Abstract
An adiabatic MEscher-GArwood (MEGA)-editing scheme, using asymmetric hyperbolic secant editing pulses, was developed and implemented in a -insensitive, 1D-semiLASER (Localization by Adiabatic SElective Refocusing) MR spectroscopic imaging (MRSI) sequence for the non-invasive mapping of γ-aminobutyric acid (GABA) over a whole brain slice. Our approach exploits the advantages of edited-MRSI at 7T while tackling challenges that arise with ultra-high-field-scans. Spatial-spectral encoding, using density-weighted, concentric circle echo planar trajectory readout, enabled substantial MRSI acceleration and an improved point-spread-function, thereby reducing extracranial lipid signals. Subject motion and scanner instabilities were corrected in real-time using volumetric navigators optimized for 7T, in combination with selective reacquisition of corrupted data to ensure robust subtraction-based MEGA-editing.
Simulations and phantom measurements of the adiabatic MEGA-editing scheme demonstrated stable editing efficiency even in the presence of ±0.15 ppm editing frequency offsets and variations of up to ±30% (as typically encountered in vivo at 7T), in contrast to conventional Gaussian editing pulses. Volunteer measurements were performed with and without global inversion recovery (IR) to study regional GABA levels and their underlying, co-edited, macromolecular (MM) signals at 2.99 ppm. High-quality in vivo spectra allowed mapping of pure GABA and MM-contaminated GABA+ (GABA + MM) along with Glx (Glu + Gln), with high-resolution (eff. voxel size: 1.4 cm3) and whole-slice coverage in 24 min scan time. Metabolic ratio maps of GABA/tNAA, GABA+/tNAA, and Glx/tNAA were correlated linearly with the gray matter fraction of each voxel. A 2.15-fold increase in gray matter to white matter contrast was observed for GABA when enabling IR, which we attribute to the higher abundance of macromolecules at 2.99 ppm in the white matter than in the gray matter.
In conclusion, adiabatic MEGA-editing with 1D-semiLASER selection is as a promising approach for edited-MRSI at 7T. Our sequence capitalizes on the benefits of ultra-high-field MRSI while successfully mitigating the challenges related to inhomogeneities, prolonged scan times, and motion/scanner instability artifacts. Robust and accurate 2D mapping has been shown for the neurotransmitters GABA and Glx.
Keywords: Magnetic resonance spectroscopic imaging, Adiabatic spectral MEGA editing, Concentric circle echo planar trajectories, Real-time motion correction, GABA, Asymmetric hyperbolic secant editing pulse
1. Introduction
1.1. MRS of neurotransmitters
Two of the major neurotransmitters in the human brain are the inhibitory neurotransmitter γ-aminobutyric acid (GABA) and the excitatory neurotransmitter glutamate (Glu). Both play essential roles in normal physiological and mental functionality (Agarwal and Renshaw, 2012; Levar et al., 2017; Northoff et al., 2007).
Altered GABAergic neurotransmission is thought to be linked to the pathophysiology of numerous neurological and neuropsychiatric disorders, including multiple sclerosis (Cawley et al., 2015), schizophrenia (Shetty et al., 2016), and Alzheimer's disease (Bai et al., 2015). GABA levels have also been studied in relation to drug treatment (Cai et al., 2012), neonatal brain development (Tomiyasu et al., 2017), aging (Gao et al., 2013), functional MRI activity (Donahue et al., 2010; Muthukumaraswam et al., 2009), and behavior (Edden et al., 2009; Stagg et al., 2011a,b). MR spectroscopy (MRS) is the only non-invasive technique for measuring in vivo GABA and Glu concentrations (Puts et al., 2012).
To date, GABA detection via MRS has mainly been conducted in single voxels within the brain, with cortical regions (e.g., occipital and prefrontal lobes) being prominent regions of interest (Brennan et al., 2017; Mahone et al., 2018; Reid et al., 2018). Reports about MRS Imaging (MRSI) are still uncommon (Bogner et al., 2014a; Hnilicová et al., 2016; Jensen et al., 2005), and all of them share the same limitations, namely, large voxel sizes, which hamper the assessment of regional, – anatomically resolved – GABA levels, and also restricted rectangular target volumes to overcome extracranial lipid artifacts, which is problematic for studies of cortical regions.
A comprehensive assessment of regional GABA levels requires measurement approaches with a) high sensitivity to overcome the low signal-to-noise ratio (SNR) associated with inherently low GABA concentrations, b) consistent high spectral quality and high spatial resolutions over the whole brain, and c) robustness against subject motion and scanner instabilities.
1.2. Ultra-high-field (7T) MRSI
MRSI at ultra-high-field (UHF) benefits from increased SNR and higher spectral resolution (Moser et al., 2012), which can be translated into higher spatial resolution to improve anatomical and chemical specificity. This requires that several technical challenges associated with accurate volume selection, stronger -field inhomogeneities, and specific absorption rate (SAR) limitations have to be overcome. Further, whole-slice MRSI needs to be robust against extracranial lipid contamination (Hangel et al., 2015). These issues can be addressed by using excitation approaches with adiabatic pulses (e.g., LASER localization instead of PRESS) to resolve signal loss and poor spatial localization (Andronesi et al., 2010; Bogner et al., 2017; Scheenen et al., 2008a,b). To fully capitalize on the increased spectral and spatial resolution at UHF, traditional MRSI sampling strategies have to be reconsidered.
1.3. Spatial-spectral sampling
Time- and SNR-efficient MRSI sampling at UHF with simultaneously increased spectral bandwidth and higher spatial resolutions is challenging. Spatial-spectral encoding (SSE) methods, such as echo-planar spectroscopic imaging (EPSI) and spiral spectroscopic imaging, are much faster than phase-encoded MRSI using parallel imaging (Strasser et al., 2017). However, at UHF and with current whole-body gradient limitations, the stated SSE methods become increasingly SNR-inefficient due to long trajectory rewinding. Self-rewinding SSE approaches, such as non-Cartesian, concentric circle echo planar trajectories (CONCEPT) (Hingerl et al., 2017), are not only more SNR-efficient at UHF, but can be intrinsically acquired with a Hamming density-weighted k-space to eliminate extracranial lipid contaminations. This is fundamental for whole-slice MRSI that includes cortical regions.
1.4. MEGA-editing
The in vivo measurement of low-abundant metabolites, such as GABA, benefits from the higher sensitivity at 7T, but remains challenging if the resonances of interest overlap with signals from more abundant metabolites (e.g., with creatine).
Several MRS techniques have been suggested to overcome this limitation, including multiple quantum filtering (Choi et al., 2006), multidimensional correlation spectroscopy (Andronesi et al., 2012), and MEscher-GArwood (MEGA)-editing (Mescher et al., 1998). Among these methods, MEGA-editing has become the most common approach due its superior editing efficiency, conceptual simplicity (Andreychenko et al., 2012; Arteaga de Castro et al., 2013), and high reproducibility (Mikkelsen et al., 2017).
MEGA-editing of GABA exploits the quantum mechanical J-coupling between the 4CH2-GABA spins at 3.01 ppm and 3CH2-GABA spins at 1.9 ppm. Two spectra (i.e., EDIT-ON and EDIT-OFF) are measured in an interleaved fashion. In the ON-acquisition, frequency-selective editing pulses applied at 1.9 ppm modulate the GABA signals at 3.01 ppm by refocusing the evolution of the scalar coupling, while, in the OFF-acquisition, the evolution remains unchanged (i.e., editing pulses applied symmetrically around water at 7.5 ppm). A difference spectrum contains a GABA signal at 3.01 ppm, and no signals that were unaffected by the editing pulses (e.g., overlying creatine signals).
Macromolecules (MM) at 1.7 ppm are partially inverted due to the editing pulse at 1.9 ppm, which results in unwanted co-editing of MM signals at 2.99 ppm. The resulting contaminated resonance, termed GABA+ (GABA + MM), complicates the interpretation. MM-suppressed GABA-editing approaches have, therefore, been proposed, including a) nulling the MM contribution via a global inversion-preparation (Rothman et al., 1993), and b) employing symmetric editing around the 1.7 ppm MM resonance with narrow-band MEGA pulses (Henry et al., 2001). While the first approach leads to inherent SNR loss, the latter approach is sensitive to temporal and spatial B0 frequency deviations, rendering it problematic for single-voxel spectroscopy (SVS), and incompatible with MRSI.
1.5. Real-time motion and scanner instability correction
As MEGA-editing is based on the subtraction of two spectra, it is particularly susceptible to patient motion or frequency drifts (Bogner et al., 2014; Evans et al., 2013). Hence, real-time corrections for any head movements and scanner instabilities are highly desirable.
Real-time correction based on 3D echo planar imaging navigators (vNav) has been introduced for MEGA-LASER at 3T and appears to be superior to retrospective approaches (e.g., frequency/phase alignment of individual averages) (Bogner et al., 2014; Hess et al., 2011). By co-registering and comparing navigator images acquired interleaved with the MRSI scan, updates (i.e., head translation, rotation, B0 frequency, and first-order shimming) are calculated in real-time on the scanner and applied before the subsequent MRSI repetition for each TR. vNavs have been shown to improve data quality by reducing subtraction artifacts, especially in patients with a predisposition for movement (Bogner et al., 2014; Heckova et al., 2018; Hnilicová et al., 2016).
vNavs have not been used for MEGA-edited MRSI at 7T as yet, but may also offer substantial improvements at UHF, especially in motion-sensitive MRSI experiments, such as MEGA-editing.
1.6. Purpose
Therefore, we propose a MEGA-edited acquisition scheme that meets the above expectations for robust and efficient high-resolution, whole-slice, edited-MRSI at 7T. Adiabatic slice excitation and MM-suppressed MEGA-editing, combined with fast non-Cartesian readout and real-time motion and B0 corrections, ensure stability against both volunteer and typical ultra-high-field artifacts.
2. Materials and methods
2.1. Subjects and hardware
Five healthy volunteers (30.8 ± 4.6 years) were included in this study. Institutional Review Board approval and written, informed consent were obtained. Measurements were performed on a 7T whole-body MR scanner (Magnetom, Siemens Healthcare, Erlangen, Germany) with a 70 mTm−1 total gradient strength and a 200 mTm−1s−1 nominal slew rate. A head coil with a 32-channel receive coil array, combined with a volume coil for transmission (Nova Medical, Wilmington, MA, USA), was used.
2.2. Sequence design
Our goal was to develop a method for high-resolution GABA mapping without MM contamination throughout a whole brain slice. This was accomplished by: a) - and chemical shift displacement error (CSDE)-insensitive, 1D-semiLASER slice selection; b) adiabatic MEGA-editing pulses combined with a global inversion-preparation to suppress macromolecules (MM) while being robust to /B0-inhomogeneities; c) rapid SNR-efficient, Hamming density-weighted, non-Cartesian spatial-spectral encoding; and d) real-time motion and B0 correction.
The resulting edited MRSI sequence combines and builds on previously published approaches (Bogner et al., 2014a; Esmaeili et al., 2017; Hingerl et al., 2017; Hwang et al., 1999), but substantially extends existing methodologies to fully exploit the benefits of whole-slice MEGA-edited MRSI at 7T.
