Abstract
Changes in the functional state of mitochondria have profound effects on other cellular compartments. Genome-wide expression analysis of Arabidopsis rps10 mutants with an RNAi-silenced expression of mitoribosomal S10 protein has revealed extensive transcriptional reprogramming. A meta-analysis comparing expression datasets of 25 mitochondrial perturbations showed a high similarity of the aox1a:rpoTmp mutant, which is defective in the alternative oxidase (AOX1a) and dual-targeted mitochondrial and plastid RNA polymerase (RPOTmp), to rps10. Both rps10 and aox1a:rpoTmp showed a significantly decreased electron flux through both the cytochrome and the alternative respiratory pathways, and a markedly decreased the expression of nuclear-encoded components of the chloroplast transcription machinery. In line with this, a decreased level of plastid transcripts was observed in rps10 and aox1a:rpoTmp, which was reflected in a reduced rate of chloroplast transcription. Chemical treatment of wild-type seedlings with respiratory inhibitors showed that only simultaneous and direct inhibition of complex IV and AOX activity decreased the level of plastid transcripts. Taken together, both chemical and genetic studies show that the limitation of the activity of two mitochondrial terminal oxidases, complex IV and AOX, negatively impacts chloroplast transcription. Salicylic acid and oxygen are discussed as putative mediators of the signalling pathway between mitochondria, nucleus and chloroplasts.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: alternative oxidase, complex IV, chloroplast transcription, NEP and PEP, Arabidopsis thaliana, low oxygen signalling
1. Introduction
Plant cells harbour two types of energy organelles, mitochondria and chloroplasts. Despite the fact that both these organelles have their own genomes, the majority of organellar proteins are nuclear encoded. Thus, to maintain organelle functions and to coordinate the expression of organellar and nuclear genes specific mechanisms have evolved, classified as anterograde and retrograde control. The anterograde control involves the regulation of organellar gene expression by nuclear-encoded proteins, whereas retrograde control involves signals originating from the organelles causing nuclear transcriptional reprogramming [1]. Moreover, additional interactions exist between mitochondria and chloroplasts involving either a retrograde signal from one of the organelles modulating the nuclear anterograde control of the other one [2,3], or simultaneous retrograde signals from both organelles that together affect nuclear gene expression [4]. Apart from the complex retrograde signalling pathways with the participation of the nucleus, there is still no convincing evidence for direct signalling between mitochondria and chloroplasts. So far, a variety of operational retrograde signalling pathways have been described that are triggered by reactive oxygen species, sugars, photosynthetic redox imbalance, tetrapyrrole intermediates and STN7-mediated signalling [5]. Moreover, several proteins mediating the response of nuclear genes to organellar dysfunction have also been identified: ABI4 (abscisic acid (ABA)-insensitive-4), cyclin-dependent kinase E1 (CDKE1/RAO1), a group of endoplasmic reticulum-bound transcription factors headed by ANAC017, RCD1 (radical-induced cell death1) and AtWRKY15/40/63 [6–14].
Mitochondria and chloroplasts are tightly linked by the exchange of metabolites and energy equivalents. Photosynthesis provides substrates for mitochondrial respiration, whereas the products of mitochondrial oxidative metabolism, particularly CO2 and ATP, can be used for photosynthetic carbon assimilation [15]. The reducing power flows from chloroplasts to mitochondria via several pathways including photorespiration [16], malate shuttles [17] and transport of photoassimilate-derived carbon-rich metabolites. Mitochondria dissipate the redox equivalents from chloroplasts, which protects them from photoinhibition [18]. Thus, the respiratory and photosynthetic electron transport chains in mitochondria and chloroplasts, respectively, act in a coordinated manner to optimize the cellular energy metabolism.
The mitochondrial electron transport chain is composed of several protein complexes that transfer electrons. In plants, it is branched into two pathways: the cyanide-sensitive cytochrome pathway (terminating at cytochrome c oxidase (COX, complex IV)) and the cyanide-resistant alternative pathway (terminating at alternative oxidase (AOX)) [19]. Both pathways obtain electrons from reduced ubiquinone, which receives them from the preceding components of the respiratory chain––NADH dehydrogenase (complex I), alternative NAD(P)H dehydrogenases, and succinate dehydrogenase (complex II). In the cytochrome pathway electrons flow from reduced ubiquinone to molecular oxygen through the cytochrome bc1 complex (complex III) and finally complex IV, generating an electrochemical proton gradient across the inner mitochondrial membrane that is then used for ATP production by ATP synthase in the process of oxidative phosphorylation (OXPHOS). The electron flow through the alternative pathway is non-phosphorylating as it bypasses the two proton-translocating complexes, III and IV, and the energy is dissipated as heat [20].
Several studies have demonstrated that changes in the functioning of the mitochondrial respiratory pathways may exert diverse effects on chloroplasts. Apart from the well-known role of the alternative and cytochrome respiratory pathways in photosynthesis optimization [21–23], it has been suggested that changes in the activity of AOX and the activity of cytochrome respiratory pathway also influence the chloroplast transcription rate [24]. Chloroplast genes are transcribed by at least three RNA polymerases: one bacterial-type plastid-encoded polymerase (PEP), composed of four basic core subunits (rpoA, rpoB, rpoC1 and rpoC2) and two nuclear-encoded polymerases (NEP): RPOTp and RPOTmp (RNA polymerases of the T3/T7 type targeted to the chloroplast and to both mitochondria and chloroplasts, respectively) [25]. PEP is mainly responsible for transcribing photosynthesis-related genes, whereas the NEPs play an especially important role in the transcription of house-keeping genes, including the rpo genes of PEP [26]. As a consequence, the abundance and activity of PEP depends on the NEP activity. The functioning of PEP requires additional nuclear-encoded factors, like sigma factors and PEP-associated proteins (PAPs) [27], while no specificity factor for the NEP enzymes has been identified to date.
Here, we demonstrate that a simultaneous restriction of the mitochondrial electron transport chain at complex IV and at AOX decreases the rate of chloroplast transcription. This effect was found using both a genetic study with two independent mutants, rps10 and aox1a:rpoTmp with reduction of complex IV and AOX activity, as well as a chemical approach using wild-type plants treated with inhibitors specific for complex IV and AOX. We propose that the diminished chloroplast transcription in response to specific mitochondrial respiratory dysfunction is owing to a negative regulation of the chloroplast transcription apparatus, including regulation at the transcriptional level of the PEP, sigma factors as well as PAPs and regulation of the NEP at the post-transcriptional level.
2. Materials and methods
(a). Growth conditions of plants
The growth conditions of RNAi-silenced rps10 mutants have been described previously [28]. All analyses were performed using the youngest leaves harvested from 9- to 10-week-old P2 and P3 phenotypes of rps10 mutant and wild-type plants of the same age growing under short-day conditions (8 h light/16 h dark) at 22°C and 100 µmol m−2 s−1. The insertional double mutants aox1a:rpoTmp were obtained from Dr. James Whelan (La Trobe University, Australia) and were previously described in [29]. For aox1a:rpoTmp, leaves harvested from six-week-old mutant and four-week-old wild-type plants growing under long-day conditions (16 h light/8 h dark) at 22°C and 100 µmol m−2 s−1 were used. For treatment with mitochondrial respiratory inhibitors, two-week-old seedlings of wild-type and rao2-1 knockout mutants of ANAC017 grown on half-strength Murashige and Skoog medium under long-day conditions were used.
(b). rps10 microarray analysis
Analysis of the global changes in transcript abundance in rps10 mutant was performed using Affymetrix ATH1 microarray gene chips. Arrays were performed using total RNA extracted from leaves of each phenotype (WT, P2 and P3) in at least three biological replicates using the GeneMATRIX Universal RNA purification kit (EURx). cDNA was prepared with the one-cycle cDNA synthesis kit followed by biotin-labelling with the 3′IVT express kit (Affymetrix, Santa Clara, CA) and hybridized to the GeneChip Arabidopsis ATH1 genome Array. Raw data were imported into the Partek Genomic Suite v 6.6 software with the use of GC robust multiarray averaging (GCRMA). Probe intensity level data were preprocessed using the RMA analysis method with background adjustment, quantile normalization and log2 transformation. GCRMA-normalized gene expression values were analysed to identify differentially expressed genes (DEGs) by a regularized t test based on a Bayesian statistical framework using the software program Cyber-T [30]. Changes were considered significant at the false discovery rate correction level of posterior probability of differential expression (ppde). p > 0.95 and twofold change. Gene ontology analysis was performed using GOrilla [31].
