The generation of gene regulatory networks from thousands of maize transcriptome data sets identifies putative transcription factor targets and candidate regulators of important metabolic pathways.
Abstract
The regulation of gene expression is central to many biological processes. Gene regulatory networks (GRNs) link transcription factors (TFs) to their target genes and represent maps of potential transcriptional regulation. Here, we analyzed a large number of publically available maize (Zea mays) transcriptome data sets including >6000 RNA sequencing samples to generate 45 coexpression-based GRNs that represent potential regulatory relationships between TFs and other genes in different populations of samples (cross-tissue, cross-genotype, and tissue-and-genotype samples). While these networks are all enriched for biologically relevant interactions, different networks capture distinct TF-target associations and biological processes. By examining the power of our coexpression-based GRNs to accurately predict covarying TF-target relationships in natural variation data sets, we found that presence/absence changes rather than quantitative changes in TF gene expression are more likely associated with changes in target gene expression. Integrating information from our TF-target predictions and previous expression quantitative trait loci (eQTL) mapping results provided support for 68 TFs underlying 74 previously identified trans-eQTL hotspots spanning a variety of metabolic pathways. This study highlights the utility of developing multiple GRNs within a species to detect putative regulators of important plant pathways and provides potential targets for breeding or biotechnological applications.
INTRODUCTION
A central goal in linking genotype to phenotype is to understand how a limited number of transcription factors (TFs) drive dynamic gene expression changes in different cell types and environmental conditions. Based on the genomes of well-characterized model systems, multicellular organisms dedicate a substantial portion of their protein-coding genes (6 to 8%) to the expression of TFs (Babu et al., 2004). TFs recognize specific cis-regulatory elements and activate or repress the transcription of specific sets of target genes by interacting with other TFs, coregulators, chromatin modifiers, and the basal transcription machinery. TFs are crucial for many important cellular processes and are widely involved in development, responses to the environment, cell cycle control, and responses to pathogens (Lee and Young, 2013). Therefore, characterizing the TF regulatory landscape within an organism is critical to expanding our knowledge of complex phenotypic traits and gene expression networks. However, genome-wide characterization of TF-target associations are only available for a handful of TFs in most crop species, such as maize (Zea mays; Ravasi et al., 2010; Nègre et al., 2011; Gerstein et al., 2012; Araya et al., 2014; Bartlett et al., 2017). In some cases, a specific TF can regulate multiple genes that are part of the same biochemical pathway, providing higher level control of cellular functions. This can make TFs an attractive target for modulation through breeding or biotechnology approaches.
A variety of approaches are available to link TFs to target genes (Springer et al., 2019). Chromatin immunoprecipitation sequencing (ChIP-seq) is a powerful approach to discover the binding sites of a particular TF (Johnson et al., 2007; Kheradpour and Kellis, 2014). In combination with transcriptome profiling of mutant stocks, ChIP-seq has been widely used to elucidate the regulatory functions of specific TFs (Robertson et al., 2007). However, ChIP-seq experiments have been generally limited in scale due to the difficulty in their execution, sensitivity to antibody quality, and inability to work when using rare or poorly expressed proteins (Kidder et al., 2011). As a result, compared to the human Encyclopedia of DNA Elements (ENCODE) project in which ChIP-seq was conducted for 694 TFs (Davis et al., 2018), even in a well-studied plant model such as Arabidopsis (Arabidopsis thaliana), there are fewer than 50 TFs with ChIP-seq/Chip data (Mathys et al., 2006; Weirauch et al., 2014; Khan et al., 2018). In maize, only eight ChIP-seq data sets have been published to date (Bolduc et al., 2012; Morohashi et al., 2012; Eveland et al., 2014; Pautler et al., 2015; Yang et al., 2016; Li et al., 2018; Zhan et al., 2018; Dong et al., 2019). The recently developed DNA affinity purification sequencing (DAP-seq) technique using an in vitro–expressed, affinity-tagged TF in combination with high-throughput genomic DNA sequencing offers a promising approach to efficiently generate genome-wide TF-target interaction maps (O’Malley et al., 2016; Bartlett et al., 2017). However, this approach has not yet been comprehensively applied to study maize regulatory landscape (Galli et al., 2018; Ricci et al., 2019) and may not reflect in vivo regulatory interactions that occur in a native chromatin context.
Homology-based attempts to computationally predict TF binding sites (TFBSs) based on existing TF binding motifs and the conservation of TFBSs among species have provided predictions of binding sites in many plants (Yilmaz et al., 2009; Chow et al., 2016; Jin et al., 2017). However, these approaches typically have a high false positive discovery rate, as there is accumulating evidence that supports the widespread contributions of sequence context in modulated sequence recognition, including flanking sequences, DNA secondary structures, and chromatin status (Siggers and Gordân, 2014). Although TFs are frequently classified as activators or repressors, gene regulation is typically controlled through the combinatorial control of different TFs, where context dependency specifies how TFs modulate the expression of target genes (Mejia-Guerra et al., 2012; Siggers and Gordân, 2014; Li et al., 2015b).
An alternative yet powerful approach to infer regulatory networks is through the use of statistical inference algorithms or machine learning techniques applied to gene expression data. The coexpression-based gene regulatory network (GRN) is an effective tool for identifying genes with essential biological functions or genes involved in a specific pathway or process (Hecker et al., 2009; Krouk et al., 2013). Inference methods that utilize transcript abundance data and reveal connections between genes have been used to construct GRNs and find important genes and regulatory relationships involved in plant growth and developmental processes, such as cell wall synthesis (Taylor-Teeples et al., 2015), regeneration (Ikeuchi et al., 2018), and root hair growth (Shibata et al., 2018). In addition, the computational inference of GRNs can help prescreen in silico potential interactions to allow focused validation of high-confidence interactions (Bassel et al., 2012). While there have been notable successes in applying coexpression-based GRNs to identify important regulatory networks in plant species, there remains a substantial gap in our knowledge of how to develop GRNs from large-scale transcriptome data sets that are currently available for optimal use in crop improvement.
Maize is an important crop species with substantial genetic and genomic resources. The B73 reference genome (AGP_v4) is a chromosome-level, high-quality assembly with well-curated gene ontology (GO)–based functional annotations (Jiao et al., 2017; Wimalanathan et al., 2018). The maize TFome project provides an invaluable resource of more than 2000 maize TF clones to facilitate high-throughput studies, including a recent yeast one-hybrid screen that identified more than a thousand TF-target interactions in the maize phenolic metabolic pathway (Burdo et al., 2014; Yang et al., 2017). Previous efforts in characterizing GRNs in maize have provided insights into regulatory networks (Li et al., 2010; Zhan et al., 2015; Walley et al., 2016; Huang et al., 2018). A maize leaf network constructed along a leaf developmental gradient revealed a dynamic transcriptome, with transcripts for basic cellular metabolism at the leaf base transitioning to transcripts for secondary cell wall biosynthesis and C4 photosynthetic development toward the tip (Li et al., 2010). Similarly, a maize endosperm network constructed using nine different endosperm, embryo, and kernel tissues/developmental stages identified an unexpected close correlation between the embryo and the aleurone layer of the endosperm (Zhan et al., 2015). Regulatory networks constructed using 23 different tissues sampled across maize developmental stages show very different topology at the mRNA level versus the protein (phosphoprotein) level, with more than 85% of regulatory hubs not conserved between the RNA network and the protein network (Walley et al., 2016). By utilizing publicly available RNA-seq data sets and the GENIE3 algorithm (Huynh-Thu et al., 2010), four tissue-specific (leaf, root, shoot apical meristem, and seed) networks were constructed, identifying well-studied key TFs in each network and revealing very different regulatory functions for many TFs (Huang et al., 2018).
In this study, we used a large number of RNA-seq data sets from maize to develop 45 different coexpression-based GRNs. The use of coexpression-based GRNs that are based upon sampling of many tissues for a single genotype (developmental atlases) or many different genotypes for a single tissue (genotype surveys) allows for widespread sampling of potential gene regulatory relationships and the detection of GRNs that are only found in some networks. We provide evidence that these networks are enriched for biologically meaningful connections and that different networks sample distinct processes or TFs. By comparing the predicted networks with data from natural variation surveys or expression quantitative trait loci (eQTLs), we identified a subset of regulatory interactions that are experimentally supported and may be important for explaining trans-eQTLs and the regulation of metabolic pathways.
RESULTS
Construction of Maize GRNs
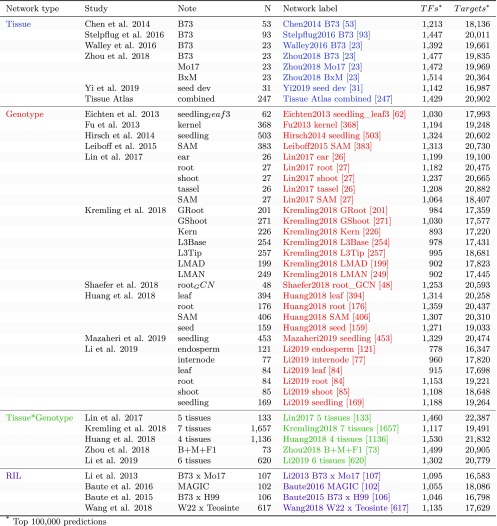
Many RNA-seq data sets are available for maize, including surveys of different tissues within a single genotype as well as surveys of expression in a single tissue across diverse germplasm. We used these data sets to generate putative GRNs based upon the expression patterns of TFs and their target genes. The set of maize TFs were obtained from PlantTFDB (Jin et al., 2017). In total, 45 putative GRNs were developed for 25 different maize RNA-seq expression data sets that were all aligned to the B73_v4 genome and normalized using consistent methods (see Methods; Figure 1). We evaluated several methods for GRN construction and found that the random forest approach (Huynh-Thu et al., 2010) provided the best performance (see Supplemental Figure 1 and Supplemental Methods for details). The expression data sets include eight developmental networks built using different tissues/developmental stages of the same genotype (five independent data sets for B73, one for Mo17, and one for B73xMo17 as well as one combined data set including 247 different tissues or stages of B73; see Methods), 28 tissue-specific networks built using the same tissue sampled from multiple inbred lines, 5 tissue-genotype networks that include multiple tissues sampled from a panel of inbred lines, and 4 networks built using recombinant inbred populations (B73xMo17, B73xH99, maize x teosinte and multiparent advanced generation intercross [MAGIC] recombinant inbred lines [RILs]; see Figure 1 for details of networks and references). For comparison purposes, we also included maize coexpression-based GRNs generated in two recent studies (Walley et al., 2016; Huang et al., 2018). These include a maize developmental network (Walley et al., 2016) as well as four tissue-specific networks (leaf, root, shoot apical meristem, and seed; Huang et al., 2018). For these previously generated networks, we downloaded and mapped the raw sequence reads and built the regulatory networks using the same pipeline used in this study. By including networks focused on specific tissues or genotypes as well as meta-networks, we were able to investigate the relative utility of focused networks as well as networks that use a more comprehensive set of data.
