Abstract
SUMOylation is a posttranslational modification (PTM) at a lysine residue and is crucial for the proper functions of many proteins, particularly of transcription factors, in various biological processes. Zinc finger homeobox 3 (ZFHX3), also known as AT motif-binding factor 1 (ATBF1), is a large transcription factor that is active in multiple pathological processes, including atrial fibrillation and carcinogenesis, and in circadian regulation and development. We have previously demonstrated that ZFHX3 is SUMOylated at three or more lysine residues. Here, we investigated which enzymes regulate ZFHX3 SUMOylation and whether SUMOylation modulates ZFHX3 stability and function. We found that SUMO1, SUMO2, and SUMO3 each are conjugated to ZFHX3. Multiple lysine residues in ZFHX3 were SUMOylated, but Lys-2806 was the major SUMOylation site, and we also found that it is highly conserved among ZFHX3 orthologs from different animal species. Using molecular analyses, we identified the enzymes that mediate ZFHX3 SUMOylation; these included SUMO1-activating enzyme subunit 1 (SAE1), an E1-activating enzyme; SUMO-conjugating enzyme UBC9 (UBC9), an E2-conjugating enzyme; and protein inhibitor of activated STAT2 (PIAS2), an E3 ligase. Multiple analyses established that both SUMO-specific peptidase 1 (SENP1) and SENP2 deSUMOylate ZFHX3. SUMOylation at Lys-2806 enhanced ZFHX3 stability by interfering with its ubiquitination and proteasomal degradation. Functionally, Lys-2806 SUMOylation enabled ZFHX3-mediated cell proliferation and xenograft tumor growth of the MDA-MB-231 breast cancer cell line. These findings reveal the enzymes involved in, and the functional consequences of, ZFHX3 SUMOylation, insights that may help shed light on ZFHX3's roles in various cellular and pathophysiological processes.
Keywords: SUMOylation, posttranslational modification (PTM), proliferation, protein stability, UBC9, zinc finger homeobox 3 (ZFHX3), protein inhibitor of activated STAT2 (PIAS2), SUMO-specific peptidase 1 (SENP1), cell proliferation, cancer, transcription factor, SUMO-conjugating enzyme UBC9, SUMO1-activating enzyme subunit 1 (SAE1)
Introduction
Posttranslational modifications (PTMs),2 including ubiquitination, acetylation, methylation, and SUMOylation of the lysine residues, are essential for a variety of cellular processes (1). Among these modifications, SUMOylation is a process in which small ubiquitin-related modifiers (SUMOs, including SUMO1 and the highly similar SUMO2 and SUMO3) are covalently attached to lysines of proteins by a specific set of activating (E1), conjugating (E2), and ligating (E3) enzymes (2, 3). SUMOylation can be reversed by specific isopeptidases referred to as sentrin/SUMO-specific proteases (SENPs) (4). The conjugation and deconjugation of SUMO is a highly dynamic and fine controlled process, and only a small fraction of a substrate is SUMOylated at a given time (5).
SUMO was initially characterized for its effects on the functions of nuclear proteins, as many transcription factors are SUMOylated. SUMOylation and ubiquitination often occur at the same lysines of a substrate protein, and SUMOylation can sometimes antagonize ubiquitination in the regulation of transcription factors (6, 7) or other substrates, such as PCNA and IκBα (8, 9). SUMOylation can also promote the ubiquitination and degradation of the modified proteins, serving as a signal for the recruitment of ubiquitin E3 ligase, as seen for PML and PARP1 (8, 10, 11). SUMOylation has also been implicated in the functions of cytoplasmic, mitochondrial, and membrane proteins (12–14). As a result, SUMOylation plays important roles in various biological processes, such as nuclear transport, transcription, chromosome segregation, and DNA repair (15). Abnormalities in the SUMO pathway thus impact various human diseases, such as Parkinson's and Huntington's diseases (16–18) and cancer (19).
Zinc finger homeobox 3 (ZFHX3), originally named ATBF1 for AT motif–binding factor 1, is a 404-kDa transcription factor that comprises four homeodomains, 23 zinc finger motifs, and several other domains. ZFHX3 was originally discovered as a negative transcriptional regulator of the AFP (α fetoprotein) gene in a hepatocellular carcinoma cell line (20), but it also plays roles in multiple pathophysiological processes, such as atrial fibrillation (21), myogenic differentiation (22), embryonic development (23), circadian regulation (24), and carcinogenesis (25, 26). For example, ZFHX3 is frequently mutated in advanced prostate cancer (27), deletion of Zfhx3 in mouse prostates induces and promotes neoplastic lesions, and ZFHX3 is essential for ERβ to inhibit cell proliferation via the down-regulation of MYC and cyclin D1 in prostate cancer cells (28). ZFHX3 can also be oncogenic in other contexts, as ZFHX3 is integral to the angiogenic activity of HIF1A/VEGFA signaling in liver cancer cells.3 ZFHX3 is rarely mutated in breast cancer (29), and ZFHX3 interacts with estrogen receptor α to modulate gene expression and cell proliferation in breast cancer cells (30, 31). During postnatal development of mouse mammary glands, Zfhx3 is essential for the progesterone signaling to induce cell proliferation, side branching, and alveologenesis (32–34). ZFHX3 has also been implicated in other types of cancers, including gastric, cervical, and head and neck (35).
Biochemically, ZFHX3 can be degraded by the ubiquitin proteasome pathway, and EFP, an estrogen-responsive RING finger ubiquitin E3 ligase, mediates the ubiquitination and degradation of ZFHX3 in breast cancer cells (29). In addition, ZFHX3 can be SUMOylated endogenously (36), and expression of ZFHX3 makes diffusely distributed nuclear SUMO1 proteins form nuclear body-like structures that are associated with PML nuclear bodies (26). Whereas SUMOylation of ZFHX3 occurs at multiple lysine residues and is nucleus-specific, the PIAS3 SUMO E3 ligase, which interacts with ZFHX3 directly, diminishes rather than enhances ZFHX3 SUMOylation (26). At present, the activating, conjugating, and ligating enzymes for ZFHX3 are unknown, and so is whether SUMOylation impacts ZFHX3 stability and function.
In this study, we identified the modifying enzymes for ZFHX3 SUMOylation and determined whether SUMOylation impacts ZFHX3 stability and function. We found that SUMO1, SUMO2, and SUMO3 can be conjugated to ZFHX3, and among the lysines that can be SUMOylated, Lys-2806 was the major SUMOylation site. In addition, Lys-2806 is evolutionarily conserved among ZFHX3 orthologous of different animal species. Interestingly, SUMOylation at Lys-2806 interfered with the ubiquitination and proteasome-mediated degradation of ZFHX3, enhancing its stability. The SAE1 E1 activating enzyme and the UBC9 E2 conjugating enzyme interacted with ZFHX3 to promote ZFHX3 SUMOylation, and PIAS2 was the only known SUMO E3 ligase responsible for ZFHX3 SUMOylation. Furthermore, the SENP1 and SENP2 deSUMOylating enzymes caused the deSUMOylation of ZFHX3. Functionally, SUMOylation of ZFHX3 at Lys-2806 promoted cell proliferation and xenograft tumor growth in a breast cancer cell line. Therefore, ZFHX3 is SUMOylated at Lys-2806 by SAE1, UBC9, and PIAS2, and the SUMOylation impacts the stability and function of ZFHX3.
Results
Lys-2806 is the major SUMOylation site of ZFHX3
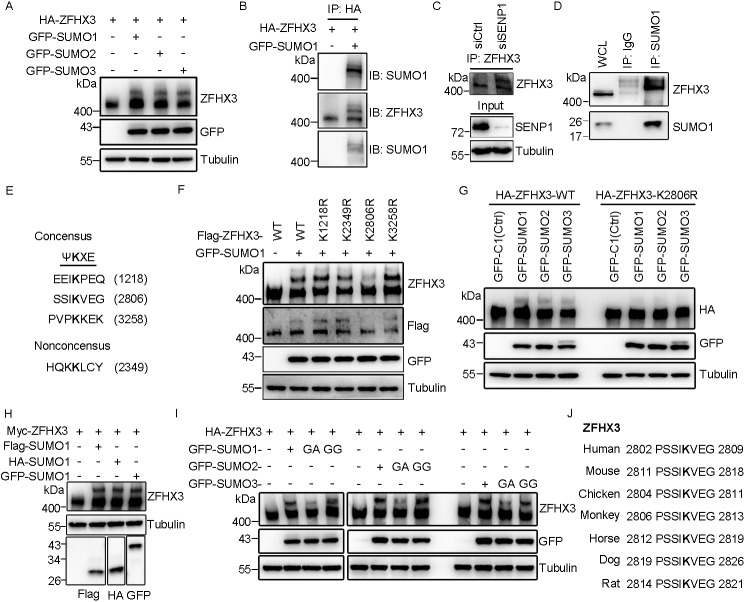
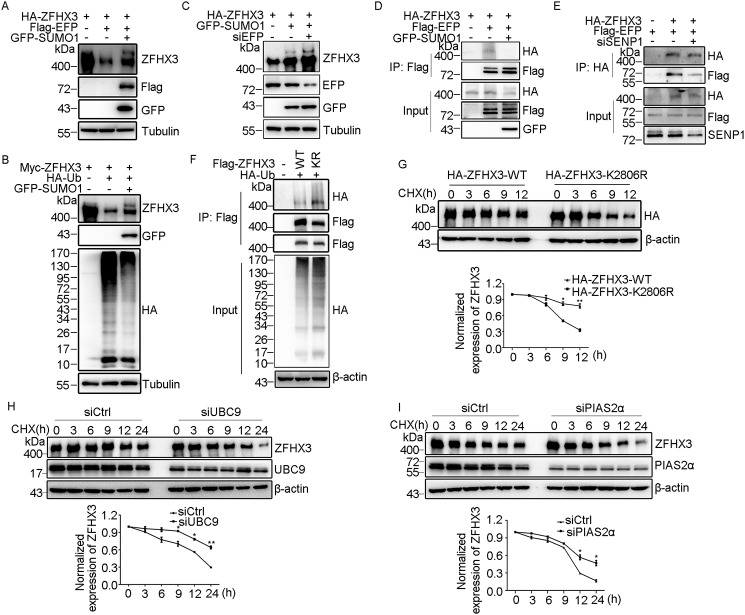
Our previous study demonstrated that ZFHX3 can be SUMOylated with SUMO1 at lysines 2349, 2806, and 3258 (26), but the enzymes for ZFHX3 SUMOylation are unknown. In this study, we were able to detect a shifted band of ZFHX3 when SUMO1, SUMO2, or SUMO3 was ectopically expressed with HA-ZFHX3 in HEK293T cells (Fig. 1A). In HA-ZFHX3 proteins precipitated with the anti-HA antibody, the shifted band was detected with both anti-SUMO1 and anti-ZFHX3 antibodies, whereas the lower unshifted band was detected only with anti-ZFHX3 antibody (Fig. 1B), confirming that the shifted band represented SUMOylated ZFHX3. In HeLa cells, RNAi-mediated silencing of SENP1 clearly increased the intensity of the upper ZFHX3 band, suggesting that SENP1 deSUMOylates endogenous ZFHX3 (Fig. 1C). Consistently, in the SUMO1-associated proteins pulled down by immunoprecipitation (IP) with anti-SUMO1 antibody in HeLa cells, ZFHX3 was also detected, and the ZFHX3 band(s) were shifted up when compared with the ZFHX3 band from cell lysate before IP (Fig. 1D). Therefore, endogenous ZFHX3 is SUMOylated.
