Abstract
Widespread loss of genes on the Y is considered a hallmark of sex chromosome differentiation. Here we show that the initial stages of Y evolution are driven by massive amplification of distinct classes of genes. The neo-Y chromosome of Drosophila miranda initially contained about 3000 protein-coding genes, but has gained over 3200 genes since its formation about 1.5 MY ago, primarily by tandem amplification of protein-coding genes ancestrally present on this chromosome. We show that distinct evolutionary processes may account for this drastic increase in gene number on the Y. Testis-specific and dosage sensitive genes appear to have amplified on the Y to increase male fitness. A distinct class of meiosis-related multi-copy Y genes independently co-amplified on the X, and their expansion is likely driven by conflicts over segregation. Co-amplified X/Y genes are highly expressed in testis, enriched for meiosis and RNAi functions, and are frequently targeted by small RNAs in testis. This suggests that their amplification is driven by X vs. Y antagonism for increased transmission, where sex chromosome drive suppression is likely mediated by sequence homology between the suppressor and distorter, through RNAi mechanism. Thus, our analysis suggests that newly emerged sex chromosomes are a battleground for sexual and meiotic conflict.
Introduction
Sex chromosomes have originated multiple times from ordinary autosomes1. After suppression of recombination, the proto-X and proto-Y-chromosomes follow separate evolutionary trajectories and differentiate2. The complete lack of recombination on Y-chromosomes renders natural selection inefficient, and Y evolution is characterized by a loss of the majority of its ancestral genes3 while the X maintains most of them. Indeed, old Y-chromosomes of various species contain only a few functional genes4,5, and Y-chromosomes of many taxa (but not all) have instead accumulated massive amounts of repetitive DNA, including transposable elements (TEs) and satellite DNA6. In some lineages, the Y-chromosome is lost entirely7.
Studies of Y-chromosomes are often hindered by a lack of high-quality reference sequences, due to the technical challenges of assembling repetitive regions. To date, the Y-chromosomes of only a handful of mammalian species have been fully sequenced8,9 and no high-quality sequences of young Y-chromosomes that have already accumulated a substantial amount of repetitive DNA have been examined.
Drosophila miranda has a pair of recently formed neo-sex-chromosomes that originated ~1.5MY ago after its split from its closely related sister species D. pseudoobscura, and has served as a model to study the initiation of sex-chromosome differentiation10. The neo-sex-chromosomes of D. miranda were formed by the fusion of a former autosome (chr3 of the pseudoobscura group11) with the ancestral, degenerate Y-chromosome of this clade12. The neo-X and neo-Y-chromosome are still homologous over much of their length, with ~98% sequence identity at homologous regions10. A previous genomic analyses using Illumina short reads confirmed the notion that genes on the Y are rapidly lost13. About 1/3 of the roughly 3000 genes ancestrally present on the neo-Y were found to be pseudogenized, and over 150 genes were entirely missing13. However, the high level of sequence similarity between the neo-X and neo-Y-chromosome, yet drastic accumulation of repeats on the neo-Y, prevented assembling the Y-chromosome using short-read data.
We recently generated a high-quality sequence assembly of the neo-Y-chromosome of D. miranda using single-molecule sequencing and chromatin conformation capture, and used extensive BAC clone sequencing and optical mapping data to confirm that our assembly is of high accuracy14. Intriguingly, instead of simply shrinking, our assembly revealed that the young neo-Y-chromosome dramatically increased in size relative to the neo-X by roughly 3-fold. We assembled 110.5 Mb of the fused ancestral Y and neo-Y-chromosome (Y/neo-Y sequence), and 25.3 Mb of the neo-X14. Most of this size increase is driven by massive accumulation of repetitive sequences —in particular TEs— which comprise over 50% of the neo-Y derived sequence14. Here, we carefully annotate the neo-sex-chromosomes using transcriptomes from multiple tissues and small RNA profiles, to study the evolution of gene content on this recently formed neo-Y-chromosome.
Results
A catalogue of genes on the neo-Y
With a comprehensive high-quality reference sequence of the neo-Y-chromosome of D. miranda, we systematically catalogued its genes. Comparison of the neo-sex-chromosome gene-content with that of D. pseudoobscura, a close relative where this chromosome pair is autosomal, allowed us to infer the ancestral gene complement and reconstruct the evolutionary history of gene gains and losses along the neo-sex-chromosomes (Figure_1A). Note that the ancestral Y-chromosome of D. pseudoobscura is not assembled and contains no annotated protein-coding genes, and our analysis focuses on neo-sex linked genes (i.e. genes present on chr3 of D. pseudoobscura). Annotation of neo-Y linked genes is a challenging task, for several reasons. Genes on the neo-Y are embedded in highly repetitive sequences, and introns often dramatically increase in size due to TE insertions15. Neo-Y genes (or pseudogenes) are also often truncated or have premature stop-codons13,16. Automated annotation thus often resulted in fragmented, split or missing gene models on the neo-Y (see Methods for details), and we used extensive manual curation of all neo-Y genes that were not simple 1:1 orthologs between species and neo-sex-chromosomes to validate and correct our gene models (see Methods). In total, we identified 6,448 genes on the neo-Y, and 3,253 genes on the neo-X, compared to 3,087 genes on the ancestral autosome that gave rise to the neo-sex-chromosome. Thus, contrary to the paradigm that Y-chromosomes undergo chromosome-wide degeneration, our analysis reveals a dramatic increase in the number of annotated genes on the neo-Y, compared to its ancestral gene complement, or that of its former homolog, the neo-X.
Figure_1. Gene content evolution of newly formed sex chromosomes. A.
Karyotype and gene content evolution on the neo-sex chromosomes of D. miranda. Shown are the karyotype of D. miranda, and its close relative D. pseudoobscura, from which it diverged about 2MY ago. In D. miranda, the fusion of an autosome (chr3) with the Y-chromosome created the neo-sex chromosomes, about 1.5 MY ago. Shown along the tree are numbers of gene amplifications (in green) and gene losses (in red) of genes ancestrally present on chr3, on the neo-X and neo-Y-chromosomes, assuming parsimony. X chromosomes are shown in red, Y-chromosomes in blue, and autosomes in black. B. Schematic representation of gene content of the D. miranda neo-Y/Y-chromosome. The D. miranda neo-Y/Y harbors different types of genes: our annotation contains 1736 ancestral single-copy genes; 1697 multi-copy Y genes (derived from 363 distinct proteins), and 2036 genes (derived from 94 distinct proteins) that co-amplified on both the X/neo-X and Y/neo-Y. Most ampliconic Y genes were ancestrally present on the autosome that formed the neo-sex chromosome (that is, they are located on chr3 of D. pseudoobscura). “Others” refers to genes not present or mapping to an unknown location in D. pseudoobscura (446 genes), or genes with complex mapping (513 genes; see Methods). C. Pie charts show the assignment of genes on the neo-Y/Y or neo-X to these different categories (using the same color scheme as in panel B), with the size of the pie scaled by the number of genes on the neo-Y/Y or neo-X. D. Co-amplified X/Y genes typically exist as tandem repeats on the X and the Y-chromosomes. Shown is a subset of 18 co-amplified X/Y gene families with meiosis and siRNA functions.
