Abstract
Introduction
Sepsis is a leading cause of mortality in burn patients. One of the major causes of sepsis in burn patients is Pseudomonas aeruginosa. We hypothesized that during dissemination from infected burn wounds and subsequent sepsis, P. aeruginosa affects the metabolome of the blood resulting in changes to specific metabolites that would serve as biomarkers for early diagnosis of sepsis caused by P. aeruginosa.
Objectives
To identify specific biomarkers in the blood after sepsis caused by P. aeruginosa infection of burns.
Methods
Gas chromatography with time-of-flight mass spectrometry was used to compare the serum metabolome of mice that were thermally injured and infected with P. aeruginosa (B–I) to that of mice that were neither injured nor infected, mice that were injured but not infected, and mice that were infected but not injured.
Results
Serum levels of 19 metabolites were significantly increased in the B–I group compared to controls while levels of eight metabolites were significantly decreased. Thymidine, thymine, uridine, and uracil (related to pyrimidine metabolism), malate and succinate (a possible sign of imbalance in the tricarboxylic acid cycle), 5-oxoproline (related to glutamine and glutathione metabolism), and trans-4-hydroxyproline (a major component of the protein collagen) were increased. Products of amino acid metabolism were significantly decreased in the B–I group, including methionine, tyrosine, indole-3-acetate, and indole-3-propionate.
Conclusion
In all, 26 metabolites were identified, including a unique combination of five metabolites (trans-4-hydroxyproline, 5-oxoproline, glycerol-3-galactoside, indole-3-acetate, and indole-3-propionate) that could serve as a set of biomarkers for early diagnosis of sepsis caused by P. aeruginosa in burn patients.
Electronic supplementary material
The online version of this article (10.1007/s11306-020-01658-2) contains supplementary material, which is available to authorized users.
Keywords: Burn, Gas chromatography with time-of-flight mass spectrometry, Infection, Metabolomics, Pseudomonas aeruginosa, Sepsis
Introduction
Sepsis is a major cause of mortality in critically ill patients, including severely burned patients (Manning 2018; Nitzschke et al. 2017). The diagnosis of sepsis in such patients is a limiting step in successful treatment. In severely burned patients, this diagnosis is particularly difficult, since the common clinical signs and symptoms of sepsis are masked by a baseline hypermetabolism (Nunez Lopez et al. 2017; Parent et al. 2016; Peltz et al. 2015), as well as an inflammatory immune response (Bortolotti et al. 2018). Laboratory testing for substantiation of sepsis is based on a combination of host reactants that are elevated during the acute phase immune response, which can be caused by infectious or immune inflammatory diseases, cancer, or trauma; and blood cells and metabolites that are altered during sepsis, septic shock, and/or organ dysfunction caused by sepsis (Biron et al. 2015; Bortolotti et al. 2018). Tests for the acute phase reactants C-reactive protein and procalcitonin have been standardized and are widely used for diagnosis of inflammation related to infection and predicting progression to sepsis, respectively (Biron et al. 2015; Fleischmann-Struzek et al. 2018; Markanday 2015). Similarly, levels of the metabolite lactate have been standardized for indication of sepsis and septic shock (Rhee et al. 2015). Levels of the proinflammatory cytokines interleukin-6 and tumor necrosis factor-alpha, which stimulate the production of the acute phase reactants, offer potential for additional markers to diagnose sepsis (Sun et al. 2015). The use of Fc-gamma receptor-1 (CD64) and high-mobility group box 1 protein are undergoing research for feasibility in assessment of sepsis in humans, but are not currently available (Hatada et al. 2005; Livaditi et al. 2006; Markanday 2015). Although blood cultures are recommended for diagnosis of the etiologic agent of sepsis, their results are negative in at least 50% of patients with sepsis (de Prost et al. 2013). Failure to recover a microorganism could be due to low microbial load in the bloodstream, prior treatment of the patient with antibiotics, or presence of an organism that cannot be grown in conventional blood culture (virus, parasite, or fastidious, slow-growing bacterium). However, sepsis can also be diagnosed by confirming an infection in the urinary tract, lower respiratory tract, a surgical site, or other body fluid (CSF, synovial fluid, peritoneal fluid, etc.) in a patient with signs and symptoms of sepsis (Comstedt et al. 2009; Heffner et al. 2010). Additionally, a noninfectious etiology, such as pancreatitis, cancer, autoimmune inflammatory disease, or burns or other trauma including surgery, is found for 18–38% of patients presenting with sepsis symptoms (Comstedt et al. 2009; de Prost et al. 2013; Heffner et al. 2010), a diagnosis often referred to as the noninfectious systemic inflammatory response syndrome (niSIRS) (Cabrera et al. 2017; Mearelli et al. 2018; Watt et al. 2015). Thus, identifying specific biomarkers for sepsis is an area of significant research interest. This will help differentiating between the various agents and conditions associated with sepsis, since numerous microbial species can be the cause (Gauer 2013). Also, the rise in multidrug resistance in certain species such as Pseudomonas aeruginosa makes it harder to empirically treat infected patients prior to knowing the cause; yet delay in initiating appropriate antibiotic treatment can substantially increase mortality in patients suffering from sepsis (Liu et al. 2017; Micek et al. 2005).
Pseudomonas aeruginosa, an opportunistic Gram-negative bacterium, is one of the major pathogens causing localized and systemic infections in different patient populations (Greenhalgh 2017). P. aeruginosa can spread systemically from localized infections at different body sites, including ventilator-associated pneumonia, urinary tract infection, or wound infection (Bassetti et al. 2018). We used a murine model of thermal injury and metabolomic analysis of serum to search for biomarkers specific for sepsis caused by dissemination of P. aeruginosa from infected burn wounds. We hypothesized that during sepsis, P. aeruginosa affects the metabolome of the blood by either consuming specific metabolites, thus decreasing their concentration; or by producing other metabolites, consequently increasing their concentration. These metabolites would be indicative of P. aeruginosa bloodstream infection and could represent potential biomarkers for early diagnosis of sepsis caused by P. aeruginosa. In this study, we identified a set of 26 metabolites related to amino acid and protein metabolism, glycolysis/gluconeogenesis, pyrimidine metabolism, fatty acid and lipid metabolism, and the tricarboxylic acid (TCA) cycle, including two unique to this study – trans-4-hydroxyproline and glycerol-3-galactoside, that have the potential to serve as novel biomarkers for P. aeruginosa causing sepsis in severely burned patients.
Materials and methods
Preparation of bacterial inoculum
The P. aeruginosa strain UCBPP-PA14 (PA14), originally isolated from an infected burn wound, was used in all the experiments (Liberati et al., 2006). Strain PA14 was grown overnight at 37 °C in Luria–Bertani (LB) broth. A 100-μL aliquot of the overnight culture was subcultured into fresh LB broth and the subcultures were grown at 37 °C for 3 h to an OD600 of 0.5. A 100-μL aliquot of the subculture was serially diluted tenfold in phosphate buffered saline (PBS) to yield an infective dose of 200 to 300 colony forming units (CFU) in 100 μL. This dose has previously been shown to produce 100% lethality in Swiss Webster mice by 2 days after thermal injury and infection with P. aeruginosa (Dzvova et al. 2018; Rahme et al. 1995).
Murine model of thermal injury
Adult female Swiss Webster mice (Charles River Laboratories, Wilmington, MA) weighing between 22 and 24 g were utilized in the murine model of thermal injury (Rumbaugh et al. 1999). Briefly, mice were anesthetized by intraperitoneal injection of 5% sodium pentobarbital (Nembutal; Diamondback Drugs, Scottsdale, AZ) at 5 mg/mL and their backs were shaved. The mice were placed in a template that exposes approximately 15% of the total body surface area and the exposed surface was placed in 90 °C water for 8 s. Such an injury is nonlethal but results in a full-thickness burn. Mice were immediately resuscitated with fluid replacement therapy. For those mice to be infected with PA14, 100 μL of PBS containing approximately 200 CFU was injected directly under the injured skin. Four groups of mice were used in these experiments: thermally injured and infected (B–I), the experimental group; and three control groups – non-injured and non-infected (NB–NI), the sham control; non-injured but infected (NB–I); and thermally injured but not infected (B–NI). After 24 h, mice in all groups were anesthetized and approximately 1 mL of blood was collected via cardiac puncture. Livers and spleens were harvested from the exsanguinated mice.
Animals were treated humanely and in accordance with the protocol approved by the Animal Care and Use Committee at the Texas Tech University Health Sciences Center (TTUHSC), Lubbock, TX.
Determination of systemic infection
To determine whether PA14 bacteremia was present, 10-μL aliquots of the blood collected from each mouse was plated on LB agar and Pseudomonas isolation agar plates (Criterion; Hardy Diagnostics, Santa Monica, CA). As the blood had begun to clot prior to plating, the numbers of CFU/mL was estimated rather than quantitated. To determine if PA14 disseminated to the livers and spleens of the animals, the individual organs were weighed, suspended in 2 mL of PBS, homogenized (Precellys tissue homogenizer; Bertin Instruments, Rockville, MD) and diluted tenfold. Ten μL of each dilution were plated on LB agar and Pseudomonas isolation agar plates to determine CFU/g of tissue. The remaining whole blood samples were allowed to stand for 1 h at room temperature for clot formation to occur (Tuck et al. 2009). The samples were centrifuged and the serum removed from each clot. The sera were stored at − 20 °C until use in the metabolomic analysis.
