Abstract
Histone H2B monoubiquitylation (H2Bub1) has central functions in multiple DNA-templated processes, including gene transcription, DNA repair, and replication. H2Bub1 also is required for the trans-histone regulation of H3K4 and H3K79 methylation. Although previous studies have elucidated the basic mechanisms that establish and remove H2Bub1, we have only an incomplete understanding of how H2Bub1 is regulated. We report here that the histone H4 basic patch regulates H2Bub1. Yeast cells with arginine-to-alanine mutations in the H4 basic patch (H42RA) exhibited a significant loss of global H2Bub1. H42RA mutant yeast strains also displayed chemotoxin sensitivities similar to, but less severe than, strains containing a complete loss of H2Bub1. We found that the H4 basic patch regulates H2Bub1 levels independently of interactions with chromatin remodelers and separately from its regulation of H3K79 methylation. To measure H2B ubiquitylation and deubiquitination kinetics in vivo, we used a rapid and reversible optogenetic tool, the light-inducible nuclear exporter, to control the subcellular location of the H2Bub1 E3 ligase, Bre1. The ability of Bre1 to ubiquitylate H2B was unaffected in the H42RA mutant. In contrast, H2Bub1 deubiquitination by SAGA-associated Ubp8, but not by Ubp10, increased in the H42RA mutant. Consistent with a function for the H4 basic patch in regulating SAGA deubiquitinase activity, we also detected increased SAGA-mediated histone acetylation in H4 basic patch mutants. Our findings uncover that the H4 basic patch has a regulatory function in SAGA-mediated histone modifications.
Keywords: histone, histone modification, gene transcription, chromatin regulation, chromatin, yeast transcription
Introduction
The nucleosome, the fundamental repeating unit of chromatin, is the first level of chromatin organization, and it is essential for the regulation of nearly all DNA-templated processes. Composed of an octamer of histone proteins, two molecules each of histones H2A, H2B, H3, and H4, the nucleosome is a partial barrier to functions such as gene transcription and DNA repair; hence, nucleosomes must be disrupted transiently for these processes to occur. One major mechanism that contributes to the transient disruption of nucleosomes is histone post-translational modifications (PTMs).2 Histone PTMs are found both in the “tail” domains, which largely influence the recruitment of effector proteins (i.e. readers), and in their globular domains, which largely influence nucleosome-DNA interactions (1).
In addition to histone PTMs being major influencers of chromatin structure and function, histones also contain basic and acidic regions or “patches” that govern nucleosome– interactions or the ability of readers to engage the nucleosome. One such region is the nucleosome acidic patch, a negatively charged cavity formed between histones H2A and H2B that contributes to chromatin function by regulating association of a multiple chromatin-modifying enzymes (2, 3). Another region, the H4 basic patch located in the H4 tail domain (between residues 16 and 20), contains multiple basic residues, i.e. arginine, histidine, and lysine (see Fig. 1A). This segment of basic residues has important functions in regulating chromatin dynamics and multiple chromatin-modifying enzymes. For example, the H4 basic patch is required for efficient Dot1-mediated histone H3 lysine 79 di- and trimethylation (H3K79me2 and H3K79me3) (4, 5). Additionally, the basic patch has a key function in maintaining the balance of heterochromatin domains in yeast by interacting with the silencing protein Sir3 when H4K16 is unacetylated (5). Acetylation of K16 within the basic patch is critical in chromatin organization (6). Recently, the H4 basic patch has been shown to function as a positive regulator of the ISWI family of chromatin-remodeling complexes (7–10) and as a regulator of Snf2 (11, 12). Chd1, another remodeler that organizes nucleosomes across the coding regions of genes, also interacts with the H4 N terminus (13).
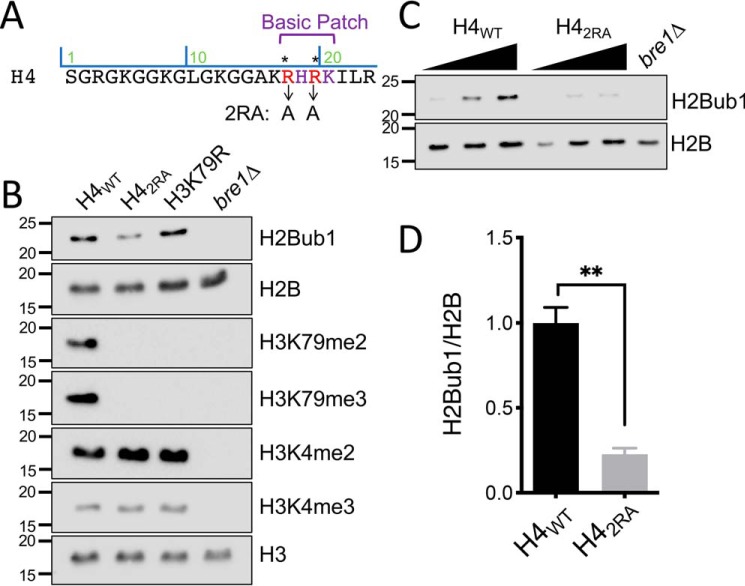
Figure 1.
The H4 basic patch is required for proper H2BK123 H2Bub1. A, sequence of the H4 N terminus with basic patch residues depicted. Arrows show the arginine residues mutated to alanine to create the basic patch mutant H42RA used throughout the study. B, immunoblotting was performed for the indicated proteins or modifications using extracts from cells that expressed WT H4 (H4WT) or the basic patch mutant (H42RA). Strains that had a mutation at H3 lysine 79 (H3K79R) or were deleted for the E3 ubiquitin ligase Bre1 (bre1Δ) were used as antibody controls for H3K79 methylation and H2Bub1, respectively. H3 was used as a loading control. C, representative immunoblot analysis of H4WT and H42RA at increasing concentrations for H2B or H2Bub1. D, relative quantification of H2Bub1/H2B from C. The means ± S.E. were analyzed from three biological replicates.
H2Bub1 is a dynamic histone PTM enriched at promoters and across the transcribed regions of genes. This modification is associated with transcription elongation and maintaining chromatin integrity through its conserved trans-histone regulation of H3K79me and H3K4me (14–20). In yeast, H2Bub1 is catalyzed by the E2/E3 ligases Rad6 and Bre1 (21–23) and is removed by the deubiquitinases Ubp8 and Ubp10. As a part of the SAGA coactivator complex, Ubp8, the catalytic subunit of the SAGA deubiquitinase (DUB) module, deubiquitinates H2Bub1 near the promoters and transcription start sites of virtually all expressed genes in eukaryotic cells to promote transcription by RNA polymerase II (24–27). This cycle of ubiquitylation and deubiquitination is important for the regulation of early elongation and the establishment of serine 2 phosphorylation on the C-terminal domain of RNA polymerase II (28, 29). Monomeric Ubp10 regulates the pattern of H2Bub1 within gene bodies and intergenic regions. Initially identified for its role in maintaining silencing of subtelomeric genes, Ubp10 has been shown to be important in maintaining global H2Bub1 levels (25, 30–32). Nune et al. (33) have described the coordinated activities of Ubp10 and the histone chaperone FACT in H2Bub1 deubiquitination and nucleosome disassembly and reassembly.
