Significance
Our findings have revealed a previously unrecognized link between circadian oscillations and intercellular variation and provide experimental evidence that stochastic transcriptional noise contributes significantly to cell-autonomous circadian periodicity. Interestingly, in separate studies, aging and cancer have been associated with increased transcriptional noise and less robust circadian rhythms. Here, we establish a direct association between transcriptional noise and circadian period. These findings may provide additional directions for researchers in the aging and cancer fields. Furthermore, circadian period may also be used as an indicator of variance in heterogeneity research and drug screening for noise control.
Keywords: circadian oscillation, transcriptional noise, period, heterogeneity/variance, single-cell imaging
Abstract
Nongenetic cellular heterogeneity is associated with aging and disease. However, the origins of cell-to-cell variability are complex and the individual contributions of different factors to total phenotypic variance are still unclear. Here, we took advantage of clear phenotypic heterogeneity of circadian oscillations in clonal cell populations to investigate the underlying mechanisms of cell-to-cell variability. Using a fully automated tracking and analysis pipeline, we examined circadian period length in thousands of single cells and hundreds of clonal cell lines and found that longer circadian period is associated with increased intercellular heterogeneity. Based on our experimental results, we then estimated the contributions of heritable and nonheritable factors to this variation in circadian period length using a variance partitioning model. We found that nonheritable noise predominantly drives intercellular circadian period variation in clonal cell lines, thereby revealing a previously unrecognized link between circadian oscillations and intercellular heterogeneity. Moreover, administration of a noise-enhancing drug reversibly increased both period length and variance. These findings suggest that circadian period may be used as an indicator of cellular noise and drug screening for noise control.
Heterogeneity exists in populations and species, within individuals, and from cell to cell. In some circumstances, cellular heterogeneity can be beneficial. For example, in preadapted subpopulations of microbial systems, heterogeneous gene expression confers adaption to fluctuating environments (1, 2). Stochastic gene expression has also been linked to cell fate decisions in higher eukaryotes (3–6), and in human cells, increasing transcriptional variation can help to eradicate latent HIV viruses (7, 8). However, in other situations, heterogeneity may be detrimental to cellular function. For example, increased nongenetic variation in gene expression is associated with aging (9–11) and cancers (12, 13) in mammalian tissues.
The origin of cell-to-cell variability is complex and may be attributed to genetic mutations (14), epigenetic modifications (15), and stochastic gene expression (16–18), although the relative contribution of each of these factors is still poorly understood (19). Stochastic gene expression is also referred to as “transcriptional noise” due to multiple rate-limiting steps and the random bursts seen in RNA polymerase II-mediated transcription (20). However, transcriptional fluctuations are further amplified by mRNA processing and protein synthesis (21), which also occur in stochastic bursts (22, 23). Studies in synthetic gene circuits suggest that negative-feedback loops may provide a way for cells to decrease transcriptional noise (24, 25).
As autonomous circadian oscillators, although cells use autoregulatory negative-feedback loops to generate precise circadian rhythms (∼24 h) (26–28), within a population, individual cells exhibit heterogeneity in circadian period length (29–31). However, the circadian period of each single cell is extremely stable over time (32), making this an ideal system for studying cellular heterogeneity. In order to investigate the origins of heterogeneity in circadian period, we analyzed and compared periods from thousands of single cells and clonal cell lines. The sources of variability were then partitioned into heritable and nonheritable components, and we estimated their contributions to phenotypic heterogeneity. Taken together, our results reveal an association between circadian period length and noise-driven intercellular heterogeneity, provide a quantitative method for assessing the contributions of heritable variance versus stochastic noise, and suggest that circadian period length is a robust indicator of cell-to-cell variability.
Results
Heterogeneous Circadian Periods from Single Cells.
