Abstract
The placenta participates in maternal insulin sensitivity changes during pregnancy; however, mechanisms remain unclear. We investigated associations between maternal insulin sensitivity and placental DNA methylation markers across the genome. We analyzed data from 430 mother-offspring dyads in the Gen3G cohort. All women underwent 75-g oral glucose tolerance tests at ∼26 weeks of gestation; we used glucose and insulin measures to estimate insulin sensitivity (Matsuda index). At delivery, we collected samples from placenta (fetal side) and measured DNA methylation using Illumina EPIC arrays. Using linear regression models to quantify associations at 720,077 cytosine-guanine dinucleotides (CpGs), with adjustment for maternal age, gravidity, smoking, BMI, child sex, and gestational age at delivery, we identified 188 CpG sites where placental DNA methylation was associated with Matsuda index (P < 6.94 × 10−8). Among genes annotated to these 188 CpGs, we found enrichment in targets for miRNAs, in histone modifications, and in parent-of-origin DNA methylation including the H19/MIR675 locus (paternally imprinted). We identified 12 known placenta imprinted genes, including KCNQ1. Mendelian randomization analyses revealed five loci where placenta DNA methylation may causally influence maternal insulin sensitivity, including the maternally imprinted gene DLGAP2. Our results suggest that placental DNA methylation is fundamentally linked to the regulation of maternal insulin sensitivity in pregnancy.
Introduction
Insulin sensitivity decreases drastically in the 2nd half of pregnancy to levels that are similar to those in individuals with early type 2 diabetes (T2D) (1). It is hypothesized that this dramatic decrease in insulin sensitivity is meant to help provide nutrients from maternal sources to the growing fetus. The placenta likely plays a role in this physiologic adaptation, but the exact mechanisms remain unclear.
The placenta is a unique organ of fetal origin that lies at the maternal and fetal interface with primary roles to optimize fetal growth, protect the fetus against infections, and produce key hormones to maintain pregnancy; these hormones profoundly influence maternal physiology. In its role of nutrient transfer, the placenta responds to both fetal demands and maternal availability of nutrients and further adapts to regulate resources allocation. Yet, the “maternal-fetal conflict” theory posits that the mother and the fetus “disagree” on an optimal level of resource allocation from the mother to the fetus to allow pregnancy to its term and a healthy baby (2). During early embryogenesis, the trophectoderm develops to form the placenta with a distinctive epigenetic process (3). The placenta demethylation process in early development is characterized by a great number of genomic regions remaining imprinted from their parent of origin. Some investigators have proposed that the maintenance of the parental origin of imprinted regions in placenta contributes to the “maternal-fetal conflict” where genes that are expressed from paternal alleles act to shunt more nutrients to the fetus while expression driven by maternal alleles helps the mother maintain her resources (2,4). Thus, it is plausible that specific DNA methylation patterns in the placenta may influence the decline in maternal insulin sensitivity that leads to the transfer of glucose and other fuels to the fetus.
Based on these hypotheses, we investigated associations between maternal insulin sensitivity estimated in the 2nd trimester and genome-wide DNA methylation in placenta collected at birth in a large prospective pregnancy cohort. We had initially hypothesized that maternal insulin sensitivity could lead to changes in placental DNA methylation (given temporality of our measures) but also tested the possibility that placenta DNA methylation may influence maternal insulin sensitivity. To untangle whether placental DNA methylation is influencing maternal insulin sensitivity or vice versa, we tested potential causality using a bidirectional Mendelian randomization (MR) framework. Additionally, we conducted pathway analyses to deepen our understanding of our findings.
Research Design and Methods
Description of Participants
This study is based on mother-child pairs in the Genetics of Glucose regulation in Gestation and Growth (Gen3G) prospective cohort. We have previously published details of recruitment and phenotypic measurements during pregnancy (5). In brief, we recruited women in the 1st trimester (V1: 5–14 weeks), inviting all women presenting for their routine prenatal laboratories at Centre Hospitalier Universitaire de Sherbrooke (CHUS). We excluded women with diabetes prior to pregnancy (known or discovered at 1st trimester) and nonsingleton pregnancies. In addition to collecting blood samples, our trained research staff collected demographics, medical history, and completed standardized anthropometric measurements. We calculated BMI using the standard formula (kg/m2).
We followed participating women in the 2nd trimester (V2: 24–30 weeks) and completed similar measurements. After overnight fasting, women completed a 75-g oral glucose tolerance testing (OGTT) for gestational diabetes mellitus (GDM) screening. We collected plasma samples at baseline (fasting) and at 1 h and 2 h during the OGTT. We measured glucose and insulin at each OGTT time point, which allowed us to derive indices of insulin sensitivity.
We followed women until delivery and collected delivery and neonatal outcomes in addition to cord blood and placenta samples. At birth, trained research staff collected placenta samples (<30 min postpartum) based on a standardized protocol: 1 cm3 placenta tissue was collected ∼5 cm from the umbilical cord insertion on both sides of the placenta. Prior studies have shown high concordance of placental DNA methylation levels at specific loci across biopsy locations (6). Placenta samples were rapidly put in RNALater (QIAGEN) and stored at 4°C for a ≥24 h and then stored at −80°C. For this study, we selected fetal side placenta samples based on availability of adequate tissue (high-quality DNA extraction) and excluded complications in late pregnancy or delivery (e.g., preeclampsia, chorioamnionitis, stillbirth).
