Four choline/ethanolamine kinases have distinct enzymatic functions in plant development and the metabolism of choline/ethanolamine in Arabidopsis.
Abstract
Phosphatidylcholine and phosphatidylethanolamine are two major phospholipid classes in eukaryotes. Each biosynthesis pathway starts with the phosphorylation of choline (Cho) or ethanolamine (Etn) catalyzed by either choline or ethanolamine kinase (CEK). Arabidopsis contains four CEK isoforms, but their isozyme-specific roles in metabolism and development are poorly described. Here, we showed that these four CEKs have distinct substrate specificities in vitro. While CEK1 and CEK2 showed substrate preference for Cho over Etn, CEK3 and CEK4 had clear substrate specificity for Cho and Etn, respectively. In vivo, CEK1, CEK2, and CEK3 exhibited kinase activity for Cho but not Etn, although the latter two isoforms showed rather minor contributions to total Cho kinase activity in both shoots and roots. The knockout mutants of CEK2 and CEK3 both affected root growth, and these isoforms had nonoverlapping cell-type-specific expression patterns in the root meristematic zone. In-depth phenotype analysis, as well as chemical and genetic complementation, revealed that CEK3, a Cho-specific kinase, is involved in cell elongation during root development. Phylogenetic analysis of CEK orthologs in Brassicaceae species showed evolutionary divergence between Etn kinases and Cho kinases. Collectively, our results demonstrate the distinct roles of the four CEK isoforms in Cho/Etn metabolism and plant development.
Phospholipids are the conserved lipid component of cellular membranes from bacteria to plants and animals. Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) are the most abundant phospholipid classes in eukaryotes. An initial reaction step for the synthesis of PC and PE is phosphorylation of choline (Cho) or ethanolamine (Etn), both of which are catalyzed by choline/ethanolamine kinase (CEK) activity. The products, phosphocholine (PCho) or phosphoethanolamine (PEtn), are subsequently converted to cytidine diphosphocholine (CDP-Cho) or cytidine diphosphoethanolamine (CDP-Etn; Cornell and Ridgway, 2015) and then incorporated into the diacylglycerol backbone to produce PC and PE, respectively (McMaster, 2018). Thus, CEK activity plays an important role in the biosynthesis of PC and PE.
Although CEK homologs have been identified and characterized in many organisms (Wu and Vance, 2010; Glunde et al., 2011; Arlauckas et al., 2016), their substrate specificity and tissue-specific roles have been of primary interest for decades (Ishidate et al., 1985a; Aoyama et al., 2004). In the 1950s, the first purified CEK from Brewer’s yeast (Saccharomyces cerevisiae) demonstrated a dual substrate specificity that phosphorylates both Cho and Etn (Wittenberg and Kornberg, 1953). In addition, the purified CEKs from rat (Rattus sp.) kidney, liver, lung, and intestinal cytosols all showed dual substrate specificities (Ishidate et al., 1985a, 1985b; Porter and Kent, 1990).
On the other hand, separate Cho and Etn kinase activities have been demonstrated in plants (Tanaka et al., 1966; Macher and Mudd, 1976; Wharfe and Harwood, 1979a, 1979b; Monks et al., 1996), mammals (Brophy et al., 1977; Upreti, 1978; Pavlidis et al., 1994; Uchida, 1997; Lykidis et al., 2001; Peisach et al., 2003), and also in yeast (Hosaka et al., 1989; Yamashita and Hosaka, 1997; Kim et al., 1998; Kim et al., 1999). For example, in rat and mouse (Mus musculus) liver and kidney, the activities of Cho kinase and Etn kinase are attributed to separate proteins (Brophy et al., 1977; Upreti, 1981). Also, the activities and ratio of these two isozymes showed variations at different stages of postnatal development (Upreti, 1978, 1981), which indicate a distinct requirement of Cho and Etn activities during the course of development. Also, two CEK isozymes in mice, CKα and CKβ, showed higher tissue distribution in testes and in heart and liver, respectively (Aoyama et al., 2004). Of note, expression of CKβ failed to complement the mutant phenotype of heterozygous CKα-knockout mice, which suggests that these isoforms may function differently in a tissue-specific manner (Wu et al., 2008; Wu and Vance, 2010).
In seed plants, Cho kinase activity is reported in the roots of barley (Hordeum vulgare) and wheat (Triticum aestivum) and the leaves of barley, wheat, tobacco (Nicotiana tabacum), spinach (Spinacia oleracea), and squash (Cucurbita pepo; Tanaka et al., 1966). We previously identified four CEKs in Arabidopsis (Arabidopsis thaliana; Lin et al., 2015). Disruption of CEK4 resulted in an embryo-lethal phenotype, and CEK4 overexpression increased both PC and PE contents (Lin et al., 2015), which indicated that CEK4 may function as an Etn kinase in vivo. Although no obvious mutant phenotype was found in knocking out any of the other three CEKs under normal conditions (Lin et al., 2015), we showed that CEK1 functions as a Cho kinase in vivo and is required for endoplasmic reticulum (ER) stress tolerance by modulating the ratio of Cho to PCho (Lin et al., 2019). However, the roles of CEK2 and CEK3 remain elusive apart from the fact that the transcription of CEK3 is induced by salt stress (Tasseva et al., 2004). How the four CEKs differentially function in vitro and in vivo in Arabidopsis remains an open question.
Here, we investigated substrate specificities of the four CEKs in vitro and in vivo. In vitro, CEK1 and CEK2 showed substrate preference to Cho over Etn, whereas CEK3 and CEK4 had clear substrate specificities to Cho and Etn, respectively. In vivo, CEK1, CEK2, and CEK3 all showed kinase activity for Cho but not for Etn, albeit the latter two isoforms showed rather minor contributions in both shoots and roots. The knockout mutants of CEK2 and CEK3, cek2-1 and cek3-1, both affected root growth under normal conditions, and in-depth phenotype observation of cek3-1 revealed that Cho kinase activity of CEK3 may be involved in cell elongation during root development. Collectively, our results demonstrate distinct roles of four CEK isoforms in Cho/Etn metabolism and plant development.
RESULTS
Transcript Levels of Four CEKs in Different Tissues
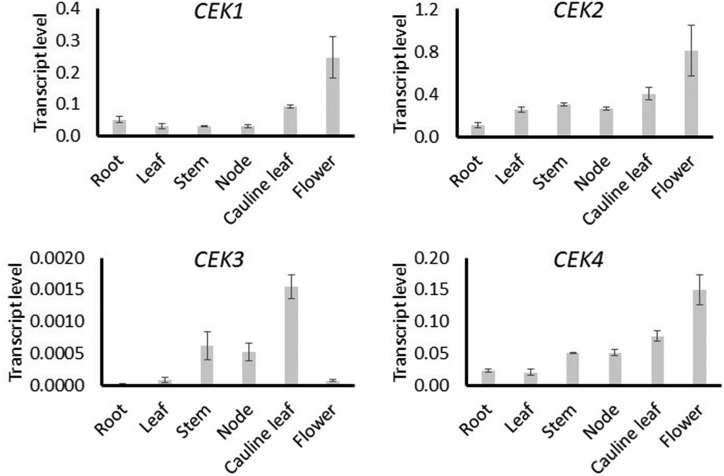
A previous in silico analysis indicated distinct tissue-specific patterns of transcript levels for four CEKs (Lin et al., 2015). To validate this, we investigated the transcript levels of four Arabidopsis CEKs in various tissues by reverse-transcription quantitative PCR (RT-qPCR) analysis using the complementary DNA (cDNA) template prepared from six different tissues (Fig. 1). The overall transcript levels of CEK1 and CEK2 were higher than the other two isoforms, particularly CEK3, which showed the lowest transcript levels among the four CEK homologs in all tissues examined. Compared with CEK1, CEK2, and CEK4, which showed higher transcript levels in reproductive tissues, the detected transcripts of CEK3 were mainly observed in the inflorescence tissues (stem, node, and cauline leaf). CEK3 transcript was barely detectable in root and flower, whereas the other three isoforms showed substantial transcript levels. In particular, CEK1, CEK2, and CEK4 showed the highest transcript levels in flower. These profiles largely support the result of the previous in silico analysis of CEKs (Lin et al., 2015). Thus, these four CEKs have distinct tissue-specific patterns of transcript level, particularly CEK3.
Figure 1.
Tissue transcript level of four CEKs by RT-qPCR. Transcript levels of CEK1, CEK2, CEK3, and CEK4 in root, leaf, stem, inflorescence node, cauline leaf, and flower. Data are averaged from three biological replicates, each with three technical replicates with sds as error bars.
