ABSTRACT
During Drosophila and vertebrate brain development, the conserved transcription factor Prospero/Prox1 is an important regulator of the transition between proliferation and differentiation. Prospero level is low in neural stem cells and their immediate progeny, but is upregulated in larval neurons and it is unknown how this process is controlled. Here, we use single molecule fluorescent in situ hybridisation to show that larval neurons selectively transcribe a long prospero mRNA isoform containing a 15 kb 3′ untranslated region, which is bound in the brain by the conserved RNA-binding protein Syncrip/hnRNPQ. Syncrip binding increases the stability of the long prospero mRNA isoform, which allows an upregulation of Prospero protein production. Adult flies selectively lacking the long prospero isoform show abnormal behaviour that could result from impaired locomotor or neurological activity. Our findings highlight a regulatory strategy involving alternative polyadenylation followed by differential post-transcriptional regulation.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Prospero, Syncrip, mRNA stability, Neuroblast, Post-transcriptional regulation
Summary: Different prospero mRNA isoforms have different stabilities, depending on an interaction with the RNA binding protein Syncrip. Isoform choice in different cell types controls the expression level of Prospero protein.
INTRODUCTION
The central nervous system (CNS) consists of a huge diversity and number of neurons that originate from a limited population of neural stem cells (NSCs) (Kelava and Lancaster, 2016), also known as neuroblasts (NBs) in Drosophila. To produce a normal CNS, NBs must divide the correct number of times and their progeny must undergo a precisely regulated programme of differentiation. Many factors and mechanisms regulating these processes have been studied in detail, and are extensively conserved between vertebrates and Drosophila (Homem and Knoblich, 2012). The emphasis in the field has been on identifying key transcription factors and their downstream transcriptional effects. However, post-transcriptional regulation, which can modulate protein expression with enhanced spatial and temporal precision, has been less well characterised.
In Drosophila embryos and larvae, NBs undergo repeated asymmetric divisions, maintaining a single large cell that retains its stem cell properties. Each division of a type I NB also produces a ganglion mother cell (GMCs), which divides only once to generate a pair of neurons (Homem and Knoblich, 2012; Knoblich, 2008). Type I NB lineage differentiation is regulated by the conserved homeodomain-containing transcription factor, Prospero (Pros)/Prox1 (Bayraktar et al., 2010). Pros activates the expression of genes required for the differentiation of type I NB progeny and suppresses the expression of genes that promote stem cell-like properties (Bello et al., 2006; Betschinger et al., 2006; Choksi et al., 2006; Doe et al., 1991; Lai and Doe, 2014; Lee et al., 2006; Matsuzaki et al., 1992; Vaessin et al., 1991). pros mRNA and Pros protein are expressed in the NBs, but are excluded from the nucleus and segregated asymmetrically into the GMC during NB division (Hirata et al., 1995; Kitajima et al., 2010; Knoblich et al., 1995; Spana and Doe, 1995). In this way, the sub-cellular localisation of Pros allows the stem cell-like properties of NBs to be maintained, while ensuring that their GMC progeny differentiate correctly. In embryonic type I NBs, Pros is expressed in GMCs and new-born neurons, but is quickly switched off as neurons mature (Srinivasan et al., 1992). In contrast, Pros expression is upregulated in larval neurons and is required to maintain neuronal identity (Carney et al., 2013), but the mechanism controlling this upregulation is not known.
Here, we examine the mechanism of upregulation of Pros expression in larval neurons using single molecule fluorescent in situ hybridisation (smFISH) and immunofluorescence (IF) in whole-mount brains (Yang et al., 2017b). We observe hugely increased pros mRNA expression in neurons, correlated with upregulated Pros protein. This highly expressed neuronal pros mRNA includes an unusually long 15 kb 3′ untranslated region (UTR) (proslong). proslong is not produced in embryos or in larval NBs, but is switched on in larval neurons. We show that proslong is stabilised by binding to the conserved RNA-Binding Protein (RBP), Syncrip (Syp)/hnRNPQ (Kuchler et al., 2014; Liu et al., 2015; McDermott et al., 2014, McDermott et al., 2012). We find that the pros 3′ UTR extension is required for the neuronal upregulation of Pros protein. Our observations highlight a novel example of alternative polyadenylation followed by differential post-transcriptional regulation through mRNA stability, which controls the level of protein expression in distinct cell types.
RESULTS
Upregulation of Pros protein in neurons is achieved through cell type-specific stabilisation of pros mRNA
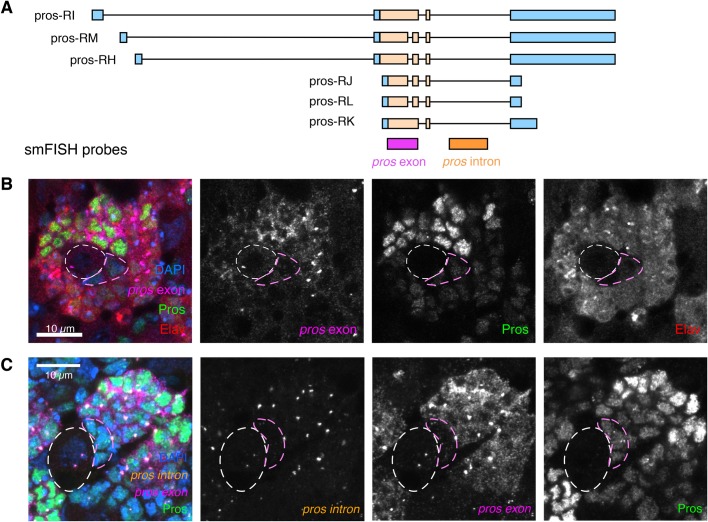
Pros protein is expressed at low levels in larval type I NBs and GMCs, where it promotes GMC differentiation, and is then upregulated in larval neurons (Carney et al., 2013; Choksi et al., 2006). To determine whether the upregulated Pros expression in neurons is driven by an increase in pros mRNA levels or by increased Pros translation, we used smFISH in whole-mount Drosophila larval brains at the wandering third instar stage (Yang et al., 2017b). We carried out four-colour imaging with smFISH against pros exon (Fig. 1A), anti-Pros antibody, anti-Elav antibody (marking differentiated neurons) and DAPI (marking DNA in all cells) (Fig. 1B). NBs were easily identified by their large cell size. As previously shown (Carney et al., 2013; Choksi et al., 2006), we found that Pros expression is low in NBs and GMCs (outlined in white and pink, respectively) and is upregulated in Elav+, post-mitotic neurons (Fig. 1B). pros exon smFISH signal is also upregulated in Elav+ cells, and correlates with the increased Pros protein expression (Fig. 1B). We conclude that increased pros transcript number is responsible for the increased Pros protein expression in the neurons compared to NBs.
Fig. 1.
Pros cell type-specific expression is regulated post-transcriptionally. (A) Illustration of smFISH probe positions on the pros mRNA transcript isoforms. (B) Third instar larval brains stained for smFISH against pros exon, with IF against Elav and Pros. pros mRNA is upregulated in post-mitotic neurons (marked with Elav), and this correlates with upregulated Pros protein. (C) smFISH against the pros intron and pros exon (probe positions shown in A) together with IF against Pros protein, shows that pros is transcribed in all cells of the Type I NB lineage, even in NBs where there are low levels of Pros protein and pros mRNA. Scale bars: 10 µm. NB, white dashed outline; GMC, pink outline.
We then asked whether the increase in pros mRNA level in neurons is regulated at the level of transcription, or post-transcriptionally through mRNA stability. We used smFISH with pros intron-specific probes to visualise the pros transcription foci (Fig. 1A,C). There are two transcription foci in each cell (these are paired in the progeny cells, thus visible as a single combined focus of higher intensity). Unexpectedly, we observed bright transcription foci in the type I NBs, but very few cytoplasmic transcripts in these cells (Fig. 1C, white outline). This result suggests that pros mRNA is more unstable in NBs than in neurons, where much higher levels of pros mRNA accumulate.
pros mRNA in larval brains contains an exceptionally long 3′ UTR
Post-transcriptional regulation is often linked to expression of distinct alternative mRNA isoforms, and neurons have been shown to selectively express transcript isoforms with unusually long 3′ UTRs (Hilgers et al., 2012, 2011; Oktaba et al., 2015). Therefore, we examined the pros isoform usage in the type I NB lineage.
