Vibralactone is a lead compound with a novel skeleton structure that shows strong inhibitory activity against pancreatic lipase. Vibralactone is not encoded by the genome directly but rather is synthesized from phenol, followed by prenylation and other enzyme reactions. Here, we used an RNA silencing approach to identify and characterize a prenyltransferase in a basidiomycete species that is responsible for the synthesis of vibralactone. The identified gene, vib-PT, was expressed in Escherichia coli to obtain a soluble and enzymatically active fusion Vib-PT protein. In vitro characterization of the enzyme demonstrated the catalytic mechanism of prenylation and broad substrate range for different aromatic acceptors and prenyl donors. These characteristics highlight the possibility of Vib-PT to generate prenylated derivatives of aromatics and other compounds as improved bioactive agents or potential prodrugs.
KEYWORDS: prenyltransferase gene, RNA silencing, heterologous expression, Stereum vibrans, vibralactone
ABSTRACT
Vibralactone, a hybrid compound derived from phenols and a prenyl group, is a strong pancreatic lipase inhibitor with a rare fused bicyclic β-lactone skeleton. Recently, a researcher reported a vibralactone derivative (compound C1) that caused inhibition of pancreatic lipase with a half-maximal inhibitory concentration of 14 nM determined by structure-based optimization, suggesting a potential candidate as a new antiobesity treatment. In the present study, we sought to identify the main gene encoding prenyltransferase in Stereum vibrans, which is responsible for the prenylation of phenol leading to vibralactone synthesis. Two RNA silencing transformants of the identified gene (vib-PT) were obtained through Agrobacterium tumefaciens-mediated transformation. Compared to wild-type strains, the transformants showed a decrease in vib-PT expression ranging from 11.0 to 56.0% at 5, 10, and 15 days in reverse transcription-quantitative PCR analysis, along with a reduction in primary vibralactone production of 37 to 64% at 15 and 21 days, respectively, as determined using ultra-high-performance liquid chromatography-mass spectrometry analysis. A soluble and enzymatically active fusion Vib-PT protein was obtained by expressing vib-PT in Escherichia coli, and the enzyme’s optimal reaction conditions and catalytic efficiency (Km/kcat) were determined. In vitro experiments established that Vib-PT catalyzed the C-prenylation at C-3 of 4-hydroxy-benzaldehyde and the O-prenylation at the 4-hydroxy of 4-hydroxy-benzenemethanol in the presence of dimethylallyl diphosphate. Moreover, Vib-PT shows promiscuity toward aromatic compounds and prenyl donors.
IMPORTANCE Vibralactone is a lead compound with a novel skeleton structure that shows strong inhibitory activity against pancreatic lipase. Vibralactone is not encoded by the genome directly but rather is synthesized from phenol, followed by prenylation and other enzyme reactions. Here, we used an RNA silencing approach to identify and characterize a prenyltransferase in a basidiomycete species that is responsible for the synthesis of vibralactone. The identified gene, vib-PT, was expressed in Escherichia coli to obtain a soluble and enzymatically active fusion Vib-PT protein. In vitro characterization of the enzyme demonstrated the catalytic mechanism of prenylation and broad substrate range for different aromatic acceptors and prenyl donors. These characteristics highlight the possibility of Vib-PT to generate prenylated derivatives of aromatics and other compounds as improved bioactive agents or potential prodrugs.
INTRODUCTION
Vibralactone was isolated from the basidiomycete Stereum vibrans (syn. Boreostereum vibrans) and identified as a lead compound with a novel skeleton structure. Importantly, vibralactone showed strong inhibitory activity (half-maximal inhibitory concentration [IC50] 0.4 μg/ml) against pancreatic lipase (1), which is comparable to that of the U.S. Food and Drug Administration-approved obesity therapeutic tetrahydrolipstatin (IC50 0.36 μg/ml). Recent structure-based optimization of vibralactones resulted in the production of a vibralactone derivative (compound C1) that exhibited the highest known inhibition of pancreatic lipase reported to date, with an IC50 of 14 nM (2). Therefore, this vibralactone derivative is considered as a potential candidate for development of a new antiobesity therapeutic.
Structure-activity relationships and molecular modeling identified prenyl as a key group in vibralactone and its derivatives that is responsible for the inhibition of pancreatic lipase activity (2). In addition, the bicyclic β-lactone vibralactone derivative emerged as a novel probe capable of labeling two isoforms of the important ClpP protease, while all other monocyclic β-lactone probes could only label ClpP2 (3). In our previous work, the vibralactone biogenesis pathway was elucidated by isotope feeding, demonstrating the involvement three main enzymes (Fig. 1A) (4). The first step involves the 3-prenylation of a phenol precursor derived from the shikimate pathway with dimethylallyl diphosphate (DMAPP) in a reaction catalyzed by an aromatic prenyltransferase. Prenyltransferases are involved in both primary and secondary metabolism in several organisms and significantly contribute to the structural diversity and bioactivities of natural products (5). Furthermore, the biological and pharmacological activities of prenylated products are usually distinct from those of nonprenylated product precursors (6–8), which makes these enzymes valuable biocatalysts in the biosynthesis of small molecules such as naphterpin (9), furaquinocin (10), sirodesmin PL (11), and clorobiocin (12). In addition to vibralactone from S. vibrans, a large number of prenylated p-hydroxybenzoic acids and their analogs are present in the fungal genus Stereum (13, 14) with diverse pharmacological activities (13, 15, 16). However, the specific role of prenyltransferase in the biosynthesis of p-hydroxybenzoate and related compounds remains unclear.
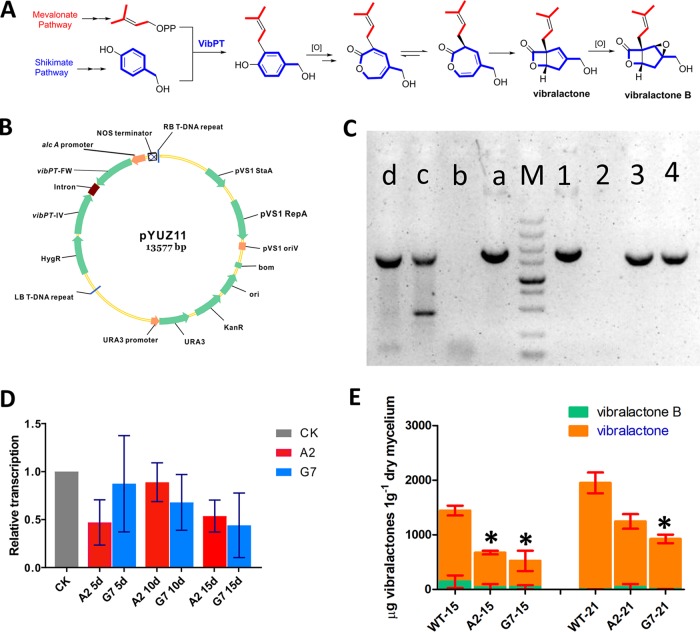
FIG 1.
Functional characterization of vib-PT by RNA silencing. (A) Biosynthetic pathway of vibralactone. (B) Plasmid map containing the RNA silencing cassette and the alcA(p), a 1,103-bp element of vib-PT-FW, the second intron of the Arabidopsis small nuclear ribonuclear protein D1, and a reverse-complemented vib-PT-IV 1,106-bp element of vib-PT. (C) PCR confirmation of the two transformants. Lane M, DL5000 marker; lanes a to d, amplification fragments from an internal segment of the hygromycin resistance gene (HygR) to PT-IV of pYUZ11, wild-type S. vibrans, A2, and G7 lines, respectively. Lanes 1 to 4 are amplification fragments from alcA(p) to intron fragments of pYUZ11, wild-type S. vibrans, and the A2 and G7 transformant lines, respectively. (D) Relative transcription levels of vib-PT quantified by RT-qPCR at 5, 10, and 15 days. CK (β-tubulin) was used as the standard (given a relative transcription level of 1) for statistical analysis of the relative transcription level of vib-PT in the transformants to that in the wild-type strain under a given condition; A2 and G7 indicate the two RNA silencing transformants. Error bars indicate the standard deviations. (E) Vibralactone and vibralactone B content in wild-type S. vibrans and the A2 and G7 transformants was determined by UPLC-MS analysis. WT-15 and WT-21 are the standards used for measuring the vibralactone content in the WT strain at days 15 and 21, respectively. A2-15, A2-21, G7-15, and G7-21 indicate the vibralactone content in the two transformants. Error bars indicate the standard deviations. Asterisks represent statistically significant differences (P < 0.001).
