Abstract
Background
This meta-analysis evaluated the association of LTF, ENAM, and AMELX polymorphisms with dental caries susceptibility.
Methods
We searched the Scopus, PubMed/Medline, Web of Science, and Cochrane Library databases to retrieve articles published by October 2019. Review Manager 5.3 software was used to estimate the odds ratios (ORs) and 95% confidence intervals (CIs). The results of publication bias tests were retrieved by Comprehensive Meta-Analysis 2.0 software.
Results
A total of 150 relevant records were identified; out of which, 16 were entered into the analysis (4 studies assessed LTF, 11 ENAM, and 11 AMELX polymorphisms). Of all polymorphisms, there was a significant association only between ENAM rs3796704 polymorphism and dental caries susceptibility. Both ENAM rs3796704 and AMELX rs17878486 polymorphisms had a significant association with dental caries risk in the Caucasian ethnicity and the studies including caries-free control group.
Conclusions
The results of this meta-analysis showed that the G allele and the GG genotype of ENAM rs3796704 were associated with an increased risk of caries in the case group compared with the control group. But there was no association between LTF rs1126478, ENAM (rs1264848 and rs3796703), and AMELX (rs946252, rs17878486, and rs2106416) polymorphisms and dental caries susceptibility.
Keywords: Dental caries, Polymorphism, Lactotransferrin, Enamelin, Amelogenin X
Highlights
1. There was an association between ENAM rs3796704 polymorphism and the risk of dental caries.
2. There was no association between polymorphisms of LTF (rs1126478), ENAM (rs1264848 and rs3796703), and AMELX (rs946252, rs17878486, and rs2106416) and dental caries susceptibility.
Introduction
Dental caries can significantly affect the general health and quality of life in the modern world [1]. Dental caries develops following demineralization of tooth structure and often results in pulpal and periapical inflammation, and subsequent pain, infection and tooth loss [1, 2]. Some environmental conditions and population groups may contribute to higher incidence of dental caries [2]. Environmental risk factors may also affect dental caries development [3]. Although exposed to the same environmental conditions, some patients may be more sensitive or more resistant to dental caries than others; such differences may be due to genetic factors in dental caries etiopathogenesis [4]. New findings raised possibilities of presence of associations between genetic factors and dental caries [5]. The etiology of dental caries involves complex interactions between genetic and environmental factors. Three prevalent genes are reportedly involved in development of dental caries namely the lactotransferrin (LTF), enamelin (ENAM), and amelogenin X (AMELX). The rs1126478 polymorphism of LTF (a saliva protein gene) produces a shift from arginine to lysine at amino acid position 47 in the antimicrobial region, and presents transcriptional activation activity [6]. LTF can decrease the lipopolysaccharide-activated innate immune response, regulate the adaptive immune system [7], and play a significant role in physiological homoeostasis, which is in turn related to disease development [8]. ENAM (a member of P/Q-rich secretory calcium-binding phosphoprotein cluster genes) is located on chromosome 4q 13.3 [9]. The gene encodes the protein enamelin, which is the largest protein found in the enamel matrix and is involved in dental enamel mineralization and its structural organization [10]. AMELX is an essential gene that produces amelogenin as the main protein of dental enamel during the secretion stage of amelogenesis [11]. AMELX polymorphisms result in distinct alternations in enamel microstructure [12]. Therefore, these polymorphisms play a critical role in regulation of mineralization and enamel thickness [13]. The association between the mutations of LTF, ENAM, and AMELX genes and dental caries susceptibility has been shown in some studies [5, 14, 15]. Therefore, we aimed to assess the association of polymorphisms of these genes and the risk of dental caries in a meta-analysis of case-control studies and therefore evaluating only the genetic influences for dental caries.
Materials and methods
Search strategy and study selection
One author systematically searched the PubMed/Medline, Web of Science, Cochrane Library, and Scopus databases to retrieve articles published by October 2019 without publication period, language, and patient’s age restrictions. The search terms or keywords were (“lactotransferrin” or “lactoferrin” or “LTF” or “enamelin” or “ENAM” or “amelogenin X” or “AMELX”) and (“dental caries” or “caries” or “decay”) and (“gene” or “polymorphism” or “variant” or “genetic”). In addition, the references of the retrieved articles related to the topic including original and review articles were searched to make sure that no study was missed. After article retrieval, another author assessed the titles and abstracts of the articles related to the topic; subsequently, the full-texts of the articles that met our eligibility criteria were downloaded and screened. After screening, the exclusion reason was recorded for any study removed, and the disagreements between the authors were resolved by another author.
Eligibility criteria
The inclusion criteria were as follows: (I) studies including two independent groups (case group with caries or high caries and caries-free control group or with low/very low caries) without age restriction, (II) studies with any defined Decayed, Missing, and Filled Teeth (DMFT) score for the two groups, (III) studies including one or more polymorphisms of LTF, ENAM, and AMELX genes with a minimum of two relevant studies for the analysis; for example, four studies assessed LTF rs1126478, ENAM rs1264848, ENAM rs3796704, ENAM rs3796703, AMELX rs946252, AMELX rs17878486, AMELX rs6639060, and AMELX rs2106416 polymorphisms; and (IV) patients and controls had to have no genetic diseases, chronic illnesses, or other disorders. We excluded irrelevant studies, studies without sufficient data for analysis, studies without a control group, studies including less than 20 individuals in each group, duplicate studies, animal studies, case reports, conference papers, reviews, and systematic reviews.
Data abstraction
Two authors independently abstracted the data of the studies analyzed in the meta-analysis. The data from each study, including first author, publication year, country of residence of the included individuals, ethnicity, mean age of individuals in the two groups, age group of individuals in each study, genotyping method, DMFT score of the two groups, and type of reported polymorphism (s) in each study, were extracted and analyzed.
Statistical analysis
Review Manager 5.3 (RevMan 5.3) software was applied to compute the odds ratios (ORs) and 95% confidence intervals (CIs). To estimate the significance of the pooled OR by the Z test, a p-value (two-sided) < 0.05 was considered significant. The I2 statistic was used to estimate heterogeneity. A p < 0.1 or I2 > 50% indicated a significant heterogeneity and we used the random-effects model for such cases; if not, the fixed-effects model was used. The publication bias across the studies was assessed using the Egger’s and Begg’s tests. If p < 0.05 (two-sided) for both tests or one, there was a significant degree of publication bias. In order to evaluate the stability/consistency of the results, the sensitivity analysis with both “the removal of one study” and “cumulative analysis” was performed. The results of these tests were retrieved by Comprehensive Meta-Analysis 2.0 (CMA 2.0) software. All authors revised the extracted data and the analyses and the disagreement between them was resolved by a discussion.
