Abstract
For centuries, flow visualization has been the art of making fluid motion visible in physical and biological systems. Although such flow patterns can be, in principle, described by the Navier-Stokes equations, extracting the velocity and pressure fields directly from the images is challenging. We addressed this problem by developing hidden fluid mechanics (HFM), a physics-informed deep-learning framework capable of encoding the Navier-Stokes equations into the neural networks while being agnostic to the geometry or the initial and boundary conditions. We demonstrate HFM for several physical and biomedical problems by extracting quantitative information for which direct measurements may not be possible. HFM is robust to low resolution and substantial noise in the observation data, which is important for potential applications.
Quantifying the flow dynamics in geophysical, living, and engineering systems requires detailed knowledge of velocity and pressure fields and has been the centerpiece of experimental and theoretical fluid mechanics for centuries. Available experimental techniques include point measurements (such as a hot wire anemometer or pitot tube) as well as smoke or dye visualization for qualitative characterization of entire flow fields. There are also quantitative flow visualizations such as particle image velocimetry and magnetic resonance imaging (1–3), but these are limited to small domains and laboratory settings. Furthermore, although experimental measurements of external flows (such as flow past a bluff object) are obtained relatively easily, albeit in small subdomains, the quantification of velocity fields for internal flows (such as blood flow in the vasculature) is very difficult or impractical. Despite substantial advances in experimental fluid mechanics, the use of measurements to reliably infer fluid velocity and pressure or stress fields is not a straight-orward task. From the theoretical standpoint, the governing equations of fluid mechanics have been derived from conservation laws (conservation of mass, momentum, and energy), leading to partial differential equations (PDEs) such as the well-known Navier-Stokes (NS) equations (4). Although accurate solutions of such equations in a “forward” setting are now available by using direct numerical simulations or other approximate forms, the computational cost is prohibitively