We developed a single-cell imaging tracking and analysis pipeline that facilitates large-scale, single-cell period analysis in a computational way (Fig. 1 A–G). Using this pipeline, we examined circadian rhythms in a primary immortalized mouse ear fibroblast cell line carrying a PER2::LUCsv bioluminescence reporter (33, 34). Analysis of 228 cells in the primary parent culture revealed a normal distribution of periods (ranging from 21.55 to 27.82 h), with a mean value of 24.38 h and SD of 1.20 h (Fig. 2 A and B and Movie S1). This average is consistent with LumiCycle recording results for the synchronized cell population (24.46 ± 0.25 h SD, n = 24), confirming that LumiCycle data represent the ensemble average of single cells. To assess the heritability of heterogeneous oscillations, 150 separate clonal cell lines were established from the parental culture. The circadian periodicity of clonal cell lines was examined as ensemble cell populations with LumiCycle analysis. These clonal cell lines showed a similar period distribution to the parental single cells (ranging from 22.76 to 27.65 h; mean, 24.81 ± 0.83 h), but smaller SD (0.83 vs. 1.20 h) and coefficient of variation (CV) (3.33% vs. 4.91%), suggesting only partially heritable variation (Fig. 2 A and B). To examine whether cell-to-cell variation remained in single cells from a more homogeneous background, we next performed single-cell imaging analysis using 10 clonal cell lines selected from the two tails of the period distribution: the short period (SP) group and the long period (LP) group (Fig. 2 B and C). Single cells within these clonal cell lines exhibited broad period distributions with consistent mean values of period for each cell population, showing that circadian period is heterogeneous even in isogenic populations (Fig. 2C, SI Appendix, Fig. S1 A–J, and Movies S2–S11).
Fig. 1.
Automated single-cell analysis reveals heterogeneous circadian periods in neighboring cells. (A) Representative bioluminescence images from immortalized mouse ear fibroblast cells carrying the PER2::LUCsv reporter, with three representative cells indicated by arrows. (B) Raw bioluminescence signals of the three representative cells. (C) Baseline-subtracted, detrended, and denoised signals of the three representative cells. (D–F) L-S periodograms for cells 1 to 3. Peak periods are indicated by dashed lines. (G) Fitted cosine trajectories of the three representative cells. Calculated periods: 24.07, 25.51, and 26.98 h for cells 1 to 3, respectively. Note the high agreement with the L-S peak periods in D–F.
Fig. 2.
Longer period is associated with increased variance in clonal cell lines. (A) Experimental scheme for establishing 150 clonal cell lines with different circadian periods from the parent culture. Different colors indicate different period length. (B) Box plot (mean ± SD, range) comparing circadian period distributions of single cells and clonal cell lines generated from the parent culture. Range for 228 single cells: 21.55 to 27.82 h. Range for 150 clonal cell lines: 22.76 to 27.65 h. (C) Violin plot visualizing period distributions of single cells for 10 clonal cell lines. The x axis indicates IDs of clonal cell lines sorted based on means of period from single-cell imaging analysis. Dots with error bar indicate mean ± SD. Number of cells is listed above each cell line. (D) Heatmap showing significance of t test between clonal cell lines based on single-cell analysis results as shown in B. (E) Heatmap showing significance of F test as in D.
Longer Circadian Period Is Associated with Increased Variance in Clonal Cell Lines.
Interestingly, clonal cell lines from the LP group had a wider period distribution (i.e., more variance in period) than the SP group (Fig. 2C and SI Appendix, Fig. S1 A–J). Statistical analysis confirmed a higher CV (average of LP vs. SP: 5.26% vs. 4.18%), and a higher SD (average of LP vs. SP: 1.40 vs. 0.98 h) for the LP group than the SP group. Similar results were also observed when number of single cells for analysis was normalized to the same value (SI Appendix, Fig. S1 A–J, light color plots). Pairwise comparisons revealed significant differences not only in period length (Fig. 2D and SI Appendix, Table S1) but also in variance between SP and LP clones (Fig. 2E and SI Appendix, Table S2). Notably, SP clone 44 and LP clone 86 were outliers compared to other group members, showing either higher CV (4.78%) with SP (23.73 ± 1.13 h), or lower CV (3.79%) with LP (26.69 ± 1.01 h) (SI Appendix, Fig. S1 E and G). In contrast to the other clonal cell lines, which were stable and maintained similar periods over time, after long-term storage and multiple passages clone 44 and clone 86 became more unstable, with periods changing to 24.72 and 24.75 h, respectively (SI Appendix, Fig. S2).
