An allelic variant of the transcription factor GAME9 reduces accumulation of toxic steroidal glycoalkaloids in ripe tomato fruit via altered binding to the promoter of a steroidal glycoalkaloid biosynthesis gene.
Keywords: GAME9, GAME17, genome-wide association study, tomato, steroidal glycoalkaloids, variation
Abstract
Steroidal glycoalkaloids (SGAs) are cholesterol-derived molecules found in the family Solanaceae. SGA content varies among different plant species and varieties. However, the genetic mechanisms regulating SGA content remain unclear. Here, we demonstrate that genetic variation in GLYCOALKALOID METABOLISM 9 (GAME9) is responsible for the variation in SGA content in tomato (Solanum lycopersicum). During a sequential analysis we found a 1 bp substitution in the AP2/ERF binding domain of GAME9. The 1 bp substitution in GAME9 was significantly associated with high SGA content and determined the binding capacity of GAME9 with the promoter of GAME17, a core SGA biosynthesis gene. The high-SGA GAME9 allele is mainly present in S. pimpinellifolium and S. lycopersicum var. cerasiforme populations and encodes a protein that can bind the GAME17 promoter. In contrast, the low-SGA GAME9 allele is mainly present in the big-fruited varieties of S. lycopersicum and encodes a protein that shows weak binding to the GAME17 promoter. Our findings provide new insight into the regulation of SGA biosynthesis and the factors that affect the accumulation of SGA in tomato.
Introduction
Steroidal glycoalkaloids (SGAs), commonly known as Solanum alkaloids, are secondary metabolites primarily found in the Solanum family (Itkin et al., 2011). These small compounds are synthesized presumably from cholesterol and typically have an oligosaccharide chain attached at the C3 position of the nitrogenous steroidal alkaloid backbone (Milner et al., 2011). Since the discovery of glycoalkaloids in Solanum nigrum by Desfososses in 1820, approximately 100 different SGAs have been identified in various tomato tissues (Mintz-Oron et al., 2008; Yamanaka et al., 2008; Itkin et al., 2013). These numerous structurally diverse metabolites are the most common biologically active compounds in the Solanaceae family and are strongly physiologically active in mammals (Chowanski et al., 2016). SGAs primarily serve as protective agents. They deter insect herbivores and help plants to resist pathogens at all levels of biological organization. SGAs are also anti-nutritional factors because they disrupt digestion and nutrient absorption in humans (James et al., 2000; Hoagland, 2009; Dolan et al., 2010; Jared et al., 2016).
In tomato, the major toxic SGAs—α-tomatine and its precursor—accumulate in green tissues (Kozukue et al., 2004). Their toxicity is due to their ability to disrupt membranes and to their acetylcholine esterase inhibitory activity (Roddick, 1990). Indeed, a dietary concentration of 1 µmol g−1 tomatine inhibits the growth of Tribolium castaneum larvae. A total SGA level that exceeds 200 mg kg−1 FW of edible tuber is considered unsafe for humans (Weissenberg et al., 1998; Korpan et al., 2004). Reducing the concentration and glycosylation of SGAs is believed to be an effective approach for decreasing the toxicity of α-tomatine during fruit ripening (Friedman, 2004; Fujiwara et al., 2004; Hoagland, 2009). Levels of this anti-nutrient were reduced or eliminated by selection and breeding during crop domestication (Itkin et al., 2013). A similar story has been reported in cucumber in that bitterness was eliminated during domestication (Shang et al., 2014). Increasing our understanding of mechanisms that influence the genetic control of SGA biosynthesis will contribute to our fundamental knowledge and provide a foundation for genetic improvement of important traits. However, the genetic mechanisms that influence this type of toxicity remain largely unknown.
GLYCOALAKLOID METABOLISM 1 (GAME1) was the first gene reported to encode an SGA biosynthetic enzyme (Itkin et al., 2011). Subsequently a pathway for SGA biosynthesis was proposed in the Solanaceae family, based on a series of GAME genes (Itkin et al., 2013; Nakayasu et al., 2017). Extensive functional characterization provides evidence that SGAs are derived from cholesterol and that cholesterol undergoes several hydroxylation, oxidation, and transamination reactions to yield unsaturated steroidal alkaloid (SA) aglycones, which are glycosylated by different UDP-glycosyltransferases to yield SGAs (Itkin et al., 2013; Sonawane et al., 2016, 2018). Unfortunately, compared with the research on the structural genes, the mechanisms that regulate SGA biosynthesis are poorly understood.
Mechanisms that coordinate the transcription of structural genes are often key regulators of complex metabolic pathways. For example, the APETALA2/Ethylene Response Factor (AP2/ERF) transcription factor ORCA2 regulates terpenoid idole alkaloid biosynthesis in Catharanthus roseus by binding an elicitor-responsive element in the promoter of the gene encoding Strictosidine Synthase (Str) (Menke et al., 1999; Li et al., 2013). Interactions between CrWRKY1 and ORCA3, a homolog of the ORCA2 gene, were reported to influence the accumulation of ajmalicine and serpentine in the roots of C. roseus (Suttipanta et al., 2011). In Nicotiana tabacum, seven AP2/ERF transcription factors encoded by the NIC2 locus can activate alkaloid-associated gene expression by binding GCC-boxes in the promoters of genes associated with nicotine biosynthesis (Hibi et al., 1994; Shoji et al., 2010). The expression of ERF189, a key transcription factor for the NIC2 locus that regulates nicotine biosynthesis, is regulated by NUP1, a plasma membrane-localized permease that takes up nicotine (Kato et al., 2014).
In tomato, GAME9 serves as a master transcriptional regulator of SGA biosynthesis by binding GCC box elements in the promoters of target genes (Cárdenas et al., 2016; Thagun et al., 2016; Nakayasu et al., 2018). In this work, we used genome-wide association study coupled with metabolomics analysis (or mGWAS) and analysed genetic variation to identify a one-base nucleotide substitution associated with variation in the SGA content in a natural population of tomato. This variation was selected during the domestication of tomato. Moreover, we confirmed that this one-base nucleotide substitution interconverts the high-SGA allele to the low-SGA allele of GAME9. The high-SGA GAME9 allele is mainly represented in Solanum pimpinellifolium (PIM) and S. lycopersicum var. cerasiforme (CER) and encodes a protein that can bind a cis-element in the promoter of GAME17—a core gene for SGA biosynthesis. In contrast, the low-SGA allele is mainly present in the big-fruited varieties of S. lycopersicum (BIG) and encodes a protein that shows weak binding to the GAME17 promoter. These findings provide insight into new strategies for regulating toxic compounds and the genetic improvement of tomato. They also provide information useful for engineering reduced levels of anti-nutritional compounds in other plants.
