Abstract
Background
The composition of the bile acid (BA) pool is closely associated with obesity and is modified by gut microbiota. Perturbations of gut microbiota shape the BA composition, which, in turn, may alter important BA signaling and affect host metabolism.
Methods
We investigated BA composition of high BMI subjects from a human cohort study and a high fat diet (HFD) obesity prone (HF-OP) / HFD obesity resistant (HF-OR) mice model. Gut microbiota was analysed by metagenomics sequencing. GLP-1 secretion and gene regulation studies involved ELISA, qPCR, Western blot, Immunohistochemistry, and Immunofluorescence staining.
Findings
We found that the proportion of non-12-OH BAs was significantly decreased in the unhealthy high BMI subjects. The HF-OR mice had an enhanced level of non-12-OH BAs. Non-12-OH BAs including ursodeoxycholate (UDCA), chenodeoxycholate (CDCA), and lithocholate (LCA) were decreased in the HF-OP mice and associated with altered gut microbiota. Clostridium scindens was decreased in HF-OP mice and had a positive correlation with UDCA and LCA. Gavage of Clostridium scindens in mice increased the levels of hepatic non-12-OH BAs, accompanied by elevated serum 7α-hydroxy-4-cholesten-3-one (C4) levels. In HF-OP mice, altered BA composition was associated with significantly downregulated expression of GLP-1 in ileum and PGC1α, UCP1 in brown adipose tissue. In addition, we identified that UDCA attenuated the high fat diet-induced obesity via enhancing levels of non-12-OH BAs.
Interpretation
Our study highlights that dysregulated BA signaling mediated by gut microbiota contributes to obesity susceptibility, suggesting modulation of BAs could be a promising strategy for obesity therapy.
Keywords: Obesity, Bile acids, Gut microbiota, Energy expenditure, GLP-1, UCP1
Research in context.
Evidence before this study
Subjects may show different metabolic status and different susceptibility to obesity even under similar diets, lifestyles, and other environmental conditions. BAs have been recognized as important signaling molecules to modulate glucose and energy metabolism. The effects of the changes of BA composition mediated by the gut microbiota on obesity susceptibility remain poorly understood, requiring continued research efforts.
Added value of this study
Our current findings support a novel BA-gut microbiota axis as a contributor to obesity susceptibility. The level of non-12-OH BAs relative to total BAs (non-12-OH BA%) was significantly increased in high BMI subjects with “metabolically healthy” phenotype from a human cohort study. Compared with the HFD obesity prone (HF-OP) mice, higher non-12-OH BA levels were observed in HFD obesity resistant (HF-OR) mice, with attenuated metabolic disorders and enhanced GLP-1 expression in ileum and PGC1α, UCP1 expression in brown adipose tissue. The changes of BA compositions were associated with altered gut microbiota. Clostridium scindens, which was significantly decreased in HF-OP mice, played a potential role in enhancing the non-12-OH BA levels. Finally, we identified that treatment with UDCA in HFD mice enhanced the non-12-OH BA% and attenuated the HFD-induced obesity phenotype.
Implications of all the available evidence
The ratio of non-12-OH BAs to 12-OH BAs is mechanistically involved in the metabolic status of obesity. Our study highlights that shifting BA synthetic pathways (between classical and alternative pathways) alters host metabolism and affects susceptibility to obesity, suggesting that modulation of BA composition could be a promising strategy for obesity management.
Alt-text: Unlabelled box
1. Introduction
Obesity is a highly prevalent health problem worldwide, and is considered a risk factor for type 2 diabetes, non-alcoholic fatty liver, hypertension, cardiovascular diseases, and cancer. In addition to genetic regulation, environmental factors such as, excessive high-calorie food intake and sedentary lifestyle play an important role in obesity development [1]. Overweight/obese individuals have different metabolic status. For example, a proportion of overweight/obese individuals can maintain healthy metabolic phenotypes (HO) and might not be at an increased risk for metabolic complications of obesity [2]. Meanwhile, susceptibility to obesity can vary considerably among individuals despite being exposed to similar diets, lifestyles, and other similar environmental conditions. This has also been observed in rodents fed the same high fat diet (HFD) which show different susceptibilities to obesity: some of them are sensitive to HFD induced obesity (obesity prone, OP), whereas others are not (obesity resistant, OR) [3]. Previous studies showed that feeding C57BL6 mice with high fat diet could generate the OP/OR model [4,5]. The heterogeneity between the OP/OR model is still largely unknown and of great significance in obesity research. Many investigators have searched for candidate factors that render individuals susceptible to diet-induced obesity. Studies comparing obesity-prone and obesity resistant subjects have been reported, the proposed anti-obesity mechanisms include: (1) expression of uncoupling protein 1 (UCP1) and uncoupling protein 2 (UCP2) in brown fat are elevated in OR mice when given HFD [6]. (2) OR mice have increased fat oxidation ability in response to increased fat intake [7]. (3) Transcriptomic and metabolomic profiling of OP and OR rats revealed that OP rats have increased activity of the Krebs cycle as well as, increased activity of the sympathetic nervous system [8]. (4) an attenuated level of central dopamine was shown to be present in animals with a predisposition to dietary obesity [9]. However, definitive reasons for obesity resistance are still largely unknown.
BAs are synthesized in hepatocytes from cholesterol, stored in the gallbladder, and released into the intestine to facilitate absorption of dietary lipids and vitamins. BAs are initially synthesized in liver via two different pathways. The classical pathway is initiated by cholesterol 7α -hydroxylase (CYP7A1), and then sterol-12α -hydroxylase (CYP8B1) to produce cholic acid (CA). CYP8B1 activity determines the ratio of CA to CDCA [10]. The alternative pathway is initiated by sterol-27-hydroxylase (CYP27A1) to form 27-hydroxycholesterol which in turn, is further hydroxylated by oxysterol 7α-hydroxylase (CYP7B1), generating CDCA, and subsequently muricholic acids (MCAs) are synthesized in the liver of mice [11]. Dysregulation of BAs has been associated with obesity [12]. Increased 12α-hydroxylated BAs levels were found to be associated with insulin resistance and type 2 diabetes (T2DM) [13,14]. Recently, BAs have been recognized as important signaling molecules that regulate numerous metabolic processes, including glucose, lipid, and energy homeostasis. The membrane G protein-coupled receptor 5 (TGR5) and the Farnesoid X receptor (FXR) are the two major receptors of BAs that regulate metabolism [15]. TGR5 and FXR signaling affect many different metabolic processes in the host. For example, TGR5 signaling induces intestinal glucagon like peptide-1 (GLP-1) release, leading to improved glucose homeostasis [16]. In addition, TGR5 is expressed in brown adipose tissue (BAT) and muscle, where its activation can promote energy expenditure and attenuate diet induced obesity by conversion of inactive thyroxine (T4) into active thyroid hormone (T3) [17]. TGR5 is also expressed in macrophages, deletion of TGR5 in macrophages contributes to inflammation and diet induced obesity [18]. FXR is involved in the regulation of BA synthesis as well as multiple glucose and lipid metabolic pathways [19]. FXR agonist obeticholic acid was found to improve obesity-related metabolic disorders [20]. FXR is activated mainly by the primary BAs, CDCA and CA, while the most potent agonists for TGR5 are LCA and deoxycholic acid (DCA). In addition, TαMCA, TβMCA, and UDCA have been identified as antagonists of FXR [11].
The intestinal microbiota has been regarded as a “metabolic organ”, which plays an important role in regulating host metabolism. In addition to facilitating nutrition absorption and energy harvesting, gut microbiota produces large amounts of small molecule metabolites. BAs are one important class of metabolites modified by gut microbiota. Germ-free mice are resistant to high fat induced obesity, and when conventionalized with normal microbiota the germ-free mice harvest more energy from the diet and increase body fat content quickly [21]. It has been shown that the gut microbiota has profound effects on BA metabolism. Primary BAs are metabolized into secondary BAs by the actions of the gut microbial enzymes through deconjugation, dihydroxylation, and dehydrogenation of BAs in the distal small intestine and colon [11]. TαMCA and TβMCA have been identified as FXR antagonists, and gut microbiota regulates intestinal FGF15 signaling through a FXR-dependent pathway [22]. Intestinal-specific deletion of FXR protected mice from HFD-induced liver steatosis and obesity [23]. Therefore, intestine-selective FXR inhibition was found to improve obesity-related metabolic dysfunction [24]. Although recent studies have revealed a significant relationship between gut microbiota and obesity, the exact molecular mechanisms are yet to be elucidated, especially the role of specific gut microbial species and their metabolites in the regulation of obesity lipid metabolism and the development of obese phenotype.
With the advent of DNA sequencing and metabolomics technologies, we are beginning to recognize the mechanistic link between intestinal microbiota and metabolic dysregulation. In the present study, we analysed serum BA profiles in high BMI subjects with or without metabolic disorders using a targeted metabolomics approach. We further analysed BA profiles in serum, liver, and feces of the HF-OP/HF-OR mice and control mice with normal diet (N). Among the three groups of mice, we identified distinct serum BA profiles which were presumably derived from different gut microbiota compositions. Therefore, gut microbiota was analysed in mice of N, HF-OP, and HF-OR groups and correlation analysis between BA and gut microbiota was performed. We also assessed the GLP-1 expression in the ileum and UCP1 and PGC1α expression in BAT as well as the effect of different BAs on GLP-1 secretion in vitro. Moreover, BA profiles were analysed after UDCA administration in HFD mice. Identification of specific BAs in combination with specific types of gut microbiota may provide new diagnostic and therapeutic strategies for obesity control.
2. Materials and methods
2.1. Study participants and definitions in the cross-sectional study
The study participants were recruited from the Shanghai Obesity Study (SHOS), which was designed to investigate the occurrence and development of metabolic syndrome and its related diseases. Subjects with high BMI were selected according to the BMI ≥ 25 kg/m2. The exclusion criteria were: type 1 diabetes, pregnancy, gallstones, severe diabetic complications (diabetic retinopathy, diabetic neuropathy, diabetic nephropathy, and diabetic foot), severe hepatic diseases including chronic persistent hepatitis, liver cirrhosis with abnormal hepatic transaminase; severe organic diseases, including cancer, coronary heart disease, renal diseases, thyroid diseases; infectious diseases; alcoholism; and continuous medication (including weight loss or psychotropic medication, use of drugs known to affect BA metabolism), acute infection or injury prior to enrolment. A total of 183 subjects with high BMI were included. Anthropometric measurements (e.g., weight and height), sex, age, personal health history, and current medications were recorded. Clinical characteristics were examined, including glucose, systolic and diastolic blood pressures (SBP and DBP), triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL), low-density lipoprotein (LDL), alanine aminotransferase (ALT), and aspartate aminotransferase (AST). “Metabolically healthy” was defined as having all of the following: fasting glucose < 6·1 mmol/L, 2 h glucose ≤ 7·8 mmol/L, and having no history of diabetes; blood pressure with SBP/DBP < 140/90 mmHg; fasting plasma TC < 5·18 mmol/L, TG < 1·7 mmol/L and HDL ≥ 0·9 mmol/L with no history of high cholesterol and no previous history of cardiovascular disease. Failure to meet all the criteria was defined as “metabolically unhealthy” [25]. According to this definition, 183 subjects with high BMI were further divided into two groups, including 121 subjects in healthy high BMI (HHB) group and 62 subjects in unhealthy high BMI (UHB) group in the study. The study was approved by the ethics committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital and all participants provided written informed consent.
