Abstract
Ubiquitin Proteasome System (UPS) is an adaptable and finely tuned system that sustains proteostasis network under a large variety of physiopathological conditions. Its dysregulation is often associated with the onset and progression of human diseases; hence, UPS modulation has emerged as a promising new avenue for the development of treatments of several relevant pathologies, such as cancer and neurodegeneration. The clinical interest in proteasome inhibition has considerably increased after the FDA approval in 2003 of bortezomib for relapsed/refractory multiple myeloma, which is now used in the front-line setting. Thereafter, two other proteasome inhibitors (carfilzomib and ixazomib), designed to overcome resistance to bortezomib, have been approved for treatment-experienced patients, and a variety of novel inhibitors are currently under preclinical and clinical investigation not only for haematological malignancies but also for solid tumours. However, since UPS collapse leads to toxic misfolded proteins accumulation, proteasome is attracting even more interest as a target for the care of neurodegenerative diseases, which are sustained by UPS impairment. Thus, conceptually, proteasome activation represents an innovative and largely unexplored target for drug development. According to a multidisciplinary approach, spanning from chemistry, biochemistry, molecular biology to pharmacology, this review will summarize the most recent available literature regarding different aspects of proteasome biology, focusing on structure, function and regulation of proteasome in physiological and pathological processes, mostly cancer and neurodegenerative diseases, connecting biochemical features and clinical studies of proteasome targeting drugs.
Keywords: Proteasome, Proteasome inhibitors, Cancer, Neurodegeneration, SARS-Cov-2
1. Introduction
Under physiological conditions human cells express approximately 10,000 proteins that must be properly folded to carry out their biological functions (Klaips, Jayaraj, & Hartl, 2018; Kulak, Geyer, & Mann, 2017). To comply with their role, some proteins retain a certain degree of structural flexibility which may render them more prone to misfolding and aggregation (Chiti & Dobson, 2006; Ciechanover & Kwon, 2017). When proteins unfold, as a consequence of aging and/or environmental stress, or else are no functionally required, they undergo degradation to limit the threat raised by their maintenance (Klaips et al., 2018). Thus, proteome fidelity (proteostasis) is achieved through a complex and multi-subcellular compartments network, which coordinates synthesis, folding, conformational upkeep and degradation (Labbadia & Morimoto, 2015; Powers, Morimoto, Dillin, Kelly, & Balch, 2009).
Finding a universal definition of the proteostasis network (PN), which would encompass the structural composition, hierarchical organization and dynamics of recruitment of main actors is particularly challenging, mostly because the expression and the activity of many PN factors are tailored depending on the different physiological stimuli the cell may experience in the context of its tissue microenvironment. PN properties can be altered by physio-pathological and multi-factorial phenomena (e.g., aging and/or environmental stress), or by mutations in PN components, which may lead to the onset/progression of different pathologies, including cancer, neurodegenerative disorders or other genetic diseases sustained by altered proteostasis (Balch, Morimoto, Dillin, & Kelly, 2008; Labbadia & Morimoto, 2015; Powers et al., 2009).
A general and widely accepted view of the PN encompasses three major branches, namely: 1) protein synthesis, which adjusts the level of bulk proteins to cell demands; 2) protein folding, which is mediated by a vast repertoire of chaperones (now referred to as “chaperome”); 3) protein degradation, which allows the proteolytic removal of undesired proteins through two main intracellular proteolytic systems, namely Ubiquitin-Proteasome-System (UPS) and autophagy (Ciechanover & Kwon, 2017; Klaips et al., 2018; Sala, Bott, & Morimoto, 2017). Furthermore, a myriad of regulatory proteins (such as transcription and metabolic factors, chromatin remodelling factors, and regulators of posttranslational modifications) act as PN auxiliary and coordinate the cross-talk between the PN compartments accounting for the afore mentioned plasticity of the PN (Klaips et al., 2018; Labbadia & Morimoto, 2015).
Therefore, unlike early scientists, who considered proteins essentially stable and prone to only a minor “wear and tear” (Schoenheimer, 1946; Schoenheimer, Ratner, & Rittenberg, 1939; Thibaudeau & Smith, 2019), it is now known that proteome is highly dynamic, and proteins constantly undergo turn over at different rates, according to their biological role (Lecker, Goldberg, & Mitch, 2006; Thibaudeau & Smith, 2019).
In the 1950s, the discovery of autophagy-lysosome system as “intracellular exergonic digestive system” by de Duve and colleagues was the first step in understanding intracellular and extracellular protein breakdown (De Duve, Gianetto, Appelmans, & Wattiaux, 1953; de Duve, Pressman, Gianetto, Wattiaux, & Appelmans, 1955; De Duve & Wattiaux, 1966; Sabatini & Adesnik, 2013). Over the same years, Simpson showed for the first time that intracellular proteolysis in mammalian cells requires energy, suggesting the existence of an additional mechanism of protein degradation (Simpson, 1953). However, this observation was considered with scepticism, since hydrolysis of the peptide bond is exergonic, and there is no apparent thermodynamic advantage in energy use (Wilkinson, 2005). However, the seminal Simpson's discovery found support in the 1970s, when Goldberg and colleagues identified a novel, cytosolic ATP-dependent proteolytic system (Bigelow, Hough, & Rechsteiner, 1981; Etlinger & Goldberg, 1977; Goldberg, 1972; Goldberg & Dice, 1974; Goldberg & St John, 1976; Thibaudeau & Smith, 2019; Wilkinson, 2005). Some years later, Wilk and Orlowski purified a 700-kDa “multicatalytic proteinase complex”, which was able to cleave peptides after hydrophobic, acidic and basic residues, suggesting the existence of multiple active sites in its structure (Wilk & Orlowski, 1980; Wilk & Orlowski, 1983). This “stacked donut ring” complex (which later was shown to be the 20S) was tnamed “proteasome”, and its orthologues were identified in all life domains (Arrigo, Tanaka, Goldberg, & Welch, 1988; Tanaka et al., 1988; Tanaka, Waxman, & Goldberg, 1983; Thibaudeau & Smith, 2019). A milestone in protein degradation field was the discovery by Ciechanover and colleagues of a 8-kDa heat-stable protein, APF-1 (later renamed “ubiquitin”), whose ATP-dependent covalent conjugation with proteins targeted them for degradation by a downstream protease, that was then identified as the 26S proteasome (Ciechanover, 2005; Ciechanover, 2013; Ciechanover, Finley, & Varshavsky, 1984; Ciechanover, Heller, Elias, Haas, & Hershko, 1980; Ciechanover, Hod, & Hershko, 2012; Hershko, Ciechanover, Heller, Haas, & Rose, 1980; Hershko, Eytan, Ciechanover, & Haas, 1982; Hough, Pratt, & Rechsteiner, 1986; Hough, Pratt, & Rechsteiner, 1987; Leestemaker & Ovaa, 2017; Varshavsky, 2006).
Over the last decade, the critical role played by UPS in the maintenance of protein homeostasis and its involvement in the pathogenesis of human diseases have been largely investigated. With respect to this, proteasome is now considered a crucial target for therapeutic intervention in many diseases, such as neurodegenerative, immune-related disorders and cancer.
In this review, we will first discuss the structure and function of proteasome under physiological conditions; then we will focus our attention on the alterations of the proteasome functionality involved in the onset and progression of neurodegeneration and cancer. Finally, we will summarize: 1) the FDA- and EMA-approved proteasome inhibitors that are used for cancer treatment as well as novel promising inhibitors currently investigated in preclinical studies and clinical trials; 2) proteasome activators as novel tools to treat neurodegenerative disorders.
2. Proteasome structure and function
2.1. General organization of UPS
The UPS is the major actor in the turn-over of most cellular soluble proteins, playing fundamental roles in several facets of cell life, such as cell cycle, apoptosis, DNA repair, antigen presentation, inflammation, cellular response to environmental stress, and morphogenesis of neuronal networks (Glickman & Ciechanover, 2002; Kunjappu & Hochstrasser, 2014).
UPS displays a hierarchical organization which encompasses two intertwined and consecutive steps: 1) the covalent attachment of ubiquitin polymers to substrates; 2) degradation by the 26S proteasome of ubiquitin-tagged substrates, followed by the release of free and recyclable ubiquitin moieties along with oligopeptides of cleared protein (Scheffner, Nuber, & Huibregtse, 1995; Glickman & Ciechanover, 2002; Grasso et al., 2017). Ubiquitin conjugation proceeds through a three-step mechanism. First, the ubiquitin-activating enzyme E1 activates ubiquitin in an ATP-dependent manner, generating a high-energy thiol ester intermediate. In the second step, activated ubiquitin is then transferred from E1 to one of several E2 enzymes (ubiquitin-conjugating enzymes), leading to the formation of another high-energy thiol ester intermediate. Finally, ubiquitin is conjugated to substrates by a ubiquitin (E3) ligase, which is responsible for substrate specificity (Ciechanover, 2013; Glickman & Ciechanover, 2002; Hough et al., 1986; Leestemaker & Ovaa, 2017; Pao et al., 2018; Pickart, 2001; Windheim, Peggie, & Cohen, 2008). The end-point of UPS is the 26S complex (hereafter referred to as 26S), a multifunctional 2500 kDa proteolytic molecular machine, composed by the 20S proteasome core particle (CP, hereafter referred to as 20S), which houses the proteolytic activity. The 20S is capped by one or two 19S regulatory particle(s) (RP) (hereafter referred to as 19S), which carry out the ATP-dependent recognition, unfolding and translocation into the 20S of the poly-ubiquitinated substrate (Ciechanover, 2005; Glickman & Ciechanover, 2002; Kunjappu & Hochstrasser, 2014; Pao et al., 2018, see also 2.2, 2.3). Over the last decades, several alternative regulators of 20S have been described, namely PA28 protein family and Blm10/PA200, whose structure, substrate specificities, and biological roles go beyond the scope of this review and are extensively reviewed elsewhere (Rechsteiner & Hill, 2005; Tanaka, 2009; Huang & Chen, 2009; Kish-Trier & Hill, 2013; Cascio, 2014; Poot et al., 2014; Schmidt & Finley, 2014; Jiang, Zhao, & Qiu, 2018; Limanaqi, Biagioni, Gaglione, Busceti, & Fornai, 2019).
Although the initial dogma on proteasome recognition mechanism states that the 26S hydrolyses only proteins tagged with at least four ubiquitin molecules, emerging evidences show that poly-ubiquitin chains are not the unique signal. In fact, in some cases, multiple or single mono-ubiquitination appears to be sufficient to label a substrate for proteasomal degradation (Kravtsova-Ivantsiv, Cohen, & Ciechanover, 2009; Shabek et al., 2012). Moreover, ornithine decarboxylase has been the first of a long series of protein substrates (i.e., Rpn4, thymidylate synthase, myelin) reported to be degraded by the 26S regardless of ubiquitination (Bercovich, Rosenberg-Hasson, Ciechanover, & Kahana, 1989; Chen, Barton, Chi, Clurman, & Roberts, 2007; Forsthoefel, Peña, Xing, Rafique, & Berger, 2004; Ju & Xie, 2004; Kudriaeva, Kuzina, Zubenko, Smirnov, & Belogurov, 2019; Li, Yuan, Pan, Liu, & Huang, 2016; Murakami et al., 1992; Rosenberg-Hasson, Bercovich, Ciechanover, & Kahana, 1989; Sheaff et al., 2000). This implies the existence of alternative molecular signals (also named “degrons”), such as specific amino acidic sequence or structural elements, that mediate proteasome recognition and degradation of substrates independently on their ubiquitination levels (Baugh, Viktorova, & Pilipenko, 2009; Kudriaeva & Belogurov, 2019).
The biological significance of ubiquitin-independent degradation of substrates by the 26S is a topic deserving great attention in order to decipher its physiological meaning in tissue homeostasis. Two proposed explanations envisage that it could be “only” a remnant of evolution, or else it could be rather a mechanism that provides, under selected circumstances, an alternative strategy to overcome the de-regulation of the canonical ubiquitin-dependent pathway (Erales & Coffino, 2014; Finley, 2009). In support of this second hypothesis, the turnover of Rpn4, a substrate and a transcriptional regulator of proteasome genes, is carried out through both ubiquitin-dependent and ubiquitin-independent pathways, providing the cell with an alternative mechanism to modulate the level of Rpn4 and of proteasome in the case of inappropriate ubiquitin conjugation (Erales & Coffino, 2014; Hanna, Meides, Zhang, & Finley, 2007; Ju & Xie, 2004).
In this regard, an intriguing example of how ubiquitin-dependent and ubiquitin-independent pathways cooperate to survey cellular homeostasis comes from the regulation of the proteome of lipid droplets (LDs), that are ubiquitous, endoplasmic reticulum-derived storage organelles from which neutral lipids are rapidly mobilized in response to cellular demands. In fact, some proteins of LDs are degraded by proteasome through the canonical ubiquitination pathway, whereas some others are processed only when the “degron” signals become unmasked upon protein insertion into the lipid monolayer (Bersuker & Olzmann, 2017). Interestingly, it has been reported that proteasome mediates ubiquitin-dependent degradation of patatin-like phospholipase domain-containing protein 3 (PNPLA3), whose sequence variant 148 M is resistant to ubiquitination and to proteasome degradation, and accumulates into LDs, contributing to non-alcoholic fatty liver disease pathogenesis (Basu Ray, 2019; Kozlitina et al., 2014; Speliotes et al., 2011).
An additional issue in deciphering the mechanisms of proteasome degradation is the ubiquitin-independent degradation in vitro of macromolecular substrates by the uncapped 20S. In fact, several studies demonstrate that the 20S is able to degrade natively unfolded as well as oxidized and misfolded proteins (Davies, 1993; Davies, 2001; Grune, Reinheckel, & Davies, 1996; Raynes, Pomatto, & Davies, 2016; Reinheckel et al., 1998; Shringarpure, Grune, Mehlhase, & Davies, 2003). Indeed, oxidative stress induces chemical alterations, bringing about conformational changes and exposure of hydrophobic residues on damaged protein surfaces (Carrard, Bulteau, Petropoulos, & Friguet, 2002; Raynes et al., 2016). These surface hydrophobic patches stimulate, in an allosteric fashion, the translocation of the substrate into the 20S proteolytic chamber (see Section 2.2 for details) (Coux, Tanaka, & Goldberg, 1996; Davies, 2001; Giulivi, Pacifici, & Davies, 1994; Kisselev, Kaganovich, & Goldberg, 2002), since under oxidative stress conditions this form is more stable than the 26S, which is quickly and reversibly inactivated likely through dissociation into free 20S and 19S particles (Reinheckel et al., 1998; Reinheckel, Ullrich, Sitte, & Grune, 2000; Shringarpure et al., 2003; Wang et al., 2010; Wang et al., 2017). Moreover, also the E1-E2-E3 cascade is transiently inactivated during oxidative stress, supporting an ubiquitin-independent degradation of oxidized proteins (Grune et al., 2011). Thus, the current view is that 20S activity on oxidized and damaged proteins might compensate for the loss of the ubiquitin-dependent activity of the 26S under redox imbalance.
Interestingly, the activity of 20S on these subsets of substrates might be assisted by PA28 which seems to increase its selectivity and activity (Fabre et al., 2014; Grune et al., 2011; Pickering & Davies, 2012). This occurrence is further discussed below in regard to the neurodegenerative disorders, since up-regulation of PA28 of immune-proteasome (i.e., an inducible proteasome subset expressed in hematopoietic cells, which plays a significant role in immunity, see Box 1) is a common feature of this class of human pathologies. Along with this, ubiquitin -independent degradation by 20S has been also demonstrated for substrates that, like oxidized proteins, have regions characterized by high hydrophobicity, such as tau protein and α-synuclein. These findings reinforce the role of 20S in the regulation of protein homeostasis independently from its association with RPs (Asher, Tsvetkov, Kahana, & Shaul, 2005; Baugh et al., 2009; David et al., 2002; Dyson & Wright, 2005; Raynes et al., 2016; Tofaris, Layfield, & Spillantini, 2001).
The herein described picture underlines the complexity of proteasome heterogeneity, since proteasome composition, specificity and activity are flexible and finely regulated at multiple steps, including post-translational modification and regulatory factors (i.e., proteasome interacting proteins) (Morozov & Karpov, 2019; Tanaka, 2009; Tundo et al., 2017). Since the proteasome pathway is extremely dynamic and reflects cellular metabolic demands (Dahlmann, 2016; Hirano, Kimura, & Kimura, 2016), it is not surprising that different proteasome forms may co-exist and fulfil different but interconnected functions that are not yet completely understood (Morozov & Karpov, 2019).
As a matter of fact, it should be emphasized that, in vertebrates, proteasome has gained considerable tissue-specificity, as supported by the existence of alternative forms of proteasome, namely (see Box 1): immuno-proteasome, thymo-proteasome, and spermato-proteasome, in which constitutive catalytic subunits of 20S are replaced by inducible/tissue-specific homologs. This is a clear-cut example of evolutionary-based mechanisms for the refinement of intracellular proteolysis (Kniepert & Groettrup, 2014; Morozov & Karpov, 2019; Murata et al., 2007; Qian et al., 2013; Tanaka, 2009; Uechi, Hamazaki, & Murata, 2014).
2.2. 20S Core particle
2.2.1. 20S structural arrangement
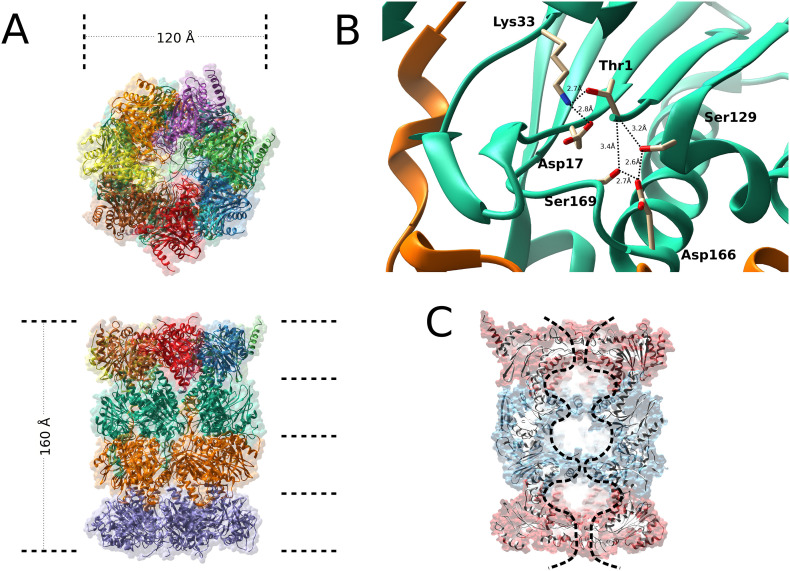
The 20S core particle, which belongs to the N-terminal nucleophilic (Ntn) hydrolase family, is a cylinder-like packed particle which contains four axial stacking heptameric rings, arranged into two outer α-rings and two inner β-rings (i.e α1–7β1–7α1–7β1–7) ( Baumeister et al., 1988; Bochtler, Ditzel, Groll, Hartmann, & Huber, 1999; Groll et al., 1997; Groll et al., 2000; Kunjappu & Hochstrasser, 2014; Tanaka, 2009). Electron micrographic studies measured its molecular dimensions that are 160 Å in length and 120 Å in diameter (Borissenko & Groll, 2007; Harris, 1968) (Fig. 1A). Eukaryotic 20S has a central channel, which houses proteolytic chambers distributed among six active β-subunits, three for each β-ring, namely: the chymotryptic-like (β5 subunit), the trypsin-like (β2) and caspase-like (β1) sites, which confer the property to preferentially cleave proteins after hydrophobic, basic and acidic residues, respectively (Groll & Huber, 2003; Tanaka, 2009; Unno et al., 2002). Historically, the 20S was the first enzyme classified as threonine protease, in which the hydroxyl group of the N-terminal Thr1 acts as nucleophile in all active subunits (Chen & Hochstrasser, 1995; Kisselev, Songyang, & Goldberg, 2000; Löwe et al., 1995). Thr1Oγ nucleophilic attack to the carbonyl carbon atom of the scissile peptide bond generates a first cleavage product, forming a covalent acyl-enzyme tetrahedral intermediate, followed by hydrolysis through the addition of a nucleophilic water molecule, which regenerates the functional active site and releases the second cleavage product (Löwe et al., 1995; Marques, Palanimurugan, Matias, Ramos, & Dohmen, 2009; Groll et al., 1999; Huber et al., 2009). Conserved residues in the proximity of Thr1, involved in the proteolysis mechanism, are Glu/Asp17 and Lys33. Lys33, which forms a salt bridge with Asp17, is positively charged at neutral pH, contributing to lower the pK a of Thr1 amino group, so that this group can work as the proton acceptor required for the activation of Thr1 hydroxyl group (Borissenko & Groll, 2007; Groll & Huber, 2003; Löwe et al., 1995) (Fig. 1B). Moreover, Ser129, Asp166, and Ser169 residues, which are close to Thr1, are required for structural stability of the proteolytic centre (Borissenko & Groll, 2007; Chen & Hochstrasser, 1995; Heinemeyer, Fischer, Krimmer, Stachon, & Wolf, 1997; Löwe et al., 1995; Seemuller, Lupas, & Baumeister, 1996). Recently, a revised interpretation of the proteasome active site architecture has been proposed, according to which proteasome can be viewed as having two triads, both essentials for an efficient proteolysis, consisting of (i) Thr1, Lys33 and Asp/Glu17 residues and (ii) Thr1, Ser129 and Asp166 residues, respectively (Huber et al., 2016). In this novel vision, Lys33 -NH2 group is expected to act as the proton shuttle, while Asp17Oγ orients Lys33 -NH2 group, making it more prone to protonation, by raising its pK a (Huber et al., 2016). The positive charge on Thr1-NH3 + group, which is essential for the binding and stabilization of the amide nitrogen of incoming peptide substrates, is favoured by the close proximity of Ser129 and Asp166 residues, which increase its pK a value, this being a crucial step for the first cleavage and for proteolytic reaction progress. In conclusion, Lys33 and Asp17 seem to be required to deprotonate the Thr1 hydroxyl side chain, whereas Ser129 and Asp166 are needed to protonate the N-terminal amine group of Thr1 (Huber et al., 2016; Vielberg, Bauer, & Groll, 2018) (Fig. 1B). It is important to emphasize that the names used to describe β-subunits catalytic activities do not reflect accurately the specificity of each active site, which is much broader, meaning that 20S function cannot be simply interpreted as the integration of the three different activities into a unique machine (Bochtler et al., 1999; Groll & Huber, 2003). Accordingly, it has been reported that the substrate specificity is modulated not only by P1 residue of the substrate, but also by the physical constraints of the substrate around proteasome active site (Bogyo, Shin, McMaster, & Ploegh, 1998; Cardozo, Vinitsky, Michaud, & Orlowski, 1994; Dick et al., 1998; Groll et al., 1997; Groll & Huber, 2003; Groll, Nazif, Huber, & Bogyo, 2002). Moreover, biochemical analysis suggested a network of intricate interconnections among the three active sites (i.e., the so called “bite and chew” mechanism), in which the chymotryptic-like site performs the first cleavage (i.e., the “bite”), followed by a series of cleavage steps at the trypsin-like and caspase-like sites (i.e., the “chewing”) (Kisselev, Akopian, Castillo, & Goldberg, 1999; Śledź et al., 2013).
Fig. 1.
Structure of 20S.
A. Structure of the 20S proteasome particle as viewed from the top (top panel) or the side (bottom panel). The protein backbone of the subunits is presented as ribbon. B. Active site of the threonine peptidase subunit (β5) of the proteasome. The protein backbone of the β5 subunit is represented as turquoise ribbon, catalytic residue (Thr1) and other residues that help to maintain the structural stability of the catalytic site (Lys33, Asp17, Ser129, Asp166 and Ser169) are represented as sticks. Polar interactions are indicated as black dashed lines together with the corresponding distances. C.
Vertical cross-section of the 20S particle, the α-subunit rings are represented as red ribbons, the β-subunit rings as blue ribbons, the outline of the internal cavity and the internal “chambers” are highlighted with a black dashed line.
All catalytically active subunits (i.e., β1, β2 and β5) are synthesized as inactive precursors, which gain their hydrolytic properties only after the proper assembly of two half-proteasome assemblies, thus generating the 20S active form (see Section 2.3 for details) (Zwickl, Kleinz, & Baumeister, 1994; Brannigan et al., 1995; Seemüller et al., 1995: Ditzel et al., 1998; Huber et al., 2016). During the final step of the proteasome maturation process, segments of the immature active sites are removed by autolysis between residues Thr1 and Gly1, a process closely related to the proteolysis mechanism (Budenholzer, Cheng, Li, & Hochstrasser, 2017; Chen & Hochstrasser, 1995; Chen & Hochstrasser, 1996; Huber et al., 2016). The pro-peptides of different β subunits act then as “co-chaperones” during 20S assembly (see Section 2.3 for details) (Budenholzer et al., 2017; Kunjappu & Hochstrasser, 2014). Noteworthy, a critical function of the pro-peptide sequence is to prevent the Nα- acetylation of catalytic Thr in β-subunits, which would block the active site function before the formation of the half-proteasome; thus, pro-peptides are removed only when the half-proteasomes are correctly assembled, an occurrence which impairs the access of Nα-acetyltransferase to active sites (Arendt & Hochstrasser, 1999; Budenholzer et al., 2017; Groll et al., 1999; Schmidtke et al., 1996; Seemuller et al., 1996). Concerning the remaining four β-subunits, it should be remarked that subunits β3, β4, and β6 subunits, which lack of the nucleophilic threonine in position 1, are catalytically inactive, and attempts to render them proteolytically active through site-directed mutations have failed (Chen et al., 1995; Groll et al., 1999; Heinemeyer et al., 1997). Conversely, the β7 subunit, which keeps conserved Thr1 and Gly-1 residues (like active subunits) remains inactive since during the maturation process the β7 pro-peptide is not cleaved at Thr 1, but at position Thr8, due to substitutions of Lys33 and Ser129 by Arg33 and Phe129, respectively. It has been also proposed that the β7 subunit shows a Ntn-hydrolase proteolytic activity at Thr8, even though the surroundings of the proposed active site differ significantly from those of other subunits, but its role is still unknown (Borissenko & Groll, 2007; Unno et al., 2002).
In proteasome architecture, while β-rings contain the proteolytic active sites (as discussed above), the outer α-rings form a nearly flat surface that binds to RPs (i.e., 19S, PA28, see Section 2.3 for details). In the free 20S (that is not engaged with the RP), N-terminal tails of the α-subunits point all inwards to the centre of the ring and neighbouring tails are tightly anchored by an intricate lattice of intra-subunits interactions, constituting “the gate”, which regulates the substrate access through a 13 Å entry pore into the antechamber (i.e, at α7-β7 interface). This passageway keeps the substrate in an unfolded state, directing it toward the catalytic chamber (i.e., at β7-β7 interface) (Bajorek & Glickman, 2004; Gaczynska & Osmulski, 2014; Groll et al., 2000; Marques et al., 2009; Ruschak, Religa, Breuer, Witt, & Kay, 2010; Unno et al., 2002) (Fig. 1C). The insertion of the substrate through this “N-terminal gate” is the rate-limiting step of proteasome activity and prevents unwanted protein degradation (Akopian, Kisselev, & Goldberg, 1997; Groll et al., 2000; Köhler et al., 2001). In fact, RP binding induces the N-terminal tails displacement and opens the gate, facilitating the substrate translocation (see Section 2.3) (Choi et al., 2016; Finley, Chen, & Walters, 2016; Marshall & Vierstra, 2019; Matyskiela, Lander, & Martin, 2013; Śledź et al., 2013). However, it is worth recalling that RP binding to 20S is not an absolute requirement for proteasome activation, since 20S can switch from an inactive “closed” conformation to an active “open” conformation spontaneously or after chemical treatment (e.g., SDS) (Bajorek & Glickman, 2004; Förster, Whitby, & Hill, 2003; Groll et al., 2000). Noteworthy, since the α3 tail points toward the centre of the channel and maintains a close interaction with all other N-termini of α subunits (Köhler et al., 2001; Köhler, Bajorek, et al., 2001), the deletion of first nine residues in α3 subunit N-tail in yeast 20S (α3Δn) induces a general disorder in the neighbouring tails, stimulating the opening of the entry pore (Köhler, Bajorek, et al., 2001; Köhler, Cascio, et al., 2001). Thus, the α3Δn mutant is in a constitutively activated “open” state and its basal proteolytic activity toward small peptides is consistently enhanced, as compared to that of wild-type (wt) 20S (Bajorek & Glickman, 2004; Köhler, Bajorek, et al., 2001; Köhler, Cascio, et al., 2001). Conversely, the double mutant α3-α7Δn more efficiently degrades macromolecular substrates with respect to either single mutant, suggesting that the interaction between these opposite tails is crucial in the regulation of gate opening (Bajorek, Finley, & Glickman, 2003; Bajorek & Glickman, 2004). Interestingly, the α3Δn mutation does not alter the assembly of 26S, as demonstrated by the evidence that the abundance and activity of mutant 26S are similar to those of wt-26S (Groll et al., 2000; Groll & Huber, 2003). Accordingly, human cell lines stably expressing α3ΔN subunits show enhanced activity of both free 20S and holoenzyme complexes. This turns out in an increase of the degradation rate of poly-ubiquitinated proteins, reinforcing the role of α3-mediated gate opening mechanism, and suggesting that the α3Δn holoenzyme could help cell to fight the proteotoxic stress (Choi et al., 2016). These data envisage that RP binding to 20S stimulates a structural rearrangement similar to that induced by the deletion of the α3 tail.
2.2.2. 20S biogenesis
Proteasome maturation refers to the process that drives the proper incorporation of individual subunits to assemble into a proteolytically active 26S. Whilst 19S assembly is largely uncovered yet (see Section 2.3.2), the 20S assembly has been uncoiled at a great molecular detail also by virtue of the extensive similarity between yeast and mammalian pathways. This similarity has widened the repertoire of methodological approaches suitable to uncover the molecular insights. In eukaryotes, the stepwise recruitment of individual α- and β-subunits to constitute a fully mature 20S requires the presence of five molecular chaperones, called Proteasome Assembly Chaperones (PAC1–4 in human, Pba1–4 in yeast) and Proteasome Maturation Protein (POMP in human, hUmp-1 in yeast) (Hirano et al., 2006; Le Tallec et al., 2007; Ramos & Dohmen, 2008). These chaperones drive the sequential insertion of the subunits preventing the formation of off-target assemblies presumably through non-catalytic activities (Burri et al., 2000; Fricke, Heink, Steffen, Kloetzel, & Krüger, 2007). First, PAC1–PAC2 and PAC3–PAC4 work as heterodimers in recruiting the α-subunits during the earliest steps of biogenesis, that is the α-ring formation (Hirano et al., 2005; Hirano et al., 2006; Le Tallec et al., 2007; Matias, Ramos, & Dohmen, 2010; Wu et al., 2018). Very recent advances in the field propose that in human cells α4, α5, α6 and α7 subunits first assemble to form a core tetrameric α-ring intermediate (α4-α7), being driven by PAC3-PAC4 heterodimers, which localize at the inner side of the nascent α-ring (Satoh et al., 2019; Wu et al., 2018). Recently, crystallographic data have allowed to identify a hydrophobic surface, surrounded by charged residues in PAC4, which is complementary to that of PAC3, thus providing a clue for the interaction between the two partners (Kurimoto et al., 2017). Notably, PACs surface was also found to display a charge complementarity with α4 and α5 subunits, envisaging the first structural basis for the binding of the heterodimer PAC3–PAC4 to the nascent 20S (Kurimoto et al., 2017).
Thereafter, the PAC1–PAC2 heterodimer binds the outer side of this assembly, favouring the recruitment of α1, α2 and α3 subunits, thus leading to the formation of a mature heptameric α-ring (Wu et al., 2018). Besides correctly introducing the α-subunit, the presence of the chaperones prevents the formation of aberrant off-pathway α-ring dimers, an occurrence potentially favoured by the sticky properties of α-subunits, in particular α7, which is prone to form high MW homo-oligomers in vitro (Kozai et al., 2017).
The α-ring is then the scaffold for subsequent insertion of the seven β-subunits through the contribution of POMP at the outer surface of the endoplasmic reticulum, which is, to date, the main intracellular localization where these events take place in human cells (Fricke et al., 2007; Hoefer, Boneberg, Grotegut, Kusch, & Illges, 2006; Krüger, Kloetzel, & Enenkel, 2001; Witt et al., 2000). The β-ring assembly starts with pro-β2, followed by β3, β4, forming the 13S complex; once these subunits are inserted, the PAC3-PAC4 heterodimer is released and pro-β5, pro-β6 and pro-β1 subunits assemble (Hirano et al., 2005; Hirano et al., 2006; Satoh et al., 2019). Remarkably, structural insights suggest that the pro-peptide is not merely involved in preventing the early activation of the catalytic Thr in the catalytically active subunit (see Section 2.1), but is necessary for further stepwise incorporation of subunits, likely through an allosteric mechanism. The pro-peptides of β2 and β5 are essential for recruitment and incorporation of β3 and β6, respectively, whereas the β5 pro-peptide is necessary for the specific interaction with POMP (Hoefer et al., 2006). The ultimate step of β-ring formation is the pro-β7 insertion and the formation of a half 20S (i.e., the 15S complex) which, upon dimerization, forms the mature 20S. Although it is proven that full activation of 20S requires a) shedding of the β-subunits pro-peptides, b) PAC1-PAC2 detachment and/or clearance and c) POMP clearance, it is not fully clear whether the degradation of the chaperones is carried out by the 20S itself or if PAC chaperones are actually cleaved or released intact to be recycled for further maturation processes. It is further widely envisaged that additional unidentified factors may take part in the maturation process with activities overlapping with those of PACs in dependence of metabolic needs of.