RNA silencing is a biological mechanism that leads to posttranscriptional gene silencing triggered by double-stranded RNA molecules to inhibit the translation of encoded proteins. RNA silencing is a powerful and reliable biotechnology, serving as a useful tool for investigating gene functions and as a rapid and convenient method for engineering strains with specific suppression of target genes (17–19). For example, use of an RNA silencing strategy demonstrated that chaetoglobosin, a PKS-NRPS hybrid produced by Penicillium expansum, is formed mainly via the core gene cheA (20). In addition, RNA silencing was used to investigate the factors involved in ganoderic acid biosynthesis (21, 22). The basidiomycete Stereum sp. has multinucleated cells; hence, it has been challenging to manipulate its genes using gene knockouts strategies (23). To overcome this limitation, in the present study, we constructed a posttranscriptional gene-silencing vector that contains a hairpin sequence to manipulate the expression of our target gene.
From our previous study, the vib-PT gene is hypothesized to encode an aromatic prenyltransferase in the S. vibrans genome (4), which is a member of the ABBA prenyltransferase superfamily. Analysis of the genome demonstrated that there are many prenyltransferase genes, but only one of them is an aromatic prenyltransferase gene. Based on the results from heterologous expression and crude enzyme activity assays (4), we hypothesized that Vib-PT catalyzes the prenyl transfer to 4-hydroxy-benzenemethanol during vibralactone biosynthesis. Therefore, in present study, we sought to gain insights into the function of vib-PT in S. vibrans. Toward this end, we applied RNA silencing technology to suppress the expression of vib-PT in vivo and then determined the expression level of vib-PT using reverse transcription-quantitative PCR (RT-qPCR) analysis. We further expressed the gene in Escherichia coli and characterized the purified protein. These findings and the detailed characterization of Vib-PT can lay the foundation for further development and application of this pathway from S. vibrans for a new antiobesity therapeutic strategy.
RESULTS
Characterization of RNA silencing transformants with suppression of vib-PT.
To investigate whether Vib-PT is responsible for the prenylation of phenols in the vibralactone biosynthesis pathway, we assembled a set of vectors that harbor genetic elements to specifically downregulate vib-PT expression. The vector pUNZ104 carries a vib-PT RNA silencing cassette under the control of the alcohol dehydrogenase promoter [alcA(p)] (Fig. 1B). The expression of this open reading frame terminates with a self-cleaving RZ that generates aberrant transcripts retained in the nuclear compartment. These transcripts are targeted by an RNA-dependent RNA polymerase and in turn processed into small interfering RNAs (24). We initially attempted the CaCl2/polyethylene glycol (PEG) method to transform S. vibrans with pUNZ104, which has been previously reported for some fungal species (25–29). Unfortunately, after many trials, no transformants were obtained using this method. Therefore, we employed an Agrobacterium tumefaciens-mediated genetic transformation approach, which resulted in two transformants, named the A2 and G7 lines, through successful transformation of the interfering fragment of pYUZ11. These two transformants were selected by hygromycin and confirmed by PCR using the total genomic DNA as a template (Fig. 1C). On the electrophoretic gel, A2 and G7 showed the expected 1,820-bp product (from an internal segment of the hygromycin resistance gene to PT-IV; see plasmid pYUZ11 in Fig. 1B) and 1,789-bp product [from alcA(p) to intron fragment; Fig. 1B], respectively, whereas wild-type (WT) S. vibrans returned negative results (Fig. 1C). Transcriptional analysis of vib-PT via RT-qPCR demonstrated that expression of vib-PT was downregulated in the two transformants compared to that detected in the WT strain. Specifically, the vib-PT mRNA level in A2 decreased by 52.9, 11.0, and 46.3% at days 5, 10, and 15, respectively, and that of G7 decreased by 12.6, 32.1, and 56.0% at days 5, 10, and 15, respectively (Fig. 1D). These results showed that vib-PT expression was successfully suppressed by our A. tumefaciens-mediated RNA silencing approach.
Although prenylphenol derivatives were previously isolated from S. vibrans (14), no direct products of Vip-PT could be obtained. We hypothesized that the direct products of Vib-PT may be a precursor for the formation of prenylphenol derivatives. Therefore, we examined the vibralactone levels in the RNA silencing lines and the control WT strains. Among the numerous vibralactone type compounds isolated from different cultural conditions (14), vibralactone and vibralactone B were the major vibralactone components obtained from S. vibrans in the culture conditions of our previous work (4). Therefore, the extracts of both RNA-silenced transformants (A2 and G7 lines) and the WT strain grown in modified potato dextrose broth (PDB) were analyzed using ultra-high-performance liquid chromatography-mass spectrometry (UPLC-MS) by monitoring the quasimolecular ion peaks of vibralactone and vibralactone B. The quasimolecular ion peaks of vibralactone and vibralactone B (at m/z 241 [M+CH3OH+H]+ and m/z 247 [M+Na]+, respectively) were observed in both the WT and RNA-silenced lines (A2 and G7), but their amounts were significantly different, with decreases of 37 and 64% at days 15 and 21, respectively, in the transformants compared to the WT (Fig. 1E). The concentrations of the two molecules were the lowest in the G7 transformant at 929.5 μg/g dry mycelium weight (DMW) at day 21. In comparison, the combined level of vibralactone and vibralactone B in the WT control was 1,950.9 μg/g DMW at day 21 (Fig. 1E).
Biochemical properties of Vib-PT.
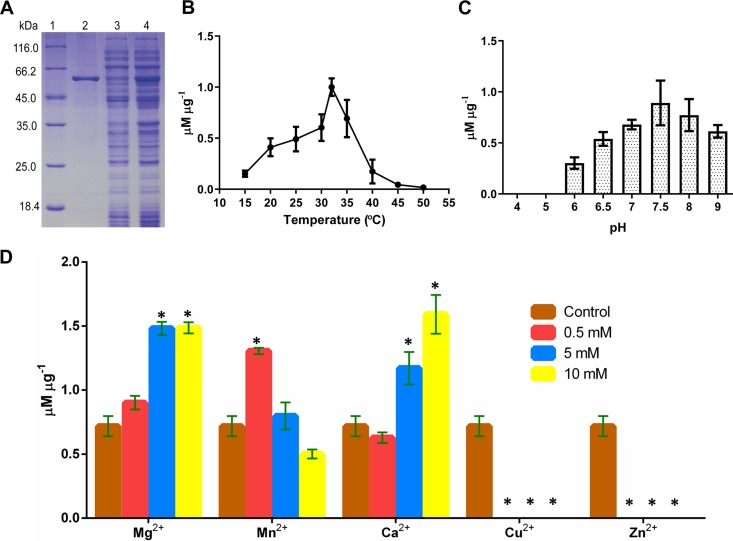
The fusion Vib-PT enzyme was expressed in E. coli, purified by Ni-NTA chromatography, and detected by SDS-PAGE. The estimated molecular mass of recombinant Vib-PT protein was determined to be 57 kDa (Fig. 2A), and the theoretical molecular weight was 56,687.35 Da. Based on the reaction profile, Vib-PT showed maximal activity at 32°C (Fig. 2B) and optimal activity at pH 7.5 in Tris-HCl (Fig. 2C). When tested with different ions, the enzyme activity did not require metal ions such as Mg2+, Mn2+, or Ca2+. However, the presence of Mg2+ or Ca2+ from 0.5 to 10 mM enhanced the enzyme activity by up to 3-fold compared to that of the control protein without a metal ion. Although 0.5 mM Mn2+ increased the activity more significantly than 0.5 mM Mg2+ or Ca2+, a decrease in the amount of the products was detected in the presence of higher concentrations of Mn2+ (5 to 10 mM). In addition, the activity of Vib-PT could not be detected in the presence of Zn2+ or Cu2+ even at a low concentration of 0.5 mM (Fig. 2D). This finding is similar to previous reports of other soluble prenyltransferases, such as CloQ from Streptomyces roseochromogenes (12), LtxC from Lyngbya majuscule (30), and CdpNPT (31), FgaPT2 (32), FtmPT1 (33), and 7-DMATS (34) from Aspergillus fumigatus but differs from those for FgaPT1 (35) and FtmPT2 (36) from A. fumigatus. Collectively, these data indicate that the enhancement of enzyme activity by Ca2+ is not a conserved property among the indole prenyltransferases.