Results
Study selection
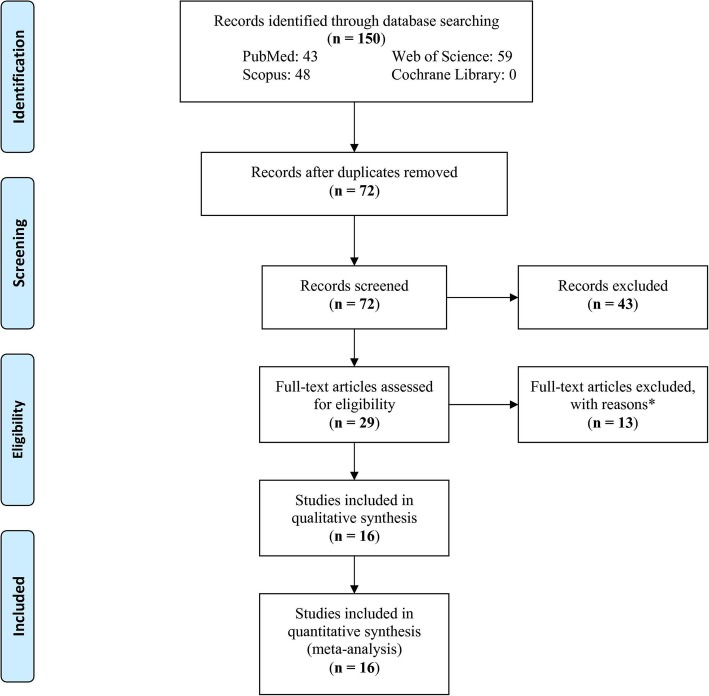
A total of 150 records were identified in the databases; after removing the duplicates and irrelevant records, 29 full-text articles were evaluated for eligibility (Fig. 1). Next, 13 articles were excluded with reasons: two studies were systematic reviews, two studies were reviews, three studies lacked sufficient data, two studies had no control group, three studies did not report any of the mentioned polymorphisms in this meta-analysis and did not have any known polymorphism either, and one study reported rs1126478 polymorphism with less than 20 individuals in each group. Finally, 16 studies were entered into the analysis.
Fig. 1.
Flow chart of study selection
Study characteristics
The characteristics of 16 studies included in this meta-analysis are presented in Table 1. The articles had been published from 2008 to 2019. Eleven studies had been conducted on Caucasians [12, 14–16, 18–23, 25], four studies had been conducted on Asians [5, 13, 17, 24], and one on mixed [11] ethnicities. Eleven studies evaluated children [5, 11, 13–16, 18–20, 22, 24], three studies evaluated adults [17, 21, 23], and two studies evaluated both adults and children [12, 25]. Four studies had assessed LTF rs1126478 in 1556 cases and 1106 controls [5, 16, 20, 24]. Five studies had assessed ENAM rs1264848 in 934 cases and 600 controls [11, 14, 19, 22, 25]. Four studies had assessed ENAM rs3796704 in 574 cases and 533 controls [11, 19, 23, 25]. Two studies had assessed ENAM rs3796703 in 585 cases and 567 controls [5, 13]. Three studies had assessed AMELX rs946252 in 151 cases and 147 controls [11, 12, 19]. Four studies had assessed AMELX rs17878486 in 249 cases and 193 controls [11, 15, 17, 19]. Two studies had assessed AMELX rs6639060 in 157 cases and 144 controls [13, 21], and two studies had assessed AMELX rs2106416 in 175 cases and 110 controls [13, 18]. One study [18] included two subsets. The genotyping method in all studies was based on polymerase chain reaction (PCR). The controls in six studies [11, 17, 18, 21–23] had low/very low rate of dental caries and others were caries-free. In addition, the two groups in the studies were introduced as the caries group versus the caries-free group, or the high-caries group versus the low/very low caries group.
Table 1.
Characteristics of the studies included in this meta-analysis
| First author, publication year | Country | Ethnicity | Mean age, years (Case/ Control) | Age group of individuals | Genotyping method | DMFT score (Case/Control) | Polymorphisms |
|---|---|---|---|---|---|---|---|
| Ouryouji, 2008 [13] | Japan | Asian | 5.4/4.8 | Children | PCR-RFLP | ≥10/0 |
ENAM rs3796703 AMELX rs6639060 AMELX rs2106416 |
| Azevedo, 2010 [16] | Brazil | Caucasian | 12 | Children | PCR-SSCP | ≥1/0 | LTF rs1126478 |
| Kang, 2011a [17] | Korea | Asian | 21.3/23.2 | Adults | PCR | > 2/≤2 | AMELX rs17878486 |
| Olszowski, 2012ab [18] | Poland | Caucasian | 5 | Children | PCR-RFLP | ≥3/< 3 | AMELX rs2106416 |
| 13 | Children | PCR-RFLP | ≥3/< 3 | AMELX rs2106416 | |||
| Gasse, 2013 [12] | France | Caucasian | 7.6/22 | Both | PCR | ≥4/0 | AMELX rs946252 |
| Jeremias, 2013a [11] | Brazil | Mixed |
Case: < 6 Control: < 20 |
Children | RT-PCR | ≥4/≤3 |
ENAM rs1264848 ENAM rs3796704 AMELX rs946252 AMELX rs17878486 |
| Ergöz, 2014 [19] | Turkey | Caucasian | 8.7/8.7 | Children | TaqMan | ≥1/0 |
ENAM rs1264848 ENAM rs3796704 AMELX rs946252 AMELX rs17878486 |
| Volckova, 2014 [20] | Czech | Caucasian | Range: 11–13 | Children | PCR-RFLP | ≥1/0 | LTF rs1126478 |
| Gerreth, 2016 [14] | Poland | Caucasian | 2.6 | Children | RT-PCR | ≥1/0 | ENAM rs1264848 |
| Yildiz, 2016a [21] | Turkey | Caucasian | Range: 20–60 | Adults | PCR-RFLP | ≥14/≤5 | AMELX rs6639060 |
| Gerreth, 2017 [15] | Poland | Caucasian | Range: 1.7–3.5 | Children | TaqMan | ≥1/0 | AMELX rs17878486 |
| Wang, 2017 [5] | China | Asian | 3.5/3.7 | Children | PCR | ≥4/0 |
LTF rs1126478 ENAM rs3796703 |
| Borilova Linhartova, 2018a [22] | Czech | Caucasian | Range: 13–15 | Children | TaqMan | ≥1/0 | ENAM rs1264848 |
| Koohpeima, 2018a [23] | Iran | Caucasian | 29.8/28.4 | Adults | ARMS-PCR | NA | ENAM rs3796704 |
| Wang, 2018 [24] | China | Asian | Range: 2–4 | Children | PCR | ≥8/0 | LTF rs1126478 |
| Devang Divakar, 2019 [25] | Saudi Arabia | Caucasian | 6.9/23.2 | Both | RT-PCR | ≥4/0 |
ENAM rs1264848 ENAM rs3796704 |
aCase group included individuals with high caries and control group included individuals with low/very low caries. b This study had two subsets: one subset was reported in 5-year-olds and another in 13-year-olds
Abbreviations: NA Not available, PCR Polymerase chain reaction, RFLP Restriction fragment length polymorphism, RT Real time, ARMS Amplification refractory mutation system, LTF Lactotransferrin, ENAM Enamelin; AMELX; Amelogenin X
The genotype prevalence of each polymorphism included in each study in both case and control groups and the p-value for the Hardy-Weinberg equilibrium (HWE) for the controls are shown in Table 2. The controls in three studies [11, 12, 21] had a deviation from the HWE (P < 0.05).
