We demonstrate that Yr15, YrG303, and YrH52 resistances are encoded by the Wtk1 locus, but express variable resistance responses to yellow rust in a genetic background-dependent manner.
Keywords: EMS mutants, phenotypic response, positional cloning, tandem kinase domains, wild emmer wheat, Wtk1, yellow rust, Yr15, YrH52, YrG303
Abstract
The wild emmer wheat (Triticum turgidum ssp. dicoccoides; WEW) yellow (stripe) rust resistance genes Yr15, YrG303, and YrH52 were discovered in natural populations from different geographic locations. They all localize to chromosome 1B but were thought to be non-allelic based on differences in resistance response. We recently cloned Yr15 as a Wheat Tandem Kinase 1 (WTK1) and show here that these three resistance loci co-segregate in fine-mapping populations and share an identical full-length genomic sequence of functional Wtk1. Independent ethyl methanesulfonate (EMS)-mutagenized susceptible yrG303 and yrH52 lines carried single nucleotide mutations in Wtk1 that disrupted function. A comparison of the mutations for yr15, yrG303, and yrH52 mutants showed that while key conserved residues were intact, other conserved regions in critical kinase subdomains were frequently affected. Thus, we concluded that Yr15-, YrG303-, and YrH52-mediated resistances to yellow rust are encoded by a single locus, Wtk1. Introgression of Wtk1 into multiple genetic backgrounds resulted in variable phenotypic responses, confirming that Wtk1-mediated resistance is part of a complex immune response network. WEW natural populations subjected to natural selection and adaptation have potential to serve as a good source for evolutionary studies of different traits and multifaceted gene networks.
Introduction
Wheat has been the basic staple food for the major civilizations of Europe, West Asia, and North Africa for at least 10 000 years (Nevo et al., 2002). Today, common wheat (Triticum aestivum L.) and durum wheat [T. turgidum ssp. durum (Desf.) Husnot] provide 20% of the calories and proteins for human consumption, as well as vitamins, dietary fibers, and phytochemicals (Shewry and Hey, 2015). Wheat annual yield reaches >>750 Mt (Food and Agriculture Organization Corporate Statistical Database, FAOSTAT); however, losses due to biotic (pathogens) and abiotic (unfavorable growth conditions) stresses prevent the maximum yield potential from being achieved.
Wheat yellow rust, also known as a stripe rust, is caused by the basidiomycete fungus Puccinia striiformis f. sp. tritici (Pst), an obligate pathogen that threatens wheat production around the globe (Chen, 2005). Yield losses due to yellow rust have ranged from 10% to 70% in susceptible varieties, and a total yield loss (100%) can occur under severe epidemics (Chen, 2005). Host resistance is considered to be the most economically and environmentally friendly strategy for yellow rust control (Chen, 2005), but widespread use of initially effective resistance genes can lead to rapid breakdown of resistance; for example, the appearance of Pst races that overcome widely deployed resistance genes (R-genes), such as Yr2, Yr9, Yr17, and Yr27, has led to destructive pandemics (Wellings, 2011; Hovmøller et al., 2015). Moreover, the rapid evolution of the pathogen facilitates an expansion of Pst into new regions, and therefore becoming an emerging issue in some countries, such as western Canada (Brar et al., 2018). Thus, resistant wheat variety breeding is a continuous process to withstand yellow rust epidemics globally, using all possible sources of Pst resistance, in order to widen and diversify the existing R-gene pool (Roelfes et al., 1992).
Wild emmer wheat (WEW), T. turgidum ssp. dicoccoides (Körn. ex Asch. & Graebner) Thell. (BBAA), discovered in 1906 in Rosh Pina, Israel by A. Aaronsohn (Aaronsohn, 1910), has been recognized as an important source for novel yellow rust resistance (Yr) genes (Fahima et al.,1998; Huang et al., 2016a; Klymiuk et al., 2019a). WEW is the undomesticated polyploid progenitor for modern cultivated tetraploid durum wheat (BBAA) and hexaploid common wheat (BBAADD), and natural populations still grow in a wide range of ecogeographical conditions distributed across the Near East Fertile Crescent (Özkan et al., 2011). These natural populations can serve as a model to study the evolutionary processes that shaped the currently observed allelic variation (Yahiaoui et al., 2009; Sela et al., 2011; Huang et al., 2016b; Lundström et al., 2017; Klymiuk et al., 2019b). Several previous studies have reported that WEW accessions exhibit high levels of resistance to inoculation with Pst isolates (Gerechter-Amitai and Stubbs, 1970; Nevo et al., 1986; Van Silfhout, 1989). Initially, Yr genes were considered novel if they expressed distinct reaction patterns to a set of Pst isolates. With this definition, Van Silfhout (1989) predicted the presence of at least 11 major Yr genes in WEW populations. Currently, the genetic position of an identified gene compared with those of previously mapped loci is considered to determine novelty (McIntosh et al., 2017). So far, six WEW-derived Yr genes have been recognized (Yr15, YrH52, Yr35, Yr36, YrTz2, and YrSM139-1B) on chromosome arms 1BS and 6BS (Huang et al., 2016a; Klymiuk et al., 2019a). Among these, only Yr36 (Fu et al., 2009) and Yr15 (Klymiuk et al., 2018) have been cloned so far.
Yr15 was discovered in WEW accession G25 (G25) by Gerechter-Amitai et al. (1989) and was shown to confer resistance against a worldwide collection of >3000 genetically diverse Pst isolates (Sharma-Poudyal et al., 2013; Ali et al., 2017; Chen and Kang, 2017; Liu et al., 2017). Only a few isolates virulent on Yr15 from Afghanistan (Van Silfhout, 1989) and Denmark (Hovmøller and Justesen, 2007) have been reported. Yr15 was localized to the short arm of chromosome 1B (Sun et al., 1997; Peng et al., 2000b; Yaniv et al., 2015), and its positional cloning revealed that it encodes a protein with 665 amino acid residues (Klymiuk et al., 2018). Yr15 protein was designated as wheat tandem kinase 1 (WTK1) since it possesses a tandem kinase–pseudokinase (TKP) domain architecture and phylogenetically groups with other proteins sharing similar TKP structure (Klymiuk et al., 2018). Functional (Wtk1) and non-functional (wtk1) WTK1 alleles differ by insertions of transposable elements, indels, and stop codons (Klymiuk et al., 2018). Functional markers detected alternative alleles in WEW, T. turgidum ssp. durum, and T. aestivum that were consistent with the phenotypic responses (Klymiuk et al., 2019b).
YrH52 was also identified in WEW, and its introgression into adapted cultivars of durum wheat and common wheat provides effective resistance to Pst (Peng et al., 1999; Klymiuk et al., 2019a). A primary genetic map of YrH52 on chromosome 1BS was developed by Peng et al. (1999, 2000a, b) using a segregating population from a cross between WEW accession H52 (the donor line of YrH52) and T. turgidum ssp. durum cv. Langdon. YrG303 was identified in the WEW donor line G303 and was shown to provide resistance to 28 Pst isolates from 19 countries tested by Van Silfhout (1989).
Yr15, YrG303, and YrH52 all localize to the distal region of chromosome 1B of wheat. In the current study, we used positional cloning and functional validation to demonstrate that Yr15, YrG303, and YrH52 are encoded by the functional Wtk1 allele. Nevertheless, WEW accessions and introgression lines that carry Yr15, YrG303, and YrH52 show different phenotypic responses after challenge with Pst. Mutations in WTK1 in the yr15, yrG303, and yrH52 susceptible mutants demonstrate conserved regions in critical kinase subdomains likely to be important for functionality.
