Editorial summary
DIVERS uses replicate sampling and spike-in sequences to quantify temporal and spatial variations and noise in microbial samples.
Metagenomic sequencing has enabled detailed investigation of diverse microbial communities, but understanding their spatiotemporal variability remains an important challenge. Here we present DIVERS, a method based on replicate sampling and spike-in sequencing. The method quantifies the contributions of temporal dynamics, spatial sampling variability, and technical noise to the variances and covariances of absolute bacterial abundances. We applied DIVERS to investigate a high-resolution time series of the human gut microbiome and a spatial survey of a soil bacterial community in Manhattan’s Central Park. Our analysis showed that in the gut, technical noise dominated the abundance variability for nearly half of the detected taxa. DIVERS also revealed substantial spatial heterogeneity of gut microbiota, and high temporal covariances of taxa within the Bacteroidetes phylum. In the soil community, spatial variability primarily contributed to abundance variance at short time scales (weeks), while temporal variability dominated at longer time scales (several months).
Introduction
Metagenomic sequencing is widely used to explore patterns of bacterial abundances and the spectrum of functions carried out by diverse microbial communities1–4. Given rapid advancements in sequencing technologies, research efforts have now moved beyond static descriptions of communities towards understanding their complex dynamics5–8. However, quantifying the sources of variability in longitudinal microbiome studies represents a key challenge in the analysis of these ecosystems. Technical sources of variability may fundamentally confound the interpretation of microbiome sequencing studies. Moreover, separating spatial sampling variability from temporal dynamics is important for understanding the underlying ecological behavior of individual bacterial taxa (Supplementary Fig. 1). Comprehensive quantification and analysis of these sources of variability also requires measurements of absolute bacterial abundances to correct for possible compositional artifacts associated with relative abundances9, 10.
Results
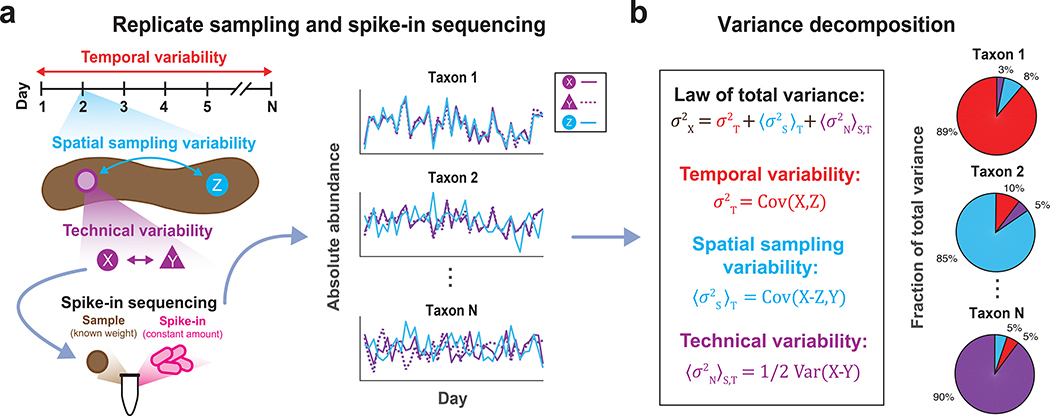
To address these key challenges, we have developed Decomposition of Variance Using Replicate Sampling (DIVERS), a broadly applicable method for metagenomic sequencing studies. DIVERS is a principled mathematical approach that utilizes the laws of total variance and covariance to separate the contributions of time, spatial sampling location, and technical noise to measured abundance variances for individual taxa and covariances for pairs of taxa:
| (A) |
| (B) |
In equations (A) and (B), Xi and Xj denote the abundances of bacterial taxa i and j, S and T are space- and time-associated random variables capturing the respective spatial and temporal processes affecting the abundances of taxa i and j, and E, Var, and Cov denote the expectation, variance, and covariance of random variables, respectively.
Naïve estimation of the terms in equations (A) and (B) requires extensive spatial sampling across the environment at every time point of a longitudinal study, and multiple technical replicates taken at every spatial location. However, such an experimental sampling approach can quickly become prohibitively laborious and expensive. To circumvent these difficulties, DIVERS uses a novel set of unbiased statistical estimators for each of the six terms in equations (A) and (B), along with a workflow to enable their exact calculation from minimal experimental measurements (Supplementary Note, Online Methods). DIVERS requires only two samples obtained from randomly chosen spatial locations at each time point of a longitudinal microbiome study, but can be generalized to accommodate more complex and unbalanced study designs (Supplementary Note). One of the two spatial replicates is then split in half to obtain two technical replicates, and bacterial absolute abundance measurements on the resulting three samples are performed using a spike-in procedure during sample processing11, 12 (Fig. 1, Supplementary Note, Online Methods). The key idea behind this approach is that bacterial taxa experiencing genuine temporal fluctuations should also exhibit large abundance covariances between spatial replicates collected across time points. In contrast, spatial variability, quantified by differences in abundances between the two random spatial locations, and technical noise will decrease temporal covariances (Supplementary Note).
Figure 1 |. DIVERS conceptual workflow.
(a) Illustration of the DIVERS workflow applied to the human fecal microbiome. Samples are collected from two random spatial locations (X and Y from the purple site, Z from the blue site, as shown on the left side of the figure) on each day of sampling and two technical replicates (X and Y) are prepared from one of these spatial locations. The resulting three samples (X, Y, and Z) are subjected to a custom spike-in procedure to estimate absolute bacterial abundances. (b) The DIVERS variance decomposition model is then applied to abundance profiles of each taxa to quantify contributions of temporal variability, spatial sampling heterogeneity and technical noise to total abundance variability.
We first assessed the performance of DIVERS on synthetic data, where the underlying temporal, spatial sampling, and technical contributions to bacterial abundance variances were known. To that end, we performed stochastic simulations of bacterial community dynamics that explicitly incorporated spatial abundance heterogeneity, as well as technical noise associated with experimental measurement error (Online Methods). Confirming our theoretical derivations, DIVERS was able to accurately quantify each of the three variability sources for all simulated species in the community (r.m.s. error = 0.02) (Supplementary Fig. 2, Supplementary Note). Computational simulations also demonstrated that DIVERS compared favorably, both in terms of speed and accuracy, to existing approaches such as the Gaussian process variance decomposition model, a method recently applied to a large human microbiome cohort5 (Supplementary Fig. 3, Online Methods).
