Significance
This study uses viral-like particle purification and subsequent unbiased genome sequencing to identify prokaryotic viruses associated with Apis mellifera. Interestingly, bacteriophages found in honey bees show a high diversity and span different viral taxa. This diversity sharply contrasts with the state-of-the-art knowledge on the relatively simple bee bacterial microbiome. The identification of multiple auxiliary metabolic genes suggests that these bacteriophages possess the coding potential to intervene in essential microbial pathways related to health and possibly also to disease. This study sheds light on a neglected part of the bee microbiota and opens avenues of in vivo research on the interaction of bacteriophages with their bacterial host, which likely has strongly underappreciated consequences on bee health.
Keywords: viral metagenomics, prokaryotic viruses, bacteriophages, Apis mellifera
Abstract
Honey bees (Apis mellifera) produce an enormous economic value through their pollination activities and play a central role in the biodiversity of entire ecosystems. Recent efforts have revealed the substantial influence that the gut microbiota exert on bee development, food digestion, and homeostasis in general. In this study, deep sequencing was used to characterize prokaryotic viral communities associated with honey bees, which was a blind spot in research up until now. The vast majority of the prokaryotic viral populations are novel at the genus level, and most of the encoded proteins comprise unknown functions. Nevertheless, genomes of bacteriophages were predicted to infect nearly every major bee-gut bacterium, and functional annotation and auxiliary metabolic gene discovery imply the potential to influence microbial metabolism. Furthermore, undiscovered genes involved in the synthesis of secondary metabolic biosynthetic gene clusters reflect a wealth of previously untapped enzymatic resources hidden in the bee bacteriophage community.
Pollination is an essential aspect for entire ecosystems, and honey bees (Apis mellifera) are considered the most economically important insect pollinators for commercial crops worldwide. Apart from the production of honey and other valuable products, honey bees contribute significantly to insect pollination, of which the economic value has been estimated at €153 billion (1). During the past decades, it has become clear that managed honey-bee colonies are under pressure from a wide variety of stressors, such as parasites (2), bacterial pathogens (3), viral pathogens (4), and others such as chemical stressors (5). Recently, more and more attention is going toward the bee microbiota, and a number of studies have attempted to characterize the honey-bee-gut microbiome (6, 7). These studies have revealed that the bacterial part of the core honey-bee-gut microbiome is dominated by 5 to 10 different bacterial species. The species that were identified belonged to three different bacterial phyla, namely the Proteobacteria, Firmicutes, and Actinobacteria (6). Transcriptome analysis further provided information on the functional potential encoded by the bacterial gut microbiome (8). From these insights a model was proposed for a microbial metabolic pathway, with different roles for different bacteria. Briefly, glycosidases and peptidases (encoded by the aforementioned core bacterial microbiome) initially break down plant polysaccharides and proteins. These products are further fermented into organic acids, gases, and alcohols, which are then further metabolized by methanogens and Clostridia species. The fact that honey bees cannot survive on unprocessed pollen alone (9) highlights the importance of microbial enzymatic digestion in honey-bee homeostasis. These findings were recently recapitulated in a study employing system-wide metabolomics (10). This study confirms that the bee-gut microbiota play a central role in the digestion and metabolization of pollen-derived components. More evidence on the existence of host–microbe interactions has revealed a positive influence of the bee-gut microbiota on weight gain in the host weight of the gut compartments, but also increasing the endogenous expression of genes involved in development and immunity, sucrose sensitivity, and insulin-like signaling (11).
Taken together, these results imply an essential role of the bee-gut microbiota in nutrition availability, bee development, and general homeostasis. This role is further strengthened by the observation that a diet-induced gut bacterial dysbiosis is associated with detrimental effects on development, mortality, and disease susceptibility (12). The fact that both the diet of honey bees and the bacterial diversity present in the honey-bee gut are much less divergent than its human counterpart led to the proposal to use honey bees as model systems for microbiota research and furthermore as a useful tool in studying the evolution and ecology of host–microbe interactions (13). Despite the recent advances in the knowledge of the honey-bee gut metagenome, the work done on honey-bee–associated bacteriophages has been based on only a few isolates and thus remains biased (14, 15). It is often postulated that (prokaryotic) viruses represent the most prevalent biological units worldwide and execute essential roles within their respective ecosystems. For example, bacteriophages play a significant role in carbon, nitrogen, and phosphorous cycling in the oceans (16) and are implied to influence soil ecology (17). The presence of auxiliary metabolic genes (AMGs: here defined as genes present in bacteriophages, but originating from bacteria with the potential to modulate microbial metabolism) within bacteriophages, and the recent discovery of a communication systems resulting in lysis and lysogeny decisions (18) reflect the important influence that these viruses play in their putative hosts and ecosystem equilibria in general. In humans, bacteriophages have been used as alternatives for antibiotics (19) and even proposed to be used as biomarkers for numerous conditions (20).
The multitude of functions that prokaryotic viruses can exert within their biosphere, combined with the fact that the honey-bee bacterial microbiome plays a crucial role in bee health, implies that the viral microbiome could play an important role in bee homeostasis as well. In this work we present an initial characterization of the prokaryotic viral microbiome associated with honey bees derived from healthy and weakened colonies using viral-like particle enrichment strategies combined with short read Illumina sequencing.
Results
Prokaryotic Virus Identification through Next-Generation Sequencing.