Table 2.
Prevalence of genotypes of the three polymorphisms (LTF, ENAM and AMELX)
| First author, publication year |
LTF rs1126478a |
ENAM s1264848a |
ENAM rs3796704a |
ENAM rs3796703b | P-value of HWE for controls | ||||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | Case | Control | ||
| Ouryouji, 2008 [13] | NA | NA | NA | NA | NA | NA | 76/4/0 | 59/8/0 | 0.603 |
| Azevedo, 2010 [16] | 16/30/16 | 19/22/7 | NA | NA | NA | NA | NA | NA | 0.877 |
| Jeremias, 2013 [11] | NA | NA | 13/44/20 | 21/41/20 | 4/11/61 | 1/19/57 | NA | NA | 0.999/0.676 |
| Ergöz, 2014 [19] | NA | NA | 4/41/55 | 1/35/64 | 0/16/78 | 1/27/69 | NA | NA | 0.108/0.351 |
| Volckova, 2014 [20] | 288/150/44 | 86/56/13 | NA | NA | NA | NA | NA | NA | 0.374 |
| Gerreth, 2016 [14] | NA | NA | 8/37/3 | 4/26/18 | NA | NA | NA | NA | 0.202 |
| Wang, 2017 [5] | 64/222/219 | 64/209/227 | NA | NA | NA | NA | 439/64/2 | 458/42/0 | 0.149/0.327 |
| Borilova Linhartova, 2018 [22] | NA | NA | 45/259/237 | 19/74/84 | NA | NA | NA | NA | 0.656 |
| Koohpeima, 2018 [23] | NA | NA | NA | NA | 0/32/204 | 0/20/146 | NA | NA | 0.409 |
| Wang, 2018 [24] | 63/224/220 | 51/168/184 | NA | NA | NA | NA | NA | NA | 0.196 |
| Devang Divakar, 2019 [25] | NA | NA | 89/58/21 | 116/68/9 | 11/58/99 | 25/76/92 | NA | NA | 0.809/0.146 |
| First author, publication year | AMELX rs946252b | AMELX rs17878486b | AMELX rs6639060b | AMELX rs2106416b | P-value of HWE for controls | ||||
| Case | Control | Case | Control | Case | Control | Case | Control | ||
| Ouryouji, 2008 [13] | NA | NA | NA | NA | 80/0/0 | 67/0/0 | 78/2/0 | 16/6/0 | NA/0.458 |
| Kang, 2011 [17] | NA | NA | 1/2/82 | 0/2/29 | NA | NA | NA | NA | 0.852 |
| Olszowski, 2012 (i) [18] | NA | NA | NA | NA | NA | NA | 24/13/0 | 21/12/1 | 0.643 |
| Olszowski, 2012 (ii) [18] | NA | NA | NA | NA | NA | NA | 26/26/6 | 28/20/2 | 0.495 |
| Gasse, 2013 [12] | 5/9/25 | 4/1/25 | NA | NA | NA | NA | NA | NA | < 0.0001 |
| Jeremias, 2013 [11] | 61/12/5 | 63/13/5 | 4/15/57 | 15/11/56 | NA | NA | NA | NA | 0.002/< 0.0001 |
| Ergöz, 2014 [19] | 5/18/11 | 10/17/9 | 8/18/14 | 3/21/8 | NA | NA | NA | NA | 0.742/0.051 |
| Yildiz, 2016 [21] | NA | NA | NA | NA | 54/11/12 | 56/9/12 | NA | NA | < 0.0001 |
| Gerreth, 2017 [15] | NA | NA | 8/10/30 | 31/14/3 | NA | NA | NA | NA | 0.422 |
Abbreviations: LTF Lactotransferrin, ENAM Enamelin, AMELX Amelogenin X, NA Not available, HWE Hardy-Weinberg equilibrium
aGenotypes: AA/AG/GG
bGenotypes: CC/CT/TT. i: 5-year-olds. ii: 13-year-olds
Pooled analysis
Table 3 shows the results of pooled analysis of each polymorphism based on five genetic models. Among the polymorphisms, only the G allele [OR = 1.38; 95%CI: 1.08, 1.76; P = 0.009; I2 = 27% (Pheterogeneity or Ph = 0.25)] and the GG genotype [OR = 1.41; 95%CI: 1.06, 1.87; P = 0.02; I2 = 18, (Ph = 0.30)]) of ENAM rs3796704 had an elevated risk in the case group compared with the control group. In addition, the funnel plots of each polymorphism are presented in the Supplementary file. Therefore, there was a significant association between ENAM rs3796704 polymorphism and dental caries susceptibility.
Table 3.
