Abstract
Long-range chromatin interactions play critical roles in genome organization and regulation of transcription. We now report a novel technique, Transposase-mediated analysis of chromatin looping (Trac-looping), for simultaneous detection of multiscale genome-wide chromatin interactions among regulatory elements and chromatin accessibility. With this technique, a bivalent oligonucleotide linker is inserted between two interacting regions such that the chromatin interactions are captured without prior chromatin fragmentation and proximity-based ligation. Application of Trac-looping to human CD4+ T cells reveals a substantial reorganization of enhancer-promoter interactions associated with changes in gene expression upon TCR stimulation.
Keywords: chromatin interaction, chromatin looping, T-cell activation, TrAC-looping
Introduction
The precise control of gene expression during cellular differentiation and disease development requires cell-type specific interactions between enhancers and promoters1–7. It is of utmost importance to define these specific chromatin interactions in order to understand the mechanisms of gene regulation associated with these processes. The development of Hi-C, the unbiased genome-wide chromatin interaction mapping technique which depends on the in vitro proximity-based ligation of chromatin ends8, has enabled the analysis of chromatin architecture from yeast to mammals9–12. Different strategies have been employed to increase the resolution of 3C-based techniques, including in situ Hi-C13, capture Hi-C14,15, ChIA-PET16,17, and Hi-ChIP18,19 which helped to increase resolution by focusing on potential regulatory regions of the genome. Here we report a novel high-resolution technique to map genome-wide interactions in multiple scales among regulatory regions without chromatin fragmentation and proximity-based ligation.
Results
Principle of Trac-looping
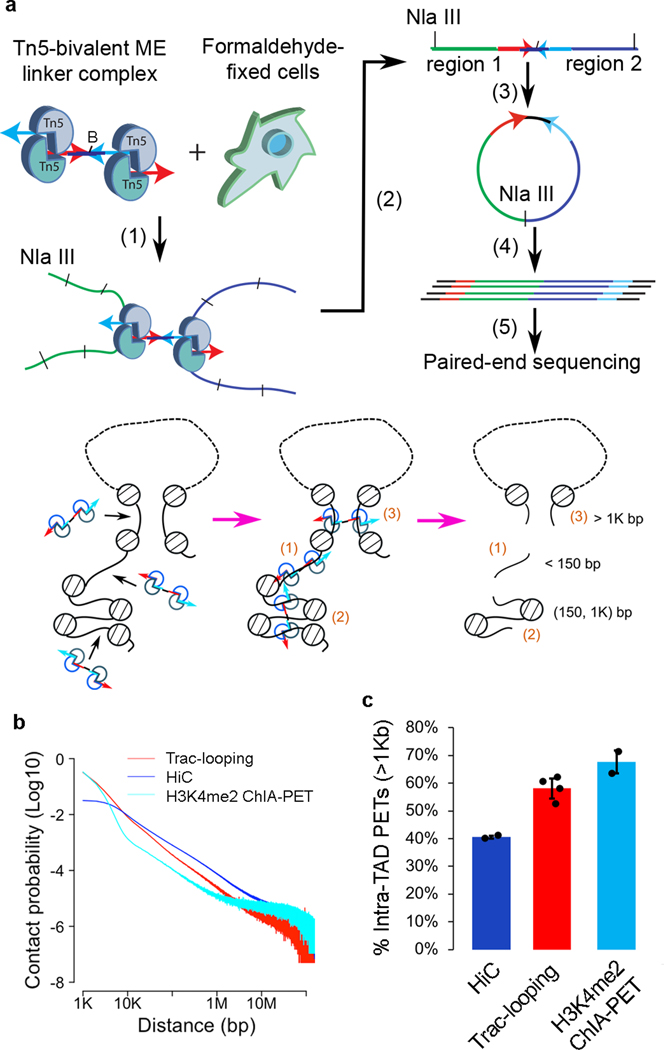
DNA transposition requires a pair of 19 bp Mosaic End (ME) sequences recognized by transposases20,21. The long DNA spacer between the MEs allows intramolecular dimerization of the transposase-ME complex (Supplementary Fig. 1a). However, when only one 19 bp ME oligonucleotide sequence (Supplementary Fig. 1b) is present, transposition leads to fragmentation and jointing of target DNA to the ME-containing adaptor, resulting in the labeling of the DNA ends by the ME sequence22. To capture genomic chromatin interactions among open chromatin regions by harnessing the activity of transposases, we designed a “bivalent ME linker” containing a pair of MEs with a 30 bp oligonucleotide spacer (Supplementary Fig. 1c). The short spacer does not have the flexibility to allow intramolecular dimerization of the transposase-ME complex and thus favors formation of a tetramer complex through the bivalent MEs in the presence of two half linkers that also contain ME sequence (Supplementary Fig. 1d–e). In addition to inserting the four MEs to the genome in cis, the tetramer can also be inserted into the genome in trans, by inserting two MEs into each of two interacting chromatin regions and resulting in “cross-linking” of the interacting chromatin regions by the bivalent oligonucleotide linker, thus identifying chromatin interactions (Fig. 1a).
Figure 1. Scheme of Trac-looping.
a.Top panel shows scheme of Trac-looping: (1) transposition of bivalent linker “in trans” into two interacting chromatin regions; (2) fragment DNA with a 4bp cutter and enrich biotinylated DNA by Strepavidin beads; (3) circulization of DNA; (4) Amplification of circular DNA; and (5) paired-end sequencing of the Trac-looping libraries. Bottom panel shows three classes of Trac-looping PETs: (1) both ends locate at the same open region (<150 bp); (2) two ends passes several nucleosomes (150 bp to 1,000 bp); (3) two ends form a loop by chromatin interaction (>1,000 bp).
b.Plot of chromatin contact probability as a function of distance (>1,000 bp) measured by Trac-looping, in situ Hi-C, and H3K4me2 ChIA-PET.
c.Percentage of intra-TAD PETs (>1,000 bp) for in situ Hi-C, Trac-looping, and H3K4me2 ChIA-PET for naïve CD4+ T cells. Data are shown as mean ± s.d. of repeated experiments for each method (HiC: n=2; TrAC-looping: n=4; ChIA-PET: n=2).
In principle, one Trac-looping assay captures multiple aspects of chromatin structure including (1) accessible regions (PETs <150 bp) similar to ATAC-seq 23,24, (2) chromatin secondary structure that includes short-range interaction spanning several nucleosomes (PETs between 150–1000 bp), and (3) chromatin interaction (PETs >1000 bp), including promoter and enhancer- associated interactions which are known to occur mostly within 200K bps19,25–29 and high-order chromatin compartmentalization30 and topologically associated domains31 (Fig. 1a).
To test the Trac-looping method, we generated libraries for resting human CD4 T cells (4 biological replicates) and stimulated CD4 T cells (2 biological replicates) (Supplementary Tables 1), with a total of 3.8 billion sequencing reads. Among the informative PETs, about 50% to 60% are <150 bp, which measure chromatin accessibility; 20% to 30% are within the range of 150bp to 1Kbp, which contain information about inter-nucleosome interaction; 15% are within the range of 1Kbp to 200Kbp, which include promoter- and/or enhancer-associated interactions; and 5% are >200Kbp, which reveal higher order chromatin organization. The distance of genomic span of PETs (>1Kbp) from Trac-looping is generally shorter than in situ Hi-C, but longer than H3K4me2 ChIA-PET (Fig. 1b). The bias to a shorter distance of Trac-looping PETs than in situ Hi-C is consistent with others’ observation that interaction among promoters and enhancers, subsets of accessible regions, preferentially occurs within short or intermediate distance19,25–29,32,33. Indeed, Trac-looping libraries have higher percentage of intra-TAD PETs than Hi-C (Fig. 1c).
