Key Points
Question
What epidemiological and clinical characteristics are associated with the development of critical illness among patients with novel coronavirus disease 2019 (COVID-19)? Can these characteristics be used to predict which patients admitted to the hospital with COVID-19 will need admission to an intensive care unit, mechanical ventilation, or will die?
Findings
In this study with a development cohort of 1590 patients and a validation cohort of 710 patients, a risk score was developed and validated to predict development of critical illness. We identified 10 independent predictors and developed a risk score (COVID-GRAM) that predicts development of critical illness. The risk score predictors included: chest radiography abnormality, age, hemoptysis, dyspnea, unconsciousness, number of comorbidities, cancer history, neutrophil-to-lymphocyte ratio, lactate dehydrogenase, and direct bilirubin.
Meaning
The COVID risk score may help identify patients with COVID-19 who may subsequently develop critical illness.
This cohort study examines the characteristics of patients admitted to the hospital with COVID-19 who will need admission to an intensive care unit, mechanical ventilation, or will die, and develops a COVID-19 risk score to predict outcomes.
Abstract
Importance
Early identification of patients with novel coronavirus disease 2019 (COVID-19) who may develop critical illness is of great importance and may aid in delivering proper treatment and optimizing use of resources.
Objective
To develop and validate a clinical score at hospital admission for predicting which patients with COVID-19 will develop critical illness based on a nationwide cohort in China.
Design, Setting, and Participants
Collaborating with the National Health Commission of China, we established a retrospective cohort of patients with COVID-19 from 575 hospitals in 31 provincial administrative regions as of January 31, 2020. Epidemiological, clinical, laboratory, and imaging variables ascertained at hospital admission were screened using Least Absolute Shrinkage and Selection Operator (LASSO) and logistic regression to construct a predictive risk score (COVID-GRAM). The score provides an estimate of the risk that a hospitalized patient with COVID-19 will develop critical illness. Accuracy of the score was measured by the area under the receiver operating characteristic curve (AUC). Data from 4 additional cohorts in China hospitalized with COVID-19 were used to validate the score. Data were analyzed between February 20, 2020 and March 17, 2020.
Main Outcomes and Measures
Among patients with COVID-19 admitted to the hospital, critical illness was defined as the composite measure of admission to the intensive care unit, invasive ventilation, or death.
Results
The development cohort included 1590 patients. the mean (SD) age of patients in the cohort was 48.9 (15.7) years; 904 (57.3%) were men. The validation cohort included 710 patients with a mean (SD) age of 48.2 (15.2) years, and 382 (53.8%) were men and 172 (24.2%). From 72 potential predictors, 10 variables were independent predictive factors and were included in the risk score: chest radiographic abnormality (OR, 3.39; 95% CI, 2.14-5.38), age (OR, 1.03; 95% CI, 1.01-1.05), hemoptysis (OR, 4.53; 95% CI, 1.36-15.15), dyspnea (OR, 1.88; 95% CI, 1.18-3.01), unconsciousness (OR, 4.71; 95% CI, 1.39-15.98), number of comorbidities (OR, 1.60; 95% CI, 1.27-2.00), cancer history (OR, 4.07; 95% CI, 1.23-13.43), neutrophil-to-lymphocyte ratio (OR, 1.06; 95% CI, 1.02-1.10), lactate dehydrogenase (OR, 1.002; 95% CI, 1.001-1.004) and direct bilirubin (OR, 1.15; 95% CI, 1.06-1.24). The mean AUC in the development cohort was 0.88 (95% CI, 0.85-0.91) and the AUC in the validation cohort was 0.88 (95% CI, 0.84-0.93). The score has been translated into an online risk calculator that is freely available to the public (http://118.126.104.170/)
Conclusions and Relevance
In this study, a risk score based on characteristics of COVID-19 patients at the time of admission to the hospital was developed that may help predict a patient’s risk of developing critical illness.
Introduction
The outbreak of the novel coronavirus disease 2019 (COVID-19) began in Wuhan, China in December 2019. Since then, it has rapidly spread around the world. As of April 16, 2020, the WHO reported a total of 1 995 983 COVID-19 cases globally, with average mortality of 6.57%.
The clinical spectrum of COVID-19 pneumonia ranges from mild to critically ill cases. Patients with mild disease present with symptoms of fever and cough, followed by sputum production and fatigue. Sepsis, respiratory failure, acute respiratory distress syndrome, heart failure, and septic shock are commonly observed in critically ill patients.1
According to the largest current report from the Chinese Center for Disease Control and Prevention with 72 314 cases, 58 574 patients (81%) were classified as mild, 10 124 (14%) were classified as severe, and 3616 (5%) were considered critical illness. The average case-fatality rate was 2.3%, but mortality was as high as 49% in patients with critical illness.2 Among 201 patients in Wuhan, Wu et al3 reported that risk factors associated with development of acute respiratory distress syndrome and death included older age, neutrophilia, organ dysfunction, coagulopathy, and elevated D-dimer levels.