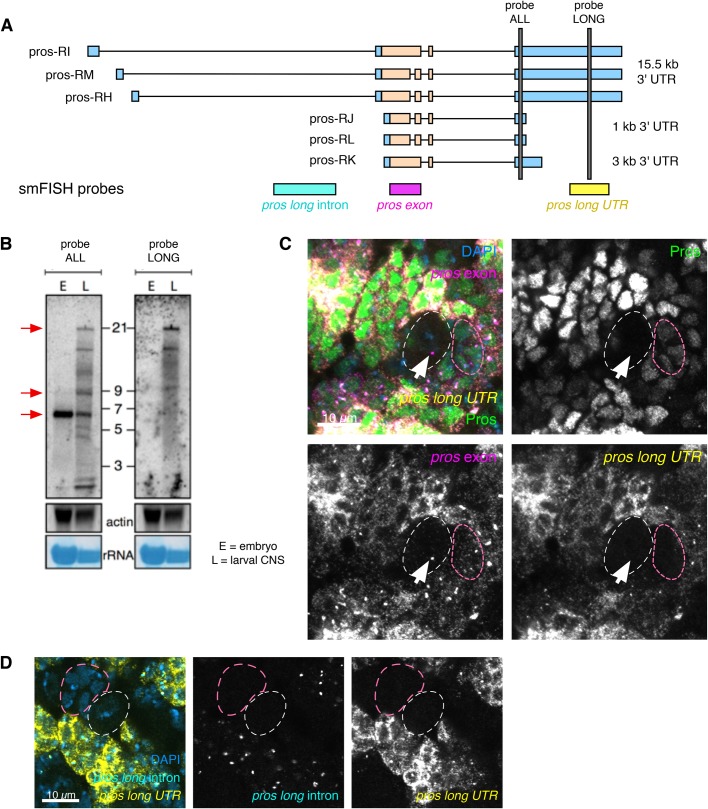
pros is annotated in FlyBase as having six isoforms with three different 3′ UTR lengths: ∼1 kb, ∼3 kb and ∼15 kb (Fig. 2A, FB2019_05, Thurmond et al., 2019). To test which isoforms are expressed in larval brains, we carried out northern blots. Our northern blot analysis used a probe that is common to all the annotated isoforms of pros transcripts (probe ALL) and detected the known ∼6 kb isoform in embryos (Fig. 2A,B) (Doe et al., 1991; Matsuzaki et al., 1992; Vaessin et al., 1991). In larval brain extracts, the northern blots show multiple-sized pros transcripts including isoforms approximately 6 kb, 9 kb and 21 kb in length (Fig. 2B), corresponding to 3′ UTR lengths of 1 kb, 3 kb and 15 kb, respectively. We also detected the 21 kb pros band in larval brains using a probe targeting the extended region of the 15 kb 3′ UTR (Fig. 2A,B). We conclude that embryos express only the shortest isoform of pros, whereas larval brains express two additional isoforms, with 3 kb and 15 kb long 3′ UTRs, respectively.
Fig. 2.
proslong isoform is expressed in larval brains and is specifically transcribed in larval neurons. (A) Diagram showing annotated pros transcript isoforms and the position of smFISH (pros long intron, pros and pros long UTR) and northern blot probes (grey bars). (B) Northern blot confirms the existence of multiple pros mRNA isoforms at 6 kb, 9 kb and 21 kb (red arrows) in the larval CNS. The 21 kb isoform corresponds to pros transcript with 15 kb 3′ UTR. E, embryo; L, larval CNS. (C) Costaining with pros exon and pros long UTR smFISH and Pros IF shows that proslong RNA is detected only in neurons. In the NB (white dotted outline), or the GMCs (pink dotted outline), pros exon is detected without pros long UTR. Pros protein is upregulated in cells expressing proslong. (D) pros long intron probe only recognises the nascent transcripts of the proslong isoform. pros long intron is detected exclusively in neurons, and not in the NB (white dotted outline) or GMCs (pink dotted outline). Scale bars: 10 µm.
To investigate the cell type-specific expression of the pros isoforms in larval brains, we performed smFISH experiments using a probe specific to the shared coding exon (pros) and a probe that is specific to the extended region of the 15 kb 3′ UTR region (pros long UTR) of the proslong transcripts (defined as the isoforms containing the 15 kb 3′ UTR), co-stained for Pros protein (Fig. 2A,C). proslong is highly expressed in the cells with highest pros exon signal, and correlates well with upregulated Pros protein. We conclude that the upregulated pros mRNA in neurons consists primarily of proslong. Co-staining with Elav to label neurons shows that proslong is expressed specifically in neurons and is absent from NBs and GMCs (Fig. S1A). However, GMCs do show low levels of pros exon smFISH signal and Pros protein (Fig. 2C, outlined in dashed pink). These observations suggest that the short isoform of pros produces the low levels of Pros seen in the GMCs and NBs.
To determine precisely where proslong is transcribed in the brain, we visualised proslong transcription using smFISH with intron probes specific for the long isoforms (Fig. 2A,D). We detected proslong-specific transcription foci only in neurons, where we also observed pros long UTR smFISH signal in the cytoplasm. In contrast, no proslong intron signal was detected in the NBs or GMCs (Fig. 2D outlined in dashed white and pink, respectively). We conclude that the proslong isoform is specifically transcribed in larval neurons. Our results support a model in which a low level of Pros protein is produced by the short pros isoform in NBs and GMCs. Transcription of proslong is switched on in the larval neurons and this isoform is stabilised, resulting in higher pros mRNA levels and upregulated Pros protein.
Syp is expressed in the type I NB lineage and stabilises pros mRNA
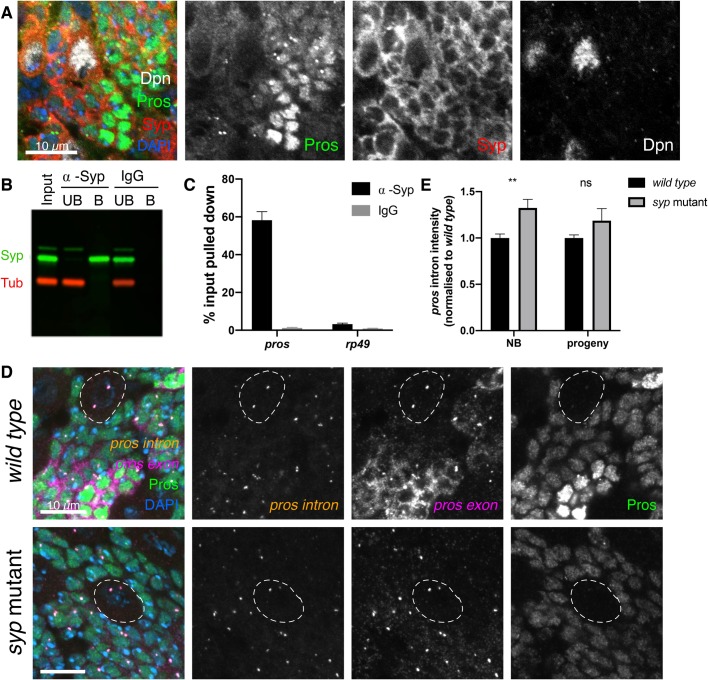
Post-transcriptional regulation, including mRNA stability, depends on the binding and action of specific RBPs. We have previously shown in larval extracts that pros mRNA associates with Syp, a highly conserved RBP (McDermott et al., 2014). Syp is a key factor in the temporal regulation of larval NBs and is known to regulate the transcription factor Chinmo to determine neuron fate (Liu et al., 2015; Ren et al., 2017; Syed et al., 2017). Syp is expressed in the larval type I NBs and their progeny (Fig. 3A) (Liu et al., 2015), and so we tested whether Syp binds pros mRNA in the larval brain. 58.3% of pros mRNA was co-immunoprecipitated with Syp in larval brain lysates, while only 3.2% of rp49, a control RNA, was pulled down (Fig. 3B,C). These results demonstrate that Syp binds specifically to pros RNA, rendering Syp a good candidate for an upstream regulator of pros mRNA stability.
Fig. 3.
