Abstract
Background and Purpose:
Selection of acute ischemic stroke (AIS) patients for endovascular treatment generally relies on dynamic susceptibility contrast (DSC) MRI or computed tomography perfusion (CTP). DSC MRI requires injection of contrast, while CTP requires high doses of ionizing radiation. The purpose of this work was to develop and evaluate a deep learning (DL)-based algorithm for assisting the selection of suitable AIS patients for endovascular treatment based on 3D pseudo-continuous arterial spin labeling (pCASL).
Methods:
A total of 167 image sets of 3D pCASL data from 137 AIS patients scanned on 1.5 and 3.0T Siemens MR systems were included for neural network training. The concurrently acquired DSC MRI was used to produce labels of hypoperfused brain regions, analyzed using commercial software. The DL and 6 machine learning (ML) algorithms were trained with 10-fold cross-validation. The eligibility for endovascular treatment was determined retrospectively based on the criteria of perfusion/diffusion mismatch in the DEFUSE 3 trial. The trained DL algorithm was further applied on 12 3D pCASL data sets acquired on 1.5 and 3T GE MR systems, without fine-tuning of parameters.
Results:
The DL algorithm can predict the DSC-defined hypoperfusion region in pCASL with a voxel-wise area under the curve (AUC) of 0.958, while the 6 ML algorithms ranged from 0.897 – 0.933. For retrospective determination for subject-level endovascular treatment eligibility, the DL algorithm achieved an accuracy of 92%, with a sensitivity of 0.89 and specificity of 0.95. When applied to the GE pCASL data, the DL algorithm achieved a voxel-wise AUC of 0.94 and a subject-level accuracy of 92% for endovascular treatment eligibility.
Conclusions:
pCASL perfusion MRI in conjunction with the DL algorithm provides a promising approach for assisting decision-making for endovascular treatment in AIS patients.
Keywords: Acute ischemic stroke (AIS), Deep learning (DL), Pseudo-continuous arterial spin labeling (pCASL), Dynamic susceptibility contrast (DSC), Perfusion imaging, Endovascular treatment, Magnetic Resonance Imaging (MRI)
Subject Terms: Ischemic Stroke, Revascularization, Treatment, MRI
Introduction
Recent clinical trials have shown that acute ischemic stroke (AIS) patients with specific patterns of penumbral tissue—the volume of brain tissue that is ischemic but not yet infarcted—can benefit from endovascular reperfusion therapy beyond the typical 6-hour treatment window from symptom onset1, 2. Computed tomography perfusion (CTP) imaging and dynamic susceptibility contrast (DSC) perfusion magnetic resonance imaging (MRI) are commonly used to estimate the penumbral tissue to select suitable AIS patients for endovascular therapies. CTP is more commonly used in emergency settings due to its speed and accessibility. However, it requires a high dose of ionizing radiation due to the continuous X-ray exposure during the passage of iodinated contrast media, which has its own risk of allergic reactions (~0.2% incidence)3. There is also debate on the accuracy of CTP in delineating the infarct core of AIS4. Diffusion-weighted MRI is regarded as the imaging standard for defining the extent of infarct core. DSC perfusion MRI relies on the injection of gadolinium-based contrast agent (GBCA) that is not suitable for patients with renal dysfunction. As most AIS patients are aged persons presenting with vascular risks (e.g. diabetes mellitus), a mandatory step (and additional time) is required to rule out contraindications for GBCAs for DSC perfusion MRI.
Arterial spin labeling (ASL) MRI techniques can provide cerebral blood flow (CBF) measures without the use of contrast agent, and it has been shown to provide largely consistent results with DSC perfusion MRI in delineating hypoperfused brain regions in AIS5, 6. ASL may be particularly suitable for patients with renal dysfunction and/or vulnerable populations such as children and pregnant women who are at risk of strokes7, 8. The scan time for ASL is generally on the order of ~5min with CBF maps generated almost immediately with vendor or custom software. However, the precise delineation of hypoperfusion lesion and penumbra in ASL images remains challenging due to the low signal-to-noise ratio (SNR) and delayed arterial transit.
Recently machine learning (ML) methods have been applied for the diagnosis and management of stroke, such as the prediction of clinical outcome of AIS patients9, 10. Machine learning models use algorithms to parse data, learn features from that data, and make informed decisions based on the learning with some user guidance. Deep learning (DL) is an advanced ML method that takes the intelligent decision-making one step further by using the neural network architecture resembling that is used for human visual perception11. Deep learning is able to capture the hierarchical and complex features of the input image automatically and can identify, classify, and quantify patterns in medical images12, 13. In this study, we developed and evaluated a DL-based algorithm to automatically identify the hypoperfusion lesion and penumbra in ASL images (in conjunction with diffusion MRI), using the perfusion lesions in DSC perfusion MRI as supervision.
Materials and Methods
The original data that support the findings of this study are available from the corresponding author upon request.
Patient selection
The present study included data from two AIS cohorts: data from the first cohort were collected from June 2010 to Sept 2013 in a registry of patients evaluated with diffusion-perfusion MRI at the University of California, Los Angeles (UCLA) Medical Center. Data from the second cohort were collected from Aug 2014 to March 2018 at the Stanford University Medical Center. Patients with AIS were included in this study if: (1) they were adults (aged >18 years); (2) they received an established clinical diagnosis of AIS; (3) ASL, DSC, and diffusion weighted imaging (DWI) data were acquired, (4) acute ischemic lesion was detected in DWI, and (5) there was an absence of previous intracranial hemorrhage, brain surgery, or large territorial lesion. The Institutional Review Board (IRB) of UCLA approved this study and waived requirement for informed consent. The IRB of Stanford University approved this study and informed consent was obtained from the participants.
MRI protocols and analysis
All MRIs of the UCLA cohort were performed on Siemens 1.5 T Avanto or 3.0 T TIM Trio systems (Erlangen, Germany). A pseudo-continuous ASL (pCASL) pulse sequence with background suppressed 3D GRASE (gradient and spin echo) readout was applied with the following parameters: TR/TE/label time=4000/22/1500ms; field-of-view=22cm; matrix size=64×64, 26×5mm slices, post-labeling delay (PLD)=2000ms, scan time 4–5min5, 6. CBF maps were generated with custom software using Interactive Data Language (IDL, Boulder, CO, USA) that included motion correction, pairwise subtraction between label and control images, averaging to generate the mean difference image and the calculation of quantitative CBF maps5, 6.
All MRIs of the Stanford cohort were acquired on GE 1.5 and 3.0T SIGNA systems (Milwaukee, WI) using a 3D pCASL sequence with fast spin echo readout and stack-of-spirals readout trajectory. Imaging parameters were: TR/TE/label time=4852/11/1500ms; PLD=2025ms, field-of-view=24cm; matrix size=64×64, 36×4mm slices, scan time ~5min. CBF maps were generated with vendor software14.
