Distinct gene repertoires are expressed during Y. pestis infection of its flea and mammalian hosts. The functions of many of these genes remain predicted or unknown, necessitating their characterization, as this may provide a better understanding of Y. pestis specialized biological adaptations to the discrete environments of its two hosts. This study provides functional context to adjacently clustered horizontally acquired genes predominantly expressed in the flea host by deciphering their fundamental processes with regard to (i) transcriptional organization, (ii) transcription activation signals, and (iii) biochemical function. Our data support a role for these genes in osmoadaptation and aromatic amino acid metabolism, highlighting these as preferential processes by which Y. pestis gene expression is modulated during flea infection.
KEYWORDS: Yersinia pestis, flea infection, hyperosmolarity stress
ABSTRACT
While alternating between insects and mammals during its life cycle, Yersinia pestis, the flea-transmitted bacterium that causes plague, regulates its gene expression appropriately to adapt to these two physiologically disparate host environments. In fleas competent to transmit Y. pestis, low-GC-content genes y3555, y3551, and y3550 are highly transcribed, suggesting that these genes have a highly prioritized role in flea infection. Here, we demonstrate that y3555, y3551, and y3550 are transcribed as part of a single polycistronic mRNA comprising the y3555, y3554, y3553, y355x, y3551, and y3550 genes. Additionally, y355x-y3551-y3550 compose another operon, while y3550 can be also transcribed as a monocistronic mRNA. The expression of these genes is induced by hyperosmotic salinity stress, which serves as an explicit environmental stimulus that initiates transcriptional activity from the predicted y3550 promoter. Y3555 has homology to pyridoxal 5′-phosphate (PLP)-dependent aromatic aminotransferases, while Y3550 and Y3551 are homologous to the Rid protein superfamily (YjgF/YER057c/UK114) members that forestall damage caused by reactive intermediates formed during PLP-dependent enzymatic activity. We demonstrate that y3551 specifically encodes an archetypal RidA protein with 2-aminoacrylate deaminase activity but Y3550 lacks Rid deaminase function. Heterologous expression of y3555 generates a critical aspartate requirement in a Salmonella enterica aspC mutant, while its in vitro expression, and specifically its heterologous coexpression with y3550, enhances the growth rate of an Escherichia coli ΔaspC ΔtyrB mutant in a defined minimal amino acid-supplemented medium. Our data suggest that the y3555, y3551, and y3550 genes operate cooperatively to optimize aromatic amino acid metabolism and are induced under conditions of hyperosmotic salinity stress.
IMPORTANCE Distinct gene repertoires are expressed during Y. pestis infection of its flea and mammalian hosts. The functions of many of these genes remain predicted or unknown, necessitating their characterization, as this may provide a better understanding of Y. pestis specialized biological adaptations to the discrete environments of its two hosts. This study provides functional context to adjacently clustered horizontally acquired genes predominantly expressed in the flea host by deciphering their fundamental processes with regard to (i) transcriptional organization, (ii) transcription activation signals, and (iii) biochemical function. Our data support a role for these genes in osmoadaptation and aromatic amino acid metabolism, highlighting these as preferential processes by which Y. pestis gene expression is modulated during flea infection.
INTRODUCTION
Plague, caused by Yersinia pestis, is a severe zoonotic disease infamous for having caused the death of millions of people in three major pandemics. This disease primarily affects rodents which become infected mainly through blood feeding by rodent-associated infected fleas. Transmission by fleas is a recent adaptation that has likely evolved in the last 1,500 to 20,000 years during the transformation of Y. pestis from its clonal ancestor, Yersinia pseudotuberculosis, which is transmitted by the fecal-oral route and causes a mild enteric self-resolving illness (1–3). Central to the biological mechanism of transmission via flea bite is Y. pestis biofilm blockage of the flea foregut proventriculus (4–6).
After acquisition from the blood of a highly bacteremic host, Y. pestis must adapt to the insect gut environment to multiply and form biofilm-mediated blockage by 7 days postinfection (7). The various physicochemical stresses Y. pestis encounters in the flea gut, such as low pH, reactive oxygen species (ROS), hyperosmolarity, nutrient limitation, and the immune response, are slowly being uncovered, along with how Y. pestis overcomes such stresses (8, 9). For example, reactive oxygen species (ROS) have been demonstrated to limit Y. pestis survival during early flea infection (10), but this oxidative stress can be counteracted by the modulation of antioxidant defenses by the Y. pestis OxyR transcriptional regulator (10). The flea gut was determined as a mildly acidic environment with a pH of ∼6.5 to 6.8 (11). Implicated in adapting to this mild acidic condition is the PhoP transcriptional regulator (12) required to maintain optimal blockage and competitive fitness in fleas and that modulates the transcription of genes required for acid stress (12–14).
Moreover, hyperosmotic stress conditions occur in the flea gut, as its content is measured to be ∼500 mOsm, much greater than that of mouse blood at 320 mOsm (11). However, genes of Y. pestis belonging to pathways involved in alleviating osmotic stress are robustly expressed during flea infection (15–17). Examples of these are (i) genes belonging to the uptake and catabolic pathways of the l-Glu amino acid group (His, Glu, Pro, and Arg) which encodes proteins that generate the osmoprotectant glutamate (17), and (ii) the betT and proVWX genes of the synthesis and transport system operons for the osmoprotectant glycine-betaine, respectively (15). In addition, the two-component signal transduction regulatory system OmpR-EnvZ that is involved in sensing and mediating adaptation to osmolarity and pH stresses in many bacterial species (18–21), including Y. pestis (22–24), is shown to be required for efficient flea blockage (11).
To further understand the flea host-specific response of Y. pestis, we turned our attention to a cluster of genes (y3555, y3551, and y3550) with lower GC content (39.8%) than that typically noted for Y. pestis (48%) genes and that ranked in the top 150 most highly expressed genes in the flea (17). Close inspection of the chromosomal context of y3555, y3551, and y3550 revealed that they are located adjacent to three other genes (y3554, y3553, and y355x) having a similarly low GC content. The y3554, y3553, and y335x genes showed robust expression in the flea gut (9, 17). The y355x gene is not annotated as such in the Y. pestis KIM10+ genome, but it is 100% identical to ypo0625 of the Y. pestis CO92 genome, the reference to which the microarrays were developed for Y. pestis flea gut infection transcriptomics studies (12, 17). All six of these low-GC-content genes were either poorly expressed or not detected under mammalian conditions. Genes sharing 99 to 100% identity to y3555, y3554, y3553, y355x, y3551, and y3550 are present in Y. pseudotuberculosis. Because genetic loss is thought to drive clonal evolution enabling the emergence of Y. pestis and its flea-borne transmissibility (25–28), we reasoned that evolutionary conservation of the encoded traits of these six genes is likely advantageous for growth and survival within new environmental niches like the flea gut. This study therefore aimed to understand the transcriptional organization and activation, as well as the roles of the y3555, y3554, y3553, y335x, y3551, and y3550 genes, in flea colonization, with a specific emphasis on the biochemical roles of the most highly expressed genes, y3555, y3550, and y3551. Our data reveal that while these preferentially expressed genes that have been evolutionarily maintained are not essential for flea colonization in the context of our experimental laboratory flea model of Y. pestis infection, they are nonetheless coordinately induced during hyperosmotic stress and contribute to aromatic amino acid metabolism, and y3555 confers enhanced growth fitness traits to Escherichia coli and Y. pestis in vitro.
RESULTS
In silico analysis of the genetic context and predicted functions of Y. pestis KIM10+ genes y3555, y3554, y3553, y355x, y3551, and y3550.
The lower GC contents of the gene cluster comprising y3555 (3,935,132 and 3,936,364), y3554 (3,933,688 and 3,935,139), y3553 (3,933,238 and 3,933,684), y355x (3,932,478 and 3,933,068), y3551 (3,932,082 and 3,932,462), and y3550 (3,931,393 and 3,931,797) suggest their acquisition by horizontal gene transfer. In silico analysis using FGENESB predicts that the continuous region of this genomic island comprises three transcriptional units, with y3555-y3554-y3553 and y355x-y3551 constituting two separate operons and y3550 representing a monocistronic transcriptional unit. Additionally, BPROM identifies distinct promoter sequences upstream of the genes y3555, y355x, and y3550 (Fig. 1A). BLAST searches were used to identify homologous proteins and conserved functional domains of genes comprising the genomic island.
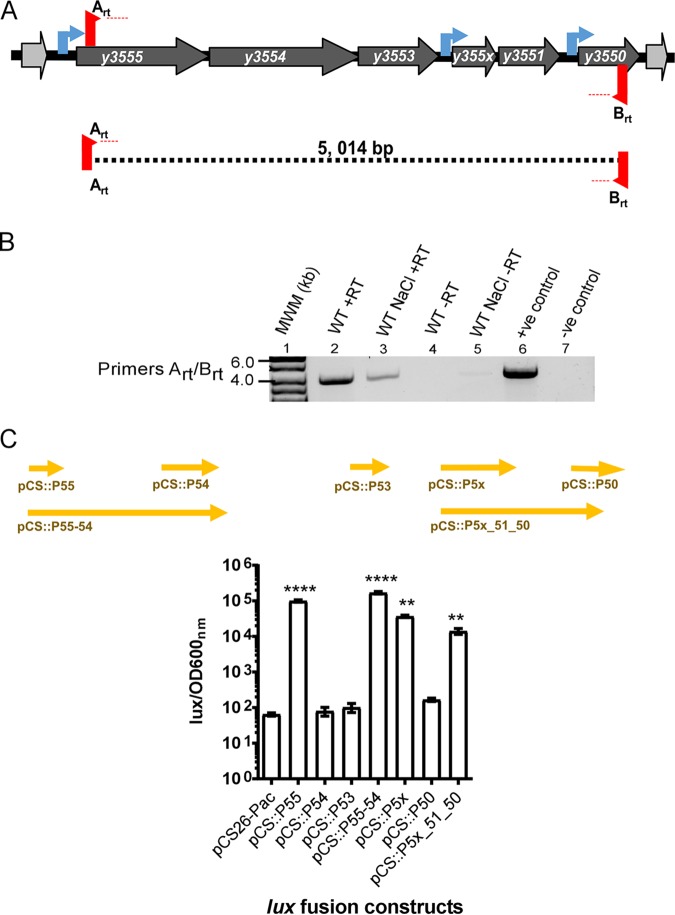
FIG 1.
