Abstract
Combinatorial inhibition of MEK1/2 and CDK4/6 is currently undergoing clinical investigation in NRAS-mutant melanoma. To prospectively map the landscape of resistance to this investigational regimen, we utilized a series of gain- and loss-of-function forward genetic screens to identify modulators of resistance to clinical inhibitors of MEK1/2 and CDK4/6 alone and in combination. First, we identified NRAS-mutant melanoma cell lines that were dependent on NRAS for proliferation and sensitive to MEK1/2 and CDK4/6 combination treatment. We then employed a genome-scale ORF overexpression screen and a CRISPR knockout screen to identify modulators of resistance to each inhibitor alone or in combination. These orthogonal screening approaches revealed concordant means of achieving resistance to this therapeutic modality, including tyrosine kinases, RAF, RAS, AKT, and PIK3CA signaling. Activated KRAS was sufficient to cause resistance to combined MEK/CDK inhibition and to replace genetic depletion of oncogenic NRAS. In summary, our comprehensive functional genetic screening approach revealed modulation of resistance to the inhibition of MEK1/2, CDK4/6 or their combination in NRAS-mutant melanoma.
Introduction
There is a lack of effective therapies for NRAS-mutant melanoma, which accounts for 20–30% of all melanomas [1]. Preclinical studies have demonstrated that oncogenic NRAS dysregulates the mitogen activated protein kinase (MAPK) signaling cascade, creating a dependency in cell lines that can be exploited with inhibitors of MEK1/2 [2, 3]. However, in clinical trials single-agent MEK inhibitors [4] had a modest impact on progression-free survival (PFS) (NCT01763164, [5]), suggesting that MEK as a single agent was insufficient to achieve durable responses.
The pre-clinical observation that CDK4/6 inhibition could attenuate NRAS oncogenic signaling when combined with MEK inhibition supported the use of combined MEK1/2 and CDK4/6 inhibitor combination in NRAS mutant melanoma [6]. A clinical trial (NCT01781572) designed to evaluate this combination in NRAS-mutant melanoma patients revealed multiple partial responses [7, 8] and is under clinical evaluation in KRAS-mutant colon (NCT02703571), lung (NCT03170206), and pancreatic (NCT02703571) cancer. Thus, understanding the resistance landscape to MEK/CDK4/6 inhibition will be imperative for improving long-term patient responses.
We employed genome-wide functional genetic screening approaches to map the landscape of resistance to MEK1/2 inhibition, CDK4/6 inhibition, and their combination in NRAS-mutant melanoma. Our analyses revealed that RTK-PI3K-AKT and RTK-RAS-RAF signaling cascades were sufficient to drive resistance to combination MEK1/2 and CDK4/6 inhibition. Our study provides an initial description of the resistance landscape to MEK1/2 and CDK4/6 combination treatment in NRAS-mutant melanoma.
Materials and Methods
Cell Lines and Reagents
Cells were maintained in DMEM (Hs936T, Hs944T; Gibco), RPMI-1640 (MELJUSO, SKMEL30, IPC298; Gibco), or EMEM (SKMEL-2; Gibco) supplemented with 10% fetal bovine serum (Sigma), and incubated at 37°C in 5% CO2 per ATCC guidelines.
Western Blot Reagents
Cells were lysed in RIPA Buffer (25mM Tris•HCl pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS; phosphatase and protease inhibitors) and resolved by Tris-Gycline SDS-PAGE. To determine the levels of activated proteins, immunoblot analyses were done with phospho-specific antibodies to AKT(S473), MEK1/2(S217/S221), RB1 (S807/811), and ERK1/2 (T202/Y204), S6 (235/236) with antibodies recognizing total AKT, RB, ERK1/2, and S6 to control for total protein expression (Cell Signaling Technologies). Antibodies to EGFR, PI3K, CCNB1, CCND1, and CCNE2 (Cell Signaling Technologies), and NRAS and phospho-AKT (Santa Cruz) were used to monitor total protein expression. Antibody to KRAS4B was obtained from Calbiochem. Antibodies for cleaved PARP (Cell Signaling Technologies) were used to monitor apoptosis. Antibody for β-actin (Sigma AC15) was used to verify equivalent loading of total cellular protein.
Small Molecule Inhibitors
Trametinib and palbociclib were purchased from Selleckchem. Trametinib was dissolved in DMSO and stored at stock concentrations of 10mM at −20 °C. Palbociclib was dissolved in water and stored at stock concentrations of 10mM at −20 °C.
siRNA Transfections
siRNA silencer select oligonucleotides against scrambled and NRAS sequences were obtained from Invitrogen and transfected into cells by using Lipofectamine RNAiMAX, according to the manufacturer’s instructions.
Lentiviral Expression Vector Infections
The pLX317 GFP, AKT1, AKT2, AKT3, PI3K H1047R, PI3K E545K, EGFR L858R, NRAS Q61L, NRAS Q61K, KRAS WT, or KRAS G13D puromycin lentivirus vector were provided by The Broad Institute Genetic Perturbation Platform, and were transiently transfected into 293T cells with a ∆8.9 and VSV-G packaging system using XtremeGENE9. Infection of melanoma cell lines was done in growth media supplemented with 5 ug/ml polybrene and selected with 2 ug/ml of puromycin for 72 hrs.
Lentiviral CRISPR Vector Infections
The pLC_V2 GFP and sgRNA RB blasticidin lentivirus vector were constructed and were transiently transfected into 293T cells with a ∆8.9 and VSV-G packaging system using XtremeGENE9. Infection of melanoma cell lines was done in growth media supplemented with 5 ug/ml polybrene and selected with 2 ug/ml of blasticidin for 72 hrs. Cells were allowed to grow for 7 days post-infection before initiation of experiments.
Anchorage-Dependent Growth Assays
To monitor proliferation, cells were plated into 96-well plates at a density of 1 x 103 (MELJUSO, Hs944T, Hs936T, and IPC298) and 2 x 103 (SKMEL30 and SKMEL2) cells per well. To quantitate cell number, after 6 days cells were stained with [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) and absorbance was measured at 490 nm. We also performed a second proliferation assay to monitor clonogenic growth. Cells were plated at 5 x 103 (MELJUSO, Hs944T, and Hs936T) and 10 x 103 (SKMEL30) cells per well, but in 12-well plates. After 7 to 10 days, paraformaldehyde-fixed cells were stained with crystal violet to visualize and quantitate colony growth.
Determination of GI50
To determine the 50% growth inhibitory concentration (GI50), 1–2 x 103 cells were seeded in 96-well plates and treated with concentrations of trametinib (from 3 nM 1μM) or palbociclib (from 30 nM to 10 μM). After 6 days of treatment, cell proliferation was analyzed with MTS and read at an absorbance of 490 nm. GI50 values were calculated with PRISM software.
Cell Cycle Assay
Cell cycle progression was quantified by flow cytometry as described (Nusse et al., 1990). Cells were treated at GI50 or siRNA for 96 hr and then washed with phosphate-buffered saline (PBS), fixed in cold 70% ethanol, and then stained with propidium iodide (PI). Modfit software was used for analyses.
Cas3/7 Glo Assay
Apoptosis assays were performed according to the manufacturer’s instructions. Staurosporine was used at 1μM for 4 h as a positive control.
STRING Analysis
ORF and CRISPR candiates were collectively uploaded to https://string-db.org/. Clusters reflect high confidence interactions.
Trametinib and Palbociclib Titration for ORF and CRISPR screens
The doses of trametinib, palbociclib, or their combination to use in these screens were determined by propagating cells in different concentrations of trametinib, palbociclib, and their combhination to determine the effect on cell proliferation. In parallel, the level of phospho-ERK or phospho-RB depletion in cells treated with different concentrations of trametinib, palbociclib, or their combination was determined. For the proliferation assay, 1 x 106 MELJUSO cells were seeded in 10 cm plates without drug. MELJUSO cells were allowed to adhere for 24 hours, after which fresh media containing different concentrations of trametinib, palbociclib, or their combination was added. MELJUSO cells were passaged or media was refreshed every 3 days, and cells were counted at each passage. For immunoblots, MELJUSO cells were treated with DMSO or the indicated concentrations of trametinib, palbociclib, or their combination for 48 hours. Phospho-ERK or phospho-RB levels were assessed by immunoblot analysis.
