Abstract
Impaired mitochondrial translation or reduced mitochondrial protein import can lead to imbalances in mitochondrial protein composition. Such mitochondrial proteotoxic stresses can trigger a nuclear transcriptional response commonly described as the mitochondrial unfolded protein response (UPRmt). Despite extensive studies of UPRmt pathways in animal and fungal systems, very little is known about how the UPRmt is regulated in plants. Through comparison of Arabidopsis thaliana whole-genome transcriptome data, it was found that most genes induced by mitochondrial ribosome inhibitor doxycycline are also induced by Complex III inhibitor antimycin A. We demonstrate that transcriptional responses to a wide range of mitochondrial proteotoxic stress-triggers are regulated by the transcription factor ANAC017, which was shown to reside in the endoplasmic reticulum (ER). By contrast, no consistent evidence was found for genes that are specifically induced by doxycycline but not antimycin A. Furthermore, ANAC017 gain- and loss-of-function mutants showed marked resistance or susceptibility, respectively, to mitochondrial stress-inducing treatments, demonstrating the physiological importance of ANAC017 during mitochondrial proteotoxic stress. Finally, it was shown that ethylene signalling promotes mitochondria-to-nucleus signalling, most likely independently of ANAC017. Overall, this study shows that in plants, the UPRmt is largely overlapping with, and perhaps identical to, ‘classical’ mitochondrial retrograde signalling, and is mediated by ER-anchored transcription factor ANAC017.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: mitochondria, unfolded protein response, retrograde signalling, UPRmt
1. Introduction
Mitochondria signal to the nucleus to regulate gene expression, allowing appropriate assembly and function of mitochondrial proteins, including the oxidative phosphorylation (OXPHOS) components [1–3]. These mitochondria-to-nucleus signalling pathways, termed retrograde signalling, contribute to cellular viability and whole-organism lifespan. Mis-regulation of mitochondrial proteostasis or function could lead to reactive oxygen species (ROS) accumulation, accumulation of damaged mtDNA, metabolic disorders and ultimately can impact on ageing and various neurodegenerative or immune system diseases [4,5]. To prevent proteotoxic stress, mitochondria use an intricate mitochondrial protein quality control system (mtPQC) of chaperones and proteases [6], which assist in protein folding and degradation of misfolded proteins, respectively. One of the most well-characterized retrograde signalling pathways is the mitochondrial unfolded protein response (UPRmt), which is induced by excessive levels of unfolded or misfolded proteins, and promotes mtPQC. The UPRmt is a transcriptional response that regulates nuclear gene expression to repair and restore dysfunctional mitochondria and adapt the organism to future stress [3].
There is strong evidence for a critical role of UPRmt in the regulation of various physiological processes, including protein-folding maintenance [7,8], recovery of the mitochondrial protein import machinery [9], increased glycolysis-related gene expression for metabolic remodelling [8], ROS detoxification [10], immune response activation [11], lifespan improvement [12,13] and delay of senescence [14]. Multiple transcription factors have been implicated as core components of mitonuclear UPRmt in invertebrates, including ATFS-1, DVE-1, and the ubiquitin-like protein UBL-5 [7,9,15]. In mammals, the activating transcription factors 4 and 5 (ATF4, ATF5), together with the C/EBP transcription factor CHOP, were identified as mediators of the UPRmt [16–18]. One possible mechanism for the regulation of UPRmt in animals is via reduced mitochondrial import efficiency of ATFS-1 or ATF5 during mitochondrial stresses. This enforces their relocation from the cytosol to the nucleus, possibly owing to the presence of mitochondrial and nuclear targeting sequences in these regulatory proteins [3,9].
In contrast with the UPRmt signalling pathways in animals, the signals and factors mediating plant UPRmt remain elusive. It was reported that the specific UPRmt response in plants can be activated through the mitochondrial ribosome inhibitor doxycycline (Dox) [19], or genetic mutation in the mitochondria ribosomal protein mrpl1 [14]. Both conditions impair mitochondrial translation and result in the upregulation of mitochondrial stress marker gene alternative oxidase AOX1a, mitochondrial HSP70 chaperone genes and genes for mitochondria ribosomal proteins (MRPs) [14]. Also, blocking of mitochondrial protein import resulted in mitochondrial proteotoxic stress in plants [14]. It has been suggested that this plant UPRmt involves an oxidative burst, and integrates hormonal signalling (mainly ethylene and auxin) with MAPK signalling to restore mitochondrial proteostasis [14]. However, genetic components that activate and execute the plant UPRmt signalling have not been identified so far. The existence of other mitochondrial retrograde regulation (MRR) pathways in plants has been demonstrated earlier using treatments that inhibit different respiratory chain complexes, such as antimycin A (AA) or rotenone [20–22], the tricarboxylic acid (TCA) cycle inhibitor monofluoroacetate [23,24], and mutants impaired in mitochondrial transcription or mitochondrial membrane protein AtPHB3 [25]. All these were found to result in the enhanced expression of a common set of nuclear genes including AOX1a, NADH dehydrogenases (NDB), AAA ATPases (OM66), mitochondrial heat shock proteins and some oxidative stress inducible genes [24–26]. The exact mechanisms of plant mitochondrial retrograde signalling are not well understood. Several regulators were initially identified as failing to upregulate the AOX1a promoter after AA treatment [22,27]. The Regulator of AOX1a 1 (RAO1) encodes a nuclear localized cyclin-dependent kinase E1 (CDKE1), presumably integrating mitochondrial energy stress signalling, with other general cellular oxidative stress signals [27]. Regulator of AOX1a 2 (RAO2) encodes a NAM/ATAF/CUC (NAC) domain containing transcription factor, ANAC017, which has been proposed to function as a more specific, and upstream, regulator of MRR pathways [22,24,28]. ANAC017 is targeted to the endoplasmic reticulum (ER) via its C-terminal transmembrane domain [22]. There is no clear agreement on the events that activate ANAC017, but it has been proposed that rhomboid proteases control NAC transcription factor proteolytic cleavage upon mitochondrial stress [22], and thereby initiate ANAC017 release from the ER and relocation to the nucleus. Other transcription factors identified through yeast one hybrid (Y1H) screens and possibly operating in plant mitonuclear retrograde signalling include ANAC013 [29], AtWRKY15 [30], AtWRKY40 and AtWRKY63 [31].
In this study, we aimed to identify novel regulators of UPRmt in plants. Our results show that there is a strong overlap between the transcriptomic responses to classic UPRmt-inducing agents and general MRR signalling. We demonstrate that mitochondrial proteotoxic stress-induced gene expression is directly regulated by the transcription factor ANAC017. In agreement with an important role of ANAC017 in mitochondrial proteotoxic stress responses, transgenic ANAC017 gain- and loss-of-function show altered tolerance and developmental responses to a range of classic UPRmt-inducing agents. In addition, we also found evidence for a regulatory role of ethylene signalling in strengthening mitochondrial retrograde responses, operating independently of ANAC017.
