Abstract
Adeno-associated virus (AAV)-based vectors have transformed into powerful elements of genetic medicine with proven therapeutic efficacy and a good safety profile. Over the years, efforts to transduce hematopoietic stem cells (HSCs) with AAV2 vectors have, however, been challenging. While there was evidence that AAV2 delivered vector genomes to primitive, quiescent, multipotential, self-renewing, in vivo engrafting HSCs, transgene expression was elusive. In this study, we review the evolution of AAV transduction of HSC, starting with AAV2 vectors leading to the isolation of a family of naturally occurring AAVs from human CD34+ HSC, the AAVHSC. The stem cell-derived AAVHSCs have turned out to have remarkable potentials for genetic therapies well beyond the hematopoietic system. AAVHSCs have tropism for a wide variety of peripheral tissues, including the liver, muscle, and the retina. They cross the blood–brain barrier and transduce cells of the central nervous system. Preclinical gene therapy studies underway using AAVHSC vectors are discussed. We review the notable ability of AAVHSCs to mediate efficient, seamless homologous recombination in the absence of exogenous nuclease activity and discuss the therapeutic implications. We also discuss early results from an AAVHSC-based clinical gene therapy trial that is underway for the treatment of phenylketonuria. Thus, the stem cell-derived AAVHSC, offer a multifaceted platform for in vivo gene therapy and genome editing for the treatment of inherited diseases.
Keywords: AAVHSC, gene therapy, gene editing, PKU, hematopoietic stem cells, AAV
Introduction
I was first introduced to adeno-associated virus (AAV) in the laboratory of James Rose at NIAID, NIH, during a postdoctoral fellowship. Intense discussions of AAV biology and its lifecycle were peppered with interesting anecdotes about the founding scientists who discovered AAV and elucidated the organization, expression, and replication, and the biology of this unique nonpathogenic, replication-deficient parvovirus. This is where I first encountered Barrie Carter, a neighbor at NIH, who played an instrumental role in deciphering every aspect of the AAV lifecycle from its genome organization, transcription, and replication to the role of the AAV-encoded proteins and the very first cloning of the AAV genome in a plasmid.1–10
It was the molecular cloning of AAV5,11–13 (Fig. 1) that jumpstarted the path that led to the advent of the role of AAV in gene therapy. Indeed, the Carter group along with the Muzyzcka group, were the first to describe the use of recombinant AAV as a gene transfer vector.12,14–16 These stalwart scientists persisted in studying this innocuous virus that survived without killing its host or leaving a significant footprint. Studies on its compact genome that encodes three capsid proteins, four replication proteins, and accessory proteins, including assembly activating protein (AAP) and potentially protein X, revealed an efficient coding mechanism of overlapping reading frames and judicious use of a splice site to encode multiple proteins.17–19 Similarly, the efficient regulation of protein expression through the use of a start codon within an alternative splice site for VP1 and an ACG start codon for VP220,21 were just some of the lessons learned from studying AAV biology. Also fascinating was the unique single-stranded DNA genome of AAV bound by palindromic, high G:C inverted terminal repeats (ITRs) at either end, which folded back on themselves and served as primers for replication, a mechanism famously termed “boustrophedonic.”22 With his foundational research on every aspect of AAV, Barrie Carter, along with a select few, laid the groundwork for our understanding of the life cycle, gene expression, and the tremendous potentials of AAV as we know it today. The first human trial of recombinant AAV vectors was initiated by Flotte and Carter in 1995, when adults with cystic fibrosis were treated with endobronchial instillation of AAV2,10 which set the stage for the phase 2a and phase 2b trials of aerosolization. These studies marked the start of the AAV genetic medicine revolution of today. In his post-NIH years, Barrie continued to shepherd AAV gene therapy through biotechnology, commercial manufacturing, and beyond. With his groundbreaking work on the early studies that defined the AAV genome, the biology of AAV-encoded proteins—to the very first human clinical trial, Barrie Carter is one of the few scientists who played an instrumental role in the evolution of the field from the very beginning. Little did we know then that this replication defective, nonpathogenic virus would have such a major impact on novel therapies for incurable diseases and would revolutionize genetic medicine. Today, there are 205 AAV-based protocols for clinical trials registered at Clinicaltrials.gov. Regulatory agencies have recently approved several AAV “drugs” that are benefitting patients and extending lives, including, Glybera,23 Luxturna,24 and Zolgensma.25
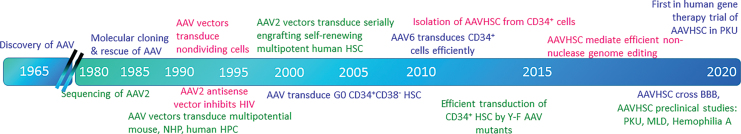
Figure 1.
Time line of major events in AAV and HSC research. AAV, adeno-associated virus; HSC, hematopoietic stem cell.
AAV Antisense Vectors for the Inhibition of HIV Infection
The early 1990s, while the incidence of HIV infection was growing exponentially, efforts to design molecular and genetic strategies to inhibit HIV replication were being initiated. We decided to test whether we could induce intracellular resistance to HIV replication by transducing cells with an AAV2 vector encoding an antisense RNA to a region common to the HIV-1 TAR/3′UTR (untranslated region), which was present in all HIV-1 transcripts and genomic RNA. We showed that T cell lines transduced with the AAV antisense vector exhibited potent reduction in HIV-1 gene expression and infectious virus production, suggesting that this was a promising approach to gene therapy for HIV-1 infection26 (Fig. 1). However, it became obvious that simply transducing differentiated T cells alone would not be sufficient for long-term efficacy. Since macrophages and T cells served as natural targets of HIV-1 infection, AAV transduction of stem cell progenitors that differentiated into HIV targets would be necessary for long-term protection. This led to our work on exploring AAV transduction of hematopoietic stem cells (HSCs).
Early work on AAV gene therapy showed that it was a promising approach for gene delivery to somatic cells.12,14–16,27,28 Stem cells, on the other hand, which comprised the ideal target for genetic correction of diseases of the hematopoietic system, were also more challenging. Successful transduction of primitive HSCs capable of both self-renewal and multilineage differentiation held the promise of permanent cure for gene-based blood diseases. However, attempts to transduce the elusive HSC were thwarted by several significant obstacles: (1) true pluripotent stem cells are rare, (2) they are difficult to identify and even more difficult to purify, and (3) in vitro manipulation resulted in loss of the key stem cell properties of self-renewal and pluripotentiality.
