A nitrate transport accessory protein interacts with two nitrilase proteins to regulate root-growth responses to nitrate and ammonium supply.
Abstract
The nitrate transport accessory protein OsNAR2 plays a critical role in root-growth responses to nitrate and nitrate acquisition in rice (Oryza sativa). In this study, a pull-down assay combined with yeast two-hybrid and coimmunoprecipitation analyses revealed that OsNAR2.1 interacts with OsNIT1 and OsNIT2. Moreover, an in vitro nitrilase activity assay indicated that indole-3-acetonitrile (IAN) is hydrolyzed to indole-3-acetic acid (IAA) by OsNIT1, the activity of which was enhanced 3- to 4-fold by OsNIT2 and in excess of 5- to 8-fold by OsNAR2.1. Knockout (KO) of OsNAR2.1 was accompanied by repressed expression of both OsNIT1 and OsNIT2, whereas KO of OsNIT1 and OsNIT2 in the osnit1 and osnit2 mutant lines did not affect expression of OsNAR2.1 or the root nitrate acquisition rate. osnit1 and osnit2 displayed decreased primary root length and lateral root density. Double KO of OsNAR2.1 and OsNIT2 caused further decreases in lateral root density under nitrate supply. Ammonium supply repressed OsNAR2.1 expression whereas it upregulated OsNIT1 and OsNIT2 expression. Both osnit1 and osnit2 showed root growth hypersensitivity to external ammonium; however, less root growth sensitivity to external IAN, higher expression of three IAA-amido synthetase genes, and a lower rate of 3H-IAA movement toward the roots were observed. Taken together, we conclude that the interaction of OsNIT1 and OsNIT2 activated by OsNAR2.1 and nitrogen supply is essential for maintaining root growth possibly via altering the IAA ratio of free to conjugate forms and facilitating its transportation.
Plants display high root growth plasticity in primary root (PR) elongation, particularly lateral root (LR) initiation and elongation, to adapt to variation in nutrient and water supply (Drew and Saker, 1975; Gojon et al., 2011; Gruber et al., 2013). Two major forms of nitrogen (N) available for plant root uptake are nitrate (NO3−) and ammonium (NH4+). Nitrate is the major form of N available for dry land crop species and is also an important source of N for paddy rice (Oryza sativa; Kirk and Kronzucker, 2005; Li et al., 2008; Xu et al., 2012). Moreover, nitrate serves as a signaling molecule in various plant developmental processes including the determination of root architecture (Perilli et al., 2012; Vidal et al., 2013; Wierzba and Tax, 2013; O’Brien et al., 2016). In Arabidopsis (Arabidopsis thaliana), several key molecules have been identified that are involved in the regulation of LR initiation and development by localized nitrate, such as the transcription factor ANR1 (Zhang and Forde, 1998), the nitrate transceptor (NRT1.1; Krouk et al., 2010), miR393, the auxin receptor AFB3, and their downstream target NAC4 (Vidal et al., 2010; 2013). The nac4 mutation results in altered LR growth but not PR growth in response to nitrate (Vidal et al., 2010). Low N in roots represses the accumulation of auxin that controls LR formation (Vidal et al., 2010; Wang et al., 2019). A gradual reduction in auxin levels is closely related to enhanced differentiation of distal stem cells in root tips in response to low nitrate (Wang et al., 2019). Moreover, NRT1.1 protein behaves like a root auxin transporter under low nitrate supply (Krouk et al., 2010).
Unlike nitrate, ammonium commonly mediates inhibition of both PR and LR elongation in most plant species (Li et al., 2010, 2014; Rogato et al., 2010). The presence of ammonium systematically alters root development, affecting processes including elongation, gravitropism, and LR branching (Lima et al., 2010; Li et al., 2014). The root lengths of the auxin-resistant mutants aux1, axr1, and axr2 are less affected by high levels of ammonium compared with wild type, indicating that auxin may be involved in ammonium-induced root length reduction (Cao et al., 1993). However, several lines of evidence have suggested that the root growth reduction induced by ammonium supply is associated with ammonium efflux in the root elongation zone, independent of the auxin pathways (Li et al., 2010; Liu et al., 2013). The combinatorial effect of nitrate on ammonium-regulated root system architecture suggests that ammonium regulates PR elongation and LR branching by distinct pathways (Liu and von Wirén, 2017).
The main form of plant auxin is indole-3-acetic acid (IAA), for which several pathways for Trp-dependent IAA biosynthesis have been proposed in plants (Woodward and Bartel, 2005; Sugawara et al., 2009; Korasick et al., 2013; Abu-Zaitoon, 2014). Indole-3-acetonitrile (IAN) has been identified as one of two key intermediates in indole-3-acetaldoxime–mediated IAA biosynthesis (Sugawara et al., 2009). IAN conversion during basal IAA production requires nitrilases (NITs) for the hydrolysis of nitriles (Müller et al., 1998; Park et al., 2003; Lehmann et al., 2017). NIT isogenes have been identified in almost all the major plant families (Piotrowski, 2008). For example, the relatively small nitrilase family in Arabidopsis, comprising four members (NIT1–NIT4), has gained diverse biological functions in nitrile metabolism (Piotrowski, 2008). Two groups of nitrilases that catalyze the conversion of IAN to IAA, namely NIT1 in Arabidopsis and ZmNIT2 in maize (Zea mays; NIT4 orthologs of Arabidopsis), have been characterized thus far. Mutation of NIT1 was shown to decrease both plant sensitivity to IAN treatment (Normanly et al., 1997) and total IAA concentration without affecting the concentration of free IAA (Lehmann et al., 2017). Overexpression of NIT1 resulted in drastic changes of both free IAA and IAN levels, resulting in a phenotype characterized by shorter PR and increased LR number (Lehmann et al., 2017). Maize kernels contain an endogenous NIT with activity toward the substrate IAN (Park et al., 2003). ZmNIT2 can hydrolyze IAN to IAA with 7- to 20-fold higher activity than AtNIT1, AtNIT2, and AtNIT3 (Park et al., 2003).
As opposed to IAA production, the proposed major role of the NIT4 subfamily of angiosperms is catalyzing the conversion of β-cyano-Ala to Asp and Asn for detoxification of hydrogen cyanide (Piotrowski et al., 2001; Jenrich et al., 2007; Piotrowski, 2008). Poaceae possesses two different NIT4 orthologs (NIT4A and NIT4B). The rice genome contains two NIT4 orthologs, namely OsNIT4A and OsNIT4B that were renamed as OsNIT1 and OsNIT2 by Ding et al. (2008), which are investigated in this study. Changes in OsNIT1 and OsNIT2 expression are similar to the changes in total IAA after bacterial infection (Ding et al., 2008), indirectly suggesting that OsNIT1 and OsNIT2 affect IAA accumulation. However, Sugawara et al. (2009) detected the presence of IAN in Arabidopsis, but not in the seedlings of maize, rice, or tobacco (Nicotiana tabacum). These data suggest a species- or organ-specific contribution of NIT to IAA production in plants.
Nitrate Assimilation Related family (NAR2) proteins have been considered as important components of NRTs in plants (Tong et al., 2005). In Arabidopsis, six of the seven AtNRT2 members require AtNAR2.1 as a component of high-affinity NRTs (Kotur et al., 2012). Disruption of AtNAR2.1 causes an almost complete loss of inducible high-affinity nitrate influx (Orsel et al., 2006). In rice, OsNAR2.1 interacts with OsNRT2.1, OsNRT2.12.2, and OsNRT2.12.3a and plays a broad role in root acquisition of nitrate in response to both low- and high-nitrate supply (Feng et al., 2011; Yan et al., 2011; Liu et al., 2014). Knockout (KO) of OsNAR2.1 causes reduced LR density and PR growth in nitrate medium (Huang et al., 2015). Notably, osnar2.1 mutants display decreased auxin distribution in the roots and the accompanying root phenotype can be largely complemented by external naphthylacetic acid (NAA; Huang et al., 2015), suggesting that OsNAR2.1 functions in both nitrate uptake and auxin-mediated nitrate signaling. Therefore, the function of the OsNAR2.1 regulatory pathway in plant auxin signaling is an intriguing question.
In this study, through a pull-down assay combined with yeast-two-hybrid (Y2H) and coimmunoprecipitation (Co-IP) analyses, we show that OsNAR2.1 directly interacts with the two NIT proteins OsNIT1 and OsNIT2 in rice. In addition to demonstrating that OsNIT1 and OsNIT2 potentially encode IAN hydrolysis enzymes, we provide a number of physiological, biochemical, and genetic lines of evidence indicating that OsNAR2.1 contributes to auxin-responsive, nitrate-regulated root growth via activation of OsNIT1 and OsNIT2.
RESULTS
OsNAR2.1 Interacts with OsNIT1 and OsNIT2 in Roots under Nitrate Supply
We first conducted a preliminary pull-down assay of nitrate-treated seedlings of rice wild type (‘Nipponbare’) using an OsNAR2.1-GST tag fusion protein (Supplemental Fig. S1A). The extracted proteins revealed several known functional proteins that potentially interact with OsNAR2.1 (Supplemental Table S1). Considering that OsNAR2.1 is involved in root responses to nitrate and auxin (Huang et al., 2015), we focused on two candidate NITs (EC 3.5.5.1), as nitrilase is the enzyme involved in IAA synthesis and detoxification in Arabidopsis and maize (Piotrowski, 2008). The predicted genes encoding NIT in the rice genome are OsNIT1 (OsNIT4A, LOC_Os02g42350) and OsNIT2 (OsNIT4B, LOC_Os02g42330), which are orthologs of Arabidopsis AtNITs and maize ZmNITs (Jenrich et al., 2007; Piotrowski, 2008).