Materials and methods
Development of mapping populations and introgression lines
YrH52
The YrH52 mapping population used in the current study consisted of 3549 F2 plants derived from a cross between resistant WEW (male) accession H52 (TD010027 from ICGB, Institute of Evolution, University of Haifa) collected in Mt. Hermon (N33°17'19'', E35°45'18''), the donor of YrH52 (Fig. 1), and a susceptible durum wheat (female) cv. Langdon (Peng et al., 1999). A homozygous resistant BC3F2 Ariel-YrH52 introgression line (LDN/H52//3*Ariel) was used to develop ethyl methanesulfonate (EMS) yrH52 mutants.
Fig. 1.
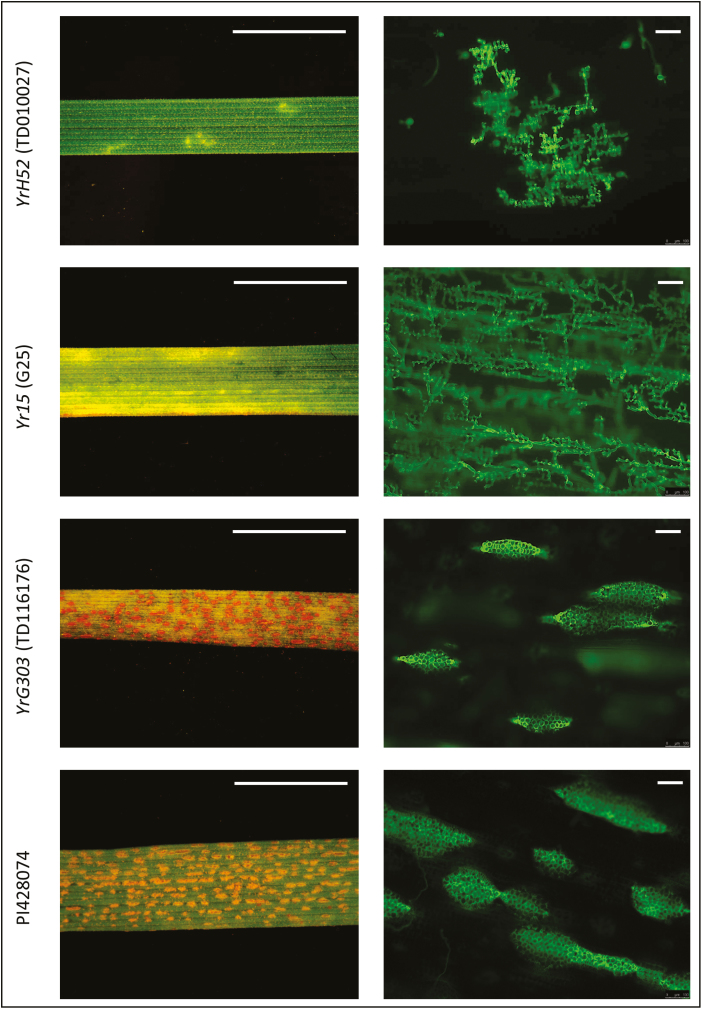
Differences in seedling phenotypic responses of WEW accessions donors of YrH52, Yr15, and YrG303, and the susceptible control PI428074 accession, at 14 dpi with Pst isolate #5006. Left panel: binocular microscopic observation of hypersensitive response and fungal sporulation (scale bar=1 cm). Right panel: sizes of fungal colonies and uredinia bags visualized by the fluorescent dye WGA–FITC (scale bar=100 µm). Note that G303 plants exhibited variable levels of resistance responses upon inoculation at the seedling stage (IT1–IT5) and only predominant IT5 is presented here. (This figure is available in color at JXB online.)
YrG303
An F2 population (1917 plants) was developed for fine mapping of YrG303 by crossing resistant WEW (male) accession G303 (TD116176 from ICGB, Institute of Evolution, University of Haifa) collected in Dishon, Israel (N33°05'09'', E35°31'03''; Fahima et al., 1998), the donor of the YrG303 gene (Fig. 1), with a susceptible T. turgidum ssp. durum accession D447 (LD393/2*Langdon ND58-322). A resistant hexaploid line 2298 (Vaskar*3/G303//Avocet) and resistant recombinant inbred lines (RILs) of the A95 mapping population derived from crossing 2298 with susceptible 2463 (Vaskar*3/G303//Avocet) were used for development of yrG303 mutants.
All parental lines were confirmed to be near homozygous using the 15K wheat single nucleotide polymorphism (SNP) array (Muqaddasi et al., 2017) (Trait Genetics GmbH, Gatersleben, Germany) consisting of 12 905 SNPs selected from the wheat 90K array (Wang et al., 2014).
Yellow rust test under controlled growth chamber conditions
Yellow rust test was carried out with Pst isolate #5006 (race 38E134) using a standard protocol for Pst inoculation described in Klymiuk et al. (2019b). Ten plants per genotype were inoculated at the seedling stage (the two-leaf stage). The yellow rust response variation was evaluated 14–18 days post-inoculation (dpi) using a 0–9 scale of infection type (IT) (Line and Qayoum, 1992) with the following interpretation of the results: IT=0–3 were considered as resistant, IT=4–6 as moderately resistant, and IT=7–9 as susceptible.
Field experiment for yellow rust assessment at the adult stage
A field experiment was conducted in order to evaluate Pst resistance of F4 recombinant lines at the adult stage. Homozygous recombinant lines from the tetraploid YrG303 mapping population, related parental lines G303, D447, 2298, and 2463, as well as susceptible control T. aestivum cv. Morocco were planted in the Institute of Evolution field located on Mt. Carmel (32°45'30.276''N, 35°1'21.7596''E) during the 2016–2017 winter growing season. Field design included three replicates per genotype randomly distributed within three 15 m long rows. Each replicate included three plants within a row planted at a distance of 30 cm between lines and 15 cm between plants. Seeds were germinated in rolled wet paper, kept in a dark cold room (4 °C) for 3 d, and then transferred to a growth room (20 °C) with a 14/10 h light/dark regime for 24 h before sowing in soil. Only seeds that showed good germination and root development were transferred to the field. Plants were inoculated at the flag leaf stage (adult inoculation) using a sprayer with fresh Pst isolate #5006 spores suspended in Soltrol® 170 light oil (Chevron Phillips Chemical Company, The Woodlands, TX, USA) 1 h before sunset at average day/night temperatures of 22/12 °C. To ensure inoculation, spreaders inoculated in controlled chamber conditions were evenly distributed in pots in the field, every second line within rows. Phenotypes were scored 14, 21, and 29 dpi for each plant; the most advanced IT was considered as the final score.
Histopathological characterization of Pst–wheat interactions within leaf tissues
Fluorescent visualization of P. striiformis structures during infection was conducted according to the protocol described by Dawson et al. (2015), with slight modifications. This protocol used wheat germ agglutinin (WGA; a lectin that binds specifically to β-(1→4)-N-acetyl-d-glucosamine, i.e. chitin) conjugated with a fluorescent dye to visualize the intercellular fungal growth and pustule formation on infected leaves. Leaf segments (10 cm long, second leaf) were sampled at 14 dpi from WEW accessions G25, G303, and H52 inoculated with urediniospores of Pst isolate #5006 as described above.
The sampled leaf segments were placed in 15 ml centrifuge tubes containing 15 ml of 1 M KOH and 2–3 drops of the surfactant alkylaryl polyether alcohol (Spreader DX) for clearing the tissue. Tubes were kept at 37 °C for 24 h, followed by three washes of samples with 50 mM Tris (pH 7.5) for neutralization of pH. After the last wash, the leaves were gently transferred to 9 cm plastic Petri dishes, excess Tris solution was removed, and 1 ml of 20 µg ml–1 WGA conjugated to the fluorophore Alexa 488 (L4895-2MG; Sigma-Aldrich) in 50 mM Tris was placed on the leaf surface. The leaves were stained with WGA for 24 h at 4 °C, and then washed with ddH2O. Stained leaf tissues were gently placed on microscope slides, immersed with antifade mounting medium for preserving fluorescence (Vectashield, Vector Laboratories), covered with a cover glass, sealed with rubber cement, and stored at 4 °C in the dark. Fluorescence microscopy was performed on an inverted fluorescence microscope, Leica DMi8 (Leica Microsystems, Wetzlar, Germany), fitted with a filter cube for the fluorescein isothiocyanate (FITC) excitation range (excitation 460–500 nm; dichroic filter 505 nm; emission 512–542 nm), and a FLUO regime to observe the WGA-stained fungal structures. Three plants of each wheat genotype were used for the investigation, and whole 10 cm long leaf segments were examined in each case. Images of the most predominant fungal colonies per field of view were recorded for each genotype.