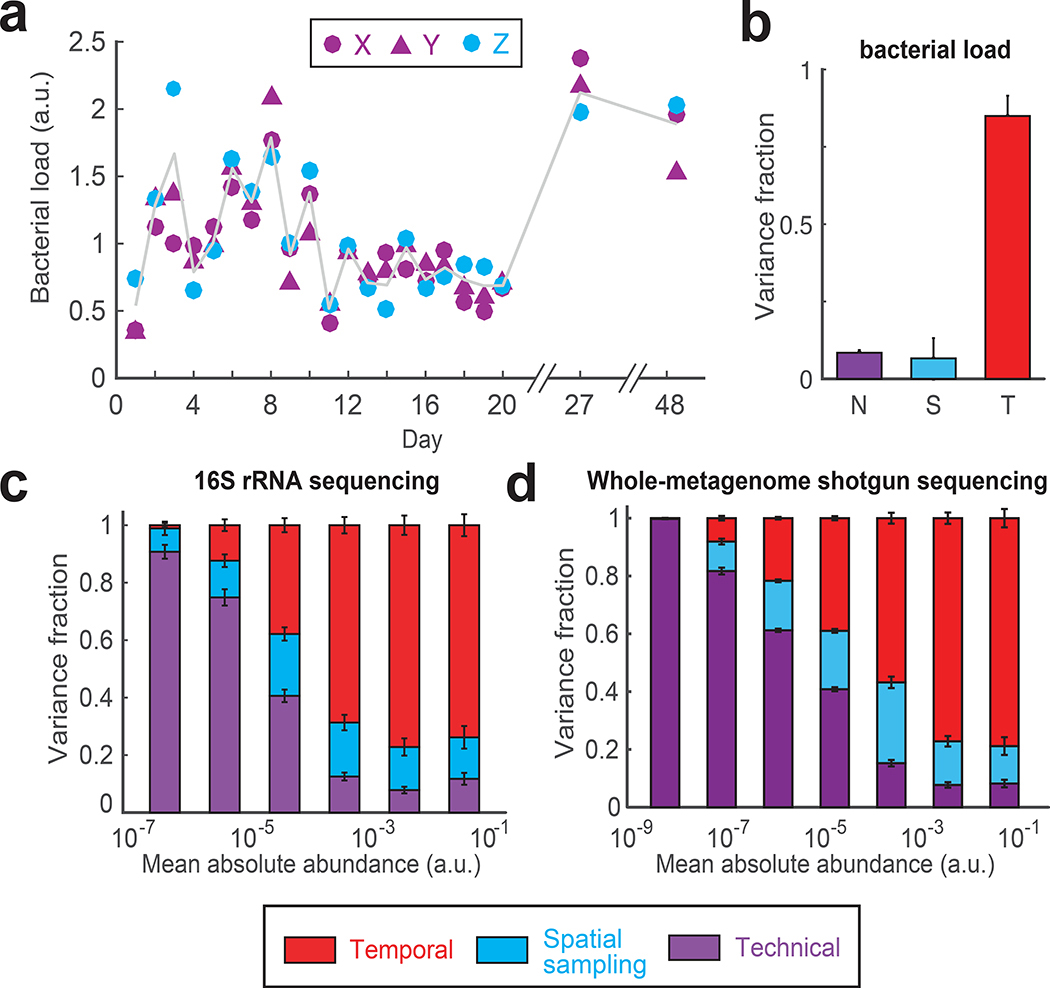
To demonstrate the utility of DIVERS in microbiome studies, we applied the approach to high-resolution time series profiling of the human fecal microbiome. Although fecal samples do not capture the full complexity of microbiota across the gastrointestinal tract13, DIVERS can be used to disentangle the effects of spatial sampling location and time across samples, an issue that is currently not well understood but fundamental to the interpretation of fecal microbiome analyses. Utilizing the DIVERS experimental sampling protocol, we performed 16S rRNA sequencing of fecal samples collected over the course of three weeks from a healthy male individual (Fig. 1, Online Methods). We first verified that our spike-in strain was not found in the human gut microbiome (Supplementary Fig. 4a). We also confirmed the accuracy of our spike-in approach to estimate fecal bacterial loads (∝ total bacterial DNA per mg of sample) using serial dilutions of input fecal matter (Supplementary Fig. 4c, Online Methods). Moreover, technical replicates from fecal samples collected over the time series showed good reproducibility (Pearson’s r = 0.9) (Supplementary Fig. 4d). We next characterized total baseline bacterial abundance variation in the human gut microbiome using DIVERS. Consistent with previous results9, we found that total bacterial abundances fluctuated substantially across samples collected on different days (coefficient of variation = ~0.5) (Fig. 2a). The observed variability was dominated by daily temporal changes, with bacterial loads remaining relatively constant across different spatial locations on each day (Fig. 2b).
Figure 2 |. Variance decomposition of gut bacterial abundances using DIVERS.
(a) Temporal profiles of bacterial loads (∝ total bacterial DNA per mg of sample) in the human gut microbiome (for definition of X, Y and Z see Fig. 1a). Gray line shows the average of spatial replicates. Bacterial loads are reported in arbitrary units and normalized to a mean of one (Online Methods). (b) Variance fraction of the bacterial load attributed to technical (N, purple), spatial sampling (S, blue), and temporal (T, red) factors as calculated by the DIVERS variance decomposition model. The averages were computed using 1000 permutations of the X/Y/Z labels. Error bars represent SEM. (c) Variance decomposition of individual OTU abundances. Absolute OTU abundances were obtained by multiplying relative abundance profiles by the bacterial load in each sample and are reported in arbitrary units (Online Methods). n = 433 OTUs were binned by their mean abundance across all samples, and stacked bars show the average variance contribution of technical, spatial sampling, and temporal sources to OTUs within each bin. Error bars represent the standard error of the mean (SEM). (d) Variance decomposition for n = 3619 individual bacterial species based on species abundances obtained by shotgun metagenomic sequencing and species profiling with Kraken28 (Online Methods).
Using measurements of bacterial loads, we calculated the absolute abundances of all operational taxonomic units (OTUs) and used DIVERS to decompose the contributions to the total abundance variances of individual OTUs (Online Methods and Supplementary Note). When OTUs were grouped by average abundance, variance profiles exhibited two regimes, with a transition occurring ~10−4 in absolute abundances (Fig. 1c). In relative abundances, this transition corresponded to a value of ~0.01% (Supplementary Fig. 5). Notably, dynamical behavior of OTUs below this abundance cutoff primarily reflected technical noise consistent with Poissonian sampling variability. Nearly half (~43%) of all OTUs detected in the fecal samples exhibited such noise-driven behavior, demonstrating that DIVERS provides a principled solution for identifying technical artifacts to be removed from subsequent analyses (Supplementary Fig. 6b,c). In contrast, the variability of OTUs above this cutoff largely reflected true temporal changes (Fig. 2c and Supplementary Fig. 6a). Differences across spatial sampling locations also contributed a substantial fraction to total variability (on average ~20% for OTUs with mean absolute abundance > 10−4), highlighting significant spatial heterogeneity of fecal samples (Fig. 2c, Supplementary Fig. 7).