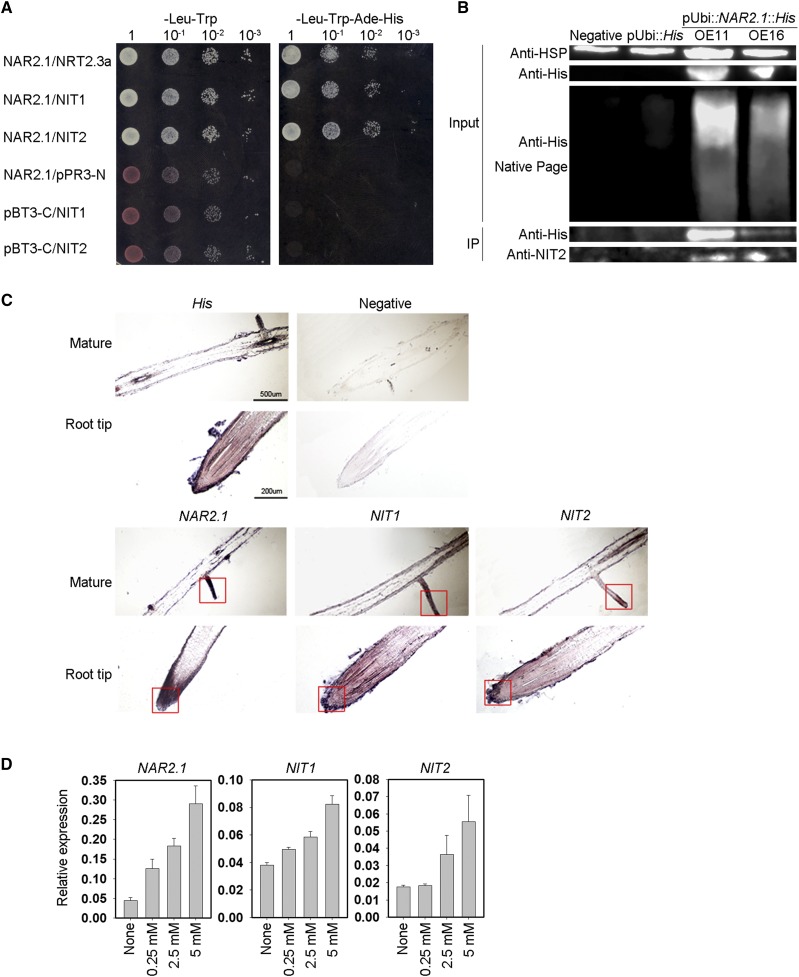
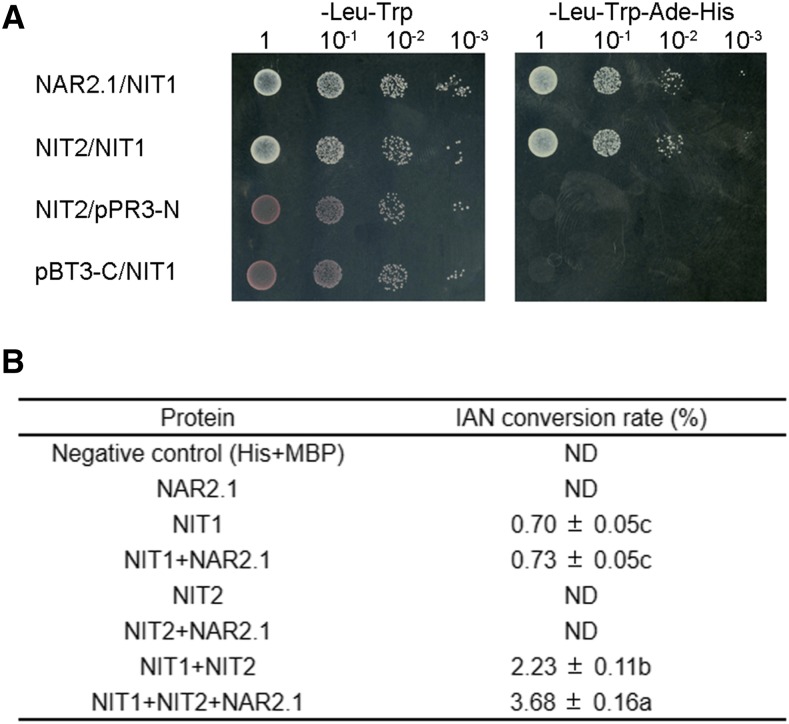
We subsequently tested for interaction between OsNAR2.1 and OsNIT1/2 through Y2H assays (Fig. 1A) as well as Co-IP assays (Fig. 1B) and mass spectrometry analysis (Supplemental Fig. S1, B and 1C). The results of these analyses confirmed that OsNAR2.1 interacts with OsNIT1 and OsNIT2 at the protein level.
Figure 1.
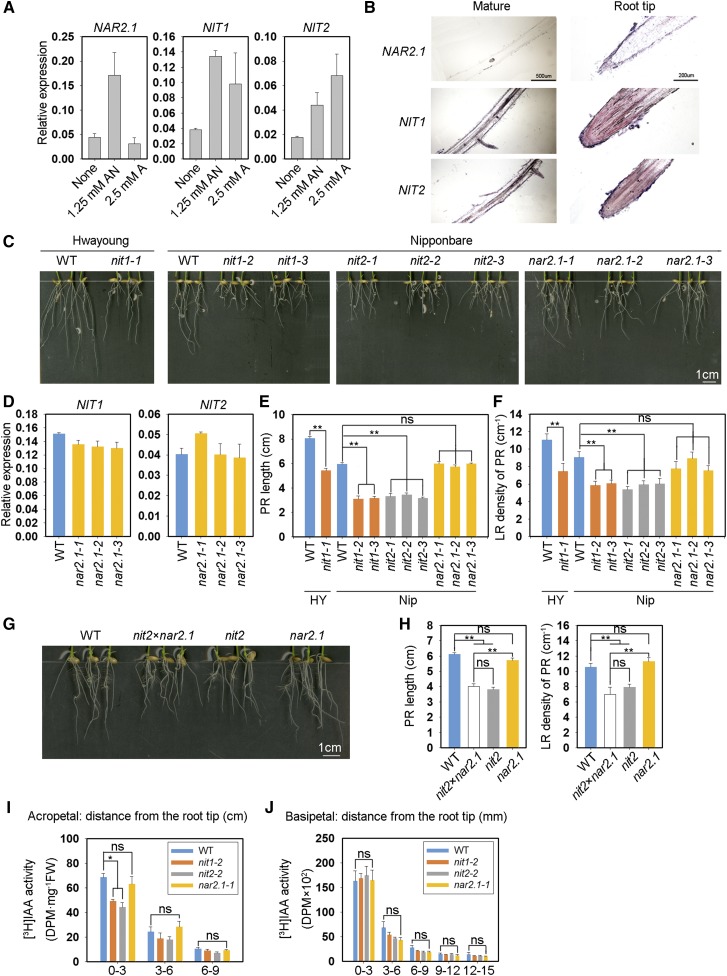
OsNAR2.1 interacts with OsNIT1 and OsNIT2 after nitrate supply to roots. A, Interaction test of OsNAR2.1 with OsNIT1 and OsNIT2 using the DUAL Membrane Pairwise Interaction Kit (Dualsystems Biotech). B, Interaction test of OsNAR2.1 with OsNIT1 and OsNIT2 using Co-IP. OE11 and OE16, two independent lines overexpressing OsNAR2.1 with 6×His tag; Negative, control line segregated from OsNAR2.1 with 6×His tag overexpression lines; pUbi::His, control line overexpressing 6×His tag only. Immunoblots were developed with anti-His antibody to detect OsNAR2.1 expression, and with anti-NIT2 to detect OsNIT2 expression. Anti-HSP used as positive control. C and D, Analysis of OsNAR2.1, OsNIT1, and OsNIT2 expression patterns in the roots of rice seedings (‘Nipponbare’) supplied with different concentrations of Ca(NO3)2. C, Expression patterns of OsNAR2.1, OsNIT1, and OsNIT2 revealed by RNA in situ hybridization in root tip and mature root area supplied with 2.5 mm of nitrate. All detection probes were based on the whole antisense strand. Negative control materials were incubated with water. D, Relative expression levels of OsNAR2.1, OsNIT1, and OsNIT2 in the roots treated with different nitrate concentrations as quantified by RT-qPCR. OsActin (LOC_Os03g50885) was used as an internal control. The values in (D) represent means ± se of three biological replicates.
To further confirm the interaction of these proteins in rice, we detected their tissue localization under the same experimental conditions. In situ hybridization analysis showed that OsNIT1, OsNIT2, and OsNAR2.1 were abundantly expressed in root tips and LRs (Fig. 1C). We have previously shown that OsNAR2.1 is expressed mainly in the roots, especially in root tips and LRs under nitrate supply (Feng et al., 2011). In this study, we found that OsNIT1, OsNIT2, and OsNAR2.1 were each transcriptionally upregulated in the roots by nitrate supply, and that gene expression induction was further enhanced by increasing nitrate supply (Fig. 1D). In addition, subcellular localization analysis showed that OsNIT1 and OsNIT2 had a wide cellular distribution (Supplemental Fig. S2), which is similar to the localization of OsNAR2.1 (Liu et al., 2014).
