Abstract
Background
Aldehyde dehydrogenase 2 (ALDH2) catalyzes the detoxification of aliphatic aldehydes, including acetaldehyde. About 45% of Han Chinese (East Asians), accounting for 8% of humans, carry a single point mutation in ALDH2*2 (E504K) that leads to accumulation of toxic reactive aldehydes.
Methods
Sequencing of a small Mexican cohort and a search in the ExAC genomic database for additional ALDH2 variants common in various ethnic groups was set to identify missense variants. These were evaluated in vitro, and in cultured cells expressing these new and common variants.
Findings
In a cohort of Hispanic donors, we identified 2 novel mutations in ALDH2. Using the ExAC genomic database, we found these identified variants and at least three other ALDH2 variants with a single point mutation among Latino, African, South Asian, and Finnish ethnic groups, at a frequency of >5/1000. Although located in different parts of the ALDH2 molecule, these common ALDH2 mutants exhibited a significant reduction in activity compared with the wild type enzyme in vitro and in 3T3 cells overexpressing each of the variants, and a greater ethanol-induced toxicity. As Alda-1, previously identified activator, did not activate some of the new mutant ALDH2 enzymes, we continued the screen and identified Alda-64, which is effective in correcting the loss of activity in most of these new and common ALDH2 variants.
Interpretation
Since ~80% of the world population consumes ethanol and since acetaldehyde accumulation contributes to a variety of diseases, the identification of additional inactivating variants of ALDH2 in different ethnic groups may help develop new ‘precision medicine’ for carriers of these inactive ALDH2.
Keywords: ALDH2 deficiency, Alda-1 and -64, Alcohol toxicity, Novel mutations, Health burden
Research in context.
Evidence before this study
A family of detoxifying enzymes called aldehyde dehydrogenases (ALDHs) has been studied with great interest due to its role in detoxifying aldehydes that accumulate through metabolism and to which we are exposed from the environment. Although the human genome has 19 ALDH genes, one ALDH emerges as a particularly important enzyme in a variety of human pathologies. This ALDH, ALDH2, is located in the mitochondrial matrix and is better known for its role in ethanol metabolism. ALDH2 dysfunction may contribute to a variety of human diseases, including cardiovascular diseases, diabetes, neurodegenerative diseases, stroke, osteoporosis, Fanconi Anemia, cancer and in the process of aging. Furthermore, an ALDH2 inactivating mutation (termed ALDH2*2) is the most common single point mutation in humans, and epidemiological studies suggest a correlation between this inactivating mutation and increased propensity for common human pathologies.
Added value of this study
We reasoned that such a common inactivating mutation must have had an evolutionary benefit and therefore may have arisen more than once. Searching the ExAC database, we now find at least 5 additional inactivating ALDH2 mutant forms in other ethnic groups, including, African, Latino, South Asian, and Finish. We characterized these 5 mutations, which represent more than 1/500 people of a given geographical or racial population, and found that each of these single point mutations caused between 50% and 90% loss of activity and some were also associated with reduced stability. Importantly, we now demonstrate the efficacy of a better ALDH2 activity booster, Alda-64.
Implications of all the available evidence
In the era of precision medicine, considering that ALDH2*2 loss of activity is associated with differences in drug metabolism and in susceptibility to a variety of chronic diseases including cancer, diabetes and neurodegeneration, determining association of these newly identified common inactivating in various other populations should help with public health and patients’ education regarding alcohol consumption and should be incorporated in decision making by physicians.
Alt-text: Unlabelled box
1. Introduction
Aldehyde dehydrogenase 2 (ALDH2) enzymatic deficiency is one of the most common race-specific human enzymopathies in the world, affecting about 560 million East Asians, or about 8% of the world population [1, 2]. ALDH2 deficiency was first characterized by symptoms of facial flushing, palpitation, headache, and vomiting with only moderate alcohol consumption among the East Asian carriers [3]. “Alcohol glow”, “alcohol flushing”, “flushing syndrome” or “alcohol intolerance” are various terminologies that describe the phenotype of this enzymatic deficiency. The unpleasant physiological responses to alcohol are caused by a rapid accumulation of acetaldehyde, due to the greatly reduced ALDH2 function [3].
The alcohol flushing syndrome is not benign. Unequivocal epidemiological data and meta-analysis have consistently shown that alcohol consumption among ALDH2*2 carriers leads to a significantly increased risk for several cancers, in particular, the upper aerodigestive tract cancers including oral, pharyngolaryngeal, and esophageal cancers [1, 4, 5]. For example, Yokoyama et al. reported that, amongst ALDH2*2 allele carriers, those who were categorized as moderate or heavy alcohol drinkers had an increased odds ratio of 72.8 for esophageal cancer compared to 44.6 for those who never drink [6]. Indeed, the International Agency for Research on Cancer has classified alcoholic beverages as a human carcinogen (Group 1) [7]. Aldehyde toxicity is not limited to cancer; it has also been implicated in many other diseases with increased vulnerability among the ALDH2*2 subjects [8, 9]. These diseases range from osteoporosis [10], cardiovascular disease [11], and Alzheimer's disease to rare genetic diseases, such as Fanconi Anemia [12, 13]. In addition, ALDH2 also plays a key role, via its reductase activity, in the conversion of nitroglycerin to nitric oxide for vasodilation [14], thus reducing the clinical benefit of traditional nitroglycerine dosing regimens among ALDH2*2 human subjects [15] when used to treat angina.
ALDH2 is a critical, NAD-dependent metabolic enzyme, responsible for the conversion of reactive endogenous and exogenous aldehydes to their corresponding, non-reactive, carboxylic acids [16]. Many commonly occurring aldehydes are relevant to human health, such as short-chain aliphatic aldehydes (e.g., acetaldehyde arising from ethanol metabolism), aromatic aldehydes (e.g., 3,4-dihydroxyphenylacetaldehyde arising from dopamine), and long-chain aliphatic aldehydes (e.g., 4-hydroxy-nonenal arising from oxidative stress lipid peroxidation). Therefore, ALDH2, a non-P450 mitochondrial detoxifying enzyme, plays a key role in protecting all organs from damage by a variety of reactive and toxic aldehydes [2].
The ALDH2*2 variant has thus far been characterized as an East Asian-specific polymorphism [17]. Extensive global geographic and population mapping based on data of 80,691 individuals from 366 population samples has confirmed that the ALDH2*2 allele is highly concentrated in areas of Southeast China, Japan, Korea, Taiwan, Singapore and Vietnam [18]. Perhaps most prominently, in Taiwan the prevalence of the ALDH2*2 carrier is as high as 49%, affecting half of the population of a single country [19].
Exhaustive literature searches on ALDH2 have found no reports or molecular characterization of any other ALDH2 variants (single amino acid substitutions or otherwise) affecting enzymatic function of this gene [18], except for a functional promoter SNP (−357G/A, rs886205) [20] that affects ALDH2 gene expression level by DNA methylation in the 5′-untranscribed region [21]. With the recent compilation of several large-scale human DNA sequencing projects worldwide, such as the 1000 genomes project [22], the human Exome Aggregation Consortium (ExAC) [23], and the Genome Aggregation Database (gnomAD) [24], we sought to better understand the worldwide distribution of ALDH2 genetic variation. Consequently, we have identified several additional common ALDH2 single amino acid variants (missense mutations) with loss of ALDH2 activity that are enriched within certain ethnic populations. In this era of precision medicine, mindful of the health implication of ALDH2*2 variant in 560 million East Asians [8,25], we recognize that the potential impact on human of common variants with reduced or loss of ALDH2 amongst other ethnic and geographical groups should be considered by policymakers, health providers, and patients.