2. Methods
(a). Plant materials and growth conditions
Arabidopsis thaliana wild-type (WT) was Columbia-0 (Col-0), and other lines used in this study were in the same genetic background, with the exception of rao2-1 mutant, which was in the Col-0 pAOX1a::LUC background [32]. Two nac017 mutant lines: rao2-1 and nac017-1 (SALK_022174) have been reported previously [22]. One ANAC017 overexpressor line (OE1) was reported before [24]. The second ANAC017 overexpressor line (OE2) was generated by cloning ANAC017 into pB7GW2 and transformed into Arabidopsis Col-0 by floral dipping. Seeds of ethylene biosynthesis mutant eto1-1 and ethylene signalling mutant ein2-1 [32] were kindly provided by Dr Kirk Overmyer (University of Helsinki). Seeds of MAPK signalling were obtained from Prof. Matthew Terry (mpk6-3; University of Southampton) and Dr Kirk Overmyer (mpk6-4).
For all gene expression analyses, seeds were first sown onto half-strength Murashige and Skoog (MS) medium supplemented with 1% (w/v) sucrose, 0.05% (w/v) 4-morpholineethanesulfonic acid (MES; Biomol, Germany) and 0.8% (w/v) agar (pH 5.8). Seeds were stratified for 3 days at 4°C and then kept for 10 days under 16/8 h light/dark cycle (100–120 µmol m−2 s−1) at 22/19°C. For mitochondrial retrograde signalling treatment, seedlings were sprayed with 50 µM antimycin A (AA; Sigma Aldrich, Sweden) in water with 0.01% Tween-20 (experimental) or water with 0.1% ethanol and 0.01% Tween-20 (mock control) and left under standard light growth conditions for 4 h and 12 h. For mitochondrial unfolded protein response treatment, seedlings were sprayed with 25 µg ml−1 Dox (Sigma Aldrich) with 0.05% MES and 0.01% Tween-20 in water (experimental) or just 0.05% MES and 0.01% Tween-20 in water (mock control). For the inhibition of mitochondrial protein import, seedlings were sprayed with 50 μM MitoBloCK-6 (MB; Tebu-bio, Denmark) in water with 0.01% Tween-20 (experimental) or water with 0.05% dimethylsulfoxide and 0.01% Tween-20 (mock control). For the inhibition of mitochondrial translation or mitochondrial membrane potential treatments, seedlings were sprayed with 100 µg ml−1 chloramphenicol (CAP; Sigma Aldrich) in water or 10 μM carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP; Sigma Aldrich), respectively (experimental), or water with 0.1% ethanol and 0.01% Tween-20 (mock control). Non-sprayed seedlings were also harvested at the beginning of each experiment as additional control (0 h). For the treatments that block or promote ethylene synthesis, seedlings were sprayed with 50 µM silver nitrate (AgNO3; Sigma Aldrich) or 1-aminocyclopropane-1-carboxylic acid (ACC; Sigma Aldrich), respectively.
For single and double treatments with AA and AgNO3 in the dark, seeds were sown onto half-strength MS medium supplemented with 1% (w/v) agar (pH 5.8) and with or without 50 µM antimycin A (AA), 50 µM AgNO3, or 0.1% ethanol as a control. After 3 days, stratification at 4°C plates were incubated in light (100–120 µmol m−2 s−1) for 2 h to induce germination and left covered with foil for 4 days under standard growth condition.
(b). Gene expression analysis by quantitative real-time polymerase chain reaction
For gene expression analysis, a pool of 10–12 whole seedlings from a single plate was collected for RNA extraction per replicate. Total RNA was extracted from up to 100 mg frozen ground tissue using the Spectrum™ Plant Total RNA Kit (Sigma Aldrich) following the manufacturer's instructions (Protocol B). To assure there was no genomic DNA contamination, additional on-column RNase free DNase I treatment step (Sigma Aldrich) was incorporated after the RNA binding step (4b) following the manufacturer's instructions. cDNA was synthesized from 500 ng of RNA with the mixture of oligo(dT) and random hexamer primers using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) following the manufacturer's instructions. Transcript levels were measured by quantitative real-time polymerase chain reaction (qRT-PCR) using SsoAdvanced™ Universal SYBR Green Supermix (Bio-Rad). Each reaction contained 20× diluted cDNA and gene specific primers at a final concentration of 0.5 µM. Samples were run on a CFX384TM Real-Time System (Bio-Rad) using the following thermal cycling conditions: 95°C for 2 min, followed by 40 cycles of 95°C for 10 s and 60°C for 10 s. The genes analysed by qRT-PCR, with their sequences, accession numbers and amplified product size, are listed in the electronic supplementary material, table S1.
(c). Genome-wide transcriptome analysis
The whole-transcriptome expression data for AA response in WT and nac017 mutants presented in figures 1 and 4 were obtained from the microarray data GSE41136 [22]. The whole-transcriptome expression data for disturbed mitochondrial translation in Dox-treated WT, or mrpl1 mutant were obtained from RNA-seq experiment GSE78862 [14]. Both datasets were reanalysed using R and Bioconductor software [33]. The R package simpleaffy [34] was used to determinate Affymetrix RMA-normalized probe level values from the raw CEL files. For the GSE78862 RNA-seq experiment, raw FASTQ reads were downloaded and mapped to the TAIR10 genome with HISAT2 [35]. The R package limma [36] or package edgeR [37] were used to determinate differently expressed genes. Only terms with the ±1 log2 fold-change in expression relative to untreated WT sample and a significant false discovery rate (FDR) value < 0.05 was considered as differently expressed. Clustering and visualization of differentially expressed genes were performed using the R function heatmap.2. Selected ethylene biosynthesis and signalling gene expression levels were extracted from GSE41136 and the calculated log2 fold changes of gene expression were visualized using the R function heatmap.2.
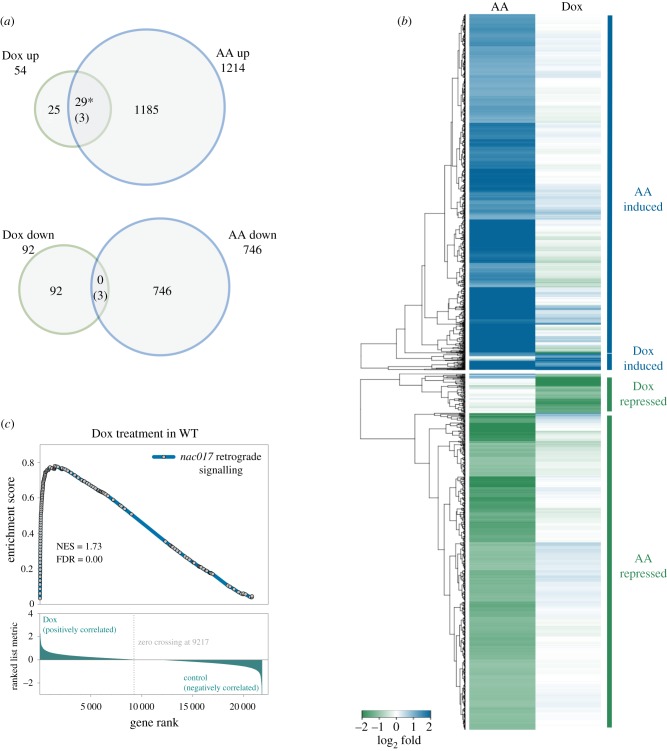
Figure 1.