high for realistic conditions and “inverse” problems. Here, we address the question of leveraging the underlying laws of physics to extract quantitative information from available flow visualizations of passive scalars, such as the transport of dye or smoke in physical systems and contrast agents in biological systems. Data assimilation techniques have been used mostly in geophysics but rely on discrete point measurements rather than images of flow visualizations. We developed an alternative approach, which we call hidden fluid mechanics (HFM), that simultaneously exploits the information available in snapshots of flow visualizations and the NS equations, combined in the context of physics-informed deep learning (5) by using automatic differentiation. In mathematics, statistics, and computer science–in particular, in machine learning and inverse problems–regularization is the process of adding information in order to prevent overfitting or to solve an ill-posed problem. The prior knowledge of the NS equations introduces important structure that effectively regularizes the minimization procedure in the training of neural networks. For example, using several snapshots of concentration fields (inspired by the drawings of da Vinci in Fig. 1A), we obtained quantitatively the velocity and pressure fields (Fig. 1, B to D).
Fig. 1. Quantifying flow visualizations.
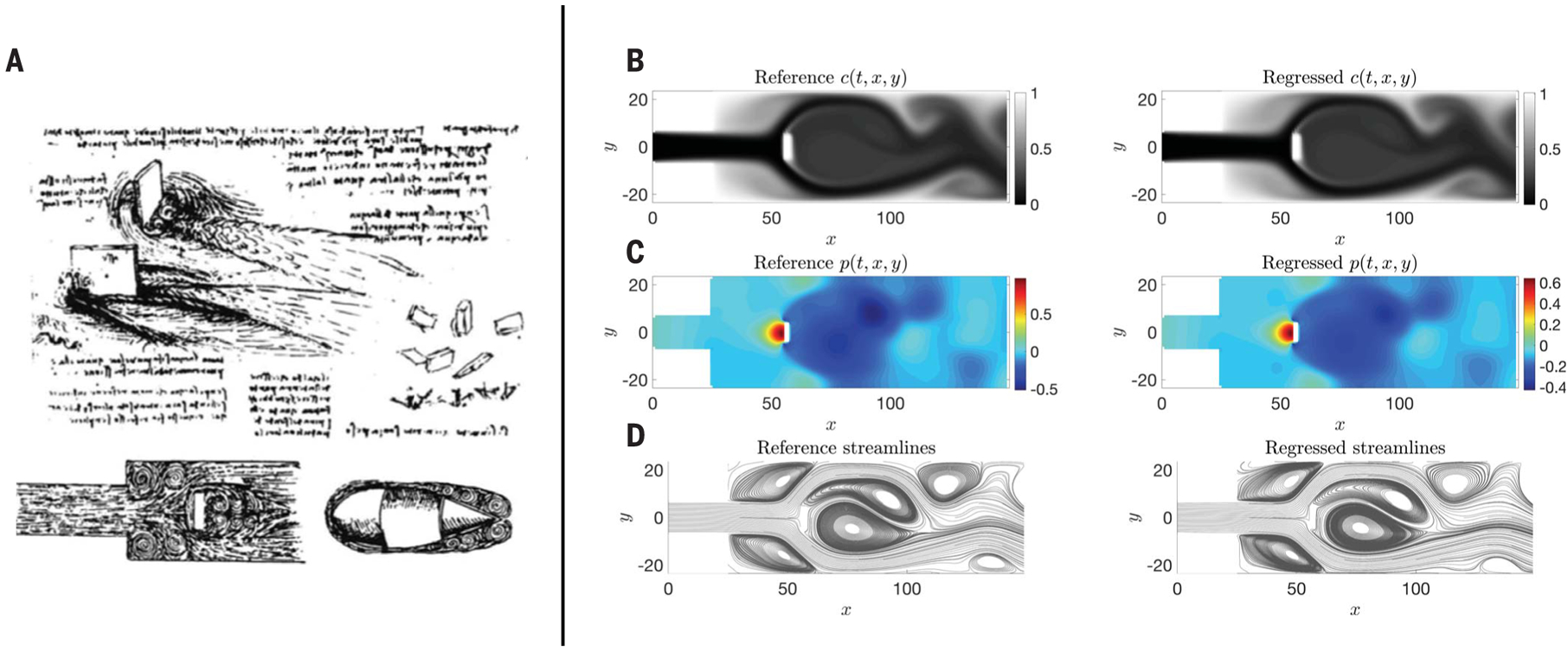
(A) Leonardo da Vinci’s scientific artistry led him to draw accurate patterns of eddies and vortices for various flow problems. [Reprinted from figure 1.4 and figure 1.5 of (17) with permission from Elsevier.] (B to D) We used HFM to quantify the velocity and pressure fields in a geometry similar to the drawing in the lower left corner of (A); our input was a point cloud of data on the concentration field c(t, x, y) shown in (B), left. In (B) to (D), left, we plot the reference concentration, pressure, and streamlines, while at right, we plot the corresponding regressed quantities of interest produced by the algorithm. [(C) and (D)] Hidden states of the system–pressure p(t, x, y) and velocity fields–obtained by using HFM based on the data on the concentration field, randomly scattered in time and space. (D) A comparison of the reference (left) and regressed (right) instantaneous streamlines. The streamlines are computed by using the velocity fields.