To further validate these results, we performed continuous wavelet transform (CWT) analysis to verify cycle-to-cycle periods of single cells. Mean peak-to-peak periods were highly consistent with cosine curve fitting results, whereas the variability within single cells was not significantly correlated with mean period length (SI Appendix, Fig. S3). There was also no significant correlation between period and goodness-of-fit across single cells from the same population, ruling out the possibility of measurement errors (SI Appendix, Fig. S4). Cell size is critical in determining period variability in stochastic simulations (35) and is also an important source of noise in gene expression (17). However, when we examined the cell size of different clones, there were no differences between the SP group and LP group in cell body or nuclear size (SI Appendix, Fig. S5 A and B). Moreover, within each clonal cell line, there was no correlation between single-cell size and period length for the majority of clones (SI Appendix, Fig. S5C). Periods of single cells were also not correlated to their physical positions (SI Appendix, Fig. S6).
Amplitude is another key parameter of circadian oscillations. Cells with various amplitudes have shown to be able to respond to stimuli differently and may affect the circadian properties of cell populations (36, 37). However, when we examined the mean peak-to-trough amplitudes of single cells, we found no (or extremely weak [R2 < 0.1]) correlation between amplitude and period length within the same cell population (SI Appendix, Fig. S7). On the population level, there is also no difference in amplitude between SP and LP groups (SI Appendix, Fig. S7C).
Period Length Is Strongly Influenced by Nonheritable Variance.
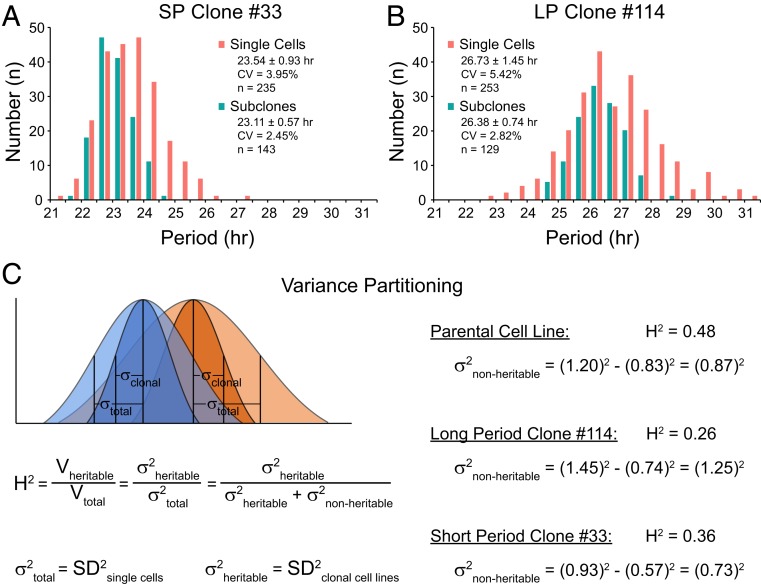
To test the heritability of heterogenous circadian period further, we used serial dilution to generate subclones from single cells of two representative parental clonal cell lines: SP clone 33 and LP clone 114 (Fig. 2C). Compared to their parental single cells, the subclones still exhibited heterogenous circadian periods with consistent mean values (23.11 vs. 23.54 h for SP clone 33; 26.38 vs. 26.73 h for LP clone 114), but decreased SD (0.57 vs. 0.93 h for SP clone 33; 0.74 vs. 1.45 h for LP clone 114) and smaller CV (2.45% vs. 3.95% for SP clone 33; 2.82% vs. 5.42% for LP clone 114), indicating partially heritable variance for both groups (Fig. 3 A and B). Subclones from SP clone 33 and LP clone 114 exhibited similar CV (2.45% vs. 2.82%); however, single cells showed robustly increased CV (3.95% vs. 5.42%) in LP clone 114, suggesting more elevated nonheritable variance than heritable variance in the LP group. F test also confirmed that single cells showed greater variance compared to subclones (1.35E-11 vs. 1.50E-3) between SP clone 33 and LP clone 114. In agreement with results from the parent culture, the difference was independent of the number of single cells examined, as similar results were observed using the same number of single cells and subclones for each group (SI Appendix, Fig. S8 A–C). Taken together, our results suggest that longer period is predominantly influenced by nonheritable rather than heritable factors.
