Abstract
Background
The severity and outcome of COVID-19 cases has been associated with the percentage of circulating lymphocytes (LYM%), levels of C-reactive protein (CRP), interleukin-6 (IL-6), procalcitonin (PCT), lactic acid (LA), and viral load (ORF1ab Ct). However, the predictive power of each of these indicators in disease classification and prognosis remains largely unclear.
Methods
We retrospectively collected information on the above parameters in 142 patients with COVID-19, stratifying them by survival or disease severity.
Findings
CRP, PCT, IL-6, LYM%, and ORF1ab Ct were significantly altered between survivors and non-survivors. LYM%, CRP, and IL-6 were the most sensitive and reliable factors in distinguishing between survivors and non-survivors. These indicators were significantly different between critically ill and severe/moderate patients. Only LYM% levels were significantly different between severe and moderate types. Among all the investigated indicators, LYM% was the most sensitive and reliable in discriminating between critically ill, severe, and moderate types and between survivors and non-survivors.
Conclusions
CRP, PCT, IL-6, LYM%, and ORF1ab Ct, but not LA, could predict prognosis and guide classification of COVID-19 patients. LYM% was the most sensitive and reliable predictor for disease typing and prognosis. We recommend that LYM% be further investigated in the management of COVID-19.
Funding
This study was supported in part by awards from the National Natural Science Foundation of China, the Foundation and Frontier Research Project of Chongqing, and the Chongqing Youth Top Talent Project.
Keywords: COVID-19, SARS-CoV-2, disease classification, prognosis, lymphocytes, lymphopenia, C-reactive protein, interlukin-6, procalcitonin, viral load
Graphical Abstract
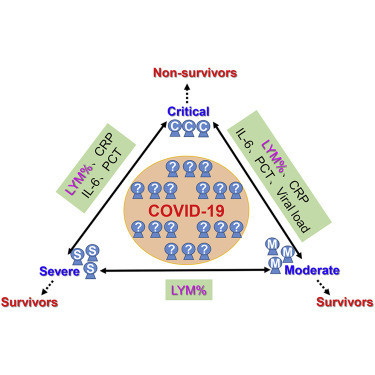
Highlights
Inflammatory cytokines, lymphocytes, and viral load predict prognosis in COVID-19 patients
Inflammatory cytokines, lymphocytes, and viral load indicate disease severity
The circulating percentage of lymphocytes distinguishes between severe and moderate types
Lymphocyte count is the most effective indicator of disease severity and prognosis
Context and Significance
There is an urgent need to identify factors that can predict outcome and disease severity in COVID-19 patients. In this study, researchers from the Third Military Medical University in Chongqing, China retrospectively enrolled 142 COVID-19 patients and measured their percentage of circulating lymphocytes as well as C-reactive protein, interleukin-6, procalcitonin, lactic acid, and viral load, factors that have been associated with the severity of the disease. The authors found that the percentage of circulating lymphocytes is the most sensitive and reliable predictor of disease severity and outcome and that levels of C-reactive protein and interleukin-6 can predict outcome. These findings may be useful in the treatment of COVID-19 patients and management of medical resources during the pandemic.
In this descriptive and retrospective study, Li Tan et al. demonstrate that the circulating percentage of lymphocytes, levels of C-reactive protein, interleukin-6, procalcitonin, and viral load can predict prognosis and guide classification of COVID-19 patients in different degrees. Among those indicators, lymphocyte percentage is the most sensitive and reliable predictor for disease typing and prognosis.
Introduction
Coronavirus disease 2019 (COVID-19) is an acute respiratory infective disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus.1 , 2 The COVID-19 pandemic has been spreading worldwide rapidly since March 2020. As of late April 2020, it was reported that more than 1,800,000 individuals had been diagnosed and this disease had caused over 200,000 deaths. The rapid increase in cases has led to heavy burdens on public healthcare resources and medical facilities.3 , 4 According to the New Coronavirus Pneumonia Diagnosis Program (5th edition), published by the National Health Commission of China, the disease severity of patients with COVID-19 can be divided into 4 categories: mild, moderate, severe, and critical.5 Current experience reveals that the majority of infected individuals (approximately >80%) are not severely affected and can recover without medical intervention, whereas a small number of cases need to be carefully treated and hospitalized.6, 7, 8, 9 The mortality rate for severe cases, particularly those that are critically ill, is quite high. It is therefore critical to identify reliable predictors for disease severity to improve outcomes and conserve medical resources.
