Abstract
Leukocyte recruitment is a universal feature of tissue inflammation and regulated by the interactions of chemokines with their G protein–coupled receptors. Activation of CC chemokine receptor 2 (CCR2) by its cognate chemokine ligands, including CC chemokine ligand 2 (CCL2), plays a central role in recruitment of monocytes in several inflammatory diseases. In this study, we used phosphoproteomics to conduct an unbiased characterization of the signaling network resulting from CCL2 activation of CCR2. Using data-independent acquisition MS analysis, we quantified both the proteome and phosphoproteome in FlpIn-HEK293T cells stably expressing CCR2 at six time points after activation with CCL2. Differential expression analysis identified 699 significantly regulated phosphorylation sites on 441 proteins. As expected, many of these proteins are known to participate in canonical signal transduction pathways and in the regulation of actin cytoskeleton dynamics, including numerous guanine nucleotide exchange factors and GTPase-activating proteins. Moreover, we identified regulated phosphorylation sites in numerous proteins that function in the nucleus, including several constituents of the nuclear pore complex. The results of this study provide an unprecedented level of detail of CCR2 signaling and identify potential targets for regulation of CCR2 function.
Keywords: phosphoproteomics, cell signaling, mass spectrometry (MS), actin, cytoskeleton, GTPase-activating protein (GAP), guanine nucleotide exchange factor (GEF), nuclear pore, chemokine, G protein-coupled receptor (GPCR), CCL2, CCR2, data-independent acquisition, C-C chemokine receptor type 2 (CCR2), C-C chemokine ligand type 2 (CCL2), signaling networks, data-independent acquisition mass spectrometry (DIA-MS)
Introduction
A universal feature of inflammatory diseases is the excessive migration of leukocytes to the affected tissues. Leukocyte recruitment is regulated by chemokines, which are secreted at the site of injury or infection and then activate chemokine receptors, G protein–coupled receptors (GPCRs)4 expressed on the target leukocytes (1–3). CCR2 is the major chemokine receptor on monocytes and macrophages, which play key roles in numerous inflammatory diseases and also express a variety of other chemokine receptors (4). The primary cognate ligands of CCR2 are considered to be the monocyte chemoattractant proteins (CCL2/MCP-1, CCL7/MCP-3, and CCL8/MCP-2 as well as CCL13/MCP-4 in humans or CCL12/MCP-5 in mice).
Animal models have consistently shown that suppression or inhibition of chemokine receptors (or chemokines) ameliorates the symptoms of inflammation in disease models. However, numerous clinical trials of chemokine receptor antagonists have failed, in many cases due to lack of efficacy in phase II or III (5, 6). Among the many factors contributing to the failure of clinical trials, one is the complexity of chemokine signaling networks (6). For example, most leukocytes express multiple chemokine receptors with interconnected intracellular signaling networks. Thus, inhibition of one receptor could be bypassed by compensatory signaling via a different receptor. Consequently, it may be advantageous to block certain intracellular effectors or pathways, perhaps in combination with receptor inhibition to avoid such compensatory effects. To identify appropriate targets for such an approach, we need a holistic understanding of the signaling networks stimulated by chemokine receptor activation.
Genetic deletion of CCR2 or silencing of CCL2 protects mice from developing atherosclerosis (7–10). Moreover, in a phase II clinical trial, inhibition of CCR2, using a humanized mAb, significantly reduced levels of C-reactive protein, an atherosclerosis biomarker (11). In type 2 diabetes, macrophage infiltration into adipose tissue is promoted by activation of CCR2. Thus, knockout or inhibition of CCR2 in mouse models reduced body mass and plasma glucose and restored insulin sensitivity (12), and CCR2 blockade lowered plasma glucose in type 2 diabetic patients (13). More than 15 clinical trials have targeted CCR2 for a broad range of indications (14), although no inhibitor has reached the clinic to date.
To gain comprehensive insights into signal transduction downstream of CCR2 activation, we have performed an unbiased systems-level phosphoproteomic analysis using data-independent acquisition (DIA) MS, which is the state-of-the-art approach to quantify thousands of peptides and proteins with superior quantitative accuracy (15–17). Specifically, we characterized the global phosphoproteome in CCR2-expressing human endothelial kidney (HEK293) cells in response to an activation time course with the principal endogenous chemokine agonist CCL2. This approach represents an advance on traditional candidate-based second-messenger assays proximal to receptor activation and offered opportunities to identify new nodes in the CCR2 signaling network. Indeed, the extensive signaling network identified from the protein phosphorylation data included expected chemokine receptor signaling pathways as well as numerous additional proteins, pointing to potential novel targets related to CCR2 signaling.
Results
Experimental design to identify the phosphorylation events underlying CCR2 activation
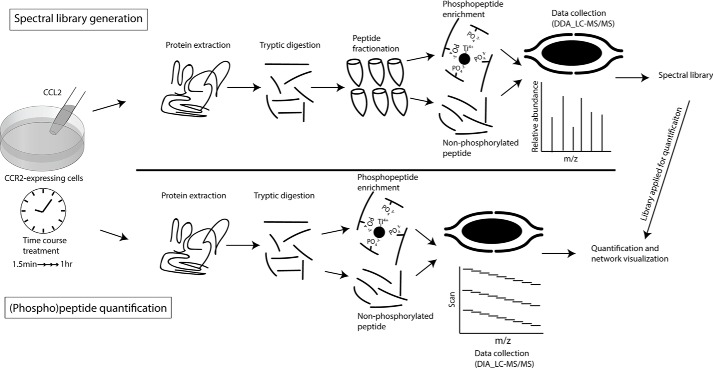
We have previously generated a CCR2-expressing cell line (FlpIn-HEK293T-CCR2) to investigate chemokine-mediated second-messenger signaling, including cAMP inhibition, β-arrestin recruitment, and internalization (18). Building on these findings, we sought to characterize the signaling networks stimulated by CCR2 activation in greater detail using phosphoproteomics. To study the kinetics of the underlying phosphorylation events, we performed a time course experiment in which FlpIn-HEK293T-CCR2 cells were treated with the full agonist CCL2 at a concentration (100 nm) that maximally activates CCR2 (18) for 1.5, 3, 6, 12, 25, and 60 min prior to harvest. Unstimulated FlpIn-HEK293T-CCR2 cells served as negative control, and each time point—including the 0-min control—was analyzed in biological triplicates (Fig. 1). The extracted proteins from each sample were digested with trypsin, and phosphopeptides were enriched using TiO2 chromatography after 10% of the tryptic digest had been removed for global proteome analysis. Both the enriched phosphopeptides and the aliquots of tryptic peptides were analyzed and quantified by DIA MS. Compared with other methods of peptide quantification, the DIA approach has improved accuracy and reproducibility and is not biased toward the most abundant proteins/peptides (19). However, quantification of peptides from DIA data must be accomplished by comparison of LC retention times and mass spectra with an independently established reference library (20).
Figure 1.
A schematic overview of (phospho)peptide spectral library generation and quantification. CCR2-expressing cells were stimulated with CCL2, and protein was extracted at multiple time points for tryptic digestion. For the generation of the spectral libraries using data-dependent acquisition MS (top half), the peptides were fractionated using basic-pH HPLC, and the phosphopeptides in each fraction were enriched using TiO2 chromatography after a small aliquot of tryptic peptides had been removed for global proteome analysis. For (phospho)peptide quantification by DIA MS (bottom half), the phosphopeptides were directly enriched with TiO2 beads after a small aliquot of tryptic peptides had been removed for global proteome analysis. The acquired spectral libraries were utilized for quantification.
To faithfully and accurately evaluate the DIA data, we generated comprehensive spectral libraries by subjecting a fourth replicate of each time point to off-line, basic-pH HPLC-C18 fractionation (21). A total of 42 fractions (7 time points × 6 fractions) for both tryptic peptides and enriched phosphopeptides were analyzed by data-dependent acquisition (DDA) MS, and the results were combined into two distinct libraries containing the spectral and chromatographic information for 115,093 peptides (derived from 8,164 proteins) for the analysis of the global proteome data and 30,586 phosphopeptides (derived from 4,831 proteins) for the analysis of the phosphoproteome data.