The low-GC-content gene cluster is transcribed as a single polycistronic mRNA. Six genes compose the low-GC-content gene cluster. (A) The three bioinformatically predicted transcriptional units are marked by the presence of predicted promoters upstream of genes y3555, y355x, and y3550 (blue arrows). (B) Reverse transcription-PCR (RT-PCR) analysis of regions (indicated by dashed lines) spanning y3555 to y3550. RT-PCRs were performed with reverse transcriptase and DNA polymerase mix (lanes 2 and 3) or only with DNA polymerase (lanes 4 to 7). PCRs in the presence of Y. pestis KIM6+ chromosomal DNA (lane 6) or absence of template (lane 7) with the same set of primers were performed as controls. MWM, molecular weight marker; +ve, positive; -ve, negative. (C) The activity of transcriptional fusion plasmids pCS::P55, pCS::P54, pCS::P53, pCS::P55_54, pCS::P5x, pCS::P50, and pCS::P5x_51_50, carrying the transcriptional fusions of the luciferase (lux) genes and predicted promoter regions (arrows) representing predicted regulatory regions of y3555, y3554, y3553, y3555-y3554, y355x, y3550, and y355x-y3551-y3550, respectively, as well as the low-copy-number empty vector pCS26-Pac. Error bars represent mean (±standard deviation [SD]) of the results from three independent experiments. Significance determined by one-way analysis of variance (ANOVA) and a Dunnett’s posttest of lux/OD values of transcriptional fusion constructs compared to the empty vector is indicated: ****, P < 0.0001, and **, P < 0.01.
The y3555 gene product exhibits homology to uncharacterized pyridoxal 5′-phosphate (PLP)-dependent transaminases, including aromatic and aspartate aminotransferases encoded in some bacteria. These PLP-dependent aminotransferases typically catalyze the interconversion of an aromatic amino acid and α-ketoglutarate to oxaloacetate and glutamate. The Y. pestis KIM10+ genome encodes two PLP-dependent aromatic aminotransferases, encoded by y2760 (aspC) and y0579 (tyrB).
The y3554 gene encodes a putative Na+/H+ antiporter (NhaC type), and there are two other antiporters (NhaA and NhaB) in the reported genome sequence of Y. pestis. A primary role for Na+/H+ cation exchange antiporters (29–31) is to extrude Na ions to counterbalance Na+ toxicity, thereby protecting against hyperosmotic saline stress. The NhaA and NhaB antiporters are essential for Y. pestis virulence in a mouse model of septicemic plague (31, 32), where they are thought to function in protecting Y. pestis against Na+ toxicity in blood.
The y3553 gene product is predicted to belong to the PYR/PYL/RCAR-like family of proteins, a part of the SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) domain protein superfamily, that are known to bind hydrophobic ligands and mediate signal transduction (33, 34). PYR/PYL/RCAR proteins are particularly well characterized in plants, where they bind phytohormones that enable the regulation of growth, development, and environmental stress responses. For example, osmotic stress responses and high salt tolerance are mediated by PYR/PYL/RCAR protein binding interactions with plant phytohormones (34–36).
The gene product of the y355x gene is predicted to be a transcriptional regulator because it contains a helix-turn-helix motif and Per-ARNT-Sim (PAS) sensory domain. PAS domains are ubiquitous in bacterial proteins involved in signal transduction and enable sensing of physical and chemical stimuli, like oxygen and redox potential (37).
The y3551 and y3550 gene products belong to the reactive intermediate deaminase (Rid) protein superfamily, originally called the YjgF/YER057c/UK114 family of proteins. Rid enzymes eliminate reactive intermediates that are produced by PLP-dependent enzymatic activity, which can alter the function of cellular targets and prevent the inhibition of transaminases (38–40). Interestingly, some Rid-encoding genes cluster with genes encoding PLP-dependent enzymes, like aminotransferases, which are susceptible to damage by the enamine 2-aminoacrylate (2AA) (41). The clustering of y3551 and y3550 with y3555, predicted to encode an aminotransferase, is consistent with this clustering pattern.
The genes y3555, y3554, y3553, y355x, y3551, and y3550 are transcribed as a polycistronic mRNA.
Gene clustering in bacteria often implies a functional association of its member genes. Previous transcriptomic analyses have shown that the y3555, y3554, y3553, y3551, and y3550 genes exhibit coordinated gene expression in the flea gut and under similar in vitro conditions (12, 17, 24). To determine if the coordinated expression of this genomic island can occur from a single long polycistronic mRNA, a two-step reverse transcription-PCR (RT-PCR) using Y. pestis parental cDNA to amplify the entire locus from y3555 to y3550 (Fig. 1B) was performed. A negative control to verify the absence of genomic DNA contamination was included by omitting reverse transcriptase during cDNA synthesis (Fig. 1B, lane 7), and a positive control using Y. pestis chromosomal DNA as the template with DNA polymerase verified the expected amplicon size (Fig. 1B, lane 6). As shown in Fig. 1B, a single 5,014-bp predicted fragment spanning all six genes from y3555 to y3550 was amplified. Together, these results confirmed that the y3555, y3554, y3553, y355x, y3551, and y3550 genes are transcribed as a long single polycistronic mRNA transcript.
Next, to determine if the predicted promoters upstream of y3555, y355x, and y3550 drive the expression of their respective downstream genes, transcriptional fusions spanning different regions of the genomic island to the luxCDABE reporter genes (Fig. 1C) were used. Luminescence directed by the plasmid carrying these transcriptional fusions was determined in the parental Y. pestis strain grown at 26°C to simulate conditions within the flea. Strains harboring fusions to the promoter of y3555 (pCS::P55) and y355x (pCS::P5x) showed ∼102- to 103-fold higher levels of luminescence expression than those of the strains carrying the empty vector (pCS26-Pac) or transcriptional fusions with potential regulatory regions of y3554 (pCS::P54), y3553 (pCS::P53), and y3550 (pCS::P50). This suggests that only the upstream regulatory regions of y3555 and y355x, and not y3550, are able to promote gene transcription using flea-matched temperature testing conditions. To confirm this observation, we generated two other transcriptional fusion constructs that allowed testing of transcription directed from the predicted upstream regulatory regions of y3555 or y355x. The transcriptional fusion construct pCS::P55-54 contains the region covered in pCS::P54 extended up to the regulatory region of y3555, and the construct pCS::P5x-51-50 contains the region of fusion pCS::P50 extended up to the regulatory region of y355x (Fig. 1C). The expression of the pCS::P55-54 and pCS::P5x-51-50 fusions was drastically increased and similar to that shown in fusions P55 and P5x, respectively. (Fig. 1C). This suggested that the promoter located upstream of y3555 can drive the expression of y3555-y3554-y3553-y355x-y3551-y3550 as a single operon and that y355x-y3551-y3550 can be transcribed as another operon from the promoter located upstream of the y355x gene.
Neither single-gene nor whole-genomic-island mutant strains exhibit fitness attenuation in competitive coinfection with the parental strain.
To test if the most highly expressed genes, y3555 and y3550, contributed to efficient colonization and formation of proventricular blockage of the flea, we constructed and tested mutants lacking the y3555 or y3550 gene alone, y3551 and y3550, y3550 and y3555, and the entire locus comprising y3555-y3554-y3553-y355x-y3551-y3550 (referred to as the “all” mutant). A previously reported y3555-y3554-y3553 mutant was also used (42). The six mutants displayed parental abilities to infect and block fleas (data not shown). Observing no difference in single flea infection, a more sensitive competitive coinfection assay (43) designed to discern the fitness of a mutant strain relative to the parental strain was performed. Fleas were coinfected with a 1:1 ratio of mutant to parental strain, harboring a kanamycin resistance cassette in the nondeleterious glmS-pstS site and called wild type (WT) glmS-pstS::Kanr, which was previously shown to exhibit equal fitness during competition with an unmarked parental strain (43). All mutants tested in coinfections with the glmS-pstS::Kanr WT strain were maintained at rates equal to that of the glmS-pstS::Kanr WT, as reflected by constant occurrence of the glmS-pstS::Kanr strain population at a mean of ∼50% of the total population of coinfecting Y. pestis immediately after infection (T0) and at 7 days (T7) postinfection (Fig. 2). These in vivo assays showed the deletion mutations did not have attenuated fitness in the flea.
FIG 2.