Four CEKs Showed Distinct Substrate Specificity in Vitro
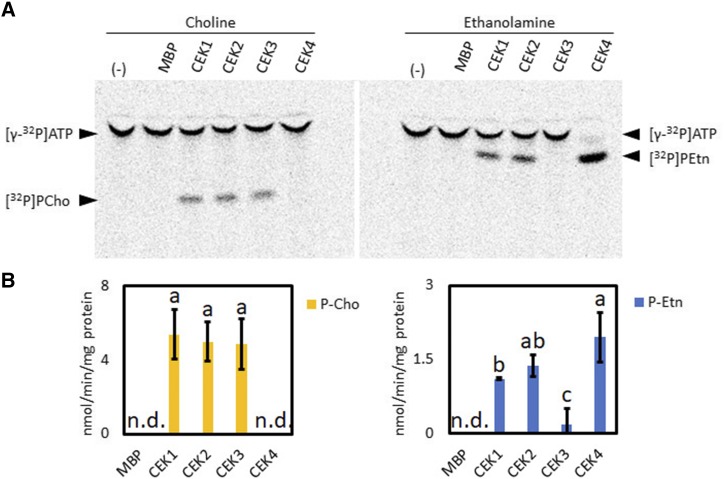
To examine the enzyme activity and substrate specificity of the four Arabidopsis CEKs, we performed in vitro enzyme activity assay with recombinant CEK proteins fused with maltose-binding protein (MBP) and expressed in Escherichia coli. To compare Cho kinase and Etn kinase activities, we used [γ-32P]ATP and non-radiolabeled Cho or Etn as substrates to detect the [32P]PCho or [32P]PEtn produced after the reaction. The result showed that incubation of Cho with CEK1, CEK2, and CEK3, but not CEK4, produced detectable amounts of [32P]PCho. On the other hand, incubation of Etn with CEK1, CEK2, and CEK4, but not CEK3, produced detectable amounts of [32P]PEtn (Fig. 2A). The quantification of enzyme activity indicated that CEK1, CEK2, and CEK3 have similar specific activities for Cho phosphorylation, while CEK3 showed much lower specific activity than CEK1, CEK2, and CEK4 in phosphorylating Etn (Fig. 2B). Based on the specific activity, CEK1 and CEK2 showed about 4-fold higher kinase activity for Cho than for Etn, whereas CEK3 showed ∼25-fold higher activity for Cho than for Etn. For CEK4, only Etn kinase activity was detected. Thus, this in vitro enzyme assay suggests that four CEKs encode functional activities with different substrate specificities: CEK1 and CEK2 prefer Cho to Etn, whereas CEK3 and CEK4 function specifically as Cho kinase and Etn kinase, respectively.
Figure 2.
In vitro enzyme activity and substrate specificity of four CEKs. A, Representative images of thin-layer chromatography plates indicating [32P]-labeled PCho or PEtn produced after incubating purified recombinant MBP-CEK1, MBP-CEK2, MBP-CEK3, and MBP-CEK4 with Cho or Etn in the presence of [γ-32P]ATP and Mg2+. The reaction with MBP but not CEK (lanes labeled MBP) or without adding protein (−) were used as negative controls. B, Specific activity of CEK isozymes in Cho or Etn kinase activity assay. Data are means ± sd from three replicates. Data obtained from at least three biologically independent experiments were analyzed by one-way ANOVA. Statistically different groups among conditions were further evaluated for significance with the Tukey’s honestly significant difference (HSD) mean separation test and displayed with lowercase letters indicating means that differ significantly. A level of P < 0.05 was considered significant. n.d., Not detected.
Pulse-Chase Metabolic Flux Analysis Dissected the Differential Contributions of CEK1, CEK2, and CEK3 to Cho and Etn Kinase Activity in Vivo
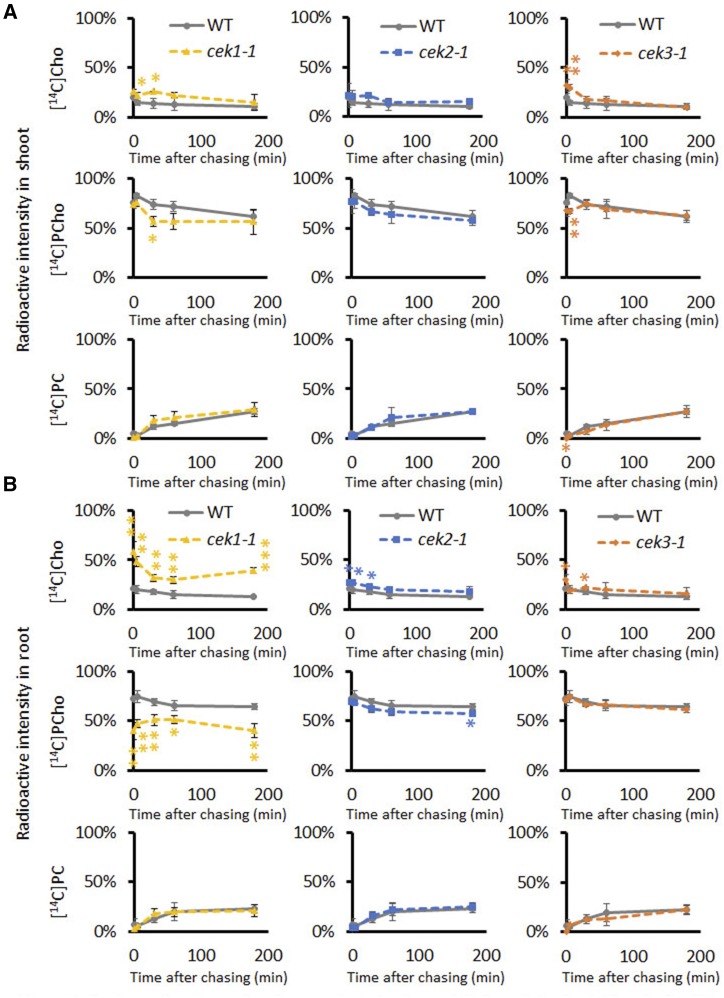
Next, we examined the role of CEKs in Cho or Etn kinase activity in vivo using a knockout mutant of each isoform except CEK4, whose knockout mutant causes an embryo-lethal phenotype (Lin et al., 2015). We recently showed that CEK1 functions as a Cho kinase in vivo (Lin et al., 2019). However, the cek1-1 mutant retained a considerable amount of PCho that could be produced by CEK2 and CEK3 harboring Cho kinase activity in vitro (Fig. 2). To dissect a differential contribution by CEK2 and CEK3 to Cho kinase activity in vivo, we performed radioactive pulse-chase metabolic flux analysis with either [14C]Cho or [14C]Etn using 14-d-old cek2-1, cek3-1, cek1-1, and wild-type seedlings (Lin et al., 2019). Following 10 min-labeling with [14C]Cho or [14C]Etn, radiolabeled metabolites in the shoots (Fig. 3A) and roots (Fig. 3B) were analyzed separately at 0, 5, 30, 60, and 180 min. The results of [14C]Cho quantification showed that Cho kinase activity was lower in cek1-1 both in shoots and roots compared with the wild type (Fig. 3), which is consistent with the previous report showing the activity in the whole seedling (Lin et al., 2019). At 30 min after chasing, the [14C]Cho level was ∼11% higher, while the [14C]PCho level was ∼17% lower in the shoots of cek1-1 than in those of the wild type (Fig. 3A). In cek2-1, Cho kinase activity decreased in roots (by ∼7%) but not in shoots, which was distinct from the result in cek3-1, where both shoots and roots showed a slight decrease in Cho kinase activity (∼16% and 6%, respectively). Although all three CEKs are committed to Cho kinase activity in vivo, none of these cek mutants affected the Etn kinase activity in either shoots or roots (Supplemental Figs. S1 and S2). Although a redundant effect on Etn kinase activity among the three CEKs is possible based on the in vitro data (Fig. 2), CEK4 may be the major Etn kinase in vivo. Thus, our results include a few important observations about the CEKs: (1) CEK1 is the major Cho kinase isoform; (2) CEK1 contributes more in roots than in shoots with respect to Cho kinase activity; and (3) none of the three CEKs individually affects Etn kinase activity and the synthesis of phospholipids PC and PE.
Figure 3.
In vivo radioactive pulse-chase analysis for the metabolism of Cho in the shoots (A) and roots (B) of cek1-1, cek2-1, and cek3-1 mutants. Fourteen-day-old wild-type (WT; gray lines), cek1-1 (yellow lines), cek2-1 (blue lines), and cek3-1 (orange lines) seedlings were labeled with [14C]Cho for 10 min and the amounts of [14C]Cho, [14C]PCho, and [14C]PC were measured at 0, 5, 30, 60, and 180 min after chasing. Ratios of [14C]Cho, [14C]PCho, and [14C]PC are shown as percentages of radioactive intensities quantified. Note that wild-type plots are in triplicate in each graph for clarity in displaying profiles in each cek mutant. Data are means ± sd from three biological replicates. Asterisks indicate statistical significance determined by Student’s t test: *P < 0.05; **P < 0.01 and ***P < 0.001.