CCR2 activation induces profound changes in the phosphoproteome
By interrogating our DIA data with these comprehensive spectral libraries using Spectronaut 10 (Biognosys), we were able to obtain quantitative information for 7,696 proteins (Table S1) and 19,931 phosphosites falling below a false discovery rate (q value) cut-off of 1% in at least one of the 21 samples (7 time points × 3 replicates; Table S2). However, to increase the confidence and stringency of the phosphoproteomics interpretation, we restricted all subsequent analyses to the 11,847 phosphopeptides that were quantified in at least 15 of the 21 samples (i.e. for which a maximum of six values had to be imputed). The associated coefficients of variation were observed to be around 10 and 25% for the proteome and phosphoproteome analysis, respectively, which demonstrates high quantitative precision and low variability between the biological replicates of each time point (Fig. S1).
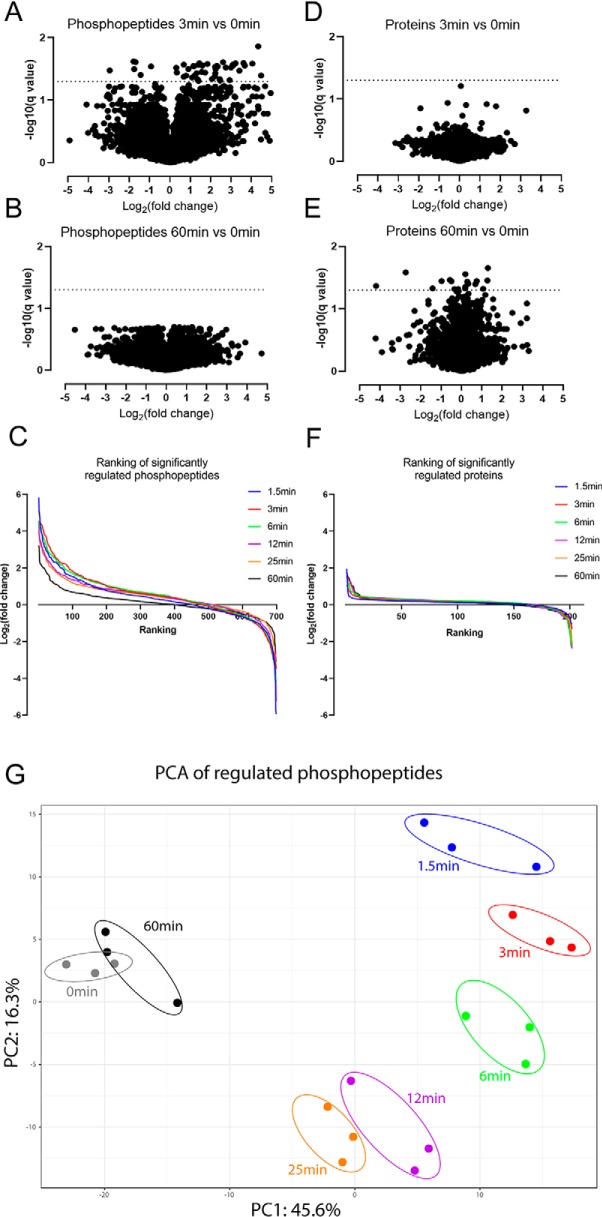
Differential expression analysis using a one-way analysis of variance (ANOVA) test revealed that the measured levels of 738 phosphosites (located on 466 proteins) and 202 proteins showed significant changes (i.e. were significantly regulated) within 60 min of CCR2 activation by CCL2 (adjusted p value <0.05; Tables S1 and S2). Importantly, only 5% of the significantly regulated phosphosites are located on proteins whose expression levels were observed to change, confirming that the majority of the observed changes in the phosphoproteome are not driven by changes in protein expression. Moreover, differential expression analyses showed that, compared with the phosphosites, the expression levels of proteins generally did not change until 60 min after CCL2 treatment, which is in agreement with the expectation that alterations in protein expression levels are much slower than changes in phosphorylation patterns (Fig. 2, A–F).
Figure 2.
CCL2-dependent phosphorylation changes arise rapidly compared with protein level changes. A and B, volcano plots representing the changes of all quantified phosphopeptides (log2(-fold change), x axis) and their statistical significance (−log10(q value), y axis) at 3 min (A) and 60 min (B) relative to unstimulated control (0 min). C, the quantified phosphopeptides at each time point have been ranked according to their log2 -fold change (relative to control) and plotted in a decreasing fashion along the x axis. D and E, volcano plots representing the changes in protein levels and their statistical significance at 3 min (D) and 60 min (E) relative to control. F, the quantified proteins at each time point have been ranked according to their log2 -fold change (relative to control) and plotted in a decreasing fashion along the x axis. G, principal component analysis (three biological repeats for each time point) for the significantly regulated phosphopeptides.
Nevertheless, phosphopeptides on significantly regulated proteins were removed from further analysis, which reduced the number of significantly regulated phosphosites to 699 (located on 441 proteins). A principal component analysis based on these 699 high-confidence phosphopeptides (Fig. 2G) revealed a time-dependent clustering pattern, which highlights the profound and rapid changes occurring in the phosphoproteome even after a short time of CCL2 exposure (1.5–3 min). These changes then gradually reversed over time to approach the untreated steady-state equilibrium after 60 min.
Gene ontology and pathway enrichment analysis
To understand the biological pathways and signal transduction networks affected by CCR2 activation, we subjected the 441 proteins that carry the 699 significantly regulated phosphosites to gene ontology analyses and to enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (22) (Table S3). As discussed below, these analyses revealed that there is a prevalence of regulated protein phosphorylation in a number of pathways known to be involved in chemokine signaling but also highlighted some groups of proteins not usually associated with chemokine receptor activation.
Signaling via canonical pathways
As expected, our set of regulated phosphoproteins is enriched for proteins in several signal transduction pathways known to be activated upon CCR2 (or other chemokine receptor) stimulation. These include the mitogen-activated protein kinase/extracellular signal–regulated kinase (MAPK/ERK) pathway, the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) pathway, and the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway (23–25).
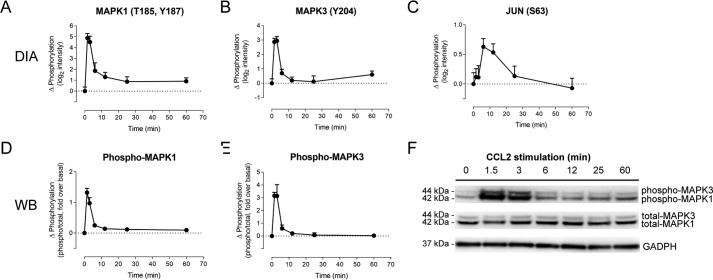
The enrichment of proteins in these pathways provides some internal validation for the accuracy of the phosphoproteomics data. However, as a means to independently validate our data set, we also sought to detect some of these phosphorylation changes using Western blotting, although such experiments were limited by the scarcity (and often questionable quality) of commercially available antibodies recognizing distinct phosphorylation sites. One of the hallmarks of chemokine activation is the phosphorylation of the two MAPK/ERK isoforms (MAPK1 and MAPK3). As shown in Fig. 3, Western blotting data are in excellent agreement with the quantitative proteomics (DIA) data, both convincingly demonstrating that MAPK1 and MAPK3 are rapidly phosphorylated upon CCR2 activation, peaking at 1.5–3 min after stimulation and then returning to baseline levels by 6–12 min after stimulation (Fig. 3, A–D and F). Also, as expected, DIA data indicated that phosphorylation of c-JUN, a downstream target of MAPK1/3 (26), peaks shortly after maximum ERK-1/2 phosphorylation (Fig. 3E).
Figure 3.
CCL2-dependent MAPK/ERK phosphorylation is quantitatively measured using DIA phosphoproteomics. CCL2-dependent changes in phosphorylation relative to unstimulated cells are shown, as a function of treatment time, for MAPK1 at position Tyr-204 (Y204), determined by DIA (A); MAPK3 at positions Thr-185 and Tyr-187 (T185/Y187) and at Tyr-187 alone (Y187), determined by DIA (B); MAPK1, at position Tyr-202 (Y202) and/or Tyr-204 (Y204), determined by Western blotting (WB) (C); and MAPK3, at positions Thr-185 and/or Tyr-187 (T185/Y187), determined by Western blotting (D). E, changes in phosphorylation of JUN at position Ser-63 (S63), determined by DIA. F, image of a representative Western blotting. Molecular weight markers have been cropped out; predicted molecular weights of the proteins are indicated. All data points for A–E are mean ± S.E. (error bars) from three biological replicates.
