Abstract
Variants associated with modulation of c-reactive protein (CRP) and plasma lipids have been investigated for polygenic overlap with Alzheimer’s Disease risk variants. We examined pleiotropic genetic effects on cognitive impairment (CI) conditioned on genetic variants (SNPs) associated with systemic inflammation as measured by CRP and with plasma lipids using data from the Health and Retirement Study (HRS). SNP enrichment is observed for CI conditioned on the secondary phenotypes of plasma CRP and lipids. Fold enrichment of 100% – 800% was observed for increasingly stringent p-value thresholds for SNPs associated with CI conditional on plasma CRP, 80%–800% for Low-Density Lipoprotein (LDL) and 80% – 600% for total cholesterol (TC). Significant associations (FDR (q) ≤ 0.05) between CI, conditional with either CRP, LDL or TC are found for the locus on chromosome 19 that contains the APOE, TOMM40, APOC1, PVRL2 genes. Relative numbers of significant SNPs in each of the genes differed by the conditional associations with the secondary phenotypes. Biological interpretation of both the genetic pleiotropy results and the individual genome-wide association results show that the variants and proximal genes identified are involved in multiple pathological processes including cholesterol metabolism, inflammation and mitochondrial transport. These findings are potentially important for AD risk prediction and development of novel therapeutic approaches.
Keywords: cognitive impairment, genetic pleiotropy, inflammation, Health and Retirement Study
1. Introduction
The association between lipid levels, inflammation, and cognitive impairment is complex. Lipid metabolism is related to inflammatory markers(Ravaglia et al., 2007) and apoE is a ligand for cholesterol transport(Poirier et al., 2014). As many reports have noted the involvement of inflammatory and lipid metabolism pathways for AD,(Akiyama et al., 2000; Di Paolo and Kim, 2011) studies that investigate cognitive impairment conditioned on phenotypes that are linked with these pathways provide the potential to identify variants associated with the earliest changes in the development of AD, vascular contributions to cognitive impairment and dementia (VCID), and other neurodegenerative diseases of aging.
Genome-wide association studies (GWAS) for late-onset Alzheimer’s disease (AD) have demonstrated associations between AD and the Apolipoprotein E (APOE) genotype as well as genetic variants involved in inflammatory and microglial activation pathways(Broussard et al., 2012; Efthymiou and Goate, 2017; Gonzalez-Reyes et al., 2017; Pimenova et al., 2018). Variants associated with modulation of c-reactive protein (CRP) and plasma lipids have been identified by GWAS and investigated for polygenic overlap with AD risk variants with the results showing that SNPs associated with CRP and plasma lipids (High-Density Lipoprotein (HDL), Low-Density Lipoprotein (LDL) and triglycerides) are also associated with increased risk for AD (e.g. genetic pleiotropy)(Desikan et al., 2015). By testing for association using conditional phenotypes, novel loci were identified that were not reported in large AD case control studies, and they provided information about the involvement of pathways related to systemic inflammation, plasma lipids (HDL, LDL and triglycerides) and AD(Desikan et al., 2015). Using multiple phenotypes for genetic studies enables investigation of shared overlap between the phenotypes, genetic variants, genes and pathways. This approach has been utilized to examine genetic pleiotropy between multiple diverse diseases and phenotypes including schizophrenia and cognitive traits(Smeland et al., 2017), bipolar disorder(Andreassen et al., 2013b), multiple sclerosis(Andreassen et al., 2015b), cardiovascular disease risk factors(Andreassen et al., 2013a), as well as schizophrenia and educational attainment.(Le Hellard et al., 2017) Other diseases and phenotypes to which the approach has been applied include Parkinson’s disease and autoimmune diseases(Witoelar et al., 2017), blood lipids and immune-related diseases(Andreassen et al., 2015a) and others(Andreassen et al., 2014a; Andreassen et al., 2014b; Liu et al., 2013).
Cognitive impairment in aging develops as a consequence of genetic and environmental factors. APOE ε4 carriers have both a higher risk of developing cognitive impairment and AD and developing symptoms earlier than APOE e4 non-carriers(Raber et al., 2004). Large, consortium GWAS studies of AD have identified SNPs in genes involved in lipid metabolism Clusterin (CLU), ATP Binding Cassette Subfamily A Member 7 (ABCA7) and inflammation Complement C3b/C4b Receptor 1 (CR1), Major Histocompatibility Complex, Class II, DR Beta 5 (HLA-DRB5)(Hollingworth et al.; Jones et al., 2010; Lambert et al., 2013; Lambert et al., 2009; Naj et al.); rare coding variants in Phospholipase C Gamma 2 (PLCG2), ABI Family Member 3 (ABI3) and Triggering Receptor Expressed On Myeloid Cells 2 (TREM2) have also been reported that support a role for innate immune response contributing directly to the development of AD(Sims et al., 2017). Genome-wide analysis to identify variants affecting the rate of age-related cognitive decline have shown a strong association with APOE (Wilson et al., 2002), a coding variant in the CR1 gene(Chibnik et al., 2011), and with a common SNP that influences the expression of Phosphodiesterase 7A (PDE7A) and Mitochondrial Fission Regulator 1 (MTFR1) which are potential regulators of inflammation and oxidative injury(De Jager et al., 2012). For environmental factors, several longitudinal studies have shown an association between inflammatory markers including CRP and Interleukin 6 (IL-6) with dementia in the elderly(Teunissen et al., 2003; Weaver et al., 2002); one study noted that increased CRP levels may precede clinical symptoms of dementia by 25 years(Schmidt et al., 2002). While genetic factors are not modifiable, control of plasma lipids, e.g. reduction of LDL levels and systemic inflammation are risk factors that are potentially modifiable by lifestyle changes and/or medication(Georgoysopoulou et al., 2016; King et al., 2003; Livingston et al., 2017; Ma et al., 2008).
In the present study, we investigate pleiotropic genetic effects on cognitive impairment conditioned on genetic variants associated with systemic inflammation and with plasma lipids (Low-Density Lipoprotein (LDL), High-Density Lipoprotein (HDL), Total Cholesterol (TC)) in a large, nationally representative longitudinal panel study of aging of older adults: the Health and Retirement Study (HRS)(Juster and Suzman, 1995).
2. Materials and methods
The HRS has been assembled from several different studies including the Asset and Health Dynamics among the Oldest Old (AHEAD) study (begun in 1993) and the HRS, which began in 1992. Other studies have been folded into the HRS including the War Baby Study and the Children of the Depression Study. Today, these studies are collectively referred to as the HRS and form a large, longitudinally followed, representative cohort of Americans aged 50 and older. All participants provide informed consent; interviews take place biennially and are conducted by the Survey Research Center at the University of Michigan. The study protocol was approved by the University of Michigan Institutional Review Board (IRB). The current project was approved by the Wake Forest School of Medicine and the Duke University Medical Center IRBs.
2.1. Participants
HRS participants are geographically dispersed across the US and are a representative sample of older Americans. Repeated biennial cognitive evaluations begin once participants turn age 65. As such the current study includes only participants’ observations once they turn age 65 and is further limited to those who participated in DNA collection in 2006 or 2008.
2.2. DNA samples, genotyping, and imputation
HRS genotype data was obtained from dbGAP (www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000428.v1.p1.). Briefly, saliva samples were collected for DNA extraction and GWA studies in 2006 and 2008. A total of 12,507 study individuals were genotyped on the Illumina HumanOmni2.5–4v1 array. The median call rate is 99.7% and the error rate estimated from 336 pairs of study sample duplicates is 6 × 10−5. Details for the genotyping procedure and the quality control approach that was applied for all of the genotypes used in the analyses in this paper are provided in the Quality Control Report for Genotypic Data for dbGaP users of the HRS genotypic data (http://hrsonline.isr.umich.edu/sitedocs/genetics/HRS_QC_REPORT_MAR2012.pdf).
Genotype imputation to dense haplotype reference panels is considered an essential tool in GWAS. We used the imputed genotypes, provided by the University of Washington Genetics coordinating Center for all GWAS analyses. Details of the imputation process are provided at http://hrsonline.isr.umich.edu/sitedocs/genetics/1000G_IMPUTE2report_HRS2_2006_2008_2010.pdf. In brief, the world-wide reference panel from the 1000 Genomes project of all 1,092 samples from the phase I integrated variant set (v3, released March 2012) was used with the IMPUTE2 software for genotype imputation, The imputation output provided a set of 21,632,048 SNPs for the downstream analysis (GWA and conditional analysis).
For APOE, 1000 Genomes imputation dosages were used for estimation of APOE genotype based on the method described in the HRS documentation (http://hrsonline.isr.umich.edu/sitedocs/genetics/candidategene/FileDescription_Longevity.pdf).
2.3. CRP and plasma lipid measurement
Extensive documentation on the sample collection, laboratory procedures for CRP and plasma lipid measurements are provided in the HRS documentation, available at http://hrsonline.isr.umich.edu/modules/meta/bio2008/desc/Biomarker2006and2008.pdf
For this study, laboratory measurements from 2008 were used if available (>95% of individuals) and from 2006 if not available in 2008. CRP and plasma lipid data from a total of 6,545 participants was available for the study. Direct measurement of LDL was not available for this cohort, therefore, to approximate LDL, the equation LDL = TC – HDL was used.
2.4. HRS Telephone Interview for Cognitive Status (TICS) for identification of Cognitive Impairment
The HRS instrument used to collect data on cognitive status is a version of the modified Telephone Interview for Cognitive Status (TICS)(Welsh et al., 1993) that has been modified further specifically for the HRS. The TICS(Brandt et al., 1988) was modeled after the Mini-Mental State Examination,(Folstein et al., 1975) a standard measure of global cognitive function, and has good sensitivity and specificity for the identification of dementia.(Manly et al., 2011) For the HRS, the TICS was modified to an abbreviated version with a total of 35 points. For the current study, we augmented TICS scores with three other approaches to identify cases of cognitive impairment. Using TICS scores, we determined cognitive impairment by using a two-step process that provides a bi-valued response (cognitively normal or cognitive impairment). First we applied a cut off of 10 points on the TICS, which is a validated cut point for cognitive impairment(Langa et al., 2008). We then added a second criteria of requiring participants to score at or below this cut off over two consecutive interviews to avoid the inclusion of false positive participants. We used informant reports of impairment based on a cutoff of 3.6 or higher on the Informant Questionnaire on Cognitive Decline in the Elderly (IQCODE)(Jorm, 1994; Jorm, 2004) indicating a severe decline in cognitive functioning. Probabilities of dementia were previously calculated in the HRS.(Hurd et al., 2015) We examined these probabilities and after evaluating concordance with other methods, we applied a cutoff of probability of impairment of 65% or higher to identify impairment. We also used two self-report indicators where participants were asked if they had been told they have a diagnosis of Alzheimer’s disease or dementia. In most cases, where the data were available, these varied methods of case ascertainment were in agreement.
2.5. Statistical and Bioinformatics Analysis
2.5.1. Genome-wide association analysis
Separate GWA analyses were run for each of the phenotypes: cognitive impairment, CRP, HDL, LDL and TC. Imputed genotypes for the 21,632,048 SNPs were used for the GWA. Cognitive impairment was run as a logistic regression for a bivariate response in PLINK while the CRP and plasma lipid phenotypes were run as linear regressions. For all GWA, an additive allelic genetic model was used.