DSC images were acquired using a gradient-echo echo-planar imaging sequence with a timed contrast-bolus passage technique (0.1mg/kg contrast administered intravenously at a rate of 5ml/s), with a TR range of 1770 to 2890ms and average TE of 44±10.4ms, matrix=128×128. Following motion correction and spatial smoothing, multiparametric perfusion maps including CBF, cerebral blood volume (CBV), time to the maximum of the tissue residue function (Tmax), and mean transit time (MTT) were generated using a method based on block-circulant singular value decomposition (cSVD) implemented in the commercial software OLEA (La Ciotat, France) for the UCLA cohort. The RAPID software (iSchemaView Inc, Menlo Park, CA)15 was used for analyzing DSC data from the Stanford cohort.
Diffusion weighted images were acquired with b value of 1000s/mm2, and range values of TR and TE were 3500–6000 and 78–118ms, respectively. The matrix size was 128×128. Apparent diffusion coefficient (ADC) images were calculated with vendor software. ASL CBF and DSC images were corregistered with DWI in each subject using SPM12 (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/), and normalized into the Montreal Neurological Institute template space using SPM12.
The flowchart for the training and evaluation of the DL algorithm is shown in Figure 1, which includes modules of data input, DL model architecture, voxel-level and subject-level evaluation. The following describes each step in detail.
Figure 1. Flowchart of the training and evaluation of DL model.
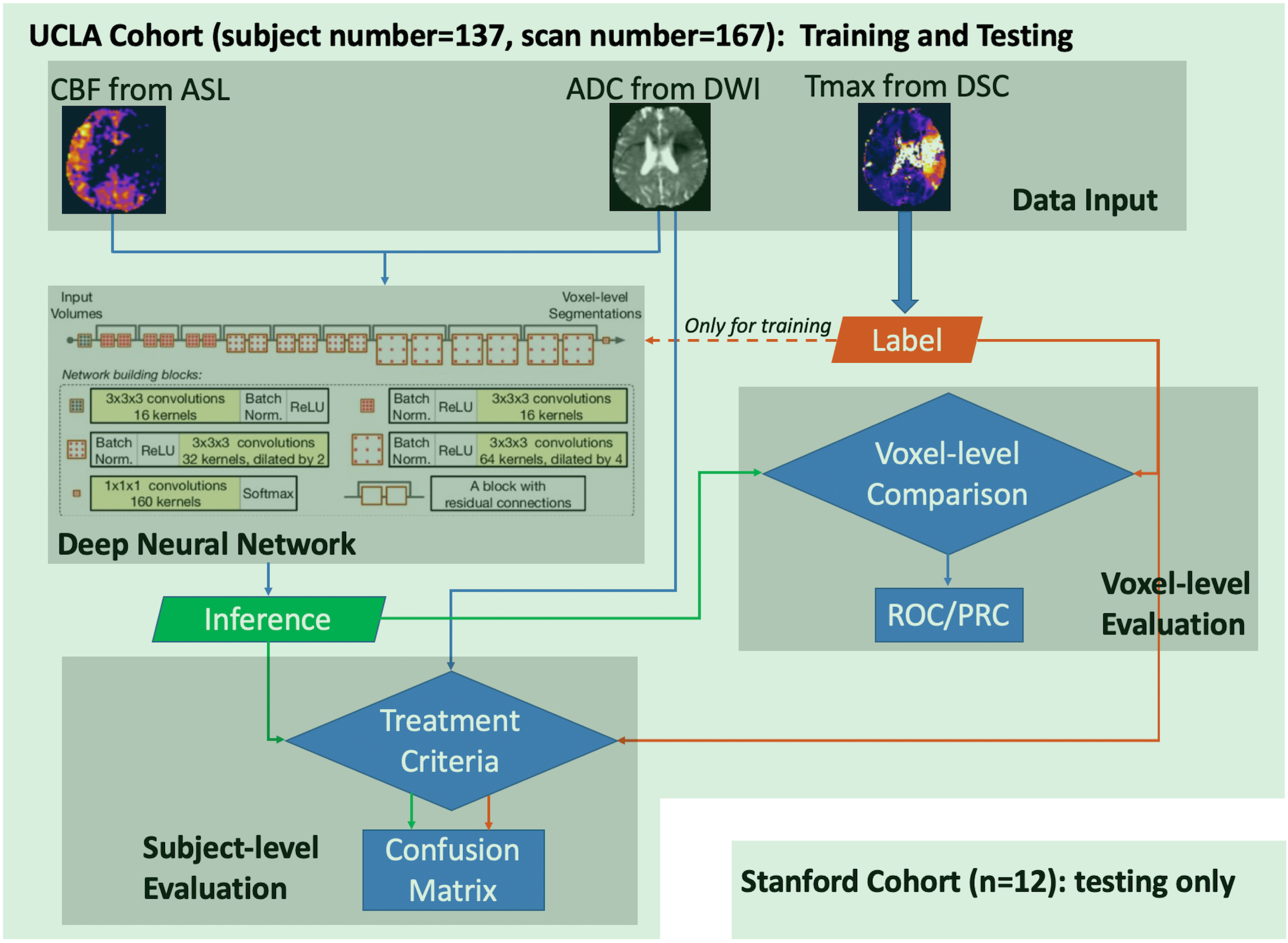
DSC MRI scans were first processed to generate parameter maps (CBF, ADC, and Tmax), then the data were used to train the Deep Neural Network. After training, inference was made and evaluated at voxel-level and subject-level. The UCLA cohort was used for both training and testing, while the Stanford cohort was only used for testing purposes.
Creating Label Images of DSC Perfusion Lesion
All calculated Tmax maps, extracted arterial input function (AIF), and venous outflow function (VOF) were inspected by an expert of perfusion MRI (DJW). Data were excluded if the AIF failed to show clear bolus passage and/or there were severe motion artifacts. The Tmax maps generated by the OLEA software for the UCLA cohort often contained speckle noise and/or residual motion artifacts on brain edges; therefore, the following denoising steps were applied to reliably identify the perfusion lesions: 1) skull-stripping using the BET program16; 2) brain segmentation using FSL FAST17 and ventricles masking with the segmented cerebrospinal fluid (CSF) regions; 3) manually setting a bounding box to include perfusion lesions; and 4) applying a threshold with Tmax≥6sec. The resultant binary maps were used as “label images” for training neural networks, with the foreground being hypoperfusion regions. For the Stanford cohort, the perfusion lesion with Tmax≥6sec was automatically generated by the RAPID software. In previous MRI and positron emission tomography studies18, 19, Tmax lesions>2sec often overestimated the volume of penumbral tissue, while stricter thresholds (>8sec) underestimated the volume of hypoperfused tissue. Based on these studies and recent clinical trials1, 18, 20, Tmax≥6sec was chosen to define hypoperfusion lesions in this study.