Syncrip selectively stabilises the proslong isoforms in neurons. (A) Syncrip (Syp) protein is expressed in the type I NB (marked with Dpn) and progeny lineage in third instar larvae. Syp is also expressed in the neurons, where Pros is upregulated. (B) Western blot confirms Syp is immunoprecipitated (IP) selectively and efficiently. α-Tubulin (Tub) was used as negative control. (C) pros mRNA is enriched by Syp IP (58.3% of input pulled down) but not by IgG control IP (1.3%). Negative control, ribosomal rp49 (α-Syp: 3.2%, IgG: 0.9%) (n=7). Error bars represent s.e.m. (D) Loss of Syp has a minimal effect on transcription levels indicated by pros intron, but pros exon signal and Pros protein are greatly reduced. NBs outlined in white. (E) Intensity of pros intron signal in wild type and syp mutant. Normalised to wild type for each experiment. Significance was calculated using t-tests. **P<0.01, ns=not significant P>0.05. NB, sum of intensity of two transcription foci; progeny, intensity of single spot including both transcription foci. Scale bars: 10 µm.
To determine whether Syp regulates the levels of pros mRNA, we compared levels of pros transcription, cytoplasmic pros transcripts and Pros protein in brains mutant for a syp allele that lacks Syp protein (see Materials and Methods). The results show that Pros protein is reduced in the neurons of syp mutant brains (Fig. 3D). Pros protein levels are maintained at low levels in the syp mutant neurons but the upregulation of Pros is lost. pros mRNA is also very substantially reduced in the syp mutants, most obviously in the neurons (Fig. 3D). We measured the intensity of pros intron signal at the transcription foci in wild type and syp mutant brains (Fig. 3E) and observed a small but significant increase in pros transcription in the syp mutant type I NBs, compared to wild type (1.3-fold increase). In progeny cells (pooled neurons and GMCs) there is no significant change in pros transcription in the syp mutant, compared to the wild type (Fig. 3E). Therefore, changes in pros transcription are not responsible for the reduction of pros transcripts in the syp mutant. We suggest that Syp mediates the upregulation of Pros protein in neurons primarily by stabilising cytoplasmic pros mRNA.
Syp negatively regulates the RBP Imp/IGF2BP in type I NBs (Liu et al., 2015) and therefore could regulate pros mRNA stability indirectly via Imp. To distinguish whether Syp regulates pros directly or indirectly, we compared the levels of pros mRNA and Pros protein in syp knockdown and imp syp double-knockdown brains (Yang et al., 2017a) (Fig. S1B). imp syp double-knockdown phenocopies the single syp knockdown (Fig. S1B). Combined with the pros pulldown in Syp immunoprecipitations, these results indicate that Syp's effects on pros are independent of its effects on Imp.
proslong is stabilised by Syp
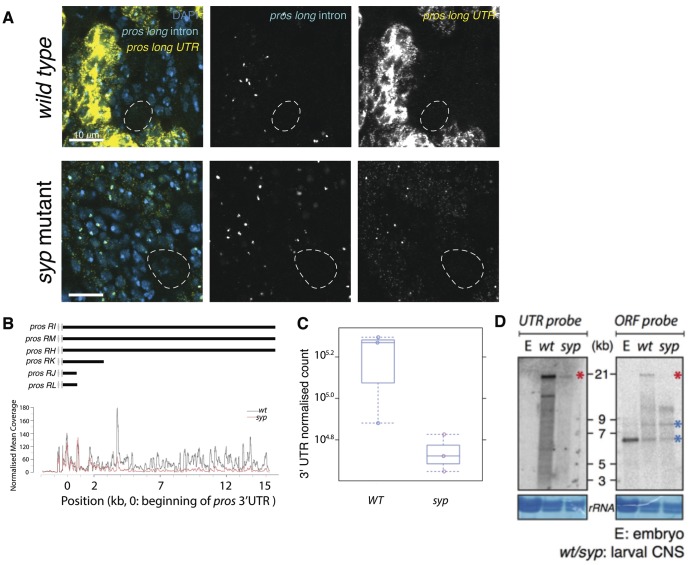
Loss of Syp has the greatest effect on pros in the neurons. In syp mutant neurons pros mRNA is substantially reduced and Pros protein upregulation is lost. We have previously shown that proslong is responsible for the neuronal pros mRNA and protein upregulation, so we asked whether Syp acts specifically on proslong. We used smFISH against the proslong 3′ UTR in syp mutant larval brains and found that it is substantially reduced in neurons, compared to wild type (Fig. 4A). We co-stained with the proslong-specific intron probe and found that proslong is transcribed normally in the syp mutant. We conclude that Syp upregulates Pros protein in neurons by stabilising proslong mRNA.
Fig. 4.
Syncrip selectively stabilises the proslong isoforms in neurons. (A) smFISH against pros long intron and pros long UTR shows that proslong is normally transcribed in the syp mutant. (B) RNA sequencing shows selective loss of proslong isoform in syp mutants. Read coverage for the first 3 kb of pros 3′ UTR is not different between wild type (wt) and syp mutant whereas the distal 12 kb of pros 3′ UTR is reduced in syp mutants. (C) Quantitative analysis of total sequencing reads of pros 3′ UTR in wild type and syp mutants. (D) Northern blot showing the selective loss of proslong isoforms in syp mutants. The intensity of the 6.5 kb and 9 kb bands are similar between wild type and syp mutants (blue asterisks) whereas the 21 kb band is reduced in syp mutants (red asterisk). Scale bars: 10 μm.
To confirm that the loss of cytoplasmic pros mRNA in syp mutants is due to specific loss of the long isoform, we carried out transcriptomic analysis of syp mutant versus wild type brains (Table S1). The results show that the abundance of proslong is specifically reduced in syp mutant brains, whereas the shorter pros forms are maintained (Fig. 4B,C). Northern blot analysis confirmed these findings (Fig. 4D).
prosshort is sufficient to maintain neuronal fate
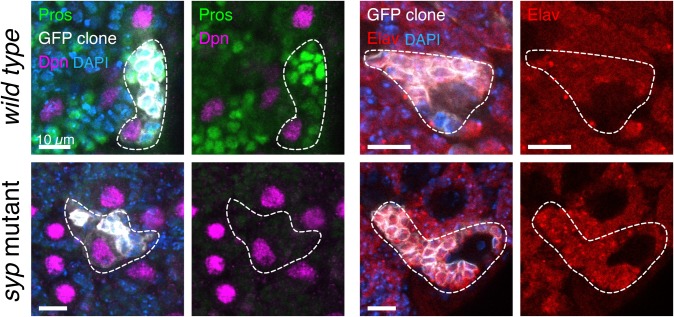
We have shown that Syp is required for the upregulation of Pros protein in larval neurons, and this occurs through the regulated mRNA stability of proslong. When Pros is depleted from neurons, but not NBs or GMCs, such as in the midlife crisis mutant, this leads to loss of neuronal differentiation and expression of NB genes (Carney et al., 2013). Therefore, we used the syp mutant as a tool to examine the specific role of the upregulated Pros protein levels in neurons, as opposed to the effect of loss of all Pros protein from the neurons. Unexpectedly, we found that neuronal differentiation progresses normally in syp mutants and neurons do not revert to NBs, as they do not switch on the NB marker, Dpn. We used clonal analysis to mark individual NB lineages in wild type and syp mutant brains and stained with Dpn, Pros and Elav (Fig. 5). Each syp mutant clone contained just one Dpn positive NB, and Elav was expressed in the progeny cells, demonstrating their neuronal identity. We conclude that the stabilisation of proslong by Syp in progeny cells is not required for the normal neuronal path of differentiation nor to prevent reversion of neurons to NBs. The remaining low level of Pros in the neurons of the syp mutant, provided by the short pros mRNA isoform, is sufficient to prevent reversion to an NB fate.
Fig. 5.
Syp and proslong are not required to maintain larval neurons in differentiated states. Clonal analysis shows normal GMC differentiation in the syp mutant. Type I NB lineage clones are marked with mCD8::GFP (outlined with white trace) and stained with IF against Pros, Dpn (wild type n=7, syp n=9) and Elav (wild type n=16, syp n=4). Both wild type and syp mutant clones have a single Dpn-expressing cell per clone and the progeny cells express Elav, marking a neuronal fate.