To further understand the function of the H4 basic patch, we investigated the possibility that there are yet-to-be elucidated aspects of histone PTM cross-talk involving this histone region. In this report, we show that the H4 basic patch is required to maintain proper H2Bub1 levels. Although the basic patch is required for Dot1-mediated H3K79me, we found that the ability of the H4 basic patch to regulate H2Bub1 is independent of Dot1 or H3K79 methylation. Furthermore, we found that the ability of the H4 basic patch to regulate H2Bub1 was also independent of its activity in regulating ATP-dependent chromatin remodelers known to be regulated by H4. We therefore examined the possibility that the H4 basic patch regulates some aspect of the H2Bub1 machinery that installs or removes this mark. Using an optogenetic tool to enable precise and rapid nuclear import or export of Bre1 (LINX-Bre1), we showed that although Bre1 installed H2Bub1 in WT and H4 basic patch mutants at comparable rates, the rate of removal of H2Bub1 by Ubp8 (but not by Ubp10) increased when the H4 basic patch was mutated. Because Ubp8 is a DUB module of the SAGA complex, we further examined and found that the H4 basic patch also contributes to the levels of H3 acetylation by SAGA. Collectively, our findings reveal an unexpected function of the H4 basic patch in negatively regulating the histone-modifying activities of SAGA.
Results
Mutation of H4 basic patch leads to reduced levels of H2B monoubiquitylation
Although studies have defined the relationship between H2Bub1 and H3K4 and H3K79 methylation, less is known about the nature of the histone cross-talk that regulates H2Bub1. In the course of our studies of the regulation of H3K79 methylation by the H4 basic patch (Fig. 1A), unexpectedly, we discovered by immunoblotting the mutation of H4 basic patch residues Arg17 and Arg19 to alanine (hereafter H42RA) also resulted in a significant reduction in H2Bub1 (Fig. 1B). As controls in these experiments, we included a deletion of BRE1 (bre1Δ), the H2Bub1 E3 ligase, and a mutation of H3K79 (H3K79R); these mutants confirmed the specificity of the histone modification-specific antibodies and verified that the H4 basic patch regulates H3K79 di- and trimethylation. We also observed the same reduction in H2Bub1 in a strain that expressed an H4 mutant lacking the entire basic patch (Δ16–20) and in a single H4-R17A mutant (data not shown). Because H4 basic patch regulation of H2Bub1 has not been reported, we sought to verify that this observation was not a property of a specific strain background. As shown in Fig. S1A, mutation of the same H4 basic patch residues in another H2A/H2B shuffle strain likewise impaired H2Bub1 levels.
Given the well-established function of the H4 basic patch in regulating Dot1-mediated H3K79 methylation, we next assessed whether the regulation of H2Bub1 by the H4 basic patch might be indirectly due to the ability of this region to regulate Dot1 and H3K79 methylation that, in turn, regulates H2Bub1. This idea was supported by a study of van Welsem et al. (34), who found that Dot1 promoted H2Bub1 formation when Dot1 levels were increased. However, we found that the deletion of DOT1 (dot1Δ) or mutation of H3K79 had no discernible effect on the levels of H2Bub1 (Fig. 1B and Fig. S1, A and B). Thus, the ability of the H4 basic patch to regulate H2Bub1 is independent of its regulation of Dot1 and H3K79 methylation. In sum, these data defined an important and previously unknown function for the H4 basic patch in regulating H2Bub1.
H4 basic patch mutant cells display phenotypes associated with the absence of H2Bub1
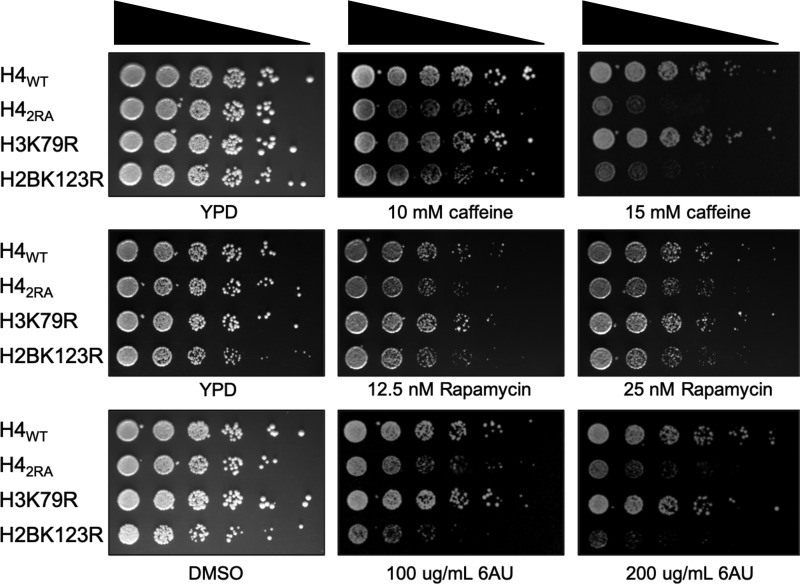
To determine the biological significance of the reduced levels of H2Bub1 in the H4 basic patch mutant, we spotted cells that expressed either H4WT, H42RA, H3K79R, or H2BK123R histones onto solid media containing various compounds to identify drug sensitivity phenotypes. As has been previously published, cells lacking all H2Bub1 (e.g. H2BK123R, rad6Δ, and bre1Δ), display sensitivity to various toxic agents including caffeine and 6-azauracil (35, 36). Interestingly, the H42RA mutant was also sensitive to these same agents as well as rapamycin (Fig. 2). In contrast, the H3K79R mutant, lacking all forms of H3K79 methylation, was not sensitive to these agents, confirming that the mechanism by which the H4 basic patch regulates H2Bub1 is independent of H3K79 methylation.
Figure 2.
Mutation of the H4 basic patch and H2BK123 share phenotypes associated with H2Bub1 loss. Overnight cultures of the indicated strains were spotted at 5-fold dilutions onto solid rich (YPD) medium or media containing the indicated chemotoxic agents and imaged after 2–3 days. 6AU, 6-azauracil.