Recent studies have shown that a variety of risk factors are associated with the prognosis of COVID-19. Patients with cardiovascular diseases, diabetes, and other comorbidities are often subject to acute respiratory distress syndrome, shock, multi-organ failure, cytokine storm, and other serious complications in COVID-19.8 , 10, 11, 12 These patients commonly have a poor prognosis. However, patients with moderate COVID-19 and no underlying diseases can also develop the complications described above and progress to the severe or critically ill types.11 , 13 According to the latest guidelines published by the National Health Commission of China, the diagnosis of severe and critically ill patients must rely on complex procedures such as imaging tests and blood gas analysis. In a pandemic situation, it is difficult to perform these diagnostic examinations on all patients. It is therefore particularly important to identify more easily detectable and earlier predictors to achieve extensive screening of patients and optimize the allocation of medical resources.
Recent studies from different cohorts of patients have identified several factors, including viral load,14 , 15 lymphocytes percentage,16 , 17 C-reactive protein (CRP),18 , 19 interleukin-6 (IL-6),17 , 20 procalcitonin (PCT),21 , 22 and lactic acid (LA)23 as warning indicators of prognosis in COVID-19 patients. However, it is still unclear which of these factors are the most sensitive and reliable indicators for predicting the prognosis of COVID-19 in the early stage. Besides, early indicators for disease classification are also urgently needed. In the present study, based on the clinical information of 142 patients with COVID-19, we compare and validate the predictive power of several reported risk factors for disease classification and prognosis in COVID-19 in a descriptive manner.
Results
Validation of Risk Factors for Prognosis in COVID-19 Patients
To investigate the mortality-associated risk factors, we initially collected clinical information from admission to discharge (or death) on a cohort of COVID-19 patients, dividing them into survivors (n = 117) and non-survivors (n = 15). First, the baseline information of these patients was analyzed. We found that older patients, especially those over 90 years of age, had a higher risk of mortality (Table S1). Female patients had a higher recovery rate and lower mortality rate than males (Table S1). The non-survivor group had a higher ratio of comorbidity, such as cardiovascular disease and diabetes, than the survivors (Table S2). In addition, relative to the cured patients, the non-survivors also had an increased incidence of complications like acute respiratory disease syndrome, septic shock, hemorrhagic shock, gastrointestinal bleeding, organ dysfunction, and multiple organ failure (Table S3).
Second, we retrospectively analyzed the serum levels of CRP, PCT, IL-6, and LA, the percentage of lymphocytes (LYM%), and Ct values of viral tests in the survivors and non-survivors. After disease onset, the levels of all the aforementioned indicators showed little change in the survivors, while they were notably altered in the non-survivors (Figures 1 A–1F). The levels of CRP and IL-6 in the peripheral blood of non-survivors increased quickly after symptom onset and further rose to a significantly higher level as compared to survivors (Figures 1A and 1C). A similar trend was observed for the PCT level, although it only began to increase in the late phase of the disease course (Figure 1B). In contrast, after symptom onset, the LYM% in non-survivors decreased quickly and remained at a significantly lower level as compared to survivors (Figure 1D). Unexpectedly, there was no obvious difference in blood LA levels between survivor and non-survivor groups (Figure 1E). In addition, non-survivors had obviously higher levels of viral load, indicated by ORF1ab Ct values, as compared to survivors (Figure 1F). Taken together, indicators CRP, PCT, IL-6, LYM%, and viral load could predict the mortality risk to varying extents. The strong predictive power of LYM% and IL-6 in mortality was also confirmed in a time-dependent ROC analysis (Figure S1).
Figure 1.
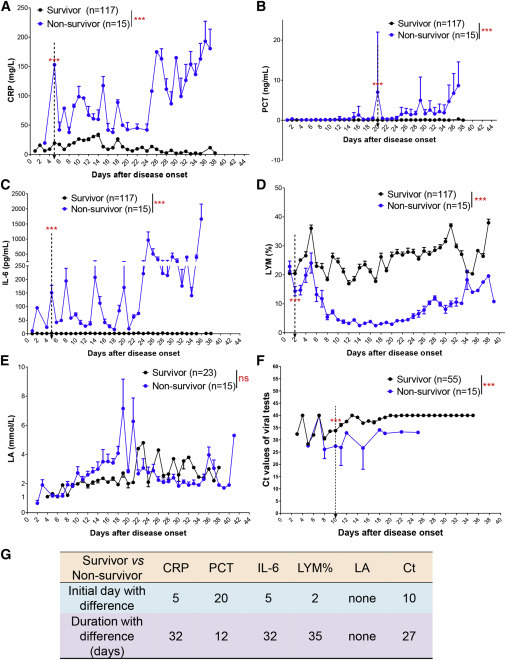
Validation of Indicators for the Prognosis of COVID-19 Patients
(A–D) Levels of CRP (A), PCT (B), IL-6 (C), and LYM% (D) in the peripheral blood of survivors (n = 117) or non-survivors (n = 15) with COVID-19 from admission to discharge.