Trametinib and Palbociclib Titration for ORF and CRISPR screens
The doses of trametinib, palbociclib, or their combination to use in these screens were determined by propagating cells in different concentrations of trametinib, palbociclib, and their combhination to determine the effect on cell proliferation.
Human ORFeome and Avana CRISPR lentiviral Libraries
Human ORFeome barcoded library was cloned in the pLX317 barcoded vector and contains 17,255 ORFs overexpressing 10,135 distinct human genes with at least 99% nucleotide and protein match (Yang et al., 2011). The creation, cloning, sequencing, and production of the Avana library have been previously described (Doench et al., 2016). The Avana library contains 73,687 barcoded guide RNAs (sgRNAS) targeting 18,675 genes and 1000 non–targeting guides (Doench et al., 2016).
Genome Scale ORF Resistance Screen
To titer the ORF library in MELJUSO cells, 3 x 106 cells were seeded per well in a 12-well plate and were infected with different amounts of virus (0, 50, 100, 150, 200, 400 µL), with a final concentration of 4 µg/mL polybrene. Cells were spun for 2 hours at 2000 rpm at 30 degrees. Approximately 6 hours after infection, 100,000 cells from each infection were seeded into duplicate wells in a 6-well plate. 24 hours after infection, one well was treated with puromycin and one with media alone. After 2–3 days of selection, cells were counted to determine the amount of virus that resulted in 30–50% infection efficiency, and this amount of virus was used in the screen. For the MELJUSO ORF resistance screen, 1 x 108 cells were infected per replicate with 40% infection efficiency, in order to obtain 1000 cells per ORF after selection (~2 x 107 surviving cells containing 17,255 ORF clones). 3 x 106 cells per well were seeded in 12-well plates and were infected with the amount of virus determined during optimization, with a final polybrene concentration of 4 µg/mL. Plates were spun for 2 hours at 2000 rpm at 30 degrees. Approximately 24 hours after infection, all wells within a replicate were pooled and were split into T225 flasks. 48 hours after infection, cells were selected in 2 µg/mL puromycin for 5 days and expanded in puromycin-free media for 4 days (MELJUSO). For MELJUSO, 4.0 x 107 cells were seeded in T225 flasks with 10 nM trametinib, 2μM palbociclib, and 10nM trametinib/2μM palbociclib on Day 0. Cells were passaged in drug or fresh media containing trametinib, palbociclib, or trametinib/palbociclib was added every 3–4 days. Cells were harvested 21 days after initiation of trametinib, palbociclib, or trametinib/palbociclib treatment. Genomic DNA was extracted using the Qiagen Blood and Cell Culture DNA Maxi Kit according to the manufacturer’s protocol. PCR of gDNA and pDNA (ORF plasmid pool used to generate virus) was performed as previously described (Doench et al., 2016).
Genome Scale CRISPR Resistance Screen
To generate cell lines stably expressing Cas9, cells were infected with the Cas9 expression vector pXPR_BRD111 and selected with 10 µg/mL blasticidin for 4–7 days. Cas9-expressing cells were maintained in 2–5 µg/mL blasticidin. To titer the Avana library in Cas9-expressing cells, 3 x 106 cells were seeded per well in a 12-well plate and were infected with different amounts of virus (0, 50, 100, 150, 200, 400 µL), with a final concentration of 4 µg/mL polybrene. Cells were spun for 2 hours at 2000 rpm at 30 degrees. Approximately 6 hours after infection, 100,000 cells from each infection were seeded into duplicate wells in a 6-well plate. 24 hours after infection, one well was treated with puromycin and one with media alone. After 2–3 days of selection, cells were counted to determine the amount of virus that resulted in 30–40% infection efficiency, and this amount of virus was used in the screen. For the MELJUSO Avana resistance screen, 1 x 108 cells were infected per replicate with 40% infection efficiency, in order to obtain 500 cells per sgRNA after selection (4.0 x 107 surviving cells containing ~77,000 sgRNAs). 3 x 106 cells per well were seeded in 12-well plates and were infected with the amount of virus determined during optimization, with a final polybrene concentration of 4 µg/mL. Plates were spun for 2 hours at 2000 rpm at 30 degrees. Approximately 6 hours after infection, all wells within a replicate were pooled and were split into T225 flasks. 24 hours after infection, cells were selected in 2 µg/mL puromycin for 5 days and expanded in puromycin-free media for 4 days (MELJUSO). For MELJUSO, 4.0 x 107 cells were seeded in T225 flasks with 10 nM trametinib, 2μM palbociclib, and 10nM trametinib/2μM palbociclib on Day 0. Cells were passaged in drug or fresh media containing trametinib, palbociclib, or trametinib/palbociclib was added every 3–4 days. Cells were harvested 21 days after initiation of trametinib, palbociclib, or trametinib/palbociclib treatment. Genomic DNA was extracted using the Qiagen Blood and Cell Culture DNA Maxi Kit according to the manufacturer’s protocol. PCR of gDNA and pDNA (sgRNA plasmid pool used to generate virus) was performed as previously described (Doench et al., 2016).
Analysis of ORF Screen
The log2(fold-change) in ORF representation between cells treated with trametinib, palbociclib, or trametinib/palbociclib for 21 days and the initial early time point of ORFs plasmid used to generate virus was calculated. Then a Z-score was calculated and a ORF was considered significant if the Z-score ≥ 2 (See Supplementary Table S1).
Analysis of CRISPR Screen
The log2 (fold-change) in sgRNA representation between cells treated with trametinib, palbociclib, or trametinib/palbociclib for 21 days and the initial pool of sgRNAs plasmid used to generate virus was calculated. Then a Z-score (x ≥ 2) was calculated. A gene was considered significant if at least half of the sgRNAs targeting the gene became enriched in the screen. STAR values were also calculated for each gene (See Supplementary Table S1).
Results
NRAS mutant melanoma cell lines are NRAS-dependent
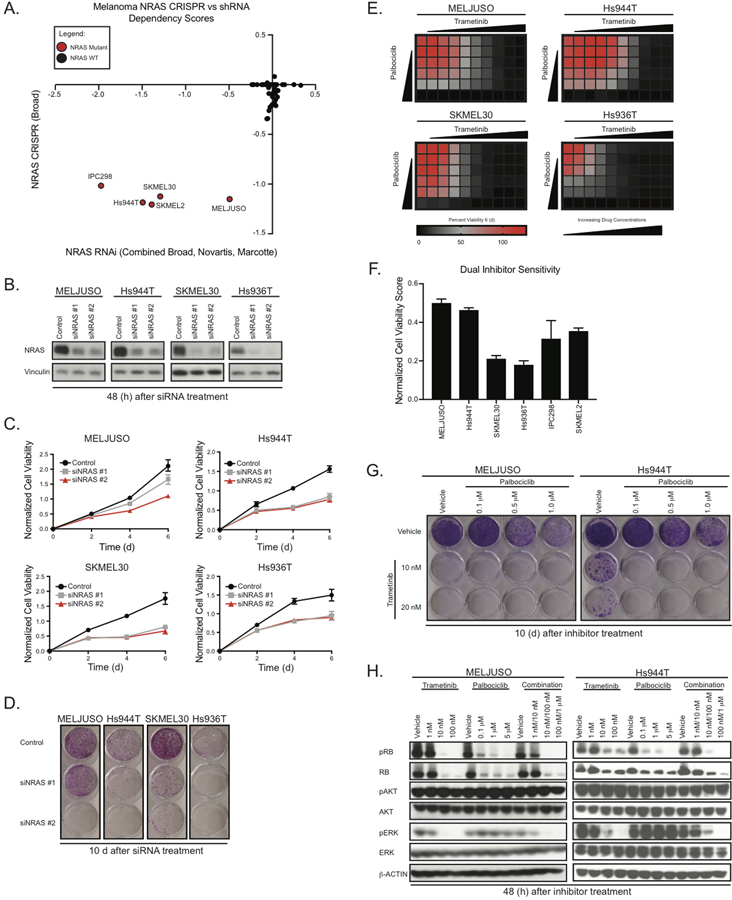
To determine the relationship between NRAS mutation and NRAS dependency in melanoma cell lines, we analyzed shRNA and CRISPR dependency data from the Broad Institute Dependency Map (https://depmap.org/portal/) [9, 10]. These analyses revealed that 5 of 5 NRAS-mutant melanoma cell lines were dependent on NRAS compared to NRAS wild-type cell lines (Fig. 1A). To validate this observation, we suppressed NRAS expression in three out of four cell lines identified in the Broad Institute Dependency Map using two NRAS targeting siRNAs. After 48 hours, we observed a greater than 90% NRAS knockdown compared to the siRNA control (Fig. 1B). NRAS depletion reduced cell viability after 4 days in an MTS assay and 10 days in an anchorage-dependent clonogenic growth assay (Fig. 1C and 1D). We observed that siNRAS #1 was mostly ineffective in the MELJUSO cell line, which harbors both activating NRAS and HRAS mutations. After performing qPCR to evaluate mRNA levels of HRAS and NRAS, we found that siNRAS #1 targets only NRAS, while siNRAS #2 targets both NRAS and HRAS.
