Abstract
The Public Health Emergency of International Concern declared the widespread outbreak of SARS-CoV-2 as a global pandemic emergency, which has resulted in 1,773,086 confirmed cases including 111,652 human deaths, as on 13 April 2020, as reported to World Health Organization. As of now, there are no vaccines or antiviral drugs declared to be officially useful against the infection. Saikosaponin is a group of oleanane derivatives reported in Chinese medicinal plants and are described for their anti-viral, anti-tumor, anti-inflammatory, anticonvulsant, antinephritis and hepatoprotective activities. They have also been known to have anti-coronaviral property by interfering the early stage of viral replication including absorption and penetration of the virus. Thus, the present study was undertaken to screen and evaluate the potency of different Saikosaponins against different sets of SARS-CoV-2 binding protein via computational molecular docking simulations. Docking was carried out on a Glide module of Schrodinger Maestro 2018-1 MM Share Version on NSP15 (PDB ID: 6W01) and Prefusion 2019-nCoV spike glycoprotein (PDB ID: 6VSB) from SARS-CoV-2. From the binding energy and interaction studies, the Saikosaponins U and V showed the best affinity towards both the proteins suggesting them to be future research molecule as they mark the desire interaction with NSP15, which is responsible for replication of RNA and also with 2019-nCoV spike glycoprotein which manage the connection with ACE2. 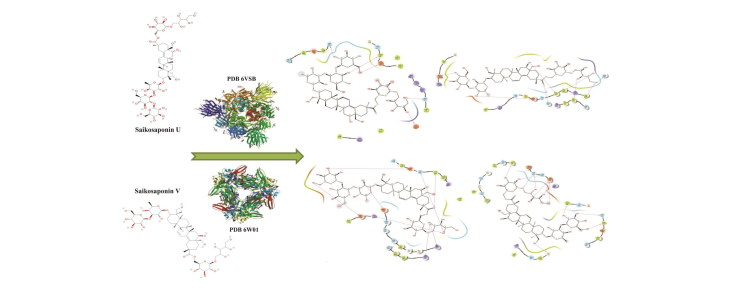
Communicated by Ramaswamy H. Sarma
Keywords: Saikosaponins, 2019-nCoV, 6W01, 6VSB, In-silico, Corona virus, virtual screening, molecular docking, Traditional Chinese Medicine
Introduction
In the last week of December 2019, Wuhan city, Hubei Province, China emerged with an outbreak of an acute resolved COVID-19 pneumonia, which has subsequently affected around 206 countries around the globe with a 6.1% fatality rate (Xu et al. 2020). The genome sequencing has revealed novel coronavirus SARS-CoV-2, formally known as 2019-nCoV as etiological agent for COVID-19 (Li et al., 2020). The genome of CoVs (enveloped, positive single-stranded RNA) is the largest genome of known RNA viruses with varying size (26 kb-32kb in length). Coronaviridae family can be divided in four genera comprising of α- and β-CoV, known to infect mammals, whereas δ- and γ- CoVs infect birds. Severe acute respiratory syndrome coronavirus i.e. SARS (2002) and the Middle East respiratory syndrome coronavirus i.e. MERS (2012) were viral pneumonia outbreaks due to β-CoVs and resulted in approximately 800 deaths (Song et al., 2019; Abdo & Eman, 2020). Whereas, this ‘Public Health Emergency of International Concern’ (PHEIC) declared due to widespread outbreak of a SARS-CoV-2 resulted in 1,773,086 confirmed cases of COVID-19, including 111,652 deaths, as of 05:00 pm CEST, 13 April 2020, reported to World Health Organization (WHO) (https://who.sprinklr.com) (World Health Organization. 2020).
The SARS-CoV-2 genome (30 kb in size) encodes a large non-structural polyprotein, open reading frame (ORF) 1a/b, which is further proteolytically cleaved to generate 15/16 proteins, five accessory proteins (ORF3a, ORF6, ORF7, ORF8 and ORF9) and four structural proteins (S, M, E and N) deliberated to be essential for SARS-CoV2 assembly and infection (Xu et al., 2020; Ortega et al., 2020; Boopathi et al., 2020).
Protrusion of the CoV spike (S) proteins from the virion envelope to cellular receptors and host cell proteases priming plays a vital role in regulation of entry of coronaviruses into cell. Key atomic-level interactions of the SARS-CoV spike protein receptor-binding domain (RBD) with its host receptor angiotensin-converting enzyme-2 (ACE-2) evidenced the regulation of human-to-human and cross-species transmissions of SARS-CoV (Chen & Du 2020). The recent report on the three-dimensional structure of the crystal structure of the 2019-nCoV main protease (PDB: 6LU7) revealed, that the protein is encoded within the viral genome (Liu et al., 2020), in complex with an inhibitor. Thus, both main protease and spike protein complexed with human ACE-2 receptor might be important targets for researchers to discover and develop vaccines and other therapeutic agents to control this new CoV.
In the absence of any validated approved therapeutic interventions this pandemic is spreading drastically all over the globe. Indeed, in current emmergent situation, the concept of drug repositioning might be a cost-effective and time-efficient option for meticulous development of possible therapeutic and/or prophylatic lead condidate(s) from well-known traditional and/or approved drugs (Li et al., 2019). Recent studies comprising reverse vaccinology and via integrated computational approaches have highlighted repurposing of FDA approved drugs and known drugs from various databases against various corona virus enzymes either individually or in combination to combat the virulence in COVID 19 (Muralidharan et al., 2020; Elmezayen et al., 2020; Khan et al., 2020; Gupta et al., 2020; Sarma et al., 2020; Pant et al., 2020; Enayatkhani et al. 2020; Khan et al., 2020). The interaction of ACE-2 and with spike protein with furin cleavage site resulting in the invasion of SARS-CoV-2 is also reported (Hasan et al. 2020). Additionally, several phytochemicals were found effective in inhibiting the 3CLpro of SARS-CoV and blocking the entry of SARS-CoV into host cells viz. phenolic compounds of Isatis indigotica root (Lin et al., 2005), baicalin (Chen et al., 2004), glycyrrhizin (Cinatl et al., 2003), quercetin (Chen et al., 2006) and Saikosaponin (Cheng et al., 2006). Moreover, ‘leads’ from natural products has also been explored for their efficacy against SARS-CoV-2 (Aanouz et al., 2020), and hesperetin, a natural citrus fruit flavonoid is found promising in suppressing the cleavage activity of the 3 C-like protease (3CLpro) of SARS-CoV in cell-free and cell-based assays (Chen et al., 2020).
