Abstract
Study Objectives
This study describes high-throughput phenotyping strategies for sleep and circadian behavior in mice, including examinations of robustness, reliability, and heritability among Diversity Outbred (DO) mice and their eight founder strains.
Methods
We performed high-throughput sleep and circadian phenotyping in male mice from the DO population (n = 338) and their eight founder strains: A/J (n = 6), C57BL/6J (n = 14), 129S1/SvlmJ (n = 6), NOD/LtJ (n = 6), NZO/H1LtJ (n = 6), CAST/EiJ (n = 8), PWK/PhJ (n = 8), and WSB/EiJ (n = 6). Using infrared beam break systems, we defined sleep as at least 40 s of continuous inactivity and quantified sleep–wake amounts and bout characteristics. We developed assays to measure sleep latency in a new environment and during a modified Murine Multiple Sleep Latency Test, and estimated circadian period from wheel-running experiments. For each trait, broad-sense heritability (proportion of variability explained by all genetic factors) was derived in founder strains, while narrow-sense heritability (proportion of variability explained by additive genetic effects) was calculated in DO mice.
Results
Phenotypes were robust to different inactivity durations to define sleep. Differences across founder strains and moderate/high broad-sense heritability were observed for most traits. There was large phenotypic variability among DO mice, and phenotypes were reliable, although estimates of heritability were lower than in founder mice. This likely reflects important nonadditive genetic effects.
Conclusions
A high-throughput phenotyping strategy in mice, based primarily on monitoring of activity patterns, provides reliable and heritable estimates of sleep and circadian traits. This approach is suitable for discovery analyses in DO mice, where genetic factors explain some proportion of phenotypic variation.
Keywords: genetics, high-throughput phenotyping, Diversity Outbred mice, sleep latency in new environment, modified Murine Multiple Sleep Latency Test, circadian period, sleep
Statement of Significance.
Diversity Outbred (DO) mice are an increasingly used resource for genetic discovery, with unique advantages. However, genetic analyses using this resource require a robust and high-throughput phenotyping strategy. This study describes such a strategy for sleep and circadian behaviors. Using DO mice and mice from their eight founder strains, we demonstrate that measures of sleep architecture, latency to sleep, sleep drive, and circadian period derived through monitoring of activity patterns are robust to the specific inactivity duration used to define sleep, are reliable in DO mice, and are significantly influenced by complex genetic effects (e.g. heritable). This phenotyping strategy can be used in future studies leveraging DO mice to discover individual or combinations of genes affecting sleep and circadian behavior.
Introduction
Many aspects of sleep and circadian behavior are heritable in humans [1, 2] and mice [3]. These heritable traits include total sleep [4], response to sleep loss [5], sleep homeostasis [6], and timing of major sleep episodes [7–10]. Data are emerging on gene variants that could explain these traits, as reviewed previously [1, 2] and evidenced by statistically significant associations with sleep phenotypes in recent large-scale genome-wide association analyses leveraging publicly available datasets, such as the UK Biobank and 23andMe [11–17]. There is also evidence that genes that may affect aspects of sleep and circadian behavior could have pleiotropic neurological consequences [2].
Identifying which genes affect sleep and circadian behavior is complicated. One strategy is to map candidate genes using association studies in outbred mouse resources, such as the Diversity Outbred (DO) mice [18–20] and then use inbred strains, in particular the Collaborative Cross [21–23], to validate the genetic effects. However, the given genetic heterogeneity of outbred mice, this strategy will require phenotyping a large number of mice to achieve adequate statistical power. The gold-standard for studying sleep in mice is implanting electrodes and recording electroencephalogram (EEG)/electromyogram (EMG) in individual mice. However, this approach is highly invasive and requires substantial technological efforts. An alternative approach is to use high-throughput phenotyping methods that do not require surgery to assess sleep and circadian rhythm [24, 25]. Toward this end, here we evaluate sleep-related phenotypes based on multiple infrared beams that are transmitted across the mouse cage, characterizing a mouse as active whenever these beams are broken. These techniques have been shown to accurately estimate sleep and wake when compared to simultaneous EEG/EMG recording in young and old mice [24, 26, 27]. Ultimately, the high-throughput phenotypes described here can be used to discover genetic associations in DO mice. As the Collaborative Cross lines have been sequenced [21–23], subsequent validation studies using gold-standard EEG/EMG in Collaborative Cross mice carrying the implicated founder alleles at specific locations can then be leveraged to derive more accurate estimates of genetic effects on sleep architecture.
Based on this strategy, we developed a high-throughput pipeline to assess sleep and circadian behaviors in individual DO mice (n = 338 total) and in mice from each of the eight founder strains of the DO population (A/J [n = 6], C57BL/6J [n = 14], 129S1/SvlmJ [n = 6], NOD/LtJ [n = 6], NZO/H1LtJ [n = 6], CAST/EiJ [n = 8], PWK/PhJ [n = 8], and WSB/EiJ [n = 6]) [18–20]. We first assessed total sleep and wake across light and dark periods, along with measurements of the number of sleep and wake bouts and average bout durations. We then performed separate experiments to capture other aspects of sleep–wake behavior, including: (1) sleep latency in a new environment as a measure of ability to maintain wakefulness [28]; (2) a modified Murine Multiple Sleep Latency Test (m-MMSLT) [29] to assess sleep drive under increasing sleep deprivation; and (3) wheel-running in constant darkness to assess circadian period. The DO mice were genotyped with a high-density genotyping array [30] and narrow-sense heritability of the sleep–circadian traits was estimated. We also quantified broad-sense heritability using data from the eight founder strains.
Our findings demonstrate that sleep and circadian phenotypes are reliable within individual mice, but also vary between founder strains and across the DO population. We found moderate to high broad-sense heritability of most traits within the founder mice, but lower estimates of narrow-sense heritability in the outbred DO mice. This provides the basis for future studies to identify gene variants and complex genetic effects that influence sleep and circadian behavior using DO mice [18–20], the Collaborative Cross [21, 23], and their founder strains.
Methods
See Supplementary material for additional details.
Animals and husbandry
Studies were done in mice at 2–3 months of age from each of the eight DO founder genotypes (A/J [n = 6], C57BL/6J [n = 14], 129S1/SvlmJ [n = 6], NOD/LtJ [n = 6], NZO/H1LtJ [n = 6], CAST/EiJ [n = 8], PWK/PhJ [n = 8], and WSB/EiJ [n = 6]) and from the DO population (n = 338). Founder strains were chosen based on community consensus in the design of the Collaborative Cross, and include five classical inbred strains (A/J, C57BL/6J, 129S1/SvlmJ, NOD/LtJ, NZO/H1LtJ) and three wild-derived strains representing different mouse sub-species (CAST/EiJ, PWK/PhJ, and WSB/EiJ) [21, 22]. As in previous studies identifying genetic influences on sleep characteristics, all mice included in the present analyses were male [6, 31]. Animals were obtained from the Jackson Laboratory at approximately 6 weeks of age; DO mice were ordered 3 times/year in groups of 50 non-siblings, following the DO production schedule (https://www.jax.org/strain/009376). All animals underwent the same protocol and testing order (see Supplementary meterial, Methods) to reduce environmental variability. Upon arrival, mice were housed in groups of five mice per cage for 3 weeks with a 12-h/12-h light/dark (L/D) cycle set at 7 am/7 pm. After this initial acclimation period, mice were individually housed for testing with the same L/D schedule. Access to chow (Laboratory Rodent Diet 5001) and water was provided ad libitum throughout the protocol. Experiments were approved by the IACUC at the University of Pennsylvania in accordance with the National Institutes of Health guidelines.
Overview of the testing protocol and phenotypes
Mice were acclimated to being singly housed with 12 h light/dark (L/D) for 5 days in a standard mouse cage, which was placed within the Accuscan IR beam system (AccuScan Instruments, Inc., Columbus, OH) that has 16 infrared beams 1 inch apart in the horizontal plane. Software records activity when two consecutive beams are broken by the mouse and activity data is stored in 10-s epochs for analysis. After acclimation, the total amount of sleep–wake and the total number of sleep–wake bouts and duration of each bout were quantified over 5 days of baseline recording. Based on previous validation studies in mice of similar age [24, 26], we operationally defined a period of inactivity (no beam breaks) of 40 s or more as sleep; other definitions are considered in sensitivity analyses. We also calculated the mouse-specific standard deviation to assess associations with day-to-day variability.
After baseline sleep–wake recording, each mouse was transferred to a new cage with fresh bedding at 11 am, a time when there is increased sleep drive. Upon transfer to a new cage, mice become more active and spend time exploring the new cage environment [28]. Latency to sleep in this new environment was determined as the onset of the first consolidated bout of sleep, defined as the first period of inactivity of 3 min or more after comparisons to gold-standard EEG-defined sleep in an independent set of C57BL/6J (n = 9) mice (see Supplementary material, Methods and Figure S1). This difference in the optimal duration of inactivity for sleep latency compared to overall sleep architecture reflects the distinction in target phenotypes. When defining latency to a single consolidated sleep bout, a longer period of inactivity (3 min) is required to avoid periods of quiet wakefulness when transferred to a new cage, whereas the shorter sleep definition (40 s) is more accurate when measuring total sleep amounts across the day [24, 26].
Following sleep latency testing in a new environment, mice were acclimated to their new cages for 5 days. Afterward, a modified Murine Multiple Sleep Latency Test (m-MMSLT) [29] was performed to provide a measure of sleep drive (homeostasis). From 10 am to 4 pm, mice were sleep deprived by gentle handling for 30 min (e.g. 10 am to 10:30 am) then allowed to sleep for 30 min (e.g. 10:30 am to 11 am), resulting in 6 total sleep opportunities where mice experienced increasing sleep deprivation. Latency to sleep onset was defined as the time to the first sleep bout, using the same 3-min inactivity definition as in new environment testing; latency to sleep was scored as 30 min if mice were active for the entire sleep opportunity. The primary measure of sleep drive was the change in latency to sleep (slope) across the six sleep opportunities. The m-MMSLT was added later in the phenotyping protocol, and therefore is only available for a subset of the DO mice (N = 174).