Materials and methods
Tomato accessions and genome-wide association mapping
A total of 2 678 533 single nucleotide polymorphisms (SNPs; minor allele frequency >5% and missing rate <10%) for 442 accessions were used in a GWAS, which included 31 PIM accessions, 124 CER accessions, and 287 BIG accessions that were from the same populations as used previously (Zhu et al., 2018). The relative quantification of the seven SGA metabolites used for mGWAS in the current work came from the previously study (Zhu et al., 2018). The association analyses were performed using the compressed mixed linear model as described previously (Zhu et al., 2018). The genome-wide suggestive thresholds (P=1/n, ≤2.4×10–7) and significance thresholds (P=0.05/n, ≤1.2×10–8) of all the SNPs were defined at a uniform threshold. VCFtools was used to perform local linkage disequilibrium (LD) analysis. The 83.9–84.2 Mb regions of SNPs (P<2.4×10–7) centered on the lead SNP (ch01:84016748) were selected to calculate the r2 of LD. The physical locations of the SNPs were identified based on tomato genomic sequence version SL2.50. All tomato plants were self-pollinated. Tomato fruit was hand-harvested at physiological maturity.
Metabolite analysis
To get more accurate phenotypic data from transgenic plants, the fruits (peel and flesh tissues without the seeds and gel) were harvested uniformly at their mature green (MG), breaker, and red ripe (RR) stages. The freeze-dried samples (100 mg) were extracted for 8 h at 4 °C with 1.0 ml 80% methanol: water (V:V) containing 0.1 mg l−1 lidocaine. The samples were centrifuged for 10 min at 12 000 g, then filtered with an organic filter with a 0.22 µm pore size (Itkin et al., 2011; Zhu et al., 2018). The extracts (2 µl) were analysed using a UPLC-ESI-qTOF instrument (Agilent 6520) that was equipped with C18 eclipse plus columns (100×2.1 mm i.d., 1.8 µm particle size), a solvent of ultrapure water (eluent A) and acetonitrile (eluent B) both containing 0.1% (v/v) formic acid at a flow rate of 0.3 ml ml−1 according to the following procedure: 5:95 v/v B:A at 0 min, 95:5 at 20 min, 95:5 at 22 min, 5:95 at 22.1 min, and 5:95 at 26 min. The column was subsequently washed for 10 min and then equilibrated before the next injection. The column temperature was 25 °C. The sample was stored at 4 °C prior to injection (Adato et al., 2009; Chen et al., 2013). Full time-of-flight (TOF) scans of HPLC effluents were used to detect the m/z range from 50 to 1500 Da in positive ion mode. For MS/MS, 5–80 collision energies were used. α-Tomatine and its precursor were identified by a comparison of their retention times and MS/MS fragments with standard compounds or to values found in the literature (Cataldi et al., 2005; Itkin et al., 2011; Iijima et al., 2013). Relative quantification of the compounds was performed with Qualitative Analysis B.04.00 (Agilent).
RNA isolation and gene expression analysis
Total RNA was isolated using the Trizol reagent (Sigma-Aldrich). cDNAs were synthesized form DNaseI-treated total RNA using a Hiscript II 1st Strand cDNA Synthesis Kit (Vazyme, China). Expression analysis of SGA-related genes in the fruits (peel and flesh tissues without the seeds and gel) of wild-type and transgenic lines was performed using three biological replicates (n=3), the actin gene (BT013524) was used as an internal control. Primer sequences are list in Supplementary Table S1 at JXB online.
Yeast one-hybrid assay
Promoter fragments from GAME17 were amplified from TS-303 genomic DNA with specific primers (see Supplementary Table S1) and cloned into the bait vector pAbAi (Clontech). The full-length GAME9135A and GAME9135V with specific primers (Supplementary Table S1) were amplified from cDNA prepared from TS-303, a CER tomato that exhibits high SGA content and AC, a BIG tomato that exhibits low SGA content, respectively. They were then fused to the GAL4 activation domain in the prey vector pGADT7. The bait plasmids were digested, linearized, integrated into the Y1HGold yeast (Saccharomyces cerevisiae) genome and cultured on SD/−Ura at 30 °C for 3 d. The bait-reporter strain was transformed with the prey vector. After 3 d, the positive yeast strains were picked and diluted with sterile water. These suspensions were spotted on an SD/−Leu medium that contained from 0 to 100 ng ml−1 aureobasidin A. The plates were incubated for 3–4 d at 30 °C. The negative (pGADT7+N-AbAi) controls were prepared as described above. The DNA–protein interactions were analysed based on the growth of the yeast strains relative to the negative controls on media with different ABA concentrations.
Transient expression assays in tobacco leaves
The two alleles GAME9135A and GAME9135V were amplified from the genomic DNA of TS-303 and AC, respectively, with specific primers (see Supplementary Table S1) and fused to pGreen II 62-SK, a CaMV 35S promoter-driven effector (Hellens et al., 2005). The fragments of the promoter were cloned into the pGreen II 0800-Luc vector harboring the firefly luciferase (LUC) reporter gene and expressed under the control of the CaMV 35S promoter (Hellens et al., 2005). Agrobacterium tumefaciens (GV2260)-mediated transformation was used to transiently express both reporters and effectors in Nicotiana benthamiana leaves. The empty pGreen II 62-SK was used as the negative control. Three days after the transfection, both fLUC and rLUC activities were measured using the dual luciferase assay reagents (Promega, USA) and an Infinite M200 plate reader (Tecan, USA). To generate β-glucuronidase (GUS) reporter proteins for observation, the two alleles GAME9135A and GAME9135V were amplified from the genomic DNA of TS-303 and AC, respectively, and fused to pHellsgate8 vector, a CaMV 35S promoter-driven effector, and the fragment of the promoter was cloned into pMV2 vector (derived from the pHellsgate8 vector with a deleted CaMV 35S promoter) harboring the GUS reporter gene. The empty pHellsgate8 was used as the negative control. Three days after the transfection, both the negative control and the fusion proteins were incubated at 37 °C for 12 h in staining buffer (100 mM sodium phosphate, pH 7, 0.1% Triton X-100, 10 mM Na2EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 1mg ml−1 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid), followed by washing with 70% (v/v) ethanol. The relative expression of GUS reporter gene and negative control was determined with the GUS primers, and the actin gene (BT013524) was used as an internal control. Primers sequences are list in Supplementary Table S1.
