Abstract
Honey bees and bumble bees belong to the same family (Apidae) and their workers exhibit a division of labor, but the style of division of labor differs between species. The molecular and neural bases of the species-specific social behaviors of Apidae workers have not been analyzed. Here, we focused on two immediate early genes, hormone receptor 38 (HR38) and early growth response gene-1 (Egr1), and late-upregulated ecdysone receptor (EcR), all of which are upregulated by foraging flight and expressed preferentially in the small-type Kenyon cells of the mushroom bodies (MBs) in the honey bee brain. Gene expression analyses in Bombus ignitus revealed that HR38 and Egr1, but not EcR, exhibited an immediate early response during awakening from CO2 anesthesia. Both premature mRNA for HR38 and mature mRNA for Egr1 were induced during foraging flight, and mRNAs for HR38 and Egr1 were sparsely detected inside the whole MB calyces. In contrast, EcR expression was higher in forager brains than in nurse bees and was expressed preferentially in the small-type Kenyon cells inside the MBs. Our findings suggest that Kenyon cells are active during foraging flight and that the function of late-upregulated EcR in the brain is conserved among these Apidae species.
Subject terms: Molecular neuroscience, Animal behaviour
Introduction
Eusocial bees, including honey bees and bumble bees, exhibit highly sophisticated sociality as represented by their caste differentiation and division of labor of workers. Their social behaviors, however, show some species-specific differences. The division of labor in honey bee workers is based on the age after eclosion1,2; older foragers convey information about the location of a food source with a unique dance called the “waggle dance” to communicate information to their nestmates about the location of a food source. On the other hand, the division of labor in bumble bees is based on their body size rather than age; smaller bumble bees remain in the hive to nurse their brood3,4 whereas larger workers are engaged in foraging outside the hive within a few days after eclosion5. In contrast to honey bees, bumble bee foragers do not communicate the location of their foraging success to their nestmates: successful foragers simply alert their nestmates to the presence of a food source by running about in the hive6,7. The molecular and neural mechanisms of honey bee foraging behavior are well investigated8–12, but few studies have examined bumble bee foraging behavior13,14.
The molecular and neural mechanisms underlying honey bee social behaviors have been investigated on the basis of the brain structure. The mushroom bodies (MBs), a higher-order center involved in learning and memory as well as in the integration of multimodal sensory information in the insect brain15, are implicated in foraging behavior in the European honey bee (A. mellifera)16–18. The MBs comprise intrinsic neurons termed Kenyon cells (KCs) that are classified into four subtypes: class I large (l)-, middle (m)-, small (s)-type KCs, and class II KCs, according to the size and position of their somata, and gene expression profiles19–24, which are conserved among Aculeate Hymenoptera25.
Previous studies used immediate early genes (IEGs), whose expression is rapidly upregulated after neuronal activation, to identify the brain regions related to certain behaviors26. Findings from these studies using a battery of IEGs, such as kakusei (noncoding RNA identified from A. mellifera)27, Egr1 (early growth response gene-1, also known as NGFI-A, Krox24, zif268, and zenk)28–31, and HR38 (hormone receptor 38, the subfamily of nuclear receptor 4A)23,32–34, suggested a possible role of the sKCs and some mKCs in the MBs in sensory processing during the foraging flight in honey bees12. In addition to HR3835, EcR (ecdysone receptor) and other ecdysone signaling genes, such as DopEcR (dopamine/ecdysteroid receptor), and Ddc (dopa decarboxylase) are also reported to be upregulated during the foraging flight in honey bees, raising the possibility that ecdysone signaling in the honey bee brain is involved in foraging behavior36.
In the present study, we analyzed two IEGs, HR38 and Egr1, and late-upregulated EcR to evaluate neural activity in the bumble bee (Bombus ignitus, Bi) and honey bee (Apis mellifera, Am) during foraging flight to disclose common and species-specific features of the neural activity related to foraging. First, we confirmed that both Bombus HR38 and Egr1 exhibit an immediate early response similar to Apis IEGs. Next, we analyzed the expression pattern of three Bombus genes, HR38, Egr1, and EcR, in the forager brain under two experimental conditions. In the first condition, the hives were set in a greenhouse, which partly resembles the natural condition, and in the second condition, the hives were set in a laboratory flight-cage, enabling us to sample foragers according to the foraging time-course. We also performed in situ hybridization to detect the expression profiles of these genes in the forager brains. Our findings indicated that both premature mRNA for HR38 and mature mRNA for Egr1 were induced in the bumble bee brains during foraging flight and both mRNAs for HR38 and Egr1 were sparsely detected inside the whole MBs. On the other hand, we showed that expression of EcR in the brain was significantly higher in nurse bees than in foragers and expressed preferentially in the sKCs of the MBs in bumble bee foragers. These results suggest that neural activity in the forager brain and the function of ecdysone signaling in the sKCs are conserved among these two species.
Materials and Methods
Animals
Bumble bee (B. ignitus) colonies at Tamagawa University (Machida-Shi, Tokyo, Japan) are usually kept under laboratory conditions (28 °C, 70% humidity, 24 h dark). For the present study, two colonies were placed in a greenhouse (Fig. 1, left panel) and three colonies were placed in a laboratory flight-cage (1 m × 50 cm × 50 cm, equipped with pollen feeding sites and absorbent cotton soaked in sugar water, Fig. 1, right panel) and maintained under laboratory conditions (25 ± 3 °C, 73 ± 5% humidity, and natural day/light hours) at Tamagawa University. A bumble bee colony was purchased from Agrisect Inc. (Inashiki-Shi, Ibaraki, Japan). Three European honey bee (A. mellifera) colonies were purchased from Kumagaya Apiary (Kumagaya-Shi, Saitama, Japan) and kept outside at the University of Tokyo (Bunkyo-Ku, Tokyo, Japan).
Figure 1.
Experimental set-up used for B. ignitus worker sampling. (Left) B. ignitus colonies were set in a greenhouse. This photo shows a hive (white box in the center of the photo) set on a block and covered with a screen to shade the hive from direct sunlight in the greenhouse (arrow). (Right) A B. ignitus colony was set in a laboratory flight-cage. A hive with its entrance (wooden box) is on the left and a small dish supplied with pollen and three brushes attached with pollen as feeders are shown at the bottom and top of the photo, respectively. Note that one forager is collecting pollen at the feeder and another is flying to search for the feeder (arrowheads).
Sampling for immediate early response analysis
A total of 70 B. ignitus workers were randomly collected from a colony and groups of 5 workers were divided into 14 insect cages (round plastic containers, 15 cm diameter and 4.5 cm high), and kept in a dark incubator at 25 °C overnight. The next morning at 8:00, all workers were set under a luminescent light in a laboratory space. Anesthesia was induced in 35 workers (7 insect cages) by supplying CO2 to the insect cages for seizure induction after hypoxia37,38, and 5 min later the CO2 in the 6 insect cages was exchanged with fresh air. The workers were collected at each time-point (0, 15, 30, 60, 120, and 180 min) after the CO2 was exchanged with fresh air (“CO2”). Workers anesthetized continuously with CO2 for 120 min were collected as a negative control (“NC”), in which we expected to detect no immediate early response. Another 35 workers (7 insect cages) that were supplied with air flow instead of CO2 were collected at the same time-points (10 min before the onset of the CO2 supply, 0, 15, 30, 60, 120, and 180 min after the CO2-fresh air exchange) as a series of positive controls (“PC”), in which we expected to detect the induction of IEGs due to the surrounding stimuli, but not due to exposure to high levels of CO2. After the bees were immediately anesthetized in iced water, the whole brains were dissected with fine tweezers and scalpels under a binocular microscope and then frozen at −80 °C for preservation.
