ALA3 and related flippases are important interactors and regulators of ADP ribosylation factor GTPase Exchange Factors regulating the polarity and trafficking of PIN auxin transporters and auxin-mediated development.
Abstract
Cell polarity is a fundamental feature of all multicellular organisms. PIN auxin transporters are important cell polarity markers that play crucial roles in a plethora of developmental processes in plants. Here, to identify components involved in cell polarity establishment and maintenance in plants, we performed a forward genetic screening of PIN2:PIN1-HA;pin2 Arabidopsis (Arabidopsis thaliana) plants, which ectopically express predominantly basally localized PIN1 in root epidermal cells, leading to agravitropic root growth. We identified the regulator of PIN polarity 12 (repp12) mutation, which restored gravitropic root growth and caused a switch in PIN1-HA polarity from the basal to apical side of root epidermal cells. Next Generation Sequencing and complementation experiments established the causative mutation of repp12 as a single amino acid exchange in Aminophospholipid ATPase3 (ALA3), a phospholipid flippase predicted to function in vesicle formation. repp12 and ala3 T-DNA mutants show defects in many auxin-regulated processes, asymmetric auxin distribution, and PIN trafficking. Analysis of quintuple and sextuple mutants confirmed the crucial roles of ALA proteins in regulating plant development as well as PIN trafficking and polarity. Genetic and physical interaction studies revealed that ALA3 functions together with the ADP ribosylation factor GTPase exchange factors GNOM and BIG3 in regulating PIN polarity, trafficking, and auxin-mediated development.
INTRODUCTION
Multicellular organisms display polarity at all levels of organization. At the cellular level, polarity is manifested by the asymmetric distribution or localization of components, typically proteins (Kania et al., 2014). In plants, directional auxin flow between cells serves as a polarizing cue that is crucial for plant morphogenesis and adaptability (Mazur et al., 2020). This polar auxin transport predominantly relies on the directional localization of PIN-FORMED (PIN) auxin efflux carriers (Wiśniewska et al., 2006; Adamowski and Friml, 2015). PIN proteins show different subcellular localization patterns during embryogenesis (Friml et al., 2003; Robert et al., 2013; Wabnik et al., 2013), root (Friml et al., 2002a) and shoot (Reinhardt et al., 2003; Heisler et al., 2005) meristem maintenance, lateral root organogenesis (Benková et al., 2003; Dubrovsky et al., 2011), and apical hook formation (Žádníková et al., 2010, 2016). PIN proteins also display dynamic changes during tropic responses (Friml et al., 2002b; Abas et al., 2006; Ding et al., 2011; Rakusová et al., 2011, 2016; Baster et al., 2013) and in response to endogenous signals such as hormones including auxin itself (Prát et al., 2018; Mazur et al., 2020), cytokinin (Marhavý et al., 2011, 2014), gibberellic acid (Löfke et al., 2013; Salanenka et al., 2018), salicylic acid (Du et al., 2013; Tan et al., 2020), strigolactone (Shinohara et al., 2013), jasmonate (Sun et al., 2011), brassinosteroids (Li et al., 2005), and danger-associated peptides (Jing et al., 2019). Thus, PINs are excellent cell polarity markers for studying the mechanism of the generation, maintenance, and rearrangement of cell polarity; nonetheless, in some developmental contexts, they can become uncoupled from genuine cell polarity.
The phosphorylation status of PIN proteins is tightly linked to their polar localization (Armengot et al., 2016). Overexpression of the Ser/Thr protein kinase PINOID (PID) leads to the apicalization of PINs, while the loss-of-function of PID together with that of two related kinases, WAVY ROOT GROWTH1 and WAVY ROOT GROWTH2, leads to the PIN polarity toward the basal side of the cell (Friml et al., 2004; Dhonukshe et al., 2015). PID kinase directly phosphorylates PINs (Michniewicz et al., 2007; Dhonukshe et al., 2015). Phosphomimic forms of PIN1 localize to the apical sides of cells, while the phospho-dead forms localize to the basal sides (Huang et al., 2010; Zhang et al., 2010). PP2A protein phosphatases function antagonistically with PID in PIN phosphorylation, and the disruption of PP2A leads to a shift in PIN polarity toward the apical side of the cell (Michniewicz et al., 2007; Dai et al., 2012). A more recent study using antibodies specifically recognizing the phosphorylated versions of PIN1 suggested that PID-mediated changes in PIN phosphorylation, which determine the polar localization of PIN, are transient, because in the steady-state situation, PIN1 appears to be phosphorylated, even at the basal side of the cell (Barbosa and Schwechheimer, 2014). Thus, despite the compelling data showing that PIN phosphorylation is necessary and sufficient for the decision about apical-basal PIN polarity, we still do not know where and how this happens.
Numerous observations also link vesicle trafficking to PIN polarity establishment and maintenance (Luschnig and Vert, 2014). A recurrent feature of PINs is that they undergo constitutive recycling between the polar domain at the plasma membrane (PM) and endosomes (Geldner et al., 2001). This allows plants to rapidly redirect auxin fluxes in response to different signals (Kleine-Vehn et al., 2008b, 2010) but also seems to play an essential role in maintaining of PIN polarity at the PM (Kitakura et al., 2011; Kleine-Vehn et al., 2011; Glanc et al., 2018; Narasimhan et al., 2020). ADP ribosylation factors (ARFs) together with their activators ARF guanine nucleotide exchange factors (ARF GEFs) are essential components in vesicle formation and thus trafficking. Arabidopsis (Arabidopsis thaliana) contains eight ARF GEFs, among which GNOM plays a major role in basal PIN polarity (Geldner et al., 2003; Kleine-Vehn et al., 2008a). The fungal toxin Brefeldin A (BFA) directly inhibits the activity of BFA-sensitive ARF GEFs. Under long-term BFA treatment or in the weak GNOM mutant gnomR5, basal localized PINs shift to the apical sides of cells (Kleine-Vehn et al., 2008b).
GNOM-LIKE1 localizes to the Golgi and regulates endoplasmic reticulum (ER)-Golgi trafficking (Richter et al., 2007; Teh and Moore, 2007), whereas it appears that GNOM, which was originally thought to localize to the trans-Golgi network/early endosome (TGN/EE), mainly localizes to the Golgi apparatus (GA) and regulates recycling to the PM but also acts at the PM itself, likely participating in endocytosis (Naramoto et al., 2010, 2014). GNOM-LIKE2 participates in pollen germination (Jia et al., 2009). The ARF GEF proteins BIG1, BIG2, BIG3, BIG4, and BIG5 localize to the TGN/EE. BIG1, BIG2, BIG3, and BIG4 are functionally redundant in the trafficking of newly synthesized and endocytosed PINs to the PM, vacuole, and cell plate (Richter et al., 2014). BIG2 and BIG5 were identified from the same forward genetic screen as BFA-VISUALIZED ENDOCYTIC TRAFFICKING DEFECTIVE3 (BEN3) and BEN1, which were implicated in endocytic trafficking (Tanaka et al., 2009; Kitakura et al., 2017). Together with BEN1, BEN2, encoding VPS45, regulates PIN Polarity (Tanaka et al., 2013). Thus, ARF GEFs are key components involved in the trafficking of PINs and other cargos, but their regulatory machinery in plants is largely elusive.
We previously aimed to identify novel components involved in cell polarity by performing a forward genetic screen for PIN polarity regulators using the Arabidopsis PIN2:PIN1-HA;pin2 line ectopically expressing PIN1 tagged with hemagglutinin in the root meristem epidermis. We demonstrated the feasibility of this screening system and identified cell wall biosynthesis components as important factors in PIN polarity maintenance (Feraru et al., 2011). Here, we report the identification of ALA3, a phospholipid flippase, as a regulator of PIN polarity. The ala3 mutants display altered auxin transport and PIN trafficking. Based on the similarity of endomembrane trafficking defects and the genetic and physical interactions of ALA3 with ARF GEFs, we propose that ALA3 is an ARF GEF regulator that is important for PIN trafficking and polarity.
RESULTS
Identification of repp12 as a PIN Polarity Regulator Mutant
To obtain a better understanding of PIN trafficking and polarity in Arabidopsis, we performed a forward genetic screen for PIN polarity regulator mutants (Feraru et al., 2011). Apically localized PIN2 in root epidermal cells is essential for the asymmetric distribution of auxin after gravistimulation and for the resulting root gravitropic response (Wiśniewska et al., 2006; Baster et al., 2013). We subjected PIN2:PIN1-HA;pin2 plants, which show predominantly basal and nonpolar-localized PIN1-HA in root epidermal cells and agravitropic root growth, to ethyl methanesulfonate (EMS) mutagenesis with the aim to identify regulator of PIN polarity (repp) mutants with apical instead of basal localization of PIN1-HA in root epidermal cells. The basic idea behind the screening was that the apically localized PIN1-HA in the repp mutants can partially function as PIN2, resulting in the restoration of gravitropic root growth, and thus mutants can be identified based on a macroscopic phenotype (Feraru et al., 2011).
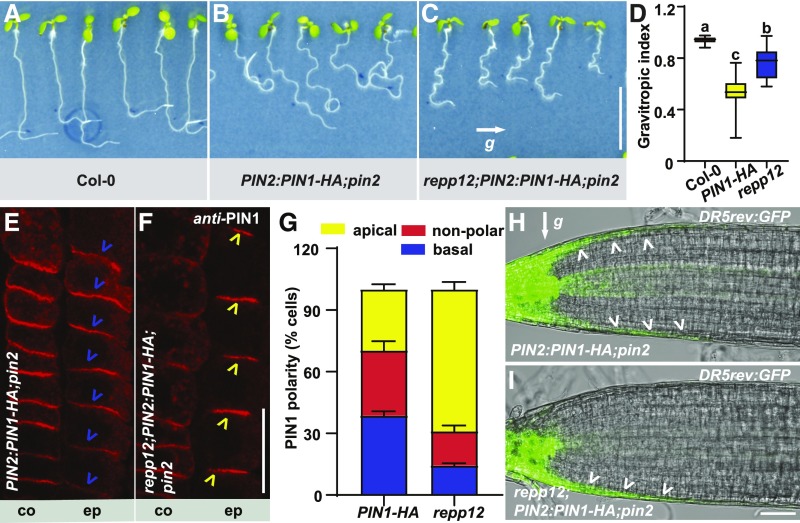
From the screen, we isolated the repp12 mutant. By contrast to the agravitropic root growth of PIN2:PIN1-HA;pin2 (Figures 1B and 1D; Supplemental Figure 1B), repp12;PIN2:PIN1-HA;pin2 displayed a positive root gravitropic response (Figures 1C and 1D; Supplemental Figure 1C) similar to that of the wild type (Figures 1A and 1D; Supplemental Figure 1A). We performed anti-PIN1 immunolocalization experiments to observe the PIN1-HA polarity switches in root epidermal cells. Indeed, compared with PIN2:PIN1-HA;pin2, with mostly basally localized PIN1-HA in root epidermal cells (Figures 1E and 1G), repp12;PIN2:PIN1-HA;pin2 showed predominantly apical PIN1-HA localization (Figures 1F and 1G). In line with the switched PIN1-HA polarity from the basal to apical side of root epidermal cells, the asymmetric distribution of auxin between the upper and lower side of the roots after gravistimulation was restored in repp12;PIN2:PIN1-HA;pin2 (Figures 1H and 1I; Supplemental Figure 1D). Taken together, these results suggest that repp12 is a PIN polarity regulator mutant.
Figure 1.
Identification of repp12 as a Mutant of a PIN Polarity Regulator.
(A) to (C) Root gravitropic responses of wild type Col-0 (A), PIN2:PIN1-HA;pin2 (B), and repp12;PIN2:PIN1-HA;pin2 (C) seedlings after 24-h gravistimulation. The letter “g” indicates the direction of the gravity vector. Scale bar = 10 mm.
(D) Box plot showing the root gravitropic index for Col-0, PIN2:PIN1-HA;pin2, and repp12;PIN2:PIN1-HA;pin2 seedlings (n > 20). For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(E) and (F) Immunolocalization analysis of PIN1-HA with PIN1 antibody in root cortex (co) and epidermal cells (ep) of PIN2:PIN1-HA;pin2 (E) and repp12;PIN2:PIN1-HA;pin2 (F) seedlings. Blue and yellow arrowheads, respectively, indicate basally and apically localized PIN1-HA. Scale bar = 20 μm.
(G) Stacked bars showing the percentage of the apical, nonpolar, or basal PIN1-HA localization in root epidermal cells of PIN2:PIN1-HA;pin2 (PIN1-HA) and repp12;PIN2:PIN1-HA;pin2 (repp12) seedlings. For each genotype, >220 root epidermal cells from >20 roots were analyzed per experiment. Error bars represent sd, calculated from three independent experiments.
(H) and (I) The auxin accumulation patterns demarcated by the auxin response indicator DR5rev:GFP in the roots of PIN2:PIN1-HA;pin2 (H) and repp12;PIN2:PIN1-HA;pin2 (I) seedlings after 10-h gravistimulation by 90°. The letter “g” indicates the direction of the gravity vector. White arrowheads indicate the distribution pattern of DR5. Scale bar = 50 μm.