Electrophoretic mobility shift assays
The whole coding sequences of GAME9135A and GAME9135V were amplified from TS-303 and AC, respectively, and then inserted into pET15d. The recombinant fusion protein with a MBP-tag was expressed in Escherichia coli DE3 (BL21) cells (Invitrogen, USA) and affinity purified using magnetic agarose (Biolabs, USA) according to the manufacturer’s instructions. To label the probes, FAM was attached to the sense oligonucleotides at their 5′ end. Sense, antisense, and mutated oligonucleotides (see Supplementary Table S1) were annealed by gradual cooling after heating at 95 °C for 5 min. An Electrophoretic mobility shift assay (EMSA) was performed with the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, USA). GAME9 protein and the probes were incubated at room temperature in 1× Binding Buffer (100 mm MgCl2, 2.5% (v/v) glycerol, 50 ng μl−1 poly (dI-dC)). After 30 min, the reaction products were separated by electrophoresis on 6% non-denaturing polyacrylamide gels. After migration, the FAM-labeled probe on the non-denaturing polyacrylamide gel was visualized using an Amersham Imager 600.
Vector construction and tomato transformation
The full-length GAME9 open reading frame (ORF) was amplified from TS-303. The GAME9-silenced fragment was amplified from AC cDNA using gene-specific primers with 5′-attB1 and 3′-attB2 extensions on the forward and reverse primers. The full-length GAME9 ORF was cloned into overexpression vector pMV2 (derived from the pHellsgate8 vector) using a CaMV 35S promoter. The GAME9-silenced fragment was recombined into the RNAi vector pHellsgate 2 using the Clonase BP reaction (Invitrogen). All vectors were introduced into the A. tumefaciens strain C58 for tomato transformation. TS-286 was transformed with the overexpression vector driven by a CaMV 35S promoter and the silencing vector. The genomic DNA from these transgenic plants was analysed with PCR-based genotyping experiments. The pertinent primers are listed in Supplementary Table S1.
Accession numbers
Sequence data from this article can be found in the Sol Genomics Network databases with the following accession numbers: ERF1 (Solyc01g090300), ERF2 (Solyc01g090310), ERF3 (Solyc01g090320), ERF5 (Solyc01g090370), GAME1 (Solyc07g043490), GAME2 (Solyc07g043410), GAME4 (Solyc12g006460), GAME6 (Solyc07g043460), GAME7 (Solyc07g062520), GAME9 (Solyc01g090340), GAME9 homolog from potato (Sotub01g029510), GAME11 (Solyc07g043420), GAME12 (Solyc12g006470), GAME17 (Solyc07g043480), GAME18 (Solyc07g043500), and MYC2 (Solyc08g076930).
Results
GAME9 is a major locus controlling SGA content in tomato
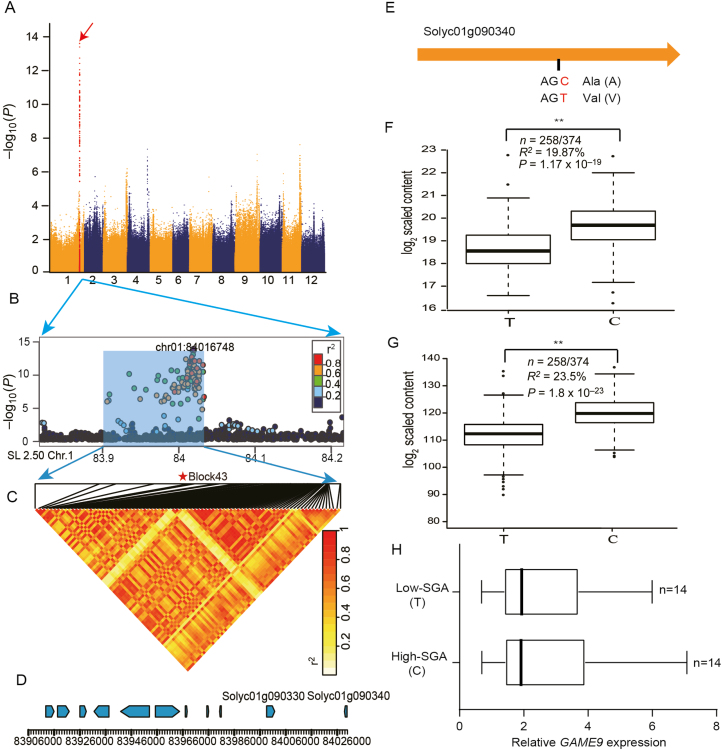
In a previous study, using a metabolic genome-wide association, we found a SNP (ch01:84016748) that showed a strong association with the levels of hydrotomatidine (SlFM1985; P=2.25×10–14), hydroxytomatidenol (SlFM0960; P=8×10–15), hydroxytomatidine (SlFM0964; P=1.78×10−10), dehydrofilotomatine (SlFM1979; P=1.11×10–7), neorickiioside A (SlFM1983; P=4.45×10–12), lycoperoside H (SlFM1984; P=5.54×10–11) and lycoperoside A (SlFM1989; P=6×10–12) (Fig. 1; Supplementary Fig. S1) (Zhu et al., 2018). Then we carefully analysed the pairwise LD distance within the 2 Mb interval centered on the SNP (ch01:84016748) from the GWAS of hydrotomatidine content (Fig. 1B). All the suggestive SNPs (P<2.4×10–7) fell into a 122.7 kb region of 83.9–84.2 Mb; there were a total of 11 genes in this region (Supplementary Table S2). A haplotype analysis of the region spanning all the significant SNPs on chromosome 1 (122.7 kb) identified 86 haploblocks (Supplementary Table S3), and many significant SNPs, including SNP ch01:84016748, can be traced back to haploblock 43 (SL2.50ch01:84014759–SL2.50ch01:84029382) (Fig. 1C). Haploblock 43 spans two genes (Fig. 1D), an unknown protein (Solyc01g090330), and an AP2/ERF (Solyc01g090340) named GAME9, which has been reported to regulate the biosynthesis of steroidal alkaloids (Cárdenas et al., 2016). Indeed, our transgenic analysis also confirmed that the overexpression and silencing of GAME9 in the high-SGA tomato cultivar TS-286 led to significantly increased or decreased levels of both α-tomatine and hydrotomatidine (Supplementary Fig. S2).