Sampling for foraging flight analysis
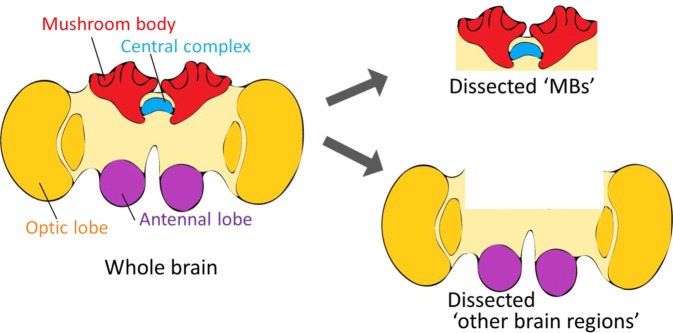
The sampling of B. ignitus foragers was performed in August 2018. For sampling in the greenhouse, “Foragers” that visited flowering fruit trees (Pouteria lucuma) with pollen loads and “Nurse bees” that were engaged in the in-hive tasks (feeding the brood, smoothing the nest combs, or warming eggs and pupae) were captured from the hives around 14:00. For sampling from the laboratory flight-cage, the day before the sampling day, all workers outside the hives were recovered in the hives and the hive entrances were closed. The next day at 8:30, the entrances were opened. The workers emerging from the entrances were immediately captured. At the same time, nurse bees in the hives were collected. Workers that were foraging around the pollen feeder at 3–7 min (8:37) and 25–30 min (9:00) after opening the entrance were collected. After they were anesthetized in ice water, the body size from the top of the head to the bottom of the abdomen of each bee was measured using a ruler with 1-mm resolution. For quantitative reverse transcription-polymerase chain reaction (qRT-PCR), the MBs and other brain regions were dissected as depicted in Fig. 2. Each dissected tissue was frozen at −80 °C for preservation.
Figure 2.
Brain dissection for qRT-PCR analysis. Schematic drawing of the brain dissection for qRT-PCR analysis. Dissected “MBs” mainly include the MBs and central complex. Dissected “other brain regions” mainly include the antennal lobes and optic lobes.
Sampling of A. mellifera foragers was performed from September to October 2019. The day before the sampling day, the bees that returned to the hive with pollen loads were caught as foragers and marked on their thorax with a non-permanent marker pen. Early the next morning at 6:30, marked bees were collected from inside the hives, i.e., foragers that had not yet begun foraging that day. At 8:30, the hive entrances were opened and the workers exiting the hive were captured immediately. Foragers returned to their hives with pollen loads were collected at 9:00, 9:30, and 12:30, respectively. Nurse bees were also collected from inside the hives at 6:30, 9:30, and 12:30 based on their behaviors of plunging their heads into honeycomb cells that contained larvae more than twice as evidence of nursing their brood39. Half of the foragers and nurse bees captured at 12:30 were incubated in a dark incubator till 22:30. All the bees were promptly anesthetized in ice water and their brains were dissected for qRT-PCR as described above. Because the hypopharyngeal glands, which synthesize royal jelly, are well developed in nurse bees whereas they are shrunken in foragers40, nurse bees collected were further screened under a stereomicroscope to collect brains only from bees with well-developed glands41.
qRT-PCR analysis
Expression analysis by qRT-PCR was performed essentially as described previously31 using TB Green Premix Ex Taq II (Tli RNaseH plus; Takara) and gene-specific primers (Table 1) with a Light Cycler 480 Instrument II (Roche Life Science, Indianapolis, IN, USA).
Table 1.
Gene-specific primers.
| Animal (analysis) | Gene name | NCBI Gene ID | Primer sequence | Size (bp) | Temp. (°C) |
|---|---|---|---|---|---|
| A. mellifera (qRT-PCR) | HR38 | 551592 | 5′-CGATTGGCTCCACAGTATTC-3′ and 5′-CTCCATGCGATGAGGCTCC-3′ | 136 | 58 |
| preHR38 | 551592 | 5′-TTATGTATGGACGTGCAGAC-3′ and 5′-ATCGGATACACGTCGATTAG-3′ | 125 | 52 | |
| EcR | 406084 | 5′-TACCACTACAACGCGCTCAC-3′ and 5′-CCTCATGTACATGTCGATCT-3′ | 120 | 56 | |
| Egr1 | 726302 | 5′-CCTCACCACCCACGTGAGAA -3′ and 5′-TGCTTGAGGTGGACTTTGGC-3′ | 117 | 58 | |
| EF1α | 408385 | 5′-TTGTGCCGTGTTAATAGTCG-3′ and 5′-GATCGGTCATGTCCATCTTG-3′ | 149 | 56 | |
| Actin | 406122 | 5′-TCCCCGAATCCCGAAAG-3′ and 5′-CGGAGGAACCAAAGGACAA-3′ | 89 | 55 | |
| B. ignitus (qRT-PCR) | HR38 | 100642535 | 5′-CGATTGGCTCCACAGTATCC-3′ and 5′-CTCCATGCGATGAGGTTCC-3′ | 136 | 58 |
| preHR38 | 100642535 | 5′-TGACGAGCCTACGACATGTC-3′ and 5′-TGAATCGTGGAAGGCGAGTT-3′ | 139 | 58 | |
| EcR | 100646757 | 5′-TATCACTACAACGCACTGAC-3′ and 5′-CCGCATGTACATATCGATCT-3′ | 120 | 55 | |
| Egr1 | 100651542 | 5′-CTTAACCACTCACGTGAGAA-3′ and 5′-TGTTTCAAGTGAACTTTCGC-3′ | 117 | 56 | |
| EF1α | 100631080 | 5′-TTGTGCCGTGTTAATAGTGG-3′ and 5′-GATCGGTCATGTCCATCTTG-3′ | 149 | 56 | |
| Actin | 100646910 | 5′-GTCTCGTTTCTCGACCATAG-3′ and 5′-ACTGATCTTCGAATGCCTAAA-3′ | 93 | 55 | |
| B. ignitus (in situ hybridzation) | HR38 | 5′-CAATCTTCTCACTACGTCCA-3′ and 5′-GGGATAGATAGTGCGCTTTC-3′ | 440 | ||
| EcR | 5′-CACTAATCAGCCCTCAGAAG-3′ and 5′-TCAAACTGAAGCACATCTCG-3′ | 566 | |||
| Egr1 | 5′-GAATCTCCTGTCCCATCATC-3′ and 5′-TGTTTCAAGTGAACTTTCGC-3′ | 573 |
NCBI, National Center for Biotechnology Information; Size, PCR product size; Temp., annealing temperature setting at Light Cycler.
The PCR conditions were as follows: 95 °C, 5 min, (95 °C, 10 s; annealing temperature of each gene is shown in Table 1, 10 s; 72 °C, 10 s) × 45 cycles, 65 °C, 1 min; 97 °C, 0 s; and 40 °C, 30 s. The selectivity of all primers was verified by agarose gel electrophoresis of the RT-PCR products amplified using Ex Taq Hot Start Version (Takara) and by analyzing melting curves of the qRT-PCR products (see Supplementary Fig. S1). The expression of each gene was normalized to that of EF1α and Actin (the expression level of each gene is shown in Supplementary Fig. S2). The relative expression was calculated using the ΔΔ Ct method (ABI user bulletin #20). The calibration samples were obtained at 0 min after the cessation of anesthesia in the CO2 group for the immediate early response validation of B. ignitus from the MBs of nurse bees in the greenhouse experiment of B. ignitus, from the MBs of nurse bees collected at 8:30 in the laboratory flight-cage experiment of B. ignitus, and from the MBs of nurse bees collected at 6:30 in the foraging flight experiment of A. mellifera.
In situ hybridization analysis
We used 7, 2, and 2 forager brains collected for the above-described experiment in B. ignitus to evaluate BiHR38, BiEcR, and BiEgr1 expression, respectively. Whole brains embedded in Tissue-Tek O.C.T. Compound (SAKURA Finetek) were frozen and sliced into 10-µm thick sections. The cDNA fragments, corresponding to the BiHR38, BiEcR, and BiEgr1 coding regions, were amplified from B. ignitus cDNA using gene-specific primers (Table 1). In situ hybridization was performed with digoxigenin-labeled riboprobes essentially as described previously42. Images of the brain slices were obtained using an optical microscope (BX-50, Olympus) and multiple photos were merged using Adobe Photoshop (CS3 EXTENDED ver.10.0, Adobe Systems) if necessary.