(c). Meta-analysis of microarray datasets
For the meta-analysis in figure 1, selected GCRMA-normalized gene expression microarray datasets were processed using Cyber-T as previously described [1,32–34]. Additionally, processed RNA-seq datasets were included [11,35]. Changes were considered significant at the false discovery rate correction level of ppde. p > 0.95 and twofold change. A full list of the transcriptome datasets used can be found in electronic supplementary material table S1. To compare similarity in gene expression patterns between different datasets, pairwise DEG overlaps were calculated via the Sørensen-Dice similarity coefficient (DSC) with formula DSC(a,b) = 2nab/(na + nb). The DSC between two datasets thus varies between 0 and 1. The complete matrix of DSC values for the rps10 and 25 additional datasets was used for hierarchical clustering in TIGR Multi-experiment Viewer 4.9.0 using Pearson correlation coefficient and represented as an unrooted tree using FigTree v. 1.4.2.
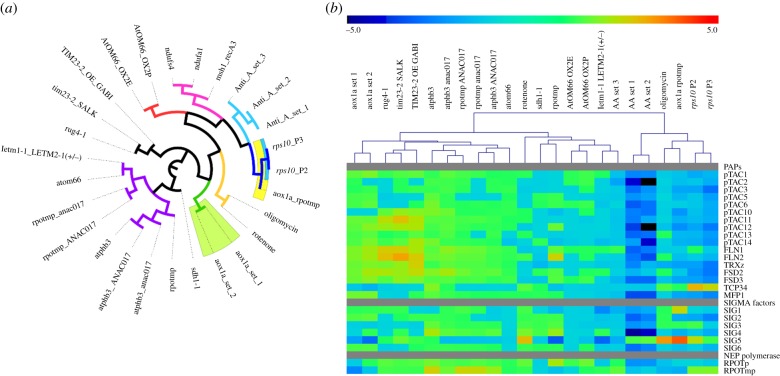
Figure 1.
Transcriptome comparison of the rps10 mutant with other mitochondrial perturbations in Arabidopsis thaliana. (a) An unrooted tree representing pairwise DEG overlaps between the datasets as calculated using the Sørensen-DSC. A complete matrix of DSC values for rps10 and 25 additional datasets was used for hierarchical clustering using Pearson correlation coefficient. Subbranches are colour coded for clarity. (b) Heat map representing gene expression of nuclear genes encoding chloroplast transcription machinery components in rps10 and 22 additional datasets representing different mitochondrial perturbations. Expression values were hierarchically clustered using Pearson correlation coefficient. Colour scale represents up (red) or down (blue) fold change values. See electronic supplementary material, table S1 for description of datasets analysed. (Online version in colour.)
(d). Real-time quantitative polymerase chain reaction and quantitative polymerase chain reaction analyses
Leaf tissue was used for DNA isolation with GeneMATRIX plant and fungi DNA purification kit (EURx). Leaf tissue or green parts of seedlings were used for RNA isolation with GeneMATRIX Universal RNA purification kit (EURx). Reverse transcription was performed using a reverse transcription kit (Applied Biosystems). The genomic DNA/cDNA was used as the template for real-time quantitative polymerase chain reaction (RT-qPCR) as described previously [28]. Relative expression was calculated using ACT2 (At3g18780) as the reference gene. The primers used are listed in electronic supplementary material, table S2a.
(e). Isolation of mitochondria and chloroplasts
Mitochondria were isolated from leaf tissue as described in [36]. Chloroplasts were isolated from leaf tissue using Chloroplast isolation kit (Sigma-Aldrich) according to the manufacturer's instruction.
(f). Respiratory activity measurements
The activity of complex IV and capacity of AOX were measured in mitochondria isolated from leaf tissue using a Clark-type oxygen electrode (Hansatech Instrument) as described in [37]. Oxygen consumption was recorded and respiration rates were calculated as nanomoles of O2 consumed per minute per milligram of mitochondrial protein.
(g). Alternative oxidase gel blot analyses
Mitochondrial proteins isolated from leaves of rps10 and wild-type plants were prepared with or without 100 mM DTT as a reductant in the sample buffer (reducing or non-reducing conditions, respectively), boiled for 5 min, and separated by SDS-PAGE on 12% polyacrylamide gels. After electrophoresis, proteins were transferred to a PVDF (polyvinylidene difluoride) membrane. Blots were probed with polyclonal rabbit antibodies against AOX1/2 (AS04 054, Agrisera). Anti-rabbit antibodies conjugated with horseradish peroxidase were used as secondary antibodies and visualized with an enhanced chemiluminescent ECL reagent, WesternBright Quantum HRP substrate (Advansta). Chemiluminescence was recorded using BioDoc-It Imaging System (UVP). Bands were quantified using QuantityOne software (Bio-Rad).
(h). Pyruvate content measurements and pyruvate uptake assays
Pyruvate was determined using the Pyruvate assay kit (MAK0711-KT, Sigma-Aldrich). For measurements, 50 µg of total extract/mitochondrial fraction isolated from the leaves of rps10 and wild-type plants were used. Briefly, 50 µl of working reagent (including enzyme mix and dye reagent) were incubated with mitochondrial fraction/extract and standards, both supplemented with pyruvate assay buffer to bring the volume to 50 µl, in darkness for 30 min at room temperature. The fluorescence intensity was measured at λex = 535/λem = 587 nm, and pyruvate content was calculated based on a standard curve.
In the pyruvate uptake assay, 100 µg of freshly isolated mitochondria from rps10 and wild-type plants were incubated in 50 µl of pyruvate transport buffer pH 7.2 (0.3 M sucrose, 5 mM KH2PO4, 10 mM TES, 10 mM NaCl, 2 mM MgSO4, 0.1% BSA) containing 10 mM pyruvate for 10 min on ice. As a control, mitochondria were incubated in the transport buffer without pyruvate. After incubation, mitochondria were re-isolated by centrifugation at 4°C for 10 min at 28 000× g and washed with mitochondria wash buffer (0.3 M sucrose, 10 mM TES, pH 7.5). Finally, the mitochondrial pellet was resuspended in wash buffer and the imported pyruvate was determined by fluorescence measurement as described above.
(i). Run-on transcription assay and dot blot hybridization
Preparation of dot blot membranes and run-on transcription assay was performed as described in [38]. The run-on assay was carried out on 107 chloroplasts in the presence of [α-32P]-UTP. DNA probes for chloroplast genes were obtained by PCR using gene-specific primers (electronic supplementary material, table S2b). Denaturated probes were dot-blotted onto nylon Hybond-N+ membrane (Amersham Pharmacia Biotech, UK) in two replicates using a Bio-Dot apparatus (Bio-Rad, USA). [32P]-labelled run-on transcripts were hybridized to the DNA probes immobilized on the membrane. Radioactive signals were analysed using Typhoon 5000 (Molecular Dynamics).
(j). Inhibitor treatments
The green part of two-week-old wild-type and rao2-1 seedlings was incubated in a solution containing appropriate mitochondrial respiratory inhibitors under light for different times, as indicated in figure legends. The final concentration of the inhibitors used was 40 µM rotenone, 10 µM antimycin A, 10 µM myxothiazol, 1 mM potassium cyanide (KCN), 10 µM oligomycin, 1 mM salicylhydroxamic acid (SHAM) or 1 mM n-propyl gallate (nPG).
