Abstract
Population-scale biobanks that combine genetic data and high-dimensional phenotyping for a large number of participants provide an exciting opportunity to perform genome-wide association studies (GWAS) to identify genetic variants associated with diverse quantitative traits and diseases. A major challenge for GWAS in population biobanks is ascertaining disease cases from heterogeneous data sources such as hospital records, digital questionnaire responses, or interviews. In this study, we use genetic parameters, including genetic correlation, to evaluate whether GWAS performed using cases in the UK Biobank ascertained from hospital records, questionnaire responses, and family history of disease implicate similar disease genetics across a range of effect sizes. We find that hospital record and questionnaire GWAS largely identify similar genetic effects for many complex phenotypes and that combining together both phenotyping methods improves power to detect genetic associations. We also show that family history GWAS using cases ascertained on family history of disease agrees with combined hospital record and questionnaire GWAS and that family history GWAS has better power to detect genetic associations for some phenotypes. Overall, this work demonstrates that digital phenotyping and unstructured phenotype data can be combined with structured data such as hospital records to identify cases for GWAS in biobanks and improve the ability of such studies to identify genetic associations.
Keywords: genetics, genetic correlation, genome-wide association study, GWAS, phenotyping, UK Biobank, population cohorts, digital phenotyping, mixture models
Introduction
Genome-wide association studies (GWAS) for binary phenotypes such as presence of a disease typically obtain cases via methods like recruitment through medical systems or archived medical samples, and they then compare these cases to controls known not to have the disease or to random population controls in which the disease is present at its population prevalence.1 However, recent studies have begun to rely on self-reported phenotypes collected via questionnaires and web or mobile phone applications.2, 3, 4, 5, 6, 7, 8, 9, 10 Such “digital phenotyping” may be faster and cheaper than standard cohort study approaches, but the extent to which this approach agrees with more traditional phenotyping approaches for GWAS is largely unknown because previous attempts to estimate the agreement between the two phenotyping approaches have focused on a small number of top associations and have not systematically assessed agreement across the hundreds or thousands of variants likely associated with complex, polygenic traits. For instance, a genome-wide study of self-reported thrombosis events found strong agreement between the top associations displayed in Manhattan plots from their self-reported thrombosis GWAS compared to previous cohort-based studies.2 Other studies have reported overlaps with genome-wide significant loci from cohort studies but have not investigated the extent to which genetic effects that did not reach genome-wide significance agree.11
In addition to self-reported phenotypes, GWAS have also been performed using family history of disease as a proxy for disease diagnosis.12,13 This genome-wide association study by proxy (GWAX) approach can be useful for childhood or late-onset diseases for which participants are difficult to recruit, and it is particularly appealing for population biobanking efforts that include questionnaires that ask about family history of disease. However, the degree to which proxy phenotyping attenuates effect sizes relative to traditional GWAS and the statistical power benefits of using GWAX in biobanks has not been explored. Estimating the agreement between digital phenotyping, GWAX, and traditional GWAS is important for understanding the extent to which these phenotyping strategies may help uncover the genetic basis of human diseases and empower the generation of therapeutic hypotheses by, for instance, identifying strong acting protein-truncating variants.14, 15, 16, 17, 18, 19
To explore the extent to which digital phenotyping or GWAX and traditional phenotyping approaches capture similar disease genetics, we developed a model, called the multivariate polygenic mixture model (MVPMM), that estimates genetic parameters such as genetic correlation, polygenicity, and scale of genetic effects and applied the model to GWAS summary statistics from phenotypes in the UK Biobank whose cases were defined using hospital records, questionnaire responses, or family history information. We applied MVPMM to GWAS summary statistics from 41 binary medical phenotypes and found that there is strong agreement between the two phenotyping methods for most complex phenotypes. We then explored the extent to which combining these two phenotyping methods improves statistical power for GWAS. We next used MVPMM to compare how well GWAX agrees with these combined case definitions for a subset of phenotypes, and we found that family history GWAS has better power to detect associations in the UK Biobank for chronic bronchitis and/or emphysema, diabetes, and Alzheimer’s disease. The results from our study demonstrate that digital phenotyping and GWAX are useful approaches for identifying cases in large biobanks and can provide increased power for identifying associations for many conditions.
Material and Methods
Quality Control of Genotype Data
We used genotype data from UK Biobank dataset release version 2 for all aspects of the study.20 To minimize the impact of cofounders and unreliable observations, we used a subset of 337,199 unrelated white British individuals that satisfied all of the following criteria: (1) self-reported white British ancestry, (2) used to compute principal components, (3) not marked as outliers for heterozygosity and missing rates, (4) do not show putative sex chromosome aneuploidy, and (5) have at most 10 putative third-degree relatives. We used PLINK v1.90b4.421 to compute the following statistics for each of 784,257 variants: (A) genotyping missingness rate, (B) p values of Hardy-Weinberg test, and (C) allele frequencies. As described previously,14 we removed variants that had (1) missingness rate greater than 1%, (2) Hardy-Weinberg disequilibrium test p value less than 1 × 10−7, (3) ambiguous cluster plots, or (4) minor allele frequencies inconsistent with gnomAD.
Hospital Record and Verbal Questionnaire Phenotype Definitions
We used the following procedure to define cases and controls for non-cancer phenotypes. For a given phenotype, ICD-10 codes (Data-Field 41202) were grouped with self-reported non-cancer illness codes from verbal questionnaires (Data-Field 20002) that were closely related. This was done by first creating a computationally generated candidate list of closely related ICD-10 codes and self-reported non-cancer illness codes, then manually curating the matches. The computational mapping was performed by using the FuzzyWuzzy python package to calculate the token set ratio between the ICD-10 code description and the self-reported illness code description. The high-scoring ICD-10 matches for each self-reported illness were then manually curated to ensure high-confidence mappings. Manual curation was required in order to validate the matches because fuzzy string matching may return words that are similar in spelling but not in meaning. For example, to create a hypertension cohort, the code description from Data-Field 20002 (“Hypertension”) was mapped to all ICD-10 code descriptions, and all closely related codes were returned (“I10: Essential (primary) hypertension” and “I95: Hypotension”). After manual curation, code I10 would be kept and code I95 would be discarded. After matching ICD-10 codes and with self-reported illness codes, cases were identified for each phenotype by using only the associated ICD-10 codes, only the associated self-reported illness codes, or both the associated ICD-10 codes and self-reported illness codes.
Questionnaire images were downloaded from the UK Biobank website (see Web Resources).