(E) LA levels in the blood of survivors (n = 23) or non-survivors (n = 15) with COVID-19.
(F) ORF1ab Ct values in viral tests with qRT-PCR in survivors (n = 55) or non-survivors (n = 15) with COVID-19.
(G) Time windows of indicators for predicting prognosis. The initial day with difference (IDD) indicates the first day when the indicator levels were significantly different between two groups. The duration with difference (DD) indicates the time frame when the indicator levels were significantly different between two groups. Comparing two groups, the time points when the values of three consecutive measurements were significantly different were counted as days of DD. The endpoint of observation was 36 days after disease onset.
The dotted arrow indicates the IDD. Data show means ± SEM. ∗∗∗p < 0.001; a mixed model with repeated measure. CRP, C-reactive protein; PCT, procalcitonin; LYM%, lymphocyte percentage; LA, lactic acid; ns, not significant.
To descriptively study the potential of these measurements as predictors, we defined two assessment indexes. The initial day with difference (IDD) after disease onset was used to indicate the sensitivity of indicators in discriminating between two groups. The duration with difference (DD) in the disease course was used to indicate the reliability of discrimination. Comparing the two groups, the time points when the values of three consecutive measurements were significantly different were counted as days of DD. The end point of the observation was 36 days after disease onset, by which time most patients had either been discharged or died. Earlier IDD and longer DD indicated a better discrimination between two continuous curves. Results showed that indicators LYM%, CRP, and IL-6 had earlier IDD (2, 5, and 5 versus 20 and 10) and longer DD (35, 32, and 32 versus 12 and 27) than PCT and ORF1ab Ct values in distinguishing between survivors and non-survivors (Figure 1G). Notably, LYM% had the earliest IDD and longest DD (Figure 1G), indicating that it might be the most sensitive and reliable predictor for the prognosis of COVID-19 patients. Interestingly, in line with their better prognosis (Table S1), female patients also had higher levels of LYM% than males in the disease course, according to a mixed model analysis (Figure S2). These findings indicate that the immune status might determine the prognosis of COVID-19 patients.
CRP Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
To validate the roles of the aforementioned indicators in disease classification, we retrospectively reclassified our cohort of patients into critically ill (n = 25), severe (n = 21), and moderate (n = 96). The critically ill patients were mainly male (76%) and older than the severe and moderate patients (76 ± 16 versus 49 ± 15 or 56 ± 16) (Table S4). A mixed model analysis indicated that the critically ill and severe/moderate types of patients had significant differences in the levels of CRP while in the hospital (Figure 2 A). According to analysis with IDD and DD, we observed that CRP could be used to discriminate between the critically ill and severe/moderate COVID-19 patients, with a better result in typing between critically ill and moderate patients (Figure 2B). However, CRP levels could not distinguish between severe and moderate types (Figures 2A and 2B).
Figure 2.
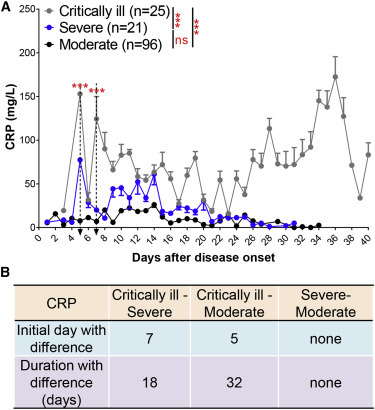
CRP Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
(A) Blood CRP levels in critically ill (n = 25), severe (n = 21), and moderate (n = 96) patients with COVID-19 from admission to discharge.
(B) Time windows of CRP for disease classification between critically ill, severe, and moderate types.
The dotted arrow indicates the IDD. Data show means ± SEM. ∗∗∗p < 0.001; a mixed model with repeated measure. ns, not significant.