Early detection of patients who are likely to develop critical illness is of great importance and may aid in delivering proper care and optimizing use of limited resources. We aimed to construct a risk prediction score based on a nationwide cohort of Chinese patients with COVID-19 to help identify patients at the time of hospital admission who are likely to develop critical illness.
Methods
Data Sources and Processing
This study was approved by the ethics committee of the First Affiliated Hospital of Guangzhou Medical University; written informed consent was waived owing to the use of deidentified retrospective data. On behalf of the National Clinical Research Center for Respiratory Disease and collaborating with the National Health Commission of the People’s Republic of China, we established a retrospective cohort to study COVID-19 cases throughout China. We obtained medical records from laboratory-confirmed hospitalized cases with COVID-19 reported to the China National Health Commission between November 21, 2019 and January 31, 2020, as previously described.4 The National Health Commission of China requested that all 1855 hospitals in China that were designated to care for COVID-19 patients submit the clinical records of all hospitalized COVID-19 cases without selection to the database by January 31, 2020. For the development cohort, we used data from the 575 hospitals that contributed clinical data by the deadline.
COVID-19 diagnoses were confirmed by positive high-throughput sequencing or real-time reverse-transcription polymerase-chain-reaction (RT-PCR) assay for nasal and pharyngeal swab specimens. A team of experienced respiratory clinicians reviewed, abstracted and cross-checked the data. Each record was checked independently by 2 clinicians. We included all patients with data on clinical status at hospitalization (laboratory findings, clinical symptoms and signs, severity, and discharge status).
Potential Predictive Variables
Potential predictive variables included the following patient characteristics at hospital admission: clinical signs and symptoms, imaging results, laboratory findings, demographic variables, and medical history. Demographic variables collected for the study included age, sex, smoking status, exposure to Wuhan (including Wuhan residency, travel history to Wuhan, or contact with people from Wuhan), residency in Hubei province, and time between onset of symptoms to admission. Medical history included number of comorbidities, chronic obstructive pulmonary disease, diabetes, hypertension, coronary artery disease, cerebrovascular disease, hepatitis B, cancer, chronic renal disease, immunodeficiency disease, and pregnancy. Clinical signs and symptoms included categorical and continuous variables: first body temperature, respiratory rate, heart rate, cardiac arrhythmia, systolic blood pressure, diastolic blood pressure, symptoms rating, fever, conjunctival congestion, nasal congestion, headache, cough, expectoration, sore throat, fatigue, hemoptysis, dyspnea, nausea and vomiting, diarrhea, arthralgia and myalgia, rigor, throat blockage, tonsillar enlargement, enlarged lymph nodes, skin rash, and unconsciousness. Imaging results included chest radiography (CXR) abnormality, the severity of CXR abnormality, chest computed tomographic (CT) imaging abnormality, and the severity of CT abnormality. Laboratory findings included partial arterial oxygen pressure, oxygen saturation, white blood cell, lymphocyte, and platelet counts, neutrophil to lymphocyte ratio, and levels of hemoglobin, C-reactive protein, procalcitonin, lactate dehydrogenase, aspartate transaminase, direct bilirubin, indirect bilirubin, total bilirubin, creatine kinase, creatinine, hypersensitive troponin I, albumin, serum sodium, serum potassium, serum chlorine, D-dimer levels, prothrombin time, and activated partial thromboplastin time.
Outcomes
We defined the severity of COVID-19 (severe vs nonsevere) based on the American Thoracic Society guidelines for community-acquired pneumonia given the extensive acceptance of this guideline.5 We defined critical COVID-19 illness as a composite of admission to the intensive care unit (ICU), invasive ventilation, or death. We adopted this composite end point because admission to ICU, invasive ventilation, and death are serious outcomes of COVID-19 that have been adopted in previous studies to assess the severity of other serious infectious diseases.5,6
Variable Selection and Score Construction
All 1590 patients hospitalized with COVID-19 in the development cohort were included for variable selection and risk score development. As described herein, 72 variables were entered into the selection process. Least Absolute Shrinkage and Selection Operator (LASSO) regression was applied to minimize the potential collinearity of variables measured from the same patient and over-fitting of variables. Imputation for missing variables was considered if missing values were less than 20%. We used predictive mean matching to impute numeric features, logistic regression to impute binary variables, and Bayesian polytomous regression to impute factor features. We used L1-penalized least absolute shrinkage and selection regression for multivariable analyses, augmented with 10-fold cross validation for internal validation. This is a logistic regression model that penalizes the absolute size of the coefficients of a regression model based on the value of λ. With larger penalties, the estimates of weaker factors shrink toward zero, so that only the strongest predictors remain in the model. The most predictive covariates were selected by the minimum (λ min). The R package “glmnet” statistical software (R Foundation) was used to perform the LASSO regression. Subsequently, variables identified by LASSO regression analysis were entered into logistic regression models and those that were consistently statistically significant were used to construct the risk score (COVID-GRAM),7 which was then used to construct a web-based risk calculator (http://118.126.104.170/). Data were analyzed between February 20, 2020 and March 17, 2020.