Family History Phenotype Definitions
We used data from Category 100034 (Family history—Touchscreen—UK Biobank Assessment Centre) to define “cases” and “controls” for family history phenotypes. This category contains data from the touchscreen questionnaire on questions related to family size, sibling order, family medical history (of parents and siblings), and age of parents (age of death if deceased). We focused on Data Coding 20107: Illness of father and 20110: Illness of mother.
Cancer Phenotype Definitions
We combined cancer diagnoses from the UK Cancer Registry with self-reported diagnoses from the UK Biobank questionnaire to define cases and controls for cancer GWAS. Individual-level ICD-10 codes from the UK Cancer Registry (Data-Field 40006) and the National Health Service (NHS; Data-Field 41202) in the UK Biobank were mapped to the self-reported cancer codes (Data-Field 20001). The mapping was performed via manual curation of ICD-10 codes for each of the self-reported cancer codes. UK Biobank field codes for self-reported cancer were created with a tree structure such that more specific cancer subtypes (e.g., “malignant melanoma”) are nested under more general categories (“skin cancer”). This tree structure was preserved in the field-code-to-ICD-10 mapping. For example, the self-reported phenotype of “lip cancer” was mapped to its field code, 1010, and the ICD-10 codes for “malignant neoplasm of lip,” C00 and C000-C009. After this mapping, individuals with an affirmative entry in one or more of the phenotype collections (self-reported cancer, cancer registry, and the NHS) were included in the case cohort for the GWAS. No secondary neoplasms were included in the cancer phenotype mappings.
Genome-Wide Association Analyses
We performed genome-wide association analyses for binary medical phenotypes in the UK Biobank across 784,257 variants genotyped by array using logistic regression with Firth-fallback as implemented in PLINK v2.00a (17 July 2017). Firth-fallback is a hybrid algorithm which normally uses the logistic regression code previously described,22 but switches to a port of logistf() in two cases: (1) if one of the cells in the 2 × 2 allele count by case/control status contingency table is empty, or (2) if logistic regression was attempted since all the contingency table cells were nonzero, but it failed to converge within the usual number of steps. We used the following covariates in our analysis: age, sex, array type, and the first four principal components, where array type is a binary variable that represents whether an individual was genotyped with UK Biobank Axiom Array or UK BiLEVE Axiom Array. For variants that were specific to one array, we did not use array as a covariate. Published summary statistics for migraine, type 2 diabetes, and rheumatoid arthritis were obtained from the GWAS Catalog and the International Headache Genetics Consortium (see Web Resources).23, 24, 25, 26
Multivariate Polygenic Mixture Model
Model Definition
We developed a two-component mixture model in order to estimate genetic parameters including correlation, scale, and proportion of non-zero genetic effects. Let be an matrix of estimated GWAS effect sizes (regression coefficient for quantitative phenotypes, log odds ratio for binary phenotypes) and let be an matrix of estimated standard errors for linkage disequilibrium (LD)-independent loci for two phenotypes. Let be a column vector with the effect sizes for the th locus and let be a column vector with standard errors for the th locus. Under the MVPMM, the estimated effect sizes are assumed to be generated from one of two mixture components. The first component is a point-mass at zero such that where . is a correlation matrix describing the correlation between the estimated effect sizes (measurement errors) at the null variants (variants that are not associated with the two phenotypes) and is a diagonal matrix with on the diagonal. The correlation matrix captures correlation in the GWAS summary statistics due to sample overlap.27,28 The second component is a multivariate normal distribution with mean zero and unknown covariance matrix such that where . is a correlation matrix describing the correlation between the genetic effects of the two phenotypes and is a length-2 vector describing the scale of the genetic effects. The correlation matrix captures correlation in the non-null GWAS summary statistics that is attributable to correlated genetic effects. The model includes a mixing parameter that describes the fraction of variants in the second component associated with the two phenotypes. The LKJ prior with was used for the correlation matrices in and . The other priors are , . We used MVPMM to estimate genetic correlations with different priors for 12 phenotypes where cases were defined using either family history of disease or diagnosis from hospital records and verbal questionnaire responses, and we found that the parameter estimates were robust to choice of priors (see Table S1). This is consistent with the large number of LD-independent variants used in the model quickly overwhelming the priors. The parameter estimates for the different priors reported in Table S1 are point estimates obtained by maximizing the joint posterior using Stan’s “optimizing” function.
Estimating Genetic Parameters Using MVPMM
We implemented MVPMM using the Stan probabilistic programming language and used MVPMM to estimate genetic parameters for a given pair of GWAS summary statistics as follows. First, we obtained GWAS summary statistics (effect size and standard error) for 361,436 LD-independent autosomal variants; these 361,436 LD-independent variants were identified using plink (−indep 50 5 2). In order to remove variants with uncertain effect size estimates, for a given pair of phenotypes, the 361,436 LD-independent variants were filtered to include only those whose standard error was less than 0.2 in both phenotypes. We then performed Markov chain Monte Carlo (MCMC) sampling using Stan with four chains for 500 iterations with 100 burn-in iterations. We calculated the statistic for the genetic correlation parameter to evaluate whether the MCMC sampling converged. For four phenotypes (Parkinson’s disease, pancreatitis, hypertension, and angina) with , we repeated MCMC sampling with four chains for 1,000 iterations with 200 burn-in iterations. We excluded bronchiectasis for the combined phenotyping versus hospital record phenotyping because MCMC sampling was extremely slow (500 iterations did not finish in 7 days).
We ran MVPMM for GWAS using cases defined based on either hospital records or questionnaire responses for 51 medical phenotypes. We then filtered out five phenotypes (peritonitis, fractured upper arm/humerus/elbow, bone disorder, stomach disorder, and fibromyalgia) which had unrealistic polygenicity estimates that indicated model failure and five phenotypes (back pain, appendicitis, endometriosis, benign breast lump, and whooping cough/pertussis) which had , indicating that the MCMC sampling did not converge.
Parameter estimates are plotted as dots that indicate the mean of the posterior distribution and bars that show the 95% highest posterior density (HPD). The 95% HPD is the smallest interval that includes 95% of the density of the posterior distribution.
Effect Size Attenuation Estimates
We calculated the attenuation for each GWAX as where is the estimated genetic correlation between the GWAS and GWAX, is the estimated scale parameter for the GWAS, and is the estimated scale parameter for the GWAX. The attenuation was calculated for each MCMC sample to obtain a posterior distribution of the attenuation for each GWAS/GWAX pair.
