Abstract
Small RNAs play an important role in plant innate immunity. However, their regulatory function in induced systemic resistance (ISR) triggered by plant growth‐promoting rhizobacteria remains unclear. Here, using Arabidopsis as a model system, one plant endogenous small RNA, miR472, was identified as an important regulator involved in the process of Bacillus cereus AR156 ISR against Pseudomonas syringae pv. tomato (Pst) DC3000. The results revealed that miR472 was down‐regulated with B. cereus AR156 treatment by comparing small RNA profiles and northern blot analysis of Arabidopsis with or without B. cereus AR156 treatment. Plants overexpressing miR472 showed higher susceptibility to Pst DC3000; by contrast, plant lines with miR472 knocked down/out showed the opposite. The transcriptome sequencing revealed thousands of differentially expressed genes in the transgenic plants. Target prediction showed that miR472 targets lots of coiled coil nucleotide‐binding site (NBS) and leucine‐rich repeat (LRR) type resistance genes and the expression of these targets was negatively correlated with the expression of miR472. In addition, transgenic plants with knocked‐out target genes exhibited decreased resistance to Pst DC3000 invasion. Quantitative reverse transcription PCR results indicated that target genes of miR472 were expressed during the process of B. cereus AR156‐triggered ISR. Taken together, our results demonstrate that the miR472‐mediated silencing pathway is an important regulatory checkpoint occurring via post‐transcriptional control of NBS‐LRR genes during B. cereus AR156‐triggered ISR in Arabidopsis.
Keywords: Arabidopsis, Bacillus cereus, induced systemic resistance, microRNA, plant innate immunity, Pseudomonas syringae pv. tomato
The plant miR472‐mediated silencing pathway is an important regulatory checkpoint occurring via post‐transcriptional control of nucleotide‐binding site and leucine‐rich repeat genes during Bacillus cereus AR156‐triggered induced systemic resistance in Arabidopsis.
Abbreviations
- CNL
CC‐NBS‐LRR
- DEG
differentially expressed gene
- ET
ethylene
- ETI
effector‐triggered immunity
- GO
gene ontology
- ISR
induced systemic resistance
- JA
jasmonic acid
- KEGG
Kyoto Encyclopaedia of Genes and Genomes enrichments
- MAMPS/PAMPs
microbial‐ or pathogen‐associated molecular patterns
- NLR
nucleotide‐binding leucine‐rich repeat receptor class
- PGPR
plant growth‐promoting rhizobacteria
- PRR
pattern recognition receptor
- PTI
PAMP‐triggered immunity
- R gene
plant disease resistance gene
- RLP/RLK
receptor‐like protein/kinase class
- RPKM
reads per kilobase of exon per million fragments
- SA
salicylic acid
- SAR
systemic acquired resistance
- TNL
TIR‐NBS‐LRR
1. INTRODUCTION
To survive in a complex and hostile soil environment, plants have evolved multiple types of inducible immune responses to attacks by pathogens (Niu et al., 2016). The plant immune system has two major branches. The first, called PAMP‐triggered immunity (PTI), uses transmembrane pattern recognition receptors (PRRs) or surface receptors, which encode transmembrane receptor‐like kinases. These kinases recognize conserved microbial‐ or pathogen‐associated molecular patterns (MAMPS or PAMPs) such as flagellin, glycoprotein, and chitin. The perception of PAMPs by PRRs triggers a cascade that includes activation of mitogen‐activated protein kinase (MAPK), oxidative burst and callose deposition, induction of defence‐related genes, and accumulation of antimicrobial compounds (Jones and Dangl, 2006; Schwessinger and Zipfel, 2008; Niu et al., 2016). Some pathogens that have successfully infected the host and caused disease can suppress PTI by secreting effectors. In turn, plants develop a second type of immune response, called effector‐triggered immunity (ETI). The ETI acts largely inside the plant cell via polymorphic nucleotide‐binding domain and leucine‐rich repeat (NB‐LRR) proteins, which are encoded by most plant disease resistance (R) genes (Dangl and Jones, 2001; Speth et al., 2007; Katiyar‐Agarwal and Jin, 2010; Kong et al., 2017). NB‐LRR‐mediated disease resistance is effective against obligate biotrophic and hemibiotrophic pathogens, which can only grow in living host tissue, but not against necrotrophic pathogens, which kill host tissue during colonization (Glazebrook, 2005; Jones and Dangl, 2006).
Depending on their protein structures, plant R proteins are divided into two major classes: receptor‐like protein/kinases (RLP/RLK) and nucleotide‐binding leucine‐rich repeat receptors (NLR) (Baker et al., 1997). RLP/RLK proteins are transmembrane immune receptors containing an N‐terminal LRR and a C‐terminal transmembrane domain with cytoplasmic kinase domain RLKs (Zhou and Yang, 2016). NLR class proteins contain an N‐terminal NB domain and C‐ terminal LRR domain. According to their N‐terminal structure, NLR proteins are divided into two subclasses: TIR‐NB‐LRRs (TNLs), which contain a toll/interleukin‐1 receptor (TIR) domain, and CC‐NB‐LRRs (CNLs), which contain a coiled‐coil domain (Austin et al., 2002). TNLs and CNLs are both intracellular immune receptors and require common signalling components. However, TNLs and CNLs possess differential genetic requirements when mediating ETI. For example, TNLs signal downstream events via EDS1/PAD4/ SAG101, while CNLs signal via nuclear Dbf2‐related kinase‐1 (Cui et al., 2015).
Small RNAs are usually 20–30 nucleotide (nt)‐long noncoding molecules, known as important regulators in plant defence against pathogens (Huang et al., 2016; Niu et al., 2016). In plants, small RNAs are divided into two major classes: small interfering RNAs (siRNA) and microRNAs (miRNA). siRNAs are generated from complementary long double‐stranded RNAs (dsRNAs) and examples consist of hc‐siRNAs, ta‐siRNAs, nat‐siRNAs, lsiRNAs, and so on (Katiyar‐Agarwal and Jin, 2010; Huang et al., 2016). miRNAs are usually 21–24 nt and are derived from RNAs with imperfectly base‐paired hairpin structures (Chen, 2009). To regulate gene expression, both miRNA and siRNA use post‐transcriptional gene silencing (PTGS) via an RNA‐induced silencing complex (RISC), which directly cleaves messenger RNAs (mRNAs) or inhibits their translation. siRNA and certain miRNAs can also regulate gene expression using transcriptional gene silencing (TGS) via DNA methylation, and histone or chromatin modification (Vaucheret, 2006; Wu et al., 2010; Cui and Cao, 2014). Numerous miRNA and siRNA are involved in plant defence against pathogens. For example, miR393, which is inducible by flg22 (a type of PAMP), was the first miRNA shown to be involved in plant innate immunity by suppressing the expression of auxin receptors (Navarro et al., 2006). nat‐siRNAATGB2, the first identified endogenous siRNA, can be specifically induced by Pseudomonas syringae pv. tomato (Pst) AvrRpt2 and positively regulates RPS2‐mediated ETI responses in Arabidopsis thaliana (Katiyar‐Agarwal et al., 2006).
Recently, miRNAs have been shown to negatively regulate the expression of plant R genes (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012). Genome‐wide analysis indicated that miRNAs can directly target numerous plant R genes. For instance, 171 out of 235 TNLs and 178 out of 290 CNLs are regulated by miRNAs in Glycine max (Zhao et al., 2015). Certain R genes with NBS‐LRR motifs could be targeted by miR482 and miR2118. In addition, further research indicated that some of these R genes with NBS‐LRR motifs were cleaved then subsequently generated series of secondary siRNAs in an RDR6‐dependent manner when they were targeted by these 22‐nt miRNAs. The newly generated secondary siRNAs can target mRNAs of several defence‐related proteins in tomato (Li et al., 2012). For example, in Arabidopsis, miR472 can modulate both PTI and ETI via post‐transcriptional control of CNL genes (Boccara et al., 2014). In tomato, miR398, miR482, and miR5300 can be significantly induced in resistant tomato cultivars on Fusarium oxysporum challenge. Ouyang and associates found that miR482/miR5300‐mediated regulation of R genes plays an important role in tomato resistance to pathogenic fungi (Ouyang et al., 2014).
In plants, local activation of PTI/ETI often triggers resistance in tissues at a site distal from the infection, which enhances resistance against a broad spectrum of pathogens (Pieterse et al., 2014; Niu et al., 2016). Examples of such induced resistance are systemic acquired resistance (SAR) and induced systemic resistance (ISR) (Jiang et al., 2016). SAR is usually triggered by exposure to virulent microbes, and is often associated with increased levels of the hormone salicylic acid (SA) and simultaneous activation of pathogenesis‐related (PR) genes such as PR1, PR2, and PR5 (Conrath et al., 2002; Niu et al., 2011, 2016; Pieterse et al., 2014). ISR is activated by nonpathogenic rhizobacteria, such as Pseudomonas and Bacillus; this mechanism is best characterized by plant growth‐promoting rhizobacteria (PGPR) (Choudhary et al., 2007; Van der Ent et al., 2009; Niu et al., 2011; Jiang et al., 2016). Distinguished from SAR, ISR normally depends on the jasmonic acid (JA)/ethylene (ET) signalling pathways. ISR is also accompanied by high expression of PR genes and plant defensin 1.2 (PDF1.2) (Van Oosten et al., 2008; Niu et al., 2011; Jiang et al., 2016). ISR, which has been demonstrated in numerous plant species such as Arabidopsis, tomato, and cucumber, can protect the host against a broad spectrum of pathogens (Van der Ent et al., 2009; Niu et al., 2011; Jiang et al., 2016). For example, the PGPR Bacillus cereus AR156 (hereafter AR156) triggers ISR by simultaneously activating SA and JA/ET signalling pathways in a manner dependent on the expression of NPR1 (Niu et al., 2011). Although ISR triggered by rhizobacteria has been shown in many plant species, few studies have examined the regulatory mechanisms. Jiang et al. identified two transcriptional factors (TFs), WRKY11 and WRKY70, that were identified as the key players in AR156‐triggered ISR to Pst DC3000 (Jiang et al., 2016). In addition to TFs, small RNAs are also involved in regulating systemic immune responses induced by rhizobacteria in plants. Using deep sequencing to analyse small RNA species in plants, Niu et al. demonstrated that miR825 and miR825* participate in modulation of AR156‐triggered ISR by negatively regulating numerous R genes; however, whether other small RNAs participate in AR156‐triggered ISR in plants remains undetermined.