After approximately 2 weeks of recovery from the new environment and m-MMSLT testing, mice were singly caged with a running wheel, where they acclimated to the change in environment for 10 days. After acclimation, voluntary wheel running in L/D 7 am/7 pm was recorded for 10 days. Subsequently, lights were turned off for 18–20 days (constant darkness; D/D) and a free-running circadian period was recorded. We used Clock Lab (Actimetrics) to estimate circadian periods from wheel-running data, employing the Lomb–Scargle algorithm [32, 33]. Thereafter, mice were returned to a 12-h L/D cycle and subsequently euthanized by cervical dislocation.
DNA/genotyping
At harvest, organs were quickly frozen in dry ice and stored at −80°C. Tail samples were collected and shipped to the Jackson Laboratory, where DNA was extracted and genotyping carried out at GeneSeek (Lincoln, NE) using the high-density Mega Mouse Universal Genotyping Array (MegaMUGA) [30]. A hidden Markov model analysis was applied to the array intensities to infer the haplotype blocks in each DO genome [20]. A total of 325 DO mice met quality standards and are included in heritability calculations.
Statistical analysis
We performed statistical analyses focused on (1) examining phenotypic variability in DO mice and comparing the phenotype values among founder mouse strains, (2) establishing reliability of the phenotype among the DO mice, and (3) calculating the proportion of phenotypic variability explained by genetic factors (i.e. heritability) within the DO mice (narrow-sense heritability; h2) and founder mice (broad-sense heritability; H2), separately. In addition to the observed scale, we created normalized phenotypes using a rank-based inverse normal transformation based on the distributions in the founder strains and DO mice, separately. Phenotypes were compared among founder strains using an analysis of variance (ANOVA). To assess reliability in DO mice, data were obtained from repeated assessments of phenotypes in the same mouse, either leveraging multiple days of recording (e.g. sleep–wake) or by repeating the experiment in a consecutive subset of animals (e.g. new environment test, m-MMSLT). Reliability was quantified using intraclass correlation coefficients (ICCs) as poor (ICC < 0.00), slight (0.00–0.20), fair (0.20–0.40), moderate (0.41–0.60), substantial (0.60–0.80), or almost perfect (0.80–1.00) [34].
To determine whether genetic architecture explains variability in measured phenotypes, we estimated the heritability (and associated confidence intervals [CIs]) of each phenotype in the founder strains and DO mice, separately. Among founder strains, we used a mixed-effects model with strain as a random effect to estimate the variance attributable to differences in strains (i.e. genetic variance). Broad-sense heritability (H2; the proportion of phenotypic variability explained by all underlying genetic factors) was calculated as this genetic variance divided by the total phenotypic variance. A non-parametric 95% CI around this estimate was calculated as the 2.5th to 97.5th percentiles from 1,000 bootstrapped samples. Among DO mice, narrow-sense heritability (h2; the proportion of phenotypic variability explained by additive genetic effects), was calculated by fitting a mixed-effects model controlling for the experimental cohort as a fixed effect and utilizing a genetic kinship matrix as a random term via the est_herit function in qtl2scan (https://github.com/rqtl/qtl2scan). A nonparametric 95% CI was calculated from the distribution of heritability estimates of 1,000 simulated phenotypes with an error covariance matrix designed to reflect the estimated covariance structure of the original phenotype by weighting the additive genetic matrix by the heritability and the identity matrix by (1-heritability) [35, 36]. Heritability estimates for which the associated 95% CI did not include zero were considered statistically significant.
Results
High-throughput phenotypes
The following sections detail the observed distributions of the high-throughput sleep and circadian characteristics, including differences among founder strains and the observed phenotypic variability among the genetically diverse DO mice. Additional details, including within-group means and standard deviations, medians and ranges, and pairwise comparisons between founder strains can be found in Supplementary material, File 1. Unless otherwise noted, data are presented as mean value ± standard deviation.
Total sleep and wake
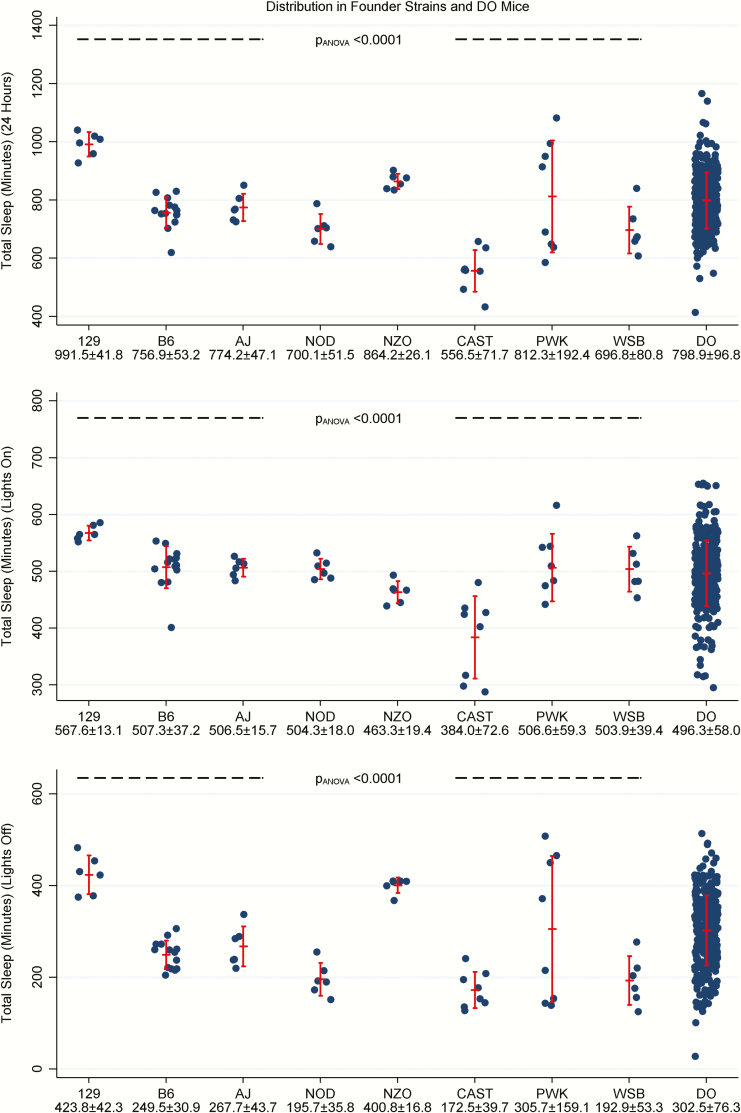
Total sleep (and wake) was estimated using our definition of sleep as 40 s or more of inactivity (Figure 1). As expected, the total amount of sleep was greater in the lights-on than the lights-off period in all mice (p = 1.86 × 10−145). Over 24-h, there were significant differences in the total amount of sleep (and wake) across founder strains (p = 2.52 × 10−10); the 129 strain had the most sleep on average (991.5 ± 41.8 min) and the CAST strain had the least (556.5 ± 71.7 min). There were significant differences in amounts of sleep (and wake) between the founder strains during both lights-on (p = 1.42 × 10−8) and lights-off (p = 2.94 × 10−9). Once again, the 129 strain had the most sleep and CAST the least in both periods (see Figure 1). The remaining strains had similar amounts of total sleep (averages between 463.3 and 507.3 min) during lights-on, while the NZO more closely resembled the 129 strain and the NOD and WSB more closely resembled the CAST during lights-off. There is some visual evidence of higher mouse-to-mouse variability in the PWK strain (see section Phenotype Variability (Standard Deviation)).
Figure 1.
Total sleep duration in founder strains and DO mice. Plots illustrate the distribution of total sleep duration over 24 h (top panel), and separately during lights-on (middle panel) and lights-off (bottom panel) in founder (N = 60) and DO mice (N = 338). Vertical error bars represent the observed mean ± standard deviation. There were significant differences in total sleep (and wake) duration across the founder strains during all time periods; the 129 strain generally slept the most and the CAST strain the least. Total sleep duration was highly variable in the DO mice, with data spanning or exceeding the full range of values observed in founder strains.
The total amount of sleep was highly variable across DO mice, spanning or exceeding the full range of values observed across founder strains (Figure 1). During the entire 24-h period, total sleep was 798.9 ± 96.8 min on average, with values ranging from 412.9 to 1,165.8 min. There was significantly more sleep on average during lights-on (496.3 ± 58.0 min; ranging from 295.0 to 655.1 min) and less sleep on average during lights-off (302.5 ± 76.3 min; ranging from 27.3 to 513.8 min) in DO mice (p = 2.01 × 10−122). This marked variation between individual DO mice is likely due, in large part, to their genetic heterogeneity.
Sleep and wake microarchitecture
To better understand sleep/wake microarchitecture, we estimated the number of sleep and wake bouts and the average duration of each bout (see Table 1 and Supplementary material, File 1). Given that bout durations were not normally distributed among the DO population, results are also presented after rank-based inverse normalization (Supplementary Figure S2); this also more clearly illustrates differences among the founder strains.
Table 1.