Fig. 3.
Longer period is predominantly associated with increased nonheritable variance. (A) Histogram comparing circadian period distributions of single cells and subclones generated from SP clone 33. Range for 253 single cells: 21.19 to 27.05 h. Range for 143 subclones: 21.95 to 24.65 h. (B) Histogram comparing circadian period distributions of single cells and subclones generated from LP clone 114. Range for 235 single cells: 22.67 to 31.27 h. Range for 129 subclones: 24.73 to 28.75 h. Statistical results were labeled as mean ± SD. All subclones in A and B were measured as whole culture with LumiCycle, and data are presented as averages from three or more experiments. (C) Variance partitioning model. Blue indicates SP clone. Orange indicates LP clone. Light color indicates single-cell data representing the total variance. Dark color indicates subclones data representing the heritable variance. The concept of broad-sense heritability H2 was adapted to measure the contributions of heritable vs. nonheritable variance to phenotypic heterogeneity.
Variance Partitioning Model for Cellular Heterogeneity.
In order to estimate the relative contributions of heritable and nonheritable (stochastic) factors, we used a variance partitioning model for cellular heterogeneity based on the experimental observations. Inspired by the genetic concept of broad sense heritability (H2) (38), the phenotypic variance Vtotal can be partitioned into heritable variance Vheritable and nonheritable variance Vnonheritable, and the index H2 is used to estimate the contribution of heritable variance Vheritable/Vtotal (Fig. 3C). Here, the Vtotal for a given cell population refers to the variance of single cells. The Vheritable refers to the variance of clonal cell lines generated from the same parent culture. The Vnonheritable is the difference between total and heritable variance.
Based on this model, in parent cultures, the comparable Vheritable (0.832) and Vnonheritable (0.872) and the H2 index value (H2: 0.48) indicate that both the heritable and nonheritable components contribute almost equally. However, in clonal cell lines, Vnonheritable is greater than Vheritable (0.732 vs. 0.572 for SP clone 33; 1.252 vs. 0.742 for LP clone 114), and there is a much lower H2 index (H2: 0.36 for SP clone 33; H2: 0.26 for LP clone 114), suggesting that nonheritable noise is the major driving force for variability in homogeneous populations. Compared to the SP group, the greater Vtotal in the LP group (0.932 vs. 1.452, SP vs. LP) indicates that more intercellular variation is associated with lengthened circadian period. Although both Vheritable and Vnonheritable increased in the LP group (0.572 vs. 0.742 and 0.732 vs. 1.252, SP vs. LP), the larger difference between Vnonheritable (1.03) compared to Vheritable (0.23), and the even smaller H2 index (0.36 vs. 0.26, SP vs. LP) in the LP group, indicates a greater contribution of nonheritable factors, further suggesting that circadian period variance is predominantly associated with nonheritable noise.
Nonheritable Transcriptional Noise Increases Variance and Lengthens Period.