Overall, we detect 1,736 ancestral single-copy orthologs between the neo-sex-chromosomes, i.e. ~56% of genes ancestrally present show a simple 1:1 relationship between species and the neo-X and neo-Y-chromosome. Furthermore, genes are degenerating on the non-recombining neo-Y. We find 143 genes that are located on chr3 in D. pseudoobscura and the neo-X of D. miranda, but are missing from our neo-Y annotation, and we fail to detect a homolog on the neo-Y by BLAST. Thus, ~5% of genes that were ancestrally present are now completely absent on the neo-Y. On the other hand, only 17 genes (~0.5% of genes ancestrally present) are absent from the neo-X but found on the neo-Y, a rate of gene loss comparable to autosomes and the ancestral X (Supplementary_Table_1). Thus, the neo-Y is indeed losing its ancestral genes at a high rate, consistent with theoretical expectation3,17 and empirical observations of gene poor ancestral Y-chromosomes4,5,8,9.
Intriguingly, however, for 457 unique single-copy D. pseudoobscura protein-coding genes, we find multiple copies in our neo-Y-chromosome annotation of D. miranda (which were all verified by manual inspection of BLAST and nucmer alignments18 and also confirmed by Illumina read-depth analysis, Supplementary_Figure_1). Genes with multiple copies on the Y/neo-Y fall into two groups. 363 unique protein-coding genes of D. pseudoobscura are also single-copy (or missing) on the X/neo-X of D. miranda, but are amplified on the neo-Y (resulting in a total of 1,697 Y-linked gene copies; two of these genes were gained from chr2; Supplementary_Table_2). The remaining 94 unique protein-coding genes of D. pseudoobscura that have amplified on the Y/neo-Y, surprisingly, have also co-amplified on the original X and neo-X chromosome of D. miranda (and harbor a total of 2,036 Y/neo-Y-linked gene copies and 647 copies on the X/neo-X; Supplementary_Table_3, Figure_1D). Most of the genes that co-amplified on the X and Y-chromosome of D. miranda were ancestrally present on the autosome that formed the neo-sex-chromosomes (i.e. chr3 in D. pseudoobscura), but some were also gained from other chromosomes (4 genes from chr2, 1 gene from chr4, and 14 genes from the ancestral X). Thus, genes on the Y/neo-Y of D. miranda fall into distinct categories (Figure_1B), and we refer to them as single-copy Y genes, multi-copy Y genes (which are single-copy on the X/neo-X), and co-amplified X/Y genes (genes that have amplified on both the X and the Y-chromosome). Genes whose ancestral location could not be determined, or genes with more complex evolutionary histories were not further analyzed (see Methods).
Properties of amplified Y genes
Many amplified gene copies –on both the X/neo-X and the Y/neo-Y-chromosome– are fragmented, and some have premature stop codons or frame shift mutations (Supplementary_Table_2, 3). We find full-length copies for 786 amplified Y genes (46%), 776 co-amplified Y-genes (38%), and 300 co-amplified X genes (46%). Thus, even if ignoring partial gene copies (which may nevertheless have function as non-coding transcripts), we still find considerably more genes on the neo-Y compared to the neo-X or the ancestral autosome that formed the neo-sex-chromosomes. Genes with truncated coding regions are less likely to produce functional proteins and may thus be pseudogenes. However, many of these amplified gene copies may instead encode functional RNAs (for example, they may be involved in RNA induced silencing, as suggested by our analysis below), and thus both full-length and fragmented copies could influence organismal fitness, if expressed. Indeed, transcriptome analysis (using only uniquely mapping RNA-seq reads) shows that most individual gene copies of amplified gene families on the Y/neo-Y are expressed in male tissues, both for partial genes and full-length transcripts. We detect expression of 71% of individual copies among multi-copy Y genes, and 94% for co-amplified X/Y genes (Supplementary_Table_4). This is consistent with many gene copies on the neo-Y indeed being functional, either as a protein or as a functional RNA.
How do genes amplify on the sex-chromosomes? The majority of multi-copy Y genes, or co-amplified X and Y genes are found in gene clusters; 89% of multi-copy Y genes are located near other copies of the same gene family (within 100 kb), and 80% of co-amplified X and 87% of co-amplified Y genes (Figure_1D, Supplementary_Figure_2,3; Supplementary_Table_5). Clustering of gene families in tandem arrays suggests that non-allelic recombination is a main factor driving gene amplification on sex-chromosomes. Additionally, phylogenetic analysis reveals that individual copies of co-amplified gene families typically cluster by chromosome, indicating independent amplification on the X and Y-chromosome, and confirming a lack of recombination between the neo-X and neo-Y (Extended_Data_Fig_1).
Multi-copy genes often show dynamic copy number evolution between individuals19. To test for variation in copy number of amplified Y/neo-Y genes in natural populations of D. miranda, we generated Y-chromosome replacement lines by backcrossing Y-chromosomes from different locations into the same genetic background (Supplementary_Table_6, Supplementary_Figure_4). This strategy avoids confounding variation at X and autosomal regions, and Y copy number polymorphisms were estimated based on Illumina read coverage (see Methods). Overall, we find relatively little variation in copy number for both multi-copy Y and co-amplified Y genes among different neo-Y-chromosomes (Extended_Data_Fig_2). Low copy number variation is consistent with reduced levels of single-nucleotide diversity on the D. miranda neo-Y-chromosome (π=0.01%; i.e. 30-fold lower than typical levels of variation in this species20), due to a recent selective sweep that completely eliminated all standing variation a few thousand years ago20.