Figure 1.
Data Sets and GRNs Developed for This Study.
Each of the coexpression-based GRNs created for this study is listed. They are separated into GRNs that utilize different tissues of B73 (blue), different genotypes (red), combinations of multiple tissues and genotypes (green), and recombinant inbred panels (purple). For each GRN, we describe the reference study, the tissue or genotype, the number of samples, and the number of TFs and targets that are classified within the top 100k edges for the GRN. The network labels from this figure are used for the remaining figures.
Evaluation of Maize GRNs Using TF Knockouts and Functional Annotations
We used several approaches to evaluate whether the putative GRNs were enriched for biologically significant edges. One approach that can be used to document the validity of GRNs is to assess the enrichment of known targets based on analysis of TF mutant RNA-seq and ChIP-seq data. Previous studies have characterized direct targets of six maize TFs using the mutant/wild-type RNA-seq combined with ChIP-seq experiments (Supplemental Table 1; Bolduc et al., 2012; Eveland et al., 2014; Pautler et al., 2015; Li et al., 2015a, 2018; Dong et al., 2019); these data sets were previously used to validate maize GRNs (Walley et al., 2016; Huang et al., 2018). We assessed the TF-target interactions in each coexpression-based GRN (rows in Figure 2) against TF-target interactions from ChIP-seq studies (columns in Figure 2A).
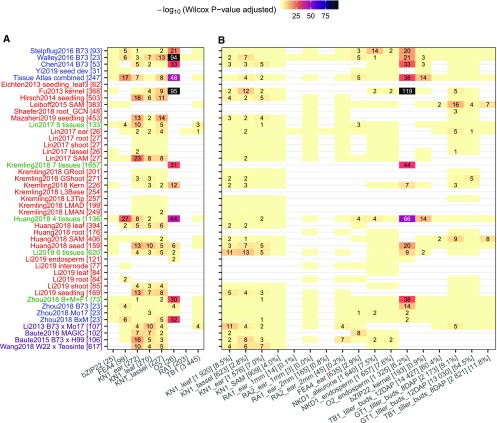
Figure 2.
TF-Target Interactions Predicted by GRNs Are Supported by Experimentally Derived TF Targets and Knockout Mutant RNA-Seq Experiments.
(A) Direct targets of published TF studies derived from ChIP-seq and mutant RNA-seq experiments.
(B) For each of the 17 maize TFs with knockout mutant RNA-seq data available, DEGs between the mutant and the wild type were identified using DESeq2 (P-value < 0.01). Wilcox rank tests were then performed using the predicted (TF-target) interaction scores (top 100k edges) between the group of true targets (DEGs) and nontargets (non_DEGs). P-values were adjusted by the Benjamini–Hochberg method implemented in R. Numbers in each cell show the adjusted test P-value (−log10 transformed). Supplemental Figure 2 provides the actual number of true targets captured by each GRN during each evaluation. Light yellow cells with no numbers indicate not significant (P > 0.05), while blank (white) cells indicate missing data where the TF being evaluated is not expressed or not variable (i.e., zero variance) in the corresponding GRN. The y axis labels correspond to the different networks listed in Figure 1. The x axis labels (e.g., KN1_ear (272) or KN1_ear [1576] [7.0%]) represent the common name for each TF, the tissue in which the TF is expressed, followed by the number of direct targets (A) or number and proportion of DEGs in TF mutant (B).
There was significant enrichment for direct targets in at least one network for four of the six TFs (Figure 2A; Supplemental Figures 2 and 3). However, the level of enrichment varied substantially, likely due to whether the TF is expressed or exhibits variable expression in each network based on the tissues or genotypes that are contained in the data set used to generate the network. It is worth noting that there are relatively few examples in which the TFs exhibit variable expression that do not show enrichment for targets (yellow cells in Figure 2A). Instead, the lack of significant enrichment often reflects either the lack of expression or lack of variable expression levels. For example, the Opaque2 (O2; Zm00001d018971) TF gene is only expressed in endosperm tissue and therefore the O2 interactions are not detected in GRNs built using only vegetative tissues. There is also enrichment for some sets of TF-targets identified by DAP-seq (Supplemental Figure 4; Galli et al., 2018; Ricci et al., 2019). While these direct targets were identified by overlaying ChIP-seq binding evidence with RNA-seq data for differentially expressed genes (DEGs) between the TF mutant and the wild type, it is likely that coexpression-based GRN predicted TF-target interactions include both direct and indirect regulatory relationships.
We used a slightly different approach to utilize all DEGs (with and without ChIP-seq binding evidence) from the TF mutants relative to the wild type to evaluate enrichment for both direct and indirect targets. We obtained paired RNA-seq data with at least two biological replicates (separate experiments) for both the mutant and the wild type from public databases for 17 maize TFs (Supplemental Table 2). For most TFs (14 of 17), at least one of the coexpression-based GRNs assigns significantly higher interaction weights to true targets (DEGs between the TF mutant and the wild type) than nontargets (non-DEGs; Figure 2B; Supplemental Figure 5). Developmental networks tend to capture regulatory relationships for many TFs, while tissue-specific networks typically generate the best performance at predicting TFs that are specific to the corresponding tissue. For example, the combined developmental network accurately predicts targets for several maize TFs including ear-expressed fasciated ear4 (fea4; Zm00001d037317), pericarp-expressed ufo1 (Zm00001d000009), and endosperm-expressed bZIP22 (Zm00001d021191), O2, and nkd (Zm00001d002654). However, O2, which is mostly expressed in kernels and endosperm (Li et al., 2015a; Zhan et al., 2018), is best predicted by a kernel network built by 368 different inbreds (Fu et al., 2013). Likewise, the Hirsch2014 seedling network only shows enrichment in the meristem-expressed KN1 (Figure 2B; Supplemental Figure 5).
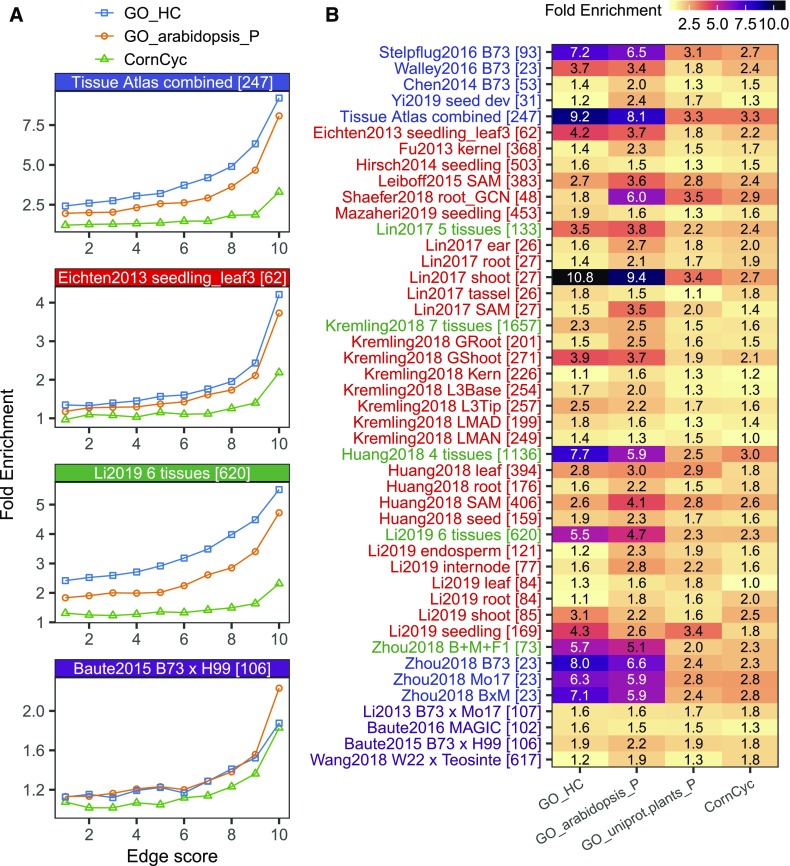
The second approach used to evaluate potential functional enrichments within the coexpression-based GRNs was to investigate how often a GRN would contain multiple genes annotated to the same GO categories or genes that are in the same metabolic pathway. For each of the networks, we documented the frequency of targets of a shared TF that are annotated with the same GO term (Wimalanathan et al., 2018) or were annotated in the same CornCyc metabolic pathway (see Methods; Andorf et al., 2016). We performed a permutation test of annotation terms to determine the expected number of shared terms among targets. Enrichment (observed shared terms/expected shared terms) was assessed for each GO/CornCyc category in each network (see Methods; Figure 3; Supplemental Figure 6). We performed enrichment analyses of shared GO terms or metabolic pathways for 10 bins of 100,000 (100k) edges in each network (based on predicted edge score). All networks exhibit significant enrichment for shared annotations of multiple targets of the same TF, and these remained significant when GO terms based on expression evidence were omitted (Supplemental Figure 6). In all cases, the enrichment was much greater for the higher ranking edges (i.e., with stronger interaction scores), with the top 100k edges showing the greatest enrichment (Figure 3A; Supplemental Figure 6). Among the developmental networks, the level of enrichment for shared annotations was generally related to the number of samples in the network, with the combined set exhibiting the greatest enrichment (Figure 2B). Within the networks that utilized diverse genotypes, there was significant variability for the enrichments for shared annotations; this variability was not clearly related to specific tissue types or the size of the networks. These observations suggest that many of these GRNs capture information that can predict pathways or biological functions that are regulated by specific TFs.