Results of pooled analysis of each polymorphism based on five genetic models
| Polymorphism, (number of studies) | G vs. A | GG vs. AA | AG vs. AA | AG + GG vs. AA | GG vs. AA + AG |
|---|---|---|---|---|---|
| OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | |
| LTF rs1126478 (n = 4) | 0.98 (0.87, 1.10), 0.68, 25, 0.26 | 1.03 (0.80, 1.33), 0.82, 4, 0.37 | 1.00 (0.80, 1.25), 0.99, 4, 0.44 | 0.99 (0.81, 1.23), 0.96, 7, 0.36 | 0.95 (0.80, 1.13), 0.58, 0, 0.45 |
| ENAM rs1264848 (n = 5) | 0.93 (0.65, 1.32), 0.68, 75, 0.003 | 0.88 (0.34, 2.30), 0.79, 76, 0.002 | 1.21 (0.88, 1.65), 0.24, 0, 0.48 | 1.24 (0.93, 1.67), 0.15, 24, 0.26 | 0.83 (0.44, 1.58), 0.58, 79, 0.0007 |
| ENAM rs3796704 (n = 4) | 1.38 (1.08, 1.76), 0.009, 27, 0.25 | 1.86 (0.96, 3.62), 0.07, 44, 0.17 | 1.27 (0.64, 2.52), 0.49, 50, 0.13 | 1.64 (0.86, 3.14), 0.13, 43, 0.17 | 1.41 (1.06, 1.87), 0.02, 18, 0.30 |
| Polymorphism, (number of studies) | T vs. C | TT vs. CC | CT vs. CC | CT + TT vs. CC | TT vs. CC + CT |
| OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | |
| ENAM rs3796703 (n = 2) | 0.92 (0.24, 3.58), 0.91, 78, 0.03 | 5.22 (0.25, 108.96), 0.29 | 0.89 (0.23, 3.48), 0.87, 77, 0.04 | 0.90 (0.23, 3.63), 0.89, 78, 0.03 | 2.90 (0.92, 9.12), 0.07, 0, 0.69 |
| AMELX rs946252 (n = 3) | 1.01 (0.68, 1.51), 0.95, 39, 0.20 | 1.27 (0.58, 2.75), 0.55, 0, 0.51 | 1.45 (0.75, 2.81), 0.27, 31, 0.23 | 1.21 (0.68, 2.14), 0.52, 0, 0.51 | 0.82 (0.43, 1.56), 0.54, 0.38, 0.20 |
| AMELX rs17878486 (n = 4) | 2.25 (0.81, 6.24), 0.12, 86, 0.0001 | 3.59 (0.55, 23.32), 0.18, 81, 0.001 | 1.45 (0.36, 5.80), 0.60, 0.67, 0.03 | 2.30 (0.51, 10.33), 0.28, 77, 0.004 | 3.10 (0.85, 11.28), 0.09, 81, 0.001 |
| AMELX rs6639060 (n = 2) | 1.08 (0.63, 1.85), 0.78 | 1.04 (0.43, 2.51), 0.94 | 1.27 (0.49, 3.30), 0.63 | 1.14 (0.56, 2.29), 0.72 | 1.00 (0.42, 2.39), 1.00 |
| AMELX rs2106416 (n = 3) | 0.59 (0.16, 2.11), 0.41, 82, 0.003 | 1.83 (0.47, 7.06), 0.38, 40, 0.20 | 0.55 (0.13, 2.25), 0.40, 80, 0.006 | 0.59 (0.15, 2.31), 0.45, 79, 0.008 | 1.67 (0.44, 6.34), 0.45, 31, 0.23 |
Abbreviations: LTF Lactotransferrin, ENAM Enamelin, AMELX Amelogenin X, NA Not available, OR Odds ratio, CI Confidence interval; Ph, Pheterogeneity
Subgroup analysis
Table 4 identifies the subgroup analyses based on the ethnicity, age group, and the control group for each polymorphism and in five genetic models. The results showed that the G allele and the GG and AG genotypes of ENAM rs3796704 had an elevated risk in the case group compared with the control group in the Caucasian ethnicity; whereas in studies with a caries-free control group, the G allele and GG genotype had an elevated risk in the case group compared with the control group. In addition, the T allele, and TT and CT genotypes of AMELX rs17878486 polymorphism had an elevated risk in the case group compared with the control group in mixed ethnicity and studies with a caries-free control group. Therefore, there was a significant association between both polymorphisms of ENAM rs3796704 and AMELX rs17878486 and dental caries susceptibility in the Caucasian ethnicity and studies including caries-free individuals as the control group.
Table 4.
Subgroup analysis of each polymorphism based on ethnicity, age group, and control group
| Polymorphism, (number of studies) | G vs. A | GG vs. AA | AG vs. AA | AG + GG vs. AA | GG vs. AA + AG |
|---|---|---|---|---|---|
| OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | |
| LTF rs1126478 | |||||
| Ethnicity | |||||
| Caucasian (2) | 1.18 (0.66, 2.11), 0.58, 73, 0.05 | 1.49 (0.58, 3.85), 0.41, 55, 0.13 | 1.02 (0.53, 1.97), 0.95, 53, 0.14 | 1.16 (0.53, 1.97), 0.95, 0.53, 0.14 | 1.33 (0.77, 2.29), 0.30, 6, 0.30 |
| Asian (2) | 0.96 (0.84, 1.09), 0.52, 0, 0.98 | 0.97 (0.73, 1.29), 0.81, 0, 0.99 | 1.07 (0.80, 1.43), 0.64, 0, 0.96 | 1.02 (0.78, 1.33), 0.91, 0, 0.97 | 0.92 (0.77, 1.10), 0.35, 0, 0.96 |
| Age group | |||||
| Children (4) | 0.98 (0.87, 1.10), 0.68, 25, 0.26 | 1.03 (0.80, 1.33), 0.82, 4, 0.37 | 1.00 (0.80, 1.25), 0.99, 4, 0.44 | 0.99 (0.81, 1.23), 0.96, 7, 0.36 | 0.95 (0.80, 1.13), 0.58, 0, 0.45 |
| Control group | |||||
| Caries free (4) | 0.98 (0.87, 1.10), 0.68, 25, 0.26 | 1.03 (0.80, 1.33), 0.82, 4, 0.37 | 1.00 (0.80, 1.25), 0.99, 4, 0.44 | 0.99 (0.81, 1.23), 0.96, 7, 0.36 | 0.95 (0.80, 1.13), 0.58, 0, 0.45 |
| ENAM rs1264848 | |||||
