Summary
This work investigates the relationship between high-glucose (HG) culture, CpG methylation of genes involved in cell signaling pathways, and the regulation of carbohydrate and lipid metabolism in hepatocytes. The results indicate that HG leads to an increase in nuclear 25-hydroxycholesterol (25HC), which specifically activates DNA methyltransferase-1 (DNMT1), and regulates gene expression involved in intracellular lipid metabolism. The results show significant increases in 5mCpG levels in at least 2,225 genes involved in 57 signaling pathways. The hypermethylated genes directly involved in carbohydrate and lipid metabolism are of PI3K, cAMP, insulin, insulin secretion, diabetic, and NAFLD signaling pathways. The studies indicate a close relationship between the increase in nuclear 25HC levels and activation of DNMT1, which may regulate lipid metabolism via DNA CpG methylation. Our results indicate an epigenetic regulation of hepatic cell metabolism that has relevance to some common diseases such as non-alcoholic fatty liver disease and metabolic syndrome.
Subject Areas: Biological Sciences, Molecular Genetics, Systems Biology
Graphical Abstract
Highlights
-
•
High glucose induces lipids accumulation via epigenetic regulation
-
•
High glucose induces lipids accumulation via DNA CpG methylation in promoter regions
-
•
High glucose induces lipid accumulation by inhibiting multiple signaling pathways
-
•
25-Hydroxycholesterol is an endogenous agonist of DNMT1, an epigenetic regulator
Biological Sciences; Molecular Genetics; Systems Biology
Introduction
A high-glucose (HG) diet has been associated with metabolic syndrome (Moreno-Fernandez et al., 2018, Zhao et al., 2016), non-alcoholic fatty liver disease (NAFLD) (Liu et al., 2018), type II diabetes (Menegazzo et al., 2015, Vallon and Thomson, 2017), and cardiovascular disease (Che et al., 2018). A HG diet can induce fatty acid accumulation in liver, elevate insulin resistance index, cause oxidative stress, and increase body weight. This has led to the association of a HG diet with the development of the metabolic syndrome (Du et al., 2010). Hepatocytes play an important role in metabolizing carbohydrates and lipids in the body. Their function changes depending on the circulating levels of insulin and the sensitivity of the insulin receptor. The inability of insulin to suppress hepatic glucose production is a key defect found in metabolic syndrome (Balland et al., 2019, O-Sullivan et al., 2015). The role of insulin has been well documented. Hepatocytes respond to elevated glucose levels by packaging it into glycogen. When carbohydrates are in excess, hepatocytes can convert this excess energy into lipids. When glucose levels drop and insulin production falls, the shortage of insulin in the blood signals the hepatocytes to utilize their storage by hydrolyzing glycogen and lipids. Dysregulation of this process can result in lipid accumulation in hepatocytes, leading to the development of metabolic disorders, including NAFLD and type II diabetes (Saltiel, 2016). HG in culture media induces lipid accumulation in hepatocytes, which has been used as an in vitro model for the study of NAFLD (Swaminathan et al., 2013). The detailed biochemical mechanisms that allow for HG to produce lipid accumulation are not fully understood.
Epigenetic modification plays a major role in the interpretation of genetic information. Methylation at position 5 of cytosine (5-methylcytosine, 5mC) in DNA is an important epigenetic modification that regulates gene expression and other functions of the genome (Moore et al., 2013). Cytosine methylation in CpG is inversely correlated with transcriptional activity of associated genes as it causes chromatin condensation and thus gene silencing (Yu et al., 2014). Recently published literature has demonstrated that dysregulation of gene expression is important in metabolism and can affect tissue function and in turn the metabolic state (Castellano-Castillo and Moreno-Indias, 2019). It has been shown that HG levels affected gene expression through DNA methylation (Wang et al., 2018). However, the molecular mechanisms by which HG levels lead to DNA methylation, the changes of gene expression patterns, and lipid accumulation in human hepatocytes are not well understood.
Oxysterols are oxidized products of cholesterol. They are short-lived but can be potent biologically active molecules involved in a plethora of functions including lipid metabolism and inflammatory responses (Cyster et al., 2014, McDonald and Russell, 2010). Oxysterols have long been known for their important role in cholesterol homeostasis, where they are involved in both transcriptional and posttranscriptional mechanisms in controlling cholesterol levels (Luu et al., 2016). Recent research with oxysterols has demonstrated their novel and sometimes surprising role associated with a wide variety of cellular functions (Vurusaner et al., 2016). They act as ligands for a growing list of receptors, including some that are of importance to the immune system and lipid metabolism. Oxysterols have also been implicated in several diseases, such as NAFLD and other metabolic syndromes. However, until now their specific binding sites and mechanism of action remain unclear. The objective of this study is to investigate the role of a specific oxysterol (25-hydroxycholesterol [25HC]) in the epigenetic regulation associated with HG induction of lipid accumulation in human hepatocytes.
Results
HG Induces Intracellular Lipid Accumulation by Increasing Gene Expression Involved in Lipid Biosynthesis in Hepatocytes
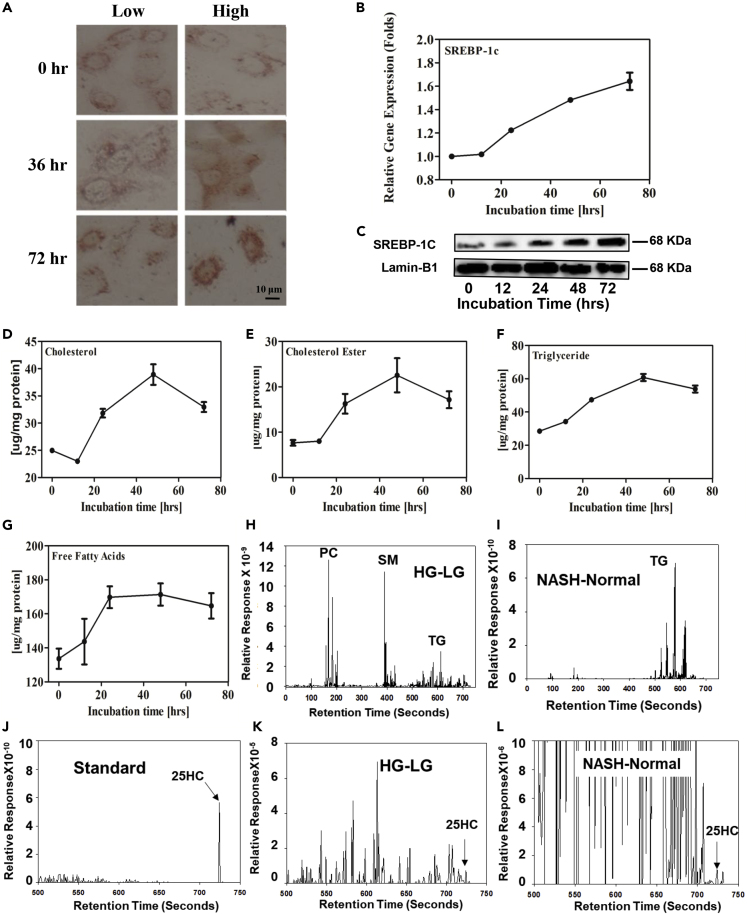
To study the effect of HG on intracellular lipid accumulation, Huh-7 cells were cultured in media with HG for different time periods and total neutral lipids were estimated by oil red O staining. As shown in Figure 1A, lipid levels started to increase at 36 h and had significantly accumulated at 72 h. HG increases expression and activation of SREBP-1C. Biosynthesis of free fatty acids and triglycerides is regulated by SREBP-1C (Horton et al., 2003). To investigate how HG increases lipid biosynthesis, total RNA was isolated from the HG-cultured Huh-7 cells. The mRNA levels of SREBP-1C were determined by real time RT-PCR. HG increased RNA levels in a time-dependent manner (Figure 1B). To confirm the results, SREBP-1C proteins were analyzed by western blot. As expected, increases in SREBP-1C protein levels (p < 0.01) in nuclear fractions were time dependent (Figure 1C). Thus, HG increases intracellular lipid levels by increasing SREBP-1C expression, which is consistent with previous reports (Pang et al., 2018, Xu et al., 2013).
Figure 1.
The Effects of HG on Lipid Accumulation in Hepatocytes
(A–L)Huh-7 cells were treated in HG media for 0, 12, 24, 48, and 72 h. The total intracellular neutral lipids were stained with 0.2% oil red O (A); inserts are shown at 400× magnification of the boxed areas (scale bar, 10 μm); mRNA levels of SREBP-1C in HG-cultured hepatocytes were determined by real-time RT-PCR analysis (B). SREBP-1C mature form protein levels in the nuclear fraction were analyzed by western blot (C); total lipids were extracted, and individual lipids were biochemically analyzed. Hepatic total cholesterol (D), cholesterol ester (E), triglyceride (F), and free fatty acid (G). Each individual level was normalized by protein concentration. Nuclear lipid composition from hepatocytes and human liver tissues was determined by lipidomic analysis. The lipid profile in nuclear fraction from Huh-7 cells cultured in HG or LG media (HG minus LG) (H); between human normal and NASH liver tissues (NASH minus normal) (I); 25HC as standard (J); the different levels of nuclear 25HC in Huh-7 cells cultured in HG and LG (K); the different levels of nuclear 25HC in human normal and NASH liver tissue (L). The lipidomic profile represents results from one of two experiments. All values are expressed as the mean ± SD.