PCT Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
We next performed a mixed-model analysis to determine whether PCT levels could discriminate between critically ill and severe/moderate patients (Figure 3 A). PCT was equally sensitive and reliable, classifying between critically ill and severe/moderate cases (Figure 3B). However, PCT levels could not distinguish between severe and moderate types.
Figure 3.
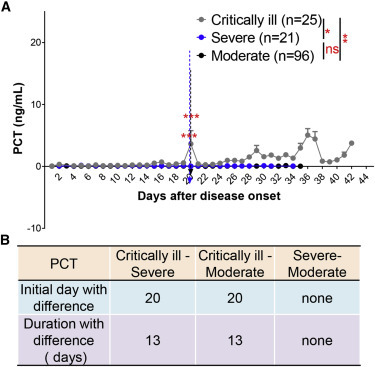
PCT Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
(A) Blood PCT levels in critically ill (n = 25), severe (n = 21), and moderate (n = 96) patients with COVID-19 from admission to discharge.
(B) Time windows of PCT for disease classification between those three types.
The IDD is marked by the dotted lines. Data show means ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; a mixed model with repeated measure. ns, not significant.
IL-6 Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
Likewise, we analyzed whether fluctuations in circulating IL-6 were significantly altered between critically ill and severe/moderate patients. Similar to CRP, we observed that IL-6 could be used to discriminate between critically ill and severe or moderate patients, but not between patients with moderate and severe COVID-19 (Figure 4 A). This indicated that IL-6 had a similar effect in typing between the critically ill and severe patients and between the critically ill and moderate ones (Figure 4B).
Figure 4.
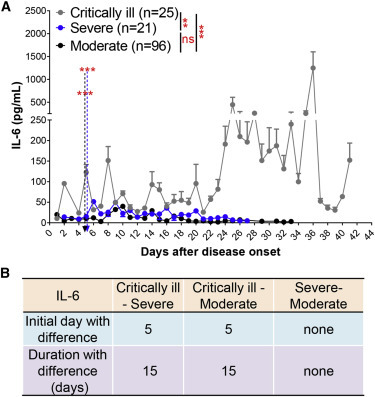
IL-6 Discriminates between Critically Ill and Severe/Moderate COVID-19 Patients
(A) Blood IL-6 levels in critically ill (n = 25), severe (n = 21), and moderate (n = 96) patients with COVID-19 from admission to discharge.
(B) Time windows of IL-6 for disease classification between those three types.
The IDD is marked by the dotted lines. Data show means ± SEM. ∗∗p < 0.01 and ∗∗∗p < 0.001; a mixed model with repeated measure. ns, not significant.
LYM% Distinguishes between Critically Ill, Severe, and Moderate COVID-19 Patients
LYM% levels were notably different between critically ill, severe, and moderate groups according to a mixed model analysis (Figure 5 A). Severe and critical patients had a rapid decrease of LYM% after disease onset. Severe patients had a subsequent restoration of LYM% 4 weeks after disease onset, whereas critical patients did not and, in some cases, had already succumbed to the disease by that time (Figure 5A). LYM% showed the earliest sensitivity and longest reliability in discriminating between groups (Figure 5B). These results indicated that LYM% could be a sensitive and reliable indicator to predict COVID-19 severity.
Figure 5.
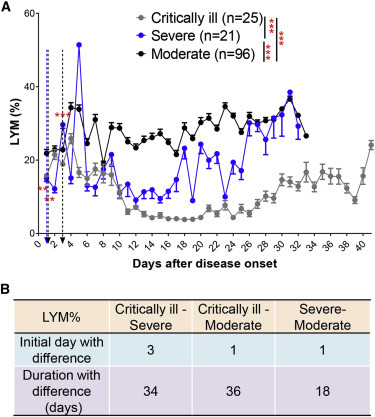
LYM% Distinguishes between Critically Ill, Severe, and Moderate COVID-19 Patients
(A) Blood LYM% in critically ill (n = 25), severe (n = 21), and moderate (n = 96) patients with COVID-19 from admission to discharge.
(B) Time windows of LYM% for disease classification between those three types.
The IDD is marked by the dotted lines. Data show means ± SEM. ∗∗∗p < 0.001; a mixed model with repeated measure.