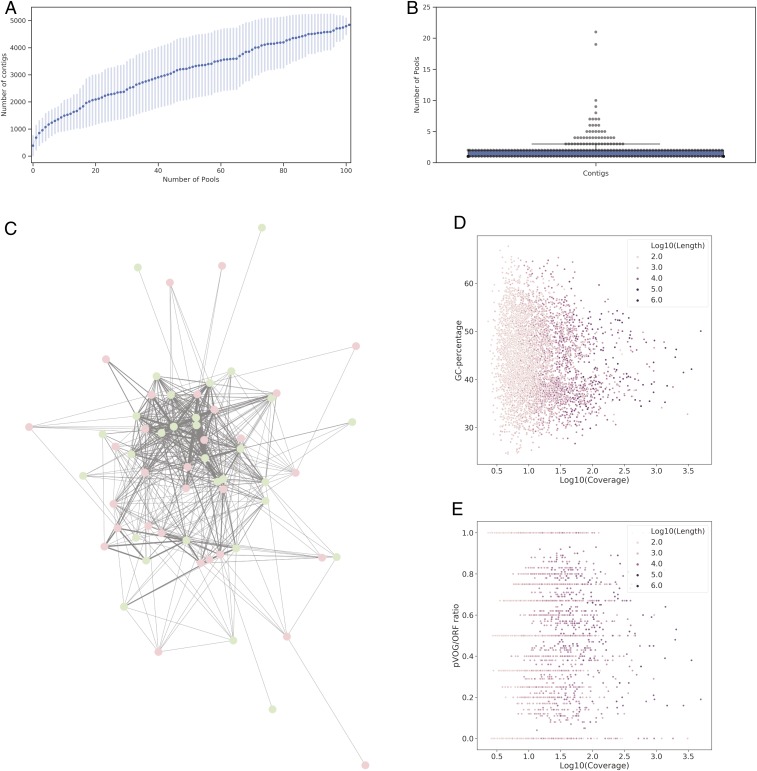
Samples comprising 300 different colonies of Flemish honey bees, collected in the framework of the EpiloBEE study (21) (SI Appendix, Fig. S1), were enriched for viruses (both DNA and RNA viruses) according to the NetoVIR protocol (22) and sequenced. These samples were initially selected to represent the Flemish population of honey bees as well as possible. In total, 102 pools containing samples from hives that were comparable (derived from healthy or weak colonies) and matched geographically and by subspecies as well as possible were analyzed (SI Appendix, Table S14, available on GitHub). Two bees from three colonies were pooled together, except for the last three pools, which contained two bees from one colony. Each pool was assigned 5 million 150-bp end reads and were sequenced using the Illumina NextSEQ platform. This approach yielded a total of 686,940,647 reads, with a median of 5,798,403 reads per pool (minimum: 2,096,600 reads; maximum: 26,307,071 reads). After de novo assembling the separate libraries, the resulting contigs were collapsed on 95% nucleotide identity over a coverage of 80%, and putative prokaryotic viral sequences were identified using VIRSorter (23) and a lowest-common-ancestor approach using DIAMOND (24). Eukaryotic viruses were omitted by using the virome decontamination mode in VIRSorter and by manually parsing the DIAMOND output. These approaches allowed the identification of 4,842 nonredundant putative prokaryotic viral contigs with a minimum length of 500 bp. Of these contigs, 20 were predicted to be circular (and thus complete genomes) (SI Appendix, Fig. S2). Of these 20 complete genomes, 11 could be assigned to known bacteriophage families (Microviridae, Siphoviridae, Myoviridae, and Podoviridae), and 7 could be assigned to a bacterial host (Bifidobacterium, Bartonella, Lactobacillus, Hafnia, and Pluralibacter) (see below). Species accumulation curves (assuming that the collapsed contigs reflect distinct viral species) reveal no plateau being reached, implying that, despite the large sampling effort and viral particle enrichment, prokaryotic viral sequence space was not fully probed (Fig. 1A). This observation is also reflected by the strong correlation between contig length and contig tpmean coverage (the average number of reads overlapping each base after removing the 10% most- and least-covered bases) (Spearman correlation coefficient = 0.74, P value < 1.10−4) (SI Appendix, Fig. S3). Reads from individual pools were aligned back to the contig representatives, and the presence of every contig in a pool was evaluated (presence being defined as tpmean coverage > 10). Most contigs larger than 5 kb were shared between less than five pools, and 20 contigs were shared between more than five pools (Fig. 1B). Pairwise comparison of the contig sharing between pools is reflected in a network that represents 70 of the 102 pools sequenced (Fig. 1C). The maximum number of contigs shared between two pools was 15. No clear clustering patterns could be observed when applying the Markov Cluster Algorithm (MCL) (25) or k-means clustering. The maximal clique observed in the network contained 21 pools. Next, the dimensionality of the coverage matrix was reduced using principal coordinate analysis (PCoA), and the clustering patterns for sample status (health vs. diseased), sampling year (2012 vs. 2013), and location (Belgian provinces) were tested using the Adonis test. No significant effect was observed for sample status, but both location and sampling year were significant, albeit with a low R-squared value (SI Appendix, Fig. S4). Retrieved putative prokaryotic virus contigs show a wide range of guanine–cytosine (GC) percentages, ranging from roughly 25 to 70% (Fig. 1D). After decorating the contigs with prokaryotic virus orthologous groups [pVOGs (26)], roughly half the contigs (2,346 contigs, or 48.5%) had a pVOG vs. open reading frame (ORF) ratio larger than 50% (Fig. 1E). Some of the contigs that fell below this ratio were more than 10 kb in size, implying that a high amount of putative viral proteins were not represented in the pVOG database. These results suggest that a large number of retrieved viral genes are not represented in the pVOG database. When plotting the average “Virusness” (frequency of a pVOG being present in viruses versus the frequency of a pVOG being present in bacteria) of the annotated pVOGs within a contig, it was shown that a slight majority of the contigs fell above 0.5 (SI Appendix, Fig. S5). The bimodal distribution reflects that a large number of contigs show a clear viral signal (average Virusness close to 1), while the enrichment of contigs with an average Virusness close to zero is a consequence of contigs carrying no detectable pVOG at all (orange dots in SI Appendix, Fig. S5).
Fig. 1.
Bee-associated prokaryotic viruses display a high interindividual diversity and contain a large number of unknown viral proteins. (A) Species accumulation curves as a function of the number of pools sequenced. Vertical lines indicate SDs based on 100 permutations. (B) Swarm plot reflecting putative viral contigs larger than 5 kb that were present in one sample or more (140 in total). Presence is defined as a coverage >10. A dot represents a single contig. The box shows the three quartile values, and the whiskers extend to 1.5 interquartile ranges of the lower and upper quartile. All 140 dots are drawn in the plot. (C) Edge-weighted spring-embedded layout network depicting the samples as nodes and edges as the number of contigs shared between them. Edge thickness reflects the number of contigs. Green nodes depict pools derived from healthy colonies; red nodes depict pools derived from weak colonies. Edge thickness ranges from 1 to 15. (D) GC percentage of all representative putative viral contigs as a function of their log10-transformed coverage in the pool of which the representative was derived. Log10-transformed length is indicated by color intensity. (E) Number of pVOGs found back in the putative viral contigs, normalized by the amount of predicted ORFs as a function of their log10-transformed coverage. Log10-transformed length is indicated by color intensity.
Host Assignment of Prokaryotic Viral Contigs.