Quantitative analysis showed that HG culture increased intracellular total cholesterol by 1.5-fold (p < 0.05) (Figure 1D), cholesterol ester by 2.9-fold (p < 0.05) (Figure 1E), triglycerides by 2.1-fold (p < 0.01) (Figure 1F), and free fatty acids by 28% (p < 0.01) at 72 h (Figure 1G). It is noted that HG culture did not significantly change free cholesterol levels (total cholesterol minus cholesterol ester). To further study the increased lipids in nuclear fractions, their composition was quantified by lipidomic analysis as shown in Figures 1H–1L and Table S2. Compared with low glucose (LG), HG increased phosphatidylcholine, sphingomyelin, and triglyceride levels with saturated fatty acids in nuclear fractions of Huh-7 cells (Figure 1H and Table S3). Compared with normal human liver tissues, non-alcoholic steatohepatitis (NASH) liver nuclear fraction showed increases mainly in triglycerides with saturated fatty acids (Figure 1I and Table S4). Interestingly, HG increased nuclear 25HC levels, whereas no change was detectable in LG (Figure 1K and Table S3); NASH liver nuclear fraction showed increases in 25HC levels by 30%, whereas decreases in 27-hydroxycholesterol (27HC) by 90% (Figure 1L and Table S4). The results imply that 25HC might play a role in the lipid accumulation.
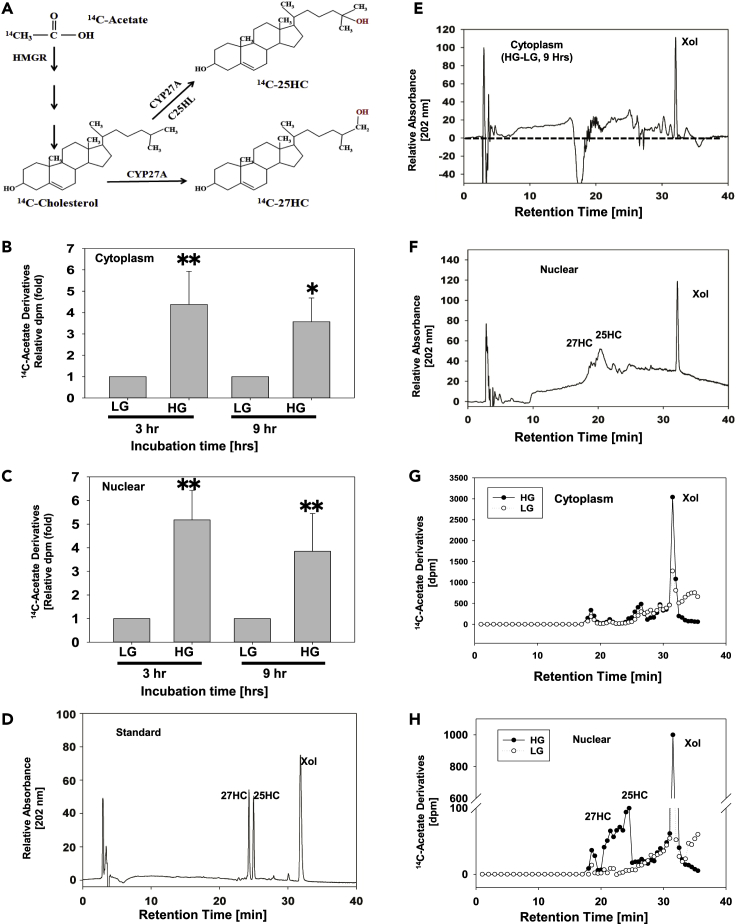
HG Increases Nuclear Levels of Newly Synthesized Cholesterol, 25-Hydroxycholesterol, and 27-Hydroxycholesterol
Oxysterols play an important role in lipid metabolism (Figure 2A). To examine the nuclear oxysterol levels in hepatocytes, Huh-7 cells were cultured in DMEM media with HG for 72 h. [14C]-Acetate was added to the media, and the cells were cultured for another 3 and 9 h (Figures 2B and 2C). Total lipids were extracted and partially purified. Individual [14C]-acetate derivatives were analyzed by high-performance liquid chromatography (HPLC). HPLC analysis showed that the product has the same retention time as 25HC and 27HC in our HPLC system (Figures 2D–2F), suggesting that the nuclear oxysterol derivative is sulfonated 25HC and 27HC. Compared with cells cultured in LG, the cells cultured in HG exhibited significantly increased levels of [14C]-cholesterol and [14C]-oxysterols, including [14C]-25HC and [14C]-27HC in nuclear fractions. About 50% of the total [14C]-25HC was detected in the nuclear fraction (Figures 2G and 2H). We also studied the distribution of exogenous 3H-25HC and found that similar percentile of the 3H-25HC will go to nuclei in 1 h (data not shown). As oxysterols have been reported to increase SREBP-1C expression and 25HC is one of the most potent regulatory oxysterols (Spann and Glass, 2013, Xu et al., 2010), cholesterol, 25HC, and 27HC were selected for further study to determine which molecule serves as an epigenetic regulator and plays a key role in lipid accumulation.
Figure 2.
HPLC Analysis of 25HC Levels in HG-Treated Human Hepatocytes
(A–H) Huh-7 cells were cultured in HG or LG media for 72 h and with [14C]-acetate for another 3 or 9 h. Nuclear and cytoplasmic fractions were isolated as described in Transparent Methods, partitioned into the chloroform phase, and analyzed by HPLC as described in Transparent Methods. The biosynthesis pathway of [14C]-sterols in the cells (A). The distribution of [14C]-acetate derivatives in cytoplasmic fraction (B) and nuclear fraction (C). The values represent means ± SD. HPLC standard elution profile of 25HC, 27HC, and cholesterol (D); elution profile of cytoplasmic extraction (E); elution profile of nuclear extraction (F); [14C]-acetate derivative profile of cytoplasmic extraction (G); and [14C]-acetate profile of HPLC products of nuclear extraction (H). The HPLC profiles represent one of three experiments. Data are represented as mean ± SD and statistic by t test. ∗Significant difference in distribution between LG and HG (p < 0.05); ∗∗p < 0.01.
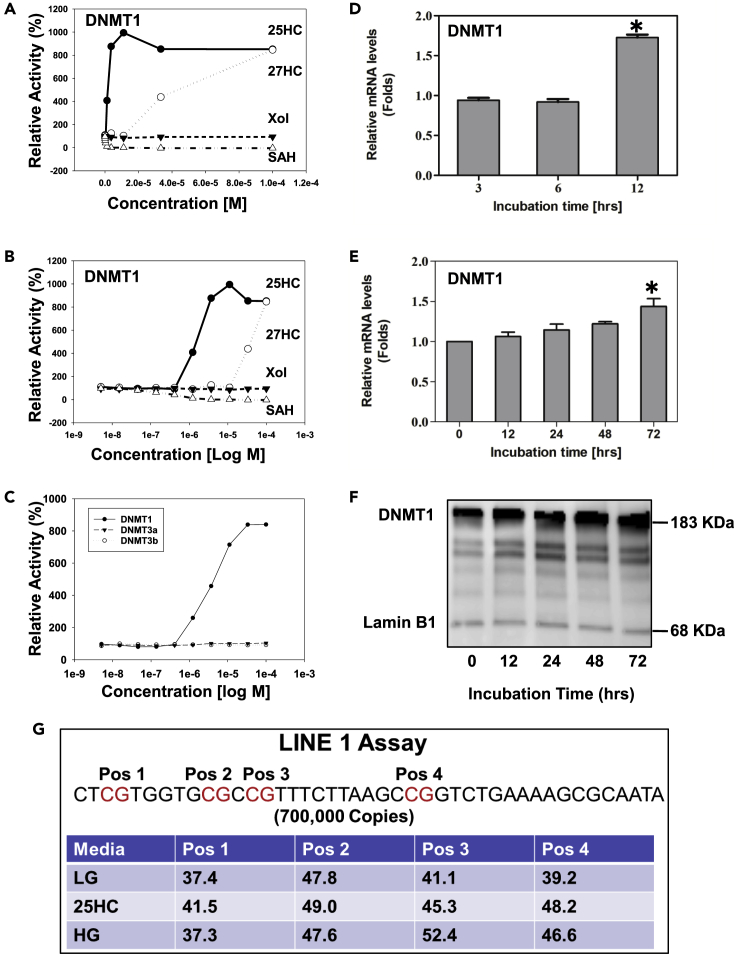
25HC Serves as an Agonist of DNMT1
Epigenetic regulators are DNA methyltransferases/demethylases and acetyltransferases/deacetylases. To study the effects of nuclear sterols on epigenetic regulation, 12 recombinant epigenetic regulators, DNA methyltransferase-1 (DNMT1), DNMT3a, DNMT3b, GCN3 (Giant congenital nevi), p300 (histone acetyltransferase), Pcaf (KAT2B lysine acetyltransferase 2B), HDAC1 (histone deacetylase 1), HDAC2 (histone deacetylase 2), HDAC3 (histone deacetylase 3), HDAC6 (histone deacetylase 6), HDAC10 (histone deacetylase 10), and KDM6B-JMJD3 (lysine demethylase 6B), were used to determine whether 25HC or cholesterol serves as their endogenous ligand(s). Interestingly, 25HC and 27HC, but not cholesterol, act on only DNMT1, and not on DNMT3a/3b (Figures 3A and 3B) and did not affect any demethylases, acetyltransferases, or deacetylases (data not shown). The kinetic study showed that 25HC increased DNMT1 by 8-fold at 3.7 μM, whereas 27HC reached the level at 100 μM, which is not physiologically significant, although both are in a dose-dependent manner. Then negative control S-adenosyl homocysteine (SAH) at 1 μM inhibited DNMT1 activity by 90% as shown in Figure 3C. Cholesterol did not have any effect on the DNMTs. The results indicate that 25HC is much more potent than 27HC (~30-fold). It is reasonable to believe that 25HC, but not 27HC, is an endogenous ligand of DNMT1, which can methylate DNA CpG and subsequently regulate gene expression. In addition, HG also increased expression of DNMT1 in hepatocytes. The RT-PCR and western blot results showed that HG and 25HC led to a time-dependent increase of DNMT1 both in mRNA and protein levels as shown in Figures 3D–3F.
