Abstract
Fucosylation of the innermost GlcNAc of N-glycans by fucosyltransferase 8 (FUT8) is an important step in the maturation of complex and hybrid N-glycans. This simple modification can dramatically affect the activities and half-lives of glycoproteins, effects that are relevant to understanding the invasiveness of some cancers, development of mAb therapeutics, and the etiology of a congenital glycosylation disorder. The acceptor substrate preferences of FUT8 are well-characterized and provide a framework for understanding N-glycan maturation in the Golgi; however, the structural basis of these substrate preferences and the mechanism through which catalysis is achieved remain unknown. Here we describe several structures of mouse and human FUT8 in the apo state and in complex with GDP, a mimic of the donor substrate, and with a glycopeptide acceptor substrate at 1.80–2.50 Å resolution. These structures provide insights into a unique conformational change associated with donor substrate binding, common strategies employed by fucosyltransferases to coordinate GDP, features that define acceptor substrate preferences, and a likely mechanism for enzyme catalysis. Together with molecular dynamics simulations, the structures also revealed how FUT8 dimerization plays an important role in defining the acceptor substrate-binding site. Collectively, this information significantly builds on our understanding of the core fucosylation process.
Keywords: glycobiology, fucosyltransferase, glycosyltransferase, substrate specificity, glycosylation, N-linked glycosylation, structural biology, enzyme mechanism, core fucose, enzyme evolution, fucosyltransferase 8 (FUT8), protein maturation
Introduction
Fucosyltransferase 8 (FUT8)3 is the mammalian α-1,6-fucosyltransferase responsible for modifying the innermost (reducing-end) GlcNAc of hybrid and complex N-glycans. This modification, referred to as core fucosylation, is ubiquitous throughout mammalian tissues and is important in the maturation of complex N-glycans within the Golgi apparatus. Core fucosylation modulates the activity of many cell surface receptors, including TGFβ1R (1, 2), epidermal growth factor receptor (3), B cell receptor (4), T cell receptor (5, 6), CD14-mediated TLR2/4 signaling (7, 8), and PD-1 (9). It also modulates the affinity of ligands for their receptors, the most notable example being the role of core fucose in decreasing the affinity of IgG for FcγRIIIa (10, 11). This latter phenomenon has inspired the development of next-generation therapeutic monoclonal antibodies that more effectively engage FcγRIIIa and demonstrate superior antibody-dependent cellular cytotoxicity (11, 12). Recently, dectin-I was identified as the first endogenous lectin that specifically recognizes core fucose (13). Collectively, these and other findings demonstrate that FUT8 plays a central role in modulating the activity of many cell surface receptors.
In mice, loss-of-function mutations in FUT8 result in severe growth retardation and development of an emphysema-like lung phenotype, purportedly because of dysregulation of TGF-β1 and epidermal growth factor receptor signaling (1, 14). These animals also exhibit behavioral abnormalities (15). Many of these phenotypes are also observed in patients with the recently described FUT8 congenital disorder of glycosylation (FUT8-CDG) (16). In contrast to FUT8-CDG, which features ablation of FUT8 activity, many cancers up-regulate FUT8 expression and this correlates with a poor prognosis (17). In melanomas, increased FUT8 activity stabilizes L1CAM to promote metastasis (18). Metastasis is also promoted by FUT8 in breast cancers, where increased core fucosylation of TGFβ1R promotes strong constitutive signaling through this receptor and tumor cell migration (19). Increased core fucosylation of α-fetoprotein is also a well-established biomarker of hepatocellular carcinoma (20).
Some have speculated that FUT8 antagonists may have therapeutic potential for the treatment of cancer (9, 18), but questions remain regarding how a hypothetical FUT8 antagonist might impact host immune responses to tumor cells. Regardless, no drug-like small-molecule inhibitors have yet been reported for FUT8 or any other human FUT. To some degree, drug discovery efforts are impeded by a limited structural understanding of this enzyme and the mechanism it employs to perform core fucosylation. The only reported FUT8 structure possesses no bound ligands (21), and our only insights into donor and acceptor substrate binding come from NMR, molecular dynamics and docking studies (22, 23). To gain a thorough understanding of how FUT8 recognizes its donor and acceptor substrates to catalyze core fucosylation, we revisited the structural biology of FUT8. The structures we obtained provide fresh insights into the conformational dynamics and molecular interactions associated with catalysis.
Results
Structural insights into nucleotide recognition by FUT8
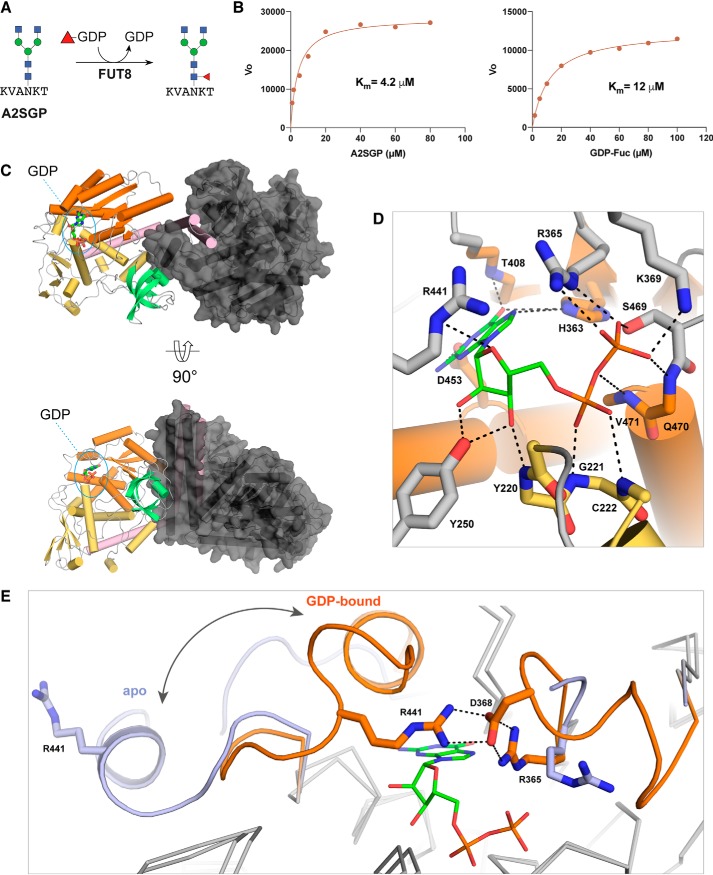
A truncated human FUT8 (HsFUT8105–575) construct missing the N-terminal transmembrane domain and unstructured region was expressed in Sf21 insect cells. The activity of the purified protein was verified using the GDP-GloTM glycosyltransferase assay with an asialo-agalacto-biantennary glycopeptide (A2SGP) derived from chicken eggs as an acceptor substrate (Fig. 1A). Using this assay, we determined a Km of 4.2 μm for GDP-fucose (GDP-Fuc) and 12 μm for A2SGP (Fig. 1B), which was in broad agreement with values reported previously (24).
Figure 1.