Genome-wide transcriptomic analysis of genes regulated by antimycin A (AA) and doxycycline (Dox). (a) Venn diagrams showing common sets of genes downregulated (down) or upregulated (up) in seedlings treated with AA for 6 h, Dox for 12 h. Numbers in parentheses correspond to the number of genes expected to overlap by chance. (*p < 6.05 × 10−23) denotes statistically significant overlap between tested sets. (b) Heatmap demonstrating gene cluster analysis of genome-wide analysis for genes induced (blue) and repressed (green) at least twofold by AA or Dox in WT. (c) Enrichment score plots analysed by GSEA using RNA sequencing data of WT plants treated with Dox (left). The list of ANAC017-dependent retrograde signalling genes was defined in Methods and was used as the gene set of interest.
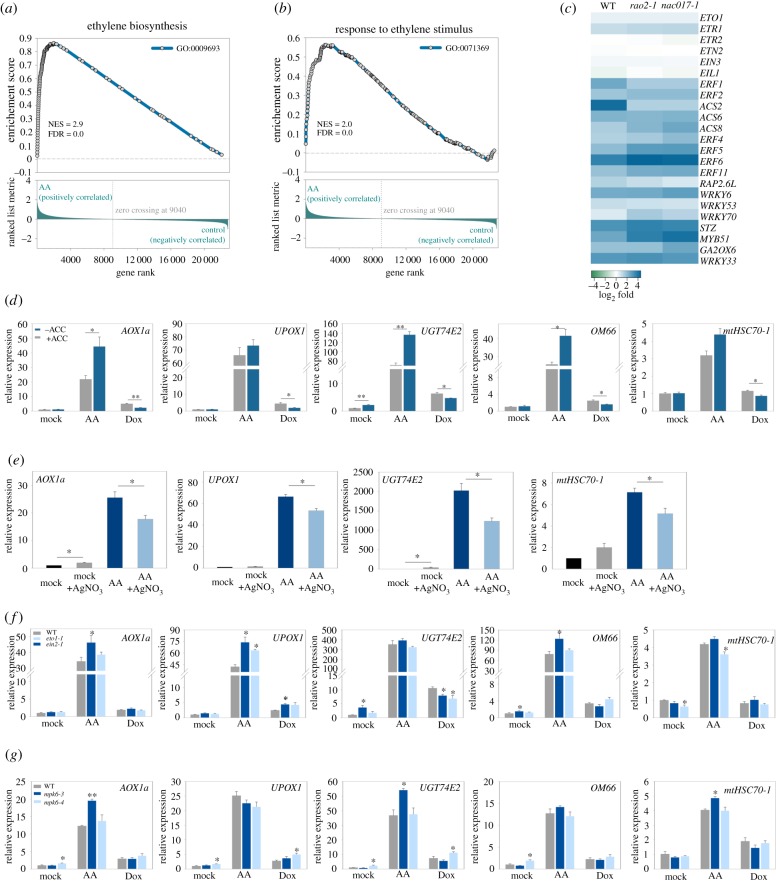
Figure 4.
ANAC017-dependent mitochondrial retrograde signalling does not require ethylene signalling and MPK6. (a,b) Enrichment score plots analysed by GSEA using microarray data of WT plants treated with antimycin A (AA). List of ethylene biosynthesis genes (GO:0071369; a) or response to ethylene stimulus genes (GO:0009693; b) was used as the gene set of interest. (c) Heat map of the expression of selected ethylene biosynthesis and signalling genes after AA treatment in WT, rao2-1 and nac017-1. High and low expressed genes are depicted in blue and green, respectively. (d,e) Expression of selected mitochondrial retrograde signalling marker genes in WT plants treated with AA or Dox and with ACC (d), or AgNO3 (e). (f,g) Expression of selected mitochondrial retrograde signalling marker genes in WT and ethylene biosynthesis (eto1-1) and ethylene-insensitive (ein2-1) mutants, or in MAPK signalling mutants (mpk6-3, mpk6-4) treated with AA or Dox. For all gene expression analysis (d–g), 10-day-old seedlings grown on half-strength MS medium were sprayed water with 50 µM AA, 25 µg ml−1 Dox or 0.1% EtOH (mock), and with or without additional supplementation with 100 µM ACC (d) or 50 µM AgNO3 (e). Seedlings were left under standard growth conditions for 4 h prior analysis. Expression was determined by qRT-PCR and is relative to mock only treated WT. Data shown are the means + s.e.m. (n = 3). Asterisks denote a significant difference versus the same treatment without ACC (d), AgNO3 (e), or versus mock-treated WT (f,g). Student's t-test (*p < 0.05, **p < 0.01).
(d). Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using GSEA software [38]. The basic settings included: number of permutations = 1000; collapse dataset = false; scoring scheme = weighted; normalization mode = meandiv. Two gene sets of ethylene biosynthesis and response to ethylene stimulus were derived from the AmiGO website (http://amigo.geneontology.org/amigo). The nac017 retrograde signalling gene set was defined in this work based on the microarray experiment from [22] (GSE41136) as genes upregulated at least two-fold on AA in WT and less induced in both rao2-1 and nac017-1 mutants at least 1.5-fold, and are listed in the electronic supplementary material, table S2. The expression input data were from RNA-seq experiment of [14] (GSE78862) or from AA response in WT (GSE41136).
3. Results
(a). Antimycin A and doxycycline treatments target a common group of mitochondrial retrograde signalling genes
A previous study by Wang & Auwerx [14] identified a novel plant UPRmt response related to chemical or genetic perturbations of mitochondrial translation. In the same work, the key role for various transcription factors in mediating UPRmt retrograde signalling was hypothesized. To further investigate the plant UPRmt response, we compared UPRmt-related full transcriptome profiles for WT (Col-0) plants treated with 25 µg ml−1 Dox for 12 h, and the mrpl1 mutant (GSE78862) with seedlings treated with 50 µM AA for 6 h (GSE41136) (figure 1a,b; electronic supplementary material, figure S1). Because these experiments were originally performed using two different approaches (22 K Affymetrix ATH1 microarray or RNA-seq), only genes detected by both platforms were included. The full list of genes identified in this study as significantly changed after Dox treatment or in mrpl1 mutant is available in the electronic supplementary material, table S3. The Dox treatment resulted in 54 upregulated and 92 downregulated genes, while the mrpl1 mutants displayed 38 up- and 29-downregulated genes. AA treatment to WT seedlings had a very strong effect on the Arabidopsis transcriptome, with 1214 genes upregulated and 746 genes downregulated (figure 1a). Twenty-nine of the 54 genes (54%) induced by the Dox treatment (figure 1a), and 17 of the 37 genes (46%) induced in the mrpl1 mutant (electronic supplementary material, figure S1), were also induced by AA. This is a much higher proportion than expected by chance (p < 0.001). However, no overlap for the downregulated genes between AA and Dox, nor mrpl1, was seen (figure 1a; electronic supplementary material, figure S1). Hierarchical clustering confirmed the similar expression of transcripts induced after AA or Dox treatments, and highlighted that while Dox triggered a very similar promotive transcriptomic response, it was also much less pronounced when compared with AA (figure 1b). The heatmap analysis additionally showed that Dox and AA trigger largely different responses for the downregulated genes. This suggests that the signalling pathways induced by the block of OXPHOS components and the UPRmt are partially similar in Arabidopsis.
