Abstract
Kidney diseases form part of the major health burdens experienced all over the world. Kidney diseases are linked to high economic burden, deaths, and morbidity rates. The great importance of collecting a large quantity of health-related data among human cohorts, what scholars refer to as “big data”, has increasingly been identified, with the establishment of a large group of cohorts and the usage of electronic health records (EHRs) in nephrology and transplantation. These data are valuable, and can potentially be utilized by researchers to advance knowledge in the field. Furthermore, progress in big data is stimulating the flourishing of artificial intelligence (AI), which is an excellent tool for handling, and subsequently processing, a great amount of data and may be applied to highlight more information on the effectiveness of medicine in kidney-related complications for the purpose of more precise phenotype and outcome prediction. In this article, we discuss the advances and challenges in big data, the use of EHRs and AI, with great emphasis on the usage of nephrology and transplantation.
Keywords: artificial intelligence, machine learning, big data, nephrology, transplantation, kidney transplantation, acute kidney injury, chronic kidney disease
1. Introduction
Kidney diseases, such as acute kidney injury (AKI) and chronic kidney disease (CKD) are major medical and public health issues worldwide, associated with high death and morbidity rates, together with great economic loss [1,2,3,4,5,6]. CKD is linked with a higher danger of argumentative outcomes, like cardiovascular complications, death, decreased quality of life, and substantial healthcare resource utilization [7,8,9,10,11], and it has been assessed that around 850 million individuals suffer different types of kidney diseases globally [12,13]. If left untreated, CKD may evolve into end-stage kidney disease (ESKD), which is associated with high mortality [14,15,16]. It is well-known that kidney diseases are very much multifactorial, with overlapping and complex clinical phenotypes, as well as morphologies [17]. The global distribution of nephrologists usually differs from one country to another, with bigger differences in its overall capacity [18]. Various nations across the world have established surveillance systems for kidney-related infections. Despite such attempts, the literature highlights that surveillance systems within third world countries are still not very strong [19]. In certain areas of some countries, basic records offices for transplantation and dialysis, as well as expert pathologists, are not even available [18,20]. Given how major gaps are always found in the main workforce in nephrology, the current eminence of kidney health management and research evidence in nephrology needs to be strengthened globally [21].
Traditionally, the randomized controlled trial (RCT) has always been used as the point of reference for offering evidence-based treatment. The numerical formulae applied in analyzing randomized control data have equally offered essential insights from numerous observational data. In the past few years, great emphasis has been placed on the pragmatic RCT, an essential component of real global research, which is applied when evaluating the great interventions within the actual clinical setting based on a great amount of samples so as to stimulate their individual practical value. A great amount of differences have been reported within nephrology, as well as some other relevant specialties. For instance, the literature indicates that nephrology trials were very limited in number and possessed minimally optimal features of high-quality designs [22]. Despite the fact that the already existing studies, as well as implemented works, have made major additions to a highly reliable prognostication, as well as an extensive understanding of the general histologic pathology, there is still a great amount of work which needs to be undertaken, as well as specific problems to be solved. The general capacity for undertaking cohort studies that involve a large sample size or Rapid Control trial is very much present across various parts of the globe, and has thereby resulted in the absence of research evidence within nephrology. In addition, limited activity in kidney research has impacted the evidence base for the treatment of kidney diseases, resulting in a lack of useful surrogate end-points for progression from the early stages of kidney disease-hindered trials [14,15]. On the same note, a great amount of cohort data could also be applied in generating relevant hypotheses and provide major insights into the etiology, pathogenesis, and prognosis of kidney diseases [23,24].
Those needs that are classified as unmet require provision of some ample spaces for the purpose of imagination in relation to leveraging the strength associated with big data, as well as relevant artificial intelligence (AI) to improve the overall status of patients with kidney diseases [25]. In this article, we discuss the big data concepts in nephrology, describe the potential use of AI in nephrology and transplantation, and also encourage researchers and clinicians to submit their invaluable research, including original clinical research studies [26,27,28,29,30], database studies from registries [31,32,33], meta-analyses [34,35,36,37,38,39,40,41,42,43,44], and artificial intelligence research [25,45,46,47,48] in nephrology and transplantation.
2. Big Data in Nephrology and Transplantation: Registries and Administrative Claims
Table 1 demonstrates known and commonly used databases that have provided big data in nephrology and transplantation [49,50,51]. For example, the United States Renal Data System (USRDS) is recognized as a state reconnaissance system that has the responsibility of collecting, analyzing, and subsequently distributing information regarding CKD and ESKD, all based on numerous big datasets. By delivering the yearly data report, the USRDS continuously tracks both the epidemiologic and economic burden linked to kidney diseases [52]. An important database in transplantation in the United States is the United Network for Organ Sharing (UNOS). The Organ Procurement and Transplantation Network (OPTN) data are linked by UNOS to the Social Security Death Master File for the purpose of augmenting ascertainment of different groups of candidates, as well as relevant deaths. The final data are attainable by different groups of researchers, and have always been applied in various studies regarding transplantation [50]. In addition to these databases in the United States, other countries worldwide also have big data within nephrology for researchers, such as the National Kidney Disease Surveillance Program in Ireland [53], the surveillance project on CKD management in Canada [54], and the China Kidney Disease Network (CK-NET), a comprehensive CKD surveillance system for China [55].
Table 1.
Nephrology and transplant databases and organizations.
| Renal and Transplant Databases | Organizations |
|---|---|
|
|
Numerous networks of international collaboration, like the International Network of CKD cohorts [56], the Therapeutic Evaluation of Steroids in IgA Nephropathy Global study [57], and the Chronic Kidney Disease Prognosis Consortium [58] have grown immensely within the last few years. There are possible advantages of introducing a traditional data element that are linked to kidney infections, like escalating the overall power of the groups which are under-represented [59]. There is, however, great need to address numerous challenges, like standardization of data, identification of the patient, plus some other additional infrastructure-related challenges. Additionally, the cadre of genomics is developing at a very rapid rate towards realizing an analysis of single cells, and subsequent great advances within metabolomics and proteomics have been developed within the past few years. A great amount of progress has equally been realized within technological developments within the areas of large-scale molecular data generation in various databases that are gene-based (Table 1). The most recent advancements in technology have made it possible for us to produce larger amounts of data, more specifically regarding the omics data. Further development of somehow less expensive genotype arrays and the subsequent presence of samples within biobanks made it possible to undertake genome-wide association studies among numerous groups of patients, offering highly essential insights into the great risk factors and the pathogenesis of multiple kidney diseases [60,61,62,63].
Within nephrology, numerous consortia-collecting biopsy biobanks of kidney tissue have been started to undertake such forms of collaborative study. Several initiatives that are aimed at extensive characterization of the relevant kidney biopsies for various groups of kidney diseases subtypes have subsequently been launched, comprising of the NEPTUNE (Nephrotic Syndrome STudy Network), ERCB (European Renal cDNA Bank), EURenOmics, C-PROBE (Clinical Phenotyping and Resource Biobank), PKU-IgAN, and more recently, TRIDENT (for diabetic nephropathy), CureGN (for glomerulopathies), and the NIDDK (National Institute of Diabetes and Digestive and Kidney Diseases) Kidney Precision Medicine Project (KPMP) [64].