Trac-looping has lower noise level compared with other methods (in situ Hi-C, ChIA-PET)
Mitochondrial DNA and genomic DNA are localized in either the cytoplasmic and nuclear compartments, respectively, and thus should not interact with each other under normal conditions. The existence of hybrid PETs consisting of mitochondrial DNA and nuclear DNA reflects false positive detection of chromatin looping. Therefore, comparing observed hybrid PETs with expected hybrid PETs under the null hypothesis could reveal the respective false positive rates of different techniques. We found that the false positive rates are 0.41 and 0.13 for the data from ChIA-PET16 and in situ Hi-C libraries generated from the same resting CD4 T cells, respectively, while the false positive rate for Trac-looping is only 0.02 (Supplementary Fig. 2a). The false positive rate based on the published in situ Hi-C data from human lymphoblastoid cells13 is 0.17. Thus, these results indicate a significant decrease in signal noise by Trac-looping compared to the other approaches for detecting chromatin interactions. Consistently, we found that PETs from the Trac-looping libraries had the highest percentage of intrachromosomal PETs (Supplementary Fig. 2b). Interestingly, cells without formaldehyde fixation gave rise to significantly more long-distance interchromosomal and inter-TAD PETs (Supplementary Fig. 2c,d), and much longer PETs (>2Mb) (Supplementary Fig. 2e). These results reveal that fixation may be critical to preserve chromatin conformation in the nucleus and to reduce random chromatin movement.
Short-distance Trac-looping PETs detect accessible chromatin regions and capture short-range nucleosome interactions
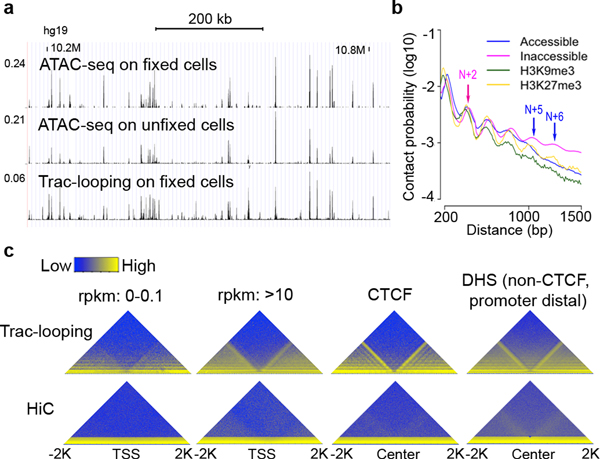
The short-distance Trac-looping PETs (<150 bps) are substantially more enriched at accessible regions than PETs from other distance ranges (Supplementary Fig. 3a) and exhibit similar profiles to that of ATAC-seq reads in both fixed and unfixed cells (Fig. 2a, Supplementary Fig. 3b–d), indicating that the short-distance Trac-looping PETs provide information on chromatin accessibility.
Figure 2. Short-distance Trac-looping PETs detect accessible chromatin regions and capture short-range nucleosome interactions.
a.Genome Browser display showing that Trac-looping and ATAC-seq detect similar chromatin accessibility profiles in fixed and unfixed cells.
b.Plots of contact probability as a function of genomic distance (150–1,500 bp) defined by Trac-looping PETs within accessible, inaccessible, H3K9me3-marked or H3K27me3-marked regions.
c.Heat maps show contact matrix at a resolution of 10 bps around TSSs of repressed and active genes, centers of CTCF binding sites, and centers of CTCF-binding-free non-promoter DHS defined by Trac-looping (upper panels). For side-by-side comparison, heat maps generated from in situ Hi-C data were also shown in the lower panels.
Chromatin may exist in two possible configurations: the “zig-zag” and the solenoid configurations 34. The “zig-zag” model predicts that nucleosome N and nucleosome N+2 are in proximal space, while the solenoid model predicts a periodicity of five to six nucleosome 35,36. Consistent with the “zig-zag” model of chromatin folding 37,38, we observed that Trac-looping PETs peaked at a distance reflecting interaction between nucleosomes N and N+2 genome-wide (Fig 2b; red arrow head), which was also observed in the yeast genome 35,39. Surprisingly, while no high level nucleosome interaction is detected in yeast 35,39, Trac-looping PETs were also enriched at a distance corresponding to interaction between nucleosome N and N+5/N+6 in the human genome (Fig. 2b; blue arrow heads), favoring the solenoid model of chromatin fiber. Interestingly, the pattern of N+5/N+6 periodic repeating fiber was only remarkable in less accessible regions (Fig. 2b; blue arrow heads) and was observed in H3K27me3-marked chromatin but not in H3K9me3-marked chromatin, consistent with a previous study that H3K9me3-marked chromatin is primarily favored in “zig-zag” while H3K27me3-marked chromatin contains mixed configurations36 (Fig. 2b). Therefore the less accessible regions, especially H3K27me3-marked region, in the human genome could contain both “zig-zag” and solenoid chromatin configurations.
Examination of chromatin interaction around functional elements such as promoters and CTCF binding sites within 2000 bp regions at a 10-bp resolution revealed a clear pattern of separation of interaction domains anchored at promoters of actively transcribed genes and at CTCF binding sites (Fig. 2c). As a control, we found no remarkable domain separation at promoters of inactive genes and at DHSs without CTCF binding (Fig. 2c). Previous Hi-C analyses reported the insulation of promoters and CTCF binding sites at large-scale with a resolution of 1,000 bp or above 12,40,41. Our results suggested that the domain insulation by promoters and CTCF occurred at mono-nucleosome level for the human genome.
Trac-looping PETs reveal high order of chromatin organization.
Trac-looping PETs also revealed similar chromatin organization detected by in situ Hi-C (Supplementary Fig. 4a). The long-distance Trac-looping PETs (>200 Kbp) recaptured the A/B compartments based on in situ Hi-C data (Supplementary Fig. 4b). Moreover, a side-by-side visualization of interaction heat maps and TAD boundaries between Trac-looping and in situ Hi-C revealed consistent TAD organizations (Supplementary Fig. 4b–d). Remarkably, interactions between CTCF-bound regions based on Trac-looping predictions were highly enriched in convergent CTCF motifs (Supplementary Fig. 4e), supporting the validity of Trac-looping in detecting structural loops at long distance. The long-range structural loops were further validated by 3D fluorescence in situ hybridization (DNA-FISH) 42 at two selected regions (Supplementary Fig. 5a,b,c).