The question remains: what is the function of proslong mRNA? A burst of nuclear Pros protein is known to have an important role in terminating NB divisions in pupae (Kohwi and Doe, 2013; Maurange et al., 2008). Furthermore, Syp is required for the final symmetric division that terminates the type I NB in the pupa (Yang et al., 2017a). In the wild type pupal brain, all type I NBs, except the mushroom body NBs, terminate by 48 h after pupal formation (APF) (Siegrist et al., 2010). Syp-depleted type I NBs persist in the pupal brain for more than 48 h APF (Yang et al., 2017a). We surmised that proslong might be required at the terminal division, stabilised by Syp to allow the upregulation of Pros in the NB. To test this hypothesis we produced a series of transgenic lines aiming to remove proslong.
Several attempts to generate flies selectively deleting the proslong transcripts were unsuccessful. These included a frame shift mutation, which did not introduce nonsense-mediated mRNA decay (NMD) of the long pros isoforms (Fig. S2; Materials and Methods), and attempting a direct deletion of the pros 3′ UTR extension (Materials and Methods). We successfully deleted the three promoters that are annotated to produce proslong (Fig. S3A), but found that the 3′ UTR extension of proslong can be transcribed from the downstream promoters (Fig. S3B) and there was no change in Pros protein expression (Fig. S3C). Finally, we used a strong SV40 transcriptional terminator to terminate transcription at the beginning of the 3′ UTR extension (see Materials and Methods). The SV40 terminator reduced but did not entirely remove the extended UTR of proslong (Fig. S4).
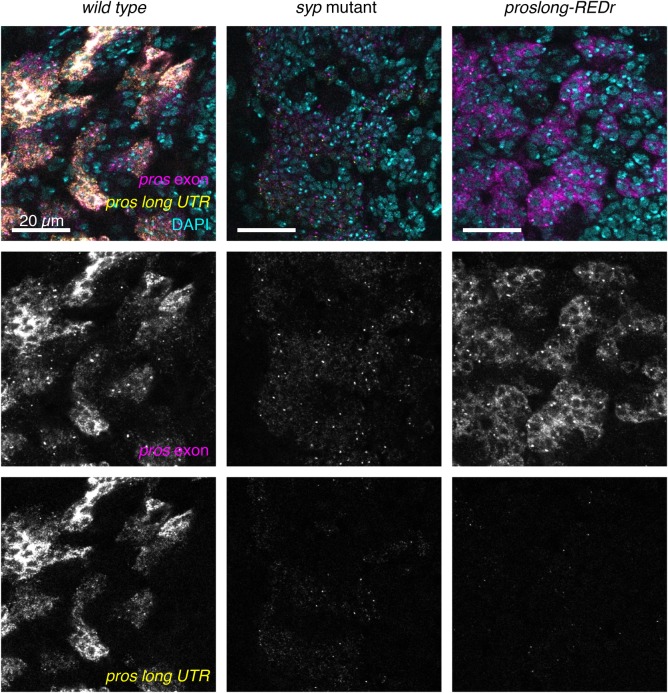
Unexpectedly, the inserted dsRED marker used to screen for integration (proslong-REDr; Materials and Methods) does terminate the 3′ UTR extension of pros (Fig. 6), allowing us to test the effect of removing the 3′ UTR extension.
Fig. 6.
proslong-REDr terminates transcription of the pros UTR extension. Insertion of a dsRED marker in reverse orientation downstream of the 3 kb UTR termination signal (proslong-REDr) results in loss of the pros UTR extension (visualised with the pros long UTR smFISH probe). The pros exon smFISH shows that proslong-REDr expresses higher levels of pros than syp mutant brains, but much less than wild type. Scale bars: 20 μm.
smFISH against the pros exon and proslong 3′ UTR showed that the long 3′ UTR is almost entirely lost in the proslong-REDr brains (Fig. 6). Therefore, regulation of pros mRNA through the 3′ UTR extension is disrupted in this line. The signal from the pros exon probes is reduced compared to the wild type, but remains higher than in the syp mutant, which could be explained by residual binding of Syp to proslong in regions outside the UTR extension. Alternatively, Syp may stabilise multiple isoforms of pros.
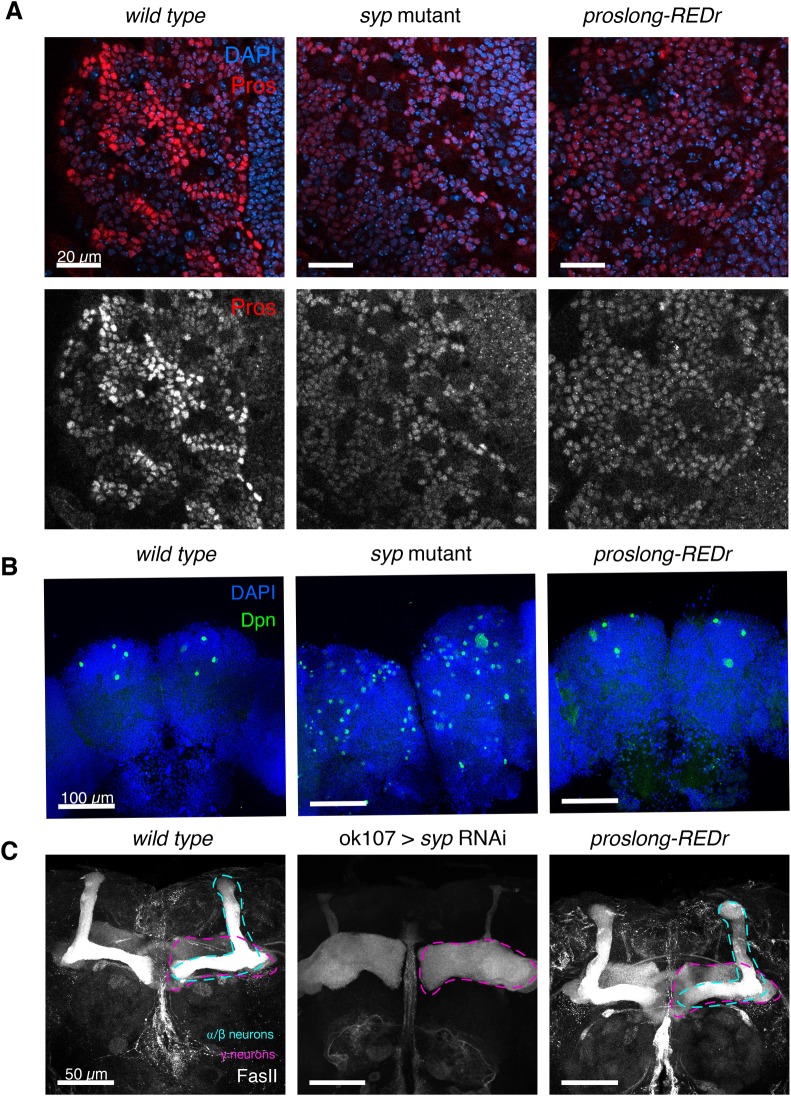
In order to understand the role of proslong in regulating Pros protein expression, we examined Pros protein level in proslong-REDr brains. We found that the cell type-specific upregulation of Pros protein in neurons is lost in proslong-REDr brains (Fig. 7A), which is consistent with the 3′ UTR extension of pros being required for the neuronal upregulation of Pros protein during larval neurogenesis.
Fig. 7.
Loss of proslong blocks Pros protein upregulation but has no effect on NB termination. (A) Pros protein level is not upregulated in the neurons of proslong-REDr larval brains. (B) Dpn marks NBs in pupal brains, 48 h APF. In wild type, only the four MB NBs remain. In the syp mutant, many NBs are long-lived in the pupal brain. In the proslong-REDr brains, type I NBs terminate normally and only the four MB NBs persist. (C) FasII IF stains γ and α/β neurons in the MB of 1-day old adult brains. In the syp RNAi knockdown brains there is loss of α/β neurons and excess γ neurons. There is no difference in MB morphology in the proslong-REDr line, compared to wild type.
To test the function of proslong in NB termination, we examined the number of NBs remaining at 48 h APF. We found that proslong is not required for normal NB termination. At 48 h APF, wild type pupae have only four NBs remaining [labelled with Dpn, the mushroom body (MB) NBs], while syp mutant pupal brains contain many persistent NBs (Fig. 7B). In the proslong-REDr brains, we observe only the four MB NBs at 48 h APF, showing that the type I NBs terminate normally, despite the loss of the proslong 3′ UTR extension.
