Abstract
Antimicrobial resistance (AMR) is a significant global threat to both public health and the environment. The emergence and expansion of AMR is sustained by the enormous diversity and mobility of antimicrobial resistance genes (ARGs). Different mechanisms of horizontal gene transfer (HGT), including conjugation, transduction, and transformation, have facilitated the accumulation and dissemination of ARGs in Gram-negative and Gram-positive bacteria. This has resulted in the development of multidrug resistance in some bacteria. The most clinically significant ARGs are usually located on different mobile genetic elements (MGEs) that can move intracellularly (between the bacterial chromosome and plasmids) or intercellularly (within the same species or between different species or genera). Resistance plasmids play a central role both in HGT and as support elements for other MGEs, in which ARGs are assembled by transposition and recombination mechanisms. Considering the crucial role of MGEs in the acquisition and transmission of ARGs, a potential strategy to control AMR is to eliminate MGEs. This review discusses current progress on the development of chemical and biological approaches for the elimination of ARG carriers.
Keywords: resistance, plasmid curing, infection, antibiotics, CRISPR
Introduction
The discovery of antibiotics and their clinical use is one of the greatest achievements in medical history. However, the acquisition and dissemination of antimicrobial resistance genes (ARGs) is a severe global problem that emerged in the post-antibiotic era (Friedman et al., 2016; Thaden et al., 2017). The acute limitation of currently available therapeutic options against common infections is responsible for increased rates of morbidity and mortality, longer treatment duration, higher hospitalization costs, and distrust in the efficacy of modern medical practices (Bennett, 2008; Shokoohizadeh et al., 2013; Jiang et al., 2017; Sultan et al., 2018). The most common antimicrobial-resistant bacterial pathogens associated with nosocomial infections were initially gathered under the acronym “ESKAPE” (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which was subsequently proposed to be updated to “ESCAPE” (E. faecium, S. aureus, Clostridium difficile, A. baumannii, P. aeruginosa, and Enterobacteriaceae) (Grundmann et al., 2006; Santajit and Indrawattana, 2016; Penes et al., 2017; World Health Organization [WHO], 2017).
The phenomenon of antimicrobial resistance (AMR) is not new as ARGs have evolved over millions of years (Shlaes et al., 1997). However, AMR is amplified in the presence of the selective pressure exerted by antibiotics (Pelgrift and Friedman, 2013). Between 2000 and 2010, global antibiotic use increased by 36%, and in the case of carbapenems reached 45% (Van Boeckel et al., 2014). Inappropriate use of antibiotics in animals also contributes to rising AMR. The global consumption of antibiotics in animal feed was estimated to be 131,109 tons in 2013, and is expected to reach 200,235 tons in 2030 (Van Boeckel et al., 2017). Incomplete microbe elimination, facilitated by microbiostatic drugs that inhibit multiplication of microbes without killing them, favors the development of drug resistance. Furthermore, incorrect administration of microbicidal drugs in terms of dosing intervals and concentration also contributes to the occurrence of AMR. The dosing interval is essential for antibiotics with a short elimination half-life, such as beta-lactams, tetracyclines, clindamycin, and the majority of macrolides; while concentration is a critical parameter for antibiotics such as vancomycin, aminoglycosides, azalides, ketolides, and quinolones (Gao et al., 2011). Genetic resistance of clinically significant pathogens is amplified by the ability of bacteria to form biofilms on viable tissues or inert substrates; these biofilms exhibit high phenotypic resistance or tolerance to high doses of antimicrobial agents (Giedraitiene et al., 2011). As a result of the selective pressure exerted by antibiotics, bacterial genomes are reshaping, and bacteria adapt and survive in the presence of antibiotics (van Elsas and Bailey, 2002). There are multiple mechanisms of adaptation of resistant bacteria to antibiotics and elucidating these mechanisms will enable the development of effective novel therapies to tackle the increasing threat of resistance.
An important strategy for combating AMR is to diminish the mobilization and persistence of ARGs in bacterial populations. This review highlights current progress in the development of chemical and biological approaches for the elimination of resistance plasmids. Such plasmids play a central role both in horizontal gene transfer (HGT) and as support for other mobile genetic elements (MGEs), in which ARGs are assembled through transposition and recombination mechanisms; the resulting MGEs can then move between chromosomes and plasmids or between plasmids. We will first describe the main MGEs involved in the global dissemination of antibiotic resistance, and then discuss current progress on the development of novel antimicrobial strategies aimed at elimination of MGEs, with a focus on resistance plasmids.
Role of MGEs in the Acquisition and Transmission of AMR
Resistance to antimicrobials can be acquired through spontaneous mutations in chromosomal genes or by HGT of ARGs. The bacterial genome includes the genomic backbone or core genome, to which a variety of MGEs, termed the accessory genome, is added, and together this comprises the bacterial pan-genome (Guimaraes et al., 2015). The term resistome refers to the part of the pan-genome that contains ARGs, both in commensal and pathogenic bacteria (D’Costa et al., 2006; Landecker, 2016). Many ARGs can move between the bacterial chromosome and plasmids, within the same species or between different species or even genera, through different mobilization mechanisms (conjugation, transduction, and transformation) (Figure 1). HGT is the primary driver of multidrug resistance (MDR) in both Gram-negative and Gram-positive bacteria. MGEs (except for gene cassettes and miniature inverted-repeat transposable elements; MITEs) are DNA fragments encoding enzymes and other proteins that mediate intracellular or intercellular mobility. Intracellular mobility occurs within the same cell, from a chromosome to a plasmid or between plasmids. This type of mobility can be achieved by non-conjugative transposons, gene cassettes, and insertion sequence common region (ISCR) elements. These MGEs are mobilized by recombination but can involve replication. Intercellular mobility is achieved by MGE elements that are capable of self-replication and conjugative transfer, such as plasmids and conjugative transposons (Frost et al., 2005; Bennett, 2008; Ilangovan et al., 2015; Partridge et al., 2018). The versatility of MGEs justified the replacement of the constant genome paradigm with that of the fluid genome (Shapiro, 1985; Pinilla-Redondo et al., 2018).
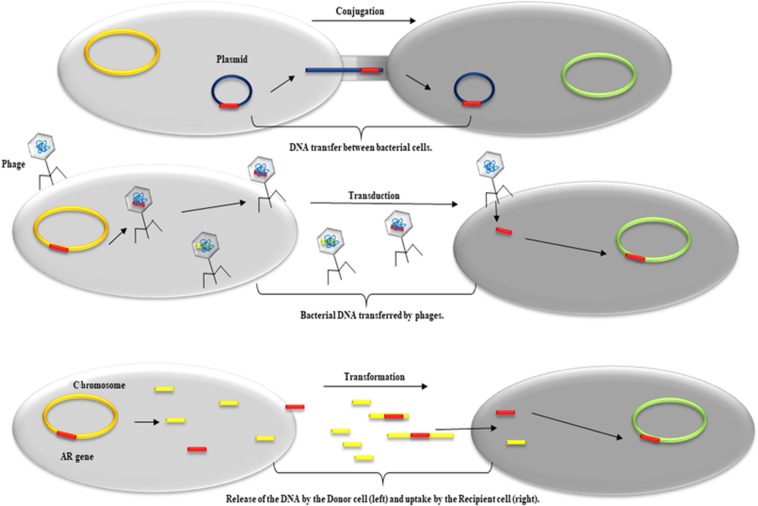
FIGURE 1.
Schematic representation of the predominant HTG mechanisms involved in the acquisition and dissemination of genetic material such as ARG. From top to bottom: Conjugation, DNA transfer between a donor cell (left) and a recipient cell (right) mediated by plasmids; Transduction, transfer of bacterial DNA between a donor cell (left) and a recipient cell (right) mediated by phages; Transformation, release of DNA by a donor cell (left) and uptake by a recipient cell (right).
The majority of clinically significant ARGs are located on MGEs. To effectively fight AMR, we need to unravel the role of MGEs in the dissemination of antibiotic resistance among clinically important pathogens.
Plasmids
Plasmids have a vital role in the accumulation and transfer of ARGs, mainly in Gram-negative bacteria, and are involved in the acquisition of resistance to most antibiotic classes, including β-lactams, aminoglycosides, tetracyclines, chloramphenicol, sulfonamides, trimethoprim, macrolides, polymyxins, and quinolones (Carattoli, 2013; Shintani et al., 2015). Plasmids, either circular or linear, are stable replicons with a complex replication apparatus (Shintani et al., 2015). Generally, plasmids are physically distinct from the primary bacterial chromosome and replicate independently; however, most of the components required for replication are provided by the host (Garcillan-Barcia and de la Cruz, 2008; Guglielmini et al., 2014). Plasmids conferring MDR are usually conjugative, capable of initiating not only their own transfer but also that of other plasmids, and possess mechanisms to control their copy-number in the cell and/or replication ability (Frost et al., 2005; Nordstrom, 2006). Plasmids guarantee transmission through different mechanisms like active partitioning systems, random segregation, or post-segregational killing (Million-Weaver and Camps, 2014). Besides conjugative plasmids, another category of plasmids are mobilizable plasmids, which are smaller in size and not self-transmissible, but they can transfer DNA to a particular host in the presence of conjugative plasmids; this transfer occurs both vertically and by HGT (Bennett, 2008).
The first classification of plasmids was based on incompatibility (Inc) groups (the mechanism that prevents the existence of plasmids with the same replication mechanism within the same bacterial cell); specific incompatibility groups were described in Enterobacteriaceae, Pseudomonas spp., and Gram-positive staphylococci (Frost et al., 2005). In MDR strains of P. aeruginosa, a series of plasmids (pS04 90, pBM41, p14057 A, and p14057 B) encoding carbapenemase resistance have been highlighted (Liu et al., 2018; Shi et al., 2018; van der Zee et al., 2018). Strains of A. baumannii show plasmid-encoded resistance to carbapenems (Cameranesi et al., 2018; Leungtongkam et al., 2018; Silva et al., 2018), aminoglycosides (armA) (Upadhyay et al., 2018), colistin (Jaidane et al., 2018), sulfonamides, or streptomycin (Hamidian et al., 2016). In addition, plasmids conferring resistance to various classes of antibiotics have been found in Staphylococcus spp. (Mugnier et al., 2009; Ruiz-Martinez et al., 2011; Hamidian et al., 2016; Holmes et al., 2016; Liu et al., 2016, 2018; Becker et al., 2018; Cameranesi et al., 2018; Fessler et al., 2018; Jaidane et al., 2018; Leungtongkam et al., 2018; Shi et al., 2018; Silva et al., 2018; Upadhyay et al., 2018; van der Zee et al., 2018). Resistance plasmids exhibit a high degree of plasticity, which is translated into an increased frequency of insertions, deletions, and changes in DNA (Kado, 2014). Plasmids may also harbor ARGs encoding efflux pumps that confer an MDR phenotype such as quinolone resistance (Jacoby et al., 2014).
Further to their direct role in HGT, plasmids can also contribute to the acquisition and dissemination of ARGs to other MGEs in which ARGs are assembled via transposition and recombination mechanisms (Stanisich, 1988; Bennett, 2004, 2008). Some of the MGE frequently involved in the acquisition of clinically relevant ARGs are briefly described below and summarized in Figure 2.
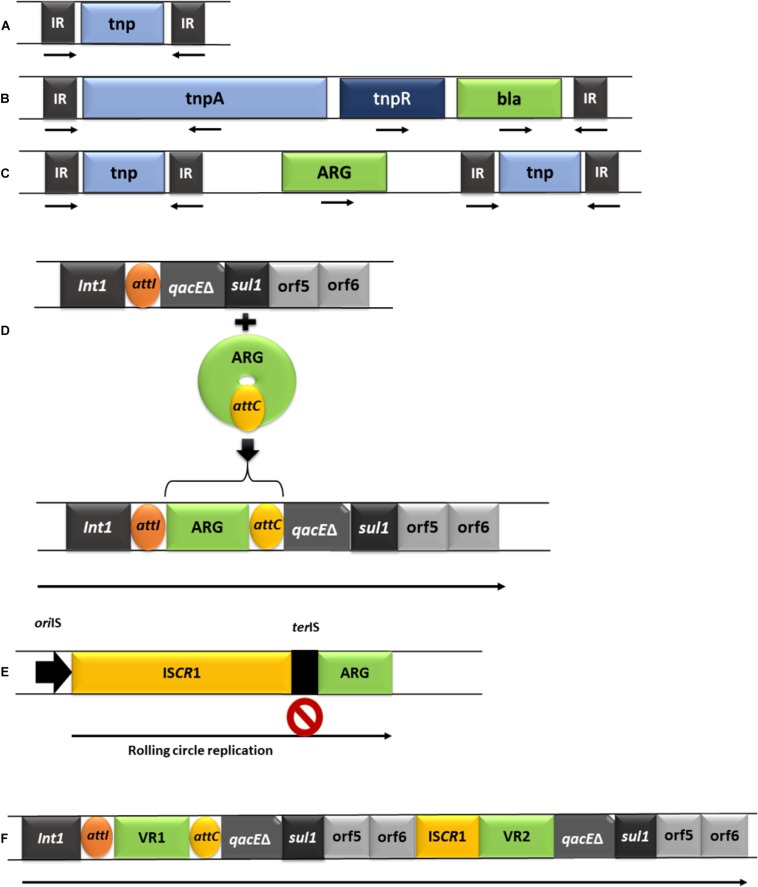
FIGURE 2.
Schematic representation of the predominant MGEs involved in acquisition and dissemination of ARGs. (A), IS element (IR: inverted repeats; tnp: transposase gene). (B), Tn3 complex transposon (tnpB: resolvase gene; ARG-antibiotic resistance gene). (C), composite transposon. (D), class I integron and the acquisition of a gene cassette (Int1: integrase gene; att1: recombination site of the integron; qacEδ: truncated segment belonging to a gene that encodes resistance to quaternary ammonium compounds; sul1: sulfonamide resistance gene; orf5/orf6: open reading frames, attC: recombination site of the gene cassette). (E), the mechanism of acquiring adjacent DNA by ISCR elements (oriIS: origin of replication; terIS: end of replication; a second stop sign is located after the ARG, allowing transposition of the entire segment by recombination). (F), complex class 1 integrons (Int1: integrase gene, followed by the attI site; VR1/VR2: variable regions e.g., ARGs, followed by the attC site).