Deep Neural Network Architecture and Training
The network used in this project was the Highres3Dnet21, which offers compact end-to-end 3D convolutional neural network (CNN) structures that maintain high-resolution multi-scale features (Figure 1). The Highres3Dnet was implemented using TensorFlow and NiftyNet22. It included 20 trainable layers with dilated convolution and dilating factors of 1, 2, and 4, respectively. Residual connections were employed for every two convolution layers. The network was trained on two Nvidia GeForce GTX 1080 Ti GPUs. CBF and ADC images were used as input, and the label image from Tmax served as the supervision. Volumes (48×48×48, batch size=4) were randomly extracted from 3D preprocessed input and label images for training. Volume level augmentation was employed, including rotation with a random angle in the range of [−10°, 10°] for each of the three orthogonal planes and spatial rescaling with a random scaling factor in the range of [0.9, 1.1]. The Dice loss function23 and Adam optimization method24 (learning rate=0.0001, β1 = 0.9, β2=0.999) were used. The total iteration numer was set to 70,000 to enable the training process to reach steady state. The hyperparameters of the network (e.g., layer number, volume size, and iteration number) were determined based on pre-training on a fixed validation dataset.
We performed the 10-fold cross-validation, in which individuals from the UCLA cohort were first divided into 10 groups. For each fold of the cross validation, 9 groups were used as the training set, and the remaining one designated as the testing set. After the process was repeated 10 times, the whole dataset was tested.
Machine Learning Models and Training
For comparison, six commonly used ML classifiers were trained on the same UCLA cohort, including linear regression classifier, ridge regression classifier, kernel ridge regression classifier, neural network classifier, Support Vector Machine (SVM) with Radial Basis Function (RBF) and random forest classifier25. The same 10-fold cross-validation scheme as used in the training of the DL model was applied: For each fold, ten thousand 13×13 patches were randomly selected from the training set, half of which the central voxel was positive voxels in the label image, and the other half of which the central voxel was negative. Once the models were trained on the ten thousand patches, inference was determined voxel by voxel on the testing set, where the classifier utilized the 13×13 patch around the central voxel to be inferred.
DL Model Performance Assessment
The DL model performance was evaluated on a subset of the UCLA cohort, herein referred to as the evaluation cohort. The inclusion criteria for the evaluation cohort were adopted from the DEFUSE 3 trial1: patients who had an occlusion of the intracranial internal carotid artery or the proximal middle cerebral artery, and the last known well time was within 16 hours of the MRI. The performance assessment was done both at voxel-level and subject-level. For voxel-level evaluation, the Dice coefficient was first calculated between the inference and the Tmax label for each subject, and the group average Dice was subsequently calculated for all subjects of the evaluation cohort. Then, 100,000 voxels were randomly selected from the DSC perfusion maps of evaluation cohort, of which 50,000 were picked from positive voxels in the label images, and the remaining 50,000 were negative voxels (to control for imbalanced positive and negative voxels in our data). Receiver Operating Characteristic (ROC) curves and Precision-Recall (PR) curves were calculated on the 100,000 voxels for the DL and 6 ML models, and the corresponding Area Under the Curve (AUC) was calculated, respectively.
Subject-level performance assessment was based on the imaging criteria for endovascular thrombectomy of the DEFUSE 3 trial1: 1) The initial infarct volume (ischemic core), identified as regions of ADC<620×10−6 mm2/s, is less than 70mL26; 2) The volume of ischemic tissue is 1.8x the volume of initial infarct volume or more; 3) The volume of potentially reversible ischemia (penumbra) is 15mL or more. For the evaluation cohort of AIS patients, the endovascular treatment eligibility was determined first with ADC and DSC Tmax label maps, then with ADC images and predicted ASL hypoperfusion lesion from the trained model. Confusion matrices, as well as sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), the total accuracy, and Cohen’s kappa coefficient were also calculated27 for the DL and 6 ML models, respectively.
The above subject-level evaluation used the optimal cut-off threshold determined by voxel-wise training/evaluation of the 6 ML methods respectively, while 0.5 was used for the DL method. We further varied the threshold for binarizing the DL/ML model output and calculated the ROC curves for selecting subjects eligible for endovascular treatment within the evaluation cohort of AIS patients.
Evaluation of the Generalizability of DL Model
To further test the generalizability of our DL model, we applied the DL model on the Stanford cohort. Inference was made based on the ADC and CBF maps, without fine-tuning model parameters. Since the ten models built through the 10-fold validation generated 10 hypoperfusion lesion probability maps for an individual, we used the consensus map of the 10 predicted lesions for evaluation, which was generated by averaging the 10 probability maps of the 10 models and subsequently binarized with threshold=0.5. Both voxel-level and subject-level evaluations with the same criteria as described above were performed, except that for voxel-level evaluation, a smaller number of voxels (10,000) was sampled because the dataset was smaller compared with the UCLA dataset.
Results
Patients
For the UCLA cohort, a total of 167 image sets from 137 patients were included for network training (1.5T: n=93; 3T: n=74). Each patient had 1–3 MRIs. For the purpose of maximizing the training datasets and improving the robustness of the DL model when dealing with different subtypes of strokes, we used all available MRIs rather than only baseline scans or patients with anterior circulation stroke. For the Stanford cohort, 12 MRI (1.5T: n=1; 3T: n=11) from 12 patients were randomly selected for the evaluation of the generalizability of DL model. The clinical and demographic information of the UCLA cohort and Stanford cohort is listed in online supplement Table I and II, respectively.
Neural network identification of ASL hypoperfusion lesions
Figure 2 shows 4 representative cases at 1.5T and 4 representative cases at 3T from the UCLA cohort, respectively. For each case, ADC, DSC Tmax, and ASL CBF maps are displayed in the first 3 columns respectively. The predicted and Tmax label of perfusion lesions are overlaid on the T2w image of DWI (the 4th column). At each field strength, 2 cases with predicted>Tmax label perfusion lesion (row 1, 2) and 2 cases with predicted<Tmax label (row 3, 4) are shown. Only the second case of 1.5T was misclassified. Overall, the network could identify the perfusion lesion defined by Tmax images although there were discrepancies between the lesion volumes of ASL and DSC MRI.
Figure 2. Four representative cases at each field strength showing the input images, label, and inference.