Statistical analysis
All statistics were performed using R statistical software (ver.3.3.3). For immediate early response analysis, two-way ANOVA (factor 1, treatment; factor 2, time) was performed. After that, for the CO2 and PC groups, one-way ANOVA, and Dunnett’s test (CO2 groups were compared with the 0 min group, the PC groups were compared with the −10 min group). To compare the NC group with the CO2 0 min group, an F test followed by Student’s t test or Welch’s t test was performed. The expression of EF1α and Actin differed slightly among the groups (Tukey-Kramer test after two-way ANOVA, CO2-NC: p < 0.01, CO2-PC: p < 0.001, see Supplementary Fig. S2a). For the greenhouse sampling of B. ignitus, two-way ANOVA (factor 1, tissue; factor 2, bee type) was performed. After the F test, Student’s t test or Welch’s t test was performed to compare each tissue between bee type. The expression of EF1α and Actin did not differ significantly for each bee type, but did differ significantly for each tissue by two-way ANOVA (also see Supplementary Fig. S2b). For the laboratory flight-cage sampling of B. ignitus, three-way ANOVA (factor 1, tissue; factor 2, bee type; factor 3, time) was performed, and then the Tukey-Kramer test was performed to compare between bee type and flight time for each tissue. The expression of EF1α was significantly different depending on the tissue (p < 0.001) and bee type (p < 0.05), and the expression of Actin was significantly different depending on the bee type (p < 0.05) (also see Supplementary Fig. S2c). For the sampling of A. mellifera, three-way ANOVA (factor 1, tissue; factor 2, bee type; factor 3, time) was performed for all samples. For each tissue, the Tukey-Kramer test was used to compare the upregulation dependence on the time-course, and Student’s t test or Welch’s t test was performed after the F test to compare nurse bees and foragers at each time-point. The expression of EF1α was significantly different depending on the tissue (p < 0.001), bee type (p < 0.001), and time (p < 0.001), and the expression of Actin was significantly different depending on the bee type (p < 0.05) (also see Supplementary Fig. S2d). The body sizes of B. ignitus were compared using Student’s t test and the Tukey-Kramer test.
Results
Validation of B. ignitus immediate early response by qRT-PCR
We first examined whether BiHR38, BiEgr1, and BiEcR show an immediate early response. We also examined premature mRNA for BiHR38 (termed as preBiHR38), because HR38 is induced a little later than Egr143. For this, B. ignitus workers were anesthetized with CO2 and seizures were induced by awakening them from anesthesia. After CO2 was supplied to the insect cages, all the bees fainted within 5 min. When the CO2 was exchanged with fresh air (cessation of anesthesia), some workers began to twitch their legs within a few minutes, which is a typical movement related to CO2-induced seizures27. Within 15 min, some bees got up and a few of them began to walk. Within 30 min, almost all workers were up, breathing with their abdomen, or grooming. Within 60 min, some workers were walking or flying, and approximately 60% of workers within 120 min and all of them within 180 min were actively walking or flying about the cage.
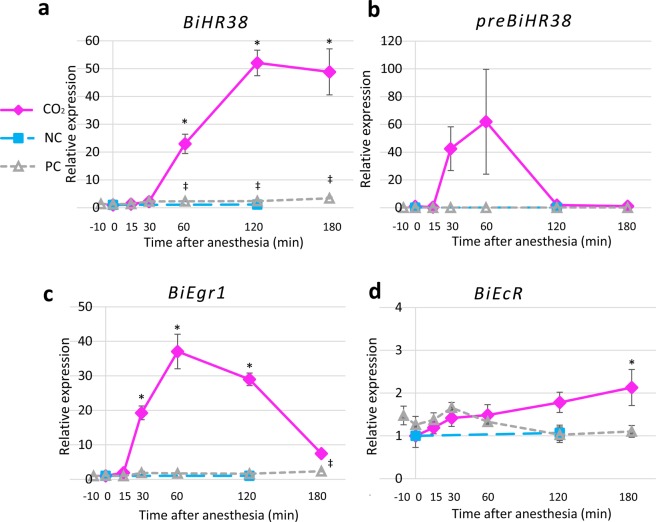
The qRT-PCR results indicated that the BiHR38 expression level normalized to that of BiEF1α changed depending on both the CO2 treatment and time after cessation of anesthesia (factor 1, 2: p < 0.001, two-way ANOVA). The relative BiHR38 expression level increased beginning 30–60 min after the cessation of CO2 anesthesia, and peaked at 120 min (Fig. 3a). In contrast, in the NC group, which was continuously anesthetized by CO2 for 120 min, the BiHR38 expression level did not change significantly compared with that at 0 min after the cessation of anesthesia as assessed by Student’s t test, indicating that the induction of BiHR38 expression is associated with awakening from anesthesia. The BiHR38 expression level in the PC group also changed slightly depending on the time after cessation of anesthesia. The BiHR38 expression level in the CO2 group was 22-fold higher than that in the PC group at the upregulation peak at 120 min (mean relative expression [normalized by EF1α], CO2: 52.1, PC: 2.4). Essentially, the same results were obtained for the BiHR38 expression level normalized with BiActin (Fig. S3a). The expression of preBiHR38 in the CO2 group also tended to increase for 15–60 min, and then rapidly decreased at 120 min after the cessation of anesthesia (Fig. 3b). Although the expression did not differ significantly between the 15–60 min time points and that at 0 min (Fig. 3b), the preBiHR38 relative expression was significantly different between 0 min and 60 min (Supplementary Fig. S3b; p < 0.05, Dunnett’s test). This finding suggests that BiHR38 is an IEG induced in the brain by seizures. The expression of BiEgr1 changed more rapidly than that of BiHR38 (Fig. 3c and Supplementary Fig. S3c). The expression of BiEgr1 increased beginning at 15–30 min, peaked at 60 min, and then decreased at 120 and 180 min. The expression changed depending on the treatment (factor 1: p < 0.001, two-way ANOVA) and time after cessation of anesthesia (factor 2: p < 0.001). The BiEcR expression level in the CO2 group differed significantly compared with that in the other two control groups (CO2-NC: p < 0.01, CO2-PC: p < 0.001, post hoc Tukey-Kramer test), as was also the case for BiHR38. In contrast, BiEcR expression increased only slightly (~2-fold) by 180 min after the cessation of anesthesia (mean of the relative expression in the CO2 group at 180 min compared with that at 0 min: 2.1; Fig. 3d and Supplementary Fig. S3d). The change in gene expression, however, was independent of both treatment and time after the cessation of anesthesia (factor 1: p = 0.16, factor 2: p = 0.42, two-way ANOVA). These findings indicated that both BiHR38 and BiEgr1, but not BiEcR, exhibit an immediate early response.
Figure 3.
Analysis of BiHR38, preBiHR38, BiEgr1, and BiEcR expression levels after seizure induction. Time-course of the expression after awakening from CO2 anesthesia of BiHR38 (a), preBiHR38 (b), BiEgr1 (c), and BiEcR (d). The expression level of each gene was analyzed by qRT-PCR and normalized with that of BiEF1α. Magenta lines indicate the group anesthetized with CO2 (“CO2”), light blue dashed lines indicate the negative control (“NC”, continuously anesthetized with CO2 for 120 min), and gray dotted lines indicate the positive control (“PC”, exposed just to air flow). All data indicate means ± SEM. Significant differences on the basis of Dunnett’s test after the ANOVA are indicated (*p < 0.05 for CO2 group, ‡p < 0.05 for PC group). Student’s t test and Welch’s t test revealed no significant difference between the NC group and the CO2 0-min group. Some errors were so low that it is difficult to see the error bars in the graph. n = 5 for each sample.