3. Results
(a). The rps10 transcriptome shows highest similarity to the transcriptome of aox1a:rpoTmp double mutant
The genome-wide transcriptional reprogramming owing to silencing of the RPS10 gene encoding mitoribosomal protein S10 in Arabidopsis was characterized using ATH1 microarray analysis (electronic supplementary material, table S3a). The transcriptomic responses were profiled for two phenotypes of the rps10 heterozygous mutant, P2 and P3, differing by the timing of the onset of the silencing [39]. P2 and P3 are independent phenotypes, but exhibit a comparable decrease in the RPS10 transcript level and consequently a similarly altered population of mitoribosomes [28]. Using the thresholds of a fold change larger than two and ppde. p > 0.95, 1923 DEGs were found in P2 compared with wild-type (1104 up- and 819 down-regulated) and 3710 DEGs (1892 up- and 1818 down-regulated) in P3. Notably, 1575 DEGs were common to P2 and P3, indicating highly similar expression patterns in the two phenotypes. The top 50 most-affected genes in both P2 and P3 are shown in electronic supplementary material, table S4. Strikingly, many defence-related genes were highly induced, including pathogenesis-related PR1, PR4, PDF1.2b and phytoalexin deficient PAD3. Also, many known target genes of ANAC017, including AOX1a, UGT74E2, At2g04050 and At2g41730 were highly induced, indicating that the mitochondrial retrograde signalling via ANAC017 was activated in the rps10 mutant [9,10,40]. A gene ontology analysis confirmed that among the up-regulated genes common to P2 and P3, those related to pathogen response, stress response, abscisic acid signalling and glucosinolate biosynthesis were overrepresented (electronic supplementary material, table S5). Also mitochondrial organization was up-regulated, reflecting the perturbations caused by silencing of RPS10. In turn, the down-regulated genes common to P2 and P3 were enriched in lipases, auxin signalling, pectin biosynthesis and microtubule-based movement. Interestingly, the most significantly down-regulated cellular component was the chloroplast, suggesting that rps10 mutants are also affected in chloroplast function.
To get a broader insight into the transcriptional changes caused by the RPS10 knock-down, the transcript profile of the rps10 P2 and P3 phenotypes was compared with the profiles caused by a range of mitochondrial perturbations in Arabidopsis owing to mutations in genes encoding other mitochondrial proteins or induced by chemical treatments affecting mitochondrial processes. Overall, 25 additional microarray or RNA-seq datasets were compared with rps10, representing 19 different types of mitochondrial perturbations. Pairwise overlaps of the DEGs were calculated for all those datasets with both rps10 phenotypes (electronic supplementary material, table S3b). The largest number of DEGs common with rps10 was found for two antimycin A treatment datasets, while the mutants with most common DEGs were aox1a:rpoTmp, a double mutant defective in the alternative oxidase (AOX1a) and the dual-targeted mitochondrial and plastid RNA polymerase (RPOTmp), and complex I mutant ndufa1. Interestingly, on average 50% of DEGs observed in the aox1a:rpoTmp double mutant were also found to be differentially expressed in rps10 P2 and P3 lines. To examine this further, the similarities between all the datasets were assessed by comparison of pairwise DEG overlaps using the Sørensen-DSC (electronic supplementary material, table S6). The complete matrix of the DSC values for the rps10 and 25 additional datasets was used for hierarchical clustering and represented as an unrooted tree (figure 1a). Also here, aox1a:rpoTmp mutant clustered most closely with rps10. Additionally, the antimycin A datasets clustered relatively close to rps10, as well as complex I mutants ndufs4 and ndufa1.
(b). rps10 and aox1a:rpoTmp loss-of-function affect chloroplast gene expression
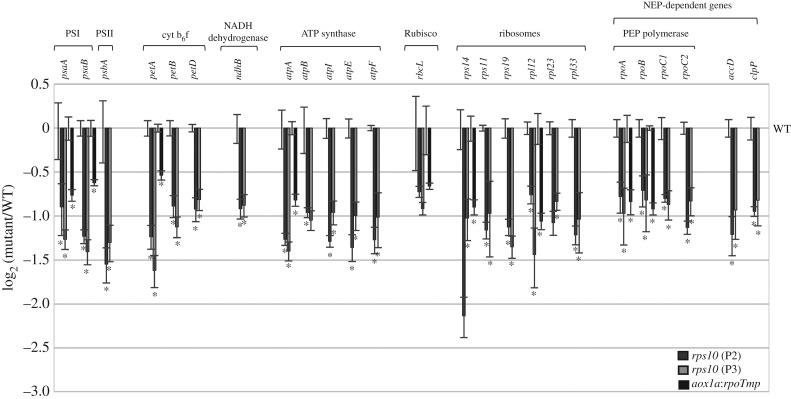
As described above, the gene ontology category ‘chloroplast’ was most significantly overrepresented among commonly down-regulated genes in rps10 (electronic supplementary material, table S5). In line with this, a clear general down-regulation of the nuclear-encoded components of chloroplast transcription machinery (including NEP polymerases, sigma factors and PEP-associated proteins) was found, with 13 genes significantly down-regulated (FC (fold change) 1.5, ppde. p > 0.95) in both rps10 phenotypes (figure 1b). Based on a meta-analysis, the most similar pattern was again observed for aox1a:rpoTmp mutants (figure 1b). The microarray results were subsequently validated using RT-qPCR, confirming down-regulation of nuclear genes related to plastid transcription for nearly all the genes tested in rps10 and aox1a:rpoTmp (electronic supplementary material, figure S1). Notably, the down-regulated expression of the nuclear genes encoding components of the plastid transcription machinery was correlated with lower chloroplast transcript level (figure 2). For all tested plastid genes belonging to different functional groups, the transcript levels measured by RT-qPCR analysis were significantly lower in the rps10 mutants compared to wild-type. Based on the similarity of the overall expression profile with rps10 mutants, plastid transcript levels were also analysed in aox1a:rpoTmp mutants. Also here the representative chloroplast transcripts were less expressed compared to wild-type (figure 2).
Figure 2.
Steady-state level of plastid transcripts in rps10 and aox1a:rpoTmp compared with wild-type plants. Data are presented as a log2 ratio of the transcript level in the mutants to those in the wild-type. The values obtained were averaged for at least three biological replicates, with error bars representing s.e.m. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05).
(c). rps10 and aox1a:rpoTmp mutants show perturbations in alternative and cytochrome mitochondrial respiratory pathways
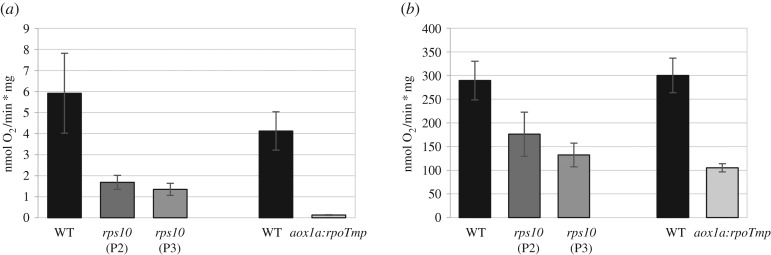
A characteristic feature of the aox1a:rpoTmp double mutant is a lack of activity of alternative oxidase and a decreased electron flux through the cytochrome pathway [29]. The first trait is caused by the lack of AOX1a expression, while the second one is owing to a dramatic reduction in the accumulation of complexes I and IV, whose subunits are mainly transcribed by the RPOTmp polymerase. Given the transcriptome similarity between rps10 and aox1a:rpoTmp, we decided to check the functionality of both the respiratory pathways in rps10 compared to their activities in aox1a:rpoTmp mitochondria. The capacity for oxygen consumption through the alternative respiratory chain was 3.5- to 4-fold decreased in both rps10 phenotypes compared with the wild-type, whereas for aox1a:rpoTmp, as expected, the alternative oxidase capacity is hardly detectable (figure 3a). The activity of the cytochrome respiratory pathway assessed by measuring complex IV activity indicated a 1.5- to 2-fold and 3-fold reduction in rps10 and aox1a:rpoTmp mitochondria, respectively, in comparison to the wild-type (figure 3b).
Figure 3.