The deepening of the molecular insights of proteasome maturation, both in terms of transcriptional regulation and of dynamics of proteins interaction, is expected to offer a new perspective for the development of therapeutic strategies based on the modulation of proteasome availability in selected tissues (Goldberg, Zhao, & Collins, 2015). Clinical and molecular studies envisage that increased POMP translation and bioavailability upon down-regulation of miR-101 (which targets POMP mRNA) is an oncogenic stimulus for breast cancer cells (Zhang, Bi, Fan, Wang, & Bao, 2015). Thus, the consequent increased proteasome intracellular content would confer protection from the proteotoxic insult to which highly proliferating cells are likely exposed, favouring cell survival (Zhang et al., 2015). Furthermore, POMP up-regulation enhanced the bulk proteasome activity under proteo-toxic conditions, providing a metabolic advantage under redox insult (Chondrogianni & Gonos, 2007). As a matter of fact, recent genetic studies on POMP promoter have identified mutations at the 3’UTR region and splicing variants in different skin inflammatory disorders, such as CANDLE syndrome (Chronic Atypical Neutrophilic Dermatosis with Lipodystrophy and Elevated temperature) or proteasome-associated autoinflammatory syndrome (PRAAS). Furthermore, increased POMP levels were observed in psoriatic skin lesions (Brehm et al., 2015; Dahlqvist et al., 2010; Ebstein, Poli Harlowe, Studencka-Turski, & Krüger, 2019; Morice-Picard et al., 2017; Poli et al., 2018; Zieba et al., 2017). In these diseases, a decrease of proteasome levels, consequent to an altered maturation, is envisaged to activate the Unfolded Protein Response (UPR) through the endoplasmic reticulum stress. Such a stressful condition is known to trigger an inflammatory stimulus, which, once chronic, would account for the pathogenesis of the disease (Dahlqvist, Törmä, Badhai, & Dahl, 2012; Ebstein et al., 2019).
Conversely, PACs involvement in pathological processes is still controversial and, probably, largely unexplored for pharmacological purposes although a functional evaluation of thielocin B analogues as protein-protein interaction inhibitors of PAC3 homodimer has been recently evaluated (Ohsawa et al., 2018). However, it must be considered that although the contribution of PACs to proteasome biogenesis (discussed above) would suggest that they are crucial for life, transgenic murine KO models for either PAC1 or PAC2 are viable with major anatomical and functional alterations limited to different brain regions (Sasaki et al., 2010). This finding underlies that the biological activity of PACs is redundant, envisaging that either additional still unknown factors can vicariate PACs activity or the self-assembling properties of free α-subunits (documented to some extent in vitro) is enough to promote the constitution of fully active 20S particles. More recently, our group reported a marked down-regulation of PAC1-PAC2 expression, along with that of α7 subunit, in primary cultures of skin fibroblasts isolated from subjects affected by Rett Syndrome (RTT), a sever neuro-developmental disorder (Sbardella et al., 2020a). These cells were characterized by two different non-sense early truncating mutations of MeCP2 (i.e., a transcriptional repressor that is mutated in the vast majority of patients affected by the syndrome) and by a concomitant severe lack of mature proteasome particles (Amir et al., 1999; Sbardella et al., 2020). Furthermore, silencing of MeCP2 expression in neuron-like cells resulted in a similar proteasome dysfunction, indicating an unprecedented role of this transcriptional regulator in proteasome biogenesis. Different approaches, including a revolutionary in vivo imaging system, suggest that only a small fraction of the intracellular proteasome particles is proteolytically active under physiological conditions (Asano et al., 2015). Thus, proteasome content appears to exceed the amount necessary to sustain life. In this view, the brain abnormalities in PAC1 KO mouse might be interpreted as the consequence of a reduced proteasome biogenesis in the tissue that is known to be more vulnerable to dysregulation of intracellular proteostasis which is primarily handled by the UPS (Sasaki et al., 2010). This suggestion would be even more fascinating if PAC1-PAC2 loss will be confirmed to occur also in the CNS of RTT, which is the tissue prevalently affected in syndrome onset and progression.
2.3. 19S regulatory particles
2.3.1. 19S structural arrangement
Gate opening is crucial in 20S function, and cells have evolved different regulators (see Box 1) which control this proteasome process (Finley et al., 2016). The predominant and best characterized 20S activator is the 19S which interacts, in the presence of ATP, with one or both ends of the 20S to form proteasome holo-complexes, 26S (i.e., single-capped) and 30S, respectively (i.e., doubly-capped) (Armon, Ganoth, & Hershko, 1990; Bard et al., 2018; Eytan, Ganoth, Armon, & Hershko, 1989; Liu et al., 2006; Marshall & Vierstra, 2019; Schmidt & Finley, 2014; Smith et al., 2005). These different proteasome assemblies (i.e., 26S and 30S) coexist, together with free 20S, in cell cytosol, and are known to cleave ubiquitinated substrates, although their substrate specificity and different biological role remain a somewhat enigmatic issue in proteasome biology. The abundance of the three main proteasome populations (i.e., 20S, 26S and 30S) seems to be finely modulated by the specific microenvironment in which the cell lives. This structural arrangement is carried out by different proteasome interacting proteins (PIPs) which can be classified either (a) extrinsic de-ubiquitinases (DUBs) (see Box 2) and/or (b) auxiliary proteasome regulators, Ecm29, HSP70 (Tanaka, 2009). In this regard, Insulin-Degrading-Enzyme (IDE), a Zn2+ protease, which behaves as a Heat-Shock Protein (Tundo et al., 2013), has been reported not only to directly bind the 20S, but also to modulate its activity through allosteric mechanisms, envisaging that it may be a novel auxiliary proteasome regulator (Sbardella et al., 2015; Tundo et al., 2017). Moreover, IDE was found to compete with 19S binding, modifying the distribution of different proteasome population in vitro (Sbardella et al., 2018). However, despite the biological relevance, the exact molecular mechanism which drives proteasome population interconversion is still far from being satisfactorily elucidated.
Once bound over the axial 20S pores, the 19S RP carries out different functions, namely (see Section 2.1): (i) recognition and unfolding of ubiquitinated substrates; (ii) opening of the 20S pore; (iii) substrates entry into the 20S catalytic chamber; (iv) release of ubiquitin moieties during substrate degradation (Collins & Goldberg, 2017; Finley & Prado, 2019; Marshall & Vierstra, 2019) (Fig. 2 ). From the structural point of view, the 19S is made up by two different sub-components, the lid and the base, which form a conformationally dynamic complex (Bajorek & Glickman, 2004; Budenholzer et al., 2017).
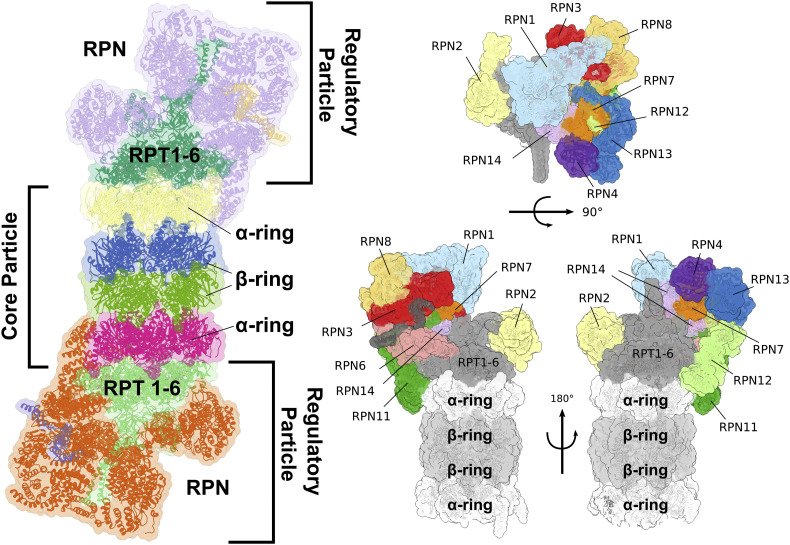
Fig. 2.
Overall organization of the proteasome 26S particle.
Left: the “Core Particle” (20S proteasome) is represented as protein ribbons, in yellow and magenta the two α-subunit rings, in blue and green the two β-subunit rings. The two regulatory particles (19S proteasome), attached on both ends of the 20S particle, are represented as protein ribbons. The group of regulatory AAA-ATPases (Rpt1–6) are coloured in dark and pale green, the non-ATPase regulators (Rpn) are coloured in violet and orange. Right: close-up of the 26S regulatory particle with the various non-ATPase subunits highlighted and labelled. The particle is shown from various point of view: on the top panel it is shown from the top; on the lower panel two opposite side views are shown.
The base binds directly to the 20S, and it is formed by a ring of six paralogous AAA-ATPases, named Rpt1-Rpt6, and three non-ATPase subunits (i.e., Rpn1, Rpn2, Rpn13), which provide multiple binding site for ubiquitin and ubiquitin-like proteins (Bard et al., 2018; He et al., 2012; Husnjak et al., 2008; Marshall & Vierstra, 2019; Saeki & Tanaka, 2012; Shi et al., 2016). Rpt subunits associate into three pairs of heterodimers (i.e., Rpt1-Rpt2, Rpt3-Rpt6, Rpt4-Rpt5), which then form the hetero-hexameric motor of proteasome. The C-terminal helical domains of Rpt1-Rpt2 subunits interact with Rpn1, while those of the heterodimer Rpt3-Rpt6 bind Rpn2. Rpn13 also interacts with the C-terminal residues of Rpn2 through its N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain, whereas the C-terminal region of Rp4-Rpt5 extends out from the base body without interaction with other proteasome subunits, at least in the resting state (Fig. 2) (Beck et al., 2012; Budenholzer et al., 2017; Djuranovic et al., 2009; Hemmis, Heard, & Hill, 2019; Husnjak et al., 2008; Tomko, Funakoshi, Schneider, Wang, & Hochstrasser, 2010; VanderLinden, Hemmis, Yao, Robinson, & Hill, 2017; Zhang et al., 2009). The first identified ubiquitin receptor was Rpn10, that is not considered part of the base, but functions as a bridge between the lid and the base, stabilizing their interaction (Aubin-Tam, Olivares, Sauer, Baker, & Lang, 2011; Beckwith, Estrin, Worden, & Martin, 2013; Erales, Hoyt, Troll, & Coffino, 2012; Maillard et al., 2011; Martin, Baker, & Sauer, 2008), as further suggested by the lid and base disassembly when Rpn10 is mutated (Deveraux, Ustrell, Pickart, & Rechsteiner, 1994; Isasa et al., 2010; Keren-kaplan et al., 2016). Importantly, mono-ubiquitination of Rpn10, which is modulated by stressful conditions, regulates its association with proteasome, and thus proteasome activity and stability (Budenholzer et al., 2017; Isasa et al., 2010; Keren-kaplan et al., 2016). An additional intrinsic ubiquitin receptor is the T1 toroidal region of the Rpn1 (Elsasser, Chandler-Militello, Müller, Hanna, & Finley, 2004; Shi et al., 2016), which, like Rpn10 and Rpn13, also recognizes ubiquitin -like domains (UBLs) of extrinsic ubiquitin receptors (i.e., HR23/Rad23, PLIC2/DsK2 and Ddi1), stimulating the proteasome-mediated degradation of ubiquitinated substrates (Leggett et al., 2002; Raasi, Varadan, Fushman, & Pickart, 2005; Saeki, Saitoh, Toh-e, & Yokosawa, 2002; Shi et al., 2016; Spyracopoulos, 2016). It remains unclear why proteasome contains such an array of ubiquitin-binding receptors, and what differential roles they might play in substrate recognition and degradation (Bard et al., 2018; Cundiff et al., 2019; Hamazaki, Hirayama, & Murata, 2015).
Upon recognition by intrinsic and extrinsic ubiquitin receptors, substrates are engaged with the AAA+ motor of the highly dynamic Rpt1–6 hexameric ring that couples ATP hydrolysis to substrate unfolding and translocation, converting chemical energy into mechanical work (de la Peña, Goodall, Gates, Lander, & Martin, 2018; Dong et al., 2019; Eisele et al., 2018). The C-terminal tails of Rpt2, Rpt3 and Rpt5 contain the conserved HbYX motif (see also Section 2.3.2) that fits into the groove between adjacent α-subunits of 20S inducing a conformational change into their N-termini which drives 20S gate opening (Rabl et al., 2008; Smith et al., 2007). Mutational studies have indicated that functions of Rpt subunits are not redundant, but they cover different roles according to the vertical position they adopt in the hexamer, such that subunits located at the top (i.e., Rpt3 and Rpt4) contribute to substrate engagement and translocation more than subunits located further down, like Rpt1 and Rpt2 (Beckwith et al., 2013; Erales et al., 2012; Lander et al., 2012; Rubin, Glickman, Larsen, Dhruvakumar, & Finley, 1998; Wehmer et al., 2017).
The peripheral lid subcomplex, which reveals significant structural and sequence similarities with COP9 signalosome and eIF3, braces one side of the base and it is composed of nine ATPase subunits, namely Rpn3, Rpn5–9, Rpn11–12 and Sem1 (also named Rpn15). Among these subunits, six are PCI (proteasome-CSN-initiation factor 3) domain containing subunits (i.e., Rpn3, Rpn5–7, Rpn9, and Rpn12), and two are MPN (Mpr1-Pad1 N-terminal) domain containing subunits (i.e., Rpn8 and Rpn11) (Beckwith et al., 2013; Erales et al., 2012; Schmidt & Finley, 2014). The most important functions of the lid subcomplex are the strengthening of 20S—19S interaction (e.g., Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together) and de-ubiquitination of substrates before their processing by the AAA-ATPase (Pathare et al., 2012; Pathare et al., 2014). Cleavage of polyubiquitin chains enables Ub recycling into the cellular pool (Budenholzer et al., 2017; Pathare et al., 2014) and it is carried out mostly by Rpn11, a Zn2+ de-ubiquitinase essential for proteasome functions and cell viability located above the translocation channel, which removes the entire ubiquitin chains of the substrates before their entry into the ATPase translocation ring (de Poot, Tian, & Finley, 2017). As a matter of fact, the close proximity of the N-terminal domain ring of the AAA+ ATPases sterically prevents the cleavage inside ubiquitin moieties by Rpn11, which, therefore, removes polyubiquitin chains by hydrolysing the isopeptide bond (located at the base of the chain) between the lysine residue of the substrate and the C-terminus of the first ubiquitin monomer (Yao & Cohen, 2002). The MPN domain of Rpn11 forms a heterodimer with the non-catalytic MPN domain of Rpn8, forming the minimal DUB-competent complex (Pathare et al., 2014; Worden, Padovani, & Martin, 2014). Even though the Rpn11/Rpn8 dimer is active when isolated, it is significantly inhibited in the free lid sub-complex through its interaction with the neighbouring lid subunit Rpn5. During lid incorporation into the 26S proteasome, conformational rearrangements occur, activating the action of the de-ubiquitinase (Dambacher, Worden, Herzik, Martin, & Lander, 2016; Verma et al., 2002; Worden et al., 2014; Yao & Cohen, 2002), which is crucial for the efficient substrate degradation. However, in order to prevent a premature ubiquitin chain removal, de-ubiquitination must be restricted to committed substrates that are engaged with the ATPase motor; therefore, the rate-limiting step in de-ubiquitination process is represented by an ubiquitin-linked conformational switch of Rpn11 Insert-1 loop from an inactive closed state to an active β-hairpin. This conformational change is activated by mechanical substrate translocation of AAA+ motor ATPase, allowing a direct coupling of substrate de-ubiquitination and degradation (Worden, Dong, & Martin, 2017). Beside Rpn11, the two extrinsic DUBs, Usp14 (see Box 2) and UchL5, which are also strongly associated with both Rpn1 and Rpn13, are involved in cleaving or editing of the ubiquitin chain from substrates (Bard et al., 2018; Guterman & Glickman, 2004; Hamazaki et al., 2006; Lam, Xu, DeMartino, & Cohen, 1997; Qiu et al., 2006; Yao et al., 2006).
2.3.2. 19S assembly
Unlike 20S, the heterogeneous and dynamic structural features of 19S demand a different and independent assembly process of the base and lid subcomplexes (Budenholzer et al., 2017; Lander et al., 2012; Marshall & Vierstra, 2019; Tomko et al., 2015; Tomko & Hochstrasser, 2014).
Base assembly has been poorly studied and two separated, but not mutually exclusive, models have been proposed up to now. According to the first model, base assembly takes place regardless of lid subunits, finding its justification mainly through studies in E. coli. On the other hand, according to the second model, 20S would act as a scaffold for 19S biogenesis, as suggested by mutagenic and immunoprecipitation studies in yeast (Beckwith et al., 2013; Funakoshi, Tomko, Kobayashi, & Hochstrasser, 2009; Li et al., 2017; Marshall & Vierstra, 2019; Park et al., 2013; Tomko et al., 2010). However, both models support the notion that an ordered recruitment of free base subunits is orchestrated by a set of dedicated chaperones, namely p27 (Nas2 in yeast), p28 (Nas6 in yeast) and S5b (Hsm3 in yeast), which join together couples of Rpts subunits, driving their correct insertion in human cells. Specifically, the interaction between the C-terminal domain of Rpt subunits with respective chaperones leads to three precursor modules formation (i.e., p27-Rpt4-Rpt5, p28-Rpt3-Rpt6, and S5bRpt1-Rpt2) (Le Tallec et al., 2007, Funakoshi et al., 2009; Roelofs et al., 2009; Saeki, Toh-e, Kudo, Kawamura, & Tanaka, 2009; Tomko et al., 2010). Remarkably, none of these chaperons is crucial for cell viability, but multiple genetic deletions become lethal under proteotoxic conditions (Budenholzer et al., 2017). Furthermore, the N-terminal domain of chaperone Adc17 binds Rpt6, mediating Rpt3-Rpt6 dimerization (Hanssum et al., 2014; Rousseau & Bertolotti, 2016); thereafter, Adc17 early dissociates by heterodimers during the assembly process, whereas Hsm3, which contacts also Rpn1, dissociates from the lid only upon completion of 19S maturation (Barrault et al., 2012; Funakoshi et al., 2009; Hanssum et al., 2014). Importantly, the p28-Rpt3-Rpt6 module also binds Rpn14, whereas the Nas6-Rpt3-Rpt6-Rpn14 module seems to form an intermediate with the p27-Rpt4-Rpt5 one. This intermediate module also interacts with Rpn2 and Rpn13, but p27 likely dissociates before the incorporation of S5b-Rpt1-Rpt2 module, since a complex displaying the two chaperones has never been observed (Funakoshi et al., 2009; Saeki et al., 2009; Tomko et al., 2010). However, it must be remarked that the ordered series of events that lead to base assembly are speculative, and differences may exist between human and yeast (Budenholzer et al., 2017), most data indicating that lid and the Rpn10 associate only when the base is completely formed (Budenholzer et al., 2017; Funakoshi et al., 2009; Roelofs et al., 2009; Saeki et al., 2009; Tomko et al., 2010).
The lid assembly proceeds through a coordinated process, characterized by an ordered series of subcomplexes interaction (Tomko et al., 2011; Fukunaga, Kudo, Toh-e, Tanaka, & Saeki, 2010; Tomko et al., 2015; Estrin, Lopez-Blanco, Chacón, & Martin, 2013). Lid assembly begins with heterodimerization of Rpn8-Rpn11, which is then followed by Rpn6, Rpn5 and Rpn9 recruitment, leading to the release of the first lid module (Estrin et al., 2013; Sharon, Taverner, Ambroggio, Deshaies, & Robinson, 2006). Unlike 20S and 19S base, no chaperones dedicated to assist the process have ever been identified, even though the intrinsically disordered Sem1 subunit of the lid seems to play a crucial role in linking Rpn3 and Rpn7 to form the heterotrimeric intermediate LP3, which is an early phase of lid biogenesis (Bohn et al., 2013; Dambacher et al., 2016; Fukunaga et al., 2010; Sone, Saeki, Toh-e, & Yokosawa, 2004). Thereafter, the first lid module and LP3 associate to form LP2, creating a complete lid that only misses the Rpn12 subunit (Estrin et al., 2013; Tomko & Hochstrasser, 2011; Yu et al., 2015). Hence, the last step is the incorporation of Rpn12, which fits its C-terminal helix into a helical bundle formed by clusters of C-termini of other Rpn subunits (Marshall & Vierstra, 2019; Tomko et al., 2015). Rpn12 binding induces a conformational change to the rest of lid, which favours the association between lid and base (Budenholzer et al., 2017; Tomko et al., 2015). During lid maturation, and possibly during lid and base connection, Rnp8/Rpn11 undergoes a conformational change, which leads to a rigid body rotation of the heterodimer, so that Rpn11 is located where it can deubiquitinate polyubiquitinated substrates before their entry in the ATPase channel (Dambacher et al., 2016; Tomko et al., 2015). As mentioned in Section 2.3.1, Rpn11 activity is inhibited by Rpn5, and further by Rpn8-Rpn9 interaction. When lid assembly is completed, the module Rpn8-Rpn11 rotates away from Rpn5, allowing Rpn11 activation (Dambacher et al., 2016; Ehlinger et al., 2013).
The last step of the holo-enzyme formation is represented by the association between 19S and 20S. The key event is the 19S-mediated gate opening, which is driven by the insertion of C-terminal HbYX motifs of Rpt2, Rpt3, and Rpt5 into the 20S α-subunit pockets (Park et al., 2013; Rabl et al., 2008; Smith et al., 2007; Tian et al., 2011). However, recently it has been shown that stable docking of HbYX motifs into the 20S is not sufficient to promote the gate opening. Accordingly, efficient gate opening has been proposed to occur only when Rpt1 and Rpt6 C-termini are engaged into the α-ring (Eisele et al., 2018; Park et al., 2013; Sokolova, Li, Polovin, & Park, 2015). Moreover, Rpn6 binding to α2 subunit facilitates 20S—19S interaction (Lander et al., 2012; Pathare et al., 2012).
Notably, 20S—19S association occurs spontaneously in vitro in the presence of ATP, whereas in cell models it seems influenced by a series of interacting proteins, such as HSP90, IDE, and Ecm29 (Imai, Maruya, Yashiroda, Yahara, & Tanaka, 2003; Sbardella et al., 2018; Tundo et al., 2017; Yamano et al., 2008). The latter protein seems to play a particularly important role under stressful conditions, since it binds structural aberrant proteasome, repressing 20S—19S interaction (De La Mota-Peynado et al., 2013; Lee et al., 2011; Lehmann, Niewienda, Jechow, Janek, & Enenkel, 2010; Panasenko & Collart, 2011; Park, Kim, Tian, Gygi, & Finley, 2011; Wang et al., 2017).
2.4. Structural conformation of the proteasome holoenzyme
A main breakthrough for understanding the structural basis of 26S came from a series of Cryo-EM studies on proteasome holoenzyme from different species, such as yeast, rat and humans (Lander et al., 2012; Matyskiela et al., 2013; Unverdorben et al., 2014; Wehmer et al., 2017; Chen, Wu, & Shen, 2016; Huang et al., 2016; Wehmer & Sakata, 2016; Bard et al., 2018). These studies have revealed the existence of at least four distinct human 26S conformational states (i.e., SA, SB, SC and SD, mirrored in yeast 26S by s1, s2, s3 and s4), that appear conserved among species. The numeric order of these main states is suggested by a structural comparison that reveals progressive and sequential movements from SA (s1) state through SB (s2) and SC (s3), to SD (s4), which is similar to SA (Bard et al., 2018; Wehmer & Sakata, 2016). In all identified conformations, the architecture and structure of 20S remains essentially unaltered, whereas the two subcomplexes of 19S, the lid and the base (see Section 2.3.1) are highly dynamic, changing the relative orientation with respect to each other and to core particles; these movements are coupled to the functional cycle of 26S (Chen et al., 2016; Unverdorben et al., 2014; Wehmer et al., 2017). However, despite the advance in knowledge on 26S structure, we have to recall that an intriguing aspect, which has never been deeply investigated, concerns the conformational transition, occurring after the binding of the first 19S, on the opposite end of 20S, where one free α-ring surface is available for the binding of a second 19S particle, which yields a double capped 30S proteasome, whose real structure, as well as the function, remains poorly understood (Tundo, Sbardella, & Coletta, 2018).
The yeast s1 is a low energy ATP-bound ground state, that is assumed to be the primary substrate-binding conformation (Matyskiela et al., 2013; Sledz et al., 2013; Unverboden et al., 2014; Wehmer et al., 2017; Ding et al., 2017; Chen et al., 2016; Zhu et al., 2017). In the s1 state, the 20S gate is closed, since the substrate translocation channel of ATPase ring is not aligned with the 20S gate, and the active site of Rpn11 is 25Ǻ away from substrate entry pore (Eisele et al., 2018; Finley & Prado, 2019; Wehmer et al., 2017). The transition toward the s2 state is driven mainly by the lid rotation, which drives Rpn11 to a position above the central processing pore of the base. On the other hand, the progression from s2 to s3 is mediated by a rearrangement of Rpt1-Rpt6, wherefore N-ring of Rpts and AAA+ domains shift toward Rpn1, thus generating a wider channel aligned with core particle axial pore (Matyskiela et al., 2013; Unverboden et al., 2014; Wehmer et al., 2017; Chen et al., 2016). Therefore, the s3 state is characterized by the axial alignment of the essential DUB Rpn11 (see also Section 2.3.1), 19S translocation channel, and 20S gate. However, in spite of these rearrangements and of the evidence that s2 and s3 states are primed for substrate degradation, the 20S gate is still mostly occluded, preventing substrate entry (Matyskiela et al., 2013; Sledz et al., 2013; Bard et al., 2018; Finley and Prado 2019). The gate becomes fully opened only during the transition from s3 to s4, inducing the entry of the substrate into the catalytic core; thus, gate opening is a consequence of the insertion of “HbYX” motif of C-termini of Rpt2-Rpt3-Rpt5 subunits into 20S pocket (see Section 2.3.1). Stable docking of HbYX motifs into the 20S is insufficient to promote gate opening, which is completed only in the s4 state upon engagement of the C-termini of Rpt6 and Rpt1 into the α-ring (Eisele et al., 2018; Finley & Prado, 2019). Besides these four states, further structural and biochemical studies have revealed recently the presence of two additional open gate states in yeast proteasome (i.e., s5 and s6) (Eisele et al., 2018). In the case of human 26S proteasome Cryo-EM studies showed that the substrate Sic1PY 26 complex (i.e., the Cdk inhibitor Sic1 from Saccharomyces cerevisiae with a Pro-Pro-Pro-Ser motif inserted into N-terminal) exists in seven conformational states, EA1, EA2, EB, EC1, EC2, ED1, ED2 (Dong et al., 2019). EA1 and EA2 states represent two initial ubiquitin recognition states; EB2 is the “de-ubiquitination” state, in which the isopeptide bond between Rpn11 and substrate is close to the zinc-active site of Rpn11; EC1 and EC2 are conformations at the onset of substrate translocation; ED1 and ED2 carry on and complete substrate translocation (Dong et al., 2019). Functional models of 26 activity, derived both from cryo-EM and biochemical analysis, couple ATP hydrolytic cycle to substrate translocation (Matyskiela et al., 2013; de la Pena et al., 2018; Dong et al., 2019). Therefore, sequential ATP hydrolysis and phosphate release, which are coordinated within the ATPase motor, seem to supply “the power” to induce conformational changes that drive the substrate through the central pore (de la Pena et al., 2018; Eisele et al., 2018; Tundo et al., 2018; Dong et al., 2019). In agreement with a “rotatory” mechanism, a hydrolytic event in a single Rpt subunit is followed by another one in the nearby subunit, thus proceeding throughout the entire ring (de la Pena et al., 2018; Eisele et al., 2018; Tundo et al., 2018; Dong et al., 2019). In fact, it has been proposed that a specific Rpt subunit binds ATP and engages substrate at the uppermost position; then, this subunit hydrolyses ATP (when at the penultimate position of the staircase), releasing the phosphate moiety and disengaging from substrate, which proceeds to the next hydrolytic step (Eisele et al., 2018; de la Pena et al., 2018). A disengaged subunit moves outward from the ring, where it can contact another segment of the translocating substrate. At the same time, the other substrate-engaged subunits carry out a coordinated and synergistic motion so that the substrate translocates by about two amino acids (~ 6 Ǻ) toward the 20S (de la Pena et al., 2018; Eisele et al., 2018; Dong et al., 2019; Finley & Prado, 2019; Majumder et al., 2019).
Importantly, cryo-electron tomography approaches have also visualized proteasome particles in their native conformation in living cells, allowing to have an insight on the percentage of different populations that harbour the cells (Asano et al., 2015; Finley & Prado, 2019; Guo, He, Li, & Le, 2017). In intact hippocampal neurons, a molecular census of proteasome conformational states showed that, in the absence of proteotoxic stress, only 20% of the 26S was engaged in substrate processing, whereas the remaining portion was in the substrate-accepting ground state. It suggests that the capacity of the proteasome system is not fully exploited by the cell under physiological conditions (Asano et al., 2015). Interestingly, poly-Gly-Ala (poly-GA) aggregates, which result from aberrant expansion of GGGGCC repeat in C9orf72 gene (i.e., the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia), recruits 26S molecules which are in the s4 state (Guo et al., 2018), unlike the general pool of proteasome. However, since poly-GA are not favourable proteasome substrates, 26S sequestration and consequent inhibition has been proposed to cover a crucial role in neurodegeneration (Finley and Prado, 2019, Guo et al., 2017).
3. Proteasome in cancer progression
3.1. Proteostasis network in cancer
Over the last decades, hallmarks of cancer cells have been described to provide a sort of universal definition which would account for the multi-step development of human tumours (Hanahan & Weinberg, 2000; Hanahan & Weinberg, 2011; Pack et al., 2014). These hallmarks, which are complementary features that enable tumour growth and metastatic dissemination, include proliferative signalling, growth suppressors inactivation, cell death resistance, replicative immortality, angiogenesis, invasiveness and dissemination, cell metabolism reprogramming and immune-surveillance evasion (Hanahan & Weinberg, 2000; Hanahan & Weinberg, 2011; Shereen, Khan, Kazmi, Bashir, & Siddique, 2020; Tundo, Sbardella, Lacal, Graziani, & Marini, 2019). Recently, resistance to proteostasis unbalance has been proposed as a new malignant hallmark of cancer, envisaging the possibility that this acquired property cooperates with the other altered circuits to allow cancer cell survival, proliferation and dissemination (Carvalho, Rodríguez, & Matthiesen, 2016; Dong & Cui, 2018; Klaips et al., 2018). Cancer cells, due to the rapid proliferation rate, are constantly under cellular stress with a consequent decrease of protein quality control (Carvalho et al., 2016; Vahid, Thaper, & Zoubeidi, 2017). However, the unbalance of protein synthesis, folding, trafficking and degradation, which usually leads normal cells to death, does not induce the same fate in cancer cells that acquire and develop, during tumour progression, novel properties to promote their survival (Calderwood, Khaleque, Sawyer, & Ciocca, 2006; Vahid et al., 2017).
In recent years, three main reasons have gained considerable insight as to why PNs are altered in human tumours, namely 1) genomic instability; 2) persistence of stressful conditions in tumour micro-environment, and 3) age-related proteome imbalance (Dong & Cui, 2018).
First, cancer cell genome is highly unstable and builds up several point mutations in protein coding sequence and/or genome mutations (e.g., large duplications, deletions, inversions, and translocations as well as altered copy numbers of entire chromosomes, such as aneuploidy). This may turn out in an inappropriate repression or activation of tumour suppressors and oncogenes, respectively, excessive protein synthesis, and/or translation of mutated proteins with altered folding, function and turn-over (Benbrook & Long, 2012; Kim & Zaret, 2015; Vogelstein et al., 2013; Weaver & Cleveland, 2006). It has been estimated that over 90% of human solid tumours harbour aneuploidies that lead to an excess in protein synthesis (Dai, Dai, & Cao, 2012; Weaver & Cleveland, 2006; Williams & Amon, 2009); indeed, this is a relevant issue mainly for proteins that become functional upon assembly in stoichiometric complexes such as in the case of ribosomes (Deshaies, 2014). Therefore, genomic alterations support a proteostasis unbalance (also referred as proteotoxic crisis) that renders cancer cells more dependent than normal cells on PNs clearance mechanisms, including UPS (Deshaies, 2014). Accordingly, yeast cells with one-third of single chromosomal aneuploidies are hypersensitive to proteasome inhibitors, and some cells “adapted” to aneuploidy harbour mutations that depress UPS activity (Torres et al., 2007; Torres et al., 2010; Torres, Williams, & Amon, 2008).
Secondly, during tumour development, tumour cells are continuously exposed to a variety of extrinsic perturbations, such as nutrient deprivation, hypoxia, and acidosis. Despite this pressure, tumour cells successfully proliferate and efficiently withstand this challenge by adapting to the fluctuations of the microenvironment, reprogramming their proteome and fully exploiting the cell defence mechanisms against proteotoxic stress. Thus, ultimately, stressful conditions lead to a disruption of the proteostasis balance, which is associated to the promotion of malignant properties (such as invasiveness, immune surveillance escape, and metabolism reprogramming), achieving a plethora of PN alterations (Oromendia & Amon, 2014; Dufey, Urra, & Hetz, 2015; Nam and Joe, 2019).
