Abstract
Background:
Adrenocortical carcinoma is a rare and aggressive malignancy with poor survival. With limited treatment options and high risk of relapse, identifying improved targets and therapies for adrenocortical carcinoma is important. We hypothesized that analysis of the database of The Cancer Genome Atlas could identify important novel biomarkers for improved therapeutic targeting of adrenocortical carcinoma.
Methods:
We utilized the University of Alabama interactive web resource to identify novel biomarkers observed in 79 adrenocortical carcinoma patients. Identified biomarkers were then examined for prognostic correlations using the cBioPortal and analyzed for statistical significance using STATA 13.0.
Results:
The Cancer Genome Atlas data mining in the University of Alabama interactive web resource for pathways associated with poor survival of patients with adrenocortical carcinoma revealed significant upregulation of genes involved in DNA damage and regulation of cell-cycle pathways, such as AURKA, AURKB, CDK1, CDK4, CDK6, PLK1, CHEK1, CHEK2, CDC7, BUB3, and MCM3 (P < .001–.05). On outcome correlation, greater expression levels of all the genes except CDK4 were associated with worse survival compared with medium or low levels of gene expression (P < .001 all) irrespective of age orsex. Consistent with our University of Alabama interactive web resource findings, data mining in the cBioPortal also revealed upregulation of genes regulating DNA-damage and cell cycle–related genes in 82% of patients (z score = 1.5).
Conclusion:
Large data mining from the The Cancer Genome Atlas and cBioPortal databases identified overexpression of genes involved in DNA damage and those regulating pathways of the cell cycle, which correlated with poorer overall survival in adrenocortical carcinoma patients.
Introduction
Adrenocortical carcinoma (ACC) is a rare and aggressive endocrine neoplasm with a poor 5-year overall survival rate of 37%–47%.1–3 Treatment options are limited for patients with locally advanced ACC, and there remains a high risk of relapse, even after major operative resection. Because of difficulties in early detection, 70% of the ACC patients present with metastases at the time of diagnosis, and for these metastatic stage IV patients, the 5-year survival rate remains especially poor at 6%–13% despite optimal treatment.4–6 Treatment options for ACC include either mitotane alone, combination multi-drug cytotoxic chemotherapy, such as etoposide, cisplatin, and doxorubicin, or other regimens, radiation therapy, or enrollment in a clinical trial. Despite these options, survival remains low, and durable complete responses for advanced disease are not expected, making it paramount to identify new biologic targets leading to improved therapies.
Recent investigations have advanced our understanding of the biology of ACC by identifying several genes that could function as potential drivers of the pathogenesis of ACC.7–11 With new target identification comes the hope for development of novel, more durable therapies for ACC that improve survival. Therefore, drugs targeting molecular pathways, such as vascular endothelial growth factor (VEGFR), epidermal growth factor receptor, insulin-like growth factor 1 receptor, mammalian target of rapamycin, and others, are being evaluated in clinical trials.7 Based on promising preclinical data, sunitinib, which targets the VEGFR pathway, entered a phase II trial in 35 patients with refractory ACC. Unfortunately, the progression-free survival and the response rate were only 2.8 months and 15%, respectively, owing to potential drug–drug interactions between sunitinib and mitotane.8 A clinical trial with sorafenib (a tyrosine kinase inhibitor which targets VEGFR and angiogenesis) in combination with weekly paclitaxel had to be suspended because of disease progression after only 8 weeks.9 The successful biologic activity of an IGF-1R inhibitor Cixutumumab (IMC-A12) in phase I clinical trials led to a phase II clinical trial of mitotane plus IMC-A12 in 20 patients with advanced or metastatic ACC. Of the 20 patients enrolled, 1 had a partial response and 7 had stable disease with median PFS of only 6 weeks, but because of limited efficacy and significant toxicity, the study was terminated.10 In addition to combination with mitotane, IMC-A12 in combination with the mammalian target of rapamycin inhibitor temsirolimus was also evaluated in 10 patients with advanced ACC in phase I clinical trial. In this study, 4 of the 10 patients had stable disease during the trial period (11). In addition to IMC-A12, phase III clinical trials of another IGF-1R inhibitor Linsitinib (OSI-906) also failed to increase the overall survival or progression-free survival of patients with ACC compared to placebo.12 Overall, these recent clinical trials demonstrate the current challenges in the treatment of advanced ACC and the lack of improved clinical outcomes with current targeted therapies, which only highlights the ongoing critical need for development of improved therapeutic targets in this disease.
Recently, The Cancer Genome Atlas (TCGA) project collected clinicopathologic data on a large global cohort of ACC samples, performing comprehensive and integrated genomic characterization of tumors matched with normal adrenal tissues.13 We hypothesized that analyzing the gene expression data of TCGA could identify important novel therapeutic and diagnostic biomarkers that correlate with poor prognosis of this disease and could present new opportunities for therapeutic targeting of ACC patients. Therefore, we mined the TCGA transcriptome data via the University of Alabama interactive web resource (UALCAN)14 and the cBioPortal resource15,16 to identify additional molecular markers that could represent novel therapeutic targets for future treatment of ACC.