In this study, we aimed to identify and characterize small RNAs that regulate systemic immune responses induced by AR156 in plants, therefore we analysed the previously obtained small RNA deep sequencing data. We found that expression of the small RNA miR472 in Arabidopsis was significantly down‐regulated by treatment with AR156. Furthermore, the expression of miR472 was significantly suppressed by AR156 upon infection with Pst DC3000. Pathogen assay results indicated that the transgenic plants overexpressing miR472 showed more susceptibility to Pst DC3000. In contrast, plants with knockdown and knockout of miR472 showed more resistance. In addition, compared with the wild type, the phenotypes of ISR triggered by AR156 shown on the above transgenic plants were also significantly affected and the ability of AR156 to trigger ISR was decreased significantly. The results of transcriptome sequencing also revealed hundreds of differentially expressed genes (DEGs) in the transgenic plants overexpressing miR472, and in plants with miR472 knockdown and knockout. We further identified five R genes, probably targeted by miR472, that were involved in the process of AR156‐triggered ISR. Taken together, our study shows that the small RNA miR472 participates in modulating AR156‐triggered ISR to Pst D3000 by negatively regulating several resistance‐related genes in plants.
2. RESULTS
2.1. Expression of miR472 is differentially regulated in B. cereus AR156‐triggered ISR in Arabidopsis
B. cereus AR156 could trigger ISR against Pst DC3000 (Niu et al., 2011); this finding agrees with our results, shown in Figure 1a,b. Transcription factors WRKY70 and WRKY11, and plant small RNAs miR825/825* serve as important regulators during this process (Jiang et al., 2016; Niu et al., 2016). To identify and characterize whether other small RNAs regulate AR156‐triggered ISR, we analysed the small RNA deep sequencing data obtained previously (Niu et al., 2016). We identified one small RNA, miR472, whose expression was significantly down‐regulated by combined treatment with AR156 and Pst DC3000, but only in Arabidopsis plants pretreated with AR156 (Figure 1c–e). The expression of miR472, assessed at 6 and 14 hr after challenge with Pst DC3000, was repressed at least 1.5‐fold in plants pretreated with AR156 compared with that pretreated with vehicle control (Figure 1c,d). Northern blotting also indicated that expression of miR472 was significantly down‐regulated in plants administered AR156/Pst DC3000 compared with that in plants administered control/Pst DC3000 (Figure 1f). These findings suggest that miR472 may play an important role in AR156‐triggered ISR.
FIGURE 1.
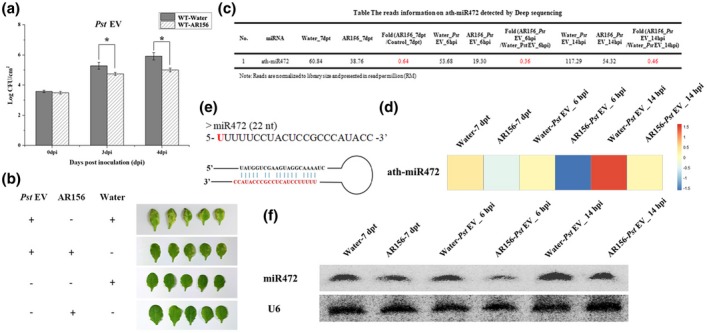
Plant endogenous small RNA miR472 is involved in regulation of Bacillus cereus AR156‐triggered induced systemic resistance (ISR) in Arabidopsis. (a) Induction of systemic resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in Arabidopsis thaliana ecotype Col‐0 wild‐type (WT) by AR156. The plants were pretreated with AR156 at 5 × 107 cfu/ml for 5 days and 0.85% NaCl. Subsequently, leaves were spray inoculated with Pst DC3000 (OD600 = 0.1). The concentration of Pst DC3000 in leaves and the effect of ISR were measured by the agar plate dilution method at 3 and 4 days post‐inoculation (dpi), respectively. (b) The phenotypic effects of AR156 ISR to Pst DC3000 in Arabidopsis. (c) The deep‐sequencing result of miR472 of AR156‐triggered ISR at different time points. The data represent reads per kilobase of exon per million fragments (RPKM) values of miR472 in different samples. (d) Heatmap showing the expression of miR472 in different samples. (e) Predicted secondary stem loop structure for miR472 with mature miR472 sequences indicated. (f) Expression of miR472 in different samples was detected by northern blotting. RNA blots were hybridized with an miR472 probe and U6 was used as a loading control
2.2. Transgenic Arabidopsis plants with overexpression or knockdown/out of miR472 show an altered defence response
To examine the role of miR472 in plant immunity, and to determine whether miR472 is required in AR156‐triggered ISR, we generated transgenic plants with overexpression or knockdown/out of miR472. We hypothesized that if miR472 indeed modulates AR156‐triggered ISR, enhancement or suppression of miR472 expression would result in an altered defence response. The schematic diagram for our transgenic plants, successfully modified to overexpress miR472 using the cauliflower mosaic virus (CaMV) 35S promoter, is shown in Figure 2a. miR472 overexpression had no major effects on the development of transgenic lines, but slightly altered growth was observed in transgenic plants (Figure 2b). Northern blotting showed that miR472 was expressed at high levels in the homozygous miR472‐overexpression (OE) lines (1#, 7#) (Figure 2c), which were subsequently used to analyse disease resistance and ISR. As expected, the miR472‐OE line was more susceptible to infection with Pst DC3000. Bacterial growth of Pst DC3000 in Arabidopsis leaves was determined at 0, 3, and 4 days post‐inoculation (dpi). The miR472‐OE lines (1#, 7#) showed a significantly higher growth of Pst DC3000 compared with that in wild‐type plants (Figure 2d). miR472 has previously been shown to negatively regulate PTI and ETI (Boccara et al., 2014), therefore Pst DC3000 (AvrPphB) (Pst DC3000 expressing AvrPphB, which activates a strong ETI) and Pst DC3000 (hrcC) (type III secretion system [TTSS] mutant of Pst DC3000, which activates PTI) were used to test resistance to disease in Arabidopsis. The growth of Pst DC3000 (AvrPphB) and Pst DC3000 (hrcC) was determined at 0, 3, and 4 dpi. Our results indicate that Pst DC3000 (AvrPphB) and Pst DC3000 (hrcC) showed high levels of proliferation in the miR472‐OE lines (1#, 7#) (Figure 2e,f). These results agree with those of previous studies (Boccara et al., 2014), and indicate that miR472 may extensively participate in PTI and ETI in Arabidopsis. We then evaluated the effects of miR472 on AR156‐induced systemic resistance in Arabidopsis. Compared with their respective controls, wild‐type Col‐0 plants pretreated with AR156 showed a significant (p < .05) reduction in disease severity, while the response of the miR472‐OE lines (1#, 7#) did not show a statistically significant difference (Figure 2g). These results indicate that miR472 may be involved in AR156‐triggered ISR.
FIGURE 2.
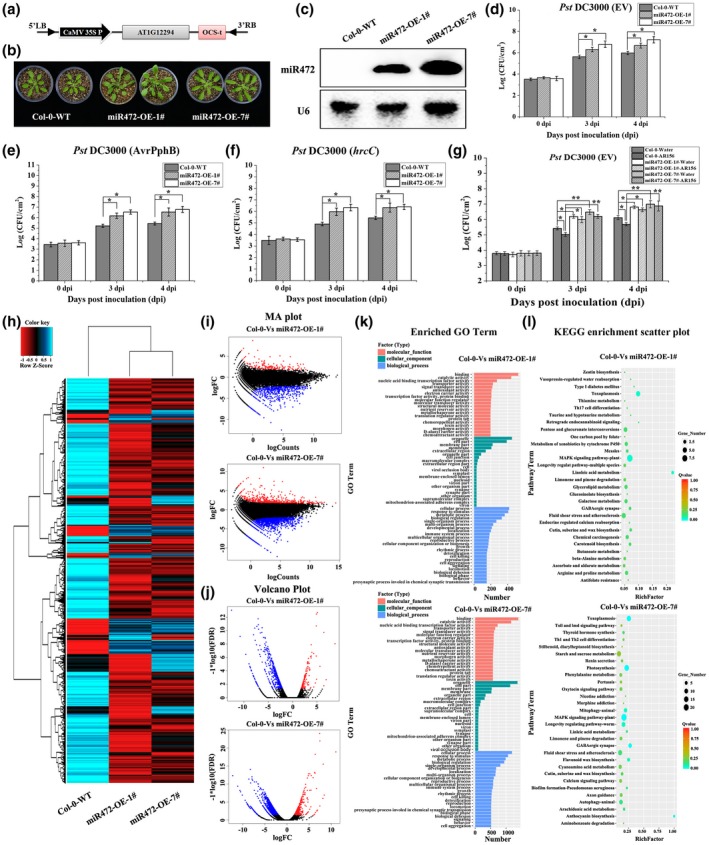
The miR472 overexpression plants are more susceptible to virulent Pseudomonas syringae pv. tomato (Pst) DC3000. The pathogen assay and induced systemic resistance (ISR) triggered by Bacillus cereus AR156 in Arabidopsis thaliana ecotype Col‐0 wild‐type (WT) and miR472 overexpression lines (miR472‐OE‐1# and miR472‐OE‐7#) were done as described in the materials and methods section. (a) Schematic diagram of miR472 overexpression constructs used to generate the transgenic plants. (b) Phenotype of miR472 overexpression transgenic plant (line 7#) was compared to ecotype Col‐0 WT, and the images show the 4‐week‐old seedlings. (c). Expression of miR472 in the overexpression lines (miR472‐OE‐1# and miR472‐OE‐7#) by northern blotting with an miR472 probe; U6 was used as a loading control. (d)–(f) Bacterial growth in 5‐week‐old plants from Col‐0 WT, miR472‐OE‐1#, and miR472‐OE‐7# infiltrated with Pst DC3000 (empty vector, EV) (OD600 = 0.0001), Pst DC3000 (AvrPphB) (OD600 = 0.0005), and Pst DC3000 (hrcC) (OD600 = 0.0001), respectively. (g) Bacterial growth assay of Pst DC3000 (EV) in Col‐0 WT, miR472‐OE‐1#, and miR472‐OE‐7# pretreated with AR156 or 0.85% NaCl. For (d)–(g), the data are means and SD (n = 24). The LSD test was performed to determine the significant differences between miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT. * and ** indicate statistically significant differences at p < .05 and p < .01, respectively. In this part, the experiments were done in three independent biological replicates and similar results were obtained. (h) Cluster analysis of differentially expressed genes (DEGs) in miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT on the expression profiles measured by RNA‐Seq. (i) Manhattan (MA) plot of the DEGs between miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT. (j) Volcano plot of the DEGs between miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT. (k) GO classifications of DEGs across one comparison between miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT. The x axis represents the number of DEGs in a category. The results of miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) versus Col‐0 WT are summarized in three main categories: molecular function (red), cellular component (green), and biological process (blue). (l) KEGG of the annotated DEGs across one comparison between miR472 OE lines (miR472‐OE‐1# and miR472‐OE‐7#) and Col‐0 WT. The y axis indicates the KEGG pathway. The x axis indicates the Rich factor. High q values are shown in red and low q values are shown in green
To further investigate the role of miR472 in AR156‐triggered ISR, we generated transgenic lines of Arabidopsis bearing a knockdown or knockout of miR472 (Figure 3a). As shown in Figure 3b, the stature of the two types of transgenic plants was smaller than that of the wild‐type plant. Moreover, the expression of miR472, assessed using northern blotting, was significantly decreased in the transgenic line STTM472‐1# and T‐DNA insertion line SALK_087945‐1# compared with that in the wild‐type control (Figure 3c). The mutants were also assessed for disease resistance and ISR. As expected, the STTM472‐1# and SALK_087945‐1# displayed enhanced resistance to Pst DC3000, Pst DC3000 (AvrPphB), and Pst DC3000 (hrcC) compared with that observed in wild‐type plants. The density of bacterial cells on the leaves of the two mutant plant lines was decreased significantly (p < .05) compared with that of the wild type (Figure 3d–f). These results indicate that miR472 can act as a negative regulator of disease resistance in plants. We also examined how AR156 affected pathogen proliferation on the leaves of plants with bearing a knockdown or knockout of miR472. The leaves of wild‐type Col‐0 plants pretreated with AR156 and inoculated with Pst DC3000 showed significantly decreased (p < .01) pathogen density at 3 and 4 dpi compared with that of respective controls inoculated with Pst DC3000 only; by contrast, the density of Pst DC3000 on the leaves of STTM472‐1# and SALK_087945‐1# remained constant irrespective of pretreatment with AR156 (Figure 3g). These results indicate that AR156‐triggered ISR was abolished in miR472 mutant lines.