2. Materials and methods
2.1. Blood sample collection, DNA purification and ALDH 2 genotyping and sequencing of human subjects
Blood samples for DNA extraction and ALDH2 genotyping were collected from volunteers under the approval of IRB protocol (INF-2938-19-19-1) with written informed consent for human subject study at the blood bank from the National Institute of Medical Sciences and Nutrition Salvador Zubirán in Mexico City. The buffy coat containing peripheral blood monocyte cells were isolated by gradient centrifugation using Lymphoprep™ (STEMCELL, Germany), density medium 1.077±0.001 g/ml. The purified peripheral blood monocytes from each sample were lyzed in lysis buffer for DNA extraction by an automatic nucleic acid extraction system (NucliSENS®easyMag®, Biomerieux, France) according to the manufacturer's instructions, and eluted in 25 μl of elution buffer. ALDH2 genotyping and DNA sequencing of all exons were carried by PCR and direct DNA sequencing of each of the exon of the entire ALDH2 genes. PCR primer sets used for each exon are listed in Table S2.
2.2. Analysis of Exome Aggregation Consortium (ExAC) database for ALDH2 variants
High-quality variants (genotype quality ≥ 20 and depth ≥ 10) in the ALDH2 transcript ENST00000261733, as of September, 2019, were collected from the ExAC Browser [23]. The ExAC database collected 60,706 unrelated individuals with a total of 490 variants recorded for the ALDH2 genomic region including 5′-, 3′-untranslated regions, intron and exon regions. The dataset was exported as an Excel file and sorted by the annotation of the variants. We focused only on variants in the amino-acid coding region of ALDH2 which comprised of 517 amino acids. The common variants were identified and defined by an allele frequency greater than 0.5% in any of the 6 major ethnic groups recorded in the database.
2.3. Cloning of ALDH2 variants
The full-length cDNA of wild type ALDH2 gene was obtained from ATCC (ATCC clone #MGC-1806) and inserted into the pET-28a (+) vector (Novagen, MA, USA) directly following the thrombin cleavage sequence for recombinant protein production in BL21 bacterial host cells. Each of the ALDH2 single amino acid variants was constructed from the wild type vector by site-directed mutagenesis using complimentary primer sets carrying the desired nucleotide substitution. The Pfu Turbo DNA polymerase was used for site-directed mutagenesis according to the manufacturer's guidelines (Agilent-Quick-change site-directed mutagenesis, #20522). After site-directed mutagenesis, each plasmid was transformed into BL21 competent E. coli (Life Science Technologies). Colonies containing the desired ALDH2 variants were selected and confirmed by DNA sequencing. The sense strand primers used in the PCR reactions were as follows: I41V, GGCATCGTGCCATTCATTGTTCACGAAAATCTGGTTGCAGAAGAC; P92T, GCGGCGCCAGGTTGAGCCCAGCTGGAAGG; T244M, 5′-GGCCCCAGCCATGGGGCCAAATCCAGG; V304M, 5′-AGTGGGCCTGTTCCATGGCCCAATCCATAT;R338W,5′-CCGGGCAACGCTCCACTCCACAAACTCAT; E504K, 5′-TGTGACAGTTTTCACTTTAGTGTATGCCTGCAGCC.
2.4. Protein expression and purification
Recombinant ALDH2 was expressed in LB broth (800 ml, with 50 μg/ml kanamycin) as previously reported [26]. In brief, culture was spun down (6000 g for 20 min at 4 °C), and the pellet was lysed with 20 ml of BugBuster Protein Extraction Reagent (Novagen, WI, USA) for 20 min at room temperature with slight agitation. GE His-Trap nickel affinity columns (GE, #11-0033-99) were first equilibrated at 4 °C by 10 ml of wash buffer. The bacterial cell lysate was added to the column and washed with another 10 ml of wash buffer. Recombinant ALDH2 protein was eluted by 3 ml of elution buffer and digested by 200 units of bovine thrombin (BioPharm, # 91-030) for 14 h at 4 °C to remove the His-tag. Sterile glycerol was added to the purified enzyme for long-term storage at −80 °C.
2.5. Co-expression of ALDH2 wildtype and ALDH2 variants
pETDuet-1 plasmid vector (Novagen, MA, USA) was used for the co-expression of ALDH2 wildtype (ALDH2*1) and each of the E504K (ALDH2*2), P92T (ALDH2*4), T244M (ALDH2*5) and R338W (ALDH2*7). A copy of a full-length wildtype ALDH2 cDNA was inserted into the His-tag cloning site and a copy of the corresponding ALDH2 variant cDNA was interested into the HA-tag cloning site. Individual ALDH2 subunits can be expressed simultaneously from two independent IPTG inducible promoters to form active ALDH2 tetramers from the pETDuet-1 vector. Co-expression of recombinant protein was achieved by 0.5 mM IPTG induction at 30 °C using BL21 bacterial host cells. Purifications of the recombinant proteins by affinity nickel columns were carried out using a standard protocol as described above according to manufacturer's instructions (Novagen, WI, USA).
2.6. Small molecule library screening for ALDH2 activators
Small molecule library screening for ALDH2 activators was carried out at the Stanford University High-Throughput Biosciences Center (Department of Chemical and Systems Biology). 63,500 compounds were screened using an established fluorescence coupled ALDH2 enzymatic assay [26]. In this assay, an increased fluorescent signal emitted by resorufin (excitation/emission, 565/590 nm) from the reduction of resazurin by diaphorase was expected from the coupled redox reaction of recombinant ALDH2 enzyme using acetaldehyde (20 mM) and NAD (10 mM) as the substrate cofactor, respectively, in the presence of an activator compound. All screening compounds were purchased from ChemBridge (San Diego, CA USA) and SPECS USA (Narragansett, RI USA) by the Stanford University High-Throughput Biosciences Center. The library compounds were dissolved in DMSO, screened and validated at 10 μM concentration. Scaled up quantifies of confirmed ALDH2 activators of Alda-1 (N-(1,3-Benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide) (MW 324.16) and Alda-64 (2-(azepane-1-carbonyl)-N-(2-chlorobenzyl)-2,3-dihydrobenzo (b) <1,4> dioxine-6-sulfonamide) (MW 464.96) were chemically synthesized by ACME Chemical (Mountain View, CA USA).
2.7. Determination of ALDH2 activity
Aldehyde dehydrogenase activity of the purified ALDH2 enzymes was measured as conversion of NAD+ to NADH using acetaldehyde as a substrate, as previously reported [26]. The assays were carried out at 25 °C in 50 mM sodium pyrophosphate buffer (pH 9.5) in the presence of 2.5 mM NAD+ and 10 mM acetaldehyde. Enzyme activities were calculated, relative to that of wildtype, as the mean values ± SD.
2.8. Expression and protein stability of ALDH2 wildtype and variants enzymes in cultured cells
3T3 cells and human-derived fibroblasts were obtained from ATCC and Coriell Institute (AG07123, RRID:CVCL_2C18), respectively. Cells were maintained in MEM supplemented with 10% (v/v) fetal bovine serum, 2 mM L-glutamine and 1% (v/v) penicillin/streptomycin at 37 °C in a humidified chamber of 95% air and 5% CO2. For experiments, cells were seeded in 96 well plates at 10,000 cells per well or 50,000 cells per well in 6 well plates for 18–24 h before transfection. Plates were transfected with 1 μg of plasmid DNA using 3 μl of Lipofectamine 2000 reagent (Life Technologies, #11668019). After 12 h, the media was replaced with fresh media to reduce toxicity of the Lipofectamine reagent. After 48 h cells were analyzed for expression and stability of the variants (under cycloheximide treatment) or treated with 50 mM ethanol for a further 48 h to determine sensitivity to oxidative stress.