Methods
Bioinformatic analysis of UALCAN
UALCAN is an interactive web portal that utilizes publicly available RNA-sequence and patient clinical data generated by TCGA consortium. Using TCGA data set, UALCAN developed a user-friendly interface that enables the analysis of profiles of relative gene expression of normal and cancer samples and gene expression based on clinicopathologic characteristics, such as race, tumor grade, cancer stage, and age. In addition, it also correlates gene expression with patient survival plotted onto Kaplan-Meier survival curves. Given this rich data resource, we utilized the UALCAN web portal to identify novel biomarkers in ACC. We initially mined genes that were associated with poor prognosis irrespective of stage and then looked at their differential regulation in various stages of ACC. The UALCAN portal calculates both high and low or medium expression for each gene based on transcript per million expression values for plotting Kaplan-Meier survival curves. By considering the top 25 percentile value, 25% of samples (20) were categorized as high expression, whereas the rest (75%) of samples (59) were considered low or medium expression.
The UALCAN web portal has an important feature that aids querying based on the gene class. The search engine has precompiled sets of genes belonging to cancer-associated pathways, such as apoptosis, cell cycle, P53 signaling, Hedgehog signaling, immune pathways, and metastasis. In addition, it has also compiled genes belonging to kinases, and genes that regulate the post-translational modifications including methylation, ubiquitination, phosphorylation, acetylation, and proteolysis that influence the pathogenesis of cancer. Using the scan genes by the class query in UALCAN, we examined the pathways and genes that are differentially expressed in advanced stage IV ACC compared to stage I and II ACC that correlated with poor patient survival. Genes whose transcripts per million expressions are low and are not differentially expressed in stage IV in comparison to stage I and II are excluded from analysis.
Data analysis in cBioPortal
The cBioPortal is an open source, web program that facilitates data mining of TCGA. Of the 92 ACC patients included in TCGA consortium data, RNA-sequencing was done for tumors and matching normal tissues from 79 patients (48 females and 31 males) at different stages of ACC. The tumor stage breakdown of the patient population involved 9 patients in stage I, 37 in stage II, 16 in stage III, and 15 in stage IV. Expression levels of genes identified as significantly altered in advanced stage ACC and correlated with patient survival from UALCAN were mined for protein as well as mRNA expressions, and a gene “heat map” was generated using the functionality built into cBioPortal.
Statistical analysis