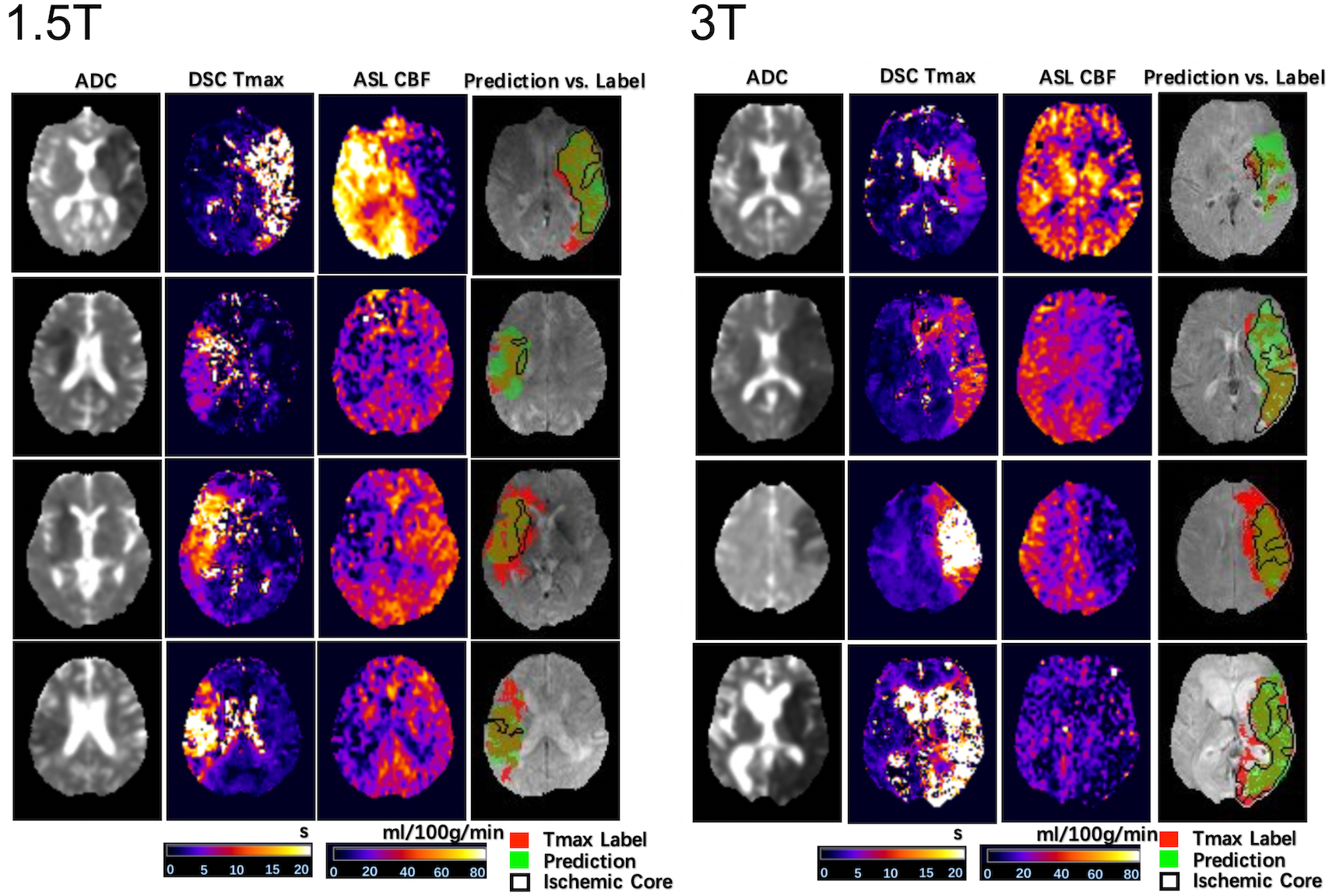
For each case, ADC, DSC Tmax, and ASL CBF are displayed in the first 3 columns, respectively. The predicted and Tmax label of perfusion lesions are overlaid on the T2w image of DWI. At each field strength, 2 cases with predicted>Tmax label perfusion lesion and 2 cases with predicted<Tmax label are shown. Only the second case of 1.5T was misclassified.
Evaluation of DL Prediction Accuracy
Thirty-eight AIS patients of the UCLA cohort with large vessel occlusion in the anterior circulation met the inclusion criteria of DEFUSE 3, which served as the evaluation cohort. For voxel-level evaluation, the group average Dice coefficient was 0.47 ± 0.23. The ROC curves showed that our DL model was able to achieve significantly superior performance compared to the 6 traditional ML methods (p<0.001). The AUC of our DL model was 0.958, while the AUC of the 6 ML models varied from 0.897 to 0.933 (Figure 3A). The PR curves showed similar results with the DL model yielding the highest AUC=0.957, while the AUC of the 6 ML models varied from 0.790 to 0.932 (p<0.001) (Figure 3B).
Figure 3. A) ROC of the DL model and ML models on the evaluation voxel set.
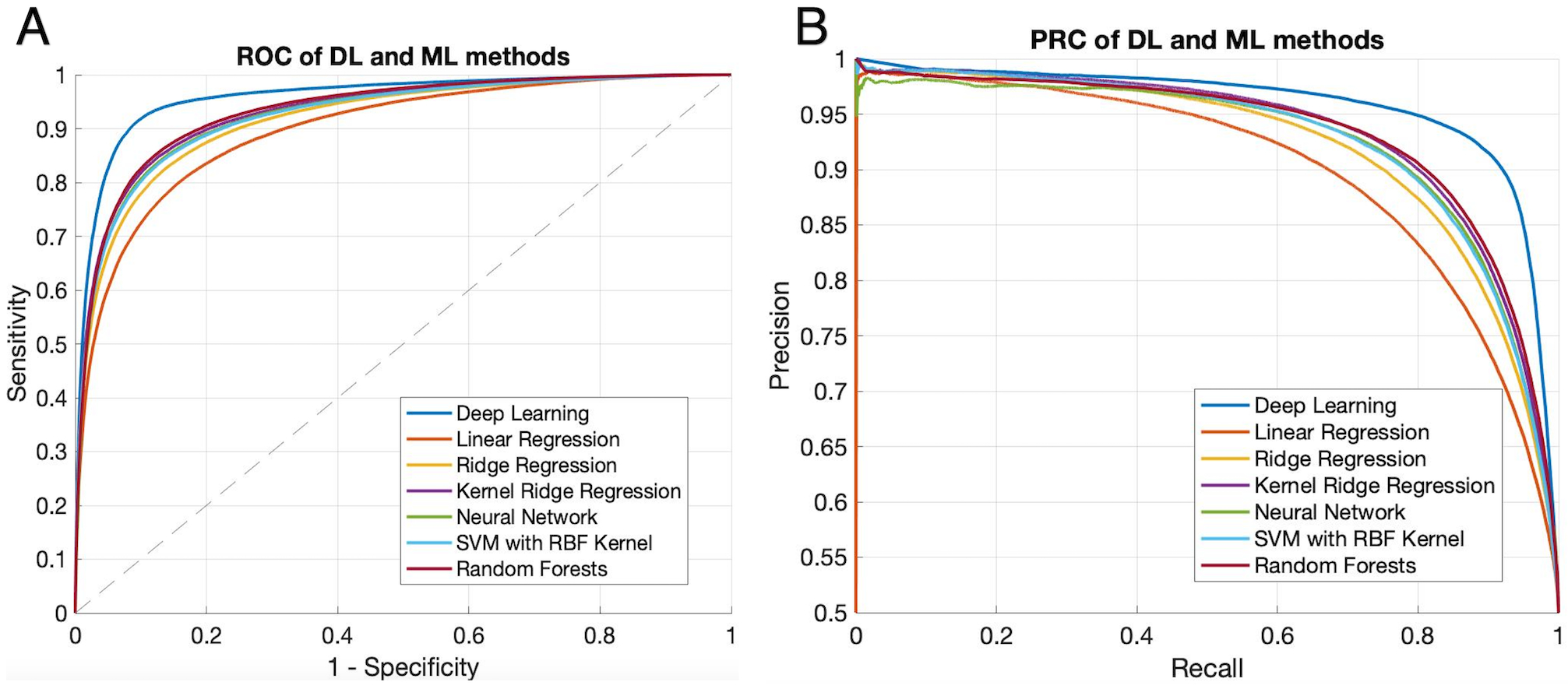
With AUC=0.958, DL model showed significantly superior performance (p<0.001) compared with traditional ML models (AUC=0.897, 0.918, 0.930, 0.925, 0.924, and 0.933 for the six methods in the legend, respectively). B) Precision-Recall curves of the DL model and ML models. Significantly superior performance of the DL model was confirmed by precision-recall curve (AUC = 0.957, p<0.001), compared to 6 ML models (AUC= 0.790, 0.802, 0.809, 0.804, 0.806, and 0.932), respectively.