LA Cannot Distinguish between Critically Ill and Severe COVID-19 Patients
LA levels were measured in blood gas assay, an analysis which is not commonly performed in moderately ill patients. We retrospectively investigated blood LA levels in the severe (n = 21) and critically ill patients (n = 25). However, there was no difference in the LA levels between these two groups of patients (Figures 6A and 6B)
Figure 6.
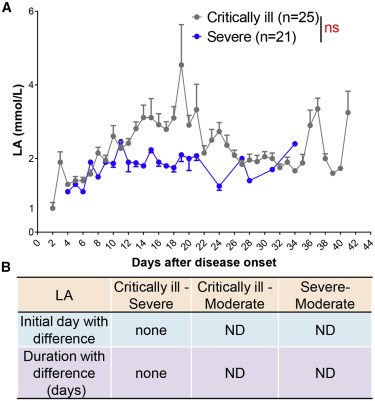
LA Cannot Distinguish between Critically Ill and Severe COVID-19 Patients
(A) Blood LA in critically ill (n = 25) and severe (n = 21) patients with COVID-19 from admission to discharge.
(B) Time windows of LA for disease classification between critically ill, severe, and moderate.
Data show means ± SEM. A mixed model with repeated measure. ns, not significant; ND, not detected.
Viral Load Distinguishes between Critically Ill and Moderate COVID-19 Patients
Unlike the indicators CRP, PCT, IL-6, or LYM%, ORF1ab Ct values were significantly altered only between critically ill and moderate groups (Figure 7 A). Our analysis also showed that ORF1ab Ct values could not distinguish between severe and moderate or critically ill patients (Figure 7B).
Figure 7.
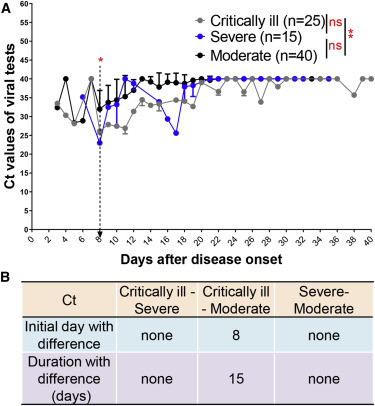
ORF1ab Ct Values Distinguish between Critically Ill and Moderate COVID-19 Patients
(A) ORF1ab Ct values in viral tests with qRT-PCR in critically ill (n = 25), severe (n = 15), and moderate (n = 40) patients with COVID-19 from admission to discharge.
(B) Time windows of ORF1ab Ct values for disease classification between those three types.
The IDD is marked by the dotted lines. Data show means ± SEM. ∗∗p < 0.01; a mixed model with repeated measure. ns, not significant.
Discussion
Given the worldwide prevalence of COVID-19, disease classification and prognostic indicators are of great significance in guiding treatment, conserving medical resources, and saving critically ill patients. In this study, we selected several reported risk factors for the prognosis of COVID-19 patients and further validated their predictive roles in disease outcome and classification. Most of the investigated factors were used for the first time to indicate the disease severity of COVID-19 patients.
We validated that the LYM% could be a sensitive and reliable predictor to distinguish between critically ill, severe, and moderate COVID-19 patients. Previous studies also supported the conclusion that lymphocyte count and function are closely related to the disease status of COVID-19.16 , 24 Patients with lowered immunity, including elderly or immunocompromised individuals, commonly presented a lower level of lymphocytes and a worse prognosis after infection with SARS-CoV-2.25 According to the present descriptive study, we show that LYM% had the earliest IDD and longest DD among all the included indicators in disease classification and prognosis prediction. Notably, we found that LYM% was the only indicator that could distinguish between the moderate and severe patients. These results indicate that LYM% was the most sensitive and reliable indicator for COVID-19.
Notably, as we described above, the serum levels of IL-6 and CRP presented with a zigzag curve during the entire course of disease in critically ill patients. We speculate that this obvious change may be caused by medical intervention. Interestingly, the changes of serum levels of these two markers in moderate and severe patients were much more stable and lower than those in critical patients. These results indicate that refractory inflammatory reaction can predict a poor outcome. Patients with this feature should be prioritized for treatment in the early stage. In clinical practice, the curve of inflammatory indicators should be drawn dynamically, similar to body temperature.