For attenuation scatterplots, GWAS summary statistics and p values were obtained for each GWAX/GWAS pair for the variants used as input to MVPMM. The following procedure was used to identify variants with reasonable effect size estimates to plot to demonstrate attenuation. Variants were filtered to include only those with in both GWAS and GWAX. If there were less than 500 variants with in both GWAS and GWAX, the p value filter threshold was increased by a factor of two until there were at least 500 variants or the threshold exceeded 0.01. This resulted in a set of variants with effect sizes that could be compared between GWAX and GWAS.
Power Calculations
Power calculations were performed using a forked version of the GeneticsDesign Bioconductor package (see Web Resources). Disease prevalence was estimated as the total number of cases identified by combining hospital records and verbal questionnaire responses and dividing by the total number of subjects. We calculated power curves for medical phenotypes with genetic correlation greater than 0.8 and GWAX phenotypes with genetic correlation greater than 0.9.
Results
Phenotyping, GWAS, and Genetic Parameter Estimation
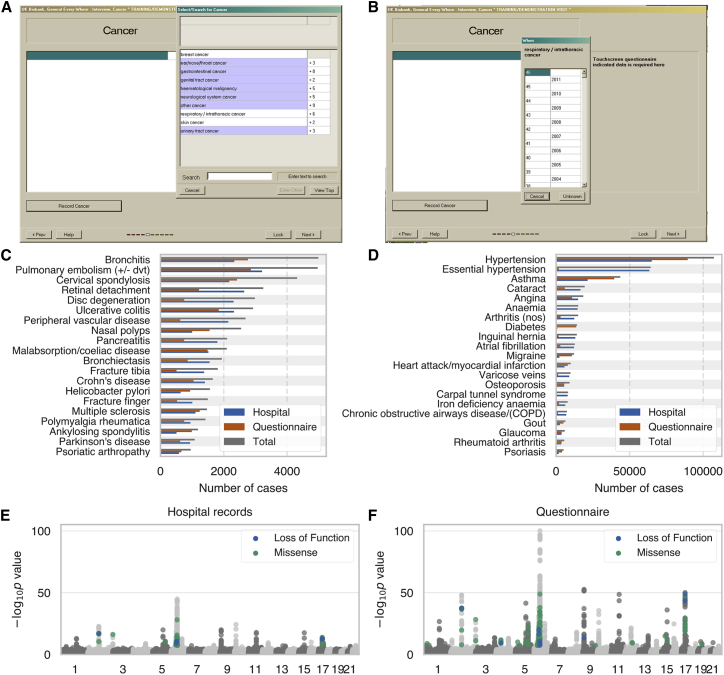
In order to perform GWAS and estimate genetic parameters, we stratified 337,199 European-ancestry UK Biobank subjects into cases and controls for 41 binary medical phenotypes through the use of hospital records or verbal questionnaire responses available from the UK Biobank (Table S2).29 The hospital records consist of hospital in-patient records (National Health Service Hospital Episode Statistics), cancer diagnoses from national cancer registries, and causes of death from national death registries. The verbal questionnaire data consisted of a computer survey that asked participants whether they had a history of several different illnesses followed by a verbal interview with a nurse to gain further confirmation of the selected diagnosis (Figure 1A–1B). The number and total fraction of cases ascertained from hospital records or questionnaire responses differed among phenotypes, although each phenotype had at least 500 cases ascertained from each method (Figure 1C–1D, Table S2). More than 80% of cases were identified using only one of the phenotyping methods for 20 of the phenotypes, and 10 phenotypes had substantial overlap (>33%) in cases identified by both hospital records and verbal questionnaire data. Overall, however, 32/41 and 20/41 phenotypes had at least 25% of cases derived solely from hospital records or questionnaire data, respectively, indicating that both phenotyping methods add a substantial proportion of cases for most diseases (Figure 1C–1D).
Figure 1.
UK Biobank Phenotype Counts and Asthma Genetic Associations
(A and B) Screenshot of UK Biobank questionnaire where participants can (A) indicate that they have been diagnosed with specific cancers or other illnesses and (B) specify at what age they were diagnosed.
(C and D) Number of cases for each of 41 medical phenotypes where cases are defined using hospital records (blue), questionnaire responses (orange), or both combined (gray).
(E and F) Manhattan plots for asthma genome-wide association studies (GWAS) with cases defined by (E) hospital records or (F) questionnaire responses. Loss-of-function and missense variants with p < 5 × 10-8 are colored blue and green, respectively. Grey dots indicate all other variants.
For each phenotype, we used cases defined based on either hospital records or verbal questionnaire responses to perform GWAS for 784,257 variants genotyped by array (see Material and Methods). For example, for asthma, we defined 21,445 cases through the use of hospital records and 39,483 cases through the use of questionnaire responses; among those cases, 17,302 were shared between both phenotyping methods. Performing GWAS for each phenotyping method yielded similar Manhattan plots, though there was less power to detect associations for hospital records, as we expected due to the lower number of cases (Figure 1E–1F). For instance, the p value for the reported association between the protein truncating variant rs146597587 in IL33 and asthma is 7.1 × 10−7 for hospital records and 2.4 × 10−14 for questionnaire responses.30 While these results illustrate the usefulness of the verbal questionnaire data, it is difficult to draw conclusions about the overall agreement between the two phenotyping methods outside of the small number of top GWAS findings.
To estimate the agreement between phenotyping using hospital records and phenotyping using verbal questionnaire responses for identifying genetic associations, we developed a Bayesian mixture model, MVPMM, and applied it to GWAS summary statistics (effect size estimate and standard error of effect size estimate) for the 41 medical phenotypes where cases were defined for each phenotype by using either hospital in-patient records or self-reported verbal questionnaire responses (Table S2). MVPMM estimates genetic parameters including genetic correlation, polygenicity, and scale of effect sizes by modeling GWAS summary statistics as drawn from either a null component where the true effect of the variant on the phenotype is zero or a non-null component where the true effect of the variant on the phenotype is non-zero. For both components, summary statistics (treated as the data) for each variant are modeled as being drawn from a multivariate normal distribution with zero mean and unknown covariance matrix. For the null component, the covariance matrix uses the standard error of the effect size estimate and estimates the correlation of errors that may be due to shared subjects. The covariance matrix for the non-null component combines the error covariance matrix from the null component with another covariance matrix that captures the genetic correlation between the phenotypes being considered. This model allows us to estimate (1) the genetic correlation between two phenotypes, (2) the fraction of loci that belong to the non-null component for both phenotypes (polygenicity), and (3) the scale of the genetic effects for each phenotype (see Material and Methods).