Structures of mouse and human FUT8 with and without GDP bound. A, the reaction catalyzed by FUT8 in our GDP-Glo assay. B, HsFUT8 is active and has Km values that are in agreement with those reported previously (24). C, the domain structure of FUT8 (coiled coil, pink; Rossman, orange and yellow; SH3, teal) and the interactions between each the two molecules in the asymmetric unit. D, the hydrogen-bonding interactions between MmFUT8 and GDP (see also Fig. S4). E, an overlay of the GDP-binding sites of apo-HsFUT8 (blue) and GDP-bound MmFUT8 (orange), illustrating the conformational changes observed for loops A and B. A salt bridge between Asp368 and Arg365/Arg441 from loops A and B, respectively, forms upon encapsulation of GDP.
Because glycosyltransferases rapidly hydrolyze their sugar nucleotide donor substrates on a protein crystallization timescale, we attempted to cocrystallize HsFUT8 with GDP rather than GDP-Fuc. These attempts failed to provide any crystals of the complex, but we did obtain crystals of the apo form that enabled redetermination of the unliganded structure at higher resolution (2.28 Å) than the existing structure (2.61 Å for PDB code 2DE0) with superior refinement statistics (Table 1) (21). As an alternative approach, we cloned and expressed mouse Fut8 (MmFUT868–575) using a similar method as for the human protein. MmFUT8 is 96.6% identical to the human homolog over the length of this truncated construct (Fig. S1). Extensive crystallization screens with this slightly different protein and GDP provided crystals that yielded a structure of MmFUT8 in its apo (1.80 Å) and GDP-bound (2.50 Å) forms (Table 1).
Table 1.
Refinement statistics for the structures reported in this study
| Structure | apo-MmFUT8 | MmFUT8–GDP | apo-HsFUT8 | HsFUT8–GDP–A2SGP |
|---|---|---|---|---|
| PDB ID | 6VLF | 6VLG | 6VLE | 6VLD |
| Space group | C 1 2 1 | P 65 2 2 | P 1 21 1 | C 1 2 1 |
| No. of protein chains in AU | 2 | 4 | 2 | 4 |
| Cell dimensions | ||||
| a, b, c (Å) | 184.87, 71.34, 126.95 | 150.82, 150.82, 472.14 | 95.02, 62.20, 109.90 | 208.31, 68.45, 249.98 |
| α, β, γ (°) | 90, 126.08, 90 | 90, 90, 120 | 90, 90.88, 90 | 90, 111.21, 90 |
| Wavelength (Å) | 0.9537 | 0.9537 | 0.9537 | 0.9537 |
| Resolution (Å)* | 49.20–1.80 (1.83–1.80) | 49.37–2.50 (2.54–2.50) | 47.50–2.28 (2.34–2.28) | 46.65–2.28 (2.32–2.28) |
| Rsym or Rmerge* | 0.047 (1.167) | 0.150 (0.664) | 0.120 (1.520) | 0.095 (1.347) |
| Rpim* | 0.044 (1.055) | 0.066 (0.366) | 0.092 (1.143) | 0.066 (0.949) |
| I/sI* | 10.6 (0.9) | 9.5 (2.0) | 8.2 (1.0) | 10.2 (1.1) |
| CC(1/2) | 0.999 (0.463) | 0.995 (0.811) | 0.997 (0.630) | 0.998 (0.435) |
| Completeness (%)* | 99.6 (99.7) | 98.8 (85.8) | 99.8 (100) | 99.9 (99.9) |
| Redundancy* | 3.4 (3.5) | 10.4 (6.2) | 5.0 (5.1) | 5.7 (5.6) |
| Wilson B-factor (Å2) | 31.78 | 33.56 | 40.13 | 47.03 |
| Refinement | ||||
| Resolution (Å) | 46.42–1.80 | 49.37–2.50 | 47.26–2.28 | 48.66–2.28 |
| No. of reflections | 123,188 | 108,979 | 58,849 | 150,217 |
| Rwork/Rfree | 0.1842 / 0.2082 | 0.1822 / 0.2273 | 0.2019 / 0.2340 | 0.1914 / 0.2239 |
| No. of non-hydrogen atoms | ||||
| Protein | 7384 | 15248 | 7492 | 15083 |
| GDP | n/a | 112 | n/a | 56 |
| A2SGP | n/a | n/a | n/a | 392 |
| Water | 626 | 925 | 376 | 628 |
| B-factors | ||||
| Protein | 43.4 | 43.3 | 50.9 | 64.2 |
| GDP | n/a | 32.0 | n/a | 56.2 |
| A2SGP | n/a | n/a | n/a | 65.6 |
| Water | 47.9 | 43.0 | 48.5 | 55.3 |
| RMSDs | ||||
| Bond lengths (Å) | 0.005 | 0.003 | 0.001 | 0.002 |
| Bond angle (°) | 0.72 | 0.61 | 0.417 | 0.482 |
| Ramachandran plot (%) | ||||
| Favored | 97.62 | 96.73 | 97.8 | 96.58 |
| Allowed | 2.38 | 3.22 | 2.2 | 3.42 |
| Disallowed | 0 | 0 | 0 | 0.05 |
* Values in parentheses are for highest-resolution shell.
The overall fold of these three FUT8 crystal structures is as described previously (21): an N-terminal coiled-coil domain followed by two Rossman folds forming a GDP substrate–binding site and a C-terminal SH3 domain of unknown function, as illustrated for MmFUT8-GDP in Fig. 1C. The backbone root mean square deviation between the four structures is low, <0.4 Å (Fig. S2). The most notable backbone perturbations observed are for two loops, Arg365-Ala375 (loop A) and Asp429-Asn446 (loop B), which are disordered or displaced in the apo-MmFUT8 and apo-HsFUT8 structure but become ordered and completely encapsulate GDP upon binding (Fig. 1, D and E). This reorganization involves the creation of several new interactions between both loops, most notably a salt bridge between Asp368 and Arg365 of loop A and Arg441 of loop B (Fig. 1E). Arg365 also forms a salt bridge with the β-phosphate of GDP, providing a link between ligand binding and organization of the encapsulating loops (Fig. 1, D and E). Mutation of Arg365 to Ala abolishes FUT8 activity (25), confirming that this residue plays a key role in organizing the encapsulating loops around the nucleotide. A detailed list of all GDP-FUT8 hydrogen bonds is provided in Fig. S3. Other noteworthy interactions include those between Asp453/His363 and the guanine base and the interactions between the ribose hydroxyl groups and Tyr250 (Fig. 1D).
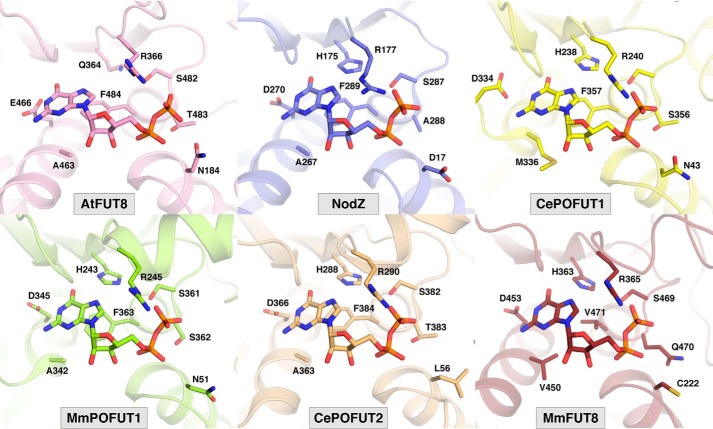
The conformational change associated with GDP encapsulation by FUT8 is unique among the FUTs studied to date. It results in burial of 96% of GDP's surface area (Fig. S3). This is comparable with or slightly higher than that observed for other FUT–GDP complexes, including AtFUT1 (95%) (26), NodZ (83%) (27), CePOFUT1 (90%) (28), MmPOFUT1 (86%) (29), and CePOFUT2 (95%) (30) (Fig. 2). Remarkably, the conformational pose of the GDP ligand and GDP–FUT interactions are nearly identical in all FUTs, including FUT8, despite significant divergence in sequence and domain architecture. Residues analogous to FUT8's Ser469, Arg365, Asp453, and His363 are conserved at a structural level across all FUTs (Fig. 2). This observation has important consequences for development of competitive inhibitors of FUTs.