Discussion
Different evolutionary processes may cause amplification of neo-Y genes
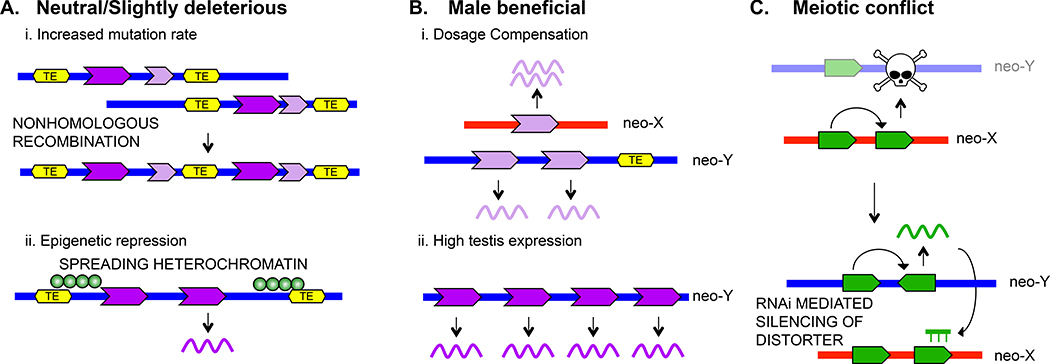
What may drive massive gene amplification on the neo-Y-chromosome? Y-chromosomes are subject to unique evolutionary forces: they lack recombination, show male-limited inheritance, and compete with the X over transmission to the next generation3,21. Indeed, our functional genomic analysis suggests that different processes appear to trigger gene-family expansion of multi-copy Y-genes versus co-amplified X/Y genes (Figure_2). Repetitive sequences, and in particular TEs are accumulating on the Y, and its high repeat content makes the Y-chromosome particularly prone to accumulate multi-copy genes for multiple reasons (Figure_2A). On one hand, repetitive sequences can provide a substrate for non-allelic homologous recombination and thereby promote gene-family expansion22. Indeed, we find several cases where repeats flank gene duplications on both the X and Y-chromosome, and may have contributed to their origination (Supplementary_Figure_5). Additionally, spreading of heterochromatin from repeats globally dampens expression of neo-Y genes23, and multi-copy gene-families may simply be more tolerated on the neo-Y (although many individual gene copies are transcribed, Supplementary_Table_4).
Figure_2. Distinct evolutionary processes may drive the accumulation of multi-copy Y genes, or co-amplified X and Y genes. A.
Amplified Y genes may have no fitness benefits or be slightly deleterious. Repeats on the neo-Y can provide a substrate for non-allelic homologous recombination and promote gene family expansion (i). Gene duplicates may be silenced by spreading heterochromatin on the neo-Y, and thus be less deleterious (ii). B. Multi-copy Y genes may provide fitness benefits to males, either through compensating for reduced gene dose of neo-Y genes (i) or by contributing to male fertility (ii). C. Co-amplified X/Y genes may be involved in an intergenomic conflict over segregation, and invoke the RNAi pathway to trigger silencing of meiotic drivers.
Gene-family expansions on the Y can also be beneficial for males (Figure_2B). Global transcription is lower from the neo-Y-chromosome of D. miranda24, and drives the evolution of dosage compensation of homologous neo-X genes25,26. D. miranda has evolved only partial dosage compensation of its neo-X-chromosome25,27, and gene amplification may help compensate for reduced gene-dose of neo-Y genes. Additionally, Y-chromosomes are transmitted from father to son, and are thus an ideal location for genes that specifically enhance male fitness28. Y-chromosomes of several species, including humans, have been shown to contain multi-copy gene-families that are expressed in testis and contribute to male fertility29–31.
Gene amplification on the Y could also be a signature of intragenomic conflicts (Figure_2C). Y-chromosomes compete with the X over transmission to the next generation32,33, and sex-chromosomes may try to cheat fair meiosis to bias their representation in functional sperm (meiotic drive). Meiotic drive on sex-chromosomes, however, reduces fertility and distorts population sex-ratios32, and creates strong selective pressure to evolve suppressors to silence selfish drivers. Suppression of sex-chromosome drive could be mediated by sequence homology between the suppressor and distorter, through RNAi mechanisms, and could result in co-amplification of genes on the sex-chromosomes. The RNAi pathway has been implicated to mediate suppression of sex-chromosome drive in Drosophila34–36.
Amplification of multi-copy neo-Y genes may increase male fitness
We find a handful of multi-copy Y gene-families that have dozens of gene-copies on the Y (six gene-families have >30 copies, and 14 gene-families have >15 copies; Figure_3, Supplementary_Figure_6), while the vast majority of multi-copy Y gene-families only have a few copies (90% of multi-copy Y gene-families have <4 copies). Dosage compensation counters ploidy differences of X-linked genes in males vs. females (one vs. two copies), and thus may contribute to amplification of multi-copy Y genes with few copies (~2–4 copies on the Y), while testis-expressed multi-copy Y genes often contain dozens of gene-copies29–31,37. Gene expression and chromatin analysis support that different evolutionary forces contribute to the accumulation of low versus high-copy number multi-copy Y gene-families.
Figure_3. Characterization of ampliconic Y genes.
Expression of multi-copy Y genes (top, purple shading) and co-amplified Y genes (bottom, green shading) in different male D. miranda samples, and tissue-specificity index (τ). Genes are sorted by their copy number on the Y. Multi-copy Y genes with high copy number are primarily expressed in testis, while gene families with low copy number are expressed in multiple tissues. Co-amplified X/Y genes show testis-biased expression, independent of copy number. The Z-Scores are calculated by scaling the rows to have 0 median and a standard deviation of 1. The data shown are presented in Data Supplement 6.
Multi-copy Y gene-families with a high copy-number (>15 copies) are expressed almost exclusively in testis (Figure_3, Supplementary_Figure_6, 7), mimicking patterns of gene-family amplification of male-fertility genes found in other species4,29,31,38,39. Their neo-X homologs, in contrast, are expressed predominantly in ovaries (Supplementary_Figure_6, 7). Gene expression profiles in D. pseudoobscura suggests that these genes were ancestrally highly expressed in testis and/or ovaries (Supplementary_Table_7), and sex-linkage may have enabled neo-Y and neo-X gametologs to specialize in their putative male- and female-specific function, respectively28.
Most multi-copy Y gene-families, in contrast, only have few copies and are ubiquitously expressed (Figure_3). Consistent with gene dosage contributing to increased copy number on the Y, the neo-X homologs of multi-copy Y genes are less likely to be dosage-compensated compared to single-copy Y genes (Extended_Data_Fig_3). In particular, male Drosophila achieve dosage compensation by recruiting the MSL-complex to their hemizygous X chromosome26, and neo-X homologs of multi-copy neo-Y genes are less likely to be targeted by the MSL complex in male D. miranda larvae27 than the neo-X homologs of single-copy neo-Y genes (p-value Fisher’s exact-test=0.007). This suggests that many multi-copy Y genes are dosage-sensitive, and additional gene-copies on the Y may contribute to dosage compensation. Truncated neo-Y genes are less likely to produce functional proteins and thus alleviate gene-dose deficiencies. Despite having many fewer copies on the Y on average (3 copies/gene), the low-copy-number multi-copy Y genes have a similar fraction of genes with at least two full-length copies (roughly half) as high-copy-number multi-copy Y genes (average 26 copies/gene), or co-amplified X/Y genes (average 22 copies/gene; see Supplementary_Table_2,3). Genes that co-amplify on the X and Y-chromosome, on the other hand, show testis-biased expression, independent of copy number (Figure_3). Gene ontology (GO) analysis found no significant enrichment of gene annotations among multi-copy Y genes, consistent with a broad category of (possibly dosage-sensitive) genes amplifying on the Y. Overall, our analysis is consistent with a considerable fraction of multi-copy Y genes having an important function in males, as supported by tissue-specific expression, or patterns of MSL-binding.