Alternative pathway respiration and complex IV activity in mitochondria isolated from rps10 and aox1a:rpoTmp compared with wild-type plants. (a) AOX capacity was measured in the presence of ATP, NADH and succinate and after successive additions of ADP, the cytochrome c oxidase inhibitor KCN and the AOX activators pyruvate and dithiothreitol. After recording oxygen consumption, the AOX inhibitor n-propyl gallate was added to control the specificity of the reaction. (b) Cytochrome c oxidase (complex IV) capacity was measured after the addition of the substrates ascorbate and cytochrome c in the presence of Triton X-100. The values obtained were averaged for three biological replicates, with error bars representing s.d. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05).
(d). Regulation of alternative oxidase activity in rps10 seems to occur at the post-translational level
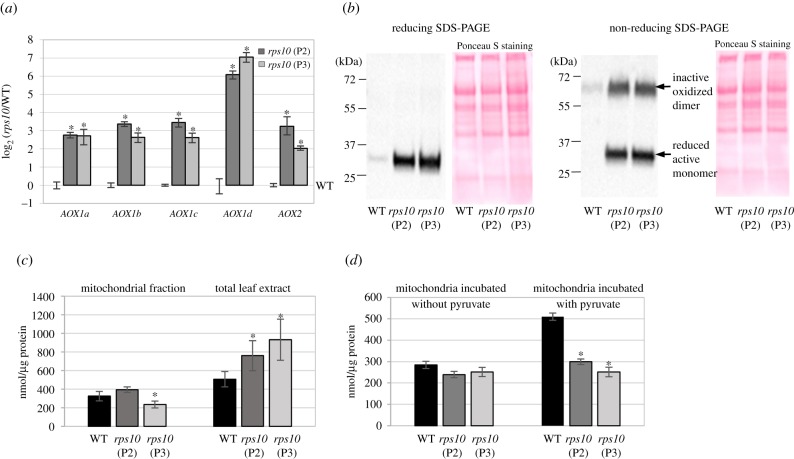
Surprisingly, the level of all five transcripts encoding different AOX isoforms as well as the abundance of the AOX protein in rps10 mitochondria were many-fold higher than in the wild-type (figure 4a,b left panel), in contradiction to the significantly decreased capacity of alternative oxidase (figure 3a). Since the redox state in isolated mitochondria could modulate AOX activity [41], we decided to determine whether AOX is present in rps10 mitochondria as an inactive oxidized dimer or a reduced active monomer. To this end, we used immunoblotting after non-reducing SDS-PAGE (figure 4b right panel). Significantly increased levels of both the inactive oxidized dimer and the reduced active monomer of AOX were detected in isolated rps10 mitochondria compared with the wild-type, indicating that the redox state of AOX itself cannot explain its decreased activity in rps10. Given that some α-keto acids, notably pyruvate, stimulate the activity of the reduced AOX form in isolated mitochondria [41], we decided to compare the levels of pyruvate in rps10 and the wild-type. Pyruvate was determined both in the mitochondrial fractions and in total leaf extracts. Whereas in the rps10 mitochondria the level of pyruvate was similar (P2 phenotype) or slightly decreased (P3 phenotype) compared with the wild-type, in the total leaf extracts, it was significantly elevated (on average, 1.6-fold) in rps10 compared with the wild-type (figure 4c). This result suggests a problem with pyruvate transport into the rps10 mitochondria. To verify it, the level of pyruvate was measured in freshly isolated mitochondria after their incubation with and without pyruvate. The external feeding of pyruvate resulted in its uptake into the wild-type (1.8-fold increase in pyruvate content), but not into the rps10 mitochondria (figure 4d).
Figure 4.
AOX expression and activity in rps10 compared with wild-type plants. (a) Steady-state level of transcripts of five AOX isoforms. Data are presented as a log2 ratio of the transcript level in the mutants to that in the wild-type. The values obtained were averaged for at least three biological replicates, with error bars representing s.e.m. Statistically significant differences from the wild-type plants are indicated by asterisks (Student's t test; p ≤ 0.05). (b) AOX protein level. Mitochondrial proteins (30 μg) from rps10 mutants and wild-type plants were separated by reducing and non-reducing SDS-PAGE (with or without the addition of 100 mM DTT, respectively) and probed with antibodies against AOX. Ponceau S staining serves as a loading control. (c) Pyruvate content in rps10 mutants compared with wild-type plants measured in mitochondrial fractions and total leaf extracts. The values obtained were averaged for three biological replicates, with error bars representing s.d. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05). (d) Pyruvate uptake into mitochondria of rps10 mutants and wild-type plants was measured as described in §2h. Control mitochondrial samples were incubated in buffer without pyruvate. The values obtained were averaged for three biological replicates, with error bars representing s.d. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05).
(e). rps10 mitochondrial dysfunction affects chloroplast transcription
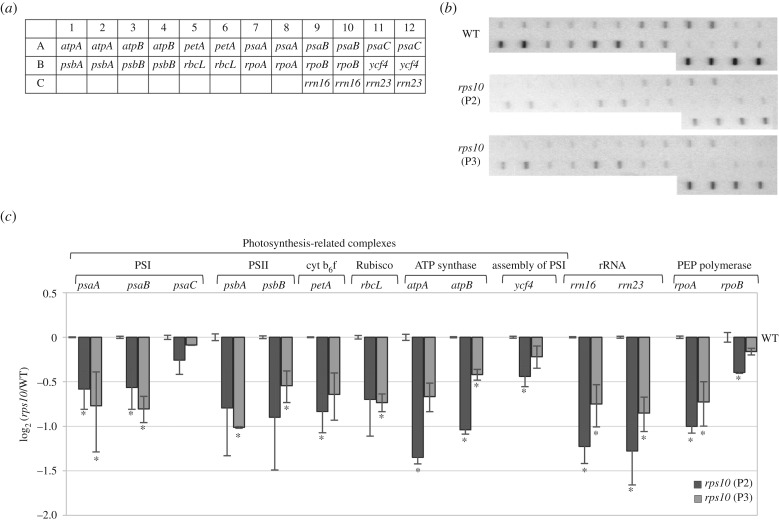
It seemed conceivable that the decreased level of chloroplast transcripts observed in rps10 and aox1a:rpoTmp could result from impairments of chloroplast transcription and/or reduced RNA stability. To distinguish between these possibilities, a run-on transcription assay was carried out on chloroplasts isolated from rps10 and wild-type plants for genes representing different functional groups (photosynthesis-related genes, rRNA genes and PEP polymerase genes). A clear reduction of transcription was observed for all the investigated genes in rps10; however, its extent was variable (figure 5). Most of the genes showed a ca. twofold decrease (slightly higher in the case of rrn16, rrn23 and atpA), only three genes showing a much weaker response: one encoding a subunit of photosystem I (PSI) (psaC), one encoding a PSI assembly protein (ycf4) and one encoding a PEP polymerase subunit (rpoB).
Figure 5.
Transcription activities of chloroplast genes in rps10 compared with wild-type plants. (a) Layout of chloroplast gene dot blots probed with run-on transcripts. Probes were applied to membranes in duplicates. (b) Representative autoradiographs of 32P-labelled run-on transcripts hybridized to chloroplast gene probes blotted on membranes. (c) Chloroplast run-on transcription rates represented as a log2 ratio of signal for rps10 to that for the wild-type. The values obtained were averaged for three biological replicates, with error bars representing s.e.m. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05).
A comparison of the transcription activity of the individual chloroplast genes with the steady-state level of their mRNA revealed a similar decrease in both parameters for the majority of genes in rps10 compared with the wild-type (electronic supplementary material, figure S2). For several genes (e.g. psaC, ycf4, rpoB), the decline of the transcript level was stronger than that of the transcription rate (electronic supplementary material, figure S2). The opposite was observed only for the rRNA genes (rrn16, rrn23), for which the quite strong reduction in transcription was associated with only a moderate decrease of the transcript level. Based on this comparison, we conclude that the decreased abundance of chloroplast transcripts in rps10 is mainly owing to their decreased transcription, although an altered stability of some transcripts contributes as well. It is worth noting that the transcriptional depression is not caused by a decreased number of chloroplast gene copies in rps10 (electronic supplementary material, figure S3). One should also note that the lower transcript abundance is accompanied by decreased levels of at least some proteins, as revealed for two chloroplast proteins: PsbC and PsbD (electronic supplementary material, figure S4).