Figure 2.
Shown are conserved residues defining the GDP-binding site across all known structures of fucosyltransferases in complex with GDP (PDB codes 5KWK (AtFUT1) (26), 3SIW (NodZ) (27), 3ZY3 (CePOFUT1) (28), 5KXQ (MmPOFUT1) (29), 5FOE (CePOFUT2) (30), and 6VLG (MmFUT8).
Structural insights into N-glycan acceptor substrate recognition by FUT8
Cocrystallization of HsFUT8105–575 with GDP and the A2SGP glycopeptide acceptor substrate provided crystals that yielded a structure with four HsFUT8 monomers in the asymmetric unit. All molecules were bound to A2SGP, but only one molecule in each dimer pair also bound GDP. Therefore, this structure provided information for the FUT8–GDP–A2SGP ternary complex and the FUT8–A2SGP binary complex. The apparent ability of GDP and A2SGP to bind FUT8 independently of each other is consistent with a rapid equilibrium random mechanism, which has been established previously for FUT8 (24).
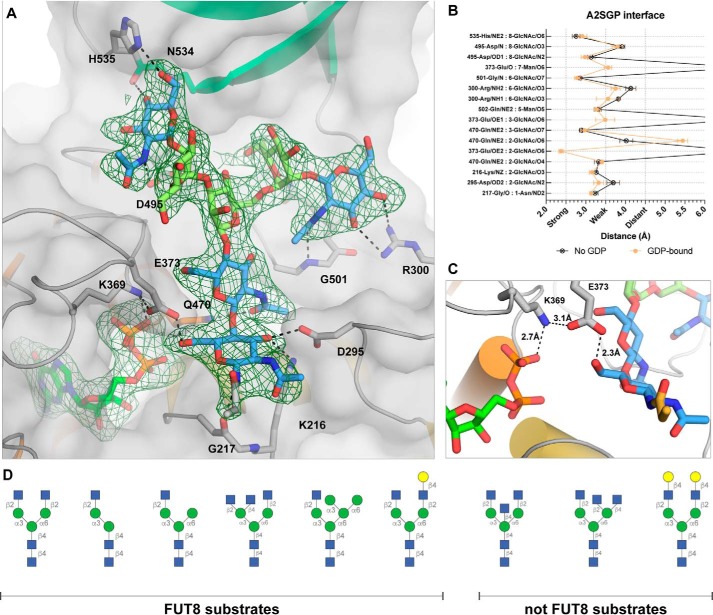
All sugars of the A2SGP substrate, and the Asn side chain to which they were attached, were resolved and modeled for each monomer (Fig. 3A). Upon binding to FUT8, the N-glycan buries 44% of its surface area. Hydrogen-bonding interactions between FUT8 and A2SGP are almost exclusively between the enzyme and the GlcNAc units comprising the core chitobiose unit and nonreducing ends of the bisected glycan (Fig. 3, A and B); the mannose units do not make any notable hydrogen-bonding interactions with FUT8. For the two protein chains in the asymmetric unit without GDP bound, loops A and B remain disordered, as for the apo structures. However, in the ternary complex with GDP bound, these loops encapsulate GDP, as observed for the MmFUT8-GDP structure. Inspection of the region between the β-phosphate of GDP and the 6-hydroxyl group of the innermost GlcNAc residue of A2SGP provides insights into which residues play a role in catalysis. Glu373 forms an intimate hydrogen bond (2.3 Å) with the 6-hydroxyl group and also interacts with Lys369, which, in turn, forms an intimate contact with the β-phosphate of GDP (Fig. 3C). This suggests that Glu273 acts as the catalytic base for catalysis and is capable of relaying a proton through Lys369 to the departing β-phosphate of GDP. In this way, the Glu273/Lys369 pair acts as proton conduit and catalytic base/acid, respectively.
Figure 3.
Acceptor substrate recognition by FUT8. A, interactions between FUT8 and the A2SGP N-glycan acceptor substrate, with an Fo-Fc omit map contoured at 1.5σ around N-glycan and GDP. Residues making hydrogen bond interactions with the N-glycan are indicated. The SH3 domain is colored in teal. B, a list of the interactions and hydrogen bond distances between A2SGP and FUT8. C, a close-up of the active site, illustrating a potential role for Glu373 and Lys369 as a proton relay to facilitate electrophilic migration of fucose from GDP-Fuc to the 6-hydroxyl group of the innermost GlcNAc. D, a selection of N-glycans known to be modified or not modified by FUT8 (31, 32), for comparison to the structure depicted in A.
Two previous studies have explored the acceptor substrate specificity of FUT8 to ascertain which features promote or impair FUT8 activity (31, 32), and these results are summarized in Fig. 3D. It is clear, through comparison with our structure, that a bisecting GlcNAc would be sterically occluded by the SH3 domain of FUT8 (Fig. 3A), consistent with this modification's ability to block core fucosylation (31, 32). Modifications of the α6-branch of the glycan are well-tolerated by FUT8, and it is clear from our structure that there is sufficient space to accommodate most types of truncation, elongation, or branching at this position. The one exception is elongation with a terminal β-1,4-GlcNAc, which would introduce steric clashes with the SH3 domain. Modification of the α3 branch is not well-tolerated by FUT8, and all typical elongation or branching residues (Fig. 3D) introduce steric clashes that would preclude binding. Notably, FUT8 activity requires the terminal β-1,2-GlcNAc of the α3 branch, suggesting that the intimate hydrogen bond between His353 and the 6-hydroxyl group of this GlcNAc is an important contributor to acceptor substrate binding.