The CHUS ethics board review committee approved this study; all women provided written consent. For this study, we included only women of European origin (self-reported) who had consented for genetics investigations.
Bioassays
We measured glucose by the glucose hexokinase method (Roche Diagnostics) immediately after collection and insulin using multiplexed particle-based flow cytometric assays (Human MILLIPLEX map kits, EMD Millipore) from plasma samples previously frozen (−80°C). We estimated insulin sensitivity using the Matsuda index, given by the following equation: 10,000/ (√[(fasting glucose × fasting insulin) × (mean glucose during OGTT × mean insulin during OGTT)]) (7). We selected the Matsuda index over other measures of insulin sensitivity because it has been validated against euglycemic-hyperinsulinemic clamps in pregnant women (7).
DNA Methylation Measurements
We purified DNA from 460 placenta samples using the AllPrep DNA/RNA/Protein Mini Kit (QIAGEN). After bisulfite conversion, the Illumina Laboratory (San Diego, CA) performed epigenome-wide DNA methylation measurements using the Infinium MethylationEPIC BeadChip. We imported methylation data into R for preprocessing using minfi. We performed quality control (QC) at the sample level, excluding samples that failed (n = 5) or had mismatched genotype based on paired cord blood data (n = 6) or sex (n = 1). Our final placenta DNA methylation data set included 448 samples. We then excluded women because of missing data (Matsuda or key covariables). Thus, our final data set for this study was composed of 430 mother-child pairs (see Supplementary Fig. 1), which fully overlap with our prior publication of maternal 2-h glucose associations with placenta DNA methylation (8).
We normalized our data as previously described (8). We applied functional normalization (9) (FunNorm) and Regression on Correlated Probes (10) (RCP) to adjust for technical variability and probe type bias, respectively. Briefly, FunNorm removes technical variability using control probes from the array and RCP corrects type II probe distributions using the distribution of proximal type I probes to increase precision. We removed cytosine-guanine dinucleotide (CpG) probes with single nucleotide polymorphisms (SNPs) at the target site (4,453), single base extension (5,552), or anywhere within the probe (71,054) with a minor allele frequency (MAF) of >5%; probes on sex chromosomes (19,129); and previously identified cross-reactive probes (40,448) (11) to analyze 720,077 high-quality probes. We used the ComBat function from the sva package to adjust for batches. We logit transformed the β-values to M values for statistical analyses, as they have been shown to be more appropriate, meeting statistical model assumptions (12). However, we also reported effect estimated on the β-value scale to ease interpretability, since it approximates the proportion of methylation (from 0 to 1).
Genotyping
We isolated DNA from maternal buffy coat using the Gentra Puregene Blood Kit (QIAGEN, Mississauga, Ontario, Canada). We performed genotyping on 598 maternal samples using Illumina MEGAEX arrays. We removed 16 samples with a call rate ≤98% or failed sample QC. All remaining samples passed additional QC measures (percent heterozygosity, sex concordance, principal components derived outliers). We removed SNPs that were monomorphic, on sex chromosomes, or had MAF <1% in our sample, Hardy-Weinburg equilibrium P value <1 × 10−8, and insertions/deletions. After the above steps, we had data available on 838,884 SNPs in 582 women. We performed genotyping imputation using ShapeIT v2.r790 phasing Haplotype Reference Consortium (HRC) r1.1 2016 reference panel and Minimac3 software provided by the Michigan Imputation Server, which resulted in a data set containing a total of 39,183,141 autosomal SNPs for the overall population. Before analyses, we excluded all monomorphic SNPs or those with an MAF <0.05.
Statistical Analyses
We described participants’ characteristics using median and interquartile range (IQR) for continuous variables and frequency and percentage for categorical variables. We used natural log (ln) transformation to obtain a normal distribution of Matsuda index and used ln values for all analyses. We conducted analyses using R, version 3.5.1.
In 430 mother-infant pairs, we performed an epigenome-wide association study (EWAS) by fitting robust linear regression models using the rlm function for each CpG site. In CpG-by-CpG models, we included DNA methylation (M values) as the response variable and Matsuda (ln) as the exposure of interest. In model 1, we adjusted for maternal age, gravidity, smoking during pregnancy, maternal BMI (1st trimester), child sex, and gestational age at delivery. To control for genomic inflation, we used ReFACTor (model 2), a reference-free method that adjusts for heterogeneity putative to cell types in EWAS from heterogenous tissues (13). We used the top-10 principal components (PCs) estimated from ReFACTor as proxy for placenta cellular heterogeneity as suggested by the scree plot (Supplementary Fig. 2). We used quantile-quantile plots and histograms for the regression P values to visually inspect genomic inflation (λ) in both models (Supplementary Fig. 3). We corrected for multiple testing using Bonferroni with significant P values <6.94 × 10−8.