H2Bub1 has a key function in the timely induction of the yeast GAL genes (37). To determine whether the H4 basic patch was also required for the induction of GAL genes like H2Bub1, we grew H4WT and H42RA cells in raffinose-containing medium prior to the addition of galactose (2% final concentration). Total RNA was isolated across the induction time course, and RT–quantitative PCR of the GAL1 locus was performed. Relative to the H4WT cells, H42RA cells displayed reduced accumulation of GAL1 transcripts during the time course (Fig. S2), suggesting that the H4 basic patch, like H2Bub1, is important for the proper regulation of GAL gene transcription. Collectively, these studies showed that the H4 basic patch is physiologically important, and its mutation phenocopies mutations in the H2Bub1 pathway.
The H4 basic patch and the ATP-dependent chromatin remodeler Chd1 regulate H2Bub1 by different mechanisms
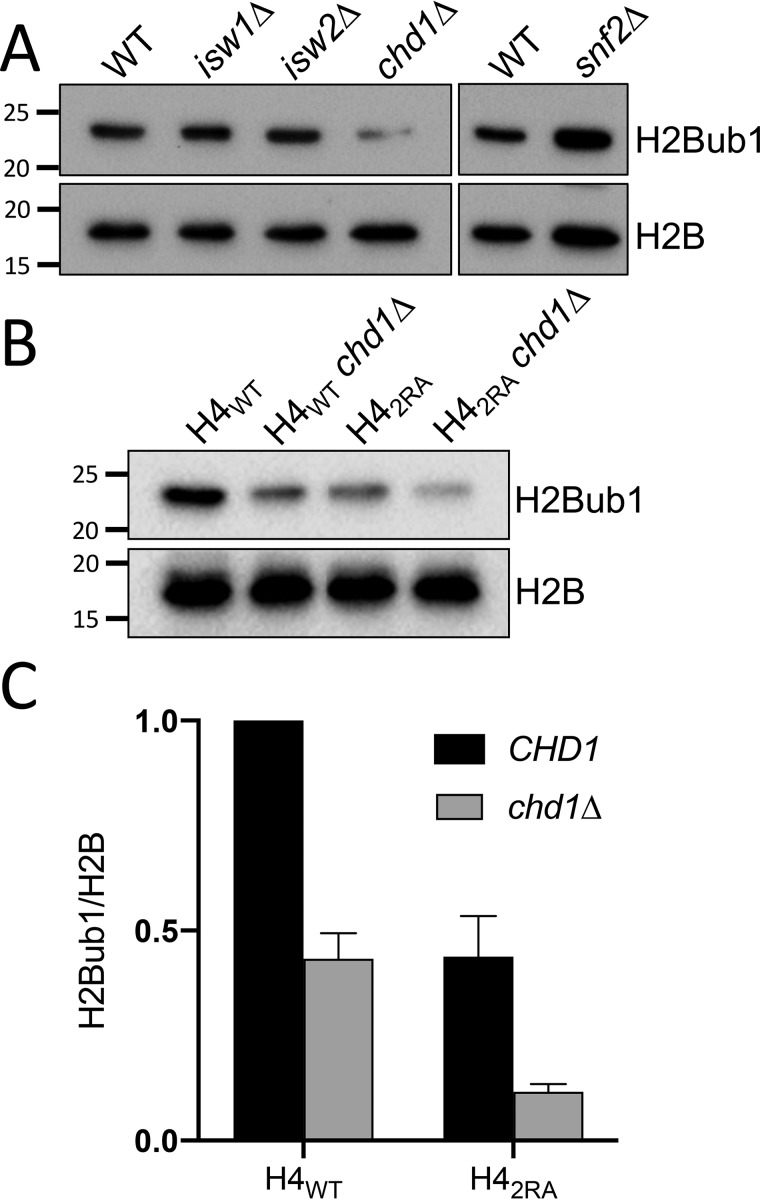
Lee et al. (38) demonstrated a function for the ATP-dependent remodeler Chd1 in regulation of H2Bub1. Additional studies show that the H4 basic patch regulates the activity of multiple chromatin remodelers including Chd1, Snf2, and Isw1. Therefore, we reasoned that a possible mechanism for how the H4 basic patch regulates H2Bub1 could be by H4 basic patch regulation of the activity of one of these remodelers, which, in turn, would regulate H2Bub1. To test this possibility, we measured global H2Bub1 levels in a panel of strains that contained individual deletions of the aforementioned chromatin remodelers (Fig. 3A). Consistent with previous studies, deletion of CHD1 (chd1Δ) resulted in a reduction of H2Bub1. In contrast to chd1Δ, however, none of the other remodeler deletions exhibited any loss of H2Bub1. To determine whether the loss of H2Bub1 in the H42RA strain was due to Chd1, we deleted CHD1 in our H4WT and H42RA strains and measured H2Bub1 levels in all strain combinations. As expected, we observed a decrease in H2Bub1 in the individual chd1Δ and H42RA mutants (Fig. 3B). Intriguingly, however, the loss of H2Bub1 in the H42RA chd1Δ double mutant was additive, suggesting that regulation of H2Bub1 by the H4 basic patch and Chd1 occurs by separate (nonepistatic) pathways.
Figure 3.
H4 basic patch regulation of H2Bub1 is independent of its interaction with chromatin remodelers. A, immunoblotting for H2Bub1 and H2B was performed using whole-cell extracts from WT or strains lacking the indicated chromatin remodelers. B, blots for H2Bub1 and H2B were derived from cells that expressed H4WT or H42RA in combination with a deletion of CHD1 (chd1Δ). C, relative quantification of H2Bub1/H2B from B. The means ± S.E. were analyzed from three biological replicates.
H4 basic patch regulation of H2B ubiquitylation is independent of the ubiquitylation machinery
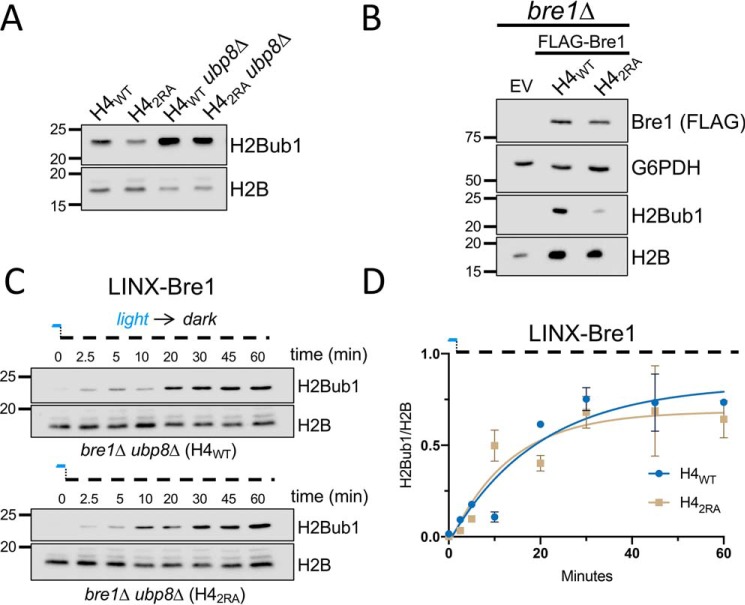
To define how the H4 basic patch regulates H2Bub1, we next considered the possibility that the region is important for Rad6/Bre1 to catalyze H2Bub1. Others have found that in the absence of one or more of the H2Bub1 DUBs, the ability of Rad6/Bre1 to properly incorporate H2Bub1 can be monitored (39, 40). More specifically, a defect in the ability of Bre1 to ubiquitylate H2B (e.g. if Lys123 is a poor substrate) resulted in reduced levels of H2Bub1 for which absence of a DUB could not compensate. Following this example, we created a deletion of UBP8 (ubp8Δ) in the context of either the H4WT or H42RA mutant and examined the levels of H2Bub1 by immunoblotting. Upon deletion of UBP8, the levels of H2Bub1 in the H42RA ubp8Δ cells was rescued to a level comparable with that in the H4WT ubp8Δ strain (Fig. 4A). These findings suggested that the ubiquitylation machinery in the H42RA mutant was functional and that the H4 basic patch was not essential for H2Bub1 catalysis.