Figure 3.
Effects of HG and 25HC on DNMT1 Expression and Enzyme Kinetics
(A–G) Effects of 25HC on DNMT1/3a/3b activities were determined by enzyme kinetics. The effects of different concentration on the enzymatic activities were tested from 0 to 0.001 M (A); effects of cholesterol, 25HC, 27HC, and S-adenosyl homocysteine (SAH, inhibitor as a negative control) on the DNMT1 activity (plots versus linear) (B); and plots versus logarithm (C). Effects of HG and 25HC on the expression of DNMT1 were determined by real-time RT-PCR. Huh-7 cells were cultured in HG media for 0, 12, 24, 48, and 72 h; mRNA levels of DNMT1 were determined by real-time RT-PCR analysis (D). Huh-7 cells were treated with 25 μM 25HC for 3, 6, and 12 h. The mRNA levels of DNMT1 were determined by real-time RT-PCR analysis (E). Protein levels of DNMT1 in the nuclei were analyzed by western blot (F). Effects of HG and 25HC on the global methylation (G). After Huh-7 cells were cultured in HG for 72 h, or treated with 25 μM of 25HC for 4 h, total DNA was purified as described in Methods. The levels of methylation were estimated by LINE-1 assay. Four CpG sites in promoter region of LINE-1 element were chosen as the target position. Data are represented as mean ± SD and statistic by t test. ∗Significant difference in expression between LG and HG (p < 0.05).
Recent studies have shown that global DNA methylation (long interspersed nucleotide element 1 [LINE-1] assay) and the methylation of specific genes are related to adipogenesis (Malodobra-Mazur et al., 2019), lipid metabolism (Chen et al., 2019, Li et al., 2020), and inflammation in visceral adipose tissues (Barajas-Olmos et al., 2018, Castellano-Castillo et al., 2018), which are in turn related to the etiology of metabolic syndrome. To study the effects of 25HC on DNA cytosine methylation in the promoter region, LINE-1 analysis was first performed to estimate CpG methylation in promoter regions. Methylation usually occurs in repetitive elements, such as LINE elements. There are 500,000 LINE elements and 40 trillion copies in total. Human LINE-1 is a retrotransposable region (promoter region) and has only 700,000 copies, which correlates to ~17% of the human genome (Lander et al., 2001). The specific sequence includes four CpG dinucleotides (Pos 1, 2, 3, and 4) as methylation targets in LINE-1 (Figure 3G). The results showed that HG significantly increases CpG methylation of LINE-1 elements in Huh-7 cells. Following culture in HG for 72 h, increases in methylation of 11% at Pos 3, and 8% at Pos 4, were observed. The results indicate that HG increases CpG methylation in promoter regions.
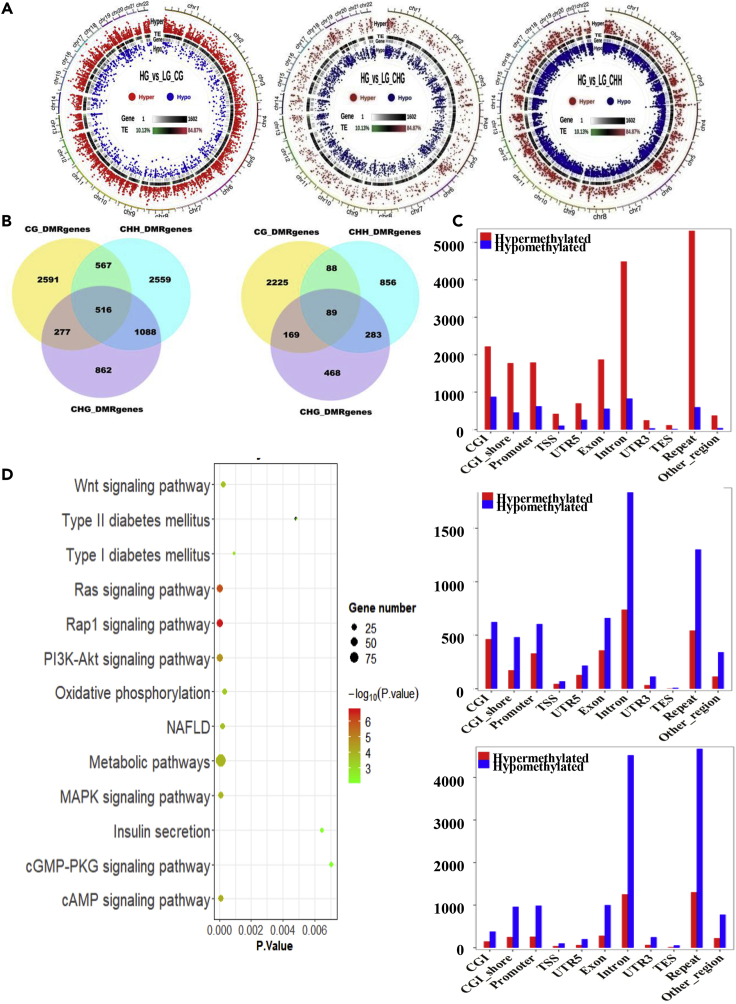
Profiles of Whole Genome-wide DNA Methylation in HG- and LG-Treated Huh-7 Cells
To understand the possible function of cytosine methylation in HG-treated Huh-7 cells, the cells were harvested for the construction of bisulfite-treated genomic DNA libraries. In total whole-genome bisulfite sequencing (WGBS) generated 401 million (LG) and 359 million (HG) raw reads from the two libraries by paired-end sequencing, respectively. Of the 394 million clean reads from LG library, 77% (305 million) were uniquely mapped to the reference genome of “human reference genome (hg38),” whereas of 354 million clean reads from HG library, 79% (279 million) were uniquely mapped to the reference genome, exhibiting an average read depth of 22 and 20, respectively. More than 82% cytosines were covered by at least 10 reads in the reference genome. The depth and density of the sequencing were enough for a high-quality genome-wide methylation analysis. Meanwhile, the bisulfite conversion efficiencies represented by the lambda DNA added into the libraries were over 99%, providing reliable and accurate results for the WGBS (Table S5)
DNA methylation levels in whole genome and differentially methylated regions (DMRs) are shown in Figure 4A. To present the global DNA methylation profiles of the two libraries, the uneven methylation levels throughout the chromosomes under CG, CHG (H represents adenosine or thymidine), and CHH contexts are shown in Figure 4B. A total of 6,012 differentially methylated genes (DMGs) were screened out among two libraries; 2,591 DMGs were identified under CG context, 862 under CHG context, and 2,559 under CHH context. In these DMGs, 277 were identified under CG and CHG contexts, 567 under CG and CHH contexts, 1,088 under CHG and CHH contexts, and only 516 under CG, CHG, and CHH contexts. Furthermore 3,549 were identified as promoter region different, and 2,225 were identified under CG context, 468 under CHG context, and 856 under CHH context. For these DMGs, 169 were identified under CG and CHG contexts, 88 under CG and CHG contexts, 283 under CHG and CHH contexts, and only 89 identified under CG, CHG, and CHH contexts.
Figure 4.
Whole-Genome Bisulfite Sequencing (WGBS) Analysis of the Effects of HG on the DNA Methylation in Hepatocytes
(A–D) Huh-7 cells were cultured in HG or LG media for 72 h. Detailed global methylation was measured by WGBS. Circos maps of DMR distribution in chromosomes (A): the first circle shows the distribution of hypermethylation DMRs, the second circle shows transposable element (TE) density, and the third circle shows the distribution of hypomethylation DMRs. Venn diagrams of DMR-associated genes (DMGs) among LG and HG libraries under CG, CHG, and CHH contexts of whole genome and promoter region (B). DNA methylation levels in different genomic functional regions of the whole genome (C), where the x axis represents the different genomic regions (CGI, CGI-shore, promoter, UTR 5, exon, intron, UTR 3 and repeat) and the y axis represents the methylation levels for LG and HG libraries under CG, CHG, and CHH contexts. Significant enrichment KEGG pathways of DMGs (D).
DNA methylation levels generally show a varied distribution across different functional regions of the genome. The methylation levels in the CGI (CG island), CGI-shore (up to 2 kbp away from the CGI), promoter (upstream 2-kb sequence from transcription starting site), 5′ UTR, exon, intron, 3′ UTR, and repeat were significantly different between HG- and LG-treated groups. It is interesting that HG has significantly higher hypermethylation levels than LG in CpG positions only (Figure 4C). In total 60,633 DMRs were identified; 4,711 (2,832 hypermethylated and 1,879 hypomethylated) were distributed in CGI, 4,986 (2,727 hypermethylated and 2,259 hypomethylated) in CGI-shore, 5,105 (2,724 hypermethylated and 2,381 hypomethylated) in exon, 15,291 (7,198 hypermethylatedand and 8,093 hypomethylated) in intron, 5,312 (2,754 hypermethylated and 2,558 hypomethylated) in promoter, 21,838 (13,023 hypermethylated and 8,815 hypomethylated) in repeat region, 210 (131 hypermethylated and 79 hypomethylated) in transcription end site elements, 811 (529 hypermethylated and 282 hypomethylated) in transcription start site (TSS) elements, 757 (350 hypermethylated and 407 hypomethylated) in 3′ UTRs, and 1,612 (916 hypermethylated and 696 hypomethylated) in 5′ UTRs. In almost all DMRs, CpGs are significantly more hypermethylated than hypomethylated. It has been reported that CG methylation in promotor regions plays a key role in silencing gene expression (Baylin, 2005).