Gene Annotation and Pathway Analyses
First, we annotated CpGs with the R package IlluminaHumanMethylationEPICmanifest (14). Second, we utilized the R package humarray (15) to find the nearest gene based on base pair distance upstream and downstream. We tested for enrichment in biologic pathways with Enrichr (16,17) Web platform using only one gene per identified CpG: at each CpG, we prioritized gene annotation from UCSC Reference extracted from the IlluminaHumanMethylationEPICmanifest (except in cases of updated gene names) and then used the closest informative gene name from humarray annotation (priority to coding genes). We focused our attention on TargetScan miRNA, 2017; ENCODE histone modifications, 2015; WikiPathways, 2019; BioCarta, 2016; GWAS Catalogue, 2019; and dbGaP-reported databases in Enrichr (16,17).
MR Analyses
We conducted MR analyses to untangle direction of effect of associations based on 401 women with complete data from genotyping arrays, placental DNA methylation, and Matsuda index. First, we used MR to test whether placental DNA methylation levels may influence maternal insulin sensitivity. We searched for SNPs in cis (within 500 kb on each side) at each of 188 CpGs identified in our model 1 and tested SNP-to-methylation associations to identify genetic instrumental variables (IVs) in each region. We removed SNPs with an MAF <0.05. We assumed a genetic additive model and chose the effect allele as associated with greater DNA methylation levels. If multiple SNPs present in the determined cis region were associated with DNA methylation levels at the CpG of interest, we used the elastic net procedure with the glmnet (18) package. We looked at models with α from 0.1 to 1 by steps of 0.1. For each model, λ was chosen as the value that gave the minimum mean cross-validated error (λ.min). Finally, the α was chosen as the value that gave the smallest mean square error. When there was more than one SNP remaining in the final model from the elastic net procedure, we built a genetic risk score (GRS) assuming an additive genetic effect and summed the number of risk alleles as a global genetic IV. If there was only one SNP associated with DNA methylation in the designated in cis region, we used additive genetic modeling for that one SNP as genetic IV. We tested associations between the genetic IVs (GRS or individual SNP) and Matsuda index (ln). To compare effect estimates, we used the two-stage least squares (TSLS) that uses the predicted DNA methylation value by its respective genetic IV as the independent variable in the linear regression to predict Matsuda index (19,20). We used the Durbin-Wu-Hausman test to test whether TSLS estimates were significantly different from the observed estimates. We corrected for the number of CpGs tested (n = 131 with genetic IV available) using false discovery rate (FDR).
Second, we used prior knowledge of SNPs known to influence insulin sensitivity (21). We selected eight SNPs (see Supplementary Table 1) that were previously established as determinants of fasting insulin in large GWAS (with P < 5 × 10−8 in Meta-Analyses of Glucose and Insulin-related traits Consortium [MAGIC] data sets) (22,23) and were also nominally associated (P < 0.05) with Matsuda index in nonpregnant individuals (24). We built a GRS assuming additive genetic effect and summed the number of risk alleles. We tested associations between the insulin sensitivity GRS and placenta DNA methylation (in M values) for the 188 CpGs identified in model 1 and corrected using FDR.
Data and Resource Availability
Data sets analyzed during the current study are not publicly available because we did not obtain consent for such public release of epigenetic data from participants. However, data are available from the corresponding author with the appropriate permission from the Gen3G study team upon reasonable request and approval of institutional review boards. Summary statistics of EWAS results for models 1 and 2 are available via https://figshare.com/s/5040ad2ece334944bf34.
Results
We present characteristics of participants in Table 1. At the beginning of pregnancy, women’s median age was 28 years (IQR 25; 31), median BMI was 23.7 kg/m2 (IQR 21.6; 27.9), one-third were primigravid, and <9% reported smoking. Median Matsuda insulin sensitivity index was estimated at 7.72 (IQR 5.69; 10.67) and appeared normally distributed after natural log transformation.
Table 1.
Characteristics of Gen3G mother-child pairs included in maternal insulin sensitivity EWAS of placenta
| Mothers (N) | 430 |
| Age (years) | 28 (25; 31) |
| Ethnicity, European descent | 430 (100.0) |
| Gravidity, 1st pregnancy | 142 (33.0) |
| Parity, 1st term pregnancy | 218 (50.7) |
| Smoking in early pregnancy | 36 (8.4) |
| BMI in early pregnancy (kg/m2) | 23.7 (21.6; 27.9) |
| Waist circumference at 1st-trimester visit (cm) | 89.1 [82.0; 97.0] |
| Blood pressure at 2nd-trimester visit (systolic/diastolic mmHg) | 107/67 (100/63; 112/72) |
| OGTT (2nd trimester) | |
| Fasting glucose (mmol/L) | 4.2 (3.9; 4.4) |
| 1-h glucose (mmol/L) | 7.1 (6; 8.2) |
| 2-h glucose (mmol/L) | 5.7 (4.8; 6.6) |
| Insulin sensitivity, Matsuda index (raw) | 7.72 (5.69; 10.67) |
| Insulin sensitivity, Matsuda index (natural log transformed) | 2.04 (1.74; 2.37) |
| GDM* | 37 (8.6) |
| Children | |
| Gestational age at birth (weeks) | 39.7 (38.9; 40.4) |
| Female sex | 204 (47.4) |
| Birth weight (kg) | 3.44 (3.17; 3.71) |
| Large for gestational age, >90th percentile | 31 (7.2) |
| Placental weight (g) | 542 (467; 642) |
Data are median (IQR) or n (%) unless otherwise indicated.
GDM was diagnosed according to International Association of the Diabetes and Pregnancy Study Groups.