All searches were performed according to the online instruction of the UALCAN and cBioPortal. P values computed by the UALCAN web portal were used to calculate significance for gene expression and overall survival. Kaplan-Meier curves were used to evaluate associations between survival and gene expression variables. All analyses were unadjusted and performed with STATA version 13.0 (StataCorp, College Station, TX), and P value < .05 was considered statistically significant.
Results
Data mining of the UALCAN indicates upregulation of genes involved in DNA damage and pathways of the cell cycle in stage IV ACC when compared to lower stage ACC.
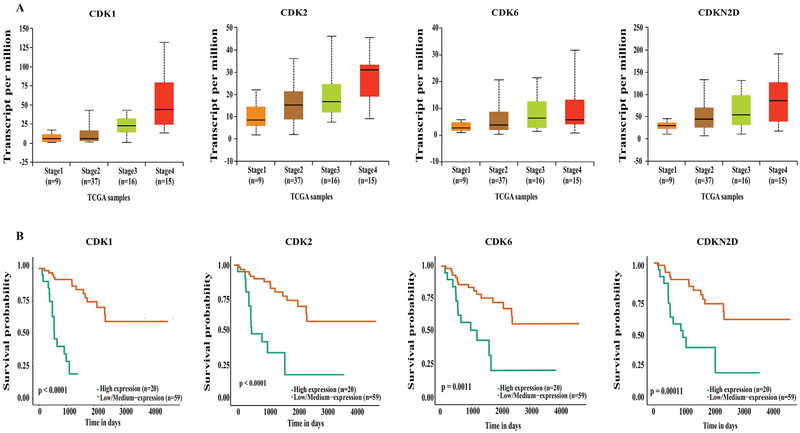
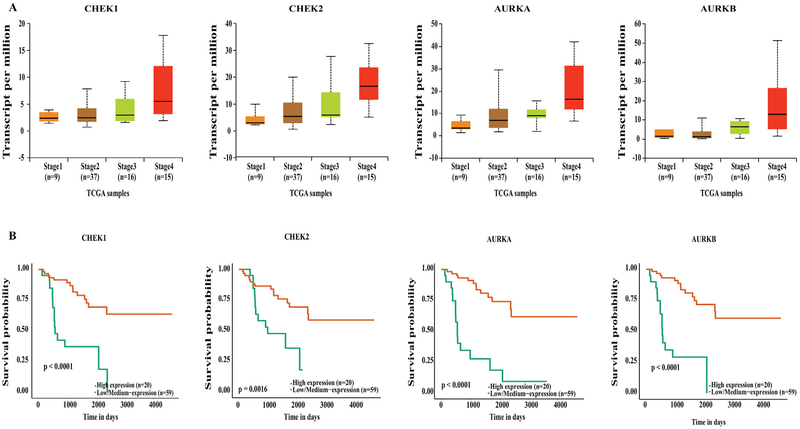
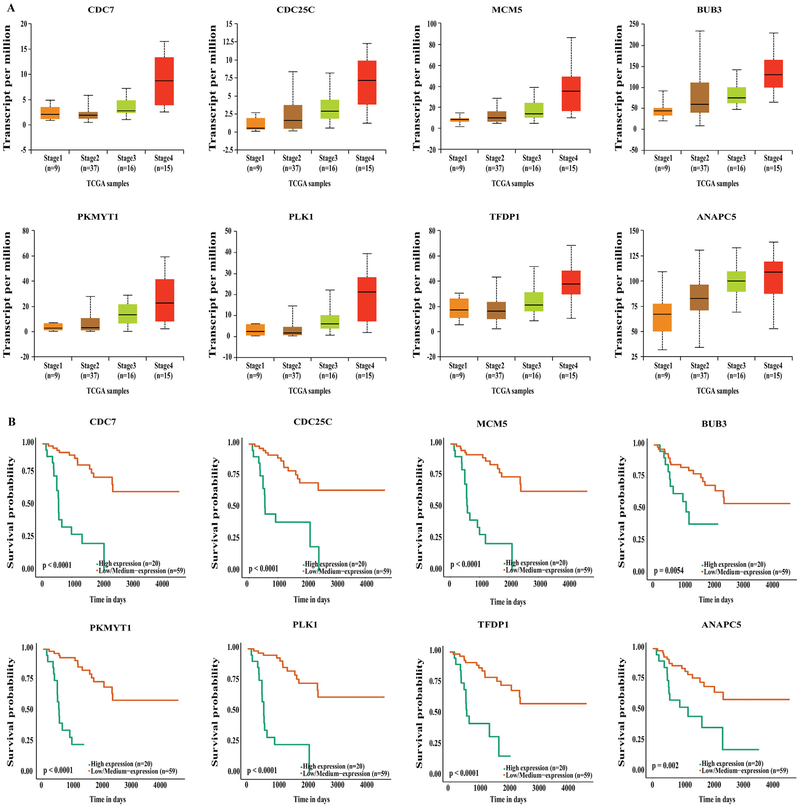
Our data mining of the UALCAN database demonstrated that genes involved in the cell cycle and DNA damage were upregulated in stage IV ACC compared to stage I and II (P < .01), independent of age or sex. The UALCAN output resulted in upregulation of cyclin-dependent kinases, such as CDK1, CDK2, CDK6, and cyclin-dependent kinase inhibitor CDKN2D, which were each associated with poorer survival (Fig 1, A and B). In addition, aurora kinases (AURKA and AURKB) and checkpoint kinases (CHEK1 and CHEK2) were also upregulated in stage IV compared to lower stage disease (P < .01), and their upregulation was associated with significantly poorer survival (Fig 2, A and B). Furthermore, genes critical for cell cycle progression (G0/G1 to S and Mitosis), DNA damage, and CDK-mediated phosphorylation of cdc6 were all upregulated in stage IV ACC (Fig 3, A), correlating with a worse survival (P < .05, P < .001, and P < .0001, respectively; Fig 3, B). These genes included the following: (1) highly conserved minichromosome maintenance protein MCM complexes (MCM 1, 2, 3, 5, and 7); (2) cell cycle–regulated E3 ubiquitin ligase, which controls progression through mitosis and the G1 phase of the cell cycle anaphase promoting complex/cyclosome (ANAPC 1, 5, 7, and 11); (3) mitotic spindle assembly checkpoint kinases (BUB1, BUB1B, and BUB3); (4) cell division cycle regulatory proteins that interact with several proteins at multiple stages of cell cycle progression (CDC7, CDC45, CDC20, CDC25A, CDC25B, and CDC25C); and (5) other proteins like PLK1, PKMYT1, and TFDP1.
Fig. 1.
(A) UALCAN output showing upregulation of cyclin-dependent kinases in stage IV ACC. (B) Kaplan-Meier survival curves showing upregulation of these genes significantly associates with poor survival compared to low and medium expression levels.
Fig. 2.
(A) UALCAN output showing upregulation of aurora and checkpoint kinases in stage IV ACC. (B) Kaplan-Meier survival curves showing upregulation of these genes significantly associates with poor survival compared to low and medium expression levels.
Fig. 3.
(A) UALCAN output showing upregulation of MCM complex genes, ANAPC genes, BUB genes and others in stage IV ACC. (B) Kaplan-Meier survival curves showing upregulation of these genes significantly associates with poor survival compared to low and medium expression levels.