As a first step toward validation of our observations in monocytes, we have used Western blotting to quantify phosphorylation of several proteins (MAPK1/3, JUN, STAT3, and RPS6KA1) in THP-1 cells, an immortalized monocyte cell line that endogenously expresses CCR2. We observed good agreement between the time courses of phosphorylation in THP-1 cells and the time courses observed for the same phosphorylation sites in our DIA phosphoproteomics data set (Fig. S2).
Phosphorylation of actin- and GTP-related proteins
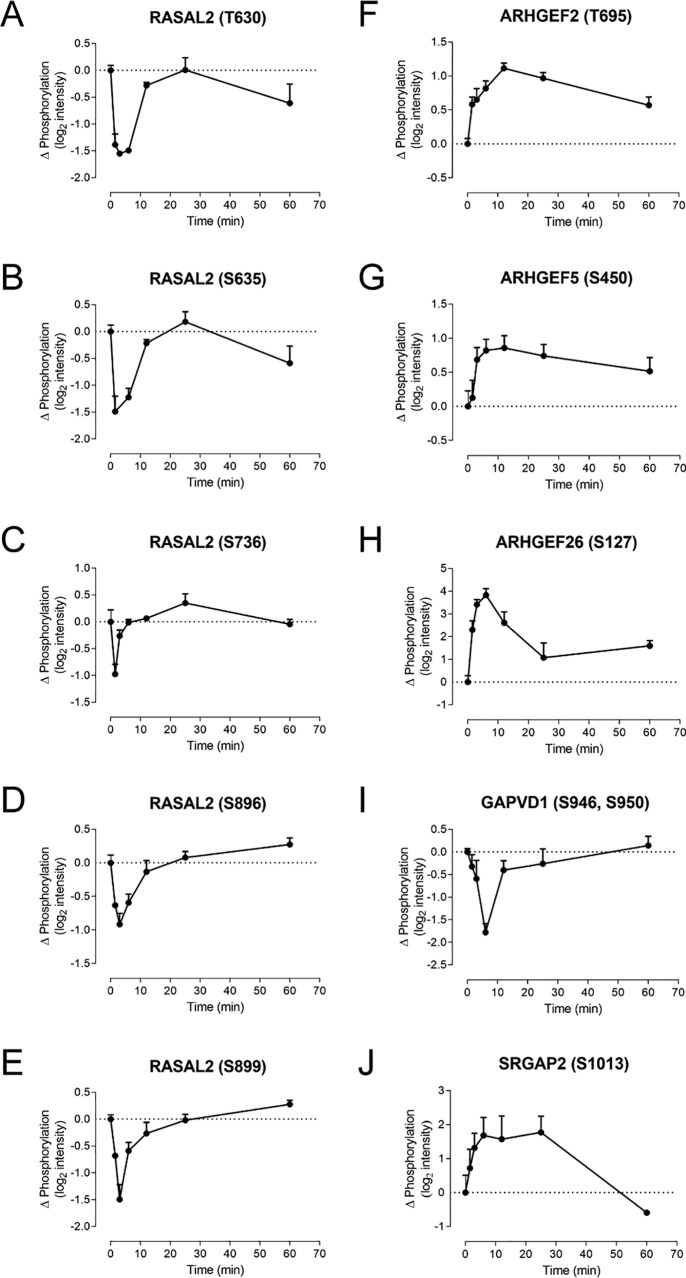
The regulated phosphoproteome was highly enriched in proteins involved in the formation of the actin cytoskeleton, cytoskeletal (re)organization, and cytoskeleton-related functions such as cell adhesion or endocytosis (Table S4). Because chemokines regulate cell migration and adhesion, these observations help to further validate the phosphoproteomics data set. Moreover, the reorganization of actin filaments is known to be controlled by the Ras family GTPases RHO, RAC, and CDC42. Although we did not directly observe phosphorylation of these proteins, we found that phosphorylation was regulated for a number of associated proteins. For example, we observed five significantly regulated phosphorylation sites in the Ras GTPase-activating protein nGAP (RASAL2), all showing similar time courses of early (1.5–6-min) decreases in phosphorylation before returning to baseline (Fig. 4, A–E). In addition, we observed significantly regulated phosphorylation for numerous guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), which are positive and negative modulators, respectively, of GTPase activity (Fig. 4 (F–J) and Fig. S3).
Figure 4.
CCL2 promotes phosphorylation of GEFs and GAPs. A–J, phosphorylation time courses in CCR2-expressing cells for selected GAPs and GEFs, including multiple phosphopeptides for RASAL2 (A–E) as well as ARHGEFs (F–H), GAPVD1 (I), and SLIT-ROBO Rho GTPase-activating protein 2 (SRGAP2) (J). Data represent CCL2-dependent changes in phosphopeptide intensity relative to unstimulated cells, mean ± S.E. (error bars) from three biological replicates.
Phosphorylation of nuclear-related proteins
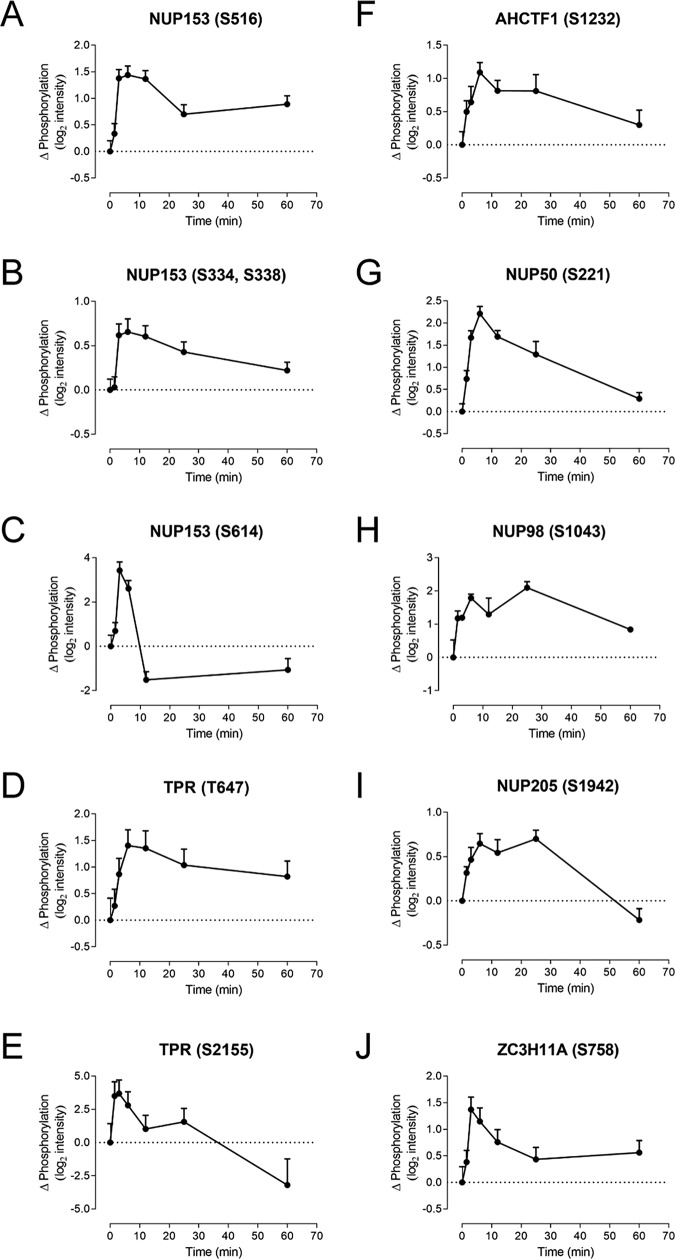
In addition to the above effects of chemokine receptor activation, we also observed phosphorylation (or dephosphorylation) for many proteins that exert functions in the nucleus, including DNA replication and repair, transcriptional regulation, RNA processing, and cell cycle regulation. Related to these, the set of regulated phosphoproteins was highly enriched in components of the nuclear pore complex and other proteins involved in nuclear export (Table S4). Among these, nuclear pore proteins NUP153 and TPR had six and four significantly regulated phosphosites, respectively (Fig. 5, A–E). In addition, whereas some of the regulated nuclear proteins underwent relatively fast changes in phosphorylation (within ∼6 min), others showed substantially slower kinetics and/or remained significantly different from baseline after 25 or 60 min (Fig. 5 and Fig. S4). To our knowledge, the regulation of nuclear protein phosphorylation after activation of CCR2 (or other chemokine receptors) has not been previously studied in detail. Its potential significance is discussed below.
Figure 5.
