Abstract
T cells engineered to express antigen-specific T cell receptors (TCRs) are potent therapies for viral infections and cancer. However, efficient identification of clinical candidate TCRs is complicated by the size and complexity of T cell repertoires and the challenges of working with primary T cells. Here, we present a high-throughput method to identify TCRs with high functional avidity from diverse human T cell repertoires. The approach uses massively parallel microfluidics to generate libraries of natively paired, full-length TCRαβ clones, from millions of primary T cells, which are then expressed in Jurkat cells. The TCRαβ-Jurkat libraries enable repeated screening and panning for antigen-reactive TCRs using peptide:MHC binding and cellular activation. We captured >2.9 million natively paired TCRαβ clonotypes from six healthy human donors and identified rare (<0.001% frequency) viral antigen–reactive TCRs. We also mined a tumor-infiltrating lymphocyte (TIL) sample from a melanoma patient and identified several tumor-specific TCRs, which, after expression in primary T cells, led to tumor cell killing.
T cells function in a broad range of immunological roles and have been used with varying degrees of success as adoptive cellular therapies (ACTs) for the treatment of cancer, autoimmunity, and infectious disease1–3. T cells engineered to express tumor-specific TCRs targeting common tumor antigens including NY-ESO-1 and MART-1 have shown promise in the clinic by directing cytotoxic T cells to kill tumor cells4–7. TCR-engineered T cell approaches are being investigated for the treatment of infectious diseases, and TCR-engineered regulatory T cells have been proposed to mitigate autoimmunity and transplant rejection8,9. Despite advances in clinical development of TCR-engineered ACTs, identification of clinically relevant TCRs has been limited by several factors. TCRs for clinical use are typically identified from human T cell repertoires using relatively low-throughput methods including ex vivo T cell expansion and single cell sorting or limited dilution schemes, which are not amenable to massively parallel analysis of TCRαβ pairs10–13. Furthermore, the primary T cells used in these TCR discovery approaches are difficult to culture and are a nonrenewable resource. Although TCRs can be engineered and affinity matured from artificial libraries14,15, these non-natural TCRs have not been subjected to endogenous thymic selection processes and can have off-target effects that are difficult to predict16. This has led to several tragic deaths caused by off-target effects17.
As such, more efficient methods for concurrently identifying and functionally validating natively-paired TCRαβ sequences would be useful. Several groups have developed strategies to sort single T cells of interest, amplify and clone the natively-paired TCRαβ chains from the single cells, and express them in primary T cells or Jurkat cell lines for functional analysis13,18. However, these approaches obtain sequences from only a few hundred single cells in parallel, complicating discovery of rare TCRαβ clonotypes. A commercial method (10X Genomics) isolates single cells into microfluidic droplets and pairs TCRαβ by fusing DNA barcodes to the TCRα and TCRβ chains19. However, single-cell barcoding methods only interrogate a small fraction of the T cell repertoire (typically no more than 20,000 cells per run), and do not generate libraries of physically-linked TCRαβ clonotypes that can be screened for functional activity.
Antibody discovery has faced many of the same challenges as TCR discovery, but antibody discovery technologies are far more advanced. For example, we and others have described methods that combine microfluidics, multiplex PCR, yeast display, and deep sequencing for ultra-high-throughput discovery of rare antibodies from human repertoires20,21. Yeast display of TCRs is possible22,23, but such methods are less useful for functional screening due to the absence of the T cell surface co-receptors CD8 and CD324, which improve TCR binding to the peptide-major histocompatibility complex (pMHC) complex and mediate downstream intracellular signaling events. Randomly-paired TCRα-TCRβ chains from primary T cells have been expressed in Jurkat cells25; however, most TCRs in such libraries are non-functional due to mis-pairing, limiting the ability to discover rare and functional TCRs.
We present an approach for massively parallel mining for rare (<0.001% frequency), antigen-reactive, natively-paired TCRαβs from human T cell repertoires (Fig. 1). We use droplet microfluidics to isolate millions of individual T cells into droplet emulsions and physically link TCRα and TCRβ chains from the single cells. These TCRαβ libraries are deep sequenced and cloned en masse into full-length lentiviral expression constructs. The full-length TCRαβ libraries are transduced into Jurkat cells, generating an immortalized, natively-paired, massively polyclonal TCRαβ expression library. The TCRαβ-Jurkat libraries are repeatedly screened and enriched for antigen reactivity by pMHC binding as well as TCR-mediated cellular activation.
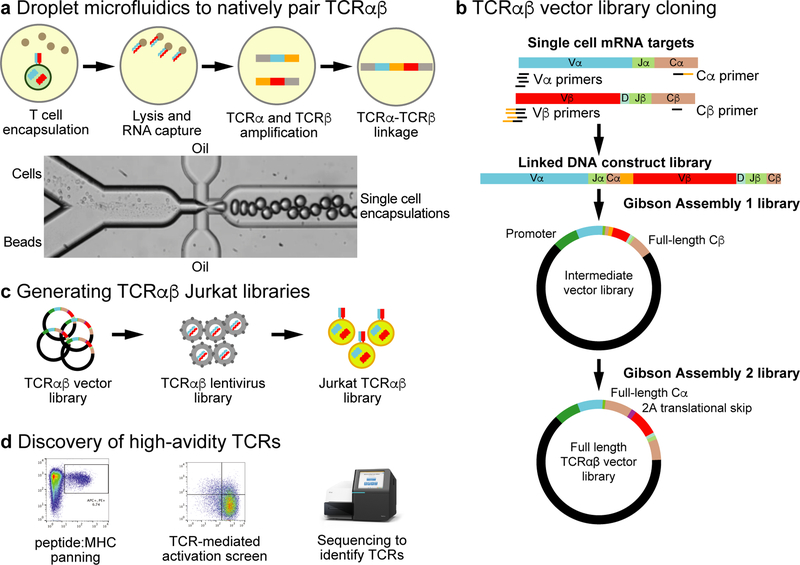
Fig. 1. Overview of the massively parallel TCRαβ repertoire mining technology.
a, T cells are encapsulated into emulsion microdroplets with lysis mixture and oligo(dT) beads. RNA-bound beads are re-injected into emulsion microdroplets with RT-PCR primers for linkage amplification of TCRα and TCRβ. b, RT-PCR primers separately amplify TCRα and TCRβ in the microdroplets. The RT-PCR introduces a region of complementarity, such that the TCRα and TCRβ amplicons are physically linked in a single amplicon. After breaking the emulsions, the linked TCRαβ amplicons are subcloned by Gibson Assembly into a vector that contains the EF1α promoter, TRAV leader sequence, and the TRBC2 (Cβ) sequence. A second Gibson Assembly introduces the TRAC (Cα) sequence, a 2A translational skip motif, and a TRBV leader sequence for full-length TCR expression. c, The full-length TCRαβ lentiviral expression library is packaged into lentiviral particles en masse and transduced into Jurkat cells creating a TCRαβ-Jurkat library. d, The TCRαβ-Jurkat library is then used to mine for TCRαβ sequences using peptide:MHC panning, TCR-mediated activation screens, and deep TCR sequencing.
Results
Analysis of Diverse, Natively-Paired TCRαβ Repertoires
Human TCRαβ repertoires comprise millions of single cells that express different TCRα and TCRβ sequences. To capture this diversity, we adapted our existing microfluidic technology20 to isolate millions of single T cells into droplets in an hour (Supplementary Fig. 1). T cells are lysed within the droplets and mRNA from single cells are captured on oligo-dT beads. The mRNA-bound beads are reinjected into droplets containing reagents for overlap extension reverse transcriptase PCR, such that the TCRα and TCRβ transcripts are amplified and physically linked into a single TCRαβ amplicon (Supplementary Fig. 2). The physically linked TCRαβ amplicon libraries are further processed for deep sequencing and cloning into expression constructs (Supplementary Fig. 3).
We used our technology to generate natively-paired TCRαβ libraries for three HLA-A*02:01 and three HLA-A*24:01 human donors that were seropositive for cytomegalovirus (CMV) and Epstein-Barr virus (EBV) (Supplementary Table 1). We ran 2.7–3.4 million T cells through the microfluidic system for each donor and processed the TCRαβ amplicons for deep sequencing (Illumina) and cloning into the lentiviral expression system.
We analyzed the six linked TCRαβ repertoires with deep sequencing (Supplementary Table 1). Illumina sequencing error can be difficult to differentiate from bona fide TCRαβ sequence diversity26,27, so we applied conservative informatics to eliminate low-quality sequence reads. We define a TCRαβ clonotype as a unique combination of CDR3α and CDR3β amino acid sequences with specific V-J gene usage. The libraries contained an average of 488,601 clonotypes (range: 339,707 to 711,589), for a total of 2,931,604 TCRαβ clonotypes, which comprehensively covers the diversity of possible TRAV-TRBV gene combinations (Fig. 2a). The TCRαβ libraries were highly diverse, with the top twenty clonotypes accounting for 10.77% of the total sequencing reads on average (range: 8.48% to 14.62%) and a long tail of low-frequency clonotypes. The average median clonotype read frequency was 0.000021% across our six libraries (Supplementary Figs. 4–5).
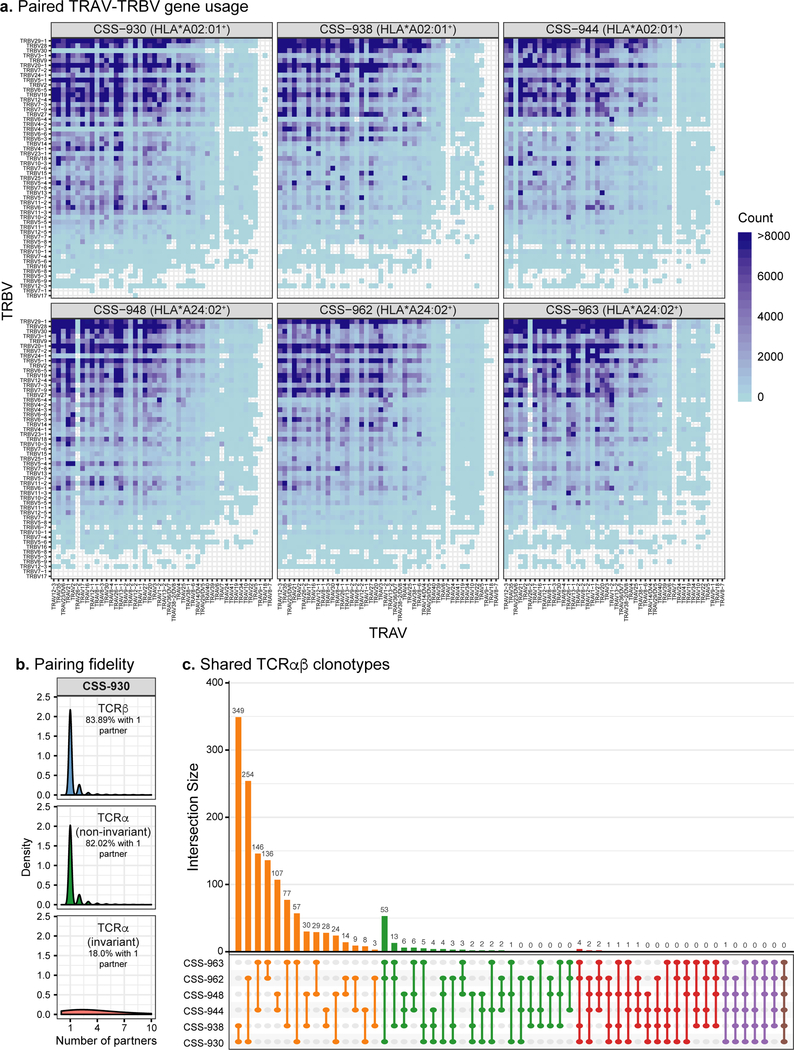
Fig. 2. Bioinformatic analysis of six paired TCRαβ repertoires.
a, Paired TRAV-TRBV gene usage across all six virus-seropositive TCRαβ repertoires. Blue shading indicates the number of unique clonotypes for each TRAV-TRBV combination for each repertoire. b, Number of partners for TCRβ, non-invariant TCRα, and known invariant TCRα sequences in the Donor CSS-930 repertoire. The density histogram indicates the relative abundance of TCR sequences comprising each category of number of partners. The percent of TCR clonotypes paired with only one partner is reported. c, UpSet plot showing counts of TCRαβ clonotypes that overlap between the six paired TCRαβ repertoires. For the Fig. 2b–c analyses, we required at least 2 sequencing reads per clonotype.