FUT8 dimerization and orientation of the SH3 domain for acceptor substrate recognition
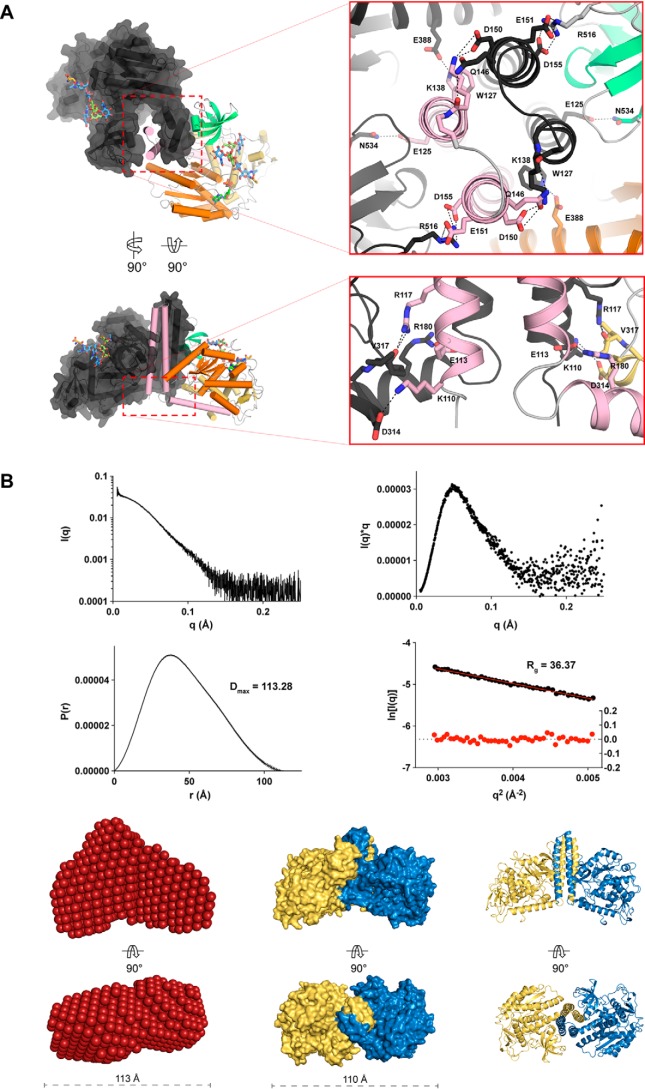
In the asymmetric units of all structures determined here, MmFUT8 and HsFUT8 formed an apparent dimer through formation of a four-helix bundle from their N-terminal coiled-coil domains. These helices interact with their neighbor's SH3 domain (Fig. 4A). This dimer could be observed in the previously determined apo structure of FUT8 (21), but the structure was reported as a monomer. Other publications have reported that HsFUT8 is a monomer in solution, based on size-exclusion chromatography experiments (33), and this has influenced the way in which molecular dynamics and docking simulations of FUT8 have been conducted (22, 23), potentially compromising the conclusions. To address this inconsistency in the literature, we performed SEC-SAXS on FUT8 (Fig. 4, B and C), which conclusively demonstrated that FUT8 exists as a dimer in solution. The buried surface area for dimerization was similar for all four structures reported here (Table S4), irrespective of which ligands were bound, and involves the same residues forming inter-chain salt bridges and hydrogen bonds (Fig. 4A).
Figure 4.
Interactions between the N-terminal coiled-coil domains drive FUT8 dimerization in solution. A, self-association of the N-terminal helices of FUT8 creates a four helix bundle that buries hydrophobic residues and creates multiple interchain salt bridges. B, intensity plot of FUT8 SAXS scattering (top left), a Kratky plot derived from FUT8 scattering showing that it forms a compact particle in solution (top right), and P(r) and Guinier plots indicating that FUT8 has a maximum dimension of 113.28 Å in solution (bottom left) and a radius of gyration of 36.37 Å (bottom right). C, a bead model of FUT8 in solution generated from solution scattering data (left) corresponds well with the dimer observed in the FUT8 crystal structure, shown as a surface (center) and cartoon (right) view.
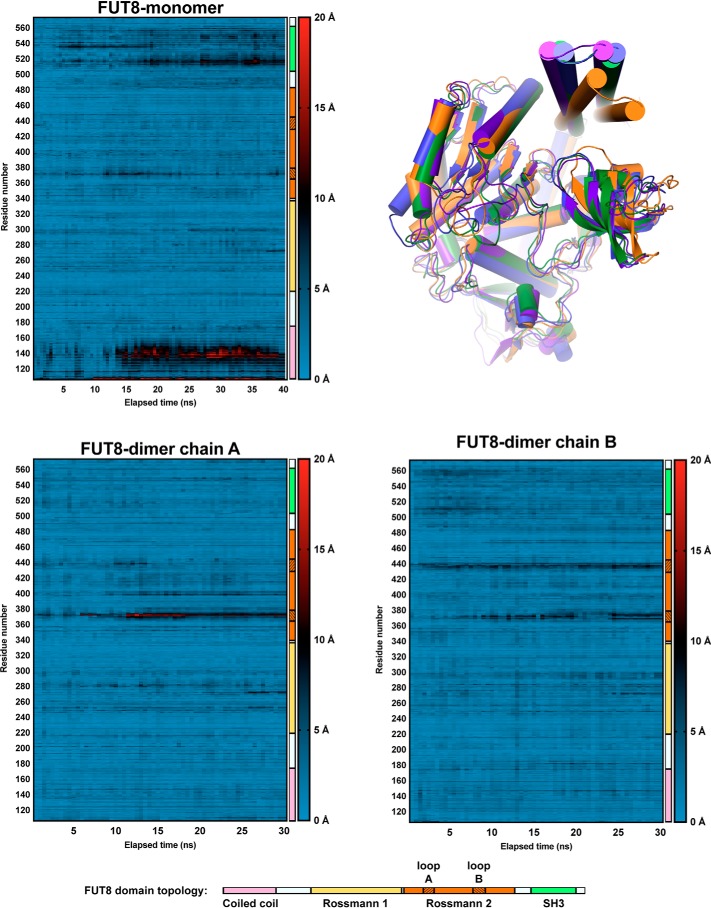
As mentioned, previous docking and molecular dynamics simulations were conducted based on the assumption that FUT8 is a monomer and suggested that the SH3 domain moves significantly after 20 ns of simulation (22). Because the SH3 domain plays an important role in recognizing the acceptor substrate, this would appear to be deleterious for catalysis. However, when considered as a dimer complex, the SH3 domain clearly binds the neighboring chain's N-terminal coiled-coil domain, which would appear to lock the SH3 domain in place (Fig. 4A). To address this discrepancy, we performed molecular dynamics simulations of unliganded FUT8 in the monomeric and dimeric state over a period of 40 and 30 ns, respectively (Fig. 5). Movements in the N-terminal coiled-coil domain and SH3 domain in the monomeric structure were replicated, with this conformational change enabling burial of hydrophobic residues and disruption of the acceptor-binding surface of the enzyme. However, in the dimeric structure, no such movements were observed (Fig. 5). In fact, for the dimer, the only significant motion observed on this timescale was in the active-site loops A and B that encapsulate GDP. These data suggest that dimerization of FUT8 is an important adaptation for buttressing the SH3 domains to maintain an extended bifurcated surface that can accommodate the bisected N-glycan acceptor substrate.
Figure 5.
Heatmaps illustrating the displacement experienced by each amino acid in FUT8 during 30–40 ns of molecular dynamics simulation. The domain to which each residue belongs is illustrated for reference, with the shaded regions of the second Rossmann domain denoting the mobile GDP-binding loops A and B of the active site. An overlay of each monomer at t = 0 ns and the end points is provided at the top right (green is at t = 0 ns, orange is the monomer at t = 40 ns, and blue and purple are dimer chains at t = 30 ns).
Discussion
This collection of structures has revealed a unique conformational change in FUT8 associated with capturing GDP and, presumably, its donor substrate GDP-Fuc. Arg365 plays a pivotal role in this process by forming a salt bridge with the β-phosphate of GDP and Asp368/Arg441 of the mobile loops. The critical importance of this residue for catalysis is supported by previous mutagenesis studies (25). Despite this unusual feature, when GDP is bound, its spatial orientation and the interactions it makes with FUT8 are largely the same as for other FUTs (26–30). This observation is particularly relevant for those seeking to develop competitive inhibitors of FUTs; selectivity may be difficult to obtain with GDP mimics or molecular scaffolds that only interact with the GDP-binding site. On the other hand, these commonalities suggest that small-molecule scaffolds may exist with pan-FUT inhibitory activity and that these might be adapted into selective inhibitors by exploring the acceptor binding site.