Gas chromatography with time-of-flight mass spectrometry (GC-TOF-MS)
Serum samples were submitted to the National Institutes of Health (NIH) West Coast Metabolomics Center at UC Davis for metabolomic analysis (https://metabolomics.ucdavis.edu/; accessed 20 Aug 2019) where untargeted metabolite profiling was done by GC-TOF-MS and the detected metabolites were identified using ChromaTOF version 2.32 and BinBase algorithm, methods based on two independent parameters: retention index and mass spectral similarity, as previously described (Fiehn et al. 2008).
Data analysis
Peak intensities of the identified metabolites were normalized using vector and median normalization methods. Relative peak intensities were log-transformed and scaled using Pareto scaling in the MetaboAnalyst program (https://www.metaboanalyst.ca; accessed 18 Aug 2019) (Chong et al. 2018). For principal component analysis (PCA) and hierarchical cluster analysis (HCA) of the metabolite data, the pre-processed and normalized dataset was imported into the ClustVis web tool (https://www.biit.cs.ut.ee/clustvis; accessed 20 Aug 2019) and further analyzed (Metsalu and Vilo, 2015). One-way analysis of variance (ANOVA) followed by post-hoc analysis using Fisher’s least significant difference (LSD) test was used for analysis of statistical significance. The alluvial plot was generated using RAWGraphs (https://rawgraphs.io/; accessed 07 Sep 2019) (Mauri et al. 2017). NetworkAnalyst software (https://www.networkanalyst.ca; accessed 20 Aug 2019) (Zhou et al. 2019) in conjunction with MetaboAnalyst was used to generate and visualize the metabolite-metabolite interaction network. The associations for the metabolite networks were extracted from STITCH (a feature of NetworkAnalyst) and only highly confident interactions were included. Receiver operating characteristic (ROC) curves and the areas under the curve (AUC) were generated using MetaboAnalyst.
Results
P. aeruginosa systemic infection occurs only in mice that are also thermally injured
We used a murine model of thermal injury to detect any differences in the serum metabolome produced by P. aeruginosa infection of the burn wound (Rumbaugh et al. 1999). For this analysis, four groups of mice were used: the B–I group of mice was thermally injured and the virulent P. aeruginosa strain PA14 was injected directly under the injured skin; and three control groups, NB–NI (sham), NB–I, and B–NI described in Materials and methods. Blood was collected from all mice 24 h post treatment. Whole blood was plated to detect bacteremia and serum was harvested for the metabolomic analysis. The livers and spleens were processed to determine systemic spread of infection. Only the B–I mice group suffered from systemic infection as indicated by recovery of PA14 from the blood (≈ 400 CFU/mL), livers (≈ 125,000 CFU/g) and spleens (≈ 100,000 CFU/g) of mice in the B–I group but not from the NB–NI, NB–I, or B–NI groups (Online Resource 1). Thus, the B–I group, the 24-h time point represents early or uncomplicated sepsis as the animals had BSI with dissemination to internal organs but were not yet moribund or dead. Without intervention PA14 has been shown to cause 100% mortality from sepsis at 48 h (Dzvova et al. 2018; Rahme et al. 1995). In contrast, the B–NI control group represents the effect of trauma without infection (similar to niSIRS).
Thermal injury plus P. aeruginosa infection specifically altered the metabolomic profile
To identify potential biomarkers present in the blood during early sepsis caused by P. aeruginosa infection of burn wounds with dissemination to the bloodstream, we utilized an untargeted metabolomic approach using GC-TOF-MS that detected metabolites related to primary metabolism, including carbohydrates and sugar phosphates, amino acids, hydroxyl acids, free fatty acids, purines, pyrimidines, aromatic compounds, and exposome-derived chemicals (Fiehn et al. 2008). We detected 531 primary metabolites among all tested groups of sera; of these, 148 metabolites were successfully identified based on their mass spectra and are reported in Online Resource 2.
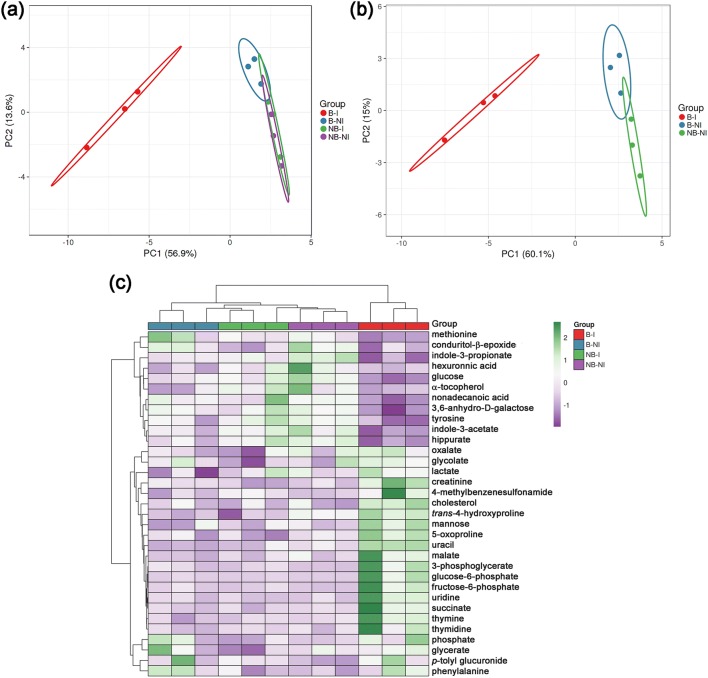
To identify metabolites that were differentially abundant among the experimental and control groups, we used the MetaboAnalyst program (Chong et al. 2018). The peak intensity levels of 34 metabolites (Online Resource 2) were found to be significantly different (P < 0.05) among the different groups by ANOVA followed post-hoc by Fisher’s LDS test. Levels of 19 metabolites were significantly increased in the B–I group compared to all the control groups, including thymidine, thymine, uridine, uracil, malate and succinate, trans-4-hydroxyproline, 5-oxoproline, and glucose-6-phosphate (Online Resource 2); while levels of eight metabolites that included glucose, indole-3-acetate, indole-3-propionate, and nonadecanoic acid were decreased (Online Resource 2). To define the metabolomic changes caused by P. aeruginosa-related sepsis following thermal injury, we used PCA and HCA, two forms of multivariate analysis. The PCA showed good separation between the samples from the B–I (Pa-sepsis) group and the control groups (Fig. 1a). The NB–NI (sham) and NB–I (infection without trauma) groups overlapped almost completely. This overlap is most likely due to the fact that uninjured mice do not become infected with PA14 following subcutaneous inoculation (Online Resource 1). Repeat PCA without the NB–I group showed that it may be possible to also identify metabolites that separate the B–I (Pa-sepsis) and B–NI (niSIRS) groups from each other (Fig. 1b). These results were confirmed by HCA, which showed two main branches on the dendrogram, with one branch representing the B–I experimental group and the other branch representing the control groups (Fig. 1c).
Fig. 1.
Metabolomic multivariate analysis of the mice plasma. a Principal component analysis (PCA) of the serum metabolomes from the 4 experimental groups (n = 3 for each group). PCA shows the distinct clustering of B–I samples separated from control groups. b PCA of the serum metabolites repeated without the NB–I group shows distinction of the B–NI group from the NB–NI group. c Heatmap and hierarchical clustering analysis of the serum metabolomes. Columns represent samples within each group and the rows show the different metabolites. Color scale implies the abundance of metabolites; green for higher abundance, purple for lower abundance
Analysis of metabolomic changes related to thermal injury and systemic infection
We used the ClassyFire tool (https://classyfire.wishartlab.com/; accessed 07 Sep 2019) (Djoumbou Feunang et al. 2016), plus the PubChem database (https://pubchem.ncbi.nlm.nih.gov/; accessed 12 Feb 2020) (Kim et al. 2019), the Human Metabolome Database (https://www.hmdb.ca/; accessed 12 Feb 2020) (Wishart et al. 2007) and the Chemical Entities of Biological Interest (ChEBI) database (https://www.ebi.ac.uk/chebi/init.do; accessed 12 Feb 2020) (Hastings et al., 2016) to broadly group the 148 metabolites into seven categories. These categories were predominated by amino acids and protein metabolites (40); sugars, sugar acids and alcohols, metabolites of glucolysis, gluconeogenesis and carbohydrate (34); and fatty acids and lipid metabolites (27); with pyrimidines and products of nucleotide/nucleoside metabolism (12); TCA cycle intermediates (11); other metabolites (7); and xenobiotics (17) also represented (Online Resource 3).
