Abstract
Background
The T-cell response to chlamydia genital tract infections in humans and mice is unusual because the majority of antigen-specific CD8 T cells are not class I restricted (referred to here as “unrestricted” or “atypical”). We previously reported that a subset of unrestricted murine chlamydia-specific CD8 T cells had a cytokine polarization pattern that included interferon (IFN)-γ and interleukin (IL)-13.
Methods
In this study, we investigated the transcriptome of CD8γ13 T cells, comparing them to Tc1 clones using microarray analysis. That study revealed that CD8γ13 polarization included IL-5 in addition to IFN-γ and IL-13. Adoptive transfer studies were performed with Tc1 clones and a CD8γ13 T-cell clone to determine whether either influenced bacterial clearance or immunopathology during Chlamydia muridarum genital tract infections.
Results
To our surprise, an adoptively transferred CD8γ13 T-cell clone was remarkably proficient at preventing chlamydia immunopathology, whereas the multifunctional Tc1 clone did not enhance clearance or significantly alter immunopathology. Mapping studies with major histocompatibility complex (MHC) class I- and class II-deficient splenocytes showed our previously published chlamydia-specific CD8 T-cell clones are MHC class II restricted.
Conclusions
The MHC class II-restricted CD8 T cells may play an important role in protection from intracellular pathogens that limit class I antigen presentation or diminish CD4 T-cell numbers or impair their function.
Keywords: CD8, Chlamydia, class II restriction, IL-5, IL-13
The CD8 response to chlamydia infections is atypical, linked both to protection and immunopathology. This article shows that CD8 complexity includes MHC class II restriction generally and protection by a CD8γ13 T-cell clone specifically, with implications for other pathogens including HIV.
Chlamydia trachomatis (Ct) urogenital serovars D–K are important because they cause immunopathology. In a subset of women, Ct infection causes fibrosis leading to infertility and ectopic pregnancies. More than 1.5 million infections are reported annually in the United States despite screening, test-and-treat programs, and effective therapy [1]. Epidemiologic data argue that test-and-treat programs arrest herd immunity leading to increased incidence of infection [2]. Women that self-clear Ct infections are less likely to be reinfected [3], which is proof of concept that vaccine-generated immunity could reduce the burden of Ct disease.
Rational vaccine development requires an immunologic goal. Mouse models predict that the most reliable mediator of protection will be class II-restricted multifunctional Th1 [4, 5]. In humans, protection from infertility [6] and susceptibility to reinfection [7] map to human leukocyte antigen (HLA) class II, which implies critical CD4 responses. In the murine Cm model, CD8 represents 5%–10% of the T-cell response in tissue [8], causing infertility [9] dependent on tumor necrosis factor (TNF)α [10]. However, in a trachoma model, monkeys protected by vaccination were susceptible to reinfection after CD8 depletion, implying CD8-mediated protection in the eye [11]. Two prominent groups investigating human Ct urogenital infections found that the majority of Chlamydia-specific CD8 clones “were not” restricted by HLA class I or CD1 [12, 13]. The role of “unrestricted” CD8 T cells is unknown.
We previously reported the CD8 response to Cm genital tract infection includes Tc1 polarized to interferon (IFN)-γ/interleukin (IL)-10/TNFα and CD8 polarized to IFN-γ/IL-10/TNFα with IL-13 [14]. Interleukin-13 knockout mice clear Cm genital infections faster with less pathology than wild-type mice [15], and IL-13 modestly enhances Cm replication in epithelial cells [14]. The CD8γ13 phenotype was originally described in humans with systemic sclerosis (SSc) [16, 17], an autoimmune disease with progressive skin and mucosal scarring. In the presence of TNFα, IL-13 triggers stromal fibroblasts to secrete transforming growth factor (TGF)β [18], the master scar formation cytokine; therefore, we hypothesized that Chlamydia-specific CD8γ13 T cells were immunopathologic and multifunctional Tc1 cells lacking IL-13 were protective. In this study, we determined the molecular fingerprints for Tc1 versus CD8γ13 T cells using expression microarray analysis, and we tested the CD8γ13 pathogenesis hypothesis by adoptive transfer.
MATERIALS AND METHODS
Mice
Four- to five-week-old female C57BL/6 mice from Harlan Laboratories (Indianapolis, IN) and Jackson Laboratories (Bar Harbor, ME) were housed in Indiana and Yale University specific pathogen-free facilities. The Institutional Animal Care and Utilization Committees at Indiana and Yale Universities approved all experimental protocols.
Cells and Bacteria
McCoy fibroblasts were cultured as previously described [19]. Mycoplasma-free Chlamydia muridarum (Nigg) were grown in McCoy cells as previously described [20]. Soluble Chlamydia antigen prepared as described [14].
Chlamydia-Specific CD8 T Cells
CD8 clones were maintained in “RPMI complete media” supplemented with cytokines as previously described [14]. For the adoptive transfer experiments, recombinant murine TGFβ1 at 10 ηg/mL was included in media for 1+ months before transfer when indicated. Murine cytokines were purchased from R&D Systems (Minneapolis, MN), except for TGFβ1 (eBioscience, San Diego, CA). Lactate dehydrogenase-release cytotoxicity assay was performed with CytoTox 96 (Pierce, Rockford) as previously described [21].
Adoptive Transfer and Genital Tract Infections
One week prior, mice were treated with 2.5 mg of medroxyprogesterone. Six days later, 1 × 106 T-cell clones were adoptively transferred via retro-orbital injection, and controls received injections of phosphate-buffered saline (PBS). T cells were purified with Ficoll-Hypaque (1083; Sigma-Aldrich, St. Louis, MO) on culture day 5 and maintained in media with 3 ng/mL IL-7 for 3 days before transfer. The day after adoptive transfer, mice were infected vaginally with 5 × 104 inclusion forming units (IFUs) of Cm in 10 µL SPG buffer, swabbed through day 49, and IFUs were determined on McCoy cells. On day 56, genital tracts were scored for pathology as previously described [22]. Each genital tract was divided into its 4 components: 2 oviducts and 2 uterine horns. For each component, macroscopically visible thinning of the organ wall with dilatation (pathologic feature) was given a score of 1 if present and 0 if absent, for a score of 0–4 per mouse; the maximal pathology score per mouse equals 4. Genital tracts were dissected and digitally imaged with a scorable pathologic feature per component and identified by an arrow if present to enable objective review.
Gene Expression Microarray Analysis
For “resting phenotype” microarrays, Chlamydia-specific CD8 clones were purified on culture day 7 and then maintained in media with 3 ηg/mL IL-7 for 48 hours without stimulation. An alloreactive CD8 clone, CD8bm1-specific for H-2Kbm1 [23], was included in the study. For the “activated” arrays, clones were purified on day 7, then activated for 14 hours in 24-well plates coated with .75 mL PBS containing .5 µg/mL anti-CD3 (145-2c11; BD Biosciences) overnight at 4⁰C, and washed before use. For resting and activated arrays, total ribonucleic acid (RNA) was isolated using an RNAse-free DNAse I step (RNeasy; QIAGEN), which was repeated 4 times to minimize false discovery. With assistance from the Indiana University Center for Medical Genomics, gene expression patterns were analyzed using the Affymetrix Mouse ST 1.0 Array (28 853 genes). Samples were labeled using the standard protocol for the WT Target Labeling and Control Reagents kit: GeneChip Whole Transcript (WT) Sense Target Labeling Assay GeneChip. Individual samples were hybridized to Mouse Gene 1.0 ST GeneChips for 17 hours, washed, stained, and scanned using the Affymetrix GeneChip Operating System (GCOS). The GCOS was used to generate data (CEL files). Arrays were visually scanned for abnormalities or defects. CEL files were imported into Partek Genomics Suite (Partek, Inc., St. Louis, MO).
