Summary
The worldwide spread of COVID-19 highlights the need for an efficient approach to rapidly develop therapeutics and prophylactics against SARS-CoV-2. The SARS-CoV-2 spike protein, containing the receptor-binding domain (RBD) and S1 subunit involved in receptor engagement, is a potential therapeutic target. We describe the development of a phage-displayed single-domain antibody library by grafting naive complementarity-determining regions (CDRs) into framework regions of a human germline immunoglobulin heavy chain variable region (IGHV) allele. Panning this library against SARS-CoV-2 RBD and S1 subunit identified fully human single-domain antibodies targeting five distinct epitopes on SARS-CoV-2 RBD with subnanomolar to low nanomolar affinities. Some of these antibodies neutralize SARS-CoV-2 by targeting a cryptic epitope located in the spike trimeric interface. Collectively, this work presents a versatile platform for rapid antibody isolation and identifies promising therapeutic anti-SARS-CoV-2 antibodies as well as the diverse immogneic profile of the spike protein.
Keywords: SARS-CoV-2, nanobody, single-domain antibody, RBD, trimeric interface, CR3022
Graphical Abstract
Highlights
-
•
A phage-displayed human single-domain antibody library is developed
-
•
Single-domain antibodies targeting five types of SARS-CoV-2 epitopes are identified
-
•
Some neutralizing antibodies target cryptic SARS-CoV-2 spike trimeric interface
-
•
Unique immunogenic profile of SARS-CoV-2 RBD is revealed
Wu et al. describe the development of a versatile platform for rapid isolation of fully human single-domain antibodies and apply this methodology to identify SARS-CoV-2-specific antibodies. These human single-domain antibodies target diverse epitopes within the SARS-CoV-2 spike protein receptor binding domain (RBD) and may yield potential therapeutic candidates for COVID-19.
Introduction
The recent outbreak of novel coronavirus disease (COVID-19) caused by SARS-CoV-2, also known as 2019-nCoV or HCoV-19 (Jiang et al., 2020), marks the third major outbreak caused by a new coronavirus in the past two decades, following severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (Li et al., 2020, Wu et al., 2020, Zhou et al., 2020, Zhu et al., 2020). Furthermore, SARS-CoV-2 is one of the most transmissible coronaviruses identified so far, with COVID-19 quickly accelerating into a global pandemic. These facts indicate that coronaviruses remain a huge threat to public health, and new prophylactic and therapeutic strategies are urgently needed.
Monoclonal antibodies (mAbs) represent the largest and fastest-growing sector in the pharmaceutical industry. During the previous SARS and MERS outbreaks, a number of neutralizing mAbs were developed and proved their therapeutic potential in the treatment of coronavirus infections (Du et al., 2009, Sui et al., 2004, ter Meulen et al., 2004, ter Meulen et al., 2006, Traggiai et al., 2004, Ying et al., 2015a, Zhu et al., 2007). Despite this, their clinical usefulness has been hampered by time-consuming and costly antibody manufacturing processes in eukaryotic systems. The large-scale production of mAbs typically takes at least 3 to 6 months, making timely production difficult in an epidemic setting.
An attractive alternative for mAbs is single-domain antibodies from camelid immunoglobulins, termed VHH, or nanobodies that are the smallest naturally occurring antigen-binding protein domains with a molecular weight of 12–15 kilodaltons (kDa) (Muyldermans, 2013). Their small size provides several advantages over conventional mAbs (150 kDa), including larger number of accessible epitopes, relatively low production costs, and ease of rapid production at kilogram scale in prokaryotic expression systems (Wu et al., 2017). More importantly, nanobodies can be administered by inhaled delivery because of their small size and favorable biophysical characteristics, making them particularly suitable for the treatment of respiratory diseases (Van Heeke et al., 2017). For instance, ALX-0171, an inhaled anti-respiratory syncytial virus (RSV) nanobody developed by Ablynx, was found to have robust antiviral effects and reduce signs and symptoms of RSV infection in animal models. Moreover, it was well tolerated at all doses when administered by inhalation in clinical trials (Larios Mora et al., 2018). These findings confirmed the feasibility of administering nanobodies via inhalation. However, the camelid origin of nanobodies limits their application as therapeutics in humans. To reduce the risk of immunogenicity, strategies for humanization of camelid nanobodies have become available in recent years but suffered from time- and labor-intensive processes (Vincke et al., 2009). Humanized nanobodies also retain a small number of camelid residues, especially several specific “hallmark” residues (F37, E44, R45, and G47) within framework region 2 (FR2), in order to maintain solubility and antigen-binding affinity of parental antibodies (Muyldermans, 2013, Wu et al., 2017).
In this study, we aimed to establish an efficient approach to rapidly develop SARS-CoV-2-specific single-domain antibodies of fully human origin, which not only could be potentially implemented in dealing with COVID-19 during the current outbreak but also could strengthen our preparedness and response capacity against emerging coronaviruses in the future.
Results
Identification of a Soluble Human Germline Immunoglobulin Heavy Chain Variable Region (IGHV) Allele
In contrast to camelid nanobodies, which are naturally devoid of light chains, heavy chain variable domains (VHs) of conventional antibodies are paired with light chain variable domains (VLs) and either are generally poorly expressed or easy to aggregate in the absence of light chains. It was proposed that the specific hallmark residues (F37, E44, R45, and G47) within FR2 could contribute to the high solubility and stability of isolated nanobodies (Muyldermans, 2013).
Interestingly, we analyzed 2391 camelid nanobody sequences from a public database and found that their FR2 regions are relatively divergent, including the hallmark residues that were considered to be strictly conserved (Figure 1 A). Furthermore, we and others have previously identified some isolated human VHs, which exhibited biophysical properties very similar to those of camelid nanobodies (Chen et al., 2008, Schneider et al., 2018). These findings inspired us to revisit the structural feature of single-domain antibodies and hypothesize that certain VH framework regions could compensate for the absence of light chain, resulting in soluble human single-domain antibodies.
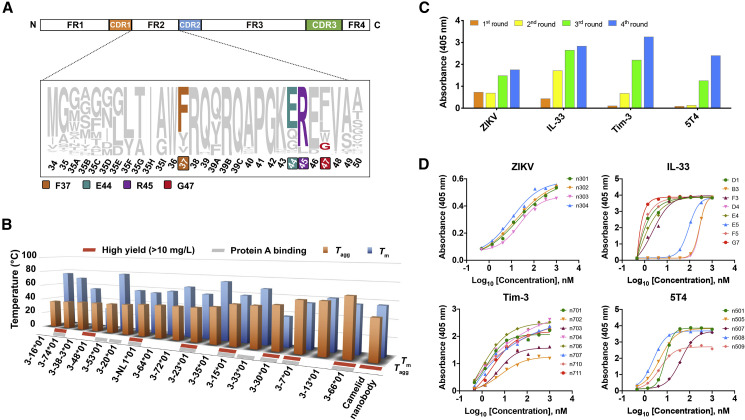
Figure 1.