Saikosaponins represent a group of oleanane derivatives, usually glucosides and are major bioactive compounds found in plants of Traditional Chinese Medicine (TCM) such as Bupleurum spp., Heteromorpha spp. and Scrophularia scorodonia (Li et al. 2018a; Li et al., 2018b). Saikosaponins have been reported for various biological activities like anti-viral, anti-tumor, anti-inflammatory, anticonvulsant, antinephritis and hepatoprotective activity both in-vivo and in-vitro (Chen et al., 2015; Yang et al., 2017; Xiao et al., 2018). Saikosaponins exhibit immunomodulatory effects with enhanced production of immunosuppressive mediators i.e. T helper (Th) 1/Th2 cells, IL-4 and IL-10 and suppression of pro-immune mediators (IFN-γ and TNF-α in splenocytes) (Huang et al., 2017). Inhibitory potential of Saikosaponins for HBV-DNA replication and HBsAg secretion, without inhibition of cell proliferation has been found to be superior to lamivudine (well-known antiviral drug) (Lin et al., 2015). Moreover, Saikosaponins (A, B2, C and D) have also been reported for their anti-coronaviral efficacy by interfering the early stage of viral replication including absorption and penetration of the virus (Cheng et al., 2006). A recent study, performed by Goswami and Bagehi (2020) reported the potential of Saikosaponins against SARS-CoV-2, showing favorable affinity towards RBD region of the spike glycoprotein using docking simulation study (Goswami & Bagchi 2020). However, it will be interesting to investigate the potency of different class of Saikosaponins towards the other protein such as NSP15 and fusion spike (S1 and S2) protein, which will give a clear indication about the selectivity of the Saikosaponins towards SARS-CoV-2. Therefore, in the current investigation attempt has been made to screen and evaluate the potency of different Saikosaponins against different sets of SARS-CoV-2 binding protein via computational molecular docking simulations.
Material and methods
Protein preparation
The docking simulation study of total 23 Saikosaponins (Supplementary file) was performed on the 1.9 A crystal structure of NSP15 Endoribonuclease from SARS CoV-2 in the complex with a citrate (PDB ID: 6W01) and prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up (PDB ID: 6VSB) which was retrieved from protein data bank (https://www.rcsb.org). The protein preparation wizard of Schrödinger maestro 2018-1 MM share version was used to prepare the structure by adding hydrogen, treating metal, deleting water molecules and assigning partial charges by using the OPLS-2005 force field, then assigning protonation states and determining restrained and further partial energy was minimized with 0.3 Å RMSD limit. The binding sites were defined after removing the ligand then grid was generated using a grid box volume of 10_10_10 Å.
Ligand preparation
The 2 D structure of ligands were downloaded from pubchem compound database (https://pubchem.ncbi.nlm.nih.gov) in SDF format and uploaded to the work space. Then ligprep module of maestro was used to generate the 3 D structure and OPLS-2005 force field was used to generate the different conformation of each ligand. The stable conformer of each Saikosaponin with minimum potential energy was further processed.
Protein ligand docking
Glide module of Schrödinger suite was used to dock the each ligand into the identified binding site of grid, and the lowest binding pose of each docking run was retained. The results of simulations were analyzed using glide XP visualizer, and the important active site interactions were analyzed along with the scoring functions.
Results
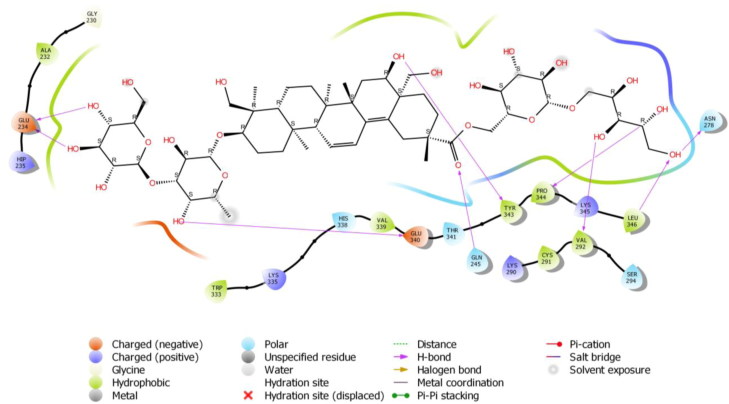
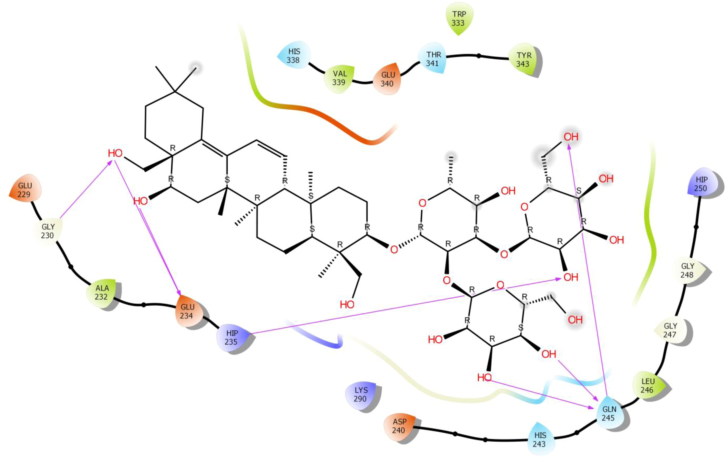
To check the potency of different Saikosaponins, the first docking simulation study was done on the crystal structure of NSP15 Endoribonuclease from SARS CoV-2 (PDB Code: 6W01). The results revealed that Saikosaponin V was properly positioned into the binding pocket constructed by polar Asn 278, Thr 341, Ser 294, Gln 245 His 338, hydrophobic Trp 333, Pro 344, Gly 230, Leu 346, Tyr 343, Val 339, Ala 232, Cys 291, Val 292 charged Glu 340, Lys 345, Lys 335, Lys 290, Hip 235 and Gln 245 amino acids with binding energy -8.35 Kcal/mol (Figure 1). Saikosaponin compounds with binding interaction parameter i.e. binding energy with the PDB: 6W01 and 6VSB of SARS-CoV-2 is depicted in Table 1 whereas details of binding interactions of top five Saikosaponins against both protein targets comprising details of protein residues involved in hydrogen bonding and hydrophobic interactions are summarized in Tables 2 and 3.
Figure 1.
Binding-interaction analysis of Saikosaponin V with NSP15 endoribonuclease (PDB ID: 6W01).
Table 1.