Cytokine Enzyme-Linked Immunosorbent Assays and Signaling Reagents
A total 2.5 × 104 purified clone T cells cultured overnight in media with 3 ηg/mL IL-7 were activated in 96-well plates with anti-CD3 (145-2c11, .5 µg/mL in PBS overnight at 4°C [washed once]), in media containing 1 ηg/mL IL-7 (R&D Systems) for 20 hours. Interferon-γ, IL-13, and IL-5 in supernatants were determined by enzyme-linked immunosorbent assay using antibody pairs with recombinant standards: IFN-γ (XMG1.2; Pierce-Thermo Fisher); IL-13 (eBio13A/eBio1316H; eBioscience); IL-5 (TRFK5/TRFK4; eBioscience). Detection was accomplished with Streptavidin-Horseradish Peroxidase (BD Biosciences) and 3,3’,5,5’-Tetramethylbenzidine (TMB) substrate (Sigma-Aldrich).
Cyclosporine A ([CsA] Sigma-Aldrich) was used at 500 ηM (2 ηg/mL). Prostaglandin D synthase (hematopoietic-type) inhibitor I in dimethyl sulfoxide ([DMSO] Cayman Chemicals, Ann Arbor, MI) was used at 2.5 µM, ~80 times the half-maximal inhibitory concentration (IC50). CrTh2 inhibitors I [(4-chloro-2-((2-methyl-5-(propylsulfonyl)phenyl)ethynyl)phenoxy)acetic acid] and II [(R)-(5-chloro-1’-(5-chloro-2-fluorobenzyl)-2,2’,5’-trioxospiro(indole-3,3→-pyrrolidin)-1(2H)-yl)acetic acid] (from EMD Millipore, Temecula, CA) were dissolved in DMSO and used at 5 µM, ~50 times the IC50.
Statistical Analysis
Summary figures are presented as “pooled” means and standard error of the mean, and single experiments are presented as pooled means and standard deviation. Figure legends indicate the number of independent experiments and mice pooled to generate the figure. Analyses were performed with Fisher’s exact test (immunopathology), Dunnett’s test (bacterial shedding), and Students t test (other data). P < .05 was considered significant. Aggregate shedding data from the 3 adoptive transfer experiments were analyzed with the Wilcoxon rank-sum test.
For the Affymetrix arrays, RMA signals were generated for core probe sets, and we used the RMA method for background correction, quantile normalization, and summarization with Median Polish. Summarized signals for each probe set were log2-transformed. Log-transformed signals wereused for principal components analysis, hierarchical clustering, and signal histograms to determine any outlier arrays. Untransformed RMA signals were used for fold-change calculations. We took a one-way analysis of variance using log2-transformed signals for all 4 experimental samples, and contrasts were made by comparing combined signals for 8sAg-1 and 8sAg-3 with the combined signals of 8uvmo-2, 8uvmo-3, and the alloreactive CD8bm1. Fold-changes were calculated using untransformed RMA signals. Genes up- or down-regulated 3-fold with P < .001 were considered in the final analysis. The microarray data presented here are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under accession number (GSE140213).
RESULTS
Transcriptome of CD8γ13 T Cells
We previously characterized Chlamydia-specific CD8 clones from mice that cleared C muridarum genital tract infections. Two CD8 clones, 8sAg-1 and 8sAg-3, polarized to IFN-γ/TNFα/IL-10/IL-13, and were not restricted by major histocompatibility complex (MHC) class Ia [14]. The novel cytokine polarization pattern and MHC-restriction prompted us to investigate the underlying gene expression patterns by comparing 2 multifunctional Chlamydia-specific Tc1 clones (8uvmo-2 and 8uvmo-3) and a Kbm1-specific alloreactive CD8 clone (CD8bm1) representing a conventional Tc1 clone. Two array analyses were performed. The first was done on T cells 9 days after passage to identify resting CD8γ13 biomarkers. The second was done 14 hours after activation with anti-CD3 to identify activated biomarkers and signaling/activation pathways. For resting and activated arrays, total RNA was isolated on 4 occasions to minimize false-positive results. Genes uniquely expressed by CD8γ13 were defined by the following: (1) same polarity for CD8γ13 (8sAg-1 + 8sAg-3 combined) versus each comparator (8uvmo-2 + 8uvmo-3, 8uvmo-2 + 8uvmo-3+CD8bm1, and CD8bm1 alone), (2) P < .01 for comparisons, (3) >3-fold difference in messenger RNA (mRNA) levels, and (4) mRNA signal for 8sAg-1 and 8sAg-3 individually exceeded all 3 Tc1 clones (eliminates genes skewed by very high or low mRNA signal by 1 CD8IL-13 clone). Table 1 shows the genes with enhanced mRNA levels in CD8γ13. Table 2 shows the genes with diminished levels. The entire table of 3-fold differences between CD8γ13 and Tc1 cells is provided in Supplemental Table 1 (resting) and Supplemental Table 2 (activated). Lineage markers are in Supplemental Table 3.
Table 1.
Genes Specifically Transcribed in CD8g13 T Cells
| CD8IL13+ vs CD8IL13− | CD8IL13+ vs CD8IL13− and alloCD8 | CD8IL13+ vs alloCD8 | |||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | P Value | Fold Change | P Value | Fold Change | P Value | Fold Change | Gene Title |
| CD8IL-13 Unique Genes Identified in “Rested” Array Only | |||||||
| Pecam1 | 1.84E-03 | 10.74 | 1.08E-03 | 11.02 | 5.66E-03 | 11.31 | platelet/endothelial cell adhesion molecule 1: marker for “central” naive CD4 T cells |
| Hpgds | 4.49E-04 | 6.86 | 1.20E-04 | 8.11 | 5.25E-04 | 9.60 | hematopoietic prostaglandin D synthase |
| Sla | 1.20E-04 | 6.39 | 1.02E-04 | 5.94 | 1.24E-03 | 5.52 | src-like adaptor: negative regulator of TCR signaling |
| Mtmr7 | 5.86E-09 | 6.24 | 8.29E-09 | 5.40 | 5.72E-07 | 4.67 | myotubularin-related protein 7: lipid phosphatase limits Th1 and Th17 differentiation |
| Dclk3 | 3.77E-11 | 5.73 | 8.80E-12 | 6.20 | 1.37E-10 | 6.72 | doublecortin-like kinase 3 |
| Ctss | 4.02E-04 | 5.33 | 1.68E-05 | 8.74 | 2.32E-05 | 14.34 | cathepsin S |
| Sulf2 | 2.14E-04 | 4.84 | 1.84E-06 | 10.10 | 1.12E-06 | 21.07 | sulfatase 2 |
| Eml1 | 2.66E-06 | 4.77 | 1.45E-07 | 6.46 | 4.21E-07 | 8.76 | echinoderm microtubule-associated protein-like 1 |
| 4930506M07Rik | 4.04E-07 | 4.15 | 2.86E-07 | 3.98 | 7.07E-06 | 3.82 | RIKEN cDNA 4930506M07 gene |