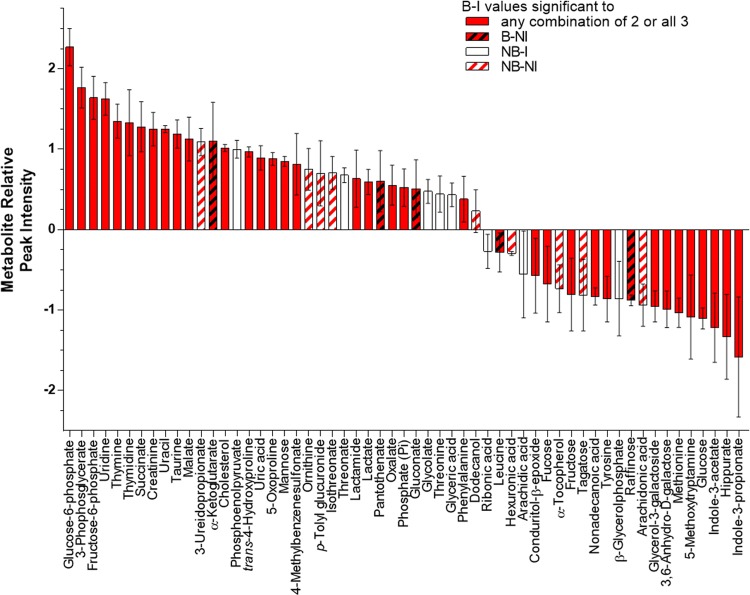
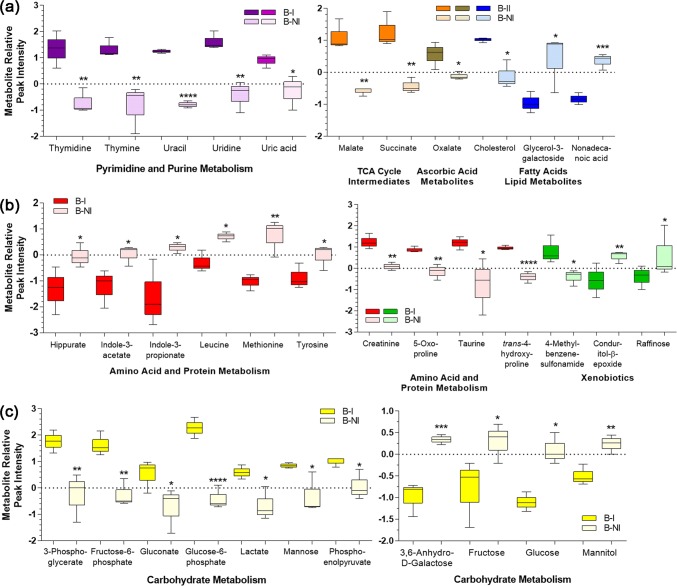
To ensure that we had selected all potentially significantly altered metabolites, further analysis using the calculated metabolite relative peak intensities and one-way ANOVA cross-comparing B–I, B–NI, and NB–NI (without the NB–I group) was performed on all 148 metabolites. In addition to the previous 34 metabolites, levels of an additional 22 metabolites were significantly increased in at least one comparison (Fig. 2, Online Resource 4). To more closely focus on those metabolites specifically altered in sepsis, we then performed one-way ANOVA on these 56 candidates using the B–I group (Pa-sepsis) as the comparator for the B–NI (niSIRS) and NB–NI groups. We then narrowed the pool of candidate biomarkers by focusing only on those that distinguished the B–I group (Pa-sepsis) from the B–NI group (niSIRS) resulting in a pool of 35 potential biomarkers. Among this group, levels of 21 metabolites were significantly reduced in the niSIRS group compared to Pa-sepsis, including seven metabolites of carbohydrate metabolism, four of pyrimidine metabolism, four of amino acid and protein metabolism, two TCA cycle intermediates, one each among fatty acid, purine, and vitamin metabolism, and one xenobiotic (Fig. 3). Levels of 14 metabolites were significantly elevated in the niSIRS group compared to Pa-sepsis: four from carbohydrate metabolism, six from amino acid and protein metabolism, two fatty acids, and two xenobiotics (Fig. 3). The remaining group of 21 metabolites did not distinguish Pa-sepsis from niSIRS (Online Resource 5). Further investigation will be necessary to determine if these metabolites would be useful in discriminating between individuals with niSIRS from trauma or other causes and those with infections who do not manifest sepsis.
Fig. 2.
Thermal injury and P. aeruginosa infection significantly increased or decreased the abundance of 58 metabolites in murine sera. Peak intensities were normalized using vector and median normalization methods and significance was determined by comparing B–I, B-NI, and NB–NI groups using one-way ANOVA followed by Fisher’s least significant difference test. Metabolites with P ≤ 0.05 are displayed from highest relative peak intensity to the lowest. Each bar represents 3 biological samples ± SEM. B–I, thermally injured/infected (Pa-sepsis); B-NI, thermally injured/not infected (niSIRS); NB–I, not injured/infected (control); NB-NI, not injured/not infected (sham control)
Fig. 3.
Thermal injury and P. aeruginosa infection significantly increased the abundance of 21 metabolites and decreased the abundance of 14 in murine sera of B–I mice compared to B–NI mice. Peak intensities were normalized using vector and median normalization methods and significance was determined using one-way ANOVA using B–I as the comparator to B–NI and NB-NI, followed by Fisher’s least significant difference test. Each box plot represents 3 biological samples and -whiskers represent their distribution. Differential abundance of a metabolites of pyrimidine and purine metabolism, intermediates of the TCA cycle, ascorbic acid metabolite, and fatty acid and lipid metabolites; b metabolites of amino acid and protein metabolism and xenobiotics; c sugars, sugar acids, and other metabolites of carbohydrate metabolism. B–I, thermally injured/infected (Pa-sepsis); B-NI, thermally injured/not infected (niSIRS); NB–NI (sham control); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Diagnostic performance of potential markers for P. aeruginosa sepsis following thermal injury
To test their diagnostic potential as markers for early sepsis with P. aeruginosa, we determined the ROC curves for all of the identified metabolites. Due to the small number of samples in our study, we applied a cut-off of ≥ 0.96 for the AUC. For 25 metabolites, the AUC were determined to be 1 (1.0) (95% confidence interval calculated using 500 bootstraps), indicating these metabolites could be considered predictive of Pa-sepsis (Table 1). Two metabolites not selected in our first analysis – tagatose and taurine (Table 1)—were included in the 25. Taurine had been selected as statistically significant in subsequent analyses of the data (Figs. 2 and 3b), but tagatose was shown not to be useful for discriminating Pa-sepsis from niSIRS (Online Resource 5). The AUC for five metabolites was 0.96 (glycerol-3-galactoside, fructose, phosphoenolpyruvate, tyrosine, and 5-methyltryptamine), suggesting that they might also be predictive (Fig. 3, Online Resource 5). However, as for tagatose, statistical analyses did not support 5-methyltryptamine as a potential biomarker for Pa-sepsis. Five of the original metabolites (glycolate, lactate, oxalate, p-tolyl glucuronide, and conduritol-β-epoxide) were eliminated by their AUC (Table 1). One metabolite with an AUC of 1.0, 3-ureidopropionate, had produced variable results in statistical analyses of the relative peak intensity data. Further analysis of the peak intensities without the NB–I group and using the B–I group as comparator revealed significant differences similar to those of uracil and uridine (Table 1). Interrogation of the Human Metabolome Database showed that 4-methylbenzenesulfonamide and 3,6-anhydro-D-galactose (as well as conduritol-β-epoxide) have been found in mice but not in humans. These metabolites were removed from consideration. The final set of 26 metabolites for consideration as potential diagnostic biomarkers of sepsis from P. aeruginosa included nine amino acids and their metabolites; seven sugars, sugar phosphates, and sugar acids; five pyrimidines and derivatives; three fatty acid and lipid metabolites; and two intermediates of the TCA cycle (Table 1).
Table 1.