We considered the transport of a passive scalar c(t, x, y, z) by a velocity field u(t, x, y, z) = [u(t, x, y, z), v(t, x, y, z), w(t, x, y, z)], which satisfies the incompressible NS equations. The passive scalar is advected by the flow and diffused but has no dynamical effect on the fluid motion itself. Smoke and dye are two typical examples of passive scalars. In this work, we assumed that the only observables are noisy data on the concentration c(t, x, y, z) of the passive scalar (Fig. 2B). This set of time-space coordinates represents a single point cloud of scattered data consisting of N data points (tn, xn, yn, zn) and their corresponding labels cn, the measured concentration value at the point (tn, xn, yn, zn). Here, the superscript n denotes the nth data point and runs from 1 to N. Given such data, scattered in space and time, we were interested in inferring the latent (hidden) quantities of interest u(t, x, y, z), v(t, x, y, z), and w(t, x, y, z) and pressure p(t, x, y, z). We aimed to develop a flexible framework that could deal with data acquired in arbitrarily complex domains such as flow around vehicles or blood flow in brain or aortic aneurysms. We approximated the function (t, x, y, z)↦(c, u, v, w, p) by means of a physics-uninformed deep neural network, which was followed by a physics-informed deep neural network (t, x, y, z)↦(e1, e2, e3, e4, e5), in which the coupled dynamics of the passive scalar and the NS equations were encoded in the outputs e1, e2, e3, e4, and e5 by using automatic differentiation (Fig. 3C and fig. S1). Here, e1 is the residual of the transport equation modeling the dynamics of the passive scalar, and e2, e3, and e4 represent the momentum equations in x, y, and z directions, respectively. Moreover, e5 corresponds to the residual of the continuity equation. In the following, we minimize the norms of these residuals to satisfy the corresponding equations that describe the underlying laws of fluid mechanics. The shared parameters of the physics-uninformed neural networks for c, u, v, w, and p and the physics-informed ones e1, e2, e3, e4, and e5 can be learned by minimizing the following mean squared error loss function
| (1) |