List of compounds with binding interaction parameter i.e. binding energy with the PDB: 6W01 and 6VSB of SARS-CoV-2.
| Sr. No. | Pubchem CID | Name | Binding energy Kcal/mol |
|
|---|---|---|---|---|
| (6W01) | (6VSB) | |||
| 1 | 100958093 | Saikosaponin V | −8.358 | −8.299 |
| 2 | 100958092 | Saikosaponin U | −7.272 | −8.429 |
| 3 | 46783811 | Saikosaponin C | −6.981 | −7.274 |
| 4 | 100962152 | Saikosaponin K | −6.79 | −6.251 |
| 5 | 124222337 | Saikosaponin 1b | −6.367 | −6.195 |
| 6 | 100962153 | Saikosaponin N | −6.192 | −6.142 |
| 7 | 132533303 | Saikosaponin R | −6.139 | −6.615 |
| 8 | 10534262 | Saikosaponin O | −5.783 | −5.538 |
| 9 | 100962151 | Saikosaponin Q | −5.728 | −5.518 |
| 10 | 101826550 | Saikosaponin G | −5.663 | −5.695 |
| 11 | 21637636 | Saikosaponin B4 | −5.587 | −5.808 |
| 12 | 21636280 | Saikosaponin H | −5.531 | −4.775 |
| 13 | 21637635 | Saikosaponin B3 | −5.175 | −6.19 |
| 14 | 102316649 | Saikosaponin 2 | −4.955 | −4.085 |
| 15 | 441945 | Saikosaponin BK1 | −4.887 | −6.195 |
| 16 | 167928 | Saikosaponin A | −4.725 | −5.143 |
| 17 | 21637642 | Saikosaponin B2 | −4.697 | −6.19 |
| 18 | 9875547 | Saikosaponin B1 | −4.61 | −6.096 |
| 19 | 21598300 | Saikosaponin F | −4.396 | −5.529 |
| 20 | 21636282 | Saikosaponin I | −4.3 | −5.794 |
| 21 | 102123130 | 16-Keto Saikosaponin A | −3.973 | −5.301 |
| 22 | 21637632 | Saikosaponin E | −3.576 | −4.905 |
| 23 | 107793 | Saikosaponin D | −3.738 | −5.638 |
Table 2.
Binding interactions of different Saikosaponins with the active site of SARS-CoV-2 NSP15 endoribonuclease (PDB ID: 6W01).
| Sr. No. | Ligand | Interaction (PDB-6W01) |
|
|---|---|---|---|
| H-Bonding | Hydrophobic Pocket | ||
| 1 | Saikosaponin V | Glu 234, Asn 278, Pro 344, Val 292, Tyr 343, Leu 346, Glu 340, Gln 245 | Trp 333, Pro 344, Gly 230, Leu 346, Tyr 343, Val 339, Ala 232, Cys 291, Val 292 |
| 2 | Saikosaponin U | Asp 240, Hip 250, Lys 290, Hip 235 , Thr 341, Glu 234 , Tyr 343, Glu 340 | Val 339, Leu 246, Pro 344, Tyr 343, Trp 333, Val 292, Cys 293, Ala 232 |
| 3 | Saikosaponin C | Lys 290, Gln 245, Hip 235, Thr 341, Asn 278, Leu 346 | Val 292, Leu 346, Pro 344, Tyr 343, Trp 333, Cys 293, Leu 246, Gly 247, Gly 248 |
| 4 | Saikosaponin K | Val 292, Hip 250, Gly 248, Tyr 343, Glu 340 | Val 292, Leu 246, Tyr 343, Trp 333, Cys 293, Gly 247, Gly 248, Cys 291, Met 331, Ala 232 |
| 5 | Saikosaponin 1b | Gly 245, Gln 245, Glu 234,Gly 230 | Val 339, Leu 246, Tyr 343, Trp 333, Gly 247, Gly 248, Ala 232, Gly 230 |
Table 3.
Binding interactions of different Saikosaponins with the active site of SARS-CoV-2 spike glycoprotein (PDB ID: 6VSB).
| Sr. No. | Ligand | Interaction (PDB-6VSB) |
|
|---|---|---|---|
| H-Bonding | Hydrophobic Pocket | ||
| 1 | Saikosaponin U | Phe 346, Ser 402, Asn 271, Glu 257, Asp 345 | Trp 270, Pro 343, Phe 346, Gly 348, Phe 403, Leu 405, Phe 371, Tyr 313 |
| 2 | Saikosaponin V | Phe 346, Ser 402, Thr 460, Ile 475, Asp 459, Thr 461, Phe 403, Gly 298 | Val 299, Leu 307, Phe 346, Phe 403, Leu 405, Phe 429, Leu 434, Ile 475, Pro 477 |
| 3 | Saikosaponin C | Phe 403, Asp 345, Lys 341, Glu 372 | Val 299, Tyr 313, Tyr 338, Leu 342, Pro 343, Phe 346, Pro 370, Phe 371, Phe 403, Leu 405 |
| 4 | Saikosaponin K | Phe 403, Ser 402, Glu 404 Asp 345 | Tyr 313, Leu 342, Pro 343, Phe 346, Pro 370, Phe 371, Phe 403, Leu 405 |
| 5 | Saikosaponin R | Thr 460, Ile 475, Ala 458, Thr 435 | Val 299, Leu 304, Leu 307, Leu 405 Leu 434, Phe 346, 429, Ile 475, Pro 477, Tyr 497, Gly 298, Ala 458, Ile 457 |
Among them, Glu 234 exhibited bi-hedral H-bonding with hydroxyl group of oxane ring (tetrahydropyran) of Saikosaponin V and other hydroxyl group of Saikosaponin V showed H-bonding with Asn 278, Pro 344, Val 292, Tyr 343 and Leu 346. Hydroxyl group of oxane ring of Saikosaponin V showed H-bonding with Glu 340 and carboxyl group of Saikosaponin V showed H-bonding with Glu 245.
Saikosaponin U was found to interact with the binding pocket constructed by polar Ser 294, Thr 341, Ser 242, His 338, His 243, Gln 245 hydrophobic Val 339, Leu 246, Pro 344, Tyr 343, Trp 333, Val 292, Cys 293, Ala 232 charged Lys 345, Glu 234, Asp 240, Glu 340, Hip 250, Lys 290 and Hip 235 amino acids with binding energy -7.27 Kcal/mol. Hydroxyl group of oxane ring of Saikosaponin U showed H-bonding with Asp 240 and Glu340 (Figure 2).
Figure 2.
Binding-interaction analysis of Saikosaponin U with NSP15 endoribonuclease (PDB ID: 6W01).
The other hydroxyl group of Saikosaponin U showed H-bonding with Hip 250, Lys 290, Hip 235 and Thr 341. The hydroxymethyl group of oxane ring of Saikosaponin U showed H-bonding with Glu 234 and carboxyl group of Saikosaponin U showed H-bonding with Tyr 343.