FIGURE 3.
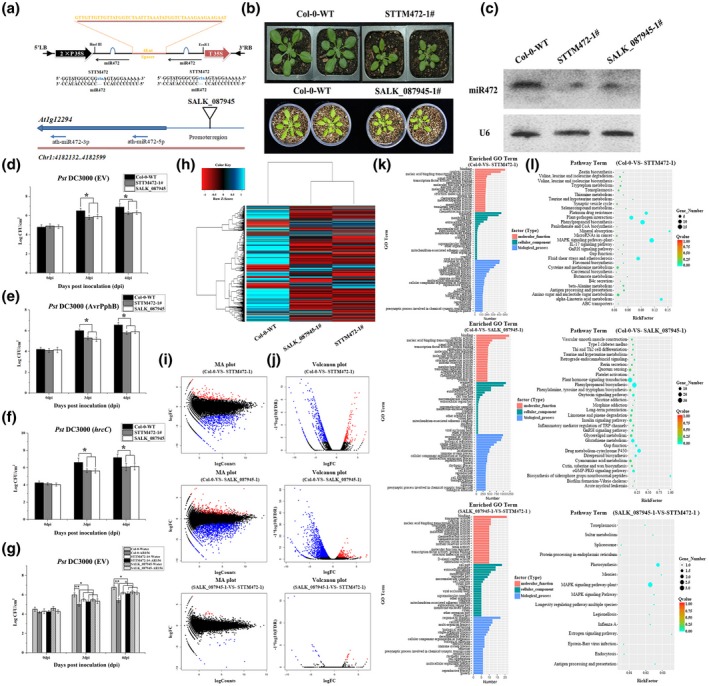
The miR472 knockdown and knockout showed more resistance to virulent Pseudomonas syringae pv. tomato (Pst) DC3000. The pathogen assay and induced systemic resistance (ISR) triggered by Bacillus cereus AR156 in Arabidopsis ecotype Col‐0 wild‐type (WT), miR472 knockdown lines (STTM472‐1#), and knockout lines (SALK_087945) were as done described in the materials and methods section. (a) Schematic diagram of the miR472 coding gene At1g12294 structure and the STTM472 structure showing the design strategy. The STTM472 construct with miR472 binding sites on flanking sides separated by a 48‐nucleotide spacer (orange) forming an imperfect weak stem‐loop. The trinucleotide bulge sequences in the miRNA binding sites are shown in blue; the complementary miR472 is shown in black. nt, nucleotides; 2 × P35S, enhanced 35S promoter; T35S, terminator. For the miR472 coding gene structure, the triangle indicates the T‐DNA insertion position. (b) Phenotype of miR472 knockdown lines (STTM472‐1#) and knockout lines (SALK_087945) were compared to ecotype Col‐0 WT, and the images show the 4‐week‐old seedlings. (c) Expression of miR472 in the miR472 knockdown lines (STTM472‐1#) and knockout lines (SALK_087945) by northern blotting with an miR472 probe; U6 was used as a loading control. (d)–(f) Bacterial growth in 5‐week‐old plants from miR472 knockdown lines (STTM472‐1#), knockout lines (SALK_087945), and Col‐0 WT infiltrated with Pst DC3000 (EV) (OD600 = 0.0001), Pst DC3000 (AvrPphB) (OD600 = 0.0005), and Pst DC3000 (hrcC) (OD600 = 0.0001), respectively. (g) Bacterial growth assay of Pst DC3000 (EV) in miR472 knockdown lines (STTM472‐1#), knockout lines (SALK_087945), and Col‐0 WT pretreated with AR156 or 0.85% NaCl. For (d)–(g) the data are means and SD (n = 24). The LSD test was performed to determine the significant differences between miR472 knockdown/out lines and Col‐0 WT. * and ** indicate statistically significant differences at p < .05 and p < .01, respectively. In this part, the experiments were done in three independent biological replicates and similar results were obtained. (h) Cluster analysis of differentially expressed genes (DEGs) in miR472 knockdown/out lines and Col‐0 WT on the expression profiles measured by RNA‐Seq. (i) Manhattan (MA) plot of the DEGs between the miR472 knockdown/out lines and Col‐0 WT. (j) Volcano plot of the DEGs between the miR472 knockdown/out lines and Col‐0 WT. (k) GO classifications of DEGs across one comparison between the miR472 knockdown/out lines and Col‐0 WT. The x axis represents the number of DEGs in a category. The results of miR472 knockdown/out lines versus Col‐0 WT are summarized in three main categories: molecular function (red), cellular component (green), and biological process (blue). (l) KEGG of the annotated DEGs across one comparison between the miR472 knockdown/out lines and Col‐0 WT. The y axis indicates the KEGG pathway. The x axis indicates the Rich factor. High q values are shown in red and low q values are shown in green
Next, we employed a comparative transcriptomic analysis using the different miR472 transgenic lines miR472‐OE‐1#, miR472‐OE‐7#, STTM472‐1#, SALK_087945‐1#, and wild‐type plant Col‐0 to examine the function of miR472 in plant defence against pathogenic invasion. The libraries constructed using different types of Arabidopsis were sequenced using a HiSeq 2500 platform in BGI. Approximately 27.1 Gb of clean reads was obtained after rigorous quality checking. As shown in Tables S5 and S6, the Q20 percentage of each library reached 96%, the Q30 percentage of each library was in the range 91.0%–92.2%, the clean reads ratio percentage of each library reached 99%, and the total mapping ratio was in the range 94.5%–95.1% (Tables S5 and S6). We then compared RNA‐Seq data obtained using transgenic lines and wild‐type plants, with each assessment being performed four times (Figure S2a). Our results show that increases and decreases in the expression level of miR472 induced changes in the expression of numerous plant genes (Table S7 and Figure S1); 2,118 DEGs showed significant changes in expression (1,521 down‐regulated and 597 up‐regulated) when we compared expression profiles of wild‐type Col‐0 and transgenic miR472‐OE‐1# and miR472‐OE‐7#. Additionally, 1,855 (1,464 down‐regulated and 391 up‐regulated) and 732 (561 down‐regulated and 171 up‐regulated) DEGs with significant changes in expression levels were identified when we compared expression profiles among wild‐type Col‐0, STTM472‐1#, and SALK_087945‐1# (Figure S1a). The fold changes in the expression of these DEGs are shown in a heat map in Figures 2h and 3h. The Manhattan and volcano plots also show the relationship between fold‐changes and miR472 overexpression in plant lines with a knockdown/out of miR472 and wild‐type plants (Figures 2i,j and 3i,j). In summary, these results indicate that differential expression of miR472 altered the expression of numerous plant genes. Moreover, analysis of the DEGs in miR472‐OE‐1# versus Col‐0, miR472‐OE‐7# versus Col‐0, STTM472‐1# versus Col‐0, and SALK_087945‐1# versus Col‐0, performed using a Venn diagram, identified 503 overlapping genes among the three groups (Figure S1b). This result indicates that these genes may be associated with miR472 regulation and expressed specifically during miR472‐regulated immune response. Otherwise, to gain insight into the potential biological processes associated with miR472 regulation process in which DEGs were involved, we performed analysis of GO and KEGG pathways. Comparison of Col‐0 and miR472‐OE lines showed that showed that 22 GO terms corresponded with “cellular component”, 20 with “molecular function”, and 93 with “biological process”. Moreover, the terms “organelle” and “cell part” were most enriched in the term “cellular component”. “Binding” was the most frequent term in “molecular function” followed by “catalytic activity” and “nucleic acid binding transcription” (Figure 2k). Furthermore, in comparison with Col‐0 and STTM472‐1# or SALK_087945‐1#, respectively, in total, 141 and 240 GO terms were assigned and significantly enriched (p < .05) to the DEGs, respectively. For Col‐0 versus STTM472‐1#, 12 GO terms corresponded to “cellular component”, 22 GO terms to “molecular function”, and 107 GO terms to “biological process”; for Col‐0 versus SALK_087945‐1#, 18 GO terms corresponded to “cellular component”, 66 GO terms to “molecular function”, and 156 GO terms to “biological process” (Figure 3k). These results indicate that miR472 could change the physiological and biochemical functions in Arabidopsis. We next conducted an analysis of KEGG pathways assigned to DEGs, and our results show that the top 30 pathways were significantly enriched (p < .05) (Figures 2l and 3l). Among these pathways, the MAPK signalling pathway, plant hormone signalling transduction pathway, and several others were related to disease resistance. These results confirm that miR472 is involved in plant defence response.