Potential biomarkers for diagnosis of P. aeruginosa sepsis
| Pathway or source/metabolite | Relative peak intensities | P value ≤ 0.05 | AUC ≥ 0.96a | Included in the set of 26 | |
|---|---|---|---|---|---|
| B–I (Pa-sepsis) | B–NI (niSIRS) | B–I:B–NI | |||
| Purine metabolism | |||||
| Uric acid | 0.893 ± 0.15 | − 0.277 ± 0.37 | 0.0042** | 0.93 | No |
| Pyrimidine metabolism | |||||
| 3-Ureidopropionate – RPI | 1.090 ± 0.17 | − 0.367 ± 0.65 | ns | 1 | Follow-upb |
| 3-Ureidopropionate – PI | 3969 ± 550.8 | 1809 ± 618.4 | 0.0314* | 1 | Yes |
| Thymidine | 1.330 ± 0.225 | − 0.687 ± 0.27 | 0.0074** | 1 | Yes |
| Thymine | 1.350 ± 0.21 | − 0.857 ± 0.525 | 0.0040** | 1 | Yes |
| Uracil | 1.250 ± 0.04 | − 0.780 ± 0.08 | < 0.0001**** | 1 | Yes |
| Uridine | 1.327 ± 0.20 | − 0.430 ± 0.345 | 0.0288* | 1 | Yes |
| Fatty acid and lipid metabolism | |||||
| Cholesterol | 1.017 ± 0.04 | − 0.110 ± 0.25 | 0.0301* | 1 | Yes |
| Glycerol-3-galactoside | − 0.953 ± 0.19 | 0.393 ± 0.52 | 0.0258* | 0.96 | Yes |
| Nonadecanoic acid | − 0.830 ± 0.11 | 0.357 ± 0.15 | 0.0007*** | 1 | Yes |
| Amino acid and protein metabolism | |||||
| 5-Oxoproline | 0.880 ± 0.08 | − 0.160 ± 0.22 | 0.0049** | 1 | Yes |
| Creatinine | 1.250 ± 0.21 | 0.087 ± 0.11 | 0.0098** | 1 | Yes |
| Hippurate | − 1.333 ± 0.53 | − 0.037 ± 0.27 | 0.0377* | 1 | Yes |
| Indole-3-acetate | − 1.220 ± 0.43 | 0.023 ± 0.23 | 0.0256* | 1 | Yes |
| Indole-3-propionate | − 1.583 ± 0.75 | 0.287 ± 0.12 | 0.0235* | 1 | Yes |
| Leucine | − 0.283 ± 0.24 | 0.710 ± 0.11 | 0.0351* | 0.81 | No |
| Methionine | − 1.033 ± 0.18 | 0.730 ± 0.41 | 0.0044** | 1 | Yes |
| Taurine | 1.190 ± 0.18 | − 0.770 ± 0.77 | 0.291* | 1 | Yes |
| trans-4-Hydroxyproline | 0.967 ± 0.06 | − 0.410 ± 0.16 | < 0.0001**** | 1 | Yes |
| Tyrosine | − 0.863 ± 0.28 | − 0.030 ± 0.29 | 0.0448* | 0.96 | Yes |
| TCA cycle | |||||
| Malate | 1.127 ± 0.27 | − 0.593 ± 0.07 | 0.0016** | 1 | Yes |
| Succinate | 1.280 ± 0.31 | − 0.433 ± 0.14 | 0.0015** | 1 | Yes |
| Glycolysis and gluconeogenesis | |||||
| 3,6-Anhydro-D-galactose | − 0.990 ± 0.23 | 0.340 ± 0.07 | 0.0005*** | 1 | Noc |
| 3-Phosphoglycerate | 1.767 ± 0.25 | − 0.267 ± 0.53 | 0.0055** | 1 | Yes |
| Fructose | − 0.807 ± 0.45 | 0.297 ± 0.26 | 0.0422* | 0.96 | Yes |
| Fructose-6-phosphate | 1.640 ± 0.45 | − 0.240 ± 0.30 | 0.0012** | 1 | Yes |
| Gluconate | 0.510 ± 0.36 | − 0.743 ± 0.49 | 0.0488* | 0.78 | Yes |
| Glucose | − 1.103 0.13 | 0.103 ± 0.21 | 0.0015** | 1 | Yes |
| Glucose-6-phosphate | 2.270 ± 0.23 | − 0.397 0.26 | < 0.0001**** | 1 | Yes |
| Lactate | 0.593 ± 0.16 | − 0.653 ± 0.37 | 0.0101* | 0.85 | No |
| Mannitol | − 0.497 ± 0.14 | 0.237 ± 0.13 | 0.0331* | 0.89 | No |
| Mannose | 0.850 ± 0.06 | − 0.273 ± 0.11 | 0.0023* | 1 | Yes |
| Phosphoenolpyruvate | 1.000 ± 0.11 | 0.070 ± 0.33 | 0.0219* | 0.96 | Yes |
| Vitamins and derivatives | |||||
| Oxalate | 0.553 ± 0.25 | − 0.113 ± 0.07 | 0.0474* | 0.85 | No |
| Xenobiotics | |||||
| 4-Methylbenzenesulfonamide | 0.813 ± 0.38 | − 0.407 ± 0.22 | 0.0169* | 1 | Noc |
| Conduritol-β-epoxide | − 0.573 ± 0.46 | 0.553 ± 0.17 | 0.0037** | 0.89 | Noc |
| Raffinose | − 0.880 ± 0.07 | 0.990 ± 0.66 | 0.0334* | 0.89 | No |
AUC area under the curve; RPI relative peak intensity; PI peak intensity; ns no significance; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
aFor AUC, the cut-off for diagnostic discrimination was set at ≥ 0.96
b3-Ureidopropionate was selected in the first and second rounds of analyses and failed the third; as the AUC was 1.0, evaluation of the peak intensity values was done
cMetabolite has not been reported in humans; information obtained from the Human Metabolome Database (https://www.hmdb.ca/; accessed 12 Feb 2020) (Wishart et al. 2007)
Pathway analysis of metabolomic changes occurring with thermal injury and systemic infection
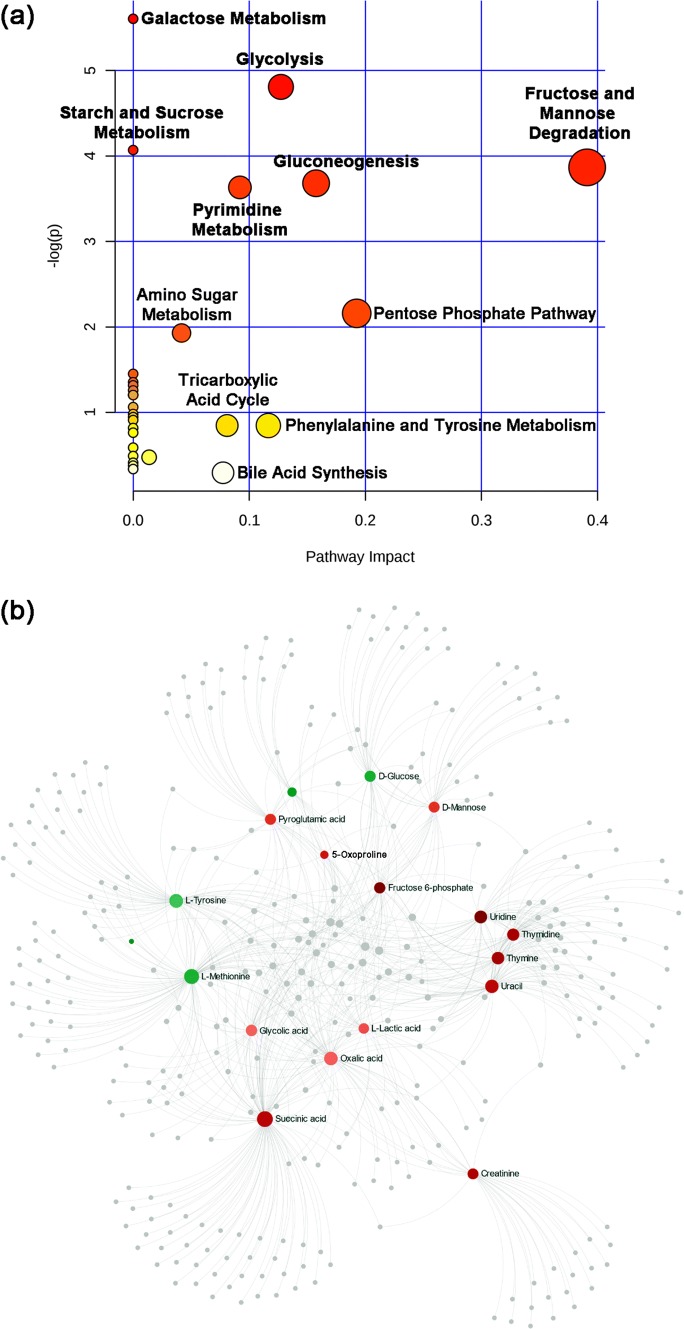
To investigate whether these metabolites would map to pathways related to the physiological changes that occur in the murine sera after thermal injury and systemic infection with P. aeruginosa, we mapped the set of metabolites using the murine Small Molecule Pathway Database within MetaboAnalyst by implementing over representation analysis with Fisher's exact test and pathway topology analysis using relative-betweenness centrality (Jewison et al. 2014). Pathway enrichment analysis for the metabolites revealed seven pathways with significant changes – six related to carbohydrate metabolism and pyrimidine metabolism (P < 0.05) (Fig. 4a). To further discover any possible metabolite-metabolite interaction, we used the network explorer function within MetaboAnalyst followed by application of NetworkAnalyst software (Zhou et al. 2019). This analysis supported the previous results, showing that the pathway for pyrimidine metabolism (metabolites thymidine, thymine, uridine, and uracil) was the most notable pathway (P < 0.001) (Fig. 4b).
Fig. 4.