Trac-looping efficiently detects interactions between accessible chromatin regions
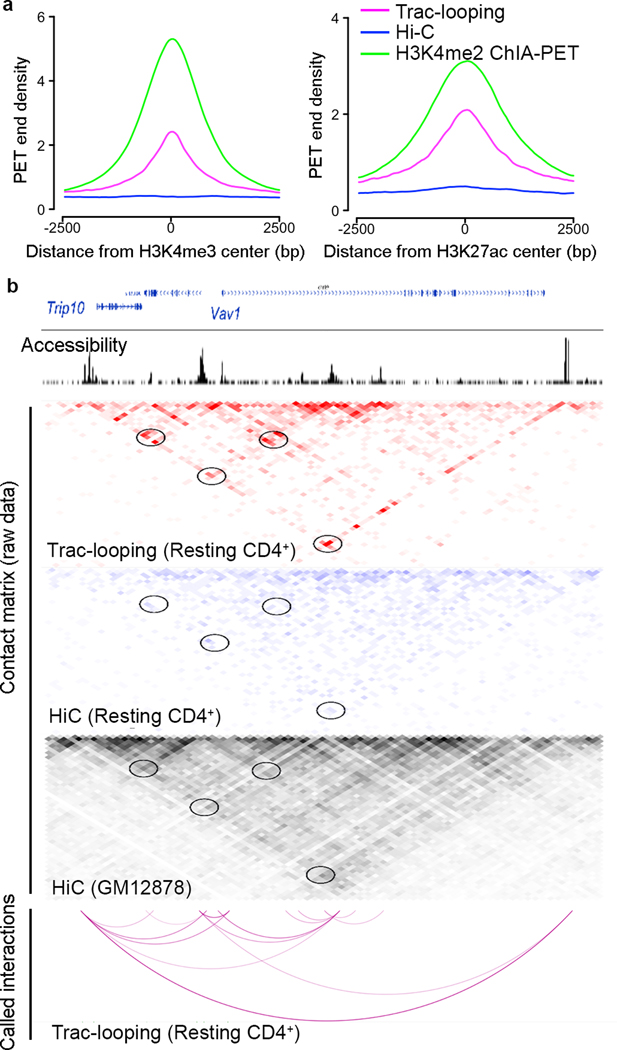
Trac-looping PETs are enriched at accessible chromatin regions and thus contain information to predict interactions among accessible regulatory elements. Indeed, the Trac-looping PETs that represent long-distance interaction events (>1,000 bp) are highly enriched at H3K4me3 and H3K27ac peaks (Fig. 3a), which represent promoter, enhancer and/or insulator regions in the genome. As expected, in situ Hi-C PETs did not exhibit notable enrichments at either promoters or enhancers, while H3K4me2 ChIA-PET reads showed significant enrichments in these regions. Consequently, Trac-looping PETs exhibited 2–3 fold more enrichment in regions associated with promoters than in situ Hi-C PETs (Supplementary Fig. 6a). As examples, we observed notable PETs enrichments between functional regions and promoters of Trip10 and Vav1, including those validated by H3K4me2 ChIA-PETs43, in the Trac-looping data (Fig. 3b). In contrast, we observed only mild enrichments (if any) from a Hi-C data set with modest sequence depth for resting CD4 (271 million informative PETs) or from very deep sequencing data for GM12878 (1.8 billion informative PETs) 33 (Fig. 3b); note that the transcription landscape of Vav1 and nearby genes was similar between resting CD4+ T cells and GM12878. Even though PETs from H3K4me2 ChIA-PET are similarly enriched at regulatory regions, our results indicate that Trac-looping has a substantially higher efficiency for detecting chromatin interactions among accessible regions (Supplementary Fig. 6b).
Figure 3. Trac-looping efficiently detects interactions between accessible chromatin regions.
a.Trac-looping PETs (>1,000 bp, magenta) are highly enriched in H3K4me3 and H3K27ac peaks. H3K4me2 ChIA-PET (green) and in situ Hi-C (blue) PETs are also plotted surrounding the H3K4me3 and H3K27ac peaks.
b.WashU genome browser showing the read density of PETs with distance less than 150 bps (for accessibility) and interaction matrices generated from Trac-looping (red) and from in-situ Hi-C (blue) for a genomic region enclosing vav1 gene locus in chromosome 19. The interaction matrices were visualized at a resolution of 2Kb. Interacting PETs longer than 200K bps were not shown. Also included for comparison is an interaction matrix from deeply sequenced in situ Hi-C data generated for GM12878 (Rao et al., 2014). Circles: interacting regions previously confirmed by H3K4me2 CHIA-PETs in resting CD4+ T cells (Chepelev et al., 2012). The predicted significant interactions for Trac-looping were also presented in the bottom panel.
We identified a total of 85,076 interactions at a resolution of 2,000 bp from a total of 261 million informative PETs of resting CD4 T cell libraries (see Methods for details; Supplementary Table 1). About 94% of the interactions between accessible regions predicted by Trac-looping were within 200 Kbp (Supplementary Fig. 6c), consistent with the observation that most of the known promoter/enhancer-associated interactions occur within this range19,25–29. Notably, 60–80% of the Trac-looping interactions were confirmed by capturing-based or antibody-assisted methods including PC-HiC44, H3K27AC Hi-ChIP45, and H3K4me2 ChIA-PET16 and in situ Hi-C data generated by third party44 and in-house, all in human CD4+ T cells (Supplementary Fig. 6d).
We evaluated the saturation of the TrAC-looping libraries by calculating how many of the interactions can be recaptured by down-sampled libraries. For the top significant interactions predicted from the whole data set (FDR < 1e-5; ~25% of all) (Supplementary Fig. 6e), >90% were still preserved as the number of sampled PETs decreased to 60% of the total (Supplementary Fig. 6f), indicating that PETs for these highly significant interactions are already saturated in the current libraries. In contrast, weak interactions (FDR between 0.05 and 0.001; ~50% of all) were far from saturation (Supplementary Fig. 6e,f). Interestingly, a substantially higher fraction of the strong interactions involved promoters and enhancers than the weak interactions (Supplementary Fig. 6g), suggesting that they are more likely to be functional interactions.
Trac-looping effectively detects promoter-enhancer interactions
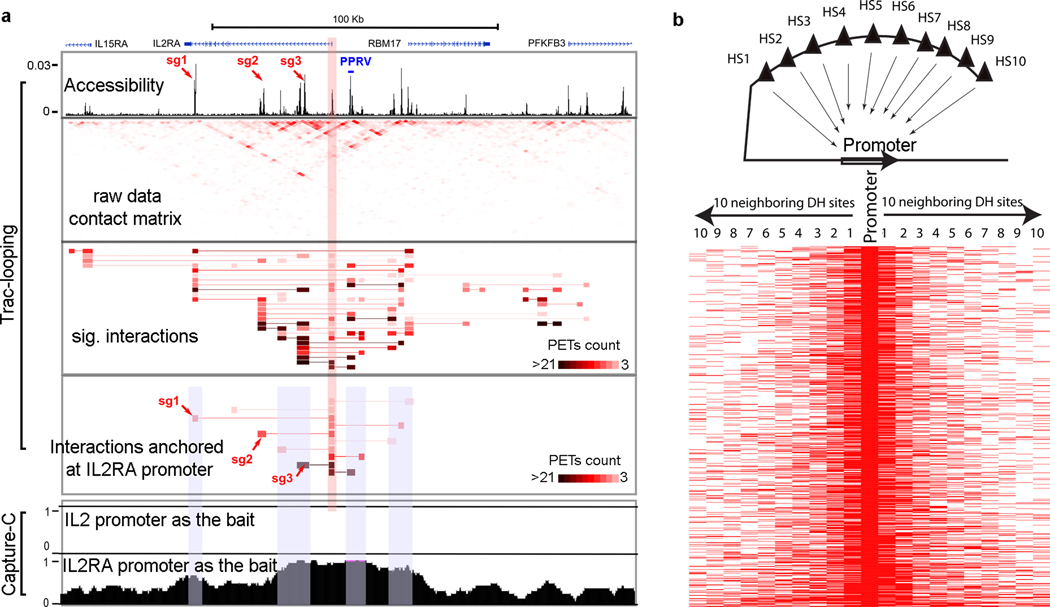
To investigate the relationship between histone modification and Trac-looping predicted chromatin interactions, we found that large proportions (40% to 70%) of active histone modifications (H3K27ac and H3K4me1/2/3) are involved in Trac-looping predicted interactions while the proportions of H3K27me3 and H3K9me3 enriched in heterochromatins are low (3%−5%) (Supplementary Fig. 7a). Moreover, we obtained consistent results by narrowing down to subsets of interacting regions validated by in situ Hi-C and sorted by distinctive chromatin status (Supplementary Fig. 7b). As expected, most of the interactions are anchored at regions enriched in histone modifications that mark promoters and enhancers (Supplementary Fig. 7c), suggesting Trac-looping effectively detects interactions between enhancers and promoters. Indeed, we identified 11,025 interactive promoters (±1kb around TSS), 26,690 interactive enhancers (see Methods for definition) and 14,138 other interactive sites (Supplementary Fig. 7d,e). Among these sites, there are 8,828 promoter-promoter, 23,232 promoter-enhancer and 34,068 enhancer-enhancer interactions. The enhancer-promoter interactions detected by Trac-looping were positively correlated with the expression of their promoters (Supplementary Fig. 7f). IL2Ra gene is critical for T cell development and has been extensively studied and annotated for its functional elements46. Our data revealed that the IL2Ra promoter interacts with a number of DHSs including the PRRV region, a well-established enhancer for IL2Ra expression (Fig. 4a), which were confirmed by (NG) Capture-C using the IL2Ra promoter as the bait (Fig. 4a). Targeting three potential enhancers of the IL2Ra gene using the dCAS9-KRAB system significantly decreased IL2Ra expression (Fig. 4a, Supplementary Fig. 7g), demonstrating that Trac-looping is capable of detecting functional enhancer-promoter interactions. Consistent with this observation, we found that active promoters and enhancers exhibit much higher interaction intensity than poised promoters and enhancers (Supplementary Fig. 7h). Super enhancers are more interactive than regular enhancers and almost all super enhancers interact with their target promoters (Supplementary Fig. 7i–k).