AAV Vectors Transduce Non-Dividing Cells
Since primitive HSCs were known to exist in a quiescent state, it became important to determine whether AAV vectors were capable of transducing nondividing cells. We tested the ability of AAV vectors to transduce cells rendered nonproliferating either by treatment with the DNA synthesis inhibitors or contact inhibition and serum starvation. Similar levels of transgene expression in both actively dividing and nondividing cells indicated that AAV vectors were capable of transducing nondividing cells29 (Fig. 1). This was the first demonstration of AAV transduction of nondividing cells and indicated that the early steps of AAV vector transduction, including virus attachment, nuclear transport, uncoating, and second-strand synthesis, occurred successfully in nondividing cells. These results also suggested that AAV transduction of primitive quiescent HSCs may be feasible.
Gene Transfer Into CD34+ Cells
The vast majority of the early work on gene delivery to HSCs was being performed with retroviral vectors. CD34+ HSC could be primed to proliferate in vitro by treatment with cytokines, rendering them good targets for retroviral transduction since retroviral genomes underwent efficient reverse transcription and integration only in dividing cells. Thus, protocols were developed to transduce ex vivo isolated CD34+ HSC with retroviral vectors. Culture conditions optimized for retrovirus transduction often employed prestimulation of cells followed by culture at high cytokine concentrations, ensuring rapid cycling of CD34+ cells and resulting in efficient retrovirus transduction.30 However, it soon became clear that efficient retrovirus transduction of CD34+ cells in vitro did not correlate with long-term multilineage engraftment in vivo, and that in vitro culture at high cytokine concentrations resulted in the loss of self-renewal and multipotential properties of CD34+ cells.
AAV Transduction of CD34+ Cell In Vitro
It was also becoming increasingly clear that culture conditions optimized for retrovirus transduction of CD34+ cells did not result in efficient AAV transduction. Episomal AAV genomes were lost from rapidly dividing cells, making them poor targets for AAV transduction. It was necessary to identify culture conditions that would enable AAV transduction, while preserving viability and the multipotential nature of CD34+ cells. Using assays for reporter transgene expression, fluorescent in situ hybridization (FISH) and Southern analyses, we showed that very low cytokine concentrations were best for AAV transduction of CD34+ cells31–33 (Fig. 1). Higher cytokine concentrations that promoted rapid cell division resulted in poor AAV transduction of CD34+ HSC from human marrow and cord blood. We also showed that prestimulation of CD34+ cells was not required and was actually deleterious to AAV transduction. Comparisons of AAV transduction of rapidly and slowly dividing CD34+ subpopulations revealed that AAV vectors persisted longer in the more quiescent CD34+ subsets, while a loss of episomal AAV genomes was observed in rapidly dividing CD34+ cells34 (Fig. 1). CD34+ cells residing in the G0 phase of the cell cycle comprised the best targets for AAV transduction. Contemporaneously, several groups showed that AAV2 vectors transduced murine, nonhuman primate (NHP) and human hematopoietic stem and progenitor cells.14,35–41
Using these culture conditions, we subsequently showed that AAV vectors transduced primitive CD34+ HSC capable of giving rise to differentiated hematopoietic colonies in long-term cultures up to 10 to 12 weeks after culture initiation.33 CD34+ cells showed chromosome-associated AAV signals by FISH. On evaluating AAV vector genomes in interphase nuclei using vector-specific fluorescent probes, we identified multiple episomal AAV vector genomes per nucleus. Interestingly, the episomal vector signals were found to be located at the periphery of the nuclei, colocalizing with heterochromatin.32
We showed that the more primitive marrow-derived CD34+CD38− subpopulation of HSCs were the most quiescent subpopulation of CD34+ cells and primarily existed in the G0 phase of the cell cycle.34 This subset of primitive HSCs was enriched for hematopoietic progenitor activity and was transduced well by AAV vectors. Notably, we also showed that, although rapidly dividing CD34+ cells were transduced by AAV, the vector genomes were rapidly lost upon cell division.34
Importantly, these experiments explained the inability to successfully transduce CD34+ cells with AAV using culture conditions optimized for retrovirus vectors. When testing AAV transduction of CD34+ HSC, many groups used culture conditions optimized for retrovirus transduction. However these culture conditions promoted rapid cell division, which led to a loss of episomal AAV genomes. As a result of transducing rapidly cycling CD34+ cells, episomal AAV vector genomes were lost and stable transduction was not observed. The use of high concentrations of cytokines, while good for retrovirus transduction, resulted in poor AAV transduction of CD34+ cells. Somewhat counterintuitively, cytokine prestimulation of CD34+ cells is often still performed before AAV transduction, resulting in a loss of episomal AAV genomes from the rapidly dividing cells. However, AAV transduction of unstimulated, quiescent HSCs, ideally followed shortly by in vivo transplantation, resulted in successful long-term gene transfer.
In contrast to the readily detectable gene marking observed after AAV2 transduction, transgene expression appeared particularly poor in vitro in primitive quiescent CD34+CD38− HSC at early time points after transduction. Several rate-limiting steps to efficient transgene expression have been demonstrated after AAV2 transduction,40–42 including donor variability in cell surface receptors for AAV, blocks to nuclear entry, uncoating and second-strand synthesis. Second-strand synthesis has been shown to be blocked when the cellular protein FKBP52 is bound to the D sequence of the AAV2 ITR. Pretreatment of cells with the receptor tyrosine kinase inhibitor tyrphostin A1 results in the phosphorylation and subsequent removal of FKBP52 from the D sequence, thereby allowing second-strand synthesis to proceed.43 We showed that second-strand synthesis is rate limiting for transgene expression in CD34+ HSC, and that tyrphostin treatment of cells leads to accelerated second-strand synthesis and augmented transgene expression.34
The Srivastava group showed that mutagenesis of surface exposed tyrosine residues to phenylalanine on AAV capsids reduced phosphorylation, ubiquitination, and proteosomal degradation of AAV virions, and resulted in increased trafficking to the nucleus and marked enhancement of transduction.44 In a series of collaborative studies, we showed that tyrosine modified AAV2 and AAV6 vectors showed significantly enhanced transduction in vitro and in vivo compared with wild-type AAV45,46 (Fig. 1). Transduction and persistence of AAV genomes in xenografted HSCs was evident long term in vivo. Similar results were observed in other tissues by other groups,47 suggesting that natural rate limitations to AAV transduction could be bypassed and would lead to further improvements in transduction.
