Abstract
Multiple systems atrophy (MSA) is a rare neurodegenerative disorder characterized by the accumulation of α-synuclein in glial cells and neurodegeneration in the striatum, substantia nigra, and cerebellum. Aberrant miRNA regulation has been associated with neurodegeneration, including alterations of specific miRNAs in brain tissue, serum, and cerebrospinal fluid from MSA patients. Still, a causal link between deregulation of miRNA networks and pathological changes in the transcriptome remains elusive. We profiled ~ 800 miRNAs in the striatum of MSA patients in comparison to healthy individuals to identify specific miRNAs altered in MSA. In addition, we performed a parallel screening of 700 transcripts associated with neurodegeneration to determine the impact of miRNA deregulation on the transcriptome. We identified 60 miRNAs with abnormal levels in MSA brains that are involved in extracellular matrix receptor interactions, prion disease, inflammation, ubiquitin-mediated proteolysis, and addiction pathways. Using the correlation between miRNA expression and the abundance of their known targets, miR-124–3p, miR-19a-3p, miR-27b-3p, and miR-29c-3p were identified as key regulators altered in MSA, mainly contributing to neuroinflammation. Finally, our study also uncovered a potential link between Alzheimer’s disease (AD) and MSA pathologies that involves miRNAs and deregulation of BACE1. Our results provide a comprehensive appraisal of miRNA alterations in MSA and their effect on the striatal transcriptome, supporting that aberrant miRNA expression is highly correlated with changes in gene transcription associated with MSA neuropathology, in particular those driving inflammation, disrupting myelination, and potentially impacting α-synuclein accumulation via deregulation of autophagy and prion mechanisms.
Keywords: Multiple systems atrophy, microRNA, Alpha-synuclein, Neurodegeneration, Inflammation, Transcription
Background
Multiple system atrophy (MSA) is a rare fatal neurodegenerative movement disorder characterized by accumulation of alpha-synuclein (α-syn) protein aggregates in oligodendroglial cells forming glial cytoplasmic inclusions (GCIs) [1–3] and triggering striatonigral degeneration or olivopontocerebellar atrophy. These pathological alterations are defined clinically by autonomic failure accompanied by parkinsonian features or cerebellar ataxia [4]. No clear familial inheritance has been defined so far and MSA is considered an orphan disease of idiopathic origin.
Multiple mechanisms may contribute to the formation of GCIs including deregulation of quality control and protein degradation, like autophagy [5–8], lower efficiency of immature oligodendrocytes to process α-syn aggregates [9], and propagation of α-syn aggregates from neurons to other glial cells [10, 11]. In addition, emerging data shows alterations in transcriptional mechanisms associated with MSA including aberrant expression of genes related to mitochondrial function, iron metabolism, immune response, and inflammation [12, 13]. Importantly, aberrant RNA processing associated with the disruption of long intervening non-coding RNA (lincRNAs) [14] and microRNA (miRNAs) were reported in MSA brains and MSA transgenic animal models, highlighting their potential involvement in pathology [15–19] and suggesting the utility of profiling miRNAs in cerebrospinal fluid (CSF) and serum as disease biomarkers [20, 21].
miRNAs are short non-coding RNA molecules that negatively impact gene expression by inducing the degradation of specific transcripts, a mechanism mediated by the binding of Argonaute proteins to the 3′ untranslated regions of target mRNAs [22]. These molecules have emerged as important regulators of biological functions and are ubiquitously expressed across tissues. Notably, the brain presents the largest diversity of miRNA species, which show specific spatial and temporal expression patterns, likely associated with their roles in neuronal development and physiology. Moreover, miRNAs have been involved in synaptic plasticity, learning and memory, and drug addiction (reviewed by Wang et al. [23]). MiRNAs fulfill important roles in sustaining myelination, as mice lacking Dicer1, implicated in miRNA biogenesis, show severe deficits in myelination and oligodendrocyte maturation failure [24]. As expected, aberrant miRNA regulation has deleterious consequences for brain function, and alterations in miRNA networks have been reported in Alzheimer’s [25], Parkinson’s [26], and Huntington’s diseases [27].
A handful of studies, including work from our group, have shown aberrant miRNA expression in MSA in association with alterations in the transcription of their known target genes [15, 17–19]. Still, a clear understanding of a causal role of aberrant miRNA networks on transcriptome disruption and neurodegeneration in MSA is lacking. Here, we report the findings from comprehensive unbiased profiling of more than 800 miRNAs from the striatum of MSA patients in comparison to healthy individuals, supported by the parallel screening of 700 transcripts previously known to be linked to neurodegeneration. We discovered novel miRNA pathways associated with MSA, identified altered expression of miRNA-mRNA targets contributing to neuroinflammation, and uncovered a potential link between AD and MSA pathologies that involve the deregulation of a miRNA and BACE1.
Methods
Human Brain Samples
Brain tissue samples were obtained from the Shiley-Marcos Alzheimer’s Disease Research Center at the University of California San Diego (UCSD-ADRC, n = 8) and from the Johns Hopkins Medical Institution Brain Resource Center (n = 10). Samples included frozen tissue from the striatum of MSA patients (n = 11) and healthy control (CT) age-matched subjects (n = 7). Case selection for this study was based on neuropathological examination and determination of a diagnosis of MSA-P [4], which is the most common MSA variant in the Western hemisphere [28]. Group demographics are presented in Supp. Table 1.
RNA Preparation and Processing
Total RNA, containing the miRNA fraction, was isolated from striatum tissue (100 mg) using the miRNeasy Mini Kit (Qiagen) as indicated by the manufacturer. Quality of the extracted RNA was further enhanced using the RNA Clean & Concentrator-5 Kit (Zymo Research). The final concentration of RNA was determined by fluorometric quantitation (Qubit, Thermo Fisher Scientific). The same RNA preparations (50 ng per panel) were used to hybridize the Human v3 miRNA assay panel and the nCounter® Human v1.0 Neuropathology panel (both from NanoString Technologies) after following the sample dilution and ligation protocols indicated by the vendor. Amplification was performed in an nCounter® SPRINT Profiler at the Stem Cell Genomics Core, Sanford Consortium for Regenerative Medicine, affiliated with UCSD.
Quantification of miRNA/mRNA by Real-Time PCR
For quantitative real-time PCR (qPCR), 1.0 μg of total RNA per sample was used for reverse transcription to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA was diluted 1:10 in ultrapure water and 5 μL of this dilution was used per reaction. qPCR was performed using TaqMan Fast Advanced Master Mix and species-specific TaqMan primers. The expression of the gene B2M (beta-2 microglobulin) was used as internal control. For miRNA qPCR, 350 ng of total RNA per sample was used for reverse transcription to cDNA using TaqMan microRNA Reverse Transcription Kit and miRNA-specific primers (Applied Biosystems), and 0.16 μL of cDNA was used per reaction. microRNA qPCR was performed using TaqMan Universal Master Mix II, no UNG, and species-specific TaqMan miRNA primers, using U6 as internal control [29]. qPCR reactions were run in duplicate in a StepOnePlus Real-Time PCR system (Applied Biosystems) and ΔCt calculations were made using StepOne software (Applied Biosystems).
Data Analysis and Statistics
Signal intensity from the array probes was exported into nSolver® analysis software v4.0 (NanoString Technologies) to normalize for probe ligation efficiency and amount of RNA used.
miRNA Analysis
miRNA abundance in each sample was normalized according to the geometric mean of the 75 most highly expressed miRNAs, as reported before [30]. The mean ± 2 standard deviations of the intensity of 5 negative control probes for each sample were used to set the background threshold for the assay. The geometric mean of 5 positive control probes in each sample was used to assess the overall quality of the assay and to measure the efficiency of probe ligation and hybridization. Only miRNAs with non-negative counts across all samples were used for downstream analysis.
mRNA Analysis
The advanced analysis module from nSolver® (run on R Bioconductor) was used to analyze the transcript abundance. mRNA signal was normalized using a set of 5 housekeeping genes (LARS, AARS, ASB7, TADA2B, CCDC127) selected from a larger set of available normalization probes and based on a stepwise exclusion of the more variable probes.
Diana mirPath v.3
KEGG pathways were generated using TarBase v.7.0, a database that catalogs published, experimentally validated miRNA-gene interactions [31]. Pathways were filtered based on P value after correction for multiple observations (false discovery rate (FDR)) and application of DAVID’s EASE score [31, 32].
miRNA-Gene Target Correlation Discovery on Ingenuity Pathway Analysis
We used IPA software to map altered miRNAs and mRNAs into functional pathways. We used core analysis to link miRNAs with their mRNA targets (experimentally validated or predicted with high confidence) based on inverse expression levels (miRNA increase and target mRNA decrease or vice versa) using fold expression, and P value data with a cutoff of P <0.05 for the difference in expression in the MSA versus CT comparison. From these relationships, we generated gene pathways to identify upstream regulators and gene interactions.
Results
Clinical and Demographic Characteristics of the Study Cohort
We analyzed postmortem brain tissue from the striatum of MSA patients (n = 11) and healthy control donors (n = 7; Supp. Table 1). Cases were selected based on neuropathological examination and confirmed the diagnosis of MSA-P [4]. For the MSA group, disease duration varied between 4 and 15 years and Braak staging for Parkinson’s (PD) pathology [33] ranged between 0 and IV. The distribution of sex, age, and PMI did not differ between control and MSA groups (P values = 0.6525, 0.4461, 0.4504 respectively, as per unpaired t test; Supp. Fig. 1A–C).
Aberrant Expression of Specific miRNAs in the Striatum of MSA Cases
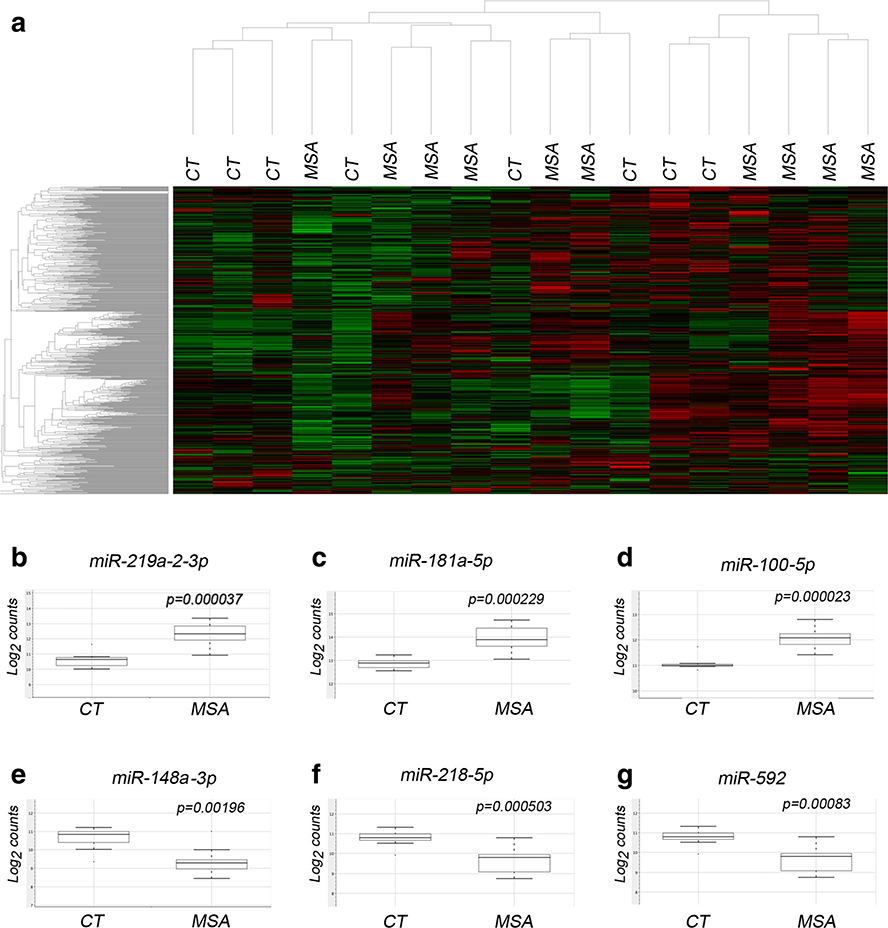
We interrogated the expression of 800 miRNAs and observed significant changes in the expression of 59 miRNAs in MSA brains at P <0.01, with the top 34 miRNAs reaching a relaxed cutoff FDR <0.2 (Table 1; Supp. Table 2 and Fig. 1b–g). Agglomerative clustering of raw miRNA expression data shows partial segregation between CT and MSA samples (Fig. 1a). Based on TargetScan classification (human release 7.0 [34]) let-7, miR-17, miR-181, and miR-25 were the top enriched families containing several miRNAs with differential expression in MSA striatum.