We tested whether proslong is required for neuronal fate specification. Syp controls α/β neuronal fate during MB development, and loss of Syp results in complete loss of α/β neurons and precocious production of γ neurons (Liu et al., 2015). Single-cell transcriptomic analysis of the adult Drosophila brain has revealed that α/β neurons express much higher levels of pros compared to γ and α’/β’ neurons (Davie et al., 2018). To test whether stabilisation of proslong by Syp is required for the specification of α/β neurons, we examined the MB morphology in 1-day old proslong-REDr adult brains (Fig. 7C). Immunostaining against FasII showed the overall morphology of α/β and γ neurons in the proslong-REDr brains are comparable to the wild type control, indicating that fate patterning of MB neurons occurs normally even in the absence of proslong.
Finally, we wondered whether proslong is required in adult flies. We found that adult flies lacking proslong show highly penetrant behavioural defects. The majority of proslong-REDr adult flies eclose normally and are morphologically normal, but display an abnormal behaviour consistent with impaired locomotor or neurological activity (Movie 1). The same phenotype is also seen in hemizygous proslong-REDr flies (Movie 2), implying that it indeed derives from depletion of the long mRNA isoform. This phenotype indicates that the proslong mRNA isoform has a function in the adult brain, although its detailed characterisation is beyond the scope of this study.
DISCUSSION
Many key regulators of NB proliferation and differentiation have been identified and characterised (Grosskortenhaus et al., 2006; Isshiki et al., 2001; Kambadur et al., 1998; Kohwi and Doe, 2013; Li et al., 2013; Narbonne-Reveau et al., 2016; Novotny et al., 2002; Pearson and Doe, 2003). Recently, an increasing number of RBPs, the key regulators of post-transcriptional processes, have been implicated in neurodevelopment (Betschinger et al., 2006; Hilgers et al., 2012; Liu et al., 2015), suggesting that the importance of post-transcriptional regulation in the brain has so far been underestimated. Here, we have applied smFISH to examine the regulation of the transcription factor (TF) Pros, a master switch promoting neuronal differentiation. We show that Pros expression is regulated by the differential stability of its mRNA isoforms depending on their 3′ UTR lengths. Unstable short mRNA isoforms produce sufficient Pros protein to prevent GMCs and their neuronal progeny from reverting back to NB identity, but a switch to the more stable proslong isoforms is required to upregulate Pros protein in larval neurons. Our findings highlight the capacity of cell type-specific alternative 3′ UTRs to mediate different modes of post-transcriptional regulation of mRNA isoforms.
Low-level prosshort has a distinct function from high-level proslong
Surprisingly, we found that neurons do not de-differentiate into NBs in syp mutants, despite the loss of neuronal Pros upregulation in the absence of proslong. Previous work has shown that Pros elimination in young or middle-aged larval neurons causes de-differentiation of neurons and their reversion to NBs (Bello et al., 2006; Betschinger et al., 2006; Carney et al., 2013; Lee et al., 2006). Our work suggests that the low levels of Pros remaining in the syp mutant (provided by prosshort) are sufficient for neurons to maintain their identity. proslong is not required to drive differentiation of GMCs to neurons or to maintain neuronal identity. Although we have not uncovered the function of proslong in larval/pupal brain development, the impaired locomotive activity of the proslong-REDr adult flies indicates a role of proslong in the adult brain, either because of an earlier neuronal specification event or due to a function of Pros in the adult brain.
The 15 kb pros UTR extension allows multiple levels of post-transcriptional regulation
Exclusion of the 3′ UTR extension from pros transcripts in the proslong-REDr brains reduces the number of pros transcripts and Pros protein in the neurons. However, the pros transcript levels are still much higher in proslong-REDr than in syp mutant brains. This residual pros signal suggests that Syp can stabilise pros mRNA through binding to additional regions of the transcript, perhaps the 5′ UTR sequence that is also unique to the proslong transcripts, or some shared sequence included in all transcripts.
The 3′ UTR extension of proslong may mediate a second regulatory step, at the level of translation. The upregulation of Pros protein in the neurons is lost in the proslong-REDr brains, despite the relatively high levels of remaining pros mRNA transcripts. While the pros exon smFISH signal is much higher in the proslong-REDr brains, compared to the syp mutant, the Pros protein levels are similar between the two genotypes. This result suggests that the proslong 3′ UTR extension includes additional regulatory sequences that promote increased pros translation, either via Syp or an unidentified second RBP. This hypothesis would explain why a moderate decrease in pros transcript levels in proslong-REDr brains leads to a loss of neural Pros protein upregulation.
Post-transcriptional and transcriptional regulation work hand-in-hand to achieve cell-type-specific gene expression
Our experiments show that Pros expression is controlled at two levels: alternative polyadenylation and then differential mRNA stability, regulated through Syp binding. The molecular mechanism underlying the cell type-specific choice of pros isoform has not yet been identified. In Drosophila embryos, Elav is recruited at the promoter of extended genes and is required to extend the 3′ UTR of brat (Hilgers, 2015; Oktaba et al., 2015). Future experiments will determine whether pros differential polyadenylation is regulated at the promotor region by a similar Elav-dependent mechanism.
Many key regulators in the brain also have complex gene structures such as multiple isoforms and long 3′ UTRs, hallmarks of post-transcriptional mechanisms (Berger et al., 2012; Hilgers et al., 2011; Stoiber et al., 2015; Tekotte et al., 2002). Such genes include the temporal regulator neuronal fate, chinmo (Liu et al., 2015), the driver of cell growth and division, myc (Samuels et al., 2020) and the mRNA-binding proteins, brat (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006) and imp (Liu et al., 2015). Quantitative smFISH approaches combined with genetics and biochemistry will allow the detailed disentanglement of the transcriptional and post-transcriptional mechanisms regulating these genes.
Regulating expression levels by differential mRNA isoform stability and long 3′ UTRs is likely to be a conserved mechanism
Mammalian SYNCRIP/hnRNPQ is an important regulator of neural development (Lelieveld et al., 2016; Liu et al., 2015; Stoiber et al., 2015) and has a number of post-transcriptional roles including regulating mRNA stability through binding at the 3′ end of transcripts (Kim et al., 2011; Kuchler et al., 2014). Prox1, the mammalian orthologue of Pros, is a tumour suppressor that regulates stem cell differentiation in the brain as well as many other organ systems (Elsir et al., 2012; Stergiopoulos et al., 2014). Like Drosophila pros, prox1 has several isoforms including alternative 3′ UTRs, and a burst of Prox1 expression is required to drive the differentiation of immature granular neurons in the adult hippocampus (Hsieh, 2012). It is plausible that post-transcriptional regulation by RBPs helps determine the expression and translation of Prox1. Application of quantitative smFISH to mammalian systems will uncover whether Prox1 expression levels, like pros, are regulated through differential stabilisation of different 3′ UTR isoforms.
MATERIALS AND METHODS
Key resources table
Contact for reagent and resource sharing
For further information and access to any reagents and fly strains used in this manuscript, please contact corresponding author Ilan Davis (ilan.davis@bioch.ox.ac.uk).
Experimental model and subject details
Fly genetics
All strains were raised on standard cornmeal-agar medium at 25°C. Oregon-R was used as the wild type strain.
Clonal analysis
For wild type, female hsFLP;Pin/CyO;Tm2,Sb/Tm6B were crossed to male act>FOT>Gal4, UAS-mCD8::GFP/CyO;Tm3,Sb/Tm6b. These lines were crossed to the syp mutant to produce parental lines to make the clones: female hsFLP;Pin/CyO;Syp[e00286]/Tm6b were crossed to male act>FOT>Gal4,UAS-mCD8::GFP/CyO;Syp[e00286]/Tm6b. Eggs were collected in a 4-h egg lay on apple juice plates with yeast paste at 25˚C. 24 h after the egg lay, newly hatched larvae were transferred to fly food vials and incubated in a 37˚C water bath for 25 min. Wandering larvae were dissected 96 h after larval hatching. For syp mutant, only non-tubby larvae were dissected. Immunofluorescence was completed as described below and clones were imaged on the ventral side.
proslong deletion mutant generation using CRISPR
sgRNA construct design and validation was performed by Dr Andrew Bassett - Genome Engineering Oxford (GEO) (Table S5). Plasmids were injected into vasa-cas9 embryos (Bloomington Stock: BL55821) as previously described (Bassett and Liu, 2014). For homologous recombination repair, the donor construct included a ds-RED marker for selection, flanked by lox sites to remove the dsRED, and an attP site to allow future insertions at the site. This inserted cassette was flanked by 1 kb homology arms to facilitate repair. The pDsRED-attP vector 51019 was used. Homology arms were amplified from purified BAC DNA (CH322-133M22).