Development of CAPS and KASP markers
Detailed protocols for the development and use of cleaved amplified polymorphic sequence (CAPS) markers for the screening of mapping populations are described in Raats et al. (2014). The sequence of the newly developed CAPS marker uhw290 is presented in Supplementary Table S1 at JXB online.
For development of new kompetitive allele-specific PCR (KASP) markers, the G303, D447, H52, and Langdon parental lines of the YrG303 and YrH52 mapping populations were genotyped using the 15K wheat SNP array. Polymorphic markers, residing on chromosome 1BS between Yr15 flanking simple sequence repeat (SSR) markers barc8 and gwm273, were identified based on their location on the consensus tetraploid wheat genetic map (Maccaferri et al. 2015). Sequences of SNP markers (RFL_Contig2160_617, IACX502, Ra_c16879_977, BS00087784_51, Excalibur_c17202_1833, wsnp_Ku_c4911_8795151, wsnp_Ex_c2111_3963161, and RAC875_c79370_378) were converted into KASP markers using the software Polymarker (Ramirez-Gonzalez et al. 2015). The primer sequences of these KASP markers are presented in Supplementary Table S2.
Detailed descriptions of the development, sequences, and conditions for amplification of KASP Yr15 functional molecular markers are provided in Klymiuk et al. (2019b). Names, primer sequences, and references of other previously published markers are presented in Supplementary Table S1.
Construction of genetic linkage maps
Primary genetic maps were developed for each of the genes, Yr15 (Klymiuk et al., 2018), YrG303, and YrH52, using the following markers: SSR markers barc8 and uhw273; KASP markers RAC875_c826_839 and BS00022902_51; and CAPS markers uhw259 and uhw264. The primary genetic maps were constructed using the MultiPoint package (http://www.multiqtl.com/). For high-resolution mapping, F2 plants from large mapping populations were screened first with two markers flanking each of the target genes. Plants that showed recombination events between the two markers were self-pollinated and progressed to F3. Ten to sixteen F3 plants of each of the F2 recombinants were analyzed with markers, and homozygous RILs were selected. The F3 RILs were then screened with molecular markers that resided within the defined YrG303 or YrH52 interval. Phenotyping of F3–4 RILs for response to Pst inoculation was performed as described above. High-resolution genetic maps were constructed using the graphical genotypes approach (Young and Tanksley, 1989). Genetic distances obtained for the low-resolution maps were used as a reference to estimate the relative genetic distances within the high-resolution maps.
Collinearity between genetic and physical maps
The best hits of a BLASTN search of the corresponding primer sequences for each genetic marker against the three genome assemblies of wheat, T. dicoccoides Zavitan (Avni et al., 2017), T. turgidum ssp. durum Svevo (Maccaferri et al., 2019), and T. aestivum Chinese Spring (Appels et al., 2018), were used to estimate average physical distances between markers. Visualization of the genetic and physical maps was performed with MapChart 2.2 software (Voorrips, 2002).
EMS mutagenesis and screening for susceptible mutants
Seeds of the homozygous resistant T. aestivum line 2298 carrying an introgression from WEW G303 that harbors the YrG303 locus and 14 homozygous resistant F6 RILs of the A95 population were treated with 0.4% EMS following the protocol described in Klymiuk et al. (2018), in order to obtain susceptible yrG303 mutants. EMS-treated M1 plants were grown in the Institute of Evolution field located on Mt. Carmel. Seedlings of M2 families of F6 RILs of the A95 population (10–20 seeds per family) were artificially inoculated with Pst under field conditions as described above, whereas seedlings of M2 families of the 2298 line (12 seeds per family) were screened under growth chamber conditions as described above.
Following the same EMS mutagenesis protocol (Klymiuk et al., 2018), seeds of T. aestivum line Ariel-YrH52 that carry an introgression from the WEW H52 line with the YrH52 locus were treated with 0.5% EMS for development of loss-of-function yrH52 mutants. EMS-treated M1 plants were grown and seedlings of M2 families (10–20 seeds per family) were screened for the response to inoculation with Pst under field conditions at the Institute of Evolution field located on Mt. Carmel as described above.
All M3 families (yrG303 and yrH52) obtained from M2 susceptible plants were inoculated in a growth chamber, as described above, to confirm homozygosity of the recessive mutations.
Sequencing of WTK1 from the mutants
DNA was isolated from freeze-dried leaves of M3 plants using the standard cetyltrimethylammonium bromide (CTAB) protocol (Doyle, 1991). Coding regions of WTK1 were sequenced from each yrG303 and yrH52 mutant using WJKDF1/WJKDR1, WJKDF2/WJKDR2, and WJKDF3/WJKDR3 primer pairs (Klymiuk et al., 2018), and screened for mutations. In order to confirm the absence of the detected mutations in the resistant background, we sequenced WTK1 regions spanning the identified mutations from two M2-derived resistant sister lines for each of the yrG303 mutants.
Structure of WTK1 protein domains
Full-length sequences of Wtk1 from G25, G303, and H52 were previously published (Klymiuk et al., 2018, 2019b) and have been deposited in NCBI GenBank under accession numbers MG649384, MK188918, and MK188919, respectively. An alignment of WTK1 KinI and KinII domains was performed with ClustalW software (Thompson et al., 1994). Secondary structures of kinase-like and pseudokinase-like domains were obtained using the web server ‘PredictProtein’ (Rost et al., 2004). Key conserved residues, the ATP-binding site, the catalytic loop, and the activation loop were defined as previously described (Klymiuk et al., 2018).
Results
YrH52 and YrG303 express distinct phenotypes in response to Pst inoculation
YrH52 in the WEW accession H52 background provides a strong resistance response (IT1) to inoculation, with the Pst isolate #5006 displaying only small dots or spots of hypersensitive response (HR) in sites of fungal penetration and initial establishment of fungal colonies (Fig. 1). The HR area corresponds to the size of fungal colonies visualized by fluorescence microscopy with WGA–FITC fluorescent dye (Fig. 1). In general, the sizes of colonies as well as the amount of HR needed to stop fungal development in YrH52 were much smaller than those of Yr15 in WEW backgrounds (Fig. 1).
YrG303 in the WEW donor background in most cases (seven out of 10 tested plants) displayed a moderate resistance response (IT5) to inoculation with Pst isolate #5006 characterized by chlorotic areas, indicating an extensive HR, accompanied by some level of fungal sporulation (Fig. 1). Such an intermediate level of resistance response of YrG303 is distinct from the strong responses of Yr15 and YrH52 (Fig. 1). Furthermore, fluorescence microscopy revealed development of a massive net of fungal hyphae and some uredinia bags with spores in WEW G303 plants inoculated with Pst (Fig. 1). However, it should be noted that some G303 plants showed IT1, IT3, and IT4 (one out of 10 tested plants for each mentioned IT).
The tetraploid YrG303 F2 mapping population showed the expected phenotypic segregation for a single dominant resistance gene 3:1 (χ 2=1.47; P=0.1) in response to Pst inoculation based on phenotyping of 843 F2 plants, and F1 plants exhibit full resistance with IT1. We used the IT scale of 0–9 (Line and Qayoum, 1992), based on which the G303 resistant parent showed IT1–IT5 and the D447 susceptible parent showed IT8–IT9. Thus, phenotypic responses of F2 plants with IT1–IT6 were classified as resistant, while those with IT7–IT9 were classified as susceptible. To confirm moderate resistance phenotypic responses (IT4–IT6), all homozygous recombinants were phenotyped in field trials at the adult stage at the F4–5 generation (Supplementary Fig. S1).