Figure 1.
NRAS mutant melanoma cell lines are NRAS dependent and sensitive to trametinib and palbociclib. A, Dependency scores (CRISPR vs shRNA) identify NRAS dependent melanoma cell lines. Red circles denote NRAS mutant and black circles denote NRAS WT [19, 20]. B, Representative immunoblots displaying the effect of NRAS knockdown after 48 h on total protein levels of NRAS. Vinculin immunoblotting was used to determine equivalent loading. C, Cell Titer MTS tetrazolium assay displaying mean cell viability of MELJUSO, SKMEL30, Hs936T, and Hs944T melanoma cell lines with or without knockdown of NRAS (six replicates), each from three independent experiments. D, Representative images of crystal violet stained clonogenic growth assays with MELJUSO, SKMEL30, Hs944T, and Hs936T melanoma cell lines with or without knockdown of NRAS. E, MTS assays showing mean cell viability of MELJUSO, Hs944T, SKMEL30, and Hs936T melanoma cell lines after 144 h of combination trametinib and palbociclib treatment, each from three independent experiments performed in triplicate. F, Normalized Cell Viability Score where DMSO is 100% cell viability as measured by MTS in MELJUSO, Hs944T, SKMEL30, Hs936T, IPC298, and SKMEL2. G, Representative images of crystal violet stained clonogenic growth assays with MELJUSO and Hs944T melanoma cell lines after 10 d of trametinib and palbociclib treatment, each from three independent experiments, performed in duplicate. H, Representative immunoblots displaying the effect of trametinib and palbociclib treatment in MELJUSO and Hs944T melanoma cell lines after 48 h on phosphorylated and total protein levels of RB, AKT, and ERK. β-actin immunoblotting was used to determine equivalent loading.
To assess the mechanism(s) through which NRAS suppression impairs growth, we investigated the modulation of cleaved PARP a well-established marker of cell death. We observed an increase in cleaved PARP total protein levels after 96 h under NRAS siRNA targeting conditions in Hs936T and Hs944T, but not MELJUSO and SKMEL30 (Supplementary Fig. 1A). Additionally, we observed increased activity of caspase-3 and caspase-7, known markers of apoptosis, in both Hs936T and Hs944T after 96 h of NRAS suppression (Supplementary Fig. 1B).
Upon NRAS suppression, a subpopulation of cells underwent programmed cell death, while a remaining population did not. To explore this observation, we evaluated the effect of NRAS depletion after 96 h on cell cycle progression using a propidium iodide (PI) stain. Our analysis revealed a significant increase in G1 arrested cells upon loss of NRAS (Supplementary Fig. S1C). This was also accompanied by a reduction in the number of cells in both S phase and G2M. Consistent with our PI staining results, we observed a reduction in the steady state protein levels of phospho-RB, cyclin B1, cyclin D1, and cyclin E2, markers of cell cycle progression (Supplementary Fig. S1D). Thus, while populations of NRAS-mutant melanoma cell lines are dependent on NRAS for growth, their response to NRAS depletion is heterogeneous, with a subset undergoing cell arrest, and others undergoing apoptosis.
NRAS-dependent melanoma cell lines are sensitive to pharmacological MEK and CDK inhibition
Next, we evaluated whether our panel of NRAS mutant melanoma cell lines were sensitive to pharmacological inhibition of MEK1/2 and CDK4/6. To test this, we employed two FDA approved inhibitors, trametinib and palbociclib and determined whether our panel of cell lines was sensitive to these agents. NRAS mutant melanoma cell line sensitivity, after 72 h and 144 h, to each single agent varied ranging from 1nM-10nM GI50 for trametinib and 1μM-10μM GI50 for palbociclib (Supplementary Fig. S1E).
Next, we evaluated the effect of combination trametinib and palbociclib treatment in our panel. We performed an MTS assay, where we treated each cell line with increasing doses of each drug and measured cell viability after 144 h. We observed that each cell line was sensitive to each drug individually as well as in combination (Fig. 1E, Supplementary Fig. S1F). By averaging the percent cell viability count of each well compared to the vehicle control, we calculated a Normalized Cell Viability Score to represent dual inhibitor sensitivity (Fig. 1F). Based on the Normalized Cell Viability Score, our panel of NRAS mutant melanoma cell lines are differentially sensitive to dual inhibition of MEK1/2 and CDK4/6. To elucidate the effect of combination trametinib and palbociclib treatment on anchorage-dependent colony formation, we treated MELJUSO, SKMEL30, Hs936T, and Hs944T with varying doses of trametinib (10nM and 20nM) and palbociclib (100nM, 0.5μM, and 1μM) as single agents as well as the combination for 10 d (Fig. 1G). Consistent with our previous observations, each cell line was sensitive to both single agents as well as the combination, though the dose response varied between cell lines.
In parallel, we assessed the levels of pathway inhibition after treatment with trametinib and palbociclib, alone and in combination after 48 h. Treatment with single agent trametinib reduced steady state levels of phospho-ERK and phospho-RB in all cell lines tested (Fig. 1H, Supplementary Fig. S1G). It has been previously shown that MAPK pharmacological inhibition can result in changes to steady state levels of phospho-AKT; in contrast, in our model, phospho-AKT levels remained constant after treatment with trametinib.
Next, we evaluated the effect of single agent palbociclib treatment in our panel of cell lines. We observed palbociclib inhibition led to a reduction in steady state levels of phospho-RB (Fig. 1H, Supplementary Fig. S1G and S1H) and an increase in CCND1 (Supplementary Fig. S1H) total protein levels, both of which are consistent with previous studies [7, 11]. Palbociclib inhibition did not alter the MAPK signaling components RAF and MEK, however we did observe an increase in phospho-ERK levels (Fig. 1H).
Finally, we examined the effects of combination trametinib and palbociclib treatment on pathway inhibition. We observed a reduction in steady state levels of phospho-ERK and phospho-RB; however, unexpectedly, combination treatment did not significantly further reduce phospho-RB levels, in 3 out of 4 NRAS mutant melanoma cell lines, with Hs936T being inconclusive (Fig. 1H).
To assess the mechanism(s) through which trametinib and palbociclib, alone and in combination, impair growth in our panel of NRAS mutant melanoma cell lines, we examined levels of cleaved PARP and cleaved caspase-3. We did not observe changes to cleaved PARP or cleaved caspase-3 total protein levels after either 48 h or 96 h of inhibitor treatment. This is in contrast to previous studies, where cell line models of NRAS-mutant melanoma underwent programmed cell death after treatment with either a MEK inhibitor alone or in combination with a CDK4/6 inhibitor [6]. However, this could be a result of using distinct melanoma cell lines, as our data support the hypothesis that NRAS-mutant melanoma cell lines are heterogeneous.