Figure 1.
Lysine 2806 is the major SUMOylation site of ZFHX3. A, ZFHX3 can be SUMOylated by SUMO1, SUMO2, and SUMO3. HEK293T cells were transfected with expression plasmids of HA-tagged ZFHX3 (HA-ZFHX3) and GFP-tagged SUMO1, SUMO2, or SUMO3, and ZFHX3 was detected by Western blotting with anti-ZFHX3 antibody. B, detection of SUMOylated ZFHX3 in HEK293T cells expressing HA-ZFHX3 and GFP-SUMO1. Cell lysates were precipitated with anti-HA-beads, and eluted proteins were immunoblotted (IB) with anti-SUMO1 (top) or anti-ZFHX3 (middle) antibody. The middle membrane was stripped and reprobed with anti-SUMO1 antibody (bottom). C and D, detection of endogenous ZFHX3 SUMOylation in HeLa cells with RNAi-mediated SENP1 knockdown by IP and Western blotting with ZFHX3 antibody (C) or in HeLa cells by IP with SUMO1 antibody and Western blotting with anti-ZFHX3 and anti-SUMO1 antibodies (D). E, four candidate SUMOylation sites in ZFHX3, including three consensuses and one nonconsensus, were predicted by the SUMOsp software. F, Lys-2806 is the major acceptor site for ZFHX3 SUMOylation. FLAG-ZFHX3, its Lys-to-Arg mutants at predicted SUMOylation sites, and GFP-SUMO1 were expressed in HEK293T cells, and ZFHX3 and its SUMOylated form were detected by Western blotting with anti-ZFHX3 and anti-FLAG antibodies. G, Lys-2806 is SUMOylated by SUMO1, SUMO2, and SUMO3. HEK293T cells expressing GFP-tagged SUMO1, SUMO2, or SUMO3 and HA-tagged ZFHX3 or its K2806R mutant were subjected to Western blotting with anti-HA and anti-GFP antibodies. H, detection of ZFHX3 SUMOylation in HEK293T cells ectopically expressing FLAG-, HA-, and GFP-tagged SUMO1 and Myc-tagged ZFHX3 by Western blotting with the indicated antibodies. I, HEK293T cells transfected with HA-ZFHX3 and different isoforms of SUMO1, SUMO2, or SUMO3 were subjected to Western blotting with the indicated antibodies. J, alignment of ZFHX3 sequences from different species surrounding the human Lys-2806 SUMOylation site, with the conserved lysine shown in boldface type.
Lysine residues undergoing SUMOylation are typically found within a SUMO modification consensus motif, ψKXE, where ψ is a large hydrophobic residue and X is any residue (37). Analysis of human ZFHX3 with the SUMOsp software (RRID:SCR_018261) identified three potential SUMOylation sites: Lys-1218, Lys-2806, and Lys-3258 (Fig. 1E), the latter two of which have been experimentally confirmed to undergo SUMOylation along with the Lys-2349 nonconsensus site (26). Each of these four lysines was mutated to arginine in the full-length ZFHX3 (FLAG-tagged, this vector is shorter than the one with an HA tag) and analyzed for SUMOylation in HEK293T cells (Fig. 1F) and HeLa cells (data not shown). Compared with WT ZFHX3, whereas mutants K1218R, K2349R, and K3258R did not cause obvious changes in ZFHX3 SUMOylation levels, mutant K2806R dramatically decreased ZFHX3 SUMOylation (Fig. 1F), indicating that Lys-2806 is the major SUMOylation site of ZFHX3. The result was similar when SUMO2 or SUMO3 was co-expressed (Fig. 1G), indicating that Lys-2806 can be SUMOylated with all three SUMO isoforms. We also constructed FLAG- and HA-tagged SUMO1 expression constructs and co-expressed each of them with Myc-tagged ZFHX3 in HEK293T cells with GFP-tagged SUMO1 as a control. Different tags of SUMO1 showed similar effects on ZFHX3 SUMOylation (Fig. 1H). The shifted band was detectable only when the WT SUMO1 or its active form SUMO1-GG was expressed and not when the SUMOylation-dead mutant SUMO1-GA was expressed. Mutation of SUMO2 or SUMO3 to the ginkgolic acid (GA) form also prevented ZFHX3 SUMOylation when co-expressed with HA-ZFHX3 (Fig. 1I). Therefore, ZFHX3 can be SUMOylated with SUMO1, SUMO2, or SUMO3. The SUMOylation motif containing Lys-2806 in ZFHX3 was evolutionarily conserved among different species of animals (Fig. 1J), suggesting that Lys-2806 is important for ZFHX3 function.
Identification of activating, conjugating, and ligating enzymes for ZFHX3 SUMOylation
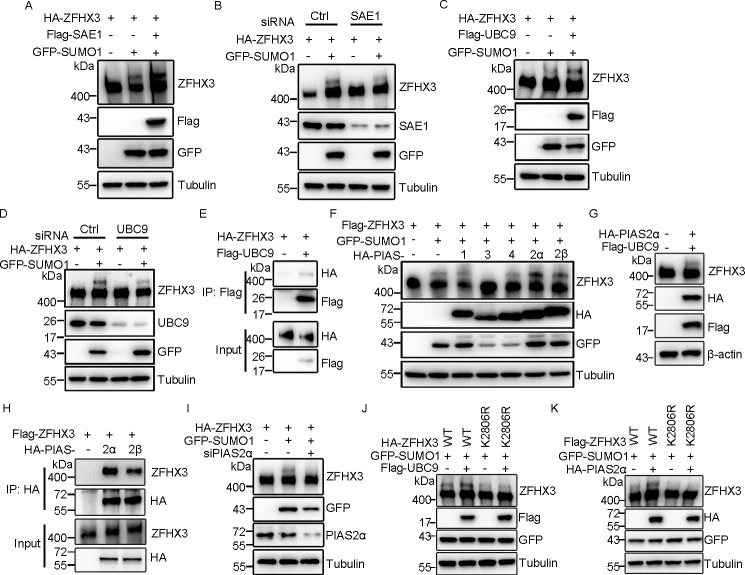
The E1 activating enzyme in the SUMO cycle of human cells is a heterodimer containing SAE1 and SAE2 subunits (37). We found that ectopic expression of SAE1 increased and its knockdown decreased SUMOylated ZFHX3, respectively, in HEK293T cells (Fig. 2, A and B), confirming that SAE1 is also the E1 for ZFHX3 SUMOylation. UBC9 is the only known E2 conjugating enzyme in the SUMO cycle (38, 39), which was also confirmed to be the case for ZFHX3, as ectopic expression and knockdown of UBC9 enhanced and reduced, respectively, ZFHX3 SUMOylation (Fig. 2, C and D). In addition, UBC9 was present in the ZFHX3 protein complex, as detected by IP and Western blotting (Fig. 2E). Therefore, SUMOylation of ZFHX3 also depends on the SAE1 E1 activating and the UBC9 E2 conjugating enzymes.
Figure 2.
Identification of SAE1 as E1 activating enzyme, UBC9 as E2 conjugating enzyme, and PIAS2 as E3 ligase for ZFHX3 SUMOylation. A and B, ectopic expression of the E1 enzyme SAE1 promotes (A), whereas its knockdown attenuates (B), ZFHX3 SUMOylation, as detected by Western blotting with the indicated antibodies in HEK293T cells expressing HA-ZFHX3 and GFP-SUMO1 in the presence or absence of FLAG-SAE1 or its siRNA. C and D, ectopic expression of UBC9 promotes (C), whereas its knockdown attenuates (D), ZFHX3 SUMOylation, as detected by Western blotting with the indicated antibodies in HEK293T cells expressing HA-ZFHX3 and GFP-SUMO1 in the presence or absence of FLAG-UBC9 or its siRNA. E, ZFHX3 physically associates with UBC9, as detected by co-IP with FLAG M2 beads and Western blotting with anti-HA antibody in HEK293T cells expressing HA-ZFHX3 in the presence or absence of FLAG-UBC9. F–I, identification of PIAS2 as the E3 ligase for ZFHX3 SUMOylation. HEK293T cells expressing FLAG-ZFHX3, GFP-SUMO1, and different HA-tagged PIASs were analyzed by Western blotting with the indicated antibodies (F). HeLa cells ectopically expressing FLAG-UBC9 and HA-PIAS2α were subjected to Western blotting with the indicated antibodies (G). HEK293T cells expressing FLAG-ZFHX3, GFP-SUMO1, and different HA-tagged PIASs were analyzed by co-IP and Western blotting with the indicated antibodies (H). HeLa cells transfected with siRNA against PIAS2α, with ectopic expression of HA-ZFHX3 and GFP-SUMO1, were analyzed by Western blotting with the indicated antibodies (I). J and K, FLAG-UBC9, HA-PIAS2α, and GFP-SUMO1 were ectopically expressed with WT ZFHX3 or its K2806R mutant in HEK293T cells, and SUMOylation of ZFHX3 was analyzed by Western blotting with anti-ZFHX3 antibody.