X/Y co-amplified genes suggest ongoing conflict over sex-chromosome transmission
Functional enrichment analysis (Figure_4), gene expression patterns and small RNA profiles (Figure_5) suggest that fundamentally different forces drive co-amplification of genes on the X and Y-chromosome.
Figure_4. Co-amplified X/Y genes are enriched for meiosis-related and RNAi functions. A.
“Interactive graph” view of enriched GO terms. Bubble color indicates the p-value; bubble size indicates the frequency of the GO term in the underlying GO database. Highly similar GO terms are linked by edges in the graph, where the line width indicates the degree of similarity. B. “TreeMap” view of enriched GO terms. Each rectangle is a single cluster representative. The representatives are joined into ‘superclusters’ of loosely related terms, visualized with different shades of gray. Size of the rectangles reflect the p-value of the GO term.
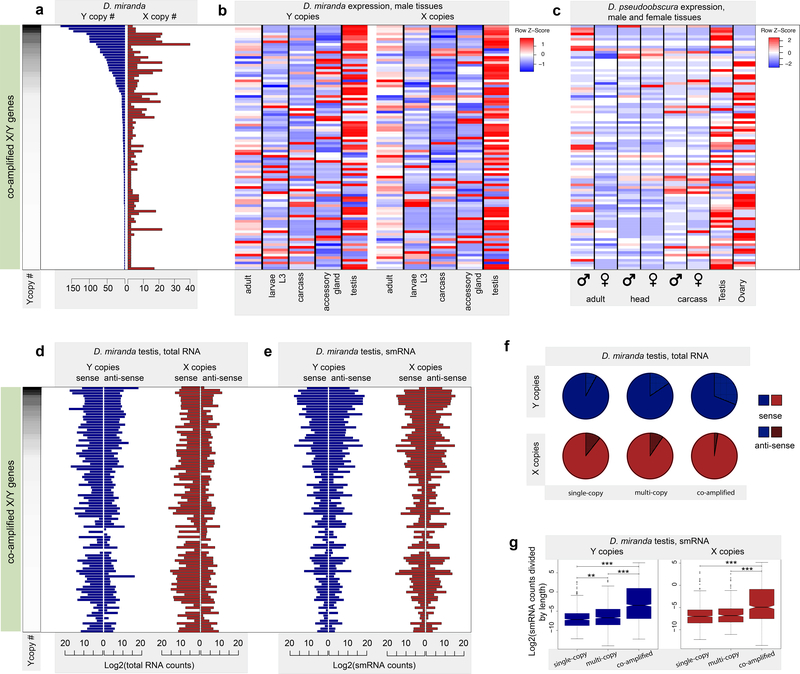
Figure_5. Co-amplified X/Y gene families produce anti-sense transcripts and small RNAs in testis.
. The 94 co-amplified X/Y gene families are sorted by copy number on the Y in panels A-E. A. Copy numbers on the Y and X chromosomes for co-amplified gene families in D. miranda. B. Tissue expression patterns for co-amplified X and Y genes in D. miranda male tissues. Co-amplified X/Y genes are highly expressed in testis. C. Tissue expression patterns of homologos of co-amplified X/Y genes in D. pseudoobscura. Homologs of co-amplified X/Y genes are highly expressed in testis and ovaries, suggesting an ancestral function in gametogenesis. D. Sense- and anti-sense transcription of total RNA for co-amplified X and Y genes in D. miranda testis. E. Sense- and anti-sense counts of small RNA for co-amplified X and Y genes in D. miranda testis. F. Fraction of sense and anti-sense transcripts produced for different categories of genes on the X/neo-X and Y/neo-Y-chromosome (i.e. ancestral single-copy Y and X genes; multi-copy Y/neo-Y genes and their neo-X gametologs; genes co-amplified on the Y/neo-Y and X/neo-X). G. Enrichment of small RNAs mapping to co-amplified X and Y genes. Shown are testis small RNA counts (normalized by total gene length for all copies of a gene family) for different categories of genes on the X/neo-X and Y/neo-Y-chromosome (as in panel F). The Wilcoxon test p-value significance is denoted by asterisks. The upper whisker and lower whisker in the boxplots show the 75th percentile + 1.5 times the interquartile range and the 25th percentile - 1.5 times the inter-quartile range, respectively. The Z-Scores are calculated by scaling the rows to have 0 mean and standard deviation 1. The data shown are presented in Data Supplement 7–13.
Overall, we identify 2683 co-amplified genes on the neo-sex-chromosomes of D. miranda (2036 Y/neo-Y and 647 X/neo-X-genes) that belong to 94 distinct proteins that were ancestrally single-copy, and phylogenetic analysis confirms their independent amplification on the X and Y (Extended_Data_Fig_1). Co-amplified X and Y-linked gene-copies are typically both highly expressed in testis (Figure_5B; Supplementary_Figure_6, 7). Testis expression of co-amplified X-linked genes is unusual, as testis-genes in Drosophila normally avoid the X-chromosome40–43, but can be understood under intragenomic conflict models21,34,35,44,45. In particular, an X-linked gene involved in chromosome segregation may evolve a duplicate that acquires the ability to incapacitate Y-bearing sperm (Figure_2C). Invasion of this segregation distorter skews the population sex-ratio and creates a selective advantage to evolve a Y-linked suppressor that is resistant to the distorter. Suppression may be achieved at the molecular level by increased copy-number of the wildtype function or by inactivation of X-linked drivers using RNAi34–36. If both driver and suppressor are dosage-sensitive, they would undergo iterated cycles of expansion, resulting in rapid co-amplification of driver and suppressor on the X and Y-chromosome32.
Consistent with meiotic conflict driving co-amplification of X/Y genes, we find that many highly co-amplified genes have well-characterized functions in meiosis (Supplementary_Table_3,8), and are ancestrally expressed in gonads (using gene expression data from D. pseudoobscura as a proxy for ancestral expression; Figure_5C, Supplementary_Figure_8). Gene ontology (GO) analysis reveals that co-amplified X/Y genes are significantly overrepresented in biological processes associated with meiosis and chromosome segregation (Figure_4). In particular, multi-copy Y genes are significantly enriched for GO categories including “nuclear division”, “spindle assembly”, “meiotic spindle midzone assembly”, “DNA packaging”, “chromosome segregation”, or “male gamete generation” (see Table_S3). Among the most highly co-amplified X/Y genes are well-studied genes with important function in meiosis, including wurstfest (145 Y and 5 X-copies), a gene involved in spindle-assembly in male meiosis; mars (48 Y and 6 X-copies), a gene involved in kinetochore-assembly and chromosome segregation, orientation disruptor (18 Y and 5 X-copies), a chromosome-localized protein required for meiotic sister chromatid cohesion, or Subito (8 Y and 11 X- copies), a gene required for spindle-organization and chromosome segregation in meiosis (Figure_4, Supplementary_Table_3,8). These important meiosis genes are typically single-copy and highly conserved across insects, but highly co-amplified on the recently evolved D. miranda X and Y-chromosome.