Big data within the medicine field might offer the opportunity to envision patients suffering from kidney diseases in a more holistic manner, using numerous lenses, each of which adequately presents the great opportunity of studying various scientific queries. Such data within the big databases might subsequently comprise of the general administrative health-related data, biometric data, biomarker data, as well as imaging, and might subsequently come from various sources, comprising of electronic health records, biobanks, reports in the internet, and various clinical registries [65]. These data from the large databases are collected and updated overtime. These data are valuable and can be used by researchers to answer numerous research questions and advance knowledge in nephrology and transplantation [66,67,68].
3. Using Electronic Health Record Data in Nephrology
Two major events have been reported within the last 10 years that seem to have changed the whole situation. To begin with is making it possible to digitalize all relevant medical information—more specifically, the initiation of EHRs that have the medical histories of the patients—and facilitate the processing of medical information using computers. This helps to make information-processing become automated by the use of given specialized software. EHRs have been greatly utilized with major regularity, clinical informatics strategies have subsequently been refined, and subsequently, the EHR field enabled [69,70].
The wide application of EHRs, when put together with the relevant novel of big data, tends to create some forms of unique opportunities for the purpose of nephrology research, as well as improvement in care for individual patients who might be suffering from kidney complications and transplantation. The data which is there within the EHR is considered big insofar as its volume is concerned. Such interventions have resulted in a new era of big data which has subsequently fueled precision medicine. These types of approach have already indicated an improved level of diagnosis, risk assessment, as well as treatment and management of numerous health conditions. With medicine getting digitalized, a great amount of data has since been developed from all aspects of health care, comprising the laboratory tests, EHR, together with medical imaging.
For instance, in the instance of electronic AKI, the automated diagnostic strategy tends to create a great opportunity to initiate predictive strategies, optimize the relevant AKI alerts, and subsequently trace AKI events across various institutions, as well as administrative datasets. The growth in the adoption of EHR and subsequent maturation of the relevant clinical informatics techniques might provide some sort of unique opportunity to advance the general predictive capabilities. Immediately, AKI has been properly diagnosed within real time, and several EHR-enabled interventions have become so viable. One of such great prospects is actually the prediction of detecting events prior to their occurrence [71]. AKI events might temporarily get anchored within the EHR, which develops a pre-disease phase of care, having the information which had accumulated before the development of AKI. With a greater amount of content, high-throughput strategies can be applied to such a group of data so as to help in identifying a form of pre-AKI signal, which can subsequently assist in discriminating between patients who are of high risk and low risk for the AKI. The capability to predict AKI risk in this manner might subsequently have some forms of dramatic impact, as presently there is no scientifically proven treatment for AKI once one develops such conditions [72]. As patients who are considered to be of high risk get identified, the extent of care can get modified, and further strategies for harm prevention implemented. In the long run, such groups of patients, institutions, and population-based techniques will result in better long- and short-term outcomes for the respective hospitals, patients, and the whole of the healthcare system. Despite the fact that potential barriers are always there, and several nuanced groups need to be taken into consideration, such approaches that are EHR-enabled have the ability to greatly improve AKI-associated knowledge and care.
Patients suffering from kidney complications are reported to have the highest level of heterogeneity in manifestation of the disease, treatment response, and overall progression. The growth in big data actually tends to stimulate the general boom of artificial intelligence that is a perfect tool that helps in handling, and also processing the big data. AI can assist in shedding light on the specific accuracy of medicines used to treat kidney diseases, for outcome prediction, and also to gain a more precise phenotype.
4. Artificial Intelligence in Nephrology and Transplantation
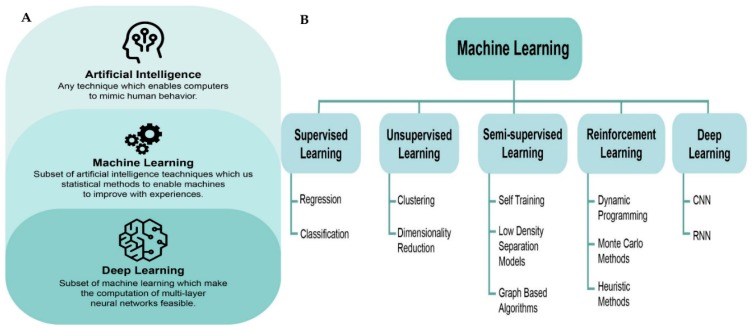
AI presently shows a very important function in nearly all areas of the day-to-day lives of human beings, as well as within different academic disciplines. Based on the fact that there has been growth in the power of computers, developments in techniques and methods, and the overall blast of the quantity of medicine, data has never been an exemption. Literature clarifies that artificial intelligence can be used in disease risk assessment. Actually, disease risk assessment has a very important influence on the general prognosis, as well as clinical intervention strategies. Accurate and rapid assessment can assist clinicians in determining the conditions of the patient, out of which optimal treatment strategies can be implemented. Links between prognosis and risk factors of the diseases are very complex. The same risk factors can be experienced within different groups of diseases, and a single disease can actually be composed of several risk factors. In such case, the links between the known risk factors and the disease has very strong correlativity, instead of simple causality. Artificial intelligence can hence be applied in doing disease risk assessment in order to understand the main factors linked to disease prognosis so as to offer effective treatment for tertiary prevention of the disease. One of the important sections of AI is machine learning, which is characterized as the study of algorithms and statistical models that computer systems utilize to learn from sample data and previous experience without being explicitly programmed to achieve particular assignments. With the ability to identify obscure patterns in the data, we can use machine learning to solve many problems, including assessing relationships of two variables, creating predictions based on baseline characteristics, identifying objects with comparable patterns, and incorporating subjects by specific criteria. Machine learning techniques have the capacities of managing complex datasets and tremendous numbers of variables that are exceeding the capability of classical statistical methods [17]—see Figure 1A. Machine learning algorithms are usually utilized without initiating as many presumptions of the underlying data. In addition, a machine-learning method can determine complex patterns of health trajectories of immense numbers of characteristics and patients, which has exhibited high predictive certainty, and been confirmed and replicated with various validation investigations [73]. Well-known machine-learning algorithms include the artificial neural network (ANN), random forest, gradient boosting trees, and support vector machine [17].
Figure 1.
(a) Relationships between artificial intelligence, machine learning, and deep learning. (b) Types of machine learning. CNN, convolutional neural network; RNN, recurrent neural network.
Inspired by the idea of mimicking the biological structure of human brains, deep learning is a subfield of machine learning based on ANN [74]. Deep-learning models can learn many levels of data design with a multiple-processing-layers model structure, attaining more powerful model performance. This cutting-edge technology has significantly changed the paradigm of visual object recognition, speech recognition, and many other domains, such as genomics and drug discovery. Deep learning techniques are increasingly being applied to biomedical data, from image processing to genomic data analysis [75]. Such methods might outperform pathologists’ fibrosis scores from histological renal biopsy images [76]. Well-known techniques include the convolutional neural network (CNN), fully connected neural network, generative adversarial network (GAN), deep reinforcement learning, and recurrent neural network (RNN) [17,77], shown in Figure 1B. AI-based clinical decision support systems (CDSS) can be implemented employing the expert system strategy, data-driven approach, or an ensemble approach by coupling both. An expert system consolidates a knowledge base containing a set of rules for specific clinical scenarios, and the initial rule set may be acquired from domain experts or learned from data through machine learning algorithms [72,78,79,80].