| Tnfrsf11b | 4.03E-06 | 3.54 | 1.34E-09 | 9.19 | 2.42E-10 | 23.83 | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| Bicd1 | 3.79E-09 | 3.14 | 1.24E-09 | 3.23 | 2.22E-08 | 3.32 | bicaudal D homolog 1 (Drosophila): movement of secretory granules in CTL |
| Mapk8 | 7.35E-04 | 3.01 | 1.91E-04 | 3.35 | 7.43E-04 | 3.73 | mitogen-activated protein kinase 8: necessary for a GITR signal --> CD4 IL-13 production |
| CD8IL-13 Unique Genes Identified in Both Rested and “Activated” Arrays | |||||||
| 1810011H11Rik | 2.12E-15 | 24.85 | 1.52E-15 | 22.31 | 8.19E-14 | 20.03 | RIKEN cDNA 1810011H11 gene |
| Arntl | 3.00E-08 | 12.80 | 5.51E-07 | 6.84 | 5.00E-04 | 3.65 | aryl hydrocarbon receptor nuclear translocator-like |
| Ccr8 | 4.58E-08 | 8.89 | 2.15E-08 | 8.90 | 4.49E-07 | 8.91 | chemokine (C-C motif) receptor 8 |
| Amelx | 1.57E-10 | 7.75 | 8.75E-12 | 10.68 | 4.13E-11 | 14.71 | amelogenin X chromosome |
| Epdr1 | 2.33E-04 | 5.60 | 1.36E-04 | 5.59 | 1.13E-03 | 5.59 | ependymin-related protein 1 (zebrafish) |
| Gm9766 | 7.61E-04 | 4.88 | 7.67E-05 | 6.79 | 1.63E-04 | 9.44 | Cep85l centrosomal protein 85-like |
| CDIL-13 Unique Genes Identified Only in the Activated Array | |||||||
| Il5 | 1.32E-13 | 30.03 | 2.74E-14 | 31.69 | 1.30E-12 | 35.28 | interleukin 5 |
| Tm4sf19 | 2.96E-17 | 25.57 | 4.13E-17 | 18.06 | 1.86E-13 | 9.01 | transmembrane 4 L 6 family member 19 |
| Il13 | 3.00E-12 | 15.71 | 4.01E-13 | 17.95 | 7.98E-12 | 23.41 | interleukin 13 |
| Gabra1 | 4.49E-17 | 14.21 | 1.09E-17 | 14.35 | 7.74E-16 | 14.64 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| I830127L07Rik | 4.32E-12 | 6.55 | 2.96E-14 | 11.16 | 1.02E-14 | 32.42 | RIKEN cDNA I830127L07 gene |
| Clgn | 1.92E-13 | 5.70 | 4.49E-16 | 10.97 | 5.42E-17 | 40.66 | calmegin |
| Mcc | 1.67E-18 | 4.28 | 2.48E-19 | 4.52 | 7.06E-18 | 5.03 | mutated in colorectal cancers |
| Cd4 | 2.34E-14 | 4.09 | 1.46E-15 | 4.72 | 9.28E-15 | 6.28 | CD4 antigen |
| Tubb6 | 6.12E-15 | 3.95 | 6.63E-18 | 7.28 | 4.11E-19 | 24.75 | tubulin, beta 6 |
| Gja5 | 3.23E-09 | 3.57 | 5.75E-11 | 4.70 | 4.89E-11 | 8.17 | gap junction protein, alpha 5 |
| Cd2ap | 2.03E-10 | 3.27 | 9.16E-12 | 3.84 | 2.95E-11 | 5.28 | CD2-associated protein: negative regulator of TCR signaling via CD2 |
| Fam169b | 1.73E-12 | 3.27 | 4.03E-14 | 4.05 | 6.02E-14 | 6.21 | family with sequence similarity 169, member B |
| Tnfsf11 | 2.48E-13 | 3.03 | 1.39E-14 | 3.42 | 7.29E-14 | 4.37 | tumor necrosis factor (ligand) superfamily, member 11: receptor for Tnfrsf11b-dendritic survival |
Bold indicates known immunological function.
Table 2.
Genes Specifically Not-transcribed in CD8g13 T Cells
| CD8IL13+ vs CD8IL13− | CD8IL13+ vs CD8IL13− and alloCD8 | CD8IL13+ vs alloCD8 | |||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | P Value | Fold Change | P Value | Fold Change | P Value | Fold Change | Gene Title |
| Non-CD8IL-13 Genes Common to the “Resting” and “Activated” Microarrays | |||||||
| Lxn | 4.33E-07 | −20.13 | 9.05E-08 | −24.69 | 8.14E-07 | −30.29 | latexin: negative regulator lymphocyte growth |
| Aldoc | 6.85E-07 | −18.72 | 1.48E-08 | −41.38 | 2.58E-08 | −91.49 | aldolase C, fructose-bisphosphate |
| A930038C07Rik | 2.40E-10 | −18.56 | 3.48E-10 | −14.70 | 3.01E-08 | −11.65 | RIKEN cDNA A930038C07 gene |
| Macc1 | 3.37E-13 | −15.30 | 1.28E-14 | −25.02 | 5.05E-14 | −40.90 | metastasis associated in colon cancer 1 |
| Mr1 | 7.15E-13 | −16.87 | 2.37E-13 | −17.81 | 5.51E-12 | −18.80 | major histocompatibility complex, class I-related |
| Phyh | 1.38E-14 | −14.62 | 2.60E-15 | −17.08 | 3.80E-14 | −19.96 | phytanoyl-CoA hydroxylase |
| Wls | 7.90E-07 | −13.15 | 1.85E-07 | −15.38 | 1.72E-06 | −17.97 | wntless homolog (Drosophila) |
| Slc27a6 | 1.64E-12 | −10.46 | 6.19E-14 | −16.05 | 2.37E-13 | −24.62 | solute carrier family 27 (fatty acid transporter), member 6 |
| Sccpdh | 1.65E-05 | −10.31 | 1.51E-06 | −15.03 | 5.23E-06 | −21.91 | saccharopine dehydrogenase (putative) |
| Atp2a3 | 5.69E-09 | −9.84 | 2.53E-10 | −14.95 | 9.07E-10 | −22.71 | ATPase, Ca++ transporting, ubiquitous |
| Peci | 2.45E-05 | −9.24 | 1.42E-05 | −9.11 | 1.74E-04 | −8.98 | peroxisomal delta3, delta2-enoyl-Coenzyme A isomerase |
| Hadh | 2.01E-09 | −8.82 | 1.45E-11 | −18.48 | 1.65E-11 | −38.75 | hydroxyacyl-Coenzyme A dehydrogenase |
| Fam38a | 3.51E-08 | −8.34 | 5.69E-08 | −6.90 | 3.87E-06 | −5.71 | family with sequence similarity 38, member A |
| Gem | 1.17E-04 | −6.54 | 2.27E-05 | −7.98 | 1.01E-04 | −9.74 | GTP binding protein: transient expression in activated T cells |
| Tmbim4 | 1.25E-04 | −5.04 | 2.77E-04 | −4.08 | 6.36E-03 | −3.30 | transmembrane BAX inhibitor motif containing 4 |
| Rgs2 | 1.06E-06 | −4.95 | 1.77E-06 | −4.26 | 8.24E-05 | −3.66 | regulator of G-protein signaling 2: Important for T-cell proliferation via IL-2 |
| Non-CD8IL-13 Genes Unique to the Activated Microarrays | |||||||
| Nrn1 | 2.86621E-19 | −30.3543 | 1.52314E-20 | −44.313 | 8.12124E-20 | −94.4417 | neuritin 1: expressed in tolerized T cells |
| Slc2a3 | 2.79463E-16 | −8.66819 | 4.67369E-16 | −6.71521 | 3.44781E-12 | −4.03027 | solute carrier family 2: necessary for CD4 but not Treg proliferation |
| Rgs18 | 2.11474E-11 | −5.14338 | 1.78358E-13 | −7.98487 | 7.48595E-14 | −19.2449 | regulator of G-protein signaling 18 |
| Sema3d | 1.29033E-12 | −4.34544 | 1.39255E-15 | −8.45173 | 8.21618E-17 | −31.9726 | sema domain, immunoglobulin domain (Ig) |
| Ccr5 | 6.33614E-12 | −3.37636 | 1.4081E-12 | −3.42894 | 6.78466E-11 | −3.53664 | chemokine (C-C motif) receptor 5 |
| Mospd1 | 2.27533E-09 | −3.21976 | 6.39868E-10 | −3.22082 | 3.4887E-08 | −3.223 | motile sperm domain containing 1 |
| Pecr | 1.19754E-11 | −3.0695 | 4.08966E-13 | −3.64045 | 9.75058E-13 | −5.12078 | peroxisomal trans-2-enoyl-CoA reductase |
aGene inclusion criteria were (1) same direction (up or down) for CD8IL-13 vs conventional CD8 clones, (2) fold change >3, (3) fold change P < .01, and (4) absolute value for messenger ribonucleic acid signal (up or down) for both CD8IL-13 T-cell clones had to be greater than for all 3 conventional T-cell clones (ie, no skewing for a large difference limited to 1 T-cell clone of any type).