KO of OsNIT1 or OsNIT2 Results in the Same Root Phenotype as the osnar2.1 Mutant under Nitrate Supply
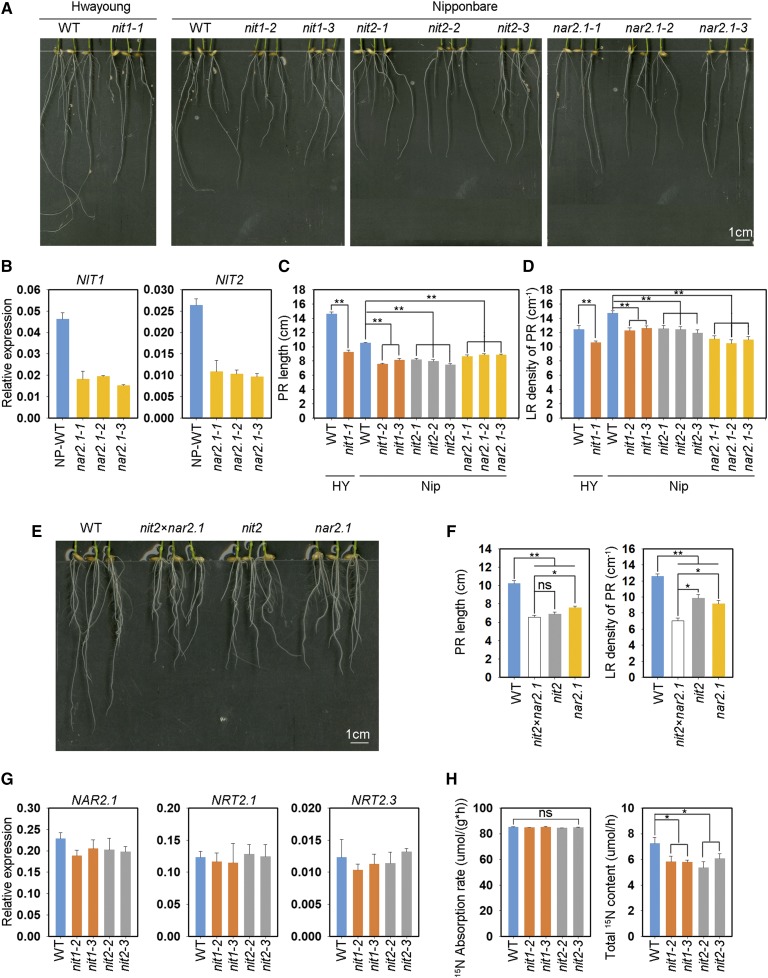
To examine the roles of OsNIT1 and OsNIT2 in root-growth responses to nitrate, we examined the root phenotypes of osnit1 and osnit2 mutants generated by CRISPR-Cas9 editing and transfer DNA (T-DNA) insertional mutagenesis. Three independent mutant lines for each gene were selected for detailed analysis (Supplemental Fig. S3). It has been shown that osnar2.1 exhibits a phenotype of short PRs and low root density under low nitrate supply (Huang et al., 2015). In this study, we observed that the root phenotypes of osnit1, osnit2, and osnar2.1 mutants in the Nipponbare background were similar under 0.25 mm nitrate supply. Under nitrate supply, inactivation of either OsNAR2.1, OsNIT1, or OsNIT2 decreased PR growth by 15% to 25% and LR density by 16% to 26% (Figs. 2A, 2C, and 2D). OsNIT1 KO in the Hwayoung background (osnit1-1) also resulted in significantly reduced PR length and LR density. Moreover, the expression levels of OsNIT1 and OsNIT2 in the roots of the osnar2.1 mutant line were largely repressed under nitrate supply (Fig. 2B) relative to their expression levels in wild type, indicating that OsNAR2.1 is necessary for nitrate-regulated expression of OsNIT1 and OsNIT2 in rice roots.
Figure 2.
KO of OsNIT1 or OsNIT2 results in the same root phenotype as the osnar2.1 mutant after nitrate supply, but does not alter root nitrate uptake rate. A to D, Seeds were first germinated in deionized water for 3 d, then were transferred to hydroponic media containing 0.125 mm Ca(NO3)2 for a further 7 d. HY, cv Hwayoung; Nip, cv Nipponbare; nit1-1, the KO mutant of OsNIT1 in the cv Hwayoung genetic background; nit1-2, nit1-3, nit2-1, nit2-2, nit2-3, nar2.1-1, nar2.1-2, and nar2.1-3, individual KO mutant lines of OsNIT1, OsNIT2, and OsNAR2.1 genes in the cv Nipponbare genetic background. A, Root phenotype of different lines. B, Effect of OsNAR2.1 KO on expression levels of OsNIT1 and OsNIT2 in roots analyzed by RT-qPCR. OsActin was used as an internal control. The values in (B to D) represent means ± se of three (B) and six (C and D) biological replicates, respectively (**P ≤ 0.01). E and F, Seeds were germinated and grown under the same conditions as in (A). nit2×nar2.1, homozygous hybrid of nit2-2 (abbreviated as nit2 in E and F) and nar2.1-1 (abbreviated as nar2.1 in E and F). The values in (F) represent means ± se of seven biological replicates (*P ≤ 0.05 and **P ≤ 0.01). G, Relative expression levels of OsNAR2.1, OsNRT2.1, and OsNRT2.3 in the roots of cv Nipponbare wild-type and the OsNIT1 and OsNIT2 KO mutants supplied with 1.25 mm Ca(NO3)2 for 1 week. H, The 15N absorption rate per root unit weight and total 15N content of wild-type (WT) plants and osnit1 and osnit2 mutants supplied with 0.125 mm Ca(15NO3)2 for 5 min. The values represent means ± se of three biological replicates (*P ≤ 0.05); ns, not significant.
To further confirm that OsNIT1 and OsNIT2 share the same function as OsNAR2.1 in root responses to nitrate, we crossed osnit2-2 and osnar2.1-1 and obtained a homozygous hybrid mutant line. Interestingly, under nitrate supply, double KO of OsNIT2 and OsNAR2.1 resulted in similar PR length inhibition as seen in osnar2.1, whereas considerably reduced LR density was observed compared to osnar2.1 or osnit2 (Figs. 2E and 2F). For unknown reasons, we could not isolate a double mutant line of osnit1 and osnar2.1 in this study. The additive effect of OsNIT2 on OsNAR2.1 function in nitrate regulation of root growth supports an interaction between OsNAR2.1 and OsNIT2, and possibly OsNIT1 (Fig. 1).
KO of OsNIT1 and OsNIT2 Does Not Alter Nitrate Uptake Rate per Root Unit Weight
We previously reported that inactivation of OsNAR2.1 limits root nitrate acquisition under a broad range of nitrate supply, which is the result of OsNAR2.1 being an essential partner protein for the function of several OsNRT2 NRTs (Yan et al., 2011). Therefore, we also investigated the roles of OsNIT1 and OsNIT2 in OsNAR2.1 expression and root nitrate uptake. In contrast to the inactivation of OsNAR2.1, which led to the repression of OsNIT1 and OsNIT2 expression (Fig. 2B), no significant changes in expression levels of OsNAR2.1, OsNRT2.1, or OsNRT2.3 were observed in either osnit1 or osnit2 mutants (Fig. 2G). Moreover, KO of OsNIT1 did not affect the expression of OsNIT2 and vice versa (Supplemental Fig. S4), indicating that both OsNIT1 and OsNIT2 are downstream components of the OsNAR2.1 regulatory pathway. We further determined that the root unit weight 15NO3− influx rate of osnit1 and osnit2 lines was comparable with wild type in a short-term (5 min) assay (Fig. 2H), even though the total amount of 15NO3− per plant in osnit1 and osnit2 was decreased by ∼20% due to the smaller root size of the mutants (Fig. 2H). These results indicate that OsNAR2.1 is upstream of OsNIT1 and OsNIT2 in a nitrate signaling pathway, and that OsNIT1 and OsNIT2 interaction with OsNAR2.1 does not directly contribute to root nitrate uptake.
KO of OsNIT1 or OsNIT2 Decreases Root Growth Sensitivity to IAN, But Not to NAA
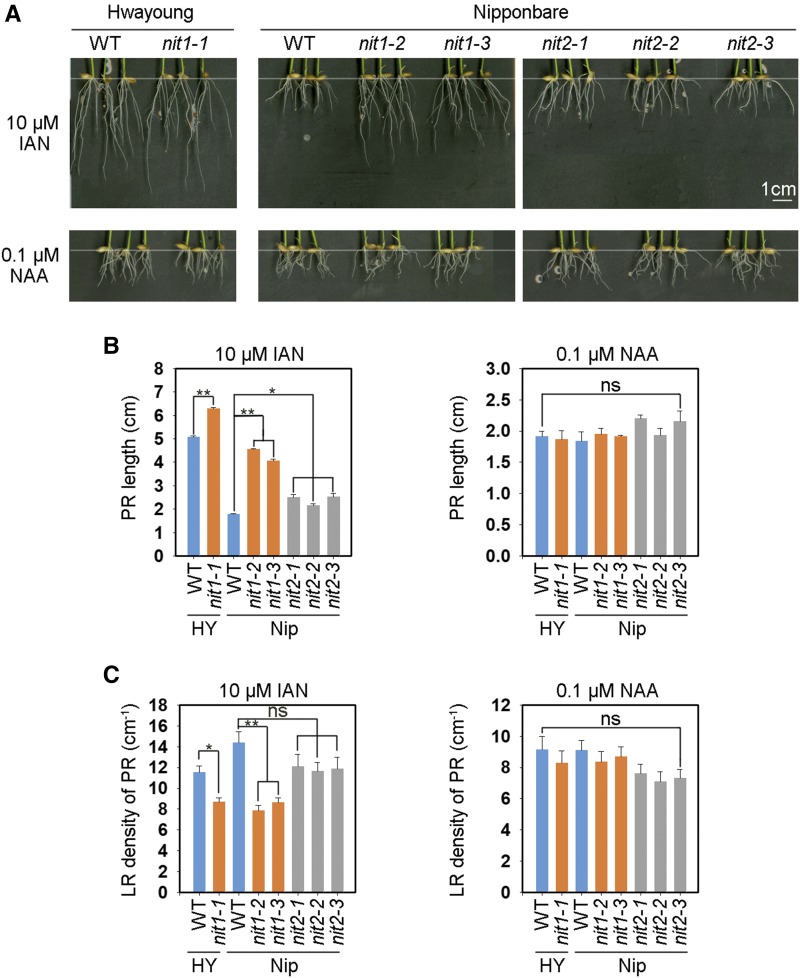
Because the presence and contribution of IAN to IAA synthesis in rice is questionable (Sugawara et al., 2009), we analyzed the effect of IAN and naphthylacetic acid (NAA, an IAA analog) on rice root growth. In wild-type plants, PR length was slightly stimulated by low (0.01 μm) IAN treatment but inhibited by high (10 μm) IAN and 0.1-μm NAA treatment, whereas LR density and the expression levels of OsNIT1 and OsNIT2 did not display distinct sensitivity to low external IAN and NAA treatment (Supplemental Fig. S5, A–C).