Assessment of Accuracy
The accuracy of COVID risk score was assessed using the area under the receiver-operator characteristic curve (AUC). We also used the AUC to compare the accuracy of the COVID-GRAM with CURB-6 models,8 which have been used in classification of the severity of community-acquired pneumonia. For internal validation of the accuracy estimates and to reduce overfit bias, we used 200 bootstrap resamples. Statistical analysis was performed with R software (version 3.6.2, R Foundation), and P < .05 was considered statistically significant.
Score Validation
To validate the generalizability of COVID risk score, we used data from hospitals that were not included in the development cohort including 710 patients. Data for the validation cohort were pooled from 4 sources: (1) a multicenter cohort of hospitals from 10 cities in Hubei province that missed the deadline for data submission, but subsequently submitted data on cases admitted before the January 31, 2020; (2) Daye Hospital (near Wuhan); (3) The First People’s Hospital of Foshan (Guangdong province), and Nanhai People’s Hospital of Foshan (Guangdong province). The later 3 hospitals reported up-to-date data as of February 28, 2020.
The variables required for calculating the COVID risk score from the validation cohort were collected and cross-checked by 2 experienced physicians (C.Z.S. and C.A.L.) and the risk score was calculated as described herein for the development cohort.
Results
Characteristics of the Development Cohort
In the development cohort, we collected data from 1590 patients from 575 hospitals in 31 provincial administrative regions between November 21, 2019 and January 31, 2020. At hospital admission, 24 of 1590 patients (1.5%) were considered to be severe and the rest (1566 [98.5%]) were considered to be mild according to the American Thoracic Society guideline.5 A total of 131 patients eventually developed critical illness (8.2%). The overall mortality was 3.2% and 1334 patients (83.9%) had a history of exposure to Wuhan.
Overall, the mean (SD) age of patients in the cohort was 48.9 (15.7) years; 904 patients (57.3%) were men and 399 (25.1%) had at least 1 coexisting condition, including hypertension (269 [16.9%]), diabetes (130 [8.2%]), and cardiovascular disease (59 [3.7%]) as the top 3 comorbidities (Table 1). Fever (1351 [88.0%]), dry cough (1052 [70.2%]), fatigue (584 [42.8%]), productive cough (513 [36.0%]), and shortness of breath (331 [23.7%]) were the most common symptoms. Most patients (1130 [71.1%]) had abnormal chest CT findings. Laboratory findings of the development cohort are presented in Table 2.
Table 1. Demographics and Clinical Characteristics Among Patients In the Development Cohort Who Did or Did Not Develop Critical Illnessa.
| Characteristic | Total, mean (SD) [range] | Critical illness | |
|---|---|---|---|
| No | Yes | ||
| No. | 1590 | 1459 | 131 |
| Age, mean (SD), y | 48.9 (15.7) [1-95] | 47.8 (15.2) | 61.6 (14.8) |
| Incubation period, mean (SD), d | 5.0 (4.1) [0-24] | 4.9 (4.1) | 5.7 (4.2) |
| Admission measures, mean (SD) | |||
| Temperature, °C | 37.3 (0.9) [35.5-40.3] | 37.4 (0.9) | 37.1 (0.9) |
| Respiratory rate, breaths/min | 21.2 (12.0) [12-65] | 21.1 (12.4) | 23.1 (5.9) |
| Heart rate, beats/min | 88.7 (14.6) [17-205] | 88.6 (14.4) | 89.7 (16.0) |
| Blood pressure, mm Hg | |||
| Systolic | 126.1 (16.4) [74-187] | 125.5 (15.6) | 131.4 (22.5) |
| Diastolic | 79.5 (25.6) [40-160] | 79 (11.3) | 84.7 (76.1) |
| Male, No./No. (%) | 904/1578 (57.3) | 816/1447 (56.4) | 88/131 (67.2) |
| Smoking status, No./No. (%) | |||
| Never | 1479/1590 (93.0) | 1366/1459 (93.6) | 113/131 (86.3) |
| Former/current | 111/1590 (7) | 93/1459 (6.4) | 18/131 (13.7) |
| Symptoms, No./No. (%) | |||
| Degree of symptoms | |||
| 0 | 73/1590 (4.6) | 67/1459 (4.6) | 6/131 (4.6) |