where the first term corresponds to the training data on the concentration of the passive scalar, whereas the last term enforces the structure imposed by the NS and transport equations at a finite set of residual points whose number and locations can be different from the actual training data. The number and locations of these points at which we penalize the equations are in our full control, whereas the data on the concentration of the passive scalar are available at the measurement points. Mini-batch gradient descent algorithms and their modern variants such as the Adam optimizer enable us to penalize the equations at virtually “infinitely” many points. Moreover, in tables S2 and S3, we demonstrate that in addition to the velocity and pressure fields, it is possible to discover other unknown parameters of the flow, such as the Reynolds and Péclet numbers, directly from the data on concentration of the passive scalar.
Fig. 2. Arbitrary training domain in the wake of a cylinder.
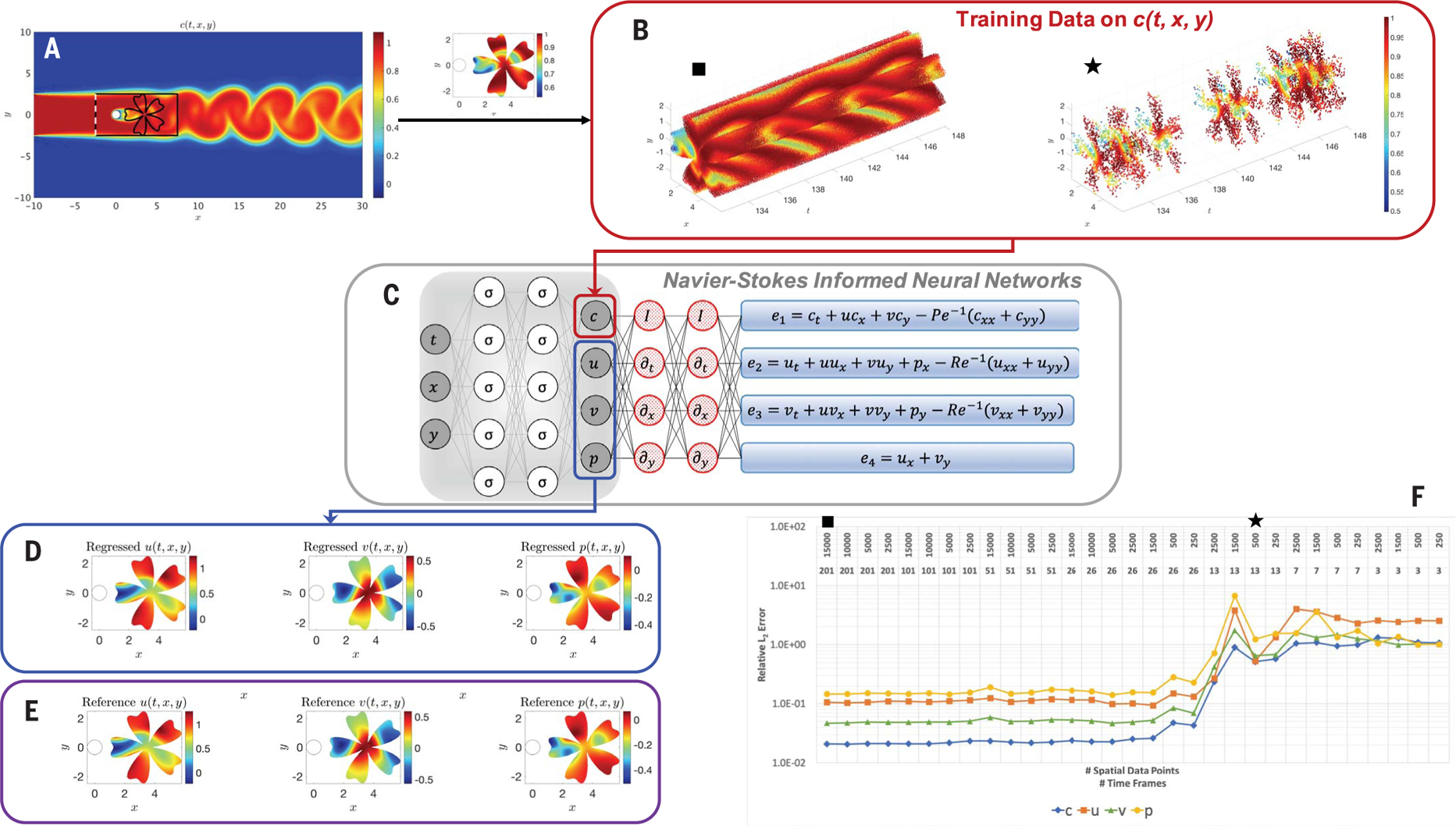
(A) Domain where the training data for concentration and reference data for the velocity and pressure are generated by using direct numerical simulation. (B) Training data on concentration c(t, x, y) in an arbitrary domain in the shape of a flower located in the wake of the cylinder. The solid black square corresponds to a very refined point cloud of data, whereas the solid black star corresponds to a low-resolution point cloud. (C) A physics-uninformed neural network (left) takes the input variables t, x, and y and outputs c, u, v, and p. By applying automatic differentiation on the output variables, we encode the transport and NS equations in the physics-informed neural networks ei, i = 1, …, 4 (right). (D) Velocity and pressure fields regressed by means of HFM. (E) Reference velocity and pressure fields obtained by cutting out the arbitrary domain in (A), used for testing the performance of HFM. (F) Relative L2 errors estimated for various spatiotemporal resolutions of observations for c. On the top line, we list the spatial resolution for each case, and on the line below, we list the corresponding temporal resolution over 2.5 vortex shedding cycles.
Fig. 3. Inferring quantitative hemodynamics in a 3D intracranial aneurysm.
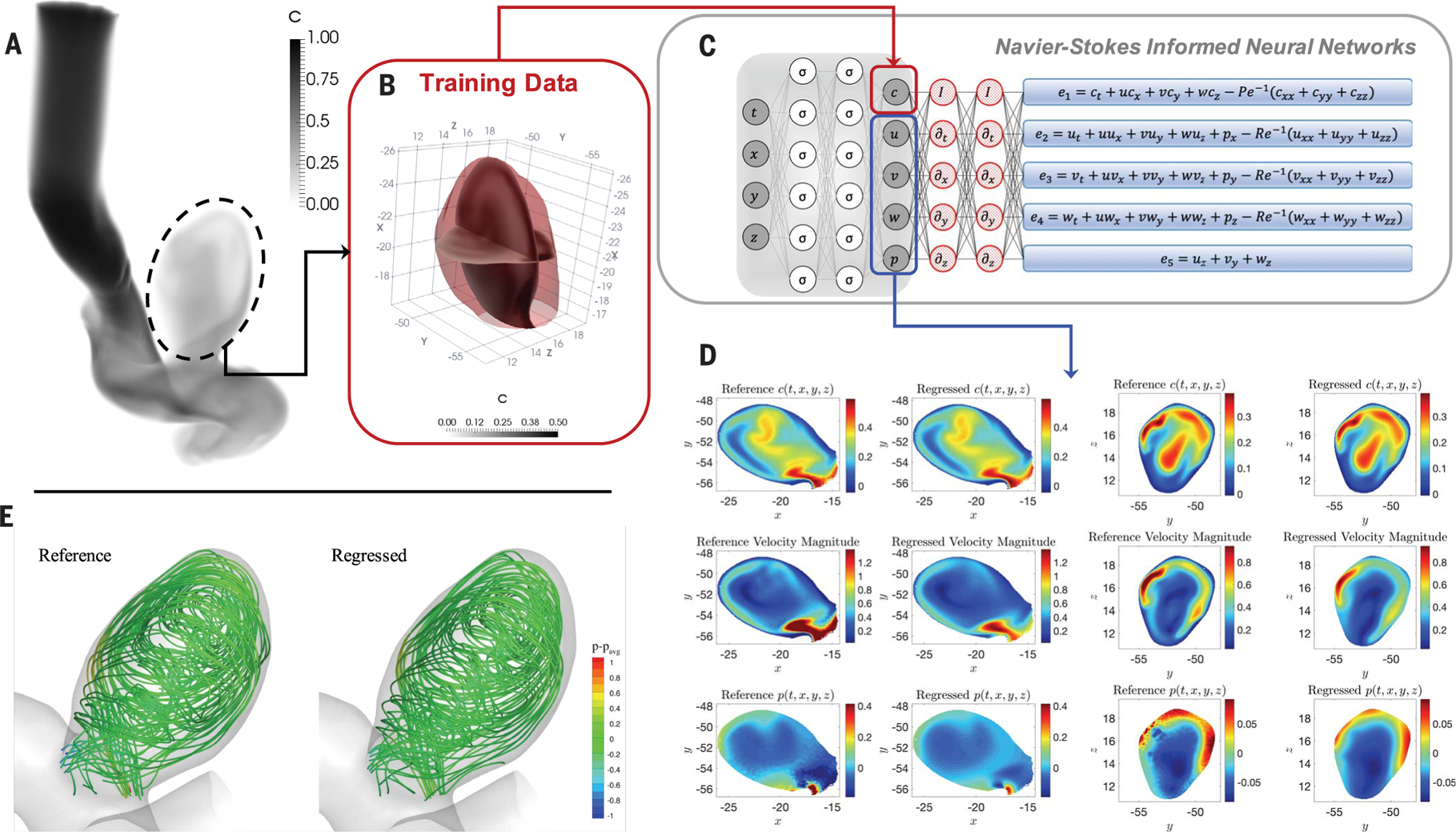