AI has recently been adopted for the prediction, diagnosis, and treatment of kidney diseases [76,81,82,83,84,85], as shown in Table 2. For example, a prediction model based on the combination of a machine learning algorithm and survival analysis has recently developed and can stratify risk for kidney disease progression among patients with IgA Nephropathy [86]. For AKI research, Tomasev et al. [83] recently used deep-learning methods to make a continuous prediction of AKI by developing a RNN model on the sequential health record data of >700,000 veterans, allowing physicians to practise with adequate data and sufficient time. In addition, regarding utilization of ANN and CNN methods, Kolachalama et al. [76] recently provided a perspicacity into the association of pathological fibrosis identified from histologic images with clinical phenotypes for patients with CKD, helping the diagnostics and prognostics of these phenotypes. Subsequently, there has been an increasing number of AI studies, with great emphasis on the usage of nephrology and transplantation [85,87,88,89].
Table 2.
Selected articles reporting the utilization of artificial intelligence, machine learning, or deep learning in the field of nephrology and kidney transplantation.
| Study | Country | Study Type | N | Subjects | Intervention |
|---|---|---|---|---|---|
| Zhou, 2020 [90] | China | R | 212 | Prediction of ARF and paraplegia after TAAAR | Machine learning classification models |
| Xu, 2020 [91] | USA | R | 37,486 | Identification the sub-phenotypes of AKI | Memory network-based deep learning approach |
| Song, 2020 [92] | USA | R | 14,039 | Longitudinal Risk Prediction of CKD in Diabetic Patients | Temporal-enhanced gradient boosting machine |
| Rashidi, 2020 [93] | USA | P, R | 101 | Early recognition of burn- and trauma-related AKI | Artificial intelligence /machine learning algorithms |
| Morid, 2020 [94] | USA | R | 22,542 | Prediction of adverse events in critical patients with AKI | Temporal pattern detection |
| Luo, 2020 [95] | China | R | 519 | Prediction of severe pneumonia during post-transplant hospitalization in recipients of a deceased-donor kidney transplant | Machine learning |
| Li, 2020 [96] | China | P | 1952 | Accuracy improvement of GFR estimation | Artificial neural network |
| Lei, 2020 [97] | China | R | 1173 | Prediction of AKI after liver cancer resection | Machine learning algorithms |
| Kate, 2020 [98] | USA | R | 44,691 | Prediction of AKI in hospitalized patients | Machine learning predictive models |
| Kang, 2020 [99] | South Korea | R | 1571 | Prediction of mortality in CRRT patients | Machine learning algorithms |
| Zimmerman, 2019 [100] | USA | R | 23,950 | Prediction of AKI following ICU admission | Machine learning models |
| Zhang, 2019 [101] | China | R | 2456 | Prediction of volume responsiveness in oliguric AKI | Machine learning models |
| Xu, 2019 [102] | USA | R | 58,976 | Prediction of mortality in patients with AKI in the ICU | Machine learning models |
| Xiao, 2019 [103] | China | R | 551 | Prediction of CKD progression | Machine learning tools |
| Mark, 2019 [104] | USA | R | 100,000 | Prediction of survival of kidney transplant recipients from UNOS | Machine learning models |
| Bae, 2019 [105] | USA | R | 120,818 | Prediction of survival after deceased donor kidney transplant from OPTN database | Machine learning methods |
AKI, acute kidney injury; ARF, acute renal failure; AUC, area under curve; CKD, chronic kidney disease; CRRT, continuous renal replacement therapy; GFR, glomerular filtration rate; ICU, intensive care unit; OPTN, Organ Procurement and Transplantation Network; P, prospective; R, retrospective; TAAAR, thoracoabdominal aortic aneurysm repair; UNOS, United Network for Organ Sharing.
5. Potential Directions and Future Scope
In order to reinforce the usage and subsequent transformation of AI as well as data–based CDSSs in nephrology, AI, as well as big data, offers the chance to actually source knowledge from expert knowledge and big data and subsequently transform it into some form of intelligent system, which can be applied in risk classification, disease diagnosis, drug discovery, and prognostic evaluation, among some other things. AI might be useful in establishing the type of kidney disease and subsequently help in solving problems related to survival analysis of the patients who have gone through kidney transplants [106,107,108,109,110,111,112,113,114]. Renal biopsy images may be a good data base for application of machine learning algorithms.
Despite having numerous imperfections, big data, as well as artificial intelligence have been applied in the field of medication from numerous parts [115,116]. There are numerous possible guidelines of using big data and artificial intelligence in nephrology that requires greater attention, as well as further consideration [74,78,117,118,119,120,121,122,123,124,125].
6. Conclusions
In summary, the present status of kidney health care, and subsequently, research evidence in nephrology requires strengthening. Big data research that is problem-driven in nephrology is very much essential in promoting the interdisciplinary incorporation and subsequent improvements in kidney disease, and it may subsequently offer some greater insights to further studies in the future. Within the present era of using big data, it is strongly believed that big data and artificial intelligence will greatly reshape research done on kidney disease and consequently improve the general clinical practice of nephrology in the near future.
Funding
This research received no external funding.
Conflicts of Interest
We do not have any financial or non-financial potential conflicts of interest.