The KO mutants of OsNIT1 showed less sensitivity to external IAN in both the cv Nipponbare and cv Hwayoung backgrounds, and exhibited phenotypes of longer PRs and reduced LR density (Fig. 3). The PR length and LR density in osnit1 was ∼140% longer and 40% less, respectively, than its cv Nipponbare wild type (Fig. 3, B and C). The osnit2 mutants showed a similar phenotype regarding changes in PR length as osnit1, but the effect was less substantial. KO of OsNIT1 or OsNIT2 did not affect root responses to external NAA (Fig. 3, B and C). These data indicate that IAN, which can be hydrolyzed by NIT proteins, may play a regulatory role in rice root growth.
Figure 3.
KO of OsNIT1 or OsNIT2 decreases root growth sensitivity to IAN, but not to NAA. A, Seeds were first germinated in deionized water for 3 d, then were transferred to the hydroponic media containing 0.125 mm Ca(NO3)2 with 10 μm IAN or 0.1 μm NAA for another 7 d before being photographed and sampled for the measurement of root growth and LR density. The genotypes are the same as those used and described in Figure 2A. The data in (B) and (C) represent means ± se of six biological replicates (*P ≤ 0.05 and **P ≤ 0.01); ns, not significant; WT, wild type.
KO of OsNIT1 or OsNIT2 Decreases Acropetal IAA Transport, But Does Not Change Root Total IAA Concentration
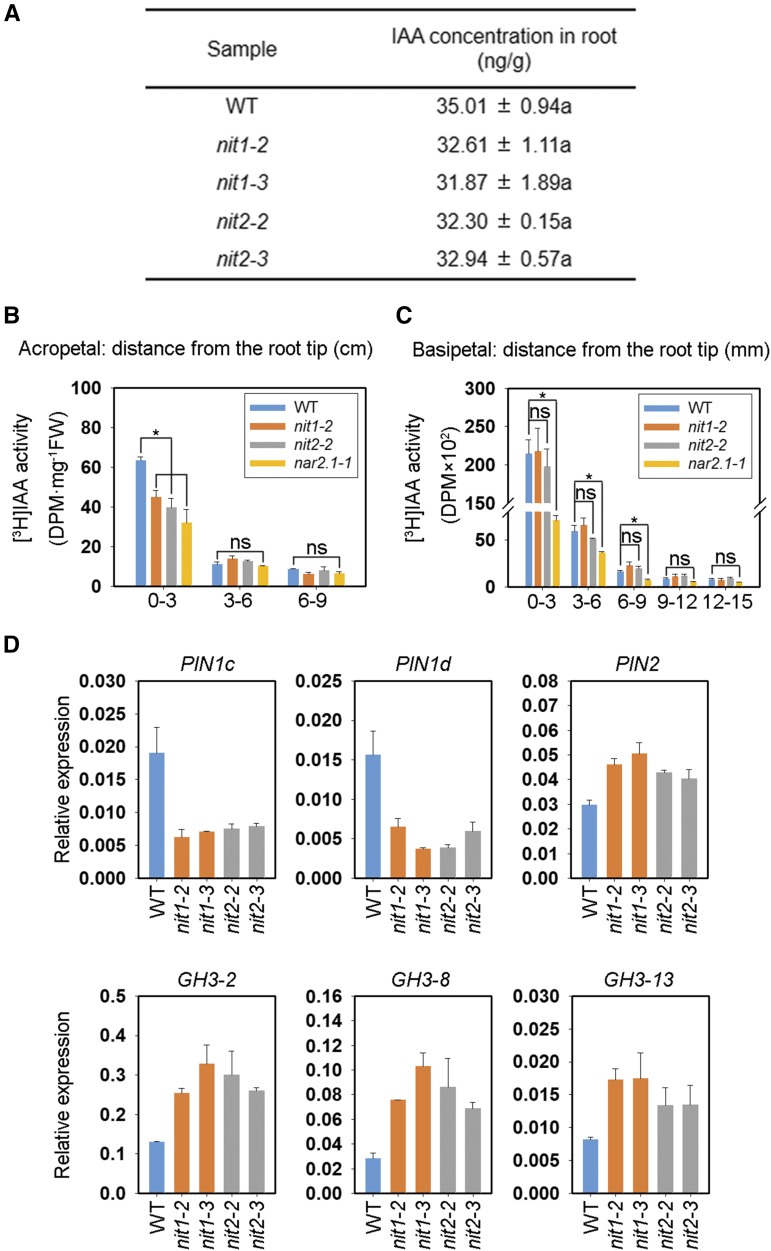
In view of the substantial responses of rice root growth to external IAN and that both OsNIT1 and OsNIT2 were shown to be involved in these responses, we attempted to analyze the effect of OsNIT1 or OsNIT2 KO on IAN and IAA concentration in rice. We did not detect IAN in wild-type cv Nipponbare seedlings (Supplemental Fig. S6A), indicating that IAN was either not present, present at levels below our detection limits, or that IAN is rapidly hydrolyzed in rice. In addition, the total concentration of IAA in the roots of osnit1 and osnit2 mutants and wild type was similar (Fig. 4A), indicating that OsNIT1 and OsNIT2 are not major influential factors for total IAA synthesis or accumulation in rice seedlings.
Figure 4.
KO of OsNIT1 or OsNIT2 alters expression of auxin efflux transporter OsPIN genes and IAA amido synthetase OsGH3 genes, and decreases acropetal IAA transport, but does not affect root total IAA concentration. A and D, Wild-type (WT; cv Nipponbare), osnit1, and osnit2 plants were grown in IRRI solution containing 1.25 mm Ca(NO3)2 for one week before being sampled for gene expression and IAA concentration analyses. A, Total IAA concentration in the roots. The roots of five seedlings were taken and mixed as one sample. B and C, wild-type, osnit1, osnit2, and osnar2.1 plants were grown in IRRI solution containing 1.25 mm Ca(NO3)2 for 10 d before being sampled for [3H]IAA transport analysis. B, Acropetal transport. FW, fresh weight. C, Basipetal transport. D, Relative expression levels of auxin efflux transporter OsPIN genes and IAA amido synthetase OsGH3 genes quantified by RT-qPCR. OsActin was used as an internal control. Data represent means ± se of three biological replicates (Asterisk represents statistically significant difference at P ≤ 0.05 level). ns, not significant.
We also compared [3H+]-IAA transport in the roots of wild-type and mutant plants under nitrate supply. Notably, as shown by the amounts of [3H+]-IAA in 0–3 cm root-tip sections, KO of OsNAR2.1 significantly restricted both root-ward and shoot-ward movement of [3H+]-IAA (Fig. 4, B and C), whereas KO of OsNIT1 or OsNIT2 caused inhibition of [3H+]-IAA root-ward movement alone (Fig. 4, B and C). Because PR length in each of the three mutant lines was shorter than that in the wild type (Fig. 2), the [3H+]-IAA transport distance from the site of application at the root–shoot junction to the root tip was in effect shorter in the mutants, and therefore, the effect of OsNIT1 or OsNIT2 disruption on acropetal transport of [3H+]-IAA in the roots is theoretically larger than that implied by the data shown in Figure 4.
To confirm that the loss of OsNIT1 or OsNIT2 function can alter auxin distribution or auxin forms, we analyzed the expression of five auxin efflux transporter-encoding genes and three IAA-amido synthetase (GH3)–encoding genes, which may prevent free IAA accumulation in rice (Xu et al., 2005; Ding et al., 2008; Wang et al., 2009, 2018; Zhang et al., 2009; Du et al., 2012. In comparison to wild type, both osnit1 and osnit2 lines exhibited repressed expression of OsPIN1c and OsPIN1d and upregulated expression of OsPIN2, OsGH3-2, OsGH3-8, and OsGH3-13 in their roots (Fig. 4D). The expression of OsPIN1a and OsPIN1b was not significantly affected in the mutants (Supplemental Fig. S6B). These data support the hypothesis that OsNIT1 or OsNIT2 regulate root growth via alteration of auxin root-ward transport and local auxin distribution.
OsNIT1 Displays IAN Hydrolysis Activity, Which Is Significantly Improved by Coexpression of OsNIT2 and OsNAR2.1
It has been shown that some NIT enzymes like sorghum SbNIT4A and SbNIT4B2 may form heteromeric complexes to achieve high catalytic activity (Jenrich et al., 2007). Therefore, we tested whether the interaction among OsNIT1, OsNIT2, and OsNAR2.1 caused enhanced IAN hydrolysis activity in vitro.
First, we observed that OsNIT1 and OsNIT2 interacted with each other in a Y2H assay (Fig. 5A), similar to the previously reported interaction between SbNIT4A and SbNIT4B2 (Jenrich et al., 2007). A short-term in vitro assay of IAN conversion to IAA quantified by liquid chromatography indicated that OsNIT1, but not OsNIT2 or OsNAR2.1, performed IAN hydrolysis (Fig. 5B). Remarkably, coexpression of OsNIT1 and OsNIT2 enhanced OsNIT1 IAN hydrolysis activity by 3.2-fold, which was further increased by 5.3-fold after the coexpression of OsNIT1 and OsNIT2 together with OsNAR2.1 (Fig. 5B; Supplemental Fig. S7). To confirm OsNIT1 function in hydrolyzing IAN to IAA and the activation of OsNIT1 enzymatic activity by OsNIT2 and OsNAR2.1, we performed the conversion rate assay using four different concentrations of IAN under the same experiment conditions except for an extension of reaction time from 1 h to 2 h. The results showed that the Michaelis–Menten equation could be used to predict the kinetics of OsNIT1 hydrolysis activity in the conversion of IAN to IAA (Supplemental Fig. S8). The additional presence of OsNIT2 or both OsNIT2 and OsNAR2.1 increased OsNIT1 enzyme affinity for IAN, resulting in a 3.8- to 4.7-fold or 5.6- to 8.2-fold increase in IAA production rate, respectively (Supplemental Table S2). These results indicate the biological significance of the interaction between OsNAR2.1 and the two NIT proteins on rice NIT activity.
