Significance
Barrier-to-autointegration factor (BAF), a highly conserved small protein in metazoans, plays multiple functions during the cell cycle; however, the regulation of BAF function is largely unclear. Here we find that the SUMOylation and de-SUMOylation cycle of BAF regulates these functions. We reveal that BAF is SUMOylated at K6, and that de-SUMOylation at this site is catalyzed by SENP1 and SENP2. SUMOylation of BAF regulates binding to lamin A/C, which is essential for nuclear retention of BAF. SUMOylation of BAF also regulates the interaction of BAF with nuclear DNA. De-SUMOylation facilitates cytoplasmic retention of BAF and loss of nuclear function. Non-SUMOylatable BAF is not efficiently retained in the nucleus, leading to structurally abnormal nuclear formation and DNA replication failure.
Keywords: cell cycle, barrier-to-autointegration factor (BAF), SUMOylation, nuclear integrity, lamin A/C
Abstract
Barrier-to-autointegration factor (BAF) is a highly conserved protein in metazoans that has multiple functions during the cell cycle. We found that BAF is SUMOylated at K6, and that this modification is essential for its nuclear localization and function, including nuclear integrity maintenance and DNA replication. K6-linked SUMOylation of BAF promotes binding and interaction with lamin A/C to regulate nuclear integrity. K6-linked SUMOylation of BAF also supports BAF binding to DNA and proliferating cell nuclear antigen and regulates DNA replication. SENP1 and SENP2 catalyze the de-SUMOylation of BAF at K6. Disrupting the SUMOylation and de-SUMOylation cycle of BAF at K6 not only disturbs nuclear integrity, but also induces DNA replication failure. Taken together, our findings demonstrate that SUMOylation at K6 is an important regulatory mechanism that governs the nuclear functions of BAF in mammalian cells.
Mutations in genes encoding nuclear periphery proteins are often associated with severe genetic disorders, diseases, and syndromes. One collective group of these disorders is laminopathy, which often carries mutations in lamin and LEM (Lap2, emerin, and MAN1) family members (1, 2). Interestingly, many of these diverse disorders affect only one or a few tissues, such as skeletal muscles, bones, adipocytes, and neuronal tissues. Hutchinson–Gilford progeria syndrome (HGPS) is a systematic laminopathy caused by point mutations of LMNA, which encodes lamin A/C. HGPS patients show severe accelerated aging and often die from atherosclerosis during adolescence (3–6). The alanine 12-to-threonine mutation (A12T) of barrier-to-autointegration factor (BAF) is associated with Néstor–Guillermo progeria syndrome (NGPS), another rare accelerated aging syndrome with clinical features similar to those of HGPS. However, in NGPS, patients lack the cardiovascular pathology characteristic of HGPS and have a longer lifespan (7). The underlying mechanism of NGPS is poorly understood.
BAF is a small multifunctional protein with roles in mitosis, nuclear dynamics, chromatin organization, gene regulation, DNA damage response, and viral infection (8). BAF is highly conserved among metazoans, and BAF depletion is lethal during embryogenesis in Caenorhabditis elegans and Drosophila melanogaster (9, 10). BAF forms homodimers and binds double-stranded DNA (dsDNA) in a sequence-independent manner (11–13). BAF functions in the maintenance of nuclear architecture during interphase (14). In mitosis, BAF is phosphorylated by vaccinia-related kinase 1 (VRK-1), which abrogates BAF interactions with other proteins to facilitate nuclear envelope (NE) disassembly (15). At the end of mitosis, BAF reassociates with chromatin and LEMs to reform the NE (16) and prevent nuclear fragmentation (17). Thus, BAF facilitates single nucleus formation.
SUMOylation of proteins by a small ubiquitin-related modifier (SUMO) regulates many cellular activities, including gene expression, signal transduction, macromolecular assembly, protein stability, nucleocytoplasmic transport, and DNA damage repair (18–20). Of the four SUMO proteins in humans—SUMO1, SUMO2, SUMO3, and SUMO4—SUMO4 remains enigmatic (19, 21, 22). Many SUMOylation proteins contain an acceptor lysine within a ΨKxE consensus sequence (where Ψ is a large hydrophobic residue and x represents any amino acid) that can be recognized by ubiquitin-conjugating enzyme 9 (Ubc9) directly. Alternatively, the SUMOylation targets without a consensus sequence recruit Ubc9 via their SUMO interaction motif (SIM), which contains a hydrophobic core with a consensus sequence V/I-X-V/I-V/I or V/I-V/I-X-V/I, or via E3 ligases (18, 19). SUMOylation can mask the interaction surface of target proteins and thus prevent their interaction with other proteins. Alternatively, SUMOylation can provide a binding site for new partners. Furthermore, if a target protein simultaneously contains an acceptor lysine for a SUMO molecule and a SIM, the intramolecular interaction between SUMO and SIM may induce a conformational change of the target (19).
Accumulating evidence shows that SUMOylation plays a pivotal role in regulation of the cell cycle (23, 24). For instance, SUMOylation promotes autophosphorylation and activation of Aurora B, which is important for localization (25, 26). Redistribution of the SUMO machinery during mitosis is essential to enable cell cycle progression (27). In this study, we demonstrate that BAF is SUMOylated, and that this modification regulates the function of BAF in nuclear integrity maintenance, DNA replication, and S phase progression.
Results
BAF Is SUMOylated at K6.
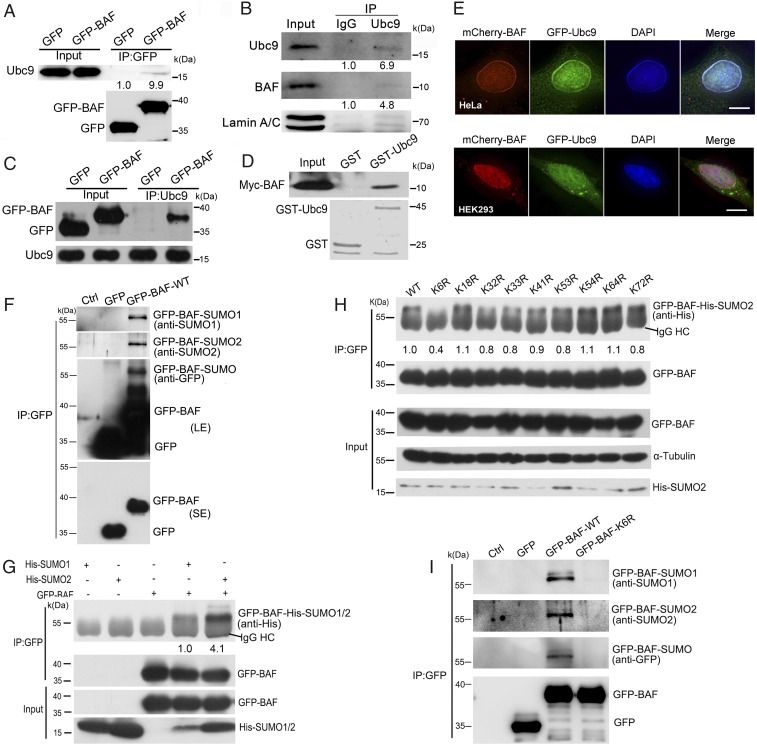
We identified proteins that interact with BAF during the cell cycle by expressing GFP-BAF in cells, followed by co-immunoprecipitation (co-IP) and Western blot analysis of the co-immunoprecipitated proteins. To our surprise, we found that Ubc9, the sole SUMO-conjugating enzyme for SUMOylation (19, 27), was co-immunoprecipitated with GFP-BAF (Fig. 1A). Using an anti-Ubc9 antibody to perform the co-IP assay, we confirmed the binding of both endogenous and exogenous BAF to endogenous Ubc9 in HEK293 cells (Fig. 1 B and C). We also expressed and purified GST-Ubc9 in bacteria and performed a GST pull-down assay in the cell lysate expressing Myc-BAF, which revealed that GST-Ubc9 pulled down the Myc-BAF (Fig. 1D). Through coexpression of GFP-Ubc9 and mCherry-BAF in HeLa and HEK293 cells, we observed that both Ubc9 and BAF colocalized in the nucleus, with enrichment at the NE (Fig. 1E). Collectively, these results demonstrate that BAF interacts and colocalizes with Ubc9 in the nucleus.