Interestingly, we further noted that the similarity between Dox and AA was mostly for the transcripts identified before as specific mitochondrial stress-responsive genes [25]. Of the 29 genes induced by both AA and Dox in WT (figure 1a), 22 (76%) were at least 1.5-fold less induced in AA-treated rao2-1 and nac017-1 mutants as compared to WT, including AOX1a, At12cys-2, UGT74E2, UPOX1, OM66, NDB2, sHSP23.5 and GSTU22. Six genes common for mrpl1 and AA (electronic supplementary material, figure S1) were also differently expressed in AA-treated nac017 mutants. Therefore, we compared the full transcriptomic responses to UPRmt (GSE78862) with nac017-dependent mitochondria signalling by performing a GSEA using a single cohort of genes, for which upregulation by AA is reduced in nac017 mutants when compared with WT, and referred here as ‘nac017 retrograde signalling’ (electronic supplementary material, table S2). Expression of ‘nac017 retrograde signalling’ genes was significantly enriched in WT seedlings treated with Dox (figure 1c), suggesting the possible role of ANAC017 in mediating at least part of the plant UPRmt signalling.
(b). ANAC017 is a major regulator of mitochondrial proteotoxic stress responses
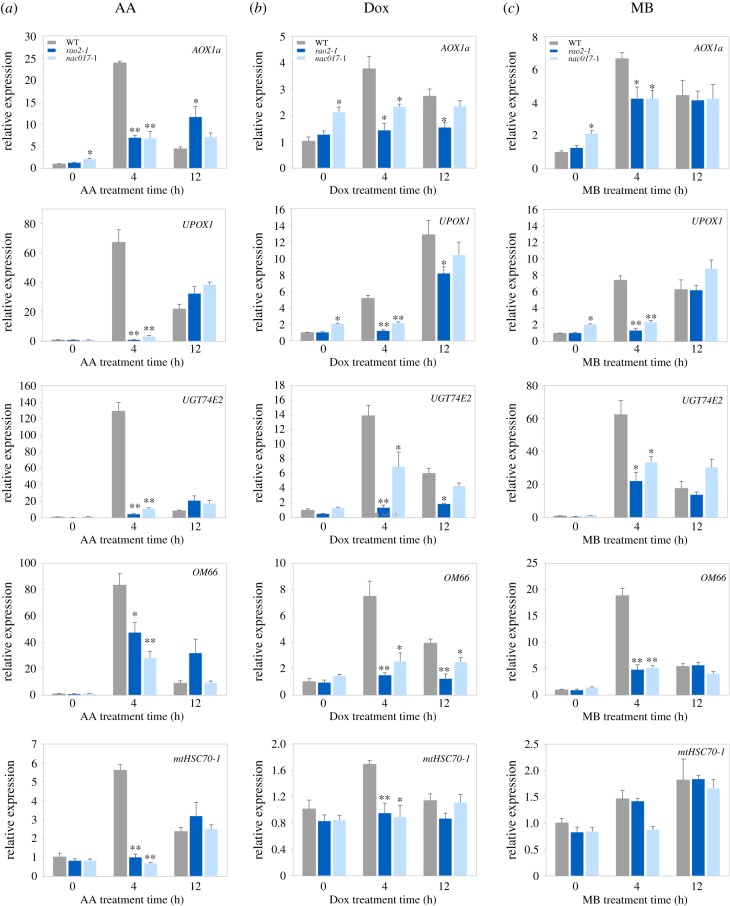
We examined whether ANAC017 signalling is required for plant UPRmt responses by assessing the expression of selected mitochondrial stress marker genes by qRT-PCR in two nac017 alleles across different treatments that perturb mitochondrial homeostasis and have classically been used to induce UPRmt in a range of systems (figure 2; electronic supplementary material, figures S2–S4) [14,19]. We first investigated expression of five nuclear-encoded genes (AOX1a, UPOX1, UGT74E2, OM66 and mtHSC70-1) which are known ANAC017-regulated genes during mitochondrial retrograde signalling (electronic supplementary material, table S2; [25]), and were also identified in this study as commonly upregulated by AA and Dox (figure 1a). As shown in figure 2a, expression of all five genes was very strongly upregulated in WT plants sprayed with 50 µM AA, with significant and consistent reduction in expression observed in both rao2-1 and nac017-1 mutants within 4 h after the treatment. At 12 h after the AA treatment, induction of all genes declined in WT, but was no further repressed in any of the nac017 alleles. Such a time-dependency for the nac017 response to AA treatment has been demonstrated previously [24] and is probably caused by activation of secondary NAC transcription factors such as ANAC013. All five mitochondrial stress marker genes were also induced by Dox in Col-0, but significantly less in rao2-1 and nac017-1 when compared with WT (figure 1b). It is worth noticing that Dox treatment activated expression of AOX1a, UPOX1, UGT74E2, OM66 and mtHSC70-1 in WT to a much lower extent than the AA treatment, which is in agreement with our genome-wide transcriptome analysis (figure 1b). Spraying with 100 µg ml−1 chloramphenicol (CAP), another inhibitor of organellar translation, showed almost the same results as Dox treatment, but the CAP treatment was overall weaker. Also here, of the two nac017 alleles, rao2-1 showed statistically significant repression of marker gene expression (electronic supplementary material, figure S3a). Treatment with MitoBloCK-6 (MB), which impairs mitochondrial protein import [39] and induces UPRmt in plants [14], also activated the same ANAC017-dependent response as AA and Dox treatments (figure 2c). A strong induction of AOX1a, UPOX1, UGT74E2 and OM66 in WT, 4 h after the MB treatment was significantly suppressed in both nac017 alleles, but not 12 h after the MB treatment (figure 2c). However, MB did not clearly induce the expression of the mitochondrial HSP70 gene (mtHSC70-1) in WT, and therefore, we also did not detect significant repression of this gene by nac017 mutation (figure 2c). The last inducer of UPRmt we tested was FCCP that alters organellar membrane potential (electronic supplementary material, figure S3b). FCCP also activated ANAC017-dependent signalling, as seen by suppressed induction of AOX1a, UPOX1, UGT74E2 and OM66, and rescued inhibition of mtHSC70-1 in two nac017 alleles when compared with WT (electronic supplementary material, figure S3b). Generally, we did not observe strong changes in expression for any of the genes tested above under the control (mock) treatment conditions at any time point, with the exception of UPOX1, for which expression was slightly, but statistically significantly, elevated in nac017-1 when compared with WT (electronic supplementary material, figure S2). To ensure the applied stress treatments did not induce significant chloroplast unfolded stress responses (UPRcp, [40,41]), we analysed the expression of several UPRcp marker genes. Neither Dox, CAP or MB induced the UPRcp genes in our own qRT-PCR experiments, or published whole transcriptomes (electronic supplementary material, figure S5), indicating that the treatments are predominantly targeting the mitochondria. Overall, these results support the conclusion that ANAC017 is a master regulator of signalling responses to mitochondrial proteotoxic stresses activated by various compounds in plants.