Table 1.
Alterations in miRNA levels in the striatum of MSA patients. Fold change expression levels were calculated for MSA cases in comparison to controls. P value as per t test. Names in italic indicate probes at false discovery rate (FDR, as per Benjamini Yekutieli method) ≤ 0.05
| Probe name | Accession # | Target sequence | Fold change | P value | FDR |
|---|---|---|---|---|---|
| Increased in MSA | |||||
| hsa-miR-219a-2-3p | MI MAT0004675 | AGAAUUGUGGCUGGACAUCUGU | 3.29 | 0.0000374 | 0.04 |
| hsa-mIR-181a-5p | MIMAT0000256 | AACAUUCAACGCUGUCGGUGAGU | 2.15 | 0.0000408 | 0.04 |
| hsa-mIR-574-3p | MIMAT0003239 | CACGCUCAUGCACACACCCACA | 2.06 | 0.0008318 | 0.17 |
| hsa- mIR-181b-5p | MIMAT0000257 | AACAUUCAUUGCUGUCGGUGGG | 2.03 | 0.0002291 | 0.08 |
| hsa-miR-181a-3p | MI MAT0000270 | ACCAUCGACCGUUGAUUGUACC | 2.00 | 0.0000403 | 0.04 |
| hsa-miR-100-5p | MI MAT0000098 | AACCCGUAGAUCCGAACUUGUG | 1.98 | 0.0000234 | 0.04 |
| hsa-miR-625-5p | MI MAT0003294 | AGGGGGAAAGUUCUAUAGUCC | 1.96 | 0.0001226 | 0.07 |
| hsa-mIR-185-Sp | MI MAT0000455 | UGGAGAGAAAGGCAGUUCCUGA | 1.74 | 0.0000717 | 0.05 |
| hsa-mIR-25-3p | MIMAT0000081 | CAUUGCACUUGUCUCGGUCUGA | 1.70 | 0.0000412 | 0.04 |
| hsa-mIR-151a-3p | MIMAT0000757 | CUAGACUGAAGCUCCUUGAGG | 1.69 | 0.0000674 | 0.05 |
| hsa-mIR-296-5p | MIMAT0000690 | AGGGCCCCCCCUCAAUCCUGU | 1.69 | 0.0000595 | 0.05 |
| hsa-let-7b-5p | MIMAT0000063 | UGAGGUAGUAGGUUGUGUGGUU | 1.66 | 0.0006138 | 0.15 |
| hsa-miR-181c-5p | MIMAT0000258 | AACAUUCAACCUGUCGGUGAGU | 1.65 | 0.0002161 | 0.08 |
| hsa-mIR-92a-3p | MIMAT0000092 | UAUUGCACUUGUCCCGGCCUGU | 1.57 | 0.0005160 | 0.15 |
| hsa-mIR-421 | MIMAT0003339 | AUCAACAGACAUUAAUUGGGCGC | 1.56 | 0.0001706 | 0.08 |
| hsa-miR-664b-3p | MIMAT0022272 | UUCAUUUGCCUCCCAGCCUACA | 1.56 | 0.0003270 | 0.11 |
| hsa-miR-590-5p | MIMAT0003258 | GAGCUUAUUCAUAAAAGUGCAG | 1.55 | 0.0009537 | 0.19 |
| hsa-miR-24-3p | MIMAT0000080 | UGGCUCAGUUCAGCAGGAACAG | 1.53 | 0.0000051 | 0.03 |
| hsa-miR-28-5p | MIMAT0000085 | AAGGAGCUCACAGUCUAUUGAG | 1.52 | 0.0006689 | 0.16 |
| hsa-miR-93-5p | MI MAT0000093 | CAAAGUGCUGUUCGUGCAGGUA | 1.49 | 0.0001367 | 0.07 |
| hsa-mIR-155-5p | MIMAT0000646 | UUAAUGCUAAUCGUGAUAGGGGGU | 1.48 | 0.0004585 | 0.14 |
| hsa-mIR-454-3p | MIMAT0003885 | UAGUGCAAUAUUGCUUAUAGGGU | 1.43 | 0.0007944 | 0.17 |
| hsa-miR-186-5p | MIMAT0000456 | CAAAGAAUUCUCCUUUUGGGCU | 1.36 | 0.0006144 | 0.15 |
| Decreased in MSA | |||||
| hsa-miR-376c-3p | MI MAT0000720 | AACAUAGAGGAAAUUCCACGU | −1.47 | 0.0004225 | 0.14 |
| hsa-mIR-539-5p | MIMAT0003163 | GGAGAAAUUAUCCUUGGUGUGU | −1.65 | 0.0001850 | 0.08 |
| hsa-mIR-3012-5p | MIMAT0000420 | UGUAAACAUCCUACACUCAGCU | −1.66 | 0.0000808 | 0.05 |
| hsa-mIR-124-3p | MIMAT0000422 | UAAGGCACGCGGUGAAUGCC | −1.78 | 0.0007659 | 0.17 |
| hsa-mIR-592 | MIMAT0003260 | UUGUGUCAAUAUGCGAUGAUGU | −1.86 | 0.0008376 | 0.17 |
| hsa-mIR-551b-3p | MI MAT0003233 | GCGACCCAUACUUGGUUUCAG | −1.93 | 0.0009979 | 0.19 |
| hsa-mIR-137 | MIMAT0000429 | UUAUUGCUUAAGAAUACGCGUAG | −2.06 | 0.0006116 | 0.15 |
| hsa-mIR-218-5p | MIMAT0000275 | UUGUGCUUGAUCUAACCAUGU | −2.20 | 0.0005030 | 0.15 |
Fig. 1.
Alterations in miRNA expression in the striatum of MSA cases. a Heat map representing the agglomerative clustering of raw miRNA abundance data plotting average Euclidean distance. Representative box plots of top upregulated (b-d) or downregulated (e-g) miRNAs in MSA showing non-parametric analysis of normalized data displaying data range and quartiles, P value as per t test
We applied mirPath v.3 analysis (DIANA tools [31]) to identify functional pathways affected by differentially expressed miRNAs (Table 2). Extracellular matrix (ECM)-receptor interaction was the top enriched pathway, involving 14 miRNAs that regulate 47 target genes. Integrins, which mediate ECM signaling events, have been largely involved in brain plasticity and pathophysiology, including oligodendrocyte differentiation and migration [35]. Aberrant miRNA expression may be related to the specific deregulation of the αv integrin subunit reported to be present in oligodendrocytes accumulating a-syn [36]. Notably, prion disease-related pathways appeared as the second most enriched category, comprised of 5 miRNAs affecting the expression of 12 genes, and potentially linking miRNAs to pathological a-syn misfolding, aggregation, and propagation in MSA. In addition, several fatty acid metabolism-related pathways were enriched in the analysis, as well as inflammation, ubiquitin-mediated proteolysis, and addiction pathways (likely indicating striatum-specific networks), thus relating global miRNA deregulation with key pathological cascades in MSA.
Table 2.
Pathway intersection query based on miRNas altered in MSA striatum. KEGG pathway enrichment by mirPath v.3 analysis based on the top 100 differentially expressed miRNAs in MSA (p > 0.05), applying Fisher’s exact test (hypergeometric distribution). Cutoff criteria: enrichment p < 0.0001, # miRNAs in pathway ≥ 5, # target genes in pathway ≥ 10. Cancer-related pathways were excluded
| KEGG pathway | P value | #genes | #mIRNAs |
|---|---|---|---|
| ECM-receptor interaction | 6.213E-124 | 47 | 14 |
| Prion diseases | 6.037E-35 | 12 | 5 |
| Biosynthesis of unsaturated fatty acids | 2.675E-14 | 10 | 6 |
| Mucin type O-glycan biosynthesis | 6.606E-14 | 24 | 22 |
| Glycosphingolipid biosynthesis | 5.101E-12 | 13 | 11 |
| TGF-beta signaling pathway | 2.567E-10 | 56 | 16 |
| Fatty acid metabolism | 4.593E-10 | 12 | 6 |
| Morphine addiction | 3.528E-09 | 49 | 10 |
| Lysine degradation | 9.419E-09 | 24 | 14 |
| Amphetamine addiction | 2.111E-08 | 43 | 19 |
| Signaling regulating stem cell pluripotency | 2.481E-08 | 103 | 25 |
| Hippo signaling pathway | 2.206E-07 | 94 | 15 |
| Focal adhesion | 8.679E-07 | 120 | 7 |
| Wnt signaling pathway | 2.195E-06 | 81 | 12 |
| Ubiquitin-mediated proteolysis | 2.195E-06 | 69 | 5 |
| Prolactin signaling pathway | 2.294E-06 | 46 | 12 |
| Glycosaminoglycan biosynthesis | 2.594E-06 | 10 | 7 |
| Xenobiotics metabolism cytochrome P450 | 7.044E-06 | 11 | 5 |
| Axon guidance | 8.688E-06 | 75 | 11 |
| Estrogen signaling pathway | 1.115E-05 | 50 | 19 |
| PI3K-Akt signaling pathway | 1.688E-05 | 126 | 7 |
| Glutamatergic synapse | 1.801E-05 | 44 | 5 |
| Long-term potentiation | 2.112E-05 | 43 | 9 |
| Circadian rhythm | 2.426E-05 | 21 | 5 |
| Nicotine addiction | 2.939E-05 | 23 | 8 |
| Thyroid hormone signaling pathway | 3.324E-05 | 73 | 14 |
| Gap junction | 4.568E-05 | 48 | 11 |
| Long-term depression | 4.943E-05 | 38 | 10 |
| N-Glycan biosynthesis | 0.0007841 | 22 | 8 |
Transcriptomic Alterations in the Striatum Impact Inflammation, Myelination, Autophagy, and Vesicle Transport in MSA
We next investigated transcriptomic alterations in MSA striatum that may contribute to disease pathology by using a focused array (nCounter® Neuropathology Panel, Nanostring) that profiles the expression of 700 genes previously associated with neuropathology. To further correlate miRNA alterations to changes in their target mRNAs, we used aliquots of the RNA samples interrogated for miRNA analysis.