2.2. Animal study
Animal experiments were approved by the ethics committee of Shanghai Jiao Tong University and performed following the guidelines of the center of Laboratory Animals at Shanghai Jiao Tong University (Shanghai, China). C57BL/6 mice (Specific pathogen-free grade, male, three weeks old) were purchased from Shanghai Laboratory Animal Co. Ltd. (SLAC, Shanghai, China) and allowed one week of acclimatization. The mice were maintained under specific pathogen-free conditions in a controlled facility with free access to chow and water (temperature 20–22 °C, humidity 45±5%, 12 h light/dark cycle).
2.2.1. Animal experiment 1
Male Mice were randomly divided into two groups with different diets for 82 weeks: (1) normal diet group (n = 8): mice were fed chow diets consisting of: 10% fat, 71% carbohydrate, and 19% protein; (2) high fat diet (HFD) group (n = 32): mice were fed with diets consisting of: 45% fat, 36% carbohydrate, and 19% protein. Mice were individually housed and body weight and food intake were measured once a week during the experiments. Average daily food intake (g/day/mouse) was calculated and converted to kcal/day/mouse according to the calorie content of the diet. Feces samples were collected at different time points (weeks 4, 18, 46, and 82) for BA assessment. After 82 weeks of feeding, mice with body weight gain in the upper quartile during the feeding period were designated as the high fat obesity prone group (HF-OP group, n = 8), while those in the lower quartile were designated as the high fat obesity resistant phenotype (HF-OR group, n = 8). The mice were fasted overnight and then anesthetized for harvesting blood. The blood was centrifuged at 3000 g for 10 min for serum isolation. Liver samples, intestinal tissues and contents, white and brown fat tissues were collected immediately after euthanasia. All samples were stored at −80 °C until used for analysis.
2.2.2. Animal experiment 2
For the UDCA supplement experiment, 15 male mice were randomly divided into three groups with different diets: normal diet group (n = 5) (10% fat, 71% carbohydrate, and 19% protein), HFD group (n = 5), and HFD+0.5%UDCA group (HFD: 60% fat, 21% carbohydrate, 19% protein). Body weight and food intake were recorded once a week during the experiments. After 8 weeks of feeding, mice were fasted overnight and then sacrificed. The harvested blood was centrifuged at 3000 g for 10 min for serum isolation. Liver, adipose tissues, intestinal tissues and contents were carefully collected and kept in liquid nitrogen before storage at −80 °C.
2.3. Biochemical analysis
For human study, the levels of serum glucose, TG, TC, HDL, LDL, AST, and ALT were measured using an automatic biochemical analyser (Hitachi 7600, Tokyo, Japan). For animal experiment, serum glucose, TG, AST, and ALT were measured using an automatic biochemical analyser (Hitachi 7600, Tokyo, Japan). Serum TC, HDL and LDL were measured using a commercial biochemical kit (ab65390, Abcam, Cambridge, United Kingdom). The liver TG and TC content were detected using a triglyceride assay kit and total cholesterol assay kit (A110-1–1, A111-1–1, Jiancheng Bioengineering Institute, Nanjing, China).
2.4. BAs analysis
BAs are amphiphilic molecules consisting of a 24-carbon steroid core and a short aliphatic side chain [26]. BAs in serum, liver, and feces were measured according to previously reported methods [27] and the detailed analytical methods are shown in the Supplemental Methods. Serum C4 was detected based on mass spectrometry methods as described previously [28] and the detailed method was shown in Supplemental Methods. BAs were quantified using ultra-performance liquid chromatography (UPLC)-triple quadrupole time-of-flight mass spectrometry (Waters Corp., Milford, MA, USA). Raw data obtained from UPLC-MS were analysed and quantified using TargetLynx version 4·1 applications manager (Waters Corp., Milford, MA). All the calibration standards solutions were mixed at appropriate concentrations and run after every ten samples for quality control. Classification of BAs and their structure information are shown in Table S1.
2.5. Sequencing of the whole microbial genome
The microbial genomic DNA samples were extracted using the DNeasyPowerSoil kit (Qiagen, Inc., Netherlands). The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The quality checked DNA sample was used to construct metagenome shotgun sequencing libraries by using the Illumina TruSeq Nano DNA LT Library Preparation Kit. Each library was sequenced by the Illumina HiSeq X-ten platform (Illumina, USA) with PE150 strategy at Personal Bio- technology Co., Ltd. (Shanghai, China). Raw sequencing reads were processed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to conduct quality control. The sequencing adapters were removed from sequencing reads using Cutadapt (v1.2.1). Low-quality reads, reads with ambiguous bases were removed using a sliding-window algorithm. The sequencing reads were aligned to the host genome and host genome reads were removed. Then quality-filtered reads were de novo assembled to construct the metagenome for each sample. Scaffolds/Scaftigs sequences were constructed by megahit (https://hku-bal.github.io/megabox/) of not less than 300 bp were selected for each sample and used to predict corresponding protein sequence by the MetaGeneMark database (http://exon.gatech.edu/GeneMark) [29]. Quality-filtered data were submitted and compared to the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database and the EggNOG database. The functions of proteins were predicted and annotated. The number of bacterial reads and relative abundances were classified by the NCBI-NT database and MEGAN (http://ab.inf.uni-tuebingen.de/software/megan5/). Lefse analysis were conducted using the online website (http://huttenhower.sph.harvard.edu/galaxy/) with α = 0·05, the threshold of LDA score was at 2·0.
2.6. Gavage of Clostridium species in conventional mice
Clostridium scindens (ATCC 35704) was purchased from the American Type Culture collection and was identified by its 16 s ribosomal sequence. The strain was cultured using Brain Heart Infusion (BHI) broth under anaerobic conditions (Bactron300 Anaerobic Chamber Glovebox, Shel Lab Inc., USA). Mice were purchased from the Laboratory Animal Services center of The Chinese University of Hong Kong, Hong Kong. The animal experiment received approval from the Committee on the Use of Human & Animal Subjects in Teaching & Research at Hong Kong Baptist University. Animal experiment followed the Animals Ordinance guidelines, Department of Health, Hong Kong SAR. Twenty-one male C57BL/J mice were divided into three groups (n = 7 per group). After confirming its activity in vitro, one treatment group was gavaged daily with 100 ul of live C.scindens PBS suspension (108 CFU/ml). One treatment group was given 100 ul heat-killed C.scindens PBS suspension (108 CFU/ml). Control mice received an equivalent volume of PBS. Mice were fed normal diet (10% fat, 71% carbohydrate, and 19% protein) throughout the experiment. After two weeks of gavage, the mice were sacrificed and liver tissue were collected for BA analysis.
2.7. RNA isolation and quantitative reverse transcription PCR
Total RNA was isolated using Trizol reagent (Invitrogen, CA, USA). The cDNA templates were obtained from 500 ng of purified RNA using iScript Reverse Transcription Supermix for RT-PCR (Bio-rad, CA). 1 × SYBR Green Master Mix buffer (Takara, Otsu, Japan) was used for quantitative RT-PCR and assays were performed on a Roche lightCycler 480 II PCR machine. Gene specific primers are listed in Table S2. Targeted gene levels were normalized to housekeeping gene levels (GAPDH) and the results were analysed using the ΔΔCT analysis method [30].
2.8. Immunohistochemistry and immunofluorescence staining
To assess the histological features of liver, epididymal adipose tissue, and brown adipose tissue, the formalin-fixed tissue samples (n = 3 per group) were embedded in paraffin and stained with routine haematoxylin and eosin (H&E). For immunofluorescence staining of ileum tissues (n = 5 per group), the fresh tissues were embedded in OCT compound (Sakura, USA) and cut into frozen intestinal sections. The slides were treated with TGR5 antibody (ab72608, Abcam, RRID:AB_2112165) and GLP-1 (ab23468, Abcam, RRID:AB_470325), and were then exposed to Alexa Fluor 488 goat Anti-mouse IgG (ab150117, Abcam, RRID:AB_2688012) and Alexa Fluor 594 goat Anti-rabbit IgG (ab150080, Abcam, RRID:AB_2650602). Nuclei were stained with 4, 6-diamidino-2-phenylindole-dihydrochloride (DAPI, Sigma chemical). After PBS washing, slides were mounted using ProlongTm Gold Antifade Mountant (Life Technologies) and were photographed with a fluorescence microscope camera (Zeiss, Oberkchen, Germany). For immunohistochemistry staining of UCP-1 in BAT (n = 3 per group), brown adipose tissues were fixed in 4% paraformaldehyde and embedded in paraffin according to the standard procedures. Then slides were deparaffinized, rehydrated, and stained with UCP-1 antibody (23673–1-AP, RRID:AB_2828003, Proteintech, China). Images were acquired using a digitalized microscope camera (Nikon, Tokyo, Japan) and results were quantified using ImageJ software.
2.9. Cell culture and ELISA
The mouse colon cell line STC-1 (RRID:CVCL_J405) and the human cecum cell line NCI-H716 (RRID:CVCL_1581) were purchased from ATCC. The cells were cultured in RPMI 1640 growth medium supplemented with 10% fetal bovine serum (Gibco, USA) at 37 °C, 5% CO2. The cells were plated into a 24 well plate with an appropriate density and then exposed to vehicle (DMSO, control, less than 0·1%), or DMSO+25uM BAs. After incubation, the culture supernatants were collected for ELISA. The secreted GLP-1 was detected according to the manufacturers’ instructions by ELISA kits (Millipore, MA, USA). Serum FGF15 levels were detected using a commercial mouse Fibroblast Growth Factor 15 ELISA Kit (CSB-EL522052MO, CUSABIO, Wuhan, China).