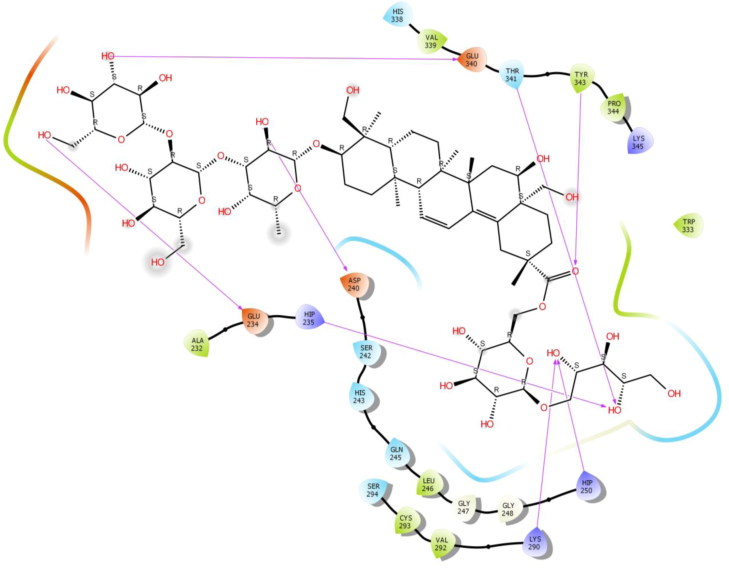
Saikosaponin C was properly positioned into the binding pocket constructed by polar Ser 244, Thr 341, Ser 294, His 243, Gln 245, Asn 278, hydrophobic Val 292, Leu 346, Pro 344, Tyr 343, Trp 333, Cys 293, Leu 246, Gly 247, Gly 248 charged Lys 345, Asp 240, Hip 250, Lys 290 and Hip 235 amino acids with binding energy -6.98 Kcal/mol. The hydroxyl group of oxane ring of Saikosaponin C exhibited bi-hedral H-bonding with Lys 290 and other hydroxyl group of oxane ring of Saikosaponin C showed H-bonding with Hip 235, Thr 341 and Asn 278 and tetrahydrofuran ring of Saikosaponin C showed H-bonding with Gln 245 (Figure 3).
Figure 3.
Binding-interaction analysis of Saikosaponin C with NSP15 endoribonuclease (PDB ID: 6W01).
Saikosaponin K was found to interact with the binding pocket constructed by polar Ser 294, Thr 341, His 338, Gln 245, hydrophobic Val 292, Leu 246, Tyr 343, Trp 333, Cys 293, Gly 247, Gly 248, Cys 291, Met 331, Ala 232 charged Lys 345, Hip 250, Lys 290, Lys 335, Glu 340 and Hip 235 amino acids with binding energy -6.79 Kcal/mol. Hydroxyl group of oxane ring of Saikosaponin K showed H-bonding with Val 292, Hip 250, Gly 248 while, hydroxymethyl group of oxane ring of Saikosaponin K showed H-bonding with Tyr 343 and other with Glu 340 (Figure 4).
Figure 4.
Binding-interaction analysis of Saikosaponin K with NSP15 endoribonuclease (PDB ID: 6W01).
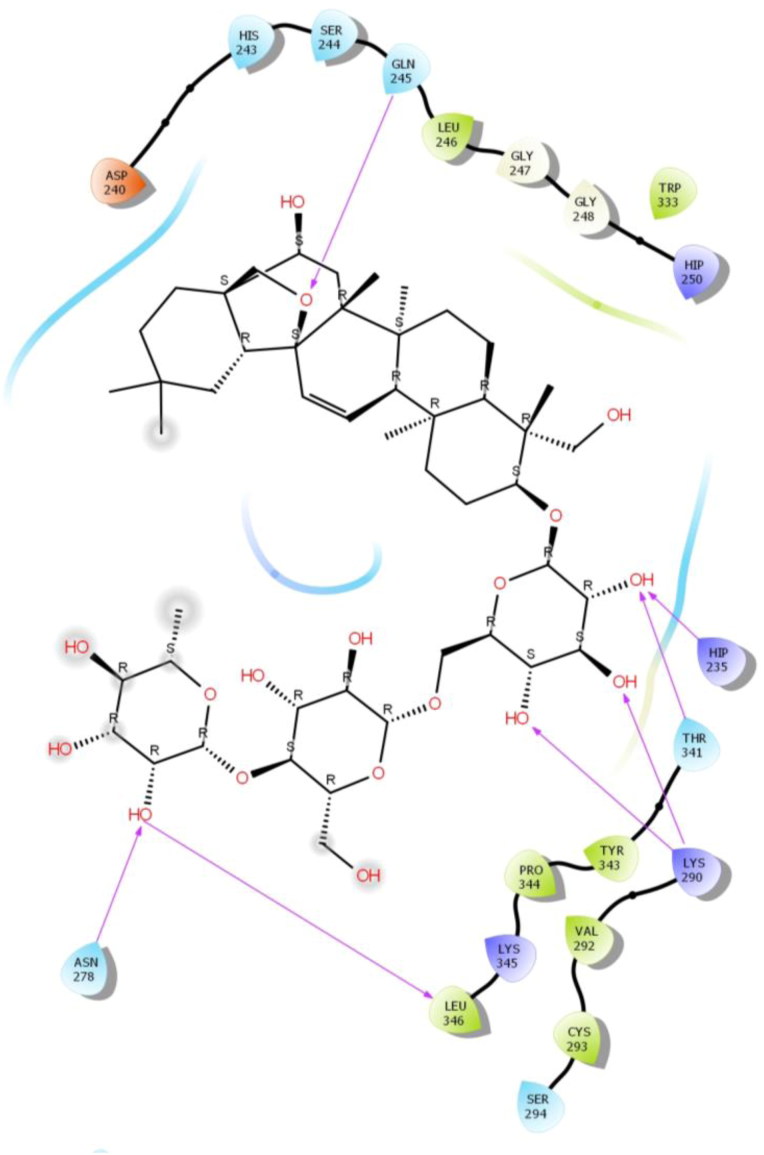
Saikosaponin 1 b was properly positioned into the binding pocket constructed by polar Thr 341, His 338, Gln 245, His 243, hydrophobic Val 339, Leu 246, Tyr 343, Trp 333, Gly 247, Gly 248, Ala 232, Gly 230 charged Lys 290, Hip 250, Glu 340, Asp 240, Glu 234, Glu 229 and Hip 235 amino acids with binding energy -6.36 Kcal/mol.
Among them, Gly 245 exhibited bi-hedral H-bonding with hydroxyl group of oxane ring of Saikosaponin 1 b and other hydroxyl group of Saikosaponin 1 b Glu 234. Hydroxymethyl group of oxane ring of Saikosaponin 1 b showed H-bonding with Gln 245 and other hydroxymethyl group with Glu 234 and Gly 230 (Figure 5).