Donor Variability
Our results indicated that most donors evaluated showed evidence of some level of AAV transduction. However, CD34+ cells from certain donors showed significantly lower levels of transduction than others.31–34 These results are in concordance with those of Ponnazhagan et al.39 who reported donor-to-donor variability of CD34 transduction by AAV vectors. Whether it is due to polymorphism of receptor expression or other factors remains unknown.
AAV Transduction of In Vivo Engrafting Self-Renewing Multipotential HSCs
Using a xenotransplant model, we showed that an AAV2 vector encoding the antisense RNA to HIV-1 transduced primitive highly purified CD34+CD38− HSCs capable of engrafting in immune-deficient mice.48 Transplantation of AAV-transduced primitive quiescent CD34+CD38− subset of HSCs led to long-term multilineage engraftment. AAV-transduced HSCs gave rise to cells of different hematopoietic lineages, including myeloid and lymphoid lineages, and CD34+ progenitors. The presence of AAV-marked cells of different hematopoietic lineages was detected to 6 months post-transplantation, confirming the multipotential nature of the AAV-transduced HSCs. Importantly, we showed that AAV-transduced HSCs were capable of serial engraftment, indicating their ability to self-renew. We additionally showed that primitive AAV-transduced HSCs harvested from the marrow of xenograft recipients retained the ability to give rise to multilineage hematopoietic colonies in vitro. These studies definitively showed that AAV2 vectors were capable of transducing primitive quiescent multipotential human HSCs capable of self-renewal and differentiation48 (Fig. 1).
AAV vectors are known to persist primarily in an episomal state following transduction. Chromosomal integration occurs at a very low frequency. This raises the question of how AAV vectors survive in HSCs. Since primitive HSCs are primarily quiescent and survive in a nondividing state for years, it is likely that AAV episomes persist for prolonged periods. It is of interest to note that stem cells are thought to possess an efficient DNA break repair system,49,50 and this may be responsible for the persistence of AAV in primitive quiescent stem cells. HSCs, in particular, have highly active DNA repair machinery, probably to guard against the accumulation of mutations and to ensure continued genomic integrity.49–51 Genes of DNA repair pathways have been found to be consistently overexpressed in stem cells. Whether these proteins or pathways play a role in AAV persistence in primitive HSCs is unknown.
Novel AAV Serotypes
Although most of the early work on AAV vectors was done with AAV2, in the early 2000s, evaluation of other AAV serotypes for gene transfer was underway.52 Guangping Gao and James Wilson reported the identification and isolation of many naturally occurring novel AAV capsids from human and NHP tissues, including the human bone marrow.53,54
AAV6 Vectors Transduce CD34+ HSC Efficiently
In a collaborative study with the Srivastava group, we evaluated AAV6 vectors for transduction of CD34+ HSC. Our results showed that AAV6 transduced CD34+ HSC more efficiently than AAV2 in both short-term in vitro as well as long-term in vivo assays46 (Fig. 1). Transduction of CD34+ cells with AAV6 resulted in ∼20-fold greater genome copies per cell at 4–6 months post-transplantation compared with AAV2. Tyrosine mutants of AAV6 showed ∼40-fold higher transduction than AAV2.46 These studies indicated that different serotypes and the use of alternate capsids had the potential to further improve AAV transduction of CD34+ HSC. AAV6 is currently used widely for the delivery of donor DNA in conjunction with nuclease-based editing platforms, including CRISPR and zinc finger nucleases.54–57
The Stem Cell-Derived AAVHSC
Novel CD34-derived AAV serotypes
Based upon the work of Gao et al.,53,54 we hypothesized that AAV may also be present in CD34+ cells. A screen of CD34+ cells from 71 healthy donors for stem cell transplantation indicated the presence of AAV in the CD34+cells of ∼70% of donors tested, although the frequency was low at ∼1/10,000 cells to 1/1,000 cells. Isolation and sequencing of AAV from CD34+ cells from two donors showed the presence of many novel naturally occurring AAV capsids. We isolated and characterized 17 of these, all of which mapped AAV Clade F58 (Fig. 1). The CD34-derived AAVs were designated AAVHSC to reflect their origin. They differed from AAV9 by 1–4 amino acids with the majority of amino acid changes being localized to VP3. Notably, frequent silent mutations were found in AAVHSCs, suggesting potential evolutionary pressure to maintain certain amino acids at specified locations.58 We reasoned that AAV derived from CD34+ cells would have tropism for HSCs, since they must have infected these cells in the first place. We also reasoned that since AAVHSCs exist naturally in human HSCs, they must have evolved and adapted to CD34+ cells over time, and we hypothesized that gene transfer vectors derived from them may elicit fewer adverse reactions.
As predicted, AAVHSC-based gene transfer vectors efficiently underwent entry and postentry processing in CD34+ cells and supported stable gene transfer into long-term in vivo engrafting human HSCs significantly better than either AAV2 or AAV8.58 AAVHSC-transduced human CD34+ cells engrafted in vivo in immune-deficient mice and gave rise to differentiated transgene-expressing progeny. Importantly, gene-marked CD34+ HSC persisted long term in xenograft recipients, indicating successful transduction of primitive progenitors. These studies indicated that AAVHSCs may serve as a new class of AAV vectors for genomic manipulation.
Biodistribution of AAVHSCs
Biodistribution studies in mice revealed that the tropism of AAVHSCs was not restricted to HSCs. Intravenous injection of AAVHSC vectors showed widespread systemic tropism. Most AAVHSCs transduced the liver efficiently. Of these, AAVHSC15 was threefold to fivefold more efficient than AAV8. Skeletal muscle, cartilage heart, and lungs were among other tissues transduced by AAVHSCs.59
Biodistribution of AAVHSCs in NHP
Recently, the biodistribution of AAVHSC7, AAVHSC15, and AAVHSC17 was reported in cynomolgus macaques following a single intravenous injection.60 As in mice, widespread tissue tropism was observed, with the liver showing the highest level of transduction. Also transduced were skeletal muscle, cardiomyocytes, kidneys, and pancreatic acinar cells.