To understand the mechanisms of growth impairment following combination drug treatment, we evaluated the effect of trametinib and palbociclib, alone and in combination on cell cycle progression. We detected a significant increase in G1 arrested cells and a reduction in the number of cells in both S phase and G2M after 96 h of inhibitor treatments (Supplementary Fig. S1I). This observation is consistent with other studies evaluating the effect of trametinib and palbociclib on cell cycle progression.
Next, we examined changes to cell cycle markers after combination treatment. Protein levels of phospho- RB, cyclin B1, and cyclin D1 were reduced after inhibitor treatments at 96 h (Supplementary Fig. S1H). Given our observations with both genetic NRAS-depletion and pharmacological inhibition of MEK1/2 and CDK4/6 in NRAS mutant melanoma cell lines, we conclude these perturbations share a similar mechanism of action as it relates to cell cycle progression. However, it is clear that a subset of cell lines undergo programmed cell death upon NRAS depletion, which is not the case with pharmacological inhibition of MEK1/2 and CDK4/6 in our model. Together, our results support the hypothesis that NRAS dependent melanoma cell lines are sensitive to pharmacological inhibition of MEK1/2 and CDK4/6.
A functional landscape of resistance to pharmacological inhibition of MEK1/2 and CDK4/6 in NRAS-mutant melanoma using an ORF expression library
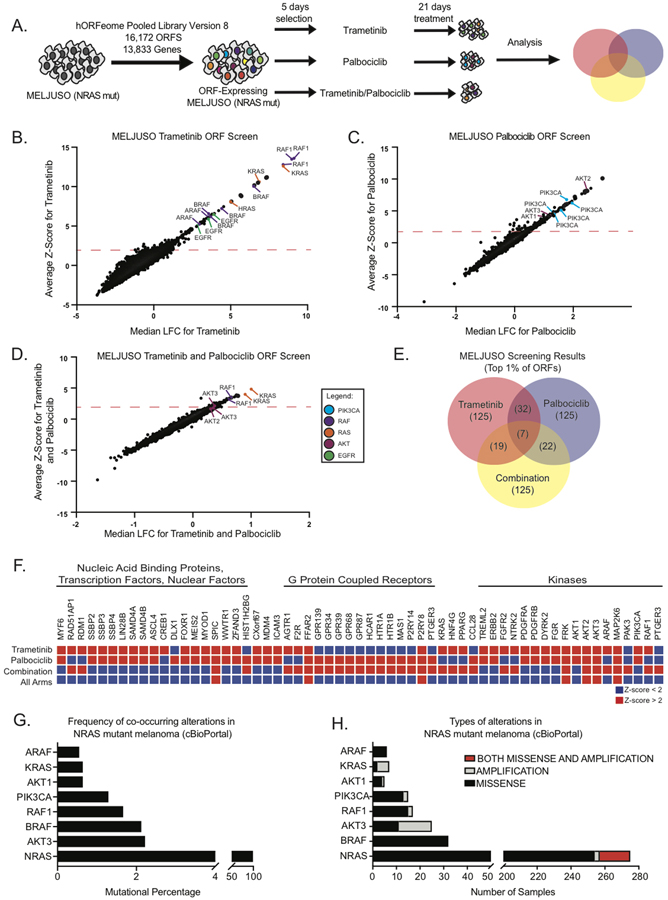
To elucidate the resistance landscape to MEK1/2 and CDK4/6 inhibition in NRAS mutant melanoma, we performed a near genome-wide pooled ORF over-expression screen to identify genetic modulators of resistance [12–16]. We performed a three-armed screen in NRAS mutant MELJUSO cell lines using trametinib (10nM), palbociclib (2μM), and trametinib/palbociclib (10nM/2μM) (Fig. 2A and Supplementary Fig. S2A, S2B, and S2C) [17].
Figure 2.
Systematic genetic ORF expression screen to understand resistance to trametinib and palbociclib in NRAS mutant melanoma cell lines. A, Screening schematics and timelines. (B, C, and D), MELJUSO cell line stably expressing the Human ORFeome Pooled Library Version 8 were treated for 21 days with trametinib (B), palbociclib (C), and trametinib/palbociclib combination. E, Venn diagram of screening candidates with a z-score greater than 2, trametinib (125), palbociclib (125), and trametinib/palbociclib (125). F, Heatmap displaying the overlap of candidates identified in each arm of the screen based on the z-score. (G and H), Identification of co-occuring mutations in NRAS-mutant melanoma from patient samples obtained from cBioPortal (www.cbioportal.org).
Our screening results revealed distinct ORFs whose over-expression caused resistance to trametinib and/or palbociclib (Fig. 2B, 2C and 2D). In the trametinib arm, we identified several genes involved in the MAPK signaling cascade, consistent with previously published screens using MAPK inhibition (Fig. 2B) [12, 18, 19]. In the palbociclib arm, we observed a significant enrichment of genes involved in the PI3K-AKT signaling cascade (Fig. 2C). In the combination trametinib/palbociclib arm, activated KRAS ORFs were the top scoring candidates (Fig. 2D). We observed some candidate overlap between trametinib and palbociclib single agents in the DNA and RNA binding proteins, G-protein coupled receptors (GPCR), tyrosine and serine/threonine protein kinases, and transcription factors (Fig. 2E and 2F). In the combination, network analysis of resistance candidates using the STRING protein-protein interaction platform identified a Ras signaling interactome and a GPCR interactome (Supplementary Fig. S2D)
Since there are limited clinical samples available for the evaluation of resistance to this combination [7, 8], we used cBioPortal (http://www.cbioportal.org/) to determine whether proteins identified in the ORF over-expression screen were altered either through point mutation or amplification in NRAS-mutant melanoma. We observed that ARAF, KRAS, AKT1, PIK3CA, RAF1, BRAF, and AKT3 were either mutated (hotspot) or amplified in NRAS-mutant melanoma (Fig. 2G and 2H), suggesting that co-occurrence of these alterations with NRAS mutation is plausible in nature.
A functional landscape of resistance to pharmacological inhibition of MEK1/2 and CDK4/6 in NRAS-mutant melanoma using a CRISP knockout library
Next, we performed a CRISPR knockout screen and assessed resistance to MEK1/2 and CDK4/6 inhibition in NRAS-mutant melanoma. We performed a three-armed screen in MELJUSO cell lines using trametinib (10nM), palbociclib (2μM), and trametinib/palbociclib (10nM/2μM) (Fig. 3A and Supplementary Fig. S3A and S3B) [20, 21]. In the trametinib arm, we observed an enrichment in negative regulators of both the EGFR-PI3K and EGFR-Ras signaling cascades. We also identified several PPP family phosphatases, transcription factors, E3 ligases, and epigenetic regulators. Overall, our CRISPR screening results support mechanisms of resistance identified in the trametinib arm of the ORF overexpression screen (Fig. 3B).
Figure 3.
Systematic CRISPR knockout screen to evaluate resistance to trametinib and palbociclib in NRAS mutant melanoma cell lines. A, Screening schematics and timelines. (B, C, and D), MELJUSO cell line stably expressing the Avana Pooled Library were treated for 21 days with trametinib (B), palbociclib (C), and trametinib/palbociclib combination. E, Venn diagram of screening candidates with a z-score and guide number greater than or equal to 2, trametinib (150), palbociclib (150), and trametinib/palbociclib (150). F, Identification of co-occuring mutations in NRAS-mutant melanoma from patient samples obtained from cBioPortal (www.cbioportal.org). G, EGFR signaling pathway displaying candidates from both the CRISPR and ORF screens.
Two independent studies evaluated the resistance landscape to trametinib in KRAS mutant (PDAC and LUAD) and NRAS mutant (LUAD) cancers [22, 23]. After analyzing all four screens, we found a subset of candidate genes associated with resistance to trametinib in two or more contexts, while a majority of candidates were unique to each cellular context (Supplementary Fig. S3C). The comparison of the melanoma NRAS screen and the PDAC KRAS screen appears informative because these screens use the same CRISPR libraries revealing CIC, KEAP1, and PPP6C to modulate MEK inhibitor sensitivity (Supplementary Fig. S3D). From these data we conclude there are likely resistance mechanisms to MEK inhibition that span disease and lineage boundaries.