REPP12 Encodes ALA3, a Phospholipid Flippase
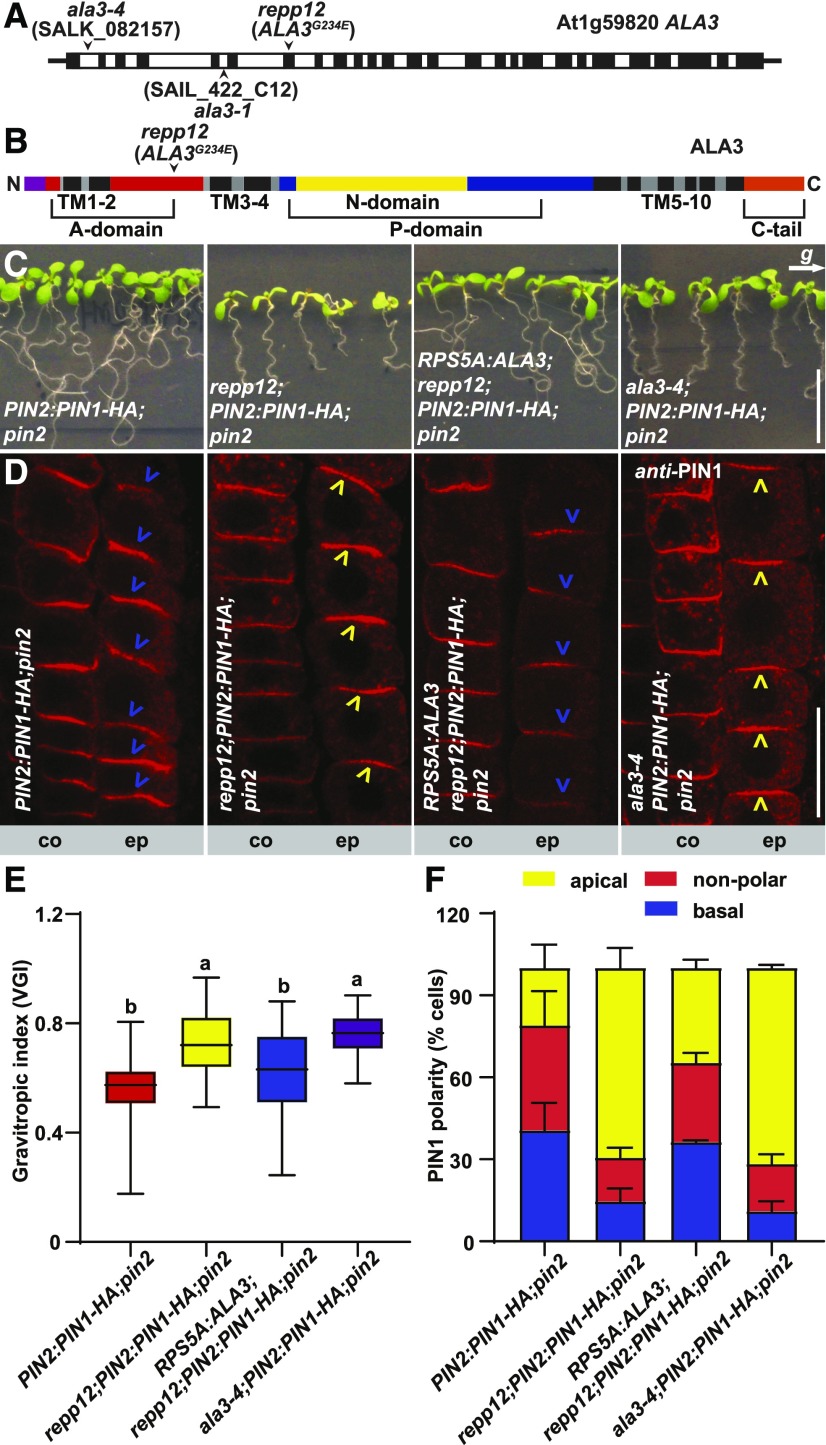
To identify the causal gene of the repp12 mutation, we performed mapping by Next Generation Sequencing of pooled genomic DNA from homozygous plants of the F2 generation with a backcross of repp12;PIN2:PIN1-HA;pin2 to PIN2:PIN1-HA;pin2. By analyzing the segregation of single nucleotide polymorphisms introduced by EMS mutagenesis, we selected a region on chromosome 1 where the causal mutation should be found. The region contained six candidate mutations (Supplemental Figures 2A and 2B). Among the six candidates, AT1G59820, which encodes AMINOPHOSPHOLIPID ATPASE3 (ALA3), carries an amino acid substitution of Gly234 to Glu (G234E; Figures 2A and 2B). ALA3 belongs to a 12-member P4-ATPase protein family in Arabidopsis that is thought to function as a phospholipid flippase. These proteins translocate phospholipids from the exoplasmic to cytoplasmic side of the two leaflets of the membrane, generating membrane curvature and contributing to vesicle formation (Poulsen et al., 2008; López-Marques et al., 2014; Nintemann et al., 2019). ALA3 is thought to participate in vesicle formation, because ala3 mutants showed defects in vesicle formation from the TGN in root peripheral columella cells (Poulsen et al., 2008). Drs2p, the ortholog of ALA3 in Saccharomyces cerevisiae, is involved in clathrin-coated vesicle formation (Muthusamy et al., 2009). Most phospholipid flippases function in a heterodimeric αβ-complex comprising α-subunit P4-ATPase and a β-subunit called Cdc50p in S. cerevisiae and ALA-Interacting Subunit (ALIS) in Arabidopsis. Cdc50p/ALIS is required for the correct localization and function of phospholipid flippase (Saito et al., 2004; López-Marqués et al., 2010, 2012; Costa et al., 2016). P4-ATPases contain 10 transmembrane helices and three cytosolic domains, including the actuator, nucleotide binding, and phosphorylation domains (Supplemental Figure 2C; Hiraizumi et al., 2019; Timcenko et al., 2019). Protein sequence analysis of P4-ATPase family members showed that G234 is located in the A domain of ALA3 protein and it is evolutionarily conserved from Arabidopsis ALA3 to ALA12 as well as all S. cerevisiae flippases, indicating the importance of this Gly for the function of flippases (Figure 2B; Supplemental Figure 2C; Supplemental Data Set 1).
Figure 2.
REPP12 Encodes ALA3, a Phospholipid Flippase.
(A) Gene model of ALA3 (At1g59820). Black boxes and white boxes represent exons and introns, respectively. Mutation site of repp12 and T-DNA insertion positions of ala3-1 and ala3-4 are indicated.
(B) Schematic diagram of ALA3. The red boxes represent the Actuator (A) domain; the yellow box represents the Nucleotide binding (N) domain; the blue boxes represent the Phosphorylation (P) domain; the black boxes represent transmembrane helixes; the purple box represents the cytoplasmic N-tail; the orange box represents cytoplasmic C-tail. The repp12 (ALA3G234E) mutation is located on the A-domain.
(C) Root gravitropic analysis of PIN2:PIN1-HA;pin2, repp12;PIN2:PIN1-HA;pin2, RPS5A:ALA3;repp12;PIN2:PIN1-HA;pin2, and ala3-4;PIN2:PIN1-HA;pin2 seedlings after 16-h gravistimulation. The letter g indicates the direction of the gravity vector. Scale bar = 10 mm.
(D) Immunolocalization of PIN1-HA with PIN1 antibody in the root cortex (co) and epidermal cells (ep) of PIN2:PIN1-HA;pin2, repp12;PIN2:PIN1-HA;pin2, RPS5A:ALA3;repp12;PIN2:PIN1-HA;pin2, and ala3-4;PIN2:PIN1-HA;pin2 seedlings. Blue and yellow arrowheads indicate basally and apically localized PIN1-HA in root epidermal cells, respectively. Scale bar = 20 μm.
(E) Box plot showing the root gravitropic index for PIN2:PIN1-HA;pin2 (n = 38), repp12;PIN2:PIN1-HA;pin2 (n = 32), RPS5A:ALA3;repp12;PIN2:PIN1-HA;pin2 (n = 55), and ala3-4;PIN2:PIN1-HA;pin2 (n = 38). For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated at least three times with similar results.
(F) Stacked bars showing the percentage of the apical, nonpolar, or basal PIN1-HA localization in root epidermal cells of PIN2:PIN1-HA;pin2, repp12;PIN2:PIN1-HA;pin2, RPS5A:ALA3;repp12;PIN2:PIN1-HA;pin2 and ala3-4;PIN2:PIN1-HA;pin2 seedlings. For each genotype, >200 root epidermal cells from >20 roots were analyzed per experiment. Error bars represent sd from three biologically independent experiments.
To determine whether the mutation in flippase ALA3 is responsible for the restoration of gravitropic root growth and apicalization of PIN1-HA in root epidermal cells of repp12;PIN2:PIN1-HA;pin2, we generated repp12;PIN2:PIN1-HA;pin2 transgenic plants expressing ALA3 under the control of the meristematic promoter RPS5A (Weijers et al., 2001). Phenotypic analysis showed that expression of wild-type ALA3 in repp12;PIN2:PIN1-HA;pin2 leads to restoration of the agravitropic root growth and basally localized PIN1-HA in root epidermal cells, as observed in PIN2:PIN1-HA;pin2 (Figures 2C to 2F). Next, we introduced a T-DNA insertion mutant of ALA3 (SALK_082157/ala3-4; Poulsen et al., 2008) into PIN2:PIN1-HA;pin2. ala3-4;PIN2:PIN1-HA;pin2 resembled repp12;PIN2:PIN1-HA;pin2, displaying a positive gravitropic response and apicalization of PIN1-HA (Figures 2C to 2F). We also used the ala3 mutant alleles to test whether the restoration of the root gravitropic growth observed in repp12;PIN2:PIN1-HA;pin2 depends on the presence of the PIN2:PIN1-HA transgene by generating ala3-4 pin2. A test using two consecutive gravistimulations showed that ala3-4 pin2 was unable to grow gravitropically, while ala3-4;PIN2:PIN1-HA;pin2 roots grew along the direction of the gravity vector, similar to the wild type (Supplemental Figure 3). These findings confirm the notion that the mutation in ALA3 rescued the agravitropic root growth of pin2, presumably through the PIN1-HA polarity shift.
To examine whether the repp12 (ALA3G234E) mutation affects the flippase activity of ALA3, we performed a phospholipid uptake assay (Poulsen et al., 2015; Jensen et al., 2017) in Arabidopsis roots using 7-nitrobenz-2-oxa-1,3-diazole-labeled phosphatidylserine (NBD-PS), which was previously shown to be a substrate of ALA3 (Poulsen et al., 2008). After incubation with 60 μM of NBD-PS for the indicated time periods, both repp12;PIN2:PIN1-HA;pin2 and ala3-4;PIN2:PIN1-HA;pin2 root tips showed less NBD-PS internalization compared with PIN2:PIN1-HA;pin2 root tips (Supplemental Figures 4A and 4B), confirming the notion that ALA3 is required for flipping PS and that the Gly G234 is essential for the function of ALA3 as a flippase.
In addition, to test whether the lipid composition is altered in the ala3 mutant, we crossed the ala3-4 mutant with plants harboring the PS biosensor 2×mCherry-C2LACT (mCH-C2LACT) and the phosphatidylinositol-4-phosphate biosensor mCitrine-1×PHFAPP1 (mCIT-1×PHFAPP1), respectively (Simon et al., 2016; Platre et al., 2018). We observed a decreased signal of mCH-C2LACT (Supplemental Figures 5A and 5C) and an increased signal of mCIT-1xPHFAPP1 at the PM of ala3-4 root epidermal cells (Supplemental Figures 5B and 5C).
In conclusion, the identified mutation in the conserved Gly G234 of the putative ALA3 flippase is the causative mutation of the PIN polarity phenotype of repp12. Furthermore, the ala3 knock-out mutation affected phospholipid uptake and membrane lipid composition, thus supporting the role of ALA3 as a flippase and confirming the importance of G234 for this activity.
ala3 Mutants Display Auxin-Related Developmental Defects and Altered Auxin Transport
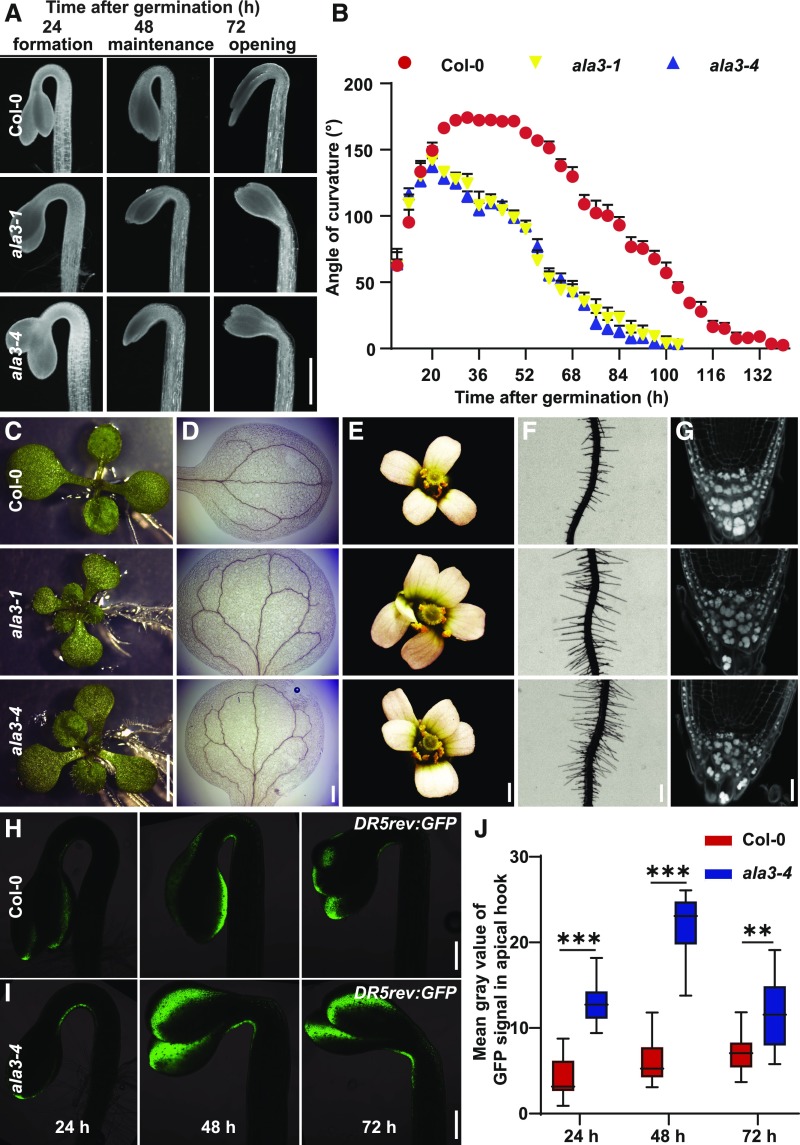
Previous studies have indicated that ala3 mutants show stunted root and shoot growth (Poulsen et al., 2008) and defects in trichome and pollen development, as well as pathogen defense (Zhang and Oppenheimer, 2009; McDowell et al., 2013; Underwood et al., 2017). However, the link between ALA3 and cell polarity/auxin-mediated development is unclear. Because we identified ALA3 as a potential PIN polarity regulator, we investigated the auxin-related developmental defects in ala3 mutants by analyzing two available T-DNA insertion mutants: ala3-1 (SAIL_422_C12) and ala3-4 (SALK_082157; Poulsen et al., 2008). Consistent with previous observations, ala3 mutants had shorter roots and smaller root meristems than the wild type (Supplemental Figures 6A, 6B, and 6E). Because the lateral root number in ala3 was comparable to that of Arabidopsis wild-type Columbia (Col-0; Supplemental Figure 6C), the lateral root density was increased in ala3 (Supplemental Figure 6D). Additionally, etiolated ala3 seedlings had slightly shorter hypocotyls than the wild type (Supplemental Figure 6F), could not form a fully closed apical hook, and showed more rapid apical hook opening than Col-0 (Figures 3A and 3B). Moreover, the ala3 mutants displayed triple cotyledons (Figure 3C), defects in cotyledon venation patterning (Figure 3D), an aberrant number of petals (Figure 3E), longer root hairs (Figure 3F; Supplemental Figure 6G), disorganized root columella cells (Figure 3G), and delayed root gravitropic responses (Supplemental Figures 6H and 6I). All of these defects in ala3 mutants are reminiscent of the phenotypes of previously reported auxin transport mutants (Friml et al., 2002a; Žádníková et al., 2010; Lampugnani et al., 2013; Prát et al., 2018), which is consistent with a role of ALA3 in auxin-regulated development.