Original investigations into FUT8 mechanism by Ihara et al. (24) indicated that FUT8 utilizes a rapid equilibrium random mechanism. This model postulates that substrates can bind independently to the enzyme in any order to form the Michaelis complex. The fact that we were able to obtain structures of FUT8 bound to acceptor or GDP alone as well as the ternary complex supports this mechanistic model. Our structures also reveal that there are no significant structural rearrangements associated with N-glycan binding and that ordering of loops A and B upon GDP binding occurs independent of the N-glycan binding site. Perhaps the greatest insights obtained from our structures with respect to enzyme mechanism is the realization that the Glu273/Lys369 residues are the key catalytic residues. Our ternary complex clearly illustrates that Glu273 forms a close contact with the 6-hydroxyl group of the innermost GlcNAc of the N-glycan acceptor substrate and that its basicity is modulated through interactions with Lys369. Therefore, Glu273 is the clear catalytic base residue. Concomitantly, Lys369 is able to shuttle a proton from Glu273 to the β-phosphate of GDP, with which it forms a salt bridge, to facilitate departure of GDP from GDP-Fuc, enabling electrophilic migration of fucose onto the hydroxyl group nucleophile of the acceptor substrate.
With core fucosylation playing such an important role in the function of proteins and maturation of N-glycans, a great deal of effort has been invested in profiling the acceptor substrate preferences of this enzyme. Our structures, and one recently reported by another laboratory (34), provide a basis for understanding the vagaries of FUT8 substrate preferences in vitro (31, 32). What is clear from our work is that the SH3 domain of FUT8 plays a defining role in recognizing N-glycan acceptor substrates. Modifications to the α3 branch of an N-glycan or a bisecting GlcNAc introduces steric clashes with the SH3 domain that prevents them from binding to FUT8. His535 of the SH3 domain also appears to form a crucial hydrogen bond with the nonreducing end GlcNAc of N-glycan substrates, which is important but not always essential for modification by FUT8 (35). The importance of this SH3 domain for defining the acceptor substrate binding site necessitates its rigidity. Our molecular dynamics simulations suggest that FUT8 dimerization has evolved to maintain this rigidity by buttressing the C-terminal SH3 domain of one chain against the N-terminal coiled-coil domain of the other chain. What this report and other structural investigations have failed to address is how the structure, hydrophobicity, and charge of glycoprotein susbtrates impacts the accessibility of their glycans to FUT8 (36); this remains an important question for future investigation.
Conclusion
FUT8 possess a unique method of capturing its GDP-Fuc donor substrate by using two mobile loops to encapsulate the nucleotide portion of this molecule; this process is largely driven by Arg365, which drives salt bridge formation between GDP and the two mobile loops. This unique feature aside, FUT8 recognizes GDP in much the same way as other FUTs, suggesting that all of these enzymes might be targetable with a common chemical scaffold. A ternary complex of FUT8, GDP, and N-glycan acceptor substrate revealed that Glu273 and Lys369 play a direct role in catalysis, with Glu273 acting as catalytic base and Lys369 relaying a proton from Glu273 to the departing phosphate of the GDP-Fuc substrate. This complex also revealed the importance of the SH3 domain in providing a bifurcated surface for N-glycan recognition and in defining the acceptor substrate scope of FUT8. The importance of the SH3 domain in substrate binding appears to have driven the evolution of FUT8 as a dimer, which restricts movement of the SH3 domain and stabilizes the acceptor-binding subsite.
Materials and methods
Cloning, expression, and purification of human and mouse FUT8
A gene encoding an N-terminal gp67 signal peptide, residues 105–585 of human FUT8 (UniProt ID Q9BYC5) and a C-terminal His10 tag (Table S1) was synthesized and cloned into the pFastBac-1 vector (Thermo Fisher) using RsrII/XhoI. A gene encoding an N-terminal gp67 signal peptide, V5 epitope tag, His10 tag, factor Xa site, and residues 68–585 of mouse FUT8 (UniProt ID Q9WTS2) (Table S1) was synthesized and cloned into the pFastBac-1 vector (Thermo Fisher) using RsrII/XhoI. Both constructs were expressed in Sf21 insect cells (Thermo Fisher) using the Bac-to-Bac Baculovirus Expression System (Thermo Fisher) following the manufacturer's protocol. Briefly, each plasmid was transformed into chemically competent DH10Bac Escherichia coli cells (Thermo Fisher), and positive clones were identified through a blue-white screen. The bacmid was prepared from these cells and transfected into Sf21 insect cells conditioned in Insect-XPRESS Protein-free Insect Cell Medium with l-glutamine (Lonza Ltd.) using Cellfectin II reagent (Thermo Fisher). The virus was passaged on Sf21 insect cells three times. For protein expression, 1 liter of Sf21 cells at a density of 1–2 × 106 cells ml−1 was infected with 30 ml of the P3 baculovirus and cultured at 27 °C for 72 h. The cells were pelleted by centrifugation (8,000 × g, 20 min, 4 °C), and the supernatant was collected. 10× buffer solution (112 ml, 500 mm Tris (pH 7.5), and 3 m NaCl) was added to the supernatant before it was filtered through a 0.22-μm membrane. The buffered and filtered supernatant was passed through a pre-equilibrated 5-ml HisTrap Excel column (GE Healthcare). The column was washed with 20 column volumes of 50 mm Tris (pH 7.5), 300 mm NaCl, followed by a one-step elution in 50 mm Tris (pH 7.5), 300 mm NaCl, and 500 mm imidazole to elute FUT8 from the column. Fractions containing protein, as judged by SDS-PAGE, were pooled and further purified by size-exclusion chromatography (SEC) using a HiLoad 16/600 Superdex 200 column (GE Healthcare) equilibrated in 50 mm Tris (pH 7.5) and 150 mm NaCl. For MmFUT868–575, the N-terminal tags were removed using Factor Xa protease (New England BioLabs) by incubation in 50 mm Tris (pH 6.5), 150 mm NaCl, and 2 mm CaCl2 overnight at room temperature. For HsFUT8105–575, the C-terminal His10 affinity tag was removed using Carboxypeptidase A (Merck) by incubation in 50 mm Tris (pH 7.5), 150 mm NaCl overnight at room temperature. Protease-treated MmFUT868–575 and HsFUT8105–575 were purified by running the reactions through a pre-equilibrated 1 ml HisTrap Excel column (GE Healthcare) and performing SEC on the column flow-through using a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated in 50 mm Tris (pH 7.5), 150 mm NaCl. Concentration of the proteins was accomplished using Amicon centrifugal filters, nominal molecular weight limit 10 kDa (Merck-Millipore). Protein yields varied between batches but were always in the region of 5–10 mg per liter of cell culture.