Filtering for identity by descent reduced the initial sample size from 12,507 to 12,484. As described in Arpawong et. al, we adjusted for population stratification by performing principal component analysis and using the first four principal components as covariates(Arpawong et al., 2017). The principal component analysis was performed on the 12,484 unrelated individuals using the R package, SNPRelate(Zheng et al., 2012). The top four principal components, age, sex and education level were included as covariates for all of the GWA (cognitive impairment, CRP and plasma lipids). For the GWA analysis, after filtering, there were 10,307 individuals available for the analysis data set, however this number was reduced to 6,545 where CRP and plasma lipid measurements were available, this set of individuals were used for all of the GWA. All genetic association analyses were performed using PLINK 1.9(Purcell et al., 2007). Since we are using imputed genotypes, the INFO criteria provided by PLINK was used for post association analysis as a quality control measure prior to the pleiotropy analysis(Purcell et al., 2007). The INFO metric is based on the ratio of empirical and expected variance in dosage. Values closer to 1 indicate better expected quality of imputation. Values can be above 1; values much greater than 1 can indicate strong departure from Hardy Weinberg equilibrium. To achieve a balance between stringency and coverage, we restricted analysis to SNPs that had INFO scores between 0.6 and 1.06. This criteria included 90% of the imputed SNPs (19,468,843 SNPs); 0.5% of SNPs had INFO scores above 1.06 and 9.5% had scores below 0.6. Manhattan plots, Q-Q plots and summary tables of the genetic association results for the individual GWA are provided in the Supplement.
2.5.2. Pleiotropy analysis
The pleiotropy analysis strategy, based on conditional false discovery rates, fold-enrichment plots and conditional quantile-quantile (q-q) plots is described in detail elsewhere.(Desikan et al., 2015) In brief, for two phenotypes A and B, pleiotropic enrichment of phenotype A conditional on phenotype B exists if the proportion of variants (SNPs) statistically significantly associated with phenotype A increases as a function of increased statistically significant SNP associations with phenotype B. The number of SNPs associated with phenotype A is determined for several thresholds of SNP association with phenotype B; the proportions are calculated relative to a baseline of all SNPs statistically significantly associated with phenotype A. For this study, phenotype A, the primary phenotype, is cognitive impairment and phenotype B, the conditional phenotypes, are the biomarkers (CRP, LDL, HDL, and TC). Fold enrichment plots graphically depict pleiotropy by showing fold enrichment in terms of numbers of SNPs on the ordinate and nominal −log10(P) values for association with cognitive impairment on the abscissa. Separate curves are shown for subsets of SNPs that reach specific levels of significance for their association with CRP, LDL, HDL and TC respectively. Conditional quantile-quantile plots for the same data shown in the fold enrichment plots provide additional assessment of genetic pleiotropy for each set of GWA results. Following the prior analysis strategy(Desikan et al., 2015), we focused the analysis for polygenic enrichment on SNPs below the standard GWAS Bonferroni-corrected p-value thresholds for association with cognitive impairment by using subsets of SNPs with a nominal −log10(P) < 9.0.
For identification of specific SNPs conditionally associated with cognitive impairment and one or more of the secondary phenotypes, a conditional false discovery rate (FDR) statistic is calculated as described in the prior analysis strategy (Desikan et al., 2015) and other publications(Andreassen et al., 2015a; Andreassen et al., 2013a; Andreassen et al., 2015b; Andreassen et al., 2014a; Andreassen et al., 2013b; Le Hellard et al., 2017; Witoelar et al., 2017). This framework is an extension of the standard analysis for FDR calculations and uses information from the secondary phenotypes (CRP and plasma lipids) to re-rank the p-values for the primary phenotype (cognitive impairment). We used a conditional FDR of 0.05 to show statistical significance; note that this level must be exceeded in both phenotypes for the overall test to be declared significant. The significance threshold of 0.05 for the conditional FDR(Hochberg and Benjamini, 1990) corresponds to 5 false positives per 100 reported associations. Manhattan plots of the FDRs for conditional association of cognitive impairment on CRP and on the plasma lipids are used to summarize the data.
2.5.3. Functional genomics bioinformatics analysis
Functional bioinformatics analysis was performed to evaluate the biological significance of the SNPs that were found to be significantly associated with cognitive impairment, conditional on the CRP and plasma lipid phenotypes. Three bioinformatics analysis tools were used to map the SNPs to genes by proximity, define the genomic context for the variant, annotate effects on phenotypes and identify relevant literature about the variant. The UCSC genome browser (http://genome.ucsc.edu/) was used to map each variant to proximate genes and to provide the first level of information about the genes and biological consequences of the genes(Kent et al., 2002). The Variant Effect Predictor (VEP) that is part of the Ensembl genome database project (http://www.ensembl.org) was used to provide information about the effects of the SNPS on genes, transcripts and regulatory regions(McLaren et al., 2016). SNPnexus (http://www.snp-nexus.org/) was used to provide additional annotation on gene/protein consequences and phenotype- and disease- association for the variants(Chelala et al., 2009; Dayem Ullah et al., 2013).
3. Results
3.1. Descriptive characteristics of the sample