| 1 | 176/1590 (11.1) | 170/1459 (11.7) | 6/131 (4.6) |
| 2 | 353/1590 (22.2) | 330/1459 (22.6) | 23/131 (17.6) |
| 3 | 409/1590 (25.7) | 378/1459 (25.9) | 31/131 (23.7) |
| 4 | 287/1590 (18.1) | 258/1459 (17.7) | 29/131 (22.1) |
| 5 | 158/1590 (9.9) | 141/1459 (9.7) | 17/131 (13.0) |
| 6 | 76/1590 (4.8) | 68/1459 (4.7) | 8/131 (6.1) |
| 7 | 36/1590 (2.3) | 29/1459 (2.0) | 7/131 (5.3) |
| 8 | 14/1590 (0.9) | 11/1459 (0.8) | 3/131 (2.3) |
| 9 | 7/1590 (0.4) | 6/1459 (0.4) | 1/131 (0.8) |
| 10 | 1/1590 (0.1) | 1/1459 (0.1) | 0/131 (0) |
| Fever | 1351/1536 (88.0) | 1237/1409 (87.8) | 114/127 (89.8) |
| Congestion | |||
| Conjunctival | 10/1345 (0.7) | 10/1235 (0.8) | 0/110 (0) |
| Nasal | 73/1299 (5.6) | 64/1191 (5.4) | 9/108 (8.3) |
| Headache | 205/1328 (15.4) | 190/1221 (15.6) | 15/107 (14) |
| Dry cough | 1052/1498 (70.2) | 959/1372 (69.9) | 93/126 (73.8) |
| Sore throat | 194/1317 (14.7) | 181/1207 (15.0) | 13/110 (11.8) |
| Productive cough | 513/1424 (36.0) | 461/1302 (35.4) | 52/122 (42.6) |
| Fatigue | 584/1365 (42.8) | 539/1250 (43.1) | 45/115 (39.1) |
| Hemoptysis | 16/1315 (1.2) | 10/1201 (0.8) | 6/114 (5.3) |
| Shortness of breath | 331/1394 (23.7) | 257/1275 (20.2) | 74/119 (62.2) |
| Nausea/vomiting | 80/1371 (5.8) | 73/1256 (5.8) | 7/115 (6.1) |
| Diarrhea | 57/1359 (4.2) | 52/1244 (4.2) | 5/115 (4.3) |
| Myalgia/arthralgia | 234/1338 (17.5) | 215/1229 (17.5) | 19/109 (17.4) |
| Chills | 163/1333 (12.2) | 151/1222 (12.4) | 12/111 (10.8) |
| Signs | |||
| Throat congestion | 21/1286 (1.6) | 21/1178 (1.8) | 0/108 (0) |
| Tonsil swelling | 31/1376 (2.3) | 30/1261 (2.4) | 1/115 (0.9) |
| Enlargement of lymph nodes | 2/1375 (0.1) | 1/1261 (0.1) | 1/114 (0.9) |
| Rash | 3/1378 (0.2) | 3/1264 (0.2) | 0/114 (0) |
| Unconsciousness | 20/1421 (1.4) | 10/1303 (0.8) | 10/118 (8.5) |
| Comorbidities, No./No. (%) | |||
| Any | 399/1590 (25.1) | 322/1459 (22.1) | 77/131 (58.8) |
| No. of comorbidities | |||
| 0 | 1191/1590 (74.9) | 1137/1459 (77.9) | 54/131 (41.2) |
| 1 | 269/1590 (16.9) | 229/1459 (15.7) | 40/131 (30.5) |
| 2 | 88/1590 (5.5) | 68/1459 (4.7) | 20/131 (15.3) |
| 3 | 34/1590 (2.1) | 20/1459 (1.4) | 14/131 (10.7) |
| 4 | 5/1590 (0.3) | 4/1459 (0.3) | 1/131 (0.8) |
| 5 | 3/1590 (0.2) | 1/1459 (0.1) | 2/131 (1.5) |
| COPD | 24/1590 (1.5) | 12/1459 (0.8) | 12/131 (9.2) |
| Diabetes | 130/1590 (8.2) | 99/1459 (6.8) | 31/131 (23.7) |
| Hypertension | 269/1590 (16.9) | 216/1459 (14.8) | 53/131 (40.5) |
| Cardiovascular disease | 59/1590 (3.7) | 46/1459 (3.2) | 13/131 (9.9) |
| Cerebrovascular disease | 30/1590 (1.9) | 20/1459 (1.4) | 10/131 (7.6) |
| Hepatitis B infection | 28/1590 (1.8) | 25/1459 (1.7) | 3/131 (2.3) |
| Malignancy | 18/1590 (1.1) | 11/1459 (0.8) | 7/131 (5.3) |
| Chronic kidney disease | 21/1590 (1.3) | 15/1459 (1.0) | 6/131 (4.6) |
| Immunodeficiency | 3/1590 (0.2) | 2/1459 (0.1) | 1/131 (0.8) |
| Abnormal chest radiography | 243/1590 (15.3) | 184/1459 (12.6) | 59/131 (45.0) |
| Severity of abnormality | |||
| 0 | 1347/1590 (84.7) | 1275/1459 (87.4) | 72/131 (55.0) |
| 1 | 138/1590 (8.7) | 104/1459 (7.1) | 34/131 (26.0) |
| 2 | 69/1590 (4.3) | 59/1459 (4.0) | 10/131 (7.6) |
| 3 | 36/1590 (2.3) | 21/1459 (1.4) | 15/131 (11.5) |
| Abnormal chest CT | 1130/1590 (71.1) | 1035/1459 (70.9) | 95/131 (72.5) |
| Severity of abnormality | |||
| 0 | 460/1590 (28.9) | 424/1459 (29.1) | 36/131 (27.5) |
| 1 | 492/1590 (30.9) | 466/1459 (31.9) | 26/131 (19.8) |
| 2 | 291/1590 (18.3) | 258/1459 (17.7) | 33/131 (25.2) |
| 3 | 248/1590 (15.6) | 224/1459 (15.4) | 24/131 (18.3) |
| 4 | 99/1590 (6.2) | 87/1459 (6.0) | 12/131 (9.2) |
| Residency in Hubei Province, yes | 647/1590 (40.7) | 552/1459 (37.8) | 95/131 (72.5) |
| Exposure to Wuhan, yes | 1334/1590 (83.9) | 1213/1459 (83.1) | 121/131 (92.4) |
Abbreviation: COPD, chronic obstructive pulmonary disease; CT, computed tomography.