Figure 3.
Enrichment of Coannotated GO/CornCyc Terms in Coregulated Network Targets.
For each network, the top 1 million predicted TF-target associations were binned to 10 bins and assessed for enrichment of GO/CornCyc functional annotation. Fold enrichment is calculated as the observed number of shared GO/CornCyc terms (by targets regulated by a common TF) divided by the expected number of shared annotation terms (determined by permutation).
(A) GO/CornCyc enrichment is shown for four selected networks.
(B) Heatmap showing enrichment of coannotated GO/CornCyc terms in the first bin (i.e., top 100k) of edges in the GRNs. See Supplemental Figure 7 for the enrichment in all bins of all newly built GRNs. A total of six sources of GO annotation were used, but only three are shown here: GO_HC (high-quality hand-curated terms transferred from maize AGP_v3 annotation), GO_arabidopsis and GO_uniprot.plants (see Supplemental Figure 6 for a complete list).
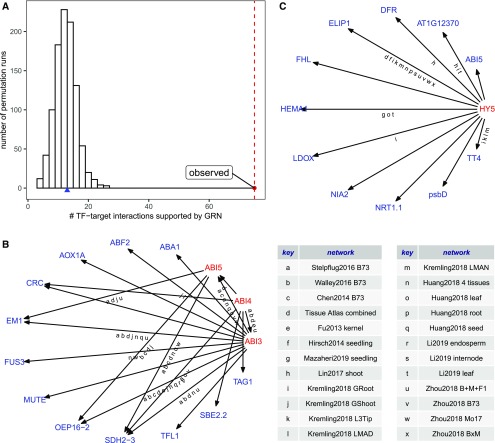
A third approach used to assess the ability of the GRNs to capture biologically relevant data was to transfer documented transcriptional regulations from Arabidopsis. We obtained 1431 high-confidence TF-target interactions in Arabidopsis from Arabidopsis Transcriptional Regulatory Map, which were collected based on systematic literature mining (Jin et al., 2015). Among these, 285 TF-target interactions could be mapped to homologous maize genes requiring both TF and target mapped to their least divergent homolog in maize. Despite the significant evolutionary distance between maize and Arabidopsis, 75 (26.5%) TF-target interactions are supported by at least 1 of the 45 GRNs (100k top edges), which is 5.8-fold higher than expected by chance (1000 permutations of the same number of 285 TF-target pairs show an average of 12.93 pairs of GRN support; Figure 4). Examples include the well-studied Elongated Hypocotyl5 (HY5; AT5G11260) TF-mediated pathway (Figure 3C; Supplemental Figure 7B) and the abscisic acid signaling pathway (Figure 3B; Supplemental Figure 8A). These two examples highlight the value of using multiple GRNs. In both cases, there is substantial variation in which TF-target interactions are detected in different GRNs (Figures 4B and 4C; Supplemental Figures 7A and 7B).
Figure 4.
GRN Predictions Show Enrichment of Documented Transcriptional Regulation Interactions from Arabidopsis.
(A) Permutation analysis showing the number of random TF-target interactions supported by at least one of the 45 GRNs (histogram) compared with the actual transcriptional regulation (transferred from Arabidopsis) with GRN support (red dashed line).
(B) Abscisic acid pathway transferred from Arabidopsis showed support for 12 of 20 edges.
(C) Six of 11 HY5 targets transferred from Arabidopsis showed support in at least one GRN. The letters along the edges of the networks in (B) and (C) indicate significant support from a specific GRN, as indicated in the key.
Together, the analysis of annotations and previously documented TF-target interactions suggests that the coexpression-based GRNs built for maize are enriched for functional interactions and provides evidence that the new set of networks generated for this study have equivalent or improved performance relative to previously published maize GRNs (Walley et al., 2016; Huang et al., 2018). We also found evidence for enrichment of predicted TF binding sites at the targets predicted by the GRNs (for details, see supplemental Methods and Supplemental Figure 8). These analyses also suggest that there is significant variability among networks in terms of which types of annotations, or which TFs, are effectively captured.
Comparison of GRNs
In this study, we developed a number of GRNs based on subsets of the data rather than simply making one larger meta-network or even a handful of meta-networks. Clearly, there are differences in GRNs that were predicted from different data sets, as some of the TFs are not expressed (or do not show variation) in some tissues and therefore will not have predicted targets. We sought to compare the information from different networks to evaluate the similarities/differences and to determine whether the meta-networks routinely provided advantages over networks focused on single tissues or genotypes.
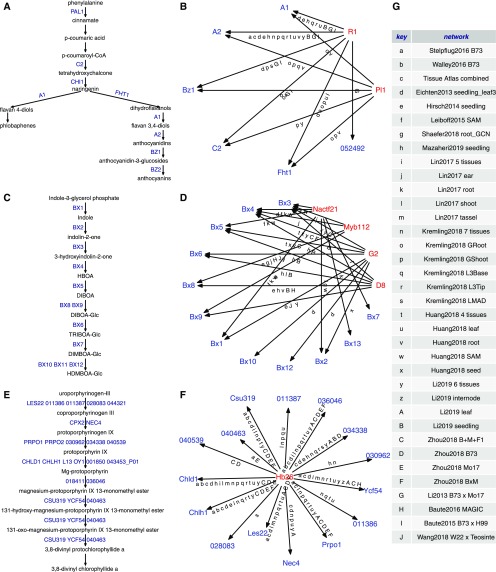
A clustering of the 45 networks identified some patterns of shared or distinct information (Supplemental Figure 9). However, there is also evidence for relatively distinct information in many of the networks, suggesting that pushing toward meta-networks may lose GRNs predicted in specific tissues or genotype panels (see supplemental Methods and Supplemental Figures 9 and 10 for details). There is also evidence that GRNs vary substantially in terms of the enrichment of specific GO terms (see supplemental Methods and Supplemental Figure 11). This suggests that distinct networks are likely capturing different information about potential biological functions. In order to highlight the potential value of using multiple GRNs, we selected several specific examples in which a TF is linked to multiple target genes within the same pathway to assess the information content in different coexpression-based GRNs (Figure 5; Supplemental Figures 12 to 14). The anthocyanin biosynthesis pathway is well characterized in maize, and there are known TFs that regulate multiple steps of this pathway (Figure 5A; Dooner et al., 1991; Petroni et al., 2014). Two of the TFs that are known to regulate structural genes in the anthocyanin biosynthesis pathway were identified in multiple GRNs (Figure 5B). However, any single network only detects a subset of these interactions (Supplemental Figure 12).
Figure 5.
Different Coexpression-Based GRNs Capture Distinct Aspects of Classic and CornCyc Metabolic Pathways.
(A) and (B) Anthocyanin biosynthesis pathway (A) regulated by R1 (Zm00001d026147) and PL1 (Zm00001d037118; see [B]).
(C) and (D) DIMBOA biosynthesis pathway (C) and four potential regulators (D): G2 (Zm00001d039260), D8 (Zm00001d033680), NACTF21 (Zm00001d036050), and MYB112 (Zm00001d046632).
(E) and (F) Chlorophyllide biosynthesis pathway (E) potentially regulated by HB26 (Zm00001d008612; see [F]).
(G) Network key mappings used in (B), (D), and (F). Mappings of reference gene IDs to aliases were obtained from MaizeGDB (https://maizegdb.org/associated_genes?type=allandstyle=table). For genes without aliases, the reference gene IDs were prefix-trimmed (Zm00001d) before displaying.
The 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) biosynthetic pathway has been well characterized in maize (Gierl and Frey, 2001), but there is little information about potential TFs that may regulate this pathway (Figure 5C). We identified four TFs with connections to multiple DIMBOA biosynthetic genes (Figure 5D). Often a single TF is linked to multiple structural genes, but the information content varies substantially between networks (Figure 5D; Supplemental Figure 13). In the case of the chlorophyllide biosynthesis pathway, there is a single TF that was identified as putatively regulating 15 genes in this pathway (Figures 5E and 5F). Often the associations were identified in many different GRNs, but in some cases, the association was only detected in a small number of GRNs (Supplemental Figure 14). Similar patterns are observed for putative TFs that may regulate multiple genes involved in zealexin biosynthesis, glycolysis, methylerythritol phosphate, and growth repression pathways (Supplemental Figure 15). These pathways provide examples of the relative value of collecting information from multiple networks to reveal potential connections between TFs and metabolic pathways.