Fig. 1.
BAF is SUMOylated at K6. (A–C) GFP-BAF interacts with Ubc9 in cells. (A) Whole lysates from HEK293 cells expressing either GFP or GFP-BAF were used for co-IP with GFP-trap beads and Western blot analysis with GFP and Ubc9 antibodies. (B) Endogenous BAF interacts with Ubc9. Whole lysates of HEK293 cells were used for co-IP with the Ubc9 antibody and Western blot analysis with BAF and lamin A/C antibodies. (C) Whole lysates of HEK293 cells expressing either GFP or GFP-BAF were used for co-IP with the Ubc9 antibody and Western blot analysis with GFP and Ubc9 antibodies. (D) BAF interacts with purified Ubc9 protein in vitro. Myc-BAF–overexpressing HEK293 cell lysates were incubated with GST or GST-Ubc9, followed by a pull-down assay using glutathione Sepharose 4B beads and Western blot analysis with Myc antibody. The loading of GST and GST-tagged Ubc9 proteins is shown by Coomassie blue staining (Bottom). (E) Ubc9 colocalizes with BAF mainly in the cell nucleus. HeLa and HEK293 cells were transfected with GFP-Ubc9 and mCherry-BAF and observed after fixation. (Scale bars: 10 μm.) (F) BAF can be SUMOylated by SUMO1 and SUMO2/3. Whole-cell lysates of HEK293 cells expressing GFP or GFP-BAF were co-immunoprecipitated with GFP-trap beads and probed with GFP, SUMO1, and SUMO2/3 antibodies. Note a slow-migrating band at approximately 57 kDa that is recognized by anti-GFP antibodies and by SUMO1 and SUMO2 antibodies. (G) BAF can be SUMOylated with both His-SUMO1 and His-SUMO2. HEK293 cells were cotransfected with GFP-BAF and His-SUMO1 or His-SUMO2. Whole-cell extracts were co-immunoprecipitated with the GFP antibody and probed with His and GFP antibodies. (H) K6 is the major SUMOylation site of BAF. GFP-BAF-WT and KR mutants coexpressed individually with SUMO2 in HEK293 cells were co-immunoprecipitated with GFP antibody, followed by Western blot analysis using the indicated antibodies. Note that K6R almost lost the positive upper bands recognized by the anti-His antibody. (I) GFP-BAF-WT, but not GFP-BAF-K6R, can be modified by endogenous SUMO1 or SUMO2/3. GFP-BAF- WT or K6R coexpressed with SUMO1 or SUMO2 in HEK293 cells were co-immunoprecipitated with GFP-trap beads, followed by Western blot analysis using the indicated antibodies. Note that only GFP-BAF-WT could be recognized by anti-SUMO1 and -SUMO2 antibodies at a Kd value of 57.
We tested whether BAF is SUMOylated using a co-IP assay in cell lysates expressing GFP-BAF. The GFP antibody recognized a slow-migrating band at ∼57 kDa and a fast-migrating band at ∼40 kDa in cell lysates expressing GFP-BAF (Fig. 1F). By serially stripping the film and reblotting with anti-SUMO1 and anti-SUMO2/3 antibodies, we found that both antibodies recognized the same slow-migrating band (Fig. 1F). The slow-migrating BAF band was precipitated with either His-SUMO1 or His-SUMO2 in reciprocal co-IP assays or Ni-NTA pull-down assays when GFP-BAF was coexpressed with His-SUMO1 or His-SUMO2 (Fig. 1G and SI Appendix, Fig. S1A). Taken together, these results demonstrate that BAF is SUMOylated by SUMO1 and SUMO2/3.
We next investigated the conjugation site(s) of BAF for SUMO. Nine conservative lysine (K) sites in the primary sequence of BAF that could be SUMOylated were revealed by multiple sequence alignments (SI Appendix, Fig. S1B). By changing each of the lysine residues to arginine (R) in the possible SUMOylation sites, we generated nine GFP-tagged non-SUMOylatable KR mutants: BAF-K6R, -K18R, -K32R, -K33R, -K41R, -K53R, -K54R, -K64R, and -K72R. These nine mutants were individually coexpressed with His-SUMO2 in cells. Co-IP revealed that only GFP-BAF-K6R lost almost all of the SUMOylation bands. The wild-type (WT) BAF and other mutants had more or fewer SUMOylation bands (Fig. 1H). WT BAF and KR mutants were coexpressed with His-SUMO1, and the His-SUMO1-conjugated proteins were isolated. The K6R mutation of BAF strongly impeded SUMOylation in cells (SI Appendix, Fig. S1C).
We then performed co-IP assays using GFP-BAF-K6R– or GFP-BAF-WT–expressing cell lysates for endogenous SUMO or coexpression of GFP-BAF-K6R or GFP-BAF-WT with either His-SUMO1 or His-SUMO3, followed by isolation of the exogenous proteins. These experiments showed that while the WT BAF was strongly SUMOylated by endogenous and exogenous SUMO1 or SUMO2/3, almost no SUMOylation on the K6R mutant was detected (Fig. 1I and SI Appendix, Fig. S1 D–G). Taken together, these findings demonstrate that BAF is SUMOylated at K6 in cells, and that the modifier can be SUMO1, SUMO2/3, or both.
Nuclear Retention and Binding of BAF to Nuclear Lamin A/C Is Regulated by SUMOylation.