Figure 5.
Binding-interaction analysis of Saikosaponin 1 b with NSP15 endoribonuclease (PDB ID: 6W01).
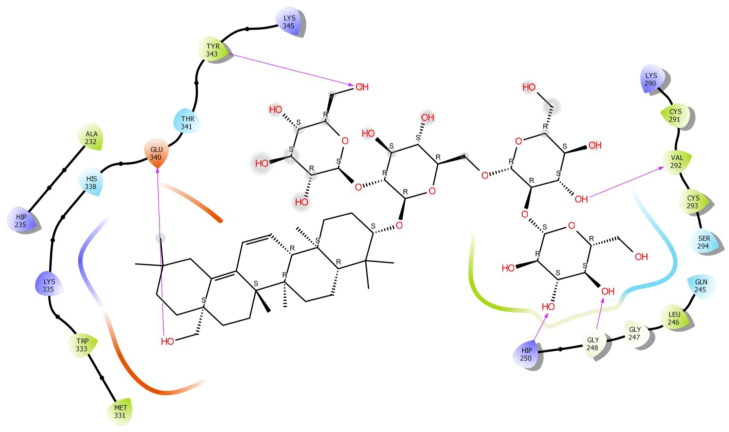
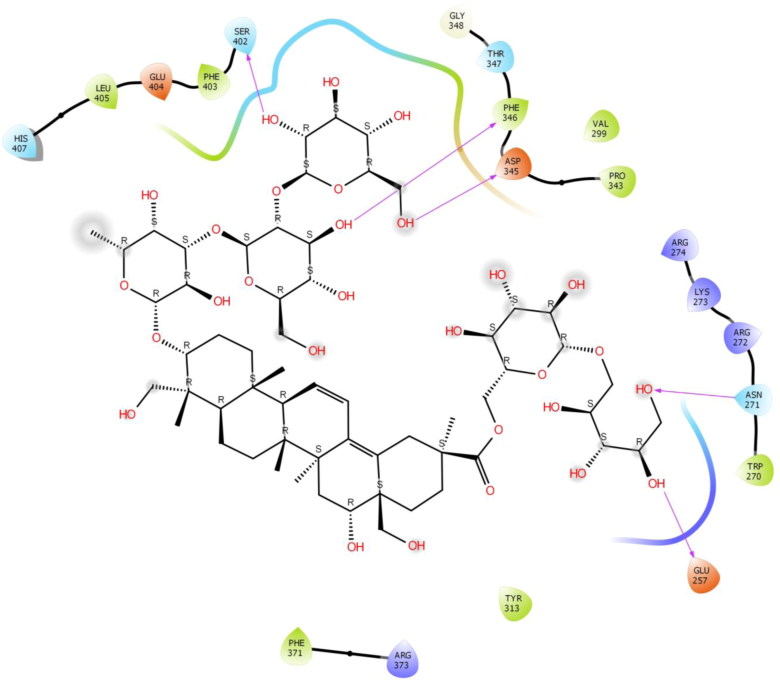
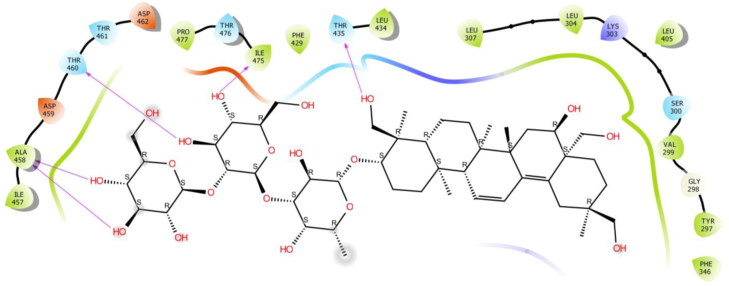
The second study was performed on the crystal structure of COVID-19 main protease enzyme (PDB Code: 6VSB). The results revealed that Saikosaponin U was properly positioned into the binding pocket constructed by polar Asn 271, Thr 347, Ser 402, His 407, hydrophobic Trp 270, Pro 343, Phe 346, Gly 348, Phe 403, Leu 405, Phe 371, Tyr 313 charged Glu 257, Arg 272, Lys 273, Arg 274, Asp 345, Arg 373 and Glu 404 amino acids with binding energy -8.42 Kcal/mol.
Hydroxyl group of oxane ring of Saikosaponin U showed H-bonding with Phe 346, Ser 402 and other hydroxyl group of Saikosaponin U showed H-bonding with Asn 271 and Glu 257, Hydroxymethyl group of oxane ring of Saikosaponin U showed H-bonding with Asp 345 (Figure 6).
Figure 6.
Binding-interaction analysis of Saikosaponin U with receptor-binding domain of spike glycoprotein (PDB ID: 6VSB).