Last, pathological and physiological senility is considered a major risk factor for protein conformational diseases, including immunological and metabolic disorders, neurodegeneration and cancer (Carrell & Lomas, 1997; Kikis, Gidalevitz, & Morimoto, 2010; van der Willik, Schagen, & Ikram, 2018). In fact, progressive exposure of stressors during aging induces accumulation of damaged and unfolded proteins which culminates in PNs alteration (Dong & Cui, 2018; Sklirou, Papanagnou, Fokialakis, & Trougakos, 2018). Thus, in a vicious circle, unbalanced PNs lead to the proteotoxic crisis, which favours tumourigenesis (Arnsburg & Kirstein-Miles, 2014; Miller, Drake, Naylor, Price, & Hamilton, 2014). As a matter of fact, in accordance with the proteotoxic crisis hypothesis, reprogramming the proteome might represent a novel therapeutic approach, since agents that target components of different PN pathways are expected to be more toxic for cancer cells than for normal cells (Deshaies, 2014; Yuan et al., 2018). In the next paragraphs, we will review the biological rationale for targeting proteasome in the context of UPS as a strategy to treat cancer.
3.2. Degradation of cancer-related proteins by proteasome
A number of preclinical studies have reported alterations of proteasome expression and activity in different type of cancers, including haematological malignancies, lung, breast, pancreatic, head and neck, and thyroid cancers (Adams, 2003; Arlt et al., 2009; Chen et al., 2005; Kumatori et al., 1990; Roeten, Cloos, & Jansen, 2018). The reason of this high proteasome activity is not well understood, even though it is likely linked to stressful conditions (e.g., hypoxia, reperfusion, alteration of growth factors and cytokines levels), which evolve in the context of a heterogeneous tumour microenvironment. Deregulation of the proteasome activity can destabilize and/or disrupt the balance between tumour suppressors and oncoproteins, promoting cancer progression (Chang & Ding, 2018; Kaplan, Torcun, Grune, Ozer, & Karademir, 2017; Ogiso, Tomida, Kim, & Tsuruo, 1999). An element of complexity in understanding the role of proteasome in carcinogenesis is also represented by the fact that most investigations are carried out in unsorted cancer cells, which do not include cancer stem cells (Voutsadakis, 2017). Thus, cancer stem cell theory states that all tumour cells derive by a small percentage of cancer stem cells capable of repopulating tumours after therapy (Hanahan & Weinberg, 2011; Simons & Clevers, 2011). Noteworthy, the proteasome function is decreased in these cells with respect to the bulk of tumour population, revealing that a better understanding of proteasome regulation in different cell sub-sets might unveil further opportunities in cancer therapy (Banno et al., 2016; Voutsadakis, 2017). Despite the criticism, there are many key proteins, degraded by proteasome, that are involved in carcinogenesis; below are listed examples of proteins, which are considered crucial in cancer progression and are reported to mediate cell death after exposure to proteasome inhibitors (Ciechanover et al., 2001; Evan & Vousden, 2001; Jang, 2018; Johnson, 2015; Soave, Guerin, Liu, & Dou, 2017) (Fig. 3 ).
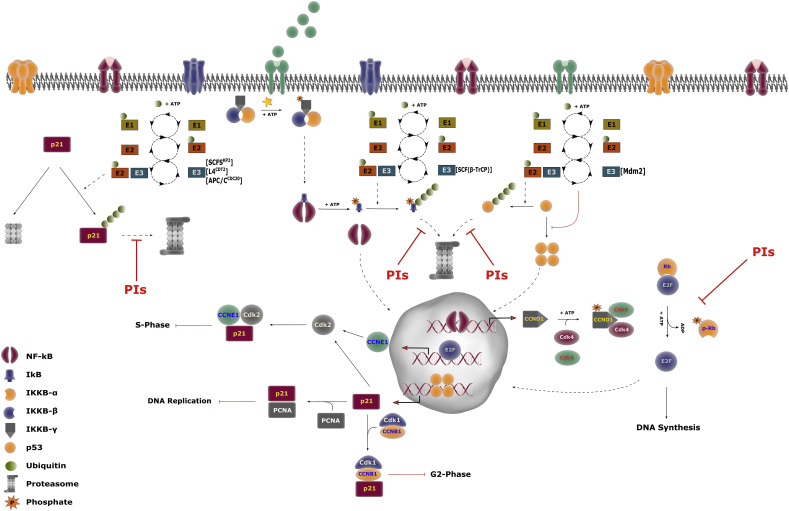
Fig. 3.
Regulation of NF-kB/E2F/Rb and p53/p21 pathways by proteasome.
NF-kB/E2F/Rb pathway: under unstimulated conditions, NF-kB is kept inactive in the cytosol by IkB inhibitor. Different stimuli (e.g., cytokines, stressors, Pathogen Associated Molecular Patterns), generically indicated as green spheres on membrane receptors, phosphorylate and activate IKKBγ subunit which thereafter phosphorylate IkB through the kinase activity held by the α and β subunits. Phosphorylated IkB is ubiquitinated and degraded by the 26S proteasome. Free NF-kB dimers translocate into the nucleus, where transcription of target genes occurs. Cyclin D1 (CCDN1) expression allows the cyclin D1-Cdk4/6 complex to form. This complex phosphorylates Rb protein, inducing its detachment from EF2 transcription factor. Free EF2 enters the nucleus and transcribes cyclin E (not shown), cyclin A and genes involved in DNA synthesis: this triggers the progression toward the S-phase.
p53/p21 pathway: under physiological conditions, p53 degradation is predominantly orchestrated by the E3-ligase MDM2, which promotes its poly-ubiquitination, and, thus, its degradation by the 26S proteasome. A number of stimuli activate the p53 pathway, inducing its tetramerization and translocation into the nucleus. Herein, p53 triggers the transcription of pro-apoptotic factors (i.e., Noxa and Bax, not shown) and CdkI p21. When expressed, p21 binds to: (i) Cdk2/cyclin E (CCNE1) complex, blocking the entry of the cell into the S phase; (ii) cyclin B (CCNB1)/Cdk1 complex, leading to a growth arrest in the G2 phase; (iii) PCNA, inhibiting DNA replication. p21 levels are also modulated through the ubiquitin-dependent degradation by the 26S, and further by a ubiquitin -independent pathway by the 20S.
Figure legend is restricted to NF-kB and p53, whose mechanisms of transcription induction is not sketched. PIs stands for Proteasome Inhibitors and the red arrows indicate the steps of NF-kB, p53 and p21 turn-over which are blocked by this class of drugs.
3.2.1. NF-kB
NF-kB is a crucial transcription factor that induces the expression of a wide range of genes involved in cell proliferation, apoptosis, inflammation and angiogenesis (Karin, Cao, Greten, & Li, 2002; Qureshi et al., 2018; Wu & Shi, 2013). Alteration of NF-kB pathway has been documented in a series of human tumours, including breast, lung, prostate, pancreatic cancer and melanoma, as well as in haematological malignancies, such as Hodgkin's/Non-Hodgkin's lymphoma and multiple myeloma (Aggarwal, 2004; Braun et al., 2006; Johnson, 2015; Karin & Greten, 2005; Kim, Hawke, & Baldwin, 2006; Perkins, 2012; Van Waes, 2007). It is generally accepted that NF-kB promotes cancer progression by inhibiting apoptosis, and that also chemo- and radiotherapy treatments activate NF-kB signalling, inducing acquired resistance to conventional cancer therapy (Baldwin, 2001; Nakanishi & Toi, 2005; Wu & Shi, 2013). Under unstimulated conditions, NF-kB homo- or hetero-dimers are sequestered in an inactive form in the cytoplasm by its inhibitor IkB. Different stimuli, including stress and chemotherapy, activate IkB kinase (i.e., IKKB) that phosphorylates IkB, leading to its ubiquitination and degradation by the proteasome. Free NF-kB dimers then translocate into the nucleus wherein they induce the transcription of target genes (Baldwin, 1996; Schwartz, Hernandez, & Mark Evers, 1999; Traenckner et al., 1995) (Fig. 3). Mammals express five NF-κB proteins, namely RelA (p65), RelB, c-Rel, p50 and p52. Proteasome is involved in the maturation process of p50 and p52, which are synthesized as large precursors of p105 and p100 respectively (Beinke & Ley, 2004; Fan & Maniatis, 1991). Treatment with proteasome inhibitors (PI), such as bortezomib, blocks p105 and p100 processing, and/or IkB degradation, thus inhibiting the NF-kB-mediated cancer promoting activity (Adams, 2004a; Johnson, 2015; Kaplan et al., 2017; Richardson et al., 2006). Indeed, NF-kB activation seems to play a major role in the antitumour effect of bortezomib, particularly in multiple myeloma and melanoma cells (Amiri, Horton, LaFleur, Sosman, & Richmond, 2004; Hideshima et al., 2002). Among the numerous proteins regulated by NF-kB signalling, cyclin D1 plays a crucial role in cancer progression, since it is a key regulator of late G1 phase of cell cycle. The cyclin D1-Cdk4/6 complexes generate the phosphorylated form of the Rb protein, resulting in the release of EF2 transcription factors, inducing its activation. This is followed by the expression of cyclin E, which interacts with Cdk2 bringing about the hyper-phosphorylation of Rb, cyclin A and genes involved in DNA synthesis. These steps anticipate the S phase progression. Downregulation of NF-kB signalling, induced by proteasome inhibition, leads instead to a decrease in cyclin D1 level, impairing the phosphorylation of Rb and, in turn, the release of E2F, thus inhibiting G1/S transition (Fig. 3) (Diehl & Ponugoti, 2010; Harbour, Luo, Dei Santi, Postigo, & Dean, 1999; Masamha & Benbrook, 2009; Rastogi & Mishra, 2012).
3.2.2. p53
p53 is a nuclear transcription factor that regulates apoptosis, DNA repair, angiogenesis, cell growth and senescence (Gupta et al., 2018; Vogelstein, Lane, & Levine, 2000); thus, regulation of its level is fundamental to guarantee cell homeostasis. This protein is characterized by a very rich functional spectrum that is the consequence of a structural complexity which renders it able to interact with a myriad of partners. p53 exists as a dynamic ensemble of different “proteoforms”, and this structural plasticity is due to the presence of intrinsically disordered regions, as well as to several modifications at transcriptional and post-translational level. Several p53 mutants form amyloid structures that aggregate in the cell through a "prion-like" fashion with a gain of function effect (Rangel et al., 2019; Rangel, Costa, Vieira, & Silva, 2014; Silva, De Moura Gallo, Costa, & Rangel, 2014). It is noteworthy that p53 unfolded mutant forms are shared in cancer and in Alzheimer's disease (AD) tissues, actually entering in the list of biomarkers that can be used for their diagnosis (Amor-Gutiérrez et al., 2020).
Under normal conditions, p53 degradation is a complex and finely regulated process, which is predominately orchestrated by the MDM2 protein, a RING-finger E3-ligase that promotes the poly-ubiquitination of p53, and, thus, its degradation by the 26S (Fig. 3) (Brown, Lain, Verma, Fersht, & Lane, 2009; Devine & Dai, 2013; Haupt, Maya, Kazaz, & Oren, 1997; Momand, Wu, & Dasgupta, 2000; Poyurovsky et al., 2007). P53 pro-apoptotic function covers a prominent role in tumour suppression, and mutations of p53 gene are among the most frequent genetic events in human tumours (Gupta et al., 2019; Kandoth et al., 2013; Niazi, Purohit, & Niazi, 2018; Walerych et al., 2016; Walerych, Lisek, & Del Sal, 2015). Additionally, tumours expressing wt-p53 often have different mechanisms to bypass its activity, such as the overexpression of MDM2 (Chène, 2003; Gupta et al., 2019; Quesnel et al., 1994). A series of studies, performed in different cancer cell models, including melanoma, head and neck and colon cancer, reveal that one of the main mechanisms of cell death induction by proteasome inhibition, is the p53 pathway stabilization (An, Hwang, Trepel, & Blagosklonny, 2000; Concannon et al., 2007; Fernández et al., 2005; Gomez-Bougie et al., 2007; Li & Johnson, 2013; Li, Li, Grandis, & Johnson, 2008; Lopes, Erhardt, Yao, & Cooper, 1997; MacLaren, Chapman, Wyllie, & Watson, 2001; Morsi, Hage-Sleiman, Kobeissy, & Dbaibo, 2018; Qin et al., 2005; Yu, Carroll, & Thomas-Tikhonenko, 2007; Zhu et al., 2005). Accordingly, pro-apoptotic factors, such as Noxa and Bax, are primary p53-responsive elements (Fig. 3) (Albert, Brinkmann, & Kashkar, 2014; Oda et al., 2000). However, controversial results are still reported, since the killing of some cancer cells was shown to involve a p53-independent mechanism of Noxa induction, providing evidence for a novel strategy to bypass the apoptotic resistance of tumour cells (Perez-Galán et al., 2006; Qin et al., 2005; Strauss et al., 2007; Devine & Dai, 2013; Yerlikaya, Okur, & Ulukaya, 2012; Xue et al., 2019).
3.2.3. p21 and p27 Cdk inhibitors
One of the main hallmarks of carcinogenesis is the loss of cell division control. Proteasome is involved in the regulation of the cell cycle, since it degrades cyclin dependent kinases (Cdk) and Cdk inhibitors (CdkIs) (Diehl & Ponugoti, 2010; Glickman & Ciechanover, 2002). Generally, the main function of CdkIs consists in the inhibition of cyclin/Cdk complexes, blocking cell division; p21 and p27 CdkIs expression is frequently suppressed in cancer, favouring the dysregulation of cell proliferation (Abbas & Dutta, 2009; Chu, Hengst, & Slingerland, 2008).
27 is a well-known negative regulator of cell cycle progression in mammalian cells which binds and suppresses the activity of two crucial complexes (i.e., Cdk2/cyclin E and Cdk2/cyclin A), mediating G1 progression and G1/S transition (Sherr & Roberts, 1999; Slingerland & Pagano, 2000). P27 is ubiquitinated by the E3-ligase complex SCFSkp2 and then degraded by the 26S (Abbas & Dutta, 2009; Chu et al., 2008; Rastogi & Mishra, 2012; Slingerland & Pagano, 2000). Low levels of p27 are reported in different cancers (including prostate, breast, and colorectal), as a consequence of an increased UPS activity, which leads to its accelerated degradation (Glickman & Ciechanover, 2002; Loda et al., 1997; Tsihlias, Kapusta, & Slingerland, 1999). Moreover, consistent with the oncogenic role of SCFSkp2, its overexpression is associated with low levels of p27, and thus with the deregulation of cell cycle progression (Gstaiger et al., 2001; Lee & Lim, 2016).
p21, whose stability is essential for cell fate decision, binds Cdk2/cyclin E complex (blocking the onset of the cell S phase) and cyclin B/Cdk1 complex (leading to a growth arrest in the G2 phase) (Abbas & Dutta, 2009; Rastogi & Mishra, 2012) (Fig. 3). Moreover, p21 binds the proliferating cell nuclear antigen (PCNA), interfering with PCNA-dependent DNA polymerase activity, inhibiting DNA replication and modulating PCNA-dependent DNA repair processes (Abbas & Dutta, 2009; Moldovan, Pfander, & Jentsch, 2007; Mortusewicz, Schermelleh, Walter, Cardoso, & Leonhardt, 2005; Walsh & Xu, 2006). Under normal conditions, p21 levels are controlled at a transcriptional level mainly by p53. In several cancer types, proteasome inhibition brings about accumulation of p53, enhancing its nuclear export, and thereby the expression of transcriptional target genes, including p21, counteracting the proliferation stimulus associated to low p21 levels (Abbas & Dutta, 2009; Brugarolas et al., 1995; Deng, Zhang, Harper, Elledge, & Leder, 1995; Eastman, 2004; Roninson, 2002). Beside a transcriptional control by p53, p21 levels are modulated through either (i) a ubiquitin-dependent degradation by the 26S, and (ii) a ubiquitin -independent pathway of degradation by the uncapped 20S, which has been proposed for the free form of p21 (Fig. 3) (Chen et al., 2007; Deng et al., 2018; Li et al., 2007; Sheaff et al., 2000; Touitou et al., 2001). In particular, case (i) requires p21 ubiquitination by three E3 ligases (i.e., SCFSKP2, L4CDT2 and APC/ CCDC20) at specific stages in an unperturbed cell cycle, which occurs only when p21 is bound to cyclin/Cdk complexes and PCNA. Consistent with these studies, proteasome inhibition has been reported to considerably increase the intracellular level of p27 and p21 in many cancers, favouring cell cycle arrest (Huang et al., 2011; Li et al., 2018; Mi, Gan, & Chung, 2011; Rastogi & Mishra, 2012; Sterz et al., 2010).
3.3. Proteasome inhibitors for cancer therapy
Although more than one thousand proteins belong to the UPS function in the ubiquitination and recognition of ubiquitinated protein substrates, the vast majority of currently available inhibitors, which have been designed and synthesized to block this pathway, target the proteolytic core of 20S. These proteasome inhibitors are broadly categorized into different groups, according to the origin (e.g., synthetic or natural products), the kinetic mechanism of inhibition (e.g., competitive or non-competitive) or else the chemical structure/reactivity. This chapter and the following one will deal with the discussion on the most promising and clinically available inhibitors, pointing out, wherever possible, their molecular action as well as their pharmacological profile and therapeutic outcome of their usage in clinic.
Proteasome inhibitors were initially developed to prevent cancer-related cachexia, in view of proteasome role in protein turnover (Manasanch & Orlowski, 2017). To date, UPS is universally considered a “bona fide” target for the development of anti-cancer drugs (Adams, 2004a, Adams, 2004b; Bullova, Cougnoux, Marzouca, Kopacek, & Pacak, 2017; Cloos et al., 2017; Gandolfi et al., 2017; King, Deshaies, Peters, & Kirschner, 1996; Landis-Piwowar et al., 2006; Narayanan et al., 2020; Niewerth et al., 2015; Roeten et al., 2018). Indeed, PIs represent the reference treatment of multiple myeloma (MM), in view of its high sensitivity to this class of anticancer agents (Chauhan et al., 2005; Fricker, 2020; Gandolfi et al., 2017; Narayanan et al., 2020; Roccaro et al., 2006). It is important to recall that MM is an aggressive and often incurable plasma cell dyscrasia characterized by uncontrolled proliferation of abnormal plasma cells, which invade the bone marrow, producing abnormal monoclonal immunoglobulins, which circulate in the blood. The poor prognosis of MM, which reflects the genomic complexity of the disease, has dramatically improved after the introduction of PIs in disease management, mainly for patients displaying a refractory MM (RMM) and relapsed and refractory MM (RRMM), as discussed in the next section (Leleu et al., 2018). As reported previously (see Section 3.2), proteasome inhibition results in multiple deleterious downstream effects in cancer cells, including down-regulation of NF-κB signalling, stabilization of p53, cell cycle arrest, which all lead to apoptosis. Moreover, PIs downregulate adhesion molecules and secretion of cytokines (Chauhan, Hideshima, & Anderson, 2005; Read et al., 1995), inhibit angiogenesis (Sunwoo et al., 2001) and induce DNA-damage (Łuczkowska, Rogińska, Ulańczyk, & Machaliński, 2020).
The effort to develop PIs has a long history and many different approaches have been adopted, ranging from the use of endogenous and/or natural compounds to the synthesis of new ones (Buac et al., 2013). Initially, proteasome targeting for cancer therapy has been viewed with scepticism, mainly because of the fundamental and crucial roles of proteasome in regulating cell homeostasis in all living cells (Park, Miller, Jun, Lee, & Kim, 2018). Although the reason for the increased cytotoxicity of PIs on proliferating tumour cells is not completely understood, it is widely reported (Chauhan, Catley, et al., 2005) that cancer cells are more dependent on proteasomal activity, likely because of the higher protein turnover they experience, thus being also more sensitive to its blockage (Almond & Cohen, 2002). Tumour cells have a proteasome pathway more active than normal cells, since an increased capability for synthesis and modification of proteins is necessary to preserve their uncontrolled cell proliferation and their high metastatic capacity (Chen et al., 2011; Chen & Madura, 2005).
Currently, three clinically approved PIs are available, namely: (i) bortezomib (Velcade, recently introduced in the market also in its generic version) (approved in 2003 and 2004, by FDA and EMA, respectively), (ii) carfilzomib (Kyprolis) (approved in 2012 and 2015, by FDA and EMA, respectively), and (iii) the first oral PI, ixazomib (Ninlaro) (approved in 2015 and 2016 by FDA and EMA, respectively) (Feling et al., 2003; Fricker, 2020; Gandolfi et al., 2017; Narayanan et al., 2020). Although the availability of PIs has led to an improvement of patients'survival rate, the therapeutic potentiality of these drugs is limited by several drawbacks, including the low potency and specificity of approved molecules, adverse effects and development of drug resistance (Assaraf et al., 2019; Cree & Charlton, 2017; Gacche & Assaraf, 2018; Gonen & Assaraf, 2012; Li, Wu, & Cheng, 2016; Wijdeven, Pang, Assaraf, & Neefjes, 2016; Zhitomirsky & Assaraf, 2016). Furthermore, the therapeutic potential of bortezomib is negatively affected by pharmacokinetic issues and by the very limited distribution to solid tumours which require exceedingly high and toxic doses (Grigoreva, Tribulovich, Garabadzhiu, Melino, & Barlev, 2015; Huang et al., 2014). The use of more recently approved carfilzomib and ixazomib has only partially allowed to overcome these issues (see Section 3.3.2.1). Therefore, there is a growing demand of novel inhibitors with different mechanisms of action and more favourable pharmacological profiles. Additionally, it emerges that the antitumour activity of PIs is markedly improved in combination with conventional therapeutic strategies or with other molecularly targeting agents, such as cell surface death receptor, autophagy, STAT3 and Histone deacetylase (HDAC) inhibitors (Li et al., 2009; Li et al., 2010; Li et al., 2012; Li & Johnson, 2012; Li, Zhou, & Chen, 2008; Pei, Dai, & Grant, 2004; Seki et al., 2010; Yoshiba et al., 2011). Accordingly, a number of preclinical and clinical studies are ongoing to evaluate further new drug combinations, and to optimize administration schedules of therapeutic protocols already used (Berenson et al., 2007; Chen, Frezza, Schmitt, Kanwar, & Dou, 2011; Chen, Retzlaff, Roos, & Frydman, 2011; Johnson, 2015; Wallington‐Beddoe, Sobieraj‐Teague, Kuss, & Pitson, 2018).
3.3.1. Chemical structure and mechanism of action
Generally, PIs are electrophilic molecular species that react covalently with the threonine residues of the proteasome active sites (Harer, Bhatia, & Bhatia, 2012). The first PIs were analogs of serine protease inhibitors, characterized by hydrophobic peptide aldehydes, mimicking substrates of the proteasome β5 active site and reacting with the nucleophilic hydroxyl group of threonine to form reversible hemiacetal adducts. However, first aldehyde inhibitors turned out to have additional targets in the cell besides the proteasome, also inhibiting cathepsin B and calpains (Kisselev & Goldberg, 2001). For this reason, other molecular scaffolds have been investigated and peptide boronates (such as bortezomib and ixazomib) as well as epoxyketones (i.e., carfilzomib and oprozomib) have been synthesized. These new and more specific PIs have experienced great success in clinics (as extensively discussed in Section 3.3.2), and most of them are currently used as therapeutic drugs, even though some of them still retain activity toward non-proteasome targets. In fact, bortezomib and most second-generation boronates also co-inhibit caspase-like sites (Kisselev, van der Linden, & Overkleeft, 2012). In the next section, the main chemical properties of PIs approved and/or ongoing in clinical trials are discussed.
3.3.1.1. First-generation proteasome inhibitors: Bortezomib
Bortezomib is a dipeptide containing phenylalanine and leucine with a boronic acid instead of a carboxylic acid, and a pyrazinoic acid moiety to protect the N-terminus. The structure of bortezomib bound to the 20S has been solved, elucidating the binding mode and mechanism of action at the molecular level (Jung et al., 2004; Groll, Berkers, Ploegh, & Ovaa, 2006) (Fig. 4B). Bortezomib binds reversibly to the chymotryptic-like (CT-L) β5 subunit of the proteasome, even though it has also been reported to bind the caspase-like (C-L) β1 and trypsin-like (T-L) β2 subunits with lower affinity (Buac et al., 2013); however, a good selectivity of bortezomib toward specific proteasome subunits is dictated by the composition of their substrate binding pockets, which differs in the three catalytic β-subunits. In the presence of bortezomib, an anti-parallel β sheet conformation is adopted by domains in the catalytic clefts, and direct hydrogen bonds are formed between the conserved residues (i.e., Gly47N, Thr21N, Thr21O, and Ala49O) of the proteasome β-type subunits and the main chain atoms of the drug, stabilizing the complex (Fig. 4B). The boronic acid is responsible for the actual inhibition, ensuring an increased specificity for the proteasome. Indeed, the boron atom covalently binds the oxygen of Thr1Oγ (the electrophilic functional group that normally reacts with peptide bonds of substrates, see Section 2.2.1), while the acidic boronate hydroxyl groups are bound to Gly47N, bringing about a stabilization of the oxyanion hole. Further stabilization of the tetrahedral boronate adduct comes from a second acidic boronate hydroxyl moiety, which works as a catalytic proton acceptor and is H-bridged to the N-terminal threonine amine atom. A wide range of specific inhibitors has been developed, but usually peptide boron esters and acids are powerful inhibitors of serine proteases, as they interact covalently but reversibly with the active hydroxyl site of this class of the enzymes (Harer et al., 2012). Furthermore, these peptide boron esters are less reactive toward circulating nucleophiles in aqueous solutions than their aldehyde counterparts (Adams et al., 1998).
Fig. 4.
Proteasome binding structures of PIs.
The structures of proteasome binding to carfilzomib (panel A), bortezomib (panel B), salinosporamide (marizomib) (panel C) and ixazomib (panel D); are reported. The β-5 subunit is represented as turquoise ribbon, the β-1 subunit is represented as purple ribbon, the inhibitors are represented as orange sticks and the protein residues interacting with the inhibitors as grey sticks.
Bortezomib induces toxicity in cancer cells through different mechanisms, including (i) inhibition of the NF-kB pathway, which has been envisaged as the main target of bortezomib clinical efficacy; (ii) stabilization of p53 pathway, which leads to apoptosis mainly by increasing the level of pro-apoptotic factors, such as NOXA and Bcl-2; (iii) modulation of CdkIs levels. Moreover, bortezomib inhibits tumour angiogenesis probably as a result of reduced vascular endothelial growth factor receptor (VEGFRs), which seems to be linked to the inhibition of NF-kB (Hideshima et al., 2001; Hideshima et al., 2003; Nunes & Annunziata, 2017; Pandit & Gartel, 2011; Sunwoo et al., 2001). Despite the plethora of mechanisms of actions responsible for the high toxicity of bortezomib toward cancer cells and the high specificity toward serine proteases, peptide boron esters containing acids, such as bortezomib, can become bio-activated to chemically reactive imine amide metabolites inducing drug toxicity (Li, Yu, Ring, & Chovan, 2013). As a matter of fact, carbinolamides metabolites have been detected after incubation with human liver proteins and the formation of GSH conjugates was also observed, both likely stemming from electrophilic reactions of the imine amides with the nucleophilic GSH. The observed metabolites seem to be produced via oxidative de‑boronation, catalyzed by hepatic cytochrome P450 enzyme, and bortezomib toxicity has been ascribed to their formation and high reactivity (see also Section 3.3.2.1). A way to reduce these adverse effects of bortezomib treatment is the use of appropriate methods for administering this agent, such as early-dose reduction and once-weekly and subcutaneous administration.
A crucial drawback, encountered when bortezomib is used as a therapeutic drug, is the rapid development of resistance in response to the treatment (Barrio et al., 2019; Lee et al., 2015). Many studies have described a plethora of strategies the cancer cells may evolve to acquire bortezomib resistance. In this regard, selective down-regulation of specific 19S subunits and the consequent reduced flux of substrates through proteasome has been reported to be a major strategy that several cancer cells may adopt to cope with proteasome inhibition (Tsvetkov et al., 2015; Tsvetkov et al., 2017). Somatic mutations in the catalytic cleft of β5 and involved in the binding to bortezomib have been further described in patients with MM who underwent prolonged therapy with PIs: in this case, resistance induction was acquired through missense mutations and resistance was effective, though at a variable extent, also to next generation PIs (see next paragraphs) (Barrio et al., 2019).
Further, a selective overexpression (up to 60-fold) of a mutant β5 protein has been proposed at the origin of the bortezomib resistance, whereas marked changes in CT-L proteasome activity are not found (Oerlemans et al., 2008). On the other hand, other studies have reported a significant decrease of the CT-L proteasome activity after 1 h in four different cell lines, maintaining such an inhibitory activity for as long as 24 h (Bonvini et al., 2007). Furthermore, an increase in the accumulation of the β5 precursor form was observed, even though no significant alteration in the expression profile of the mature form was detected (Yerlikaya & Okur, 2019). Analogously, it has been also reported that α5 promotes the tumourigenic process of prostate cancer cells and is linked to bortezomib resistance (Fu et al., 2018). Other changes in bortezomib resistant cell lines, such as increased expression of β1 and β5 proteasome subunits, upregulation of pro-apoptotic proteins of the Bcl-2 protein family, Bax and Noxa have been also reported (Wu, Yang, & Saitsu, 2016). Moreover, the lack of some proteins, such as XBP1, which decreases the endoplasmic reticulum burden and affects the unfolded protein response, has also been proposed as a possible cause of bortezomib resistance (Fall et al., 2014). Interestingly, certain factors have been proposed as predictive markers of response to bortezomib treatment. Among others, KLF9, CDK5, Nampt and accumulation of unfolded proteins in the endoplasmic reticulum (ER stress) and UPR-associated markers (XBP1, ATF3, and AFT4) have been identified to play an important role in bortezomib sensitivity. It is therefore clear that further studies are demanded in order to better understand the underlying mechanisms which limit the use of this compound for cancer treatment.
3.3.1.2. Second-generation proteasome inhibitors
The socalled second generation of PIs includes drugs with different chemical features that reflect their different pharmacological profile (see Section 3.3.2.2); they can be either epoxyketones (e.g., carfilzomib and oprozomib), peptide boronate (e.g., ixazomib and delazomib) or else nonpeptide proteasome inhibitor, such as marizomib.
In general, epoxyketone PIs are characterized by a short peptide core, and a terminal α,β-epoxyketone dual electrophilic reactive warhead, which determines their activity (Schrader et al., 2016). The most important representative of this PI class is carfilzomib, a tetrapeptide with a terminal epoxyketone group, which seems to be highly specific for the proteasome (see 3.3.1.1, 3.3.2.2.2) (Muz et al., 2016). It displays an inhibitory power equivalent to that of bortezomib for CT-L subunits of the proteasome (IC50 = 6 nM), whereas C-L and T-L sites are only very weakly inhibited by carfilzomib (IC50 = 2400 and 3600 nM, respectively); thus it is considered a selective inhibitor of CT-L activity (Demo et al., 2007; Boccon-Gibod et al., 2020). Carfilzomib forms a covalent adduct between its C-terminal ketone moiety and Thr1O of each inhibited subunit (Fig. 4A). Additionally, unlike peptide boronates, such as bortezomib and ixazomib, carfilzomib forms a stable morpholine ring between Thr1 N-terminal amino group and epoxide α carbon. These further covalent interactions dramatically enhance the specificity of epoxyketones for proteasome with respect to other proteases, making this PI class essentially irreversible under physiological-treatment conditions (Groll et al., 2000). Crystallographic studies on human 20S proteasome in complex with carfilzomib clarify the structural basis for the high in vivo drug's selectivity for CT-L activity. The high specificity for CT-L activity can be attributed to van der Waals interactions between carfilzomib and S1, S3, and S4 pockets of β5 subunits, whereas in the T-L β2 subunit, carfilzomib forms favourable van der Waals interactions only with S3 and S4 pockets, but not with S1 (Fig. 4A). In fact, the main differences between CT-L and T-L sites mostly reside in the S1 pocket size, which is much wider in the T-L subunit than in the CT-L one, rendering less effective the van der Waals contacts between the P1 leucyl group of carfilzomib and the S1 pocket of the T-L subunit. In addition, His116 of β7 subunit sterically blocks the entry of carfilzomib P4 phenyl group into the S4 pocket of the T-L subunit, shifting P4 up to 3.7 Ǻ away from the S4. Furthermore, the polarity of the C-L S1 pocket impairs an interaction with the carfilzomib hydrophobic P1 leucyl group (Fig. 4A). As a whole, these differences lead to a disordered N-terminus of carfilzomib, likely contributing to the higher IC50 value of C-L activity (Harshbarger, Miller, Diedrich, & Sacchettini, 2015).
The mechanisms through which carfilzomib induces cell death are less known than for bortezomib, even though several studies demonstrated that it elicits programmed cell death acting in different ways, such as (i) activating c-Jun-N-terminal kinase, (ii) promoting mitochondrial membrane depolarization and favouring cytochrome c release, (iii) increasing the levels of pro-apoptotic factor Noxa, and activating caspase-3 and caspase-7 (Narayanan et al., 2020; Parlati et al., 2009). The introduction of carfilzomib in clinical therapy has allowed to overcome some criticisms related to bortezomib administration, like a reduced incidence of adverse effects (e.g., severe peripheral neuropathy) and acquired resistance; therefore, it has become a key option for the treatment of RMM patients (see Section 3.3.2.2.1). However, a number of patients also display intrinsic resistance or develop resistance to carfilzomib treatment (Shah et al., 2018). The reasons for this might stem from mutations or overexpression of proteasome catalytic subunits, but a likely contributor to carfilzomib resistance could also be the overexpression of the efflux pump P-glycoprotein (P-gp), reducing the drug intracellular concentration, since carfilzomib is a recognized substrate of this enzyme (Ao et al., 2012; Besse et al., 2018; Lee et al., 2019; Zang, Kirk, & Johnson, 2014; Zheng, Liu, Zheng, & Hu, 2017).