Nonsense-Mediated mRNA-Decay mutant
We took advantage of the unique coding sequence that is specific to the three long isoforms of pros, contributing an additional 300 amino acids to the N-terminus of Pros protein. We used CRISPR/Cas9 with non-homologous end joining to induce a frame shift, which introduces a stop codon 80 bp downstream (Fig. S2A). This stop codon would be expected to produce a truncated protein and induce Nonsense mediated mRNA decay (NMD) of the proslong transcripts. However, this line (pros-longSTOP) showed no reduction in either proslong-specific smFISH signal (Fig. S2B), or Pros protein (Fig. S2C). This result suggests that the upstream coding sequence is not actually translated in proslong in the neurons. Instead, Pros protein is most likely produced from the downstream translation start site.
UTR extension deletion
We aimed to delete the pros UTR extension directly using a CRISPR strategy with homologous recombination (HR), but this was unsuccessful. We speculate that the mutant line was embryonic dominant lethal, implying unknown roles for the UTR extension or non-coding transcription from this region.
Upstream promoter deletion
We used an HR strategy to delete the three promoters that are annotated to produce the UTR extended isoform, collectively termed proslong (Fig. S3A). We found that this line (prosΔpromUP) successfully abolished transcription from the upstream promoters, but the proslong-specific UTR probe showed substantial remaining expression, albeit reduced compared to wild type (Fig. S3B). Furthermore, the level of Pros protein was unchanged compared to wild type (Fig. S3C). We conclude that the UTR extension of proslong can be transcribed from the downstream promoters. The lower levels of proslong in this line are sufficient to produce the wild type upregulation of Pros protein in neurons.
Transcriptional termination
We designed a strategy to terminate transcription at the beginning of the 3′ UTR extension using a strong SV40 transcriptional terminator. This strategy uses a two-step process to insert an attP site and then integrate the SV40 terminator. The SV40 terminator (proslong-SV40) did not prevent the formation of the extended UTR of proslong (Fig. S4). Unexpectedly, the inserted dsRED marker used to screen for integration does terminate the UTR extension of pros (Fig. 6). We tested lines with the dsRED marker inserted in both orientations, which had the same phenotype. The dsRED inserted in the opposite orientation to pros (proslong-REDr) showed the lowest expression of dsRED allowing use of the red channel for smFISH staining, and so this line was used for further experiments.
Method details
RNA extraction
Third instar larval brains were homogenised in IP buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mini tablet of Complete EDTA-free protease inhibitor and 2 μl RNAse inhibitor (RNAsin Plus RNase Inhibitor, Promega). RNA extraction was performed using the RNAspin Mini kit (GE Healthcare).
Western blot
NuPage Novex 4–12% Bis-Tris gels (Invitrogen) were used for all SDS-PAGE experiments. Following SDS-PAGE, proteins were transferred to nitrocellulose membrane using the XCell II™ Blot Module (Invitrogen) following the manufacturer's protocol. Protein bands were visualised with a quantitative infrared imaging system (LI-COR Odyssey).
Northern blot
5 µl of purified RNA samples was mixed with 3.5 µl 37% formaldehyde and 10 µl 100% formamide, 2 µl supplemented 10x MOPS buffer (400 mM MOPS buffer, 100 mM NaOAc, 10 mM Na2EDTA). Sample mixture was incubated at 55°C for 15 min and 4 µl of loading dye (1 mM EDTA, 0.25% BromoPhenolBlue, 50% Glycerol) was added and run on an 1.5% agarose gel at 120 V in 1× MOPS buffer. ssRNA ladder (NEB) was used as size marker for all northern blot experiments.
RNA was transferred to pre-wet Nylon membrane (Hybond-N, Amersham). Blotted RNA was crosslinked to the membrane with either a UV crosslinker or by baking at 80°C for >1 h. The membrane was hybridised in hybridisation buffer (0.4 M Na2HPO4, 6% SDS and 1 mM EDTA) for 1 h at 57°C with RNA probes prepared using the Prime-a-Gene kit (Promega) (for list of primers used to generate RNA probes, see Table S2), washed twice with wash buffer 1 (40 mM Na2HPO4, 5% SDS and 1 mM EDTA) and twice with wash buffer 2 (40 mM Na2HPO4, 5% SDS and 1 mM EDTA). RNA was detected overnight in PhosphorImager (Molecular Dynamics).
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was performed using a real time PCR detection system [CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad)] and in 25 µl consisted of 12.5 µl 2× SYBRGreen Mastermix (Thermo Fisher Scientific), 0.75 µl of gene-specific primer (10 µM stock, forward and reverse, Table S3), 4 µl of cDNA and 7 µl of nuclease-free water. Cycle threshold value was calculated by the Bio-Rad CFX software using a second differential maximum method. For each of three biological replicates, a dilution series of the input sample was produced (10%, 2%, 0.5%. 0.01% of input). qPCR for each set of primers was performed on this series and the Cq values were plotted against log10 dilution to find the formula of the line. The Cq value of each pulldown sample (anti-Syp and anti-IgG) was inputted into this formula to calculate the % of input pulled down. For each set of primers, a two-tailed Student's t-test was used to compare the % input pulldown in the test (anti-Syp) and control (anti-IgG) samples.
Immunoprecipitation
Guinea pig anti-Syp antibody and IgG antibody were cross linked to ProteinA magnetic beads (Abcam) following the manufacturer's protocol. For each replicate, 90 third instar larval brains dissected in Schneider's medium were homogenised in immunoprecipitation (IP) buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mini tablet of Complete EDTA-free protease inhibitor and 2 µl RNAse inhibitor [RNAsin® Plus RNase Inhibitor (Promega)] and topped up to 100 µl in IP buffer. For each reaction, 20 µl of lysate was taken as a 50% input sample, which was taken directly to RNA extraction. 40 µl of lysate was incubated with 25 µl of each antibody cross-linked beads over night at 4°C on rotator wheel. For each reaction, 100 µl of lysate was incubated with 20 µl of 50% bead slurry over night at 4°C on rotator wheel. Next day, supernatant was transferred to fresh tubes and beads were washed five times for 5 min each with 200 µl cold IP buffer at 4°C. After final wash, beads were resuspended in 40 µl extraction buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA and 1.3% SDS, 1:100 RNAsin) and incubated at 65°C, 1000 rpm for 30 min on thermomixer. The elution step was repeated and the supernatant were pooled. RNA was then extracted from the IP eluates and the input sample and used for cDNA library synthesis.
RNA sequencing
Three biological replicates (n=3), each replicate consists of a pool of 100 larval brains. Following RNA extraction, mRNA was enriched using NEB Next® Poly(A) mRNA Magnetic Isolation Module (NEB). Briefly, extracted RNA sample was mixed with Oligo d(T)25 beads and heated to 65°C for 5 min followed by incubation at 4°C for 1 h to allow binding. Following incubation, the beads were washed 5 times for 5 min each at 4°C and RNA was eluted by heating the beads at 80°C for 5 min. Poly(A) enriched RNA was then used for library production using the Ion Total RNA-Seq Kit v2 for Whole Transcriptome Libraries (Life Technologies). Following quality-control steps, adaptors were hybridised to the RNA fragments and RT reaction was performed followed by cDNA amplification with Ion Xpress RNA Barcode primers. Prior to sequencing, quality of cDNA libraries were assessed using Agilent High Sensitivity DNA Kit with the Agilent 2100 Bioanalyser. Libraries were pooled to a total concentration of 100 pM, with three samples multiplexed per chip. Sequencing was performed on an Ion Proton Sequencer, using the Ion PI IC 200 Kit (Life Technologies). Ion PI chips were prepared following the manufacturer's instructions and loaded using the Ion Chef System.
