Capsule Summary
We report that MAIT cells repress group 2 innate lymphoid cell activation and restrict allergen-induced airway inflammation and airway hyperresponsiveness.
Keywords: mucosal associated invariant T cells, group 2 innate lymphoid cells, allergic airway inflammation, airway hyperresponsiveness
To the editor:
Asthma is a complicated airway inflammatory disorder that involves dysregulation of multiple adaptive and innate immune cell types. Mucosal-associated invariant T cells (MAIT) are innate-like T cells that recognize microbial vitamin B metabolites presented by MR1, but can function through both TCR dependent and independent pathways.1, 2 MAIT cells are abundant in healthy human individuals, accounting for a significant proportion of human CD8+ T cells. Recent studies have indicated that MAIT cells may be implicated in asthma. A recent study demonstrated that increased frequency of MAIT cells in one year-old infants was associated with a reduced risk of asthma by age 7.3 Another study by an independent research group revealed that adult severe asthma patients had decreased numbers of MAIT cells in peripheral blood, induced sputum, bronchoalveolar lavage, and endobronchial biopsy tissues.4 However, it remains unclear whether MAIT cell deficiency in asthma patients is a result of increased airway inflammation or corticosteroid therapy.4 The precise role of MAIT cells in asthma pathogenesis is also largely unknown.
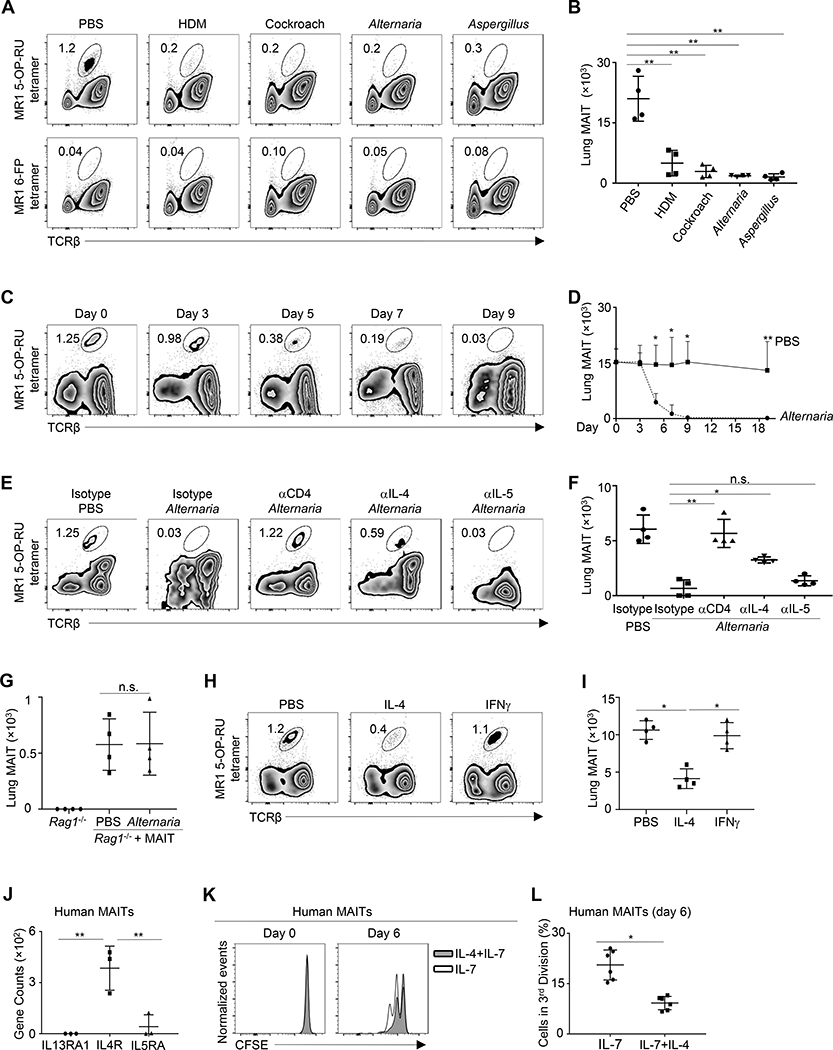
This study aimed to understand the function and regulation of MAIT cells in allergic airway inflammation. We first hypothesized that allergen-induced airway inflammation may directly lead to MAIT cell deficiency, independent of steroid therapy. To test this, we intranasally administered C57BL/6 mice with a variety of different common allergens, including extracts from Alternaria, Aspergillus, house dust mite (HDM), and cockroach. Strikingly, the numbers of lung-resident MAIT cells were greatly reduced in mice with repeated intranasal exposure to all the allergens we tested (Fig 1, A and B). None of these mice received steroid treatment. Thus, MAIT cells are diminished during allergic airway inflammation, independent of steroid treatment.