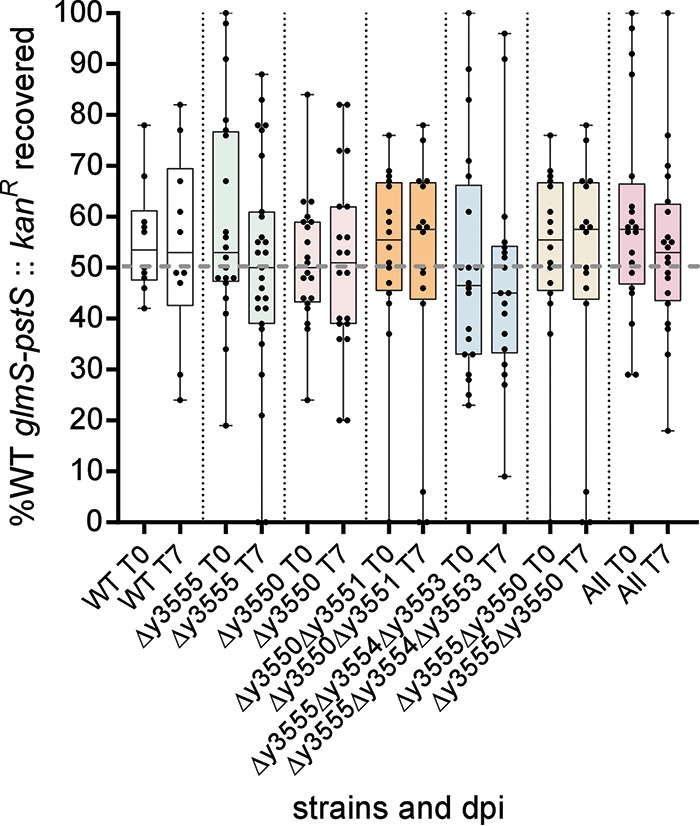
Y. pestis low-GC-content gene mutants coinfected with the parental strain do not exhibit competitive fitness defects in fleas. Fleas were infected using a 1:1 ratio of mutant to parental strains harboring a kanamycin resistance cassette in the chromosome (WT glmS-pstS::Kanr). The percent WT glmS-pstS::Kanr from the total coinfecting Y. pestis population per flea (dots) at days 0 and 7 is reflected. The error bars represent the mean plus the minimum and maximum percent WT glmS-pstS::Kanr per flea. The data were analyzed using an unpaired Student t test. dpi, days postinfection.
Predicted promoters located upstream of y3555, y355x, and y3550 are responsive to hyperosmotic salinity stress.
In a previous study, the genome-wide transcriptional responses of the enzootic strain Y. pestis bv. Microtus to hyperosmotic stress mediated by high salt found that genes homologous to the Y. pestis KIM6+ y3555, y3554, y3553, y355x, y3551, and y3550 genes are upregulated (24). This study suggested that the genomic island is responsive to hyperosmotic stress. To determine if the y3555, y3554, y3553, y355x, y3551, and y3550 genes were induced in response to hyperosmolarity stress in the KIM6+ epidemic strain, we exposed this strain to 0.5 M NaCl (reflects a high osmolarity of 1,000 mOsm) and determined the steady-state mRNA expression levels of these genes. The expression levels for the genes y3555, y3554, and y3553 showed between 1.5- and 2.0-fold increases, while the expression of the genes determined to compose the second operon, i.e., y355x-y3551-y3550, showed between 4- and 6-fold increases in gene expression (Fig. 3A).
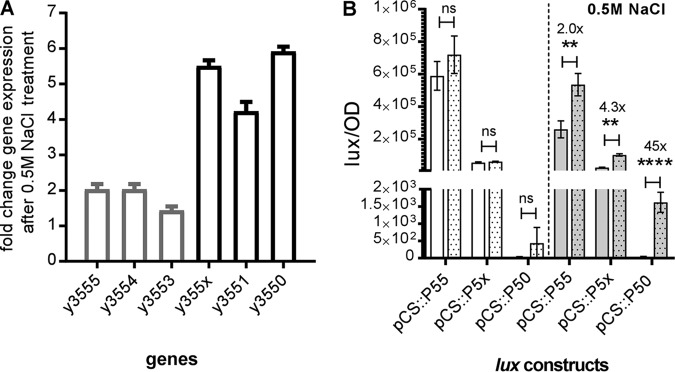
FIG 3.
Gene expression of low-GC-content genes is induced during exposure to saline hyperosmotic stress. (A) Steady-state mRNA transcript levels of low-GC-content genes in a Y. pestis KIM6+ parental strain treated or not treated with 0.5 M NaCl were determined. The data are represented as the fold change in gene expression of treated versus untreated mRNA transcript levels. Error bars represent the mean (±SD) of the results from 3 independent experiments. (B) Activities of the transcriptional fusion plasmids pCS::P55, pCS::P5x, and pCS::P50 carrying the transcriptional fusions of the luciferase genes and predicted promoter regions of y3555, y355x, y3550, respectively, pre-treatment with vehicle (white bars) or 0.5 M NaCl (gray bars) and 100 min post-treatment with vehicle (white dotted bars) or 0.5 M NaCl (gray dotted bars). Error bars represent the mean (±SD) of the results from three independent experiments. Data were analyzed using Student's t test; **, P < 0.01; ****, P < 0.0001; ns, not significant.
Additionally, the induction of lux transcriptional reporter fusion activity was assessed for the pCS::P55, pCS::P5x, and pCS::P50 transcriptional fusion reporter constructs to determine if reporter activity increased after NaCl exposure. This time readings were taken 100 min after exposure to 0.5 M NaCl (Fig. 3B). Interestingly, while the Lux reporter activity from pCS::P55 and pCS::P5x showed 2- and 4-fold increases, respectively, a remarkable 45-fold increase in Lux reporter activity from pCS::P50 was observed following exposure to 0.5 M NaCl. These data indicated that, while the promoters upstream of y3555 and y355x promote transcription at flea-matched temperatures, they exhibit increased transcriptional activity under hyperosmotic/saline conditions, whereas the promoter upstream of y3550 is active only under hyperosmotic/saline conditions. RT-PCR analysis (Fig. 1B) indicates that the genomic island continues to be expressed as a single long transcript following exposure to 0.5 M NaCl.
Y3555 generates a critical requirement for aspartate in a Salmonella enterica aspC mutant.
The ridA and aspC genes, encoding aspartate aminotransferases, cluster in many bacterial chromosomes (41). Some online genome resources, including KEGG, Phyre2 (44), and BLAST, predict that Y3555 is an aspartate aminotransferase, and for this reason, aspartate was considered as a possible substrate for the enzyme. However, based on structural alignments, it appears that Y3555 has a glutamate instead of an aspartate residue in the active site to coordinate the pyridine nitrogen of PLP. These initial alignments suggest Y3555 may be a fold type IV aminotransferase (45), not a fold type I aminotransferase like AspC (45) (see Fig. S1 in the supplemental material).
Nonetheless, to experimentally query if Y3555 had indeed predicted aspartate aminotransferase activity, the ability of y3555 to restore the growth of an S. enterica aspC789::Km mutant strain was tested. Growth of the S. enterica aspC789::Km mutant containing either the empty pTRc99A plasmid or plasmid pTRc_55, carrying an isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible y3555 gene, was determined (Fig. 4). The S. enterica aspC789::Km mutant has a growth defect in glucose minimal medium, since it relies on low-level promiscuous aspartate aminotransferase activity of the tyrosine aminotransferase TyrB. The S. enterica aspC789::Km mutant strains containing either the empty pTRc99A plasmid or plasmid pTRc_55 had similar growth on minimal glucose medium with aspartate. Expression of the aspC gene from S. enterica allows full growth of the mutant strain (Fig. S2). Surprisingly, in the absence of aspartate, the addition of 100 μM IPTG, presumed to induce the expression of y3555, inhibited the growth of the S. enterica aspC789::Km mutant (Fig. 4). This growth defect was eliminated with the addition of aspartate, suggesting that the critical aspartate requirement was facilitated by Y3555 expression. There are a few possibilities for this effect that cannot be distinguished by the data here, including that Y3555 could affect TyrB flux or produce tyrosine and phenylalanine that shut down TyrB expression and/or activity (46–48).
FIG 4.
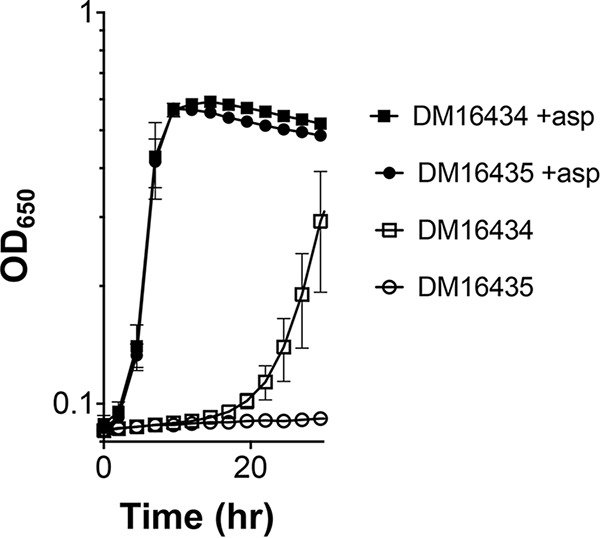
Induction of y3555 generates an aspartate requirement for an S. enterica aspC789::Km mutant. S. enterica aspC789::Km mutants expressing y3555 DM16435 or the empty vector control DM16434 were grown in minimal glucose (11 mM) medium plus IPTG (100 μM), with (+asp) or without aspartate (1 mM). Error bars represent the mean (±standard error of the mean [SEM]) values from three biological replicates.
Y3551, but not Y3550, is a functional RidA deaminase.