Table 1 summarizes the descriptive characteristics of the sample. The sample consisted of 6,545 individuals interviewed from 1992 to 2010 who provided both DNA and biomarker samples. Demographic data, TICS scores, biomarker (CRP, HDL, LDL and TC) and APOE genotype are summarized for the entire sample, impaired individuals (classified as cognitively impaired as defined in Methods) and unimpaired individuals. For the entire sample the mean age was 72.2(sd 7.5) with a higher proportion (58%) of females. There was a statistically significant (P<0.0001) difference of 7.0 years in age between the impaired (78.9 years, sd 7.5) and the unimpaired (71.8 years, sd 7.3) groups. There was no statistically significant difference in the proportion of females between the impaired and unimpaired groups. There were highly statistically significant differences (P < 0.0001) in education between the impaired (11.3 years, sd 3.8) and unimpaired (12.6 years, sd 3.0) groups. There were highly statistically-significant differences in APOE genotype frequencies between the impaired and unimpaired groups with higher APOE ε4 frequencies in the impaired group (Table 1).
Table 1.
Summary statistics for demographic, TICS, APOE genetics and biomarkers for the sample.
| Measurement | Full cohort | Impaired | Unimpaired | P value for difference between affected and Unaffected |
|---|---|---|---|---|
| n | 6545 | 355 | 6190 | |
| Age, mean (SD), y | 72.22 (7.48) | 78.85 (7.46) | 71.84 (7.30) | < 0.0001 |
| % Female | 57.51 | 57.69 | 54.37 | ns |
| Education | 12.57 (3.05) | 11.32 (3.78) | 12.64 (2.99) | < 0.0001 |
| CRP mean (SD) | 2.12 (3.75) | 1.96 (3.72) | 2.13 (3.75) | ns |
| HDL mean (SD) | 55.36 (15.37) | 53.38 (13.70) | 55.48 (15.46) | 0.033 |
| LDL mean (SD) | 139.06 (42.42) | 138.15 (43.37) | 139.11 (42.37) | ns |
| TC mean (SD) | 191.55 (44.36) | 189.49 (46.50) | 191.67 (44.23) | ns |
| APOE genotypes | ||||
| APOE ε2/ε2 | 0.98% | 0.28% | 1.02% | ns |
| APOE ε2/ε3 | 16.24% | 14.93% | 16.32% | ns |
| APOE ε2/ε4 | 2.31% | 4.23% | 2.20% | 0.013 |
| APOE ε3/ε3 | 61.57% | 51.55% | 62.15% | P < 0.0001 |
| APOE ε3/ε4 | 17.82% | 24.79% | 17.42% | 0.0004 |
| APOE ε4/ε4 | 1.08% | 4.23% | 0.90% | P < 0.0001 |
| APOE ε4 carrier | 21.21% | 33.24% | 20.52% | P < 0.0001 |
There were no statistically significant differences between the impaired and unimpaired groups for the plasma CRP, LDL or TC biomarkers. There was a marginally statistically-significant (p=0.03) difference for HDL between the impaired and unimpaired groups. Histograms of the plasma CRP and lipid biomarkers are shown in Fig 1. The distributions of the CRP and lipid biomarkers showed a slight departure from normality (p=0.01 (for all of the biomarkers)) based on the Komologorov-Smirnov-Lilliefors test. A log transformation did not correct for normality, therefore the untransformed data was used.
Fig 1. Histograms of the plasma CRP and lipid biomarkers.
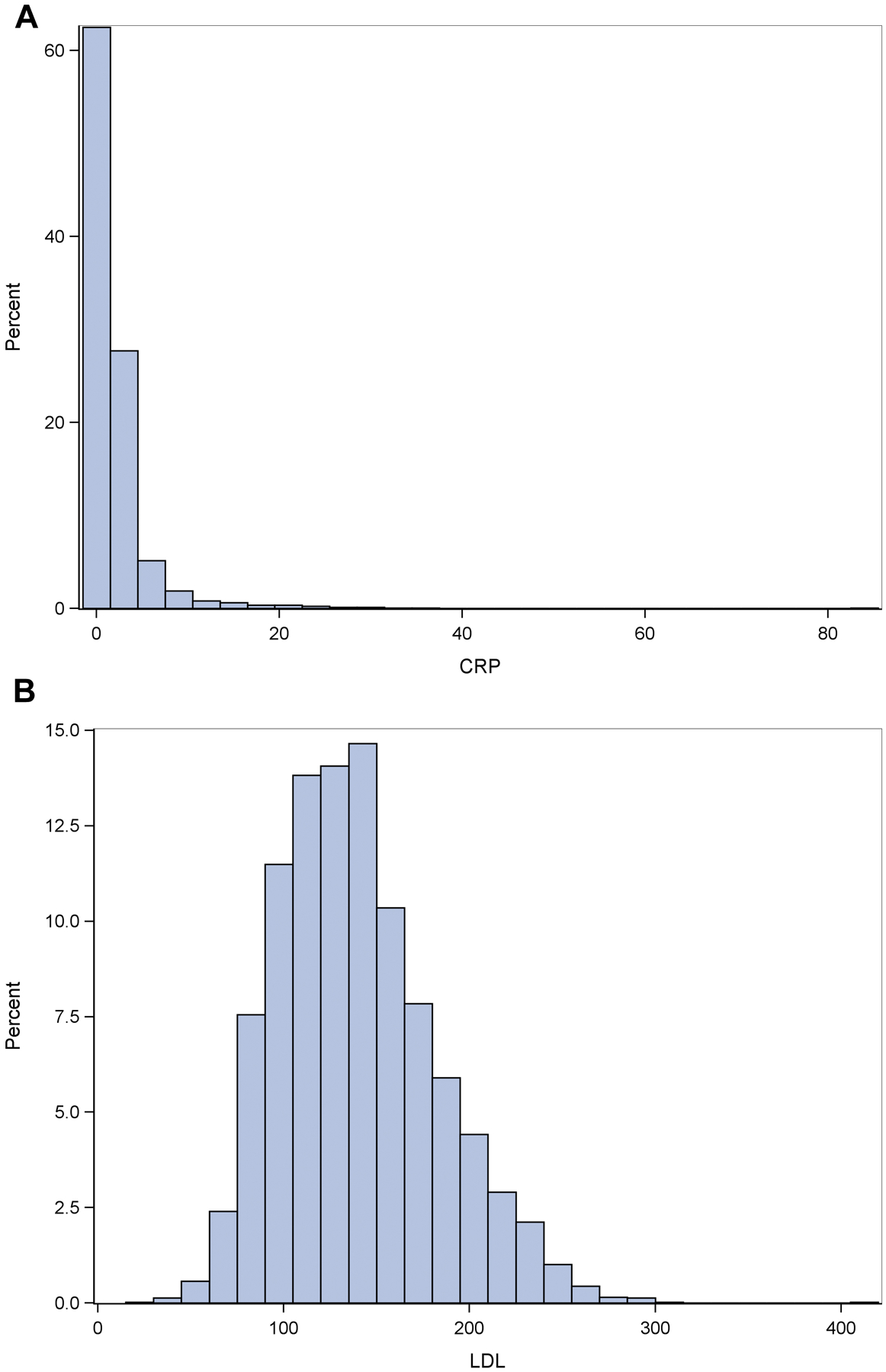
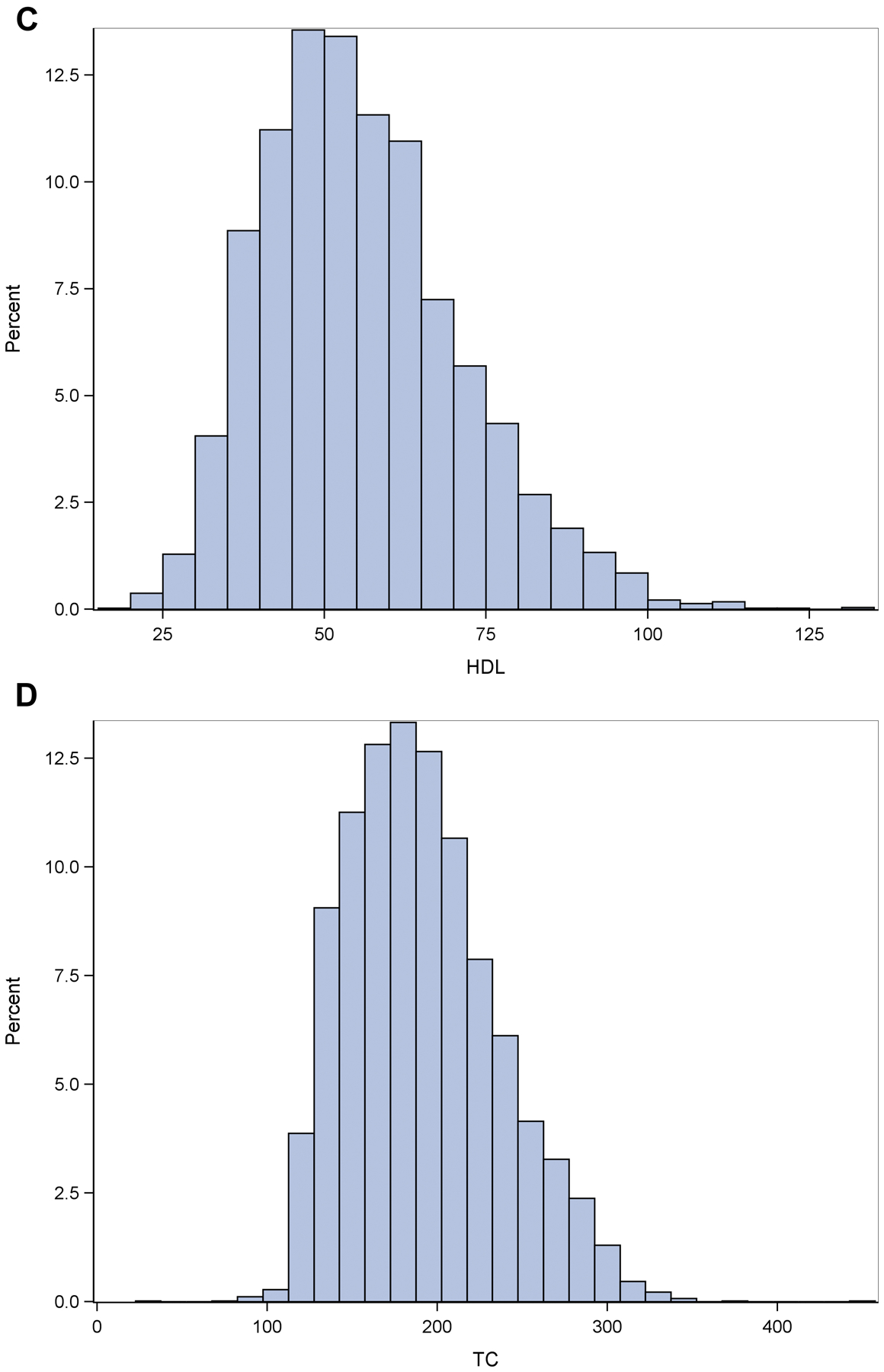
Ordinate shows the proportion of values represented by the histogram bars for CRP (Fig 1A), LDL (Fig 1B), HDL (Fig 1C),TC (Fig 1D and CI(1E)).
3.2. Genome-wide association summary results for the individual phenotypes
Prior to assessment of polygenetic overlap between cognitive impairment and CRP/plasma lipids, the individual genome-wide association studies for each phenotype were analyzed for quality control and overall genetic association statistics. The supplement contains summaries for each phenotype: Manhattan plots (Supplementary Fig. S1), q-q plots (Supplementary Fig. S2) and Tables of association statistics (Supplementary Table S1). The genome-wide significance level was 8.3*10−9 based on a Bonferroni correction for the number of SNPs. For the primary cognitive impairment phenotype, numerous genome-wide significant SNPs in the APOE-TOMM40 region of chromosome 19 were observed (Supplementary Fig. S1E), as was a single SNP near the SMYD3 gene on chromosome 1. For the HDL phenotype, SNPs in the genomic region on chromosome 16 overlapping and near the Cholesteryl Ester Transfer Protein (CETP) gene reached genome wide significance, with p values as low as 1*10−13. An objective of this study to test for polygenic effects below the standard GWAS significance threshold. Therefore the Tables of results (Supplementary Table S1) report on SNPs with a nominal level of significance of −log10(P) ≥ 6 corresponding to P≤1*10−6. Inspection of the Manhattan plots (Supplementary Fig. S1) show several regions of the genome with nominal levels of association (P≤1*10−6) for the different phenotypes. The q-q plots (Supplementary Fig. S2) show that population stratification was accounted for in the association analysis. A notable result for the CRP phenotype is the identification of the APOE coding SNP, rs429358 at an association significance level of 2.38*−7. The minor allele (C) for this variant, has a frequency of 14% in the HRS cohort which is comparable to the 15% reported for the 1000 Genomes Phase 3 combined population.
3.3. Assessment of polygenic overlap between cognitive impairment and CRP/plasma lipids
The fold enrichment plot (Fig 2A) demonstrates SNP enrichment for cognitive impairment across different levels of significance with CRP association. This result and the associated conditional FDR Manhattan plot (Fig 3A) support polygenic overlap between CRP and cognitive impairment. Notably, the fold enrichment plot for CRP is monotonic increasing and support fold enrichment of 100% – 800% for increasingly stringent p value thresholds for the SNPs associated with cognitive impairment (CI); this relationship is observed for all p value thresholds for the CRP association; these associations are largely driven by the high level of enrichment of SNPs in the APOE-TOMM40 region of chromosome 19. For LDL and TC, selection of SNPs with the highest threshold for association (-log10(P HDL) ≥ 3.0) showed fold enrichment of approximately 80% –800% for LDL and approximately 80%-600% for TC for increasingly stringent p value thresholds associated with CI. As with CRP, the high level of enrichment of SNPs in the APOE-TOMM40 region of chromosome 19 was the primary region driving the enrichment. HDL did not demonstrate fold enrichment above a level of 2.0% and therefore the hypothesis of polygenic overlap between CI and HDL is not supported by the data.
Fig 2. Fold-enrichment plots.
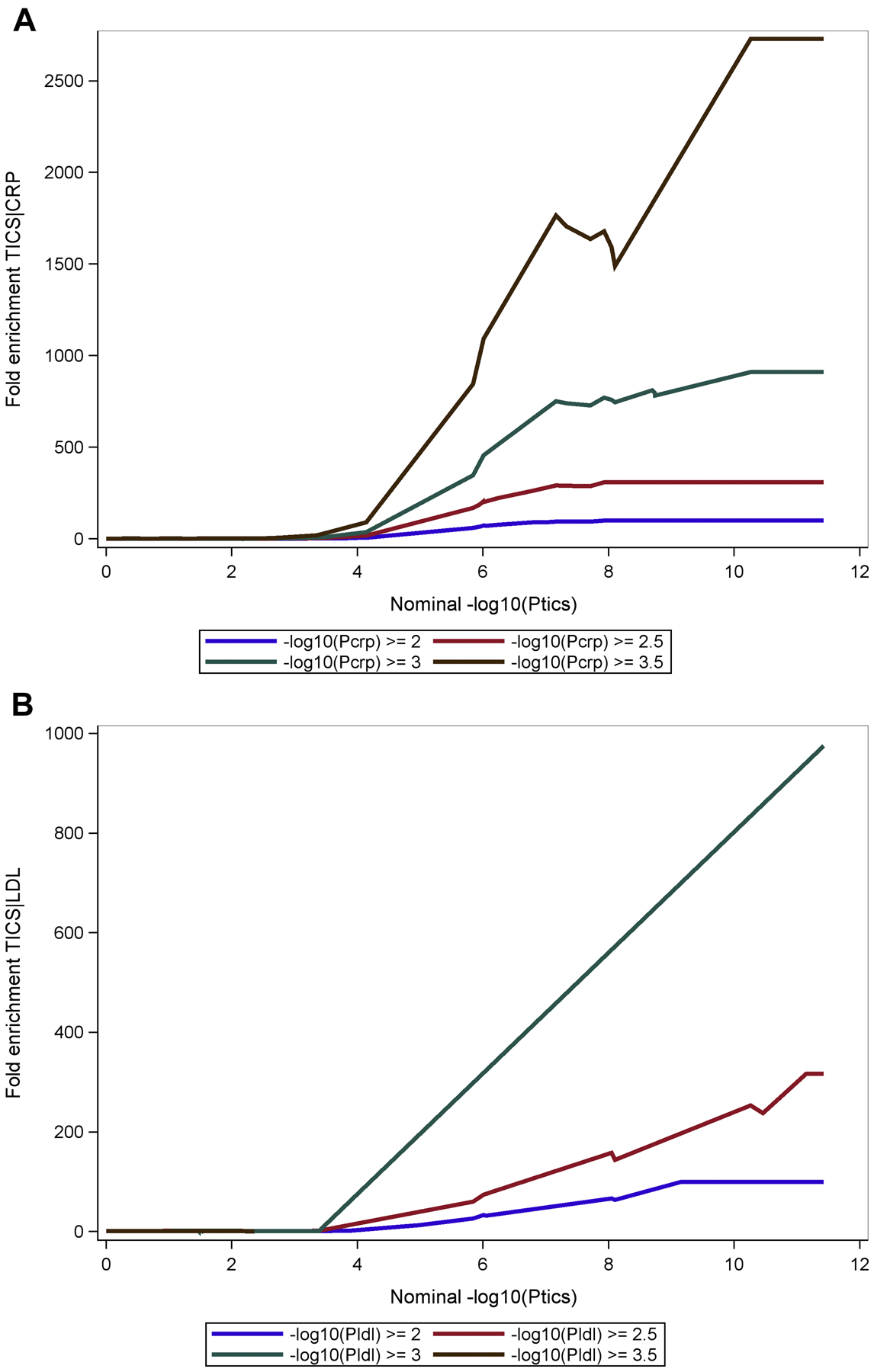
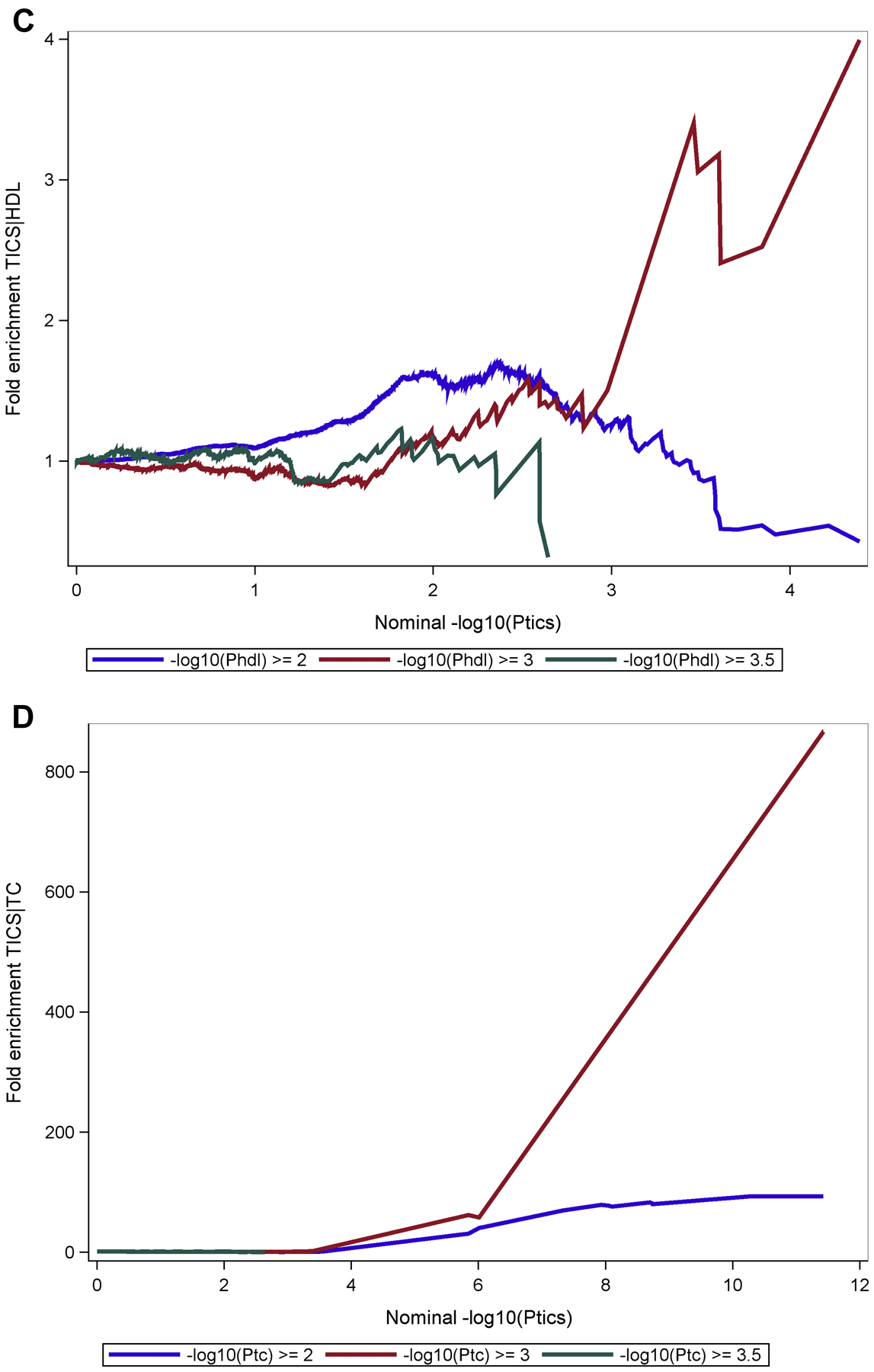
Ordinate is fold-enrichment, abscissa is nominal −log10(p) for cognitive impairment below the standard genome-wide association study threshold of P < 1 × 10−9 as a function of significance of association with CRP (Fig 2A), LDL (Fig 2B), HDL (Fig 2C) and TC (Fig 2D). Curves are differentiated by the threshold for level of statistical significance in the secondary phenotype (CRP and plasma lipids).