Random fluctuations in gene expression are inevitable at the single-cell level because of the stochastic nature of biochemical reactions (16). However, this intrinsic noise can be modulated (8, 39). Therefore, we asked whether circadian period is altered with increased intrinsic noise using a thymidine analog, idoxuridine, that acts as a transient transcriptional noise enhancer (8). As expected, idoxuridine increased the intrinsic cell-to-cell variance in the expression of ∼3,300 housekeeping genes without changing mean expression (SI Appendix, Fig. S9A). When we treated our clonal cell lines with idoxuridine, periods were significantly lengthened (∼1.5 h on average) (Fig. 4 A–C) (SI Appendix, Table S3). As both thymidine and idoxuridine can inhibit DNA replication, we wanted to rule out this potential side effect. We treated cells with thymidine and found no effect on circadian periodicity, suggesting that the effects of idoxuridine are not due to inhibited DNA replication (SI Appendix, Fig. S9B). Although LumiCycle data suggested reduced amplitude (Fig. 4A), single-cell analysis revealed even higher mean value from idoxuridine treatment (5.97 ± 3.45 vs. 4.23 ± 1.70 photons/min) (SI Appendix, Fig. S9C), possibly due to an effect of cell proliferation inhibition on amplitude at the population level. Furthermore, single-cell analysis revealed that idoxuridine also increased the variance of period heterogeneity (Fig. 4D and Movies S12 and S13). It is important to note that, due to the transient effect of the noise enhancer, only the first 6 d of single-cell imaging after drug treatment were analyzed. Beyond that window, the drug-treated cell population had comparable heterogeneity to controls. Because enhancing transcriptional noise increased both nonheritable variance and circadian period length, these findings suggest that lengthened circadian period is likely caused by increased stochastic noise.
Fig. 4.
Noise enhancement lengthens period and increases variation. (A and B) Two representative experiments showing effects of the noise enhancer idoxuridine on SP clone 128 and LP clone 114. LumiCycle traces are baseline subtracted. (C) Histogram showing period change for all 10 clonal cell lines. Results are average of three or more experiments. Error bars indicate SD. P < 0.001 for all tests. (D) Histogram showing period distributions of single cells from SP clone 128 with either vehicle control or idoxuridine treatment for 48 h. Values indicate mean ± SD. Control cells: 20.43 to 27.49 h, 5.75% (range, CV). Idoxuridine-treated cells: 20.16 to 30.90 h, 8.03%. P value of t test and F test between two conditions: 4.49E-07, 1.86E-04.
Discussion
Using fully automated single-cell tracking and precise analysis methods, we revealed an association between circadian period length and cell-to-cell variation in clonal cell lines: Longer period was associated with greater heterogeneity. When there was a discrepancy, such as clone 44 with shorter period but larger variance, or clone 86 with longer period but smaller variance, the population became more unstable and period changed remarkably to become longer or shorter, respectively, to be consistent with their heterogeneity status. Interestingly, in a stochastic model of circadian oscillators, the variability in period of simulated single cells increased from 1.03 h (SD) to 1.54 h when the variation in biochemical parameters increased to 5% of the mean values of the rate constant (40), consistent with our experimental observation of an average of 0.98 h SD in SP groups and 1.40 h SD in LP groups.
We also developed a method to estimate the contributions of different sources to the total phenotypic heterogeneity, by partitioning them into two categories: heritable and nonheritable. Because of mitotic inheritance, both genetic mutations and epigenetic modifications serve as heritable sources (14, 15). The nonheritable sources accounting for heterogeneity are more complex, including transcriptional, translational, and environmental fluctuations (16, 17). Although the underlying mechanisms are not clear, our findings suggest that period heterogeneity in the parent culture was equally influenced by heritable components and stochastic noise, while the heterogeneity in clonal cell lines was predominantly driven by noise. This relationship between circadian oscillation and stochastic noise is extremely interesting. In synthetic gene circuits, autoregulatory negative-feedback loops have been demonstrated as a way to control and decrease transcriptional noise and provide stability (24, 25). On the other hand, mathematical simulations suggest that increasing stochastic noise can increase heterogeneity of single-cell circadian oscillations (40, 41). However, it is less clear whether circadian transcriptional–translational negative-feedback loops play such a stabilizing role under physiological conditions (40). Nonetheless, our experiments using small molecules to manipulate transcriptional noise demonstrate that increasing transcriptional noise enhances heterogeneity and also significantly lengthens circadian period.