Figure 4. Trac-looping efficiently detects interactions between accessible chromatin regions.
a.Genome browser displays the Trac-looping detected interactions around IL2RA gene locus. Accessible regions, raw contact matrix (resolution = 2kb), all significant interactions (color indicates PET count) and the interactions linked to IL2RA promoter were shown. The (NG) Capture-C data using IL2 promoter (negative control) as the bait and the IL2RA promoter as the bait were also included. The IL2RA promoter region was highlighted in red bar. Three dCAS9-KRAB repression targets were indicated by red arrows. The PPRV region was also indicated in the accessibility panel.
b.Promoters tend to interact with nearby accessible enhancers. The interaction between the top 1,000 most interactive promoters and the 10 nearest DHSs on each side of the promoter was examined as indicated by the cartoon on the top. +1 and −1 DHSs are closest to the promoters; +10 and −10 DHSs are most distant to the promoters. Red color indicates interactions.
Chromatin accessibility is essential for transcriptional regulation and chromatin interactions. To test whether Trac-looping detected interactions can solely be explained by proximal chromatin accessibility, we compiled the 10 nearest accessible sites on each side of the top 1000 most interactive promoters and examined their interaction with the promoter. Interestingly, 64% of promoters interacted with the closest accessible sites (+1 or −1 site) (Fig. 4b, Supplementary Fig. 8a), suggesting that promoters tend to interact with nearby regulatory DHSs. However, we also found that 36% of the interactions take place by looping out at least one nearest accessible site and 23% of interactions loop out at least two nearest accessible sites (Fig. 4b, Supplementary Fig. 8a). Next, we examined a random set of 3000 DHSs in the genome and found that 32%, 18%, 12% and 7% of them loop out at least 1, 2, 3 and 4 DHSs, respectively, before the first interaction (Extended Data Fig. 8b). In fact, 24% of all enhancers skip their nearest promoters to form enhancer-promoter interactions (Extended Data Fig. 8c,d). These results indicate that chromatin accessibility and spatial proximity are not sufficient to allow detection of chromatin interaction by Trac-looping.
Reorganization of enhancer-promoter interaction upon TCR stimulation of CD4+ T cells
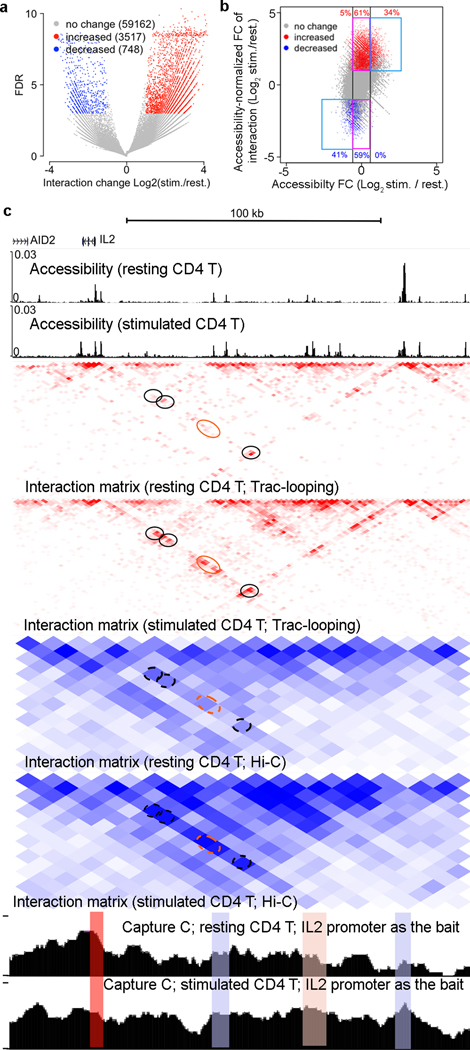
To investigate whether Trac-looping can detect the promoter-enhancer interaction changes induced by T cell activation, we called differential interactions between resting and activated CD4 T cells. To remove the potential bias caused by accessibility change, the interaction changes were normalized against accessibility change (Supplementary Fig. 9a,b) (see also Methods). We predicted 3517 (5.5%) pairs of HSs and 748 (1.2%) pairs of HSs that showed an increase and a decrease in interaction during T cell stimulation, respectively (Fig. 5a,b). Of the sites predicted with an increase in interaction, 66% showed no substantial increase in accessibility (FC<1.5; Fig. 5b). Specific examples included the interactions between the il2 promoter and three putative enhancers located 46.5, 51.4, and 121.8 Kbp in the upstream region of its TSS (Fig. 5c; black circles), which were independently confirmed by capture-C (Fig. 5c) and in situ Hi-C (Fig. 5c; dashed black circles). For the remaining 34%, an increase in accessibility was notable but insufficient to explain the increase in interaction. A specific example was shown for the interaction between the il2 promoter and an enhancer cluster located 83.1 to 96.4Kbp in the upstream region (Fig. 5c; orange circles); notably, the increase in the interaction were also confirmed by the accessibility-insensitive in situ Hi-C. At a genome-wide scale, the increase in interaction predicted by Trac-looping at HSs also exhibited an increase in interacting PETs from the in situ Hi-C data (Supplementary Fig. 9c). Relevant to T cell biology, genes associated with increases in interaction were associated with increases in gene expression upon T cell stimulation (Supplementary Fig. 9d), and were enriched in biological processes related to T cell activation (Supplementary Fig. 9e). Similarly, 59% of the sites predicted with a decrease in interaction showed no substantial decrease in accessibility (FC<1.5; Fig. 5b), while for the remaining 41%, the decrease in interacting PETs significantly exceeded the decrease in accessibility (Fig. 5b). Interacting HSs predicated with a decrease in interaction from Trac-looping also exhibited a decrease in the in situ Hi-C PETs (Supplementary Fig. 9f). Genes associated with a decrease in interaction from Trac-looping tended to be downregulated in expression level (Supplementary Fig. 9g) and were enriched in biological process including DNA packaging and chromatin assembly (Supplementary Fig. 9h).
Figure 5. Reorganization of enhancer-promoter interaction upon TCR stimulation of CD4+ T cells.
a.Volcano plot shows increased (red) and decreased (blue) interactions (FDR<1e-3, accessibility-normalized FC>2).
b.Plots of accessibility-normalized fold change of interacting PETs vs. accessibility change at anchors for all the identified interactions. Significant increased (FDR<1e-3, FC>2, red) or decreased (FDR<1e-3, FC>2, blue) interactions were highlighted.
a-b: Data shown represents total 63,427 merged interactions identified from resting and activated CD4 T cells. Details of identifying significantly changed interactions are presented in the online Methods.
c.Heat maps show the contact intensity defined by Trac-looping PETs (red, bin size=2kb) and in situ Hi-C PETs (blue, bin size=10kb because of low sequence depth) in resting and stimulated T cells. Tracks for the accessibility were shown at the top. Black circles: examples of interaction increase with no accessibility increase observed at anchor regions. Orange circle: example of interaction increase with also accessibility increase at anchor regions. The bottom two panels show the (NG) Capture-C data using IL2 promoter as the bait. The anchors (including IL2 promoter) of the highlighted interaction examples in the matrix were also highlighted for the Capture-C data.