Mapping the metabolites to pathways and networks. a Pathway enrichment analysis of the 35 metabolites showing significant changes in the B–I group. Pathways labeled in red contain the highest number of mapped metabolites (P < 0.05). The map was produced by analyzing the metabolites through over-representation analysis with Fisher’s exact test and pathway topology using the murine Small Molecule Pathway Database within MetaboAnalyst. b Metabolite-metabolite interaction network highlighting potential functional relationships among the annotated metabolites. Metabolites in shades of red are those with higher concentration in thermally injured and P. aeruginosa infected mice; those in shades of green are those with lower concentration. All gray circles are metabolites linked to, but not present in, our dataset
Discussion
In this study, we identified a set of 26 potential biomarkers for early sepsis related to P. aeruginosa infection of thermal injury in mice. We demonstrated that only thermally injured mice infected with P. aeruginosa developed sepsis within 24 h (Online Resource 1). The GC-TOF-MS analysis of murine sera showed a difference in the abundance of numerous metabolites during P. aeruginosa-related sepsis compared to control groups. Similar to previously reported studies, we found differential abundance of amino acids, metabolites of glycolysis and gluconeogenesis, pyrimidines, fatty acids and lipids, and the TCA cycle (Figs. 2 and 3) (Hoerr et al. 2012; Lin et al. 2016; Liu et al. 2010; Ludwig and Hummon 2017; Wei et al. 2016). Differences in models, including the protocol used to induce sepsis (infection of burn wound, intraperitoneal injection, or cecal ligation and puncture [CLP]), the organism(s) used (P. aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, E. coli, or mixed infection from CLP), timing of harvest of samples (4 h post infection, 12 h, or 24 h), and methods used to detect metabolites (LC/MS versus 1H NMR versus GC/MS) makes it difficult to directly compare the entire sets of results from these different studies. In several studies, the majority of reported metabolites constituted acylcarnitines and lipid molecules best discovered by LC/MS (Lin et al. 2016; Liu et al. 2010; Wei et al. 2016), which makes it difficult to compare the metabolites we detected by GC-TOF/MS. However, Hoerr et al. (Hoerr et al. 2012), who used 1H- NMR, found 43 metabolites differentially abundant in mice infected with P. aeruginosa by intraperitoneal injection compared to the 56 we found in mice with P. aeruginosa burn wound infections (Fig. 2) (Hoerr et al. 2012). We found 35 metabolites in common with their study, but only 10 were similar in levels; notably, glucose was reduced in both studies and amino acids and protein metabolites were found in similar abundance: 2-hydroxybutyrate, creatinine, taurine and phenylalanine were elevated while asparagine was reduced and isoleucine, alanine and glutamine were detected but levels were the same in sepsis and controls (Online Resource 4) (Hoerr et al. 2012). Thus, comparison of functional groups of metabolites or even individual metabolites provides a way to determine which metabolites might be used for the diagnosis of sepsis related to P. aeruginosa.
Among the metabolites that were increased in the blood of thermally-injured mice septic with P. aeruginosa were thymidine, thymine, uridine, and uracil, which are involved in pyrimidine metabolism (Fig. 3a). These nucleobases and nucleosides serve fundamental roles in the replication of genetic material, gene transcription, protein synthesis, and cellular metabolism. Thymine and uracil are incorporated into DNA and RNA, respectively, during their synthesis. Uracil was among nine metabolites identified by Liu et al. that were changed when thermally injured rats became septic (Liu et al. 2010). While we did not identify all nine of their markers, two –gluconate and uric acid – followed the same pattern of increase in the septic animals and decrease in the animals that were thermally injured only (Online Resource 4) (Liu et al. 2010). It has been demonstrated that uracil influences quorum sensing and biofilm formation in P. aeruginosa by enhancing transcription of quorum sensing genes with subsequent increase in biofilm formation (Ueda et al. 2009). Since uracil was only increased in the B–I group, PA14 dissemination preceded the increase in uracil. Whether the increase in serum uracil would then help those PA14 organisms remaining in the wound to establish biofilm, leading to more dissemination, is not known at this time. P. aeruginosa can also utilize pyrimidines as a carbon and/or nitrogen source (West and Chu 1986). Interestingly, 3-ureidopropionate, which is a metabolite of uracil, was identified by ROC analysis as a distinguishing marker in septic mice (Table 1, Online Resource 3).
Trans-4-hydroxyproline is a major constituent of the collagen protein found in connective tissue, muscle, and skin (Ananthanarayanan 1983). This molecule is released due to collagen breakdown (Wu et al. 2011). While the release of trans-4-hydroxyproline could occur with tissue injury, the level was reduced in the B–NI group compared to the NB–NI group (Online Resource 4). P. aeruginosa is known to be able to degrade collagen (Hao et al. 1999). The increase in trans-4-hydroxyproline in the B–I group, compared to all control groups, is most likely a result of the collagen degradation happening when P. aeruginosa invaded the tissue and disseminated. Werthén et al. (2004) showed that P. aeruginosa degrades the collagen in skin biopsies within hours of inoculation (Werthén et al. 2004). Moreover, P. aeruginosa, which can use trans-4-hydroxyproline as a carbon and nitrogen source, has an uptake system for it that is induced by the presence of the molecule (Li and Lu 2016).
The increase in 5-oxoproline, also known as pyroglutamic acid, in septic mice could be indicative of imbalance in glutamine and glutathione metabolism as previously documented in critically ill patients that experience low glutathione levels (Liu et al. 2014). Known as pyroglutamic acidosis, this can also occur with other conditions besides sepsis such as chronic therapeutic paracetamol (acetaminophen) use, malnutrition, antibiotics and renal impairment (Hunter et al. 2016). However, 5-oxoproline could be used in conjunction with other potential diagnostic markers to indicate Pa-sepsis.
The amino acids methionine and tyrosine were significantly decreased in septic mice compared to the control group, while taurine was increased (Fig. 3b, Online Resource 4). Although the specific etiologic agent of the sepsis in each patient was not indicated, a previous study in humans showed that compared to nonseptic patients (n = 18), the levels of methionine and tyrosine were significantly decreased in patients with sepsis (n = 35) on the day of admission to intensive care while the level of taurine was significantly increased (Su et al. 2015). Recently, Lawal et al. (2018) used analysis of volatile organic compounds to determine whether compounds specific to Enterobacter cloacae or P. aeruginosa would be produced when the organisms were grown individually or in co-culture in artificial sputum medium (Lawal et al. 2018). They found some compounds such as cyclopentanone were produced by both organisms individually and in co-culture; some were produced by P. aeruginosa alone and in co-culture (1-undecene, pyrrole, methyl 2-ethylhexanoate) and some by E. cloacae alone but greatly reduced in co-culture (2-methyl-1-propanol, 3-methyl-1-butanol, 2-phenylethanol) (Lawal et al., 2018). Other compounds were found elevated only in co-culture—methyl 2-methylbutyrate, isoamyl butyrate, 2-methylbutyl acetate (Lawal et al. 2018). Since other organisms besides P. aeruginosa can use amino acids as sources of nitrogen and/or carbon, we would expect similar findings—some metabolites unique to specific organisms and some metabolites in common—if other organisms such as Staphylococcus aureus or E. cloacae were tested.
Indole-3-acetate and indole-3-propionate, which were significantly decreased in septic thermally-injured mice, are products of tryptophan metabolism by the gut microbiota (Gao et al. 2018). Further, some studies showed that with sepsis the gut microbiome function is disrupted (McDonald et al. 2016; Stadlbauer et al. 2019), which could lead to the decrease of these metabolites. P. aeruginosa does not possess the enzymes to produce either indole metabolite from tryptophan (Bortolotti et al. 2016). Why the reduction in indole-3-acetate and indole-3-propionate occurred primarily in the B–I group is not known at this time. Their decrease could be a sign of dysbiosis that requires further study.
Organic acids including malic, succinic, oxalic and lactic acids were at higher levels in septic mice, which could be a sign of a disturbance in the TCA cycle (malic and succinic acids) and the presence of oxidative stress (oxalic and lactic acids). The TCA cycle is used by aerobic organisms to release stored energy. Meylan et al. (2017) showed that TCA cycle activity drives the lethality of the aminoglycoside antibiotic tobramycin in P. aeruginosa (Meylan et al. 2017). They found that supplementation of media with fumarate and succinate, components of the lower part of the TCA cycle, sensitized stationary-phase cells to killing by tobramycin (Meylan et al. 2017). In contrast, components of the upper TCA cycle such as citrate had little effect (Meylan et al. 2017). Numerous metabolites found as components of the TCA cycle were detected in our metabolomics profiles, including significantly increased levels of succinate and malate (Figs. 2 and 3), as well as the presence of citrate, isocitrate, cis-aconitate, α-ketoglutarate, and fumarate (Online Resources 2 and 4). Besides serving as markers for sepsis, the increased levels of specific TCA cycle metabolites might be useful in predicting alterations in tobramycin susceptible phenotype.