CCL2 promotes phosphorylation of nuclear-related proteins. Shown are phosphorylation time courses in CCR2-expressing cells for selected nuclear pore complex proteins and proteins involved in nuclear export. These include multiple phosphosites for nuclear pore complex protein NUP153 (A–C) and nucleoprotein TPR (D and E) as well as individual sites for additional nuclear pore complex proteins (F–I) and the mRNA export protein zinc finger CCCH domain–containing protein 11A (ZC3H11A) (J). Data represent CCL2-dependent changes in phosphopeptide intensity relative to unstimulated cells, mean ± S.E. (error bars) from three biological replicates.
A proposed CCL2-CCR2 signaling network
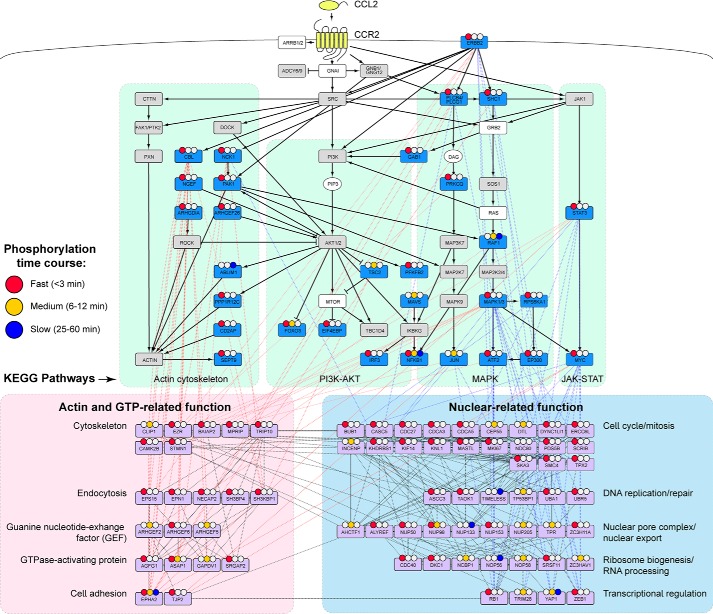
To provide further insights into the signaling pathways resulting from CCR2 activation, we have generated a network of interconnected proteins whose phosphorylation status changes between any of the measured time points (Fig. 6). This network was assembled in several steps. First, the enriched KEGG pathways were connected with each other and with known CCR2 proximal signaling events. Second, we added regulated phosphoproteins that were identified by interrogation of the STRING protein-protein interaction database to have at least three connections to members of the initial network; many of these were also connected to each other. Subsequently, the network was manually curated and organized to indicate the functional clusters discussed above.
Figure 6.
CCL2-CCR2 phosphoproteomic signaling network. Proteins with significantly regulated phosphorylation in response to CCL2 stimulation (one-way ANOVA, FDR < 0.05) were manually curated into a signaling network. KEGG pathways, indicated by green shaded areas and black lines, include proteins with significantly regulated phosphosites (blue boxes), proteins with phosphosites that were observed but not regulated (gray boxes), proteins with no observed phosphosites (white boxes), and nonprotein components (white ovals). The black solid lines connecting these proteins represent activating (arrowhead) or inhibitory (blunt head) interactions. Many additional proteins (light purple boxes) that have not been associated with chemokine receptor signaling but have known interactions with at least three other proteins in the network are broadly grouped into actin-related and nuclear-related biological processes (pink and blue shaded areas, respectively). Interactions between such proteins and KEGG pathway proteins are shown as red and blue dashed lines. Inter- and intramolecular interactions between nuclear-related and actin/GTP-related proteins shown as black dashed lines. Regulated phosphorylation events were classified as fast, medium, or slow, based on the timing of the maximal phosphorylation response relative to baseline (colored circles).
To assist in illustrating the temporal dynamics of phosphorylation across this network, we classified all 699 regulated phosphosites into three groups (fast, medium, and slow responders) based on the time when the changes reached their maximal amplitudes compared with the baseline control (colored circles in Fig. 6; see Table S2). The majority of the phosphosites (74%) reached their maximal amplitude within 3 min (fast responders), 20% showed maximal changes after 6–12 min (medium responders), and only a very small proportion (7%) reached maximal changes after 25–60 min. Fast, medium, and slow responders were fairly evenly distributed throughout the network without any apparent spatial-temporal pattern, and some proteins even carry multiple phosphosites that reach their maximal (de)phosphorylation levels after substantially different time periods; examples include transcription factor NFκB1, pleiotropic Ser/Thr kinase PRKD1, and cell adhesion–associated kinase EPHA2 (Fig. 6). These observations accentuate the complexity of the CCR2 signal transduction network.
Discussion
Signaling via pro-inflammatory chemokine receptors is critical for the stimulation of leukocyte migration in numerous inflammatory diseases. However, most studies of chemokine receptor signal transduction have focused on detection of well-characterized pathways and effectors (e.g. activated G proteins, cAMP levels, phosphorylated MAPK1/3, Ca2+ mobilization, and β-arrestin recruitment) or cellular properties (e.g. chemotaxis and proliferation) as readouts for receptor activation. A smaller number of studies have investigated specific features of the pathways that connect receptor activation to cellular outcomes (27–30). In this study, to gain new insights into the mechanisms of CCR2-activated signal transduction, we have taken the unbiased phosphoproteomics approach to identify numerous phosphorylation (and dephosphorylation) events resulting from CCR2 activation. This approach has the potential to corroborate prior observations as well as reveal new aspects of the CCR2 signaling network. Nonetheless, although the discovery proteomics approach used here provides valuable insights into the dynamics of CCL2-stimulated phosphorylation events, it should be noted that any information regarding the subcellular localization of these signaling events is lacking. This will be an area of interest for future investigation.
We identified nearly 700 phosphopeptides that are significantly regulated in response to CCR2 activation. In addition to well-known signal transduction pathways, our data clearly demonstrate that CCR2 activation substantially affects actin-, GTP-, and nuclear-related proteins. Importantly, a significant proportion of these identified phosphopeptides have not previously been linked to chemokine signaling, and some may be viable targets to interfere with or disrupt chemokine signaling.
It is noteworthy that we observed phosphorylation of the epidermal growth factor receptor (EGFR) family member ERBB2 and several proteins known to be phosphorylated downstream of EGFRs (Fig. 6). This is consistent with previous reports of cross-talk between chemokine receptors and EGFRs (31–34). We therefore speculate that such cross-talk also occurs for CCR2, although the details are likely to be cell type–specific.
In a previous phosphoproteomic study of chemokine signaling in breast cancer stem cells, Yi et al. (30) described the effects of the chemokine CXCL12 (stromal cell–derived factor-1, SDF-1), which activates the receptor CXCR4 to promote cancer metastasis. They reported phosphorylation changes for numerous kinases and used these to construct a putative MAPK signaling cascade. Despite the differences in the cell types, the chemokine-receptor pair, and the experimental and data analysis methods used, the results of this previous study are broadly consistent with the observations described herein. Notably, several proteins with regulated phosphorylation were identified in both studies, including MAP1A and MAP1B (involved in regulation of microtubule dynamics), protein kinase A–anchoring proteins (involved in chemotaxis), and catenin (involved in cell adhesion). Moreover, Yi et al. (30) also reported phosphorylation changes for some GEFs and NUPs, albeit substantially fewer than observed in our data set. The similarities between the findings of these studies suggest that the common pathways identified have general significance for mediating the responses of diverse cell types to chemokine receptor stimulation.
Our phosphoproteomics data set highlighted the involvement of GTP-related proteins in CCR2 signaling. Several GAPs and GEFs have been previously implicated in chemokine signaling. In one well-characterized example, stimulation of T lymphocytes by the chemokine CXCL12, the ligand for CXCR4, induced tyrosine phosphorylation of three Rho guanine nucleotide exchange factors (ARHGEFs)—SOS1, ARHGEF1, and DOCK2—giving rise to integrin presentation and consequent cell adhesion and arrest (35). In another study, CCL2 was found to induce tyrosine phosphorylation of p115 RhoGEF (ARHGEF1), which was implicated in the migration and proliferation of human aortic smooth muscle cells in response to CCL2 (27). Although the current study did not detect significant regulation of these specific factors, it identified a number of additional GAPs and GEFs that responded to CCL2 stimulation (Fig. 4 and Fig. S3) but have not previously been associated with chemokine signaling. Importantly, several of these GAPs and GEFs are expressed in monocytes, based on RNA-Seq data reported for THP-1 monocytes (36), including ARHGAP21, ARHGAP45, ARHGEF2, ARHGEF5, ARHGEF6, and ARHGEF11, suggesting that the regulated phosphorylation observed here may also occur in cells that endogenously express CCR2. Interestingly, the majority of the GTPase regulatory proteins underwent rapid changes in phosphorylation after CCL2 stimulation and remained in their modified state until 12 or 25 min after stimulation before returning to their basal phosphorylation levels. This is consistent with the typical time course of leukocyte chemotaxis, in which initial responses may be detected within ∼30 min but continue for several hours (37).