Insertion Sequences
Insertion sequences (IS) are the smallest (0.7–2.5 Kb) and simplest transposable elements found in bacteria (Mahillon and Chandler, 1998; Aminov, 2011). These elements are usually flanked by short, mostly inverted repeats, which sometimes generate direct target duplications (DR) when they are integrated into the target DNA (Siguier et al., 2015). IS differ from transposons by the absence of cargo or passenger genes, which are responsible for functions other than mobilization. Currently, there are more than 4500 IS listed in dedicated databases like ISFinder (Siguier et al., 2015; Vandecraen et al., 2017). IS are involved in AMR through their ability to transfer ARGs, but also by their ability to modulate the expression of ARGs; this occurs following integration of IS within the ARGs, or by the IS providing an active promoter for ARGs (Siguier et al., 2006; Partridge et al., 2018). For example, IS can increase expression of efflux pumps (Olliver et al., 2005; Siguier et al., 2015). The role of IS in antibiotic resistance has been highlighted by numerous studies, particularly those related to resistance to colistin and carbapenem. The most common mechanism for the development of colistin resistance is inactivation of the gene mgrB in K. pneumoniae, following the transposition of different types of IS, such as IS903, ISKpn26, IS10R, and IS5 (Cannatelli et al., 2014; Berglund et al., 2018). In colistin-resistant strains of Klebsiella sp., alteration of mgrB and phoP gene sequences by different IS, such as ISKpn14, ISKpn28, IS903, IS5, and IS3, can sometimes induce a pandrug-resistance phenotype (Giordano et al., 2018; Uz Zaman et al., 2018). IS also play a vital role in carbapenem resistance through a mechanism similar to that of colistin resistance but involving the inactivation of oprD and omp genes (Lev et al., 2017; Bocharova et al., 2019). The oprD gene is inactivated by the insertion of ISPpu-21 (Shariati et al., 2018). In addition to the IS themselves, there are other similar transposable elements (TEs) that harbor transposase genes (autonomous) or depend on host cell elements (non-autonomous) (Siguier et al., 2015). When IS elements are carrying passenger genes, they are termed IS transporters (tISs) (Siguier et al., 2006). In contrast to complex transposons that exist only as a single copy in a specific replicon, IS can be present as multiple copies, thus contributing to the accumulation of ARGs (Rankin et al., 2011).
Resistance Transposons
Transposons (Tn) are a category of MGEs that carry ARGs. Many Tn have the ability to jump from/to different locations in the genome, and are capable of mediating the mobility of both intramolecular and intermolecular ARG (Bennett, 2004, 2008; Babakhani and Oloomi, 2018). Bacterial Tn can be divided into two types, composite (two IS elements flanking a central gene) and complex (containing the tnpA gene encoding transposase, the tnpR gene encoding resolvase, as well as one or more cargo genes) (Genilloud et al., 1988; Bennett, 2008; Partridge, 2011). MITEs and palindrome-associated transposable elements (PATEs) are included in the category of non-autonomous derivatives (Siguier et al., 2015). The predominant ARG-containing Tn whose transmission is a challenge when treating infections are Tn5 (encoding resistance to neomycin and kanamycin in A. baumannii and P. aeruginosa), Tn10 (encoding tetracycline resistance), Tn9, Tn903, Tn1525, and Tn2350 (Genilloud et al., 1988; Partridge, 2011).
Integrons
Integrons are MGEs that have the ability to accumulate gene cassettes, including ARGs, and to disseminate them through other MGEs. Sedentary integrons are DNA elements found in the chromosomes of many species and were initially discovered due to their association with AMR (Mazel, 2006; Partridge et al., 2009). In contrast to sedentary integrons, mobile resistance integrons are often found in plasmids (Ponce-Rivas et al., 2012). The role of these elements in the acquisition and dissemination of ARGs is crucial, especially in Gram-negative bacteria (Ponce-Rivas et al., 2012), but they are also present in Gram-positive bacteria (Nandi et al., 2004). Integrons contain the gene encoding integrase (IntI), an enzyme that allows the incorporation of circular DNA segments by site-specific recombination (Cambray et al., 2010). They also harbor a specific integration site, at which one or more gene cassettes can be inserted by the integrase (Recchia and Hall, 1995; Bennett, 2008; Partridge et al., 2009). Gene cassettes are usually small DNA fragments of 500–1000 base pairs, which can be mobilized by integrase. Generally, the gene cassettes comprise a single open reading frame (ORF) followed by a short recombination site termed attC (formerly “59 bases element”). Since the majority of these cassettes are promoterless, expression of their genes depends on the integron promoter (Bennett, 1999, 2008). Gene cassettes contain ARGs encoding resistance to different antibiotic classes (Recchia and Hall, 1995; Nordmann and Poirel, 2002), as well as antiseptics and disinfectants (Recchia and Hall, 1995; Bennett, 2008). Integrons are divided into several classes (class 1, class 2, and class 3) depending on the amino acid sequence of the IntI enzyme. Class 1 integrons, which are typically associated with plasmids, are most commonly encountered in clinical isolates from hospitals and elderly care facilities, but have also been found in food production chain isolates (e.g., cattle farm isolates) (Belaynehe et al., 2018; Faghri et al., 2018; Rajpara et al., 2018). ISCRs are transposable elements that are a similar size to IS elements, are often associated with class 1 integrons, and are capable of mobilizing adjacent DNA via a rolling-circle mechanism (Bennett, 2008). When ISCR elements are associated with class 1 integrons, they form complex class 1 integrons (Bennett, 2008; Toleman and Walsh, 2008, 2011).
Genomic Islands
In addition to classical MGEs, such as conjugative plasmids or resistance transposons, an additional category of MGEs is a series of genomic islands that are capable of mediating their own excision, called integrative and conjugative elements (ICE) (Burrus et al., 2002; Burrus and Waldor, 2004; Dobrindt et al., 2004; Juhas et al., 2009; Wozniak and Waldor, 2010). The concept of pathogenicity islands (PAIs) was first described in 1980 by Hacker et al. (1983) who analyzed the virulence mechanisms of strains of E. coli isolated from urine cultures and observed the presence of unstable chromosomal regions bearing different virulence characteristics. Studies on multiple genomic islands have identified several common and essential features of these chromosomal regions: they are DNA segments with a size of 10–200 kb; they insert within tRNA genes; they contain directly repeated recognition sequences; and they contain cryptic genes encoding factors involved in integration, insertion, or transfer (Hacker et al., 1990).
Integrative and Conjugative Elements
Integrative and conjugative elements (ICE) were first described in 1946 by Lederberg and Tatum (1946), and are responsible for HGT of most resistance and virulence factors (Llosa et al., 2002; Burrus and Waldor, 2004; Fernandez-Lopez et al., 2006; de la Cruz et al., 2010; Smillie et al., 2010). ICE are 18–600 kbp in size and share several common characteristics with genomic islands, including insertion at a specific site, association with phage integrase genes, and being flanked by inverted repeats (Toleman and Walsh, 2011). Excision and integration of ICE are accomplished through a recombinase, often termed an integrase. The integrases associated with ICE are tyrosine or serine recombinases, and are homologous to the integrases found in temperate phages (Wozniak and Waldor, 2010). The insertion site for ICE in the bacterial chromosome is attB and is usually located in the gene encoding tRNA, hence the ICE attachment site is termed att (Grindley et al., 2006). Some ICE have low specificity for the att site, and thus may have an affinity for other sites (Bedzyk et al., 1992; Roberts and Mullany, 2009). Similar to conjugative plasmids, the excision and transfer of ICE are mediated by a type IV secretion system, but in contrast to the conjugative plasmids, which are capable of autonomous replication, ICE integrate into the chromosome and replicate with it (Burrus, 2017). However, some ICE are capable of autonomous plasmid-like replication (Johnson and Grossman, 2015). When ICE are mobilizing bacterial DNA, such as genomic islands, they are termed integrative mobilization elements (IME) (Gonzalez-Candelas and Francino, 2012). In terms of ICE conjugation, the transfer mechanism is similar to that encountered in plasmids. In the case of plasmids, the relaxase enzyme binds to the DNA and introduces a break in oriT to initiate rolling-circle replication. Relaxase remains bound to the single-stranded DNA and forms a complex with a specific coupling protein that allows the translocation of DNA into the recipient cell (Lanka and Wilkins, 1995). ICE mediate the acquisition of genes conferring selective advantages such as resistance to antibiotics or heavy metals, degradation of some compounds, increased bacterial fitness, ability to achieve symbiosis, use of alternative carbon sources, expression of virulence factors such as type III and IV secretion systems, which play an essential role in regulating contact with host cells, disruption of signal transduction, or promotion of apoptosis (Roberts and Smith, 1980; Shoemaker et al., 1980; Mays et al., 1982; Magot, 1983; Hochhut et al., 1997; Ravatn et al., 1998; Nishi et al., 2000; Dobrindt et al., 2004; Schmidt and Hensel, 2004).
Integrative and conjugative elements play a vital role in the acquisition and intercellular transmission of ARGs. Through their own integration and mobilization apparatus, these elements have the ability to mobilize adjacent sequences, including genomic islands or composite transposons carrying ARGs (Delavat et al., 2017). Examples include Tn10 found in ICEHpaT3T1 from Haemophilus parainfluenzae and ICEHin1056 from H. influenzae, containing tetracycline and chloramphenicol resistance genes (Juhas et al., 2007); R391, a plasmid of the SXT ICE family that carries kanamycin resistance genes (Pembroke et al., 2002), and ICEPmiJpn1 described in Proteus mirabilis and encoding resistance to broad-spectrum beta-lactamases (Harada et al., 2010; Mata et al., 2011). There are also a number of ICE encountered in H. influenzae (ICEHin1056, ICEHin299, ICEHin2866, ICEHpa8f, ICEHin028, ICEHinB) (Juhas et al., 2007). ICEEc2, identified in E. coli, contains Tn7, which can be mobilized independently, and class 2 integrons. Tn7 carries dfrA1, sat2, and aadA1, which are responsible for resistance to trimethoprim, streptothricin, and streptomycin/spectinomycin, respectively (Roche et al., 2010). Another large ICE family is ICETn4371 found in Beta- and Gamma-Proteobacteria. Members of this ICE family, such as ICETn43716061 found in P. aeruginosa, display transfer mechanisms similar to IncP plasmids and carry different ARGs (Castanheira et al., 2007). The Tn21 transposon subfamily, containing pKLC102/PAPI-1 and PAGI-2/PAGI-3 (P. aeruginosa-pathogenicity island-type ICE) carbapenem resistance genes are integrated into tRNALys and tRNAGly (Klockgether et al., 2007). The Tn4371 family in P. aeruginosa (e.g., ICETn43716061) also carries carbapenem ARGs (Fonseca et al., 2015). Tn916-type elements that encode tetracycline or minocycline resistance, via the tet(M) gene, may embed additional ARGs for other antibiotics such as macrolides, lincosamides, and streptogramins (MLS) and kanamycin/neomycin in the case of Tn1545 (Cochetti et al., 2008). Roberts et al. (2008) observed that the majority of transposable elements, including composite transposons, mobilizable transposons, ICE, and genomic islands, possess similar transposition mechanisms (serine or tyrosine recombinases). Consequently, it was suggested that all these elements capable of integration and conjugation should be called conjugative transposons, even though most of them integrate into a single specific site (Roberts et al., 2008). The term “conjugative transposon” was first used by Franke and Clewell (1981) when characterizing the Tn916 element from E. faecalis (Franke and Clewell, 1981; Brochet et al., 2009).
Targeting MGEs to Combat Antibiotic Resistance
The ability of bacteria to adapt to all currently available antibiotics has led to an acute need for new, more effective antibiotics or the development of alternative therapeutic strategies (Seal et al., 2018). MGEs, especially those containing resistance plasmids, transposons, and integrons, play a crucial role in the accumulation and dissemination of ARGs in both the clinical and environmental sectors. Consequently, there is a strong argument for considering that one potential strategy to control AMR is through the elimination of these MGEs. In the field of medicine, the concept of “curing” refers to various clinical techniques applied to repair a defective system (Kennedy, 1981; Dow, 1990). In terms of AMR, “curing” is predominantly used to describe the process of removing ARGs from bacterial populations, and compounds used for this purpose are called “curing agents.” Considering that most ARGs and virulence factors are located on plasmids, the term “curing” has been associated with the removal of plasmids since 1971 (Bouanchaud and Chabbert, 1971). Over the past half-century, several studies have focused on testing antibacterial compounds, such as detergents, biocides, intercalary agents, and nanoparticles (Table 1), bacteriophage- and microbiota-based therapies, or the CRISPR system for curing resistance plasmids (Buckner et al., 2018).
TABLE 1.