Figure 3.
ala3 Mutants Exhibit Auxin-Related Developmental Defects.
(A) Representative images of apical hooks of Col-0, ala3-1, and ala3-4 seedlings during three phases of apical hook development. The photographs were taken during the apical hook formation phase (24-h time after germination), the maintenance phase (48 h), and the opening phase (72 h). Scale bar = 1 mm.
(B) Kinetics of apical hook development in Col-0–, ala3-1–, and ala3-4–etiolated seedlings. Angles of apical hooks from six seedlings for each genotype were measured (starting from ∼8 h after germination, every 4 h). Error bars represent sd. The experiment was repeated three times with similar results.
(C) Triple cotyledons of the ala3-1 and ala3-4 mutants. Frequencies of triple cotyledons’ appearance: Col-0 (0% ± 0%), ala3-1 (11.3% ± 1.8%), ala3-4 (6.8% ± 1.5%). Values represent means ± sd, calculated from two biologically independent experiments (n > 60 seedlings of each genotype per experiment). Scale bar = 4 mm.
(D) More complex cotyledon venation pattern in the ala3-1 and ala3-4 mutants compared with Col-0. Frequencies of cotyledons with defects in venation pattern: Col-0 (6% ± 2.8%), ala3-1 (58% ± 12.4%), and ala3-4 (61% ± 9.7%). Values represent means ± sd, calculated from three biologically independent experiments (n > 37 cotyledons of each genotype per experiment). Scale bar = 200 μm.
(E) Flowers with aberrant numbers of petals in the ala3-1 and ala3-4 mutants. Frequencies of flowers with aberrant numbers of petals: Col-0 (0% ± 0%), ala3-1 (15.2% ± 5%), and ala3-4 (10% ± 2.9%). Percentage of flowers with aberrant numbers of petals from each 47-d–old plant was calculated. Values represent means ± sd (n > 7 plants). The experiment was repeated three times with similar results. Scale bar = 2 mm.
(F) Longer root hairs of the ala3-1 and ala3-4 mutants compared with Col-0. Scale bar = 200 μm.
(G) Disorganized root columella cells were observed in the ala3-1 and ala3-4 mutants compared with Col-0. Frequencies of roots showing disorganized root columella cells: Col-0 (0%), ala3-1 (100%), and ala3-4 (100%). For each genotype, more than nine roots have been checked. Every root of ala3 mutants exhibits disorganized columella cells. The experiment was repeated three times with similar results. Scale bar = 40 μm.
(H) and (I) The DR5rev:GFP signal in the 24-h–, 48-h–, and 72-h–apical hooks of Col-0 (H) and the ala3-4 mutant (I) Scale bar = 200 μm.
(J) Quantification of DR5rev:GFP signal in the 24-h–, 48-h–, and 72-h–apical hooks of Col-0 and ala3-4 mutant seedlings. For each time point, >12 seedlings were evaluated per genotype. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Asterisks indicate significant differences according to multiple t tests using the Holm–Sidak method (***P < 0.001, **P < 0.01). The experiment was repeated at least 3 times with similar results.
To investigate whether auxin distribution is perturbed in the ala3 mutants, we introduced the DR5rev:GFP auxin reporter into the ala3-4 mutant background. A maximum DR5 signal was detected in the concave side of the apical hook of etiolated Col-0 and ala3-4 seedlings, but the signal was much stronger in ala3-4 than the wild type (Figures 3H to 3J; Supplemental Figures 7A and 7B). DR5 signal in the epidermal cells was also stronger in ala3-4 hypocotyls compared with Col-0 (Supplemental Figure 7C). Moreover, slightly enhanced DR5 signal was observed in ala3-4 versus Col-0 root caps (Supplemental Figures 7D and 7E). All these observations are consistent with auxin transport defects in ala3-4 mutants. To further test this notion, we performed an auxin transport assay in hypocotyls using 3H-IAA and observed slightly enhanced auxin transport in both ala3-1 and ala3-4 versus Col-0 (Supplemental Figure 7F). Collectively, these observations are consistent with the notion that ALA3 functions in auxin-mediated development, particularly some aspect of auxin transport.
ala3 Mutants Show Defects in BFA-Sensitive Trafficking
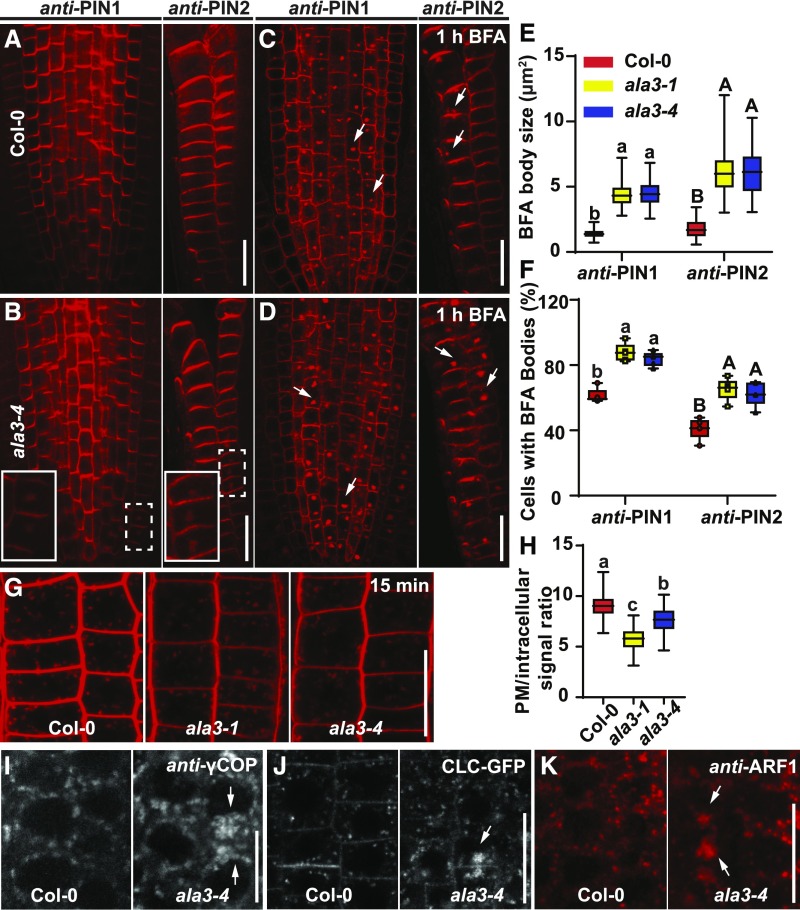
As PIN1-HA polarity was switched from the basal to apical domain in ala3 roots, we were interested in determining whether there are also polarity changes in endogenous PIN proteins in the mutants. To address this question, we performed immunostaining experiments of ala3-1 and ala3-4 using anti-PIN1 and anti-PIN2 antibodies. We did not observe pronounced polarity changes in the localization of endogenous PIN1 or PIN2 in roots of the ala3 mutants (Figures 4A and 4B; Supplemental Figure 8A). However, we observed intracellular aggregations of PIN1 and PIN2 in these roots, indicating that ala3 mutations perturb the trafficking of PIN proteins (Figures 4A and 4B; Supplemental Figure 8A).
Figure 4.
The Trafficking Defects in ala3 Mutants.
(A) to (D) Immunolocalization of PIN1 and PIN2 in 4-d–old roots of Col-0 and ala3-4 seedlings treated with DMSO (A) and (B) and 50 μM of BFA for 1 h (C) and (D). Aggregations were observed in the indicated cells (by the dashed line rectangle), which were magnified (full line rectangle). White arrows indicate PIN1- and PIN2-labeled BFA bodies. Scale bar = 20 μm.
(E) Quantification of BFA body size. Eighty BFA bodies from five roots per genotype were measured. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(F) Quantification of the percentage of cells with BFA bodies per root. Five roots per genotype were evaluated. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum showing all points. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(G) Two micromoles of FM4-64 staining of Col-0, ala3-1, and ala3-4 seedlings for 15 min. Scale bar = 20 μm.
(H) Quantification of FM4-64 internalization (PM/intracellular signal). More than 40 cells from 10 roots were measured. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated at least three times with similar results.
(I) Immunolocalization of Golgi marker γCOP in Col-0 and ala3-4. White arrows indicate the aggregations of γCOP in ala3-4. Frequencies of cells with aggregations: Col-0 (0% ± 0%) and ala3-4 (2.4% ± 0.8%). Values represent means ± sd. For each genotype, >10 single-scanned images were collected and analyzed per experiment. Two biologically independent experiments were done with similar results. Scale bar = 10 μm.
(J) CLC-GFP in Col-0 wild type and ala3-4 roots. Aggregations were observed in ala3-4. Frequencies of cells with aggregations: Col-0 (0% ± 0%) and ala3-4 (2% ± 0.88%). Values are means ± sd. For each genotype, six single-scanned images were collected and analyzed per experiment. White arrows indicate the aggregations. Three biologically independent experiments were done with similar results. Scale bar = 20 μm.
(K) Immunolocalization of ARF1 in Col-0 and ala3-4 roots. Aggregations were observed in ala3-4. Frequencies of cells with aggregations: Col-0 (0% ± 0%) and ala3-4 (2.9% ± 0.92%). Values represent means ± sd. For each genotype, nine single-scanned images per experiment were collected and analyzed. White arrows indicate the aggregations of ARF1 in ala3-4. Three biologically independent experiments were performed with similar results. Scale bar = 20 μm.
The fungal toxin BFA targets ARF GEF regulators of vesicle budding, thereby inhibiting protein recycling and causing the accumulation of PM-localized proteins into intracellular aggregates known as BFA bodies (Geldner et al., 2003). This effect is reversible; after BFA washout, the BFA bodies disappear and membrane proteins go back to the PM (Feraru et al., 2012). Thus, BFA is a convenient tool to study protein trafficking. We used this approach to analyze PIN trafficking defects in the ala3 mutants. After treatment with 50-μM BFA for 1 h, PIN1- and PIN2-labeled BFA bodies were detected in Col-0, ala3-1, and ala3-4 (Figures 4C and 4D; Supplemental Figure 8B). Nonetheless, both BFA body size and the percentage of cells with BFA bodies were significantly higher in the ala3 mutants than Col-0 (Figures 4C to 4F; Supplemental Figure 8B). After BFA washout for 90 min, both PIN1- and PIN2-labeled BFA bodies disappeared in Col-0, whereas they persisted in >20% root cells in ala3-1 and ala3-4 (Supplemental Figures 8C to 8F). These data suggest that the mutation of ALA3 perturbs BFA-sensitive trafficking of PIN proteins, which is consistent with the PIN polarity- and auxin-related developmental defects observed in the ala3 mutants.
Interestingly, in another fluorescence imaging-based forward genetic screen for mutants in which we looked for plants with altered sensitivity to the effect of auxin on endocytosis, we identified the auxin resistant endocytosis2 (are2) and are3 mutants. Auxin inhibits endocytosis of PINs and other proteins from the PM domain of root cells (Paciorek et al., 2005). Concomitant treatment of 10-μM NAA/25-μM BFA results in the absence of BFA-induced PIN internalization in wild-type seedlings (Supplemental Figure 9A). By contrast, the are2 and are3 mutants displayed persistent internalization of PIN1-GFP in the BFA bodies (Supplemental Figures 9B and 9C). Next Generation Sequencing and further data analysis using a predecessor of the now improved and publicly available artMAP tool (Javorka et al., 2019) showed that the are3 mutant harbors a mutation resulting in an early stop codon in ALA3. Moreover, sequencing of ALA3 in the are2 mutant uncovered a mutation at the splice site between the first exon and first intron of ALA3 (Supplemental Figure 9D). Immunostaining of PIN1 and PIN2 in F1 individuals from the crosses are2 × ala3-4 and are3 × ala3-4 showed that PIN1 and PIN2 were internalized in BFA bodies after a concomitant treatment of 10-μM NAA/25-μM BFA for 1 h (Supplemental Figures 9E to 9G), demonstrating the allelism of these mutants. The identification of two additional ala3 alleles from an independent screen focused on BFA-sensitive PIN trafficking further supports a role for ALA3 in ARF GEF-mediated processes.
ala3 Mutations Disrupt Vesicular Trafficking Related to the PM, TGN, and Golgi
To further characterize the trafficking defects in the ala3 mutants, we traced the endocytic pathway using the amphiphilic styryl dye FM4-64, which is incorporated into membrane lipid bilayers and can be used as a tracer of endocytosis (Jelínková et al., 2010; Dejonghe et al., 2019). After staining the roots with 2 μM of FM4-64 for 15 min, the ala3 mutants showed much weaker signal on the PM compared with Col-0 (Figure 4G; Supplemental Figure 10A). The reduced FM4-64 staining on ala3 root epidermal cells might be related to a change in the charge on the PM or altered lipid composition (Supplemental Figure 5). However, it is not clear whether a change in the charge or lipid composition would decrease the efficiency of FM4-64 incorporation into the membrane (McDowell et al., 2015). Further quantification of the PM/cytosol FM4-64 signal intensity ratio suggested that endocytosis increased in the ala3 mutants (Figures 4G and 4H). Additionally, after staining the roots with FM4-64 for 30 min, an aggregation of FM4-64 staining signals was observed in the ala3 mutants (Supplemental Figure 10B). These findings, together with the abnormal BFA bodies observed in the ala3 mutants, indicate that these mutants have defects in the endosomal compartments that are sensitive to BFA, such as the Golgi or TGN.