To characterize proper TCRα-TCRβ pairing within our linked TCRαβ libraries, we spiked Jurkat cells into healthy donor primary T cells at 0.1% or 0.001% frequency and ran the samples through our microfluidic platform. We found that 92.9% and 98.6% of Jurkat TCRα paired with Jurkat TCRβ, for the 0.1% and 0.001% mixtures, respectively.
To broadly assess the TCRα-TCRβ pairing fidelity generated by our microfluidic system, we compared the number of unique TCRβ partners paired to known invariant and non-invariant TCRα sequences within our linked TCRαβ libraries (Fig. 2b; Supplementary Fig. 6). Invariant TCRα sequences are expressed by specialized T cell populations, such as natural killer T (NKT) and mucosal-associated invariant T (MAIT) cells28,29. These T cell populations express a constrained set of invariant TCRα CDR3 sequences, such that repertoire diversity is driven by the TCRβ chain30–32. Previously, single cell analysis has demonstrated that identical TCRα CDR3 sequences pair with numerous TCRβ sequences33. We therefore reasoned that known invariant TCRα sequences would pair with a variety of TCRβ, whereas non-invariant TCRα would generally pair with only one TCRβ, providing a method to quantify our TCRαβ pairing fidelity. In agreement with this, we observed that a high proportion (81.45%) of invariant TCRα sequences paired with more than one TCRβ, whereas a significantly smaller fraction (18.90%; one-sided binomial proportion test, z-statistic, p < 2.2×10−16) of non-invariant TCRα paired with more than one TCRβ (Fig. 2b; Supplementary Fig. 6). Similarly, the majority of TCRβ sequences (79.88%) paired with a single TCRα. These data suggest that our cloned TCR libraries retain high TCRαβ pairing fidelity. The TCRβ and non-invariant TCRα sequences that were found to pair to multiple partners could represent natural features of these T cell repertoires or could have resulted from mispairing during TCRαβ linkage due to multiple cells in droplets, PCR-based artifacts, or other technical errors.
Recent work using single chain TCR sequencing methods has shown considerable overlap between TCR repertoires from unrelated individuals, both in terms of shared TCR sequences and common sequence motifs34,35. Globally, we observed very little overlap between donors’ TCRαβ repertoires, with only one TCRαβ clonotype shared among five donors and twelve shared among at least four donors (Fig. 2c). No clear global distinctions were observed between HLA-A*02:01 and HLA-A*24:02 genotypes, even though all six donors are CMV+ and EBV+, and therefore are likely to have some anti-viral T cells in the periphery. Notably, we observed higher cross-donor sharing of unlinked TCRs with 1735 TCRα and 17 TCRβ sequences shared among at least five repertoires, and 4866 TCRα and 128 TCRβ sequences shared among at least four repertoires (Supplementary Figs. 7–9). This suggests that conventional single chain TCR sequencing methods may over-estimate functional repertoire overlap between unrelated individuals.
Generation of Natively-Paired TCRαβ Jurkat Expression Libraries
To understand how well our method captures TCR diversity, we compared the TCRβ clonotype diversity present in our natively-paired TCRαβ amplicon libraries to the TCRβ clonotype diversity obtained using single chain TCRβ amplification of RNA from a matched aliquot of primary T cells (Supplementary Table 2). We controlled for the number of input cells and sequenced to similar depth (average of 5.4 million and 5.0 million reads from single chain TCRβ and natively-paired TCRαβ libraries, respectively) across all samples. We identified an average of 303,599 unique TCRβ clonotypes from single chain amplification and 210,249 TCRβ clonotypes from the paired TCRαβ libraries, suggesting a clonotype yield of about 71%. Furthermore, the natively-paired TCRαβ libraries captured, on average, 97% of the top 100 and 91% of the top 250 clonotypes identified in the single chain TCRβ libraries (Supplementary Fig. 10). These data confirmed that our natively-paired TCRαβ libraries efficiently capture the clonotypes present in the primary T cell population, and, as expected, the rate of clonotype overlap between single chain TCRβ and paired TCRαβ libraries decreased with decreasing clonotype frequency.
We next developed a workflow to clone these natively-paired TCRαβ amplicon libraries into expression libraries. An important innovation was to optimize Gibson Assembly for cloning the libraries of TCRαβ amplicons en masse into a lentiviral expression vector, followed by another library-wide Gibson Assembly to introduce the TCRα constant domain, a ribosomal skip motif (P2A), and a TCRβ leader sequence, for full-length TCRαβ expression (Supplementary Fig. 3). We generated over 5 million bacterial transformants at each cloning step to maintain the starting clonotype diversity (Supplementary Table 2). The full-length TCRαβ expression libraries were then packaged into lentiviral particles for transduction into Jurkat cells, generating immortalized, natively-paired TCRαβ-Jurkat expression libraries. Unlike primary cells, the TCRαβ-Jurkat libraries can be efficiently processed through multiple rounds of panning, across multiple antigens, using both binding to pMHC multimers and activation by artificial antigen-presenting cells (aAPCs).
To improve Jurkat TCR-mediated cellular activation in response to HLA class I presented peptides, we engineered the TCRβ deficient (ΔTCRβ) Jurkat cell line [J.RT3-T3.5] to express human CD8 (Supplementary Fig. 11). We then transduced the six full-length TCRαβ expression libraries into the CD8+ ΔTCRβ Jurkat cells at low infection rates (estimated 10–20%) to obtain single integration events. We obtained CD3+/TCRαβ+ surface expression in about 10% of pre-selected cells, which increased to 42–56% after selection for stable lentiviral integration (Supplementary Fig. 12). Assuming a Poisson probability distribution, we calculated that a 20% transduction efficiency should result in about 90% of our selected cells expressing a single TCR36. To determine the actual frequency of cells expressing single TCRs, we sorted single CD3+/TCRαβ+ cells from two post-selected TCRαβ-Jurkat libraries and amplified the expressed TCRβ mRNA. This analysis confirmed that >80% of the TCRαβ-Jurkat library cells express a single TCR (Supplemental Fig. 13).
Finally, we confirmed that the TCRαβ-Jurkat expression library build maintained the clonotype diversity present in the starting natively-paired TCRαβ amplicon libraries. Deep TCRβ sequencing demonstrated that the various molecular cloning steps and the fully selected TCRαβ-Jurkat libraries were highly correlated with their respective starting TCRαβ amplicon library, with Pearson correlation coefficients ranging from 0.77 to 0.97 (Supplementary Figs. 14–15). In addition, >95% of the 100 most frequent TCRβ clonotypes were maintained through the full process (Supplementary Fig. 16). In total, the six healthy donor TCRαβ-Jurkat libraries express over one million TCRβ clonotypes with an average of 186,117 unique TCRβ clonotypes per library (Supplementary Table 2).
Identification of Rare Anti-Viral TCRs
Antigen-specific T cells are conventionally identified by staining primary T cells with pMHC multimers, but any further functional analysis requires single cell cloning. Unlike primary T cells, Jurkat cells are easy to culture and expand indefinitely, enabling us to apply a “panning” approach. In this method, TCRαβ-Jurkat libraries are stained with dextramer and sorted by FACS. These “Round 1 dextramer” cells are expanded, re-stained with dextramer, and sorted a second time (“Round 2 dextramer”); this cycle can be repeated as necessary to enrich for rare antigen-specific clones. To test the method, we first performed control experiments with a previously reported HLA-A*02:01/MART-1(ELA) reactive TCRαβ clone, MEL537, spiked into the Donor CSS-930 TCRαβ-Jurkat library at a range of 0.001–1% frequency. Panning recovered the clone quantitatively across all levels tested (Supplementary Figs. 17–18).
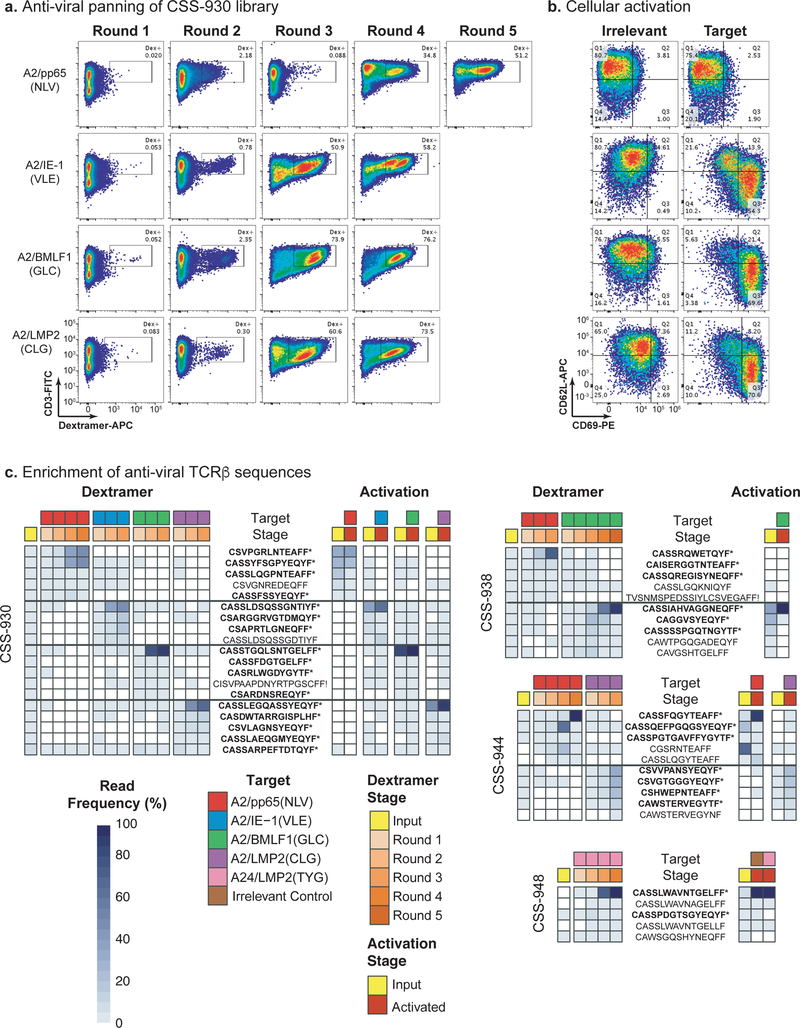
To identify virus-specific TCRαβ clones, we panned the six TCRαβ-Jurkat libraries with fluorescently-labeled pMHC dextramers against eight well-characterized antigens from CMV and EBV (Supplementary Table 3). Multiple rounds of panning enriched for dextramer-binding cells to four A*02:01 targets and one A*24:02 target across our donor libraries (Fig. 3a; Supplementary Figs. 19–21). Two of the A*24:02 donors yielded no binders. Dextramer-binding cells were rare (<0.1%) in the starting TCRαβ-Jurkat libraries and increased to 47–98% with panning. The Jurkat cell expression system also provided the ability to test these dextramer enriched TCRs for functional activity by staining for activation markers CD69 and CD62L (non-activated cells are CD69−/CD62L+ and activated cells are CD69+/CD62L−). We used an aAPC activation assay with peptide-pulsed T2 cells to functionally validate the fully panned Jurkat samples (Fig. 3b; Supplementary Figs. 19–21). This confirmed strong cell activation in 6 of 8 samples, weak activation for the A24/LMP2(TYG) sample, and no activation for the CSS-930 Jurkat library enriched for A2/pp65(NLV) binding.