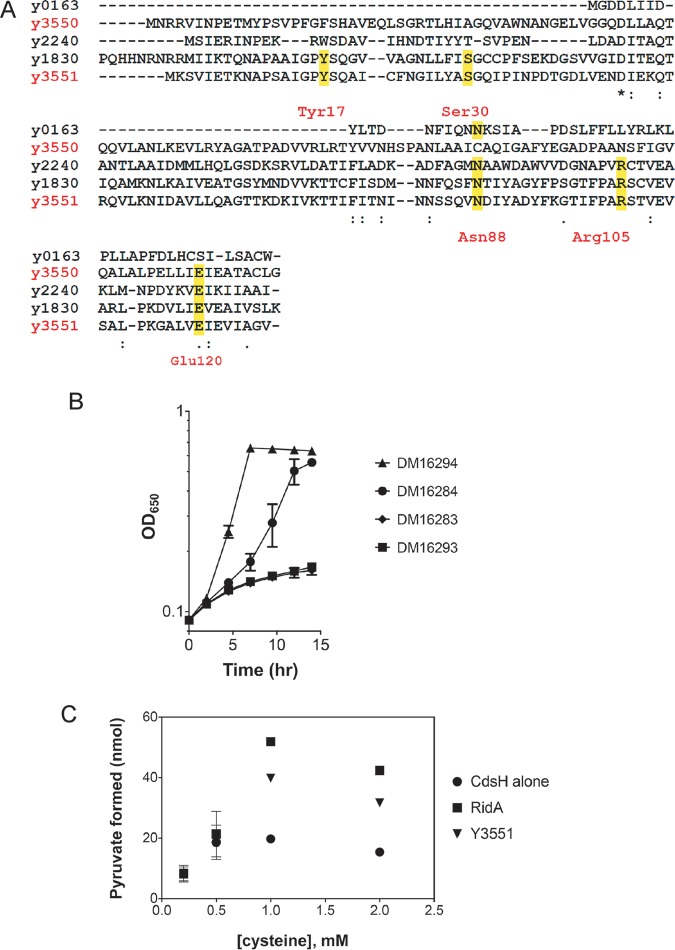
Proteins of the Rid superfamily are defined as belonging to the archetypal RidA subfamily or Rid1 to -7 subfamilies based on phylogenetic analysis (41). RidA enzymes hydrolyze reactive imine/enamine metabolic intermediates (i.e., 2-aminoacrylate [2AA]) (40), which are generated as obligate catalytic intermediates by some PLP-dependent enzymes. An active-site arginine (Arg105) is critical for RidA imine hydrolysis activity (40). Rid4 to -7 subfamily members lack the active-site arginine and an ascribed biochemical function. Amino acid alignment of the Y3550 and Y3551 sequences with Rid family homologs (Fig. 5A) revealed that Y3551 had the requisite arginine in the active site. Y3550 is missing an active-site arginine, contains a phenylalanine at position 17 rather than the typical tyrosine, and has a variable asparagine at position 88. These features of Y3550 conform to those described previously for Rid5 or Rid6 proteins (41).
FIG 5.
Y3551 is a RidA enzyme. (A) Multiple-sequence alignment of all Y. pestis KIM6+ Rid family homologs. Amino acids highlighted in yellow represent conserved residues found in archetypal RidA subfamily members. y3551 complements an S. enterica ridA mutant. (B) The S. enterica ridA1::Tn10 (Tc) mutant carried the empty vector control DM16293 or plasmid pET28b expressing E. coli ridA DM16294, y3551 DM16284, or y3550 DM16283. Strains were grown in a 96-well plate at 37°C shaking in minimal NCE glucose (11 mM) with serine (5 mM). Error bars represent the mean (±SEM) from three biological replicates. (C) Y3551 deaminates 2-aminoacrylate in vitro. Purified S. enterica cysteine desulfhydrase (CdsH) was used to generate 2AA from cysteine. Reactions with or without the addition of Y3551 or S. enterica RidA measured coupled pyruvate formation and NADH oxidation, as previously described (Ernst et al. [67] and Kredich [69]). NADH oxidation was used as a measurement of pyruvate formation. RidA enzymes deaminate 2AA in vitro, increasing the reaction rate. The initial reaction rate of pyruvate formation is plotted versus the concentration of cysteine. As a control, CdsH alone or S. enterica RidA was used. Y3551 deaminated 2AA derived from cysteine. Error bars represent the standard error of the mean from technical triplicates.
To test if Y3551 and/or Y3550 had the deaminase activity conserved for RidA enzymes, the S. enterica ridA1::Tn10 (Tc) mutant strain was transformed with the plasmid constructs pET28b::ridA, pET28a::y3551, and pET28a::y3550. Minimal defined medium supplemented with serine was used for growth analysis. In the S. enterica ridA1::Tn10 (Tc) mutant, 2AA is generated from serine by the serine/threonine dehydratase IlvA (EC 4.3.1.19). In the absence of RidA, 2AA accumulates and damages some PLP-dependent enzymes, which can result in growth defects (49). Growth was monitored to determine if Y3551 and/or Y3550 could restore growth to the S. enterica ridA1::Tn10 (Tc) mutant. The S. enterica ridA1::Tn10 (Tc) mutant strain carrying the empty vector control failed to grow in this medium, and as expected, it was complemented by the pET28b::ridA plasmid. Significantly, the pET28a::y3551 plasmid restored significant growth to the strain, indicating that Y3551 had 2AA deaminase activity (Fig. 5B). However, the pET28a::y3550 plasmid failed to restore growth to the S. enterica ridA1::Tn10 (Tc) mutant strain (Fig. 5B). Control experiments confirmed that the addition of isoleucine restored full growth to all strains, confirming that the weak growth was due to 2AA accumulation (data not shown).
To confirm that Y3551 had 2AA hydrolysis activity, as indicated by the complementation analyses, assays were done with pure proteins. Cysteine desulfhydrase (CdsH) converts cysteine to 2AA, which can be deaminated to pyruvate by solvent water or a RidA enzyme. Thus, RidA activity is detected as an accelerated rate of pyruvate formation in a CdsH assay. The assay was performed using purified Y3551 and S. enterica RidA enzymes, and the data are shown in Fig. 5C. In the presence of either purified S. enterica RidA or Y3551, the initial rate of pyruvate formation was accelerated over the control lacking RidA when the cysteine concentration was 1 mM or greater (Fig. 5C). These data confirmed that Y3551 is a RidA family enzyme with 2AA hydrolysis activity.
One other disparate biochemical activity previously assigned to Rid enzymes is that of an endoribonuclease (41, 50). The Y3551 and Y3550 proteins were in fact previously designated l-PSP family endonucleases by BLAST analyses (12, 17). However, our purified Y3551 and Y3550 did not show any RNase activity (data not shown), supporting the idea that these proteins do not encode such a function in Y. pestis.
Overexpression of y3555 enhances the growth of E. coli and Y. pestis.
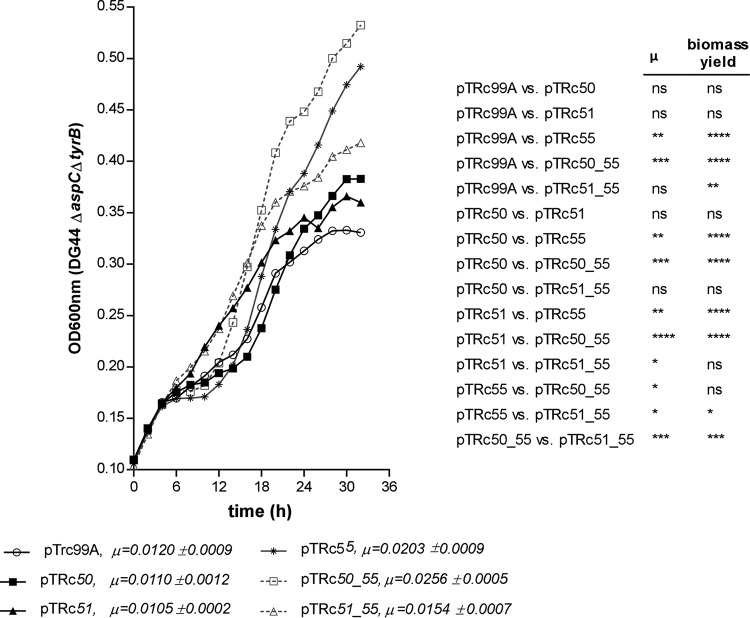
The uncharacterized protein Y3555 enhanced the growth of Y. pestis strains grown in rich medium. The Y. pestis y3555 and y3550 mutants expressing Y3555 from a high-copy-number plasmid (pCR4-TOPO) grew significantly faster than did the wild-type strain grown under the same conditions (Fig. 6). A similar observation was made in an E. coli aspartate (aspC) and tyrosine (tyrB) aromatic aminotransferase double mutant that requires either aspartate or tyrosine for growth (46, 51). This strain, called DG44, was used in early trial experiments replicating those in the S. enterica aspC359::Km mutant, but those experiments were abandoned due to a lack of genetic complementation tools. It has been suggested that Rid enzymes may be encoded by genes for PLP-dependent enzymes that are targets of 2AA damage, including branched-chain amino acid transaminases and aspartate aminotransferases (41). These reports, along with the gene synteny of y3550, y3551, and y3555, suggest that either y3551 or y3550 may prevent the accumulation of a toxic metabolic intermediate of y3555. The improvement in growth observed with the overexpression of y3555 presented an opportunity to test if y3550 or y3551 expression combined with that of y3555 could further improve the growth kinetics of the DG44 strain. The DG44 strain was transformed with inducible plasmids carrying a single y3555, y3550, or y3551 gene and either the y3555 and y3550 genes or the y3555 and y3551 genes. Significantly higher growth rate and biomass yield were observed for the DG44 strain expressing y3555 (μ = 0.0203 ± 0.0009) but not y3550 (μ = 0.0110 ± 0.0012) and y3551 (μ = 0.0105 ± 0.0002) relative to the empty vector-containing strain (μ = 0.0120 ± 0.0009) (Fig. 7). The coexpression of y3555 with y3550 (μ = 0.0256 ± 0.0005) or y3551 (μ = 0.0154 ± 0.0007) increased the growth rate and biomass yield similar to those of the y3555-expressing strain, indicating that the increased growth rate and yield were y3555 dependent (Fig. 7). However, the highest growth rates occurred in the strain coexpressing y3550 and y3555, followed by the strain expressing y3555 alone, and next, the strain coexpressing y3555 and y3551. The y3550- and y3555-coexpressing strain achieved overall higher biomass yield than did all other strains tested except the y3555-expressing strain. Strains expressing y3550 or y3551 alone had growth rates and biomass yields similar to those of the empty vector-containing strain.