Figure 1. MAIT cells are diminished during allergic airway inflammation.
A, Lung MAIT cells in C57BL/6 mice challenged with the indicated allergen extracts every other day for 19 days. B, Numbers of lung MAIT cells in allergen-challenged mice. C, Lung MAIT cells in mice challenged with Alternaria extracts every other day for the indicated time points. D, Numbers of lung MAIT cells in Alternaria-challenged mice. E, Lung MAIT cells in mice challenged with Alternaria extracts and treated with the indicated antibodies every other day for 7 days. F, Numbers of lung MAIT cells in mice challenged with Alternaria and treated with the indicated antibodies. G, Numbers of lung MAIT cells in Rag1−/− mice that received MAIT cell transfer and challenged with Alternaria or PBS every other day for 7 days. H, Lung MAIT cells in mice treated with IL-4 or IFNγ daily for 7 days. I. Numbers of MAIT cells in cytokine-trmice treated with the indicated cytokines. J. Gene expression by RNA-seq. K, CFSE levels in MAIT cells before and after culturing with IL-7, or together with IL-4. L, Percentage of MAIT cells in the third division at day 6 of culture. N= 4 mice or 6 human samples per group, 2–3 independent experiments. *p < 0.05; **p < 0.01.
To examine the mechanisms underlying allergen-induced MAIT cell deficiency, we examined the kinetics of MAIT cell reduction throughout the time course of Alternaria-allergen induced airway inflammation. MAIT cell reduction occurred after 6 days of exposure to Alternaria allergen, a time point at which T cell inflammation becomes dominant in the lung (Fig 1, C and D). Indeed, depletion of CD4+ T cells by anti-CD4 mAbs restored MAIT cell numbers, suggesting that proinflammatory T cells mediate MAIT cell deficiency in the lung (Fig 1, E and F). We next purified lung MAIT cells by fluorescence activated cell sorting (FACS), and then transferred MAIT cells into Rag1−/− mice followed by 8 days of Alternaria challenge. The number of MAIT cells that were transferred into Rag1−/− mice remained intact during Alternaria-induced airway inflammation (Fig. 1, G). These results thus highlight an important role for adaptive immune cells, especially CD4+ T cells, in mediating MAIT cell decline during allergic airway inflammation. Notably, neutralization of IL-4, but not IL-5, partially alleviated MAIT cell deficiency in mice that inhaled Alternaria, indicating that IL-4 but not IL-5 regulates MAIT cell maintenance (Fig 1, E and F). In addition, administration of IL-4, but not IFNγ, impaired the maintenance of pulmonary MAIT cells (Fig 1, H and I). Indeed, RNA-seq revealed that human MAIT cells (see Figure E1 in this article’s Online Repository) expressed IL4R but not IL5R or IL13R (Fig 1, J). To clarify whether IL-4 may directly regulate human MAIT cell biology, we purified MAIT cells from human peripheral blood by FACS, labelled them with CFSE, and cultured them in the presence or absence of IL-4. Exposure to IL-4 inhibited the proliferation of purified human MAIT cells, demonstrated by reduced dilution of CFSE (Fig 1, K and L). Together, these data indicate that pro-inflammatory T cells may impair the maintenance of MAIT cells during allergic airway inflammation, in part through direct effects of IL-4.