Fig. 3. Identification of virus-specific TCRαβ clones by library panning and cellular activation screens.
a, Panning the Donor CSS-930 TCRαβ-Jurkat library with pMHC dextramers for four different viral antigens: A2/pp65(NLV), A2/IE-1(VLE), A2/BMLF1(GLC), and A2/LMP2(CLG). pMHC dextramer signal is on the x-axis and CD3 staining is on the y-axis. The inset box indicates the gate used to sort for pMHC dextramer-positive cells. Panning experiments were completed one time. b, Cellular activation of the fully pMHC dextramer-enriched cell populations co-cultured with peptide-pulsed T2 cells. T2 cells were loaded with four different viral antigens, right column (target), from top to bottom: pp65(NLV), IE-1(VLE), BMLF1(GLC), and LMP2(CLG). The left column comprises irrelevant peptide targets loaded onto T2 cells. CD69 staining is on the x-axis and CD62L staining is on the y-axis. Cellular activation experiments were independently repeated twice with similar results. c, Enrichment of anti-viral TCRβ sequences for Donors CSS-930, CSS-938, CSS-944, and CSS-948. The legend indicates color coding for each target, pMHC dextramer panning, and cellular activation stage. TCRβ frequencies are indicated by a heat map, from light to dark blue. TCRβ clonotypes labeled with a star (*) and in bold font were engineered into monoclonal Jurkat cell lines.
To benchmark our dextramer panning method against conventional sorting of primary T cells, we stained PBMCs from donor CSS-930 with the four viral A*02:01 dextramers, sorted for CD8+/Dextramer+ cells, and conducted TCRβ clonotype analysis. The primary PBMCs displayed a low frequency of viral-pMHC binding T cells (range: 0.055–0.32%), in agreement with our TCRαβ-Jurkat libraries (Supplementary Fig. 22). The dextramer-binding TCRβ clonotypes were typically quite rare in the primary T cell population (Supplementary Table 4) with a median read frequency of 0.0003%, suggesting that many of these clonotypes would have limited overlap across independent cell aliquots. Six of the TCRβ clonotypes were present at >0.01% in the primary T cell repertoire, and three (50%) of these were also identified by panning of the TCRαβ-Jurkat library, indicating that the TCRαβ-Jurkat libraries are able to capture common dextramer-binding TCRs.
A major advantage of the TCRαβ-Jurkat library system is the ability to quickly assess the cellular activity of the dextramer-enriched TCRs and reduce our false positive rate. To identify functional TCRs, we performed in vitro aAPC activation screens on partially-enriched TCRαβ-Jurkat populations and sorted for activated (CD69+/CD62L−) cells (Supplementary Fig. 23). We used deep sequencing to quantify clonotype enrichment in the activated cell population versus the starting population and integrated these data with the pMHC dextramer-panning data (Fig. 3c). We found that only 19 of the 44 (43.2%) clonotypes most enriched by pMHC dextramer panning were also enriched in the activation screen. For example, A2/pp65(NLV) dextramer panning of the CSS-930 library strongly enriched for two TCRs; however, these TCRs did not display cellular activation in response to NLV-pulsed T2 cells. These data suggest that combining an activation screen with pMHC dextramer panning could reduce false positive rates.
Characterization of Individual Anti-Viral TCRs
To quantify pMHC dextramer panning false positive rates, we selected 34 of the most frequent TCRαβ clonotypes following pMHC dextramer panning (mean: 24.55%; median: 7.3%; range: 0.2% - 97.6%) for monoclonal expression in Jurkat (Table 1; Supplementary Table 5). These clonotypes were generally very rare in the initial TCRαβ-Jurkat libraries (mean: 0.036%; median: 0.003%), with multiple clonotypes present at <0.0001%. On average, pMHC dextramer panning enriched the clonotypes 136,672-fold (median: 3,640), with two clonotypes, NLV.1 and NLV.8, enriching more than one million-fold. Of note, the NLV.8 clonotype has previously been identified by other groups and shown to have high functional avidity and cytotoxic activity38,39.
Table 1.
Overview of our TCRαβ-Jurkat mining and functional validation. Clone and donor identifiers, pMHC targets, pMHC dextramer panning TCRβ sequence frequencies, cellular activation TCRβ frequencies, and monoclonal validation data for TCRαβ clones identified with our technology. “ND” stands for not determined. “+” indicates a binder, “-“ indicates a non-binder.
| TCRD Read Frequency (%) | TCRD Read Frequency (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clone ID | Donor | Target | Starting Library | Post-Panning | Dextramer Enrichment | Input | Activated Cells | Activation Enrichment | Monoclonal Binding | Monoclonal EC50 (nM) |
| NLV.1 | CSS-930 | pp65(NLV) | 0.000035% | 47.61% | 1,359,118 | 33.47% | 35.08% | 1.048 | Not expressed | ND |
| NLV.2 | CSS-930 | pp65(NLV) | 0.090000% | 46.54% | 517 | 28.89% | 23.89% | 0.827 | + | No activation |
| NLV.3 | CSS-930 | pp65(NLV) | 0.000035% | 2.64% | 75,364 | 16.70% | 0.04% | 0.002 | − | ND |
| NLV.4 | CSS-930 | pp65(NLV) | 0.000035% | 0.54% | 15,415 | 0.95% | 0.003% | 0.003 | − | ND |
| NLV.5 | CSS-938 | pp65(NLV) | 0.090000% | 72.86% | 810 | ND | 1 ND I | ND | − | ND |
| NLV.6 | CSS-938 | pp65(NLV) | 0.000557% | 6.45% | 11,577 | ND | ND | ND | − | ND |
| NLV.7 | CSS-938 | pp65(NLV) | 0.010000% | 3.98% | 398 | ND | 1 ND 1 | ND | − | ND |
| NLV.8 | CSS-944 | pp65(NLV) | 0.000043% | 96.76% | 2,254,385 | 6.59% | 79.74% | 12.100 | + | 6.616 |
| NLV.9 | CSS-944 | pp65(NLV) | 0.000172% | 1.53% | 8,912 | 21.36% | 1.65% | 0.077 | + | No activation |
| NLV.10 | CSS-944 | pp65(NLV) | 0.000300% | 0.97% | 3,229 | 0.12% | 0.75% | 6.250 | + | 0.267 |
| VLE.1 | CSS-930 | IE-1 (VLE) | 0.030000% | 44.54% | 1,485 | 36.51% | 63.22% | 1.732 | + | 4.650 |
| VLE.2 | CSS-930 | IE-1 (VLE) | 0.000035% | 15.93% | 454,752 | 9.16% | 4.35% | 0.475 | − | ND |
| VLE.3 | CSS-930 | IE-1 (VLE) | 0.000841% | 14.92% | 17,747 | 6.98% | 3.94% | 0.564 | Not expressed | ND |
| VLE.4 | CSS-930 | IE-1 (VLE) | 0.160000% | 5.49% | 34 | 3.45% | 2.92% | 0.846 | − | ND |
| GLC.1 | CSS-930 | BMLF1 (GLC) | 0.160000% | 85.49% | 534 | 78.56% | 88.64% | 1.128 | + | 38.895 |
| GLC.2 | CSS-930 | BMLF1 (GLC) | 0.000035% | 3.61% | 103,054 | 3.25% | 2.43% | 0.748 | − | ND |
| GLC.3 | CSS-930 | BMLF1 (GLC) | 0.002697% | 0.85% | 315 | 2.95% | 0.27% | 0.092 | − | ND |
| GLC.4 | CSS-930 | BMLF1 (GLC) | 0.000490% | 0.51% | 1,040 | 0.84% | 0.06% | 0.071 | − | ND |
| GLC.5 | CSS-930 | BMLF1 (GLC) | 0.030000% | 0.38% | 13 | 0.50% | 0.02% | 0.040 | Not expressed | ND |
| GLC.6 | CSS-938 | BMLF1 (GLC) | 0.002340% | 92.46% | 39,513 | 35.47% | 76.86% | 2.167 | Weak | 591.950 |
| GLC.7 | CSS-938 | BMLF1 (GLC) | 0.000260% | 2.95% | 11,346 | 27.76% | 0.03% | 0.001 | − | ND |
| GLC.8 | CSS-938 | BMLF1 (GLC) | 0.003714% | 1.77% | 477 | 14.30% | 0.005% | 0.000 | − | ND |
| CLG.1 | CSS-930 | LMP2(CLG) | 0.020000% | 64.75% | 3,238 | 46.44% | 83.95% | 1.808 | + | 0.355 |
| CLG.2 | CSS-930 | LMP2(CLG) | 0.001857% | 12.03% | 6,480 | 9.35% | 1.75% | 0.187 | − | ND |
| CLG.3 | CSS-930 | LMP2(CLG) | 0.000035% | 8.14% | 232,372 | 4.73% | 0.82% | 0.173 | − | ND |
| CLG.4 | CSS-930 | LMP2(CLG) | 0.002522% | 3.27% | 1,297 | 1.26% | 6.60% | 5.238 | Weak | 25.595 |
| CLG.5 | CSS-930 | LMP2(CLG) | 0.008477% | 0.73% | 86 | 0.75% | 0.06% | 0.080 | − | ND |
| CLG.6 | CSS-944 | LMP2(CLG) | 0.460000% | 29.03% | 63 | 0.27% | 18.54% | 68.667 | − | No activation |
| CLG.7 | CSS-944 | LMP2(CLG) | 0.003863% | 33.35% | 8,633 | 0.29% | 26.06% | 89.862 | Weak | 0.668 |
| CLG.8 | CSS-944 | LMP2(CLG) | 0.001245% | 25.27% | 20,302 | 0.25% | 15.32% | 61.280 | − | No activation |
| CLG.9 | CSS-944 | LMP2(CLG) | 0.002833% | 11.45% | 4,042 | 0.04% | 5.84% | 146.000 | − | No activation |
| TYG.1 | CSS-948 | LMP2(TYG) | 0.009477% | 97.58% | 10,297 | 8.99% | 92.71% | 10.313 | Weak | 2.516 |
| TYG.2 | CSS-948 | LMP2(TYG) | 0.060000% | 0.20% | 3 | 0.01% | 0.0065% | 0.653 | − | ND |
| TYG.3 | CSS-948 | LMP2(TYG) | 0.060000% | 0.26% | 4 | 3.67% | 0.0065% | 0.00178 | − | ND |
| PMEL.1 | CSS-799 | PMEL(KTW) | 0.120000% | 87.35% | 728 | 4.26% | 64.23% | 15.077 | + | 14.505 |
| PMEL.2 | CSS-799 | PMEL(KTW) | 0.001963% | 1.43% | 728 | 0.44% | 6.23% | 14.159 | + | 13.547 |
| PMEL.3 | CSS-799 | PMEL(KTW) | 0.001963% | 0.03% | 15 | 0.004% | 0.56% | 158.222 | + | 11.967 |
| PMEL.4 | CSS-799 | PMEL(KTW) | 0.300000% | 1.77% | 6 | 0.58% | 5.43% | 9.362 | + | 12.840 |
| PMEL.5 | CSS-799 | PMEL(KTW) | 0.300000% | 4.62% | 15 | 0.46% | 0.94% | 2.043 | + | 5.148 |
ND = Not Determined
− = No Dextramer Binding
+ = Dextramer Binding
Three of the 34 monoclonal TCRαβ constructs were poorly expressed in Jurkat cells, leaving 31 clones for downstream analysis. We demonstrated that 11/31 (35.5%) of the monoclonal TCRαβ cell lines bound their cognate pMHC and 9/11 (81.8%) of the validated binders induced cellular activation (Fig. 4a; Supplementary Figs. 24–28; Table 1; Supplementary Table 5). We also built two negative control monoclonal TCRαβ cell lines from high frequency clonotypes in the starting Donor CSS-930 TCRαβ library which persisted throughout pMHC dextramer panning. These high frequency negative control cell lines did not bind any of the four HLA-A*02:01 pMHC dextramers, as expected (Supplementary Fig. 29).