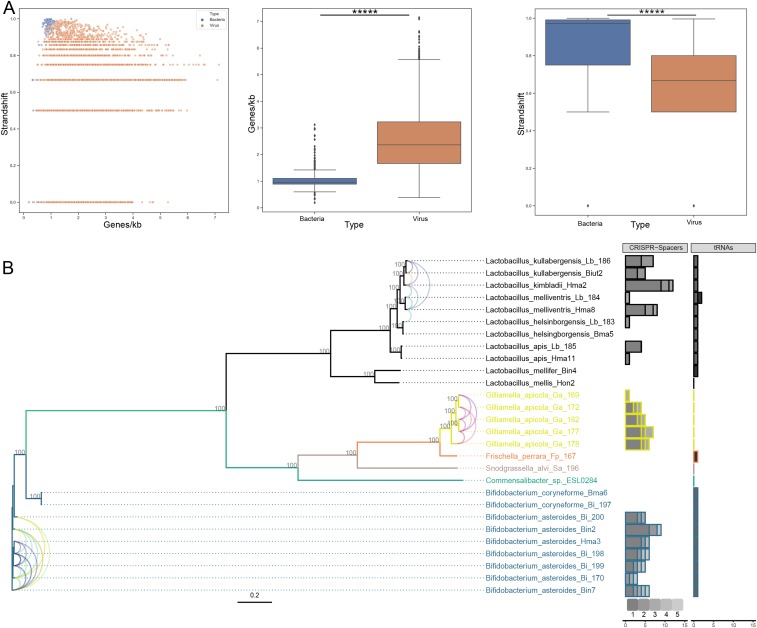
Putative bacteriophage genomic sequences can be linked to their specific host by taking advantage of the CRISPR-spacer sequences that they encode and by transfer RNA (tRNA) similarity. We constructed a bee-specific gut bacterial microbiome dataset by collating data available on IMG/M and from Ellegaard et al. (27) (SI Appendix, Table S15). This collated dataset includes bacterial sequences from six different genera, including Lactobacillus, Bifidobacterium, Commensalibacter, Gilliamella, Snodgrassella, and Frischella. To minimize the possibility that any of the putative prokaryotic viral contigs were of bacterial origin, the coding density and frequency of strand shift were calculated for both the bacterial contig set and the bacteriophage contig set (Fig. 2A). Both these parameters were significantly different between both sets. On average, the coding density (defined as number of predicted genes/kilobase) was higher in the virus dataset (2.50) than the bacterial dataset (1.07) (Mann–Whitney U test, P value = 5.10−164). The average frequency of strand shift (defined as the frequency that two neighboring genes start in different frames) was higher in the bacterial dataset (0.84) than in the viral dataset (0.63) (Mann–Whitney U test, P value = 4.10−87). These observations, together with the fact that no single copy bacterial marker genes [as defined by Lee et al. (28)] could be identified in the putative prokaryotic viral contig set, suggest that the viral contig set contains no or very little bacterial contamination. In total, 76 putative bacteriophage contigs could be linked to specific bacteria using these approaches. These contigs were within the length range of 1 to 107 kb, and four were predicted to be circular (and thus depict full-length genomes). Of these 76 contigs, 32 could be assigned to the genus Lactobacillus, 17 to the genus Gilliamella, and 27 to the genus Bifidobacterium. No viral contigs could be linked to the Frischella, Snodgrassella, and Commensalibacter group of bacteria since these bacterial genomes did not contain any detectible CRISPR array. However, one tRNA hit was found against the genus Frischella (Fig. 2B). The majority of host-called viral contigs were linked to a single bacterium, but 17 of them displayed CRISPR-spacer hits against more than one bacterial species. One putative viral contig could even be linked to five different bacteria, but none of the host-linked viral contigs could be assigned to more than one genus, suggesting a restricted host range. Only five of the putative viral contigs contained a tRNA signature that could be linked to specific bacteria. Two of those contigs gave hits against nearly all of the Bifidobacterium or Lactobacillus species included in this analysis. One of those contigs also gave a hit to the only Frischella species included, although there was no CRISPR-spacer evidence found to confirm this. Since bees sample the environment, it cannot be excluded that some of the retrieved viral sequences reflect environmental bacteriophages rather than true bee-gut viruses. To this extent, an additional CRISPR-spacer search was ran by using the spacers present in the CRISPR database (CRISPRdb) (29). These results confirm 19 of the 76 previous hits against the bee-gut–specific bacteria. Furthermore, 50 additional hits were found, of which 32 were for bacterial genera present in the bee gut (6 Lactobacillus hits, 3 Bifidobacterium hits, 18 Bartonella hits, 5 Gilliomella hits). The 18 remaining putative hosts identified potentially reflect environmental bacteria (SI Appendix, Table S19, available on GitHub).
Fig. 2.
Retrieved prokaryotic viruses display a significant difference in genomic variables and infect a wide range of known bee-gut bacteria. (A) Frequency of strand shift in function of coding density (number of ORFs per kilobase). Data from the bacterial dataset are indicated in blue; data from the viral dataset are indicated in orange. Boxplots for individual parameters are also denoted, and asterisks designate significance (Mann–Whitney U test; P value for coding density = 5.10−164; P value for strand shift frequency = 4.10−87). The box shows the three quartile values, and the whiskers extend to 1.5 interquartile ranges of the lower and upper quartile. Dots independently drawn fall outside of this range. (B) Maximum-likelihood phylogenetic tree for bacterial sequences included in the host-calling effort. Gray integers indicate bootstrap values. The tree is colored according to bacterial genera. Number of contigs linked to a specific bacterial species are indicated by the stacked horizontal bar plots (CRISPR-spacer counts and tRNA similarity). Shades of gray indicate the number of specific bacterial species that gave hits to a single contig (CRISPR spacers) or indicate a specific viral contig (tRNA similarity). Single contigs displaying CRISPR-spacer hits to multiple bacteria are indicated with colored tax links between the tips. A single color corresponds to a single contig.
Classification of Prokaryotic Viral Contigs.