Total DMGs, 6,012 genes, were significantly enriched in 288 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (189 hypermethylated and 129 hypomethylated). DMGs in promoter regions were significantly enriched in 169 KEGG pathways (83 hypermethylated and 51 hypomethylated) (Table S5), of which 79 (57 hypermethylated and 5 hypomethylated) were under CG context. Seventy-two DMGs (25 hypermethylated and 34 hypomethylated) were significantly enriched under CHG context and 18 (1 hypermethylated and 12 hypomethylated) under CHH context.
To analyze the function of DMGs in promoter regions under CG context, 79 KEGG pathways (p < 0.05) were identified. In these pathways 57 were identified as hypermethylation and 5 as hypomethylation. Thirteen of them are related to metabolic syndrome (Figure 4D). Six of these pathways including PI3K-Akt signaling pathway (Table 1), cAMP signaling pathway (Table 2), NAFLD (Table 3), type II diabetes mellitus (Table 4), and insulin secretion (Table 5) are directly related to carbohydrate, lipid, and energy metabolism. The most significant genes involved in these signaling pathways include insulin (INS), retinoid X receptor alpha (RXRα), insulin receptor substrate 1 (IRS1), insulin receptor (INSR), phosphoinositide-3-kinase regulatory subunit 5 (PIK3R5), mitogen-activated protein kinase 1 (MAPK1), cyclin-dependent kinase inhibitor 1B (CDKN1B), protein kinase N2 (PKN2), calcium/calmodulin-dependent protein kinase II beta (CAMK2B), peroxisome proliferator-activated receptor alpha (PPAR), mitogen-activated protein kinase 1 (MAPK1), potassium calcium-activated channel subfamily N member 3 (KCNN3), glucagon-like peptide 1 receptor (GLP1R), potassium calcium-activated channel subfamily U member 1 (KCNU1), potassium calcium-activated channel subfamily N member 4 (KCNN4), and calcium voltage-gated channel subunit alpha1D (CACNA1D). It has been well believed that insulin resistance is the major mechanism in the development and progression of NAFLD/NASH (Engin, 2017). Interestingly, DMRs in the promoter regions of key genes in these signaling pathways are hypermethylated, indicating that the gene expressions are down-regulated, which fits well with the mechanism of NAFLD mechanism.
Table 1.
Methylation of DMR in Gene Promoter Regions of PI3K-Akt Signaling Pathway (p = 0.000523)
| Gene Name | DMR Location in Promoter Region |
DMR (Methylation %) |
||||
|---|---|---|---|---|---|---|
| Chromosome | Start | End | HG | LG | HG-LG | |
| COL11A1 | Chr1 | 1.03 × 108 | 1.03 × 108 | 92.12 | 51.66 | 40.46 |
| GNG5P2 | ChrX | 1.1 × 108 | 1.1 × 108 | 47.26 | 6.76 | 40.5 |
| KRAS | Chr12 | 25250819 | 25250967 | 17.42 | 5.25 | 12.18 |
| INS | Chr11 | 2162594 | 2162810 | 79.13 | 23.5 | 55.62 |
| JAK1 | Chr1 | 64967244 | 64967529 | 67.24 | 15.6 | 51.65 |
| LAMA5 | Chr20 | 62368047 | 62368299 | 31.14 | 8.64 | 22.5 |
| BCL2 | Chr18 | 63320785 | 63321134 | 63.22 | 35.11 | 28.1 |
| HRAS | Chr11 | 536242 | 537214 | 61.26 | 18.46 | 42.8 |
| FGFR3 | Chr4 | 1792656 | 1792887 | 60.41 | 18.3 | 42.11 |
| CD19 | Chr16 | 28935890 | 28936133 | 88.21 | 47.13 | 41.08 |
| LAMB1 | Chr7 | 1.08 × 108 | 1.08 × 108 | 71.14 | 24.24 | 46.9 |
| RAC1P2 | Chr4 | 46724260 | 46724573 | 73.99 | 47.16 | 26.82 |
| MAPK1 | Chr22 | 21867333 | 21867621 | 23.71 | 3.56 | 20.15 |
| CDKN1B | Chr12 | 12714273 | 12715037 | 93.4 | 23.71 | 69.69 |
| PIK3R5 | Chr17 | 8888466 | 8888776 | 89.6 | 36.73 | 52.87 |
| BCL2L11 | Chr2 | 1.11 × 108 | 1.11 × 108 | 72.75 | 42.65 | 30.1 |
| PDGFB | Chr22 | 39242292 | 39242477 | 83.66 | 54.37 | 29.29 |
| ANGPT1 | Chr8 | 1.07 × 108 | 1.07 × 108 | 46.11 | 5.16 | 40.96 |
| IL6 | Chr7 | 22726101 | 22726775 | 80.53 | 30.36 | 50.18 |
| MDM2 | Chr12 | 68806524 | 68806756 | 75.71 | 33.96 | 41.74 |
| INSR | Chr19 | 7295041 | 7295182 | 57.84 | 14.92 | 42.92 |
| FGF11 | Chr17 | 7437822 | 7438331 | 50.31 | 19.51 | 30.81 |
| LPAR3 | Chr1 | 84894412 | 84894734 | 90.41 | 45.1 | 45.31 |
| FGF12 | Chr3 | 1.93 × 108 | 1.93 × 108 | 85.93 | 40.52 | 45.41 |
| FGFR2 | Chr10 | 1.22 × 108 | 1.22 × 108 | 24.32 | 8.01 | 16.3 |
| COL2A1 | Chr12 | 48004804 | 48005182 | 83.53 | 12.91 | 70.62 |
| FGF7P3 | Chr9 | 40106680 | 40106914 | 32.35 | 7.07 | 25.28 |
| CSF3 | Chr17 | 40014561 | 40014935 | 73.05 | 18.59 | 54.46 |
| FOXO3 | Chr6 | 1.09 × 108 | 1.09 × 108 | 25.28 | 4.11 | 21.17 |
| NR4A1 | Chr12 | 52051741 | 52052037 | 61.95 | 14.22 | 47.73 |
| F2R | Chr5 | 76715992 | 76716157 | 27.58 | 4.89 | 22.69 |
| PKN2 | Chr1 | 88684561 | 88684789 | 37.22 | 4.97 | 32.25 |
| EIF4E1B | Chr5 | 1.77 × 108 | 1.77 × 108 | 42.38 | 11.45 | 30.93 |
| RXRα | Chr9 | 1.34 × 108 | 1.34 × 108 | 36.99 | 6.91 | 30.08 |
| AC113189.4 | Chr17 | 7437822 | 7438331 | 50.31 | 19.51 | 30.81 |
| CSF3R | Chr1 | 36482185 | 36482512 | 90.94 | 41.71 | 49.23 |
| IRS1 | Chr2 | 2.27 × 108 | 2.27 × 108 | 34.01 | 5.95 | 28.06 |
| PDGFA | Chr7 | 521434 | 521719 | 83.23 | 59.88 | 23.35 |
Table 2.
Methylation of DMR in Gene Promoter Regions of cAMP Signaling Pathway (p = 0.001699)
| Gene Name | DMR Location in Promoter Region |
DMR (Methylation %) |
||||
|---|---|---|---|---|---|---|
| Chromosome | Start | End | HG | LG | HG-LG | |
| PDE4C | Chr19 | 18220846 | 18221310 | 85.7 | 24.91 | 60.79 |
| PPARα | Chr22 | 46149487 | 46149878 | 94.42 | 59.47 | 34.95 |
| HCAR2 | Chr12 | 1.23 × 108 | 1.23 × 108 | 87.48 | 45.91 | 41.57 |
| GNAS | Chr20 | 58891431 | 58891578 | 35.11 | 5.71 | 29.4 |
| CACNA1D | Chr3 | 53493469 | 53494019 | 74.84 | 25.81 | 49.03 |
| CAMK2B | Chr7 | 44221409 | 44221698 | 86.07 | 45 | 41.07 |
| RAC1P2 | Chr4 | 46724260 | 46724573 | 73.99 | 47.16 | 26.82 |
| PLD2 | Chr17 | 4806842 | 4806937 | 18.7 | 5.64 | 13.06 |
| MAPK1 | Chr22 | 21867333 | 21867621 | 23.71 | 3.56 | 20.15 |
| PIK3R5 | Chr17 | 8888466 | 8888776 | 89.6 | 36.73 | 52.87 |
| GLP1R | Chr6 | 39048762 | 39048955 | 86.25 | 59.95 | 26.3 |
| ADCY1 | Chr7 | 45574683 | 45574877 | 59.55 | 33.76 | 25.79 |
| PDE4B | Chr1 | 66331798 | 66332539 | 86.73 | 37.91 | 48.82 |
| NFATC1 | Chr18 | 79399054 | 79399517 | 78.35 | 33.06 | 45.29 |
| PDE4D | Chr5 | 59215742 | 59215941 | 94.05 | 54.39 | 39.66 |
| VAV3 | Chr1 | 1.08 × 108 | 1.08 × 108 | 88.49 | 24.15 | 64.34 |
| F2R | Chr5 | 76715992 | 76716157 | 27.58 | 4.89 | 22.69 |
| ADCY4 | Chr14 | 24334404 | 24334719 | 86.49 | 53.89 | 32.6 |
| HCN2 | Chr19 | 587907 | 588099 | 56.37 | 20.45 | 35.92 |
| GIPR | Chr19 | 45669075 | 45669744 | 83.03 | 39.41 | 43.62 |
| HTR4 | Chr5 | 1.49 × 108 | 1.49 × 108 | 95.66 | 78.7 | 16.96 |
| AL590635.1 | Chr6 | 91816380 | 91819294 | 93.85 | 28.92 | 64.92 |
| GRIN1 | Chr9 | 1.37 × 108 | 1.37 × 108 | 37.91 | 13.55 | 24.36 |
| PPP1R1B | Chr17 | 39630003 | 39630266 | 65.33 | 17.07 | 48.25 |
| NPR1 | Chr1 | 1.54 × 108 | 1.54 × 108 | 76.11 | 53.96 | 22.15 |
Table 3.