Figure 4.
The H4 basic patch does not interfere with Bre1 ubiquitylation of H2B. A, immunoblot analysis of H2Bub1 and H2B in H4WT and H42RA in the presence or absence of UBP8 (ubp8Δ). The ability of Bre1 to increase H2Bub1 in the H42RA ubp8Δ strain suggested that the H4 basic patch mutant does not impair Bre1 catalysis. B, H4WT bre1Δ and H42RA bre1Δ cells were transformed with empty vector (EV) or ADH1-driven FLAG-BRE1 and subjected to immunoblot analysis with the indicated antibodies. Visualizing normal levels of Bre1 protein in the H42RA strain indicated that Bre1 function in catalysis was normal; Bre1 protein stability is normally diminished when Bre1 is unable to catalyze H2Bub1 (41). C, optogenetic-mediated control of the E3 ligase Bre1 (LINX-Bre1) reveals similar kinetics of H2B ubiquitylation in H4WT and H42RA. D, quantification of data from C. The means ± S.E. were analyzed from two biological replicates.
In addition to the above approach, another outcome that we observed for the absence of Bre1 catalysis was reduced Bre1 protein stability (41). To determine whether Bre1 protein level was reduced in the H42RA mutant, we expressed a FLAG-BRE1 construct in H4WT bre1Δ and H42RA bre1Δ cells and analyzed Bre1 levels (FLAG) and H2Bub1 levels by immunoblot (Fig. 4B). As expected, we did not detect H2Bub1 in the bre1Δ cells. In contrast, and upon plasmid rescue with our FLAG-BRE1 construct, we observed a restoration of H2Bub1 levels in H4WT that was reduced in the H42RA strain. This result indicated that our rescue experiment recapitulated our original observations on H2Bub1 regulation by the H4 basic patch. Importantly, we did not find any significant difference in Bre1 protein levels between the H4WT and H42RA strains, suggesting that Bre1 catalytic activity, specifically, and the ubiquitylation machinery, generally, were unaffected by the H42RA mutation.
Although the foregoing studies were informative, they were limited by the fact that they captured only final, steady-state levels of H2Bub1. Recently, for the study of dynamic epigenetic regulation, optogenetic tools have emerged that enable rapid and reversible nucleocytoplasmic shuttling to overcome the steady-state limitation. We used a previously characterized optogenetic tool developed in our lab, light-inducible nuclear export (LINX), that uses blue light to control the cellular localization of Bre1 (42). Briefly, when cells lacking BRE1 and UBP8 are transformed with the LINX-Bre1 construct and grown in blue light, LINX-Bre1 is rapidly sequestered in the cytoplasm and phenocopies a bre1Δ strain. Removal of blue light results in import of LINX-Bre1 into the nucleus, rendering the cells BRE1+ and enabling precise measurement of the in vivo kinetics of ubiquitylation and deubiquitination with unprecedented control and resolution. Thus, we measured the kinetics of LINX-Bre1–mediated ubiquitylation in H4WT and H42RA cells that lacked endogenous BRE1 and UBP8. These cells were grown to log phase in the presence of blue light before being switched to the dark to release Bre1 into the nucleus. As shown in Fig. 4C, H2B ubiquitylation occurred rapidly, consistent with published results (42). Significantly, we did not observe any significant difference in the kinetics of ubiquitylation between the H4WT and H42RA cells, providing strong, in vivo evidence that the ubiquitylation machinery in the H42RA mutant was fully functional and that the H4 basic patch regulates H2Bub1 independent of Bre1.
The H4 basic patch regulates SAGA-associated Ubp8 deubiquitination
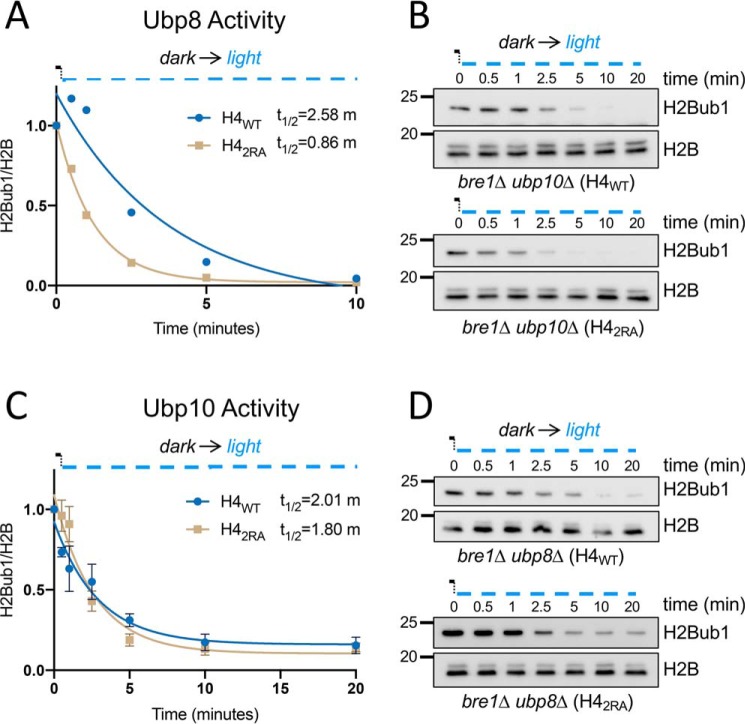
We next turned our attention to the effect of the H4 basic patch on H2Bub1 deubiquitination. We took advantage of the LINX-Bre1 system to measure the dynamics of deubiquitination. To distinguish between the activities of the two H2Bub1 DUBs, Ubp8 and Ubp10, we created H4WT bre1Δ and H42RA bre1Δ strains that lacked either UBP8 (bre1Δ ubp8Δ) or UBP10 (bre1Δ ubp10Δ). The cells were grown to log phase in the dark to maintain normal H2Bub1 by LINX-Bre1. Then we exposed the cells to blue light to rapidly remove LINX-Bre1 from the nucleus, preventing any subsequent ubiquitylation and enabling us to measure the kinetics of DUB activity in vivo. In the H4WT cells, the half-lives of the H2Bub1 mark in the presence of Ubp8 or Ubp10 were ∼2.58 and 2 min, respectively. The half-life of the H2Bub1 mark in H42RA cells in the presence of Ubp10 was similar to that in H4WT, ∼1.8 min. Strikingly, the half-life of the H2Bub1 mark in H42RA cells in the presence of Ubp8 decreased 3-fold (to less than 1 min; Fig. 5, A and C). This result suggested that the H4 basic patch regulates Ubp8, and it may limit the activity or accessibility of Ubp8. Given these findings, we also employed an in vitro DUB assay; we used cell extracts from yeast strains doubly deleted for UBP8 and UBP10 to generate high levels of H2bub1 as a substrate for recombinant Ubp8 and Ubp10. We assayed an affinity-purified Upb8-DUB module previously shown to interact with the nucleosome and affinity-purified Ubp10. These in vitro assays did not reveal a difference in the rates of deubiquitination between H4WT and H42RA cells (Fig. S3). Thus, the regulation of Ubp8 by the H4 basic patch may involve either a regulatory mechanism that is not captured in in vitro assays or that depends on the full SAGA complex and not the SAGA-associated DUB module alone.