Development of a Versatile Platform for Rapid Isolation of Fully Human Single-Domain Antibodies
(A) Representation of camelid nanobody framework (FR) and CDR, showing the four hydrophilic amino acids (Phe37, Glu44, Arg45, Gly47) in the FR2 region that could contribute to high solubility and stability of isolated nanobodies.
(B) Characterization of biophysical properties (protein yield, protein A binding capacity, stability, and aggregation) of 17 isolated human germline IGHV alleles, along with a camelid nanobody.
(C) Polyclonal phage ELISA showing the binding of the first to fourth rounds of phages to target antigens.
(D) Binding activity of purified single-domain antibodies against target antigens evaluated by ELISA.
Therefore, we searched the IMGT database for human immunoglobulin heavy chain variable region (IGHV) alleles that have the same germline framework regions (FR1, FR2, or FR3) with m36, an HIV-1-neutralizing VH that was found to be highly soluble and stable. As a result, 17 human germline IGHV alleles, along with a camelid nanobody (VHH#3) as control (Beirnaert et al., 2017), were cloned, expressed in Escherichia coli, and characterized for their biophysical properties (Figure 1B). Eight out of 17 alleles could be highly expressed with yields of over 10 mg/L bacterial culture, and 10 out of 17 possessed protein-A-binding capabilities. Notably, germline 3-66∗01 exhibited the most advantageous properties, including midpoint transition temperature (T m) comparable to that of camelid nanobody and the highest aggregation temperature (T agg) among all tested antibodies. These results confirmed the feasibility of using human single-domain antibodies as ideal alternatives to camelid nanobodies in therapeutic applications.
Development of a Fully Human Single-Domain Antibody Library
Next, we aimed to establish a generalizable platform for rapid development of human single-domain antibodies. We used germline 3-66∗01 framework regions as the scaffold for grafting of heavy-chain complementarity-determining regions CDR1, CDR2, and CDR3 cloned from several naive antibody libraries (Table S1). These libraries were previously constructed from the blood of healthy adult donors, and their effectiveness had already been proved by the successful isolation of potent germline-like human monoclonal antibodies against a variety of targets such as H7N9 avian influenza virus (Yu et al., 2017), MERS-CoV (Ying et al., 2015b), and Zika virus (Wu et al., 2017). Consequently, such CDR grafting resulted in a very large and highly diverse phage-displayed single-domain antibody library.
To validate the quality of the library, several rounds of parallel biopanning selection were performed against a set of representative antigens, including viral antigen, cytokine, and surface antigen on immune or tumor cells. In all tests, potent phage enrichments were observed (Figure 1C), and panels of single-domain antibodies could be identified with binding affinities in the low-nanomolar/subnanomolar range (Figure 1D). These antibodies are monomeric and could be solubly expressed at high levels in Escherichia coli with yields ranging from 15 to 65 mg/L culture (Figure S1). Moreover, their sequences are of fully human origin with minimal divergence from the germline predecessors.
Identification of SARS-CoV-2-Specific Single-Domain Antibodies
This technology enabled us to rapidly develop fully human single-domain antibodies against SARS-CoV-2. To this end, the receptor-binding domain (RBD) of SARS-CoV-2 was first used as the target antigen during bio-panning. Significant enrichment was achieved after two rounds of panning, and a panel of 18 unique single-domain antibodies were selected for further studies (Figure 2 A). They bound potently and specifically to the SARS-CoV-2 RBD and could be divided into three competition groups (A, B, or C) by competition binding assays (Figures 2A and 2B). Most of the antibodies belonged to competition group A represented by n3021, which was also the most enriched clone with subnanomolar affinity (0.6 nM) to RBD (Figure 2C; Table S2). The group A antibodies showed moderate competition with ACE2 for the binding to RBD (Figures 2A and S2) and had no binding to a RBD variant (T500A/N501A/G502A) with mutation of ACE2-binding residues (Figure S3), indicating that their epitope overlaps with ACE2-binding motifs of RBD. To our surprise, none of these antibodies showed efficient neutralization at 50 μg/mL in a well-established SARS-CoV-2 pseudovirus infection assay (data not shown) (Xia et al., 2020a, Xia et al., 2020b). These results suggest that some non-neutralizing epitopes are relatively immunogenic in the isolated SARS-CoV-2 RBD, in contrast to that of SARS-CoV and MERS-CoV, in which the neutralizing subregion was found to be highly immunogenic (Berry et al., 2010).
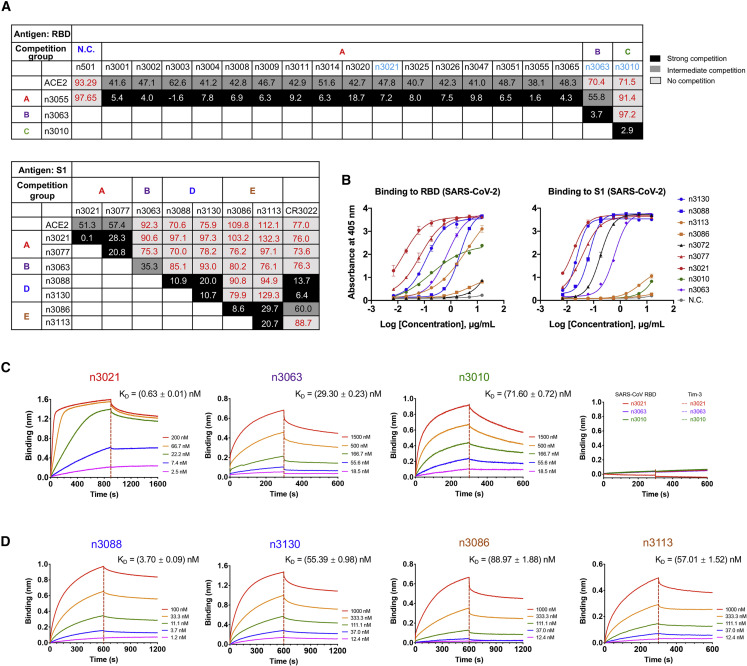
Figure 2.
Characterization of Single-Domain Antibodies Identified from Antibody Library Using SARS-CoV-2 RBD and S1 as Panning Antigens
(A) Eighteen single-domain antibodies identified by panning against SARS-CoV-2 RBD and 5 antibodies by using SARS-CoV-2 S1 as panning antigens were tested in competition binding assay. Competition of these antibodies with each other, or ACE2, or the antibody CR3022 for RBD binding were measured by BLI. The antibodies are displayed in 5 groups (A, B, C, D, or E). The values are the percentage of binding that occurred during competition in comparison with non-competed binding, which was normalized to 100%, and the range of competition is indicated by the box colors. Black-filled boxes indicate strongly competing pairs (residual binding <30%), gray-filled boxes indicate intermediate competition (residual binding 30%–69%), and white-filled boxes indicate non-competing pairs (residual binding ≥ 70%).