Data are mean (SD), No./No. (%), where No. is the total number of patients with available data.
Table 2. Laboratory Findings Among Patients Who Did or Did Not Develop Critical Illness.
| Variable | Total, mean (SD) [range] | Critical illness, mean (SD) | |
|---|---|---|---|
| No | Yes | ||
| No. | 1590 | 1459 | 131 |
| Total urine volume, mL/d | 794.6 (901.6) [0-3810] | 622.1 (855.4) | 1155.1 (907.8) |
| Pao2 (with oxygen inhalation), mm Hg | 84.3 (36.2) [10.2-242.0] | 86.5 (36.0) | 67.2 (33.3) |
| FiO2, % | 27.5 (16.6) [21.0-99.7] | 26.6 (14.7) | 33.2 (25.2) |
| Pao2 (without oxygen inhalation), mm Hg | 92.7 (13.5) [6.12-254.0] | 93.6 (12.5) | 85.8 (18.2) |
| Neutrophil cell count, ×109/L | 4.14 (2.2) [0.69-24.0] | 3.9 (1.9) | 6.4 (3.6) |
| Lymphocyte count, ×109/L | 1.4 (3.1) [0-45.0] | 1.5 (3.3) | 0.7 (0.4) |
| Platelet count, ×109/L | 179.5 (70.7) [0.1-602.0] | 180.1 (70.4) | 173.4 (73.7) |
| Hemoglobin, g/L | 123.5 (43.9) [3.33-414.0] | 124.2 (43.9) | 115.5 (43.5) |
| C-reactive protein, mg/L | 34.8 (49.2) [0-624.0] | 30.6 (43.8) | 84.5 (76.3) |
| Procalcitonin, ng/mL | 0.7 (9.8) [0-252.7] | 0.8 (10.3) | 0.6 (1.4) |
| Lactate dehydrogenase, U/L | 314.3 (693.7) [1.0-1411.0] | 273.6 (135.2) | 723.6 (2239.5) |
| Aminotransferase, U/L | |||
| Aspartate | 49.7 (451.9) [5.1-203.0] | 34.1 (20.9) | 205.1 (1493.4) |
| Alanine | 43.1 (242.5) [2.0-435.0] | 34.4 (37.2) | 130.1 (795.9) |
| Bilirubin, mmol/L | |||
| Direct | 4 (2.7) [0-22.3] | 3.7 (2.3) | 6.5 (4.1) |
| Indirect | 7.4 (4.5) [0-41.2] | 7.2 (4.3) | 8.9 (5.3) |
| Total | 11.8 (14.2) [0.1-97.0] | 11.4 (14.7) | 15.2 (8.4) |
| Creatine kinase, U/L | 135.5 (246.7) [0.05-1013.0] | 123 (125.3) | 258.9 (702.8) |
| Creatinine, μmol/L | 76 (71.4) [4.05-1441.0] | 71.8 (54.2) | 118.7 (158.4) |
| Hypersensitive troponin I, pg/mL | 76.3 (586.4) [0-8622.0] | 42.7 (439.0) | 288.1 (1124.2) |
| Albumin, g/L | 38.7 (9.1) [0-135.0] | 39.3 (8.9) | 32.6 (8.9) |
| Sodium, mmol/L | 140.5 (50.4) [124.4-151.0] | 139.7 (41.2) | 148.7 (103.1) |
| Potassium, mmol/L | 4.4 (6.4) [2.3-6.7] | 4.4 (6.7) | 4.1 (0.8) |
| Chlorine, mmol/L | 103.8 (28.2) [89.8-119.0] | 103.7 (29.5) | 105 (4.8) |
| D-dimer level, mg/L | 25.5 (138.2) [0-2660.0] | 26.3 (144.8) | 19.1 (70.1) |
| Prothrombin time, s | 17.4 (48.2) [0-183.0] | 17.6 (50.2) | 15.9 (24.1) |
| Activated partial thromboplastin time, s | 42.5 (143.7) [3.0-499.0] | 43.3 (150.6) | 34.8 (50.9) |
| Neutrophil-lymphocyte ratio | 5.1 (5.6) [0.06-78.2] | 4.4 (3.8) | 12.7 (12.4) |
Abbreviations: FiO2, fraction of inspired oxygen; Pao2, partial pressure of oxygen.
Predictor Selection
Seventy-two variables measured at hospital admission (Table 1 and Table 2) were included in the LASSO regression. After LASSO regression selection (eFigure 1 in the Supplement), 19 variables remained significant predictors of critical illness, including clinical features and blood test results, CXR abnormality, age, exposure to Wuhan, first and highest body temperature, respiratory rate, systolic blood pressure, hemoptysis, dyspnea, skin rash, unconsciousness, number of comorbidities, chronic obstructive pulmonary disease (COPD), cancer, oxygen saturation levels, neutrophils, neutrophil to lymphocyte ratio, lactate dehydrogenase, direct bilirubin, and creatinine levels.