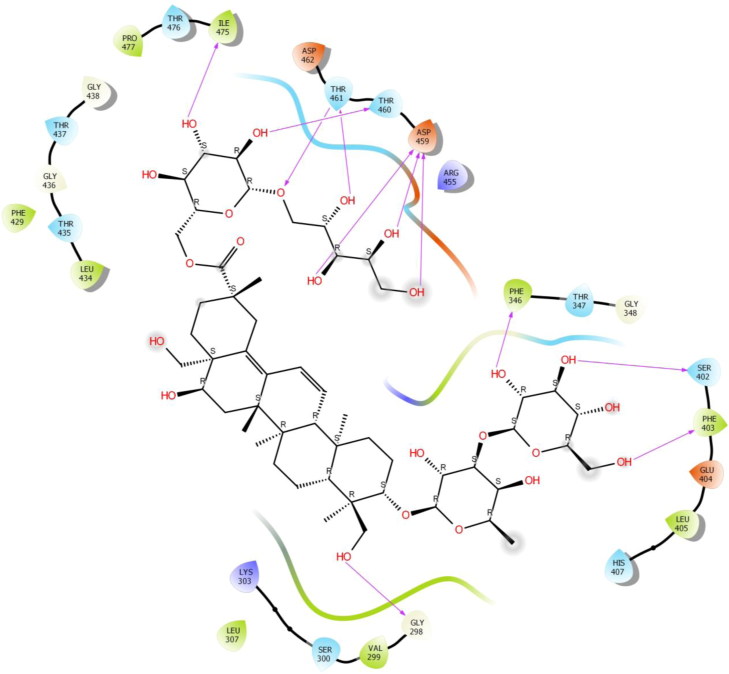
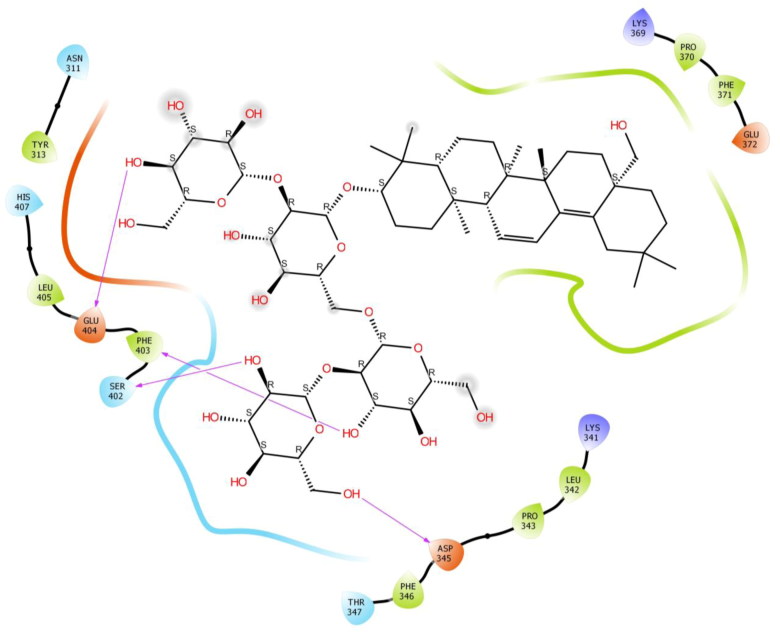
Saikosaponin V was found to interact with the binding pocket constructed by polar Ser 300, Thr 347, Ser 402, His 407, Thr 435, Thr 437, Thr 460, Thr 461, Thr 476 hydrophobic Val 299, Leu 307, Phe 346, Phe 403, Leu 405, Phe 429, Leu 434, Ile 475, Pro 477 charged Lys 303, Glu 404, Arg 455, Asp 459 and Asp 462 amino acids with binding energy -8.29 Kcal/mol. Phe 346, Ser 402, Thr 460, Ile 475 exhibited H-bonding with Hydroxyl group of oxane ring of Saikosaponin V and other hydroxyl group of Saikosaponin V showed H-bonding with Asp 459 and Thr 461, Hydroxymethyl group of oxane ring of Saikosaponin V showed H-bonding with Phe 403, Hydroxymethyl group of other ring showed H-bonding with Gly 298 and ether linkage showed H-bonding with Thr 461 (Figure 7).
Figure 7.
Binding-interaction analysis of Saikosaponin V with receptor-binding domain of spike glycoprotein (PDB ID: 6VSB).
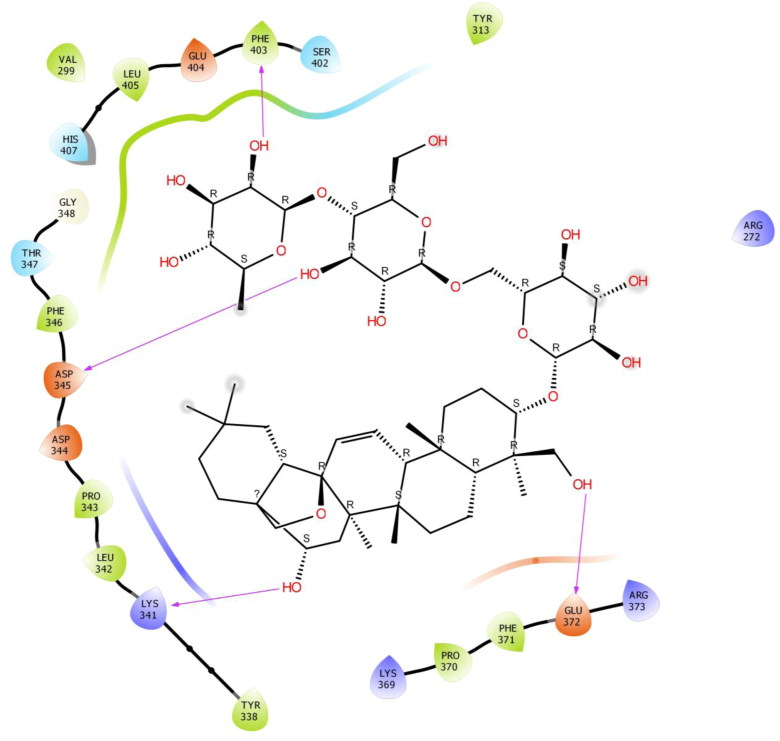
Saikosaponin C was properly positioned into the binding pocket constructed by polar Thr 347, Ser 402, His 407, hydrophobic Val 299, Tyr 313, Tyr 338, Leu 342, Pro 343, Phe 346, Pro 370, Phe 371, Phe 403, Leu 405 charged Lys 341, Asp 344, Asp 345, Lys 369, Glu 372, Arg 373 and Glu 404 amino acids with binding energy -7.27 Kcal/mol. Hydroxyl group of oxane ring of Saikosaponin C showed H-bonding with Phe 403 and Asp 345, other hydroxyl group of Saikosaponin C showed H-bonding with Lys 341 and Glu 372 (Figure 8).
Figure 8.
Binding-interaction analysis of Saikosaponin C with receptor-binding domain of spike glycoprotein (PDB ID: 6VSB).
Saikosaponin K was found to interact with the binding pocket constructed by polar Asn 311, Thr 347, Ser 402, His 407 hydrophobic Tyr 313, Leu 342, Pro 343, Phe 346, Pro 370, Phe 371, Phe 403, Leu 405 charged Lys 341, Asp 345, Lys 369, Glu 372 and Glu 404 amino acids with binding energy -6.25 Kcal/mol. The hydroxyl group of oxane ring of Saikosaponin K exhibited H-bonding with Phe 403, Ser 402, Glu 404. Hydroxymethyl group of oxane ring of Saikosaponin V showed H-bonding with Asp 345 (Figure 9).
Figure 9.
Binding-interaction analysis of Saikosaponin K with receptor-binding domain of spike glycoprotein (PDB ID: 6VSB).
Saikosaponin R was properly positioned into the binding pocket constructed by polar Ser 300, Thr 435, Thr 460, Thr 461, Thr 476, hydrophobic Val 299, Leu 304, Leu 307, Leu 405 Leu 434, Phe 346, 429, Ile 475, Pro 477, Tyr 497, Gly 298, Ala 458, Ile 457 charged Lys 303, Asp 459 and Asp 462 amino acids with binding energy -6.19 Kcal/mol. Hydroxyl group of oxane ring of Saikosaponin R showed H-bonding with Thr 460, Ile 475 and bi-hedral H-bonding with Ala 458, hydroxymethyl group of oxane ring of Saikosaponin R showed H-bonding with Thr 435 (Figure 10).