2.9. ALDH2 enzyme activity in cells
Enzymatic activity of ALDH2 was determined by measuring the conversion of NAD+ to NADH, as described previously [26]. The assays were carried out at 25 °C in 50 mM sodium pyrophosphate buffer (pH 9.5) in the presence of 2.5 mM NAD+ and 10 mM acetaldehyde. Measurement of ALDH2 activity was determined by directly adding 100 μg of the total lysate to the reaction mix and reading absorbance at 340 nm for 15 min. Activity was measured in the absence and presence of disulfiram (1 μM) and the difference between the two rates was attributed to the ALDH2.
2.10. ROS production
For cellular ROS detection, cells were incubated with 2,7-dichloro-fluorescein diacetate (DCFDA) (Abcam, #ab113851) 100 μM for 30 min at 37 °C in the dark, and fluorescence was analyzed with excitation/emission at 495/529 nm, using SpectraMax M2e (Molecular devices). Fluorescence intensity was then normalized for cell number.
2.11. ATP measurements
Relative intracellular ATP levels were determined using ATP-based CellTiter-Glo Luminescent Cell Viability kit (Promega, #G7570), which causes cell lysis and generates a luminescent signal proportional to the amount of ATP present. For intracellular ATP levels, opaque-walled 96-well plates with cell lysate (50 μl) were prepared. An equal volume of the single-one-step reagent provided by the kit was added to each well and incubated for 30 min at room temperature. ATP content was measured using a luminescent plate reader SpectraMax M2e (Molecular devices).
2.12. Western blot analysis
Protein concentrations were determined using the Bradford assay (Thermo Fisher Scientific). Proteins were resuspended in Laemmli buffer containing 2‐mercaptoethanol, loaded on SDS–PAGE, and transferred on to nitrocellulose membrane, 0.45 μm (Bio‐Rad) as previously described [27, 28]. Cell supernatant was cleared of cellular debris by centrifugation at 1000 g for 10 min. The total lysate was resuspended in Laemmli buffer containing 2‐mercaptoethanol, loaded on SDS–PAGE, and transferred on to nitrocellulose membrane, 0.45 μm (Bio‐Rad). Membranes were cut at appropriate molecular weights and then probed with a mouse monoclonal antibody against β-actin (Cell Signaling Technology, # 12262, RRID:AB_2566811) and a rabbit monoclonal antibody against ALDH2 (Abcam Cat# ab108306, RRID:AB_10862581), and visualized by ECL (0.225 mM p‐coumaric acid; Sigma), 1.25 mM 3‐aminophthalhydrazide (Luminol; Fluka) in 1 M Tris pH 8.5. Scanned images of the exposed X‐ray film or images acquired with Azure Biosystems C600 were analyzed with ImageJ to determine relative band intensity. Quantification was performed on samples from independent cultures for each condition.
2.13. 4-HNE protein adduct measurements
4-Hydroxynonenal (4HNE) protein adducts were determined by Elisa according to manufacturer's protocol (Abcam, #ab118970). Briefly, 50 μg of protein from human-derived fibroblasts lysate were loaded onto 4-HNE conjugate coated wells, blocked using the provided blocking solution and incubated with anti-4-HNE antibody at room temperature for 1 h. After washing three times with washing buffer, HRP-peroxidase was added to each well and incubated for 1 h at room temperature with gentle shaking. Wells were washed again and incubated with the provided substrate solution. Color density was measured at 450 nm after adding stop solution and quantified by comparison with a 4-HNE-BSA standard curve.
2.14. Statistical analysis
Prism 8.0 (GraphPad Software) was used for the statistical analysis. Data shown are the mean ± s.d. with P < 0.05 considered statistically significant. Group differences were analyzed with one-way analysis of variance (ANOVA) followed by Fishers Least Significant Difference (LSD) test. Data distribution was assumed to be normal, but this was not formally tested. No statistical methods were used to predetermine sample sizes.
3. Results
3.1. A survey for the presence of the Latino variants of ALDH2 in a small cohort from Mexico City, Mexico
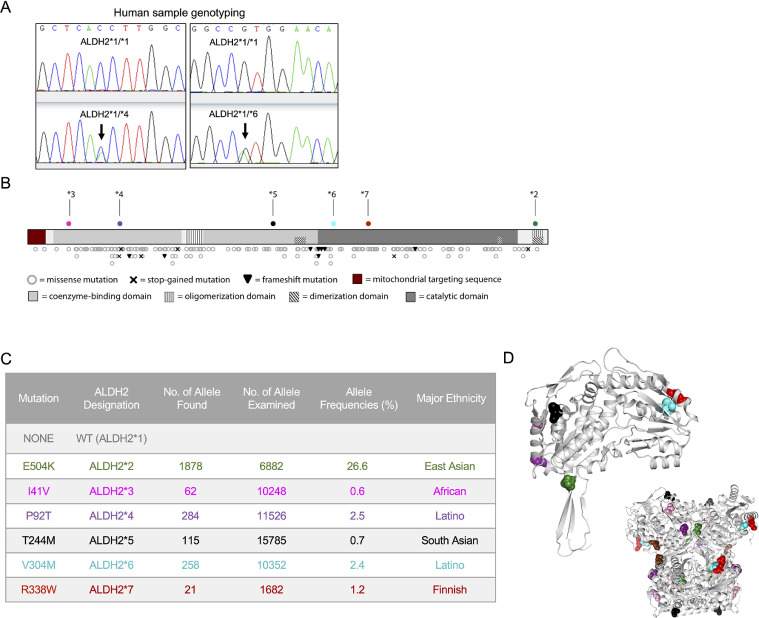
ALDH2*2 mutation has been extensively studied for its contribution to various human diseases [1, 4, 5]. Since our publication on Alda-126, we received numerous correspondences from people of non-Asian heritage suffering from “alcohol flushing” like syndrome. To start determining whether this or other mutations exist in non-East Asians, we obtained 150 human DNA samples from healthy donors from a local blood bank in Mexico City, Mexico and sequenced the entire ALDH2 gene. None had the East Asian mutation (E504K). However, we identified six subjects (4%) with a heterozygous genotype of P92T and five subjects (3.3%) with V304M (Fig. 1(A)). These results indicated that non-East Asian ALDH2 variants could be relatively common in other human populations.
Fig. 1.
Unstudied, high-frequency variants of ALDH2 have been extracted from the ExAC database (A). Sequence analysis from human samples showing examples of the Latino ALDH2*4 (P92T) and ALDH2*6 (V304M) variants. (B) All mutations identified in the Exome Aggregation Consortium (ExAC) database are shown on a linearized map of the ALDH2 protein structure. (C) Previously uncharacterized mutations in ALDH2 were identified using the ExAC database. Those variants with allele frequency percentages greater than 0.1% in a given ethnic population are shown in Table S1, along with the previously studied mutant E504K. Mutation numbering refers to the amino acid sequence of the mature protein with the 17-amino acid mitochondrial targeting sequence. Color code assignments are used consistently in the reporting of this work. Ethnicity assignments were made based off of available information in the ExAC database and might not represent all affected populations. (D) The mutated residues are shown with their respective color codes on the crystal structure (PDB: 1O05) of the ALDH2 monomer (top) and tetramer (bottom, inset).