All the samples passed QC parameters based on positive control probes, binding density, hybridization, and imaging steps. Unsupervised hierarchical clustering of normalized data showed disease status (CT or MSA) as the main clustering factor (Supp. Figure 1G). Analysis of the four principal components showed no association of sex, age, or PMI with the prominent signal in the gene expression dataset (Supp. Figure 1D–F). Braak staging scores were available for only 7/11 MSA samples; therefore, we only used the disease group as the analysis variable. Analysis of neuronal, glial, and endothelial markers contained in the array to measure cell-specific abundance showed significant changes in oligodendroglial markers (P value < 0.001), and overall, gene expression was increased in glial-types and decreased in neurons in MSA samples (Supp. Figure 1H), aligning with the pathological hallmarks of MSA.
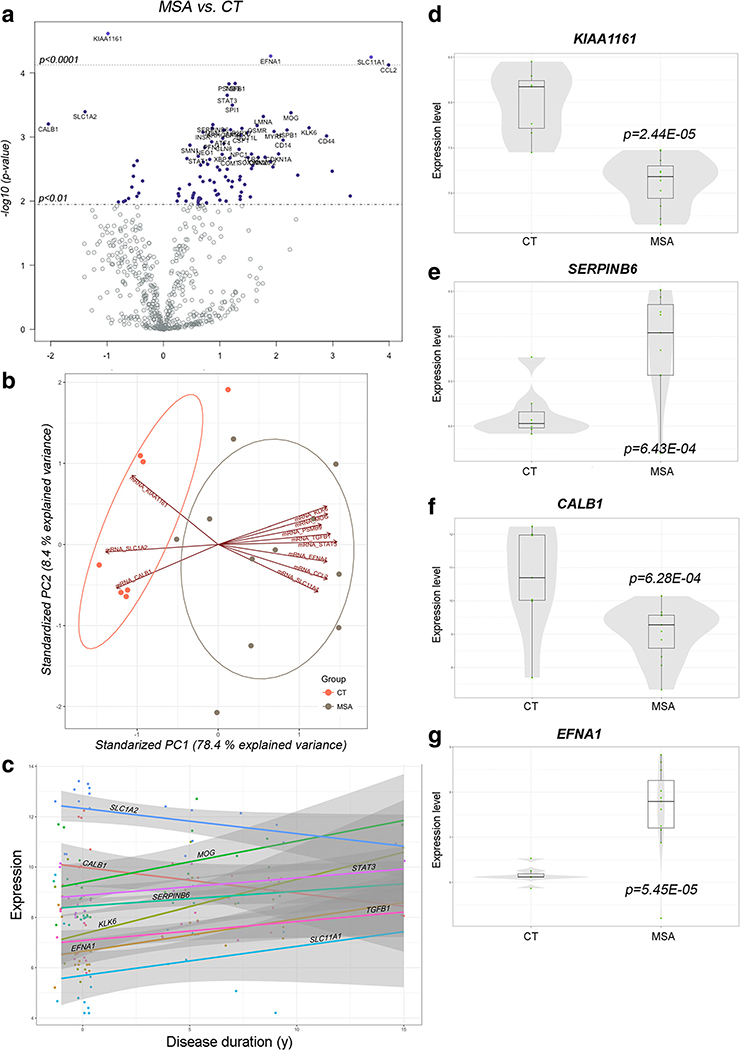
We identified 96 genes as differentially expressed (DE) in MSA samples at P <0.01, with the top 29 mRNAs reaching a relaxed cutoff at FRD <0.2 (Table 3, Supp. Table 3 and Fig. 2a, d–g). We further validated the array findings by profiling the abundance of selected targets, including 4 miRNAs and 6 mRNAs, differentially expressed in MSA by qPCR (Supp. Figure 2). All the tested miRNAs and mRNAs showed similar changes to those determined by array analysis.
Table 3.
Transcriptomic alterations in MSA striatum
| Gene | Accession # | Log2 fold change | P value | FDR | Functional pathway |
|---|---|---|---|---|---|
| CCL2 | NM_002982.3 | 3.99 | 7.51E-05 | 0.085 | Cytokines, UPR |
| SLC11A1 | NM_000578.2 | 3.68 | 5.66E-05 | 0.085 | Autophagy |
| CD44 | NM_001001392 | 2.89 | 9.66E-04 | 0.176 | Act. microglia |
| KLK6 | NM_001012964 | 2.58 | 7.19E-04 | 0.172 | Myelination |
| MOG | NM_001008228 | 2.26 | 4.19E-04 | 0.172 | Myelination |
| HSPB1 | NM_001540.3 | 2.19 | 7.80E-04 | 0.172 | Oxidative stress, UPR |
| CD14 | NM_000591.2 | 2.12 | 1.12E-03 | 0.188 | Growth factor signaling |
| MYRF | NM_001127392 | 1.96 | 8.21E-04 | 0.172 | Myelination |
| EFNA1 | NM_004428.2 | 1.90 | 5.45E-05 | 0.085 | Growth factor signaling |
| LMNA | NM_005572.2 | 1.77 | 4.79E-04 | 0.172 | Act. microglia, UPR |
| OSMR | NM_003999.2 | 1.66 | 6.64E-04 | 0.172 | Cytokines |
| DOT1L | NM_032482.2 | 1.48 | 8.68E-04 | 0.172 | Chromatin modification |
| P2RX7 | NM_002562.5 | 1.39 | 7.35E-04 | 0.172 | Vesicle trafficking |
| CSF1 | NM_000757.4 | 1.37 | 9.37E-04 | 0.176 | Act. microglia, cytokines |
| TGFB1 | NM_000660.4 | 1.27 | 1.46E-04 | 0.112 | Cytokines, myelination |
| SPI1 | NM_003120.1 | 1.22 | 3.18E-04 | 0.172 | Chromatin modification |
| JAM3 | NM_032801.3 | 1.19 | 7.73E-04 | 0.172 | Act. microglia, myelination |
| PSMB9 | NM_002800.4 | 1.16 | 1.48E-04 | 0.112 | Act. microglia, UPR |
| STAT3 | NM_003150.3 | 1.13 | 2.23E-04 | 0.145 | Growth factor signaling |
| ARHGEF10 | NM_014629.2 | 1.07 | 7.82E-04 | 0.172 | Myelination |
| CLN8 | NM_018941.3 | 1.07 | 1.27E-03 | 0.198 | Oxidative stress |
| ATF4 | NM_001675.2 | 1.05 | 1.05E-03 | 0.183 | Oxidative stress, UPR |
| SERPINB6 | NM_004568.4 | 0.88 | 6.43E-04 | 0.172 | Act. microglia |
| MSN | NM_002444.2 | 0.87 | 7.29E-04 | 0.172 | Neuronal cytoskeleton |
| PFN1 | NM_005022.2 | 0.85 | 1.19E-03 | 0.193 | Neural connectivity |
| INSR | NM_000208.2 | 0.70 | 8.45E-04 | 0.172 | Neural connectivity |
| CALB1 | NM_004929.2 | −2.04 | 6.28E-04 | 0.172 | Vesicle trafficking |
| SLC1A2 | NM_004171.3 | −1.39 | 4.04E-04 | 0.172 | Vesicle trafficking |
| KIAA1161 | NM_020702.3 | −0.98 | 2.44E-05 | 0.085 |
Fold change expression levels were calculated for MSA cases in comparison to controls; normalized to 5 reference genes present in the array. P value as per t test. FDR, false discovery rate as per Benjamini-Yekutieli method. Text in italics denote functional groups and pathways that contribute to MSA pathology. UPR, unfolded protein response; Act Microglia, activated microglia
Fig. 2.
Differential expression of genes involved in neurodegeneration in the MSA striatum. a Volcano plot displaying genes differentially transcribed in MSA with high statistical significance. Labels for the 40 top genes are indicated in the plot. b Principal component analysis biplot including the 15 most significant differentially expressed genes. Projections in opposite directions in PC1 indicate high-degree contribution of the indicated genes in clustering. c Trend plot for visualization of changes in expression as a function of disease progression. Color lines indicate trajectory. Transcription showing higher degree of change is labeled in the graph. d-g Univariate plots showing expression of select genes. Box plots depict median and second quartile of expression. Overlying violin plots (gray) show log2 expression quartiles and the estimated distribution of expression levels. P values indicated as per Student t test
PCA biplots showed that transcriptional alterations of the top 15 DE genes (based on significance) explain the majority of the variance in our analysis and segregate the MSA samples from the CT samples (Fig. 2b), suggesting an important contribution of these genes to pathology. Furthermore, these genes are functionally associated with inflammation, myelination, autophagy, and vesicle transport, which appear to be the most affected pathways in MSA.
Preliminary visualization of transcript abundance as a function of disease duration shows a trend for increased transcription of MOG—encoding myelin oligodendrocyte protein— and KLK6—encoding kallikrein 6—as the disease progresses, likely associated with aberrant myelination (Fig. 2c). Importantly, KLK6 shows a ~ 2.6-fold increased expression in MSA and is specifically involved in a-syn degradation. KLK6 is also known to be altered in PD and dementia with Lewy bodies (DLB) [37]. These transcriptional alterations may represent cellular responses to a-syn accumulation. On the other hand, the progressive decay in CALB1 and SCL1A2 expression, both encoding proteins with functions in glutamate transport, may contribute to excitotoxicity (Fig. 2c).
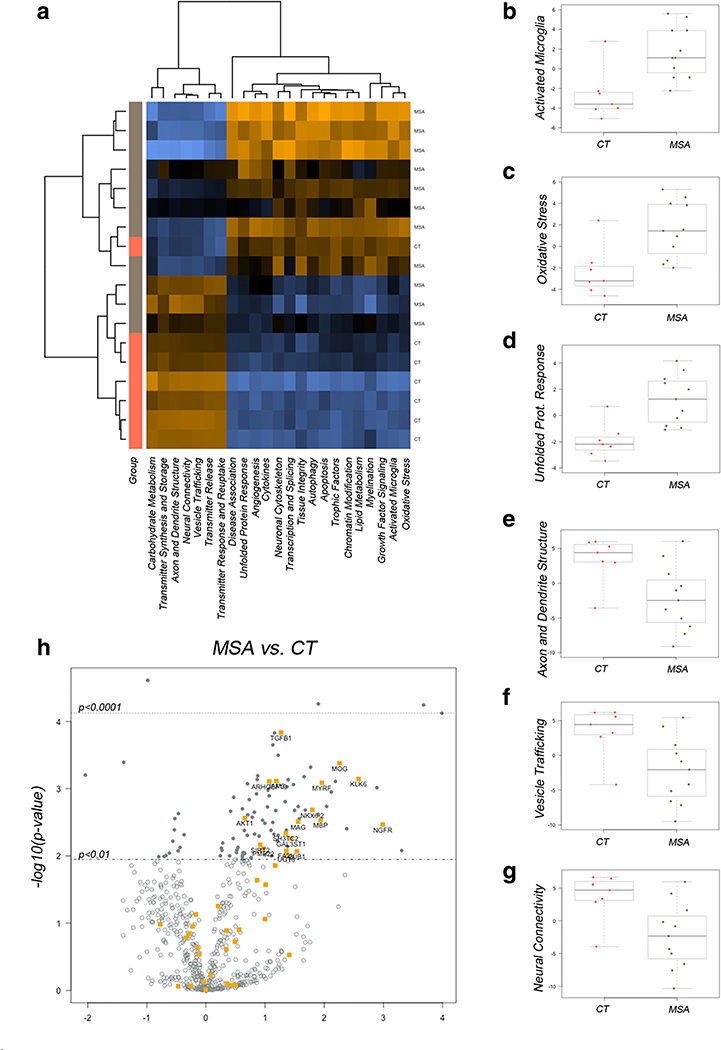
To better understand the potential contribution of these transcriptional alterations to MSA pathology, we visualized the pathway scores obtained from principal component analysis based on the weighted, averaged expression of genes assigned to a particular pathway in a heat map [38]. We observed clustering of MSA and CT samples (Fig. 3a). Overall, scores for pathological functions including microglial activation, oxidative stress, and unfolded protein response were increased in the MSA striatum (Fig. 3b–d); while pathways associated with neuronal functions, like axonal structure, connectivity, and vesicle transport appeared with lower scores in MSA samples (Fig. 3e–g). Importantly, genes associated with myelination in pathway analysis showed differential expression in MSA (Fig. 3h), in agreement with oligodendrocyte alterations and the signature pathological changes that characterize MSA.