(f). Chemical inhibition of complex IV and alternative oxidase decreases the level of chloroplast transcripts
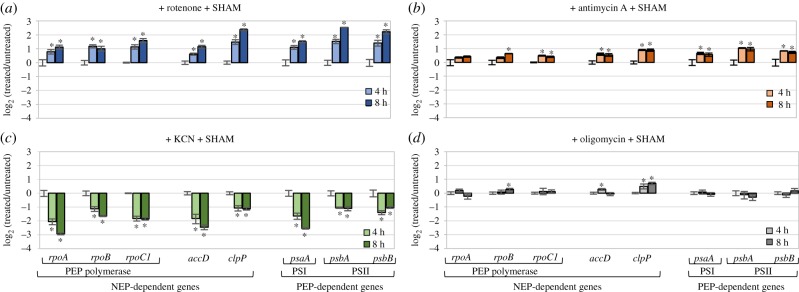
The results obtained so far for the rps10 and aox1a:rpoTmp mutants suggested that a simultaneous restriction of the electron flux through both the cytochrome and alternative mitochondrial respiratory pathways decreases the abundance of chloroplast transcripts. To define which mitochondrial electron transport chain complex/es has/have to be inhibited to prompt this specific chloroplast response, RT-qPCR analysis was performed after incubation of two-week-old wild-type seedlings with the AOX inhibitor SHAM in combination with one of OXPHOS complex-specific inhibitors: rotenone (inhibitor of complex I), antimycin A (inhibitor of complex III), KCN (inhibitor of complex IV) or oligomycin (inhibitor of ATP synthase). The analysis revealed that only simultaneous inhibition of AOX and complex IV by KCN causes a drop in the levels of chloroplast transcripts (figure 6). We observed a ca. two- to fourfold decrease in the steady-state level of these transcripts after 4 h of treatment, whereas after prolonged incubation (8 h), the decrease was even more pronounced, up to eightfold compared to untreated plants (figure 6c). By contrast, treating the seedlings with mixtures of SHAM with any of the other OXPHOS-specific inhibitors did not change or even increased the chloroplast transcript abundance (figure 6a,b,d).
Figure 6.
Steady-state level of plastid transcripts in wild-type seedlings treated for 4 and 8 h with mitochondrial respiratory inhibitors compared with untreated seedlings. Changes in the level of transcripts in seedlings treated with a mixture of (a) rotenone and SHAM, (b) antimycin A and SHAM, (c) KCN and SHAM, (d) oligomycin and SHAM. Data are presented as a log2 ratio of the transcript level seedlings treated with the inhibitors to that in water-treated control. The values obtained were averaged for four biological replicates, with error bars representing s.e.m. Statistically significant differences from the untreated seedlings are indicated by asterisks (Student's t test; p ≤ 0.05). (Online version in colour.)
The requirement for a simultaneous inhibition of complex IV and AOX activity to decrease the level of chloroplast transcripts was also observed when we used KCN in combination with an another inhibitor of AOX––n-propyl gallate (nPG) (electronic supplementary material, figure S5a). Additionally, we checked the level of chloroplast transcripts after a complete inhibition of complex III using a mixture of antimycin A and myxothiazol. Antimycin A blocks the reduction of cytochrome c1, whereas myxothiazol inhibits the reduction of both cytochromes b. nPG in combination with both these inhibitors of complex III still failed to decrease the steady-state level of chloroplast transcripts (electronic supplementary material, figure S5b).
To investigate the nature of the hypothetical mitochondrial retrograde signal(s) involved in the chloroplast response to a simultaneous inhibition of complex IV and AOX, we determined the level of three transcripts (rpoA, accD and psbA) in seedlings of a rao2-1 knockout mutant of ANAC017, a well-known regulator of the mitochondrial retrograde signalling pathway, after treatment with KCN and SHAM. As for the wild-type seedlings, the level of the plastid transcripts was significantly reduced in rao2-1 in response to a simultaneous inhibition of complex IV and AOX (electronic supplementary material, figure S6), which excludes ANAC017 as the sole mediator in the mitochondrial signalling leading to the down-regulation of chloroplast transcription.
(g). Both nuclear-encoded polymerases- and plastid-encoded polymerase-dependent genes are down-regulated in response to chemical inhibition of complex IV and alternative oxdiase
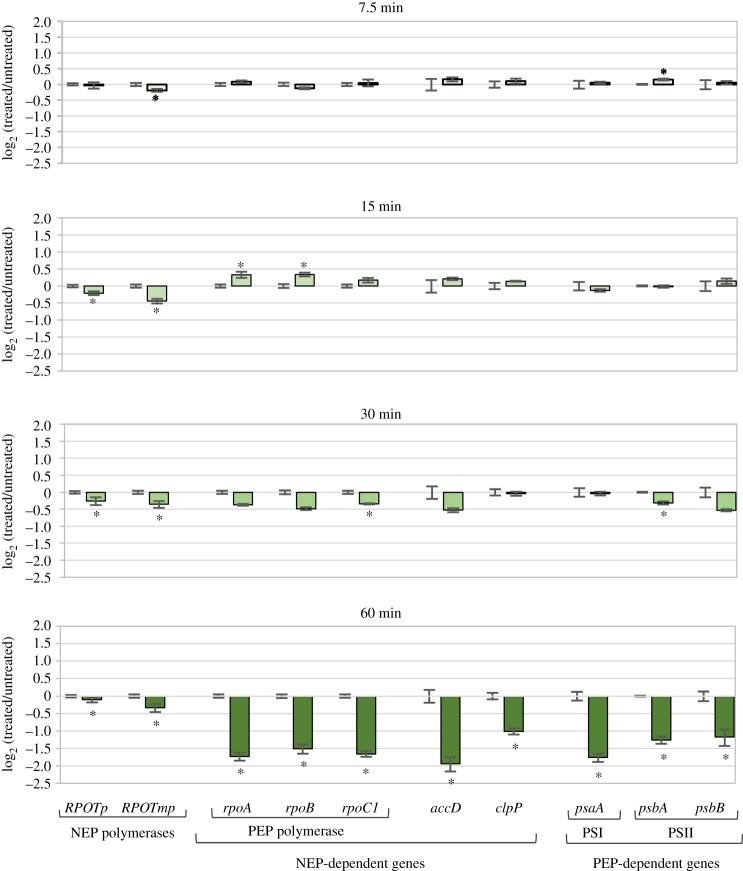
Next, we evaluated the effect of a simultaneous inhibition of complex IV (with KCN) and AOX (with SHAM) on the transcript level of the genes encoding the chloroplast RNA polymerases, NEP (RPOTp and RPOTmp) and PEP, as well as the transcripts dependent on their activities. The measurements were performed after treatment of the seedlings with the inhibitors for different time periods: 7.5, 15, 30 and 60 min. A small but statistically significant decrease was already observed for the RPOTmp polymerase transcript after 7.5 min, while a similar change for RPOTp took 15 min, but these were maintained at similar levels for both enzymes upon longer treatment (figure 7). By contrast, a clear drop in the PEP transcript level was found only after 60 min of treatment, but it was much more pronounced (ca. threefold) than that for the NEPs. Notably, at this time point, but not earlier, a significant reduction in the level of transcripts of both NEP- and PEP-dependent genes was detected. Taken together, these results indicate that the inhibition of the two terminal mitochondrial oxidases strongly reduces the abundance of the PEP-encoding transcripts and, to a similar extent, of both the PEP- and NEP-dependent transcripts even though the level of the NEP-encoding transcripts is reduced only slightly. It should be emphasized that the PEP transcripts behaved similarly in rps10 and aox1a:rpoTmp mutants, showing a nearly twofold lower abundance than in the wild-type (figure 2), while the NEP-encoding transcripts behaved differently from each other and inconsistently in the two mutants (electronic supplementary material, figure S1).