Analysis of gene expression of B. ignitus during foraging flight by qRT-PCR
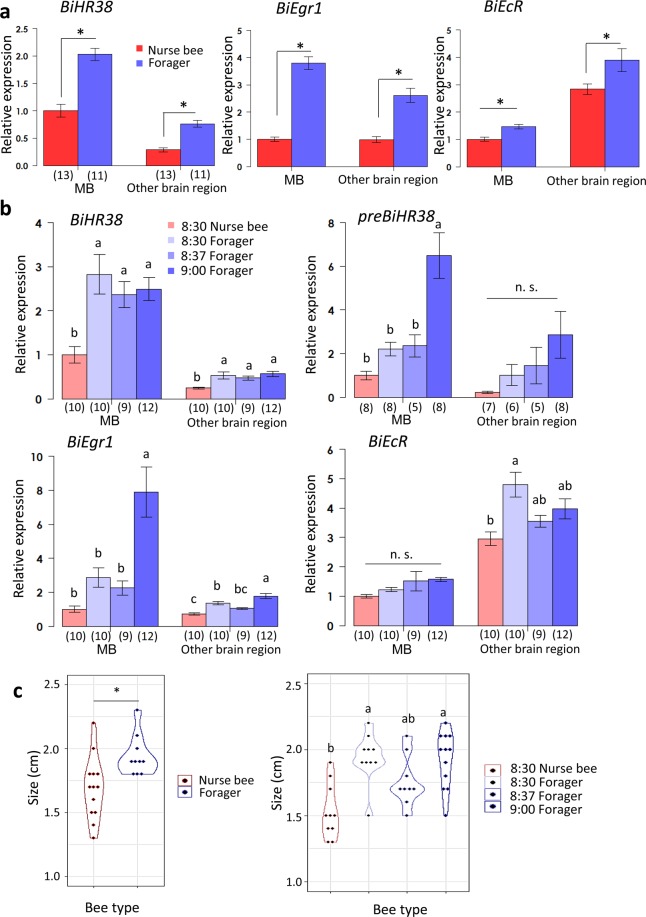
First, we analyzed the expression levels of BiHR38, BiEgr1, and BiEcR of B. ignitus nurse bees and foragers captured in the greenhouse. We examined not only the MBs but also other brain regions to analyze the major brain regions that are active during the foraging flight. The gene expression level was normalized with that of either BiEF1α (Fig. 4a and Table 2) or BiActin (Supplementary Fig. S4a and Supplementary Table S1). The expression of BiHR38 was significantly higher in foragers than in nurse bees in both the MBs (~2.0-fold) and the other brain regions (~2.7-fold; p < 0.05, Student’s t test in Fig. 4a, the expression folds were calculated from the mean of each group in Table 2). The expression of BiEgr1, which notably increases during the honey bee foraging flight31,36,44 was also significantly higher in foragers than in nurse bees in both MBs (~3.8-fold) and the other brain regions (~2.6-fold). The expression of BiEcR was also slightly but significantly higher in foragers than in nurse bees in both the MBs (~1.5-fold) and the other brain regions (~1.4-fold). Moreover, the expression levels of BiHR38, BiEgr1, and BiEcR differed significantly between the MBs and other brain regions (factor 1: p < 0.05, two-way ANOVA on each gene). Interestingly, however, whereas the expression of BiHR38 and BiEgr1 was higher in the MBs than in the other brain regions, BiEcR expression was higher in brain regions other than the MBs. Essentially, the same results were obtained when the gene expression level was normalized with BiActin (Supplementary Fig. S4a and Supplementary Table S1).
Figure 4.
Analysis of BiHR38, preBiHR38, BiEgr1, and BiEcR expression during foraging flight by qRT-PCR. Expression analysis for the greenhouse experiment (a) and the laboratory flight-cage experiment (b). The expression level of each gene was analyzed by qRT-PCR and normalized with that of BiEF1α. Each bar represents the mean ± SEM. Significant differences are indicated by asterisks (p < 0.05, Student’s t test or Welch’s t test after the F test) on the error bars in (a), or different letters (p < 0.05, Tukey-Kramer test in (b), respectively). The sample size is indicated by the number in parentheses below the horizontal axis. (c) The body sizes of B. ignitus workers captured in the greenhouse (left) and those from the flight-cage (right). Significant differences are indicated by asterisks in the left panel (p < 0.05, Student’s t test), and different letters in the right panel (p < 0.05, Tukey-Kramer test) n.s., not significant.
Table 2.
Gene expression of B. ignitus in the greenhouse sampling normalized using EF1α.
| Mean ± SEM | MB | Other brain region | ||
|---|---|---|---|---|
| Nurse bee | Forager | Nurse bee | Forager | |
| BiHR38 | 1 ± 0.12 | 2.03 ± 0.11 | 0.28 ± 0.04 | 0.76 ± 0.06 |
| BiEgr1 | 1 ± 0.08 | 3.80 ± 0.23 | 0.99 ± 0.10 | 2.62 ± 0.26 |
| BiEcR | 1 ± 0.08 | 1.47 ± 0.08 | 2.83 ± 0.19 | 3.89 ± 0.41 |
Relative gene expression in each brain tissue of workers normalized to BiEF1α in the B. ignitus greenhouse sampling compared with the MBs of nurse bees.
Next, we analyzed whether the expression levels of BiHR38, BiEgr1, and BiEcR in the MBs and other brain regions of foragers change during the foraging flight. B. ignitus foragers kept in the laboratory flight-cage exited the bee hive as soon as the hive entrance was open at 8:30. Because the feeder was so close to the hive entrance it took less than 30 min to complete a single foraging flight (i.e., searching for the feeder, obtaining pollen loads, and returning to the hive). The gene expression level was normalized with that of either BiEF1α (Fig. 4b and Table 3) or BiActin (Supplementary Fig. S4b and Supplementary Table S2). The expression level of BiHR38 normalized with that of BiEF1α changed depending on the tissue and bee type, but did not change depending on the flight time (factor 1, 2: p < 0.001, factor 3: p = 0.57, three-way ANOVA). BiHR38 expression in both the MBs and other brain regions was significantly higher (~2.4–2.8-fold in the MBs and 1.9–2.3-fold in other brain regions) in foragers than in nurse bees at any time after the onset of foraging, while no significant change in BiHR38 expression was detected in relation to the time after the onset of a foraging flight (p < 0.05, Tukey-Kramer [Fig. 4b], the expression folds were calculated from the mean of each group in Table 3). Significant upregulation of preBiHR38 was observed in the MBs at 30 min after the onset of a foraging flight (p < 0.05, Tukey-Kramer test). The expression level of BiEgr1 changed depending on the tissue and flight time, but did not change in relation to the bee type (factor 1, 3: p < 0.001, factor 2: p = 0.10, three-way ANOVA). BiEgr1 expression in both the MBs and other brain regions was significantly higher in foragers at 30 min after the onset of foraging (~2.7-fold in the MBs and 1.3-fold in other brain regions comparing foragers at 8:30 and those at 9:00 in Table 3). Whereas, the expression level of BiEcR changed depending on the tissue and bee type (factor 1, 2: p < 0.001, factor 3: p = 0.22, three-way ANOVA), there was no significant upregulation in either tissue at any time-point after the onset of the foraging flight (Fig. 4b). Additionally, the BiEcR expression level in the MBs was higher (~1.6-fold) in foragers at 9:00 than in nurse bees, and that in the other brain regions was higher (~1.3-fold) in foragers at 9:00 than in nurse bees (Table 3). Essentially, the same results were obtained for gene expression levels normalized with that of BiActin (Supplementary Fig. S4b).
Table 3.
Gene expression of B. ignitus in the laboratory flight-cage sampling normalized using EF1α.
| Mean ± SEM | MB | Other brain region | ||||||
|---|---|---|---|---|---|---|---|---|
| 8:30 | 8:37 | 9:00 | 8:30 | 8:37 | 9:00 | |||
| Nurse bee | Forager | Nurse bee | Forager | |||||
| BiHR38 | 1 ± 0.19 | 2.83 ± 0.45 | 2.37 ± 0.29 | 2.49 ± 0.26 | 0.25 ± 0.02 | 0.53 ± 0.08 | 0.48 ± 0.05 | 0.57 ± 0.06 |
| preBiHR38 | 1 ± 0.20 | 2.21 ± 0.32 | 2.36 ± 0.51 | 6.48 ± 1.05 | 0.21 ± 0.05 | 1.02 ± 0.48 | 1.46 ± 0.83 | 2.85 ± 1.07 |
| BiEgr1 | 1 ± 0.19 | 2.87 ± 0.56 | 2.25 ± 0.43 | 7.90 ± 1.46 | 0.73 ± 0.06 | 1.35 ± 0.10 | 1.05 ± 0.06 | 1.78 ± 0.14 |
| BiEcR | 1 ± 0.06 | 1.22 ± 0.07 | 1.51 ± 0.33 | 1.57 ± 0.07 | 2.95 ± 0.23 | 4.80 ± 0.42 | 3.54 ± 0.20 | 3.97 ± 0.34 |
Relative gene expression in each brain tissue of workers normalized to BiEF1α in the B. ignitus laboratory flight-cage sampling compared with the MBs of nurse bees collected at 8:30.