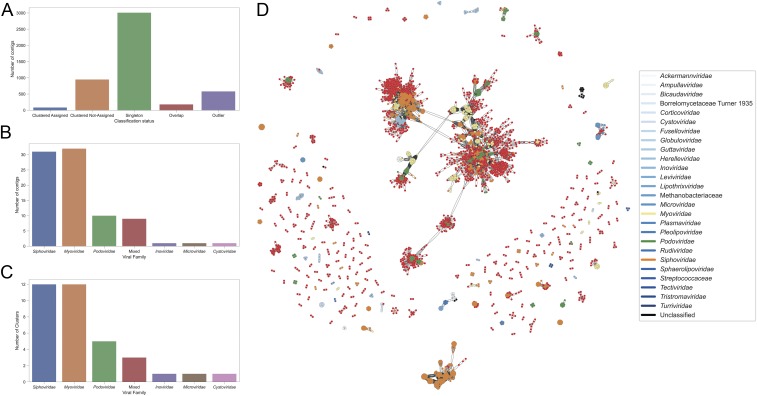
In an attempt to classify the newly discovered sequences, we ran vConTACT2 (30) on the putative prokaryotic viral sequences retrieved, using the Prokaryotic viral REFSEQ 88 database. This method uses gene-sharing networks to taxonomically assign prokaryotic viruses solely based on their sequence. The algorithm classifies sequences either as “singletons” (no shared gene content), “outliers” (weakly connected with a cluster of sequences), or as part of a cluster (30). Of the 4,842 nonredundant prokaryotic viral contigs (>500 bp), 3,010 were singletons, 582 were outliers, 181 showed strong overlap between more than one established cluster (not allowing for their unambiguous classification), and 1,034 could be unambiguously clustered (Fig. 3A). The clustered contigs are represented by 403 viral genome clusters (which are said to be equivalent to the genus taxonomic level). Of these viral genome clusters, 368 clusters contained no REFSEQ sequences at all. The remaining 35 clusters (representing 85 contigs) were mostly related to the families Siphoviridae and Myoviridae, although the families Podoviridae, Inoviridae, Microviridae, and Cystoviridae were also represented (Fig. 3 B and C and SI Appendix, Table S16). The resulting network of clustered genome sequences reveals the newly discovered sequences as widely dispersed throughout known REFSEQ sequences (Fig. 3D), despite the fact that this network reflects only about 20% of the recovered sequences (the remaining sequences are singletons). Of the 537 viral contigs that were larger than 5 kb, 71 (13.2%) were singletons, 126 (23.5%) were outliers, 67 (12.5%) showed too much overlap to be unambiguously classified, and 273 (50.1%) could be unambiguously clustered. Of the clustered sequences, 73 could be assigned to a viral family. Although the relative amount of assigned contigs versus the other categories was higher in the dataset with large viral contigs (SI Appendix, Fig. S6A) compared to the small contigs (Fig. 3A), the assignment to different viral families remained comparable to the full dataset (Fig. 3 B and C vs. SI Appendix, Fig. S6 B and C). The network projection also revealed that, despite the loss of a substantial number of small clusters, the viral diversity remained widespread. The relative increase in clustered viral sequences seen only when looking at contigs above 5 kb, combined with the observation that most of the singleton contigs are shorter in length than the other groups (SI Appendix, Fig. S7), reflect that a shorter sequence length hampers the classification process in this dataset. Because viruses lack universal marker genes, and very few of the recovered proteins could be clustered together (see below), phylogenetic trees were drawn for the five largest protein clusters (PCs), as identified by vConTACT2. These five largest protein clusters contained reference proteins annotated as “Ribonucleotide reductase” (PC1), “Endonuclease” (PC2), “ssDNA binding protein” (PC3), “Endonuclease” (PC4), and “Thymidilate synthase” (PC5). The number of proteins (identified in this study) in each protein cluster was highly variable. PC1 contained 27 identified proteins (400 proteins in total), PC2 contained 70 identified proteins (389 proteins in total), PC3 contained 83 identified proteins (331 proteins in total), PC4 contained 34 identified proteins (302 proteins in total), and PC5 contained 8 identified proteins (283 proteins in total). The identified sequences do not fall into distinct clades and seem to be dispersed over the entire phylogenetic spectrum of their respective trees (SI Appendix, Fig. S8A). Given the lack of large protein clusters containing many of the identified sequences, the branch lengths in between all tips on the protein cluster trees were calculated and linked to the minimum path length between the corresponding genomes in the vConTACT2 network. These minimum path lengths were calculated using the Bellman–Ford algorithm. Both metrics were significantly positively correlated (Spearman rank correlation coefficient = 0.43; P value = 0.0), although when breaking up between the types (bee-associated viral contig, reference or bee-associated viral contig, and references combined) the correlation coefficients ranged from 0.44 to 0.82 (SI Appendix, Fig. S8B). These results imply that the distances between connected nodes within the vConTACT2 network can also be interpreted (to some degree) as phylogenetic distances.
Fig. 3.
The vast majority of retrieved prokaryotic viruses cannot be classified confidently. (A) Counts indicating the classification status of the putative viral contigs using vConTACT2. “Clustered Assigned” denotes retrieved contigs falling into clusters containing reference sequences; “Clustered Not-Assigned” denotes retrieved contigs falling in clusters without reference sequences. (B) Number of clusters that contained both confidently clustered large contigs and reference sequences. (C) Number of clusters that contained both confidently clustered contigs and reference sequences. (D) Scalable force directed placement layout genome network containing the retrieved clustered putative prokaryotic viruses (red) and the viral family of reference sequences (other colors). The most prevalent viral families are indicated in yellow (Myoviridae), green (Podoviridae), and orange (Siphoviridae).
Functional Potential and Selection Signatures of Honey-Bee–Associated Prokaryotic Viral Genes.