Fig 3. Conditional Manhattan plots of the conditional −log10 (FDR) values for cognitive impairment given CRP (Fig 3A), LDL (Fig 3B), HDL (Fig 3C) and TC (Fig 3D).
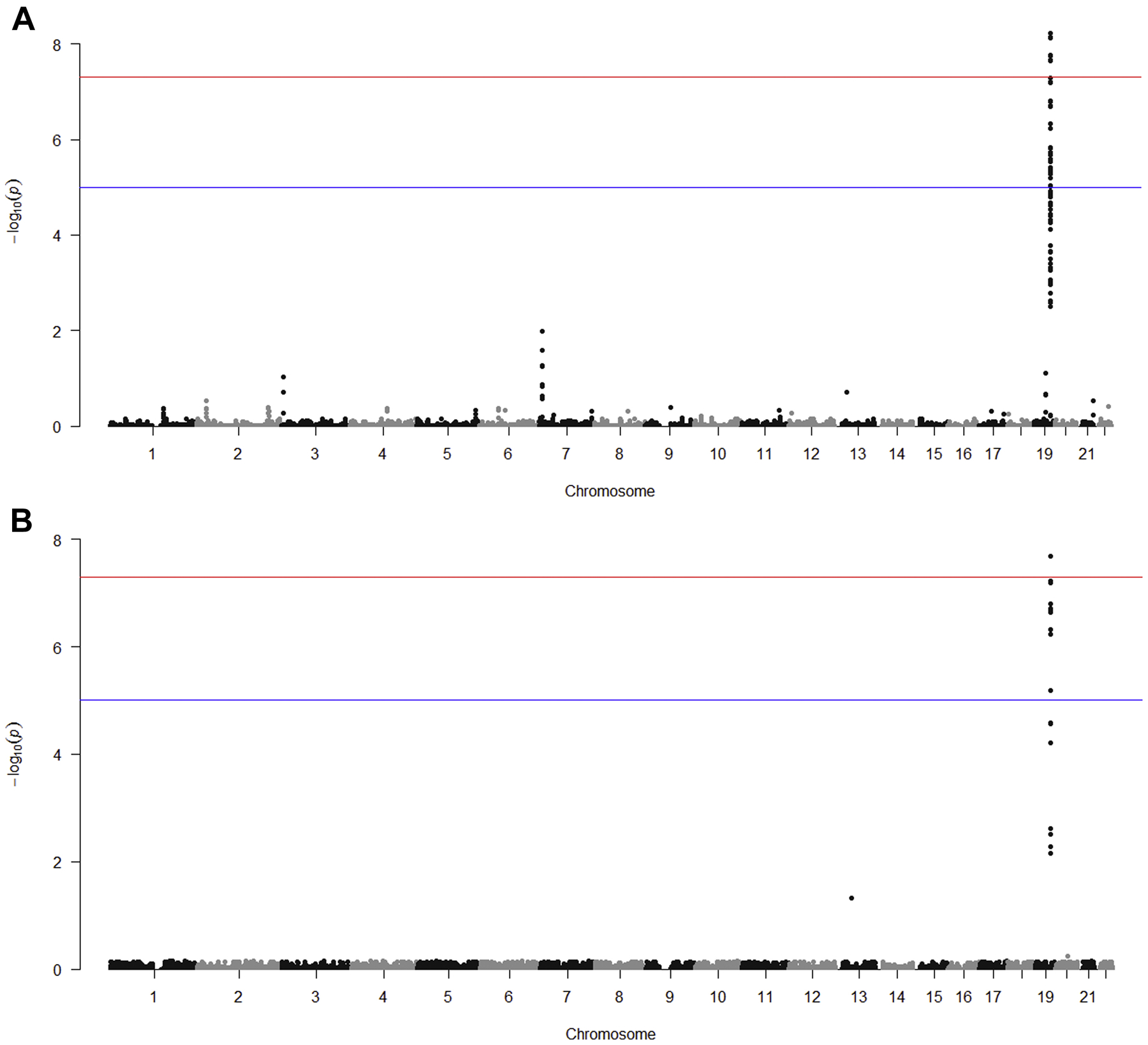
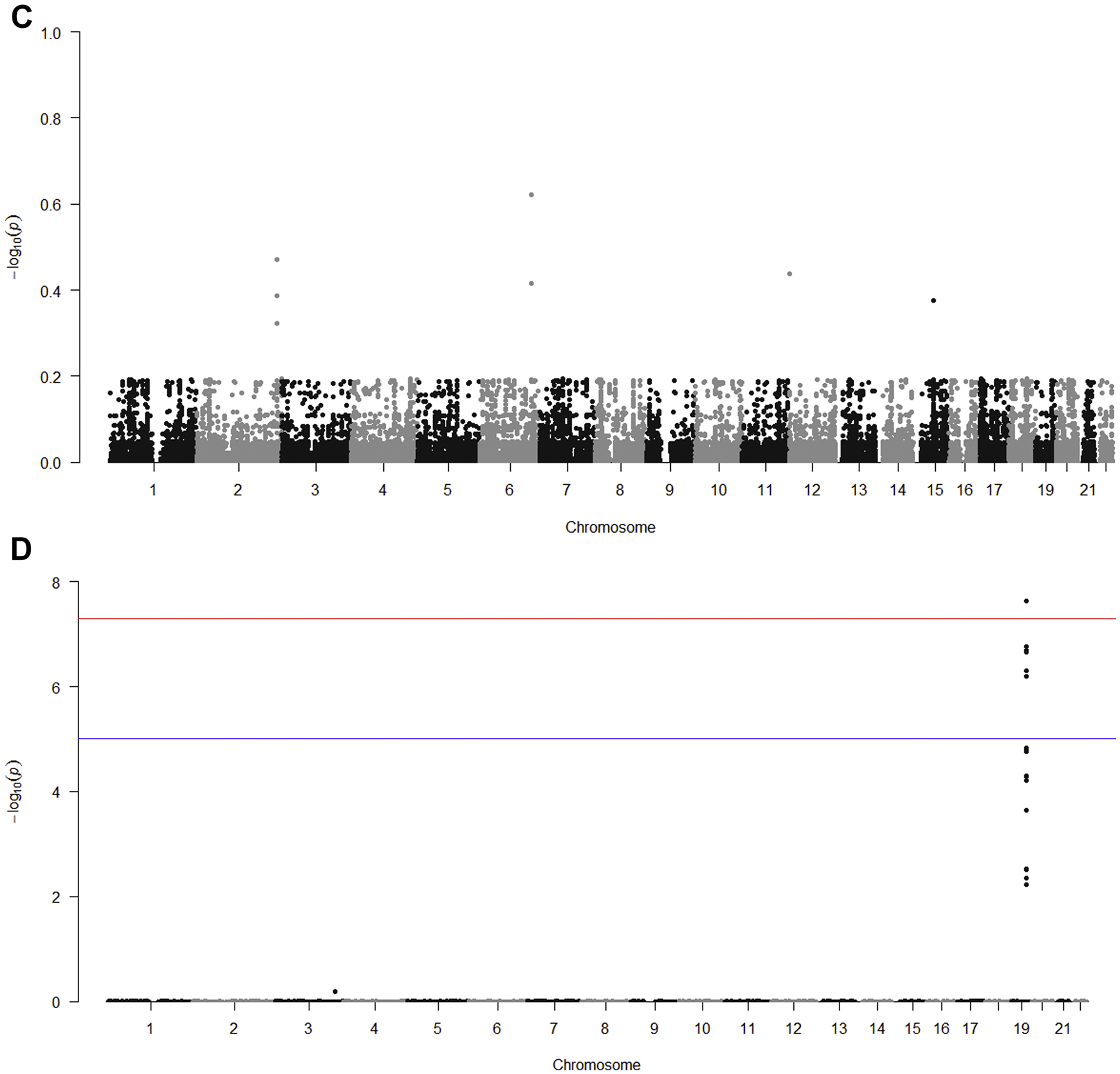
Conditional Manhattan plots show the FDR q value for cognitive impairment conditional on each of the four secondary phenotypes: CRP (Fig 3A), LDL (Fig 3B), HDL (Fig 3C) and TC (Fig 3D). Genome-wide significant line (red) is drawn at −log10(5 × 10−8), suggestive line (blue) is drawn at −log10(1 × 10−5).