References
- 1.Sutherland S.M., Goldstein S.L., Bagshaw S.M. Leveraging Big Data and Electronic Health Records to Enhance Novel Approaches to Acute Kidney Injury Research and Care. Blood Purif. 2017;44:68–76. doi: 10.1159/000458751. [DOI] [PubMed] [Google Scholar]
- 2.Kashani K., Cheungpasitporn W., Ronco C. Biomarkers of acute kidney injury: The pathway from discovery to clinical adoption. Clin. Chem. Lab. Med. 2017;55:1074–1089. doi: 10.1515/cclm-2016-0973. [DOI] [PubMed] [Google Scholar]
- 3.Srivali N., Thongprayoon C., Cheungpasitporn W., Ungprasert P., Caples S.M. Unusual cause of pleural effusion: Ovarian hyperstimulation syndrome. QJM. 2016;109:197–198. doi: 10.1093/qjmed/hcv182. [DOI] [PubMed] [Google Scholar]
- 4.Sanguankeo A., Upala S., Cheungpasitporn W., Ungprasert P., Knight E.L. Effects of Statins on Renal Outcome in Chronic Kidney Disease Patients: A Systematic Review and Meta-Analysis. PLoS ONE. 2015;10:e0132970. doi: 10.1371/journal.pone.0132970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheungpasitporn W., Thongprayoon C., Kittanamongkolchai W., Sakhuja A., Mao M.A., Erickson S.B. Impact of admission serum potassium on mortality in patients with chronic kidney disease and cardiovascular disease. QJM. 2017;110:713–739. doi: 10.1093/qjmed/hcx118. [DOI] [PubMed] [Google Scholar]
- 6.Jadlowiec C., Smith M., Neville M., Mao S., Abdelwahab D., Reddy K., Moss A., Aqel B., Taner T. Acute Kidney Injury Patterns Following Transplantation of Steatotic Liver Allografts. J. Clin. Med. 2020;9:954. doi: 10.3390/jcm9040954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheungpasitporn W., Thongprayoon C., O’Corragain O.A., Edmonds P.J., Kittanamongkolchai W., Erickson S.B. Associations of sugar-sweetened and artificially sweetened soda with chronic kidney disease: A systematic review and meta-analysis. Nephrology. 2014;19:791–797. doi: 10.1111/nep.12343. [DOI] [PubMed] [Google Scholar]
- 8.Wijarnpreecha K., Thongprayoon C., Chesdachai S., Panjawatanana P., Ungprasert P., Cheungpasitporn W. Associations of Proton-Pump Inhibitors and H2 Receptor Antagonists with Chronic Kidney Disease: A Meta-Analysis. Dig. Dis. Sci. 2017;62:2821–2827. doi: 10.1007/s10620-017-4725-5. [DOI] [PubMed] [Google Scholar]
- 9.Wijarnpreecha K., Thongprayoon C., Thamcharoen N., Panjawatanan P., Cheungpasitporn W. Association of coffee consumption and chronic kidney disease: A meta-analysis. Int. J. Clin. Pr. 2017;71 doi: 10.1111/ijcp.12919. [DOI] [PubMed] [Google Scholar]
- 10.Wijarnpreecha K., Thongprayoon C., Nissaisorakarn P., Jaruvongvanich V., Nakkala K., Rajapakse R., Cheungpasitporn W. Association of Helicobacter pylori with Chronic Kidney Diseases: A Meta-Analysis. Dig. Dis. Sci. 2017;62:2045–2052. doi: 10.1007/s10620-017-4516-z. [DOI] [PubMed] [Google Scholar]
- 11.Wijarnpreecha K., Thongprayoon C., Scribani M., Ungprasert P., Cheungpasitporn W. Noninvasive fibrosis markers and chronic kidney disease among adults with nonalcoholic fatty liver in USA. Eur. J. Gastroenterol Hepatol. 2018;30:404–410. doi: 10.1097/MEG.0000000000001045. [DOI] [PubMed] [Google Scholar]
- 12.Glassock R.J., Warnock D.G., Delanaye P. The global burden of chronic kidney disease: Estimates, variability and pitfalls. Nat. Rev. Nephrol. 2017;13:104–114. doi: 10.1038/nrneph.2016.163. [DOI] [PubMed] [Google Scholar]
- 13.Jha V., Garcia-Garcia G., Iseki K., Li Z., Naicker S., Plattner B., Saran R., Wang A.Y., Yang C.W. Chronic kidney disease: global dimension and perspectives. Chronic kidney disease: Global dimension and perspectives. Lancet. 2013;382:260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 14.Kaewput W., Thongprayoon C., Rangsin R., Ruangkanchanasetr P., Bathini T., Mao M.A., Cheungpasitporn W. Association between serum uric acid and chronic kidney disease in patients with hypertension: A multicenter nationwide cross-sectional study. J. Evid. Based Med. 2019;12:235–242. doi: 10.1111/jebm.12364. [DOI] [PubMed] [Google Scholar]
- 15.Piccoli G.B., Breuer C., Cabiddu G., Testa A., Jadeau C., Brunori G. Where Are You Going, Nephrology? Considerations on Models of Care in an Evolving Discipline. J. Clin. Med. 2018;7:199. doi: 10.3390/jcm7080199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaewput W., Thongprayoon C., Chewcharat A., Rangsin R., Satirapoj B., Kaewput C., Suwannahitatorn P., Bathini T., Mao M.A., Cato L.D., et al. Rate of kidney function decline and factors predicting progression of kidney disease in type 2 diabetes mellitus patients with reduced kidney function: A nationwide retrospective cohort study. Ther. Apher. Dial. 2020 doi: 10.1111/1744-9987.13480. [DOI] [PubMed] [Google Scholar]
- 17.Xie G., Chen T., Li Y., Chen T., Li X., Liu Z. Artificial Intelligence in Nephrology: How Can Artificial Intelligence Augment Nephrologists’ Intelligence? Kidney Dis. 2020;6:1–6. doi: 10.1159/000504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin A. Global challenges in kidney diseases. Nephrol Dial. Transpl. 2018;33:371–372. doi: 10.1093/ndt/gfy037. [DOI] [PubMed] [Google Scholar]
- 19.Yang C., Kong G., Wang L., Zhang L., Zhao M.-H. Big data in nephrology: Are we ready for the change? Nephrology. 2019;24:1097–1102. doi: 10.1111/nep.13636. [DOI] [PubMed] [Google Scholar]
- 20.Kolachalama V.B., Singh P., Lin C.Q., Mun D., Belghasem M.E., Henderson J.M., Francis J.M., Salant D.J., Chitalia V.C. Association of Pathological Fibrosis With Renal Survival Using Deep Neural Networks. Kidney Int. Rep. 2018;3:464–475. doi: 10.1016/j.ekir.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bello A.K., Levin A., Tonelli M., Okpechi I.G., Feehally J., Harris D., Jindal K., Salako B.L., Rateb A., Osman M.A., et al. Assessment of Global Kidney Health Care Status. JAMA. 2017;317:1864–1881. doi: 10.1001/jama.2017.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inrig J.K., Califf R.M., Tasneem A., Vegunta R.K., Molina C., Stanifer J.W., Chiswell K., Patel U.D. The landscape of clinical trials in nephrology: A systematic review of Clinicaltrials.gov. Am. J. Kidney Dis. 2014;63:771–780. doi: 10.1053/j.ajkd.2013.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulsen T., Jamuar S.S., Moody A.R., Karnes J.H., Varga O., Hedensted S., Spreafico R., Hafler D.A., McKinney E.F. From Big Data to Precision Medicine. Front. Med. 2019;6:34. doi: 10.3389/fmed.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shilo S., Rossman H., Segal E. Axes of a revolution: Challenges and promises of big data in healthcare. Nat. Med. 2020;26:29–38. doi: 10.1038/s41591-019-0727-5. [DOI] [PubMed] [Google Scholar]