CEK2 and CEK3 Exhibited Disparate Tissue-Specific Expression Patterns
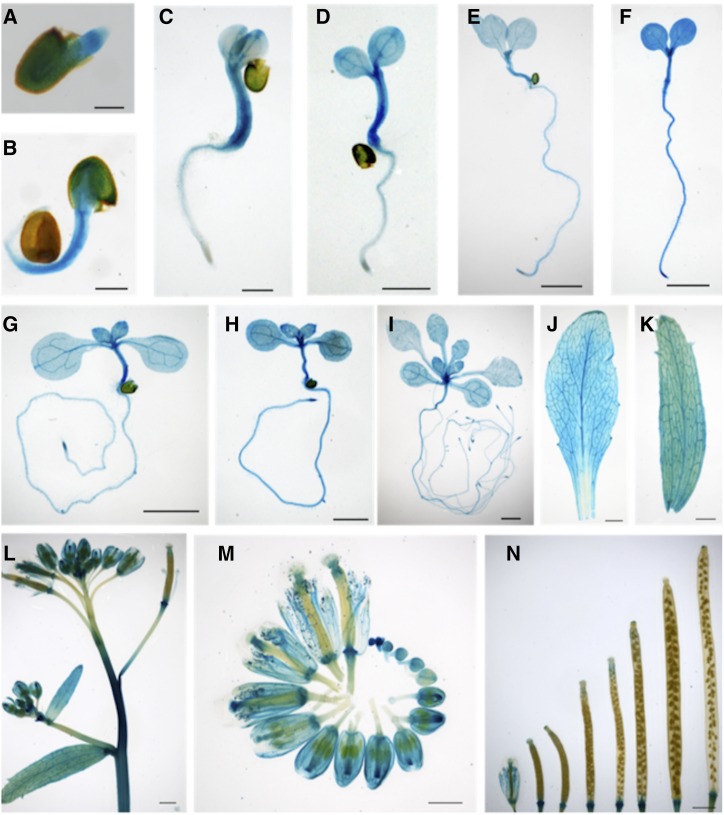
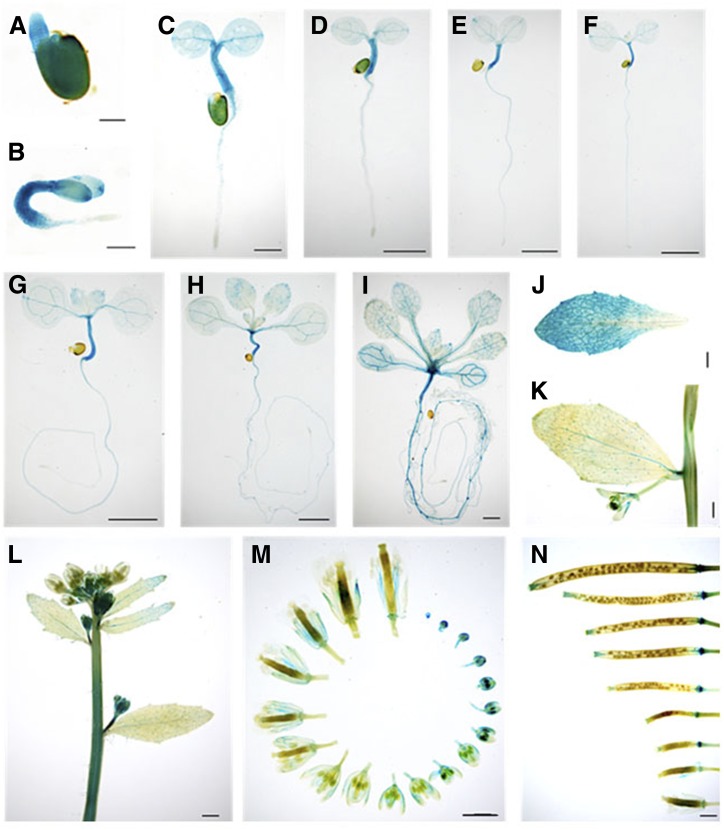
Previous reports showed that CEK1 and CEK4 have different tissue-specific expression patterns (Lin et al., 2015, 2019), possibly related to their distinct substrate specificities (Fig. 2). Since CEK2 and CEK3 function as the Cho kinases in vitro (Fig. 2) and in vivo (Fig. 3), we examined whether the expression patterns overlap with that of CEK1, which is preferentially expressed in vegetative tissues, particularly in root (Lin et al., 2019). To perform histochemical GUS staining assay for both CEK2 and CEK3, we transformed wild-type plants with a plasmid vector containing the whole genomic sequence of CEK2 or CEK3 with a GUS reporter in the C-terminal end of their protein-coding sequences (ProCEK2:CEK2-GUS and ProCEK3:CEK3-GUS). In the transgenic plants harboring these transgenic reporters, GUS staining was first observed in the entire embryos of CEK2 (Fig. 4, A and B) and CEK3 (Fig. 5, A and B), then in the hypocotyls, shoot apical meristems, and vasculature (Figs. 4, C–I, and 5, C–I, for CEK2 and CEK3, respectively). Furthermore, CEK2 and CEK3 expressions were confined in the vasculatures of rosette and cauline leaves (Figs. 4, J and K, and 5, J and K). In reproductive tissues, high GUS expression was observed in the inflorescence stem and basal part of developing siliques in both CEK2 (Fig. 4, L and N) and CEK3 (Fig. 5, L and N). Since the inflorescence showed relatively higher expression levels, we then examined their flowers closely at different developmental stages. In CEK2, strong GUS staining patterns were observed in all stages of flower development, particularly in young stigma, sepals and petals, and filaments (Fig. 4M). In CEK3, the strongest expression was noticeable only in developing buds, specifically in young anthers, and moderate expression was only evident in the filament of mature flowers (Fig. 5M). These staining patterns were confirmed in other transgenic plant lines harboring CEK2-GUS (Supplemental Fig. S3) and CEK3-GUS (Supplemental Fig. S4). Taken together, our results show that CEK2 and CEK3 have distinct tissue specificities, particularly during flower development.
Figure 4.
Tissue-specific expression of CEK2-GUS by histochemical GUS staining of transgenic plants harboring ProCEK2:CEK2-GUS (line 11). A to C, Time course of the GUS staining profile of germinating seeds at 1 (A), 2 (B), and 3 (C) d after planting. Seeds were stratified in sterile water for 1 d and then placed on MS-agar plates. D to I, Young seedlings at 4 (D), 5 (E), 6 (F), 7 (G), 10 (H), and 14 (I) DAG. J to N, Images of various plant tissues, including rosette leaf (J), cauline leaf (K), inflorescence (L), flowers at different developmental stages (M), and developing siliques (N). Scale bars = 200 μm (A and B), 1 mm (C, L, and N), and 2 mm (D–K and M).
Figure 5.
Tissue-specific expression of CEK3-GUS by histochemical GUS staining of transgenic plants harboring ProCEK3:CEK3-GUS (line 4). A to C, Time course of the GUS staining profile of germinating seeds at 1 (A), 2 (B), and 3 (C) d after planting. Seeds were stratified in sterile water for 1 d and then placed on MS-agar plates. D to I, Young seedlings at 4 (D), 5 (E), 6 (F), 7 (G), 10 (H), and 14 (I) DAG. J to N, Images of various plant tissues, including rosette leaf (J), cauline leaf (K), inflorescence (L), flowers at different developmental stages (M), and developing siliques (N). Scale bars = 200 μm (A and B), 1 mm (C, L, and N), and 2 mm (D–K and M).
CEK2 and CEK3 Were Localized at the ER But Showed Distinct Cell-Type Expression Patterns in the Root Meristematic Zone
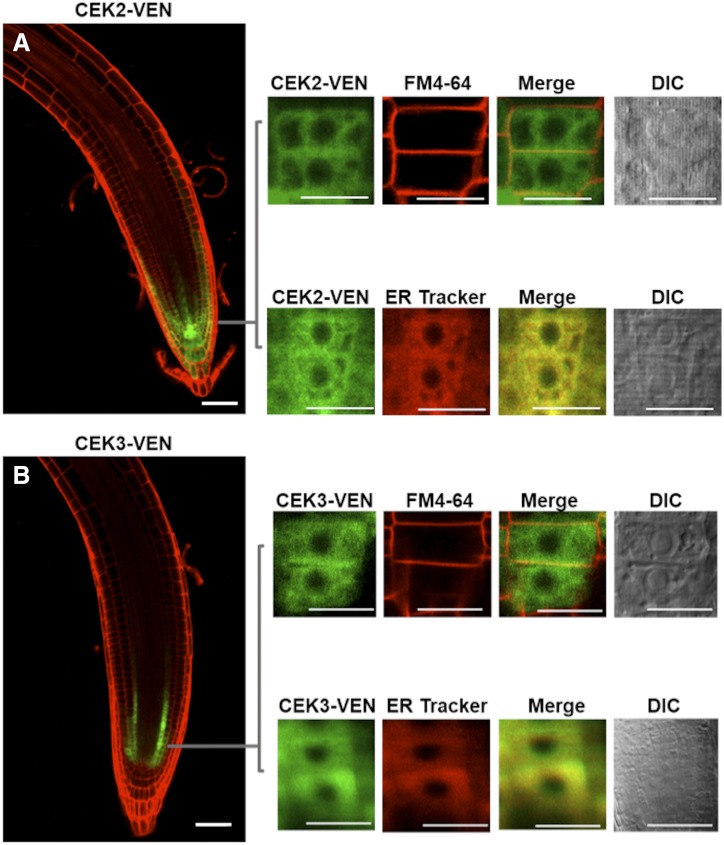
A consistent effect of CEK2 and CEK3 on Cho phosphorylation in roots (Fig. 3B) prompted us to investigate the expression patterns of these CEKs in root cells. Transgenic Arabidopsis plants were produced that express a triple repeat of a Venus fluorescent reporter construct (VEN) C-terminally fused to the protein coding sequence of CEK2 and CEK3 (ProCEK2:CEK2-VEN and ProCEK3:CEK3-VEN). Observation of their expression patterns under the confocal microscope showed that expression of both CEK2-VEN and CEK3-VEN was specific to the root meristematic zone (Fig. 6). However, we noted distinct cell type-specific expression patterns: CEK2-VEN was expressed in the columella, lateral root cap, and epidermis (Fig. 6A), while CEK3-VEN was observed exclusively in the initial few cells of cortex and endodermis (Fig. 6B). We magnified the Venus signal and overlapped it with staining of FM4-64 (as the plasma membrane marker) and ER Tracker. Both CEK2 and CEK3 colocalized with the ER Tracker but not with FM4-64. These results suggest that CEK2 and CEK3 are both localized mainly in the ER but still exhibit distinct nonoverlapping cell-type-specific expression patterns in the root meristematic zone.