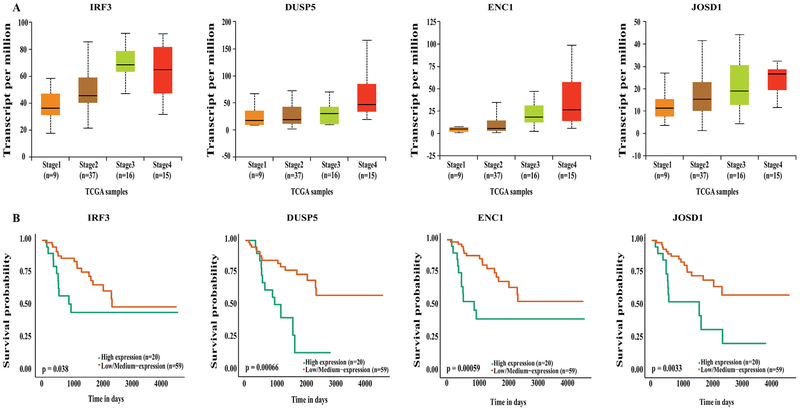
Genes involved in several other pathways, like immune modulation, metabolism, Wnt and RET signaling, were also significantly upregulated in stage IV ACC compared to stage I and II ACC, and this upregulation also correlated with a poorer survival (Figs 4, A and B). The genes that were overexpressed in stage IV ACC and correlated with a significantly worse survival included the following: (1) genes involved in the immune pathway: NF-kB, interferon, and TLR signaling genes (RelB, IRF3, and HMBG3), (2) genes involved in innate immunity: TRIM11 and LRSAM1, and (3) genes involved with RET signaling, metabolism, and Wnt signaling: DUSP5, CBL, ENC1, JOSD1, and several others.
Fig. 4.
(A) UALCAN output showing upregulation of immune pathway, RET signaling, metabolism and Wnt pathway representative genes in stage IV ACC. (B) Kaplan-Meier survival curves showing upregulation of these genes significantly associates with poor survival compared to low and medium expression levels.
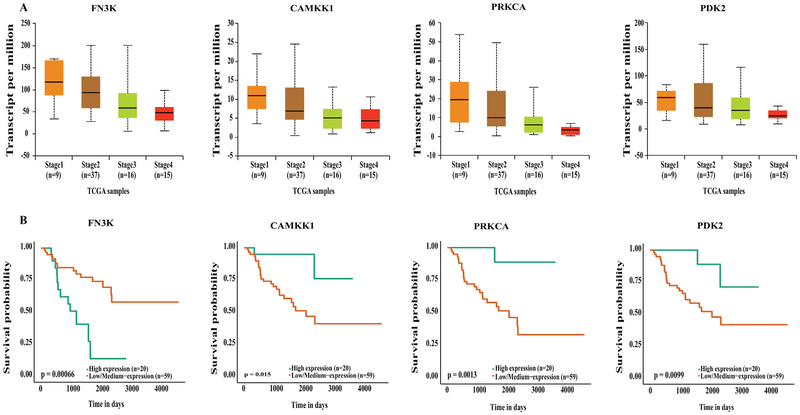
Significant downregulation was only observed in 4 genes (fructosamine 3 kinase [FNK3], calcium/calmodulin-dependent kinase 1 [CAMKK1], protein kinase C alpha [PRKCA], and pyruvate dehydrogenase kinase 2 [PDK2]) when comparing stage IV ACC to the stage I and II ACC, and each was significantly associated with poorer overall survival (Figs 5, A and B). Of these 4 downregulated genes, FN3K and PDK2 regulate metabolism, whereas CAMKK1 is involved in cell survival. The protein kinase C (PKC) family of proteins (including PRKCA) phosphorylate a wide variety of proteins and hence are involved in the regulation of several signaling pathways.
Fig. 5.
(A) UALCAN output showing downregulation of genes in stage IV ACC. (B) Kaplan-Meier survival curves showing upregulation of these genes significantly associates with poor survival compared to low and medium expression levels.
cBioPortal analysis
To further validate our UALCAN findings, we next queried for the expression of various gene signatures that were found to be upregulated in stage IV ACC compared to lower stage I, II, and III ACC. The genes associated with DNA damage and cell cycle pathway whose expression was significantly associated with worse survival of ACC patients were included in the query. Overall expressions of differentially expressed genes at the mRNA or protein levels were assessed using cBioPortal. Differentially expressed genes were evaluated using either mRNA z scores of RNAseq V2 RSEM or protein level z scores by reverse phase protein array (RPPA). The z score threshold was set at ±1.5, and this represents the mean expression values between altered and unaltered groups.