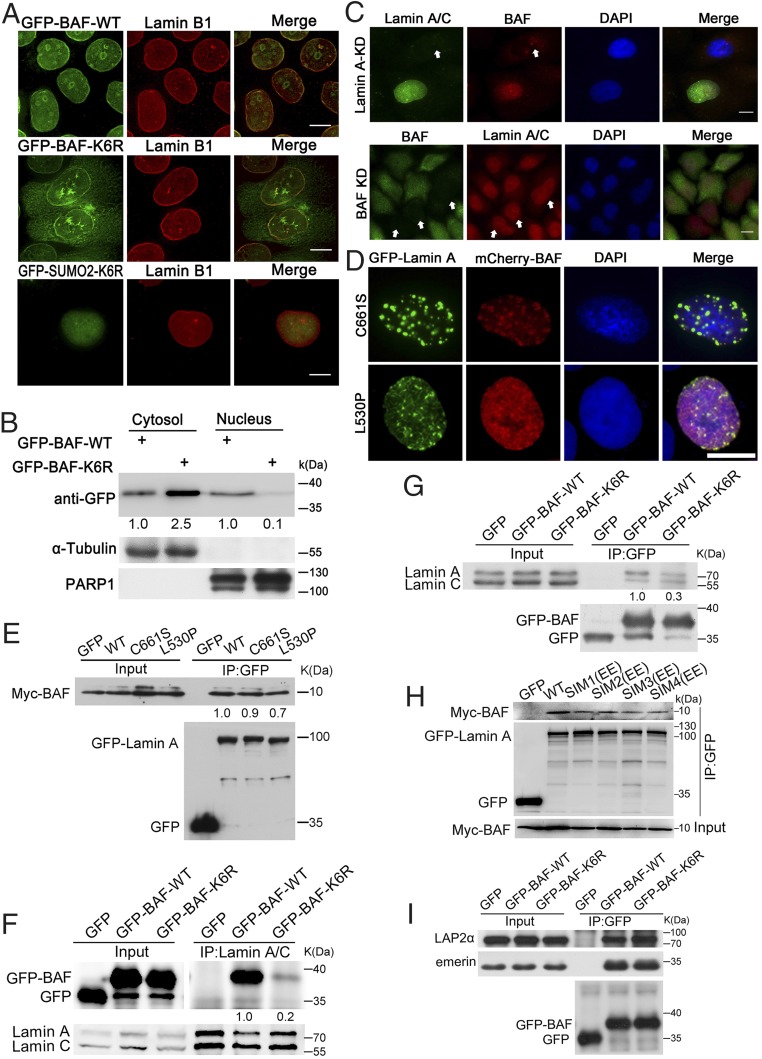
Previous studies have shown that GFP-fused BAF localizes to the nuclear envelope and the nucleoplasm similar to endogenous BAF (28, 29). We performed immunofluorescence experiments in Myc-BAF–, GFP-BAF–, and mCherry-BAF–overexpressing HeLa cells and found all the tagged constructs used in this study (GFP, mCherry and Myc tags) showed consistent localization with previous reports (SI Appendix, Fig. S1H). To determine the function of BAF SUMOylation, we first investigated the subcellular localization of both GFP-BAF and K6R mutants in cells. The results showed that GFP-BAF was localized predominantly at the NE and nucleoplasm, whereas GFP-BAF-K6R exhibited a dramatic reduction in nuclear localization in most cells (Fig. 2A). Cytoplasmic and nuclear fractionation assays in cells overexpressing GFP-BAF and GFP-BAF-K6R showed that GFP-BAF was significantly higher in the nuclear fraction compared with GFP-BAF-K6R, and that GFP-BAF-K6R exhibited more cytoplasmic retention (Fig. 2B). Importantly, when a SUMO2 molecule was fused to GFP-BAF-K6R (GFP-SUMO2-K6R) to mimic SUMO conjugation (30–32), the expressed fusion protein, GFP-SUMO2-K6R, was localized predominantly to the nucleus (Fig. 2A). In other words, the addition of a SUMO molecule rescued the nuclear localization failure of K6R.
Fig. 2.
SUMOylation of BAF enhances its binding to nuclear lamin A/C and nuclear retention. (A) Distinct localization of GFP-BAF-WT, GFP-BAF-K6R, and GFP-SUMO2-K6R. HeLa cells expressing GFP-BAF-WT, GFP-BAF-K6R, or GFP-SUMO2-K6R were stained with the lamin B1 antibody and DAPI. (B) BAF-K6R failed to localize to the nucleus. The protein levels of GFP-BAF in nuclear and cytoplasmic fractions were determined by immunoblotting with an anti-GFP antibody. The relative purity of the nuclear and cytoplasmic fractions was confirmed by sequential probing for the nuclear marker PARP1 and the cytoplasmic marker α-tubulin. (C) BAF failed to localize to the nucleus in lamin A/C knockdown cells. HeLa cells were transfected with lamin A/C siRNA (Upper) or BAF shRNA vector (Lower) to deplete endogenous lamin A/C and BAF, respectively, followed by immunostaining with BAF and lamin A/C antibodies. White arrows point to lamin A/C or BAF in relevant knockdown cells. (D) Distribution of BAF is affected by prelamin A/C nuclear accumulation. HeLa cells were transfected with GFP-lamin A-C661S or GFP-lamin A-L530P and mCherry-BAF. Note that when lamin A localized to intranuclear aggregates, BAF was found predominantly at these structures. (E) Both C661S and L530P mutants of lamin A interact with BAF. HEK293 cell lysates coexpressing GFP, GFP-lamin A-WT, GFP-lamin A-C661S, or GFP-lamin A-L530P with Myc-BAF were co-immunoprecipitated with GFP-trap beads and probed with Myc and GFP antibodies. (F) The interaction between BAF-K6R and endogenous lamin A/C is very weak. HEK293 cells were transfected with GFP-BAF or GFP-BAF-K6R. The total cell lysates were used for co-immunoprecipitation with lamin A/C antibody and probed with lamin A/C and GFP antibodies. (G) BAF SUMOylation is important for its interaction with lamin A/C. HEK293 cells expressing GFP-BAF or GFP-BAF-K6R were co-immunoprecipitated with the GFP antibody and analyzed by Western blot analysis using lamin A/C and GFP antibodies. (H) The interaction between BAF and SIM-deficient mutants of lamin A is reduced. HEK293 cell lysates coexpressing Myc-BAF with GFP-lamin A or its SIM mutants were co-immunoprecipitated with GFP-trap beads and analyzed by Western blot analysis using GFP and Myc antibodies. SIM (EE) mutants were obtained by substitution of two hydrophobic amino acids with glutamic acid residues in each respective SIM. SIM1(EE): 226EEEI229; SIM2(EE):362EEDI365; SIM3(EE): 494EETI497; SIM4(EE): 547LTEE550. (I) SUMOylation of BAF does not affect its binding to LEM domain proteins. HEK293 cell lysates individually expressing GFP-BAF and GFP-BAF-K6R were used for co-immunoprecipitation with GFP antibody and were probed with emerin, LAP2α, and GFP antibodies. DNA was stained with DAPI. (Scale bars: 10 μm.)