Bold indicates known immunological function.
Transforming Growth Factor-β, Calcineurin, and Hpgds in CD8γ13 T Cells
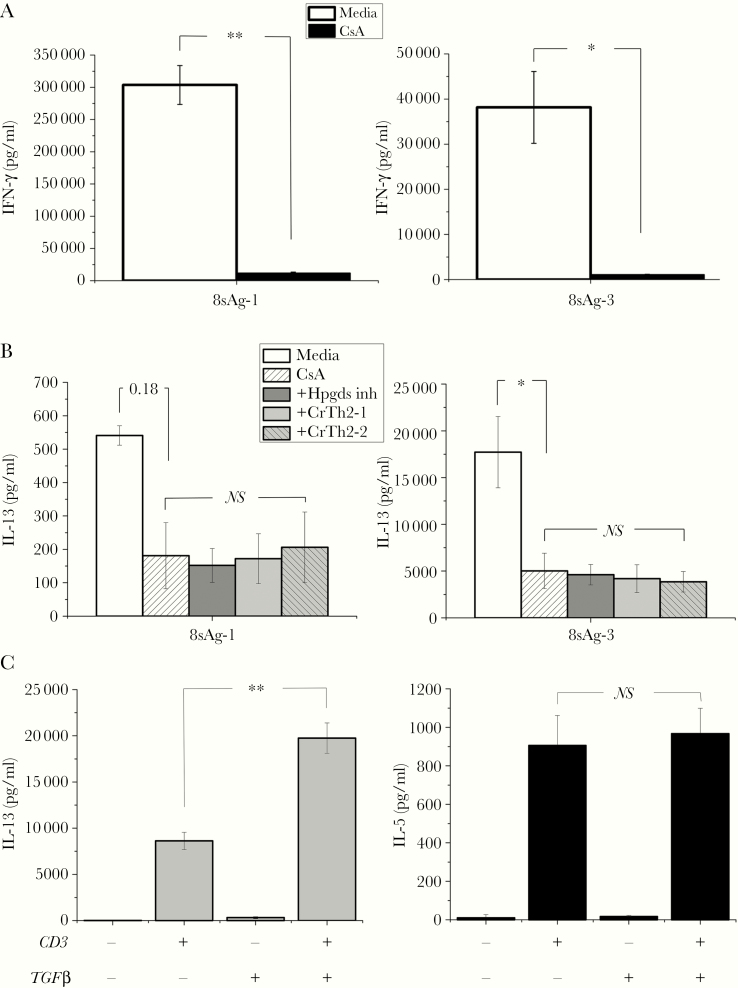
The array identified hematospecific prostaglandin D synthetase (Hpgds) mRNA as uniquely transcribed by resting CD8γ13. Hpgds is interesting because its enzymatic product prostaglandin D binds Crth2 resulting in T-cell receptor (TCR)-independent IL-13 production in Th2 cells [24]; all of the CD8 clones had positive Crth2 mRNA signals (data not shown). To test whether Hpgds played a role in cytokine production, CD8γ13 8sAg-1 and 8sAg-3 were activated with anti-CD3 in the absence and presence of calcineurin, Hpgds, and CrTh2 inhibitors; then, IL-13 and IFN-γ were measured in culture supernatants (Figure 1A and B). Interferon-γ was dramatically inhibited, whereas IL-13 was partially calcineurin independent (CsA resistant). The CsA-resistant portion of IL-13 production was not inhibited by an Hspgds inhibitor nor 2 CrTh2 small molecule inhibitors. CD8γ13TCR signaling includes a calcineurin-independent pathway that accounts for 25%–30% of IL-13 production that was not explained by CrTh2. In a previous study, we found that maintenance of IL-13 production by CD4γ13 T cells required TGFβ1 [21]. To test for a CD8γ13 TGFβ1 requirement, 8sAg-3 was passaged under usual conditions with and without TGFβ1 for 6 weeks and then tested for the ability to produce IL-13 and IL-5 (Figure 1C). The 6-week experiment showed that 8sAg-3 IL-13 production was enhanced by exogenous TGFβ1 without affecting IL-5. Although readily detectable after CD3-activation, unlike IL-13, IL-5 was not produced by 8sAg-3 activated by infected epithelial cells (data not shown).
Figure 1.
Interferon (IFN)-γ and interleukin (IL)-13 in CD8γ13 T cells. A total of 2.5 × 104 T cells, 8sAg-1 and 8sAg-3, were activated by immobilized anti-CD3 antibody without and with inhibitors for 24 hours; supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA). (A) Interferon-γ in the absence and presence of cyclosporine A (CsA). (B) Interleukin-13 in the absence and presence of CsA and in the presence of CsA combined with hematospecific prostaglandin D synthetase (Hpgds) and CrTh2 inhibitors (eg, +Hpgds inh). Aggregate data from 2 independent experiments. For B, the comparisons are Media vs CsA, then CsA vs CsA plus the 3 inhibitors (Hspgds, CrTh2-1, CrTh2-2). 8sAg-1 and 8sAg-2 without CD3 activation made no detectable IFN-γ or IL-13 vs CD3-Media (P < .01, not graphed). (C) Effect of transforming growth factor (TGF)β1 on IL-13 and IL-5. CD8γ13 clone 8sAg-3 was maintained in (1) usual culture and (2) usual culture plus TGFβ1 for 6 weeks then tested for production of IL-13 and IL-5. A total of 2.5 × 104 T cells was added to wells without and with anti-CD3; supernatants were collected at 24 hours and analyzed by ELISA. A single experiment was done with triplicate wells. Student’s t test analysis: *, P < .05 and **, P <.005. NS, not significant.