3.4. Specific variants and genes identified by conditional false discovery rate analysis
Table 2 shows the FDR analysis to identify SNPs associated with cognitive impairment conditional on association for each secondary phenotype with a conditional FDR (q) ≤ 0.05. The locus on chromosome 19 that contains the APOE, TOMM40, APOC1, and PVRL2 genes is shown to have a strong, statistically-significant association of cognitive impairment conditional with either CRP, LDL or TC. The odds ratios for the association of the SNPs in these loci with cognitive impairment ranged from 1.4 to 1.9, consistent with a moderate effect size for the APOE ε4 allele. Relative numbers of significant SNPs in each of the genes: APOE, TOMM40, APOC1 and PVRL2 differed by the secondary phenotypes. Two additional loci showed modest, statistically-significant associations of cognitive impairment conditional on a secondary phenotype: conditional with CRP, a locus on chromosome 7 (near gene AC009500.2) (p=0.01) and conditional with LDL, a locus on chromosome 13 (near gene SPERT) (p=0.05). The same direction of allelic effects was observed for all SNPs associated with cognitive impairment conditional on association with CRP, LDL or TC.
Table 2.
Loci reaching statistical significance at FDR ≤0.05 for association with cognitive impairment conditional on association for the plasma lipids and CRP phenotypes.
| Biomarker | SNP | Chr | Position | Nearest Gene | Minor allele | MAF | Conditional FDR | Cognitive Impairment P Value | Cognitive Impairment OR (95% CI) | Biomarker Beta (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| CRP | rs73058377 | 7 | 9,080,297 | AC009500.2 | T | 0.40 | 1.03E-02 | 7.28E-05 | 1.31 (1.12 – 1.60) | −0.26 (−0.39 – −0.13) |
| CRP | rs1541411 | 7 | 9,081,364 | AC009500.2 | A | 0.46 | 5.78E-02 | 4.38E-04 | 1.25 (1.08 – 1.49) | −0.24 (−0.37 – −0.11) |
| CRP | rs11561890 | 7 | 9,086,743 | AC009500.2 | G | 0.42 | 9.54E-02 | 2.90E-04 | 1.28 (1.09 – 1.55) | −0.23 (−0.36 – −0.10) |
| CRP | rs118110581 | 19 | 31,450,347 | CTC-400I9.3 | C | 0.06 | 7.92E-02 | 1.13E-04 | 1.63 (1.16 – 2.74) | −0.44 (−0.71 – −0.17) |
| CRP | rs12972156 | 19 | 45,387,459 | PVRL2 | C | 0.13 | 1.81E-06 | 1.83E-09 | 1.72 (1.32 – 2.47) | 0.35 (0.16 – 0.54) |
| CRP | rs12972970 | 19 | 45,387,596 | PVRL2 | G | 0.13 | 1.57E-06 | 1.85E-09 | 1.72 (1.32 – 2.46) | 0.35 (0.16 – 0.54) |
| CRP | rs34342646 | 19 | 45,388,130 | PVRL2 | G | 0.13 | 1.49E-06 | 2.02E-09 | 1.71 (1.32 – 2.45) | 0.35 (0.16 – 0.54) |
| CRP | rs283812 | 19 | 45,388,568 | PVRL2 | T | 0.21 | 3.22E-04 | 1.02E-07 | 1.50 (1.23 – 1.94) | 0.24 (0.07 – 0.41) |
| CRP | rs283815 | 19 | 45,390,333 | PVRL2 | A | 0.24 | 8.92E-04 | 9.69E-07 | 1.41 (1.18 – 1.75) | 0.25 (0.09 – 0.40) |
| CRP | rs6857 | 19 | 45,392,254 | PVRL2 | C | 0.14 | 7.42E-09 | 3.75E-12 | 1.79 (1.38 – 2.53) | 0.48 (0.30 – 0.65) |
| CRP | rs71352238 | 19 | 45,394,336 | TOMM40 | T | 0.12 | 4.32E-06 | 1.97E-08 | 1.65 (1.28 – 2.32) | 0.38 (0.20 – 0.57) |
| CRP | rs184017 | 19 | 45,394,969 | TOMM40 | T | 0.24 | 1.64E-04 | 1.59E-07 | 1.45 (1.21 – 1.82) | 0.24 (0.08 – 0.40) |
| CRP | rs2075650 | 19 | 45,395,619 | TOMM40 | A | 0.13 | 1.23E-05 | 6.86E-08 | 1.57 (1.25 – 2.11) | 0.37 (0.18 – 0.56) |
| CRP | rs157581 | 19 | 45,395,714 | TOMM40 | T | 0.24 | 9.33E-04 | 1.17E-06 | 1.41 (1.18 – 1.75) | 0.24 (0.08 – 0.40) |
| CRP | rs34404554 | 19 | 45,395,909 | TOMM40 | C | 0.13 | 2.92E-06 | 1.18E-08 | 1.62 (1.28 – 2.21) | 0.37 (0.18 – 0.56) |
| CRP | rs11556505 | 19 | 45,396,144 | TOMM40 | C | 0.13 | 9.33E-06 | 4.72E-08 | 1.58 (1.26 – 2.14) | 0.37 (0.18 – 0.56) |
| CRP | rs157582 | 19 | 45,396,219 | TOMM40 | C | 0.24 | 8.85E-04 | 1.06E-06 | 1.41 (1.18 – 1.75) | 0.24 (0.08 – 0.40) |
| CRP | rs59007384 | 19 | 45,396,665 | TOMM40 | G | 0.22 | 5.58E-04 | 5.75E-07 | 1.43 (1.19 – 1.80) | 0.26 (0.10 – 0.42) |
| CRP | rs769449 | 19 | 45,410,002 | APOE | G | 0.10 | 1.74E-08 | 3.51E-11 | 1.87 (1.39 – 2.85) | 0.42 (0.22 – 0.63) |
| CRP | rs429358 | 19 | 45,411,941 | APOE | T | 0.14 | 2.16E-08 | 5.47E-11 | 1.69 (1.34 – 2.31) | 0.50 (0.31 – 0.69) |
| CRP | rs10414043 | 19 | 45,415,713 | APOE | G | 0.12 | 1.62E-04 | 9.84E-07 | 1.53 (1.21 – 2.07) | 0.42 (0.22 – 0.63) |
| CRP | rs7256200 | 19 | 45,415,935 | APOE | G | 0.12 | 2.19E-04 | 1.44E-06 | 1.52 (1.21 – 2.06) | 0.42 (0.22 – 0.63) |
| CRP | rs73052335 | 19 | 45,420,082 | APOC1 | A | 0.10 | 5.92E-09 | 8.97E-12 | 1.94 (1.42 – 3.06) | 0.41 (0.20 – 0.62) |
| CRP | rs12721046 | 19 | 45,421,254 | APOC1 | G | 0.12 | 2.07E-06 | 7.10E-10 | 1.78 (1.34 – 2.63) | 0.30 (0.10 – 0.50) |
| CRP | rs12721051 | 19 | 45,422,160 | APOC1 | C | 0.15 | 7.07E-09 | 7.15E-12 | 1.79 (1.38 – 2.54) | 0.38 (0.19 – 0.56) |
| CRP | rs56131196 | 19 | 45,422,846 | APOC1 | G | 0.16 | 2.62E-06 | 7.94E-09 | 1.59 (1.27 – 2.12) | 0.38 (0.19 – 0.56) |
| CRP | rs4420638 | 19 | 45,422,946 | APOC1 | A | 0.16 | 2.56E-06 | 9.06E-09 | 1.59 (1.27 – 2.11) | 0.38 (0.19 – 0.56) |
| CRP | rs111789331 | 19 | 45,427,125 | APOC1 | T | 0.12 | 1.20E-05 | 6.87E-09 | 1.75 (1.32 – 2.62) | 0.34 (0.13 – 0.54) |
| CRP | rs66626994 | 19 | 45,428,234 | APOC1 | G | 0.13 | 2.39E-03 | 8.92E-07 | 1.59 (1.23 – 2.24) | 0.31 (0.10 – 0.52) |
| LDL | rs11620068 | 13 | 46,300,824 | SPERT | A | 0.14 | 4.71E-02 | 1.04E-05 | 1.45 (1.17 – 1.90) | −3.39 (−5.81 – −0.98) |
| LDL | rs6857 | 19 | 45,392,254 | PVRL2 | C | 0.14 | 2.08E-08 | 3.75E-12 | 1.79 (1.38 – 2.53) | −4.02 (−6.41 – −1.63) |
| LDL | rs769449 | 19 | 45,410,002 | APOE | G | 0.10 | 1.99E-07 | 3.51E-11 | 1.87 (1.39 – 2.85) | −4.31 (−7.07 – −1.55) |
| LDL | rs429358 | 19 | 45,411,941 | APOE | T | 0.14 | 2.33E-07 | 5.47E-11 | 1.69 (1.34 – 2.31) | −4.10 (−6.64 – −1.57) |
| LDL | rs10414043 | 19 | 45,415,713 | APOE | G | 0.12 | 2.40E-03 | 9.84E-07 | 1.53 (1.21 – 2.07) | −4.57 (−7.33 – −1.82) |
| LDL | rs7256200 | 19 | 45,415,935 | APOE | G | 0.12 | 3.06E-03 | 1.44E-06 | 1.52 (1.21 – 2.06) | −4.56 (−7.32 – −1.80) |
| LDL | rs73052335 | 19 | 45,420,082 | APOC1 | A | 0.10 | 1.62E-07 | 8.97E-12 | 1.94 (1.42 – 3.06) | −4.25 (−7.09 – −1.42) |
| LDL | rs12721046 | 19 | 45,421,254 | APOC1 | G | 0.12 | 6.41E-06 | 7.10E-10 | 1.78 (1.34 – 2.63) | −3.75 (−6.40 – −1.10) |
| LDL | rs12721051 | 19 | 45,422,160 | APOC1 | C | 0.15 | 6.09E-08 | 7.15E-12 | 1.79 (1.38 – 2.54) | −3.86 (−6.34 – −1.39) |
| LDL | rs56131196 | 19 | 45,422,846 | APOC1 | G | 0.16 | 2.71E-05 | 7.94E-09 | 1.59 (1.27 – 2.12) | −3.98 (−6.45 – −1.51) |
| LDL | rs4420638 | 19 | 45,422,946 | APOC1 | A | 0.16 | 2.57E-05 | 9.06E-09 | 1.59 (1.27 – 2.11) | −3.98 (−6.45 – −1.51) |
| LDL | rs66626994 | 19 | 45,428,234 | APOC1 | G | 0.13 | 5.37E-03 | 8.92E-07 | 1.59 (1.23 – 2.24) | −3.78 (−6.58 – −0.98) |
| TC | rs12972156 | 19 | 45,387,459 | PVRL2 | C | 0.13 | 1.77E-05 | 1.83E-09 | 1.72 (1.32 – 2.47) | −3.22 (−5.62 – −0.83) |
| TC | rs12972156 | 19 | 45,387,459 | CTB-129P6.4 | C | 0.13 | 1.77E-05 | 1.83E-09 | 1.72 (1.32 – 2.47) | −3.22 (−5.62 – −0.83) |
| TC | rs12972970 | 19 | 45,387,596 | PVRL2 | G | 0.13 | 1.53E-05 | 1.85E-09 | 1.72 (1.32 – 2.46) | −3.22 (−5.62 – −0.83) |
| TC | rs12972970 | 19 | 45,387,596 | CTB-129P6.4 | G | 0.13 | 1.53E-05 | 1.85E-09 | 1.72 (1.32 – 2.46) | −3.22 (−5.62 – −0.83) |
| TC | rs34342646 | 19 | 45,388,130 | PVRL2 | G | 0.13 | 1.46E-05 | 2.02E-09 | 1.71 (1.32 – 2.45) | −3.22 (−5.60 – −0.83) |
| TC | rs34342646 | 19 | 45,388,130 | CTB-129P6.4 | G | 0.13 | 1.46E-05 | 2.02E-09 | 1.71 (1.32 – 2.45) | −3.22 (−5.60 – −0.83) |
| TC | rs6857 | 19 | 45,392,254 | PVRL2 | C | 0.14 | 2.34E-08 | 3.75E-12 | 1.79 (1.38 – 2.53) | −3.83 (−6.08 – −1.58) |
| TC | rs6857 | 19 | 45,392,254 | CTB-129P6.4 | C | 0.14 | 2.34E-08 | 3.75E-12 | 1.79 (1.38 – 2.53) | −3.83 (−6.08 – −1.58) |
| TC | rs34404554 | 19 | 45,395,909 | TOMM40 | C | 0.13 | 6.21E-05 | 1.18E-08 | 1.62 (1.28 – 2.21) | −3.19 (−5.55 – −0.83) |
| TC | rs11556505 | 19 | 45,396,144 | TOMM40 | C | 0.13 | 2.28E-04 | 4.72E-08 | 1.58 (1.26 – 2.14) | −3.18 (−5.54 – −0.82) |
| TC | rs769449 | 19 | 45,410,002 | APOE | G | 0.10 | 5.08E-07 | 3.51E-11 | 1.87 (1.39 – 2.85) | −4.17 (−6.76 – −1.59) |
| TC | rs429358 | 19 | 45,411,941 | APOE | T | 0.14 | 6.33E-07 | 5.47E-11 | 1.69 (1.34 – 2.31) | −3.87 (−6.24 – −1.49) |
| TC | rs10414043 | 19 | 45,415,713 | APOE | G | 0.12 | 3.06E-03 | 9.84E-07 | 1.53 (1.21 – 2.07) | −4.62 (−7.20 – −2.04) |
| TC | rs7256200 | 19 | 45,415,935 | APOE | G | 0.12 | 2.98E-03 | 1.44E-06 | 1.52 (1.21 – 2.06) | −4.60 (−7.18 – −2.02) |
| TC | rs73052335 | 19 | 45,420,082 | APOC1 | A | 0.10 | 1.73E-07 | 8.97E-12 | 1.94 (1.42 – 3.06) | −3.84 (−6.48 – −1.19) |
| TC | rs12721051 | 19 | 45,422,160 | APOC1 | C | 0.15 | 2.07E-07 | 7.15E-12 | 1.79 (1.38 – 2.54) | −3.07 (−5.39 – −0.76) |
| TC | rs56131196 | 19 | 45,422,846 | APOC1 | G | 0.16 | 5.11E-05 | 7.94E-09 | 1.59 (1.27 – 2.12) | −3.17 (−5.48 – −0.86) |
| TC | rs4420638 | 19 | 45,422,946 | APOC1 | A | 0.16 | 5.24E-05 | 9.06E-09 | 1.59 (1.27 – 2.11) | −3.17 (−5.48 – −0.86) |