The top scoring guides from the palbociclib arm of the CRISPR screen were RB1 targeting guides, which is consistent with the known mechanism of action for palbociclib (Fig. 3C) [24, 25]. We utilized two sgRNAs that efficiently knocked out RB1 as measured by protein expression (Supplementary Fig. S3E) and led to palbociclib resistance (Supplementary Fig. S3F). In addition, we identified multiple guides targeting NF2, PTEN, RBL2, and RALGAPB, all of which are regulatory components of the PI3K-AKT-mTOR signaling cascade [26] and are consistent with our results from the ORF expression screen.
Similar to our observations in the ORF screen, there were far fewer genes whose deletion could drive resistance to combination MEK/CDK inhibition relative to those discovered for single agents (Fig. 3D and 3E). In addition, many of the top scoring genes only had a single scoring guide in the presence of the combination, technically limiting our ability to identify genes that drive resistance to the combination. However, we did observe RB1, which has been linked to Ras signaling and has been implicated in resistance to CDK4/6-directed therapeutics. Together with the ORF screening data, these findings suggest that few genes beyond Ras, itself, can overcome combination trametinib/palbociclib treatment.
Similar to the ORF over expression screen, we observed several CRISPR candidates that were either truncated or deleted in patient samples (NF2, TSC1, FBXW7, RB1, PTEN, TSC2, PPP6C, and TP53) (Fig. 3F), again suggesting that co-occurrence with NRAS is feasible in human tumors (cbioportal). Together the ORF and CRISPR screening results highlight modulators to MAPK and PI3K signaling cascades that are clinically plausible (Fig. 3G).
After performing both the ORF over-expression and the CRISPR depletion screens, we gathered potential candidates based on the aforementioned statistical measurements and conducted a STRING analysis, which is an unbiased pathway network analysis tool. From these analyses, we found that the RTK-RAS-RAF and RTK-PI3K-AKT pathways were amongst the most highly enriched pathways (Supplementary Fig. S3G). Given the tractable clinical targets, we focused on validating various members of these two signaling cascades.
EGFR-PI3K-AKT signaling modulates resistance to trametinib/palbociclib treatment in NRAS mutant melanoma cell lines
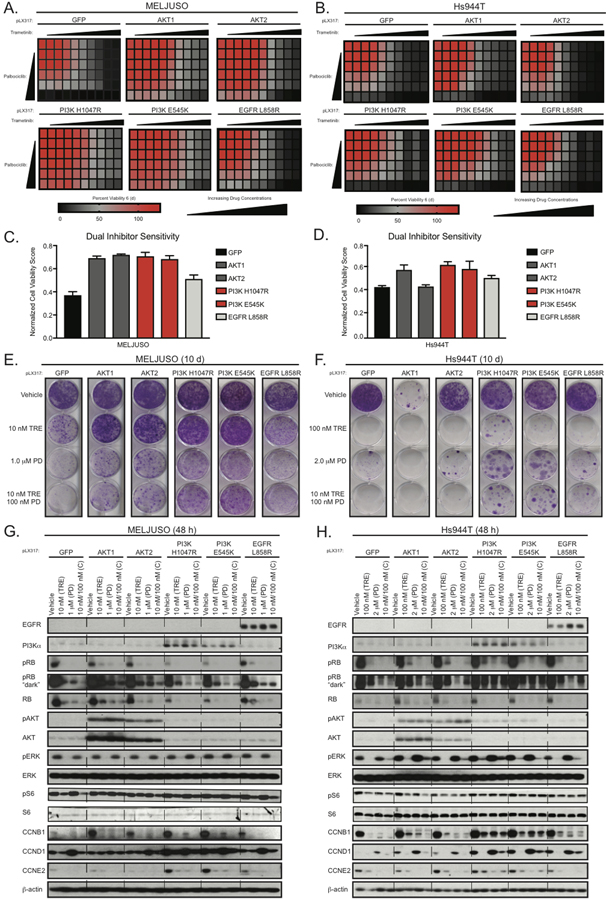
Next, we validated if the EGFR-PI3K-AKT signaling cascade can impact response to trametinib and/or palbociclib. The ORF over-expression screen revealed that wild-type AKT, activated PI3K, and activated EGFR all conferred resistance, thus we chose validation constructs representing this observation (Supplemental Table S1). We infected MELJUSO, Hs944T, SKMEL30, and Hs936T cell lines with GFP, AKT1, AKT2, PI3KH1047R, PI3KE545K, and EGFRL858R, and then treated them with trametinib and/or palbociclib in a 6 d cell viability assay. Consistent with our previous observation that NRAS mutant melanoma cell lines are heterogeneous, our screening validation data highlight distinct modulators of resistance amongst the cell lines assayed. We observed that AKT1, AKT2, PI3KH1047R, PI3KE545K, or EGFRL858R expression led to resistance to trametinib and/or palbociclib in MELJUSO and Hs936T, (Fig. 4A and 4B, Supplementary Fig. S4A and S4B), whereas only PI3KH1047R or PI3KE545K mediated resistance in SKMEL30 and Hs944T (Fig. 4C and 4D, Supplementary Fig. S4C and S4D). AKT3 caused resistance to both inhibitors alone and in combination in the MELJUSO, SKMEL30, and Hs936T cell lines (Supplementary Fig. S4E and S4F).
Figure 4.
EGFR-PI3K-AKT signaling modulates resistance to trametinib/palbociclib treatment in NRAS mutant melanoma cell lines. (A and C), Cell Titer MTS tetrazolium assays showing mean cell viability of MELJUSO (A) and Hs944T (C) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR after 144 h of combination trametinib and palbociclib treatment, each from three independent experiments performed in triplicate. (B and D), Normalized Cell Viability Score where DMSO is 100% cell viability as measured by MTS in MELJUSO (B) and Hs944T (D). (E and F), Representative images of crystal violet stained clonogenic growth assays with MELJUSO (E) and Hs944T (F) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR after 10 d of trametinib, palbociclib, and trametinib/palbociclib combination treatment, each from three independent experiments performed in triplicate. (G and H), Representative immunoblots displaying the effect of trametinib, palbociclib, and trametinib/palbociclib combination treatment in MELJUSO (G) and Hs944T (H) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR after 48 h on phosphorylated and/or total protein levels of RB, AKT, ERK, S6, CCNB1, CCND1, and CCNE2. β-actin immunoblotting was used to determine equivalent loading.
Next, we determined the effect of the EGFR-PI3K-AKT signaling cascade on clonogenic growth. In MELJUSO we observed that expression of AKT1, AKT2, PI3KH1047R, and PI3KE545K caused resistance to trametinib (Fig. 4E), whereas EGFRL858R had a modest effect. Notably, PI3KH1047R and PI3KE545K only mediated trametinib resistance in Hs944T (Fig. 4F). Expression of AKT1 alone in the Hs944T cell line caused an unexpected, yet reproducible, growth defect. In MELJUSO and Hs944T, expressing AKT1 (only MELJUSO), AKT2, PI3KH1047R, PI3KE545K or EGFRL858R drove resistance to palbociclib (Fig. 4E and 4F). We observed a similar pattern of insensitivity in cells treated with the combination, though resistance phenotypes were reduced for AKT2 and EGFR. These results are consistent with our cell viability findings. Overall, our data suggests that the EGFR-PI3K-AKT signaling cascade can modulate resistance to trametinib and/or palbociclib, though the strength of resistance is cell line dependent.
Next, we assessed pathway inhibition in the MELJUSO, SKMEL30, Hs936T, and Hs944T cell lines expressing AKT1, PI3K, or EGFR constructs to identify potential mechanisms overcoming the inhibitor block. First, we checked total protein levels of our AKT, PI3K, and EGFR constructs in our panel of cell lines (Fig. 4G and 4H, Supplementary Fig. S4G–I). We then treated each cell line with single agent trametinib and/or palbociclib for 48 h. We observed a reduction of phospho-ERK (trametinib and combination) and phospho-RB (trametinib, palbociclib, and combination) in our GFP expressing cell lines, suggesting target inhibition. In MELJUSO, SKMEL30, and Hs936T expressing AKT, PI3K, and EGFR, we did not observe any changes to phospho-ERK after inhibitor treatments, suggesting that these genes drive resistance independent of MAPK signaling (Fig. 4G, Supplementary Fig. S4H and S4I). This was in contrast to the Hs944T cells where we observed that expression of AKT1, AKT2, and PI3KH1047R lead to increased levels of phospho-ERK (Fig. 4H).