(B) Binding capacities of single-domain antibodies to SARS-CoV-2 RBD or S1 measured with ELISA. Data are shown as mean ± SD.
(C) Binding kinetics of representative antibodies from competition groups A, B, and C to SARS-CoV-2 RBD and binding specificity to SARS-CoV RBD or Tim-3, as measured by BLI.
(D) Binding kinetics of competition groups D and E antibodies to SARS-CoV-2 S1.
Interestingly, we also found that the group C antibody n3010 bound potently to SARS-CoV-2 RBD but did not show any binding to S1 protein, indicating that it recognized a cryptic epitope hidden in S1 (Figure 2B). Therefore, we performed another set of biopanning selection with SARS-CoV-2 S1 protein instead of RBD as the target antigen, and a substantially different spectra of antibodies were identified (Figure 2A). Most antibodies showed evident binding to both S1 and RBD, whereas only one antibody, n3072, had strong binding to S1 but no binding to RBD (Figure 2B). In contrast to the dominant enrichment of group A antibodies from RBD panning, the antibodies identified from S1 panning were very diverse, covering four distinct epitopes on RBD, including competition groups A (n3021, n3077), B (n3063), and two additional competition groups D (n3088, n3130) and E (n3086, n3113) (Figures 2A and 2D; Table S2). H3 loops of the identified single-domain antibodies from five competition groups are diverse in sequence and length, and no preferential occurrence of particular amino acids was observed (Table S3).
Neutralizing Antibodies Recognize Two Distinct Epitopes on SARS-CoV-2 RBD
We further measured the neutralization activities of these antibodies with the pseudovirus neutralizing assay. Group E antibodies n3086 and n3113 showed moderate neutralization activities, inhibiting SARS-CoV-2 pseudovirus infection in a dose-dependent manner with half-maximal inhibitory concentration (IC50) values of 26.6 and 18.9 μg/mL, respectively (Figure 3 A). Group D antibodies n3088 or n3130 had IC50 values of 3.3 and 3.7 μg/mL and IC80 values of 10.5 and 11.5 μg/mL, respectively (Figure 3B). The mixture of n3088 or n3130 with group E antibody n3113 showed neutralization of SARS-CoV-2 in a synergistic fashion, with IC50 values of 0.51 and 0.70 μg/mL and IC80 values of 3.2 and 3.6 μg/mL, respectively (Figure 3B). Furthermore, no cytopathic effect (CPE) was observed for group D antibodies, whereas evident CPE was detected in the wells containing live SARS-CoV-2 virus and group E antibodies at 20 μg/mL (Figure 3C). The group D antibodies n3130 and n3088 neutralized live SARS-CoV-2 with an IC50 of 4.0 and 2.6 μg/mL, respectively (Figure 3D).
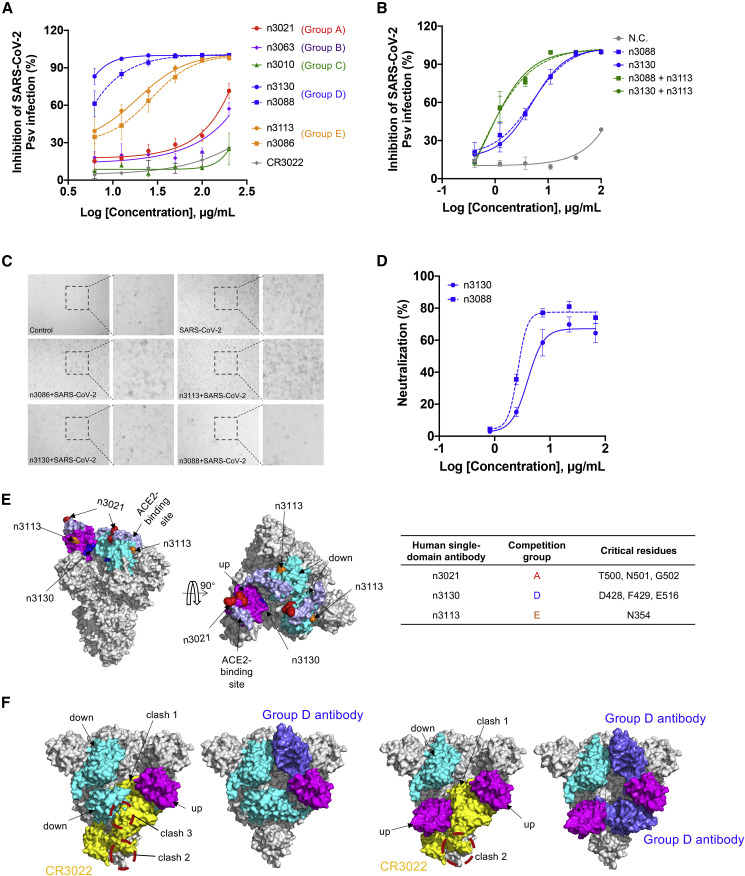
Figure 3.
Neutralization and Epitope Mapping of Single-Domain Antibodies
(A) Antibody-mediated neutralization against luciferase-encoding pseudotyped virus with spike protein of SARS-CoV-2. Pseudotyped viruses preincubated with antibodies at indicated concentrations were used to infect Huh-7 cells, and inhibitory rates (%) of infection were calculated by luciferase activities in cell lysates. Error bars indicate mean ± SD from three independent experiments.
(B) Neutralization of SARS-CoV-2 pseudotyped virus by single-domain antibody cocktails. Group D antibody n3088 or n3130 was combined with an equimolar amount of n3113. For cocktails, the concentration on the x axis indicates that of the individual single-domain antibody. Data are shown as mean ± SD.
(C) Neutralization of live SARS-CoV-2 (clinical isolate nCoV-SH01) by 20 μg/mL of single-domain antibodies.
(D) Neutralization activities of group D antibodies n3088 and n3130 against live SARS-CoV-2. Data are shown as mean ± SD.
(E) Representative single-domain antibody from competition groups A, D, and E are listed with residues critical for binding. The critical residues are highlighted as spheres from epitope mapping experiments and shown by colors which correspond to the competition group designation as (A). The ecto-domian of SARS-CoV-2 spike glycoprotein (PBD entry 6VSB) is shown as surface with RBD colored in magenta or cyan for up or down conformation. ACE2-binding site is shown as slate spheres.
(F) Comparison of the binding model of n3088 and CR3022 Fab to homotrimeric S protein with the RBD protomers adopt a single “up” or double “up” conformation. CR3022 Fab and single-domain antibody n3088 were represented as yellow and blue surface, respectively.