To further characterize this defect, we performed immunostaining experiments using the antibody against the γ-subunit of coat protein complex (γ-COP; Movafeghi et al., 1999), which labels the GA (Feraru et al., 2012). We observed aggregates of γCOP-positive Golgi compartments in ala3-4 (Figure 4I). In wild-type Col-0, CLC2-GFP is localized to the PM, cell plate, and TGN/EE (Kitakura et al., 2011; Ito et al., 2012), while in ala3-4, we detected aggregated CLC-GFP (Figure 4J), indicating the formation of TGN/EE aggregates. ARF1, a key regulator of vesicle formation, is localized to both the Golgi and TGN (Naramoto et al., 2010). By immunostaining with anti-ARF1 antibody (Pimpl et al., 2000), we also observed ARF1 aggregates in the ala3-4 mutant (Figure 4K). The aggregates of γCOP-, CLC2-, and ARF1-labeled Golgi or TGN/EE in ala3-4 resembled BFA-induced changes in endomembrane compartments (Naramoto et al., 2014). Together, these findings suggest that ala3 is impaired in BFA-sensitive trafficking processes that take place at the PM, TGN, and Golgi.
ALA3 Localizes to the PM, TGN, and Golgi
Various efforts to study the function of ALA3 have focused on investigating its subcellular localization. However, neither N-terminal– nor C-terminal–tagged GFP-ALA3 fusion proteins showed any visible signal in Arabidopsis seedlings when expressed under the control of the 35S promoter (Poulsen et al., 2008; Zhang and Oppenheimer, 2009), even though expressing Pro35S:GFP-ALA3 in the ala3-4 mutant background complemented the root and vegetative growth defects of this mutant (Poulsen et al., 2008). GFP-ALA3 colocalized with the Golgi marker ST-YFP when transiently expressed in Nicotiana tabacum (tobacco) leaves (Poulsen et al., 2008).
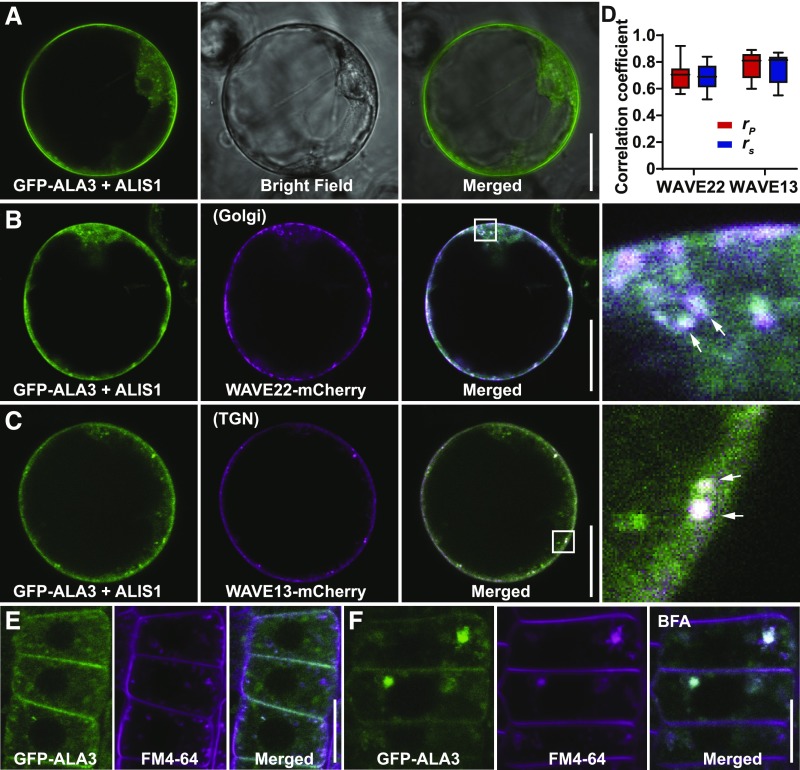
To study the subcellular localization of ALA3, we used Arabidopsis protoplasts transiently expressing the Pro35S:GFP-ALA3 (Poulsen et al., 2008) and Pro35S:YFP-ALA3 constructs. After transiently co-expressing YFP-ALA3 with the ER marker RFP-p24δ5 (Langhans et al., 2008) in protoplasts, we observed colocalization of YFP-ALA3 with RFP-p24δ5, suggesting that ALA3 is localized to the ER (Supplemental Figure 11A). ALIS1, the β-subunit of the heterodimeric flippase, is essential for the localization and function of ALA3 (Poulsen et al., 2008; López-Marqués et al., 2010). Transient co-expression of GFP/YFP-ALA3 with untagged ALIS1 in protoplasts resulted in GFP/YFP-ALA3 localization at both the PM and endomembrane compartments, partially colocalizing with FM4-64 (Figure 5A; Supplemental Figure 11C). To determine which endomembrane compartment ALA3 localizes to, we transiently co-expressed GFP-ALA3 and untagged ALIS1 with the Golgi marker WAVE22-mCherry or the TGN marker WAVE13-mCherry in protoplasts (Geldner et al., 2009). Consistent with a previous report (Poulsen et al., 2008), GFP-ALA3 colocalized with the Golgi marker WAVE22-mCherry (Figures 5B and 5D). Besides the Golgi localization of ALA3, we also observed colocalization of GFP-ALA3 with the TGN marker WAVE13-mCherry (Figures 5C and 5D). Taken together, our data suggest that ALA3 localizes to the PM, TGN, and Golgi, which is in agreement with the corresponding trafficking defects of ala3 mutants.
Figure 5.
Subcellular Localization of ALA3 to the PM, Golgi, and TGN.
(A) Transient co-expression of GFP-ALA3 and untagged ALIS1 in protoplasts. PM and endomembrane compartment signals were observed. Scale bar = 20 μm.
(B) and (C) Transient co-expression of GFP-ALA3, untagged ALIS1 with the Golgi marker WAVE22-mCherry (B) or TGN marker WAVE13-mCherry (C) in protoplasts. The right representations for (B) and (C) were enlarged from the regions highlighted by the white boxes in the merged representations. The arrows in the right panels for (B) and (C) indicate the colocalization of GFP-ALA3 with WAVE22-mCherry (B) and WAVE13-mCherry (C), respectively. Scale bars = 20 μm.
(D) Evaluation of the colocalization of GFP-ALA3 with Golgi marker WAVE22-mCherry and TGN marker WAVE13-mCherry when co-expressed with untagged ALIS1 in protoplasts. The Pearson correlation coefficient (rP) and Spearman correlation coefficient (rS) were calculated after analysis with 24 manually selected regions of interest from 12 individual protoplast cells. The values of rP and rS range from +1 (perfect correlation) to −1 (negative correlation). Error bars represent sd. The experiment was repeated three times with similar results.
(E) The colocalization of GFP-ALA3 and FM4-64. After induction with 10 μM of β-estradiol for 12 h, 4-d–old XVE:GFP-ALA3;repp12 seedlings were stained with 2 μM of FM4-64 for 15 min. Scale bar = 20 μm.
(F) Representative images of BFA body formation of GFP-ALA3 and FM4-64. After 10 μM of β-estradiol induction for 12 h, 4-d–old XVE:GFP-ALA3;repp12 seedlings were stained with 2 μM of FM4-64 and treated with 50 μM of BFA for 1 h. Scale bar = 20 μm.
To test whether the repp12 (ALA3G234E) mutation affects the subcellular localization of ALA3, we transiently co-expressed YFP-ALA3G234E and the ER marker RFP-p24δ5 in protoplasts. Confocal imaging of the transfected protoplasts revealed colocalization between the markers both expressed alone and in ALIS1-cotransfected cells (Supplemental Figures 11B and 11D). This is markedly different from the subcellular localization of wild-type ALA3 in the presence of ALIS1 (Figure 5A; Supplemental Figure 11C). These results indicate that the ALA3G234E mutation interferes with the localization of ALA3, presumably by affecting its exit from the ER.
To further determine the subcellular localization of ALA3, we constructed the β-estradiol-inducible line XVE:GFP-ALA3 in the repp12 background. The XVE:GFP-ALA3 construct was not able to complement the gravitropic root growth defect of repp12 even though the position of the tag was the same as for the functional Pro35S:GFP-ALA3 construct (Supplemental Figures 11E and 11F), perhaps because the expression of GFP-ALA3 is not homogenous after β-estradiol induction (Supplemental Figure 11G). After 10 μM of β-estradiol induction for 12 h, GFP-ALA3 signal was observed on the PM and intracellular compartments, which partially colocalized with FM4-64 (Figure 5E). This observation is consistent with what we observed when expressing GFP-ALA3 and untagged ALIS1 in Arabidopsis protoplasts. BFA/FM4-64 treatment of XVE:GFP-ALA3;repp12 seedlings led to the formation of intracellular GFP-ALA3 aggregates (Figure 5F).
ALAs Act Redundantly in PIN Trafficking and Polarity
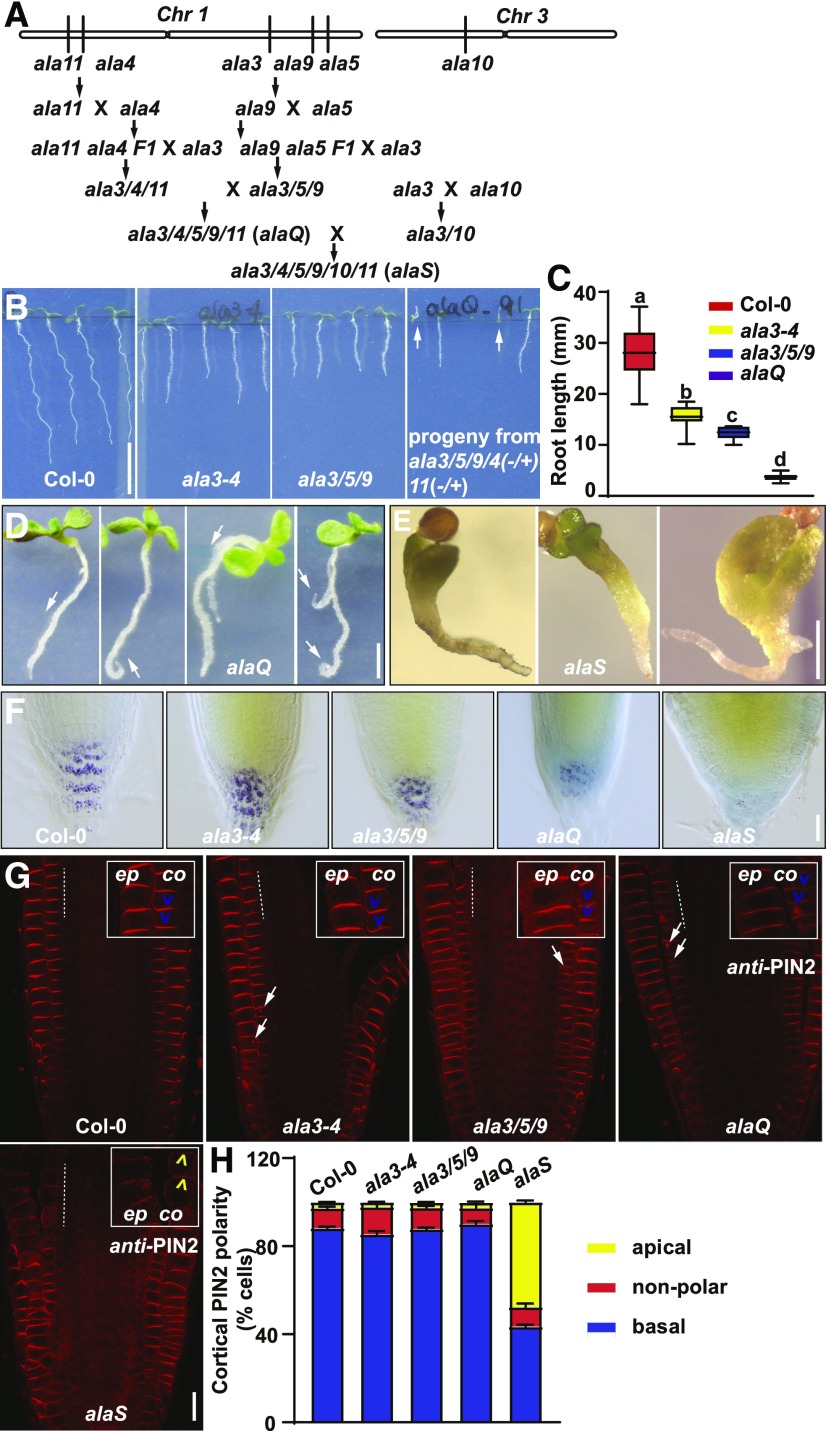
ALA3 belongs to the P4-ATPase gene family, which contains 12 members in Arabidopsis. Therefore, functional redundancy might occur among ALA gene family members. Based on gene expression profiles during root development (Supplemental Figure 12A) and the location of ALA3 in the ALA3-12 subclade (Nintemann et al., 2019), we selected mutants of ALA4, ALA5, ALA9, ALA10, and ALA11 to construct multiple mutants with ala3-4. We obtained T-DNA insertion lines ala4-1 (WiscDsLox435D3), ala5-1 (SALK_049232), ala9-1 (SALK_128495), ala10-1 (SALK_024877C), and ala11-1 (SAIL_90_A12). All these mutants carry insertions in the exons of the respective genes, and RT-PCR analysis showed that ala4-1 (ala4), ala5-1 (ala5), ala9-1 (ala9), ala10-1 (ala10), and ala11-1 (ala11) are knockout mutants (Supplemental Figure 12B). We generated multiple combinations including the double mutant ala3/10, the triple mutants ala3/4/11 and ala3/5/9, the quintuple mutant ala3/4/5/9/11 (alaQ), and the sextuple mutant ala3/4/5/9/10/11 (alaS; Figure 6A). The mutant ala3/5/9 had shorter shoots and roots than ala3 (Figure 6B and 6C). The root lengths of ala3/10 and ala3/4/11 were comparable to ala3 (Supplemental Figure 12C), whereas ala3/4/11 shoots displayed severe developmental defects (Supplemental Figure 12E). The homozygous mutants alaQ and alaS were sterile (Supplemental Figures 12D and 12E; Figures 6D and 6E). Notably, alaQ and alaS seedlings had significantly shorter primary roots and much weaker Lugol’s staining patterns at the root tip compared with ala3 (Figures 6D to 6F). Both alaQ and alaS seedlings were agravitropic (Figure 6D and 6E). To further test whether the agravitropic root growth of alaQ and alaS resulted from defects in PIN polarity or trafficking, we performed immunolocalization using anti-PIN2 antibodies. In ala3 and ala3/5/9 seedlings, PIN2 localized to the basal side of the PM in root cortex cells, similar to that in Col-0 (Figures 6G and 6H). However, PIN2 formed aggregates inside the cells of ala3 and ala3/5/9 (Figure 6G). By contrast, PIN2 was present in larger aggregations in alaQ (Figure 6G) and, interestingly, the basal PIN2 localization switched to the apical side of cortex cells in alaS roots (Figures 6G and 6H). These findings suggest that ALA gene family members play functionally redundant roles in regulating PIN trafficking and the apical–basal polarity of PIN.