FIG 2.
Properties of purified Vib-PT. (A) SDS-PAGE analysis of the heterologous expression and purification of Vib-PT. Lane 1, protein molecular weight marker; lane 2, purified fusion Vib-PT; lane 3, soluble protein; lane 4, cell extract of total protein after induction with 0.1 mM IPTG at 16°C for 20 h. (B) Optimal temperature for the prenylation of 4-hydroxy-benzenemethanol catalyzed by Vib-PT. Error bars indicate the standard deviations. (C) Determination of optimal pH for the prenylation of 4-hydroxy-benzenemethanol catalyzed by Vib-PT. Error bars indicate the standard deviations. (D) Effect of metal ions on the activity of Vib-PT. The activity of Vib-PT was assayed in the absence (as a control) or presence of various metal ions at 0.5, 5, and 10 mM. Error bars indicate the standard deviations. Asterisks represent statistically significant differences (P < 0.001).
In vitro prenylation of phenols by Vib-PT.
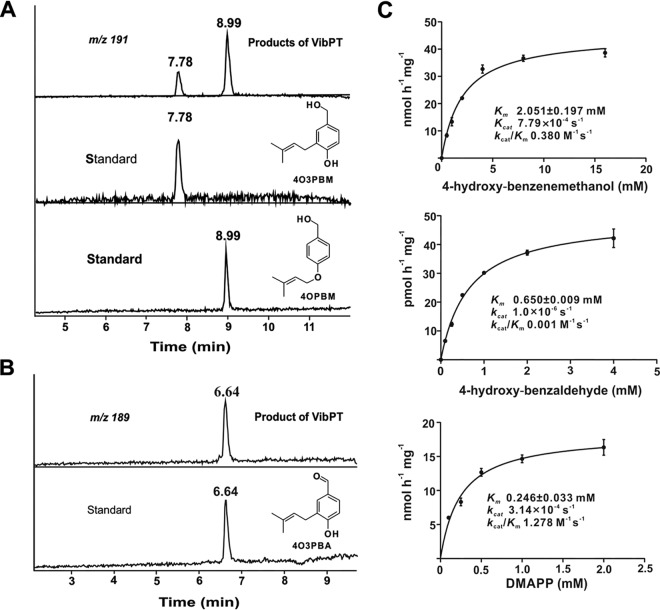
In our previous study, Vib-PT expressed through the vector pET32a+ in E. coli Rosetta 2(DE3)pLysS showed activity but poor solubility (4). To characterize the catalytic activity of Vib-PT, we optimized the conditions for the heterologous expression of the vib-PT gene. First, vib-PT was cloned in a different vector, pET28a, and transformed into E. coli BL21(DE3) cells. Second, the expression was induced at different temperatures and/or with different concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside). To measure the enzymatic activity, Vib-PT was incubated with 4-hydroxy-benzenemethanol or 4-hydroxy-benzaldehyde in the presence of DMAPP, and UPLC-MS was used to analyze the formation of enzymatic products in the incubation mixtures. As shown in Fig. 3A, products were detected in the incubation mixture of 4-hydroxy-benzenemethanol with DMAPP. Two extracted ion chromatogram peaks of m/z 191 [M–H]− were detected at 7.78 and 8.99 min (Fig. 3A) and were absent in the control assay containing 4-hydroxy-benzenemethanol and DMAPP with inactivated Vib-PT that was boiled for 20 min. This result demonstrated that the reaction depended on the presence of an active enzyme. The product obtained at 7.78 min was confirmed by comparing the retention times with an authentic reference sample, synthesized according to 4-hydroxy-3-(3-methylbut-2-en-1-yl)-benzenemethanol (4O3PBM) (Fig. 3A), while the product at 8.99 min was identified as 4-(3-methylbut-2-en-1-yl)oxy-benzenemethanol (4OPBM) (Fig. 3A). In addition, one peak of m/z 189 [M–H]− at 6.64 min was detected in the incubation mixture of 4-hydroxy-benzaldehyde with DMAPP (Fig. 3B). Through the same analytical method, the product was verified to be 4-hydroxy-3-(3-methylbut-2-en-1-yl)-benzaldehyde (4O3PBA) in comparison to the inactivated control (Fig. 3B) and the authentic reference sample (Fig. 3B).
FIG 3.
Activity assays and prenylation reaction catalyzed by purified Vib-PT with Michaelis-Menten kinetics. (A) UPLC-MS of the prenylation of 4-hydroxy-benzenemethanol catalyzed by Vib-PT. Extracted ion chromatographs of UPLC-MS analysis of catalytic activity with Vib-PT and with authentic 4O3PBM and 4OPBM from chemical synthesis. (B) UPLC-MS analysis of the prenylation of 4-hydroxy-benzaldehyde catalyzed by Vib-PT. Extracted ion chromatographs from UPLC-MS analyses of the catalytic activity with Vib-PT and with authentic 4O3PBA from chemical synthesis are shown. (C) Enzyme kinetic parameters were determined with different concentrations of 4-hydroxy-benzenemethanol, 4-hydroxy-benzaldehyde, and DMAPP. Error bars indicate the standard deviations.
Kinetics of Vib-PT activity with DMAPP and phenols as cosubstrates.
The common Michaelis-Menten kinetic parameters for Vib-PT activity were determined at pH 7.5 and 32°C to maintain physiological relevance with respect to this species. After a 1-h incubation period, which was within the linear range for product formation, the reactions were quenched by the addition of methanol and centrifuged to remove the precipitated protein prior to analysis of the diluted supernatant by UPLC-MS. Kinetic studies of the reaction catalyzed by the fusion protein Vib-PT were performed using DMAPP as the isopentenyl donor and 4-hydroxy-benzenemethanol and 4-hydroxy-benzaldehyde as the acceptors. When the initial rate was measured by varying the concentrations of 4-hydroxy-benzenemethanol and 4-hydroxy-benzaldehyde with DMAPP at fixed concentrations, a set of intersecting lines was obtained by regression analysis of the data at each DMAPP concentration (Fig. 3C and see Fig. S1 in the supplemental material). Vib-PT exhibited an apparent Km of 0.246 ± 0.033 mM for DMAPP using 4-hydroxy-benzenemethanol as the acceptor, a Km of 2.051 ± 0.197 mM for 4-hydroxy-benzenemethanol, and a Km of 0.650 ± 0.009 mM for 4-hydroxy-benzaldehyde (Fig. 3C). The maximum reaction velocity (Vmax) with the individual substrates ranged from 0.049 nmol h−1 mg−1 for 4-hydroxy-benzaldehyde to 45.380 nmol h−1 mg−1 for 4-hydroxy-benzenemethanol. The corresponding apparent turnover numbers (kcat) were 7.79 × 10−4 s−1 and 1.0 × 10−6 s−1 for 4-hydroxy-benzenemethanol and 4-hydroxy-benzaldehyde, respectively.
Substrate specificity of Vib-PT.
