Abstract
Purpose
The brain-derived neurotrophic factor (BDNF) gene has been shown to be important for synaptic plasticity in animal models. Human research has suggested that BDNF genotype may influence stroke recovery. Some studies have suggested a genotype-specific motor-related brain activation in stroke recovery. However, recovery from aphasia in relation to BDNF genotype and language-related brain activation has received limited attention. We aimed to explore functional brain activation by BDNF genotype in individuals with chronic aphasia. Consistent with findings in healthy individuals and individuals with poststroke motor impairment, we hypothesized that, among individuals with aphasia, the presence of the Met allele of the BDNF gene is associated with reduced functional brain activation compared to noncarriers of the Met allele.
Method
Eighty-seven individuals with chronic stroke-induced aphasia performed a naming task during functional magnetic resonance imaging scanning and submitted blood or saliva samples for BDNF genotyping. The mean number of activated voxels was compared between groups, and group-based activation maps were directly compared. Neuropsychological testing was conducted to compare language impairment between BDNF genotype groups. The Western Aphasia Battery Aphasia Quotient (Kertesz, 2007) was included as a covariate in all analyses.
Results
While lesion size was comparable between groups, the amount of activation, quantified as the number of activated voxels, was significantly greater in noncarriers of the Met allele (whole brain: 98,500 vs. 28,630, p < .001; left hemisphere only: 37,209 vs. 7,000, p < .001; right hemisphere only: 74,830 vs. 30,630, p < .001). This difference was most strongly expressed in the right hemisphere posterior temporal area, pre- and postcentral gyrus, and frontal lobe, extending into the white matter. Correspondingly, the atypical BDNF genotype group was found to have significantly less severe aphasia (Western Aphasia Battery Aphasia Quotient of 64.2 vs. 54.3, p = .033) and performed better on a naming task (Philadelphia Naming Test [Roach, Schwartz, Martin, Grewal, & Brecher, 1996] score of 74.7 vs. 52.8, p = .047). A region of interest analysis of intensity of activation revealed no group differences, and a direct comparison of average activation maps across groups similarly yielded null results.
Conclusion
BDNF genotype mediates cortical brain activation in individuals with chronic aphasia. Correspondingly, individuals carrying the Met allele present with more severe aphasia compared to noncarriers. These findings warrant further study into the effects of BDNF genotype in aphasia.
Supplemental Material
Presentation Video
Approximately a third of stroke patients experience aphasia, a language impairment that affects functional communication abilities (Brady, Kelly, Godwin, Enderby, & Campbell, 2016; Engelter et al., 2006). Current estimates suggest there are approximately 2 million stroke survivors living with chronic aphasia in North America (Simmons-Mackie, 2018). Persons with aphasia (PWA) are at high risk for depression, activity limitations, and social isolation (Cruice, Worrall, & Hickson, 2006; Kauhanen et al., 2000; Parr, 2007); the presence of aphasia severely affects health-related quality of life (Lam & Wodchis, 2010). Although some PWA experience spontaneous recovery in the months following stroke (Cramer, 2008), the vast majority of individuals with chronic aphasia never fully recover (Duffy, 1995). Speech and language therapy has been shown to be effective in treating aphasia (Brady et al., 2016; Breitenstein et al., 2017); however, there is great individual variability in responsiveness to treatment (Fridriksson, 2010; Helm-Estabrooks, 2002; Lazar & Antoniello, 2008). It is still somewhat unclear how specific factors relate to treatment-induced recovery. Social support has consistently been associated with recovery (Hilari, Needle, & Harrison, 2012; Lanyon, Rose, & Worrall, 2013; Vickers, 2010). Biographical and physiological factors that have been found to contribute to treatment-induced recovery include initial severity of aphasia (Laska, Hellblom, Murray, & Kahan, 2001; Pedersen, Jørgensen, Nakayama, Raaschou, & Skyhoj, 1995), age (Laska et al., 2001), lesion size, and location (Henseler, Regenbrecht, & Obrig, 2014; Laska et al., 2001; Naeser et al., 1998). However, the extent to which these biographical and physiological factors contribute to language recovery is debated (e.g., El Hachioui et al., 2013; Lazar, Speizer, Festa, Krakauer, & Marshall, 2008). Recently, some evidence has emerged suggesting that molecular and neurobiological factors may influence recovery (Balkaya & Cho, 2019). While mixed results have been reported (de Boer et al., 2017; Fridriksson et al., 2018; Marangolo et al., 2014), this line of research may offer crucial evidence to further understand the processes involved in spontaneous and treatment-based recovery from aphasia after stroke.
Neuroplastic processes that mediate cognitive recovery after stroke are guided by multiple molecular factors (Dancause & Nudo, 2011). One of which, brain-derived neurotrophic factor (BDNF), has recently been shown to be an important factor for neuroplastic processes at the level of synaptic activity (Cowansage, LeDoux, & Monfils, 2010; Fritsch et al., 2010). BDNF represents a family of neurotrophins that regulate cortical synaptic plasticity (Akeneya, Tsumoto, & Hatanaka, 1997; Lu, 2003), for example, survival, growth, and differentiation of cells in the nervous system (Huang & Reichardt, 2001). Unlike most other growth factors, secretion of BDNF is both constitutive and activity dependent, and the BDNF protein is secreted both from axons and dendrites in response to neuronal activity (Balkaya & Cho, 2019; Lessmann & Brigadski, 2009). Activity-dependent secretion of BDNF plays a crucial role in synaptic plasticity (Binder & Scharfman, 2004). This knowledge has been applied in several animal studies that have shown that BDNF is involved in long-term potentiation (LTP) processes, which may be crucial for learning (Fritsch et al., 2010; Lamb et al., 2015; Minichiello et al., 1999). The BDNF protein is encoded by the BDNF gene. A common single-nucleotide polymorphism (SNP) identified at nucleotide 196 (rs6265), resulting in a switch from valine to methionine at codon position 66 (Val66Val to Val66Met or Met66Met), has been associated with impairments in intracellular trafficking. The Met allele variants have been shown to result in 18%–30% less activity-dependent secretion of BDNF (Val66Met: 18%, Met66Met: 30%; Z. Y. Chen et al., 2004; Egan et al., 2003). The Met allele variation is present in one (Val66Met) or both alleles (Met66Met) in approximately 30% of people in the United States (Egan et al., 2003; Shimizu, Hashimoto, & Iyo, 2004), whereas the prevalence ranges between 0% and 70% in various racial groups (Petryshen et al., 2010). The Met66Met variant is rare compared with the other two variants: 3% in a Caucasian sample and virtually absent in other populations. Therefore, the Met allele variant groups are frequently pooled together in experimental designs (e.g., de Boer et al., 2017; Frodl et al., 2007) to increase statistical power of the analysis.
The presence of the Met allele variant has functional consequences. In studies on healthy individuals, the polymorphism has been associated with poorer memory performance (Hariri et al., 2003; Ho et al., 2006), decreased learning, and decreased activity-related cortical plasticity (Cheeran et al., 2008; Fritsch et al., 2010; Kleim et al., 2006; McHughen et al., 2010). McHughen et al. (2010) found smaller activation volume within several brain regions in subjects with the Met allele compared to those without the allele, as well as decreased motor learning and poorer retention over 4 days. Similarly, studies in healthy individuals have consistently reported greater functional activation in several brain areas in noncarriers of the Met allele compared to carriers (W. Chen et al., 2016; Frielingsdorf et al., 2010). In particular, carriers of a Met allele show a smaller hippocampal volume and greater deficits in motor learning and memory tasks (Z. Y. Chen et al., 2006; Cowansage et al., 2010; Egan et al., 2003; Hariri et al., 2003; Pang et al., 2004; Pezawas et al., 2004). BDNF levels increase in response to brain damage (Schäbitz et al., 2004; Zhang & Pardridge, 2006; Zhao et al., 2001), and findings from stroke studies correspondingly indicate that carriers of a Met allele have worse functional recovery after stroke (Johansson, 2011; Shimizu et al., 2004), as well as decreased region-specific brain activation in motor tasks (Kim, Quinlan, Gramer, & Cramer, 2016).
The effects of the Val66Met/Met66Met polymorphism in aphasia recovery remain to be thoroughly investigated. Similar to other cognitive abilities after stroke, language recovery is mediated by neuroplastic processes (Cowansage et al., 2010; Fridriksson et al., 2018; Thompson, den Ouden, Bonakdarpour, Garibaldi, & Parrish, 2010). Past studies on aphasia have either focused on BDNF serum levels, BDNF genotype, or both. With this in mind, it should be noted that neurotrophic factors are produced by many different cell types, including some in situ in the brain (Bejot et al., 2011), which may not be reflected in BDNF serum levels (Yoshimura et al., 2010). De Boer et al. (2017) found no significant difference in language recovery between carriers and noncarriers of the Met allele in 53 individuals with aphasia in the acute phase (< 3 months poststroke) receiving individualized impairment-based treatment. A study by Mirowska-Guzel et al. (2013) found no differences in BDNF serum levels between patients (n = 20, ≤ 3 months poststroke) who improved and those who did not improve after receiving repetitive transcranial magnetic stimulation adjuvant to behavioral treatment. However, they did report a decrease in BDNF serum levels for individuals receiving repetitive transcranial magnetic stimulation. No effects of BDNF genotype were found (Mirowska-Guzel et al., 2013). Marangolo et al. (2014) investigated the role of bihemispheric transcranial direct current stimulation (tDCS) on language recovery and BDNF serum levels in seven PWA (> 6 months poststroke). Participants underwent an intensive language therapy while receiving either active tDCS stimulation or a sham condition. They found no significant differences in BDNF serum levels after tDCS stimulation but did find a significant positive correlation in the active stimulation condition between percent change in BDNF levels and performance on a verb naming task. Lastly, Harnish et al. (2018) studied BDNF serum levels in patients (n = 5, > 6 months poststroke) receiving aphasia treatment and following an aerobic exercise plan and reported a decrease in BDNF serum levels for the first 6 weeks of therapy, followed by a retreat to baseline levels for the last 6 weeks of therapy.