2.10. Western blot analyses
Brown adipose tissues and liver tissues were lysed in RIPA buffer with protease inhibitors. The concentrations of the protein were quantified with the BCA protein assay kit (#23225, Thermo, California, USA). Equal amounts of protein were electrophoresed on 10% SDS-page gels and transferred to a PVDF membrane. The membranes were blocked at room temperature with 5% non-fat milk and the membranes were incubated with antibodies against UCP1 (1:1000, #14670, RRID:AB_2687530, Cell Signalling Technology, Beverly, MA), peroxisome-proliferator-activated receptor γ coactivator 1alpha (PGC1α) (1:1000, 66369–1-Ig, RRID:AB_2828002, Proteintech, China), CYP7B1 (1:1000, ab138497, RRID:AB_2828001, Abcam, Cambridge, United Kingdom), CYP8B1 (1:1000, ab191910, RRID:AB_2828000, Abcam, Cambridge, United Kingdom), FXR (ab28480, RRID:AB_726991, Abcam, Cambridge, United Kingdom), FGF15 (sc398338, RRID:AB_2827999, Santa Cruz, CA) and beta-actin (1:1000, #3700, RRID:AB_2242334, Cell Signalling Technology, Beverly, MA) at 4 °C overnight. The membranes were washed three times with TBST buffer and followed by a 2 h incubation at room temperature with HRP conjugated secondary antibody (1:5000, #7074, RRID:AB_2099233, #7076, RRID:AB_330924, Cell Signalling Technology, Beverly, MA). The signals were detected using an ECL kit (Bio-Rad, CA).
2.11. Statistical analysis
Results are presented as mean ± SD. Statistical significance was analysed using the unpaired Student's t-test. The Mann-Whitney U test was used for BA data analysis because most of the data were not normally distributed. Statistical analyses were calculated using GraphPad prism 8.0 (GraphPad software, La Jolla, CA, USA) and SPSS version 17.0 (IBM SPSS, Chicago, IL, USA). Correlations between BAs and microbiome abundances were performed using Spearman's correlation analysis with the post hoc correction using the FDR method. The (OPLS-DA) was performed using SIMCA version 13.0 (Umetrics, Umea, Sweden). Partial least squares-discriminant analysis (PLS-DA), Principal component analysis (PCA) and heatmaps depicting spearman correlation or metabolic profile were generated using R studio (RStudio, Boston, MA, USA). Differences between experimental groups were considered significant at p<0·05.
3. Results
3.1. Non-12-OH BAs were significantly decreased in the unhealthy high BMI subjects
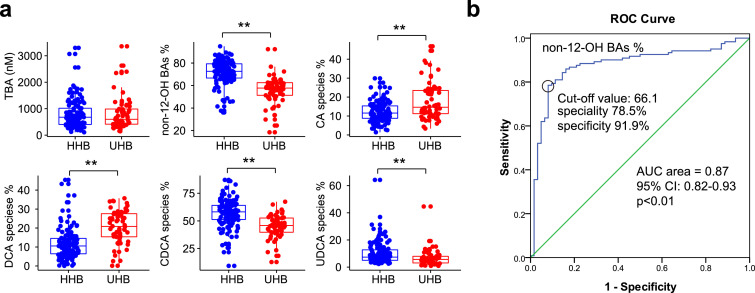
In the cross-sectional study, including 121 healthy high BMI subjects (HHB) and 62 unhealthy high BMI subjects (UHB), we observed that there were significant differences in the clinical characteristics between the HHB and UHB group. Briefly, BMI, glucose, TC, and TG were significantly increased in UHB group (Table S3). Serum BA profiles were examined in both groups. The levels of HCA, HDCA, GHDCA, UDCA, CDCA, and GUDCA were significantly decreased in the UHB group, and all these BAs belonged to non-12-OH BAs. Meanwhile, the levels of DCA, GDCA, TLCA, TDCA, and TCA were elevated in the UHB group, and most of these BAs belonged to 12-OH BAs. (Table S4). The proportions of BA species were further analysed and the results showed that UHB subjects had significantly lower non-12-OH BAs proportion while total BA concentrations were not changed between the two groups (Fig. 1a). The proportions of CA species and DCA species were obviously increased in UHB group, whereas the proportions of CDCA species and UDCA species were decreased compared with HHB subjects (Fig. 1a). Logistic regression analysis showed that the proportion of non-12-OH BAs was significantly associated with the presence of healthy high BMI after adjustment for age and gender (OR 1·129, 95% CI: 1·083–1·176, p<0·01) (logistic regression). In addition, we performed a receiver-operating characteristic (ROC) analysis and found that the proportion of non-12-OH BAs had good performance in predicting healthy high BMI subjects (AUC area = 0·87, 95% CI: 0·82–0·93, p<0·01, a cut-off value of 66·1 with a sensitivity of 78·5% and a specificity of 91·9%) (ROC curve analysis) (Fig. 1b). These results suggested that altered non-12-OH BA composition may be involved in different obesity metabolic status.
Fig. 1.
Serum BA analysis from HHB and UHB group. (a) Total BA concentrations and major BA ratio of the subjects from the HHB and UHB group. BA species include unconjugated, taurine-conjugated, and glycine-conjugated BAs. The proportions of BA species were calculated as follows: concentrations of BA species / Total BA × 100%. n = 121 in HHB group, n = 62 in UHB group. HHB: healthy high BMI group; UHB: unhealthy high BMI group. **p<0•01 (Mann–Whitney U test). (b) ROC of non-12-OH BAs% in predicting healthy high BMI. AUC: area under curve.
3.2. The different physiological features of HF-OP and HF-OR mice
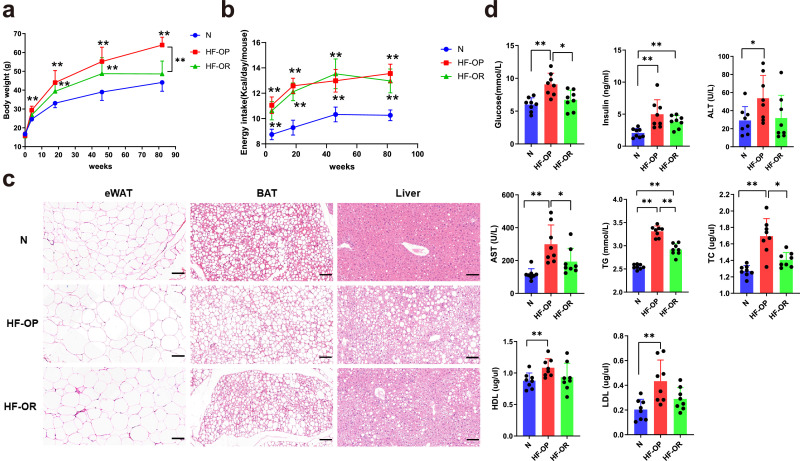
To determine the role of non-12-OH BAs in different obesity status, we established an animal model. After feeding with HFD, the body weights of HF-OP group mice were significantly different from that of the HF-OR group (Fig. 2a), while there were no significant differences in food intake between HF-OP and HF-OR groups (Fig. 2b). Compared with normal diet fed mice at each time point, HF-OP mice showed bodyweight gain progressively from the 4th week to the 82nd week of HFD. However, HF-OR mice showed a slower rate of weight gain and the body weight was significantly lower at the 82nd week compared with HF-OP group. Relative to normal diet mice group, HF-OP mice showed hypertrophy of adipocytes from epididymal white adipose tissue, higher lipid content in BAT, and more lipid droplets accumulation in liver, while the changes were mild in the HF-OR group (Fig. 2c). In addition, the HF-OP mice had significantly higher serum fasting glucose, insulin, ALT, AST, TG, TC, HDL-c, and LDL-c levels than the normal diet mice (Fig. 2d). However, lower serum glucose, AST, TG, and TC levels were observed in HF-OR group compared with HF-OP mice, suggesting HF-OR mice had a healthier overweight/obesity phenotype (HO status). Similarly, the liver TG and TC content was elevated in HF-OP group and HF-OR group, with a higher level in HF-OP group (Fig. S1).
Fig. 2.
The physiological changes in the N, HF-OP and HF-OR group. (a) Body weights and daily energy intake (b) at different time points. Data are expressed as mean ± SD (n = 8 per group). **p<0•01 (unpaired Student's t-test) compared with N group. (c) Representative images of H&E staining of eWAT, BAT and Liver sections. Scale bars, 100 μm. eWAT: epididymal white adipose tissue. (d) Serum parameters. Data are expressed as mean ± SD (n = 8 per group). *p<0•05 and **p<0•01 (unpaired Student's t-test).