The higher levels of inflammatory indicators and the lower level of LYM% lead to another interesting question: whether the inflammatory cytokine storm is due to impaired lymphocytes, such as T cells. As we know, some T cell subsets, i.e., regulatory T cells (Tregs), are responsible for inflammatory regulation.26 , 27 The impaired function of Tregs may contribute to the uncontrolled inflammation in critical patients. It should be noted that, in the late stage of non-survivors, all the levels of proinflammatory indicators, LYM%, and LA displayed an acute increase (Figures 1A–1E). This phenomenon indicated a significant dysregulation of inflammation, immunity, and metabolism in these dying patients.
Persistently low levels of ORF1ab Ct values in viral tests indicated sustained high levels of viral load in the critically ill patients.15 However, it should be noted that there were no obvious differences in the duration of viral shedding between the severe and non-severe patients, which was consistent with a previous study.8 Recently, we reported a moderate patient with long duration of viral shedding for 49 days.28 This phenomenon was in line with evidence of asymptomatic infection reported in some populations.29
Conclusions
In the present study, based on the information obtained from 142 patients with COVID-19, we analyzed the predictive power of several reported indicators for disease severity and prognosis. We found that CRP, PCT, IL-6, LYM%, and viral load could predict the prognosis and guide classification of COVID-19 patients to different extents. LYM% was the most sensitive and reliable predictor for disease outcome and classification, especially for the typing of severe and moderate cases. During the COVID-19 pandemic, precise classification and prognosis prediction are critical for saving insufficient medical resources, stratifying treatment, and improving the survival rate of critically ill patients. We recommend that LYM% be used independently or in combination with other indicators in the management of COVID-19.
Limitations of Study
First, this study is a retrospective study in a single center and the conclusions obtained need to be further verified by other centers. Second, the sample size of the non-survivor group in this study is not large enough to exclude bias in the results of the analysis. Third, this study only suggests the potential of a series of indicators in disease typing and prognosis, and their validation and implementation in the clinic need to be further explored.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Patients blood samples and nasal swabs | This study | N/A |
| Critical Commercial Assays | ||
| C-Reactive Protein Reagents | Lifotronic Technology Co., Ltd | 8319178 |
| Elecsys® BRAHMS PCT | Roche Diagnostics GmbH | 05056888 200 |
| Elecsy® IL-6 | Roche Diagnostics GmbH | 05109442 190 |
| Fluorocell WDF | Sysmex Corporation | CV377552 |
| Lysercell WNR | Sysmex Corporation | AN577063 |
| Lysercell WDF | Sysmex Corporation | CV377552 |
| Lysercell WPC | Sysmex Corporation | CS412800 |
| SULFOLYSER | Sysmex Corporation | 83401621 |
| CELLPACK DFL | Sysmex Corporation | BN337547 |
| Radiometer ABL800(Lactic acid) | Radiometer | N/A |
| SARS-CoV-2 RT-qPCR Kit | DAAN Gene Corporation | DA0930 |
| Nucleic Acid Isolation Kit | Tianlong Science and Technology Co., Ltd | T183 |
| Oligonucleotides | ||
| Primer sequences for the ORF1ab: Forward primer (F): CCCTGTGGGTTTTACACTTAA; Reverse primer (R): ACGATTGTGCATCAGCTGA; Fluorescent probe (P): 5′-FAM-CCGTCTGCGGTATGT GGAAAGGTTATGG-BHQ1-3′ |
DAAN Gene Corporation | DA0930 |
| Software and Algorithms | ||
| GraphPad 8.01.244 | GraphPad | https://downloadly.win/graphpad-prism-8-0-1-244/ |
| SPSS Statistics for Windows, version 18.0 (SPSS 18.0) | SPSS Inc., Chicago, Ill., USA | https://www.ibm.com/support/pages/downloading-ibm-spss-modeler-180 |
| R 3.6.3 | Bell Laboratories (formerly AT&T, now Lucent Technologies) by John Chambers and colleagues. | https://www.r-project.org/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hongming Miao (hongmingmiao@sina.com).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate or analyze datasets.
Experimental Model and Subject Details
Patient information
All cases were taken from the General Hospital of Central Theater Command (Wuhan, Hubei province, People’s Republic of China). This study was approved by the Ethics Committee of the hospital. All subjects signed informed consent forms at admission to hospital. A total of 142 patients hospitalized between January 14, 2020 and March 14, 2020 were investigated, of which 96 cured cases were moderate type, 21 cured cases were severe type and 25 death cases were critically ill type. Among the critically ill, 15 patients died. The information on gender, age, comorbidities and complications in these patients were presented in Table S1–S4. All patients were confirmed by viral detections of SARS-CoV-2 via oropharyngeal swabs using quantitative RT-PCR for ORF1ab, which ruled out infection by other respiratory viruses such as influenza virus A, influenza virus B, coxsackie virus, respiratory syncytial virus, parainfluenza virus and enterovirus.