Fig. 1.
Loci associated with hydrotomatidine content. (A) Manhattan plot for a GWAS of SGA (hydrotomatidine) content. (B) Detailed plot for 83.9–84.2 Mb on chromosome 1 (x-axis). Lead SNP is indicated by darker shading. A representation of pairwise r2 values (a measure of LD) among all SNPs in region 83.9–84.2 Mb, where the shading of each box corresponds to the r2 value as shown by the key. (C) A representation of the pairwise r2 values among all polymorphic sites in the 122.7 kb genomic region corresponding to (B), where the shade of each box corresponds to the r2 value according to the key. Haploblock 43 (marked by star) contains lead SNP associated with fruit SGA content. (D) Gene structure of 11 genes in region 83.9–84.2 Mb, an unknown protein (Solyc01g090330), and an APETALA2/Ethylene Response Factor (Solyc01g090340) in haploblock 43. (E) Model and mutation for GAME9. (F) The effect of different alleles on the content of hydrotomatidine (SlFM1985). The data were plotted as a function of a particular SNP (ch01:84029382). The middle line of the box indicates the median, the box indicates the range of the 25th to 75th percentiles of the total date, the whiskers indicate the interquartile range, and the outer dots are outliers. (G) Total SGA content in different genotypes. The box plot (detailed information same as for (F)) includes measurements for hydrotomatidine (SlFM1985), hydroxytomatidenol (SlFM0960), hydroxytomatidine (SlFM0964), dehydrofilotomatine (SlFM1979), neorickiioside A (SlFM1983), lycoperoside H (SlFM1984), and lycoperoside A (SlFM1989). The data were plotted as a function of a particular SNP (ch01:84029382). (H) The relative expression of GAME9 in red ripe tomato fruit. n refers to the number of appropriate genotypes of tomato accessions used in this study. (This figure is available in color at JXB online.)
A natural variant of GAME9 associated with SGA content in tomato
To investigate functional allelic variation at the GAME9 locus, we analysed the nucleotide sequence of GAME9 in 374 tomato accessions with diverse SGA content. The resequencing of the GAME9 coding region indicated that a polymorphism (SNP G/A, ch01:84029382) that causes an alanine to valine substitution at position 135 (A135V) in the N-terminus of AP2/ERF binding domain explained 19.87% and 23.5% of the variation in the level of hydrotomatidine and the seven SGAs in tomato fruit, respectively (Fig. 1E, F, G). A subsequent analysis indicated a significant difference (P=1.8×10–23, n=374) between the levels of the seven SGAs in the two alleles of GAME9 (Fig. 1G). Thus, the GAME9 genotype can be classified into two different haplotypes in tomato. We refer to the ‘low-SGA’ content phenotype as GAME9135V, which is mainly present in the BIG population (86.5%), and the ‘high-SGA’ content phenotype as GAME9135A, which is mainly present in the PIM (100%) and CER (71.2%) populations. Further, we randomly selected 14 accessions of high SGA and 14 accessions of low SGA and measured expression of GAME9 in red ripe fruit by quantitative RT-PCR. The expression of GAME9 showed no significant difference in red ripe fruit between high- and low-SGA accessions (Fig. 1H). These findings indicate that this allelic variant of GAME9 may play a unique role in the transcriptional regulation of SGA.
GAME9 specifically binds a GC-rich element in the GAME17 promoter
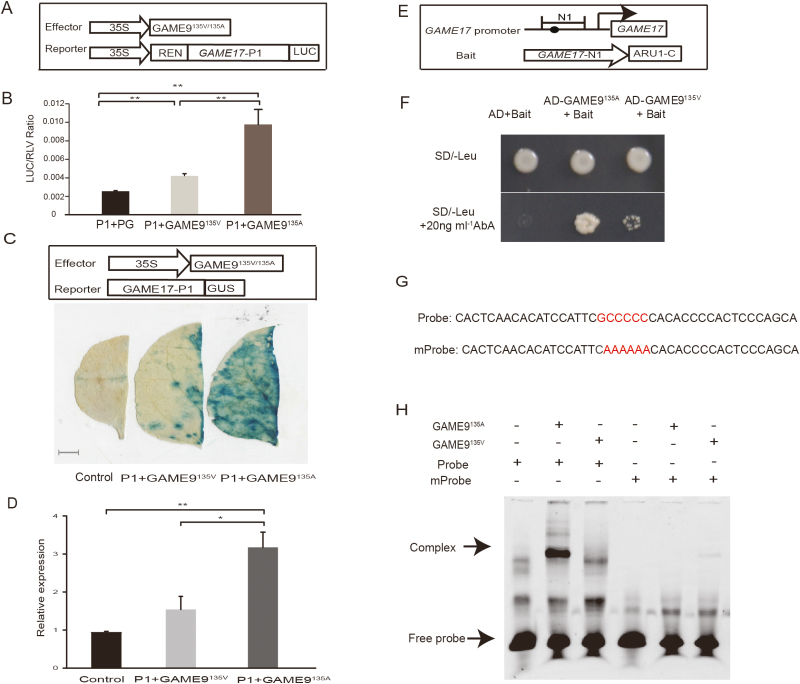
Previous study showed that ERF189, ORCA3, and GAME9 can bind to a GC-rich box (5′-CCGCCCTCCA-3′) in the tobacco PMT2 promoter (Shoji et al., 2013). We discovered similar elements in the promoters of GAME7, GAME4, and GAME17 (see Supplementary Table S4). Furthermore, expression analysis revealed that GAME4 and GAME17 were significantly down-regulated in the GAME9-RNAi lines and GAME17 was down-regulated to a greater extent than the other GAME genes (Supplementary Fig. S3). Thus, we tested whether GAME9135A can directly affect the transcription of GAME17. A yeast one hybrid (Y1H) assay indicated that GAME9135A can bind to the N1 motif (−898 to −378 bp) of the GAME17 promoter and that GAME9135A cannot bind the N2 motif (−666 to −378 bp) of the GAME17 promoter (Supplementary Fig. S4A, B). To independently test the idea that GAME9135A can bind the GAME17 promoter, we generated two promoter deletion constructs and tested whether GAME9135A could induce the transcription of these genes in tobacco (N. benthamiana) leaves. We found that the P1 sequence (from −898 bp to the translational start codon) was essential to activate a luciferase (LUC) reporter gene. In contrast, the P2 sequence (from −671 bp to the translational start codon) was not necessary (Supplementary Fig. S4C, D). Thus, we conclude that the GAME9135A protein can bind the core sequence between −898 and −671 bp in the GAME17 promoter. We tested whether the GC-rich element (5′-CGCCCCCC-3′) in this core sequence could serve as a GAME9135A binding site using an EMSA. We compared the ability of GAME9135A to bind probes containing this GC-rich sequence with probes containing mutations in this GC-rich sequence (Supplementary Fig. S4E). We found that the GAME9135A fusion protein can bind the wild-type cis-element (5′-CGCCCCCC-3′) but not the mutant cis-element (Supplementary Fig. S4F). These data indicate that GAME9135A can directly bind the promoter of GAME17 to positively activate the transcription of genes required for SGA biosynthesis.