2.3. miR472 alters defence response to Pst DC3000 by targeting a group of plant resistance genes in Arabidopsis
miRNAs regulate post‐transcriptional gene expression by mRNA degradation and/or translational repression, therefore identification of miRNAs targets is important to determine the mechanisms involved in miRNA activity. As previously shown, miR472 targets dozens of CC‐NB‐LRRs (CNLs) to initiate the production of RDR6‐dependent secondary siRNAs (Boccara et al., 2014). Therefore, we next aimed to find and validate more targets of miR472, and to determine which of these targets are involved in AR156‐triggered ISR. In total, 37 targets were predicted by the plant small RNA target analysis server, psRNATarget; 26 of these targets were CNLs (Table S8). In Figure 4a, the large red box indicates the predicted target region of miR472 in CNLs. Moreover, phylogenetic analyses based on amino acid sequences of the 26 predicted CNLs indicated the presence of three groups (Figure 4b). Subsequently, we analysed the differential expression of 26 predicted targets, as well as RNA‐Seq data. As shown in Figure 4c and Table S9, 20 CNLs were down‐regulated in miR472‐OE‐1# and miR472‐OE‐7#, and seven CNLs were up‐regulated in both STTM472‐1# and SALK_087945‐1# compared with the profile of wild‐type Col‐0 plants. Finally, five CNLs identified as possible targets of miR472 were identified as At1g15890, At1g61300, At1g12220, At5g43730, and At5g43740. To confirm the transcriptomic data, expression of these five CNLs was analysed by quantitative reverse transcription PCR (RT‐qPCR), which indicated that the patterns of gene expression in these five CNLs were consistent with those shown by RNA‐Seq (Figure S2). To confirm that these five genes were indeed true targets of miR472, we performed Agrobacterium‐mediated transient co‐expression in Nicotiana benthamiana to express green fluorescent protein (GFP):CNL fusion protein alone and co‐expressing miR472. The results of western blotting indicate that co‐expression of miR472 with the five CNLs significantly decreased the mRNA levels of the CNLs (Figure 4d), which was not observed in the case of the GFP:CNLs fusion protein. Confocal imaging of N. benthamiana epidermal cells revealed that accumulation of CNLs in the cells was suppressed via co‐expression with miR472 (Figure 5a). Moreover, analysis of fluorescence intensity showed that co‐expression of miR472 with the five CNLs significantly decreased accumulation of GFP compared with the levels of respective controls (Figure 5b). We then performed Agrobacterium‐mediated transient co‐expression assays to express a β‐glucuronidase (GUS):CNL fusion protein alone and in co‐expression with miR472 in N. benthamiana. GUS histochemical staining was used to verify the results. As shown in Figure S3, the results of GUS histochemical staining indicated that GUS expression decreased significantly in the leaves of plants in which miR472 was co‐expressed with the five CNLs (Figure S3). So far, these findings indicate that these five CNLs are the true targets of miR472.
FIGURE 4.
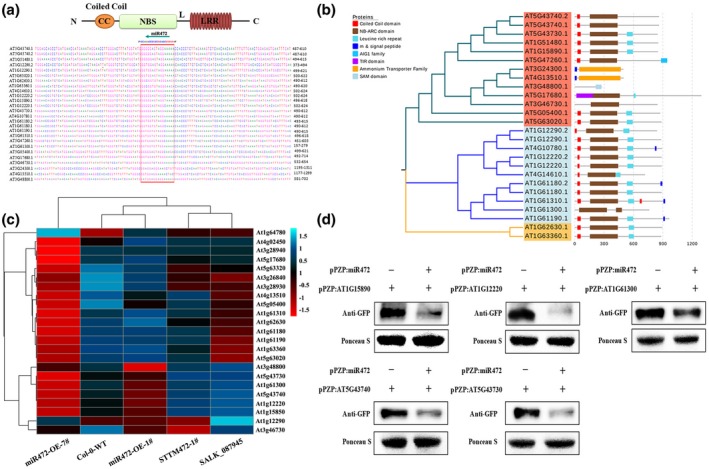
The prediction and validation of miR472 target genes. (a) The predicted information of miR472 target genes. Nucleotide sequences targeted by miR472 were aligned to corresponding genes, with the CC‐NBS‐LRR gene structure depicted at the top. CC, coiled‐coil domain; NBS, nucleotide‐binding site domain; LRR, leucine‐rich repeat domain. Red boxes on gene sequences represent the target binding site by miR472. (b) Phylogenetic tree of predicted targets based on the amino acid sequence. Sequences were aligned using ClustalX, and the tree was generated using the online software EvolView v. 3 (Subramanian et al., 2019). (c) Cluster analysis of predicted miR472 target genes in miR472 overexpression lines, knockdown/out lines and Arabidopsis thaliana Col‐0 wild‐type (WT) on the expression profiles measured by RNA‐Seq. (d) Expression levels of miR472 targets fused with green fluorescent protein (GFP) in Nicotiana benthamiana by western blotting with a commercial GFP antibody that crossreacts with GFP tag. Ponceau S staining was used as protein loading control. The experiments were done in three independent biological replicates and similar results were obtained
FIGURE 5.
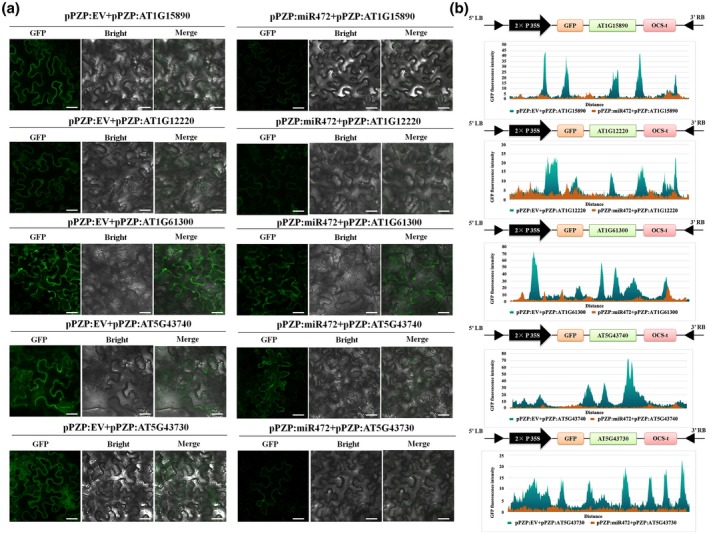
Five targets of miR472 were certificated by green fluorescent protein (GFP) fluorescence fusion labelling in vitro. Confocal imaging of Nicotiana benthamiana epidermal cells expressing miR472 and its targets. (a) Epidermal cells of N. benthamiana leaves transiently expressing miR472 target coding genes alone, when fused with a GFP tag or co‐expressing with miR472 were observed using confocal microscopy and the photographs were taken 48 hr after infiltration. Scale bars = 20 μm. (b) The expression levels of target genes were quantified through detecting the fluorescence intensity. The values on the y axis indicate the relative intensity of the GFP fluorescence signal. The experiments were done in three independent biological replicates and similar results were obtained
We evaluated the role of these five CNLs in plant defence response to Pst DC3000. For this, we generated transgenic lines bearing a knockout of these five CNLs and used these lines for a pathogen challenge assay. The five T‐DNA insertion mutant lines, obtained from TAIR, were designated as SALK_060360C, SALK_061926C, SALK_015294C, SALK_025605C, and SALK_013860C, and contained T‐DNA insertions in At1g15890, At1g61300, At1g12220, At5g43740, and At5g43730, respectively. On challenge with Pst DC3000, all five CNL lines exhibited significantly higher bacterial growth (Figure 6a). Compared with Col‐0 wild‐type plants, the five CNL lines showed increased susceptibility to disease, which was manifested as stronger yellowing or chlorosis in leaves (Figure 6b). These results indicate that the five identified targets of miR472 positively regulate disease resistance, thereby further clarifying the role of miR472 in regulating the defence response.
FIGURE 6.