Figure 2.
ANAC017 regulates mitochondrial stress responses. WT, rao2-1 and nac017-1 seedlings were grown on half-strength MS medium. Seedlings were sprayed with: (a) 50 µM antimycin A (AA), (b) 25 µg ml−1 doxycycline (Dox) or (c) 50 µM MitoBloCK-6 (MB) and left for 4 and 12 h under white light. Gene expression was determined by qRT-PCR and is relative to untreated WT (0 h) (n = 3). Asterisks denote a significant difference versus WT for the same time point, Student's t-test (*p < 0.05, **p < 0.01).
We also wanted to test whether there is a specific UPRmt response in plants that is different from ‘classic’ MRR signalling, and whether it also shows the ANAC017-dependency. Based on the transcriptomic analysis, we chose three genes (ULP1B, GDS2-like and DIN2) that were induced by Dox and two genes (XTH21 and RHS15) that were repressed by Dox, but not induced after AA treatment in WT and non-differently expressed in nac017 mutants. We then examined their expression by qRT-PCR under the same AA, Dox or MB conditions as described above (electronic supplementary material, figure S4). Our qRT-PCR results were largely inconsistent with the previously published whole-transcriptome study, as reflected by lack, or low induction of ULP1B, GDS2-like and DIN2 in WT treated with Dox (electronic supplementary material, figure S4b), as well elevated DIN2 expression in AA-treated WT. Only two genes XTH21 and RHS15 were repressed in WT consistently across three treatments and under both time points, but only XTH21 expression showed ANAC017-dependence as evidenced by more pronounced XTH21 repression in both nac017 alleles 12 h after AA, or Dox treatments (electronic supplementary material, figure S4a,b). Therefore, we were unable to define additional UPRmt signalling responses that are clearly independent of ANAC017.
(c). ANAC017 regulates physiological alterations to mitochondrial protein imbalance stresses
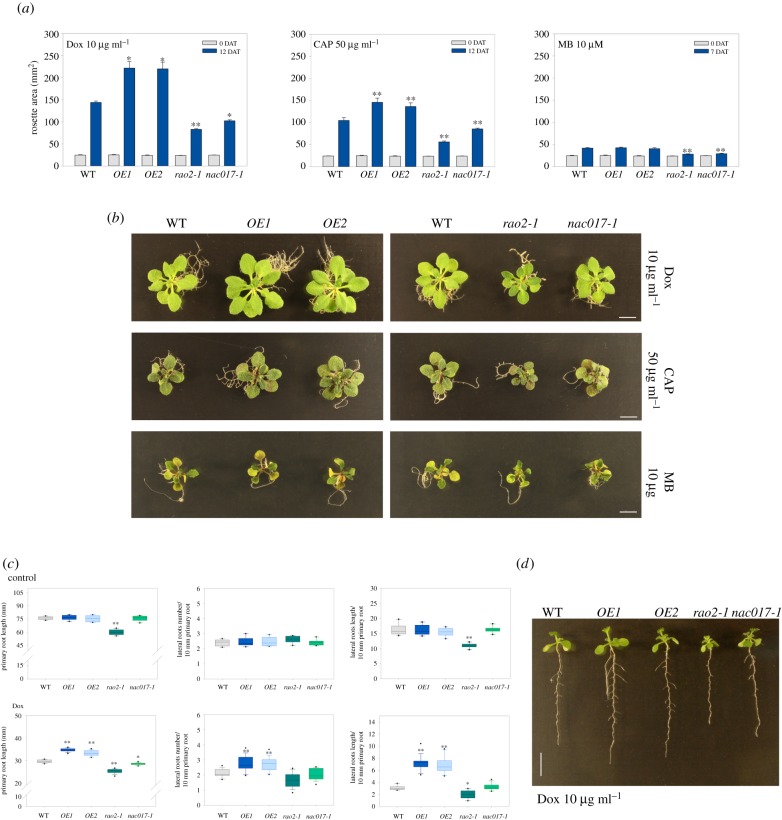
To determine whether the ability of ANAC017 to control nuclear gene expression has physiological significance in plant UPRmt, we treated nac017 mutants and ANAC017 overexpressor lines with chemical agents that disturb mitochondrial proteostasis. ANAC017 OE1 plants look similar to WT, while ANAC017 OE2 plants had an overall smaller rosette size, as previously reported [24,42]. This appears to be correlated with the ANAC017 expression levels of the plants at this age (electronic supplementary material, figure S6). No difference in root growth was observed in either overexpression line compared to Col-0. We first transferred 10-day-old seedlings onto half-strength MS agar plates supplemented with 10 µg ml−1 Dox, 50 µg ml−1 CAP or 10 µM MB and allowed them to grow under standard photoperiodic conditions for 12 days (Dox, CAP), or 7 days (MB) (figure 3a,b). Consistent with previous reports, all agents inhibited plant growth [14], with MB having the most severe effect (figure 3a,b). When mitochondrial translation was blocked by Dox or CAP, both ANAC017 overexpressor lines (OE1 and OE2) showed increased rosette area size (ca 50–60%), while rao2-1 and nac017-1 mutants showed significantly more pronounced growth retardation (ca 7–43%) when compared with the WT. In a parallel experiment, both nac017 mutants also showed a hypersensitive response to MB, but no significant difference between WT and ANAC017 OE1/OE2 rosette sizes were observed (figure 3a,b). The inhibitory effect of Dox on lateral roots in Arabidopsis is well documented [14,19], so we tested whether this response is also controlled by ANAC017. In agreement with previous reports, Dox reduced primary root length and lateral root length in WT seedlings transferred to Dox for 7 days (figure 3c). Under these conditions, both OE lines showed statistically significant higher length of primary and lateral rootss and two nac017 mutants had significantly shorter root length than the WT (figure 3c,d). However, the rao2-1 allele displayed shorter root length already in control experimental conditions (figure 3c), as observed previously [24]. We did not observe significant effects of Dox on lateral root number in WT or nac017 mutants, but it was slightly increased in both OE lines grown on Dox (figure 3c). Together, these results are supportive of an important role of ANAC017 in controlling plant responses to mitochondrial proteotoxic stress.