Comparisons of sleep architecture phenotypes in founder strains and DO mice
| Measure* | Diversity Outbred | Founder strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 129/SvlmJ | C57BL/6J | A/J | NOD/LtJ | NZO/H1LtJ | CAST/EiJ | PWK/PhJ | WSB/EiJ | ANOVA P-value† | ||
| Number of sleep bouts | ||||||||||
| 24 h | 234.0 ± 53.4 | 308.5 ± 26.3 | 230.6 ± 40.8 | 234.0± 31.0 | 242.4 ± 27.0 | 277.9 ± 24.1 | 188.8 ± 34.3 | 229.9 ± 32.3 | 210.8 ± 46.8 | 2.58 × 10−6 |
| Lights-on | 117.4± 29.5 | 139.3 ± 16.1 | 115.2 ± 18.6 | 130.4 ± 9.8 | 130.3 ± 14.8 | 137.8 ± 15.2 | 118.6 ± 21.0 | 139.2 ± 14.9 | 128.5 ± 15.2 | 0.0148 |
| Lights off | 116.9 ± 34.0 | 169.6 ± 12.1 | 115.6 ± 25.6 | 103.7 ± 28.2 | 112.4 ± 15.6 | 140.2 ± 11.7 | 70.2 ± 21.6 | 91.0 ± 33.6 | 82.3 ± 37.6 | 3.38 × 10−8 |
| Average sleep bout duration (min) | ||||||||||
| 24 h | 3.70 ± 1.28 | 3.26 ± 0.30 | 3.42 ± 0.59 | 3.40 ± 0.38 | 2.92 ± 0.30 | 3.15 ± 0.35 | 3.03 ± 0.37 | 3.60 ± 0.75 | 3.41 ± 0.47 | 0.1749 |
| Lights-on | 5.16 ± 3.59 | 4.34 ± 0.75 | 4.69 ± 0.84 | 3.94 ± 0.28 | 3.97 ± 0.48 | 3.45 ± 0.54 | 3.31 ± 0.48 | 3.88 ± 0.89 | 4.01 ± 0.50 | 0.0011 |
| Lights-off | 2.77 ± 0.88 | 2.51 ± 0.27 | 2.25 ± 0.44 | 2.81 ± 0.52 | 1.75 ± 0.23 | 2.91 ± 0.23 | 2.60 ± 0.43 | 3.29 ± 0.96 | 2.61 ± 0.72 | 0.0001 |
| Average wake bout duration (min) | ||||||||||
| 24 h | 2.99 ± 1.15 | 1.47 ± 0.20 | 3.09 ± 0.74 | 2.96 ± 0.59 | 3.11 ± 0.53 | 2.09 ± 0.18 | 4.92 ± 1.20 | 2.90 ± 1.21 | 3.75 ± 1.10 | 5.22 × 10−8 |
| Lights-on | 2.07 ± 0.81 | 1.10 ± 0.08 | 1.94 ± 0.61 | 1.64 ± 0.21 | 1.71 ± 0.27 | 1.89 ± 0.16 | 3.02 ± 1.22 | 1.57 ± 0.50 | 1.73 ± 0.47 | 3.00 × 10−5 |
| Lights-off | 4.54 ± 6.46 | 1.77 ± 0.31 | 4.36 ± 1.33 | 5.22 ± 2.00 | 4.88 ± 1.06 | 2.32 ± 0.28 | 8.95 ± 2.93 | 6.02 ± 4.24 | 8.18 ± 4.91 | 2.27 × 10−5 |
*Data presented as mean ± standard deviation.
† p-Value from analysis of variance (ANOVA) comparing values across founder strains.
We first examined the total number of sleep bouts (see Table 1 and Supplementary Figure S2); results for wake bouts are nearly identical since each wake bout precedes a corresponding sleep bout. Significant differences in the number of bouts were observed over the entire day (p = 2.58 × 10−6), with fewer sleep bouts in the CAST strain (188.8 ± 34.3 bouts) and a larger number of sleep bouts in the 129 strain (308.5 ± 26.3). Differences among founder strains were observed in both light and dark periods, although stronger differences were seen in lights-off (p = 3.38 × 10−8) than lights-on (p = .015). All founder strains had an average number of sleep bouts between 115.2 and 139.3 during lights-on. However, during the active lights-off period, the CAST strain had the fewest sleep bouts (70.2 ± 21.6) and the 129 strain the largest number of sleep bouts (169.6 ± 12.1). Among the DO mice, we observed an average of 234.0 ± 53.4 sleep bouts across the entire day (range = 84.0–391.0), with a similar average number of sleep bouts in both lights-on (117.4 ± 29.5; range = 27.4–219.2 bouts) and lights-off (116.9 ± 34.0; range = 10.4–211.0 bouts). Observed values spanned and exceeded the distribution observed across founders, reflecting genetic heterogeneity and likely epistasis (gene–gene interactions).
When we assessed the average duration of sleep and wake bouts over 24 h among founder strains, there was a difference in average wake bout duration (p = 5.22 × 10−8), but not average sleep bout duration (p = .175). All strains had a mean average sleep bout durations ranging from 2.92 to 3.60 min. The CAST mice had the longest wake bouts (4.92 ± 1.20 min) and 129 strain the shortest (1.47 ± 0.20 min). On the other hand, average bout durations of sleep and wake were significantly different among founder strains during both the lights-on (p = .0011 and p = 3.00 × 10−5 for sleep and wake, respectively) and lights-off (p = .0001 and p = 2.27 × 10−5) periods (see Table 1). During lights-on, the CAST strain displayed the shortest average sleep bout duration and longest average wake bout duration. The NZO also displayed relatively short sleep bout duration during lights-on, while the 129 strain had the shortest average wake bout duration during this period. Similarly, the 129 strain showed the shortest wake bout duration and the CAST strain the longest during lights-off; the NOD mice had the shortest average sleep bout duration.
Overall, observed values of average sleep and wake bout durations in DO mice encompass and exceed variation among the founder strains (see Supplementary material, File 1 and Figure S2). This includes individual mice with extremely long bout durations that extend well beyond the range observed in founders, which may represent an extreme genetic phenotype. DO mice had an average sleep bout duration of 3.70 ± 1.28 min (range: 2.15–12.14 min) and average wake bout duration of 2.99 ± 1.15 min (range: 1.47–11.32 min). In general, more variability was observed in the bout durations corresponding to the expected behavior in the specific time period, that is, sleep bout duration during lights-on (sleep period) and wake bout duration during lights-off (active period). We observed a mean average sleep bout duration of 5.16 ± 3.59 min, with estimates ranging from 2.11 to 28.34 min during lights-on. In contrast, the average wake bout duration during lights-on was 2.07 ± 0.81 min, with a range of only 0.936.84 min. During lights-off, average wake bout duration was 4.23 ± 2.69 min, with a range of 1.31–32.44 min, after excluding a single outlier of 112.18 min (average wake bout duration was 4.54 ± 6.46 min when including this point). Sleep bout duration during lights-off demonstrated a mean of 2.77 ± 0.88 and a range of 1.22–6.42 min. Thus, we again found large heterogeneity in the DO mice.
Sleep latency in a new environment
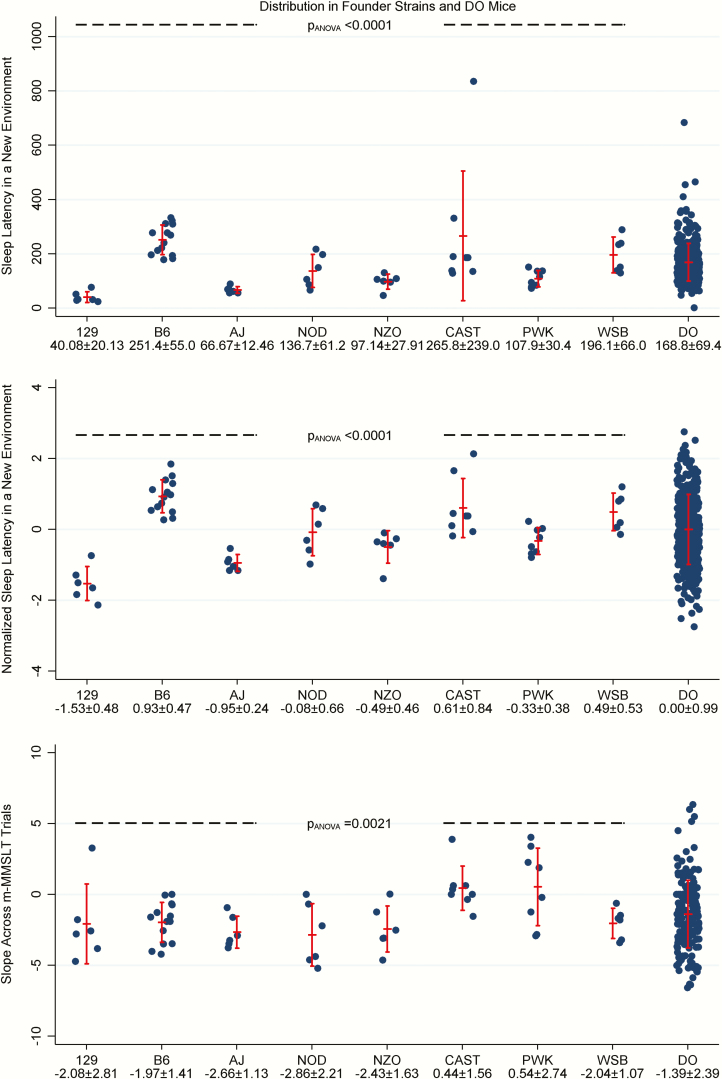
Estimates of latency to sleep onset in a new environment were obtained for founder strains and DO mice (see Figure 2). When examining the values among founder strains, we observed significant differences between mice for both observed (p = 3.49 × 10−5) or rank-based inverse normalized (p = 2.11 × 10−12) values; one CAST mouse was an apparent outlier, with an extremely long sleep latency of 835.3 min. On average, we observed higher sleep latencies in a new environment in the B6, CAST, and WSB strains, intermediate values in the NOD, NZO, and PWK strains, and the lowest average values among the 129 and AJ mice. When assessing the distribution in the DO mouse population, mean sleep latency was 168.8 ± 69.4 min, with a wide range from 0.83 to 683.7 min.
Figure 2.
Sleep latency in a new environment and modified Murine Multiple Sleep Latency Test phenotypes in founder strains and DO mice. Estimates of latency to sleep in a new environment and the slope of latency to sleep across m-MMSLT trials are illustrated within the founder and DO mice. Vertical error bars represent the observed mean ± standard deviation. For sleep latency in a new environment, significant differences were seen among founders (N = 60) on both the observed scale (p = 3.49 × 10−5; top panel) and after rank-based inverse normalization to correct for outliers in the distributions (p = 2.11 × 10−12; middle panel). In general, longer sleep latency is observed for the B6, CAST, and WSB strains, while the shortest sleep latency was seen in the 129 strain. Values in DO mice (N = 338) span the full range seen in the founder strains. We also observed significant differences among the founder strains (N = 60) for the slope of latency to sleep across m-MMSLT trials (p = .0021; bottom panel). In general, the CAST and PWK strain exhibit slope values close to zero (indicating relative resistance to increased sleep deprivation), whereas other founders demonstrated negative slopes (shorter latency to sleep for later trials). We observe broad variability among DO mice (N = 174), with a negative slope on average, but values ranging from −6.59 (indicating high sleep drive) to 6.34 (indicating longer latency to sleep over trials).