In order to further experimentally validate the developed workflow and the variance decomposition model, we performed a set of control experiments that specifically eliminated either temporal or spatial variability from fecal samples (Online Methods). First, we collected fecal samples from ten independent spatial locations of the same stool specimen. This procedure effectively simulated five consecutive time points of the DIVERS protocol, but without any temporal contribution to microbiota variability. Second, to remove spatial variability, we carried out eight consecutive days of sampling with spatial replicates that were homogenized on each day before sequencing (Online Methods). Reassuringly, the model correctly predicted no temporal or spatial contributions to OTU abundance variability when the corresponding signals were removed from the data (Supplementary Fig. 8).
The observed patterns of variability in the human gut microbiome may be influenced by factors specific to 16S rRNA library preparation and sequencing, such as differential 16S rRNA copy number, 16S primer and PCR biases, and OTU clustering approaches14, 15. We therefore carried out whole-metagenome shotgun sequencing (WMGS) of the same fecal samples and estimated species absolute abundances in each sample (Online Methods). Applying the DIVERS variance decomposition model to WMGS species abundances, we obtained results highly similar to those obtained using 16S rRNA sequencing (Fig. 2d, Supplementary Fig. 9). Specifically, the behavior of over half of all detected species was predominantly explained by technical noise, with spatial sampling variability contributing ~20% to the total variance for abundant species (mean absolute abundance > 10−5). This demonstrates that overall patterns of individual taxa variability uncovered by DIVERS are robust and the method is applicable to another rapidly expanding sequencing methodology.
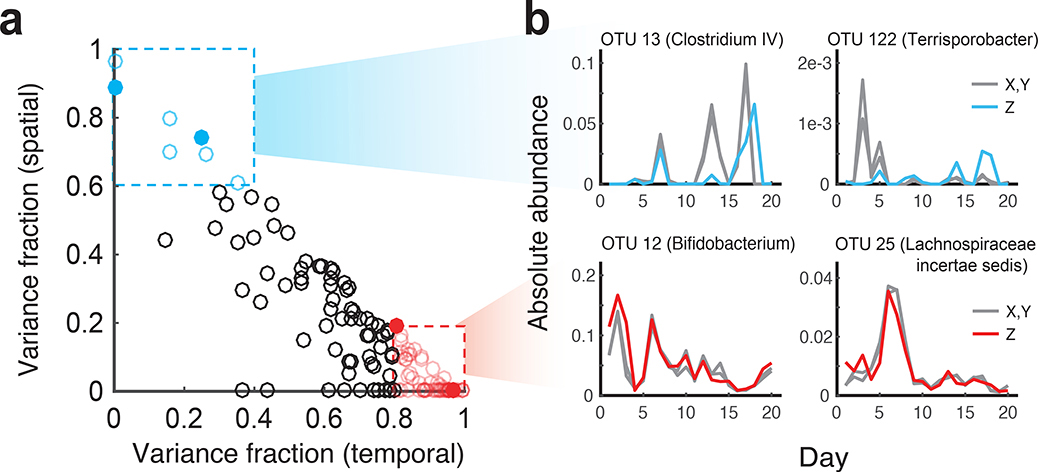
Based on the DIVERS variance decomposition, we identified several abundant OTUs whose time series were primarily shaped by either temporal (OTU 12, Genus: Bifidobacterium and OTU 25, Genus: Lachnospiracea incertae sedis) or spatial variation (OTU 13, Genus: Clostridium IV and OTU 122, Genus: Terrisporobacter) (Fig. 3a,b). Thus, in addition to revealing OTUs dominated by technical noise, DIVERS demonstrates substantial temporal and spatial variability contributing to the abundance changes of individual fecal bacteria.
Figure 3 |. Identifying individual taxa with high temporal or spatial sampling variance.
(a) Identification of specific OTUs with either high temporal variance (red points, variance fraction > 0.8) or high spatial sampling variance (blue points variance fraction > 0.6) contributions. Only abundant OTUs (mean absolute abundance > 10−4) are shown. (b) Time series of individual OTUs, corresponding to filled points in a, whose abundance variation is predominantly attributed to temporal (red) or spatial sources (blue). Gray lines correspond to abundances of technical replicates (X,Y) obtained from the same spatial location, and colored lines correspond to abundances from the second spatial replicate (Z).
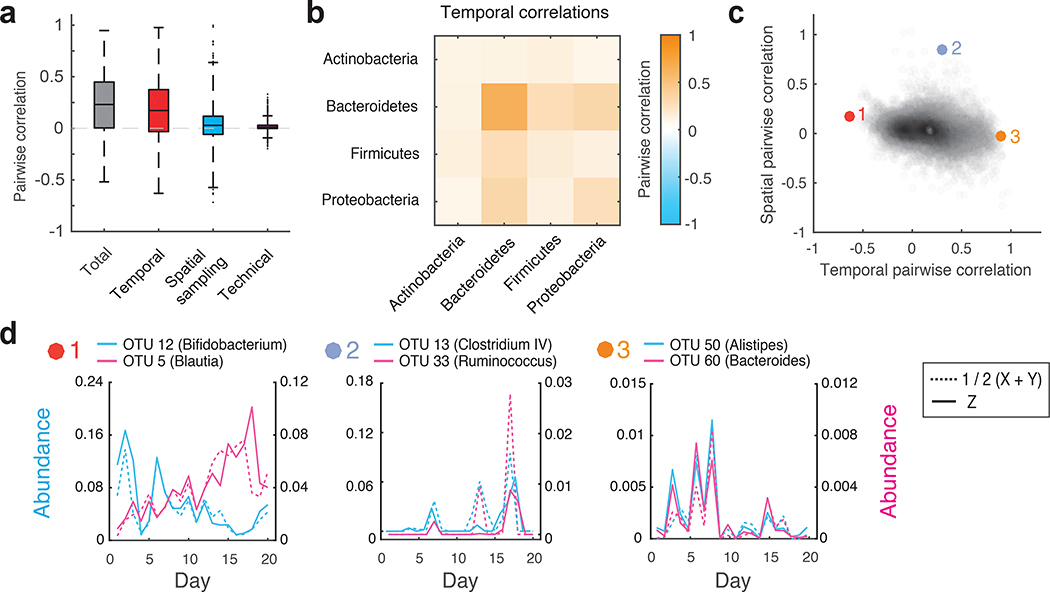
Fluctuations in bacterial abundances often result from the collective behavior of multiple different taxa, whose interactions are reflected in correlated abundance changes16. DIVERS can also be used to quantify the factors contributing to abundance correlations between pairs of OTUs in a microbial community (Online Methods and Supplementary Note). Applying this analysis to human fecal samples, we found that the majority of pairwise abundance correlations were due to temporal sources, with relatively smaller contributions arising from spatial sampling location and technical noise (Fig. 4a and Supplementary Fig. 10a,b). Consistent with previous results9, we also found that total correlations based on absolute abundance measurements were generally larger than correlations calculated using relative abundances, an effect primarily caused by the variance in bacterial loads across samples (Supplementary Fig. 10c,d and Supplementary Note).