Figure 5.
The H4 basic patch contributes to H2B deubiquitination by influencing Ubp8 but not Ubp10. A, H4WT and H42RA cells lacking UBP10 were grown to log phase in the dark to maintain LINX-Bre1 in the cytoplasm. Time point 0 was taken in the dark, and subsequent time points were collected after exposure to blue light. Immunoblot analysis of H2Bub1 and H2B was performed and quantified. The half-life measured from each curve was determined from single exponential fits of the data. B, representative immunoblots from A. C, H4WT and H42RA cells lacking UBP8 were grown to log phase in the dark to maintain LINX-Bre1 in the cytoplasm. Time point 0 was taken in the dark, and subsequent time points were collected after exposure to blue light. The data were processed as above. D, representative immunoblots from C.
The H4 basic patch regulates SAGA-dependent H3 acetylation
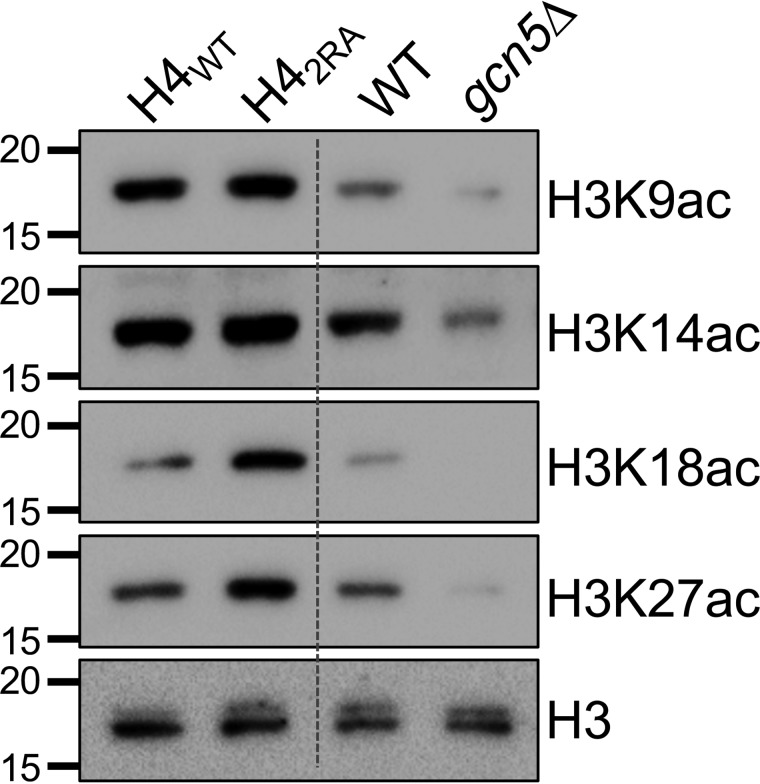
The SAGA complex consists of four distinct modules and two enzymatic functions: deubiquitination and lysine acetyltransferase (KAT) activities (43). The main target of the KAT module is the histone H3 N terminus, specifically lysine residues 9, 14, 18, 23, 27, and 36 (44). Although the KAT and DUB modules are distinct structurally, they are in close proximity to one another, and the DUB module modestly stimulates KAT activity (45). To determine whether the H4 basic patch also influences SAGA-dependent histone acetylation, we analyzed the histone acetylation levels at H3K9, K14, K18, and K27 in H4WT and H42RA cells alongside a gcn5Δ strain as a control (Fig. 6). Intriguingly, in the H42RA cells, we observed an increase in H3K18 and H3K27 acetyl levels but little change in H3K9 and H3K14 acetyl levels. However, H3K9 and H3K14 are also targets of other KATs (NuA3 and Rtt109, respectively). Of the acetylation sites tested, H3K18ac and H3K27ac are almost exclusively catalyzed by SAGA, as demonstrated by the complete loss of acetylation at these two sites. Thus, the selective increase in H3K18 and H3K27 acetylation in H42RA cells was likely due to SAGA-specific acetylation changes caused by the loss of H4 basic patch function. To eliminate the possibility that the H42RA mutant alters KAT activity more generally, we analyzed H3K56ac levels in the H4WT and H42RA cells; we did not find any difference between the strains (Fig. S4). Taken together, these data suggest that, in addition to regulating the DUB model in SAGA, the basic patch also regulates the SAGA KAT module. Thus, these findings document an important inhibitory function of the H4 basic patch in SAGA-associated histone modification functions.
Figure 6.
The H4 basic patch regulates SAGA-dependent H3 acetylation in addition to H2B deubiquitination. The results of immunoblot analysis of H3 N-terminal acetyl-lysine residues in H4WT and H42RA cells are shown. gcn5Δ and matched WT cells were included as controls.
Discussion
We have shown that, by negatively regulating SAGA-associated activities, the H4 basic patch ensures proper levels of H2Bub1 and H3 acetylation. The H4 basic patch regulates many other chromatin-modifying activities, e.g. Dot1-mediated H3K79 methylation and ISWI chromatin remodeling. However, we found that regulation of H2Bub1 and histone acetylation by the H4 basic patch were independent of its regulation of Dot1 or other chromatin remodelers that influence H2Bub1. Thus, these studies document a new function for the H4 tail and reveal another layer of cross-talk regulation of the SAGA complex.