The current study also revealed phosphorylation changes for numerous proteins with nuclear-related functions. This is also expected, considering that several of the known (and observed) signaling pathways regulate the activation of transcription factors and that chemokine stimulation is known to up-regulate transcription and protein synthesis, as well as cell proliferation (38). Thus, we observed regulated phosphorylation of proteins related to transcriptional regulation, RNA processing, and aspects of cell division, such as cell cycle regulation, DNA replication or repair, and ribosome biogenesis. Interestingly, among the 47 proteins with regulated phosphorylation that are categorized as having nuclear-related functions in our network (Fig. 6, blue box), to the best of our knowledge, only one, MKI67, has previously been reported to be regulated by chemokine signaling. MKI67 is a marker of cell proliferation (39) whose expression has been reported to be induced by treatment of myoblasts with the CCR2 ligand CCL7 (40). On the other hand, two of the identified transcriptional regulators, YAP1 and ZEB1, have been reported to promote expression of chemokines, including CCL2 and CCL8 (41, 42), suggesting that they may contribute to amplification of chemokine-stimulated responses through positive feedback loops. The identification of so many proteins not previously known to be regulated by chemokines highlights the level of detail provided by the phosphoproteomics approach and its potential to yield novel information.
Among the nuclear-related proteins that were particularly prevalent in the CCL2-regulated phosphoproteome were proteins involved in the nuclear pore complex and nuclear export (Fig. 5 and Fig. S4). The nuclear pore complex is comprised of seven subcomplexes (Y-complex, NUP93, NUP62, NDC1, NUP214, NUP98, and TPR), each consisting of proteins with high affinity for each other (43). Phosphorylation of nuclear pore complex proteins has previously been reported to regulate nuclear translocation, the number of nuclear pore complexes per nucleus, and nuclear envelope reorganization during mitosis (44–47). We observed phosphopeptides derived from proteins in all but one of the nuclear pore subcomplexes, the NUP62 complex, which forms the central pore, and significant phosphorylation changes for proteins in four subcomplexes. Notably, the significantly regulated proteins included all three proteins comprising the TPR complex (TPR, NUP50, and NUP153), which forms the nuclear basket, projecting into the nucleoplasm and apparently contributing to nuclear organization (48). The prevalence of nuclear pore proteins among the significantly regulated proteins observed here strongly suggests that CCR2 activation gives rise to reorganization of the nuclear pore and that this is one mechanism by which chemokine receptor activation regulates transcription and translation.
There is little previous literature regarding the influence of chemokine receptor signaling on nuclear translocation. In one notable exception, Balan and Pal (49) reported that signaling of receptor CXCR3-B in breast cancer cell lines increased nuclear localization of BACH-1 and increased nuclear export of NFE2L2 (also called NRF1). BACH-1 and NFE2L2 regulate transcription of the anti-apoptotic protein heme oxygenase-1 in a positive and negative manner, respectively. Thus, the nuclear redistribution of these factors resulted in decreased expression of heme oxygenase-1 and enhanced apoptosis.
Previous studies have also suggested a more direct role of one nuclear pore protein linking chemokine signaling to actin dynamics and cell migration. Terashima et al. (50) identified a novel protein in THP-1 monocytes that associated with the intracellular, C-terminal region of CCR2 after CCL2 stimulation and was required for CCR2-dependent activation of PI3K/AKT and GTP-Rac, leading to actin reorganization and cell migration. This protein was named “FROUNT” but was later shown to be identical to a component of the nuclear pore complex named NUP85 (51, 52). The same group found that FROUNT also colocalized with chemokine receptor CCR5 and with actin after chemokine (CCL4) stimulation, promoting pseudopodial protrusion toward high chemokine concentrations (53). FROUNT was not identified in our data set, possibly indicating differences in FROUNT expression between HEK293 cells and THP-1 cells. Moreover, none of the other nuclear pore proteins has significant sequence identity to FROUNT, so it seems unlikely that they would regulate actin in the same manner. On the other hand, we observed four other components of the same nuclear pore subcomplex in which FROUNT is located (the Y-complex). Among these, NUP133, NUP107 and ELYS were significantly regulated, and Nup107 phosphosites also showed kinetic profiles suggestive of regulated phosphorylation (Fig. S4). Thus, it is possible that phosphorylation changes of these proteins alter the localization or availability of FROUNT for regulation of actin and chemotaxis.
This phosphoproteomics study was performed using a stably transfected CCR2 cell line, which we have previously used as a model system to study chemokine receptor signaling (18). These receptor overexpression systems offer several experimental advantages, including uniform expression of the transgene (54), which is a distinct advantage for proteomics studies as it is likely to reduce variability in signaling across the population of cells. Moreover, we and others have used HEK293 cells transfected with CCR2 to investigate nuances in chemokine-dependent signaling, such as partial and biased agonism (18, 55). Although these studies generally focus on molecular events at or proximal to the receptor rather than downstream pathways, HEK293 cells transfected with CCR2 have been reported to undergo chemotaxis in response to CCL2 stimulation (56). Nevertheless, given the importance of the CCL2-CCR2 signaling axis in monocyte recruitment in the context of inflammation, it will be important to follow up the current study by phosphoproteomics analyses of CCR2 signaling in monocytic cells, which we are currently pursuing in our laboratory.
Experimental procedures
Sample preparation
The generation of a HEK293 cell line stably expressing CCR2 (FlpIn-HEK293T-CCR2) was described previously (18). FlpIn-HEK293T-CCR2 cells were cultured for 16 h in fetal bovine serum–free Dulbecco's modified Eagle's medium containing 10 μg/ml tetracycline to induce the expression of CCR2 before stimulation with 100 nm CCL2 for 1.5, 3, 6, 12, 25, or 60 min. Untreated cells served as the 0-min control. THP-1 monocytes were cultured in RPMI with 10% heat-inactivated fetal bovine serum (Thermo Fisher Scientific) and were serum-starved for 4 h prior to the CCL2 stimulation time course. After stimulation, cells were washed three times with ice-cold PBS and lysed in 1% sodium deoxycholate, 100 mm Tris (pH 8.0) using a probe sonicator (Soniprep 150, MSE). The lysate was reduced with 10 mm tris(2-carboxyethyl)phosphine (Thermo Fisher Scientific) and alkylated with 40 mm chloroacetamide (Sigma), and the protein concentration was determined using the bicinchoninic acid (BCA) assay kit (Pierce). A total of four independent biological replicates were collected.
For DIA quantification, 2 mg of protein was digested overnight at 37 °C with trypsin at a ratio of 1:100 (w/w) (Trypsin Gold, Promega), and sodium deoxycholate was removed by extraction with an equal volume of water-saturated ethyl acetate prior to desalting the peptides with 30-mg HLB cartridges (Waters). 10% of each sample was directly subjected to LC-MS/MS for global proteome analysis. The remaining 90% was used for phosphopeptide enrichment by TiO2 as described previously (57, 58), followed by LC-MS/MS analysis.
For the generation of the spectral libraries, 10 mg of protein was digested overnight at 37 °C with trypsin at a ratio of 1:100 (w/w) (Trypsin Gold, Promega). The peptides were fractionated using basic pH fractionation as described previously (21). A total of 42 fractions were collected and pooled in a noncontiguous manner to reduce the number of fractions to six. 10% of each fraction was directly analyzed by LC-MS/MS to generate the proteome library. The remaining 90% was used for phosphopeptide enrichment by TiO2 and analyzed by LC-MS/MS to generate the phosphopeptide library.
LC-MS/MS
All samples were analyzed by LC-MS/MS using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled online to an RSLC nano-HPLC (Ultimate 3000, UHPLC, Thermo Fisher Scientific). Samples were loaded onto a 100-μm, 2-cm nanoviper Pepmap100 trap column, eluted, and separated on an RSLC nano column (75 μm × 50 cm), Pepmap100 C18 analytical column (Thermo Fisher Scientific). The tryptic peptides or phosphopeptides were separated by increasing concentrations of 80% acetonitrile, 0.1% formic acid at a flow of 250 nl/min for 150 min.