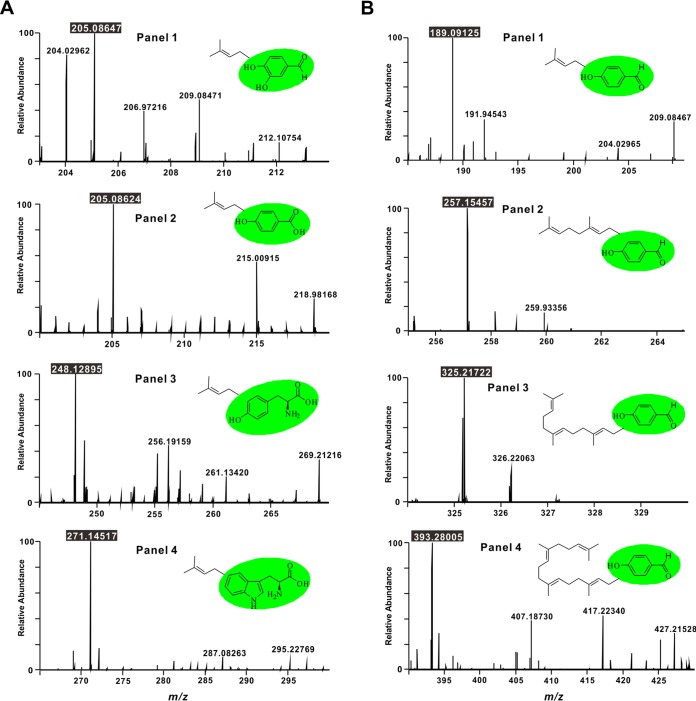
In general, prenyltransferases have broad substrate specificity. To further evaluate the substrate range of Vib-PT, 3,4-dihydroxy-benzaldehyde, 4-hydroxy-benzoic acid, l-phenylalanine, l-tyrosine, and l-tryptophan were used as acceptors at 1 mM each in standard assays with DMAPP. Four aromatic derivatives, including 3,4-dihydroxy-benzaldehyde, 4-hydroxy-benzoic acid, l-tyrosine, and l-tryptophan, could be prenylated with DMAPP by Vib-PT; however, no prenylated product was detected in the reaction of l-phenylalanine and DMAPP as the substrates. Prenylation products corresponding to the aromatic derivatives were detected by UPLC-high-resolution (HR)-electrospray ionization (ESI)-MS, demonstrating a quasimolecular ion peak for the expected prenylated product of 3,4-dihydroxy-benzaldehyde at m/z 205.08647 [M–H]− (calculated for C12H13O3, 205.08592) (Fig. 4A, panel 1). Similarly, in the UPLC-HR-ESI-MS spectrum of the incubation with 4-hydroxy-benzoic acid, the prenylated product was detected at m/z 205.08624 [M–H]− (calculated for C12H13O3, 205.08592) (Fig. 4A, panel 2). Moreover, the corresponding prenylated products of l-tyrosine and l-tryptophan were observed at m/z 248.12895 [M–H]− (calculated for C14H18NO3, 248.12812) and 271.14517 [M–H]− (calculated for C16H19N2O2, 271.14410), respectively (Fig. 4A, panels 3 and 4).
FIG 4.
HR-ESI-MS spectra of prenylation on different substrates by Vib-PT. (A) HR-ESI-MS spectra of the reaction products of Vib-PT with different aromatic derivatives as acceptors. (Panels 1 to 4) 3,4-Dihydroxy-benzaldehyde, 4-hydroxy-benzoic acid, l-tyrosine, and l-tryptophan were used as acceptors at 1 mM, respectively. (B) HR-ESI-MS spectra of the reaction products of Vib-PT with DMAPP, GPP, FPP, and GGPP as donors (panels 1 to 4, respectively) and 4-hydroxy-benzaldehyde as the acceptor. All reactions were carried out in standard assays at least twice.
The donor specificity of Vib-PT was probed using DMAPP, geranyl diphosphate (GPP), farnesyl diphosphate (FPP), or geranylgeranyl diphosphate (GGPP) and 4-hydroxy-benzaldehyde as an acceptor. All expected prenylated products could be detected by UPLC-HR-ESI-MS in the assay (Fig. 4B). In the HR-ESI-MS spectrum, a quasimolecular ion peak for the expected prenylated product was observed at m/z 189.09125 [M–H]− (calculated for C12H13O2, 189.09101) (Fig. 4B, panel 1), and the geranylated product was detected at m/z 257.15457 [M–H]− (calculated for C17H21O2, 257.15361) (Fig. 4B, panel 2) from incubation of 4-hydroxy-benzaldehyde with GPP. Moreover, the corresponding products of FPP and GGPP were observed at m/z 325.21722 [M–H]− (calculated for C22H29O2, 325.21621) and 393.28005 [M–H]− (calculated for C27H37O2, 393.27881), respectively (Fig. 4B). The retention times of these products were 12.7, 16.4, 18.2, and 19.5 min, consistent with their polarity (see Fig. S2 in the supplemental material). Therefore, we conclude that Vib-PT catalyzes the in vitro prenylation of 4-hydroxy-benzaldehyde using DMAPP, GPP, FPP, and GGPP as prenyl donor substrates.
DISCUSSION
Vibralactone is a lead compound as the most potent drug candidate for inhibition of pancreatic lipase in antiobesity treatment. Our previous analyses of the vibralactone biogenesis pathway showed that the first step is prenylation of the phenol derivative precursor. In this study, we expressed and characterized the vib-PT gene from S. vibrans and demonstrated its function by biochemical characterization.
The enzymatic products of 4-hydroxy-benzenemethanol were identified as 4-(3-methylbut-2-en-1-yl)oxy-benzenemethanol and 4-hydroxy-3-(3-methylbut-2-en-1-yl)-benzenemethanol by UPLC-MS analysis and comparison with chemically synthetic compounds. Interestingly, we found that Vib-PT catalyzed the formation of an O–C bond and C–C bond between 4-hydroxy-benzenemethanol and the prenyl donor DMAPP. The reaction mechanism of aromatic prenyltransferases is expected to involve the formation of a carbocation on the prenyl donor (37). This cation then carries out an electrophilic attack on the aromatic ring of the prenyl acceptor. Mechanistically, this is similar to Friedel-Crafts alkylation, in which the energy barrier to carbocation formation is reduced by catalysts (38, 39). In enzyme-catalyzed aromatic prenyl transfer reactions, a hydrogen bond with an aromatic ring helps to lower the positive charge of the intermediary σ complex. Similarly, in our in vitro experiment, the prenyl group was connected with the 4-OH of 4-hydroxy-benzenemethanol and then transferred slowly to 3-C via the prenyltransferase activity. Interestingly under acidic (such as 0.1% formic acid) and nonenzymatic conditions, 4OPBM could be quickly converted to 4O3PBM (Fig. S3).
Many prenyltransferases can catalyze the formation of both O- and N-prenyl adducts, which is similar to the activity of the known ABBA prenyltransferase family (11, 40, 41) and that observed for other prenyltransferases. For example, NphB and Fnq26a from the Gram-positive bacteria Streptomyces spp. have been reported to catalyze both C- and O-prenyl of various phenolic substances (42, 43) and be N-prenyl prenylated by FtmPT2 (36), FtmPT1 (33), and CdpNPT (31). Liu and coworkers systematically investigated the secondary metabolites of S. vibrans but only found derivatives of the C–C bond between the aromatic ring and the prenyl group (14, 44, 45). Vib-PT could catalyze the formation of an O–C bond between 4-hydroxy-benzenemethanol and the prenyl donor DMAPP in vitro, but no prenylated aromatic derivatives with an O–C bond between the aromatic ring and the prenyl group had been reported in S. vibrans until now. In addition, the pH of the cultural system decreased to about 3 after culture for 21 days, so we speculated that this is an important factor.
To date, fungal ABBA family prenyltransferases have been found to catalyze six different kinds of prenyl transfer reactions to various positions: the “regular” C-prenylation catalyzed by FtmPT1 (46), 7-DMATS (34), and FgaPT2 (47, 48) and their orthologues MaPT (49), NotC (50), and VrtC (51); O-prenylation catalyzed by XptB (52), TyrPT (53), and SirD (11); N-prenylation catalyzed by FtmPT2 (36); the “reverse” C-prenylation catalyzed by BrePT (54) and AnaPT (55); the “reverse” O-prenylation catalyzed by LynF (56); and the “reverse” N-prenylation catalyzed by FtmPT1 (33) and CdpNPT (31). Based on sequence comparison, it is not possible to predict the prenylation position of an ABBA family prenyltransferase or to distinguish a C- and an N- or a “regular” and a “reverse” prenyltransferase, and no conserved domain has yet been identified for any groups of the fungal ABBA family prenyltransferases.
BLAST analysis of Vib-PT showed 91% identity to a hypothetical protein from genomic data of S. hirsutum (57) and 67% similarity to BYPB (a prenyltransferase) from Stereum sp. strain BY1 (58) at the amino acid level. BYPB was recently reported in a stereaceous basidiomycete, which could prenylate orsellinic acid. Many prenylated compounds have been isolated and identified from S. hirsutum (59, 60), and the present characterization of Vib-PT provides direct insights into the biosynthesis of these metabolites.
MATERIALS AND METHODS
Synthesis of standard compounds.
The synthesis of 4-hydroxy-3-(3-methylbut-2-en-1-yl)-benzenemethanol (4O3PBM) was performed according to our previous work (4).
4-Hydroxybenzaldehyde (0.61 g) and 3,3-dimethylallyl bromide (1.12 g) (both from J&K) were dissolved in dimethylformamide (10 ml) at room temperature (23 to 25°C), and then K2CO3 (1.04 g) was added in one portion. After stirring for 2 h, the initial exothermic reaction was stopped, and the reaction mixture was diluted with water (20 ml). The aqueous phase was extracted by using ethyl acetate (EtOAc), and the organic extracts were combined, washed with brine, dried over Na2SO4, and then filtered and concentrated under reduced pressure. The residue was purified by using silica gel column chromatography (petroleum ether/EtOAc, 50:1 to 20:1) to afford 4-hydroxy-3-(3-methylbut-2-en-1-yl) benzaldehyde (0.86 g, 91%) as a colorless oil (61).