4. Discussion
This study showed that genetic variants associated with cognitive impairment are also associated with CRP and plasma lipids (LDL, and TC) in a large study of older adults. The fold enrichment and conditional q-q plots show similarity to previously published results for conditional association with AD as a function of stringency for levels of significance of association with CRP or plasma lipids(Desikan et al., 2015); with the results for CRP being the most consistent and replicative in terms of congruency of the fold enrichment curves. Enrichment was observed for the plasma lipids LDL and TC but not for HDL. A locus on chromosome 19 that contains the APOE, TOMM40, APOC1, PVRL2 genes show a statistically-significant conditional association with cognitive impairment for three secondary phenotypes, CRP, LDL and TC. SNPs in this locus were found to show a strong, genome-wide significant association in the GWA for cognitive impairment, however, the strength of this association was greatly amplified when the conditional analysis was performed. While this locus shows a strong effect in the conditional analysis where all of the phenotypes were measured in the same individuals, larger cohorts and/or meta analyses of GWAS data have the potential to identify additional loci with common variants that demonstrate pleiotropic genetic effects on cognitive impairment, systemic inflammation and plasma lipids.
Considering that the HRS cohort is a large, nationally representative longitudinal panel study of aging, the fact that few genome-wide significant results are identified for the individual GWA is not surprising; as a comparator, a GWAS meta-analysis of general cognitive function report from the COGENT consortium identified two genome-wide significant SNPs(Trampush et al., 2017). For the present study, the odds-ratios reported for the nominally-significant loci are consistent with the low effect size common SNPs, while the odds-ratio reported for the chromosome 19 SNPs are consistent with those reported for the association between APOE and cognitive impairment (OR=1.68, 95% CI:1.03–2.75)(Jefferson et al., 2015). The current study utilized stringent selection criteria for SNPs based on the imputed genotypes. High stringency was used to maximize the likelihood that the results will replicate. The borderline significance of the association of the APOE coding SNP, rs429358 with CRP merits discussion. Prior large studies have shown that plasma CRP levels are influenced by the common genetic polymorphisms within the APOE gene, specifically, that the APOE ε4 allele is associated with low levels of plasma CRP(Hubacek et al., 2010). The MAF for this SNP in our study was also consistent with the frequency reported for the 1000 Genomes Phase 3 combined population, an important result considering the moderate effect size of this variant on many diseases of aging including AD. The finding of genome-wide significant SNPs in the genomic region on chromosome 16 overlapping and near the CETP gene that are identified for the HDL phenotype is consistent with the biological function of this gene. The CETP gene encodes cholesteryl ester transfer protein. This protein is found in plasma, where it is involved in the transfer of cholesteryl ester from high density lipoprotein (HDL) to other lipoproteins.
There are several aspects of our findings that are important in consideration of the role of inflammation with the development of cognitive impairment. First, the strong conditional associations between cognitive impairment and the biomarkers are observed only for CRP, LDL and TC but not HDL. This finding points to the involvement of inflammation-related biological processes that are tagged by these biomarkers with the development of cognitive impairment. Although APOE is clearly a genetic factor which is common to cognitive impairment(Jefferson et al., 2015; Qian et al., 2017; Welsh-Bohmer et al., 2009), LDL(Radwan et al., 2014) and CRP(Kahri et al., 2006), it is interesting to note that numerous (8) SNPs are observed in the TOMM40 gene for the CRP conditional association, but not for LDL. Only two TOMM40 SNPs are identified in the conditional association of cognitive impairment with TC. Recent literature has suggested that haplotypes of APOE-TOMM40 have complex effects of multiple late-onset disease related phenotypes(Bekris et al., 2010; Bekris et al., 2012; Chiba-Falek et al., 2018; Gottschalk et al., 2014; Li et al., 2013; Linnertz et al., 2014). Interestingly, in another study of the HRS, TOMM40 effects independent of APOE were identified for aging-related verbal memory(Arpawong et al., 2017). The finding that the CRP phenotype alone has the most number of associated TOMM40 SNPs warrants further investigation to understand whether there is a biological basis rooted in metabolic or mitochondrial dysfunction for the genetic association.
The locus on chromosome 19 that contains the CTB-129P6.4 that is statistically-significant for the cognitive impairment association conditional with TC is located close to the TOMM40 and PVRL2 genes and therefore the association is likely a consequence of linkage disequilibrium. The locus on chromosome 7 containing AC009500.2 constitutes a pseudogene of unknown biological function, however, the conditional FDR for this locus (q=0.013) is several orders of magnitude greater than observed for the chromosome 19 loci containing APOE. The locus on chromosome 13 containing the SPERT (spermatid associated) has unclear biological relevance to the phenotypes and has a conditional FDR (q=0.047) that is narrowly significant at the q=0.05 level.
The availability of cognitive and CRP/plasma lipid biomarker data for the sample allowed the genetic pleiotropy analysis to be performed within the same set of individuals. Although genetic pleiotropy analysis can be performed on GWAS summary-level statistics (beta coefficients and p values) the present study utilized independent GWA for cognitive impairment, CRP and the plasma lipids. As noted in Desikan et al., the conditional FDR framework can detect genetic pleiotropy independent of directionality and that both phenotypes are utilized to identify genetic association that may not be detected with a single phenotype GWA (Desikan et al., 2015). The results of this study set the stage to examine how genetic variation in genes and pathways involved in inflammation as measured by CRP and plasma lipid metabolism may identify individuals at risk to develop cognitive impairment, mild cognitive impairment and AD later in life.
This study has several limitations. The conditional FDR framework controls for the likelihood of false positive results and is based on the number of statistical tests; however, replication in additional cohorts of the conditional associations would strengthen statistical support. Future planned work will replicate the analysis in other cohorts; specifically the Women’s Health Initiative Memory Study (WHIMS) which has both the cognitive phenotype and the biomarker measurements available. The cognitive phenotype considered in the study is comparable to other studies of cognitive impairment but cannot be applied to a specific differential diagnosis for dementia. Further work will assess genetic pleiotropy using alternative measures of cognitive impairment and cognitive decline based on the HRS and other studies.
The overall approach used in this study has the potential to provide biologically-relevant information about the relationship of inflammation and lipid metabolism with the development of cognitive impairment during the process of aging. Inflammation and lipid metabolism are potentially modifiable risk factors that can be controlled through lifestyle interventions and/or medications and therefore have the potential to delay the onset of cognitive impairment.
Supplementary Material
Highlights.
We examined pleiotropic genetic effects on cognitive impairment (CI) with HRS data.
We tested for SNP enrichment for CI conditional with plasma CRP and lipid levels.
SNP enrichment was observed for CI conditioned on CRP levels, LDL levels and total cholesterol levels.
Significant associations between cognitive impairment, conditional with either CRP, LDL or TC were found for the locus on chromosome 19 that contains the APOE, TOMM40, APOC1, PVRL2 genes.
Variants and proximal genes identified are involved in multiple pathological processes including cholesterol metabolism, inflammation and mitochondrial transport.
Acknowledgments
Funding provided by NIH- R01AG042633. The work was supported by access to equipment (MiCDA Enclave VDI Environment) made possible by the University of Michigan.
Footnotes
Disclosure statement
The authors report no actual or potential conflict of interest.