Evaluation of Maize GRNs Based on Differential Gene Expression among Maize Genotypes
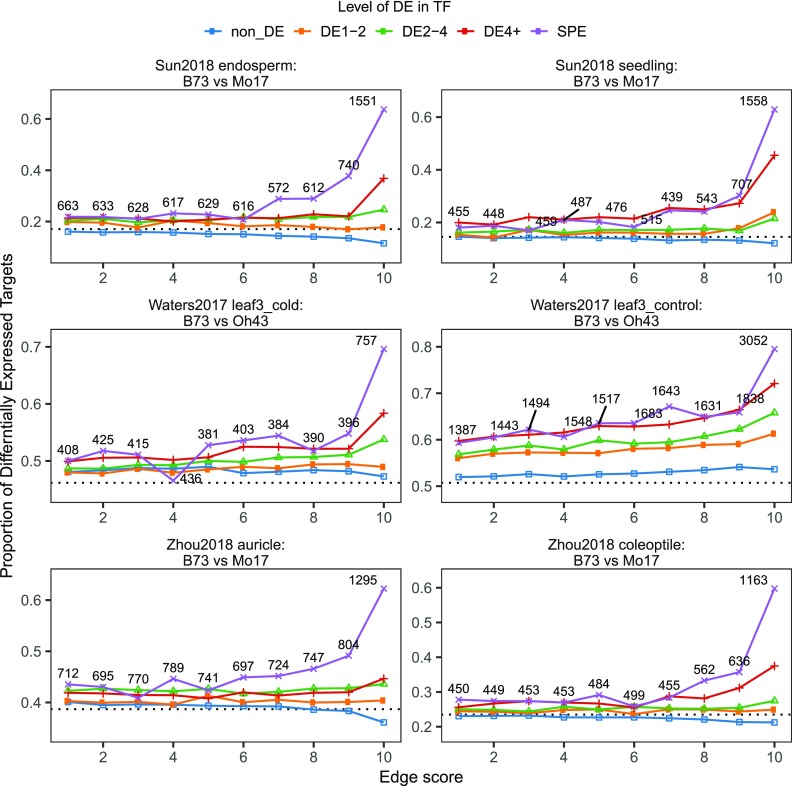
One goal in generating coexpression-based GRNs is to predict key TFs that could be modulated in order to affect changes in expression for sets of target genes. This would enable approaches that could control traits in a fashion that is often not achievable through changes in single enzymes (Grotewold, 2008). We sought to assess how often coexpression-based GRN-predicted associations would be supported using natural variation in gene expression based on comparisons of paired genotypes of maize. We selected four published transcriptomic studies that included at least two maize genotypes. These studies explored differences in transcriptome response between seedling leaf tissues of B73 and five other genotypes under control, cold, and heat conditions (Waters et al., 2017), between B73 and Mo17 root tissues under control and drought conditions (Marcon et al., 2017), between five tissues of B73 and Mo17 under standard growth conditions (Sun et al., 2018), and a tissue atlas spanning 23 paired tissues between B73 and Mo17 (Supplemental Table 3; Zhou et al., 2019). We obtained data for a total of 42 paired tissue/treatment samples with at least three biological replicates per sample between B73 and a different genotype (Supplemental Table 3) and identified DEGs between the two genotypes in each of the 42 paired tissue/treatment samples. In theory, if a regulator (TF) exhibits differential expression (DE) between the two genotypes in a certain tissue/condition, we anticipate that the targets of this TF (as predicted by coexpression-based GRNs) will also exhibit DE in the same tissue/condition. For every comparison, we classified each TF into different DE categories (non_DE, DE1-2, DE2-4, and DE4+ and single parent expression [SPE], where one genotype is not expressed) and assessed the proportion of predicted targets also exhibiting DE (Figure 6; Supplemental Figure 16). In addition, we binned the TF-target predictions in each network into 10 groups according to their interaction score predicted by the random forest regression model with the assumption that stronger TF-target interactions may receive stronger support from the paired genotype data sets (see Methods).
Figure 6.
TF-Target Validation of the Combined Tissue Network in Three Selected Natural Variation Data Sets.
Each panel shows the proportion of differentially expressed targets regulated by TFs showing different DE levels between two genotypes in one tissue/treatment condition. For each network, the top 1 million TF-target predictions were binned to 10 groups based on the interaction score in the GRN. Each TF-target pair is classified according to the DE level of the TF (non_DE, DE1-2, DE2-4, and DE4+, or SPE) in each network. The proportion of TF-target pairs with the target al.so showing DE was then determined for each category. Within each panel, the actual numbers of TF-target pairs falling into the SPE category (i.e., purple line) are labeled next to each point. Dashed line in each panel represents the genome-wide (background) proportion of DE genes in each tissue/treatment setting.
Our initial analysis focused on testing the predictions of the coexpression-based GRNs built using the combined tissue atlas representing 247 samples of B73. As expected, for TFs that do not exhibit DE (non_DE) or only show minor changes (DE1-2) between the two tested genotypes, the predicted targets show very little or no enrichment for DE (Figure 6; Supplemental Figure 9, blue lines). However, when the TF exhibits high levels of DE (e.g., DE4+) or SPE between the two genotypes, there is a markedly increased likelihood that the putative targets exhibit DE levels (Figure 6; Supplemental Figure 16, red and purple lines). In addition, the TF-target interaction score has a strong impact on the validation rate in paired genotype data sets, as observed for the functional annotation enrichments. The TF-target predictions with rank 10 (top 10% of predictions) exhibit a much higher proportion of targets being DE (60 to 80%) for TFs in the SPE groups (Figure 6; Supplemental Figure 16). These observations suggest that (1) presence/absence (i.e., SPE) rather than quantitative (i.e., DE1-2, DE2-4) changes in TF gene expression are much more likely to result in changes in target gene expression levels and (2) only the top network predictions with the highest interaction scores have high predictive power in paired genotype data sets. Therefore, we only utilized the rank 10 edges (TF-target interactions) in each network and focused on the group of TFs showing SPE patterns to compare the validation performance of different networks in each of the paired genotype data sets (by calculating the enrichment P-value of target DE proportion for TFs showing SPE pattern versus the background genome-wide DE rate; see Methods).
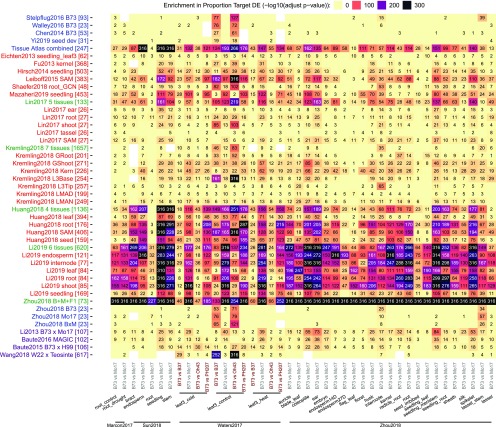
Several patterns emerge when comparing the levels of support for different GRNs based on paired genotype data sets. Developmental networks built within a single genotype generally have low power in predicting targets that are DE, with the exception of the combined atlas data set including 247 different tissues/stages (Figure 7, networks in blue). Despite being built using only tissues from the reference B73 genotype, the combined tissue network shows significant predictive power in almost all paired genotype data sets regardless of the two genotypes being compared (Figure 7). Not surprisingly, the strongest enrichment was observed using the paired B73 and Mo17 developmental atlas network (tissue atlas combined in Figure 7; Zhou et al., 2019) to predict DEGs between B73 and Mo17 samples, likely because the variation between these two genotypes was included within the information used to generate the GRNs. Indeed, this B73-Mo17 paired developmental network shows poorer predictive power whenever the genotypes compared in the validation data set include a different genotype such as Oh43 or PH207 (Figure 7). The same is true for the four RIL networks, with the B73xMo17 RIL network doing the best on comparisons between B73 and Mo17 samples and the B73xH99 RIL network showing the greatest performance on comparisons between B73 and non-Mo17 samples. The MAGIC RIL network, which is based on seven inbred parents, showed enrichment in a wider range of comparisons spanning more genotypes, while the maize W22-teosinte RIL network rarely predicts the transcriptional variation within maize populations with the exception of some B37 and Oh43 samples (Figure 7).
Figure 7.
Enrichment in Differentially Expressed Targets Regulated by TFs That Show SPE Patterns.
The color and number in each cell represent the enrichment P-value (−log10 transformed, Benjamini–Hochberg adjusted hypergeometric test P-value) of (SPE TF regulated) the proportion of the DE target relative to the genome-wide proportion of DEGs for each GRN (row-wise) evaluated against a tissue/treatment condition in a natural variation data set (column-wise). Only edges in the first bin (top 100k) of each network were taken.
Single-tissue networks generally show low predictive power unless the two genotypes in the validation data set being examined are among the genotype panel that was used to build the network (e.g., Li2019 endosperm network at predicting endosperm samples, Leiboff2015 shoot apical meristem network at predicting seedling and meristem samples). Meta-networks built from multiple tissues and multiple genotypes sometimes perform better than individual single-tissue networks (such as Li2017 networks), but in other cases show worse performance (such as the Huang2018 and Li2019 networks). Surprisingly, the largest network (Kremling2018 and subnetworks) rarely shows any predictive power in surveyed paired genotype data sets. This could be due to the different library creation methods used compared to the rest of the networks [3′ RNA-seq versus poly(A) RNA-seq], the relatively low sequencing depth (on average, three to five times), or the dilution of B73-Mo17 variation in the larger genotype panel (Figure 7).
Overlap with Trans-eQTL Hotspots
Three eQTL studies in maize have been published to date: one using 105 B73xMo17 RILs (Li et al., 2013), one using an association panel of 368 inbred lines (Fu et al., 2013; Liu et al., 2017), and one using 623 maize W22xteosinte RILs (Wang et al., 2018). These studies identified 96, 518, and 125 significant trans-eQTL hotspots, respectively, that each remotely regulate the expression levels of multiple target genes (Li et al., 2013; Liu et al., 2017; Wang et al., 2018). We asked whether the target genes associated with each hotspot tend to share a common TF regulator based on the TF-target predictions made by each coexpression-based GRN. In most cases, target genes from the same trans-eQTL hotspot are more likely to be regulated by a common TF (as predicted by GRNs) than a simulated data set with randomly assigned eQTL-eGene associations (see Methods; Supplemental Figures 17 and 18). In addition, the top 100,000 edges of each network consistently show much greater enrichment for coregulating trans-eQTL targets than the rest of the edges (i.e., top 200,000 to 1,000,000 edges; Supplemental Figure 17), which is consistent with the results of GO/CornCyc functional validation. We thus only focused on the top 100k edges in each network for subsequent analyses. Interestingly, the two biparental RIL networks (Li2013 B73xMo17 and Baute2015 B73xH99) show much stronger enrichment for regulating the trans-eQTL targets than the W22xTeosinte RIL network and the multi-parent MAGIC network (Supplemental Figure 18). Fifteen nonredundant networks showing the highest enrichment among the coexpression-based GRNs were chosen for subsequent analysis (Supplemental Figure 18).