Modified Murine Multiple Sleep Latency Test (m-MMSLT)
To assess sleep drive under increasing amount of sleep deprivation, we compared the slope of change in sleep latency across m-MMSLT trials within founder strains and DO mice (see Figure 2). There was a significant difference in the m-MMSLT slope among founder strains (p = .0021), with the CAST (0.44 ± 1.56) and PWK (0.54 ± 2.74) strains demonstrating average slopes close to zero (indicating relative resistance to increasing sleep deprivation), while all other strains showed negative slopes ranging from −1.97 to −2.86, indicating shorter latency to sleep with increasing sleep deprivation. We observed wide variability in this measurement among the DO mice, with a mean slope across m-MMSLT trials of −1.39 ± 2.39 and values ranging from −6.59 (indicating high sleep drive) to 6.34 (indicating increasing sleep latency over trials). Overall, 79.9% of DO mice tested had decreasing latency across trials (i.e. negative slopes).
Circadian period
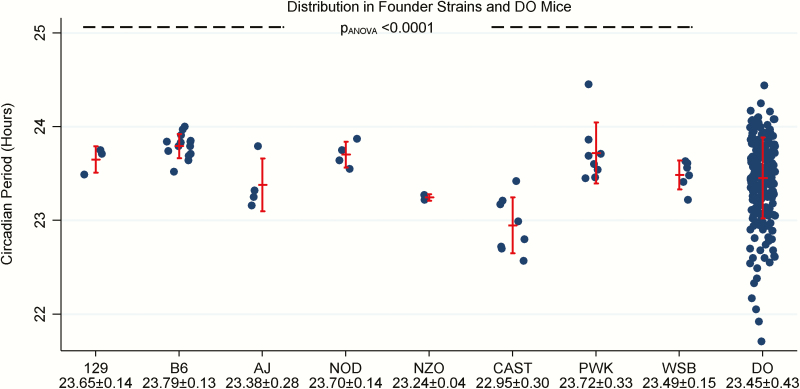
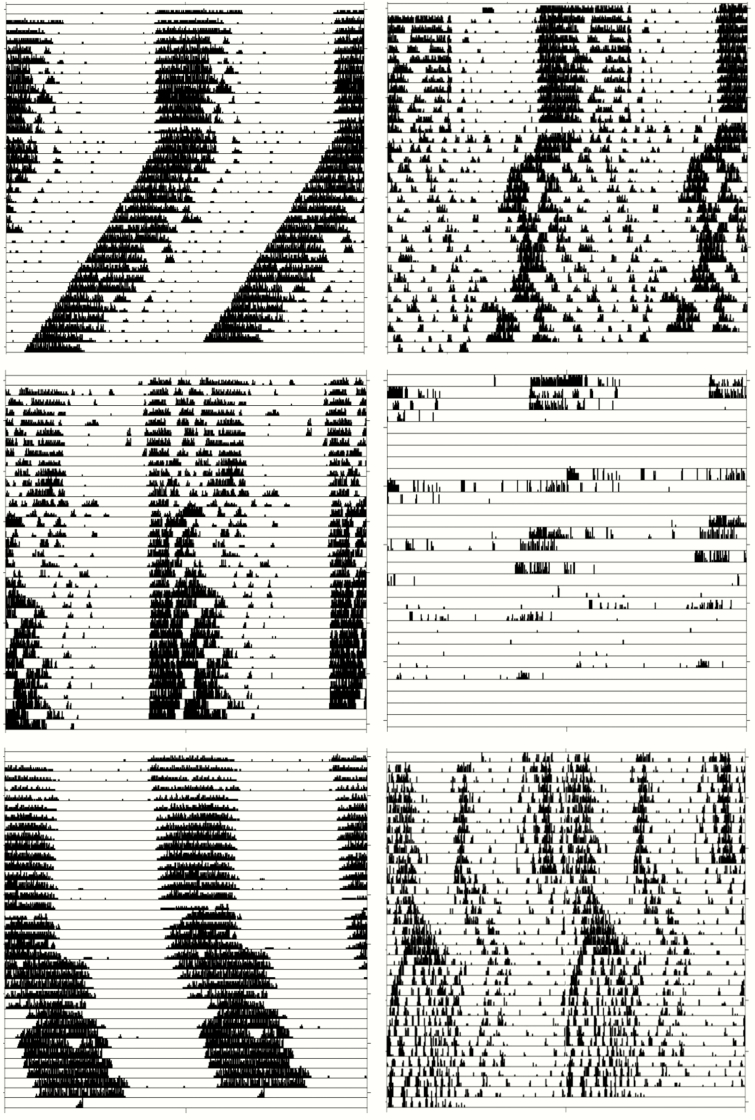
The circadian period was examined during 18–20 days of constant darkness (see Figure 3). The circadian period was quantifiable in 49 of 58 founder mice studied (84%), compared to 228 of 332 DO mice (69%) that underwent the wheel running paradigm. In mice where a circadian period could not be determined, mice either showed no clear circadian period in wheel running or had a general lack of running on the wheel (see Figure 4 for examples of data from wheel running experiments in individual mice). Among the founder mice, average values of the circadian period ranged from 22.95 to 23.79 h and there were differences among the strains (p = 1.52 × 10−8). The wild-derived CAST mice exhibited the shortest average period of 22.95 ± 0.30 h. Among the DO mice with available data, we observed a mean period of 23.45 ± 0.43 h, with values covering and exceeding the spread observed in the founder strains (range from 21.71 to 24.44 h), including some mice with notably short circadian periods (<22.5 h).
Figure 3.
Circadian period in founder strains and DO mice. The circadian period is shown in the founder strains (N = 49) and DO mice (N = 228). Vertical error bars represent the observed mean ± standard deviation. Among these mice with a measurable circadian period, we found significant differences across the founder strains (p = 1.52 × 10−8), with the CAST strain exhibiting the shortest period and several strains observed to have a period closer to 24 h. Among the DO mice, the average circadian period was 23.5 ± 0.4 h, with a range of 21.7 to 24.4 h. Thus, the circadian phenotype in the DO population fully covers and extends beyond the spread observed in the founders.
Figure 4.
Examples of data from circadian wheel-running experiments. Examples of data obtained from wheel running experiments utilized to estimate circadian period among the DO mice, including scenarios where the period was and was not able to be estimated. Each row represents a double-plotted day of wheel running activity, used to estimate circadian period. Panels on the left illustrate animals with valid estimates of the circadian period either <24 h (top left), =24 h (middle left), or > 24 h (bottom left). Panels on the right illustrate scenarios where we are unable to estimate the circadian period due to non-consolidated activity (top right), lack of sufficient wheel running (middle right), or a combination of both factors (bottom right). These plots illustrate the broad diversity in the circadian period and wheel-running behavior in the DO mouse population.
Phenotype variability (standard deviation)
Finally, for sleep and wake characteristics we calculated the standard deviation of the measurements across 5 days of recording within each individual mouse. Detailed results are presented in Supplementary material, File 1.
For baseline measures of sleep and wake over the entire 24 h, there were significant differences in within-animal standard deviation among founder strains for total sleep (p = .004), number of sleep bouts (p = .018), and average sleep bout (p = .029) and wake bout (p = .0002) duration. The PWK strain demonstrated the highest average SD for total sleep time (69.0 ± 46.7 min), with all other founder strains showing similar SD between 23.1–39.4 min. Similarly, the PWK showed the largest SD in a number of sleep bouts (35.0 ± 8.5 bouts) and average sleep bout duration (0.63 ± 0.33 min), as well as the second-highest SD in average wake bout duration (0.59 ± 0.35 min), just below the CAST (0.68 ± 0.35 min). Results were similar when evaluating variability in sleep characteristics in lights-on and lights-off, except for no significant difference across founder strains for either total sleep in lights-on (p = .201) or average wake bout duration during lights-on (p = .167). PWK again showed greater variability for most measurements, except the number of sleep bouts during lights-off, where AJ showed a higher SD (24.2 ± 11.3 min) than other strains. As with other phenotypes, the DO mice showed a wide range in SD values across traits, with some mice exhibiting SD values extending well above those in founders (see Supplementary material, File 1).
Robustness of inactivity-based sleep definitions
We evaluated the robustness of sleep and wake traits, sleep latency in a new environment, and slope of latency to sleep across m-MMSLT trials to alternative definitions of sleep using different durations of inactivity.
Sleep and wake characteristics
To examine the robustness of sleep phenotypes to alternative inactivity definitions, we conducted sensitivity analyses altering the definition of sleep to 20, 60, and 80 s of inactivity in a sample of 24 sequentially tested DO mice and all founder mice. Average estimates of each trait across different definitions are shown in Supplementary Table S1. The mean estimates of total sleep demonstrate expected changes using the different definitions—there is more sleep using a definition of at least 20 s and less sleep with a definition of at least 80 s. All estimates were highly correlated during 24-h (Spearman’s rho ≥ 0.97), lights-on (Spearman’s rho ≥ 0.97) and lights-off (Spearman’s rho ≥ 0.97), indicating that the different definitions give consistent relative estimates of total sleep, with the rank order of mice from the most to the least sleep remaining nearly identical across definitions (see Supplementary Figure S3).