GWAS Based on Hospital Records or Questionnaire Responses
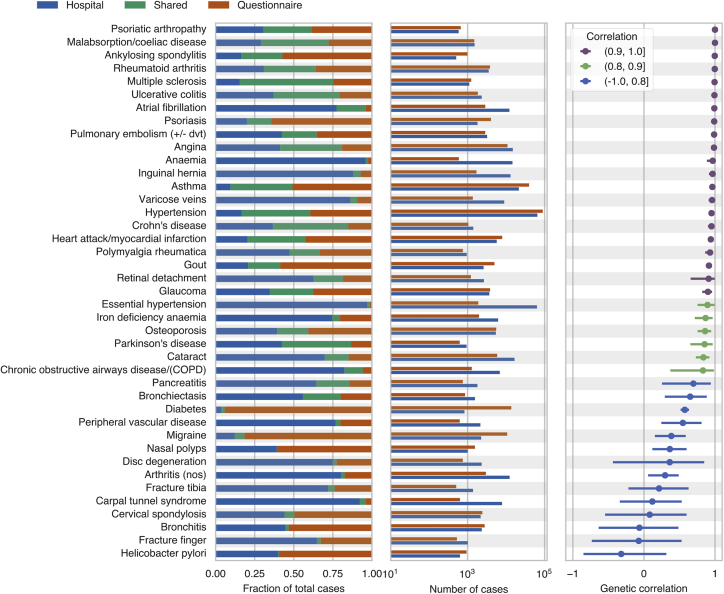
To systematically examine whether the GWAS results for phenotyping using hospital records or questionnaire responses agreed across a broader range of associations, we applied MVPMM to the GWAS summary statistics for the 41 phenotypes in order to estimate the genetic correlation between the results of both phenotyping methods (Figure 2, Table S3, Materials and Methods). The genetic correlation estimates from MVPMM were robust according to the statistic (Figure S1) and agreed in large part with those from LD score regression (Figure S2).31 We found that 21/41 phenotypes had genetic correlations greater than 0.9; this result indicates strong agreement of genetic effects between cases identified using hospital records and those identified using verbal questionnaire data. Another 6/41 phenotypes had genetic correlations greater than 0.8; this indicates moderate agreement between the two phenotyping methods. For instance, the genetic correlation for asthma as defined by the two phenotyping methods was 0.96 (95% HPD 0.95–0.98, see Material and Methods). We identified 43,626 total asthma cases between both phenotyping methods, 40% of which were identified by both methods and 51% of which were identified only based on the verbal questionnaire responses. These results indicate that the large number of asthma cases contributed by the verbal questionnaire responses capture similar disease genetics to those of the cases indicated by hospital records. We observed similar results, where a large number of cases were identified from questionnaire responses, for several other diseases such as ankylosing spondylitis, psoriasis, myocardial infarction, gout, and others (Figure 2); this demonstrates that the two phenotyping methods agree for a range of phenotypes including both chronic and acute conditions.
Figure 2.
Estimated Genetic Correlations for Hospital Record and Verbal Questionnaire GWAS
The first panel from left indicates the fraction of cases that were ascertained from hospital records only (blue), questionnaire responses only (orange), or both phenotyping methods (green). The second panel shows the number of cases ascertained from hospital records (blue) and questionnaire responses (orange). The third panel shows the estimated genetic correlation from the multivariate polygenic mixture model (MVPMM); the dots represent the means of the posterior distributions, and the error bars are the 95% highest posterior density (see Material and Methods).
There were 14 phenotypes that had genetic correlations less than 0.8, a result that indicates less agreement between cases defined based on hospital records and those defined based on questionnaire data, though notably several of these pairs were predicted to have positive, non-zero correlations (Figure 2). For instance, the genetic correlations for migraine (0.38, 95% HPD: 0.15–0.59), diabetes (0.58, 95% HPD: 0.52–0.63), peripheral vascular disease (0.55, 95% HPD: 0.25–0.81), and carpal tunnel syndrome (0.19, 95% HPD: −0.34–0.53) were all less than 0.8; this indicates that there may be differences in the case populations captured by the phenotyping methods for these diseases. The Manhattan plots for these phenotypes are also different for the two phenotyping methods, a result which demonstrates that even the top associations are not necessarily consistent between the two methods for these phenotypes (Figure S3).
To further explore these genetic correlations of less than 0.8, we compared the UK Biobank hospital record and questionnaire GWAS summary statistics to GWAS summary statistics from published studies that did not use the UK Biobank for migraine and diabetes as well as rheumatoid arthritis, which had a high genetic correlation (0.996, 95% HPD: 0.993–0.999).23, 24, 25, 26 We subsetted the published GWAS associations to LD-independent variants that were used to estimate genetic correlations with MVPMM and that had p < 1 × 10−5 in the published studies, and we compared the estimated effect sizes from the published studies to the effect sizes from GWAS performed using hospital records or questionnaire data from the UK Biobank. We found that there was good agreement in the direction of estimated effect sizes between hospital record and questionnaire GWAS and the published effect sizes for rheumatoid arthritis, and these results were consistent with the high estimated genetic correlation between hospital record and questionnaire GWAS (Figure S4). The effect sizes for verbal questionnaire GWAS for migraine and diabetes also agreed with the published GWAS, but the effect sizes for hospital record GWAS differed in direction for more loci. Additionally, the hospital record and questionnaire effect sizes were consistent for rheumatoid arthritis but were less consistent for migraine and diabetes (Figure S4). These results indicate that for migraine and diabetes, the power to detect associations in the hospital record GWAS may be affected by the small number of hospital record cases and relatively large number of cases identified from questionnaire data that are included as controls for the hospital record GWAS (Figure 2), and these results also demonstrate that it is particularly important to consider cases derived from questionnaire data for some phenotypes that have a small number of hospital record cases.