- 25.Gameiro J., Branco T., Lopes J.A. Artificial Intelligence in Acute Kidney Injury Risk Prediction. J. Clin. Med. 2020;9:678. doi: 10.3390/jcm9030678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gore E.J., Gomes-Neto A.W., Wang L., Bakker S., Niesters H., de Joode A., Verschuuren E., Westra J., Leer-Buter C.V. Torquetenovirus Serum Load and Long-Term Outcomes in Renal Transplant Recipients. J. Clin. Med. 2020;9:440. doi: 10.3390/jcm9020440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swarte J.C., Douwes R.M., Hu S., Vich Vila A., Eisenga M.F., van Londen M., Gomes-Neto A.W., Weersma R.K., Harmsen H., Bakker S. Characteristics and Dysbiosis of the Gut Microbiome in Renal Transplant Recipients. J. Clin. Med. 2020;9:386. doi: 10.3390/jcm9020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thölking G., Gillhaus N.H., Schütte-Nütgen K., Pavenstädt H., Koch R., Suwelack B., Reuter S. Conversion to Everolimus was Beneficial and Safe for Fast and Slow Tacrolimus Metabolizers After Renal Transplantation. J. Clin. Med. 2020;9:328. doi: 10.3390/jcm9020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheungpasitporn W., Kremers W.K., Lorenz E., Amer H., Cosio F.G., Stegall M.D., Gandhi M.J., Schinstock C.A. De novo donor-specific antibody following BK nephropathy: The incidence and association with antibody-mediated rejection. Clin. Transpl. 2018;32:e13194. doi: 10.1111/ctr.13194. [DOI] [PubMed] [Google Scholar]
- 30.Chewcharat A., Thongprayoon C., Cheungpasitporn W., Mao M.A., Thirunavukkarasu S., Kashani K.B. Trajectories of Serum Sodium on In-Hospital and 1-Year Survival among Hospitalized Patients. Clin. J. Am. Soc. Nephrol. 2020 doi: 10.2215/CJN.12281019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaewput W., Thongprayoon C., Boonpheng B., Ungprasert P., Bathini T., Chewcharat A., Srivali N., Vallabhajosyula S., Cheungpasitporn W. Inpatient Burden and Mortality of Goodpasture’s Syndrome in the United States: Nationwide Inpatient Sample 2003–2014. J. Clin. Med. 2020;9:455. doi: 10.3390/jcm9020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheungpasitporn W., Thongprayoon C., Ungprasert P., Wijarnpreecha K., Kaewput W., Leeaphorn N., Bathini T., Chebib F.T., Kröner P.T. Subarachnoid Hemorrhage in Hospitalized Renal Transplant Recipients with Autosomal Dominant Polycystic Kidney Disease: A Nationwide Analysis. J. Clin. Med. 2019;8:524. doi: 10.3390/jcm8040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeaphorn N., Thongprayoon C., Chon W.J., Cummings L.S., Mao M.A., Cheungpasitporn W. Outcomes of kidney retransplantation after graft loss as a result of BK virus nephropathy in the era of newer immunosuppressant agents. Am. J. Transpl. 2019 doi: 10.1111/ajt.15723. [DOI] [PubMed] [Google Scholar]
- 34.Lertjitbanjong P., Thongprayoon C., Cheungpasitporn W., O’Corragain O.A., Srivali N., Bathini T., Watthanasuntorn K., Aeddula N.R., Salim S.A., Ungprasert P., et al. Acute Kidney Injury after Lung Transplantation: A Systematic Review and Meta-Analysis. J. Clin. Med. 2019;8:1713. doi: 10.3390/jcm8101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thongprayoon C., Kaewput W., Thamcharoen N., Bathini T., Watthanasuntorn K., Lertjitbanjong P., Sharma K., Salim S.A., Ungprasert P., Wijarnpreecha K., et al. Incidence and Impact of Acute Kidney Injury after Liver Transplantation: A Meta-Analysis. J. Clin. Med. 2019;8:372. doi: 10.3390/jcm8030372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wongboonsin J., Thongprayoon C., Bathini T., Ungprasert P., Aeddula N.R., Mao M.A., Cheungpasitporn W. Acetazolamide Therapy in Patients with Heart Failure: A Meta-Analysis. J. Clin. Med. 2019;8:349. doi: 10.3390/jcm8030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez Suarez M.L., Thongprayoon C., Mao M.A., Leeaphorn N., Bathini T., Cheungpasitporn W. Outcomes of Kidney Transplant Patients with Atypical Hemolytic Uremic Syndrome Treated with Eculizumab: A Systematic Review and Meta-Analysis. J. Clin. Med. 2019;8:919. doi: 10.3390/jcm8070919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chewcharat A., Thongprayoon C., Bathini T., Aeddula N.R., Boonpheng B., Kaewput W., Watthanasuntorn K., Lertjitbanjong P., Sharma K., Torres-Ortiz A., et al. Incidence and Mortality of Renal Cell Carcinoma after Kidney Transplantation: A Meta-Analysis. J. Clin. Med. 2019;8:530. doi: 10.3390/jcm8040530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheungpasitporn W., Thongprayoon C., Craici I.M., Sharma K., Chesdachai S., Khoury N.J., Ettore A.S. Reactivation of BK polyomavirus during pregnancy, vertical transmission, and clinical significance: A meta-analysis. J. Clin. Virol. 2018;102:56–62. doi: 10.1016/j.jcv.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Thongprayoon C., Cheungpasitporn W., Lertjitbanjong P., Aeddula N.R., Bathini T., Watthanasuntorn K., Srivali N., Mao M.A., Kashani K. Incidence and Impact of Acute Kidney Injury in Patients Receiving Extracorporeal Membrane Oxygenation: A Meta-Analysis. J. Clin. Med. 2019;8:981. doi: 10.3390/jcm8070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thongprayoon C., Kaewput W., Thamcharoen N., Bathini T., Watthanasuntorn K., Salim S.A., Ungprasert P., Lertjitbanjong P., Aeddula N.R., Torres-Ortiz A., et al. Acute Kidney Injury in Patients Undergoing Total Hip Arthroplasty: A Systematic Review and Meta-Analysis. J. Clin. Med. 2019;8:66. doi: 10.3390/jcm8010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanduri S.R., Cheungpasitporn W., Thongprayoon C., Bathini T., Kovvuru K., Garla V., Medaura J., Vaitla P., Kashani K.B. Incidence and Mortality of Acute Kidney Injury in Patients Undergoing Hematopoietic Stem Cell Transplantation: A Systematic Review and Meta-analysis. QJM. 2020 doi: 10.1093/qjmed/hcaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thongprayoon C., Cheungpasitporn W., Mao M.A., Sakhuja A., Erickson S.B. Admission calcium levels and risk of acute kidney injury in hospitalised patients. Int. J. Clin. Pr. 2018;72:e13057. doi: 10.1111/ijcp.13057. [DOI] [PubMed] [Google Scholar]
- 44.Thongprayoon C., Khoury N.J., Bathini T., Aeddula N.R., Boonpheng B., Leeaphorn N., Ungprasert P., Bruminhent J., Lertjitbanjong P., Watthanasuntorn K., et al. BK polyomavirus genotypes in renal transplant recipients in the United States: A meta-analysis. J. Evid. Based Med. 2019;12:291–299. doi: 10.1111/jebm.12366. [DOI] [PubMed] [Google Scholar]