We next investigated the molecular basis for nuclear localization of BAF after SUMOylation. BAF interacts with many important binding partners inside the nucleus, including lamin A/C and LEM domain proteins. We first knocked down lamin A/C in cells, and found that this almost totally abolished the nuclear localization of BAF (Fig. 2C). Treating these cells with leptomycin B, a protein nuclear export inhibitor, or MG132, a proteasome inhibitor, did not rescue the nuclear localization failure (SI Appendix, Fig. S2 A and B), suggesting that nuclear disappearance of BAF in lamin A/C knockdown cells was not due to nuclear export or protein degradation. On the other hand, we found that BAF knockdown did not affect the nuclear pool of lamin A/C (Fig. 2C). Specifically, for endogenous BAF observation, 4% paraformaldehyde was used for fixation at room temperature, followed by Triton X-100 permeation, different from cold methanol fixation for tagged-BAF observation, since endogenous BAF was difficult to observe under methanol fixation. Next, we generated two GFP-tagged lamin A mutants—C661S, a nonfarnesylated form of prelamin A, and L530P, a permanently farnesylated prelamin A (33)—and coexpressed them individually with mCherry-tagged BAF in cells. We observed that both lamin A mutants localized to the nucleus with distinct patterns, and that, interestingly, BAF colocalized with both, as reported previously (Fig. 2D) (34). Furthermore, we found that both of the lamin A mutants interacted with BAF (Fig. 2E). Importantly, through reciprocal immunoprecipitation assays using cell lysates expressing BAF-WT and BAF-K6R, we found that the binding of BAF-K6R to lamin A/C was severely reduced (Fig. 2 F and G and SI Appendix, Fig. S2 C and D). These data suggest that the interaction between lamin A/C and BAF is dependent on BAF K6 SUMOylation. To confirm this, we generated a panel of GFP-tagged SIM-defective lamin A mutants, SIM1(EE) to SIM4(EE) (35), and individually coexpressed them with Myc-BAF in cells. Interactions between BAF and the SIM-deficient lamin A mutants were decreased compared with WT lamin A (Fig. 2H). These data indicate that the interaction between lamin A/C and BAF is SUMOylation-dependent. In contrast, we found that BAF SUMOylation did not affect its interaction with the LEM proteins, emerin, LAP2α, and LAP2β (Fig. 2I and SI Appendix, Fig. S2 E and F) or its dimerization status (SI Appendix, Fig. S2G). Thus, we conclude that the interaction between BAF and lamin A/C occurs through the SUMO molecule on BAF and the SIM domain on lamin A/C, and that this interaction results in the retention of BAF in the nucleus.
SUMOylation of BAF Also Regulates Its Binding to Nuclear DNA and Proliferating Cell Nuclear Antigen in S Phase.
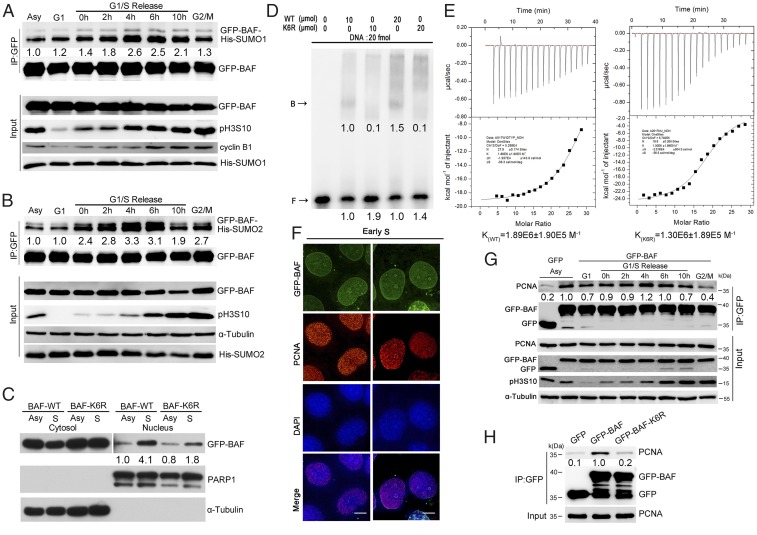
Given the importance of BAF for cell cycle regulation, we investigated its SUMOylation kinetics during the cell cycle. We found that SUMOylation of BAF by SUMO proteins occurred mainly in S phase (Fig. 3 A and B), as demonstrated by coexpressing GFP-BAF with His-SUMO1 or His-SUMO2. Since SUMOylation of BAF is required for its nuclear accumulation, we examined the relationship between SUMOylation and nuclear accumulation of BAF in S phase. When GFP-BAF or GFP-BAF-K6R was stably expressed in cells, GFP-BAF was significantly enriched in S phase nuclear fraction, as expected, but GFP-BAF-K6R did not efficiently accumulate in S phase nucleus (Fig. 3C).
Fig. 3.
SUMOylation of BAF also regulates its binding to nuclear DNA and PCNA in S phase. (A) SUMOylation of BAF by SUMO1 peaks in S phase. HEK293 cells coexpressing GFP-BAF and His-SUMO1 were synchronized at G1, G1/S transition, S, G2, and G2/M phases. Cells were collected for immunoprecipitation using GFP-trap beads and analyzed by Western blot analysis using the indicated antibodies. (B) SUMOylation of BAF by SUMO2 also peaks in S phase. HEK293 cells coexpressing GFP-BAF and His-SUMO2 were synchronized at different phases, collected for immunoprecipitation using GFP-trap beads, and analyzed by Western blot analysis using the indicated antibodies. (C) BAF SUMOylation promotes its nuclear accumulation in S phase. Asynchronous and S-phase–arrested HEK293 cells stably expressing GFP-BAF or GFP-BAF-K6R were subjected to subcellular fractionation. Protein levels of GFP-BAF in the nuclear and cytoplasmic fractions were determined by immunoblotting with an anti-GFP antibody. The relative purity of the nuclear and cytoplasmic fractions was confirmed by sequential probing for the nuclear marker PARP1 and the cytoplasmic marker α-tubulin, respectively. (D) SUMOylation is required for BAF binding to dsDNA. EMSA results of BAF-WT or BAF-K6R protein binding to a 5′-biotin–labeled dsDNA probe are shown. B represents the BAF-bound DNA, and F represents the free DNA. (E) The binding of BAF-WT and BAF-K6R proteins to dsDNA was measured by ITC, with association equilibrium constants (KA) as indicated. The association equilibrium constant of BAF-WT protein with dsDNA (KWT) is 1.89 ± 0.19 μM, whereas the association equilibrium constant of BAF-K6R protein with dsDNA (KK6R) is 1.30 ± 0.189 μM. (F) BAF colocalizes with PCNA during S phase. HeLa cells expressing GFP-BAF were synchronized at the early S phase by releasing the G1/S-arrested cells for 2 h and then stained with PCNA antibody and DAPI. (Scale bars: 10 μm.) (G) BAF interacts with PCNA during S phase. HEK293 cells expressing GFP-BAF synchronized at different phases were subjected to immunoprecipitation using GFP-trap beads and Western blot analysis using the indicated antibodies. (H) The K6R mutation reduces the binding of BAF to PCNA. HEK293 cells expressing GFP-BAF or GFP-BAF-K6R were co-immunoprecipitated with GFP-trap beads and analyzed by Western blot analysis using PCNA and GFP antibodies.