Methylation of DMR in Gene Promoter Regions of Non-alcoholic Fatty Liver Disease (NAFLD) (p = 0.003206)
| Gene Name | DMR Location in Promoter Regions |
DMR (Methylation %) |
||||
|---|---|---|---|---|---|---|
| Chromosome | Start | End | HG | LG | HG-LG | |
| RAC1P2 | Chr4 | 46724260 | 46724573 | 73.99 | 47.16 | 26.82 |
| MTCYBP22 | Chr5 | 1 × 108 | 1 × 108 | 90.01 | 52.51 | 37.50 |
| RXRα | Chr9 | 1.34 × 108 | 1.34 × 108 | 36.99 | 6.91 | 30.08 |
| MTCO3P22 | Chr5 | 1 × 108 | 1 × 108 | 88.87 | 45.71 | 43.16 |
| MTCO1P46 | Chr2 | 2.12 × 108 | 2.12 × 108 | 83.83 | 26.28 | 57.54 |
| MTCO2P22 | Chr5 | 1 × 108 | 1 × 108 | 88.87 | 45.71 | 43.16 |
| INSR | Chr19 | 7295041 | 7295182 | 57.84 | 14.92 | 42.92 |
| AC234779.1 | Chrx | 1.41 × 108 | 1.41 × 108 | 89.36 | 19.05 | 70.31 |
| COX6CP2 | Chr20 | 50478286 | 50478554 | 81.66 | 43.62 | 38.04 |
| BID | Chr22 | 17774417 | 17774536 | 20.06 | 5.89 | 14.17 |
| MAP3K5 | Chr6 | 1.37 × 108 | 1.37 × 108 | 81.00 | 40.19 | 40.81 |
| PIK3R5 | Chr17 | 8888466 | 8888776 | 89.60 | 36.73 | 52.87 |
| BCL2L11 | Chr2 | 1.11 × 108 | 1.11 × 108 | 72.75 | 42.65 | 30.10 |
| COX6B2 | Chr19 | 55354472 | 55354665 | 85.45 | 65.5 | 19.95 |
| PPARα | Chr22 | 46149487 | 46149878 | 94.42 | 59.47 | 34.95 |
| INS | Chr11 | 2162594 | 2162810 | 79.13 | 23.5 | 55.62 |
| IL6 | Chr7 | 22726101 | 22726775 | 80.53 | 30.36 | 50.18 |
| IRS1 | Chr2 | 2.27 × 108 | 2.27 × 108 | 34.01 | 5.95 | 28.06 |
| DDIT3 | Chr12 | 57521695 | 57521932 | 68.03 | 14.01 | 54.02 |
| NDUFA4L2 | Chr12 | 57240882 | 57241348 | 72.33 | 37.01 | 35.32 |
Table 4.
Methylation of DMR in Gene Promoter Regions of Type II Diabetes Mellitus (p = 0.032118)
| Gene Name | DMR Location in Promoter Regions |
DMR (Methylation %) |
||||
|---|---|---|---|---|---|---|
| Chromosome | Start | End | HG | LG | HG-LG | |
| INSR | Chr19 | 7295041 | 7295182 | 57.84 | 14.92 | 42.92 |
| INS | Chr11 | 2162594 | 2162810 | 79.13 | 23.5 | 55.62 |
| SOCS2 | Chr12 | 93572989 | 93573585 | 87.30 | 53.22 | 34.08 |
| MAPK1 | Chr22 | 21867333 | 21867621 | 23.71 | 3.56 | 20.15 |
| HK1 | Chr10 | 69318270 | 69318400 | 63.47 | 43.17 | 20.30 |
| PIK3R5 | Chr17 | 8888466 | 8888776 | 89.60 | 36.73 | 52.87 |
| CACNA1D | Chr3 | 53493469 | 53494019 | 74.84 | 25.81 | 49.03 |
| IRS1 | Chr2 | 2.27 × 108 | 2.27 × 108 | 34.01 | 5.95 | 28.06 |
Table 5.
Methylation of DMR in Gene Promoter Regions of Insulin Secretion (p = 0.035573)
| Gene Name | DMR Location in Promoter Regions |
DMR (Methylation %) |
||||
|---|---|---|---|---|---|---|
| Chromosome | Start | End | HG | LG | HG-LG | |
| ADCY4 | Chr14 | 24334404 | 24334719 | 86.49 | 53.89 | 32.6 |
| INS | Chr11 | 2162594 | 2162810 | 79.13 | 23.5 | 55.62 |
| KCNN3 | Chr1 | 1.55 × 108 | 1.55 × 108 | 83.22 | 46.68 | 36.54 |
| CHRM3 | Chr1 | 2.4 × 108 | 2.4 × 108 | 58.07 | 21.85 | 36.22 |
| GLP1R | Chr6 | 39048762 | 39048955 | 86.25 | 59.95 | 26.3 |
| KCNU1 | Chr8 | 36928346 | 36930601 | 64.67 | 11.97 | 52.7 |
| KCNN4 | Chr19 | 43774123 | 43774347 | 31.44 | 10.69 | 20.76 |
| GNAS | Chr20 | 58888560 | 58888756 | 41.48 | 18.28 | 23.2 |
| CACNA1D | Chr3 | 53493469 | 53494019 | 74.84 | 25.81 | 49.03 |
| ADCY1 | Chr7 | 45574683 | 45574877 | 59.55 | 33.76 | 25.79 |
| CAMK2B | Chr7 | 44221409 | 44221698 | 86.07 | 45 | 41.07 |
Target Next-Generation Bisulfite Sequencing to Confirm the WGBS Results
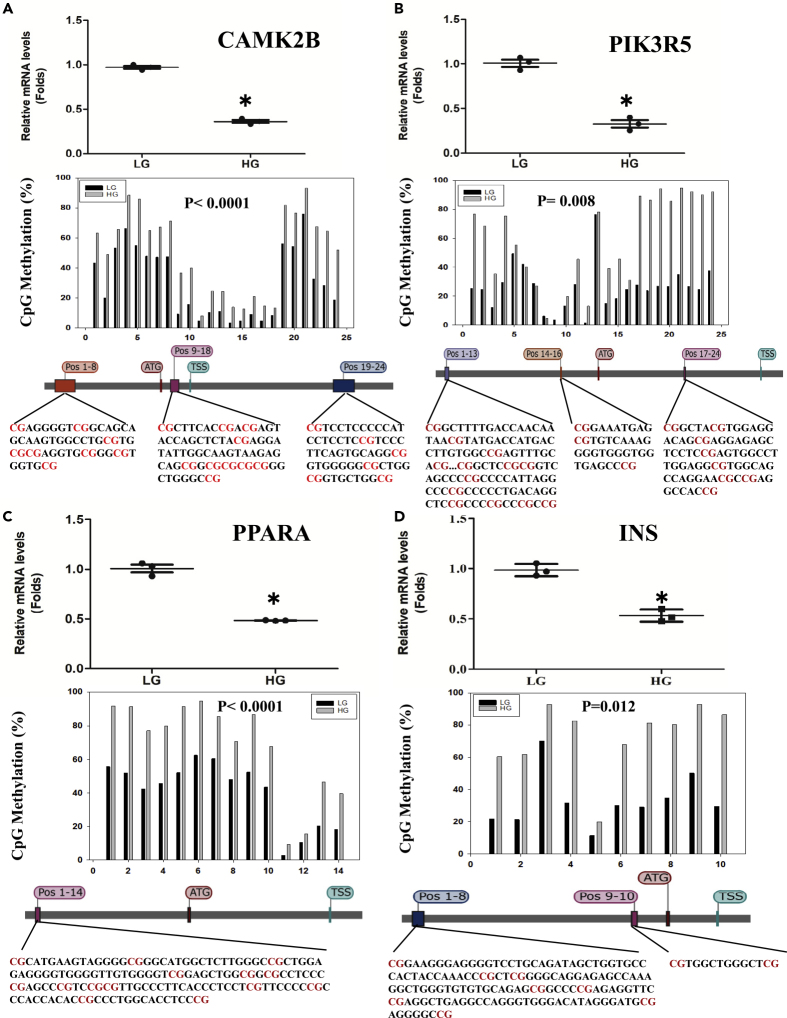
To confirm the results from WGBS, the methylation levels of CpG sites in promoter regions of PPARα, INS, CAMK2B, and PIK3R5, key genes in NAFLD, type II diabetes mellitus, insulin secretion, and cAMP signaling pathways, were determined using target next-generation bisulfite sequencing (tNGBS) (Figure 5). The results showed that HG increases the methylation levels of every CpG site not only in promoter region but also in exon and intron regions. For CAMK2B gene (Figure 5A), 24 CpG sites were identified including 8 (Pos 1–8) in promoter region, 4 (Pos 9–12) in exon region, and 12 (Pos 13–24) in intron region. HG increases the methylation levels of Pos 1–8 by 22%, Pos 9–12 by 18%, and Pos 13–24 by 19%. For PIK3R5 gene (Figure 5B), 24 CpG sites were identified including 8 (Pos1–8) in exon region, 13 (Pos 9–21) in promoter region, and 3 (Pos 22–24) in intron region. HG increases the methylation level of Pos 1–8 by 60% on average, Pos 9–21 by 15%, and Pos 22–24 by 19%. For PPARα gene (Figure 5C), HG increases the methylation levels of 14 CpG sites (Pos 1–14) in promoter region by 27%. For INS gene (Figure 5D), HG increases the methylation levels of 10 CpG sites (Pos 1–10) by 40% on average.