Figure 5.
Copresence of OsNAR2.1, OsNIT1m, and OsNIT2 improves OsNIT1-mediated IAN hydrolysis. A, Interaction test between OsNIT1 and OsNIT2 using the DUAL Membrane Pairwise Interaction Kit (Dualsystems Biotech). B, OsNIT1 and OsNIT2 enzyme activity. IAN conversion rate (consumed IAN content/total IAN content × 100%) was used as a measure of enzymatic activity by HPLC. Reaction time was 1 h at 28°C. Both the 6×His and MBP tag proteins were used as negative controls. The data represent means ± se of four biological replicates (different lowercase letters represent statistically significant difference at P ≤ 0.05 level). ND, not detectable.
Unlike OsNAR2.1, OsNIT1, and OsNIT2 Are Upregulated by Ammonium and Function in Ammonium Root-growth Responses
We previously reported that expression of OsNAR2.1 is activated by nitrate whereas it is repressed by ammonium (Feng et al., 2011). Moreover, there are no significant root phenotypic differences between OsNAR2.1-mutant and wild-type plants under ammonium supply (Yan et al., 2011; Liu et al., 2014; Huang et al., 2015). However, we noticed that the expression levels of both OsNIT1 and OsNIT2 were upregulated in the roots by ammonium (Fig. 6A). To further confirm these expression dynamics, we detected OsNIT1 and OsNIT2 tissue localization in rice under the same experimental conditions. In situ hybridization analyses showed that OsNIT1 and OsNIT2 were abundantly expressed in root tips and LRs under ammonium supply, whereas OsNAR2.1 expression was very faint in comparison (Fig. 6B).
Figure 6.
KO of OsNIT1 and OsNIT2 decreases both PR and LR growth after supply of ammonium. A and B, Expression levels and patterns of OsNAR2.1, OsNIT1, and OsNIT2 in the roots of rice seedings (‘Nipponbare’) in response to growth in IRRI solution containing 1.25 mm NH4NO3 (AN) or 1.25 mm (NH4)2SO4 (A). Relative gene expression levels under the above experimental conditions quantified by RT-qPCR (A). OsActin was used as an internal control. The values in (A) represent means ± se of three biological replicates. B, Expression patterns of OsNAR2.1, OsNIT1, and OsNIT2 determined by RNA in situ hybridization in root tips and mature root area supplied with 2.5 mm ammonium. All detection probes were based on the whole antisense strand. ScC to F, Seeds were first germinated in deionized water for 3 d, then transferred to hydroponic media containing 0.125 mm (NH4)2SO4 for another 7 d. Genotypes are the same as those used and described in Figure 2A. D, Effect of OsNAR2.1 KO on expression levels of OsNIT1 and OsNIT2 in roots determined by RT-qPCR. OsActin was used as an internal control. The values in (D to F) represent means ± se of three (D) and six (E and F) biological replicates (**P ≤ 0.01). G and H, Seeds were germinated and grown under the same conditions as described for (C). Genotypes are the same as those used and described in Figure 2E. The values in (H) represent means ± se of seven biological replicates (**P ≤ 0.01). I and J, Wild-type (WT; cv Nipponbare) plants and KO mutant plants were grown in IRRI solution containing 1.25 mm of (NH4)2SO4 for 10 d before being sampled for [3H]IAA transport analysis. I, Acropetal transport. J, Basipetal transport. Data represent means ± se of three biological replicates (P ≤ 0.05). ns, not significant.
We subsequently supplied different genotypes with 0.25 mm of NH4+ and observed that the osnit1 and osnit2 mutants in the cv Nipponbare background consistently displayed a phenotype of shorter PRs and lower LR root density, both of which were ∼35% to 45% less than in wild type (Fig. 6C). By contrast, KO of OsNAR2.1 in both the cv Hwayoung and cv Nipponbare backgrounds did not alter the expression of OsNIT1 and OsNIT2 (Fig. 6D) or root growth under ammonium supply (Fig. 6, E and F). Moreover, double mutation of OsNAR2.1 and OsNIT2 resulted in a similar root phenotype as osnit2 (Fig. 6, G and H), confirming that OsNAR2.1 is not involved in the role that NIT proteins play in root responses to ammonium supply.
To further verify that OsNIT1 and OsNIT2 function in regulating root responses to ammonium, we investigated acropetal transport of [3H+]-IAA in the corresponding mutant lines. In agreement with the repression of OsNAR2.1 expression by ammonium supply, there was no significant difference in either acropetal or basipetal transport of IAA in osnar2.1 compared to wild type (Fig. 6, I and J). Moreover, inactivation of OsNIT1 or OsNIT2 did not significantly affect IAA basipetal (shoot-ward) transport in roots under ammonium supply (Fig. 6J), comparable to that seen in roots under nitrate supply (Fig. 4C). Remarkably, in osnit1 and osnit2, the amount of [3H+]-IAA transported from its site of application at the root–shoot junction to the 0- to 3-mm root tip section was less than that in wild type (Fig. 6I). Because the ammonium-supplied mutant lines showed much shorter PR length than wild type (Fig. 6, C and E), these data confirm that OsNIT1 and OsNIT2 play regulatory roles in root responses to both ammonium and nitrate.
DISCUSSION
For efficient acquisition of N with varied forms and concentrations, plants develop sophisticated regulatory pathways in altering root morphology, architecture, N transport, and assimilation. We have previously reported that OsNAR2.1 in rice is activated by nitrate and inhibited by ammonium (Feng et al., 2011). As a component of NRTs, OsNAR2.1 contributes to both root nitrate acquisition and nitrate-regulated root growth (Yan et al., 2011; Liu et al., 2014; Huang et al., 2015). In this study, we identified that OsNIT1 and OsNIT2 are two new interacting proteins of OsNAR2.1 and found that they are the down-stream components of the OsNAR2.1 regulation pathway in the root response to nitrate supply. Moreover, ammonium supply could activate the interaction of OsNIT1 and OsNIT2 for maintaining root growth in rice.
The Role of OsNIT1 and OsNIT2 as the Components of NIT Enzyme in Catalyzing IAN Conversion and IAA Distribution
Previously, two groups of NITs that can catalyze the conversion of IAN to IAA, NIT1 orthologs in Brassicaceae and NIT4 orthologs in maize have been characterized (Wajant and Effenberger, 2002; Park et al., 2003; Ishikawa et al., 2007; Kriechbaumer et al., 2007). In this study, we detected that OsNIT1 alone belonging to NIT4 subfamily could also have the activity to hydrolyze IAN (Fig. 5B; Supplemental Fig. S8; Supplemental Table S2), whereas the velocity was slow and similar to that catalyzed previously by NIT1 and NIT4 in Arabidopsis, maize, Brassica rapa, and Sorghum bicolor (Wajant and Effenberger, 2002; Park et al., 2003; Ishikawa et al., 2007; Jenrich et al., 2007; Kriechbaumer et al., 2007). In addition, the Km value of SbNIT4A/B2 with IAN was 0.16 mm (Jenrich et al., 2007), which is comparable to Km value (0.65 mm) of OsNIT1/2/OsNAR2.1 (Supplemental Fig. S8), indicating the similar NIT enzyme activity in rice and S. bicolor. Moreover, as showed in Arabidopsis, nit1 mutants were resistant to external IAN, indicating that the slow IAN hydrolysis activity of NIT1 was sufficient to produce an auxin-overproduction phenotype (Normanly et al., 1997). Similarly, we observed that osnit2, in particular osnit1, displayed a stimulated PR growth and a reduced LR growth as compared to wild type in the presence of exogenous IAN (Fig. 3, A and B), indicating that inactivation of OsNIT1 or OsNIT2 could prevent auxin-overproduction in rice (Supplemental Fig. S5B). Notably, IAN was not detectable in the seedlings of rice (‘Nipponbare’) in this study (Supplemental Fig. S6A), the same as reported by Sugawara et al. (2009). In addition, KO of OsNIT1 or OsNIT2 did not significantly affect total IAA content in the roots of seedlings (Fig. 4A). These results indicated that NITs may not be the key enzymes in IAA biosynthesis as proposed by Piotrowski (2008), but affect auxin-related regulatory pathways in plants.
S. bicolor contains three NIT4 isoforms—SbNIT4A, SbNIT4B1, and SbNIT4B2 (Jenrich et al., 2007). Interestingly, each isoform of SbNIT4 did not possess the enzymatic activity in hydrolyzing β-cyano-Ala, whereas the heteromeric complexes of SbNIT4A/B1 and SbNIT4A/B2 showed high activity in catalyzing the hydrolysis of β-cyano-Ala (Jenrich et al., 2007). The SbNIT4A/B2 complex could also catalyze the conversion of IAN to IAA (Jenrich et al., 2007). The assay of site-specific mutagenesis of the active Cys residue demonstrates that hydrolysis of β-cyano-Ala is catalyzed by the SbNIT4A isoform in both complexes whereas hydrolysis of IAN occurs at the SbNIT4B2 isoform (Jenrich et al., 2007). In maize, ZmNIT2 is expressed in auxin synthesizing tissues and shows efficient activity in hydrolyzing IAN to IAA (Kriechbaumer et al., 2007). Notable, ZmNIT2 could have an additional enzymatic function in turnover of β-cyano-Ala when it forms heteromers with the orthologs ZmNIT1 (Kriechbaumer et al., 2007). Interestingly, the IAN hydrolysis activity of OsNIT1 was also strongly enhanced by presence of OsNIT2 (Fig. 5, A and B). Taken together, we predict that the basic function of Poaceae NIT in catalyzing either IAN to IAA or hydrolyzing β-cyano-Ala is determined by one NIT component and enhanced by the other.