Inclusion of these 19 variables in a logistic regression model resulted in 10 variables that were independently statistically significant predictors of critical illness and were included in risk score. These variables included CXR abnormality (OR, 3.39; 95% CI, 2.14-5.38; P < .001), age (OR, 1.03; 95% CI, 1.01-1.05; P = .002), hemoptysis (OR, 4.53; 95% CI, 1.36-15.15; P = .01), dyspnea (OR, 1.88; 95% CI, 1.18-3.01; P = .01), unconsciousness (OR, 4.71; 95% CI, 1.39-15.98; P = .01), number of comorbidities (OR, 1.60; 95% CI, 1.27-2.00; P < .001), cancer history (OR, 4.07; 95% CI, 1.23-13.43; P = .02), neutrophil-to-lymphocyte ratio (OR, 1.06; 95% CI, 1.02-1.10; P = .003), lactate dehydrogenase (OR, 1.002; 95% CI, 1.001-1.004, P < .001), and direct bilirubin (OR, 1.15; 95% CI, 1.06-1.24; P = .001) (Table 3).
Table 3. Multivariable Logistic Regression Model for Predicting Development of Critical Illness in 1590 Patients Hospitalized With COVID-19 in Wuhan.
| Variables | Odds ratio (95% CI) | P value |
|---|---|---|
| X-ray abnormality (yes vs no) | 3.39 (2.14-5.38) | <.001 |
| Age, per y | 1.03 (1.01-1.05) | .002 |
| Hemoptysis (yes vs no) | 4.53 (1.36-15.15) | .01 |
| Dyspnea (yes vs no) | 1.88 (1.18-3.01) | .01 |
| Unconsciousness (yes vs no) | 4.71 (1.39-15.98) | .01 |
| No. of comorbidities | 1.60 (1.27-2.00) | <.001 |
| Cancer history (yes vs no) | 4.07 (1.23-13.43) | .02 |
| Neutrophil to lymphocyte ratio | 1.06 (1.02-1.10) | .003 |
| Lactate dehydrogenase, U/L | 1.002 (1.001-1.004) | <.001 |
| Direct bilirubin, μmol/L | 1.15 (1.06-1.24) | .001 |
| Constant | 0.001 |
Abbreviation: COVID-19, coronavirus disease 2019.
Construction of the Risk Score and Web-Based Calculator
The COVID risk score was constructed based on the coefficients from the logistic model. We used the following formulas for the logistic model to calculate the probability and 95% confidence intervals9: probability = exp( ∑ β × X)/[1+ exp( ∑ β × X)], lower limit of 95% CI = exp[ ∑ Xn × βn− ∑ z × SE(β)]/{1+exp[ ∑ Xn × βn- ∑ z × SE(β)]}, upper limit of 95% CI = exp[ ∑ Xn × βn+ ∑ z × SE(β)]/{1+exp[ ∑ Xn × βn+ ∑ z × SE(β)]}.