3.3. Dysregulated BAs profiles in HF-OP group and a relative expansion of non-12-OH BAs proportion in the HF-OR group
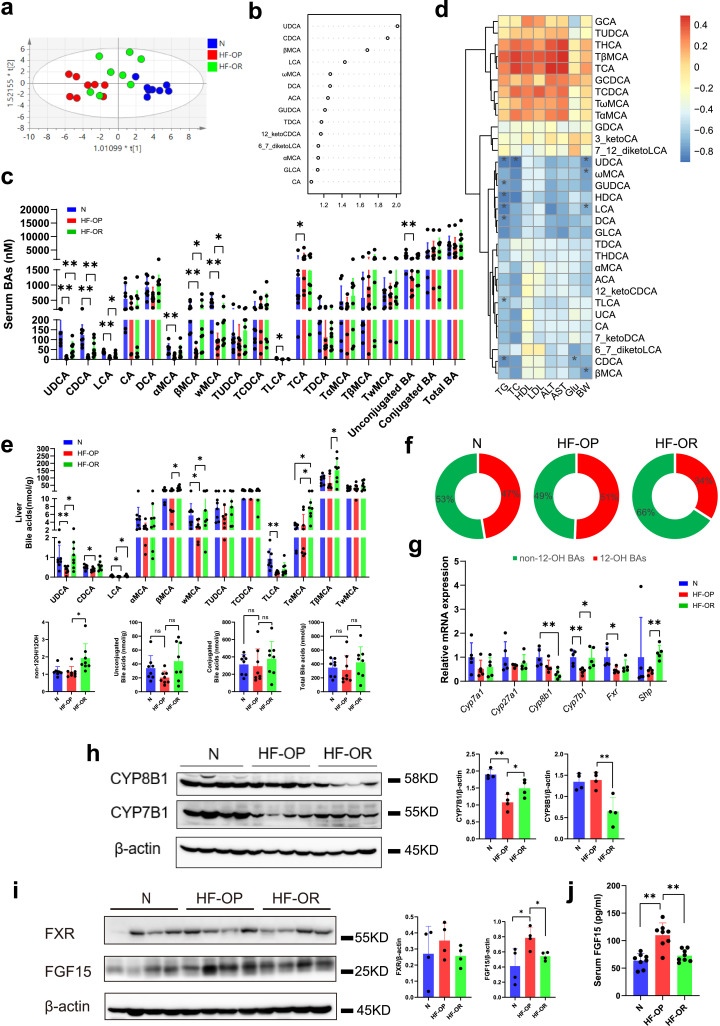
BAs play an important role in obesity as signaling molecules. To assess the BAs profile in the three groups of mice, ultra-performance liquid chromatography quadruple time of flight mass spectrometry (UPLC/QTOFMS) was applied to acquire their serum BAs profiles. We observed a clear separation between the normal diet group and HF-OP group mice from the orthogonal partial least squared-discriminant analysis (OPLS-DA) model established with the identified serum BAs. Similarly, a separation trend between the HF-OP group and HF-OR group was also observed (Fig. 3a). The variable importance in projection (VIP) scores from OPLS-DA model indicated that UDCA was the top BA that resulted in the group separation between the HF-OP and HF-OR group (Fig. 3b, 95%CI shown in Table S5). The concentrations of serum BAs are summarized in Table S6. We found that UDCA, CDCA, LCA, βMCA and ωMCA were significantly decreased in the HF-OP group and furthermore, all these differential BAs belonged to the non-12-OH BA class. In addition, TCA was increased in HF-OP group and unconjugated BAs were decreased in HF-OP group compared with N group. However, there were no significant differences in conjugated and total BAs among the three groups (Fig. 3c). To elucidate the association of serum BAs and blood biochemical parameters, Spearman correlation heatmap analyses were performed for the three groups. We found that most of the differential BAs had a significantly negative correlation with body weight or serum TG. Moreover, UDCA had a negative correlation with serum TC, and CDCA had a negative correlation with fasting glucose (Fig. 3d). Similar results were observed in the livers of the HF-OP group mice, where we found that UDCA, CDCA, LCA, and ωMCA were significantly decreased in HF-OP, whereas βMCA, TαMCA, and TβMCA were significantly increased in HF-OR group (Fig. 3e and Table S7). Interestingly, although there were no significant changes of non-12-OH/12-OH ratios between N and HF-OP group, we found that the ratios of non-12-OH/12-OH BAs in liver were significantly increased in the HF-OR group compared with the HF-OP group, while total BAs showed no significant differences (Fig. 3e). The proportion of non-12-OH BA was increased in the HF-OR group (Fig. 3f). To determine the basis for the altered BAs profile, we performed mRNA analysis of liver tissue to evaluate the expression of enzymes responsible for both classical and alternative BA synthesis. Whereas gene expression for both Cyp7a1 and Cyp27a1 were not significantly altered, Cyp8b1 expression was significantly decreased in the HF-OR group compared with the normal diet (N) group. Meanwhile, Cyp7b1 expression in the HF-OP group was decreased by half compared with the N group and the HF-OR group (Fig. 3g). Consistently, western blot results showed that CYP7B1expression was reduced in HF-OP group while CYP8B1 expression was downregulated in the HF-OR group (Fig. 3h). BA synthesis is not only regulated by the hepatic FXR/SHP pathway, but also by intestinal FXR/FGF15 signaling. We detected Fxr/Shp mRNA expression in liver and found that Fxr expression was significantly downregulated in HF-OP group compared with N group, suggesting that impaired FXR pathway might be involved in HF-OP group. However, Shp expression was increased in HF-OR group compared with HF-OR group, implying hepatic FXR activation (Fig. 3g). There were no significant changes of Pxr and Lxr mRNA expression in the liver of mice in both HF-OP and HF-OR groups (Fig. S2). In addition, we examined the impact of intestinal FXR signaling and found that the expression of FGF15 was significantly increased in the ileum tissue of the HF-OP group mice, whereas there were no significant changes in FXR expression (Fig. 3i). Accordingly, serum FGF15 was also significantly increased in the HF-OP group mice (Fig. 3j).
Fig. 3.
Dysregulated BA profiles in the HF-OP group and relative expansion of non-12-OH BAs composition in the HF-OR group. (a) Orthogonal partial least squared-discriminant analysis (OPLS-DA) scores plot of serum BA profiles showing the groupings of N (blue) group, HF-OP (red) and HF-OR (green). (b) VIP scores of OPLS-DA based on the serum BA profiles between the HF-OP and HF-OR group. A BA with VIP more than 1 was considered important in the discrimination between the groups. (c) Dysregulated BAs in the HF-OP group. The data are presented as the mean ± SD. *p<0•05 and **p<0•01 (Mann–Whitney U test). (d) Heatmap of spearman correlation coefficients between serum BAs and blood biochemical parameters from all samples in the three groups (n = 24, 8 samples per group). The gradient colours represent the correlation coefficients, with red color being more positive and blue color indicating more negative. *p<0•05 (Spearman's correlation with the post hoc correction using the Holm method). (e) Differential BAs in liver tissue with a relative expansion of non-12-OH BA composition in HF-OR group. The data are presented as the mean ± SD. *p<0•05 and **p<0•01 (Mann–Whitney U test), ns showed no significance. (f) Pie graphs show the mean percentage of non-12-OH BAs in the liver among the three groups. (g) The relative mRNA levels of Fxr, Shp and critical enzymes responsible for BA synthesis in liver among the three groups. The data are presented as the mean ± SD. **p<0•01 (unpaired Student's t-test). n = 5 per group. (h) The expression of CYP8B1 (n = 4 per group) and CYP7B1 (n = 4 per group) in liver were detected by western blot. Data are presented as the mean ± SD. *p<0•05 and **p<0•01 (unpaired Student's t-test). (i) The expression of FXR and FGF15 in ileum were detected by western blot. Data are presented as the mean ± SD. *p<0•05 (unpaired Student's t-test). (j) Serum FGF15 levels were detected by ELISA. Data are presented as the mean ± SD. **p<0•01 (unpaired Student's t-test).
3.4. Dynamic change of BAs profile in feces
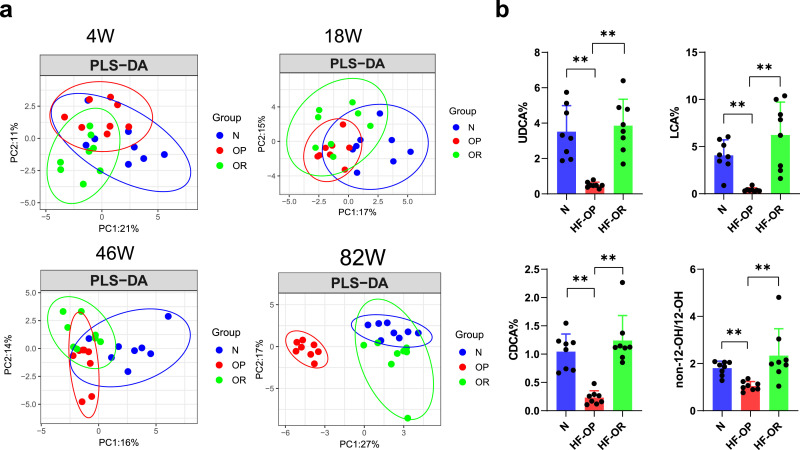
To explore the dynamic changes of fecal BAs, we collected the feces at the chosen time points of 4, 18, 46, and 82 weeks and measured the BA profile in the stool samples from the N, HF-OP and HF-OR groups. Partial least-square discriminant analysis (PLS-DA) of the fecal BA profiles were performed. The results showed that there was a large overlap of BA profiles among the three groups at week 4. HFD (HF-OP group+ HF-OR group) and normal diet group began to separate at week 18. At week 46, the BA profile from the HF-OP group was gradually separated from that of the HF-OR group. Finally, the HF-OP group displayed an obvious separation from both the N and HF-OR groups at week 82. In contrast, the scatter plot distribution of HF-OR group was close to that of the N group at week 82 (Fig. 4a). The PLS-DA model at week 82 explained the 44% variation (PC1: 27%, and PC2: 17%). Fecal BA concentrations at week 82 were analysed, and the results showed that the levels of LCA, UDCA, HDCA, and CDCA were significantly decreased in the HF-OP group compared with N and HF-OR group, while CA was increased in the HF-OP group compared with the HF-OR group (Table S8). In addition, we found that the composition percentage of UDCA, LCA, and CDCA in the HF-OP group were significantly decreased compared with the N and HF-OR groups at week 82 (Fig. 4b). Of note, we also observed that ratios of non-12-OH/12-OH BAs in HF-OR group were significantly increased at week 82 compared with the HF-OP group.
Fig. 4.
The time-dependent alteration of the fecal BAs profile. (a) Partial least squares-discriminant analysis (PLS-DA) analysis of the fecal BAs profile from different time points of the three groups (n = 8 per group). (b) Dysregulated BAs in feces of N, HF-OP and HF-OR groups at week 82. The data are presented as the mean ± SD. **p<0•01 (Mann–Whitney U test). n = 8 per group.