FIG 6.
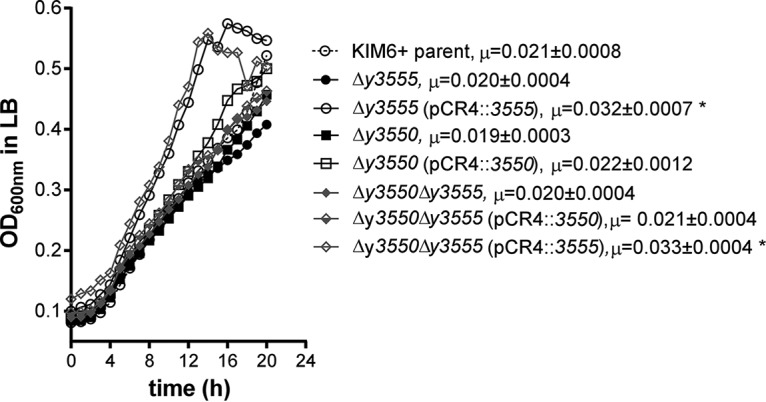
Overexpression of y3555 increases the growth fitness of Y. pestis strains. Shown are the growth kinetics of the Y. pestis KIM6+ parental strain, Δy3550 mutant, Δy3555 mutant, and Δy3550 Δy3555 mutant alone or overexpressing either y3555 (pCR4::3555) or y3550 (pCR4::3550) in LB medium. Data represent the mean values of the results from three independent experiments. The growth rate (μ) calculated from regression analysis is given next to each strain name. Data were analyzed using one-way ANOVA and a Tukey post hoc test. Asterisks mark significantly different growth rates of mutant strains versus that of the parental strain.
FIG 7.
Heterologous expression of y3555 increases the growth rate of the Escherichia coli ΔaspC ΔtyrB (DG44) mutant. Shown are the growth kinetics of E. coli DG44 in M9 minimal medium supplemented with 0.4% glucose, 0.1% Casamino Acids, succinate (2.5 mg/ml), malate (2.5 mg/ml), and α-ketoglutarate (1 mg/ml), as well as 100 μg/ml each of asparagine, glutamine, and glutamic acid. The growth rate (μ) is given next to each plasmid name. Error bars represent mean values of the results from seven independent replicates for the E. coli DG44 studies. One-way ANOVA and Tukey’s multiple-comparison test were used to determine significant differences in growth rate and final biomass yield. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.000; ns, not significant.
DISCUSSION
In this work, we have determined that the coordinated expression of an uncharacterized cluster of low-GC-content genes, y3555-y3554-y3553-y355x-y3551-y3550, that show exceptionally robust expression during flea infection, is accounted for by their cotranscription as a single polycistronic mRNA transcript. Adjacently clustered genes are often grouped transcriptionally as an operon and frequently encode functionally related proteins. This feature allows protein synthesis to be controlled coordinately in response to the needs of the bacterium only when and where the proteins are required. While the genomic island is expressed only at flea-matched temperatures (16, 17), the further induction of these genes following exposure to hyperosmotic saline stress implicates these genes as being functionally concerned with osmoadaptation during the Y. pestis flea life stage. Indeed, protein homology for the y3553 and y3554 gene products clearly predicts that these proteins have direct roles in hypersaline osmotic stress adaptation. In the case of Y3553, active exchange of a sodium ion for a proton is predicted, whereas Y3554 is likely involved in transducing osmotic stress signals to mediate osmoadaptation. However, disruption of the genomic island in the “all” mutant did not result in any survival defects after exposure to 0.5 M NaCl during in vitro LB broth culture, as this mutant survived at levels comparable to those of the parental Y. pestis strain (data not shown). This suggested that absence of the genomic island is not critical for adaptation to osmotic stress in vitro. Nonetheless, y3555, a metabolic gene showing putative roles in amino acid metabolism, confers a striking ability to enhance the growth fitness of E. coli, a trait that is also made apparent during the overexpression of y3555 in Y. pestis strains. These two findings allude to a role of the genomic island in optimizing amino acid metabolism in the context of hyperosmotic stress. This is fitting since (i) Y. pestis primarily metabolizes amino acids during flea infection (15–17) and (ii) the flea gut has a demonstrated physicochemical status of high osmolarity (11), while several Y. pestis metabolic pathways involved in osmoprotection are transcriptionally induced during flea gut infection (15, 17).
To begin to understand the function of Y3555, it was expressed in trans in a well-characterized S. enterica aspC mutant. An S. enterica aspC mutant has a growth defect that is restored with the addition of aspartate or aspC expressed in trans. However, when Y3555 was expressed in the S. enterica aspC359::Km mutant, growth was not restored but in fact further retarded. This surprising result may, however, hint at the function of Y3555. There is evidence to show that in an aspC mutant of S. enterica or E. coli, TyrB produces minimal aspartate that allows for some growth (48, 52), but the addition of tyrosine or phenylalanine inhibits tyrB expression and growth (51). As such, one explanation for the growth defect of the S. enterica aspC359::Km mutant expressing Y3555 is that Y3555 increases tyrosine and/or phenylalanine pools that lead to tyrB inhibition. Thus, our findings suggest that while Y3555 is not an aspartate aminotransferase, it could instead be an aromatic aminotransferase. Alignments of Y3555 with AspC and TyrB from both S. enterica and Y. pestis suggest that Y3555 proteins are fairly dissimilar (see Fig. S1 in the supplemental material). Instead, Y3555 aligns with over 85% identity to uncharacterized PLP-dependent aminotransferases from foodborne isolates of S. enterica sequenced by the Centers for Disease Control and Prevention (Fig. S3). The crystallization of Y3555 may help determine the substrates or products of the protein, and their characterization is ongoing.
During active metabolism, cellular function can be compromised by an anomalous accumulation of endogenously produced reactive intermediates of metabolism, e.g., 2AA. The elimination of such metabolic intermediates is therefore needed to optimize cellular function, growth, and fitness. The highly conserved Rid family proteins prevalent in all domains of life (41, 49) are among the first enzymes characterized to support the optimization of metabolic processes by neutralizing obligatory reactive metabolic intermediates. Currently, RidA enzymes are well characterized as detoxifiers of 2AA in several bacterial species (38, 40, 41, 49, 53). Our data contribute to this list by establishing that in Y. pestis, the y3551 gene encodes an archetypal RidA protein that can deaminate the reactive enamine intermediate 2AA. Additionally, the Y3550 gene product likely belongs to the Rid5-6 subclass of the Rid family proteins whose functions have not yet been characterized. Further adding to its distinctive status is that while the promoters predicted immediately upstream of y3555 and y355x can drive the transcription of y3550, a dedicated promoter predicted immediately upstream of the Y3550 coding sequence is exclusively activated by hyperosmotic stress to inclusively drive y3550 transcription.
It remains elusive how Y3555, Y3550, and Y3551 operate collaboratively to achieve optimal metabolic activity in response to osmotic stress. The achievement of increased growth rate in the DG44 strain coexpressing y3555 and y3550 suggests a synergistic activity of these two proteins. Such a case can be explained by the specific Rid activity meted out by Y3550 being able to potentially eliminate a metabolic by-product that is toxic and/or produced by Y3555. One complementary observation within a context of Y3555 serving in aromatic amino acid metabolic processes is that BPROM predicts a binding box of the transcriptional factor TyrR (54–56) in the promoter sequence of y3550 (the predicted promoter region spans nucleotide positions −120 to −87). In many enteric bacteria, TyrR regulates genes involved in the uptake and biosynthesis of aromatic amino acids in response to the presence or absence of each of the three aromatic amino acids (tyrosine, tryptophan, and phenylalanine) which function as cofactors for TyrR (54, 55). For example, in E. coli, TyrR is responsive to tyrosine and able to regulate the expression of tyrosine biosynthesis genes tyrB, aroF-tyrA, and aroLM and genes encoding aromatic amino acid transporters, aroP and mtrR (55).
Conversely, the suppressed growth rate in the DG44 strain coexpressing y3555 and y3551 versus that of y3555 expression alone suggests antagonistic activity of these two encoded proteins. Such a scenario can be explained by Y3551 RidA deaminase activity protecting an unknown enzyme whose activity either competes with or generates reactive intermediates to which Y3555 is susceptible. The coexpression of Y3551 RidA and Y3550 in Y. pestis might therefore accomplish a balance in metabolic fitness as it pertains to aromatic amino acid metabolic activities.
Our experimental data demonstrate that mutations in the genomic island afford no compromise in the growth and survival or competitive fitness of Y. pestis in fleas. This is confounding because highly elevated expression of unrequired genes would be energetically expensive if not required in the flea gut environment. It is possible that controlled lab-reared flea experiments cannot discern a crucial function of gene products that in wild fleas feeding on wild rodent blood sources (different from lab mouse species) is otherwise apparent. It has been demonstrated that the infectious blood source influences Y. pestis infection in fleas (57) and that the gut microbiota of wild fleas is more diverse than that of lab-reared fleas (58). Perhaps in the wild, a competitive coinfection arises between Y. pestis and diverse gut microbes necessitating genomic island gene product function to heighten its competitive fitness.