We next performed binding competition assay by biolayer interferometry (BLI) and found that all of the competition groups D and E antibodies are incapable of competing with ACE2 for SARS-CoV-2 RBD binding (Figures 2A and S2). The group E antibody n3113 did not exhibit any binding to the RBD of SARS-CoV-2 isolate SZTH-004 that had two mutations (N354D/D364Y) to the most prevelant isolate, but it bound potently with isolate IDF0372 that has the V367F mutation, suggesting that the epitope of group E antibodies was located at a region surrounding N354, distinct from the ACE2 binding site (Figures 3E and S3).
Furthermore, we found that the group D antibodies potently competed with CR3022 (Figures 2A and S2), a human mAb cross-reactive to SARS-CoV (ter Meulen et al., 2006) and SARS-CoV-2 (Tian et al., 2020). The critical residues (D428, F429, E516) for the binding of group D antibody n3130 were located within the epitope of CR3022 as indicated by alanine scanning mutagenesis of SARS-CoV-2 RBD (Figure S3). Recent studies indicated that CR3022 recognizes a cryptic epitope that is only accessible when S protein is in “open” conformations (Yuan et al., 2020). This epitope could be less accessible in SARS-CoV-2 than in SARS-CoV, explaining why CR3022 was capable of neutralizing SARS-CoV but not SARS-CoV-2 (Wrapp et al., 2020, Yuan et al., 2020). As shown in the recent research (Yuan et al., 2020) and our modeling results (Figure 3F), the binding of CR3022 to homotrimeric SARS-CoV-2 spike is sterically hindered even when the RBD protomers adopt a double “up” conformation. In contrast, despite binding to the same cryptic epitope, the small-size single-domain antibody is very well tolerated in the lateral space between two adjacent RBD protomers.
Discussion
It is very intriguing that the panning with SARS-CoV-2 S1 or RBD protein as antigen resulted in a substantially different spectra of antibodies. The antibodies identified from S1 panning were very diverse, covering four distinct epitopes on SARS-CoV RBD (competition groups A, B, D, and E). In contrast, most of the antibodies from RBD panning belonged to competition group A represented by n3021, which was also the most dominant clone after two rounds of panning. Furthermore, group A antibodies showed moderate competition with ACE2 for RBD binding but proved to be ineffective viral neutralizers. This phenomenon is quite different from that of SARS-CoV, in which the dominance of an antigenic loop within RBD made it relatively easy to isolate potent SARS-CoV neutralizing antibodies independent of repertoire, species, quaternary structure, and the technology used to derive the antibodies (Berry et al., 2010). Similarly, we previously used MERS-CoV S1 or RBD to isolate antibodies from a naive antibody library, and panning that used either of the two antigens led to dominant enrichment of antibodies that precisely targeted 90% of the receptor binding site within RBD and neutralized the virus potently (Ying et al., 2015b). It was proposed that viruses like SARS-CoV have not yet evolved such that their membrane glycoproteins could avoid direct immune recognition of a single site critical to virus pathogenesis (Berry et al., 2010). It should be pointed out that the difference in the immunogenicity of RBD was observed solely based on in vitro experiments and might not correlate with humoral immune responses in vivo. In this regard, it is imperative to investigate the immunogenic characteristics of SARS-CoV-2 RBD with special attention to potentially antigenic and non-neutralizing epitopes.
It is also striking that the SARS-CoV-2-specific neutralizing antibodies from competition groups D and E are incapable of competing with ACE2 for SARS-CoV-2 RBD binding. This phenomenon was rarely observed in SARS-CoV, confirming the unique immunogenic profile of SARS-CoV-2. Further investigations are needed to understand the underlying mechanisms that govern these diverse sets of neutralizing and non-neutralizing SARS-CoV-2 antibodies, which could have important implications for the development of effective vaccines.
More interestingly, the group D antibodies n3088 and n3130 were found to neutralize SARS-CoV-2 by targeting a “cryptic” epitope located in the spike trimeric interface. The mAb CR3022 also recognizes this epitope but cannot neutralize SARS-CoV-2, implying the advantage of small-size single-domain antibodies to target cryptic eptiopes. Such hidden trimeric interface epitope was also observed recently in the study of influenza antibodies by our group (Yu et al., 2017) and several other groups (Bajic et al., 2019, Bangaru et al., 2019, Watanabe et al., 2019). It is probable that such an epitope would also be important in the study of coronaviruses, and n3088 or n3130 could be ideal for synergistically used with other SARS-CoV-2 neutralizing antibodies, especially the ACE2-competing neutralizing antibodies.
Fully human single-domain antibodies offer the potential for prevention and treatment of COVID-19. First, antibodies derived entirely from human sequences would be less immunogenic than camelid or humanized nanobodies, leading to improved safety and efficacy when used in humans. Indeed, despite humanization, caplacizumab, the first nanobody approved by FDA, still contains multiple camelid residues to maintain the antigen binding affinity (Ulrichts et al., 2011). Second, small size and favorable biophysical properties allow for large-scale production of single-domain antibodies within a few weeks in prokaryotic expression systems, thus enabling rapid implementation in an outbreak setting. Furthermore, single-domain antibodies could be delivered to the lung via inhalation, which could offer considerable advantages for treatment of COVID-19, including fast onset of action, low systemic exposure, and high concentration of therapeutics at the site of disease. Lastly, single-domain antibodies can be used alone or synergistically with other neutralizing antibodies. Their small size makes them ideal building blocks for generation of bispecific or multi-specific antibodies to prevent the appearance of viral escape mutants. They can also be easily engineered to further increase neutralization activity by increasing binding moieties. For instance, the trivalent nanobody ALX-0171 was found to have 6,000-fold increased neutralization potency against RSV-A and >10,000-fold against RSV-B compared to its monovalent format (Detalle et al., 2015).