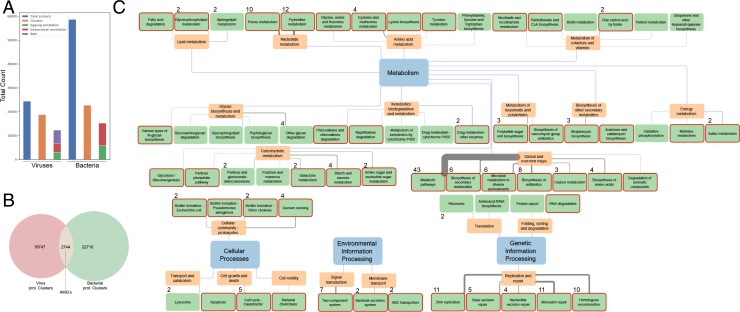
To gain insights into the functional potential encoded by the retrieved bacteriophages, InterProScan (31) and eggNOG-mapper (32, 33) were utilized for domain annotations. To reduce computational burden, protein sequences from predicted genes were collapsed on 50% amino acid identity before analysis. This procedure reduced the amount of putative proteins from 24,420 to 18,747, although the vast majority of clusters comprised less than five protein sequences (Fig. 4A and SI Appendix, Fig. S9A). In an attempt to identify AMGs, we blasted the viral protein sequences against the proteins encoded in the same bee-gut bacterial microbiome dataset used for host calling (SI Appendix, Table S15). Prior to blasting, the bacterial protein dataset was clustered using the same parameters as for the viral proteins. A viral protein was considered a genuine AMG when the alignment had an e-value smaller than 1e-5. Of the 18,747 viral protein clusters, 2,744 were identified as AMGs (Fig. 4B). To estimate the proportion of the identified AMGs originating from prophage regions, PHASTER (34) was run on the bacterial contigs (SI Appendix, Table S17). The bacterial proteins found in the AMG search were evaluated whether they fell in these regions or not. In total, 45 prophage regions were discovered, and 95 of the 1,506 (roughly 6%) bacterial counterparts of the identified AMGs fell inside these regions. Roughly 65% of the cluster representatives of the viral proteins (12,286) showed significant hits against the EggNOG database, the different databases used by InterProScan, or both (Fig. 4A). Of all of the Clusters of Orthologous Groups (COG) categories that the viral proteins could be assigned to, category S had the highest number of clusters assigned (“Function Unknown,” 4,293 clusters), followed by typical viral replication signatures (“Replication, recombination, and repair” [468 clusters], “Cell wall/membrane/envelope biogenesis” [212 clusters], and “Transcription” [166 clusters]) (SI Appendix, Fig. S9B). A complete enumeration of Gene Ontology (GO) accessions plotted into treemaps using REVIGO (35) reveals similar characteristics as the COG categories and a general lack of in-depth annotation of the retrieved viral proteins (SI Appendix, Fig. S10). In an attempt to further elucidate functional potential, the GO accessions were projected onto the pathways of which they are a part of using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapper (36) (Fig. 4C). Some of the retrieved pathways reflect functions that could influence bacteria directly and are involved in biofilm formation, quorum sensing, and bacterial chemotaxis. Other represented pathways reflect a wide range of basic metabolic functions, including lipid-, carbohydrate-, nucleotide-, and amino acid metabolism. Interestingly, also xenobiotic degradation, glycan biosynthesis, and terpenoid and polyketide metabolism were represented. The bacterial annotations for the previously defined AMGs were also projected into pathways, and overlapping pathway annotations were identified (Fig. 4C, red-outlined rectangles). Interestingly, nearly all of the identified pathways in the viral protein clusters were represented by the bacterial annotations for the AMG set. This observation further confirms the idea that the prokaryotic viral contigs contain the coding potential to influence bacterial metabolic state and homeostasis.
Fig. 4.
Functional annotation reveals a large metabolic overlap between bacterial and prokaryotic virus proteins. (A) Number of viral and bacterial proteins included in the analysis (blue), number of clusters remaining after collapsing on 50% AA identity (orange), and amount of protein clusters with a hit through either eggnog-mapper or InterProScan. (B) Venn diagram depicting the number of protein clusters and the number of putative AMGs identified. (C) Functional network depicting the KEGG pathways represented in the viral protein clusters. Edge weight reflects the number of GO accessions associated with each pathway, ranging from 1 to 43. All edge weights larger than 1 are indicated with a number on the terminal node. Pathways outlined with a red rectangle depict functional pathways found encoded by viral genes that are reflected by the bacterial representatives of the AMGs as well.
In an attempt to further characterize the signatures of secondary metabolites, as well as the other pathway functions that reflect the role of secondary metabolites, antiSMASH (37) was run. In total, four gene clusters were identified, all containing one gene with a bacteriocin signature (SI Appendix, Fig. S11). The four genes containing the bacteriocin signature had amino acid similarities with GenBank proteins ranging from 34 to 97%. Of the gene clusters identified, 13 to 53% of neighboring genes showed similarity to other genes represented in the antiSMASH database. Finally, an attempt was made to characterize selection signatures within the encoded genes. To achieve this, single-nucleotide polymorphisms (SNPs) were called per representative contig present in every pool, and nonsynonymous vs. synonymous substitution rates were calculated using SNPgenie (38). The majority of genes had a πN/πS ratio lower than 1, but 52 proteins revealed a positive selection signature in at least one pool (SI Appendix, Fig. S12). Of those 52 proteins, 11 were functionally annotated. These functions included mostly capsid and tail domains, but also transglycosylase and endopeptidase functions (see SI Appendix, Table S18, available on GitHub).
Discussion
This study provides an unbiased look at the prokaryotic viral communities associated with honey bees. The fact that the species accumulation curve did not reach a plateau phase implies that the full prokaryotic virus diversity has not been probed, despite the large sampling effort in combination with viral-like particle enrichment. The fact that some pools contained a high number of eukaryotic viral reads [also replicating in the honey-bee gut (39)] might have resulted in a suboptimal probing of the bacteriophages in these pools. The retrieved prokaryotic viral sequences display a large diversity, reflected by the number of annotated genes and the number of pVOGs that could be described. The general lack of classification of the contigs, and the fact that the majority of encoded genes could not have a function described, reflects this observation. Furthermore, the nonflattened species accumulation curve in combination with the strong positive correlation between coverage and length implies that many of the contigs described are fragmented genomes. In contrast to the gut bacterial microbiome, the prokaryotic viral communities display a high level of individualism where very few of the sequences are found back in many pools. Whether this observation could be a consequence of the sampling effort, or truly reflects the lack of a core gut virome, remains enigmatic. No significant correlation could be described between the bacteriophage communities derived from healthy and weak bees, but both location and sampling year had a significant effect (albeit with a low R-squared value). This observation reinforces the idea of high individuality and implies a dynamic nature of the bacteriophage communities. Host assignment of the viral sequences resulted in the assignment of only 1% of the contigs to their respective bacteria. Because both the methods (CRISPR-spacer sequences and tRNA similarity) used for host assignment do not have a very high sensitivity, it is likely that the number of viral sequences infecting members of the core gut bacterial microbiome is much higher. On the other hand, since entire bees were used for viral discovery, and not dissected guts, it cannot be excluded that some sequences represent soil- or plant-associated phage communities. The overlapping results from the CRISPRdb search and the bee bacterium-specific search revealed that the majority of sequences are indeed true gut-specific bacteriophages, but that environmental “contamination” cannot