Figure 4 |. Decomposition of contributions to pairwise OTU abundance correlations in the human gut microbiome.
(a) Boxplots of total, temporal, spatial, and technical correlations for all pairs of abundant OTUs (average absolute abundance > 10−4). Boxes denote the median and interquartile ranges, with maximum whisker lengths three times the interquartile range. (b) Temporal correlations of OTU abundances within and between different phyla; colors reflect average temporal correlations between pairs of OTUs from the indicated phyla. Data are shown for all highly abundant OTUs (mean absolute abundance >10−4) from the Actinobacteria (n = 10), Bacteroidetes (n=15), Firmicutes (n=103), and Proteobacteria (n=5). (c) Temporal and spatial correlations for all pairs of abundant OTUs (average absolute abundance > 10−4). Colored points (1–3) indicate pairs of OTUs with temporal profiles shown in d. (d) Temporal abundance profiles for pairs of OTUs highlighted in c. Pairs exhibit (from left to right): 1) Substantial negative temporal (ρT = −0.63, p = 4×10−4), 2) substantial positive spatial (ρS = 0.85, p = 3×10−4), and 3) substantial positive temporal (ρT = 0.90, p < 10−4) correlations. For every OTU pair, blue and pink solid lines show abundances of each OTU measured from the same spatial location (Z). Blue and pink dashed lines show the average between technical replicates (1/2(X+Y)) of each OTU measured from the second spatial location. See online methods Eq. 9 for definition of the temporal correlation; p values were estimated by generating 104 random abundance series for the pairs of OTUs with known temporal variances, and then using these time series to compute the temporal covariances.
Next, we examined factors contributing to the correlations of OTU abundances within and between the four most prevalent gut bacterial phyla. Interestingly, the Bacteroidetes exhibited significantly larger intra-phyla temporal abundance correlations compared to the rest of the community (p < 10−10, Wilcoxon rank sum test) (Fig. 4b, Supplementary Fig. 11). This result was also observed at the family level, and was not due to differences in 16S rRNA sequence similarity across taxa (Supplementary Fig. 12, Supplementary Fig. 13). The coordinated temporal changes of the Bacteroidetes may reflect fluctuations in the availability of dietary polysaccharides on each day that are specifically metabolized by these bacteria17, 18, as well as previously observed cross-feeding interactions between these taxa19, 20. In addition, our analysis revealed several interesting examples of OTU pairs with positive and negative correlation contributions from temporal and spatial factors (Fig. 4c,d). For example, OTU 12 (blue) and OTU 5 (pink) display substantial negative temporal correlation (Fig. 4d, panel 1) independent of spatial locations. In contrast, OTU 50 (blue) and OTU 60 (pink) display substantial positive temporal correlation (Fig. 4d, panel 3). The substantial positive spatial correlation between OTU 13 and OTU 33 (Fig. 4d, panel 2) is reflected by their similar abundance profiles at two independent spatial locations; dashed lines of different colors represent the two OTUs at one location, and solid lines of different colors represent the two OTUs at the other location. These examples highlight the diversity of bacterial dynamics, and demonstrate the ability of DIVERS to disentangle factors contributing to the abundance correlations between different taxa.
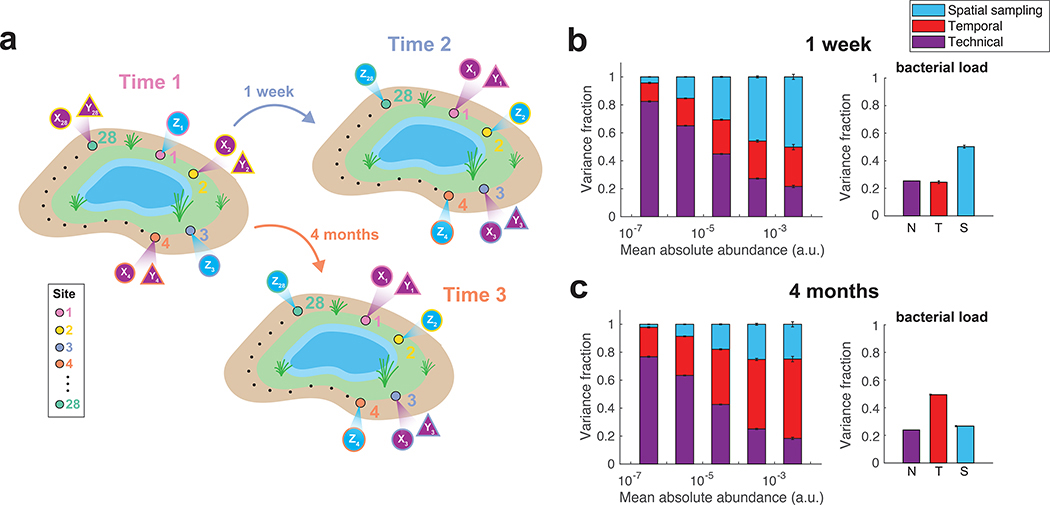
The DIVERS variance decomposition approach is not limited to the human gut microbiome, and can be used to investigate the contributions to bacterial abundance variability in diverse ecological communities. To demonstrate this, we performed an analysis of spatial variation of a soil microbial community in Manhattan’s Central Park in New York City. Urban microbiomes, including soil communities in Central Park, have been previously shown to exhibit substantial microbial diversity21. To explore spatial abundance variability, we utilized a modified protocol that inverted the hierarchy of spatial and temporal sampling replicates (Supplementary Note). Specifically, we collected soil samples from twenty-eight sites spread uniformly around a small man-made pond in the northwest section of Central Park (Fig. 5a, Supplementary Fig. 14, Supplementary Table 3, Online Methods). Samples were collected from identical locations on three different days, two of which were one week apart and the third over four months later. This experimental design allowed us to compare overall patterns of spatial and temporal variation at the timescales of one week and several months. Following the DIVERS protocol, a single time point from each spatial location was subjected to two independent rounds of sample preparation and sequencing (Fig. 5a), and a spike-in approach was used to estimate bacterial loads in each soil sample (Fig. 5b,c and Supplementary Fig. 4b, Online Methods).