The level of the sugar mannose was increased in sera of mice with sepsis. Due to the abundance of mannose on host cell surfaces, this increase could be a result of cell damage and the release of mannose into the bloodstream; however, levels of mannose in the controls was considerably lower. It was recently shown that the mannose level also increases in the blood of trauma patients and that P. aeruginosa can sense that increase and upregulate its mannose transport system to use the sugar as a carbon source (Elmassry et al. 2019). Compared to controls, fructose-6-phosphate (F6P) and glucose-6-phosphate (G6P) were found at significantly higher levels in B–I mice, 4.7-fold and 18-fold, respectively; while glucose was found to be significantly lower (− 2.3-fold). Moreover, the average ratio of glucose to G6P in septic mice was 1:23, a drop of 47-fold compared to the average ratio of 1:1090 in the three control groups. These results suggest disturbances occurred within the glycolytic/gluconeogenic pathway.
Interest in, and understanding of, the fluidity of the human metabolome in response to different conditions has led to analyses looking for changes in metabolite abundance related to general demographics such as age, sex, and race (Lawton et al. 2008) as well as studies focused on specific metabolic responses present in a specific disease; such as examination of the metabolome to study pathophysiology of oxidative stress in cancer (Andrisic et al. 2018). Because of the lethal nature of sepsis and the rapidity with which it can kill, and because current markers such as serum lactate, C-reactive protein, and procalcitonin have been shown to be insufficiently discriminating (Mann et al. 2011; Rhee et al. 2015), there has been much interest in metabolomics as a means to identify markers that could better predict which patients with sepsis are likely to progress to severe sepsis, septic shock, and/or succumb (Ferrario et al. 2016; Holder et al. 2016; Langley et al. 2014, 2013; Seymour et al. 2013; Su et al. 2014). Additionally, analysis of the metabolome has been used to determine which patients with signs and symptoms of sepsis are truly septic or are suffering from noninfectious SIRS (Schmerler et al. 2012). As with the studies in animal models, differences in types of infections (bloodstream versus pneumonia), timing (< 4 h post admittance to 24 h to 3–7 days), and methods of detection of metabolites make comparison of the studies difficult. Comparing sepsis survivors and nonsurvivors to patients with niSIRS, Langley et al. (Langley et al. 2014) found that patients who survived sepsis had lower levels of malate, phosphate, and ketogenic amino acids, while nonsurvivors had higher levels of lactate, malate, phosphate, and glucogenic amino acids and lower levels of glucose. In our study the B–I mice will become sepsis nonsurvivors while the B–NI mice represent the niSIRS group. Compared to B–NI mice, the B–I mice had significantly higher levels of malate and lactate and lower levels of glucose, but the phosphate levels were elevated in both over sham (normal) controls similar to the nonsurvivors (Fig. 3, Online Resource 5). Levels of the ketogenic amino acids leucine and tyrosine were significantly reduced as were levels of the glucogenic amino acid methionine (Fig. 3, Table 1), similar to their survivors (Langley et al. 2014). Whether testing at 48 h (near the usual time of death in the model) would result in a shift in the levels of phosphate and amino acids closer to those observed in the nonsurvivors is not known at this time. Although Langley et al. (Langley et al. 2014) reported no differences in metabolomic profiles for the types of pathogens present in their sepsis patients, it is also possible that the changes in amino acid levels could reflect the growth of P. aeruginosa in the B–I mice. Ferrario et al. (2016) also compared sepsis survivors at day 7 to their profiles on day 1, and nonsurvivors at day 7 to survivors at day 7. We detected three of the metabolites considered significant in the survivors at day 7— “sugars,” serine, and creatinine—with the exact opposite results; sugars and serine were reduced while creatinine was elevated in the B–I mice (Figs. 2, 3, Online Resource 4) (Ferrario et al. 2016). Amino acids were the major metabolites significantly different in the nonsurvivors at day 7 (Ferrario et al. 2016). We detected all six of the amino acids, but the levels for the two amino acids that were significantly different in our septic mice (asparagine and tyrosine) were opposite – reduced rather than elevated (Figs. 2, 3, and Online Resource 4) (Ferrario et al. 2016). Since there was no mention of specific pathogens isolated from the septic patients, the levels of metabolites we observed may reflect the growth of P. aeruginosa. Unfortunately, many of the metabolites reported in the human metabolome studies on sepsis were detected by LC/MS and constitute phosphocholines, glycerophospholipids, sphingomyelins, and acylcarnitines, which are not detected by GC/MS, the method we used (Ferrario et al. 2016; Schmerler et al. 2012; Su et al. 2014). Despite the similarity of methodology within those studies, it is interesting that few, if any, of the metabolites identified as potential biomarkers were the same from one study to the next (Ferrario et al. 2016; Schmerler et al. 2012; Su et al. 2014). What we found in common with these studies is that, despite the differences in approaches, we identified metabolites from the same metabolic pathways.
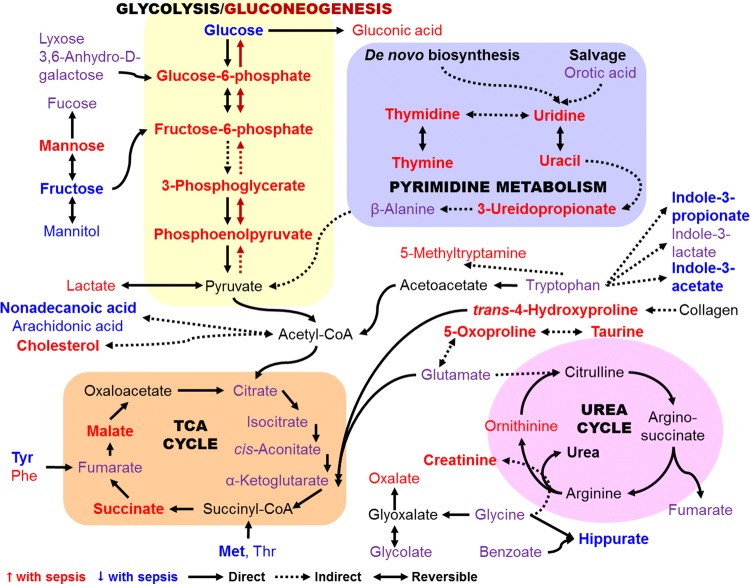
In summary, 25 of the 26 metabolites with levels significantly altered by thermal injury plus P. aeruginosa sepsis were mapped to four metabolic pathways—glycolysis/gluconeogenesis, the TCA cycle, the urea cycle, and pyrimidine metabolism plus related amino acid metabolic links (Fig. 5). Additionally, 24 intermediate metabolites within these pathways were also detected among the 148 identified metabolites, although the changes in their levels were not found to be significant enough to be considered among the diagnostic biomarkers (Fig. 5). Certainly, a substantial disturbance in metabolism occurs during sepsis from dissemination of P. aeruginosa from local infection, which emphasizes the value of this approach to identify new potential markers of sepsis.
Fig. 5.
Schematic overview of the metabolites with serum levels significantly altered with thermal injury and P. aeruginosa sepsis. Metabolites with increased levels are in red and those with decreased levels are in blue; bold metabolites are included in the set of potential biomarkers for Pa-sepsis—25 of the 26 metabolites in the set map to these four interconnected pathways. Metabolites in purple were among the 148 identified metabolites detected in the sera but changes in their levels were not considered significant; however, they are intermediates in the pathways shown. Metabolites in black were not detected in our analysis. The rounded shapes labeled in all capitals contain the four metabolic pathways most interlinked. Solid lines, direct reactions; dotted lines, indirect reactions; arrowhead indicates direction of the reaction; double arrowhead, reversible reactions
Within our set of 26 potential biomarkers, we identified a group of five metabolites that constitute a unique set of biomarkers: trans-4-hydroxyproline (a marker of collagen degradation), 5-oxoproline (a marker of oxidative stress), indole-3-acetate (dysbiosis), indole-3-propionate (dysbiosis; recently implicated as a potential marker of oxidative stress in cancer (Andrisic et al. 2018; Ke et al. 2016)), and glycerol-3-galactoside (possibly reflecting glycerolipid synthesis following stress (Grapov et al. 2019)). This unique group, together with selected metabolites from pyrimidine, glucose, and fatty acid metabolism and the TCA cycle (e.g., uracil, glucose-6-phosphate, nonadecanoic acid, malate, and succinate (Fig. 3) that were the most discriminatory for Pa-sepsis from niSIRS may have the potential to serve as diagnostic biomarkers for sepsis caused by P. aeruginosa in severely burned patients.
Conclusion
Metabolomics studies are integral to the identification of new markers to assist the diagnosis of sepsis in critically ill patients. Among the analyzed metabolites, 26 metabolites were shown to have high predictive power for P. aeruginosa sepsis following thermal injury. It is very clear that these changes in the metabolome occurred only with thermal injury plus infection. Examination of sepsis following infection of burn wounds by other microbes such as the Gram-positive coccus S. aureus and the Gram-negative bacillus Acinetobacter baumannii would help establish the specificity of these metabolites as markers for bacterial sepsis in general, Gram-negative bacterial sepsis, or for the individual organism. The latter would be most helpful in early diagnosis as it would assist in the determination of appropriate antibiotic therapy. While we used an untargeted metabolomics approach to detect as many metabolites as possible, once a set of markers is established and validated in humans, shifting to a targeted approach of identifying just those specific “P. aeruginosa sepsis” metabolites would be the next step. Alternatively, should a specific pathway be confirmed as predictive, such as the pyrimidine metabolic pathway, then detection and quantitation of those metabolites could be sufficient.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Joanna E. Swickard for critical reading of this manuscript.