be ruled out completely. The observation that nearly all of the members of the core gut bacterial microbiome now have viral sequences associated with them reinforces the idea that at least a part of the viral community described here is truly part of the bee-gut virome. Since viral-like particle enrichment techniques never perform perfectly, the question arises that some of the retrieved sequences could have originated from bacteria. Since bacteriophages can also be integrated into bacterial genomes as prophages, the identification process can be prone to errors. To ensure that the sequences retrieved in this study originate from viruses, and not from bacterial contamination, coding densities and strand shift were calculated and differed significantly from the bacterial dataset. Both the parameters were chosen since a new bacteriophage identification algorithm identified these as the most informative in the discrimination between viruses and bacteria (40). Additionally, no bacterial marker genes could be identified with Anvi’o (41), using the single-copy gene bacterial Hidden Markov Model (HMM) profiles defined by Lee et al. (28). Furthermore, a large number of the GO terms associated with the putative viral proteins contain virus-specific signatures, and the overlap between bacterial—and viral—protein clusters (defined here as AMGs) remains relatively small. One would expect a much larger overlap between these cluster sets if contaminated by bacterial sequences. Taken together, this evidence supports the idea that very few to none of the sequences used in this study are of bacterial origin. Classification of the putative viral sequences resulted in roughly 20% of all of the sequences being clustered into 403 putative viral genera (genome clusters), but many of the clusters contained only two sequences or did not contain a reference genome from an established (International Committee on Taxonomy of Viruses [ICTV]-recognized) virus genus or family. The fact that roughly 50% (273 of 537) of contigs larger than 5 kb could be clustered reflects that a short sequence length can hamper the ability to classify these sequences. Since the accuracy of the vConTACT2 algorithm is estimated at more than 95% (based on ICTV genera) (30), the confidence in the classification performance of the large sequences is high. The proportion of clustered sequences (roughly 20% of all sequences and roughly 50% of sequences larger than 5 kb) is higher than in a similar study on permafrost viruses (17% sequences larger than 10 kb clustered) (42) and in human gut datasets (18% of sequences larger than 10 kb clustered) (43). Despite the relatively high proportion of clustered sequences, the classification results reflect the strikingly large diversity of prokaryotic viral communities associated with bees and how much of the viral diversity still remains untapped. This is reinforced by the fact that only very few putative bee-associated phage sequences were present in the largest protein clusters created for classification. The same patterns of diversity are also reflected in the protein annotation. Most of the predicted protein clusters remain unannotated. Of the proteins predicted to be under strong directional selection, only 20% could be assigned a function. Since it is probable that these proteins fulfill cornerstone functions in viral replication or important functions in the viral life cycle, the lack of annotation of these proteins reflects how little is known about these processes. Of the proteins that could be annotated in a meaningful way, the vast majority was specific for nucleic acid processing/metabolism and are most often derived from polymerase sequences, which are often the easiest to identify with very specific domains. Represented pathways contained a plethora of metabolic functions, including carbohydrate, protein, and lipid processing pathways. Many of these functions are also represented by the bacterial counterpart of the bee microbiome, implying that the bee-gut virome contains the coding potential for a vast range of metabolic functions and could directly intervene within the gut ecosystem. The best-represented pathways, such as genetic information processing and nucleotide metabolism, most likely reflect the rewiring strategy of phages to tune the bacterial cell metabolism toward virus replication, which has been described before (44). The lipid and nucleotide metabolism pathways most likely point in the same direction. The presence of environmental information-processing pathways suggests that some of the retrieved bacteriophages have the potential to probe the environment. It has been shown that the two-component system can be exploited by viruses to provide an environmental sensor system (45). The presence of more basal metabolic pathways, such as energy metabolism and carbohydrate metabolism, implies that also in the bee microbiome bacteriophages can modulate the metabolic state rather than hijack the microbial cell and deplete it for resources, as has been shown before (46). Biofilm formation, quorum sensing, and chemotaxis pathways were also represented within the retrieved viral communities, suggesting the potential of the viral communities to interfere in microbial processes on the bacterial population level. The presence of metabolic pathways involved in secondary metabolites and even terpenoids and polyketides raised the question if any other genes could be involved in bacteria–bacteria interactions. The discovery of four bacteriocin gene clusters implies that these bacteriophages do not directly influence only their own host and their metabolism but encode the potential to exert an effect on other bacteria in the same ecosystem throughout their host. Some of the identified bacteriocin genes were rather divergent, and very few of the neighboring genes within the cluster gave any hit at all. These findings imply that, while the essential host–microbe interactions in honey bees are known, the virus–bacteria interactions in the bee gut are highly intertwined. Finally, we can highlight the potential role that the prokaryotic viral community can play in the gut and microbial metabolism and thus indirectly influence bee development, health, and homeostasis.
Materials and Methods
Library Preparation and Next Generation Sequencing.
Samples were taken from the Flemish section of the EpiloBEE project from both sampling years (autumn 2012 and 2013) from different hives. In the framework of this study, colony health was determined retrospectively by assessing which colonies survived the winter or not. Two honey bees per colony were taken and homogenized for 1 min in phosphate-buffered saline, using ceramic beads (Precellys, Bertin Technologies) at 4,000 hz using a tissue homogenizer (Minilys, Bertin Technologies). Homogenates from three colonies were pooled together using equal volumes for feasibility reasons. Samples were pooled based on status (weak or healthy colonies), subspecies, and location (see SI Appendix, Table S14, available on GitHub). After pooling, the homogenates were prepared for sequencing using the NetoVIR protocol (22). Briefly, homogenates were centrifuged (17,000 × g for 3 min), filtered (0.8 μm), and treated with a mixture of nucleases (Benzonase, Novagen) and micrococcal nuclease (New England Biolabs).
Next, nucleic acids (both DNA and RNA) were extracted using the RNA viral extraction kit (Qiagen), reverse-transcribed, and amplified using the WTA2 kit (Sigma-Aldrich) and prepared for sequencing using the Nextera XT kit (Illumina). Libraries were quantified using a qubit fluorometer, and insert sizes were asserted using a bioanalyzer (Agilent). Only libraries that had molarities above 4 nM and an average size of 300 bp or more were considered for sequencing. Paired-end sequencing was performed using the Illumina NextSEQ platform, assigning 5 million clusters per pool (10 million reads with a base length of 150). In total, 102 pools were sequenced (representing 300 colonies).
Read Processing, Bacteriophage Contig Identification, and Classification.