In summary, we report here the development of a versatile platform for rapid isolation of fully human single-domain antibodies and their application for screening of antibodies against SARS-CoV-2. These antibodies could represent promising candidates for prophylaxis and therapy of COVID-19 and serve as reagents to facilitate vaccine development.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody | Sigma-Aldrich | Cat# A8592-1MG; RRID: AB_439702 |
| Anti-M13-horseradish peroxidase (HRP) polyclonal antibody | Pharmacia | Cat# 27-9421 |
| CR3022 scFv | This paper | N/A |
| Bacterial and Virus Strains | ||
| TG1 Electrocompetent Cells | Lucigen | Cat# 60502-1 |
| HB2151 Escherichia coli Strains | This paper | N/A |
| SARS-CoV-2 strain nCoV-SH01 | Fudan University | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphate Buffered Saline solution | HyClone | Cat# SH30256.01B |
| Ni Sepharose High Performance | GE Healthcare | Cat# 17526802 |
| Protein G Sepharose 4 Fast Flow resin | GE Healthcare | Cat# 17061805 |
| BirA500 Kit | Avidity | N/A |
| Polymyxin B sulfate salt | Sigma-Aldrich | Cat# P1004-5MU |
| High Fidelity PCR Master | Roche | Cat# 12140314001 |
| SfiI | New England Biolabs | Cat# R0123L |
| Dynabeads™ MyOne™ Streptavidin T1 | Thermo Fisher | Cat# 65601 |
| ABTS | Thermo Fisher | Cat# 002024 |
| EZ-Link™ Sulfo-NHS-LC-LC-Biotin | Thermo Fisher | Cat# A35358 |
| Biotinylated ZIKV DIII | This paper | N/A |
| Biotinylated Human TIM-3 / HAVCR2 | ACROBiosystems | Cat# TM3-H82E7 |
| Human IL-33 | ACROBiosystems | Cat# IL3-H52H7 |
| Human TPBG / 5T4 | ACROBiosystems | Cat# TPG-H52E5 |
| Human TNF-α | Sino Biological Inc. | Cat# 10602-HNAE |
| SARS-CoV-2 Spike S1-His | Sino Biological Inc. | Cat# 40591-V08H |
| SARS-CoV-2 Spike RBD-His | Sino Biological Inc. | Cat# 40592-V08B |
| ACE2-His Protein | Novoprotein Scientific Inc. | Cat# C419 |
| Critical Commercial Assays | ||
| Bright-Glo™ Luciferase Assay System | Promega | Cat# E2620 |
| Cell Couting Kit-8 Assay Kit | Dojindo | Cat# CK04-20 |
| Experimental Models: Cell Lines | ||
| Expi293 Expression System | Thermo Fisher | Cat# A14635 |
| Vero E6 cells | ATCC | Cat# CRL-1586 |
| Recombinant DNA | ||
| Germline IGHV alleles genes | Genscript | N/A |
| VHH#3 gene | Genscript | N/A |
| RBD gene: SARS-CoV-2 (strain WIV04, GISAID accession number EPI_ISL_402124) | Genscript | N/A |
| RBD gene: SARS-CoV-2 (strain SZTH-004, GISAID accession number EPI_ISL_406595) | Genscript | N/A |
| RBD gene: SARS-CoV-2 (strain IDF0372, GISAID accession number EPI_ISL_406596) | Genscript | N/A |
| RBD gene: SARS-CoV (strain WH20) | Genscript | N/A |
| pComb3x vector | Addgene | Cat# 63891 |
| pSecTag2B expression vector | Thermo Fisher | Cat# V90020 |
| Software and Algorithms | ||
| IMGT | IMGT | www.imgt.org |
| ForteBio Data Analysis software | Pall ForteBio LLC | N/A |
| SWISS-MODEL | SWISS-MODEL | https://swissmodel.expasy.org |
| Z-dock | Z-dock | http://zdock.umassmed.edu |
| Pymol | Pymol | N/A |
| Other | ||
| M13KO7 helper phages | Thermo Fisher | Cat# 18311019 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact, Tianlei Ying (tlying@fudan.edu.cn).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate anny unique datasets or code.
Experimental Models and Subject Details
Cells and Viruses
Vero E6 cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in 37°C, 5% CO2 atmosphere. The authentic SARS-CoV-2 virus hCoV-SH01 used in the neutralization assay was obtained from Shanghai Medical College and stored at −80°C in a BSL-3 laboratory (Fudan University).
Method Details
Expression and Purification of Single-Domain Antibodies
A panel of 17 human germline IGHV alleles have been identified from IMGT database (http://www.imgt.org/) by searching for sequences that have one of these FRs: FR1, EVQLVESGGGLVQPGGSLRLSCAAS, FR2: MSWVRQAPGKGLEWVS, and FR3: YYADSVKGRFTISRDNSKNTLYLQMNSLRAEDTAVYYC. The allele genes joined with the identical CDR3 and FR4 (ARLRDGFNNGFDYWGQGTLVTVSS), as well as a previously reported camelid-derived nanobody that targets human TNF-α (12), were synthesized (Genscript, Nanjing, China), and then subcloned into the pComb3x vector with N-terminal OmpA signal peptide (MKKTAIAIAVALAGFATVAQA) and C-terminal tags comprising a hexahistidine (His6) tag and a Flag tag in tandem. Their expression was performed in Escherichia coli HB2151 bacterial culture at 30°C for 14 h accompanied with 1 mM IPTG. The cells were harvested and lysed by Polymyxin B (Sigma-Aldrich) at 30°C for 0.5 h. Supernatant was obtained by centrifugation at 8000 rpm for 10 min and loaded over Ni-NTA as manual described (GE Healthcare). Resin was washed by washing buffer [10 mM Na2HPO4, 10 mM NaH2PO4 (pH 7.4), 500 mM NaCl and 20 mM imidazole] and protein were eluted in elution buffer [10 mM Na2HPO4, 10 mM NaH2PO4 (pH 7.4), 500 mM NaCl and 250 mM imidazole]. The collected pure fractions were immediately buffer-exchanged into phosphate buffered saline solution (PBS) (Hyclone) and concentrated using an Amicon ultra centrifugal concentrator (Millipore) with a molecular weight cut-off of 3 kDa. Purity was estimated to be over 95% by SDS-polyacrylamide gel electrophoresis, and protein concentration was measured using the NanoDrop 2000 spectrophotometer (Thermo Fisher).
Determination of Melting and Aggregation Temperatures
Thermal stability and colloidal stability were assessed by using an Uncle/UNit system (Unchained Labs, Pleasanton, CA). Briefly, the static light scattering (SLS) at 473 nm was used as an indicator for colloidal stability, reporting the onset of aggregation temperature (T agg), which can be defined as the temperature at which the measured scatter reaches a threshold that is approximately 10% of its maximum value. The changes in the SLS signal represented changes in the average molecular mass observed due to protein aggregation. Thermal stability was evaluated at an intrinsic fluorescence intensity ratio (350/330 nm) by measuring the temperature of the on-set of melting. The samples at a concentration of 0.5 mg/mL were heated from 20°C to 95°C using 1°C increments, with an equilibration time of 60 s before each measurement. Measurements were made in duplicates.
Size-exclusion-high-performance liquid chromatography (SEC-HPLC)
Fifty μg of purified human germline IGHV allele proteins, or purified SARS-CoV-2-specific single domain neutralizing antibodies, were applied to a TSK-Gel Super SW3000 (TSK-GEL) using Waters AQUITY UPLC H-class system. The mobile phase was PBS buffer (pH 7.4) run at a flow rate of 0.4 mL/min. Absorbance was monitored at 280 nm. The UV trace was analyzed and integrated by area under the curve to determine percent aggregation, monomer and degradants.