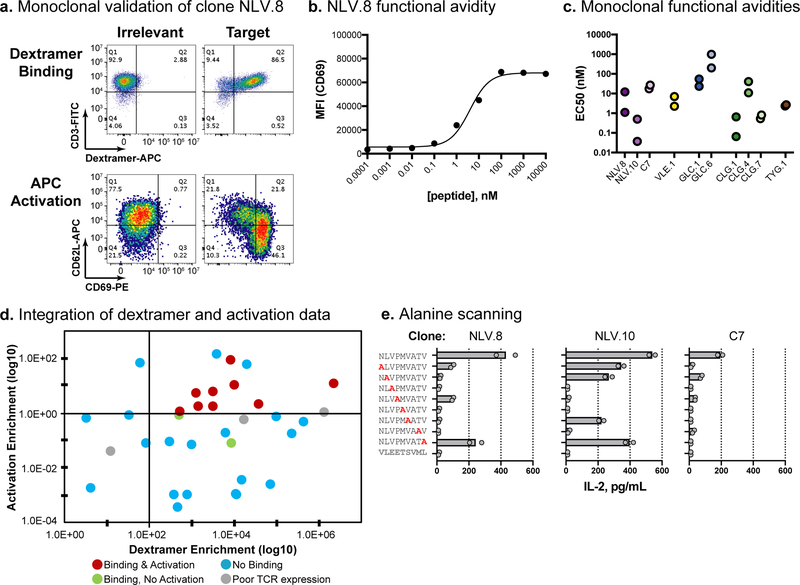
Fig. 4. Experimental validation of viral-specific TCRαβ clones.
a, Validation data for the NLV.8 monoclonal TCR. The top panels show dextramer binding; pMHC dextramer signal is on the x-axis and CD3 staining is on the y-axis. The bottom panels show Jurkat cell activation upon co-culture with peptide-loaded aAPCs; CD69 staining is on the x-axis and CD62L staining is on the y-axis. The left column is irrelevant and the right column is target peptide. Dextramer binding and cellular activation experiments were independently repeated twice with similar results. b, Functional avidity analysis of clone NLV.8 using T2 cells pulsed with a dilution series of peptide. Peptide loading concentration is on the x-axis and median fluorescence intensity (MFI) for CD69 staining is on the y-axis. Means are plotted from n=2 independent experiments. c, Functional avidity EC50 values calculated using peptide-pulsed T2 cells as described in b. Two independent experiments were conducted for each clone and the calculated EC50 values from each experiment are plotted. d, Integration of pMHC dextramer panning and activation data. Each point represents a unique clone, with clones colored according to their respective validation result. The dextramer enrichment of each clone is on the x-axis, and the activation enrichment (ratio of activated:input TCRβ read count frequency) is on the y-axis (based on data from Table 1). e, Functional specificity of viral-specific clones, using alanine scanning and IL-2 secretion. Clones NLV.8 and NLV.10 and the positive control (C7) are shown. T2 cells were loaded with wild type, single alanine mutation, or irrelevant peptides, and IL-2 secretion was measured by ELISA (x-axis). Replicate data (circles) and means (bars) are plotted for n=2 independent experiments.
We further characterized the functional avidities of the eleven TCRαβ monoclonal cell lines that displayed target-specific pMHC binding. We co-cultured each TCRαβ-Jurkat line with peptide-pulsed T2 cells (dilution series: 0.0001–10,000nM) and quantified cell activation by flow cytometry for surface CD69 staining (Fig. 4b; Supplementary Fig. 30). The previously-reported αA2/pp65(NLV) “C7” clone was used as a positive control40. The NLV.2 and NLV.9 cell lines failed to induce cellular activation, while functional avidity EC50s for the other nine lines ranged from 0.3nM to 565nM (median: 5.62nM) (Fig. 4c; Table 1; Supplementary Table 5).
We retrospectively assessed the benefit of combining pMHC dextramer panning with activation enrichment a priori in the initial screen (Fig. 4d). If we had applied a 100-fold pMHC dextramer enrichment cutoff, 15/26 clones (58%) would have been false positives for pMHC dextramer binding, and 17/26 clones (65%) would have been false positives for activation. If we had additionally applied an activation enrichment requirement of >1-fold, the overall false positive rate would have decreased to 2/11 (18%). Seven of the nine monoclonal TCRαβ cell lines that validated as activators were the highest frequency clonotypes following pMHC dextramer panning, indicating that dextramer panning can efficiently enrich for functional TCRs. However, this approach also enriched for two strong pMHC dextramer-binding clones (NLV.2 and NLV.9) that did not induce cellular activation, a phenomenon reported previously for some high affinity TCRs41. Including the activation enrichment requirement would have eliminated these non-activating TCRs from further analysis. Additionally, the activation enrichment would have verified functional activity for two low frequency (<4%) post-panning TCRs, NLV.10 and CLG.4. Given these results, combining dextramer and cellular activation enrichment measurements decreases the false positive rate and could also decrease the false negative rate.
Finally, we investigated the specificity of eight TCR clones by measuring IL-2 secretion upon incubation with a series of peptides with single alanine substitutions (Fig. 4e; Supplementary Fig. 31). Alanine mutations that reduced peptide presentation or TCR interaction induced lower IL-2 secretion than wild type cognate peptides. These data provide some evidence of contact residues and antigen specificity, which varied between TCRs and pMHC targets. Though a larger library of pMHC would better characterize specificity42, these alanine scanning data provide evidence that we have identified TCRs with varied specificity toward their cognate pMHC, rather than spurious TCRs that are artifacts of our methodology.
Identification and Characterization of Anti-Tumor TCRs from TILs
We obtained informed consent from an HLA-A*02:01+ Stage IV melanoma patient who was unsuccessfully treated with conventional therapy but who achieved a partial response to autologous tumor-inflitrating lymphocyte (TIL) adoptive cell therapy (ACT)43. We ran 1.36 million of the post-expanded therapeutic TILs through the microfluidic system and generated sequencing libraries from the natively-paired TCRαβ amplicons. Deep sequencing revealed a diverse TCRαβ repertoire with 395,464 unique TCRαβ clonotypes in our natively-paired TCRαβ amplicon library (Supplementary Table 1; Supplementary Fig. 32; Fig. 5a). We also confirmed a high estimated TCRβ clonotype yield of 96.6% with 86,068 unique TCRβ clonotypes present in the starting cell population, 83,172 in the linked TCRαβ amplicon library, and 71,753 in the Jurkat expression library (Supplementary Table 2). Furthermore, the linked TCRαβ amplicon library captured 100% of the top 250 TCRβ clonotypes present in the expanded TIL cell population (Supplementary Fig. 32e).
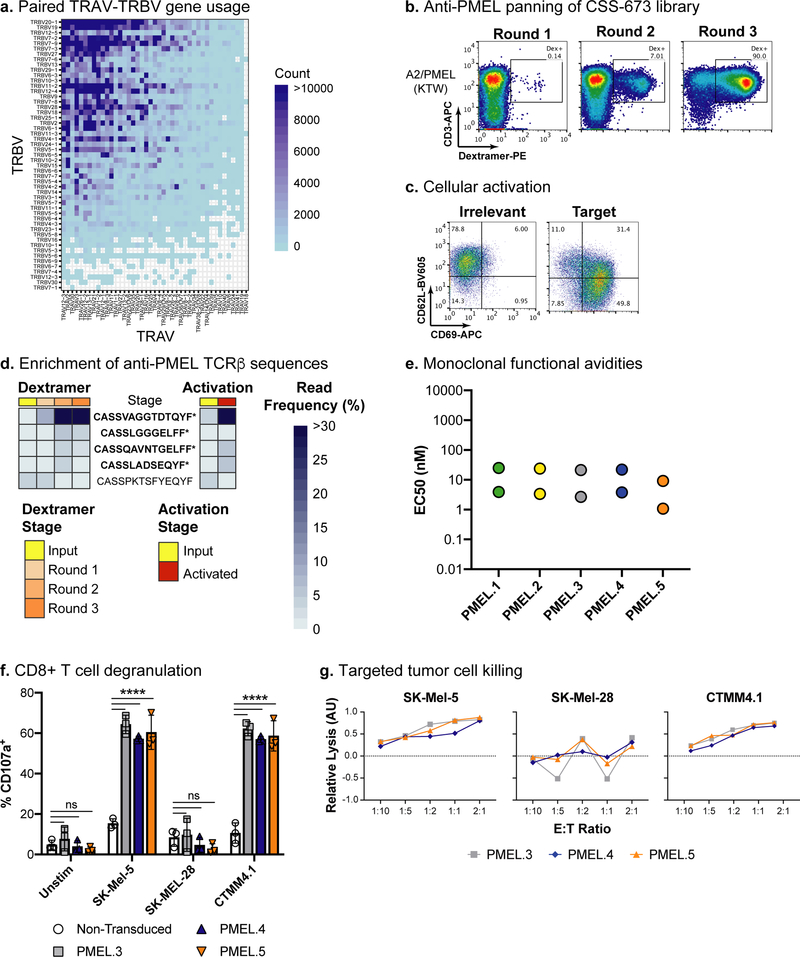
Fig. 5. Identification and experimental validation of anti-tumor antigen TCRαβ clones from tumor-infiltrating lymphocytes.
a, Paired TRAV-TRBV gene usage across the TIL TCRαβ repertoire. Blue shading indicates the number of unique clonotypes for each TRAV-TRBV combination. b, Panning the TIL (CSS-673) CD8− TCRαβ-Jurkat library with a pMHC dextramer for A2/PMEL(KTW). pMHC dextramer signal is on the x-axis and CD3 staining is on the y-axis. The inset box indicates the gate used to sort for pMHC dextramer-positive cells. The panning experiment was completed one time. c, Cellular activation data using the fully pMHC dextramer-enriched cell populations co-cultured with peptide-pulsed T2 cells. T2 cells were loaded with irrelevant peptide (left) and the PMEL(KTW) target (right). CD69 staining is on the x-axis and CD62L staining is on the y-axis. The cellular activation experiment was independently repeated twice with similar results. d, Enrichment of anti-PMEL TCRβ sequences from the TIL sample. The legend indicates color coding for pMHC dextramer panning and cellular activation stage. TCRβ frequencies are indicated by a heat map, from light to dark blue. TCRβ clonotypes labeled with a star (*) and in bold font were engineered into monoclonal Jurkat cell lines. e, Functional avidity EC50 values calculated using CD8+ monoclonal TCRαβ Jurkat cell lines co-cultured with T2 cells pulsed with a dilution series of peptide (see Supplementary Fig. 36). Two independent experiments were conducted for each clone and the calculated EC50 values from each experiment are plotted. f, T cell degranulation assay for three anti-A2/PMEL(KTW) TCRαβ clones transduced into primary human T cells. TCR-transduced T cells (i.e., Effector cells, E) were incubated with SK-Mel-5 (HLA-A*02:01+; PMEL+), SK-Mel-28 (HLA-A*02:01−; PMEL+), or CTMM4.1 (HLA-A*02:01+; PMEL+) tumor cells (i.e., Target cells, T) at an E:T ratio of 1:2 and stained for the degranulation marker, CD107a. The percentage of CD8+ T cells that were CD107a+ was measured for TCR-transduced T cells from three independent human donors and compared to non-transduced control T cells. Means and standard deviations are plotted from n=3 independent experiments. A two-way ANOVA with Dunnett’s test statistic was conducted. ns = not significant; **** = <0.0001 adjusted P value. g, Tumor cell killing data was measured using a WST-1 assay for the three anti-A2/PMEL(KTW) TCRαβ clones transduced into primary human T cells. TCR-transduced T cells were incubated with tumor cells at various E:T ratios as indicated on the x-axis, and the Relative Lysis (Arbitrary Units; AU) is reported on the y-axis. Mean values are plotted from triplicate wells for one experiment. AU data were transformed to show lysis mediated by TCR-transduced T cells relative to non-transduced T cells.
We built a TIL TCRαβ-Jurkat expression library and screened for pMHC dextramer binding to four well-characterized melanoma antigens expressed in patient matched tumor tissue (Supplementary Tables 2–3; Supplementary Fig. 33). The TIL TCRαβ-Jurkat library only displayed clear binding to the A2/PMEL(KTW) dextramer. This binding was enriched with panning (Fig. 5b), and cellular activation was confirmed by co-culture with peptide-pulsed T2 cells (Fig. 5c). We used deep sequencing to identify five TCRαβ clonotypes that were enriched by dextramer panning and aAPC-induced cellular activation (Fig. 5d; Supplementary Fig. 34; Table 1; Supplementary Table 5). As we observed previously in the virus-seropositive libraries, the anti-A2/PMEL(KTW) binders were initially rare (≤0.3%) in the TIL TCRαβ-Jurkat library. This low level of anti-tumor antigen reactivity in TILs is consistent with recent reports using analogous methods11.