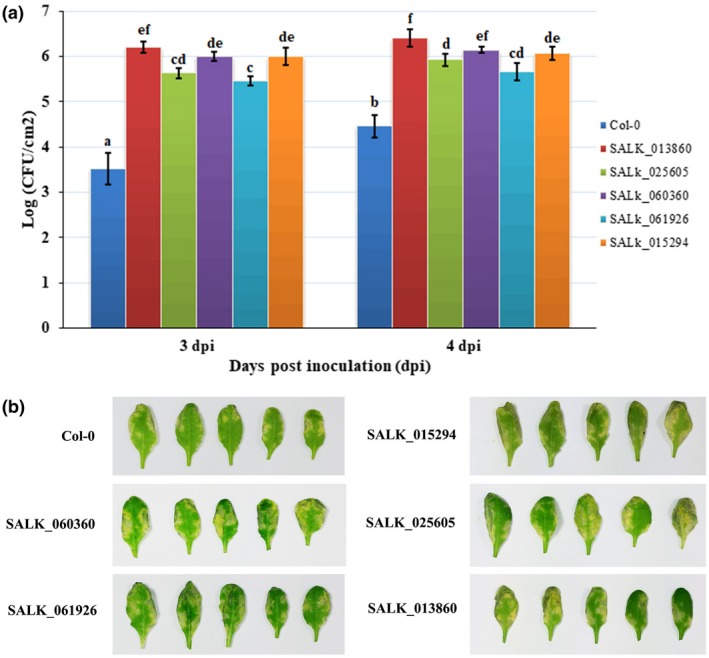
Reduced basal resistance against virulent Pseudomonas syringae pv. tomato (Pst) DC3000 observed in the miR472 target genes mutant lines. (a) Bacterial growth in 5‐week‐old Arabidopsis thaliana plants from T‐DNA insert lines of miR472 target genes and Col‐0 wild‐type (WT) infiltrated with Pst DC3000 (EV) (OD600 = 0.0001), the T‐DNA insert lines being SALK_060360C (At1g15890), SALK_061926C (At1g61300), SALK_015294C (At1g12220), SALK_025605C (At5g43740), and SALK_013860C (At5g43730). The data are means and SD (n = 24). The LSD test was performed to determine the significant differences between T‐DNA insert lines and Col‐0 wild‐type (WT). Letters above the bars indicate statistically significant differences between treatments (LSD test, p < .05). (b) Disease symptoms caused by Pst DC3000 (empty vector, EV) infection in different T‐DNA insert lines and Col‐0 WT plants. A representative plant from each treatment was photographed at 5 days post‐inoculation. All the experiments were done in three independent biological replicates and similar results were obtained
2.4. Target genes of miR472 are coordinately expressed with B. cereus AR156‐triggered ISR
Subsequently, we examined whether miR472 may modulate AR156‐triggered ISR by regulating its five putative CNL targets. For this we used RT‐qPCR to analyse the expression levels of these five CNLs during AR156‐triggered ISR. Our results show that all five targets showed significantly altered expression levels in AR156‐treated Arabidopsis compared with mock‐treated controls; this expression pattern was not dependent on inoculation with Pst DC3000. As shown in Figure 7, the expression of At1g15890, At1g61300, At1g12220, At5g43740, and At5g43730 was up‐regulated in the leaves of plants treated only with AR156 for 3 and 5 days compared with that of plants administered mock treatment. These target genes were also up‐regulated in plants pretreated with AR156 and then challenged with Pst DC3000. Increases in the expression of these genes were significantly greater compared with those in plants treated only with Pst DC3000 (Figure 7). In summary, our results indicate that the target genes of miR472 were coordinately expressed and involved in the modulation of AR156‐triggered ISR against the invasion of Pst DC3000.
FIGURE 7.
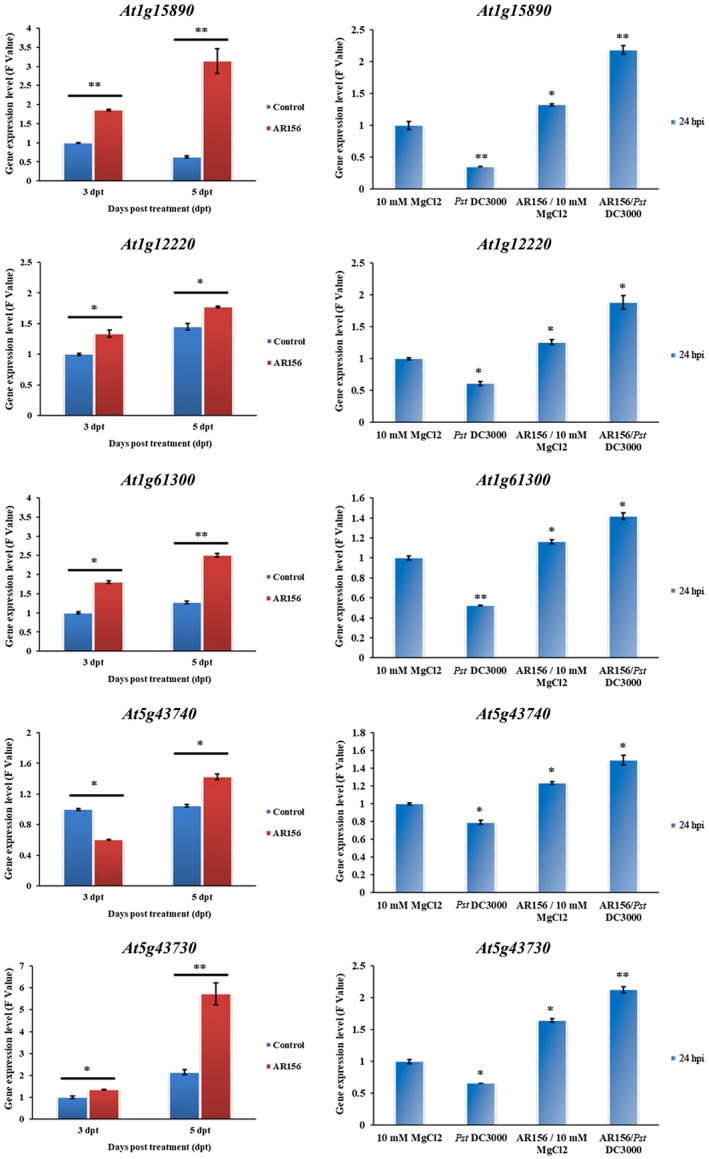
Target genes of miR472 were coordinately expressed with Bacillus cereus AR156‐triggered induce systemic resistance (ISR). The expression profiles of miR472 target genes At1g15890, At1g12220, At1g61300, At5g43740, and At5g43730 in Arabidopsis thaliana seedlings with AR156‐triggered ISR were detected. The plants were pretreated with AR156 at 5 × 107 cfu/ml for 5 days and 0.85% NaCl. Subsequently, leaves were spray inoculated with Pseudomonas syringae pv. tomato (Pst) DC3000 (empty vector, EV) (OD600 = 0.1). The leaves in different treatments were harvested at different time points and total RNA was extracted (dpt, days post‐treatment; hpi, hours post‐inoculation). Expressions of miR472 target genes were examined by quantitative reverse transcription PCR. The expression values of the individual genes were normalized using β‐Tubulin 4 as an internal standard. The data are means and SD (n = 24). The LSD test was performed to determine the significant differences between T‐DNA insert lines and A. thaliana Col‐0 wild‐type. * and ** indicate statistically significant differences at p < .05 and p < .01, respectively. The experiments were done in three independent biological replicates and similar results were obtained
3. DISCUSSION
3.1. Plant endogenous small RNA miR472 plays an essential role in B. cereus AR156‐triggered ISR
ISR triggered by PGPRs is an important mechanism in broad‐spectrum biocontrol against various pathogens. The molecular mechanisms underlying ISR have been studied extensively in various plant species such as A. thaliana. Many signalling components and molecular processes are involved in the interactions between ISR elicitors and the host (Pieterse et al., 2014). For instance, B. cereus AR156 can trigger ISR by simultaneously activating the SA and JA/ET signalling pathways in an NPR1‐dependent manner and the transcription factors WRKY70 and WRKY11 were identified as “biological switches” involved in the regulation of these processes (Niu et al., 2011; Jiang et al., 2016). Furthermore, the regulatory mechanisms involved in PGPR‐triggered ISR are gradually being revealed. Small RNAs in plants are important regulators of plant defence against pathogens (Huang et al., 2016; Niu et al., 2016). miRNA can negatively regulate the expression of plant R genes (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012). The expression of miR398, miR482, and miR5300 is increased by targeting a group of R genes via challenge with F. oxysporum in resistant tomato cultivars (Ouyang et al., 2014). As for small RNAs involving in regulating PGPR‐triggered ISR, we reported that expression of the miR825/825* pair was significantly down‐regulated by infection with Pst DC3000 in plants pretreated with AR156 (Niu et al., 2016). We used small RNA deep sequencing data obtained previously (Niu et al., 2016) to seek more small plant RNAs participating in AR156‐triggered ISR systemically, and to further explore how they regulate the process of AR156‐triggered ISR in this study. For this, we evaluated the differential expression levels and function of target genes such as transcription factors and R genes. Our results indicate that the expression of miR472 was significantly down‐regulated by pretreatment with AR156 and challenged with Pst DC3000, while it was slightly induced by Pst DC3000 infection only (Figure 1c–e). Northern blotting confirmed that accumulation of miR472 in Arabidopsis was modulated by B. cereus AR156 (Figure 1f). The effect of miR472 on AR156‐triggered ISR was also analysed using plant lines with miR472 knockdown and knockout. As shown in Figure 2g, treatment with AR156 significantly reduced (p < .05) the density of Pst DC3000 in the leaves of wild‐type Col‐0 at 3 and 4 dpi compared with that on the leaves of respective controls. However, AR156‐induced reduction of Pst DC3000 density in the leaves of the miR472‐OE lines (1#, 7#) was not significant (p < .05) (Figure 2g). Conversely, the density of Pst DC3000 in the leaves of miR472 mutant lines STTM472‐1# and SALK_087945‐1# remained constant with or without AR156 treatment (Figure 3g). These findings indicate that miR472 plays an essential role in AR156‐triggered ISR. In future studies we will investigate whether this miR472‐mediated pathway represents a new mechanism in ISR regulation.
3.2. Arabidopsis miR472 negatively regulates PTI and ETI responses to the infection of Pst DC3000
miR472‐mediated post‐transcriptional regulation was found to be involved in plant defensive responses against Pst DC3000. Boccara et al. discovered that miR472 could target several NBS‐LRR genes in an RDR6/DCL4‐dependent manner. The targeting of mRNA by 22‐nt miRNAs was assisted by a cascade of secondary siRNAs that were generated in an RDR6‐dependent manner (Boccara et al., 2014). In our present study, it was found that the miR472‐OE lines (1#, 7#) were more susceptible to infection with Pst DC3000, while plants with knockdown and knockout of miR472 were more resistant to this pathogen. The miR472‐OE lines (1#, 7#) displayed a significantly higher growth of Pst DC3000 compared with that of wild‐type plants; however, the growth of Pst DC3000 declined sharply in miR472 mutant lines STTM472‐1# and SALK_087945‐1# (Figures 2d and 3d). miR472 negatively regulated PTI responses to the infection of Pst DC3000. Boccara et al. found that the induction of RSG1and RPS5 mRNA via flg22 was significantly impaired in miR472‐OE lines (1#, 7#) compared with that in wild‐type controls. However, production of reactive oxygen species triggered by flg22 remained normal, while callose deposition was reduced in miR472‐OE lines (1#, 7#) compared with that in wild‐type controls (Boccara et al., 2014). In our present study, the miR472 transgenic plants were inoculated with Pst DC3000 (hrcC), and the growth of Pst DC3000 (hrcC) was then determined in Arabidopsis leaves at 0, 3, and 4 dpi. We found that Pst DC3000 (hrcC) showed high growth levels in the miR472‐OE lines (1#, 7#), but low growth levels in the miR472 mutant lines STTM472‐1# and SALK_087945‐1#, compared with that in wild‐type control (Figures 2f and 3f). These results indicate that miR472 is involved in the regulation of the PTI process.