Understanding and having a readout of noise in cellular systems has implications both for basic scientific studies and translational research. For example, gene editing using the CRISPR-Cas9 system requires isolating clonal cell lines to obtain the desired genotype (42). Thus, clonal heterogeneity may represent a more serious obstacle to precise gene editing than nuclease-mediated off-target effects (43). Our findings suggest that using isogenic cells will improve the precision of gene editing since there is less heritable clonal heterogeneity from homogeneous cell populations, especially from those with shorter circadian period length. Our study also indicates that circadian period may be a useful indicator of variation in heterogeneity research and drug screening for noise control.
Intriguingly, circadian clock function declines with both aging and cancer (44–46). Considering that both of these processes are also associated with increased transcriptional variation (9–11, 13), a better understanding of the relationship between the circadian clock and cellular heterogeneity is warranted. Here, we establish a direct link between circadian oscillations and intercellular variability and provide experimental evidence that stochastic transcriptional noise contributes significantly to cell-autonomous circadian periodicity. Thus, our study may provide additional directions for researchers in the aging and cancer fields.
Materials and Methods
Bioluminescence Recording and Data Analysis.
To measure bioluminescence rhythms, we used an immortalized fibroblast cell line carrying a PER2::LUCsv fusion bioluminescence reporter (33, 34). For cell cultures, confluent cells from 35-mm culture dishes were synchronized with 100 nM dexamethasone for 2 h, then changed to Hepes-buffered recording medium containing 2% fetal bovine serum (FBS) (29), and loaded into a LumiCycle luminometer for at least 7 d recording (Actimetrics). The period was analyzed with LumiCycle Analysis program (Actimetrics). All LumiCycle period analysis results shown in this paper were averages of three or more experiments. Baseline-subtracted signals were exported to Excel to generate bioluminescence traces. To test the stability of clonal cell lines, two-way ANOVA with Bonferroni’s multiple comparisons was performed using GraphPad Prism.
For single-cell imaging, cells were changed to recording medium containing 2% B27 and 1% FBS without dexamethasone synchronization. An inverted microscope (Leica DM IRB) in a heated lucite chamber custom-engineered to fit around the microscope stage (Solent Scientific) kept the cells at a constant 36 °C and was mounted on an anti-vibration table (TMC) equipped with a 10× objective. A cooled CCD camera with backside illuminated E2V CCD 42-40, 2,048 × 2,048 pixel, F-mount adapter, −100 °C cooling (Series 600; Spectral Instruments) was used to capture the luminescence signal at 30-min intervals, with 29.6-min exposure duration, for at least 12 d. To increase the signal-to-noise ratio, 8 × 8 binning was used.
Single-Cell Tracking and Analysis.
The bioluminescence signal of each single cell, outlined with a region of interest, was tracked using ImageJ (47, 48) with the Trackmate plugin (49). Only cells which could be continuously tracked for at least 288 frames (6 d) were used for downstream analysis (except for noise enhancer treatment). For noise enhancer treatment experiment, only cells that could be continuously tracked for at least 168 frames during the first 7 d were used for downstream analysis. For video presentations, cosmic ray artifacts were removed by pixelwise comparison of two consecutive images using the minimum value of each pixel (ImageJ). Statistical analysis of single-cell imaging was performed with a Python code. Our code is publicly available (https://github.com/johnabel/per2py) and freely available for automation of data processing. Briefly, cosmic ray artifacts were removed by using a top-hat filter (image J → remove outliers). After detrending via Hodrick–Prescott filter and denoising via eigendecomposition and signal reconstruction, we used Lomb–Scargle (L-S) spectral periodogram analysis to estimate period length in the range of 14 to 40 h. Then for rhythmic cells, we calculated the circadian period length by fitting a decaying sinusoid curve, using a least-squares fit with constraint of within ±1 h of the period peak from L-S spectral analysis. The L-S power and goodness of sinusoid fitting can also serve as criteria to eliminate mis-tracking of neighboring migrating cells due to loss of signal in dark phase. Only cells with both power of L-S peak >0.7 and goodness of fit >0.7 were used for downstream periodicity analysis (50). Criteria for noise enhancer treatment experiment is power of L-S peak >0.7 and goodness of fit >0.9 due to fewer cycles analyzed. Our tracking and analysis pipeline can record the position information of each cell, allowing examination of the correlation between position and periodicity.