Figure 7.
Steady-state level of transcripts encoding NEP, PEP and transcripts dependent on NEP and PEP activities. Changes in the steady-state level of transcripts in wild-type seedlings treated with a mixture of KCN and SHAM compared with untreated seedlings. Incubation with the inhibitors was for 7.5, 15, 30 and 60 min. Data are presented as a log2 ratio of the transcript level seedlings treated with the inhibitors to that in water-treated control. The values obtained were averaged for at least three biological replicates, with error bars representing s.e.m. Statistically significant differences from the untreated seedlings are indicated by asterisks (Student's t test; p ≤ 0.05). (Online version in colour.)
(h). Salicylic acid and oxygen limitation negatively modulate the expression of nuclear-encoded components of the chloroplast transcription machinery
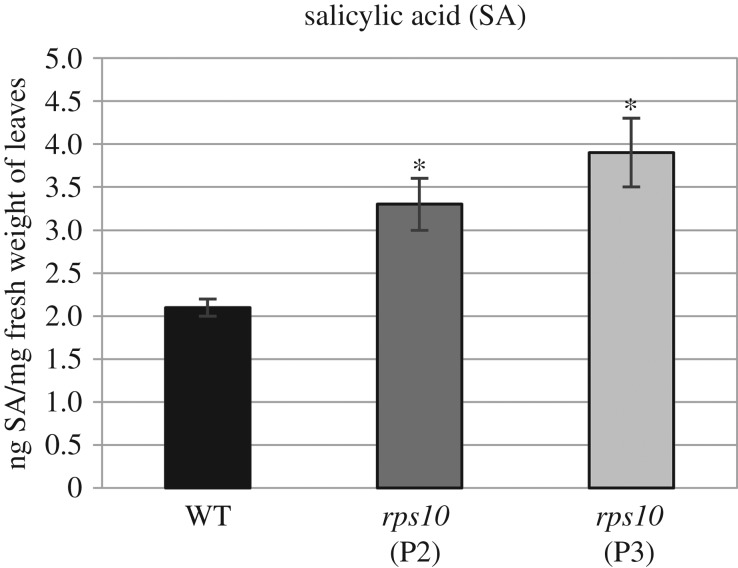
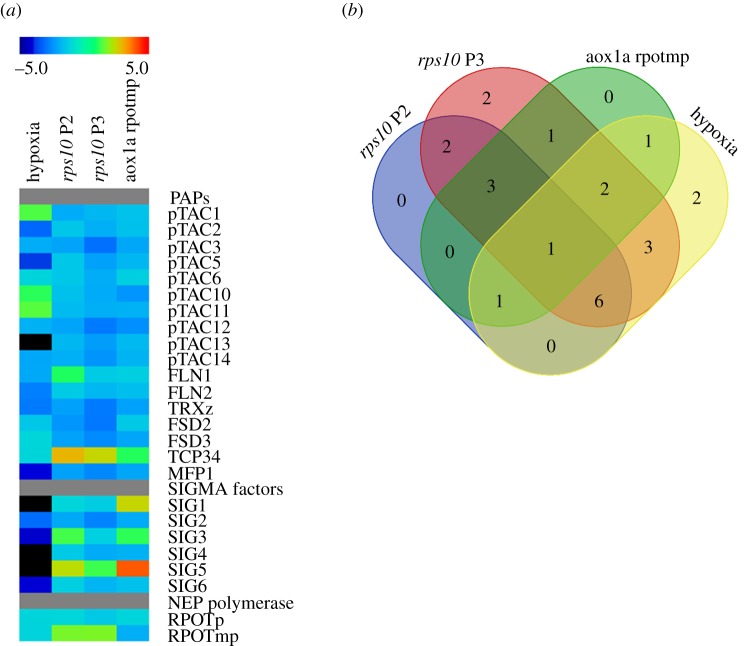
To gain insight into the signal transduction cascade leading to a decreased rate of chloroplast transcription, we looked for factors that down-regulate the expression of nuclear-encoded components of chloroplast transcription machinery, similarly to rps10 and aox1a:rpoTmp plants. Notably, a recent study showed that some of phytohormones, like abscisic acid (ABA), methyl jasmonate (MJ) and salicylic acid (SA), decrease the expression of nuclear genes encoding the components of plastid transcription apparatus [42]. Interestingly, one of the above-mentioned phytohormones, SA, was 1.5- to 2-fold increased in leaf extracts of both rps10 phenotypes compared with the wild-type (figure 8). Moreover, a strong down-regulation of nuclear genes encoding components of the chloroplast transcription machinery, similar to or even in some cases stronger than that observed in rps10 and aox1a:rpoTmp, was revealed in plants growing under hypoxic conditions (figure 9).
Figure 8.
Salicylic acid level in rps10 and wild-type plants. Salicylic acid was quantified by ultra-high performance liquid chromatography-electrospray ionization-tandem mass spectrometry (UHPLC-ESI-MS/MS) method at The Franciszek Górski Institute of Plant Physiology of the Polish Academy of Science, Krakow, as described in [43,44]. The values obtained were averaged for at least four biological replicates, with error bars representing s.d. Statistically significant differences from the wild-type are indicated by asterisks (Student's t test; p ≤ 0.05).
Figure 9.
Expression level of nuclear genes encoding components of the chloroplast transcription machinery in selected mitochondrial mutants and plants growing under hypoxic conditions. (a) Colour scale represents up (red) or down (blue) fold change values relative to the wild-type. Publicly available dataset GSE 14420 [45] representing three-week-old wild-type plant exposed to 4 h of hypoxia was analysed. (b) Common differentially expressed nuclear genes encoding chloroplast transcription machinery components (ppde. p > 0.95, FC (fold change) 1.5) in representative microarray datasets. (Online version in colour.)
4. Discussion
In this study, we provide evidence for a signalling pathway linking mitochondria, the nucleus and chloroplasts. We have discovered that a simultaneous inhibition of mitochondrial AOX together with mitochondrial complex IV, but not with other complexes of the mitochondrial oxidative phosphorylation system, causes a decrease in the rate of chloroplast transcription and consequently the steady-state levels of chloroplast transcripts in Arabidopsis.
A meta-analysis of published transcriptomic data obtained for different mitochondrial perturbations revealed a high similarity of two seemingly unrelated Arabidopsis thaliana mutants: rps10 and aox1a:rpoTmp, the first suffering from a deficit of the mitoribosomal S10 protein [28] and the second from a lack of the AOX1a protein and the dual-targeted RPOTmp polymerase [29]. We showed here that both these mutants have a significantly restricted activity in two mitochondrial respiratory complexes, complex IV and AOX (figure 3). The strongly reduced activity of AOX in aox1a:rpoTmp results from the lack of a functional AOX enzyme, whereas the low activity of complex IV is caused by the absence of the RPOTmp polymerase responsible for the transcription of the cox1 gene encoding a subunit of complex IV [46]. In rps10, the mutant with altered mitochondrial translation, the decline in complex IV activity is a consequence of a reduced synthesis of its mitochondrially encoded subunits [28], whereas the decreased activity of AOX seems to result from its post-translational regulation and is not connected with the redox state of the AOX protein but rather with the limited availability of pyruvate needed to activate AOX (figure 4d). However, since the relevance of pyruvate stimulation in vivo was questioned because of its high intramitochondrial concentration [47], the explanation of the decreased activity of AOX in rps10 requires a future research.