Oprozomib is an orally bioavailable peptide epoxyketone, currently tested in ongoing clinical trials. It is a tripeptide structural analogue of carfilzomib, which was synthesized to improve drug absorption. In fact, it is thought that smaller peptides are absorbed more effectively across the small intestine wall (Hamman, Enslin, & Kotzé, 2005; Zhou et al., 2009). Like carfilzomib, oprozomib primarily exhibits irreversible binding kinetics to CT-L subunit (Rajan & Kumar, 2016), and the co-crystal structure of human 20S and oprozomib enhanced the knowledge of how proteasome active sites interact with peptide epoxyketone inhibitors (Schrader et al., 2016). Oprozomib, like other epoxyketone inhibitors, forms a seven-membered, 1,4-oxazepano adduct with the catalytic Thr within the β5 active site, whereas (as also reported for carfilzomib) previous findings reported the formation of a 1,4-morpholino adduct. Therefore, these novel solved structures have indicated that, during the second step of the inhibitory reaction, the Thr N-terminal amino group attacks the β carbon rather than the α carbon of the inhibitor's epoxide (Carmony, Lee, & Kim, 2016; Schrader et al., 2016). Concerning the mechanism through which oprozomib mediates cancer cell death, it has been demonstrated that oprozomib induces apoptosis through the activation of caspase 3, 8 and 9 (Chauhan et al., 2010), PARP cleavage, and, interestingly, it seems to block angiogenesis that it is known to play a key role in MM progression (Chauhan et al., 2010a; Giuliani, Storti, Bolzoni, Palma, & Bonomini, 2011; Podar et al., 2001; Zhu et al., 2019).
Ixazomib and delazomib are both orally available structural analogues of bortezomib with a boronic acid as pharmacofore. In particular, ixazomib is a dipeptidyl leucine boronic acid, that was developed through a large-scale screening of boron-containing PIs in the search of compounds with an increased efficacy and reduced side effects with respect to bortezomib (Chauhan et al., 2011; Kupperman et al., 2010; Offidani et al., 2014). Since it belongs to the same chemical class of bortezomib, it is not surprising that its acts through a similar mechanism of action. In fact, proteasome subunit inhibition occurs when boric acid group forms a covalent bond with the hydroxyl group of the catalytic N-terminal threonine residue (Muz et al., 2016). Like bortezomib, ixazomib reversibly blocks the CT-L of the β5 subunit (IC50 = 3.4 nmol/L for ixazomib vs 2.7 nmol/L for bortezomib) (Chauhan et al., 2011; Lee, De la Mota-Peynado, & Roelofs, 2011) (Fig. 4D). Noteworthy, proteasome dissociation half-life for ixazomib is relatively shorter than for bortezomib (18 min for ixazomib and 110 for bortezomib), improving its general tissue distribution and rendering this drug more “re-available” (see Section 3.3.2.2.2) (Kupperman et al., 2010; Narayanan et al., 2020). Biochemical analysis and in vitro studies showed that at high concentrations ixazomib (like bortezomib) inhibits also other proteolytic sites of 20S proteasome (Chauhan et al., 2011). However, the most important advancement of ixazomib with respect to bortezomib is the possibility of an oral administration; thus, ixazomib can be formulated as an ester citrate prodrug (MLN2238), which is rapidly hydrolyzed in aqueous solution (e.g., plasma) to the pharmacologically active metabolite MLN2238 with free boric acid (Chauhan et al., 2011; Gupta et al., 2019; Kupperman et al., 2010; Okazuka & Ishida, 2018). Like bortezomib, the apoptotic activity of ixazomib is mediated by caspase 3–8-9 activation through a stabilization of the p53 pathway (see 3.2.2, 3.2.3) (Muz et al., 2016). Interestingly, microRNA studies in MM cells revealed that ixazomib induces upregulation of the tumour suppresso miR33b, leading to apoptosis by blocking proto-oncogene PIM-1 (Tian et al., 2012).
Another peptide boronate is delazomib, which reversibly inhibits CT-L subunit with a magnitude and a mechanism of action similar to bortezomib and ixazomib (Dorsey et al., 2008; Piva et al., 2008). Moreover, like bortezomib, in vitro studies have revealed that delazomib primary target is the inhibition of NF-kB pathway, with a consequent alteration of the expression of several NF-kB downstream effectors (Kubiczkova, Pour, Sedlarikova, Hajek, & Sevcikova, 2014; Piva et al., 2008).
The main representative of the PI third class is marizomib, also named salinosporamide A, which is a natural product deriving from a sediment obligate marine actinomycete identified as Salinospora tropica (strain CNB-392) (Potts & Lam, 2010; Pereira et al., 2019). It is characterized by a different non-peptide-based structure with respect to other PIs so far described. Its unique structure displays a fused γ-lactam-β-lactone ring system containing a cyclohexenyl carbinol and chloroethyl functional groups. Marizomib is an irreversible inhibitor of all catalytic subunits of 20S, with IC50 values ranging from the low pM to mid nM range (Fenical et al., 2009), and it produces a prolonged 20S inhibition (≥72 h) (Potts et al., 2011). Crystallographic structure of the complex between Salinosporamide A and yeast 20S showed that the drug perfectly occupies the active sites of all three pairs of catalytic subunits of 20S (Fenical et al., 2009; Groll et al., 2006) (Fig. 4C). These findings provided a detailed understanding of the proteasome-ligand interactions at the molecular level, revealing a unique mechanism of action that renders the inhibitor irreversible. The first step of interaction is represented by the formation of a covalent ester bound between the catalytic N-terminal Thr1Oγ of each 20S subunit and the carbonyl of the β-lactone ring of the inhibitor (Fig. 4C). β-lactone ring opening is followed by chlorine elimination, giving rise to a stable 5-membered cyclic ether (Groll et al., 2006). One of the main downstream effect observed after proteasome inactivation by marizomib is the inhibition of NF-κB activation, in a fashion similar to other PIs (Narayanan et al., 2020). Remarkably, as also discussed in Section 3.3.2.2.3, the main advantage of marizomib is the capability to overcome the blood-brain barrier, which opened a novel therapeutic potential for this inhibitor, eliciting the research in attempt to improve its pharmacological profile (Park et al., 2018; Singh et al., 2010).
An intriguing point on proteasome biology, which reflects on the identification of more clinically effective PIs and/or novel combination therapy, concerns the individual role and the functional meaning of different proteasome subunits. Although these crucial aspects are poorly understood, β5 subunit was initially identified as the rate limiting protease for proteasome-dependent protein turnover (Heinemeyer et al., 1997; Arendt & Hochstrasser, 1999; Groll et al., 1999). Consequently, PIs were designed to target β5 subunits, as it comes from the above reported chemical features of main PIs (Kubiczkova et al., 2014; Kisselev et al., 2012). However, advanced chemical manipulations, which allowed to monitor the activity of each individual proteolytic subunit, have pointed out that, at higher concentrations, all β5-targeted PIs lose their subunit selectivity and inhibit also the β1 and/or β2 types of proteasome subunits (Kraus et al., 2015; Bruin et al., 2016). These co-inhibition patterns differ among individual PIs and seem to be responsible for the overcoming of drug resistance observed at higher concentrations. In this respect, recent investigations have revealed that β5 and β2 co-inhibition, exclusively achieved by high levels of carfilzomib, is the most effective proteasome inhibition profile in MM (Besse et al., 2019). Therefore, it has been proposed that a better comprehension of the significance of different coinhibitory patterns should help to understand the differential activity and toxicity observed during treatment with different PIs as well as with different doses of the same drug (Besse et al., 2019). Moreover, these findings could provide the rationale for preclinical and clinical investigations of a novel treatment schedules. These results showed that differences on the functional proteasome-interacting groups of the PI (i.e., epoxyketone, β-lactone, or boronate) cannot account for the observed differences in the clinical efficacy of the various drugs (reversible versus irreversible proteasome binding); differences in the PIs affinity toward the various proteasome subunits should be considered instead. The observed differences in PIs affinities are mainly due to different interactions of the specific PI side chains with each of the proteasome subunits. The chemical interpretation of the different PIs inhibition capability for the various proteasome subunits (Gozzetti et al., 2017) also explains why drugs combination is more effective than monotherapy.
3.3.2. Clinical pharmacology
3.3.2.1. First-generation proteasome inhibitors: bortezomib
In the 1990s, the reversible PI bortezomib (formerly named as PS-341) PI, was initially developed as anti-inflammatory and anti-cachectic agent. However, preclinical studies soon unravelled that bortezomib was highly effective against different tumours, in particular MM, inducing growth arrest and apoptosis and inhibiting angiogenesis (Mitch & Goldberg, 1996; Adams et al., 1998; Adams et al., 1999; Teicher, Ara, Herbst, Palombella, & Adams, 1999; Hideshima et al., 2001; LeBlanc et al., 2002; Ma et al., 2003; Mitsiades et al., 2003; Sánchez-Serrano, 2006; Caravita, de Fabritiis, Palumbo, Amadori, & Boccadoro, 2006). Additionally, in vitro studies revealed that bortezomib increased in vitro tumour chemosensitivity and overcame chemoresistance to dexamethasone, doxorubicin, and melphalan, suggesting also its use in combination therapies (Hideshima et al., 2001; Ma et al., 2003; Mitsiades et al., 2003). The overall bulk of in vitro and in vivo studies supported clinical investigations of bortezomib in patients with MM, who had received at least two prior therapies and have demonstrated disease progression after the last therapy (Caravita et al., 2006; Park et al., 2018; Robak & Robak, 2019), leading to the first global approval of PI for cancer treatment.
A pivotal early phase I study investigated the maximum-tolerated dose, dose-limiting toxicity, and pharmacodynamics of bortezomib in patients with refractory haematological malignancies, showing activity against RMM (Orlowski et al., 2002). Subsequently, a phase 2 study (CREST) showed the efficacy of the PI, as single agent or in combination with dexamethasone, in patients with relapsed MM after frontline therapy (Jagannath et al., 2004). These observations provided the rationale for a phase 2 open-label, single-arm (SUMMIT) trial, which included 202 patients with RRMM receiving at least two prior therapies, in which bortezomib (1.3 mg/m2) was administered by intravenous bolus twice weekly for 2 weeks, followed by 1 week without treatment, for up to eight cycles (24 weeks). This study reported 27.7% complete or partial response rate, a median response of 12 months and manageable adverse effects (Richardson et al., 2003). Moreover, bortezomib increased the time to progression to a higher extent (2–4 folds) compared to the last treatment patients received before entering the clinical trial. These impressive results led to the accelerated approval of bortezomib for the treatment of patients with RRMM who had received at least two prior therapies, a particularly difficult-to-treat patient population. An extended follow-up of the SUMMIT study reported a median time to progression of 13.9 months for responding patients, whereas of 1.3 months for those with progressive disease or not evaluable (Richardson, Mitsiades, Hideshima, & Anderson, 2006). The phase 3 trial, APEX, comparing bortezomib with high-dose dexamethasone for RRMM after one to three previous lines of treatment, showed a significant increased survival in patients treated with the PI (one-year survival rates of 80% versus 66%, P = .003; the hazard ratio for overall survival (OS) = was 0.57, P = .001) (Richardson et al., 2005; Cengiz Seval & Beksac, 2018) and these results led in 2005 to the regular approval to bortezomib. The superiority of bortezomib was further confirmed after an extended follow-up, in which the reported median OS was 29.8 months for bortezomib and 23.7 months for high-dose dexamethasone, despite crossover from dexamethasone to bortezomib arm (Richardson et al., 2007). Thereafter, in 2007 FDA approved label expansion of bortezomib to include patients with impaired kidney function, without the requirement of dose adjustment).
Bortezomib was also tested in multidrug regimens, since the shift in clinical practice to a more aggressive approach, including PIs (as also discussed in Section 3.2), has improved survival outcomes (Leleu et al., 2019). In the DOXIL-MMY-3001 phase 3 study the safety and efficacy of bortezomib in combination with pegylated liposomal doxorubicin were compared to those of bortezomib as a single agent in patients with RRMM who had received at least one prior treatment. The doublet therapy was more effective than monotherapy, even though associated with a higher incidence of grade 3/4 (80 vs 64%, respectively) myelosuppression, gastrointestinal events, and hand-foot syndrome (Orlowski et al., 2007). However, despite the significant increase in time to progression observed in the group treated with the drug combination, the final results of OS analysis after a median follow-up of 103 months indicated no significant differences between the two treatments (Orlowski et al., 2016). The triple combination bortezomib-thalidomide-dexamethasone resulted in increased median time to progression (19.5 versus 13.8 months; hazard ratio, 0.59; P < .001) compared to the dual combination of the immunomodulatory agent thalidomide plus dexamethasone, in patients with MM progressing or relapsing after autologous stem-cell transplantation (ASCT), as demonstrated in a phase 3 study (MMVAR/IFM 2005–04) (Garderet et al., 2012). Although a direct comparison between trials is not possible, the observed time to progression was higher than that observed in the APEX trial where bortezomib was tested as single agent (6.2 months) or in the DOXIL-MMY-3001 trial where the PI was combined with liposomal doxorubicin (9.3 month). The addition of bortezomib to thalidomide-dexamethasone was associated with a substantial increase of cumulative, dose-related grade 3 peripheral sensory neuropathy (Garderet et al., 2012). Since thalidomide is also neurotoxic, in the triple combination this agent was replaced by lenalidomide, which is a better-tolerated analogue. Indeed, a phase 2 trial demonstrated a similar median time to progression of 19.5 months, but a markedly lower rate of grade 3 peripheral neuropathy compared to the triple combination including thalidomide (2% versus 29%) (Richardson et al., 2014). The current guidelines recommend the triple combination of bortezomib, lenalidomide and dexamethasone as a preferred salvage regimen for previously treated multiple myeloma and as first-line therapy irrespective of transplantation eligibility (National Cancer Comprehensive Network, NCCN Clinical Practice Guidelines–Multiple-Myeloma.Version3.2020 https://www.nccn.org/professionals/physician_gls/pdf/myeloma.pdf. Accessed April 4, 2020). For RRMM patients who previously received lenalidomide, bortezomib and dexamethasone have also been evaluated in combination with the other immunomodulatory agent pomalidomide. In particular, the phase 3 clinical trial OPTIMISMM showed a significant increase in median progression-free survival (PFS) compared to the doublet bortezomib-dexamethasone (11.2 months versus 7.1 months, HR = 0.61, P 〈0,0001) (Richardson et al., 2019). Two other interesting therapeutic triplet regimen in RRMM, which included bortezomib, are: (i) bortezomib-dexamethasone and the anti-CD38 monoclonal antibody daratumumab approved in 2016 by FDA and EMA for the treatment of patients who had received at least one prior therapy; (ii) bortezomib-dexamethasone and HDAC inhibitor panobinostat approved in 2015 by FDA and EMA for the treatment of patients who had received at least two prior therapies, including bortezomib and an immunomodulatory drug.
In the phase 3 CASTOR trial, the addition of darutumumab resulted in significantly longer PFS (median PFS 16.7 vs. 7.1 months, HR = 0.31) compared to bortezomib plus dexamethasone, but it was associated to infusion-related reactions and higher rates of thrombocytopenia and neutropenia (Palumbo et al., 2016; Spencer et al., 2018).
Panabinostat was the first pan-HDAC inhibitor approved to treat MM, which acts via epigenetic modification and inhibition of the aggresome pathway (i.e. a proteasome-independent pathway that eliminates misfolded proteins). The approval for RRMM was based on the results from the pivotal phase 3 PANORAMA-1 clinical trial, which demonstrated an improvement in PFS of 7.8 months for the three-drug combination compared with placebo plus bortezomib and dexamethasone in this patient population (11.99 vs. 8.08, P < .0001), even though several adverse events were more frequently observed in the panobinostat group (San et al., 2014; San-Miguel et al., 2014; San et al., 2016).
A phase 2 study has also investigated the combination of bortezomib plus dexamethasone with the immunomodulatory agent elotuzumab, a monoclonal antibody against SLAMF7 (signalling lymphocytic activation molecule F7), reporting encouraging results (median PFS 9.7 vs. 6.9 months) (Jakubowiak et al., 2016). Based on the results of this study, the National Comprehensive Cancer Network (NCCN) panel guidelines have included also this triple combination among the therapeutic option for RRMM who have previously received at least one prior therapy (National Cancer Comprehensive Network, NCCN Clinical Practice Guidelines – Multiple Myeloma. Version 3.2020. https://www.nccn.org/professionals/physician_gls/pdf/myeloma.pdf. Accessed April 4, 2020).
Once bortezomib efficacy was established for RRMM in the early 2000s, attention turned to patients with newly diagnosed disease (NDMM), in whom its efficacy was tested with or without dexamethasone, showing that the combined treatment was associated with improved response rate without additional severe toxicities compared to PI monotherapy (Mateos et al., 2006; Mateos et al., 2008; Harousseau et al., 2010; Jagannath et al., 2005; Okazuka & Ishida, 2018). In 2008, the key phase 3 trial VISTA led to the approval of bortezomib, in combination with melphalan and prednisone, by FDA for previously untreated MM and by EMA for previously untreated MM not eligible for high dose chemotherapy and stem-cell transplantation (SCT) (San Miguel et al., 2008). Melphalan plus prednisone, was the standard of care for NDMM patients over 65 years old, being instead high-dose chemotherapy followed by SCT the preferred treatment for patients under the age of 65 years (Barlogie et al., 1997; Alexanian et al., 1969; San-Miguel et al., 2016). In the VISTA trial, 682 patients were randomized to receive either melphalan and prednisone or the same schedule with the addition of bortezomib (at the dose of 1.3 mg/ml). At the initial analysis, the time to disease progression was 24.0 months in the bortezomib group and 16.6 months in the control group (HR for the bortezomib group = 0.48; P < .001) (San-Miguel et al., 2008). The interim and final analyses confirmed the efficacy of the triplet regimen: after median follow-ups of 36.7 months and 60.1 months, 35% and 31% risk of death reduction were reported, respectively, in the bortezomib-containing group versus the control group (HR = 0.653; P < .001 and 0.695; P < .001, respectively) (Mateos et al., 2010; San Miguel et al., 2013). The final reported median OS was 56.4 vs 43.1 months (San Miguel et al., 2013). Even in this case, peripheral neuropathy events were more frequently documented in the bortezomib group. Interestingly, an Italian phase 3 study introduced thalidomide in the triplet regimen of the VISTA trial: 511 patients were randomly assigned to receive nine cycles of the four-drug combination bortezomib-melphalan-prednisone-thalidomide followed by continuous bortezomib-thalidomide as maintenance (VMPT-VT), or bortezomib-melphalan-prednisone (VMP, control group) at the same doses. In the initial analysis, the median PFS was not reached in the VMPT-VT arm and was 27.3 months in the VMP group; the 3-year PFS was 56% in patients receiving VMPT-VT and 41% in those receiving VMP (HR = 0.67; P = .008) (Palumbo et al., 2010). A longer follow up confirmed the higher survival benefit of the VMPT-VT protocol compared to the triplet combination (median PFS 35.3 months versus 24.8 months; HR = 0.58; P < .001) (Palumbo et al., 2014). Thereafter, several randomized, open label, phase 3 clinical trials (IFM 2005–01; GIMEMA; PETHEMA/GEM and MMY-3006) tested bortezomib-dexamethasone doublet regimen or bortezomib-dexamethasone-thalidomide triplet regimen in patients with previously untreated MM as induction therapy before SCT. These and other clinical trials demonstrated a statistically significant improvement in the post-transplantation complete response rate as a consequence of bortezomib inclusion in the induction regimens, even though the incidence of peripheral neuropathy was increased (Horousseau et al., 2010; Cavo et al., 2010; Rosiñol et al., 2012; Sonneveld et al., 2013). The results of these studies led to EMA approval in 2013 of bortezomib with dexamethasone or with dexamethasone plus thalidomide for the induction treatment of patients with previously untreated MM, eligible for high dose chemotherapy followed by SCT. Thereafter, the bortezomib-lenalinomide-dexamethasone triplet regimen has become one of the standard induction therapies before SCT (Okazuka & Ishida, 2018). In two different phase 2 studies (i.e., IFM and IFM/DFCI 2009) this therapeutic regimen was tested in patients with NDMM eligible for SCT as induction therapy, and as induction and consolidation therapy, respectively, with encouraging results in terms of PFS (Richardson et al., 2010; Roussel et al., 2014). Additionally, results from a phase 3 clinical trial demonstrated that the lenalidomide-containing triplet therapy (RVD) followed by high-dose chemotherapy plus SCT was associated with significantly longer PFS than the RVD therapy alone, even though OS did not differ significantly between the two approaches (Attal et al., 2017). A more recent study also confirmed the efficacy of the RVD regimen in the pre-transplant induction therapy so that it has to be considered as a standard of care in this clinical setting (Rosiñol et al., 2019). Recently, a phase 3 trial has evaluated this regimen with respect to lenalidomide-dexamethasone in patients with previously untreated MM, who were not planned for immediate SCT, demonstrating a significant improvement in terms of PFS (43 months vs 30 months, HR = 0.712, P = .0018) and OS (75 months vs 64 months, HR = 0,709, P = .025) with an acceptable risk-benefit profile (Durie et al., 2017). Therefore, although, as above mentioned, high-dose chemotherapy plus autologous SCT is the standard treatment for NDMM in adults up to 65 years of age, the use of combination therapy with lenalidomide, bortezomib and dexamethasone has raised questions about the role and timing of transplantation (Attal et al., 2017; Okazuka & Ishida, 2018).
As in the case of RRMM treatment, bortezomib is always the backbone for therapeutic regimens for NDMM in combination with other novel targeted agents (Seval and Beksac, 2018). Accordingly, two therapeutic regimens, including the monoclonal antibody daratumumab, have been approved by both FDA and EMA:
1) in 2018, bortezomib, daratumumab, melphalan and prednisone combination for NDMM patients, who are ineligible for ASCT. This approval was based on the results of the open-label, multicentre phase 3 ALCYONE (MMY3007) study, in which patients ineligible for high-dose chemotherapy followed by ASCT (age > 65 years or comorbidities), were randomized to receive either daratumumab-bortezomib-melphalan-prednisone (D-VMP) or bortezomib-melphalan-prednisone (VMP, control group). The 3-year OS was significantly higher in the D-VMP group than in the VMP group (78% vs 67.9%); the PFS was also improved in the D-VMP arm (HR = 0.42 for daratumumab group; P < 0·0001) (Mateos et al., 2020);
2) in 2019 (FDA) and 2020 (EMA), daratumumab, bortezomib, thalidomide and dexamethasone for NDMM patients, who are eligible for ASCT, based on the results of the open-label, phase 3 CASSIOPEIA trial. In this study, patients were randomly assigned to receive four pre-transplantation induction and two post-transplantation consolidation cycles of bortezomib, thalidomide and dexamethasone alone or the same regimen plus daratumumab. At day 100 after transplantation, 39% patients in the daratumumab group achieved a complete response or better versus 26% in the control group (p < .0001), with acceptable safety (Moreau et al., 2019).
Initially, intravenous injection was the standard administration route for bortezomib. Thereafter, a large randomized phase 3 clinical trial, compared the efficacy and safety of subcutaneous versus intravenous treatment, at the approved 1.3 mg/ml dose and twice per week schedule in patients with RRMM, showing that subcutaneous administration induced similar effects in terms of overall response rate, but with improved tolerability and reduction of the incidence of peripheral neuropathy. Thus, currently subcutaneous injection is the preferred method of bortezomib administration, since this route is also more convenient for patients (Arnulf et al., 2012; Moreau et al., 2011).
Common adverse effects associated with bortezomib administration are fatigue, gastrointestinal toxicity, trombocytopenia, anorexia, and peripheral neuropathy (i.e., hyperesthesia, hypoesthesia, neuropathic pain, weakness), which is one of the most important complications that negatively affects the patient's quality life and daily activity (Seval and Beksac, 2018). Peripheral neuropathy has been regarded as an off-target effect, since it is due to inhibition of HtrA2/Omi, a serine protease involved in neuronal survival with potency near or equivalent to that for the proteasome (Arastu-Kapur et al., 2011; Park et al., 2018).
Moreover, other adverse events described so far are: (i) tumour lysis syndrome (Sanagawa et al., 2020); (ii) cardiovascular toxicities (Enrico et al., 2007; Grandin, Ky, Cornell, Carver, & Lenihan, 2015), (iii) acute interstitial nephritis and rarely (iv) a severe syndrome of inappropriate anti-diuresis (SIAD) (Cheungpasitporn et al., 2015; Peng, Chen, & Lou, 2017). Furthermore, treatment with bortezomib is associated with an increased risk of Varicella Zoster Virus (VZV) infection, and a continuous prophylaxis with antiviral agents, such as acyclovir and valacyclovir, is recommended (Aoki, Nishiyama, Imahashi, & Kitamura, 2011; Chanan-Khan et al., 2008; Robak & Robak, 2019; Teh, Harrison, Worth, & Slavin, 2016).
The pharmacokinetics of bortezomib is poorly documented mostly due to analytical difficulties (Leveque et al., 2007). Two different studies on patients with prostate cancer and MM suggested that its kinetic profile is characterized by a large distribution volume (Vd), 721-1270 L, high systemic clearance, ranging from 1095 mL/min to 1866 mL/min, and terminal half-life ranging between 10 h and 31 h (calculated over a 24 h period) (Papandreou et al., 2004; Levêque, Carvalho, & Maloisel, 2007).
For what concerns bortezomib clearance, the drug is converted into inactive de‑boronated metabolites by different cytocrome P450 enzymes (CYPs) (e.g., 1A2, 2C9, 2C19, 2D6, and 3A4) (Pekol et al., 2005; Uttamsingh, Lu, Miwa, & Gan, 2005), as also confirmed by studies in which the co-administration of ketoconazole, a CYP3A4 inhibitor, and rifampicin, a CYP3A4 inducer, increased and decreased patients' exposure to bortezomib, respectively (Hellmann et al., 2011; Venkatakrishnan et al., 2009). Since bortezomib undergoes oxidative metabolism in the liver, a study was carried out on whether patients with a reduced hepatic functionality require dose adjustment (LoRusso et al., 2012; Robak & Robak, 2019). In a first phase 1 clinical trial, the pharmacokinetics and safety of bortezomib in patients with varying degrees of hepatic impairment were evaluated, revealing that patients with mild hepatic impairment did not require a starting dose adjustment, whereas patients with moderate or severe hepatic impairment required a reduced dose of 0.7 mg/m2, and constantly monitoring during treatment (LoRusso et al., 2012). These data were also confirmed by the VISTA trial, and specific dosing recommendations for patients with hepatic impairment are inserted into the drug label.
Bortezomib efficacy has been also investigated in other haematological malignancies, and several trials are currently ongoing, including Light Chain Amyloidosis (ALA), Waldenstrom Macroglobulinemia, Acute Lymphoblastic and Myeloid Leukemia, Indolent B-cell non-Hodgkin Lymphoma, Diffuse Large B-cell Lymphoma, T-cell lymphomas, as also recently reviewed elsewhere (Robak & Robak, 2019). Moreover, bortezomib is used off-label in refractory or relapsed T-cell lymphomas, Waldenström Macroglobulinemia and ALA (Du, Yang, & Zhang, 2016; Robak & Robak, 2019). The most promising results have been reported for ALA, in which bortezomib, dexamethasone and cyclophosphamide, or bortezomib, dexamethasone and melphalan represent the most commonly used first-line treatments, although several opened questions demand further investigation (Venner et al., 2012; Mikhael et al., 2012; Palladini et al., 2004; Kastritis et al., 2019; Robak & Robak, 2019). In recent years, the introduction of bortezomib has had a great impact in the cure of Mantle cell Lymphoma (MCL), a non-Hodgkin lymphoma with a short remission duration to standard therapies, and a median OS of approximately 6–7 years (Banks et al., 1992; Fisher et al., 1995; Teodorovic et al., 1995; Weisenburger et al., 2000; Vose, 2017). Therefore, there is a great need of therapeutic strategies directed against novel molecular targets. The chromosomal translocation t(11;14)-(q13;q32) is the molecular hallmark of MCL, resulting in overexpression of cyclin D1, which is not typically expressed in normal lymphocytes (Vose, 2017), and the constitutive activation of NF-kB, which also plays a key role in MCL growth and survival (by controlling cyclin D1 expression, as reported in a previous section (see Section 3.2.1) (Pham, Tamayo, Yoshimura, Lo, & Ford, 2003; Rosenberg et al., 1991). Therefore, proteasome inhibition has been envisaged as an achievable therapeutic strategy, which was confirmed by in vitro studies showing that NF-kB inhibition mediated by bortezomib leads to cell cycle arrest and apoptosis in MCL cells (Pham et al., 2003).
Based on preclinical studies and a phase 1 trial in patients with refractory hematologic malignancies, including, besides MM (see above), MCL and follicular lymphomas (Orlowski et al., 2002), the clinical efficacy of bortezomib as single agent was investigated in patients with relapsed and refractory MCL (Goy et al., 2005; O'Connor et al., 2005; Goy et al., 2009; O'Connor et al., 2009). In 2006, the results of a pivotal open-label, single arm, multicentre phase 2 trials (PINNACLE) led to bortezomib approval by FDA for the treatment of MCL in patients who had received at least one prior treatment. In this trial, 155 patients with progressive MCL, who had undergone at least one prior therapy received, 1.3 mg/m2 of bortezomib on day 1, 2, 5, and 11 of each 3-weeks cycle. The results showed an OR rate of 31% (median duration, 9.3 months), a complete response rate (CR + unconfirmed CR) of 8% (median duration, 15.4 months), a median time to response of 40 days (range, 31 to 204 days), and a median time to progression of 6.2 months. Adverse events were similar to those reported in other studies with bortezomib, such as peripheral neuropathies and gastrointestinal symptoms (Fisher et al., 2006). An extended follow-up confirmed the positive trend, reporting a median OS of 35.4 months in responding patients (Goy et al., 2009). Combined regimens, including bortezomib, have been evaluated in phase 2 clinical trials with a small number of patients, showing improved efficacy that needs to be confirmed by further investigation: bendamustine-bortezomib-rituximab (of 29 patients evaluable for efficacy, 83% achieved an objective response), and bortezomib plus cyclophosphamide-doxorubicin-vincristine-prednisone (CHOP) (OS = 36.6 for patients treated with bortezomib plus CHOP and 11.6 months for those treated with CHOP alone) (Weigert et al., 2009; Orciuolo, Buda, Pelosini, & Petrini, 2010; Agathocleous et al., 2010; Furtado, Johnson, Kruger, Turner, & Rule, 2015; Kouroukis et al., 2011; Lamm et al., 2011; Friedrberg et al., 2011).
Bortezomib as single agent was tested also in previously untreated MCL, demonstrating clinical activity (Belch et al., 2007; Robak & Robak, 2019). However, more promising results were obtained when bortezomib was combined with rituximab-CHOP (R-CHOP), as demonstrated by: (i) phase 1/2 studies on previously untreated patients with MCL (Ruan et al., 2011; Till et al., 2016; Vose, 2017); (ii) a phase 2 study on patients with newly diagnosed MCL, who received also bortezomib as maintenance therapy (Ruan et al., 2011; Till et al., 2016). These trials demonstrated that the combination of R-CHOP with bortezomib followed by bortezomib maintenance improves PFS, as compared to R-CHOP alone, with acceptable toxicity, suggesting further investigation (Ruan et al., 2011; Till et al., 2016). In a large, randomized phase 3 trial, 487 patients with untreated, newly diagnosed MCL, who were not eligible for transplantation, were randomly assigned to two groups, one receiving R-CHOP and a modified R-CHOP regimen with bortezomib in place of vincristine (VR-CAP). After a median follow-up of 40 months, PFS was 14.4 months in the R-CHOP group versus 24.7 months in the VR-CAP group (HR = 0.63; P < .001). Moreover, the final analysis after a median follow-up of 82 months revealed a significantly longer OS in the VR-CAP group than in the R-CHOP group (90.7 months vs 55.7 months; HR =0.66, P = .001), with a manageable toxicity profile. These data led to FDA and EMA approval of bortezomib in combination with rituximab, cyclophosphamide, doxorubicin and prednisone for the treatment of adult patients with previously untreated MCL, who are unsuitable for SCT (Drach et al., 2018; Robak et al., 2015; Robak et al., 2018; Robak et al., 2019). Another strategy, combining bortezomib or lenalidomide with bendamustine and rituximab, has shown efficacy in both first-line and salvage therapy for MCL (Albertsson-Lindblad et al., 2016; Campo & Rule, 2015). Accordingly, in a prospective, multicentre phase 2 study evaluating rituximab, bendamustine, bortezomib and dexamethasone as first-line treatment for patients with MCL aged 65 years or older, at median follow-up of 52 months, the 2-year PFS was 70%, clearly demonstrating that this regimen is active and demands further evaluation (Gressin et al., 2019). Despite other investigations have reported promising data, the results of a recent phase 2 trial for newly diagnosed MCL, in which bortezomib was administered as maintenance treatment after induction therapy with three cycles of R-CHOP, two cycles of high-dose cytarabine, BEAM (carmustine, etoposide, cytarabine, melphalan) and ASCT, demonstrated no positive effects of bortezomib as maintenance therapy (Doorduijn et al., 2020).