Intravenous delivery of AAVHSCs resulted in widespread transduction of the central nervous system (CNS), indicating that AAVHSCs were capable of crossing the blood–brain barrier (BBB). Widespread transduction of glial cells was observed throughout the CNS. AAVHSCs were also found to transduce neurons, astrocytes, oligodendrocytes, and satellite cells in the brain, spinal cord, dorsal root ganglia, as well as the peripheral nervous system. AAVHSC7 and AAVHSC15 transduced the retinal ganglion cell layer and the inner cell layer after intravenous injection.60 These results support the use of AAVHSC vectors for gene therapy of both peripheral organs as well as the CNS. Systemic treatment of inherited diseases often does not address CNS manifestations of disease and vice versa, leading to insufficiently effective therapies. As such, AAVHSCs may be ideal for the treatment of diseases with sequelae in both compartments, which have historically been difficult to address.
Pre-existing neutralizing antibodies to AAVHSCs
Neutralizing antibodies to AAV are prevalent in many species, including humans. A recent study by Ellsworth et al. queried the presence of pre-existing neutralizing antibodies to AAVHSCs in a large sampling of human sera.61 Eighty percent of serum donors were found to be seronegative for neutralizing antibodies to AAVHSCs. Of the seropositive donors, only 6% were found to have high titers. Since pre-existing neutralizing antibodies ablate AAV transduction and render systemic therapy ineffective, this study indicates that a large fraction of patients could potentially be treated with AAVHSCs.61
AAVHSCs mediate seamless, accurate genome editing in the absence of exogenous nucleases
Previous studies had demonstrated that AAV is capable of mediating genome editing without the requirement for double-stranded DNA breaks by exogenous nucleases.62–67 However, the reported editing frequencies were low, generally well below 0.5%. Upon testing the capacity of AAVHSC vectors to mediate genome editing, we observed that AAVs mapping to Clade F, including AAVHSC and AAV9, mediated efficient genome editing without the need for prior nuclease-mediated DNA breaks68 (Fig. 2).
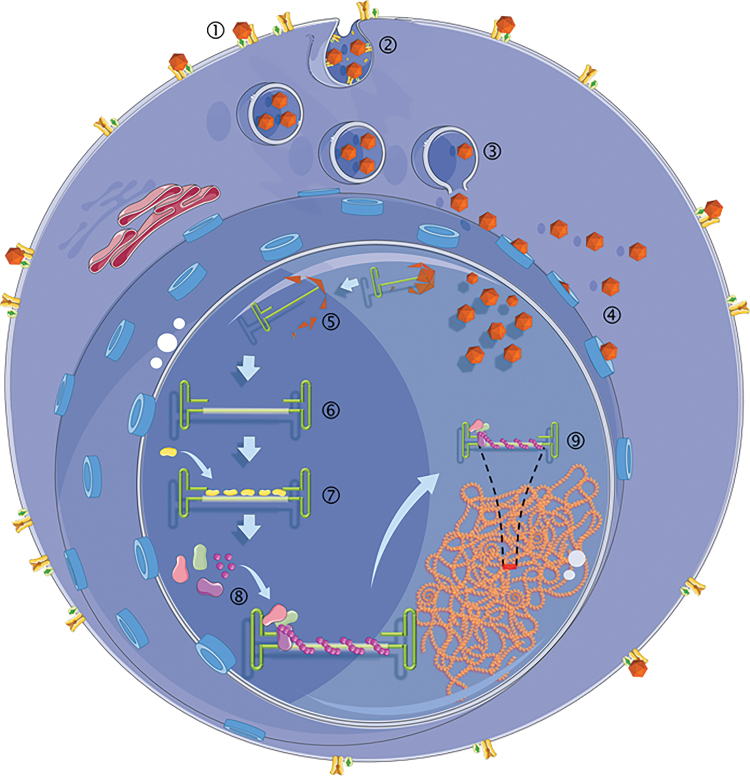
Figure 2.
Events in the AAVHSC life cycle leading to genome editing: (1) binding of AAVHSC virions to cell surface receptor and co-receptors. (2) Endosomal entry of AAVHSCs. (3) Release of AAVHSCs from endosomes. (4) Entry of AAV through the nuclear pore complex.76,77 (5) Uncoating of AAVHSC virions and release of the single-stranded vector genome. (6) The single-stranded AAV vector genome with homology region and ITRs. (7) Recruitment of DNA repair proteins to the AAV editing genome. (8) Formation of nucleoprotein filament complex. (9) Homology search between editing vector genome and chromosomal DNA. ITRs, inverted terminal repeats.
Interestingly, we found that the lower efficiency of editing of AAV2, AAV6, and AAV8 was not due to their inability to transduce cells or deliver genomes to the nuclei, as AAV6 is known to transduce CD34+ and other cells efficiently.46 Since the genomes of all editing vectors were identical and based on AAV2, we concluded that capsid proteins play a role in determining editing efficiency. The role of AAV capsid proteins in modulation editing efficiency is under investigation. Figure 2 summarizes the potential sequence of events leading to AAV editing.
Notably, efficient in vitro editing was dependent upon high-quality, high-titered vector stocks and the use of high multiplicities of infection (MOIs), at or above 150,000 vector genomes/cells.68 Attempts to replicate AAVHSC editing at low MOIs with poor-quality AAV vector batches that cause toxicity have predictably failed.67–69 Rogers et al.70 and Dudek and Porteus71 failed to observe Clade F AAV-mediated non-nuclease editing. However, their experiments were primarily performed at MOI: 5,000–10,000, due to toxicity of their vector stocks. These results are actually in concordance with Smith et al.,68,69 where we showed that high MOIs were required to observe efficient editing. Vector preparations used for editing must be of sufficiently high quality such that no toxicity due to contaminants is observed at the high multiplicities required for editing.69
At the molecular level, editing was found to be seamless and highly accurate, with no evidence of on-target mutations.68 AAVHSC-based editing was guided by homology arms complementary to the genomic target site and operated exclusively by homologous recombination (HR), as evidenced by the absolute requirement for BRCA2, an essential mediator of HR. The complete absence of any evidence of NHEJ-based repair, including insertion/deletions and incorporation of AAV ITR, supported these conclusions. We showed editing at multiple genomic loci, including AAVS1 and Rosa26, and in a variety of cell types. AAVHSC-based editing was observed to be most efficient in primary cells and in vivo in mice. Remarkably, editing appeared to also occur efficiently in postmitotic cells, such as the adult liver in vivo, freshly isolated human liver sections, myocytes, and unstimulated primary CD34+ cells. The AAVHSC platform can be designed to mediate single-nucleotide changes in the genome, indicating its potential to correct point mutations.68
AAVHSC vectors mediated in vivo editing following intravenous delivery. We showed targeted insertion of a promoterless firefly luciferase open reading frame cassette into Intron 1 of the murine Rosa26 locus.68 Serial in vivo imaging of injected mice showed luciferase expression early after injection and was stable long term. Seamless editing was confirmed by sequencing and Southern analyses. As with other chromosomal loci, no ITR sequences were observed as being inserted into the targeted insertion site.