Next, we examined changes to levels of phospho-RB. In SKMEL30 and Hs936T cell lines expressing our AKT, PI3K, and EGFR constructs, we observed a significant rebound of phospho-RB in the presence of trametinib and/or palbociclib (Supplementary Fig. S4H and S4I). In MELJUSO cells expressing the same constructs there was a slight rebound of phospho-RB in the presence of trametinib and the combination, but not uniformly in the presence of palbociclib (Fig. 4G). This was in contrast to Hs944T cells, where expression of PI3KH1047R and PI3KE545K resulted in slight rebounding of phospho-RB in the presence of trametinib and/or palbociclib (Fig. 4H). These data indicate that resistance to trametinib and/or palbociclib in NRAS mutant melanoma cell lines is partially associated with re-activation of phospho-RB, which is consistent with the notion of pathway re-activation.
Next, we assessed phospho-S6 levels, as they have been previously associated with resistance to trametinib and/or palbociclib treatment in other NRAS mutant melanoma models. Treatment with trametinib alone and in combination with palbociclib reduced phospho-S6 levels in all cell lines examined (Fig. 4G, 4H, and Supplementary Fig. S4H and S4I). In the palbociclib arm, we observed a slight decrease in phospho-S6 levels. In MELJUSO, SKMEL30, and Hs936T cell lines expressing our AKT, PI3K, and EGFR constructs we observed a significant rebounding of phospho-S6 levels in the presence of trametinib alone or in combination (Fig. 4G and Supplementary Fig. S4H and S4I). However, only in the Hs944T cells expressing either PI3KH1047R, PI3KE545K or EGFRL858R caused a rebound of phospho-S6 levels (Fig. 4H). These data suggest that reengagement of mTOR signaling is associated with resistance to trametinib and palbociclib treatment in NRAS mutant melanoma, which is consistent with known in vivo and clinical mechanisms of inhibitor resistance [7, 11].
Next, we investigated the effect of trametinib and/or palbociclib treatment on levels of cell cycle markers. In the MELJUSO and Hs944T cell lines expressing AKT, PI3K, and EGFR constructs, we found that cyclin E2 levels were restored in the presence of trametinib and/or palbociclib, which correlated with the observed resistance phenotype (Fig. 4G and 4H). In Hs944T cells expressing either PI3KH1047R or PI3KE545K, we observed that cyclin B1 and cyclin D1 levels rebounded in the presence either/both drugs. Together, these results suggest that NRAS mutant melanoma cell lines can overcome inhibitor sensitivity to trametinib and/or palbociclib by increasing the levels of phospho-RB and phospho-S6, as well as cell cycle markers.
KRAS signaling modulates resistance to trametinib and trametinib/palbociclib treatment in NRAS mutant melanoma cell lines
Alleles of activated KRAS were amongst the top scoring hits in the trametinib single agent screen and comprised the top two scoring ORFs identified in the combination arm. We expressed GFP or activated KRASG13D in the MELJUSO, SKMEL30, Hs936T, and Hs944T cell lines and assayed cell viability and clonogenic growth. Similar to our screening results, we observed that activated KRAS caused strong resistance to trametinib and trametinib/palbociclib in all cell lines evaluated in cell viability and clonogenic growth (Fig. 5A–F and Supplementary Fig. S5A–D). However, KRAS WT could not overcome sensitivity to trametinib/palbociclib treatment (Supplmentary Fig. S5D–I). When these cell lines were treated with palbociclib alone, we observed minimal outgrowth in the MELJUSO, Hs936T, and Hs944Tcell lines, but not the SKMEL30 cell line, where there was no measurable advantage to having activated KRAS mutations.
Figure 5.
RAS signaling modulates resistance to trametinib/palbociclib treatment in NRAS mutant melanoma cell lines. (A and C), Cell Titer MTS tetrazolium assay showing mean cell viability of MELJUSO (A) and Hs936T (C) cell lines expressing GFP, KRAS, or NRAS after 144 h of combination trametinib and palbociclib treatment, each from three independent experiments performed in triplicate. (B and D), Normalized Cell Viability Score where DMSO is 100% cell viability as measured by MTS in MELJUSO (B) and Hs936T (D). (E and F), Representative images of crystal violet stained clonogenic growth assays with MELJUSO (E) and Hs936T (F) cell lines expressing GFP, KRAS or NRAS after 10 d of trametinib, palbociclib, and trametinib/palbociclib combination treatment, each from three independent experiments performed in triplicate. (G and H), Representative immunoblots displaying the effect of trametinib, palbociclib, and trametinib/palbociclib combination treatment in MELJUSO (G) and Hs936T (H) cell lines expressing GFP [51], KRAS, or NRAS after 48 h on phosphorylated and/or total protein levels of NRAS, RB, AKT, ERK, S6, CCND1, and KRAS. β-actin immunoblotting was used to determine equivalent loading.
NRAS clones are not present in the ORFeome Library used here. Thus, we investigated whether over-expression of activated NRASQ61L/K could drive resistance to trametinib and/or palbociclib in NRAS-mutant cells. As observed with KRAS, mutant NRAS drove resistance to trametinib and trametinib/palbociclib, with minimal impact on palbociclib resitance (Fig. 5A–F, Supplementary Fig. S5A). Together, these data imply that activated Ras isoforms are sufficient to drive resistance to overcome MEK1/2 and CDK4/6 inhibition.
Consistent with the resistance phenotypes observed with expression of activated Ras isoforms, expression of mutant NRAS or KRAS led to an increase in cyclin D1 levels and a rebound of phospho-RB and phospho-S6 levels in the presence of trametinib, alone and in combination with palbociclib, whereas no rebound of phospho-RB levels was seen with palbociclib alone (Fig. 5G and 5H, Supplementary Fig. S5J). Additionally, phospho-ERK levels were elevated in the presence of trametinib and the combination in cell lines expressing activated KRAS and NRAS. Together these data support the idea that activated RAS is the strongest driver of resistance to combination trametinib and palbociclib in NRAS mutant melanoma cell lines.