References
- 1.Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WST, Hampel H, Hull M, Landreth G, Lue LF, Mrak R, Mackenzie IR, McGeer PL, O’Banion MK, Pachter J, Pasinetti G, Plata–Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss–Coray T, 2000. Inflammation and Alzheimer’s disease. Neurobiology of Aging 21(3), 383–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreassen OA, Desikan RS, Wang Y, Thompson WK, Schork AJ, Zuber V, Doncheva NT, Ellinghaus E, Albrecht M, Mattingsdal M, Franke A, Lie BA, Mills IG, Aukrust P, McEvoy LK, Djurovic S, Karlsen TH, Dale AM, 2015a. Abundant genetic overlap between blood lipids and immune-mediated diseases indicates shared molecular genetic mechanisms. PLoS One 10(4), e0123057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, Rujescu D, Werge T, van de Bunt M, Morris AP, McCarthy MI, International Consortium for Blood Pressure, G., Diabetes Genetics, R., Meta-analysis, C., Psychiatric Genomics Consortium Schizophrenia Working, G., Roddey JC, McEvoy LK, Desikan RS, Dale AM, 2013a. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet 92(2), 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, Zuber V, Bettella F, Ripke S, Kelsoe JR, Kendler KS, O’Donovan MC, Sklar P, Psychiatric Genomics Consortium Bipolar, D., Schizophrenia Work, G., International Multiple Sclerosis Genetics, C., McEvoy LK, Desikan RS, Lie BA, Djurovic S, Dale AM, 2015b. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry 20(2), 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreassen OA, McEvoy LK, Thompson WK, Wang Y, Reppe S, Schork AJ, Zuber V, Barrett-Connor E, Gautvik K, Aukrust P, Karlsen TH, Djurovic S, Desikan RS, Dale AM, International Consortium for Blood Pressure Genome-Wide Association Studies, G.F.f.O.C., 2014a. Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension 63(4), 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, Kendler KS, O’Donovan MC, Rujescu D, Werge T, Sklar P, Psychiatric Genomics C, Bipolar D, Schizophrenia Working G, Roddey JC, Chen CH, McEvoy L, Desikan RS, Djurovic S, Dale AM, 2013b. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet 9(4), e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreassen OA, Zuber V, Thompson WK, Schork AJ, Bettella F, Consortium P, Cruk G, Djurovic S, Desikan RS, Mills IG, Dale AM, 2014b. Shared common variants in prostate cancer and blood lipids. Int J Epidemiol 43(4), 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreone BJ, Chow BW, Tata A, Lacoste B, Ben-Zvi A, Bullock K, Deik AA, Ginty DD, Clish CB, Gu C, 2017. Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron 94(3), 581–594 e585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arpawong TE, Pendleton N, Mekli K, McArdle JJ, Gatz M, Armoskus C, Knowles JA, Prescott CA, 2017. Genetic variants specific to aging-related verbal memory: Insights from GWASs in a population-based cohort. PLoS One 12(8), e0182448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekris LM, Galloway NM, Montine TJ, Schellenberg GD, Yu CE, 2010. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet 153B(2), 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekris LM, Lutz F, Yu CE, 2012. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet 57(1), 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, Gu C, 2014. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509(7501), 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett S, Thomas AJ, 2014. Depression and dementia: cause, consequence or coincidence? Maturitas 79(2), 184–190. [DOI] [PubMed] [Google Scholar]
- 14.Brandt J, Spencer M, Folstein M, 1988. The Telephone Interview for Cognitive Status. Neuropsychiatry, Neuropsychology, and Behavioral Neurology 1, 111–117. [Google Scholar]
- 15.Broussard GJ, Mytar J, Li RC, Klapstein GJ, 2012. The role of inflammatory processes in Alzheimer’s disease. Inflammopharmacology 20(3), 109–126. [DOI] [PubMed] [Google Scholar]
- 16.Byers AL, Yaffe K, 2011. Depression and risk of developing dementia. Nat Rev Neurol 7(6), 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, Maurage CA, Huot L, Bensemain F, Laumet G, Ayral AM, Fievet N, Hauw JJ, DeKosky ST, Lemoine Y, Iwatsubo T, Wavrant-Devrieze F, Dartigues JF, Tzourio C, Buee L, Pasquier F, Berr C, Mann D, Lendon C, Alperovitch A, Kamboh MI, Amouyel P, Lambert JC, 2009. Transcriptomic and genetic studies identify IL-33 as a candidate gene for Alzheimer’s disease. Mol Psychiatry 14(11), 1004–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chelala C, Khan A, Lemoine NR, 2009. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics 25(5), 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Tang J, Huang L, Lin J, 2015. Expression and prognostic value of Mycl1 in gastric cancer. Biochem Biophys Res Commun 456(4), 879–883. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Zheng Z, Tang J, Lin X, Wang X, Lin J, 2013. Association of polymorphisms and haplotype in the region of TRIT1, MYCL1 and MFSD2A with the risk and clinicopathological features of gastric cancer in a southeast Chinese population. Carcinogenesis 34(5), 1018–1024. [DOI] [PubMed] [Google Scholar]
- 21.Chiba-Falek O, Gottschalk WK, Lutz MW, 2018. The effects of the TOMM40 poly-T alleles on Alzheimer’s disease phenotypes. Alzheimers Dement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, Hardy JA, Huentelman MJ, Corneveaux JJ, Reiman EM, Evans DA, Bennett DA, De Jager PL, 2011. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol 69(3), 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook SE, Marsiske M, McCoy KJ, 2009. The use of the Modified Telephone Interview for Cognitive Status (TICS-M) in the detection of amnestic mild cognitive impairment. J Geriatr Psychiatry Neurol 22(2), 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dayem Ullah AZ, Lemoine NR, Chelala C, 2013. A practical guide for the functional annotation of genetic variations using SNPnexus. Brief Bioinform 14(4), 437–447. [DOI] [PubMed] [Google Scholar]
- 25.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, Yu L, Leurgans SE, Tran D, Aubin C, Anderson CD, Biffi A, Corneveaux JJ, Huentelman MJ, Alzheimer’s Disease Neuroimaging, I., Rosand J, Daly MJ, Myers AJ, Reiman EM, Bennett DA, Evans DA, 2012. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiology of aging 33(5), 1017 e1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desikan RS, Schork AJ, Wang Y, Thompson WK, Dehghan A, Ridker PM, Chasman DI, McEvoy LK, Holland D, Chen CH, Karow DS, Brewer JB, Hess CP, Williams J, Sims R, O’Donovan MC, Choi SH, Bis JC, Ikram MA, Gudnason V, DeStefano AL, van der Lee SJ, Psaty BM, van Duijn CM, Launer L, Seshadri S, Pericak-Vance MA, Mayeux R, Haines JL, Farrer LA, Hardy J, Ulstein ID, Aarsland D, Fladby T, White LR, Sando SB, Rongve A, Witoelar A, Djurovic S, Hyman BT, Snaedal J, Steinberg S, Stefansson H, Stefansson K, Schellenberg GD, Andreassen OA, Dale AM, Inflammation Working G, International Genomics of Alzheimer’s Disease, P., DemGene I, 2015. Polygenic Overlap Between C-Reactive Protein, Plasma Lipids, and Alzheimer Disease. Circulation 131(23), 2061–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Paolo G, Kim T-W, 2011. Linking Lipids to Alzheimer’s Disease: Cholesterol and Beyond. Nature Reviews. Neuroscience 12(5), 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efthymiou AG, Goate AM, 2017. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol Neurodegener 12(1), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folstein MF, Folstein SE, McHugh PR, 1975. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12(3), 189–198. [DOI] [PubMed] [Google Scholar]
- 30.Georgoysopoulou EN, Panagiotakos DB, Pitsavos C, Kalogeropoulou A, Ntertimani M, Pitaraki E, Chrysohoou C, Skoumas I, Tousoulis D, Stefanadis C, 2016. Adherence to Mediterranean diet has a mediating effect inflammation as regards cardiovascular disease risk: The 10-year (2002–12) follow-up of ATTICA study. Clin Nutr ESPEN 13, e67. [Google Scholar]
- 31.Gonzalez-Reyes RE, Nava-Mesa MO, Vargas-Sanchez K, Ariza-Salamanca D, Mora-Munoz L, 2017. Involvement of Astrocytes in Alzheimer’s Disease from a Neuroinflammatory and Oxidative Stress Perspective. Front Mol Neurosci 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottschalk WK, Lutz MW, He YT, Saunders AM, Burns DK, Roses AD, Chiba-Falek O, 2014. The Broad Impact of TOM40 on Neurodegenerative Diseases in Aging. J Parkinsons Dis Alzheimers Dis 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He R, Kidder BL, 2017. H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochberg Y, Benjamini Y, 1990. More powerful procedures for multiple significance testing. Stat Med 9(7), 811–818. [DOI] [PubMed] [Google Scholar]
- 35.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J, Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43(5), 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubacek JA, Peasey A, Pikhart H, Stavek P, Kubinova R, Marmot M, Bobak M, 2010. APOE polymorphism and its effect on plasma C-reactive protein levels in a large general population sample. Hum Immunol 71(3), 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurd MD, Martorell P, Langa K, 2015. Future Monetary Costs of Dementia in the United States under Alternative Dementia Prevalence Scenarios. J Popul Ageing 8(1–2), 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferson AL, Beiser AS, Seshadri S, Wolf PA, Au R, 2015. APOE and mild cognitive impairment: the Framingham Heart Study. Age Ageing 44(2), 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, Pahwa JS, Jones N, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Peters O, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Ruther E, Carrasquillo MM, Pankratz VS, Younkin SG, Hardy J, O’Donovan MC, Owen MJ, Williams J, 2010. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One 5(11), e13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorm A, 1994. A short form of the informant questionnaire on cognitive decline in the elderly (IQCODE): development and cross-validation [published erratum appears in Psychol Med 1995 Mar;25(2):437]. Psychological Medicine 24, 145–153. [DOI] [PubMed] [Google Scholar]
- 41.Jorm AF, 2004. The Informant Questionnaire on cognitive decline in the elderly (IQCODE): a review. Int Psychogeriatr 16(3), 275–293. [DOI] [PubMed] [Google Scholar]
- 42.Juster FT, Suzman R, 1995. An overview of the health and retirement study. Journal of Human Resources 30, S7–S56. [Google Scholar]
- 43.Kahri J, Soro-Paavonen A, Ehnholm C, Taskinen MR, 2006. ApoE polymorphism is associated with C-reactive protein in low-HDL family members and in normolipidemic subjects. Mediators Inflamm 2006(3), 12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaup AR, Byers AL, Falvey C, Simonsick EM, Satterfield S, Ayonayon HN, Smagula SF, Rubin SM, Yaffe K, 2016. Trajectories of Depressive Symptoms in Older Adults and Risk of Dementia. JAMA Psychiatry 73(5), 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keaney J, Campbell M, 2015. The dynamic blood-brain barrier. FEBS J 282(21), 4067–4079. [DOI] [PubMed] [Google Scholar]
- 46.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, 2002. The human genome browser at UCSC. Genome Res 12(6), 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kidder BL, Hu G, Zhao K, 2014. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol 15(2), R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King DE, Egan BM, Geesey ME, 2003. Relation of dietary fat and fiber to elevation of C-reactive protein. Am J Cardiol 92(11), 1335–1339. [DOI] [PubMed] [Google Scholar]
- 49.Klaassen RV, Stroeder J, Coussen F, Hafner AS, Petersen JD, Renancio C, Schmitz LJ, Normand E, Lodder JC, Rotaru DC, Rao-Ruiz P, Spijker S, Mansvelder HD, Choquet D, Smit AB, 2016. Shisa6 traps AMPA receptors at postsynaptic sites and prevents their desensitization during synaptic activity. Nat Commun 7, 10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin C-F, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau M-T, Choi S-H, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MCD, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer’s Disease, I., Genetic, Environmental Risk in Alzheimer’s, D., Alzheimer’s Disease Genetic, C., Cohorts for, H., Aging Research in Genomic, E., Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RFAG, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JSK, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang L-S, Dartigues J-F, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P, 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41(10), 1094–1099. [DOI] [PubMed] [Google Scholar]