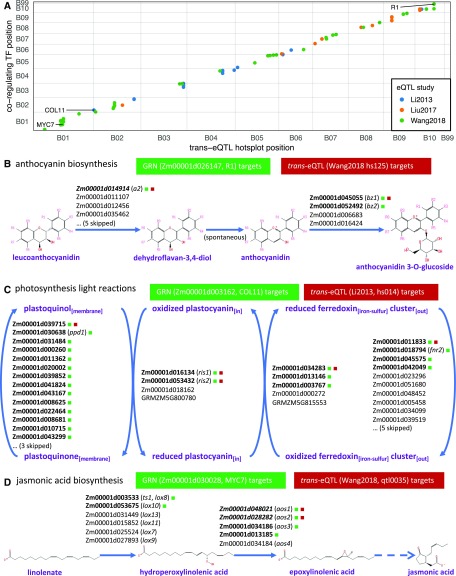
In total, 372 TFs were identified in at least 2 of the 15 high-quality networks that show significant coregulation with at least one trans-eQTL hotspot (hypergeometric test enrichment P-value < 0.01; see Methods). When plotting the physical positions of these TFs against the corresponding trans-eQTL hotspot locations (coordinates from older assembly versions were all lifted over to the AGP_v4 assembly), we found frequent colocalization of these two coordinates (Supplemental Figure 19). This suggests that many TFs fall within previous trans-eQTL hotspots and are predicted by multiple GRNs to regulate the same set of target genes (as eQTL targets; i.e., eGenes). Sixty-eight TFs were found to colocalize with 74 previously identified trans-eQTL hotspots (within 50 Mbp on the same chromosome), each showing significant overlap between their GRN-predicted targets and eQTL targets (see Methods; Figure 8A; Supplemental Data Set). In other words, GRN predictions led to the identification of the candidate regulator TF underlying 74 trans-eQTL hotspots.
Figure 8.
Identification of TFs Underlying Trans-eQTL Hotspots Identified in Previous Studies.
(A) Colocalization of TFs predicted by GRNs in this study and trans-eQTL hotspots identified in previous studies that regulate the same set of targets. Each dot represents a TF that is supported by at least two high-quality networks to show significant coregulation with at least one trans-eQTL hotspot and is within 50-Mbp distance from the trans-eQTL hotspot location.
(B) to (D) Identification of R1, COL11, and MYC7, which colocalize with previously identified trans-eQTL hotspots and act as master regulators of the maize anthocyanin biosynthesis pathway (B), photosynthesis light reaction pathway (C), and JA biosynthesis pathway (D), respectively.
Inspecting these 68 TF regulators validated by trans-eQTL hotspots, we found both well-studied TFs with characterized functions and a number with yet unknown functions. One well-studied example is the colored1 gene (R1; Zm00001d026147) that regulates the anthocyanin biosynthesis pathway (Figure 8B). A crucial step in this pathway is the oxidation of the colorless leucoanthocyanidins to generate the colored anthocyanidins, which play important roles as pigments in flowers and fruits in numerous plants across the plant kingdom to attract insects for pollination and act as protectants against UV-B irradiation (Koes et al., 2005; Grotewold, 2006). A previous study identified a trans-eQTL hotspot on chromosome 10 regulating the expression levels of eight genes in the flavonoid biosynthesis pathway and suggested that the basic helix-loop-helix (bHLH) family TF gene R1 is the underlying regulator (Wang et al., 2018). Indeed, R1 is predicted by at least three independent GRNs in this study to regulate a set of target genes that are significantly enriched not only in flavonoid and anthocyanin biosynthesis pathways but also in the targets of the previously identified trans-eQTL hotspot (Figure 8B; Supplemental Data Set). Among the shared targets are anthocyaninless2 (a2; Zm00001d014914), bronze1 (bz1; Zm00001d045055), and bronze2 (bz2; Zm00001d052492), which is consistent with previous findings from functional characterizations (Figure 8B; Grotewold et al., 1998; Hernandez et al., 2004).
A number of TFs were found to be involved in photosynthesis-related pathways (Supplemental Data Set). One such example is the C2C2-CO-like-TF 11 (COL11; Zm0001d003162), which coregulates a similar set of targets with a previously identified trans-eQTL hotspot (hs014, Li2013; Figure 8C). Regulated genes of the trans-eQTL showed enrichment for GO terms photosynthesis, photosystem I, and photosynthetic membrane categories (Li et al., 2013). Targets of COL11 in our newly built GRNs are also enriched in similar GO categories as well as the photosynthesis light reaction pathways (Supplemental Data Set). Although there is no direct evidence that COL11 plays a role in this pathway, the closest Arabidopsis ortholog (AtCOL3; AT2G24790, BLAST E-value of 8e-43) is annotated as a positive regulator of photomorphogenesis/flower development (Lamesch et al., 2012). Furthermore, COL11 is among the candidate genes associated with flowering time, according to a previous genome-wide nested association mapping study (Dong et al., 2012; Hung et al., 2012; Jamann et al., 2017). Therefore, COL11 is a strong candidate underlying the trans-eQTL (hs014) identified in a previous study (Li et al., 2013).
Another less-studied TF (myc transcription factor7 [MYC7]; Zm0001d030028) was predicted to regulate many of the same target genes as a previously identified trans-eQTL hotspot (Figure 8D; Supplemental Table 3). Among the targets regulated by MYC7 are two lipoxygenase genes (TS1 or LOX8, and LOX10) and three allene oxide synthase genes (AOS1, AOS2, and AOS3), all of which are involved in the jasmonic acid (JA) biosynthesis pathway (Figure 8D). Although no previous study has suggested that MYC7 plays a role in the JA biosynthesis pathway, one study showed that MYC7 mRNA levels increased in response to iron (Fe) starvation and that homologous expression of MYC7 restored the growth of the yeast fet3 fet4 mutant, which is defective in both low- and high-affinity Fe transport (Loulergue et al., 1998). Interestingly, JAs play an important role in the response to Fe-deficiency stress (Maurer et al., 2011; Hindt and Guerinot, 2012; Kobayashi and Nishizawa, 2012). Therefore, it is possible that MYC7 is induced in Fe-deficient environments to activate the JA biosynthesis pathway. Indeed, the closest ortholog of ZmMYC7 in Arabidopsis, ATMYC2 (AT1G32640, BLAST E-value of 8e-142), is induced by dehydration stress and abscisic acid treatment and regulates diverse JA-dependent functions (Lamesch et al., 2012). Moreover, it should be noted that although previous eQTL analysis mapped two of the pathway genes (aos1 and aos2) to an eQTL hotspot (qtl0035) spanning the MYC7 gene, it was not able to fine-map the actual TF regulator, nor was it possible to identify the involved pathway due to lack of power to map other pathway genes to this hotspot (Figure 7C; Wang et al., 2018). Therefore, GRNs built in this study offer a powerful and efficient approach to pinpoint the actual regulator underlying the trans-eQTL hotspot and complement previous eQTL studies in finding true TF-target associations.
DISCUSSION
The Value of Multiple Networks
One approach often used to generate coexpression networks and coexpression-based GRNs is to include as many different samples as possible in a single large network. We were interested in determining whether we would obtain distinct insights from different networks developed from specific subsets of the larger available data sets. By consistently analyzing a large number of maize RNA-seq data sets, we generated many different coexpression-based GRNs. The comparison of GRNs generated from many tissues for a single genotype (developmental atlases) or many different genotypes for a single tissue (genotype surveys) can allow for widespread sampling of potential gene regulatory relationships. While these networks are all enriched for biologically meaningful connections, different networks capture distinct TF-target associations (Figure 1) and show enrichment in distinct processes and functions (Figures 2 and 4). Increasing sample size was reported to generally have a positive effect on network performance (Ballouz et al., 2015; Huang et al., 2017), consistent with our comparison of tissue-specific or genotype-specific (i.e., developmental atlas) networks of different sizes. Interestingly, this is not always the case when comparing networks constructed from samples spanning multiple tissues of the same genotype panel (i.e., meta-networks) to the various tissue-specific networks (i.e., using only samples from the same tissue for the genotype panel). For most ubiquitously expressed TFs and general processes, meta-networks perform better than individual tissue-specific networks (Figures 2, 3, and 5). However, for certain TFs or processes with tissue-specific expression patterns (Figure 2, KN1 or O2), the transcriptional variation specific to a tissue will be diluted in the meta-network with a much larger sample size, leading to a weaker signal of true TF-target associations, which was also observed in a previous study comparing different coexpression networks in rice (Oryza sativa; Childs et al., 2011). Thus, which type of network (a tissue-specific network with large sample size, or a meta-network spanning multiple tissues with moderately large sample size) should be used depends on the goal of the analysis (general interactions or tissue-specific interactions).
Network Predictions Contain Many False Positives
While these coexpression networks contain useful information and show significant enrichments for specific annotations or pathways, they are also very prone to false positive predictions, especially when sample sizes are small. By binning network predictions according to their interaction scores, we found that top-scoring network predictions (top 100,000) consistently show better performance (enrichment in functional annotation, validation rate in paired genotype data sets, and so on) than the rest of the predictions (Figures 2 and 5). Across networks built in this study, we found that the top 100,000 predictions (or top 0.2 to 0.3% predictions from each network, assuming a total of 2000 TFs by 20,000 targets = 40 million possible edges) is a good cutoff to control for false positives while also keeping a high true positive rate (Supplemental Figure 6), although a previous study comparing network topologies in yeast (Saccharomyces cerevisiae), Caenorhabditis elegans, and fruitfly (Drosophila melanogaster) suggests that 3.5 to 11.7% (i.e., 1.4 to 4.7 million) of all possible edges should be used (Ouma et al., 2018). While this works well for most small- to moderate-sized networks where a steep drop of fold enrichment is typically observed from rank-10 to rank-9 predictions, it is not true for some well-powered networks with large sample size, such as the tissue atlas combined (Huang2018 four tissues, Li2019 six tissues, and Zhou2018 B+M+F1 networks), in which a strong enrichment is observed for rank-9 or even rank-8 network predictions (Supplemental Figure 6). Therefore, the level of functional enrichment in different-sized networks can be used as an empirical indicator to inform proper network filtering.
Presence/Absence Rather Than Quantitative Changes in TF Expression Are More Likely to Result in Changes in Target Gene Expression
By examining the power of our coexpression-based GRNs to predict covarying TF-target relationships in paired genotype data sets that include natural variation for gene expression, we found that presence/absence expression changes of a TF (i.e., no expression versus moderately expressed) are very likely to result in significant expression changes in target genes (Figure 6). On the other hand, more subtle quantitative changes in TF expression (e.g., differentially expressed but less than fourfold change) are less likely to be associated with measurable changes (e.g., significant DE) in the targets. This finding has potential implications for how to best manipulate TFs in order to affect downstream pathways and ultimately traits: overexpressing or underexpressing a TF that is already moderately expressed will not be as effective as complete knockout of an actively expressing TF or activating a TF that is normally repressed. However, changing tissue-specific expression patterns of a TF may result in novel changes to target gene expression in specific tissues.