Globally, we detected 1,591 (17.7% of all interacting promoters) and 589 (6.6% of all) promoters with significantly increased and decreased interactions, respectively, upon TCR stimulation. Among all the interacting promoters, about 1.6% have enhancers switched during T cell activation (Supplementary Fig. 10a,b).
To test the changes in interaction networks upon TCR signaling, we identified 7,820 and 8,930 interaction networks in resting and activated CD4 T cells, respectively, mainly within TADs (Supplementary Fig. 11a). The connectivity of these networks can either increase or decrease upon TCR signaling (Supplementary Fig. 11b), which is correlated with gene expression changes (Supplementary Fig. 11c; p-value=0.007; Supplementary Fig. 11d; p-value=1.7e-16). Our analysis revealed that 94 and 36 gene promoters in resting and activated T cells, respectively, interact with five or more promoters (Supplementary Fig. 11e–g), which may represent hubs of interaction.
FOS family motifs are enriched in chromatin regions with increased interaction upon T cell activation
Our analysis revealed that the TGAGTCA motif for the FOS family of transcription factors, which were highly upregulated by T cell stimulation (Supplementary Fig. 12a), was significantly enriched in regions with increased interaction (Supplementary Fig. 12b). Footprinting analysis using the reads of both short and long PETs from the Trac-looping libraries revealed that the DNA sequence surrounding the FOS motifs exhibit footprint patterns in stimulated but not in resting CD4 T cells (Supplementary Fig. 12b), suggesting the binding of the FOS family factors is associated with increased interactions. Indeed, our ChIP-Seq assays confirmed that the binding of FOSL1/Fra-1 to these regions is enriched after T cell activation (Supplementary Fig. 12c–e). In summary, our results suggest that the Trac-looping data can be used to predict TFs involved in chromatin interactions and accessibility.
Discussion
TrAC-looping is a novel technology for mapping genome-wide interactions among transcriptional regulatory elements and unlike 3C-based methods such as Hi-C and ChIA-PET, it does not require prior chromatin fragmentation and proximity-based ligation. We show that TrAC-looping PETs contain information for (1) chromatin accessibility and TF footprints (PETs <150 bp); (2) short-range nucleosome to nucleosome interactions (PETs between 150 to 1000 bp); (3) intermediate to long-range regulatory element interactions and high-order chromatin organizations including TADs and A/B compartments (PETs > 1000 bp). Application of Trac-looping to T cell activation led to the detection of changed interaction at thousands of enhancers and promoters, which contribute to the altered gene expression in activated CD4 T cells. Our integrated analysis of Trac-looping data with ChIP-seq data suggested a potential role of the FOS family of transcription factors in the reorganization of enhancer-promoter interactions and gene activation. In summary, we have developed a novel technique for defining genome-wide enhancer-promoter interactions, which may be used to characterize dynamic regulation of 3D nuclear structure at high-resolution in a variety of biological systems.
Methods
Expression construct of Hyperactive Tnp
The coding sequence of Tn5 transposase Tnp was amplified from the pfd-Tn5[pfdA31-Tn5] (ATCC cat# 77330) as 3 PCR fragments while introducing 3 mutations: E54K, M56A and L372P to generate the hyperactive Tn5 22,47. The PCR fragments were assembled by NEB Gibson builder (NEB cat# E5510S) into pET-15b (Novagen cat# 69661–3) to make the hyperactive Tnp expression construct pET15b-His6Tnp (available now at addgene, plasmid#79807).
Expression and purification of hyperactive Tnp
Competent cells BL21 Gold (DE3) (Agilent cat# 230132) were transformed with pET15b-His6Tnp. One single colony was inoculated and cultured in 60ml LB + Carbenicillin (100 μg/ml) at 37°C, 200rpm for overnight, which was diluted to 1.2 liter of the same media and continued to culture till OD600 at 0.8. To induce Tnp expression, IPTG was added to 0.5mM (final concentration) and shaken at 200rpm at room temperature for 4 hours. Following cooling down in ice water, the cells were harvested by spinning at 3700rpm for 15min. The cell pellets were resuspended in 30 ml lysis buffer (50mM Tris-HCl pH8.0, 300 mM NaCl, 20mM Imidazole, 0.1% Triton X-100, 10 μg/ml Pepstatin A (calbiochem cat# 516481), 10 μg/ml Leupeptin Hemisulfate (calbiochem cat# 108975), 10 μg/ml Chymostatin (calbiochem cat# 230790), 6 μg/ml Antipain Dihydrochloride (Sigma cat#A6191), 1mg/ml lysozyme), put on ice for 30 minutes and sonicated with a microtip to lyse the cells. The lysates were spun at 15,000rpm for 10 min at 4°C before adding 5mM 2ME (2-mecapotoethenal), 1 μM PMSF and 1M NaCl.
To purify the Tnp, the cell lysates were added to 0.8 ml 50% Ni-NTA agarose bead slurry (Qiagen cat#1018244), which was washed with 50mM Tris-HCl pH8.0, 300 mM NaCl and 20mM Imidazole, and incubated for one hour at 4°C with rotation. The beads were spun down at 1320rpm for 5min at 4°C and washed once with 15ml wash buffer (50mM Tris-HCl pH8.0, 1M NaCl , 20mM Imidazole, 0.1% Triton® X-100). The beads were transferred to a 1ml syringe with glass wool on the bottom and washed with 20ml wash buffer. The bound Tnp was eluted using 3 ml of elution buffer (50mM Tris-HCl pH8.0, 1M NaCl , 250mM Imidazole, 0.1% Triton® X-100) and collected into 6 Eppendorf tubes.
Transposome complex assembly
To anneal the primers for TrAC-looping, 250μl 20μM Bio67F (sequence in the primer list, all primers are ordered from Integrated DNA Technologies, Inc.) and 250 μl 20 μM 67bpR in EB were annealed in 55 μl 10x buffer (0.5 M Tris-acetate pH 7.5, 1.5 M potassium acetate, 40 mM spermidine) by heating at 98°C for 5min followed by slow cooling to room temperature. This bivalent ME linker is called “67bpR-Bio67F”. Similarly, ME3ddCTPtop and 5NH2MEbottom were annealed to form NHddCTP. To a mixture of 16 μl 100% glycerol, 4.5 μl 50 μM annealed NHddCTP, 12.5 μl 9 μM 67bpR-Bio67F was added 30 μl 1mg/ml HisTnp enzyme and incubated at room temperature for 20min to form transposome complex.
To anneal for primers for ATAC-seq, pMENTS and i5Top primer are anneal for pMi5. pMENTS and i7Bottom primer are anneal for pMi7. To assembly ATAC-seq complex, 8 μl H2O, 16 μl 100% glycerol, 4.5 μl 50 μM annealed pMi5, 4.5 μl 50 μM annealed pMi7 was mix and added to 30 μl 1mg/ml HisTnp enzyme. Incubated at room temperature for 20min to form transposome complex before use.