Our study outlines a negative function of the basic patch on SAGA-associated histone-modifying activities. However, it remains unknown precisely how the H4 basic patch restrains the SAGA coactivator complex. Detailed cryo-EM studies of SAGA and the DUB module in SAGA showed that the histone acetylation and DUB modules are in close proximity to the nucleosomal core (45–49); however, the H4 tail was not resolved, and thus these studies do not provide insight into how the H4 basic patch interacts. Although our in vitro DUB assay with the Upb8-DUB module did not reveal a change in the H2Bub1 turnover rate when the H4 basic patch was altered (Fig. S3), our in vivo and time-resolved optogenetic studies did reveal a change in H2Bub1 turnover rate (Fig. 5). Thus, the H4 basic patch may be involved in modulating an upstream regulatory pathway during transcription that regulates SAGA activity, perhaps by an indirect mechanism, instead of regulating enzymatic activity by a physical interaction that was perturbed in the mutant. Interestingly, both H4-R17A and SAGA DUB module member Sgf11-R78 interact with acidic patch residue H2A-E64, and DUB activity in the Sgf11-R78A mutant was severely reduced (49, 50). We hypothesize that the H4 basic patch regulates H2Bub1 activity by competing with Sgf11 for association with the H2A acidic patch.
An additional, perhaps related possibility to explain this regulation is the fact that the H4 basic patch–H2A acidic patch interaction affects higher-order chromatin structure that may influence the accessibility of chromatin to chromatin-modifying enzymes. In this scenario, the mutation of the H4 basic patch would relieve its stabilizing effect on chromatin structure and “free” additional nucleosomes for SAGA binding, thereby increasing the overall H2Bub1 and H3 acetylation activities of the SAGA enzyme complex. The precise explanation for how this mechanism operates will be interesting to determine.
In sum, these studies reveal a novel regulatory mechanism for the activity of SAGA that likely is important for proper gene regulation and other functions associated with SAGA. Given the highly conserved nature of yeast, we expect that this function of the H4 tail is conserved in more complex eukaryotes.
Experimental procedures
Yeast strains
Strains and plasmids used in this study are listed in Tables S1 and S2. Strains used in Figs. 1 and 3A were derived from the parental histone shuffle strain, YAA524. All other histone shuffle mutants used were derived from the parental histone shuffle strain, yDT51. Gene disruptions and endogenous overexpression were performed as described (51) and verified by PCR and immunoblotting. Plasmids were generated using a standard site-directed mutagenesis protocol, and primers were designed with the Agilent QuikChange Primer Design tool.
Preparation of whole-cell extracts and immunoblots
Cells were collected by centrifugation and stored at −80 °C. Cell pellets were thawed on ice, resuspended in 200 μl of ice-cold TCA buffer (10 mm Tris 8, 10% TCA, 25 mm NH4OAc, 1 mm EDTA), moved to a microcentrifuge tube, and incubated on ice for 10 min. The samples were centrifuged at 13,000 rpm for 5 min at room temperature to collect precipitated protein. After aspirating the supernatant, the pellet was resuspended 70–100 μl of 0.1 m Tris, pH 11, 3% SDS and heated at 95 °C for 10 min followed by centrifugation at 13,000 rpm for 1 min to collect cellular debris. Protein concentration was measured using the Bio-Rad DC protein assay kit (catalog no. 5000112) and diluted to normalize concentrations across samples. The samples were diluted in 2× loading dye, and 10–15 μl of sample was subjected to SDS-PAGE followed by transfer to polyvinylidene difluoride membrane for immunoblot analyses.
Yeast spotting assays
Saturated overnight yeast cultures were diluted to an A600 of 0.5, and 5-fold serial dilutions of the cells were plated onto YPD or SC medium containing the indicated chemotoxic agents. The plates were imaged after 2–4 days at 30 °C.
Optogenetic and in vivo DUB assays
LINX-Bre1 optogenetic time courses were performed as described (42). Briefly, colonies of LINX-Bre1 transformed in bre1Δ, bre1Δubp8Δ, or bre1Δubp10Δ strains were grown overnight in SC-LEU–containing medium in the dark (Fig. 5) or in the presence of blue light (Fig. 4). Cell density was measured, diluted to an A600 of 0.5, and grown for 4 h in the same light condition as the overnight culture. Time courses began when cultures were moved from light to dark or vice versa. At each time point, the same volume of culture was collected and added to the appropriate volume of 100% TCA to yield a final TCA concentration of 20%, followed immediately by mixing and centrifugation at 5000 rpm for 5 min at 4 °C. The supernatant was aspirated, and the pellets were stored at −80 °C until processed for Western blotting analysis as detailed above.
Antibodies
Immunoblots were developed using ECL Prime (Amersham Biosciences catalog no. RPN2232). The following antibodies were used: ubiquityl-histone H2B (Cell Signaling Technologies catalog no. 5546; 1:5000), H2B (Active Motif catalog no. 39237; 1:5000), H3K79me3 (Abcam catalog no. 2621; 1:2500), H3K79me2 (Active Motif catalog no. 39143; 1:2500), H3K4me3 (EpiCypher catalog no. 13-0004; 1:5000), H3K9ac (Active Motif catalog no. 39917, 1:2500), H3K14ac (Millipore catalog no. 07-353; 1:2500), H3K18ac (Millipore catalog no. 07-354; 1:2500), H3K27ac (Abcam catalog no. ab4729; 1:2500), H3K56ac (Active Motif catalog no. 39281; 1:1000), H3 (EpiCypher catalog no. 13-0001; 1:5000), G6PDH (Sigma–Aldrich catalog no. A9521; 1:100,000), FLAG-M2 (Sigma-Aldrich catalog no. F1804; 1:5000), and Myc (Millipore; 1:2500). Rabbit (Amersham Biosciences catalog no. NA934; donkey anti-rabbit) and mouse (Amersham Biosciences catalog no. NA931; sheep anti-mouse) secondary antibodies were used at 1:10,000.
Galactose induction
Yeast strains that expressed either the WT or mutant histone H4 were grown overnight in medium containing 2% raffinose. Cell density was measured and diluted to an A600 of 0.25 and grown for 4 h in medium containing 2% raffinose. The induction time course was begun by adding 20% galactose to a final concentration of 2%. At each time point the same volume of each culture was harvested and centrifuged at 4500 rpm for 5 min. The supernatant was aspirated, and the cell pellets were stored at −80 °C.