We also explored different options to identify potential TF-target associations in the context of natural variation (Figure 7). The genotype panel used to build a cross-genotype network in large determines the performance of this network in predicting expression variation in a validation data set. In theory, the more genotypes that go into network construction, the wider the range of expression variation that will be captured by the network. However, genotype-specific signals for rare alleles may be diluted in such large genotype networks as the panel size continues increasing (e.g., the poor performance of Hirsch2014 and Kremling2018). The ideal network for a specific validation data set is always a network spanning the compared genotypes or their close relatives (phylogenetic neighbors). On the other hand, a comprehensive developmental network—although built solely with B73 samples—had strikingly good performance in almost all validation data sets, indicating that developmental covariation is a good predictor of allelic variation in TFs that will affect GRNs among genotypes. However, it should be noted that when the genotype for which the developmental network is made (i.e., B73) lacks a functional TF that is present in other lines, the GRN for this TF will not be predicted in the B73 developmental network.
Identification of TFs That May Regulate Important Metabolic Pathways
By integrating information from our network predictions with known metabolic pathways or previous eQTL mapping results, we were able to link TFs with potential pathways. Using information on metabolic pathways in maize, we identified many examples of TFs that regulate multiple structural genes in these pathways. In some cases, this identified TFs known to regulate the pathway, but in other cases this resulted in prediction of TFs that may regulate these pathways. By comparing the edges between the TF and putative targets in the pathway in the different GRNs, it became clear that the compilation of information from many different networks provided a much more complete set of linkages of TFs to pathways than using single GRNs. The analysis of trans-eQTL hotspots identified 68 TFs as the putative sources underlying 74 previously identified trans-eQTL hotspots. These putative trans-regulators span a variety of metabolic pathways, including the well-studied bHLH TF R1, which regulates the anthocyanin biosynthesis pathway, as well as the less-studied CONSTANS-LIKE TF COL11 and the bHLH TF MYC7, which might act as crucial regulators for the photosynthesis light reaction pathway and JA biosynthesis pathway, respectively. This highlights the utility of a large set of coexpression-based GRNs for identifying TFs that may underlie trans-eQTL hotspots. These are potential targets for breeding or biotechnology applications to influence specific pathways or traits.
In summary, we compiled a comprehensive resource of maize regulatory networks using a diverse collection of public RNA-seq data sets. These networks are supported by previous characterizations of TF knockout mutants (Figure 1), show different levels of enrichment in functional annotation including GO and CornCyc metabolic pathways (Figure 3), and greatly expand the breadth and depth of previous work on maize GRNs (Walley et al., 2016; Huang et al., 2018). When evaluated against external data sets of natural variation, some of the newly built GRNs achieve high predictive power in predicting transcriptional changes in targets (Figures 6 and 7). In addition, GRN predictions show significant overlap with the results of previous eQTL studies, in many cases allowing the fine mapping/pinpointing of the master regulators underlying trans-eQTL hotspots (Figure 8). Among these potential master regulators are well-studied TFs such as R1 as well as TFs with uncharacterized functions yet promising external evidence (Figure 8). These validated regulators, as well as other high-confidence network predictions, provide excellent candidates for accurate and efficient manipulation of valuable traits and pathways. We have constructed a dedicated web portal (maizeGRN, https://maizeumn.github.io/maizeGRN) for sharing these predicted TF-target interactions with the community.
METHODS
RNA-Seq Data Sets, Mapping, and Normalization
Raw sequencing reads from 21 published RNA-seq studies (9 developmental atlas/tissue time-course studies [Liu et al., 2013; Chen et al., 2014; Chettoor et al., 2014; Li et al., 2014; Yu et al., 2015; Stelpflug et al., 2016; Walley et al., 2016; Yi et al., 2019; Zhou et al., 2019], 11 population studies [Eichten et al., 2013; Fu et al., 2013; Hirsch et al., 2014; Leiboff et al., 2015; Lin et al., 2017; Kremling et al., 2018; Schaefer et al., 2018; Li et al., 2019; Mazaheri et al., 2019; Zhou et al., 2019), and 4 RIL studies [Li et al., 2013; Baute et al., 2015, 2016; Wang et al., 2018]) were downloaded from the National Center for Biotechnology Information Sequence Read Archive, trimmed using fastp (Chen et al., 2018), and mapped to the maize B73 AGP_v4 genome (Jiao et al., 2017) using hisat2 (Kim et al., 2015). Uniquely mapped reads were assigned to and counted for the 46,023 reference gene models (Ensembl Plants v41) using FeatureCounts (Liao et al., 2014). Raw read counts were then normalized using the trimmed mean of M-values normalization approach (Robinson et al., 2010) to give counts per million (CPM) reads and then further normalized by gene coding sequence lengths to give fragments per kilobase of exon per million reads values. Hierarchical clustering and principal component analysis/t-distributed stochastic neighbor embedding analysis was used to explore sample clustering patterns. Outlier replicates with low mapping rate or poor correlation with other replicates from the same sample were discarded. Replicates (technical or biological) were merged into a single sample with the resulting expression matrix re-normalized. All tissue/developmental atlas data sets (Liu et al., 2013; Chen et al., 2014; Chettoor et al., 2014; Li et al., 2014; Yu et al., 2015; Stelpflug et al., 2016; Walley et al., 2016; Yi et al., 2019; Zhou et al., 2019) were combined and re-normalized to create a larger developmental expression data set. The three population studies spanning multiple tissues in panels of inbred lines (Lin et al., 2017; Kremling et al., 2018; Zhou et al., 2019) were separated by tissue to create five, seven, and six tissue-specific networks, respectively. Pipeline scripts, normalization code, and expression matrices are available at Github (https://github.com/orionzhou/rnaseq).
GRN Construction
Normalized CPM matrices from the 21 aforementioned RNA-seq data sets were filtered to remove silent (CPM < 1 in all samples) and invariable (sd of CPM = 0) genes. A set of 2289 maize TFs were obtained from PlantTFDB (Jin et al., 2017) and converted to 2211 AGP_v4 gene models using the v3_to_v4 mapping table from maizeGDB (Andorf et al., 2016). All GRNs were built using the Python machine learning library scikit-learn and XGBoost (Pedregosa et al., 2011; Chen and Guestrin, 2016). Transformed CPM matrices and the list of putative TFs were used to train three regression models (random forest, extra trees, and xgboost) for each data set using the RandomForestRegressor(), ExtraTreesRegressor(), and XGBRegressor() classes, respectively. RandomForest and ExtraTrees regression models were built using the parameters “–n_estimators=1000–criterion=mse–max_features=sqrt” and XGBoost regression models were built using parameters “–n_estimators=1000–max_depth=3–learning_rate=0.0001–reg_alpha=0–reg_lambda=1” (Pedregosa et al., 2011; Chen and Guestrin, 2016). For each regression approach, 44 GRNs in total were constructed, including four developmental networks built using different tissues/developmental stages of the B73 line (three independent data sets and one combined data set including 237 different tissues or stages of B73), 23 tissue-specific networks built using the same tissue sampled from multiple inbred lines, 4 tissue-genotype networks that include multiple tissues sampled from a panel of inbred lines, and 1 network built using shoot apical meristem sampled from 108 B73xMo17 RILs (Figure 1). Five GRNs generated in two recent studies including a maize developmental network (Walley et al., 2016) and four tissue-specific networks (leaf, root, shoot apical meristem, and seed; Huang et al., 2018) were also included (Figure 1).
GRN Evaluation Using Results from Existing TF Functional Studies
Previous studies have characterized several maize TFs including KNOTTED1 (KN1; Bolduc et al., 2012), RAMOSA1 (RA1; Eveland et al., 2014), FEA4 (Pautler et al., 2015), O2 (Li et al., 2015a), HDA101 (Yang et al., 2016), and bZIP22 (Li et al., 2018) through ChIP-seq and/or knockout mutant RNA-seq analysis. The predicted targets of these known TFs serve as good candidates to evaluate the biological relevance of the GRNs built in this study. The performance of each GRN was evaluated using the receiver operating characteristic (ROC) curve space, defined over quantities derived from a confusion matrix that consists of four basic numbers that represent the correctness of link predictions: the number of correctly recognized true network links (true positives [TP]), number of correctly recognized absent links in the true network (true negatives [TN]), and links that either have been incorrectly predicted to be present (false positives [FP]) or true network links that were predicted as absent (false negatives [FN]). The ROC curve then depicts the relative trade-offs between TP rate (i.e., ) and FP rate (i.e., ). Area under the curve was then evaluated for each GRN, which integrates the area below the two-dimensional ROC curve. Area under the receiver operating characteristic (AUROC) values range from 0 to 1, with a value of 1 representing a perfect classifier, values of ∼0.5 indicating that the classifier is no better than a default (random) classifier, and values below 0.5 indicating even worse performance than a random classifier.
Since ROC curve can be misinterpreted if the problem under consideration is characterized with an imbalanced distribution of class values, that is, there are far more TN than TP, which is often the case in GRN reconstruction, we used a partial AUROC score that only considers the first part of the curve before an FP rate of 10% is reached. Similarly, a random classifier will have a score of 0.005 for the partial AUROC, with a higher score (>0.005) indicating a better classifier.
Instead of using the direct targets for each TF, a slightly different approach was also taken to utilize all DEGs from the TF mutant relative to the wild type to evaluate enrichment for both direct and indirect targets. Paired RNA-seq data with at least two biological replicates for both the mutant and the wild type were collected for 17 maize TFs (Supplemental Table 1). DEGs between the TF mutant and the wild type were then identified using DESeq2 (P-value < 0.01; Love et al., 2014). Wilcox rank test was then performed using the predicted (TF-target) interaction scores between the group of true targets (DEGs) and nontargets (non_DEGs), with test P-value −log10 transformed and missing data suggesting the TF being tested (knocked out) is not expressed in the corresponding GRN.