(A) Domain (right internal carotid artery with an aneurysm) where the training data for concentration and reference data for the velocity and pressure are generated by using a direct numerical simulation. (B) The training domain containing only the ICA sac is shown, in which two perpendicular planes have been used to interpolate the reference data and the predicted outputs for plotting 2D contours. (C) Schematic of the NS-informed neural networks, which take c(t, x, y, z) data as the input and infer the velocity and pressure fields. (D) Contours of instantaneous reference and regressed fields plotted on two perpendicular planes for concentration c, velocity magnitude, and pressure p in each row. The first two columns show the results interpolated on a plane perpendicular to the z axis, and the next two columns are plotted for a plane perpendicular to the y axis. (E) Flow streamlines are computed from the reference and regressed velocity fields colored by the pressure field. The range of contour levels is the same for all fields for better comparison [movies S1 and S2, which correspond to (A) and (E)].
We first considered external flows, starting with the prototypical problem of a two-dimensional (2D) flow past a circular cylinder at Reynolds number Re = 100 and Péclet number for the passive scalar Pe = 100 (Fig. 2). We have performed a direct numerical simulation [using the spectral element method (6)] to generate the training data and also the reference velocity and pressure fields in order to investigate the accuracy of HFM. The passive scalar is injected at the inlet on the left boundary. As illustrated in Fig. 2, the shape and extent of the boundaries of the training domains that we chose for our analysis could be arbitrary. However, there are two important factors that need to be considered when choosing the training domain. First, the concentration field of the passive scalar must be present within the training domain so that its information can be used to infer other flow variables. Second, to avoid the need for specifying appropriate boundary conditions for velocities, there must be sufficient gradients of concentration normal to the boundaries (∂c/∂n ≠ 0) in order for the method to be able to infer a single solution for the velocity field. In regions of the training domain in which the concentration profile alone does not carry sufficient information to guarantee a single velocity or pressure field, one could provide the algorithm with additional information such as data on the velocities or pressure (for example, the no-slip boundary condition imposed for velocity on the wall boundaries).