| Ethnicity | |||||
| Caucasian (4) | 0.86 (0.56, 1.32), 0.49, 80, 0.002 | 0.67 (0.18, 2.45), 0.54, 82, 0.0009 | 1.13 (0.80, 1.59), 0.48, 0, 0.45 | 1.18 (0.86, 1.62), 0.31, 35, 0.20 | 0.76 (0.34, 1.72), 0.51, 84, 0.0003 |
| Mixed (1) | 1.23 (0.79,1.91), 0.36 | 1.62 (0.64, 4.09), 0.31 | 1.73 (0.77, 3.90), 0.18 | 1.69 (0.78, 3.68), 0.18 | 1.09 (0.53, 2.23), 0.82 |
| Age group | |||||
| Children (4) | 0.82 (0.57, 1.18), 0.28, 67, 0.03 | 0.58 (0.19, 1.82), 0.35, 74, 0.01 | 1.30 (0.84, 2.01), 0.26, 3, 0.36 | 1.16 (0.76, 1.75), 0.49, 42, 0.16 | 0.66 (0.37, 1.17), 0.15, 70, 0.02 |
| Control group | |||||
| Caries free (3) | 0.79 (0.39, 1.63), 0.53, 86, 0.0006 | 0.42 (0.03, 5.41), 0.51, 88, 0.0003 | 1.00 (0.66, 1.51), 1.00, 0, 0.44 | 0.74 (0.27, 2.02), 0.56, 55, 0.11 | 0.66 (0.15, 2.95), 0.59, 89, < 0.0001 |
| Low caries (2) | 1.03 (0.83, 1.29), 0.78, 0, 0.37 | 1.30 (0.79, 2.14), 0.30, 0, 0.59 | 1.57 (0.97, 2.53), 0.07, 0, 0.76 | 1.45 (0.92, 2.28), 0.11, 0, 0.62 | 0.90 (0.66, 1.23), 0.51, 0, 0.57 |
| ENAM rs3796704 | |||||
| Ethnicity | |||||
| Caucasian (3) | 1.43 (1.10, 1.85), 0.007, 45, 0.16 | 2.49 (1.19, 5.24), 0.02, 0, 0.85 | 1.74 (0.81, 3.74), 0.16, 0, 0.98 | 2.16 (1.05, 4.45), 0.04, 0, 0.85 | 1.41 (1.04, 1.91), 0.03, 45, 0.16 |
| Mixed (1) | 1.11 (0.57, 2.15), 0.77 | 0.27 (0.03, 2.47), 0.24 | 0.14 (0.01, 1.46), 0.10 | 0.24 (0.03, 2.17), 0.20 | 1.43 (0.67, 3.05), 0.36 |
| Age group | |||||
| Children (2) | 1.46 (0.92, 2.32), 0.11, 22, 0.26 | 0.61 (0.13, 2.93), 0.54, 38, 0.20 | 0.35 (0.07, 1.74), 0.20, 35, 0.22 | 0.54 (0.11, 2.56), 0.44, 37, 0.21 | 1.71 (1.02, 2.85), 0.04, 0, 0.53 |
| Control group | |||||
| Caries free (2) | 1.62 (1.21, 2.17), 0.001, 0, 0.59 | 2.499 (1.19, 5.24), 0.02, 0, 0.85 | 1.74 (0.81, 3.74), 0.16, 0, 0.98 | 2.16 (1.05, 4.45), 0.04, 0, 0.85 | 1.67 (1.17, 2.39), 0.0050, 0.58 |
| Low caries (2) | 0.97 (0.63, 1.50), 0.89, 0, 0.61 | 0.27 (0.03, 2.47), 0.24 | 0.14 (0.01, 1.46), 0.10 | 0.24 (0.03, 2.17), 0.20 | 1.05 (0.66, 1.68), 0.83, 0, 0.32 |
| Polymorphism, (number of studies) | T vs. C | TT vs. CC | CT vs. CC | CT + TT vs. CC | TT vs. CC + CT |
| OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | OR (95%CI), P-value, I2 (%), Ph | |
| ENAM rs3796703 | |||||
| Ethnicity | |||||
| Asian (2) | 0.92 (0.24, 3.58), 0.91, 78, 0.03 | 5.22 (0.25, 108.96), 0.29 | 0.89 (0.23, 3.48), 0.87, 77, 0.04 | 0.90 (0.23, 3.63), 0.89, 78, 0.03 | 2.90 (0.92, 9.12), 0.07, 0, 0.69 |
| Age group | |||||
| Children (2) | 0.92 (0.24, 3.58), 0.91, 78, 0.03 | 5.22 (0.25, 108.96), 0.29 | 0.89 (0.23, 3.48), 0.87, 77, 0.04 | 0.90 (0.23, 3.63), 0.89, 78, 0.03 | 2.90 (0.92, 9.12), 0.07, 0, 0.69 |
| AMELX rs946252 | |||||
| Ethnicity | |||||
| Caucasian (2) | 0.95 (0.35, 2.55), 0.92, 69, 0.07 | 1.42 (0.54, 3.76), 0.48, 17, 0.27 | 2.80 (0.93, 8.43), 0.07, 0, 0.38 | 1.63 (0.67, 4.01), 0.28, 0, 0.42 | 0.73 (0.19, 2.87), 0.66, 67, 0.08 |
| Mixed (1) | 0.99 (0.53, 1.86), 0.98 | 1.03 (0.28, 3.75), 0.96 | 0.95 (0.40, 2.25), 0.91 | 0.98 (0.46, 2.07), 0.95 | 1.04 (0.29, 3.75), 0.95 |
| Age group | |||||
| Children (2) | 1.21 (0.77,1.91), 0.42, 0, 0.37 | 1.54 (0.61, 3.92), 0.36, 0, 0.37 | 1.24 (0.61, 2.49), 0.56, 5, 0.31 | 1.24 (0.66, 2.33), 0.50, 24, 0.25 | 1.26 (0.56, 2.83), 0.25 |
| Control group | |||||
| Caries free (2) | 0.95 (0.35, 2.55), 0.92, 69, 0.07 | 1.42 (0.54, 3.76), 0.48, 17, 0.27 | 2.80 (0.93, 8.43), 0.07, 0, 0.38 | 1.63 (0.67, 4.01), 0.28, 0, 0.42 | 0.73 (0.19, 2.87), 0.66, 67, 0.08 |
| Low caries (1) | 0.99 (0.53, 1.86), 0.98 | 1.03 (0.28, 3.75), 0.96 | 0.95 (0.40, 2.25), 0.91 | 0.98 (0.46, 2.07), 0.95 | 1.04 (0.29, 3.75), 0.95 |
| AMELX rs17878486 | |||||
| Ethnicity | |||||
| Caucasian (2) | 2.87 (0.35, 23.23), 0.32, 95, < 0.0001 | 5.12 (0.09, 278.85), 0.42, 93, 0.0002 | 1.00 (0.12, 8.23), 1.00, 0.81, 0.02 | 2.03 (0.10, 42.44), 0.65, 92, 0.0004 | 6.17 (0.41, 92.05), 0.19, 91, 0.001 |
| Asian (1) | 1.38 (0.25, 7.75), 0.71 | 0.93 (0.04, 23.52), 0.97 | 0.33 (0.01, 12.82), 0.56 | 0.89 (0.04, 22.53), 0.95 | 1.89 (0.30, 11.85), 0.50 |
| Mixed (1) | 1.87 (1.06, 3.30), 0.03 | 3.82 (1.19, 12.21), 0.02 | 5.11 (1.33, 19.72), 0.02 | 4.03 (1.27, 12.75), 0.02 | 1.39 (0.69, 2.80), 0.35 |
| Age group | |||||
| Children (3) | 2.48 (0.76, 8.05), 0.13, 90, < 0.0001 | 4.69 (0.55, 40.16), 0.16, 86, 0.0007 | 1.72 (0.37, 7.92), 0.48, 75, 0.02 | 2.65 (0.49, 14.35), 0.26, 84, 0.002 | 3.55 (0.72, 17.60), 0.12, 87, 0.0004 |