Moreover, it is known that more than a single pathway contributes to metabolic robustness, and metabolic versatility is tolerated in the repertoire of metabolic gene pathways that might be switched on for a common physiological purpose, e.g., sustaining Y. pestis growth and survival in the flea gut environment under hyperosmotic stress conditions. For example, the osmoprotectant glycine-betaine is produced by proVW and betI, and these genes have been shown to be expressed by Y. pestis during flea infection (15). Additionally, the Y. pestis genome encodes homologs of some members of the genomic island. Two PLP-dependent aromatic aminotransferase genes, y2760 (aspC) and y0579 (tyrB), which convert aspartate and tyrosine to glutamate, respectively, are induced at robust levels during a Y. pestis flea infection (17). The y3554 gene is one of three genes encoding a Na+/H+ antiporter (nhaA, nhaB, and nhaC). Two ridA homologs, y2240 and y1830, and a putative Rid1 to -7 superfamily-encoding gene, y0163, may be able to compensate for the functions of their homologs in the genomic island (Fig. 5A).
Overall, our data reinforce a model wherein once Y. pestis enters the flea in the blood meal, it transitions into an environment of elevated osmotic stress (11) likely exacerbated by water removal from the blood meal during flea digestive processes (59). Osmotic pressure in the blood meal is contributed by the salts it contains, the concentrations of which are expected to increase after water removal. The requirement for NhaB and NhaC Na+/H+ antiporters for the virulence of Y. pestis during septicemic infection supports the idea that this bacterium faces hyperosmotic stress in mammalian blood (31, 32). Simultaneously, Y. pestis must optimize its growth fitness to establish a transmissible infection in the hyperosmotic flea gut environment. The mechanisms by which Y. pestis counterbalances osmotic stress in the flea, however, remain unclear, although our current data suggest that this is linked to amino acid metabolism shifts that support osmoadaptation within the flea gut. Interestingly, Y. pestis encodes and expresses several biochemical and regulatory pathways that function in alleviating osmotic stress, emphasizing (11, 15) that osmotic stress is a perilous stress to which Y. pestis must adapt in the flea gut. Preservation of compensatory/redundant osmotic stress resistance pathways that allow simultaneous optimization of Y. pestis growth fitness to establish a transmissible infection in fleas as such is critical for the maintenance of disease cycles.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this work are listed in Table S1 in the supplemental material. Bacterial cultures were grown at 28°C with shaking in brain heart infusion (BHI) medium. When necessary, media were supplemented with carbenicillin (100 μg/ml) or kanamycin (50 μg/ml). For the osmolarity assays, overnight cultures of parental KIM6+ strains were used to inoculate fresh LB medium and were incubated at 28°C to reach mid-logarithmic phase; thereafter, cultures were exposed to 0.5 M NaCl. Bacteria were immediately mixed with RNAprotect bacterial reagent (Qiagen) to minimize RNA degradation and then harvested at 4°C and stored at −80°C until RNA isolation.
Salmonella enterica serovar Typhimurium LT2 was the parental strain for all S. enterica mutants. Overnight cultures of S. enterica were grown in 8 g/liter Difco nutrient broth (NB) with NaCl (5 g/liter) and the appropriate antibiotic (150 μg/ml ampicillin or 50 μg/ml kanamycin). Minimal no-carbon E (NCE) was supplemented with MgSO4 (1 mM), trace minerals, glucose (11 mM), and, where indicated, serine (5 mM), isoleucine (0.3 mM), aspartate (1 mM), and/or isopropyl-β-d-1-thiogalactopyranoside (IPTG; 100 μM) (60, 61). All chemicals were purchased from the Sigma-Aldrich Chemical Company in St. Louis, MO.
Construction of plasmids.
The plasmids and primers used in this study are listed in Tables S1 and S2, respectively.
Lux transcriptional fusion constructs.
The Lux transcriptional fusions were generated by cloning a PCR product containing the corresponding regulatory region into the low-copy-number vector pCS26-Pac that carries the promoterless luxCDABE operon (62). PCR products were obtained with the 50R-BHI/50F-XhoI, 51R-BHI/51F-XhoI, 50R-BHI/51F-XhoI, 53R-BHI/53F-XhoI, 54R-BHI/54F-XhoI, 55R-BHI/55F-XhoI, and 54R-BHI/55F-XhoI primer pairs (Table S2). They were then digested with BamHI and XhoI restriction enzymes (NEB) and cloned into the pCS26-Pac plasmid previously digested with the same restriction enzymes, generating plasmids pCS::P50, pCS::P5x, pCS::P5x_51_50. pCS::P53, pCS::P54, pCS::P55, and pCS::P55_54, respectively (Table S1). Lux transcriptional fusion plasmids were transformed into the Y. pestis KIM6+ parental strain.
pTRc99A and pET28a constructs.
The pTRc99A- and pET28a-inducible expression plasmids were generated by cloning PCR products obtained for the y3550, y3551, and y3555 genes from the p270/p271, p272/p273, and p274/p275 primer pairs, respectively. The PCR products were digested with BamHI and XhoI and ligated into pTrc99A or pET28a previously digested with BamHI and SalI restriction enzymes. The plasmid constructs generated from cloning y3550, y3551, and y3555 into pTRc99A are referred to as pTRc55, pTRc50, and pTRc51, and for pET28a cloning, they are referred to as pET28a::y3550, pET28a::y3551, and pET28a::y3555, respectively. To clone y3555 together with y3551 or y3550 into pTrc99A, y3555 was amplified with primers p508 and p509, digested with HindIII, and ligated into pTRc50 or pTRc51 such that y3555 was inserted downstream of y3550 or y3551. These plasmids were referred to as pTRc50_55 and pTRc51_55, respectively. Plasmids were transformed into transformed into E. coli DG44 and selected for on LB–100 μg/ml carbenicillin.
Tn7-based chromosomal integration of a kanamycin resistance cassette into Y. pestis KIM6+ strain.
The kanamycin resistance cassette from the plasmid pKD4 was amplified with primers p395 and p396 (Table S2). The PCR product was digested with BamHI and EcoRI, ligated into pUC18R6K-mini-Tn7T previously digested with the same restriction enzymes, transformed into DH5α lambda pir chemical competent E. coli, and selected for on LB–50 μg/ml kanamycin. The plasmid, referred to as pUC18R6K-mini-Tn7T::kanr, was isolated and verified for sequence. Electrocompetent Y. pestis KIM6+ was transformed with pUC18R6K-mini-Tn7T::kanr and pTNS2 and selected for on heart infusion broth (HIB)–50 μg/ml kanamycin plates. Colonies were screened with the pstSup2 and pTn7R primers for insertion at the glmS-pstS site. This Y. pestis KIM6+ strain with the kanamycin cassette integrated at the glmS-pstS site was referred to as Y. pestis KIM6+ glm-pstS::Kanr.
Lux transcriptional reporter assay.
Y. pestis KIM6+ carrying plasmids containing the lux fusions was grown overnight in LB supplemented with appropriate antibiotics. An aliquot was used to inoculate fresh LB medium the next day. The cultures were grown on a shaker at 28°C to mid-logarithmic phase and then treated or not with 0.5 M NaCl for 100 min. Aliquots of 1 ml were taken out to determine the optical density of the culture by measuring the optical density at 600 nm (OD600). One-tenth of a milliliter of each culture was dispensed into a 96-well plate (Costar), and the luminescence of each well was measured in the Infinite M1000 reader (Tecan), with an integration time of 1 s and using the Tecan i-control software version 1.7.1.12. Plates were first shaken with a 2-mm linear amplitude 5-s pulse. Data were normalized by dividing relative light units (RLU) by the OD600 to obtain the final RLU corrected for bacterial growth.
RT-PCR.
The Y. pestis KIM6+ strain was grown in Miller’s LB broth at 26°C to mid-logarithmic phase; the culture was split in two, and one-half received 0.5 M NaCl and the other distilled water vehicle control treatment. After 20 min, culture RNA was stabilized immediately with RNAprotect bacterial reagent (Qiagen). Total RNA was purified using the Quick-RNA miniprep kit (Zymo). Purified RNA was subject to Turbo DNase I (Ambion) treatment, according to the manufacturer’s instructions. Then, 800 to 1,600 ng of RNA was used in the first-strand cDNA synthesis reaction using Maxima H Minus reverse transcriptase (Thermo Fisher), according to the manufacturer’s instructions. Negative controls were performed by omitting the reverse transcriptase. The cDNA was then treated with RNase H (NEB) at 37°C for 30 min. The PCR amplification step was performed using Phusion polymerase (Thermo Fisher), 100 ng of cDNA, and the Y3555Set2Fw/5051RT-R primer pair (each primer at a final concentration of 0.5 μM) (Table S2); this pair amplifies a 5,014-bp region upstream of y3555 to 226 bp within y3550. RT-PCR cycles were as follows: one cycle of 1 min at 98°C for initial denaturation, and 31 cycles of 10 s at 98°C, 20 s at 68°C, and 1 min 40 s at 72°C for PCR amplification with the Y3555Set2Fw/5051RT-R primer pair. A final elongation step of 5 min at 72°C was included. Controls for the expected size of the PCR products were performed by using chromosomal DNA from Y. pestis KIM6+ as the template. The PCR products were analyzed by 1% agarose gel electrophoresis gels. The GeneRuler 1-kb DNA ladder (Thermo Scientific) was run alongside the PCR products.
Construction of mutant and complementation strains.