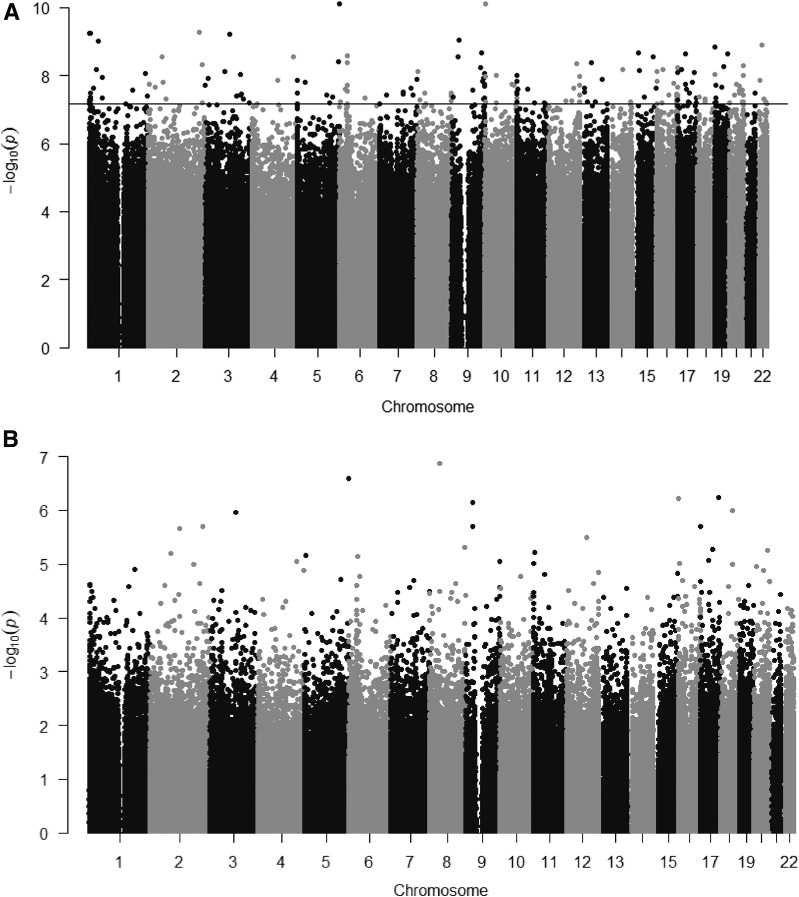
In model 1, we found that maternal insulin sensitivity was associated with placental DNA methylation at 188 CpGs (P values <6.94 × 10−8; adjustment for maternal age, gravidity, smoking, maternal BMI, child sex, and gestational age at delivery). Adding GDM status as covariate had minimal impact on association estimates at identified CpGs (0.02%–9.9% changes in β-coefficients). These 188 individual CpGs were distributed across the genome (Fig. 1A); in 14 regions, multiple CpGs were in close genomic vicinity and were annotated to the same gene (Supplementary Table 2). Among annotated genes, we identified 12 genes known to be imprinted in the placenta (9 maternally imprinted, SPHKAP, CNTN6, KCNIP4, PODXL, DLGAP2, KCNQ1, DSCAML1, GPC6, and OCA2, and 3 paternally imprinted, H19/MIR675, MCF2L, and LINC01056) (25). We also noted that specific miRNAs were listed at 11 loci (Supplementary Table 2). We examined our findings and found no CpGs overlapping with the list of CpGs that we had previously identified for associations with maternal 2-h glucose in the same cohort (8), despite moderate correlation (r = −0.44) between Matsuda and 2-h glucose. In model 2, adjusting bioinformatically for cell type heterogeneity, we did not find any individual CpGs that reached Bonferroni (Fig. 1B). Examining PCs generated by ReFACTor that reflect the cellular heterogeneity of tissue samples, we observed that PC1, PC2, and PC5 were strongly associated with Matsuda index, suggesting that cell type–specific placental DNA methylation profile is strongly related to maternal insulin sensitivity (Supplementary Table 3).
Figure 1.
Manhattan plots representing the results of the epigenome-wide association analyses between maternal insulin sensitivity (Matsuda index, ln transformed) and placenta methylation (in M values). A: Model 1 adjusted for maternal age, gravidity, smoking, maternal BMI, sex, and gestational age at delivery (genomic inflation = 2.884). The horizontal line indicates the Bonferonni level of statistical significance (P values <6.94 × 10−8). B: Model 2 adjusted for maternal age, gravidity, smoking, maternal BMI, sex, and gestational age at delivery and 10 PCs from ReFACTor (genomic inflation = 1.158).
MR 1: Does Placental Methylation Affect Maternal Insulin Sensitivity?