We first engineered the anti-A2/PMEL(KTW) TCRαβ clonotypes into Jurkat cells that lacked the CD8 coreceptor. All five properly paired clones bound pMHC dextramer and showed CD8-independent cellular activation against peptide-loaded T2 cells as well as endogenously expressed antigen on the HLA-A*02:01+ melanoma tumor cell line, SK-Mel-5 (Supplementary Fig. 35). We also tested mis-paired TCRα-TCRβ cell lines as negative controls, and none of these showed strong dextramer binding or cellular activation (PMEL.5–4 showed weak dextramer binding). These results suggest that the anti-PMEL TCR clones have high functional activity. We then re-engineered the anti-PMEL TCRαβ clones into CD8 positive Jurkat cells and determined their functional avidity EC50s, which ranged from 5.1 to 14.5nM (Table 1; Fig. 5e; Supplementary Fig. 36). To test TCR-mediated cytotoxicity, we transduced the anti-PMEL TCRs into primary human T cells. Two of the TCRs (PMEL.1 and PMEL.2) expressed poorly in primary T cells. However, the three well-expressed clones induced strong CD107a staining, a marker of cytotoxic degranulation, in CD8+ T cells from three human donors when co-cultured with the HLA-A*02:01+ melanoma cell line SK-Mel-5 and unrelated HLA-A*02:01+ Stage IV melanoma patient derived CTMM4.1 tumor cells, but not when co-cultured with the HLA-A*02:01− melanoma cell line SK-Mel-28 (Fig 5f; Supplementary Figs. 37–38). These TCRs also demonstrated targeted tumor cell killing of HLA-A*02:01+ SK-Mel-5 and CTMM4.1 cells (Fig. 5g; Supplementary Fig. 39), suggesting that cell killing was A2-restricted.
Discussion
In a recent query of the VDJdb website, we found fewer than one thousand high-confidence paired TCRαβ sequences with known binding specificity and functional avidity44, even though a single person’s T cell repertoire comprises millions of clonotypes. Here we introduce a method to greatly accelerate the pace of identifying natively-paired TCRαβ sequences and their cognate antigen specificities. Each immortalized T cell repertoire can be mined for TCRs against large panels of pMHC targets. Our method has the potential to scale, providing an opportunity to comprehensively profile human TCR reactivity to a diverse set of antigens.
We captured over 2.9 million natively paired TCRαβ clonotypes from six healthy virus-seropositive human donors. Compared with prior published reports using single-cell microfluidic and other methods10,36–39, we obtained 10 to 100-fold more natively paired TCRαβ sequences. Similarly, two recently published single cell studies have each profiled a total of about 30,000 TILs across multiple patient samples obtaining 2,000 – 10,000 unique TCRαβ clonotypes per patient19,45, whereas our study profiled 1.36 million post-expanded TIL cells used for a therapeutic infusion product and we identified 395,464 unique TCRαβ clonotypes. We then took the seven libraries of TCRαβ amplicons and built highly diverse, natively-paired expression libraries in Jurkat cells. We identified and validated nine high-avidity anti-viral TCRs and five high-avidity anti-tumor TCRs from these seven libraries. We also showed that integrating activation-based screening with dextramer binding is critical for reducing the false positive rate and could also be useful for decreasing the false negative rate, differentiating our system from other TCR discovery methods. Additionally, we identified three anti-PMEL TCRs that induced strong HLA-restricted CD8+ T cell degranulation and cell killing in response to endogenously presented antigen on the established melanoma cell line SK-Mel-5 and unrelated patient derived cancer cells in vitro, demonstrating that this approach can efficiently identify functional TCRs from natively-paired T cell repertoires.
Despite our progress, there remain many areas for improvement and further investigation. Though we benchmarked our technology against pMHC dextramer sorting of primary T cells, we could not compute our false negative rate, since the number of “true positives” in the repertoires is unknown. Further, our technology still requires low-throughput methods to validate functional avidity for each individual TCR identified in a screen. Also, the pMHC dextramer panning approach described here led to a highly oligoclonal enrichment of a small number of TCRs, whereas the cellular activation enrichment was able to identify several lower frequency TCRs in partially pMHC dextramer-panned populations. Thus, a key area of continued optimization is in our flow sorting approach. For example, conducting the cellular activation enrichment after a single round of pMHC dextramer binding could improve our false positive and negative rates. Alternatively, implementing an initial selection step based on activation followed by pMHC dextramer sorting may be beneficial. To identify even rarer clones, future work could make TCRαβ-Jurkat libraries from T cells expanded ex vivo with peptide-loaded APCs. The Jurkat activation system could also be improved, for example, by using artificial reporter systems or alternative endogenous markers for activation. Also, our methods were assessed for identification of TCRs against only twelve pMHC targets across only two HLA types. Different targets and HLA types will certainly require further protocol development, for example by panning with activation rather than pMHC binding, when pMHC multimer reagents are not available. Finally, to simultaneously screen for novel pMHC targets and their cognate TCRs, we are actively working on methods for screening large pMHC target libraries against TCRαβ-Jurkat libraries.
The functional TCRs identified in this study have potential for translation to clinical applications. Pioneering groups use virus-specific primary T cells as a way to restore long-term viral immunity after hematopoietic stem cell transplantation46,47. The EBV and CMV TCRs identified in our study, plus TCRs for adenovirus, BK virus, and respiratory syncytial virus (RSV) could one day be used for this clinical ACT application, perhaps offering a more streamlined manufacturing protocol and higher potency. Recently, chimeric antigen receptor T cell (CAR-T) therapies were approved by the US Food and Drug Administration for leukemia and lymphoma48,49; however, no CAR-T or ACTs have been approved for solid tumors. The anti-PMEL TCRs identified in this study could be assessed for potential development into TCR-engineered T cells for advanced melanoma. Because our TCRs are not artificially affinity-matured, they may be less likely to show off-target toxicity17. In conclusion, our technology has removed a substantial bottleneck in functional TCR discovery and the development of novel ACT.
Online Methods
Sourcing and Processing Human Materials
De-identified peripheral blood mononuclear cells (PBMCs) in leukopaks were obtained from HLA-A*02:01 and HLA-A*24:02 healthy donors (AllCells, Alameda, CA, USA), under an Institutional Review Board (IRB) approved protocol. T cells were isolated from the PBMCs using the EasySep™ Human T Cell Enrichment Kit (Stemcell Technologies, Vancouver, BC, CA). Following isolation, T cells were cryopreserved using CryoStor® CS10 (Stemcell Technologies, Vancouver, BC, CA). For downstream single cell TCRα-TCRβ linkage, cells were thawed, washed, and resuspended at 5,000–6,000 cells per μl in cold DPBS+0.5% BSA with 12% OptiPrep™ Density Gradient Medium (Sigma, St. Louis, MO, USA). The resuspended cells were then used for microfluidic encapsulation as described in the next section.
Human TILs were generated from tumor biopsies from consenting individuals undergoing TIL clinical therapy for metastatic melanoma using a proprietary method which in brief involves the following. Tumor biopsies were disaggregated to a single cell suspension using collagenase and DNAse, then plated in tissue culture plates in the presence of IL-2 for up to 21 days. Following this initial outgrowth, the TIL underwent a rapid expansion protocol (REP) by mixing the derived TIL cells with donor-mixed irradiated PBMC feeder cells with addition of OKT3 and IL-2, for 14 days prior to cryopreservation. Patient matched tumor lines were grown by plating digested tumor material onto 24-well tissue culture plates in RPMI supplemented with 10% FBS (Sigma, St. Louis, MO, USA), 0.01 M HEPES (Sigma, St. Louis, MO, USA), and 1% Penicillin/streptomycin (Sigma, St. Louis, MO, USA). After 24 hours the supernatant was removed from the plate and fresh media was added to the original well. Wells were observed every few days and media was changed and wells were split as necessary. The CTMM4.1 patient-derived cell line was derived from an adherent cell population of an HLA-A*02:01+ melanoma patient. The CTMM4.1 melanoma cells and CSS-673 TIL samples were from unrelated patients.
For TCR-mediated tumor cell killing assays, human CD8+ T cells were isolated from normal buffy coats, obtained from the National Health Service Blood Transfusion (NHSBT) service. PBMCs were isolated by diluting 50:50 in PBS with 0.5% FBS and 2mM EDTA, then layering onto Ficoll-Paque (GE Healthcare, Chicago, IL, USA) before centrifuging for 20 minutes. Red blood cells were removed using 1× PharmLyse RBC lysing buffer (BD, San Jose, CA, USA) and CD8+ T cells were isolated from PBMCs using the EasySep™ Human CD8+ T Cell Isolation Kit (Stemcell Technologies, Vancouver, BC, CA).
Generating Paired TCRα-TCRβ Linkage Libraries
Library generation is divided into three steps: (i) poly(A)+ mRNA capture, (ii) multiplexed overlap extension reverse transcriptase polymerase chain reaction (OE-RT-PCR), and (iii) nested PCR to remove artifacts and add adapters for deep sequencing or expression libraries20.
For poly(A)+ mRNA capture, we used a custom designed co-flow emulsion droplet microfluidic chip fabricated from glass (Dolomite, Royston, UK). The microfluidic chip has two input channels for fluorocarbon oil (Dolomite, Royston, UK), one input channel for the cell suspension mix, and one input channel for oligo-dT beads (New England Biolabs, Ipswich, MA, USA) in 0.5M NaCl, 0.5% Tween-20, and 20mM DTT. The input channels are etched to 50μm × 150μm for most of the chip’s length, narrow to 55μm at the droplet junction, and are coated with hydrophobic Pico-Glide (Dolomite, Royston, UK). Three Mitos P-Pump pressure pumps (Dolomite, Royston, UK) are used to pump the liquids through the chip. Droplet size depends on pressure, but typically we find that droplets of ~45μm diameter are optimally stable. Emulsions were collected into 1.5mL microcentrifuge tubes and incubated at 40°C for 30 minutes to capture mRNA onto oligo-dT beads. Emulsions were then broken using Pico-Break (Dolomite, Royston, UK) and mRNA-bound beads were magnetically isolated.
For multiplex OE-RT-PCR, mRNA-bound beads were re-encasuplated into droplets with an OE-RT-PCR mix. The OE-RT-PCR mix contains 2× one step RT-PCR buffer (ThermoFisher, Waltham, MA, USA), 2.0mM MgSO4, SuperScript III reverse transcriptase (ThermoFisher, Waltham, MA, USA), and Platinum Taq (ThermoFisher, Waltham, MA, USA), plus a mixture of primers directed against the TRAC, TRBC, and all V-gene regions (Supplementary Fig. 2). TCRα and TCRβ chains are physically linked by overlapping primer sequences included on the TRAC and TRBV primers. The amplified DNA was recovered from the droplets using a proprietary droplet breaking solution (GigaMune Inc., South San Francisco, CA, USA) and purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany).
For nested PCR, the OE-RT-PCR product was first run on a 1.7% agarose gel and a band at 800–1200bp was excised and purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Bethlehem, PA, USA). Nested PCR was performed using NEBNext amplification mix (New England Biolabs, Ipswich, MA, USA) to add adapters for Illumina sequencing or cloning into a mammalian expression construct. PCR products were run on a 1.2% agarose gel, and the 800–1100bp band was excised and purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Bethlehem, PA, USA).
We ran 2.7–3.4 million live T cells from each of our six healthy PBMC donors through this workflow to generate our six viral-seropositive TCRαβ libraries (Supplementary Table 1). We ran 1.4 million live T cells from the post-REP TIL sample to generate the TIL TCRαβ library (Supplementary Table 1).