GRN Evaluation Using GO and CornCyc
GO annotations for maize AGP_v4 were obtained from maize-GAMER (Wimalanathan et al., 2018), and maize metabolic pathway information was downloaded from CornCyc at maizeGDB (Andorf et al., 2016). We evaluated how often two genes annotated to the same GO term are predicted to be regulated by the same TF in a certain GRN. Specifically, for each GRN, we counted the number of gene pairs coannotated to the same GO term that are also coregulated by the same TF. To obtain the background level of GO/regulator sharing, we shuffled the GO labels and counted the number of such gene pairs again. This process was repeated 100 times to create a distribution of coregulated gene pairs for each GO term in random networks, which enables the calculation of a significance P-value for the observed level of coregulation. A P-value threshold of 0.05 was used to assess significance level. If a GO term was found to be significantly enriched, an enrichment score (fold change) was calculated using the observed number of coregulated gene pairs divided by the average number of coregulated gene pairs in 1000 permutations. Only GO or CornCyc terms with more than two members predicted as targets in each GRN were included in the analysis. The global enrichment fold change and significance level were determined by summing all coregulated gene pairs over different functional categories for each GRN as well as the permuted network.
GRN Evaluation Using Natural Variation Data Sets
Data for a total of 42 paired tissue/treatment samples with at least three biological replicates per sample between B73 and a different genotype were obtained from four published transcriptomic studies (Supplemental Table 2; Marcon et al., 2017; Waters et al., 2017; Sun et al., 2018; Zhou et al., 2019). In order to test the assumption that a regulator (TF) with DE between the two genotypes in a certain tissue/condition would result in DE for the targets of this TF (as predicted by GRNs) in the same tissue/condition, we implemented the following assessment. DEGs between the two genotypes were first identified in each pair of tissue/treatment samples using DESeq2 (Love et al., 2014). In each comparison, a TF was classified into different DE categories (non_DE, DE1-2, DE2-4, DE4+, and SPE, where one genotype has completely lost expression) and the proportion of predicted targets also exhibiting DE was assessed. In addition, the TF-target predictions in each network were binned into 10 groups according to their interaction score predicted by the regression model with the assumption that stronger TF-target interactions may receive stronger support from the natural variation data sets.
eQTL Validation
eQTL-eGene associations were downloaded from three previous maize eQTL studies (see Supplemental Methods; Li et al., 2013; Liu et al., 2017; Wang et al., 2018) and converted to AGP_v4 genome coordinates and gene IDs. cis- and trans-interactions were determined based on whether the eQTL and eGene (i.e., target gene) are on the same chromosome and within 1-Mbp physical distance. eQTLs that regulate more than 10 target eGenes in trans were then tagged as trans-eQTL hotspots and kept for further evaluation. eGenes associated with each trans-eQTL hotspot were checked for enrichment of being coregulated by common TFs as predicted by each GRN, using an identical permutation approach described in the GO/CornCyc enrichment section. In general, target genes within the same trans-eQTL hotspot are much more likely to be regulated by a common TF (as predicted by each GRN) than a randomly generated GRN. Whenever an enrichment was detected (i.e., a TF as predicted by the GRN sharing a significant number of target genes with a previously identified trans-eQTL hotspot), the physical location of the TF was obtained for a colocalization test with the trans-eQTL hotspot (on the same chromosome and within 50 Mbp).
Accession Numbers
All network predictions are available for download at https://maizeumn.github.io/maizeGRN. Code used to build and evaluate networks is available at GitHub (https://github.com/orionzhou/grn). The processed data sets used to create networks and predicted interactions are deposited at http://hdl.handle.net/11299/212030.
Supplemental Data
Supplemental Figure 1. Comparison of GRNs built using different methods according to the enrichment of functional annotations (Gene Ontology, CornCyc, and so on).
Supplemental Figure 2. Number of true TF targets captured by the top one million predictions and the top 100K predictions in each GRN.
Supplemental Figure 3. Evaluation of GRNs using support from direct targets of eight known TFs.
Supplemental Figure 4. Evaluation of GRNs using support from 31 maize TF DAP-Seq data sets.
Supplemental Figure 5. Evaluation of GRNs using support from 17 maize TF knockout mutant RNA-Seq data sets.
Supplemental Figure 6. Enrichment of co-annotated GO/CornCyc terms in co-regulated network targets.
Supplemental Figure 7. Different GRNs capture distinct parts of documented transcriptional regulation interactions from Arabidopsis for the abscisic acid (ABA) pathway and HY5 (Elongated Hypocotyl 5) regulated pathway.
Supplemental Figure 8. Evaluation (AUROC and Wilcox P-value) of constructed GRNs using four sets of predicted TF-target interactions based on TF binding site motif, conserved element of TFBS motif, or FunTFBS.
Supplemental Figure 9. Hierarchical clustering of 45 GRNs.
Supplemental Figure 10. T-SNE clustering of 45 GRNs.
Supplemental Figure 11. Hierarchical clustering of 98 Gene Ontology (Uniprot.Plants, level 6) terms using fold enrichment in different GRNs.
Supplemental Figure 12. Different GRNs support different parts of the anthocyanin biosynthesis pathway.
Supplemental Figure 13. Different GRNs support different parts of the DIMBOA pathway.
Supplemental Figure 14. Different GRNs support different parts of the chlorophyllide biosynthesis pathway regulated by homeobox-transcription factor 26 (HB26, Zm00001d008612).
Supplemental Figure 15. Different coexpression-based GRNs capture distinct parts of classic and CornCyc metabolic pathways.
Supplemental Figure 16. TF-target validation of the combined tissue network in all six selected natural variation data sets.
Supplemental Figure 17. Enrichment of co-regulated targets between previously identified trans-eQTL hotspots and TF-target associations predicted by GRNs.
Supplemental Figure 18. Enrichment of co-regulated targets between previously identified trans-eQTL hotspots and TF-target associations predicted by GRNs.
Supplemental Figure 19. Co-localization of TFs predicted by GRNs in this study and trans-eQTL hotspots identified in previous studies that regulate the same set of targets.
Supplemental Table 1. ChIP-Seq and DAP-Seq data sets used in this study.
Supplemental Table 2. TF knockout mutant RNA-Seq data sets used in this study.
Supplemental Table 3. Natural variation data sets used for validation in this study.
Supplemental Data Set. GRN-predicted TFs supported by trans-eQTL hotspots.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Sarah N. Anderson, Maria Katherine Mejía-Guerra, and Peter Hermanson for reading through the article and providing valuable feedback. We thank the Minnesota Supercomputing Institute at the University of Minnesota (http://www.msi.umn.edu) for providing resources that contributed to the research results reported within this article. This study was funded by the National Science Foundation (grants IOS-1546899 and IOS-1733633). This work is supported in part by Michigan State University and the National Science Foundation Research Traineeship Program, Division of Graduate Education (grant DGE-1828149 to F.A.G.C.).
AUTHOR CONTRIBUTIONS
P.Z., C.N.H., S.P.B., and N.M.S. conceived the experiments. E.G., S.P.B., and N.M.S. secured funding. P.Z. and N.M.S. performed the experiments and analyzed data. Z.L. and F.G.C. contributed additional RNA-seq data to the pipeline. P.Z. and N.M.S. wrote the article. E.M., P.A.C., J.M.N., E.G., and C.N.H. provided expertise, ideas, and feedback.
Footnotes
Articles can be viewed without a subscription.
References
- Andorf C.M., et al. (2016). MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 44 (D1): D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C.L., et al. (2014). Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M.M., Luscombe N.M., Aravind L., Gerstein M., Teichmann S.A.(2004). Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 14: 283–291. [DOI] [PubMed] [Google Scholar]
- Ballouz S., Verleyen W., Gillis J.(2015). Guidance for RNA-seq co-expression network construction and analysis: Safety in numbers. Bioinformatics 31: 2123–2130. [DOI] [PubMed] [Google Scholar]
- Bartlett A., O’Malley R.C., Huang S.C., Galli M., Nery J.R., Gallavotti A., Ecker J.R.(2017). Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12: 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Gaudinier A., Brady S.M., Hennig L., Rhee S.Y., De Smet I.(2012). Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baute J., Herman D., Coppens F., De Block J., Slabbinck B., Dell’Acqua M., Pè M.E., Maere S., Nelissen H., Inzé D.(2015). Correlation analysis of the transcriptome of growing leaves with mature leaf parameters in a maize RIL population. Genome Biol. 16: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baute J., Herman D., Coppens F., De Block J., Slabbinck B., Dell’Acqua M., Pè M.E., Maere S., Nelissen H., Inzé D.(2016). Combined large-scale phenotyping and transcriptomics in maize reveals a robust growth regulatory network. Plant Physiol. 170: 1848–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S.(2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo B., et al. (2014). The Maize TFome--Development of a transcription factor open reading frame collection for functional genomics. Plant J. 80: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., Hauck A., Lai J.(2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J.(2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Guestrin C. (2016). XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM), pp. 785–794. [Google Scholar]
- Chettoor A.M., Givan S.A., Cole R.A., Coker C.T., Unger-Wallace E., Vejlupkova Z., Vollbrecht E., Fowler J.E., Evans M.M.(2014). Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 15: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.L., Davidson R.M., Buell C.R.(2011). Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS One 6: e22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.-N., Zheng H.-Q., Wu N.-Y., Chien C.-H., Huang H.-D., Lee T.-Y., Chiang-Hsieh Y.-F., Hou P.-F., Yang T.-Y., Chang W.-C.(2016). PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44 (D1): D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.A., et al. (2018). The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46 (D1): D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Danilevskaya O., Abadie T., Messina C., Coles N., Cooper M.(2012). A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One 7: e43450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Xiao Y., Govindarajulu R., Feil R., Siddoway M.L., Nielsen T., Lunn J.E., Hawkins J., Whipple C., Chuck G.(2019). The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nat. Commun. 10: 3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A.(1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2013). Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland A.L., et al. (2014). Regulatory modules controlling maize inflorescence architecture. Genome Res. 24: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., et al. (2013). RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 4: 2832. [DOI] [PubMed] [Google Scholar]
- Galli M., Khakhar A., Lu Z., Chen Z., Sen S., Joshi T., Nemhauser J.L., Schmitz R.J., Gallavotti A.(2018). The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 9: 4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M.B., et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Frey M.(2001). Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213: 493–498. [DOI] [PubMed] [Google Scholar]