PIASs are the major class of SUMO E3 ligases, and five mammalian PIAS proteins, including PIAS1, PIAS2 (commonly known as PIASX, which has two isoforms, α and β), PIAS3, and PIAS4 (also known as PIASY), have been identified, and they function as SUMO E3 ligases with substrate specificity (37, 40, 41). To determine the E3 ligase for ZFHX3 SUMOylation, we co-expressed each of the five PIAS E3 ligases with ZFHX3 and SUMO1 in HEK293T cells and examined ZFHX3 SUMOylation by Western blotting. PIAS2α and -2β clearly increased, PIAS1 and PIAS4 did not change, and PIAS3 decreased ZFHX3 SUMOylation (Fig. 2F), indicating that both isoforms of PIAS2 are E3 ligases for ZFHX3 SUMOylation. Furthermore, when FLAG-tagged UBC9 and HA-tagged PIAS2α were simultaneously expressed in HeLa cells, an upper band of endogenous ZFHX3 was detected (Fig. 2G), supporting both UBC9 and PIAS2α as SUMOylation enzymes for ZFHX3.
Both PIAS2 isoforms were associated with ZFHX3, as demonstrated by IP and Western blotting (Fig. 2H), and knockdown of PIAS2 (α isoform) by siRNA clearly decreased the SUMOylation of ZFHX3 (Fig. 2I). Ectopic expression of PIAS3 diminished rather than enhanced ZFHX3 SUMOylation, which is consistent with a previous study (26). These findings demonstrate that PIAS2 functions as an E3 ligase for ZFHX3 SUMOylation.
To test whether UBC9 and PIAS2 act on lysine 2806 of ZFHX3, we ectopically expressed FLAG-tagged UBC9 or HA-tagged PIAS2α with GFP-tagged SUMO1 and HA- or FLAG-tagged WT ZFHX3 and ZFHX3-K2806 mutant in HEK293T cells. Western blotting demonstrated that, whereas UBC9 and PIAS2α obviously promoted SUMOylation of WT ZFHX3, neither UBC9 nor PIAS2α had a detectable effect on the ZFHX3-K2806R mutant (Fig. 2, J and K). Therefore, UBC9 and PIAS2 indeed can act on Lys-2806 in ZFHX3 SUMOylation.
SENP1 is a major deSUMOylating enzyme for ZFHX3
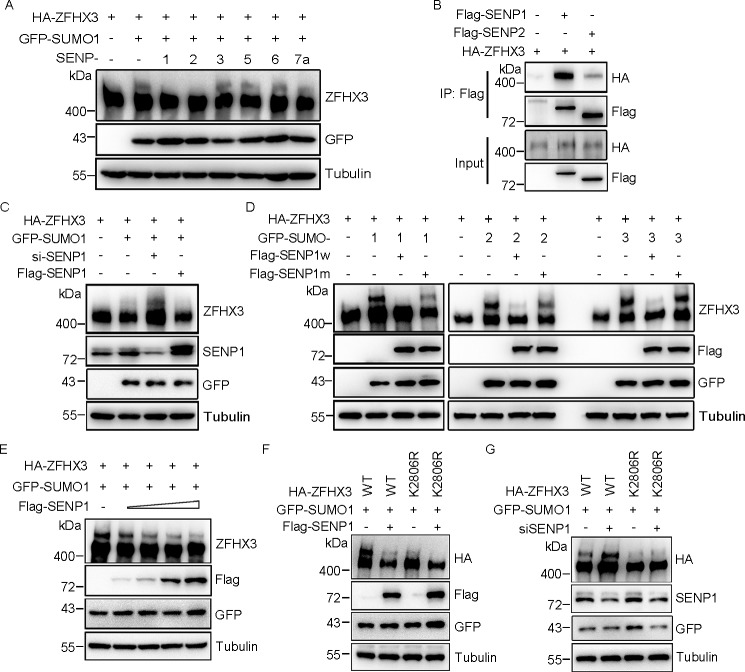
SUMOylation is a dynamic process that can be reversed by deSUMOylating enzymes known as SENPs. Six SENPs have been identified in humans, and they have different cellular localization and substrate specificity (42). To identify the SENP(s) for ZFHX3, SUMO1 and each of the six SENPs were co-transfected with HA-ZFHX3 into HEK293T cells, and ZFHX3 SUMOylation was assessed. SENP1 and SENP2 decreased ZFHX3 SUMOylation, whereas other SENPs did not (Fig. 3A). Co-IP and Western blotting demonstrated that SENP1 had a stronger interaction with ZFHX3 than SENP2 (Fig. 3B), so we focused on SENP1 for additional analyses. Consistent with its deSUMOylating effect on ZFHX3, silencing the endogenous SENP1 by RNAi increased (Fig. 3C) and ectopic expression of SENP1 abrogated ZFHX3 SUMOylation. The C603A mutant of SENP1, which no longer has a catalytic activity, failed to decrease ZFHX3 SUMOylation. Similar results were observed when SUMO2 or SUMO3 was co-expressed with SENP1 and its mutant (Fig. 3D). Ectopic expression of increasing amounts of SENP1 plasmids resulted in a dose-dependent decrease in ZFHX3 SUMOylation (Fig. 3E), further confirming the deSUMOylating activity of SENP1 for ZFHX3.
Figure 3.
SENP1 deSUMOylates ZFHX3. A, SENP1 and SENP2, but not SENP3, -5, -6, and -7a, deSUMOylate ZFHX3. HEK293T cells were transfected with HA-ZFHX3, GFP-SUMO1, and the indicated isoforms of SENPs, and SUMOylation of ZFHX3 was analyzed by Western blotting with antibody against ZFHX3. B, ZFHX3 interacts with SENP1 and SENP2. HEK293T cells expressing HA-ZFHX3 and FLAG-SENP1 or FLAG-SENP2 were subjected to immunoprecipitation and Western blotting with the indicated antibodies. C, depletion of SENP1 enhanced SUMOylation of ZFHX3. HEK293T cells were transfected with control or siRNA against SENP1, with ectopic expression of HA-ZFHX3 and GFP-SUMO1 or FLAG-SENP1 for Western blotting detection as indicated. D, SENP1 deSUMOylates ZFHX3 dependent on its catalytic activity. HA-ZFHX3 and GFP-SUMO1/SUMO2/SUMO3 were ectopically expressed in the presence of WT SENP1 or the SENP1 C603A mutant in HEK293T cells, and SUMOylation of ZFHX3 was examined by Western blotting with the indicated antibodies. E, SENP1 reduces SUMOylation of ZFHX3 in a dose-dependent manner. HEK293T cells were transfected with HA-ZFHX3 and GFP-SUMO1 with different amounts of FLAG-SENP1, and SUMOylation of ZFHX3 was analyzed by Western blotting with the indicated antibodies. F and G, SENP1-mediated ZFHX3 deSUMOylation mainly occurs at Lys-2806 of ZFHX3. WT ZFHX3 or its K2806R mutant was transfected with GFP-SUMO1 and FLAG-SENP1 into HEK293T cells (F), and siRNAs against SENP1 were transfected into HeLa cells with expression plasmids of GFP-SUMO1 and WT ZFHX3 or its K2806R mutant (G), and SUMOylation of ZFHX3 was analyzed by Western blotting with anti-HA antibody.
In addition, whereas ectopic expression of SENP1 significantly decreased the SUMOylation of WT ZFHX3, it had little effect on the SUMOylation-deficient ZFHX3-K2806R mutant (Fig. 3F). Similarly, whereas knockdown of SENP1 significantly increased the SUMOylation of WT ZFHX3, it did not cause a similar change when the ZFHX3-K2806R mutant was expressed (Fig. 3G), even though other lysines of ZFHX3 can also be SUMOylated. These results firmly establish SENP1 as a deSUMOylating enzyme for ZFHX3.
SENP1 interacts with ZFHX3
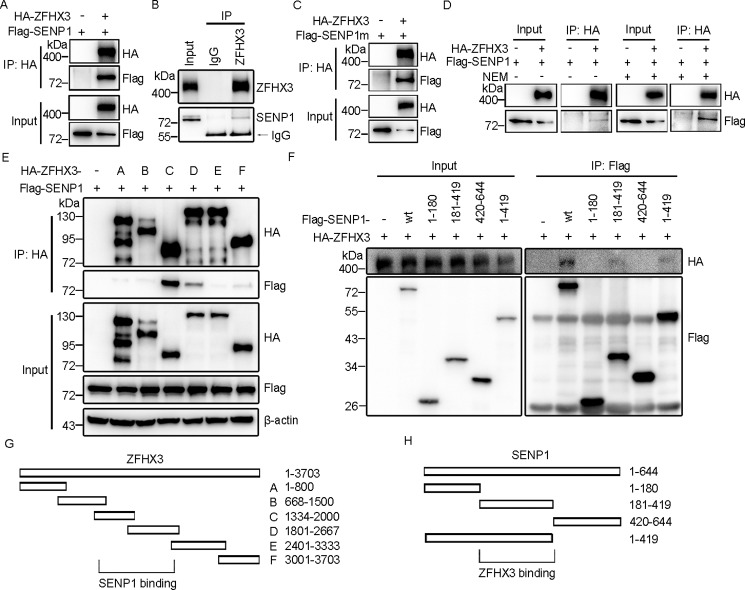
We then characterized the interaction between ZFHX3 and SENP1. Both ectopically expressed SENP1 and ZFHX3 (Fig. 4A) and the endogenous SENP1 and ZFHX3 showed a protein-protein interaction, as demonstrated by IP and Western blotting (Fig. 4B). The interaction was independent of SENP1's catalytic activity, as ZFHX3 also interacted with the catalytically inactive C603A mutant of SENP1 (Fig. 4C), and treatment with the N-ethylmaleimide (NEM) inhibitor of SENP1 did not decrease the interaction (Fig. 4D). To map the interacting domains of SENP1 and ZFHX3, different fragments of both SENP1 and ZFHX3 were co-expressed in HEK293T cells, and co-IP and Western blotting were performed. A fragment of ZFHX3 (residues 1334–2667) showed an interaction with SENP1 (Fig. 4, E and G), whereas a fragment of SENP1 (residues 181–419) interacted with ZFHX3 (Fig. 4, F and H).
Figure 4.