To the best of our knowledge, only one study has investigated the effects of BDNF genotype in individuals with chronic aphasia. A recent double-blinded randomized clinical trial by our group examined whether responses to anodal tDCS during aphasia treatment was genotype specific in 66 PWA (> 6 months poststroke). Participants underwent an aphasia treatment (five 45-min sessions per week for 3 weeks) and were randomized to receive adjuvant anodal tDCS or a sham tDCS condition. We observed an interaction between BDNF genotype and tDCS condition for improvement in naming, where participants with Val66Val receiving anodal tDCS improved significantly more than participants with Val66Val receiving sham tDCS and Met allele carriers regardless of condition (Fridriksson et al., 2018).
The current study relied on data from the same individuals reported in the study of Fridriksson et al. (2018), in addition to more recently acquired data from a separate, ongoing study of aphasia recovery. We aimed to explore functional brain activation by BDNF genotype in individuals with chronic aphasia. In addition, we explored BDNF genotype–specific differences in common language measures. Consistent with previous literature on the relationship between BDNF genotype and task-related brain activation in healthy individuals (W. Chen et al., 2016; Frielingsdorf et al., 2010; Hariri et al., 2003; McHughen et al., 2010) and stroke (Kim et al., 2016), we tested the hypothesis that the presence of the BDNF Met allele polymorphism is associated with reduced functional brain activation and worse performance on language measures.
Method
Participants
Eighty-seven participants were included in the current study; 65 of those had previously been included in the study of Fridriksson et al. (2018). All participants were recruited at the University of South Carolina and the Medical University of South Carolina. Inclusion criteria for participants recruited for the Fridriksson et al. study were as follows: single ischemic stroke to the left hemisphere, greater than 6 months poststroke, between the ages of 21 and 80 years, previously right-handed, aphasia as confirmed using the Western Aphasia Battery–Revised (WAB-R; Kertesz, 2007), no magnetic resonance imaging (MRI) contraindications, and were able to provide informed consent for participation. Inclusion criteria for the additional participants were identical, except individuals who had suffered hemorrhagic strokes or multiple strokes were also eligible. Exclusion criteria for participation in the study of Fridriksson et al.'s (2018) randomized clinical trial included previous history of brain surgery, seizures during the previous 12 months, sensitive scalp (as per patient report), greater than 80% naming accuracy on the Philadelphia Naming Test (PNT; Roach, Schwartz, Martin, Grewal, & Brecher, 1996), and being unable to overtly name at least five of 40 items during the functional MRI (fMRI) session. Other participants were excluded if they had severely limited verbal output (WAB-R Spontaneous Speech score of 0–1), severely impaired auditory comprehension (WAB-R Comprehension score of 0–1), or bilateral stroke. Fifteen individuals did not meet the inclusion criteria and were thus not included in the analysis (see Fridriksson et al., 2018). We did not collect information about whether or how much previous aphasia treatment participants received. All participants provided written consent for study inclusion.
Among the participants, 61% (53) had typical BDNF genotype (Val66Val), and 39% (34) had the atypical genotype (Val66Met/Met66Met). The mean age of participants with typical genotype was 59.6 years (SD = 11.2, range: 29–77) and 57.7 years (SD = 10.9, range: 30–76) for participants with atypical genotype. In the typical genotype group, 17 participants were female, and the racial distribution was as follows: 41 Caucasian and 12 African American. In the atypical genotype group, 12 participants were female, and the racial distribution was as follows: 31 Caucasian, two African American, and one Asian. The groups were not significantly different in terms of age (mean difference = 1.9 years, p = .433), racial distribution (Caucasian vs. non-Caucasian; χ2(1, N = 87) = 2.772, p = .096), time poststroke (mean difference = 9.5 months, p = .253), education (mean difference = 0.1 year, p = .834), lesion size (mean difference = 20.8 cc, p = .257), amount of exercise (per patient report; mean difference before stroke = 0.5 day, p = .388, after stroke = 0.1 day, p = .802), or severity of stroke symptoms as indexed by the National Institutes of Health Stroke Scale (NIHSS; mean difference = 1.2, p = .147). Participants' characteristics are presented in Table 1.
Table 1.
Participants' biographical characteristics for the typical and atypical genotype groups.
| Characteristic | Typical BDNF genotype (Val66Val), n = 53 |
Atypical BDNF genotype (Val66Met, Met66Met), n = 34 |
95% CI of the difference | Two-sided p value | ||
|---|---|---|---|---|---|---|
| M | SD | M | SD | |||
| Age | 59.6 | 11.2 | 57.7 | 10.9 | [–2.9, 6.7] | .433 |
| Time poststroke (months) | 44.0 | 38.7 | 34.5 | 36.9 | [–6.9, 26.0] | .253 |
| Education (years) | 15.1 | 2.4 | 15.2 | 2.9 | [–1.3, 1.0] | .834 |
| Lesion size (cc) | 121.4 | 73.2 | 142.2 | 88.4 | [–57.2, 15.5] | .257 |
| Exercise before stroke (days/week) | 3.0 | 2.6 | 3.5 | 2.5 | [–1.6, 0.6] | .388 |
| Exercise now (days/week) | 3.0 | 2.4 | 3.1 | 2.2 | [–1.2, 0.9] | .802 |
| NIHSS | 5.1 | 3.2 | 6.3 | 3.8 | [–2.7, 0.4] | .147 |
Note. BDNF = brain-derived neurotrophic factor; CI = confidence interval; NIHSS = National Institutes of Health Stroke Scale.
WAB-R was used to characterize aphasia types. In the typical genotype group, 25 participants had Broca's aphasia, 12 had anomic aphasia, 11 had conduction aphasia, two had Wernicke's aphasia, two had transcortical motor aphasia, and one had global aphasia. In the atypical genotype group, 16 had Broca's aphasia, six had anomic aphasia, six had conduction aphasia, three had Wernicke's aphasia, and three had global aphasia. There was no significant difference in the distribution of nonfluent (Broca's, transcortical motor, and global) and fluent (anomic, conduction, and Wernicke's) aphasia types across groups, χ2(1, N = 87) = 0.078, p = .780. In terms of biographical measures, proportionally more subjects reported having a history of depression in the atypical genotype group, although this difference did not reach statistical significance (nine vs. six cases), χ2(1, N = 87) = 3.332, p = .068. A somewhat opposite pattern was observed for diabetes, where 11 individuals with typical genotype had diabetes, compared to two in the atypical group. However, this difference was not statistically significant, χ2(1, N = 87) = 3.605, p = .058.
Behavioral Measure
All neuropsychological tests, except for the NIHSS, were administered by American Speech-Language-Hearing Association–certified speech-language pathologists. A trained neurologist administered the NIHSS. All tests were conducted within 2 days of fMRI data acquisition upon study entry. Language testing included the WAB-R, the PNT, and the Pyramids and Palm Trees Test (Howard & Patterson, 1992). We also administered the Matrix Reasoning subtest of the Wechsler Adult Intelligence Scale–Third Edition (Wechsler, 1997) as an index of nonverbal cognitive skills. Furthermore, participants who were included in the study of Fridriksson et al. (2018) were also assessed with the Boston Naming Test (Kaplan, Goodglass, & Weintraub, 1983), the Apraxia of Speech Rating Scale (Strand, Duffy, Clark, & Josephs, 2014), and the Apraxia Battery for Adults–Second Edition (Dabul, 2000).
BDNF Genotyping
For the participants recruited for the study of Fridriksson et al. (2018), a 2-ml whole blood sample was collected at the same date as behavioral testing and neuroimaging. The blood sample was labeled with each participant's de-identified study number and frozen. Saliva samples (0.75 ml) were collected from the remaining participants by stroking the inside of their cheeks with individual, nonreusable Oragene sponges. Saliva samples were collected at a later date than behavioral testing and neuroimaging sessions and stored at room temperature. DNA genotyping is equally accurate for blood and saliva samples (Van Oene, Alic, Jackson, & Lem, 2006). Samples were transported to DNA Genotek (https://www.dnagenotek.com/) in two batches, where DNA extraction and SNP genotyping were completed on rs6265. Blood samples were extracted using Qiagen PureGene reagents and a validated extraction protocol. Saliva samples were extracted using SNPstream and a validated extraction protocol. Genotyping for the SNP was accomplished using a TaqMan single-tube genotyping assay. The TaqMan assay is an allele discrimination assay using amplification and a pair of fluorescent dye detectors that target the SNP. One fluorescent dye is attached to the detector that is a perfect match to the first allele (e.g., an “A” nucleotide), and a different fluorescent dye is attached to the detector that is a perfect match to the second allele (e.g., a “C” nucleotide). During polymerase chain reaction, the polymerase releases the fluorescent probe into solution where it is detected using end point analysis in a Life Technologies 7900HT Real-Time instrument. Primers and probes were obtained through Life Technologies design and manufacturing. Genotypes were determined using Life Technologies' Taqman Genotyper v1.0.1 software. Participants with a Val66Val (e.g., “C/C”) expression were considered typical BDNF genotype, and those with Val66Met (e.g., “C/A”) or Met66Met (e.g., “A/A”) were considered to have atypical BDNF genotype. Samples were then destroyed as per protocol.