Possible involvement of RNAi in sex-chromosome drive
Additionally, GO-analysis reveals a significant overrepresentation of co-amplified X/Y genes associated with piRNA metabolism and generation of small-RNA’s (Figure_4). Again, this is expected under recurring sex-chromosome drive where silencing of distorters is achieved by RNAi, since compromising the small RNA pathway would release previously silenced drive systems36. Noteworthy genes in the RNAi pathway that are typically single-copy in insects but co-amplified on the X and Y of D. miranda include Dicer-2 (26 Y and 6 X- copies), a endonuclease that cuts long double-stranded RNA into siRNAs, cutoff (7 Y and 9 X- copies), a gene involved in transcription of piRNA-clusters, or shutdown (50 Y and 22 X-copies), a co-chaperone necessary for piRNA biogenesis (Supplementary_Table_3,8). Thus, functional enrichment supports a model of meiotic conflict driving co-amplification of X/Y genes.
We gathered stranded RNA-seq and small-RNA profiles from wildtype D. miranda testis, to obtain insights into the molecular mechanism of putative sex-chromosome drive. Consistent with meiotic drive and suppression through RNAi causing co-amplification of X/Y genes, the vast majority of co-amplified X/Y genes produce both sense and antisense transcripts and small-RNA’s (Figure_5D–G,6). Globally, co-amplified Y genes show significantly higher levels of anti-sense transcription and small-RNA production than single-copy Y genes, or multi-copy Y genes. Likewise, small-RNA levels are higher for co-amplified X-linked genes, compared to single-copy X genes, or X homologs of multi-copy Y genes (Figure_5D–G; Wilcoxon-test p-value<10−16, Supplementary_Table_9). Anti-sense transcription of many co-amplified X/Y genes suggests that they may function not as proteins, but instead as functional RNA by generating double-stranded RNA and triggering the RNAi silencing pathway. Targeting of co-amplified X/Y genes by small-RNA’s in testis demonstrates that small-RNA production is not simply a consequence of the repeat-rich environment of the neo-Y but instead a property of co-amplified X/Y genes. Overall, our data are consistent with sex-chromosome drive having repeatedly led to characteristic patterns of gene amplification of homologous genes on both the X and the Y-chromosomes that are targeted by small-RNAs (Figure_6).
Figure_6.
Examples of co-amplified X and Y genes. Shown are the genomic architecture of co-amplified gene families on the neo-X and neo-Y (repetitive regions are displayed in yellow, and co-amplified genes in green, other genes in gray), and expression profiles from testis (stranded RNA-seq and small RNA profiles in red for the neo-X and blue for the neo-Y). For each gene (A. fest; B. mars; C. zip), only a representative subset of copies is shown.
Conclusions
Contrary to the paradigm that Y-chromosomes undergo global degeneration, we document a high rate of gene gain on the recently formed neo-Y-chromosome of D. miranda, mainly through amplification of genes that were ancestrally present on the autosome (chr3) that became the neo-Y. Our comparative genomic analysis reveals different types of amplified Y genes, and we show that their acquisition likely is driven by different selective pressures. Multi-copy genes exclusive to the Y presumably increase male fitness, while genes that are co-amplified on the X and Y likely reflect intragenomic conflict. Multi-copy Y genes come in two flavors, and our analysis suggests that they are selected and amplifying on the Y either because of their testis-specific function, or to compensate for gene-dosage deficiencies. Genes with testis-biased expression often have dozens of copies on the Y, and their neo-X homologs are often expressed in ovaries, and sex linkage may have allowed these former homologs to specialize in their sex-specific roles36. Ubiquitously expressed housekeeping genes also duplicate on the Y, possibly to mitigate gene-dose deficiencies of partially silenced neo-Y genes; these genes are present at a much lower copy number, and are targeted less often by the dosage-compensation-complex on the X.
Co-amplified X/Y genes are highly expressed in testis and often have functions in chromosome segregation and RNAi, and their parallel amplification on the X and Y may be a consequence of ongoing X-Y interchromosomal conflicts over segregation. Sequence homology between putative drivers and their suppressors on the sex-chromosomes, and their widespread targeting by small-RNAs suggests that RNAi is involved in silencing rampant sex-chromosome drive. If amplified Y-genes are involved in a battle with the X over fair transmission, changes in gene copy-number may bias inclusion into functional sperm, and trigger repeated co-amplification of distorters and suppressors on the sex-chromosomes.
Either sex chromosome could initiate this evolutionary tug-of-war over transmission, but the X chromosome is a priori more likely to acquire segregation distorters, creating strong selection to evolve suppressors on the Y. On one hand, natural selection is impaired on the non-recombining Y3, making drivers more likely to originate on the X. Additionally, the heterochromatic nature of a Y-chromosome may render it especially vulnerable to be exploited by selfish elements during meiosis46.
Rampant sex-chromosome drive can have important evolutionary consequences. Strong selective pressure to amplify Y-linked suppressors of meiotic drive may indirectly account for the complete genetic decay of the Y. Since the Y-chromosome lacks recombination, strong positive selection for meiotic drive suppressors can propel linked deleterious mutations to fixation17, and the ongoing degeneration of ancestral Y genes may thus be a by-product of silencing recurrent meiotic drivers arising on the X. Patterns of molecular variation are suggestive of episodes of recurrent positive selection shaping neo-Y evolution of D. miranda20, and natural lines of D. miranda show a wide range of sex-ratio bias (with typically female-biased sex ratios12). These observations are consistent with recurrent and ongoing conflicts over segregation affecting the genomic architecture of sex-chromosomes in this species.
Genetic conflict between X-Y ampliconic genes may also contribute to hybrid sterility and consequent reproductive isolation33,47,48. Segregation distortion can result in male hybrid sterility in Drosophila49, and further functional characterization of co-amplified, lineage-specific X-Y gene families will be needed to test the proposed link between X-Y genetic conflict and hybrid sterility.
X-Y interchromosomal conflict, and its consequent impact on gene amplification on sex chromosomes, may be widespread. In both human and mouse, the X and Y have co-acquired and amplified genes, and meiotic drive has been invoked to explain this co-amplification9,50–52,53. Co-amplified genes have also been found in D. melanogaster54, and RNAi-mechanisms mediate suppression of sex-ratio drive in flies34–36. Highly amplified gene families have been detected in other mammals55 and across fruit flies56, suggesting that sex chromosome drive may be prevalent in evolution; to determine the true phylogenetic range of lineage-specific acquisition and amplification of X-Y genes, high-quality sex chromosome assemblies across more taxa are needed.