3.5. Dysregulated BAs were associated with gut microbiota
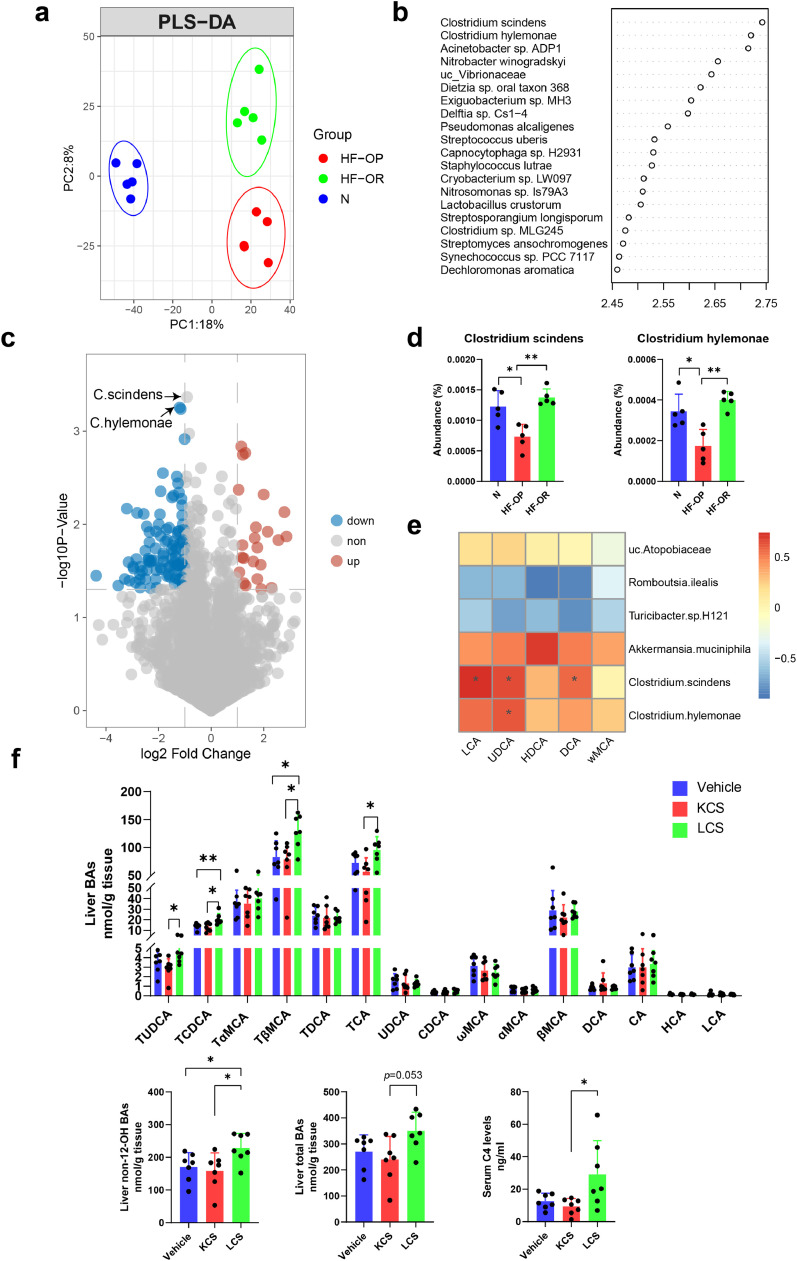
The gut microbiota is tightly associated with metabolic disorders, including obesity. Additionally, the gut microbiota has a great impact on BA composition. We speculated that the dysregulated BA profile was associated with differences in the gut microbiota. Whole-genome shotgun metagenomics sequencing was performed to compare the fecal microbiota compositions of mice in the N, HF-OP, and HF-OR groups at week 82. Whole-gene counts and α diversity (including Simpson, Chao1, ACE, and Shannon index) were found to be unchanged (Fig. S3a, b). The phyla relative abundances did not show any significant differences between the three groups (Fig. S3c). The plot from the PLS-DA analysis at the species level showed that the gut microbiota composition was significantly different among the three groups (Fig. 5a). There was an obvious separation between the HF-OR and HF-OP groups (Fig. S3d). The top 20 VIP scores from PLS-DA analysis between the HF-OP and HF-OR groups indicated that the species Clostridium scindens and Clostridium hylemonae were the top 2 gut microbiota that resulted in the group separation between the HF-OP and HF-OR groups (Fig. 5b). In addition, the species C. scindens and C. hylemonae were labelled in the volcano plots (Fig. 5c). C. scindens and C. hylemonae, both members of the Clostridium XIVa cluster, are important bacterial species for BA metabolism because they have high levels of BA bioconversion activity [31]. We found that C. scindens and C. hylemonae were significantly decreased in the HF-OP group compared with the N and HF-OR group (Fig. 5d). To further elucidate the main altered microbes, a Linear discriminant analysis effect size (Lefse) method was performed to compare gut microbiota between the HF-OP and HF-OR groups. Akkermansia muciniphila has been reported to be involved in diet-induced obesity and inversely correlates with body weight [32]. The results showed that species Akkermansia muciniphila and uc_Atopobiaceae were increased in HF-OR group, whereas the species Turicibacter H121 and Romboutsia ilealis were increased in the HF-OP group (Fig. S4). To visualize the correlation of gut microbiota and BA abundances, Spearman correlation was conducted between the main secondary BA abundances (% total) in feces and the relative abundances of differential bacteria species identified above. We found that C. scindens had significantly positive correlations with UDCA, LCA, and DCA, and C. hylemonae had a significant positive correlation with UDCA (Fig. 5e). These results suggested that gut microbiota dysbiosis may impose a substantial impact on BA composition. We further performed KEGG pathway analysis of gut microbial gene sequencing. There was an obvious separation of KEGG function as noted by the PLS-DA model among the three groups (Fig. S5a). To identify differential functional pathways, Lefse analysis of KEGG functional pathways was performed. The results showed that regulation of lipolysis in adipocytes, biosynthesis of unsaturated fatty acids, and secondary BA biosynthesis were enriched in the normal diet group, while olfactory transduction, nitrogen metabolism, and glutathione metabolism were enriched in the HF-OR group. Glutathione is involved in improving obesity related oxidative stress [33], and might be associated with glycine conjugated BA metabolism. Further, the HF-OP group gained more function of protein digestion and absorption and was prone to Cushing's syndrome as well as, endocrine and metabolic diseases, all of which suggests a link to obesity (Fig. S5b). KEGG pathway enrichment analysis showed that baiB, a key enzyme of the BA 7a-dehydroxylation pathway, was significantly enriched in the HF-OR group compared with the HF-OP group (Fig. S5c). To identify the role of intestinal C. scindens in BA regulation, we analysed the BA profile of mice that underwent C. scindens gavage after confirming its activity in vitro (Fig. S6). Interestingly, we found that C. scindens significantly enhanced the levels of TUDCA, TCDCA, TβMCA, and TCA in liver, especially the levels of non-12-OH BAs (Fig. 5f and Table S9). In addition, serum 7α‑hydroxy-4-cholesten-3-one (C4), the BA precursor, was significantly elevated in C. scindens treated mice, suggesting that it played an important role in regulation BA de novo synthesis.
Fig. 5.
Altered BA profiles were associated with gut microbiota. (a) Partial least-square discriminant analysis (PLS-DA) at species level identified by metagenomic sequencing. The HF-OP group is shown in red, the HF-OP group is shown in green, and the N group is shown in blue. PC1 and PC2 account for 18% and 8% respectively. n = 5 per group. (b) VIP scores of top 20 species in PLS-DA model between HF-OP and HF-OR group. (c) Visualization of differentially expressed species-level bacteria by volcano plots. Red dots represent significantly high abundance in the HF-OP group compared with the HF-OR group; blue dots represent significantly low abundance in the HF-OP group compared with the HF-OR group. (d) The abundance of Clostridium scindens and Clostridium hylemonae were significantly decreased in the HF-OP group. Data are presented as the mean ± SD. **p<0•01 (unpaired Student's t-test). (e) Spearman correlations of the relative BA abundance (% total) in feces with the relative abundance of differential microbial species from the samples in the group of N (n = 5), HF-OP (n = 5) and HF-OR (n = 5). The gradient colours represent the correlation coefficients, with red color being more positive and blue color indicating more negative. *p<0•05 (Spearman's correlation after the post hoc correction using the FDR method). (f) The BA profile in the mouse liver and serum C4 levels after 2 weeks Clostridium scindens gavage (n = 7 per group). The data are presented as the mean ± SD. Vehicle: PBS group; KCS: heat-killed C. scindens group; LCS: live C. scindens group. *p<0•05 and **p<0•01 (Mann–Whitney U test).
3.6. Altered BA composition was accompanied by reduction of GLP-1 levels and energy expenditure in HF-OP group mice
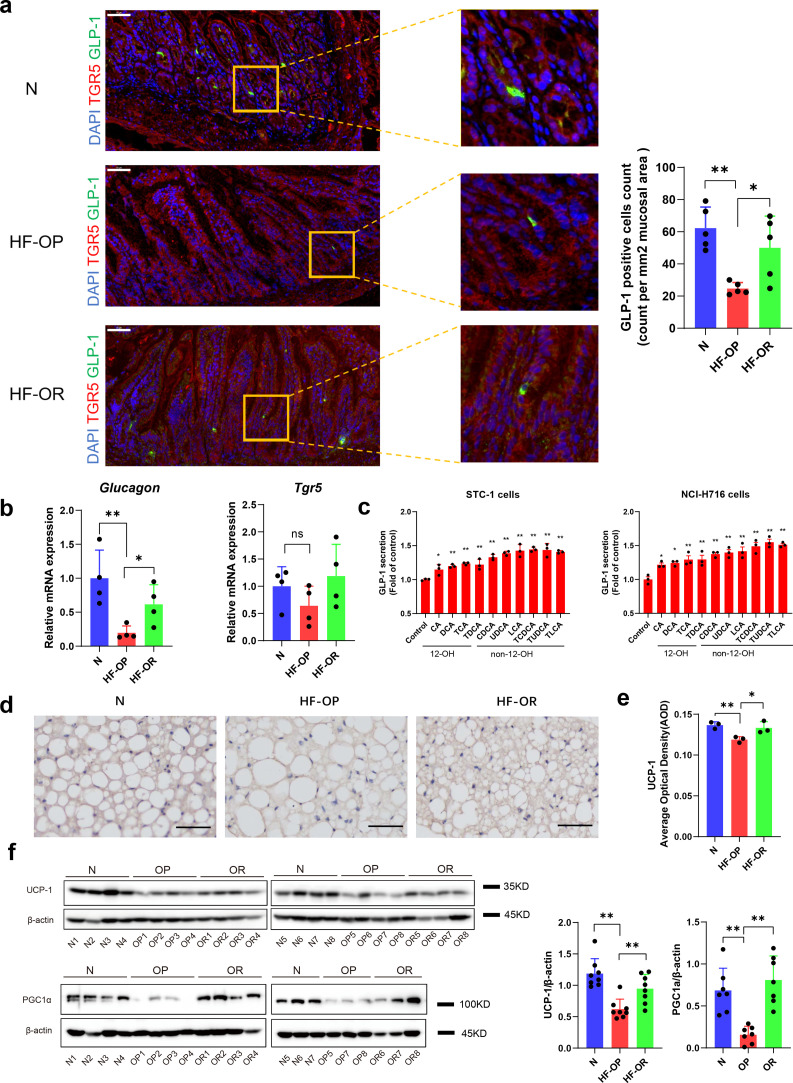
TGR5, a G protein-coupled receptor, can be activated by several BAs, and LCA is the most potent natural agonist for TGR5 [34,35]. Activation of TGR5 in enteroendocrine cells resulted in enhanced GLP-1 release, which helps to improve pancreatic β cell functions and insulin resistance. Next, we showed that ileum GLP-1 was decreased significantly in the HF-OP group using immunofluorescent staining techniques (Fig. 6a). Further, the results were confirmed using real-time PCR. The Glucagon (the precursor of GLP-1) mRNA levels were significantly downregulated in the HF-OP group, whereas the Tgr5 mRNA expression did not differ in the ileum among the three groups (Fig. 6b). Moreover, we performed an ELISA assay to assess the effects of BAs on GLP-1 secretion in vitro. The results showed that non-12-OH BAs such as CDCA, UDCA, LCA, and their taurine conjugates displayed a stronger potential to enhance GLP-1 secretion significantly in the enteroendocrine cell lines STC-1 and NCI-H716 compared with 12-OH BAs (Fig. 6c). TGR5 is expressed in BAT and it is reported that activation of TGR5 by BAs triggers an increase in energy expenditure and attenuates diet-induced obesity. We observed that the BAT marker, UCP1, was significantly decreased in the BAT of HF-OP mice as noted by immunohistochemical staining (Fig. 6d, e). Confirmed by western blot, expression of key thermogenic gene expressions in BAT, including UCP1 and PGC1α, were markedly downregulated in mice from the HF-OP group (Fig. 6f).