GDP-Glo assay of FUT8 activity
FUT8 activity was assayed using the GDP-GloTM glycosyltransferase assay (Promega) with 3-μl reactions being conducted in a 1536-well microtiter plate. Reactions contained assay buffer (50 mm Tris (pH 7), 100 mm NaCl, 0.01% Triton X-100, and 0.1% BSA), 5 μm A2SGP (Fushimi Pharmaceutical Co.), 10 μm GDP-Fuc, and 5 nm FUT8 unless stated otherwise. After incubation for 20 min at room temperature, the reactions were stopped by addition of 1 μl of 4% acetic acid prepared in assay buffer for 10 min. The resulting decrease in pH completely inactivated FUT8. To bring the pH back to neutral, 1 μl of 700 mm NaOH prepared in assay buffer was added to the reaction for 2 min. To detect the GDP product, 2.5 μl of GDP-GloTM glycosyltransferase assay (Promega) nucleotide detection reagent was added. Plates were sealed and incubated at room temperature for 60 min. Chemiluminescence was quantitated on an EnVision Multimode plate reader (PerkinElmer Life Sciences). The readout time was 0.1 s per well. To determine the Km of the A2SGP acceptor substrate under these conditions, reactions were conducted with serial dilutions of A2SGP starting at 80 μm, with 15 μm GDP-Fuc and 5 nm FUT8. To determine the Km of the GDP-Fuc donor substrate under these conditions, reactions were conducted with serial dilutions of GDP-Fuc starting at 100 μm, with 5 μm A2SGP and 5 nm FUT8. All data were fit to the appropriate model using Prism 8 (GraphPad).
Crystallization of apo-MmFUT868–575
MmFUT868–575 in 50 mm Tris (pH 7.4) and 300 mm NaCl was concentrated to 15 mg ml−1. A single crystal was grown over 2 weeks in a sitting drop at room temperature by mixing 0.5 μl of well solution containing 0.25 m (NH4)2SO4 and 10% PEG 3350 with 0.5 μl of MmFUT8 solution. The crystal was cryoprotected by supplementing the mother liquor with 25% glycerol and cryocooled using liquid nitrogen.
Crystallization of MmFUT868–575 in complex with GDP
MmFUT868–575 in 50 mm Tris (pH 7.4), 300 mm NaCl, and 1 mm GDP at 0.1 mg ml−1 was incubated at 4 °C overnight prior to a final concentration of 15 mg ml−1 MmFUT868–575. A single crystal was grown over a week in a sitting drop at room temperature by mixing 0.5 μl of well solution containing 0.25 m (NH4)2SO4 and 10% PEG 3350 with 0.5 μl of MmFUT8:GDP solution. The crystal was cryoprotected by supplementing the mother liquor with 25% glycerol and cryocooled using liquid nitrogen.
Crystallization of apo-HsFUT8105–575
HsFUT8105–575 in 50 mm Tris (pH 7.4) and 50 mm NaCl was concentrated to 2 mg ml−1. Crystals were grown over 3 days at 20 °C by mixing 1 μl of well solution containing 12% (w/v) PEG 20000, 2.5% (v/v) DMSO, and 0.1 m HEPES (pH 7.5) with 1 μl of protein solution. The crystal was cryoprotected by supplementing the mother liquor with 25% ethylene glycol (v/v) and cryocooled using liquid nitrogen.
Crystallization of HsFUT8 in complex with A2SGP and GDP
HsFUT8105–575 in 50 mm Tris (pH 7.4) and 50 mm NaCl at 3 mg ml−1 was mixed with A2SGP and GDP (in water) to final concentrations of 2 mg ml−1 HsFUT8105–575, 0.5 mm A2SGP, and 2 mm GDP. The mixture was incubated on ice for 30 min prior to setting up crystallization experiments. A single crystal was grown over 10 weeks in sitting drops at 8 °C by mixing 1 μl of well solution containing 2 m NH4SO4, 0.2 m NaCl, and 0.1 m sodium cacodylate (pH 6.5) with 1 μl of FUT8:A2SGP:GDP solution. The crystal was cryoprotected by supplementing the mother liquor with 3 m NH4SO4 and cryocooled using liquid nitrogen.
Data collection and structure determination
Data were collected at the Australian Synchrotron (MX2 beamline) and processed using XDS (37). All structures were solved by molecular replacement using PHASER (38) using the apo structure of human FUT8 as a search model (PDB code 2DE0) (21). The final models were built in Coot (39) and refined with Phenix (40). Data collection and refinement statistics are summarized in Table 1. Figures were prepared using PyMOL.
Small-angle X-ray scattering and modeling
SEC-SAXS was performed using a Coflow apparatus at the Australian Synchrotron (41, 42). Purified MmFUT8 was analyzed at a preinjection concentration of 100 μm. Chromatography for SEC-SAXS was performed at 22 °C with a 5/150 Superdex S200 Increase column at a flow rate of 0.4 ml min−1 in 50 mm Tris (pH 7.9), 100 mm NaCl, 5% glycerol, and 0.2% sodium azide. Inclusion of glycerol and azide was essential to prevent capillary fouling because of photo-oxidation of buffer components. Scattering data were collected for 1-s exposures over a q range of 0.01–0.51 Å−1. A buffer blank for each SEC-SAXS run was prepared by averaging 10–20 frames pre- or post-protein elution. Scattering curves from peaks corresponding to MmFUT8 were then buffer-subtracted, scaled across the elution peak, and compared for interparticle effects. Identical curves (five to ten) from each elution were then averaged for analysis. Data were analyzed using the ATSAS package, Scatter, and SOMO solution modeler (43).
Molecular dynamics
The FUT8 monomeric and dimeric systems were created using either one or two copies of chain A from the HsFUT8 substrate bound structure (PDB code 6VLD). Each system was solvated in an orthorhombic box, expanding 12 Å in each direction from the protein chain(s), and neutralized with Na+ and Cl− at 150 mm. These steps were carried out with the AutoPSF Builder in VMD 1.9.4 (44). Molecular dynamics were simulated using the CHARMM36 force field for proteins (45), the TIP3P (46) water model, and sodium and chloride ion parameters from Beglov and Roux (47). Both systems were minimized with NAMD version 2.13 (48) using a conjugate gradient for 10,000 steps. Next, the systems were annealed by heating from 60 K to 300 K at a rate of 10 K/12 ps. After annealing, both systems were allowed to equilibrate at 300 K for 1,000 ps. The annealing and equilibration phases were carried out in a constant pressure/temperature (NPT) ensemble using the Langevin piston barostat set to 1 atm and with harmonic constraints on all nonhydrogen protein atoms. After this, the harmonic restraints on the protein were removed, and the monomeric system was simulated for 40 ns and the dimeric system for 30 ns. All simulations were performed using a time step of 2 fs and using periodic boundary conditions with the particle mesh Ewald method to determine the electrostatics of the system. The aligned backbone root mean square deviations of the trajectories were calculated with the RMSDVT Visualizer Tool in VMD and plotted in Prism 8 (GraphPad).
Author contributions
M. A. J., M. D., and E. D. G.-B. data curation; M. A. J., M. D., and E. D. G.-B. formal analysis; M. A. J. and E. D. G.-B. validation; M. A. J., M. D., J. P. L., R. M., A. J., K. E. J., R. G., and E. D. G.-B. investigation; M. A. J. and E. D. G.-B. visualization; M. A. J., M. D., J. P. L., R. M., A. J., K. E. J., R. G., and E. D. G.-B. methodology; M. A. J., M. D., and E. D. G.-B. writing-original draft; M. A. J. and E. D. G.-B. project administration; E. D. G.-B. conceptualization; E. D. G.-B. resources; E. D. G.-B. supervision; E. D. G.-B. funding acquisition; E. D. G.-B. writing-review and editing.