However, in all of the examples considered in this work except for one (figs. S8 and S9), we have relied solely on the information encapsulated within the data on the concentration of the passive scalar. As shown in Fig. 2, good quantitative agreement can be achieved between the predictions of the algorithm and the reference data within a completely arbitrary training domain downstream of the cylinder. The input to the algorithm is essentially a point cloud of data on the concentration of the passive scalar, scattered in space and time (Fig. 2B). We performed a systematic study (Fig. 2F and fig. S5) with respect to the spatiotemporal resolution of the training data for the concentration profile of the passive scalar and verified that the proposed algorithm is very robust with respect to the spatiotemporal resolution of the point cloud of data. Specifically, the algorithm breaks down if for training there are fewer than five time snapshots per vortex shedding cycle or fewer than 250 points in the spatial domain. More details for this case–including quantifying the fluid forces on the cylinder, a benchmark problem for 3D external flow past a circular cylinder, and specifically an analysis of HFM robustness to significant noise levels in the concentration data–are given in figs. S3 to S14. Moreover, we demonstrate the effectiveness of HFM in using information from streaklines (fig. S11 and table S4) and show its robustness irrespective of the boundary conditions imposed at the cylinder wall (c = 0 or ∂c/∂n = 0) to generate the training data.
Next, we focused on internal flows, first demonstrating the effectiveness of HFM for a 2D channel with a stenosis for which we aimed to infer the wall shear stresses (figs. S15 to S18). We studied a realistic biomedical application of HFM for 3D physiologic blood flow in a patient-specific intracranial aneurysm (ICA) (Fig. 3). The aneurysm was located in the cavernous segment of the right internal carotid artery at the level of the eye and beneath the brain (7). Exact concentration fields were generated numerically by using patient-specific boundary conditions, a physiologic flow waveform at the inlet along with a uniform concentration for the passive scalar. Because of the characteristics of our algorithm, we could focus only on the regions where the velocity and pressure fields were needed, reducing substantially the size of data and the cost of training. We first cropped the aneurysm sac out from the rest of geometry and then used only the passive scalar data within the ICA sac (Fig. 3B) for training, whereas no information was used for the boundary conditions. The reference and regressed concentration, velocity, and pressure fields within the ICA sac at a sample time instant were then projected on two separate planes perpendicular to the y and z axes (Fig. 3D). We observed very good agreement between the reference and regressed fields, given the complexity of the flow field. More details on this case, including the predictions for wall shear stress components on the wall of the ICA sac, are given in figs. S19 to S21.
The algorithm we developed is agnostic to the geometry, initial, and boundary conditions, hence providing flexibility in choosing the domain of interest for data acquisition as well as subsequent training and predictions. Moreover, the current methodology allows us to construct computationally efficient and fully differentiable surrogates for velocity and pressure fields that can be further used to estimate other quantities of interest, such as shear stresses and vorticity fields. Transport of scalar fields in fluid flow has been studied in many applications, such as aerodynamics, biofluid mechanics, and nonreactive flow mixing, to name a few. The use of smoke in wind tunnels or dye in water tunnels for flow visualization and quantification has long been practiced in experimental fluid mechanics (8). Moreover, recent techniques in planar laser-induced fluorescence imaging combined with particle image velocimetry have been developed to assess the relationships between scalar and velocity-vorticity fields (9, 10). The use of scalar transport in conjunction with advanced imaging modalities to quantify blood flow in the vascular networks is now a common practice. For example, coronary computed tomography (CT) angiography is typically performed on multidetector CT systems after the injection of a nondiffusible iodine contrast agent, which allows coronary artery visualization and the detection of coronary stenoses (11). Another example is the quantification of cerebral blood flow, either in the prognostic assessment of strokes in patients by using a contrast agent and perfusion CT (12) or in cognitive neuro-science with the use of functional magnetic resonance imaging that only relies on the blood-oxygen-level–dependent contrast to measure brain activity (13). As demonstrated in this work, a direct implication of the current method is quantifying the hemodynamics in the vasculature. This could potentially have a substantial impact on the clinical diagnosis (especially with the noninvasive methods) of vascular diseases associated with important pathologies such as heart attack and stroke. Blood flow shear stresses acting on the vascular wall are crucial in the prognosis of a vascular disease, and their quantification is clinically important (14, 15). Using the proposed method, it is possible to estimate the wall shear stresses at no extra cost. This will simplify the complexities of the state-of-the-art methods in which extracting the exact boundaries of the vessels from the clinical images is required (16).