Materials and methods
Genome and data availability
A 150-kb fragment of the Y-chromosome was found to be missing in the previous genome assembly14 and the Y fragment was correctly reinserted before all downstream analyses. The updated genome assembly has been submitted to GenBank. All the data that were used and generated for this project are given in Supplementary_Table_10.
De novo transcriptome assembly
To mask repeats in the genome, we used RepeatMasker57 with custom de novo repeat libraries, generated using RepeatModeler58 and Repdenovo59, along with the Drosophila repeat library from Repbase60. The de novo repeat libraries are given in Data Supplement 1, and a repeat-masked gff file is given in Data Supplement 2. Paired end RNA-seq reads from several male and female tissues (heads, carcass, whole body, testis, ovary, accessory gland, spermatheca, 3rd instar larvae) were then aligned to the repeat-masked genome using HiSat261 with the --dta parameter on default settings. The resulting alignment file was used to assemble the transcriptome using the software StringTie62 with default parameters. Fasta sequences of the transcripts were extracted from the gtf output produced by StringTie using the gffread utility.
Gene annotation with Maker
We ran Maker63 three times to iteratively build and improve the gene annotation of the neo-sex chromosomes. For the first Maker run, we used annotated protein sequences for D. melanogaster and D. pseudoobscura from flybase.org, our de novo assembled D. miranda transcripts (see above), and the gene predictors Augustus64 and SNAP65 to get the initial set of predictions. The parameters est2genome and protein2genome were set to 1 to allow Maker to directly build gene models from the transcript and protein alignments, and we used the Augustus fly gene model and the SNAP D. melanogaster hmm file for this first run. The predictions from the first round were then used to train Augustus using BUSCO66 and also to train SNAP. The new Augustus gene model and SNAP hmm file were then used during the second Maker run, with the parameters est2genome and protein2genome set to 0. The maximum intron size was increased to 20000bp (default 10000bp). The results from the second round were then used to train Augustus and SNAP again, before the final round of Maker. This process resulted in a total of 21,524 annotated genes in D. miranda.
Orthology detection
Transcript sequences for D. pseudoobscura were downloaded from flybase.org and only the largest transcript per gene was retained for downstream analyses. De novo annotated D. miranda transcripts were then aligned to this filtered D. pseudoobscura transcript set using BLAST67. Alignments with percentage identity <60% were discarded and the best alignment was calculated based on the e-value, score, % identity and alignment lengths. Each D. miranda transcript was thus assigned the ortholog that was its best BLAST hit. We identified paralogous genes in the D. pseudoobscura genome as those for which at least 80% of the sequence of one aligned to the other and vice versa. Paralogous genes in the D. miranda – D. pseudoobscura orthology calls were replaced by a single gene name from the duplicated gene family.
Identifying multicopy genes
The gene annotation produced by Maker had roughly 2,500 more genes annotated on the Y/neo-Y compared to the neo-X, and hundreds of genes had multiple annotated copies on the Y/neo-Y-chromosome (and also the X chromosome and autosomes in some cases). Based on the orthology calls from BLAST, 822 Maker annotated genes had more than 2 copies on the Y/neo-Y, and 209 of those genes had more than two copies on both the X/neo-X and Y/neo-Y. In our initial Maker annotation, 366 genes were missing on the neo-Y and 155 genes were missing on the neo-X. However, closer inspection revealed that the annotation was often fragmented, especially on the Y/neo-Y-chromosome, which led to an overestimation of the number of distinct genes that had duplicated, but subsequent BLAST searches also revealed that Maker often failed to annotate individual copies of gene families. On the other hand, several genes in the annotation were “chimeras”, where two genes were collapsed into one by Maker and thus one of the genes appeared to be missing from the gene annotation, if it got assigned to the other D. pseudoobscura gene during orthology assignment. Thus, the actual number of missing genes is much smaller than our initial Maker annotation suggested. We thus manually verified, and if necessary fixed, each gene model that was annotated by Maker, and inferred to be either duplicated on the Y/neo-Y or missing from the neo-X and/or Y/neo-Y annotations. We used nucmer18 from the mummer package to individually align (one gene at a time) the sequences of their corresponding D. pseudoobscura orthologs to the D. miranda genome with the parameters --maxmatch and --nosimplify. Alignment coordinates were manually stitched together to get full gene coordinates. Only fragments that were at least 25% the length of the corresponding D. pseudoobscura ortholog, or at least 1000-bp long, were counted as duplicates/paralogs in the D. miranda genome. We also performed BLAST searches to identify the genes that had been lost from the neo-sex chromosomes.
In total, we annotate 6,448 genes on the neo-Y. Of these, 1,736 are ancestral single-copy Y genes (i.e. they were present on the ancestral autosome that formed the neo-Y). 1,105 of these genes were readily identified on both the neo-X and neo-Y by our Maker annotation, and are used as the single-copy orthology gene set in our analysis. 631 ancestral single-copy Y genes were initially missed or mis-qualified by our Maker annotation (i.e. 347 neo-Y genes were wrongly annotated as multi-copy by Maker, but our manual inspection revealed that they were present only as single-copy genes, and 114 neo-X genes and 170 neo-Y genes were missing from the Maker annotation, but found to be present on the neo-X and neo-Y, respectively, after manual checking using nucmer18.
In addition, we identify 457 genes (with 3,733 gene copies) that have become amplified on the neo-Y: 1,697 multi-copy Y genes (with a single copy on the neo-X), and 2,036 co-amplified neo-Y genes (which also amplified on the X/neo-X). In addition, we detect 959 genes in our Maker annotation that were not further considered. These “other” genes are comprised of 159 neo-Y genes that lack a homolog in D. pseudoobscura, 287 neo-Y genes that are present on an unknown location in D. pseudoobscura, 189 single-copy neo-Y genes that are present at multiple other locations in the genome (based on the Maker annotation), and 324 genes (from 49 unique proteins) with complicated mapping which could not be included in any categories of our analysis (i.e. genes for which the number of copies were ambiguous based on alignments such as for nested/overlapping genes; genes for which many alignments of variable identity were observed; genes which were amplifying on autosomes and the Y-chromosome; chimeric genes).
Thus, after manual verification, we identified 94 genes that have co-amplified on the X/neo-X and the Y/neo-Y, with 647 copies on the X/neo-X and 2036 copies on the Y/neo-Y (and 58 copies on the autosomes). We also identified 363 genes that have only amplified on the Y/neo-Y-chromosome, with a total of 1697 copies on the Y/neo-Y. Thus, the Y/neo-Y-chromosome has gained at least 3200 gene copies.
We identified 17 genes that are present on chr3 in D. pseudoobscura but are missing on the neo-X in D. miranda; 6 of those genes are found on other chromosomes in the D. miranda genome and 6 are still present on the Y-chromosome in D. miranda. We identified 143 genes that are present on chr3 in D. pseudoobscura but absent on the neo-Y in D. miranda and 138 of those are still present on the neo-X in D. miranda. However, 5 genes have been lost from both the neo-X and neo-Y-chromosomes and BLAST searches failed to identify other chromosomal locations that those genes could have moved to. Genome annotations for all genes, multi-copy Y genes and their homologs, and co-amplified X and Y genes are given in Data Supplements 3–5.