Author contributions
ANH, SD, JAG conceived and designed research. MME and NM conducted experiments. MME and JACH analyzed data. MME, MJS, and JACH interpreted the results. SD and JAG provided consultation on clinical relevance. MME, JACH, and ANH wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was funded in part by the Burn Center of Research Excellence (BCoRE) in the Department of Surgery at TTUHSC, Lubbock, TX, USA. MME was funded by a grant-in-aid from the Texas Tech University Association of Biologists and a summer dissertation research award, and a graduate student research support award from the Texas Tech University Graduate School.
Data availability
The metabolomics and metadata reported in this paper are available via the Metabolomics Workbench public repository (Sud et al., 2016) (https://www.metabolomicsworkbench.org/data/DRCCMetadata.php?Mode=Study&StudyID=ST001101) under study identifier ST001101. These data include the 383 unidentified metabolites as well as the 148 identified metabolites.
Compliance with ethical standards
Conflict of interest
Moamen M. Elmassry, Nithya Mudaliar, Jane A. Colmer-Hamood, Michael J. San Francisco, John A. Griswold, Sharmila Dissanaike, and Abdul N. Hamood declare that they have no conflict of interest.
Research involving animals and human participants
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Animals were treated humanely and in accordance with the protocol approved by the Animal Care and Use Committee at the Texas Tech University Health Sciences Center (TTUHSC), Lubbock, TX. This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ananthanarayanan VS. Structural aspects of hydroxyproline-containing proteins. Journal of Biomolecular Structure & Dynamics. 1983;1(3):843–855. doi: 10.1080/07391102.1983.10507485. [DOI] [PubMed] [Google Scholar]
- Andrisic L, Dudzik D, Barbas C, Milkovic L, Grune T, Zarkovic N. Short overview on metabolomics approach to study pathophysiology of oxidative stress in cancer. Redox Biology. 2018;14:47–58. doi: 10.1016/j.redox.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs in Context. 2018;7:212527. doi: 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron BM, Ayala A, Lomas-Neira JL. Biomarkers for sepsis: What is and what might be? Biomarker Insights. 2015;10(Suppl 4):7–17. doi: 10.4137/BMI.S29519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti P, Faure E, Kipnis E. Inflammasomes in tissue damages and immune disorders after trauma. Frontiers in Immunology. 2018;9:1900. doi: 10.3389/fimmu.2018.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti P, Hennart B, Thieffry C, Jausions G, Faure E, Grandjean T, et al. Tryptophan catabolism in Pseudomonas aeruginosa and potential for inter-kingdom relationship. BMC Microbiology. 2016;16(1):137. doi: 10.1186/s12866-016-0756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera CP, Manson J, Shepherd JM, Torrance HD, Watson D, Longhi MP, et al. Signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: A prospective cohort study. PLoS Medicine. 2017;14(7):e1002352. doi: 10.1371/journal.pmed.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Research. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstedt P, Storgaard M, Lassen AT. The systemic inflammatory response syndrome (SIRS) in acutely hospitalised medical patients: A cohort study. Scandinavian Journal of Trauma, Resuscitation and Emergency Medicine. 2009;17:67. doi: 10.1186/1757-7241-17-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Prost N, Razazi K, Brun-Buisson C. Unrevealing culture-negative severe sepsis. Critical Care. 2013;17(5):1001. doi: 10.1186/cc13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou Feunang Y, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. Journal of Cheminformatics. 2016;8:61. doi: 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzvova N, Colmer-Hamood JA, Griswold JA, Hamood AN. Heparinase is essential for Pseudomonas aeruginosa virulence during thermal injury and infection. Infection and Immunity. 2018;86(1):e00755-17. doi: 10.1128/iai.00755-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmassry MM, Mudaliar NS, Kottapalli KR, Dissanaike S, Griswold JA, San Francisco MJ, et al. Pseudomonas aeruginosa alters its transcriptome related to carbon metabolism and virulence as a possible survival strategy in blood from trauma patients. mSystems. 2016;4(4):e00312-18. doi: 10.1038/srep20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario M, Cambiaghi A, Brunelli L, Giordano S, Caironi P, Guatteri L, et al. Mortality prediction in patients with severe septic shock: A pilot study using a target metabolomics approach. Scientific Reports. 2016;6:20391. doi: 10.1038/srep20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, et al. Quality control for plant metabolomics: Reporting MSI-compliant studies. The Plant Journal. 2008;53(4):691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- Fleischmann-Struzek C, Mikolajetz A, Schwarzkopf D, Cohen J, Hartog CS, Pletz M, et al. Challenges in assessing the burden of sepsis and understanding the inequalities of sepsis outcomes between National Health Systems: Secular trends in sepsis and infection incidence and mortality in Germany. Intensive Care Medicine. 2018;44(11):1826–1835. doi: 10.1007/s00134-018-5377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauer, R. L. (2013). Early recognition and management of sepsis in adults: The first six hours. American Family Physician, 88(1), 44–53. https://www.ncbi.nlm.nih.gov/pubmed/23939605. [PubMed]
- Grapov D, Fiehn O, Campbell C, Chandler CJ, Burnett DJ, Souza EC, et al. Exercise plasma metabolomics and xenometabolomics in obese, sedentary, insulin-resistant women: Impact of a fitness and weight loss intervention. American journal of physiology. Endocrinology and metabolism. 2019;317(6):E999–E1014. doi: 10.1152/ajpendo.00091.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh DG. Sepsis in the burn patient: a different problem than sepsis in the general population. Burns Trauma. 2017;5:23. doi: 10.1186/s41038-017-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JL, Nagano T, Nakamura M, Kumagai N, Mishima H, Nishida T. Effect of galardin on collagen degradation by Pseudomonas aeruginosa. Experimental Eye Research. 1999;69(6):595–601. doi: 10.1006/exer.1999.0755. [DOI] [PubMed] [Google Scholar]
- Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Research. 2016;44(D1):D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada T, Wada H, Nobori T, Okabayashi K, Maruyama K, Abe Y, et al. Plasma concentrations and importance of High Mobility Group Box protein in the prognosis of organ failure in patients with disseminated intravascular coagulation. Thrombosis and Haemostasis. 2005;94(5):975–979. doi: 10.1160/TH05-05-0316. [DOI] [PubMed] [Google Scholar]
- Heffner AC, Horton JM, Marchick MR, Jones AE. Etiology of illness in patients with severe sepsis admitted to the hospital from the emergency department. Clinical Infectious Diseases. 2010;50(6):814–820. doi: 10.1086/650580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerr V, Zbytnuik L, Leger C, Tam PP, Kubes P, Vogel HJ. Gram-negative and Gram-positive bacterial infections give rise to a different metabolic response in a mouse model. Journal of Proteome Research. 2012;11(6):3231–3245. doi: 10.1021/pr201274r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder AL, Gupta N, Lulaj E, Furgiuele M, Hidalgo I, Jones MP, et al. Predictors of early progression to severe sepsis or shock among emergency department patients with nonsevere sepsis. International Journal of Emergency Medicine. 2016;9(1):10. doi: 10.1186/s12245-016-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RW, Lawson C, Galitsiou E, Gifford F, Neary JJ. Pyroglutamic acidosis in association with therapeutic paracetamol use. Clinical Medicine (Lond) 2016;16(6):524–529. doi: 10.7861/clinmedicine.16-6-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, et al. SMPDB 2.0: Big improvements to the small molecule pathway database. Nucleic Acids Research. 2014;42(D1):D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke C, Li A, Hou Y, Sun M, Yang K, Cheng J, et al. Metabolic phenotyping for monitoring ovarian cancer patients. Scientific Reports. 2016;6:23334. doi: 10.1038/srep23334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Research. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley RJ, Tipper JL, Bruse S, Baron RM, Tsalik EL, Huntley J, et al. Integrative "omic" analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. American Journal of Respiratory and Critical Care Medicine. 2014;190(4):445–455. doi: 10.1164/rccm.201404-0624OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Science Translational Medicine. 2013;5(195):195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal O, Knobel H, Weda H, Nijsen TME, Goodacre R, Fowler SJ, et al. TD/GC-MS analysis of volatile markers emitted from mono- and co-cultures of Enterobacter cloacae and Pseudomonas aeruginosa in artificial sputum. Metabolomics. 2018;14(5):66. doi: 10.1007/s11306-018-1357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Li G, Lu CD. Molecular characterization of LhpR in control of hydroxyproline catabolism and transport in Pseudomonas aeruginosa PAO1. Microbiology. 2016;162(7):1232–1242. doi: 10.1099/mic.0.000300. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proceedings of the Natlional Academy of Sciences USA. 2006;103(8):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Liu X, Sun L, Li J, Hu Z, Xie H, et al. Comparison of sepsis rats induced by caecal ligation puncture or Staphylococcus aureus using a LC-QTOF-MS metabolomics approach. Infection Genetics and Evolution. 2016;43:86–93. doi: 10.1016/j.meegid.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Liu VX, Fielding-Singh V, Greene JD, Baker JM, Iwashyna TJ, Bhattacharya J, et al. The timing of early antibiotics and hospital mortality in sepsis. American Journal of Respiratory and Critical Care Medicine. 2017;196(7):856–863. doi: 10.1164/rccm.201609-1848OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XR, Zheng XF, Ji SZ, Lv YH, Zheng DY, Xia ZF, et al. Metabolomic analysis of thermally injured and/or septic rats. Burns. 2010;36(7):992–998. doi: 10.1016/j.burns.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hyde AS, Simpson MA, Barycki JJ. Emerging regulatory paradigms in glutathione metabolism. Advances in Cancer Research. 2014;122:69–101. doi: 10.1016/B978-0-12-420117-0.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livaditi O, Kotanidou A, Psarra A, Dimopoulou I, Sotiropoulou C, Augustatou K, et al. Neutrophil CD64 expression and serum IL-8: sensitive early markers of severity and outcome in sepsis. Cytokine. 2006;36(5–6):283–290. doi: 10.1016/j.cyto.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Ludwig KR, Hummon AB. Mass spectrometry for the discovery of biomarkers of sepsis. Molecular BioSystems. 2017;13(4):648–664. doi: 10.1039/c6mb00656f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EA, Wood GL, Wade CE. Use of procalcitonin for the detection of sepsis in the critically ill burn patient: A systematic review of the literature. Burns. 2011;37(4):549–558. doi: 10.1016/j.burns.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Manning J. Sepsis in the burn patient. Critical Care Nursing Clinics North America. 2018;30(3):423–430. doi: 10.1016/j.cnc.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Markanday A. Acute phase reactants in infections: Evidence-based review and a guide for clinicians. Open Forum Infectious Diseases. 2015;2(3):ofv098. doi: 10.1093/ofid/ofv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri, M., Elli, T., Caviglia, G., Uboldi, G., & Azzi, M. (2017) 'RAWGraphs: A visualisation platform to create open outputs' 12th Biannual Conference on Italian SIGCHI. September 18 - 20, 2017. Cagliari, Italy ACM, pp. 28:1–28:5. (Accessed: 09/07/2019).