Figure 6.
Functional Redundancy of ALA Gene Family Members.
(A) The locations of ALA3, ALA4, ALA5, ALA9, and ALA11 on Chromosome 1 (Chr1), as well as ALA10 on Chromosome 3 (Chr3). The strategy of crossing was aimed at combining the most closely linked mutations first, so that they later segregated almost like a single locus. Mutants with the genes lying closest (ALA4 with ALA11, and ALA5 with ALA9) were crossed. After obtaining double mutants of ala4/11 and ala5/9 in the F1 generation, ala3 and ala4/11 F1, ala3 and ala5/9 F1, and ala3 and ala10 were crossed. Subsequently, ala3/4/11 with ala3/5/9 were combined to generate quintuple mutants ala3/4/5/9/11 (alaQ). Lastly, ala3/4(+/−)/5(+/−)/9(+/−)/11(+/−) was crossed with ala3/10 to obtain sextuple mutants ala3/4/5/9/10/11 (alaS).
(B) Phenotypic analysis of the roots from 6-d–old Col-0, ala3-4, ala3/5/9, and ala3/5/9/4(+/−)/11(+/−) progeny seedlings. White arrows indicate the alaQ mutants. Scale bar = 10 mm.
(C) Quantification of root length in Col-0, ala3-4, ala3/5/9, and alaQ seedlings shown in (B) (n > 10). For the box plots, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(D) Phenotypes of 10-d–old alaQ mutant seedlings. White arrows indicate the agravitropic root growth of alaQ mutants. Scale bar = 2 mm.
(E) Phenotypes of 7-d–old alaS mutant seedlings. The arrested embryo development and agravitropic root growth of alaS mutants. Scale bar = 1 mm.
(F) Lugol’s staining of root tips of 7-d–old Col-0, ala3-4, ala3/5/9, alaQ, and alaS seedlings. Scale bar = 40 μm.
(G) Immunolocalization of PIN2 in Col-0, ala3-4, ala3/5/9, alaQ, and alaS roots. The indicated cells (by a dashed line) were magnified, as shown at the top of each representation. “ep” indicates the root epidermal cell and “co” indicates root cortex cell. Blue and yellow arrowheads, respectively, indicate the basally and apically localized PIN2 in cortex cells. White arrows indicate the aggregations of PIN2. Scale bar = 20 μm.
(H) Evaluation of PIN2 polarity in Col-0, ala3-4, ala3/5/9, alaQ, and alaS root cortex cells. Numbers of root cortex cells with apically, basally, and nonpolarly localized PIN2 from each root were summed up and represented as a percentage of the total number of evaluated cells. For each genotype, >100 root cortex cells from >10 roots were evaluated per experiment. Error bars represent sd, calculated from three independent experiments.
ALA3 Interacts Genetically and Physically with ARF GEFs GNOM and BIG3
We demonstrated that ALA proteins are important for PIN polarity and trafficking, as has been shown for the ARF GEFs (Geldner et al., 2003; Kleine-Vehn et al., 2008a; Tanaka et al., 2009, 2013; Richter et al., 2014). Additionally, the S. cerevisiae orthologs of ALAs, DRS2p and Neo1p, are associated with S. cerevisiae ARF GEFs Gea2p and Mon2p (Chantalat et al., 2004; Wicky et al., 2004; Tsai et al., 2013). In line with the notion that ALA3 participates in ARF GEF activity, inhibiting ARF GEF activity with BFA led to the same apicalization of PIN1-HA and rescued the gravitropic root growth in PIN2:PIN1-HA;pin2 plants (Supplemental Figure 13), which was the original basis for the identification of ALA3 in our screen.
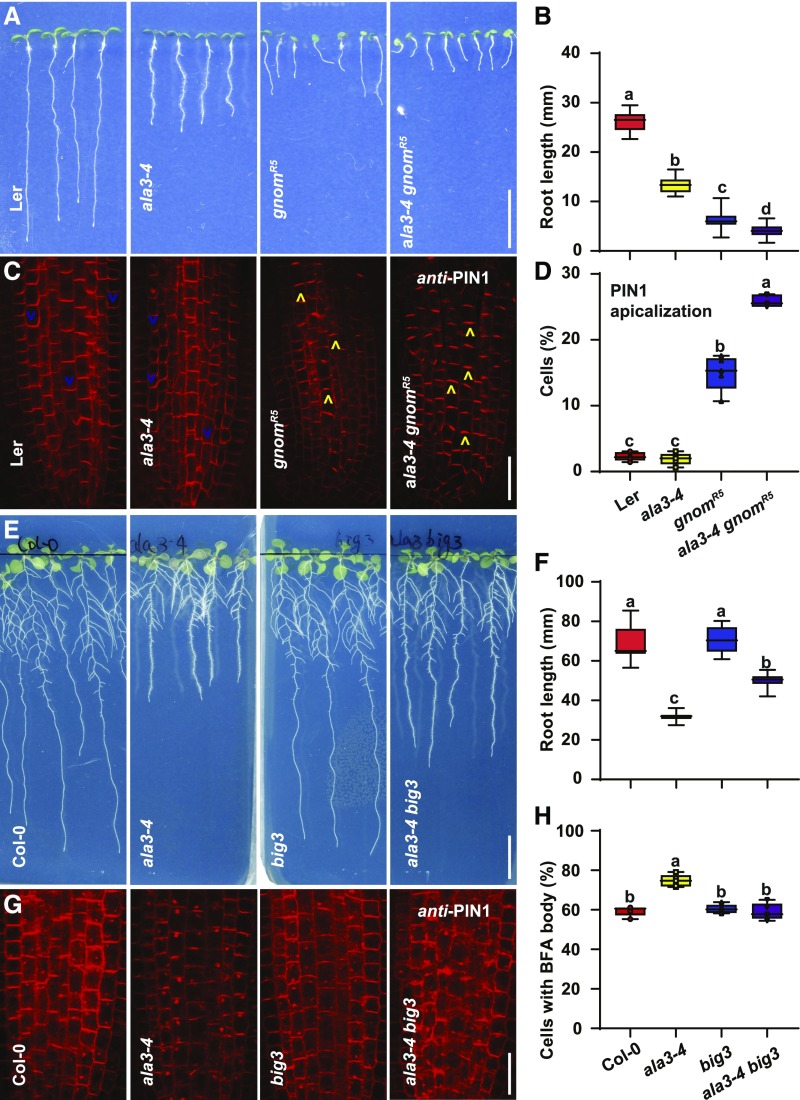
To further explore the functional links between ALA3 and ARF GEFs, we crossed ala3-4 with gnomR5 and big3. gnomR5 is a partial loss-of-function mutant of GNOM that shows collapsed root apical meristems and apicalization of PIN1 in the stele (Kleine-Vehn et al., 2008a). The ala3-4 gnomR5 double mutant exhibits shorter root compared with gnomR5 (Figures 7A and 7B). Moreover, we observed more apicalization of PIN1 in the stele cells of ala3-4 gnomR5 compared with gnomR5 (Figures 7C and 7D). In accordance with previous findings (Kleine-Vehn et al., 2008a), the roots of the gnomR5 mutant did not form as many BFA bodies as the corresponding wild type after BFA treatment (Supplemental Figures 14A and 14B). Notably, the ala3-4 gnomR5 double mutant formed abnormally large BFA bodies more similar to those in ala3-4 (Supplemental Figures 14A and 14B). These findings suggest that the ala3 and gnom mutations act synergistically on PIN polarity, whereas in terms of BFA body formation, ala3 is rather epistatic to gnom.
Figure 7.
Genetic Interactions between ALA3 and ARF GEFs.
(A) Root phenotypes of 7-d–old Ler, ala3-4, gnomR5, and ala3-4 gnomR5seedlings. Ler (control) and gnomR5 seedlings were obtained from the segregating population of heterozygous gnomR5 (+/−). ala3-4 and ala3-4 gnomR5 seedlings were from the segregating population of heterozygous ala3-4 (−/−) gnomR5 (+/−). Scale bar = 10 mm.
(B) Box plot showing the root length for the seedlings as in (A), n > 15 seedlings. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(C) Immunolocalization of PIN1 in Ler, ala3-4, gnomR5, and ala3-4 gnomR5. Blue and yellow arrowheads, respectively, indicate basally and apically localized PIN1. Scale bar = 20 μm.
(D) Box plot with individual data points showing the percentage of apically localized PIN1 in roots of Ler, ala3-4, gnomR5, and ala3-4 gnomR5 (n = 5 roots). For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum showing all points. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(E) Twelve-d–old Col-0, ala3-4, big3, and ala3-4 big3 seedlings. Scale bar = 10 mm.
(F) Box plot showing the root length of seedlings shown in (E), n > 11 seedlings. big3 partially rescues the root growth phenotype of ala3-4. For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
(G) Immunolocalization of PIN1 in Col-0, ala3-4, big3, and ala3-4 big3 seedlings treated with 50 μM of BFA for 1 h. Scale bar = 20 μm.
(H) Box plot with individual data points showing the percentage of root stele cells with PIN1-labeled BFA bodies (n = 5 roots). For the box plot, the box defines the first and third quartiles, and the central line in the box represents the median. Whiskers, from minimum to maximum showing all points. Different letters represent significant differences at P < 0.05 by one-way ANOVA with a Tukey multiple comparisons test. The experiment was repeated three times with similar results.
We next analyzed the ala3-4 big3 double mutant to examine the functional relationship of ALA3 with BIG ARF GEFs. BIG1 to BIG4 regulate the late secretory trafficking pathway (Richter et al., 2014). BFA treatment blocks the late secretion of newly synthesized PINs to the PM and prevents the establishment of PIN polarity in the big3 mutant (Richter et al., 2014; Glanc et al., 2018). Notably, crossing with the big3 mutant partially rescued the root growth defect of ala3-4 (Figures 7E and 7F). Unlike ala3-4 gnomR5, ala3-4 big3 had basally localized PIN1 in the stele and PIN1 aggregates (Supplemental Figure 14C). However, after 1 h of BFA treatment, the formation of abnormal BFA bodies in ala3-4 was blocked by introducing the big3 mutation (Figures 7G and 7H). After 12 h of BFA treatment, like the big3 mutant, PIN1 lost its polarity and was retained in dispersed BFA bodies in the cytosol of the ala3-4 big3 double mutant (Supplemental Figure 14D). Taken together, these observations indicate that big3 is epistatic to ala3.
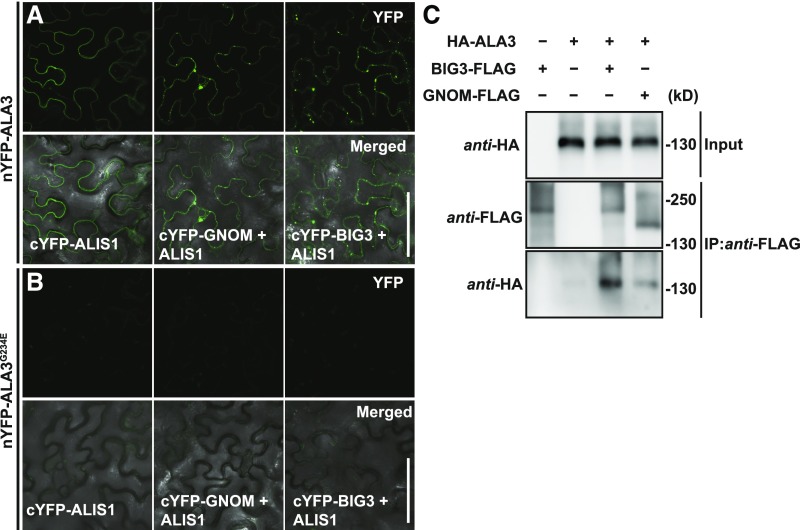
Because genetic interaction analysis showed that ALA3 collaborates with ARF GEFs to regulate PIN polarity and trafficking, we asked whether ALA3 and ARF GEFs physically interact. To investigate this notion, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaf epidermal cells. YFP signal was observed when nYFP-ALA3 (nYFP, n-terminal fragment of YFP) was co-infiltrated with cYFP-ALIS1 (cYFP, c-terminal fragment of YFP) or with combinations containing untagged ALIS1 and cYFP-GNOM or cYFP-BIG3 (Figure 8A), indicating that ALA3 physically interacts with ARF GEFs GNOM and BIG3. However, YFP signal was not detected by confocal microscopy when nYFP-ALA3G234E was cotransformed into N. benthamiana with cYFP-ALIS1, cYFP-GNOM and untagged ALIS1, or cYFP-BIG3 and untagged ALIS1 (Figure 8B), suggesting that the repp12 (ALA3G234E) mutation disrupts the physical interactions of ALA3 with ALIS1, GNOM, or BIG3. Co-immunoprecipitation (Co-IP) assays performed in N. benthamiana transiently co-expressing HA-ALA3 and CFP-ALIS1 with either GNOM-FLAG or BIG3-FLAG verified the interactions of ALA3 with ARF GEFs GNOM and BIG3 (Figure 8C).
Figure 8.
ALA3 Physically Interacts with ARF GEFs.
(A) and (B) BiFC analysis of the interactions of ALA3 (A) or ALA3G234E (B) with ALIS1, with GNOM and with BIG3 in N. benthamiana leaf epidermal cells. The split nYFP fusions nYFP-ALA3 or nYFP-ALA3G234E were co-expressed with cYFP-ALIS1, or with cYFP-GNOM and untagged ALIS1, or with cYFP-BIG3 and untagged ALIS1 in N. benthamiana leaf epidermal cells. YFP signals were checked at ∼72 h after infiltration. The images were collected with the same settings used for confocal microscopy, including the laser power, pinhole size, and gain level. The experiment was repeated three times with similar results. Scale bars = 100 μm.
(C) Co-IP assays testing the interactions of ALA3 with GNOM or BIG3 in N. benthamiana. GNOM-FLAG or BIG3-FLAG was transiently co-expressed with HA-ALA3 and CFP-ALIS1 in N. benthamiana leaf epidermal cells. Plants transiently expressing BIG3-FLAG were used as a positive control, and plants transiently co-expressing HA-ALA3 and CFP-ALIS1 were used as a negative control. Proteins were extracted from infiltrated leaves and analyzed using anti-FLAG and anti-HA antibodies. Input levels of HA-ALA3 protein in the crude protein extracts were analyzed by immunoblotting with anti-HA antibody. Immunoprecipitated (IP) BIG3-FLAG or GNOM-FLAG proteins were probed with anti-HA antibody to detect Co-IP of HA-ALA3.