Allelic variation in GAME9 determines its binding capacity with GAME17
To learn more about the influence of the SNP (ch01:84029382) that affects the N-terminal AP2/ERF domain of GAME9, we used a CaMV 35S promoter-driven GAME9135V effector (pGreen II 62-SK) to perform transient transactivation and promoter binding assays. The infiltrated plants revealed that the P1 sequence in the GAME17 promoter could support the transactivation activity of both GAME9135V and GAME9135A. However, the transient overexpression of GAME9135A could activate transcription of the luciferase reporter gene (3.8-fold) more than could the transient overexpression of GAME9135V (1.6-fold) (Fig. 2A, B). This result was independently verified with a GAME17 promoter-driven GUS reporter gene. The histochemical staining of GUS activity in tobacco leaves showed that with GUS expression driven by the GAME9135V effector there was little expression relative to the control, while GUS expression driven by the GAME9135A effector was significant relative to the control (Fig. 2C, D). Additionally, the GAME9135V positive yeast strains grew more slowly and showed weak binding activity relative to the GAME9135A positive yeast strains (Fig. 2E, F). Finally, EMSAs were performed to compare the binding activities of GAME9135V and GAME9135A to its binding site in the GAME17 promoter in vitro. Although the GAME9135A protein was able to retard the mobilities of GAME17 promoter fragments containing the GC-rich element, the GAME9135V protein was not able to bind the same DNA fragments (Fig. 2G, H). Taken together, GAME9135A is a more potent activator of the GAME17 promoter than is GAME9135V and the protein encoded by GAME9135V—an allelic variant of GAME9135A—appears to have weak ability to bind the GAME17 promoter.
Fig. 2.
Transactivation of GAME17 promoters by GAME9135A and GAME9135V. (A) Schematic diagrams of the constructs used for the dual luciferase assay. The reporter constructs were created by cloning promoter fragment of GAME17, P1 (−898 bp to the translational start codon), into pGreen II 0800-Luc. The effector constructs were created by cloning the full-length open reading frames of GAME9135A and GAME9135V into pGreen II 62-SK. (B) Transactivation activity of GAME9135A and GAME9135V on the GAME17 promoter in tobacco leaves. LUC, firefly luciferase activity; RLU, Renilla luciferase activity. The P1 and the pGreen II 62-SK empty (PG) vector were used as controls. Bars represent mean values ±SE (n>6). Asterisks indicate statistically significant differences determined using a t-test: **P<0.01. (C) GUS activity of GAME9135A and GAME9135V on the GAME17 promoter in tobacco leaves. Schematic diagrams of the constructs used for the GUS activity assay are shown at the top. P1: the promoter fragment of GAME17 (−898 bp to the translational start codon). P1 and pHellsgate8 empty vector were used as controls. Scale bar, 1 cm. (D) GUS expression level driven by GAME9135V and GAME9135A effector. The control and fragment of GAME17 promoter correspond to those shown in (C). A validation experiment (n=4) was performed. Asterisks indicate statistically significant differences determined using a t-test: *P<0.05, **P<0.01. (E) Schematic diagrams of the GAME17 promoter and reporter constructs used for the Y1H assay. The circles indicate the cis-acting GC-rich element (−724 to −717 bp) within the GAME17 promoter. N1 (−898 to −378 bp) indicates the promoter fragments of GAME17 that were cloned into the bait plasmid pAbAi. (F) GAME17 promoter-binding activity of GAME9135A and GAME9135V. The promoter binding activities were determined using a Y1H assay. The bait vector and the empty pGADT7 vector were co-transformed into Y1Gold as a negative control. All transformants were grown on a selective medium containing (top) or lacking (bottom) 20 ng ml−1 antibiotic (AbA). (G) Wild-type and mutant probes were used for EMSAs. The wild-type probe was synthesized based on the GAME17 promoter sequence. The cis-element sequence was replaced with AAAAAA in the mutant probe (mProbe). The hypothetical cis-element is indicated with shaded letters. (H) In vitro binding of GAME9135A and GAME9135V to the promoter of GAME17. −, absence; +, presence. The protein–DNA complex and free probe are indicated. (This figure is available in color at JXB online.)