Reads were clipped using Trimmomatic (version 0.38) (47), removing WTA2 and Nextera XT adapters and the leading 19 bases, and tailing 15 bases were cropped. Reads were trimmed using a sliding window of 4 with a PHRED score cutoff of 20 with a minimum size of 50 bp. Trimmed reads were assembled using SPAdes (version 3.12.0) (48) on metagenomic setting with kmer sizes of 21, 33, 55, and 77. Resulting contigs larger than 500 bp were clustered on 95% nucleotide identity over a coverage of 80% using ClusterGenomes (https://bitbucket.org/MAVERICLab/docker-clustergenomes). Putative prokaryotic viral sequences were identified using VIRSorter (version 1.05) (23) and by including sequences that had a lowest-common ancestor [as assigned by KronaTools (version 2.7.1) (49)] to any prokaryotic viral family, after alignment with the nonredundant protein database (downloaded September 30, 2018) from National Center for Biotechnology Information (NCBI). Reads were mapped back to these representative putative prokaryotic viral sequences using bwa-mem (50), and the resulting bam files were postprocessed using BamM (https://github.com/Ecogenomics/BamM), allowing only alignments with 95% nucleotide identity over 90% of the length. Coverages were calculated from these postprocessed bam files with BamM using the tpmean counting option. Dimension reduction was performed on the coverage matrix using the PCoA function implemented in the ape package in R (51) and formally tested using the adonis test implemented in the vegan package in R (52). Predicted viral sequences were classified using the BLASTP mode incorporated in vConTACT2 (30), using the Prokaryotic Viral RefsEq. 88 database MCL (25) for protein clustering and ClusterONE (53) for genome clustering. The resulting vConTACT2 network was processed using the graph-tool library (54) in python. Phylogenetic trees for the five biggest protein clusters were created by aligning the protein sequences with MAFFT (l-INS-i setting) and trimming the resulting alignment with trimAL (version 1.2) using the gappyout preset. Trees were subsequently created with RaxML (version 8.2.12) (55) using automatic model selection. Statistics from the phylogenetic trees were processed using the ete3 toolkit (56), implemented in python.
Host Calling.
Bacterial sequences were retrieved from IMG/M database (JGI) by using the query “honey bee” and complemented with the sequences from Ellegaard et al. (27) (SI Appendix, Table S15). CRISPR spacers were predicted from these bacterial sequences using MINCED (version 0.2.0) (57). This CRISPR-spacer collection was subsequently blasted on the nucleotide level against a database containing the retrieved bacteriophage sequences, using the blastN algorithm with the additional settings -ungapped and -perc_identity 100. These settings are more conservative than usual (58), but were selected to achieve the highest possible specificity at the expense of sensitivity. In parallel, tRNA genes were predicted from the retrieved bacteriophage sequences using Aragorn (version 1.2.38) (59) and blasted against the bacterial sequences using the blastN algorithm with an e-value cutoff of 1e-5. To estimate how many of the retrieved viral sequences are derived from the environment rather than reflecting true bee-gut bacteriophages, an additional analysis using CRISPR spacers from the CRISPRdb (29) was run using the same blastN parameters as before. A concatenated protein alignment for the bacterial sequences was created with Anvi’o (version 5) (41), using the “phylogenomics” workflow. The resulting alignment was trimmed using trimAl (version 1.2), using the gappyout preset. Protein models were calculated with ProtTest3 (version 3.4.2) (60), and the phylogeny was created using RAxML (version 8.2.12) (55) under the LG + I + G + F model. The resulting tree was visualized using ggtree (version 3.10) (61).
Functional Analysis.
Putative viral genes were predicted with prodigal, using the bacterial genetic code (11). Resulting proteins were clustered using CD-HIT (version 4.8.1) (62) with a threshold of 50% amino acid (AA) similarity. Bacterial genes (from the aforementioned bacterial dataset) were predicted and clustered via the same pipeline. Representative protein sequences were analyzed using InterProScan (version 5.30–69.0) (31) and eggNOG-mapper (version 1.0.2) (32). Downstream analysis was performed with REVIGO (35) and the KEGG Pathway Maps (36). Prophage regions were identified in the bacterial contigs using PHASTER (34). Antimicrobial functions were extracted from the InterProScan and eggNOG-mapper output and complemented using antiSMASH (version 5.0.0) (37) output. SNPs were called using freebayes (version 1.2.0) (63) on the filtered bam files using flags -X, -u and -p1. Resulting VCF files were filtered for a quality threshold of 20. SNP statistics were subsequently calculated using SNPgenie (38).
Statistics.
The species accumulation curve was calculated using the “specaccum” function within vegan, R version 3.5.3 (52), using all 102 sequenced pools. The difference between coding density and strand shift frequency, as well as the difference in contig length between clustered contigs and singleton contigs was calculated with Python using the two-tailed Mann–Whitney U test implemented in the SciPy library. For the coding density and strand shift frequency analysis, 24,420 predicted viral genes were used and 58,704 predicted bacterial genes. For the clustered-contig versus singleton-contig length difference, 1,034 clustered contigs were used and 3,010 singleton contigs were used. Correlations were calculated with the Spearman’s rank-order correlation implemented in the SciPy library. For the correlation between coverage and length, 4,842 putative viral contigs were used. Correlations between branch length distance and node distances in the network were calculated using 224,714 pairs.
Data Accessibility.
Retrieved prokaryotic viral sequences larger than 5 kb were submitted to NCBI GenBank (accession numbers available in SI Appendix, Table S19, available on GitHub). Raw reads were deposited in NCBI’s Sequence Read Archive (SRA) database under project accession no. PRJNA579886 (SRA accession numbers are also available in SI Appendix, Table S19, available on GitHub). Analysis notebooks have been deposited on GitHub (https://github.com/Matthijnssenslab/beevir). All intermediate result files and outputs generated, as well as the fasta sequences for nucleotides and proteins, are also available through the GitHub repository. A complete overview of the wet laboratory work and data-processing pipeline is given in SI Appendix, Fig. S13.
Supplementary Material
Acknowledgments
This project was funded by the Belgian Federal Public Service Health, Food Chain Safety and Environment (grant RF 16/6306 ViroBee). We thank the EpiloBEE study (European Commission) for providing access to the samples. L.B. was funded by Research Foundation Flanders (FWO). C.K.Y was funded by the Interfaculty Council for the Development Cooperation (IRO) from the KU Leuven. The computing power for this work was provided by the Flemish Supercomputer Centre (VSC) and financed by the FWO and the Department of Economy, Science and Inovation (EWI) of the Flemish government.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Retrieved prokaryotic viral sequences larger than 5 kb were deposited in the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers available in SI Appendix, Table S18, available on GitHub). Raw reads were deposited in NCBI’s Sequence Read Archive (SRA) database under accession no. PRJNA579886 (SRA accession numbers are also available in SI Appendix, Table S18, available on GitHub). Analysis notebooks are available at GitHub (https://github.com/matthijnssenslab/beevir). All intermediate results files and outputs that were generated, as well as the fasta sequences for nucleotides and proteins, are also available through the GitHub repository.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921859117/-/DCSupplemental.