Karyotype plots showing co-amplified X/Y genes were produced using karyoploteR68. Plots showing multi-copy gene locations on the neo-X and neo-Y (Y1 and Y2) were created using genoPlotR69.
Y-chromosome replacement lines
Y-chromosomes from seven D. miranda lines (Supplementary_Table_6) were moved into an MSH22 (reference) background by repeated backcrossing of hybrid males (8 generations) with virgin MSH22 females. We then extracted DNA from a single male from each Y-chromosome replacement line using a Qiagen DNeasy kit following the standard extraction protocol. DNA libraries were prepared using the Illumina TruSeq Nano Prep kit and sequenced on a Hiseq 4000 with 100bp PE reads. Raw reads from these seven Y-chromosome replacement lines along with sequencing data for three isofemales lines (MA03.4, 0101.7, MA03.2) were initially mapped to the reference MSH22 genome using BWA mem70. The resulting files were processed using Samtools71 and PCR duplicates were removed using Picard Tools. We called SNPs using GATK’s UnifiedGenotyper72 and filtered SNPs using VCFtools73 and retained biallelic SNPs, positions with no more than 50% of individuals missing a call, and individual genotypes with GQ > 30 and depth > 3 and < 80.
To confirm Y replacement, we first estimated nucleotide diversity (π) using VCFtools across each chromosome with the expectation that it should be uniformly low given that MSH22 is an inbred line. We noted a few regions that showed peaks of elevated heterozygosity (Supplementary_Figure_4A), which is indicative of either residual heterozygous regions in the MSH22 line or suggests that our backcrossing failed to replace all chromosomes with MSH22 chromosomes. Further, the MSH22 YBB51 line appeared to be heterozygous across all of chr4 while all its other chromosomes appeared to be replaced with MSH22 chromosomes. However, given that variation on chr4 of line BB51 likely contributes little to Y-chromosome gene amplification estimates, we retained MSH22 YBB51 in our analyses. Phylogenetic networks using SplitsTree74 for chr2 (17,189 SNPs) and the Y-chromosome (1,543 SNPs) confirmed that chr2 of Y-chromosome replacement lines appeared identical to MSH22 while the seven Y-chromosomes all appeared genetically distinct (Supplementary_Figure_4B, C).
Read coverage analysis to infer gene copy number
We used DIAMOND75 to align raw Illumina reads from each Y-chromosome replacement line to the longest isoform for each D. miranda protein (n=12,180). Only the top hit for each read was retained, and mean coverage over each protein was estimated using Bedtools76. To estimate the copy number for co-amplified Y, multi-copy Y, and multi-copy autosome and X genes we normalized estimates using median coverage over 98 randomly selected X-linked single copy genes.
Phylogenetic analysis of co-amplified X/Y genes
Co-amplified X/Y gene regions and their D. pseudoobscura ortholog were aligned using MAFFT77. Due to the fragmented nature of some Y copies, a small number of copies were removed to maximize the number of informative sites while retaining most gene copies. We created rooted maximum-likelihood phylogenetic trees using RAxML 8.2.978 with 200 bootstrap replicates and a GTR + gamma model of sequence evolution. Phylogenetic trees were visualized using FigTree version 1.4.3 (https://github.com/rambaut/figtree/).
Gene expression analysis
Kallisto79 was used to quantify gene expression and calculate TPM values for each gene in our annotation using several male and female tissues (whole body, carcass, 3rd instar larvae, gonads, spermatheca and accessory gland) using default parameters and 100 bootstraps. The R function heatmap.2 from the gplots package (https://cran.r-project.org/web/packages/gplots/index.html) was used to plot heatmaps to visualize tissue-specific differences in gene expression. Each row in the heatmap is a different gene and the different columns represent different tissues. The heatmap was row-normalized (each row was scaled to have mean 0 and standard deviation 1) to indicate the tissue with the highest expression for each gene. The tissue-specificity index, τ was calculated in the following manner:
where, xi is the expression of the gene (TPM) in tissue i, n is the number of tissues, and
GO analysis
GO analysis was done using GOrilla80 and the D. melanogaster orthologs of multi-copy Y genes or genes co-amplifying on the X and Y were used as the target set. The GO terms that were enriched and had a p-value less than 10−3 and FDR less than 0.05 were visualized using the software Revigo81.
Testis RNA libraries
We dissected testes from 3–8 day old virgin males of D. miranda (strain MSH22) reared at 18°C on Bloomington food. We used Trizol (Invitrogen) and GlycoBlue (Invitrogen) to extract and isolate total RNA. We resolved 20 μg of total RNA on a 15% TBE-Urea gel (Invitrogen) and size selected 19–29 nt long RNA, and used Illumina’s TruSeq Small RNA Library Preparation Kit to prepare small RNA libraries, which were sequenced on an Illumina HiSeq 4000 at 50 nt read length (single-end). We used to Ribo-Zero to deplete ribosomal RNA from total RNA, and used Illumina’s TruSeq Stranded Total RNA Library Preparation Kit to prepare stranded testis RNA libraries, which were sequenced on an Illumina HiSeq 4000 at 100 nt read length (paired-end).
Analysis of testes smRNA and testes totalRNA data
Stranded total RNA paired-end reads were mapped to the D. miranda genome using HiSat261 with default parameters and the --rna-strandness parameter set to RF. Single-end small RNA-seq reads were aligned to the genome using bowtie282 and default parameters. BamCoverage from the deeptools package83 was used to convert bam alignment files to bigwig format in both cases to be visualized using IGV. Sense and antisense transcription estimates were obtained based on the alignment and the orientation of genes using bedtools84. The number of small RNA and total RNA reads mapping to the co-amplified X/Y genes were summed for each gene family, based on sense or antisense transcription, and barplots of counts were plotted in log2 scale using R (Figure_5D, E). The number of small RNA reads mapping to each annotated gene (sense and antisense counts) were divided by the gene/fragment length and boxplots were plotted in R for single copy Y genes, genes that have only amplified on the Y and genes that have co-amplified on the X and Y (Figure_5G).
Extended Data
Extended Data Fig. 1. Phylogenetic relationships of co-amplified X/Y genes in D. miranda.
Maximum-likelihood trees of D. miranda X and Y gene copies with nodes showing >70 bootstrap support highlighted with black circles. X-linked copies are shown in red, Y-linked copies shown in blue, with distinct X and Y groupings collapsed. Fasta alignments are in Data Supplement 14.
Extended Data Fig. 2. Copy number estimates for co-amplified Y genes, multi-copy Y genes, and multi-copy autosome and X genes.