2.2.1. Spatial localization using 1D-semiLASER
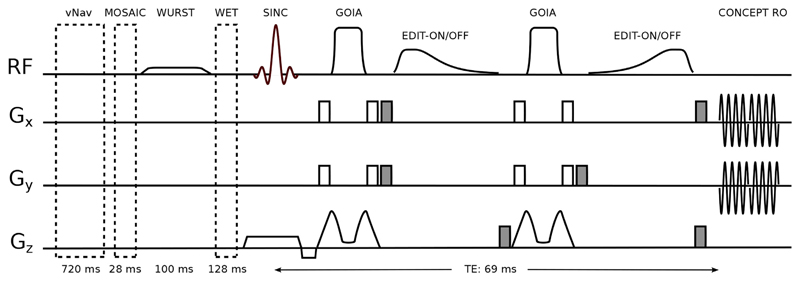
Volume of interest (VOI) localization was achieved by a -insensitive 1D-semiLASER sequence (Fig. 1). Slice-selective excitation was performed by a Hamming-filtered, three-lobe sinc pulse with a 0.9 ms duration and a 4.44 kHz bandwidth, exciting a 50% thicker slice than the target thickness in order to ensure a flat-top excitation profile within the target slice while eliminating possible contaminations from outside the slice (e.g., from the nasal cavity or extracranial lipids) that might occur with non-selective (adiabatic) excitation. One pair of Gradient Offset Independent Adiabatic (GOIA) pulses (W16,4 modulation, 8 ms duration, and 10 kHz bandwidth) refocused only the target slice thickness spoiling other coherences from outside. GOIA pulses provide accurate -insensitive localization with low-power requirements and negligible CSDEs (Andronesi et al., 2010; Tannús and Garwood, 1997). A safety margin of 40% above the adiabatic threshold was used for the adiabatic GOIA pulses to account for the inhomogeneities that are expected at 7T over the whole slice (Supplementary Figure 1).
Fig. 1.
A 1D-semiLASER sequence (i.e., Hamming-filtered Gaussian excitation pulse and a pair of GOIA-refocusing pulses) including an adiabatic MEGA-editing scheme. Prior to MRSI excitation, volumetric navigators for real-time motion correction, MOSAIC prescans for coil combination, WURST inversion recovery for MM suppression and WET water suppression modules are played out. MEGA-editing elements include asymmetric adiabatic editing pulses and surrounding spoiler gradients (shaded in gray). CONCEPT spatial-spectral encoding readout starts after a TE of 69 ms.
2.2.2. Adiabatic MEGA-editing
Editing pulses and spoiler gradients (30 mT/m amplitude with a 2 ms duration) were arranged in a way similar to that originally proposed for MEGA-PRESS (Mescher et al., 1996). However, to account for the strong B0 and inhomogeneities at 7T across the slice, we introduced a modified MEGA scheme that incorporated adiabatic editing pulses (Fig. 1). For this, we replaced the conventional Gaussian editing pulses by adiabatic editing pulses, which are more suitable for whole-slice MEGA-editing at 7T. Adiabatic MEGA lipid/water suppression was shown previously at 3T (Esmaeili et al., 2017) for full slab imaging using asymmetric hypergeometric pulses (Rosenfeld et al., 1996). Here, we extend this approach to homonuclear adiabatic MEGA J-difference editing at 7T. Due to larger spectral dispersion at 7T compared to 3T, the editing pulses at 7T can have a larger inversion transition band (defined as the frequency range in which −0.95 < Mz/M0 < 0.95).
Hence, instead of using hypergeometric pulses which have very sharp transition bands (50 Hz) but require higher peak voltage, we designed asymmetric adiabatic pulses based on stretched hyperbolic secant modulations (Hwang et al., 1999; Xin et al., 2009), which provide a sufficiently narrow transition band for 7T, but with lower peak voltage. The design of our asymmetric adiabatic pulses was also constrained by their duration to yield an overall echo time (TE) close to the optimum value (68 ms) for GABA editing. With this design a pulse sequence with total TE of 69 ms was obtained as shown in Fig. 1. Detailed pulse parameters are described in Section 2.3.
In the ON-acquisition, the editing pulses were placed such that the 1.9 ppm GABA resonance remained well within the flat-top of the MEGA inversion profile, even in the presence of B0 variations of about ±0.15 ppm (Supplementary Figure 1). Due to the broad inversion flat-top of the MEGA pulse, the MM resonance at 1.7 ppm (i.e., J-coupled to MM2.99ppm) was fully co-edited, but, unlike with narrow-band Gaussian pulses, the MM contribution was stable, even in the presence of substantial frequency offsets (i.e., temporal frequency drift or spatial B0 inhomogeneities) (Harris et al., 2014; Near et al., 2011; van der Veen et al., 2017). In the OFF-acquisition, instead of setting the editing pulse far up-field (e.g., 10 ppm as for GSH editing), to reduce SAR no editing pulse was applied while keeping the overall sequence timing unchanged.
To suppress these unwanted MM contributions and extract the pure GABA signal, a global inversion-preparation was applied to null co-edited MM signals at 2.99 ppm (Prinsen et al., 2017). A 100-ms-long, 40th-order WURST (wideband, uniform rate and smooth truncation) pulse with a bandwidth of 1300 Hz centered at 3.0 ppm was used for inversion. The global inversion also efficiently suppressed any remaining extracranial lipid signals, thereby removing any remaining baseline distortions.
2.2.3. Spatial-spectral sampling
Time-efficient MRSI sampling was achieved by using Hamming density-weighted, concentric circle echo planar trajectories (CONCEPT), as described by Hingerl et al. at 7T. This approach optimizes the SNR per unit time and the point-spread-function (PSF) compared to constant-density approaches (Hingerl et al., 2017; Strasser et al., ISMRM, 2017, No. 1251). This PSF improvement further mitigates extracranial lipid artifacts.
2.2.4. Real-time motion and scanner instability correction
Dynamic motion and B0-field changes were tracked with dual-contrast, multi-shot 3D-EPI navigators (vNav) inserted prior to the water suppression module and coil combination prescans of the MRSI sequence. In each TR, the required translation and rotation updates, as well as carrier frequency changes, were calculated online and applied in real-time on the MR scanner. These vNavs have been used previously in various MR experiments at 3T, including MRI (Alhamud et al., 2016; Reuter et al., 2015; Stout et al., 2017), MRS (Hess et al., 2013, 2011), and MRSI (Bogner et al., 2014a, 2014b; Saleh et al., 2016).
Real-time motion updates were applied in each TR, while frequency updates were applied only on pairs of EDIT-ON/EDIT-OFF acquisitions, as this was shown to be superior to individual corrections (Bogner et al., 2014; Zhu et al., 2011). Selective reacquisition of motion-corrupted data was employed and triggered if the head translation or rotation updates were larger than three times the standard deviation of the motion that was normally observed during an average volunteer scan (i.e., 0.4 mm or 0.40°). The affected data were discarded and immediately reacquired in the following TR, and a maximum of 25% of the originally estimated data were allowed to be reacquired.
As previously presented (Moser et al., ISMRM, 2017, No. 1253), vNavs were adjusted to deal with ghosting and lipid artifacts that are significantly worse at 7T. The main steps included local phase correction to eliminate N/2-ghosting (Feiweier et al., 2011), water-selective excitation to eliminate fat artifacts, and a reduction of the EPI factor from 32 to 16 (i.e., 2-shot instead of single-shot EPI) to reduce geometrical brain distortions.
2.3. Simulations – adiabatic MEGA-editing
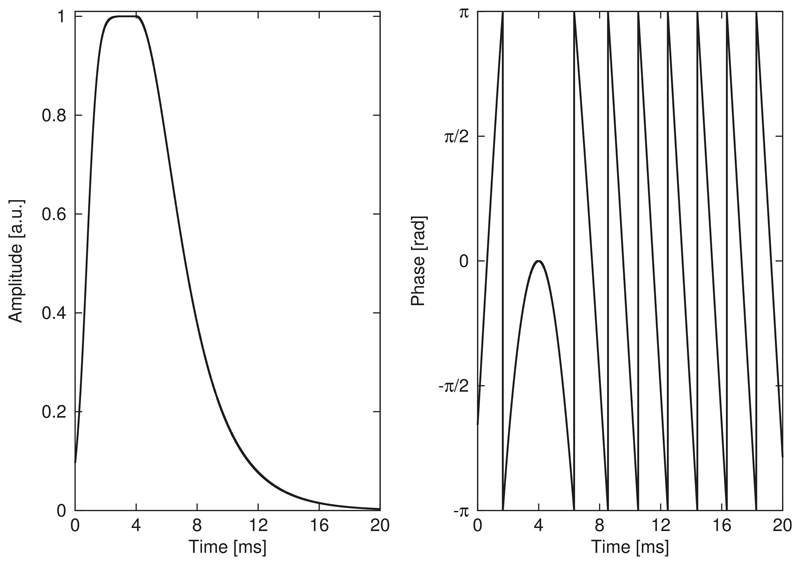
A highly efficient, asymmetric, adiabatic full-passage pulse was synthesized in Matlab (R2013a, MathWorks, Natick, MA, USA) from a combination of two adiabatic half-passage pulses with different modulation functions. The pulse was composed of one-half of a 32-ms-long HS-1 pulse and half of an 8-ms-long HS-4 pulse, resulting in a total pulse length of 20 ms (Fig. 2). The second editing pulse (Fig. 1) was created by time-reversing the first editing pulse with additionally inverted phase modulation to obtain a frequency inversion profile identical to the first editing pulse.
Fig. 2.
The asymmetric adiabatic full-passage pulse for spectral MEGA-editing was designed from a combination of one-half of a 32-ms-long hyperbolic secant (HS) pulse of order 1 and the other half of an 8-ms-long HS-4. Amplitude and phase modulation functions exhibit a smooth transition at the connecting point of the two individual components.
The frequency inversion profile, amplitude, and phase modulation functions were simulated via the MATPULSE/VeSPA toolkit (Matson, 1994). Immunity, with regard to frequency offset and variations, was simulated and compared to a conventional sinc-Gaussian pulse (8.3 ms duration, FWHM = 120 Hz, full width at 95% maximum = 30 Hz) as previously used in SVS experiments at 7T (Chen et al., 2017).
We performed quantum mechanical simulations of this new 1D-semi-LASER sequence of the 3.01 ppm GABA resonance using NMRScope-B (Starvčuk et al., 2009) with J-coupling constants from Govind et al. (2015). The editing efficiency of the asymmetric editing pulse was compared to the above-mentioned Gaussian pulse with respect to sensitivity to ±30% variations and a frequency offset of 0.15 ppm.
2.4. Phantom measurements
The inversion profile of the asymmetric editing pulse was obtained from repeated measurements of the signal intensity of the 3.03 ppm creatine singlet resonance in a phantom (10 mM creatine) while subsequently sweeping over editing frequencies from −1.0 ppm to 5.0 ppm in 0.15 ppm increments. The center frequencies of measured and simulated profiles were matched by frequency shifting and the simulated profile from Section 2.3 was experimentally validated by comparing transition and flat-top bands. Further, a phantom containing 20 mM GABA was measured and the 3.01 ppm GABA resonance shapes in the EDIT-ON/EDIT-OFF/DIFF-spectra were qualitatively compared to simulated results from Section 2.3.
2.5. Volunteer measurements
The in vivo protocols started with acquiring fast, anatomical 3D T1-weighted images using an MP2RAGE (magnetization-prepared 2 rapid acquisition gradient echoes) sequence (Marques et al., 2010). The MRSI slice was positioned transversally above the ventricles. Second-order, field map-based B0-shimming was used with a shim volume of 220 × 220 × 18 mm3. The reference MRSI transmit voltage was calculated from a pre-saturation, turbo-FLASH-based -mapping sequence (Chung et al., 2010; Klose, 1992). In total, all MRI prescans, B0 shimming, and slice positioning took ~15 min.
Five volunteers were scanned with two MRSI sequences using our proposed adiabatic editing sequence, one with and one without applying the global inversion-preparation to study the MM contamination to the 3.01 ppm signal. All MRSI protocols shared the following settings: TR 2.8 s; TE 69 ms; if applicable TI 300 ms (Prinsen et al., 2017); FOV 220 × 220 mm2; 32 × 32 matrix; slice thickness 16 mm; effective voxel size, 1.4 cm3 bandwidth 2778 Hz; FA 83° (Ernst angle for GABA); 2 averages with 2-step phase cycling; 902 FID points; 2 temporal interleaves; 64 k-space rings; TA 24:12 min.