Plasmid curing compounds.
| Curing agent | Species of interest | Plasmid target | References |
| SDS | S. aureus | Penicillinase plasmid | Sonstein and Baldwin, 1972 |
| E. coli | pBR322;pBR325 | Keyhani et al., 2006 | |
| E. coli | 212587, 212973, 208366, and 207940 isolates carrying plasmids | Zaman et al., 2010 | |
| P. aeruginosa | pBC15 plasmid | Raja and Selvam, 2009 | |
| Ethidium bromide | Streptomycetes | pIJ303 and pIJ61 plasmids | Crameri et al., 1986 |
| E. coli | 212587, 212973, 208366, and 207940 isolates carrying plasmids | Zaman et al., 2010 | |
| L. acidophilus | 20.3 bp chloramphenicol resistant plasmid | Karthikeyan and Santosh, 2010 | |
| E. aerogenes | pKpQIL carbapenem resistant plasmid | Pulcrano et al., 2016 | |
| Acridin-orange Acriflavine | Salmonella | Chloramphenicol resistant plasmid | Adetosoye and Rotilu, 1985 |
| Shigella | |||
| Lactobacillus | pDR101 | Chassy et al., 1978 | |
| O. oeni | pRS1, pRS2, and pRS3 | Mesas et al., 2004 | |
| E. coli | 212587, 212973, 208366, and 207940 isolates carrying plasmids | Zaman et al., 2010 | |
| S. aureus | Beta-lactam resistance plasmid | Ojo et al., 2014 | |
| Triclosan (irgasan) Fusidic acid | S. aureus | Mupirocin resistance 48 Md plasmid | Irish et al., 1998 |
| E. coli | pMIB4 plasmid | Riber et al., 2016 | |
| Nitric oxide nanoparticles | P. aeruginosa | Plasmid carrying antibiotic-resistance genes | Jones et al., 2010 |
| S. aureus | |||
| E. coli | |||
| Trichophyton mentagrophytes | |||
| T. rubrum | |||
| A. baumannii | |||
| Chitosan | E. coli | Plasmid carrying antibiotic-resistance genes | Bavya et al., 2019 |
| S. aureus | |||
| Silver nanoparticles | S. aureus | methicillin-resistant plasmid | Huang et al., 2011 |
Chemical Strategies for Removing MGEs
Chemical agents used for the elimination of resistance elements in bacteria act through several mechanisms, including replication blockage, DNA breaks, or inhibition of conjugation (Tables 1, 2). The effectiveness of the agent varies depending on the bacterial strain, presence of plasmids, and growth conditions.
TABLE 2.
Conjugation inhibitors and their targets for the elimination of antibiotic resistance.
| Conjugative inhibitor | Species of interest | Target | Results | References |
| Intercalating agents | Salmonella typhimurium | Plasmid resistance determinants | Inhibition of plasmid DNA replicons | Hahn and Ciak, 1976; Adetosoye and Rotilu, 1985 |
| Nitrofuran derivatives | Enterobacteriaceae | Plasmid DNA replication | DNA replication blocking | Michel-Briand and Laporte, 1985 |
| Unsaturated fatty acids | E. coli | R388 and the F-plasmid derivative pOX38 | Plasmid conjugation inhibition | Fernandez-Lopez et al., 2005 |
| Bisphosphonates | E. coli | Relaxase enzyme | Disrupting conjugative DNA transfer | Lujan et al., 2007 |
| Antibodies | E. coli | Relaxase activity | Relaxase blocking, inhibition of conjugative transfer | Garcillan-Barcia et al., 2007 |
| Chemical inhibitors of transposons recombination | E. coli | Tn3 recombinase | Tn3 transposition blocking | Fennewald and Capobianco, 1984 |
Detergents have been used to remove resistance plasmids since 1972. Sodium dodecyl sulfate (SDS) has shown excellent efficiency in removing resistance plasmids in both Gram-positive (e.g., the penicillin resistance plasmid from S. aureus) and Gram-negative (e.g., E. coli and P. aeruginosa) bacteria (Sonstein and Baldwin, 1972; Keyhani et al., 2006; Raja and Selvam, 2009; Zaman et al., 2010). However, high concentrations of SDS are required, which result in gastrointestinal side effects and thus prohibit the use of SDS in humans and animals (Buckner et al., 2018). Another class of compounds used to remove resistance plasmids are the intercalating agents, such as ethidium bromide, acridine-orange, and acriflavine. Elimination of resistance plasmids by ethidium bromide has been demonstrated in Gram-positive (Lactobacillus acidophilus) and Gram-negative (E. coli, Enterobacter aerogenes) bacteria, as well as in actinomycetes (Streptomycetes) (Crameri et al., 1986; Karthikeyan and Santosh, 2010; Zaman et al., 2010; Pulcrano et al., 2016). Acridine-orange and acriflavine have successfully cured resistance plasmids in E. coli (Zaman et al., 2010), Salmonella spp. and Shigella spp. (Adetosoye and Rotilu, 1985), Lactobacillus spp. (Chassy et al., 1978), Oenococcus oeni (Mesas et al., 2004), and S. aureus (Ojo et al., 2014). However, the use of intercalating agents is associated with the risk of mutagenic effects. Furthermore, intercalating agents appear to be inefficient in eliminating large plasmids, such as those found in Rhizobium spp. and Agrobacterium spp. (Rosenberg et al., 1981). Biocides such as triclosan (irgasan) or fusidic acid have been used since 1998 for the successful removal of resistance plasmids in Gram-negative (E. coli) and Gram-positive (methicillin-resistant S. aureus) bacteria (Irish et al., 1998; Riber et al., 2016).
Recently, nanoparticles have been proposed as potential tools to combat bacterial resistance (Jones et al., 2010; Bavya et al., 2019). An advantage of using nanoparticles is that they simultaneously target multiple structures, decreasing the risk of selecting/acquiring resistance to them (Zhang et al., 2010). Nanoparticles exert their antibacterial effects through multiple mechanisms, including destruction of the bacterial membrane with elimination of cytoplasmic components, inactivation of DNA or protein binding, and release of reactive oxygen species. Blocking the function of cellular components leads to oxidative stress, electrolyte imbalance, enzyme inhibition, and finally, cell death (Huang et al., 2011; Knetsch and Koole, 2011; Wang et al., 2017). Due to their effects on DNA integrity (double-strand breaks, deaminations, alkylating agent formation, and inhibition of DNA repair enzymes) (Schairer et al., 2012; Nejdl et al., 2017), different nanoparticles might also be regarded as MGE curing agents. Another advantage of using nanoparticles is that their antibacterial action can be maintained for an extended period of time with no loss in stability (Cheow and Hadinoto, 2014). Platinum and copper nanoparticles are instrumental in the elimination of resistance plasmids as they interact with the supercoiled plasmid DNA or with topoisomerases involved in replication, transcription, and recombination processes, ultimately leading to elimination of the plasmids (Lakshmi et al., 1988; Lakshmi and Polasa, 1991; Antonoglou et al., 2019). Copper nanoparticles have also been used in plasmid DNA degradation experiments as well as for blocking plasmid conjugation (Chatterjee et al., 2014; Klumper et al., 2017). Despite the potential for metal nanoparticles to be used as weapons against AMR, bacteria are capable of developing resistance to the nanoparticles themselves, probably facilitated by the global use of metals in fields such as agriculture, animal feed supplements, and disinfectant production (Gupta and Silver, 1998; Rai et al., 2012). Bacterial resistance to silver, copper, and zinc nanoparticles has been highlighted in both Gram-negative and Gram-positive bacteria isolated from inert surfaces, soil, or the intestinal contents of animals fed with zinc and copper supplements (Cason et al., 1966; Santo et al., 2010; Altimira et al., 2012; Yazdankhah et al., 2014; Poole, 2017; Xu et al., 2017). Genes conferring resistance to metals are usually located on plasmids, posing a significant risk of very rapid dissemination through HGT (Dupont et al., 2011). Furthermore, the co-existence of antibiotic resistance and metal resistance genes within the same MGE is a possible mechanism for selecting antibiotic resistance (Poole, 2017). Another type of nanoparticle, the organic nanoparticles such as chitosan nanoparticles, demonstrate resistance curing activity by affecting the integrity of plasmid DNA and the conjugation capacity (Bozkir and Saka, 2004).
Conjugation is involved in the dissemination of plasmids and other MGEs such as conjugative transposons and ICE. Since most MGEs use the same proteins for their transfer, HGT could be blocked by conjugation inhibitors (COINs) (de la Cruz et al., 2010; Baquero et al., 2011; Table 1). Many experiments have demonstrated that intercalating agents, heterocyclic compounds, acridine dyes, quinolones, and unsaturated fatty acids such as linoleic and linolenic acid can act as COINs (Hahn and Ciak, 1976; Adetosoye and Rotilu, 1985; Michel-Briand and Laporte, 1985; Molnar et al., 1992; Fernandez-Lopez et al., 2005). A possible target of COINs is relaxase, the most critical enzyme in the conjugation process as it cuts the plasmid at the oriT origin. Inhibiting conjugation by targeting relaxase has been demonstrated for bisphosphonates (etidronate, clodronate) and for specifically designed antibodies (Garcillan-Barcia et al., 2007; Lujan et al., 2007). Another potential target of COINs is to limit or block the site-specific recombinase enzymes that have a central role in the transposition process (Fennewald and Capobianco, 1984). However, some COINs are unable to translocate the bacterial cell membrane, hence research has been directed toward the discovery of new classes of permeable compounds (Wigle et al., 2009).
Chemical agents have been successfully used to remove resistance plasmids, but use of these compounds to limit antibiotic resistance in humans is problematic. As stated previously, high concentrations of SDS are required to remove resistance plasmids and this results in gastrointestinal side effects such as colitis; consequently, SDS use is banned in humans and animals (Raja and Selvam, 2009). Studies have demonstrated that intercalating agents are effective in eliminating resistance plasmids (Chassy et al., 1978; Mesas et al., 2004; Ojo et al., 2014; Pulcrano et al., 2016), but the risk of mutagenic effects must be considered. Nanoparticles are another weapon against bacterial resistance, but the main impediment to their use in humans is the lack of information regarding their safety and how they affect the biological integrity of organisms, particularly in terms of producing toxicological, cytotoxic, and genotoxic effects (Li T. et al., 2018). Nanoparticles are predominantly used in doses below the threshold concentrations; thus, they are not considered harmful to the body. However, bioaccumulation of nanoparticles in the body following long-term exposure is well known (Hasan et al., 2018). Therefore, further research on long-term nanoparticle toxicity and carcinogenesis is needed. Quinolones inhibit bacterial DNA replication by interfering with DNA-gyrase activity, and numerous studies have highlighted the plasmid-curing effect of quinolones in E. coli in vitro and in vivo assays (Weisser and Wiedemann, 1985; Michel-Briand et al., 1986; Courtright et al., 1988; Fu et al., 1988; Selan et al., 1988). Despite these results, the use of quinolones to eliminate plasmids containing ARGs may lead to a fitness advantage in plasmid-containing cells and would therefore select for plasmid maintenance. Phenothiazines, such as chlorpromazine, also have plasmid-curing activity in E. coli (Molnar et al., 1976) and methicillin-resistant S. aureus (Costa et al., 2010). Although the role of these chemical agents has been demonstrated in vitro, further studies are needed to clarify the efficacy of these compounds in vivo. In the case of chlorpromazine, the concentration required in the intestine to remove resistance plasmids is considerable (Maier et al., 2018). For this reason, it is necessary to develop strategies that allow targeted delivery of these chemical agents and avoid oral administration in order to increase their efficiency and reduce the risk of toxicity.
Biological Strategies for Removing MGEs
Bacteriophages
Bacteriophages are viral parasites capable of infecting bacteria by recognizing surface receptors, injecting their genetic material into the host, and replicating using the host cellular machinery. Phages exert ecological and genetic effects on bacteria at the population level, and these effects can impact plasmid stability (Thompson, 1994). This is due to epistatic interactions between the cost of chromosomal phage-resistant mutations and the cost of maintaining plasmids (Buckling and Rainey, 2002). Although phage-mediated transfer of ARGs between bacteria has been demonstrated for numerous bacterial species, the transduction occurs at a low rate (between 10–6 and 10–9 transductants/pfu). One exception is methicillin-resistant S. aureus that contains a category of MGEs called phage-inducible chromosomal islands (PICIs), which are associated with the highest transduction frequency (10–1 transductants/pfu) (Calero-Caceres and Muniesa, 2016; Torres-Barcelo, 2018). Phages may enhance the persistence of ARGs as an adaptation strategy to restrictive environmental conditions, e.g., wastewater aggressively treated using UV, temperature, or pH. However, genetically modified phages could be used to increase antibiotic susceptibility of resistant strains. The alarming increase in resistance has also led to the revival of phage therapy in order to sensitize resistant bacteria by eliminating resistance and virulence factors (Lin et al., 2017). Jalasvuori et al. (2011) showed that the PRD1 phage determined the loss of RP4 and RN3 resistance plasmids from strains of E. coli and Salmonella spp. and inhibited the conjugation ability of the remaining resistant bacteria. Another study demonstrated that the M13KE filamentous phage could block plasmid conjugation by interacting with the conjugative F pilus in E. coli. Furthermore, addition of the M13 phage g3p minor protein results in complete inhibition of conjugation, suggesting this protein has an essential role in the process (Lin et al., 2011). Harrison et al. (2015) eliminated the pQBR103 megaplasmid in P. fluorescens using the SBW252 lytic phage. Recently, Chan et al. (2016) revealed that the OMKO1 phage isolated from P. aeruginosa could sensitize antibiotic-resistant strains to erythromycin, ceftazidime, tetracycline, and ciprofloxacin. Together, these studies demonstrate the possibility of using phages to reduce the prevalence of resistance plasmids in bacterial populations as well as to block plasmid conjugation. In addition, phages can be successfully used to increase the sensitivity of bacterial strains to antibiotics.