Subject-level evaluation was performed according to the imaging criteria for endovascular thrombectomy of the DEFUSE 3 trial. The sensitivity, specificity, PPV, NPV, and Cohen’s kappa coefficient were 0.89, 0.95, 0.94, 0.90, and 0.84, respectively (the corresponding confusion matrix is shown in online supplement Table III). The accuracy was 0.92 (95% CI: [0.79, 0.98]). In comparison, the accuracy, sensitivity, specificity, PPV, NPV, and Cohen’s kappa coefficient (online supplement Table IV), were also calculated for the 6 ML methods. The highest accuracy among the 6 ML models was 0.84 (95% CI: [0.69, 0.94]). When the cutoff threshold was varied to generate ROC curves, although without significant differences, the DL model still yielded the highest AUC of 0.950, while the AUC of ML algorithms ranged from 0.915 to 0.949.
Cross-validation on Stanford cohort
Our pretrained DL models were tested on the 12 patients of the Stanford cohort scanned on GE 1.5 and 3T MR scanners, without any fine-tuning of parameters. The results from the consensus maps of the 10 models are reported here. Three cases are shown in Figure 4 with the same layout as Figure 2 (two correctly and one incorrectly classified for treatment). The average Dice coefficient was 0.43 ± 0.25. Voxel-level evaluation showed that the AUC of the ROC and PR curve was 0.942 (Figure 5A) and 0.931 (Figure 5B) respectively. Only one among the 12 tested subjects was classified incorrectly (the corresponding confusion matrix is shown in online supplement Table V). Subject-level evaluation yielded an accuracy of 0.92 (95% CI: [0.62, 0.99]), with sensitivity, specificity, PPV, and NPV of 0.75, 1.00, 1.00 and 0.89, respectively. Prediction of subject-level treatment eligibility in the Stanford cohort was highly reproducible as all the 10 DL models trained on the UCLA cohort yielded the same accuracy of 0.92.
Figure 4. Three representative cases of the Stanford cohort showing the input images, inference, and label.
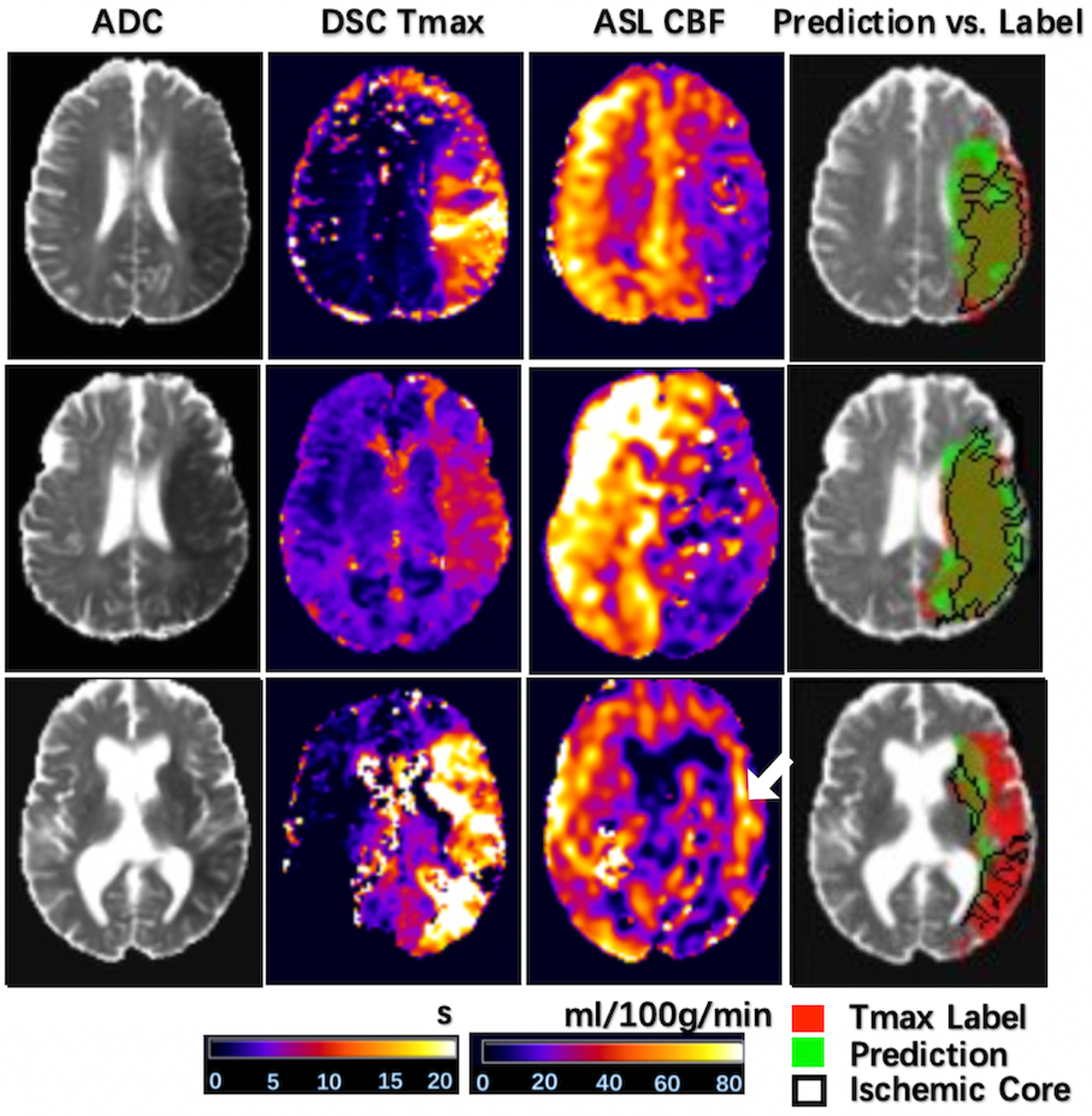
The same layout is used as in Figure 2. The first two were correctly classified, while the third one was misclassified. The arrow indicates hyperperfusion signal likely arising from delayed arterial transit effects.