Mapping of the Restriction Elements of Chlamydia-Specific CD8 T-Cell Clones
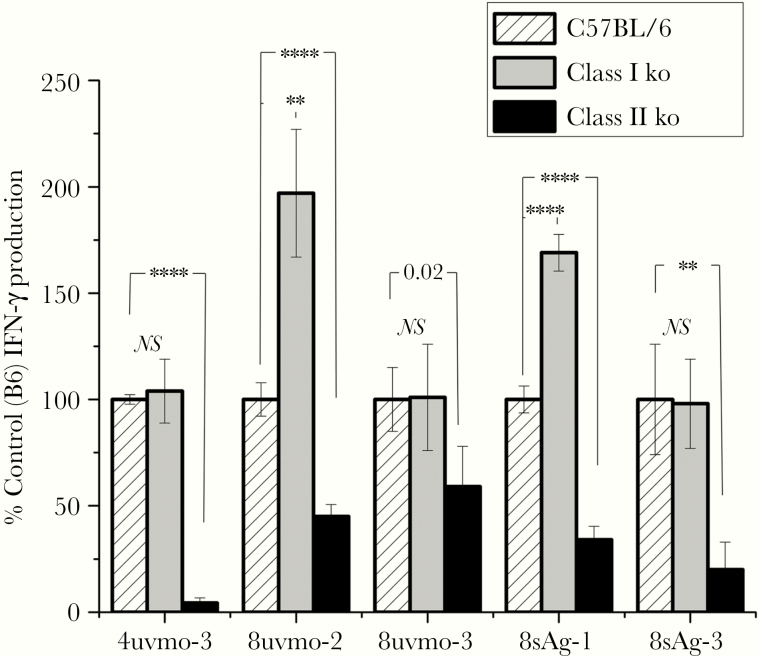
We previously showed that CD8γ13 clones 8sAg-1 and 8sAg-3 were not restricted by class Ia because they equally recognized antigen-pulsed splenocytes from C57BL/6 and KbDb knockout (class Ia deficient) mice. In the same study, mapping of Tc1 clones 8uvmo-2 and 8uvmo-3 was indeterminate due to bystander activation (IFN-γ-enhancing cytokines released by irradiated antigen-presenting cells [APC] exposed to UV-Cm). To get around those issues, we used low-dose UV-Cm-pulsed C57BL/6, β 2-microglobulin knockout (deficient in class Ia, Ib, CD1, MR1), and Ciita knockout (no class II) splenocytes as APC. A well characterized Chlamydia-specific CD4 clone, 4uvmo-3, that recognizes infected epithelial cells in CD4-dependent fashion (MHC class II restricted) [19] was included as a control (Figure 2). CD8 T cells conventionally need class Ia, Ib, MR1, or CD1 molecules to present antigen, which are absent in β 2-microglobulin knockout splenocytes. The 4 CD8 clones and CD4 clone recognized UV-Cm-pulsed splenocytes from C57BL/6 mice and MHC class I-deficient β 2-microglobulin knockout mice as expected for the CD4 clone, and this is anomalous but consistent with our published data for the CD8 clones. It is curious that 8uvmo-2 and 8sAg-1 activation was enhanced with class I-deficient splenocytes, suggesting they have killer inhibitory receptor (Ly-49 family) arrays favoring activation over inhibition in the absence of class I. Critically, all clones, CD4 and CD8, showed significantly diminished activation by class II-deficient UV-Cm-pulsed splenocytes consistent with MHC class II restriction, although 8uvmo-3 was marginally significant, and we consider that result indeterminate.
Figure 2.
Major histocompatibility complex restriction mapping with class I-deficient (β2 micro ko) and class II-deficient (Ciita ko) splenocyte antigen-presenting cells (APC). A total of 2.5 × 105 indicated APC preincubated with 2.8 inclusion forming units UV-Cm per cell for 1 hour, then cocultured with 2.5 × 104 T-cell clones for 48 hours; supernatants were collected and analyzed for interferon (IFN)-γ by enzyme-linked immunosorbent assay. Data normalized to IFN-γ level with wild-type C57BL/6 APC were set as 100% for each experiment. Data were aggregated from 2 independent experiments. Student’s t test analysis: **, P < .005; ****, P < .00005. ko, knockout; NS, not significant comparison of C57BL/6 to class I ko (β2 micro ko).
Adoptive Transfer of CD8 T-Cell Clones
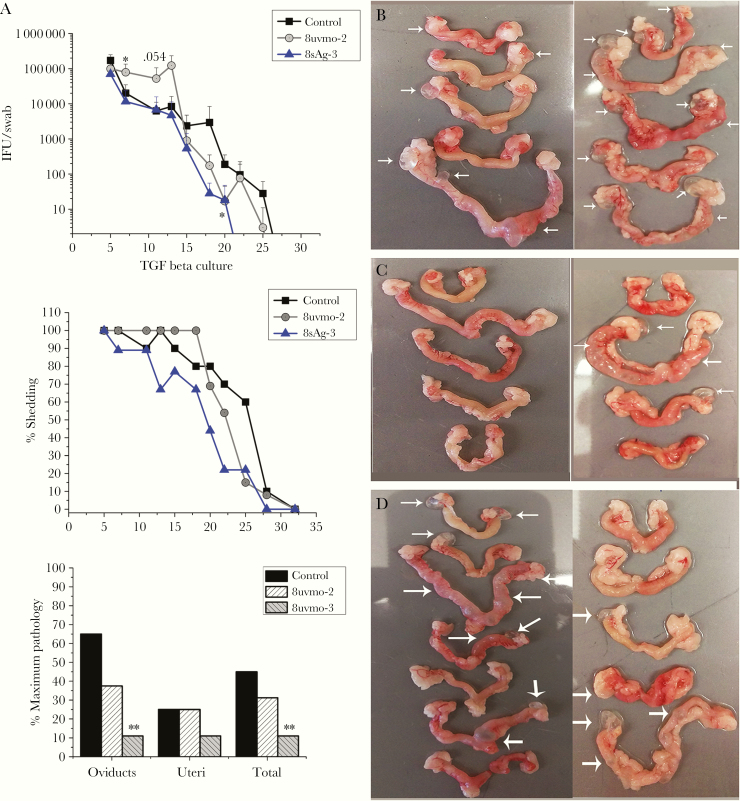
To test the hypothesis that multifunctional Tc1 were protective and CD8γ13 was immunopathologic, we chose the following representative clones: multifunctional, Tc1 = 8uvmo-2; “pathologic”, CD8γ13= 8sAg-3. Both CD8 clones are activated by C muridarum-infected epithelial cells and terminate replication in them [14]. For initial adoptive transfer experiments, TGFβ1 was included in 8uvmo-2 and 8sAg-3 cultures for 1+ months before transfer. One day before genital tract challenge, 1 million 8uvmo-2 or 8sAg-3 T cells were adoptively transferred into C57BL/6 mice, whereas control mice received PBS. Bacterial shedding was monitored through day 49, and immunopathology was scored on day 56. A staggered (1 month later) repeat experiment was done, and data were aggregated for analysis. The Tc1 clone 8uvmo-2 trended toward enhancing bacterial shedding without significantly influencing immunopathology; similarly, another Tc1 clone, 8uvmo-3, showed no protection from shedding or pathology (Supplemental Figure 1). In contrast, the CD8γ13 8sAg-3 clone significantly reduced oviduct immunopathology with a trend toward lower shedding (Figure 3).
Figure 3.
Adoptive transfer of T-cell clones maintained in transforming growth factor (TGF)β cultures. Medroxyprogesterone-treated C57BL/6 mice received phosphate-buffered saline (control) or 1 million CD8 clone cells, 8uvmo-2 (Tc1) or 8sAg-3 (CD8γ13), 24 hours before vaginal challenge with 5 × 104 inclusion forming units (IFU) Chlamydia muridarum. (A) Shedding as IFU/swab, percentage of mice shedding over time, and immunopathology score on day 56. Genital tracts from (B) control mice, (C) 8sAg-3 mice, and (D) 8uvmo-2 mice. White arrows indicate damage to uterine horn or hydrosalpinx. Data are from 2 independent experiments; number of mice = number of genital tracts. Shedding was analyzed with Dunnett’s test, and pathology was analyzed with Fisher’s exact test; *, P < .05 and **, P < .005.