Construction of Human Single-Domain Antibody Library
For the construction of the fully human single-domain antibody library, grafting of CDR1, CDR2, and CDR3 regions of heavy chain from several naive antibody libraries was carried out by PCR using specific oligonucleotides (Table S1) and combined with framework sequences from the germline IGHV3-66∗01 subfamily by a series of overlap extension PCR (OE-PCR). To obtain maximal diversity, twenty PCRs for each library template were carried out separately to obtain CDR repertoires using multiple primer combinations. PCR was performed in a volume of 50 μL using High Fidelity PCR Master (Roche), 200 pM concentration of each primer, and 0.1 μg of templates (library plasmid DNA) for 30 cycles (45 s at 94°C, 45 s at 55°C, and 1 min at 72°C). All products from the source libraries were pooled at a molarity ratio calculated by counting the number of donors for construction of these libraries. The primers used to amplify CDR fragments contained identical sequences that are homologous to framework sequences and serve as the overlap for OE-PCR. Finally, the entire single-domain antibody fragments were digested with SfiI (New England Biolabs) and cloned into phagemid pComb3x allowing the expression of C-terminal His6-Flag tagged single-domain antibodies. The recombinant vector was electro-transformed into competent TG1 bacteria (Lucigen) at 1.8 kV, 25 μF, and 200 Ω. Pre-warmed 2 × YT medium was added and incubated at 37°C with shaking at 250 rpm for 1 h. Ten μl of the culture was 10-fold serially diluted in 100 μL of 2 × YT medium and plated on 2 × YT agar plates containing 2% glucose (w/v) and 100 μg/mL ampicillin. The plates were incubated overnight at 37°C, and the diversity of the library was calculated next day by counting the number of colonies, multiplying by the culture volume and dividing by the plating volume. For preparation of single-domain antibody library, the cultures were incubated for additional 2 h at 37°C by adding 100 μg/mL ampicillin and 2% (w/v) glucose. TG1 cells were infected with M13KO7 helper phages (Invitrogen) when the optical density at 600 nm (OD600) reached 0.6, and incubated for 1 h at 37°C. The TG1 were harvested and resuspended in 2 × YT medium with ampicillin and kanamycin (100 μg/mL), and the bacteria were cultured overnight at 30°C. Next day, the cultures were centrifuged and phages were precipitated from the supernatant by adding 5% (w/v) polyethylene glycol (PEG) 8000-NaCl (20% PEG8000, 2.5 mol/L NaCl). Precipitated phages were collected by centrifugation, resuspended in sterile PBS, and frozen in aliquots of 1013 phage particles per tube at −80°C.
Preparation of Panning Antigens and Proteins
The biotinylated human Tim-3, human IL-33 and human 5T4 were purchased from ACROBiosystems Inc. The baculovirus-expressed SARS-CoV-2 Spike S1-His and RBD-His were purchased from Sinobiological Inc. (Beijing, China). The biotinylated ZIKV envelope DIII from the MR766 strain was stored in our laboratory. The recombinant RBD sequences encoding amino acids R319-F541 of SARS-CoV-2 (isolate WIV04 from GISAID accession number EPI_ISL_402124, isolate SZTH-004 from GISAID accession number EPI_ISL_406595, isolate IDF0372 from GISAID accession number EPI_ISL_406596), amino acids R306-F527 of SARS-CoV (isolate WH20) were cloned into pSecTag2B expression vector with an additional C-terminal Fc fragment of human IgG1 and AviTag in tandem and transiently expressed in Expi293 cells (Thermo Fisher) as secreted proteins. After 3 days, supernatants were harvested by centrifuging the culture at 2500 rpm for 15 min and filtering the supernatant with a 0.22 μm vacuum filter. The protein was purified using protein G resin (GE Healthcare). Equilibration and wash steps were performed with PBS and proteins were eluted in 0.1 M glycine pH 2.7. The eluates were equilibrated to pH 7.4 using 1 M Tris-HCl (pH 9.0) and immediately buffer-exchanged into PBS and concentrated using an Amicon ultra centrifugal concentrator (Millipore) with a molecular weight cut-off of 10 kDa. Purity was estimated as 95% by SDS–polyacrylamide gel electrophoresis, and protein concentration was measured using the NanoDrop 2000 spectrophotometer (Thermo Fisher). The biotinylated proteins used in bio-panning and BLI experiment were prepared by the BirA biotin-protein ligase in PBS for 30 min at 30°C according to the manufacturer’s instructions (Avidity), which adds biotin covalently to AviTag in a highly specific manner, or by non-specific labeling using sulfo-NHS-LC-Biotin (Thermo Fisher).
Antibody Screening from Phage Library
Panning protocols were carried out essentially as described previously (13, 15). Briefly, one aliquot of the frozen library phage stocks was precipitated with 5% PEG8000-NaCl and re-suspended in PBS. Phages from library were pre-blocked in 3% milk powder (w/v) in PBS (MPBS) and incubated with the biotinylated antigen in 1% MPBS for 30 min, and then incubated with streptavidin-coated magnetic beads (Invitrogen) for 1.5 h. After washes with PBST (PBS buffer supplemented with 0.05% Tween 20), bound phages were used to infect mid-log phase TG1 bacteria at 37°C for 1 h. Then, TG1 bacteria were grown in 2 × YT medium with 100 μg/mL ampicillin and 2% (w/v) glucose at 37°C and 250 rpm. After 2 h, the cells were infected with M13KO7 helper phages (Invitrogen) for 30 min at room temperature. The infected cells were harvested and re-suspended into 2 × YT medium supplemented with 100 μg/mL ampicillin and kanamycin, followed by incubation overnight at 30°C and 180 rpm. The phages were precipitated from culture supernatant with PEG8000-NaCl and re-suspended in PBS for subsequent panning. About 1 × 1012 phage particles were used for the 2nd, 3rd, and 4th rounds of bio-panning. The enrichment for antigen-specific phages after each round of panning was assessed by polyclonal phage enzyme-linked immunosorbent assay. Positive clones expressing human single-domain antibodies were identified from the enriched rounds of panning by using soluble expression-based enzyme-linked immunosorbent assay. The identified clones were analyzed and classified into different families prominently based on their amino acid sequences similarity.
Enzyme-linked Immunosorbent Assay (ELISA)
Costar half-area high binding assay plates (Corning #3690) were coated with purified protein at 100 ng/well in PBS overnight at 4°C, and blocked with PBS buffer containing 3% milk powder (w/v) at 37°C. For polyclonal phage ELISA, phages from each round of panning were incubated with immobilized antigen and bound phages were detected with anti-M13-horseradish peroxidase (HRP) polyclonal antibody (Pharmacia). For the purified antibody binding assay, serially diluted antibody solutions were added and incubated for 1.5 h at 37°C, and bound antibodies were detected with monoclonal anti-Flag-HRP antibody (Sigma-Aldrich). The enzyme activity was measured with the subsequent addition of substrate ABTS (Invitrogen) and signal reading was carried out at 405 nm using a Microplate Spectrophotometer (Biotek).