Figure 5. ROC Curve (A) and Precision-Recall Curve (B) on evaluation voxel set of the Stanford cohort.
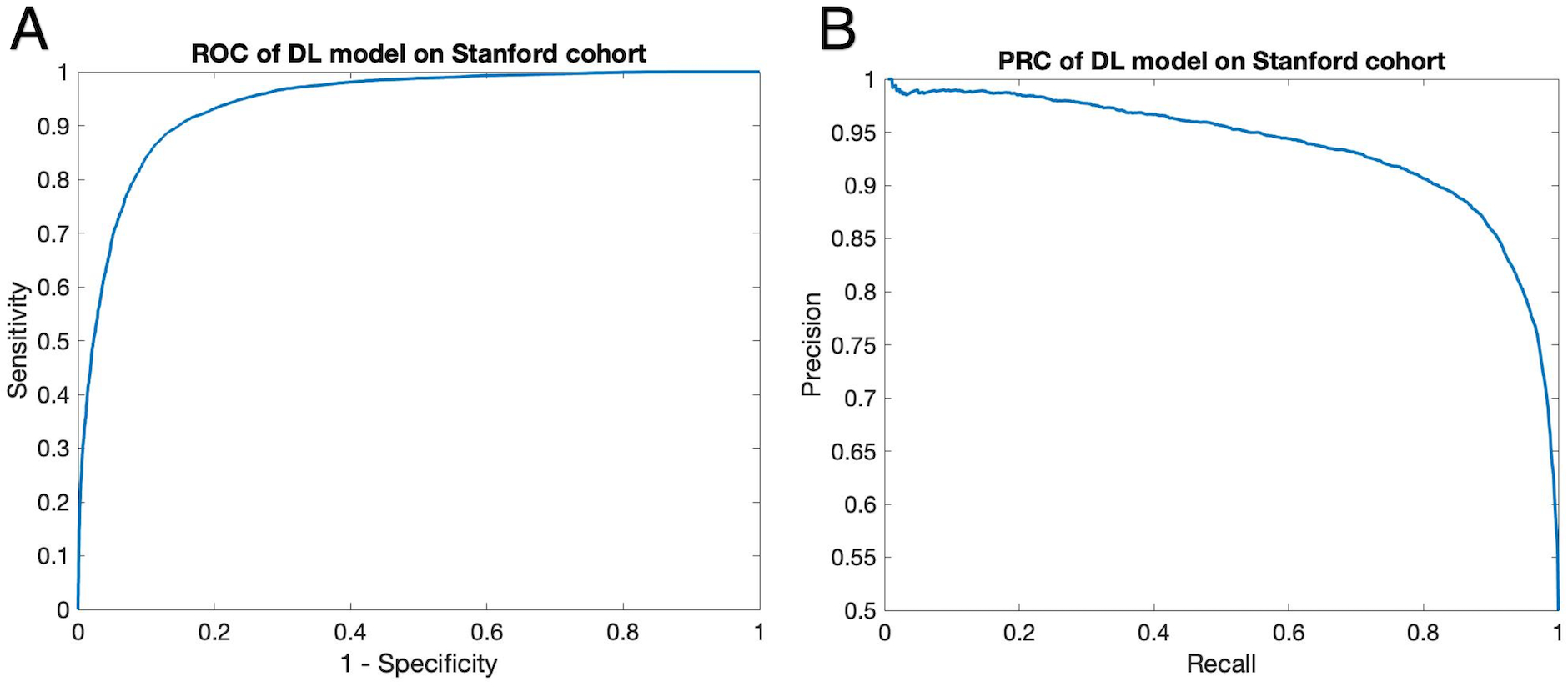
Without any parameter fine-tuning, our DL model achieved reasonable performance on the 10,000-voxel evaluation set, with AUC=0.942 for the ROC curve and 0.931 for the precision-recall curve.
The bottom row of Figure 4 shows the case that was incorrectly classified for treatment eligibility. The DL model still identified hypoperfusion lesions on ASL CBF, however the hyperperfusion signals (arrow) likely arising from delayed arterial transit effects restricted the predicted lesion volume to be below the threshold for perfusion/diffusion mismatch.
Discussion
Recent randomized clinical trials have consistently reported positive outcomes by utilizing CTP and/or DSC MRI to identify and quantify the penumbral tissue, using automated post-processing software such as RAPID1, 2, 28–30. In this project, we trained DL models to assist the decision-making for endovascular thrombectomy with non-contrast ASL images, using the hypoperfusion lesion observed on DSC MRI as the label. The DL models were able to consistently achieve an accuracy of 92% for imaging-based criteria for endovascular therapy in two independent cohorts of AIS patients scanned on Siemens and GE MRI scanners at 1.5 and 3T. If verified in randomized clinical trials, DL based algorithms may have several advantages compared to existing methods using CTP and DSC MRI. With the proposed method, any AIS patients including the elderly, children, and pregnant women can be evaluated, as long as they do not have contraindications for MRI. As diffusion MRI is considered the gold standard for delineating the infarct core, an entirely non-invasive MRI protocol including ASL perfusion and diffusion MRI could potentially improve the diagnostic accuracy and ischemic tissue characterization for AIS patients compared to CTP. Past studies have shown that the infarct cores identified by diffusion MRI and CTP often do not match each other31, 32. Since AIS patients regularly undergo multiple MRIs without concerns of ionizing radiation, ASL scans can be repeated as often as required to follow disease progression. With sufficient clinical evidence, the proposed method may extend the treatment window to subacute stages and may assist in the management of reperfusion injury33, 34.