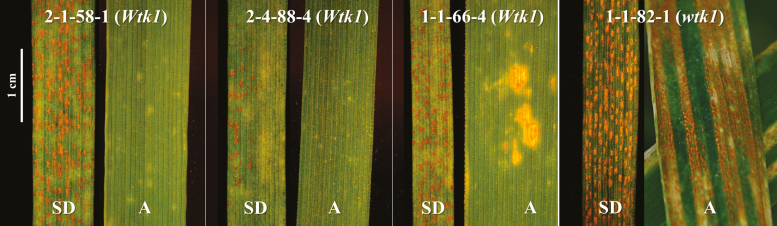
The fine mapping of YrG303 demonstrated that three out of the 124 homozygous recombinant lines were susceptible at the seedling stage even though they were expected to be resistant based on their genotype (Supplementary Fig. S1). We repeated seedling inoculation experiments in multiple generations (F3–F5) with similar results. However, artificial field inoculation with the same Pst isolate at the adult stage resulted in resistance response of these lines (Fig. 2). Furthermore, G303 plants exhibited different levels of resistance responses upon inoculation at the seedling stage (IT1–IT5) as compared with complete resistance (IT0–IT1) at the adult stage. In addition, after introgression of YrG303 to the hexaploid common wheat cultivar Avocet (introgression line 2298), this gene provided full resistance at the seedling stage to Pst inoculation with IT1 (Fig. 3).
Fig. 2.
Comparisons of seedling and adult responses of four recombinant lines from the YrG303 segregating mapping population (G303×D447) to inoculation with Pst isolate #5006 at 14 and 21 dpi, respectively. SD, seedling inoculation under controlled growth chamber conditions; A, adult plant inoculation under field conditions. RILs 2-1-58-1, 2-4-88-4, and 1-1-66-4 harbor the functional Wtk1 allele, while 1-1-82-1 harbors the non-functional wtk1 allele, and was used as a susceptible control. (This figure is available in color at JXB online.)
Fig. 3.
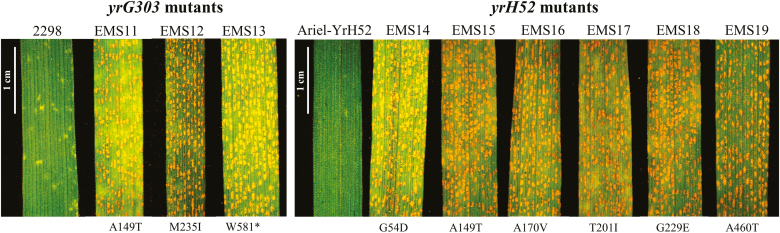
Susceptible reaction of yrG303 and yrH52 mutants to Pst inoculation at 14 dpi with Pst isolate #5006. 2298 and Ariel-YrH52 are wild-type YrG303 and YrH52 hexaploid introgression lines, respectively, used to develop mutants. (This figure is available in color at JXB online.)
Fine mapping of YrH52 and YrG303
YrH52
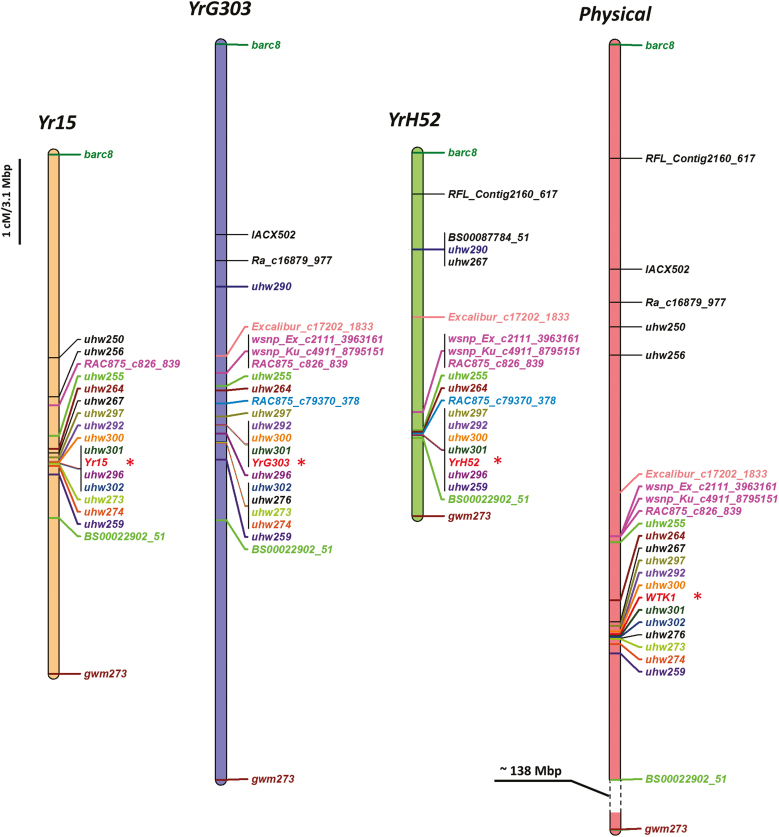
Previously, YrH52 was mapped using low-resolution mapping as proximal to Yr15 (Peng et al., 2000a, b). A high-resolution genetic map of the YrH52 genetic region was constructed using an F2 mapping population. In total, 3549 F2 plants of the YrH52 mapping population were screened for recombinants using the YrH52 flanking markers, of which 3211 F2 plants were screened using wmc406 and gwm413 markers (218 recombination events were detected) and 338 F2 plants were screened with barc8 and gwm273 markers (12 recombination events were detected). In total, 194 homozygous F3–4 RILs were developed and used for high-resolution mapping based on the graphical genotypes approach (Fig. 4). The genetic region between barc8 and gwm273 markers was saturated with 10 CAPS and eight KASP markers (Supplementary Fig. S2). Fine mapping of YrH52 revealed that its genetic position co-segregated with three dominant markers (uhw292, uhw300, and uhw301) and three co-dominant markers (uhw297, uhw296, and uhw259), and overlaps with the location of the Yr15 locus (Klymiuk et al., 2018). Sequencing of the full-length genomic WTK1 from WEW H52 revealed that it is identical to Wtk1 from WEW G25.
Fig. 4.
Genetic and physical maps of Yr15, YrG303, and YrH52 showing the same position for all three genes that correspond to WTK1. The consensus physical map represents three reference genomes, based on 1BS pseudomolecules of WEW Zavitan, T. durum Svevo, and T. aestivum Chinese Spring. The consensus physical map contains only collinear markers between the genetic and the physical maps. (This figure is available in color at JXB online.)
YrG303
We conducted preliminary experiments and localized YrG303 distal to marker gwm413, suggesting that YrG303 is different from Yr15, which was mapped proximal to gwm413 (Yaniv et al., 2015). In order to localize YrG303 more precisely relative to Yr15, we performed fine mapping of this locus. A high-resolution genetic map was developed by screening of 1381 F2 plants, from the YrG303 tetraploid mapping population, with SSR markers gwm273 and barc8 flanking a chromosome interval of 8.5 cM, and then 536 F2 plants with internal SNP markers RAC875_c826_839 and BS00022902_51, flanking a 1.7 cM interval that harbors the target gene (Fig. 4). These screenings identified heterozygous F2 recombinant lines within the region spanning YrG303. From them, a total of 124 homozygous F3–4 recombinants were developed and used for graphical genotyping of YrG303. These recombinants were genotyped with 23 PCR markers (SSR, CAPS, KASP, and dominant gene-specific markers) that showed polymorphisms between the parental lines (G303 and D447) (Fig. 4; Supplementary Fig. S1) and phenotyped by Pst inoculation. Three dominant gene-specific markers (uhw292, uhw300, and uhw301), previously mapped to chromosome 1BS (Klymiuk et al., 2018), were found to be co-segregating with the YrG303 Pst resistance phenotype and verified the location of YrG303 on the short arm of chromosome 1B. The fine mapping of the gene revealed that the position of YrG303 was the same as the Yr15 locus (Klymiuk et al., 2018) (Fig. 4). Sequencing of the full-length genomic WTK1 from a WEW G303 individual plant showing IT1, and another one showing IT5, revealed full identity between the two sequences and Wtk1 from WEW G25.