SENP1 interacts with ZFHX3. A and B, ZFHX3 and SENP1 interact with each other. HEK293T cells expressing HA-ZFHX3 and FLAG-SENP1 were subjected to immunoprecipitation and Western blotting with the indicated antibodies (A). HeLa cells were subjected to immunoprecipitation with ZFHX3 antibody in the presence of NEM and Western blotting with anti-ZFHX3 and SENP1 antibodies to detect the interaction of endogenous ZFHX3 and SENP1 (B). C and D, SENP1 binds to ZFHX3 independent of its catalytic activity. HEK293T cells transfected with HA-ZFHX3 and FLAG-SENP1 C603A mutant (C) or the same cells transfected with HA-ZFHX3 and FLAG-SENP1 in the presence or absence of NEM (D) were subjected to immunoprecipitation and Western blotting with the indicated antibodies. E, SENP1 interacts with the central region of ZFHX3. HEK293T cells transfected with FLAG-SENP1 and WT HA-ZFHX3 or its deletion mutants were subjected to immunoprecipitation and Western blotting with the indicated antibodies. F, ZFHX3 interacts with the central region of SENP1. HEK293T cells transfected with HA-ZFHX3 and WT FLAG-SENP1 or its deletion mutants were subjected to immunoprecipitation and Western blotting with the indicated antibodies. G and H, diagrams of ZFHX3 and its deletion mutants (G) as well as SENP1 and its deletion mutants (H).
SUMOylation of ZFHX3 enhances its protein stability
Although SUMOylation itself does not directly mediate the degradation of a protein as ubiquitination does through the proteasome pathway, it can affect the ubiquitination of proteins and thus indirectly modulate protein stability. For example, SUMOylation enhances the stability of Smad4 (43) and PCNA (44). EFP is a ubiquitin E3 ligase that mediates the ubiquitination and subsequent degradation of ZFHX3 via the ubiquitin proteasome pathway (29). In testing whether SUMOylation alters the ubiquitination and degradation of ZFHX3, HEK293T cells were transfected with ZFHX3 and EFP in the presence of SUMO1, and protein levels were measured. Overexpression of SUMO1, which enhanced ZFHX3 SUMOylation, attenuated EFP-mediated degradation of ZFHX3 (Fig. 5A). Consistently, ectopic expression of ubiquitin increased ZFHX3 ubiquitination, which was also decreased by SUMO1 overexpression (Fig. 5B). As expected, knockdown of EFP significantly increased both SUMOylated and total ZFHX3 (Fig. 5C), further suggesting that SUMOylation protects ZFHX3 from proteasome-mediated degradation. Ectopic expression of SUMO1 also weakened the interaction of ZFHX3 with EFP (Fig. 5D), and consistently, knockdown of SENP1, which enhanced ZFHX3 SUMOylation, significantly attenuated the ZFHX3-EFP interaction (Fig. 5E). These results further indicate that SUMOylation of ZFHX3 protects ZFHX3 from EFP-mediated ubiquitination and degradation.
Figure 5.
SUMOylation of ZFHX3 enhances its protein stability. A, degradation of ZFHX3 by the EFP E3 ligase is partially rescued by SUMO1 expression in HeLa cells transfected with HA-ZFHX3, FLAG-SENP1, and SUMO1, as detected with anti-ZFHX3 antibody. B, SUMOylation prevents the degradation of ZFHX3 induced by ubiquitin. HeLa cells transfected with Myc-ZFHX3, HA-Ub, and SUMO1 were subjected to Western blotting with the indicated antibodies. C, knockdown of endogenous EFP increases the levels of ZFHX3 and SUMOylated ZFHX3. HeLa cells were transfected with siRNA against EFP with HA-ZFHX3 and GFP-SUMO1, and ZFHX3 and SUMOylated ZFHX3 were detected with anti-ZFHX3 antibody. D, SUMO1 weakens the ZFHX3-EFP interaction. HEK293T cells transfected with HA-ZFHX3 and FLAG-EFP in the presence or absence of GFP-SUMO1 were treated with MG132 (20 μm, 6 h) and then subjected to immunoprecipitation and Western blotting with the indicated antibodies. E, SENP1 enhances the interaction between ZFHX3 and EFP. HeLa cells were transfected with siRNA against SENP1 with HA-ZFHX3 and FLAG-EFP, treated with MG132, and then subjected to immunoprecipitation with HA-beads and Western blotting with the indicated antibodies. F, loss of SUMOylation enhances the ubiquitination of ZFHX3. WT ZFHX3 or its K2806R mutant was ectopically expressed with HA-Ub in HEK293T cells, which were treated with MG132 and then subjected to immunoprecipitation and Western blotting with anti-FLAG and anti-HA antibodies, respectively. G, half-life of ZFHX3-K2806R is shorter than that of ZFHX3-WT. HEK293T cells were transfected with WT ZFHX3 or the ZFHX3-K2806R mutant, treated with 100 μg/ml CHX for the indicated times, and then subjected to Western blotting with anti-HA antibody. Representative blots are shown at the top, and the quantitation of relative protein levels is shown at the bottom (data were from three independent experiments). *, p < 0.05; **, p < 0.01. H and I, half-life of endogenous ZFHX3 was determined by the CHX assay in HeLa cells transfected with siRNA against UBC9 (H) or PIAS2α (I) and treated with CHX for the indicated times. Western blotting, protein quantification, and statistical analysis were performed the same way as in G except that data were from two independent experiments. Error bars, S.E.
To further test the role of SUMOylation in ZFHX3 stability, we analyzed whether SUMOylation of ZFHX3 affects its ubiquitination. Mutation of the major SUMOylation site (i.e. Lys-2806) significantly enhanced the ubiquitination of ZFHX3 (Fig. 5F, top), and the global ubiquitination level also appeared to be increased (Fig. 5F, bottom). As expected, the half-life of ZFHX3 was significantly reduced by the K2806R mutation (Fig. 5G). We also tested whether SUMOylation affects the stability of endogenous ZFHX3. UBC9 and PIAS2α was knocked down by RNAi in HeLa cells, and the cycloheximide (CHX) assay was performed. Knockdown of either UBC9 or PIAS2α significantly reduced the half-life of endogenous ZFHX3 (Fig. 5, H and I). These findings support a role for SUMOylation in protein stability of ZFHX3.
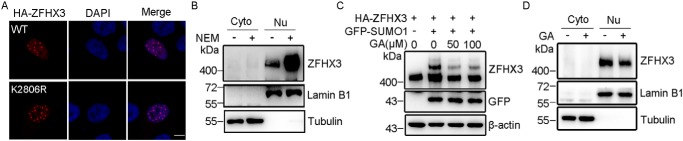
In addition to stability, SUMOylation is also known to regulate substrate subcellular distribution (4, 45), so we also tested whether ZFHX3's cellular localization is affected by SUMOylation. In HeLa cells transfected with WT ZFHX3 or its K2806R mutant, immunofluorescence staining demonstrated that both ZFHX3 and ZFHX3-K2806R massed to form nuclear body-like dots in the nucleus, and no apparent differences were noticeable (Fig. 6A). However, elevated SUMOylation, as induced by the SENP1 inhibitor NEM, increased the amount of nuclear ZFHX3, as detected by Western blotting in the cytoplasmic and nuclear fractions of cells (Fig. 6B). Consistently, the intervention of ZFHX3 SUMOylation by different concentrations of GA (Fig. 6C), an inhibitor of protein SUMOylation (46, 47), reduced the amount of nuclear ZFHX3 (Fig. 6D).
Figure 6.
SUMOylation does not change the subcellular localization of ZFHX3. A, subcellular localization of ZFHX3. WT ZFHX3 or its K2806R mutant plasmid was transfected into HeLa cells, and ZFHX3 was immunostained with anti-HA antibody (red) and visualized by fluorescence microscopy. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 5 μm. B, nuclear expression of ZFHX3 was increased by NEM treatment. Cytoplasmic and nuclear fractions were collected from HeLa cells treated with or without NEM and then subjected to Western blotting with antibodies against the lamin B1 nuclear protein, the tubulin cytoplasmic protein, and ZFHX3. C, GA attenuates ZFHX3 SUMOylation. Hela cells were transfected with HA-ZFHX3 and GFP-SUMO1, treated with different concentrations of GA for 4 h, and then subjected to Western blotting with the indicated antibodies. D, GA attenuates the nuclear localization of ZFHX3. HeLa cells were treated with 50 μm GA for 4 h and then analyzed as in B.
SUMOylation at Lys-2806 is necessary for ZFHX3 to promote cell proliferation and tumor growth
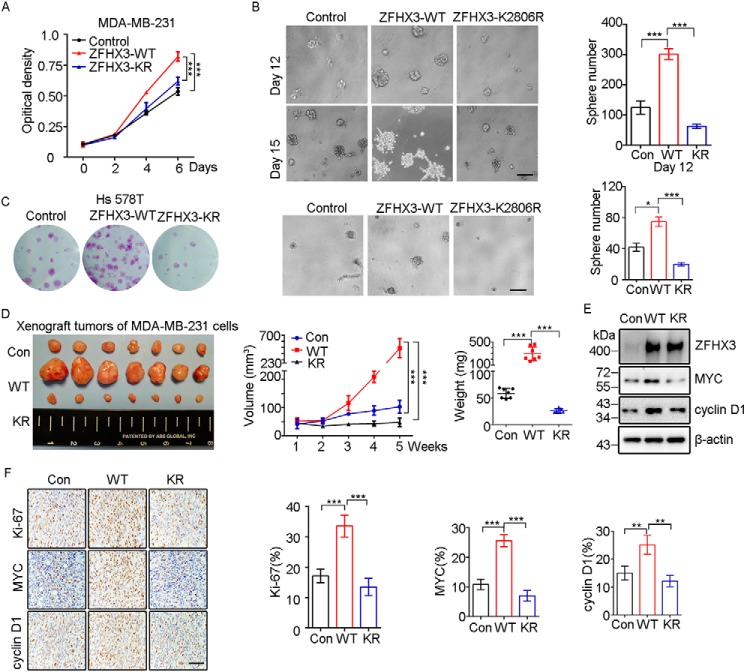
Given that Lys-2806 is the major SUMO modification site of ZFHX3 and SUMOylation often modulates molecular functions, we determined whether SUMOylation modulates ZFHX3 function. In prostate cancer cells, ZFHX3 coordinates with ERβ to inhibit cell proliferation via the down-regulation of MYC and cyclin D1 (48). Therefore, we stably expressed ZFHX3 and its K2806R mutant in several prostate cancer cell lines (PC-3, 22Rv1, and C4-2B) and breast cancer ones (MCF-7, T-47D, Hs 578T, and MDA-MB-231) to prepare cells that stably express ZFHX3 and its K2806R mutant. Unfortunately, we were only able to obtain such cells for MDA-MB-231 and Hs 578T triple-negative breast cancer cell lines. Compared with parental cells, whereas ectopic expression of WT ZFHX3 in MDA-MB-231 cells significantly increased cell proliferation, as indicated by the SRB and colony formation (data not shown) assays in two-dimensional culture and the sphere formation assay in Matrigel (Fig. 7, A and B), expression of the K2806R mutant showed opposing effects. In addition, the larger spheres in Matrigel from the WT ZFHX3 formed branches, an indicator of higher motility (Fig. 7B, bottom panels), whereas the K2806R mutant did not. Similar results were obtained in the Hs 578T breast cancer cell line (Fig. 7C). Consistent with these in vitro assays, subcutaneous injection of MDA-MB-231 cells expressing different forms of ZFHX3 into nude mice demonstrated that whereas the WT ZFHX3 significantly promoted tumor growth, as indicated by tumor images, growth curves, and tumor weights at 35 days after injection (Fig. 7D), the K2806R mutant significantly suppressed tumor growth. These results indicate that SUMOylation is necessary for ZFHX3 to promote the proliferation of breast cancer cells.