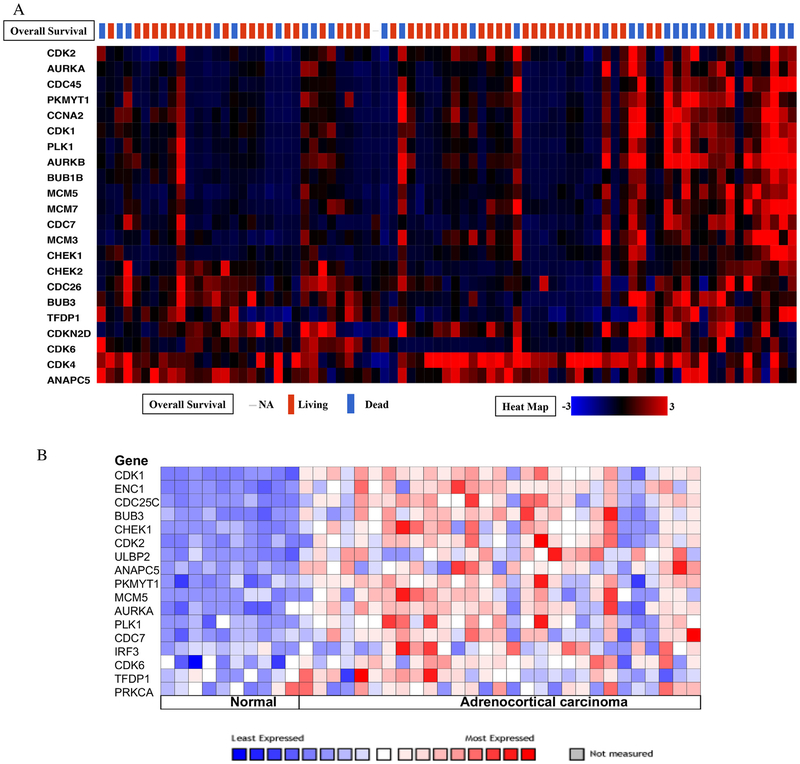
The results from cBioPortal data mining indicated that the genes involved in DNA damage and the cell cycle pathway were altered in 65 of the 79 sequenced patient samples (82%; Fig 6, A). Expression levels of less than 10% were not included in the figure given their low likelihood of contribution. Expression levels of all the queried genes, except the highly upregulated CDK4 gene (51% upregulated), was found to be correlated with poor survival in the UALCAN (each with P < .05). Of the 21 genes modulated in 68% of the patients, CDK1, AURKB, CDKN2D, and ANAPC5 were altered by 23%, 22%, 29%, and 32%, respectively, whereas the rest of the genes were altered between 10% and 20%. The heat map also indicated that approximately 80% of the patients who had several of these cell cycle genes upregulated (as clusters) had significantly worse overall survival than the 20% that did not cluster and were deceased. Additional, Oncomine Giordano ACC data analysis also revealed upregulation of genes involved in the DNA damage and the cell cycle pathway in ACC compared to normal adrenal (Fig 6, B).17 Together these results suggest that upregulated DNA damage and the cell cycle pathways may play a critical role in ACC biology and prognosis.
Fig. 6.
(A) The results from cBioPortal data mining indicated that cell cycle and DNA damage pathway related genes are altered in 82% of the sequenced patient samples. (B) Oncomine Giordano Data analysis revealed upregulation of cell cycle and DNA damage pathway genes in ACC compared to normal adrenal gland.
Discussion
ACC is a very rare disease with an annual incidence of 0.7 to 2 cases per million.18,19 Despite the recent identification and targeting of several important molecular pathways associated with its aggressiveness and biology, early clinical trials with these novel agents have not improved prognosis in advanced stage patients, warranting the search for better targets in this disease. Given the heterogeneity of ACC tumors across the population, there is a justifiable rationale that analyzing comprehensive genomic data across a large cohort of patients with the disease through TCGA network might be able to identify and unmask several genetic alterations that correlate well with prognosis and provide a new resource for biomarker identification. Through a better understanding of this relationship between genetic alterations in ACC and survival outcome, identification of these prognostic and diagnostic targets could then be translated more effectively into novel improved therapies for ACC patients. In this regard, we analyzed in the present study available data from TCGA and showed that genes involved in both DNA damage and the cell cycle pathway are highly upregulated in patients with advanced stage ACC.
Overall, 21 genes belonging to the DNA damage and cell cycle pathways were identified as potential biomarkers for highgrade, stage IV ACC, which were significantly modulated in their expression compared to early stage disease tumors. In addition, the overexpression of these biomarkers significantly correlated with poorer survival outcome. Transcriptome profiling of 33 ACC patients using Affymetrix Human Genome U133 plus 2.0 oligonucleotide arrays had identified 2,875 differentially regulated genes.20 Gene signature of the upregulated genes involved pathways of DNA damage and the cell cycle from our study included BUB1, CDC25A, and MCM5. Upregulation of these 3 cell cycle genes was also observed in the DNA microarray data, which corroborated our findings.20 Our use of data mining of differentially expressed genes in advanced stage IV ACC with poor survival is more clinically focused compared to global transcriptome profiling of the entire cohort (which is more commonly performed) without incorporation of survival outcome and staging for correlation. This approach allowed us to have a more clinically focused data analysis to the study and resulted in the identification of specific gene signatures that are uniquely upregulated only in advanced stage IV ACC. Because small molecule inhibitors targeting several genes that participate in DNA damage and cell cycle pathways are already in clinical trials, our findings contribute to a better understanding of the key drivers of poor prognosis in the disease; hopefully, this approach may improve the selection and rationale of new therapeutic strategies and combinations that have potential to enhance response and outcomes in ACC patients with more advanced disease.