Validation of WTK1 as a candidate gene for YrG303 and YrH52 resistance genes by EMS mutagenesis
A total of ~2500 seeds of the YrG303 introgression lines and ~1000 seeds of the YrH52 introgression line were treated with EMS. The germination rate of M1 seeds ranged between 73% and 80%. For YrG303, we screened ~800 M1-derived families generated from F6 RILs of the hexaploid A95 population, and only one susceptible mutant was identified. Furthermore, we screened ~1200 M1-derived families generated from hexaploid line 2298, and two additional susceptible mutants were identified. Sequencing of WTK1 from these susceptible lines revealed that they all carried independent mutations in the coding region of the WTK1 sequence (Fig. 3; Supplementary Table S3). For YrH52, we screened 724 M1-derived families generated from the hexaploid introgression line Ariel-YrH52 and identified six mutants that all contained independent mutations in WTK1 (Fig. 3; Supplementary Table S3).
Point mutations result in disruption of WTK1 function
WTK1 possesses two distinct kinase domains, with the first kinase domain (KinI) predicted to encode a functional kinase, while the second (KinII) lacks several critical residues and therefore is predicted to be a pseudokinase (Klymiuk et al., 2018). All 19 susceptible EMS mutants developed based on Yr15 (Klymiuk et al., 2018), YrH52, and YrG303 introgression lines were analyzed for the positions of specific mutations within Wtk1 (Fig. 5). Five susceptible yr15 mutants carried SNP changes in the KinI kinase domain and five carried SNP changes in the KinII pseudokinase domain (Klymiuk et al., 2018). The majority of the non-functional yrG303 and yrH52 mutants developed within the current study carried mutations in KinI; only one yrG303 mutant and one yrH52 mutant carried mutations in the KinII pseudokinase domain (Supplementary Table S3). Resistant sister lines of all yrG303 mutants deriving from the same M1 families showed the presence of the wild-type allele of Wtk1, therefore validating that the mutations in KinI and KinII domains are responsible for the loss of function, and that the resistance conferred by Yr15, YrG303, and YrH52 is encoded by only one gene.
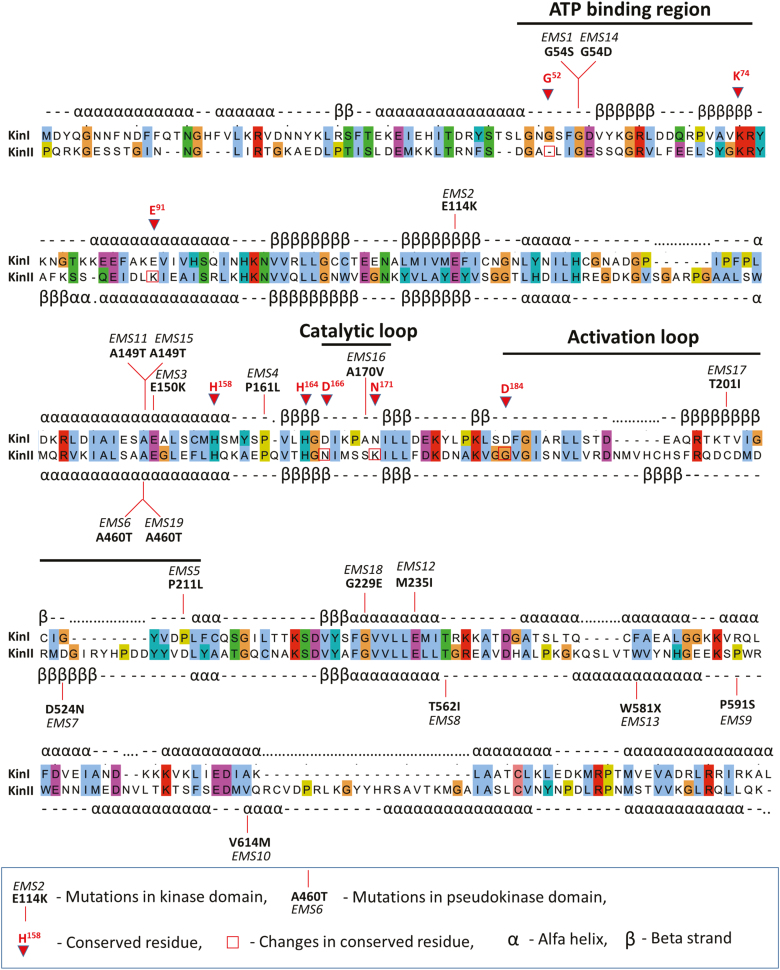
Fig. 5.
Primary and secondary structures of WTK1 kinase and pseudokinase domains alongside positions of knockout EMS mutations in yr15, yrG303, and yrH52 susceptible mutants. The diagram of WTK1 domain architecture highlights eight key conserved residues in the kinase domain (with numbers that correspond to their positions in cAPK (Hanks et al., 1988)) and the absence of five of them in the pseudokinase domain. Vertical lines indicate EMS mutations that block resistance. KinI, kinase domain; KinII, pseudokinase domain. (This figure is available in color at JXB online.)
The EMS mutations in WTK1 that affect recognition in yr15, yrG303, and yrH52 susceptible mutants have not been identified in key conserved residues, but do map to conserved regions in critical kinase subdomains such as the ATP-binding region, catalytic loop, and activation loop (Fig. 5). Three amino acids (Gly54, Ala149, and Ala460) were disrupted in different wheat genetic backgrounds and all inhibited WTK1-mediated immunity to yellow rust, confirming that these residues are critical for WTK1 responses (Fig. 5). Gly54 is located in KinI’s ATP-binding region and may directly affect WTK1’s kinase activity (Fig. 5). Ala149 and Ala460 map to the same α-helical region present in KinI and KinII domains. Neither Ala149 nor Ala460 maps to catalytic residues, but their presence in the same region of KinI and KinII indicates that these alanine residues may be required for proper WTK1 folding and/or Pst effector binding.
Discussion
Recent advances in the plant innate immunity system have shed light on possible mechanisms and organization of resistance networks. However, identification of R-genes and an understanding of their allelic series serve as an essential initial step to dissect complex plant–pathogen interactions. Many studies have been performed to discover novel Pst R-genes. However, the deployment of novel R-genes from WEW into common wheat is a long process due to ploidy differences, negative linkage drag, etc. For R-genes that map to similar chromosome regions, it is important to determine if they are in fact different genes or alleles of the same genes because this will impact strategies for their effective deployment.
YrG303, YrH52, and Yr15 R-genes derived from WEW
Yr15, YrG303, and YrH52 genes originated from different WEW accessions and were all mapped to the short arm of chromosome 1B. However, they were initially considered distinct R-genes, due to differences in geographic distribution of their donor lines and different resistance reaction patterns in response to inoculation with the same Pst races. Yr15 originated in WEW accession G25 collected in Rosh Pinna, Israel (Gerechter-Amitai et al., 1989), while WEW accession G303 harboring YrG303 was collected in Dishon (Israel), and WEW accession H52, the donor line of YrH52, was collected in Mt. Hermon (Israel), that represent different habitats. These three WEW accessions showed different level of resistance to Pst isolate #5006 from Israel, with YrH52 (H52) displaying IT1, Yr15 (G25) IT3, and YrG303 (G303) IT5 all proved to have corresponding differences at the level of development of pathogenic fungal colonies (Fig. 1). Fine genetic mapping of the three genes demonstrated that all three map to the same genetic interval on chromosome arm 1BS (Fig. 4; Supplementary Figs S1, S2). Moreover, sequencing of WTK1 from the susceptible yrG303 and yrH52 EMS mutants showed that all of them contain loss-of-function mutations in Wtk1 (Figs 3, 5; Supplementary Table S3). Thus, other genes reported to be located on chromosome 1BS, especially those originating from WEW, such as YrSM139-1B (Zhang et al., 2016) and YrTz2 (Wang et al., 2018), should be tested for allelism/identity to Wtk1.