We report here that the mitochondrial dysfunctions found in rps10 and aox1a:rpoTmp, namely the inhibition of complex IV and AOX, cause a down-regulation of the abundance of plastid transcripts belonging to different functional groups (figure 2). Notably, the same effect is observed when complex IV and AOX are inhibited chemically in wild-type plants (figure 6c). Based on the comparison of the transcription activity of the chloroplast genes with the steady-state level of their mRNA in rps10, we conclude that the changes in the plastid transcript abundance are largely owing to a decline in their synthesis rate (electronic supplementary material, figure S2; figure 5). The negative regulation of chloroplast transcription was observed for genes dependent on the PEP and the NEP polymerases, including PEP polymerase genes. Since PEP is transcribed by NEP [26], we postulate instead that the NEP activity is modulated first in response to simultaneous inhibition of complex IV and AOX. Given that the function of one of the NEP (RPOTmp) is limited mostly to mitochondria [46,48], we believe that RPOTp, but not RPOTmp, is involved in reducing the rate of chloroplast transcription in response to the reduced activity of complex IV and AOX. Based on a very small decrease in the transcript level of NEP, at least in a chemical assay (figure 7), it seems that the regulation of NEP occurs at the post-transcriptional level. Notably, the reduced plastid transcriptional activity in rps10 coincides well with the down-regulation of the transcript level of numerous nuclear genes encoding components of the chloroplast transcription machinery, like sigma factors and PEP-associated proteins that are required for PEP activity (figure 1b; electronic supplementary material, figure S1). A similarly reduced level of these components was also found for aox1a:rpoTmp. To conclude, we postulate that the inhibited activity of mitochondrial complex IV and AOX affects the chloroplast transcription, most likely by a negative regulation of numerous components of the chloroplast transcription apparatus, including NEP at the post-transcriptional level and PEP with nuclear-encoded sigma factors and PAPs at the transcriptional level.
In this context, a recent finding concerning the effects of phytohormones on the expression of nuclear genes of the plastid transcription machinery is noteworthy [42]. It was shown that ABA, MJ and SA decrease the expression of nuclear genes of the chloroplast transcription machinery, including sigma factors and PAPs [42], so their action results in a similar effect to that observed here for rps10 and aox1a:rpoTmp. A significantly increased level of SA in rps10 leaf extracts (figure 8) supports the hypothesis that SA could act as a mediator in the signal transduction cascade from mitochondria to the nucleus to chloroplasts leading to a decline in the level of chloroplast transcription machinery components.
An involvement of phytohormones in the retrograde response to mitochondrial proteotoxic stress has also been suggested [49]. In that case, an impaired mitochondrial translation owing to a loss of mitoribosomal protein MRPL1 activated hormone signalling (involving ethylene and auxin), which in turn led to a strong transcriptional induction of the nuclear genes encoding mitochondrial HSP70 chaperones and mitoribosomal proteins. Since this retrograde response was induced by a loss of a mitoribosomal protein, it seems likely that a similar response could be activated in rps10 plants also deficient in a mitoribosomal protein. However, the transcript level of the nuclear-encoded components of mitoribosomes is not up-regulated in the rps10 mutant [28]. Thus, the signalling cascade initiated by the inhibition of the two terminal mitochondrial oxidases is different from that induced by mitochondrial proteotoxic stress. Moreover, our data indicate that the signal transduction pathway from mitochondria via the nucleus to chloroplasts is not mediated by ANAC017 [11], since we observed a decreased level of plastid transcripts after chemical inhibition of complex IV and AOX even in the absence of ANAC017 (electronic supplementary material, figure S6).
Given that both complex IV and AOX as terminal mitochondrial oxidases reduce oxygen to water, we propose that the signal to reduce the rate of chloroplast transcription is related to a drop in oxygen consumption. It should be stressed that regardless of whether the oxygen is not consumed or withdrawn from the cell, electron transport is blocked at the terminal oxidases (complex IV, AOX) [1]. In line with this assumption, a strong down-regulation of many nuclear genes encoding components of the chloroplast transcription machinery, similar to that in rps10 and aox1a:rpoTmp, was revealed in plants growing under low oxygen stress (hypoxia) (figure 9). Based on this finding, we speculate that a signal related to oxygen metabolism could lead to a decline of chloroplast transcription. At least in animal systems, it has been shown that complex IV may act as an oxygen sensor [50]. An overlap between low oxygen signalling and mitochondrial retrograde signalling (potentially controlled by the ANAC017 pathway) induced by inhibition of mitochondrial electron transport chain pathway at complex III by antimycin A has been reported [1]. As antimycin A treatment also results in a down-regulation of the nuclear-encoded components of the chloroplast transcription machinery, as observed during hypoxia and in the rps10 and aox1a:rpoTmp mutants (figures 1b and 9), it appears that two independent mitochondria-sensed low oxygen signalling pathways are active.
An involvement of both mitochondrial electron transport pathways (alternative and cytochrome) in the generation of signals influencing the transcription of plastid genes has been suggested in [24]. However, they emphasized the role of AOX activity, disregarding the importance of individual complexes of the cytochrome pathway in this response. By contrast, our results show unequivocally the necessity of the cytochrome respiratory pathway being inhibited at complex IV, but not at the other oxidative phosphorylation complexes, to bring about a reduction of chloroplast transcription. Thus, it is not the inhibition of the two mitochondrial electron transport chain pathways per se, but the restricted function of complex IV and AOX that affects the chloroplast function. Moreover, our results are based both on chemical treatment and studies of independent mitochondrial mutants (rps10 and aox1a:rpoTmp), providing much-needed in vivo confirmation of this pathway.
Taken together, we provide evidence from chemical and genetic studies that the limitation of the activity of two mitochondrial terminal oxidases down-regulates chloroplast transcription. Our data highlight the key role of complex IV in this response. We propose that oxygen and/or salicylic acid act as mediators in the signalling linking the restricted activity of complex IV and AOX with chloroplast transcription. However, the underlying molecular regulatory components of this signalling pathway remain to be established.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
The datasets supporting the conclusions of this article are included within the article and its electronic supplementary material files. Microarray data of rps10 have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under the accession dataset E-MTAB-8557.
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
H.J. and M.K.-O. designed the study. A.A.-S., M.K.-O. and U.K. performed the experiments. O.V.A. performed analysis of transcriptomic data. All authors analysed the data. M.K.-O., H.J. and O.V.A. wrote the manuscript with contribution from A.A.-S.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grant no. 2013/11/D/NZ1/00288 from the National Science Centre, Poland, to M.K.-O. O.V.A. was supported by the Swedish Research Council (Vetenskapsrådet 2017-03854).