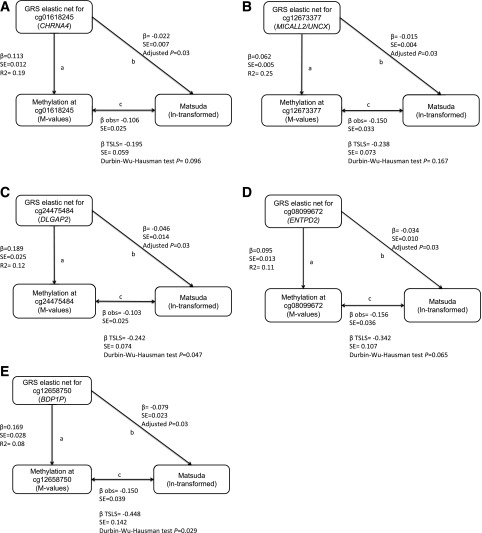
We identified genetic IVs in 131 of the 188 CpGs identified in model 1 (Supplementary Table 4). Specific GRS built with selected cis-SNPs captured respective CpG methylation levels with r2 ranging from 1% to 32%. We found 28 GRS capturing methylation at their respective CpG that were nominally associated (P < 0.05) with Matsuda index: 5 of these were statistically significant at FDR <0.05. These five GRS represented methylation levels at cg01618245 (CHRNA4), cg12673377 (MICALL2/UNCX), cg24475484 (DLGAP2), cg08099672 (ENTPD2), and cg03699074 (BDP1P). In all five cases, higher DNA methylation levels (as represented by GRS) were associated with lower Matsuda index and were in line with the direction of effect detected in our primary observational analyses (Fig. 2). TSLS estimates were also all in the same direction as that of the observed associations and significant (FDR <0.05). In two of the five CpGs (cg03699074 at BDP1P and cg24475484 at DLGAP2), the Durbin-Wu-Hausman test suggested that observed associations between DNA methylation and Matsuda index might be confounded. The fact that MR estimates were larger than the observational estimates suggests that observational estimates were negatively confounded and that “true” causal effects may be larger than the “observed.”
Figure 2.
MR supporting direction of effect at five loci where placenta DNA methylation may influence maternal insulin sensitivity: cg01618245 (CHRNA4) (A), cg12673377 (MICALL2/UNCX) (B), cg24475484 (DLGAP2) (C), cg08099672 (ENTPD2) (D), and cg03699074 (BDP1P) (E). In each panel, the a arrow indicates the association between genetic IV representing the fetal placental DNA methylation levels at CpG site (using GRS from maternal genotypes), the b arrow indicates the association with the build genetic IV and Matsuda index, and the c arrow (with β and SE below) indicates the observed (obs) association between methylation levels at the CpG and Matsuda index (reverse of original EWAS, to allow comparison of βs). TSLS estimates, SE, and Durbin-Wu-Hausman test P values are presented under observed estimates for the c arrows. All estimates are unadjusted (no covariates); the adjusted P values for b association results are FDR adjusted for number of tests performed (n = 131 with a genetic IV available).
MR 2: Does Maternal Insulin Sensitivity Affect Placenta?
The insulin sensitivity GRS build with eight SNPs captured ∼1.5% of Matsuda index variance (r2 = 0.015). Among our 188 identified CpGs (model 1), we did not identify any CpGs at which the insulin sensitivity GRS was associated with placental DNA methylation levels (Supplementary Table 5).
Pathways Analyses
Among transcription pathway databases, we noticed a strong enrichment in the TargetScan miRNA database (Supplementary Table 6): we found 34 miRNA target terms with adjusted P values <0.05 including hsa-miR-3180(-3p), hsa-miR-3196, hsa-miR1538, and hsa-miR-4745-3p at the top of the list. We also observed enrichment in the ENCODE histone modifications database, mainly driven by H3K27me3 in a variety of tissues and cell types (Supplementary Table 7).
The top term emerging from the GWAS Catalog (Supplementary Table 8) was “DNA methylation (parent-of-origin)” with H19, MIR675, and SEPT5 (P = 3.76 × 10−5; adjusted P = 0.017). Among the top 10 terms from the GWAS Catalog, we also observed “hemoglobin A1c” (driven by TCF7L2, KCNQ1, and PIEZO1) and “fasting insulin” (driven by TCF7L2 and CNTN6), which is the most common proxy of insulin resistance in large GWAS analyses (22) (both adjusted P value >0.05). From dbGaP, we identified 10 terms that reached adjusted P values <0.05 including “body mass index” and “body height” and other cardiovascular traits (triglycerides and blood pressure [Supplementary Table 9]).
In BioCarta, the top pathway was “role of PPAR-γ coactivators in obesity and thermogenesis,” with MED1 and RXRA leading the emergence of this pathway (Supplementary Table 10). The top pathways emerging from WikiPathways (Supplementary Table 11) were “adipogenesis” (6 of 130 genes: MEF2A, WWTR1, MBNL1, RXRA, GATA4, and PRLR) and “genes targeted by miRNAs in adipocytes,” driven by KCNQ1 and HCN2 (2 of13 genes), yet neither pathway had adjusted P values <0.05.