Fig. 6.
GLP-1 levels in ileum and energy expenditure were decreased significantly in HF-OP group. (a) Representative immunofluorescent images showing TGR5(red), GLP-1(green) in the ileum tissue of N, HF-OP and HF-OR groups. Scale bars, 50 μm. GLP-1 positive cells count was expressed as counts per mm2 of mucosal area. Five fields of view were chosen randomly per sample to calculate mean count and area, values are presented as mean ± SD. n = 5 per group. The count and area were analysed using image J software. *p<0•05 and **p<0•01 (unpaired Student's t-test). (b) The GLP-1 precursor Glucagon and Tgr5 mRNA in ileum tissue were measured using real-time PCR assay. Data are presented as the mean ± SD. *p<0•05 and **p<0•01 (unpaired Student's t-test). n = 4 per group. (c) GLP-1 secretion was detected in STC-1 and NCI-H716 cells using ELISA with treatment of BAs, and data were obtained from 3 independent experiments. *p<0•05 and **p<0•01 (unpaired Student's t-test). (d) Representative UCP1 immunostaining of BAT sections from the three groups. Scale bars, 50 μm. (e) Quantification of UCP1 immunostaining average optical density. Data are presented as the mean ± SD. n = 3 per group. *p<0•05 and **p<0•01 (unpaired Student's t-test). (f) The expression of UCP1 (n = 8 per group) and PGC-1α (n = 7 per group) in BAT were detected by western blot. Data are presented as the mean ± SD. **p<0•01 (unpaired Student's t-test).
3.7. UDCA treatment attenuated diet-induced obesity
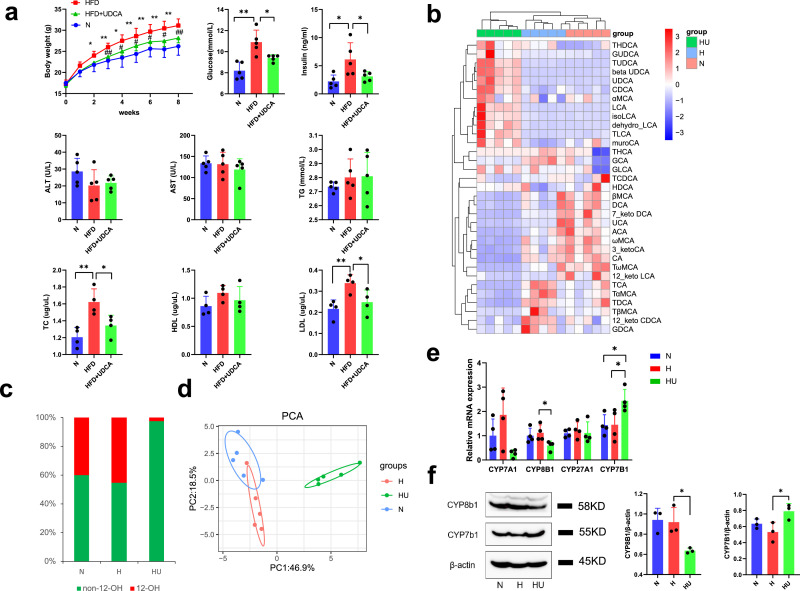
To further confirm the beneficial effects of non-12-OH BAs mediated by gut microbiota, we fed HFD mice with 0·5% UDCA (w/w), which was the most decreased BA detected in HF-OP group. Compared with mice in the HFD group, mice in the HFD+UDCA group showed a greater reduction in body weight gain (Fig. 7a). In addition, serum biochemical parameters were also improved in the HFD+UDCA group. Mice in the HFD group had higher fasting glucose, insulin, TC, HDL-c, and LDL-c levels compared with the N group, whereas the HFD+UDCA group displayed lower fasting glucose, insulin, TC, and LDL-c levels compared with HFD group (Fig. 7a). Levels of AST and ALT revealed no significant differences between the HFD group and the HFD+UDCA group, which suggested that the UDCA dose did not provoke liver inflammation or damage. The heatmap of serum BA profiles showed that oral supplementation with UDCA significantly increased non-12-OH BAs, CDCA, TLCA, LCA, and TUDCA levels, while the amounts of 12-OH BAs, CA, TCA, and TDCA were decreased (Fig. 7b). We found that the HFD+UDCA group had a significantly increased serum non-12-OH BA proportion (Fig. 7c). The principal component analysis (PCA) scores plot showed that HFD changed serum BA profiles and that the HFD+UDCA group displayed a distinct serum BA profile (Fig. 7d). To determine the basis for the altered BA profile, liver enzyme mRNAs responsible for BA synthesis including Cyp7a1, Cyp8b1, Cyp27a1, and Cyp7b1 were detected by qPCR. Compared with the HFD group, the HFD+UDCA group had a substantial reduction of Cyp8b1 mRNA expression while there was a significant increase in Cyp7b1 mRNA expression (Fig. 7e). The results were further confirmed by western blot (Fig. 7f). CYP8B1 is a critical enzyme for synthesis of CA, one of the main subtypes in the class of 12-OH BAs. CYP7B1 is a critical enzyme contributing to non-12-OH BA synthesis. Therefore, our data suggested that UDCA treatment mainly increased the alternative BAs synthesis pathway and suppressed the classical BA synthesis pathway and thus, contributed to the non-12-OH BAs proportion expansion.
Fig. 7.
UDCA treatment attenuated obesity induced by high fat diet. (a) UDCA improved the metabolic profile: changes of body weight and serum paraments. Data are presented as the mean ± SD. n = 5 per group. *p<0•05 and **p<0•01 (unpaired Student's t-test), HFD group vs N group; #p <0•05 and ##p<0•01 (unpaired Student's t-test), HFD+UDCA group vs HFD group. (b) Heatmap of serum bile acids profile of normal diet group (N), high fat diet group (H) and high fat diet + UDCA group (HU). The gradient colours in the heatmap depicted the z-scale value of serum bile acids concentration. (c) The mean percentage of 12-OH bile acids and non-12 bile acids in serum from all the samples of the three group (n = 5 per group). (d) The principal component analysis (PCA) analysis of serum bile acids. n = 5 per group. N: normal diet group; H: high fat diet group; HU: high fat diet group+0•5%UDCA group. (e) Relative mRNA levels of Cyp7a1, Cyp8b1, Cyp27a1 and Cyp7b1 in liver detected by q-PCR assay. All data are presented as the mean ± SD. n = 4 per group. *p<0•05 (unpaired Student's t-test). (f) The expression of CYP8B1 and CYP7B1 in liver of three group mice were detected by western blot. Data are presented as the mean ± SD. n = 3 per group. *p<0•05 (unpaired Student's t-test).
4. Discussion
In the study, we examined the role of BA signaling mediated by the gut microbiota and how changes in BA composition affect host metabolism. There are many enzymes in the liver involved in the synthesis of BA from cholesterol. Among them, CYP7A1 is the rate-limiting enzyme for BA synthesis, whereas CYP8B1 determines the ratio of CA to CDCA [11]. We found that mice in the HF-OR had a significant reduction of CYP8B1, thereby causing relative expansion of the non-12-OH BAs proportion in the HF-OR group. Recently, several studies have reported that depletion or downregulation of CYP8B1 may exert beneficial effect to host metabolic status. CYP8B1−/- mice are resistant to western diet-induced obesity and hepatic steatosis and insulin resistance by reducing lipid absorption [36]. It has been shown that loss of CYP8B1 in mice improved glucose tolerance by increasing GLP-1 secretion [37]. Moreover, Li. et al. showed that depletion of a liver-enriched long non-coding RNA could downregulate CYP8B1 expression and increased the conjugated MCA/CA ratio, thus enhancing apoC2 expression and improving lipid metabolism [38]. In mouse liver, CDCA is converted to α- and β-MCA. Our results also showed that TαMCA and TβMCA, as potential FXR antagonists, were significantly increased in the liver of the mice from HF-OR group compared with the mice from HF-OP group, leading to intestinal FGF15 signaling inhibition once they were released into intestine. CYP8B1 has been shown to be downregulated in mice that have undergone vertical sleeve gastrectomy (VSG), a popular weight-loss surgery to combat morbid obesity, and this TGR5 dependent regulation contributes to BA pool shifts after VSG [39]. Depletion or downregulation of CYP8B1 expression would eliminate or decrease the amount of 12-OH BAs formed and thus increase the non-12-OH/12‑hydroxy BAs ratio. CA is a highly efficient BA in mixed micelle formation for dietary fat and cholesterol absorption, altered BA composition consisting of increased MCA and UDCA make the BA pool more hydrophilic, thus resulting in reduced fat absorption [40]. A previous study using germ-free and conventional mice showed that altered BA hydrophobicity through BSH deconjugation induced the transcriptional expression of key genes involved in lipid metabolism and transportation [41]. It has also been reported that TDCA, derived from CA, is more likely to increase micelle size and aggregation number, due to a lowering of the relative hydrophilic-lipophilic balance (HLB) [42]. Another study showed that trihydroxy-TCA proved superior in promoting cholesterol uptake compared to dihydroxy-TCDCA [43]. BAs promote lipid absorption by increasing paracellular transport, epithelial membrane fluidity, and opening of tight junctions [44]. Furthermore, altered BA profiles were associated with cholesterol metabolism, as evidenced by the fact that some secondary BAs were positively correlated with plasma simvastatin concentration, predicting individual responses to statin treatment [45].
Additionally, CYP7B1, a key enzyme in the alternate pathway of BA synthesis, was increased in the HF-OR group compared with the HF-OP group. Cold exposure has been reported to trigger a metabolic program and enhance energy expenditure, and this process is partially mediated by CYP7B1 [46]. Lower hepatic CYP7B1 expression was observed in obese patients with T2DM, and knockout of CYP7B1 significantly downregulated UCP-1 expression in BAT, suggesting that CYP7B1 derived BAs might exhibit higher TGR5 receptor activation ability. Conversely, AAV-mediated CYP7B1 overexpression in the liver caused higher energy expenditure in BAT and more O2 consumption in vivo [46]. Similarly, in the present study, UCP-1 and GLP-1 expression were downregulated in the HF-OP group, and we showed that non-12-OH BAs showed a stronger ability to enhance GLP-1 secretion in vitro.