Figure 5 |. Decomposition of contributions to the variance of soil bacterial abundances.
(a) Illustration of the DIVERS sampling protocol applied to a Central Park soil microbial community. Samples were collected one week and four months apart from twenty-eight spatial sites spread uniformly around a small pond in the northwest area of the park. At each site i, two technical replicates were collected using samples from one time point (Xi and Yi), whereas a single measurement was made at the other time point (Zi) (b,c) DIVERS variance decomposition of n = 24667 individual OTU abundances (left panels) and n = 27 bacterial loads (right panels). Left panels: OTUs were binned by their mean abundance across all samples, and stacked bars show the average variance contribution of technical, spatial sampling, and temporal sources to OTUs within each bin. Right panels: variance contribution of technical, spatial sampling, and temporal sources to bacterial load. Error bars represent the SEM. Temporal variability reflects average changes in the community at the two time scales (1 week in b, 4 months in c).
Similar to the human gut, DIVERS revealed pervasive technical noise in OTUs with low abundance (log10 mean absolute abundance < −4.5). Spatial sampling location was the major source of variability when comparing time points separated by one week (Fig. 5b, Supplementary Fig. 15). However, when comparing time points separated by four months, temporal variability predominated, demonstrating the ability of DIVERS to quantify differences in contributions to abundance variability at various timescales in the community (Fig. 5c). Applying the DIVERS covariance decomposition model to soil bacteria, we observed a relatively low degree of abundance correlations between all OTU pairs, as well as within and between different bacterial phyla (Supplementary Fig. 16). These results indicate relatively weak patterns of co-occurrences between soil bacteria22, but significant spatial and temporal abundance variability of individual taxa22–24.
Discussion
While current sequencing technologies make it possible to profile bacterial communities at high temporal resolution, novel approaches are required for the proper interpretation and in-depth analyses of collected data. The pervasive contributions of technical and spatial factors to the bacterial abundance variability revealed in our analysis underscores the need for principled approaches like DIVERS. Future studies can employ the DIVERS hierarchical sample collection and analysis framework to quantify and minimize the biases due to technical noise. Importantly, researchers can use DIVERS to estimate noise contributions for species of interest, instead of relying on arbitrary abundance cutoffs to consider data for individual species. When applying DIVERS in future studies it will be important to understand how the contribution of various factors to bacterial abundance fluctuations vary at different spatial and temporal scales. Identifying and understanding unusual patterns of spatial and temporal variances and co-variances may also serve as biomarkers of disease and perturbed microbiota states.
Another crucial challenge for future studies is understanding microbiota variances and variance contributions in absolute rather than relative terms. This will minimize potential biases due to compositional nature of many current metagenomic datasets. In the present study we used a microbial spike-in technique with a single species to evaluate factors contributing to absolute bacterial abundance variability. Multiple other approaches, such as measurement of microbial DNA content11, quantitative PCR25, and flow cytometry9, can also be used with DIVERS to evaluate changes in absolute bacterial abundances. Comparing the performance of these techniques, and future improvements in the spike-in technology (e.g. optimization of spike-in amounts, use of multiple spike-in species, automated weighing of samples) will minimize technical noise associated with the spike-in process, improve accuracy, and allow more efficient utilization of sequencing coverage.
Although we focus on human gut and soil microbial communities, DIVERS can be readily applied to explore patterns of variation and technical noise in any bacterial ecosystem across different hosts and environments (Supplementary Note, online tutorial of DIVERS is available at https://github.com/hym0405/DIVERS). Moreover, given the flexibility of the developed quantitative framework, it can be easily extended to other sequencing-based applications, such as the characterization of human immune cell repertoires26 and gene expression changes in tumors27.
Online Methods
Ethical review
This study was approved and conducted under Columbia University Medical Center Institutional Review Board protocol AAAR0753. Written informed consent was obtained from the subject in the study, a healthy male adult.
Fecal sample collection and storage
Fecal samples were collected daily over the course of twenty days, with two additional samples taken on days 27 and 48 of the study. After defecation, inverted sterile 200 μL pipette tips (Rainin RT-L200F) were used to core out a small sample from the stool specimen, which was placed immediately in a sterile cryovial (Sarstedt 72.694.106). Samples were then immediately placed in a −20 °C freezer and transferred to a −80 °C freezer for long-term storage.
Replicate fecal sampling experimental protocol
To enable decomposition of gut bacterial abundance variability into temporal, spatial and technical contributions, a replicate sampling approach was utilized. Specifically, on each day of the time series, two fecal samples were collected from random spatial locations of the same stool specimen. For one of these samples, two technical replicates were prepared in parallel by splitting the individual fecal core. Thus, a total of three samples were processed for each day of the time series: two technical replicates from a single spatial location (denoted samples X and Y) and a second spatial replicate (denoted sample Z). To further characterize technical noise, a single fecal sample was also subjected to 12 independent rounds of sample processing and sequencing. Metadata associated with all fecal samples are provided in Supplementary Table 1. Theoretical details associated with the DIVERS approach are described in the Supplementary Note.
Soil site description, sample collection and storage
Soil samples were collected in June and October of 2018 from The Pool in Central Park, Manhattan (approximately 40.795oN, 73.960oW), a man-made body of water located in the northwest area of the park. Soil cores were collected on two days exactly one week apart, and a third day roughly four months after the initial sampling time point, from twenty-eight sites located on the periphery of the water’s edge. The average distance between adjacent sites was ~8 meters. Photographs were taken at each site to ensure sampling accuracy at the same location from different time points (Supplementary Fig. 14). Following soil collection, samples were transferred to a −80 °C freezer for long-term storage.
Replicate soil sampling experimental protocol
Similar to our fecal sampling protocol, a replicate sampling approach was utilized to collect soil bacteria. For a given pair of time points, technical replicates (denoted samples X and Y) were prepared from a single sample collected from one of the two time points by splitting the individual soil core. The time points for which technical replicates were prepared were alternated between neighboring spatial sampling sites. Following the DIVERS protocol, a single measurement was made from samples collected the remaining time point (denoted sample Z). Metadata associated with all soil samples are provided in Supplementary Table 3. Due to technical error associated with sample preparation, site 25 was excluded from any further downstream analyses. Theoretical details associated with the DIVERS approach are described in the Supplementary Note.