Analysis of the transposome complexes by chromatography
The mixture of the Tn5 complexes was resolved by a Superdex 200 10/300 GL column (Cat. 17–5175-01, GE Healthcare) controlled by an AKTA chromatography system. The column was pre-equilibrated with 20mM TrisHCl, pH7.4, 250mM NaCl, 20% glycerol. Approximately 250 μl of the reaction mixture was applied onto the column and eluted by same buffer at a flow rate of 0.4ml/min. Fractions were collected at 0.5ml/tube. The DNA purified from peak fractions A7 and A11 by phenol: chloroform extraction and ethanol precipitation was run on a 2% E-gel. The formation of Tn5 tetramer complex is confirmed by the presence of both the bivalent linker and the half linker in fraction A7. The dimer formation is confirmed by the presence of only the half linker in fraction A11.
TrAC-looping (Transposition-mediated Analysis of Chromatin Looping)
Human CD4+ T cells were purified from human blood using human CD4 T cell enrichment Kit from Stemcell Technologies (cat# 19052) according to the manufactures manual. Cells used for biological repeats are from different individual donors. For the TRAC-looping protocol improvement, cells were purified using human naive CD4 T cell isolation kit II (Milteyi Biotec, order no. 130–094-131). The cells were used directly as resting T cells or activated using anti-CD3/CD28 beads (Invitrogen Dynabeads Human T-Activator CD3/CD28, Cat# 11131D) for 16 hours at 37°C as activated or stimulated cells. To perform the TrAC-looping assay, 100 million cells in 100 ml culture media were cross-linked by adding 16% formaldehyde (Thermo Scientific prod# 28908) to 1% and incubated at room temperature for 5 minutes with gentle shaking, followed by adding glycine to 0.125M to quench the reaction for 5min and washing twice with PBS. The cell pellets were resuspended in 100 ml cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.5% IGEPAL CA-630) and incubated on ice for 15min. The cells were gently spun down and resuspended in 4.5 ml of the same cold lyse buffer, followed by addition of 500 μl 10x reaction buffer (0.5 M Tris-acetate pH 7.5, 1.5 M potassium acetate, 100 mM magnesium acetate, 40 mM spermidine) and 80μl transposon complex. The reaction mixture was split into 10 eppendorf tubes and incubated at 50°C 1.5 hours. The reaction was stopped by adding 25 μl 0.5M EDTA, 16μl 10% SDS and 30 μl Protease K (Roche cat# 03115828001) to each tube. Following incubate at 65°C for overnight, the genomic DNA was purified by phenol-chloroform extraction and ethanol precipitation. The DNA pellets were resuspended in 1 ml of elution buffer (EB).
To repair the 9bp gap generated in the transposition reaction, 200 μg gDNA was incubated with T4 DNA polymerase (NEB cat# M0203L) in NEBuffer2.1 in the presence of 0.1mM dNTP at room temperature for one hour. The reaction was stopped by adding EDTA to 25mM and the mixture was added to 2772 μl prewarmed Ampure XP beads (Agencourt part number: A63881) to purify >300bp DNA fragments. Following washes with 70% ethanol twice, the bound DNA was eluted with 1100μl of 1x cutsmart buffer. The eluates were split into 2 tubes and digested with 33 μl of Nla III (NEB, R0125L) and MluCI (NEB, R0538L), respectively, at 37°C for 3 hours. To each of the reactions were added 50μl Streptavidin C1 beads (Invitrogen cat# 65001) that is prewashed and resuspended in 533μl 2x B/W buffer and incubated for 30min at room temperature.
Following three washes with 1ml 1x B/W +0.1% TritonX-100, the beads were resuspended in 50 μl of elution buffer (870 μl EB+30 μl 10% SDS+100 μl Protease K) and incubated at 55°C for 4h hours. The eluted DNA was purified by phenol-chloroform extraction and ethanol precipitation. For intramolecular circulization, the DNA was incubated in the presence of T7 DNA ligase in 50ml of ligation buffer (66 mM Tris-HCl pH 7.4, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 7.5% Polyethylene glycol (PEG 6000) at room temperature for overnight. The ligated DNA was precipitated by ethanol and then resuspended in 55 μl sampling buffer from templify100 kit (GE Hearlthcare cat# 25–6400-10). The DNA was denatured at 97°C for 3min and then chilled on ice. RCA was performed by adding 50 μl Reactions buffer +2 μl enzyme mix to the denatured DNA mix and incubated at 30°C for overnight. Use 1 μl of RCA product for PCR by mixing with 25 μl NEB Phusion HF master mix, 23 μl H2O, 1 μl 10 μM PCR primer N501, 1 μl 10 μM PCR primer N701 with the cycling conditions: 98 °C for 30 s; 11 cycle of 98 °C for 10 s, 65 °C for 30 s and 72 °C for 8 s; 72 °C for 5 min; 4C hold. The PCR products were resolved on E-gel and the DNA with a size range of 250–650bp was isolated and sequenced by paired-end sequencing on Illumina Hiseq 2500.
ATAC-seq
A total of 100k human CD4+ T resting cells, fixed or native (not fixed) were washed with PBS. The cell pellets were washed with RSB buffer (10mM Tris-HCl, pH 7.5, 10mM NaCl, 3mM MgCl2) once and resuspended in 52 μl cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.5% IGEPAL CA-630) and incubated on ice for 15min. add 6 ul 10x reaction buffer and 2 μl ATAC-seq Tnp complex. Put in PCR machine for 37 °C 30 min. Add 3 μl 0.5M EDTA to stop the reaction. Add 40 μl H2O + 3 μl 10% SDS +5 μl proteinase K. put the samples in a 65 °C incubator overnight. Transfer samples to a new Eppendorf and extract DNA by phenol-chloroform extraction and ethanol precipitation. The DNA pellets were resuspended in 10 μl of elution buffer (EB). Set up PCR by adding 25 μl of master Phusion HF mix, 22 μl H2O, 1 μl of one of the 10uM N501-N508 and 1 μl of one of the 10uM N701-N712. PCR: 72°C for 5min, 98°C for 45 sec. 19 cycles of ( 98°C, 15”; 65°C, 30”; 72°C, 30”), 72°C 5min. 4°C hold. Gel purify the 200–500 bp smear for sequencing.
3D fluorescence in situ hybridization (FISH)
CD4+ T cells were purified and/or activated as described as above for resting cells and activated cells. The cells were then adhered on cell-tak™ (Corning cat# 354240) coated slides before being fixed for 10 minutes in 4% paraformaldehyde. For the slide pretreatment, we followed a previously published protocol 42.
Bacterial artificial chromosome (BACs) DNA were chosen ~75kb upstream and downstream of the DNaseI hypersensitive sites to confirm whether two HSs on each of the two areas that show interaction by Trac-looping looped together on chromosome 5 (RP11–101N10 and RP11–1130A7) and chromosome 17 (RP11–133J18 and RP11–138B14). Two additional sets of BAC clones were chosen on chromosome 17 (RP11–138J18, RP11–12F14 and RP11–138J18, RP11–139I13) to serve as a negative control. Each BAC DNA was extracted using the plasmid maxi kit (Qiagen cat# 12162) and labeled using nick translation with either Spectrum Orange (Abbott Molecular cat# 02N33–050) or Dy505 (Dyomics cat# Dy-505), and their chromosomal location confirmed by chromosome FISH. The labeled DNA was then precipitated overnight before being co-denatured with the pretreated slides for FISH hybridization at 72°C for five minutes. The slides were then incubated at 37°C for two nights. Hybridized slides were detected using our standard online lab protocol without ethanol dehydration (https://ccr.cancer.gov/Genetics-Branch/thomas-ried under resources).
The hybridized slides were imaged using a 100X objective on a DeltaVision microscope (Applied Precision) and processed using image decovolution. Approximately 15 fields were imaged with approximately 25 cells analyzed for each slide. Nuclei were individually cropped, surface rendered and subjected to 3D measurements using 3D-constructor and Image-Pro Plus 6.3 (Media Cybernetics). The distance between the red and green probes was calculated using the geometric centers of each probe.