Acquisition of Neuroimaging Data
Each participant underwent an MRI session that included T1- and T2-weighted structural MRI and fMRI. The fMRI task utilized a simple picture-naming paradigm designed to allow us to effectively isolate activation associated with naming. During 10 min of fMRI scanning, participants completed a picture-naming task in which 40 colored pictures of high-frequency nouns were back-projected on an MRI-compatible screen and seen via a mirror mounted on the scanner head coil. For the purpose of establishing a baseline for the fMRI data analysis, 20 colored abstract images were shown at random among the real picture presentation. Pictures were presented for 2 s each. Participants were instructed to overtly name target pictures representing nouns once they appeared on the screen and to stay silent when abstract images appeared on the screen. All naming attempts were recorded using a nonferrous microphone and were subsequently scored off-line.
To improve clarity of the audio recordings and to minimize speech-related head motion, a sparse imaging sequence was utilized where a single full brain volume was collected every 10 s. Each volume acquisition lasted 2 s, allowing for 8 s of scanner silence until the next volume was collected; these 8 s of silence were utilized for stimulus presentation and response, in which a single picture was shown for 2 s and a naming attempt was recorded. To better model the hemodynamic response in the fMRI data analysis, the interval between picture presentation was jittered (i.e., different time points following each stimulus presentation were sampled). The interstimulus interval varied between 6 and 8 s. To minimize the chance that participants would speak during fMRI data collection, pictures were always presented during the silent period between scans, appearing at least 3 s prior to acquisition of the subsequent scan.
MRI data were collected on a 3T Siemens Trio scanner with a 12-element head coil or a Siemens Prisma 3T with a 20-channel head coil. fMRI acquisition sequence was kept consistent across scanners. Upon visual examination, we did not encounter a difference in the sensitivity for detecting activation in images acquired with the 12- and 20-channel head coils. Task fMRI was acquired using T2* MRI echoplanar imaging with sparse sampling: 64 full brain volumes, 90° flip angle, repetition time = 10 s, acquisition time = 2 s, echo time = 30 ms, in-plane resolution = 3.25 × 3.25 mm, slice thickness = 3.2 mm (no gap), and 33 axial slices collected in planes aligned parallel to the anterior commissure–posterior commissure line.
Preprocessing of Structural Neuroimaging Data
Image preprocessing was performed blinded to genetic and clinical data. Each individual's fMRI data were preprocessed using Statistical Parametric Mapping software (SPM12, Wellcome Department of Imaging Neuroscience). Lesions were manually drawn on the T2 weighted image by a neurologist who was blind to all participant testing information. The T2-image was coregistered to the T1 image, and these parameters were used to reslice the lesion into the native T1 space. The resliced lesion maps were smoothed with a 3-mm full-width half-maximum Gaussian kernel to remove jagged edges associated with manual drawing. We then performed enantiomorphic segmentation–normalization (Nachev, Coulthard, Jäger, Kennard, & Husain, 2008) using SPM12 and MATLAB scripts we developed (Rorden, Bonilha, Fridriksson, Bender, & Karnath, 2012) as follows: First, a mirrored image of the T1 scan (reflected around the midline) was created, and this mirrored image was coregistered to the native T1 image. We then created a chimeric image based on the native T1 scan with the lesioned tissue replaced by tissue from the mirrored scan (using the smoothed lesion map to modulate this blending). SPM12's unified segmentation–normalization (Ashburner & Friston, 2005) was used to warp this chimeric image to standard space, with the resulting spatial transform applied to the native T1 scan as well as the lesion map and the T2 scan (which used the T1 segmentation parameters to mask nonbrain signal). Note that this high-resolution T2 scan has identical features and similar contrast to the individual's low-resolution T2* fMRI scan (a property leveraged for normalizing the fMRI data). The normalized lesion map was then binarized, using a 50% threshold.
Preprocessing of Functional Neuroimaging Data
fMRI data were corrected for motion using the SPM12 “realign and unwarp” procedure with default settings. We then performed brain extraction on the T2-weighted images using the SPM12 script spm_brain_mask with default settings. Slice time correction was also done using SPM12. Stimulus onsets were convolved with the canonical hemodynamic response function (HRF) and its temporal derivative in SPM12. The mean extracted fMRI volume for each participant was nonlinearly normalized to the individual's brain-extracted and normalized T2 image (correcting for both individual differences and spatial distortions observed in echoplanar imaging sequences) and resliced to 2 mm isotropic. All fMRI data were then spatially smoothed with a Gaussian kernel with full-width half-maximum of 6 mm. The voxelwise fMRI time courses were detrended using the following regressors: mean signal from the white matter, obtained from the chimeric T1 image; time courses of the six motion parameters estimated at the motion correction step; and linear trends. In addition, we used independent component analysis to identify and remove lesion-driven artifacts in the data (Yourganov, Fridriksson, Stark, & Rorden, 2018).Following the aforementioned preprocessing steps, we used SPM12's default functions to model and estimate the effects for our two conditions (overt naming and silent abstract images), producing t maps demonstrating voxels brighter for picture naming > abstract (Ashburner et al., 2012).
Data Analysis
To examine our hypothesis that the presence of atypical BDNF genotype is associated with reduced brain activation in individuals with chronic aphasia, we utilized general linear modeling and a standard HRF to analyze contrast maps containing naming-related activation. For first-level analysis, we used SPM12's default functions to model and estimate the effects for our two conditions (overt naming and silent viewing of abstract images). The HRF was modeled using a Gamma function and a temporal derivative. This procedure yielded whole-brain contrast maps showing brain locations where BOLD response was significantly different between the two conditions. For second-level analysis, we analyzed the group-level differences between the typical and atypical genotype groups using a two-sample t test. The same procedure was utilized to compare activation maps separately for the left and right hemispheres by applying explicit masks to the maps. We corrected for multiple comparisons using the SPM default method of familywise error (FWE; p = .05) correction based on random field theory. WAB-R Aphasia Quotient (AQ) score was used as a covariate in all analyses to account for differences in aphasia severity. In addition, to compute the number of significantly active voxels for each group, we conducted one-sample t tests (within a group) to obtain significant activation maps at a statistical threshold of 0.05 with FWE correction to control for multiple comparisons. We used the Batch Descriptives function of MRIcron (Rorden & Brett, 2000) to quantify the number of significantly activated voxels and an independent-samples t test for group comparison.
Results
Clear differences in language impairment were observed between the typical BDNF genotype group and the atypical BDNF genotype group in overall aphasia severity on the WAB-R AQ (typical vs. atypical BDNF; 64.2 vs. 54.3, p = .033) and naming accuracy measured on the PNT (74.7 vs. 52.8, p = .047). In contrast, no observable difference in performance was found for the Pyramids and Palm Trees Test (45.7 vs. 46.2, p = .598) and the Wechsler Adult Intelligence Scale–Third Edition (12.4 vs. 11.8, p = .635), indicating that the groups only differed in terms of their language impairment. Although performance on the naming task in the scanner was slightly better on average in the typical BDNF genotype group, this difference was not significant (16.0 vs. 13.6, p = .336). In a subset of 65 participants described in the study of Fridriksson et al. (2018), the typical genotype group performed significantly better on the Boston Naming Test (22.8 vs. 14.4, p = .042), lending further support to the evident group difference in anomia severity. Furthermore, no group difference in the same subset of participants was revealed for severity of apraxia of speech measured on the Apraxia of Speech Rating Scale (2.9 vs. 2.9, p = .988), but the atypical genotype group was found to have more severe limb apraxia (46.6 vs. 41.1, p = .006) and oral apraxia (43.3 vs. 37.9, p = .016) on the Apraxia Battery for Adults–Second Edition (see Supplemental Material S1). Behavioral testing results for the full sample are shown in Table 2.
Table 2.
Results from behavioral testing on participants with typical and atypical brain-derived neurotrophic factor (BDNF) genotype.
| Characteristic | Typical BDNF genotype (Val/Val), n = 53 |
Atypical BDNF genotype (Val/Met, Met/Met), n = 34 |
95% CI of the difference | Two-sided p value | ||
|---|---|---|---|---|---|---|
| M | SD | M | SD | |||
| WAB-R AQ | 64.2 | 20.3 | 54.3 | 21.0 | (0.81, 18.98) | .033* |
| Spont. Sp. | 12.2 | 4.4 | 10.4 | 4.6 | (–0.21, 3.75) | .079 |
| Aud. Comp. | 8.1 | 1.6 | 7.4 | 1.7 | (0.04, 1.49) | .040* |
| Repetition | 5.8 | 2.9 | 4.6 | 2.9 | (–0.10, 2.43) | .071 |
| Naming | 6.1 | 2.7 | 4.8 | 2.7 | (0.06, 2.44) | .040* |
| PNT | 74.7 | 51.1 | 52.8 | 43.3 | (0.26, 43.60) | .047* |
| PPTT | 45.7 | 4.2 | 46.2 | 4.6 | (–2.47, 1.43) | .598 |
| WAIS-III Matrix Score | 12.4 | 5.6 | 11.8 | 5.1 | (–1.76, 2.87) | .635 |
Note. CI = confidence interval; WAB-R AQ = Western Aphasia Battery–Revised Aphasia Quotient; Spont. Sp. = Spontaneous Speech; Aud. Comp. = Auditory Comprehension; PNT = Philadelphia Naming Test; PPTT = Pyramids and Palm Trees Test; WAIS-III = Wechsler Adult Intelligence Scale–Third Edition.
p < .05.