One additional volunteer was scanned to compare our proposed method (CONCEPT readout, adiabatic MEGA-editing, and real-time corrections, IR-ON) to a more conventional approach including spiral readout (8 angular and 3 temporal interleaves) (Adalsteinsson et al., 1998), narrow-band Gaussian editing pulses (FWHM = 120 Hz) as suggested by Chen et al. for MM suppression via symmetric editing around the 1.7 MM resonance at 7T (Chen et al., 2017), and no real-time corrections. The main MRSI parameters described above were employed for both scans.
An optimized WET (Water suppression Enhanced through T1 effects) scheme (Ogg et al., 1994) using four suppression pulses at 7T was used as previously described (Hangel et al., 2016). The MOSAIC coil combination was employed (Moser et al., ISMRM, 2018, No. 1306), which extends the MUSICAL coil combination (Strasser et al., 2013) to spatial-spectral encoded MRSI by acquiring reference data in an interleaved fashion.
2.6. Data processing
Offline MRSI data reconstruction and processing was performed using an in-house-developed software package (Povavžan et al., ISMRM, 2015, No. 1973) based on Matlab (R2013a, MathWorks, Natick, MA, USA), Bash (version 4.2.25, Free Software Foundation, Boston, MA, USA), and MINC (MINC tools, v2.0, McConnell Brain Imaging Center, Montreal, QC, Canada). Data reconstruction included a modified Pipe-Menon pre-gridding density compensation (Pipe and Menon, 1999), an off-resonance correction (Mayer et al., 2006), and convolution gridding (Jackson et al., 1991) using a Kaiser-Bessel kernel (width of 3).
Spectral fitting was performed via the LCModel (Provencher, 2001) with metabolic basis sets simulated using NMRScope-B. In vivo EDIT-OFF spectra were fitted in the spectral range of 1.8–4.2 ppm, with a basis set that included N-acetyl-aspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphocholine (PCh), glycerophosphocholine (GPC), creatine (Cr), phosphocreatine (PCr), Glu, glutamine (Gln), glutathione (GSH), myo-inositol (m-Ins), and GABA. Apodization by an 8 Hz Gaussian filter was used to display DIFF-spectra. DIFF-spectra were fitted with a basis set that contained only GABA and Glu between 2.65 and 4.2 ppm. The spectral range between 3.17 and 3.65 ppm was excluded from fitting to improve the LCModel results.
Maps of spectral quality measures (i.e., SNR and linewidth), as well as spectral fitting quality (i.e., CRLBs) were obtained. For each voxel, the SNR was calculated with an adapted pseudo-replica method (Robson et al., 2008) and the linewidth was determined based on the NAA resonance with an in-house developed MATLAB routine. Moreover, maps of metabolites and their ratios were obtained. In particular, GABA/tNAA maps were acquired from DIFF-spectra, where MM contributions were nulled via inversion recovery. GABA+/tNAA maps were obtained from DIFF-spectra without inversion recovery. Glx/tNAA maps were acquired in both cases. In all cases, tNAA (NAA + NAAG) maps obtained from IR-ON EDIT-OFF spectra were used for normalization (Stagg et al., 2009), thereby reducing lipid and MM contributions to the tNAA signal while avoiding to introduce significant T1 weighting (Xin et al., 2013).
2.7. Data evaluation
T1-weighted images were segmented into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) voxels via BET (Jenkinson et al., 2005) and FSL/FAST (Zhang et al., 2001). For statistical analysis, the GM and WM maps were down-sampled to the MRSI resolution and the PSF was matched via Hamming-filtering. The GM/WM fraction was derived for each MRSI voxel.
The mean and standard deviation of the SNR (i.e., GABA, GABA+, and tNAA), linewidth (i.e., FWHM of tNAA), and Cramér-Rao lower bounds (CRLBs of all fitted metabolites) over all brain voxels were assessed and compared between IR-ON/OFF using paired t-tests. Differences in metabolite concentrations between GM and WM were assessed via correlation analysis between metabolic ratio maps (i.e., for GABA/tNAA, GABA+/tNAA, Glx/tNAA, and tCr/tNAA) and GM/WM tissue composition.
3. Results
3.1. Simulations
Amplitude and phase modulation functions of the asymmetric 20-ms-long adiabatic HS pulse are provided in Fig. 2.
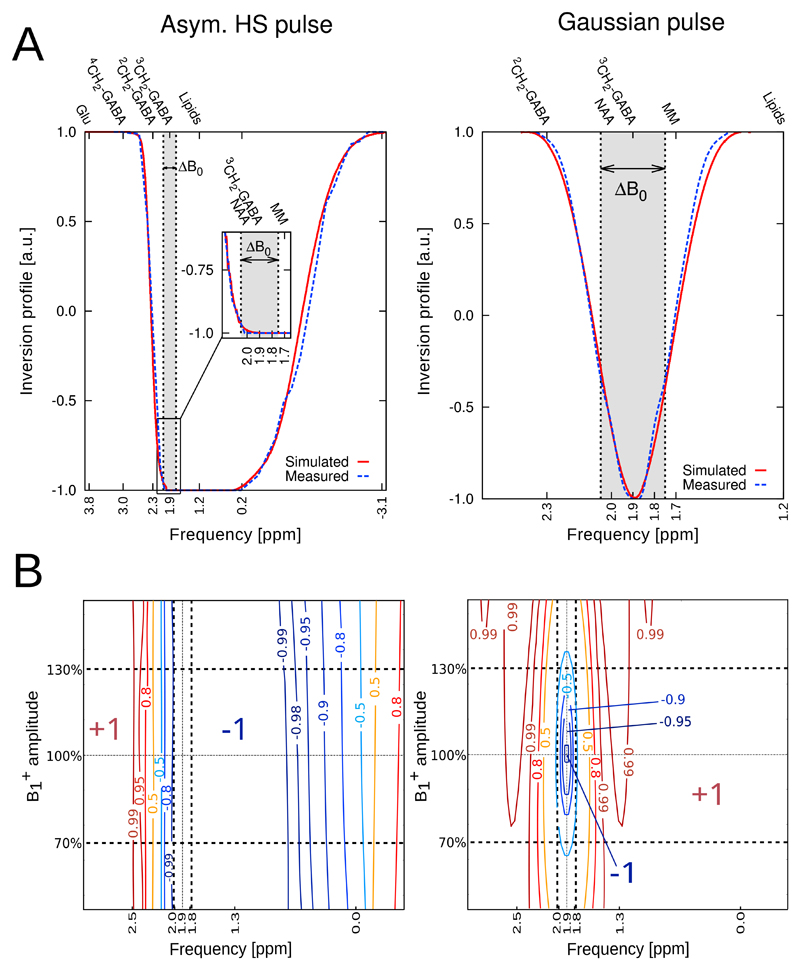
Fig. 3(A) illustrates the simulated inversion profiles for the two editing pulse types. The narrow transition band of the asymmetric HS pulse was 162 Hz, which was lower than that provided by a symmetric HS pulse of equal duration (i.e., 183 Hz). The broad transition band on the other side of the frequency profile of the asymmetric HS pulse was 818 Hz. The flat-top covered a bandwidth of 534 Hz. The flat-top of the asymmetric HS pulse included important resonances, such as GABA at 1.9 ppm, NAA at 2.0 ppm, and macromolecules (MM) at 1.7 ppm. The FWHM of the Gaussian pulse was 120 Hz. The Gaussian inversion profile was centered at the 1.9 ppm GABA resonance affecting the MM resonances at 1.7 ppm only by partial inversion leading to a smaller amount of co-edited MM signal compared to the asymmetric HS pulse.
Fig. 3.
(A) Inversion profiles for our asymmetric adiabatic HS editing pulse compared to a conventional Gaussian pulse (120 Hz FWHM). Measured and simulated profiles are matched in center frequency and graphically overlaid. A frequency band of ΔB0 = ±0.15 ppm is indicated to visualize the frequency offset sensitivity of the Gaussian pulse, while the adiabatic pulse yields robust, full inversion over ΔB0. For the asymmetric HS pulse, stable inversion of the 3CH2-GABA resonance at 1.9 ppm, macromolecules (MM) at 1.7 ppm, and lipids at 1.2 ppm is achieved, while the Gaussian pulse affects MM at 1.7 ppm only by partial inversion. (B) Immunity plot summarizing the inversion behavior in the presence of variations of ±30% and ±0.15 ppm editing frequency offsets of the asymmetric HS pulse and the Gaussian pulse. While the Gaussian pulse exhibits only a small region of full inversion (−1), the inversion efficiency of the HS pulse is invariant to variations. Also, full inversion is achieved in the presence of a ±0.15 ppm editing frequency offset for the asymmetric HS pulse, which is important to compensate for in vivo B0 field inhomogeneities. Dashed lines indicate the regions of = ±30% and ΔB0 = ±0.15 ppm.
Simulated 4CH2-GABA resonance shapes from the EDIT-ON/EDIT-OFF/DIFF-spectra are provided in Supplementary Figure 2. Immunity against B0 and variations is summarized in a 2D inversion efficiency contour plot in Fig. 3(B).
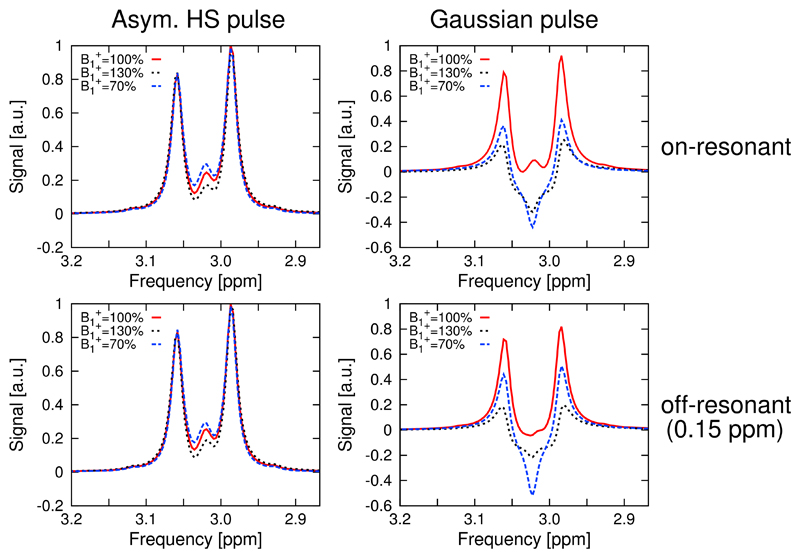
Simulations of the 4CH2-GABA 3.01 ppm resonance in Fig. 4 showed high editing stability using the asymmetric HS editing pulse, with less than 1.8% integrated GABA signal amplitude variation, even in the presence of a ±30% offset. For the HS editing pulse, a 0.15 ppm editing frequency offset showed negligible loss (of 1%) of the edited GABA signal. Also, the shape of the GABA multiplet was preserved in all cases for the HS pulse. At = 100%, the Gaussian editing pulse yielded a 10% and 29% less integrated signal of the edited GABA multiplet compared to the HS pulse for the on-resonance and off-resonance case, respectively. A severe signal drop by factors of 5 and 15.8, together with a strongly deteriorated triplet structure (i.e., outer multiplet peaks were significantly decreased and the inner peak was negative), was obtained for the Gaussian editing pulse compared to the HS editing pulse at = 70% and = 130%, respectively.
Fig. 4.
Simulations of the 4CH2-GABA triplet at 3.01 ppm. The resonance shapes showed high editing stability in case of the adiabatic editing pulse in the presence of variations of ±30% and 0.15 ppm editing frequency offsets (<1.8% integrated signal loss). At = 100%, the conventional Gaussian pulse yielded up to 29% less integrated signal in the off-resonance case, while a 15.8-fold lower signal integral, together with a strongly deteriorated triplet structure, was obtained for the Gaussian pulse compared to the HS pulse at = 70% and = 130%.
3.2. Phantom measurements
The measured 4CH2-GABA resonances from the EDIT-ON/EDIT-OFF/DIFF-spectra are provided in Supplementary Figure 2 and support the simulated results from Section 3.1. The experimentally determined transition bands were 160 Hz downfield and 920 Hz upfield, with a flat-top of 538 Hz, which is in good agreement with simulated values.