To further validate the periodicity measurement using another method, we reanalyzed all single-cell data with CWT using WAVOS toolkit in MATLAB (51). Fed with the detrended and denoised signal, after removing edge-affected data, we calculated peak-to-peak periods for each single cell and defined the mean value as the period of each cell. The amplitude was defined as the mean peak-to-trough difference.
Student’s t test and two-tailed F test were performed in Excel. P values were adjusted using Benjamini–Hochberg (BH) method. GraphPad Prism was used to perform Pearson correlation coefficient analysis and to generate heatmaps for t test and F test based on log-transformed q value. Violin plot and box plot were generated in R using ggplot2 (52).
Noise Enhancer Characterization and Treatment.
Mouse E14 embryonic stem cells (mESCs) (male) (53) were cultured in feeder-free conditions on gelatin-coated plates with ESGRO-2i medium (Millipore; catalog #SF016-200). For noise enhancer testing, mESCs were treated with either 10 μM idoxuridine (Sigma; catalog #I7125) or DMSO for 24 h. Afterward, cells were trypsinized with trypLE and spun down for 5 min at 90 × g. Single-cell RNA-seq libraries were prepared according to the 10× Genomics user guide and sequenced on an Illumina HiSeq 4000 platform. Data were aligned to mm10 using 10× Cell Ranger v2. The gene-barcode matrices were then filtered and normalized in Seurat using the “LogNormalize” gene scaling method (54). A list of human housekeeping genes was generated and converted to their mouse orthologs (55). Gene expression noise for housekeeping genes was quantified using the fano factor to minimize the effect of mean-variance dependency (21). For noise enhancer treatment experiment, fibroblast cells were seeded at ∼25% confluence and treated with 20 μM idoxuridine for 48 h before loading to LumiCycle or microscopy. DMSO was used as a vehicle control. The same dosage of thymidine was given as another control.
For details about the generation of clonal cell lines and cell size measurements, please see SI Appendix, Materials and Methods.
Contact for Reagent and Resource Sharing.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author.
Data Availability.
The noise enhancer RNA-seq data have been deposited in GEO (accession no. GSE147386). All other data are included with the manuscript.
Supplementary Material
Acknowledgments
This research was supported by the Howard Hughes Medical Institute. The noise enhancer molecule was developed with the support of NIH grant AI109593. We thank Dr. John H. Abel and Dr. Francis J. Doyle III from Harvard Medical School for their help with the single-cell imaging analysis pipeline, Gokhul Kilaru for bioinformatics support, and all J.S.T. laboratory members, Dr. Carla B. Green, and Dr. Shin Yamazaki for helpful discussions. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
Data deposition: The noise enhancer RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE147386). Statistical analysis of single-cell imaging was performed with a Python code. Our code has been deposited in GitHub (https://github.com/johnabel/per2py) and is freely available for automation of data processing.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922388117/-/DCSupplemental.