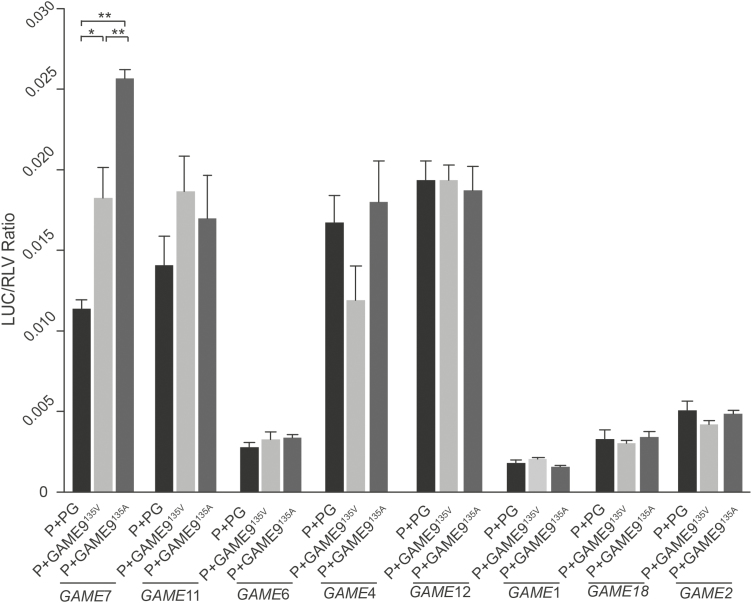
To test whether this regulatory strategy is used for other GAME genes, the transactivation of GAME7 (−682 bp to the translational start codon), GAME11 (−1581 bp to the translational start codon), GAME6 (−1112 bp to the translational start codon), GAME4 (−1201 bp to the translational start codon), GAME12 (−1612 bp to the translational start codon), GAME1 (−1220 bp to the translational start codon), GAME18 (−517 bp to the translational start codon), and GAME2 (−1551 bp to the translational start codon) by GAME9135A and GAME9135V was tested in tobacco leaves. We found that GAME9135A and GAME9135V are capable of transactivating the GAME7 promoter (Fig. 3). More importantly, a similar trend was observed with GAME7 in that GAME9135A was a more potent activator than GAME9135V (Figs 2, 3). In addition, we randomly selected 14 accessions of high SGA and 14 accessions of low SGA and measured expression of GAME17 in red ripe fruit by quantitative RT-PCR. The expression of GAME17 showed a significant difference in red ripe fruit between high- and low-SGA accessions (see Supplementary Fig. S5). So these findings showed that GAME9135A can bind the GC-rich element (5′-GCCNNCC-3′) in the promoters of GAME17 and GAME7, and relative to GAME9135A, GAME9135V weakly binds and activates the transcription of genes that contribute to SGA biosynthesis. Taken together, it is clear that the binding activity of GAME9 depends on allelic variation that influences the N-terminal AP2/ERF domain.
Fig. 3.
Transactivation assays of putative downstream gene promoters by GAME9135V and GAME9135A. The capacity of GAME9135V and GAME9135A to transactivate eight different promoters of candidate downstream genes was evaluated in tobacco leaves. P, promoter fragment of candidate downstream genes; PG, pGreen II 62-SK empty vector. A validation experiment (n>8) was performed. Asterisks indicate statistically significant differences determined using a t-test: *P<0.05, **P<0.01.
Discussion
Alkaloids are a major and extensively studied class of plant metabolites with diverse biological activities. They are widely reported in important crop plants, such as tomato, potato (Solanum tuberosum), and eggplant (Solanum melongena) (Eich, 2008; Buckingham et al., 2010). Understanding the genetic mechanisms that control the accumulation of SGAs will help us to improve the nutritional quality and consumption value of important crops, but until now, most of the research on SGAs has focused on their regulation by jasmonate, the elucidation of compound structures, and SGA biosynthesis (van der Fits and Memelink, 2000; De Boer et al., 2011; Itkin et al., 2013; Schwahn et al., 2014; Umemoto et al., 2016; Sonawane et al., 2018). The genetic control of SGA content—especially the core SGA biosynthesis pathway that acts between cholesterol and α-tomatine—is not entirely clear and requires further investigation. In this study, we provide multiple lines of experimental evidence that help to clarify the impact of genetic variation on regulation of SGAs in tomato.
As described above, we found that natural variation in GAME9 had a role in regulating the accumulation of the seven SGAs during domestication. Genetically, PIM belongs to the wild tomato group that originated in the Andean region of South America and is the ancestor of the CER and BIG groups that were later dispersed to other parts of the world and subjected to selection by local farmers and breeders (Lin et al., 2014). In general, because of their anti-nutritional activities, there was a strong selection against SGAs during the development of BIG from PIM (Zhu et al., 2018). Changes to the GAME9 protein acquired during evolution possibly affected the accumulation of SGA metabolites because GAME9 is a major regulator of SGA biosynthesis. Other than a distinct genetic factor, one possible reason for the variation of SGA content during domestication could lie in the base pair substitution in the coding sequence of GAME9 differentially regulating SGA biosynthesis. Beyond that, our analysis revealed that GAME9 is located inside a tandem duplication of an ERF gene and that among the natural populations we analysed, GAME9 is the only gene in the tandem duplication encoding a protein with a serine-rich C-terminal domain (see Supplementary Fig. S6). This finding indicates that lack of the serine-rich domain in tomato GAME9-like proteins prevents these proteins from directly participating in SGA biosynthesis but does not exclude a possible indirect contribution to SGA biosynthesis.
Functional characterization by transient luciferase expression assays, Y1H assay and EMSA showed that GAME9 can directly regulate transcription from the GAME7 and GAME17 promoters (Figs 2, 3). Furthermore, a similar element in the promoters of GAME7 and GAME17 was discovered (see Supplementary Table S4). These finding indicated that GAME9 can directly bind a GC-rich sequence that is a highly conserved functional element (5′-GCCNNCC-3′) in the promoter regions of GAME17 and GAME7 rather than the canonical GCC box to regulate the core SGA biosynthetic pathway, although GAME9 has substantial GCC box-binding activity in vitro. More importantly, the allelic variation located in the region of GAME9 encoding the N-terminal AP2/ERF domain has a decisive effect on the binding activity of GAME9 (Fig. 2). The GAME9-based regulatory strategy that controls SGA biosynthesis in tomato with the two alleles of GAME9 may be suitable not only to regulate the core SGA biosynthetic pathway but also to regulate the upstream biosynthetic genes of the cholesterol pathway and possibly other genes that act upstream of this pathway.