We also examined how the number of sleep bouts and estimates of average sleep and wake bout durations changed based on the duration of inactivity used to define sleep (see Supplementary Table S1 and Figure S3). We again observed the expected differences in average values, with fewer bouts and longer bout durations as we increased the duration of inactivity for defining sleep–wake (see Supplementary Table S1). There were also high pairwise correlations compared to the primary 40-s rule. Specifically, for the number of sleep bouts, Spearman’s correlations were at least 0.92 during 24-h, at least 0.88 during lights-on, and at least 0.92 during lights-off when comparing alternative definitions to the 40-s rule. For sleep and wake bout duration, correlations with the 40-s definition were at least 0.92 over 24-h, at least 0.94 during lights-on, and at least 0.89 during lights-off. Correlations remained reasonably high when comparing more extreme differences in the rules, with the lowest observed Spearman correlation equal to 0.69 between the number of sleep bouts during lights-on defined using 20 or 80 s. Thus, rankings are robust across definitions, particularly in comparison to the primary sleep definition.
Sleep latency in a new environment
Next, we evaluated the robustness of latency to sleep onset in a new environment to alternative definitions of the first consolidated sleep bout as 2, 2.5, 3.5, and 4 min of inactivity; the primary definition based on comparison to EEG was 3 min of inactivity. As the definition of the first sleep bout increases, we see expected increases in sleep latency, ranging from 160.7 ± 79.8 min based on 2 min of inactivity to 172.0 ± 80.6 min based on 4 min of inactivity. As shown in Supplementary Figure S4, there is a high correlation across definitions, with Spearman’s rank correlations of at least 0.93 for all measurements. Thus, the ordering of animals with respect to sleep latency in a new environment is insensitive to the chosen definition within this range.
Modified Murine Multiple Sleep Latency Test
As with sleep latency in a new environment, we evaluated the robustness of the latency to sleep slope across m-MMSLT trials using definitions of 2, 2.5, 3 (primary), 3.5, and 4 min of inactivity. Average slope values became more positive as the sleep bout definition increased, with values of −1.65 ± 2.50 for 2 min, −1.52 ± 2.38 for 2.5 min, −1.42 ± 2.33 for 3 min, −1.35 ± 2.33 for 3.5 min, and −1.31 ± 2.27 for 4 min of inactivity. In all cases, the slope remains negative on average, demonstrating the expected shorter latency to sleep with increasing sleep deprivation. As shown in Supplementary Figure S4, we observed high correlations among measures with alternative definitions. Compared to the primary definition of 3 min of inactivity, all Spearman rank correlations are at least 0.91. For more extreme comparisons, the smallest correlation was equal to 0.83 when comparing the 2- and 4-min definitions. Thus, the relative ordering of the data remains robust.
Reliability of high-throughput phenotypes
In addition to evaluating alternative definitions of sleep, we evaluated the reliability of measured sleep traits across repeated measurements in DO mice. Reliability of sleep and wake characteristics were evaluated across multiple days of recording, while the reliability of latency to sleep in a new environment and slope across m-MMSLT trials was assessed by repeat experiments in subsets of DO mice. Results for all measures assessed are shown in Figure 5.
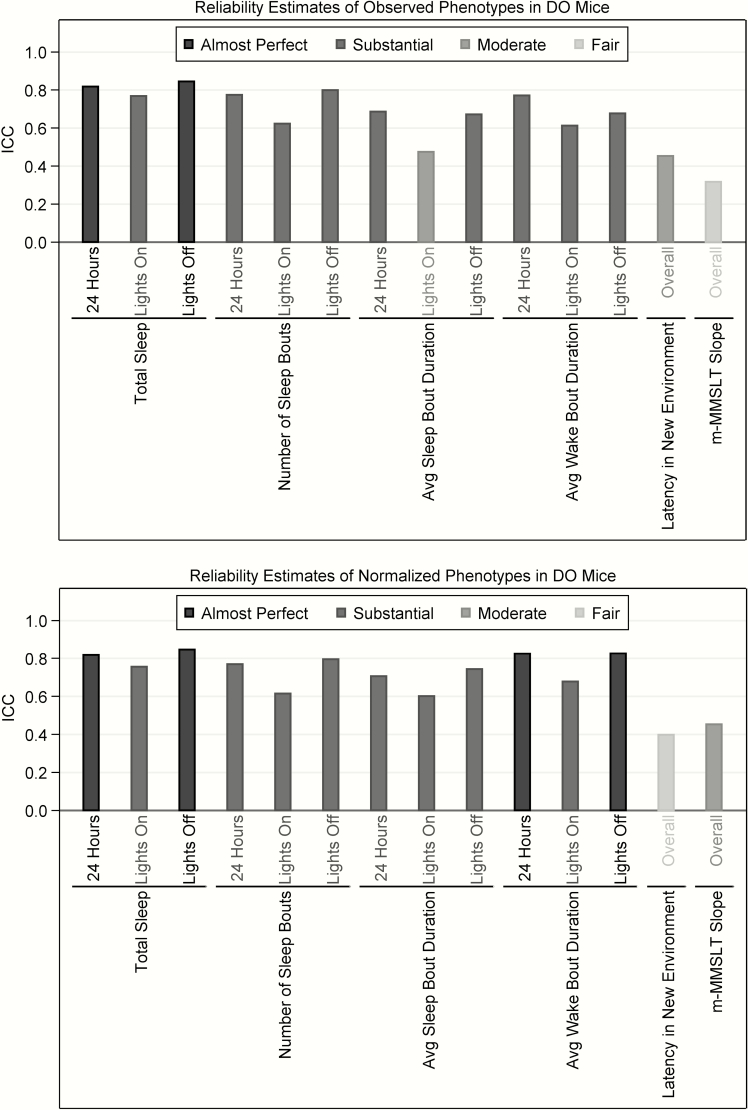
Figure 5.
Reliability of observed and normalized phenotypes in DO mice. Estimates of reliability are illustrated based on repeated measurements of sleep and wake characteristics, sleep latency in a new environment, and the slope across m-MMSLT trials. Reliability can be interpreted based on ranges of ICC values as almost perfect (ICC = 0.80–1.00), substantial (0.60–0.80), moderate (0.40–0.60), fair (0.20–0.40), slight (0.00–0.20), or poor (<0.00). We find that nearly all phenotypes are at least substantially reliable, apart from moderate reliability of average sleep bout duration in lights-on and sleep latency in a new environment, as well as fair reliability for the slope across m-MMSLT trials. Similarly, after rank-based inverse normalization nearly all phenotypes are at least substantially reliable, apart from moderate reliability of latency to sleep slope across m-MMSLT trials and fair reliability for sleep latency in a new environment.
Sleep and wake characteristics
Sleep and wake parameters were estimated across 5 days, which allowed us to evaluate reliability in the DO mice. Across 24 h, all measurements of sleep amounts, bout numbers and bout durations were at least substantially reliable (ICCs ranging from 0.686 to 0.819). When comparing lights-on and lights-off separately, similar results were observed. During lights-on, all traits were substantially reliable (ICCs ranging from 0.613 to 0.769), except average sleep bout duration, which showed moderate reliability on the observed scale (ICC = 0.475), but reliability in the substantial range after rank-based inverse normalization (ICC = 0.601). During the lights-off period, all phenotypes showed at least substantial reliability (ICCs ranging from 0.672 to 0.847).
Sleep latency in a new environment
Repeated new environment tests were conducted for a set of 24 DO mice that were sequentially tested to assess reliability. Sleep latency in a new environment was moderately reliable with an ICC (95% CI) of 0.453 (0.076, 0.719) on the observed scale and at the upper end of the fairly reliable range with an ICC of 0.398 (0.009, 0.684) after normalization. This lower estimate of reliability is likely partially confounded by differences in stress between the first and second days of the experiment, as nearly all latency estimates (21 of 24 [87.5%]) in the second experiment were shorter than those in the first experiment (median [range] difference = −37.8 [−133.5, 4.67] min; see Supplementary Figure S5). Thus, there is moderate reliability of the latency to sleep in a new environment.
Modified Murine Multiple Sleep Latency Test
Similarly, we evaluated the reliability of the m-MMSLT data by repeating the experiment within a set of 24 sequentially tested DO mice. Results demonstrated fair reliability on the observed scale, with an ICC of 0.317 (−0.084, 0.632), but moderate reliability after normalization (ICC [95% CI] = 0.453 [0.076, 0.719]). Unlike data on sleep latency in a new environment, we observed no meaningful difference between the values on the two repeated runs (median [range] difference = 0.00 [−4.02, 4.77]). Thus, there is evidence of fair to moderate reliability of the slope of the latency to sleep across m-MMSLT trials in DO mice.
Heritability of high-throughput phenotypes
To estimate the proportion of phenotypic variance explained by genetic factors, we calculated the heritability (and associated 95% CIs) for each trait within the founder strains and DO mice, separately. Given differences in the underlying genetic models, calculations in the founder mice can be interpreted as broad-sense heritability (H2), which estimates the proportion of phenotypic variability due to all underlying genetic factors (e.g. additive and dominance single gene effects and epistasis), whereas estimates in DO mice correspond to narrow-sense heritability (h2), which estimates the proportion of phenotypic variability due to additive genetic effects only. Heritability estimates for which the 95% CI did not include 0% were considered significant. Overall, we observe lower or nonsignificant estimates of narrow-sense heritability in the DO mice, compared to significant broad-sense heritability in founders (Figure 6 and Supplementary Figure S6 and Supplementary material, File 1); differences in these estimates suggest important non-additive genetic effects.