Spike-in strain for calculation of bacterial absolute abundances
A spike-in approach was utilized during sample processing to allow for calculation of total bacterial abundances per mass of fecal or soil matter. Sporosarcina pasteurii (ATCC 11859), an environmental bacterium that was confirmed to be absent in our fecal and soil samples, was grown to saturation in NH4-YE medium (ATCC medium 1376). It was then concentrated by centrifugation, resuspended in ~0.1X volume phosphate buffered saline (PBS) with 20% glycerol, and stored in cryovials at −80 °C for subsequent use during genomic DNA extraction.
Sample genomic DNA extraction
Genomic DNA (gDNA) extraction was performed using a custom liquid handling protocol based on the Qiagen MagAttract PowerMicrobiome DNA/RNA Kit (Qiagen 27500–4-EP) adapted for lower volumes. Briefly, a 96 well plate (Axygen P-DW-20-C) was loaded with 1 mL of 0.1 mm Zirconia Silica beads (Biospec 11079101Z) using a loading device (Biospec 702L). During sample processing, appropriate negative controls were run on each plate (i.e. water control). 10 uL of thawed and concentrated spike-in strain was added to each well; for soil samples, the spike-in strain was diluted 1:25. 10–500 mg of each sample (average 45.9 mg, standard deviation 14.7 mg for fecal samples; average 298.5 mg, standard deviation 62.8 mg for soil samples) was added to the plate using a sterile plastic spatula, and the weight added for each sample was determined via an analytical balance. 750 μL of lysis solution was then added to each well (90 mL master mix, 9 mL 1M Tris HCl pH 7.5, 9 mL 0.5M EDTA pH 8.0, 11.25 mL 10% SDS, 22.5 mL Qiagen lysis reagent, 38.25 mL nuclease free water). The plate was centrifuged down for 1 min at 4500xg and a bead sealing mat was affixed to the plate (Axygen AM-2ML-RD). The plate was then placed on a bead beater (Biospec 1001) and subjected to bead beating for 5 min followed by 10 min for cooling. This bead beating cycle was repeated, for a total of 10 min of bead beating. The plate was centrifuged down for 5 min at 4500xg and 200uL of supernatant was transferred to a V-bottom microplate. 35 μL of Qiagen inhibitor removal solution was added to each well and mixed by vortexing, incubated 4 °C for 5 min, and the plate was again centrifuged down for 5 min at 4500xg. 100 μL of supernatant was removed from the plate and placed in a round-bottom plate (Corning 3795). The plate was then placed on a robotic liquid handler (Biomek 4000) for magnetic bead purification of the supernatant per the manufacturers recommendations but at a scaled volume; magnetic beads in binding solution were mixed in each well, and subjected to 3 washes with wash solution and elution in 100 uL of nuclease free water into a new plate.
16S rRNA amplicon sequencing
16S sequencing of the V4 region was performed utilizing a custom protocol and a dual indexing scheme adapted from Kozich et al1. Briefly, dual indexing sequencing primers were adapted from the previous study, but we utilized Illumina Nextera barcode sequences and altered 16S primers to match updated 505f and 806rB primers (see Table S2 for sequences). A 20 μL PCR amplification was set up in a 96 well skirted PCR microplate: 1 μM forward 5XX barcoded primer, 1 μM reverse 7XX barcoded primer, 1 μL prepared gDNA, 10 uL NEBNext Q5 Hot Start HiFi Master Mix (NEB M0543L), 0.2X final concentration SYBR Green I. A quantiative PCR amplification (98°C 30s; cycle: 98°C 20s, 55°C 20s, 65°C 60s, 65°C 5m) was performed and cycling was stopped during exponential amplification (typically 12–20 cycles) and the reaction was advanced to the final extension step.
The resulting PCRs were quantified utilizing a SYBR Green I dsDNA assay; 2 μL of PCR product was added to 198 μL of TE with 1X final concentration SYBR Green I and fluorescence was quantified on a microplate reader. Samples were pooled based on this quantification on a robotic liquid handler (Biomek 4000). The resulting ~390 bp amplicon from the pool was then gel-purified utilizing a 2% E-gel (Invitrogen) and Wizard SV gel extraction kit.
Final libraries were then quantified by Qubit dsDNA HS assay and sequenced on the Illumina MiSeq platform (V2 500 or 300 cycle kit) according to the manufacturers instructions with modifications. Specifically, the library was loaded at 10 pM with 20% PhiX spike-in, and custom sequencing primers were spiked into the MiSeq reagent cartridge (6 uL of 100 μM stock; well 12: read1, well 13: index1, well 14: read2).
Sequence analysis and OTU clustering
Resulting sequence data was analyzed using USEARCH1 version 9.2.64. Specifically, raw reads were merged using the –fastq_mergepairs command (for 2×250 reads, the options –fastq_maxdiffs 10 –fastq_maxdiffpct 10 were utilized). Merged sequences were filtered using the –fastq_filter command with options –fastq_maxee 1.0 and –fastq_minlen 240. Resulting sequences were dereplicated (–derep_fulllength), clustered into OTUs (–cluster_otus) and the merged reads were searched against OTUs sequences (–usearch_global) at 97% identity. Taxonomic assignments of OTUs were made using the RDP classifier2.
Whole-metagenome shotgun sequencing
The same genomic DNA utilized for 16S rRNA sequencing was subjected to metagenomic shotgun sequencing following a published protocol for low-volume Nextera library preparation3. Barcoded samples were pooled and sequencing was performed on the Illumina HiSeq platform (2×150 reads). Coverage was 4.25 ± 2.08 million reads (average ± s.d.) per sample.
Abundance estimation from metagenomic sequencing
We used Kraken4 to assign taxonomies to individual short reads, using a database of complete NCBI RefSeq bacterial genomes as well as the genome of Sporosarcina pasteurii, our spike-in strain. To estimate species level abundances, the fraction of total reads directly assigned to each reference genome was normalized by the total assembly length of that genome. Normalized read abundances were then summed over all reference genomes belonging to a given species. These summed abundances were then renormalized such that the total abundance of all detected species was equal to one.