It has been shown that inactivation of NIT1 resulted in decrease of total IAA, but not free IAA in the seedlings of Arabidopsis (Lehmann et al., 2017). In this study, we found that KO of OsNIT1 or OsNIT2 did not change the concentration of total IAA (Fig. 4A), but confined the acropetal transport of 3H-IAA in the roots (Fig. 4B). In comparison to wild type, osnit1 and osnit2 mutants showed upregulated expression of GH3 genes (Fig. 4D) and downregulation of two putative IAA efflux transporter genes OsPIN1c and OsPIN1d expression in the nitrate supplied roots (Fig. 4D). Notably, OsPIN1c and 1d are orthologs genes of AtPIN1 expressed in Arabidopsis vascular tissue (Gälweiler et al., 1998). Double mutation of OsPIN1c and OsPIN1d resulted in a short PR phenotype (Li et al., 2019). Therefore, we hypothesize that OsPIN1c and OsPIN1d are also involved in rice root IAA distribution. It can be deduced that the activation of OsNIT1 and OsNIT2 mediates root growth partially via altering the IAA ratio of free to conjugate forms and transportation.
Interaction between OsNIT1 and OsNIT2 and Activation by OsNAR2.1 Contribute to Nitrate-regulated Root Growth in Rice
Root architecture is shaped through N interactions with PR and LRs. It is known that interactions with auxin signaling are important to N regulation of root branching (Lavenus et al., 2013; Forde, 2014). In this study, we found that KO of either OsNIT1 or OsNIT2 resulted in shorter length of PR and lower LR density at nitrate supply condition (Fig. 2). In comparison to their wild type, the mutants showed less sensitivity of the root growth to external IAN (Fig. 3, A and B) and lower rate of root-ward movement of 3H-IAA (Fig. 4B). However, the contribution of OsNIT1 and OsNIT2 to maintain root growth seems different to their putative orthologs in Arabidopsis. KO of AtNIT1 and AtNIT2 did not affect the length of PR whereas overexpression of AtNIT1 resulted in shorter PR and higher LR density (Lehmann et al., 2017).
Notably, the impaired root growth of both osnit1 and osnit2 mutants was similar to that of osnar2.1 mutant (Fig. 2,A, C, and D). Previously, we have shown that knockdown of OsNAR2.1 inhibited LR formation under nitrate supply (Huang et al., 2015). The osnar2.1 mutant showed significantly fewer LRs than wild type co-occurring with decreased auxin transport from shoots to roots even at similar N concentrations in their roots (Huang et al., 2015), suggesting that OsNAR2.1 probably functions in nitrate-signaling in addition to nitrate uptake (Yan et al., 2011; Liu et al., 2014). In this study, we found that OsNAR2.1, OsNIT1, and OsNIT2 are colocalized in the root tissues (Fig. 1C) and all of them showed nitrate-enhanced expression in the roots (Fig. 1D). We provide robust evidence that the three proteins can interact with each other. The presence of OsNAR2.1 and OsNIT2 enhanced the enzyme activity of OsNIT1 (Fig. 5B). The preliminary pull-down assay (Supplemental Fig. S1), together with the Y2H assay (Fig. 1A) and the Co-IP assay (Fig. 1B) as well as the mass spectrometry analysis (Supplemental Fig. S1, B and 1C), all support this notion.
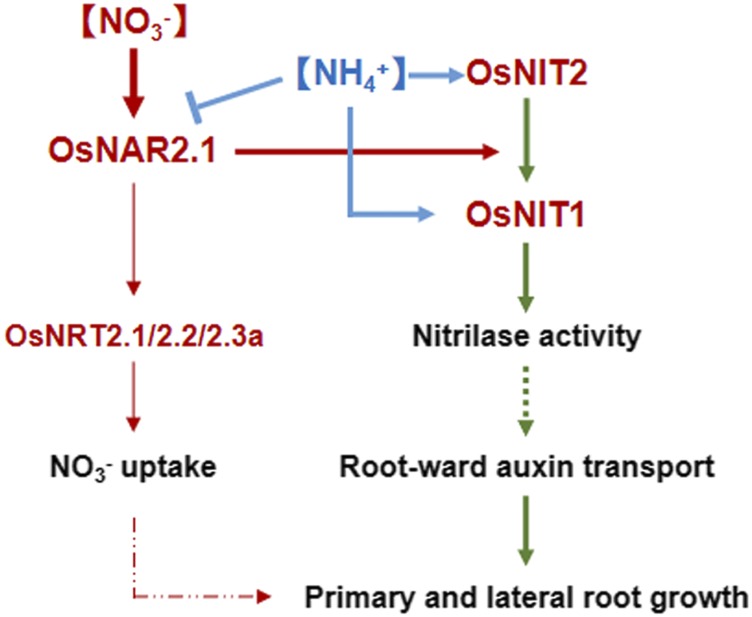
We also found that OsNAR2.1 is the regulatory upstream of OsNIT1 and OsNIT2 at nitrate supply. KO of OsNAR2.1 repressed the expression of OsNIT1 and OsNIT2 (Fig. 2B), whereas inactivation of OsNIT1 or OsNIT2 did not affect the expression of OsNAR2.1 (Fig. 2G). OsNAR2.1 enhanced the enzyme activity of OsNIT1 and OsNIT2 (Fig. 5B) and functions in root nitrate acquisition at wide range (Yan et al., 2011; Liu et al., 2014), whereas OsNIT1 and OsNIT2 mutations did not affect root nitrate uptake rate (Fig. 2H). Nevertheless, the double mutation of OsNAR2.1 and OsNIT2 (osnit2-2 × osnar2.1-1) showed a smaller LR density than the two parental mutants under nitrate conditions (Fig. 2F). Because OsNAR2.1 is essential to activate the nitrate uptake functions of OsNRT2.1, OsNRT2.2 and OsNRT2.3a in rice (Yan et al., 2011; Liu et al., 2014), it is not so surprising that the regulatory role of OsNAR2.1 in the occurrence of LRs is broader than that of OsNIT2, and probably OsNIT1 as well. Therefore, our results demonstrated that OsNAR2.1 could regulate root formation not only by interacting with OsNIT1 and OsNIT2, but also through the nitrate uptake (Fig. 7).
Figure 7.
Model for the role of OsNAR2.1, OsNIT1, and OsNIT2 in mediating root growth in rice. Interaction of OsNAR2.1 with OsNRT2.1, OsNRT2.2, and OsNRT2.3a, and their effect on root nitrate uptake, were previously reported in Feng et al. (2011), Yan et al. (2011), and Liu et al. (2014). All other noted functions are demonstrated in this study.
OsNIT1 and OsNIT2 also play a regulatory role on root growth that is partially independent from OsNAR2.1. In osnar2.1 mutant, both the activities of NIT1 and the NIT1+NIT2 complex might be impaired but remain significant (Fig. 2B). Because OsNIT1 alone showed an IAN to IAA conversion activity and OsNIT2 could enhance the OsNIT1 activity in hydrolyzing IAN (Fig. 5A), it is not surprising that osnar2.1 or osnit2 single mutant has a weaker phenotype than their double osnit2 × osnar2.1 mutant (Fig. 2F).
Remarkably, the role of OsNAR2.1 in nitrate uptake was independent from OsNIT1 and OsNIT2 (Fig. 2, G and H), whereas the role of OsNIT1 and OsNIT2 in the regulation of root growth can be enhanced by OsNAR2.1 (Fig. 5B; Supplemental Fig. S7; Supplemental Table S2). As suggested in the pull-down and Co-IP assays (Supplemental Fig. S1; Supplemental Table S1), OsNAR2.1 might have more than just NIT and NRTs for potential interacting protein candidates. Therefore, the nitrate signaling role of OsNAR2.1 not only relies on its interaction with the OsNITs and NRTs, but also other unknown regulators.
Independent Role of OsNIT1 and OsNIT2 in Maintaining Ammonium-supplied Root Growth
It has been shown that ammonium triggered LR branching and inhibition of PR and LR elongation (Liu et al., 2013; Zou et al., 2013), which was the same as that we observed in this study (Fig. 6). The repression of PIN2-altered auxin distribution in the root apices exposed to ammonium suggested the auxin involvement in the ammonium repression of root growth (Liu et al., 2013; Zou et al., 2013). Because OsNAR2.1 was repressed when ammonium was provided as the only N source (Fig. 6A; Feng et al., 2011) it was not surprising that KO of OsNAR2.1 did not significantly affect the root phenotypes in the ammonium supply condition (Fig. 6, C–H). However, ammonium supply enhanced expression of both OsNIT1 and OsNIT2, and their inactivation repressed acropetal transport of 3H-IAA and limited both PR length and LR densities (Fig. 6). The data clearly demonstrated that OsNIT1 and OsNIT2 have independent roles via activation of NIT in maintaining root growth that is basically not regulated by OsNAR2.1. There might be other ammonium-induced proteins that activate the enzyme activity of OsNIT1 and/or OsNIT2 for altering the form and transportation of IAA. This speculation is worth future investigation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp. japonica) of the ‘Nipponbare’ background was used for physiological experiments and rice transformation (mutant constructed by CRISPRCas9 system). The osnit1-1 T-DNA insertion mutant (Line ID PFG_1C-01739.R) with a genetic background of japonica cv Hwayoung was obtained from RiceGE in Korea (http://signal.salk.edu/cgi-bin/RiceGE).. For hydroponic experiments of nearly 1-month duration, which formed the basis of most of the experiments in this article (except for the 10-d hydroponic experiment described below), rice seeds were surface-sterilized in a 30% (v/v) NaClO solution for 30 min, washed, and germinated on half-strength Murashige and Skoog medium for 3 d at 25°C in darkness and for another 5 d in growth-chamber conditions as follows: 14-h light/10-h dark photo cycle, day/night temperatures of 30°C/24°C, and relative humidity of ∼60%. The air in the growth room was refreshed every 6 h. Hydroponic experiments were performed using the rice normal nutrient solution from the International Rice Research Institute (IRRI). Nitrate treatment used Ca(NO3)2 instead of NH4NO3, whereas ammonium treatment used (NH4)2SO4 instead of NH4NO3. Twenty seedlings were grown in each culture vessel containing 7.5 L nutrient solution, and the solution was changed every 2 d. After germination for 8 d, rice seedlings were initially treated with half-strength nutrient solution for 6 d, then transferred to full-strength culture solution containing either 1.25 mm NH4NO3, 1.25 mm Ca(NO3)2, or 1.25 mm (NH4)2SO4 for another 7 d before treatments or sampling. For 10-d hydroponic experiments, rice seeds were surface-sterilized in a 30% (v/v) NaClO solution for 30 min, washed, and germinated in deionized water for 3 d at 25°C in darkness. Seeds displaying a comparable extent of germination (just had a white tip) were transferred to hydroponic media containing either 0.125 mm Ca(NO3)2, 0.125 mm (NH4)2SO4, or other experimental treatments as described in the figure legends for another 7 d, and maintained in the growth-chamber conditions described above. Thirty seedlings were grown in each culture vessel containing nearly 3 L of solution, and the solution was changed every 2 d.