Figure 6.
Cell type-specific expression pattern and subcellular localization in the seedling roots of transgenic plants harboring ProCEK2:CEK2-VEN (line 14; A) or ProCEK3:CEK3-VEN (line 24; B). Magnified images of VEN fluorescence were merged with FM4-64 and ER Tracker staining to observe the subcellular localization. Scale bars = 50 μm for the overview and 10 μm for the magnified images. DIC, Differential interference contrast.
CEK2 and CEK3 Mutants Displayed Abnormal Root Growth
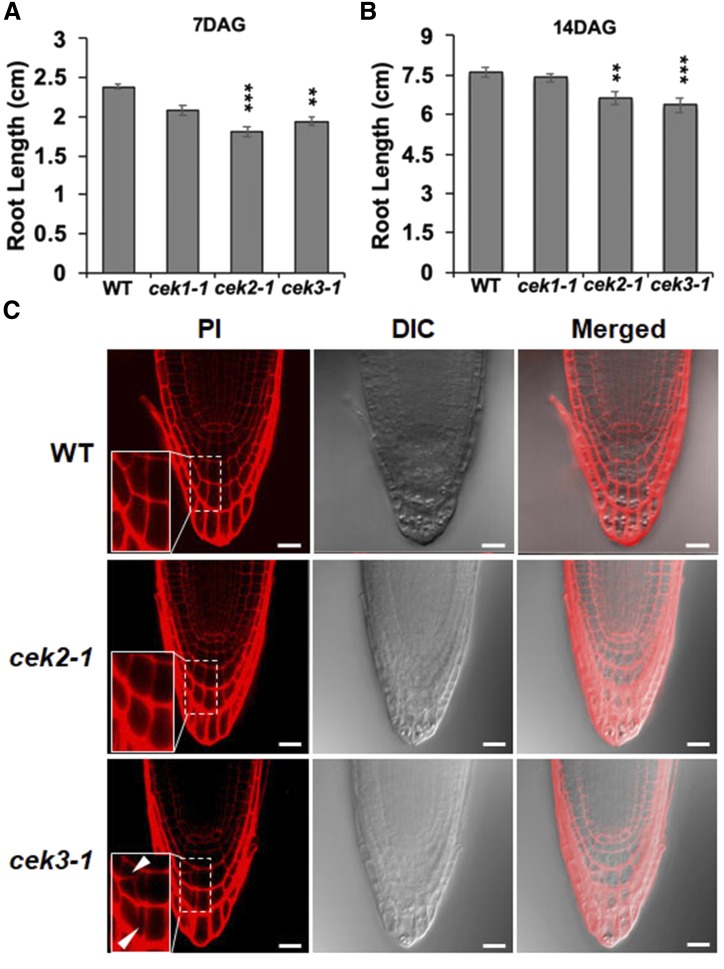
Since root cell-type-specific expression patterns of CEK2-VEN and CEK3-VEN suggested a specific role of these CEKs in root development, we observed the growth phenotype of seedling roots in transfer DNA-tagged knockout mutants cek1-1, cek2-1, and cek3-1 (Lin et al., 2015, 2019). Whereas cek1-1 did not show any significant difference in root length compared with the wild type, both cek2-1 and cek3-1 displayed significantly shorter root length in seedlings both 7 d-after-germination (DAG) and 14 DAG (Fig. 7, A and B). We observed the root tips of cek2-1 and cek3-1 by propidium iodide (PI) staining to check whether the short root phenotype is associated with a defective cellular architecture in the root apices of the mutants. In 7 DAG seedlings, we found some irregular shape and extra division in the columella cells of the root tips (Fig. 7C, white arrows) and an abnormal root cell architecture in cek3-1 but not in cek2-1 or the wild type. As observed in the magnified image of the root tip in cek3-1, the root tip contained odd-shaped columella cells possibly due to abnormal cell division (Fig. 7C, white arrows). We did not observe any morphological phenotype in any organs of cek1-1, cek2-1, and cek3-1 other than seedling roots. These observations denote the important role of CEK3 in the proper development of the columella cells for normal root growth. Thus, we further characterized cek3-1 for defective root growth.
Figure 7.
Root development in the seedlings of cek2-1 and cek3-1 mutants. A and B, Measurement of root length at 7 (A) and 14 (B) DAG. Data are means ± se of three biological replicates (n > 25). Asterisks indicate statistical significance by Student’s t test: **P < 0.01 and ***P < 0.001. C, Cellular architecture of root tips in 7-DAG wild-type, cek2-1, and cek3-1 seedlings as detected by PI stain. Inset pictures are magnified images of the columella cells in the dashed rectangle. White arrows indicate abnormally shaped columella cells. Scale bars = 20 µm. DIC, Differential interference contrast.
The cek3-1 Mutant Affected the Elongation of Root Cells
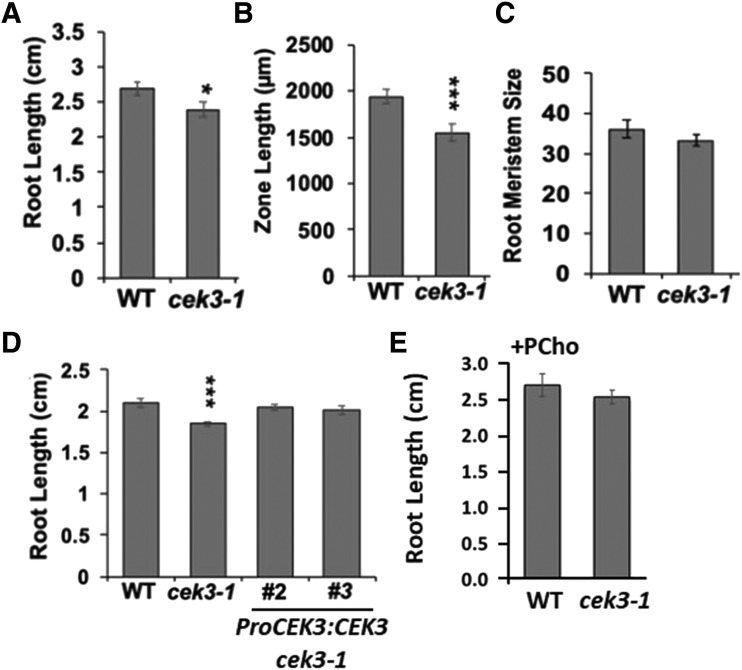
To address what cellular defect caused the short root phenotype in cek3-1, we observed the roots of 7 DAG seedlings. In the aerial part, we did not observe any abnormality, as previously reported (Lin et al., 2015). In the root, however, knockout of CEK3 caused not only shorter root growth (Fig. 8A) but also a shorter distance from the root tip to the first root hair of the maturation zone (length of elongation zone; Fig. 8B). To discern whether these phenotypes were caused by peculiarities in either cell division or cell elongation in the roots, we checked the root meristem size of both the wild type and cek3-1, which was expressed as the number of meristematic cortex cells from the quiescent center to the first elongated root cortex cell (Fig. 8C). However, we observed no significant difference in the root meristem size of cek3-1 compared to the wild type. Thus, the short root length and elongation zone observed in the cek3-1 mutant suggest that CEK3 may be involved in maintaining the normal cell elongation process in Arabidopsis roots.
Figure 8.
Knockout of CEK3 affected the elongation of root cells. A to C, Root length (A), zone length (B), and root meristem size (C) of 7 DAG wild-type and cek3-1 seedlings. D, Root length of 7 DAG wild-type and cek3-1 seedlings and genetically complemented lines (ProCEK3:CEK3 cek3-1, lines 2 and 3). E, Root length of 7 DAG wild-type and cek3-1 seedlings supplemented with PCho (100 μm). Data are means ± se of three biological replicates (n > 30). Asterisks indicate statistical significance determined by Student’s t test: *P < 0.05 and ***P < 0.001.
Genetic and Chemical Complementation Rescued the Root Phenotype in cek3-1
To test whether the cellular anomalies observed in the cek3-1 mutant were due to genetic ablation of CEK3, we transduced ProCEK3:CEK3 in the cek3-1 background to perform a genetic complementation test. The root length of two independent genetic complementation lines (ProCEK3:CEK3 cek3-1 lines 2 and 3) was not significantly different from that of the wild type, while that of cek3-1 was significantly shorter (Fig. 8D), which indicates that the root phenotype in cek3-1 is due to knockout of CEK3. Next, to examine whether the short root phenotype is due to compromised Cho kinase activity, we exogenously supplemented the reaction product PCho and measured the root length. As shown in Figure 8E, the root length of cek3-1 supplemented with PCho was not significantly different from that of the wild type. Overall, these data suggest that CEK3 is a functional Cho kinase localized at the ER in cortex/endodermal cells of the root meristematic zone and may be involved in root cell elongation.