We next investigated the effects of SUMOylation on BAF binding to nuclear DNA. First, we performed an in vitro protein-DNA binding assay using purified BAF proteins from HEK293 cells and dsDNA, followed by an electrophoretic mobility shift assay (EMSA). We found that BAF-K6R bound significantly less dsDNA compared with WT BAF (Fig. 3D). Using the same proteins and DNA, we confirmed the foregoing results by isothermal titration calorimetry (ITC) (Fig. 3E). Nuclear localization of BAF has been reported to correlate with S phase progression (36); thus, we investigated the nuclear localization of BAF during the cell cycle via GFP-BAF expression in cells, followed by immunofluorescence labeling. The results not only confirmed the nuclear localization of BAF, but also demonstrated that GFP-BAF was oriented at the nuclear foci, bound and colocalized with proliferating cell nuclear antigen (PCNA) in S phase (Fig. 3 F and G).
PCNA is a cofactor of DNA polymerases that encircles DNA, which recruits crucial players to the replication fork and marks sites of DNA synthesis during S phase (37–39). A previous study showed that localization of PCNA has cell cycle-dependent properties (37). In G1 and G2 phases, it equally distributes throughout the nucleus. In early S phase, it agglomerates to small and equally distributed foci, which are located at the nuclear periphery in mid S phase. In late S phase, PCNA forms large foci near the center of the nuclei. When GFP-BAF or GFP-BAF-K6R were expressed followed by co-IP, we found that endogenous PCNA co-immunoprecipitated with GFP-BAF, but not with GFP-BAF-K6R (Fig. 3H). These data indicate that BAF interacts with PCNA under the regulation of SUMOylation. Taken together, these results show that SUMOylation-regulated nuclear accumulation of BAF in S phase enhances its binding to dsDNA and PCNA.
SENP1 and SENP2 Are Responsible for de-SUMOylation of BAF at K6.
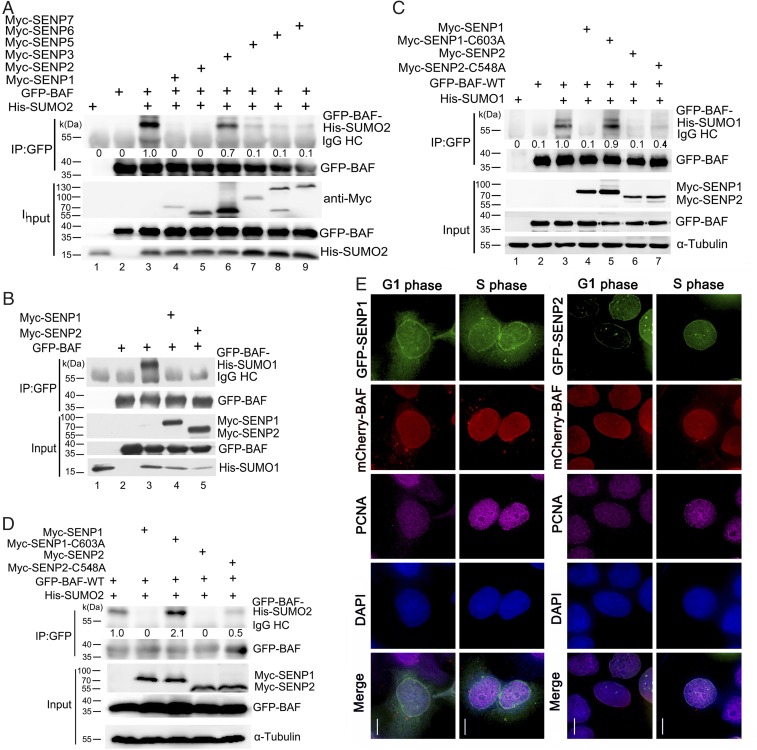
SUMOylation of proteins is a reversible process, and the SUMOylation/de-SUMOylation cycle is highly dynamic. The regulated deconjugation of SUMO from its substrates ensures the plasticity of protein interaction networks (18). Deconjugation of SUMO is catalyzed by cysteine proteases, termed SUMO isopeptidases, and the sentrin-specific protease (SENP) family of isopeptidases is responsible for most deconjugations of SUMO molecules from their target proteins (22). To uncover the isopeptidases responsible for the de-SUMOylation of BAF, we generated Myc-tagged constructs for all six human SENPs: Myc-SENP1, Myc-SENP2, Myc-SENP3, Myc-SENP5, Myc-SENP6, and Myc-SENP7. These six SENPs were individually coexpressed with both His-SUMO2 and GFP-BAF in cells, followed by detection of de-SUMOylation. We found that while the SUMOylated band of GFP-BAF was significantly increased in cells expressing both GFP-BAF and His-SUMO2 (Fig. 4A), it was reduced or absent in the cells coexpressing SENP1 or SENP2 (Fig. 4A). In contrast, coexpressing GFP-BAF and His-SUMO2 with SENP3, SENP5, SENP6, or SENP7 did not significantly reverse the SUMO2 modification, although a reduced deconjugation effect was identified (Fig. 4A). Strikingly, when GFP-BAF and His-SUMO1 were coexpressed with SENP1 and SENP2, both SENPs together efficiently deconjugated SUMO1 from GFP-BAF (Fig. 4B). We also generated two catalytically inactive mutants, SENP1 C603A and SENP2 C548A (40, 41). The mutant and WT SENPs were individually coexpressed with GFP-BAF and His-SUMO1 in cells. We found that both mutants failed to deconjugate SUMO from BAF (Fig. 4 C and D).
Fig. 4.