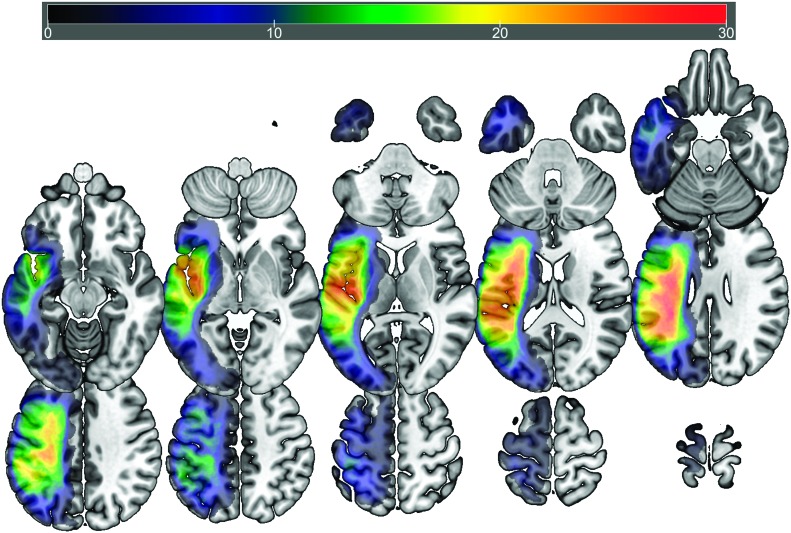
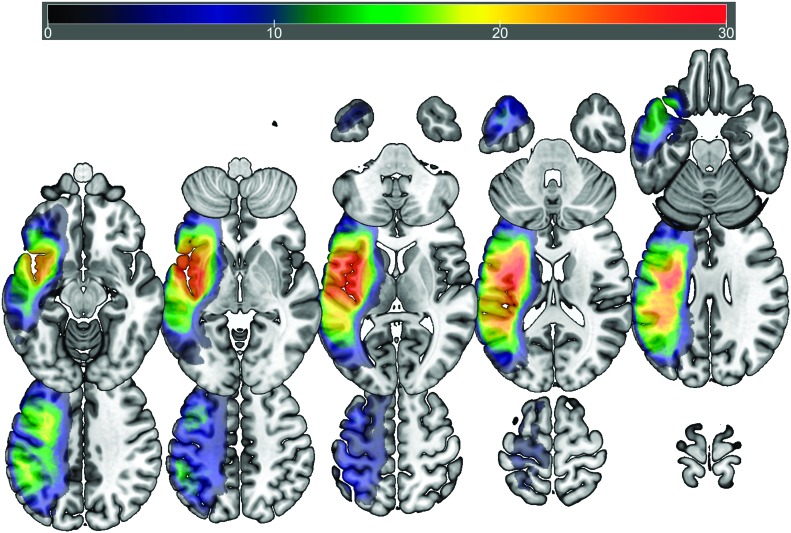
Consistent with the primary aim of the study, we sought out to explore whether BDNF genotype influences functional brain activation during language processing in PWA. Participants in the typical and atypical BDNF genotype groups presented with distributed cortical and subcortical lesions that covered the middle cerebral artery territory, extending from the posterior occipital and parietal lobes to anterior temporal and frontal areas. The greatest overlap at a voxelwise level was identified in the superior longitudinal fasciculus (MNI coordinates: −34 −36 28) in the typical genotype group and the superior longitudinal fasciculus (MNI: −36 −8 25) and the insula (MNI: −44 −9 3) in the atypical group. Figures 1 and 2 present overlap images for the typical and atypical genotype groups, respectively.
Figure 1.
Lesion overlay map (n = 53; maximum overlap, n = 39) showing lesion distribution of participants in the typical genotype group. Warmer colors indicate greater overlap.
Figure 2.
Lesion overlay map (n = 34; maximum overlap, n = 29) showing lesion distribution of participants in the atypical genotype group. Warmer colors indicate greater overlap.
We used a first-level specification to prepare and analyze individual contrast maps of picture naming versus abstract image viewing. This procedure allowed us to isolate naming-related activation for each individual. Then, we used a second-level specification to create average activation maps for each group, that is, typical and atypical genotype groups. Subsequently, we performed a two-sample t test to directly compare whole-brain activation maps between groups. The same procedure was utilized to compare activation maps separately for the left and right hemispheres by applying explicit masks to the maps. When controlling for WAB-R AQ and using an FWE threshold of 0.05, no statistically significant differences were observed in intensity of cortical activation across the typical and atypical BDNF genotype groups.
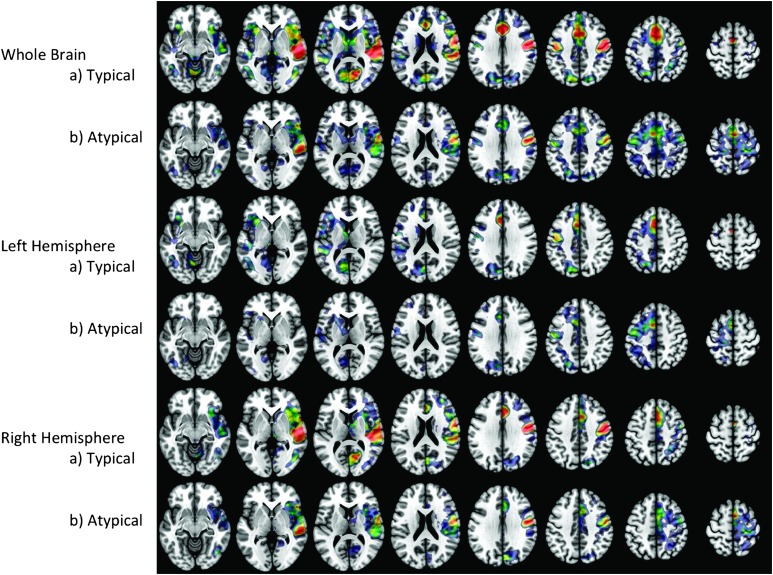
Using one-sample t tests, we obtained significant naming-related activation maps for each group. Figure 3 presents average activation maps at an FWE of 0.05 for the whole brain and separately for each hemisphere. The overall activation pattern was similar in both groups, with the greatest intensity of activation present in the bilateral posterior temporal gyrus, pre- and postcentral gyrus, and the longitudinal fissure. We observed greater distribution and intensity of activation in the right hemisphere in the typical genotype group, particularly in the posterior temporal lobe, pre- and postcentral gyrus, and frontal lobe white matter, while these areas were only minimally activated in the atypical BDNF group. Although the pattern of activation was similar between groups, the intensity of activation was greater across most areas in the typical BDNF genotype group.
Figure 3.
Significant activation during picture naming over abstract image viewing in the typical and atypical genotype groups. Warmer colors indicate greater activation intensity (t = 3.0–8.0).
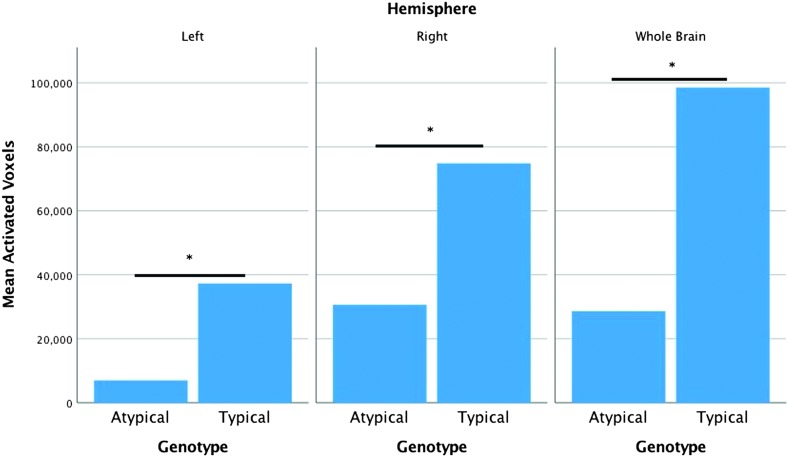
In order to quantify the observed difference, we obtained the number of voxels where functional naming-related activation was significantly greater than zero when factoring out the variance explained by the WAB-R AQ for each genotype group at the whole-brain level and separately for either hemisphere. Using an independent-samples t test, we found that the number of activated voxels was greater in the typical genotype group compared to the atypical group at the whole-brain level (98,500 vs. 28,630), t(85) = 18.63, p < .001, in the left hemisphere (37,290 vs. 7,000), t(85) = 8.33, p < .001, and in the right hemisphere (74,830 vs. 30,630), t(85) = 11.29, p < .001 (see Figure 4). In terms of the interpretation of this finding, it is important to note that the number of significantly activated voxels in the left and right hemisphere does not and should not add up to the number of activated voxels at the whole-brain level. This stems from the fact that the analyses are conducted separately for the whole-brain level and each hemisphere, and therefore, the number of voxels included in the calculations for t values is different across analyses, that is, roughly twice as many voxels go into the calculation for the whole-brain level compared to hemispheric-level analyses.
Figure 4.
Number of activated voxels at the whole-brain level and for the left and right hemispheres separately for the typical and atypical brain-derived neurotrophic factor genotype groups. *p < .001.