Linked TCRαβ Repertoire Sequencing
Deep TCRαβ sequencing libraries were quantified using a quantitative PCR Illumina Library Quantification Kit (Kapa Biosystems, Wilmington, MA, USA) and diluted to 8.5–10pM. Libraries were sequenced on a MiSeq (Illumina, San Diego, CA, USA) using v3 600-cycle MiSeq Reagent Kits, according to the manufacturer’s instructions. To identify the paired sequences from the TCRαβ libraries, we obtain forward reads of 357 cycles that cover the TCRα V gene and CDR3, and reverse reads of 162 cycles that cover the TCRβ CDR3 and enough of the TCRβ V gene for accurate calling. Repertoire sequencing data are available in the Short Read Archive, under BioProject ID PRJNA541985.
To exclude base call errors, we use the expected error filtering method50. The expected number of errors (E) for a read is calculated from its Phred scores. By default, reads with E>3 are discarded. Except where noted, statistics for TCR clonotype counts include singleton sequencing reads. We classify a unique clonotype by the CDR3 amino acid sequence plus V and J gene usage using International Immunogenetics (IMGT) nomenclature. Exceptions include the pairing fidelity density plots (Fig. 2b; Supplementary Figs. 6 and 32c), the cross sample comparison UpSet (Fig 2c; Supplementary Figs. 7–8) and percent sharing (Supplementary Fig 9) plots, and the library build cloning correlation and heat maps (Supplementary Figs. 14 and 16), for which we exclude singleton sequence reads from the analysis, to reduce the risk of Illumina barcode sequence miscalls.
To identify reading frame and CDR3 amino acid sequences generated by V(D)J rearrangements, we first processed a database of well-curated TCR sequences51 (IMGT, http://www.imgt.org/download/LIGM-DB/) to generate position-specific sequence matrices (PSSMs) for the 5’ and 3’ CDR3 junctions (Supplementary Fig. 40). Each nucleotide sequence from the Illumina sequencing runs was translated into all reading frames. We then used the PSSMs to identify the FR3-CDR3 (5’) and CDR3-FR4 (3’) junctions and the appropriate protein reading frame for each of the nucleotide sequences. To report a CDR3 sequence, we required 5’ and 3’ PSSM hits in the same reading frame. Additionally, sequences that had low PSSM identity scores were marked with an exclamation point. These steps allowed us to predict valid, functional, CDR3 sequences with high confidence. We queried TCRα and TCRβ nucleotide sequences against the IMGT database of reference V and J gene germline sequences using UBLAST52 (https://www.drive5.com/usearch/manual/ublast_algo.html); V and J genes were identified based on the UBLAST alignments with the best alignment (lowest E-values).
Single Chain TCRα and TCRβ Repertoire Sequencing
For TCRβ analysis of primary T cells, RNA was isolated from 3 million T cells from healthy donors CSS-930, 938, 944, 948, 962, and 963 and from 2 million T cells from the TIL infusion product, CSS-673, using the NucleoSpin RNA Plus kit (Macherey-Nagel, Bethlehem, PA, USA). TCRβ V(D)J regions were amplified using a mixture of primers directed against the V-genes and a common primer within the TRBC region (Supplementary Fig. 2); these primers contained adapters for Illumina sequencing. RT-PCR was conducted using SuperScript III reverse transcriptase (ThermoFisher, Waltham, MA, USA) and Platinum Taq (ThermoFisher, Waltham, MA, USA). These amplicons were run on 1.7% agarose gels and the about 450bp bands were excised and purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Bethlehem, PA, USA). Samples were separately quantified using an Illumina Library Quantification Kit (Kapa Biosystems, Wilmington, MA, USA). After diluting to 9pM, libraries were sequenced on a MiSeq (Illumina, San Diego, CA, USA) using v2 300-cycle and 500-cycle MiSeq Reagent Kits, according to the manufacturer’s instructions.
For single chain sequencing of the recombinant TCRα-TCRβ expression libraries, TCRα and TCRβ V(D)J regions were amplified separately using universal primers that contained adapters for Illumina sequencing, within the TRAV.Leader and TRAC regions for TCRα and within the TRBV.Leader and TRBC regions for TCRβ. We conducted RT-PCR off RNA samples using SuperScript III reverse transcriptase (ThermoFisher, Waltham, MA, USA) and Platinum Taq (ThermoFisher, Waltham, MA, USA). We used NEBNext amplification mix (New England Biolabs, Ipswich, MA) for PCR off plasmid DNA. These amplicons were run on 1.7% agarose gels and the 500–600bp band was excised and purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Bethlehem, PA, USA). Samples were separately quantified using an Illumina Library Quantification Kit (Kapa Biosystems, Wilmington, MA, USA). After diluting to 9pM, libraries were sequenced on a MiSeq (Illumina, San Diego, CA, USA) using v2 300-cycle and 500-cycle MiSeq Reagent Kits, according to the manufacturer’s instructions.
TCRα and TCRβ sequences were analyzed separately using the same methods as described above for paired sequences.
Visualization of TCRα and TCRβ Repertoires
To generate the Vα-Vβ gene usage heatmaps, we tallied the number of unique clonotypes (CDR3 amino acid sequence + V + J gene calls) for each alpha chain variable gene and beta chain variable gene pair. The heatmaps were generated in R 3.4.2 using ggplot2 3.1.0.
To evaluate TCRαβ pairing precision, we first annotated each TCRα as non-invariant or invariant, based on previously published invariant TCRα sequences53–58. For each TCRα and TCRβ clonotype, we calculated the number of unique partners they were found with and generated density plots (Fig. 2b; Supplementary Fig. 6 and 32c).
To compare TCR clonotype sharing among donors, we first curated a list of all TCRα, TCRβ, and TCRαβ clonotypes (defined as unique CDR3 amino acid sequences + V+J gene usage) across libraries from all donors. We then asked if each clonotype was observed with at least two reads in a given library. The UpSet plots were generated using in R 3.4.2 using UpSetR 1.3.3.
To illustrate clonotype enrichment through the panning rounds, we selected the five most abundant clones from the fully dextramer enriched population and display their sequencing read frequency as heatmaps for each round of dextramer panning. Similarly, we present the sequencing read frequency heatmaps for these clonotypes from the activation assays.
Generating Recombinant TCRαβ-Jurkat Expression Libraries
We developed a subcloning workflow to convert the linked TCRαβ amplicons into full-length lentiviral expression constructs (Supplementary Fig. 3). In this workflow, we first used nested PCR to add overhang adapters to the 5’ and 3’ ends of the linked TCRαβ amplicons for downstream Gibson assembly. Then, we used NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA) to insert the linked TCRαβ library into a pReceiver-based lentiviral vector (GeneCopoeia, Rockville, MD, USA) that contains the EF1α promoter, a TCRα leader sequence, the TCRβ constant region, and the Puromycin resistance gene. We transformed this intermediate library into Endura electrocompetent E. coli (Lucigen, Middleton, WI, USA), plated onto LB Lennox carbenicillin plates (Teknova, Hollister, CA, USA), and scraped and pooled >5 million colonies from each library for plasmid purification. Plasmids were purified using the endotoxin free ZymoPURE II Plasmid Maxiprep Kit (Zymo Research, Irvine, CA, USA). These intermediate libraries were linearized with a NheI-HF (New England Biolabs, Ipswich, MA, USA) restriction digest present within the linker region, run on a 0.8% agarose gel, and gel extracted using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Bethlehem, PA, USA). Note: we selected the NheI restriction site for this linearization because it is not present in the TCRα or TCRβ V genes, and, on average, it is present in less than 3% of the linked TCRαβ amplicons (Supplementary Table 1). To create the full-length TCRαβ lentiviral libraries, we performed a second Gibson assembly to insert the TCRα constant region, a ribosomal skip motif59 (P2A), and a TCRβ leader sequence. These full-length TCRαβ lentiviral libraries were transformed into Endura electrocompetent cells and purified using the endotoxin free maxiprep kit as described above.
Engineering Human CD8 Expressing Jurkat Cells
To screen our natively-paired TCRαβ libraries for TCRs reactive to MHC-class I presented peptides, we engineered the TCRβ deficient (ΔTCRβ) Jurkat cell line J.RT3-T3.5 (ATCC TIB-153) to stably express human CD860. We built a lentiviral vector with the PGK promoter driving expression of human CD8A, a P2A ribosomal skip motif, CD8B(M-1) 61, and an IRES-Blasticidin resistance gene cassette (Supplementary Fig. 11) in the pReceiver (GeneCopoeia, Rockville, MD, USA) backbone. Lentivirus was packaged and transduced into TCRβ deficient Jurkat cells as described below. Blasticidin selection was started on day two post-transduction and continued for 14 days to select for stable integration. CD8 surface expression was confirmed by flow cytometry (Supplementary Fig. 11) and this CD8+ ΔTCRβ Jurkat cell line was cryopreserved for future use.
Lentiviral Transduction of Jurkat Cells
We optimized our lentiviral transduction protocol to obtain a low transduction efficiency to ensure that we expressed only one TCRαβ pair per cell. We packaged lentivirus into VSV-G pseudotyped lentiviral particles using the 3rd generation ViraSafe Lentiviral Packaging System (Cell Biolabs, San Diego, CA, USA) and Lenti-Pac 293Ta cells (GeneCopoeia, Rockville, MD, USA) as previously described62. 21 million Lenti-Pac 293Ta cells per 10cm plate were transfected with 4.3μg pCMV-VSV-G, 4.3μg pRSV-Rev, 4.3μg pCgpV, and 4.3μg of the TCRαβ expression libraries using Lipofectamine 3000 (ThermoFisher, Waltham, MA, USA) following the manufacturer’s protocol. Lentiviral supernatant was collected at 48 hours post-transfection, spun down at 500×g for 10 minutes to eliminate cellular debris, and clarified through a 0.45μm syringe filter. RNA was isolated from fresh lentiviral supernatant using the NucleoSpin RNA Virus kit (Macherey-Nagel, Bethlehem, PA, USA) and viral titer was measured using the Lenti-X qRT-PCR Titration Kit (Takara, Mountain View, CA, USA) following the manufacturers’ protocols. Fresh lentiviral supernatant was used to transduce CD8+ ΔTCRβ Jurkat cells. Clarified lentiviral supernatant was added at a 1:10 ratio with Jurkat cells in RPMI media with 10% FBS and 8μg/ml Polybrene (EMD Millipore, Burlington, MA, USA). See Supplementary Table 2 for the number of viral particles in each transduction. For the healthy donor libraries, 40 million CD8+ ΔTCRβ Jurkat cells were incubated with 20mL of lentiviral particles in a total volume of 200mL for 6 hours and then media was exchanged. Two days after viral transduction, Jurkat cells were analyzed for cell surface CD3 and TCRαβ expression to measure viral transduction efficiency. Surface CD3 and TCRαβ expression was seen on 8–14% of transduced cells compared to 4% on the parental ΔTCRβ Jurkat cells. Cells were then cultured for 14 days with puromycin to select for stable integration and again assessed for CD3 and TCRαβ surface expression. TCRαβ surface expression increased to 42–56% following selection. For monoclonal TCRαβ cell line generation, 800,000 CD8+ ΔTCRβ Jurkat cells were transduced and selected with puromycin for 14 days. CD3 and TCRαβ surface expression was measured following selection.
The TIL TCRαβ expression library (CSS-673) was transduced into 5 million parental CD8− ΔTCRβ Jurkat cell line J.RT3-T3.5 (ATCC TIB-153) using 2.5mL of fresh lentiviral supernatant in 25ml total volume and selected for stable integration as described above.
TCRαβ-Jurkat libraries can be made available to non-profit researchers, subject to approval by ATCC.
Verifying Recombinant TCRαβ-Jurkat Libraries
To confirm that the stably selected TCRαβ-Jurkat libraries express one TCR clone per cell, we sorted single CD3+/TCRαβ+ cells into 96-well plates as described in Supplementary Fig. 13. We amplified the TCRβ chain using a modified multistep RT-PCR and nested PCR approach based on Han et al. using universal primers within the TRBV.Leader and TRBC regions63. Wells that gave >90% of sequence reads with the same TCRβ clonotype (i.e., CDR3 amino acid sequence and V+J gene usage) were classified as having one TCR clone. All other wells were manually inspected and the number of unique clonotypes were counted. Control well positions were sorted for 0, 2, 5, and 10 cells to verify proper cell sorting and TCR clonotype counts.