Next-generation (NG) Capture-C
The (NG) Capture-C assays were performed according to published procedures15 with the following modification. The human CD4 T cells were purified and fixed in the same way as for Trac-looping. Following digestion of chromatin and 3C ligation, multiple oligonucleotides covering the IL2RA and IL2 promoter regions, respectively, were hybridized to the 3C libraries without using the blockers. The biotin-labeled oligo probe sequences are provided in Supplementary Table 2.
Inhibition of enhancer activity by dCAS9-KRAB
Three sgRNA targets from the IL2RA genomic region and one control sequence from another chromosome were selected (see Supplementary Table 2 for sequences). These above target sequences were cloned into Origene’s pGuide (cat# GE100042) plasmid for constructing the sgRNA expression plasmids sgRNA1, sgRNA2, sgRNA3 and sgRNA Control. The dCas9-KRAB plasmid (Addgene plasmid # 84245, pSLQ2815 pPB: CAG-Puro-WPRE PGK-KRAB-tagBFP-SpdCas9) was a gift from Stanley Qi. For inhibiting enhancers, human rested CD4+ T cell were purified using human CD4+ T cell enrichment kit (stem cell cat#19052) and transfected with 1.5μg dCas9 KRAB plasmid, 1.5μg sgRNA plasmid and 2μg pmaxGFPTM by following the instruction of amaxaTM human T cell nucleofectorTM kit. The GFP+ positive cells were sorted by AIRE II after 24 hours of transfection. The sorted cells were activated by beads bound antiCD3 and antiCD28 antibodies for 24 hours. The RNAs were isolated from the cells and analyzed by qRT-PCR. The IL2RA TaqMan® Gene Expression Assay and eukaryotic 18S rRNA Endogenous Control (cat#: Hs00907782_m1, 4352930E, respectively) were from Thermo Fisher. The IL2RA expression level was normalized against 18S rRNA as input control. The efficiency of transcriptional repression by sgRNA1, sgRNA2, sgRNA3 is presented as compared to the sgRNA control.
Data analysis
TrAC-looping PETs mapping
The paired-end tags (PETs) from TraAC-looping libraries were mapped to the human reference genome (hg19) using an iterative mapping approach with bowtie2 48 and the PETs with the low mapping quality were filtered out (MAPQ<10). Because the mapped PETs with the same coordinate on the genome were considered to be PCR replicates derived from the same original DNA fragments, only one uniquely mapped PET was kept at each coordinate. The uniquely aligned PETs were subjected to further analysis and basic information of the PETs was provided (Supplementary Table 1).
False Positive Rate (FPR) of TrAC-looping, HiC and CHIA-PET
Mitochondrial DNA and nuclear DNA were segregated to mitochondrion and nucleus in cells, respectively. And we used the hybrid of the two as a measure for noise or false positive rate. We denoted nuclear DNA by N and mitochondrial DNA by M. For each paired end tag (PET), there are three possible scenarios: PETs with both ends from nuclear DNA (NN), PETs with both ends from mitochondrial DNA (MM), and hybrid PETs with one end from nuclear DNA while the other end from mitochondrial DNA (NM). We could infer the proportion of nuclear DNA and mitochondrial DNA based on the number of PETs in each category. The frequency of nuclear DNA and frequency of mitochondrial DNA could be denoted and , respectively, in which . Furthermore, we could calculate and .
If nuclear DNA and mitochondrial DNA randomly formed the PETs, the frequency of PETs categories will follow binomial theorem (p + q)2 = p2 + 2pq + q2 = 1. Therefore, the expected frequency of NN PET and expected frequency of MM PET is and , respectively. Especially, we got the expected frequency of NM PET and of expected number of NM PET under the null hypothesis . We could calculate the false positive rates (FPR) of the library by , in which any observed NM PET is treated as noise.
PET classification
We classified PETs into three groups according to their lengths. The PETs <150 bp were used to define genome-wide hypersensitive sites (HS). The PETs within 150 bp to 1000 bp were used to investigate nucleosomal interactions. The PETs >1000 bp were used to predict chromatin interactions and characterize higher-order chromatin organization, such as TAD and A/B compartments. We conducted peak calling using short reads (<150 bp) for genome-wide hypersensitive sites (HSs) by MACS 49 using default parameters. The HSs smaller than 2kb were extended to 2kb and the HSs were merged if they overlapped after extension.
Identifying significant interactions
We used the model in Mango50 to calculate the statistical significance of an “interaction” between two HS. In short, we first estimated the probability of observing an PET between two HSs separated by a given distance and characterized by a given joint accessibility. The accessibility of HS was measured by all PET-end tag density. Then, we calculated the P value of observing k or more PETs between two HSs based on binomial distribution. Multiple testing correction (Benjamini-Hochberg) was conducted and only significant interaction were kept for further analysis (FDR ≤0.05 and strength ≥3PET).
Validating interaction by alternative technologies
To validate the predicted interaction in resting CD4 T cells by TrAC-looping, we used data from different chromatin interaction technologies downloaded from previous publications or generated in house. Interactions for in situ Hi-C were identified using Homer (FDR<0.05, bin size=2K bp). Interactions for H3K4me2 ChIP-PET and H3K27ac HiChIP were made using the same approach as did for TrAC-looping. Interactions for PCHiC were directly downloaded from publication.
Identifying differential interactions
We identified significantly changed interactions between stimulated and resting CD4+ T cells. We down-sampled PETs from resting CD4 T cell library to the same number as the stimulated CD4 T cell library. Significant interactions in resting and stimulated CD4 were merged as an interaction union set. Interacting PETs from the two conditions were compared after normalized against accessibility levels (measured by PETs <150bp). Specifically, suppose one interaction I has two anchors A and B. The interacting PETs from two conditions are I1 and I2, the library sizes are L1 and L2, and the accessibility levels are A1, B1, A2, and B2. Suppose two conditions have the same interaction level, the expected interacting PETs in condition 2 is estimated as If observed PETs I2 is significantly larger than , we predicted this interaction as increased in condition 2. The P value of observing I2 or more PETs was calculated based on binomial distribution. Multiple testing correction (Benjamini-Hochberg) was conducted and FDR<0.001 was used as a cut-off. Decreased interaction was identified similarly.
Analysis of capture Hi-C data
Significant interactions (score ≥5) detected by promoter capture Hi-C data in resting CD4 T cells were downloaded from Javierre et al.51.
Analysis of ChIP-seq data
The ChIP-seq for FOSL1 was performed with an antibody from Santa Cruz Biotechnoloy, Inc. (cat#: sc-183) using the protocol as described 52. The ChIP-seq data of H3K4me3, H3K4me1, H3K4me2, H3K27me3, H2A.Z and CTCF for CD4 resting T cell were downloaded from Barski et al. 52. The ChIP-seq data of H3K27ac for CD4 resting T were downloaded from Wang et al. 53. The ChIP-seq data of H3K9ac for CD4 resting T were downloaded from Bernstein et al. 54. The reads were mapped to hg18 using bowetie2 and the reads with MAPQ<10 were removed. The ChIP-seq peaks were called using MACS with default parameters. The distributions of tags from TrAC-looping (treated both ends of PET separately) around these H3K4me3, H3K27ac and CTCF peaks were checked.
Comparing TrAC-looping with in situ Hi-C and ChIA-PET
We compared TrAC-looping with in situ Hi-C and H3K4me2 ChIP-PET data in the resting CD4 T cells. In situ Hi-C library in resting CD4 T cells was newly generated and H3K4me2 ChIP-PET data from Chepelev et al.16 were downloaded. To compare the efficiency of identifying interactions between TrAC-looping and H3K4me2 ChIA-PET, the same number of PETs (100M PETs) were sampled from each library and the same method as described above were conducted with the same parameters (bin size = 2K).