Discussion
The current study explored functional brain activation in relation to BDNF genotype in participants with chronic aphasia as a result of left-hemisphere stroke. While we did not observe a significant difference in activation intensity in specific brain regions across groups, fMRI during a naming task revealed a greater number of activated voxels at a whole-brain level and in both hemispheres in individuals with typical BDNF genotype compared to those with atypical BDNF genotype. This difference was particularly prevalent in the right-hemisphere posterior temporal lobe, pre- and postcentral gyrus, and frontal lobe. Correspondingly, the atypical genotype group was found to be more severely affected by aphasia as indicated by poorer performance on the WAB-R and the PNT.
These results are consistent with studies in healthy individuals (W. Chen et al., 2016; Jabbi et al., 2017; McHughen et al., 2010) and the general stroke population (Kim et al., 2016) where decreased task-related functional brain activation has been reported for individuals carrying the BDNF Met allele polymorphism. Notably, McHughen et al. (2010) observed smaller activation volume in a broad sensorimotor network in individuals with the Met allele present in response to right index finger movement. Furthermore, carriers of the Met allele showed activation volume reduction while noncarriers showed activation volume expansion in response to 25 min of right index finger training. Previous studies have linked the polymorphism to abnormal modulation of hippocampal function during a memory task and reduced gray matter in several areas of frontal cortex (Egan et al., 2003), experience-dependent plasticity of the motor cortex (Kleim et al., 2006), and abnormal motor cortex plasticity after various forms of inhibitory and excitatory repetitive transcranial magnetic stimulation protocols (Cheeran et al., 2008). Therefore, McHughen et al. inferred that the Met allele polymorphism alters neuronal processes in a manner that produces differences in brain response to short-term training. Additional evidence suggests that the observed genotype-specific difference in functional neural activation may relate to differences in long-term cortical development and plasticity, particularly in terms of hippocampal (Bueller et al., 2006; Frodl et al., 2007; Pezawas et al., 2004; Szeszko et al., 2005), prefrontal cortex (Pezawas et al., 2004), and temporal and occipital gray matter volume (Ho et al., 2006).
While the effects of BDNF genotype on functional brain activation in healthy individuals are well documented, little is known about how genotype impacts brain activation poststroke. However, our results—and similarly the results of Kim et al. (2016)—indicate that the modulatory effects of genetic factors on the normal brain remain important after stroke. This effect seems to be stable despite the functional (Hamilton, Chrysikou, & Coslett, 2011; Teasell, Bayona, & Bitensky, 2015) and structural (Berthier et al., 2011; Grefkes & Fink, 2011) reorganization that takes place as a result of brain damage. Specifically, our results show increased activation in both hemispheres in the typical compared to the atypical genotype group. Given the exploratory nature of the current study, these results may be interpreted in several different ways in terms of the mechanisms of language recovery after stroke. First, recovery has been related to the structure and function of perilesional regions in the damaged left hemisphere (Fridriksson, Richardson, Fillmore, & Cai, 2012; Hillis et al., 2006; Meinzer & Breitenstein, 2008; Szaflarski, Allendorfer, Banks, Vannest, & Holland, 2013; Warburton, Price, Swinburn, & Wise, 1999). Although we did not perform a region-specific analysis, our findings may be interpretable under these assumptions in that naming-related activation in the left hemisphere was considerably greater (5.3-fold) in the typical versus the atypical group. Notably, Fridriksson et al. (Fridriksson, 2010; Fridriksson et al., 2012) reported that activation in perilesional frontal lobe areas predicted treatment-related improvement in naming in PWA, and they concluded that these areas are important for recovery. Second, contralateral right hemisphere homotopic regions have been suggested to mediate recovery (Blasi et al., 2002; Hartwigsen & Saur, 2019; Leff et al., 2002; Lukic et al., 2017; Musso et al., 1999; Saur et al., 2006; Turkeltaub et al., 2012; Winhuisen et al., 2005). Although we failed to identify a significant difference in specific language-related areas in the right hemisphere, we found that naming-related activation in the right hemisphere in the typical group was 2.4-fold that of the atypical group. The distributed activation in the right hemisphere may certainly contribute to language-related recovery according to this line of evidence. Lastly, contralateral right hemisphere homotopic regions have also been found to deter recovery through transcallosal disinhibition of the lesioned left hemisphere (Belin et al., 1996; Naeser, Martin, Nicholas, Baker, Seekins, Helm-Estabrooks, et al., 2005; Naeser, Martin, Nicholas, Baker, Seekins, Kobayashi, et al., 2005; Thiel et al., 2006; for further discussion, see Turkeltaub, 2015). While our results do not provide ground to support or invoke theories of interhemispheric disinhibition, further study into genotype-specific brain activation may be a prospective area to study neural reorganization after stroke.
To our knowledge, this is the first large-scale study investigating the role of the BDNF gene in chronic aphasia. Previous studies have included participants in the acute phase of aphasia (de Boer et al., 2017; Mirowska-Guzel et al., 2013) or a small sample size (Harnish et al., 2018; Marangolo et al., 2014). The novelty of the current study thus lies in a larger sample of individuals in the chronic phase of aphasia, and as such, our data present a new perspective to the role of BDNF in language recovery. To this end, the observed group difference in brain activation, together with distinctive baseline difference in language measures, can be interpreted in light of reorganization of the brain after stroke. Presence of the Met allele results in 18%–30% less activity-dependent secretion of BDNF (Z. Y. Chen et al., 2004; Egan et al., 2003), which affects the potential for cortical synaptic plasticity and LTP processes crucial for learning (Fritsch et al., 2010; Lamb et al., 2015). Therefore, the presence of the Met allele may affect the potential for recovery at the molecular level. The effects are likely too subtle to be detected shortly after stroke, but our results indicate that these effects may play an important role in long-term recovery in aphasia. The finding by Fridriksson et al. (2018) that typical BDNF genotype enhances the effects of anodal tDCS through LTP lends further support to this conclusion. However, further study into the exact mechanism by which typical BDNF genotype mediates recovery is needed to decipher whether and how this knowledge can be used to improve clinical practice.
The current study had several limitations. First, we failed to find a significant difference in specific brain regions when we directly compared activation maps at the whole-brain level and separately for each hemisphere across groups. This null finding may suggest that the evident increase in activation is not strongly region specific, but rather that a distributed network of regions contributed in the naming task. It should be noted that we used a stringent criteria of FWE = .05, and we did not explore a priori hypotheses about the contribution of certain regions of the brain. Thus, our data do not enable us to make decisive inferences concerning the mechanism of functional reorganization of the language network in aphasia. Second, we did not control for exposure to rehabilitation services and treatment participants may have received before study entry. It is conceivable that the typical BDNF genotype group has received more treatment, and this is being reflected in our results. Similarly, factors such as motivation and social support were not considered. Lastly, although the number of participants in each group was relatively large compared to most studies in aphasia, the difference in group size across groups may impact the power of the analyses conducted. These limitations should be considered for further studies into the effects of BDNF genotype in aphasia.
The results of the current study suggest that BDNF genotype may play an important role in language recovery after stroke. Effects of the Met allele polymorphism appear to lead to poorer language-related prognosis, similar to functional cognitive (Cramer & Procaccio, 2012; Egan et al., 2003; Johansson, 2011) and motor-related (Helm, Tyrell, Pohlig, Brady, & Reisman, 2016; Kim et al., 2016; van der Vliet, Ribbers, Vandermeeren, Frens, & Selles, 2017) recovery reported in the stroke literature. While our results are somewhat limited by the exploratory nature of the study and lack of control for possible confounding variables (e.g., time in treatment), they do suggest a promising venue to further our knowledge of language recovery after stroke. These results do need to be replicated, and an area of particular interest might be to investigate more thoroughly the causational relationship between functional brain activation and language outcome measures by genotype. Considering the variability in response to language-based treatment in aphasia, our results warrant further exploration of the underlying molecular mechanisms guiding recovery.