Taken together, these findings indicated that the expression of both preBiHR38 and BiEgr1, but not BiEcR, significantly increased in association with the foraging flight (Fig. 4b). Moreover, the expression level of BiHR38 and BiEgr1 was higher in the MBs than in the other brain regions, whereas the expression level of BiEcR was higher in brain regions other than the MBs (Fig. 4a,b).
The body sizes of the foragers were significantly larger than those of nurse bees in both the greenhouse and laboratory experiments (Fig. 4c, p < 0.05, Student’s t test and the Tukey-Kramer test), which is consistent with a previous observation that relatively larger bumble bee workers tend to be engaged in foraging and smaller workers tend to engage in the in-hive tasks.
Reexamination of gene expression in A. mellifera during foraging flight by qRT-PCR
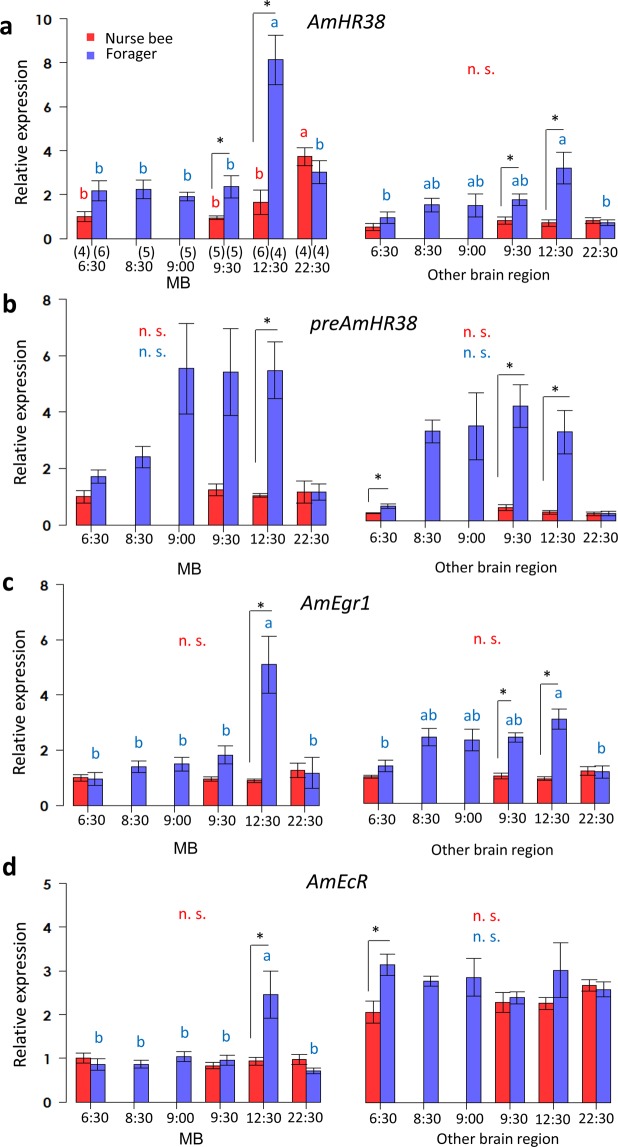
To compare our results for B. ignitus with those for A. mellifera, we reexamined the neural activity of A. mellifera during foraging flight using the same sampling protocol. As seen in the laboratory flight-cage experiment for B. ignitus, A. mellifera workers came out from the bee hive as soon as the hive entrance was open at 8:30. The expression level of each gene was normalized with that of either AmEF1α (Fig. 5 and Table 4) or AmActin (Supplementary Fig. S5 and Supplementary Table S3). The relative expression levels of AmHR38 normalized with that of AmEF1α changed depending on the tissue (the MBs or other brain regions), bee type (nurse bees or foragers), and time (factor 1, 2, 3: p < 0.001, three-way ANOVA). The expression level of AmHR38 was not significantly different from that of nurse bees until 60 min after the onset of the foraging flight (9:30), and was significantly higher in active foragers at 12:30 than in nurse bees in both the MBs and other brain regions (~4.9-fold in the MBs and 4.5-fold in other brain regions, calculated with the data in Table 4). The expression decreased at 22:30 in foragers that were captured after the foraging flight and kept in a dark incubator for 10 h. Together, these findings indicate that AmHR38 was induced by the foraging flight. preAmHR38 was markedly upregulated from 30–60 min after the onset of the foraging flight and the expression level was maintained in foragers at 12:30 and again decreased in foragers at 22:30 (Fig. 5b). The change in AmEgr1 expression also depended on the bee type and flight time (factor 2, 3: p < 0.01, factor 1: p = 0.61, three-way ANOVA). The expression level of AmEgr1 at 12:30 was significantly higher in foragers than in nurse bees in both the MBs and other brain regions (Fig. 5c, ~5.7-fold in the MBs and 3.5-fold in other brain regions), and again decreased in foragers at 22:30. AmEcR expression in the MBs was also significantly higher (~2.6-fold) in foragers than in nurse bees at 12:30, but there was no significant difference in the AmEcR expression levels in the other brain regions between nurse bees and foragers at 12:30 (Fig. 5d, the same trend as shown in Supplementary Fig. S5d). Although gene expression levels in nurse bees also changed significantly at some sampling points (AmHR38, as seen in Fig. 5a at 22:30, AmEgr1, as seen in Supplementary Fig. S5c at 12:30 and 22:30, and AmEcR, as seen in Supplementary Fig. S5d between 6:30 and 12:30), significant differences were not consistently detected for the expression levels normalized with that of either AmEF1α or AmActin.
Figure 5.
Analysis of AmHR38, preAmHR38, AmEgr1, and AmEcR expression during foraging flight by qRT-PCR. Expression levels of AmHR38 (a), preAmHR38 (b), AmEgr1 (c), and AmEcR (d) were analyzed by qRT-PCR and normalized with that of AmEF1α. Each bar represents the mean ± SEM. Significant differences are indicated using different letters (p < 0.05, Tukey-Kramer test for nurse bees [red] and for foragers [blue] during the time-course, in each brain tissue respectively) or asterisks (p < 0.05, Student’s t test or Welch’s t test after the F test) on the error bars (black). The sample size is shown below the horizontal axis in parentheses. n.s., not significant.
Table 4.
Gene expression of A. mellifera normalized using EF1α.
| Mean ± SEM | MB | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 6:30 | 8:30 | 9:00 | 9:30 | 12:30 | 22:30 | |||||
| Nurse bee | Forager | Forager | Forager | Nurse bee | Forager | Nurse bee | Forager | Nurse bee | Forager | |
| AmHR38 | 1 ± 0.23 | 2.17 ± 0.47 | 2.25 ± 0.42 | 1.91 ± 0.20 | 0.95 ± 0.08 | 2.36 ± 0.50 | 1.65 ± 0.55 | 8.13 ±1.13 | 3.73 ± 0.40 | 3.01 ± 0.52 |
| preAmHR38 | 1 ± 0.23 | 1.71 ± 0.24 | 2.40 ± 0.37 | 5.54 ± 1.60 | 1.25 ± 0.20 | 5.41 ± 1.54 | 1.04 ± 0.07 | 5.48 ± 1.01 | 1.15 ± 0.39 | 1.16 ± 0.30 |
| AmEgr1 | 1 ± 0.12 | 0.98 ± 0.25 | 1.40 ± 0.22 | 1.50 ± 0.25 | 0.95 ± 0.07 | 1.82 ± 0.32 | 0.90 ± 0.06 | 5.11 ± 1.03 | 1.27 ± 0.26 | 1.17 ± 0.56 |
| AmEcR | 1 ± 0.11 | 0.86 ± 0.13 | 0.86 ± 0.09 | 1.04 ± 0.12 | 0.83 ± 0.07 | 0.96 ± 0.11 | 0.93 ± 0.09 | 2.46 ± 0.54 | 0.97 ± 0.12 | 0.71 ± 0.06 |
| Mean ± SEM | Other brain region | |||||||||
| AmHR38 | 0.50 ± 0.16 | 0.92 ± 0.26 | 1.51 ± 0.31 | 1.50 ± 0.52 | 0.80 ± 0.16 | 1.75 ± 0.28 | 0.70 ± 0.15 | 3.17 ± 0.72 | 0.81 ± 0.13 | 0.70 ± 0.13 |
| preAmHR38 | 0.26 ± 0.02 | 0.51 ± 0.07 | 3.17 ± 0.40 | 3.35 ± 1.18 | 0.45 ± 0.10 | 4.07 ± 0.76 | 0.29 ± 0.07 | 3.14 ± 0.78 | 0.24 ± 0.06 | 0.24 ± 0.08 |
| AmEgr1 | 0.95 ± 0.06 | 1.34 ± 0.21 | 2.39 ± 0.32 | 2.29 ± 0.40 | 0.97 ± 0.11 | 2.38 ± 0.17 | 0.88 ± 0.06 | 3.04 ± 0.37 | 1.16 ± 0.15 | 1.13 ± 0.21 |
| AmEcR | 2.07 ± 0.26 | 3.15 ± 0.25 | 2.78 ± 0.11 | 2.87 ± 0.44 | 2.29 ± 0.23 | 2.40 ± 0.13 | 2.27 ± 0.14 | 3.03 ± 0.62 | 2.68 ± 0.13 | 2.60 ± 0.17 |
Relative gene expression in each brain tissue of workers normalized to AmEF1α in the A. mellifera sampling compared with the MBs of nurse bees collected at 6:30.