| Adults (1) | 1.38 (0.25, 7.75), 0.71 | 0.93 (0.04, 23.52), 0.97 | 0.33 (0.01, 12.82), 0.56 | 0.89 (0.04, 22.53), 0.95 | 1.89 (0.30, 11.58), 0.50 |
| Control group | |||||
| Caries free (2) | 2.87 (0.35, 23.23), 0.32, 95, < 0.0001 | 5.12 (0.09, 278.85), 0.42, 93, 0.0002 | 1.00 (0.12, 8.23), 1.00, 0.81, 0.02 | 2.03 (0.10, 42.44), 0.65, 92, 0.0004 | 6.17 (0.41, 92.05), 0.19, 91, 0.001 |
| Low caries (2) | 1.82 (1.06, 3.11), 0.03, 0, 0.74 | 3.28 (1.14, 9.43), 0.03, 0, 0.42 | 3.42 (1.04, 11.28), 0.04, 47, 0.17 | 3.43 (1.21, 9.75), 0.02, 0, 0.39 | 1.44 (0.75, 2.77), 0.27, 0, 0.76 |
| AMELX rs6639060 | |||||
| Ethnicity | |||||
| Caucasian (1) | 1.08 (0.63, 1.85), 0.78 | 1.04 (0.43, 2.51), 0.94 | 1.27 (0.49,3.30), 0.63 | 1.14 (0.56, 2.29), 0.72 | 1.00 (0.42, 2.39), 1.00 |
| Age group | |||||
| Adults (1) | 1.08 (0.63, 1.85), 0.78 | 1.04 (0.43, 2.51), 0.94 | 1.27 (0.49,3.30), 0.63 | 1.14 (0.56, 2.29), 0.72 | 1.00 (0.42, 2.39), 1.00 |
| AMELX rs2106416 | |||||
| Ethnicity | |||||
| Caucasian (2) | 1.25 (0.77, 2.03), 0.37, 30, 0.23 | 1.83 (0.47, 7.06), 0.38, 40, 0.20 | 1.20 (0.65, 2.22), 0.56, 0, 0.54 | 1.25 (0.69, 2.27), 0.46, 0, 0.35 | 1.67 (0.44, 6.34), 0.45, 31, 0.23 |
| Age group | |||||
| Children (3) | 0.59 (0.16, 2.11), 0.41, 82, 0.003 | 1.83 (0.47, 7.06), 0.38, 40, 0.20 | 0.55 (0.13, 2.25), 0.40, 80, 0.006 | 0.59 (0.15, 2.31), 0.45, 79, 0.008 | 1.67 (0.44, 6.34), 0.45, 31, 0.23 |
Abbreviations: LTF Lactotransferrin, ENAM Enamelin, AMELX Amelogenin X, OR Odds ratio, CI Confidence interval; Ph, Pheterogeneity
Sensitivity analysis
One study [19] was omitted from the analysis of AMELX rs17878486 because the outlier data and the results illustrated that by deleting this study, the CT [OR = 3.07; 95%CI: 1.36, 6.94; P = 0.007; I2 = 0% (Ph = 0.37)] and CT + TT [OR = 5.72; 95%CI: 2.83, 11.59; P = < 0.00001; I2 = 21% (Ph = 0.0.28)] genotypes in dental caries patients were significantly superior to controls and with a low heterogeneity, respectively (Table 3). In addition, other sensitivity analyses including “one study excluded” and “cumulative analysis” were performed and the previous results did not change qualitatively. Although the genotype distribution of the controls in three studies [11, 12, 21] did not follow the HWE, these analyses reported that the pooled ORs based on all genetic models were steady.
Publication bias
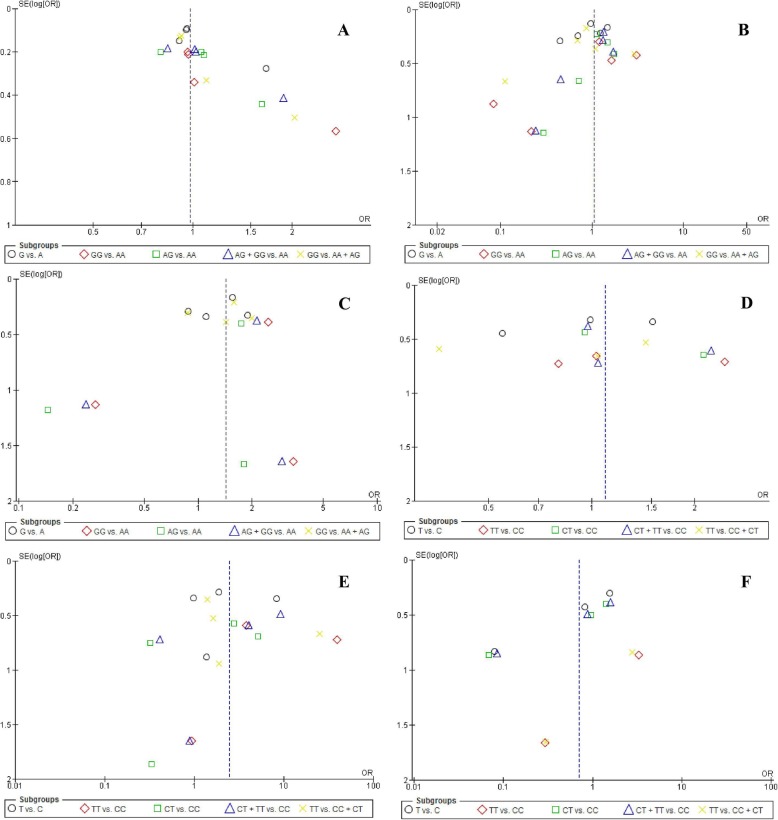
Both Egger’s and Begg’s tests were done on the previous pooled analyses with a minimum of three studies (Fig. 2). The results revealed a publication bias regarding GG vs. AA, AG vs. AA, and AG + GG vs. AA models of LTF rs1126478 polymorphism (Begg’s test: P < 0.05) and also T vs. C and CT + TT vs. CC models of AMELX rs2106416 polymorphism (Begg’s test: P < 0.05).
Fig. 2.
Funnel plots of the analysis of each polymorphism with a minimum of three studies: A) LTF rs1126478, B) ENAM s1264848, C) ENAM rs3796704, D) AMELX rs946252, E) AMELX rs17878486, and F) AMELX rs2106416
Discussion
The effect of environmental risk factors [26] and genetic predisposition [27] on development of caries has been well identified. This meta-analysis assessed the association between the common polymorphisms of LTF, ENAM, and AMELX and the risk of dental caries. The findings showed that ENAM rs3796704 polymorphism had an increased risk in the case group compared with the control group especially in the Caucasian ethnicity and studies with caries-free individuals as the control group.
The prevalence of LTF polymorphisms differs among various ethnicities [28]. The genotypes related to LTF level are associated with decreased function and this may lead to decreased defense against infection with stronger stimuli for granulocyte activation and desorption, leading to greater LTF release [28]. Four studies [5, 16, 20, 24] included in this meta-analysis assessed LTF rs1126478 polymorphism. However, none of them found any association between this polymorphism and dental caries susceptibility. In addition, the subgroup analysis in this meta-analysis did not find any association between this polymorphism and dental caries risk. Thus, we can exclude LTF rs1126478 polymorphism as a risk factor for dental caries; however, more accurate confirmation of results requires further research through larger studies on different ethnicities.