3.3. Volunteer measurements
A graphical representation of motion and B0-field changes, as measured by the vNavs during a typical MRSI scan, is provided in Supplementary Figure 3.
Fitted spectra (DIFF and EDIT-OFF) from different brain regions are shown in Fig. 5, including voxels with typically good spectral quality and voxels where fitting is challenging due to baseline distortions and B0 inhomogeneities (frontal lobe). Doublets were observed for 4CH2-GABA signals at 3.01 ppm and co-edited Glx signals at 3.74 ppm in the DIFF-spectra, while EDIT-OFF spectra included the full set of brain metabolites. Supplementary Figure 4 shows EDIT-OFF and DIFF spectra over the full metabolite range.
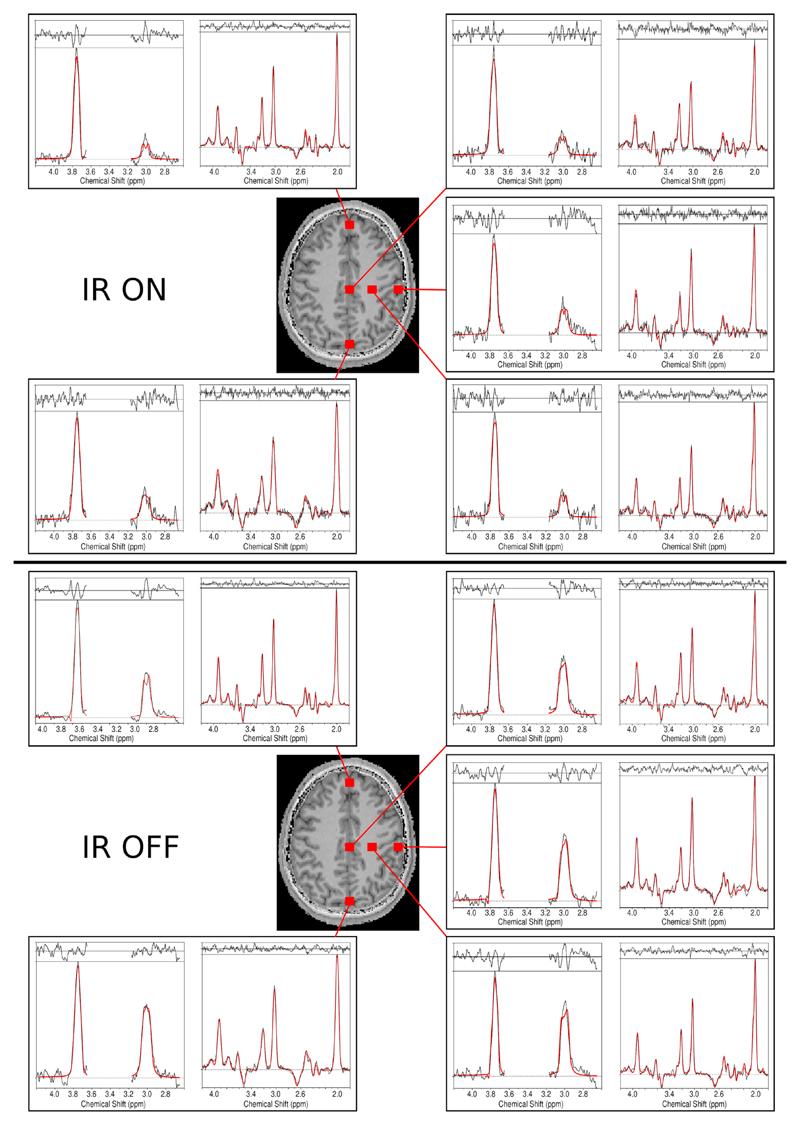
Fig. 5.
Sample spectra including the LCModel fits (in red) for volunteer 1. Spectra from different brain regions that included voxels with typically good spectral quality (mesial gray matter and left frontoparietal white matter) and lower spectral quality (frontal and occipital lobes) are shown. For each voxel, DIFF and EDIT-OFF spectra are grouped together. For comparison, spectra with inversion recovery ON and OFF are provided. A decrease in overall SNR, as well as a significantly reduced GABA peak, is observed in the IR-ON case. The effective voxel size was 1.4 cm3.
Applying IR reduced the mean SNR calculated over all volunteers and all voxels by about 50% from 241.1 ± 99.0 to 119.6 ± 47.1 (p < 0.005). The mean linewidth of NAA changed from 12.1 ± 2.9 to 11.5 ± 3.2 when applying IR (p < 0.005). The CRLBs of tNAAOFF increased from 2.0 ± 1.5 to 2.3 ± 1.2 (p = 0.21), without and with IR, respectively. Similar CRLB changes were observed for tCrOFF, with CRLBs increasing from 2.2 ± 1.3 to 2.6 ± 1.2 (p = 0.62). The mean GABA CRLBs increased from 4.2 ± 2.8 to 8.0 ± 2.3 when applying IR (p < 0.005), while for Glu the CRLBs increased from 3.2 ± 2.2 to 3.6 ± 2.6 (p = 0.015).
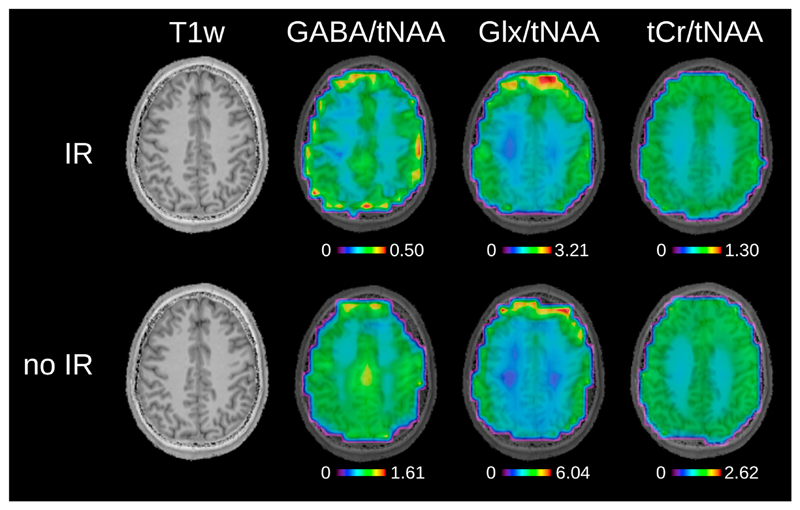
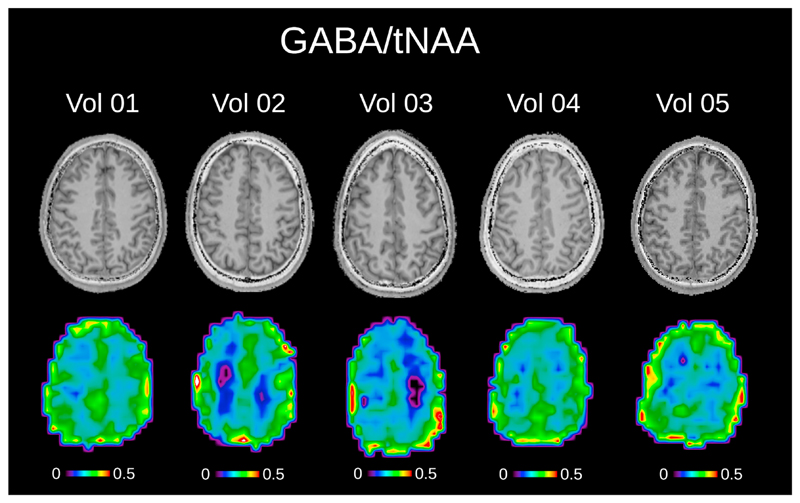
Fig. 6 depicts metabolic ratio maps (i.e., GABA/tNAA, GABA+/tNAA, GlxDIFF/tNAA, and tCr/tNAA) of one volunteer including anatomical MP2RAGE images (maps without anatomical overlay are provided in Supplementary Figure 5). Fig. 7 shows the pure GABA maps (with IR-ON) obtained for all five volunteers.
Fig. 6.
Metabolic ratio maps for inversion recovery ON and OFF for volunteer 1. T1-weighted images are shown, as well as GABA/tNAA and Glx/tNAA maps from DIFF spectra. tCr/tNAA maps are also provided and were obtained from EDIT-OFF spectra. The tNAA map from EDIT-OFF IR-ON spectra was used as normalization in all cases. Metabolic maps were acquired using the following MRSI settings: FOV 220 × 220 mm2; 32 × 32 matrix; slice thickness 16 mm; TA 24:12 min. The maps were interpolated to a 64 × 64 matrix for display.
Fig. 7.
The pure GABA/tNAA metabolic maps without MM contamination, as well as the corresponding T1-weighted images are shown for all five volunteers. The tNAA map from EDIT-OFF IR-ON spectra was used as normalization in all cases. Metabolic maps were acquired using the following MRSI settings: FOV 220 × 220 mm2; 32 × 32 matrix; slice thickness 16 mm; TA 24:12 min. The maps were interpolated to a 64 × 64 matrix for display.
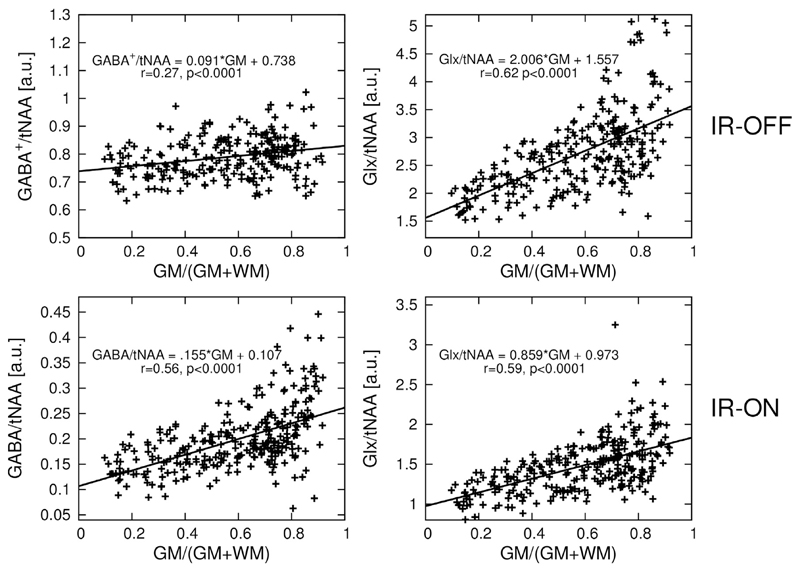
The linear regression result of GABA/tNAA and Glx/tNAA versus the GM fraction is provided in Fig. 8. GM/WM ratios calculated from linear regression over all volunteers for all metabolic maps are listed in Table 1. The GM/WM ratio for tCr/tNAA was similar when applying IR and not applying IR, with values around 1.5. Also, for GlxOFF/tNAA, the GM/WM ratio was decreased only a small amount (i.e., <6% difference between IR-ON and IR-OFF). GABA/tNAA showed a 2.15-fold increase in GM/WM contrast when applying IR, while GlxDIFF/tNAA experienced a slight contrast reduction of about 17%.
Fig. 8.
Linear regression plot of GABA/tNAA and Glx/tNAA versus gray matter tissue composition. tNAA obtained from EDIT-OFF IR-ON spectra was used as normalization in all cases. Linear regression equation, as well as r-value and p-values are provided. GABA/tNAA shows a signal decrease by a factor of ~4 in the case of IR-ON compared to IR-OFF (IR reduces the GABA signal by a factor of ~2 and eliminates MM contributions), while a factor of only ~1.7 in signal decrease is observed in the case of Glx/tNAA.
Table 1.