Seemingly successful experimental trials using phages to treat pediatric dysentery (Summers, 2004), cholera and skin infections (Abedon et al., 2011), and bubonic plague (Summers, 2004) sparked interest in phage therapy both in Europe and the United States. However, attempts to repeat these trials and achieve positive results failed; this was due to an incomplete understanding of phage biology, and because of the large-scale development of a wide range of antibiotics that could be used to treat these infections. Experimental data obtained from the use of phage therapy in animals, as well as data from observational studies conducted in humans, were not followed by clinical studies to confirm the therapeutic value of phages. However, in recent years, the abusive use of broad-spectrum antibiotics (Ventola, 2015), as well as the rapid evolution and dissemination of resistant bacteria (Kumarasamy et al., 2010), has stimulated research into phage therapy (Lin et al., 2017), and data from promising clinical trials have been published. Schooley et al. (2017) have used phagotherapy in a patient with necrotic pancreatitis caused by a MDR strain of A. baumannii. Other studies have obtained favorable results for the phagotherapy of an aortic graft infection with P. aeruginosa (Chan et al., 2018), pneumonia caused by a MDR strain of P. aeruginosa in a cystic fibrosis patient (Law et al., 2019), a Mycobacterium abscessus infection in a patient with cystic fibrosis (Dedrick et al., 2019), and periprosthetic, musculoskeletal, and lung infections (Maddocks et al., 2019; Onsea et al., 2019; Tkhilaishvili et al., 2019). Contrary to these studies, there are reports of the inefficiency of phages in treating bacterial infections (Sarker et al., 2016; Jault et al., 2019), which suggests that the clinical use of phages requires standardization. One of the greatest challenges in phage therapy is the selection of bacterial strains that are resistant to phage action (Azam and Tanji, 2019; Taylor et al., 2019; Yuan et al., 2019). Further studies are required to clearly understand phage biology and elucidate the mechanisms leading to the emergence of phage resistance.
Incompatibility-Based Plasmid Curing and Toxin/Antitoxin Systems
Plasmid incompatibility is generally defined as the inability of two co-resident plasmids to be stably inherited in the absence of outside selection. Thus, if the introduction of a second plasmid destabilizes transmission of the first plasmid, the two plasmids are incompatible. This occurs because the two plasmids share the same replication and partitioning mechanisms. Consequently, under the influence of selective pressure, the resident plasmid can be eliminated (Novick, 1987). Elimination of plasmids based on incompatibility has historically been used to elucidate the mechanisms involved in elimination, and to study the interactions between the plasmid and the host (Uraji et al., 2002). One of the main disadvantages of incompatibility-based plasmid curing methods is the need for repeated cloning and detailed knowledge of the target plasmid. Also a significant problem in the construction of interference plasmids is the requirement to know the replication and partition control region before curing, as well as the need to include additional plasmid genes (Ni et al., 2008). This incompatibility-based strategy has been employed in a variety of bacteria. In L. acidophilus, L. plantarum, and L. pentosus it was used to eliminate approximately 2.3-kb resident plasmids (Bringel et al., 1989; Posno et al., 1991). Ni et al. (2008) used plasmid incompatibility to study the role of plasmids in the pathogenesis of Yersinia pestis. The technique has also been used to remove strains of Bacillus anthracis carrying high pathogenicity plasmids, thus allowing observation of their role in capsule formation and toxin production (Wang et al., 2011; Liu et al., 2012). Hale et al. (2010) constructed a plasmid incompatibility system called pCURE to eliminate F-like plasmids and IncP-1α from E. coli. This system comprises elements of repression (transcriptional repressor, antisense RNA), the origin of replication to compete for essential steps, as well as an antitoxin repressor to control the toxin/antitoxin system (Hale et al., 2010). Toxin/antitoxin systems, also known as post-segregational cell killing or addiction systems, are components of natural plasmids that ensure their persistence in bacterial populations by blocking the growth of daughter cells that do not inherit the plasmid. These systems consist of a labile antitoxin that quenches the activity of the stable toxin. Blocking antitoxin gene expression upon plasmid loss leads to faster depletion of the antitoxin than the toxin, which de-represses toxin activity, and ultimately results in programmed cell death (Hayes, 2003). Recently, Kamruzzaman et al. (2017) constructed incompatibility plasmids in combination with genes encoding antitoxins and replicons, in order to eliminate the blaIMP–4 and blaCMY–2 genes, in both in vitro and in vivo experimental models. Target plasmids were eliminated in the presence of antibiotics for selecting for the interference plasmid (Kamruzzaman et al., 2017).
Successful in vitro elimination of plasmids through incompatibility systems suggests that this strategy could be applied in vivo, to both humans and animals. However, in order to achieve this desideratum, in-depth research is necessary to overcome current limitations of the system, such as the need for repeated cloning, detailed characterization of target plasmids, and prior knowledge of replication and segregation control regions (Ni et al., 2008). Moreover, the requirement to use antibiotics to select eliminated plasmids may be a significant disadvantage to the method. Another aspect that needs further research is the interaction between the interference and resistance plasmids, including the reduction of antibiotic selection.
Utilization of CRISPR/Cas System to Eliminate MGEs Involved in AMR
As stated above, strategies to remove MGEs based on chemical compounds, phages, or incompatibility based-curing plasmid systems have many limitations. All previously described strategies require several stages of bacterial growth in the presence of stressor agents, such as high temperature or intercalating agents, which may lead to unwanted mutations. Therefore, novel approaches for the elimination of MGEs involved in AMR have been proposed.
An attractive alternative strategy for combating bacterial resistance uses the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) system, which was initially described in 1987 by Ishino et al. (1987). CRISPR/Cas is an immune defense system in bacteria that is capable of recognizing foreign nucleic acids and destroying them through associated caspases. One of the significant advantages of this system is its high specificity. This is due to the existence of short repetitive sequences in CRISPR loci that are separated from each other by single sequences of 26–72 pairs derived from MGEs such as plasmids or transposons (Li H. Y. et al., 2018). The CRISPR/Cas mechanism of defense against foreign genetic elements is accomplished in three stages: acquisition, expression, and interference (Crawley et al., 2018). The acquisition stage comprises the insertion of single sequences (spacers) derived from MGEs into repetitive loci of the host chromosome; these sequences are separated from each other by repetitive sequences. The expression stage involves transcribing the complex of repetitive and spacer sequences into a single RNA transcript that will be further processed by caspases in short CRISPR RNAs. In the final stage, the interference phase, foreign nucleic acids are identified based on complementarity with CRISPR RNAs, and their degradation is executed by caspases (Walker and Hatoum-Aslan, 2017). Discrimination between self and non-self is accomplished through sequences from the foreign nucleic acid called protospacers. These sequences are positioned between short DNA sequences (2–6 bp) called protospacer adjacent motifs (PAMs). Cas9 (CRISPR-associated protein 9) will not cleave to a protospacer sequence unless there is a neighboring PAM. CRISPR loci do not contain PAMs, hence direct target recognition is achieved by the CRISPR system without the risk of degrading its own nucleic acid (Marraffini and Sontheimer, 2010; Figure 3). The CRISPR system is classified into six main types and 33 subtypes. Each type has several structural and functional characteristics, but the most distinctive feature is the cas genes and proteins they encode, which play an essential role in recognizing and degrading invading nucleic acids. The number of cas genes ranges from 4 to 20, and the diversity of the corresponding Cas proteins form an ensemble of properties that are essential to the CRISPR immune mechanism (Makarova and Koonin, 2015).
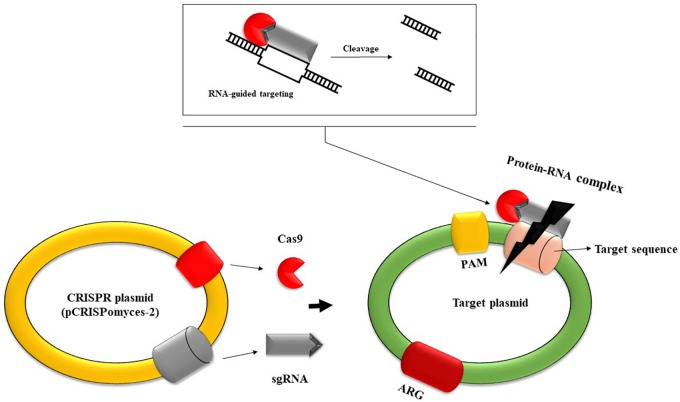
FIGURE 3.
Schematic representation of CRISPR-based plasmid system capable of removing MGE-like resistance plasmids. This system contains two sgRNA transcripts, the cas9 nuclease, and other structural elements. Firstly, sgRNA forms a complex with cas nuclease. The sgRNA transcripts guide cas9 nuclease to introduce double-stranded breaks at the ends of the target DNA, leading to cleavage. Direct target recognition is achieved through recognition of protospacer adjacent motifs (PAM), short DNA sequences that are not found in CRISPR loci, so there is no risk of self-degradation (So et al., 2017). Subsequently, the gap is filled through homologous recombination by an editing template. This system can be used to edit the genome of several antibiotic-resistant bacterial strains, leading to the removal of resistance determinants.
The idea that the CRISPR system could acquire new repetitive nucleic acid sequences of extrachromosomal origin, mainly from phages and plasmids, significantly increased interest in using the system to limit HGT by blocking plasmid conjugation (Bolotin et al., 2005; Pourcel et al., 2005). Subsequently, there have been numerous studies on this topic. The first study of this type was conducted by Marraffini and Sontheimer (2008). The authors attempted to use the CRISPR system interference to block the conjugation of plasmids in S. epidermidis. S. epidermidis strains contain a CRISPR locus containing a homolog spc1 spacer with a region encoding the nickase gene found in conjugative plasmids from this strains (Diep et al., 2006). Silent mutations were introduced into the target gene of the pG0400 conjugate plasmid, generating a mutant plasmid (pG0). Both wild-type and mutant plasmids were tested for conjugation ability. In the control strain, which lacked the CRISPR locus and spc1 complementary to the nickase gene, the conjugation frequency was similar for both plasmids. In the strain harboring the CRISPR locus, only the mutated plasmid with the modified sequence was transferred by conjugation. This demonstrated that the CRISPR system could block plasmid conjugation in a site-specific manner. Furthermore, based on the complementarity between the spc1 spacer and the nickase gene region, the authors showed that CRISPR interference could block plasmid transformation (Marraffini and Sontheimer, 2008). Jiang et al. (2013) showed that the CRISPR system could edit the genome of E. coli. An A to C transversion was introduced in the rpsL gene and a pCRISPR:rpsL plasmid harboring a spacer that would guide dual-RNA:Cas9 cleavage of the wild-type rpsL gene was constructed. Following incubation of the strain of interest with the plasmid, deletion of the rpsL gene was observed (Jiang et al., 2013). Removal of ARGs was also demonstrated by Citorik et al. (2014), using a variant of the CRISPR system encountered in S. pyogenes. The authors built plasmids in which they introduced the CRISPR elements as well as a copy of the blaSHV–18 and blaNDM–1 target genes, conferring resistance to extended-spectrum beta-lactam antibiotics. Elimination of blaSHV–18 and blaNDM–1 plasmids was achieved by packaging the CRISPR elements into a bacteriophage. Following treatment of clinical isolates of E. coli bearing the target plasmids with the constructed phage, a significant reduction in viable bacterial cells was observed (Citorik et al., 2014). Removal of kanamycin resistance genes through the CRISPR system has been demonstrated by Bikard et al. (2014) for strains of S. aureus.
Yosef et al. (2015) introduced the CRISPR elements (cas genes, and spacer sequences targeting the blaNDM-1 and blaCTX-M-15 genes) into a lysogenic phage, and following lysogenization of the resistant bacteria with the constructed phage, elimination of resistance plasmids was observed. The CRISPR system had exhibited the ability to not only remove resistance plasmids, but also to block their HGT (Yosef et al., 2015). Kim et al. (2016) designed a CRISPR plasmid capable of recognizing the blaTEM and blaSHV genes from strains of E. coli producing extended-spectrum β-lactamases. Following transformation of bacterial cells with the CRISPR plasmid, elimination of plasmid-encoding beta-lactamase production was observed, demonstrating the action of caspase at the level of the blaTEM and blaSHV target regions. Furthermore, after elimination of the resistance plasmid, the bacterial strains became sensitive to a series of other antibiotics to which they have previously shown resistance (Kim et al., 2016). Wang et al. (2019) constructed a pMBLcas9 plasmid expressing Cas9, used to clone target single-guide RNAs (sgRNAs) for plasmid curing. The recombinant plasmid pMBLcas9-sgRNA was transferred by conjugation into two clinical isolates of E. coli. In this study, four native plasmids in isolate 14EC033 and two native plasmids in isolate 14EC007 were successfully eliminated in a stepwise manner using pMBLcas9. In addition, two native plasmids in 14EC007 were simultaneously eliminated by tandemly cloning multiple sgRNA in pMBLcas9, sensitizing isolate 14EC007 to polymyxin and carbenicillin (Wang et al., 2019). In Zymomonas mobilis strain ZM4 the resistance plasmids pZZM402 and pZZM403 were eliminated by targeting the replicase-encoding gene which, once inactivated, impairs plasmid replication and leads to subsequent elimination (Cao et al., 2017). The CRISPR system was also used to target some conserved regions within the ColE1 replicons encountered in 91% of the plasmids found in the databases. Lauritsen et al. (2017) constructed a vector in which they introduced all required CRISPR elements as well as two complementary RNA sequences with the conserved regions that guide the cascade nine nuclease to introduce double-strand breaks. This induced elimination of resistance plasmids in E. coli and other bacteria possessing replicons with conserved regions that are targets for the CRISPR system (Lauritsen et al., 2017). CRISPR systems have been designed and delivered in E. coli via transformation and conjugation to eliminate the plasmid-borne mcr-1 gene (Sun et al., 2017; Dong et al., 2019). Efficient editing of a target locus using a CRISPR-based system was achieved in S. aureus (Liu et al., 2017), B. subtilis (So et al., 2017), E. faecalis (Hullahalli et al., 2017), and E. coli (Lauritsen et al., 2018). In summary, this array of studies conducted on various bacterial strains demonstrates the utility of the CRISPR system to eliminate resistance plasmids, as well as blocking HGT of the plasmids. The CRISPR system can also be used for antibiotic sensitization of resistant strains.