Plants have evolved an extensive repertoire of immune receptors, and R proteins monitor vital host proteins for signs of pathogenic activity. miR472 and RDR6‐dependent secondary siRNAs are involved in RPS5‐mediated resistance (Simonich and Innes, 1995; Dangl and Jones, 2001; Boccara et al., 2014). In our present study, we also observed that Pst DC3000 (AvrPphB) multiplied to high levels in the miR472‐OE lines (1#, 7#), but remained at low levels in the miR472 mutant lines STTM472‐1# and SALK_087945‐1# compared with the levels observed in the wild‐type plant (Figures 2e and 3e). Taken together, we proved that miR472 is involved in both ETI and PTI via the use of mutants of Pst DC3000 that activate different branches of plant immune system. Although these results were consistent with those obtained in previous studies (Boccara et al., 2014), our approach and methodology differed.
3.3. An altered expression level of miR472 activates a large number of DEGs
Transcriptome sequencing is a widely used, highly efficient, molecular biological research method used to obtain detailed transcriptome information (Chen et al., 2017; Jiang et al., 2019). To further detail how miR472 regulates plant disease resistance, we used the transgenic plants miR472‐OE‐1#, miR472‐OE‐7#, STTM472‐1#, SALK_087945‐1#, and wild‐type Col‐0 for transcriptome sequencing. In this study, the results inform us that an altered expression level of miR472, regardless of whether it was enhanced or decreased, activated a large number of DEGs (Table S7 and Figure S1). Thousands of DEGs were identified in miR472 transgenic plants compared with the profile of wild‐type plant Col‐0 (Figures 2h–j and 3h–j). However, only a few DEGs were identified in data generated using STTM472‐1# and SALK_087945‐1# (Figure 3h–j). The comparison of miR472 knockdown line STTM472‐1# and knockout line SALK_087945‐1# indicates that miR472 mutants were generated successfully and were a reliable model for analysis using transcriptome sequencing. MAPKs are involved in plant growth, development, and interactions between plants and pathogens (Yu and Tang, 2004; Bi and Zhou, 2017). MAPKs also play important roles in plant response to pathogen infections, and many pathogens secrete effectors that can inhibit the MAPK cascade (Bi and Zhou, 2017). In our present study, we also analysed the GO and KEGG pathways, and found that miR472 was involved in the MAPK signalling pathway that responds to plant pathogens (Figures 2k,l and 3k,l). Here, by using transcriptome analysis techniques, we discovered that endogenous miR472 plays an important role in PTI mediated by the MAPK cascade.
3.4. miR472 modulates the defence response to Pst DC3000 by targeting a group of CNLs in Arabidopsis
miRNAs can regulate the plant defence response by targeting numerous R genes. In our study, 37 genes were identified as targets of miR472 via bioinformatics prediction. Twenty‐six of these genes were CNLs and included the previously reported targets At1g12220, At1g51480, and At5g43730 (Table S8; Boccara et al., 2014). Subsequently, the five CNLs At1g15890, At1g61300, At1g12220, At5g43730, and At5g43740 were identified as possible targets of miR472 via RNA‐Seq and RT‐qPCR validation (Figures 4c and S2, and Table S9). To confirm the five identified targets, we also performed Agrobacterium‐mediated transient co‐expression assays. Using western blotting, GFP fluorescence, and GUS histochemical staining, we found that miR472 can inhibit the accumulation of proteins encoded by these five target genes (Figures 4d, 5, and S3). To clarify the role of these five CNLs in the defence response to Pst DC3000, we generated transgenic lines bearing a knockout of these five CNLs and used them for a pathogen challenge with Pst DC3000. Following infection with Pst DC3000, all five CNL T‐DNA insertion lines showed significantly higher pathogenic bacterial growth (Figure 6a). Compared with that of wild‐type Col‐0 plants, all five CNL mutants showed increased susceptibility to disease, manifested as stronger yellowing or chlorosis of the leaves (Figure 6b). These results show that the five identified targets of miR472 positively regulated disease resistance in Arabidopsis. We also identified four new CNL genes that were targeted by miR472 and could mediate plant defence response. Our results confirm that miR472‐mediated plant defence depends not only on RPS5‐mediated ETI, but also on other CNLs. This finding also complements previous studies reported by Boccara et al. (2014).
3.5. B. cereus AR156‐triggered ISR against Pst DC3000 depends on CNL‐mediated basal immunity in Arabidopsis
Studies examining the molecular mechanisms of ISR have mainly focused on the roles of the crucial signalling pathways and key regulated proteins. Pieterse et al. demonstrated that WCS417r‐triggered ISR was dependent on the JA/ET signalling pathway and NPR1 in Arabidopsis (Pieterse et al., 1996). In our previous study, we found that AR156 triggered ISR by simultaneously activating the SA and JA/ET signalling pathways in a manner dependent on the expression of the NPR1 gene, and the transcription factors WRKY70 and WRKY11 were identified as “biological switches” involved in the regulation of these processes (Niu et al., 2011; Jiang et al., 2016). In this study, we wanted to examine whether miR472 is also involved in AR156‐triggered ISR. For this, we used RT‐qPCR to analyse the expression of miR472 targets during the process of AR156‐triggered ISR. Our results revealed that all of the targets showed significantly altered expression levels in AR156‐treated Arabidopsis compared with those in mock‐treated seedlings; this pattern of expression was not dependent on inoculation with Pst DC3000. The target genes of miR472 were up‐regulated in plants pretreated with AR156 and challenged with Pst DC3000. Increases in the expression of these genes were significantly greater than those in plants treated with Pst DC3000 alone (Figure 7). These findings indicate that miR472 performs a regulatory function in AR156‐triggered ISR by targeting of the expression levels of CNL genes involved in disease resistance. However, how these CNLs mediate plant immunity and ISR remains to be detailed in our future studies.
3.6. A proposed model for miR472 functions in regulating AR156‐triggered ISR
In summary, we identified miR472 as an important regulator of AR156‐triggered ISR. Arabidopsis roots and leaves were used to examine the regulatory mechanisms of endogenous small RNA in AR156‐triggered ISR (Figure 8). In Arabidopsis, MAMPs of B. cereus AR156 are recognized via their respective receptors, leading to the generation and release of a distinct ISR signal in the roots, which is then transferred to the leaves. To trigger ISR, AR156 suppresses the expression of miR472, which is involved in plant basal immunity, then increases the accumulation of proteins encoded by the CNL genes targeted by miR472. Therefore, the expression of R genes, involved in mediation of PTI and ETI, is considerably enhanced, thereby increasing the expression of PR proteins, activating hypersensitive responses, and increasing host resistance against pathogens such as Pst DC3000. In our present study, we determined how B. cereus AR156 triggers ISR by using miR472 to enhance the expression of R genes and increase the accumulation of their protein products.
FIGURE 8.
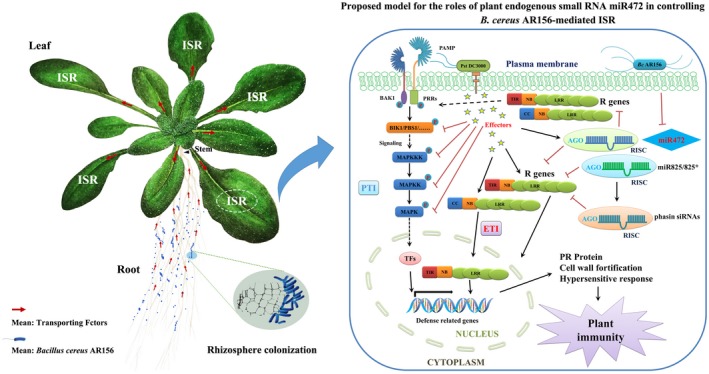
Proposed model for the roles of plant endogenous small RNA miR472 in controlling Bacillus cereus AR156‐mediated induced systemic resistance (ISR)
To the best of our knowledge, this study is the first to determine that miR472 plays an important role in regulating PGPR‐triggered ISR in Arabidopsis via targeting five CNLs and, more importantly, provides a new way to study the mechanism of regulation on PGPR ISR to pathogens. Moreover, it provides theoretical guidance for the development and application of biopesticides.
4. EXPERIMENTAL PROCEDURES
4.1. Plants, bacterial strains, and growth conditions
All the plants used in this study are listed in Table S1. The seeds for each line were sown in vermiculite, then 3‐week‐old seedlings were transferred into 200‐ml pots filled with a mixture of sterilized vermiculite and potting soil at a volume ratio of 2:1, with one seedling per pot. The transplanted seedlings were cultivated in a growth chamber with a 10 hr day (200 μmol⋅m−2⋅s−1 at 22 °C) and a 14 hr night (20 °C) cycle at 70% relative humidity and supplied with modified half‐strength Hoagland nutrient solution (Hoagland and Arnon, 1938) weekly.
All the bacterial strains used in this study are listed in Table S2. The pathogens were grown in liquid King's B medium containing 50 mg/L rifampicin at 28 °C overnight. The cultured Pst DC3000 cells were pelleted by centrifugation at 5,000 × g for 10 min, and then resuspended in 10 mM MgSO4 containing 0.01% (vol/vol) of surfactant Silwet L‐77 (Sigma). Cell concentration was adjusted with respect to experimental purposes, using OD600 = 0.01 for spray inoculation and OD600 = 0.0001 for inoculation via syringe infiltration (as described by Jiang et al., 2016) with several modifications; the concentrations of Pst DC3000 and Pst DC3000 (hrc C) were adjusted to OD600 = 0.0005 for use.
B. cereus AR156 was grown on Luria Bertani (LB) agar at 28 °C for 24 hr. Subsequently, the AR156 cells were pelleted by centrifugation at 5,000 × g for 10 min and resuspended in sterile 0.85% NaCl; cell concentration was adjusted to 5 × 107 cfu/ml for experimental purposes.