4-Hydroxy-3-(3-methylbut-2-en-1-yl) benzaldehyde (0.38 g) was dissolved in methanol (10 ml) and then cooled in an ice bath, followed by the addition of sodium borohydride (0.114 g). After the reaction mixture was stirred for 2 h at 0°C, the solution was removed under reduced pressure, and the residue was dissolved in water (10 ml) and extracted by using EtOAc. The organic layers were washed with brine and dried over Na2SO4 and then purified by silica gel column chromatography (petroleum ether/EtOAc, 10:1 to 4:1) to give 4-(3-methylbut-2-en-1-yl)oxy-benzenemethanol (4OPBM; 0.32 g, 83%) (62).
4-(3-Methylbut-2-en-1-yl)oxy-benzenemethanol (4OPBM) was obtained as colorless oil. ESI-MS m/z: 191 [M–H]−. 1H-NMR (500 MHz, CDCl3) δ: 7.24 (2H, dd, J = 2.0, 8.3 Hz), 6.87 (2H, dd, J = 2.0, 8.3 Hz), 5.45 (1H, m), 4.87 (2H, s), 4.50 (2H, d, J = 4.1 Hz), 1.77 (3H, s), 1.73 (3H, s); 13C-NMR (125 MHz, CDCl3) δ: 18.2 (q), 25.9 (q), 64.9 (t), 65.8 (t), 115.6 (d), 121.3 (d), 129.6 (d), 134.7 (s), 138.4 (s), 159.7 (s).
4-Hydroxy-3-(3-methylbut-2-en-1-yl)benzaldehyde (4O3PBA) was obtained as a colorless oil. ESI-MS m/z: 189 [M–H]−. The NMR data were consistent with those reported in the literature (63).
RNA silencing plasmid construction for PEG-mediated transformation.
The vector pUCH2-8, containing the hygromycin B resistance gene, was used as a plasmid for expression in basidiomycetes, and the vector pMCB17apx was used to clone the alcA(p). First, the alcA(p) was cloned into the expression vector pUCH2-8. The alcA(p) primers AlcA-F and AlcA-R were designed contained HindIII and BamHI digestion sites (Table S1), and pMCB17apx was used as the PCR template. The gene was then cloned between the HindIII and BamHI sites of pUCH2-8 to construct the vector pUNZ101. The vib-PT gene was isolated via RT-PCR from mRNA as described in our previous work (4). To improve RNA silencing efficiency, the two arms of the target gene vib-PT were connected by introns from plants using the construction principle of an efficient intron-spliced hairpin RNA (ihpRNA) expression vector. A pUC18-RNAi plasmid (gift from Heriberto Cerutti at the University of Nebraska—Lincoln) that harbors the second intron of the Arabidopsis small nuclear ribonuclear protein D1 (locus at 4g02840) was used to construct the RNA silencing cassette. A 1,103-bp element derived from the PCR product was subsequently reamplified to incorporate convenient cloning sites at the 5′ and 3′ ends containing HpaI and BamHI cleavage sites by PCR using the primer set P-2UP-BamHI and PT-2DN-HpaI. A reverse-complemented vib-PT 1,106-bp element derived from the PCR product was then reamplified by PCR using the primer set PT-2iv-UP-sac1 and PT-2iv-DN-xho1-NotI containing SacI and XhoI-NotI cleavage sites. The two PCR products were purified, digested with corresponding endonucleases, and cloned into the two ends of the intron of the pUC18-RNAi vector to construct the vector pUNZ103. The 2,456-bp RNA silencing cassette was digested with BamHI and NotI and cloned into pUNZ101 to obtain pUNZ104, which was used for PEG-mediated transformation.
RNA silencing plasmid construction for Agrobacterium-mediated transformation.
To obtain a plasmid for Agrobacterium-mediated transformation, the RNA silencing cassette, including the alcA(p), was reamplified into two fragments by PCR using the pUNZ104 vector as the template and primer pairs PT-FW F/PT-FW R and PT-IVF/PT-IV R. One fragment was a 1,479-bp element containing the alcA(p) and vib-PT forward sequence, and the other fragment was a 1,357-bp element containing the intron and vib-PT reverse-complemented sequence. After digestion with BglII and dephosphorylation, a 1,479-bp element was subcloned into the pC-HYG-YR vector (Addgene, plasmid 61765) to achieve pYUZ10 by In-Fusion ligation (In-Fusion HD cloning kit; TaKaRa Biological Company, Dalian, China) (64). The pYUZ10 vector and a 1,357-bp element were digested with SalI, dephosphorylated, and then linked by In-Fusion ligation to obtain pYUZ11.
Culture of S. vibrans mycelia and protoplast preparation.
Stereum vibrans YMF1.05754, a basidiomycete of the Stereaceae family, was preserved in the State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, People’s Republic of China. Mycelia of S. vibrans were cultured in potato dextrose agar for 3 days, and the fresh mycelia were cut into small pieces and cultured in TG liquid medium (10 g liter−1 tryptone and 10 g liter−1 glucose) for 3 days. The strains were filtered and collected in bottles containing enzymatic hydrolysates (0.8 mg ml−1 cellulase, 0.8 mg ml−1 snailase, and 0.4 mg ml−1 driselase) for 6 h. Enzymatic hydrolysates were filtered, and protoplasts were collected and placed at 4°C for 2 h.
Culture of A. tumefaciens.
Agrobacterium competent cell lines AGL1, LBA4404, EHA105, and GV3101 were obtained from Biomed (Beijing, China) (see Table S2 in the supplemental material). The pYUZ11 plasmid was introduced into the four strains of A. tumefaciens according to previously described methods (65). A. tumefaciens strains LBA4404, GV3101, EHA105, and AGL-1 harboring the binary vector pYUNZ11 were grown at 28°C on a rotatory shaker (180 rpm) in 50 ml of Luria-Bertani (LB) broth supplemented with 50 μg ml−1 kanamycin to an optical density at 600 nm (OD600) of 0.5 to 0.6. Induction medium (IM) was used for transformation using A. tumefaciens, and modified potato dextrose agar (205.2 g liter−1 sucrose, 200 g liter−1 peeled potato, 10 g liter−1 molasses, and 15 g liter−1 agar) for the protoplast recovery (66). The bacterial solution was diluted in 50 ml of IM containing 0.3 mM acetosyringone (AS) to an OD600 of 0.15 and then grown at 28°C on a rotatory shaker (180 rpm) to an OD600 of 0.5 to 0.6 in the dark. The cells were harvested by centrifugation at 5,000 rpm for 5 min.
S. vibrans infected by A. tumefaciens .
The induced A. tumefaciens solution was mixed with the prepared protoplast suspension at a ratio of 1:1, and then 100 to 150 μl of the mixed solution was coated on an IM solid plate in the dark at 22°C, followed by coculture for 48 h or until the mycelium grew out. Two validation steps were performed in the screening transformants. The first step was to cover the medium with a layer containing 500 μg ml−1 cefotaxime to inhibit the growth of A. tumefaciens. After the growth of the mycelium, a layer containing 150 μg ml−1 hygromycin B was added to cover the medium for the second screening, and transformants that could grow on the resistance selection plate were obtained and named S. vibrans A2 and G7 (67).
RT-qPCR analysis of vib-PT expression.
Extraction of fungal genomic DNA was accomplished using a genomic DNA extraction kit (Tsingke, Kunming, China) and validated by 1% agarose gel electrophoresis. The total RNA of the fungi was extracted according to the Axygen kit procedure (Axygen Biotech Company, Hangzhou, China) and verified by 1% agarose gel electrophoresis with 0.1% formaldehyde.
RNA was extracted from the mycelia at 5, 10, and 15 days according to the metabolic period of vibralactone, and the transformants were fermented for two time periods (15 and 21 days). A TaKaRa reverse transcription kit was used for the synthesis of cDNA, which was used as the template for qPCR. β-Tubulin was used as an internal reference, and qPCR was performed using the primer pairs β-tubulin F/β-tubulin R and PT F/PT R (Table S1) according to the Roche-LightCycler 480 relative quantitative method (Roche Center, China). The relative transcription level of each gene was calculated as the ratio of the transcription level in the transformants to that in the WT strain at a given time point according to the 2−ΔΔCT method (68). All assays were repeated seven times. PCR product purification kits were purchased from Biological Company (Sangon Biotech, Shanghai, China).