In summary, the genetic and physical interactions of ALA3 with both BIG3 and GNOM, together with the finding that these proteins play similar roles in PIN polarity and trafficking, suggest that ALA3 and ARF GEFs act together to regulate these processes.
DISCUSSION
Flippases Function As Regulators of Polarity and Trafficking in Plants
Here, we performed a forward genetic screen for PIN polarity regulator mutants and identified the repp12 mutant, which is defective in the ALA3 gene encoding the P4-ATPase/phospholipid flippase. Flippases are thought to translocate phospholipids from the exoplasmic to the cytoplasmic side of the two leaflets of a membrane, thereby generating and maintaining the asymmetric distribution of phospholipids, which is essential for vesicle formation during endomembrane trafficking (Panatala et al., 2015; Poulsen et al., 2015; Best et al., 2019).
GFP-ALA3 localizes to the GA when transiently expressed in the tobacco leaves, and the Golgi localization of ALA3 does not depend on the presence of the β-subunit ALIS1 (Poulsen et al., 2008). Our ALA3 localization study using Arabidopsis protoplasts revealed that ALIS1 is required for the correct localization of ALA3 to the PM and endomembrane compartments. By performing a GFP-ALA3 colocalization study, we observed that ALA3 not only localizes to the Golgi but also to the PM and TGN. These observations are supported by the results of a proteomics study in which ALA3 was detected in the PM, Golgi, and TGN fractions (Li et al., 2012; Heard et al., 2015). In addition, transmission electron microscopy showed that ala3 mutants have obvious defects including hypertrophied trans-Golgi stacks and secretory vesicle formation in peripheral columella cells of roots (Poulsen et al., 2008). These observations support the hypothesis that ALA3 regulates trafficking from the Golgi/TGN to the PM and back to the TGN. Consistent with this notion, ala3 mutants showed defects in endocytosis as well as punctate aggregations of GA and TGN/EE, indicating secretion defects.
Newly synthesized PIN proteins are secreted from the Golgi to PM and undergo recycling between the TGN and PM (Geldner et al., 2003; Łangowski et al., 2016). ala3 mutations perturb the trafficking of PIN proteins and interfere with the basal polarity of ectopically expressed PIN1-HA. However, ala3 mutants do not show defects in the polarity of endogenous PIN proteins, primarily because the polarity of ectopically expressed PIN1-HA in root epidermal cells is more sensitive and more easily perturbed than the polarity of endogenous PIN proteins (Kleine-Vehn et al., 2008a). Analysis of the quintuple and sextuple mutants of ALA gene family members revealed strong defects in the polarity and trafficking of endogenous PIN2 protein. However, a previous study showed that PIN2 localizes to the apical side of older root cortex cells (Kleine-Vehn et al., 2008b). Could the shorter root meristem size of alaS lead to the change in cortical PIN2 polarity? Many mutants or transgenic lines with changes in cortical PIN2 polarity have smaller root meristems than the wild type, such as the ARF GEF mutant gnomR5 (Geldner et al., 2004), protein phosphatase mutants pp2aa1,3 (Michniewicz et al., 2007) and fypp1 fypp3 (Dai et al., 2012), and plants overexpressing PID (Michniewicz et al., 2007). In all of these cases, it is difficult to dissect cause from effect but given the data on inducible PID (Friml et al., 2004) and BFA-based inhibition of ARF GEFs (Kleine-Vehn et al., 2008a), both lead to cortical PIN2 apicalization without meristem shortening, suggesting that polarity switch is the primary cause of this phenotype. Furthermore, the ala3 gnomR5 double mutant showed enhanced changes in PIN polarity (Figures 7C and 7D). This finding, together with other results, links ALA3 activity with the GNOM-regulated PIN polarity pathway, again supporting the direct, primary effect of the ala3 mutation on PIN polarity. In agreement with the PIN trafficking defects, the ala3 mutants displayed altered auxin distribution and multiple auxin-related developmental defects. Nonetheless, ALA3 does not specifically regulate the polarity and trafficking of PINs, as indicated by the finding that the trafficking of PEN3, a transporter that plays a role in pathogen defense, is perturbed in ala3 leaves (Underwood et al., 2017) and by the defective internalization of generic endocytic markers. In addition, trichome development and pollen tube growth, which rely on polarized endomembrane trafficking, are compromised in ala3 mutants (Zhang and Oppenheimer, 2009; McDowell et al., 2013). Overall, these observations indicate that ALA proteins play redundant, crucial roles in polarity and trafficking processes in Arabidopsis.
The Flippase/ARF GEF Module Plays an Evolutionarily Conserved Role in Subcellular Trafficking
In the S. cerevisiae and the nematode Caenorhabditis elegans, flippases physically interact with ARF GEFs to regulate membrane remodeling (Chantalat et al., 2004; Wicky et al., 2004; Tsai et al., 2013; McGough et al., 2018). Our finding that ALA3 physically interacts with both GNOM and BIG3 suggests that the cooperative action of flippases with ARF GEFs in endomembrane trafficking is evolutionarily conserved.
Our results also indicate that ALA3 has different genetic relationships with GNOM and BIG3. The ala3 gnomR5 double mutant showed synergistic effects on the apicalization of PIN1 and root growth. By contrast, ala3 and big3 showed epistatic interactions in root growth and BFA-sensitive trafficking. Interestingly, different types of genetic interactions were also observed among different yeast flippases (Drs2p, Neo1p), ARFs (Arf1, Arl1), and ARF GEFs (Gea2, Ysl2; Chen and Graham, 1998; Chantalat et al., 2004; Wicky et al., 2004). In the S. cerevisiae, both the drs2△ gea2△ and drs2△ arf1△ double mutants show additive growth defects, but overexpression of another yeast flippase, Neo1p, rescued the ARF GEF ysl2△ mutant defects and deletion of ARL1 rescued neo1-37 (Chen and Graham, 1998; Chantalat et al., 2004; Wicky et al., 2004). Our observations from Arabidopsis suggest that the ALA3 flippase has acquired similarly divergent relationships with ARF GEFs.
In the S. cerevisiae, the ARF GEF Gea2p stimulates the activity of the flippase Drs2p (Natarajan et al., 2009; Tsai et al., 2013). By contrast, our genetic studies indicated that the flippase ALA3 regulates ARF GEFs, not the other way around. These findings suggest that the mechanisms of vesicle formation by flippase/ARF GEFs are different in Arabidopsis and S. cerevisiae or, presumably, in both systems, the activities of these components are mutually regulated. Regardless, more detailed biochemical and cell biological characterization of this machinery in Arabidopsis is needed.
Potential Roles of Flippases in Vesicle Formation
Apart from the conserved flippase/ARF GEF module, studies in other organisms have proposed more models for the involvement of flippases in vesicle formation and trafficking. One typical model is that flippases generate an imbalance in the quantities of phospholipids between the two leaflets of a membrane, causing it to bend toward the cytosol side of the cells and thus providing membrane curvature for vesicle budding (Muthusamy et al., 2009; López-Marques et al., 2014; Panatala et al., 2015; Best et al., 2019). The concentration of specific phospholipids on the cytosolic leaflet as well as membrane curvature can affect the membrane association of soluble proteins that are crucial for vesicle formation, such as small GTPases, Arf GTPase-activating protein (Arf GAP), clathrin, COP coats, or Bin/amphiphysin/Rvs-domain proteins. Multiple lines of evidence support this scenario. The drs2p mutant exhibits late Golgi defects and decreased numbers of clathrin-coated vesicles (Chen et al., 1999; Gall et al., 2002). The translocation of the phospholipid PS by budding yeast Drs2p generates membrane curvature and anionic surface charge, which is required for TGN/EEs recruitment of Arf GAP Gcs1, as it contains a curvature-sensing Arf GAP Lipid Packing Sensor motif (Xu et al., 2013). In addition, the translocation of PS by the human flippase ATP8A1 is needed for the association of membrane fission protein EHD1 to recycling endosomes (Lee et al., 2015). Moreover, the S. cerevisiae flippase complex Dnf1- or Dnf2-Lem3/Cdc50p promotes the rapid membrane disassociation of Cdc42, a small GTPase that functions as a key regulator of cell polarity establishment (Das et al., 2012).
Interestingly, a recent study in Arabidopsis showed that, ROP6, an ortholog of the S. cerevisiae Cdc42, is stabilized by PS into the nanodomains on the PM in response to auxin. Changes in PS biosynthesis altered the numbers of ROP6 clusters on the PM, thus affecting endocytosis and root gravitropism (Platre et al., 2019). We speculate that similar to S. cerevisiae and human flippases, Arabidopsis flippases mediate the asymmetric distribution of phospholipids, a process that is also required for the recruitment of soluble lipid binding proteins involved in vesicle formation to the membrane.
METHODS
Plant Material
Previously published Arabidopsis (Arabidopsis thaliana) lines were used in this study: PIN2:PIN1-HA;pin2 (Wiśniewska et al., 2006), pin2 (Luschnig et al., 1998), DR5rev:GFP (Benková et al., 2003), PIN2:PIN1-HA;pin2;DR5rev:GFP (Feraru et al., 2011), ala3-1 (Poulsen et al., 2008), ala3-4 (Poulsen et al., 2008), ala10-1 (Poulsen et al., 2015), Pro35S:CLC2-GFP (Konopka et al., 2008), gnomR5 (Geldner et al., 2004), and big3 (Richter et al., 2014). pUBQ10:2×mCherry-C2LACT (Platre et al., 2018), and pUBQ10:mCitrine-1×PHFAPP1 (Simon et al., 2016).
The following Arabidopsis lines were generated in this study: ala3-4 pin2, repp12;PIN2:PIN1-HA;pin2;DR5rev:GFP, ala3-4;DR5rev:GFP, repp12;PIN2:PIN1-HA;pin2;RPS5A:ALA3, ala3-4;PIN2:PIN1-HA;pin2, repp12;PIN2:PIN1-HA;pin2;XVE:GFP-ALA3, ala3-4;Pro35S:CLC2-GFP, are2, are3, are2 ala3-4, are3 ala3-4, ala4-1, ala5-1, ala9-1, ala11-1, ala3/4/11, ala3/5/9, ala3/10, ala3/4/5/9/11 (alaQ), ala3/4/5/9/10/11 (alaS), ala3 gnomR5, ala3 big3, ala3-4;2×mCherry-C2LACT, and ala3-4;mCitrine -1×PHFAPP1. All information about seeds can be found in Supplemental Table 1.
Vector Construction and Transformation
The entry vectors used in this study include ALA3-pENTR/D-TOPO (Poulsen et al., 2008), ALIS1-pDONR221, GNOM-pDONR221, GNOM-TGA-pDONR221, BIG3-pDONR221, BIG3-TGA-pDONR221, ALA3G234E-pENTR/D-TOPO, ProUBQ10-pDONRP4P1r (Adamowski et al., 2018), and ProRPS5A-pDONRP4P1r. The ALIS1, GNOM, and BIG3 sequences were amplified from cDNA. The ALA3G234E mutation was generated by site-directed mutagenesis of ALA3-pENTR/D-TOPO using a QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The primers are listed in Supplemental Table 2.
The expression vectors were constructed using the Gateway system (Karimi et al., 2002, 2005; Bensmihen et al., 2004; Belda-Palazón et al., 2012). ProRPS5A:ALA3 in pK7m24GW,3, XVE:GFP-ALA3 in pMDCGWm42, Pro35S:HA-ALA3 in pAlligator 2, Pro35S:CFP-ALIS1 in pB7WGC2, Pro35S:ALIS1 in p2GW7, Pro35S:ALIS1 in pB2GW7, Pro35S:YFP-ALA3 in p2YGW7, Pro35S:YFP-ALA3G234E in p2YGW7, ProUBQ:GNOM-FLAG in pB7m34GW, ProUBQ:BIG3-FLAG in pB7m34GW, Pro35S:nYFP-ALA3 in YFN43, Pro35S:nYFP-ALA3G234E in pYFN43, Pro35S:cYFP-ALIS1 in pYFC43, Pro35S:cYFP-GNOM in pYFC43, and Pro35S:cYFP-BIG3 in pYFC43. The plasmids used in this study are listed in Supplemental Table 3.
XVE:GFP-ALA3 and ProRPS5A:ALA3 was transformed into repp12:PIN2:PIN1-HA;pin2 plants using the floral dip method (Clough and Bent, 1998).
Plant Growth Conditions
Arabidopsis seedlings were grown on Murashige and Skoog (MS) medium supplemented with 1% (w/v) Suc and 0.8% (w/v) Phytoagar (pH 5.9) at 21°C under a long-day photoperiod (16-h light/8-h dark). Seven-d–old seedlings were transferred to soil and grown under a long-day photoperiod (16 h light/8 h dark). The light sources used were Philips GreenPower light emitting diode production modules in a deep red, far red, blue combination with a photon density of 140 µmol/m2/s ± 20%.
Forward Genetic Screen and Next-Generation Sequencing
The M2 generation of 5-d–old EMS-mutagenized PIN2:PIN1-HA;pin2 seedlings (progenies of 2,600 M1 plants) were gravistimulated three to four times (10 h to 16 h of 90° gravistimulation) and scored for individuals with positive responses. In the M3 generation, the candidates showing positive root gravitropic responses and changes in PIN1-HA polarity were confirmed.
For Next Generation Sequencing, the population used for whole-genome sequencing was selected from a F2-segregating population in the repp12;PIN2:PIN1-HA;pin2 mutant backcrossed to PIN2:PIN1-HA;pin2. DNA from 100 seedlings was isolated (DNeasy Plant Mini Kit, QIAGEN; MagJET Plant Genomic DNA Kit, Thermo Fisher Scientific) and sequenced as described by Kitakura et al. (2017) and Depuydt et al. (2013).
Fluorescence Imaging-based Forward Genetic Screen and Next-Generation Sequencing
M2 generation of 5-d–old EMS-mutagenized PIN1:PIN1-GFP seedlings (progenies of 1,920 M1) were cotreated with 10 μM of NAA and 25 μM of BFA for 1 h and screened under a fluorescent microscope for individuals with PIN1-GFP signal accumulated in BFA bodies and therefore insensitive to the effect of auxin on endocytosis. In the M3 generation, the candidates were confirmed, the alleles were backcrossed to the parental line three times, and homozygous lines showing a clear cellular phenotype were selected.
For Next Generation Sequencing, genomic DNA from three homozygous backcrossed individuals and the parental PIN1:PIN1-GFP plant were submitted to the Vienna Biocenter Core Facility (https://www.viennabiocenter.org/facilities/) for whole-genome sequencing. The interactive tool artMAP (Javorka et al., 2019) was used to map EMS-induced mutations.