Taken together, these results suggested that both HR38 and Egr1 were significantly upregulated by foraging flight in both B. ignitus and A. mellifera, and a slight upregulation was also observed for late-induced gene EcR.
Detection of activated cells by in situ hybridization
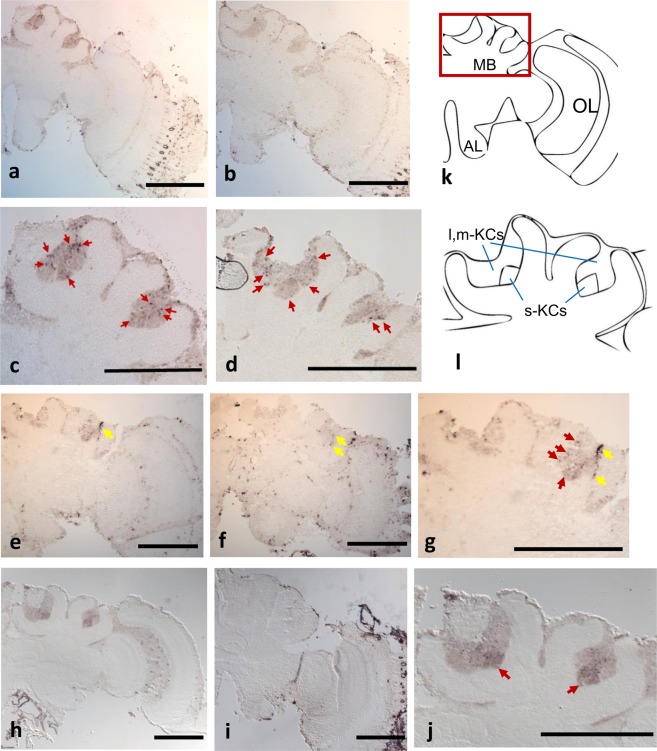
We performed in situ hybridization analysis using brain sections of B. ignitus foragers to detect the cells activated in the forager brains. Both BiHR38 and BiEgr1 were strongly expressed in the MBs (Fig. 6a–d and e–g). Cells activated by BiHR38 and BiEgr1 were detected sparsely in the whole KCs (Fig. 6c,d,g). On the other hand, BiEcR was detected preferentially and locally in the small-type KCs in the MBs (Fig. 6h–j). BiEcR was also weakly detected in the whole brain cortex (Fig. 6h). Thus, consistent with our qRT-PCR results, these findings suggested that both BiHR38 and BiEgr1 were induced mainly in the MBs, and that BiEcR was expressed both in the sKCs and other brain regions, as previously reported in A. mellifera23,31,45.
Figure 6.
In situ hybridization of BiHR38, BiEcR, and BiEgr1 in forager brain sections. Expression of BiHR38 (a–d), BiEgr1 (e–g), and BiEcR (h–j) was analyzed by in situ hybridization. Sections of brain hemispheres hybridized with antisense (a,e,h) or sense probes (b,f,i) are shown. Panel (k) indicates schematic drawing of the brain hemisphere and panel (l) indicates the magnified view of the MB enclosed by the red square in panel (k). Panels (c,d,g,j) indicate magnified views of the MB area in panels (a,e,h), respectively. (a–c) Forager collected at 9:00 in the laboratory flight-cage; (d–j) forager collected as “Forager” in the greenhouse. Representative signals are indicated by red arrows. Processing-induced damage to the tissue is indicated by yellow arrows (e–g, respectively). The s (m,l)-KCs: small-type (middle-, large-) Kenyon cells, AL: antennal lobe, OL: optic lobe. Scale bars = 500 µm.
Discussion
After awakening from CO2 anesthesia, the expression of preBiHR38, BiHR38, and BiEgr1 was prominently induced, indicating that both HR38 and Egr1 exhibit an immediate early response in the brains of B. ignitus. BiEgr1 was induced earlier (~30 min after the cessation of anesthesia) than BiHR38 (~60 min; Fig. 3a,c), which is consistent with previous studies in moths34, flies34, honey bees31, and mammals43. Expression of BiHR38 and BiEgr1 was very low, but significantly upregulated in the PC group, suggesting that novel surroundings, for example, light exposure or novel visual objects, stimulated the bees44,46. Expression of preBiHR38 was also induced transiently within 30 min, earlier than BiHR38 expression (Fig. 3a,b), which likely reflects the time needed for premature RNA to be processed into mature RNA. A previous study showed that induction of the expression of honey bee IEGs such as kakusei and AmEgr1 is much more rapid – within 15–30 min after the cessation of anesthesia27,31. In contrast, our present results showed that the increased expression of both preBiHR38 and BiEgr1 was significant and prominent within 30 min after the cessation of CO2 anesthesia (Fig. 3a,c). These findings give rise to the possibility that CO2 treatment (~5 min), which was long enough to fully anesthetize the large B. ignitus workers, caused deep anesthesia and that it took a relatively long time for the workers to awake from the anesthesia, which resulted in a time-lag between the cessation of anesthesia and IEG expression. On the other hand, BiEcR expression was not significantly induced till 120 min (Fig. 3d), and was expected to be higher at 180 min after the awakening from anesthesia, indicating that BiEcR did not exhibit an immediate early response. Unexpectedly, expression of both BiHR38 and BiEgr1 was only slightly induced in bees treated with air instead of CO2 and exposed to the same luminescent light, which we originally expected to act as a positive control. It is possible that bees that had been kept in a dark incubator were exposed to some light and thus light exposure did not fully act as a stimulus to activate the IEGs.
In this experiment, we set colonies in a greenhouse or in a laboratory flight-cage for the sampling of B. ignitus workers, but we used colonies maintained outside for the sampling of A. mellifera workers. This was because honey bee workers tended to gather around the fluorescent lamp, making it difficult for them to forage normally in the restricted laboratory space. In contrast, the foraging of bumble bee workers did not seem to be affected in the greenhouse or in the laboratory flight-cage. In this small flight-cage, bumble bees workers tended to complete one foraging flight in less than 30 min, and therefore we collected workers at three time-points within 30 min after the onset of the foraging flight. We found that expression of preBiHR38 and BiEgr1 was upregulated 30 min after the onset of the foraging flight more prominently in the MBs than in the other brain regions, suggesting that the MB neural activity increased in the bumble bee during the foraging flight. It is plausible that induction of preBiHR38 preceded that of BiHR38 and eventually resulted in increased expression of BiHR38 in foragers at 14:00. (Fig. 4a). In contrast, BiEcR was not significantly upregulated by the foraging flight at 30 min (Fig. 4b), which is consistent with our previous finding that BiEcR did not show an immediate early response until 120 min after awakening from anesthesia (Fig. 3d). The expression level of BiEcR, however, was also higher in foragers than in nurse bees collected at 14:00, like BiHR38 and BiEgr1 (Fig. 4a). This might be explained by the fact that the expression level of BiEcR was slightly, but significantly, higher in the CO2-treated group 180 min after awakening from anesthesia (Fig. 3d). It might be that BiEcR was upregulated later by the foraging experience, as reported previously36.