Overall, the AAVHSCs represent a single component editing platform that functions efficiently in vivo without the need for nuclease-mediated double-stranded DNA breaks and is being developed for therapeutic in vivo editing for inherited diseases, such as hemophilia and phenylketonuria (PKU).72
Clinical gene therapy of PKU with AAVHSC15
In 2019, Homology Medicines initiated a Phase1/2 open-label, randomized, concurrently controlled, dose escalation clinical gene therapy trial for PKU using an AAVHSC15 vector (pheNIX study, Clinicaltrials.gov Identifier NCT03952156). This trial, based upon Ahmed et al.,73 is designed to test the safety and efficacy of a single intravenous injection of an AAVHSC15 vector encoding a functional copy of the phenylalanine hydroxylase (PAH) gene in adults with PKU. PKU is an autosomal recessive inborn error of metabolism due to an absence of functional PAH, a hepatic enzyme that converts phenylalanine to tyrosine. PAH deficiency leads to an inability to metabolize phenylalanine, resulting in developmental neurologic and metabolic problems. There is currently no adequate treatment for PKU that addresses the underlying genetic mutation. Management of PKU requires a highly restrictive diet, which leads to compliance failures.
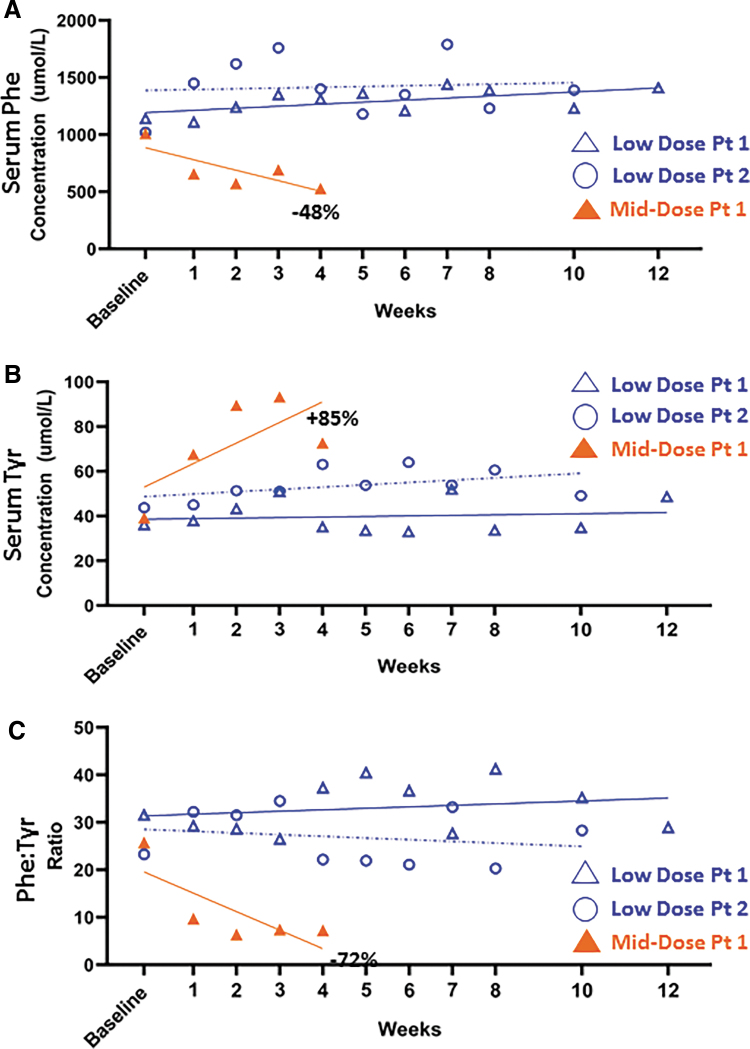
Early results of the pheNIX gene therapy showed that intravenous injection of AAVHSC15 was well tolerated at both doses tested, which ranged from low to mid 1e13 vector genomes/kg (Homology Medicines, Inc. Webcast, December 17, 2019). No treatment-related adverse effects were observed at either dose, indicating safety of the in vivo use of AAVHSC15. One patient treated with the higher of the two doses showed a significant decline, by 48%, in serum phenylalanine levels (Fig. 3A) and a concurrent rise in serum tyrosine levels, by 85%, within 4 weeks after treatment initiation (Fig. 3B). The phenylalanine: tyrosine ratio declined by 72% by 4 weeks after injection (Fig. 3C). Thus, while the trial is still ongoing and data collection continues, early results support evidence of safety and efficacy of AAVHSC-PAH gene therapy for the treatment of PKU (Homology Medicines, Inc. Webcast, December 17, 2019. Data disclosed through December 2, 2019).
Figure 3.
Evaluation of AAVHSC15-PAH gene therapy in adults with PKU. Early results of pheNIX study, Clinicaltrials.gov Identifier NCT03952156. Data shown are from initiation of the trial until December 2, 2019. (A) Serum phenylalanine levels are shown. Each symbol represents a unique subject. The two patients shown in blue were treated at the low does. Open blue triangle, low dose, Patient 1. Open blue circle, low dose, Patient 2. Closed orange triangle, mid-dose, Patient 1. (B) Serum tyrosine levels. (C) Ratio of serum levels of phenylalanine to tyrosine. Low dose and mid dose represent low to mid 1e13 vector genomes/kg. Baseline and weekly measurements are shown. PAH, phenylalanine hydroxylase; PKU, phenylketonuria.