Our framework is general and can be extended to other disciplines; for example, in electromagnetics, when given data on the electric field and knowing the Maxwell’s equations, we can infer the magnetic field. We have also verified the robustness of HFM to low resolution and substantial noise in the observed concentration fields (Fig. 2 and figs. S5 and S6), which suggests that HFM may find applications in engineering and biomedicine.
Supplementary Material
ACKNOWLEDGMENTS
We thank the anonymous referees for their very helpful suggestions.
Funding: This work received support from the Defense Advanced Research Projects Agency (DARPA) Enabling Quantification of Uncertainty in Physical Systems (EQUiPS)grant N66001-15-2-4055 and DARPA- Artificial Intelligence Research Associate (AIRA) grant HR00111990025, the Air Force Office of Scientific Research (AFOSR) grant FA9550-17-1-0013, the NIH grant U01HL142518, and the U.S. Department of Energy grant DE-AC05-76RL01830.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: All data and codes used in this manuscript are in the supplementary materials and are publicly available on GitHub (18, 19).
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/367/6481/1026/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S21
Tables S1 to S4
References (20–37)
Movies S1 and S2
REFERENCES AND NOTES
- 1.Westerweel J, Dabiri D, Gharib M, Exp. Fluids 23, 20–28 (1997). [Google Scholar]
- 2.Pereira F, Lu J, Castano-Graff E, Gharib M, Exp. Fluids 42, 589–599 (2007). [Google Scholar]
- 3.Shattuck M, Behringer R, Johnson G, Georgiadis J, j.Fluid MechJ 332, 215–245 (1997). [Google Scholar]
- 4.Batchelor GK, An Introduction to Fluid Dynamics (Cambridge Univ. Press, 2000). [Google Scholar]
- 5.Raissi M, Perdikaris P, Karniadakis GE, J. Comput. Phys 378, 686–707 (2019). [Google Scholar]
- 6.Karniadakis GE, Sherwin S, Spectral/hp Element Methods for Computational Fluid Dynamics (Oxford Univ. Press, ed. 2, 2013). [Google Scholar]
- 7.Baek H, Karniadakis GE, J. Comput. Phys 231, 629–652 (2012). [Google Scholar]
- 8.Barlow JB, Rae WH Jr., A. Pope, INCAS Bull. 7, 133 (2015). [Google Scholar]
- 9.Koochesfahani M, Dimotakis P, AIAA J 23, 1700–1707 (1985). [Google Scholar]
- 10.Crimaldi J, Exp. Fluids 44, 851–863 (2008). [Google Scholar]
- 11.Voros S et al. , JACC Cardiovasc. Imaging 4, 537–548 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Wintermark M et al. , Ann. Neurol 51, 417–432 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Aron AR, Behrens TE, Smith S, Frank MJ, Poldrack RA, J. Neurosci 27, 3743–3752 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaaban AM, Duerinckx AJ, AJR Am. J. Roentgenol 174, 1657–1665 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Zarins CK et al. , Circ. Res 53, 502–514 (1983). [DOI] [PubMed] [Google Scholar]
- 16.Boussel L et al. , Magn. Reson. Med 61, 409–417 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama Y, Introduction to Fluid Mechanics (Butterworth-Heinemann, 2018). [Google Scholar]
- 18.Raissi M, maziarraissi/HFM: Hidden fluid mechanics (version v1.0). Zenodo (2019); 10.5281/zenodo.3566161. [DOI]
- 19.Yazdani A, alirezayazdani1/HFM: HFM - Synthetic data generators (version v1.0). Zenodo (2019); 10.5281/zenodo.3567215. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