3.2. Survey of the human Exome Aggregation Consortium (ExAC) database reveals novel high-frequency ALDH2 variants
To expand our findings from the limited donor study, and determine whether these and other variants are common, we analyzed the human Exome Aggregation Consortium (ExAC) database; which provides harmonized exome DNA sequencing data from 60,706 unrelated individuals [23]. We identified 490 variants, spanning the 42 kb of the ALDH2 genomic region. Within the 515 amino acid coding sequence, which consists of 12 exons, a total of 280 variants were identified. Amongst these, we identified 185 non-synonymous exon variants, including the ALDH2*2 variant, 6 stop-gained mutations, and 8 frameshift mutations (Fig. 1(B), and Table S1). The data compiled in the ExAC were grouped into six different major human ethnic categories: European (non-Finnish), Finnish, Latino, African, East Asian, South Asian. The 185 non-synonymous exon variants that we identified occurred in a wide range of allele frequencies. Five missense variants were present in a frequency greater than 0.5% of the corresponding ethnic and geographical groups, which include the two we found in the small cohort from Mexico City, P92T and V304M.
Following the established nomenclature (ALDH2*1 for the common active, so called wildtype, variant and ALDH2*2 for the common East Asian E504K mutant variant) we designated these new variants as ALDH2*3 (I41V), ALDH2*4 (P92T), ALDH2*5 (T244M), ALDH2*6 (V304M), and ALDH2*7 (R338W) (Fig. 1(C)). As expected, the ALDH2*2 (E504K) variant is the most prevalent genetic variant worldwide, with an allele frequency of 26.6% among East Asians, which is 10 times higher than the next most two common variants, ALDH2*4 (P92T) and ALDH2*6 (V304M), with allele frequencies of 2.5% and 2.4%, respectively, among Latinos (Fig. 1(B)). As identified in the ExAC database, other high-frequency variants are also present in Finnish (ALDH2*7), South Asian (ALDH2*5) and African (ALDH2*3) cohorts.
3.3. Recombinantly expressed ALDH2 variants have diminished activity in vitro
The ALDH2 tetramer functions as a dimer of dimers, where each of the four monomeric units contains a dinucleotide (NAD+) binding domain and a catalytic domain featuring three consecutive cysteine residues and a glutamate, which is required for the catalysis at the active site. Dimerization occurs via key contacts between two monomers, and subsequent tetramerization is assisted by contacts between three-stranded β-sheets in each monomer. The common variants span the full length of the ALDH2 primary structure and are interspersed throughout the dimerization, oligomerization, coenzyme-binding domain, and catalytic domain [29] (Fig. 1(C) and (D)). Based on location alone, the functional consequences of these common variants are not clear. Therefore, we studied the in vitro enzymatic activity of these new variants.
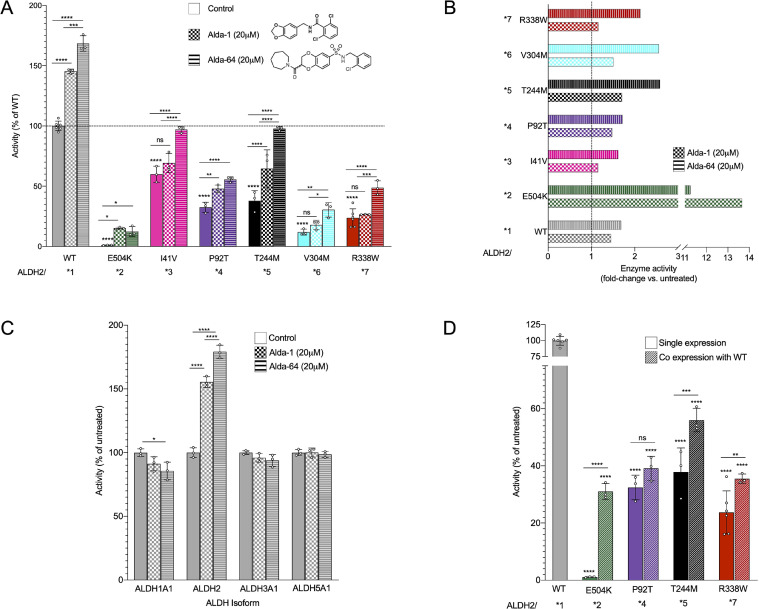
We focused on the five variants with relatively high allele-frequency (>0.5% in their population) and those that present with homozygotic individuals in the genomic datasets (Fig. 1(C), Table S1). In addition to ALDH2*2, these five unreported ALDH2 variants had a significant reduction in activity as compared with the ALDH2*1 variant (referred to as wild type from here on; Fig. 2(A)). The most active variant of those studied, ALDH2*3 (I41V), which was identified in the African population, had a mean activity of only 60% relative to the untreated wild type enzyme, even though this variant results from only a single-carbon change in the amino acid sidechain.
Fig. 2.
Newly characterized ALDH2 mutants show decreased activity in vitro compared to wildtype (WT) enzyme. (A)ALDH2 activity of the recombinant mutants upon acetaldehyde treatment was measured using the conversion of NAD+ to NADH, without treatment (solid bars) and with the ALDH2 activators Alda-1 and Alda-64 (checkered and striped bars, respectively). n = 3–6; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05. (B) Fold change of ALDH2 activity improvement in each individual ALDH2 variants by Alda-1 and Alda-64. (C) Dehydrogenase activity measured in other ALDH isoforms after treatment with Alda-1 and Alda-64. n = 3; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; * p-value <0.05. (D)ALDH2 activity measured in enzyme produced in a co-expression system of WT and ALDH2 variants to mimic heterozygous carriers. n = 3; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; ** p-value <0.01; * p-value <0.05.
The two Latino variants, ALDH2*4 (P92T) and ALDH2*6 (V304M) had only 32% and 11% activity of wildtype, respectively (Fig. 2(A)), and together potentially represent about 40 million people in the world [30]. Of importance from an epidemiological perspective is also the Finnish variant ALDH2*7 (R338W), with an activity 23% of wildtype (Fig. 2(A)). Although the population represented by this allele is much smaller than the African, Latino, or Asian counterparts, the ability to harness the unprecedented data available from the FinnGen initiative will allow us to search out direct correlations between this Finnish variant and health outcomes. Finally, although the ALDH2*5 variant (T244M) has 38% of wildtype activity (Fig. 2(A)), this mutation, like the Latino variants, represents a significant global population of potentially 14 million South Asians (0.7% of the total 2 billion South Asians) [31].
3.4. ALDH2 isozyme-specific activators, Alda-1 and Alda-64, and their effects on ALDH2 variants
The first ALDH2-specific enzyme activator, Alda-1, was identified previously by a high throughput screening of 65,000 compounds, using purified recombinant ALDH2*2 [26]. Since then, an additional 65,000 small-molecule compounds were added to the library, and an additional screen of that library identified 380 hits that increased wildtype ALDH2 activity by 12−160% at 20 μM. Of these, we confirmed 21 new ALDH2 activator compounds. Racemic Alda-64 (2-(azepane-1-carbonyl)-N-(2-chlorobenzyl)-2,3-dihydrobenzo (b) <1,4> dioxine-6-sulfonamide) (MW 464.96; Fig. 2(B)) was the most potent activator of ALDH2 and was therefore characterized here further.