The cek2-1 cek3-1 Double Mutant Did Not Further Enhance the Short Root Phenotype
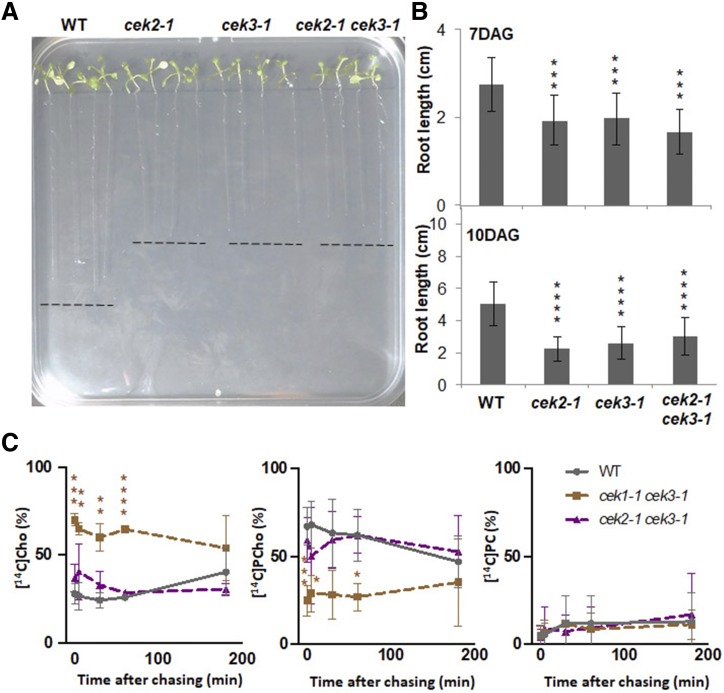
Since both cek2-1 and cek3-1 single mutants showed a similar short root phenotype (Fig. 7), we created a double mutant to test whether the double knockout mutant shows an enhanced root phenotype. Although seedlings of cek2-1 cek3-1 produced a shorter root length, it was not significantly shorter than that of cek2-1 or cek3-1 either at 7 or 10 DAG (Fig. 9, A and B). Also, no morphological phenotype was observed in the aerial part of the double mutant. These results suggest that the role of CEK2 and CEK3 in root growth may overlap despite their distinct cell-type expression patterns.
Figure 9.
Characterization of cek double mutants. A, Wild-type, cek2-1, cek3-1, and cek2-1 cek3-1 seedlings at 10 DAG on MS-agar media. B, Root length of seedlings at 7 and 10 DAG. Data are means ± sd from 12 seedlings for each genotype, and are from four biological replicates. Asterisks indicate statistical significance examined by Student’s t test: ***P < 0.001 and ****P < 0.0001. C, In vivo radioactive pulse-chase analysis of the metabolism of Cho in wild-type, cek1-1 cek3-1, and cek2-1 cek3-1 seedlings. Fourteen-day-old seedlings of the wild type (gray lines), cek1-1 cek3-1 (brown lines), and cek2-1 cek3-1 (purple lines) were labeled with [14C]Cho for 10 min and the amounts of [14C]Cho, [14C]PCho, and [14C]PC were measured at 0, 5, 30, 60, and 180 min after chasing. The ratios of [14C]Cho, [14C]PCho, and [14C]PC are shown as percentages of the radioactive intensities quantified. Data are means ± sd from three biological replicates. Asterisks indicate statistical significance examined by Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
cek1-1 cek3-1 But Not cek2-1 cek3-1 Considerably Decreased Cho Kinase Activity in Vivo
The results of pulse-chase analysis (Fig. 3; Supplemental Figs. S1 and S2) suggest that CEK1, CEK2, and CEK3 function as Cho kinases, whereas only CEK4 functions as an Etn kinase in vivo. To further examine how these three Cho kinases contribute differentially to the total Cho kinase activity in vivo, we produced multiple mutants by genetic crossing of cek1-1, cek2-1, and cek3-1. In F2 progeny, we obtained cek1-1 cek3-1 and cek2-1 cek3-1 by PCR-based genotyping. However, we isolated only cek1-1/+ cek2-1/+ but not cek1-1/− cek2-1/−. Because no morphological change was observed in gametophytes or embryos of cek1-1/+ cek2-1/+ plants, we genotyped the offsprings of double heterozygous mutants, which again did not produce any homozygous mutants. As CEK1 and CEK2 are both on the same chromosome, we speculated that cek1-1/− cek2-1/− cannot be obtained due to high genetic linkage. Thus, we abandoned the production of cek1-1/− cek2-1/− and cek1-1/− cek2-1/− cek3-1/−, and focused on cek1-1 cek3-1 and cek2-1 cek3-1. We first examined whether any of these Cho kinases is transcriptionally upregulated in the mutant background. We found no such upregulation except CEK2 in the cek1-1 background, which showed a >3-fold increase (Supplemental Fig. S5). Next, we performed radioactive pulse-chase metabolic flux analysis with [14C]Cho using 14-d-old cek1-1 cek3-1, cek2-1 cek3-1, and wild-type seedlings to test whether Cho kinase activity is further decreased in these double mutants. Although cek2-1 cek3-1 did not show any significant difference from the wild type in the profiles of [14C]Cho, [14C]PCho, and [14C]PC, we observed a significant decrease in Cho kinase activity in cek1-1 cek3-1 (Fig. 9C). Considering that a major part of seedling biomass is attributable to the shoot, the level of [14C]Cho detected in cek1-1 cek3-1 was considerably higher than that of cek1-1 (Fig. 3). These results suggest that CEK3 rather than CEK2 plays an additive role to CEK1 in total Cho kinase activity in vivo.
Phylogenetic Analysis of the CEK Family in Brassicaceae Species
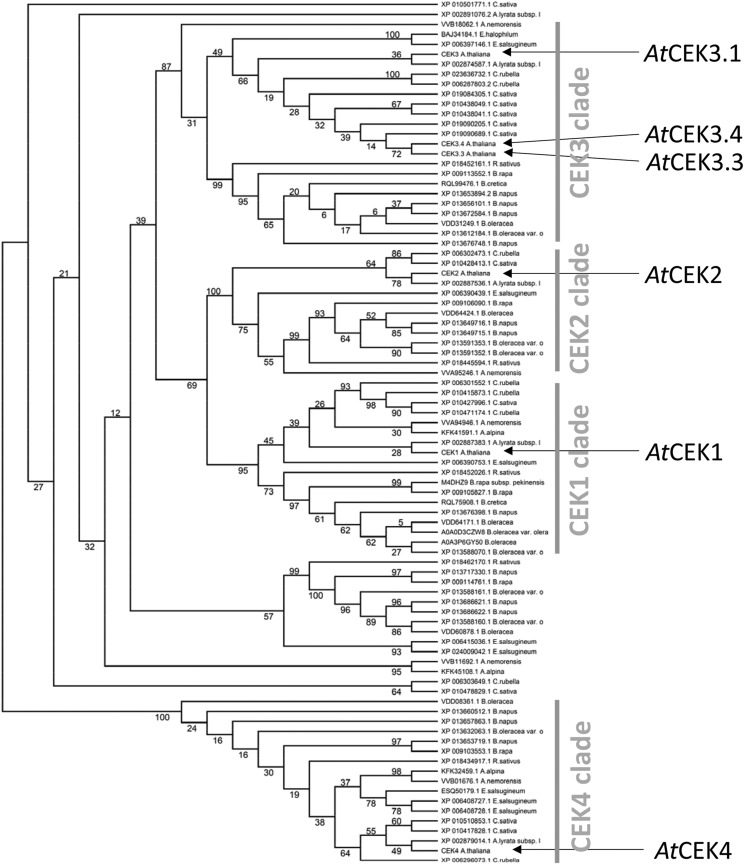
To give relevance to the functions and evolution of the four Arabidopsis CEK paralogs in terms of their differences and similarities, we created a phylogenetic tree of the four paralogs in Arabidopsis and 87 orthologs in 15 fully/semisequenced Brassicaceae species available at the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/) and The Universal Protein Resource (https://www.uniprot.org/). As shown in Figure 10, the tree clearly separated AtCEK4 from the other AtCEKs, and the clade for Cho kinase (i.e. AtCEK1, AtCEK2, and AtCEK3) was more complicated than that for Etn kinase (i.e. AtCEK4). Among AtCEK1, AtCEK2, and AtCEK3 (including three predicted splice variants, AtCEK3.1, AtCEK3.3, and AtCEK3.4), AtCEK1 and AtCEK2 were more closely related. Regarding the other Brassicaceae species analyzed, most of them possess CEK1 (except Brassica napus and Eutrema salsugineum), CEK2 (except Arabis alpina, B. cretica, and E. salsugineum), CEK3 (except A. alpina), and CEK4 (except B. cretica and E. salsugineum). Besides, some species possess an additional copy of CEK that is more distantly related to CEK1, CEK2, and CEK3. Thus, Etn kinase and Cho kinase show different evolutionary divergence in Brassicaceae species.