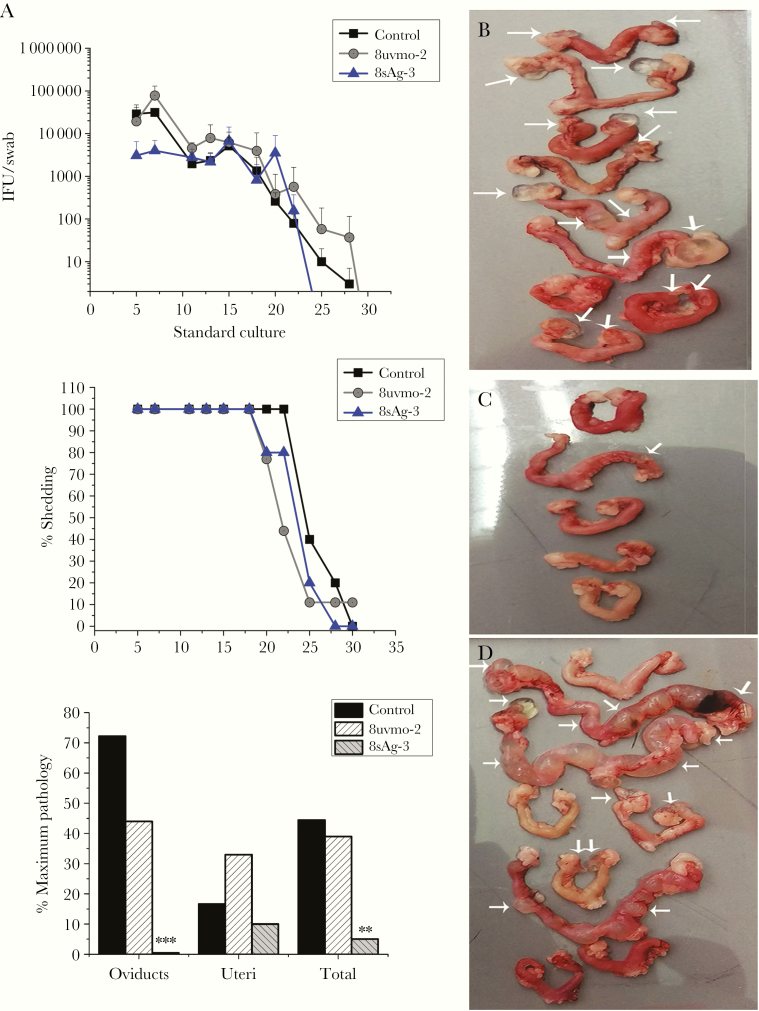
After analyzing first and second experiments, we were surprised to find high bacterial shedding in 8uvmo-2 mice. We considered the possibility that extended culture of Tc1 8uvmo-2 in TGFβ1 may have undermined its ability to clear infection. We repeated the experiment using 8uvmo-2 and 8sAg-3 that were never exposed to TGFβ1. 8uvmo-2 again did not enhance bacterial clearance or significantly alter immunopathology. In contrast, 8sAg-3 remained solidly protective against development of oviduct pathology with a trend toward lower bacterial shedding (Figure 4). In C57BL/6 mice, an adoptively transferred clone was added to an endogenous immune response that effectively cleared C muridarum. Others have shown, using doxycycline to interrupt infection-dependent adaptive immunity maturation, that T cell-mediated protective immunity is functionally established by postinfection on day 7 and B-cell immunity (antibody titers) is established by day 10 [25]. The window for an adoptively transferred monoepitope-specific T-cell clone to single-handedly reduce bacterial shedding (a tall order) is in days 1 to 10 postinfection. Analysis of aggregate data from day 5 and 7 of our 3 adoptive-transfer experiments revealed that the CD8γ13 clone 8sAg-3 reduced bacterial shedding on day 5 (mean IFU/swab 46 000 vs control 124 000 with P = .007) and day 7 (mean IFU 8900 vs control 24 000 with P = .006).
Figure 4.
Adoptive transfer of T-cell clones maintained without transforming growth factor β in cultures. Medroxyprogesterone-treated C57BL/6 mice received phosphate-buffered saline (control) or 1 million CD8 T-cell clones, 8uvmo-2 (Tc1) or 8sAg-3 (CD8γ13), 24 hours before vaginal challenge with 5 × 104 inclusion forming units (IFU) Chlamydia muridarum. (A) Shedding by IFU/swab, percentage of mice shedding over time, and immunopathology scores on day 56. Genital tracts from (B) control mice, (C) 8sAg-3 mice, and (D) 8uvmo-2 mice. White arrows indicate damage to uterine horn or hydrosalpinx. Data are from 1 experiment; number of mice = number of genital tracts. Shedding was analyzed with Dunnett’s test, and pathology was analyzed with Fisher’s exact test; **, P < .005 and ***, P < .0005.
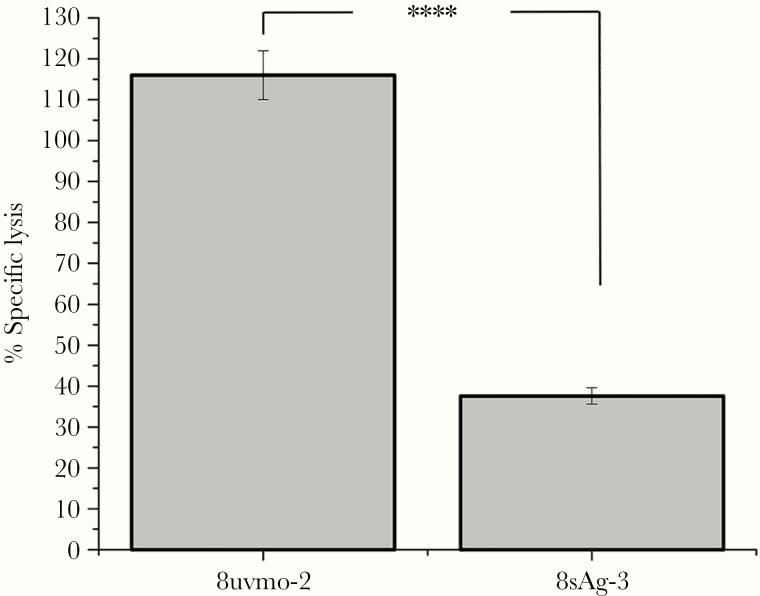
Cytolytic Potential of 8sAg-3 and 8uvmo-2
Perforin-mediated cytolysis by Chlamydia-specific CD4 T cells is detrimental to termination of Cm replicating in epithelial cells [26]. In the setting of protective and nonprotective phenotypes for 8sAg-3 and 8uvmo-2, respectively, we hypothesized that 8sAg-3 may be a noncytolytic CD8 T cell. Cytolytic assays with infected epithelial cells are problematic due to their fragility. We compared intrinsic cytolytic capacity using a redirected-lysis assay, coating FcR-bearing P815 cells with anti-CD3 antibody and measuring release of intracellular enzyme lactose dehydrogenase in a 4-hour assay (Figure 5). 8uvmo-2 had greater intrinsic killing capacity than 8sAg-3. It is plausible that the reduced cytolytic capability of 8sAg-3 contributes to its protective phenotype.
Figure 5.
Cytolytic capacity of 8uvmo-2 (Tc1) and 8sAg-3 (CD8γ13) were determined in a redirected lysis assay. A total of 10 000 CD8 clone cells was cocultured with 10 000 P815 cells in the presence of anti-CD3 antibody for 4 hours. Lactate dehydrogenase release from damaged P815 cells compared with P815 controls without CD3 antibody and used to determine percentage-specific lysis. Single experiment with quadruplicate wells. Student’s t test analysis; ****, P < .00005.
DISCUSSION
The murine response to Cm genital tract infection includes Chlamydia-specific unrestricted CD8 T cells polarized to IFN-γ/IL-13 [14]. In this study, we document a CD8γ13 transcriptome and found that adoptive transfer of a CD8γ13 clone protected oviducts from immunopathology. Arguments against nonspecific effects, in parallel a multifunctional Tc1 clone, did not enhance bacterial clearance or significantly alter immunopathology. Those results profoundly upended our SSc-based hypothesis that CD8γ13 were pathologic and multifunctional Tc1 cells were protective. Although it is not definitive, CD8γ13 seems to be protective in the context of Ct genital tract infections.