Figure 3.
ANAC017 regulates Arabidopsis growth during mitochondrial proteotoxic stress. (a) Rosette areas of WT, ANAC017 overexpressor lines (OE1, OE2) and nac017 mutants (rao2-1, nac017-1) pre-grown for 10 days on half-strength MS medium, and transferred for an additional 12 days on medium supplemented with 10 µg ml−1 doxycycline (Dox) or 50 µg ml−1 chloramphenicol (CAP), or for 7 days on medium supplemented with 10 µM MitoBloCK-6 (MB) (n = 3 × 15). Data shown are the means + s.e.m. Asterisks denote a significant difference versus WT before transfer or 7–12 days after transfer to plates with translation inhibitors (one-way ANOVA and Tukey HSD post hoc test, *p < 0.05, **p < 0.01). (b) Phenotypes of 17–22-day-old plants analysed in (a). Scale bar indicates 6 mm. (c) Root phenotypes of WT, OE1, OE2, rao2-1 and nac017-1. Five-day-old plants were transferred on medium, with (lower panel) or without (top panel) 10 µg ml−1 Dox and left to grow vertically for 7 days. Horizontal lines in the box plots indicate the median and the 5th/95th percentiles of outliers are indicated as black dots. Data shown are the means ± s.e.m. (n = 9–15 × 3). Asterisks denote a significant difference versus WT (one-way ANOVA and Tukey HSD post hoc test, *p < 0.05, **p < 0.01). (d) Phenotypes of 12-day-old Dox-treated plants analysed in (c). Scale bar indicates 7 mm.
(d). Ethylene signalling interacts with mitochondria retrograde singling
Previous reports have highlighted the potential importance of phytohormone signalling during mitochondrial stress [14,43,44]. An increase in ethylene biosynthesis, expression of ethylene marker genes and genes encoding mitochondrial ribosomal proteins was previously observed during UPRmt. This suggested a possibly important role for ethylene signalling in the control of UPRmt in plants [14]. Therefore, we tested whether ANAC017-mediated MRR also requires anterograde feedback from ethylene signalling. As shown in figure 4a,b, GSEA using AA treatment to WT array dataset evidenced significant over-representation of Gene Ontology categories involved in ethylene biosynthesis and response to ethylene stimulus. This suggests that the involvement of ethylene in mitochondrial signalling might not be restricted to UPRmt, but is a common factor controlling MRR. Analysis of the microarray dataset for AA treatment (GSE41136) showed that many ethylene biosynthesis and signalling genes are induced in WT 6 h after AA treatment, similarly to what has been reported before for Dox treatment and mrpl1 mutation [14] (figure 4c). However, expression of these genes was induced in two nac017 alleles mostly to the same level as in the WT (figure 4c), arguing that the ethylene-related genes are not part of ANAC017-dependent mitochondria retrograde regulation. To gain more understanding on the interaction between ANAC017 and ethylene, we studied phenotypes of nac017 mutants and ANAC017 overexpressor lines in 4 days dark grown seedlings fed with AA and silver nitrate (AgNO3), a well-established antagonist of ethylene signalling (electronic supplementary material, figure S7a,b). AA reduced hypocotyl length in a dose-dependent manner in WT, and had much more pronounced inhibitory effect in rao2-1 and nac017-1 mutants, while hypocotyls of ANAC017 OE1 and OE2 lines on AA were significantly longer than the WT (electronic supplementary material, figure S7a,b). In support of a role of ethylene in mitochondrial retrograde signalling, blocking of ethylene signalling by AgNO3 partially restored hypocotyl inhibition by AA, and this rescue was statistically significant in all tested genotypes (electronic supplementary material, figure S7a). This result also suggests that there is no requirement for ANAC017 signalling to mediate ethylene responses. In agreement with the AgNO3 impact on the AA response in the dark, ethylene overproducing mutant eto1-1 showed significantly shorter hypocotyls, while ethylene-insensitive mutant ein2-1 showed significantly longer hypocotyls, when compared with the WT when fed with AA in the dark for 4 days (electronic supplementary material, figure S6d).
To examine whether the ethylene impact on physiological responses to AA is also important for the transcriptomic responses, we analysed expression of five mitochondrial stress-responsive marker genes in response to AA and Dox in the presence or absence of ACC, an ethylene precursor (figure 4d). Incubation with ACC alone did not induce MRR marker genes (only a weak induction was observed in UGT74E2), suggesting that ethylene cannot induce MRR responses on its own. Co-incubation with ACC resulted in enhanced responses to AA for several MRR markers, as observed previously [44]. Conversely, blocking ethylene signalling by addition of AgNO3 partially inhibited MRR gene induction by AA (figure 4e). This stimulatory effect of ethylene could be confirmed genetically, as the ethylene overproducing mutant eto1-1 also showed stronger AA-induction of MRR genes (figure 4f). Interestingly, the addition of ACC partially repressed Dox-induced gene expression, suggesting that there might be a complex interplay between ethylene signalling and various mitochondrial stress signalling pathways.
The mitogen activated protein kinase MPK6 was shown to have increased phosphorylation in response to a wide range of mitochondrial stress inducers [14] and is a known stimulator of ethylene signalling and biosynthesis [45]. Therefore, it was suggested to play a central role in UPRmt signalling in plants [14]. To test a potential involvement of MPK6 in mitochondrial stress-induced signalling of ANAC017 target genes, two alleles of mpk6 were exposed to AA or Dox for 6 h (figure 4g). Overall, no consistent effects were observed in both mpk6 alleles that would suggest a positive role of MPK6 in the induction of ANAC017-regulated genes under mitochondrial stress conditions. Together these results are supportive for a promoting role of ethylene in mediating responses to mitochondrial stresses in an ANAC017-independent way, but ethylene itself is not sufficient for inducing MRR-target gene expression.
4. Discussion
While mitochondria-to-nucleus signalling in plants has originally been demonstrated by blocking OXPHOS protein complexes [46], components of retrograde communication in animals were identified through inhibiting a wide range of mitochondrial processes, which all lead to organellar proteotoxic stress [3,47]. The current models in plants suggest that mitochondrial signals trigger the proteolytic cleavage of ANAC017 from the ER, leading to altered nuclear gene expression [28]. By analysing the transcriptomic and physiological responses to five different mitochondrial stressors, which interfere with mitochondrial translation, protein import, membrane potential and OXPHOS, we show here that ANAC017 plays a role in coordinating the general mitochondrial retrograde response. Because most of these treatments are known to result in mitochondrial protein imbalance and proteotoxic stress [14,19], we propose that ANAC017 is also a key regulator of the UPRmt in plants.