On the basis of bortezomib success in haematological malignancies, its potential application in the treatment of solid tumours has been explored (Roeten et al., 2018). A large amount of data has been collected in vitro and in vivo aiming at characterizing the possible activity of bortezomib in different models, such as pancreatic and breast cancers, hepatocellular and anaplastic thyroid carcinoma, with contradictory results (Chen et al., 2016; Huang et al., 2019; Roeten et al., 2018). One of the most promising strategy is the combination of bortezomib with radiotherapy, which results in synergistic effects as a consequence of the bortezomib-induced cell accumulation at the G2/M radiosensitive phase of cell cycle and modulation of radio-resistance mechanisms (i.e., NF-kB activation, loss of p53 and DNA double-strand break repair) (Cron et al., 2013; Zhu et al., 2015; Roeten et al., 2018). Additionally, bortezomib might act as chemosensitizer in combination with standard chemotherapy as demonstrated in models of chemo-resistant small cell lung cancer xenografts (Taromi et al., 2017). An impressive number of clinical trials on bortezomib (897) are reported in ClinicalTrial.gov. A number of these trials have been also performed in different solid tumours (see Table 1 ), revealing, as for preclinical studies, conflicting results. Due to their poor prognosis, two types of solid tumours have been mainly investigated, namely:
Table 1.
Clinical trials with bortezomib in non-haematological cancers.a
| Combined agent | NCT identifier | Phase | Status | Setting |
|---|---|---|---|---|
| Carboplatin | NCT00028912 | 1 | Terminated | Recurrent or progressive ovarian epithelial, primary peritoneal, or fallopian tube cancer |
| Tanespimycin | NCT00096005 | 1 | Terminated | Advanced solid tumours |
| Docetaxel | NCT00064636 | 1 | Terminated | Solid tumours |
| / | NCT00004002 | 1 | Completed | Advanced solid tumours |
| / | NCT00091117 | 1 | Completed | Advanced malignancies |
| / | NCT00021216 | 1 | Completed | Pediatric advanced solid tumours |
| / | NCT02220049 | 1 | Completed | Solid tumours |
| / | NCT00054483 | 1 | Completed | Advanced cancers |
| Vorinostat | NCT01132911 | 1 | Completed | Refractory and recurrent solid tumours |
| Dacarbazine | NCT00580320 | 1 | Completed | Melanoma and sarcoma |
| Temozolomide | NCT00544284 | 1 | Completed | Solid tumours |
| Celecoxib | NCT00290680 | 1 | Completed | Solid tumours |
| Chemotherapy | NCT00424840 | 1 | Terminated | Lung cancer |
| Lapatinib | NCT01497626 | 1 | Terminated | Solid tumours |
| / | NCT02220049 | 1 | Completed | Solid tumours |
| Topotecan | NCT00388089 | 1 | Completed | Solid tumours |
| Carboplatin | NCT00059618 | 1 | Completed | Ovarian, abdominal, or fallopian tube cancer. |
| 5-Fluorouracil, leucovorin, oxaliplatin | NCT00098982 | 1 | Completed | Advanced or metastatic colorectal cancer |
| Cetuximab, radiation with or without cisplatin | NCT00629226 | 1 | Completed | Head and neck cancer |
| 5-Fluorouracil, external-beam radiation therapy | NCT00280176 | 1 | Completed | Rectal cancer |
| Chemoradiation | NCT00329589 | 1 | Completed | Brain, head and neck, and cervix cancer |
| Erlotinib | NCT00895687 | 1 | Completed | Advanced cancer |
| Mitoxantrone | NCT00059631 | 1 | Completed | Prostate cancer |
| Vorinostat | NCT00731952 | 1 | Completed | NSCLC |
| Cetuximab with or without cisplatinum | NCT01445405 | 1 | Completed | Head and neck cancer |
| Placlitaxel | NCT00030368 | 1 | Completed | Advanced or metastatic solid tumours |
| Trastuzumab | NCT00199212 | 1 | Completed | Overexpressing Her-2 breast cancer |
| Bevacizumab | NCT00428545 | 1 | Completed | Advanced malignancies |
| Gemcitabine, carboplatin | NCT00052338 | 1 | Completed | NSCLC |
| Carboplatin, etoposide | NCT00027898 | 1 | Completed | Advanced solid tumours |
| 5-Fluorouracil, leucovorin | NCT00007878 | 1 | Completed | Metastatic solid tumour |
| Radiotherapy | NCT00011778 | 1 | Completed | Head and neck cancer |
| Topotecan | NCT00068484 | 1 | Completed | Advanced malignancies |
| Topotecan | NCT00541359 | 1 | Completed | Advanced solid tumours |
| Pegylated liposomal doxorubicin, gemcitabine | NCT00500422 | 1 | Completed | Advanced solid tumours |
| Omeprazole | NCT00298779 | 1 | Completed | Advanced solid tumours or Non-Hodgkin's lymphoma |
| Cetuximab | NCT00622674 | 1 | Completed | Advanced solid tumours |
| Gemcitabine | NCT00620295 | 1 | Completed | Advanced solid tumours |
| Paclitaxel | NCT00667641 | 1 | Completed | Metastatic or unresectable malignant solid tumours |
| Varinostat | NCT00227513 | 1 | Completed | Metastatic or unresectable solid tumours |
| Paclitaxel, carboplatin | NCT00028587 | 1 | Completed | Advanced solid tumours |
| Carboplatin | NCT01074411 | 1 | Completed | Ovarian epithelial, fallopian tube or primary peritoneal cancer |
| Belinostat | NCT00348985 | 1 | Completed | Advanced solid tumours |
| Vorinostat | NCT00994500 | 1 | Completed | Advanced solid tumours |
| Doxorubicin | NCT00023855 | 1 | Completed | Advanced solid tumours |
| Irinotecan | NCT00644696 | 1 | Completed | Neuroblastoma |
| Interferon α-2b | NCT01462773 | 1 | Completed | Melanoma |
| Sorafenib | NCT01078961 | 1 | Completed | Melanoma |
| Temozolomide, bevacizumab | NCT01435395 | 1 | Completed | Recurrent glioblastoma |
| Clofarabine | NCT02211755 | 1 | Recruiting | Relapsed solid tumours |
| NK cells | NCT00720785 | 1 | Recruiting | Advanced cancers |
| Gemcitabine, doxorubicin | NCT00479128 | 1 | Active, not recruiting | Solid tumours |
| DFMO | NCT02139397 | 1 | Active, not yet recruiting | Relapsed and refractory neuroblastoma |
| Pembrolizumab, cisplatin | NCT04265872 | 1 | Not yet recruiting | Breast cancer |
| Panitumumab | NCT01504477 | 1/2 | Terminated | Advanced colorectal cancer |
| Carboplatin, docetaxel | NCT00714246 | 1/2 | Terminated | Non small cell lung cancer (NSCLC) |
| Temozolomide | NCT00512798 | 1/2 | Terminated | Solid tumours or melanoma |
| Vandetanib | NCT00923247 | 1/2 | Terminated | Medullary Thyroid Carcinoma |
| Docetaxel | NCT00064610 | 1/2 | Completed | Androgen-indipendent prostate cancer |
| Carboplatin, paclitaxel | NCT00093756 | 1/2 | Completed | NSCLC |
| Temozolomide | NCT03643549 | 1/2 | Recruiting | Glioblastoma |
| Cannabidiol, leucovorin, oxaliplatin, bevacizumab, irinotecan, gemcitabine, temozolomide | NCT03607643 | 1/2 | Not yet recruiting | Glioblastoma, gastrointestinal malignancies, MM |
| Bevacizumab | NCT00411593 | 1/2 | Withdrawn | NSCLC |
| Docetaxel, cisplatin | NCT00313690 | 1/2 | Withdrawn | NSCLC |
| / | NCT00200382 | 2 | Terminated | NSCLC |
| / | NCT00346645 | 2 | Terminated | NSCLC |
| / | NCT00085410 | 2 | Terminated | Bile duct or gallbladder carcinoma |
| / | NCT00346645 | 2 | Terminated | NSCLC |
| / | NCT00117351 | 2 | Terminated | Bronchioloalveolar carcinoma and adenocarcinoma |
| Panobinostat | NCT01056601 | 2 | Terminated | Pancreatic cancer progressing after gemcitabine therapy |
| LH-RH agonist drug and androgen receptor antagonists | NCT00103376 | 2 | Terminated | Relapsed prostate cancer |
| Irinotecan | NCT00106262 | 2 | Terminated | Progressive, recurrent or metastatic cervical, vulvar, or vaginal cancer |
| Sorafenib | NCT01100242 | 2 | Terminated | Renal carcinoma |
| Carboplatin | NCT00416793 | 2 | Terminated | Metastatic pancreatic cancer |
| Doxorubicin | NCT00574236 | 2 | Terminated | Metastatic breast cancer |
| Fluorouracil, leucovorin | NCT00103103 | 2 | Terminated | Metastatic or unresectable gastric or gastroesophageal junction adenocarcinoma |
| Erlotinib | NCT00283634 | 2 | Terminated | RRNSCLC |
| / | NCT00025584 | 2 | Completed | Metastatic breast cancer |
| / | NCT00028639 | 2 | Completed | Breast cancer |
| / | NCT00068289 | 2 | Completed | NSCLC |
| / | NCT00023712 | 2 | Completed | Recurrent ovarian epithelial or primary peritoneal cancer |
| / | NCT00425503 | 2 | Completed | Prostate cancer |
| / | NCT00017329 | 2 | Completed | Metastatic kidney cancer |
| / | NCT00051987 | 2 | Completed | Relapsed and refractory colorectal cancer |
| / | NCT00513877 | 2 | Completed | Mesothelioma |
| / | NCT00024011 | 2 | Completed | Metastatic melanoma |
| / | NCT00027716 | 2 | Completed | Metastatic sarcoma |
| / | NCT00051987 | 2 | Completed | Relapsed and refractory colorectal cancer |
| / | NCT00077441 | 2 | Completed | Liver cancer |
| Docetaxel | NCT00183937 | 2 | Completed | Hormoine refractory prostate cancer |
| Docetaxel | NCT00064012 | 2 | Completed | RRNSCLC |
| Docetaxel | NCT00193232 | 2 | Completed | Advanced hormone, refractory prostate cancer |
| Acyclovir | NCT01833143 | 2 | Completed | KRAS mutant NSCLC |
| Gemcitabine, cisplatin | NCT01633645 | 2 | Completed | NSCLC |
| Carboplatin, paclitaxel | NCT00107341 | 2 | Completed | Unresectable, metastatic esophagus or gastroesophageal junction cancer |
| Gemcitabine, carboplatin | NCT00075751 | 2 | Completed | NSCLC |
| Pemetrexed | NCT00343720 | 2 | Completed | NSCLC |
| Doxorubicin | NCT03509246 | 2 | Recruiting | Ovarian cancer |
| Gemcitabine | NCT00305734 | 2 | Completed | Recurrent or metastatic nasopharyngeal cancer |
| Docetaxel | NCT00425750 | 2 | Completed | Head and neck cancer |
| Docetaxel | NCT00362882 | 2 | Completed | Recurrent NSCLC |
| With or without docetaxel | NCT00051974 | 2 | Completed | NSCLC |
| Doxorubicin | NCT00610792 | 2 | Withdrawn | Ovarian cancer |
| Vorinostat | NCT00798720 | 2 | Completed | NSCLC |
| With or without gemcitabine | NCT00052689 | 2 | Completed | Metastatic pancreatic cancer |
| Doxorubicin | NCT00083226 | 2 | Completed | Liver cancer |
| Temozolomide | NCT00990652 | 2 | Completed | Glioma |
| Cetuximab, docetaxel | NCT00118183 | 2 | Completed | NSCLC |
| / | NCT00104871 | 2 | Completed | Metastatic thyroid cancer |
| Irinotecan | NCT00061932 | 2 | Completed | Gastroesophageal junction or stomach cancer |
| / | NCT00118144 | 2 | Completed | Lung Cancer |
| Doxorubicin | NCT00077428 | 2 | Completed | Recurrent, or metastatic adenoid cystic carcinoma |
| Temozolomide, radiotherapy | NCT00998010 | 2 | Completed | Glioblastoma |
| Tamoxifen | NCT00108069 | 2 | Completed | Brain cancers |
| / | NCT00072150 | 2 | Completed | Urothelial transitional cell carcinoma |
| Cisplatin | NCT00458913 | 2 | Completed | Mesothelioma. |
| Irinotecan | NCT00103259 | 2 | Completed | Head and neck cancer. |
| Vorinostat | NCT00641706 | 2 | Completed | Glioblastoma |
| Linsitinib, erlotinib, paclitaxel, dexamethasone | NCT02057380 | 2 | Completed | Advanced solid tumours |
| Vorinostat | NCT00937495 | 2 | Completed | Advanced sarcoma |
| Avastin | NCT00611325 | 2 | Completed | Recurrent glioma |
| Carboplatin, placlitaxel | NCT00288041 | 2 | Completed | Metastatic melanoma |
| Doxorubicin | NCT03509246 | 2 | Recruiting | Ovarian cancer |
| / | NCT00367718 | 2 | Not yet recruiting | Recurrent nasopharyngeal carcinoma |
| / | NCT03345303 | 3 | Recruiting | Intrahepatic cholangiocarcinoma patients |
See https://clinicaltrials.gov/, accessed April 27, 2020. (/ no drug)
(a) Small cell lung cancer and non-small lung cancer, in which bortezomib as single agent has revealed limited efficacy, whereas combination therapy with paclitaxel, carboplatin, and concurrent thoracic radiation seemed more encouraging (Gatti, Zuco, Zaffaroni, & Perego, 2013; Zhao, Zhai, Gygi, & Goldberg, 2015);
(b) Head and neck squamous cell carcinoma, in which, despite promising preclinical studies, clinical trials revealed poor results (Lin, Chen, Chen, Cheng, & Chen, 2012; Li et al., 2013; Gilbert et al., 2013);
Different mechanisms have been proposed to justify the different bortezomib activity between solid tumours and haematological malignancies, including: (i) alterations and mutations of the proteasome subunit composition; (ii) drug penetration; (iii) activation of compensatory mechanisms, such as autophagy; (iv) resistance to apoptosis induction, even though the exact role of each one of these mechanisms needs to be further investigated. Interestingly, to overcome the poor penetration of bortezomib in solid tumours, an alternative strategy currently investigated consists in a delivery system based on nanoparticles or micelle formulation, as also studied for other PIs (Ao et al., 2015; Coelho, Almeida, Santos-Silva, Pereira, & Coelho, 2016; Shen et al., 2014).
3.3.2.2. Second-generation proteasome inhibitors
The strategy of proteasome activity inhibition and the introduction of bortezomib in clinical practice have dramatically changed the battle against MM, even though it was immediately evident that this drug suffers from several drawbacks that needed to be overcome. For example, (i) mutation in the β5 subunits, (ii) induction of drug efflux from cells and (iii) activation of signalling cascades promoting cell survival are all resistance mechanisms that have been identified in bortezomib-resistant cell lines (Bringhen, De Wit, & Dimopoulos, 2017; Oerlemans et al., 2008; Sherman & Li, 2020). Therefore, second-generation PIs have been designed and tested, including carfilzomib, ixazomib, delanozomib, oprozomib and marizomib, whose properties will be discussed in the next sections. Particular emphasis will be given to carfilzomib and ixazomib, which are both FDA- and EMA-approved.
3.3.2.2.1. Carfilzomib
As mentioned in the previous section, two main limitation in the clinical use of bortezomib are (i) the extent of proteasome inhibition and (ii) proteasome recovery after inhibition (Deshaies, 2014). These issues stimulated an intense research, which culminated with the discovery of the natural compound epoxomicin, a covalent and irreversible inhibitor of the β5 subunit (Hanada et al., 1992; Meng et al., 1999; Myung, Kim, Lindsten, Dantuma, & Crews, 2001; Deshaies, 2014), which was isolated by an unidentified Actinomycetes strain (see also Section 3.3.1.2). Importantly, epoxomicin owned unprecedented and exceptional selectivity for proteasome with respect to PI already available (Kim & Crews, 2013; Sin et al., 1999). This high selectivity was supposed to guarantee more physiological tolerability, and a more favourable pharmacological profile than bortezomib. Moreover, the ability to irreversibly bind the β5 subunit implies that the only way to recover proteasome activity is the induction of novel synthesis of functional proteasome particles (Demo et al., 2007; Deshaies, 2014; O'Connor et al., 2009). Thus, through a medicinal chemistry approach, epoxomicin became the scaffold for the synthesis of a more potent tetrapeptide epoxyketone, YU-101 (the parent lead compound of PR-171 or carfilzomib), an injectable drug, which is quickly cleared from plasma (Demo et al., 2007; Myung et al., 2001; Kim & Crews, 2013; Deshaies, 2014). As a matter of fact, for carfilzomib an antitumor activity greater than that of bortezomib was reported in a human tumour xenograft model (Demo et al., 2007).
Thereafter, based on the results of in vitro and in vivo preclinical studies, carfilzomib (Kyprolis) safety profile and clinical efficacy were evaluated in two pioneering studies, in which two different schedules were tested in patients with relapsed or refractory haematological malignancies. In the first phase 1 dose-escalating clinical trial (PX-171–001), carfilzomib doses ranging from 1.2 to 20 mg/m2 were administered intravenously on five consecutive days following 9 days of rest in 14-day cycles, until occurrence of unacceptable toxicity or disease progression. This study revealed that the drug was well tolerated (a dose of 15 mg/m2 was established as the maximum tolerated dose [MTD]), and was active in patients with MM or Waldenström macroglobulinemia that were treatment-refractory or relapsed after at least two lines of treatment (Connor et al., 2009b). In the second phase 1 trial (PX-171-002), carfilzomib (1.2–27 mg/m2) was administered twice a week on two consecutive days for 3 weeks in a 4-week cycle; the results showed that this treatment schedule was well-tolerated, being the majority of adverse events manageable and low-grade in severity, and showed activity in particular against RRMM (Alsina et al., 2012). This regimen was then selected for the subsequent clinical studies, including the PX-171-003A1 and ASPIRE trials. In 2012, in consideration of the results of an open-label, single-arm phase 2 study (PX-171-003-A1), which enrolled 266 patients, carfilzomib received accelerated approval by FDA as single agent for the treatment of MM in patients with clinical evidence of disease progression after at least two prior therapies, including bortezomib and an immunomodulatory agent. In this study, bortezomib was administered at the dose of 20 mg/m2 to a patient population in the majority (89%) resistant to bortezomib and the reported overall response rate was 23.7% with median duration of response of 7.8 months (Herndon et al., 2013). Over the last years, several phase 1 and 2 studies were run both on NDMM and, mainly on RRMM, by using different schedules of carfilzomib as monotherapy, or in combination with other agents, such as low-dose of dexamethasone and lenalidomide (being lenalidomide plus dexamethasone a reference treatment for RRMM). These trials generated a bulk of clinical data which demonstrated the efficacy and tolerability of this PI (see ClinicalTrials.gov) (Vij et al., 2012; Wang et al., 2013; Siegel et al., 2013; Moreau et al., 2015; Papadopoulos et al., 2015; Berenson et al., 2016). The results of these phase 2 studies supported the progression into phase 3 trials, which have further led in 2015 to the EMA approval of carfilzomib. In particular, based on the results of the open label ASPIRE phase 3 study (NCT01080391), EMA approved carfilzomib in combination with lenalidomide and dexamethasone, for the treatment of adult patients with MM, who have received at least one prior therapy. Furthermore, in 2015, FDA approved carfilzomib in combination with lenalidomide and dexamethasone for the treatment of patients with MM, who had received one to three prior lines of therapy (Stewart et al., 2015; Stewart et al., 2016). In the ASPIRE trial, 792 patients with RRMM were randomly assigned in a 1:1 ratio to the carfilzomib group, in which the drug was part of a triple combination therapy with lenalidomide and dexamethasone, or to the lenalidomide plus dexamethasone control group (Stewart et al., 2015). Carfilzomib was administrated for 18 cycles, at a starting dose of 20 mg/m2 on the first cycle with subsequent escalation to reach the target dose of 27 mg/m2 in the following cycles. The addition of carfilzomib significantly increased the median PFS as compared with lenalidomide and dexamethasone alone (26.3 months vs. 17.6 months; HR for progression or death, 0.69; P = .0001). Furthermore, the triplet therapy showed a favourable risk-benefit profile and improved the health-related quality of life of RRMM patients (Stewart et al., 2015; Stewart et al., 2016). Moreover, in 2016 FDA and EMA extended carfilzomib approval, in combination with dexamethasone, to patients with RRMM, who had received one to three lines of therapy (FDA) or at least one prior therapy (EMA), on the basis of the results of the phase 3 randomized, open label, ENDEAVOR study (NCT01568866). In this head-to head comparative study of bortezomib and carfilzomib, 929 patients with RMM were randomly assigned to receive carfilzomib plus low-dose dexamethasone or bortezomib plus low-dose dexamethasone (Dimopoulos et al., 2016). In this study, carfilzomib regimen was 27 mg/m2 in the first cycle and 56 mg/m2 thereafter, infused over 30 min, which is the maximum tolerated dose of carfilzomib tested in combination with dexamethasone in phase 1/2 clinical trials (Papadopulos et al., 2015; Dimopoulos et al., 2016). The primary endpoint of the trial was PFS that was reported to be longer for the carfilzomib group, as compared to the bortezomib one (18.7 versus 9.4 months, HR = 0.53; P < .0001) (Dimopoulos et al., 2016). In another interim analysis aimed at comparing the OS between the two PI, it has been shown that patients treated with carfilzomib had a statistically significant and clinically meaningful improvement in OS than those treated with bortezomib (median 47.6 months versus 40.0 months; HR 0.791; 95%, P = .010) (Dimopoulos et al., 2017; Ludwig et al., 2019; Dimopoulos et al., 2019; O'Connor et al., 2009). In 2018, FDA extended the use of carfilzomib to include a once-weekly dosing option in combination with dexamethasone (once-weekly Kd70) for patients with RRMM. This approval was based on data from the randomized, open-label, phase 3 A.R.R.O.W. trial, in which 478 patients with RRMM, who had received at least two (but no more than three) prior therapies (including bortezomib and an immunomodulatory drug), were assigned to receive a 30-min infusion of once-weekly (70 mg/m2) carfilzomib vs a 10-min infusion of twice-weekly (27 mg/m2). All patients also received the same dose of dexamethasone (Moreau et al., 2018). The primary endpoint of the trial, PFS, was 11.2 months for the once-weekly regimen versus 7.6 months for the twice-weekly one (HR = 0.69; P = .0014). The ORR in patients treated with the once-weekly regimen was 62.9% versus 40.8% for those treated with twice-weekly (p < .0001) (Moreau et al., 2018; Moreau et al., 2019; Moreau et al., 2020). Thus, the once-weekly carfilzomib was safe and more effective as compared to the twice weekly schedule.
As mentioned above, early phase 1/2 trials suggested that carfilzomib in combination with other agents, such as melphalan and prednisone (Moreau et al., 2015), lenalidomide and low-dose dexamethasone (Jakubowiak et al., 2013), or thalidomide and low-dose dexamethasone (Wester et al., 2019), could be a therapeutic opportunity also for NDMM patients, although the outcomes and the related adverse events are still not convincing. Carfilzomib seems to have a distinct pattern of adverse effects with respect to bortezomib. In fact, the rate of peripheral neuropathy is lower than for bortezomib, whereas some patients are affected by cardiovascular complication, such as hypertension and heart failure, rendering the ecocardiography assessment advisable before the onset of treatment. Additionally, unlike bortezomib, carfilzomib can lead to renal failure (Siegel et al., 2013; Chari & Hajje, 2014; Korde et al., 2015; Manasanch & Orlowski, 2017). Adverse events in common with bortezomib are fatigue, anemia, nausea and thrombocytopenia (Siegel et al., 2012). Currently, a great number of clinical studies on carfilzomib are listed in ClinicalTrials.gov. Besides MM, carfilzomib is evaluated in clinical trials for solid tumours, including lung, refractory renal, and metastatic prostate cancers (Table 2 ). However, like bortezomib, its therapeutic potential is limited by the low distribution within the tumour mass, thus requiring very high and toxic doses to elicit a response (Huang et al., 2014; Grigoreva et al., 2015; Johnson, 2015).
Table 2.
Clinical trials with carflizomb and ixazomib in non-haematological cancers.a
| Drug | Combined agent | NCT identifier | Phase | Status | Setting |
|---|---|---|---|---|---|
| Carfilzomib | |||||
| Dexamethasone | NCT02257476 | 1 | Completed | Advanced solid tumours | |
| Cyclophosphamide, etoposide | NCT02512926 | 1 | Recruiting | Pediatric relapèsed and refractory solid tumours | |
| INCB052793, gemcitabine, nab-placlitaxel, dexamethasone, bortezomib, lenalidomide, azacitidine, pomalidomide, INCB050465, INCB039110 | NCT02265510 | 1/2 | Terminated | Advanced solid tumours | |
| Carboplatin, etoposide | NCT01987232 | 1/2 | Completed | Small cell lung cancer | |
| Dexamethasone | NCT00531284 | 1/2 | Completed | Solid tumours, lymphoma, RRMM | |
| / | NCT01775930 | 2 | Completed | Refractory renal cell carcinoma | |
| / | NCT00884312 | 2 | Completed | Solid tumours and MM | |
| Dexamethasone, acyclovir | NCT02047253 | 2 | Completed | Prostate cancer | |
| / | NCT02318784 | 2 | Active non recruiting | Neuroendocrine cancers | |
| Ixazomib | |||||
| Fulvestrant | NCT02384746 | 1 | Terminated | Breast cancer | |
| / | NCT00830869 | 1 | Completed | Advanced non haematological malignancies | |
| / | NCT01830816 | 1 | Completed | Advanced solid tumours and RRMM | |
| / | NCT01912222 | 1 | Completed | Solid tumours and haematological malignancies | |
| / | NCT02630030 | 1 | Completed | Glioblastoma | |
| Ketoconazole, rifampin, clarithromycin | NCT01454076 | 1 | Completed | Advanced non haematological malignancies lymphoma | |
| Vorinostat | NCT02042989 | 1 | Active, non yet recruiting | Advanced cancers | |
| Erlotinib | NCT02942095 | 1 | Active, non yet recruiting | Solid tumours | |
| Selinexor | NCT03880123 | 1 | Recruiting | Advanced sarcoma | |
| Nelfinavir | NCT03422874 | 1 | Withdrawn | Advanced solid tumours and lymphoma | |
| Pegylated (IFN)α2b | NCT02447887 | 1/2 | Terminated | Renal cell carcinoma | |
| Gemcitabine, doxorubicin | NCT02420847 | 1/2 | Active, non yet recruiting | Urothelial cancer | |
| Carboplatin | NCT02993094 | 1/2 | Recruiting | Triple negative breast cancer | |
| Doxorubicin, gemcitabine | NCT03587662 | 2 | Recruiting | Advanced kidney cancer | |
See https://clinicaltrials.gov/, accessed April 27, 2020. (/ no drug).
For what concerns pharmacokinetics, preclinical and clinical studies have shown that carfilzomib has an extremely short half-life of about 12–40 min (Demo et al., 2007; Park et al., 2018; Yang et al., 2011). Furthermore, it displays a rapid systemic clearance (116–123 lLh) and a large distribution volume at steady state (9–28 L) (Alsina et al., 2012; Wang et al., 2013; Papadopoulos et al., 2013). At all doses tested, carfilzomib clearance exceeded hepatic blood flow, envisaging a contribution of extra-hepatic mechanisms, via peptidase cleavage and epoxide hydrolysis, to its overall elimination. Cytochrome P450-mediated metabolism plays a minor role, suggesting that carfilzomib pharmacokinetic profile is poorly altered by administration of CYP inducers or inhibitors. Furthermore, no meaningful differences in carfilzomib pharmacokinetics are detected between patients with normal renal function and those with renal impairment (Badros et al., 2013; Wang, Martin, et al., 2013; Quach et al., 2017). This is of particular relevance, taking into account that renal insufficiency is a common and often severe complication occurring in MM patients.
3.3.2.2.2. Ixazomib
Both bortezomib and carfilzomib require parental administration, and are associated to the development of specific toxic effects (see 3.3.1.1, 3.3.2.1a), mainly peripheral neuropathy and cardiovascular adverse events (carfilzomib) (Moreau et al., 2016; Richardson et al., 2006; Dimopoulus et al., 2010; Richardson et al., 2012; Richardson et al., 2013; Dimopoulos et al., 2017; Waxman et al., 2018). In fact, although the weekly dosing and subcutaneous administration of bortezomib have attenuated the risk of peripheral neuropathy with bortezomib, this adverse effect is still an important concern. Moreover, bortezomib has a limited tissue distribution, due to its slow dissociation rate from red blood cells. Thus, development of an orally available PIs with improved pharmacokinetics and better tolerability profile was required (Gupta et al., 2019). Accordingly, ixazomib (Ninlaro) represents the first orally administered PI approved for clinical use by FDA and EMA. This agent is a reversible inhibitor that preferentially binds the β5 site of the 20S proteasome. In vitro and in vivo preclinical studies reported for ixazomib a therapeutic efficacy greater than for bortezomib in different cancer models, including MM, as well as improved pharmacokinetics, pharmacodynamics, and antitumour activity in xenograft models. In particular, in MM xenograft models, mice treated with ixazomib presented a significant longer survival time than those treated with bortezomib (Chauhan, Catley, et al., 2005; Kupperman et al., 2010; Chauhan et al., 2011; Lee, De la Mota-Peynado, & Roelofs, 2011). In addition, ixazomib induced apoptosis in MM cells resistant to bortezomib without affecting the viability of normal cells, suggesting a potential efficacy in patients with disease relapse after treatment with bortezomib-containing regimens (Chauhan et al., 2011). Therefore, based on the encouraging results observed in these preclincal studies, together with its more convenient oral administration, ixazomib rapidly advanced in phase 1 clinical trials on RRMM to evaluate its safety and tolerability, as single-agent administered once-weekly (NCT00963820) or twice-weekly (NCT00932698) (Kumar et al., 2014 ; Richardson, Baz, et al., 2014). In the once-weekly dosing study, the maximum tolerated dose was determined to be 2.97 mg/m2, whereas in the twice-weekly dosing was 2 mg/m2. Overall, ixazomib was generally well tolerated; no severe neuropathy was reported and most of the observed toxicities were manageable. These studies also indicated that ixazomib absorption is rapid, with a maximum plasma concentration at approximatively 1 h post-dose. After multiple dosing, the terminal half-life was 3.3–7.4 days and 3.6–11.3 days in the once-weekly and twice-weekly regimes, respectively. The ORR were 18% and 15% for once weekly and twice-weekly treatments, respectively, supporting the use of both schedules (Kumar, Bensinger, et al., 2014; Richardson, Baz, et al., 2014). The efficacy of ixazomib as a single agent (5.5 mg/m2 weekly for 3–4 weeks) was confirmed in the first part of a phase 2 trial recruiting 33 patients with relapsed MM who were PI naïve or previously exposed to bortezomib but were not refractory to this agent (NCT01415882) (Kumar et al., 2015). Moreover, in a second phase of this trial the efficacy and tolerability of ixazomib were evaluated in combination with dexamethasone in patients showing lack of adequate response or disease progression. The ORR was 34% and the main toxic effects observed were nausea, thrombocytopenia and fatigue that were in line with the ixazomib toxicity profile (Kumar et al., 2015). The efficacy of the ixazomib (weekly doses of 4 or 5.5 mg/m2) and dexamethasone combination was further investigated in another phase 2 trial recruiting patients with MM that had relapsed after at least 1 previous therapy but not refractory to bortezomib. The results of this study revealed that the ixazomib-dexamethasone doublet had promising efficacy and acceptable tolerability (Richardson et al., 2018) (ORR = 31% and 51% with 4.0 mg and 5.5 mg, respectively). However, the combination with the higher dose of ixazomib was more toxic albeit, indicating the potential requirement of dose reductions to attenuate adverse effects (Kumar et al., 2016).
In preclinical studies ixazomib was shown to exert synergistic effects with lenalidomide, and the results of these studies provided the rationale for the clinical testing of the PI with lenalidomide plus desamethasone (Chauhan et al., 2011). The clinical efficacy and manageability of adverse events reported in early trials (Richardson et al., 2014), were confirmed in the phase 3, randomized, double-blind, placebo-controlled TOURMALINE-MM1 trial (NCT01564537), whose results led to FDA (2015) and EMA (2016) approval of the triplet regimen combining ixazomib with lenalidomide and dexamethasone in MM patients, who had received at least one prior therapy (Moreau et al., 2016). In this trial, 722 patients, who had RMM or RRMM, were randomly assigned to receive ixazomib plus lenalidomide–dexamethasone (ixazomib group) or placebo plus lenalidomide–dexamethasone (placebo group). Interim results demonstrated that the addition of ixazomib significantly prolonged PFS compared to the control group (median 20.6 vs. 14.7 months; HR = 0.74; P = .01). Importantly, a benefit in terms of PFS was observed with ixazomib regimen in all patient subgroups, including subjects with high-risk cytogenetic abnormalities (del(17p), t(4;14), and/or t(14;16)), who are known to be burdened by a very severe prognosis (Avet-Loiseau et al., 2016; Moreau et al., 2016). An arm of the TOURMALINE-MM1 trial also included patients previously treated with PI therapy and thalidomide/ lenalidomide combination; results demonstrated a substantial clinical benefit in terms of prolonged PFS with the ixazomib-lenalidomide/dexamethasone triplet regardless of prior administered therapy (Mateos et al., 2017). A regional expansion of TOURMALINE-MM1 study in China population supported the clinical benefit of the ixazomib-containing triplet therapy, further reporting a significantly increase in OS (ixazomib-lenalidomide-dexamethasone vs placebo-lenalidomide-dexamethasone: median OS 25.8 vs 15.8 months, after median follow-up of 20.2 and 19.1 months, respectively; HR = 0.419; P = .001) (Hou et al., 2017). Importantly, the combination of ixazomib with the lenalidomide-dexamethasone regimen was associated with a limited additional toxicity, and had no adverse impact on patient-reported quality of life. Commonly reported grade ≥ 3 adverse events with ixazomib include gastrointestinal symptoms, rash, thrombocytopenia, and arrhythmia (Hari et al., 2018; Hou et al., 2017; Leleu et al., 2018; Moreau et al., 2016). For what concerns the peripheral neuropathy, this adverse event never exceeded 3% in all studies (Bonnet & Moreau, 2017; Richardson et al., 2018). The low risk of peripheral neuropathy associated with ixazomib use is probably due to the high specificity of ixazomib in inhibiting the chymotrypsin-like (CT-L) site of the proteasome (Muz et al., 2016).