Supplementary Material
Acknowledgments
This article stems from the 2018 Research Symposium at the American Speech-Language-Hearing Association Convention, which was supported by the National Institute on Deafness and Other Communication Disorders of the National Institutes of Health under Award R13DC003383. The study was supported by the National Institute on Deafness and Other Communication Disorders (Award P50 DC014664 and U01 DC011739 (PI: Fridriksson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding Statement
This article stems from the 2018 Research Symposium at the American Speech-Language-Hearing Association Convention, which was supported by the National Institute on Deafness and Other Communication Disorders of the National Institutes of Health under Award R13DC003383. The study was supported by the National Institute on Deafness and Other Communication Disorders (Award P50 DC014664 and U01 DC011739 (PI: Fridriksson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Akeneya Y., Tsumoto T., & Hatanaka H. (1997). Brain-derived neurotrophic factor enhances long-term potentiation in rat visual cortex. Journal of Neuroscience, 17, 6707–6716. https://doi.org/10.1523/JNEUROSCI.17-17-06707.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J., Chen C., Moran R., Henson R., Glauche V., & Phillips C. (2012). SPM12 manual the FIL methods group (and honorary members). Retrieved from http://www.fil.ion.ucl.ac.uk/spm/doc/spm12_manual.pdf
- Ashburner J., & Friston K. J. (2005). Unified segmentation. NeuroImage, 26(3), 839–851. https://doi.org/10.1016/j.neuroimage.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Balkaya M., & Cho S. (2019). Genetics of stroke recovery: BDNF val66met polymorphism in stroke recovery and its interaction with aging. Neurobiology of Disease, 126, 36–46. https://doi.org/10.1016/j.nbd.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejot Y., Prigent-Tessier A., Cachia C., Giroud M., Mossiat C., Bertrand N., … Marie C. (2011). Time-dependent contribution of non-neuronal cells to BDNF production after ischemic stroke in rats. Neurochemistry International, 53, 102–111. https://doi.org/10.1016/j.neuint.2010.10.019 [DOI] [PubMed] [Google Scholar]
- Belin P., Van Eeckhout P., Zilbovicius M., Remy P., François C., Guillaume S., … Samson Y. (1996). Recovery from nonfluent aphasia after melodic intonation therapy: A PET study. Neurology, 47(6), 1504–1511. https://doi.org/10.1212/wnl.47.6.1504 [DOI] [PubMed] [Google Scholar]
- Berthier M. L., García-Casares N., Walsh S. F., Nabrozidis A., Ruíz de Mier R. J., Green C., … Pulvermüller F. (2011). Recovery from post-stroke aphasia: Lessons from brain imaging and implications for rehabilitation and biological treatments. Discovery Medicine, 12(65), 275–289. [PubMed] [Google Scholar]
- Binder D. K., & Scharfman H. E. (2004). Brain-derived neurotrophic factor. Growth Factors, 22(3), 123–131. https://doi.org/10.1080/08977190410001723308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi V., Young A. C., Tansy A. P., Petersen S. E., Snyder A. Z., & Corbetta M. (2002). Word retrieval learning modulates right frontal cortex in patients with left frontal damage. Neuron, 36, 159–170. https://doi.org/10.1016/S0896-6273(02)00936-4 [DOI] [PubMed] [Google Scholar]
- Brady M. C., Kelly H., Godwin J., Enderby P., & Campbell P. (2016). Speech and language therapy for aphasia following stroke. Cochrane Database of Systematic Reviews, 6 https://doi.org/10.1002/14651858.CD000425.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenstein C., Grewe T., Flöel A., Ziegler W., Springer L., Martus P., … Baumgaertner A. (2017). Intensive speech and language therapy in patients with chronic aphasia after stroke: A randomised, open-label, blinded-endpoint, controlled trial in a health-care setting. Lancet, 389(10078), 1528–1538. https://doi.org/10.1016/S0140-6736(17)30067-3 [DOI] [PubMed] [Google Scholar]
- Bueller J. A., Aftab M., Sen S., Gomez-Hassan D., Burmeister M., & Zubieta J.-K. (2006). BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biological Psychiatry, 59, 812–815. https://doi.org/10.1016/j.biopsych.2005.09.022 [DOI] [PubMed] [Google Scholar]
- Cheeran B., Talelli P., Mori F., Koch G., Suppa A., Edwards M., … Rothwell J. C. (2008). A common polymorphism in the brain derived neurotrophic factor (BDNF) gene modulates human cortical plasticity and the response to rTMS. The Journal of Physiology, 586(23), 5717–5725. https://doi.org/10.1113/jphysiol.2008.159905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Chen C., Xia M., Wu K., Chen C., He Q., … Dong Q. (2016). Interaction effects of BDNF and COMT genes on resting-state brain activity and working memory. Frontiers in Human Neuroscience, 10, 540 https://doi.org/10.3389/fnhum.2016.00540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Jing D., Bath K. G., Ieraci A., Khan T., Siao C. J., … Lee F. S. (2006). Genetic variant BDNF (val66met) polymorphism alters anxiety-related behaviour. Science, 314, 140–143. https://doi.org/10.1126/science.1129663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Patel P. D., Sant G., Ment C. X., Teng K. K., Hempstead B. L., & Lee F. S. (2004). Variant brain-derived neurotropic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. Journal of Neuroscience, 24(18), 4401–4411. https://doi.org/10.1523/JNEUROSCI.0348-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowansage K. K., LeDoux J. E., & Monfils M. H. (2010). Brain-derived neurotrophic factor: A dynamic gatekeeper of neural plasticity. Current Molecular Pharmacology, 3, 12–29. https://doi.org/10.2174/1874467211003010012 [DOI] [PubMed] [Google Scholar]
- Cramer S. C. (2008). Repairing the human brain after stroke: I. Mechanisms of spontaneous recovery. Annals of Neurology, 63, 272–287. https://doi.org/10.1002/ana.21393 [DOI] [PubMed] [Google Scholar]
- Cramer S. C., & Procaccio V. (2012). Correlation between genetic polymorphisms and stroke recovery: Analysis of the GAIN Americas and GAIN international studies. European Journal of Neurology, 19(5), 718–724. https://doi.org/10.1111/j.1468-1331.2011.03615.x [DOI] [PubMed] [Google Scholar]
- Cruice M., Worrall L., & Hickson L. (2006). Quantifying aphasic people's social lives in the context of non-aphasic peers. Aphasiology, 20(12), 1210–1225. https://doi.org/10.1080/02687030600790136 [Google Scholar]
- Dabul B. (2000). Apraxia Battery for Adults–Second Edition (ABA-2). Austin, TX: Pro-Ed. [Google Scholar]
- Dancause N., & Nudo R. J. (2011). Shaping plasticity to enhance recovery after injury. Progress in Brain Research, 192, 273–295. https://doi.org/10.1016/B978-0-444-53355-5.00015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer R. G. A., Spielmann K., Heijenbrok-Kal M. H., van der Vliet R., Ribbers G. M., & van de Sandt-Koenderman W. M. E. (2017). The role of the BDNF Val66Met polymorphism in recovery of aphasia after stroke. Neurorehabilitation and Neural Repair, 31(9), 851–857. https://doi.org/10.1177/1545968317723752 [DOI] [PubMed] [Google Scholar]
- Duffy J. (1995). Motor speech disorders. St. Louis, MO: Mosby. [Google Scholar]
- Egan M. F., Kojima M., Callicott J. H., Goldberg T. E., Kolachana B. S., Bertolino A., … Weinberger D. R. (2003). The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell, 112(2), 257–269. https://doi.org/10.1016/S0092-8674(03)00035-7 [DOI] [PubMed] [Google Scholar]
- El Hachioui H., Lingsma H. F., van de Sandt-Koenderman M. W., Dippel D. W., Koudstaal P. J., & Visch-Brink E. G. (2013). Long-term prognosis of aphasia after stroke. Journal of Neurology, Neurosurgery, & Psychiatry, 84, 310–315. https://doi.org/10.1136/jnnp-2012-302596 [DOI] [PubMed] [Google Scholar]
- Engelter S. T., Gostynski M., Papa S., Frei M., Born C., Ajdacic-Gross V., … Lyrer P. A. (2006). Epidemiology of aphasia attributable to first ischemic stroke. Stroke, 37(6), 1379–1384. https://doi.org/10.1161/01.STR.0000221815.64093.8c [DOI] [PubMed] [Google Scholar]
- Fridriksson J. (2010). Preservation and modulation of specific left hemisphere regions is vital for treated recovery from anomia in stroke. Journal of Neuroscience, 30(35), 11558–11564. https://doi.org/10.1523/JNEUROSCI.2227-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridriksson J., Elm J., Stark B. C., Basilakos A., Rorden C., Sen S., … Bonilha L. (2018). BDNF genotype and tDCS interaction in aphasia treatment. Brain Stimulation, 11(6), 1276–1281. https://doi.org/10.1016/j.brs.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridriksson J., Richardson J. D., Fillmore P., & Cai B. (2012). Left hemisphere plasticity and aphasia recovery. NeuroImage, 60, 854–863. https://doi.org/10.1016/j.neuroimage.2011.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frielingsdorf H., Bath K. G., Soliman F., DiFede J., Casey B. J., & Lee F. S. (2010). Variant brain-derived neurotrophic factor Val66Met endophenotypes: Implications for posttraumatic stress disorder. Annals of the New York Academy of Sciences, 1208, 150–157. https://doi.org/10.1111/j.1749-6632.2010.05722.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch B., Reis J., Martinowich K., Schambra H. M., Ji Y., Cohen L. G., & Lu B. (2010). Direct current stimulation promotes BDNF-dependent synaptic plasticity: Potential implications for motor learning. Neuron, 66, 198–204. https://doi.org/10.1016/j.neuron.2010.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T., Schüle C., Schmitt G., Born C., Baghai T., Zill P., … Meisenzahl E. (2007). Association of the brain-derived neurotrophic factor Val66Met polyorphism with reduced hippocampal volumes in major depression. Archives of General Psychiatry, 64, 410–416. https://doi.org/10.1001/archpsyc.64.4.410 [DOI] [PubMed] [Google Scholar]