Figure 10.
Binding-interaction analysis of Saikosaponin R with receptor-binding domain of spike glycoprotein (PDB ID: 6VSB).
Discussion
The corona viruses are said to be enveloped RNA viruses having the largest genome among all RNA viruses and containing a replicase gene covered with nonstructural proteins (NSP). Among these NSPs, the NSP15 is a nidoviral RNA uridylate-specific Endoribonuclease (NendoU) which carries a catalytic C-terminal domain, which is specific for uridine (U). A recent study has reported that the NSP15 NendoU activity is mainly responsible for the interference of protein replication and innate immune response (Deng et al., 2017) and therefore, NSP15 is said to be very essential for biological progression of coronavirus.
The studies conducted on the structures of SARS- and MERS-CoV NSP15 revealed that the virus protein constitutes of hexamers i.e. dimers of trimers. Further, each monomeric unit is comprised of ∼345 amino acids which are folded into three domains viz. N-terminal, middle and NendoU C-terminal catalytic domain, where the NendoU C-terminal catalytic domain comprises two β-sheets which are antiparallel. Study have reported that the NSP15 is responsible for cutting the double-stranded (ds) RNA substrates with specificity via the Mn + 2-dependent Endoribonuclease activity. The main active sites, conserved among SARS-CoV-2, SARS-CoV and MERS-CoV proteins are located in a shallow groove between the two β-sheets that contain six key amino acids viz. His235, His250, Lys290, Thr341, Tyr343, and Ser294.Among them, His235, His250, Lys290 constitute the catalytic triad, His235 serves as general acid, His250 acts a base, while Ser294 together withTyr343 have been found to govern U specificity.
Our observations revealed that, the Saikosaponin V having two oxane rings substituted with 3 hydroxyl group and the side chain contains 4 hydroxyl and an ester linkage showed very high binding with active site having a furrow between the two β-sheets, carries amino acids Lys290, Thr341, Tyr343 and Ser29. Among these Lys290 have been proposed to comprise the catalytic triad and Ser294 with Tyr343 are believed to govern U specificity. Four hydroxyl group of side chain provides a proper grip between hydrophobic pockets. Saikosaponin U having 2 hydroxyl and hydroxymethyl group of oxane ring and other 2 hydroxyl group and an ester linkage in side chain showed very high binding with active site carries key residues Lys290, Thr341, Tyr343, and Ser294. An ester linkage of side chain provides a proper grip between hydrophobic pockets. Saikosaponin C having two oxane ring substituted with 4 hydroxyl groups and the other chain containing furan ring, showed very high binding with active site carries residues Lys290 Thr341, Tyr343, and Ser294. Hydroxyl group of oxane ring provides a proper grip between hydrophobic pockets. Saikosaponin K is having 3 hydroxyl and hydroxymethyl group of oxane ring and other hydroxymethyl group in other chain showed very high binding with active site carrier key residues Lys290, Thr341, Tyr343, and Ser294. Hydroxyl and hydroxymethyl group of oxane ring provides a proper grip between hydrophobic pockets. Saikosaponin 1 b having two oxane ring substituted with 3 hydroxyl and hydroxymethyl group and the other chain contains hydroxyl and hydroxymethyl group showed very high binding with active site carries residues Lys290 Thr341and Tyr343. Hydroxymethyl group of other ring provides a proper grip between hydrophobic pocket.
It has been reported that, 2019-nCoV uses a densely glycosylated, homotrimeric class I fusion spike protein to make entry into the host cells. This S protein occurs in a metastable prefusion conformation which undergoes structural rearrangement so that the viral membrane gets fuses with the host cell membrane (Li, 2016; Bosch et al., 2003). This fusion process is accelerated by binding of S1 subunit to the host-cell receptor and by transition of the S2 subunit to a highly stable postfusion conformation (Walls et al., 2017). The receptor-binding domain (RBD) of S1 subunit undergoes hinge-like conformational movement to engage the host-cell receptor, which transiently hide/expose the determinants of receptor binding. Further, the S1/S2 subunits have been reported to possess arginine-arginine-alanine-arginine (RRAR) a protease cleavage site. Recent studies have also reported that, 2019-nCoV S and SARS-CoV S shares the same functional host cell receptor i.e. angiotensin-converting enzyme 2 (ACE2). In addition, the 2019-nCoV RBD-SD1 106 fragment (S residues from 319–591) have reported to have ACE2 binding site. The affinity with which ACE2 bounds to 2019-nCoV S ectodomain is 10- to 20-fold higher than ACE2 binding to SARS-CoV S. Due to the higher binding affinity of 2019-nCoV S for human ACE2, the spread of 2019-nCoV became apparently much prevalent from human-to-human (Chan et al., 2020).
Simulation study on crystal structure of 2019-nCoV spike glycoprotein revealed that all the Saikosaponin buried properly into the RBD of S1 subunit and also positioned into the N-terminal domain. Due to the help of RBD this virus makes connection with ACE 2 receptor and gets entry into the host cell. Based on the binding energy, Saikosaponin U and V were the top ranked ligand. The Saikosaponin U has octadecahydropicene ring in which one side is substituted with 3 oxane ring and other side with one oxane ring. The Saikosaponin V having the same ring substituted with 2 oxane ring at one side whereas other side with one oxane rings. The extra oxane ring in Saikosaponin U provides better grip into the binding and hydrophobic pocket. The Saikosaponin U showed good interaction with triple arginine, which is responsible for recognition and cleavage of furin, this step facilitates the 2019-nCoV spike with ACE 2 receptor.
Saikosaponin R also having two oxane ring substituted with 4 hydroxyl and the side chain contains one hydroxymethyl group showed very high binding affinity and buried into the domain of binding site, out of which one hydroxyl group of oxane ring which provides a proper grip between hydrophobic pocket.
Conclusion
In a nutshell, from all the simulation studies on both proteins, it can be concluded that Saikosaponin U and V have octadecahydropicene ring with substituted oxane ring but one more extra oxane ring of Saikosaponin U provide better grip into the wide spread binding pocket (residues from 319-519) of spike glycoprotein. The smaller structure of Saikosaponin V makes it best fit ligand into the narrow binding pocket of NSP 15. Thus we can say that Saikosaponin U and V would be the future research interest ligand as they mark the desire interaction with NSP15, which is responsible for replication of RNA and also with 2019-nCov spike glycoprotein which manage the connection with ACE2.