Fig. 3.
Gene set analysis identifies MSA-associated pathways in the striatum. a Heat map showing distribution of scores summarizing the data from all pathways’ genes into a single score and clustering across disease groups. Orange indicates high and blue indicates low scores respectively. Box plots showing median scores with quartile ranges in MSA and CT samples for selected pathways associated with inflammation and cell death (b-d) and neuronal physiology (e-g). h Volcano plot displaying genes differentially transcribed in MSA with high statistical significance and involved in myelination. Labels for the top genes are indicated in the plot
Gene set enrichment analysis (GSEA) showed high enrichment for the frequency of SAF-1-canonical binding sites in 20 DE genes (LMNA, PFN1, DDIT3, EFNA1, CDKNA1, STAT3, BACE1, NFKBIA, PHF21A, JUN, HSPB1, SOX10, NEO1, GSN, TNFRSF1A, MAG, MSN, SH3TC2, CD44, and SPI1) with FDR q value = 1.96E-05. In addition, NFAT binding was enriched in 16 genes (CDKN1A, STAT3, NFKBIA, PHF21A, SOX10, GSN, TNFRSF1A, MECP2, PMP22, CREB1, SOX9, CCL2, XBP1, CUL3, CALB1, and CAST) with FDR q value = 1.47E-04. SAF-1 is a transcription factor that responds to inflammatory stimuli and contributes to the development of insoluble amyloid A aggregates in amyloidosis [39]. NFAT is a complex transcription factor that is implicated in neurodegenerative changes like the induction of dystrophic neurites and dendritic spine loss, due to the activation of calcineurin signaling by amyloid-beta [40]. Therefore, upstream deregulation of these transcription factors may also add to striatal transcriptomic alterations in response to a-syn aggregation during the course of MSA. GSE analysis of deregulated genes in MSA also identified miR-124 (predicted to target 10 genes: LMNA, STAT3, BACE1, MECP2, CREB1, SOX9, CCL2, LCLAT1, FA2H, and KIAA1161) with FDR q value = 4.75E-05 and miR-27a and b (predicted to target 9 genes: NEO1, CREB1, NOVA1, INSR, SLC1A2, CSF1, NGFR, EGFR, andDOT1L) with FDR q value = 7.63E-05 as enriched regulators of the DE gene set. Both miRNAs showed differential expression in our array analysis, thus validating our findings and further highlighting their role in MSA pathology (Tables 1 and 5).
Table 5.
Alterations in selected miRNAs in MSA is associated with inverse changes in transcription in target genes. In silico analysis of miRNAs deregulated in MSA brains and their target mRNAs that also showed differential expression in MSA. Coefficients (r) correspond to Pearson’s correlation.
| ID | Fold change | P value | Symbol | Log2 fold | P value | Correlat. r | P value |
|---|---|---|---|---|---|---|---|
| hsa-let-7b-5p | 1.66 | 0.0006 | AP1S1 | −0.86 | 0.0131 | −0.223 | 0.3738 |
| GNPTAB | −0.54 | 0.0198 | −0.176 | 0.4831 | |||
| RASGRP1 | −1.03 | 0.0271 | −0.200 | 0.4253 | |||
| hsa-miR-100-5p | 1.98 | 0.0001 | PPP3CA | −1.17 | 0.0470 | −0.688** | 0.0016 |
| hsa-miR-10a-5p | 1.26 | 0.0074 | CAMK2B | −0.77 | 0.0430 | −0.039 | 0.8756 |
| hsa-miR-1185-5p | −1.36 | 0.0092 | GNG2 | 1.51 | 0.0288 | −0.291 | 0.2411 |
| hsa-miR-124-3p | −1.78 | 0.0007 | CCL2 | 3.99 | 7.5E-05 | −0.343 | 0.1625 |
| STAT3 | 1.13 | 0.0002 | −0.647** | 0.0037 | |||
| SERPINB6 | 0.87 | 0.0006 | −0.695** | 0.0014 | |||
| DNM2 | 1.68 | 0.0020 | −0.598** | 0.0087 | |||
| CERS2 | 1.22 | 0.0049 | −0.753*** | 0.0003 | |||
| GSN | 1.21 | 0.0059 | −0.632** | 0.0049 | |||
| FA2H | 1.36 | 0.0084 | −0.710*** | 0.0010 | |||
| STX2 | 0.63 | 0.0184 | −0.695** | 0.0014 | |||
| GNG2 | 1.51 | 0.0288 | −0.339 | 0.1685 | |||
| hsa-miR-1249-3p | 1.48 | 0.0102 | SOX9 | −0.47 | 0.0084 | −0.490* | 0.0389 |
| hsa-miR-128-3p | −1.66 | 0.0120 | IL13RA1 | 0.89 | 0.0050 | −0.711*** | 0.0009 |
| NOVA1 | 0.27 | 0.0082 | −0.474* | 0.0465 | |||
| hsa-miR-132-3p | −1.61 | 0.0331 | MECP2 | 0.24 | 0.0099 | −0.439 | 0.0679 |
| NFE2L2 | 0.49 | 0.0113 | −0.659** | 0.0029 | |||
| hsa-miR-148a-3p | −2.39 | 0.0019 | UGT8 | 1.36 | 0.0093 | −0.693** | 0.0014 |
| hsa-miR-149-5p | −1.30 | 0.0420 | CSF1 | 1.37 | 0.0009 | −0.562* | 0.0151 |
| hsa-miR-154-5p | −1.47 | 0.0024 | JAM3 | 1.19 | 0.0007 | −0.603** | 0.0080 |
| hsa-miR-155-5p | 1.48 | 0.0004 | LCLAT1 | −0.53 | 0.0040 | −0.430 | 0.0749 |
| hsa-miR-15a-5p | 1.43 | 0.0058 | EGFR | −0.52 | 0.0027 | −0.249 | 0.3191 |
| PPP2R5C | −0.56 | 0.0060 | −0.548* | 0.0184 | |||
| AKT3 | −0.41 | 0.0271 | −0.391 | 0.1086 | |||
| hsa-miR-181a-5p | 2.15 | 0.0001 | GRIA2 | −0.59 | 0.0414 | −0.521* | 0.0266 |
| hsa-miR-19a-3p | 1.59 | 0.0248 | SLC1A2 | −1.39 | 0.0004 | −0.645** | 0.0038 |
| CDS1 | −0.76 | 0.0162 | −0.681** | 0.0018 | |||
| SLC8A1 | −0.54 | 0.0280 | −0.592** | 0.0096 | |||
| hsa-miR-218-5p | −2.20 | 0.0005 | TNC | 2.38 | 0.0039 | −0.546* | 0.0190 |
| GNAI2 | 0.62 | 0.0097 | −0.601** | 0.0083 | |||
| hsa-miR-222-3p | −2.01 | 0.0424 | SOD2 | 1.36 | 0.0041 | −0.484* | 0.0418 |
| hsa-miR-23b-3p | 1.56 | 0.0010 | SEC23A | −0.49 | 0.0251 | −0.473* | 0.0471 |
| hsa-miR-25-3p | 1.70 | 0.0001 | SLC12A5 | −0.66 | 0.0345 | −0.727*** | 0.0006 |
| hsa-miR-27b-3p | 1.57 | 0.0098 | CDS1 | −0.76 | 0.0162 | −0.681** | 0.0018 |
| PLCL2 | −0.68 | 0.0251 | −0.603** | 0.0080 | |||
| PDPK1 | −0.36 | 0.0397 | −0.570* | 0.0133 | |||
| hsa-miR-296-5p | 1.69 | 0.0001 | GBA | −0.46 | 0.0023 | −0.482* | 0.0425 |
| PDPK1 | −0.36 | 0.0397 | −0.535* | 0.0220 | |||
| hsa-miR-299-5p | −1.83 | 0.0103 | GNPTG | 0.60 | 0.0122 | −0.785*** | 0.0001 |
| hsa-miR-29c-3p | −1.52 | 0.0048 | BACE1 | 0.82 | 0.0047 | −0.737*** | 0.0005 |
| TNFRSF1A | 1.15 | 0.0047 | −0.460 | 0.0548 | |||
| PMP22 | 0.97 | 0.0077 | −0.724*** | 0.0007 | |||
| GPR37 | 0.89 | 0.0439 | −0.558* | 0.0160 | |||
| hsa-miR-301a-3p | 1.52 | 0.0375 | CDS1 | −0.76 | 0.0162 | −0.669** | 0.0024 |
| CALM1 | −0.76 | 0.0279 | −0.703** | 0.0011 | |||
| hsa-miR-30b-5p | −1.66 | 0.0001 | JUN | 0.65 | 0.0092 | −0.533* | 0.0225 |
| GNAI2 | 0.62 | 0.0097 | −0.475* | 0.0463 | |||
| C9orf72 | 0.68 | 0.0119 | −0.388 | 0.1108 | |||
| STX2 | 0.63 | 0.0184 | −0.503* | 0.0331 | |||
| BECN1 | 0.19 | 0.0403 | −0.148 | 0.5560 | |||
| hsa-miR-376c-3p | −1.47 | 0.0004 | HIF1A | 0.42 | 0.0401 | −0.160 | 0.5238 |
| hsa-miR-423-5p | 1.51 | 0.0031 | CALM1 | −0.76 | 0.0279 | −0.285 | 0.2515 |
| THY1 | −0.75 | 0.0293 | −0.309 | 0.2109 | |||
| hsa-miR-495-3p | −1.59 | 0.0033 | CD9 | 0.86 | 0.0231 | −0.693** | 0.0014 |
| hsa-miR-505-3p | −1.47 | 0.0458 | HMGB1 | 0.54 | 0.0074 | −0.599** | 0.0086 |
| C9orf72 | 0.68 | 0.0119 | −0.595** | 0.0091 | |||
| hsa-miR-520f-3p | 1.37 | 0.0197 | AKT3 | −0.41 | 0.0271 | −0.122 | 0.6295 |
| hsa-miR-582-5p | −1.38 | 0.0141 | NOVA1 | 0.27 | 0.0082 | −0.121 | 0.6321 |
| NFE2L2 | 0.49 | 0.0113 | −0.241 | 0.3340 | |||
| hsa-miR-625-5p | 1.96 | 0.0001 | CHRNB2 | −0.86 | 0.0403 | −0.720*** | 0.0007 |
| hsa-miR-744-5p | 1.31 | 0.0224 | THY1 | −0.75 | 0.0293 | −0.095 | 0.7050 |
| SLC12A5 | −0.66 | 0.0345 | −0.082 | 0.7450 | |||
| CAMK2B | −0.77 | 0.0430 | −0.041 | 0.8703 | |||
| MAP2K2 | −0.25 | 0.0449 | −0.018 | 0.9430 | |||
| hsa-miR-9-5p | −1.22 | 0.0173 | TNC | 2.38 | 0.0039 | −0.068 | 0.7872 |
| BACE1 | 0.82 | 0.0047 | −0.372 | 0.1274 | |||
| PMP22 | 0.97 | 0.0077 | −0.329 | 0.1814 |
*P < 0.05, **P < 0.01, and ***P < 0.001 for the correlation between miRNA and mRNA levels. Gene names in italic indicate experimental verified effect of miRNA on target; gene names on regular font indicate targets predicted with high confidence (according to TargetScan, Tarbase, and/or miRecords)
We next applied ingenuity pathway analysis (IPA) to predict canonical pathways affected in MSA. To visualize the extent of these alterations and the potential crosstalk with other pathways, we used a more relaxed cutoff for DE genes at P < 0.05 and we allowed the software to include genes from the IPA database that modulate, interact, or associate with those present in the array. Neuroinflammation was the top activated pathway (30 genes identified in the dataset, P value = 4.87E-20).