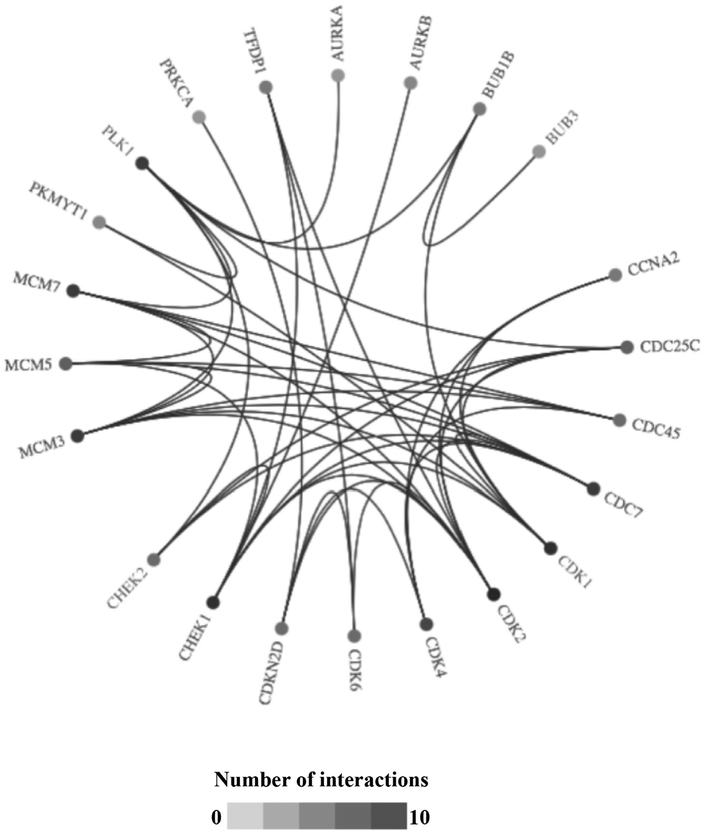
Uncontrolled cellular proliferation is directed to a great extent by cell cycle checkpoint proteins that play a key role in cancer development. Our analysis demonstrated an upregulation of DNA damage and cell cycle–related proteins like cyclin-dependent kinase, aurora kinase, polo-like kinase, and checkpoint kinase. Checkpoint kinases like CHEK1 and CHEK2 not only regulate cell cycle but also play an important role in DNA repair. There are several inhibitors that are currently in clinical trials in other diseases that target proteins involved in these proliferation modulating pathways. Comprehensive analysis of the literature and the Open Target bioinformatics platform resulted in several drugs that are currently undergoing clinical trials targeting the identified genes from the DNA damage and cell cycle pathways (Table 1).21 Additional connectivity analysis of the target gene sets in Open Target showed the interaction between these genes (Fig 7). Translating these genetic expression findings to ACC patients, there is rationale to consider use of inhibitors of cell cycle proteins currently in clinical trials in other cancers for advanced ACC patients. Given that uncontrolled growth and DNA repair alterations appear to be key genetic perturbations associated with poor prognosis in advanced ACC, there appears to be molecular rationale to utilize inhibitors of these cell cycle proteins (already in clinical use) in an attempt to improve prognosis and outcomes in ACC.
Table 1.
Cell cycle and DNA repair pathway proteins targeting inhibitors in clinical trials for cancer21,22,23
| Inhibitor | Targets | Clinical trials |
|---|---|---|
| Flavopiridol | ||
| AT7519 | Phase I | |
| MK7965 | Phase I | |
| AZD 5438 | Phase II | |
| R-roscovitine | Phase I | |
| PHA-848125/PHA-848125AC | Pan cdk | Phase I Phase I |
| PD0332991 | Phase III | |
| LY2835219 | Phase III | |
| LEE011 | CDK-4 and CDK-6 | Phase III |
| PHA-739358 | Phase IIand III | |
| MLN8237 | Pan AURK AURKA AURKB | Phase II |
| AZD1152 | ||
| Rigosertib | Phase II and III Phase II and III |
|
| Volasertib | PLK | Phase II |
| TKM-080301 | ||
| AZD1775 | Phase I and II | |
| MK8776 | Phase I and II Phase I and II |
|
| LY2606368 | CHEK1/CHEK2 | |
| ALISERTIB | AURKA | Phase III |
| ENMD-981693 | Phase II | |
| AT-9283 | Phase II | |
| TOZASERTIB | Phase II | |
| Danusertib | Phase II | |
| ILORASERTIB | Phase II | |
| TAS-119 | Phase I | |
| MK-5108 | Phase I | |
| RG-1530 | Phase I | |
| CYC-116 | Phase I | |
| MLN-8054 | Phase I | |
| TTP-607 | Phase I | |
| PF-03814735 | Phase I | |
| Barasertib | AURKB | Phase III |
| BI-811283 | Phase II | |
| ENMD-2076 | Phase II | |
| Chiauranib | Phase I | |
| GSK-1070916 | Phase I | |
| MK-6592 | Phase I | |
| KW-2449 | Phase I | |
| TAK-901 | Phase I | |
| AMG-900 | Phase I | |
| SNS-314 | Phase I | |
| BMS-863233 | CDC7 | Phase II |
| NMS-1116354 | Phase I | |
| DINACICLIB | CDK1 | Phase III |
| Milciclib | Phase II | |
| AG-24322 | Phase I | |
| RGB-286638 | Phase I | |
| TG-02 | Phase I | |
| AZD-5438 | Phase I | |
| SELICICLIB | CDK2 | Phase II |
| RG-547 | Phase II | |
| BMS-387032 | Phase I | |
| Ribociclib | CDK4 | Phase IV |
| ABEMACICLIB | Phase III | |
| Voruciclib | Phase II | |
| PALBOCICLIB | CDK6 | Phase IV |
| ALVOCIDIB | Phase III | |
| Roniciclib | Phase II | |
| AT-7519 | Phase II | |
| UCN-01 | Phase II | |
| PHA-793887 | Phase I | |
| LY-2606368 | CHEK1 | Phase II |
| RABUSERTIB | Phase II | |
| PREXASERTIB | Phase II | |
| RG-7602 | Phase I | |
| RG-7741 | Phase I | |
| SCH-900776 | Phase I | |
| XL-844 | Phase I | |
| AZD-7762 | Phase I | |
| PF-00477736 | Phase I | |
| SODIUM DICHLOROACETATE | PDK1 | Phase IV |
| VOLASERTIB | Phase III | |
| BI-2536 | Phase II | |
| MK-1496 | Phase I | |
| Cafusertib | Phase I | |
| NMS-1286937 | Phase I | |
| TAK-960 | Phase I | |
| GSK-461364 | Phase I | |
| MIDOSTAURIN | PRKCA | Phase III |
| SOTRASTAURIN | Phase II | |
| GSK-690693 | Phase I |
Fig. 7.
Connectivity pathway analysis of cell cycle and DNA damage genes using Open Target platform.23