Figure 7.
SUMOylation is required for ZFHX3 to promote cell proliferation and tumor growth of MDA-MB-231 breast cancer cells. A, the K2806R mutation of ZFHX3 dramatically decreases cell growth, as measured by the SRB assay. B, interruption of ZFHX3 SUMOylation prevents ZFHX3 from promoting sphere formation in Matrigel. The data are presented as the number of spheres (>75 μm) per well (1600 seeded cells) after culture for 12 days (top) or 15 days (bottom). Representative bright field images for spheres are shown for each group. Scale bar, 200 μm. ***, p < 0.001. C, SUMOylation deficiency of ZFHX3 significantly reduces cell proliferation in Hs 578T cells. Growth of cells with ectopic expression of control vector, WT ZFHX3, or ZFHX3 K2806R mutant plasmid were measured by crystal violet staining and Matrigel assays described as above. Scale bar, 200 μm. *, p < 0.05; ***, p < 0.001. D, interruption of SUMOylation prevents ZFHX3 from promoting xenograft tumor growth. Cells stably expressing control vector, WT ZFHX3, and the K2806R mutant were inoculated into nude mice subcutaneously (1.5 × 106 cells/site). Tumor volumes were measured at the indicated times (middle), and tumors were isolated at 35 days (images are shown at the left) and weighted (right). ***, p < 0.001. E, detection of the indicated molecules in cells stably transfected with control vector, WT ZFHX3, or its K2806R mutant by Western blotting with respective antibodies. F, detection of Ki-67, MYC, and cyclin D1 by IHC staining in xenograft tumors from D (left), and the rates of staining positivity for each of the molecules are shown at the right. Scale bar, 100 μm. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
MYC and cyclin D1 are involved in the suppressive function of ZFHX3 in the proliferation of prostate cancer cells (48), so we determined whether they are also involved in ZFHX3 function in breast cancer cells. Ectopic expression of WT ZFHX3 in MDA-MB-231 cells up-regulated MYC and cyclin D1, but the K2806R mutant did not (Fig. 7E). In xenograft tumors, immunohistochemical (IHC) staining further revealed that WT ZFHX3 increased the expression of the cell proliferation marker Ki-67 and proliferation-promoting cyclin D1 and MYC, but the K2806R mutant did not (Fig. 7F). Collectively, these data suggest that SUMOylation of ZFHX3 at Lys-2806 is necessary for ZFHX3 to promote cell proliferation and tumor growth.
Discussion
The ZFHX3 transcription factor modulates multiple pathophysiological processes, such as embryonic development, carcinogenesis, and atrial fibrillation (23–25), and can be posttranslationally modified by SUMOylation (26) and ubiquitination (29). In this study, we determined how ZFHX3 is SUMOylated and deSUMOylated, and whether SUMOylation impacts the stability and function of ZFHX3.
SUMOylation is an enzymatic process that covalently conjugates SUMOs to the lysines of proteins by enzymes analogous to those of ubiquitination, including the E1 activating enzyme, the E2 conjugating enzyme, and the specific SUMO E3 ligase. For the SUMOylation of ZFHX3, we demonstrated that SUMO1, SUMO2, and SUMO3 can each be conjugated to the protein (Fig. 1). In addition, whereas multiple lysines in ZFHX3, including Lys-2349, Lys-2806, and Lys-3258, can be SUMOylated, which is consistent with a previous study (26), we demonstrated that Lys-2806 is the major acceptor site because the K2806R mutation significantly reduced SUMO1/2/3-modified ZFHX3 (Fig. 1). As expected, SUMOylation of ZFHX3 required the established E1 and E2 enzymes, including the SAE1 E1 activating enzyme and the UBC9 E2 conjugating enzyme, because their ectopic expression and knockdown clearly increased and decreased, respectively, the level of SUMOylated ZFHX3 (Fig. 2).
Among the common E3 ligases (i.e. the PIAS family members PIAS1, PIAS2, PIAS3, and PIAS4), only PIAS2 has the ligase activity for ZFHX3, because its expression modulates ZFHX3 SUMOylation level, and it interacts with ZFHX3 (Fig. 2). Interestingly, the PIAS3 E3 ligase in fact decreased ZFHX3 SUMOylation, which is consistent with previous studies where PIAS3 physically interacts with ZFHX3 to reduce its SUMOylation (26, 49).
Among the common deSUMOylating enzymes, SENP1 and SENP2 have deSUMOylating activities for ZFHX3, because their ectopic expression reduced ZFHX3 SUMOylation, and both SENP1 and SENP2 interacted with ZFHX3, even though SENP2 was weaker than SENP1 in the interactions (Fig. 3). Further supporting this conclusion, knockdown of SENP1 or mutation of its catalytic domain increased ZFHX3 SUMOylation (Fig. 3), endogenous ZFHX3-SENP1 interaction also occurred (Fig. 4), and the ZFHX3-SENP1 interaction regions were mapped to residues 1334–2667 of ZFHX3 and 181–419 of SENP1 (Fig. 4). Notably, the catalytic domain of SENP1 is dispensable for its interaction with ZFHX3, as neither the C603A inactivating mutation nor the NEM inhibitor of SENP1 affected the ZFHX3-SENP1 interaction (Fig. 4).
Similar to ubiquitin in both structure and biochemistry, SUMO commonly conjugates to lysine residues of its substrate proteins (2). In addition, SUMOylation of lysines can prevent the same residue's ubiquitination and subsequent protein degradation, as seen for Mdm2, PCNA, IκBα, and other proteins (8, 37, 44, 50). On the other hand, SUMO can also act as a signal for the recruitment of a ubiquitin E3 ligase to a substrate to promote the substrate's ubiquitination and degradation, as seen for PML (15). For ZFHX3, SUMOylation prevents its ubiquitination and subsequent degradation via the ubiquitin proteasome pathway, which is based on multiple lines of evidence. For example, SUMOylation reduced the ubiquitination and degradation of ZFHX3 by EFP (Fig. 5, A–E), a known ubiquitin E3 ligase for ZFHX3, and interruption of SUMOylation at Lys-2806 by the K2806R mutation increased the ubiquitination of ZFHX3 and led to more degradation of ZFHX3 (Fig. 5, F and G).
At present, it remains unknown which lysines are responsible for the ubiquitination and degradation of ZFHX3, even though the EFP E3 ubiquitin ligase is clearly involved. In addition, it is unknown whether Lys-2806 can be ubiquitinated, although its lack of SUMOylation enhanced the overall ubiquitination and degradation of ZFHX3 (Fig. 5).
SUMOylation often occurs in the nucleus, and its interruption can lead to cytoplasmic translocation of a target protein by changing its inter- or intramolecular interactions (15, 51). For ZFHX3, its nuclear localization is required for its SUMOylation, as failure to enter into the nucleus prevents its SUMOylation (26), and ZFHX3 is normally localized in the nucleus to regulate gene transcription via promoter binding (52). Interestingly, deSUMOylation does not appear to cause detectable cytoplasmic translocation of ZFHX3, because the K2806R mutation, which attenuated ZFHX3 SUMOylation to a large extent, did not change ZFHX3's nuclear localization (Fig. 6A), and neither did chemical activation or inhibition of ZFHX3 SUMOylation (Fig. 6, B–D). On the other hand, cytoplasmic translocation likely alters the function of ZFHX3, as its cytoplasmic localization often occurs in different types of cancers, including gastric cancer and those of the skin, head and neck, and bladder, and cytoplasmic translocation correlates with worse survival in head and neck cancer patients (53–55). In addition, multiple nuclear localization signals (NLSs) have been identified for ZFHX3, including NLS1387, NLS2947, and NLS2987 defined in one study (56) and NLS KRK2615-2617 defined in another study (26). Currently, it is unknown how ZFHX3 shuttles between the cytoplasm and the nucleus.
SUMOylation clearly plays important roles in a variety of biological processes, such as development and carcinogenesis (19). SUMOylation prevents the ubiquitination and subsequent degradation of ZFHX3 (Fig. 5), and quantitative reduction in ZFHX3 has functional consequences because ZFHX3 is haploinsufficient (23, 35). More specifically, SUMOylation is essential for ZFHX3 to promote cell proliferation and tumor growth at least in some breast cancer cell lines (i.e. MDA-MB-231 and Hs 578T), as the K2806R mutation, which clearly interrupts ZFHX3 SUMOylation, significantly attenuated the promoting effects of ZFHX3 on colony and sphere formation and xenograft tumor growth (Fig. 7).
Whereas SUMOylation at Lys-2806 is crucial for the function of ZFHX3 in the proliferation of some breast cancer cell lines and this SUMOylation site is evolutionarily conserved among different animal species, it is unknown whether the promoting effect of SUMOylated ZFHX3 on cell proliferation is also there for normal mammary epithelial cells, where Zfhx3 is essential for the progesterone signaling to induce ductal cell proliferation during side branching (34). In addition, it is also unknown whether SUMOylation modulates the function of ZFHX3 in prostate cancer cells, where ZFHX3 clearly possesses a tumor suppressor activity based on its frequent inactivating mutations in advanced human prostate cancer and the induction of neoplastic lesions by its deletion (35). We are in the process of generating a transgenic mouse line in which the K2806R mutation can be induced by Cre expression, which will enable us to examine whether Zfhx3 SUMOylation affects its functions in mammary gland development, prostate tumorigenesis, and likely other pathophysiological processes.