Supplementary Material
Acknowledgments
We thank the beamline staff at the Australian Synchrotron for help with X-ray data collection as well as Dr. Janet Newman and Dr. Bevan Marshall at the Commonwealth Scientific and Industrial Research Organization Collaborative Crystallization Centre (C3) for assistance with protein crystallization. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation detector.
This work was supported by The Walter and Eliza Hall Institute of Medical Research, National Health and Medical Research Council of Australia Project Grant GNT1139549, the Australian Cancer Research Fund, and a Victorian State Government Operational Infrastructure support grant. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Table S1.
The atomic coordinates and structure factors (codes 6VLD, 6VLE, 6VLF, and 6VLG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FUT
- fucosyltransferase
- TGF
- transforming growth factor
- A2SGP
- asialo-agalacto-biantennary glycopeptide
- Fuc
- fucose
- SH
- Src homology
- SEC
- size-exclusion chromatography
- SAXS
- small-angle X-ray scattering.
References
- 1. Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., Mizuno-Horikawa Y., Nakano M., Asahi M., Takahashi M., Uozumi N., Ihara S., Lee S. H., Ikeda Y., Yamaguchi Y., et al. (2005) Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 102, 15791–15796 10.1073/pnas.0507375102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin H., Wang D., Wu T., Dong C., Shen N., Sun Y., Sun Y., Xie H., Wang N., and Shan L. (2011) Blocking core fucosylation of TGF-β1 receptors downregulates their functions and attenuates the epithelial-mesenchymal transition of renal tubular cells. Am. J. Physiol. Renal Physiol. 300, F1017–F1025 10.1152/ajprenal.00426.2010 [DOI] [PubMed] [Google Scholar]
- 3. Matsumoto K., Yokote H., Arao T., Maegawa M., Tanaka K., Fujita Y., Shimizu C., Hanafusa T., Fujiwara Y., and Nishio K. (2008) N-glycan fucosylation of epidermal growth factor receptor modulates receptor activity and sensitivity to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 99, 1611–1617 10.1111/j.1349-7006.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li W., Yu R., Ma B., Yang Y., Jiao X., Liu Y., Cao H., Dong W., Liu L., Ma K., Fukuda T., Liu Q., Ma T., Wang Z., Gu J., et al. (2015) Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J. Immunol. 194, 2596–2606 10.4049/jimmunol.1402678 [DOI] [PubMed] [Google Scholar]
- 5. Fujii H., Shinzaki S., Iijima H., Wakamatsu K., Iwamoto C., Sobajima T., Kuwahara R., Hiyama S., Hayashi Y., Takamatsu S., Uozumi N., Kamada Y., Tsujii M., Taniguchi N., Takehara T., and Miyoshi E. (2016) Core fucosylation on T cells, required for activation of T-cell receptor signaling and induction of colitis in mice, is increased in patients with inflammatory bowel disease. Gastroenterology 150, 1620–1632 10.1053/j.gastro.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 6. Liang W., Mao S., Sun S., Li M., Li Z., Yu R., Ma T., Gu J., Zhang J., Taniguchi N., and Li W. (2018) Core fucosylation of the T cell receptor is required for T cell activation. Front. Immunol. 9, 78 10.3389/fimmu.2018.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iijima J., Kobayashi S., Kitazume S., Kizuka Y., Fujinawa R., Korekane H., Shibata T., Saitoh S. I., Akashi-Takamura S., Miyake K., Miyoshi E., and Taniguchi N. (2017) Core fucose is critical for CD14-dependent Toll-like receptor 4 signaling. Glycobiology 27, 1006–1015 10.1093/glycob/cwx075 [DOI] [PubMed] [Google Scholar]
- 8. Nakayama K., Wakamatsu K., Fujii H., Shinzaki S., Takamatsu S., Kitazume S., Kamada Y., Takehara T., Taniguchi N., and Miyoshi E. (2019) Core fucose is essential glycosylation for CD14-dependent Toll-like receptor 4 and Toll-like receptor 2 signalling in macrophages. J. Biochem. 165, 227–237 10.1093/jb/mvy098 [DOI] [PubMed] [Google Scholar]
- 9. Okada M., Chikuma S., Kondo T., Hibino S., Machiyama H., Yokosuka T., Nakano M., and Yoshimura A. (2017) Blockage of core fucosylation reduces cell-surface expression of PD-1 and promotes anti-tumor immune responses of T cells. Cell Rep. 20, 1017–1028 10.1016/j.celrep.2017.07.027 [DOI] [PubMed] [Google Scholar]
- 10. Okazaki A., Shoji-Hosaka E., Nakamura K., Wakitani M., Uchida K., Kakita S., Tsumoto K., Kumagai I., and Shitara K. (2004) Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcγRIIIa. J. Mol. Biol. 336, 1239–1249 10.1016/j.jmb.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 11. Shields R. L., Lai J., Keck R., O'Connell L. Y., Hong K., Meng Y. G., Weikert S. H., and Presta L. G. (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- 12. Niwa R., Natsume A., Uehara A., Wakitani M., Iida S., Uchida K., Satoh M., and Shitara K. (2005) IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J. Immunol. Methods 306, 151–160 10.1016/j.jim.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 13. Manabe Y., Marchetti R., Takakura Y., Nagasaki M., Nihei W., Takebe T., Tanaka K., Kabayama K., Chiodo F., Hanashima S., Kamada Y., Miyoshi E., Dulal H. P., Yamaguchi Y., Adachi Y., et al. (2019) Fucose on an IgG antibody is an endogenous ligand of Dectin-1. Angew. Chem. Int. Ed. Engl. 58, 18697–18702 10.1002/anie.201911875 [DOI] [PubMed] [Google Scholar]
- 14. Wang X., Fukuda T., Li W., Gao C.-X., Kondo A., Matsumoto A., Miyoshi E., Taniguchi N., and Gu J. (2009) Requirement of Fut8 for the expression of vascular endothelial growth factor receptor-2: a new mechanism for the emphysema-like changes observed in Fut8-deficient mice. J. Biochem. 145, 643–651 10.1093/jb/mvp022 [DOI] [PubMed] [Google Scholar]
- 15. Fukuda T., Hashimoto H., Okayasu N., Kameyama A., Onogi H., Nakagawasai O., Nakazawa T., Kurosawa T., Hao Y., Isaji T., Tadano T., Narimatsu H., Taniguchi N., and Gu J. (2011) α1,6-fucosyltransferase-deficient mice exhibit multiple behavioral abnormalities associated with a schizophrenia-like phenotype: importance of the balance between the dopamine and serotonin systems. J. Biol. Chem. 286, 18434–18443 10.1074/jbc.M110.172536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng B. G., Xu G., Chandy N., Steyermark J., Shinde D. N., Radtke K., Raymond K., Lebrilla C. B., AlAsmari A., Suchy S. F., Powis Z., Faqeih E. A., Berry S. A., Kronn D. F., and Freeze H. H. (2018) Biallelic mutations in FUT8 cause a congenital disorder of glycosylation with defective fucosylation. Am. J. Hum. Genet. 102, 188–195 10.1016/j.ajhg.2017.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrera H., Dilday T., Uber A., Scott D., Zambrano J. N., Wang M., Angel P. M., Mehta A. S., Drake R. R., Hill E. G., and Yeh E. S. (2019) Core-fucosylated tetra-antennary N-glycan containing a single N-acetyllactosamine branch is associated with poor survival outcome in breast cancer. Int. J. Mol. Sci. 10.3390/ijms20102528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agrawal P., Fontanals-Cirera B., Sokolova E., Jacob S., Vaiana C. A., Argibay D., Davalos V., McDermott M., Nayak S., Darvishian F., Castillo M., Ueberheide B., Osman I., Fenyö D., Mahal L. K., and Hernando E. (2017) A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell 31, 804–819.e7 10.1016/j.ccell.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tu C.-F., Wu M.-Y., Lin Y.-C., Kannagi R., and Yang R.-B. (2017) FUT8 promotes breast cancer cell invasiveness by remodeling TGF-β receptor core fucosylation. Breast Cancer Res. 19, 111–111 10.1186/s13058-017-0904-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aoyagi Y., Isemura M., Suzuki Y., Sekine C., Soga K., Ozaki T., and Ichida F. (1985) Fucosylated α-fetoprotein as marker of early hepatocellular carcinoma. Lancet 2, 1353–1354 [DOI] [PubMed] [Google Scholar]