Immunofluorescence
Third instar larval brains were dissected in Schneider's medium and fixed in 4% formaldehyde solution (4% formaldehyde in 0.3% PBTX), for 25 min at room temperature. Following incubation with primary and secondary antibody, samples were mounted in VECTASHIELD anti-fade mounting medium (Vector Laboratories). All imaging experiments were performed with three replicates and representative images are shown.
RNA smFISH for larval brains
Dissected third instar larval brains were fixed in 4% paraformaldehyde solution for 25 min at room temperature followed by three rinses in 0.3% PBTX (PBS and 0.3% triton-X). Samples were washed three times for 15 min each in 0.3% PBTX at room temperature and incubated in pre-hybrdisation buffer (10% deionised formamide in prepared in 2× SSC) for 5 min at 37°C. Hybridisation was performed by incubating samples overnight at 37°C in the dark with gentle shaking in hybridisation buffer [10% deionised formamide, 5% dextran sulphate (Sigma-Aldrich), 2× SSC] containing 250 nM gene-specific fluorescently labelled Stellaris® DNA probe set (BioSearch Technologies). Following hybridisation, samples were rinsed three times in pre-hybridisation buffer and washed a further three times, for 15 min each time in pre-hybridisation buffer at 37°C. DAPI was included during the second 15 min wash. Samples were mounted in VECTASHIELD anti-fade mounting medium (Vector Laboratories) and imaged. Samples were protected from light for all steps including hybridisation.
smFISH with immunofluorescence for larval brains
For smFISH with anti-Dpn antibody, the smFISH was performed identically as above, then, after washing three times 30 min in pre-hybridisation buffer on Day 2, samples were blocked in blocking buffer (1% BSA in 0.3% PBTX) for 1 h at room temperature. Samples were incubated with primary antibody in blocking buffer for 3 h at room temperature, washed, and then incubated with secondary antibody for 1 h at room temperature. Final washes were performed using 0.3% PBTX, once including DAPI.
For all other antibodies, the protocol is identical to the smFISH protocol with the following modifications. Before hybridisation, samples were blocked in blocking buffer (1% BSA in 0.3% PBTX) for 1 h at room temperature. Antibody at the appropriate dilution was included with the Stellaris® DNA probes during the overnight hybridisation step. Counter-stain with secondary antibody was performed on the second day following hybridisation. After final wash, samples were mounted in VECTASHIELD anti-fade mounting medium (Vector Laboratories) and immediately imaged.
Image acquisition
Larval brains were imaged on the ventral side. Fixed imaging of larval brains was performed using an inverted Olympus FV3000 Laser Scanning Microscope with Becker and Hickel FLIM system and with an inverted Olympus FV1200 Laser Scanning Microscope with high sensitivity galium arsenide phosphide (GaAsP) detectors (Olympus). Images were acquired using ×20 0.75 NA UPlanSApo, ×40 1.3 NA Oil UPlan FLN, ×60 1.4 NA and ×100 1.4 NA Oil UPlanSApo objective lenses. The laser units used were solid state 405 and 488 lasers, argon 488, 515, 568 and 633 lasers.
Quantification and statistical analysis
Bioinformatic analysis of RNA sequencing data
Base calling, read trimming and sample de-multiplexing were done using the standard Ion Torrent Suite. The reads were aligned to the Drosophila genome (ENSEMBL assembly BDGP5, downloaded 8 Jan 2015) using the TopHat aligner (v2.0.13) (Kim et al., 2013). To quantitate gene expression, uniquely aligned reads were assigned to the Drosophila exome (BDGP5) using htseq-count. Differential was assessed using negative binomial generalised linear models implemented in R/Bioconductor package DESeq2 (Love et al., 2014). Differential expression analysis is presented in Table S1.
Image analysis for smFISH
pros intron probes were used to compare pros transcription rates in wild type and syp mutant brains (Fig. 3E). Transcription rates were compared separately for type I NBs and for progeny cells (including GMCs, immature and mature neurons). pros intron transcription foci were identified in 3D with the spot detection feature of Imaris Image Analysis software (Imaris, Bitplane), using a 1.2 μm diameter spot. Spots overlapping the edge of the image were removed. Average intensity was measured for each spot.
In type I NBs, pros transcription foci are found spatially separated in the nucleus and therefore the intensity of two spots were added together for each NB. In progeny cells, the transcription foci are close in the nucleus and are measured in a single spot. Intensity measurements were normalised to the average wild type intensity for each experimental replicated (i.e. the average of wild type measurements from a single day of staining/imaging).
Supplementary Material
Acknowledgements
We are grateful to Andrew Bassett (Genome Engineering Oxford, GEO) for assistance in designing and validating CrispR guide RNAs, to Tomek Dobrzycki for initial work that lead to the project, to MICRON Oxford (http://micronoxford.com), supported by a Wellcome Strategic Awards to I.D. (091911/B/10/Z and 107457/Z/15/Z) providing access to equipment and advice on advanced imaging techniques, to Alan Wainman and Richard M. Parton for advice on advanced microscopy and to Darragh Ennis for help and advice with fly husbandry and administration. We are also indebted to Alfredo Castello, Lidia Vasilieva, Jordan Raff and Neil Brockdorff for their comments. We would also like to thank the Cambridge Fly Facility for transgenic production and the Bloomington Drosophila Stock Centre.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.J.S., L.Y., D.I.-H., I.D.; Methodology: T.J.S., Y.A., A.I.J., F.R., J.Y.L., L.Y., C.-P.Y., T.L., D.I.-H., I.D.; Software: A.I.J.; Formal analysis: T.J.S., Y.A., J.Y.L.; Investigation: T.J.S., Y.A., J.Y.L.; Writing - original draft: T.J.S., L.Y.; Writing - review & editing: T.J.S., D.I.-H, I.D.; Visualization: T.J.S., A.I.J.; Supervision: I.D.; Funding acquisition: I.D., T.L.
Funding
T.J.S. was funded by Wellcome Trust Four-Year PhD Studentship (105363/Z/14/Z). L.Y. was funded from the Clarendon Trust and Goodger Fund. F.R. was funded by a Marie Curie Postdoctoral Fellowship. C.-P.Y. and T.L. were supported by Howard Hughes Medical Institute. J.Y.L. was funded by the Clarendon Trust. D.I.-H. was funded by University College London. A.I.J. was funded by a Wellcome Senior Research Fellowship to I. D. (096144/Z/17/Z). F. R. was funded by a Wellcome Investigator Award to I.D. (209412/Z/17/Z).