SENP1 and SENP2 are responsible for BAF de-SUMOylation. (A) The deconjugation efficiencies of SUMO2 conjugated-BAF by different SENP family members are different. HEK293 cells expressing GFP-BAF and His-SUMO2 were cotransfected with various Myc-tagged SENP members. Whole-cell extracts were co-immunoprecipitated with a GFP antibody and probed with His and GFP antibodies. Note that while SENP1 and SENP2 completely reversed the SUMO2 modification of BAF, SENP3, SENP5, SENP6, and SENP7 reduced this SUMO2 modification only slightly. (B) Both SENP1 and SENP2 efficiently deconjugate the SUMO1 modification of BAF. HEK293 cells expressing GFP-BAF and His-SUMO1 were cotransfected with Myc-SENP1 or Myc-SENP2. Whole-cell extracts were co-immunoprecipitated with GFP antibody and probed with His and GFP antibodies. (C) The catalytically inactive SENP1-C603A and SENP2-C548A mutants are unable to deconjugate the SUMO1 modification of BAF. HEK293 cells expressing GFP-BAF and His-SUMO1 were transfected with Myc-SENP1, Myc-SENP1-C603A, Myc-SENP2, or Myc-SENP2-C548A. Whole-cell extracts were immunoprecipitated with the GFP antibody and probed with His and GFP antibodies. (D) Catalytically inactive SENP1-C603A and SENP2-C548A are unable to deconjugate the SUMO2 modification of BAF. HEK293 cells coexpressing GFP-BAF and His-SUMO2 were cotransfected with Myc-SENP1, Myc-SENP1-C603A, Myc-SENP2, or Myc-SENP2-C548A. Whole-cell extracts were immunoprecipitated with GFP antibody and probed with His and GFP antibodies. (E) SENP1 and SENP2 colocalize with BAF. HeLa cells coexpressing GFP-SENP1 or GFP-SENP2 and mCherry-BAF were fixed and stained with a PCNA antibody and DAPI. (Scale bars: 10 μm.)
SENP1 and SENP2 localize to the nucleoplasm and concentrate at the NE through their interaction with components of the nuclear pore complex (42–45). Here we not only confirmed their nuclear localization patterns, but also found that both SENP1 and SENP2 colocalized with BAF in interphase (Fig. 4E and SI Appendix, Fig. S3A). We also observed that SENP3 and SENP5 were localized mainly in the nucleolus, as reported previously (46–48), and did not colocalize with BAF (SI Appendix, Fig. S3 B–E). SENP6 and SENP7 have been reported to localize to the nucleoplasm (22, 45, 49, 50). Here we confirmed this localization and found small fractions of SENP6 and SENP7 concentrated at the NE (SI Appendix, Fig. S3 F–I). Taken together, these results demonstrate that SENP1 and SENP2 colocalize with BAF at the nucleoplasm and NE and catalyze de-SUMOylation of SUMOylated BAF.
The K6 SUMOylation and de-SUMOylation Cycle of BAF Is Essential for Nuclear Integrity and DNA Replication.
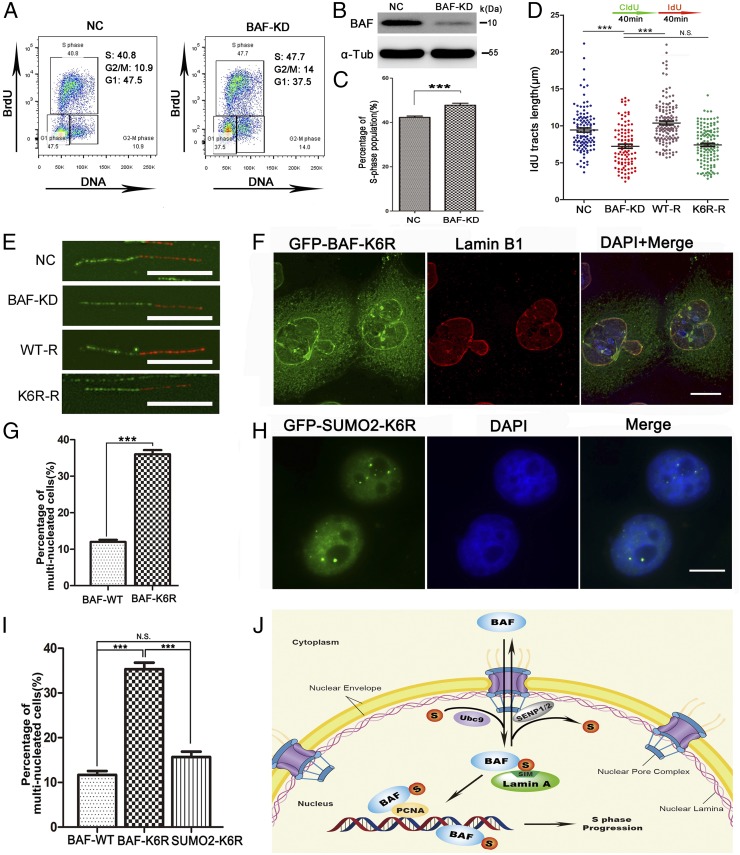
Finally, we investigated the function of the SUMOylation/de-SUMOylation cycle of BAF at K6 in cells. We first observed a significant S phase retention (∼16.9%) in BAF knockdown cells (Fig. 5 A–C). We then performed a DNA fiber assay to investigate the replication fork dynamics in control and BAF-depleted cells. Cells were transfected with control or BAF shRNA vectors, followed by pulse-labeling with chlorodeoxyuridine (CldU) for 40 min and then incubation with iododeoxyuridine (IdU) for 40 min. We examined the replication rate under normal conditions by measuring IdU tract lengths. We found that BAF-depleted cells exhibited significantly decreased average tract lengths (on average) compared with control cells (7.24 μm vs. 9.44 μm) (Fig. 5 D and E), indicating that BAF is required for DNA replication. When the IdU tract length was converted to kilobases using a common conversion factor, the DNA fibers were 2.59 kb/μm using the spreading methods (Materials and Methods) (51, 52). The replication rate was ∼0.61 kb/min in control cells and 0.46 kb/min in BAF-depleted cells, an ∼26% slowdown. Importantly, this DNA replication delay could be efficiently rescued by expression of BAF-WT, but not by expression of BAF-K6R (Fig. 5 D and E). Collectively, these results indicate that SUMOylation of BAF is directly involved in DNA replication.
Fig. 5.