Ratios for gray/white matter contrast for different metabolic ratio maps averaged over all five volunteers. GABA/tNAA and GlxDIFF/tNAA were obtained from DIFF-spectra, while GlxOFF/tNAA and tCr/tNAA were obtained from EDIT-OFF spectra. The GM/WM ratios are presented with inversion recovery (IR-ON) and without (IR-OFF). tNAA obtained from EDIT-OFF IR-ON spectra was used as normalization in all cases.
| GM/WM contrast | ||
|---|---|---|
| IR-OFF | IR-ON | |
| GABA/tNAA | 1.11 ± 0.13 | 2.39 ± 0.06 |
| GlxDIFF/tNAA | 2.21 ± 0.05 | 1.84 ± 0.17 |
| GlxOFF/tNAA | 2.23 ± 0.12 | 2.11 ± 0.11 |
| tCr/tNAA | 1.53 ± 0.05 | 1.50 ± 0.04 |
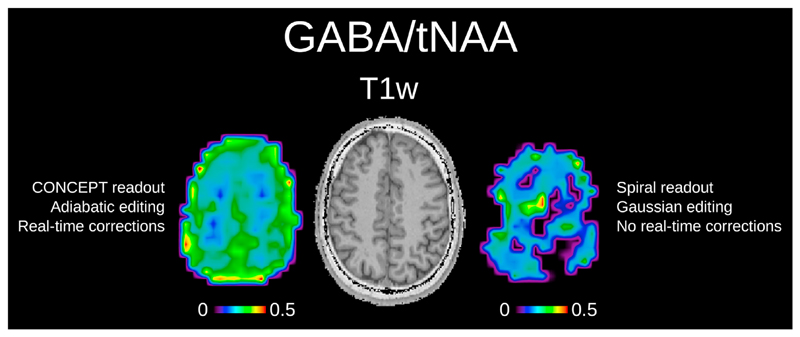
Fig. 9 shows the qualitative in vivo comparison of the pure GABA/tNAA maps obtained with our proposed approach compared to a conventional approach including spiral readout, Gaussian editing pulses and no real-time corrections.
Fig. 9.
Qualitative comparison pure GABA/tNAA maps obtained via our new editing approach (adiabatic editing CONCEPT readout, real-time corrections, and IR-based MM suppression) compared to a more conventional approach (narrow-band Gaussian editing pulses symmetric editing around MM at 1.7 ppm, spiral readout, no real-time corrections). Lower SNR, -related editing efficiency loss and the lack of real-time corrections resulted in low-quality GABA maps for the conventional approach and render any reasonable quantitative evaluation impossible.
4. Discussion
In this study, an adiabatic MEGA-editing scheme was developed and incorporated into a -insensitive MRSI sequence, which enabled whole-slice metabolic imaging of neurotransmitters in the human brain with unprecedented high-resolution at 7T.
Our approach exploits the advantages of edited-MRSI at 7T while tackling challenges that arise with an ultra-high-field. Our approach was validated by simulations, phantom, and in vivo measurements and included: a) adiabatic 1D-semiLASER selection that yielded improved -insensitive localization with negligible CSDE; b) asymmetric adiabatic MEGA-editing pulses that mitigated regionally different editing efficiencies while keeping the overall echo time optimal, i.e., ~1/(2J); c) spatial-spectral encoding that used the time-efficient CONCEPT readout with substantially improved PSF; d) real-time updating of the MRSI sequence based on vNavs, in combination with selective reacquisition of corrupted data, for robust, subtraction-based MEGA-editing.
4.1. Spatial distribution of GABA
Previous MRSI studies provided indications of higher GABA concentrations in cortical GM compared to WM, with quite heterogeneous results of GM/WM ratios ranging from 1.48 to 8.12 (Choi et al., 2006; Jensen et al., 2005; Zhu et al., 2011). These reports are encouraging from a biochemical perspective, as GABA exerts its effects via GABAergic synapses that are predominantly located in the gray matter (Petroff et al., 1995). Also, GABA is mainly metabolized from Glu via glutamate decarboxylase (Stagg et al., 2011a,b) and Glu is known to exhibit a GM/WM contrast (Marsman et al., 2013). This highlights the significance of tissue composition in GABA measurements, but all previous MRSI reports suffer from low spatial resolution, which makes the regional assessment of GABA levels difficult. In all reports, voxels from brain regions with typically poor spectral quality were discarded (e.g., anterior to the ventricular system due to problems linked to susceptibility and off-resonance effects from the sinus-cavity), thereby reducing the number of gray matter voxels for further analysis. Also, the regional distribution of macromolecular contributions to the measured GABA signals needs to be addressed, which may bias the GM/WM contrast of GABA.
We present the first study to investigate regional GABA and GABA+ levels based on whole-slice metabolic maps with an effective voxel size of 1.4 cm3 in about 24 min, while Zhu et al. were mapping GABA+ levels with voxel sizes of 7 cm3 in 17 min at 3T (Zhu et al., 2011).
The majority of published GABA-edited DIFF spectra contained fitted resonances of NAA (2.01 ppm), GABA (3.01 ppm), and Glx (3.74 ppm). We achieved more stable LCModel results by fitting a tighter spectral range that contained only GABA and Glx and also spared the spectral range in-between. Spectral quality was high throughout the FOV, with only little differences in the linewidth for the IR-ON/IR-OFF scan. A noticeable SNR reduction, by up to 50%, was observed when enabling inversion recovery for all metabolites, while the GABA resonance was affected by IR in two ways: a) underlying MM contributions were fully nulled; and b) the GABA signal itself was reduced due to incomplete signal recovery.
Metabolic maps showed distinct anatomical contrast and were used to calculate metabolite abundance differences in GM and WM. Linear regression showed a clear elevation of tCr levels in GM and was in the range of previously published reports (Gasparovic et al., 2009; Wang and Li, 1998). No significant changes induced by inversion recovery were found, which is explained by the predominance of Cr compared to other signals found around 3 ppm. Without IR, GABA+/tNAA showed only little GM/WM contrast, but this contrast increased 2.15-fold when IR (i.e., GABA/tNAA) was enabled. We attribute this to an elevated abundance of MM2.99 ppm in the white matter compared to the gray matter, which is in accordance with the results of Povavžan et al. at 3T (Povavžan et al., ISMRM, 2017, No. 1058). However, the characterization and distribution of MM are an on-going field of research, with age-specific, pathologically altered, and even inter-subject differences being described (Chong et al., 2011; Hofmann et al., 2001; Mader et al., 2001). GM/WM contrasts obtained for Glx/tNAA showed similar results for EDIT-OFF and DIFF spectra and for IR turned ON/OFF, and were also in the range described in the literature (Gasparovic et al., 2009). Only for GlxDIFF/tNAA with enabled IR was an unexpected reduction in contrast of 17% observed. This may have been caused by T1 effects or by unanticipated co-editing due to as yet unknown metabolite couplings, as our adiabatic editing pulses exhibited broad editing bands. This should be further investigated.
4.2. Spatial localization using 1D-semiLASER
Inhomogeneities in the radio frequency field are a known challenge for many MR experiments at 7T, especially when targeting low abundant metabolites in MRSI. Up to 50% variation has been reported for 7T in the human brain (Singh et al., 2013). Severe flip angle variations and CSDE, as well as poor selection profiles, make conventional PRESS localization almost impossible at 7T. Localization based on -insensitive adiabatic refocusing (LASER or semiLASER) can mitigate these challenges (Scheenen et al., 2008a,b). The high SAR demand of full LASER sequences at 7T has been tackled by using either gradient offset modulated adiabatic (GOIA) pulses (Bhattacharyya et al., ISMRM, 2017, No. 3012), or by using semi-LASER sequences, a combination of a non-adiabatic excitation pulse and high-bandwidth adiabatic Frequency Offset Corrected Inversion (FOCI) pulses (Arteaga de Castro et al., 2013).
Our aim was to perform MRSI over a whole brain slice without being restricted to a confined LASER box inside the brain and including even cortical areas. Our proposed 1D-semiLASER sequence is an extension of the semi-LASER sequence using only one pair of GOIA pulses. The use of a non-adiabatic sinc excitation pulses leaves some -sensitivity, but is a good compromise between -immunity and SAR demand. The use of only one pair of GOIA pulses allowed for significant SAR reduction and longer and more sophisticated editing pulses to be incorporated into the TE of 69 ms.
4.3. Adiabatic MEGA-editing
All MEGA-editing reports thus far have employed an editing scheme very similar to the one initially proposed by Mescher et al. (1998). If B0 and inhomogeneities are sufficiently small in the volume of interest (e.g., SVS in small voxels placed in the occipital lobe), such a non-adiabatic editing approach is certainly adequate. However, this approach may become increasingly problematic even for SVS with medium/large sized voxels in regions known to be difficult to shim (e.g., hippocampus), and unfeasible for MRSI at 7T. While most reports for 7T used conventional Gaussian pulses for MEGA-editing (Cai et al., 2012; Chen et al., 2017; Terpstra et al., 2002), there are only a limited number of studies that used more sophisticated but longer (~30–44 ms) pulses, such as Shinnar Le-Roux (SLR) pulses (Andreychenko et al., 2012; Henry et al., 2001; Kaiser et al., 2007). SLR pulses provide substantially improved inversion profiles (i.e., smaller inversion bandwidth combined with narrow transition bands) compared to Gaussian pulses, but are also more sensitive to errors. This makes their use less attractive at 7T.
Due to their -insensitivity adiabatic pulses have been used for MEGA lipid/water suppression pulses at 3T in full slab 3D MRSI (Esmaeili et al., 2017) and for pre-excitation CHESS water/fat suppression (Zhu et al., 2010). Asymmetric adiabatic pulses have been also used at 7T to perform CHESS water/fat suppression for full slice MRSI (Zhu et al., 2013). Adiabatic MEGA-editing was already employed for heteronuclear spectral editing (Garwood and Merkle, 1991), but to our knowledge, in this paper we present the first homonuclear J-difference editing sequence to use adiabatic editing pulses. In contrast to symmetric editing pulses, asymmetric pulses feature narrower transition bands, while being sufficiently short to remain within an optimal TE of 1/(2J)~69 ms. Broadband adiabatic pulses with one narrow transition band are very well-suited for MEGA-editing at 7T, thereby ensuring stable, full inversion in the presence of substantial B0 frequency offsets as encountered in whole-slice MRSI. The flat-top of our designed asymmetric pulses covered a range of more than 500 Hz, broad enough to include the 1.9 ppm GABA resonance over ΔB0 variations of ±0.15 ppm and fully suppress lipid signals at 1.25 ppm. This might lead to lipid signals in the DIFF spectra (Supplement Figure 4) as lipids are suppressed in the ON-acquisition, but not in the OFF-acquisition. However, this did not cause problems with our tight fitting range. The baseline was adequately estimated by LCModel in the GABA region. Hence, we decided to deactivate the editing pulses in the OFF-acquisition (the editing pulse amplitude was set to zero) in order to save SAR. If in future studies (e.g., 3D-MRSI) lipids will be more problematic, they could be reduced by activating the editing pulses in the OFF-acquisition. Their frequency would need to be set such that the main lipid resonances will be suppressed in both acquisitions, while affecting the 1.9 ppm GABA resonance only in the ON-acquisition.
Our 1D-semiLASER sequence with only one pair of GOIA refocusing pulses is well suited for incorporating the asymmetric, adiabatic, 20-ms-long editing pulses within a TE of 69 ms, which would not be feasible in a full LASER or semiLASER sequence with more pulses used for volume selection. As adiabatic editing pulses are undoubtedly more SAR-demanding than conventional Gaussian pulses, a TR of 1600 ms as frequently used in GABA-edited MRS at 3T was not feasible, but the 1D-semiLASER sequence helped to minimize the required TR and keep it far below other reported TR values at 7T (Andreychenko et al., 2012; Chen et al., 2017).