At 20 μM, Alda-64 increased ALDH2 catalytic activity by 80% as compared to 55% by Alda-1. Similar to Alda-1, Alda-64 exhibited specificity for the ALDH2 family; it did not activate ALDH1, ALDH3A1 or ALDH5A1 (Fig. 2(A), (C) and [26]). Alda-1 increased the activity of the E504K variant (ALDH2*2 homo-tetramer) by ~14-fold and Alda-64 activated by ~11-fold (Fig. 2(A) and (B)), but this activity was still only about ~20% of absolute wild type activity. In contrast, Alda-64 brought the activity of ALDH2*3 (I41V) and ALDH2*5 (T244M) to wildtype levels, effects that were substantially greater than the effect of Alda-1 on these variants. There was >2-fold activation of ALDH2*7 (R338W), ALDH2*6 (V304M) by Alda-64 and a ~70% increase in activity of ALDH2*4 (P92T; Fig. 2(A) and (B)). Surprisingly, only the Latino variant P92T (ALDH2*4) and South Asian variant T244M (ALDH2*5) exhibited a significant activation by Alda-1 (~70% increase each; Fig. 2(B)); there was no significant effect of Alda-1 on the activity of the other three new variants tested, suggesting that the reduced enzyme activity caused by these mutations might cause a structural change in a way that is fundamentally different from that of E504K and the Alda-1-responsive P92T and T244M variants.
3.5. Dominant negative effect of the ALDH2 variants
As discussed, ALDH2 is a tetramer, consisting of four monomeric subunits of 55 kDa encoded by the ALDH2 gene [16]. The ALDH2*2 (E504K) subunit exerts a dominant-negative effect on the wildtype ALDH2*1 subunit in the heterotetramer, due to structural changes in the NAD co-factor binding site [9, 32]. The ALDH2 activity of human carriers of the heterozygous ALDH2*1/*2 genotype is 25–40% of wild type enzyme, which explains the alcohol flushing reaction in ALDH2 heterozygote carriers [33]. It is therefore important to determine whether human carriers of a heterozygous genotype of any of the newly identified ALDH2 variants also have reduced ALDH2 catalytic activity, as is the case of ALDH2*1/*2 heterozygous East Asians. Therefore, we next co-expressed wild type and each the four ALDH2 variants, E504K, P92T, T244M and R338W, on a bacterial dual expression vector, pETDuet-1, to evaluate the dominant-negative effect of the new variants on the wild type subunit. Co-expressed wild type/E504K (ALDH2*1/*2) heterotetramer had 31% of the enzyme activity relative to the wild type/wild type (ALDH2*1/*1) activity, corroborating previously published co-expression results [26], and reflecting, for example, the dominant negative effect in human liver tissue [33, 34]. The P92T, T244M and R338W all appear to have a greater degree of dominant-negative effect on the enzymatic activity of ALDH2, relative to ALDH2*2 (Fig. 2(D)). The activity of homozygote-equivalent mutants was indistinguishable from the activity of the heterozygotes, ALDH2*1/*4 (wildtype/P92T) and only slightly elevated for ALDH2*1/*5 (wild type/T244M) and ALDH2*1/*7 (wild type/R338M); the activity of all the ALDH2 heterozygotic variants ranged from 30% to 55% of the WT activity (Fig. 2(D)).
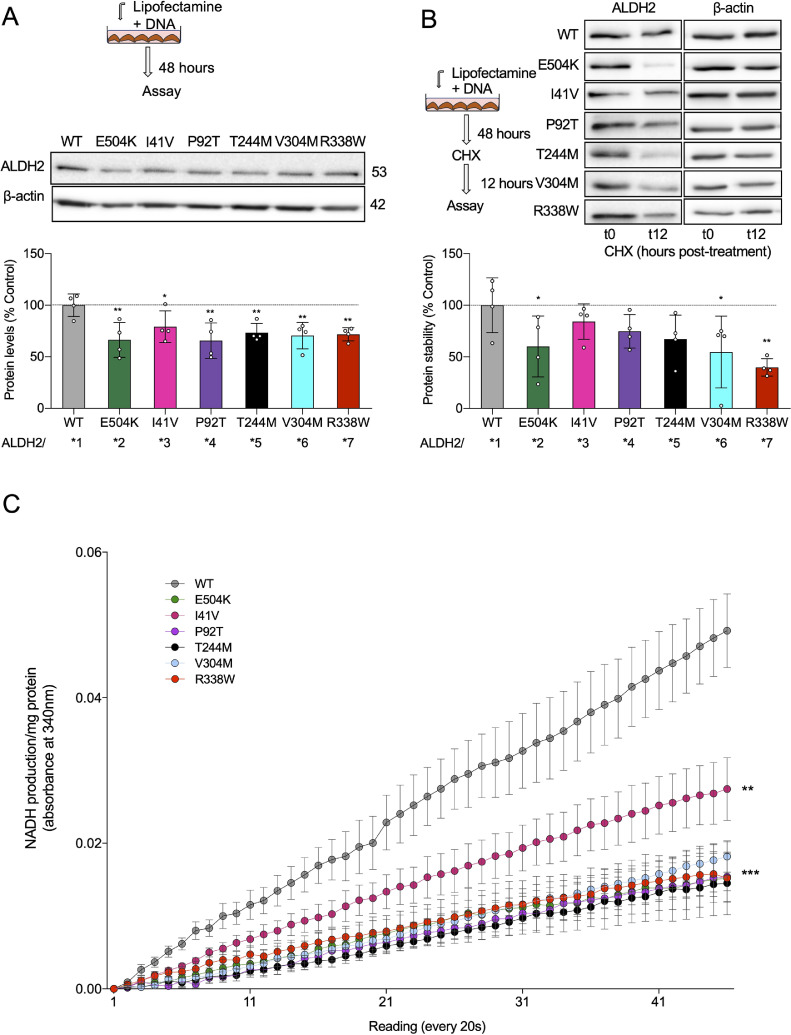
3.6. Expression and stability of ALDH2 variants in cultured cells
To understand the effects on protein activity and stability of these new variants, we transiently transfected 3T3 cells. Surprisingly, when transfected with the same amount of DNA, the expression of all the mutants was significantly reduced as compared to the overexpressed WT (ALDH2*1) (Fig. 3(A)). Interestingly, the stability of protein, when measured following a 12-h cycloheximide treatment, showed that only ALDH*2, *6 and *7 (E504K, V304M and R338W), had reduced protein stability whereas ALDH2*3, *4 and *5 (I41V, P92T, T244) variants had similar stability to that of WT (Fig. 3(B)). We also measured the effect on ALDH2 activity and found similarly low activity in all the 3T3 lines expressing the inactive variants, with the ALDH2*3 overexpressing cell line showing the highest activity and stability (Fig. 3(B) and (C)).
Fig 3.
Novel ALDH2 variants show lower stability and activity in transiently transfected cells. (A) Experimental paradigm; ALDH2 protein levels were determined in transiently transfected 3T3 cells by immunoblotting of total cell lysate. Levels were quantified and presented as ratio vs β−actin (loading control). n = 4; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) ** p-value <0.01; * p-value <0.05. (B) Experimental paradigm; ALDH2 protein stability was determined in transiently transfected 3T3 cells by immunoblotting of total cell lysate after 100 μm cycloheximide for 12 h. Levels were quantified and presented as ratio vs β-actin (loading control). n = 4; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) ** p-value <0.01; * p-value <0.05. (C)ALDH2 specific activity was determined in transiently transfected 3T3 cells. n = 4; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001 vs other groups.