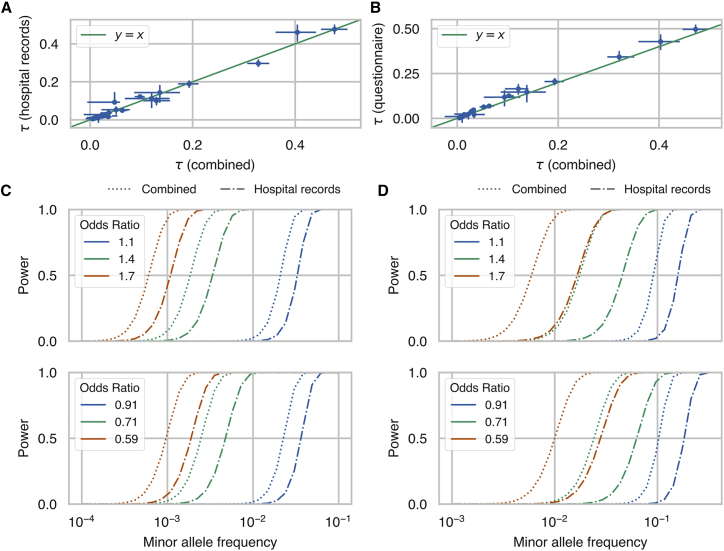
Given the high genetic correlation between the two phenotyping methods for many of the phenotypes tested here, we combined together cases from both phenotyping methods, performed GWAS analysis using the combined cases, and used MVPMM to estimate genetic parameters between GWAS summary statistics from combined cases and those from questionnaire cases or hospital record cases (Table S3). We found a high correlation between the combined GWAS and GWAS performed using either questionnaire cases or hospital record cases. 29 phenotypes had genetic correlations greater than 0.8 for the hospital record GWAS, and 33 phenotypes had correlations greater than 0.8 for the verbal questionnaire GWAS. We compared the estimates from MVPMM for the scale of effects, which captures how strong the genetic effects are for each phenotype definition, and we found that the scale of effects generally agreed between the combined GWAS and the questionnaire or hospital record GWAS (Figure 3A–3B); this finding indicates that there is not a large amount of effect size attenuation due to combining the phenotyping methods. In order to investigate the practical impact of including cases ascertained from questionnaire responses, we calculated the power to detect associations using the combined cases compared to using either the questionnaire or hospital record cases, and we found an increase in power for detecting associations for both risk and protective rare variants (Figure 3C–3D, Figures S5 and S6). The increase in power differs across phenotypes depending on the fraction of total cases that are added by including cases ascertained from questionnaire data. Notably, identifying additional cases caused a larger increase in the power to detect rare protective variants which are especially useful for identifying therapeutic targets.14,17,18,32
Figure 3.
Scale of Genetic Effects and Power Estimates
(A and B) Estimates of scale of genetic effects from the multivariate polygenic mixture model (MVPMM) for genome-wide association studies (GWAS) summary statistics generated using cases ascertained from hospital records and verbal questionnaire (combined) versus summary statistics generated using only cases ascertained from (A) hospital records or (B) verbal questionnaires; the dots represent the means of the posterior distributions and the error bars are the 95% highest posterior density (HPD). Phenotypes whose 95% HPD size for was less than 0.1 for both hospital record and verbal questionnaire comparisons are plotted.
(C and D) Statistical power to detect associations between rare genetic variants at different minor allele frequencies for (C) asthma and (D) psoriasis in the UK Biobank. Dot-dash lines show power for GWAS performed using only cases ascertained from hospital records, and dotted lines show power for GWAS performed using cases ascertained from both hospital records and verbal questionnaire data. Top panel shows power for rare risk variants and bottom panel shows power for rare protective variants. Different colors indicate power for different association effect sizes. The only parameters that differ between the dot-dash lines and dotted lines of a given color are the numbers of cases and controls; data represented by the dot-dash lines include cases that were identified from hospital record data, and data represented by the dotted lines include cases identified from either hospital record or verbal questionnaire data.
GWAS Using Disease Diagnosis or Family History of Disease
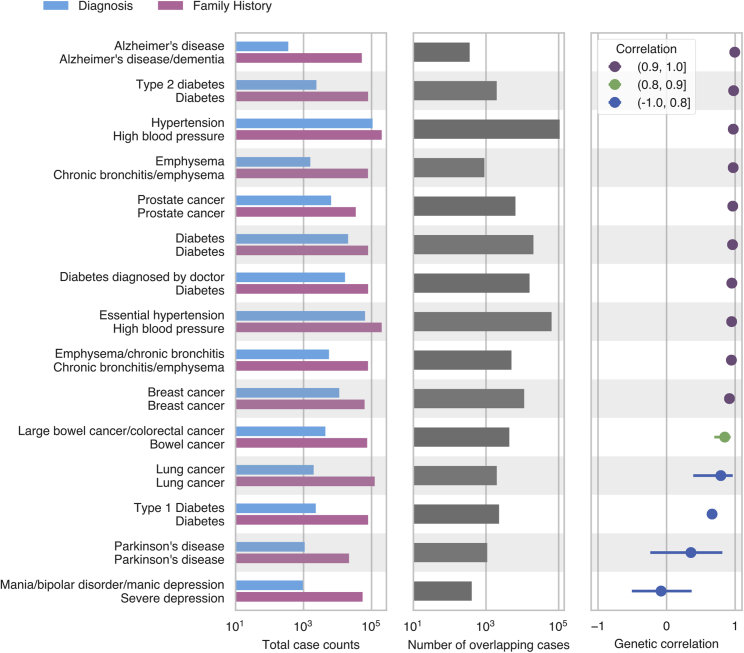
Another approach for identifying loci associated with disease is a GWAX, in which cases are defined as biobank participants that have a relative with a particular disease.12,13 We estimated genetic parameters for 15 diseases by using summary statistics from a traditional GWAS in which cases were identified from hospital records and/or from questionnaire responses and summary statistics for the same disease from a GWAX based on the presence of disease in the parents of the subject (ascertained from questionnaire data). We included multiple disease definitions for diabetes and emphysema that rely on different aspects of the UK Biobank phenotyping data. We restricted our analysis to diseases with at least 1,000 GWAS cases (except for Alzheimer’s disease), though notably, the number of cases is generally much larger for GWAX than for GWAS. We found that the genetic correlation was greater than 0.9 for 10/15 comparisons and that four comparisons had genetic correlations less than 0.8 (Figure 4, Table S3). One of the comparisons with genetic correlation less than 0.8 was family history of “severe depression” and “mania/bipolar disorder/manic depression.” In this case, these two case definitions were matched due to the word “depression,” but they actually capture two different diseases, depression and bipolar disorder, and the low genetic correlation reflects this. Another comparison with genetic correlation less than 0.8 is type 1 diabetes and family history of diabetes. However, family history of diabetes has a high correlation with other diabetes definitions that likely include type 2 diabetes cases, indicating that family history of diabetes mostly captures cases for type 2 diabetes; this is consistent with the higher prevalence of type 2 diabetes in the UK Biobank.33
Figure 4.