DDA MS was used to analyze the fractions for the generation of the spectral libraries. In detail, the LC eluent was nebulized and ionized using a nano-electrospray source (Thermo Fisher Scientific) with a distal coated fused silica emitter (New Objective). The capillary voltage was set at 1.7 kV. The Q Exactive mass spectrometer operated in DDA mode was set to switch between full MS scans and subsequent MS/MS acquisitions. Survey full-scan MS spectra (m/z 375–1800) were acquired with 70,000 resolution (at m/z 200) after accumulation of ions to a 3 × 106 target value with a maximum injection time of 30 ms. Dynamic exclusion was set to 20 s. The 10 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the collision cell by higher-energy collisional dissociation with a fixed injection time of 60 ms, 17,500 resolution, and automatic gain control (AGC) target of 5 × 104.
The following settings were applied to the mass spectrometer operated in DIA mode: MS1 scans were acquired between 370 m/z and 2,000 m/z with a resolution of 70,000 (at m/z 200) using an AGC target of 1 × 106 and a maximum ion injection time of 50 ms. Subsequent MS2 scans were acquired with a resolution of 17,500 (at m/z 200) using an AGC target of 2 × 105 with automatic injection time control and a loop count of 25. To minimize the number of co-fragmented peptides and phosphopeptides in a given DIA window and to increase quantitative precision and specificity, we narrowed the DIA isolation windows from the conventionally used 25 m/z to 12 m/z. As a consequence, each sample had to be injected two or three times to consecutively cover a mass range of 375–975 m/z (for global proteome analyses) and 375–1275 m/z (for phosphoproteome analyses) while still adhering to a duty cycle of 2 s (Fig. S5).
Data analysis
MaxQuant and its implemented search engine Andromeda (version 1.5.2.8) (59) was used to interrogate all DDA files against a human protein sequence database, which was downloaded from Uniprot/SwissProt in May 2016, containing 20,199 entries. Cysteine carbamidomethylation was specified as a fixed modification, and methionine oxidation, N-terminal acetylation, and phosphorylation at serine, threonine, or tyrosine were set as variable modifications. Up to two missed cleavages and a mass tolerance of 20 ppm for the first search were permitted. Only proteins and peptides falling below a false discovery rate (FDR) of 1% were considered for subsequent analyses.
Spectronaut 10 (Biognosys) was used for both the generation of the spectral libraries based on the MaxQuant result files and the quantification of the acquired DIA files. The parameters for the generation of the spectral libraries were kept as default. In brief, (i) mass tolerance was set as dynamic, (ii) the identification FDR threshold for MaxQuant results was set at 1%, (iii) protein inference was performed and specific trypsin digestion was used, (iv) fragment ions with an amino acid length of less than 3 were removed, (v) the m/z range was specified between 300 and 1800, (vi) the empirical iRT database was used for iRT calibration, and (vii) the minimum relative fragment intensity was set to 5, and the best number of fragments per peptide was set between 3 and 6.
For quantification of the acquired DIA files, the raw files belonging to individual injections of the same sample were merged using the HTRMS Converter (Biognosys). All parameters for phosphopeptide or protein quantification were kept as default. In brief, (i) intensity extraction for MS1 and MS2 was set as maximum and mass tolerance was set as dynamic, (ii) retention time extraction and calibration were set as dynamic using an automatic/nonlinear regression, (iii) a mutated database was used as decoy, (iv) the Kernel density estimator was used for p value estimator, (v) the MS2 area and sum precursor quantity were used for generating protein quantities, (vi) the q value was used for data filtering, and (vii) cross-run and global normalization strategies were used for intensity normalization, and protein inference was set as automatic. The normalized protein quantities or phosphopeptide intensities were exported for further analysis.
Statistical analysis
The DIA quantification results were analyzed using Perseus (60) and the R statistical framework. Observations with more than six missing values across 21 data points (7 time points × 3 biological replicates each) were excluded to maximize data quality. Missing values for the remaining data were imputed based on random draws from a left-centered Gaussian distribution in each sample 1.8 S.D. away from the mean and a width of 0.3 (60). A one-way ANOVA test was performed, and the p values were adjusted using the Benjamini–Hochberg method. An adjusted p value cut-off of less than 0.05 was used to identify significantly regulated phosphopeptides or proteins upon CCL2 treatment.
For the phosphoproteome data, all phosphopeptides with a localization probability of <0.75 (based on the phosphoproteome library created from the MaxQuant search results) were removed. In addition, phosphosites were excluded if the underlying protein itself had been observed to be significantly regulated based on the proteome data. Significantly regulated phosphopeptides were classified into three groups: fast (1.5 or 3 min), medium (6 or 12 min), and slow (25 or 60 min) responders based on the time when the changes reached their maximal amplitude compared with the baseline control.
Network generation
The list of unique proteins with significantly regulated phosphopeptides was submitted to the DAVID Bioinformatics Database (22) for KEGG pathway and gene ontology analyses. The significantly enriched pathways (Benjamini–Hochberg, FDR < 0.05, Table S3) were first manually examined and curated into a preliminary CCL2-dependent phosphoproteomic network using Cytoscape (61). The network was further expanded based on proteins that fell under broad categories of actin/GTPase-related activity and nuclear-related biological processes. These additional proteins were input into the STRING database (62) to investigate protein-protein interactions, and new proteins with more than three interactions were added to the network.
SDS-PAGE and Western blotting
Proteins were separated by SDS-PAGE (4–12% BisTris protein gels (Life Technologies, Inc.)) and transferred onto polyvinylidene difluoride membranes (GE Healthcare) using a Mini Trans-Blot Cell system (Bio-Rad). The membrane was washed three times with TBS-T buffer for 5 min and blocked for 1 h at room temperature with 5% BSA in TBS-T. The blocked membrane was incubated overnight at 4 °C with the primary antibody followed with a horseradish peroxidase–conjugated secondary antibody for 1 h at room temperature. The polyvinylidene difluoride membrane was developed with Western Lightning Plus ECL reagent (PerkinElmer Life Sciences), and images were captured and quantified with a ChemiDoc and ImageLab software (Bio-Rad). The following antibodies were used: rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (2118, Cell Signaling Technology (CST)); rabbit anti-phospho-p44/42 MAPK (ERK1/2) (Thr-202/Tyr-204) (9101, CST); rabbit anti-p44/42 MAPK (ERK1/2) (9102, CST); mouse anti-STAT3 (9139, CST); rabbit anti-phospho-STAT3 (Ser-727) (9134, CST); rabbit anti-c-JUN (9165, CST); rabbit anti-phospho-c-JUN (Ser-63) (2361, CST); rabbit anti-RSK1/RSK2/RSK3 (9355, CST); rabbit anti-phospho-p90RSK (Thr-359) (8753, CST); rabbit anti-ATF2 (ab32160, Abcam); rabbit anti-phospho-ATF2 (Thr-71) (ab32019, Abcam); goat anti-mouse IgG (M8642, Sigma–Aldrich), and goat anti-rabbit IgG (ab6721, Abcam) as the horseradish peroxidase–labeled secondary antibodies.
Data availability
All raw data have been deposited at PRIDE with the ProteomeXchange data set identifier PXD015845.
Author contributions
C. H., S. R. F., O. K., M. C., R. B. S., and M. J. S. conceptualization; C. H., S. R. F., A. D. S., O. K., R. B. S., and M. J. S. formal analysis; C. H., O. K., M. C., R. B. S., and M. J. S. supervision; C. H., O. K., M. C., R. B. S., and M. J. S. funding acquisition; C. H. and S. R. F. validation; C. H., S. R. F., A. D. S., O. K., M. C., R. B. S., and M. J. S. investigation; C. H., S. R. F., A. D. S., O. K., R. B. S., and M. J. S. visualization; C. H., S. R. F., R. B. S., and M. J. S. methodology; C. H., S. R. F., A. D. S., O. K., R. B. S., and M. J. S. writing-original draft; C. H. and M. J. S. project administration; C. H., S. R. F., A. D. S., O. K., M. C., R. B. S., and M. J. S. writing-review and editing; A. D. S., O. K., M. C., R. B. S., and M. J. S. data curation; A. D. S., O. K., and M. C. software.