Figure 10.
Unrooted maximum likelihood tree of CEK homologs in Brassicaceae species. This tree was calculated with the raxmlGUI 2.0 program. The numbers on each branch indicate the confidence level in percentage of maximum likelihood method.
DISCUSSION
Distinct Roles of the Four CEKs in Cho/Etn Metabolism
Earlier enzymological studies in soya bean demonstrated two distinct CEK activities: a Cho-preferring kinase (with substrate preference for Cho over Etn of 3:1; Wharfe and Harwood, 1979b) and an Etn-specific kinase (Wharfe and Harwood, 1979a). Although this evidence suggests that plants possess multiple copies of CEK activity with different substrate specificity, a comprehensive study on the substrate specificity of the CEK family has not been conducted previously in any plant species. Here, results of the in vitro enzyme activity assay demonstrated that four CEK isozymes in Arabidopsis have clear substrate specificities (Fig. 2). Whereas CEK2 showed similar substrate specificity to CEK1 (Lin et al., 2019), CEK3 showed a clearer specificity for Cho than for Etn. Remarkably, CEK4 showed substrate specificity for Etn. Thus, four CEKs showed different substrate preference in vitro. For the three Cho kinases, we tested in vivo activities by pulse-chase radiolabeling experiments in the mutants. Previously, cek1-1 was shown to have a significant defect in phosphorylating Cho in whole seedlings (Lin et al., 2019). Our data elaborated that the defect was more pronounced in roots than in shoots (Fig. 3). Compared with cek1-1, both cek2-1 and cek3-1 showed a rather limited effect on Cho phosphorylation. Also, none of these cek mutants affected Etn kinase activity in vivo (Supplemental Figs. S1 and S2). To investigate a possible functional redundancy among Cho kinases, we produced double mutants and observed an enhanced Cho kinase defect in cek1-1 cek3-1 but not in cek2-1 cek3-1 (Fig. 9C). Regarding the effect of Cho kinase activity on PC biosynthesis, in vivo pulse-chase flux analysis showed that none of the single and double mutants examined affected PC production (Figs. 3 and 9C; Lin et al., 2019). Although functional redundancy among these three isozymes cannot be ruled out until a triple mutant is characterized, a contribution of Cho kinase activity to PC biosynthesis may be minor in Arabidopsis. Taken together, we suggest that (1) CEK4 may be the sole Etn kinase in vivo; (2) CEK1 may be the major Cho kinase isoform, whereas CEK3, rather than CEK2, may play a minor role in total Cho kinase in vivo; and (3) Cho kinase activity may have no major impact on PC biosynthesis in Arabidopsis.
Distinct Roles of the Four CEKs in Plant Growth and Development
Seedling phenotype observation indicated that both cek2-1 and cek3-1 mutants show reduced root length (Fig. 7, A and B). Since CEK2 and CEK3 are both Cho kinases in vivo (Fig. 3), the root phenotype may be associated with reduced production of PCho. Indeed, exogenous supplementation of PCho rescued the root phenotype in cek3-1 (Fig. 8E). PCho is required for root growth, as a knockout of phospho-base N-methyltransferase1 (PMT1), which catalyzes the major pathway for PCho biosynthesis, showed the short-root phenotype (Cruz-Ramírez et al., 2004), and the phenotype was enhanced in pmt1 pmt2 double mutants (Chen et al., 2019; Liu et al., 2019). Also, another pathway that produces PCho from PC catalyzed by nonspecific phospholipase C (NPC) 2 and NPC6 is involved in root growth (Ngo et al., 2019). Regarding PCho production from Cho by Cho kinase, CEK1, CEK2, and CEK3 may be responsible based on the in vivo data (Fig. 3). However, cek1-1 mutant did not show any root growth defect under normal growth conditions (Fig. 7, A and B) despite the fact that CEK1 has a major contribution to the Cho kinase activity in both shoots and roots (Fig. 3; Lin et al., 2019). Interestingly, CEK2-VEN and CEK3-VEN showed somewhat complementary cell-type-specific expression patterns in the root meristematic zone (Fig. 6; Supplemental Fig. S6). Although CEK2-VEN was more predominantly expressed in root columella cells, a more evident defect in the columella cell structure was found in the cek3-1 mutant (Fig. 7C). Since CEK3-VEN was expressed specifically in juvenile cortex/endodermal cells in root, and defective cell elongation was associated with the short root phenotype in cek3-1, CEK3 may produce PCho in this type of cell to maintain cell elongation in roots. It is possible that the three CEK isoforms produce PCho at different cell types for different purposes in roots.
Our substrate specificity assays provide a clue in addressing the embryonic lethal phenotype of cek4-1 (Lin et al., 2015). Since CEK4 is an Etn-specific kinase (Fig. 2), the lethal phenotype may be caused by the loss of Etn kinase activity in vivo. Since neither CEK1, CEK2, nor CEK3 likely functions as an Etn kinase in vivo, based on the result of pulse-chase analysis (Fig. 3; Supplemental Figs. S1 and S2), the lethality of cek4-1 may be due to lack of Etn kinase activity that cannot be compensated by any of the remaining CEK isoforms. Unlike Cho kinase, Etn kinase activity takes part in the metabolic pathway of de novo PC and PE biosynthesis (Lin et al., 2015). Starting from the conversion of Ser to Etn by Ser decarboxylase1 (SDC1; Rontein et al., 2001; Yunus et al., 2016), CEK4 phosphorylates Etn to produce PEtn, which is a common precursor for the biosynthesis of PC and PE. It should be noted that the knockout of SDC1 causes embryonic lethal phenotype (Yunus et al., 2016), similar to the effect with knockout of CEK4 (Lin et al., 2015). Triple knockout mutants of three PMTs, which block the conversion of PEtn to PCho in the PC biosynthesis pathway, did not cause embryonic lethality (Chen et al., 2019; Liu et al., 2019). However, a knockout of CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE (PECT1), which converts PEtn to CDP-Etn in the PE biosynthesis pathway, caused an embryonic lethal phenotype (Mizoi et al., 2006). These pieces of evidence suggest that the embryonic lethality may be due to a defect in PE biosynthesis.
An Evolutionary Insight into the CEK Family
Phylogenetic analysis of the CEKs in Brassicaceae species indicated that the Cho kinase family (CEK1, CEK2, and CEK3 clades) is more diversified than the Etn kinase family (CEK4 clade; Fig. 10). Most species included in the analysis possess homologs for each Cho kinase isozyme, suggesting that functional divergence among the three Cho kinases may be conserved in Brassicaceae. However, of note, A. alpina possesses CEK1 and another distantly related isoform, but not CEK2 or CEK3. The A. alpina genome is fully sequenced (Willing et al., 2015) and the species is known to grow on a rocky alpine mountain under harsh growth conditions (Toräng et al., 2015). By contrast, the phylogenetic tree for the CEK4 clade showed a less diverse pattern. Whereas PC is an abundant phospholipid class in most eukaryotes, this lipid class is absent from most prokaryotes. In contrast, PE is widely present in both prokaryotes and eukaryotes. Heterotrophic eukaryotes such as animals obtain Cho through diet intake, which is readily used for the biosynthesis of PC (Wu and Vance, 2010). In plants, however, Cho is synthesized from Etn by Etn kinase (Lin et al., 2015), PMTs (Chen et al., 2018, 2019; Liu et al., 2018, 2019), and PCho phosphatase (May et al., 2012; Angkawijaya and Nakamura, 2017; Hanchi et al., 2018). Since knockout of all PMTs makes the mutant unable to synthesize PCho and PC (Chen et al., 2019; Liu et al., 2019), it follows that this pathway may be essential for the production of Cho-containing compounds. Thus, in Arabidopsis, as a photoautotrophic plant, Etn kinase plays a crucial role in synthesizing both PC and PE, while Cho kinase has less commitment to PC biosynthesis compared with heterotrophic organisms that can take up exogenous Cho through diet. The facts that Etn compounds—but not Cho compounds—are widely found in prokaryotes, and that Etn kinase is absolutely required for PC and PE biosynthesis might have exerted strong selection pressure for the CEK4 clade through evolution. It is possible that the contribution of Cho kinase activity to PC biosynthesis may differ between autotrophic and heterotrophic organisms.
In conclusion, we revealed the isozyme-specific roles of the four CEKs in vitro and in vivo in Arabidopsis. Our results demonstrate the divergent roles of these four CEKs in phospholipid metabolism and plant development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants (Columbia-0 ecotype) were grown under a 16-h light/8-h dark photoperiodic condition at 22°C with light intensity of 150 μmol m−2 s−1. Murashige and Skoog (MS) medium was used at one-half strength for plant culture (Murashige and Skoog, 1962). The mutant seeds are as previously reported (Lin et al., 2015). Double mutants were produced by genetic crossing. For root observation, plants were grown vertically on a MS-agar plate.
Vector Construction and Plant Transformation
ProCEK2:CEK2-GUS
A 3,430-bp genomic sequence for CEK2 was amplified by PCR with the primers PK072 and JL249 (Supplemental Table S1). The fragment was cloned into the pENTR/D-TOPO plasmid vector (Invitrogen, Thermo Fisher Scientific) to obtain pPK33. Then, the SfoI site was added immediately before the stop codon of pPK33 by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000) with the primer JL273 to obtain pPK39. The GUS cassette was inserted into the SfoI site of pPK39 to obtain pPK45.