Pull Down Assay
Details of OsNAR2.1-GST tag fusion protein generation are provided below in the “Preparation of Recombinant Proteins” section. Target proteins were purified using ProteinIso GST Resin (DP201; Transgen Biotech). Total protein of rice (‘Nipponbare’) roots that were treated with nitrate was extracted with a Plant Protein Extraction Kit (CW0885S; CWBio). Total protein was incubated with the OsNAR2.1-GST fusion protein for 4 h at 4°C. Bound proteins were collected and subjected to two-dimensional electrophoresis. The gel was recovered and analyzed for each gel point protein.
Y2H Assay
The interactions between OsNAR2.1 and either OsNIT1 or OsNIT2 were tested using the DUAL Membrane Pairwise Interaction Kit (Dualsystems Biotech; Yan et al., 2011). HIS3 and ADE2 were used as reporter genes in the yeast strain NMY51, with each strain carrying a pair of bait and prey plasmids (pBT3-C and pPR3-N are the control vectors with no cloned complementary DNA (cDNA).
Full-length cDNA of OsNAR2.1 was cloned into pBT3-C (LEU2, KanR; Liu et al., 2014) and OsNIT1 and OsNIT2 cDNAs were cloned into pPR3-N (TRP1, AmpR; primers detailed in Supplemental Table S3), and expression vectors were cotransformed into yeast strain NMY51 (MATa his3 trp1 leu2 ade2 LYS2::HIS3 ura3::lacZ ade2::ADE2 GAL4) using the DS Yeast Transformation Kit (Dualsystems Biotech). Transformed colonies were selected in synthetic dropout medium without Leu and Thr (SD-LW) medium and incubated for growth of positive transformants. For growth assays, several independent positive transformants were selected and grown in SD-LW liquid medium at 30°C overnight. Culture concentrations were adjusted to OD546 = 0.8 and diluted 10, 100, and 1,000 times. Five microliters of each dilution were spotted onto SD-LW and SD without Ala, His, Leu, and Thr solid media, respectively, and incubated at 30°C for 2.5 d.
Co-IP Assay
Root protein was extracted from rice (‘Nipponbare’) OsNAR2.1+His and His overexpression lines using a Plant Protein Extraction Kit (CW0885S; CWBio). Total root protein was passed through ProteinIso Ni-NTA Resin (DP101; Transgen Biotech) to obtain binding protein (Elution buffer: 300 mm NaCl, 50 mm NaH2PO4, 400 mm imidazole, 10 mm Tris base, pH 8.0). Anti-His analysis for testing OsNAR2.1 interaction with other proteins after immunoprecipitation was performed by Native PAGE and all other analyses were SDS-PAGE. anti-NIT2 was purified from a rabbit injected with a specific peptide chain of OsNIT2 (amino acid sequence: EKNSAAKSDGISRT). A portion of the sample was subjected to immunoblot analysis using indicated antibodies, and the remaining sample was incubated with 10 mm dithiothreitol, 55 mm of ammonium iodoacetate, and 1 μg trypsin for enzymatic hydrolysis overnight. Afterward, the polypeptide was desalted by a C18 column, then drained and dissolved with 15 μL of Loading buffer (0.1% [v/v] formic acid, 3% [v/v] acetonitrile). The peptide was analyzed by liquid chromatography-tandem mass spectrometry (ekspert nanoLC 400 system, Triple TOF 5600-Plus; AB Sciex).
RNA In Situ Hybridization
Longitudinal sections of root tips and mature roots of wild-type (cv Nipponbare) seedlings with a length of ∼10 mm were fixed in FAA solution (1.85% [v/v] formaldehyde, 5% [v/v] acetic acid, and 63% [v/v] ethanol), dehydrated with a mixture of ethanol and 1-butanol, and then embedded in paraffin. The embedded sections were sliced (10-µm thickness) using a microtome (model no. RM2235; Leica). The full-length cDNA sequences of OsNAR2.1, OsNIT1, and OsNIT2 were cloned into pENTR-d-TOPO (primers detailed in Supplemental Table S3). Digoxin (DIG)-labeled RNA probes in antisense orientation were synthesized using T7 RNA polymerase, with each linearized plasmid DNA as a template, using the DIG RNA Labeling Kit as described in Ishiyama et al. (2004). RNA in situ hybridization with DIG-labeled RNA probes was performed as described in Ishiyama et al. (2004).
RNA Extraction, cDNA Synthesis, and Reverse Transcription Quantitative PCR
Total RNA was extracted from plant samples using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using the HIScript II Reverse Transcriptase with gDNA wiper (cat. no. R223-01; Vazyme). Reverse transcription quantitative PCR (RT-qPCR) was performed using the AceQ qPCR SYBR Green Master Mix (cat. no. Q111-01; Vazyme) on the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. Relative expression level of each sample was determined by normalizing it to the amount of OsActin1 (LOC_Os03g50885) detected in the same sample and presented as 2−ΔCT. All primers used for RT-qPCR are detailed in Supplemental Table S3.
Determination of 15N-NO3− Uptake Rate
Rice seedlings were first planted in IRRI solution containing 1.25 mm NH4NO3, then they were deprived of N for 3 d. Next, plants were first transferred into 0.1 mm of CaSO4 for 1 min, then to a complete nutrient solution containing 0.25 mm 15NO3− (Ca[15NO3−]2) for 5 min, and finally to 0.1 mm CaSO4 for 1 min. Then, we used paper to blot excess water from the plants. The shoots and roots were separated. Root samples were placed in an oven at 105°C for 30 min to inactivate the enzymes, and further dried to a constant weight at 70°C. After recording their dry weights, the samples were ground into powder using a ball mill. The Isotope Ratio Mass Spectrometer System (model no. Flash 2000 HT; Thermo Fisher Scientific) was used to determine the 15N content of the samples.
Determination of Total IAA and IAN
Samples were ground to a power with liquid N and freeze-dried. IAA measurement was performed as described below. The above dry powder was added with 1 ng of [13C]6-IAA and extracted three times with 80% (v/v) methanol. After the extraction was concentrated, ethyl acetate containing 5% (v/v) acetic acid was added, and the extraction was collected again. Extractions were freeze-dried once more, after which 10 μL of pyridine and 40 μL of Bis(trimethylsilyl)trifluoroacetamide were added and reacted at 80°C for 30 min. Finally, 50 μL of N-hexane was added for mass spectrometry using the method described in Novák et al. (2012). The instrument was a GC-QqQ MS (model no. 7890a-5975b; Agilent) and the column was a DB-5MS (30 m × 0.25 mm × 0.10 μm; Agilent; Novák et al., 2012). For IAN measurement, the above dry powder was extracted three times with methanol, and the extraction was concentrated and then subjected to liquid phase analysis by the standard of GB/T 16631-2008 HPLC. The instrument used was a model no. 1200 High Performance Liquid Chromatograph (Agilent; Sugawara et al., 2009).
3H-IAA Transport Assay
Acropetal and basipetal auxin transport was assayed in excised seminal roots as described by Lewis and Muday (2009) with minor modifications. For acropetal auxin transport, agar mixtures containing 0.7% (w/v) agar, 0.04% (v/v) [3H]IAA (26.0 Ci/mmol), 10 μm cold IAA, 2% DMSO, and 25 mm MES(pH 5.5) were prepared in a scintillation vial. After shoot excision at 1 cm above the root-shoot junction, 20-μL agar droplets of the [3H]IAA solution were applied to the cut surface. After a 6-h incubation in 60% to 70% humidity at 25°C in darkness, root segments were excised at distances of 0–1, 1–2, and 2–3 cm from the root apex and weighed. Then the root segments were immediately placed in scintillation solution (3 mL) for 12 h. For basipetal auxin transport, 20-μL agar droplets (agar mixtures as described above) of the [3H]IAA solution were applied to root segments excised at 0- to 3-mm, 3- to 6-mm, 6- to 9-mm, 9- to 12-mm, and 12- to 15-mm distances from the root apex. The root segments were digested with perchloric acid and then immediately placed in scintillation solution (3 mL) for 12 h. The amount of radioactivity of [3H] in each sample was determined using a model no. LS6500 (Beckman).
Preparation of Recombinant Proteins
To obtain recombinant proteins, the plasmids MBP-OsNIT1, OsNIT2-6×His, and OsNAR2.1-6×His (primers detailed in Supplemental Table S3) were transformed into Escherichia coli strain Transetta (DE3; cat no. CD801-01; Transgen Biotech). The bacterial cells were cultivated at 37°C shaking at 150 to 200 rpm. At an A600 of 0.6–0.8, the bacterial cells were induced by 0.5 mm IPTG at 16°C for 16 h. Cells were collected and disrupted using ultrasonic cell breakers (model no. 120; Thermo Fisher Scientific) on ice. The recombinant proteins were affinity-purified with Amylose Resin High Flow (cat. no. E8022; New England Biolabs) or ProteinIso Ni-NTA Resin (cat. no. DP101; Transgen Biotech) according to the manufacturers’ instructions.