Nonpolar gene deletion mutant strains were generated by the lambda (λ) red recombinase system (63, 64) and gene-specific primer pairs shown in Table S2. The y3550 and y3555 genes and the entire continuous region spanning genes y3555, y3554, y3553, y355x, y3551, and y3550 (referred to as “all”) and the tandem genes y3551 and y3550 were replaced with a selectable kanamycin (Km) resistance cassette in the Y. pestis KIM6+ strain (Table S1). The Δy3550 Δy3555 double mutant was constructed similarly, but for this, the Δy3550 mutant was used. Then, the FLP recombination target (FRT)-flanked Km cassette was excised from these strains after transformation with the helper plasmid pFLP3 (Table S1), expressing the FLP recombinase that acts on the directly repeated FRT (65). Both pKOBEG-sacB and pFLP3 were cured by sucrose counterselection by growing the strains on LB without NaCl and supplemented with 10% sucrose. The strains generated were Δy3550 (VV150), Δy3555 (VV151), Δy3550 Δy3551 (VV278), Δy3550 Δy3555 (VV279), and “all” (VV280) mutants (Table S1) and retained an FRT scar. To construct plasmids expressing y3550 or y3555, a DNA fragment containing the y3550 gene alone or y3555 with its native promoter sequence was amplified by PCR using the 3550-F/3555-R or 3550-F/3555-R primer pair (Table S2) and cloned into vector pCR4-TOPO, generating plasmids pCR4::3550 and pCR4::3555, respectively. All mutant strains were verified by PCR and sequencing.
Reverse transcriptase qPCR.
Total RNA of Y. pestis KIM6+ was purified using the Quick-RNA miniprep kit (Zymo). Chromosomal DNA was removed by incubating 5 μg of RNA from each strain with Turbo DNase I (Ambion), according to the manufacturer’s instructions. cDNA was synthesized in a reaction mixture containing 2 μg of each DNase-treated RNA, 100 pmol random hexamers, and 0.5 mM dinucleoside triphosphates (dNTPs), using RiboLock RNase inhibitor (Thermo Fisher) and Maxima H minus reverse transcriptase (Thermo Fisher), according to the manufacturer’s protocol. The obtained cDNA was used as the template for quantitative PCR (qPCR) assays, with 0.5 μM primer pairs for y3550, y3551, y3553, y3554, y3555, or gyrB (Table S2) and the SsoAdvanced Universal SYBR green Supermix (Bio-Rad). Real-time PCRs were performed with the c1000 Touch thermal cycler and the CFX96 real-time system (Bio-Rad), and data were collected using the CFX Manager software 2.1 (Bio-Rad). The reaction conditions were 95°C for 3 min, 95°C for 10 s, and 58°C for 30 s during 40 cycles. The level of gyrB mRNA was used as an internal control to normalize the results. The quantification technique used to analyze data from cultures was the 2–ΔΔCT method (66). All qPCRs were performed in triplicate and were repeated using RNA purified from three independent bacterial cultures or flea infections.
Flea infections.
For coinfections, Xenopsylla cheopis fleas were infected with blood containing a 1:1 mix of parental KIM6+ marked at the attn7 site (65), called KIM6+ glmS-pstS::Kanr, and a mutant strain, as previously described (43). The infected fleas were fed twice weekly on a live neonatal mouse. To compare the bacterial load over the infection, 20 fleas were individually triturated and plated on BHI agar and BHI agar containing kanamycin to select for the KIM6+ glmS-pstS::Kanr at 2 h (time zero) and 7 days postinfection. All experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Washington State University, Pullman, WA, and were conducted in strict accordance with institutional guidelines based on the U.S. National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Growth fitness of E. coli strains DH5α and DG44 expressing Y. pestis low-GC-content genes.
E. coli strains harboring IPTG-inducible constructs were grown overnight in LB containing 100 μg/ml carbenicillin with shaking at 37°C. For the E. coli DH5α strains, the overnight inoculum was diluted 1:200 into fresh M9 medium supplemented with 0.4% glucose and 0.1% Casamino Acids medium (Difco). For the E. coli D44 strains, the overnight inoculum was diluted 1:200 into fresh M9 medium supplemented with 0.4% glucose and 0.1% Casamino Acids medium (Difco), succinate (2.5 mg/ml), malate (2.5 mg/ml), α-ketoglutarate (1 mg/ml), and 100 μg/ml each asparagine, glutamine, and glutamic acid, as previously described (46). Growth curves were generated at 37°C using a Bioscreen C shaking incubator (Growth Curves USA, Piscataway, NJ).
Growth quantification of S. enterica strains.
The growth of S. enterica in liquid culture was assessed using a BioTek EL808 microtiter plate reader by measuring the optical density at 650 nm at 37°C at a low shaking speed. Overnight cultures of S. enterica in biological triplicate were grown in rich medium with the appropriate antibiotic at 37°C. Cultures were pelleted and resuspended in an equal volume of sterile NaCl (8.5 g/liter). The cell suspension was used to inoculate growth medium (2% inoculum), and growth was monitored. The growth data were plotted in a log10 format using GraphPad Prism 7.0c, and the error bars represent the standard error of the mean for three biological replicates.
Protein purification.
Proteins were purified as previously described (53, 67, 68). EcRidA and Y3551 were purified from E. coli strain BL21-AI harboring either pET20-ridA (DM12740) or pET28b-y3551 (DM16279). Overnight cultures grown in superbroth with ampicillin were used to inoculate 3 liters of superbroth with ampicillin. Cultures were grown for ∼3 h at 37°C with shaking until an OD650 of 0.7 was reached. Fresh arabinose was added to a final concentration of 0.2%, and cultures were incubated with shaking at 37°C overnight. A clarified supernatant was injected onto a nickel-nitrilotriacetic acid (Ni-NTA) Superflow resin, and proteins were purified according to the manufacturer’s protocol (Qiagen). Protein aliquots were frozen in liquid nitrogen and stored at −80°C. Purified CdsH was a gift from Dustin Ernst and was purified similarly from E. coli strain BL21-AI containing pET14b-cdsH, as previously described (67).
CdsH assay.
2AA deaminase activity was determined using a coupled assay with purified cysteine desulfhydrase (CsdH), RidA, and Y3551, as previously described (67, 69). Assay mixtures (100 μl) contained 100 mM Tris-HCl (pH 8), NADH (250 μM), 5′-phosphate (PLP)-phosphate (30 μM), lactate dehydrogenase (5 U), and purified CdsH (0.27 μM). The reaction mixtures contained RidA or Y3551 (0.19 μM) or an equal volume buffer as a control. The reactions were initiated in triplicate with the addition of freshly prepared l-cysteine (final concentration, 0.5 to 2.5 mM). The reactions were monitored continuously in a 96-well quartz plate by measuring the absorbance at 340 nm for 2 min using a SpectraMax M2 microplate reader. The initial rate of pyruvate formation was calculated from the rate of NADH oxidation in the first 30 s, along with the molar extinction coefficient (ε = 6,200 M−1 cm−1).
In silico analyses.
In silico predictions of genomic island genetic organization and prediction of promoters were achieved using FGENESB and BPROM (Softberry, Inc., Mt. Kisco, NY) freeware. Alignment of amino acid sequences of Rid proteins was accomplished using the EMBL-MUSCLE tool (https://www.ebi.ac.uk/Tools/msa/muscle/). The protein secondary structure prediction and structural alignments of Y3555 were made using Phyre2 (44).
Statistical analysis.
Statistical analysis was accomplished using GraphPad Prism 7.0, the details for which are provided in “Growth quantification of S. enterica strains,” above, or the legends to Fig. 1 to 3, 6, and 7.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kameron Gravelle, Stephanie Baker, and Veronica Harris for technical assistance.