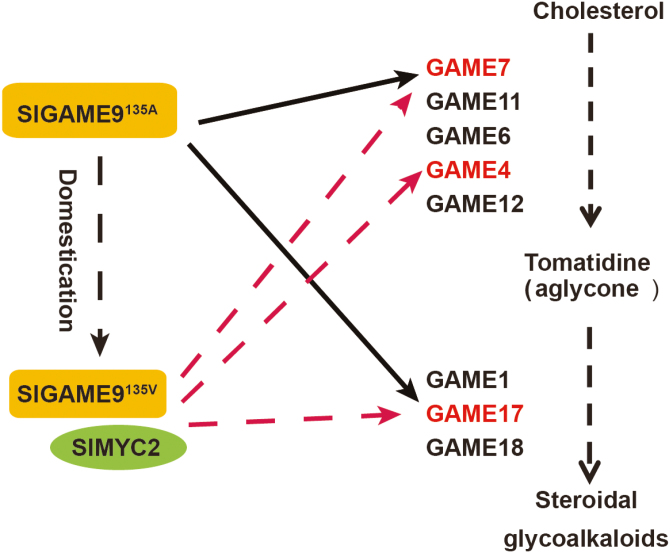
In contrast to GAME9135A, which can closely bind the GAME17 promoter, GAME9135V showed weak binding activity with the GAME17 promoter. However, how GAME9135V ensures the flux of precursors in times of SGA production in the BIG population is still unclear. A previous study reported that GAME9135V must bind another transcription factor, such as the jasmonate signaling component SlMYC2 (Solyc08g076930), to up-regulate GAME4 and GAME7 expression in a different scenario (Cárdenas et al., 2016) (Fig. 4). Meanwhile, in C. roseus and N. tabacum, the AP2/ERFs ORCA3 and ERF189 are regulated by the basic helix–loop–helix (bHLH) transcription factors CrMYC2 and NtMYC2, respectively, which potentially function as activators of gene expression by binding G-boxes in the ORCA3 and ERF189 promoters (Shoji and Hashimoto, 2011; Zhang et al., 2011). Previously, other studies reported that a jasmonate-inducible bHLH1 from Nicotiana enhanced the activity of ORCA1 (De Sutter et al., 2005; De Boer et al., 2011). Based on previous reports, we speculate that this cooperation may be more helpful and perhaps necessary to produce SGAs in the ‘low-SGA’ varieties where GAME9135V has lower transactivation activity relative to the ‘high-SGA’ material, where bHLH-like transcription factors may be helpful but not necessary.
Fig. 4.
Influence of two haplotypes of GAME9 on the steroidal alkaloid pathway. GAME9 activates the synthesis of SGAs in tomato. GAME9135A is mainly present in the PIM and CER populations and encodes a protein that can bind the GAME7 and GAME17 promoter. In contrast, GAME9135V is mainly present in the BIG population and encodes a protein that shows weak binding to the GAME17 promoter, and which might use an intermediate bHLH-like transcription factor, such as SlMYC2, to up-regulate SGA biosynthesis. (This figure is available in color at JXB online.)
In general, a high concentration of glycoalkaloids and their byproducts adversely affects the nutritional value of tomato fruit and human health (Roddick, 1996). Thus, breeding tomato varieties that accumulate low levels of alkaloids and elucidating the underlying molecular mechanism that promotes the accumulation of SGAs to facilitate these breeding efforts are significantly important. In this study, we determined that natural variation in GAME9 plays an important role in regulating SGA biosynthesis. Furthermore, in tomato, GAME9135A can bind target gene promoters and regulate SGA biosynthesis. In contrast, GAME9135V showed a weak binding activity with target gene promoters and may need to interact with an additional factor, such as SlMYC2, to enhance the binding activity with target gene promoters. Compared with GAME9135A, GAME9135V appears to have relatively less influence on the production of SGAs. This molecular mechanism is entirely different from the previously reported mechanisms that lower the toxicity of SGAs and clearly explains the regulatory mechanism of toxicity. Moreover, this control strategy is probably one of the most crucial for up-regulating the flux of precursors in times of SGA production and for balancing essential phytosterol biosynthesis and the breeding of low alkaloid tomato varieties during their domestication. In tomato, the practical value of this work is that we identified the key allelic variant (SNP G/A, ch01:84029382) in GAME9 that can be used to develop a CAPS maker that will be useful for breeding low alkaloid varieties of tomato with better nutritional value.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Manhattan plots for GWAS on six SGA contents.
Fig. S2. In vivo function of GAME9.
Fig. S3. SGA pathway-related gene expression at the mature green fruit stage in GAME9-RNAi lines.
Fig. S4. GAME9135A binds the GAME17 promoter and activates its expression.
Fig. S5. The relative expression of GAME17 in red ripe tomato fruit.
Fig. S6. Alignment of GAME9 amino acid sequences.
Table S1. Primers used in this study.
Table S2. Genes within 122.7 kb of the SNP (ch01:84016748) most highly associated with fruit hydrotomatidine content
Table S3. The SNP and their P-value (associated with fruit hydrotomatidine content) in the 86 haploblocks.
Table S4. The GC-rich motifs found in the promoters of GAME4, GAME7, and GAME17.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (31991182, 31572125 and 31872118). We thank Professor Robert M. Larkin and Professor Hanhui Kuang for critical reading of our manuscript.
Author contributions
ZY, GY, HL, and JZ planned and designed the research. GY, CL, CS, HZ, SG, GZ, HL, and ZY performed experiments and conducted fieldwork. GY, CL, LZ, GA, GZ, and ZY analysed data. GY wrote the manuscript and ZY, HL, JZ, SM, and CY revised the manuscript.