References
- 1.Wagner S, Van Aken O, Elsässer M, Schwarzländer M. 2018. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiol. 176, 1156–1170. ( 10.1104/pp.17.01387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuo M, Obokata J. 2006. Remote control of photosynthetic genes by the mitochondrial respiratory chain. Plant J. 47, 873–882. ( 10.1111/j.1365-313X.2006.02839.x) [DOI] [PubMed] [Google Scholar]
- 3.Emanuel C, Weihe A, Graner A, Hess WR, Börner T. 2004. Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 38, 460–472. ( 10.1111/j.0960-7412.2004.02060.x) [DOI] [PubMed] [Google Scholar]
- 4.Pesaresi P, Masiero S, Eubel H, Braun H-P, Bhushan S, Glaser E, Salamini F, Leister D. 2006. Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18, 970–991. ( 10.1105/tpc.105.039073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleine T, Leister D. 2016. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta Bioenerg. 1857, 1313–1325. ( 10.1016/j.bbabio.2016.03.017) [DOI] [PubMed] [Google Scholar]
- 6.Giraud E, Van Aken O, Ho LHM, Whelan J.. 2009. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296. ( 10.1104/pp.109.139782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco NE, Guinea-Díaz M, Whelan J, Strand Å. 2014. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Phil. Trans. R. Soc. B 369, 20130231 ( 10.1098/rstb.2013.0231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ. 2019. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 179, 18–23. ( 10.1104/pp.18.01047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng S, et al. 2013. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. ( 10.1105/tpc.113.113985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Clercq I, et al. 2013. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490. ( 10.1105/tpc.113.117168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Aken O, Ford E, Lister R, Huang S, Millar AH.. 2016. Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 88, 542–558. ( 10.1111/tpj.13276) [DOI] [PubMed] [Google Scholar]
- 12.Shapiguzov A. et al. 2019. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife 8, e43284 ( 10.7554/eLife.43284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderauwera S, et al. 2012. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 20 113–20 118. ( 10.1073/pnas.1217516109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Aken O, Zhang B, Law S, Narsai R, Whelan J.. 2013. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162, 254–271. ( 10.1104/pp.113.215996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghavendra AS, Padmasree K. 2003. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8, 546–553. ( 10.1016/j.tplants.2003.09.015) [DOI] [PubMed] [Google Scholar]
- 16.Watanabe CKA, Yamori W, Takahashi S, Terashima I, Noguchi K. 2016. Mitochondrial alternative pathway-associated potoprotection of photosystem II is related to the photorespiratory pathway. Plant Cell Physiol. 57, pcw036 ( 10.1093/pcp/pcw036) [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. 2018. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 28, 448–461. ( 10.1038/s41422-018-0024-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niyogi KK. 2000. Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3, 455–460. ( 10.1016/S1369-5266(00)00113-8) [DOI] [PubMed] [Google Scholar]
- 19.Rasmusson AG, Møller IM. 2015. Multiple energy conservation bypasses in oxidative phosphorylation of plant mitochondria. In A companion to plant physiology and development, 6th Edition (eds Taiz L, Zeiger E, Møller IM, Murphy A), Chapter 12.3. Sunderland, MA: Sinauer. [Google Scholar]
- 20.Siedow JN, Umbach AL. 2000. The mitochondrial cyanide-resistant oxidase: structural conservation amid regulatory diversity. Biochim. Biophys. Acta Bioenerg. 1459, 432–439. ( 10.1016/S0005-2728(00)00181-X) [DOI] [PubMed] [Google Scholar]
- 21.Yoshida K, Terashima I, Noguchi K. 2006. Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol. 47, 22–31. ( 10.1093/pcp/pci219) [DOI] [PubMed] [Google Scholar]
- 22.Strodtkötter I, et al. 2009. Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with Antimycin A. Mol. Plant 2, 284–297. ( 10.1093/mp/ssn089) [DOI] [PubMed] [Google Scholar]
- 23.Vishwakarma A, Tetali SD, Selinski J, Scheibe R, Padmasree K. 2015. Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann. Bot. 116, 555–569. ( 10.1093/aob/mcv122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zubo YO, Potapova TV, Tarasenko VI, Börner T, Konstantinov YM. 2014. The rate of transcription in Arabidopsis chloroplasts depends on activity of alternative electron transfer pathway in mitochondria. Dokl. Biochem. Biophys. 455, 76–79. ( 10.1134/S1607672914020094) [DOI] [PubMed] [Google Scholar]
- 25.Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S. 2015. Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 66, 6957–6973. ( 10.1093/jxb/erv415) [DOI] [PubMed] [Google Scholar]
- 26.Hajdukiewicz PTJ, Allison LA, Maliga P. 1997. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. ( 10.1093/emboj/16.13.4041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfalz J, Pfannschmidt T. 2013. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. ( 10.1016/j.tplants.2012.11.003) [DOI] [PubMed] [Google Scholar]
- 28.Kwasniak M, Majewski P, Skibior R, Adamowicz A, Czarna M, Sliwinska E, Janska H. 2013. Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in Arabidopsis mitochondria. Plant Cell 25, 1855–1867. ( 10.1105/tpc.113.111294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kühn K, et al. 2015. Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol. 167, 228–250. ( 10.1104/pp.114.249946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kayala MA, Baldi P. 2012. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 40, W553–W559. ( 10.1093/nar/gks420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 10, 48 ( 10.1186/1471-2105-10-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B, et al. 2012. LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects. J. Biol. Chem. 287, 41 757–41 773. ( 10.1074/jbc.M112.383836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, et al. 2014. The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J. 80, 709–727. ( 10.1111/tpj.12665) [DOI] [PubMed] [Google Scholar]
- 34.Van Aken O, Whelan J.. 2012. Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific sesponses in Arabidopsis. Front. Plant Sci. 3, 281 ( 10.3389/fpls.2012.00281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, et al. 2017. The transcription factor MYB29 is a regulator of ALTERNATIVEOXIDASE1a. Plant Physiol. 173, 1824–1843. ( 10.1104/pp.16.01494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyu W, Selinski J, Li L, Day DA, Murcha MW, Whelan J, Wang Y. 2018. Isolation and respiratory measurements of mitochondria from Arabidopsis thaliana. J. Vis. Exp. 131, e56627 ( 10.3791/56627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacoby RP, Millar AH, Taylor NL.. 2015. Assessment of respiration in isolated plant mitochondria using Clark-type electrodes. In Plant mitochondria. Methods in molecular biology, vol. 1305 (eds Whelan J, Murcha M), pp. 165–185. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- 38.Zubo YO, Börner T, Liere K. 2011. Measurement of transcription rates in Arabidopsis chloroplasts. In Chloroplast research in Arabidopsis. Methods in Molecular Biology (Methods and Protocols), vol. 774 (ed. Jarvis R.), pp. 171–182. New York, NY: Humana Press. Humana Press, New York, USA. [DOI] [PubMed] [Google Scholar]
- 39.Majewski P, Wołoszyńska M, Jańska H. 2009. Developmentally early and late onset of RPS10 silencing in Arabidopsis thaliana: genetic and environmental regulation. J. Exp. Bot. 60, 1163–1178. ( 10.1093/jxb/ern362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lama S, Broda M, Abbas Z, Vaneechoutte D, Belt K, Säll T, Vandepoele K, Van Aken O.. 2019. Neofunctionalization of mitochondrial proteins and incorporation into signaling networks in plants. Mol. Biol. Evol. 36, 974–989. ( 10.1093/molbev/msz031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millenaar FF, Lambers H. 2003. The alternative oxidase: in vivo regulation and function. Plant Biol. 5, 2–15. ( 10.1055/s-2003-37974) [DOI] [Google Scholar]
- 42.Danilova MN, Andreeva AA, Doroshenko AS, Kudryakova NV, Kuznetsov VV, Kusnetsov VV. 2018. Phytohormones regulate the expression of nuclear genes encoding the components of the plastid transcription apparatus. Dokl. Biochem. Biophys. 478, 25–29. ( 10.1134/S1607672918010076) [DOI] [PubMed] [Google Scholar]
- 43.Dziurka M, et al. 2016. Local and systemic hormonal responses in pepper leaves during compatible and incompatible pepper–tobamovirus interactions. Plant Physiol. Biochem. 109, 355–364. ( 10.1016/j.plaphy.2016.10.013) [DOI] [PubMed] [Google Scholar]
- 44.Płażek A, Słomka A, Kopeć P, Dziurka M, Hornyák M, Sychta K, Pastuszak J, Dubert F. 2019. Effects of high temperature on embryological development and hormone profile in flowers and leaves of common Buckwheat (Fagopyrum esculentum Moench). Int. J. Mol. Sci. 20, 1705 ( 10.3390/ijms20071705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. 2009. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149, 1724–1738. ( 10.1104/pp.108.131912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kühn K, et al. 2009. Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21, 2762–2779. ( 10.1105/tpc.109.068536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Del-Saz NF, Ribas-Carbo M, McDonald AE, Lambers H, Fernie AR, Florez-Sarasa I. 2018. An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends Plant Sci. 23, 206–219. ( 10.1016/j.tplants.2017.11.006) [DOI] [PubMed] [Google Scholar]
- 48.Tarasenko VI, Katyshev AI, Yakovleva TV, Garnik EY, Chernikova VV, Konstantinov YM, Koulintchenko MV. 2016. RPOTmp, an Arabidopsis RNA polymerase with dual targeting, plays an important role in mitochondria, but not in chloroplasts. J. Exp. Bot. 67, 5657–5669. ( 10.1093/jxb/erw327) [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Auwerx J. 2017. Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell 68, 540–551. ( 10.1016/j.molcel.2017.10.006) [DOI] [PubMed] [Google Scholar]
- 50.Sommer N, et al. 2017. Mitochondrial complex IV subunit 4 isoform 2 is essential for acute pulmonary oxygen sensing. Circ. Res. 121, 424–438. ( 10.1161/CIRCRESAHA.116.310482) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its electronic supplementary material files. Microarray data of rps10 have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under the accession dataset E-MTAB-8557.
The datasets supporting this article have been uploaded as part of the electronic supplementary material.