Figure 5.
The Relationship between DNA CpG Methylation and Gene Expression
(A–D) Huh-7 cells were cultured in HG or LG media for 72 h, and the methylation levels of the CpG sites of key genes involved in signaling pathways were analyzed by tNGBS. Huh-7 cells were cultured in HG media for 12 h, and the mRNA levels of these genes were determined by real-time RT-PCR analysis. The CpG methylation of the CAMK2B gene is shown in (A), where the top image shows the mRNA levels, the middle shows the methylation levels of 5′ upstream (Pos 1–14) region, and the bottom shows the sequence. CpG methylation of the PIK3R5 gene is shown in (B), where the top image shows the mRNA levels; the middle shows the methylation levels of 5′ upstream (Pos 1–13), intron (Pos 14–16), and exon (Pos 17–24) regions; and the bottom shows the gene sequence. CpG methylation of the PPARα gene is shown in (C), where the top shows the mRNA levels; the middle, the methylation levels of 5′ upstream (Pos 1–8), exon (Pos 9–12), and intron (Pos 13–24) regions; and the bottom, the gene sequence. CpG methylation of the INS gene is shown in (D), where the top image shows the mRNA levels; the middle, the methylation levels of 5′ UTR (Pos 1–2) and 5′ upstream (Pos 3–10) regions; and the bottom, the gene sequence. Data are represented as mean ± SD and statistic by t test. ∗Significant difference in expression between LG and HG (p < 0.05).
To explore whether DNA methylation in promoter region is involved in down-regulation of gene expression, four multiple function genes were selected for confirmation (Figure 5). Gene expressions were estimated by RT-PCR, and CpG methylations were confirmed by tNGBS. The relationship of gene expression with CpG methylation of CAMK2B is shown in Figures 5A; with PIK3R5, in 5B; with PPARα, in 5C; and with INS, in 5D. The results showed that all these gene expressions were significantly decreased when Huh-7 cells were cultured in HG, which is highly consistent with the results from the tNGBS analysis. The results confirm that the CpG methylation in the genes is responsible for the down-regulation of their expression.
Discussion
This study proposes a regulatory pathway for HG to lead to induction of lipid accumulation through 25HC and epigenetic regulation through hypermethylation. HG induces lipid accumulation through increased nuclear 25HC levels. The potent regulatory molecule 25HC, but not 27HC, activates DNMT1, which methylates cytosine of CpG in promoter regions, suppresses their expression, and down-regulates the associated signaling pathways. This proposed mechanism sheds new light on understanding how consuming excess sugar can lead to an increase in lipid biosynthesis, lipid accumulation, and risk of weight gain. These results propose some of the epigenetic mechanisms by which high-sugar diet may lead to insulin resistance and inflammation, both of which are risk factors for metabolic syndrome, through epigenetic regulation.
Sugars, such as fructose and glucose, can be catabolized to acetyl-CoA, which is the major substrate for producing energy, ATP, in vivo. When ATP concentrations reach high levels, excess acetyl-CoA is shunted into synthesis of cholesterol, free fatty acids, and triglycerides as a means of energy storage. The correlation between cholesterol and triglyceride biosynthesis has not been elucidated. In the present report, we demonstrate evidence that HG increases nuclear 25HC levels and subsequently increases triglyceride biosynthesis. We also may have explored for the first time that 25HC serves as an activator of DNMT1 and plays an important role in global regulation of metabolic pathways, cell survivals, and cell death by regulating critical signaling pathways. It has been reported that 25HC as a ligand of liver X receptor (LXRs) plays an important role in the control of lipid metabolism (Janowski et al., 1996). LXR ligands up-regulate expression of cholesterol reverse transporters such as ABCA1 and ABCG1 via activation of LXR/RXR heterodimers. ABCA1/G1 is known to mediate efflux of cellular cholesterol and phospholipids, which is the target for therapy of anti-atherosclerosis (Zhu et al., 2012). Unfortunately, administration of the synthetic ligands to mice triggers induction of the lipogenic pathway and elevates plasma triglyceride levels (Schultz et al., 2000). Addition of 25HC to human hepatocytes increases gene expression of key enzymes, ACC and FAS, in lipid biosynthesis and increases intracellular lipids levels via SREBP-1C pathway (Ma et al., 2008). 25HC has also been described to function in immune system, suppressing immunoglobulin production in B cells (Bauman et al., 2009) and inducing expression of the inflammatory cytokine interleukin-8 (Wang et al., 2004). Recently, 25HC has been shown to directly restrict target cell entry of enveloped viruses by inhibiting fusion of virus and cell membranes (Doms et al., 2018, Ke et al., 2017, Li et al., 2017, Liu et al., 2013, Wang et al., 2017). More interestingly, it has been reported that 25HC can be sulfated to 25-hydroxycholesterol 3-sulfate, 25HC3S, which has been shown as a distinct yet potent regulator of cellular functions (Li et al., 2007, Ren et al., 2007). Both 25HC3S and 25HC play important roles in lipid metabolism, inflammatory responses, and cell survival but act in a direction opposite to each other (Ren and Ning, 2014). The observation of global regulation is consistent with the present conclusion that 25HC serves as an agonist of epigenetic regulator, DNMT1. Whether the sulfation of 25HC to 25HC3S is the fundamental regulatory mechanism needs to be further investigated.
It is also interesting to notice that HG hyper-methylates CpG but hypomethylates CHG and CHH in whole gene regions including CpG island, CpG island shore, promoter, and TSS region as shown in Figure 4C. It is well-believed that CpG methylation in promoter regions is directly related with silencing gene expression (Lewis et al., 1992). However, the physiological significance of hypomethylation of CHG and CHH is unknown. DNA methylation is established and maintained by three essential DNA methyltransferases, DNMT1, DNMT3a, and DNM3b, in mammals (Koerner and Barlow, 2010, Long et al., 2017). Methylated DNA is then recognized by methyl CpG-binding proteins along with associated co-repressors that lead to silencing of the associated promoter (Clouaire and Stancheva, 2008). The in vivo role of DNMT1 in carcinogenesis has been widely studied using hypomorphic mice instead of DNMT1−/− mice that die during embryogenesis (Subramaniam et al., 2014). DNMT1 hypomorphic mice exhibit resistance to alcohol-induced hepatic steatosis compared with the wild-type mice (Kutay et al., 2012). It has been reported that metabolic syndrome has an impact on the global methylation pattern in visceral adipose tissue (Castellano-Castillo and Moreno-Indias, 2019). The results indicate that the steatosis is most likely dependent on the DNA methylation in related genes, which silences gene expression. In this study, we report that HG incubation induces DNA CpG methylation in promoter regions of the key signaling pathways, which induces lipid accumulation in human hepatocytes. We found that HG incubation increases 25HC levels in hepatocyte nuclei. 25HC activates DNMT1 and increases 5mCpG in promoter regions of 2,225 genes.
Furthermore, the HG-induced CpG methylation in promoter regions covers 79 KEGG pathways. Fifty-seven pathways were identified as significantly hypermethylated. To our knowledge, 13 of these pathways are directly related to lipid metabolism, including metabolic signaling pathway, Ras signaling pathway (Ye et al., 2017), cAMP signaling pathway (Ravnskjaer et al., 2016), PI3K-Akt signaling pathway (Zhou et al., 2019), MAPK signaling pathway (Yang et al., 2018), Wnt signaling pathway (Yao et al., 2018), NAFLD (Engin, 2017), type 1 diabetes mellitus (Ritchie et al., 2017), cGMP-PKG signaling pathway (Schwarz et al., 2018), FoxO signaling pathway (Cheng and White, 2011), type II diabetes mellitus (Samuel and Shulman, 2012), insulin secretion (Eto et al., 2002), and insulin signaling pathway (Consitt et al., 2009) as shown in Table 1. As hypermethylation in promoter regions is believed to silence the gene expression, it is reasonable to conclude that HG induces metabolic disorders via methylation of their key genes in the pathways, which were confirmed by analysis of mRNA levels as shown in Figure 5. Only one (p < 0.05) tight junction pathway was identified as hypomethylation. However, its physiological significance is unknown.