AMELX is the most significant factor for development of normal enamel [17] and AMELX aberration predominantly causes mineralization defects and congenital disorders such as amelogenesis imperfecta [29]. Therefore, some researchers suggest that genetic variations contribute to structural changes of the enamel and may create more levels of mineral loss, bacterial extension or biofilm deposition [30]. In our meta-analysis on AMELX rs946252, AMELX rs17878486, AMELX rs6639060, and AMELX rs2106416 polymorphisms, none of them was associated with the risk of dental caries. However, the sensitivity analysis revealed that AMELX rs17878486 polymorphism could be a risk factor for dental caries but the ethnicity and type of selected controls were the effective factors on the association between AMELX rs17878486 polymorphism and risk of dental caries. In addition, two studies included in our meta-analysis [11, 15] showed an association between AMELX rs17878486 polymorphism and dental caries susceptibility. Therefore, it may point to the role of AMELX rs17878486 polymorphism in development of dental caries more than others.
ENAM gene may also play a role in enamel formation [31] and any change in genes that encode enamel proteins may lead to enamel malformation [32]. Among studies on ENAM polymorphisms included in this meta-analysis, one study on Saudi patients [25] reported an elevated risk of ENAM rs1264848 polymorphism and another study on Polish children [14] showed the protective role of ENAM rs1264848 polymorphism in dental caries, which was confirmed by Abbasoğlu et al., in their study on Turkish children [33]. Also, one study [25] reported an elevated risk of ENAM rs3796704 polymorphism and another study [5] showed an elevated risk of ENAM rs3796703 polymorphism in dental caries patients compared with controls. One research [22] reported that there were significant differences in the minor allele frequency between the Poles and Czechs populations. In addition, the sensitivity analysis confirmed the effect of ethnicity and type of control group on the association of ENAM rs3796704 and risk of dental caries. Bayram and colleagues [34] showed that ENAM polymorphisms may affect the development of enamel and these effects may be different between primary and permanent dentitions.
Small number of studies included in each analysis and different DMFT scores for selecting the cases and controls between the studies were two important limitations of the present meta-analysis. However, in most analyses, the heterogeneity was low and there was no publication bias.
Conclusions
The findings of this meta-analysis confirmed that the G allele and the GG genotype of ENAM rs3796704 polymorphism were associated with an elevated risk of caries in the case group compared with the control group. But there was no association between LTF rs1126478, ENAM rs1264848, ENAM rs3796703, AMELX rs946252, AMELX rs17878486, and AMELX rs2106416 polymorphisms and dental caries susceptibility. However, subgroup analysis showed an association between ENAM rs3796704 and AMELX rs17878486 polymorphisms and dental caries susceptibility in the Caucasian ethnicity and studies including caries-free individuals as the control group. In addition, sensitivity analysis showed an increased risk of AMELX rs17878486 polymorphism in the case group compared with the control group. The future analyses with more cases in various areas should have focused on possible effects of gene-environmental interactions on caries experience.
Supplementary information
Acknowledgements
This work was performed in partial fulfillment of the requirements for a doctorate degree in General Dentistry (Sajjad Jahedi), in Faculty of Dentistry, Kermanshah University of Medical Sciences, Kermanshah, Iran.
Abbreviations
- LTF
Lactotransferrin
- ENAM
Enamelin
- AMELX
Amelogenin X
- OR
Odds ratio
- HWE
Hardy-Weinberg equilibrium
- CI
Confidence interval
- DMFT
Decayed, Missing, and Filled Teeth
Authors’ contributions
RS and MaS contributed to the conception and design of the work. SJ contributed to the acquisition and analysis of data. HRM, MMI, AG, HM, and MoS contributed to interpretation of data and created new software used in the work. MaS drafted the manuscript and substantively revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Research Council of Kermanshah University of Medical Sciences (Grant Number: 980711). The funding body did not contribute to the design of the study, the collection, analysis, and interpretation of data or in writing of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Kermanshah University of Medical Sciences, Kermanshah, Iran (approval number: IR.KUMS.REC.1398.795). In addition, consent to participate form was signed by each participant in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12903-020-01121-7.
References
- 1.Hunter PB. Risk factors in dental caries. Int Den J. 1988;38:211–217. [PubMed] [Google Scholar]
- 2.Zander A, Sivaneswaran S, Skinner J, Byun R, Jalaludin B. Risk factors for dental caries in small rural and regional Australian communities. Rural Remote Health. 2013;13:2492. [PubMed] [Google Scholar]
- 3.Slayton RL, Cooper ME, Marazita ML. Tuftelin, mutans streptococci, and dental caries susceptibility. J Dent Res. 2005;84:711–714. doi: 10.1177/154405910508400805. [DOI] [PubMed] [Google Scholar]
- 4.Renuka P, Pushpanjali K, Sangeetha R. Review on “Influence of host genes on dental caries”. J Dent Med Sci. 2013;4:86–92. [Google Scholar]
- 5.Wang M, Qin M, Xia B. The association of Enamelin, Lactoferrin, and tumour necrosis factor alpha gene polymorphisms with high caries susceptibility in Chinese children under 4 years old. Arch Oral Biol. 2017;80:75–81. doi: 10.1016/j.archoralbio.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Velliyagounder K, Kaplan JB, Furgang D, Legarda D, Diamond G, Parkin RE, Fine DH. One of two human lactoferrin variants exhibits increased antibacterial and transcriptional activation activities and is associated with localized juvenile periodontitis. Infect Immun. 2003;71:6141–6147. doi: 10.1128/IAI.71.11.6141-6147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Actor JK, Hwang SA, Kruzel ML. Lactoferrin as a natural immune modulator. Curr Pharm Des. 2009;15:1956. doi: 10.2174/138161209788453202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruzel ML, Zimecki M, Actor JK. Lactoferrin in a context of inflammation-induced pathology. Front Immunol. 2017;8:1438. doi: 10.3389/fimmu.2017.01438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hashimi N, Sire J-Y, Delgado S. Evolutionary analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 32-kDa peptide and reveals selective adaptation in rodents and primates. J Mol Evol. 2009;69:635–656. doi: 10.1007/s00239-009-9302-x. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett JD. Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent. 2013;2013:684607. doi: 10.1155/2013/684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeremias F, Koruyucu M, Küchler EC, Bayram M, Tuna EB, Deeley K, Pierri RA, Souza JF, Fragelli CM, Paschoal MA, Gencay K, Seymen F, Caminaga RM, dos Santos-Pinto L, Vieira AR. Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol. 2013;58:1434–1442. doi: 10.1016/j.archoralbio.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasse B, Grabar S, Lafont AG, Quinquis L, Opsahl Vital S, Davit-Béal T, Moulis E, Chabadel O, Hennequin M, Courson F, Droz D, Vaysse F, Laboux O, Tassery H, Al-Hashimi N, Boillot A, Carel JC, Treluyer JM, Jeanpierre M, Beldjord C, Sire JY, Chaussain C. Common SNPs of AmelogeninX (AMELX) and dental caries susceptibility. J Dent Res. 2013;92:418–424. doi: 10.1177/0022034513482941. [DOI] [PubMed] [Google Scholar]