The sequences of Wtk1 from G25, G303, and H52 have been published previously (Klymiuk et al., 2018, 2019b); however, in G303 and H52 genetic backgrounds, we did not prove that the resistance phenotype is conferred by the Wtk1 allele. Here, based on fine genetic mapping, we show that the genetic positions of YrG303 and YrH52 indeed coincide with the physical position of Wtk1 (Fig. 4), and that mutations in Wtk1 lead to susceptibility in both YrG303 and YrH52 introgression lines (Figs 3, 5; Supplementary Table S3). Furthermore, YrG303 and YrH52 both possess a Wtk1 functional allele that is identical to the Yr15 sequence of Wtk1. We hypothesize that such high sequence conservation represents a specific character of TKP proteins as compared with the high level of variability observed for the known nucleotide-binding domain and leucine-rich repeat (NLR) proteins (MacQueen et al., 2019). Clustering, rapid evolution, and the presence of multiple alleles is a known character of many NLRs (Bhullar et al., 2009; Marchal et al., 2018; Adachi et al., 2019). TKPs also tend to cluster together, probably reflecting the evolutionary mechanisms by which they evolved via duplication or fusion of two kinase domains (Klymiuk et al., 2018). For example, WTK1 has three tandem copies on each of chromosomes 6A and 6B (Klymiuk et al., 2019b). However, in contrast to NLRs, we did not detect diverse Wtk1 alleles, and differences in phenotypic responses are likely to be associated with differences in the genetic backgrounds as discussed below.
The spectrum of Wtk1 phenotypic responses
Three components, host, pathogen, and environment, serve as a basis for the concept of the disease triangle and determine the degree of severity of the disease (Agrios, 2005). According to the gene for gene model, host–pathogen interactions result in co-evolution of virulence genes (e.g. pathogen effectors) and resistance genes (e.g. host receptors) (Flor, 1971). However, even in the case of compatible interaction between host and pathogen, disease may not occur or symptoms will be limited due to unfavorable environmental conditions for pathogen development (Agrios, 2005). The plant immunity system is multifaceted and comprises recognition (via receptors) and response (a transduction network of multiple genes deployed) parts (Jones and Dangl, 2006). Different alleles of various R-genes, such as wheat Yr5/YrSP (Marchal et al., 2018), wheat Pm3 (Srichumpa et al., 2005), barley Mla (Seeholzer et al., 2010), flax L locus (Ellis et al., 1999), tomato Pto (Rose et al., 2005), Arabidopsis Rpm1 locus (Stahl et al., 1999), and Arabidopsis RPP13 (Rose et al., 2004), possess distinct resistance specificities and responses. These are based on the possibility of allelic variation in R-genes to recognize pathogen effectors with different efficiencies. Clearly, this does not apply for Yr15, YrG303, and YrH52 because their sequences are identical.
One possible explanation for the observed variation in phenotypic responses of Wtk1 carriers could be related to differences in expression levels. However, we have shown previously (Klymiuk et al., 2018) that differences in expression levels of Yr15 (Wtk1) in transgenic and introgression lines did not relate to phenotypic expression differences. Also the basal level of Yr15 (Wtk1) expression is low and did not change significantly after inoculation with Pst (Klymiuk et al., 2018).
Introgression of these genes into different genetic backgrounds shows repeatable differences in resistance response upon Pst inoculation. We have previously shown that tetraploid (T. turgidum ssp. durum) and hexaploid (T. aestivum) Yr15 introgression lines all exhibit diverse phenotypic responses to Pst inoculation (Klymiuk et al., 2018). We demonstrated that the introgression of YrG303 from WEW G303 to T. aestivum 2298 improved the resistance phenotype from IT5 to IT1 (Figs 1, 3). This could be possible due to variable regulation of Wtk1 expression, perhaps because of variation in the flanking regulatory sequences (which were not obtained in the current study). However, this seems unlikely given that the introgression lines mentioned here were developed through conventional breeding and are likely to carry relatively large introgressed segments, including those flanking Wtk1 up- and downstream (Yaniv et al., 2015). Similar situations occur during introgression or transformation of other R-genes. For example, transformation of the LR34res allele into two genetic backgrounds of wheat resulted in variation in resistance of transgenic lines, which was explained by differences in genes modifying LR34res activity (Risk et al., 2012). Moreover, diverse phenotypic responses are typical not only for wheat–rusts interactions, but also for other pathosystems. In particular, transfer of R-genes/quantitative resistance loci for resistance to rice blast resulted in variation in the resistance reaction of improved lines to inoculation with Magnaporthe oryzae (Hasan et al., 2016). A more likely explanation of unexpected phenotypic results is that the effectiveness of R-genes is altered following introgression into a new background (Adachi et al., 2019). According to current understanding of the plant immunity system, some response networks may be shared between different receptors, while others are very specific. Faulty connections between nodes in the NLR network may arise after a cross between distinct plant genotypes that can result in NLR misregulation and autoimmunity of progeny, although in parental lines the immune system worked well (Adachi et al., 2019).
Although Wtk1 provides resistance at both seedling and adult stages, YrG303 exhibited a resistance response in three independent tetraploid recombinant lines only at the adult stage under field inoculation (Fig. 2). Taking into account that controlled conditions in the dew chamber used for seeding inoculation are more favorable for Pst development than field conditions used for adult stage inoculation, our results suggest that the environment plays an important role in the resistance response of these genes. Moreover, WEW accessions that carry functional Wtk1 alleles and were subjected to natural selection and adaptation (Klymiuk et al., 2019b) show different levels of resistance within the same experiment. An analogous situation was shown for the Pto locus in wild tomato (Lycopersicon spp.) populations, where susceptible phenotypes were detected even in the presence of alleles conferring AvrPto recognition, suggesting non-functionality of some of the genes from the Pto resistance response network (Rose et al., 2005). Taking all of the above into account, we hypothesize that phenotypic differences in the presence of Wtk1 probably originate from differences in the genetic background, rather than from the presence of different Wtk1 resistance alleles. Nevertheless, we do not exclude that other factors, such as the presence of genes–suppressors of R-genes in some genetic backgrounds, may influence phenotypic responses of Wtk1 carrier lines (Kema et al., 1995).
Positions of loss-of-function mutations in WTK1
Both the kinase and pseudokinase domains of WTK1 are necessary to provide resistance to Pst (Klymiuk et al., 2018). Plant pseudokinases are important players in diverse biological processes and represent ~10% of the kinase domains in higher eukaryotes (Castells and Casacuberta, 2007; Reiterer et al., 2014; Niu et al., 2016). Emerging evidence indicates that pseudokinases themselves act as signaling molecules and modulate the activity of catalytically active kinase partners (Müller et al., 2008; Reiterer et al., 2014). Consistent with this hypothesis and the previous study (Klymiuk et al., 2018), EMS mutations blocking resistance were mapped to both pseudokinase and kinase domains of WTK1 (Fig. 5). In the current study, we identified informative EMS mutations in conserved kinase motifs such as the ATP-binding region, the catalytic loop, and the activation loop (Kornev and Taylor, 2010). Nevertheless, these mutations did not affect previously defined key conserved residues involved in kinase activity (Kannan et al., 2007; Klymiuk et al., 2018). It seems that these mutations are still able to affect WTK1’s kinase activity possibly by disrupting the correct folding of the protein or by preventing binding of pathogen effectors. Interestingly, we identified single nucleotide mutations caused by EMS treatment in three residues, Gly54, Ala149, and Ala460, that each blocked resistance in different genetic backgrounds (Fig. 5). Gly54 maps to KinI’s ATP-binding region and corresponds to Gly55 of the α cAMP-dependent protein kinase catalytic subunit (cAPK) (Hanks et al., 1988) known to be part of a glycine-rich loop that coordinates the ATP phosphates during binding (Taylor and Kornev, 2011). Ala149 and Ala460 occur in the same α-helical region in both KinI and KinII domains of WTK1, respectively, and correspond to Ala151 of cAPK (Hanks et al., 1988). Although the functions of Ala151 itself remain unclear, it is located close to the LXXLH158 motif that is involved in providing the link between the catalytically important DFG motif and substrate-binding regions as a part of the hydrogen-bonding network (Kannan et al., 2007). Thus, it seems that the Ala149/Ala460 region of WTK1 is important for KinI/II associations, effector binding, or target binding.