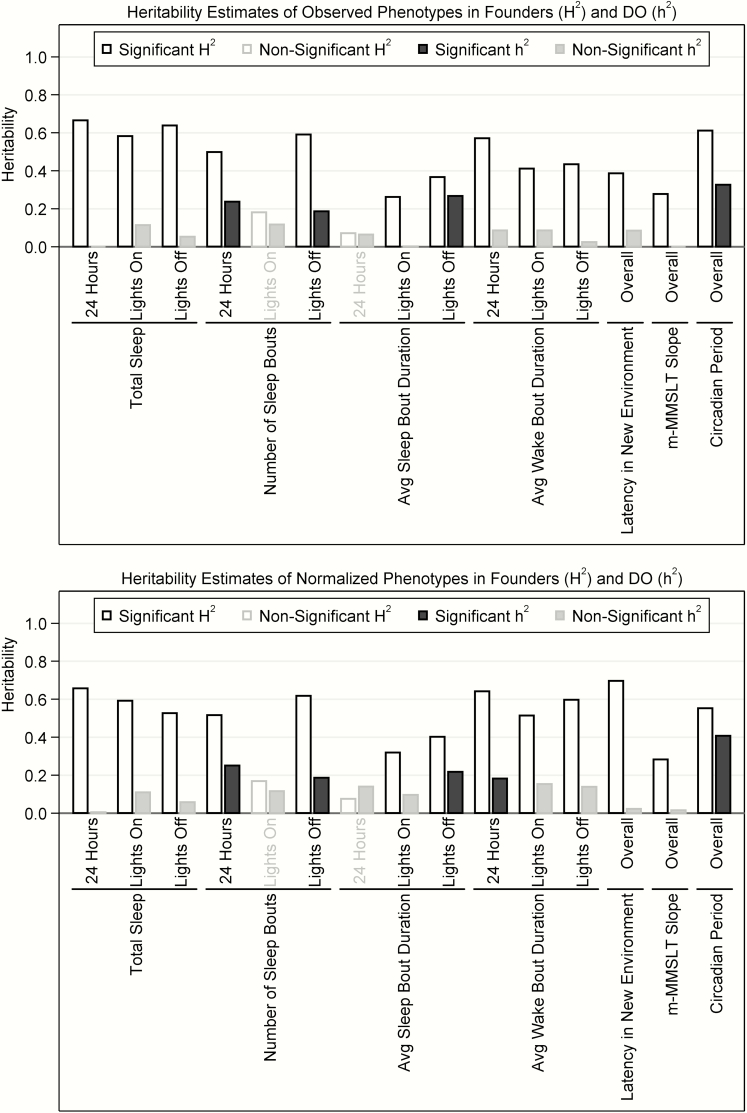
Figure 6.
Heritability of observed and normalized phenotypes in founder strains and DO mice. The estimated broad-sense (H2) and narrow-sense (h2) heritability of measured phenotypes within the founder and DO mice are illustrated for traits on both the observed (top panel) and rank-based inverse normal transformed (bottom panel) scales. Heritability estimates with 95% CIs that did not include 0 were considered significant and are shown in black (hollow bars for H2 or solid bars for h2), while estimates where the CI included 0 are considered non-significant and presented in gray. Overall, considerably larger estimates of heritability were observed within the founder mice compared to the DO mice. This difference in broad-sense (within founders) and narrow-sense (within DO mice) heritability likely reflects the importance of non-additive genetic effects in determining the variability in measured sleep and circadian traits.
Sleep and wake characteristics
For estimates of total sleep over 24-h and during lights-on and lights-off, results among the founders showed high heritability, indicating that 66.5% (95% CI: 51.4%, 81.6%) of variance in total sleep during the entire day, 58.3% (41.5%, 75.1%) of variability in total sleep during lights-on and 63.9% (46.5%, 81.3%) of variability in lights-off is explained by genetic differences. However, among the DO mice, we did not observe significant heritability for total sleep in any of the time periods, with h2 estimates between 0% and 11.4% and all 95% CIs including 0%.
Significant heritability was seen for some measures of sleep–wake microarchitecture in both the founder and DO mice, although higher heritability was again seen among founders. In particular, during lights-on we estimated that 26.2% (5.3%, 47.0%) of variability in sleep bout duration and 41.1% (18.8%, 63.5%) of variability in wake bout duration was explained by genetic factors in founder mice. In lights-off, H2 was increased slightly to 36.7% (15.8%, 57.6%) for sleep bout duration and 43.5% (22.1%, 64.8%) for wake bout duration. Interestingly, the significant H2 for average wake bout duration (57.2% [41.9%, 72.4%]), but not average sleep bout duration (7.1% [0.0%, 26.8%]), was maintained over the entire 24 h. There was considerably higher heritability for the number of sleep bouts during lights-off (59.1% [43.4%, 74.8%]), compared to non-significant estimates during lights-on (18.1% [0.0%, 37.2%]). In this case, heritability was also observed when summarizing over the entire 24-h period (49.9% [32.5%, 67.3%]). Among the DO mice, significant narrow-sense heritability was observed for the sleep bout duration during lights-off (26.7% [9.3%, 51.0%]), as well as the number of sleep bouts during both lights-off (18.6% [1.6%, 41.1%]) and 24 h (23.8% [4.3%, 47.7%]). No measures during the lights-on period had significant h2 estimates in DO mice (see Figure 6). Similar results were seen for rank-based inverse normalized phenotypes (Figure 6); a significant h2 emerged for normalized wake bout duration during 24 h in DO mice (18.3% [1.1%, 40.6%]). Thus, sleep–wake bout characteristics show evidence of heritability in founder mice, with a few modest heritability estimates among DO mice, mainly in lights-off. This likely highlights the importance of complex genetic influences on variability in sleep microarchitecture.
Sleep latency in a new environment
We next examined the evidence for heritability of latency to sleep in a new environment. There was significant broad-sense heritability for this trait among the founders. Specifically, genetic factors were estimated to explain 38.6% (6.1%, 71.2%) of variability in latency to sleep based on observed data, which increased to an H2 (95% CI) of 69.7% (59.8%, 79.6%) after rank-based inverse normalization to account for a large outlier observed in the CAST mice. Among the DO mice, we observed a non-significant heritability estimate of 8.5% (0.0%, 26.5%) that decreased to 2.3% (0.0%, 18.9%) for the normalized phenotype. Thus, results again support complex genetic influences on sleep latency in a new environment.
Modified Murine Multiple Sleep Latency Test
Next, we examined the heritability of the slope of latency to sleep across m-MMSLT trials. Consistent with prior phenotypes, we observed significant heritability among founders for the observed (27.8% [2.5%, 53.1%]) and rank-based inverse normalized (28.3% [4.6%, 52.0%]) phenotype. However, no evidence of narrow-sense heritability was observed for this trait among the DO population.
Circadian period
The circadian period was one of the most heritable traits examined across both founder and DO mouse populations. In particular, the circadian period showed a heritability of 61.2% (43.8%, 78.6%) among the founder mice and a heritability of 32.6% (5.4%, 71.3%) among the DO mice. Estimates were generally similar for rank-based inverse normalized data, with a slight decrease among founders (55.3% [40.7%, 69.9%]) and an increase among DO mice (40.8% [12.7%, 84.7%]). Thus, there is strong evidence that both additive and nonadditive genetic factors significantly influence variability in the circadian period.
Phenotype variability (standard deviation)
Finally, we evaluated genetic influences on within animal day-to-day variability (standard deviation) of baseline sleep characteristics over 5 days of recording. In general, lower heritability estimates were seen for these measures in both founders and DO mice, suggesting an increased role of nongenetic influences on day-to-day variability (Supplementary Figure S6).
More specifically, no significant heritability was observed for SD of total sleep time in either founders or DO mice. For sleep microarchitecture, significant broad-sense heritability was observed for variability of the number of sleep bouts during lights-on (H2 [95% CI] = 33.4% [7.9%, 58.9%]), average sleep bout duration in lights-on (23.7% [3.7%, 43.7%]) and lights-off (25.6% [3.6%, 47.6%]), and average wake bout duration during lights-off (33.5% [10.2%, 56.8%]) and summarized over 24 h (36.1% [15.7%, 56.6%]). Among the DO mice, only the variability in average sleep bout duration over 24 h demonstrated significant heritability (h2 [95% CI] = 16.4% [0.3%, 37.5%]).
Discussion
We have described a comprehensive, high-throughput, and noninvasive approach to assessing multiple phenotypes related to sleep and circadian behavior in mice. We have shown that there are highly significant differences among the eight founder strains of the Collaborative Cross [21, 23] and DO mice [18–20] in nearly all the phenotypes assessed. Importantly, our study also demonstrated the robustness and reliability of these phenotypes in DO mice. Furthermore, using data in the eight founder strains, we have shown the measured phenotypes have evidence of significant broad-sense heritability. While estimates of narrow-sense heritability calculated in DO mice were lower, significant heritability was still observed for multiple traits. In particular, the circadian period demonstrated the largest heritability in DO mice, and was one of the more heritable traits in founders; thus, it appears both additive and non-additive genetic effects play a role in explaining variance. Ultimately, larger heritability estimates in founders than DO mice suggest that non-additive genetic effects (e.g. dominance and epistasis) play an important role in explaining phenotypic variability, as may be expected given the complex nature of the traits. Overall, our results indicate that genetic factors play a moderate role in determining variability in the high-throughput traits examined here; non-genetic factors may also play a role, although these differences are expected to be minimal given our experimental design.
Challenges and approaches to identifying genes affecting sleep and circadian behavior
Almost all aspects of sleep and circadian rhythm in humans are heritable, as previously reviewed [1, 2], including total sleep [4], response to sleep deprivation [5, 6], and timing of sleep [7–10]. Many common sleep disorders are also heritable [1, 2]. Moreover, sleep abnormalities are common in neurological and psychiatric disorders, suggesting possible pleiotropic genetic associations [2]. Thus, there are many opportunities for the discovery of genetic factors driving variation in sleep and circadian biology.
Studying sleep and circadian genetics in humans
Although there are many sleep and circadian phenotypes to study in humans, our current toolkit for uncovering gene variants underlying these quantitative phenotypes is limited. This is largely due to feasibility. Large genetic studies have identified gene variants using self-reported phenotypes to assess sleep duration and chronotype within datasets such as the UK Biobank and 23andMe [11–14]. However, self-report phenotypes are problematic because there are significant variations when compared to results from objective data [37–39], making subjective assessment of sleep a “noisy” phenotype that requires a very large number of subjects. Studies leveraging objective sleep data from accelerometry within subsets of these populations are emerging and have identified associated variants [15–17], but results are not always consistent with self-reported phenotypes. Ultimately, human studies that use objective in-depth or invasive phenotyping necessarily have smaller sample sizes given the increased burden in obtaining these endpoints. Moreover, human studies do not typically involve interventions to assess different aspects of the behavior, as we have described here. Due to these challenges in obtaining robust phenotypes, it will be difficult to conduct human studies with sufficient sample size to find gene variants associated with important sleep and circadian traits, such as response to sleep loss (i.e. sleep drive).