4.2. Plant inoculations and bacterial counts
Six‐week‐old Arabidopsis seedlings were challenged with the indicated pathogens and then assessed for ISR using methods published previously (Katiyar‐Agarwal and Jin, 2007; Niu et al., 2011; Jiang et al., 2016). For the pathogen challenge procedure, we used a suspension containing Pst DC3000 (OD600 = 0.0001), which was inoculated via syringe infiltration. Leaf samples of each treated plant were collected by acock borer. Bacterial titres were determined by plating the organisms and counting the number of colonies at 0, 3, and 4 dpi. At least 24‐leaf discs were collected for each treatment. The density of Pst DC3000 in the Arabidopsis leaves was determined and expressed as cfu/g fresh leaf. For the ISR assay, 10 ml AR156 cell suspension (5 × 107 cfu/ml) was poured onto the soil around the root of each seedling in each pot; an equivalent volume of 0.85% NaCl solution was used as control. Half of the seedlings receiving each treatment were challenged after 5 days by spraying the leaves with Pst DC3000 cell suspension (OD600 = 0.01) until all the Arabidopsis leaves were covered with fine droplets. The remaining seedlings were sprayed with 10 mM MgSO4 as control treatment. All the seedlings were maintained in a dew chamber at 100% relative humidity for 3 days and transferred to a growth chamber after inoculation. As described above, bacterial titres were determined at 0, 3, and 4 dpi. Each experiment was performed three times.
4.3. Genetic cloning, vector construction, and generation of transgenic plants
All the plasmids and primers used in this study are listed in Tables S3 and S4, respectively. To generate an miR472 overexpression construct, miR472 precursors were cloned into the Gateway destination vector pEG100 (Earley et al., 2006). The miR472 knockdown vector, STTM‐472, used to inactivate miR472 in Arabidopsis, was generated according to the method of Yan et al. (2012). Next, we generated the overexpression construct of miR472 target genes according to methods described by Tzfira et al. (2005). Full‐length cDNA sequences of these R genes were cloned via PCR amplification. The PCR fragments of full‐length R‐protein coding sequences were inserted into the pPZP‐RCS1 vector (Tzfira et al., 2005; Wang et al., 2017).
All the constructs described above were electroporated into Agrobacterium tumefaciens GV3101 according to the methods described by Clough and Bent (1998). Generation of the transgenic Arabidopsis lines was performed using a floral dip method as reported previously (Koornneef and Pieterse, 2008); transformed seedlings were selected as described by Harrison et al. (2006) and at least two transgenic lines of each gene were selected.
4.4. RNA extraction, RT‐qPCR, and hybridization
For RNA extraction, 0.5 g fresh weight Arabidopsis leaves was collected. Total RNA for each sample was prepared using TRIzol (Invitrogen) as described in the manufacturer's recommendations. RT‐PCR was performed using 0.5 mg total RNA. Reverse transcription was conducted using HiScript Q Select RT Super Mix (Vazyme) (Jiang et al., 2016). RT‐qPCR was conducted using a 7500 Real‐Time PCR System (ABI) with a SYBR Premix Ex Taq II (Takara) and 200 nM final primer concentration. The conditions were as follows: 50 °C for 10 min, predenaturation at 95 °C for 10 min, then 40 cycles of denaturation at 95 °C for 15 s, and annealing and elongation at 60 °C for 30 s. Relative expression levels were calculated as described previously (Niu et al., 2016). Expression of At‐β‐TUB4 was used as internal standard. All the primers used for RT‐qPCR are listed in Table S4. Each experiment was repeated three times.
Northern blotting was performed as described previously (Park et al., 2002). In brief, miR472 hybridization was performed using 100 μg total RNA extracted previously, which was separated at 100 V for 6 hr on a denaturing 15% polyacrylamide gel containing 8 M urea. After electroblotting, RNA was transferred to Nytran N nylon blotting membranes (Whatman/GE Healthcare) using a trans‐blot electrophoretic transfer cell (Bio‐Rad). RNA was then bound to the membrane using UV cross‐linking and baking at 80 °C for 1 hr in a UVP HL‐2000 Hybri Linker hybridization oven and crosslinker (Analytik Jena UVP). The α‐32P dCTP‐labelled probes were generated using a Prime‐It II Random Primer Labelling Kit (Agilent Technologies). The primers used for probe labelling are listed in Table S4. Hybridization was performed in a hybridization oven at 65 °C overnight and washed according to Hybond's instructions. Radioactive signals were detected using Phosphor Imager screens and developed on a Typhoon Imager (GE Healthcare).
4.5. Protein extraction and western blotting
For protein extraction, leaves were collected from seedlings and stored in liquid nitrogen. Total protein was extracted using RIPA lysis buffer (Beyotime). Protein concentration was quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). For western blotting, 10 μg total protein was size‐fractionated using 12% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis, then transferred to a Hybond‐P polyvinylidene fluoride (PVDF) membrane (GE Healthcare) using a trans‐blot electrophoretic transfer cell (Bio‐Rad) containing 10% methanol protein transfer buffer. The membrane was probed with a monoclonal anti‐GFP antibody (Roche) and a secondary horseradish peroxidase‐conjugated antibody (anti‐mouse‐HRP) (Beyotime). The horseradish peroxidase was detected using Clarity Western ECL Substrate (Bio‐Rad) on a Bio‐Rad VersaDoc 5000 (Bio‐Rad). Protein expression was quantified using Quantity One 1‐D software (Bio‐Rad). Total proteins on the membrane were stained with 0.1% (wt/vol) Ponceau S, as described previously (Janes, 2015).
4.6. Target genes of miR472 prediction and verification
To predict the target genes of miR472, we used psRNATarget (http://plantgrn.noble.org/psRNATarget/), a server for conducting target analysis of plant small RNAs. The guidelines for predicting miR472 targets were set as described by Allen et al. (2005). In brief, only one gap with a double penalty was allowed when miR472 was matched with its targets. Inclusion parameters were (a) the nucleotides at positions 10 and 11 of miR472 must match the target completely; and (b) a maximum of three continuous mismatches was allowed if the mismatch region contained at least two G/U pairs. Additionally, the penalty score for the region used for miR472 target selection was less than 3.0 (Allen et al., 2005; Zhang et al., 2011). To verify the actual target genes of miR472, the expression of predicted target genes in the constructed miR472 transgenic lines was analysed using RNA‐Seq data. The expression levels of these genes were detected by RT‐qPCR. An Agrobacterium‐mediated tobacco transient expression system was used to simultaneously express miR472 and its targets genes in tobacco. Subsequently, the expression of each target gene and its respective protein product was quantitatively detected using western blotting, GFP fluorescence, and GUS staining.
4.7. Construction of RNA‐Seq library and RNA sequencing
In this study, a total of eight samples was obtained from: A. thaliana ecotype Col‐0, a line overexpressing miR472, a line bearing a knockdown of miR472, and T‐DNA insertion lines. All samples were obtained using the same growth period, and each seedling was sampled twice. Construction of RNA sequencing libraries was performed using 20 μg total RNA for each sample via a TruSeq RNA Sample Prep Kit v. 2 (Illumina). First, mRNA isolated from the leaves of different lines was purified, dissociated into short fragments, and used to synthesize first‐strand cDNA via a Transcriptor First Strand cDNA Synthesis Kit (Roche). Subsequently, double‐stranded DNA was synthesized using DNA polymerase I and RNase H (Thermo Fisher Scientific), followed by end repair via single‐adenine residue and adapter ligation. The final cDNA library was generated using Phusion High‐Fidelity DNA polymerase (Thermo Scientific). Illumina sequencing was carried out using a HiSeq 2500 platform (Illumina). All sequencing data were saved as FASTQ files and deposited into the NCBI database (BioProject accession number: PRJNA548632; BioSample accession numbers: SAMN12045474, SAMN12045475, SAMN12045476, SAMN12045477, SAMN12045478, SAMN12045479, SAMN12045480, SAMN12045481, SAMN12054380, and SAMN12054381).
4.8. Bioinformatic analysis
4.8.1. Processing of raw reads, de novo assembly, and functional annotation
To obtain sequencing information, raw reads were initially processed and optimized as described previously (Zhang et al., 2015). De novo assembly of the RNA‐Seq library was performed using the Trinity platform (Grabherr et al., 2011). Clean reads were then mapped onto the corresponding contigs, and unigenes with no extension on either end were generated by assembling all the contigs. The assembled unigenes were subjected to similarity search against the NCBI nonredundant protein database via BlastX. Subsequently, Blast annotations were mapped back onto the Uniprot protein database and the GO terms were extracted. To examine putative functions, the assembled unigenes were searched against other databases such as the Swiss‐Prot protein database, GO database, and KEGG pathway database (Wu et al., 2016).
4.8.2. Identification of DEGs and GO enrichment
To quantify the expression of the genes characterized in this study, we mapped RPKM and fragments per kilobase of transcript per million mapped reads. In brief, differential expression analysis was performed using the DESeq2 R package v. 1.24.0, and DEGs were determined via log‐fold expression change (log FC) less than −2 or greater than 2. Venn diagrams were drawn using Venny v. 2.1.0 as described by Oliveros (2007). The KEGG pathway and GO enrichment analyses for all DEGs were conducted using the GOseq R v. 1.36.0 package and KOBAS v. 2.0 plate (Jiang et al., 2019).
4.9. Data analysis
All bioassays used in this study were performed at least three times using 48 seedlings per treatment. Analysis of variance was carried out using IBM SPSS Statistics software v. 21.0 (SPSS Inc.). Mean comparison was conducted using a least significant difference (LSD) test ± SE or SD; p < .05 was considered statistically significant.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
We want to thank Professor Hong‐Wei Zhao (Nanjing Agricultural University) for kind advice and help. We also want to thank Jing‐Da Xu and Hong‐Li Wang for experimental assistance. This study was supported by the Natural Science Foundation of Jiangsu Province (Young Scholars Program, BK20170709), the National Natural Science Foundation of China (Young Scholars Program, 31701829), the National Natural Science Foundation of China (31672075, 31972322), the China Agriculture Research System (CARS‐21), the National Postdoctoral Program for Innovative Talents (BX201600074), and the China Postdoctoral Science Foundation (2017M611839).