Although we first expected that AmHR38, AmEgr1, and AmEcR were upregulated in the brain within 30 min from the onset of foraging on the basis of a previous report, they were not upregulated even at the end of one foraging bout (Fig. 5a–d) except for preAmHR38 (Fig. 5b). The expression of AmHR38 did not change for 30 min after the onset of foraging when preAmHR38 was already upregulated (Fig. 5b), suggesting that AmHR38 could be induced by a single foraging flight longer than 30 min. This could be account for the finding that the AmHR38 expression level was higher in foragers than in nurse bees collected at 12:30 (Fig. 5a). Also, neither AmEgr1 nor AmEcR was upregulated in the MBs within 30 min after the onset of the foraging flight, but the expression of both was higher in foragers than in nurse bees collected at 12:30 and downregulated in foragers captured and kept in a dark incubator for 10 h (Fig. 5c,d). It might be that AmEgr1 induction could not be detected 30 min after the onset of the foraging flight because we did not analyze the foraging flight time of individual workers: we might have collected workers that had been engaged in foraging for less than 30 min, even 30 min after the entrance was open to allow them to forage freely.
Also, in the present study, BiHR38 and BiEgr1 expression levels were high (~2-3-fold) in foragers at 8:30 before the onset of the foraging flight compared with that in nurse bees at the same time, and these genes were preferentially expressed in the MBs over the other brain regions (Fig. 4b). This was also at least partly true for the honey bees: the AmHR38 expression level was higher in foragers than in nurse bees at 6:30 (~2-fold; Fig. 5a). We assume that the MB neural activity of foragers at that time was already upregulated, although the foragers inside the hive had not yet engaged in foraging. Eusocial bee foragers (A. mellifera and B. terrestris) have a steady circadian rhythm as they work outside in daylight and are influenced by temperature, whereas the circadian rhythm in nurse bees is attenuated as they work all day in a dark hive with a constant temperature10,47,48. Bumble bee (B. terrestris) foragers in the wild become active at almost 6:00 when they are turning out from the hive by ones and twos49. In our study, the hive entrance was shut until 8:30, which is the usual start time for bumble bees to forage. We assume that the foragers recalled the start time for foraging and their foraging-related neural activity was induced, as previously reported36,50. Egr1 is suggested to be an IEG that is upregulated before the onset of foraging by the reward learning associated with time51. Social bee foragers must deal with changes in the good flowering locations according to the time of day. At the end of the day, the foragers must remember the best feeding locations at the different times to be ready for the next day. Thus, for effective foraging, the induction of HR38 in the MBs – the higher center for memory and learning – in the early morning may reflect neural activity needed to recall their foraging experience.
In the B. ignitus brain, both BiHR38 and BiEgr1 were expressed sparsely in the entire MBs of the foragers captured at 14:00 in the greenhouse, contrary to a previous report that both genes were expressed preferentially in the sKCs in the brains of honey bee foragers23,31 (Fig. 6d,g). It is thus possible that the differences in the expression patterns in the MBs of HR38 and Egr1 are related to the species-specific traits of the honey bee and bumble bee foraging behaviors. It is also possible that they only reflect differences in the experimental conditions set for the honey bees and bumble bees (e.g., flight distance, flight speed, or feeder). It is necessary to discriminate these two possibilities in future studies.
It is noteworthy that EcR was expressed preferentially in the sKCs in the MBs in both B. ignitus and A. mellifera (Fig. 6j). Ecdysone signaling is suggested to be involved not only in molting or metamorphosis, but also in various social behaviors in insects52. Although three types of class I KCs in the MBs are reported to be conserved among Aculeate insects25, EcR protein is distributed in the whole MBs in Camponotus japonicus53. It might be that the functions of EcR in the brain differ between Formicidae and Apoidea, and are conserved among two Apidae species: the bumble bees and honey bees.
In conclusion, our findings suggested that the brain neural activity evoked by foraging flight are at least partly conserved among two social bee species; honey bees and bumble bees. Especially, it is possible that the functions of late-upregulated EcR in the sKCs is conserved among these two species. Further studies focusing on the signaling cascade that involves these IEGs are needed to confirm the above hypothesis.
Supplementary information
Author contributions
S.I. and T.K. designed the experiments. S.I., M.N., S.A. and M.O. collected the bumble bees (Bombus ignitus) and S.I. collected the honey bees (Apis mellifera). S.I. and Y.S. designed gene-specific primers and performed subcloning. S.I. performed all the other investigations (experiments and curation of the data). S.I., M.N., S.A., M.O. and T.K. discussed on the results. S.I. drafted the manuscript and S.I., M.N., S.A., M.O. and T.K. wrote the paper. T.K. supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64701-1.
References
- 1.von Frisch, K., Chadwick, L. E. & Seeley, T. D. The dance language and orientation of bees. (Harvard Univ. Press, 1967).
- 2.Seeley, T. D. The wisdom of the hive: the social physiology of honey bee colonies. (Harvard Univ. Press, 1995).
- 3.Goulson D, et al. Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Anim. Behav. 2002;64:123–130. doi: 10.1006/anbe.2002.3041. [DOI] [Google Scholar]
- 4.Cameron SA. Temporal patterns of division of labor among workers in the primitively eusocial bumble bee, Bombus griseocollis (Hymenoptera: Apidae) Ethology. 1989;80:137–151. doi: 10.1111/j.1439-0310.1989.tb00735.x. [DOI] [Google Scholar]
- 5.Riveros AJ, Gronenberg W. Olfactory learning and memory in the bumblebee Bombus occidentalis. Naturwissenschaften. 2009;96:851–856. doi: 10.1007/s00114-009-0532-y. [DOI] [PubMed] [Google Scholar]
- 6.Dornhaus, A. & Chittka, L. Food alert in bumblebees (Bombus terrestris): possible mechanisms and evolutionary implications. Behav. Ecol. Sociobiol.50, 570–576 (2001).