Although this data analysis is very encouraging and sheds new light on some unexplored targets and mechanisms of disease progression and poor survival, there are several limitations to this approach. First, although TCGA database is by far the largest ACC cohort with full genetic analysis data, the total number of cases is < 100 patients, which limits the statistical power of these findings given the heterogeneity of the disease across the spectrum. Despite this limitation, the clustering of data even in this relatively small cohort demonstrates significant separation of gene targets between stage IV and earlier stage patients, which also correlates highly with prognosis differences. This is also the reason we utilized 2 separate genetic analysis platforms to validate our data and to limit signal to noise ratio given the large number of genetic expression signatures included in TCGA database. Another limitation of this study is that identification of novel gene targets that correlate with poorer prognosis may not translate into successful novel therapies that inhibit those targets. Therefore it would be prudent to validate the therapeutic potential of these novel ACC target genes and pathway with solid preclinical in vivo studies before clinical applications.
In conclusion, data mining of TCGA gene expression identified a unique set of gene alterations in advanced ACC patients related to genes involved in DNA damage and cell cycle pathways, which correlated significantly with poorer survival. These data suggest that targeting the DNA damage and cell cycle pathways either alone or in combination with current therapies could represent a novel therapeutic adjuvant approach for ACC patients. Because some of these pathway inhibitors are already in clinical trials for other malignancies, preclinical validation of the therapeutic potential of these novel ACC therapeutics would provide a strong rationale for rapid clinical translation to patients with advanced ACC who currently lack promising new therapeutic options for their disease.
Discussion
Dr Quan-Yang Duh (San Francisco, CA): Very nice presentation and very exciting stuff. I sort of see this as similar to the thyroid cancer genome study.
Mitotane has been around for a long time. Our experience has been that there are some patients for whom it worked well, but in most patients it actually didn’t work. I assume that some of the patients that you have in this series had Mitotane. Have you looked at those who have taken Mitotane, and then compared responders to non-responders, to see where they actually come out different?
Dr Mark Cohen: For the TCGA, actually, the tumors were taken at the time of initial surgery, so it was likely prior to any Mitotane exposure. We have not looked at that specifically to see the effects of Mitotane on these pathways, especially in patients who have recurred after Mitotane therapy. That’s something we certainly could do.
Dr Quan-Yang Duh (San Francisco, CA): It’s more of a question of whether you can predict which patient you should put on Mitotane.
Dr Mark Cohen: I think that’s certainly a possibility. In other cancers we have shown that doing the genetic analysis pre-treatment allows you to see which pathways are going to be susceptible. If it is not susceptible where Mitotane is targeting, then it’s probably not going to work. We have seen that with melanoma and some other types of tumors. That’s certainly something that we will plan to do at some point.
Dr James Howe (Iowa City, IA): Mark, this is great work, and hopefully this will lead to some translational improvements in the care of these patients. You focused on 10 or 20 genes that you showed us, but I’m sure you had hundreds or even thousands of differentially expressed genes. How did you decide to just look at those?
Dr Mark Cohen: What we initially did was to look at level of expression as some cutoff for what we are interested in, in terms of genes of interest. So all of these that were of interest had modulation of at least 5- to 10-fold over normal. That narrowed it down from a larger pool to a much smaller pool. Then we looked at these high-modulation genes and whether they were increased or decreased as our key targets to follow.