The triplet regimen ixazomib-lenalidomide-dexamethasone followed, when feasible, by single-agent ixazomib as maintenance therapy, was investigated also in patients with NDMM in different trials. In in a phase 1/2 study (NCT01217957), the combination therapy was well tolerated and associated with high ORR (92%) (Kumar et al., 2014). Furthermore, analysis of the long-term efficacy and safety of this regimen, confirmed that ixazomib-lenalidomide-dexamethasone followed by ixazomib maintenance was highly active and caused manageable toxicity in this clinical setting. In particular, out of 65 enrolled patients, 23 patients discontinued induction for SCT, whereas in the remaining 42 patients, the ORR was 80%, including 63% very good partial response and 32% complete responses; these data underscore the feasibility of long-term maintenance treatment with single-agent ixazomib (Kumar et al., 2019). Furthermore, in NDMM patients, a phase 1/2 dose-escalation study investigated the all-oral ixazomib-melphalan-prednisone regimen, followed by single-agent ixazomib maintenance, in transplant ineligible patients with encouraging results (San-Miguel et al., 2018). Recently, the TOURMALINE-MM3 trial (NCT02181413) investigated the ixazomib suitability versus placebo as a maintenance therapy in NDMM to delay disease progression and prolong patients' survival following ASCT. The results of this study revealed that ixazomib induced a 28% reduction in the risk of progression or death compared to placebo (median PFS 26.5 months vs 21.3 months; HR = 0.72; P = .0023), thus representing an additional therapeutic option for these patients (Dimopoulos et al., 2019b). Promising results come also from an ongoing phase 3 trial where ixazomib is administered in patients with RMM as post-ASCT maintenance strategy (Striha et al., 2018).
Several studies are currently investigating the activity of ixazomib in patients with immunoglobulin light chain (AL) amyloidosis, Waldestrom Macroglobulinemia, bone plasmocytome and other non-haematological malignancies (Smith et al., 2015; Smolewski & Rydygier, 2019; ClinicalTrials.gov). Until now the best results were obtained in a phase 1/2 study which evaluated the safety, tolerability, and preliminary efficacy of ixazomib in patients with relapsed/refractory AL amyloidosis, paving the road to a phase 3 study which is currently ongoing (NCT01659658) (Sanchorawala et al., 2017; Smolewski & Rydygier, 2019).
Using all collected clinical data, ixazomib pharmacokinetics was characterized by an absolute oral bioavailability of 58%, terminal long half-life of 9.5 days, large distribution volume of 543 L, and systemic clearance of approximately 1.86 L/h (Gupta, Hanley, et al., 2018; Park et al., 2018; Richardson, Hofmeister, et al., 2018; Gupta et al., 2019). The faster dissociation rate of ixazomib compared to bortezomib, which allows it to associate and dissociate consecutively with more than one proteasome particle, likely contributes to the improved drug distribution into tissues (Kupperman et al., 2010). Moreover, it has been shown that plasma exposure increases linearly with higher administered dose, and no dose adjustment is required on the basis of race, age, sex, body weight, mild-moderate renal impairment, and mild hepatic impairment (Gupta, Zhao, Hui, Esseltine, & Venkatakrishnan, 2015; Gupta et al., 2016; Gupta et al., 2019). At clinical doses, ixazomib is mainly metabolized by non-CYP enzymes; in fact, no significant effect on its pharmacokinetics has been reported after the concomitant administration of CYP3A inhibitors, such as ketoconazole and clarithromycin, in patients with advanced solid tumours and lymphoma (NCT01454076) (Gupta, Hanley, et al., 2018; Gupta et al., 2018). However, the concomitant administration of the CYP3A-inducer rifampin causes a clinically relevant reduction in ixazomib activity, supporting the advice to avoid this combined treatment schedule, and underlying the complexity of ixazomib metabolism (Gupta, Singh, Varshney, & Khan, 2018a; Gupta et al., 2019). In all clinical trials so far described, ixazomib was administered on an empty stomach (Gupta et al., 2019). However, since the absorption and metabolism of an oral drug can change with food, according to the US regulatory guidance (Singh & Malhotra, 2004; United States Food and Drug Administration), a phase 1 study in adult patients with advanced solid tumours or lymphoma was carried out to evaluate whether pharmacokinetics of ixazomib might be altered when administered after a high-calorie, high-fat meal (Gupta, Herzlich, Sauer, & Chan, 2016; Gupta et al., 2019). The results of this study showed that a high-fat meal reduces the rate and extent of absorption of ixazomib, supporting its administration on empty stomach, at least 1 h before or at least 2 h after food intake. These recommendations are inserted in the ixazomib prescribing information (Gupta, Herzlich, et al., 2016; Gupta et al., 2019). Currently, 139 clinical studies on ixazomib are reported in ClinicalTrials.gov, also including the studies on solid tumurs (Table 2).
3.3.2.2.3. Investigational PIs: oprozomib, marizomib and delanzomib
To overcome the clinical limitations of FDA/EMA approved PIs, a number of novel compounds have been identified over the last years. However, the only three drugs currently under evaluation in clinical trials are: oprozomib (PER-047 and ONX 0912); marizomib (NPI-0052, salinosporamide A) and delanzomib (CEP-18770) (Table 3 ).
Table 3.
Clinical trials with oprozomib, marizomib and delazomib.a
| Drug | Combined agent | NCT identifier | Phase | Status | Setting |
|---|---|---|---|---|---|
| Oprozomib | |||||
| / | NCT01129349 | 1 | Completed | Advanced solid tumours | |
| Midazolam | NCT02244112 | 1 | Completed | Advanced malignancies | |
| Dexamethasone, pomalidomide | NCT01999335 | 1 | Completed | MM | |
| Dexamethasone, pomalidomide | NCT02939183 | 1 | Active, not yet recruiting | RRMM | |
| / | NCT01416428 | 1/2 | Completed | Haematological malignancies | |
| Dexamethasone | NCT01832727 | 1/2 | Completed | RRMM | |
| Lenalidomide, dexamethasone, cyclophosphamide | NCT01881789 | 1/2 | Completed | NDMM | |
| Melphalan, prednisone | NCT02072863 | 1/2 | Completed | NDMM | |
| Sorafenib | NCT02227914 | 1/2 | Withdrawn | Haepatocellular carcinoma | |
| Marizomib | |||||
| / | NCT00396864 | 1 | Completed | Advanced solid tumours, lymphoma | |
| Dexamethasone | NCT00629473 | 1 | Completed | Advanced tumours | |
| Vorinostat | NCT00629473 | 1 | Completed | Pancreatic cancer, melanoma, lymphoma, NSCLC | |
| Pomalidomide, dexamethasone | NCT02103335 | 1 | Completed | RRMM | |
| Panobinostat | NCT04341311 | 1 | Not yet recruiting | Pediatric diffuse intrinsic pontine glioma | |
| Temozolomide, radiotherapy | NCT02903069 | 1 | Active, not yet recruiting | Brain cancer | |
| Bevacizumab | NCT02330562 | 1/2 | Active, not yet recruiting | Glioma, glioblastoma | |
| / | NCT00461045 | 2 | Completed | RRMM | |
| / | NCT03727841 | 2 | Not yet recruiting | Ependymoma | |
| Temozolomide, radiotherapy, bevacizumab, lomustine, ABI-009 | NCT03463265 | 2 | Recruiting | Glioblastoma | |
| Temozolomide, radiotherapy | NCT03345095 | 3 | Recruiting | Glioblastoma | |
| Delanzomib | |||||
| / | NCT00572637 | 1 | Completed | Solid tumours and NHL | |
| / | NCT01023880 | 1/2 | Terminated | RRMM | |
| Dexamethasone, lenalidomide | NCT01348919 | 1/2 | Terminated | RRMM | |
See https://clinicaltrials.gov/, accessed April 27, 2020. (no drug)
Oprozomib is an oral drug designed to improve the absorption rate, dosing flexibility, and to overcome two established bortezomib-resistance mechanisms, such as mutations in the proteasome β5 subunit, and drug efflux mediated by ATP-binding cassette transporters (Verbrugge et al., 2012; Zhou et al., 2009). Preclinical studies showed that oprozomib has an antitumour activity comparable to that of carfilzomib, stimulating early clinical investigation (Chauhan et al., 2010; Park et al., 2018). Some phase 1b/2 trials have evaluated oprozomib efficacy and safety profile as single agent or as a component of combined regimens in patients with RRMM. In particular, its combination with dexamethasone provided encouraging results. However, grade ≥ 3 adverse events occurred in approximately 80% of patients and the most common adverse events of any grade were gastrointestinal disorders (up to 84.8%), underscoring the requirement of novel oprozomib formulations to improve its gastrointestinal tolerability (Hari et al., 2019). Recently, based on preclinical studies that have indicated greater efficacy of oprozomib in combination with dexamethasone and pomalidomide compared to oprozomib monotherapy, a phase 1b trial has evaluated this triplet regimen in 31 patients with RRMM (Sanchez et al., 2017; Shah et al., 2019). The results of this study showed encouraging results in terms of efficacy but also confirmed the toxicity profile and the high pharmacokinetic variability of the original bortezomib formulation tested (Shah et al., 2019), stimulating further clinical trials in order to evaluate novel drug formulations with improved gastrointestinal tolerability. Moreover, a phase 1b/2 study where oprozomib was tested as single agent revealed promising results in terms of ORR and tolerability also in patients with Waldenström Macroglobulinemia (Ghobrial et al., 2019). Recently, two multicentre open-label 1b/2 phase studies evaluated three oprozomib-based regimens in transplant-ineligible NDMM patients: oprozomib-dexamethasone plus lenalidomide or cyclophosphamide (oprozomib003) and oprozomib-melphalan-prednisone (oprozomib006) (Hari, Matous, et al., 2019). Although anti-myeloma activity was reported also in NDMM patients, gastrointestinal toxicities and the inter-individual pharmacokinetic variability limit oprozomib efficacy and clinical use, reinforcing the need of novel formulations (Hari et al., 2019). Accordingly, a phase 1b dose-exploration study (NCT02939183) in RRMM is currently ongoing in the attempt to evaluate two new oprozomib formulations (Hari, Matous, et al., 2019; ClinicalTrials.gov). Despite encouraging clinical results in MM and promising data obtained in preclinical models of solid tumours, in a first dose-escalation study in patients with advanced refractory or recurrent NSCLC and colorectal cancer, oprozomib as single agent showed minimal antitumour activity, with clinically relevant gastrointestinal toxicity (Infante et al., 2016; Zang et al., 2012).
The pharmacokinetic profile of oprozomib was first investigated in preclinical models: the drug was rapidly absorbed (2–3 min) in duodenum and jejunum with an estimated absolute oral bioavailability of approximately 39% (Park et al., 2018; Zhou et al., 2009). Moreover, phase 1 studies revealed a plasma half-life of about 1 h and a clearance that exceeded the hepatic blood flow, indicating extra-hepatic contribution to its metabolism. Accordingly, the epoxide hydrolase, which seems to be the primary enzyme involved in oprozomib metabolism, is expressed in many other tissues beyond the liver (Fang et al., 2015; Wang, Chemmama, et al., 2017). Therefore, though developed to improve carfilzomib pharmacokinetics properties, oprozomib still displays a high systemic clearance and a short half-life (Wang, Martin, et al., 2013, Wang, Yang, et al., 2013; Fang et al., 2015).
Marizomib is different from the structural point of view with respect to other PIs, and this translates into a different mechanism of proteasome inhibition, efficacy and toxicity profile (Gozzetti et al., 2017). In vitro and in vivo studies demonstrated that marizomib induces apoptosis in MM and other haematological and solid malignancies with a lower toxicity compared to bortezomib (Ruiz et al., 2006; Potts et al., 2011). Importantly, marizomib induced apoptosis even in tumour cells from MM patients relapsing after various prior therapies including bortezomib and/or thalidomid (Potts et al., 2011; Chauhan, Catley, et al., 2005; Singh et al., 2010). Early phase clinical trials testing different treatment schedules of the PI as single agent in patients with advanced malignancies, reported marizomib activity mainly in patients with RRMM. Remarkably, marizomib did not exhibit severe peripheral neuropathy, warranting further evaluation (Harrison et al., 2016; Levin et al., 2016). The most important adverse events observed in the phase 1 trials were fatigue, nausea, diarrhoea, and infusion site pain (Harrison et al., 2016). In accordance with preclinical models in which marizomib was found to synergistically act with immunomodulatory agents (Chauhan, Singh, Aujay, et al., 2010; Das et al., 2015), a phase 1 clinical trial demonstrated that the triplet combination of marizomib, pomalidomide and low-dose dexamethasone was well tolerated and endowed with promising activity in heavily pre-treated, high-risk RRMM patients, without increasing the incidence of adverse events (Spencer et al., 2018). As mentioned in previous sections, PIs are relatively ineffective in treating solid tumours. Thanks to its more lipophilic structure, an additional differential feature of marizomib compared to other PIs is the ability to cross the blood-brain barrier in different species. Accordingly, preclinical studies demonstrated that oral administration of marizomib inhibits proteasome activity in the brain, and displays a greater activity than bortezomib in a range of solid tumour xenograft models (Potts et al., 2011; Shabaneh et al., 2013; Di et al., 2016). In fact, marizomib was found to induce apoptosis in glioma cells, with minimal toxic effect on normal neurons (Di et al., 2016; Manton et al., 2016). Based on these studies, this PI is currently evaluated in a clinical trial (NCT03345095) for treating newly diagnosed glioblastoma, the most common aggressive malignant primary brain tumour in adults, having a median survival of about 12 months, after debulking surgery and radiotherapy (Weller, Le Rhun, Preusser, Tonn, & Roth, 2019).
The pharmacokinetic profile of intravenously administered marizomib was investigated in a phase 1 clinical trial on patients with advanced solid malignancies, indicating a short half-life (Chauhan, Singh, Ciccarelli., 2010) (lower than 30 min), rapid clearance (0.9–22 L/min), and a large volume of distribution (15–416 L) (Harrison et al., 2016; ). Although the involvement of extra-hepatic clearance in the overall marizomib elimination has been proposed, detailed studies on excretion, metabolism, and in general pharmacokinetic-pharmacodinamic profiles are not available (Potts et al., 2011; Harrison et al., 2016; Park et al., 2018).
Delanzomib is an oral PI that in vitro has shown significant activity on MM and a panel of solid tumours. Furthermore, both intravenous and oral administration resulted in complete tumour regression in MM xenograft models, and increased mice survival in a systemic model of human MM (Piva et al., 2008). Moreover, administration of delanzomib in combination with other conventional anti-MM therapies, such as melphalan plus bortezomib and dexamethasone plus lenalidomide, was more effective than treatment with either agent alone (Sanchez et al., 2010; Sanchez et al., 2012). Nevertheless, the results of early phase trials were not so encouraging (Gallerani et al., 2013; Vogl et al., 2017). In a phase 1 trial, delanzomib showed a linear plasma pharmacokinetic profile, lack of peripheral neuropathy, but a very high incidence of severe skin toxicity in (53% of patients) (Gallerani et al., 2013). In a second multicentre phase 1/2 study delanzomib as single-agent was investigated in patients with RRMM, but no efficacy was reported, whereas severe adverse events, such as rash and thrombocytopenia were reported. Thus, development of delanzomib for myeloma was discontinued (Vogl et al., 2017). To date, it is not clear the rationale of different clinical and preclinical investigations (Park et al., 2018).
3.4. New concepts for proteasome inhibitors
Beside the inhibitors, described in Section 3.3, which switch off directly the enzymatic activity of the proteasome by locking down the active site(s) through a chemical bond with specific residues, new class(es) of molecules is/are emerging, which act(s) instead as modulator(s) of the proteasome activity rather than directly inhibiting it. Obviously, they are not alternative to other inhibitors, which often inhibit more efficiently the proteasome enzymatic action, but they rather affect the UPS activity to a variable extent and fashion, interfering at a different level, such as the interaction with 19S RP and/or conformational changes of the 20S. Their investigation is very recent and obviously almost no pharmacological studies have been carried out on these new class(es), but nonetheless it is very important to report on them because they might represent in the near future a relevant implementation of present therapeutic approaches.
3.4.1. Porphyrins
Porphyrins are an old class of antitumour agents which is now again at the center of renewed scientific interests for the possible role as multifunctional (anticancer) drugs. They are organic heterocyclic macrocycles with an extended π system that on one hand makes them highly hydrophobic and, on the other hand, provides porphyrins with a remarkably high extinction coefficient together with additional photo-physical properties. The latter properties make them well-suited to accomplish, for example, both clinical phototherapy (PDT) and cancer imaging, rendering them suitable to be employed in a multi-tasking role as theranostic tools (Tsolekile, Nelana, & Oluwafemi, 2019). Apparently, their poor aqueous solubility might represent a major restriction to their application in the clinical use. Yet, their very versatile chemistry has allowed to get formulations for topical and systemic treatments thanks to the possibility to synthesize an almost unlimited number of water-soluble derivatives. Easy functionalization of the core (in particular in the meso- position) allows to tune their solubility, aggregation tendency and electronic properties by only choosing the nature, number and reciprocal topology of substituents. In addition to the chemistry related to the periphery, the central core also has a manifold role in determining the steric and physicochemical behavior (e.g., absorption and emission properties) of porphyrins.
It is difficult to overstate the physiological relevance of porphyrins, since their biological role (such as oxygen carrier or storage and redox balance among others) indeed depends on the correct cell localization and on the matching between their structural features and those of the hosting cavity/compartment. For example, anionic porphyrins (e.g., Uro, Copro, PP-IX, with their carboxylates) tend to localize in the acidic compartment of the lysosome. The role of porphyrins (natural or synthetic) and their relevance in affecting biological processes is underlined by the evidence that uncontrolled endogenous porphyrins exposure, caused by hemoproteins release (i.e., ectopic porphyrins) and their subsequent binding to essential proteins may impair protein function, inducing an oxidative damage and altering cellular functions.
In addition to the well-known cellular damage, resulting by reactive oxygen species (ROS), photogenerated by porphyrins catalyzed reactions, several experimental observations have recently remarked porphyrin toxicity in the dark, that seems to be correlated to inhibition phenomena involving HSP90, (Lee, Lee, Lim, Kim, & Kim, 2013) telomerase (Masood et al., 2003), (Cory et al., 2002) and proteasome. Other observations (Szokalska et al., 2009) demonstrate that the cytotoxic effects, caused by a porphyrin photosensitizer in PDT, could be potentiated through inhibition of proteasome, encouraging to screen the ability of porphyrins to inhibit the proteasome activity (Chauhan, et al., 2005; Chauhan et al., 2010).
The first evidence that porphyrins, in particular hemin, could reduce “the protein degradation ATP dependent”, not yet known as proteasome, dates back to 40 years ago (Etlinger & Goldberg, 1980). Afterwards, many reports showed that hemin effect on ubiquitin-dependent proteolysis is not restricted to erythroid cells but hemin is a negative UPS modulator in all eukaryotic cells (Haas & Rose, 1981; Vierstra & Sullivan, 1988).
The first molecular investigation on purified 20S has demonstrated that micromolar amounts of cationic porphyrins inhibit reversibly all three main protease activities of proteasome (Santoro et al., 2012). Quite interestingly, porphyrins activity is finely modulated (tuned) both by the nature and reciprocal topology of peripheral substituents and by the stereochemistry of the macrocyclic ring center. Thus, the inhibitory efficiency of the cationic macrocycles increases with the number of positive substituents in the meso position. As far as the porphyrin core is concerned, it is evident from the experimental data that among the various metallo-derivatives, the most active ones are those with no axial ligands, the activity decreasing going from penta- to hexacoordinated metals. In particular, the naked cationic porphyrins are the most active ones, indicating that the molecule should be flat in order to interact effectively with the proteasome. Interestingly, thanks to their high extinction coefficient these molecules are “visible inhibitors”, and in this sense they behave as very efficient spectroscopic “probes” for UV–Vis stopped-flow kinetic analysis. The latter studies, combined with NMR and computational study, helped in defining the tetra-cationic H2T4 as a competitive inhibitor which binds the gate area on the α-ring, hindering the substrate access to catalytic chamber (Santoro et al., 2016).
Starting from the first evidence, an accurate kinetic and computational analysis of the surface of the α-subunit ring revealed then that the positive charges play a critical role in the inhibition of the 20S, showing that cationic porphyrins may act as tuneable gatekeepers of the 20S (see Fig. 9 in Santoro, Cunsolo, et al., 2016). Indeed, the α-ring represents a receptor-like region physiologically involved in ionic interactions with canonical RPs; Ss a matter of fact, the regular arrangement of aminoacidic residues in these surfaces has been found to represent a sort of electrostatic code, exploited by the 19S, and regulating the gating phenomena (see 2.2.1, 2.3.1). The charges of porphyrins represent the keys able to interfere with this “electrostatic code”, and, depending on their spatial distribution, a high variety of binding modes and inhibition mechanisms have been observed (Di Dato et al., 2017). Furthermore, some functional effects, characterized by cooperative phenomena, are the resulting of conformational rearrangements that can reverberate onto the β5 subunit (Arba et al., 2018). Finally, additional binding modes involve interactions with both the α- and β-rings regions, acting directly on the β5 catalytic subunit.
In conclusion, porphyrins are excellent candidates for multi-tasking biological active molecules. As an example, quite recently it was shown (Vallelian et al., 2015) that high levels of intracellular heme disrupt cellular homeostasis through the combined activities of oxidative damage and proteasome inhibition, thus resulting in the accumulation of damaged proteins that contribute to the triggering of cell death. As far as it concerns their interactions with proteasome, porphyrins can either induce a partial competitive occlusion by hindering the substrate access into the catalytic chamber or else behave as an allosteric modulator, regulating the occurrence of conformational change(s) that affect(s) the dynamic equilibrium between the open and the closed state of the proteasome gates.
3.4.2. Metal complexes
The use of metal complexes as anticancer drugs has been adopted since early 1960s with the discovery and development of cisplatin and its derivatives (Alderden, Hall, & Hambley, 2006). The first metal complexes used as anticancer agents were designed to interact with the cancerous cell DNA, inducing apoptosis of cancer cells, but it has been also demonstrated that they can alter the cellular redox chemistry through binding either the metal or other ligand redox centers of biomolecules involved in cellular redox pathways. Since tumour cells have a more reducing environment than normal cells, due to the accelerated metabolic activity, high rates of cell growth and proliferation, selectivity can be reached by using metal complexes which contain redox active metal ions. These are then reduced in the reducing environment of the cancer cell and metal complex drugs become activated. Therefore, the reduced metal ion (Co2+, Pt2+, Fe2+, Cu+, etc.) exerts its anticancer activity with a marked selectivity for tumour cells, as the unique ability of metal complexes to undergo redox activation processes involve both metal and ligand redox centers and it can be tuned to specific potentials ( Zhang & Sadler, 2017 ). However, the idea of using metal complexes to disrupt proteasome activity in order to have an anticancer effect is relatively recent (Shagufta & Ahmad, 2020). Disulfiram was the first metal complex containing copper capable of inducing apoptosis in cancer cells by inhibiting proteasome activity (Chen, Cui, Yang, & Dou, 2006). It was later demonstrated that disulfiram rapidly converts in vivo to its reduced metabolite diethyldithiocarbamate (DDTC) before exerting its anticancer activity (Pang, Chen, Cui, & Ping Dou, 2007). Interestingly, in most cases the presence of the metal ion turned out to be fundamental to have proteasome activity inhibition, since the non-metallated ligand has been demonstrated to be ineffective for this purpose. For example, the asymmetric ligands, containing pyridine and 4,6-substituted phenol moieties alone, do not have any influence on proteasome activity (Shakya, Peng, Liu, Heeg, & Verani, 2006); conversely, the copper (II) chloride salt of this compound inhibits the proteasome activity in cell free conditions and, for this reason, it has been proposed that the copper complex works as a carrier to cross the cell membrane. Such an assumption would imply that the effectiveness of the metal complex should strongly depend on the metal-ligand species formed and, once inside the cell, the copper shuttling complex should induce proteasome inhibition by releasing the copper ions, which become available to coordinate with proteasome, involving amino acids capable of forming Cu—N, Cu—S, or Cu—O bonds (Hindo et al., 2009). However, elucidating the molecular mechanisms by which copper ions are able to inhibit proteasome activity is a very challenging task, due to the very complex cellular environment and the difficulty in monitoring the fate of intracellular copper ions (Satriano et al., 2013). Indeed, it has been reported in cell-free conditions that Cu (II) ions promote conformational changes associated to an impaired channel gating, without catalyzing redox reactions nor disrupting the assembly of the 20S proteasome (Santoro et al., 2016). On the contrary, HeLa cells, grown in a Cu (II)-supplemented medium, exhibit a decreased proteasome activity, which was then restored in the presence of an antioxidant. For this reason, it has been proposed that, although the Cu(II)-inhibited 20S activities may be associated to proteasome conformational changes, favouring the closed state of the core particle, other effects may occur, such as ROS-mediated proteasome flooding and disassembly of the 26S proteasome into 20S and 19S.
Beside copper, other metal ions have been used in complex with various ligands to inhibit proteasome activity. For example, cadmium, though carcinogenic for humans, has been tested in complex with several organic ligands, such as indole-3-butyrric acid and indole-3-propionic acid. Strikingly, proteasomal inhibition, as well as accumulation of ubiquitinated proteins and induction of apoptosis, were observed in MDA MB 231 breast cancer cells, whereas non-tumourigenic breast MCF10A cells were much less sensitive to the cadmium complexes, indicating cell-specific apoptotic death (Zhang et al., 2013). Cadmium complexes with heterocycle-L-tryptophan Schiff base ligands such as 2-acetylpyrazine-L-tryptophan,5-methylfurfural-L-tryptophan and 5-bromo-2-thiophenecarbaldehyde-L-tryptophan have also been synthesized and tested for cancer specific proteasome inhibitory and apoptosis -inducing activities. Results show that the inhibition of the proteasomal CT-L activity is strongly depending on the ligand; thus, while the Cd complex with 2-acetylpyrazine-L-tryptophan and 5-methylfurfural-L-tryptophan were powerful inhibitors, the use of 5-bromo-2-thiophenecarbaldehyde-L-tryptophan as ligand produced an inactive complex (Zhang, Li, Huang, Guan, & Zhu, 2017).
Manganese and gold complexes have also been investigated for their inhibitory activity on proteasome in cancer. In particular, the cefepime-Mn complex has been demonstrated to inhibit the proteasomal CT-L activity and to induce the apoptosis of breast cancer cells in a dose-and time-dependent manner (Zhang, Schulz, et al., 2015).
Gold(III) and gold(I) dithiocarbamate complexes have also been reported to be strong proteasomal CT-L activity inhibitors with IC50 values ~1.1 μM. Interestingly, the different oxidation states of the gold ion seem to affect the mechanism of inhibition, as only the higher gold oxidation state has been reported to produce significant levels of ROS (Zhang et al., 2010). Several additional gold complexes have been reported to have IC50 in the μM range toward all three main proteasome catalytic activities, such as mononuclear gold, dinuclear(III) complexes and gold(I) phosphine complexes, whereas aurofin, an established gold(I) drug (currently in clinical use for the treatment of rheumatoid arthritis), has been demonstrated to be completely inactive in the modulation of proteasome (Micale et al., 2014). The gold(III) complex AuL12 (dibromo[ethyl-N-(dithiocarboxy-kS,kS’)-N-methylglycinate])has shown attractive properties in terms of anticancer activity and toxicity. AuL12 was found to inhibit proteasome activity in living cells with an efficiency comparable to that of bortezomib (Tomasello et al., 2017). Furthermore, AuL12 also inhibits Lys48-linked poly-ubiquitination in vitro at a concentration of about 7 μM, interfering with Ub activation reactions catalyzed by E1 enzymes. Another approach, based on the use of metal complexes to inhibit the function of the UPS system in cancerous cells, is to target deubiquitinases instead of the proteasome. As an example, nickel, as well as gold(I), pyrithione complexes were tested for their anticancer activity and it was found that both complexes are able to inhibit the UPS by targeting the 19S-associated deubiquitinases without directly affecting proteasome activity (Li et al., 2019; Zhao et al., 2016).
Finally, it is worth mentioning that metal ions, such as copper, have a strong inhibitory effect also on other different enzymes (Grasso et al., 2011, Grasso et al., 2012), which have been found to be associated with the proteasome and to be able to modulate its activity (Sbardella et al., 2015; Sbardella et al., 2018). For this reason, the use of metal complexes to modulate proteasome activity should consider all possible mechanisms and actors involved in the UPS system, including the interaction of the metal complexes with regulatory proteins, such as IDE (Tundo et al., 2017).
4. Proteasome alteration in neurodegeneration
Although proteins are generally found in the right folded and functional state in healthy cells, unfolded configurations are present, mostly occurring upon exposure to environmental stressors; furthermore, they may originate from multi-faceted alterations in translation, folding and intracellular trafficking. Under physiological conditions, the misfolded variants of proteins are either: (i) tagged for degradation via UPS or autophagy pathways, or (ii) correctly refolded back to the native state by chaperones, or else (iii) sequestered into intracellular compartments, such as aggresomes, which preserve them for the following refolding or degradation (Hartl & Hayer-Hartl, 2009; Chen, Retzlaff, et al., 2011; Escusa-Toret, Vonk, & Frydman, 2013; Hipp, Park, & Hartl, 2014; Sontag, Vonk, & Frydman, 2014; Sweeney et al., 2017). Misfolded proteins can aggregate to form high-molecular weight species of different nature, such as soluble oligomers, prefibrillar species and highly ordered amyloid structure, which often consist of different aggregated-prone and normally folded proteins (Olzscha et al., 2011; Hong, Han, Fink, & Uversky, 2011; Brettschneider, Del Tredici, Lee, & Trojanowski, 2015; Sweeney et al., 2017; Olzscha, 2019). Although there is compelling evidence that many proteins (if not all) can form amyloid-like structures under stressful conditions, nonetheless disease-associated amyloidogenic proteins are characterized by intrinsic structurally disordered elements in their free soluble form (Guijarro, Sunde, Jones, Campbell, & Dobson, 1998; Tzotzos & Doig, 2010). Thus, a common hallmark of neurodegenerative diseases is the accumulation of misfolded protein aggregates into affected tissues, leading to a derangement of PN, and, ultimately, to progressive death of neurons (Bredesen, Rao, & Mehlen, 2006; Goedert, Clavaguera, & Tolnay, 2010; McAlary, Plotkin, & Cashman, 2019; Chiti & Dobson, 2017). In general, the major component of insoluble deposits is a specific-disease related protein, such as β-amyloid and tau in Alzheimer's disease (AD), α-synuclein in Parkinson's disease (PD), and huntingtin in Huntington Disease (HD), even though overlapping similarities between syndromes are reported (Jellinger, 2012; McAlary et al., 2019).
Neurons, as well as each post-mitotic cell, are very susceptible to proteostasis unbalance mainly due to their long lifespan, morphology and enhanced metabolism (Tai & Schuman, 2008). In particular, UPS is crucial in synaptic protein turnover, calcium flux dynamics, long-terminal plasticity and memory (Bingol & Schuman, 2004; Bingol & Schuman, 2006; Colledge et al., 2003; Djakovic et al., 2012; Djakovic, Schwarz, Barylko, DeMartino, & Patrick, 2009; Fonseca, Vabulas, Hartl, Bonhoeffer, & Nägerl, 2006; Guo & Wang, 2007; Lopez-Salon, Alonso, Vianna, Viola, & Mello e Souza, Izquierdo, Pasquini, & Medina, 2001; Tai & Schuman, 2008; Wu et al., 2009). Furthermore, besides the intracellular proteasome, a membrane-associated proteasome complex, specific for the nervous system, has been recently discovered (Ramachandran & Margolis, 2017). This complex seems to be involved in the modulation of neuronal function by degrading intracellular proteins into peptides that are then released into synaptic cleft, where they stimulate postsynaptic N-methyl-d-aspartate neuronal signalling (Ramachandran & Margolis, 2017). Impaired proteasome activity is reported in idiopathic neurodegenerative diseases, and some hereditary form of neurodegeneration is due to mutations in UPS components, such as PARK1 and PINK (see next paragraphs) (Ciechanover & Brundin, 2003; McKinnon & Tabrizi, 2014; Ortega, Díaz-Hernández, & Lucas, 2007; Ortega & Lucas, 2014; Thibaudeau & Smith, 2019). A wide range of studies support the notion that a decrease of proteasome activity with age positively correlates with misfolded protein accumulation. This culminates then, in the presence of other pathological stressors at which aged people are exposed, with the progressive development of neurodegeneration (Smith, 2018; Vigouroux, Briand, & Briand, 2004; Mattson & Magnus, 2006; Chondrogianni & Gonos, 2010; Zabel et al., 2010; Tomaru et al., 2012). Accordingly, targeted proteasome inhibition in brain of animal models of neurodegeneration reproduces some clinical and neuropathological signatures of human diseases (McNaught et al., 2002; McNaught, Perl, Brownell, & Olanow, 2004; Ciechanover & Brundin, 2003; Bedford et al., 2008; Li et al., 2010). Despite this evidence, it is still unclear whether reduced proteasome functionality is a primary event in neurodegeneration onset or the consequence of misfolded protein aggregation (Ciechanover & Brundin, 2003; Dantuma & Bott, 2014; Ortega & Lucas, 2014; Thibaudeau & Smith, 2019). Soluble oligomers, which are believed to be the most toxic and pathologically significant species among the different forms of aggregates, have been shown to impair proteasome activity (Cecarini et al., 2008; Tseng, Green, Chan, Blurton-Jones, & LaFerla, 2008; Díaz-Hernández et al., 2006; Bence, Sampat, & Kopito, 2001; Deriziotis et al., 2011; Dantuma & Bott, 2014). Recently, a common mechanism has been proposed, according to which oligomers of different diseases-related proteins inhibit the 20S proteasome activity through an allosteric-driven interaction (Smith, 2018; Thibaudeau, Anderson, & Smith, 2018; Ciehanover, Hod, & Hershko, 1978). Specifically, the soluble oligomers, which adopt a similar three-dimensional conformation, were reported to bind the 20S, stabilizing the gate in the closed configuration. Oligomers-induced proteasome impairment seems then to be counteracted by HbYX peptides, which mimic the gate physiological opening induced by HbYX motifs of 19S ATPase (see also Section 2.3.2) (Thibaudeau et al., 2018). Accordingly, α3Δn-HEK293 cells exhibit increased degradation of proteasome substrates, including neurodegenerative disease-related proteins (see also Section 2.3.2) (Choi et al., 2016). These results support the scientific hypothesis that drugs, which directly open the 20S gate, might have a relevant therapeutic potential in the management of neurodegenerations (Smith, 2018; Thibaudeau et al., 2018; Thibaudeau & Smith, 2019; VerPlank, Lokireddy, Zhao, & Goldberg, 2019). In this respect, it should be pointed out that the action of porphyrins (see Section 3.4.1) looks very promising (Di Di Dato et al., 2017).