References
- 1.Vanengelsdorp D., Meixner M. D., A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 103, S80–S95 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Forfert N., et al. , Parasites and pathogens of the honeybee (Apis mellifera) and their influence on Inter-Colonial Transmission. PLoS One 10, e0140337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fünfhaus A., Ebeling J., Genersch E., Bacterial pathogens of bees. Curr. Opin. Insect Sci. 26, 89–96 (2018). [DOI] [PubMed] [Google Scholar]
- 4.McMenamin A. J., Flenniken M. L., Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 26, 120–129 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Mitchell E. A. D., et al. , A worldwide survey of neonicotinoids in honey. Science 358, 109–111 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Ellegaard K. M., Engel P., Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10, 446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regan T., et al. , Characterisation of the British honey bee metagenome. Nat. Commun. 9, 4995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee F. J., Rusch D. B., Stewart F. J., Mattila H. R., Newton I. L. G., Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol. 17, 796–815 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Haydak M. H., Honey bee nutrition. Annu. Rev. Entomol. 15, 143–156 (1970). [Google Scholar]
- 10.Kešnerová L., et al. , Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, e2003467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H., Powell J. E., Steele M. I., Dietrich C., Moran N. A., Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 4775–4780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maes P. W., Rodrigues P. A. P., Oliver R., Mott B. M., Anderson K. E., Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 25, 5439–5450 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Engel P., et al. , The bee microbiome: Impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 7, e02164–e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady T. S., et al. , Bacteriophages as an alternative to conventional antibiotic use for the prevention or treatment of Paenibacillus larvae in honeybee hives. J. Invertebr. Pathol. 150, 94–100 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Merrill B. D., Grose J. H., Breakwell D. P., Burnett S. H., Characterization of Paenibacillus larvae bacteriophages and their genomic relationships to firmicute bacteriophages. BMC Genomics 15, 745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitbart M., Bonnain C., Malki K., Sawaya N. A., Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Trubl G., et al. , Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3, e00076-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erez Z., et al. , Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dedrick R. M., et al. , Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakhshinejad B., Ghiasvand S., Bacteriophages in the human gut: Our fellow travelers throughout life and potential biomarkers of health or disease. Virus Res. 240, 47–55 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Jacques A., et al. ; EPILOBEE Consortium , A pan-European epidemiological study reveals honey bee colony survival depends on beekeeper education and disease control. PLoS One 12, e0172591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conceição-Neto N., et al. , Modular approach to customise sample preparation procedures for viral metagenomics: A reproducible protocol for virome analysis. Sci. Rep. 5, 16532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Van Dongen S., Graph clustering via a discrete uncoupling process. SIAM J. Matrix Anal. Appl. 30, 121–141 (2008). [Google Scholar]
- 26.Grazziotin A. L., Koonin E. V., Kristensen D. M., Prokaryotic Virus Orthologous Groups , Prokaryotic virus orthologous groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 45, D491–D498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellegaard K. M., Engel P., New reference genome sequences for 17 bacterial strains of the honey bee gut microbiota. Microbiol. Resour. Announc. 7, e00834-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M. D., GToTree: A user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grissa I., Vergnaud G., Pourcel C., The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8, 172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bin Jang H., et al. , Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mitchell A. L., et al. , InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huerta-Cepas J., et al. , Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huerta-Cepas J., et al. , eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arndt D., et al. , PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blin K., et al. , antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson C. W., Moncla L. H., Hughes A. L., SNPGenie: Estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31, 3709–3711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boncristiani H. F. Jr, Di Prisco G., Pettis J. S., Hamilton M., Chen Y. P., Molecular approaches to the analysis of deformed wing virus replication and pathogenesis in the honey bee, Apis mellifera. Virol. J. 6, 221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amgarten D., Braga L. P. P., da Silva A. M., Setubal J. C., MARVEL, a tool for prediction of bacteriophage sequences in metagenomic bins. Front. Genet. 9, 304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eren A. M., et al. , Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emerson J. B., et al. , Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 3, 870–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory A. C., Zablocki O., Howell A., Bolduc B., Sullivan M. B., The human gut virome database, bioRxiv:10.1101/655910 (2 July 2019).
- 44.Enav H., Mandel-Gutfreund Y., Béjà O., Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2, 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Q., Chisholm S. W., Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr. Biol. 22, 124–128 (2012). [DOI] [PubMed] [Google Scholar]
- 46.De Smet J., et al. , High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 10, 1823–1835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ondov B. D., Bergman N. H., Phillippy A. M., Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12, 385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradis E., Schliep K., Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Oksanen J., et al. , vegan: Community Ecology Package. https://cran.r-project.org/web/packages/vegan/index.html. Accessed 30 September 2019.
- 53.Nepusz T., Yu H., Paccanaro A., Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 9, 471–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peixoto T. P., The Graph-Tool Python Library (2017). https://figshare.com/articles/graph_tool/1164194. Accessed 8 October 2019.
- 55.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bland C., et al. , CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edwards R. A., McNair K., Faust K., Raes J., Dutilh B. E., Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 40, 258–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laslett D., Canback B., ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darriba D., Taboada G. L., Doallo R., Posada D., ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu G., Lam T. T.-Y., Zhu H., Guan Y., Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 35, 3041–3043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (21 September 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Retrieved prokaryotic viral sequences larger than 5 kb were submitted to NCBI GenBank (accession numbers available in SI Appendix, Table S19, available on GitHub). Raw reads were deposited in NCBI’s Sequence Read Archive (SRA) database under project accession no. PRJNA579886 (SRA accession numbers are also available in SI Appendix, Table S19, available on GitHub). Analysis notebooks have been deposited on GitHub (https://github.com/Matthijnssenslab/beevir). All intermediate result files and outputs generated, as well as the fasta sequences for nucleotides and proteins, are also available through the GitHub repository. A complete overview of the wet laboratory work and data-processing pipeline is given in SI Appendix, Fig. S13.