In this work, we were able to define common gene expression signatures shared between two mitochondrial stresses, AA and Dox (figure 1), which were consistent with the previously defined ANAC017-dependent regulon (electronic supplementary material, table S2; [25]). Interestingly, many ANAC017 target genes are also the classic targets of UPRmt in animals, which function in various physiological processes summarized by Melber & Haynes [47]. These include genes for mitochondrial protein homeostasis and import, including chaperone proteins like heat shock proteins (sHSP23.5, mtHSC70-1), a GrpE family protein MITOCHONDRIAL GRPE 1 (MG1), AtOM66 (BCS1), which has a possible AAA protease function, and mitochondrial import protein TIM17-1. Under ANAC017 control are also genes for OXPHOS protein components like AOX1a, NDB2, NDB4 and STOMATIN-LIKE 1/2 protein; proteins that regulate metabolism like ALANINE AMINOTRANSFERASE 2 (ALAAT2), ASPARTATE AMINOTRANSFERASE 2 (ASP2), 2,3-BIPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE (IPGAM2); genes for mitochondrial dynamics proteins like OPTIC ATROPHY 3 (OPA3) and finally proteins with a role in ROS reduction and detoxification like AOX1a, Multidrug And Toxin Efflux carriers (At2g04050), glutathione S-transferase TAU 25 and SULPHOTRANSFERASE 12 (SOT12). We therefore suggest that the classical (ANAC017-dependent) mitochondrial retrograde response underlies—and is probably identical to—the UPRmt in plants. In agreement, we could not find consistent evidence for genes that are specifically induced by, for example, Dox but not AA (electronic supplementary material, figure S4). Interestingly, the set of downregulated genes are partially overlapping between AA and Dox (electronic supplementary material, figure S4), but also contain many distinct components (figure 1). The importance of ANAC017 in control of proteotoxic stress responses in plants is strongly supported by its ability to improve growth resistance to a wide range of mitochondrial dysfunctions induced by AA, Dox, CAP or MB that are also model systems for UPRmt in animals [12,13,19,47]. Many of the classic inducers of UPRmt (FCCP, CAP, Dox) used in non-plant systems could potentially affect chloroplast function in plants. Our analysis, however, suggests that plant UPRcp genes are not induced by these treatments, indicating that the effects are predominantly targeting mitochondria also in plants. To what extent, it is the imbalanced accumulation of mitochondrial proteins caused by the inhibition of mitochondrial translation or import itself, or alternatively the indirect effects this would have on the OXPHOS chain, that triggers the nuclear transcriptional responses, also remains unclear.
Despite our understanding of the downstream targets of ANAC017-retrograde signalling, the events that initiate its release from the ER are indeed still unknown. One possible hypothesis suggests hydrogen peroxide (H2O2) generated during mitochondrial stresses could induce ANAC017 cleavage. In both animals and plants, treatments that block OXPHOS complexes, mitochondrial translation, protein import or proton gradient were all shown to induce ROS (e.g. superoxide and H2O2) production [14,17,22]. Another possibility is that in plants, UPRmt involves induction of the UPRER [48]. However, based on the array dataset for AA treatment to WT (GSE41136), we do not see induction of the classic UPRER marker genes [49] including IRE1A, IRE1B, TMS1, NAC089. Some of these genes are even repressed by AA, e.g. PDI6, PDI9, BIP1, CRT1 or SDF2. Therefore, it is possible that the UPRmt in plants requires the UPRER to be maintained at a low level. Moreover, UPRmt in plants seems directed specifically towards the induction of mitochondrial heat shock protein genes, like mtHSC70-1 and sHSP23.5, rather than the chloroplastic (cpHsc70-1, cpHsc70-1), the cytosolic (e.g. HSP70T-1, HSC70-2, HSP70-15) or the ER (BIP1, BIP3) counterparts [14]. Despite the apparent physical separation between mitochondria and ER, ANAC017 release from the ER may be facilitated at contact sites between the mitochondria and the ER, which have been demonstrated also in plants [50].
Finally, it appears that the UPRmt is integrated with hormonal signalling in plants [14]. Previous work showed that auxin signalling is antagonistic to retrograde signalling, keeping check on the growth versus stress response balance [43,44]. Also ethylene was pointed out to play a role during UPRmt [14]. Here, we confirm that ethylene signalling plays a more general role in retrograde signalling. Its precise role and how it is controlled is not entirely clear yet, but our data suggest that ethylene is required for a fully extended retrograde response at a transcriptional and physiological level, indicating a promoting effect. Blocking ethylene signalling during UPRmt/retrograde signalling partially rescues hypocotyl extension in the dark. This suggests that the reduced growth caused by mitochondrial inhibition is not merely a result of energetic inhibition of the plants, but at least in part a regulated phenomenon with input from ethylene signalling. One study identified the mitochondrial prohibitin atphb3 mutant in a screen for lines with enhanced sensitivity to ethylene [51]. It was shown that atphb3 mutants show strongly inhibited hypocotyl elongation under ethylene treatment. By contrast, suppression of ethylene signalling with AgNO3 partially rescued the shorter hypocotyl phenotype of the atphb3 mutant, also indicating that the reduced growth of the mutant is partially controlled by ethylene signalling. The atphb3 mutant has constitutively induced ANAC017-dependent retrograde signalling [25,52] and produces higher levels of ethylene [51]. Treatment with Dox, CAP or MB also activates ethylene-responsive genes and promoter constructs, indicating ethylene production is probably increased [14]. It thus seems that mitochondrial inhibition induces ethylene production and sensitizes the plants to ethylene effects. The induction of ethylene-responsive genes seems unaffected in anac017 mutants (figure 4), suggesting the ethylene production and/or signalling are ANAC017-independent. In conclusion, there are at least three signalling pathways active during UPRmt/retrograde signalling: ANAC017-dependent induction of gene expression, ethylene signalling that enhances the growth reduction and gene expression induction (but operates outside of ANAC017 itself) and auxin signalling that represses retrograde signalling and helps restore the balance towards growth.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
The NGS/microarray data used are a reanalysis of previously published, publicly available data (accession numbers provided in the paper). Reanalysed data are provided in the electronic supplementary material, tables.
Authors' contributions
S.M.K. participated in the design of the study, carried out laboratory work, participated in data analysis, conceived experiments and wrote the manuscript; A.D. carried out laboratory work and critically revised the manuscript; O.V.A. conceived and designed the study, participated in data analysis and participated in writing the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
S.M.K. was supported by a fellowship from the Carl Trygger Foundation (grant no. CTS 17: 487). O.V.A. was supported by the Swedish Research Council (Vetenskapsrådet 2017-03854), the Australian Research Council (grant no. DP160103573), Crafoord Foundation (grant no. 20170862), Carl Trygger Foundation (grant no. CTS 17: 487) and Carl Tesdorpf Stiftelse.