EGFR-PI3K-AKT signaling differentially modulates sensitivity of NRAS mutant melanoma cell lines to genetic loss of activated NRAS
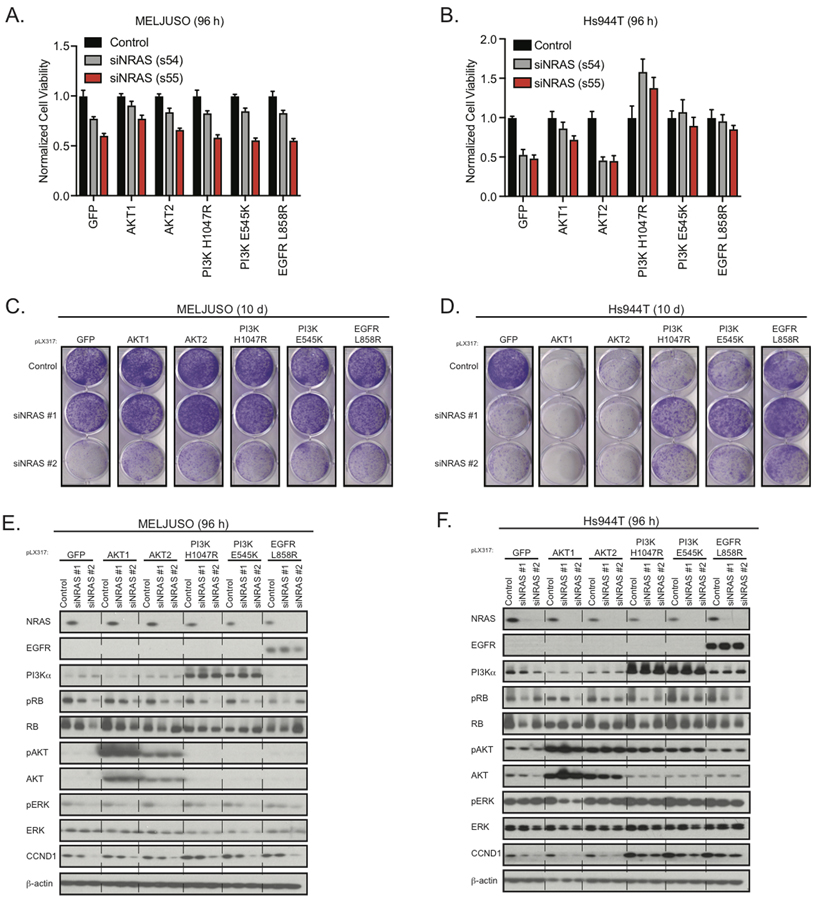
Our data suggest that expression of single components found in the EGFR-PI3K-AKT signaling cascade can partly overcome sensitivity to trametinib and/or palbociclib. This led us to evaluate whether any of these components could overcome genetic ablation of activated NRAS. To address this question, we expressed AKT1, AKT2, PI3KH1047R, PI3KE545K or EGFRL858R in MELJUSO and Hs944T cell lines, treated with siRNAs and assessed cell viability and clonogenic growth. Expression of AKT1, PI3K, or EGFR was sufficient to partially rescue NRAS depletion in MELJUSO (Fig. 6A and 6B, Supplementary Fig. S6A). However, rescue was much more pronounced in the Hs944T cell line in cell viability and clonogenic assays (Fig. 6C and 6D, Supplementary Fig. S6A). Expression of AKT1, PI3K, or EGFR constructs in SKMEL30 and Hs936T was unable to overcome genetic ablation of NRAS (Supplementary Fig. S6A–G). These differences did not appear to be a consequence of the expression levels of components of the EGFR-PI3K-AKT pathway or the result of co-occurring mutation. These data highlight the heterogeneity of NRAS-mutant melanoma cell lines and suggest that mutant NRAS ablation in some contexts may not be enough to prevent re-occurrence.
Figure 6.
EGFR-PI3K-AKT signaling cascade differentially effects loss of genetic NRAS in NRAS mutant melanoma cell lines. (A and B), Cell Titer MTS tetrazolium assay displaying mean cell viability of MELJUSO (A) and Hs944T (B) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR with or without knockdown of NRAS (six replicates) after 96 h. (C and D), Representative images of crystal violet stained clonogenic growth assays with MELJUSO (C) and Hs944T (D) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR with or without knockdown of NRAS (duplicate), each from three independent experiments. (E and F), Representative immunoblots displaying the effect of MELJUSO (E) and Hs944T (F) cell lines expressing GFP, AKT1, AKT2, PI3K, or EGFR with or without knockdown of NRAS, after 96 h on phosphorylated and/or total protein levels of NRAS, EGFR, PI3K, RB, AKT, ERK, and CCND1. β-actin immunoblotting was used to determine equivalent loading.
Next, we assessed the effects of genetic loss of NRAS on signaling output. Genetic loss of NRAS resulted in a reduction in phospho-ERK and phospho-RB levels in the GFP control, which could not be rescued by expression of AKT1, PI3K, or EGFR constructs in MELJUSO and SKMEL30 cells (Fig. 6E, Supplementary Fig. S6F and S6H). The Hs944T cell line expressing GFP did not yield a reduction in levels of phospho-ERK after NRAS siRNA treatment, whereas phospho-RB levels displayed minimal changes. Consistent with their ability to rescue NRAS ablation in Hs944T, PI3KH1047R, PI3KE545K, or EGFRL858R expression caused an increase in cyclin B1 and cyclin D1 suggesting these cell lines can rescue NRAS ablation via re-engaging the cell cycle (Fig. 6F, Supplementary Fig. S6I). These data imply that single components in the EGFR-PI3K-AKT signaling cascade may partially, but not fully replace genetic loss of NRAS.
Activated KRAS can compensate for genetic loss of activated NRAS
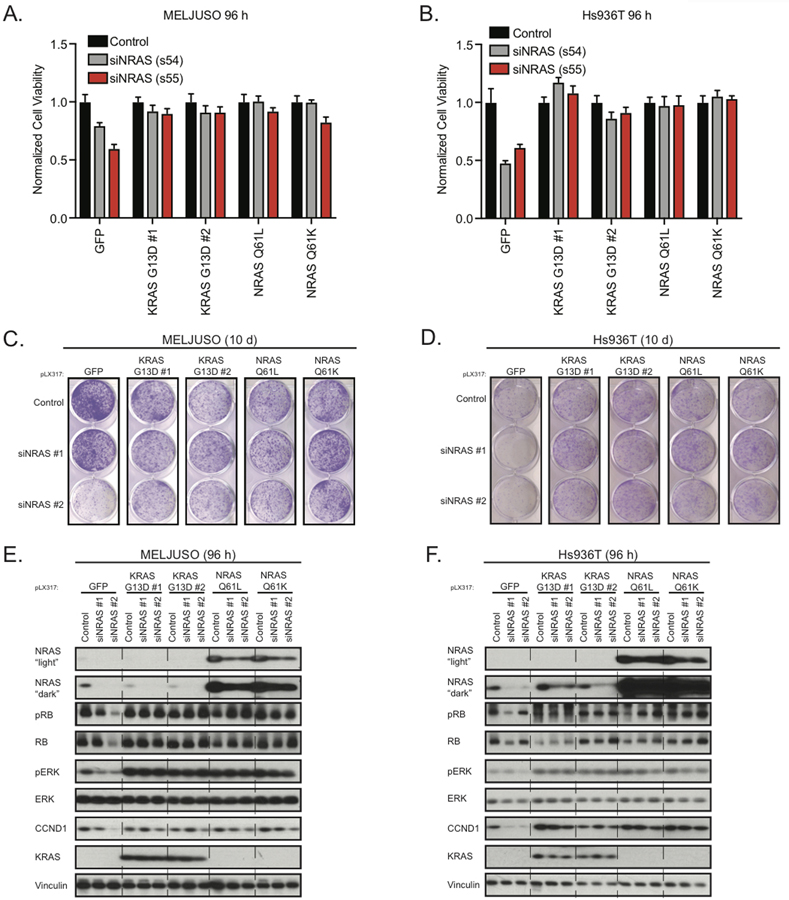
Next, we examined whether activated KRAS could compensate for genetic loss of NRAS. We first expressed activated KRAS constructs in the MELJUSO, SKMEL30, Hs936T, and Hs944T cell lines. Next, we treated cell lines expressing activated KRAS with NRAS-targeted siRNA and control siRNA. After 96 h and 144 h post siRNA treatment, we assessed cell viability and observed that cell lines expressing activated KRAS grew equally well in the presence of NRAS-targeted siRNA compared to control siRNA (Fig. 7A and 7B, Supplementary Fig. S7A–C). This observation was further extended in clonogenic growth assays, where activated KRAS uniformly restored growth to the same levels as the control (Fig. 7C and 7D, Supplementary S7D and S7E). When treated with NRAS-targeted siRNA, cell lines expressing activated NRAS were able to grow to similar levels as cell lines treated with the siRNA control. This observation suggests that our siRNAs in fact target NRAS.
Figure 7.
Active KRAS can replace loss of genetic NRAS. (A and B), Cell Titer MTS tetrazolium assay displaying mean cell viability of MELJUSO (A) and Hs936T (B) cell lines expressing GFP, KRAS, or NRAS with or without knockdown of NRAS (six replicates) after 96 h, each from three independent experiments. (C and D), Representative images of crystal violet stained clonogenic growth assays with MELJUSO (C) and Hs936T (D) cell lines expressing GFP, KRAS, or NRAS with or without knockdown of NRAS (duplicate), after 10 d each from three independent experiments. (E and F), Representative immunoblots displaying the effect of MELJUSO (E) and Hs936T (F) cell lines expressing GFP, KRAS, or NRAS with or without knockdown of NRAS, after 96 h on phosphorylated and/or total protein levels of NRAS, RB, ERK, CCND1, and KRAS. Vinculin immunoblotting was used to determine equivalent loading.