Supplementary Material
Glossary
Abbreviations
- ACE2
Angiotensin Converting Enzyme 2
- WHO
World Health Organization
- SARS
Severe Acute Respiratory syndrome
- MERS
Middle East Respiratory Syndrome
- SARS-COV-2
Coronavirus Disease Strain
- ORF
Open Reading Frame
- CoV
Corona Virus
- N Protein
Nucleocapsid protein
- 2019-nCoV
2019 Novel Coronavirus
- COVID-19
Coronavirus Disease 2019
- SDF
Structure Data File
- 3CLpro
Chymotrypsin-like protease
- TCM
Traditional Chinese Medicine
- Th
T helper
- IL
Interleukin
- IFN
Interferon
- TNF
Tumor Necrosis Factor
- RBD
Receptor Binding Domain
- RMSD
Root Mean Square Deviation
- OPLS
Optimized Potentials for Liquid Simulations
- NSP
Nonstructural Protein
Acknowledgements
All authors are thankful to Dr. Prajwal Nandekar, Senior Scientist, Schrodinger for helping in computational studies in present investigation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., & Hao P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. 10.1007/s11427020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19:Computational investigations. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdo A. E., & Eman B. A. (2020). Novel Guanosine Derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 Coronavirus Structure, Mechanism of Action, Antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B. J., Van der Zee R., de Haan C. A., & Rottier P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801–8811. 10.1128/JVI.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., Xing F., Liu J., Yip C. C.-Y., Poon R. W.-S., Tsoi H.-W., Lo S. K.-F., Chan K.-H., Poon V. K.-M., Chan W.-M., Ip J. D., Cai J.-P., Cheng V. C.-C., Chen H., Hui C. K.-M., & Yuen K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Chan K. H., Jiang Y., Kao R. Y., Lu H. T., Fan K. W., Cheng V. C., Tsui W. H., Hung I. F., Lee T. S., Guan Y., Peiris J. S., & Yuen K. Y. (2004). In-vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. Journal of Clinical Virology, 31(1), 69–75. 10.1016/j.jcv.2004.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., & Du Q. (2020). Potential natural compounds for preventing 2019-nCoV infection. Preprints, 2020, 2020010358 10.20944/preprints202001.0358.v3 [DOI] [Google Scholar]
- Chen J., Duan M., Zhao Y., Ling F., Xiao K., Li Q., Li B., Lu C., Qi W., Zeng Z., Liao M., Liu Y., & Chen W. (2015). Saikosaponin A inhibits influenza A virus replication and lung immunopathology. Oncotarget, 6(40), 42541–42556. 10.18632/oncotarget.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O. W., Zhu W., Puah C. M., Shen X., & Jiang H. (2006). Binding interaction of quercetin-3-beta-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): Structure-activity relationship studies reveal salient pharmacophore features. Bioorganic and Medicinal Chemistry, 14(24), 8295–8306. 10.1016/j.bmc.2006.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., & Zhao Z. J. (2020). Structure analysis of the receptor binding of 2019-nCoV. Biochemical and Biophysical Research Communications, 17(20), 30339–30339. 10.1016/j.bbrc.2020.02.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P. W., Ng L. T., Chiang L. C., & Lin C. C. (2006). Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clinical and Experimental Pharmacology and Physiology, 33(7), 612–216. 10.1111/j.1440-1681.2006.04415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., & Doerr H. W. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. The Lancet, 361(9374), 2045–2046. 10.1016/S0140-6736(03)13615-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R. C., O’Brien A., Mielech A. M., Yi G., Kao C. C., & Baker S. C. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National Academy of Sciences, 114(21), E4251–E4260. 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): Insilico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami T., & Bagchi B. (2020). Molecular docking study of receptor binding domain of SARS-CoV-2 spike glycoprotein with saikosaponin, a triterpenoid natural product. Preprints 2020. 10.26434/chemrxiv.12033774.v1 [DOI]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zheng X., Yang X., & Fan S. (2017). Stimulation of osteogenic differentiation by saikosaponin-A in bone marrow stromal cells Via WNT/beta-catenin pathway. Calcified Tissue International, 100(4), 392–401. 10.1007/s00223-017-0242-y [DOI] [PubMed] [Google Scholar]
- Khan R. J., Kumar Jha R., Muluneh Amera G., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Li C. C., Wang X. J., & Wang H. R. (2019). Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discovery Today, 24(3), 726–736. 10.1016/j.drudis.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K. S. M., Lau E. H. Y., Wong J. Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., … Feng Z. (2020). Early transmission dynamics in Wuhan, China, of novel Coronavirus-infected pneumonia. New England Journal of Medicine, 382(13), 1199–1207. 26 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Q., Song Y. N., Wang S. J., Rahman K., Zhu J. Y., & Zhang H. (2018. b). Saikosaponins: A review of pharmacological effects. Journal of Asian Natural Products Research, 20(5), 399–411. 10.1080/10286020.2018.1465937 [DOI] [PubMed] [Google Scholar]
- Li X., Li X., Huang N., Liu R., & Sun R. (2018. a). A comprehensive review and perspectives on pharmacology and toxicology of saikosaponins. Phytomedicine, 50, 73–87. 15 10.1016/j.phymed.2018.09.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. W., Tsai F. J., Tsai C. H., Lai C. C., Wan L., Ho T. Y., Hsieh C. C., & Chao P. D. (2005). Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Research, 68(1), 36–42. 10.1016/j.antiviral.2005.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. T., Chung C. Y., Hsu W. C., Chang S. P., Hung T. C., Shields J., Russell R. S., Lin C. C., Li C. F., Yen M. H., Tyrrell D. L., Lin C. C., & Richardson C. D. (2015). Saikosaponin b2 is a naturally occurring terpenoid that efficiently inhibits hepatitis C virus entry. Journal of Hepatology, 62(3), 541–548. 10.1016/j.jhep.2014.10.040 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang B., Jin Z., Yang H., & Rao Z. (2020). The crystal structure of COVID-19 main protease in complex with an inhibitor N3. 10.2210/pdb6LU7/pdb [DOI]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Ortega J. T., Serrano M. L., Pujol F. H., & Rangel H. R. (2020). Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target. EXCLI Journal, 19, 400–409. 10.17179/excli2020-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Prasad Dhibar D., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., & Qin C. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses, 11(1), 59 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C., Tortorici M. A., Snijder J., Xiong S., Bosch B. J., Rey F. A., & Veesler D. (2017). Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proceedings of the National Academy of Sciences, 114(42), 11157–11162. 10.1073/pnas.1708727114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). Coronavirus Disease (COVID-2019) https://who.sprinklr.com
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., & Wang F.-S. (2020). Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet Respiratory Medicine, 8(4), 420–422. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen X., Jiang C., He K., & Hu Y. (2017). Antiviral and immunoregulatory role against PCV2 in-vivo of Chinese herbal medicinal ingredients. Journal of Veterinary Research, 61(4), 405–410. 10.1515/jvetres-2017-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.