Many protocols for plasmid curing/ARG elimination using CRISPR have been proposed (Table 3). However, there are some limitations of this strategy. These limitations include: a known target plasmid replication mechanism is required,; there is a risk of chromosomal ARG acquisition in the interfering plasmid (Kamruzzaman et al., 2017); and the majority of the studies demonstrating the ability of the CRISPR system to eliminate resistance plasmids, as well as to block the dissemination of ARGs by HTG, were performed in vitro. The efficiency of the CRISPR system to eliminate ARGs has been demonstrated in vivo in different mammalian models. Price et al. (2019) revealed that the E. faecalis CRISPR system could block dissemination of resistance plasmids in the mouse gut. However, it remains to be established how much can be extrapolated from these studies to other mammalian organisms. Successful oral administration of phages for targeting bacteria in the intestinal tract (Corbellino et al., 2019) has led to the proposal that phages could be used as a vehicle for delivering the CRISPR system into intestinal microbiota to eliminate ARGs. However, this would require a collection of phages specially designed to target ARGs, the optimal concentration would need to be established, and knowledge of several barriers that occur in vivo would be required, such as inactivation of bacteriophages by gastric acid, and neutralization of phages by the spleen and the immune system (Merril et al., 2003).
TABLE 3.
Protocols used for plasmid curing/ARGs elimination via CRISPR.
| Curing/Elimination strategy | Species of interest | Plasmid/Genes target | Delivery | Results | References |
| Plasmid conjugation blocking by CRISPR system via complementarity between spc1 CRISPR locus and a nes gene region from staphylococcal plasmids | S. aureus S. epidermidis | pG0400 plasmid nes gene | Conjugation Electroporation | Plasmid conjugation and transformation blocking Limiting the spread of antibiotic resistance | Marraffini and Sontheimer, 2008 |
| S. pneumoniae: introducing the ermAM gene together with a premature stop codon in the srtA locus. E. coli pCRISPR:rpsL plasmid construct with a spacer that would guide Cas9 cleavage | S. pneumoniae E. coli | ermAM, erythromycin resistance gene rpsL, streptomycin resistance gene | Transformation | Killing of transformed cells CRISPR selection of non-edited cells | Jiang et al., 2013 |
| CRISPR plasmid constructs bearing the copy of target genes, introduction of double-strand breaks by complementary RNA-guided nucleases in targets CRISPR bacteriophage constructs using for targeting plasmids of interest | E. coli Galleria mellonella | blaSHV–18 blaNDM–1 pZE-blaNDM–1–gfp | Conjugation Viral transduction | Sequence-specific cytotoxicity Excluding high-copy antibiotic resistance plasmids Re-sensitizing a resistant population to antibiotics. | Citorik et al., 2014 |
| Insertion of CRISPR array in a staphylococcal vector to obtain pDB114, programmed to target kanamycin resistant gene Antimicrobial CRISPR cas phagemid to target the methicillin resistance gene CRISPR array that target plasmids of interest | S. aureus | aph-3, kanamycin resistance gene mecA, methicillin resistance gene pUSA01 pUSA02 pUSA03 | Transformation Transduction | Sequence-specific killing of staphylococci resistant to kanamycin or methicillin Loss of pUSA02 plasmid Immunization of staphylococci against pUSA02 transfer. | Bikard et al., 2014 |
| Phage transferable CRISPR cas system | E. coli | blaNDM–1 and blaCTX–M–15 encoding resistance to carbapenems pNDM and pCTX antibiotic resistance plasmid | Lysogenization Transformation | Resistance plasmid curing Prevention of horizontal gene transfer Sensitizing bacteria to multiple antibiotic resistance genes | Yosef et al., 2015 |
| CRISPR plasmid construct, able to recognize target sequences from ESBL strains | E. coli | bla, beta-lactamase genes tet, tetracycline resistance gene pUC19 mediating Amp resistance pET21b pBR322 | Transformation Conjugation | Re-sensitization to antibiotics of E. coli carrying ESBL plasmids CRISPR mediated clearance of whole plasmids | Kim et al., 2016 |
| CRISPR plasmid designed to target a sequence from the replicase gene | Zymomonas mobilis | pZZM402 pZZM403 | Transformation Electroporation | Elimination of native plasmid of Z. mobilis | Cao et al., 2017 |
| Targeting conserved regions from colE1 replicons via CRISPR | E. coli P. putida | pZE-GFP, pZA-GFP pZS-GFP | Transformation | Efficient plasmid curing | Lauritsen et al., 2017 |
| mcr-1 knockout via pCas: mcr CRISPR plasmid | E. coli | Plasmid-borne mcr-1 gene, colistin resistant. | Electroporation BMAP-27 antimicrobial peptide | Sensitization of E. coli strains to colistin following mcr-1 elimination | Sun et al., 2017 |
| pLQ-Pxyl/tet-cas9-Pspac-sgRNA construct, designed to target plasmid of interest | S. aureus | pLQ-KO-tgt-50 bp pLQ-KO-rocA | Transformation | Efficient editing of the target locus | Liu et al., 2017 |
| CRISPR -based plasmid pHCas9 targeting pHT01 and pB0A | B. subtilis | pHT01 pB0A | Transformation | Both plasmids cured by serial culture in antibiotic-free conditions | So et al., 2017 |
| CRISPR2 locus manipulation to obtain pCR2-ermB and pCR2-Phage1 | E. faecalis | PRP pTEF1 pAM771 | Conjugation Electroporation | Decrease in conjugation frequency of the plasmids harboring ARGs | Hullahalli et al., 2017 |
| CRISPR Cas 9 – based plasmid curing system (pFREE) targeting all major plasmid replicon in molecular biology | E. coli | SEVA vectors | Transformation | Efficient curing of target plasmids | Lauritsen et al., 2018 |
| pMCas9- mcr-1 CRISPR construct able to eliminate mcr-1 gene pMob-Cas9 construct, delivered by conjugation | E. coli | Plasmid-borne mcr-1 gene, conferring colistin resistance | Transformation Conjugation | Elimination of plasmid-borne mcr-1 via CRISPR system delivered via transformation and conjugation assay | Dong et al., 2019 |
| Metal stressors exposure | E. coli | pKJK5 broad range plasmid | Conjugation Filter mating experiments | Plasmid elimination | Klumper et al., 2017 |
Conclusion
The global increase in antibiotic resistance is a significant challenge in the fields of medicine and microbial ecology. Rapid development of effective strategies to reduce and control bacterial resistance is required. MGEs have a pivotal role in the acquisition and transmission of ARGs in clinical and environmental sectors, and one approach to control resistance is through elimination of these MGEs. Different chemical (biocides, nanoparticles, antibodies) and biological (engineered phages, commensal microbiota) strategies have been developed, with most of the strategies being directed toward curing the resistance plasmids or inhibiting the conjugation process. However, despite the potential array of approaches directed toward elimination of MGEs, these strategies need refining to overcome the challenges identified in this literature survey. These challenges include the cost-efficiency ratio, the narrow bacterial host spectrum, resistance to phages or chemical agents, the need for a known target plasmid replication mechanism, the risk of chromosomal ARG acquisition in the interfering plasmid (CRISPR technology), and the inability to remove big plasmids. Future work should focus on tackling these challenges to develop a successful strategy to combat antibiotic resistance.
Author Contributions
MC conceived and corrected the manuscript. MC, CB, LP, and CV contributed to the literature survey and revised the manuscript. CV drafted the manuscript. LP designed the figures.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Zong Zhiyong for critical reading of the revised manuscript and helpful suggestions.
Footnotes
Funding. The authors have been supported by the Executive Unit for Financing Higher Education, Research, Development and Innovation (UEFISCDI) under grants PN-III-P4-ID-PCCF-2016-0114 and PN-III-P3-3.1-PM-RO-CN-2018-0147.
References
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. (2011). Phage treatment of human infections. Bacteriophage 1 66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adetosoye A. I., Rotilu I. O. (1985). Infectious drug resistance and antibiotic resistance curing in Salmonella and Shigella isolates from cases of diarrhoea. Revue d’elevage Med. Vet. Pays Trop. 38 433–437. [PubMed] [Google Scholar]
- Altimira F., Yanez C., Bravo G., Gonzalez M., Rojas L. A., Seeger M. (2012). Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 12:193. 10.1186/1471-2180-12-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. 10.3389/fmicb.2011.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonoglou O., Lafazanis K., Mourdikoudis S., Vourlias G., Lialiaris T., Pantazaki A., et al. (2019). Biological relevance of CuFeO2 nanoparticles: antibacterial and anti-inflammatory activity, genotoxicity. DNA and protein interactions. Mater. Sci. Eng. C Mater. Biol. Appl. 99 264–274. 10.1016/j.msec.2019.01.112 [DOI] [PubMed] [Google Scholar]
- Azam A. H., Tanji Y. (2019). Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 103 2121–2131. 10.1007/s00253-019-09629-x [DOI] [PubMed] [Google Scholar]
- Babakhani S., Oloomi M. (2018). Transposons: the agents of antibiotic resistance in bacteria. J. Basic Microbiol. 58 905–917. 10.1002/jobm.201800204 [DOI] [PubMed] [Google Scholar]
- Baquero F., Coque T. M., de la Cruz F. (2011). Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob. Agents Chemother. 55 3649–3660. 10.1128/AAC.00013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavya M. C., Vimal Rohan K., Gaurav G. B., Srivasatava R. (2019). Synergistic treatment strategies to combat resistant bacterial infections using Schiff base modified nanoparticulate - hydrogel system. Mater. Sci. Eng. C Mater. Biol. Appl. 95 226–235. 10.1016/j.msec.2018.10.080 [DOI] [PubMed] [Google Scholar]
- Becker K., van Alen S., Idelevich E. A., Schleimer N., Seggewiss J., Mellmann A., et al. (2018). Plasmid-Encoded Transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 24 242–248. 10.3201/eid2402.171074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedzyk L. A., Shoemaker N. B., Young K. E., Salyers A. A. (1992). Insertion and excision of Bacteroides conjugative chromosomal elements. J. Bacteriol. 174 166–172. 10.1128/jb.174.1.166-172.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaynehe K. M., Shin S. W., Yoo H. S. (2018). Interrelationship between tetracycline resistance determinants, phylogenetic group affiliation and carriage of class 1 integrons in commensal Escherichia coli isolates from cattle farms. BMC Vet. Res. 14:340. 10.1186/s12917-018-1661-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M. (1999). Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. 43 1–4. [PubMed] [Google Scholar]
- Bennett P. M. (2004). “Genome plasticity Methods,” in Molecular Biology Vol 266 Genomics, Proteomics and Clinical Bacteriology, eds Woodford N., Johnson A. (Totowa: Human Press Inc; ), 71–113. [Google Scholar]
- Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153(Suppl. 1), S347–S357. 10.1038/sj.bjp.0707607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund B., Hoang N. T. B., Tarnberg M., Le N. K., Svartstrom O., Khu D. T. K., et al. (2018). Insertion sequence transpositions and point mutations in mgrB causing colistin resistance in a clinical strain of carbapenem-resistant Klebsiella pneumoniae from Vietnam. Int. J. Antimicrob. Agents 51 789–793. 10.1016/j.ijantimicag.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Bikard D., Euler C. W., Jiang W., Nussenzweig P. M., Goldberg G. W., Duportet X., et al. (2014). Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32 1146–1150. 10.1038/nbt.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharova Y., Savinova T., Shagin D. A., Shelenkov A. A., Mayanskiy N. A., Chebotar I. V. (2019). Inactivation of the oprD porin gene by a novel insertion sequence ISPa195 associated with large deletion in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate. J. Glob. Antimicrob. Resistance 17 309–311. 10.1016/j.jgar.2019.01.016 [DOI] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151(Pt 8), 2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Bouanchaud D. H., Chabbert Y. A. (1971). The problems of drug-resistant pathogenic bacteria. Practical effectiveness of agents curing R factors and plasmids. Ann. N. Y. Acad. Sci. 182 305–311. 10.1111/j.1749-6632.1971.tb30666.x [DOI] [PubMed] [Google Scholar]
- Bozkir A., Saka O. M. (2004). Chitosan-DNA nanoparticles: effect on DNA integrity, bacterial transformation and transfection efficiency. J. Drug Target. 12 281–288. 10.1080/10611860410001714162 [DOI] [PubMed] [Google Scholar]
- Bringel F., Frey L., Hubert J. C. (1989). Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid 22 193–202. 10.1016/0147-619x(89)90002-4 [DOI] [PubMed] [Google Scholar]
- Brochet M., Da Cunha V., Couve E., Rusniok C., Trieu-Cuot P., Glaser P. (2009). Atypical association of DDE transposition with conjugation specifies a new family of mobile elements. Mol. Microbiol. 71 948–959. 10.1111/j.1365-2958.2008.06579.x [DOI] [PubMed] [Google Scholar]
- Buckling A., Rainey P. B. (2002). Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269 931–936. 10.1098/rspb.2001.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner M. M. C., Ciusa M. L., Piddock L. J. V. (2018). Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. FEMS Microbiol. Rev. 42 781–804. 10.1093/femsre/fuy031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V. (2017). Mechanisms of stabilization of integrative and conjugative elements. Curr. Opin. Microbiol. 38 44–50. 10.1016/j.mib.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Burrus V., Pavlovic G., Decaris B., Guedon G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46 601–610. 10.1046/j.1365-2958.2002.03191.x [DOI] [PubMed] [Google Scholar]
- Burrus V., Waldor M. K. (2004). Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155 376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- Calero-Caceres W., Muniesa M. (2016). Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 95 11–18. 10.1016/j.watres.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Cambray G., Guerout A. M., Mazel D. (2010). Integrons. Ann. Rev. Genet. 44 141–166. 10.1146/annurev-genet-102209-163504 [DOI] [PubMed] [Google Scholar]