Preclinical Studies of AAVHSCs
Gene therapy for metachromatic leukodystrophy
Preclinical studies of AAVHSC vectors for gene therapy for other inherited diseases are also underway, including metachromatic leukodystrophy (MLD). Intravenous injection of an AAVHSC15 vector encoding a functional copy of the arylsulfatase-A (ARSA) lysosomal enzyme, missing in MLD, resulted in rapid and sustained therapeutic levels of ARSA expression in the brain and spinal cord in an ARSA knockout mouse model.74 Supraphysiologic levels of ARSA were observed within 2 weeks after intravenous AAVHSC15 injection. Expression was stable over time. AAVHSC-based gene therapy for MLD is currently in investigational new drug (IND)-enabling studies at Homology Medicines, Inc.
Gene editing for PKU
As with most inborn errors of metabolism, in PKU, developmental sequelae due to the inability to metabolize phenylalanine begin early in life. Elevated levels of serum phenylalanine and a reduced availability of tyrosine for synthesis of neurotransmitters have severe effects on PKU patients.75 Thus, early genetic intervention is highly desirable and also feasible due to the existence of a nationwide newborn screening program for PKU. However, there is a risk of loss of episomal AAV vector genomes in infants following gene therapy due to growth during childhood. Thus, genome editing is an attractive treatment strategy to treat inherited diseases early in life. Targeted genomic insertion of the wild-type PAH gene in the liver would be expected to result in a permanent correction of the disease. Thus, preclinical studies on therapeutic genome editing for PKU with an AAVHSC15 editing vector are underway. Targeted insertion of a promoterless PAH complementary DNA (cDNA) at the PAH locus with AAVHSC15 resulted in rapid reduction of serum phenylalanine levels and restoration of normal levels of PAH expression in both a murine model of PKU (enu2) as well as in human/mouse xenograft model.72 Gene editing for PKU is currently in IND-enabling studies.
Gene editing for hemophilia A
Preclinical studies of gene editing to correct the I22I inversion mutation in hemophilia A are also underway. This inversion mutation accounts for almost half of hemophilia A patients and results in an inversion of exons 23–26 relative to the rest of the FVIII gene. Preliminary results showed that AAVHSC editing of the FVIII gene was successful and stable over time (unpublished data). Sequence analysis confirmed that before, editing was seamless with no insertion/deletion mutations or ITR insertions.
Lessons learned and the path forward
Almost two and a half decades of research on AAV and HSC have led to some interesting and unexpected revelations. AAV vectors were found to efficiently transduce primitive quiescent HSCs residing in the G0 phase of the cell cycle, a primitive cell population that has been challenging to transduce with other vectors.31–34 Although AAV transduction of HSCs resulted in efficient delivery of the vector genomes to the nuclei, transgene expression was more difficult to detect. Several limitations to transgene expression have since been identified and strategies to overcome these have been designed.40–44
The identification of naturally occurring AAV in CD34+ human HSCs was a surprising and impactful finding.58 The finding that AAV, which was originally thought to be a respiratory virus traveling with its adenovirus helper, would find its way to marrow-resident HSCs was unexpected. Also unexpected was the high frequency of healthy stem cell donors that harbored AAV in their CD34+ cells. Somewhat surprisingly, AAVHSCs turned out to have broad tropism beyond HSCs, including the liver, muscle, and the CNS.59,60 Their ability to cross the BBB supports important clinical utility. The low seroprevalence of neutralizing antibodies to AAVHSCs may be a corollary of their HSC origins.61 Since T cells develop immune tolerance to proteins recognized as “self,” perhaps, exposure of progenitor cells to AAVHSCs results in lower immunogenicity. Like other AAVs, AAVHSCs were found to mediate HR. However, perhaps the most surprising property of the AAVHSC is their significantly increased HR efficiency in the absence of exogenous nuclease activity.68 The use of HR instead of the nonhomologous end joining editing pathways commonly utilized by the nuclease-based editing platforms lead to seamless and precise genome editing by AAVHSCs. The elucidation of mechanisms by which AAVHSC capsid proteins mediate efficient editing will surely lead to more interesting revelations.
Meanwhile, AAVHSCs are being advanced into clinical trials. Early data from clinical trials of human gene therapy for PKU with AAVHSCs showed safety and evidence of dose-dependent efficacy, even as IND-enabling genome editing studies are underway. The option to treat inherited diseases either by episomal AAVHSC-mediated gene transfer or the permanent correction of the genome by precise and accurate editing offers attractive future strategies for genetic medicines.
Acknowledgments
The authors are indebted to Albert Seymour, Arthur Tzianabos, Theresa McNeely, and Cara Mayfield for sharing data from the early clinical trial from their Webcast and preclinical data presented at conferences. We thank also Lakshmi Bugga and Ka Ming Pang for consulting on the article.
Author Disclosure
S.C. is a co-founder of, has equity in, and serves on the scientific advisory board of Homology Medicines Inc., K.K.W. has equity in Homology Medicines, Inc.
Funding Information
This work was supported by the National Institutes of Health grants P01CA59308, R01AI40001, PO1HL60898, R01HL087285, and P30CA033572, and a grant from the California Institute of Regenerative Medicine, CIRM DISC2–10524.