Conclusions and future perspectives
Here we show that Yr15, YrG303, and YrH52 all represent one tandem kinase gene, Wtk1. Wtk1 sequences of YrG303 and YrH52 are identical to those of Yr15. Differences in phenotypic responses of WEW donor lines and introgression lines of Yr15, YrG303, and YrH52 are probably related to differences due to genetic backgrounds rather than to the presence of different alleles. Our results indicate that YrG303 and YrH52 are synonymous to Yr15. However, we cannot exclude the option that the three WEW donor lines (G25, H52, and G303) harbor additional Yr genes. The positions of mutations in EMS-treated lines that led to disruption of Wtk1 function were associated with conserved regions in critical kinase subdomains, although key conserved residues were not affected. This information will be useful for future work on the possible molecular mechanism of Wtk1 and its role in plant innate immunity. The Wtk1-mediated resistance network is diverse in WEW natural populations subjected to natural selection and adaptation, confirming that WEW natural populations have potential to serve as a good source for evolutionary studies of different traits and multifaceted gene networks.
Supplementary data
Supplementary data are available at JXB online.
Table S1. A list of SSR, CAPS, and KASP markers used in the current study.
Table S2. A list of KASP markers from the Yr15 region developed based on SNPs from the wheat 15K SNP array.
Table S3. Molecular characterization of the yr15, yrG303, and yrH52 EMS mutants.
Fig. S1. Graphical genotype of selected recombinant inbred lines (RILs) from the YrG303 tetraploid mapping population.
Fig. S2. Graphical genotype of selected RILs from the YrH52 mapping population.
Acknowledgements
This research was supported by the US–Israel Binational Agricultural Research and Development Fund (IS-4628-13, US-4916-16) and the Israel Science Foundation (1719/08, 1366/18). We thank S. Barinova, J. Cheng, T. Kis-Papo, S. Khalifa, T. Krugman, D. Lewinsohn, I. Manov, O. Rybak, and I. Shams for their support, advice, and discussions.
Glossary
Abbreviations
- cAPK
α cAMP-dependent protein kinase catalytic subunit
- CAPS
cleaved amplified polymorphic sequence
- dpi
days post-inoculation
- EMS
ethyl methanesulfonate
- HR
hypersensitive response
- KASP
kompetitive allele-specific PCR
- NLR
nucleotide-binding domain and leucine-rich repeat
- Pst
Puccinia striiformis f. sp. tritici
- R-gene
resistance gene
- SNP
single nucleotide polymorphism
- SSR
simple sequence repeat
- TKP
tandem kinase–pseudokinase
- WEW
wild emmer wheat
- WGA
wheat germ agglutinin
- wtk1
wheat tandem kinase 1
- Wtk1
functional WTK1 allele
- WTK1
non-functional WTK1 allele
- Yr gene
yellow rust resistance gene
References
- Aaronsohn A. 1910. Agricultural and botanical explorations in Palestine (No. 180). Washington, DC: US Government Printing Office. [Google Scholar]
- Adachi H, Derevnina L, Kamoun S. 2019. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Current Opinion in Plant Biology 50, 121–131. [DOI] [PubMed] [Google Scholar]
- Agrios GN. 2005. Plant pathology. New York: Academic Press. [Google Scholar]
- Ali S, Rodriguez-Algaba J, Thach T, et al. 2017. Yellow rust epidemics worldwide were caused by pathogen races from divergent genetic lineages. Frontiers in Plant Science 8, 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels R, Eversole K, Feuillet C, et al. 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O, et al. 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. [DOI] [PubMed] [Google Scholar]
- Bhullar NK, Street K, Mackay M, et al. 2009. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proceedings of the National Academy of Sciences, USA 106, 9519–9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GS, Ali S, Qutob D, Ambrose S, Lou K, Maclachlan R, Pozniak CJ, Fu YB, Sharpe AG, Kutcher HR. 2018. Genome re-sequencing and simple sequence repeat markers reveal the existence of divergent lineages in the Canadian Puccinia striiformis f. sp. tritici population with extensive DNA methylation. Environmental Microbiology 20, 1498–1515. [DOI] [PubMed] [Google Scholar]
- Castells E, Casacuberta JM. 2007. Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. Journal of Experimental Botany 58, 3503–3511. [DOI] [PubMed] [Google Scholar]
- Chen X, Kang Z. 2017. Stripe rust. Berlin: Springer Science+Business Media B.V. [Google Scholar]
- Chen XM. 2005. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Canadian Journal of Plant Pathology 27, 314–337. [Google Scholar]
- Dawson AM, Bettgenhaeuser J, Gardiner M, et al. 2015. The development of quick, robust, quantitative phenotypic assays for describing the host–nonhost landscape to stripe rust. Frontiers in Plant Science 6, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. 1991. DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW, eds. Molecular techniques in taxonomy. NATO ASI Series (Series H: Cell Biology) 57. Berlin: Springer, 283–293. [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. The Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahima T, Roder MS, Grama A, et al. 1998. Microsatellite DNA polymorphism divergence in Triticum dicoccoides accessions highly resistant to yellow rust. Theoretical and Applied Genetics 96, 187–195. [Google Scholar]
- Flor HH. 1971. Current status of the gene-for-gene concept. Annual Review of Phytopathology 9, 275–96. [Google Scholar]
- Fu D, Uauy C, Distelfeld A, et al. 2009. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerechter-Amitai ZK, Stubbs RW. 1970. A valuable source of yellow rust resistance in Israeli populations of wild emmer, Triticum dicoccoides Koern. Euphytica 19, 12–21. [Google Scholar]
- Gerechter-Amitai ZK, van Silfhout CH, Grama A, et al. 1989. Yr15—a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G25. Euphytica 43, 187–190. [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52. [DOI] [PubMed] [Google Scholar]
- Hasan MM, Rafii MY, Ismail MR, Mahmood M, Alam MA, Abdul Rahim H, Malek MA, Latif MA. 2016. Introgression of blast resistance genes into the elite rice variety MR263 through marker-assisted backcrossing. Journal of the Science of Food and Agriculture 96, 1297–1305. [DOI] [PubMed] [Google Scholar]
- Hovmøller MS, Justesen AF. 2007. Appearance of atypical Puccinia striiformis f. sp. tritici phenotypes in north-western Europe. Australian Journal of Agricultural Research 58, 518–524. [Google Scholar]
- Hovmøller MS, Walter S, Bayles RA, et al. 2015. Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathology 65, 402–411. [Google Scholar]
- Huang L, Raats D, Sela H, et al. 2016. a Evolution and adaptation of wild emmer wheat populations to biotic and abiotic stresses. Annual Review of Phytopathology 54, 276–301. [DOI] [PubMed] [Google Scholar]
- Huang L, Sela H, Feng L, et al. 2016. b Distribution and haplotype diversity of WKS resistance genes in wild emmer wheat natural populations. Theoretical and Applied Genetics 129, 921–934. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, et al. 2007. Structural and functional diversity of the microbial kinome. PLoS Biology 5, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema GHJ, Lange W, Van Silfhout CH. 1995. Differential suppression of stripe rust resistance in synthetic wheat hexaploids derived from Triticum turgidum subsp. dicoccoides and Aegilops squarrosa. Phytopathology 85, 425–429. [Google Scholar]
- Klymiuk V, Fatiukha A, Fahima T. 2019b Wheat tandem kinases provide insights on disease-resistance gene flow and host–parasite co-evolution. The Plant Journal 98, 667–679. [DOI] [PubMed] [Google Scholar]
- Klymiuk V, Fatiukha A, Huang L, et al. 2019. a Durum wheat as a bridge between wild emmer wheat genetic resources and bread wheat. In: Miedaner T, Korzun V, eds. Application of genetic and genomic research in cereals. Cambridge: Woodhead Publishing, 201–230. [Google Scholar]