Root Gravitropic Index
Six-day–old seedlings were grown vertically on MS medium. We evaluated root gravitropism using the parameter vertical growth index (vertical length/total root length), as previously described by Grabov et al. (2005).
Genotyping and RT-PCR
All primers used for genotyping are listed in Supplemental Table 2. The primers were used for genotyping and RT-PCR of ala4, ala5, ala9, ala10, and ala11. RNA was extracted from 4-d–old seedlings with an RNeasy kit (QIAGEN), and cDNA was synthetized with an iScript cDNA kit (Bio-Rad). PCR was performed with gene-specific primers spanning the T-DNA insertions, and the housekeeping gene TUB2 was used as the internal control.
Imaging via Confocal Laser Scanning Microscopy
Fluorescence imaging was performed with a model no. LSM800 confocal laser-scanning microscope (Zeiss) with the following parameters: Cy3 (excitation wavelength: 548 nm), GFP (excitation wavelength: 488 nm), mCITRINE (excitation wavelength: 488 nm), RFP (excitation wavelength: 587 nm), mCherry (excitation wavelength: 587 nm), FM4-64 (excitation wavelength: 506 nm), PI staining (excitation wavelength: 536 nm), and NBD (excitation wavelength: 459 nm).
Immunostaining
Immunostaining was performed with 4-d–old seedlings as previously described by Sauer et al. (2006). The primary antibodies used were rabbit anti-PIN1 (Paciorek et al., 2005) diluted 1:1,000 (v/v), rabbit anti-PIN2 (Abas et al., 2006) diluted 1:1,000 (v/v), rabbit anti-ARF1 (cat. no. AS08 325; Agrisera; Pimpl et al., 2000) diluted 1:1,000 (v/v), and rabbit anti-γCOP (cat. no. AS08 327; Agrisera; Movafeghi et al., 1999) diluted 1:1,000 (v/v).
The secondary antibody used was sheep anti-rabbit conjugated with Cy3 (cat. no. C2306; Sigma-Aldrich) diluted 1:600 (v/v).
BFA Treatment
For immunostaining, 4-d–old seedlings were incubated in liquid MS medium at a final concentration of 50 μM of BFA for 1 h. As controls, 4-d–old seedlings were incubated in liquid MS medium supplemented with DMSO.
To evaluate BFA-rescued agravitropic root growth of PIN2:PIN1-HA;pin2 seedlings, 5-d–old seedlings were transferred to MS medium with 10 μM of BFA or DMSO and vertically grown for 16 h.
NBD-PS Uptake Assay in Arabidopsis
A quantity of 1-palmitoyl-2-(6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl)-sn-glycero-3-phosphoserine (NBD-PS) was obtained from Sigma-Aldrich. The NBD-PS uptake assay was performed as previously described by Poulsen et al. (2015) and Jensen et al. (2017). Briefly, 5-d–old seedlings were incubated in liquid MS medium supplemented with a final concentration of 60 μM of NBD-PS for the indicated time periods. After incubation, the samples were washed twice with liquid MS medium and visualized under an LSM800 confocal laser-scanning microscope (Zeiss).
Analysis of Cotyledon Vasculature
Cotyledons of 11-d–old seedlings were harvested and incubated overnight in 70% ethanol. The cotyledons were transferred to 4% (v/v) HCl/20% (v/v) methanol solution and incubated at 65°C for 15 min, followed by incubation in 7% (v/v) NaOH/70% (v/v) ethanol at room temperature for 15 min. Next, the cotyledons were incubated in a 70% (v/v), 50% (v/v), 25% (v/v), and 10% (v/v) ethanol series (5 min each), followed by 25% (v/v) glycerol/5% (v/v) ethanol solution. Finally, the cotyledons were mounted in 50% (v/v) glycerol and imaged by differential interference contrast microscopy (Olympus BX53).
FM4-64 Staining
Four-d–old seedlings were incubated in MS liquid medium containing 2 μM of FM4-64 dye for 15 min and 30 min, followed by immersion in MS liquid medium for a few seconds before imaging under an LSM800 confocal laser-scanning microscope (Zeiss).
Propidium Iodide, modified Pseudo-Schiff Propidium Iodide, and Lugol’s Staining
For propidium iodide (PI) staining, 4-d–old seedlings were stained with 10 mg/L of PI for 1 min, washed briefly in water for a few seconds to remove excess PI dye, and imaged under a model no. LSM800 confocal microscope (Zeiss).
For modified pseudo-Schiff propidium iodide staining, 7-d–old seedlings were stained as previously described by Truernit et al. (2008). Samples were imaged by confocal microscopy (Zeiss LSM800 confocal microscope).
For Lugol’s staining, 4-d–old seedlings were dipped in Lugol’s solution for 1 min, rinsed in liquid MS medium, and imaged by differential interference contrast microscopy (Olympus BX53).
Auxin Transport Assay
The auxin transport assay was performed as previously described by Lewis and Muday (2009). Briefly, one droplet (5 μL) of MS medium/1.25% (w/v) agar with a final concentration of 500 μM of 3H-IAA was placed on the top of an intact hypocotyl of a 6-d–old etiolated seedling (15 hypocotyls per sample, six replicates), followed by incubation in the dark for 6 h. Roots and the upper parts of the hypocotyls were cut off, and the rest of the hypocotyls were harvested and stored in liquid nitrogen. Samples were ground and 1 mL of scintillation solution (PerkinElmer, 6,013,199) was added to the samples. The samples were incubated in scintillation solution overnight and measured with a scintillation counter (model no. 300XL; Hidex). A sample with only scintillation solution was used as a background control.
Protoplast Preparation and PEG-Mediated Transformation
Protoplasts were prepared and transformed as previously described by Mathur and Koncz (1998). Plasmids were prepared with an E.Z.N.A. Plasmid Maxi Kit I (Omega Bio-Tek). Ten micrograms of each plasmid was transformed into the protoplasts. The transformed protoplast cells were incubated in the dark at room temperature for 12 h to 16 h before imaging under an LSM800 confocal microscope (Zeiss).
Colocalization Analysis
To evaluate the colocalization of GFP-ALA3 with ER, FM4-64, TGN, or Golgi markers, 24 regions of interest from 12 individual transfected protoplasts were selected. Pearson and Spearman correlation coefficients were calculated using the Pearson–Spearman correlation colocalization plugin (French et al., 2008) in the software ImageJ (https://imagej.nih.gov/ij/download.html).
Transient Expression in Nicotiana benthamiana
N. benthamiana plants were grown at 21°C under a long-day photoperiod (16-h light/8-h dark) for 6 weeks. Agrobacterium tumefaciens strain GV3101 harboring plasmids with the targeted gene and p19 strain were grown in Super Optimal broth with Catabolite repression medium with the appropriate antibiotics at 28°C overnight. The agrobacterial cells were harvested and resuspended in infiltration solution (10 mM of MES at pH 5.6, 10 mM of MgCl2, and 200 μM of acetosyringone) until OD600 to 0.3, followed by incubation in the dark for 3 h before infiltration into plant leaves.
BiFC Assays
The genes encoding GNOM, BIG3, ALIS1, ALA3, and ALA3G234E were cloned into BiFC Gateway vectors (Belda-Palazón et al., 2012). The corresponding binary vectors were co-infiltrated with p19 strain into N. benthamiana leaf epidermal cells. After infiltration for 72 h, BiFC fluorescence was visualized under a model no. LSM800 confocal microscope (Zeiss).
Protein Extraction and Co-IP
N. benthamiana leaves co-expressing the indicated combination of proteins were harvested at 72 h after infiltration. Total proteins were extracted from N. benthamiana leaves with extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 1 mM of dithiothreitol, 10 mM of MgCl2, 1% [v/v] Triton, 1 mM of EDTA, one protease inhibitor cocktail [Sigma-Aldrich], and one protein phosphatase inhibitor tablet [Sigma-Aldrich]). The immunoprecipitation was performed using a μMACS DYKDDDDK Isolation Kit (Miltenyi Biotec) following the manufacturer’s instructions. The immunoprecipitates were separated by 10% (v/v) SDS-PAGE and detected with anti-FLAG-HRP antibody (cat. no. A8592; Sigma-Aldrich) or anti-HA-Peroxidase antibody (cat. no. 12013819001; Sigma-Aldrich).
Statistical Analysis
All data were analyzed using one-way ANOVA with a Tukey multiple comparisons test, multiple t tests with the Holm–Sidak method, or two-tailed t test in the software Prism v8.3.0 (GraphPad). Detailed summaries of statistical analysis are presented in Supplemental Data Set 2.
Accession Numbers
Sequences data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ALA1 (At5g04930), ALA2 (At5g44240), ALA3 (At1g59820), ALA4 (At1g17500), ALA5 (At1g72700), ALA6 (At1g54280), ALA7 (At3g13900), ALA8 (At3g27870), ALA9 (At1g68710), ALA10 (At3g25610), ALA11 (At1g13210), ALA12 (At1g26130), ALIS1 (At3g12740), PIN1 (At1g73590), PIN2 (At5g57090), CLC2 (At2g40060), ARF1 (At2g47170), γCOP (At4g34450), GNOM (At1g13980), BIG3 (At1g01960), Drs2p (CAY77625.1), Neo1p (AJR52924), Dnf1p (AJV36506), Dnf2p (AJV18765), and Dnf3p (AJS99534).
The next generation sequencing data have been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) under Gene Expression Omnibus Series accession no. GSE146989.
Supplemental Data
Supplemental Figure 1. The repp12;PIN2:PIN1-HA;pin2 mutant shows a positive root gravitropic response.
Supplemental Figure 2. The results of Next Generation Sequencing of the repp12 mutant.
Supplemental Figure 3. ala3 cannot restore the root gravitropic response of the pin2 mutant without PIN2:PIN1-HA.
Supplemental Figure 4. The repp12 and ala3-4 mutants show reduced NBD-PS uptake.
Supplemental Figure 5. Lipid homeostasis is disturbed in ala3 mutants.
Supplemental Figure 6. ala3 mutants show auxin-related developmental defects.
Supplemental Figure 7. Auxin distribution is altered in ala3 mutants.
Supplemental Figure 8. ala3 mutants exhibit trafficking defects.
Supplemental Figure 9. Identification of are2 and are3 mutants.
Supplemental Figure 10. FM4-64 staining of the roots of ala3 mutants.
Supplemental Figure 11. Subcellular localization of ALA3 and ALA3G234E.
Supplemental Figure 12. ALA gene family members play functionally redundant roles in plant development.
Supplemental Figure 13. Inhibition of ARF GEF activity has similar effects as the ala3 mutation on PIN-HA polarity and gravitropism.
Supplemental Figure 14. Genetic interaction between ALA3 and ARF GEFs.
Supplemental Table 1. List of transgenic lines and mutants used in this study.
Supplemental Table 2. List of primers used in this study.
Supplemental Table 3. List of plasmids used in this study.
Supplemental Data Set 1. Alignment of 5 P4-ATPases from S. cerevisiae and 12 P4-ATPases from A. thaliana.
Supplemental Data Set 2. Detailed statistical analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Rosa Laura López-Marqués for kindly providing plasmids ALA3-pENTR/D-TOPO and Pro35S:GFP-ALA3, and Luca Santuari for helping with Next Generation Sequencing analysis. We thank the Nottingham Arabidopsis Stock Center for providing T-DNA insertional mutant lines; we also thank the Bioimaging and Life Science facilities of the Institute of Science and Technology Austria, the Cellular Imaging Core Facility of the Central European Institute of Technology supported by the Czech-BioImaging large Research Infrastructure project (LM2018129 funded by the Ministry of Education, Youth and Sports of the Czech Republic), and the Plant Sciences Core Facility of the Central European Institute of Technology, Masaryk University, for their support with obtaining a part of the scientific data. This work was supported by the European Research Council (grant 742985), the Austrian Science Fund (grant I 3630-B25), the China Scholarship Council (PhD scholarship to X.Z.), the Czech Science Foundation (to M.Z., V.P., A.S.S., grant GA 17-17966Y to M.Z. and GA18-26981S to J.F.), the European Regional Development Fund (Project “REMAP” CZ.02.1.01/0.0/0.0/15_003/0000479 to V.K.R.), and the European Molecular Biology Organization (ALTF 723-2015 to S.T.).
AUTHOR CONTRIBUTIONS
X.Z., M.A., P.M., S.T., M.Z., T.N., and J.F. designed the experiments; X.Z., M.A., M.Z., S.T., L.R., V.P., A.S.S., Y.Z., and V.K.R. performed the experiments; C.S.H. contributed to NGS analysis of the repp12 mutant; X.Z. and J.F. wrote the article, and all authors revised it.