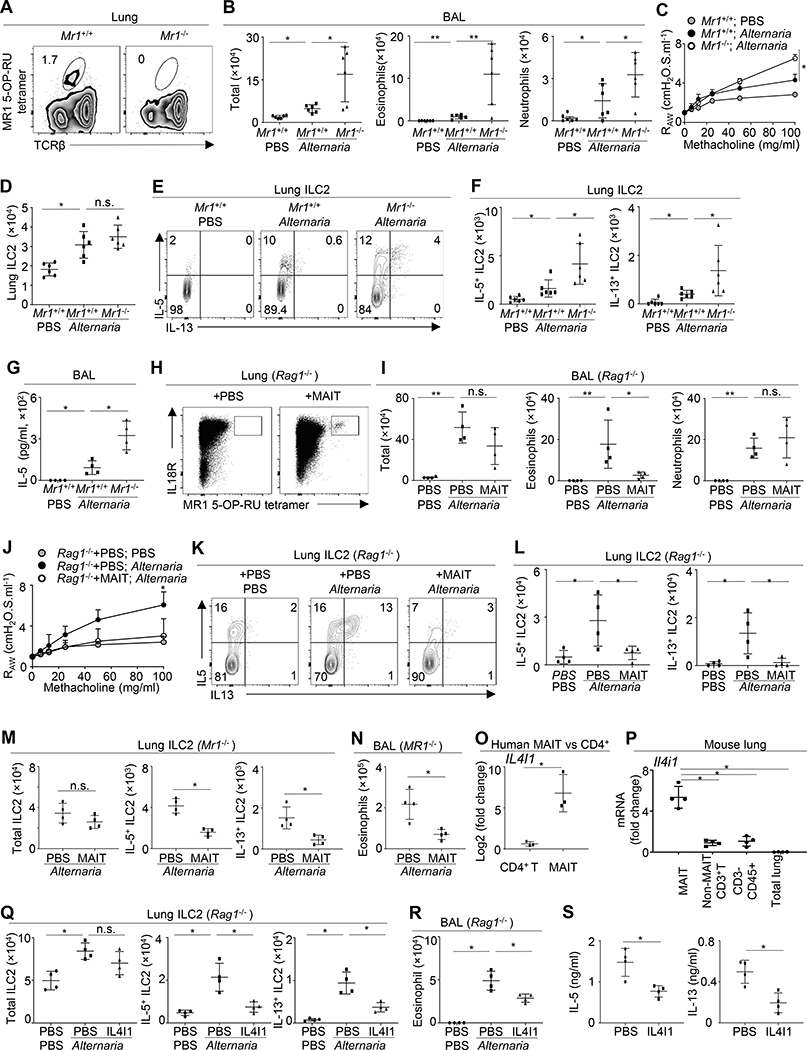
We sought to understand the specific role of MAIT cells in allergic airway inflammation and airway hyperresponsiveness (AHR). Alternaria is a fungal allergen that can trigger acute and severe asthma attacks in humans. Studies with mouse models have shown that Alternaria can rapidly and potently activate lung-resident group 2 innate lymphoid cells (ILC2), eliciting airway inflammation and AHR.5–7 Using newly generated Mr1−/− mice (see Figure E2 in this article’s Online Repository), we examined the role of MAIT cells in Alternaria-induced acute airway inflammation and AHR (Fig 2, A). Alternaria inhalation elicited eosinophilic and neutrophilic inflammation and moderate AHR in wildtype C57BL/6 mice within 3 days (Fig 2, B). Compared with wildtype mice, Mr1−/− mice exhibited greatly increased eosinophilic inflammation and also enhanced neutrophilic inflammation in response to Alternaria challenge (Fig 2, B). AHR was also moderately increased in Mr1−/− mice (Fig 2, C). We then examined ILC2 response in the lungs of Alternaria-challenged mice. Lung-resident ILC2 were identified as CD45+Lin−Thy1+T1/ST2+ cells (see Figure E3, A in this article’s Online Repository). As previously described,5 Alternaria inhalation led to increased proliferation and enhanced expression of IL-13 and IL-5 by lung-resident ILC2 in wildtype mice (Fig 2, D–F, and Figure E3, B). Although ILC2 number and proliferation were not significantly changed in Mr1−/− mice (see Figure E3, B in this article’s Online Repository), the expression of both IL-13 and IL-5 by ILC2 was greatly increased in Mr1−/− mice after Alternaria challenge, suggesting that MAIT cells inhibit ILC2 responses during allergic airway inflammation (Fig 2, D–F). Lung IL-5 and IL-13 concentrations were significantly increased in Mr1−/− mice, compared to wildtype mice after Alternaria challenge (see Figure E3, C in this article’s Online Repository). Compared to wildtype mice, Alternaria-challenged Mr1−/− mice also exhibited higher concentrations of IL-5 in the bronchoalveolar lavage fluid (Fig 2, G). We next examined ILC2 activity during house dust mite (HDM)-induced airway inflammation. Compared to wildtype mice, Mr1−/− mice exhibited markedly increased expression of IL-5 and IL-13 in lung ILC2, after 5 daily doses of HDM challenge (see Figure E3, D in this article’s Online Repository). BAL eosinophilic infiltration was also greatly increased in Mr1−/− mice upon HDM challenge (see Figure E3, E in this article’s Online Repository). Thus, MAIT cells also restrain HDM-induced allergic airway inflammation. To further verify the role of MAIT