- 21. Ihara H., Ikeda Y., Toma S., Wang X., Suzuki T., Gu J., Miyoshi E., Tsukihara T., Honke K., Matsumoto A., Nakagawa A., and Taniguchi N. (2007) Crystal structure of mammalian α1,6-fucosyltransferase, FUT8. Glycobiology 17, 455–466 10.1093/glycob/cwl079 [DOI] [PubMed] [Google Scholar]
- 22. Kötzler M. P., Blank S., Bantleon F. I., Wienke M., Spillner E., and Meyer B. (2013) Donor assists acceptor binding and catalysis of human α1,6-fucosyltransferase. ACS Chem. Biol. 8, 1830–1840 10.1021/cb400140u [DOI] [PubMed] [Google Scholar]
- 23. Kötzler M. P., Blank S., Bantleon F. I., Spillner E., and Meyer B. (2012) Donor substrate binding and enzymatic mechanism of human core α1,6-fucosyltransferase (FUT8). Biochim. Biophys. Acta 1820, 1915–1925 10.1016/j.bbagen.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 24. Ihara H., Ikeda Y., and Taniguchi N. (2006) Reaction mechanism and substrate specificity for nucleotide sugar of mammalian α1,6-fucosyltransferase: a large-scale preparation and characterization of recombinant human FUT8. Glycobiology 16, 333–342 10.1093/glycob/cwj068 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi T., Ikeda Y., Tateishi A., Yamaguchi Y., Ishikawa M., and Taniguchi N. (2000) A sequence motif involved in the donor substrate binding by α1,6-fucosyltransferase: the role of the conserved arginine residues. Glycobiology 10, 503–510 10.1093/glycob/10.5.503 [DOI] [PubMed] [Google Scholar]
- 26. Urbanowicz B. R., Bharadwaj V. S., Alahuhta M., Peña M. J., Lunin V. V., Bomble Y. J., Wang S., Yang J.-Y., Tuomivaara S. T., Himmel M. E., Moremen K. W., York W. S., and Crowley M. F. (2017) Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism. Plant J. 91, 931–949 10.1111/tpj.13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brzezinski K., Dauter Z., and Jaskolski M. (2012) Structures of NodZ α1,6-fucosyltransferase in complex with GDP and GDP-fucose. Acta Crystallogr. D Biol. Crystallogr. 68, 160–168 10.1107/S0907444911053157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lira-Navarrete E., Valero-González J., Villanueva R., Martínez-Júlvez M., Tejero T., Merino P., Panjikar S., and Hurtado-Guerrero R. (2011) Structural insights into the mechanism of protein O-fucosylation. PLoS ONE 6, e25365 10.1371/journal.pone.0025365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z., Han K., Pak J. E., Satkunarajah M., Zhou D., and Rini J. M. (2017) Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 13, 757–763 10.1038/nchembio.2381 [DOI] [PubMed] [Google Scholar]
- 30. Valero-González J., Leonhard-Melief C., Lira-Navarrete E., Jiménez-Osés G., Hernández-Ruiz C., Pallarés M. C., Yruela I., Vasudevan D., Lostao A., Corzana F., Takeuchi H., Haltiwanger R. S., and Hurtado-Guerrero R. (2016) A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat. Chem. Biol. 12, 240–246 10.1038/nchembio.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tseng T. H., Lin T. W., Chen C. Y., Chen C. H., Lin J. L., Hsu T. L., and Wong C. H. (2017) Substrate preference and interplay of fucosyltransferase 8 and N-acetylglucosaminyltransferases. J. Am. Chem. Soc. 139, 9431–9434 10.1021/jacs.7b03729 [DOI] [PubMed] [Google Scholar]
- 32. Calderon A. D., Liu Y., Li X., Wang X., Chen X., Li L., and Wang P. G. (2016) Substrate specificity of FUT8 and chemoenzymatic synthesis of core-fucosylated asymmetric N-glycans. Org. Biomol. Chem. 14, 4027–4031 10.1039/C6OB00586A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kamińska J., Glick M. C., and Kocielak J. (1998) Purification and characterization of GDP-L-Fuc: N-acetyl β D-glucosaminide α1→6fucosyltransferase from human blood platelets. Glycoconj. J. 15, 783–788 10.1023/A:1006959915435 [DOI] [PubMed] [Google Scholar]
- 34. García-García A., Ceballos-Laita L., Serna S., Artschwager R., Reichardt N. C., Corzana F., and Hurtado-Guerrero R. (2020) Structural basis for substrate specificity and catalysis of α1,6-fucosyltransferase. Nat. Commun. 11, 973 10.1038/s41467-020-14794-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Q., Zhang R., Cai H., and Wang L.-X (2017) Revisiting the substrate specificity of mammalian α1,6-fucosyltransferase reveals that it catalyzes core fucosylation of N-glycans lacking α1,3-arm GlcNAc. J. Biol. Chem. 292, 14796–14803 10.1074/jbc.M117.804070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thaysen-Andersen M., and Packer N. H. (2012) Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 10.1093/glycob/cws110 [DOI] [PubMed] [Google Scholar]
- 37. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Storoni L. C., McCoy A. J., and Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 10.1107/S0907444903028956 [DOI] [PubMed] [Google Scholar]
- 39. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 40. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kirby N., Cowieson N., Hawley A. M., Mudie S. T., McGillivray D. J., Kusel M., Samardzic-Boban V., and Ryan T. M. (2016) Improved radiation dose efficiency in solution SAXS using a sheath flow sample environment. Acta Crystallogr. D Struct. Biol. 72, 1254–1266 10.1107/S2059798316017174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kirby N. M., Mudie S. T., Hawley A. M., Cookson D. J., Mertens H. D. T., Cowieson N., and Samardzic-Boban V. (2013) A low-background-intensity focusing small-angle X-ray scattering undulator beamline. J. Appl. Crystallogr. 46, 1670–1680 10.1107/S002188981302774X [DOI] [Google Scholar]
- 43. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 10.1107/S0021889803012779 [DOI] [Google Scholar]
- 44. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 45. Huang J., and MacKerell A. D. Jr. (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 47. Beglov D., and Roux B. (1994) Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys. 100, 9050–9063 10.1063/1.466711 [DOI] [Google Scholar]
- 48. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.