While in vivo variations nearly reach ±28% in transverse slices above the ventricles, even higher inhomogeneities of up to ±47% can be observed in more central slices (Supplementary Figure 1). The advantage of our adiabatic editing approach is that inhomogeneities can be compensated for by increasing the pulse amplitude to fulfil the adiabatic condition throughout the slice/slab even in such cases. Furthermore, stronger B0 inhomogeneities in central brain areas (Supplementary Figure 1) can be compensated for via adiabatic MEGA-editing. The only “drawback” is that higher B0 variations lead to increased extension of the MEGA inversion profile into the metabolite spectral region to cover all B0 variations. This is not a limitation unless the B0 inhomogeneities become large enough that the transition band of the inversion profile reaches the 3.0 ppm GABA resonance.
MM-suppressed detection of GABA was achieved by B0 and -insensitive adiabatic inversion recovery. Although IR inherently leads to metabolite signal loss, other MM-suppressive approaches, such as symmetric editing around 1.7 ppm (Henry et al., 2001), are not feasible in the presence of large B0 frequency variations across the FOV.
Phantom measurements showed a high agreement of the EDIT-ON, EDIT-OFF, and DIFF GABA resonance shapes, not only with our simulations, but also with the literature reports (Near et al., 2013).
Our simulations proved that adiabatic MEGA-editing pulses successfully mitigated editing efficiency loss caused by inhomogeneities, as can be inferred from Figs. 3 and 4. In the presence of variations of ±30%, negligible reductions (<1.8%) of integrated, edited signal intensity were observed, while conventional Gaussian editing pulses yielded a signal loss of up to a factor of 15 when also considering a 0.15 ppm frequency offset. Even without frequency and offsets the adiabatic editing pulses yielded about a 10% more integrated signal compared to Gaussian editing pulses, which we attribute to adiabatic spin-locking effects.
4.4. Spatial-spectral sampling
Robust MEGA-editing requires a minimum of four measurement repetitions (two-step phase cycling and acquisition of EDIT-ON/EDIT-OFF spectra). This leads to prolonged scan times for conventional phase-encoded MRSI. Non-Cartesian sampling based on concentric circle trajectories offers acceleration factors of up to two orders of magnitude compared to phase-encoded MRSI (Hingerl et al., 2017). By intrinsically sampling a Hamming-weighted k-space, substantial PSF improvements and an SNR gain of +24% and +40% could be achieved compared to elliptical phase-encoding and parallel imaging accelerated MRSI, respectively (Hingerl et al., 2017; Strasser et al., 2017).
It is well known that artifacts from extracranial lipids may cause severe baseline distortions, especially when the FOV extends over the whole brain slice. In previous GABA-edited MRSI reports, Bogner et al. were restricted to a cuboid LASER selection box within the brain, and hence, could avoid lipid contaminations (Bogner et al., 2014), while Zhu et al. employed highly optimized, hyper geometric dual-band pulses at 3T with outer-volume suppression for water and lipid suppression (Zhu et al., 2011). Both approaches would face significant SAR problems at 7T.
4.5. Real-time motion and scanner instability correction
Uncorrected subject head motion and B0 field changes degrade edited MRSI data (Evans et al., 2013; Heckova et al., 2018; Puts et al., 2012) and are a substantial source of measurement bias in MRI in general (Zaitsev et al., 2015). In addition to the SNR loss caused by line broadening, subtraction errors of misaligned spectra lead to improperly subtracted Cr signals, and hence, an overestimation and variability of GABA signals. Older, healthy subjects and, in particular, patients with Parkinson's disease or mild cognitive impairment showed up to 2.5-fold more involuntary head movement than young healthy volunteers (Heckova et al., 2018).
In contrast to SVS, MRSI does not allow for simple frequency and phase alignment during post-processing without an additional navigator due to its spatial encoding. Our real-time correction approach is similar to other studies from 3T and is based on volumetric EPI navigators inserted prior to water suppression in each MRSI TR (Bogner et al., 2014; Hnilicová et al., 2016). These vNavs were capable of monitoring head motion, frequency, and shim changes in real-time with sub-millimeter precision via two low-resolution 3D magnitude/phase images of different TE. Following Bogner et al. and Zhu et al., frequency updates were applied only to pairs of EDIT-ON/EDIT-OFF acquisitions to prevent unfavorable rounding, as only integer update values were allowed on our scanner hardware (Bogner et al., 2014; Zhu et al., 2011). To further compensate for head movement between EDIT-ON and EDIT-OFF, we implemented selective reacquisition of corrupted data. If the rejection criterion (i.e., >0.4 mm translation or >0.4° rotation) was triggered between an EDIT-ON/EDIT-OFF pair, this specific pair was reacquired immediately. However, if the criterion was triggered before an EDIT-ON/EDIT-OFF pair, the previous pair was reacquired. A maximum of 25% of the total scan time was imposed as maximum scan time prolongation.
While vNavs have been shown to reduce spatial variability and subtraction artifacts, and thereby increasing the reproducibility of GABA-edited MRSI at 3T (Hnilicová et al., 2016), to our knowledge, no previous report has implemented this real-time correction approach for edited MRSI at 7T. As reported previously for non-edited MRSI (>Moser et al., ISMRM, 2017, No. 1253), several optimizations to the EPI navigator protocol used on 3T had to be made to exploit the potential of vNavs at 7T. As Nyquist N/2-ghosting artifacts were significantly worse at 7T (Beisteiner et al., 2011), a local phase correction (Feiweier et al., 2011; Lima Cardoso et al., 2018) was implemented. Fat artifacts were eliminated by using water-selective excitation, which allowed shorter TRs compared to dedicated fat suppression pulses. vNavs at 3T have been employed with slab-selective excitation, which was switched to non-selective excitation due to water-selective excitation at 7T. Possible fold-in artifacts were eliminated via increasing the FOV in the z-direction. Spatial image distortions were reduced by lowering the EPI factor from 32 to 16 (i.e., dual-shot EPI rather than single-shot EPI).
4.6. Limitations
Adiabatic MEGA-editing is a promising approach for ultra-high field scanning, but requires significantly longer editing pulses compared to conventional Gaussian pulses. We achieved a TE of 69 ms by asymmetric editing pulses combined with only one pair of GOIA pulses in a 1D-semi-LASER localization. Employing different volume selections might lead to prolonged echo times.
Our asymmetric HS editing pulses have a broad inversion profile, with one narrow and one broad transition band. This approach works well if the resonance of interest (in our case GABA at 3.01 ppm) is downfield of the co-edited resonance targeted by the editing pulses (in our case GABA at 1.9 ppm). In case of GSH, however, the resonance of interest (i.e., 2.95 ppm) is upfield of the co-edited resonance (4.56 ppm). A simple flip of the inversion profile by inverting the phase modulation function of the asymmetric HS pulse would resolve this issue.
Our approach tackles multiple challenges of whole-slice edited-MRSI on a software basis. However, modern hardware based approaches (i.e., lipid crusher coils (Boer et al., 2015), B1 shimming (Hetherington et al., 2010), B0 shim arrays (Stockmann et al., 2016)) might further improve data quality.
In this study, we investigated only the distribution of the neurotransmitters GABA and Glx using the MEGA-editing technique. However, our edited MRSI approach could further be applied to other highly interesting metabolites, including the onco-metabolite 2-hydroxyglutarate (Andronesi et al., 2016), lactate (Arteaga de Castro et al., 2013) in brain tumors, and the antioxidant GSH in neurodegenerative and psychiatric disorders (Terpstra et al., 2003).
5. Conclusion
Adiabatic MEGA-editing incorporated into a 1D-semiLASER-MRSI sequence is feasible at 7T and can account for the severe B0 and variations frequently encountered in whole-slice MRSI at ultra-high field. Integrating time-efficient, spatial-spectral CONCEPT encoding, and realtime motion and scanner instability correction makes this approach a promising technique for whole-brain, spectrally edited mapping of neurotransmitters, antioxidants, and onco-metabolites.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuroimage.2018.09.039.
Acknowledgments
This study received support from the Austrian Science Fund (FWF): KLI 646, P 30701 and J 4124; the FFG Bridge Early Stage Grant #846505.
Abbreviations
- CSDE
chemical shift displacement error
- CONCEPT
concentric circle echo planar trajectories
- CRLB
Cramér-Rao lower bound
- Cr
creatine
- CSF
cerebrospinal fluid
- EPI
echo planar imaging
- EPSI
echo-planar spectroscopic imaging
- FA
flip angle
- FID
free induction decay
- FOV
field of view
- FWHM
full width at half maximum
- GABA
γ-aminobutyric acid
- GABA+
γ-aminobutyric acid + macromolecules
- Gln
glutamine
- Glu
glutamate
- Glx
glutamate + glutamine
- GM
gray matter
- GOIA
gradient offset independent adiabatic
- GSH
glutathione
- HS
hyperbolic secant
- IR
inversion recovery
- m-Ins
myo-inositol
- MEGA
Mescher-Garwood
- MM
macromolecule
- MP2RAGE
magnetization-prepared 2 rapid acquisition gradient echoes
- MOSAIC
multi-channel data of spectroscopic imaging assembled via interleaved calibrations
- MRSI
magnetic resonance spectroscopic imaging
- NAA
N-acetyl-aspartate
- NAAG
N-acetyl-aspartyl glutamate
- PCr
phosphocreatine
- PSF
point-spread function
- PRESS
point-resolved spectroscopy
- SAR
specific absorption rate
- semi-LASER
semi-localized adiabatic selective refocusing
- SLR
Shinnar-Le Roux
- SNR
Signal-to-noise ratio
- SSE
spatial-spectral encoding
- SVS
single-voxel spectroscopy
- TA
acquisition time
- tCr
Cr + PCr
- TE
echo time
- TI
inversion time
- tNAA
NAA + NAAG
- TR
repetition time
- UHF
ultra-high field
- WET
water suppression enhanced through T1 effects
- vNav
volumetric navigator
- WM
white matter
- WURST
wideband uniform rate and smooth truncation
Footnotes
Declarations of interest
None.
Contributor Information
Philipp Moser, Email: philipp.a.moser@meduniwien.ac.at.
Lukas Hingerl, Email: lukas.hingerl@meduniwien.ac.at.
Bernhard Strasser, Email: bstrasser@mgh.harvard.edu.
Michal Považan, Email: mpovaza1@jhmi.edu.
Gilbert Hangel, Email: gilbert.hangel@meduniwien.ac.at.
Ovidiu C. Andronesi, Email: ovidiu@nmr.mgh.harvard.edu.
Andre van der Kouwe, Email: andre@nmr.mgh.harvard.edu.
Stephan Gruber, Email: stephan@nmr.at.
Siegfried Trattnig, Email: siegfried.trattnig@meduniwien.ac.at.