Calculation of absolute taxa abundances
Bacterial load in each sample was calculated using the following formula:
where, Ri is the sequenced relative abundance of the spike-in strain in sample i, C0 is the constant amount of spike-in strain (units of total DNA copies) added to each sample, Wi is the weight of the fecal or soil sample i (mg), and ρi is the bacterial load per fecal/soil mass (DNA copies/mg). Solving for ρi,
where we have measured Ri and Wi experimentally. Note that relative changes in ρi are independent of the constant C0. We therefore scaled the bacterial loads within fecal or soil samples to a mean of unity. Relative abundance profiles (with the spike-in strain excluded) were then multiplied by this scaled quantity to obtain absolute OTU or species abundances in arbitrary units that were used for the analyses.
Assessment of the DIVERS spike-in sequencing approach to estimate absolute bacterial abundances
To assess the accuracy of the DIVERS spike-in approach in estimating absolute abundances, we performed a spike-in dilution series. Specifically, two fecal samples from different individuals were homogenized in 5X volume sterile PBS by vortexing, and passed through a 40 micron sterile filter. The fecal filtrate was then serially diluted 1:2 in sterile PBS to generate samples with exponentially decreasing fecal matter. Constant volumes (100 uL) of the undiluted and diluted samples were then subjected to the DIVERS spike-in sequencing approach as described previously.
Based on the above formula used to calculate bacterial loads, we derived a single relationship that described the expected behavior of sequenced spike-in strain abundances across the dilution series:
where R0 is the sequenced relative abundance of the spike-in strain in the original, undiluted fecal sample, and Ri is the relative abundance of the spike-in strain in the ith sample of the dilution series (i.e. sample i = 1 contains one half of the input fecal matter of the original sample). We show excellent agreement between expected and observed behavior in Supplementary Fig. 4c.
Variance decomposition of taxa abundances and bacterial loads
DIVERS utilizes the replicate sampling and sequencing protocol described above to decompose measured bacterial abundance variances. Let X denote the abundance of an individual species or OTU. Using the law of total variance, the variance of X can be written as a sum of three components representing temporal, spatial, and technical sources of variability:
| (1) |
where, S and T are space and time-associated random variables capturing the spatial and temporal processes affecting the abundance of X across samples. Following the notation in Fig. 1, each of the terms in equation (1) is estimated as follows (see Supplementary Note for full derivations):
| (2) |
| (3) |
| (4) |
where X, Z and Y, Z denote pairs of spatial replicate measurements of either bacterial load or individual OTU/species abundances. As described earlier, spatial replicates are obtained from two independent spatial locations in the environment at every time point. In contrast, X and Y denote technical replicates that are measured from the same spatial location.
Covariance decomposition of taxa abundances
Using the law of total covariance, the covariance between the abundances of any two taxa i and j, denoted Xi and Xj, can also be written as a sum of temporal, spatial and technical contributions:
| (5) |
Each of the terms in (5) is estimated using the replicate sampling and sequencing protocol as follows (see Supplementary Note for full derivations):
| (6) |
| (7) |
| (8) |
where, Xi, Zi and Yi, Zi denote spatial replicate measurements of the abundance of taxa i, and Xi, Yi denote technical replicates. To obtain temporal, spatial and technical correlations shown in Fig. 4, we normalize each covariance contribution by the respective standard deviations of individual taxa:
| (9) |
Variances and covariances of taxa abundances were calculated using data obtained across the twenty consecutive days of fecal sampling and twenty-seven soil sites. The variance decomposition of bacterial loads also included samples taken from days 27 and 48 of the times series. To minimize artifacts due to technical noise, only OTUs with a log10 mean absolute abundance >−4 and >−3.5 were included in the covariance decomposition analysis of fecal and soil samples respectively. These cutoffs was chosen based on the observed variance profiles of individual OTUs. To compare contributions across gut bacterial phyla, 16S rRNA sequence-based phylogenetic distances were calculated using the pairwise2 module of Biopython.
Two component variance and covariance decompositions
In certain cases, it may be useful to separate technical from non-technical (biological) sources of variability in microbiome studies. DIVERS can also perform a two component variance and covariance decomposition using the laws of total variance and covariance:
| (10) |
| (11) |
Note here that B is a random variable that now simultaneously captures the temporal and spatial factors affecting the abundance of taxon i.
Each term in equation (10) can be written as follows:
| (12) |
| (13) |
Terms in equation (11) can be written as:
| (14) |
| (15) |
As before, X and Y reflect technical measurements of the same biological sample. Not that the biological sources of variance and covariance now reflect both temporal and spatial factors. However, this interpretation is subject to change depending on the exact study design of experiments. See the Supplementary Note for full derivations and more in-depth discussion of the interpretation of each of the terms in equations (10) and (11).
Stochastic simulations of microbiota dynamics
To assess the performance of the DIVERS variance decomposition model, we carried out stochastic simulations of bacterial dynamics and measurement noise. We considered a community of interacting species on a 2D lattice, where at each time point, species were allowed to increase their abundance through birth, decrease their abundance through death, or migrate randomly to a neighboring location. These dynamics were governed by the following set of reactions:
| (16) |
| (17) |
| (18) |
| (19) |
where represents the abundance of species i at grid location (x,y), and , and are the respective per-capita birth rates, death rates and migration rates of species i at location (x,y). Migration rates for each species were chosen to be independent of spatial location. Per-capita birth and death rates were given by the following density-dependent logistic equation5:
| (20) |
| (21) |
| (22) |
where ri is the intrinsic growth rate of species i, is the carrying capacity of species i at location (x,y), and A is a matrix encoding interactions between community members (elements of A may be both positive or negative). To incorporate environmental stochasticity into our model, we multiplied species abundances by a Gaussian random variable at each time step: , with Finally, to simulate technical noise associated with experimental measurement error6, 7, we modeled final observed abundances as a Poisson random variable with mean and variance equal to
Simulations were carried out using the Gillespie algorithm on a 10 × 10 lattice with continuous boundary conditions. The following parameters were used for simulations: nspecies = 10, Ki ~ unif(100,500), νi ~ unif(0.5,2), ri ~ unif(0.2,0.5), Aij ~ unif(−0.2,0.5), ϵ = 3 * 10−4. The true temporal abundance variance for each species was calculated empirically as: , where is the average abundance of species i at time t across spatial locations, is the average abundance of species i over all time points and all spatial locations, and T is the length of the simulation. Similarly, the spatial abundance variance of each species was calculated empirically as: , where S is the number of considered spatial locations in the environment. Finally, technical variance was calculated empirically as (Supplementary Note).
Taking and to be technical replicates draw from the same spatial location at each time point t, and to be a single technical replicate drawn from a different spatial location, we then used equations (2–4) of the DIVERS variance decomposition model to estimate the temporal, spatial sampling and technical abundance variances of each species in the simluated community. In Supplementary Fig. 2, we compare these estimated variances using DIVERS to the quantities calculated empirically as described above.