- 45.Lin S.Y., Hsieh M.H., Lin C.L., Hsieh M.J., Hsu W.H., Lin C.C., Hsu C.Y., Kao C.H. Artificial Intelligence Prediction Model for the Cost and Mortality of Renal Replacement Therapy in Aged and Super-Aged Populations in Taiwan. J. Clin. Med. 2019;8:995. doi: 10.3390/jcm8070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Díez-Sanmartín C., Sarasa Cabezuelo A. Application of Artificial Intelligence Techniques to Predict Survival in Kidney Transplantation: A Review. J. Clin. Med. 2020;9:572. doi: 10.3390/jcm9020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azuaje F., Kim S.Y., Perez Hernandez D., Dittmar G. Connecting Histopathology Imaging and Proteomics in Kidney Cancer through Machine Learning. J. Clin. Med. 2019;8:1535. doi: 10.3390/jcm8101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsiao C.C., Tu H.T., Lin C.H., Chen K.H., Yeh Y.H., See L.C. Temporal Trends of Severe Hypoglycemia and Subsequent Mortality in Patients with Advanced Diabetic Kidney Diseases Transitioning to Dialysis. J. Clin. Med. 2019;8:420. doi: 10.3390/jcm8040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gout A.M., Martin N.C., Brown A.F., Ravine D. PKDB: Polycystic Kidney Disease Mutation Database—A gene variant database for autosomal dominant polycystic kidney disease. Hum. Mutat. 2007;28:654–659. doi: 10.1002/humu.20474. [DOI] [PubMed] [Google Scholar]
- 50.Massie A.B., Kucirka L.M., Segev D.L. Big data in organ transplantation: Registries and administrative claims. Am. J. Transpl. 2014;14:1723–1730. doi: 10.1111/ajt.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papadopoulos T., Krochmal M., Cisek K., Fernandes M., Husi H., Stevens R., Bascands J.L., Schanstra J.P., Klein J. Omics databases on kidney disease: Where they can be found and how to benefit from them. Clin. Kidney J. 2016;9:343–352. doi: 10.1093/ckj/sfv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Port F.K., Held P.J. The US Renal Data System at 30 Years: A Historical Perspective. Am. J. Kidney Dis. 2019;73:459–461. doi: 10.1053/j.ajkd.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Stack A.G., Casserly L.F., Cronin C.J., Chernenko T., Cullen W., Hannigan A., Saran R., Johnson H., Browne G., Ferguson J.P. Prevalence and variation of Chronic Kidney Disease in the Irish health system: Initial findings from the National Kidney Disease Surveillance Programme. Bmc Nephrol. 2014;15:185. doi: 10.1186/1471-2369-15-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bello A.K., Ronksley P.E., Tangri N., Singer A., Grill A., Nitsch D., Queenan J.A., Lindeman C., Soos B., Freiheit E., et al. A national surveillance project on chronic kidney disease management in Canadian primary care: A study protocol. BMJ Open. 2017;7:e016267. doi: 10.1136/bmjopen-2017-016267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saran R., Steffick D., Bragg-Gresham J. The China Kidney Disease Network (CK-NET): “Big Data-Big Dreams”. Am. J. Kidney Dis. 2017;69:713–716. doi: 10.1053/j.ajkd.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Dienemann T., Fujii N., Orlandi P., Nessel L., Furth S.L., Hoy W.E., Matsuo S., Mayer G., Methven S., Schaefer F., et al. International Network of Chronic Kidney Disease cohort studies (iNET-CKD): A global network of chronic kidney disease cohorts. BMC Nephrol. 2016;17:121. doi: 10.1186/s12882-016-0335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lv J., Zhang H., Wong M.G., Jardine M.J., Hladunewich M., Jha V., Monaghan H., Zhao M., Barbour S., Reich H., et al. Effect of Oral Methylprednisolone on Clinical Outcomes in Patients With IgA Nephropathy: The TESTING Randomized Clinical Trial. JAMA. 2017;318:432–442. doi: 10.1001/jama.2017.9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsushita K., Ballew S.H., Astor B.C., Jong P.E.d., Gansevoort R.T., Hemmelgarn B.R., Levey A.S., Levin A., Wen C.-P., Woodward M., et al. Cohort profile: The chronic kidney disease prognosis consortium. Int. J. Epidemiol. 2013;42:1660–1668. doi: 10.1093/ije/dys173. [DOI] [PubMed] [Google Scholar]
- 59.Levin A., Tonelli M., Bonventre J., Coresh J., Donner J.-A., Fogo A.B., Fox C.S., Gansevoort R.T., Heerspink H.J.L., Jardine M., et al. Global kidney health 2017 and beyond: A roadmap for closing gaps in care, research, and policy. Lancet. 2017;390:1888–1917. doi: 10.1016/S0140-6736(17)30788-2. [DOI] [PubMed] [Google Scholar]
- 60.O’Seaghdha C.M., Fox C.S. Genome-wide association studies of chronic kidney disease: What have we learned? Nat. Rev. Nephrol. 2011;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wuttke M., Köttgen A. Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol. 2016;12:549–562. doi: 10.1038/nrneph.2016.107. [DOI] [PubMed] [Google Scholar]
- 62.Ahlqvist E., van Zuydam N.R., Groop L.C., McCarthy M.I. The genetics of diabetic complications. Nat. Rev. Nephrol. 2015;11:277–287. doi: 10.1038/nrneph.2015.37. [DOI] [PubMed] [Google Scholar]
- 63.Mohan C., Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat. Rev. Nephrol. 2015;11:329–341. doi: 10.1038/nrneph.2015.33. [DOI] [PubMed] [Google Scholar]
- 64.Lindenmeyer M.T., Kretzler M. Renal biopsy-driven molecular target identification in glomerular disease. Pflug. Arch. 2017;469:1021–1028. doi: 10.1007/s00424-017-2006-y. [DOI] [PubMed] [Google Scholar]
- 65.Rumsfeld J.S., Joynt K.E., Maddox T.M. Big data analytics to improve cardiovascular care: Promise and challenges. Nat. Rev. Cardiol. 2016;13:350–359. doi: 10.1038/nrcardio.2016.42. [DOI] [PubMed] [Google Scholar]
- 66.Cheungpasitporn W., Thongprayoon C., Ungprasert P., Wijarnpreecha K., Raimondo M., Kroner P.T. Acute pancreatitis in end-stage renal disease patients in the USA: A nationwide, propensity score-matched analysis. Eur. J. Gastroenterol. Hepatol. 2019;31:968–972. doi: 10.1097/MEG.0000000000001449. [DOI] [PubMed] [Google Scholar]
- 67.Thongprayoon C., Kaewput W., Boonpheng B., Ungprasert P., Bathini T., Srivali N., Vallabhajosyula S., Castaneda J.L., Monga D., Kanduri S.R., et al. Impact of ANCA-Associated Vasculitis on Outcomes of Hospitalizations for Goodpasture’s Syndrome in the United States: Nationwide Inpatient Sample 2003-2014. Medicina. 2020;56:103. doi: 10.3390/medicina56030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ungprasert P., Koster M.J., Cheungpasitporn W., Wijarnpreecha K., Thongprayoon C., Kroner P.T. Inpatient epidemiology and economic burden of granulomatosis with polyangiitis: A 10-year study of the national inpatient sample. Rheumatology. 2020 doi: 10.1093/rheumatology/keaa069. [DOI] [PubMed] [Google Scholar]
- 69.Evans R.S. Electronic Health Records: Then, Now, and in the Future. Yearb. Med. Inform. 2016;25(Suppl. 1):S48–S61. doi: 10.15265/IYS-2016-s006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutherland S.M., Goldstein S.L., Bagshaw S.M. Acute Kidney Injury and Big Data. Contrib. Nephrol. 2018;193:55–67. doi: 10.1159/000484963. [DOI] [PubMed] [Google Scholar]
- 71.Sutherland S.M. Electronic Health Record-Enabled Big-Data Approaches to Nephrotoxin-Associated Acute Kidney Injury Risk Prediction. Pharmacotherapy. 2018;38:804–812. doi: 10.1002/phar.2150. [DOI] [PubMed] [Google Scholar]