References
- 1. Carter BJ, Rose JA. Transcription in vivo of a defective parvovirus: sedimentation and electrophoretic analysis of RNA synthesized by adenovirus-associated virus and its helper adenovirus. Virology 1974;61:182–199 [DOI] [PubMed] [Google Scholar]
- 2. Carter BJ, Fife KH, de la Maza LM, et al. Genome localization of adeno-associated virus RNA. J Virol 1976;19:1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myers MW, Carter BJ. Assembly of adeno-associated virus. Virology 1980;102:71–82 [DOI] [PubMed] [Google Scholar]
- 4. de la Maza LM, Carter BJ. Molecular structure of adeno-associated virus variant DNA. J Biol Chem 1980;255:3194–3203 [PubMed] [Google Scholar]
- 5. Laughlin CA, Tratschin JD, Coon H, et al. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 1983;23:65–73 [DOI] [PubMed] [Google Scholar]
- 6. Tratschin JD, West MH, Sandbank T, et al. A human parvovirus, adeno-associated virus, as a eucaryotic vector: transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol Cell Biol 1984;4:2072–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tratschin JD, Miller IL, Smith MG, et al. Adeno-associated virus vector for high-frequency integration, expression, and rescue of genes in mammalian cells. Mol Cell Biol 1985;5:3251–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flotte TR, Afione SA, Conrad C, et al. Stable in vivo expression of the cystic fibrosis transmembrane conductance regulator with an adeno-associated virus vector. Proc Natl Acad Sci U S A 1993;90:10613–10617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Owens RA, Weitzman MD, Kyöstiö SR, et al. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol 1993;67:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flotte TR, Zeitlin PL, Reynolds TC, et al. Phase I trial of intranasal and endobronchial administration of a recombinant adeno-associated virus serotype 2 (rAAV2)-CFTR vector in adult cystic fibrosis patients: a two-part clinical study. Hum Gene Ther 2003;14:1079–1088 [DOI] [PubMed] [Google Scholar]
- 11. Samulski RJ, Berns KI, Tan M, et al. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci U S A 1982;79:2077–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci U S A 1984;81:6466–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Senapathy P, Carter BJ. Molecular cloning of adeno-associated virus variant genomes and generation of infectious virus by recombination in mammalian cells. J Biol Chem 1984;259:4661–4666 [PubMed] [Google Scholar]
- 14. LaFace D, Hermonat P, Wakeland E, et al. Gene transfer into hematopoietic progenitor cells mediated by an adeno-associated virus vector. Virology 1988;162:483–486 [DOI] [PubMed] [Google Scholar]
- 15. Chejanovsky N, Carter BJ. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology 1989;171:239–247 [DOI] [PubMed] [Google Scholar]
- 16. Srivastava CH, Samulski RJ, Lu L, et al. Construction of a recombinant human parvovirus B19: adeno-associated virus 2 (AAV) DNA inverted terminal repeats are functional in an AAV-B19 hybrid virus. Proc Natl Acad Sci U S A 1989;86:8078–8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tratschin JD, Miller IL, Carter BJ. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol 1984;51:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hermonat PL, Labow MA, Wright R, et al. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol 1984;51:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 2010;107:10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarty DM, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol 1991;65:2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becerra SP, Koczot F, Fabisch P, et al. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol 1988;62:2745–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Straus SE, Sebring ED, Rose JA. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci U S A 1976;73:742–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burnett JR, Hooper AJ. Alipogene tiparvovec, an adeno-associated virus encoding the Ser(447)X variant of the human lipoprotein lipase gene for the treatment of patients with lipoprotein lipase deficiency. Curr Opin Mol Ther 2009;11:681–691 [PubMed] [Google Scholar]
- 24. Russell S, Bennett J, Wellman JA, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 2017;390:849–860. DOI:10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoy SM. Onasemnogene abeparvovec: first global approval. Drugs 2019;79:1255–1262 [DOI] [PubMed] [Google Scholar]
- 26. Chatterjee S, Johnson PR, Wong KK Jr.. Dual-target inhibition of HIV-1 in vitro by means of an adeno-associated virus antisense vector. Science 1992;258:1485–1488 [DOI] [PubMed] [Google Scholar]
- 27. Kessler PD, Podsakoff GM, Chen X, et al. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci U S A 1996;93:14082–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koeberl DD, Alexander IE, Halbert CL, et al. Persistent expression of human clotting factor IX from mouse liver after intravenous injection of adeno-associated virus vectors. Proc Natl Acad Sci U S A 1997;94:1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Podsakoff G, Wong KK Jr., Chatterjee S. Efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. J Virol 1994;68:5656–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoban MD, Romero Z, Cost GJ, et al. Delivery of genome editing reagents to hematopoietic stem/progenitor cells. Curr Protoc Stem Cell Biol 2016;36:5B.4.1–5B.4.10 [DOI] [PubMed] [Google Scholar]
- 31. Chatterjee S, Lu D, Podsakoff G, et al. Strategies for efficient gene transfer into hematopoietic cells. The use of adeno-associated virus vectors in gene therapy. Ann N Y Acad Sci 1995;770:79–90 [DOI] [PubMed] [Google Scholar]
- 32. Fisher-Adams G, Wong KK Jr., Podsakoff G, et al. Integration of adeno-associated virus vectors in CD34+ human hematopoietic progenitor cells after transduction. Blood 1996;88:492–504 [PubMed] [Google Scholar]
- 33. Chatterjee S, Li W, Wong CA, et al. Transduction of primitive human marrow and cord blood-derived hematopoietic progenitor cells with adeno-associated virus vectors. Blood 1999;93:1882–1894 [PubMed] [Google Scholar]
- 34. Paz H, Wong CA, Li W, et al. Quiescent subpopulations of human CD34-positive hematopoietic stem cells are preferred targets for stable recombinant adeno-associated virus type 2 transduction. Hum Gene Ther 2007;18:614–626 [DOI] [PubMed] [Google Scholar]
- 35. Schimmenti S, Boesen J, Claassen EA, et al. Long-term genetic modification of rhesus monkey hematopoietic cells following transplantation of adenoassociated virus vector-transduced CD34+ cells. Hum Gene Ther 1998;9:2727–2734 [DOI] [PubMed] [Google Scholar]
- 36. Kurpad C, Mukherjee P, Wang XS, et al. Adeno-associated virus 2-mediated transduction and erythroid lineage-restricted expression from parvovirus B19p6 promoter in primary human hematopoietic progenitor cells. J Hematother Stem Cell Res 1999;8:585–592 [DOI] [PubMed] [Google Scholar]
- 37. Zhou SZ, Broxmeyer HE, Cooper S, et al. Adeno-associated virus 2-mediated gene transfer in murine hematopoietic progenitor cells. Exp Hematol 1993;21:928–933 [PubMed] [Google Scholar]
- 38. Zhou SZ, Cooper S, Kang LY, et al. Adeno-associated virus 2-mediated high efficiency gene transfer into immature and mature subsets of hematopoietic progenitor cells in human umbilical cord blood. J Exp Med 1994;179:1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ponnazhagan S, Mukherjee P, Wang XS, et al. Adeno-associated virus type 2-mediated transduction in primary human bone marrow-derived CD34+ hematopoietic progenitor cells: donor variation and correlation of transgene expression with cellular differentiation. J Virol 1997;71:8262–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hansen J, Qing K, Srivastava A. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J Virol 2001;75:4080–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhong L, Li W, Yang Z, et al. Impaired nuclear transport and uncoating limit recombinant adeno-associated virus 2 vector-mediated transduction of primary murine hematopoietic cells. Hum Gene Ther 2004;15:1207–1218 [DOI] [PubMed] [Google Scholar]