- Grefkes C., & Fink G. R. (2011). Reorganization of cerebral networks after stroke: New insights from neuroimaging with connectivity approaches. Brain, 134(5), 1264–1276. https://doi.org/10.1093/brain/awr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R. H., Chrysikou E. G., & Coslett B. (2011). Mechanisms of aphasia recovery after stroke and the role of noninvasive brain stimulation. Brain and Language, 118(1–2), 40–50. https://doi.org/10.1016/j.bandl.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri A. R., Goldberg T. E., Mattay V. S., Kolachana B. S., Callicott J. H., Egan M. F., & Weinberger D. R. (2003). Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. Journal of Neuroscience, 23(17), 6690–6694. https://doi.org/10.1523/JNEUROSCI.23-17-06690.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnish S. M., Rodriguez A. D., Blackett D. S., Gregory C., Seeds L., Boatright J. H., & Crosson B. (2018). Aerobic exercise as an adjuvant to aphasia therapy: Theory, preliminary findings, and future directions. Clinical Therapeutics, 40(1), 38.e6–48.e6. https://doi.org/10.1016/j.clinthera.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwigsen G., & Saur D. (2019). Neuroimaging of stroke recovery from aphasia—Insights into plasticity of the human language network. NeuroImage, 190, 14–31. https://doi.org/10.1016/j.neuroimage.2017.11.056 [DOI] [PubMed] [Google Scholar]
- Helm E. E., Tyrell C. M., Pohlig R. T., Brady L. D., & Reisman D. S. (2016). The presence of a single-nucleotide polymorphism in the BDNF gene affects the rate of locomotor adaptation after stroke. Experimental Brain Research, 234, 341–351. https://doi.org/10.1007/s00221-015-4465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm-Estabrooks N. (2002). Cognition and aphasia: A discussion and a study. Journal of Communication Disorders, 35(2), 171–186. https://doi.org/10.1016/S0021-9924(02)00063-1 [DOI] [PubMed] [Google Scholar]
- Henseler I., Regenbrecth F., & Obrig H. (2014). Lesion correlates of patholinguistic profiles in chronic aphasia: Comparisons of syndrome-, modality- and symptom-level assessment. Brain, 137(Pt. 3), 918–930. https://doi.org/10.1093/brain/awt374 [DOI] [PubMed] [Google Scholar]
- Hilari K., Needle J. J., & Harrison K. L. (2012). What are the important factors in health-relates quality of life for people with aphasia? A systematic review. Archives of Physical Medicine and Rehabilitation, 93(Suppl. 1), S86–S95. https://doi.org/10.1016/j.apmr.2011.05.028 [DOI] [PubMed] [Google Scholar]
- Hillis A. E., Kleinman J. T., Newhart M., Heidler-Gary J., Gottesman R., Barker P. B., … Chaudhry P. (2006). Restoring cerebral blood flow reveals neural regions critical for naming. Journal of Neuroscience, 26(31), 8069–8073. https://doi.org/10.1523/JNEUROSCI.2088-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. C., Milev P., O'Leary D. S., Librant A., Andreasen N. C., & Wassink T. H. (2006). Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor Val66Met gene polymorphism in patients with schizophrenia and healthy volunteers. Archives of General Psychiatry, 63(7), 731–740. https://doi.org/10.1001/archpsyc.63.7.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D., & Patterson K. (1992). The Pyramids and Palm Trees Test: A test of semantic access from words and pictures. Cambridge, United Kingdom: Thames Valley Test Company. [Google Scholar]
- Huang E. J., & Reichardt L. F. (2001). Neurotrophins: Roles in neuronal development and function. Annual Review Neuroscience, 24, 677–736. https://doi.org/10.1146/annurev.neuro.24.1.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbi M., Cropp B., Nash T., Kohn P., Kippenhan J. S., Masdeu J. C., … Berman K. F. (2017). BDNF Val66Met polymorphism tunes frontolimbic circuitry during affective contextual learning. NeuroImage, 162, 373–383. https://doi.org/10.1016/j.neuroimage.2017.08.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B. B. (2011). Current trends in stroke rehabilitation. A review with focus on brain plasticity. Acta Neurologica Scandinavica, 123, 147–159. https://doi.org/10.1111/j.1600-0404.2010.01417.x [DOI] [PubMed] [Google Scholar]
- Kaplan E., Goodglass H., & Weintraub S. (1983). Boston Naming Test. Philadelphia, PA: Lea & Fabiger. [Google Scholar]
- Kauhanen M. L., Korpelainen J. T., Hiltunen P., Määttä R., Mononen H., Brusin E., … Myllylä V. V. (2000). Aphasia, depression, and non-verbal cognitive impairment in ischemic stroke. Cerebrovascular Diseases, 10(6), 455–461. https://doi.org/10.1159/000016107 [DOI] [PubMed] [Google Scholar]
- Kertesz A. (2007). Western Aphasia Battery–Revised (WAB-R). San Antonio, TX: Pearson. [Google Scholar]
- Kim D. Y., Quinlan E. B., Gramer R., & Cramer S. C. (2016). BDNF Val66Met polymorphism is related to motor system function after stroke. Physical Therapy, 96, 533–539. https://doi.org/10.2522/ptj.20150135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleim J. A., Chan S., Pringle E., Schallert K., Procaccio V., Jimenez R., & Cramer S. C. (2006). BDNF val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nature Neuroscience, 9(6), 735–737. https://doi.org/10.1038/nn1699 [DOI] [PubMed] [Google Scholar]
- Lam J. M., & Wodchis W. P. (2010). The relationship of 60 disease diagnoses and 15 conditions to preference-based health-related quality of life in Ontario hospital-based long-term care residents. Medical Care, 48(4), 380–387. https://doi.org/10.1097/MLR.0b013e3181ca2647 [DOI] [PubMed] [Google Scholar]
- Lamb Y. N., McKay N. S., Thompson C. S., Hamm J. P., Waldie K. E., & Kirk I. J. (2015). Brain-derived neurotrophic factor val66met polymorphism, human memory, and synaptic neuroplasticity. Wiley Interdisciplinary Reviews: Cognitive Science, 6, 97–108. https://doi.org/10.1002/wcs.1334 [DOI] [PubMed] [Google Scholar]
- Lanyon L. E., Rose M. L., & Worrall L. (2013). The efficacy of outpatient and community-based aphasia group interventions: A systematic review. International Journal of Speech-Language Pathology, 15(4), 359–374. https://doi.org/10.3109/17549507.2012.752865 [DOI] [PubMed] [Google Scholar]
- Laska A. C., Hellblom A., Murray V., Kahan T., & Von Arbin M. (2001). Aphasia in acute stroke and relation to outcome. Journal of Internal Medicine, 249(5), 413–422. https://doi.org/10.1046/j.1365-2796.2001.00812.x [DOI] [PubMed] [Google Scholar]
- Lazar R. M., & Antoniello D. (2008). Variability in recovery from aphasia. Current Neurology and Neuroscience Reports, 8(6), 497–502. https://doi.org/10.1007/s11910-008-0079-x [DOI] [PubMed] [Google Scholar]
- Lazar R. M., Speizer A. E., Festa J. R., Krakauer J. W., & Marshall R. S. (2008). Variability in language recovery after first-time stroke. Journal of Neurology, Neurosurgery, & Psychiatry, 79, 530–534. https://doi.org/10.1136/jnnp.2007.122457 [DOI] [PubMed] [Google Scholar]
- Leff A., Crinion J., Scott S., Turkheimer F., Howard D., & Wise R. (2002). A physiological change in the homotopic cortex following left posterior temporal lobe infarction. Annals of Neurology, 51(5), 553–558. https://doi.org/10.1002/ana.10181 [DOI] [PubMed] [Google Scholar]
- Lessmann V., & Brigadski T. (2009). Mechanisms, locations, and kinetics of synaptic BDNF secretion: An update. Neuroscience Research, 65, 11–22. https://doi.org/10.1016/j.neures.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Lu B. (2003). BDNF and activity-dependent synaptic modulation. Learning and Memory, 10, 86–98. https://doi.org/10.1101/lm.54603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic S., Barbieri E., Wang X., Caplan D., Kiran S., Rapp B., … Thompson C. K. (2017). Right hemisphere grey matter volume and language functions in stroke aphasia. Neural Plasticity, 2017, 14 https://doi.org/10.1155/2017/5601509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangolo P., Fiori V., Gelfo F., Shofany J., Razzano C., Caltagirone C., & Angelucci F. (2014). Bihemispheric tDCS enhances language recovery but does not alter BDNF levels in chronic aphasic patients. Restorative Neurology and Neuroscience, 32, 367–379. https://doi.org/10.3233/RNN-130323 [DOI] [PubMed] [Google Scholar]
- McHughen S. A., Rodriguez P. F., Kleim J. A., Kleim E. D., Marchal Crespo L., Procaccio V., & Cramer S. C. (2010). BDNF val66met polymorphism influences motor system function in the human brain. Cerebral Cortex, 20(5), 1254–1262. https://doi.org/10.1093/cercor/bhp189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinzer M., & Breitenstein C. (2008). Functional imaging studies of treatment-induced recovery in chronic aphasia. Aphasiology, 22(12), 1251–1268. https://doi.org/10.1080/02687030802367998 [Google Scholar]
- Minichiello L., Korte M., Wolfer D., Kühn R., Unsicker K., Cestari C., … Klein R. (1999). Essential role for TrkB receptors in hippocampus-mediated learning. Neuron, 24, 401–414. https://doi.org/10.1016/S0896-6273(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Mirowska-Guzel D., Gromadzka G., Seniow J., Lesniak M., Bilik M., Waldowski K., … Czlonkowska A. (2013). Association between BDNF-196 G>A and BDNF-270 C>T polymorphism, BDNF concentration, and rTMS-supported long-term rehabilitation outcome after ischemic stroke. NeuroRehabilitation, 32(3), 573–582. https://doi.org/10.3233/NRE-130879 [DOI] [PubMed] [Google Scholar]