Next, we next examined how activated KRAS and NRAS were able to overcome NRAS-targeted siRNA treatment. We observed a greater than 50% knockdown in cell lines treated with NRAS-target siRNA compared to control siRNA (Fig. 7E and 7F, Supplementary Fig. S7F and S7G). Our analyses revealed a significant rebound in phospho-ERK and phospho-RB levels (Fig. 7E and 7F). We did not observe a change in the levels of phospho-AKT in either KRAS or NRAS expressing cell lines suggesting that in this context both NRAS and KRAS signal through the MAPK cascade. Consistent with our growth phenotype, we observed a rebound in levels of the cell cycle markers cyclin B1, cyclin D1, and cyclin E2 in activated KRAS and NRAS expressing cell lines (Fig. 7E and 7F, Supplementary S7H and S7I). Based on these data, it seems that re-engagement of the cell cycle is the mechanism by which activated NRAS and KRAS overcome genetic loss of activated NRAS in NRAS-mutant melanoma.
Discussion
There is no Ras-specific approved therapy for the treatment of advanced NRAS-mutant melanoma; however, there are several therapeutic avenues under pre-clinical and clinical evaluation. The modest-but-clear clinical effectiveness of MEK inhibition as a single agent in NRAS-mutant melanoma drove interest in designing rational therapeutic combination strategies to enhance MEK inhibition (NCT01763164) [4, 27–29]. This idea resulted in the identification of CDK4/6 as a modulator of MEK inhibition in NRAS-mutant melanoma. Consequently, this combination is under clinical evaluation in several Ras-mutant cancers.
In our study, we present the first resistance landscape to MEK1/2 and CDK4/6 single agent and combination treatment in NRAS-mutant melanoma. Using both ORF gain-of-function and CRISPR loss-of-function screens we provide evidence that modifying levels of components of the RTK-PI3K-AKT and RAS-RAF signaling cascades alters sensitivity to trametinib and/or palbociclib. These discoveries are particularly relevant for the rational design of future combinatorial therapeutics for patients with NRAS-mutant melanoma and other RAS-driven cancers. One clinical trial is evaluating binimetinib/LEE011 (NCT01781572), and enrolled patients have experienced either a partial response (34%), stable disease (52%) or progressive disease (14%) [8].
Our screening results are highly complementary to recent studies, as we identified both ORFs and CRISPRs that represent components of the RTK-PI3K-AKT-mTOR signaling cascade that modulate sensitivity to trametinib and/or palbociclib (Fig. 2 and 3) [7, 11]. These findings will be of particular significance information regarding trametinib/palbociclib efficacy in Ras-mutant cancer in clinical trials becomes available. Together, our findings highlight resistance mechanisms to single agent and combination MEK1/2 and CDK4/6 in NRAS-mutant melanoma and reveal insights into RAS biology.
Supplementary Material
Statement of Significance.
Findings reveal that NRAS-mutant melanomas can acquire resistance to genetic ablation of NRAS or combination MEK1/2 and CDK4/6 inhibition by upregulating activity of the RTK-RAS-RAF and RTK-PI3K-AKT signaling cascade.
Acknowledgements
Grant Support: This work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation in Mexico. This work was supported by a Career Development Award from the Melanoma Research Foundation (C.M.J.). We acknowledge additional support from the National Cancer Institute to L.A. Garraway (3R35CA197737) and M. Meyerson (1R35CA197568) and Damon Runyon Cancer Research Foundation award to T.K. Hayes (David Livingston Fellow). M. Meyerson is an American Cancer Society Research Professor.
COI: Matthew Meyerson is a scientific advisory board member for OrigiMed and receives consulting fees for this role. He was a consultant for Foundation Medicine and held equity, some of which was sold to Roche. He receives research support from Bayer and is an inventor of several joint patents and patent applications, none of which have been licensed at the time of this disclosure (12–17-18). In addition, he is an inventor on several patents on EGFR mutations in lung cancer diagnosis, licensed to LabCorp, for which he receives royalties. He is also an inventor on several patent applications on Fusobacterium, none issued or licensed, and the inventor on a patent for pathogen discovery, not licensed, for none of which he receives royalties. Levi Garraway is a co-founder and equity holder of Tango Therapeutics. He was a consultant for Foundation Medicine and held equity, some of which was sold to Roche.
References
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2018. CA Cancer J Clin, 2018. 68(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Lin WM, et al. , Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res, 2008. 68(3): p. 664–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solit DB, et al. , BRAF mutation predicts sensitivity to MEK inhibition. Nature, 2006. 439(7074): p. 358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascierto PA, et al. , MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol, 2013. 14(3): p. 249–56. [DOI] [PubMed] [Google Scholar]
- 5.Dummer R, et al. , Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol, 2017. 18(4): p. 435–445. [DOI] [PubMed] [Google Scholar]
- 6.Kwong LN, et al. , Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med, 2012. 18(10): p. 1503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano G, et al. , A Preexisting Rare PIK3CA(E545K) Subpopulation Confers Clinical Resistance to MEK plus CDK4/6 Inhibition in NRAS Melanoma and Is Dependent on S6K1 Signaling. Cancer Discov, 2018. 8(5): p. 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosman JA, K.M., Lolkema MPJ, Postow MA, Schwartz G, Franklin C, et. al., A phase 1b/2 study of LEE011 in combination with binimetinib (MEK162) in patients with NRAS-mutant melanoma: early encouraging clinical activity. Journal of Clinical Oncology, 2014. 32: p. 9009. [Google Scholar]
- 9.Tsherniak A, et al. , Defining a Cancer Dependency Map. Cell, 2017. 170(3): p. 564–576 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald ER 3rd, et al. , Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell, 2017. 170(3): p. 577–592 e10. [DOI] [PubMed] [Google Scholar]
- 11.Teh JLF, et al. , In Vivo E2F Reporting Reveals Efficacious Schedules of MEK1/2-CDK4/6 Targeting and mTOR-S6 Resistance Mechanisms. Cancer Discov, 2018. 8(5): p. 568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johannessen CM, et al. , COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature, 2010. 468(7326): p. 968–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johannessen CM, et al. , A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature, 2013. 504(7478): p. 138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le X, et al. , Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov, 2016. 6(10): p. 1134–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao DD, et al. , KRAS and YAP1 converge to regulate EMT and tumor survival. Cell, 2014. 158(1): p. 171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson FH, et al. , A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell, 2015. 27(3): p. 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, et al. , A public genome-scale lentiviral expression library of human ORFs. Nat Methods, 2011. 8(8): p. 659–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes TK, et al. , Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell, 2016. 29(1): p. 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharifnia T, et al. , Genetic modifiers of EGFR dependence in non-small cell lung cancer. Proc Natl Acad Sci U S A, 2014. 111(52): p. 18661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doench JG, et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016. 34(2): p. 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde M, et al. , Uncoupling of sgRNAs from their associated barcodes during PCR amplification of combinatorial CRISPR screens. PLoS One, 2018. 13(5): p. e0197547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krall EB, et al. , KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, et al. , ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Rep, 2017. 18(6): p. 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condorelli R, et al. , Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol, 2018. 29(3): p. 640–645. [DOI] [PubMed] [Google Scholar]
- 25.Konecny GE, et al. , Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res, 2011. 17(6): p. 1591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin TD, et al. , Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell, 2014. 53(2): p. 209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatzivassiliou G, et al. , Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature, 2013. 501(7466): p. 232–6. [DOI] [PubMed] [Google Scholar]
- 28.Ribas A, et al. , Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol, 2014. 15(9): p. 954–65. [DOI] [PubMed] [Google Scholar]
- 29.Zimmer L, et al. , Phase I expansion and pharmacodynamic study of the oral MEK inhibitor RO4987655 (CH4987655) in selected patients with advanced cancer with RAS-RAF mutations. Clin Cancer Res, 2014. 20(16): p. 4251–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.