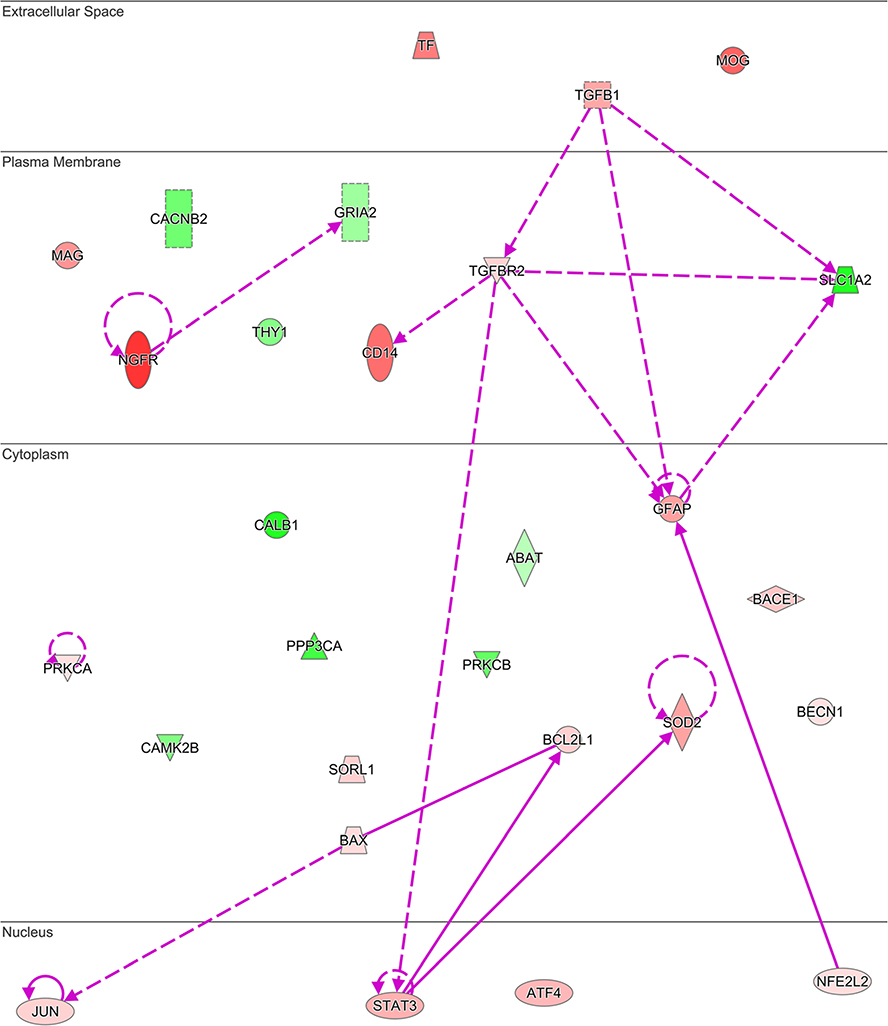
Furthermore, the disease enrichment module in IPA identified 20 DE genes clustering with neuromuscular disease (P value interaction 4.23E-9), and also overlapping with disorders of basal ganglia (20 genes, P value = 9.68E-10), movement disorders (19 genes, P value = 6.07E-9), and progressive motor neuropathy (14 genes, P value = 1.30E-8). Unexpectedly, Alzheimer’s disease (29 genes, P value = 4.55E-21; Fig. 4 and Table 4) and amyloidosis (30 genes, P value = 5.52E-22) were the top enriched disorders involving a different set of genes deregulated in MSA than those contributing to basal ganglia disorders. This finding suggests previously unreported mechanisms of neurodegeneration shared between MSA and AD.
Fig. 4.
Genes with altered expression in MSA striatum overlap networks associated with Alzheimer’s disease neuropathology. Network built using 29 genes showing differential transcription in MSA that were associated to AD in the disease enrichment analysis module of IPA. Red shades indicate increased and green shades indicate decreased expression in the data with intensity of shading representing fold change magnitude. Solid lines denote direct and dash lines indicate indirect interactions between gene products. Arrowheads denote reaction direction
Table 4.
Transcripts altered in the striatum of MSA patients overlapping networks affected in Alzheimer’s disease pathology. Genes were identified by disease enrichment analysis in IPA with a P value for the group interaction = 5.52E-22. Fold change expression levels were calculated for MSA cases in comparison to controls, P value as per t test
| Gene ID | Accession # | Log2 fold change | P value |
|---|---|---|---|
| NGFR | NM_002507.3 | 2.99 | 0.00341 |
| MOG | NM_001008228.2 | 2.26 | 0.00041 |
| CD14 | NM_000591.2 | 2.12 | 0.00112 |
| TF | NM_001063.2 | 1.9 | 0.00240 |
| MAG | NM_001199216.1 | 1.56 | 0.00310 |
| GFAP | NM_002055.4 | 1.39 | 0.01190 |
| SOD2 | NM_000636.2 | 1.36 | 0.00414 |
| TGFB1 | NM_000660.4 | 1.27 | 0.00014 |
| STAT3 | NM_003150.3 | 1.13 | 0.00022 |
| ATF4 | NM_001675.2 | 1.05 | 0.00105 |
| BACE1 | NM_012104.3 | 0.823 | 0.00473 |
| BCL2L1 | NM_138578.1 | 0.695 | 0.01150 |
| SORL1 | NM_003105.5 | 0.676 | 0.03580 |
| JUN | NM_002228.3 | 0.657 | 0.00922 |
| TGFBR2 | NM_001024847.1 | 0.646 | 0.00901 |
| BAX | NM_138761.3 | 0.517 | 0.00884 |
| NFE2L2 | NM_006164.3 | 0.498 | 0.01130 |
| PRKCA | NM_002737.2 | 0.262 | 0.00766 |
| BECN1 | NM_003766.2 | 0.196 | 0.04030 |
| ABAT | NM_020686.5 | −0.434 | 0.02630 |
| GRIA2 | NM_001083620.1 | −0.594 | 0.04140 |
| THY1 | NM_006288.2 | −0.754 | 0.02930 |
| CAMK2B | NM_001220.3 | −0.778 | 0.04300 |
| CACNB2 | NM_000724.3 | −0.889 | 0.04170 |
| PRKCB | NM_212535.1 | −0.963 | 0.04430 |
| PPP3CA | NM_000944.4 | −1.17 | 0.04700 |
| SLC1A2 | NM_004171.3 | −1.39 | 0.00040 |
| CALB1 | NM_004929.2 | −2.04 | 0.00062 |
Concordant Changes Between miRNA Effectors and mRNA Targets in MSA
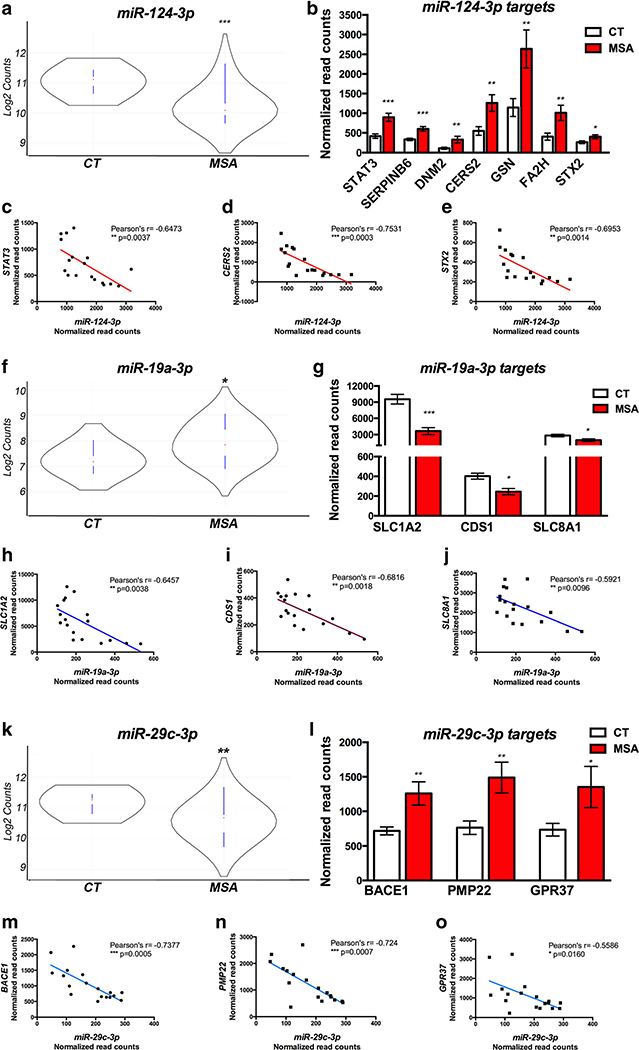
To determine the causal role of miRNA deregulation in driving MSA pathology, we used the miRNA target filtering function in IPA to identify miRNA and target mRNA pairs among those showing significant alterations in MSA striatum. Analysis thresholds were set as follows: (a) differential expression of miRNA and mRNA (independent analysis) in MSA at P < 0.05; (b) inverse relation between miRNA and target-mRNA abundance (increased miRNA-decreased mRNA and vice versa); (c) target validated experimentally or predicted with high confidence. We identified 34 miRNAs targeting 54 genes altered in MSA (Table 5). We further refined our analysis by investigating the correlation between miRNA abundance and target mRNA levels in each sample which narrowed down the list to 24 miRNAs and 38 genes showing significant correlation by Pearson’s analysis (at P < 0.05) and with r scores ranging between − 0.473 and − 0.785 (Table 5). MiR-124–3p, miR-19a-3p, miR-27b-3p, and miR-29c-3p were highly correlated with the expression levels of multiple DE target genes (Fig. 5)
Fig. 5.