NIT Activity Assays
Assays were carried out in a volume of 50 μL solution containing 50 mm KPi (pH 8.0), 1 mm dithiothreitol, 3 mm ATP, 3 mm substrate IAN, and 2 μg for each purified enzyme except for OsNIT1, for which 6 μg was used because the western blot result showed many bands. IAN conversion rate (consumed IAN content/total IAN content × 100%) was used as a measure of enzymatic activity as analyzed by HPLC. Reaction time was 1 to 2 h at 28°C, with 200 μL methanol added to stop reactions. An aliquot (15 μL) of the diluted sample was injected into the HPLC system (model no. 1200LC; Agilent) equipped with a ZORBAX C18 SB-Aq column (Agilent). The flow rate was 0.8 mL/min, and the sample was eluted with 0.1% (v/v) H3PO4 (5 min), followed by a linear methanol gradient to 40% (v/v) methanol in 7 min, and held at this composition for an additional 18 min. The column effluent was monitored at 280 nm. Under these conditions, the retention times of IAA and IAN were 23.161 and 28.684 min, respectively.
Subcellular Localization
For subcellular localization constructs, the full-length open reading frames of OsNIT1 and OsNIT2 were amplified and subcloned into the intermediate vectors pSAT6A-EGFP-N1 and pSAT6-EGFP-C1 to generate OsNIT1-GFP, GFP-OsNIT1, OsNIT2-GFP, and GFP-OsNIT2 vectors. All vectors were introduced into the final expression vector pRCS2-ocs-nptII with the restriction endonuclease PI-PspI. The constructs were transformed into rice protoplasts by the polyethylene-glycol–mediated method. The isolation and transformation of rice protoplast was performed as described in Jia et al. (2011). In brief, 10-μg plasmid DNA of each construct was transformed into 0.2-mL protoplast suspension. HDEL fusion mCherry served as an endoplasmic reticulum marker. After incubation at 28°C in darkness for 12 to 15 h, fluorescence signals in rice protoplasts were detected. Confocal microscopy images were taken using a model no. TCS SP8X Confocal Laser Scanning Microscope (Leica). Excitation/emission wavelengths were 488 nm/495 to 556 nm for GFP and 587 nm/600 to 650 nm for mCherry.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: LOC_Os02g42350 (OsNIT1), LOC_Os02g42330 (OsNIT2), LOC_Os02g38230 (OsNAR2.1), LOC_Os02g02170 (OsNRT2.1), LOC_Os01g50820 (OsNRT2.3), LOC_Os06g12610 (OsPIN1a), LOC_Os02g50960 (OsPIN1b), LOC_Os11g04190 (OsPIN1c), LOC_Os12g04000 (OsPIN1d), LOC_Os06g44970 (OsPIN2), LOC_Os01g55940 (OsGH3-2), LOC_Os07g40290 (OsGH3-8), LOC_Os11g32520 (OsGH3-13).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. OsNAR2.1 interacts with OsNIT1 and OsNIT2.
Supplemental Figure S2. Subcellular localization analysis of OsNIT1 and OsNIT2.
Supplemental Figure S3. Characterization of different genotypes used in this article.
Supplemental Figure S4. Gene expression pattern of OsNIT1 in osnit2 and OsNIT2 in osnit1.
Supplemental Figure S5. Selection of appropriate IAN treatment concentration.
Supplemental Figure S6. Extracted IAN was not detectable in the seedlings of Nipponbare wild-type and expression levels of OsPIN1a and OsPIN1b were unchanged in osnit1 or osnit2.
Supplemental Figure S7. The enzyme activity of OsNIT1 and OsNIT2 for IAN hydrolysis.
Supplemental Figure S8. The kinetics of IAN conversion rate to IAA by NIT enzyme.
Supplemental Table S1. List of proteins identified by pull-down assay.
Supplemental Table S2. The enzyme activity for conversion of IAN to IAA at four substrate concentration ranges after a 2-h reaction time.
Supplemental Table S3. The primers used in this article.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0100700), the National Natural Science Foundation of China (grant no. 31930101), the Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource Utilization, the Innovative Research Team Development Plan of the Ministry of Education of China (grant nos. IRT_17R56 and KYT201802), and Fundamental Research Funds for the Central Universities (KYZ201869).
Articles can be viewed without a subscription.
References
- Abu-Zaitoon YM.(2014) Phylogenetic analysis of putative genes involved in the tryptophan-dependent pathway of auxin biosynthesis in rice. Appl Biochem Biotechnol 172: 2480–2495 [DOI] [PubMed] [Google Scholar]
- Cao Y, Glass AD, Crawford NM(1993) Ammonium inhibition of Arabidopsis root growth can be reversed by potassium and by auxin resistance mutations aux1, axr1, and axr2. Plant Physiol 102: 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S(2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Saker LR(1975) Nutrient supply and the growth of the seminal root system in barley: II. Localized, compensatory increases in lateral root growth and rates op nitrate uptake when nitrate supply is restricted to only part of the root system. J Exp Bot 26: 79–90 [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L(2012) A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63: 6467–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G(2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Forde BG.(2014) Nitrogen signalling pathways shaping root system architecture: An update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K(1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E(2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N(2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen S, Liang Z, Zhang C, Yan M, Chen J, Xu G, Fan X, Zhang Y(2015) Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci Rep 5: 18192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Okazaki K, Kuroda H, Itoh K, Mitsui T, Hori H(2007) Molecular cloning of Brassica rapa nitrilases and their expression during clubroot development. Mol Plant Pathol 8: 623–637 [DOI] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H(2004) Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J Biol Chem 279: 16598–16605 [DOI] [PubMed] [Google Scholar]
- Jenrich R, Trompetter I, Bak S, Olsen CE, Møller BL, Piotrowski M(2007) Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc Natl Acad Sci USA 104: 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G(2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GJD, Kronzucker HJ(2005) The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: A modelling study. Ann Bot 96: 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC(2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotur Z, Mackenzie N, Ramesh S, Tyerman SD, Kaiser BN, Glass ADM(2012) Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol 194: 724–731 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E(2007) Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J Exp Bot 58: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, Benkova E, et al. (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L(2013) Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lehmann T, Janowitz T, Sánchez-Parra B, Alonso MP, Trompetter I, Piotrowski M, Pollmann S(2017) Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front Plant Sci 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK(2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Li B, Li G, Kronzucker HJ, Baluška F, Shi W(2014) Ammonium stress in Arabidopsis: Signaling, genetic loci, and physiological targets. Trends Plant Sci 19: 107–114 [DOI] [PubMed] [Google Scholar]
- Li Q, Li BH, Kronzucker HJ, Shi WM(2010) Root growth inhibition by NH4+ in Arabidopsis is mediated by the root tip and is linked to NH4+ efflux and GMPase activity. Plant Cell Environ 33: 1529–1542 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu J, Wu L, Shao Y, Wu Y, Mao C(2019) Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol 60: 2720–2732 [DOI] [PubMed] [Google Scholar]
- Li YL, Fan XR, Shen QR(2008) The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant Cell Environ 31: 73–85 [DOI] [PubMed] [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wirén N(2010) Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22: 3621–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang D, Tao J, Miller AJ, Fan X, Xu G(2014) Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol 204: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lai N, Gao K, Chen F, Yuan L, Mi G(2013) Ammonium inhibits primary root growth by reducing the length of meristem and elongation zone and decreasing elemental expansion rate in the root apex in Arabidopsis thaliana. PLoS One 8: e61031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, von Wirén N(2017) Ammonium as a signal for physiological and morphological responses in plants. J Exp Bot 68: 2581–2592 [DOI] [PubMed] [Google Scholar]
- Müller A, Hillebrand H, Weiler EW(1998) Indole-3-acetic acid is synthesized from l-tryptophan in roots of Arabidopsis thaliana. Planta 206: 362–369 [DOI] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B(1997) Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K(2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA(2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9: 837–856 [DOI] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ(2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiol 142: 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WJ, Kriechbaumer V, Möller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E(2003) The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol 133: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S(2012) Growth and development of the root apical meristem. Curr Opin Plant Biol 15: 17–23 [DOI] [PubMed] [Google Scholar]
- Piotrowski M.(2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69: 2655–2667 [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schönfelder S, Weiler EW(2001) The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode β-cyano-l-alanine hydratase/nitrilase. J Biol Chem 276: 2616–2621 [DOI] [PubMed] [Google Scholar]
- Rogato A, D’Apuzzo E, Barbulova A, Omrane S, Parlati A, Carfagna S, Costa A, Lo Schiavo F, Esposito S, Chiurazzi M(2010) Characterization of a developmental root response caused by external ammonium supply in Lotus japonicus. Plant Physiol 154: 784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H(2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ(2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA(2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA(2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Effenberger F(2002) Characterization and synthetic applications of recombinant AtNIT1 from Arabidopsis thaliana. Eur J Biochem 269: 680–687 [DOI] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P(2009) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2: 823–831 [DOI] [PubMed] [Google Scholar]
- Wang L, Guo M, Li Y, Ruan W, Mo X, Wu Z, Sturrock CJ, Yu H, Lu C, Peng J, Mao C(2018) LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J Exp Bot 69: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong Z, Friml J, Zhang J(2019) Nitrate modulates the differentiation of root distal stem cells. Plant Physiol 180: 22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzba MP, Tax FE(2013) Notes from the underground: Receptor-like kinases in Arabidopsis root development. J Integr Plant Biol 55: 1224–1237 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B(2005) Auxin: Regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ(2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou H, Wu P(2005) A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46: 1674–1681 [DOI] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G(2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34: 1360–1372 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG(1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y(2009) Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol 151: 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N, Li B, Chen H, Su Y, Kronzucker HJ, Xiong L, Baluška F, Shi W(2013) GSA-1/ARG1 protects root gravitropism in Arabidopsis under ammonium stress. New Phytol 200: 97–111 [DOI] [PubMed] [Google Scholar]