However, although it was generally accepted that, once formed, amyloid aggregates are resistant to proteasome degradation (Sweeney et al., 2017), it has been recently reported that proteasome holoenzyme seems to possess a “fibril-fragmenting activity”, being able to reduce the size of large tau and α-synuclein fibrils into smaller entities in vitro, thus opening a novel perspective in understanding proteasome role in neurodegeneration (Cliffe et al., 2019; Ye, Klenerman, & Finley, 2020). In the rest of this chapter, the contribution of proteasome to the onset and progression of main neurodegenerative diseases is reported. Additionally, we will focus on strategies developed so far to enhance proteasome activity.
4.1. Alzheimer's disease
In the late 1901, the German neuropathologist Alois Alzheimer reported about the presence of amyloid plaques and neurofibrillary tangles (NFTs) in the brain of a woman suffering from progressive cognitive decline (Stelzma, Schnitzlein, & Murllagh, 1995). This was the very first paper reporting a case of senile dementia, a neurodegenerative disease that will be later commonly recognized as Alzheimer's Disease (AD). Almost 80 years after this ground-breaking report, protein aggregates present in amyloid senile plaques (i.e. amyloid peptides) (Glenner & Wong, 1984) and NFTs (i.e., hyperphosphorylated tau) (Lee, Balin, Otvos, & Trojanowski, 1991) were fully characterized. However, only over the last two decades, attention in the area of protein aggregation has increased considerably, transforming it into a key subject of study in diverse research areas ranging from chemistry to biology and medicine. The most important reason for the rising attention in this field is that most of the disorders are associated with amyloid aggregation (Chiti & Dobson, 2006) and neurodegeneration, which are becoming more and more expensive in terms of health care and social cost worldwide (Alzheimer's Disease International, 2010; Chiti & Dobson, 2017; Ciehanover et al., 1978). After an initial enthusiasm in targeting amyloid protein aggregation as a possible therapeutic approach to treat AD, other protective mechanisms associated with properties of the cellular environment, such as the existence of molecular chaperones and degradation mechanisms have attracted increasing attention (Hartl, Bracher, & Hayer-Hartl, 2011; Morimoto, 2008). Substantial shreds of evidence point to UPS malfunction as an important factor playing a key role in Aβ amyloid growth and AD pathogenesis. This is not surprising if one bears in mind that UPS surveillance is needed for a tightly regulated maintenance of all proteome components involved in memory formation, as well as synaptic plasticity and functioning (Djakovic et al., 2012; Lopez-Salon et al., 2001; Tai & Schuman, 2008).
However, a deeper understanding of all components of the proteome quality control network is needed to allow us to envisage a successful regulation of all pathogenic pathways. As an example, it is critical to single out all the key components of the UPS, including the upstream processes, involved in AD pathogenesis to allow the design of small molecules with higher efficacy and less severe side effects (Cao, Zhong, Toro, Zhang, & Cai, 2019). In particular, proteasome function (if compared to age-matched controls) declines in AD brains, whereas other proteasome isoforms, such as immunoproteasome, are overexpressed in astrocytes (Keller, Hanni, & Markesbery, 2000; Nijholt et al., 2011). As a matter of fact, several studies underscored that immunoproteasome is upregulated in glial cells surrounding Aβ plaques present in affected brains (Yeo et al., 2019). The same work reported that YU102, a specific PI, abolished the production of inflammation cytokines from glial cells and improved cognitive performance in AD mice without any evident effect on Aβ plaques deposition. Hence, the proteasome is now emerging as a major target in the treatment of memory loss and cognitive impairment in AD (Al Mamun et al., 2020; Hegde, Smith, Duke, Pourquoi, & Vaz, 2019). A very recent study, involving 48 CEpatients and 50 healthy volunteers, has clearly shown that proteasome levels are significantly decreased in erythrocytes of patients affected by AD. Moreover, the same study revealed that ubiquitin is overexpressed in red blood cells of AD individuals, thus suggesting that (i) the UPS is heavily involved in the pathogenesis of the disease and (ii) both Ub and proteasome may be investigated as AD biomarkers (Lv et al., 2020). The role of proteasome inactivation in AD development, was addressed in vivo by studies using transgenic mice. In particular, APPswe/PS1de9 AD mice were crossed with mice expressing a green fluorescent protein (GFP) fused to a degradation signal (CL-1) targeted by the proteasome; these studies revealed that GFP protein-linked proteasome substrates build up in the hippocampus and cortex of AD mice at 4 weeks of age, and they were also confirmed by a concomitant accumulation of p53, an endogenous proteasome substrate, and of poly-ubiquitinated proteins. Altogether, these results suggest that the proteasome function is altered in AD mice even at a very young age, well before cognitive impairment and amyloid fibril deposition (Liu, Fung, Chong, Shukla, & Hilgenfeld, 2014).
Although it is known that Aβ is a proteasome substrate, it may also, in turn, inhibit 20S peptidase activity; in this respect, scanning transmission electron microscopy (SEM) experiments have shown that Aβ peptide binds the 20S and inhibits its proteolytic activity (Gregori, Hainfeld, Simon, & Goldgaber, 1997). These findings also reconcile with experiments outlining that neuronal cells incubated with amyloid Aβ peptides do exhibit inhibitory effect on proteasome activity (Cecarini et al., 2008; Lopez Salon, Pasquini, Besio Moreno, Pasquini, & Soto, 2003; Tseng et al., 2008). Impaired tau metabolism has been also associated with abnormal UPS activity; thus, tau may be ubiquitinated at diverse sites and UPS impairment is involved in tauopathies (Cripps et al., 2006; David et al., 2002; Han et al., 2014; Keck, Nitsch, Grune, & Ullrich, 2003; Lee, Lee, & Rubinsztein, 2013; Metcalfe, Huang, & Figueiredo-Pereira, 2012; Morris et al., 2015; Tai et al., 2012; Thomas, Cripps, & Yang, 2009). Hyperphosphorylated tau oligomers build up at synaptic and postsynaptic junctions in AD, (Tai et al., 2012) and tau insoluble assemblies inhibit proteasome activity, leading to an accumulation of poly-ubiquitinated proteins (Myeku et al., 2016). It has also been shown that CHIP (C-terminus of Hsp70-interacting proteins) E3 ligase, is overexpressed in AD patients and its downregulation brings about the accumulation of ubiquitinated tau proteins (Dickey et al., 2007; Petrucelli et al., 2004; Shimura, Schwartz, Gygi, & Kosik, 2004). Recently, it has been demonstrated that toxic Aβ oligomeric assemblies may bind proteasome, impairing its activity by interfering with gating phenomena. Moreover, as mentioned previously, other different proteins (e.g., α-synuclein and huntingtin), known to self-assemble into similar 3D structures, have the potential to inhibit 20S activity by a similar mechanism, thus pointing to a general oligomer-driven model of proteasome inhibition (Thibaudeau et al., 2018). Notably, clearance of full-length monomeric tau was ATP-independent, whereas on the contrary, fibrillar tau hydrolysis was strictly related to the ATPase activity of the proteasome (Cliffe et al., 2019).
Neurotoxic 30-mer Aβ assemblies (termed amylo-spheroids) are present in AD brains, showing to be the main responsible of neuronal cells death. Although it is still unclear how amylo-spheroids form in the brain and activate neurodegenerative processes, proteasome inhibition was shown to dramatically promote their accumulation in the trans-Golgi network of excitatory neurons, altering dendritic transport (Komura et al., 2019).
Besides Aβ/tau aggregation in toxic oligomers, other causes are known to come into play for proteasome impairment in AD, including abnormal generation of ROS (de Vrij, Fischer, van Leeuwen, & Hol, 2004). Indeed, high intracellular levels of the redox-active metal ions Cu(II) are known to be associated to both Aβ and tau pathologies, even though it is known that only Cu(I) is internalized into the cells by the plasma transporter Ctr1(Maryon, Molloy, Zimnicka, & Kaplan, 2007). In this respect, it should be outlined that Cu(II) ions are present at relatively high concentration in the brain and their levels are known to increase with age (Morita, Kimura, & Itokawa, 1994; Tarohda, Yamamoto, & Amamo, 2004). As a matter of fact, Cu(II) ions are also found in amyloid plaques present in the AD brain (Lovell, Robertson, Teesdale, Campbell, & Markesbery, 1998; Suh et al., 2000) co-purifying with Aβ from AD brain tissues (Opazo et al., 2002). Intriguingly, several Cu(II) complexes exhibit remarkable proteasome inhibitory capacities (Daniel, Gupta, Harbach, Guida, & Dou, 2004; Marzano, Pellei, Tisato, & Santini, 2009). All these findings reconcile with recent reports that demonstrate that Cu (II) ions inhibit all proteolytic activities of the 20S (Grasso et al., 2017; Santoro, Monaco, et al., 2016; Bellia et al., 2019).
Human neuroblastoma cell lines SH-SY5Y, differentiated with retinoic acid, reproduce the neuronal morphology and function and are for this reason widely employed to mimic primary neurons. Low doses of MG-115, a known PI, brought about tau hyperphosphorylation, microtubule destabilization, and an impaired dendritic development in SH-SY5Y cell lines. Moreover, an inhibited proteasome activity increased the levels of signal proteins involved in AD pathogenesis, such as c-Jun N-terminal kinase, c-Jun and extracellular signal-regulated protein kinase (ERK). Notably, in the same study it came out evident that an inhibition of c-Jun and ERK was able to nullify the effects of proteasome inhibition. These results suggest that a reduced proteasome activity may induce an activation of c-jun/ERK signalling and, in turn, a cascade of adverse events leading to neuronal death (Agholme et al., 2014).
Indirect proteasome inactivation may also play a significant role in AD development. For example, ubiquitin hydrolase (Uch-L1) is known to raise cytosolic mono-ubiquitin levels, thus enhancing protein degradation by the proteasome; not surprisingly, low Uch-L1 levels are correlated with amyloid accumulation. Furthermore, a neuron F-box protein Fbx2 may act as an E3 ligase, facilitating the ubiquitination of the β-secretase and its degradation by the proteasome, leading to a decrease of amyloid Aβ generation; notably, both Fbx2 and Uch-L1 increase synaptic plasticity and memory function in AD mice (Gong, Radulovic, Figueiredo-Pereira, & Cardozo, 2016). It was also reported that UBB+1, a ubiquitin form arising from a pathogenic mutation in the Ub gene through addiction of 19 residues to the C-terminus of the protein, inhibits proteasome activity and is present in neurofibrillary tangles of AD patients (Fischer et al., 2003). Proteasome activation by exogenous agents may represent a promising strategy for AD therapy; in fact, feeding wild-type Caenorhabditis elegans with 18α-glycyrrhetinic acid (18α-GA), a known proteasome activator, resulted in an increased proteasome activitiy and increased lifespan of worms. Moreover, higher proteasome activity was related to lower paralysis rates in C. elegans AD models. Notably, analogous promising results were confirmed also when murine and human neuronal cells were treated with 18α-GA (Papaevgeniou et al., 2016).
A defective proteasome activity has been also related to increased levels of the APP-derived intracellular C-terminal membrane fragment β (CTFβ), a neurotoxic peptide with significant roles in AD pathogenesis (Bustamante et al., 2018).
4.2. Parkinson's disease
Parkinson's Disease (PD) is a multi-factorial neurodegenerative disease, which primarily affects the nigro-striatal dopaminergic motor neurons. The pathognomonic histological lesion of PD, though not shared by all disease variants, is the formation of peri-nuclear intracellular aggregates, called Lewy Bodies (LB) (extremely rich in a proto-typical amyloidogenic protein, i.e. α-synuclein, and ubiquitinated proteins) through a process referred to as aggresome formation. It is still debated whether aggresome formation is a pathogenic event in PD progression or it rather represents a protective pathway activated to limit the high toxicity of the soluble amyloidogenic oligomers, which chronologically precedes aggregates formation (Raiss et al., 2016; Wakabayashi et al., 2013; Wakabayashi, Tanji, Mori, & Takahashi, 2007). In support of this second hypothesis, which is the prevalent one, aggresomes formation is a phenomenon orchestrated by several proteins (including HSP10, p62, HDAC6, GRB1, NUB1, c-Abl among the others), which either selectively bind α-synuclein or modulate its solubility and interacting properties through insertion of post-translational modifications, such as phosphorylation (further discussed below) (Anisimov et al., 2019; Mahul-Mellier et al., 2014; Takahashi et al., 2018; Tanji et al., 2010). Finally, under healthy conditions an extensive crosstalk between the UPS and autophagy is expected to tightly balance aggresome growth and to avoid the protein imbalance of neurons (Yang et al., 2013). According to this PD pathogenesis framework, inherited and acquired conditions which predispose to the disease onset would impair this sophisticated mechanism of proteostasis maintenance.
Although very recently it has been reported that Ub-positive inclusions in LB mostly deal with K63 (i.e., autophagy-related) rather than UPS-specific K48 linkages, extensive studies on the genetically inherited familiar variants and idiopathic forms of the disease indicate that PD is probably the neurodegenerative disorder displaying the highest degree of association with a multi-faceted dysregulation of E3-ligases and of proteasome proteolytic activities early at disease onset (Bentea, Verbruggen, & Massie, 2017; Krebiehl et al., 2010; Savolainen, Albert, Airavaara, & Myöhänen, 2017; Ugras et al., 2018).
Remarkably, although α-synuclein is a natural substrate of 20S proteasome, widely used to monitor its proteolytic activity under different experimental conditions, structural conformational changes of α-synuclein have been reported to transform it in an inhibitor of the 20S proteolytic activity (Dächsel et al., 2005; Zondler et al., 2017). On the other hand, since IDE was shown to halt in vitro the oligomerisation of α-synuclein, as well as that of other amyloidogenic substrates (Sharma et al., 2015; Sharma, Chorell, & Wittung-Stafshede, 2015; de Tullio et al., 2013), an important still unexplored aspect is the role of IDE and, possibly, of IDE::20S complexes; thus, interaction of the enzyme with the 20S might stimulate further information on the functionality of these pathways in fighting the proteotoxic stress in PD.
By analogy with other neurodegenerative disorders, the metabolic dysregulation, which stimulates neuron loss in PD, deals with redox imbalance, proteotoxicity and metalions dys-homeostasis (Bentea et al., 2017; Zondler et al., 2017; Kumar et al., 2018; Le, 2014; Janda, Isidoro, Carresi, & Mollace, 2012). Nonetheless, extensive redox imbalance chemically induced by rotenone and paraquat (i.e, mitochondrial respiratory chain inhibitors), leads to the development of specific forms of parkinsonism.
Interestingly, α-synuclein in LB is often phosphorylated at Ser129, a post-translational modification which reduces the aggregating propensity of the protein; impairment of the phosphorylation at Ser129, brough about by redox nitrosylation of neighbouring Tyr residues together with di-tyrosine cross-linking, especially at the C-terminus of the protein, induces pathological structural alterations of the protein (Kleinknecht et al., 2016).
Furthermore, exceedingly high concentrations of metal ions, such as Zn2+, Cu2+ and iron have been long detected in post-mortem brain biopsies (Le, 2014); in particular, in murine models it has been observed that, besides metal-related toxicity, Zn2+ ions also trigger the expression and oligomerization-propensity of α-synuclein in nigrostriatal tissues and the selective loss of specific proteasome subunits, such as β5 and Rpt6 (Kumar et al., 2018). Additionally, copper metabolism appears to be implicated in inducing cell death, since over-expression of α-synuclein and copper transporters stimulated loss of proteasome function, regardless of the tendency to form aggregates (Anandhan et al., 2015; Lan, Chen, Chai, & Hu, 2016).
A bulk of molecular evidences on PD pathogenesis comes from studies on genetic inheritance of the disease. Mutations of at least six genes have been linked with hereditary PD, namely α-synuclein (SNCA or PARK1), Parkin (PARK2), ubiquitin carboxyhydroxylase L1 (UCH-L1 or PARK5), PTEN-induced putative kinase 1 (PINK-1 or PARK6), DJ-1 (PARK7), and leucine-rich repeat kinase 2 (LRRK2 or PARK8) (Janda et al., 2012; Nuytemans, Theuns, Cruts, & Van Broeckhoven, 2010). Interestingly, all proteins, encoded by these genes, were found to interact at some level with proteasome or ubiquitin-conjugating enzymes.
In particular, DJ-1 is an intracellular protein with pleiotropic activities which encompass cell morphology, functionality of mitochondria and ROS balance (Irrcher et al., 2010); most notably, it has been shown to positively regulate the transcriptional activity of Nrf-2 upon inactivation of PTEN and activation of PI3kinase/Akt/mTOR pathway. Nrf2 is a master regulator of anti-oxidant defense systems including transcription of proteasome genes (Niki et al., 2020). A tight link between DJ-1 and Akt/mTOR pathway has been reported also in Drosophila melanogaster, underscoring the degree of conservation across evolution of such a relevant pathway for neuron homeostasis (Yang et al., 2005). Interestingly, detection of hyper-stimulated autophagy in DJ-1 deficient neurons is another indirect proof of the pivotal role of DJ-1 in stimulating the Akt/mTOR signalling, since this kinase is the major autophagy inhibitor. In this framework, it looks relevant to clarify the potentially controversial inhibitory activity of DJ-1 on the 20S catalytic activity both on synthetic and natural substrates, recently described (Moscovitz et al., 2015). However, the authors propose that DJ-1 up-regulates the expression of proteasome subunits whilst repressing the catalytic activity of 20S assemblies, thus finely tuning the 20S proteasome bulk proteolytic activity. This activity would be necessary to address the cell need in balancing the clearance of oxidatively damaged proteins and that of native intrinsically unstructured proteins, which coordinate regulatory and signalling events (Moscovitz et al., 2015).
Another gene, critically involved in PD, is LKKR2 and the clinical features of LRRK2-PD are often indistinguishable from idiopathic PD, with accumulation of α-synuclein and/or tau and/or ubiquitin in intraneuronal aggregates (Lichtenberg, Mansilla, Zecchini, Fleming, & Rubinsztein, 2011). Although the mechanisms through which LKKR2 mediates toxicity are unknown, its mutation brings about a gain of functional mechanisms by means of an increased kinase activity, which was shown to stimulate α-synuclein aggregation and cytotoxicity (Lin et al., 2009). Furthermore, LRRK2 overexpression in cells and in vivo down-regulates UPS activity which turns out into the accumulation of intracellular substrates (Lichtenberg et al., 2011).
Probably, the most studied protein in PD is the ring-finger E2-dependent E3 ubiquitin–protein ligase parkin, which plays a plethora of intracellular functions linked to PD pathogenesis. Mutations in the UBL domain of parkin, as well as those observed in inherited PD cases, render the protein highly unstable, making it possible parkin detection in the cell lines only in the presence of PI. However, it is debated whether parkin genetic mutations impair the ability of carrying out ubiquitin conjugation of substrates or else disruption of the UBL domain enhances auto-catalytic self-ubiquitination of parkin promoting its self-clearance (Chaugule et al., 2011). As a matter of fact, integrity of UBL is crucial for the regulation of parkin function in cells, and it has been proposed that parkin mutations lead to its altered sub-cellular localization and may further increase the aggregating-propensity of parkin (Santos, Morais, Pereira, Sequeiros, & Alonso, 2019).
Parkin binds concurrently substrates and proteasome through interaction with 19S subunits, especially Rpn1, even though additional receptors have been identified (Chaugule et al., 2011; Kabayama et al., 2017; Um et al., 2010; Um et al., 2006). Specifically, parkin was found to further bind Rpn13 through the UBL domain. On its turn, Rpn13 may regulate protein turnover of parkin, since Rpn13 silencing increases parkin bio-availability in cell (Aguileta et al., 2015).
Although the affinity of parkin for 19S subunits is not very high, being likely in the μM range, it has been proposed that this interaction regulates an allosteric mechanism which activates parkin ubiquitin ligase activity (Aguileta et al., 2015; Chaugule et al., 2011). Conversely, it looks still controversial whether parkin binds also the 20S, and the divergent findings are probably dependent on the experimental models used (Dachsel et al., 2005).
Biological function of parkin deals with turn-over of a plethora of intracellular substrates.
A direct clue between parkin and α-synuclein was originally provided by identifying the selective ubiquitination of a specialized form of α-synuclein expressed in neurons, α22SYn (Shimura et al., 2001).
Additional substrates have been then identified, supporting a broader relevance of parkin activity in neuron homeostasis. A non-exhaustive list of substrates includes (i) CDCrel-1, which is the synaptic vesicle associated protein, (ii) p62, which is ubiquitinated at K13 by parkin, and (iii) p62 intracellular abundance, which follows an inverse linearity with parkin expression (Okatsu et al., 2010; Song et al., 2016). Being p62 involved in aggresome formation, this finding elicits additional considerations on the cross-talk between the contribution of the key players of proteostasis in PD.
Another substrate of parkin is STEP6, which builds up in the striatum of PD subjects and also in murine models of the disease. The increase in STEP6, which follows parkin loss, is associated with a decrease in the phosphorylation of ERK1/2 and its downstream target, pCREB [phospho-CREB (cAMP response element-binding protein)] (Kurup et al., 2015). Interestingly, dopamine signalling affects the de-phosphorylation of STEP6 and its catalytic activity; thus, in dopamine-deficient neurons STEP6 would be more active, depressing the ERK1/2 signalling pathway mentioned above, and grossly impacting on neuron homeostasis. Furthermore, STEP6 and BDNF are regulated through a reciprocal feedback mechanism and this may outline the loss of the neuron growth factor in PD.
Remarkably, one of the most relevant items about parkin biological role is its interaction with PINK, a Ser/Thr protein kinase encoded by the PINK1 gene, a major surveyor of mitochondria quality control, by targeting damaged mitochondria to autophagy-mediated clearance (i.e., mitophagy) (Greene et al., 2012; Gao et al., 2016; Fedorowicz et al., 2014; Rosen et al., 2010; Sun & Büeler, 2019; Gao et al., 2016).
In healthy mitochondria, PINK1 is quickly degraded by several mitochondrial peptidase, whereas in de-polarized mitochondria it is no longer cleaved and becomes exposed to the mitochondrial membrane, recruiting parkin for ubiquitination of various mitochondrial proteins thereby labelling the organelles for mitophagy (Greene et al., 2012). Thus, down-regulation in parkin bioavailability turns out into a reduced clearance of damaged mitochondria which become a major source of ROS, dramatically contributing to the redox imbalance.
Furthermore, upon enzymatic shedding, PINK1 is further released as a soluble cytosolic form, called PINK1-s, which assists recruitment of parkin to the mitochondrial membrane and further contributes to the delivery of aggregating-prone proteins to the forming aggresomes during proteasome inhibition. Thus, PINK1-s works as a sensor that links the proteasomal deficiency signal to the aggresome formation process (Fedorowicz et al., 2014; Gao et al., 2016).
As a whole, molecular insights on PD strongly support that tailored strategies of UPS modulation would provide a significant therapeutic efficacy in delaying disease progression.
4.3. Huntigton's disease
Huntington disease (HD) is a neuro-degenerative disorder with autosomal dominant inheritance caused by a triplet expansion (i.e., CAG) in the huntingtin gene (Htt) which induces the protein to hold an exceedingly long poly-glutamine (polyQ) stretch at the N-terminus. A polyQ length > 35 residues render pathogenic Htt (hereafter referred to as mHtt), which then acquires aggregating-prone properties (Di Figlia et al., 1997; Boland et al., 2018); the removal of the poly-Q stretch appears to be sufficient to rescue neuron homeostasis and to milder neuro-degeneration in murine models of HD (Zheng et al., 2010). From the clinical point of view, the disease is characterized by the anticipation phenomenon, that is an increase in the CAG triplet repetitions, and thus of polyQ length, over generations, bringing about an early onset of the disease through a more severe phenotype which further strengthens the pathogenic relevance of the polyQ stretch length (Labbadia & Morimoto, 2013).
Although several neuronal cells are affected during disease progression, medium-spiny neurons of striatum are those displaying the highest degree of alteration. This feature might be due to the glutamergic cytotoxicity through an exceedingly high N-methyl d-aspartate receptor (NMDAR) activity (Labbadia & Morimoto, 2013). However, differences in synaptic and extra-synaptic activities of these receptors have suggested that dysfunction of neuronal circuits might derive from a primary dysregulation of intracellular pathways, including those regulating proteostasis, through largely unknown mechanisms.
Molecular research on Htt has clarified that the toxicity of the mutated protein is given by the release of N-terminal fragments, which hold the polyQ stretch, upon cleavage of the full-length protein by intracellular proteases, most likely caspase 6 (Soares, Reis, Pinho, Duchen, & Oliveira, 2019; Tebbenkamp et al., 2011).
From the early research on HD, multi-faceted interactions between mHtt fragments and intracellular proteolytic pathways have been sketched and therapeutic strategies proposed to improve the clearance of the proteins (Harding & Tong, 2018; Zhang et al., 2019). Scherzinger first reported that poly-glutamine expansion had an amyloidogenic-like behaviour (Scherzinger et al., 1997). As a matter of fact, cellular aggregates in cell-based models and, most notably, in post-mortem CNS explants of human subjects display positivity to either ubiquitin, proteasome subunits and several E3 ligases staining, thus envisaging a specific recruitment of proteasome complexes in growing aggregates (DiFiglia et al., 1997; Juenemann, Wiemhoefer, & Reits, 2015; Dasgupta et al., 2015). However, soluble mHtt is not efficiently targeted by the 26S proteasome and the lack of efficient ubiquitination for proteasomal degradation leads to intracellular aggregation driven by the intrinsic disordered structure of mHtt (Juenemann et al., 2015; Schipper-Krom, Juenemann, & Reits, 2012; Juenemann et al., 2013). Conversely, mHtt fragments are extensively targeted by E3 ligases and aggregates formation lead to the progressive sequestration of them with severe consequences for cell metabolism. More recently, the isolated mutated exon-1 of Htt appears to be a ubiquitin-independent substrate of free 20S, even though a direct proteasome inhibition by exon1-Htt, as occurring for other amyloidogenic proteins, is not expected to be a major mechanism of proteo-toxicity in HD (Juenemann et al., 2018). It is instead more likely that Htt perturbs UPS functionality through the modulation of metabolic pathways and through transcriptional and post-translational events mostly involving the Ubiquitin-Conjugating Enzymes which are discussed below. In fact, it is well known that expression of mHtt stimulates a deep re-arrangement of the genes being transcribed, which are, among others, mitochondrial membranes, chromatin remodelling, lipid binding proteins, protein folding and a plethora of E3 ligases (Tang et al., 2011). Furthermore, mHtt expression was reported to affect cell cycle, which obviously is not an issue for neurons, but it may affect glial cells and their mechanisms of preservation of CNS homeostasis (van Hagen et al., 2017).
Abnormal synaptic transmission was reported to induce proteasome impairment in murine models of HD and a molecular mechanism through an increased cAMP signalling and the concomitant decreased of PKA activity was proposed to explain this feature in neurons. Nonetheless, the cAMP/PKA pathway has been long considered central to HD pathogenesis by virtue of the pivotal role played by the two molecular pathways in neuron homeostasis and plasticity. The PKA holoenzyme, which is catalytically inactive in the absence of cAMP, is made up of two PKA-Rs and two catalytic subunits (See section 4.5 and Box 3) (Lin et al., 2013); in the presence of cAMP, cAMP-bound PKA-Rs dissociate from the catalytic subunits, which are then degraded by the proteasome. As a consequence, an impaired proteasome activity would allow the PKA-Rs to gather up reducing the amounts of free PKA catalytic subunits and thereby impairing PKA activity. On the other hand, PKA carries out the phosphorylation of Rpt6 at Ser120, a post-translational modification which enhances the activity of the proteasome and the constitution of capped particles (Lin et al., 2013). Furthermore, during synaptic sprouting Rpt6 can also be phosphorylated by Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) in a neuronal activity-dependent manner (Jarome, Kwapis, Ruenzel, & Helmstetter, 2013). Therefore, the HD onset might be correlated to a vicious cycle wherefore proteasome content drops down, due to its seizure, and the PKA-mediated stimulation of proteolysis fades out.
A significant contribution to HD pathogenesis further comes from the iPSC model of HD. Remarkably, HD-iPSCs display constitutive increased proteasome activity, which was found to regulate the levels of both normal and mutated Htt, contributing to suppress polyQ-expanded Htt aggregation (Koyuncu et al., 2018; Liu et al., 2017). As a matter of fact, HD iPSCs do not accumulate polyQ-expanded Htt aggregates even after multiple passages. Accordingly, a dysfunction in proteasome activity results in impaired Htt levels and aggregation of mHtt also in HD-iPSCs.
In iPSC, a major role in facilitating Htt clearance seems to be played by the E3 ligase UBR5. Although loss of UBR5 did not impair pluripotency markers in human control iPSCs, it induced instead the formation of misfolded protein aggregates (i.e., aggresomes) (Koyuncu et al., 2018).
The iPSCs experimental model has allowed to cast further light on the transcriptional activity of FOXOs, a subgroup of the Forkhead transcription factors (human cells encode four FOXO proteins, FOXO1, FOXO3a, FOXO4 and FOXO6), which are central regulators of cell metabolism (Liu et al., 2017). Remarkably, HD iPSCs start lacking proteasome activity and expression upon differentiation into neural cells, an occurrence which appears to be dependent on FOXO4, which is downregulated during differentiation. Such a downregulation appeared to be dependent on the enhanced activation of Akt, a serine-threonine kinase which acts as an upstream regulator of FOXOs by mediating their clearance, in HD iPSC-derived neurons (Liu et al., 2017). However, activated and total Akt levels strongly vary among different HD experimental models adopted and they were reported to be reduced in a rat HD model and in post-mortem brain extracts from patients with HD (Colin et al., 2005). Thus, further studies are demanded to address the relevance of this key metabolic pathway in preserving the PN in HD.
As mentioned above, several E3-ligases tag mutant Htt and are sequestered in aggregates. In different cell lines, mHtt clearance is usually carried out by canonical E3 ligases, such as UBR1, UBE3A, HSP and also non-canonical E3 ligases, such as Herp which, however, contain(s) a UBL domain (Luo et al., 2018). Conversely, a role of de-ubiquitinase ataxin-3, involved in the pathogenesis of other neurodegenerative disorders (such as spino-cerebellar ataxia type 3), has not been confirmed in HTT progression by studies in murine models (Zeng, Tallaksen-Greene, Wang, Albin, & Paulson, 2013). Moreover, atypical ubiquitination of mHtt by some E3 ligases, such as WWP1, may favour disease progression (Lin et al., 2016).
Conversely, ubiquilin-1, a highly conserved family of proteins which facilitate protein disposal through autophagy and UPS and which is down-regulated in early HD, improves the clearance of Htt (Safren et al., 2014).
Similarly, Usp14 has a favourable effect in cells expressing mutant Htt cells by decreasing the aggregate load and by enhancing cell viability (Hyrskyluoto et al., 2014).
Finally, although UBE3A overexpression is known to promote UPS-mediated degradation of transfected mHtt in cultured cells, it is still unclear how UBE3A expression levels impact HD pathology. Remarkably, when the E3-ligase was up-regulated a drop in K63 ubiquitination of mHtt was observed (Bhat, Yan, Wang, Li, & Li, 2014). In this study, the presence of the pathological polyQ stretch was proposed to alter the overall folding of mHtt favouring the formation of K63 Ub linkages, also through cooperation of p62/SQSTM1, which are more prone to aggregation (Lim et al., 2015). Thus, to stimulate the UBE3A activity would be relevant to limit the toxicity of mHtt.