Discussion
Our results suggest that placental DNA methylation is fundamentally linked to maternal insulin sensitivity regulation. Using MR, we identified five loci where placental DNA methylation may be modulating the pregnancy-associated decrease in insulin sensitivity. To our knowledge, this is the 1st study to suggest that placental DNA methylation may causally influence maternal insulin sensitivity. Other identified loci are within known placenta-specific imprinted regions, consistent with the theory of maternal-fetal conflict. In addition, our assessment of cellular heterogeneity showed that the two 1st components of overall placenta DNA methylation profile are strongly associated with maternal insulin sensitivity. On one hand, it is possible that insulin sensitivity influences cell repertoire in the placenta as well as cell lineage commitment early during development. This cellular model has been termed “polycreodism,” or systematic variability in cell fate, which is relevant during embryonic development (26). Our MR 2 analyses did not support this direction, but our IV for insulin sensitivity was limited (r2 = 0.015). On the other hand, placental DNA methylation at delivery might also reflect DNA methylation stability across gestation at some loci. Despite the well-known global increase in placenta DNA methylation throughout gestation, Novakovic et al. (27) showed that substantial changes in methylation (β ≥ 0.2) were observed in 954 CpG sites between the 1st and 3rd trimesters and in only 157 CpG sites between the 2nd and 3rd trimesters (out of >26,000 CpG sites). Moreover, Schroeder et al. (28) demonstrated that partially methylated domains are common in placenta (37% of placental genome) and stable across gestation. Furthermore, by definition, imprinted loci remain stably methylated during fecundation and throughout in utero development (4).
miRNAs are suspected to have key roles in placenta development and function (29). Many of our findings implicated miRNAs as a potential link between placental DNA methylation and maternal insulin sensitivity. First, among the 188 identified CpGs, 11 were annotated to an miRNA as one of the closest genes. Second, TargetScan miRNA showed that our list of genes was greatly enriched for targets of multiple human miRNAs. In addition, we identified CpGs near genomic imprinted regions that contain miRNAs known to play an important role in placenta, e.g., at the H19/MIR675 and DIO3OS loci. DIO3OS is located near the placenta-specific miRNA cluster on Chr14q32 known as C14MC in the imprinted Dlk1-Dio3 domain. DIO3 is paternally imprinted during fetal development, suggesting that DIO3OS is a noncoding gene that may have a role in maintaining monoallelic maternal expression of DIO3 (30). The locus H19/MIR675 is paternally imprinted and thus maternally expressed (29). miR-675 is a highly conserved miRNA, located in the 1st exon of H19, and is specifically expressed by the placenta, with expression rising as the gestation advances (31). A putative role of miR-675 is to limit placental growth, likely via reducing the expression of IGF1R (main receptor of IGF2 key placental growth factor) (31).
It is notable that many imprinted genes are predominantly or solely expressed in the placenta (29). The different parental origin of DNA methylation patterns led to the maternal-fetal conflict hypothesis whereby paternal expression should favor fetal growth by deriving more maternal resources, while the maternally expressed genes should act to conserve maternal resources. We identified three loci at which the annotated gene is a known paternally imprinted (maternally expressed) gene in the placenta (including H19/MIR675) and nine loci at which the annotated gene is maternally imprinted (paternally expressed) in placental tissue (25) including KCNQ1. Our MR investigations suggested that methylation levels at cg24073146 in KCNQ1 could causally influence maternal insulin sensitivity, yet our MR estimate at this locus was nonsignificant after accounting for multiple testing. Loss of maternal-specific methylation of KCNQ1 causes Beckwith-Wiedemann syndrome, characterized by prenatal overgrowth and hypoglycemia in infancy (32). During normal fetal development, fetal pancreas shows monoallelic expression of KCNQ1, suggesting an important role of imprinting at this locus during pancreatic development, while adult pancreas shows biallelic expression (33). Genetic variants at KCNQ1 are well-established T2D risk variants with evidence of parent-of-origin effect where the transmission of maternal allele shows a very strong association for risk of T2D in comparison with paternal transmission (34).
Another identified CpG (cg24475484) is located within a known placenta-specific maternally imprinted region annotated to DLGAP2. DLGAP2 is biallelically expressed in the brain, but only paternally expressed in the testis (35), and was differentially methylated in spermatozoal DNA of infertility studies (36). Our MR analyses supported that methylation at cg24475484 (DLGAP2) causally influences maternal insulin sensitivity (FDR <0.05), in line with the maternal-fetal conflict hypothesis where the paternally expressed gene would reduce maternal insulin sensitivity to drive more nutrients toward the fetus.
Based on our MR analyses, we found that placenta DNA methylation levels at some identified CpGs may be causal in influencing maternal insulin sensitivity: four loci passed FDR significance threshold in addition to cg24475484 (DLGAP2). These CpGs were not located near genes with known placental function; this highlights the importance of agnostic approaches to discover new biologic candidates. Some of these loci deserve further functional studies in placenta and/or other relevant tissues. For example, cg08099672 is located near ENTPD2 expressed by the placenta, in addition to ovaries and testis, nervous system, and islets of Langerhans (37). The placenta expresses MICALL2 (located at ∼80 kb of cg12673377), and the protein is also highly detected in pancreas, adrenal, and stomach tissues (38). One intriguing finding is cg01618245, located near CHRNA4, which encodes for cholinergic receptor, nicotinic, α4, associated with epilepsy and nicotine addiction (39). It is noteworthy that two miRNAs (MIR3674 and MIR596) are located nearby cg24475484 at DLGAP2 (<150 kb apart) and the MIR4326 is located ∼72 kb from cg01618245 at CHRNA4, again implicating placental miRNAs as potential biologic mediators of pregnancy-associated changes in maternal insulin sensitivity.