Luary C. Martínez-Chavarría was supported by a postdoctoral fellowship from CONACYT (grants 186381 and 203527). The study was supported by competitive grants from the NIH with numbers 1R21AI097974 (to V.V.), 5R01AI117016-02 (to V.V.), and GM095837 (D.M.D.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A 101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen RJ, Gage KL. 2009. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res 40:1. doi: 10.1051/vetres:2008039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch BJ, Erickson DL. 2008. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol 322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinnebusch BJ, Perry RD, Schwan TG. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch BJ, Jarrett CO, Bland DM. 2017. “Fleaing” the plague: adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu Rev Microbiol 71:215–232. doi: 10.1146/annurev-micro-090816-093521. [DOI] [PubMed] [Google Scholar]
- 8.Chouikha I, Hinnebusch BJ. 2012. Yersinia–flea interactions and the evolution of the arthropod-borne transmission route of plague. Curr Opin Microbiol 15:239–246. doi: 10.1016/j.mib.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinnebusch BJ, Sebbane F, Vadyvaloo V. 2012. Transcriptional profiling of the Yersinia pestis life cycle, p 1–19. In Carniel E, Hinnebusch BJ (ed), Yersinia systems biology and control. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 10.Zhou W, Russell CW, Johnson KL, Mortensen RD, Erickson DL. 2012. Gene expression analysis of Xenopsylla cheopis (Siphonaptera: Pulicidae) suggests a role for reactive oxygen species in response to Yersinia pestis infection. J Med Entomol 49:364–370. doi: 10.1603/me11172. [DOI] [PubMed] [Google Scholar]
- 11.Bontemps-Gallo S, Fernandez M, Dewitte A, Raphael E, Gherardini FC, Pradel E, Koch L, Biot F, Reboul A, Sebbane F. 2019. Nutrient depletion may trigger the Yersinia pestis OmpR-EnvZ regulatory system to promote flea-borne plague transmission. Mol Microbiol 112:1471–1482. doi: 10.1111/mmi.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vadyvaloo V, Viall AK, Jarrett CO, Hinz AK, Sturdevant DE, Hinnebusch BJ. 2015. Role of the PhoP-PhoQ gene regulatory system in adaptation of Yersinia pestis to environmental stress in the flea digestive tract. Microbiology 161:1198–1210. doi: 10.1099/mic.0.000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PC, Hinnebusch BJ. 2013. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J Bacteriol 195:1920–1930. doi: 10.1128/JB.02000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuto HS, Vadyvaloo V, McPhee JB, Poinar HN, Holmes EC, Bliska JB. 2018. A single amino acid change in the response regulator PhoP, acquired during Yersinia pestis evolution, affects PhoP target gene transcription and polymyxin B susceptibility. J Bacteriol 200:e00050-18. doi: 10.1128/JB.00050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chouikha I, Sturdevant DE, Jarrett C, Sun YC, Hinnebusch BJ. 2019. Differential gene expression patterns of Yersinia pestis and Yersinia pseudotuberculosis during infection and biofilm formation in the flea digestive tract. mSystems 4:e00217-18. doi: 10.1128/mSystems.00217-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vadyvaloo V, Jarrett C, Sturdevant D, Sebbane F, Hinnebusch BJ. 2007. Analysis of Yersinia pestis gene expression in the flea vector. Adv Exp Med Biol 603:192–200. doi: 10.1007/978-0-387-72124-8_16. [DOI] [PubMed] [Google Scholar]
- 17.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernardini ML, Fontaine A, Sansonetti PJ. 1990. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J Bacteriol 172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty S, Kenney LJ. 2018. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front Microbiol 9:2656. doi: 10.3389/fmicb.2018.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty S, Winardhi RS, Morgan LK, Yan J, Kenney LJ. 2017. Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat Commun 8:1587. doi: 10.1038/s41467-017-02030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao H, Zhang Y, Han Y, Yang L, Liu X, Guo Z, Tan Y, Huang X, Zhou D, Yang R. 2011. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol 11:39. doi: 10.1186/1471-2180-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Zhang Y, Tan Y, Wang L, Xiao X, Guo Z, Zhou D, Yang R. 2011. Transcriptional regulation of ompF2, an ompF paralogue, in Yersinia pestis. Can J Microbiol 57:468–475. doi: 10.1139/w11-028. [DOI] [PubMed] [Google Scholar]
- 24.Han Y, Zhou D, Pang X, Zhang L, Song Y, Tong Z, Bao J, Dai E, Wang J, Guo Z, Zhai J, Du Z, Wang X, Wang J, Huang P, Yang R. 2005. Comparative transcriptome analysis of Yersinia pestis in response to hyperosmotic and high-salinity stress. Res Microbiol 156:403–415. doi: 10.1016/j.resmic.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Sun YC, Hinnebusch BJ, Darby C. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A 105:8097–8101. doi: 10.1073/pnas.0803525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun YC, Jarrett CO, Bosio CF, Hinnebusch BJ. 2014. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 15:578–586. doi: 10.1016/j.chom.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wren BW. 2003. The yersiniae–a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- 28.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. 2008. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol 190:8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlowski J, Grinstein S. 2011. Na+/H+ exchangers. Compr Physiol 1:2083–2100. doi: 10.1002/cphy.c110020. [DOI] [PubMed] [Google Scholar]
- 30.Vaish M, Price-Whelan A, Reyes-Robles T, Liu J, Jereen A, Christie S, Alonzo F III, Benson MA, Torres VJ, Krulwich TA. 2018. Roles of Staphylococcus aureus Mnh1 and Mnh2 antiporters in salt tolerance, alkali tolerance, and pathogenesis. J Bacteriol 200:e00611-17. doi: 10.1128/JB.00611-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minato Y, Ghosh A, Faulkner WJ, Lind EJ, Schesser Bartra S, Plano GV, Jarrett CO, Hinnebusch BJ, Winogrodzki J, Dibrov P, Häse CC. 2013. Na+/H+ antiport is essential for Yersinia pestis virulence. Infect Immun 81:3163–3172. doi: 10.1128/IAI.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Häse CC, Fedorova ND, Galperin MY, Dibrov PA. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65:353–370, table of contents. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer LM, Koonin EV, Aravind L. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43:134–144. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, Lu T, Miao W, Sun L, Tian M, Wang J, Hao F. 2017. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 5:e4126. doi: 10.7717/peerj.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Ge H, Wang X, Tang R, Wang Y, Zhao F, Lan W, Luan S, Yang L. 2017. Overexpression of pyrabactin resistance-like abscisic acid receptors enhances drought, osmotic, and cold tolerance in transgenic poplars. Front Plant Sci 8:1752. doi: 10.3389/fpls.2017.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Y, Zhang X, Sun T, Tian Q, Zhang WH. 2018. Glutamate receptor homolog3.4 is involved in regulation of seed germination under salt stress in Arabidopsis. Plant Cell Physiol 59:978–988. doi: 10.1093/pcp/pcy034. [DOI] [PubMed] [Google Scholar]
- 37.Möglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz G, Downs DM. 2004. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J Bacteriol 186:803–810. doi: 10.1128/jb.186.3.803-810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christopherson MR, Schmitz GE, Downs DM. 2008. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J Bacteriol 190:3057–3062. doi: 10.1128/JB.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrecht JA, Flynn JM, Downs DM. 2012. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J Biol Chem 287:3454–3461. doi: 10.1074/jbc.M111.304477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niehaus TD, Gerdes S, Hodge-Hanson K, Zhukov A, Cooper AJ, ElBadawi-Sidhu M, Fiehn O, Downs DM, Hanson AD. 2015. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics 16:382. doi: 10.1186/s12864-015-1584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuto HS, Svetlanov A, Palmer LE, Karzai AW, Bliska JB. 2010. Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect Immun 78:3700–3715. doi: 10.1128/IAI.00062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemon AK, Silva-Rohwer A, Sagawa J, Vadyvaloo V. 2019. Co-infection assay to determine Yersinia pestis competitive fitness in fleas. Methods Mol Biol 2010:153–166. doi: 10.1007/978-1-4939-9541-7_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg M. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Percudani R, Peracchi A. 2009. The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 10:273. doi: 10.1186/1471-2105-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gelfand DH, Steinberg RA. 1977. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol 130:429–440. doi: 10.1128/JB.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Camakaris H, Pittard J. 2002. Molecular analysis of tyrosine-and phenylalanine-mediated repression of the tyrB promoter by the TyrR protein of Escherichia coli. Mol Microbiol 45:1407–1419. doi: 10.1046/j.1365-2958.2002.03108.x. [DOI] [PubMed] [Google Scholar]
- 48.Powell JT, Morrison JF. 1978. Role of the Escherichia coli aromatic amino acid aminotransferase in leucine biosynthesis. J Bacteriol 136:1–4. doi: 10.1128/JB.136.1.1-4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambrecht JA, Schmitz GE, Downs DM. 2013. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 4:e00033-13. doi: 10.1128/mBio.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morishita R, Kawagoshi A, Sawasaki T, Madin K, Ogasawara T, Oka T, Endo Y. 1999. Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J Biol Chem 274:20688–20692. doi: 10.1074/jbc.274.29.20688. [DOI] [PubMed] [Google Scholar]
- 51.Gelfand DH, Rudo N. 1977. Mapping of the aspartate and aromatic amino acid aminotransferase genes tyrB and aspC. J Bacteriol 130:441–444. doi: 10.1128/JB.130.1.441-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Dyk TK, LaRossa RA. 1986. Sensitivity of a Salmonella Typhimurium aspC mutant to sulfometuron methyl, a potent inhibitor of acetolactate synthase II. J Bacteriol 165:386–392. doi: 10.1128/jb.165.2.386-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irons J, Hodge-Hanson KM, Downs DM. 2018. PA5339, a RidA homolog, is required for full growth in Pseudomonas aeruginosa. J Bacteriol 200:e00434-18. doi: 10.1128/JB.00434-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Argaet VP, Wilson TJ, Davidson BE. 1994. Purification of the Escherichia coli regulatory protein TyrR and analysis of its interactions with ATP, tyrosine, phenylalanine, and tryptophan. J Biol Chem 269:5171–5178. [PubMed] [Google Scholar]
- 55.Pittard J, Camakaris H, Yang J. 2005. The TyrR regulon. Mol Microbiol 55:16–26. doi: 10.1111/j.1365-2958.2004.04385.x. [DOI] [PubMed] [Google Scholar]
- 56.Smith HQ, Somerville RL. 1997. The tpl promoter of Citrobacter freundii is activated by the TyrR protein. J Bacteriol 179:5914–5921. doi: 10.1128/jb.179.18.5914-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bland DM, Jarrett CO, Bosio CF, Hinnebusch BJ. 2018. Infectious blood source alters early foregut infection and regurgitative transmission of Yersinia pestis by rodent fleas. PLoS Pathog 14:e1006859. doi: 10.1371/journal.ppat.1006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones RT, Vetter SM, Montenieiri J, Holmes J, Bernhardt SA, Gage KL. 2013. Yersinia pestis infection and laboratory conditions alter flea-associated bacterial communities. ISME J 7:224–228. doi: 10.1038/ismej.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lehane M. 2005. Managing the blood meal, p 84–115. In Lehane M. (ed), The biology of blood-sucking in insects, 2nd ed Cambridge University Press, New York, NY. [Google Scholar]
- 60.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 61.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized [sic] atmosphere. Appl Environ Microbiol 32:781–791. doi: 10.1128/AEM.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bjarnason J, Southward CM, Surette MG. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J Bacteriol 185:4973–4982. doi: 10.1128/jb.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 65.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 66.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Ernst DC, Lambrecht JA, Schomer RA, Downs DM. 2014. Endogenous synthesis of 2-aminoacrylate contributes to cysteine sensitivity in Salmonella enterica. J Bacteriol 196:3335–3342. doi: 10.1128/JB.01960-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lambrecht JA, Browne BA, Downs DM. 2010. Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J Biol Chem 285:34401–34407. doi: 10.1074/jbc.M110.160515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kredich NM, Foote LJ, Keenan BS. 1973. The stoichiometry and kinetics of the inducible cysteine desulfhydrase from Salmonella Typhimurium. J Biol Chem 248:6187–6196. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.