In summary, we have identified the enzymes for ZFHX3 SUMOylation, which include the SAE1 E1 activating enzyme, the UBC9 E2 conjugating enzyme, the PIAS2 E3 ligase, and the SENP1 and SENP2 deSUMOylating enzymes. We have also demonstrated that among the lysines of ZFHX3 that can be SUMOylated, Lys-2806 is the major SUMOylation site, and SUMOylation at Lys-2806 enhances ZFHX3 stability by preventing its ubiquitination and proteasome-mediated degradation. While not affecting ZFHX3's nuclear localization, SUMOylation is essential for ZFHX3 to promote cell proliferation and xenograft tumor growth in the MDA-MB-231 breast cancer cell line. These findings will be helpful for understanding how ZFHX3 functions in different pathophysiological processes.
Experimental procedures
Cell lines
All cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA). HEK293T, HeLa, and Hs 578T cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). MDA-MB-231 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. All cells were maintained at 37 °C with 5% CO2.
Plasmid preparation and transfection
Expression plasmids for ZFHX3, ZFHX3-NLSm, SUMO1, EFP, and ubiquitin were described previously (26, 29). PCR and cloning of PCR products were used to generate expression plasmids for SUMO1 in pcDNA3.0-FLAG and pCMV-HA, for SUMO2 and SUMO3 in pEGFP-C1, and for SENP1-C603A and UBC9 in p3xFLAG-CMV-10 following standard procedures. Mutants of ZFHX3 and SENP1 plasmids were generated by PCR-mediated site-directed mutagenesis. PCR primers for gene cloning and site-directed mutagenesis are listed in Table 1. Expression plasmids for all PIASs and SENPs were kindly provided by Dr. Jinke Cheng of Shanghai Jiao Tong University School of Medicine (57). Identities of all expression plasmids were confirmed by DNA sequencing. Plasmid was transfected into cells using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Table 1.
Primer sequences used in gene cloning
| Name | Sequence (5′–3′) |
|---|---|
| FLAG-SUMO1 | |
| Forward | GAAGATCTGATGTCTGACCAGGAGGCA |
| Reverse | GGGGTACCGACTAAACTGTTGAATGACCCCC |
| HA-SUMO1 | |
| Forward | GAAGATCTCTATGTCTGACCAGGAGGCA |
| Reverse | GGGGTACCCTAAACTGTTGAATGACCCCC |
| SUMO2 | |
| Forward | GAAGATCTATGGCCGACGAAAAGCC |
| Reverse | GGGGTACCGTAGACACCTCCCGT |
| SUMO3 | |
| Forward | GAAGATCTATGTCCGAGGAGAAGCC |
| Reverse | GGGGTACCGAAACTGTGCCCTG |
| SUMO2-GA | |
| Forward | GAAGATCTATGGCCGACGAAAAGCC |
| Reverse | GGGGTACCTCCCGTCTGCTGTTGGAAC |
| SUMO2-GG | |
| Forward | GAAGATCTATGGCCGACGAAAAGCC |
| Reverse | GGGGTACCACCTCCCGTCTGCTGTTG |
| SUMO3-GA | |
| Forward | GAAGATCTATGTCCGAGGAGAAGCC |
| Reverse | GGGGTACCTCCCGTCTGCTGCTG |
| SUMO3-GG | |
| Forward | GAAGATCTATGTCCGAGGAGAAGCC |
| Reverse | GGGGTACCACCTCCCGTCTGCTG |
| SAE1 | |
| Forward | CGGAATTCAATGGTGGAGAAGGAGGAGGCTGG |
| Reverse | GGGGTACCGATCACTTGGGGCCAAGGCACTC |
| UBC9 | |
| Forward | CGGAATTCAATGTCGGGGATCGCCCTCAG |
| Reverse | GGGGTACCGATTATGAGGGCGCAAACTTCTTGGC |
| ZFHX3-K1218R | |
| Forward | TGAGGAGATCAGGCCGGAGCAGATGTAC |
| Reverse | CCTGATCTCCTCAGCTGTTTTTGTCGC |
| ZFHX3-K2349R | |
| Forward | GCACCAGAAGAGGCTGTGTTACAAGGAT |
| Reverse | CACAGCCTCTTCTGGTGCTTGATGAGATC |
| ZFHX3-K2806R | |
| Forward | TTCCTCCATTAGGGTGGAAGGGATTGAA |
| Reverse | CCTAATGGAGGAAGGGCTTAGAAGAGTT |
| ZFHX3-K3258R | |
| Forward | GCCTGTCCCCAGGAAGGAGAAAGGAGAG |
| Reverse | CCTGGGGACAGGCAGGGGTTCCCCTTTC |
| SENP1(1–180) | |
| Forward | CGGAATTCAATGGATGATATTGCTGATAGG |
| Reverse | GGGGTACCGAACTAACATGTCGCCTCTGA |
| SENP1(181–419) | |
| Forward | CGGAATTCAACAGCAGAAGAGACAGTTCAA |
| Reverse | GGGGTACCGATTCATCTTCACTATCAGTTAAT |
| SENP1(420–644) | |
| Forward | CGGAATTCATTTCCTGAAATTACAGAGGAAATG |
| Reverse | GGGGTACCGACTACAAGAGTTTTCGGTGGA |
| SENP1(1–419) | |
| Forward | CGGAATTCAATGGATGATATTGCTGATAGG |
| Reverse | GGGGTACCGATTCATCTTCACTATCAGTTAAT |
| SENP1-C603A | |
| Forward | ATGGAAGTGACGCTGGGATGTTTGCCTGC |
| Reverse | AGCGTCACTTCCATTCATCTGCTGAGGAA |
Antibodies and reagents
Monoclonal anti-FLAG M2 and anti-HA affinity gels, NEM, MG132, CHX, and ginkgolic acid C15:1 were purchased from Sigma. Normal rabbit IgG was from Santa Cruz Biotechnology, Inc. (Dallas, TX). Protein A– and G–agarose were from GE Healthcare. Complete protease inhibitor mixture tablet was from Roche Applied Science. Details of primary antibodies are listed in Table 2. Secondary antibodies, including horseradish peroxidase–conjugated goat anti-rabbit IgG (catalogue no. 7074) and goat anti-mouse IgG (catalogue no. 7076), were from Cell Signaling (Danvers, MA).
Table 2.
Antibodies used in the study
| Antibody | Company | Catalog no. | Dilutiona |
|---|---|---|---|
| ZFHX3 | Previously prepared | Ref. 28 | WB 1:800 |
| FLAG | Sigma | F1804 | WB 1:3000; IF 1:1000 |
| HA | Cell Signaling | 3724S | WB 1:3000; IF 1:1000 |
| GFP | Cell Signaling | 2555 | WB 1:3000 |
| cyclin D1 | Abcam | ab134175 | WB 1:5000; IHC 1:2000 |
| MYC | Cell Signaling | 9402S | WB 1:1000 |
| MYC | Abcam | ab32072 | IHC 1:200 |
| SENP1 | Abcam | ab108981 | WB 1:1000 |
| Ki-67 | Abcam | ab15580 | IHC 1:2000 |
| Lamin B1 | Abcam | ab16048 | WB 1:1000 |
| UBC9 | Abcam | ab75854 | WB 1:1000 |
| SAE1 | Abcam | ab185552 | WB 1:1000 |
| PIASXα | Santa Cruz Biotechnology | sc-166494 | WB 1:500 |
| PIAS3 | Cell Signaling | 9042 | WB 1:1000 |
| SUMO1 | Abcam | ab32058 | WB 1:1000 |
| EFP | BD Biosciences | 610570 | WB 1:500 |
| β-actin | Sigma | A1978 | WB 1:8000 |
| α-Tubulin | Sigma | T6199 | WB 1:8000 |
a WB, Western blotting; IF, immunofluorescence; IHC, immunohistochemistry.
Establishment of cell lines ectopically expressing ZFHX3
MDA-MB-231 and Hs 578T breast cancer cells were transfected with pcDNA3.0-FLAG and FLAG-tagged WT ZFHX3 (ZFHX3-WT) or its K2806R mutant (ZFHX3-K2806R) and selected with 1 mg/ml G418 for 14 days. Clones were isolated and subjected to Western blotting with anti-FLAG antibody to identify those expressing FLAG-tagged ZFHX3.
Immunofluorescence staining
Cells were transfected with the indicated plasmids for 24 h and then seeded onto coverslips in 6-well plates overnight, washed with PBS, fixed in 4% paraformaldehyde under room temperature for 30 min, permeabilized with 0.2% Triton X-100 for 10 min, and then blocked with 5% bovine serum in PBS. Cells on coverslips were then incubated with primary antibody at 4 °C overnight, washed three times with PBS, incubated with secondary antibody conjugated with TRITC (Life Technologies; A11008) at room temperature for 2 h in a lightproof box, counterstained with 4′,6-diamidino-2-phenylindole (Sigma) for 5 min, rinsed with PBS, mounted to a glass slide with a drop of mounting medium, and sealed with nail polish. The immunofluorescence images were taken by a laser-scanning confocal microscope (Zeiss, LSM710, Jena, Thuringen, Germany).
Immunoprecipitation and Western blotting
For IP, cells were washed twice with cold PBS after various treatments, lysed at 4 °C for 30 min by gentle shaking in IP buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.5, and 1% Nonidet P-40, supplemented with 1% protease inhibitor mixture). For the detection of SUMOylated proteins, NEM was added to the lysis buffer (20 mm) to preserve SUMOylation of ZFHX3 during the experiment. After centrifugation at 12,000 × g for 15 min, cell lysates were incubated with anti-FLAG or anti-HA-agarose beads at 4 °C for 2 h. For endogenous co-IP, cell lysates were incubated with anti-ZFHX3 antibody or normal IgG for 4 h at 4 °C. Protein A/G-agarose beads were then added and incubated for 2 h at 4 °C. After washing with cold IP buffer five times, immune complexes were collected by centrifugation at 2000 rpm for 2 min, boiled in 2× SDS sample buffer for 10 min, and subjected to Western blotting.
For Western blotting, cell lysates or immunoprecipitates were separated by 4% (for full-length ZFHX3) or 10% (for all other proteins) SDS-PAGE, and proteins were then transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA). Membranes were then blocked with 5% nonfat milk, incubated with the primary antibody overnight at 4 °C, and then incubated with horseradish peroxidase–linked secondary antibody for 2 h at room temperature. After adding Western Bright ECL reagents (Advansta, Menlo Park, CA), protein signals were detected with a luminescent image analyzer (Jun Yi Dong Fang, Beijing, China).