A proper K6 SUMOylation and de-SUMOylation cycle of BAF is essential for nuclear integrity maintenance and DNA replication. (A) Down-regulation of BAF delays S phase progression. HEK293 cells were transfected with control shRNA vector (NC) or BAF shRNA vector (BAF-KD) and stained with anti-BrdU antibody and DAPI, and the cell cycle distribution was determined by flow cytometry analysis. (B) Expression levels of BAF in cells with control shRNA vector (NC) or BAF shRNA vector (BAF-KD) in A were analyzed by Western blot analysis. (C) Quantitation of BrdU-positive S phase cells in control (NC) or BAF knockdown (BAF-KD) HEK293 cells. Six independent experiments were conducted. (D) BAF SUMOylation is required for normal DNA replication. HEK293 cells were transfected with control shRNA vector (NC), BAF shRNA vector (BAF-KD), and BAF shRNA vector together with GFP-BAF-WT resistant to BAF shRNA (WT-R) or BAF shRNA together with GFP-BAF-K6R resistant to BAF shRNA (K6R-R). Cells were pulse-labeled as outlined at the top of the panel. DNA fibers were stained with antibodies recognizing IdU (red) and CldU (green). The lengths of the IdU tracts were measured and presented. Approximately 100 fibers were counted per sample. (E) Representative images of DNA fibers in control (NC), BAF knockdown (BAF-KD), BAF-WT rescue (WT-R), or BAF-K6R rescue (K6R-R) cells in D. (F) Overexpression of GFP-BAF-K6R increases abundance of the distorted nuclei and multinucleated cells. (G) Quantitation of multinucleated cells in GFP-BAF-WT and GFP-BAF-K6R-overexpressing HeLa cells is shown. Approximately 200 cells were counted per sample, and three independent experiments were conducted. (H) Conjugation of SUMO2 to the N terminus of BAF-K6R rescued the nuclear accumulation failure and the nuclear structure of BAF-K6R. (I) Quantitation of multinucleated HeLa cells overexpressing GFP-BAF-WT, GFP-BAF-K6R, or GFP-SUMO2-K6R. Approximately 200 cells were counted per sample and three independent experiments were conducted. The statistical data in G and I are presented as mean ± SD ***P < 0.001; N.S., no significant difference (Student’s t test). DNA was stained with DAPI. (Scale bars: 10 μm.) (J) A working model. BAF is a small protein that shuttles between the cytoplasm and the nucleus through the nuclear pore complexes in interphase cells. BAF is SUMOylated mainly at K6, and this modification promotes its nuclear localization by enhancing its binding to the SIM motifs of lamin A/C to physically tether chromatin to the nuclear lamina and/or other nuclear structures to maintain nuclear integrity. Both the SUMOylation level and the nuclear accumulation of BAF peak in S phase to promote localization to the DNA replication sites and interaction with PCNA and dsDNA for the regulation of DNA replication and S phase progression. De-SUMOylation of BAF is catalyzed by SENP1 and SENP2, which facilitates its nuclear export through the nuclear pore complexes.
We investigated the effects of the SUMOylation/de-SUMOylation cycle of BAF at K6 on nuclear integrity. As shown in SI Appendix, Fig. S4A, BAF-K6R–overexpressing cells exhibited two patterns. When the expression level of BAF-K6R was low, BAF-K6R showed similar localization as BAF-WT, without nuclear abnormalities; however, when the expression level of BAF-K6R was high, BAF-K6R mutant showed a dramatic loss of nuclear accumulation and localized mainly to the cytoplasm. In contrast, BAF-WT–overexpressing cells exhibited only one pattern (Fig. 2A). When the mutant (GFP-BAF-K6R) but not the WT (GFP-BAF) was overexpressed, the cells showed high percentages of distorted nuclei and multinuclei (Fig. 5 F and G). These results suggest that it was the loss of nuclear BAF in BAF-K6R–highly overexpressing cells that led to the nuclear abnormality. When GFP-SUMO2-K6R was expressed, the fusion protein localized predominantly to the nucleus and efficiently rescued the nuclear structure, similar to GFP-BAF-WT (Fig. 5 H and I).
To examine the effects of BAF SUMOylation in other cell types, we transfected human osteosarcoma U2OS cells and monkey kidney COS7 cells with GFP-BAF-WT, GFP-BAF-K6R, or GFP-SUMO2-K6R and performed immunofluorescence assays. We found that BAF-K6R expression induced a high percentage of distorted nuclei and multinuclei, similar to HeLa cells. SUMO2-K6R significantly rescued the nuclear structure, indicating that the effect of K6R on nuclear integrity is not limited to HeLa cells, and that BAF SUMOylation at K6 has a general function in other mammalian cell lines (SI Appendix, Fig. S4 B–E). Thus, we conclude that SUMOylation is essential for nuclear localization of BAF, and that this localization regulates nuclear integrity.
Collectively, these results demonstrate that the SUMOylation/de-SUMOylation cycle of BAF ensures its timely interaction with lamin A/C and DNA, thus contributing to its nuclear distribution and function. When this SUMOylation/de-SUMOylation cycle is disrupted, BAF tends to lose its nuclear accumulation and function.
Discussion
The nuclear periphery forms a selective barrier between the nucleus and cytoplasm and serves as a structural domain supporting nuclear organization and function. Numerous nuclear periphery proteins are involved in the regulation of nuclear integrity, NE dynamics, genome stability, and various cellular biological processes. BAF, a small inner nuclear membrane-associated protein, has multiple roles during the cell cycle. In this work, we found that SUMOylated BAF regulates nuclear integrity and DNA replication.
Posttranslational modifications of proteins play vital roles in the functional regulation of target proteins in multiple pathways (27, 53). Here we found that reversible SUMOylation of BAF at K6 plays a pivotal role in its functional regulation. Since lamin A/C is indispensable for BAF nuclear localization regulated by SUMOylation, the disruption of nuclear integrity caused by disturbing BAF SUMOylation may be due to the nuclear structural function of lamin A/C, which needs the assistance of SUMOylated BAF. SUMOylation studies have shown that SUMO frequently modifies entire functional groups of proteins (27). In the present case, SUMOylated BAF and lamin A/C might act as a complex to maintain nuclear integrity.
The mechanism by which BAF contributes to DNA replication is unclear, although we have demonstrated that BAF localizes to DNA replication sites and interacts with PCNA during S phase. Furthermore, the down-regulation of BAF impedes DNA replication. Since expression of the non-SUMOylatable mutant BAF-K6R distorts the nuclear structure, BAF likely influences chromatin remodeling; lack of SUMOylation disrupts the interaction of BAF with PCNA and dsDNA during DNA replication. SUMOylation of BAF might contribute to DNA replication at multiple levels, including structural support and direct participation in the DNA replication process. In addition, SUMOylation of BAF may influence the regulation of gene expression in the nucleus.
Overall, this study reveals previously unanticipated roles of SUMOylation in modulating BAF functions in interphase. Based on our present data, we propose a working model for the functions of SUMOylation and de-SUMOylation of BAF at K6 (Fig. 5J). BAF is a small protein that shuttles between the cytoplasm and nucleus through nuclear pore complexes. SUMOylation may regulate BAF subcellular localization and function. In interphase, BAF is SUMOylated mainly at K6. This modification promotes SUMOylated BAF binding to lamin A/C through the SIM domains of lamins, maintaining the nuclear localization and function of BAF in the maintenance of nuclear integrity. SUMOylation of BAF also regulates its binding to DNA and DNA replication regulators, which is important for DNA replication. Disrupting SUMOylation of BAF not only disturbs nuclear integrity, but also induces DNA replication failure. SENP1 and SENP2 catalyze the de-SUMOylation of BAF and may induce its nuclear export.
In conclusion, our findings shed light on the crucial roles of BAF in the maintenance of nuclear integrity, nuclear dynamics, and the cell cycle. These findings may have important implications for understanding how SUMOylation and de-SUMOylation regulate functions of the nuclear periphery proteins.