- 13.Ouryouji K, Imamura Y, Fujigaki Y, Oomori Y, Yanagisawa S, Miyazawa H, Wang P-L. Analysis of mutations in the amelogenin and the enamelin genes in severe caries in Japanese pediatric patients. Pediatric Dent J. 2008;18:79–85. doi: 10.1016/S0917-2394(08)70126-5. [DOI] [Google Scholar]
- 14.Gerreth K, Zaorska K, Zabel M, Borysewicz-Lewicka M, Nowicki M. Association of ENAM gene single nucleotide polymorphisms with dental caries in polish children. Clin Oral Investig. 2016;20:631–636. doi: 10.1007/s00784-016-1743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerreth K, Zaorska K, Zabel M, Borysewicz-Lewicka M, Nowicki M. Chosen single nucleotide polymorphisms (SNPs) of enamel formation genes and dental caries in a population of polish children. Adv Clin Exp Med. 2017;26:899–905. doi: 10.17219/acem/63024. [DOI] [PubMed] [Google Scholar]
- 16.Azevedo LF, Pecharki GD, Brancher JA, Cordeiro CA, Jr, Medeiros KG, Antunes AA, Arruda ES, Werneck RI, de Azevedo LR, Mazur RF, Moysés SJ, Moysés ST, Faucz FR, Trevilatto PC. Analysis of the association between lactotransferrin (LTF) gene polymorphism and dental caries. J Appl Oral Sci. 2010;18:166–170. doi: 10.1590/S1678-77572010000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang SW, Yoon I, Lee HW, Cho J. Association between AMELX polymorphisms and dental caries in Koreans. Oral Dis. 2011;17:399–406. doi: 10.1111/j.1601-0825.2010.01766.x. [DOI] [PubMed] [Google Scholar]
- 18.Olszowski T, Adler G, Janiszewska-Olszowska J, Safranow K, Kaczmarczyk M. MBL2, MASP2, AMELX, and ENAM gene polymorphisms and dental caries in polish children. Oral Dis. 2012;18:389–395. doi: 10.1111/j.1601-0825.2011.01887.x. [DOI] [PubMed] [Google Scholar]
- 19.Ergöz N, Seymen F, Gencay K, Tamay Z, Deeley K, Vinski S, Vieira AR. Genetic variation in Ameloblastin is associated with caries in asthmatic children. Eur Arch Paediatr Dent. 2014;15:211–216. doi: 10.1007/s40368-013-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volckova M, Linhartova PB, Trefna T, Vlazny J, Musilova K, Kukletova M, Kukla L, Holla LI. Lack of association between lactotransferrin polymorphism and dental caries. Caries Res. 2014;48:39–44. doi: 10.1159/000351689. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz G, Ermis RB, Calapoglu NS, Celik EU, Türel GY. Gene-environment interactions in the etiology of dental caries. J Dent Res. 2016;95:74–79. doi: 10.1177/0022034515605281. [DOI] [PubMed] [Google Scholar]
- 22.Borilova Linhartova P, Deissova T, Musilova K, Zackova L, Kukletova M, Kukla L, Izakovicova HL. Lack of association between ENAM gene polymorphism and dental caries in primary and permanent teeth in Czech children. Clin Oral Investig. 2018;22:1873–1877. doi: 10.1007/s00784-017-2280-2. [DOI] [PubMed] [Google Scholar]
- 23.Koohpeima F, Hashemi-Gorji F, Mokhtari MJ. Evaluation of caries experience in two genders and ENAM polymorphism in Iranian adults. Meta Gene. 2018;17:78–81. doi: 10.1016/j.mgene.2018.05.002. [DOI] [Google Scholar]
- 24.Wang M, Qin M. Lack of association between LTF gene polymorphisms and different caries status in primary dentition. Oral Dis. 2018;24:1545–1553. doi: 10.1111/odi.12939. [DOI] [PubMed] [Google Scholar]
- 25.Devang Divakar D, Alanazi SAS, Assiri MYA, Mohammed Halawani S, Zaid Alshehri S, Ahmed Saeed Al-Amri S, Mustafa M. Association between ENAM polymorphisms and dental caries in children. Saudi J Biol Sci. 2019;26:730–735. doi: 10.1016/j.sjbs.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peres KG, Peres MA, Araujo CL, Menezes AM, Hallal PC. Social and dental status along the life course and oral health impacts in adolescents: a population-based birth cohort. Health Qual Life Outcomes. 2009;7:95. doi: 10.1186/1477-7525-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werneck RI, Mira MT, Trevilatto PC. A critical review: an overview of genetic influence on dental caries. Oral Dis. 2010;16:613–623. doi: 10.1111/j.1601-0825.2010.01675.x. [DOI] [PubMed] [Google Scholar]
- 28.Videm V, Dahl H, Wålberg LE, Wiseth R. Functional polymorphisms in the LTF gene and risk of coronary artery stenosis. Hum Immunol. 2012;73:554–559. doi: 10.1016/j.humimm.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Greene SR, Yuan ZA, Wright JT, Amjad H, Abrams WR, Buchanan JA, Trachtenberg DI, Gibson CW. A new frameshift mutation encoding a truncated amelogenin leads to x-linked amelogenesis imperfecta. Arch Oral Biol. 2002;47:211–217. doi: 10.1016/S0003-9969(01)00111-X. [DOI] [PubMed] [Google Scholar]
- 30.Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, Vieira AR. Enamel formation genes are associated with high caries experience in turkish children. Caries Res. 2008;42:394–400. doi: 10.1159/000154785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley JL, Swanson WJ. Dietary change and adaptive evolution of enamelin in humans and among primates. Genetics. 2008;178:1595–1603. doi: 10.1534/genetics.107.077123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babbitt CC, Warner LR, Fedrigo O, Wall CE, Wray GA. Genomic signatures of diet-related shifts during human origins. Proc Biol Sci. 2011;278:961–969. doi: 10.1098/rspb.2010.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbasoğlu Z, Tanboğa İ, Küchler EC, Deeley K, Weber M, Kaspar C, Korachi M, Vieira AR. Early childhood caries is associated with genetic variants in enamel formation and immune response genes. Caries Res. 2015;49:70–77. doi: 10.1159/000362825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayram M, Deeley K, Reis MF, Trombetta VM, Ruff TD, Sencak RC, Hummel M, Dizak PM, Washam K, Romanos HF, Lips A, Alves G, Costa MC, Granjeiro JM, Antunes LS, Küchler EC, Seymen F, Vieira AR. Genetic influences on dental enamel that impact caries differ between the primary and permanent dentitions. Eur J Oral Sci. 2015;123:327–334. doi: 10.1111/eos.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.