ProCEK2:CEK2-VEN
The triple (3×) repeat of a Venus fluorescent reporter construct was inserted into the SfoI site of pPK39 to obtain pPK44.
ProCEK3:CEK3
A 5,452-bp genomic sequence for CEK3 was amplified by PCR with the primers JL269 and JL254. The fragment was cloned into the pENTR/D_TOPO plasmid vector (Invitrogen, Thermo Fisher Scientific) to obtain pLH1.
ProCEK3:CEK3-GUS
The SfoI site was inserted before the stop codon of pLH1 by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000) with the primer JL302 to obtain pLH2. Next, the GUS cassette was inserted into the SfoI site of pLH2 to obtain pPK35.
ProCEK3:CEK3-VEN
The triple (3×) repeat of a Venus fluorescent reporter construct was inserted into the SfoI site of pLH2 to obtain pPK34.
The obtained entry vector plasmids pPK45, pPK44, pLH1, pPK35, and pPK34 were recombined into the pBGW destination vector by Gateway LR reaction (Karimi et al., 2002) to obtain pPK48, pPK52, pGA006, pPK43, and pPK42, respectively. These plant binary vectors were transduced into cek2-1 (for pPK48 and pPK52) or cek3-1 (for pGA006, pPK43, and pPK42) via Agrobacterium tumefaciens-mediated gene transformation. For each transformation, a total of 24 T1 plants were genotyped and T2 seeds from those carrying transgenic lines were harvested individually. To distinguish the transgene from endogenous CEK2, the following primers were designed: JL252 and KK098 for ProCEK2:CEK2-GUS, JL251 and KK104 for ProCEK2:CEK2-VEN, YN896 and YN748 for ProCEK3:CEK3, PK073 and KK098 for ProCEK3:CEK3-GUS, and PK073 and KK104 for ProCEK3:CEK3-VEN. Lines used for observation were ProCEK2:CEK2-GUS, lines 11 and 22; ProCEK2:CEK2-VEN, lines 14 and 19; ProCEK3:CEK3 cek3-1, lines 3 and 9; ProCEK3:CEK3-GUS, lines 4 and 21; and ProCEK3:CEK3-VEN, lines 18 and 24.
RNA Extraction and RT-qPCR
Total RNA was isolated from samples as follows: 14-d-old root and leaf, 25-d-old stem, inflorescence node, cauline leaf, and flowers in stages 1–14. RNA was extracted by TRI Reagent (AM9738, Invitrogen, Thermo Fisher Scientific) including DNase treatment, and cDNA was synthesized using the SuperScriptIII First-Strand Synthesis Kit (11752050, Invitrogen, Thermo Fisher Scientific). RT-qPCR was performed using the QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). The comparative threshold cycle method was used to determine relative gene expression, with the expression of ACT2 (KK129/KK130) as an internal control. Data are means ± sd from three biological replicates for each tissue sample, which involves three technical replicates. The primer sets for RT-qPCR are as reported (Nakamura et al., 2014).
Recombinant Protein Production and Enzyme Assay
Recombinant proteins of CEK2, CEK3, and CEK4 were produced, purified and used for Cho or Etn kinase activity assay as reported for CEK1 (Lin et al., 2019). For cloning of CEK2, CEK3, and CEK4, their open reading frames (1,053 bp, 1,041 bp, and 1,125 bp, respectively) were amplified by PCR from Arabidopsis cDNA, using Phusion high-fidelity DNA polymerase (M0530S, New England Biolabs) with the primer sets JL329/JL330, JL354/JL332, and JL333/JL334, respectively. PCR products were digested with restriction enzymes NdeI and BamHI for CEK2 and CEK4, and EcoRV and EcoRI for CEK3. The PCR fragments were ligated into the pMAL-c5x vector (N8108S, New England Biolabs) to produce pMAL-CEK2 (pJL103), pMAL-CEK3 (pJL104), and pMAL-CEK4 (pJL105) for expression of recombinant proteins N-terminally tagged with MBP in Escherichia coli strain C41 (DE3; 60442, Lucigen).
Radioisotope Pulse-Chase Labeling Assay
The pulse-chase labeling assay was conducted as follows: 14-d-old wild-type, cek1-1, cek2-1, cek3-1, cek1-1 cek3-1, and cek2-1 cek3-1 seedlings were labeled with either 192.4 kBq of [14C]Cho chloride (52 mCi/mmol, PerkinElmer) or 40.7 kBq of [14C]Etn (55 mCi/mmol, American Radiolabeled Chemicals) for 10 min; then the seedlings were washed twice with water and the labeling of metabolites was chased at 0, 5, 30, 60, and 180 min. The extraction and quantification of the labeled compounds from root or shoot are as previously described (Lin et al., 2019). For each sample, data are means ± sd from three biological replicates.
Histochemical GUS Staining
GUS staining of different tissues from various development stages was performed as previously described (Lin et al., 2015).
Confocal Microscopy
Subcellular localization of both ProCEK2:CEK2-VEN and ProCEK3:CEK3-VEN was observed by confocal microscopy (LSM 510 Meta; Carl Zeiss) equipped with Plan-Apochromat 20×/0.8-NA, and Plan-Apochromat 10×/0.45-NA. To stain the plasma membrane or the ER, samples were immersed in 5 μg mL−1 of FM4-64 (F34653, Thermo Fisher Scientific) for 5 min or 2 μm of the ER-Tracker Red dye (E34250, Thermo Fisher Scientific) for 30 min, respectively, before confocal observation. Images were captured using the LSM 510 version 3.2 (Carl Zeiss) with filters for Venus (514 nm laser, 520–555 nm band pass), FM4-64 (543 nm laser, 560–615 nm band pass), and ER-Tracker Red dye (543 nm laser, 560 nm long pass).
Root Cell Architecture Observation
The architecture of the root cells was observed by PI staining according to the previous report (Cruz-Ramírez et al., 2004) and was performed under the confocal microscope. Root meristem size was observed as previously described (Berger et al., 1998).
Phylogenetic Analysis of the CEK Family in Brassicaceae
Sequences of CEK homologs in Brassicaceae were acquired from the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/) and The Universal Protein Resource (https://www.uniprot.org/) using the protein BLAST search with Arabidopsis CEKs as queries. Arabidopsis CEK sequences used were according to The Arabidopsis Information Resource (http://www.arabidopsis.org/), including CEK1, 2, 4, and 3 splice variants of CEK3. Amino acid sequence alignment was performed using Multiple Sequence Comparison by Log-Expectation (MUSCLE; https://www.ebi.ac.uk/Tools/msa/muscle/). Four rounds of modifications according to the first round of structural modeling were performed as described (Sato, 2010). Jalview (Clamp et al., 2004) was used to remove the sites having >20% of gaps in sequences. The retrieved alignment (87 operational taxonomic unit, 380 sites) was used for estimating the optimal model on MEGA X (Kumar et al., 2018). The optimal model was determined with the lowest Bayesian Information Criterion score, as shown in Supplemental Dataset S1. The tree was estimated on raxmlGUI 2.0 (Edler et al., 2019) using a maximum likelihood statistical method with 1,000 replications of bootstrap. The alignment files used for phylogenetic analysis are shown in Supplemental Dataset S2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g71697 (CEK1), At1g74320 (CEK2), At4g09760 (CEK3), and At2g26830 (CEK4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. In vivo radioactive pulse-chase analysis for the metabolism of Etn in the shoots of cek1-1, cek2-1, and cek3-1 mutants.
Supplemental Figure S2. In vivo radioactive pulse-chase analysis for the metabolism of Etn in the roots of cek1-1, cek2-1, and cek3-1 mutants.
Supplemental Figure S3. Tissue-specific expression of CEK2-GUS by histochemical GUS staining of transgenic plants harboring ProCEK2:CEK2-GUS (line 22).
Supplemental Figure S4. Tissue-specific expression of CEK3-GUS by histochemical GUS staining of transgenic plants harboring ProCEK3:CEK3-GUS (line 21).
Supplemental Figure S5. Transcript levels of CEK1, CEK2 and CEK3 in 14-d-old wild-type, cek1-1, cek2-1, cek3-1, cek1-1 cek3-1, and cek2-1 cek3-1 seedlings.
Supplemental Figure S6. Cell type-specific expression pattern and subcellular localization in the seedling roots of transgenic plants harboring ProCEK2:CEK2-VEN (line 19) or ProCEK3:CEK3-VEN (line 18).
Supplemental Table S1. List of oligonucleotide sequences used in this study.
Supplemental Dataset S1. Maximum Likelihood fits of 56 different amino acid substitution models.
Supplemental Dataset S2. Sequences of CEK homologs in Brassicaceae used for phylogenetic analysis.
Acknowledgments
We thank Pannada Kungwon and Lydia Hangelmann for making DNA constructs.
Footnotes
This work was supported by the Ministry of Science and Technology, Taiwan (grant nos. 107–2628–B–001–001 and 108–2628–B–001–011 to Y.N).
Articles can be viewed without a subscription.