- 72.Sutherland S.M. Big Data and Pediatric Acute Kidney Injury: The Promise of Electronic Health Record Systems. Front. Pediatr. 2019;7:536. doi: 10.3389/fped.2019.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shipp M.A., Ross K.N., Tamayo P., Weng A.P., Kutok J.L., Aguiar R.C., Gaasenbeek M., Angelo M., Reich M., Pinkus G.S., et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 74.Saez-Rodriguez J., Rinschen M.M., Floege J., Kramann R. Big science and big data in nephrology. Kidney Int. 2019;95:1326–1337. doi: 10.1016/j.kint.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 75.Angermueller C., Pärnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.SGarcelon N., Burgun A., Salomon R., Neuraz A. Electronic health records for the diagnosis of rare diseases. Kidney Int. 2020;97:676–686. doi: 10.1016/j.kint.2019.11.037. [DOI] [PubMed] [Google Scholar]
- 77.Stead W.W. Clinical Implications and Challenges of Artificial Intelligence and Deep Learning. JAMA. 2018;320:1107–1108. doi: 10.1001/jama.2018.11029. [DOI] [PubMed] [Google Scholar]
- 78.Obermeyer Z., Emanuel E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 80.Pennington J., Socher R., Manning C.D., editors. Glove: Global vectors for word representation; Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); Doha, Qatar. 25–29 October 2014. [Google Scholar]
- 81.Hermsen M., de Bel T., den Boer M., Steenbergen E.J., Kers J., Florquin S., Roelofs J., Stegall M.D., Alexander M.P., Smith B.H., et al. Deep Learning-Based Histopathologic Assessment of Kidney Tissue. J. Am. Soc. Nephrol. 2019;30:1968–1979. doi: 10.1681/ASN.2019020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ginley B., Lutnick B., Jen K.Y., Fogo A.B., Jain S., Rosenberg A., Walavalkar V., Wilding G., Tomaszewski J.E., Yacoub R., et al. Computational Segmentation and Classification of Diabetic Glomerulosclerosis. J. Am. Soc. Nephrol. 2019;30:1953–1967. doi: 10.1681/ASN.2018121259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomašev N., Glorot X., Rae J.W., Zielinski M., Askham H., Saraiva A., et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572:116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Escandell-Montero P., Chermisi M., Martínez-Martínez J.M., Gómez-Sanchis J., Barbieri C., Soria-Olivas E., Mari F., Vila-Francés J., Stopper A., Gatti E., et al. Optimization of anemia treatment in hemodialysis patients via reinforcement learning. Artif. Intell. Med. 2014;62:47–60. doi: 10.1016/j.artmed.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 85.Barbieri C., Molina M., Ponce P., Tothova M., Cattinelli I., Ion Titapiccolo J., Mari F., Amato C., Leipold F., Wehmeyer W., et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int. 2016;90:422–429. doi: 10.1016/j.kint.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 86.Chen T., Li X., Li Y., Xia E., Qin Y., Liang S., Xu F., Liang D., Zeng C., Liu Z. Prediction and Risk Stratification of Kidney Outcomes in IgA Nephropathy. Am. J. Kidney Dis. 2019;74:300–309. doi: 10.1053/j.ajkd.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 87.Samal L., D’Amore J.D., Bates D.W., Wright A. Implementation of a scalable, web-based, automated clinical decision support risk-prediction tool for chronic kidney disease using C-CDA and application programming interfaces. J. Am. Med. Inf. Assoc. 2017;24:1111–1115. doi: 10.1093/jamia/ocx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tangri N., Stevens L.A., Griffith J., Tighiouart H., Djurdjev O., Naimark D., Levin A., Levey A.S. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 2011;305:1553–1559. doi: 10.1001/jama.2011.451. [DOI] [PubMed] [Google Scholar]
- 89.Ravizza S., Huschto T., Adamov A., Böhm L., Büsser A., Flöther F.F., Hinzmann R., König H., McAhren S.M., Robertson D.H., et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat. Med. 2019;25:57–59. doi: 10.1038/s41591-018-0239-8. [DOI] [PubMed] [Google Scholar]
- 90.Zhou C., Wang R., Jiang W., Zhu J., Liu Y., Zheng J., et al. Machine learning for the prediction of acute kidney injury and paraplegia after thoracoabdominal aortic aneurysm repair. J. Card. Surg. 2020;35:89–99. doi: 10.1111/jocs.14317. [DOI] [PubMed] [Google Scholar]
- 91.Xu Z., Chou J., Zhang X.S., Luo Y., Isakova T., Adekkanattu P., Ancker J.S., Jiang G., Kiefer R.C., Pacheco J.A., et al. Identifying sub-phenotypes of acute kidney injury using structured and unstructured electronic health record data with memory networks. J. Biomed. Inform. 2020;102:103361. doi: 10.1016/j.jbi.2019.103361. [DOI] [PubMed] [Google Scholar]
- 92.Song X., Waitman L.R., Yu A.S., Robbins D.C., Hu Y., Liu M. Longitudinal Risk Prediction of Chronic Kidney Disease in Diabetic Patients Using a Temporal-Enhanced Gradient Boosting Machine: Retrospective Cohort Study. JMIR Med. Inform. 2020;8:e15510. doi: 10.2196/15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rashidi H.H., Sen S., Palmieri T.L., Blackmon T., Wajda J., Tran N.K. Early Recognition of Burn- and Trauma-Related Acute Kidney Injury: A Pilot Comparison of Machine Learning Techniques. Sci. Rep. 2020;10:205. doi: 10.1038/s41598-019-57083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morid M.A., Sheng O.R.L., Del Fiol G., Facelli J.C., Bray B.E., Abdelrahman S. Temporal Pattern Detection to Predict Adverse Events in Critical Care: Case Study With Acute Kidney Injury. JMIR Med. Inform. 2020;8:e14272. doi: 10.2196/14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo Y., Tang Z., Hu X., Lu S., Miao B., Hong S., Bai H., Sun C., Qiu J., Liang H., et al. Machine learning for the prediction of severe pneumonia during posttransplant hospitalization in recipients of a deceased-donor kidney transplant. Ann. Transl. Med. 2020;8:82. doi: 10.21037/atm.2020.01.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li N., Huang H., Qian H.-Z., Liu P., Lu H., Liu X. Improving accuracy of estimating glomerular filtration rate using artificial neural network: Model development and validation. J. Transl. Med. 2020;18:120. doi: 10.1186/s12967-020-02287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lei L., Wang Y., Xue Q., Tong J., Zhou C.-M., Yang J.-J. A comparative study of machine learning algorithms for predicting acute kidney injury after liver cancer resection. PeerJ. 2020;8:e8583. doi: 10.7717/peerj.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kate R.J., Pearce N., Mazumdar D., Nilakantan V. A continual prediction model for inpatient acute kidney injury. Comput. Biol. Med. 2020;116:103580. doi: 10.1016/j.compbiomed.2019.103580. [DOI] [PubMed] [Google Scholar]