Screening Recombinant TCRαβ-Jurkat Libraries
We screened the virus-seropositive donor TCRαβ-Jurkat libraries with pMHC dextramers (Immudex, København, Denmark) targeting common viral antigens from CMV and EBV (Supplementary Table 3). The TIL TCRαβ-Jurkat library was stained with pMHC dextramers (Immudex, København, Denmark) targeting common melanoma antigens. We stained 2–5 million TCRαβ-Jurkat library cells with 10μl of APC- or PE-conjugated dextramer at room temperature for 10 minutes. Cells were then stained with an anti-CD3 antibody (FITC or APC conjugated; clone: UCHT1; BioLegend, San Diego, CA, USA) for 30 minutes at 4ºC and DAPI (BioLegend, San Diego, CA, USA) to assess cell viability. Cells were then sorted on a FACSMelody or BD Influx (BD Biosciences, San Jose, CA, USA) for Live CD3+/dextramer+ cells. The sorted Jurkat cells were recovered and expanded in RPMI media with 10% FBS and 100U/ml Pen/Strep (ThermoFisher, Waltham, MA, USA). Once cells reached high viability (>85%) and appropriate cell numbers, 2 million cells were lysed, and RNA was extracted using the NucleoSpin RNA Plus kit (Macherey-Nagel, Bethlehem, PA, USA) for single chain TCRα and TCRβ repertoire sequencing as described above. Multiple rounds of dextramer staining, FACS sorting, and cell expansion were conducted to enrich for populations of pMHC-binding TCRs.
Following pMHC-binding enrichment, we co-cultured TCRαβ-Jurkat cell populations with T2 peptide-pulsed aAPCs to assess cell activation. We pulsed T2 cells with 10μM peptide, mixed 200,000 peptide-pulsed T2 cells with 200,000 TCRαβ-Jurkat cells per well in 96-well round-bottom plates (Falcon; Fisher Scientific, Hampton, NH, USA), and cultured for 16–20 hours. Cells were harvested out of the round-bottom plates and stained for HLA-A2 (clone: BB7.2; BioLegend, San Diego, CA, USA), CD69 (clone: FN50; BioLegend, San Diego, CA, USA), CD62L (clone: DREG-56; Bio-Legend, San Diego, CA, USA), and cell viability with DAPI. Cells were analyzed on a FACSMelody, BD Influx, or CytoFLEX LX (Beckman Coulter, Brea, CA, USA) for activation (HLA-A2−/CD69+/CD62L−). We used 1× Cell Stimulation Cocktail (ThermoFisher, Waltham, MA, USA) as a positive control and irrelevant peptide-pulsed T2 cells as a negative control. T2 cells natively express HLA-A*02:01, and we generated a stable T2 cell line that expresses HLA-A*24:02 and GFP (data not shown) for additional peptide presentation.
To identify TCRs present in peptide-activated Jurkat cells, we co-cultured partially enriched TCRαβ-Jurkat cell populations with peptide-pulsed T2 cells, stained with the activation markers described above and sorted for activated (HLA-A2−/CD69+/CD62L−) cells. These sorted cells were lysed and RNA isolated using the NucleoSpin RNA Plus XS kit (Macherey-Nagel, Bethlehem, PA, USA) for single chain TCRα and TCRβ repertoire sequencing. For the anti-viral input TCR repertoires, we collected RNA from 2 million TCRab-Jurkat cells from the cell population used in the activation assay. For the anti-PMEL input TCR repertoire, we sorted 30,000 Jurkat cells (HLA-A2−) directly from the co-cultured cell population prior to sorting for the activated (HLA-A2−/CD69+/CD62L−) Jurkat cells.
Peptides were synthesized at >90% purity (ELIM Biopharm, Hayward, CA, USA), resuspended in DMSO to 4mg/ml, aliquoted for single use, and stored at −20˚C.
Peptide-MHC Dextramer Staining of PBMCs and TCRβ Repertoire Analysis
To detect primary T cells that bind viral-pMHC dextramers, we stained about 4 million PBMCs. First, we incubated PBMCs with human FcR Blocking Reagent (Miltenyi Biotec, Auburn, CA, USA) at room temperature for 10 minutes and then stained with 10μl of APC-conjugated pMHC dextramer at room temperature for 10 minutes. Following this, cells were stained with anti-CD3-FITC (clone: UCHT1; BioLegend, San Diego, CA, USA), anti-CD4-PE/Cy7 (clone: OKT4; BioLegend, San Diego, CA, USA), and anti-CD8-APC/H7 (clone: SK1; BD Pharmingen, San Diego, CA, USA) for 30 minutes at 4ºC and DAPI (BioLegend, San Diego, CA, USA) to assess cell viability. CD8+/Dextramer+ cells were sorted on a FACSMelody (BD Biosciences, San Jose, CA, USA) in purity mode. Cells were lysed and RNA was isolated using the NucleoSpin RNA Plus XS kit (Macherey-Nagel, Bethlehem, PA, USA). Single chain TCRβ repertoire analysis was conducted using a previously described multistep RT-PCR plus two round nested PCR approach to build Illumina sequencing libraries63. We then compared the TCRβ sequences identified in the CD8+/Dextramer+ cell populations with those present in (i) a separate aliquot of primary T cells and (ii) enriched by dextramer panning and cellular activation in the TCRαβ-Jurkat library (Supplementary Table 4).
Monoclonal TCRαβ Characterization in Jurkat Cells
Enriched TCRα and TCRβ single chain sequences were identified from the pMHC-binding and cell activation screens. We used this enrichment data and the natively paired TCRα-TCRβ sequencing data to identify candidate antigen-reactive TCR clones. We designed full-length TCRα-TCRβ lentiviral expression constructs using the Illumina sequencing data, specifically the CDR3 nucleotide sequences and V-gene calls. We synthesized full-length anti-viral TCRα-TCRβ lentiviral expression constructs using the BioXp 3200 system (SGI-DNA, La Jolla, CA, USA) and full-length anti-PMEL TCRα-TCRβ lentiviral constructs using gBlocks (IDT, Coralville, IA, USA). These monoclonal TCRαβ expression constructs follow the same layout as the TCRαβ libraries. Lentiviral plasmids were sequence verified by Sanger sequencing, packaged into VSV-G pseudotyped lentiviral particles, transduced into ΔTCRβ Jurkat cells, and stable cell lines were selected. Anti-viral TCRαβ clones were introduced into CD8+ ΔTCRβ Jurkat cells, and anti-PMEL TCRαβ clones were first introduced into CD8− ΔTCRβ Jurkat cells and then into CD8+ ΔTCRβ Jurkat cells. TCRαβ-Jurkat clones can be made available to non-profit researchers, subject to approval by ATCC.
Monoclonal TCRαβ-Jurkat cell lines were assessed for pMHC binding and cellular activation. We stained 0.5–1 million cells with 5μl of pMHC dextramer and anti-CD3 antibodies and analyzed for binding as described above. We then ran co-culture assays with the monoclonal TCRαβ-Jurkat cell lines that showed pMHC binding. As described above, we pulsed T2 cells with 10μM peptide, mixed 200,000 peptide-pulsed T2 cells with 200,000 TCRαβ-Jurkat cells per well, and measured cell activation by staining for CD69 and CD62L. We then assessed TCRαβ-mediated cell activation in response to tumor cells by co-culturing 500,000 anti-PMEL TCRαβ-Jurkat cells with a confluent monolayer of melanoma cells for SK-MEL-5 (HLA-A*02:01+, PMEL+; ATCC: HTB-70) and SK-MEL-28 (HLA-A*02:01−, PMEL+; ATCC: HTB-72) in a 24-well plate.
We measured functional avidity of the monoclonal TCRs that showed cellular activation using T2 cells pulsed with a dilution series of peptide (0.1pM–10μM). Cells were co-cultured in duplicate wells for 20 hours, harvested, pooled, and stained for CD69 and CD62L activation markers as described above. Cells were run on the CytoFLEX LX and Jurkat cell CD69 Median Fluorescence Intensity (MFI) was calculated using FlowJo (Treestar/BD Biosciences, San Jose, CA, USA) and analyzed in Prism (GraphPad, San Diego, CA, USA). A nonlinear three parameter curve was fit to the dose response data. Two independent experiments were conducted, and the mean and standard deviation values were plotted for the dose response curves. EC50 values were calculated for each independent experiment and plotted.
We analyzed a set of viral antigen-reactive TCRs for peptide specificity using alanine scanning mutagenesis as previously described64. Synthetic peptides with alanine substitutions were obtained from Pepscan (purity = crude), resuspended in DMSO to 5mg/ml, aliquoted, and stored at −20˚C. T2 cells were pulsed with 10μg/ml of peptide and co-cultured with monoclonal CD8+ TCRαβ-Jurkat cell lines for 24 hours as described above. Following co-culture, cell supernatant was removed and stored at −80˚C. IL-2 levels were measured in cell supernatant by sandwich ELISA (BioLegend, San Diego, CA, USA) following the manufacturer’s protocol. MaxiSorp ELISA plates were coated with antibody at 4˚C overnight. ELISA wells were blocked with PBS + 1% BSA, 50μl of cell supernatant was added to duplicate wells and incubated at room temperature for 2 hours, and IL-2 levels were measured using an anti-IL-2 detection antibody followed by Avidin-HRP incubation and signal development with aTMB substrate solution. Signal absorbance was read at 450nm on a SpectraMax i3x (Molecular Devices, Sunnyvale, CA, USA). IL-2 levels were calculated using a standard curve run on the same plate. Two independent experiments were conducted and data points were plotted with the mean in Prism (GraphPad).
Monoclonal TCRαβ Characterization in Human T Cells
For enhanced expression, detection and proper pairing of the recombinant TCRαβ clones in human T cells, we generated human-mouse fusion TCRs65. The variable regions of the α- and β-chains of the PMEL specific TCRs were gene synthesized (Genewiz, South Plainfield, NJ, USA) and cloned in frame with the murine TCRα and TCRβ constant domains. A P2A cleavage sequence was added between the TCR chains and this expression cassette was cloned into the pSF.Lenti plasmid (Oxford Genetics, Oxford, England, UK) under the control of an EF1α promoter.
Lentiviral production was performed using a three-plasmid packaging system (Cell Biolabs, San Diego, CA, USA) by mixing 10μg of each plasmid, plus 10μg of the pSF.Lenti lentiviral plasmid containing the recombinant TCRs, together in serum free RPMI containing 50 mM CaCl2. The mixture was added dropwise to a 50% confluent monolayer of 293T cells in 75 cm2 flasks. The viral supernatants were collected at 48 and 72 hours post transfection, pooled and concentrated using LentiPac lentiviral supernatant concentration solution (GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s instructions. Lentiviral supernatants were concentrated 10-fold and used to directly infect primary human T cells in the presence of 4μg/ml polybrene (Sigma-Aldrich, Dorset, England, UK). Human T cells were activated for 24 hours with Human T-Activator CD3/CD28 Dynabeads (Invitrogen, ThermoFisher, Waltham, MA, USA) at a 3:1 bead to cell ratio according to the manufacturer’s instructions before addition of lentiviral supernatant.
After transduction of the T cells, flow sorting was conducted to isolate the TCR-positive populations by staining with anti-mouse TCRβ (H57–597; BioLegend, San Diego, CA, USA) antibody, on a BD Influx flow sorter. The sorted cells were then expanded by REP (described above) and the resulting post-expanded cells were used fresh or cryopreserved for later use in freezing media (FBS + 10% DMSO) and frozen by controlled rate freezing in a Mr Frosty™ (ThermoFisher, Waltham, MA, USA). When cryopreserved cells were used, the transduced T cells were thawed and recovered overnight at 5% CO2 and 37˚C in fresh RPMI-1640 media with 10% FBS, 0.01 M HEPES, and 200 IU/mL IL-2. Recombinant TCR surface expression was confirmed i) after the REP expansion of mouse TCRβ+ sorted cells and ii) upon thawing of cryopreserved cells for use in the WST-1 assay using the anti-mouse TCRβ [clone: H57–597] antibody and ran on a MACSquantVYB (Miltenyi Biotec, Bisley, UK) (Supplementary Fig. 39). Data was analyzed using FlowJo.