RNAi
For gene silencing by RNAi, siRNAs (details are shown in Table 3) were synthesized by Sangon Biotech (Shanghai, China) and transiently transfected into cells using the Lipofectamine RNAiMAX reagent according to the manufacturer's instructions. Cells were harvested after transfection for 48–72 h and analyzed by Western blotting.
Table 3.
Sequences of siRNAs used in the study
| Name | Sense sequence (5′–3′) |
|---|---|
| SAE1 | AGCGAGCUCAGAAUCTCAATT |
| UBC9 | GGGAUUGGUUUGGCAAGAATT |
| SENP1 | GGACCAGCUUUCGCUUUCUTT |
| EFP | GCACUGGAUGAUGUGAGAATT |
| PIAS2α | AAGAUACUAAGCCCACAUUUGTT |
| PIASX3 | CAAACAGACAGGUGGAAAATT |
CHX assay
To examine protein stability, cells were seeded into 12-well plates at a density of 2 × 105 cells/well, cultured for 24 h, and transfected with the indicated plasmids. After treatment with CHX (100 μg/ml) for the indicated times, cells were then harvested and subjected to Western blotting.
Subcellular fractionation
Cells were collected, resuspended in lysis buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT, and 1% protease inhibitor mixture), incubated for 5 min on ice, and centrifuged at 450 × g at 4 °C for 5 min. Cell pellets were then resuspended and incubated in lysis buffer with 0.6% IGEPAL CA-630 for 15 min on ice. After centrifugation at 12,000 × g and 4 °C for 30 s, the supernatant was collected as the cytoplasmic fraction. The pellets were washed at least three times in lysis buffer and lysed in extraction buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 0.42 m NaCl, 0.2 mm EDTA, 25% glycerol, 1 mm DTT, and 1% protease inhibitor mixture) for 30 min on ice. After centrifugation at 20,000 × g for 5 min, the supernatant was collected as the nuclear fraction. Western blotting was applied to detect cytoplasmic and nuclear proteins.
SRB and colony formation assays
The SRB assay was performed to monitor cell proliferation and death, as described previously (27). Briefly, cells were plated into 24-well plates with 4 replicates/group, incubated for different times, fixed, stained with SRB, and measured for optical intensities, which indicated cell numbers.
For the colony formation assay, 1000 cells were plated into 6-well plates in triplicate. Cells were cultured for 10 or more days until colonies were clearly visible, fixed in 4% paraformaldehyde, and stained with 0.25% crystal violet. The numbers of colonies were scored by using the ImageJ program. Each experiment was repeated at least twice.
Matrigel assay
Eight-well chamber slides (FALCON, Corning, Inc., catalogue no. 353097) were precoated with 40 μl of growth factor–reduced Matrigel (BD Biosciences, catalogue no. 354230) and set in the cell culture incubator for 30 min to allow Matrigel solidification. A total of 1600 cells were overlaid onto the gel in medium supplemented with 2% Matrigel in a well. The medium was replenished every 3 days. 12 or 15 days later, images of spheres were taken, and the number of spheres at each well was counted by using the ImageJ program. Each experiment was repeated twice.
Mouse xenograft tumor growth assay
The animal studies were approved by the Nankai University School of Life Science Animal Care and Use Committee. For MDA-MB-231 cells expressing vector, FLAG-ZFHX3-WT, or FLAG-ZFHX3-K2806R, 1.5 × 106 cells in 100 μl of PBS/Matrigel (2:1) (catalogue no. 354234) were injected into the flanks of a 4-week-old nude mouse. Tumor volumes were recorded every 7 days using a Vernier caliper, and the tumor volume was estimated as follows: V = (length × width × height × 0.5) mm3. Five weeks after injection, mice were sacrificed, and the tumors were isolated, photographed, weighed, and sectioned for IHC staining.
IHC staining
Tissue sections of xenograft tumors (4 μm thick) were immunostained with anti-MYC, anti-cyclin D1, or anti-Ki-67 antibodies overnight at 4 °C. The peroxidase-conjugated streptavidin method was performed, followed by the 3,3′-diaminobenzidine procedure according to the manufacturer's protocols (Dako, Agilent Pathology Solutions). Hematoxylin was used for counterstaining.
Statistical analysis
All in vitro experiments were repeated at least twice unless stated otherwise, and results from one experiment are shown. Prism 6 software (GraphPad Software, San Diego, CA) was used for all statistical analyses. All quantitative data are expressed as mean ± S.E. Differences were analyzed using Student's t test or one-way analysis of variance for multiple group comparisons. p < 0.05 was considered as statistically significant difference.
Data availability
All of the data are contained within the article.
Author contributions
R. W., J. F., M. L., J. A., J. Liu, W. C., J. Li., and G. M. investigation; R. W. and J. A. formal analysis; R. W. visualization; R. W. writing-original draft; L. F. project administration; Z. Z., B. Z., and J.-T. D. writing-review and editing; J.-T. D. conceptualization; J.-T. D. funding acquisition; J.-T. D. supervision.
Acknowledgments
We thank Drs. Ang Gao and Qingxia Hu for valuable comments, suggestions, and help during the study.
This work was supported by National Natural Science Foundation of China (NSFC) Grants 81472464 and 31871466. The authors declare that they have no conflicts of interest with the contents of this article.
C. Fu, N. An, J. Liu, J. A, B. Zhang, M. Liu, Z. Zhang, L. Fu, X. Tian, D. Wang, and J.-T. Dong (2020) J. Biol. Chem. 295, in press.
- PTM
- posttranslational modification
- ZFHX3
- zinc finger homeobox 3
- ERβ
- estrogen receptor β
- SUMO
- small ubiquitin-like modifier
- SENP
- SUMO-specific peptidase
- HA
- hemagglutinin
- IP
- immunoprecipitation
- PIAS
- protein inhibitor of activated STAT
- PCNA
- proliferating cell nuclear antigen
- GA
- ginkgolic acid
- SRB
- sulforhodamine B
- IHC
- immunohistochemical
- PML
- promyelocytic leukemia protein
- NLS
- nuclear localization signal
- NEM
- N-ethylmaleimide
- CHX
- cycloheximide
- TRITC
- tetramethylrhodamine isothiocyanate.
References
- 1. Chen C. H., Chang C. C., Lee T. H., Luo M., Huang P., Liao P. H., Wei S., Li F. A., Chen R. H., Zhou X. Z., Shih H. M., and Lu K. P. (2013) SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. 73, 3951–3962 10.1158/0008-5472.CAN-12-4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson S. P., and Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 3. Knipscheer P., Flotho A., Klug H., Olsen J. V., van Dijk W. J., Fish A., Johnson E. S., Mann M., Sixma T. K., and Pichler A. (2008) Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31, 371–382 10.1016/j.molcel.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 4. Kim Y. H., Choi C. Y., and Kim Y. (1999) Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. U.S.A. 96, 12350–12355 10.1073/pnas.96.22.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hay R. T. (2013) Decoding the SUMO signal. Biochem. Soc. Trans. 41, 463–473 10.1042/BST20130015 [DOI] [PubMed] [Google Scholar]
- 6. Ji Z., Degerny C., Vintonenko N., Deheuninck J., Foveau B., Leroy C., Coll J., Tulasne D., Baert J. L., and Fafeur V. (2007) Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 26, 395–406 10.1038/sj.onc.1209789 [DOI] [PubMed] [Google Scholar]
- 7. Aillet F., Lopitz-Otsoa F., Egaña I., Hjerpe R., Fraser P., Hay R. T., Rodriguez M. S., and Lang V. (2012) Heterologous SUMO-2/3-ubiquitin chains optimize IκBα degradation and NF-κB activity. PLoS ONE 7, e51672 10.1371/journal.pone.0051672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Creton S., and Jentsch S. (2010) SnapShot: the SUMO system. Cell 143, 848–848.e1 10.1016/j.cell.2010.11.026 [DOI] [PubMed] [Google Scholar]
- 9. Ran Y., Liu T. T., Zhou Q., Li S., Mao A. P., Li Y., Liu L. J., Cheng J. K., and Shu H. B. (2011) SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol. Cell Biol. 3, 283–292 10.1093/jmcb/mjr020 [DOI] [PubMed] [Google Scholar]
- 10. Hendriks I. A., and Vertegaal A. C. O. (2016) A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- 11. Jalal D., Chalissery J., and Hassan A. H. (2017) Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases. Nucleic Acids Res. 45, 2242–2261 10.1093/nar/gkw1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harder Z., Zunino R., and McBride H. (2004) Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr. Biol. 14, 340–345 10.1016/j.cub.2004.02.004 [DOI] [PubMed] [Google Scholar]
- 13. Barry R., John S. W., Liccardi G., Tenev T., Jaco I., Chen C. H., Choi J., Kasperkiewicz P., Fernandes-Alnemri T., Alnemri E., Drag M., Chen Y., and Meier P. (2018) SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 9, 3001 10.1038/s41467-018-05321-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang T., Cao Y., Zheng Q., Tu J., Zhou W., He J., Zhong J., Chen Y., Wang J., Cai R., Zuo Y., Wei B., Fan Q., Yang J., Wu Y., et al. (2019) SENP1-Sirt3 signaling controls mitochondrial protein acetylation and metabolism. Mol. Cell 75, 823–834.e5 10.1016/j.molcel.2019.06.008 [DOI] [PubMed] [Google Scholar]
- 15. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eckermann K. (2013) SUMO and Parkinson's disease. Neuromolecular Med. 15, 737–759 10.1007/s12017-013-8259-5 [DOI] [PubMed] [Google Scholar]
- 17. Henley J. M., Craig T. J., and Wilkinson K. A. (2014) Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 94, 1249–1285 10.1152/physrev.00008.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., and Marsh J. L. (2004) SUMO modification of huntingtin and Huntington's disease pathology. Science 304, 100–104 10.1126/science.1092194 [DOI] [PubMed] [Google Scholar]
- 19. Seeler J. S., and Dejean A. (2017) SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197 10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
- 20. Yasuda H., Mizuno A., Tamaoki T., and Morinaga T. (1994) ATBF1, a multiple-homeodomain zinc finger protein, selectively down-regulates AT-rich elements of the human α-fetoprotein gene. Mol. Cell Biol. 14, 1395–1401 10.1128/MCB.14.2.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng W., Kao Y., Chao T., Lin Y., Chen S., and Chen Y. (2019) MicroRNA-133 suppresses ZFHX3-dependent atrial remodelling and arrhythmia. Acta Physiol. (Oxf.) 227, e13322 10.1111/apha.13322 [DOI] [PubMed] [Google Scholar]