Our pathway analyses highlighted many genes and pathways involved in adipose tissue regulation. From WikiPathways, “adipogenesis” (40) emerged from genes such as MEF2A, WWTR1, MBNL1, RXRA, and GATA4 and, notably, PRLR, which encodes for the prolactin receptor. MED1 and RXRA (±MEF2A) also highlighted the role of “PPAR-γ coactivators in obesity and thermogenesis” (BioCarta) and “energy metabolism” (WikiPathways), with PPARGC1A central to this pathway (41). We also identified two probes annotated to PRDM16, a key regulator of brown adipose tissue differentiation. Our group previously demonstrated in candidate gene studies that maternal hyperglycemia is associated with placental DNA methylation at PRDM16 and PPARGC1A (42); our current analyses using an agnostic approach validate our previous findings. On the other hand, none of the 188 identified CpGs in the current analyses overlapped with loci identified in our prior EWAS of maternal 2-h glucose and placenta DNA methylation (8). The absence of overlapping findings may be due to different biological phenomena or indicate that we would need a larger sample size to observe associations between placenta DNA methylation and both of these two moderately correlated glycemic traits.
Strengths and Limitations
Among our strengths, we have investigated a large number of placenta samples using the most comprehensive DNA methylation array, covering >720,000 CpGs across the genome, in a prospective cohort of pregnant women with well-characterized phenotypes, including a validated measure of insulin sensitivity during pregnancy. We were able to account for many potential confounders and applied an MR approach to test potential causality. Despite our attempts at untangling direction of effect, we found only a handful of CpGs from which MR supported causality. It is likely that the MR 2 analyses were limited in power given that our IV captured only 1.5% of the variance in Matsuda, resulting in a weak IV—one of the major limitations of our MR analyses in this direction. After adjustment for heterogeneity using ReFACTor, none of the CpGs reached P values <6.94 × 10−8, so it is possible that our signals from model 1 reflect placental cell-specific DNA methylation. Top ReFACToR PCs, reflecting cell type heterogeneity, were associated with insulin sensitivity; this might suggest that early DNA methylation programming might be driving a distinctive repertoire of cells in the placenta, also known as polycreodism (26). We feel this is highly biologically relevant and that future studies should investigate whether specific cells are responsible for the signals that we found and potential causal biological effects on maternal insulin sensitivity. Finally, our cohort is composed of women of European descent and thus findings may not be generalizable to other ethnicities.
Conclusion
In summary, our findings support a placental DNA methylation signature fundamentally linked to maternal insulin sensitivity. We identified CpGs at which our MR investigations supported that placental DNA methylation has a causal influence on maternal insulin sensitivity. The enrichment in miRNA targets and identification of specific miRNAs add to recent literature implicating miRNAs in placenta biology, either as paracrine or endocrine actors. Stimulation of insulin responsive cells (adipocytes, hepatocytes, myocytes) or trophoblasts by exposure to identified miRNAs could reveal potential functions. Finally, the identification of both maternally and paternally imprinted genes is in line with the maternal-fetal conflict hypothesis and yet also suggests that imprinted genes from both parents regulate maternal insulin sensitivity during pregnancy.
Supplementary Material
Article Information
Funding. This work was supported by American Diabetes Association Pathways Accelerator award 1-15-ACE-26 (to M.-F.H), Fonds de recherche du Québec en santé (FRSQ) 20697 (to M.-F.H), Canadian Institutes of Health Research MOP 115071 (to M.-F.H), and Diabète Québec grants (to P.P. and L.B.). L.B. is a senior research scholar from FRSQ. L.B., M.-F.H. and P.P. are members of the FRSQ-funded Centre de Recherche du Centre Hospitalier Universitaire de Sherbrooke. C.E.P. is supported by career development awards from the National Institute of Diabetes and Digestive and Kidney Diseases (K23DK113218) and the Robert Wood Johnson Foundation’s Harold Amos Medical Faculty Development Program (74256).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.-F.H. conceived the original study design and analyses plan, supervised the analyses, interpreted the results, and wrote the manuscript. M.D. contributed to sample and data collection. A.C. and C.A. carried out analyses, contributed to interpretation of data, and reviewed and critically edited the manuscript. M.D., C.E.P., P.M.C., P.P., and L.B. reviewed and critically edited the manuscript. All authors approved the final version. M.-F.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, 22–26 June 2018.
Footnotes
This article contains Supplementary Data online at https://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0798/-/DC1.