- 7.Barron AB, Plath JA. The evolution of honey bee dance communication: a mechanistic perspective. J. Exp. Biol. 2017;220:4339–4346. doi: 10.1242/jeb.142778. [DOI] [PubMed] [Google Scholar]
- 8.Whitfield CW, Cziko A, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 9.Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends Genet. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Bloch, G. The social clock of the honeybee. J. Biol. Rhythms25, 307–317 (2010). [DOI] [PubMed]
- 11.Milkos GLG, Maleszka R. Epigenomic communication systems in humans and honey bees: From molecules to behavior. Horm. Behav. 2011;59:399–406. doi: 10.1016/j.yhbeh.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko K, Suenami S, Kubo T. Gene expression profiles and neural activities of Kenyon cell subtypes in the honeybee brain: identification of novel ‘middle-type’ Kenyon cells. Zool. Lett. 2016;2:14. doi: 10.1186/s40851-016-0051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodaira Y, Ohtsuki H, Yokoyama J, Kawata M. Size-dependent foraging gene expression and behavioral caste differentiation in Bombus ignitus. BMC Res. Notes. 2009;2:184. doi: 10.1186/1756-0500-2-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobback J, Mommaerts V, Vandersmissen HP, Smagghe G, Huybrechts R. Age-and task-dependent foraging gene expression in the bumblebee Bombus terrestris. Arch. Insect Biochem. Physiol. 2011;76:30–42. doi: 10.1002/arch.20401. [DOI] [PubMed] [Google Scholar]
- 15.Heisenberg M. What do the mushroom bodies do for the insect brain? an introduction. Learn. Mem. 1998;5:1–10. [PMC free article] [PubMed] [Google Scholar]
- 16.Fahrbach SE, Moore D, Capaldi EA, Farris SM, Robinson GE. Experience-expectant plasticity in the mushroom bodies of the honeybee. Learn. Mem. 1998;5:115–123. [PMC free article] [PubMed] [Google Scholar]
- 17.Farris SM, Robinson GE, Fahrbach SE. Experience- and age-related outgrowth of intrinsic neurons in the mushroom bodies of the adult worker honeybee. J. Neurosci. 2001;21:6395–6404. doi: 10.1523/JNEUROSCI.21-16-06395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobrin SE, Herlihy JD, Robinson GE, Fahrbach SE. Muscarinic regulation of Kenyon cell dendritic arborizations in adult worker honey bees. Arthropod Struct. Dev. 2011;40:409–419. doi: 10.1016/j.asd.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamikouchi A, Takeuchi H, Sawata M, Natori S, Kubo T. Concentrated expression of Ca2+/calmodulin-dependent protein kinase II and protein kinase C in the mushroom bodies of the brain of the honeybee Apis mellifera L. J. Comp. Neurol. 2000;417:501–510. doi: 10.1002/(SICI)1096-9861(20000221)417:4<501::AID-CNE8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi H, et al. Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honeybee brain. Insect Mol. Biol. 2001;10:487–494. doi: 10.1046/j.0962-1075.2001.00288.x. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi H, et al. Prepro-tachykinin gene expression in the brain of the honeybee Apis mellifera. Cell Tissue Res. 2004;316:281–293. doi: 10.1007/s00441-004-0865-y. [DOI] [PubMed] [Google Scholar]
- 22.Paul RK, Takeuchi H, Kubo T. Expression of two ecdysteroid-regulated genes, Broad-Complex and E75, in the brain and ovary of the honeybee (Apis mellifera L.) Zool. Sci. 2006;23:1085–1092. doi: 10.2108/zsj.23.1085. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki Y, et al. Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor. FEBS Lett. 2006;580:2667–2670. doi: 10.1016/j.febslet.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko K, et al. Novel middle-type Kenyon cells in the honeybee brain revealed by area-preferential gene expression analysis. PLoS One. 2013;8:e71732. doi: 10.1371/journal.pone.0071732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oya S, Kohno H, Kainoh Y, Ono M, Kubo T. Increased complexity of mushroom body Kenyon cell subtypes in the brain is associated with behavioral evolution in hymenopteran insects. Sci. Rep. 2017;7:13785. doi: 10.1038/s41598-017-14174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommerlandt FMJ, Brockmann A, Rössler W, Spaethe J. Immediate early genes in social insects: a tool to identify brain regions involved in complex behaviors and molecular processes underlying neuroplasticity. Cell. Mol. Life Sci. 2019;76:637–651. doi: 10.1007/s00018-018-2948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiya T, Kunieda T, Kubo T. Increased neural activity of a mushroom body neuron subtype in the brains of forager honeybees. PLoS One. 2007;2:e371. doi: 10.1371/journal.pone.0000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 29.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc. Natl. Acad. Sci. USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein Y, Halachmi N, Egoz-Matia N, Toder M, Salzberg A. The proprioceptive and contractile systems in Drosophila are both patterned by the EGR family transcription factor Stripe. Dev. Biol. 2010;337:458–470. doi: 10.1016/j.ydbio.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 31.Ugajin A, Kunieda T, Kubo T. Identification and characterization of an Egr ortholog as a neural immediate early gene in the European honeybee (Apis mellifera L.) FEBS Lett. 2013;587:3224–3230. doi: 10.1016/j.febslet.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell MA, Muscat GEO. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velarde RA, Robinson GE, Fahrbach SE. Nuclear receptors of the honey bee: annotation and expression in the adult brain. Insect Mol. Biol. 2006;15:583–595. doi: 10.1111/j.1365-2583.2006.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujita N, et al. Visualization of neural activity in insect brains using a conserved immediate early gene, Hr38. Curr. Biol. 2013;23:2063–2070. doi: 10.1016/j.cub.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 35.Baker KD, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/S0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- 36.Singh AS, Shah A, Brockmann A. Honey bee foraging induces upregulation of early growth response protein 1, hormone receptor 38 and candidate downstream genes of the ecdysteroid signalling pathway. Insect Mol. Biol. 2018;27:90–98. doi: 10.1111/imb.12350. [DOI] [PubMed] [Google Scholar]
- 37.Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- 38.Rodgers, C. I. et al. Stress preconditioning of spreading depression in the locust CNS. PLoS One2, e1366 (2007). [DOI] [PMC free article] [PubMed]
- 39.Ugajin A, et al. Identification and initial characterization of novel neural immediate early genes possibly differentially contributing to foraging-related learning and memory processes in the honeybee. Insect Mol. Biol. 2018;27:154–165. doi: 10.1111/imb.12355. [DOI] [PubMed] [Google Scholar]
- 40.Huang ZY, Robinson GE, Borst DW. Physiological correlates of division of labor among similarly aged honey bees. J. Comp. Physiol. A. 1994;174:731–739. doi: 10.1007/BF00192722. [DOI] [PubMed] [Google Scholar]
- 41.Ueno, T., Kawasaki, K. & Kubo, T. Preparation of single-cohort colonies and hormone treatment of worker honeybees to analyze physiology associated with role and/or endocrine system. J. Vis. Exp. e54240, 10.3791/54240 (2016). [DOI] [PMC free article] [PubMed]
- 42.Suenami S, et al. Analysis of the differentiation of Kenyon cell subtypes using three mushroom body-preferential genes during metamorphosis in the honeybee (Apis mellifera L.) PLoS One. 2016;11:e0157841. doi: 10.1371/journal.pone.0157841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kendall G, Ensor E, Brar-Rai A, Winter J, Latchman DS. Nerve growth factor induces expression of immediate-early genes NGFI-A (Egr-1) and NGFI-B (nur 77) in adult rat dorsal root ganglion neurons. Mol. Brain Res. 1994;25:73–79. doi: 10.1016/0169-328X(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 44.Lutz CC, Robinson GE. Activity-dependent gene expression in honey bee mushroom bodies in response to orientation flight. J. Exp. Biol. 2013;216:2031–2038. doi: 10.1242/jeb.084905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi, H., Paul, R. K., Matsuzaka, E. & Kubo, T. EcR-A expression in the brain and ovary of the honeybee (Apis mellifera L.). Zool. Sci.24, 596–603 (2007). [DOI] [PubMed]
- 46.Sommerlandt FMJ, Rössler W, Spaethe J. Impact of light and alarm pheromone on immediate early gene expression in the European honeybee, Apis mellifera. Entomol. Sci. 2017;20:122–126. doi: 10.1111/ens.12234. [DOI] [Google Scholar]
- 47.Bloch G, Toma DP, Robinson GE. Behavioral rhythmicity, age, division of labor and period expression in the honey bee brain. J. Biol. Rhythms. 2001;16:444–456. doi: 10.1177/074873001129002123. [DOI] [PubMed] [Google Scholar]
- 48.Yerushalmi S, Bodenhaimer S, Bloch G. Developmentally determined attenuation in circadian rhythms links chronobiology to social organization in bees. J. Exp. Biol. 2006;209:1044–1051. doi: 10.1242/jeb.02125. [DOI] [PubMed] [Google Scholar]
- 49.Stelzer RJ, Chittka L. Bumblebee foraging rhythms under the midnight sun measured with radiofrequency identification. BMC Biol. 2010;8:93. doi: 10.1186/1741-7007-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naeger NL, Robinson GE. Transcriptomic analysis of instinctive and learned reward-related behaviors in honey bees. J. Exp. Biol. 2016;219:3554–3561. doi: 10.1242/jeb.144311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah A, Jain R, Brockmann A. Egr-1: A candidate transcription factor involved in molecular processes underlying time-memory. Front. Psychol. 2018;9:865. doi: 10.3389/fpsyg.2018.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey A, Bloch G. Juvenile hormone and ecdysteroids as major regulators of brain and behavior in bees. Curr. Opin. Insect Sci. 2015;12:26–37. doi: 10.1016/j.cois.2015.09.006. [DOI] [Google Scholar]
- 53.Nemoto M, Hara K. Ecdysone receptor expression in developing and adult mushroom bodies of the ant Camponotus japonicus. Dev. Genes Evol. 2007;217:619–627. doi: 10.1007/s00427-007-0172-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.