Drosophila and forward genetics approaches
Beyond human studies, various genetic discovery approaches have been applied in animal models to identify genes relevant to sleep and circadian traits. In Drosophila, unbiased forward genetics approaches have been particularly useful [3, 40]. For example, the screening of established mutants has identified multiple genes that affect amounts of sleep and wake [41, 42]. In recent analyses, Toda et al. performed an unbiased screen of over 12,000 Drosophila genetic lines to identify a single gene that induces sleep [43]. In addition to these large-scale genetics screens, Harbison et al. leveraged an artificial selection procedure—breeding short- and long-sleeping flies over multiple generations—to produce extreme short- and long-sleep phenotypes [44]. This approach identified 126 polymorphisms on 80 candidate genes that influence sleep in outbred fly populations [44]. This work has been extended to the creation of the Drosophila Sleep Inbred Panel (SIP), which allows evaluations of candidate polymorphisms across consistent genetic backgrounds [45]. In translating this forward genetics approach to mouse models, Funato et al. screened over 8,000 randomly mutagenized mice and identified only two dominant mutations affecting sleep and wake based on EEG/EMG recording [46]. As more recently described by Miyoshi et al., this number has been increased to approximately 10,000 mice, and approximately 10 heritable loci have been identified [47]. While these approaches are able to identify genes, forward genetics techniques are ultimately less practical in mice given their longer lifespan compared to Drosophila, as well as the labor-intensive and costly nature of obtaining EEG/EMG recording and scoring in many thousands of animals [47]. Instead, analyses have typically relied on classical approaches within inbred mouse strains [31, 48, 49].
Genetic analyses within inbred and outbred mice
Total sleep and wake, as well as the duration of different sleep stages, vary between inbred mouse strains [4]. Multiple quantitative trait loci (QTLs) have been associated with different amounts of sleep stages in the lights-on and lights-off periods [48, 49]. Although a number of QTLs has been identified, it has been difficult to identify the relevant genes within the QTL. For example, a QTL has been identified for sleep homeostasis (i.e. increase in delta power following sleep deprivation) using recombinant inbred mice [6]. However, the identified QTL region contained over 200 genes. Thus, determining the causative gene variants remains challenging. Informatics examination of the QTL region suggested that Homer1a might be the responsible gene [50], although studies of Homer1a knockout mice did not exhibit differences in sleep homeostasis [51]. Instead, mice lacking Homer1a exhibited an inability to sustain long bouts of wakefulness in the lights-off period when compared to wild-type mice [51].
Thus, a clear limitation of analyses using inbred mice is that the broad genomic regions identified by linkage peaks, encompassing hundreds of genes, make it difficult to identify the specific gene(s) responsible for a phenotype. DO mice provide a key benefit in this regard. As a result of increased recombination and smaller haplotype blocks through outbreeding, the DO mice allow the identification of smaller QTL regions that may contain only a few genes (rather than hundreds) [18–20]. The utility of analyses in DO mice is evidenced by a number of examples of genetic factors being identified using this approach [18, 19, 52–59]. Thus, it seems reasonable to propose that finding genes underlying sleep and circadian phenotypes using DO mice is likely to be a productive strategy. It requires, however, phenotyping of hundreds of mice to provide sufficient statistical power. Thus, the ideal phenotyping strategy needs to be high-throughput, as with the approach described here.
Robust, reliable and heritable high-throughput sleep and circadian traits
To establish the utility of the high-throughput phenotypes described here for future genetic analyses in DO mice, it is key to demonstrate their robustness and reliability, as well as show that underlying genetic factors are likely to influence trait variability.
Robustness
Our noninvasive, high-throughput phenotyping strategy for sleep and wake is based on activity monitoring and defining sleep as 40 s or more of continuous inactivity [24]. This definition has been validated previously in young [24] and old [26] B6 mice. It is, however, conceivable that for different inbred strains or DO mice an alternative definition of sleep could be more appropriate. To assess this, we evaluated the robustness of sleep and wake phenotypes using different durations of inactivity to define sleep (i.e. 20, 60, or 80 s). While, not surprisingly, the actual amounts of sleep changed across these definitions, the rank ordering of mice with respect to the different phenotypes remains consistent. Similarly, we validated an optimal inactivity-based definition of the first consolidated sleep bout by comparing data against EEG-defined sleep latency in an independent sample of B6 mice. Our optimal definition for latency to consolidated sleep was 3 min of inactivity; data on the sleep latency in a new environment and slope across m-MMSLT trials were again robust to deviations from this definition (ranging from 2 to 4 min). Thus, we believe that utilizing one specific duration of inactivity to defined sleep will not impede the discovery of gene variants using the DO mouse strategy. Ultimately, if specific gene variants are found for certain phenotypes, these can be assessed in replication studies using Collaborative Cross mice with the relevant alleles at the specific location identified, combined with gold-standard EEG/EMG recording to quantify the phenotype of interest.
Reliability
We have also shown that most phenotypes are reliable in DO mice. Analyses leveraged repeated experiments (for sleep latency in a new environment and m-MMSLT) or multiple days of recording (for sleep architecture). In general, nearly all measured sleep and wake phenotypes showed at least substantial reliability (ICC values of 0.6 and above). While all measures were reliable, we also found that reliability was higher in the lights-off period for the majority of measures. Thus, it is conceivable that when DO mice are used to identify novel gene variants, data from the lights-off period will prove more informative. In contrast, measurements during m-MMSLT and new environment testing showed fair/moderate reliability. Given that these latencies to sleep tests involve some amount of stress to the animals (either movement to a new environment or interaction with experimenters during sleep deprivation for m-MMSLT), this lower reliability likely reflects some interplay between true sleep latency and the influence of stress (which affects the ability to sleep). This was particularly evident in repeated testing of sleep latency in a new environment, where nearly all animals demonstrated faster latency to sleep on the second experiment compared to the first. Future studies should carefully consider the potential confounding effects of stress on these phenotypes, which may include the use of alternative experimental approaches to minimize this influence and increase the reliability of the traits.
Heritability
Importantly for discovering genetic associations, for most phenotypes assessed we observed significant broad-sense heritability, with 95% CIs that did not include zero. Estimates of broad-sense heritability (or the proportion of variability explained by additive, dominant, and epistasis genetic effects) were derived from data in the eight founder strains, while narrow-sense heritability (or the proportion of variability explained by additive genetic effects only) was calculated in DO mice. Thus, while the analysis methods differ, comparisons of the heritability values in the two mouse samples provide insight into potential genetic mechanisms of effect; larger broad-sense heritability suggests important non-additive genetic effects. Emphasizing the likely complex genetic influences on sleep and circadian phenotypes, heritability estimates in the founder strains were indeed larger and more often statistically significant than those obtained from the data from the DO mice. Despite these differences, we did find that both approaches gave robust estimates of heritability for a circadian period (61.2% in founders and 32.6% in DO mice). Thus, additive genetic effects may be particularly important for this trait. However, we could not assess the circadian period in all mice we studied, due to some mice either not wheel-running or having a disrupted circadian rhythm of wheel-running after switching to constant darkness. It will be of interest to determine whether DO mice can be used to identify novel clock genes despite this limitation. Interestingly, measures of sleep and wake microarchitecture showed more heritability in DO mice than total sleep–wake amounts; thus, these phenotypes may be more promising from a genetic standpoint. Ultimately, the development of new analysis approaches that can evaluate nonadditive genetic effects within DO mice will be particularly important.
While the narrow-sense heritability estimates of sleep architecture in DO mice were generally low compared to broad-sense heritability estimates, results were similar to those observed in large-scale human analyses performed in many thousands of individuals [12, 14–17]. The median narrow-sense (or additive) heritability estimate for sleep architecture measures in our DO mice was 8.5%, with values ranging from 0.0% to 26.7%. In recent human studies utilizing self-reported sleep duration, corresponding additive heritability estimates fall between 7.0% and 10.3% [12, 14, 17]. Similarly, recent studies using accelerometry have estimated additive heritability between 2.8% and 22.3% for sleep-related variables [16] and between 10% and 21% for sleep and activity behaviors [15]. Therefore, the modest evidence for additive genetic heritability observed in our sample of DO mice is supported by similar analyses in human samples that leverage genome-wide genotype data. It is likely that our comparatively small sample of DO mice is not sufficiently powered to detect significant narrow-sense heritability estimates in these lower ranges.
In addition to mean estimates, recent analyses in Drosophila have identified and validated multiple genes that influence day-to-day variability in sleep phenotypes [60]. Thus, we also examined the heritability of within-mouse standard deviations of sleep and wake phenotypes across multiple days of recording. While there was some evidence of significant broad-sense heritability for these traits, in general, there appeared to be a less genetic influence on day-to-day variability in both founders and DO mice when compared to the average values of traits themselves. Recent studies in humans using accelerometry have similarly suggested low heritability of the variability of sleep duration (h2 [95% CI] = 2.8% [2.0%, 3.6%]) [16]. Thus, additional studies may be required to determine the role of genetic influences on the day-to-day variability of sleep phenotypes in mice.
Strengths, limitations and future directions
Strengths of this study include the ability to perform reliable, non-invasive screening of a large number of DO mice, the inclusion of mice from each of the eight founder strains to quantify broad-sense heritability, the carefully designed phenotyping paradigms that minimize environmental variance, and the robust analysis methodologies.
There are also limitations. Our high-throughput phenotyping approach using beam-breaks does not allow assessment of stage-specific sleep characteristics (e.g. non-rapid eye movement [NREM] and rapid eye movement [REM]); video-based analysis coupled with machine learning approaches using simultaneous EEG is a promising future direction to capture these characteristics [61]. Despite prior validation of our inactivity definition against EEG/EMG recording [24, 26], estimates of total sleep obtained using our inactivity-based method are higher than external, strain-specific estimates using either EEG/EMG or a Piezoelectric system [31, 62]. While a number of factors could influence these differences, including environment or periods of quiet wakefulness scored as sleep, we have demonstrated that the specific inactivity definition does not influence the relative ordering of individual mice; thus, results of within and between strain comparisons are expected to be robust. Validation studies of genetic associations identified using the approaches described herein should be performed with gold-standard EEG/EMG in the Collaborative Cross lines carrying the implicated founder alleles at specific genetic locations to more accurately estimate effects on sleep amounts.