References
- Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998;39:889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- Agarwal N, Renshaw PF. Proton MR spectroscopy-detectable major neurotransmitters of the brain: biology and possible clinical applications. Am J Neuroradiol. 2012;33:595–602. doi: 10.3174/ajnr.A2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhamud A, Taylor PA, van der Kouwe AJW, Meintjes EM. Real-time measurement and correction of both B0 changes and subject motion in diffusion tensor imaging using a double volumetric navigated (DvNav) sequence. Neuroimage. 2016;126:60–71. doi: 10.1016/j.neuroimage.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DWJ. Efficient spectral editing at 7 T: GABA detection with MEGA-sLASER. Magn Reson Med. 2012;68:1018–1025. doi: 10.1002/mrm.24131. [DOI] [PubMed] [Google Scholar]
- Andronesi OC, Gagoski BA, Adalsteinsson E, Sorensen AG. Correlation chemical shift imaging with low-power adiabatic pulses and constant-density spiral trajectories. NMR Biomed. 2012;25:195–209. doi: 10.1002/nbm.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi OC, Loebel F, Bogner W, Marjanska M, Vander Heiden MG, Iafrate AJ, Dietrich J, Batchelor TT, Gerstner ER, Kaelin WG, Chi AS, et al. Treatment response assessment in IDH-mutant glioma patients by noninvasive 3D functional spectroscopic mapping of 2-hydroxyglutarate. Clin Canc Res. 2016;22:1632–1641. doi: 10.1158/1078-0432.CCR-15-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi OC, Ramadan S, Ratai E-M, Jennings D, Mountford CE, Sorensen AG. Spectroscopic imaging with improved gradient modulated constant adiabaticity pulses on high-field clinical scanners. J Magn Reson. 2010;203:283–293. doi: 10.1016/j.jmr.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga de Castro CS, Boer VO, Andreychenko A, Wijnen JP, van der Heide UA, Luijten PR, Klomp DWJ. Improved efficiency on editing MRS of lactate and γ-aminobutyric acid by inclusion of frequency offset corrected inversion pulses at high fields. NMR Biomed. 2013;26:1213–1219. doi: 10.1002/nbm.2937. [DOI] [PubMed] [Google Scholar]
- Bai X, Edden RAE, Gao F, Wang G, Wu L, Zhao B, Wang M, Chan Q, Chen W, Barker PB. Decreased γ-aminobutyric acid levels in the parietal region of patients with Alzheimer's disease. J Magn Reson Imag. 2015;41:1326–1331. doi: 10.1002/jmri.24665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisteiner R, Robinson S, Wurnig M, Hilbert M, Merksa K, Rath J, Höllinger I, Klinger N, Marosi C, Trattnig S, Geissler A. Clinical fMRI: evidence for a 7T benefit over 3T. Neuroimage. 2011;57:1015–1021. doi: 10.1016/j.neuroimage.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya P, Lowe M, Andronesi OC. GABA Editing with Reduced Sensitivity to B1 Inhomogeneity and Improved Detectability at 7T Using MEGA-LASER. Proceedings of the 25th Annual Meeting of the ISMRM; Hawaii. 2017. p. 3012. [Google Scholar]
- Boer VO, van de Lindt T, Luijten PR, Klomp DWJ. Lipid suppression for brain MRI and MRSI by means of a dedicated crusher coil. Magn Reson Med. 2015;73:2062–2068. doi: 10.1002/mrm.25331. [DOI] [PubMed] [Google Scholar]
- Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJW, Strasser B, Marjańska M, Trattnig S, Grant E, Rosen B, Andronesi OC. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014a;103:290–302. doi: 10.1016/j.neuroimage.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner W, Hangel G, Esmaeili M, Andronesi OC. 1D-spectral editing and 2D multispectral in vivo 1H-MRS and 1H-MRSI - methods and applications. Anal Biochem. 2017;529:48–64. doi: 10.1016/J.AB.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Bogner W, Hess AT, Gagoski B, Tisdall MD, van der Kouwe AJW, Trattnig S, Rosen B, Andronesi OC. Real-time motion- and B0-correction for LASER-localized spiral-accelerated 3D-MRSI of the brain at 3T. Neuroimage. 2014b;88:22–31. doi: 10.1016/j.neuroimage.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan BP, Admon R, Perriello C, LaFlamme EM, Athey AJ, Pizzagalli DA, Hudson JI, Pope HG, Jensen JE. Acute change in anterior cingulate cortex GABA, but not glutamine/glutamate, mediates antidepressant response to citalopram. Psychiatry Res Neuroimaging. 2017;269:9–16. doi: 10.1016/j.pscychresns.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Nanga RP, Lamprou L, Schinstine C, Elliott M, Hariharan H, Reddy R, Epperson CN. The impact of gabapentin administration on brain GABA and glutamate concentrations: a 7T 1H-MRS study. Neuropsychopharmacology. 2012;37:2764–2771. doi: 10.1038/npp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley N, Solanky BS, Muhlert N, Tur C, Edden RAE, Wheeler-Kingshott CAM, Miller DH, Thompson AJ, Ciccarelli O. Reduced gamma-aminobutyric acid concentration is associated with physical disability in progressive multiple sclerosis. Brain. 2015;138:2584–2595. doi: 10.1093/brain/awv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sigurdsson HP, Pépées SE, Auer DP, Morris PG, Morgan PS, Gowland PA, Jackson SR. Activation induced changes in GABA: functional MRS at 7 T with MEGA-sLASER. Neuroimage. 2017;156:207–213. doi: 10.1016/J.NEUROIMAGE.2017.05.044. [DOI] [PubMed] [Google Scholar]
- Choi I-Y, Lee S-P, Merkle H, Shen J. In vivo detection of gray and white matter differences in GABA concentration in the human brain. Neuroimage. 2006;33:85–93. doi: 10.1016/j.neuroimage.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Chong DGQ, Kreis R, Bolliger CS, Boesch C, Slotboom J. Two-dimensional linear-combination model fitting of magnetic resonance spectra to define the macromolecule baseline using FiTAID, a Fitting Tool for Arrays of Interrelated Datasets. Magn Reson Mater Physics, Biol Med. 2011;24:147–164. doi: 10.1007/s10334-011-0246-y. [DOI] [PubMed] [Google Scholar]
- Chung S, Kim D, Breton E, Axel L. Rapid B1þ mapping using a preconditioning RF pulse with TurboFLASH readout. Magn Reson Med. 2010;64:439–446. doi: 10.1002/mrm.22423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue MJ, Near J, Blicher JU, Jezzard P. Baseline GABA concentration and fMRI response. Neuroimage. 2010;53:392–398. doi: 10.1016/j.neuroimage.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Edden RAE, Muthukumaraswamy SD, Freeman TCA, Singh KD. Orientation discrimination performance is predicted by GABA concentration and gamma oscillation frequency in human primary visual cortex. J Neurosci. 2009;29:15721–15726. doi: 10.1523/JNEUROSCI.4426-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili M, Bathen TF, Rosen BR, Andronesi OC. Three-dimensional MR spectroscopic imaging using adiabatic spin echo and hypergeometric dual-band suppression for metabolic mapping over the entire brain. Magn Reson Med. 2017;77:490–497. doi: 10.1002/mrm.26115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Puts NAJ, Robson SE, Boy F, McGonigle DJ, Sumner P, Singh KD, Edden RAE. Subtraction artifacts and frequency (mis-)alignment in J-difference GABA editing. J Magn Reson Imag. 2013;38:970–975. doi: 10.1002/jmri.23923. [DOI] [PubMed] [Google Scholar]
- Feiweier T, Huwer S, Huwer T, Kim H, Porter DA, Thorsten Speckner. Method and Magnetic Resonance System to Reduce Distortions in Diffusion Imaging. 2011 [Google Scholar]
- Gao F, Edden RAE, Li M, Puts NAJ, Wang G, Liu C, Zhao B, Wang H, Bai X, Zhao C, Wang X, Barker PB. Edited magnetic resonance spectroscopy detects an age-related decline in brain GABA levels. Neuroimage. 2013;78:75–82. doi: 10.1016/j.neuroimage.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood M, Merkle H. Heteronuclear spectral editing with adiabatic pulses. J Magn Reson. 1991;94:180–185. doi: 10.1016/0022-2364(91)90307-F. [DOI] [PubMed] [Google Scholar]
- Gasparovic C, Yeo R, Mannell M, Ling J, Elgie R, Phillips J, Doezema D, Mayer AR. Neurometabolite concentrations in gray and white matter in mild traumatic brain injury: an 1H-magnetic resonance spectroscopy study. J Neurotrauma. 2009;26:1635–1643. doi: 10.1089/neu.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind V, Young K, Maudsley AA. Corrigendum: proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, editors. NMR Biomed. 2000;13:129–153. doi: 10.1002/nbm.3336. NMR Biomed. 2015; 28: 923–924. [DOI] [PubMed] [Google Scholar]
- Hangel G, Strasser B, Povavžan M, Gruber S, Chmelík M, Gajdovsík M, Trattnig S, Bogner W. Lipid suppression via double inversion recovery with symmetric frequency sweep for robust 2D-GRAPPA-accelerated MRSI of the brain at 7 T. NMR Biomed. 2015;28:1413–1425. doi: 10.1002/nbm.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangel G, Strasser B, Povavzan M, Heckova E, Hingerl L, Boubela R, Gruber S, Trattnig S, Bogner W. Ultra-high resolution brain metabolite mapping at 7 T by short-TR Hadamard-encoded fiD-MRSI. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.10.043. [DOI] [PubMed] [Google Scholar]
- Harris AD, Glaubitz B, Near J, John Evans C, Puts NAJ, Schmidt-Wilcke T, Tegenthoff M, Barker PB, Edden RA. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72:941–948. doi: 10.1002/mrm.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckova E, Povazan M, Strasser B, Krumpolec P, Hnilicov P, Hangel GJ, Moser PA, Andronesi OC, Van Der Kouwe AJ, Valkovic P, Ukropcova B, et al. Real-time correction of motion and imager instability artifacts during 3D γ-aminobutyric acid-edited MR spectroscopic imaging. Radiology. 2018;286 doi: 10.1148/radiol.2017170744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45:517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Hess AT, Dylan Tisdall M, Andronesi OC, Meintjes EM, Van Der Kouwe AJW. Real-time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66:314–323. doi: 10.1002/mrm.22805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess AT, Van Der Kouwe AJW, Mbugua KK, Laughton B, Meintjes EM. Quality of 186 child brain spectra using motion and b0 shim navigated single voxel spectroscopy. J Magn Reson Imag. 2013;40:958–965. doi: 10.1002/jmri.24436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington HP, Avdievich NI, Kuznetsov AM, Pan JW. RF shimming for spectroscopic localization in the human brain at 7 T. Magn Reson Med. 2010;63:9–19. doi: 10.1002/mrm.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerl L, Bogner W, Moser P, Povavzan M, Hangel G, Heckova E, Gruber S, Trattnig S, Strasser B. Density-weighted concentric circle trajectories for high resolution brain magnetic resonance spectroscopic imaging at 7T. Magn Reson Med. 2017 doi: 10.1002/mrm.26987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicová P, Povavzan M, Strasser B, Andronesi OC, Gajdovsík M, Dydak U, Ukropec J, Dobrota D, Trattnig S, Bogner W. Spatial variability and reproducibility of GABA-edited MEGA-LASER 3D-MRSI in the brain at 3 T. NMR Biomed. 2016;29:1656–1665. doi: 10.1002/nbm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann L, Slotboom J, Boesch C, Kreis R. Characterization of the macromolecule baseline in localized (1)H-MR spectra of human brain. Magn Reson Med. 2001;46:855–863. doi: 10.1002/mrm.1269. [DOI] [PubMed] [Google Scholar]
- Hwang T-L, van Zijl PCM, Garwood M. Asymmetric adiabatic pulses for NH selection. J Magn Reson. 1999;138:173–177. doi: 10.1006/jmre.1999.1713. [DOI] [PubMed] [Google Scholar]
- Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding (computerised tomography application) IEEE Trans Med Imag. 1991;10:473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- Jenkinson Pechaud M, Smith S. {BET2: MR}-Based Estimation of Brain. Skull and Scalp Surfaces. 2005 [Google Scholar]
- Jensen JE, deB Frederick B, Renshaw PF. Grey and white matter GABA level differences in the human brain using two-dimensional,J-resolved spectroscopic imaging. NMR Biomed. 2005;18:570–576. doi: 10.1002/nbm.994. [DOI] [PubMed] [Google Scholar]
- Kaiser LG, Young K, Matson GB. Elimination of spatial interference in PRESS-localized editing spectroscopy. Magn Reson Med. 2007;58:813–818. doi: 10.1002/mrm.21407. [DOI] [PubMed] [Google Scholar]