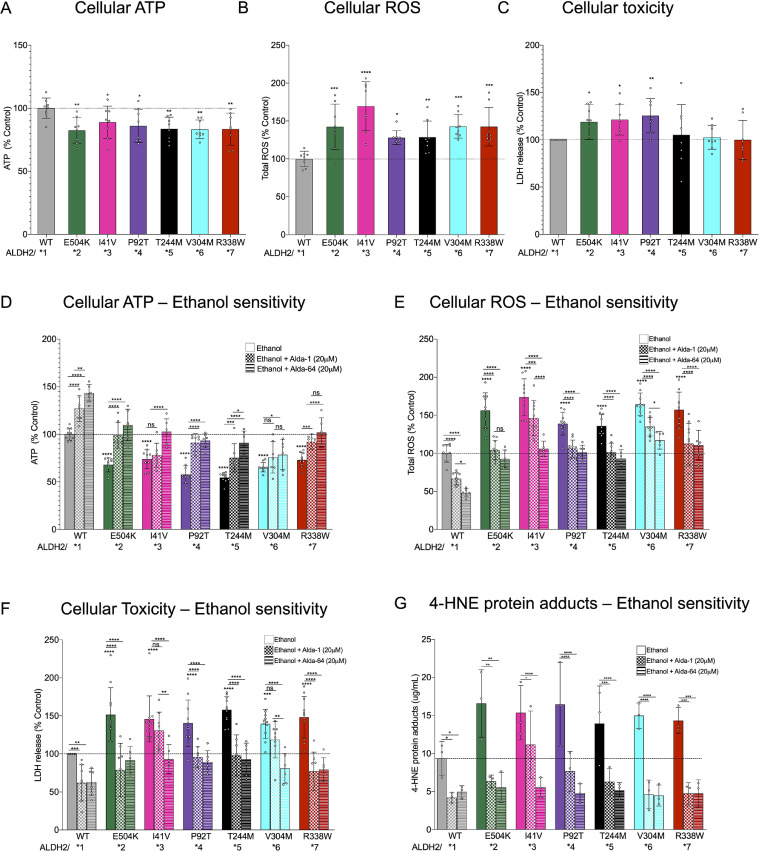
We next measured the cellular ATP levels, cell ROS, and cell death in the 3T3 cell lines overexpressing WT, as well as in each of the ALDH2 variants. We observed a mild reduction in ATP 25–75% as well as an increase in cellular ROS accumulation across all the mutants similar to the defects in E504K (Fig. 4(A) and (B)). However, only three mutations, ALDH2*2, *3 and *4 (E504K, I41V, P92T), showed a significantly increased cell death under basal levels (Fig. 4(C)). We observed a similar response of increased cell death along with accumulation of 4-HNE protein adducts in human-derived fibroblasts expressing ALDH2*2, *3 and *4 (E504K, I41V, P92T) variants relative to cells expressing WT (Figs. S1A and S1B). 4-HNE is an aldehyde generated during lipid peroxidation and cleared by ALDH2.
Fig 4.
Novel ALDH2 mutant are sensitive to ethanol toxicity. (A) Cellular ATP levels were measured in transiently transfected 3T3 cells using ATP-based CellTiter-Glo Luminescent Cell Viability kit. n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) ** p-value <0.01; * p-value <0.05. (B) Total cellular ROS levels were measured with 2,7 dichloro- fluorescein diacetate in cells treated as in (A). n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) ** p-value <0.01; * p-value <0.05. (C) Lactate dehydrogenase activity levels in supernatant in cells treated as in (A). n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) ** p-value <0.01; * p-value <0.05. (D) Cellular ATP levels were measured in transiently transfected 3T3 cells in the presence or absence of Alda-1/Alda-64 (20 μM/48 h; 50 mM Ethanol) using ATP-based CellTiter-Glo Luminescent Cell Viability kit. n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; ** p-value <0.01; * p-value <0.05. (E) Total cellular ROS levels were measured with 2,7 dichloro- fluorescein diacetate in 3T3 cells treated as in (D). n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; ** p-value <0.01; * p-value <0.05. (F) Lactate dehydrogenase activity levels in supernatant in 3T3 cells treated as in (D). n = 8; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; ** p-value <0.01; * p-value <0.05. (G) 4-HNE protein adducts in transiently transfected human-derived fibroblasts treated as in (D). n = 3; Mean, standard deviation, probability by one‐way ANOVA (with Fischer's LSD post hoc test) **** p-value <0.0001; *** p-value <0.001; ** p-value <0.01; * p-value <0.05.
Since ALDH2 activity is required for alcohol detoxification, we assessed the consequence of ethanol injury in 3T3 cells expressing the various ALDH2 variants. Exposure to ethanol (50 mM) for 48 h significantly reduced cellular ATP levels and increased ROS levels in each of the cell lines expressing mutant ALDH2 variants relative to the cell line expressing WT ALDH2 (Fig. 4(D) and (E)). There was also a corresponding increase in ethanol-induced cell death in both 3T3 cells and human-derived fibroblasts individually expressing ALDH2 variants relative to the cell line expressing WT ALDH2 (Figs. 4(E), (F) and S1C). Both Alda-1 and Alda-64 rescued the defects significantly, but the effect of Alda-64 appeared to be superior in terms of restoring cellular ATP levels, reducing ROS, and importantly, reducing cell death as measured by LDH release (Fig. 4(D)–(F)), especially in the case of ALDH2*3 and ALDH2*5. Alda-1 and Alda-64 treatment also reduced the levels of 4-HNE protein adducts (Fig. 4(G)) and cell death (Fig. S1C) in human-derived fibroblasts expressing the different ALDH2 variants exposed to ethanol (50 mM) for 48 h, with exception of ALDH2*3 and *6 (I41V, V304M) variants, which did not improve survival upon Alda1 treatment.
4. Discussion
560 million people, or 8% of the world population, are affected by the enzymopathy of ALDH2 deficiency. Despite affecting such a large percentage of the world population, all current ALDH2 deficient subjects are reported to carry one identical single amino acid variant (ALDH2*2, rs671), which appears to have originated from a single founder in East Asia 2000–3000 years ago [35]. No other ALDH2 single amino acid polymorphism has been reported or characterized thus far. In contrast, another very common human enzymopathy, glucose-6-phosphate dehydrogenase (G6PD) deficiency (also known as favism), which affects 400 million people or 5.7% of the world population, has been well-documented with more than 160 single amino acid variants [36]. Carriers of many of these variants have reduced G6PD enzyme activity and are susceptibility to anemia, newborn jaundice, or acute hemolysis induced by oxidative stress [36]. Both ALDH2 and G6PD are multimeric metabolic enzymes, consisting of similarly sized protein subunits of 517 and 515 amino acids, respectively. We reasoned that both enzymopathies have, at some point in time, provided some health benefits to humans, perhaps in the presence of bacterial, viral or parasite-induced diseases [37]. Regardless of the potential evolutionary advantage for these common enzymopathies in humans, we predicted that other inactivating mutations in ALDH2 may also be common among other ethnic and geographical groups.
In this study, we confirmed the presence of at least five new common variants that cause a substantial loss of ALDH2 activity. As all of these mutants are dominant (Fig. 2(D)), and given the allele frequencies and 2019 world population, we estimate that 80 million Latinos (10% of the world 800 million Latinos carrying the P92T and V304M variants), 28 million South Asians (1.4% of the 2 billion South Asians carrying the T224M variant) and 14 million Africans (1.2% of the 1.2 billion Africans carrying the I41V variant) may be affected by ALDH2 enzyme deficiency. Thus, there could be an additional ~120 million individuals who are ALDH2 enzyme deficient worldwide, in addition to the 560 million East Asians carrying the ALDH2*2 mutation. Of particular consequence to human health, the two very common Latino variants, ALDH2*4 (P92T) and ALDH2*6 (V304M), together affecting ~5% of Latinos, had only 32% and 11% activity of wildtype, respectively (Fig. 2(A)).