Comparison of DIVERS to the Gaussian process variance decomposition model
Using synthetic data, we compared the performance of DIVERS to a recently described approach wherein a Gaussian process8 was used to model variability in measured bacteral abundances9.
We first simulated a time series of bacterial abundances using a generative statistical model, in which the true contibutions of temporal, spatial and technical contributions were inputs into the simulation. Specifically, average bacterial abundances across spatial locations in the environment at each time point {⟨xT=1⟩,⟨xT=2⟩…⟨xT=L⟩} were first drawn from a gamma distribution10 with mean equal to and variance . Note that the parameter is the true temporal variance that DIVERS and the Gaussian process model attempt to estimate. To model spatial abundance heterogeneity at any given time point t, we defined an additional gamma distribution with an average abundance equal to ⟨xT=t⟩, with variance equal to . This spatial abundance variance at each time point was itself drawn from a distribution with mean . Note again that the quantity is exactly the spatial abundance variance that both models estimate. We then generated a synthetic time series wherein we sampled twice from each of the spatial abundance distributions defined at every time point to simulate abundances at two different spatial locations . Finally, for a given time point t and spatial location s, we modeled technical variability associated with experimental measurement error using a Poisson random variable31 with mean and variance equal to xT=t,S=s. The true technical variability is then given by , which is equal to the technical variance at a given time point and spatial location, averaged over all time points and spatial locations. We sampled two technical replicates from each spatial location to represent two sequenced abundances from location s. Notably, the specific choice of distributions to model temporal, spatial, and technical variances is arbitrary.
We then used the DIVERS variance decomposition model to estimate the temporal, spatial, and technical contributions to simulated abundance variances. We used a simulation length of L = 20, the number of time points for which human fecal data was collected in this study. We also used the Gaussian process decomposition procedure to estimate the corresponding temporal, spatial, and technical variance contributions in simulations using publicly available code provided by the study of Lloyd-Price et. al.9. In the referenced study, the Gaussian process procedure was applied to a large cohort of individuals with sparsely sampled time points, while our simulations reflected a single, densely sampled time series. We therefore modified several of the terms in the Gaussian process covariance function used by Lloyd-Price et. al. Specifically, we set the interindividual variability contribution to zero and treated the “biological variablity” in the covariance function utlized in Lloyd-Price et. al. as the spatial sampling variablity of interest. Other terms remained unchanged. All MCMC sampling parameters used in the Gaussian process inference procedure were taken directly from Lloyd-Price et. al.9
Removal of temporal or spatial variability from fecal samples
We conducted two sets of control experiments to remove either temporal or spatial variability of OTU abundances from fecal samples. Specifically, to eliminate temporal contributions, we re-sampled a single stool specimen ten times total to simulate five consecutive days of time series sampling. To eliminate spatial variability, replicate sampling was conducted for eight consecutive days; on each day, fecal samples obtained from random spatial locations were homogenized together by combining fecal samples, and then mechanically homogenizing in 1X PBS with a P200 pipette tip. The resulting homogenized sample was then split into technical triplicates and processed following the normal DIVERS protocol.
Code availability
MATLAB scripts to perform all variance and covariance decomposition analyses from original OTU abundance tables are available on GitHub at https://github.com/brianwji/DIVERS. Implementation of DIVERS in R is available on GitHub at https://github.com/hym0405/DIVERS.
Data availability
Sequencing data is available at NCBI SRA under PRJNA541083.
Supplementary Material
Acknowledgments
H.H.W. acknowledges funding from the NIH (R01AI132403, R01DK118044), Burroughs Wellcome Fund (PATH 1016691), Bill & Melinda Gates Foundation (INV-000609), and the Schaefer Research Scholars Program for this work. R.U.S. is supported by a Fannie and John Hertz Foundation Fellowship and a NSF Graduate Research Fellowship (DGE-1644869). BJ is supported in part by the NIH under Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (T32GM007367) and by the MD-PhD program at Columbia University. DV acknowledges funding from the NIH (R01GM079759, R01DK118044).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenhower C et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segata N et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9, 811–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson LR et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt DE et al. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Faust K, Lahti L, Gonze D, de Vos WM & Raes J Metagenomics meets time series analysis: unraveling microbial community dynamics. Curr Opin Microbiol 25, 56–66 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Martin-Platero AM et al. High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat Commun 9, 266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandeputte D et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Friedman J & Alm EJ Inferring correlation networks from genomic survey data. PLoS Comput Biol 8, e1002687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stammler F et al. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 4, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tkacz A, Hortala M & Poole PS Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zmora N et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 174, 1388–1405 e1321 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Gohl DM et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34, 942–949 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Langille MG et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faust K & Raes J Microbial interactions: from networks to models. Nat Rev Microbiol 10, 538–550 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Sonnenburg ED et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg ED et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakoff-Nahoum S, Foster KR & Comstock LE The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakoff-Nahoum S, Coyne MJ & Comstock LE An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24, 40–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez KS et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc Biol Sci 281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien SL et al. Spatial scale drives patterns in soil bacterial diversity. Environ Microbiol 18, 2039–2051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carini P a.D.-B., M and Hinckley, E and Brewer, TE and Rue, G and Vanderburgh, C and McKnight, D and Fierer, N Unraveling the effects of spatial variability and relic DNA on the temporal dynamics of soil microbial communities. bioRxiv, 402438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fierer N & Jackson RB The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contijoch EJ et al. Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wargo JA, Reddy SM, Reuben A & Sharma P Monitoring immune responses in the tumor microenvironment. Curr Opin Immunol 41, 23–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood DE & Salzberg SL Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15, R46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 1.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Garrity GM, Tiedje JM & Cole JR Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baym M et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood DE & Salzberg SL Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15, R46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilpatrick AM & Ives AR Species interactions can explain Taylor’s power law for ecological time series. Nature 422, 65–68 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Grun D, Kester L & van Oudenaarden A Validation of noise models for single-cell transcriptomics. Nat Methods 11, 637–640 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Marioni JC, Mason CE, Mane SM, Stephens M & Gilad Y RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18, 1509–1517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams C.a.R., CE Gaussian processes for machine learning, Vol. 2 (MIT Press, Cambridge, MA; 2006). [Google Scholar]
- 9.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sala C et al. Stochastic neutral modelling of the Gut Microbiota’s relative species abundance from next generation sequencing data. BMC Bioinformatics 17 Suppl 2, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is available at NCBI SRA under PRJNA541083.