cells in Alternaria-induced airway inflammatory responses, we sorted MAIT cells from IL-7-treated mice and transferred purified MAIT cells into Rag1−/− mice (Fig 2, H). Consistent with previous reports,6 Alternaria inhalation activated ILC2 and induced innate eosinophilia and AHR in Rag1−/− mice that lacked T/B lymphocytes (Fig 2, I and J). Notably, transfer of MAIT cells alleviated eosinophilic infiltration and repressed AHR (Fig 2, I and J). Transfer of MAIT cells markedly reduced both IL-5 and IL-13 production by lung ILC2, verifying that MAIT cells can inhibit ILC2 responses (Fig 2, K and L). Of note, MAIT cell transfer did not significantly alter the number of total lung ILC2, but specifically repressed cytokine expression by ILC2 (see Figure E3, F in this article’s Online Repository). Transfer of MAIT cells into Alternaria-challenged Mr1−/− mice also repressed the expression of both IL-5 and IL-13 in lung ILC2, without significantly affecting the number of total ILC2 (Fig 2, M, and Figure E3, G in this article’s Online Repository). MAIT cell transfer alleviated airway eosinophilic infiltration in Mr1−/− mice (Fig 2, N). Thus, MAIT cells are capable of repressing ILC2 function even in the absence of MR1-expressing antigen-presenting cells. Together, these data reveal a novel role for MAIT cells in repressing ILC2 responses and restricting allergen-induced airway inflammation and AHR.
Figure 2. MAIT cells restrict Alternaria-induced airway inflammation and hyperresponsiveness.
A, Lung MAIT cells in Mr1+/+ and Mr1−/− mice. B, Bronchoalveolar lavage (BAL) cells in mice challenged with PBS or Alternaria for 3 days. C, FlexiVent analysis. D, Numbers of lung ILC2. E. Cytokine expression by ILC2. F, Numbers of IL-5+ or IL-13+ ILC2. G, BAL IL-5 concentrations. H, Lung MAIT cells in Rag1−/− mice with or without MAIT cell transfer. I, BAL cell numbers in Rag1−/− mice challenged with Alternaria or PBS, with or without MAIT cell transfer. J, FlexiVent analysis. K, Cytokine expression by lung ILC2. L, Numbers of IL-5+ or IL-13+ ILC2. M, Numbers of ILC2 in Mr1−/− mice challenged with Alternaria, with or without MAIT cell transfer. N, BAL eosinophilic numbers. O, IL4I1 expression in human blood CD4+ T cell and MAIT cells. P, Il4i1 expression in lung MAIT cells, non-MAIT T cells, non-T immune cells, and the whole lung tissue of naïve mice. Q, Numbers of ILC2 in Rag1−/− mice challenged with PBS or Alternaria, with or without IL4I1 treatment. R, BAL eosinophilic numbers. S, IL-5 and IL-13 production in mouse lung ILC2 cultured with or without IL4I1. N= 3–6 mice per group, 3–4 independent experiments. *p < 0.05; **p < 0.01.