Method Details
Disease classification and discharge criteria
All cases were diagnosed and classified according to the New Coronavirus Pneumonia Diagnosis Program (5th edition) published by the National Health Commission of China. Clinical manifestations consist of four categories, mild, moderate, severe and critically ill. The mild clinical symptom were mild with no pulmonary inflammation on imaging. The moderate is the overwhelming majority, showing symptoms of respiratory infections such as fever, cough, and sputum, and pulmonary inflammation on imaging; when symptoms of dyspnea appear, including any of the following: shortness of breath, RR ≥ 30bpm, blood oxygen saturation ≤ 93% (at rest), PaO2 / FiO2 ≤ 300 mmHg, or pulmonary inflammation that progresses significantly within 24 to 48 hours > 50%, it was classified as severe; respiratory failure, shock, and organ failures that require intensive care were critically ill. Among them, mild patients were not admitted in this designated hospital.
The patient can be discharged if he/she simultaneously meets the following conditions recommended in the guidelines: 1) The body temperature returns to normal for more than 3 days. 2) Respiratory symptoms improve significantly. 3) Pulmonary imaging shows significant improvement in acute exudative lesions. 4) Negative nucleic acid test for two consecutive sputum and nasopharyngeal swabs and other respiratory tract specimens (at least 24 hours apart).
Laboratory examination of blood samples
Serum CRP levels were determined by the turbidimetry (Lifotronic PA900, Shenzhen, China). Serum PCT and IL-6 concentrations were measured by the electrochemiluminescence immunoassay (Roche COBAS e411, Mannheim, Germany). Routine blood tests including LYM% were performed by a hematology analyzer (Sysmex XN-9000, Kobe, Japan). LA levels were determined by a blood gas analyzer (Radiometer ABL800, Copenhagen, Denmark).
Quantitative RT-PCR assay for SARS-CoV-2
The pharyngeal swab of patients were collected and the total RNA was extracted using the nucleic acid extraction kit (Tianlong, Xi’an, China). The quantitative RT-PCR assay was performed using a SARS-CoV-2 nucleic acid detection kit (Chinese Center for Disease Control and Prevention recommended) according to the manufacturer’s protocol (DAAN Gene, Guangzhou, China). Open reading frame 1ab (ORF1ab) was simultaneously amplified and tested during the quantitative RT-PCR assay. A cycle threshold value (Ct-value) less than 40 was defined as a positive test result, and a Ct-value of 40 or more was defined as a negative test.
Data collection
In this study, the basic information, complete blood count, serum biochemical test, inflammatory indicators, viral load and disease outcome of all included patients were collected from admission to discharge or death. This study was approved by the Ethics Committee of the hospital. All subjects signed informed consent forms at admission to hospital.
Quantification and Statistical Analysis
In this study, GraphPad 8.01.244, SPSS 18.0 and R 3.6.3 were used for data mapping and statistics. Mann-whitney U test was used for comparison between two groups of continuous data, and kruskal-wallis H was used for comparison between multiple groups. Group comparisons for categorical variables were performed by using the Chi-square or Fisher exact probability methods. The laboratory examination indexes were expressed as the means ± s.e.ms. and were analyzed using a mixed model with repeated-measure. If the difference between two groups was significant, a descriptive study would be performed. Initial day with difference and duration with difference between two groups were used to respectively indicate the sensitivity and reliability of predictors for discrimination roughly. To calculate the hazard ratios (HR) of IL-6 and LYM%, the univariable Cox proportional hazard ratio regression model was performed. The sensitivity and specificity of prognostic indicators were evaluated by the time-dependent receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) was quantified by the timeROC package. All tests were bilateral, and p < 0.05 was considered statistically significant. For each parameter of all data presented, ∗ indicates p < 0.05, ∗∗ indicates p < 0.01 and ∗∗∗ indicates p < 0.001.
Acknowledgments
We thank all of the medical and scientific workers for the efforts in fighting against SARS-CoV-2. This study was supported in part by award numbers 81872028 and 81672693 (to H.M.) from the National Natural Science Foundation of China, cstc2017jcyjBX0071 (to H.M.) from the Foundation and Frontier Research Project of Chongqing, and T04010019 (to H.M.) from the Chongqing Youth Top Talent Project.