Correlation between altered miRNAs and gene targets in MSA ► striatum. a, f, k Violin plots showing abundance of selected miRNAs in CT and MSA brains. Red dot indicates median count and black curve indicates frequency of counts based on log2; blue lines indicate lower and upper values and extend over first and third quartiles. b, g, l Bar graphs showing transcript abundance for gene targets of miRNAs depicted in (a-f-k) that showed significant correlation in the paired analysis. c-e, h-j, m-o Plots showing top correlated miR-gene target pairs as per Pearson’s analysis. *P<0.05, **P<0.01, and ***P<0.001 as per t test in the group comparisons (MSA vs. CT) or per Pearson’s in the correlation miR-gene target expression
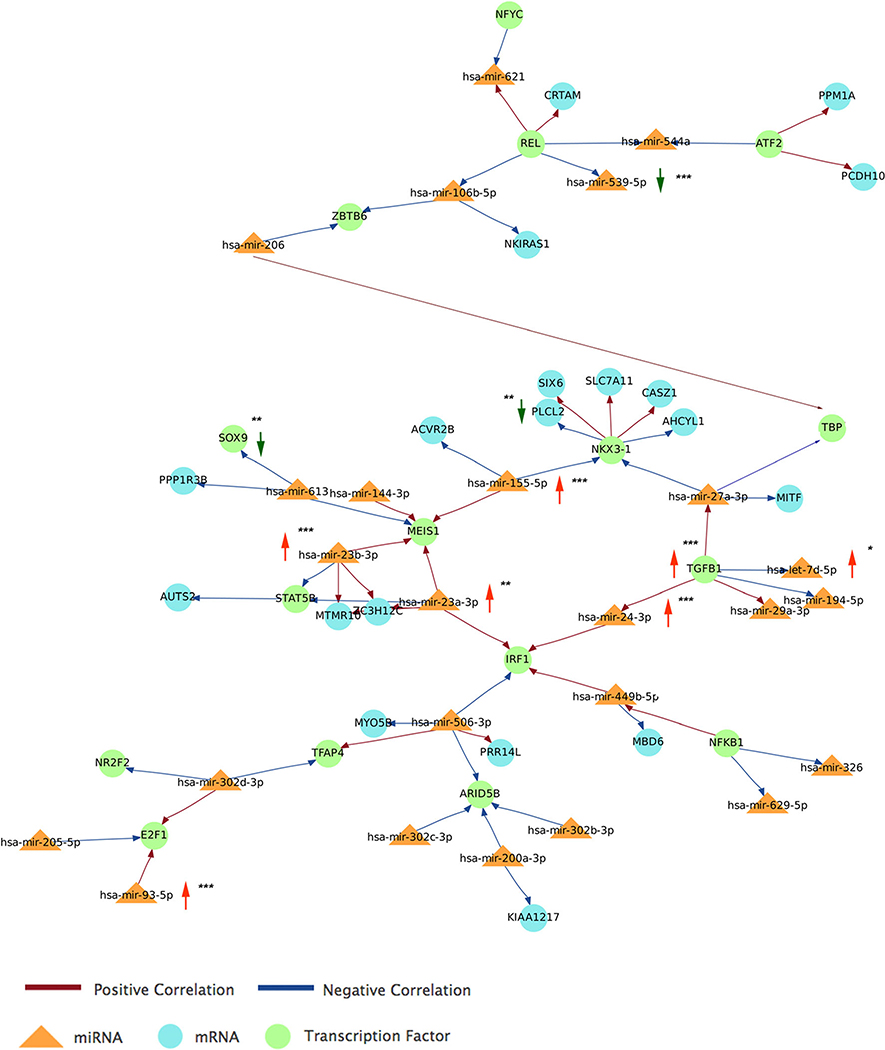
Finally, we ran an integrative analysis of miRNA and gene expression targets using Magia2 web tools [41] and including transcription factors to visualize extended networks that may contribute to MSA pathology. We performed a paired analysis using the matched miRNAs and target mRNAs altered in MSA striatum that showed significant correlation by Pearson analysis (Table 5). We used TargetScan as the prediction tool with a stringency of 0.8, which restricted the analysis to the top 20% interactions. The resulting main network was built upon miR-93–5p, miR-24–3p, let-7d-5p, miR-23a-3p, miR-23b-3p, miR-155–5p, and miR-539–5p from our array analysis and further expanded by the inclusion of 18 additional miRNAs and transcription factor nodes that interacted with those miRNAs according to previously published reports (Fig. 6). Pathway analysis using the molecules involved in the abovementioned network showed the top enriched pathways as neuroinflammation signaling (n = 31 molecules, P = 3.4E-20), opioid signaling (n = 27 molecules, P = 4.8E-18), Huntington’s disease (n = 26 molecules, P = 1.26E-16), and CREB signaling in neurons (n = 24 molecules, P = 6.1E-16); once more supporting a prominent role of inflammation in MSA pathology and validating the specificity of our in silico analysis with enrichment of striatum-associated functions.
Fig. 6.
Generation of expanded networks linking miRNAs altered in MSA, transcription factors, and additional regulatory molecules. Main network obtained by integrative analysis of paired miRNAs and gene targets affected in MSA and expanded by Pearson’s correlation and TargetScan prediction tools in Magic2. Vertical arrows indicate miRNAs and transcripts altered in MSA brains (red = increased, green = decreased abundance). *P <0.05, **P < 0.01, and ***P <0.001 as per t test in the group comparisons (MSA vs. CT)
Discussion
Transcriptional deregulation plays an important role in neurodegeneration and emerging evidence shows that mechanisms that modulate gene expression—including miRNAs—may present novel therapeutic and biomarker value. While a great deal of attention has been placed into miRNA profiling in AD and PD, much less is known about the role of this mechanism in MSA pathology. Here, we report the results from comprehensive profiling of miRNA and mRNA levels in the striatum, a region severely affected by pathology [4], in MSA cases and healthy control donors. We interrogated the abundance of 800 miRNAs (unbiased regarding function) in parallel to 700 transcripts previously associated with neurodegeneration using Nanostring™ technology.
Overall, this integrative analysis supports the involvement of miRNA deregulation in MSA, with more than 60 miRNAs showing aberrant expression in affected brains. Altered miRNAs mapped to functions like ECM-receptor interactions, prion disorders, autophagy, and inflammation. In addition, we observed deregulation of multiple transcripts, including GBA and BACE1 previously unreported, suggestive of shared mechanisms of neurodegeneration between MSA and Alzheimer’s disease.
In agreement with our previous study using qPCR [18], we found let-7b and another three members of the let-7 family increased in MSA brains. Let-7 is the most studied family of miRNAs and the first to be discovered in human tissues and has been largely implicated in cancer and inflammation [42]. Let-7 is highly abundant in the CNS [43], is implicated in autophagy [44], and was reported to induce neurodegeneration via activation of toll-like receptor (TLR) 7 in mouse neurons in vitro and to be increased in cerebrospinal fluid (CSF) from Alzheimer’s patients [45]. Interestingly, TLR7 signaling aggravates pathology in a model of multiple sclerosis, by activation of the myelin oligodendrocyte glycoprotein-specific T cell [46]. Since a-syn is known to activate TLR receptors, including TLR7 [47, 48], upregulation of the let-7 family may have a direct role on MSA pathology. Moreover, members of this family regulate glucose metabolism in several organs, and overexpression of let-7 in mice alters glucose metabolism, reducing glucose tolerance and insulin secretion [49]. Although the potential role of let-7 on insulin resistance and aberrant insulin signaling reported in MSA brains [50] remains to be investigated, this family of miRNAs appears to be a node that links multiple disease pathways.
In addition, we observed the miR-17 family to be altered in MSA brains, with 4 members showing differential expression. Interestingly, these miRNAs are downregulated in the brains of AD patients and are associated with suppression of amyloid precursor protein [51, 52], supporting the idea of potential pathology overlap between AD and MSA at the miRNA level, as observed in the pathway analyses we present here.
Upregulation of miR-181a, b, and c-5p (all members of the miR-181 family) could be highly relevant to MSA pathology, as miR-181b has been shown to be involved in autophagy inhibition by regulating PTEN/Akt/mTOR signaling. Mir-181b has been reported to be downregulated in a model of PD in association with MPP+ toxicity and increased cell death [53]. In our study, we found miR-181 elevated in MSA striatum, which potentially may reduce autophagy, further aggravating a-syn accumulation. Furthermore, miR-181 family members are linked to enhanced expression of pro-inflammatory cytokines in response to inflammatory agents in astrocytes [54], a function that may also contribute to neurodegeneration in MSA.
The search for signaling pathways that may be affected by aberrant miRNA expression in MSA highlights the role of these regulatory molecules in pathology. One of the top enriched pathways was related to prion diseases, involving miR-148a, miR-495, miR-548n, miR-410, and miR-543 and target genes PRNP, HSPA5, NCAM1, NCAM2, FYN, NOTCH1, PRKX, ELK1, PRKACB, BAX, IL1A, and MPAK1. Notably, recent studies report that, while the prion protein Prp (encoded by the PRNP gene) is not necessary for a-syn propagation, it can modulate the effect of a-syn fibrils accelerating spreading [55] and contributing to internalization of a-syn aggregates [56]. The relation between Prp and amyloid-β and its involvement in Alzheimer’s pathology has been largely described [57–59]. Furthermore, Prp infected cells show deregulation of miRNA profiles [60]. Here, we present evidence supporting that miRNA-mediated mechanisms are potentially implicated in regulating Prp and may further contribute to a-syn propagation in MSA pathology.
Pathway analysis also showed enrichment of three addiction-related pathways, including morphine, amphetamine, and nicotine. These pathways share multiple genes associated with a neural transmission that converge into specific functions of the striatum, a brain region that mediates acute and chronic effects of drugs of addiction [61]. Our results point to a striatum-specific miRNA network that becomes altered in MSA. Further studies may elucidate whether other cerebellar-specific pathways are deregulated in MSA-C cases and the degree of concordant changes between these two disease variants.
Accumulation of misfolded a-syn has been largely associated with neuroinflammation and recognized as a major driver of neuronal death in Parkinson’s disease and MSA [62, 63]. In agreement, we found altered expression of cytokines—including CCL2, the top increased transcript in MSA striatum—as well as molecules involved in microglia activation. Moreover, genes involved in autophagy, like SLC11A1 and ATP6V1H also appeared to be deregulated in MSA brains, potentially contributing to reduced clearance of protein aggregates, a common failure associated with proteinopathies [18, 64, 65].
Another interesting finding from our transcriptomic analysis is the significant decrease of GBA in MSA striatum. GBA encodes for glucocerebrosidase, a lysosomal enzyme implicated in glycolipid metabolism; mutations in this gene cause Gaucher’s disease. Genetic variants of glucocerebrosidase have also been associated with PD and DLB, and some studies suggest a higher frequency of glucocerebrosidase variations in MSA brains [66, 67]. To the best of our knowledge, this is the first report of decreased levels of GBA transcripts in MSA and particularly in association with miR-24–3p as a likely regulator of GBA expression.
We also observed a significant increase in myelin oligodendrocyte glycoprotein (MOG) transcripts in MSA brains, along with KLK6, MYRF, NKX6, FA2H, and SH3TC2, indicating myelin alterations. With the exception of KLK6, for which a recent study showed that ablation in mice attenuates symptoms of inflammatory demyelinating disease in transgenic models [68], all the other enzymes are needed for myelin maintenance and their increased expression may be a compensatory mechanism in response to myelin loss and oligodendrocyte degeneration as MSA progresses [69–72]. The alteration in MOG transcription we observed in MSA brains has never previously been reported.