As in previous studies, analyses were performed only in male mice; future analyses should evaluate the performance and heritability of phenotypes in female mice. To understand any gender differences, genetic associations discovered in male DO mice should be validated using both male and female mice from the implicated Collaborative Crosslines, as well as male and female knock-out mice when appropriate. Analyses of the reliability of sleep and wake architecture utilized data from five consecutive days of recording. While results provide insight into the stability of the target phenotypes, future studies examining the reproducibility of these traits with repeated testing over a longer time period (e.g. 1 month apart) would be insightful. A recent study on EEG/EMG sleep parameters measured 4 weeks apart suggests that some, but not all, are correlated on first and second measurement [47]. Finally, the broad-sense heritability estimates derived in founder mice attribute differences between strains solely to genetic effects; it is possible that unknown non-genetic effects could influence differences and bias H2 estimates. However, the pipeline was designed to diminish environmental variability and, thus, we expect these non-genetic differences to be minimal.
Conclusions
We have implemented and assessed a high-throughput phenotyping strategy for evaluating different sleep and circadian phenotypes in mice. The majority of the phenotypes, we were assessed both reliable among DO mice and have evidence of broad-sense heritability in founder strains. Comparatively, fewer traits demonstrated narrow-sense (or additive) heritability in DO mice, underscoring the importance of complex, nonadditive genetic effects for determining variability in these traits. Ultimately, DO mice have unique advantages when compared to other model systems. The genetic complexity created by random outbreeding results in clear phenotypic heterogeneity among DO mice, with phenotype values extending beyond ranges observed in the original founder strains. This heterogeneity is more akin to that seen in human population studies, and increases the likelihood of large genetic effects. Importantly, the increased recombination in DO mice also allows the identification of narrow genetic linkage regions, facilitating identification of individual genes affecting traits of interest. Thus, when combined with appropriate analysis approaches and large enough samples, this strategy is likely to lead to the identification of novel sleep and circadian genes.
Supplementary Material
Funding
The work described herein was supported by the National Institutes of Health (NIH) grants R01 HL111725, R01 GM070683 and P01 HL094307.
Conflict of interest statement. Dr. Pack is The John L. Miclot Professor of Medicine at the University of Pennsylvania; funds for this endowment were provided by the Philips Respironics Foundation. Dr. Churchill, Dr. Svenson, Dr. Gatti and Dr. Simecek disclose that The Jackson Laboratory produces and sells a wide variety of mice for research, including Diversity Outbred mice. All other authors report no financial conflicts of interest related to the present manuscript. Authors report no non-financial conflicts of interest related to the present manuscript.
Author’s contributions
Design: GC, KLS, AIP; Mouse phenotyping: LZ, JL, RJG; Statistical analysis: BTK, PS, DG; Writing: BTK, RJG, DCL, KLS, GC, AIP.
References
- 1. Gehrman PR, et al. Genetics of sleep disorders. Psychiatr Clin North Am. 2015;38(4):667–681. [DOI] [PubMed] [Google Scholar]
- 2. Veatch OJ, et al. Pleiotropic genetic effects influencing sleep and neurological disorders. Lancet Neurol. 2017;16(2):158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci. 2009;10(8):549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tafti M, et al. Quantitative trait loci approach to the genetics of sleep in recombinant inbred mice. J Sleep Res. 1999;8(Suppl 1):37–43. [DOI] [PubMed] [Google Scholar]
- 5. Kuna ST, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35(9):1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franken P, et al. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21(8):2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aguiar GF, et al. Patterns of daily allocation of sleep periods: a case study in an Amazonian riverine community. Chronobiologia. 1991;18(1):9–19. [PubMed] [Google Scholar]
- 8. Hur Y-M, et al. Genetic and environmental influence on morningness-eveningness. Pers Individ Dif. 1998;25(5):917–925. [Google Scholar]
- 9. Vink JM, et al. Genetic analysis of morningness and eveningness. Chronobiol Int. 2001;18(5):809–822. [DOI] [PubMed] [Google Scholar]
- 10. Klei L, et al. Heritability of morningness-eveningness and self-report sleep measures in a family-based sample of 521 hutterites. Chronobiol Int. 2005;22(6):1041–1054. [DOI] [PubMed] [Google Scholar]
- 11. Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doherty A, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones SE, et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun. 2019;10(1):1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Churchill GA, et al. The diversity outbred mouse population. Mamm Genome. 2012;23(9–10):713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Svenson KL, et al. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190(2):437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gatti DM, et al. Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda). 2014;4(9):1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Churchill GA, et al. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–1137. [DOI] [PubMed] [Google Scholar]
- 22. Collaborative Cross Consortium. The genome architecture of the collaborative cross mouse genetic reference population. Genetics. 2012;190(2):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava A, et al. Genomes of the mouse collaborative cross. Genetics. 2017;206(2):537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pack AI, et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28(2):232–238. [DOI] [PubMed] [Google Scholar]
- 25. Mang GM, et al. Evaluation of a piezoelectric system as an alternative to electroencephalogram/electromyogram recordings in mouse sleep studies. Sleep. 2014;37(8):1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Naidoo N, et al. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neurosci. 2008;28(26):6539–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikonova EV, et al. Transcriptional profiling of cholinergic neurons from basal forebrain identifies changes in expression of genes between sleep and wake. Sleep. 2017;40(6). doi:10.1093/sleep/zsx059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parmentier R, et al. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci. 2002;22(17):7695–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Veasey SC, et al. Murine multiple sleep latency test: phenotyping sleep propensity in mice. Sleep. 2004;27(3):388–393. [DOI] [PubMed] [Google Scholar]
- 30. Morgan AP, et al. The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda). 2015;6(2):263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franken P, et al. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22(2):155–169. [PubMed] [Google Scholar]
- 32. Lomb LR. Least-squares frequency analysis of unequally spaced data. Astrophys Space Sci. 1976;39:447–462. [Google Scholar]
- 33. Zielinski T, et al. Strengths and limitations of period estimation methods for circadian data. PLoS One. 2014;9(5):e96462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Landis JR, et al. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 35. Schweiger R, et al. Fast and accurate construction of confidence intervals for heritability. Am J Hum Genet. 2016;98(6):1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Genz A, et al. Computation of Multivariate Normal and t Probabilities. Dordrecht, the Netherlands: Springer; 2009. [Google Scholar]
- 37. McCall WV, et al. Subjective estimates of sleep differ from polysomnographic measurements in obstructive sleep apnea patients. Sleep. 1995;18(8):646–650. [DOI] [PubMed] [Google Scholar]
- 38. Harvey AG, et al. (Mis)perception of sleep in insomnia: a puzzle and a resolution. Psychol Bull. 2012;138(1):77–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castillo J, et al. Sleep-wake misperception in sleep apnea patients undergoing diagnostic versus titration polysomnography. J Psychosom Res. 2014;76(5):361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Axelrod S, et al. Studying circadian rhythm and sleep using genetic screens in Drosophila. Methods Enzymol. 2015;551:3–27. [DOI] [PubMed] [Google Scholar]
- 41. Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434(7037):1087–1092. [DOI] [PubMed] [Google Scholar]
- 42. Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321(5887):372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toda H, et al. A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila. Science. 2019;363(6426):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harbison ST, et al. Selection for long and short sleep duration in Drosophila melanogaster reveals the complex genetic network underlying natural variation in sleep. PLoS Genet. 2017;13(12):e1007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Serrano Negron YL, et al. The sleep inbred panel, a collection of inbred drosophila melanogaster with extreme long and short sleep duration. G3 (Bethesda). 2018;8(9):2865–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Funato H, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539(7629):378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyoshi C, et al. Methodology and theoretical basis of forward genetic screening for sleep/wakefulness in mice. Proc Natl Acad Sci USA. 2019;116(32):16062–16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tafti M. Quantitative genetics of sleep in inbred mice. Dialogues Clin Neurosci. 2007;9(3):273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Summa KC, et al. The genetics of sleep: insight from rodent models. Sleep Med Clin. 2011;6(2):141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mackiewicz M, et al. Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol Genomics. 2008;33(1):91–99. [DOI] [PubMed] [Google Scholar]
- 51. Naidoo N, et al. Role of Homer proteins in the maintenance of sleep-wake states. PLoS One. 2012;7(4):e35174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Logan RW, et al. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013;12(4):424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Recla JM, et al. Precise genetic mapping and integrative bioinformatics in diversity outbred mice reveals hydin as a novel pain gene. Mamm Genome. 2014;25(5–6):211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smallwood TL, et al. High-resolution genetic mapping in the diversity outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3 (Bethesda). 2014;4(12):2353–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. French JE, et al. Diversity outbred mice identify population-based exposure thresholds and genetic factors that influence benzene-induced genotoxicity. Environ Health Perspect. 2015;123(3):237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tyler AL, et al. Epistatic networks jointly influence phenotypes related to metabolic disease and gene expression in diversity outbred mice. Genetics. 2017;206(2):621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yuan JT, et al. Genome-wide association for testis weight in the diversity outbred mouse population. Mamm Genome. 2018;29(5–6):310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shorter JR, et al. Quantitative trait mapping in diversity outbred mice identifies two genomic regions associated with heart size. Mamm Genome. 2018;29(1–2):80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gatti DM, et al. Genetic background influences susceptibility to chemotherapy-induced hematotoxicity. Pharmacogenomics J. 2018;18(2):319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu KJ, et al. Genotype influences day-to-day variability in sleep in Drosophila melanogaster. Sleep. 2017;41(2). doi:10.1093/sleep/zsx205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McShane BB, et al. Assessing REM sleep in mice using video analysis. Sleep. 2012;35(3):433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bogue MA, et al. Mouse phenome database: an integrative database and analysis suite for curated empirical phenotype data from laboratory mice. Nucleic Acids Res. 2018;46(D1):D843–D850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.