Biolayer Interferometry (BLI) Binding Assays
BLI was carried out on an OctetRED96 device (Pall FortéBio) and assays were carried out in 96-well black plates. For the binding kinetics of single-domain antibodies with SARS-CoV-2 S1, the recombinant S1-His protein (Sinobiological Inc) at 15 μg/mL buffered in sodium acetate (pH 5.0) was immobilized onto activated AR2G biosensors (Pall FortéBio) and incubated with threefold serial dilutions of single-domain antibodies in kinetics buffer (PBS buffer supplemented with 0.02% Tween 20). The experiments included the following steps at 37°C: (1) equilibration (60 s); (2) activation of AR2G by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride/N-hydroxysucci-nimide (300 s); (3) immobilization of S1 protein onto sensors (100 s); (4) quenching with ethanolamine (300 s); (5) baseline in kinetics buffer (120 s); (6) association of antibodies for measurement of k on (300-600 s); and (7) dissociation of antibodies for measurement of k off (300-600 s). For measuring binding kinetics of single-domain antibodies with SARS-CoV-2 RBD, Avi-tagged recombinant RBD was biotinylated with the BirA biotinylation kit (Avidity), diluted in kinetics buffer and immobilized on streptavidin (SA) coated biosensors (Pall FortéBio) at ~50% of the sensor maximum binding capacity. Baseline was established in kinetics buffer and loaded biosensors were dipped into wells containing serial dilutions of single-domain antibodies for 300-600 s. RBD:antibody complexes were then allowed to dissociate in kinetics buffer. All the curves were fitted by a 1:1 binding model using the Data Analysis software (FortéBio). Mean k on, k off, and KD values were determined by averaging binding curves within a dilution series having R2 values of greater than 95% confidence level. Real-time interactions between purified SARS-CoV RBD or an irrelevant protein (Tim-3) as control and single-domain antibodies were determined. The streptavidin-coated biosensors were loaded with biotinylated SARS-CoV RBD or Tim-3, and then incubated with 15 μg/mL of single-domain antibodies in kinetics buffer and binding was measured after 300 s of association.
Binding Competition Assays
The epitopes of anti-SARS-CoV-2 single-domain antibodies were initially mapped by binding competition assays using BLI. Sensor tips loaded with SARS-CoV-2 RBD, as described above, were immersed into wells containing the first competing single-domain antibody at a concentration (15 μg/mL) necessary to reach binding saturation after 300 s. Next, biosensors were dipped into wells containing the second single-domain antibody with the same concentration at 15 μg/mL, or 17 μg/mL ACE2 (Novoprotein Scientific Inc., Shanghai), in the presence of the first competing single-domain antibody, and binding was measured after 300 s of association. For competition of single-domain antibodies with the antibody CR3022, 6 μg/mL of CR3022 in scFv format were incubated with sensors loaded with SARS-CoV-2 RBD for 300 s to reach binding saturation. Then, the sensors were immersed into wells containing the competition single-domain antibody at 15 μg/mL in the presence of the 6 μg/mL CR3022, and binding was measured for 300 s. The signal obtained for binding of the second antibody in the presence of the first antibody was expressed as a percentage of the uncompeted binding of the second antibody that was derived independently. The antibodies were defined as competing if the presence of first antibody reduced the signal of the second antibody to less than 30% of its maximal binding capacity, and non-competing when binding was greater than 70%. A level of 30%–69% was considered intermediate competition.
Pseudotyped Virus Neutralization
To determine the neutralization activity of single-domain antibodies, the pseudotyped virus neutralization assay was performed. Briefly, 293 T cells were co-transfected with expression vectors of pcDNA3.1-SARS-CoV-2-S (encoding SARS-CoV-2 S protein) and pNL4-3.luc.RE bearing the luciferase reporter-expressing HIV-1 backbone, as previously described (Xia et al., 2020a). The supernatants containing SARS-CoV-2 pseudotyped virus were harvested. Serial dilutions of single-domain antibodies in DMEM supplemented with 10% fetal calf serum were incubated with pseudoviruses at 37°C for 1 h and then the mixtures were added to monolayer Huh-7 cells (104 per well in 96-well plates). Twelve h after infection, culture medium was refreshed and then incubated for an additional 48 h. The luciferase activity was calculated for the detection of relative light units using the Bright-Glo Luciferase Assay System (Promega). A nonlinear regression analysis was performed on the resulting curves using Prism (GraphPad) to calculate half-maximal inhibitory concentration (IC50) values.
Live Virus Microneutralization Assay
SARS-CoV-2 strain nCoV-SH01 was obtained from a biosafety level 3 (BSL-3) laboratory of Fudan University (Zhang et al., 2020). Viral cytopathic effect (CPE)-based neutralization assay was conducted to determine the neutralization of human single-domain antibodies against live SARS-CoV-2 virus. In brief, 1 × 104 Vero E6 cells were seeded in 96-well plates and cultured for overnight at 37°C. Single-domain antibodies were diluted at a final concentration of 20 μg/mL in culture medium and mixed with 200 PFU of SARS-CoV-2. Following incubation for 1 h at 37°C, the virus and antibody mixtures were added to the Vero E6 cells, and medium was replaced with fresh DMEM 2 h later. CPE was observed daily and recorded on Day 3 post-infection. To calculate the IC50 of neutralizing single-domain antibodies, the live virus microneutralization assay was used as previously described (Sui et al., 2004, Zhu et al., 2007). Briefly, the antibodies were serially diluted 3-fold and incubated with 200 PFU of SARS-CoV-2 infectious strain (nCoV-SH01) for 1 h at 37°C. The virus and antibodies were then added to a 96-well plate with 4 × 104 Vero E6 cells/well, and checked for CPE at 3 days postinfection. Cell viability was assessed using CCK-8 kit (Dojindo). All assays were performed at least four times.
Site-Directed Mutagenesis for Epitope Mapping of Single-Domain Antibodies
To determine the epitopes of anti-SARS-CoV-2 single-domain antibodies, a panel of SARS-CoV-2 RBD variants were generated. The recombinant expressing vector pSecTag2B-RBD encoding wild-type SARS-CoV-2 RBD from strain WIV04 was subjected to alanine scanning mutagenesis using a site-directed mutagenesis kit (Yeasen, Shanghai, China). A single residue or sequential residues were changed to alanine. Then, variants of RBD were expressed in Expi293 cells and purified from the conditioned culture media using protein G resin (GE Healthcare) as described above. Purified wild-type and mutant RBD were coated on 96-well ELISA plates for overnight at 4°C. After blocking with 3% BSA in PBS, single-domain antibodies n3021, n3113, and n3130 with 3-fold serial dilutions were added and incubated for 1.5 h at 37°C. Plates were washed by PBST and HRP-conjugated anti-Flag was used for detection. Enzyme activity was measured with the subsequent addition of substrate ABTS, and signal reading was carried out at 405 nm.