References
- 1.Acar M., Mettetal J. T., van Oudenaarden A., Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40, 471–475 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Carey J. N., et al. , Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell 173, 196–207.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernet M. F., et al. , Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440, 174–180 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang H. H., Hemberg M., Barahona M., Ingber D. E., Huang S., Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vassar R., Ngai J., Axel R., Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74, 309–318 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Chambers I., et al. , Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Weinberger L. S., Burnett J. C., Toettcher J. E., Arkin A. P., Schaffer D. V., Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 tat fluctuations drive phenotypic diversity. Cell 122, 169–182 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Dar R. D., Hosmane N. N., Arkin M. R., Siliciano R. F., Weinberger L. S., Screening for noise in gene expression identifies drug synergies. Science 344, 1392–1396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Jimenez C. P., et al. , Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355, 1433–1436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enge M., et al. , Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahar R., et al. , Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441, 1011–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Spencer S. L., Gaudet S., Albeck J. G., Burke J. M., Sorger P. K., Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaffer S. M., et al. , Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrell R. A., McGranahan N., Bartek J., Swanton C., The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kelsey G., Stegle O., Reik W., Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Raser J. M., O’Shea E. K., Noise in gene expression: Origins, consequences, and control. Science 309, 2010–2013 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj A., van Oudenaarden A., Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel J., Widmer L., St. John P., Stelling J., Doyle F. III, A coupled stochastic model explains differences in cry knockout behavior. IEEE Life Sci. Lett. 1, 10.1109/LLS.2015.2439498 (2015). [DOI] [Google Scholar]
- 19.Eling N., Morgan M. D., Marioni J. C., Challenges in measuring and understanding biological noise. Nat. Rev. Genet. 20, 536–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenstra T. L., Rodriguez J., Chen H., Larson D. R., Transcription dynamics in living cells. Annu. Rev. Biophys. 45, 25–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen M. M. K., Desai R. V., Simpson M. L., Weinberger L. S., Cytoplasmic amplification of transcriptional noise generates substantial cell-to-cell variability. Cell Syst. 7, 384–397.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J., Xiao J., Ren X., Lao K., Xie X. S., Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Cai L., Friedman N., Xie X. S., Stochastic protein expression in individual cells at the single molecule level. Nature 440, 358–362 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Dublanche Y., Michalodimitrakis K., Kümmerer N., Foglierini M., Serrano L., Noise in transcription negative feedback loops: Simulation and experimental analysis. Mol. Syst. Biol. 2, 41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becskei A., Serrano L., Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Mohawk J. A., Green C. B., Takahashi J. S., Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L., The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S., Kay S. A., Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb A. B., Angelo N., Huettner J. E., Herzog E. D., Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 16493–16498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagoshi E., et al. , Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Leise T. L., Wang C. W., Gitis P. J., Welsh D. K., Persistent cell-autonomous circadian oscillations in fibroblasts revealed by six-week single-cell imaging of PER2:LUC bioluminescence. PLoS One 7, e33334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z., et al. , Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc. Natl. Acad. Sci. U.S.A. 109, 101–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo S. H., et al. , Period2 3′-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. U.S.A. 114, E8855–E8864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forger D. B., Peskin C. S., Stochastic simulation of the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 102, 321–324 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb A. B., Taylor S. R., Thoroughman K. A., Doyle F. J. 3rd, Herzog E. D., Weakly circadian cells improve resynchrony. PLoS Comput. Biol. 8, e1002787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu C., Liang X., Yang H., Rohling J. H., Heterogeneity induces rhythms of weakly coupled circadian neurons. Sci. Rep. 6, 21412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plomin R., DeFries J. C., McClearn G. E., Behavioral Genetics: A Primer (W. H. Freeman, New York, ed. 2, 1990). [Google Scholar]
- 39.Hansen M. M. K., et al. , A post-transcriptional feedback mechanism for noise suppression and fate stabilization. Cell 173, 1609–1621.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko C. H., et al. , Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 8, e1000513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St John P. C., Doyle F. J. 3rd, Quantifying stochastic noise in cultured circadian reporter cells. PLoS Comput. Biol. 11, e1004451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veres A., et al. , Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Musiek E. S., Holtzman D. M., Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 354, 1004–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Z., et al. , Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 9, 1556–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rueden C. T., et al. , ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tinevez J. Y., et al. , TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Ruf T., The Lomb-Scargle periodogram in biological rhythm research: Analysis of incomplete and unequally spaced time-series. Biol. Rhythm Res. 30, 178–201 (1999). [Google Scholar]
- 51.Harang R., Bonnet G., Petzold L. R., WAVOS: A MATLAB toolkit for wavelet analysis and visualization of oscillatory systems. BMC Res. Notes 5, 163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham H., ggplot2: Elegant Graphics for Data Analysis, Use R! (Springer, New York, 2016). [Google Scholar]
- 53.Sokolik C., et al. , Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Syst. 1, 117–129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg E., Levanon E. Y., Human housekeeping genes, revisited. Trends Genet. 29, 569–574 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The noise enhancer RNA-seq data have been deposited in GEO (accession no. GSE147386). All other data are included with the manuscript.