Quantification of vibralactones by UPLC-MS.
The transformants were inoculated into 300 ml of modified PDB (20 g liter−1 glucose, 200 g liter−1 peeled potato, 1.5 g liter−1 MgSO4, 3.0 g liter−1 KH2PO4, 10 mg liter−1 thiamine hydrochloride) at 25°C and 140 rpm, and WT S. vibrans was used as the control group. The fermentation was conducted for two stages: 15 and 21 days. After fermentation, the mycelia were filtered and dried and then extracted three times by using ethyl acetate and dried under a vacuum. The remaining residues were dissolved in methanol and transferred to a 2-ml volumetric flask to analyze the vibralactone contents. All assays were repeated three times.
Cloning, expression, and purification of the recombinant Vib-PT.
vib-PT was cloned into pET28a in-frame with a C-terminal His tag sequence using EcoRI and NotI (Invitrogen). The recombinant plasmid was transformed into E. coli BL21(DE3) cells. Transformants were grown in LB medium supplemented with kanamycin (50 μg ml−1) at 37°C and 220 rpm for 12 h and then diluted to 1:100 with fresh LB medium. The diluted cultures were grown to an OD600 of approximately 0.4 to 0.6, induced with 0.1 mM IPTG, and incubated for another 20 h at 15°C with shaking at 90 rpm. The induced E. coli BL21(DE3) cells were centrifuged at 6,000 rpm for 10 min, and the pellet was suspended in binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM imidazole) and then purified by one-step Ni-NTA affinity chromatography to yield homogeneous protein. The protein was eluted with 200 mM imidazole (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 200 mM imidazole) and was desalted with Sephadex G-25 against 50 mM Tris-HCl buffer (pH 7.5) with 5% glycerol to remove the imidazole. The protein homogeneity was assessed by 12% SDS-PAGE, followed by Coomassie brilliant blue R-250 staining. The protein concentration was determined by the Bradford method using bovine serum albumin as a standard without glycerol (69).
Vib-PT activity assay.
The enzyme assays (100 μl) contained 1 mM 4-hydroxy-benzenemethanol or 1 mM 4-hydroxy-benzaldehyde, 1 mM DMAPP (Cayman Chemical), 5.0% (vol/vol) glycerol, 1% (vol/vol) dimethyl sulfoxide (DMSO), 50 mM Tris-HCl (pH 7.5), and purified recombinant protein (30 to 50 μg). The reaction mixtures were incubated at 37°C for 4 h, and the products were extracted three times with ethyl acetate. The ethyl acetate phases were concentrated to dryness after collection by centrifugation for 5 min at 12,000 rpm.
Characterization of the biochemical properties of Vib-PT.
The activity of purified Vib-PT was determined in a reaction mixture (100 μl) containing 50 mM Tris-HCl (pH 7.5, 5% glycerol), 1 mM 4-hydroxy-benzenemethanol (or 1 mM 4-hydroxy-benzaldehyde), 1 mM DMAPP, and 5 to 8 μM purified fusion Vib-PT at 37°C for 4 h. The effect of temperature on enzymatic activity was tested after incubation of 8.1 μM Vib-PT in 50 mM Tris-HCl (pH 7.5) buffer at various temperatures ranging from 15 to 50°C for 4 h. To determine the optimal pH for Vib-PT, the prenylation activity was analyzed at an optimal temperature of 32°C in each buffer at various pH values (4.0, 5.0, 6.0, 6.5, 7.0, 7.5, 8.0, and 9.0) for 4 h with 2.7 μM Vib-PT. The effect of metals on Vib-PT activity was tested in the presence of 0.5, 5, and 10 mM (final concentration) MgCl2·6H2O, CaCl2, MnCl2, CuCl2, or ZnCl2 for 4 h at the optimal temperature (32°C) and pH (7.5) with 6.2 μM Vib-PT. Each experiment was performed in triplicate.
Kinetic analysis of fusion Vib-PT.
The assays for kinetic parameter analysis contained various 4-hydroxy-benzenemethanol concentrations (0.5, 1, 2, 4, 8, and 16 mM) and 4-hydroxy-benzaldehyde concentrations (0.1, 0.25, 0.5, 1, 2, and 4 mM) with 5.6 and 6.8 μM purified Vib-PT, 4% (vol/vol) DMSO for solubility, 1 mM DMAPP, 1.5 mM TCEP, and 10 mM Ca2+ in 50 mM Tris-HCl (pH 7.5, 5% glycerol). To determine the kinetic parameters of DMAPP, 4.0 μM Vib-PT, 4 mM 4-hydroxy-benzenemethanol, 1.5 mM TCEP, 10 mM Ca2+ in 50 mM Tris-HCl (pH 7.5, 5% glycerol), and DMAPP at final concentrations of 0.1, 0.25, 0.5, 1.0, and 2 mM were used. Since this reaction provides two products, we measured the levels of C- and O-prenylation, which were converted to molar concentration, respectively, and finally considered the data together as the Michaelis constant. The reactions were incubated for 1 h at 32°C and then quenched exactly as described above. Each experiment was performed in triplicate.
Assay of substrate specificity.
Reactions of Vib-PT toward different substrates were conducted in a reaction mixture (100 μl) containing 50 mM Tris-HCl (pH 7.5, 5% glycerol), 4.0 μM Vib-PT, 1 mM l-tryptophan (or 3,4-dihydroxy-benzaldehyde, 4-hydroxy-benzoic acid, l-phenylalanine, l-tyrosine, and l-tryptophan), 4% (vol/vol) DMSO for solubility, 1 mM DMAPP (or GPP, FPP, or GGPP), 1.5 mM TCEP, and 10 mM CaCl2. Incubations were carried out at 32°C for 4 h. Each assay was repeated at least twice.
Quantification and statistical analysis.
The enzyme reactions were stopped by adding 3 × 100 μl of EtOAc. The EtOAc phases were concentrated to dryness after collection by centrifugation for 5 min at 12,000 rpm. The product mixtures were dissolved in 50 μl of methanol and analyzed by an Agilent series UPLC 1290 instrument with an Agilent 6500 Accurate-Mass Q-TOF system (Agilent Technologies). A Zorbax Eclipse Plus C18 column (Rapid Resolution HD; 2.1 mm × 50 mm, 1.8 μm; Agilent) was applied for analysis at a flow rate of 300 μl min−1 with gradient elution (the mobile phase consisted of solvent A [water] and solvent B [methanol]). The gradient program was as follows: 0 to 2 min (8% B), 3 min (8 to 45% B), 4 to 6 min (45 to 55% B), 7 to 10 min (55 to 100% B), and 11 to 14 min (100% B). This method was used to analyze the products of 4-hydroxy-benzenemethanol and 4-hydroxy-benzaldehyde reactions catalyzed by Vib-PT.
The prenylated products of 3,4-dihydroxy-benzenemethanol, 4-hydroxy-benzoic acid, l-tryptophan, l-tyrosine, and l-phenylalanine were detected on a Thermo Scientific Dionex Ultimate 3000 UHPLC system with a Thermo high-resolution Q Exactive focus mass spectrometer (Thermo, Bremen, Germany). The injection volume was 2 μl, and the chromatographic column was Hypersil Gold (100 mm × 2.1 mm; Thermo Fisher Scientific), with a particle size of 1.9 μm. The mobile phase was a gradient prepared from a 0.5% formic acid aqueous solution (A) and methanol (B). The elution began with 35% B from 2 to 15 min, and the proportion of B was increased linearly to 98% at 10 min, kept for 5 min, brought back to 35% B at 20.1 min, and kept for 5 min for a total of 25 min. The flow rate of the mobile phase was maintained at 200 μl/min. Mass spectrometry was performed on a Thermo Quadrupole Exactive Focus system (Thermo Fisher Scientific). The products were analyzed under negative-ion mode. The optimized conditions were as follows: sheath gas, 35 liters min−1; auxiliary gas (Ar), 10 liters min−1; and capillary potential, 2.5 kV.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Heriberto Cerutti at University of Nebraska—Lincoln for providing the pUC18-RNAi plasmid, to Ji-Kai Liu at Kunming Institute of Botany for providing Stereum vibrans, and to Jianping Xu of McMaster University for his valuable comments and critical discussion. We also thank Yan-Long Yang for synthesizing three compounds as authentic reference samples [(4-hydroxy-3-(3-methylbut-2-en-1-yl)-benzenemethanol, 4-(3-methylbut-2-en-1-yl)oxy-benzenemethanol and 4-hydroxy-3-(3-methylbut-2-en-1-yl)benzaldehyde)].