Data availability
The presented RNA sequencing data has been deposited with Gene Expression Omnibus (GEO), with accession number GSE145670.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.049684.supplemental
References
- Awasaki T., Kao C., Lee Y., Yang C., Huang Y., Pfeiffer B. D., Luan H., Jing X., Huang Y., He Y. et al. (2014). Making Drosophila lineage-restricted drivers via patterned recombination in neuroblasts. Nature Neuroscience, 17, 631-7 [DOI] [PubMed] [Google Scholar]
- Bassett A. and Liu J.-L. (2014). CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69, 128-136. 10.1016/j.ymeth.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Bayraktar O. A., Boone J. Q., Drummond M. L. and Doe C. Q. (2010). Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 5, 26 10.1186/1749-8104-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B., Reichert H. and Hirth F. (2006). The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639-2648. 10.1242/dev.02429 [DOI] [PubMed] [Google Scholar]
- Berger C., Harzer H., Burkard T. R., Steinmann J., van der Horst S., Laurenson A.-S., Novatchkova M., Reichert H. and Knoblich J. A. (2012). FACS purification and transcriptome analysis of drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2, 407-418. 10.1016/j.celrep.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K. and Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241-1253. 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Carney T. D., Struck A. J. and Doe C. Q. (2013). midlife crisis encodes a conserved zinc-finger protein required to maintain neuronal differentiation in Drosophila. Development 140, 4155-4164. 10.1242/dev.093781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E., Fischer B. E., van Steensel B., Micklem G. and Brand A. H. (2006). Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775-789. 10.1016/j.devcel.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft Ł., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C. et al. (2018). A single-cell transcriptome atlas of the aging drosophila brain. Cell 174, 982-998.e920. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Q., Chu-LaGra Q., Wright D. M. and Scott M. P. (1991). The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451-464. 10.1016/0092-8674(91)90463-9 [DOI] [PubMed] [Google Scholar]
- Elsir T., Smits A., Lindström M. S. and Nistér M. (2012). Transcription factor PROX1: its role in development and cancer. Cancer Metastasis Rev. 31, 793-805. 10.1007/s10555-012-9390-8 [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R., Robinson K. J. and Doe C. Q. (2006). Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 20, 2618-2627. 10.1101/gad.1445306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. M., Pfeiffer B. D., Rubin G. M. and Truman J. W., (2015). Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. eLife. 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V. (2015). Alternative polyadenylation coupled to transcription initiation: Insights from ELAV-mediated 3′ UTR extension. RNA Biol. 12, 918-921. 10.1080/15476286.2015.1060393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Perry M. W., Hendrix D., Stark A., Levine M. and Haley B. (2011). Neural- specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. USA 108, 15864-15869. 10.1073/pnas.1112672108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Lemke S. B. and Levine M. (2012). ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259-2264. 10.1101/gad.199653.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata J., Nakagoshi H., Nabeshima Y. and Matsuzaki F. (1995). Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377, 627-630. 10.1038/377627a0 [DOI] [PubMed] [Google Scholar]
- Homem C. C. F. and Knoblich J. A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297-4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Hsieh J. (2012). Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 26, 1010-1021. 10.1101/gad.187336.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S. and Doe C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511-521. 10.1016/S0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J. and Odenwald W. F. (1998). Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12, 246-260. 10.1101/gad.12.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I. and Lancaster M. A. (2016). Stem cell models of human brain development. Cell Stem Cell 18, 736-748. 10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- Kim D.-Y., Kwak E., Kim S.-H., Lee K.-H., Woo K.-C. and Kim K.-T. (2011). hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res. 39, 8901-8914. 10.1093/nar/gkr605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima A., Fuse N., Isshiki T. and Matsuzaki F. (2010). Progenitor properties of symmetrically dividing Drosophila neuroblasts during embryonic and larval development. Dev. Biol. 347, 9-23. 10.1016/j.ydbio.2010.06.029 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y. and Jan Y. N. (1995). Asymmetric segregation of numb and prospero during cell division. Nature 377, 624-627. 10.1038/377624a0 [DOI] [PubMed] [Google Scholar]
- Kohwi M. and Doe C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823-838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler L., Giegerich A. K., Sha L. K., Knape T., Wong M. S., Schröder K., Brandes R. P., Heide H., Wittig I., Brune B. et al. (2014). SYNCRIP-dependent Nox2 mRNA destabilization impairs ROS formation in M2-polarized macrophages. Antioxid Redox Signal. 21, 2483-2497. 10.1089/ars.2013.5760 [DOI] [PubMed] [Google Scholar]
- Lai S. L. and Doe C. Q. (2014). Transient nuclear Prospero induces neural progenitor quiescence. Elife 3, e03363 10.7554/eLife.03363.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Wilkinson B. D., Siegrist S. E., Wharton R. P. and Doe C. Q. (2006). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441-449. 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Lelieveld S. H., Reijnders M. R. F., Pfundt R., Yntema H. G., Kamsteeg E.-J., de Vries P., de Vries B. B. A., Willemsen M. H., Kleefstra T., Lohner K. et al. (2016). Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 19, 1194-1196. 10.1038/nn.4352 [DOI] [PubMed] [Google Scholar]
- Li X., Erclik T., Bertet C., Chen Z., Voutev R., Venkatesh S., Morante J., Celik A. and Desplan C. (2013). Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456-462. 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yang C.-P., Sugino K., Fu C.-C., Liu L.-Y., Yao X., Lee L. P. and Lee T. (2015). Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 350, 317-320. 10.1126/science.aad1886 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F., Koizumi K., Hama C., Yoshioka T. and Nabeshima Y. (1992). Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem. Biophys. Res. Commun. 182, 1326-1332. 10.1016/0006-291X(92)91878-T [DOI] [PubMed] [Google Scholar]
- Maurange C., Cheng L. and Gould A. P. (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891-902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- McDermott S. M., Meignin C., Rappsilber J. and Davis I. (2012). Drosophila syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during Axis specification. Biology Open 1, 488-497. 10.1242/bio.2012885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott S. M., Yang L., Halstead J. M., Hamilton R. S., Meignin C. and Davis I. (2014). Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. RNA 20, 1593-1606. 10.1261/rna.045849.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne-Reveau K., Lanet E., Dillard C., Foppolo S., Chen C. H., Parrinello H., Rialle S., Sokol N. S. and Maurange C. (2016). Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila. Elife 5, e13463 10.7554/eLife.13463.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T., Eiselt R. and Urban J. (2002). Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development 129, 1027-1036. [DOI] [PubMed] [Google Scholar]
- Oktaba K., Zhang W., Lotz T. S., Jun D. J., Lemke S. B., Ng S. P., Esposito E., Levine M. and Hilgers V. (2015). ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol. Cell 57, 341-348. 10.1016/j.molcel.2014.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J. and Doe C. Q. (2003). Regulation of neuroblast competence in Drosophila. Nature 425, 624-628. 10.1038/nature01910 [DOI] [PubMed] [Google Scholar]
- Ren Q., Yang C.-P., Liu Z., Sugino K., Mok K., He Y., Ito M., Nern A., Otsuna H. and Lee T. (2017). Stem cell-intrinsic, seven-up-triggered temporal factor gradients diversify intermediate neural progenitors. Curr. Biol. 27, 1303-1313. 10.1016/j.cub.2017.03.047 [DOI] [PubMed] [Google Scholar]
- Samuels T. J., Järvelin A. I., Ish-Horowicz D. and Davis I. (2020). Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. eLife Sci. 9, e51529 10.7554/eLife.51529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist S. E., Haque N. S., Chen C.-H., Hay B. A. and Hariharan I. K. (2010). Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr. Biol. 20, 643-648. 10.1016/j.cub.2010.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E. P. and Doe C. Q. (1995). The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121, 3187-3195. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Peng C. Y., Nair S., Skeath J. B., Spana E. P. and Doe C. Q. (1992). Biochemical analysis of prospero protein during asymmetric cell division: cortical prospero is highly phosphorylated relative to nuclear prospero. Dev. Biol. 204 10.1006/dbio.1998.9079 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos A., Elkouris M. and Politis P. K. (2014). Prospero-related homeobox 1 (Prox1) at the crossroads of diverse pathways during adult neural fate specification. Front. Cell Neurosci. 8, 454 10.3389/fncel.2014.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber M. H., Olson S., May G. E., Du M. O., Manent J., Obar R., Guruharsha K. G., Bickel P. J., Artavanis-Tsakonas S., Brown J. B. et al. (2015). Extensive cross-regulation of post-transcriptional regulatory networks in Drosophila. Genome Res. 25, 1692-1702. 10.1101/gr.182675.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed M. H., Mark B. and Doe C. Q. (2017). Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. Elife 6, e26287 10.7554/eLife.26287.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekotte H., Berdnik D., Torok T., Buszczak M., Jones L. M., Cooley L., Knoblich J. A. and Davis I. (2002). Dcas is required for importin-α3 nuclear export and mechano-sensory organ cell fate specification in Drosophila. Dev. Biol. 244, 396-406. 10.1006/dbio.2002.0612 [DOI] [PubMed] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S., Marygold S. J., Matthews B. B., Millburn G., Antonazzo G., Trovisco V. et al (2019). FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759-D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaessin H., Grell E., Wol E., Bier E., Jan L. Y. and Jan Y. N. (1991). prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, 941-953. 10.1016/0092-8674(91)90367-8 [DOI] [PubMed] [Google Scholar]
- Yang C.-P., Samuels T. J., Huang Y., Yang L., Ish-Horowicz D., Davis I. and Lee T. (2017a). Imp and Syp RNA-binding proteins govern decommissioning of Drosophila neural stem cells. Development 144, 3454-3464. 10.1242/dev.149500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Titlow J., Ennis D., Smith C., Mitchell J., Young F. L., Waddell S., Ish-Horowicz D. and Davis I. (2017b). Single molecule fluorescence in situ hybridisation for quantitating post-transcriptional regulation in Drosophila brains. Methods 126, 166-176. 10.1016/j.ymeth.2017.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.