The CD8γ13 transcriptome includes interesting biomarkers. The cell surface protein mRNA that is most highly and uniquely transcribed by CD8γ13 T cells is 1810011H11Rik, an uncharacterized 95 amino acid (aa) protein with a predicted signal sequence, transmembrane domain, and cytoplasmic domain, suggesting a signaling scaffold. 1810011H11Rik’s highly conserved human homolog C10orf128 has been validated at the protein level [27].
CD8γ13 have unique TCR signaling/activation pathways. T-cell receptor activation of 8sAg-3 led to the production of IL-13 and IL-5; 25%–30% of IL-13 was calcineurin independent based on CsA resistance. The core transcriptome includes reduced Rsg2 and Gem [28, 29] with enhanced Amelx [30], genes associated with proliferation, and the absence of mRNA for latexin (lxn), a negative regulator of T-cell proliferation [31]. At rest, CD8γ13 have enhanced Sla, a negative regulator of TCR signaling [32], and Dclk3, a serine/threonine kinase of unknown function. Sla has a 104aa cytoplasmic domain that may serve as a proximal TCR SH3 and SH2 signaling scaffold to temper TCR activation.
The IL-13/IL-5/IFN-γ cytokine polarization of the 2 chlamydia-specific CD8γ13 T-cell clones is not synonymous with MHC class II restriction because HLA-E (class Ib)-restricted IL-13/IL-5 T cells described in tuberculosis patients have the same polarization pattern [33]. CD8γ13 T cells were originally described in SSc. In SSc subjects, but not healthy controls, CD8 T cells had elevated Gata3 mRNA and Gata3-dependent IL-13 production without differences in T-bet [34]. In our CD8γ13 resting and activated arrays, Gata3 mRNA was elevated in CD8γ13 clones (2.02-fold and 2.15-fold, P < .00001), without making the arbitrary 3-fold cutoff for inclusion (see Table 1), whereas T-bet (Tbx21) was not significantly different (−1.19-fold; not significant; rested) or slightly lower (−1.25-fold; P < .0001; activated). Other investigators showed that TGFβ1 increased Gata3-dependent IL-13 production by SSc T cells [35], consistent with our finding that 8sAg-3 IL-13 production was enhanced by TGFβ1. The microarray data provide preliminary insight into CD8γ13 differentiation with enhanced mRNA levels of mitogen-activated protein kinase 8 (Mapk8), a signaling molecule critical for natural Treg differentiation into pathologic IL-13-secreting CD4 T cells [36], and for myotubularin-related protein 7 (Mmtr7), a lipid phosphatase that prevents Th0 differentiation into Th2 and Th17 [37]. The combination of Gata3 with enhanced Mmtr7/Mapk8 mRNA suggests an IL-13 polarization configuration that differs from the Gata3/Eomes/Flh2 pattern seen in chlamydia-specific CD4γ13 T cells [21].
Adoptively transferred CD8γ13 8sAg-3 protected mice from immunopathology. It is interesting to note that the more cytolytic Tc1 8uvmo-2 did not facilitate clearance and trended toward delaying it. We have shown that, with Chlamydia-specific CD4 T cells, perforin-mediated killing is detrimental to Cm termination in epithelial cells [26], and others have shown that perforin-deficient mice clear infections with normal kinetics [38]. Cervical CD8 T cells in women with prior Chlamydia infection have low perforin levels, which possibly reflect an attenuated phenotype that balances microbial protection with fetal tolerance [39]; however, perforin-low CD8 may be a protective T-cell phenotype. Cytolysis seems to be unhelpful due to enhanced inflammation, reduced termination of intracellular chlamydia, and/or killing of APC, particularly B cells [40], possibly delaying assembly of memory lymphocyte clusters anchoring chlamydia-specific Trm [41].
CONCLUSIONS
We previously reported that unrestricted CD8 clones recognized antigen-pulsed wild-type and KbDb knockout splenocytes equally. In this report, at least 3 of those Chlamydia-specific CD8 clones, including 2 of 2 CD8γ13 clones, were found to be class II restricted. That result reasonably explains “uncomfortable” published data including the following: (1) MHC class II knockout mice are unable to clear Cm genital tract infections, but CD4 knockout mice clear infections with only a 1-week delay [42], “consistent with class II-restricted CD8 T cells filling the void”; (2) vaccine-generated trachoma protection in macaques genetically maps to class II [43] but was shown to be dependent on CD8 T cells by depletion [11], “consistent with class II-restricted CD8”; and (3) the unrestricted CD8 phenotype based on framework antibodies against HLA class I, II, and CD1 [12, 13] could not block unrestricted CD8 “because CD8αβ does not bind MHC class II, and class II-restricted CD8s are either coreceptor independent or use an unknown coreceptor”. The MHC class II-restricted CD8 T cells are not new. They are present in mixed lymphocyte reactions [44], they recognize soluble antigen presented by antigen-specific B cells [40], they are seen in viral infections [45, 46], and they may be a principal driver of chronic transplant rejection [47, 48]. Human and mouse data support class II-restricted CD8 T cells, including a CD8γ13 subset, and play a role in protection and immunopathology. They may have evolved under selective pressure from intracellular pathogens that block or limit class I antigen presentation (eg, chlamydia [49]) or diminish CD4 T-cell numbers or impair their function (eg, human immunodeficiency virus). A better understanding of them may provide new insights into pathogenesis and vaccine development.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Jeanette McClintick and the Indiana University Center for Medical Genomics provided critical assistance with microarray data analysis.
Financial support. This research was funded by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (Grant R21AI114953; to R. M. J.).