An online calculator based on COVID-GRAM was developed to allow clinicians to enter the values of the 10 variables required for the risk score with automatic calculation of the likelihood (with 95% CIs) that a hospitalized patient with COVID-19 will develop critical illness (http://118.126.104.170/) (Figure)
Figure. The Online Web-Based Calculatora for Predicting Critical Illness Among Patients With COVID-19.
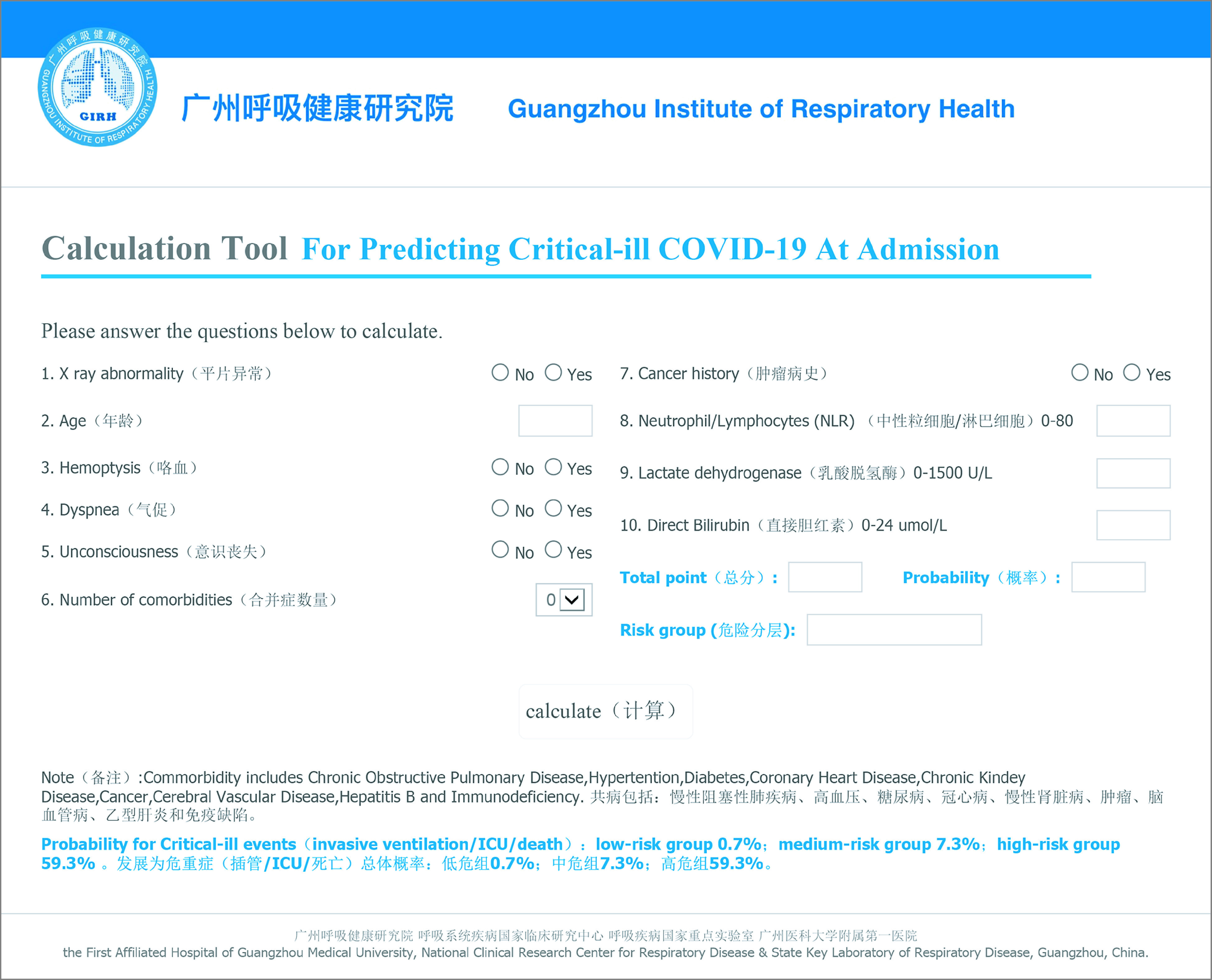
aGuangzhou Institute of Respiratory Health is responsible for the web-based calculator (http://118.126.104.170/).
The Performance of COVID Risk Score
By internal bootstrap validation, the mean AUC based on data from the development cohort was 0.88 (95% CI, 0.85-0.91) (eFigure 2 in the Supplement). The AUC of COVID risk score for patients in the epicenter at Hubei was 0.87 (95% CI, 0.83-0.91) and outside Hubei was 0.82 (95% CI, 0.73-0.90). The predictive value of COVID-GRAM was higher than the CURB-6 model, which had an AUC of 0.75 (95% CI, 0.70-0.80) for correct prediction of development of critical illness (P < .001).
Validation of COVID-GRAM
The validation cohort included 710 patients with a mean (SD) age of 48.2 (15.2) years, 382 (53.8%) were men and 172 (24.2%) had at least 1 coexisting condition. Critical illness eventually developed in 87 (12.3%) of these patients and 8 (1.1%) died. Variables used in COVID risk score for the validation cohort are shown in Table 4; eTable 1 in the Supplement. The accuracy of COVID risk score in the validation cohort was similar to that of the development cohort with an AUC in the validation cohort of 0.88 (95% CI, 0.84-0.93) (eFigures 3 and 4 and eTable 2 in the Supplement).
Table 4. Demographics and Clinical Characteristics of Patients in Validation Cohorts.