- 99.Kang M.W., Kim J., Kim D.K., Oh K.-H., Joo K.W., Kim Y.S., Han S.S. Machine learning algorithm to predict mortality in patients undergoing continuous renal replacement therapy. Crit. Care. 2020;24:42. doi: 10.1186/s13054-020-2752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zimmerman L.P., Reyfman P.A., Smith A.D.R., Zeng Z., Kho A., Sanchez-Pinto L.N., Luo Y. Early prediction of acute kidney injury following ICU admission using a multivariate panel of physiological measurements. Bmc Med. Inform. Decis. Mak. 2019;19(Suppl. 1):16. doi: 10.1186/s12911-019-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Z., Ho K.M., Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit. Care. 2019;23:112. doi: 10.1186/s13054-019-2411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Z., Luo Y., Adekkanattu P., Ancker J.S., Jiang G., Kiefer R.C., Pacheco J.A., Rasmussen L.V., Pathak J., Wang F. Stratified Mortality Prediction of Patients with Acute Kidney Injury in Critical Care. Stud. Health Technol. Inform. 2019;264:462–466. doi: 10.3233/SHTI190264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao J., Ding R., Xu X., Guan H., Feng X., Sun T., Zhu S., Ye Z. Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J. Transl. Med. 2019;17:119. doi: 10.1186/s12967-019-1860-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mark E., Goldsman D., Gurbaxani B., Keskinocak P., Sokol J. Using machine learning and an ensemble of methods to predict kidney transplant survival. PLoS ONE. 2019;14:e0209068. doi: 10.1371/journal.pone.0209068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bae S., Massie A.B., Thomas A.G., Bahn G., Luo X., Jackson K.R., Ottmann S.E., Brennan D.C., Desai N.M., Coresh J., et al. Who can tolerate a marginal kidney? Predicting survival after deceased donor kidney transplant by donor-recipient combination. Am. J. Transpl. 2019;19:425–433. doi: 10.1111/ajt.14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoo K.D., Noh J., Lee H., Kim D.K., Lim C.S., Kim Y.H., Lee J.P., Kim G., Kim Y.S. A Machine Learning Approach Using Survival Statistics to Predict Graft Survival in Kidney Transplant Recipients: A Multicenter Cohort Study. Sci. Rep. 2017;7:8904. doi: 10.1038/s41598-017-08008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Niel O., Bastard P. Artificial Intelligence in Nephrology: Core Concepts, Clinical Applications, and Perspectives. Am. J. Kidney Dis. 2019;74:803–810. doi: 10.1053/j.ajkd.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 108.Improta G., Mazzella V., Vecchione D., Santini S., Triassi M. Fuzzy logic-based clinical decision support system for the evaluation of renal function in post-Transplant Patients [published online ahead of print, 2019 Nov 12] J. Eval. Clin. Pract. 2019 doi: 10.1111/jep.13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Atallah D.M., Badawy M., El-Sayed A., Ghoneim M.A. Predicting kidney transplantation outcome based on hybrid feature selection and KNN classifier. Multimed. Tools Appl. 2019;78:20383–20407. doi: 10.1007/s11042-019-7370-5. [DOI] [Google Scholar]
- 110.Nematollahi M., Akbari R., Nikeghbalian S., Salehnasab C. Classification Models to Predict Survival of Kidney Transplant Recipients Using Two Intelligent Techniques of Data Mining and Logistic Regression. Int. J. Organ. Transpl. Med. 2017;8:119–122. [PMC free article] [PubMed] [Google Scholar]
- 111.Tapak L., Hamidi O., Amini P., Poorolajal J. Prediction of Kidney Graft Rejection Using Artificial Neural Network. Healthc Inf. Res. 2017;23:277–284. doi: 10.4258/hir.2017.23.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shahmoradi L., Langarizadeh M., Pourmand G., Fard Z.A., Borhani A. Comparing Three Data Mining Methods to Predict Kidney Transplant Survival. Acta Inf. Med. 2016;24:322–327. doi: 10.5455/aim.2016.24.322-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luck M., Sylvain T., Cardinal H., Lodi A., Bengio Y. Deep learning for patient-specific kidney graft survival analysis. arXiv. 2017170510245 [Google Scholar]
- 114.Topuz K., Zengul F.D., Dag A., Almehmi A., Yildirim M.B. Predicting graft survival among kidney transplant recipients: A Bayesian decision support model. Decis. Support. Syst. 2018;106:97–109. doi: 10.1016/j.dss.2017.12.004. [DOI] [Google Scholar]
- 115.Lyell D., Coiera E. Automation bias and verification complexity: A systematic review. J. Am. Med. Inf. Assoc. 2017;24:423–431. doi: 10.1093/jamia/ocw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Agarwal R., Sinha A.D. Big data in nephrology-a time to rethink. Nephrol. Dial. Transpl. 2018;33:1–3. doi: 10.1093/ndt/gfx330. [DOI] [PubMed] [Google Scholar]
- 117.Lee C.H., Yoon H.-J. Medical big data: Promise and challenges. Kidney Res. Clin. Pr. 2017;36:3–11. doi: 10.23876/j.krcp.2017.36.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calvert J., Saber N., Hoffman J., Das R. Machine-Learning-Based Laboratory Developed Test for the Diagnosis of Sepsis in High-Risk Patients. Diagnostics. 2019;9:20. doi: 10.3390/diagnostics9010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lim E.-C., Park J.H., Jeon H.J., Kim H.-J., Lee H.-J., Song C.-G., Hong S.K. Developing a Diagnostic Decision Support System for Benign Paroxysmal Positional Vertigo Using a Deep-Learning Model. J. Clin. Med. 2019;8:633. doi: 10.3390/jcm8050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heo S.-J., Kim Y., Yun S., Lim S.-S., Kim J., Nam C.-M., Park E.-C., Jung I., Yoon J.-H. Deep Learning Algorithms with Demographic Information Help to Detect Tuberculosis in Chest Radiographs in Annual Workers’ Health Examination Data. Int. J. Env. Res. Public Health. 2019;16:250. doi: 10.3390/ijerph16020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kooman J.P., Wieringa F.P., Han M., Chaudhuri S., van der Sande F.M., Usvyat L.A., Kotanko P. Wearable health devices and personal area networks: can they improve outcomes in haemodialysis patients? Nephrol. Dial. Transplant. 2020;35(Suppl. 2):ii43–ii50. doi: 10.1093/ndt/gfaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shortliffe E.H., Sepúlveda M.J. Clinical Decision Support in the Era of Artificial Intelligence. JAMA. 2018;320:2199–2200. doi: 10.1001/jama.2018.17163. [DOI] [PubMed] [Google Scholar]
- 123.Santo B.A., Rosenberg A.Z., Sarder P. Artificial intelligence driven next-generation renal histomorphometry. Curr. Opin. Nephrol. Hypertens. 2020;29:265–272. doi: 10.1097/MNH.0000000000000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Na L., Yang C., Lo C.C., Zhao F., Fukuoka Y., Aswani A. Feasibility of Reidentifying Individuals in Large National Physical Activity Data Sets From Which Protected Health Information Has Been Removed With Use of Machine Learning. JAMA Netw. Open. 2018;1:e186040. doi: 10.1001/jamanetworkopen.2018.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Obermeyer Z., Lee T.H. Lost in Thought—The Limits of the Human Mind and the Future of Medicine. N. Engl. J. Med. 2017;377:1209–1211. doi: 10.1056/NEJMp1705348. [DOI] [PMC free article] [PubMed] [Google Scholar]