Author Contributions
Conceptualization, H.M.; Methodology, G.L., H.M., X.J., and X.K.; Investigation, L.T., Qiongshu Wang, Qi Wang, and X.J.; Writing–Original Draft, H.M. and X.K.; Writing–Review & Editing, H.M. and Y.L.; Funding Acquisition, H.M.; Resources, L.T., Qiongshu Wang, and Qi Wang; Supervision, Y.L., Qiongshu Wang, and H.M.
Declaration of Interests
The authors declare no competing interests.
Published: May 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.medj.2020.05.002.
Supplemental Information
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Rio C., Malani P.N. COVID-19-New Insights on a Rapidly Changing Epidemic. JAMA. 2020;323:1339–1340. doi: 10.1001/jama.2020.3072. [DOI] [PubMed] [Google Scholar]
- 3.Nicoli F., Gasparetto A. Italy in a Time of Emergency and Scarce Resources: The Need for Embedding Ethical Reflection in Social and Clinical Settings. J. Clin. Ethics. 2020;31:92–94. [PubMed] [Google Scholar]
- 4.Yan Y., Shin W.I., Pang Y.X., Meng Y., Lai J., You C., Zhao H., Lester E., Wu T., Pang C.H. The First 75 Days of Novel Coronavirus (SARS-CoV-2) Outbreak: Recent Advances, Prevention, and Treatment. Int. J. Environ. Res. Public Health. 2020;17:2323. doi: 10.3390/ijerph17072323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490. Published online February 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L.Q., Huang T., Wang Y.Q., Wang Z.P., Liang Y., Huang T.B., Zhang H.Y., Sun W., Wang Y. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korean Society of Infectious Diseases and Korea Centers for Disease Control and Prevention Analysis on 54 Mortality Cases of Coronavirus Disease 2019 in the Republic of Korea from January 19 to March 10, 2020. J. Korean Med. Sci. 2020;35:e132. doi: 10.3346/jkms.2020.35.e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K., Chen Y., Lin R., Han K. Clinical features of COVID-19 in elderly patients: A comparison with young and middle-aged patients. J. Infect. 2020;80:e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du R.H., Liang L.R., Yang C.Q., Wang W., Cao T.Z., Li M., Guo G.Y., Du J., Zheng C.L., Zhu Q. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020;55:2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC COVID-19 Response Team Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19) - United States, February 12-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa345. Published online March 28, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. Published online April 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan C., Huang Y., Shi F., Tan K., Ma Q., Chen Y., Jiang X., Li X. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J. Med. Virol. 2020 doi: 10.1002/jmv.25871. Published online April 13, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:1. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 21.Lippi G., Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chim. Acta. 2020;505:190–191. doi: 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., Li Q., Jiang C., Zhou Y., Liu S. Risk factors of critical & mortal COVID-19 cases: A systematic literature review and meta-analysis. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.021. Published online April 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang P., Ding Y., Xu Z., Pu R., Li P., Yan J., Liu J., Meng F., Huang L., Shi L. Epidemiological and clinical features of COVID-19 patients with and without pneumonia in Beijing, China. medRxiv. 2020 doi: 10.1101/2020.02.28.20028068. 2020.2002.2028.20028068. [DOI] [Google Scholar]
- 24.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. Published online March 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Applegate W.B., Ouslander J.G. COVID-19 Presents High Risk to Older Persons. J. Am. Geriatr. Soc. 2020;68:681. doi: 10.1111/jgs.16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okeke E.B., Uzonna J.E. The Pivotal Role of Regulatory T Cells in the Regulation of Innate Immune Cells. Front. Immunol. 2019;10:680. doi: 10.3389/fimmu.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhry A., Rudensky A.Y. Control of inflammation by integration of environmental cues by regulatory T cells. J. Clin. Invest. 2013;123:939–944. doi: 10.1172/JCI57175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan L., Kang X., Zhang B., Zheng S., Liu B., Yu T., Yang F., Wang Q., Miao H. A special case of COVID-19 with long duration of viral shedding for 49 days. medRxiv. 2020 doi: 10.1101/2020.03.22.20040071. 2020.2003.2022.20040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimball A., Hatfield K.M., Arons M., James A., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Chisty Z., Public Health – Seattle & King County. CDC COVID-19 Investigation Team Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility - King County, Washington, March 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:377–381. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets.