- Klymiuk V, Yaniv E, Huang L, et al. 2018. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase–pseudokinase family. Nature Communications 9, 3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Taylor SS. 2010. Defining the conserved internal architecture of a protein kinase. Biochimica et Biophysica Acta 1804, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Line RF, Qayoum A. 1992. Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America, 1968–1987. Technical Bulletin 1788 . Washington, DC: United States Department of Agriculture. [Google Scholar]
- Liu T, Wan A, Liu D, Chen X. 2017. Changes of races and virulence genes in Puccinia striiformis f. sp. tritici, the wheat stripe rust pathogen, in the United States from 1968 to 2009. Plant Disease 101, 1522–1532. [DOI] [PubMed] [Google Scholar]
- Lundström M, Leino MW, Hagenblad J. 2017. Evolutionary history of the NAM-B1 gene in wild and domesticated tetraploid wheat. BMC Genetics 18, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Harris NS, Twardziok SO, et al. 2019. Durum wheat genome highlights past domestication signatures and future improvement targets. Nature Genetics 51, 885–895. [DOI] [PubMed] [Google Scholar]
- Maccaferri M, Ricci A, Salvi S, et al. 2015. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnology Journal 13, 648–663. [DOI] [PubMed] [Google Scholar]
- MacQueen A, Tian D, Chang W, et al. 2019. Population genetics of the highly polymorphic RPP8 gene family. Genes 10, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Zhang J, Zhang P, et al. 2018. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nature Plants 4, 662–668. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Dubcovsky J, Rogers WJ, et al. Catalogue of gene symbols for wheat: 2017 Supplement https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf.
- Müller R, Bleckmann A, Simon R. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell 20, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqaddasi QH, Brassac J, Börner A, et al. 2017. Genetic architecture of anther extrusion in spring and winter wheat. Frontiers in Plant Sciences 8, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E, Gerechter-Amitai Z, Beiles A, et al. 1986. Resistance of wild wheat to stripe rust: predictive method by ecology and allozyme genotypes. Plant Systematics and Evolution 153, 13–30. [Google Scholar]
- Nevo E, Korol AB, Beiles A, et al. 2002. Evolution of wild emmer and wheat improvement: population genetics, genetic resources, and genome organization of wheats progenitor, Triticum dicoccoides. Berlin: Springer. [Google Scholar]
- Niu D, Lii YE, Chellappan P, et al. 2016. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nature Communications 7, 11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan H, Willcox G, Graner A, et al. 2011. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genetic Resources and Crop Evolution 58, 11–53. [Google Scholar]
- Peng JH, Fahima T, Röder MS, et al. 1999. Microsatellite tagging of the stripe-rust resistance gene YrH52 derived from wild emmer wheat, Triticum dicoccoides, and suggestive negative crossover interference on chromosome 1B. Theoretical and Applied Genetics 98, 862–872. [Google Scholar]
- Peng JH, Fahima T, Röder MS, et al. 2000. a Microsatellite high-density mapping of the stripe rust resistance gene YrH52 region on chromosome 1B and evaluation of its marker-assisted selection in the F2 generation in wild emmer wheat. New Phytologist 146, 141–154. [Google Scholar]
- Peng JH, Fahima T, Röder MS, et al. 2000. b High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica 109, 199–210. [DOI] [PubMed] [Google Scholar]
- Raats D, Yaniv E, Distelfeld A, et al. 2014. Application of CAPS markers for genomic studies in wild emmer wheat. In: Savrukov Y, ed. Cleaved amplified polymorphic sequences (CAPS) markers in plant biology. New York: Nova Science Publishers, 31–61. [Google Scholar]
- Ramirez-Gonzalez RH, Uauy C, Caccamo M. 2015. PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31, 2038–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V, Eyers PA, Farhan H. 2014. Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends in Cell Biology 24, 489–505. [DOI] [PubMed] [Google Scholar]
- Risk JM, Selter LL, Krattinger SG, et al. 2012. Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnology Journal 10, 477–487. [DOI] [PubMed] [Google Scholar]
- Roelfes AP, Singh RP, Saari EE. 1992. Rust diseases of wheat: concepts and methods of disease management. Mexico: CIMMYT. [Google Scholar]
- Rose LE, Bittner-Eddy PD, Langley CH, et al. 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LE, Langley CH, Bernal AJ, Michelmore RW. 2005. Natural variation in the Pto pathogen resistance gene within species of wild tomato (Lycopersicon). I. Functional analysis of Pto alleles. Genetics 171, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. 2004. The PredictProtein server. Nucleic Acids Research 32, W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer S, Tsuchimatsu T, Jordan T, et al. 2010. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Molecular Plant-Microbe Interactions 23, 497–509. [DOI] [PubMed] [Google Scholar]
- Sela H, Loutre C, Keller B, et al. 2011. Rapid linkage disequilibrium decay in the Lr10 gene in wild emmer wheat (Triticum dicoccoides) populations. Theoretical and Applied Genetetics 122, 175–187. [DOI] [PubMed] [Google Scholar]
- Sharma-Poudyal D, Chen XM, Wan AM, et al. 2013. Virulence characterization of international collections of the wheat stripe rust pathogen, Puccinia striiformis f. sp. tritici. Plant Disease 97, 379–386. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Hey SJ. 2015. The contribution of wheat to human diet and health. Food and Energy Security 4, 178–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srichumpa P, Brunner S, Keller B, Yahiaoui N. 2005. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiology 139, 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, et al. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400, 667–671. [DOI] [PubMed] [Google Scholar]
- Sun GL, Fahima T, Korol AB, et al. 1997. Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat, Triticum dicoccoides. Theoretical and Applied Genetics 95, 622–628. [Google Scholar]
- Taylor SS, Kornev AP. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends in Biochemical Sciences 36, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Silfhout CH. 1989. Identification and characterization of resistance to yellow rust and powdery mildew in wild emmer wheat and their transfer to bread wheat. PhD Thesis, Agricultural University, Wageningen. [Google Scholar]
- Voorrips RE. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93, 77–78. [DOI] [PubMed] [Google Scholar]
- Wang S, Wong D, Forrest K, et al. 2014. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnology Journal 12, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZZ, Xie JZ, Guo L, et al. 2018. Molecular mapping of YrTZ2, a stripe rust resistance gene in wild emmer accession TZ-2 and its comparative analyses with Aegilops tauschii. Journal of Integrative Agriculture 17, 1267–1275. [Google Scholar]
- Wellings CR. 2011. Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. [Google Scholar]
- Yahiaoui N, Kaur N, Keller B. 2009. Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. The Plant Journal 57, 846–856. [DOI] [PubMed] [Google Scholar]
- Yaniv E, Raats D, Ronin Y, et al. 2015. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Molecular Breeding 35, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Tanksley SD. 1989. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theoretical and Applied Genetics 77, 95–101. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang L, Wang C, et al. 2016. Molecular mapping and marker development for the Triticum dicoccoides-derived stripe rust resistance gene YrSM139-1B in bread wheat cv. Shaanmai 139. Theoretical and Applied Genetics 129, 369–376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.