DL based prediction of hypoperfusion lesions is instantaneous (<1sec per subject), while the post-processing of CTP and DSC MRI takes a few minutes using RAPID software35; Although the net time for CTP and DSC MRI scanning is only 1–2min, additional time and staffing is required for the preparation of contrast injection and in the case of DSC MRI, to rule out contraindications for GBCAs. Therefore, the total time for ASL scanning (~5min), CBF calculation (seconds) and DL prediction of penumbra (<1sec) could be shorter than that required for CTP and DSC MRI scanning and post-processing. It is worth noting that fast ASL scanning within 1–2min is feasible with DL-based denoising36, 37. The potentially lower overhead for time and management of patient safety with the proposed method could be clinically significant in AIS treatment, given that “time is brain”.
Machine learning and in particular DL algorithms are ideally suited for ASL imaging in stroke. Although ASL has been shown to provide largely consistent results with DSC MRI in delineating hypoperfused brain regions in AIS5, 6, in practice ASL at a single PLD (e.g. 2 seconds) tends to overestimate hypoperfusion lesion volume due to delayed arterial transit effects. On the other hand, delayed arterial transit effects in ASL images may indicate collateral flow38. However, there are no objective criteria for their radiographic definition and interpretation. The low SNR of ASL images also makes it difficult to determine the boundary of the hypoperfusion lesion. Although often criticized as a “black box”, advanced ML and DL algorithms offer powerful tools to extract complex features from multi-dimensional data without explicit characterization of these features. In the present study, our DL model apparently realized multiple functions including denoising of ASL images, integration with diffusion MRI, and segmentation of hypoperfusion lesions. The DL algorithm outperformed 6 ML methods which were trained on the same dataset acquired on Siemens MR scanners, and it was demonstrated that the DL algorithm can be generalized to an independent dataset collected on GE MR scanners. Although different commercial software packages (OLEA and RAPID) were used to generate the Tmax label of hypoperfusion lesions in the two cohorts respectively, our DL model was able to achieve a high accuracy of 92% in both cohorts. As a further validation, the mean pCASL CBF values increase from the infarct core, to the predicted penumbral tissue, and to the contralateral region at both 1.5 and 3T (see online supplement Table VI). These results strongly indicate that the proposed DL model is well suited and valid for identifying hypoperfusion lesions on ASL images in AIS patients, for two main field strengths and MRI platforms used clinically.
In the present study, we trained 10 models using the 167 image sets of the UCLA cohort with 10-fold validation. When evaluated on the Stanford cohort, the consensus or averaged probability map from the 10 models yielded a significantly higher AUC of ROC (0.942) compared with the 10 individual models (AUC of ROC: 0.897±0.050), while the subject-level treatment eligibility results remain the same (0.92) for all the models. Therefore, although additional time (~8sec per subject) and storage (~140MB in total) is needed for 10 models, we recommend using the averaged result of the 10 models for future applications.
It is important to note that the clinical decision to proceed with endovascular thrombectomy is not only based on the imaging criteria of diffusion-perfusion mismatch, but also on the individual patient’s medical history and clinical condition. While the ASL perfusion DL model cannot and should not be used independently for treatment decision-making, interventionists may consider utilizing it to supplement their assessment of stroke and treatment planning, when imaging with completely non-invasive MRI.
There are a few limitations and future directions of the present study. The detected hypoperfusion lesions on ASL did not match completely with that of DSC MRI with a moderate Dice coefficient of 0.47 in the UCLA cohort and 0.43 in the Stanford cohort. We noticed that the Dice coefficients were lower in patients with smaller hypoperfusion lesions due to their sensitivity to the mismatch between ASL and DSC MRI. There were 4 out of a total of 50 cases that were incorrectly classified for treatment eligibility, as shown in the bottom row of Figure 4. This discrepancy may arise from different mechanisms of ASL and DSC PWI, which merits further research in larger cohorts. The performance of our DL model is likely to be further improved with larger training datasets, as well as by integrating other clinical information such as demographics, electronic medical records, biomarkers, and real-time continuous monitoring using wearables39. Although we did not adjust our DL model to test the Stanford data (due to small sample size and also to avoid bias), it is also possible to fine-tune the model with a few cases when dealing with MRI data from different vendors. Finally, future clinical studies are needed to evaluate how well the DL penumbra can predict follow-up infarct in AIS patients when there is lack of reperfusion.
Conclusion
With a high accuracy of 92% for imaging-based criteria for endovascular treatment in two independent cohorts of AIS patients and superior performance compared to ML methods, the proposed ASL perfusion DL model may provide a promising approach for assisting decision-making for endovascular treatment in AIS patients.
Supplementary Material
Acknowledgements:
The authors thank Drs. Yonggang Shi and Ben Duffy for helpful discussions.
Sources of Funding:
This work was supported by National Institute of Health (NIH) grants UH2-NS100614, R01EB028297, R01NS066506.
Disclosures:
Dr. Zaharchuk received research support from GE Healthcare, Bayer, Nvidia, and has equity interest in Subtle Medical. Dr. Wang received consulting fees from Bioclinica, Parexel, Translational MRI, and has equity interest in Translational MRI. Dr Liebeskind received consulting fees from Cerenovus, Genentech, Medtronic, and Stryker. Dr. Kim is funded by the BrightFocus Foundation grant (A2019052S).