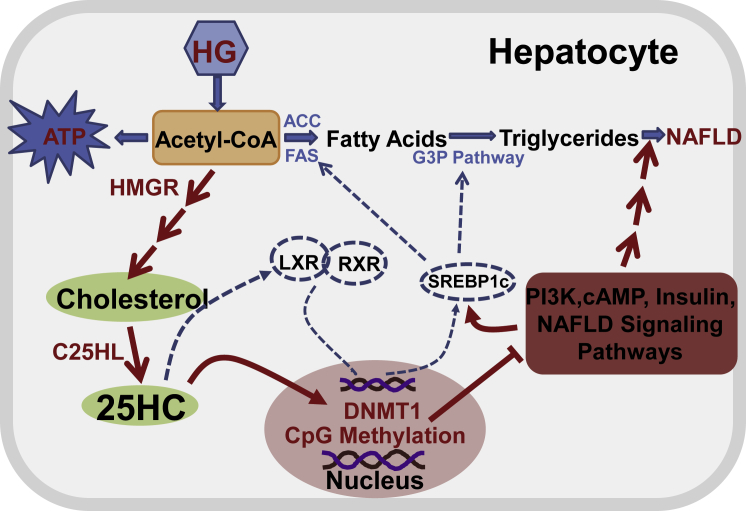
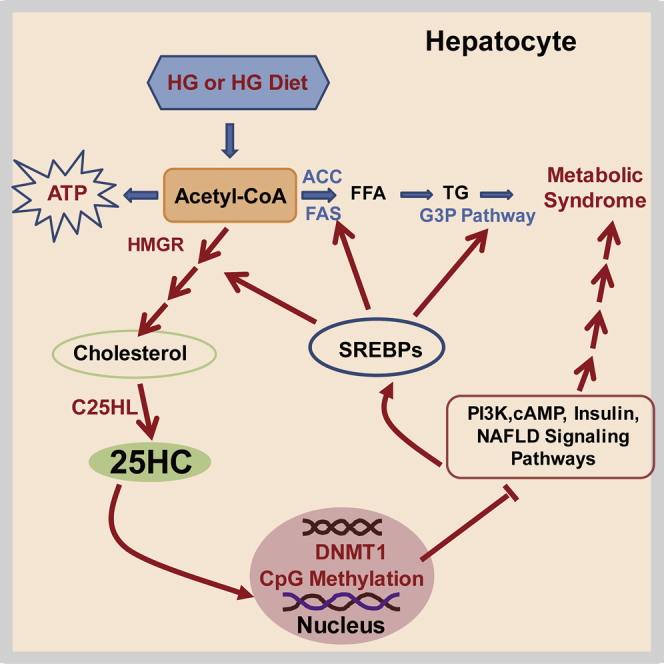
The current results most likely represent the etiology for the development of high sugar diet-induced fatty liver diseases. Based on the results, we summarize the regulatory pathway, which may play an important role in the occurrence of NAFLD and metabolic syndrome. When cells are incubated in HG media, high sugar consumption will increase intracellular 25HC, which in turn will activate DNMT1, as shown in Figure 6.
Figure 6.
Proposed Regulatory Pathway of Intracellular Lipid Homeostasis
A high-sugar diet produces an excess of acetyl-CoA, which is the energy source for generation of ATP. The excess of acetyl-CoA can also be used for synthesis of cholesterol and subsequently for synthesis of oxysterols such as 25-hydroxycholesterol by key enzymes, 3-hydroxy-3-methyl-glutaryl-CoA reductase and cholesterol 25-hydroxylase (C25HL) (Lund et al., 1998). 25HC activates LXR and upregulates SREBP-1C expression, which results in increasing biosynthesis of free fatty acids and triglycerides in a known pathway. On the other hand, 25HC enters nuclei and activates DNMT1, which selectively methylates CpG in promoter regions and silences gene expression, leading to down-regulation of many signaling pathways including PI3K, cAMP, insulin, NAFLD, and metabolic pathways. The dashed blue lines represent known pathways, and the red solid lines represent the proposed pathways. The proposed pathways play an important role in the occurrence of NAFLD and metabolic syndrome.
Limitations of the Study
We are investigating the effect of knockout and knockdown of 25HC syntheses on the activity of DNA methyltransferases and considering this as a limitation of this study. The enzyme kinetic results provide strong advance of this question. All the materials are available to all interested scientists per request. Subsection lead contact to Yaping Wang.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The lipidomic data were generated by the VCU Massey Cancer Center Lipidomics Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30 CA016059. This work was supported by VA Merit Review grant and DURECT Research Agreement to VCU. Jim Brown and WeiQi Lin, DURECT Inc., San Francisco, CA, edited the manuscript.
Author Contributions
Y.W. designed and conducted the experiments and wrote the paper; S.R. designed experiments, wrote the paper, and provided response to the project; and L.C., W.M.P., D.H., and P.B.H gave a suggestion and edited the manuscript.
Declaration of Interests
S.R. and Virginia Commonwealth University obtain license-related payments from DURECT Corporation, Cupertino, CA.
Published: May 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101102.
Supplemental Information
References
- Balland E., Chen W., Dodd G.T., Conductier G., Coppari R., Tiganis T., Cowley M.A. Leptin signaling in the arcuate nucleus reduces insulin's capacity to suppress hepatic glucose production in obese mice. Cell Rep. 2019;26:346–355.e3. doi: 10.1016/j.celrep.2018.12.061. [DOI] [PubMed] [Google Scholar]
- Barajas-Olmos F., Centeno-Cruz F., Zerrweck C., Imaz-Rosshandler I., Martinez-Hernandez A., Cordova E.J., Rangel-Escareno C., Galvez F., Castillo A., Maydon H. Altered DNA methylation in liver and adipose tissues derived from individuals with obesity and type 2 diabetes. BMC Med. Genet. 2018;19:28. doi: 10.1186/s12881-018-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman D.R., Bitmansour A.D., McDonald J.G., Thompson B.M., Liang G., Russell D.W. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci U S A. 2009;106:16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Castellano-Castillo D., Moreno-Indias I. Altered adipose tissue DNA methylation status in metabolic syndrome: relationships between global DNA methylation and specific methylation at adipogenic, lipid metabolism and inflammatory candidate genes and metabolic variables. J. Clin. Med. 2019;8:87. doi: 10.3390/jcm8010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Castillo D., Morcillo S., Clemente-Postigo M., Crujeiras A.B., Fernandez-Garcia J.C., Torres E., Tinahones F.J., Macias-Gonzalez M. Adipose tissue inflammation and VDR expression and methylation in colorectal cancer. Clin. Epigenetics. 2018;10:60. doi: 10.1186/s13148-018-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che Y., Wang Z.P., Yuan Y., Zhang N., Jin Y.G., Wan C.X., Tang Q.Z. Role of autophagy in a model of obesity: a longterm high fat diet induces cardiac dysfunction. Mol. Med. Rep. 2018;18:3251–3261. doi: 10.3892/mmr.2018.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shi Y., Liu N., Wang Z., Yang R., Yan B., Liu X., Lai W., Liu Y., Xiao D. DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism. Epigenetics Chromatin. 2019;12:59. doi: 10.1186/s13072-019-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., White M.F. Targeting Forkhead box O1 from the concept to metabolic diseases: lessons from mouse models. Antioxid. Redox Signal. 2011;14:649–661. doi: 10.1089/ars.2010.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T., Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol. Life Sci. 2008;65:1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consitt L.A., Bell J.A., Houmard J.A. Intramuscular lipid metabolism, insulin action, and obesity. IUBMB Life. 2009;61:47–55. doi: 10.1002/iub.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Dang E.V., Reboldi A., Yi T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 2014;14:731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- Doms A., Sanabria T., Hansen J.N., Altan-Bonnet N., Holm G.H. 25-Hydroxycholesterol production by the cholesterol-25-hydroxylase interferon-stimulated gene restricts mammalian reovirus infection. J. Virol. 2018;92 doi: 10.1128/JVI.01047-18. e01047–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., Shi Y.H., Le G.W. Oxidative stress induced by high-glucose diet in liver of C57BL/6J mice and its underlying mechanism. Mol. Biol. Rep. 2010;37:3833–3839. doi: 10.1007/s11033-010-0039-9. [DOI] [PubMed] [Google Scholar]
- Engin A. Non-alcoholic fatty liver disease. Adv. Exp. Med. Biol. 2017;960:443–467. doi: 10.1007/978-3-319-48382-5_19. [DOI] [PubMed] [Google Scholar]
- Eto K., Yamashita T., Matsui J., Terauchi Y., Noda M., Kadowaki T. Genetic manipulations of fatty acid metabolism in beta-cells are associated with dysregulated insulin secretion. Diabetes. 2002;51:S414–S420. doi: 10.2337/diabetes.51.2007.s414. [DOI] [PubMed] [Google Scholar]
- Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., Goldstein J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski B., Willy P., Devi T., Falck J., Mangelsdorf D. An oxysterol signaling pathway mediated by the nuclear receptor Lxr-alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Ke W., Fang L., Jing H., Tao R., Wang T., Li Y., Long S., Wang D., Xiao S. Cholesterol 25-hydroxylase inhibits porcine reproductive and respiratory syndrome virus replication through enzyme activity-dependent and -independent mechanisms. J. Virol. 2017;91 doi: 10.1128/JVI.00827-17. e00827–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner M.V., Barlow D.P. Genomic imprinting-an epigenetic gene-regulatory model. Curr. Opin. Genet. Dev. 2010;20:164–170. doi: 10.1016/j.gde.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay H., Klepper C., Wang B., Hsu S.H., Datta J., Yu L., Zhang X., Majumder S., Motiwala T., Khan N. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PLoS One. 2012;7:e41949. doi: 10.1371/journal.pone.0041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li X., Pandak W.M., Erickson S.K., Ma Y., Yin L., Hylemon P., Ren S. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta,25-diol 3-sulfate, in hepatocytes. J. Lipid Res. 2007;48:2587–2596. doi: 10.1194/jlr.M700301-JLR200. [DOI] [PubMed] [Google Scholar]
- Li C., Deng Y.Q., Wang S., Ma F., Aliyari R., Huang X.Y., Zhang N.N., Watanabe M., Dong H.L., Liu P. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang J., Wang L., Feng G., Li G., Yu M., Li Y., Liu C., Yuan X., Zang G. Impaired lipid metabolism by age-dependent DNA methylation alterations accelerates aging. Proc Natl Acad Sci U S A. 2020;117:8660. doi: 10.1073/pnas.1919403117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu S.Y., Aliyari R., Chikere K., Li G., Marsden M.D., Smith J.K., Pernet O., Guo H., Nusbaum R., Zack J.A. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jiang S., Zhao Y., Sun Q., Zhang J., Shen D., Wu J., Shen N., Fu X., Sun X. Geranylgeranyl diphosphate synthase (GGPPS) regulates non-alcoholic fatty liver disease (NAFLD)-fibrosis progression by determining hepatic glucose/fatty acid preference under high-fat diet conditions. J. Pathol. 2018;246:277–288. doi: 10.1002/path.5131. [DOI] [PubMed] [Google Scholar]