| Characteristic | No./No. (%) | ||
|---|---|---|---|
| Validation cohort, total | Critical illness | ||
| No | Yes | ||
| No. | 729 | 642 | 87 |
| Age, mean (SD) [range], y | 48.2 (15.2) [4-88] | 46.2 (14.3) | 63.1 (13.1) |
| Neutrophil-lymphocyte ratio | 5.8 (8.7) [0.08-109.2] | 4.3 (3.8) | 17.1 (20.0) |
| Lactate dehydrogenase, mean (SD) [range], U/L | 288.3 (151.2) [106-1390] | 264.1 (119.6) | 479.5 (223.8) |
| Direct bilirubin, mean (SD) [range], umol/L | 9.7 (9.6) [0-79] | 9.1 (9.5) | 14.1 (9.8) |
| Abnormal chest radiograph | 355/723 (49.1) | 277/639 (43.3) | 78/84 (92.9) |
| Hemoptysis | 8/724 (1.1) | 7/642 (1.0) | 1/82 (1.2) |
| Shortness of breath | 118/724 (16.3) | 70/642 (10.9) | 48/82 (58.5) |
| Unconsciousness | 6/724 (0.8) | 0/642 (0) | 6/82 (0.7) |
| Comorbidities | |||
| 0 | 550/722 (76.2) | 521/642 (81.2) | 29/80 (36.3) |
| 1 | 103/722 (14.3) | 83/642 (12.9) | 20/80 (25.0) |
| 2 | 56/722 (7.8) | 33/642 (5.1) | 23/80 (28.8) |
| 3 | 10/722 (1.4) | 3/642 (0.4) | 7/80 (8.8) |
| 4 | 3/722 (0.4) | 2/642 (0.3) | 1/80 (1.3) |
| Malignant disease | 9/723 (1.2) | 9/642 (1.4) | 0/81 (0) |
Discussion
In this study, we developed and validated a clinical risk score and a web-based risk calculator to predict the development of critical illness among hospitalized COVID-19 infected patients. The performance of this risk score was satisfactory with accuracy based on AUCs in both the development and validation cohorts of 0.88. The web-based calculator can be used by clinicians to estimate an individual hospitalized patient’s risk of developing critical illness. The 10 variables required for calculation of the risk of developing critical illness are generally readily available at hospital admission, and the web-based calculator is easy to use. If the patient’s estimated risk for critical illness is low, the clinician may choose to monitor, whereas high-risk estimates might support aggressive treatment or admission to the ICU. We deliberately did not categorize risk into low-, moderate-, and high-risk groups, as we believe that clinicians are better informed by calculating the risk estimate for each individual patient and making decisions based on local or regional conditions. For example, in areas with good access to clinical and supportive care, patient outcomes might be optimized by deciding to provide more aggressive care to moderate risk patients. In contrast, in areas with high case volume and/or limited resources, the decision might be to provide less aggressive care to moderate-risk patients to maximize availability of ICU beds and ventilators.
Chest radiography abnormality, age, hemoptysis, dyspnea, unconsciousness, number of comorbidities, cancer history, neutrophil-to-lymphocyte ratio, lactate dehydrogenase, and direct bilirubin were included in the COVID risk score. Previous studies have found several of these variables to be risk factors for severe illness related to COVID-19. Wu et al3 found that older age and more comorbidities were associated with a higher risk of developing ARDS in patients infected with COVID-19. A previous study10 from our group found that patients with COVID-19 with cancer had higher risk of severe events compared with patients without cancer (39% vs 18%). Zhou and colleagues1 found lower lymphocyte count, higher lactate dehydrogenase, and more imaging abnormalities in patients who died from COVID-19 disease.
Limitations
Potential limitations of this study include a modest sample size for constructing the risk score and a relatively small sample for validation. The data for score development and validation are entirely from China, which could potentially limit the generalizability of the risk score in other areas of the world. Additional validation studies of the COVID risk score from areas outside China should be completed.
Conclusions
In this study, we developed a risk score and web-based calculator to estimate the risk of developing critical illness among patients with COVID-19 based on 10 variables commonly measured on admission to the hospital. Estimating the risk of critical illness could help identify patients who are and are not likely to develop critical illness, thus supporting appropriate treatment and optimizing the use of medical resources.
eTable 1. Demographics and clinical characteristics of patients in validation cohorts.
eTable 2. External validation of the COVID-GRAM.
eFigure 1. Feature selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model. (A) LASSO coefficient profiles of the 72 baseline features. (B) Tuning parameter (λ) selection in the LASSO model used 10-fold cross-validation via minimum criteria.
eFigure 2. The area under the receiver-operator characteristic (ROC) curve (AUC) of predicting critical illness among patients with COVID-19.
eFigure 3. External validation of COVID-GRAM in an independent cohort.
eFigure 4. External validation of COVID-GRAM in four independent cohorts.
eAppendix. Acknowledgements.
Reference
- 1.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;(Feb):24. doi: 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 3.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;(Mar):13. doi: 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y, et al. ; China Medical Treatment Expert Group for Covid-19 . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;(Feb):28. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia. an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45-e67. doi: 10.1164/rccm.201908-1581ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368(24):2277-2285. doi: 10.1056/NEJMoa1305584 [DOI] [PubMed] [Google Scholar]
- 7.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364-1370. doi: 10.1200/JCO.2007.12.9791 [DOI] [PubMed] [Google Scholar]
- 8.Neill AM, Martin IR, Weir R, et al. Community acquired pneumonia: aetiology and usefulness of severity criteria on admission. Thorax. 1996;51(10):1010-1016. doi: 10.1136/thx.51.10.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Long JS. Confidence intervals for predicted outcomes in regression models for categorical outcomes. Stata Journal. 2005;5(4):537-559. doi: 10.1177/1536867X0500500405 [DOI] [Google Scholar]
- 10.Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335-337. doi: 10.1016/S1470-2045(20)30096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Demographics and clinical characteristics of patients in validation cohorts.
eTable 2. External validation of the COVID-GRAM.
eFigure 1. Feature selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model. (A) LASSO coefficient profiles of the 72 baseline features. (B) Tuning parameter (λ) selection in the LASSO model used 10-fold cross-validation via minimum criteria.
eFigure 2. The area under the receiver-operator characteristic (ROC) curve (AUC) of predicting critical illness among patients with COVID-19.
eFigure 3. External validation of COVID-GRAM in an independent cohort.
eFigure 4. External validation of COVID-GRAM in four independent cohorts.
eAppendix. Acknowledgements.