Footnotes
Articles can be viewed without a subscription.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C.(2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. [DOI] [PubMed] [Google Scholar]
- Adamowski M., Friml J.(2015). PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M., Narasimhan M., Kania U., Glanc M., De Jaeger G., Friml J.(2018). A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30: 700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengot L., Marquès-Bueno M.M., Jaillais Y.(2016). Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 67: 4015–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa I.C.R., Schwechheimer C.(2014). Dynamic control of auxin transport-dependent growth by AGCVIII protein kinases. Curr. Opin. Plant Biol. 22: 108–115. [DOI] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCFTIR1/AFB-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B., Ruiz L., Martí E., Tárraga S., Tiburcio A.F., Culiáñez F., Farràs R., Carrasco P., Ferrando A.(2012). Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J.(2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F.(2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131. [DOI] [PubMed] [Google Scholar]
- Best J.T., Xu P., Graham T.R.(2019). Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 59: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S., Park S.K., Hua Z., Liu K., Gobin R., Peyroche A., Rambourg A., Graham T.R., Jackson C.L.(2004). The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 117: 711–722. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Graham T.R.(1998). An arf1Δ synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics 150: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Ingram M.F., Rosal P.H., Graham T.R.(1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147: 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costa S.R., Marek M., Axelsen K.B., Theorin L., Pomorski T.G., López-Marqués R.L.(2016). Role of post-translational modifications at the β-subunit ectodomain in complex association with a promiscuous plant P4-ATPase. Biochem. J. 473: 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Zhang C., Kania U., Chen F., Xue Q., McCray T., Li G., Qin G., Wakeley M., Terzaghi W., Wan J., Zhao Y., et al. (2012). A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Slaughter B.D., Unruh J.R., Bradford W.D., Alexander R., Rubinstein B., Li R.(2012). Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat. Cell Biol. 14: 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejonghe W., et al. (2019). Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 15: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L., Hardtke C.S.(2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. USA 110: 7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R.(2015). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 142: 2386–2387. [DOI] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski Ł., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J.(2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452. [DOI] [PubMed] [Google Scholar]
- Du Y., Tejos R., Beck M., Himschoot E., Li H., Robatzek S., Vanneste S., Friml J.(2013). Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. USA 110: 7946–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky J.G., Napsucialy-Mendivil S., Duclercq J., Cheng Y., Shishkova S., Ivanchenko M.G., Friml J., Murphy A.S., Benková E.(2011). Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 191: 970–983. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E.(2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Asaoka R., Paciorek T., De Rycke R., Tanaka H., Nakano A., Friml J.(2012). BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 24: 3074–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J.(2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343. [DOI] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P.(2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628. [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wiśniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K.(2002a). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G.(2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K.(2002b). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Gall W.E., Geething N.C., Hua Z., Ingram M.F., Liu K., Chen S.I., Graham T.R.(2002). Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 12: 1623–1627. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G.(2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J.(2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K.(2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G.(2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400. [DOI] [PubMed] [Google Scholar]
- Glanc M., Fendrych M., Friml J.(2018). Mechanistic framework for cell-intrinsic re-establishment of PIN2 polarity after cell division. Nat. Plants 4: 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A., Ashley M.K., Rigas S., Hatzopoulos P., Dolan L., Vicente-Agullo F.(2005). Morphometric analysis of root shape. New Phytol. 165: 641–651. [DOI] [PubMed] [Google Scholar]
- Heard W., Sklenář J., Tomé D.F.A., Robatzek S., Jones A.M.E.(2015). Identification of regulatory and cargo proteins of endosomal and secretory pathways in Arabidopsis thaliana by proteomic dissection. Mol. Cell. Proteomics 14: 1796–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M.(2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hiraizumi M., Yamashita K., Nishizawa T., Nureki O.(2019). Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365: 1149–1155. [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R.(2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., Nakano A.(2012). Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69: 204–216. [DOI] [PubMed] [Google Scholar]
- Javorka P., Raxwal V.K., Najvarek J., Riha K.(2019). artMAP: A user-friendly tool for mapping ethyl methanesulfonate-induced mutations in Arabidopsis. Plant Direct 3: e00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelínková A., Malínská K., Simon S., Kleine-Vehn J., Parezová M., Pejchar P., Kubeš M., Martinec J., Friml J., Zazímalová E., Petrásek J.(2010). Probing plant membranes with FM dyes: Tracking, dragging or blocking? Plant J. 61: 883–892. [DOI] [PubMed] [Google Scholar]
- Jensen M.S., Costa S.R., Duelli A.S., Andersen P.A., Poulsen L.R., Stanchev L.D., Gourdon P., Palmgren M., Günther Pomorski T., López-Marqués R.L.(2017). Phospholipid flipping involves a central cavity in P4 ATPases. Sci. Rep. 7: 17621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D.J., Cao X., Wang W., Tan X.Y., Zhang X.Q., Chen L.Q., Ye D.(2009). GNOM-LIKE 2, encoding an adenosine diphosphate-ribosylation factor-guanine nucleotide exchange factor protein homologous to GNOM and GNL1, is essential for pollen germination in Arabidopsis. J. Integr. Plant Biol. 51: 762–773. [DOI] [PubMed] [Google Scholar]
- Jing Y., Zheng X., Zhang D., Shen N., Wang Y., Yang L., Fu A., Shi J., Zhao F., Lan W., Luan S.(2019). Danger-associated peptides interact with PIN-dependent local auxin distribution to inhibit root growth in Arabidopsis. Plant Cell 31: 1767–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania U., Fendrych M., Friml J.(2014). Polar delivery in plants; commonalities and differences to animal epithelial cells. Open Biol. 4: 140017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., de Meyer B., Hilson P.(2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A.(2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Adamowski M., Matsuura Y., Santuari L., Kouno H., Arima K., Hardtke C.S., Friml J., Kakimoto T., Tanaka H.(2017). BEN3/BIG2 ARF GEF is involved in Brefeldin A-Sensitive trafficking at the trans-Golgi network/early endosome in Arabidopsis thaliana. Plant Cell Physiol. 58: 1801–1811. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J.(2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., et al. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J.(2008a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Ding Z., Jones A.R., Tasaka M., Morita M.T., Friml J.(2010). Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 107: 22344–22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Łangowski Ł., Wiśniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008b). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y.(2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani E.R., Kilinc A., Smyth D.R.(2013). Auxin controls petal initiation in Arabidopsis. Development 140: 185–194. [DOI] [PubMed] [Google Scholar]
- Langhans M., Marcote M.J., Pimpl P., Virgili-López G., Robinson D.G., Aniento F.(2008). In vivo trafficking and localization of p24 proteins in plant cells. Traffic 9: 770–785. [DOI] [PubMed] [Google Scholar]
- Łangowski Ł., Wabnik K., Li H., Vanneste S., Naramoto S., Tanaka H., Friml J.(2016). Cellular mechanisms for cargo delivery and polarity maintenance at different polar domains in plant cells. Cell Discov. 2: 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Uchida Y., Wang J., Matsudaira T., Nakagawa T., Kishimoto T., Mukai K., Inaba T., Kobayashi T., Molday R.S., Taguchi T., Arai H.(2015). Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34: 669–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.R., Muday G.K.(2009). Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4: 437–451. [DOI] [PubMed] [Google Scholar]
- Li B., Takahashi D., Kawamura Y., Uemura M.(2012). Comparison of plasma membrane proteomic changes of Arabidopsis suspension-cultured cells (T87 Line) after cold and ABA treatment in association with freezing tolerance development. Plant Cell Physiol. 53: 543–554. [DOI] [PubMed] [Google Scholar]
- Li L., Xu J., Xu Z.H., Xue H.W.(2005). Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C., Zwiewka M., Heilmann I., Van Montagu M.C.E., Teichmann T., Friml J.(2013). Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. USA 110: 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués R.L., Poulsen L.R., Hanisch S., Meffert K., Buch-Pedersen M.J., Jakobsen M.K., Pomorski T.G., Palmgren M.G.(2010). Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA α-subunit. Mol. Biol. Cell 21: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marqués R.L., Poulsen L.R., Palmgren M.G.(2012). A putative plant aminophospholipid flippase, the Arabidopsis P4 ATPase ALA1, localizes to the plasma membrane following association with a β-subunit. PLoS One 7: e33042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marques R.L., Theorin L., Palmgren M.G., Pomorski T.G.(2014). P4-ATPases: Lipid flippases in cell membranes. Pflugers Arch. 466: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R.A., Grisafi P., Fink G.R.(1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12: 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Vert G.(2014). The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141: 2924–2938. [DOI] [PubMed] [Google Scholar]
- Marhavý P., Bielach A., Abas L., Abuzeineh A., Duclercq J., Tanaka H., Pařezová M., Petrášek J., Friml J., Kleine-Vehn J., Benková E.(2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21: 796–804. [DOI] [PubMed] [Google Scholar]
- Marhavý P., Duclercq J., Weller B., Feraru E., Bielach A., Offringa R., Friml J., Schwechheimer C., Murphy A., Benková E.(2014). Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 24: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Mathur J., Koncz C.(1998). PEG-mediated protoplast transformation with naked DNA. Methods Mol. Biol. 82: 267–276. [DOI] [PubMed] [Google Scholar]
- Mazur E., Kulik I., Hajný J., Friml J.(2020). Auxin canalization and vascular tissue formation by TIR1/AFB-mediated auxin signaling in Arabidopsis. New Phytol.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell S.C., López-Marqués R.L., Cohen T., Brown E., Rosenberg A., Palmgren M.G., Harper J.F.(2015). Loss of the Arabidopsis thaliana P4-ATPases ALA6 and ALA7 impairs pollen fitness and alters the pollen tube plasma membrane. Front Plant Sci 6: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell S.C., López-Marqués R.L., Poulsen L.R., Palmgren M.G., Harper J.F. (2013). Loss of the Arabidopsis thaliana P4-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS One 8: e62577.23667493 [Google Scholar]
- McGough I.J., de Groot R.E.A., Jellett A.P., Betist M.C., Varandas K.C., Danson C.M., Heesom K.J., Korswagen H.C., Cullen P.J.(2018). SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat. Commun. 9: 3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Movafeghi A., Happel N., Pimpl P., Tai G.H., Robinson D.G.(1999). Arabidopsis Sec21p and Sec23p homologs. Probable coat proteins of plant COP-coated vesicles. Plant Physiol. 119: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy B.P., Natarajan P., Zhou X., Graham T.R.(2009). Linking phospholipid flippases to vesicle-mediated protein transport. Biochim. Biophys. Acta 1791: 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Kleine-Vehn J., Robert S., Fujimoto M., Dainobu T., Paciorek T., Ueda T., Nakano A., van Montagu M.C.E., Fukuda H., Friml J.(2010). ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl. Acad. Sci. USA 107: 21890–21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Otegui M.S., Kutsuna N., de Rycke R., Dainobu T., Karampelias M., Fujimoto M., Feraru E., Miki D., Fukuda H., Nakano A., Friml J.(2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan M., Johnson A., Prizak R., Kaufmann W.A., Tan S., Casillas-Pérez B., Friml J.(2020). Evolutionarily unique mechanistic framework of clathrin-mediated endocytosis in plants. eLife 9: e52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P., Liu K., Patil D.V., Sciorra V.A., Jackson C.L., Graham T.R.(2009). Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat. Cell Biol. 11: 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nintemann S.J., Palmgren M., López-Marqués R.L.(2019). Catch you on the flip side: A critical review of flippase mutant phenotypes. Trends Plant Sci. 24: 468–478. [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J.(2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. [DOI] [PubMed] [Google Scholar]
- Panatala R., Hennrich H., Holthuis J.C.M.(2015). Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 128: 2021–2032. [DOI] [PubMed] [Google Scholar]
- Pimpl P., Movafeghi A., Coughlan S., Denecke J., Hillmer S., Robinson D.G.(2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre M.P., et al. (2018). A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev. Cell 45: 465–480.e11. [DOI] [PubMed] [Google Scholar]
- Platre M.P., et al. (2019). Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62. [DOI] [PubMed] [Google Scholar]
- Poulsen L.R., López-Marqués R.L., McDowell S.C., Okkeri J., Licht D., Schulz A., Pomorski T., Harper J.F., Palmgren M.G.(2008). The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20: 658–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L.R., López-Marqués R.L., Pedas P.R., McDowell S.C., Brown E., Kunze R., Harper J.F., Pomorski T.G., Palmgren M.(2015). A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat. Commun. 6: 7649. [DOI] [PubMed] [Google Scholar]
- Prát T., Hajný J., Grunewald W., Vasileva M., Molnár G., Tejos R., Schmid M., Sauer M., Friml J.(2018). WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet. 14: e1007177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H., Abbas M., Han H., Song S., Robert H.S., Friml J.(2016). Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr. Biol. 26: 3026–3032. [DOI] [PubMed] [Google Scholar]
- Rakusová H., Gallego-Bartolomé J., Vanstraelen M., Robert H.S., Alabadí D., Blázquez M.A., Benková E., Friml J.(2011). Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67: 817–826. [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C.(2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.D., Rios G., Koncz C., Robinson D.G., Jürgens G.(2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492. [DOI] [PubMed] [Google Scholar]
- Richter S., Kientz M., Brumm S., Nielsen M.E., Park M., Gavidia R., Krause C., Voss U., Beckmann H., Mayer U., Stierhof Y.D., Jürgens G.(2014). Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3: e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Grones P., Stepanova A.N., Robles L.M., Lokerse A.S., Alonso J.M., Weijers D., Friml J.(2013). Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23: 2506–2512. [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K.(2004). Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanenka Y., Verstraeten I., Löfke C., Tabata K., Naramoto S., Glanc M., Friml J.(2018). Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc. Natl. Acad. Sci. USA 115: 3716–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benková E., Friml J. (2006). Immunocytochemical Techniques for Whole-Mount in Situ Protein Localization in Plants. Nat. Protoc. 1: 98–103. [DOI] [PubMed] [Google Scholar]
- Shinohara N., Taylor C., Leyser O.(2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.L., Platre M.P., Marquès-Bueno M.M., Armengot L., Stanislas T., Bayle V., Caillaud M.C., Jaillais Y. (2016). A PtdIns(4)P-driven Electrostatic Field Controls Cell Membrane Identity and Signalling in Plants. Nat. Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen Q., Qi L., Jiang H., Li S., Xu Y., Liu F., Zhou W., Pan J., Li X., Palme K., Li C.(2011). Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol. 191: 360–375. [DOI] [PubMed] [Google Scholar]
- Tan S., Abas M., Verstraeten I., Glanc M., Molnár G., Hajný J., Lasák P., Petřík I., Russinova E., Petrášek J., Novák O., Pospíšil J., et al. (2020). Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr. Biol. 30: 381–395.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., Rakusová H., Uemura T., Feraru M.I., De Rycke R., Robert S., Kakimoto T., Friml J.(2013). Cell polarity and patterning by PIN trafficking through early endosomal compartments in Arabidopsis thaliana. PLoS Genet. 9: e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.K., Moore I.(2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496. [DOI] [PubMed] [Google Scholar]
- Timcenko M., et al. (2019). Structure and autoregulation of a P4-ATPase lipid flippase. Nature 571: 366–370. [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C.(2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P.C., Hsu J.W., Liu Y.W., Chen K.Y., Lee F.J.S.(2013). Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc. Natl. Acad. Sci. USA 110: E668–E677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W., Ryan A., Somerville S.C.(2017). An Arabidopsis lipid flippase is required for timely recruitment of defenses to the host-pathogen interface at the plant cell surface. Mol. Plant 10: 805–820. [DOI] [PubMed] [Google Scholar]
- Wabnik K., Robert H.S., Smith R.S., Friml J.(2013). Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 23: 2513–2518. [DOI] [PubMed] [Google Scholar]
- Weijers D., Franke-van Dijk M., Vencken R.J., Quint A., Hooykaas P., Offringa R.(2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wicky S., Schwarz H., Singer-Krüger B.(2004). Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol. Cell. Biol. 24: 7402–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Růžička K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J.(2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Xu P., Baldridge R.D., Chi R.J., Burd C.G., Graham T.R.(2013). Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J. Cell Biol. 202: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Žádníková P., Wabnik K., Abuzeineh A., Gallemi M., van der Straeten D., Smith R.S., Inzé D., Friml J., Prusinkiewicz P., Benková E.(2016). A model of differential growth-guided apical hook formation in plants. Plant Cell 28: 2464–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzyński T., Pěnčík A., Rolčík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Oppenheimer D.G.(2009). IRREGULAR TRICHOME BRANCH 2 (ITB2) encodes a putative aminophospholipid translocase that regulates trichome branch elongation in Arabidopsis. Plant J. 60: 195–206. [DOI] [PubMed] [Google Scholar]