- Long M.D., Smiraglia D.J., Campbell M.J. The genomic impact of DNA CpG methylation on gene expression; relationships in prostate cancer. Biomolecules. 2017;7:E15. doi: 10.3390/biom7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E.G., Kerr T.A., Sakai J., Li W.P., Russell D.W. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 1998;273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- Luu W., Sharpe L.J., Capell-Hattam I., Gelissen I.C., Brown A.J. Oxysterols: old tale, new twists. Annu. Rev. Pharmacol. Toxicol. 2016;56:447–467. doi: 10.1146/annurev-pharmtox-010715-103233. [DOI] [PubMed] [Google Scholar]
- Ma Y., Xu L., Rodriguez-Agudo D., Li X., Heuman D.M., Hylemon P.B., Pandak W.M., Ren S. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1369–E1379. doi: 10.1152/ajpendo.90555.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malodobra-Mazur M., Cierzniak A., Dobosz T. Oleic acid influences the adipogenesis of 3T3-L1 cells via DNA Methylation and may predispose to obesity and obesity-related disorders. Lipids Health Dis. 2019;18:230. doi: 10.1186/s12944-019-1173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.G., Russell D.W. Editorial: 25-Hydroxycholesterol: a new life in immunology. J. Leukoc. Biol. 2010;88:1071–1072. doi: 10.1189/jlb.0710418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzo L., Ciciliot S., Poncina N., Mazzucato M., Persano M., Bonora B., Albiero M., Vigili de Kreutzenberg S., Avogaro A., Fadini G.P. NETosis is induced by high glucose and associated with type 2 diabetes. Acta Diabetol. 2015;52:497–503. doi: 10.1007/s00592-014-0676-x. [DOI] [PubMed] [Google Scholar]
- Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez S., Garces-Rimon M., Vera G., Astier J., Landrier J.F., Miguel M. High fat/high glucose diet induces metabolic syndrome in an experimental rat model. Nutrients. 2018;10:E1502. doi: 10.3390/nu10101502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O-Sullivan I., Zhang W., Wasserman D.H., Liew C.W., Liu J. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J., Cui J., Xi C., Shen T., Gong H., Dou L., Lin Y., Zhang T. Inhibition of poly(ADP-ribose) polymerase increased lipid accumulation through SREBP1 modulation. Cell Physiol. Biochem. 2018;49:645–652. doi: 10.1159/000493028. [DOI] [PubMed] [Google Scholar]
- Ravnskjaer K., Madiraju A., Montminy M. Role of the cAMP pathway in glucose and lipid metabolism. Handbook Exp. Pharmacol. 2016;233:29–49. doi: 10.1007/164_2015_32. [DOI] [PubMed] [Google Scholar]
- Ren S., Ning Y. Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 2014;306:E123–E130. doi: 10.1152/ajpendo.00552.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Li X., Rodriguez-Agudo D., Gil G., Hylemon P., Pandak W.M. Sulfated oxysterol, 25HC3S, is a potent regulator of lipid metabolism in human hepatocytes. Biochem. Biophys. Res. Commun. 2007;360:802–808. doi: 10.1016/j.bbrc.2007.06.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie R.H., Zerenturk E.J., Prakoso D., Calkin A.C. Lipid metabolism and its implications for type 1 diabetes-associated cardiomyopathy. J. Mol. Endocrinol. 2017;58:R225–R240. doi: 10.1530/JME-16-0249. [DOI] [PubMed] [Google Scholar]
- Saltiel A.R. Insulin signaling in the control of glucose and lipid homeostasis. Handbook Exp. Pharmacol. 2016;233:51–71. doi: 10.1007/164_2015_14. [DOI] [PubMed] [Google Scholar]
- Samuel V.T., Shulman G.I. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J.R., Tu H., Luk A., Repa J.J., Medina J.C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D.J. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K.R.L., de Castro F.C., Schefer L., Botigelli R.C., Paschoal D.M., Fernandes H., Leal C.L.V. The role of cGMP as a mediator of lipolysis in bovine oocytes and its effects on embryo development and cryopreservation. PLoS One. 2018;13:e0191023. doi: 10.1371/journal.pone.0191023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann N.J., Glass C.K. Sterols and oxysterols in immune cell function. Nat. Immunol. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- Subramaniam D., Thombre R., Dhar A., Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K., Kumar S.M., Clemens D.L., Dey A. Inhibition of CYP2E1 leads to decreased advanced glycated end product formation in high glucose treated ADH and CYP2E1 over-expressing VL-17A cells. Biochim. Biophys. Acta. 2013;1830:4407–4416. doi: 10.1016/j.bbagen.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Vallon V., Thomson S.C. Targeting renal glucose reabsorption to treat hyperglycaemia: the pleiotropic effects of SGLT2 inhibition. Diabetologia. 2017;60:215–225. doi: 10.1007/s00125-016-4157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurusaner B., Leonarduzzi G., Gamba P., Poli G., Basaga H. Oxysterols and mechanisms of survival signaling. Mol. Aspects Med. 2016;49:8–22. doi: 10.1016/j.mam.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Wang F., Ma Y., Barrett J.W., Gao X., Loh J., Barton E., Virgin H.W., McFadden G. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 2004;5:1266–1274. doi: 10.1038/ni1132. [DOI] [PubMed] [Google Scholar]
- Wang J., Zeng L., Zhang L., Guo Z.Z., Lu S.F., Ming S.L., Li G.L., Wan B., Tian K.G., Yang G.Y. Cholesterol 25-hydroxylase acts as a host restriction factor on pseudorabies virus replication. J. Gen. Virol. 2017;98:1467–1476. doi: 10.1099/jgv.0.000797. [DOI] [PubMed] [Google Scholar]
- Wang Q., Tang S.B., Song X.B., Deng T.F., Zhang T.T., Yin S., Luo S.M., Shen W., Zhang C.L., Ge Z.J. High-glucose concentrations change DNA methylation levels in human IVM oocytes. Hum. Reprod. 2018;33:474–481. doi: 10.1093/humrep/dey006. [DOI] [PubMed] [Google Scholar]
- Xu L., Bai Q., Rodriguez-Agudo D., Hylemon P.B., Heuman D.M., Pandak W.M., Ren S. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45:821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- Xu L., Kim J.K., Bai Q., Zhang X., Kakiyama G., Min H.K., Sanyal A.J., Pandak W.M., Ren S. 5-cholesten-3beta,25-diol 3-sulfate decreases lipid accumulation in diet-induced nonalcoholic fatty liver disease mouse model. Mol. Pharmacol. 2013;83:648–658. doi: 10.1124/mol.112.081505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Suh W.I., Kang N.K., Lee B. MAPK/ERK and JNK pathways regulate lipid synthesis and cell growth of Chlamydomonas reinhardtii under osmotic stress, respectively. Sci. Rep. 2018;8:13857. doi: 10.1038/s41598-018-32216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Sun S., Wang J., Fei F., Dong Z., Ke A.W., He R., Wang L., Zhang L., Ji M.B. Canonical Wnt signaling remodels lipid metabolism in Zebrafish hepatocytes following Ras oncogenic insult. Cancer Res. 2018;78:5548–5560. doi: 10.1158/0008-5472.CAN-17-3964. [DOI] [PubMed] [Google Scholar]
- Ye X., Li M., Hou T., Gao T., Zhu W.G., Yang Y. Sirtuins in glucose and lipid metabolism. Oncotarget. 2017;8:1845–1859. doi: 10.18632/oncotarget.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., McIntosh C., Lister R., Zhu I., Han Y., Ren J., Landsman D., Lee E., Briones V., Terashima M. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24:1613–1623. doi: 10.1101/gr.172015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Guo X., Wang O., Zhang H., Wang Y., Zhou F., Liu J., Ji B. Fructose and glucose combined with free fatty acids induce metabolic disorders in HepG2 cell: a new model to study the impacts of high-fructose/sucrose and high-fat diets in vitro. Mol. Nutr. Food Res. 2016;60:909–921. doi: 10.1002/mnfr.201500635. [DOI] [PubMed] [Google Scholar]
- Zhou M., Ren P., Li S., Kang Q., Zhang Y., Liu W., Shang J., Gong Y., Liu H. Danhong injection attenuates high-fat-induced atherosclerosis and macrophage lipid accumulation by regulating the PI3K/AKT insulin pathway. J. Cardiovasc. Pharmacol. 2019;74:152–161. doi: 10.1097/FJC.0000000000000691. [DOI] [PubMed] [Google Scholar]
- Zhu R., Ou Z., Ruan X., Gong J. Role of liver X receptors in cholesterol efflux and inflammatory signaling (review) Mol. Med. Rep. 2012;5:895–900. doi: 10.3892/mmr.2012.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.