Relative to WT ALDH2 (ALDH2*1), the impairment in ALDH2 activity in vitro was much greater for the ALDH2*2 variant as compared with the new ALDH2 mutant variants (Fig. 2(A)). When overexpressed in 3T3 cells, except for the African variant, ALDH2*3, the loss of ALDH2 activity for all the variants were similar, showing about 30% lower protein levels, with ALDH2*3 showing the least effect on levels or stability (Fig. 3(A) and (B)). All the variants show about 60% lower ALDH2 activity, again with ALDH2*3 being the least impaired. Whether these data reflect the stability and activity of the variants in patient-derived cells remains to be determined. However, these data are quite consistent with the relative impairment of ALDH2 activity in the in vitro assays.
Our studies comparing the resistance of cells overexpressing each of the mutants to ethanol-induced toxicity (Figs. 4 and S1) suggest that the in vivo consequences of these variants might be comparable. We found similar increases in cell death (~50%), an increase in cellular ROS levels (~50%) and reduced cellular ATP levels (~35%) following exposure to ethanol (Fig. 4(D)–(F)). Even under basal conditions, increase in cell ROS and cellular ATP levels were similar (Fig. 4(A) and (B)). Moreover, we observed a pronounced accumulation of 4-hydroxynonenal (4-HNE)-protein adducts in human-derived fibroblasts overexpressing ALDH2 variants (which have reduced ALDH2 activity) compared with cells overexpressing ALDH2 WT. These findings suggest an impaired aldehyde detoxification capacity in cells overexpressing ALDH2 variants.
The difference between the effect of the two small molecule activators of ALDH2, Alda-1 and Alda-64, on the various variants suggests a different molecular mechanism for their activation. Alda-1 was not as effective as Alda-64 in compensating for the loss of activity on cellular ATP levels, cellular ROS levels and accumulation of 4-HNE protein adducts in the presence of ethanol for ALDH2*3 and ALDH2*6, translating to less protection from ethanol-induced cell death in both 3T3 cells and human-derived fibroblasts overexpressing these variants versus the other ALDH2 variants (Fig. 4(D)–(G)). These data underscore the opportunity for further exploration of both the structural origin of the new variants’ dysfunction and also for developing of new, more efficacious classes of ALDH2 activators. Moreover, ALDH2 activation using either Alda-1 or Alda-64 was sufficient to counteract the excessive 4-HNE accumulation and protecting cells against aldehydic load. These new findings along with our previous data demonstrate the critical role of ALDH2 in metabolizing toxic aldehydes.
If the functional deficits of ALDH2*3, *4, *5, *6 and *7 are confirmed in cells derived from individual carriers of these variants, there are new implications to our findings for these common variants in specific ethnic and geographic populations; our very small sample of 150 Latinos in Mexico City, identifying six subjects carrier of ALDH2*4 and five carriers of ALDH2*6 (Fig. 4(G)), supports the need for larger genotyping studies in the corresponding populations to confirm the prevalence of these variants. As discussed, epidemiological studies suggest an association between ALDH2*2 deficiency, alcohol consumption, and certain common and rare diseases [9]. Previous genome-wide association studies have implied that several SNPs near the ALDH2 chromosome region (12q24) might be associate with variation in alcohol consumption [38]. However, a more recent and thorough genome-wide study examining 86,627 individuals from 4 different major ethnic groups (non-Hispanic white, Hispanic/Latino, East Asian and African Americans) ruled out that any of these nearby SNPs in the non-coding sequence of ALDH2 were causal or contributed to variation in alcohol consumption [39]. New consideration should be given to educating people at risk of these diseases regarding the consumption of alcoholic beverages.
Our study provides the first molecular evidence, based on a human genomic database search together with in vitro enzymatic assay and cellular characterization, that there are at least five more ALDH2 deficiencies in humans, caused by mutations other than the common and well-characterized E504K substitution (ALDH2*2). We also showed that it is possible to correct these enzymopathies with small molecules, Alda-1 and Alda-64, identified in a small molecule screen. Confirmation of the health consequences of the ALDH2 variants in humans is required, as are epidemiological studies to determine the extent of their impact on global populations. The contribution of other rarer ALDH2 variants should also be considered. Our current research efforts focus on enzyme structural studies, and drug development using new chemotypes, to identify compounds that specifically target each ALDH2 variant more effectively. Finally, considering that at least 80% of the world's population consumes alcoholic beverages and that even moderate consumption poses a health risk, as has been reported for increased risk of upper aerodigestive tract cancer and other diseases in ALDH2*2 East Asians,1,5,8,9 the health impact of the newly identified common ALDH2 mutations in other populations should be considered. In the era of precision medicine, the health risks for carriers of the common ALDH2 variants in each of the ethnic groups should, therefore, be evaluated.
5. Caveat and limitations
The novel and prevalent non-East Asian ALDH2 variants identified and characterized in this manuscript are limited to the database collection by ExAC. Although the ExAC database has included the six major ethnic groups worldwide, it still represents only a small fraction of the world population and does not exclude the possibility that other distinct, common and undescribed ALDH2 variants may exist in other races and subethnic groups in different regions of the world. Our survey of the Latino ALDH2 variant is based on 150 human samples collected from Mexico. This is a small number of sample size from a single location for the Latino ethnic group. Future and more extensive collections and characterizations are needed to confirm our observations and the frequencies reported by the human genomic database. Finally, clinical studies to correlate the genotype of the carriers of the new ALDH2 variant with reduced ALDH2 activity and their phenotype of alcohol flushing reactions are needed.
6. Author contributions
C-HC and DM-R conceived the research idea and designed the research plan. C-HC, JCBF, AUJ, MCS, SJL, JH-MH, NDF and RM performed the experiments. PRC, DDM, FLB and GHRQ prepared IRB approval, collected human blood samples in Mexico and participated in manuscript discussion. C-HC, JCBF, AUJ and DM-R wrote the manuscript. All authors read and approved the manuscript.
7. Funding
C-HC & AUJ are supported by R37 MERIT award NIH AAA11147 to DM-R. JCBF is supported by FAPESP (#2018/18627-3) and CNPq (307934/2018-7). S-JL is supported by the Ministry of Science and Technology of Taiwan, under Grant No. MOST 107-2917-I-002-001. JH is supported by Graduate Student Study Abroad Program, Ministry of Science and Technology, Taiwan. None of the funding organization had any involvement in the study design, execution of the experiments, analysis and interpretation of data, the writing and submission of the manuscript.
Declaration of Competing Interest
Daria Mochly-Rosen and Che-Hong Chen hold patents related to Alda-1 activation of ALDH2*1 and ALDH2*2. One of the patents is licensed to Foresee Pharmaceuticals, a company that DM-R consults. However, these authors do not own stocks of the company and none of this research is supported by the company.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102753.