Materials and Methods
Human BAF was cloned from a HeLa cell cDNA library, by RT-PCR and inserted into related vectors. To obtain recombinant Flag-tagged BAF-WT and BAF-K6R proteins, BAF-WT and BAF-K6R were cloned into the pCAG vector. SUMO1 was cloned from a cDNA library by RT-PCR and inserted into PEF1-HisB. More FACS data for identifying cell cycle phases in Fig. 3 A–C and G are provided in SI Appendix, Figs. S5 and S6, respectively.
Detailed information on cell culture, cell cycle synchronization, plasmids and antibodies, plasmid DNA transfection, RNA interference, viral transduction, protein purification, immunofluorescence, subcellular protein fractionation, co-IP, GST fusion protein pull-down assays, Ni-NTA pull-down assays, DNA fiber assays, electrophoretic mobility shift assays, and ITC is provided in SI Appendix, Materials and Methods.
Data Availability Statement.
All pertinent data are provided in the main text and SI Appendix. A list of the reagents included in this study is available on request from the corresponding author.
Supplementary Material
Acknowledgments
We thank Dr. Jing Yi (Shanghai Jiao Tong University) and Dr. Li Yu (Tsinghua University) for providing reagents and other members of our laboratory for valuable comments. We also thank our colleagues Drs. Hongxia Lu, Liying Du, Dong Liu, Hui Li, Xiaochen Li, and Guilan Li at the National Center for Protein Science at Peking University for assistance with microscopic imaging, mass spectrometry, flow cytometry and protein preparation and identification. B.Y. is a visiting student from Shanghai Jiao Tong University School of Medicine. This work was supported by grants from the Ministry of Science and Technology of China and the National Natural Science Foundation of China (31520103906, 2016YFA0500201, 2016YFA0100501, and 31430051).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912984117/-/DCSupplemental.
References
- 1.Worman H. J., Bonne G., “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 313, 2121–2133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capell B. C., Collins F. S., Human laminopathies: Nuclei gone genetically awry. Nat. Rev. Genet. 7, 940–952 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Bione S., et al. , Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8, 323–327 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Bonne G., et al. , Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21, 285–288 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Butin-Israeli V., Adam S. A., Goldman A. E., Goldman R. D., Nuclear lamin functions and disease. Trends Genet. 28, 464–471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber K. H., Kennedy B. K., When lamins go bad: Nuclear structure and disease. Cell 152, 1365–1375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puente X. S., et al. , Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 88, 650–656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamin A., Wiebe M. S., Barrier to autointegration factor (BANF1): Interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol. 34, 61–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa K., et al. , Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116, 3811–3823 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Segura-Totten M., Kowalski A. K., Craigie R., Wilson K. L., Barrier-to-autointegration factor: Major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158, 475–485 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai M., et al. , Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat. Struct. Biol. 5, 903–909 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Umland T. C., Wei S. Q., Craigie R., Davies D. R., Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39, 9130–9138 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Bradley C. M., Ronning D. R., Ghirlando R., Craigie R., Dyda F., Structural basis for DNA bridging by barrier-to-autointegration factor. Nat. Struct. Mol. Biol. 12, 935–936 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Margalit A., Brachner A., Gotzmann J., Foisner R., Gruenbaum Y., Barrier-to-autointegration factor—A BAFfling little protein. Trends Cell Biol. 17, 202–208 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Molitor T. P., Traktman P., Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol. Biol. Cell 25, 891–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haraguchi T., et al. , Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121, 2540–2554 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Samwer M., et al. , DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170, 956–972.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flotho A., Melchior F., Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Geiss-Friedlander R., Melchior F., Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Seeler J. S., Dejean A., SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Wang L., et al. , SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 15, 878–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz K., Piller T., Müller S., SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 131, jcs21190 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Wan J., Subramonian D., Zhang X. D., SUMOylation in control of accurate chromosome segregation during mitosis. Curr. Protein Pept. Sci. 13, 467–481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watts F. Z., The role of SUMO in chromosome segregation. Chromosoma 116, 15–20 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Ban R., Nishida T., Urano T., Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells 16, 652–669 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Miranda G., et al. , SUMOylation modulates the function of Aurora-B kinase. J. Cell Sci. 123, 2823–2833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eifler K., Vertegaal A. C. O., SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimi T., et al. , Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 147, 31–41 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Halfmann C. T., et al. , Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol. 218, 2136–2149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmstrom S., Van Antwerp M. E., Iñiguez-Lluhi J. A., Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. U.S.A. 100, 15758–15763 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross S., Best J. L., Zon L. I., Gill G., SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10, 831–842 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wen D., Wu J., Wang L., Fu Z., SUMOylation promotes nuclear import and stabilization of polo-like kinase 1 to support its mitotic function. Cell Rep. 21, 2147–2159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mounkes L. C., Kozlov S., Hernandez L., Sullivan T., Stewart C. L., A progeroid syndrome in mice is caused by defects in A-type lamins. Nature 423, 298–301 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Capanni C., et al. , Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle 9, 2600–2610 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Moriuchi T., Kuroda M., Kusumoto F., Osumi T., Hirose F., Lamin A reassembly at the end of mitosis is regulated by its SUMO-interacting motif. Exp. Cell Res. 342, 83–94 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Haraguchi T., et al. , Nuclear localization of barrier-to-autointegration factor is correlated with progression of S phase in human cells. J. Cell Sci. 120, 1967–1977 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Schönenberger F., Deutzmann A., Ferrando-May E., Merhof D., Discrimination of cell cycle phases in PCNA-immunolabeled cells. BMC Bioinformatics 16, 180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moldovan G. L., Pfander B., Jentsch S., PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Mailand N., Gibbs-Seymour I., Bekker-Jensen S., Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Bawa-Khalfe T., Cheng J., Lin S. H., Ittmann M. M., Yeh E. T., SENP1 induces prostatic intraepithelial neoplasia through multiple mechanisms. J. Biol. Chem. 285, 25859–25866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garvin A. J., et al. , The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev. 33, 333–347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow K. H., Elgort S., Dasso M., Ullman K. S., Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus 3, 349–358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goeres J., et al. , The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol. Biol. Cell 22, 4868–4882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hang J., Dasso M., Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Nayak A., Müller S., SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15, 422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong L., Yeh E. T., Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281, 15869–15877 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Haindl M., Harasim T., Eick D., Muller S., The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun C., et al. , Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183, 589–595 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maison C., et al. , The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat. Struct. Mol. Biol. 19, 458–460 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Shen L. N., Geoffroy M. C., Jaffray E. G., Hay R. T., Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem. J. 421, 223–230 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Daigaku Y., Davies A. A., Ulrich H. D., Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465, 951–955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson D. A., Pombo A., Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z., Lu W., Roles of ubiquitination and SUMOylation on prostate cancer: Mechanisms and clinical implications. Int. J. Mol. Sci. 16, 4560–4580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All pertinent data are provided in the main text and SI Appendix. A list of the reagents included in this study is available on request from the corresponding author.