- 42. Zhong L, Chen L, Li Y, et al. Self-complementary adeno-associated virus 2 (AAV)-T cell protein tyrosine phosphatase vectors as helper viruses to improve transduction efficiency of conventional single-stranded AAV vectors in vitro and in vivo. Mol Ther 2004;10:950–957 [DOI] [PubMed] [Google Scholar]
- 43. Mah C, Qing K, Khuntirat B, et al. Adeno-associated virus type 2-mediated gene transfer: role of epidermal growth factor receptor protein tyrosine kinase in transgene expression. J Virol 1998;72:9835–9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhong L, Li B, Mah CS, et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci U S A 2008;105:7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kauss MA, Smith LJ, Zhong L, et al. Enhanced long-term transduction and multilineage engraftment of human hematopoietic stem cells transduced with tyrosine-modified recombinant adeno-associated virus serotype 2. Hum Gene Ther 2010;21:1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song L, Kauss MA, Kopin E, et al. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy 2013;15:986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiao C, Zhang W, Yuan Z, et al. Adeno-associated virus serotype 6 capsid tyrosine-to-phenylalanine mutations improve gene transfer to skeletal muscle. Hum Gene Ther 2010;21:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Santat L, Paz H, Wong C, et al. Recombinant AAV2 transduction of primitive human hematopoietic stem cells capable of serial engraftment in immune-deficient mice. Proc Natl Acad Sci U S A 2005;102:11053–11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myllyperkio MH, Vilpo JA. Increased DNA single-strand break joining activity in UV-irradiated CD34 versus. Mutat Res 1999;425:169–176 [DOI] [PubMed] [Google Scholar]
- 50. Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annu Rev Med 2005;56:495–508 [DOI] [PubMed] [Google Scholar]
- 51. Bender CF, Sikes ML, Sullivan R, et al. Cancer predisposition and hematopoietic failure in Rad50S/S mice. Genes Dev 2002;16:2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong L, Li W, Li Y, et al. Evaluation of primitive murine hematopoietic stem and progenitor cell transduction in vitro and in vivo by recombinant adeno-associated virus vector serotypes 1 through 5. Hum Gene Ther 2006;17:321–333 [DOI] [PubMed] [Google Scholar]
- 53. Gao G, Alvira MR, Somanathan S, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A 2003;100:6081–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao G, Vandenberghe LH, Alvira MR, et al. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 2004;78:6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang J, DeClercq JJ, Hayward SB, et al. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res 2016;44:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen S, Sun S, Moonen D, et al. CRISPR-READI: efficient generation of knockin mice by CRISPR RNP electroporation and AAV donor infection. Cell Rep 2019;27:3780.e4–3789.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ling C, Bhukhai K, Yin Z, et al. High-efficiency transduction of primary human hematopoietic stem/progenitor cells by AAV6 vectors: strategies for overcoming donor-variation and implications in genome editing. Sci Rep 2016;6:35495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smith LJ, Ul-Hasan T, Carvaines SK, et al. Gene transfer properties and structural modeling of human stem cell-derived AAV. Mol Ther 2014;22:1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith LJ, Kauss A, Wong KK, et al. Enhanced hepatic transgene expression following intravenous delivery of novel stem cell-derived recombinant AAV vectors. Mol Ther 2010;18:S1–S220514033 [Google Scholar]
- 60. Ellsworth JL, Gingras J, Smith LJ, et al. Clade F AAVHSCs cross the blood brain barrier and transduce the central nervous system in addition to peripheral tissues following intravenous administration in nonhuman primates. PLoS One 2019;14:e0225582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ellsworth JL, O'Callaghan M, Rubin H, et al. Low seroprevalence of neutralizing antibodies targeting two clade F AAV in humans. Hum Gene Ther Clin Dev 2018;29:60–67 [DOI] [PubMed] [Google Scholar]
- 62. Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet 1998;18:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Porteus MH, Cathomen T, Weitzman MD, et al. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol 2003;23:3558–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vasileva A, Linden RM, Jessberger R. Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Res 2006;34:3345–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barzel A, Paulk NK, Shi Y, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 2015;517:360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller DG, Wang PR, Petek LM, et al. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol 2006;24:1022–1026 [DOI] [PubMed] [Google Scholar]
- 67. Khan IF, Hirata RK, Russell DW. AAV-mediated gene targeting methods for human cells. Nat Protoc 2011;6:482–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith LJ, Wright J, Clark G, et al. Stem cell-derived clade F AAVs mediate high-efficiency homologous recombination-based genome editing [published correction appears in Proc Natl Acad Sci U S A. 2019 Jan 2;116(1):337]. Proc Natl Acad Sci U S A 2018;115:E7379–E7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chatterjee S. Efficient nuclease-free HR by clade F AAV requires high MOIs with high quality vectors. Mol Ther 2019;27:2058–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rogers GL, Chen HY, Morales H, et al. Homologous recombination-based genome editing by clade F AAVs is inefficient in the absence of a targeted DNA break. Mol Ther 2019;27:1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dudek AM, Porteus MH. AAV6 is superior to clade F AAVs in stimulating homologous recombination-based genome editing in human HSPCs. Mol Ther 2019;27:1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wright J, Ellsworth J, St. Martin T, et al. Nuclease-free and promoter-less AAVHSC-mediated genome editing in vivo corrects the disease phenotype in a mouse model of phenylketonuria. Mol Ther 2019;27:S462 [Google Scholar]
- 73. Ahmed S, Rubin H, Wang M, et al. Sustained correction of a murine model of phenylketonuria following a single intravenous administration of AAVHSC15-PAH. Mol Ther Methods Clin Dev 2020;17:568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gingras J, St-Martin T, Gall K, et al. HMI-202: investigational gene therapy for treatment of metachromatic leukodystrophy (MLD). Mol Genet Metab 2020;129:S62 [Google Scholar]
- 75. Harding CO, Winn SR, Gibson KM, et al. Pharmacologic inhibition of l-tyrosine degradation ameliorates cerebral dopamine deficiency in murine phenylketonuria (PKU). J Inherit Metab Dis 2014;37:735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nicolson SC, Samulski RJ. Recombinant adeno-associated virus utilizes host cell nuclear import machinery to enter the nucleus. J Virol 2014;88:4132–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kelich JM, Ma J, Dong B, et al. Super-resolution imaging of nuclear import of adeno-associated virus in live cells. Mol Ther Methods Clin Dev 2015;2:15047. [DOI] [PMC free article] [PubMed] [Google Scholar]