- 52.Langa KM, Chernew ME, Kabeto MU, Herzog AR, Ofstedal MB, Willis RJ, Wallace RB, Mucha LM, Straus WL, Fendrick AM, 2001. National estimates of the quantity and cost of informal caregiving for the elderly with dementia. J Gen Intern Med 16(11), 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langa KM, Larson EB, Karlawish JH, Cutler DM, Kabeto MU, Kim SY, Rosen AB, 2008. Trends in the prevalence and mortality of cognitive impairment in the United States: is there evidence of a compression of cognitive morbidity? Alzheimers Dement 4(2), 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Hellard S, Wang Y, Witoelar A, Zuber V, Bettella F, Hugdahl K, Espeseth T, Steen VM, Melle I, Desikan R, Schork AJ, Thompson WK, Dale AM, Djurovic S, Andreassen OA, Schizophrenia Working Group of the Psychiatric Genomics, C., 2017. Identification of Gene Loci That Overlap Between Schizophrenia and Educational Attainment. Schizophr Bull 43(3), 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li G, Bekris LM, Leong L, Steinbart EJ, Shofer JB, Crane PK, Larson EB, Peskind ER, Bird TD, Yu CE, 2013. TOMM40 intron 6 poly-T length, age at onset, and neuropathology of AD in individuals with APOE epsilon3/epsilon3. Alzheimers Dement 9(5), 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, Liu L, Yang S, Song N, Zhou X, Gao J, Yu N, Shan L, Wang Q, Liang J, Xuan C, Wang Y, Shang Y, Shi L, 2014. Histone demethylase KDM5B is a key regulator of genome stability. Proceedings of the National Academy of Sciences of the United States of America 111(19), 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linnertz C, Anderson L, Gottschalk W, Crenshaw D, Lutz MW, Allen J, Saith S, Mihovilovic M, Burke JR, Welsh-Bohmer KA, Roses AD, Chiba-Falek O, 2014. The cis-regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement 10(5), 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu JZ, Hov JR, Folseraas T, Ellinghaus E, Rushbrook SM, Doncheva NT, Andreassen OA, Weersma RK, Weismuller TJ, Eksteen B, Invernizzi P, Hirschfield GM, Gotthardt DN, Pares A, Ellinghaus D, Shah T, Juran BD, Milkiewicz P, Rust C, Schramm C, Muller T, Srivastava B, Dalekos G, Nothen MM, Herms S, Winkelmann J, Mitrovic M, Braun F, Ponsioen CY, Croucher PJ, Sterneck M, Teufel A, Mason AL, Saarela J, Leppa V, Dorfman R, Alvaro D, Floreani A, Onengut-Gumuscu S, Rich SS, Thompson WK, Schork AJ, Naess S, Thomsen I, Mayr G, Konig IR, Hveem K, Cleynen I, Gutierrez-Achury J, Ricano-Ponce I, van Heel D, Bjornsson E, Sandford RN, Durie PR, Melum E, Vatn MH, Silverberg MS, Duerr RH, Padyukov L, Brand S, Sans M, Annese V, Achkar JP, Boberg KM, Marschall HU, Chazouilleres O, Bowlus CL, Wijmenga C, Schrumpf E, Vermeire S, Albrecht M, Consortium U-P, Rioux JD, Alexander G, Bergquist A, Cho J, Schreiber S, Manns MP, Farkkila M, Dale AM, Chapman RW, Lazaridis KN, International, P.S.C.S.G., Franke A, Anderson CA, Karlsen TH, International, I.B.D.G.C., 2013. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet 45(6), 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, Ballard C, Banerjee S, Burns A, Cohen-Mansfield J, Cooper C, Fox N, Gitlin LN, Howard R, Kales HC, Larson EB, Ritchie K, Rockwood K, Sampson EL, Samus Q, Schneider LS, Selbaek G, Teri L, Mukadam N, 2017. Dementia prevention, intervention, and care. Lancet. [DOI] [PubMed] [Google Scholar]
- 60.Ma Y, Hebert JR, Li W, Bertone-Johnson ER, Olendzki B, Pagoto SL, Tinker L, Rosal MC, Ockene IS, Ockene JK, Griffith JA, Liu S, 2008. Association between dietary fiber and markers of systemic inflammation in the Women’s Health Initiative Observational Study. Nutrition 24(10), 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manly JJ, Schupf N, Stern Y, Brickman AM, Tang M-X, Mayeux R, 2011. Telephone-Based Identification of Mild Cognitive Impairment and Dementia in a Multicultural Cohort. Arch Neurol 68(5), 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F, 2016. The Ensembl Variant Effect Predictor. Genome Biol 17(1), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen LN, Ma D, Shui G, Wong P, Cazenave-Gassiot A, Zhang X, Wenk MR, Goh EL, Silver DL, 2014. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509(7501), 503–506. [DOI] [PubMed] [Google Scholar]
- 65.Nho K, Ramanan VK, Horgusluoglu E, Kim S, Inlow MH, Risacher SL, McDonald BC, Farlow MR, Foroud TM, Gao S, Callahan CM, Hendrie HC, Niculescu AB, Saykin AJ, Alzheimer’s Disease Neuroimaging, I., 2015. Comprehensive gene- and pathway-based analysis of depressive symptoms in older adults. J Alzheimers Dis 45(4), 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pimenova AA, Raj T, Goate AM, 2018. Untangling Genetic Risk for Alzheimer’s Disease. Biol Psychiatry 83(4), 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poirier J, Miron J, Picard C, Gormley P, Theroux L, Breitner J, Dea D, 2014. Apolipoprotein E and lipid homeostasis in the etiology and treatment of sporadic Alzheimer’s disease. Neurobiol Aging 35 Suppl 2, S3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian J, Wolters FJ, Beiser A, Haan M, Ikram MA, Karlawish J, Langbaum JB, Neuhaus JM, Reiman EM, Roberts JS, Seshadri S, Tariot PN, Woods BM, Betensky RA, Blacker D, 2017. APOE-related risk of mild cognitive impairment and dementia for prevention trials: An analysis of four cohorts. PLoS Med 14(3), e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raber J, Huang Y, Ashford JW, 2004. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging 25(5), 641–650. [DOI] [PubMed] [Google Scholar]
- 71.Radwan ZH, Wang X, Waqar F, Pirim D, Niemsiri V, Hokanson JE, Hamman RF, Bunker CH, Barmada MM, Demirci FY, Kamboh MI, 2014. Comprehensive evaluation of the association of APOE genetic variation with plasma lipoprotein traits in U.S. whites and African blacks. PLoS One 9(12), e114618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ravaglia G, Forti P, Maioli F, Chiappelli M, Montesi F, Tumini E, Mariani E, Licastro F, Patterson C, 2007. Blood inflammatory markers and risk of dementia: The Conselice Study of Brain Aging. Neurobiol Aging 28(12), 1810–1820. [DOI] [PubMed] [Google Scholar]
- 73.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma’ayan A, 2016. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt R, Schmidt H, Curb JD, Masaki K, White LR, Launer LJ, 2002. Early inflammation and dementia: a 25-year follow-up of the Honolulu-Asia Aging Study. Ann Neurol 52(2), 168–174. [DOI] [PubMed] [Google Scholar]
- 75.Sillen A, Brohede J, Lilius L, Forsell C, Andrade J, Odeberg J, Ebise H, Winblad B, Graff C, 2010. Linkage to 20p13 including the ANGPT4 gene in families with mixed Alzheimer’s disease and vascular dementia. J Hum Genet 55(10), 649–655. [DOI] [PubMed] [Google Scholar]
- 76.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nothen MM, White CC, Hamilton-Nelson KL, Epelbaum J, Maier W, Choi SH, Beecham GW, Dulary C, Herms S, Smith AV, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sanchez-Juan P, Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank-Garcia A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henandez I, Barber RC, Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel-Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Jr WTL, Fairchild TJ, Bossu P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris J, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairen MD, Hodges A, Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleo A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J, Consortium A, Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin LW, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Morris JC, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult-Stahlbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu CK, Gerad/Perades, C.A.E., Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Mann D, Tsolaki M, Clarimon J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Goate AM, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues JF, DeStefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, Van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Launer LJ, Escott-Price V, Mayeux R, Deleuze JF, Amin N, Holmans PA, Pericak-Vance MA, Amouyel P, van Duijn CM, Ramirez A, Wang LS, Lambert JC, Seshadri S, Williams J, Schellenberg GD, 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, Krull F, Bettella F, Eriksen JA, Witoelar A, Davies G, Fan CC, Thompson WK, Lam M, Lencz T, Chen CH, Ueland T, Jonsson EG, Djurovic S, Deary IJ, Dale AM, Andreassen OA, Neuro CCWG, 2017. Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teunissen CE, van Boxtel MP, Bosma H, Bosmans E, Delanghe J, De Bruijn C, Wauters A, Maes M, Jolles J, Steinbusch HW, de Vente J, 2003. Inflammation markers in relation to cognition in a healthy aging population. J Neuroimmunol 134(1–2), 142–150. [DOI] [PubMed] [Google Scholar]
- 79.Trampush JW, Yang MLZ, Yu J, Knowles E, Davies G, Liewald DC, Starr JM, Djurovic S, Melle I, Sundet K, Christoforou A, Reinvang I, DeRosse P, Lundervold AJ, Steen VM, Espeseth T, Raikkonen K, Widen E, Palotie A, Eriksson JG, Giegling I, Konte B, Roussos P, Giakoumaki S, Burdick KE, Payton A, Ollier W, Horan M, Chiba-Falek O, Attix DK, Need AC, Cirulli ET, Voineskos AN, Stefanis NC, Avramopoulos D, Hatzimanolis A, Arking DE, Smyrnis N, Bilder RM, Freimer NA, Cannon TD, London E, Poldrack RA, Sabb FW, Congdon E, Conley ED, Scult MA, Dickinson D, Straub RE, Donohoe G, Morris D, Corvin A, Gill M, Hariri AR, Weinberger DR, Pendleton N, Bitsios P, Rujescu D, Lahti J, Le Hellard S, Keller MC, Andreassen OA, Deary IJ, Glahn DC, Malhotra AK, Lencz T, 2017. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry 22(11), 1651–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ueki M, Kawasaki Y, Tamiya G, for Alzheimer’s Disease Neuroimaging, I., 2017. Detecting genetic association through shortest paths in a bidirected graph. Genet Epidemiol 41(6), 481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vinkers DJ, Gussekloo J, Stek ML, Westendorp RG, van der Mast RC, 2004. Temporal relation between depression and cognitive impairment in old age: prospective population based study. BMJ 329(7471), 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vinkers DJ, van der Mast RC, 2011. Temporal course of depressive symptoms during the development of Alzheimer disease: blowing hot and cold over depression and cognitive impairment. Neurology 76(17), 1531; author reply 1531, discussion 1531–1532. [DOI] [PubMed] [Google Scholar]
- 83.Weaver JD, Huang MH, Albert M, Harris T, Rowe JW, Seeman TE, 2002. Interleukin-6 and risk of cognitive decline: MacArthur studies of successful aging. Neurology 59(3), 371–378. [DOI] [PubMed] [Google Scholar]
- 84.Welsh-Bohmer KA, Ostbye T, Sanders L, Pieper CF, Hayden KM, Tschanz JT, Norton MC, Cache Country Study G, 2009. Neuropsychological performance in advanced age: influences of demographic factors and Apolipoprotein E: findings from the Cache County Memory Study. Clin Neuropsychol 23(1), 77–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Welsh KA, Breitner JCS, Magruderhabib KM, 1993. Detection of Dementia in the Elderly Using Telephone Screening of Cognitive Status. Neuropsy Neuropsy Be 6(2), 103–110. [Google Scholar]
- 86.Wilson RS, Schneider JA, Barnes LL, Beckett LA, Aggarwal NT, Cochran EJ, Berry-Kravis E, Bach J, Fox JH, Evans DA, Bennett DA, 2002. The apolipoprotein E epsilon 4 allele and decline in different cognitive systems during a 6-year period. Arch Neurol 59(7), 1154–1160. [DOI] [PubMed] [Google Scholar]
- 87.Witoelar A, Jansen IE, Wang Y, Desikan RS, Gibbs JR, Blauwendraat C, Thompson WK, Hernandez DG, Djurovic S, Schork AJ, Bettella F, Ellinghaus D, Franke A, Lie BA, McEvoy LK, Karlsen TH, Lesage S, Morris HR, Brice A, Wood NW, Heutink P, Hardy J, Singleton AB, Dale AM, Gasser T, Andreassen OA, Sharma M, International Parkinson’s Disease Genomics Consortium, N.A.B.E.C., United Kingdom Brain Expression Consortium, I., 2017. Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases. JAMA Neurol 74(7), 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiong F, Wu C, Chang J, Yu D, Xu B, Yuan P, Zhai K, Xu J, Tan W, Lin D, 2011. Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res 71(15), 5175–5181. [DOI] [PubMed] [Google Scholar]
- 89.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS, 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24), 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu B, Chen C, Xue G, Moyzis RK, Dong Q, Chen C, Li J, He Q, Lei X, Wang Y, Lin C, 2014. The SEMA5A gene is associated with hippocampal volume, and their interaction is associated with performance on Raven’s Progressive Matrices. Neuroimage 88, 181–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