Analysis of topologically associating domains (TADs)
Each chromosome was separated into 40Kb bins and interaction matrix of each chromosome was generated for either Hi-C data or TrAC-looping data from CD4+ T cell. The interaction matrix was normalized by Hi-Corrector 55. The normalized interaction matrix was used as input for identifying TAD by TopDom 56. The TADs identified by TrAC-looping and Hi-C are highly consistent with each other.
Analysis of RNA-Seq
The reads from RNA-Seq libraries were mapped to the human genome (hg18) using bowtie2 48. The gene expression level was measured by RPKM (Reads Per Kilobase per million mapped reads) and number of reads in a gene. The significant increased and decreased genes after T cell activation were identified using EdgeR (FDR < 0.05) 57.
Annotation of HSs and interactions
The HS that contains transcription start site (TSS) in refSeq or H3K4me3 peaks is classified as promoter. The HS that overlaps with H3K27ac/H3K4me1/H3K4me2 peaks but not promoters is classified as enhancer. The HS that neither is a promoter nor an enhancer is classified as other which includes insulator. Based on the category of HS, the interactions were classified into 6 categories: Promoter-Promoter, Promoter-Enhancer, Promoter-Other, Enhancer-Enhancer, Enhancer-Other, Other-Other. Super-enhancers were determined using H3K27ac ChIP-seq data and ROSE58.
Analysis of CTCF loop orientation
The CTCF position weight matrix (JASPAR motif: MA0139.1) was downloaded from JASPAR database. We extended ±100bp of each HS summit to obtain potential transcription factor (TF) binding regions. Then we searched the CTCF motif in all the potential TF binding regions using fimo, a module of meme 59. The HS contained very strong CTCF motif was considered as CTCF binding sites (p value <1×106). We kept 1Kb as the interaction resolution and identified the interaction among the CTCF binding sites, which is a subset of the total interaction among HSs.
GO Enrichment analysis
To examine whether particular gene categories/pathways were enriched in certain gene lists, the GO enrichment analysis was performed using DVAID 60. The GO categories with FDR <0.05 were consider as significant.
Statistical Analysis
All the statistical tests were executed using the R statistical package (http://www.r-project.org/). One-sided t-tests were performed to compare the values between two groups for the data in Supplementary Figs. 5c, 7g, 9c,d,f,g, and 11c,d.
Supplementary Material
Supplementary Table 1. PETs information of Trac-looping libraries from human CD4 T cells.
Supplementary Table 2. Oligonucleotides sequences used in the study.
Acknowledgements
We thank the NHLBI DNA Sequencing Core facility for sequencing the libraries; the NIH Biowulf High Performance Computing Systems for computing service; the Department of Transfusion Medicine for preparing lymphocyte cells procured by apheresis from donor blood. We thank Drs. Wai Lim Ku for critical reading of the manuscript. The work was supported by the Division of Intramural Research of NHLBI, NIH and CCR, NCI, NIH.
Footnotes
Competing Financial Interests Statements
The authors declare no competing interests.
Data availability and Accession Code Availability Statements
The Trac-looping data and RNA-seq date are available from GSE87254.
References
- 1.Vakoc CR. et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell 17, 453–462, doi: 10.1016/j.molcel.2004.12.028 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504, 306–310, doi: 10.1038/nature12716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei Z. et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13, 36–47, doi: 10.1016/j.stem.2013.05.010 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Phillips-Cremins JE. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295, doi: 10.1016/j.cell.2013.04.053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolhuis B, Palstra RJ, Splinter E, Grosveld F. & de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular cell 10, 1453–1465 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Palstra RJ. et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35, 190–194, doi: 10.1038/ng1244 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Drissen R. et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes & development 18, 2485–2490, doi: 10.1101/gad.317004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M. & Kleckner N. Capturing chromosome conformation. Science 295, 1306–1311, doi: 10.1126/science.1067799 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Duan Z. et al. A three-dimensional model of the yeast genome. Nature 465, 363–367, doi: 10.1038/nature08973 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanizawa H. et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic acids research 38, 8164–8177, doi: 10.1093/nar/gkq955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieberman-Aiden E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293, doi: 10.1126/science.1181369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380, doi: 10.1038/nature11082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao SP. et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell in-press, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mifsud B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47, 598–606, doi: 10.1038/ng.3286 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Davies JO. et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods 13, 74–80, doi: 10.1038/nmeth.3664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chepelev I, Wei G, Wangsa D, Tang Q. & Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell research 22, 490–503, doi: 10.1038/cr.2012.15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z. et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627, doi: 10.1016/j.cell.2015.11.024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumbach MR. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mumbach MR. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reznikoff WS. Tn5 as a model for understanding DNA transposition. Mol Microbiol 47, 1199–1206 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Reznikoff WS. The Tn5 transposon. Annu Rev Microbiol 47, 945–963, doi: 10.1146/annurev.mi.47.100193.004501 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Adey A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome biology 11, R119, doi: 10.1186/gb-2010-11-12-r119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangadharan S, Mularoni L, Fain-Thornton J, Wheelan SJ. & Craig NL. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proceedings of the National Academy of Sciences of the United States of America 107, 21966–21972, doi: 10.1073/pnas.1016382107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buenrostro JD, Giresi PG, Zaba LC, Chang HY. & Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218, doi: 10.1038/nmeth.2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Q. et al. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat. Genet. 49, 1428–1436 (2017). [DOI] [PubMed] [Google Scholar]
- 26.He B, Chen C, Teng L. & Tan K. Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. U S A 111, E2191–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma W. et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 12, 71–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z. et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman-Aiden E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JR. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes JR. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Rao SS. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luger K, Dechassa ML. & Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13, 436–447, doi: 10.1038/nrm3382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh TH. et al. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell 162, 108–119, doi: 10.1016/j.cell.2015.05.048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risca VI, Denny SK, Straight AF. & Greenleaf WJ. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature 541, 237–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorigo B. et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573, doi: 10.1126/science.1103124 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Song F. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380, doi: 10.1126/science.1251413 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Hsieh TS, Fudenberg G, Goloborodko A. & Rando OJ. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat Methods 13, 1009–1011, doi: 10.1038/nmeth.4025 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Nora EP. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385, doi: 10.1038/nature11049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonev B. et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572 e524, doi: 10.1016/j.cell.2017.09.043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cremer M. et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol 463, 205–239, doi: 10.1007/978-1-59745-406-3_15 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Chepelev I, Wei G, Wangsa D, Tang Q. & Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javierre BM. et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384 e1319, doi: 10.1016/j.cell.2016.09.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumbach MR. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet, doi: 10.1038/ng.3963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HP, Imbert J. & Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev 17, 349–366, doi: 10.1016/j.cytogfr.2006.07.003 (2006). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 47.Goryshin IY. & Reznikoff WS. Tn5 in vitro transposition. J Biol Chem 273, 7367–7374 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Langmead B. & Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phanstiel DH, Boyle AP, Heidari N. & Snyder MP. Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics 31, 3092–3098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Javierre BM. et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837, doi: 10.1016/j.cell.2007.05.009 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Wang Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40, 897–903, doi: 10.1038/ng.154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein BE. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Gong K, Li Q, Alber F. & Zhou XJ. Hi-Corrector: a fast, scalable and memory-efficient package for normalizing large-scale Hi-C data. Bioinformatics 31, 960–962, doi: 10.1093/bioinformatics/btu747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin H. et al. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic acids research 44, e70, doi: 10.1093/nar/gkv1505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MD, McCarthy DJ. & Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whyte WA. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL. et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT. & Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. PETs information of Trac-looping libraries from human CD4 T cells.
Supplementary Table 2. Oligonucleotides sequences used in the study.