Recombinant TCR-mediated T cell degranulation was measured by culturing TCR-transduced human T cells with melanoma cell lines (SK-Mel-5, SK-Mel28, and CTMM4.1) and staining for CD107a. Transduced T-cells were co-cultured with tumor cells at a 1:2 effector to target ratio with 1:1000 monensin and brefeldin, and 1:50 anti-human anti-CD107a-PE (clone: REA792, Miltenyi Biotec, Bisley, UK) antibody for 4 hours. The cells were then stained with eFluor®780 Fixable Viability Dye (eBioscience, Waltham, MA, USA), along with extracellular staining with CD8-APC (clone: SK1, BioLegend, London, UK) and CD4-FITC (Clone: SK3, BD Biosciences, San Jose, CA, USA), before fixing in 4% PFA. Cells were resuspended in 200μL DPBS + 2mM EDTA + 0.5% FBS before the samples were run on the MACSQuant Flow Cytometer. Triplicate wells were run for each condition per human T cell donor. Three independent experiments were conducted using three different human T cell donors. Data were analyzed using FlowJo and Prism (GraphPad) software, and statistical analysis was conducted using a two-way ANOVA with Dunnett’s multiple comparisons test. Data from the three independent experiments are plotted with mean and standard deviation values in Figure 5f. The three technical replicates (dots) for each of the three human donors are plotted with mean (bars) values in Supplementary Figure 37.
TCR-directed tumor cell killing was measured using water-soluble tetrazolium salt (WST-1) assays on co-cultured tumor and T cell samples. 20,000 tumor cells were co-cultured with varying amounts of T cells at Effector to Target (E:T) ratios of 1:10, 1:5, 1:2, 1:1, and 2:1 in a total volume of 200μl per well in 96-well round-bottom plates and incubated overnight. The following day, cells were spun down, 100μL of supernatant was removed and 10μL Cell Proliferation Reagent WST-1 (Roche, Basel, Switzerland) was added to the cells. After 30 minutes incubation, 80μL of supernatant was transferred to a flat-bottom plate for analysis on a FLUOStar Omega plate reader (BMG Labtech, Ortenberg, Germany). For the TCR-specific killing signal, we performed the following transformation:
Triplicate wells were run for each co-culture condition and mean and standard deviation values from the resulting WST-1 measurements were plotted for one experiment using Prism (GraphPad).
Supplementary Material
Acknowledgements
This work was partially funded by NIAID grant R43AI120313-01, to D.S.J., and NCI grant R43CA232942, to M.J.S. N.O. is supported with funding from the MRC, Engineering and Physical Sciences Research Council (EPSRC) Centre for Doctoral Training (EP/L014904/1). Dr. Rena Mizrahi, Mr. Matthew Adams, Ms. Renee Leong, and Mr. Jackson Leong (GigaGen, Inc., South San Francisco, CA, USA) assisted with development of Gibson Assembly protocols. Drs. Erica Stone and Sheila Keating (GigaGen Inc., South San Francisco, CA, USA) provided useful discussions around general T cell immunology. Dr. Ryan Guest (Immetacyte Ltd., Manchester, England, UK) provided assistance with isolation and expansion of TILs.
Footnotes
Competing interests
M.J.S., A.L.N., E.K.W., A.S.A., M.A.A., Y.W.L., R.C.E., and D.S.J. are salaried employees of GigaGen Inc., which is an affiliate of GigaMune Inc. GigaMune Inc. pays cash to GigaGen Inc. for research services. M.J.S., A.L.N., E.K.W., J.M.H., A.S.A., M.A.A., R.C.E., Y.W.L., E.H.M., M.C., and D.S.J. are holders of equity shares of GigaMune Inc. J.M.H. and M.C. hold research positions at Massachusetts General Hospital. Massachusetts General Hospital has entered into a research collaboration with GigaMune Inc. M.C. is currently an employee of AstraZeneca. M.C. owns equity in Revitope Oncology and Gritstone Oncology. M.C. received consultant fees from Merck Laboratories. J.S.B. and R.E.H. are salaried employees of Immetacyte Ltd. R.E.H. is a holder of equity shares of Immetacyte Ltd. The viral TCRs and TCR repertoire mining methods are described in USPTO provisional patent application 62/821808, assigned to GigaMune (M.J.S., A.L.N., E.K.W., Y.W.L., A.S.A., M.A.A., and D.S.J). The PMEL TCRs are described in USPTO provisional patent application 62/842691, assigned to GigaMune (M.J.S., A.S.A., M.A.A., and D.S.J). Methods for generating TCR libraries are described in patents WO2012083225A2, US20160362470A1, US20170247684A1, and US20170247683A1, assigned to GigaGen or GigaMune (M.J.S., A.S.A., E.H.M., and D.S.J.).
Data availability
Pre-sort TCRα-TCRβ repertoire fastq sequence files are deposited at the Short Read Archive (http:www.ncbi.nlm.nih.gov/sra/), BioProject ID PRJNA541985. All other data are available from the corresponding author upon reasonable request.
References
- 1.Yee C, Adoptive T cell therapy: points to consider. Curr. Opin. Immunol 51, 197–203 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ, Prockop S, & Bollard CM Virus-Specific T Cells: Broadening Applicability. Biol. Blood Marrow Transplant 24, 13–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romano M, Fanelli G, Albany CJ, Giganti G, & Lombardi G, Past, Present, and Future of Regulatory T Cell Therapy in Transplantation and Autoimmunity. Front Immunol 10, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chodon T, et al. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin Cancer Res 20, 2457–2465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapoport AP et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med 21, 914–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tendeiro Rego R, Morris EC & Lowdell MW T-cell receptor gene-modified cells: past promises, present methodologies and future challenges. Cytotherapy 21, 341–357 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, & Wang L, The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review. Technol. Cancer Res. Treat 18, 1533033819831068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadelain M, Rivière I, & Riddell S, Therapeutic T cell engineering. Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson NAJ & Levings MK Antigen-specific regulatory T cells: are police CARs the answer? Transl Res 187, 53–58 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Linnemann C, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med 19, 1534–1541 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Scheper W, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 25, 89–94 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Yossef R, et al. Enhanced detection of neoantigen-reactive T cells targeting unique and shared oncogenes for personalized cancer immunotherapy. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, et al. A cloning and expression system to probe T-cell receptor specificity and assess functional avidity to neoantigens. Blood 132, 1911–1921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol 23, 349–354 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Wagner EK et al. Human cytomegalovirus-specific T cell receptor engineered for high affinity and soluble expression using mammalian cell display. Journal of Biological Chemistry (2019). doi: 10.1074/jbc.RA118.007187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Border EC, Sanderson JP, Weissensteiner T, Gerry AB & Pumphrey NJ Affinity-enhanced T-cell receptors for adoptive T-cell therapy targeting MAGE-A10: strategy for selection of an optimal candidate. Oncoimmunology 8, e1532759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linette GP et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122, 863–871 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X-ZJ et al. Rapid cloning, expression, and functional characterization of paired αβ and γδ T-cell receptor chains from single-cell analysis. Mol Ther Methods Clin Dev 3, 15054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azizi E, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler AS et al. Rare, high-affinity anti-pathogen antibodies from human repertoires, discovered using microfluidics and molecular genomics. MAbs 9, 1282–1296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, et al. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat Biotechnol 36, 152–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieke MC et al. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc Natl Acad Sci USA 96, 5651–5656 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith SN, Harris DT & Kranz DM in Yeast Surface Display 1319, 95–141 (Springer; New York, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhns MS, Davis MM & Garcia KC Deconstructing the form and function of the TCR/CD3 complex. Immunity 24, 133–139 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Tsuji T, et al. Rapid Construction of Antitumor T-cell Receptor Vectors from Frozen Tumors for Engineered T-cell Therapy. Cancer Immunology Research 6, 594–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat Methods 11, 653–655 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Heather JM, Ismail M, Oakes T, & Chain B, High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Brief. Bioinformatics 19, 554–565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Exley M, Porcelli S, Furman M, Garcia J, & Balk S, CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant V alpha 24 J alpha Q T cell receptor alpha chains. J Exp Med 188, 867–876 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Gold MC et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med 211, 1601–1610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellabona P, Padovan E, Casorati G, Brockhaus M, & Lanzavecchia A, An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4–8- T cells. J Exp Med 180, 1171–1176 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porcelli S, Yockey CE, Brenner MB & Balk SP Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med 178, 1–16 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Held K, Beltrán E, Moser M, Hohlfeld R, & Dornmair K, T-cell receptor repertoire of human peripheral CD161hiTRAV1–2+ MAIT cells revealed by next generation sequencing and single cell analysis. Human Immunology 76, 607–614 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Dash P, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature Publishing Group 547, 89–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature Publishing Group 547, 94–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, & Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole DK et al. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. Journal of Biological Chemistry 284, 27281–27289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trautmann L, et al. Selection of T Cell Clones Expressing High-Affinity Public TCRs within Human Cytomegalovirus-Specific CD8 T Cell Responses. J.I 175, 6123–6132 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Day EK et al. Rapid CD8+ T Cell Repertoire Focusing and Selection of High-Affinity Clones into Memory Following Primary Infection with a Persistent Human Virus: Human Cytomegalovirus. J Immunol 179, 3203–3213 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Yang X, et al. Structural Basis for Clonal Diversity of the Public T Cell Response to a Dominant Human Cytomegalovirus Epitope. Journal of Biological Chemistry 290, 29106–29119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sibener LV et al. Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding. Cell 174, 672–687.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birnbaum ME et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shugay M, et al. VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res 46, D419–D427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yost KE et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerdemann U, et al. Rapidly generated multivirus-specific cytotoxic T lymphocytes for the prophylaxis and treatment of viral infections. Mol Ther 20, 1622–1632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett AJ & Bollard CM The coming of age of adoptive T-cell therapy for viral infection after stem cell transplantation. Ann Transl Med 3, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Leary MC et al. FDA Approval Summary: Tisagenlecleucel for Treatment of Patients with Relapsed or Refractory B-cell Precursor Acute Lymphoblastic Leukemia. Clin Cancer Res 25, 1142–1146 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Bouchkouj N, et al. FDA Approval Summary: Axicabtagene Ciloleucel for Relapsed or Refractory Large B-cell Lymphoma. Clin Cancer Res 25, 1702–1708 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Edgar RC & Flyvbjerg H, Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Lefranc M-P et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 37, D1006–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Exley M, Garcia J, Balk SP & Porcelli S, Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J Exp Med 186, 109–120 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han M, et al. Invariant or highly conserved TCR alpha are expressed on double-negative (CD3+CD4-CD8-) and CD8+ T cells. J Immunol 163, 301–311 (1999). [PubMed] [Google Scholar]
- 55.Démoulins T, Gachelin G, Bequet D, & Dormont D, A biased Valpha24+ T-cell repertoire leads to circulating NKT-cell defects in a multiple sclerosis patient at the onset of his disease. Immunol. Lett 90, 223–228 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Greenaway HY et al. NKT and MAIT invariant TCRα sequences can be produced efficiently by VJ gene recombination. Immunobiology 218, 213–224 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol 14, 706–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepore M, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun 5, 3866 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Funston GM, Kallioinen SE, de Felipe P, Ryan MD & Iggo RD Expression of heterologous genes in oncolytic adenoviruses using picornaviral 2A sequences that trigger ribosome skipping. J. Gen. Virol 89, 389–396 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Lyons GE et al. Influence of human CD8 on antigen recognition by T-cell receptor-transduced cells. Cancer Res. 66, 11455–11461 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Thakral D, Dobbins J, Devine L, & Kavathas PB Differential expression of the human CD8beta splice variants and regulation of the M-2 isoform by ubiquitination. J Immunol 180, 7431–7442 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Zufferey R, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72, 9873–9880 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han A, Glanville J, Hansmann L, & Davis MM Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol (2014). doi: 10.1038/nbt.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chheda ZS et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med 215, 141–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goff SL et al. Enhanced receptor expression and in vitro effector function of a murine-human hybrid MART-1-reactive T cell receptor following a rapid expansion. Cancer Immunology, Immunotherapy 59, 1551–1560 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.