Potential conflicts of interest. R. M. J. has a U.S. Patent (No. 9,683,262 B2) derived, in part, from this work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers for Disease Control and Prevention. Sexually transmitted disease surveillance 2015. Atlanta: US Department of Health and Human Services; 2016. [Google Scholar]
- 2. Brunham RC, Rekart ML. The arrested immunity hypothesis and the epidemiology of chlamydia control. Sex Trans Dis 2008; 35:53–4. [DOI] [PubMed] [Google Scholar]
- 3. Geisler WM, Lensing SY, Press CG, Hook EW 3rd. Spontaneous resolution of genital Chlamydia trachomatis infection in women and protection from reinfection. J Infect Dis 2013; 207:1850–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu H, Jiang X, Shen C, et al. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun 2010; 78:2272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu H, Karunakaran KP, Kelly I, et al. Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model: correlation with MHC class II peptide presentation and multifunctional Th1 cells. J Immunol 2011; 186:3615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen CR, Gichui J, Rukaria R, Sinei SS, Gaur LK, Brunham RC. Immunogenetic correlates for Chlamydia trachomatis-associated tubal infertility. Obstet Gynecol 2003; 101:438–44. [DOI] [PubMed] [Google Scholar]
- 7. Olson KM, Tang J, Brown L, Press CG, Geisler WM. HLA-DQB1*06 is a risk marker for chlamydia reinfection in African American women. Genes Immun 2019; 20:69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang J, Maxion H, Champion CI, Liu G, Kelly KA. Expression of CXCR3 on adaptive and innate immune cells contributes oviduct pathology throughout Chlamydia muridarum infection. J Mucosal Immunol Res 2017; 1:104. [PMC free article] [PubMed] [Google Scholar]
- 9. Igietseme JU, He Q, Joseph K, et al. Role of T lymphocytes in the pathogenesis of chlamydia disease. J Infect Dis 2009; 200:926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vlcek KR, Li W, Manam S, et al. The contribution of chlamydia-specific CD8(+) T cells to upper genital tract pathology. Immun Cell Biol 2016; 94:208–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olivares-Zavaleta N, Whitmire WM, Kari L, Sturdevant GL, Caldwell HD. CD8+ T cells define an unexpected role in live-attenuated vaccine protective immunity against Chlamydia trachomatis infection in macaques. J Immunol 2014; 192:4648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gervassi AL, Probst P, Stamm WE, Marrazzo J, Grabstein KH, Alderson MR. Functional characterization of class Ia- and non-class Ia-restricted Chlamydia-reactive CD8+ T cell responses in humans. J Immunol 2003; 171:4278–86. [DOI] [PubMed] [Google Scholar]
- 13. Matyszak MK, Gaston JS. Chlamydia trachomatis-specific human CD8+ T cells show two patterns of antigen recognition. Infect Immun 2004; 72:4357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson RM, Kerr MS, Slaven JE. An atypical CD8 T-cell response to Chlamydia muridarum genital tract infections includes T cells that produce interleukin-13. Immunology 2014; 142:248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asquith KL, Horvat JC, Kaiko GE, et al. Interleukin-13 promotes susceptibility to chlamydial infection of the respiratory and genital tracts. PLoS Pathog 2011; 7:e1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuschiotti P, Medsger TA Jr, Morel PA. Effector CD8+ T cells in systemic sclerosis patients produce abnormally high levels of interleukin-13 associated with increased skin fibrosis. Arthritis Rheum 2009; 60:1119–28. [DOI] [PubMed] [Google Scholar]
- 17. Fuschiotti P, Larregina AT, Ho J, Feghali-Bostwick C, Medsger TA Jr. Interleukin-13-producing CD8+ T cells mediate dermal fibrosis in patients with systemic sclerosis. Arthritis Rheum 2013; 65:236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006; 12:99–106. [DOI] [PubMed] [Google Scholar]
- 19. Jayarapu K, Kerr MS, Katschke A, Johnson RM. Chlamydia muridarum-specific CD4 T-cell clones recognize infected reproductive tract epithelial cells in an interferon-dependent fashion. Infect Immun 2009; 77:4469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson RM. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect Immun 2004; 72:3951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson RM, Yu H, Strank NO, Karunakaran K, Zhu Y, Brunham RC. B cell presentation of Chlamydia antigen selects out protective CD4gamma13 T cells: implications for genital tract tissue-resident memory lymphocyte clusters. Infect Immun 2018; 86:e00614–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson RM, Kerr MS, Slaven JE. Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J Immunol 2012; 188:1896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jayarapu K, Kerr M, Ofner S, Johnson RM. Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms. J Immunol 2010; 185:6911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue L, Gyles SL, Wettey FR, et al. Prostaglandin D2 causes preferential induction of proinflammatory Th2 cytokine production through an action on chemoattractant receptor-like molecule expressed on Th2 cells. J Immunol 2005; 175:6531–6. [DOI] [PubMed] [Google Scholar]
- 25. Su H, Morrison R, Messer R, Whitmire W, Hughes S, Caldwell HD. The effect of doxycycline treatment on the development of protective immunity in a murine model of chlamydial genital infection. J Infect Dis 1999; 180:1252–8. [DOI] [PubMed] [Google Scholar]
- 26. Johnson RM, Kerr MS, Slaven JE. Perforin is detrimental to controlling [corrected] C. muridarum replication in vitro, but not in vivo. PLoS One 2013; 8:e63340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 28. Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, et al. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci U S A 2000; 97:12272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, Kelly K. Gem: an induced, immediate early protein belonging to the Ras family. Science 1994; 265:241–4. [DOI] [PubMed] [Google Scholar]
- 30. Fukuda T, Sanui T, Toyoda K, et al. Identification of novel amelogenin-binding proteins by proteomics analysis. PLoS One 2013; 8:e78129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y, Howard D, Rector K, et al. Latexin is down-regulated in hematopoietic malignancies and restoration of expression inhibits lymphoma growth. PLoS One 2012; 7:e44979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (SLAP) is a negative regulator of T cell receptor signaling. J Exp Med 2000; 191:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Meijgaarden KE, Haks MC, Caccamo N, Dieli F, Ottenhoff TH, Joosten SA. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS Pathog 2015; 11:e1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Medsger TA Jr, Ivanco DE, Kardava L, Morel PA, Lucas MR, Fuschiotti P. GATA-3 up-regulation in CD8+ T cells as a biomarker of immune dysfunction in systemic sclerosis, resulting in excessive interleukin-13 production. Arthritis Rheum 2011; 63:1738–47. [DOI] [PubMed] [Google Scholar]
- 35. Baraut J, Farge D, Jean-Louis F, et al. Transforming growth factor-β increases interleukin-13 synthesis via GATA-3 transcription factor in T-lymphocytes from patients with systemic sclerosis. Arthritis Res Ther 2015; 17:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joetham A, Schedel M, Takeda K, et al. JNK2 regulates the functional plasticity of naturally occurring T regulatory cells and the enhancement of lung allergic responses. J Immunol 2014; 193:2238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo L, Martens C, Bruno D, et al. Lipid phosphatases identified by screening a mouse phosphatase shRNA library regulate T-cell differentiation and protein kinase B AKT signaling. Proc Natl Acad Sci U S A 2013; 110:E1849–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perry LL, Feilzer K, Hughes S, Caldwell HD. Clearance of Chlamydia trachomatis from the murine genital mucosa does not require perforin-mediated cytolysis or Fas-mediated apoptosis. Infect Immun 1999; 67:1379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ibana JA, Myers L, Porretta C, et al. The major CD8 T cell effector memory subset in the normal and Chlamydia trachomatis-infected human endocervix is low in perforin. BMC Immunol 2012; 13:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shinohara N, Watanabe M, Sachs DH, Hozumi N. Killing of antigen-reactive B cells by class II-restricted, soluble antigen-specific CD8+ cytolytic T lymphocytes. Nature 1988; 336:481–4. [DOI] [PubMed] [Google Scholar]
- 41. Johnson RM, Brunham RC. Tissue-resident T cells as the central paradigm of chlamydia immunity. Infect Immun 2016; 84:868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morrison RP, Feilzer K, Tumas DB. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 1995; 63:4661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kari L, Whitmire WM, Olivares-Zavaleta N, et al. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 2011; 208:2217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shinohara N, Hozumi N, Watanabe M, Bluestone JA, Johnson-Leva R, Sachs DH. Class II antigen-specific murine cytolytic T lymphocytes (CTL). II. Genuine class II specificity of Lyt-2+ CTL clones. J Immunol 1988; 140:30–6. [PubMed] [Google Scholar]
- 45. Hioe CE, Hinshaw VS. Induction and activity of class II-restricted, Lyt-2+ cytolytic T lymphocytes specific for the influenza H5 hemagglutinin. J Immunol 1989; 142:2482–8. [PubMed] [Google Scholar]
- 46. Morrison LA, Braciale VL, Braciale TJ. Expression of H-2I region-restricted cytolytic activity by an Lyt-2+ influenza virus-specific T lymphocyte clone. J Immunol 1985; 135:3691–6. [PubMed] [Google Scholar]
- 47. Schnickel GT, Whiting D, Hsieh GR, et al. CD8 lymphocytes are sufficient for the development of chronic rejection. Transplantation 2004; 78:1634–9. [DOI] [PubMed] [Google Scholar]
- 48. Vessie EL, Hirsch GM, Lee TD. Aortic allograft vasculopathy is mediated by CD8(+) T cells in cyclosporin A immunosuppressed mice. Transpl Immunol 2005; 15:35–44. [DOI] [PubMed] [Google Scholar]
- 49. Karunakaran KP, Yu H, Jiang X, et al. Discordance in the epithelial cell-dendritic cell MHC class II immunoproteome: implications for Chlamydia vaccine development. J Infect Dis 2019; 221:841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.