This study was financially supported by the National Natural Science Foundation of China (31760018, 31560016, and 31970060), the Applied Basic Research Foundation of Yunnan Province (2018FA006), and the Joint Fund from the Ministry of Education and Yunnan University (C176280101). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare that they have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Chen Y, Li G, Yin S, Xu J, Ji Z, Xiu X, Liu L, Ma D. 2007. Genetic polymorphisms involved in toxicant-metabolizing enzymes and the risk of chronic benzene poisoning in Chinese occupationally exposed populations. Xenobiotica 37:103–112. doi: 10.1080/00498250601001662. [DOI] [PubMed] [Google Scholar]
- 2.Wei K, Wang GQ, Bai X, Niu YF, Chen HP, Wen CN, Li ZH, Dong ZJ, Zuo ZL, Xiong WY, Liu JK. 2015. Structure-based optimization and biological evaluation of pancreatic lipase inhibitors as novel potential antiobesity agents. Nat Prod Bioprospect 5:129–157. doi: 10.1007/s13659-015-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiler E, Braun N, Bottcher T, Kastenmuller A, Weinkauf S, Sieber SA. 2011. Vibralactone as a tool to study the activity and structure of the ClpP1P2 complex from Listeria monocytogenes. Angew Chem Int Ed Engl 50:11001–11004. doi: 10.1002/anie.201104391. [DOI] [PubMed] [Google Scholar]
- 4.Zhao P-J, Yang Y-L, Du L, Du L, Liu J-K, Zeng Y. 2013. Elucidating the biosynthetic pathway for vibralactone: a pancreatic lipase inhibitor with a fused bicyclic β-lactone. Angew Chem Int Ed Engl 52:2298–2302. doi: 10.1002/anie.201208182. [DOI] [PubMed] [Google Scholar]
- 5.Heide L. 2009. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol 13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G. 2005. Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem 12:717–739. doi: 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- 7.Li SM. 2010. Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat Prod Rep 27:57–78. doi: 10.1039/b909987p. [DOI] [PubMed] [Google Scholar]
- 8.Wollinsky B, Ludwig L, Hamacher A, Yu X, Kassack MU, Li SM. 2012. Prenylation at the indole ring leads to a significant increase of cytotoxicity of tryptophan-containing cyclic dipeptides. Bioorg Med Chem Lett 22:3866–3869. doi: 10.1016/j.bmcl.2012.04.119. [DOI] [PubMed] [Google Scholar]
- 9.Kuzuyama T, Noel JP, Richard SB. 2005. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumano T, Tomita T, Nishiyama M, Kuzuyama T. 2010. Functional characterization of the promiscuous prenyltransferase responsible for furaquinocin biosynthesis: identification of a physiological polyketide substrate and its prenylated reaction products. J Biol Chem 285:39663–39671. doi: 10.1074/jbc.M110.153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer A, Li SM. 2010. A tyrosine O-prenyltransferase catalyses the first pathway-specific step in the biosynthesis of sirodesmin PL. Microbiology 156:278–286. doi: 10.1099/mic.0.033886-0. [DOI] [PubMed] [Google Scholar]
- 12.Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li SM, Heide L. 2003. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci U S A 100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi QY, Ren JW, Sun LW, He LW, Bao L, Yue W, Sun QM, Yao YJ, Yin WB, Liu HW. 2015. Structurally diverse sesquiterpenes produced by a Chinese Tibet fungus Stereum hirsutum and their cytotoxic and immunosuppressant activities. Org Lett 17:3098–3101. doi: 10.1021/acs.orglett.5b01356. [DOI] [PubMed] [Google Scholar]
- 14.Chen HP, Liu JK. 2017. Secondary metabolites from higher fungi. Prog Chem Org Nat Prod 106:1–201. doi: 10.1007/978-3-319-59542-9_1. [DOI] [PubMed] [Google Scholar]
- 15.Ito-Kobayashi M, Aoyagi A, Tanaka I, Muramatsu Y, Umetani M, Takatsu T. 2008. Sterenin A, B, C, and D, novel 11β-hydroxysteroid dehydrogenase type 1 inhibitors from Stereum sp. SANK 21205. J Antibiot (Tokyo) 61:128–135. doi: 10.1038/ja.2008.121. [DOI] [PubMed] [Google Scholar]
- 16.Ma K, Bao L, Han J, Jin T, Yang X, Zhao F, Li S, Song F, Liu M, Liu H. 2014. New benzoate derivatives and hirsutane type sesquiterpenoids with antimicrobial activity and cytotoxicity from the solid-state fermented rice by the medicinal mushroom Stereum hirsutum. Food Chem 143:239–245. doi: 10.1016/j.foodchem.2013.07.124. [DOI] [PubMed] [Google Scholar]
- 17.Nakayashiki H, Hanada S, Quoc NB, Kadotani N, Tosa Y, Mayama S. 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42:275–283. doi: 10.1016/j.fgb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Timmons L, Tabara H, Mello CC, Fire AZ. 2003. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol Biol Cell 14:2972–2983. doi: 10.1091/mbc.e03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 20.Schumann J, Hertweck C. 2007. Molecular basis of cytochalasan biosynthesis in fungi: gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J Am Chem Soc 129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 21.Li CY, Shi L, Chen DD, Ren A, Gao T, Zhao MW. 2015. Functional analysis of the role of glutathione peroxidase (GPx) in the ROS signaling pathway, hyphal branching and the regulation of ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet Biol 82:168–180. doi: 10.1016/j.fgb.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Liu YN, Zhang TJ, Lu XX, Ma BL, Ren A, Shi L, Jiang AL, Yu HS, Zhao MW. 2017. Membrane fluidity is involved in the regulation of heat stress induced secondary metabolism in Ganoderma lucidum. Environ Microbiol 19:1653–1668. doi: 10.1111/1462-2920.13693. [DOI] [PubMed] [Google Scholar]
- 23.Vasiliauskas R, Stenlid J. 1998. Influence of spatial scale on population structure of Stereum sanguinolentum in northern Europe. Mycol Res 102:93–98. doi: 10.1017/S0953756297004516. [DOI] [Google Scholar]
- 24.Dang Y, Yang Q, Xue Z, Liu Y. 2011. RNA interference in fungi: pathways, functions, and applications. Eukaryot Cell 10:1148–1155. doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Li R, Liu Q, Wang Q, Chen M, Li B. 2006. A highly efficient polyethylene glycol-mediated transformation method for mushrooms. FEMS Microbiol Lett 256:203–208. doi: 10.1111/j.1574-6968.2006.00110.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Zong H, Zhuge B, Lu XY, Fang HY, Zhu JL, Zhuge J. 2016. Protoplast preparation and polyethylene glycol (PEG)-mediated transformation of Candida glycerinogenes. Biotechnol Bioproc E 21:95–102. doi: 10.1007/s12257-015-0686-8. [DOI] [Google Scholar]
- 27.Robinson HL, Deacon JW. 2001. Protoplast preparation and transient transformation of Rhizoctonia solani. Mycol Res 105:1295–1303. doi: 10.1017/S0953756201005159. [DOI] [Google Scholar]
- 28.Curragh HJ, Mooibroek H, Wessels JGH, Marchant R, Mullan E. 1993. Protoplast formation and DNA-mediated transformation of Fusarium culmorum to hygromycin B resistance. Mycol Res 97:313–317. doi: 10.1016/S0953-7562(09)81127-1. [DOI] [Google Scholar]
- 29.Alhawatema MS, Gebril S, Cook D, Creamer R. 2017. RNAi-mediated downregulation of a melanin polyketide synthase (pks1) gene in the fungus Slafractonia leguminicola. World J Microb Biot 33:179. doi: 10.1007/s11274-017-2346-y. [DOI] [PubMed] [Google Scholar]
- 30.Edwards DJ, Gerwick WH. 2004. Lyngbyatoxin biosynthesis: sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc 126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 31.Yin WB, Ruan HL, Westrich L, Grundmann A, Li SM. 2007. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. Chembiochem 8:1154–1161. doi: 10.1002/cbic.200700079. [DOI] [PubMed] [Google Scholar]
- 32.Unsold IA, Li SM. 2005. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 33.Grundmann A, Li SM. 2005. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 151:2199–2207. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 34.Kremer A, Westrich L, Li SM. 2007. A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. Microbiology 153:3409–3416. doi: 10.1099/mic.0.2007/009019-0. [DOI] [PubMed] [Google Scholar]