Supplementary Material
Acknowledgments
This study used NCRIS (National Collaborative Research Infrastructure Strategy)-enabled infrastructure through the Monash Proteomics and Metabolomics Facility. We thank Robert J. A. Goode from the Monash Proteomics and Metabolomics Facility for assistance in data analysis.
This work was supported by National Health and Medical Research Council Project Grant APP1140874 (to M. J. S., M. C., and R. B. S.), the Monash Institute of Pharmaceutical Sciences Large Grant Support Scheme (to M. C.), and Monash University Joint Medicine-Pharmacy Grant JMP16-18 (to M. J. S. and M. C.). In addition, this study was further supported by the Office of the Vice-Provost for Research and Research Infrastructure (VPRRI) at Monash University and Bioplatforms Australia (BPA). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S5.
- GPCR
- G protein–coupled receptor
- ANOVA
- analysis of variance
- ARHGAP
- Rho GTPase-activating protein
- ARHGEF
- Rho guanine nucleotide exchange factor
- CCR2
- chemokine receptor 2
- CCL2
- C-C chemokine ligand type 2
- DIA
- data-independent acquisition
- DDA
- data-dependent acquisition
- EGFR
- epidermal growth factor receptor
- ERK1/2
- extracellular signal-regulated kinase 1/2
- FDR
- false discovery rate
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- MAPK
- mitogen-activated protein kinase
- NUP
- nucleoporin
- TPR
- tetratricopeptide repeat
- RASAL2
- Ras GTPase-activating protein nGAP
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- DAVID
- Database for Annotation, Visualization, and Integrated Discovery
- PI3K
- phosphoinositide 3-kinase/protein kinase B
- AKT
- protein kinase B
- JAK
- Janus kinase
- STAT
- signal transducer and activator of transcription
- AGC
- automatic gain control
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CST
- Cell Signaling Technology.
References
- 1. Baggiolini M. (2001) Chemokines in pathology and medicine. J. Intern. Med. 250, 91–104 10.1046/j.1365-2796.2001.00867.x [DOI] [PubMed] [Google Scholar]
- 2. Gerard C., and Rollins B. J. (2001) Chemokines and disease. Nat. Immunol. 2, 108–115 10.1038/84209 [DOI] [PubMed] [Google Scholar]
- 3. Moser B., Wolf M., Walz A., and Loetscher P. (2004) Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 25, 75–84 10.1016/j.it.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 4. Sandblad K. G., Jones P., Kostalla M. J., Linton L., Glise H., and Winqvist O. (2015) Chemokine receptor expression on monocytes from healthy individuals. Clin. Immunol. 161, 348–353 10.1016/j.clim.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 5. Proudfoot A. E. (2002) Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115 10.1038/nri722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russo R. C., Garcia C. C., and Teixeira M. M. (2010) Anti-inflammatory drug development: broad or specific chemokine receptor antagonists? Curr. Opin. Drug Discov. Devel. 13, 414–427 [PubMed] [Google Scholar]
- 7. Boring L., Gosling J., Cleary M., and Charo I. F. (1998) Decreased lesion formation in CCR2−/− mice reveals a role for chemokines in the initiation of atherosclerosis. Nature 394, 894–897 10.1038/29788 [DOI] [PubMed] [Google Scholar]
- 8. Dawson T. C., Kuziel W. A., Osahar T. A., and Maeda N. (1999) Absence of CC chemokine receptor-2 reduces atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis 143, 205–211 10.1016/S0021-9150(98)00318-9 [DOI] [PubMed] [Google Scholar]
- 9. Gu L., Okada Y., Clinton S. K., Gerard C., Sukhova G. K., Libby P., and Rollins B. J. (1998) Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol. Cell 2, 275–281 10.1016/S1097-2765(00)80139-2 [DOI] [PubMed] [Google Scholar]
- 10. Gosling J., Slaymaker S., Gu L., Tseng S., Zlot C. H., Young S. G., Rollins B. J., and Charo I. F. (1999) MCP-1 deficiency reduces susceptibility to atherosclerosis in mice that overexpress human apolipoprotein B. J. Clin. Invest. 103, 773–778 10.1172/JCI5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert J., Lekstrom-Himes J., Donaldson D., Lee Y., Hu M., Xu J., Wyant T., Davidson M., MLN1202 Study Group (2011) Effect of CC chemokine receptor 2 CCR2 blockade on serum C-reactive protein in individuals at atherosclerotic risk and with a single nucleotide polymorphism of the monocyte chemoattractant protein-1 promoter region. Am. J. Cardiol. 107, 906–911 10.1016/j.amjcard.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 12. Brown M. F., Avery M., Brissette W. H., Chang J. H., Colizza K., Conklyn M., DiRico A. P., Gladue R. P., Kath J. C., Krueger S. S., Lira P. D., Lillie B. M., Lundquist G. D., Mairs E. N., McElroy E. B., et al. (2004) Novel CCR1 antagonists with improved metabolic stability. Bioorg. Med. Chem. Lett. 14, 2175–2179 10.1016/j.bmcl.2004.02.022 [DOI] [PubMed] [Google Scholar]
- 13. White G. E., Iqbal A. J., and Greaves D. R. (2013) CC chemokine receptors and chronic inflammation—therapeutic opportunities and pharmacological challenges. Pharmacol. Rev. 65, 47–89 10.1124/pr.111.005074 [DOI] [PubMed] [Google Scholar]
- 14. Struthers M., and Pasternak A. (2010) CCR2 antagonists. Curr. Top. Med. Chem. 10, 1278–1298 10.2174/156802610791561255 [DOI] [PubMed] [Google Scholar]
- 15. Lawrence R. T., Searle B. C., Llovet A., and Villén J. (2016) Plug-and-play analysis of the human phosphoproteome by targeted high-resolution mass spectrometry. Nat. Methods 13, 431–434 10.1038/nmeth.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosenberger G., Liu Y., Röst H. L., Ludwig C., Buil A., Bensimon A., Soste M., Spector T. D., Dermitzakis E. T., Collins B. C., Malmström L., and Aebersold R. (2017) Inference and quantification of peptidoforms in large sample cohorts by SWATH-MS. Nat. Biotechnol. 35, 781–788 10.1038/nbt.3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bekker-Jensen D. B., Bernhardt O. M., Hogrebe A., Martinez-Val A., Verbeke L., Gandhi T., Kelstrup C. D., Reiter L., and Olsen J. V. (2020) Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 11, 787 10.1038/s41467-020-14609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huma Z. E., Sanchez J., Lim H. D., Bridgford J. L., Huang C., Parker B. J., Pazhamalil J. G., Porebski B. T., Pfleger K. D. G., Lane J. R., Canals M., and Stone M. J. (2017) Key determinants of selective binding and activation by the monocyte chemoattractant proteins at the chemokine receptor CCR2. Sci. Signal. 10, eaai8529 10.1126/scisignal.aai8529 [DOI] [PubMed] [Google Scholar]
- 19. Ludwig C., Gillet L., Rosenberger G., Amon S., Collins B. C., and Aebersold R. (2018) Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 14, e8126 10.15252/msb.20178126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Searle B. C., Pino L. K., Egertson J. D., Ting Y. S., Lawrence R. T., MacLean B. X., Villén J., and MacCoss M. J. (2018) Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 9, 5128 10.1038/s41467-018-07454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Batth T. S., Francavilla C., and Olsen J. V. (2014) Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 13, 6176–6186 10.1021/pr500893m [DOI] [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 23. Biswas S. K., and Sodhi A. (2002) Tyrosine phosphorylation-mediated signal transduction in MCP-1-induced macrophage activation: role for receptor dimerization, focal adhesion protein complex and JAK/STAT pathway. Int. Immunopharmacol. 2, 1095–1107 10.1016/S1567-5769(02)00055-3 [DOI] [PubMed] [Google Scholar]
- 24. Ko J., Yun C. Y., Lee J. S., Kim J. H., and Kim I. S. (2007) p38 MAPK and ERK activation by 9-cis-retinoic acid induces chemokine receptors CCR1 and CCR2 expression in human monocytic THP-1 cells. Exp. Mol. Med. 39, 129–138 10.1038/emm.2007.15 [DOI] [PubMed] [Google Scholar]
- 25. Jiménez-Sainz M. C., Fast B., Mayor F. Jr., and Aragay A. M. (2003) Signaling pathways for monocyte chemoattractant protein 1-mediated extracellular signal-regulated kinase activation. Mol. Pharmacol. 64, 773–782 10.1124/mol.64.3.773 [DOI] [PubMed] [Google Scholar]
- 26. Werle M., Schmal U., Hanna K., and Kreuzer J. (2002) MCP-1 induces activation of MAP-kinases ERK, JNK and p38 MAPK in human endothelial cells. Cardiovasc. Res. 56, 284–292 10.1016/S0008-6363(02)00600-4 [DOI] [PubMed] [Google Scholar]