- Klose U. Mapping of the radio frequency magnetic field with a MR snapshot FLASH technique. Med Phys. 1992;19:1099–1104. doi: 10.1118/1.596828. [DOI] [PubMed] [Google Scholar]
- Levar N, van Leeuwen JMC, Denys D, van Wingen GA. Divergent influences of anterior cingulate cortex GABA concentrations on the emotion circuitry. Neuroimage. 2017;158:136–144. doi: 10.1016/j.neuroimage.2017.06.055. [DOI] [PubMed] [Google Scholar]
- Lima Cardoso P, Dymerska B, Bachratá B, Fischmeister FPS, Mahr N, Matt E, Trattnig S, Beisteiner R, Robinson SD. The clinical relevance of distortion correction in presurgical fMRI at 7 T. Neuroimage. 2018;168:490–498. doi: 10.1016/J.NEUROIMAGE.2016.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader I, Seeger U, Weissert R, Klose U, Naegele T, Melms A, Grodd W. Proton MR spectroscopy with metabolite-nulling reveals elevated macromolecules in acute multiple sclerosis. Brain. 2001;124:953–961. doi: 10.1093/brain/124.5.953. [DOI] [PubMed] [Google Scholar]
- Mahone EM, Puts NA, Edden RAE, Ryan M, Singer HS. GABA and glutamate in children with Tourette syndrome: a 1H MR spectroscopy study at 7 T. Psychiatry Res Neuroimaging. 2018 doi: 10.1016/J.PSCYCHRESNS.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele P-F, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 2010;49:1271–1281. doi: 10.1016/j.neuroimage.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Marsman A, Mandl RCW, van den Heuvel MP, Boer VO, Wijnen JP, Klomp DWJ, Luijten PR, Hilleke EHP. Glutamate changes in healthy young adulthood. Eur Neuropsychopharmacol. 2013;23:1484–1490. doi: 10.1016/J.EURONEURO.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Matson GB. An integrated program for amplitude-modulated RF pulse generation and re-mapping with shaped gradients. Magn Reson Imaging. 1994;12:1205–1225. doi: 10.1016/0730-725x(94)90086-7. [DOI] [PubMed] [Google Scholar]
- Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized13C imaging. Magn Reson Med. 2006;56:932–937. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(SICI)1099-1492(199810)11:6<266::AID-NBM530=3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Mescher M, Tannus a, Johnson MO, Garwood M. Solvent suppression using selective echo dephasing. J Magn Reson, Ser A. 1996;123:226–229. doi: 10.1006/jmra.1996.0242. [DOI] [Google Scholar]
- Mikkelsen M, Barker PB, Bhattacharyya PK, Brix MK, Buur PF, Cecil KM, Chan KL, Chen DY-T, Craven AR, Cuypers K, Dacko M, et al. Big GABA: edited MR spectroscopy at 24 research sites. Neuroimage. 2017;159:32–45. doi: 10.1016/J.NEUROIMAGE.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser E, Stahlberg F, Ladd ME, Trattnig S. 7-T MR-from research to clinical applications? NMR Biomed. 2012;25:695–716. doi: 10.1002/nbm.1794. [DOI] [PubMed] [Google Scholar]
- Moser P, Strasser B, Hingerl L, Povavzan M, Hangel G, Andronesi OC, Gagoski B, Hess A, Tisdall D, van der Kouwe A, Trattnig S, et al. Spiral-Accelerated Short-TE MRSI with B1-Insensitive 1D-SemiLASER Localization and Real-Time Motion Correction at 7T. Proceedings of the 25th Annual Meeting of the ISMRM; Hawaii. 2017. No. 1253. [Google Scholar]
- Moser P, Strasser B, Hingerl L, Povavzan M, Hangel G, Heckova E, Gruber S, Trattnig S, Bogner W. MOSAIC - A Generalized Multi-Channel Coil Combination for 1H-MRSI via Interleaved Calibration Scans. Proceedings of the 26th Annual Meeting of the ISMRM; Paris. 2018. No. 1306. [Google Scholar]
- Muthukumaraswamy SD, Edden RAE, Jones DK, Swettenham JB, Singh KD. Resting GABA concentration predicts peak gamma frequency and fMRI amplitude in response to visual stimulation in humans. Proc Natl Acad Sci USA. 2009;106:8356–8361. doi: 10.1073/pnas.0900728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Evans CJ, Puts NAJ, Barker PB, Edden RAE. J-difference editing of gamma-aminobutyric acid (GABA): simulated and experimental multiplet patterns. Magn Reson Med. 2013;70:1183–1191. doi: 10.1002/mrm.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Simpson R, Cowen P, Jezzard P. Efficient γ-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24:1277–1285. doi: 10.1002/nbm.1688. [DOI] [PubMed] [Google Scholar]
- Northoff G, Walter M, Schulte RF, Beck J, Dydak U, Henning A, Boeker H, Grimm S, Boesiger P. GABA concentrations in the human anterior cingulate cortex predict negative BOLD responses in fMRI. Nat Neurosci. 2007;10:1515–1517. doi: 10.1038/nn2001. [DOI] [PubMed] [Google Scholar]
- Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson. 1994;B 104:1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- Petroff OA, Pleban LA, Spencer DD. Symbiosis between in vivo and in vitro NMR spectroscopy: the creatine, N-acetylaspartate, glutamate, and GABA content of the epileptic human brain. Magn Reson Imaging. 1995;13:1197–1211. doi: 10.1016/0730-725x(95)02033-p. [DOI] [PubMed] [Google Scholar]
- Pipe JG, Menon P. Sampling density compensation in MRI: rationale and an iterative numerical solution. Magn Reson Med. 1999;41:179–186. doi: 10.1002/(sici)1522-2594(199901)41:1<179::aid-mrm25>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Prinsen H, de Graaf RA, Mason GF, Pelletier D, Juchem C. Reproducibility measurement of glutathione, GABA, and glutamate: towards in vivo neurochemical profiling of multiple sclerosis with MR spectroscopy at 7T. J Magn Reson Imag. 2017;45:187–198. doi: 10.1002/jmri.25356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- Puts NAJ, Edden RAE. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Spectrosc. 2012;60:1–26. doi: 10.1016/j.pnmrs.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MA, Salibi N, White DM, Gawne TJ, Denney TS, Lahti AC. 7T proton magnetic resonance spectroscopy of the anterior cingulate cortex in first-episode schizophrenia. Schizophr Bull. 2018 doi: 10.1093/schbul/sbx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Tisdall MD, Qureshi A, Buckner RL, van der Kouwe AJW, Fischl B. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. Neuroimage. 2015;107:107–115. doi: 10.1016/j.neuroimage.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med. 2008;60:895–907. doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld D, Panfil SL, Zur Y. Analytic solutions of the Bloch equation involving asymmetric amplitude and frequency modulations. Phys Rev. 1996;54:2439–2443. doi: 10.1103/PhysRevA.54.2439. [DOI] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci USA. 1993;90:5662–5666. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Alhamud A, Near J, van der Kouwe AJW, Meintjes EM. Volumetric navigated MEGA-SPECIAL for real-time motion and shim corrected GABA editing. NMR Biomed. 2016;29:248–255. doi: 10.1002/nbm.3454. [DOI] [PubMed] [Google Scholar]
- Scheenen TWJ, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. Magma. 2008a;21:95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. Short echo time1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008b;59:1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- Shetty AK, Bates A. Potential of GABA-ergic cell therapy for schizophrenia, neuropathic pain, and Alzheimer's and Parkinson's diseases. Brain Res. 2016;1638:74–87. doi: 10.1016/j.brainres.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Cai K, Haris M, Hariharan H, Reddy R. On B1 inhomogeneity correction of in vivo human brain glutamate chemical exchange saturation transfer contrast at 7T. Magn Reson Med. 2013;69:818–824. doi: 10.1002/mrm.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg CJ, Bachtiar V, Johansen-Berg H. The role of GABA in human motor learning. Curr Biol. 2011a;21:480–484. doi: 10.1016/j.cub.2011.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg CJ, Bachtiar V, Johansen-Berg H. What are we measuring with GABA magnetic resonance spectroscopy? Commun Integr Biol. 2011b;4:573–575. doi: 10.4161/cib.4.5.16213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg CJ, Wylezinska M, Matthews PM, Johansen-Berg H, Jezzard P, Rothwell JC, Bestmann S. Neurochemical effects of theta burst stimulation as assessed by magnetic resonance spectroscopy. J Neurophysiol. 2009;101:2872–2877. doi: 10.1152/jn.91060.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starvčuk Z, Starvčuková J, švtrbák O, Graveron-Demilly D. Simulation of coupled-spin systems in the steady-state free-precession acquisition mode for fast magnetic resonance (MR) spectroscopic imaging. Meas Sci Technol. 2009;20 doi: 10.1088/0957-0233/20/10/104033. 104033. [DOI] [Google Scholar]
- Stockmann JP, Witzel T, Keil B, Polimeni JR, Mareyam A, LaPierre C, Setsompop K, Wald LL. A 32-channel combined RF and B0 shim array for 3T brain imaging. Magn Reson Med. 2016;75:441–451. doi: 10.1002/mrm.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JN, Tisdall MD, McDaniel P, Gagoski B, Bolar DS, Grant PE, Adalsteinsson E. Assessing the effects of subject motion on T 2 relaxation under spin tagging (TRUST) cerebral oxygenation measurements using volume navigators. Magn Reson Med. 2017;78:2283–2289. doi: 10.1002/mrm.26616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Chmelik M, Robinson SD, Hangel G, Gruber S, Trattnig S, Bogner W. Coil combination of multichannel MRSI data at 7 T: MUSICAL. NMR Biomed. 2013;26:1796–1805. doi: 10.1002/nbm.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Povavzan M, Hangel G, Hingerl L, Chmelik M, Gruber S, Trattnig S, Bogner W. (2 + 1)D-CAIPIRINHA accelerated MR spectroscopic imaging of the brain at 7T. Magn Reson Med. 2017;78:429–440. doi: 10.1002/mrm.26386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannús A, Garwood M. Adiabatic pulses. NMR Biomed. 1997;10:423–434. doi: 10.1002/(sici)1099-1492(199712)10:8<423::aid-nbm488>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Terpstra M, Henry P-G, Gruetter R. Measurement of reduced glutathione (GSH) in human brain using LCModel analysis of difference-edited spectra. Magn Reson Med. 2003;50:19–23. doi: 10.1002/mrm.10499. [DOI] [PubMed] [Google Scholar]
- Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47:1009–1012. doi: 10.1002/mrm.10146. [DOI] [PubMed] [Google Scholar]
- Tomiyasu M, Aida N, Shibasaki J, Umeda M, Murata K, Heberlein K, Brown MA, Shimizu E, Tsuji H, Obata T. In vivo estimation of gamma-aminobutyric acid levels in the neonatal brain. NMR Biomed. 2017;30:e3666. doi: 10.1002/nbm.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen JW, Marenco S, Berman KF, Shen J. Retrospective correction of frequency drift in spectral editing: the GABA editing example. NMR Biomed. 2017;30:e3725. doi: 10.1002/nbm.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li SJ. Differentiation of metabolic concentrations between gray matter and white matter of human brain by in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 1998;39:28–33. doi: 10.1002/mrm.1910390107. [DOI] [PubMed] [Google Scholar]
- Xin L, Frenkel H, Mlynárik V, Morgenthaler FD, Gruetter R. Selective resonance suppression 1H-[13C] NMR spectroscopy with asymmetric adiabatic RF pulses. Magn Reson Med. 2009;61:260–266. doi: 10.1002/mrm.21829. [DOI] [PubMed] [Google Scholar]
- Xin L, Schaller B, Mlynarik V, Lu H, Gruetter R. Proton T 1 relaxation times of metabolites in human occipital white and gray matter at 7 T. Magn Reson Med. 2013;69:931–936. doi: 10.1002/mrm.24352. [DOI] [PubMed] [Google Scholar]
- Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: a complex problem with many partial solutions. J Magn Reson Imag. 2015;42:887–901. doi: 10.1002/jmri.24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhu H, Edden RAE, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med. 2011;65:603–609. doi: 10.1002/mrm.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Ouwerkerk R, Barker PB. Dual-band water and lipid suppression for MR spectroscopic imaging at 3 Tesla. Magn Reson Med. 2010;63:1486–1492. doi: 10.1002/mrm.22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Soher BJ, Ouwerkerk R, Sch€ar M, Barker PB. Spin-echo magnetic resonance spectroscopic imaging at 7 T with frequency-modulated refocusing pulses. Magn Reson Med. 2013;69:1217–1225. doi: 10.1002/mrm.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.