Estimated Genetic Correlations for Family History and Combined Hospital Record and Verbal Questionnaire GWAS
The first panel from left indicates the number of cases ascertained from combined hospital records and questionnaire responses (blue) or family history of disease (pink). The second panel shows the number of cases overlapping between both phenotyping methods. The third panel shows the estimated genetic correlation between genome-wide association studies (GWAS) and genome-wide association study by proxy (GWAX) from multivariate polygenic mixture model (MVPMM); the dots represent the means of the posterior distributions and the error bars are the 95% highest posterior density.
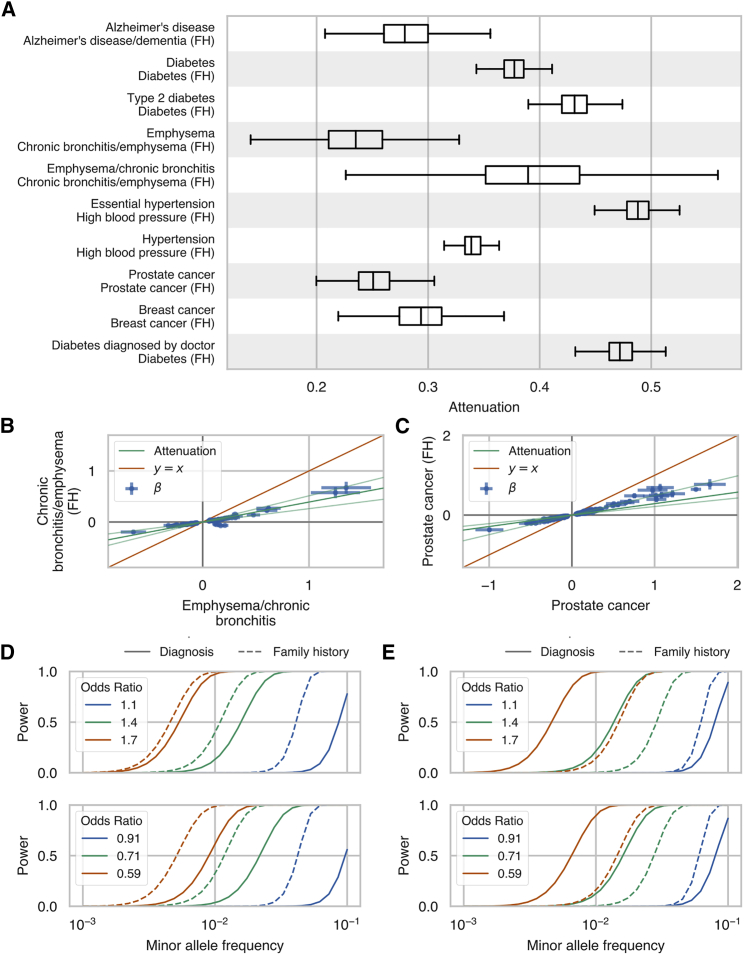
Because our GWAX uses subjects whose parents had a particular disease, we expect that the effect sizes of the associated variants identified by GWAX will be attenuated relative to the effect sizes estimated from GWAS.13 We used the scale of effects estimates for each phenotype to estimate the attenuation for GWAX compared to combined hospital record and/or verbal questionnaire GWAS for the 10 phenotypes with genetic correlations greater than 0.9. We found that the estimated attenuation factors ranged from 0.24–0.54 and that estimated effect sizes were generally scaled consistently with the estimated attenuation factor (Figure 5A–5C, Figure S7). Although the smaller effect sizes of GWAX may decrease the power to detect genetic associations compared to GWAS in the UK Biobank, we find that, in practice, this decrease in power is offset by much larger case sizes in GWAX for some phenotypes (Figure 5D and 5E, Figures S8 and S9). For instance, the power to detect associations for chronic bronchitis and/or emphysema, diabetes, and Alzheimer’s disease is higher when using GWAX, whereas the power to detect associations is higher with combined hospital record and verbal questionnaire GWAS for other phenotypes, such as prostate cancer.
Figure 5.
GWAS Effect Size Attenuation, Scale Effects, and Power Estimates Family History GWAS
(A) Boxplots of the posterior distribution of effect size attenuation (see Material and Methods) from genome-wide association study by proxy (GWAX) using cases ascertained based on family history of disease versus genome-wide association studies (GWAS) using cases ascertained using combined hospital records and verbal questionnaire responses.
(B and C) Effect sizes and standard errors (error bars) for family history GWAX (y axis) versus combined hospital records and verbal questionnaire responses GWAS (x axis) for (B) chronic bronchitis and/or emphysema and (C) prostate cancer. The dark green line indicates the mean of the posterior distribution of attenuation from the multivariate polygenic mixture model (MVPMM) and the light green lines indicate lower and upper bounds of 95% highest posterior density of attenuation (see Material and Methods).
(D and E) Statistical power to detect association between rare genetic variants at different minor allele frequencies for (D) chronic bronchitis and/or emphysema and (E) prostate cancer in the UK Biobank. Solid lines show power for GWAS performed using cases ascertained from hospital records and questionnaire responses, and dashed lines show power for GWAS performed using cases ascertained from family history of disease. Top panel shows power for rare risk variants and bottom panel shows power for rare protective variants. Different colors indicate power for different association effect sizes. The only parameters that differ between the solid lines and dashed lines of a given color are the numbers of cases and controls.
Discussion
In this study, we present a method, called the MPVMM, for estimating genetic parameters from GWAS summary statistics, and we use the method to evaluate the extent to which GWAS using cases ascertained from hospital records, verbal questionnaire responses, and family history of disease agree across 41 diverse medical phenotypes. We found that GWAS using cases ascertained from hospital records or questionnaire responses had genetic correlation greater than 0.8 for 27 of the 41 phenotypes; this indicates that the two phenotyping methods identify similar disease genetics for many complex diseases. Combining both phenotyping methods for GWAS does not greatly alter effect size estimates relative to using either method individually, but due to the increased number of cases, it does increase power to identify genetic associations. We also showed that GWAX, where family history of disease is used to identify cases, has genetic correlation greater than 0.8 with combined hospital record and questionnaire GWAS for 11 of 16 pairs of traits analyzed; this demonstrates that the GWAX approaches based on digital phenotyping can also be used to identify variant-disease associations. Finally, we showed that the power to detect genetic associations in the UK Biobank is greater for GWAX than GWAS for chronic bronchitis and/or emphysema, diabetes, and Alzheimer’s disease.