References
- 1.Catalano PM, Huston L, Amini SB, Kalhan SC. Longitudinal changes in glucose metabolism during pregnancy in obese women with normal glucose tolerance and gestational diabetes mellitus. Am J Obstet Gynecol 1999;180:903–916 [DOI] [PubMed] [Google Scholar]
- 2.Fowden AL, Moore T. Maternal-fetal resource allocation: co-operation and conflict. Placenta 2012;33(Suppl. 2):e11–e15 [DOI] [PubMed] [Google Scholar]
- 3.Januar V, Desoye G, Novakovic B, Cvitic S, Saffery R. Epigenetic regulation of human placental function and pregnancy outcome: considerations for causal inference. Am J Obstet Gynecol 2015;213(Suppl.):S182–S196 [DOI] [PubMed] [Google Scholar]
- 4.Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M. Imprinted genes and the epigenetic regulation of placental phenotype. Prog Biophys Mol Biol 2011;106:281–288 [DOI] [PubMed] [Google Scholar]
- 5.Guillemette L, Allard C, Lacroix M, et al. . Genetics of Glucose regulation in Gestation and Growth (Gen3G): a prospective prebirth cohort of mother-child pairs in Sherbrooke, Canada. BMJ Open 2016;6:e010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen BG, Byun HM, Cox B, et al. . Variation of DNA methylation in candidate age-related targets on the mitochondrial-telomere axis in cord blood and placenta. Placenta 2014;35:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22:1462–1470 [DOI] [PubMed] [Google Scholar]
- 8.Cardenas A, Gagné-Ouellet V, Allard C, et al. . Placental DNA methylation adaptation to maternal glycemic response in pregnancy. Diabetes 2018;67:1673–1683 [DOI] [PubMed] [Google Scholar]
- 9.Fortin JP, Labbe A, Lemire M, et al. . Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 2014;15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics 2016;32:2659–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidsley R, Zotenko E, Peters TJ, et al. . Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du P, Zhang X, Huang CC, et al. . Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmani E, Zaitlen N, Baran Y, et al. . Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 2016;13:443–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen KD. IlluminaHumanMethylationEPICmanifest: Manifest for Illumina's EPIC methylation arrays. R package version 0.3.0, 2016
- 15.Cooper N, humarray: Simplify Analysis and Annotation of Human Microarray Datasets R package version 1.2, 2017
- 16.Chen EY, Tan CM, Kou Y, et al. . Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22 [PMC free article] [PubMed] [Google Scholar]
- 19.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163 [DOI] [PubMed] [Google Scholar]
- 20.Angrist JD, Imbens GW. Two-stage least squares estimation of average causal effects in models with variable treatment intensity. J Am Stat Assoc 1995;90:431–442 [Google Scholar]
- 21.Powe CE, Nodzenski M, Talbot O, et al. . Genetic determinants of glycemic traits and the risk of gestational diabetes mellitus. Diabetes 2018;67:2703–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning AK, Hivert MF, Scott RA, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012;44:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prokopenko I, Poon W, Mägi R, et al. . A central role for GRB10 in regulation of islet function in man. PLoS Genet 2014;10:e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Delgado M, Court F, Vidal E, et al. . Human oocyte-derived methylation differences persist in the placenta revealing widespread transient imprinting. PLoS Genet 2016;12:e1006427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 2017;18:441–451 [DOI] [PubMed] [Google Scholar]
- 27.Novakovic B, Yuen RK, Gordon L, et al. . Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 2011;12:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder DI, Blair JD, Lott P, et al. . The human placenta methylome. Proc Natl Acad Sci USA 2013;110:6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malnou EC, Umlauf D, Mouysset M, Cavaillé J. Imprinted MicroRNA gene clusters in the evolution, development, and functions of mammalian placenta. Front Genet 2019;9:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez A, Martinez ME, Croteau W, St Germain DL. Complex organization and structure of sense and antisense transcripts expressed from the DIO3 gene imprinted locus. Genomics 2004;83:413–424 [DOI] [PubMed] [Google Scholar]
- 31.Keniry A, Oxley D, Monnier P, et al. . The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MP, DeBaun MR, Mitsuya K, et al. . Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 1999;96:5203–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Travers ME, Mackay DJ, Dekker Nitert M, et al. . Insights into the molecular mechanism for type 2 diabetes susceptibility at the KCNQ1 locus from temporal changes in imprinting status in human islets. Diabetes 2013;62:987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyssenko V, Groop L, Prasad RB. Genetics of type 2 diabetes: it matters from which parent we inherit the risk. Rev Diabet Stud 2015;12:233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DLGAP2 gene [Internet] , 2019. Available from https://www.genecards.org/cgi-bin/carddisp.pl?gene=DLGAP2&keywords=DLGAP2. Accessed 29 October 2019
- 36.Sujit KM, Sarkar S, Singh V, et al. . Genome-wide differential methylation analyses identifies methylation signatures of male infertility. Hum Reprod 2018;33:2256–2267 [DOI] [PubMed] [Google Scholar]
- 37.ENTPD2 gene [Internet] , 2019. https://www.genecards.org/cgi-bin/carddisp.pl?gene=ENTPD2&keywords=ENTPD2. Accessed 29 October 2019
- 38.MICALL2 gene [Internet] , 2019. Available from https://www.genecards.org/cgi-bin/carddisp.pl?gene=MICALL2&keywords=MICALL2. Accessed 29 October 2019
- 39.CHRNA4 gene [Internet] , 2019. https://www.genecards.org/cgi-bin/carddisp.pl?gene=CHRNA4&keywords=CHRNA4. Accessed 29 October 2019
- 40.Patti M-E, Hanspers K, Salomonis N, et al. Adipogenesis (Homo sapiens) [Internet] , 2018. Available from https://www.wikipathways.org/index.php/Pathway:WP236. Accessed 29 October 2019
- 41.Kosho M, Pico A, Hanspers K, et al. Energy metabolism [Internet] , 2017. Available from https://www.wikipathways.org/index.php/Pathway:WP1541. Accessed 29 October 2019
- 42.Côté S, Gagné-Ouellet V, Guay SP, et al. . PPARGC1α gene DNA methylation variations in human placenta mediate the link between maternal hyperglycemia and leptin levels in newborns. Clin Epigenetics 2016;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.