We next examined the mechanisms by which MAIT cells may repress ILC2 responses. RNA-seq results demonstrated that human MAIT cells expressed a high amount of an anti-inflammatory molecule interleukin-4-induced gene 1 (IL4I1) (Fig 2, O). Using qPCR, we verified that mouse lung MAIT cells expressed markedly higher amounts of Il4i1, compared to other T cells and other immune cells in the lung (Fig 2, P). The expression of Il4i1 in total lung tissue, which consists chiefly of non-immune cells, was barely detectable (Fig 2, P). IL4I1 is a L-amino-acid oxidase that converts phenylalanine to phenylpyruvate, hydrogen peroxide, and ammonia.8 IL4i1 has been found to repress T cell inflammation during auto-immune diseases and may also repress B1 cell function.8 The immune-repressive effects of IL4I1 were thought be meditated in part through hydrogen peroxide toxicity.8 Whether IL4I1 may regulate ILC2 function remains unknown. We administrated IL4I1 intravenously into Alternaria-challenged Rag1−/− mice at the dose of 1.5 μg per day. A previous study indicated that this dose can repress T cell inflammation and reduce disease severity in a mouse model of EAE without causing obvious toxic effects.9 Notably, treatment with IL4i1 markedly repressed IL-5 and IL-13 expression in lung ILC2 in Alternaria-challenged Rag1−/− mice, indicating a novel role for IL4I1 in repressing ILC2 function (Fig 2, Q). IL4I1 treatment also reduced eosinophilic infiltration in Rag1−/− mice (Fig 2, R). We next sorted lung ILC2 from C57BL/6 mice and cultured purified ILC2 in the presence or absence of recombinant Il4I1, together with a cocktail of activating cytokines. Exposure to IL4I1 led to decreased production of IL-13 and IL-5 from cultured ILC2, indicating that IL4I1 may directly repress ILC2 activation (Fig 2, S). IL4I1 did not significantly affect the proliferation of ILC2, but repressed cytokine production of ILC2 at per cell level (Figure E3, H–J, in this article’s Online Repository). Together, MAIT cells may inhibit ILC2 responses in part through the expression of IL4I1.
Here we report a novel role for MAIT cells in repressing ILC2 responses and restricting allergic airway inflammation and AHR. We demonstrated that Mr1−/− mice that lacked MAIT cells had exacerbated ILC2 responses, increased airway inflammation and elevated AHR in response to acute Alternaria inhalation. Adoptive transfer of MAIT cells repressed ILC2 responses and alleviated airway inflammation and AHR. We found that both human and mouse MAIT cells expressed high amounts of the anti-inflammatory molecule IL4I1 that inhibited ILC2 activation. Further, MAIT cells were diminished after repeated allergen exposure due to the effects of pro-inflammatory T cells, suggesting a negative feedback loop that may contribute to sustained airway inflammation. The deficiency of MAIT cells in some asthma patients might contribute to an exacerbated inflammatory response to allergens and other stimuli.
Materials and Methods
Human blood samples and RNA sequencing
Blood samples from 20–45 years old healthy controls were collected. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll gradient centrifugation. MAIT cells were sorted by flow cytometric cell sorting followed by in vitro culture or RNA seq. Informed consent was obtained from all subjects. The study was approved by the Albany Medical Center Institutional Review Board.
Mice, Allergen Challenge, Flexivent Analysis, and Adoptive Transfer
Mr1+/− mice on C57BL/6 background were generated by Taconic bioscience using CRISPR technique and bred in the specific pathogen-free animal facility of Albany Medical Center (see Figure E2 in this article’s Online Repository). Rag1−/− mice were purchased from the Jackson laboratory. 6–10 weeks-old female and male Mr1−/− mice and wildtype littermate controls were used. All mouse procedures were approved by the Institutional Animal Care and Use Committee of Albany Medical College.