Quantitative real-time PCR
RNA was isolated by a hot acid phenol method as described (52). Crude RNA was treated with DNase (Promega catalog no. M6101) followed by RNA cleanup (Qiagen RNeasy mini kit, catalog no. 74106). cDNA was synthesized from 500 ng to 1 μg of total RNA using random hexamer primers and Superscript reverse transcriptase III (Thermo Fisher Scientific, catalog no. 108-80044). The cDNA was diluted 1:25 before being subjected to real-time PCR (primers listed in Table S3). Quantitative RT-PCR was performed using the iTaq Universal SYBR Green Master mix according to the manufacturer's instructions (Bio-Rad, catalog no. 1725125), and the relative quantities of transcripts were calculated using the ΔΔCt method (53) with ACT1 as a control. The data shown are the replicates of three independent experiments with three technical replicates in each experiment.
In vitro DUB assays
Yeast strains that expressed either WT or mutant histone H4, FLAG-tagged histone H2B and lacking both UBP8 and UBP10 were grown overnight in 25 ml of YPD and harvested by centrifugation at 4500 rpm for 5 min. Cell pellets were frozen and stored at −80 °C. The pellets were resuspended in 500 μl of 10 mm Tris-Cl, pH 7.4, 300 mm sorbitol, 100 mm NaCl, 5 mm MgCl2, 5 mm EDTA, 10% glycerol, 0.1% Igepal-30 and split into two microcentrifuge tubes before being subjected to standard glass bead lysis by vortexing at 4 °C. The tubes were punctured with a push pin and placed into a clean tube and spun to 3000 rpm three times to separate the lysate from the beads. Final lysate was clarified by centrifugation at 13,000 rpm for 10 min at 4 °C. Protein concentration was determined using the Bio-Rad Bradford assay. To perform the assay, 50–100 μg of total protein was diluted to a final volume of 20 μl. A master mix containing enough sample to complete the experiment was prepared and preincubated at 30 °C with mixing for 30 min before addition of enzyme. The time course was begun by addition of recombinant Ubp8 DUB module (rDUBm) or Ubp10 enzyme. At the indicated time points, 20 μl of sample was removed and added to a microcentrifuge tube containing 5 μl of 5× loading dye and heated at 95 °C immediately. 20 μl of the sample was subjected to 12% SDS-PAGE, which was blotted and probed with an M2-FLAG primary antibody to detect both the FLAG-H2Bub1 and FLAG-H2B signal from the same membrane.
Author contributions
H. A. M., M. B. C., and B. D. S. conceptualization; H. A. M. data curation; H. A. M. validation; H. A. M. and A. M. L. investigation; H. A. M. visualization; H. A. M., A. M. L., and M. B. C. methodology; H. A. M. and B. D. S. writing-original draft; H. A. M., M. B. C., and B. D. S. writing-review and editing; A. M. L. and M. B. C. resources; B. D. S. supervision; B. D. S. funding acquisition; B. D. S. project administration.
Supplementary Material
Acknowledgments
We thank Strahl lab members, especially Raghuvar Dronamraju, Julia Difiore, and Glenn Wozniak, for technical support and Howard Fried for editorial suggestions. We thank Michael Morgan, Melesse Nune, and CynthiaWolberger for providing us with purified recombinant Ubp10 and Ubp8-DUB module enzyme. We also thank Jef Boeke, Mary Bryk and Fred Winston for yeast shuffle strains.
This work was supported by National Institutes of Health Grant GM126900 (to B. D. S.) and Cancer Cell Biology Training Program Grant T32CA071341 (to H. A. M.). B. D. S. acknowledges that he is a cofounder of EpiCypher, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1–S4.
- PTM
- post-translational modification
- H2Bub1
- histone H2B monoubiquitylation
- SAGA
- Spt-Ada-Gcn5 acetyltransferase coactivator
- LINX
- light-inducible nuclear exporter
- KAT
- lysine acetyltransferase
- DUB
- deubiquitinase.
References
- 1. Lawrence M., Daujat S., and Schneider R. (2016) Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32, 42–56 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 2. Kalashnikova A. A., Porter-Goff M. E., Muthurajan U. M., Luger K., and Hansen J. C. (2013) The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface. 10, 20121022 10.1098/rsif.2012.1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGinty R. K., and Tan S. (2015) Nucleosome structure and function. Chem. Rev. 115, 2255–2273 10.1021/cr500373h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fingerman I. M., Li H. C., and Briggs S. D. (2007) A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21, 2018–2029 10.1101/gad.1560607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altaf M., Utley R. T., Lacoste N., Tan S., Briggs S. D., and Côté J. (2007) Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28, 1002–1014 10.1016/j.molcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M. J., Davie J. R., and Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 10.1126/science.1124000 [DOI] [PubMed] [Google Scholar]
- 7. Clapier C. R., Längst G., Corona D. F., Becker P. B., and Nightingale K. P. (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell Biol. 21, 875–883 10.1128/MCB.21.3.875-883.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamiche A., Kang J. G., Dennis C., Xiao H., and Wu C. (2001) Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. U.S.A. 98, 14316–14321 10.1073/pnas.251421398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clapier C. R., Nightingale K. P., and Becker P. B. (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 30, 649–655 10.1093/nar/30.3.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clapier C. R., and Cairns B. R. (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492, 280–284 10.1038/nature11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Racki L. R., Naber N., Pate E., Leonard J. D., Cooke R., and Narlikar G. J. (2014) The histone H4 tail regulates the conformation of the ATP-binding pocket in the SNF2h chromatin remodeling enzyme. J. Mol. Biol. 426, 2034–2044 10.1016/j.jmb.2014.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X., Li M., Xia X., Li X., and Chen Z. (2017) Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 10.1038/nature22036 [DOI] [PubMed] [Google Scholar]
- 13. Sundaramoorthy R., Hughes A. L., El-Mkami H., Norman D. G., Ferreira H., and Owen-Hughes T. (2018) Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. Elife 7, e35720 10.7554/eLife.35720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., and Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 10.1074/jbc.C200348200 [DOI] [PubMed] [Google Scholar]
- 15. Sun Z. W., and Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- 16. Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., and Strahl B. D. (2002) trans-Histone regulatory pathway in chromatin. Nature 418, 498 10.1038/nature00970 [DOI] [PubMed] [Google Scholar]
- 17. Ng H. H., Xu R. M., Zhang Y., and Struhl K. (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 10.1074/jbc.C200433200 [DOI] [PubMed] [Google Scholar]
- 18. Fleming A. B., Kao C. F., Hillyer C., Pikaart M., and Osley M. A. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- 19. Chandrasekharan M. B., Huang F., and Sun Z. W. (2009) Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. U.S.A. 106, 16686–16691 10.1073/pnas.0907862106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weake V. M., and Workman J. L. (2008) Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 10.1016/j.molcel.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 21. Robzyk K., Recht J., and Osley M. A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–504 10.1126/science.287.5452.501 [DOI] [PubMed] [Google Scholar]