Jiang C, Fan Z, Li Z, et al. Bacillus cereus AR156 triggers induced systemic resistance against Pseudomonas syringae pv. tomato DC3000 by suppressing miR472 and activating CNLs‐mediated basal immunity in Arabidopsis . Molecular Plant Pathology. 2020;21:854–870. 10.1111/mpp.12935
Chunhao Jiang and Zhihang Fan contributed equally to this study.
DATA AVAILABILITY STATEMENT
Sequencing data were deposited in the NCBI BioProject database at https://www.ncbi.nlm.nih.gov/bioproject/, accession number PRJNA548632. Sample information is in the NCBI BioSample database at https://www.ncbi.nlm.nih.gov/biosample/, accession numbers SAMN12045474, SAMN12045475, SAMN12045476, SAMN12045477, SAMN12045478, SAMN12045479, SAMN12045480, SAMN12045481, SAMN12054380, and SAMN12054381.
REFERENCES
- Allen, E. , Xie, Z. , Gustafson, A.M. and Carrington, J.C. (2005) microRNA‐directed phasing during trans‐acting siRNA biogenesis in plants. Cell, 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Austin, M.J. , Muskett, P. , Kahn, K. , Feys, B.J. , Jones, J.D.G. and Parker, J.E. (2002) Regulatory role of SGT1 in early R gene‐mediated plant defenses. Science, 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Baker, B. , Zambryski, P. , Staskawicz, B. and Dinesh‐Kumar, S.P. (1997) Signaling in plant–microbe interactions. Science, 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bi, G. and Zhou, J.M. (2017) Map kinase signaling pathways: a hub of plant–microbe interactions. Cell Host & Microbe, 21, 270–273. [DOI] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiébeauld, O. , Jay, F. and Colot, V. (2014) The Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐ and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Pathogens, 10, e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.M. (2009) Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology, 25, 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Shen, Q. , Lin, R. , Zhao, Z. , Shen, C. and Sun, C. (2017) De novo transcriptome analysis in Dendrobium and identification of critical genes associated with flowering. Plant Physiology and Biochemistry, 119, 319–327. [DOI] [PubMed] [Google Scholar]
- Choudhary, D.K. , Prakash, A. and Johri, B.N. (2007) Induced systemic resistance (ISR) in plants: mechanism of action. Indian Journal of Microbiology, 47, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Pieterse, C.M. and Mauch‐Mani, B. (2002) Priming in plant–pathogen interactions. Trends in Plant Science, 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Cui, X. and Cao, X. (2014) Epigenetic regulation and functional exaptation of transposable elements in higher plants. Current Opinion in Plant Biology, 21, 83–88. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology, 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D.G. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K.M. et al (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. et al (2011) Full length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology, 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, S.J. , Mott, E.K. , Parsley, K. , Aspinall, S. , Gray, J.C. and Cottage, A. (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods, 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland, D.R. and Arnon, D.I. (1938) The water culture method for growing plants without soil. California Agricultural Experiment Station, 347, 36–39. [Google Scholar]
- Huang, J. , Yang, M. and Zhang, X. (2016) The function of small RNAs in plant biotic stress response. Journal of Integrative Plant Biology, 58, 312–327. [DOI] [PubMed] [Google Scholar]
- Janes, K.A. (2015) An analysis of critical factors for quantitative immunoblotting. Science Signaling, 8, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.H. , Huang, Z.Y. , Xie, P. , Gu, C. , Li, K. , Wang, D.C. et al (2016) Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156‐induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis . Journal of Experimental Botany, 67, 157–174. [DOI] [PubMed] [Google Scholar]
- Jiang, C.H. , Yao, X.F. , Mi, D.D. , Li, Z.J. , Yang, B.Y. , Zheng, Y. et al. (2019) Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis F21 against Fusarium wilt on watermelon. Frontiers in Microbiology, 10, 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , Morgan, R. , Dahlbeck, D. , Borsani, O. , Villegas, A.J. , Zhu, J.K. et al (2006) A pathogen‐inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences of the United States of America, 103, 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. and Jin, H. (2007) Discovery of pathogen‐regulated small RNAs in plants. Methods in Enzymology, 427, 215–227. [DOI] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. and Jin, H. (2010) Role of small RNAs in host–microbe interactions. Annual Review of Phytopathology, 48, 225–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Qiu, X. , Kang, J. , Wang, Y. , Chen, H. , Huang, J. et al (2017) A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Current Biology, 27, 981–991. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiology, 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Pignatta, D. , Bendix, C. , Brunkard, J.O. , Cohn, M.M. , Tung, J. et al (2012) MicroRNA regulation of plant innate immune receptors. Proceedings of the National Academy of Sciences of the United States of America, 109, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. et al (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Niu, D.D. , Liu, H.X. , Jiang, C.H. , Wang, Y.P. , Wang, Q.Y. , Jin, H.L. et al (2011) The plant growth‐promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate‐ and jasmonate/ethylene‐dependent signalling pathways. Molecular Plant‐Microbe Interactions, 24, 533–542. [DOI] [PubMed] [Google Scholar]
- Niu, D.D. , Xia, J. , Jiang, C.H. , Qi, B.B. , Ling, X.Y. , Lin, S.Y. et al (2016) Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825* and activating defense‐related genes in Arabidopsis . Journal of Integrative Plant Biology, 58, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros, J.C. (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html [Accessed 20 March 2020].
- Ouyang, S. , Park, G. , Atamian, H.S. , Han, C.S. , Stajich, J.E. , Kaloshian, I. et al (2014) MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum . PLoS Pathogens, 10, e1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W. , Li, J. , Song, R. , Messing, J. and Chen, X. (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Current Biology, 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van Wees, S.C. , Hoffland, E. , Van Pelt, J.A. and Van Loon, L.C. (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis‐related gene expression. The Plant Cell, 8, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology, 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Zipfel, C. (2008) News from the frontline: recent insights into PAMP‐triggered immunity in plants. Current Opinion in Plant Biology, 11, 389–395. [DOI] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Chen, H.M. , Patel, K. , Bond, D.M. , Santos, B.A. and Baulcombe, D.C. (2012) A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. The Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonich, M. and Innes, R.W. (1995) A disease resistance gene in Arabidopsis with specificity for the AvrPph3 gene of Pseudomonas syringae pv. phaseolicola . Molecular Plant‐Microbe Interactions, 8, 637–640. [DOI] [PubMed] [Google Scholar]
- Speth, E.B. , Lee, Y.N. and He, S.Y. (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Current Opinion in Plant Biology, 10, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, B. , Gao, S. , Lercher, M.J. , Hu, S. and Chen, W. (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Research, 47, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira, T. , Tian, G.W. , Lacroix, B. , Vyas, S. , Li, J. , Leitner‐Dagan, Y. et al (2005) pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Molecular Biology, 57, 503–516. [DOI] [PubMed] [Google Scholar]
- Van der Ent, S. , Van Wees, S.C. and Pieterse, C.M. (2009) Jasmonate signaling in plant interactions with resistance‐inducing beneficial microbes. Phytochemistry, 70, 1581–1588. [DOI] [PubMed] [Google Scholar]
- Van Oosten, V.R. , Bodenhausen, N. , Reymond, P. , Van Pelt, J.A. , Van Loon, L.C. , Dicke, M. et al (2008) Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis . Molecular Plant‐Microbe Interactions, 21, 919–930. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2006) Post‐transcriptional small RNA pathways in plants: mechanisms and regulations. Genes & Development, 20, 759–771. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Lacroix, B. , Guo, J. and Citovsky, V. (2017) The Agrobacterium VirE2 effector interacts with multiple members of the Arabidopsis VIP1 protein family. Molecular Plant‐Microbe Interactions, 19, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , Zhou, H. , Zhang, Q. , Zhang, J. , Ni, F. , Liu, C. et al (2010) DNA methylation mediated by a microRNA pathway. Molecular Cell, 38, 465–475. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Wu, J. , Li, S.S. , Zhang, H.J. , Feng, C.Y. , Yin, D.D. et al (2016) Transcriptome sequencing and metabolite analysis for revealing the blue flower formation in waterlily. BMC Genomics, 17, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Gu, Y. , Jia, X. , Kang, W. , Pan, S. , Tang, X. et al (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis . The Plant Cell, 24, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.W. and Tang, K.X. (2004) MAP kinase cascades responding to environmental stress in plants. Acta Botanica Sinica, 46, 127–136. [Google Scholar]
- Zhai, J. , Jeong, D.H. , De Paoli, E. , Park, S. , Rosen, B.D. , Li, Y. et al (2011) MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased trans‐acting siRNAs. Genes & Development, 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhao, H. , Gao, S. , Wang, W.C. , Katiyaragarwal, S. , Huang, H.D. et al (2011) Arabidopsis argonaute 2 regulates innate immunity via miRNA393∗‐mediated silencing of a golgi‐localized snare gene, MEMB12 . Molecular Cell, 42, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.C. , Li, W.J. , Dou, Y.J. , Zhang, J.X. , Jiang, G.H. , Miao, L.X. et al (2015) Transcript quantification by RNA‐Seq reveals differentially expressed genes in the red and yellow fruits of Fragariavesca . PLoS ONE, 10, e0144356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Cai, C. , Zhai, J. , Lin, F. , Li, L. , Shreve, J. et al (2015) Coordination of microRNAs, phasiRNAs, and NB‐LRR genes in response to a plant pathogen: insights from analyses of a set of soybean Rps gene near‐isogenic lines. Plant Genome, 8. doi: 10.3835/plantgenome2014.09.0044. [DOI] [PubMed] [Google Scholar]
- Zhou, J.M. and Yang, W.C. (2016) Receptor‐like kinases take center stage in plant biology. Science China Life Sciences, 59, 863–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited in the NCBI BioProject database at https://www.ncbi.nlm.nih.gov/bioproject/, accession number PRJNA548632. Sample information is in the NCBI BioSample database at https://www.ncbi.nlm.nih.gov/biosample/, accession numbers SAMN12045474, SAMN12045475, SAMN12045476, SAMN12045477, SAMN12045478, SAMN12045479, SAMN12045480, SAMN12045481, SAMN12054380, and SAMN12054381.