For co-amplified Y genes we show all genes that were identified as co-amplified. For the multi-copy Y genes we only show genes with >3 copies on the Y. For multi-copy autosome and X genes we show only genes with >4 total copies. Multi-copy autosome and X estimates are predicted to be highly similar given that the autosome and X background in each Y-chromosome replacement line is nearly identical. Slight deviations are probably due to stochasticity in sequencing and read mapping, residual heterozygosity in the MSH22 line, or unique Y-chromosome gene amplifications.
Extended Data Fig. 3. Dosage compensation status of neo-X homologs of single-copy (left) and multi-copy (right) Y genes.
Shown are the relative numbers of neo-X genes that are bound by the MSL-complex (and are thus dosage compensated), and those not bound (and thus not dosage compensated). MSL-binding data were generated for male D. miranda larvae. Genes with multiple copies on the Y are less likely to be dosage compensated on the X. The data are presented in Data Supplement 15.)
Supplementary Material
Acknowledgments
Funded by NIH grants (R01GM076007, GM101255 and R01GM093182) to DB. We thank L. Gibilisco for generating small RNA libraries and K. Chatla and A. Tran for generating genomic libraries.
Footnotes
Competing interests
The authors declare that no competing interests exist.
Data availability
BioProject ID PRJNA545539 and NEE webpage.
References
- 1.Bachtrog D et al. Are all sex chromosomes created equal? Trends Genet. 27, 350–357 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth B Model for evolution of Y chromosomes and dosage compensation. Proc. Natl. Acad. Sci. U.S.A. 75, 5618–5622 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachtrog D Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahajan S & Bachtrog D Convergent evolution of Y chromosome gene content in flies. Nat Commun 8, 785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellott DW et al. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat. Genet. 49, 387–394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatti M & Pimpinelli S Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 26, 239–275 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Blackmon H, Ross L & Bachtrog D Sex Determination, Sex Chromosomes, and Karyotype Evolution in Insects. J. Hered. 108, 78–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes JF et al. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature 437, 100–103 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Soh YQS et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachtrog D & Charlesworth B Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416, 323–326 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Muller HJ in The new Systematics (ed. JS H) [Google Scholar]
- 12.Dobzhansky T Drosophila Miranda, a New Species. Genetics 20, 377–391 (1935). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q & Bachtrog D Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337, 341–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan S, Wei KH-C, Nalley MJ, Gibilisco L & Bachtrog D De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 16, e2006348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho AB, Lazzaro BP & Clark AG Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl. Acad. Sci. U.S.A. 97, 13239–13244 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachtrog D, Hom E, Wong KM, Maside X & de Jong P Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 9, R30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charlesworth B & Charlesworth D The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 355, 1563–1572 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz S et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucotte EA et al. Dynamic Copy Number Evolution of X- and Y-Linked Ampliconic Genes in Human Populations. Genetics 209, 907–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachtrog D Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36, 518–522 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Meiklejohn CD & Tao Y Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. (Amst.) 25, 215–223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konkel MK & Batzer MA A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 20, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q et al. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11, e1001711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachtrog D Expression profile of a degenerating neo-y chromosome in Drosophila. Curr. Biol. 16, 1694–1699 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ellison CE & Bachtrog D Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 342, 846–850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchesi JC & Kuroda MI Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol 7, a019398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alekseyenko AA et al. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 27, 853–858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice WR Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Skaletsky H et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Bellott DW et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez D et al. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Jaenike J Sex Chromosome Meiotic Drive. 113958 32, 25–49 (2003). [DOI] [Google Scholar]
- 33.Frank SA Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45, 262–267 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Tao Y, Masly JP, Araripe L, Ke Y & Hartl DL A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 5, e292 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao Y et al. A sex-ratio Meiotic Drive System in Drosophila simulans. II: An X-linked Distorter. PLoS Biol. 5, e293 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C-J et al. The hpRNA/RNAi Pathway Is Essential to Resolve Intragenomic Conflict in the Drosophila Male Germline. Dev. Cell 46, 316–326.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brashear WA, Raudsepp T & Murphy WJ Evolutionary conservation of Y Chromosome ampliconic gene families despite extensive structural variation. Genome Res. 28, 1841–1851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellott DW et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho AB, Dobo BA, Vibranovski MD & Clark AG Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 98, 13225–13230 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturgill D, Zhang Y, Parisi M & Oliver B Demasculinization of X chromosomes in the Drosophila genus. Nature 450, 238–241 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assis R, Zhou Q & Bachtrog D Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol 4, 1189–1200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meiklejohn CD, Landeen EL, Cook JM, Kingan SB & Presgraves DC Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 9, e1001126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vibranovski MD, Zhang Y & Long M General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19, 897–903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller JL et al. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 40, 794–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller JL et al. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 45, 1083–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helleu Q et al. Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc. Natl. Acad. Sci. U.S.A. 113, 4110–4115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurst LD & Pomiankowski A Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128, 841–858 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larson EL, Keeble S, Vanderpool D, Dean MD & Good JM The Composite Regulatory Basis of the Large X-Effect in Mouse Speciation. Mol. Biol. Evol. 34, 282–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phadnis N & Orr HA A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323, 376–379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lahn BT & Page DC A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum. Mol. Genet. 9, 311–319 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Cocquet J et al. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8, e1002900 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cocquet J et al. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7, e1000244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larson EL, Kopania EEK & Good JM Spermatogenesis and the Evolution of Mammalian Sex Chromosomes. Trends Genet. 34, 722–732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balakireva MD, YuYa Shevelyov, Nurminsky DI, Livak KJ & Gvozdev VA Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster. Nucleic Acids Res. 20, 3731–3736 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy WJ et al. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2, e43 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellison CE & Bachtrog D Recurrent gene amplification on Drosophila Y chromosomes suggests cryptic sex chromosome drive is common on young sex chromosomes. doi: 10.1101/324368 [DOI]
- 57.Smith A, Hubley R & Green P RepeatMasker Open-4.0. RepeatMasker Open-4.0. Available at: (Accessed: 30 August 2017)
- 58.Smith A & Hubley R RepeatModeler Open-1.0. RepeatMasker Open-4.0. Available at: (Accessed: 30 August 2017)
- 59.Chu C, Nielsen R & Wu Y REPdenovo: Inferring De Novo Repeat Motifs from Short Sequence Reads. PLoS ONE 11, e0150719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bao W, Kojima KK & Kohany O Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pertea M et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campbell MS, Holt C, Moore B & Yandell M Genome Annotation and Curation Using MAKER and MAKER-P. Curr Protoc Bioinformatics 48, 411.1–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanke M & Waack S Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 Suppl 2, ii215–25 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Korf I Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV & Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Camacho C et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gel B & Serra E karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guy L, Kultima JR & Andersson SGE genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danecek P et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huson DH & Bryant D Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Buchfink B, Xie C & Huson DH Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Supek F, Bošnjak M, Škunca N & Šmuc T REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramírez F, Dündar F, Diehl S, Grüning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quinlan AR BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47, 1112.1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.