Prediction of the Conformation of n3088 and Its Complex with SARS-CoV-2 Spike
The homology model of single-domain antibody n3088 was built on the SWISS-MODEL online server (https://swissmodel.expasy.org) using the searched templates. The complex model of SARS-CoV-2 RBD (Lan et al., 2020) and n3088 was prepared on Z-dock (http://zdock.umassmed.edu) (Pierce et al., 2014). The binding model of n3088 or CR3022 to homotrimeric S were generated by Pymol.
Quantification and Statistical Analysis
Binding experiments are presented as the mean values ± SD calculated from two independent experiments. Neutralization is the geometric mean of the IC50 values calculated using four-parameter logistic regression from at least two independent experiments performed in triplicate. The nanobody sequences were collected from an online database (http://ican.ils.seu.edu.cn). All data were graphed using Prism software (version 8, GraphPad Software).
Acknowledgments
We thank C. Qin from Beijing Institute of Microbiology and Epidemiology; A. Huang and S. Zhou from our group; Y. Wu and Y. Wang from Fudan University BSL-3 laboratory; and Q. Wang and other staff at the Core Facility of Microbiology and Parasitology, Shanghai Medical College, for help with experiments. This work was supported by grants from the National Key R&D Program of China (2019YFA0904400), National Natural Science Foundation of China (81822027, 81630090), National Megaprojects of China for Major Infectious Diseases (2018ZX10301403), and Chinese Academy of Medical Sciences (2019PT350002).
Author Contributions
T.Y., Y.W., and C.T. conceived and designed the study. Y.W., T.Y., and C.L. performed most of the experiments with assistance from X.T., Y.K., Z.W., and Z.Y.. S.X., L.L., and S.J. performed pseudovirus neutralization assay. C.G., R.Z., and Y.X. performed live SARS-CoV-2 neutralization assay. T.Y. and Y.W. integrated the data and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Declaration of Interests
Y.W. and T.Y. are listed as inventors on two patent applications related to this work.
Published: May 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.04.023.
Contributor Information
Yanling Wu, Email: yanlingwu@fudan.edu.cn.
Tianlei Ying, Email: tlying@fudan.edu.cn.
Supplemental Information
References
- Bajic G., Maron M.J., Adachi Y., Onodera T., McCarthy K.R., McGee C.E., Sempowski G.D., Takahashi Y., Kelsoe G., Kuraoka M., Schmidt A.G. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe. 2019;25:827–835.e6. doi: 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Lang S., Schotsaert M., Vanderven H.A., Zhu X., Kose N., Bombardi R., Finn J.A., Kent S.J., Gilchuk P. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell. 2019;177:1136–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirnaert E., Desmyter A., Spinelli S., Lauwereys M., Aarden L., Dreier T., Loris R., Silence K., Pollet C., Cambillau C., de Haard H. Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions. Front. Immunol. 2017;8:867. doi: 10.3389/fimmu.2017.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.D., Hay K., Rini J.M., Yu M., Wang L., Plummer F.A., Corbett C.R., Andonov A. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2:53–66. doi: 10.4161/mabs.2.1.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu Z., Feng Y., Dimitrov D.S. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. USA. 2008;105:17121–17126. doi: 10.1073/pnas.0805297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K. Generation and Characterization of ALX-0171, a Potent Novel Therapeutic Nanobody for the Treatment of Respiratory Syncytial Virus Infection. Antimicrob. Agents Chemother. 2015;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Shi Z., Shu Y., Song J., Gao G.F., Tan W., Guo D. A distinct name is needed for the new coronavirus. Lancet. 2020;395:949. doi: 10.1016/S0140-6736(20)30419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Larios Mora A., Detalle L., Gallup J.M., Van Geelen A., Stohr T., Duprez L., Ackermann M.R. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. MAbs. 2018;10:778–795. doi: 10.1080/19420862.2018.1470727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Pierce B.G., Wiehe K., Hwang H., Kim B.-H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D., Xiong Y., Hu P., Wu D., Chen W., Ying T., Zhu Z., Dimitrov D.S., Dropulic B., Orentas R.J. A Unique Human Immunoglobulin Heavy Chain Variable Domain-Only CD33 CAR for the Treatment of Acute Myeloid Leukemia. Front. Oncol. 2018;8:539. doi: 10.3389/fonc.2018.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S.C., Olurinde M., Choe H. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B.H., van den Brink E.N., Weverling G.J., Martina B.E.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrichts H., Silence K., Schoolmeester A., de Jaegere P., Rossenu S., Roodt J., Priem S., Lauwereys M., Casteels P., Van Bockstaele F. Antithrombotic drug candidate ALX-0081 shows superior preclinical efficacy and safety compared with currently marketed antiplatelet drugs. Blood. 2011;118:757–765. doi: 10.1182/blood-2010-11-317859. [DOI] [PubMed] [Google Scholar]
- Van Heeke G., Allosery K., De Brabandere V., De Smedt T., Detalle L., de Fougerolles A. Nanobodies® as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- Vincke C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- Watanabe A., McCarthy K.R., Kuraoka M., Schmidt A.G., Adachi Y., Onodera T., Tonouchi K., Caradonna T.M., Bajic G., Song S. Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell. 2019;177:1124–1135.e16. doi: 10.1016/j.cell.2019.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Jiang S., Ying T. Single-Domain Antibodies As Therapeutics against Human Viral Diseases. Front. Immunol. 2017;8:1802. doi: 10.3389/fimmu.2017.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Li H., Lu L., Dimitrov D.S., Jiang S. Development of human neutralizing monoclonal antibodies for prevention and therapy of MERS-CoV infections. Microbes Infect. 2015;17:142–148. doi: 10.1016/j.micinf.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D.S., Zhou T. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Song H., Wu Y., Chang S.Y., Wang L., Li W., Hong B., Xia S., Wang C., Khurana S. A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin. Cell Host Microbe. 2017;22:471–483.e5. doi: 10.1016/j.chom.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020:eabb7269. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Yi Z., Wang Y., Teng Z., Xu W., Song W., Cai X., Sun Z., Gu C., Zhou Y. Isolation of a 2019 novel coronavirus strain from a coronavirus disease 2019 patient in Shanghai. J. Microbes Infect. 2020;15:16–21. [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate anny unique datasets or code.