- 22. Berry F. B., Miura Y., Mihara K., Kaspar P., Sakata N., Hashimoto-Tamaoki T., and Tamaoki T. (2001) Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 276, 25057–25065 10.1074/jbc.M010378200 [DOI] [PubMed] [Google Scholar]
- 23. Sun X., Fu X., Li J., Xing C., Martin D. W., Zhang H. H., Chen Z., and Dong J. T. (2012) Heterozygous deletion of Atbf1 by the Cre-loxP system in mice causes preweaning mortality. Genesis 50, 819–827 10.1002/dvg.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parsons M. J., Brancaccio M., Sethi S., Maywood E. S., Satija R., Edwards J. K., Jagannath A., Couch Y., Finelli M. J., Smyllie N. J., Esapa C., Butler R., Barnard A. R., Chesham J. E., Saito S., et al. (2015) The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell 162, 607–621 10.1016/j.cell.2015.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun X., Xing C., Fu X., Li J., Zhang B., Frierson H. F. Jr., and Dong J. T. (2015) Additive effect of Zfhx3/Atbf1 and Pten deletion on mouse prostatic tumorigenesis. J. Genet. Genomics 42, 373–382 10.1016/j.jgg.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun X., Li J., Dong F. N., and Dong J. T. (2014) Characterization of nuclear localization and SUMOylation of the ATBF1 transcription factor in epithelial cells. PLoS ONE 9, e92746 10.1371/journal.pone.0092746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun X., Frierson H. F., Chen C., Li C., Ran Q., Otto K. B., Cantarel B. M., Vessella R. L., Gao A. C., Petros J., Miura Y., Simons J. W., and Dong J. T. (2005) Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat. Genet. 37, 407–412 10.1038/ng1528 [DOI] [PubMed] [Google Scholar]
- 28. Hu Q., Zhang B., Chen R., Fu C., A J., Fu X., Li J., Fu L., Zhang Z., and Dong J. T. (2019) ZFHX3 is indispensable for ERβ to inhibit cell proliferation via MYC downregulation in prostate cancer cells. Oncogenesis 8, 28 10.1038/s41389-019-0138-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong X. Y., Fu X., Fan S., Guo P., Su D., and Dong J. T. (2012) Oestrogen causes ATBF1 protein degradation through the oestrogen-responsive E3 ubiquitin ligase EFP. Biochem. J. 444, 581–590 10.1042/BJ20111890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dong X. Y., Sun X., Guo P., Li Q., Sasahara M., Ishii Y., and Dong J. T. (2010) ATBF1 inhibits estrogen receptor (ER) function by selectively competing with AIB1 for binding to the ER in ER-positive breast cancer cells. J. Biol. Chem. 285, 32801–32809 10.1074/jbc.M110.128330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong X. Y., Guo P., Sun X., Li Q., and Dong J. T. (2011) Estrogen up-regulates ATBF1 transcription but causes its protein degradation in estrogen receptor-α-positive breast cancer cells. J. Biol. Chem. 286, 13879–13890 10.1074/jbc.M110.187849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M., Zhao D., Ma G., Zhang B., Fu X., Zhu Z., Fu L., Sun X., and Dong J. T. (2013) Upregulation of ATBF1 by progesterone-PR signaling and its functional implication in mammary epithelial cells. Biochem. Biophys. Res. Commun. 430, 358–363 10.1016/j.bbrc.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 33. Zhao D., Ma G., Zhang X., He Y., Li M., Han X., Fu L., Dong X. Y., Nagy T., Zhao Q., Fu L., and Dong J. T. (2016) Zinc finger homeodomain factor Zfhx3 is essential for mammary lactogenic differentiation by maintaining prolactin signaling activity. J. Biol. Chem. 291, 12809–12820 10.1074/jbc.M116.719377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma G., Gao A., Yang Y., He Y., Zhang X., Zhang B., Zhang Z., Li M., Fu X., Zhao D., Wu R., Qi L., Hu Q., Li J., Fu L., et al. (2019) Zfhx3 is essential for progesterone/progesterone receptor signaling to drive ductal side-branching and alveologenesis in mouse mammary glands. J. Genet. Genomics 46, 119–131 10.1016/j.jgg.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 35. Sun X., Fu X., Li J., Xing C., Frierson H. F., Wu H., Ding X., Ju T., Cummings R. D., and Dong J. T. (2014) Deletion of atbf1/zfhx3 in mouse prostate causes neoplastic lesions, likely by attenuation of membrane and secretory proteins and multiple signaling pathways. Neoplasia 16, 377–389 10.1016/j.neo.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lumpkin R. J., Gu H., Zhu Y., Leonard M., Ahmad A. S., Clauser K. R., Meyer J. G., Bennett E. J., and Komives E. A. (2017) Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 8, 1171 10.1038/s41467-017-01271-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 10.1016/j.molcel.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 38. Gong L., Kamitani T., Fujise K., Caskey L. S., and Yeh E. T. H. (1997) Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272, 28198–28201 10.1074/jbc.272.45.28198 [DOI] [PubMed] [Google Scholar]
- 39. Bassi C., Ho J., Srikumar T., Dowling R. J. O., Gorrini C., Miller S. J., Mak T. W., Neel B. G., Raught B., and Stambolic V. (2013) Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341, 395–399 10.1126/science.1236188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bischof O., Schwamborn K., Martin N., Werner A., Sustmann C., Grosschedl R., and Dejean A. (2006) The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell 22, 783–794 10.1016/j.molcel.2006.05.016 [DOI] [PubMed] [Google Scholar]
- 41. Yang S. H., and Sharrocks A. D. (2005) PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 24, 2161–2171 10.1038/sj.emboj.7600690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng J., Kang X., Zhang S., and Yeh E. T. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shimada K., Suzuki N., Ono Y., Tanaka K., Maeno M., and Ito K. (2008) Ubc9 promotes the stability of Smad4 and the nuclear accumulation of Smad1 in osteoblast-like Saos-2 cells. Bone 42, 886–893 10.1016/j.bone.2008.01.009 [DOI] [PubMed] [Google Scholar]
- 44. Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., Sung P., and Ulrich H. D. (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 10.1016/j.molcel.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 45. Du J. X., Bialkowska A. B., McConnell B. B., and Yang V. W. (2008) SUMOylation regulates nuclear localization of Kruppel-like factor 5. J. Biol. Chem. 283, 31991–32002 10.1074/jbc.M803612200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fukuda I., Ito A., Hirai G., Nishimura S., Kawasaki H., Saitoh H., Kimura K., Sodeoka M., and Yoshida M. (2009) Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 16, 133–140 10.1016/j.chembiol.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 47. Luo H. B., Xia Y. Y., Shu X. J., Liu Z. C., Feng Y., Liu X. H., Yu G., Yin G., Xiong Y. S., Zeng K., Jiang J., Ye K., Wang X. C., and Wang J. Z. (2014) SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 111, 16586–16591 10.1073/pnas.1417548111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du Y., Hou G., Zhang H., Dou J., He J., Guo Y., Li L., Chen R., Wang Y., Deng R., Huang J., Jiang B., Xu M., Cheng J., Chen G. Q., Zhao X., and Yu J. (2018) SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 46, 5195–5208 10.1093/nar/gky156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nojiri S., Joh T., Miura Y., Sakata N., Nomura T., Nakao H., Sobue S., Ohara H., Asai K., and Ito M. (2004) ATBF1 enhances the suppression of STAT3 signaling by interaction with PIAS3. Biochem. Biophys. Res. Commun. 314, 97–103 10.1016/j.bbrc.2003.12.054 [DOI] [PubMed] [Google Scholar]
- 50. Nishida T., and Yamada Y. (2011) The nucleolar SUMO-specific protease SMT3IP1/SENP3 attenuates Mdm2-mediated p53 ubiquitination and degradation. Biochem. Biophys. Res. Commun. 406, 285–291 10.1016/j.bbrc.2011.02.034 [DOI] [PubMed] [Google Scholar]
- 51. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1016/j.bbrc.2011.02.034 [DOI] [PubMed] [Google Scholar]
- 52. Li M., Zhang C., Zhong Y., and Zhao J. (2017) Cellular localization of ATBF1 protein and its functional implication in breast epithelial cells. Biochem. Biophys. Res. Commun. 490, 492–498 10.1016/j.bbrc.2017.06.068 [DOI] [PubMed] [Google Scholar]
- 53. Sun X., Li J., Sica G., Fan S. Q., Wang Y., Chen Z., Muller S., Chen Z. G., Fu X., Dong X. Y., Guo P., Shin D. M., and Dong J. T. (2013) Interruption of nuclear localization of ATBF1 during the histopathologic progression of head and neck squamous cell carcinoma. Head Neck 35, 1007–1014 10.1002/hed.23077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mabuchi M., Kataoka H., Miura Y., Kim T. S., Kawaguchi M., Ebi M., Tanaka M., Mori Y., Kubota E., Mizushima T., Shimura T., Mizoshita T., Tanida S., Kamiya T., Asai K., and Joh T. (2010) Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-β signal transduction. Biochem. Biophys. Res. Commun. 398, 321–325 10.1016/j.bbrc.2010.06.090 [DOI] [PubMed] [Google Scholar]
- 55. Kawaguchi M., Hara N., Bilim V., Koike H., Suzuki M., Kim T. S., Gao N., Dong Y., Zhang S., Fujinawa Y., Yamamoto O., Ito H., Tomita Y., Naruse Y., Sakamaki A., et al. (2016) A diagnostic marker for superficial urothelial bladder carcinoma: lack of nuclear ATBF1 (ZFHX3) by immunohistochemistry suggests malignant progression. BMC Cancer 16, 805 10.1186/s12885-016-2845-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kataoka H., Miura Y., Kawaguchi M., Suzuki S., Okamoto Y., Ozeki K., Shimura T., Mizoshita T., Kubota E., Tanida S., Takahashi S., Asai K., and Joh T. (2017) Expression and subcellular localization of AT motif binding factor 1 in colon tumours. Mol. Med. Rep. 16, 3095–3102 10.3892/mmr.2017.7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li R., Wei J., Jiang C., Liu D., Deng L., Zhang K., and Wang P. (2013) Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 73, 5742–5753 10.1158/0008-5472.CAN-13-0538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data are contained within the article.