BA synthesis is regulated not only by intestinal FXR/FGF15 signaling, but also by negative feedback of hepatic FXR/SHP pathway. We therefore examined intestinal and hepatic FXR signaling. FGF15, produced by ileal enterocytes after FXR activation, is secreted into the portal vein to reach the liver, where it binds to FGFR4/β-Klotho receptor complex to inhibit CYP7A1 activity [47]. In the study, we observed that HF-OP mice had increased FGF15 levels. However, the expression of Cyp7a1 only had a downregulated trend in HF-OP group without statistical significance. One possible mechanism to explain this phenomenon is that inhibition of intestinal FGF15 signaling on CYP7A1 is partially abolished by dysregulated hepatic FXR signaling, which leads to less feedback inhibition of CYP7A1. It has been reported that levels of FXR protein and mRNA were decreased in patients with NAFLD [48]. Accordingly, we observed that Fxr was decreased in the HF-OP group mice with obvious hepatic steatosis. FXR activation lowers hepatic lipids accumulation, due to reducing the lipogenic genes and their regulators including fatty acid synthase (FASN), stearoyl-COA desaturase (SCD), and sterol regulatory element-binding protein-1c (SREBP-1c) [49]. FXR plays an essential role in glucose homeostasis because FXR-null mice develop severe fatty liver and elevated circulating free fatty acids, leading to peripheral insulin resistance [50]. Reduced Fxr expression and its most potential agonist CDCA might have a dual effect in aggravating metabolic disorder in HF-OP mice, thus contributing to the obesity prone phenotype.
In this study, we found that UDCA treatment could upregulate the expression of CYP7B1 and downregulated CYP8B1. Different BAs have differential effects on BA signaling. Consistent with our results, it has been reported that low dose (0.1% and 0.3%) UDCA administration increased CYP7B1 expression and downregulated CYP8B1 expression [51], suggesting that UDCA might play an important role in promoting changes in non-12-OH/12-OH bile BA ratio. Another study showed that UDCA administration stimulated BA synthesis by reducing FGF15, accompanied by elevated serum C4 [52]. UDCA and LCA are derived from CDCA via gut microbiota metabolism. We found that two members of the Clostridium XIVa cluster, C. scindens and C. hylemonae species were approximately twofold lower in the HP-OP group compared with the N and HF-OR group. In addition to 7a/β-dehydroxylating activity, 7α/β-hydroxysteroid dehydrogenases were detected among members of the genus Clostridium including C. scindens [53], suggesting that the epimerization of CDCA to UDCA might be mediated by these bacteria. In addition, 7α/β-hydroxysteroid dehydrogenases have higher affinities for dihydroxy-BAs (CDCA) than for trihydroxy-BAs (CA) [53]. These studies supported our finding that there was a significantly positive correlation between bacteria with UDCA, LCA, and DCA. Furthermore, baiB, encodes BA CoA ligase, a key enzyme for 7α-dehydroxylation to produce LCA from CDCA [53]. Oral supplementation of CDCA increased human brown adipose tissue activity with increased energy expenditure via TGR5 [54]. Among the natural endogenous BAs, LCA is considered the most potent ligand for TGR5 [55]. Binding of an agonist to the TGR5 receptor in enteroendocrine L cells stimulated GLP-1 release, thereby affecting metabolic homeostasis [56]. GLP-1, is an incretin secreted by enteroendocrine L cells and plays a critical role in obesity by alleviating hepatic steatosis via induction of fatty acid oxidation and reduction of lipogenesis [57]. UDCA has been proven to have clinical effects for treating various hepatic diseases including non-alcoholic steatohepatitis [58]. We found that UDCA not only enhanced GLP-1 release in vitro, but also alleviated HFD-induced obesity by oral administration. In agreement with our study, Murakami et al. previously showed that UDCA stimulated GLP-1 secretion in healthy subjects [59]. In addition to exhibiting affinity for TGR5, UDCA has been considered an FXR antagonist, and inhibition of FXR in enteroendocrine cells could also enhance GLP- 1 secretion [60,61]. Gavage with a mixture of UDCA and LCA reduced hyperlipidaemia, and treatment with a bacterial strain capable of elevating UDCA, LCA and succinate levels, could alleviate obesity, hyperglycaemia, and hepatic steatosis in HFD-fed mice [62]. C. scindens has been shown to be associated with resistance to Clostridium difficile infection in a secondary BA dependent manner [63]. Meanwhile, C. scindens has also been shown to be associated with ameliorated chronic inflammatory enteropathy [64]. And in strain background obesity prone or obesity resistant mice, C. scindens showed strong negative correlation with body weight [65]. Our previous study showed that oral gavage of C. scindens in C57BL/6 J mice reduced FGF15 expression in the ileum and increased serum C4 levels as well as CYP7A1 expression [28]. It has been shown that inhibition of intestinal FXR/FGF15 signaling exerts beneficial effects on obesity associated metabolic dysfunction [24]. Additionally, CYP7A1 is regulated more strongly by intestinal FXR/FGF15 pathway while CYP8B1 is more sensitive to FXR activation in liver [66]. Injection of FGF15 in wild-type mice significantly suppressed CYP7A1 expression but did not affect CYP8B1 [66]. A previous study showed that germ free (GF) mice had reduced FGF15 levels and predominantly increased non-12-OH BAs [22]. Meanwhile, CYP7A1 and CYP7B1 were increased in GF mice compared with conventionally raised mice whereas CYP8B1 was not [22]. As reported in our previous study [28], after C. scindens gavage, CYP7A1 expression was significantly increased while there was no obvious change in CYP8B1 expression. Taken together, these studies suggest that inhibition of intestinal FGF15 signaling enhances hepatic BA synthesis, especially via the alternative pathway. We also observed significantly increased serum C4 concentrations, and BAs, especially non-12-OH BAs in the liver of the C. scindens treated mice model. In addition, reduced Clostridia has been observed in the intestines of humans with obesity and type 2 diabetes [67]. A recent important finding has been reported that colonization with Clostridia protects against obesity via T cell-mediated regulation of the microbiota and downregulation of CD36, a receptor responsible for uptake of long-chain fatty acids (LCFs) [68].
When energy intake exceeds energy expenditure, causing an imbalance of energy metabolism, obesity is likely to develop. Therefore, increasing energy expenditure is a potential strategy for controlling obesity. We observed that UCP-1 expression was significantly decreased in the HF-OP group. It has been shown previously that UCP1 activity had a determinative role for obesity development in mice [69]. Thermogenesis mediated by UCP-1 protein in brown adipose tissues can combat obesity and UCP1 expression can be regulated by BA. For example, CDCA, LCA, and other TGR5 agonists increase UCP1 expression in brown adipocytes, implying TGR5 is critical to mediate the process [54]. Another study showed that adding BA to HFD reduced adiposity and hepatic lipogenesis and improved glucose tolerance in WT but not in UCP-1 KO mice [70]. Moreover, the GLP-1 receptor is expressed in BAT, and the GLP-1 receptor agonist liraglutide stimulates BAT thermogenesis and adipocyte browning by enhancing UCP-1 expression [71]. In the present study, we found that UCP1 and PGC1α were significantly downregulated in the HF-OP group mice. The PGC1α → UCP1 signaling pathway is mediated by TGR5 as evidenced by the fact that administration of BA mimetic (INT-777, specific agonist for TGR5) increased mitochondrial counts and induced UCP-1 exclusively in TGR5+/+ mice but not TGR5 knockout mice [72], thus, illustrating that this process was TGR5 dependent. BAs metabolized by bacterial 7a-dehydroxylation were found to have a much stronger activation effect on TGR5 than primary BAs, and TGR5 activation is one of the proposed mechanisms to ameliorate obesity. Interestingly, TGR5 knockout had decreased CYP7B1 expression [73], suggesting that TGR5 signaling might regulate the expression of CYP7B1.
There are several limitations in this study. First, metabolic cages were not used to measure calorimetry to calculate energy expenditure. Second, we did not measure short chain fatty acids in the intestinal contents, which may also affect GLP-1 secretion. In addition, lipids excretion in feces were not investigated. Further studies would be required to fully understand the mechanistic link between the gut microbiota dysbiosis-induced changes of BA composition with metabolic status.
To summarize, in this study we found that changes in BA signaling mediated by gut microbiota altered host metabolism and contributed to obesity status. The close links between BAs and metabolic status raises the possibility that modulation of BAs could be a promising strategy for obesity therapy.
Declaration of Competing Interest
The authors declare that they have no conflicts of interests.
Acknowledgments
Acknowledgement
We thank all participants who provided written informed consent and donated specimens for this study. We thank Prof. Zhaoxiang Bian and Dr. Ling Zhao (Hong Kong Baptist University) for providing C.scindens manipulation mice specimens and valuable support.
Funding sources
This work was supported by National Key R and D Program of China (2017YFC0906800), National Natural Science Foundation of China (81772530, 81974073), the National Institutes of Health/National Cancer Institute Grant 1U01CA188387-01A1, and the China Scholarship Council (No. 201706230089). The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report.
Author contributions
Wei Jia was principal investigator of this study. Zhaoxiang Bian provided valuable support for C. scindens gavage animal experiment. Wei Jia, Aaihua Zhao, Xiaojiao Zheng, and Guoxiang Xie designed the study. Meilin Wei conducted key experiments of the study and perform the data analysis and drafted the manuscript. Fengjie Huang, Yunjing Zhang, Wei Yang, and Ling Zhao conducted the animal experiments. Kun Ge, Chun Qu, Mengci Li, Shouli Wang, and Xiaolong Han helped to perform the experiments and collected the data. Wei Jia and Cynthia Rajani revised the manuscript.
Data sharing
The metagenomic sequences were provided and available at NCBI Sequence Read Archive (SRA) repository with accession code PRJNA616396. Datasets and R-code related to this study can be found at https://github.com/mweixq/OPOR.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102766.
Contributor Information
Zhaoxiang Bian, Email: bzxiang@hkbu.edu.hk.
Wei Jia, Email: wjia@cc.hawaii.edu, xie26guo@gmail.com.