- McDonald, D., Ackermann, G., Khailova, L., Baird, C., Heyland, D., Kozar, R., et al. (2016). Extreme dysbiosis of the microbiome in critical illness. mSphere, 1(4). doi:10.1128/mSphere.00199–16. [DOI] [PMC free article] [PubMed]
- Mearelli F, Fiotti N, Giansante C, Casarsa C, Orso D, De Helmersen M, et al. Derivation and validation of a biomarker-based clinical algorithm to rule out sepsis from noninfectious systemic inflammatory response syndrome at emergency department admission: A multicenter prospective study. Critical Care Medicine. 2018;46(9):1421–1429. doi: 10.1097/CCM.0000000000003206. [DOI] [PubMed] [Google Scholar]
- Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research. 2015;43(W1):W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chemical Biology. 2017;24(2):195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrobial Agents and Chemotherapy. 2005;49(4):1306–1311. doi: 10.1128/AAC.49.4.1306-1311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzschke S, Offodile AC, 2nd, Cauley RP, Frankel JE, Beam A, Elias KM, et al. Long term mortality in critically ill burn survivors. Burns. 2017;43(6):1155–1162. doi: 10.1016/j.burns.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Nunez Lopez O, Cambiaso-Daniel J, Branski LK, Norbury WB, Herndon DN. Predicting and managing sepsis in burn patients: current perspectives. Therapeutics and Clinical Risk Management. 2017;13:1107–1117. doi: 10.2147/TCRM.S119938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent BA, Seaton M, Sood RF, Gu H, Djukovic D, Raftery D, et al. Use of metabolomics to trend recovery and therapy after injury in critically ill trauma patients. JAMA Surgery. 2016;151(7):e160853. doi: 10.1001/jamasurg.2016.0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz ED, D'Alessandro A, Moore EE, Chin T, Silliman CC, Sauaia A, et al. Pathologic metabolism: an exploratory study of the plasma metabolome of critical injury. The Journal of Trauma and Acute Care Surgery. 2015;78(4):742–751. doi: 10.1097/TA.0000000000000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Rhee C, Murphy MV, Li L, Platt R, Klompas M. Lactate testing in suspected sepsis: Trends and predictors of failure to measure levels. Critical Care Medicine. 2015;43(8):1669–1676. doi: 10.1097/CCM.0000000000001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh, K. P., Griswold, J. A., & Hamood, A. N. (1999). Contribution of the regulatory gene lasR to the pathogenesis of Pseudomonas aeruginosa infection of burned mice. The Journal of Burn Care & Rehabilitation 20(1), 42–49. https://www.ncbi.nlm.nih.gov/pubmed/9934636. [DOI] [PubMed]
- Schmerler D, Neugebauer S, Ludewig K, Bremer-Streck S, Brunkhorst FM, Kiehntopf M. Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. Journal of Lipid Research. 2012;53(7):1369–1375. doi: 10.1194/jlr.P023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour CW, Yende S, Scott MJ, Pribis J, Mohney RP, Bell LN, et al. Metabolomics in pneumonia and sepsis: An analysis of the GenIMS cohort study. Intensive Care Medicine. 2013;39(8):1423–1434. doi: 10.1007/s00134-013-2935-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer V, Horvath A, Komarova I, Schmerboeck B, Feldbacher N, Klymiuk I, et al. Dysbiosis in early sepsis can be modulated by a multispecies probiotic: A randomised controlled pilot trial. Benef Microbes. 2019;10(3):265–278. doi: 10.3920/BM2018.0067. [DOI] [PubMed] [Google Scholar]
- Su L, Huang Y, Zhu Y, Xia L, Wang R, Xiao K, et al. Discrimination of sepsis stage metabolic profiles with an LC/MS-MS-based metabolomics approach. BMJ Open Respiratory Research. 2014;1(1):e000056. doi: 10.1136/bmjresp-2014-000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Li H, Xie A, Liu D, Rao W, Lan L, et al. Dynamic changes in amino acid concentration profiles in patients with sepsis. PLoS ONE. 2015;10(4):e0121933. doi: 10.1371/journal.pone.0121933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Research. 2016;44(D1):D463–D470. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Liang J, Yao X, Lu C, Zhong T, Hong X, et al. The activation of EGFR promotes myocardial tumor necrosis factor-alpha production and cardiac failure in endotoxemia. Oncotarget. 2015;6(34):35478–35495. doi: 10.18632/oncotarget.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, et al. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. Journal of Proteome Research. 2009;8(1):113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Attila C, Whiteley M, Wood TK. Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microbial Biotechnology. 2009;2(1):62–74. doi: 10.1111/j.1751-7915.2008.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DG, Horgan PG, McMillan DC. Routine clinical markers of the magnitude of the systemic inflammatory response after elective operation: a systematic review. Surgery. 2015;157(2):362–380. doi: 10.1016/j.surg.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Wei Y, Gao N, Zhang Z, Zu X, Hu Z, Zhang W, et al. Metabolic changes at the early stage of sepsis induced by cecal ligation and puncture in rats and the interventional effects of Huang-Lian-Jie-Du-Tang. Journal of Chromatography B. 2016;1026:176–182. doi: 10.1016/j.jchromb.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Werthén M, Davoudi M, Sonesson A, Nitsche DP, Morgelin M, Blom K, et al. Pseudomonas aeruginosa-induced infection and degradation of human wound fluid and skin proteins ex vivo are eradicated by a synthetic cationic polymer. Journal of Antimicrobial Chemotherapy. 2004;54(4):772–779. doi: 10.1093/jac/dkh407. [DOI] [PubMed] [Google Scholar]
- West, T. P., & Chu, C. P. (1986). Utilization of pyrimidines and pyrimidine analogues by fluorescent pseudomonads. Microbios, 47(192), 149–157. https://www.ncbi.nlm.nih.gov/pubmed/3097460. [PubMed]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Research. 2007;35(suppl 1):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Bazer FW, Burghardt RC, Johnson GA, Kim SW, Knabe DA, et al. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids. 2011;40(4):1053–1063. doi: 10.1007/s00726-010-0715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Research. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics and metadata reported in this paper are available via the Metabolomics Workbench public repository (Sud et al., 2016) (https://www.metabolomicsworkbench.org/data/DRCCMetadata.php?Mode=Study&StudyID=ST001101) under study identifier ST001101. These data include the 383 unidentified metabolites as well as the 148 identified metabolites.