A remarkable result from our study is the identification of overlapping networks between MSA and AD. Although our analysis is limited by the use of a biased array centered on neuropathology which may overstate the significance of gene-network associations, we eventually anticipated a potential overlap with Parkinson’s disease and Lewy body disease (due to common mechanisms elicited by a-syn aggregation). Instead, the transcriptomic alterations shared with Alzheimer’s disease are notable. We report an increase in BACE1 gene expression; BACE1 is responsible for the cleavage of the amyloid precursor protein (APP) into amyloid-β fragments in a crucial step for the initiation of amyloid pathology [73]. Amyloid-β has not been detected as a component of GCIs in MSA brains, although neuropathological examination has shown amyloid plaque deposition in some cases, mostly MSA-C presentation, to be associated with cerebellar degeneration [28, 74]. Our study based on a sensitive platform for transcript quantification has revealed these alterations in the striatum of MSA cases. While further studies are needed to rule out that our observations are not due to concurrent AD pathology in the cases analyzed, the fact that we detected alterations in the transcription of multiple genes that are germane to AD neurodegeneration is likely an indication for the crosstalk of pathological pathways. As AD pathology is highly frequent in the elderly population and with the increased momentum in research and drug development for AD, these shared mechanisms may potentially benefit MSA patients if reposition of novel AD drugs becomes suitable. Interestingly, a recent study pointed at miR-99, miR-132, and miR-129 to be highly associated with AD pathology [25]. We found these same miRNAs to be altered in MSA brain samples, supporting that the overlapping transcriptome alterations may be mediated by common miRNA networks operating in both pathologies.
Overall, our findings highlight a potential role for miRNAs on initiating and/or sustaining neuroinflammation. We detected a significant decrease of miR-124–3p—this miRNA is highly abundant in the brain and a key player in microglial activation, regulating the polarization of microglia between M1 pro-inflammatory and M2 anti-inflammatory phenotypes [75]. Intracerebral injection of miR-124 in a mice model for cerebral stroke shifts microglia towards M2 phenotypes, reducing inflammation [76]. Therefore, reduced miR-124 levels in MSA brains may sustain pro-inflammatory microglia fueling neuronal death in the striatum. Furthermore, miR-124 is expressed in dopaminergic neurons and reduced in MPTP-mouse models of PD, where overexpression of miR-124 partially rescued neuronal loss in the striatum. Notably, the mechanism of neuroprotection involves changes in the transcription of miR-124 targets and modulation of autophagy [77]. The role of miR-124 in autophagy is mediated by one of its targets: BACE1. Whereas several studies point to the deregulation of miR-124 in AD cases, some data is contradictory regarding whether there is an increase or decrease of this miRNA [78]; these differences may be a result of the region- and stage-specific expression. Nonetheless, the effects of miR-124 imbalance in AD are likely mediated by the combination of altered processing of amyloid precursor protein and increased microglial activation. Our study also reveals that the reduction of miR-29c-3p may contribute to increased BACE1 levels in MSA brains, potentially feeding into a deleterious pathway common to MSA to AD.
The study of miRNA deregulation in MSA is still limited, with a handful of reports addressing the question from cellular and animal models and a few on human samples, ranging from postmortem brain to serum. Though limited by the analysis of a small cohort, a common factor in the field, our results partially validate those reported previously by other groups, including downregulation of miR-130a, miR-376a, miR-377-3p, miR-87b-3p, and miR-379–5 in the striatum of a transgenic mouse model overexpressing α-syn under the PLP promoter [16]. Importantly, our results replicate findings in MSA brains, including decreased levels of miR-129–2-3p, miR-123–3p, miR-128–3p, miR-149–5p, miR-124–3p, and miR-379–5p and increased levels of miR-1290 and miR-23a-3p profiled in both the pons and cerebellum of MSA samples [79], and thus highlights the relevance of these miRNAs in pathology. The interest in miRNA profiling is also based on their potential as disease biomarkers. In this respect, our results from MSA striatum are in agreement with the observation of increased levels of miR-24, miR-185, miR-25, miR-454, miR-186, and let-7b in the serum of MSA patients [20], as well as with increased miR-223 and miR-24 in the circulating miRNA fraction of MSA patients’ serum [19], as well as increased miR-24 levels reported in CSF from MSA patients [21].
Conclusions
Taken in all, our results provide a comprehensive appraisal of miRNA alterations in MSA and their effect on the striatal transcriptome. Our results partially overlap with previous reports applying different methods in independent human samples or animal models, offering robust evidence that aberrant miRNA expression in MSA is highly correlated with altered transcription of genes associated with neuropathology, has fundamental roles in driving inflammation, disrupting myelination, and potentially impacting α-syn accumulation via deregulation of autophagy and prion mechanisms. High overlap between networks deregulated in AD and MSA point to a complex pathological cascade operating in MSA that may involve alterations in amyloid processing proteins. As research in AD gains momentum, insights from this disease can greatly benefit MSA patients in the future, if therapies can be repositioned for this devastating disorder.
Supplementary Material
Acknowledgments
We are grateful to the University of California, San Diego Shiley-Marcos AD Research Center, and the Johns Hopkins Medical Institution Brain Resource Center for the provision of brain tissue. The authors want to thank Dr. Elsa Molina, Director of the Sanford Stem Cell Clinical Center, and UCSD-Sanford Consortium for Regenerative Medicine for technical assistance with array processing.
Funding information This work was supported by the NIH grant NS092803 from NINDS to P.D. The UCSD Shiley-Marcos Alzheimer’s Disease Research Center is supported by the NIH grant AG05131.
Abbreviations
- MSA
multiple system atrophy
- α-syn
alpha-synuclein
- GCIs
glial cytoplasmic inclusions
- lincRNAs
long intervening non-coding RNA
- miRNAs
microRNA
- CSF
cerebrospinal fluid
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- CT
control cases
- ECM
extracellular matrix
- DLB
dementia with Lewy bodies
- GSEA
gene set enrichment analysis
- DE
differentially expressed genes
- IPA
ingenuity pathway analysis
- APP
amyloid precursor protein
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12035–019-1577–3) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spillantini MG, Crowther RA, Jakes R, Cairns NJ, Lantos PL, Goedert M (1998) Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett 251(3):205–208 [DOI] [PubMed] [Google Scholar]
- 2.Wakabayashi K, Hayashi S, Kakita A, Yamada M, Toyoshima Y, Yoshimoto M, Takahashi H (1998) Accumulation of alpha-synu- clein/NACP is a cytopathological feature common to Lewy body disease and multiple system atrophy. Acta Neuropathol 96(5):445–452 [DOI] [PubMed] [Google Scholar]
- 3.Wakabayashi K, Yoshimoto M, Tsuji S, Takahashi H (1998) Alpha- synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci Lett 249(2–3):180–182 [DOI] [PubMed] [Google Scholar]
- 4.Gilman S, Wenning GK, Low PA, Brooks DJ, Mathias CJ, Trojanowski JQ, Wood NW, Colosimo C et al. (2008) Second consensus statement on the diagnosis of multiple system atrophy. Neurology 71(9):670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miki Y, Tanji K, Mori F, Utsumi J, Sasaki H, Kakita A, Takahashi H, Wakabayashi K (2016) Alteration of upstream autophagy- related proteins (ULK1, ULK2, Beclin1, VPS34 and AMBRA1) in Lewy body disease. Brain Pathol 26(3):359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz L, Goldbaum O, Bergmann M, Probst-Cousin S, RichterLandsberg C (2012) Involvement of macroautophagy in multiple system atrophy and protein aggregate formation in oligodendrocytes. J Mol Neurosci 47(2):256–266 [DOI] [PubMed] [Google Scholar]
- 7.Vijayakumaran S, Wong MB, Antony H, Pountney DL (2015) Direct and/or indirect roles for SUMO in modulating alpha- synuclein toxicity. Biomolecules 5(3):1697–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winslow AR, Rubinsztein DC (2011) The Parkinson disease protein alpha-synuclein inhibits autophagy. Autophagy 7(4):429–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaji S, Maki T, Kinoshita H, Uemura N, Ayaki T, Kawamoto Y, Furuta T, Urushitani M et al. (2018) Pathological endogenous alpha- synuclein accumulation in oligodendrocyte precursor cells potentially induces inclusions in multiple system atrophy. Stem Cell Rep 10(2):356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konno M, Hasegawa T, Baba T, Miura E, Sugeno N, Kikuchi A, Fiesel FC, Sasaki T et al. (2012) Suppression of dynamin GTPase decreases alpha-synuclein uptake by neuronal and oligodendroglial cells: a potent therapeutic target for synucleinopathy. Mol Neurodegener 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woerman AL, Watts JC, Aoyagi A, Giles K, Middleton LT, Prusiner SB (2018) Alpha-synuclein: multiple system atrophy prions. Cold Spring Harb Perspect Med. Cold Spring Harb Perspect Med 2;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Mills JD, Halliday GM, Janitz M (2015) Role of transcriptional control in multiple system atrophy. Neurobiol Aging 36(1): 394–400 [DOI] [PubMed] [Google Scholar]
- 13.Langerveld AJ, Mihalko D, DeLong C, Walburn J, Ide CF (2007) Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov Disord 22(6):766–777 [DOI] [PubMed] [Google Scholar]
- 14.Mills JD, Ward M, Kim WS, Halliday GM, Janitz M (2016) Strand- specific RNA-sequencing analysis of multiple system atrophy brain transcriptome. Neuroscience 322:234–250 [DOI] [PubMed] [Google Scholar]
- 15.Lee ST, Chu K, Jung KH, Ban JJ, Im WS, Jo HY, Park JH, Lim JY et al. (2015) Altered expression of miR-202 in cerebellum of multiple-system atrophy. Mol Neurobiol 51 (1): 180–186 [DOI] [PubMed] [Google Scholar]
- 16.Schafferer S, Khurana R, Refolo V, Venezia S, Sturm E, Piatti P, Hechenberger C, Hackl H et al. (2016) Changes in the miRNA- mRNA regulatory network precede motor symptoms in a mouse model of multiple system atrophy: clinical implications. PLoS One 11(3):e0150705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubhi K, Rockenstein E, Kragh C, Inglis C, Spencer B, Michael S, Mante M, Adame A et al. (2014) Widespread microRNA dysregulation in multiple system atrophy - disease-related alteration in miR- 96. Eur J Neurosci 39(6):1026–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valera E, Spencer B, Mott J, Trejo M, Adame A, Mante M, Rockenstein E, Troncoso JC et al. (2017) MicroRNA-101 modulates autophagy and oligodendroglial alpha-synuclein accumulation in multiple system atrophy. Front Mol Neurosci 10:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallelunga A, Ragusa M, Di Mauro S, Iannitti T, Pilleri M, Biundo R, Weis L, Di Pietro C et al. (2014) Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front Cell Neurosci 8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kume K, Iwama H, Deguchi K, Ikeda K, Takata T, Kokudo Y, Kamada M, Fujikawa K et al. (2018) Serum microRNA expression profiling in patients with multiple system atrophy. Mol Med Rep 17(1):852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques TM, Kuiperij HB, Bruinsma IB, van Rumund A, Aerts MB, Esselink RAJ, Bloem BR, Verbeek MM (2017) MicroRNAs in cerebrospinal fluid as potential biomarkers for Parkinson’s disease and multiple system atrophy. Mol Neurobiol 54(10):7736–7745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J (2014) The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 21(9):743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Kwon EJ, Tsai LH (2012) MicroRNAs in learning, memory, and neurological diseases. Learn Mem 19(9):359–368 [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X et al. (2010) MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65(5):612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick E, Rajagopal S, Wong H-K, McCabe C, Xu J, Tang A, Imboywa SH, Schneider JA et al. (2017) Dissecting the role of non-coding RNAs in the accumulation of amyloid and tau neuropathologies in Alzheimer’s disease. Mol Neurodegener 12(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz J, Takousis P, Wohlers I, Itua I, Dobricic V, Binder H, Middleton L, Ioannidis J et al. (2018) Systematic meta-analyses identify differentially expressed microRNAs in Parkinson’s disease. 10.1101/253849 [DOI] [PubMed] [Google Scholar]