For allergen challenge, 50μg of extracts from house dust mite, or cockroach, or Alternaria alternata, or Aspergillus Fumigatus were administered intranasally every other day for the indicated time periods. All allergen extracts were purchased from the Greer laboratory. Bronchoalveolar immune cell infiltration, MAIT and ILC2 response were examined at 24 hours after the last dose of allergen administration. Airway hyperresponsiveness was measured by a FlexiVent system (SCIREQ) as we previously described.E1
For antibody treatment, mice were intranasally challenged with 50μg of Alternaria extracts on day 0, 2, 4 and 6. 1500ug of anti-IL-4 (11B11, Bio X Cell), 250ug of anti-IL-5 (TRFK5, Bio X Cell), or isotype control (TNP6A7, Bio X Cell) were injected intraperitoneally on day −1, 1, 3, 5. 100ug of anti-CD4 (GK1.5, Bio X Cell) was injected intraperitoneally on day −1, 1, 5. Mice were euthanized and MAIT cells were examined at day 7. For cytokine treatment, mice were intraperitoneally injected with 400ng IL-4 or IFNγ, or PBS control daily for 7 days. Mice were euthanized and MAIT cells were examined on the day after the last challenge.
For adoptive transfer, donor wildtype C57BL/6 mice that were treated with 400ng IL-7 daily for 7 days for expansion of MAITs. MAITs were sorted from the lungs of IL-7-treated mice and transferred intravenously into Rag1−/− mice. Mice were rested for 4 weeks, followed examination of reconstitution efficiency with flow cytometry analysis. Alternaria-allergen challenge was also performed at 4 weeks post adoptive transfer.
Flow Cytometry and Cell Sorting
APC conjugated human and mouse MR1 5-OP-RU tetramers and control MR1 6-FP tetramers were obtained from NIH Tetramer Core Facility. The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne.E2 Tetramer staining was performed at room temperature for 30mins, together with antibodies for surface staining. Antibodies used to help identify human MAIT included anti-CD3 (OKT3), anti-CD45 (HI30), and anti-CD8 (SK1). Antibodies used to help identify mouse lung MAIT included anti-TCRβ (H57–597), anti-Thy1.2 (53–2.1), and anti-CD45 (104).
Mouse ILC2 were identifies as CD45+Lin−Thy1.2+ST2+ cells as we previously described.E1, 3–5 Anti-lineage (Lin) antibodies for mouse ILC2 included anti-B220 (RA3–6B2), anti-CD3 (2C11), anti-TCRβ (H57), anti-TCRγδ (GL-3), anti-CD11b (M1/70), anti-Nk1.1 (PK136). Other antibodies used include anti-Siglec F (E50–2440), anti-CD11c (N418), anti-Gr-1 (RB6–8C5), anti-ST2 (DJ8), anti-IL-13 (eBio13A), anti-IL-5 (TRFK5). Antibodies were purchased from BD bioscience, eBioscience, Biolegend or MD Bioproducts. Flow cytometric analysis was performed on a 3-laser FACSCanto (BD Biosciences) and cell sorting was performed on a FACSAria II (BD Biosciences).
Cell culture
For human MAIT culture, MAITs were sorted by FACS, and labelled with CFSE using a the CellTrace™ CFSE Cell Proliferation Kit (Invitrogen) according to the manufacturer’s instructions. CFSE-labelled cells were cultured with α-MEM medium containing 20% FCS and 30ng/ml recombinant human IL-7, in the presence or absence of 10ng/ml recombinant human IL-4. CFSE levels were examined at different time points.
For mouse ILC2 culture, ILC2 were sorted from the lungs of naïve wildtype C57BL/6 mice. Sorted ILC2 were cultured with α-MEM medium containing 20% FCS and 10ng/ml of recombinant mouse IL-2, IL-7 and IL-33. 100ng/ml Recombinant mouse IL4I1 (R&D) or control PBS was added in some wells. Cytokines were purchased from Biolegend or R&D.
Assessment of cytokine production
Mice were euthanized and lungs were perfused by 10ml cold PBS. Lungs were minced by a scissors and digested in Hank’s balanced salt solution with 0.1mg/ml Liberase TL (Roche Diagnostics) and 10U/ml DNase I (Roche Diagnostics). Cells were filtered through a 100μm cell strainer and re-stimulated with PMA/ Ionomycin in the presence of 1μg/ml monensin at 37°C for 2.5 hrs. Intracellular cytokine staining was performed using the Cytofix/Cytoperm Kit (BD) according to the manufacturer’s instructions.