Reference:
- 1.Albers GW, Marks MP, Kemp S, Christensen S, Tsai JP, Ortega-Gutierrez S, et al. Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. N Engl J Med. 2018;378:708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouslama M, Haussen DC, Grossberg JA, Dehkharghani S, Bowen MT, Rebello LC, et al. Computed tomographic perfusion selection and clinical outcomes after endovascular therapy in large vessel occlusion stroke. Stroke. 2017;48:1271–1277 [DOI] [PubMed] [Google Scholar]
- 3.Bottinor W, Polkampally P, Jovin I. Adverse reactions to iodinated contrast media. Int j angiol. 2013:149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer PW, Souza L, Kamalian S, Hirsch JA, Yoo AJ, Gonzalez RG, et al. Limited reliability of computed tomographic perfusion acute infarct volume measurements compared with diffusion-weighted imaging in anterior circulation stroke. Stroke. 2015;46:419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang DJ, Alger JR, Qiao JX, Gunther M, Pope WB, Saver JL, et al. Multi-delay multi-parametric arterial spin-labeled perfusion mri in acute ischemic stroke - comparison with dynamic susceptibility contrast enhanced perfusion imaging. NeuroImage. Clinical 2013;3:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang DJ, Alger JR, Qiao JX, Hao Q, Hou S, Fiaz R, et al. The value of arterial spin-labeled perfusion imaging in acute ischemic stroke: Comparison with dynamic susceptibility contrast-enhanced mri. Stroke. 2012;43:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moatti Z, Gupta M, Yadava R, Thamban S. A review of stroke and pregnancy: Incidence, management and prevention. Eur J Obstet Gynecol Reprod Biol. 2014;181:20–27 [DOI] [PubMed] [Google Scholar]
- 8.Kupferman JC, Zafeiriou DI, Lande MB, Kirkham FJ, Pavlakis SG. Stroke and hypertension in children and adolescents. J Child Neurol. 2017;32:408–417 [DOI] [PubMed] [Google Scholar]
- 9.Asadi H, Dowling R, Yan B, Mitchell P. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLOS ONE. 2014;9:e88225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Os HJA, Ramos LA, Hilbert A, van Leeuwen M, van Walderveen MAA, Kruyt ND, et al. Predicting outcome of endovascular treatment for acute ischemic stroke: Potential value of machine learning algorithms. Frontiers in Neurology. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444 [DOI] [PubMed] [Google Scholar]
- 12.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol. 2018;39:1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaharchuk G, El Mogy IS, Fischbein NJ, Albers GW. Comparison of arterial spin labeling and bolus perfusion-weighted imaging for detecting mismatch in acute stroke. Stroke. 2012;43:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straka M, Albers GW, Bammer R. Real-time diffusion-perfusion mismatch analysis in acute stroke. J Magn Reson Imaging. 2010;32:1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang YY, Brady M, Smith S. Segmentation of brain mr images through a hidden markov random field model and the expectation-maximization algorithm. Ieee T Med Imaging. 2001;20:45–57 [DOI] [PubMed] [Google Scholar]
- 18.Olivot J-M, Mlynash M, Thijs VN, Kemp S, Lansberg MG, Wechsler L, et al. Optimal tmax threshold for predicting penumbral tissue in acute stroke. Stroke. 2009;40:469–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasawa M, Jones PS, Guadagno JV, Christensen S, Fryer TD, Harding S, et al. How reliable is perfusion mr in acute stroke? Stroke. 2008;39:870–877 [DOI] [PubMed] [Google Scholar]
- 20.Davis SM, Donnan GA, Parsons MW, Levi C, Butcher KS, Peeters A, et al. Effects of alteplase beyond 3 h after stroke in the echoplanar imaging thrombolytic evaluation trial (epithet): A placebo-controlled randomised trial. The Lancet Neurology. 2008;7:299–309 [DOI] [PubMed] [Google Scholar]
- 21.Li WQ, Wang GT, Fidon L, Ourselin S, Cardoso MJ, Vercauteren T. On the compactness, efficiency, and representation of 3d convolutional networks: Brain parcellation as a pretext task. Lect Notes Comput Sc. 2017;10265:348–360 [Google Scholar]
- 22.Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, et al. Niftynet: A deep-learning platform for medical imaging. Comput Methods Programs Biomed. 2018;158:113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milletaride F, Navab N, Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. Int Conf 3d Vision. 2016:565–571 [Google Scholar]
- 24.Diederik P Kingma JB. Adam: A method for stochastic optimization Proceedings of the 3rd International Conference on Learning Representations (ICLR); 2015. May 7–9; San Diego, California (CA): ICLR; 2015. [Google Scholar]
- 25.McKinley R, Hung F, Wiest R, Liebeskind DS, Scalzo F. A machine learning approach to perfusion imaging with dynamic susceptibility contrast mr. Front Neurol. 2018;9:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purushotham A, Campbell BCV, Straka M, Mlynash M, Olivot JM, Bammer R, et al. Apparent diffusion coefficient threshold for delineation of ischemic core. Int J Stroke. 2015;10:348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stehman SV. Selecting and interpreting measures of thematic classification accuracy. Remote Sens Environ. 1997;62:77–89 [Google Scholar]
- 28.Saver JL, Goyal M, Bonafe A, Diener HC, Levy EI, Pereira VM, et al. Stent-retriever thrombectomy after intravenous t-pa vs. T-pa alone in stroke. N Engl J Med. 2015;372:2285–2295 [DOI] [PubMed] [Google Scholar]
- 29.Campbell BC, Mitchell PJ, Kleinig TJ, Dewey HM, Churilov L, Yassi N, et al. Endovascular therapy for ischemic stroke with perfusion-imaging selection. N Engl J Med. 2015;372:1009–1018 [DOI] [PubMed] [Google Scholar]
- 30.Goyal M, Demchuk AM, Menon BK, Eesa M, Rempel JL, Thornton J, et al. Randomized assessment of rapid endovascular treatment of ischemic stroke. N Engl J Med. 2015;372:1019–1030 [DOI] [PubMed] [Google Scholar]
- 31.Copen WA, Morais LT, Wu O, Schwamm LH, Schaefer PW, Gonzalez RG, et al. In acute stroke, can ct perfusion-derived cerebral blood volume maps substitute for diffusion-weighted imaging in identifying the ischemic core? PLoS One. 2015;10:e0133566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copen WA, Yoo AJ, Rost NS, Morais LT, Schaefer PW, Gonzalez RG, et al. In patients with suspected acute stroke, ct perfusion-based cerebral blood flow maps cannot substitute for dwi in measuring the ischemic core. PLoS One. 2017;12:e0188891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S, Ma SJ, Liebeskind DS, Yu D, Li N, Qiao XJ, et al. Aspects-based reperfusion status on arterial spin labeling is associated with clinical outcome in acute ischemic stroke patients. Journal of Cerebral Blood Flow and Metabolism. 2018;38:382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S, Liebeskind DS, Dua S, Wilhalme H, Elashoff D, Qiao XJ, et al. Postischemic hyperperfusion on arterial spin labeled perfusion mri is linked to hemorrhagic transformation in stroke. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2015;35:630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell BC, Parsons MW. Imaging selection for acute stroke intervention. Int J Stroke. 2018;13:554–567 [DOI] [PubMed] [Google Scholar]
- 36.Kim KH, Choi SH, Park SH. Improving arterial spin labeling by using deep learning. Radiology. 2017:171154. [DOI] [PubMed] [Google Scholar]
- 37.Gong E, Pauly J, Zaharchuk G. Boosting snr and/or resolution of arterial spin label (asl) imaging using multi-contrast approaches with multi-lateral guided filteranddeepnetworks. Proc ISMRM. 2017;25:3983 [Google Scholar]
- 38.Zaharchuk G, Do HM, Marks MP, Rosenberg J, Moseley ME, Steinberg GK. Arterial spin-labeling mri can identify the presence and intensity of collateral perfusion in patients with moyamoya disease. Stroke. 2011;42:2485–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saber H, Somai M, Rajah GB, Scalzo F, Liebeskind DS. Predictive analytics and machine learning in stroke and neurovascular medicine. Neurol Res. 2019:1–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