References
- Adato A, Mandel T, Mintz-Oron S, et al. 2009. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genetics 5, e1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham J, Baggaley KH, Roberts AD, Szabo LF. 2010. Dictionary of Alkaloids with CD-ROM, 2nd edn. Boca Raton: CRC Press. [Google Scholar]
- Cárdenas PD, Sonawane PD, Pollier J, et al. 2016. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nature Communications 7, 10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi TR, Lelario F, Bufo SA. 2005. Analysis of tomato glycoalkaloids by liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Communications in Mass Spectrometry 19, 3103–3110. [DOI] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J. 2013. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molecular Plant 6, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Chowanski S, Adamski Z, Marciniak P, et al. 2016. A review of bioinsecticidal activity of solanaceae alkaloids. Toxins 8, E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A. 2011. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. The Plant Journal 66, 1053–1065. [DOI] [PubMed] [Google Scholar]
- De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, Karimi M, Inzé D, Goossens A, Hilson P. 2005. Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. The Plant Journal 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Dolan LC, Matulka RA, Burdock GA. 2010. Naturally occurring food toxins. Toxins 2, 2289–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eich E. 2008. Solanaceae and Convolvulaceae: secondary metabolites: Biosynthesis, chemotaxonomy, biological and economic significance (a handbook). Berlin, Heidelberg: Springer Verlag. [Google Scholar]
- Friedman M. 2004. Analysis of biologically active compounds in potatoes (Solanum tuberosum), tomatoes (Lycopersicon esculentum), and jimson weed (Datura stramonium) seeds. Journal of Chromatography A 1054, 143–155. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Takaki A, Uehara Y, Ikeda T, Okawa M, Yamauchi K, Ono M, Yoshimitsu H, Nohara T. 2004. Tomato steroidal alkaloid glycosides, esculeosides A and B, from ripe fruits. Tetrahedron 60, 4915–4920. [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y. 1994. Gene expression in tobacco low-nicotine mutants. The Plant Cell 6, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland RE. 2009. Toxicity of tomatine and tomatidine on weeds, crops and phytopathogenetic fungi. Allelopathy Journal 23425–436. [Google Scholar]
- Iijima Y, Watanabe B, Sasaki R, Takenaka M, Ono H, Sakurai N, Umemoto N, Suzuki H, Shibata D, Aoki K. 2013. Steroidal glycoalkaloid profiling and structures of glycoalkaloids in wild tomato fruit. Phytochemistry 95, 145–157. [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, et al. 2013. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, et al. 2011. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. The Plant Cell 23, 4507–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G, Roddick MW, Leonard AL. 2000. Membrane disruption and enzyme inhibition by naturally-occurring and modified chacotriose-containing Solanum steroidal glycoalkaloids. Phytochemistry 56, 603–610. [DOI] [PubMed] [Google Scholar]
- Jared JJ, Murungi LK, Wesonga J, Torto B. 2016. Steroidal glycoalkaloids: chemical defence of edible African nightshades against the tomato red spider mite, Tetranychus evansi (Acari: Tetranychidae). Pest Management Science 72, 828–836. [DOI] [PubMed] [Google Scholar]
- Kato K, Shoji T, Hashimoto T. 2014. Tobacco nicotine uptake permease regulates the expression of a key transcription factor gene in the nicotine biosynthesis pathway. Plant Physiology 166, 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpan YI, Nazarenko EA, Skryshevskaya IV, Martelet C, Jaffrezic-Renault N, El’skaya AV. 2004. Potato glycoalkaloids: true safety or false sense of security? Trends in Biotechnology 22, 147–151. [DOI] [PubMed] [Google Scholar]
- Kozukue N, Han JS, Lee KR, Friedman M. 2004. Dehydrotomatine and α-tomatine content in tomato fruits and vegetative plant tissues. Journal of Agricultural and Food Chemistry 52, 2079–2083. [DOI] [PubMed] [Google Scholar]
- Li CY, Leopold AL, Sander GW, Shanks JV, Zhao L, Gibson SI. 2013. The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biology 13, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, et al. 2014. Genomic analyses provide insights into the history of tomato breeding. Nature Genetics 46, 1220–1226. [DOI] [PubMed] [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink J. 1999. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. The EMBO Journal 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner SE, Brunton NP, Jones PW, O’Brien NM, Collins SG, Maguire AR. 2011. Bioactivities of glycoalkaloids and their aglycones from Solanum species. Journal of Agricultural and Food Chemistry 59, 3454–3484. [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, et al. 2008. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiology 147, 823–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu M, Shioya N, Shikata M, et al. 2018. JRE4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. The Plant Journal 94, 975–990. [DOI] [PubMed] [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M. 2017. A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiology 175, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddick JG. 1990. The acetylcholinesterase-inhibitory activity of steroidal glycoalkaloids and their aglycones. Phytochemistry 29, 1513–1518. [Google Scholar]
- Roddick JG. 1996. Steroidal glycoalkaloids: nature and consequences of bioactivity. Advances in Experimental Medicine and Biology 404, 277–295. [DOI] [PubMed] [Google Scholar]
- Schwahn K, de Souza LP, Fernie AR, Tohge T. 2014. Metabolomics-assisted refinement of the pathways of steroidal glycoalkaloid biosynthesis in the tomato clade. Journal of Integrative Plant Biology 56, 864–875. [DOI] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, et al. 2014. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088. [DOI] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. 2011. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant & Cell Physiology 52, 1117–1130. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. 2010. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. The Plant Cell 22, 3390–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Mishima M, Hashimoto T. 2013. Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiology 162, 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane PD, Heinig U, Panda S, et al. 2018. Short-chain dehydrogenase/reductase governs steroidal specialized metabolites structural diversity and toxicity in the genus Solanum. Proceedings of the National Academy of Sciences, USA 115, E5419–E5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane PD, Pollier J, Panda S, et al. 2016. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nature Plants 3, 16205. [DOI] [PubMed] [Google Scholar]
- Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. 2011. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiology 157, 2081–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thagun C, Imanishi S, Kudo T, et al. 2016. Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant & Cell Physiology 57, 961–975. [DOI] [PubMed] [Google Scholar]
- Umemoto N, Nakayasu M, Ohyama K, Yotsu-Yamashita M, Mizutani M, Seki H, Saito K, Muranaka T. 2016. Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiology 171, 2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Weissenberg M, Levy A, Svoboda AJ, Ishaaya I. 1998. The effect of some Solanum steroidal alkaloids and glycoalkaloids on larvae of the red flour beetle, Tribolium castaneum, and the tobacco hornworm, Manduca sexta. Phytochemistry 47, 203–209. [DOI] [PubMed] [Google Scholar]
- Yamanaka T, Vincken JP, De Waard P, Sanders M, Takada N, Gruppen H. 2008. Isolation, characterization, and surfactant properties of the major triterpenoid glycosides from unripe tomato fruits. Journal of Agricultural and Food Chemistry 56, 11432–11440. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J. 2011. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. The Plant Journal 67, 61–71. [DOI] [PubMed] [Google Scholar]
- Zhu G, Wang S, Huang Z, et al. 2018. Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.