- Cameranesi M. M., Moran-Barrio J., Limansky A. S., Repizo G. D., Viale A. M. (2018). Site-Specific Recombination at XerC/D Sites Mediates the Formation and Resolution of Plasmid Co-integrates Carrying a blaOXA-58- and TnaphA6-Resistance Module in Acinetobacter baumannii. Front. Microbiol. 9:66. 10.3389/fmicb.2018.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli A., Giani T., D’Andrea M. M., Di Pilato V., Arena F., Conte V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58 5696–5703. 10.1128/AAC.03110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q. H., Shao H. H., Qiu H., Li T., Zhang Y. Z., Tan X. M. (2017). Using the CRISPR/Cas9 system to eliminate native plasmids of Zymomonas mobilis ZM4. Biosci. Biotechnol. Biochem. 81 453–459. 10.1080/09168451.2016.1189312 [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. IJMM 303 298–304. [DOI] [PubMed] [Google Scholar]
- Cason J. S., Jackson D. M., Lowbury E. J., Ricketts C. R. (1966). Antiseptic and aseptic prophylaxis for burns: use of silver nitrate and of isolators. Br. Med. J. 2 1288–1294. 10.1136/bmj.2.5525.1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M., Gales A., Walsh T. R., Campana E. H., Tolema M. A. (2007). “Association of blaSPM-1 with the transposon Tn4371,” in Proceedings od the 47th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy 17-20 September 2007, Chicago, IL. [Google Scholar]
- Chan B. K., Sistrom M., Wertz J. E., Kortright K. E., Narayan D., Turner P. E. (2016). Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6:26717. 10.1038/srep26717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B. K., Turner P. E., Kim S., Mojibian H. R., Elefteriades J. A., Narayan D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018 60–66. 10.1093/emph/eoy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy B. M., Gibson E. M., Guiffrida A. (1978). Evidence for plasmid-associated lactose metabolism inLactobacillus casei subsp.casei. Curr. Microbiol. 1 141–144. 10.1007/BF02601666 [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K., Chakraborty R., Basu T. (2014). Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology 25:135101. 10.1088/0957-4484/25/13/135101 [DOI] [PubMed] [Google Scholar]
- Cheow W. S., Hadinoto K. (2014). Antibiotic polymeric nanoparticles for biofilm-associated infection therapy. Methods Mol. Biol. 1147 227–238. 10.1007/978-1-4939-0467-9_16 [DOI] [PubMed] [Google Scholar]
- Citorik R. J., Mimee M., Lu T. K. (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32 1141–1145. 10.1038/nbt.3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochetti I., Tili E., Mingoia M., Varaldo P. E., Montanari M. P. (2008). erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52 1285–1290. 10.1128/AAC.01457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbellino M., Kieffer N., Kutateladze M., Balarjishvili N., Leshkasheli L., Askilashvili L., et al. (2019). Eradication of a multi-drug resistant, carbapenemase-producing Klebsiella pneumoniae isolate following oral and intra-rectal therapy with a custom-made, lytic bacteriophage preparation. Clin. Infect. Dis. 70 1998–2001. 10.1093/cid/ciz782 [DOI] [PubMed] [Google Scholar]
- Costa S. S., Ntokou E., Martins A., Viveiros M., Pournaras S., Couto I., et al. (2010). Identification of the plasmid-encoded qacA efflux pump gene in meticillin-resistant Staphylococcus aureus (MRSA) strain HPV107, a representative of the MRSA Iberian clone. Int. J. Antimicrob. Agents 36 557–561. 10.1016/j.ijantimicag.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Courtright J. B., Turowski D. A., Sonstein S. A. (1988). Alteration of bacterial DNA structure, gene expression, and plasmid encoded antibiotic resistance following exposure to enoxacin. J. Antimicrob. Chemother. 21(Suppl. B), 1–18. 10.1093/jac/21.suppl_b.1 [DOI] [PubMed] [Google Scholar]
- Crameri R., Davies J. E., Hutter R. (1986). Plasmid curing and generation of mutations induced with ethidium bromide in streptomycetes. J. Gen. Microbiol. 132 819–824. 10.1099/00221287-132-3-819 [DOI] [PubMed] [Google Scholar]
- Crawley A. B., Henriksen E. D., Stout E., Brandt K., Barrangou R. (2018). Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci. Rep. 8:11544. 10.1038/s41598-018-29746-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa V. M., McGrann K. M., Hughes D. W., Wright G. D. (2006). Sampling the antibiotic resistome. Science 311 374–377. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Frost L. S., Meyer R. J., Zechner E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34 18–40. 10.1111/j.1574-6976.2009.00195.x [DOI] [PubMed] [Google Scholar]
- Dedrick R. M., Guerrero-Bustamante C. A., Garlena R. A., Russell D. A., Ford K., Harris K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25 730–733. 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavat F., Miyazaki R., Carraro N., Pradervand N., van der Meer J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41 512–537. 10.1093/femsre/fux008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367 731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2 414–424. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- Dong H., Xiang H., Mu D., Wang D., Wang T. (2019). Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int. J. Antimicrob. Agents 53 1–8. 10.1016/j.ijantimicag.2018.09.017 [DOI] [PubMed] [Google Scholar]
- Dow K. H. (1990). The enduring seasons in survival. Oncol. Nurs. Forum 17 511–516. [PubMed] [Google Scholar]
- Dupont C. L., Grass G., Rensing C. (2011). Copper toxicity and the origin of bacterial resistance–new insights and applications. Metallomics 3 1109–1118. 10.1039/c1mt00107h [DOI] [PubMed] [Google Scholar]
- Faghri J., Nouri S., Jalalifar S., Zalipoor M., Halaji M. (2018). Investigation of antimicrobial susceptibility, class I and II integrons among Pseudomonas aeruginosa isolates from hospitalized patients in Isfahan. Iran. BMC Res. Notes 11:806. 10.1186/s13104-018-3901-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennewald M. A., Capobianco J. (1984). Isolation and analysis of inhibitors of transposon Tn3 site-specific recombination. J. Bacteriol. 159 404–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lopez R., Garcillan-Barcia M. P., Revilla C., Lazaro M., Vielva L., de la Cruz F. (2006). Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30 942–966. 10.1111/j.1574-6976.2006.00042.x [DOI] [PubMed] [Google Scholar]
- Fernandez-Lopez R., Machon C., Longshaw C. M., Martin S., Molin S., Zechner E. L., et al. (2005). Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151(Pt 11), 3517–3526. 10.1099/mic.0.28216-0 [DOI] [PubMed] [Google Scholar]
- Fessler A., Kadlec K., Wang Y., Zhang W. J., Wu C., Shen J., et al. (2018). Small antimicrobial resistance plasmids in livestock-associated methicillin-resistant Staphylococcus aureus CC398. Front. Microbiol. 9:2063. 10.3389/fmicb.2018.02063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca E. L., Marin M. A., Encinas F., Vicente A. C. (2015). Full characterization of the integrative and conjugative element carrying the metallo-beta-lactamase bla SPM-1 and bicyclomycin bcr1 resistance genes found in the pandemic Pseudomonas aeruginosa clone SP/ST277. J. Antimicrob. Chemother. 70 2547–2550. 10.1093/jac/dkv152 [DOI] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. (1981). Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. D., Temkin E., Carmeli Y. (2016). The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 22 416–422. 10.1016/j.cmi.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3 722–732. 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- Fu K. P., Grace M. E., Hsiao C. L., Hung P. P. (1988). Elimination of antibiotic-resistant plasmids by quinolone antibiotics. Chemotherapy 34 415–418. 10.1159/000238601 [DOI] [PubMed] [Google Scholar]
- Gao P., Nie X., Zou M., Shi Y., Cheng G. (2011). Recent advances in materials for extended-release antibiotic delivery system. J. Antibiot. 64 625–634. 10.1038/ja.2011.58 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia M. P., de la Cruz F. (2008). Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60 1–18. 10.1016/j.plasmid.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia M. P., Jurado P., Gonzalez-Perez B., Moncalian G., Fernandez L. A., de la Cruz F. (2007). Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 63 404–416. 10.1111/j.1365-2958.2006.05523.x [DOI] [PubMed] [Google Scholar]
- Genilloud O., Blazquez J., Mazodier P., Moreno F. (1988). A clinical isolate of transposon Tn5 expressing streptomycin resistance in Escherichia coli. J. Bacteriol. 170 1275–1278. 10.1128/jb.170.3.1275-1278.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedraitiene A., Vitkauskiene A., Naginiene R., Pavilonis A. (2011). Antibiotic resistance mechanisms of clinically important bacteria. Medicina 47 137–146. [PubMed] [Google Scholar]
- Giordano C., Barnini S., Tsioutis C., Chlebowicz M. A., Scoulica E. V., Gikas A., et al. (2018). Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of ISL3 on plasmids. Int. J. Antimicrob. Agents 51 260–265. 10.1016/j.ijantimicag.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Candelas F., Francino M. P. (2012). “Barriers to horizontal gene transfer: fuzzy and evolvable boundaries Horizontal gene transfer,” in Microorganisms, ed. Francino M. (UnitedKingdom: Caister Academic Press; ), 47–75. [Google Scholar]
- Grindley N. D., Whiteson K. L., Rice P. A. (2006). Mechanisms of site-specific recombination. Ann. Rev. Biochem. 75 567–605. [DOI] [PubMed] [Google Scholar]
- Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. (2006). Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368 874–885. 10.1016/S0140-6736(06)68853-3 [DOI] [PubMed] [Google Scholar]
- Guglielmini J., Neron B., Abby S. S., Garcillan-Barcia M. P., de la Cruz F., Rocha E. P. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42 5715–5727. 10.1093/nar/gku194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes L. C., Florczak-Wyspianska J., de Jesus L. B., Viana M. V., Silva A., Ramos R. T., et al. (2015). Inside the Pan-genome - methods and software overview. Curr. Genomics 16 245–252. 10.2174/1389202916666150423002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Silver S. (1998). Silver as a biocide: will resistance become a problem? Nat. Biotechnol. 16:888. 10.1038/nbt1098-888 [DOI] [PubMed] [Google Scholar]
- Hacker J., Bender L., Ott M., Wingender J., Lund B., Marre R., et al. (1990). Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 8 213–225. 10.1016/0882-4010(90)90048-u [DOI] [PubMed] [Google Scholar]
- Hacker J., Knapp S., Goebel W. (1983). Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F. E., Ciak J. (1976). Elimination of resistance determinants from R-factor R1 by intercalative compounds. Antimicrob. Agents Chemother. 9 77–80. 10.1128/aac.9.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L., Lazos O., Haines A., Thomas C. (2010). An efficient stress-free strategy to displace stable bacterial plasmids. Biotechniques 48 223–228. 10.2144/000113366 [DOI] [PubMed] [Google Scholar]
- Hamidian M., Ambrose S. J., Hall R. M. (2016). A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 8 43–50. 10.1016/j.plasmid.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Harada S., Ishii Y., Saga T., Tateda K., Yamaguchi K. (2010). Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 54 3545–3550. 10.1128/AAC.00111-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E., Wood A. J., Dytham C., Pitchford J. W., Truman J., Spiers A., et al. (2015). Bacteriophages limit the existence conditions for conjugative plasmids. mBio 6:e00586-15. 10.1128/mBio.00586-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Morshed M., Memic A., Hassan S., Webster T. J., Marei H. E. (2018). Nanoparticles in tissue engineering: applications, challenges and prospects. Int. J. Nanomed. 13 5637–5655. 10.2147/IJN.S153758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301 1496–1499. 10.1126/science.1088157 [DOI] [PubMed] [Google Scholar]
- Hochhut B., Jahreis K., Lengeler J. W., Schmid K. (1997). CTnscr94, a conjugative transposon found in enterobacteria. J. Bacteriol. 179 2097–2102. 10.1128/jb.179.7.2097-2102.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. H., Moore L. S., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387 176–187. 10.1016/S0140-6736(15)00473-0 [DOI] [PubMed] [Google Scholar]
- Huang L., Dai T., Xuan Y., Tegos G. P., Hamblin M. R. (2011). Synergistic combination of chitosan acetate with nanoparticle silver as a topical antimicrobial: efficacy against bacterial burn infections. Antimicrob. Agents Chemother. 55 3432–3438. 10.1128/AAC.01803-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullahalli K., Rodrigues M., Palmer K. L. (2017). Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. eLife 6:e26664. 10.7554/eLife.26664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangovan A., Connery S., Waksman G. (2015). Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 23 301–310. 10.1016/j.tim.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Irish D., Eltringham I., Teall A., Pickett H., Farelly H., Reith S., et al. (1998). Control of an outbreak of an epidemic methicillin-resistant Staphylococcus aureus also resistant to mupirocin. J. Hospital Infect. 39 19–26. 10.1016/s0195-6701(98)90239-0 [DOI] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Strahilevitz J., Hooper D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2. 10.1128/microbiolspec.PLAS-0006-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaidane N., Naas T., Mansour W., Radhia B. B., Jerbi S., Boujaafar N., et al. (2018). Genomic analysis of in vivo acquired resistance to colistin and rifampicin in Acinetobacter baumannii. Int. J. Antimicrob. Agents 51 266–269. 10.1016/j.ijantimicag.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Jalasvuori M., Friman V. P., Nieminen A., Bamford J. K., Buckling A. (2011). Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol. Lett. 7 902–905. 10.1098/rsbl.2011.0384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault P., Leclerc T., Jennes S., Pirnay J. P., Que Y. A., Resch G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19 35–45. 10.1016/S1473-3099(18)30482-1 [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Ellabaan M. M. H., Charusanti P., Munck C., Blin K., Tong Y., et al. (2017). Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8:15784. 10.1038/ncomms15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. M., Grossman A. D. (2015). Integrative and Conjugative Elements (ICEs): what they do and how they work. Ann. Rev. Genet. 49 577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. L., Ganopolsky J. G., Labbe A., Prakash S. (2010). A novel nitric oxide producing probiotic patch and its antimicrobial efficacy: preparation and in vitro analysis. Appl. Microbiol. Biotechnol. 87 509–516. 10.1007/s00253-010-2490-x [DOI] [PubMed] [Google Scholar]