Concentrations of IL-5 and IL-13 from lung homogenate, bronchoalveolar lavage, and culture supernatant were examined by ELISA kits according to the manufacturer’s instructions (Thermo).
RNA-seq and qPCR
For RNA-seq, RNA was extracted and cDNA libraries was generated using the SMART-Seq v4 Ultra Low Input RNA kit (Takara) and Nextera XT DNA Library Prep kit (Illumina). High throughout single-end 75 bp sequencing was performed with NextSeq 500 (Illumina) at the Microarray and Nexgen Sequencing core at University at Albany. RNA-seq data were deposited at GEO (accession number: GSE132799).
For qPCR, RNA was extracted using RNeasy (Qiagen). cDNA was generated using reverse transcriptase superscript II kit (Invitrogen). qPCR was performed with the following Taqman probes (Applied Biosytems™): Hs00541746_m1 (human IL4I1); Mm00515786_m1 (Mouse Il4i1).
Statistical analysis
Student’s t-test was used to compare the difference between two groups. Anova was used to compare the difference among three or more groups. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01HL137813, R01AG057782, and K22AI116728 (to Q.Y.), the Alexandrine and Alexander L. Sinsheimer Scholar Award (to Q.Y.), and R01HL110951, R01HL130304 (to D.D.T.).
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012; 491:717–23. [DOI] [PubMed] [Google Scholar]
- 2.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 2003; 422:164–9. [DOI] [PubMed] [Google Scholar]
- 3.Chandra S, Wingender G, Greenbaum JA, Khurana A, Gholami AM, Ganesan AP, et al. Development of Asthma in Inner-City Children: Possible Roles of MAIT Cells and Variation in the Home Environment. J Immunol 2018; 200:1995–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinks TS, Zhou X, Staples KJ, Dimitrov BD, Manta A, Petrossian T, et al. Innate and adaptive T cells in asthmatic patients: Relationship to severity and disease mechanisms. J Allergy Clin Immunol 2015; 136:323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty TA, Khorram N, Chang JE, Kim HK, Rosenthal P, Croft M, et al. STAT6 regulates natural helper cell proliferation during lung inflammation initiated by Alternaria. Am J Physiol Lung Cell Mol Physiol 2012; 303:L577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snelgrove RJ, Gregory LG, Peiro T, Akthar S, Campbell GA, Walker SA, et al. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol 2014; 134:583–92 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol 2011; 186:4375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano F, Molinier-Frenkel V. An Overview of l-Amino Acid Oxidase Functions from Bacteria to Mammals: Focus on the Immunoregulatory Phenylalanine Oxidase IL4I1. Molecules 2017; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Psachoulia K, Chamberlain KA, Heo D, Davis SE, Paskus JD, Nanescu SE, et al. IL4I1 augments CNS remyelination and axonal protection by modulating T cell driven inflammation. Brain. 2016;139(Pt 12):3121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Yang Q, Ge MQ, Kokalari B, Redai IG, Wang X, Kemeny DM, et al. Group 2 innate lymphoid cells mediate ozone-induced airway inflammation and hyperresponsiveness in mice. J Allergy Clin Immunol 2016; 137:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014; 509:361–5. [DOI] [PubMed] [Google Scholar]
- 3.Shen X, Pasha MA, Hidde K, Khan A, Liang M, Guan W, et al. Group 2 innate lymphoid cells promote airway hyperresponsiveness through production of VEGFA. J Allergy Clin Immunol 2018; 141:1929–31 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Xu X, Pasha MA, Siebel CW, Costello A, Haczku A, et al. Cutting Edge: Notch Signaling Promotes the Plasticity of Group-2 Innate Lymphoid Cells. J Immunol 2017; 198:1798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen X, Liang M, Chen X, Pasha MA, D’Souza SS, Hidde K, et al. Cutting Edge: Core Binding Factor beta Is Required for Group 2 Innate Lymphoid Cell Activation. J Immunol 2019; 202:1669–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.