References
- Angkawijaya AE, Nakamura Y(2017) Arabidopsis PECP1 and PS2 are phosphate starvation-inducible phosphocholine phosphatases. Biochem Biophys Res Commun 494: 397–401 [DOI] [PubMed] [Google Scholar]
- Aoyama C, Liao H, Ishidate K(2004) Structure and function of choline kinase isoforms in mammalian cells. Prog Lipid Res 43: 266–281 [DOI] [PubMed] [Google Scholar]
- Arlauckas SP, Popov AV, Delikatny EJ(2016) Choline kinase α—Putting the ChoK-hold on tumor metabolism. Prog Lipid Res 63: 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Hung C-Y, Dolan L, Schiefelbein J(1998) Control of cell division in the root epidermis of Arabidopsis thaliana. Dev Biol 194: 235–245 [DOI] [PubMed] [Google Scholar]
- Brophy PJ, Choy PC, Toone JR, Vance DE(1977) Choline kinase and ethanolamine kinase are separate, soluble enzymes in rat liver. Eur J Biochem 78: 491–495 [DOI] [PubMed] [Google Scholar]
- Chen W, Salari H, Taylor MC, Jost R, Berkowitz O, Barrow R, Qiu D, Branco R, Masle J(2018) NMT1 and NMT3 N-methyltransferase activity is critical to lipid homeostasis, morphogenesis, and reproduction. Plant Physiol 177: 1605–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Taylor MC, Barrow RA, Croyal M, Masle J(2019) Loss of phosphoethanolamine N-methyltransferases abolishes phosphatidylcholine synthesis and is lethal. Plant Physiol 179: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ(2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Cornell RB, Ridgway ND(2015) CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog Lipid Res 59: 147–171 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, González-Ortega E, Herrera-Estrella L(2004) The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler D, Klein J, Antonelli A, Silvestro D (2019) raxmlGUI 2.0 β: A graphical interface and toolkit for phylogenetic analyses using RAxML. bioRxiv 800912 doi: 10.1101/800912 [Google Scholar]
- Glunde K, Bhujwalla ZM, Ronen SM(2011) Choline metabolism in malignant transformation. Nat Rev Cancer 11: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchi M, Thibaud MC, Légeret B, Kuwata K, Pochon N, Beisson F, Cao A, Cuyas L, David P, Doerner P, et al. (2018) The phosphate fast-responsive genes PECP1 and PPsPase1 affect phosphocholine and phosphoethanolamine content. Plant Physiol 176: 2943–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Kodaki T, Yamashita S(1989) Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli. J Biol Chem 264: 2053–2059 [PubMed] [Google Scholar]
- Ishidate K, Furusawa K, Nakazawa Y(1985a) Complete co-purification of choline kinase and ethanolamine kinase from rat kidney and immunological evidence for both kinase activities residing on the same enzyme protein(s) in rat tissues. Biochim Biophys Acta 836: 119–124 [PubMed] [Google Scholar]
- Ishidate K, Iida K, Tadokoro K, Nakazawa Y(1985b) Evidence for the existence of multiple forms of choline (ethanolamine) kinase in rat tissues. Biochim Biophys Acta 833: 1–8 [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A(2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim K, Kim KH, Storey MK, Voelker DR, Carman GM(1999) Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J Biol Chem 274: 14857–14866 [DOI] [PubMed] [Google Scholar]
- Kim KH, Voelker DR, Flocco MT, Carman GM(1998) Expression, purification, and characterization of choline kinase, product of the CKI gene from Saccharomyces cerevisiae. J Biol Chem 273: 6844–6852 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K(2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Kanehara K, Nakamura Y(2019) Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol 223: 1904–1917 [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Liu YC, Nakamura Y(2015) The choline/ethanolamine kinase family in Arabidopsis: Essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27: 1497–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Lin Y-C, Kanehara K, Nakamura Y(2018) A pair of phospho-base methyltransferases important for phosphatidylcholine biosynthesis in Arabidopsis. Plant J 96: 1064–1075 [DOI] [PubMed] [Google Scholar]
- Liu YC, Lin Y-C, Kanehara K, Nakamura Y(2019) A methyltransferase trio essential for phosphatidylcholine biosynthesis and growth. Plant Physiol 179: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykidis A, Wang J, Karim MA, Jackowski S(2001) Overexpression of a mammalian ethanolamine-specific kinase accelerates the CDP-ethanolamine pathway. J Biol Chem 276: 2174–2179 [DOI] [PubMed] [Google Scholar]
- Macher BA, Mudd JB(1976) Partial purification and properties of ethanolamine kinase from spinach leaf. Arch Biochem Biophys 177: 24–30 [DOI] [PubMed] [Google Scholar]
- May A, Spinka M, Köck M(2012) Arabidopsis thaliana PECP1: Enzymatic characterization and structural organization of the first plant phosphoethanolamine/phosphocholine phosphatase. Biochim Biophys Acta 1824: 319–325 [DOI] [PubMed] [Google Scholar]
- McMaster CR.(2018) From yeast to humans—roles of the Kennedy pathway for phosphatidylcholine synthesis. FEBS Lett 592: 1256–1272 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Nakamura M, Nishida I(2006) Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks DE, Goode JH, Dewey RE(1996) Characterization of soybean choline kinase cDNAs and their expression in yeast and Escherichia coli. Plant Physiol 110: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F(1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamura Y, Teo NZW, Shui G, Chua CHL, Cheong W-F, Parameswaran S, Koizumi R, Ohta H, Wenk MR, Ito T(2014) Transcriptomic and lipidomic profiles of glycerolipids during Arabidopsis flower development. New Phytol 203: 310–322 [DOI] [PubMed] [Google Scholar]
- Ngo AH, Kanehara K, Nakamura Y(2019) Non-specific phospholipases C, NPC2 and NPC6, are required for root growth in Arabidopsis. Plant J 100: 825–835 [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Ramaswami M, Tanouye MA(1994) The Drosophila easily shocked gene: A mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79: 23–33 [DOI] [PubMed] [Google Scholar]
- Peisach D, Gee P, Kent C, Xu Z(2003) The crystal structure of choline kinase reveals a eukaryotic protein kinase fold. Structure 11: 703–713 [DOI] [PubMed] [Google Scholar]
- Porter TJ, Kent C(1990) Purification and characterization of choline/ethanolamine kinase from rat liver. J Biol Chem 265: 414–422 [PubMed] [Google Scholar]
- Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu W-I, Voelker DR, Basset G, Hanson AD(2001) Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276: 35523–35529 [DOI] [PubMed] [Google Scholar]
- Sato N.(2010) Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol Phylogenet Evol 56: 176–186 [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A(2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 28: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Tolbert NE, Gohlke AF(1966) Choline kinase and phosphorylcholine phosphatase in plants. Plant Physiol 41: 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseva G, Richard L, Zachowski A(2004) Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett 566: 115–120 [DOI] [PubMed] [Google Scholar]
- Toräng P, Wunder J, Obeso JR, Herzog M, Coupland G, Ågren J(2015) Large-scale adaptive differentiation in the alpine perennial herb Arabis alpina. New Phytol 206: 459–470 [DOI] [PubMed] [Google Scholar]
- Uchida T.(1997) A novel high-molecular mass mammalian ethanolamine kinase. Biochim Biophys Acta 1349: 13–24 [DOI] [PubMed] [Google Scholar]
- Upreti RK.(1978) Search for sex-dependent and gestation-induced changes in choline and ethanolamine phosphorylating activities. Experientia 34: 166–167 [DOI] [PubMed] [Google Scholar]
- Upreti RK.(1981) Choline and ethanolamine phosphorylation in rodents as a function of age. Age (Omaha) 4: 52–56 [Google Scholar]
- Wharfe J, Harwood JL(1979a) Lipid metabolism in germinating seeds. Purification of ethanolamine kinase from soya bean. Biochim Biophys Acta 575: 102–111 [DOI] [PubMed] [Google Scholar]
- Wharfe J, Harwood JL(1979b) Purification of choline kinase from soya bean In Appelqvist LA, and Liljenberg C, eds, Advances in the Biochemistry and Physiology of Plant Lipids. Elsevier, Amsterdam, pp 443–447 [Google Scholar]
- Willing EM, Rawat V, Mandáková T, Maumus F, James GV, Nordström KJ, Becker C, Warthmann N, Chica C, Szarzynska B, et al. (2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants 1: 14023. [DOI] [PubMed] [Google Scholar]
- Wittenberg J, Kornberg A(1953) Choline phosphokinase. J Biol Chem 202: 431–444 [PubMed] [Google Scholar]
- Wu G, Aoyama C, Young SG, Vance DE(2008) Early embryonic lethality caused by disruption of the gene for choline kinase α, the first enzyme in phosphatidylcholine biosynthesis. J Biol Chem 283: 1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Vance DE(2010) Choline kinase and its function. Biochem Cell Biol 88: 559–564 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Hosaka K(1997) Choline kinase from yeast. Biochim Biophys Acta 1348: 63–69 [DOI] [PubMed] [Google Scholar]
- Yunus IS, Liu YC, Nakamura Y(2016) The importance of SERINE DECARBOXYLASE1 (SDC1) and ethanolamine biosynthesis during embryogenesis of Arabidopsis thaliana. Plant J 88: 559–569 [DOI] [PubMed] [Google Scholar]