- Grotewold E.(2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57: 761–780. [DOI] [PubMed] [Google Scholar]
- Grotewold E.(2008). Transcription factors for predictive plant metabolic engineering: Are we there yet? Curr. Opin. Biotechnol. 19: 138–144. [DOI] [PubMed] [Google Scholar]
- Grotewold E., Chamberlin M., Snook M., Siame B., Butler L., Swenson J., Maddock S., St Clair G., Bowen B.(1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740. [PMC free article] [PubMed] [Google Scholar]
- Hecker M., Lambeck S., Toepfer S., van Someren E., Guthke R.(2009). Gene regulatory network inference: Data integration in dynamic models-a review. Biosystems 96: 86–103. [DOI] [PubMed] [Google Scholar]
- Hernandez J.M., Heine G.F., Irani N.G., Feller A., Kim M.-G., Matulnik T., Chandler V.L., Grotewold E.(2004). Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 279: 48205–48213. [DOI] [PubMed] [Google Scholar]
- Hindt M.N., Guerinot M.L.(2012). Getting a sense for signals: Regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Vendramin S., Shi L., McGinnis K.M.(2017). Construction and optimization of a large gene coexpression network in maize using RNA-seq data. Plant Physiol. 175: 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zheng J., Yuan H., McGinnis K.(2018). Distinct tissue-specific transcriptional regulation revealed by gene regulatory networks in maize. BMC Plant Biol. 18: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L.M., Tian F., Bradbury P.J., Chen C., Flint-Garcia S.A., McMullen M.D., Ware D., Buckler E.S., Doebley J.F., Holland J.B.(2012). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 109: E1913–E1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu V.A., Irrthum A., Wehenkel L., Geurts P.(2010). Inferring regulatory networks from expression data using tree-based methods. PLoS One 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Shibata M., Rymen B., Iwase A., Bågman A.-M., Watt L., Coleman D., Favero D.S., Takahashi T., Ahnert S.E., Brady S.M., Sugimoto K.(2018). A gene regulatory network for cellular reprogramming in plant regeneration. Plant Cell Physiol. 59: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamann T.M., Sood S., Wisser R.J., Holland J.B.(2017). High-throughput resequencing of maize landraces at genomic regions associated with flowering time. PLoS One 12: e0168910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., He K., Tang X., Li Z., Lv L., Zhao Y., Luo J., Gao G.(2015). An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 32: 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G.(2017). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 (D1): D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B.(2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502. [DOI] [PubMed] [Google Scholar]
- Khan A., et al. (2018). JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46 (D1): D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P., Kellis M.(2014). Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42: 2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B.L., Hu G., Zhao K.(2011). ChIP-Seq: Technical considerations for obtaining high-quality data. Nat. Immunol. 12: 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L.(2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N.K.(2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152. [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F.(2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Kremling K.A.G., Chen S.-Y., Su M.-H., Lepak N.K., Romay M.C., Swarts K.L., Lu F., Lorant A., Bradbury P.J., Buckler E.S.(2018). Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523. [DOI] [PubMed] [Google Scholar]
- Krouk G., Lingeman J., Colon A.M., Coruzzi G., Shasha D.(2013). Gene regulatory networks in plants: Learning causality from time and perturbation. Genome Biol. 14: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Young R.A.(2013). Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiboff S., Li X., Hu H.-C., Todt N., Yang J., Li X., Yu X., Muehlbauer G.J., Timmermans M.C.P., Yu J., Schnable P.S., Scanlon M.J.(2015). Genetic control of morphometric diversity in the maize shoot apical meristem. Nat. Commun. 6: 8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W.(2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Li C., Qiao Z., Qi W., Wang Q., Yuan Y., Yang X., Tang Y., Mei B., Lv Y., Zhao H., Xiao H., Song R.(2015a). Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell 27: 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yue Y., Chen H., Qi W., Song R.(2018). The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 30: 2402–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2014). Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc. Natl. Acad. Sci. USA 111: 7582–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Petsch K., Shimizu R., Liu S., Xu W.W., Ying K., Yu J., Scanlon M.J., Schnable P.S., Timmermans M.C.P., Springer N.M., Muehlbauer G.J.(2013). Mendelian and non-Mendelian regulation of gene expression in maize. PLoS Genet. 9: e1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Li Y., Varala K., Coruzzi G.M.(2015b). From milliseconds to lifetimes: Tracking the dynamic behavior of transcription factors in gene networks. Trends Genet. 31: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., et al. (2019). Highly genotype- and tissue-specific single-parent expression drives dynamic gene expression complementation in maize hybrids. bioRxiv •••: 668681. [DOI] [PubMed] [Google Scholar]
- Lin H.-Y., Liu Q., Li X., Yang J., Liu S., Huang Y., Scanlon M.J., Nettleton D., Schnable P.S.(2017). Substantial contribution of genetic variation in the expression of transcription factors to phenotypic variation revealed by eRD-GWAS. Genome Biol. 18: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Luo X., Niu L., Xiao Y., Chen L., Liu J., Wang X., Jin M., Li W., Zhang Q., Yan J.(2017). Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol. Plant 10: 414–426. [DOI] [PubMed] [Google Scholar]
- Liu W.-Y., et al. (2013). Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc. Natl. Acad. Sci. USA 110: 3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulergue C., Lebrun M., Briat J.-F.(1998). Expression cloning in Fe2+ transport defective yeast of a novel maize MYC transcription factor. Gene 225: 47–57. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S.(2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon C., Paschold A., Malik W.A., Lithio A., Baldauf J.A., Altrogge L., Opitz N., Lanz C., Schoof H., Nettleton D., Piepho H.-P., Hochholdinger F.(2017). Stability of single-parent gene expression complementation in maize hybrids upon water deficit stress. Plant Physiol. 173: 1247–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34: D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer F., Müller S., Bauer P.(2011). Suppression of Fe deficiency gene expression by jasmonate. Plant Physiol. Biochem. 49: 530–536. [DOI] [PubMed] [Google Scholar]
- Mazaheri M., et al. (2019). Genome-wide association analysis of stalk biomass and anatomical traits in maize. BMC Plant Biol. 19: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Guerra M.K., Pomeranz M., Morohashi K., Grotewold E.(2012). From plant gene regulatory grids to network dynamics. Biochim. Biophys. Acta 1819: 454–465. [DOI] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., et al. (2011). A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.(2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma W.Z., Pogacar K., Grotewold E.(2018). Topological and statistical analyses of gene regulatory networks reveal unifying yet quantitatively different emergent properties. PLOS Comput. Biol. 14: e1006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler M., Eveland A.L., LaRue T., Yang F., Weeks R., Lunde C., Je B.I., Meeley R., Komatsu M., Vollbrecht E., Sakai H., Jackson D.(2015). FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27: 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F., et al. (2011). Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12: 2825–2830. [Google Scholar]
- Petroni K., Pilu R., Tonelli C.(2014). Anthocyanins in corn: A wealth of genes for human health. Planta 240: 901–911. [DOI] [PubMed] [Google Scholar]
- Ravasi T., et al. (2010). An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci W.A., et al. (2019). Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., et al. (2007). Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K.(2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer R.J., Michno J.-M., Jeffers J., Hoekenga O., Dilkes B., Baxter I., Myers C.L.(2018). Integrating co-expression networks with GWAS to prioritize causal genes in maize. Plant Cell 30: 2922–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Breuer C., Kawamura A., Clark N.M., Rymen B., Braidwood L., Morohashi K., Busch W., Benfey P.N., Sozzani R., Sugimoto K.(2018). GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 145: dev159707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T., Gordân R.(2014). Protein-DNA binding: Complexities and multi-protein codes. Nucleic Acids Res. 42: 2099–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N., de León N., Grotewold E.(2019). Challenges of translating gene regulatory information into agronomic improvements. Trends Plant Sci. 24: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Stelpflug S.C., Sekhon R.S., Vaillancourt B., Hirsch C.N., Buell C.R., de Leon N., Kaeppler S.M.(2016). An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: plantgenome2015.04.0025. [DOI] [PubMed] [Google Scholar]
- Sun S., et al. (2018). Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50: 1289–1295. [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J.W., Sartor R.C., Shen Z., Schmitz R.J., Wu K.J., Urich M.A., Nery J.R., Smith L.G., Schnable J.C., Ecker J.R., Briggs S.P.(2016). Integration of omic networks in a developmental atlas of maize. Science 353: 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen Q., Wu Y., Lemmon Z.H., Xu G., Huang C., Liang Y., Xu D., Li D., Doebley J.F., Tian F.(2018). Genome-wide analysis of transcriptional variability in a large maize-teosinte population. Mol. Plant 11: 443–459. [DOI] [PubMed] [Google Scholar]
- Waters A.J., Makarevitch I., Noshay J., Burghardt L.T., Hirsch C.N., Hirsch C.D., Springer N.M.(2017). Natural variation for gene expression responses to abiotic stress in maize. Plant J. 89: 706–717. [DOI] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalanathan K., Friedberg I., Andorf C.M., Lawrence-Dill C.J.(2018). Maize GO annotation-methods, evaluation, and review (maize-GAMER). Plant Direct 2: e00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., et al. (2017). A maize gene regulatory network for phenolic metabolism. Mol. Plant 10: 498–515. [DOI] [PubMed] [Google Scholar]
- Yang H., Liu X., Xin M., Du J., Hu Z., Peng H., Rossi V., Sun Q., Ni Z., Yao Y.(2016). Genome-wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., et al. (2019). High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31: 974–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A., Nishiyama M.Y. Jr., Fuentes B.G., Souza G.M., Janies D., Gray J., Grotewold E.(2009). GRASSIUS: A platform for comparative regulatory genomics across the grasses. Plant Physiol. 149: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-P., et al. (2015). Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proc. Natl. Acad. Sci. USA 112: E2477–E2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J., Li G., Ryu C.-H., Ma C., Zhang S., Lloyd A., Hunter B.G., Larkins B.A., Drews G.N., Wang X., Yadegari R.(2018). Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 30: 2425–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J., Thakare D., Ma C., Lloyd A., Nixon N.M., Arakaki A.M., Burnett W.J., Logan K.O., Wang D., Wang X., Drews G.N., Yadegari R.(2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27: 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Hirsch C.N., Briggs S.P., Springer N.M.(2019). Dynamic patterns of gene expression additivity and regulatory variation throughout maize development. Mol. Plant 12: 410–425. [DOI] [PubMed] [Google Scholar]