- Juhas M., Power P. M., Harding R. M., Ferguson D. J., Dimopoulou I. D., Elamin A. R., et al. (2007). Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8:R237. 10.1186/gb-2007-8-11-r237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., van der Meer J. R., Gaillard M., Harding R. M., Hood D. W., Crook D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33 376–393. 10.1111/j.1574-6976.2008.00136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I. (2014). Historical events that spawned the field of plasmid biology. Microbiol. Spectr. 2:10.1128/microbiolspec.PLAS-0019-2013. 10.1128/microbiolspec.PLAS-0019-2013 [DOI] [PubMed] [Google Scholar]
- Kamruzzaman M., Shoma S., Thomas C. M., Partridge S. R., Iredell J. R. (2017). Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS One 12:e0172913. 10.1371/journal.pone.0172913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan V., Santosh S. W. (2010). Comparing the efficacy of plasmid curing agents in Lactobacillus acidophilus. Benef. Microb. 1 155–158. 10.3920/BM2009.0038 [DOI] [PubMed] [Google Scholar]
- Kennedy I. (1981). The Unmasking of Medicine. London: Paladin, Granada Publishmg. [Google Scholar]
- Keyhani J., Keyhani E., Attar F., Haddadi A. (2006). Sensitivity to detergents and plasmid curing in Enterococcus faecalis. J. Indus. Microbiol. Biotechnol. 33 238–242. 10.1007/s10295-005-0261-y [DOI] [PubMed] [Google Scholar]
- Kim J. S., Cho D. H., Park M., Chung W. J., Shin D., Ko K. S., et al. (2016). CRISPR/Cas9-Mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum beta-lactamases. J. Microbiol. Biotechnol. 26 394–401. 10.4014/jmb.1508.08080 [DOI] [PubMed] [Google Scholar]
- Klockgether J., Wurdemann D., Reva O., Wiehlmann L., Tummler B. (2007). Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J. Bacteriol. 189 2443–2459. 10.1128/JB.01688-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumper U., Dechesne A., Riber L., Brandt K. K., Gulay A., Sorensen S. J., et al. (2017). Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 11 152–165. 10.1038/ismej.2016.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch M., Koole L. (2011). New strategies in the development of antimicrobial coatings: the example of increasing usage of silver and silver nanoparticles. Polymers 3 340–366. [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India. Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi V. V., Polasa H. (1991). Curing of pBR322 and pBR329 plasmids in Escherichia coli by cis-dichlorodiamine platinum(II) chloride (Cis-DDP). FEMS Microbiol. Lett. 62 281–284. 10.1016/0378-1097(91)90171-6 [DOI] [PubMed] [Google Scholar]
- Lakshmi V. V., Sridhar P., Khan B. T., Polasa H. (1988). Mixed-ligand complexes of platinum(II) as curing agents for pBR322 and pBR329 (ColE1) plasmids in Escherichia coli. J. Gen. Microbiol. 134 1977–1981. 10.1099/00221287-134-7-1977 [DOI] [PubMed] [Google Scholar]
- Landecker H. (2016). Antibiotic resistance and the biology of history. Body Soc. 22 19–52. 10.1177/1357034X14561341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Wilkins B. M. (1995). DNA processing reactions in bacterial conjugation. Ann. Rev. Biochem. 64 141–169. 10.1038/sj.emboj.7601806 [DOI] [PubMed] [Google Scholar]
- Lauritsen I., Kim S. H., Porse A., Norholm M. H. H. (2018). Standardized cloning and curing of plasmids. Methods Mol. Biol. 1772 469–476. 10.1007/978-1-4939-7795-6_28 [DOI] [PubMed] [Google Scholar]
- Lauritsen I., Porse A., Sommer M. O. A., Norholm M. H. H. (2017). A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb. Cell Fact. 16:135. 10.1186/s12934-017-0748-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law N., Logan C., Yung G., Furr C. L., Lehman S. M., Morales S., et al. (2019). Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 47 665–668. 10.1007/s15010-019-01319-0 [DOI] [PubMed] [Google Scholar]
- Lederberg J., Tatum E. L. (1946). Gene recombination in Escherichia coli. Nature 158:558. [DOI] [PubMed] [Google Scholar]
- Leungtongkam U., Thummeepak R., Tasanapak K., Sitthisak S. (2018). Acquisition and transfer of antibiotic resistance genes in association with conjugative plasmid or class 1 integrons of Acinetobacter baumannii. PLoS One 13:e0208468. 10.1371/journal.pone.0208468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev A. I., Astashkin E. I., Shaikhutdinova R. Z., Platonov M. E., Kartsev N. N., Volozhantsev N. V., et al. (2017). Identification of IS1R and IS10R elements inserted into ompk36 porin gene of two multidrug-resistant Klebsiella pneumoniae hospital strains. FEMS Microbiol. Lett. 364:fnx072. 10.1093/femsle/fnx072 [DOI] [PubMed] [Google Scholar]
- Li H. Y., Kao C. Y., Lin W. H., Zheng P. X., Yan J. J., Wang M. C., et al. (2018). Characterization of CRISPR-Cas systems in clinical Klebsiella pneumoniae isolates uncovers its potential association with antibiotic susceptibility. Front. Microbiol. 9:1595. 10.3389/fmicb.2018.01595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liang W., Xiao X., Qian Y. (2018). Nanotechnology, an alternative with promising prospects and advantages for the treatment of cardiovascular diseases. Int. J. Nanomed. 13 7349–7362. 10.2147/IJN.S179678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Jimenez J., Derr J., Vera P., Manapat M. L., Esvelt K. M., et al. (2011). Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS One 6:e19991. 10.1371/journal.pone.0019991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. M., Koskella B., Lin H. C. (2017). Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Therapeut. 8 162–173. 10.4292/wjgpt.v8.i3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang L., Chen D., Peters B. M., Li L., Li B., et al. (2018). Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from pseudomonas aeruginosa. Int. J. Antimicrob. Agents 51 145–150. 10.1016/j.ijantimicag.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Liu Q., Jiang Y., Shao L., Yang P., Sun B., Yang S., et al. (2017). CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Acta Biochim. Biophys. Sin. 49 764–770. 10.1093/abbs/gmx074 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang D., Wang H., Feng E., Zhu L., Wang H. (2012). Curing of plasmid pXO1 from Bacillus anthracis using plasmid incompatibility. PLoS One 7:e29875. 10.1371/journal.pone.0029875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Llosa M., Gomis-Ruth F. X., Coll M., de la Cruz F. F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45 1–8. 10.1046/j.1365-2958.2002.03014.x [DOI] [PubMed] [Google Scholar]
- Lujan S. A., Guogas L. M., Ragonese H., Matson S. W., Redinbo M. R. (2007). Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc. Natl. Acad. Sci. U.S.A. 104 12282–12287. 10.1073/pnas.0702760104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S., Fabijan A. P., Ho J., Lin R. C. Y., Ben Zakour N. L., Dugan C., et al. (2019). Bacteriophage therapy of ventilator-associated Pneumonia and Empyema caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 200 1179–1181. 10.1164/rccm.201904-0839LE [DOI] [PubMed] [Google Scholar]
- Magot M. (1983). MTransfer of antibiotic resistances from Clostridium innocuum to Clostridium perfringens in the absence of detectable plasmid DNA. FEMS Microbiol. Lett. 18 149–151. [Google Scholar]
- Mahillon J., Chandler M. (1998). Insertion sequences. Microbiol. Mol. Biol. Rev. 62 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L., Pruteanu M., Kuhn M., Zeller G., Telzerow A., Anderson E. E., et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555 623–628. 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Koonin E. V. (2015). Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 1311 47–75. 10.1007/978-1-4939-2687-9_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11 181–190. 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata C., Navarro F., Miro E., Walsh T. R., Mirelis B., Toleman M. (2011). Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J. Antimicrob. Chemother. 66 2266–2270. 10.1093/jac/dkr286 [DOI] [PubMed] [Google Scholar]
- Mays T. D., Smith C. J., Welch R. A., Delfini C., Macrina F. L. (1982). Novel antibiotic resistance transfer in Bacteroides. Antimicrob. Agents Chemother. 21 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4 608–620. 10.1038/nrmicro1462 [DOI] [PubMed] [Google Scholar]
- Merril C. R., Scholl D., Adhya S. L. (2003). The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2 489–497. 10.1038/nrd1111 [DOI] [PubMed] [Google Scholar]
- Mesas J. M., Rodriguez M. C., Alegre M. T. (2004). Plasmid curing of Oenococcus oeni. Plasmid 51 37–40. [DOI] [PubMed] [Google Scholar]
- Michel-Briand Y., Laporte J. M. (1985). Inhibition of conjugal transfer of R plasmids by nitrofurans. J. Gen. Microbiol. 131 2281–2284. 10.1099/00221287-131-9-2281 [DOI] [PubMed] [Google Scholar]
- Michel-Briand Y., Uccelli V., Laporte J. M., Plesiat P. (1986). Elimination of plasmids from Enterobacteriaceae by 4-quinolone derivatives. J. Antimicrob. Chemother. 18 667–674. 10.1093/jac/18.6.667 [DOI] [PubMed] [Google Scholar]
- Million-Weaver S., Camps M. (2014). Mechanisms of plasmid segregation: have multicopy plasmids been overlooked? Plasmid 75 27–36. 10.1016/j.plasmid.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar J., Foldeak S., Nakamura M. J., Rausch H., Domonkos K., Szabo M. (1992). Antiplasmid activity: loss of bacterial resistance to antibiotics. APMIS Suppl. 30 24–31. [PubMed] [Google Scholar]
- Molnar J., Mandi Y., Kiraly J. (1976). Antibacterial effect of some phenothiazine compounds and R-factor elimination by chlorpromazine. Acta Microbiol. Acad. Sci. Hungar. 23 45–54. [PubMed] [Google Scholar]
- Mugnier P. D., Poirel L., Nordmann P. (2009). Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191 2414–2418. 10.1128/JB.01258-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Maurer J. J., Hofacre C., Summers A. O. (2004). Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U.S.A. 101 7118–7122. 10.1073/pnas.0306466101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejdl L., Kudr J., Moulick A., Hegerova D., Ruttkay-Nedecky B., Gumulec J., et al. (2017). Platinum nanoparticles induce damage to DNA and inhibit DNA replication. PLoS One 12:e0180798. 10.1371/journal.pone.0180798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B., Du Z., Guo Z., Zhang Y., Yang R. (2008). Curing of four different plasmids in Yersinia pestis using plasmid incompatibility. Lett. Appl. Microbiol. 47 235–240. 10.1111/j.1472-765x.2008.02426.x [DOI] [PubMed] [Google Scholar]
- Nishi A., Tominaga K., Furukawa K. (2000). A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 182 1949–1955. 10.1128/jb.182.7.1949-1955.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Poirel L. (2002). Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8 321–331. [DOI] [PubMed] [Google Scholar]
- Nordstrom K. (2006). Plasmid R1–replication and its control. Plasmid 55 1–26. 10.1016/j.plasmid.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Novick R. P. (1987). Plasmid incompatibility. Microbiol. Rev. 51 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojo S. K., Sargin B. O., Esumeh F. I. (2014). Plasmid curing analysis of antibiotic resistance in beta-lactamase producing Staphylococci from wounds and burns patients. PJBS 17 130–133. 10.3923/pjbs.2014.130.133 [DOI] [PubMed] [Google Scholar]
- Olliver A., Valle M., Chaslus-Dancla E., Cloeckaert A. (2005). Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob. Agents Chemother. 49 289–301. 10.1128/AAC.49.1.289-301.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onsea J., Soentjens P., Djebara S., Merabishvili M., Depypere M., Spriet I., et al. (2019). Bacteriophage application for difficult-to-treat musculoskeletal infections: development of a standardized multidisciplinary treatment protocol. Viruses 11:891. 10.3390/v11100891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35 820–855. 10.1111/j.1574-6976.2011.00277.x [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Kwong S. M., Firth N., Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Tsafnat G., Coiera E., Iredell J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33 757–784. 10.1111/j.1574-6976.2009.00175.x [DOI] [PubMed] [Google Scholar]
- Pelgrift R. Y., Friedman A. J. (2013). Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv. Rev. 65 1803–1815. 10.1016/j.addr.2013.07.011 [DOI] [PubMed] [Google Scholar]
- Pembroke J. T., MacMahon C., McGrath B. (2002). The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 59 2055–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penes N. O., Muntean A. A., Moisoiu A., Muntean M. M., Chirca A., Bogdan M. A., et al. (2017). An overview of resistance profiles ESKAPE pathogens from 2010-2015 in a tertiary respiratory center in Romania. Roman. J. Morphol. Embryol. 58 909–922. [PubMed] [Google Scholar]
- Pinilla-Redondo R., Cyriaque V., Jacquiod S., Sorensen S. J., Riber L. (2018). Monitoring plasmid-mediated horizontal gene transfer in microbiomes: recent advances and future perspectives. Plasmid 99 56–67. 10.1016/j.plasmid.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Ponce-Rivas E., Munoz-Marquez M. E., Khan A. A. (2012). Identification and molecular characterization of class 1 integrons in multiresistant Escherichia coli isolates from poultry litter. Appl. Environ. Microbiol. 78 5444–5447. 10.1128/AEM.00660-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25 820–832. 10.1016/j.tim.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Posno M., Leer R. J., van Luijk N., van Giezen M. J., Heuvelmans P. T., Lokman B. C., et al. (1991). Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57 1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C., Salvignol G., Vergnaud G. (2005). CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151(Pt 3), 653–663. 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- Price V. J., McBride S. W., Hullahalli K., Chatterjee A., Duerkop B. A., Palmer K. L. (2019). Enterococcus faecalis CRISPR-Cas is a robust barrier to conjugative antibiotic resistance dissemination in the murine intestine. mSphere 4:e00464-19. 10.1128/mSphere.00464-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulcrano G., Pignanelli S., Vollaro A., Esposito M., Iula V. D., Roscetto E., et al. (2016). Isolation of Enterobacter aerogenes carrying blaTEM-1 and blaKPC-3 genes recovered from a hospital Intensive Care Unit. APMIS 124 516–521. 10.1111/apm.12528 [DOI] [PubMed] [Google Scholar]