- Musso M., Weiller C., Kiebel S., Müller S. P., Bülau P., & Rijntjes M. (1999). Training-induced brain plasticity in aphasia. Brain, 122, 1781–1790. https://doi.org/10.1093/brain/122.9.1781 [DOI] [PubMed] [Google Scholar]
- Nachev P., Coulthard E., Jäger H., Kennard C., & Husain M. (2008). Enantiomorphic normalization of focally lesioned brains. NeuroImage, 39(3-3), 1215–1226. https://doi.org/10.1016/j.neuroimage.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeser M. A., Baker E. H., Palumbo C. L., Nicholas M., Alexander M. P., Samaraweera R., … Weissman T. (1998). Lesion site patterns in severe, nonverbal aphasia to predict outcome with a computer-assisted treatment program. Archives of Neurology, 55(11), 1438–1448. https://doi.org/10.1001/archneur.55.11.1438 [DOI] [PubMed] [Google Scholar]
- Naeser M. A., Martin P. I., Nicholas M., Baker E. H., Seekins H., Helm-Estabrooks N., … Pascual-Leone A. (2005). Improved naming after TMS treatments in a chronic, global aphasia patient—Case report. Neurocase, 11(3), 182–193. https://doi.org/10.1080/13554790590944663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeser M. A., Martin P. I., Nicholas M., Baker E. H., Seekins H., Kobayashi M., … Pascual-Leone A. (2005). Improved picture naming in chronic aphasia after TMS to part of right Broca's area: An open-protocol study. Brain and Language, 93(1), 95–105. https://doi.org/10.1016/j.bandl.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Pang P. T., Teng H. K., Zaitsev E., Woo N. T., Sakata K., Zhen S., … Lu B. (2004). Cleavage of proBDNF by tPA/plasmin is essential for long-term hippocampal plasticity. Science, 306, 487–491. https://doi.org/10.1126/science.1100135 [DOI] [PubMed] [Google Scholar]
- Parr S. (2007). Living with severe aphasia: Tracking social exclusion. Aphasiology, 21(1), 98–123. https://doi.org/10.1080/02687030600798337 [Google Scholar]
- Pedersen P. M., Jørgensen H. S., Nakayama H., Raaschou H. O., & Skyhoj T. (1995). Aphasia in acute stroke: Incidence, determinants, and recovery. Annals of Neurology, 38(4), 659–666. https://doi.org/10.1002/ana.410380416 [DOI] [PubMed] [Google Scholar]
- Petryshen T. L., Sabeti P. C., Aldinger K. A., Fry B., Fan J. B., Schaffner S. F., … Sklar P. (2010). Population genetic study of the brain-derived neurotrophic factor (BDNF) gene. Molecular Psychiatry, 15(8), 810–815. https://doi.org/10.1038/mp.2009.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L., Verchinski B. A., Mattay V. S., Callicott J. H., Kolachana B. S., Straub R. E., … Weinberger D. R. (2004). The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. Journal of Neuroscience, 24(45), 10099–10102. https://doi.org/10.1523/JNEUROSCI.2680-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach A., Schwartz M. F., Martin N., Grewal R. S., & Brecher A. (1996). The Philadelphia Naming Test: Scoring and rationale. Clinical Aphasiology, 24, 121–133. Retrieved from http://psycnet.apa.org/record/2017-27183-001 [Google Scholar]
- Rorden C., Bonilha L., Fridriksson J., Bender B., & Karnath H. O. (2012). Age-specific CT and MRI templates for spatial normalization. NeuroImage, 61(4), 957–965. https://doi.org/10.1016/j.neuroimage.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden C., & Brett M. (2000). Stereotaxic display of brain lesions. Behavioural Neurology, 12, 191–200. https://doi.org/10.1155/2000/421719 [DOI] [PubMed] [Google Scholar]
- Saur D., Lange R., Baumgaertner A., Schraknepper V., Willmes K., Rijntjes M., & Weiller C. (2006). Dynamics of language reorganization after stroke. Brain, 129, 1371–1384. https://doi.org/10.1093/brain/awl090 [DOI] [PubMed] [Google Scholar]
- Schäbitz W. R., Berger C., Kollmar R., Seitz M., Tanay E., Kiessling M., … Sommer C. (2004). Effect of brain-derived neurotrophic factor treatment and forced arm use on functional motor recovery after small cortical ischemia. Stroke, 35, 992–997. https://doi.org/10.1161/01.STR.0000119754.85848.0D [DOI] [PubMed] [Google Scholar]
- Shimizu E., Hashimoto K., & Iyo M. (2004). Ethnic difference of the BDNF 196 G/A (Val66Met) polymorphism frequencies: The possibility to explain ethnic mental traits. American Journal of Medical Genetics: Part B: Neuropsychiatric Genetics, 126, 122–123. https://doi.org/10.1002/ajmg.b.20118 [DOI] [PubMed] [Google Scholar]
- Simmons-Mackie N. (2018). Aphasia in North America. Moorestown, NJ: Aphasia Access. [Google Scholar]
- Strand E. A., Duffy J. R., Clark H. M., & Josephs K. (2014). The Apraxia of Speech Rating Scale: A tool for diagnosis and description of apraxia of speech. Journal of Communication Disorders, 51, 43–50. https://doi.org/10.1016/j.jcomdis.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaflarski J. P., Allendorfer J. B., Banks C., Vannest J., & Holland S. K. (2013). Recovered vs. not-recovered from post-stroke aphasia: The contributions from the dominant and non-dominant hemispheres. Restorative Neurology and Neuroscience, 31(4), 347–360. https://doi.org/10.3233/RNN-120267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeszko P. R., Lipsky R., Mentschel C., Robinson D., Gunduz-Bruce H., Sevy S., … Malhotra A. K. (2005). Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Molecular Psychiatry, 10, 631–636. https://doi.org/10.1038/sj.mp.4001656 [DOI] [PubMed] [Google Scholar]
- Teasell R., Bayona N. A., & Bitensky J. (2015). Plasticity and reorganization of the brain post stroke. Topics in Stroke Rehabilitation, 12(3), 11–26. https://doi.org/10.1310/6AUM-ETYW-Q8XV-8XAC [DOI] [PubMed] [Google Scholar]
- Thiel A., Habedank B., Herholz K., Kessler J., Winhuisen L., Haupt W. F., & Heiss W. D. (2006). From the left to the right: How the brain compensates progressive loss of language function. Brain and Language, 98(1), 57–65. https://doi.org/10.1016/j.bandl.2006.01.007 [DOI] [PubMed] [Google Scholar]
- Thompson C. K., den Ouden D. B., Bonakdarpour B., Garibaldi K., & Parrish T. B. (2010). Neural plasticity and treatment-induced recovery of sentence processing in agrammatism. Neuropsychologia, 48(11), 3211–3227. https://doi.org/10.1016/j.neuropsychologia.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub P. E. (2015). Brain stimulation and the role of the right hemisphere in aphasia recovery. Current Neurology and Neuroscience Reports, 15(11), 72 https://doi.org/10.1007/s11910-015-0593-6 [DOI] [PubMed] [Google Scholar]
- Turkeltaub P. E., Coslett H. B., Thomas A. L., Faseyitan O., Benson J., Norise C., & Hamilton R. H. (2012). The right hemisphere is not unitary in its role in aphasia recovery. Cortex, 48, 1179–1186. https://doi.org/10.1016/j.cortex.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet R., Ribbers G. M., Vandermeeren Y., Frens M. A., & Selles R. W. (2017). BDNF Val66Met but not transcranial direct current stimulation affects motor learning after stroke. Brain Stimulation, 10(5), 882–892. https://doi.org/10.1016/j.brs.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Van Oene M., Alic S., Jackson A., & Lem P. (2006). SNP genotyping of DNA from Oragene®/saliva samples with SNPstream®. Retrieved from http://www.dnagenotek.com/ROW/pdf/MK-AN-014.pdf
- Vickers C. P. (2010). Social networks after the onset of aphasia: The impact of aphasia group attendance. Aphasiology, 24(6–8), 902–913. https://doi.org/10.1080/02687030903438532 [Google Scholar]
- Warburton E., Price C. J., Swinburn K., & Wise R. J. (1999). Mechanisms of recovery from aphasia: Evidence from positron emission tomography studies. Journal of Neurology, Neurosurgery, & Psychiatry, 66, 155–161. https://doi.org/10.1136/jnnp.66.2.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. (1997). Wechsler Adult Intelligence Scale–Third Edition (WAIS-III). San Antonio, TX: NCS Pearson. [Google Scholar]
- Winhuisen L., Thiel A., Schumacher B., Kessler J., Rudolf J., Haupt W. F., & Heiss W. D. (2005). Role of the contralateral inferior frontal gyrus in recovery of language function in poststroke aphasia: A combined repetitive transcranial magnetic stimulation and positron emission tomography study. Stroke, 36, 1759–1763. https://doi.org/10.1161/01.STR.0000174487.81126.ef [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Ochi H., Isobe N., Matsushita T., Motomura K., Matsuoka T., … Kira J. (2010). Altered production of brain-derived neurotrophic factor by peripheral blood immune cells in multiple sclerosis. Multiple Sclerosis, 16, 1178–1188. https://doi.org/10.1177/1352458510375706 [DOI] [PubMed] [Google Scholar]
- Yourganov G., Fridriksson J., Stark B., & Rorden C. (2018). Removal of artifacts from resting-state fMRI data in stroke. NeuroImage Clinical, 17, 297–305. https://doi.org/10.1016/j.nicl.2017.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., & Pardridge W. M. (2006). Blood-brain barrier targeting of BDNF improves motor function in rats with middle cerebral artery occlusion. Brain Research, 1111, 227–229. https://doi.org/10.1016/j.brainres.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Zhao L. R., Risedal A., Wojcik A., Hejzlar J., Johansson B. B., & Kokaia Z. (2001). Enriched environment influences brain-derived neurotrophic factor levels in rat forebrain after focal stroke. Neuroscience Letters, 305, 169–172. https://doi.org/10.1016/S0304-3940(01)01837-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.