Appendix. Supplementary materials
References
- 1.Brooks P.J., Enoch M.A., Goldman D., Li T.K., Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6(3):e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchitti S.A., Brocker C., Stagos D., Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Exp Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida A., Huang I.Y., Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81(1):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Q., Wu J., Cai Q., Chen E.Z., Jiang Z.Y. Association between Glu504Lys polymorphism of ALDH2 gene and cancer risk: a meta-analysis. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama A., Omori T., Yokoyama T. Alcohol and aldehyde dehydrogenase polymorphisms and a new strategy for prevention and screening for cancer in the upper aerodigestive tract in East Asians. Keio J Med. 2010;59(4):115–130. doi: 10.2302/kjm.59.115. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama T., Yokoyama A., Kato H., Tsujinaka T., Muto M., Omori T. Alcohol flushing, alcohol and aldehyde dehydrogenase genotypes, and risk for esophageal squamous cell carcinoma in Japanese men. Cancer Epidemiol Biomarkers Prev. 2003;12(11 Pt 1):1227–1233. [PubMed] [Google Scholar]
- 7.Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Bouvard V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8(4):292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 8.Gross E.R., Zambelli V.O., Small B.A., Ferreira J.C., Chen C.H., Mochly-Rosen D. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol. 2015;55:107–127. doi: 10.1146/annurev-pharmtox-010814-124915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C.H., Ferreira J.C., Gross E.R., Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev. 2014;94(1):1–34. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshi H., Hao W., Fujita Y., Funayama A., Miyauchi Y., Hashimoto K. Aldehyde-stress resulting from ALDH2 mutation promotes osteoporosis due to impaired osteoblastogenesis. J Bone Miner Res. 2012;27(9):2015–2023. doi: 10.1002/jbmr.1634. [DOI] [PubMed] [Google Scholar]
- 11.Pang J., Wang J., Zhang Y., Xu F., Chen Y. Targeting acetaldehyde dehydrogenase 2 (ALDH2) in heart failure-Recent insights and perspectives. Biochim Biophys Acta Mol Basis Dis. 2017;1863(8):1933–1941. doi: 10.1016/j.bbadis.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Van Wassenhove L.D., Mochly-Rosen D., Weinberg K.I. Aldehyde dehydrogenase 2 in aplastic anemia, Fanconi anemia and hematopoietic stem cells. Mol Genet Metab. 2016;119(1–2):28–36. doi: 10.1016/j.ymgme.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabe M., Koike T., Ohtsubo K., Imai E., Morimoto T., Takakura H. Associations of complementation group, ALDH2 genotype, and clonal abnormalities with hematological outcome in Japanese patients with Fanconi anemia. Ann Hematol. 2019;98(2):271–280. doi: 10.1007/s00277-018-3517-0. [DOI] [PubMed] [Google Scholar]
- 14.Opelt M., Eroglu E., Waldeck-Weiermair M., Russwurm M., Koesling D., Malli R. Formation of nitric oxide by aldehyde dehydrogenase-2 is necessary and sufficient for vascular bioactivation of nitroglycerin. J Biol Chem. 2016;291(46):24076–24084. doi: 10.1074/jbc.M116.752071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Zhang D., Jin W., Shao C., Yan P., Xu C. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116(2):506–511. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmetz C.G., Xie P., Weiner H., Hurley T.D. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5(5):701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 17.Goedde H.W., Agarwal D.P., Fritze G., Meier-Tackmann D., Singh S., Beckmann G. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88(3):344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Borinskaya S., Yoshimura K., Kal'ina N., Marusin A., Stepanov V.A. Refined geographic distribution of the oriental ALDH2*504Lys (nee 487Lys) variant. Ann Hum Genet. 2009;73(Pt 3):335–345. doi: 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo H.R., Wu G.S., Pakstis A.J., Tong L., Oota H., Kidd K.K. Origin and dispersal of atypical aldehyde dehydrogenase ALDH2487Lys. Gene. 2009;435(1–2):96–103. doi: 10.1016/j.gene.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 20.Harada S., Okubo T., Nakamura T., Fujii C., Nomura F., Higuchi S. A novel polymorphism (-357G/A) of the ALDH2 gene: linkage disequilibrium and an association with alcoholism. Alcohol Clin Exp Res. 1999;23(6):958–962. [PubMed] [Google Scholar]
- 21.Pathak H., Frieling H., Bleich S., Glahn A., Heberlein A., Haschemi Nassab M. Promoter polymorphism rs886205 genotype interacts with DNA methylation of the ALDH2 regulatory region in alcohol dependence. Alcohol Alcohol. 2017;52(3):269–276. doi: 10.1093/alcalc/agw106. [DOI] [PubMed] [Google Scholar]
- 22.Genomes Project C., Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lappalainen T., Scott A.J., Brandt M., Hall I.M. Genomic analysis in the age of human genome sequencing. Cell. 2019;177(1):70–84. doi: 10.1016/j.cell.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto A. Importance of an aldehyde dehydrogenase 2 polymorphism in preventive medicine. Nihon Eiseigaku Zasshi. 2018;73(1):9–20. doi: 10.1265/jjh.73.9. [DOI] [PubMed] [Google Scholar]
- 26.Chen C.H., Budas G.R., Churchill E.N., Disatnik M.H., Hurley T.D., Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi A.U., Minhas P.S., Liddelow S.A., Haileselassie B., Andreasson K.I., Dorn G.W., 2nd Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nat Neurosci. 2019;22(10):1635–1648. doi: 10.1038/s41593-019-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi A.U., Saw N.L., Shamloo M., Mochly-Rosen D. Drp1/Fis1 interaction mediates mitochondrial dysfunction, bioenergetic failure and cognitive decline in Alzheimer's disease. Oncotarget. 2018;9(5):6128–6143. doi: 10.18632/oncotarget.23640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marie-Cardine A., Bruyns E., Eckerskorn C., Kirchgessner H., Meuer S.C., Schraven B. Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J Biol Chem. 1997;272(26):16077–16080. doi: 10.1074/jbc.272.26.16077. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez F., Hu J., Kershaw K., Hastings K.G., Lopez L., Cullen M.R. County-level Hispanic ethnic density and cardiovascular disease mortality. J Am Heart Assoc. 2018;7(19) doi: 10.1161/JAHA.118.009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volgman A.S., Palaniappan L.S., Aggarwal N.T., Gupta M., Khandelwal A., Krishnan A.V. Atherosclerotic cardiovascular disease in South Asians in the United States: epidemiology, risk factors, and treatments: a scientific statement from the American Heart Association. Circulation. 2018;138(1):e1–e34. doi: 10.1161/CIR.0000000000000580. [DOI] [PubMed] [Google Scholar]
- 32.Chen C.H., Joshi A.U., Mochly-Rosen D. The role of mitochondrial aldehyde dehydrogenase 2 (ALDH2) in neuropathology and neurodegeneration. Acta Neurol Taiwan. 2016;25(4):111–123. [PMC free article] [PubMed] [Google Scholar]
- 33.Lai C.L., Yao C.T., Chau G.Y., Yang L.F., Kuo T.Y., Chiang C.P. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38(1):44–50. doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Q., Weiner H., Johnston T., Crabb D.W. The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced Hela cells. J Clin Invest. 1995;96(5):2180–2186. doi: 10.1172/JCI118272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S., Fritze G., Fang B.L., Harada S., Paik Y.K., Eckey R. Inheritance of mitochondrial aldehyde dehydrogenase: genotyping in Chinese, Japanese and South Korean families reveals dominance of the mutant allele. Hum Genet. 1989;83(2):119–121. doi: 10.1007/BF00286702. [DOI] [PubMed] [Google Scholar]
- 36.Hwang S., Mruk K., Rahighi S., Raub A.G., Chen C.H., Dorn L.E. Correcting glucose-6-phosphate dehydrogenase deficiency with a small-molecule activator. Nat Commun. 2018;9(1):4045. doi: 10.1038/s41467-018-06447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cappadoro M., Giribaldi G., O'Brien E., Turrini F., Mannu F., Ulliers D. Early phagocytosis of glucose-6-phosphate dehydrogenase (G6PD)-deficient erythrocytes parasitized by Plasmodium falciparum may explain malaria protection in G6PD deficiency. Blood. 1998;92(7):2527–2534. [PubMed] [Google Scholar]
- 38.Baik I., Cho N.H., Kim S.H., Han B.G., Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93(4):809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann T.J., Passarelli M.N., Graff R.E., Emami N.C., Sakoda L.C., Jorgenson E. Genome-wide association study of prostate-specific antigen levels identifies novel loci independent of prostate cancer. Nat Commun. 2017;8:14248. doi: 10.1038/ncomms14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.