- Rai M. K., Deshmukh S. D., Ingle A. P., Gade A. K. (2012). Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 112 841–852. 10.1111/j.1365-2672.2012.05253.x [DOI] [PubMed] [Google Scholar]
- Raja C., Selvam G. (2009). Plasmid profile and curing analysis of Pseudomonas aeruginosa as metal resistant. Int. J. Environ. Sci. Technol. 6 259–266. [Google Scholar]
- Rajpara N., Nair M., Chowdhury G., Mukhopadhyay A. K., Ramamurthy T., Niyogi S. K., et al. (2018). Molecular analysis of multidrug resistance in clinical isolates of Shigella spp. from 2001-2010 in Kolkata. India: role of integrons, plasmids, and topoisomerase mutations. Infect. Drug Resist. 11 87–102. 10.2147/IDR.S148726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin D. J., Rocha E. P., Brown S. P. (2011). What traits are carried on mobile genetic elements, and why? Heredity 106 1–10. 10.1038/hdy.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R., Studer S., Springael D., Zehnder A. J., van der Meer J. R. (1998). Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. Strain B13. J. Bacteriol. 180 4360–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchia G. D., Hall R. M. (1995). Gene cassettes: a new class of mobile element. Microbiology 141(Pt 12), 3015–3027. 10.1099/13500872-141-12-3015 [DOI] [PubMed] [Google Scholar]
- Riber L., Burmolle M., Alm M., Milani S. M., Thomsen P., Hansen L. H., et al. (2016). Enhanced plasmid loss in bacterial populations exposed to the antimicrobial compound irgasan delivered from interpenetrating polymer network silicone hydrogels. Plasmid 8 72–78. 10.1016/j.plasmid.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Roberts A. P., Chandler M., Courvalin P., Guedon G., Mullany P., Pembroke T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60 167–173. 10.1016/j.plasmid.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P. (2009). A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17 251–258. 10.1016/j.tim.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Roberts M. C., Smith A. L. (1980). Molecular characterization of “plasmid-free” antibiotic-resistant Haemophilus influenzae. J. Bacteriol. 144 476–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche D., Flechard M., Lallier N., Reperant M., Bree A., Pascal G., et al. (2010). ICEEc2, a new integrative and conjugative element belonging to the pKLC102/PAGI-2 family, identified in Escherichia coli strain BEN374. J. Bacteriol. 192 5026–5036. 10.1128/JB.00609-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Denarie J., Casse-Delbart F. (1981). Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184 326–333. 10.1007/bf00272926 [DOI] [PubMed] [Google Scholar]
- Ruiz-Martinez L., Lopez-Jimenez L., d’Ostuni V., Fuste E., Vinuesa T., Vinas M. (2011). A mechanism of carbapenem resistance due to a new insertion element (ISPa133) in Pseudomonas aeruginosa. Int. Microbiol. 14 51–58. 10.2436/20.1501.01.135 [DOI] [PubMed] [Google Scholar]
- Santajit S., Indrawattana N. (2016). Mechanisms of Antimicrobial Resistance in ESKAPE pathogens. Biomed Res. Int. 2016:2475067. 10.1155/2016/2475067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo C. E., Morais P. V., Grass G. (2010). Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76 1341–1348. 10.1128/AEM.01952-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker S. A., Sultana S., Reuteler G., Moine D., Descombes P., Charton F., et al. (2016). Oral phage therapy of acute bacterial diarrhea with two Coliphage preparations: a randomized trial in children from Bangladesh. EBiomedicine 4 124–137. 10.1016/j.ebiom.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schairer D. O., Chouake J. S., Nosanchuk J. D., Friedman A. J. (2012). The potential of nitric oxide releasing therapies as antimicrobial agents. Virulence 3 271–279. 10.4161/viru.20328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Hensel M. (2004). Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17 14–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R. T., Biswas B., Gill J. J., Hernandez-Morales A., Lancaster J., Lessor L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61 10.1128/AAC.02221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal B. S., Drider D., Oakley B. B., Brussow H., Bikard D., Rich J. O., et al. (2018). Microbial-derived products as potential new antimicrobials. Vet. Res. 49:66. 10.1186/s13567-018-0563-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selan L., Scazzocchio F., Oliva B., Schippa S., Allocati N., Renzini G. (1988). Plasmid “curing” by some recently synthetized 4-quinolone compounds. Chemioterapia 7 292–294. [PubMed] [Google Scholar]
- Shapiro J. A. (1985). Mechanisms of DNA reorganization in bacteria. Int. Rev. Cytol. 93 25–56. [DOI] [PubMed] [Google Scholar]
- Shariati A., Azimi T., Ardebili A., Chirani A. S., Bahramian A., Pormohammad A., et al. (2018). Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran. Iran. New Microbes New Infect. 21 75–80. 10.1016/j.nmni.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Liang Q., Feng J., Zhan Z., Zhao Y., Yang W., et al. (2018). Coexistence of two novel resistance plasmids, blaKPC-2-carrying p14057A and tetA(A) -carrying p14057B, in Pseudomonas aeruginosa. Virulence 9 306–311. 10.1080/21505594.2017.1372082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani M., Sanchez Z. K., Kimbara K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6:242. 10.3389/fmicb.2015.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M., Gerding D. N., John J. F., Jr., Craig W. A., Bornstein D. L., Duncan R. A., et al. (1997). Society for Healthcare Epidemiology of America and Infectious Diseases Society of America Joint Committee on the Prevention of Antimicrobial Resistance: guidelines for the prevention of antimicrobial resistance in hospitals. Clin. Infect. Dis. 25 584–599. 10.1086/513766 [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Smith M. D., Guild W. R. (1980). DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in Pneumococcus. Plasmid 3 80–87. 10.1016/s0147-619x(80)90036-0 [DOI] [PubMed] [Google Scholar]
- Shokoohizadeh L., Mobarez A. M., Zali M. R., Ranjbar R., Alebouyeh M., Sakinc T., et al. (2013). High frequency distribution of heterogeneous vancomycin resistant Enterococcous faecium (VREfm) in Iranian hospitals. Diagn. Pathol. 8:163. 10.1186/1746-1596-8-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Gourbeyre E., Varani A., Ton-Hoang B., Chandler M. (2015). Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr. 3:MDNA3-0030-2014. 10.1128/microbiolspec.MDNA3-0030-2014 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L., Mourao J., Grosso F., Peixe L. (2018). Uncommon carbapenemase-encoding plasmids in the clinically emergent Acinetobacter pittii. J. Antimicrob. Chemother. 73 52–56. 10.1093/jac/dkx364 [DOI] [PubMed] [Google Scholar]
- Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P., de la Cruz F. (2010). Mobility of plasmids. Microbiology and molecular biology reviews. MMBR 74 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So Y., Park S. Y., Park E. H., Park S. H., Kim E. J., Pan J. G., et al. (2017). A Highly efficient CRISPR-Cas9-mediated large genomic deletion in Bacillus subtilis. Front. Microbiol. 8:1167. 10.3389/fmicb.2017.01167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonstein S. A., Baldwin J. N. (1972). Nature of the elimination of the penicillinase plasmid from Staphylococcus aureus by surface-active agents. J. Bacteriol. 111 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A. (1988). “Identification and analysus of plasmids at the genetic level,” in Methods in Microbiology Vol 21 Plasmid Technology, 2nd Edn, eds Grinsted J., Bennett P. M. (London: Academic Press; ), 11–47. [Google Scholar]
- Sultan I., Rahman S., Jan A. T., Siddiqui M. T., Mondal A. H., Haq Q. M. R. (2018). Antibiotics, resistome and resistance mechanisms: a bacterial perspective. Front. Microbiol. 9:2066. 10.3389/fmicb.2018.02066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers W. C. (2004). “Bacteriophage research: early history,” in Bacteriophages: Biology And Applications, 1 Edn, eds Kutter E., Sulakvelidze A. (Boca Raton, FL: CRC Press; ), 5–27. [Google Scholar]
- Sun L., He T., Zhang L., Pang M., Zhang Q., Zhou Y., et al. (2017). Generation of newly discovered resistance gene mcr-1 Knockout in Escherichia coli using the CRISPR/Cas9 system. J. Microbiol. Biotechnol. 27 1276–1280. 10.4014/jmb.1611.11021 [DOI] [PubMed] [Google Scholar]
- Taylor V. L., Fitzpatrick A. D., Islam Z., Maxwell K. L. (2019). The diverse impacts of phage morons on bacterial fitness and virulence. Adv. Virus Res. 103 1–31. 10.1016/bs.aivir.2018.08.001 [DOI] [PubMed] [Google Scholar]
- Thaden J. T., Li Y., Ruffin F., Maskarinec S. A., Hill-Rorie J. M., Wanda L. C., et al. (2017). Increased costs associated with bloodstream infections caused by multidrug-resistant gram-negative bacteria are due primarily to patients with hospital-acquired infections. Antimicrob. Agents Chemother. 61:e0170916. 10.1128/AAC.01709-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. N. (1994). The Coevolutionary Process. Chicago, IL: University of Chicago Press. [Google Scholar]
- Tkhilaishvili T., Winkler T., Muller M., Perka C., Trampuz A. (2019). Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64:AAC.00924-19. 10.1128/AAC.00924-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Walsh T. R. (2008). Evolution of the ISCR3 group of ISCR elements. Antimicrob. Agents Chemother. 52 3789–3791. 10.1128/AAC.00479-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Walsh T. R. (2011). Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35 912–935. 10.1111/j.1574-6976.2011.00294.x [DOI] [PubMed] [Google Scholar]
- Torres-Barcelo C. (2018). The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg. Microb. Infect. 7:168. 10.1038/s41426-018-0169-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S., Khyriem A. B., Bhattacharya P., Bhattacharjee A., Joshi S. R. (2018). High-level aminoglycoside resistance in Acinetobacter baumannii recovered from Intensive Care Unit patients in Northeastern India. Indian J. Med. Microbiol. 36 43–48. 10.4103/ijmm.IJMM_17_225 [DOI] [PubMed] [Google Scholar]
- Uraji M., Suzuki K., Yoshida K. (2002). A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet. Syst. 77 1–9. 10.1266/ggs.77.1 [DOI] [PubMed] [Google Scholar]
- Uz Zaman T., Albladi M., Siddique M. I., Aljohani S. M., Balkhy H. H. (2018). Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect. Drug Resist. 11 1183–1187. 10.2147/IDR.S161146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel T. P., Gandra S., Ashok A., Caudron Q., Grenfell B. T., Levin S. A., et al. (2014). Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 14 742–750. 10.1016/S1473-3099(14)70780-7 [DOI] [PubMed] [Google Scholar]
- Van Boeckel T. P., Glennon E. E., Chen D., Gilbert M., Robinson T. P., Grenfell B. T., et al. (2017). Reducing antimicrobial use in food animals. Science 357 1350–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee A., Kraak W. B., Burggraaf A., Goessens W. H. F., Pirovano W., Ossewaarde J. M., et al. (2018). Spread of carbapenem resistance by transposition and conjugation among Pseudomonas aeruginosa. Front. Microbiol. 9:2057. 10.3389/fmicb.2018.02057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas J. D., Bailey M. J. (2002). The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42 187–197. 10.1111/j.1574-6941.2002.tb01008.x [DOI] [PubMed] [Google Scholar]
- Vandecraen J., Chandler M., Aertsen A., Van Houdt R. (2017). The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 43 709–730. 10.1080/1040841X.2017.1303661 [DOI] [PubMed] [Google Scholar]
- Ventola C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. PT 40 277–283. [PMC free article] [PubMed] [Google Scholar]
- Walker F. C., Hatoum-Aslan A. (2017). Conjugation assay for testing CRISPR-Cas anti-plasmid immunity in Staphylococci. Bio Protoc. 7:e2293. 10.21769/BioProtoc.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu X., Feng E., Zhu L., Wang D., Liao X., et al. (2011). Curing the plasmid pXO2 from Bacillus anthracis A16 using plasmid incompatibility. Curr. Microbiol. 62 703–709. 10.1007/s00284-010-9767-2 [DOI] [PubMed] [Google Scholar]
- Wang L., Hu C., Shao L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 12 1227–1249. 10.2147/IJN.S121956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., He D., Li B., Guo Y., Wang W., Luo X., et al. (2019). Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J. Antimicrob. Chemother. 74 2559–2565. 10.1093/jac/dkz246 [DOI] [PubMed] [Google Scholar]
- Weisser J., Wiedemann B. (1985). Elimination of plasmids by new 4-quinolones. Antimicrob. Agents Chemother. 28 700–702. 10.1128/aac.28.5.700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle T. J., Sexton J. Z., Gromova A. V., Hadimani M. B., Hughes M. A., Smith G. R., et al. (2009). Inhibitors of RecA activity discovered by high-throughput screening: cell-permeable small molecules attenuate the SOS response in Escherichia coli. J. Biomol. Screen. 14 1092–1101. 10.1177/1087057109342126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2017). Global Priority list of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: WHO. [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Xu Y., Xu J., Mao D., Luo Y. (2017). Effect of the selective pressure of sub-lethal level of heavy metals on the fate and distribution of ARGs in the catchment scale. Environ. Pollut. 220(Pt B), 900–908. 10.1016/j.envpol.2016.10.074 [DOI] [PubMed] [Google Scholar]
- Yazdankhah S., Rudi K., Bernhoft A. (2014). Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 25:25862. 10.3402/mehd.v25.25862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I., Manor M., Kiro R., Qimron U. (2015). Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. U.S.A. 112 7267–7272. 10.1073/pnas.1500107112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wang L., Li X., Tan D., Cong C., Xu Y. (2019). Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii. Virus Res. 272:197734. 10.1016/j.virusres.2019.197734 [DOI] [PubMed] [Google Scholar]
- Zaman M., Pasha M., Akhter M. (2010). Plasmid curing of Escherichia coli cells with ethidium bromide, sodium dodecyl sulfate and acridine orange. Bangladesh J. Microbiol. 1 28–31. [Google Scholar]
- Zhang L., Pornpattananangku D., Hu C. M., Huang C. M. (2010). Development of nanoparticles for antimicrobial drug delivery. Curr. Med. Chem. 17 585–594. 10.2174/092986710790416290 [DOI] [PubMed] [Google Scholar]