Appendix. Supplementary materials
References
- 1.Castillo J.J., Orlando R.A., Garver W.S. Gene-nutrient interactions and susceptibility to human obesity. Genes Nutr. 2017;12(1):29. doi: 10.1186/s12263-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stefan N., Häring H.-.U., Hu F.B., Schulze M.B. Metabolically healthy obesity: epidemiology, mechanisms, and clinical implications. Lancet Diabetes & Endocrinol. 2013;1(2):152–162. doi: 10.1016/S2213-8587(13)70062-7. [DOI] [PubMed] [Google Scholar]
- 3.Levin B.E., Dunn-Meynell A.A., Balkan B., Keesey R. Selective breeding for diet-induced obesity and resistance in Sprague-Dawley rats. Am J Physiol. 1997;273(2):R725–RR30. doi: 10.1152/ajpregu.1997.273.2.R725. [DOI] [PubMed] [Google Scholar]
- 4.Gu Y., Liu C., Zheng N., Jia W., Zhang W., Li H. Metabolic and gut microbial characterization of obesity-prone mice under a high-fat diet. J Proteome Res. 2019;18(4):1703–1714. doi: 10.1021/acs.jproteome.8b00945. [DOI] [PubMed] [Google Scholar]
- 5.Huang X.-.F., Zavitsanou K., Huang X. Dopamine transporter and D2 receptor binding densities in mice prone or resistant to chronic high fat diet-induced obesity. Behav Brain Res. 2006;175(2):415–419. doi: 10.1016/j.bbr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 6.Surwit R.S., Wang S., Petro A.E. Diet-induced changes in uncoupling proteins in obesity-prone and obesity-resistant strains of mice. Proc Natl Acad Sci U S A. 1998;95(7):4061–4065. doi: 10.1073/pnas.95.7.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang S., Graham B., Yakubu F., Lin D., Peters J., Hill J. Metabolic differences between obesity-prone and obesity-resistant rats. Am J Physiol Regul Integrat Compar Physiol. 1990;259(6):R1103–R1R10. doi: 10.1152/ajpregu.1990.259.6.R1103. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Xie Z., Lin J. Transcriptomic and metabonomic profiling of obesity-prone and obesity-resistant rats under high fat diet. J Proteome Res. 2008;7(11):4775–4783. doi: 10.1021/pr800352k. [DOI] [PubMed] [Google Scholar]
- 9.Geiger B.M., Behr G.G., Frank L.E. Evidence for defective mesolimbic dopamine exocytosis in obesity-prone rats. The FASEB J. 2008;22(8):2740–2746. doi: 10.1096/fj.08-110759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang J.Y. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlström A., Sayin S.I., Marschall H.-.U., Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Ma H., Patti M.E. Bile acids, obesity, and the metabolic syndrome. Best Pract Res Clin Gastroenterol. 2014;28(4):573–583. doi: 10.1016/j.bpg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haeusler R.A., Astiarraga B., Camastra S., Accili D., Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes. 2013;62(12):4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brufau G., Stellaard F., Prado K. Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology. 2010;52(4):1455–1464. doi: 10.1002/hep.23831. [DOI] [PubMed] [Google Scholar]
- 15.Fiorucci S., Mencarelli A., Palladino G., Cipriani S. Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharmacol Sci. 2009;30(11):570–580. doi: 10.1016/j.tips.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Thomas C., Gioiello A., Noriega L. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe M., Houten S.M., Mataki C. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 18.Perino A., Pols T.W.H., Nomura M., Stein S., Pellicciari R., Schoonjans K. TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation. J Clin Invest. 2014;124(12):5424–5436. doi: 10.1172/JCI76289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubara T., Li F., Gonzalez F.J. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2013;368(1–2):17–29. doi: 10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cipriani S., Mencarelli A., Palladino G., Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res. 2010;51(4):771–784. doi: 10.1194/jlr.M001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bäckhed F., Ding H., Wang T. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayin S.I., Wahlström A., Felin J. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Jiang C., Xie C., Li F. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C., Xie C., Lv Y. Intestine-selective farnesoid x receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6(1):1–18. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni Y., Zhao L., Yu H. Circulating unsaturated fatty acids delineate the metabolic status of obese individuals. EBioMedicine. 2015;2(10):1513–1522. doi: 10.1016/j.ebiom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S., Maitra U. Chemistry and biology of bile acids. Curr Sci. 2004;87(12):1666–1683. [Google Scholar]
- 27.Xie G., Zhong W., Li H. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013;27(9):3583–3593. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., Yang W., Chen Y. A Clostridia-Rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J Clin Invest. 2019;130(1) doi: 10.1172/JCI130976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Ridlon J.M., Harris S.C., Bhowmik S., Kang D-J, Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7(1):22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everard A., Belzer C., Geurts L. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mytilineou C., Kramer B.C., Yabut J.A. Glutathione depletion and oxidative stress. Parkinsonism Relat Disord. 2002;8(6):385–387. doi: 10.1016/s1353-8020(02)00018-4. [DOI] [PubMed] [Google Scholar]
- 34.Porez G, Prawitt J, Gross B, Staels BJJolr. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease. J Lipid Res. 2012:jlr. R024794. [DOI] [PMC free article] [PubMed]
- 35.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7(8):678. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 36.Bertaggia E., Jensen K.K., Castro-Perez J. Cyp8b1 ablation prevents Western diet-induced weight gain and hepatic steatosis because of impaired fat absorption. Am J Physiol Endocrinol Metabolism. 2017;313(2):E121–EE33. doi: 10.1152/ajpendo.00409.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur A., Patankar J.V., de Haan W. Loss of CYP8B1 improves glucose homeostasis by increasing GLP-1. Diabetes. 2015;64(4):1168–1179. doi: 10.2337/db14-0716. [DOI] [PubMed] [Google Scholar]
- 38.Li P., Ruan X., Yang L. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21(3):455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGavigan A.K., Garibay D., Henseler Z.M. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut. 2017;66(2):226–234. doi: 10.1136/gutjnl-2015-309871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D.Q.-.H., Tazuma S., Cohen D.E., Carey M.C. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: studies in the gallstone-susceptible mouse. Am J Physiol Gastrointestinal Liver Physiol. 2003;285(3):G494–G502. doi: 10.1152/ajpgi.00156.2003. [DOI] [PubMed] [Google Scholar]
- 41.Joyce S.A., MacSharry J., Casey P.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci. 2014;111(20):7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuoka K., Suzuki M., Honda C., Endo K., Moroi Y. Micellization of conjugated chenodeoxy-and ursodeoxycholates and solubilization of cholesterol into their micelles: comparison with other four conjugated bile salts species. Chem Phys Lipids. 2006;139(1):1–10. doi: 10.1016/j.chemphyslip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Nordskog B.K., Phan C.T., Nutting D.F., Tso P. An examination of the factors affecting intestinal lymphatic transport of dietary lipids. Adv. Drug Deliv. Rev. 2001;50(1–2):21–44. doi: 10.1016/s0169-409x(01)00147-8. [DOI] [PubMed] [Google Scholar]
- 44.Pavlović N., Goločorbin-Kon S., Ðanić M. Bile acids and their derivatives as potential modifiers of drug release and pharmacokinetic profiles. Front Pharmacol. 2018;9:1283. doi: 10.3389/fphar.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaddurah-Daouk R., Baillie R.A., Zhu H. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS ONE. 2011;6(10) doi: 10.1371/journal.pone.0025482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worthmann A., John C., Ruhlemann M.C. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med. 2017;23(7):839–849. doi: 10.1038/nm.4357. [DOI] [PubMed] [Google Scholar]
- 47.Inagaki T., Choi M., Moschetta A. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z.-.X., Shen W., Sun H. Effects of nuclear receptor FXR on the regulation of liver lipid metabolism in patients with non-alcoholic fatty liver disease. Hepatol Int. 2010;4(4):741–748. doi: 10.1007/s12072-010-9202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe M., Houten S.M., Wang L. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113(10):1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma K., Saha P.K., Chan L., Moore D.D. Farnesoid x receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116(4):1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song P., Rockwell C.E., Cui J.Y., Klaassen C.D. Individual bile acids have differential effects on bile acid signaling in mice. Toxicol Appl Pharmacol. 2015;283(1):57–64. doi: 10.1016/j.taap.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller M., Thorell A., Claudel T. Ursodeoxycholic acid exerts farnesoid x receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol. 2015;62(6):1398–1404. doi: 10.1016/j.jhep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridlon J.M., Kang D-J, Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Broeders E.P., Nascimento E.B., Havekes B. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 2015;22(3):418–426. doi: 10.1016/j.cmet.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Sato H., Macchiarulo A., Thomas C. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure− activity relationships, and molecular modeling studies. J Med Chem. 2008;51(6):1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- 56.Pols T.W., Noriega L.G., Nomura M., Auwerx J., Schoonjans K. The bile acid membrane receptor TGR5: a valuable metabolic target. Dig Dis. 2011;29(1):37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding X., Saxena N.K., Lin S., Gupta N.A., Anania F.A. Exendin-4, a glucagon-like protein-1 (GLP-1) receptor agonist, reverses hepatic steatosis in ob/ob mice. Hepatol. 2006;43(1):173–181. doi: 10.1002/hep.21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratziu V., De Ledinghen V., Oberti F. A randomized controlled trial of high-dose ursodesoxycholic acid for nonalcoholic steatohepatitis. J Hepatol. 2011;54(5):1011–1019. doi: 10.1016/j.jhep.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 59.Murakami M., Une N., Nishizawa M., Suzuki S., Ito H., Horiuchi T. Incretin secretion stimulated by ursodeoxycholic acid in healthy subjects. Springerplus. 2013;2(1):20. doi: 10.1186/2193-1801-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trabelsi M.S., Daoudi M., Prawitt J. Farnesoid x receptor inhibits glucagon-like peptide-1 production by enteroendocrine l cells. Nat Commun. 2015;6:7629. doi: 10.1038/ncomms8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun L., Xie C., Wang G. Gut microbiota and intestinal fxr mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang K., Liao M., Zhou N. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26(1):222–235. doi: 10.1016/j.celrep.2018.12.028. e5. [DOI] [PubMed] [Google Scholar]
- 63.Buffie C.G., Bucci V., Stein R.R. Precision microbiome reconstitution restores bile acid mediated resistance to clostridium difficile. Nature. 2015;517(7533):205. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S., Martins R., Sullivan M.C. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7(1):126. doi: 10.1186/s40168-019-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ussar S., Griffin N.W., Bezy O. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015;22(3):516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim I., Ahn S.-.H., Inagaki T. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48(12):2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Qin J., Li Y., Cai Z. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 68.Petersen C., Bell R., Klag K.A. T cell–mediated regulation of the microbiota protects against obesity. Science. 2019;365(6451):eaat9351. doi: 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feldmann H.M., Golozoubova V., Cannon B., Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9(2):203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 70.Zietak M., Kozak L.P. Bile acids induce uncoupling protein 1-dependent thermogenesis and stimulate energy expenditure at thermoneutrality in mice. Am J Physiol Endocrinol Metab. 2016;310(5):E346–E354. doi: 10.1152/ajpendo.00485.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beiroa D., Imbernon M., Gallego R. GLP-1 agonism stimulates brown adipose tissue thermogenesis and browning through hypothalamic AMPK. Diabetes. 2014;63(10):3346–3358. doi: 10.2337/db14-0302. [DOI] [PubMed] [Google Scholar]
- 72.Velazquez-Villegas L.A., Perino A., Lemos V. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-017-02068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donepudi A.C., Boehme S., Li F., Chiang J.Y. G-protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis in mice. Hepatology. 2017;65(3):813–827. doi: 10.1002/hep.28707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.