- 27.Jin J, Cheng Y, Zhang Y, Wood W, Peng Q, Hutchison E, Mattson MP, Becker KG et al. (2012) Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J Neurochem 123(4):477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jellinger KA (2014) Neuropathology of multiple system atrophy: new thoughts about pathogenesis. Mov Disord 29(14):1720–1741 [DOI] [PubMed] [Google Scholar]
- 29.Peltier HJ, Latham GJ (2008) Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14(5):844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogasawara R, Akimoto T, Umeno T, Sawada S, Hamaoka T, Fujita S (2016) MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol Genomics 48(4):320–324 [DOI] [PubMed] [Google Scholar]
- 31.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43(W1):W460–W466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA (2003) Identifying biological themes within lists of genes with EASE. Genome Biol 4(10):R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E (2003) Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24(2): 197–211 [DOI] [PubMed] [Google Scholar]
- 34.Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 12:4. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WuX Reddy DS (2012) Integrins as receptor targets for neurological disorders. Pharmacol Ther 134(1):68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuboi K, Grzesiak JJ, Bouvet M, Hashimoto M, Masliah E, Shults CW (2005) Alpha-synuclein overexpression in oligodendrocytic cells results in impaired adhesion to fibronectin and cell death. Mol Cell Neurosci 29(2):259–268 [DOI] [PubMed] [Google Scholar]
- 37.Miners JS, Renfrew R, Swirski M, Love S (2014) Accumulation of alpha-synuclein in dementia with Lewy bodies is associated with decline in the alpha-synuclein-degrading enzymes kallikrein-6 and calpain-1. Acta Neuropathol Commun 2:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomfohr J, Lu J, Kepler TB (2005) Pathway level analysis of gene expression using singular value decomposition. BMC Bioinf 6:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray A, Shakya A, Kumar D, Benson MD, Ray BK (2006) Inflammation-responsive transcription factor SAF-1 activity is linked to the development of amyloid A amyloidosis. J Immunol 177(4):2601–2609 [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee A, Soto C (2011) Role of calcineurin in neurodegeneration produced by misfolded proteins and endoplasmic reticulum stress. Curr Opin Cell Biol 23(2):223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bisognin A, Sales G, Coppe A, Bortoluzzi S, Romualdi C (2012) MAGIA(2): from miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update). Nucleic Acids Res 40(Web Server issue):W13–W21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roush S, Slack FJ (2008) The let-7 family of microRNAs. Trends Cell Biol 18(10):505–516 [DOI] [PubMed] [Google Scholar]
- 43.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12(9):735–739 [DOI] [PubMed] [Google Scholar]
- 44.Ham O, Lee SY, Lee CY, Park JH, Lee J, Seo HH, Cha MJ, Choi E et al. (2015) Let-7b suppresses apoptosis and autophagy of human mesenchymal stem cells transplanted into ischemia/reperfusion injured heart 7by targeting caspase-3. Stem Cell Res Ther 6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G et al. (2012) An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci 15(6):827–835 [DOI] [PubMed] [Google Scholar]
- 46.Lalive PH, Benkhoucha M, Tran NL, Kreutzfeldt M, Merkler D, Santiago-Raber ML (2014) TLR7 signaling exacerbates CNS autoimmunity through downregulation of Foxp3+ Treg cells. Eur J Immunol 44(1):46–57 [DOI] [PubMed] [Google Scholar]
- 47.Beraud D, Twomey M, Bloom B, Mittereder A, Ton V, Neitzke K, Chasovskikh S, Mhyre TR et al. (2011) Alpha-synuclein alters toll-like receptor expression. Front Neurosci 5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim C, Ho DH, Suk JE, You S, Michael S, Kang J, Joong Lee S, Masliah E et al. (2013) Neuron-released oligomeric alpha-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun 4:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frost RJ, Olson EN (2011) Control of glucose homeostasis and insulin sensitivity by the let-7 family of microRNAs. Proc Natl Acad Sci U S A 108(52):21075–21080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassil F, Canron MH, Vital A, Bezard E, Li Y, Greig NH, Gulyani Kapogiannis D et al. (2017) Insulin resistance and exendin-4 treatment for multiple system atrophy. Brain 140(5):1420–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert SS, De Strooper B (2009) Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci 32(4): 199–206 [DOI] [PubMed] [Google Scholar]
- 52.Schonrock N, Matamales M, Ittner LM, Gotz J (2012) MicroRNA networks surrounding APP and amyloid-beta metabolism- implications for Alzheimer’s disease. Exp Neurol 235(2):447–454 [DOI] [PubMed] [Google Scholar]
- 53.Li W, Jiang Y, Wang Y, Yang S, Bi X, Pan X, Ma A, Li W (2018) MiR-181b regulates autophagy in a model of Parkinson’s disease by targeting the PTEN/Akt/mTOR signaling pathway. Neurosci Lett 675:83–88 [DOI] [PubMed] [Google Scholar]
- 54.Hutchison ER, Kawamoto EM, Taub DD, Lal A, Abdelmohsen K, Zhang Y, Wood WH 3rd, Lehrmann E et al. (2013) Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 61(7): 1018–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urrea L, Segura-Feliu M, Masuda-Suzukake M, Hervera A, Pedraz L, Garcia Aznar JM, Vila M, Samitier J et al. (2018) Involvement of cellular prion protein in alpha-synuclein transport in neurons. Mol Neurobiol 55(3):1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aulic S, Masperone L, Narkiewicz J, Isopi E, Bistaffa E, Ambrosetti E, Pastore B, De Cecco E et al. (2017) Alpha- synuclein amyloids hijack prion protein to gain cell entry, facilitate cell-to-cell spreading and block prion replication. Sci Rep 7(1): 10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calella AM, Farinelli M, Nuvolone M, Mirante O, Moos R, Falsig J, Mansuy IM, Aguzzi A (2010) Prion protein and Abeta-related synaptic toxicity impairment. EMBO Mol Med 2(8):306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas LT, Salazar SV, Kostylev MA, Um JW, Kaufman AC, Strittmatter SM (2016) Metabotropic glutamate receptor 5 couples cellular prion protein to intracellular signalling in Alzheimer’s disease. Brain 139(Pt 2):526–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kostylev MA, Kaufman AC, Nygaard HB, Patel P, Haas LT, Gunther EC, Vortmeyer A, Strittmatter SM (2015) Prion-proteininteracting amyloid-beta oligomers of high molecular weight are tightly correlated with memory impairment in multiple Alzheimer mouse models. J Biol Chem 290(28):17415–17438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellingham SA, Coleman BM, Hill AF (2012) Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 40(21):10937–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lobo MK, Nestler EJ (2011) The striatal balancing act in drug addiction: distinct roles of direct and indirect pathway medium spiny neurons. Front Neuroanat 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caplan IF, Maguire-Zeiss KA (2018) Toll-like receptor 2 signaling and current approaches for therapeutic modulation in synucleinopathies. Front Pharmacol 9:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joshi N, Singh S (2018) Updates on immunity and inflammation in Parkinson disease pathology. J Neurosci Res 96(3):379–390 [DOI] [PubMed] [Google Scholar]
- 64.Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, Hansen L, Adame A et al. (2010) Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One 5(2):e9313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Wong E, Cuervo AM (2010) Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13(7):805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitsui J, Matsukawa T, Sasaki H, Yabe I, Matsushima M, Durr A, Brice A, Takashima H et al. (2015) Variants associated with Gaucher disease in multiple system atrophy. Ann Clin Transl Neurol 2(4): 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sklerov M, Kang UJ, Liong C, Clark L, Marder K, Pauciulo M, Nichols WC, Chung WK et al. (2017) Frequency of GBA variants in autopsy-proven multiple system atrophy. Mov Disord Clin Pract 4(4):574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bando Y, Hagiwara Y, Suzuki Y, Yoshida K, Aburakawa Y, Kimura T, Murakami C, Ono M et al. (2018) Kallikrein 6 secreted by oligodendrocytes regulates the progression of experimental autoimmune encephalomyelitis. Glia 66(2):359–378 [DOI] [PubMed] [Google Scholar]
- 69.Duncan GJ, Plemel JR, Assinck P, Manesh SB, Muir FGW, Hirata R, Berson M, Liu J et al. (2017) Myelin regulatory factor drives remyelination in multiple sclerosis. Acta Neuropathol 134(3): 403–422 [DOI] [PubMed] [Google Scholar]
- 70.Meixner M, Jungnickel J, Grothe C, Gieselmann V, Eckhardt M (2011) Myelination in the absence of UDP-galactose: ceramide galactosyl-transferase and fatty acid 2 -hydroxylase. BMC Neurosci 12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Southwood C, He C, Garbern J, Kamholz J, Arroyo E, Gow A (2004) CNS myelin paranodes require Nkx6–2 homeoprotein transcriptional activity for normal structure. J Neurosci 24(50):11215–11225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vijay S, Chiu M, Dacks JB, Roberts RC (2016) Exclusive expression of the Rab11 effector SH3TC2 in Schwann cells links integrin- alpha6 and myelin maintenance to Charcot-Marie-tooth disease type 4C. Biochim Biophys Acta 1862(7):1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peters F, Salihoglu H, Rodrigues E, Herzog E, Blume T, Filser S, Dorostkar M, Shimshek DR et al. (2018) BACE1 inhibition more effectively suppresses initiation than progression of beta-amyloid pathology Acta Neuropathol 135(5):695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bujan B, Hofer MJ, Oertel WH, Pagenstecher A, Burk K (2013) Multiple system atrophy of the cerebellar type (MSA-C) with concomitant beta-amyloid and tau pathology. Clin Neuropathol 32(4): 286–290 [DOI] [PubMed] [Google Scholar]
- 75.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL (2011) MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP- alpha-PU.1 pathway. Nat Med 17(1):64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamzei Taj S, Kho W, Aswendt M, Collmann FM, Green C, Adamczak J, Tennstaedt A, Hoehn M (2016) Dynamic modulation of microglia/macrophage polarization by miR-124 after focal cerebral ischemia. JNeurolmmune Pharmacol 11(4):733–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, Wang H, Gong X et al. (2016) MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to Bim. Brain Pathol 26(2):167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sonntag KC, Woo TU, Krichevsky AM (2012) Converging miRNA functions in diverse brain disorders: a case for miR-124 and miR-126. Exp Neurol 235(2):427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wakabayashi K, Mori F, Kakita A, Takahashi H, Tanaka S, Utsumi J, Sasaki H (2016) MicroRNA expression profiles of multiple system atrophy from formalin-fixed paraffin-embedded samples. Neurosci Lett 635:117–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.