Although we observed genetic correlation greater than 0.8 for 27 of 41 phenotypes for GWAS using cases ascertained from hospital records or verbal questionnaire, we did find low genetic correlation in some cases. Some of the phenotypes with low genetic correlation likely are mainly driven by environmental factors or have low heritability; these phenotypes include finger and tibia fractures, Helicobacter pylori infection, and carpal tunnel syndrome.34 We did identify some common diseases, such as peripheral vascular disease, migraine, and diabetes, that had low, though positive, genetic correlations. These lower correlations could be caused by differences in the case populations captured by the two phenotyping methods. For instance, peripheral vascular disease patients identified from hospital records may have more severe disease that has required a hospital visit compared to cases that only reported the disease in their questionnaire. Differences in disease subtypes between cases identified by the two phenotyping methods, as well as incorrect ICD-10 code assignment or self-reporting of diagnoses, may also drive differences between GWAS using the two phenotyping methods.35 In the cases of diabetes and migraines, the relatively small number of cases identified from hospital records compared to the number of cases identified from questionnaire responses may underlie the low genetic correlation observed between the two GWAS approaches. Future studies that perform higher-resolution or longitudinal phenotyping, which can more accurately identify cases and controls, can be used to further investigate the causes underlying heritable diseases that have low genetic correlation in this study.
Because we independently ascertained cases based on hospital records or verbal questionnaire responses for this study, there are varying levels of overlap between the cases used for GWAS for the two phenotyping methods (Figure 2). For instance, nearly 60% of multiple sclerosis cases were identified using both phenotyping methods while only 1% of essential hypertension cases were identified by both. Because overlapping samples may bias genetic correlation estimates derived from GWAS summary statistics, the MVPMM model includes a term in the covariance matrix for the non-null component of the mixture model; this captures the contribution of shared samples to the GWAS effect sizes. In practice, we observed that this parameter was highly correlated with sample overlap as expected, although we did find that an increase of 10% in the percentage of shared cases corresponded to ∼1% increase in genetic correlation for phenotypes with genetic correlation greater than 0.8 (= 0.116, 95% confidence interval: −0.02–0.251, p = 0.091) in a model that also accounted for the total number of cases. It is possible that this may represent bias in the genetic correlation estimates from MVPMM due to sample overlap, though the effect on genetic correlation estimates is likely negligible for the purpose of broadly assessing genetic correlation.
This work demonstrates that power to detect genetic associations in population biobanks is improved by using diverse phenotyping approaches to improve the classification of subjects into cases and controls. The power to detect associations in biobanks can be affected in different ways when phenotyping is inaccurate. For instance, phenotyping quality may be associated with disease severity: it may be more likely for subjects with more severe disease to be identified as cases. In this scenario, GWAS may identify variants that are associated with severe forms of disease. Because diseases are observed at population prevalence in biobanks, and most biobank subjects do not have a given disease, the power to detect associations is most improved by identifying cases that are incorrectly labeled as controls. This was observed for diabetes and migraine, for which the number of hospital record cases was low compared to the number of questionnaire cases, impacting the power of the hospital record GWAS. It is also possible that phenotyping quality may vary across different phenotypes. Some phenotypes may be difficult to ascertain or confused with other phenotypes, such as migraines and headaches. While it is clear that incorrect classification of cases as control is problematic for GWAS, these results indicate that the extent of incorrect classification can vary across phenotypes in a biobank study.
The high genetic correlation between GWAS based on questionnaire data and GWAS based on hospital records shows that the two methods capture similar disease genetics for many diseases, though some diseases have low genetic correlation for GWAS using the two phenotyping approaches. In the UK Biobank, participants completed a touchscreen questionnaire and had a follow-up interview with a nurse to discuss any diagnoses for major illnesses and procedures. Future studies will explore to what extent other digital or questionnaire phenotyping approaches such as phone or internet applications, waiting room surveys, or features extracted by natural language processing also identify similar disease genetics to those identified by GWAS that ascertain cases using more traditional recruitment methods. Such comparisons may be aided by the adoption of standardized questionnaire approaches across datasets or biobanks so that digital phenotyping methods can easily be shared in the way that phenotyping based on structured medical data is now shared.36 The results from this study illustrate how such efforts will benefit GWAS in population-scale biobanks by improving power to detect novel genetic associations.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This research has been conducted using the UK Biobank resource. We thank all the participants in the UK Biobank study. M.A.R. is supported by Stanford University and a National Institutes of Health (NIH) Center for Multi- and Trans-ethnic Mapping of Mendelian and Complex Diseases grant (5U01 HG009080). C.D. is supported by a postdoctoral fellowship from the Stanford Center for Computational, Evolutionary, and Human Genomics and a seed grant from the Stanford ChEM-H Institute. Y.T. is supported by a Funai Overseas Scholarship from Funai Foundation for Information Technology and the Stanford University Biomedical Informatics Training Program. The primary and processed data used to generate the analyses presented here are available in the UK Biobank access management system (see Web Resources) for application 24983, “Generating effective therapeutic hypotheses from genomic and hospital linkage data” (see Web Resources), and the results are displayed in the Global Biobank Engine (see Web Resources). Research reported in this publication was supported by the National Human Genome Research Institute of the NIH under Award Number R01HG010140. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: April 9, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.03.007.