References
- 1.Ryan MT, Hoogenraad NJ. 2007. Mitochondrial–nuclear communications. Annu. Rev. Biochem. 76, 701–722. ( 10.1146/annurev.biochem.76.052305.091720) [DOI] [PubMed] [Google Scholar]
- 2.Kleine T, Leister D. 2016. Retrograde signalling: organelles go networking. Biochim. Biophys. Acta. 1857, 1313–1325. ( 10.1016/j.bbabio.2016.03.017) [DOI] [PubMed] [Google Scholar]
- 3.Quirós PM, Mottis A, Auwerx J. 2016. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226. ( 10.1038/nrm.2016.23) [DOI] [PubMed] [Google Scholar]
- 4.Jovaisaite V, Mouchiroud L, Auwerx J. 2014. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 217, 137–143. ( 10.1242/jeb.090738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moehle EA, Shen K, Dillin A. 2018. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J. Biol. Chem. 294, 5396–5407. ( 10.1074/jbc.TM117.000893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker BM, Haynes CM. 2011. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem. Sci. 36, 254–261. ( 10.1016/j.tibs.2011.01.004) [DOI] [PubMed] [Google Scholar]
- 7.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. 2006. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–239. ( 10.1534/genetics.106.061580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol. Cell 58, 123–133. ( 10.1016/j.molcel.2015.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. ( 10.1126/science.1223560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papa L, Germain D. 2014. SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell. Biol. 34, 699–710. ( 10.1128/MCB.01337-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Samuel BS, Breen PC, Ruvkun G. 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410. ( 10.1038/nature13204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durieux J, Wolff S, Dillin A. 2011. The cell-non-autonomous nature of electron transport chain mediated longevity. Cell 144, 79–91. ( 10.1016/j.cell.2010.12.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houtkooper RH, et al. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. ( 10.1038/nature12188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Auwerx J. 2017. Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell. 68, 540–551. ( 10.1016/j.molcel.2017.10.006) [DOI] [PubMed] [Google Scholar]
- 15.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480. ( 10.1016/j.devcel.2007.07.016) [DOI] [PubMed] [Google Scholar]
- 16.Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26, 2037–2043. ( 10.1016/j.cub.2016.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quirós PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Auwerx J. 2017. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045. ( 10.1083/jcb.201702058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horibe T, Hoogenraad NJ. 2007. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE 2, e835 ( 10.1371/journal.pone.0000835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moullan N, et al. 2015. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691. ( 10.1016/j.celrep.2015.02.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dojcinovic D, Krosting J, Harris AJ, Wagner DJ, Rhoads DM. 2005. Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol. Biol. 58, 159–175. ( 10.1007/s11103-005-5390-1) [DOI] [PubMed] [Google Scholar]
- 21.Schwarzlander M, Konig AC, Sweetlove LJ, Finkemeier I. 2012. The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 63, 1735–1750. ( 10.1093/jxb/err374) [DOI] [PubMed] [Google Scholar]
- 22.Ng S, et al. 2013b. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signalling in Arabidopsis. Plant Cell 25, 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umbach AL, et al. 2012. Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS ONE 7, e44339 ( 10.1371/journal.pone.0044339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, Whelan J. 2016. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 171, 2150–2165. ( 10.1104/pp.16.00273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Aken O, Ford E, Lister R, Huang S, Millar AH. 2016. Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 88, 542–558. ( 10.1111/tpj.13276) [DOI] [PubMed] [Google Scholar]
- 26.Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J. 2009. Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2, 1310–1324. ( 10.1093/mp/ssp053) [DOI] [PubMed] [Google Scholar]
- 27.Ng S, et al. 2013. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 288, 3449–3459. ( 10.1074/jbc.M112.416727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J. 2014. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 7, 1075–1093. ( 10.1093/mp/ssu037) [DOI] [PubMed] [Google Scholar]
- 29.De Clercq I, et al. 2013. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490. ( 10.1105/tpc.113.117168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanderauwera S, et al. 2012. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 20 113–20 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Aken O, Zhang B, Law S, Narsai R, Whelan J. 2013. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162, 254–271. ( 10.1104/pp.113.215996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzman P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 ( 10.1186/gb-2004-5-10-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson CL, Miller CJ. 2005. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics 21, 3683–3685. ( 10.1093/bioinformatics/bti605) [DOI] [PubMed] [Google Scholar]
- 35.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. ( 10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (eds Gentleman R, Carey V, Dudoit S, Irizarry R, Huber H), pp. 397–420. New York, NY: Springer. [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15 545–15 550. ( 10.1073/pnas.0506580102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dabir DV, et al. 2013. A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev. Cell 25, 81–92. ( 10.1016/j.devcel.2013.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dogra V, Duan J, Lee KP, Kim C. 2019. Impaired PSII proteostasis triggers a UPR-like response in the var2 mutant of Arabidopsis. J. Exp. Bot. 70, 3075–3088. ( 10.1093/jxb/erz151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llamas E, Pulido P, Rodriguez-Concepcion M. 2017. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet. 13, e1007022 ( 10.1371/journal.pgen.1007022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng X, et al. 2019. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 180, 634–653. ( 10.1104/pp.18.01603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F. 2014. Mitochondrial perturbation negatively affects auxin signalling. Mol. Plant 7, 1138–1150. ( 10.1093/mp/ssu071) [DOI] [PubMed] [Google Scholar]
- 44.Ivanova A, et al. 2014. A functional antagonistic relationship between auxin and mitochondrial retrograde signalling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiol. 165, 1233–1254. ( 10.1104/pp.114.237495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn A, Harter K. 2009. Mitogen-activated protein kinase cascades and ethylene: signaling, biosynthesis, or both? Plant Physiol. 149, 1207–1210. ( 10.1104/pp.108.132241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanlerberghe GC, McIntosh L. 1994. Mitochondrial electron transport regulation of nuclear gene expression. Studies with the alternative oxidase gene of tobacco. Plant Physiol. 105, 867–874. ( 10.1104/pp.105.3.867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melber A, Haynes CM. 2018. UPRmt regulation and output: a stress response mediated by mitochondrial–nuclear communication. Cell Res. 28, 281–295. ( 10.1038/cr.2018.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howell SH. 2013. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64, 477–499. ( 10.1146/annurev-arplant-050312-120053) [DOI] [PubMed] [Google Scholar]
- 49.Kim JS, Yamaguchi-Shinozaki K, Shinozaki K. 2018. ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176, 2221–2230. ( 10.1104/pp.17.01414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller SJ, Reski R. 2015. Mitochondrial dynamics and the ER: the plant perspective. Front. Cell Dev. Biol. 3, 78 ( 10.3389/fcell.2015.00078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christians MJ, Larsen PB. 2007. Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J. Exp. Bot. 58, 2237–2248. ( 10.1093/jxb/erm086) [DOI] [PubMed] [Google Scholar]
- 52.Van Aken O, et al. 2007. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52, 850–864. ( 10.1111/j.1365-313X.2007.03276.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS/microarray data used are a reanalysis of previously published, publicly available data (accession numbers provided in the paper). Reanalysed data are provided in the electronic supplementary material, tables.