- 22. Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., and Shilatifard A. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 10.1016/S1097-2765(02)00802-X [DOI] [PubMed] [Google Scholar]
- 23. Kao C. F., Hillyer C., Tsukuda T., Henry K., Berger S., and Osley M. A. (2004) Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18, 184–195 10.1101/gad.1149604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Batta K., Zhang Z., Yen K., Goffman D. B., and Pugh B. F. (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 10.1101/gad.177238.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schulze J. M., Hentrich T., Nakanishi S., Gupta A., Emberly E., Shilatifard A., and Kobor M. S. (2011) Splitting the task: Ubp8 and ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 25, 2242–2247 10.1101/gad.177220.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonnet J., Wang C. Y., Baptista T., Vincent S. D., Hsiao W. C., Stierle M., Kao C. F., Tora L., and Devys D. (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28, 1999–2012 10.1101/gad.250225.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baptista T., Grünberg S., Minoungou N., Koster M. J. E., Timmers H. T. M., Hahn S., Devys D., and Tora L. (2017) SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell 68, 130–143.e5 10.1016/j.molcel.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., and Berger S. L. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 10.1101/gad.1144003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyce A., Xiao T., Whelan K. A., Kosman C., Walter W., Eick D., Hughes T. R., Krogan N. J., Strahl B. D., and Berger S. L. (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27, 275–288 10.1016/j.molcel.2007.01.035 [DOI] [PubMed] [Google Scholar]
- 30. Emre N. C., Ingvarsdottir K., Wyce A., Wood A., Krogan N. J., Henry K. W., Li K., Marmorstein R., Greenblatt J. F., Shilatifard A., and Berger S. L. (2005) Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell 17, 585–594 10.1016/j.molcel.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 31. Gardner R. G., Nelson Z. W., and Gottschling D. E. (2005) Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell Biol. 25, 6123–6139 10.1128/MCB.25.14.6123-6139.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orlandi I., Bettiga M., Alberghina L., and Vai M. (2004) Transcriptional profiling of ubp10 null mutant reveals altered subtelomeric gene expression and insurgence of oxidative stress response. J. Biol. Chem. 279, 6414–6425 10.1074/jbc.M306464200 [DOI] [PubMed] [Google Scholar]
- 33. Nune M., Morgan M. T., Connell Z., McCullough L., Jbara M., Sun H., Brik A., Formosa T., and Wolberger C. (2019) FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics. Elife 8, e40988 10.7554/eLife.40988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Welsem T., Korthout T., Ekkebus R., Morais D., Molenaar T. M., van Harten K., Poramba-Liyanage D. W., Sun S. M., Lenstra T. L., Srivas R., Ideker T., Holstege F. C. P., van Attikum H., El Oualid F., Ovaa H., et al. (2018) Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism. Nucleic Acids Res. 46, 11251–11261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song Y. H., and Ahn S. H. (2010) A bre1-associated protein, large 1 (Lge1), promotes h2b ubiquitylation during the early stages of transcription elongation. J. Biol. Chem. 285, 2361–2367 10.1074/jbc.M109.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klucevsek K. M., Braun M. A., and Arndt K. M. (2012) The Paf1 complex subunit rtf1 buffers cells against the toxic effects of [PSI+] and defects in rkr1-dependent protein quality control in Saccharomyces cerevisiae. Genetics 191, 1107–1118 10.1534/genetics.112.141713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao T., Kao C.-F., Krogan N. J., Sun Z.-W., Greenblatt J. F., Osley M. A., and Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell Biol. 25, 637–651 10.1128/MCB.25.2.637-651.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee J. S., Garrett A. S., Yen K., Takahashi Y. H., Hu D., Jackson J., Seidel C., Pugh B. F., and Shilatifard A. (2012) Codependency of H2B monoubiquitination and nucleosome reassembly on Chdl. Genes Dev. 26, 914–919 10.1101/gad.186841.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chandrasekharan M. B., Huang F., Chen Y.-C., and Sun Z.-W. (2010) Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol. Cell Biol. 30, 3216–3232 10.1128/MCB.01008-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cucinotta C. E., Young A. N., Klucevsek K. M., and Arndt K. M. (2015) The nucleosome acidic patch regulates the H2B K123 monoubiquitylation cascade and transcription elongation in Saccharomyces cerevisiae. PLoS Genet. 11, e1005420 10.1371/journal.pgen.1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wozniak G. G., and Strahl B. D. (2014) Catalysis-dependent stabilization of Bre1 fine-tunes histone H2B ubiquitylation to regulate gene transcription. Genes Dev. 28, 1647–1652 10.1101/gad.243121.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yumerefendi H., Lerner A. M., Zimmerman S. P., Hahn K., Bear J. E., Strahl B. D., and Kuhlman B. (2016) Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Nat. Chem. Biol. 12, 399–401 10.1038/nchembio.2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weake V. M., and Workman J. L. (2012) SAGA function in tissue-specific gene expression. Trends Cell Biol. 22, 177–184 10.1016/j.tcb.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cieniewicz A. M., Moreland L., Ringel A. E., Mackintosh S. G., Raman A., Gilbert T. M., Wolberger C., Tackett A. J., and Taverna S. D. (2014) The bromodomain of gcn5 regulates site specificity of lysine acetylation on histone H3. Mol. Cell Proteomics 13, 2896–2910 10.1074/mcp.M114.038174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Han Y., Luo J., Ranish J., and Hahn S. (2014) Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546 10.15252/embj.201488638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., Cohen R. E., and Wolberger C. (2010) Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328, 1025–1029 10.1126/science.1190049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Samara N. L., Ringel A. E., and Wolberger C. (2012) A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity. Structure 20, 1414–1424 10.1016/j.str.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Köhler A., Zimmerman E., Schneider M., Hurt E., and Zheng N. (2010) Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 10.1016/j.cell.2010.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morgan M. T., Haj-Yahya M., Ringel A. E., Bandi P., Brik A., and Wolberger C. (2016) Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 10.1126/science.aac5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koehler C., Bonnet J., Stierle M., Romier C., Devys D., and Kieffer B. (2014) DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt–Ada–Gcn5–acetyltransferase (SAGA). J. Biol. Chem. 289, 8989–8999 10.1074/jbc.M113.500868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- 52. Collart M. A., and Oliviero S. (2001) Preparation of yeast RNA. Curr. Protoc. Mol. Biol. Chapter 13, Unit 13.12 10.1002/0471142727.mb1312s23 [DOI] [PubMed] [Google Scholar]
- 53. Livak K. J., Wills Q. F., Tipping A. J., Datta K., Mittal R., Goldson A. J., Sexton D. W., and Holmes C. C. (2013) Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods 59, 71–79 10.1016/j.ymeth.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.