Acknowledgments
We would like to thank Dr Thomas Giordano, University of Michigan Department of Pathology, for his expertise and direction for our TCGA data analysis.
Footnotes
Presented at the 39th annual meeting of the American Association of Endocrine Surgeons in Durham, North Carolina, May 6–8, 2018.
Conflicts of interest
This work was partly funded by the National Institutes of Health (T32 CA009672, R01 CA173292, R01 CA120458), the University of Michigan Comprehensive Cancer Center Support Grant (P30-CA-046592), and the University of Michigan Department of Surgery. The authors have indicated that they have no other conflicts of interest regarding the content of this article.
References
- 1.Fassnacht M, Libe R, Kroiss M, Allolio B. Adrenocortical carcinoma: a clinician’s update. Nat Rev Endocrinol. 2011;7:323–335. [DOI] [PubMed] [Google Scholar]
- 2.Erdogan I, Deutschbein T, Jurowich C, Kroiss M, Ronchi C, Quinkler M, et al. The role of surgery in the management of recurrent adrenocortical carcinoma. J Clin Endocrinol Metab. 2013;98:181–191. [DOI] [PubMed] [Google Scholar]
- 3.Icard P, Goudet P, Charpenay C, Andreassian B, Carnaille B, Chapuis Y, et al. Adrenocortical carcinomas: surgical trends and results of a 253-patient series from the French Association of Endocrine Surgeons study group. World J Surg. 2001;25:891–897. [DOI] [PubMed] [Google Scholar]
- 4.Else T, Williams AR, Sabolch A, Jolly S, Miller BS, Hammer GD. Adjuvant therapies and patient and tumor characteristics associated with survival of adult patients with adrenocortical carcinoma. J Clin Endocr Metab. 2014;99:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassnacht M, Johanssen S, Quinkler M, Bucsky P, Willenberg HS, Beuschlein F, et al. Limited prognostic value of the 2004 International Union Against Cancer Staging Classification for adrenocortical carcinomas. Cancer. 2009;115:243–250. [DOI] [PubMed] [Google Scholar]
- 6.Fassnacht M, Kroiss M, Allolio B. Update in adrenocortical carcinoma. J Clin Endocr Metab. 2013;98:4551–4564. [DOI] [PubMed] [Google Scholar]
- 7.Fay AP, Elfiky A, Telo GH, McKay RR, Kaymakcalan M, Nguyen PL, et al. Adrenocortical carcinoma: the management of metastatic disease. Crit Rev Oncol Hematol. 2014;92:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroiss M, Quinkler M, Johanssen S, van Erp NP, Lankheet N, Pollinger A, et al. Sunitinib in refractory adrenocortical carcinoma: a phase II, single-arm, open-label trial. J Clin Endocrinol Metab. 2012;97:3495–3503. [DOI] [PubMed] [Google Scholar]
- 9.Berruti A, Sperone P, Ferrero A, Germano A, Ardito A, Priola AM, et al. Phase II study of weekly paclitaxel and sorafenib as second/third-line therapy in patients with adrenocortical carcinoma. European Journal of Endocrinology. 2012;166:451–458. [DOI] [PubMed] [Google Scholar]
- 10.Lerario AM, Worden FP, Ramm CA, Hasseltine EA, Stadler WM, Else T, et al. The combination of insulin-like growth factor receptor 1 (IGF-1R) antibody cixutumumab and mitotane as a first-line therapy for patients with recurrent/metastatic adrenocortical carcinoma: a multi-institutional NCI-sponsored trial. Horm Cancer-Us. 2014;5:232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naing A, Kurzrock R, Burger A, Gupta S, Lei X, Busaidy N, et al. Phase I trial of cixutumumab combined with temsirolimus in patients with advanced cancer. Clin Cancer Res. 2011;17:6052–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fassnacht M, Berruti A, Baudin E, Demeure MJ, Gilbert J, Haak H, et al. Linsitinib (OSI-906) versus placebo for patients with locally advanced or metastatic adrenocortical carcinoma: a double-blind, randomised, phase 3 study. Lancet Oncol. 2015;16:426–435. [DOI] [PubMed] [Google Scholar]
- 13.Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;29:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu JJ, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. Onocomine: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Else T, Kim AC, Sabolch A, Raymond VM, Kandathil A, Caoili EM, et al. Adrenocortical carcinoma. Endocr Rev. 2014;35:282–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilimoria KY, Shen WT, Elaraj D, Bentrern DJ, Winchester DJ, Kebebew E, et al. Adrenocortical Carcinoma in the United States: treatment utilization and prognostic factors. Cancer. 2008;113:3130–3136. [DOI] [PubMed] [Google Scholar]
- 20.Giordano TJ, Kuick R, Else T, Gauger PG, Vinco M, Bauersfeld J, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K Development of cell-cycle checkpoint therapy for solid tumors. Jpn J Clin Oncol. 2015;45:1097–1102. [DOI] [PubMed] [Google Scholar]
- 23.Koscielny G, An P, Carvalho-Silva D, Cham JA, Fumis L, Gasparyan R, et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45 D985–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]