Web Resources
GeneticsDesign Bioconductor package, https://bioconductor.org/packages/devel/bioc/html/GeneticsDesign.html
Global Biobank Engine, https://biobankengine.stanford.edu
GWAS Catalog, www.ebi.ac.uk/gwas/
International Headache Genetics Consortium, www.headachegenetics.org/
logistf(), https://cran.r-project.org/web/packages/logistf/index.html
Modified GeneticsDesign package, https://github.com/cdeboever3/GeneticsDesign
UK Biobank access management system, https://amsportal.ukbiobank.ac.uk/
UK Biobank application 24983, http://www.ukbiobank.ac.uk/wp-content/uploads/2017/06/24983-Dr-Manuel-Rivas.pdf
UK Biobank source for Figure 1A, http://biobank.ctsu.ox.ac.uk/crystal/crystal/images/vs_review_2.png
UK Biobank source for Figure 1B, http://biobank.ctsu.ox.ac.uk/crystal/crystal/images/vs_when_1.png
Supplemental Data
References
- 1.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinds D.A., Buil A., Ziemek D., Martinez-Perez A., Malik R., Folkersen L., Germain M., Mälarstig A., Brown A., Soria J.M., METASTROKE Consortium, INVENT Consortium Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum. Mol. Genet. 2016;25:1867–1874. doi: 10.1093/hmg/ddw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang D., Nalls M.A., Hallgrímsdóttir I.B., Hunkapiller J., van der Brug M., Cai F., Kerchner G.A., Ayalon G., Bingol B., Sheng M., International Parkinson’s Disease Genomics Consortium. 23andMe Research Team A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McConnell M.V., Shcherbina A., Pavlovic A., Homburger J.R., Goldfeder R.L., Waggot D., Cho M.K., Rosenberger M.E., Haskell W.L., Myers J. Feasibility of Obtaining Measures of Lifestyle From a Smartphone App: The MyHeart Counts Cardiovascular Health Study. JAMA Cardiol. 2017;2:67–76. doi: 10.1001/jamacardio.2016.4395. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y., Shmygelska A., Tran D., Eriksson N., Tung J.Y., Hinds D.A. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson N., Macpherson J.M., Tung J.Y., Hon L.S., Naughton B., Saxonov S., Avey L., Wojcicki A., Pe’er I., Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinds D.A., McMahon G., Kiefer A.K., Do C.B., Eriksson N., Evans D.M., St Pourcain B., Ring S.M., Mountain J.L., Francke U. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira M.A.R., Matheson M.C., Tang C.S., Granell R., Ang W., Hui J., Kiefer A.K., Duffy D.L., Baltic S., Danoy P., Australian Asthma Genetics Consortium Collaborators Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol. 2014;133:1564–1571. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onnela J.-P., Rauch S.L. Harnessing Smartphone-Based Digital Phenotyping to Enhance Behavioral and Mental Health. Neuropsychopharmacology. 2016;41:1691–1696. doi: 10.1038/npp.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny J.C., Bastarache L., Ritchie M.D., Carroll R.J., Zink R., Mosley J.D., Field J.R., Pulley J.M., Ramirez A.H., Bowton E. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S., Sham P., Daly M.J. Parental phenotypes in family-based association analysis. Am. J. Hum. Genet. 2005;76:249–259. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J.Z., Erlich Y., Pickrell J.K. Case-control association mapping by proxy using family history of disease. Nat. Genet. 2017;49:325–331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 14.DeBoever C., Tanigawa Y., Lindholm M.E., McInnes G., Lavertu A., Ingelsson E., Chang C., Ashley E.A., Bustamante C.D., Daly M.J., Rivas M.A. Medical relevance of protein-truncating variants across 337,205 individuals in the UK Biobank study. Nat. Commun. 2018;9:1612. doi: 10.1038/s41467-018-03910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emdin C.A., Khera A.V., Chaffin M., Klarin D., Natarajan P., Aragam K., Haas M., Bick A., Zekavat S.M., Nomura A. Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat. Commun. 2018;9:1613. doi: 10.1038/s41467-018-03911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBoever C., Aguirre M., Tanigawa Y., Spencer C.C.A., Poterba T., Bustamante C.D., Daly M.J., Pirinen M., Rivas M.A. Bayesian model comparison for rare variant association studies of multiple phenotypes. bioRxiv. 2018 doi: 10.1101/257162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivas M.A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C.K., Boucher G., Ripke S., Ellinghaus D., Burtt N., National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC) United Kingdom Inflammatory Bowel Disease Genetics Consortium. International Inflammatory Bowel Disease Genetics Consortium Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivas M.A., Graham D., Sulem P., Stevens C., Desch A.N., Goyette P., Gudbjartsson D., Jonsdottir I., Thorsteinsdottir U., Degenhardt F., UK IBD Genetics Consortium. NIDDK IBD Genetics Consortium A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat. Commun. 2016;7:12342. doi: 10.1038/ncomms12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 doi: 10.1101/166298. [DOI] [Google Scholar]
- 21.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill Andres, Loh Pu-Ru, Bharadwaj Ragu B., Pons Pascal, Shang Jingbo, Guinan Eva. Stepwise distributed open innovation contests for software development: acceleration of genome-wide association ana lysis. Gigascience. 2017;6:1–10. doi: 10.1093/gigascience/gix009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gormley P., Anttila V., Winsvold B.S., Palta P., Esko T., Pers T.H., Farh K.-H., Cuenca-Leon E., Muona M., Furlotte N.A., International Headache Genetics Consortium Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 2016;48:856–866. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turchin M.C., Stephens M. Bayesian multivariate reanalysis of large genetic studies identifies many new associations. PLoS Genet. 2019;15:e1008431. doi: 10.1371/journal.pgen.1008431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cichonska A., Rousu J., Marttinen P., Kangas A.J., Soininen P., Lehtimäki T., Raitakari O.T., Järvelin M.-R., Salomaa V., Ala-Korpela M. metaCCA: summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics. 2016;32:1981–1989. doi: 10.1093/bioinformatics/btw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D., Helgason H., Sulem P., Bjornsdottir U.S., Lim A.C., Sveinbjornsson G., Hasegawa H., Brown M., Ketchem R.R., Gavala M. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLoS Genet. 2017;13:e1006659. doi: 10.1371/journal.pgen.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan, B., Loh, P.-R., Finucane, H., Ripke, S., and Yang, J., Schizophrenia Working Group Psychiatric Genomics Consortium (2014). Patterson, N., Daly, M.J., Price, A.L., and Neale, B.M (LD Score Regression Distinguishes Confounding from Polygenicity in Genome-Wide Association Studies). [DOI] [PMC free article] [PubMed]
- 32.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 33.Thomas N.J., Jones S.E., Weedon M.N., Shields B.M., Oram R.A., Hattersley A.T. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol. 2018;6:122–129. doi: 10.1016/S2213-8587(17)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiberg A., Ng M., Schmid A.B., Smillie R.W., Baskozos G., Holmes M.V., Künnapuu K., Mägi R., Bennett D.L., Furniss D. A genome-wide association analysis identifies 16 novel susceptibility loci for carpal tunnel syndrome. Nat. Commun. 2019;10:1030. doi: 10.1038/s41467-019-08993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsky J., Drucker E.A., Ramelson H.Z. AMIA Annu Symp Proc. 2018. Accuracy and Completeness of Clinical Coding Using ICD-10 for Ambulatory Visits; pp. 912–920. [PMC free article] [PubMed] [Google Scholar]
- 36.Kirby J.C., Speltz P., Rasmussen L.V., Basford M., Gottesman O., Peissig P.L., Pacheco J.A., Tromp G., Pathak J., Carrell D.S. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J. Am. Med. Inform. Assoc. 2016;23:1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.