- 27. Singh N. K., Janjanam J., and Rao G. N. (2017) p115 RhoGEF activates the Rac1 GTPase signaling cascade in MCP1 chemokine-induced vascular smooth muscle cell migration and proliferation. J. Biol. Chem. 292, 14080–14091 10.1074/jbc.M117.777896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Hayre M., Salanga C. L., Kipps T. J., Messmer D., Dorrestein P. C., and Handel T. M. (2010) Elucidating the CXCL12/CXCR4 signaling network in chronic lymphocytic leukemia through phosphoproteomics analysis. PLoS ONE 5, e11716 10.1371/journal.pone.0011716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wojcechowskyj J. A., Lee J. Y., Seeholzer S. H., and Doms R. W. (2011) Quantitative phosphoproteomics of CXCL12 (SDF-1) signaling. PLoS ONE 6, e24918 10.1371/journal.pone.0024918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yi T., Zhai B., Yu Y., Kiyotsugu Y., Raschle T., Etzkorn M., Seo H. C., Nagiec M., Luna R. E., Reinherz E. L., Blenis J., Gygi S. P., and Wagner G. (2014) Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, E2182–E2190 10.1073/pnas.1404943111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kodali R., Hajjou M., Berman A. B., Bansal M. B., Zhang S., Pan J. J., and Schecter A. D. (2006) Chemokines induce matrix metalloproteinase-2 through activation of epidermal growth factor receptor in arterial smooth muscle cells. Cardiovasc. Res. 69, 706–715 10.1016/j.cardiores.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 32. Bolitho C., Hahn M. A., Baxter R. C., and Marsh D. J. (2010) The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr. Relat. Cancer 17, 929–940 10.1677/ERC-10-0107 [DOI] [PubMed] [Google Scholar]
- 33. Marsigliante S., Vetrugno C., and Muscella A. (2013) CCL20 induces migration and proliferation on breast epithelial cells. J. Cell Physiol. 228, 1873–1883 10.1002/jcp.24349 [DOI] [PubMed] [Google Scholar]
- 34. Cheng Y., Qu J., Che X., Xu L., Song N., Ma Y., Gong J., Qu X., and Liu Y. (2017) CXCL12/SDF-1α induces migration via SRC-mediated CXCR4-EGFR cross-talk in gastric cancer cells. Oncol. Lett. 14, 2103–2110 10.3892/ol.2017.6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toffali L., Montresor A., Mirenda M., Scita G., and Laudanna C. (2017) SOS1, ARHGEF1, and DOCK2 rho-GEFs mediate JAK-dependent LFA-1 activation by chemokines. J. Immunol. 198, 708–717 10.4049/jimmunol.1600933 [DOI] [PubMed] [Google Scholar]
- 36. Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., et al. (2015) Proteomics: tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 37. Ward P. A. (1968) Chemotoxis of mononuclear cells. J. Exp. Med. 128, 1201–1221 10.1084/jem.128.5.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Hayre M., Salanga C. L., Handel T. M., and Allen S. J. (2008) Chemokines and cancer: migration, intracellular signalling and intercellular communication in the microenvironment. Biochem. J. 409, 635–649 10.1042/BJ20071493 [DOI] [PubMed] [Google Scholar]
- 39. Yang C., Su H., Liao X., Han C., Yu T., Zhu G., Wang X., Winkler C. A., O'Brien S. J., and Peng T. (2018) Marker of proliferation Ki-67 expression is associated with transforming growth factor β1 and can predict the prognosis of patients with hepatic B virus-related hepatocellular carcinoma. Cancer Manag. Res. 10, 679–696 10.2147/CMAR.S162595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee J., Park J., Kim Y. H., Lee N. H., and Song K. M. (2019) Irisin promotes C2C12 myoblast proliferation via ERK-dependent CCL7 upregulation. PLoS ONE 14, e0222559 10.1371/journal.pone.0222559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen X. J., Deng Y. R., Wang Z. C., Wei W. F., Zhou C. F., Zhang Y. M., Yan R. M., Liang L. J., Zhong M., Liang L., Wu S., and Wang W. (2019) Hypoxia-induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis. 10, 508 10.1038/s41419-019-1748-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Young K., Tweedie E., Conley B., Ames J., FitzSimons M., Brooks P., Liaw L., and Vary C. P. (2015) BMP9 crosstalk with the Hippo pathway regulates endothelial cell matricellular and chemokine responses. PLoS ONE 10, e0122892 10.1371/journal.pone.0122892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kabachinski G., and Schwartz T. U. (2015) The nuclear pore complex–structure and function at a glance. J. Cell Sci. 128, 423–429 10.1242/jcs.083246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nardozzi J. D., Lott K., and Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32 10.1186/1478-811X-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajanala K., Sarkar A., Jhingan G. D., Priyadarshini R., Jalan M., Sengupta S., and Nandicoori V. K. (2014) Phosphorylation of nucleoporin Tpr governs its differential localization and is required for its mitotic function. J. Cell Sci. 127, 3505–3520 10.1242/jcs.149112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hazawa M., Lin D. C., Kobayashi A., Jiang Y. Y., Xu L., Dewi F. R. P., Mohamed M. S., Hartono, Nakada M., Meguro-Horike M., Horike S. I., Koeffler H. P., and Wong R. W. (2018) ROCK-dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 19, 73–88 10.15252/embr.201744523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCloskey A., Ibarra A., and Hetzer M. W. (2018) Tpr regulates the total number of nuclear pore complexes per cell nucleus. Genes Dev. 32, 1321–1331 10.1101/gad.315523.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duheron V., Chatel G., Sauder U., Oliveri V., and Fahrenkrog B. (2014) Structural characterization of altered nucleoporin Nup153 expression in human cells by thin-section electron microscopy. Nucleus 5, 601–612 10.4161/19491034.2014.990853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balan M., and Pal S. (2014) A novel CXCR3-B chemokine receptor-induced growth-inhibitory signal in cancer cells is mediated through the regulation of Bach-1 protein and Nrf2 protein nuclear translocation. J. Biol. Chem. 289, 3126–3137 10.1074/jbc.M113.508044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Terashima Y., Onai N., Murai M., Enomoto M., Poonpiriya V., Hamada T., Motomura K., Suwa M., Ezaki T., Haga T., Kanegasaki S., and Matsushima K. (2005) Pivotal function for cytoplasmic protein FROUNT in CCR2-mediated monocyte chemotaxis. Nat. Immunol. 6, 827–835 10.1038/ni1222 [DOI] [PubMed] [Google Scholar]
- 51. Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., and Forbes D. J. (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11, 853–864 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- 52. Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., and Matunis M. J. (2002) Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Toda E., Terashima Y., Sato T., Hirose K., Kanegasaki S., and Matsushima K. (2009) FROUNT is a common regulator of CCR2 and CCR5 signaling to control directional migration. J. Immunol. 183, 6387–6394 10.4049/jimmunol.0803469 [DOI] [PubMed] [Google Scholar]
- 54. O'Gorman S., Fox D. T., and Wahl G. M. (1991) Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251, 1351–1355 10.1126/science.1900642 [DOI] [PubMed] [Google Scholar]
- 55. Berchiche Y. A., Gravel S., Pelletier M. E., St-Onge G., and Heveker N. (2011) Different effects of the different natural CC chemokine receptor 2b ligands on β-arrestin recruitment, Gαi signaling, and receptor internalization. Mol. Pharmacol. 79, 488–498 10.1124/mol.110.068486 [DOI] [PubMed] [Google Scholar]
- 56. Röhrl J., Yang D., Oppenheim J. J., and Hehlgans T. (2010) Human β-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 184, 6688–6694 10.4049/jimmunol.0903984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jensen S. S., and Larsen M. R. (2007) Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645 10.1002/rcm.3254 [DOI] [PubMed] [Google Scholar]
- 58. Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., and Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell Proteomics 6, 1103–1109 10.1074/mcp.T600060-MCP200 [DOI] [PubMed] [Google Scholar]
- 59. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 60. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 61. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., and Mering C. V. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data have been deposited at PRIDE with the ProteomeXchange data set identifier PXD015845.