- 35.Unsold IA, Li SM. 2006. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem 7:158–164. doi: 10.1002/cbic.200500318. [DOI] [PubMed] [Google Scholar]
- 36.Grundmann A, Kuznetsova T, Afiyatullov S, Li SM. 2008. FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. Chembiochem 9:2059–2063. doi: 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
- 37.Metzger U, Keller S, Stevenson CE, Heide L, Lawson DM. 2010. Structure and mechanism of the magnesium-independent aromatic prenyltransferase CloQ from the clorobiocin biosynthetic pathway. J Mol Biol 404:611–626. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 38.Gebler JC, Woodside AB, Poulter CD. 1992. Dimethylallyltryptophan synthase: an enzyme-catalyzed electrophilic aromatic-substitution. J Am Chem Soc 114:7354–7360. doi: 10.1021/ja00045a004. [DOI] [Google Scholar]
- 39.Haug-Schifferdecker E, Arican D, Brückner R, Heide L. 2010. A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyl-transferase reaction. J Biol Chem 285:16487–16494. doi: 10.1074/jbc.M110.113720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wunsch C, Zou HX, Linne U, Li SM. 2015. C7-prenylation of tryptophanyl and O-prenylation of tyrosyl residues in dipeptides by an Aspergillus terreus prenyltransferase. Appl Microbiol Biotechnol 99:1719–1730. doi: 10.1007/s00253-014-5999-6. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Liebhold M, Xie X, Li SM. 2015. Tyrosine O-prenyltransferases TyrPT and SirD displaying similar behavior toward unnatural alkyl or benzyl diphosphate as their natural prenyl donor dimethylallyl diphosphate. Appl Microbiol Biotechnol 99:7115–7124. doi: 10.1007/s00253-015-6452-1. [DOI] [PubMed] [Google Scholar]
- 42.Haagen Y, Unsold I, Westrich L, Gust B, Richard SB, Noel JP, Heide L. 2007. A soluble, magnesium-independent prenyltransferase catalyzes reverse and regular C-prenylations and O-prenylations of aromatic substrates. FEBS Lett 581:2889–2893. doi: 10.1016/j.febslet.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumano T, Richard SB, Noel JP, Nishiyama M, Kuzuyama T. 2008. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg Med Chem 16:8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding JH, Feng T, Li ZH, Li L, Liu JK. 2012. Twelve new compounds from the basidiomycete Boreostereum vibrans. Nat Prod Bioprospect 2:200–205. doi: 10.1007/s13659-012-0060-x. [DOI] [Google Scholar]
- 45.Chen HP, Jiang MY, Zhao ZZ, Feng T, Li ZH, Liu JK. 2018. Vibralactone biogenesis-associated analogues from submerged cultures of the fungus Boreostereum vibrans. Nat Prod Bioprospect 8:37–45. doi: 10.1007/s13659-017-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jost M, Zocher G, Tarcz S, Matuschek M, Xie X, Li SM, Stehle T. 2010. Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity. J Am Chem Soc 132:17849–17858. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- 47.Metzger U, Schall C, Zocher G, Unsold I, Stec E, Li SM, Heide L, Stehle T. 2009. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A 106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan AL, Zocher G, Stec E, Stehle T, Li SM. 2015. Site-directed mutagenesis switching a dimethylallyl tryptophan synthase to a specific tyrosine C-3-prenylating enzyme. J Biol Chem 290:1364–1373. doi: 10.1074/jbc.M114.623413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Y, Williams RM, Sherman DH. 2008. Molecular analysis of a 4-dimethylallyltryptophan synthase from Malbranchea aurantiaca. J Biol Chem 283:16068–16076. doi: 10.1074/jbc.M801991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Y, de Wet JR, Cavalcoli J, Li S, Greshock TJ, Miller KA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM, Sherman DH. 2010. Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J Am Chem Soc 132:12733–12740. doi: 10.1021/ja1049302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chooi YH, Cacho R, Tang Y. 2010. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol 17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CC, Oakley BR. 2011. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc 133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan A, Chen H, Wu R, Xu H, Li SM. 2014. A new member of the DMATS superfamily from Aspergillus niger catalyzes prenylations of both tyrosine and tryptophan derivatives. Appl Microbiol Biotechnol 98:10119–10129. doi: 10.1007/s00253-014-5872-7. [DOI] [PubMed] [Google Scholar]
- 54.Yin S, Yu X, Wang Q, Liu XQ, Li SM. 2013. Identification of a brevianamide F reverse prenyltransferase BrePT from Aspergillus versicolor with a broad substrate specificity towards tryptophan-containing cyclic dipeptides. Appl Microbiol Biotechnol 97:1649–1660. doi: 10.1007/s00253-012-4130-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou K, Ludwig L, Li SM. 2015. Friedel-crafts alkylation of acylphloroglucinols catalyzed by a fungal indole prenyltransferase. J Nat Prod 78:929–933. doi: 10.1021/np5009784. [DOI] [PubMed] [Google Scholar]
- 56.McIntosh JA, Donia MS, Nair SK, Schmidt EW. 2011. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc 133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 58.Braesel J, Fricke J, Schwenk D, Hoffmeister D. 2017. Biochemical and genetic basis of orsellinic acid biosynthesis and prenylation in a stereaceous basidiomycete. Fungal Genet Biol 98:12–19. doi: 10.1016/j.fgb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Duan YC, Meng XX, Yang YL, Yang YH, Zhao PJ. 2015. Two new phenol derivatives from Stereum hirsutum FP-91666. J Asian Nat Prod Res 17:324–328. doi: 10.1080/10286020.2014.959439. [DOI] [PubMed] [Google Scholar]
- 60.Yun BS, Cho Y, Lee IK, Cho SM, Lee TH, Yoo ID. 2002. Sterins A and B, new antioxidative compounds from Stereum hirsutum. J Antibiot (Tokyo) 55:208–210. doi: 10.7164/antibiotics.55.208. [DOI] [PubMed] [Google Scholar]
- 61.Miege F, Meyer C, Cossy J. 2011. Rhodium-catalyzed cycloisomerization involving cyclopropenes: efficient stereoselective synthesis of medium-sized heterocyclic scaffolds. Angew Chem Int Ed Engl 50:5932–5937. doi: 10.1002/anie.201101220. [DOI] [PubMed] [Google Scholar]
- 62.Hanashima S, Manabe S, Ito Y. 2005. Divergent synthesis of sialylated glycan chains: combined use of polymer support, resin capture-release, and chemoenzymatic strategies. Angew Chem Int Ed Engl 44:4218–4224. doi: 10.1002/anie.200500777. [DOI] [PubMed] [Google Scholar]
- 63.Moriarty RM, Grubjesic S, Surve BC, Chandersekera SN, Prakash O, Naithani R. 2006. Synthesis of abyssinone II and related compounds as potential chemopreventive agents. Eur J Med Chem 41:263–267. doi: 10.1016/j.ejmech.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 64.Sleight SC, Bartley BA, Lieviant JA, Sauro HM. 2010. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res 38:2624–2636. doi: 10.1093/nar/gkq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flowers JL, Vaillancourt LJ. 2005. Parameters affecting the efficiency of Agrobacterium tumefaciens-mediated transformation of Colletotrichum graminicola. Curr Genet 48:380–388. doi: 10.1007/s00294-005-0034-1. [DOI] [PubMed] [Google Scholar]
- 66.Liu L, Long LK, An Y, Yang J, Xu XX, Hu CH, Liu G. 2013. The thioredoxin reductase-encoding gene ActrxR1 is involved in the cephalosporin C production of Acremonium chrysogenum in methionine-supplemented medium. Appl Microbiol Biotechnol 97:2551–2562. doi: 10.1007/s00253-012-4368-6. [DOI] [PubMed] [Google Scholar]
- 67.Sidhu YS, Cairns TC, Chaudhari YK, Usher J, Talbot NJ, Studholme DJ, Csukai M, Haynes K. 2015. Exploitation of sulfonylurea resistance marker and non-homologous end joining mutants for functional analysis in Zymoseptoria tritici. Fungal Genet Biol 79:102–109. doi: 10.1016/j.fgb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.