4.4. Involvement of UPS in retinal diseases
By virtue of its anatomical localization retina is also called the “window to the brain” (London, Benhar, & Schwartz, 2013). Clinical and molecular investigation into this highly specialized nervous tissue has been long considered promising to search for early diagnostic and prognostic bio-markers of CNS disorders, such as AD, through non-invasive approaches. The identification of either quantifiable metabolic by-products, released in main eye fluids (e.g., vitreous, humour aqueous, tears), or of ultra-structural alterations pathognomonic of a given diseases would fulfil this clinical opportunity. With respect to the second point, non-invasive imaging approaches, such as (i) retinal fiber layer imaging with spectral domain optical coherence tomography (OCT), (ii) Visual Evoked Potentials (VEPs) and (iii) Doppler haemodynamic parameters of retinal veins (Berisha, Feke, Trempe, McMeel, & Schepens, 2007; Dehabadi, Davis, Wong, & Cordeiro, 2014; Iseri, Altinaş, Tokay, & Yüksel, 2006; Parisi et al., 2001; Nag & Wadhwa, 2012), have been already used to detect early morphological alterations in AD subjects and the advancement in imaging techniques will certainly allow to improve the sensibility and specificity of the proposed diagnostic tools.
Conversely, the identification of biomarkers in eye fluids is still devoid of solid evidences and a much greater knowledge of the retina metabolism is required to address this task. However, research on this topic is limited by technical and histological issues, namely (a) the difficulty in isolating vital retina tissue from human post-mortem explants, and, mostly, (b) the technical inability in obtaining homogeneous primary cultures of individual retina cells. In fact, retina is arranged into complex three-dimensional layers in the posterior segment of the eye populated by (i) photoreceptor cells (i.e., rod and cones), which carry out from the physical to chemical transduction of the light stimulus during vision, (ii) bi-polar cells, which transduce rod and cones signal to (iii) Retinal Ganglion Cells (RGCs) which, finally, convey this information to the visual cortex through the optic nerve their axons generate (Belenky, Smeraski, Provencio, Sollars, & Pickard, 2003; Lobanova et al., 2018). In addition, optimization of the light stimulus perception and transduction along with cells nourishment and survey of tissue homeostasis is carried out by a multitude of different cell lineages (i.e., amacrine cells, horizontal cells, retinal pigment epithelium, muller glia cells), which lie in the retina layers. An extensive ultrastructural organization of the retina and other segments of the eye is provided elsewhere (Gupta, Hanley, et al., 2016; Nag & Wadhwa, 2012).
Being terminally differentiated, photoreceptors, bi-polar cells and RGC, like any other post-mitotic cell, are vulnerable to proteostasis unbalance which may originate from either inherited or acquired disorders. A clearcut example are the metabolic complications of glaucoma and diabetic retinopathy (DR), discussed below to a greater detail, and the inheritance of mutated alleles of proteins, either tissue specific (e.g., rhodopsin, a protein which senses light in photoreceptors) or not (i.e., optineurin), which pose an amyloidogenic threat in retinal degenerative diseases and genetic variants of glaucoma, respectively (Lobanova et al., 2018; Swarup & Sayyad, 2018; Minegishi et al., 2016; Shen et al., 2011; Yao et al., 2018; Fernandez-Godino & Pierce, 2018; Piippo et al., 2018; Felszeghy et al., 2019; Shen et al., 2019; Sirohi & Swarup, 2016; Ying et al., 2015; Shen, Li, Chen, Chern, & Tu, 2015; Li et al., 2015; Caballero, Liton, Challa, Epstein, & Gonzalez, 2004; Caballero, Liton, Epstein, & Gonzalez, 2003). Furthermore, the physiological enhanced metabolism of this tissue is sustained through the maintenance of a lipid profile extremely rich in long-chain poly-unsaturated fatty acids and very long chain poly-unsaturated fatty acids for membrane stability, and a high O2 tension which are both sources of ROS production, proteo-toxicity, lipid peroxidation and membrane damage (Gorusupudi, Liu, Hageman, & Bernstein, 2016) .
Whilst autophagy activation shows controversial issues, being also an apoptosis inducer in photoreceptors after prolonged proteo-toxicity, such as in models of inherited retinal degeneration, UPS activation appears to bring about only metabolic benefits (Blasiak, Pawlowska, Szczepanska, & Kaarniranta, 2019; Yao et al., 2018) inasmuch proteasome loss leads to pathogenic events. Accordingly, proteasome pharmacological inhibition in rodents quickly turns out in retina degeneration (Kageyama, Ota, Sasaoka, Katsuta, & Shinomiya, 2019). Nonetheless, a deepening on the molecular insights of UPS into the retina would help addressing several unresolved issues regarding UPS biology. In fact, highly specific mechanisms of regulation of protein turn-over are supposed to have evolved since metabolic activity follows a circadian rhythm with alternance of light and dark hours. Such an alternance is mirrored by a specular overall proteolytic burden and the molecular clock(s), which are finalized to turn the UPS off once light hours are over are unknown. No evidence for recruitment of specific PIPs and/or post-translational modification of UPS members is reported, but their identification might pave the road to novel mechanisms of regulation of proteasome proteolytic activity (Fan et al., 2013; Fukuhara, Dirden, & Tosini, 2001; Knowles et al., 2009; Naash, Al-Ubaidi, & Anderson, 1997).
Moreover, surprisingly, murine transgenic models and human ex-vivo models of eye diseases point toward a major role of non-canonical proteasome assemblies in retina development and homeostasis. A predominant contribution seems to be played by the immunoproteasome, by means of either PA28 expression and incorporation of inducible proteasome subunits in 20S assemblies (See Box 1), which is over-expressed to an exceptionally high degree in synaptic terminals and in photoreceptors (Hussong, Kapphahn, Phillips, Maldonado, & Ferrington, 2010;Hussong et al., 2011; Shang & Taylor, 2012; Lobanova et al., 2018; Aghdam and Mahmoudpour, 2016; Basler et al., 2015). In detail, differentiation of murine retinal progenitor cells into their mature lineages requires the mTORC1-dependent STAT1 activation, which triggers the transcriptional up-regulation of PSMB9 gene (which encodes for a catalytic subunit of immunoproteasome) but not of PSMB6 or PSMB7 (which encode for canonical subunits with trypsin-like and caspase-like proteolytic activities) (Choi et al., 2018). Assembly of functional immunoproteasome is supposed to assist the 26S in clearing the bulk of short half-life proteins that accumulate in highly replicating cells (Choi et al., 2018). Nonetheless, this finding envisages that the immunoproteasome might have a higher affinity than 26S for cell cycle substrates and for oxidized and unfolded proteins which may accumulate during replication. This last possibility is consistent with biochemical properties reported to date for the immunoproteasome and with the finding on a murine model of an inherited retinal degeneration, wherefore photoreceptors carry a rhodopsin allele mutation (i.e., P23H). This mutation renders the protein unfolded and aggregating-prone, while the overexpression of PA28 counteracts the degeneration improving photoreceptor survival in such a murine model (Raule et al., 2014; Raule, Cerruti, & Cascio, 2014; Lobanova et al., 2018).
The role of immunoproteasome and its substrate specificities gain further relevance when we consider that retina is an immune-privileged organ so that the processing of antigenic peptides, as well as their presentation, follow highly specific dynamics to regulate local immune response and immune-surveillance of this tissue, mostly concerning the maintenance of immune tolerance versus retinal self-antigens (Lipski et al., 2017; McPherson, Heuss, Pierson, & Gregerson, 2014; Schuld et al., 2015; Voigt et al., 2017).
Although indirectly, the Rett Syndrome (RTT) case (see also Section 2.2.2) might further offer a clue for studying the specific regulation of proteasome biogenesis in retinal cells. RTT is a neurodevelopmental disorder classified as rare X-linked genetic disease (Amir et al., 1999). In >95% of cases, girls, affected by the syndrome harbour a de-novo mutation in the MeCP2 gene, which encodes for an epigenetic transcriptional regulator with largely unknown biological functions (Chahrour et al., 2008).
Whilst several brain areas display neuroanatomical abnormalities, retina, as well as vision, appear to be unaffected during RTT onset and progression (Jain et al., 2010; Rose, Wass, Jankowski, Feldman, & Djukic, 2019). Interestingly, whilst MeCP2 is ubiquitous in human tissues and maximally abundant in the CNS, it is poorly expressed in the retina and this may outline the absence of morphological and functional alterations of the retina and the visual pathways in this disease. It has been recently unveiled that primary fibroblasts, isolated from skin biopsies of RTT subjects harbouring non-sense MeCP2 mutations, suffer from a defective proteasome biogenesis (see Section 2.2.2) due to the MeCP2 dependent down-regulation of PAC1 and PAC2 along with the α7 subunit of 20S (Sbardella et al., 2020). Upon MeCP2 silencing, this defect shows up in human neuron-derived cells envisaging a general contribution of this transcription regulator in proteasome biogenesis. Thus, molecular investigation into retina might help addressing unresolved issues about the contribution of individual proteasome assemblies in PN and in immune responses and further clarify the transcriptional and molecular dynamics of proteasome biogenesis.
Among the neuro-degenerative eye disorders leading to irreversible blindness, which display the highest prevalence in western countries, there are (a) diabetic retinopathy (DR), that is a micro-vascular complication of diabetes, and (b) glaucoma. The clinical and epidemiological features underscoring onset and progression of these diseases are extensively discussed elsewhere and will not be discussed herein (Lombardo et al., 2013; Parravano et al., 2013; Picconi et al., 2018; Tarr, Kaul, Chopra, Kohner, & Chibber, 2013). However, they represent two eye disorders wherefore UPS and alteration of the PN appear to follow a specular pattern and for which pharmacological strategies targeting the UPS might provide a valid therapeutic opportunity.
Contribution of the UPS in DR onset is poorly studied, even though several lines of research support a pivotal role played by the proteasome in regulating the nuclear activity of key transcription factors and the release of cytokines in the early vascular response to the hyper-glycemic insult (Aghdam, Gurel, Ghaffarieh, Sorenson, & Sheibani, 2013; Campello, Esteve-Rudd, Cuenca, & Martín-Nieto, 2013; Rahimi, 2012).
Although choroidal endothelial cells seem to contribute only marginally to this phenomenon, an aberrant proliferation of retinal endothelial cells (which ultimately lead to micro-haemorragic lesion, vessel leakage and irreversible fibrosis) follows the increased secretion and bio-availability of VEGFs. Several independent research teams suggest that the retinal cell type, which first senses the hyper-glycemia is the Muller glia, and indeed metabolism of this cell type appears to be sensitive, through unknown mechanisms, to fluctuations in glucose concentration, as those occurring in vivo in diabetic subjects (Wang, Xu, Elliott, Zhu, & Le, 2010; Le, 2017; Picconi et al., 2019; Picconi et al., 2017; Voigt et al., 2017; Matteucci et al., 2014; Sbardella et al. 2017). This insult stimulates the secretion of VEGFs as well as of other pro-inflammatory cytokines. VEGFs synthesis is mostly regulated by the transcriptional activity of NF-kB and HIF-1α whose nuclear translocation is regulated by proteasome proteolytic activity (see Section 3.2.1) (Alkalay et al., 1995; Traenckner, Wilk, & Baeuerle, 1994; Chen et al., 1995; Ferrara, 2004).
Therefore, a better understanding of the molecular mechanisms of enhanced protein turn-over upon hyper-glycemia in Muller glia and, possibly, in additional retina cell types would allow to envisage selective therapeutic approaches to target VEGF synthesis rather than its biological cascade once secreted. In fact, clinical regimens for DR treatment mostly deal with VEGF inhibitors which however do not distinguish between the physiological and pathological angiogenesis with the former being as much relevant for retina homeostasis as blocking the latter one would be for DR progression (Ferrara, 2004; Lacal & Graziani, 2018). Thus, selective targeting of factors, that regulate the proteasome-mediated turn-over of players in DR progression, would help to overcome the limitation of traditional proteasome inhibition strategies, which stop bulky proteolytic burden thereby compromising PN.
The involvement of proteasome in the onset of glaucoma appears to follow an opposite path to that observed in Muller glia cells in DR. In fact, there is a compelling evidence that PN might be dys-regulated in both main clinical forms of glaucoma, namely (i) primary open-angle glaucoma (POAG) and (ii) normal-tension glaucoma (Oddone et al., 2016; Quaranta et al., 2016; Agarwal, Gupta, Agarwal, Saxena, & Agrawal, 2009; Weinreb, Aung, & Medeiros, 2014; Wunderlich, Golubnitschaja, Pache, Eberle, & Flammer, 2002; Caballero et al., 2003).
In this regard, the term glaucoma encompasses an heterogeneous group of neurodegenerative disorders characterized by the loss of retinal ganglion cells (RGC) and atrophy of the optic nerve their axons generate (Minegishi et al., 2016; Oddone et al., 2016; Swarup & Sayyad, 2018).
Disease etiology displays a multi-factorial profile, wherefore genetic and acquired factors concur in determining its onset and progression. Among the acquired factors, redox imbalance and increased Intraocular Pressure (IOP) are likely the most relevant ones. In POAG, IOP increase is sustained especially through a pathogenic mechanism linked to the altered metabolism of an endothelial-like cell histo-type called Trabecular Meshwork Cell (TMC). TMCs synthesize and secrete the trabecular meshwork (TM), a specialized form of extracellular matrix localized in the anterior segment of the eye (at the sclero-corneal limit), which drains the aqueous humour, a fluid which shapes the eye-globe and nourishes the lining cells (Agarwal et al., 2009; Weinreb et al., 2014). Upon metabolic dysregulation, TMs acquire a senescent-like phenotype and display enhanced apoptosis which leads to a pathological remodelling of TM associated to an obstruction for the outflow of aqueous humour (Agarwal et al., 2009; Weinreb et al., 2014; Micera et al., 2016; Vernazza et al., 2019). This brings about the increase in (IOP) thereby exerting a mechanical compression of the retina and optic nerve, localized in the posterior eye, ultimately leading to RGCs loss, optic nerve degeneration and visual decline. Therapy with glucocorticoids (GC) has been long known to induce acute iatrogenic form of glaucoma by affecting the TMC metabolism, but the primary alteration of such a cell after GC administration is unknown (Roberti et al., 2020). Interestingly, TMCs express a ubiquitous protein, called myocilin (i.e., from the MYOC gene), whose expression is up-regulated in TMCs when exposed to GC, but also oxidative stress and cytokines (Qiu, Shen, Shyam, Yue, & Ying, 2014; Wang et al., 2019; Resch & Fautsch, 2009; Micera et al., 2016); when over-expressed, this protein is supposed to pose a metabolic threat to TMCs through unexplored gain of function mechanisms (Jain et al., 2017; Kim et al., 2001). This pathogenic effect may occur either in the intracellular or extracellular compartments, in accordance with the broad localization of the protein. Furthermore, mutations in MYOC gene are the most studied cause of the juvenile form of glaucoma and, among the different point mutations described so far, the most prevalent ones (e.g., Pro370Leu) render myocilin amyloidogenic and aggregating-prone (Wang et al., 2019; Yam, Gaplovska-Kysela, Zuber, & Roth, 2007). Myocilin is a proteasome substrate, and, in the absence of GC therapy, proteasome activity appears to decline in TMCs culture isolated from patients suffering from glaucoma in an age-dependent manner; further, myocilin expression in HeLA cells was found to decrease the bioavailability of some 20S subunits (Qiu et al., 2014). Conversely, the GC effect on MYOC processing and proteasome regulation is unknown. Nonetheless, the role of proteasome impairment in driving cell senescence and the role of proteasome re-activation in delaying this phenomenon is well studied in several cell types but not in TMCs (Chondrogianni & Gonos, 2004; Deschênes-Simard, Lessard, Gaumont-Leclerc, Bardeesy, & Ferbeyre, 2014).
Even though transgenic models do not always support an unequivocal role of MYOC in glaucoma onset, the interactome of this protein is worth being studied to address the metabolism of TMC (Jain et al., 2017; Joe, Nakaya, Abu-Asab, & Tomarev, 2015; Kim et al., 2001; Senatorov et al., 2006; Zhou, Grinchuk, & Tomarev, 2008). Therefore, to study the dynamics of myocilin digestion in TMCs and how proteasome might undergo dysregulation under metabolic conditions, that are commonly seen in glaucoma subjects, might help to explore the molecular insights of pharmacological strategies based on UPS rescue.
Differently from POAG, normal-tension glaucoma is not supported by an increased intra-ocular pressure and the degeneration of the optic nerve likely depends on a primary insult on RGCs. Even in this case, a tight involvement of intracellular proteolytic pathways is largely envisaged. Optineurin gene encodes for a protein involved in intracellular vesicle trafficking, and expression of mutated forms of optineurin induces a severe dysregulation of the UPS and of autophagy (Shen et al., 2015; Shen et al., 2011; Sirohi & Swarup, 2016).
Besides the molecular findings discussed above, a major suggestion for the relevance of proteasome in handling the PN in retina comes indirectly from biochemical pharmacology (Sbardella, Tundo, et al., 2020). Several clinical trials worldwide support the therapeutic efficacy of citicoline for glaucoma treatment (Carnevale et al., 2019; Parisi et al., 2008; Parisi et al., 2018; Parisi et al., 2019; Roberti et al., 2015). Citicoline, also known as CDP choline, is a drug made up by choline and cytidine diphosphate which displays optimal bioavailability and easily crosses the blood brain barrier (Faiq, Wollstein, Schuman, & Chan, 2019). Although the mechanisms of action of citicoline have never been identified at molecular detail, its wide usage in clinical regimens is based on its outstanding safety profile and on the efficacy also in neurological disorders, including neurodegenerative disease, such as the early phases of AD and PD onset, though the trials having been run are still limited (Eberhardt, Birbamer, Gerstenbrand, Rainer, & Traegner, 1990).
Our group has very recently reported that citicoline is an allosteric modulator of proteasome in vitro and in vivo, wherefore citicoline binds the 20S with a very high affinity (i.e., in the low nanomolar range), stimulating the clearance of synthetic substrates as well as of α-synuclein (Sbardella, Tundo, et al., 2020). Surprisingly, in neuron-derived cells, citicoline was found to both stimulate 20S activity and to promote the assembly of proteolytic active capped assemblies (i.e., 26S and 30S, see Section 2.3.2) (Sbardella, Tundo, et al., 2020). As a matter of fact, cells stimulated with citicoline experience a very significant increase in the overall proteolytic burden by the UPS. Therefore, although it is reasonable that the proteasome stimulation is not the only therapeutic effect, the neuro-protective role of citicoline highlights the relevance of proteasome functionality in maintaining the post-mitotic cells homeostasis. Nonetheless, citicoline experience in clinical trials might be looked as a proof of concept that the activation of UPS is a valid strategy to delay the progression of pathologies sustained by proteo-toxicity.
4.5. Targeting proteasome as novel tool against proteotoxic diseases
Neurodegenerative diseases are clinically heterogeneous proteinopathies sustained by the accumulation of aggregates of misfolded disease associated proteins (see also Section 4.1). The prevalence and incidence of neurodegeneration increases dramatically with age, and, since people life expectancy rises worldwide, also the number of individuals suffering from these pathologies is expected to dramatically increase in the next years (Jones & Tepe, 2019; McAlary et al., 2019). Despite social and economic relevance, no effective cure still exists; therefore, the development of novel therapeutic approaches is essential.
As discussed above (see Section 4.1), UPS alteration contributes to disease onset and progression, leading to an intense research effort with the purpose of identifying therapeutic strategies targeting UPS. Different approaches to enhance UPS functionality have been proposed, spanning from stimulation of ubiquitination and/or inhibition of de-ubiquitination (See Box 2), or inhibition of protein aggregation, being this last strategy founded on evidences that monomeric proteins are better degraded by proteasome than oligomers (Dantuma & Bott, 2014; Wertz & Murray, 2019). In addition, modern strategies envisage the direct proteasome stimulation by either (i) drugs which specifically target proteasome particles increasing their bulk proteolytic activities (see Section 2.2.1) or (ii) phosphorylation of proteasome subunits (Ottobelli et al., 2013; Parisi et al., 2015; Myeku & Duff, 2018).
According to the first point, the identification of “drug-like” molecules, which directly activate proteasome, is challenging. Notably, a chemical genetics screening of over 2750 compounds using a proteasome activity probe as a readout in a high-throughput live-cell fluorescence-activated cell sorting-based assay has led to the identification of more than ten compounds that increase proteasome activity (Leestemaker et al., 2017). A promising, but still poorly explored strategy, is the development of therapeutic peptides and/or peptidomimetics, designed on the basis of specific binding regions of natural proteasome regulators (Wilk & Chen, 1997; Fosgerau & Hoffmann, 2015; Jones & Tepe, 2019). In general, the advantage of peptide usage seems to deal with the higher specificity and selectivity with respect to molecular target; however, beside the complex synthesis, they suffer from low metabolic stability and poor membrane permeability (Fosgerau and Hoffmann, 2015, Lau and Dunn, 2018). The most common class of synthetic peptides acting as proteasome activators are based on the HbYX motif (see Section 2.3.2) (Lau & Dunn, 2018). In this context, it has been shown that peptides derived from C-termini Rpt2 and Rp5, and a 14-mer peptide, based on the C-terminal fragment of Blm10 (the yeast ortholog of PA200, see Box 1) containing the HbYX motif, efficiently stimulated proteasome activity in vitro (Sadre-Bazzaz, Whitby, Robinson, Formosa, & Hill, 2010; Smith et al., 2007; Karpowicz et al., 2015). Recently, it has been reported that upon introduction of the HbYX sequence, the proline- and arginine-rich peptide (PR11), which is a 20S allosteric inhibitor, turned to be a proteasome activator in vitro and in cell model (Giżyńska et al., 2019; Osmulski et al., 2020). Another example of a “drug-like” molecule with a different unknown mechanism of action is the proteasome-activating peptide 1 (PAP1), which increases the chymotrypsin-like proteasomal catalytic activity in vitro and in cell models and is further able to halt protein aggregation (Dal Vechio, Cerqueira, Augusto, Lopes, & Demasi, 2014). Among proteasome activators, natural compounds, such as oleuropein, betulinic acid and fatty acids deserve particular attention, and their features will be discussed in the next sections.
Concerning the phosphorylation strategy, a bulk of studies has shown that reversible phosphorylation of proteasome subunits positively regulates its function (Myeku & Duff, 2018; VerPlank et al., 2019; VerPlank & Goldberg, 2017; VerPlank & Goldberg, 2018). Accordingly, protein kinase A (PKA)-dependent phosphorylation is involved in the regulation of multiple aspects of proteasome functionality, such as: (i) enhancement of Rpt6 ATPase activity through phosphorylation which further stimulates the association of 20S and 19S in vitro (See Box 3), (ii) increase in the proteasome capacity to clear out ubiquitinated proteins, peptides and ATP as well as the degradation of aggregation-prone proteins in cells upon phosphorylation of Rpn6 at serine14 (Box 3) (Asai et al., 2009; Jarome et al., 2013; Lokireddy, Kukushkin, & Goldberg, 2015; Lu et al., 2008; Zhang et al., 2007). Moreover, phosphorylation of Rpt6 by kinase CaMKIIα induces 26S translocation into dendritic spines in primary neurons, promoting local protein breakdown and driving the formation of synaptic connections (Bingol et al., 2010; Djakovic et al., 2009; Hamilton et al., 2012; Jarome et al., 2013). Therefore, proteasome subunit phosphorylation has been suggested to rescue proteasome function and it could represent a promising strategy to treat neurodegeneration (Myeku & Duff, 2018; VerPlank & Goldberg, 2017). Recently, it has been reported that, in hyppocampal neurons, only 20% of proteasome seems to be in an (see Section 2.4) “active” substrate-engaged state, whereas the remaining part is in an “inactive” substrate-accepting ground state. Therefore, it has been speculated that phosphorylation increases the percentage of active forms of proteasome, recruiting “idle” particles as well as directly stimulating their activities (Asano et al., 2015; Myeku & Duff, 2018). As a matter of the fact, a promising strategy should be the stimulation of PKA activity through the modulation of the amplitude of cAMP signal. The cAMP level is curtailed by cyclic nucleotide phosphor-di-esterases (PDE), which act negatively by regulating PKA signals. Inhibition of PDE stimulates cAMP/PKA axis and activates proteasome, opening to a novel potential use of PDE inhibitors in the CNS diseases treatment (Myeku & Duff, 2018; VerPlank & Goldberg, 2017). Accordingly, it has been shown that PDE4 selective inhibition by rolipram induces phosphorylation of several subunits of 26S, leading to an increase in mouse models of UPS-mediated clearance of tau and amyloid aggregate, accompanied by a reduced cognitive impairment (Myeku et al., 2016; Smith, Pozueta, Gong, Arancio, & Shelanski, 2009; Vitolo et al., 2002). Moreover, cAMP/PKA axis activation, which follows PDE10 inhibition, reduces Htt aggregation through a proteasome-dependent mechanism, and ameliorates motor and cognitive deficit in Htt mouse model (Beaumont et al., 2016; Giampà et al., 2010; Harada, Suzuki, & Kimura, 2017; Lin et al., 2013). Furthermore, the administration of the FDA approved PDE3 inhibitor, cilostazol, to a mouse model of tauopathy enhanced proteasome function and attenuated the tauopathy and cognitive decline in rTg4510 mice, suggesting that this drug could be potentially repurposed for the treatment of patients with early-stage tauopathy (Schaler & Myeku, 2018). As a whole, despite some early encouraging results in mouse model, it seems clear that the broad range of biological functions, mediated by cAMP, can reduce the clinical efficacy of PDE inhibitors, due to their adverse effects (Heckman, Blokland, Bollen, & Prickaerts, 2018; Myeku & Duff, 2018). It is worth recalling that cAMP/PKA pathway transduces the intracellular signalling of a number of hormones: thus, in such a way, proteasome function can be regulated by hormonal and metabolic stimuli (see Box 3) (VerPlank and Goldberg, 2015).
4.5.1. Direct enhancing proteasome activity by natural compounds
Many organisms have developed a large number of small molecules, which modulate the activity of UPS components (Rousseau & Bertolotti, 2016). Those of nutraceutical origin (contained in fruits, in vegetables and their extracts) are very attractive for their positive effects as antiaging and in the treatment and the prevention of a wide range of pathologies. In fact, dietary phytochemicals exhibit broad and different biological activities, including antioxidant action, free radical scavenging, anti-inflammatory and metal-chelating properties, that represent the evolutive result of the vegetable system defense. All of them are secondary metabolites that plants produce to counteract against various stresses (Murakami, 2013) thus, they can be considered “multifunctional drug-like molecules” (see Section 4.5), and it is not surprising such a high variety of targets, since they are small molecules with very simple chemical structures.
Concerning the activity of these compounds on proteasome, those able to activate/enhance proteasome activity are rare (Bonfili et al., 2008; Dahlmann et al., 1993; Huang & Chen, 2009) and they are often characterized by an ambivalent behaviour, acting alternatively as inhibitors and/or activators according to diverse conditions. As an example, the effects of curcumin (Cuanalo-Contreras & Moreno-Gonzalez, 2019) (1E,6E)-1,7-bis-(4-idrossi-3-metossifenil)-epta-1,6-dien-3,5-dione) on the UPS reflects the hormesis principle (i.e., the biphasic dose-response to an environmental agent characterized by a low dose stimulation or beneficial effect and a high dose inhibitory or toxic effect), being characterized by an inverted U shape dose-response (Ali & Rattan, 2006); thus, curcumin treatment (up 1 μM for 24 h) increases proteasome activity in keratinocytes, but it displays an inhibitory effect at 10 μM (Murakami, 2013). In particular, curcumin induces 26S perturbation, leading to an impairment of cell proliferation in various cancer cells and reduction of cancer burden in mice (Banerjee et al., 2018).
By analogy, quercetin (3,3′,4′,5,7-pentahydroxyflavonethe), the most abundant flavonoid found in fruits and vegetables, which has been initially reported to be a 20S inhibitor (Chen et al., 2005) (IC50 = 3.5 μM), it has been shown to enhance the proteasome activity in vivo and to reduce the Aβ-induced toxicity in a dose-dependent manner when administered to a Caenorhabditis elegans AD model (Chondrogianni et al., 2010). Likewise, the polyphenol resveratrol, that was previously described as a natural direct PI (Yang, Landis-Piwowar, Chen, Milacic, & Dou, 2008; Qureshi et al., 2012), recently has been reported to enhance proteasome activity recovering the impaired proteostasis in a C. elegans AD model, and in AD transgenic mice (Regitz, Fitzenberger, Mahn, Dußling, & Wenzel, 2016); in addition, resveratrol has been shown to enhance cognitive activity by increasing 20S proteasome subunits levels and stimulating proteasome activity (Corpas, Griñán-Ferré, Rodríguez-Farré, Pallàs, & Sanfeliu, 2019).
Hereafter, we focus only on bioactive compounds that directly target the naked catalytic particle, 20S, thus enhancing the ubiquitin-ATP-independent proteolysis, the main pathway degrading the oxidatively damaged and intrinsically disordered proteins (Ben-Nissan & Sharon, 2014).
Oleuropein, the most abundant phenolic compound extracted from Olea europaea (leaf and olives), enhances all three proteasome activities in vitro and promotes cellular resistance to oxidants, prolonging human fibroblasts lifespan (Katsiki, Chondrogianni, Chinou, Rivett, & Gonos, 2007). Systemic administration of oleuropein in pigs increased 20S activity in the subcortical white matter, reducing the damaged proteins accumulation after hypoxia and hypothermia and protecting the myelin.
The triterpene betulinic acid, extracted from the lipid fraction of the algae Phaeodactylum tricornutum and of many other medicinal plants, activates preferentially the CT-L activity (Huang, Ho, & Chen, 2007) with minor effects on T-L and C-L activities. Some neuroprotective effects are reported in the transgenic C. elegans PD model, where betulinic acid decreased α-synuclein accumulation and the 6-hydroxydopamine-induced dopaminergic neuron degeneration (Mullauer, Kessler, & Medema, 2010). Furthermore, it indirectly promoted the enhancement of proteasome activity by regulating rpn1 expression and downregulation of the apoptosis pathway gene, egl-1 (Tsai et al., 2017). Interestingly, betulinic acid has shown a neuroprotective effect in vascular dementia rat models, re-establishing the cerebral blood flow, restoring behaviour parameters and significantly improving the BDNF levels, with a restrain of the oxidative stress and of inflammatory parameters (Kaundal, Zameer, Najmi, Parvez, & Akhtar, 2018).
Besides the proteasomal effect, oleuropein and betulinic acid could be also considered as pleiotropic small molecules, for their anti-HIV (Mayaux et al., 1994; Yang, Gong, Zhang, & Lu, 2016) and anti-tumour activity toward some cancer cell lines (Pisha et al., 1995; Saeed, Mahmoud, Sugimoto, Efferth, & Abdel-Aziz, 2018). Although both oleuropein and betulinic acid have been previously reported to be natural proteasome activators (Katsiki et al., 2007), some authors recently clarified that the stimulatory activity is restricted to fluorogenic substrates, and no effect has been reported on the turnover of mis-folded proteins in vitro or in living cells.
Among naturally occurring activators of 20S proteasome there are some physiological cellular components, such as mucopolisaccarides (e.g., heparin), glycolipids (e.g., ceramides, lysophosphatidyl-inositol and cardiolipin) (Matsumura & Aketa, 1991; Ruiz de Mena, Mahillo, Arribas, & Castaño, 1993), and some proteins (not included in the physiological UPS control), such as the arginine-rich histone H3, a chromatin binding protein able to selectively enhance protein degradation by the proteasome (Orlowski, 2001).
The detailed mechanism by which these compounds regulate 20S degradation is still largely undetermined, but it might be related to the gate opening, mimicking the RP interaction (see 2.2.1, 2.3.1).
4.5.2. Fatty acids
Fatty acids are likely the first small molecules that were first described as 20S proteasome modulators. Indeed, our basic knowledge of proteasome function is rooted on the pioneering studies conducted throughout the 80s and 90s (Orlowski & Wilk, 1981; Ishiura et al., 1986; Folco, Busconi, Martone, & Sanchez, 1988), using fatty acids and SDS as activators. The first systematic study on proteasome peptidase activity reported the effects of several saturated and unsaturated fatty acids examining various carbon-chain lengths and underlining that the optimal 20S activation potency may be achieved by using fatty acids with a C18-C20 chain carbon; thus, the oleic acid resulted the most active compound, with an activating effect 50-fold higher than SDS (Dahlmann, Rutschmann, Kuehn, & Reinauer, 1985). Although our knowledge of proteasome structure and function was still in its infancy, in those early reports it was proposed that activation mechanism could be basically related to conformational changes occurring in the enzyme. Furthermore, they also proposed that fatty acids, abundant in muscle, could participate in the physiological regulation of proteasome-mediated protein degradation. Later on, Orlowski et al. performed a detailed kinetic analysis, reporting that lauric acid activates the C-L activity like SDS; increasing concentrations of lauric acid caused a shift in the apparent Km toward lower substrate concentrations with a concomitant increase in Vmax (Orlowski, Cardozo, Hidalgo, & Michaud, 1991). However, unlike the SDS-mediated one, this activation occurred with a sigmoidal shape of the velocity curve, suggesting the presence of two (or more) substrate binding sites interacting cooperatively. In other words, in the absence of an external activator only part of this activity is manifested, thus underlining the cooperative control of allosteric sites and the concepts of “latency” and of “multi-proteasic complex”; however, there is still some controversy concerning the ability of most fatty acids to enhance the three main 20S proteolytic activities. Actually, fatty acids often exhibit a double-faced nature, behaving as activators and/or inhibitors, according to the type of activity measured. In ostrich liver the C-L proteasome activity was found to be activated by all (with the exception of decanoic acid) types of fatty acids in a concentration-dependent fashion, whereas the CT- and T-L activities were differentially inhibited (Klinkradt, Naudé, Muramoto, & Oelofsen, 1997). An attempt to clarify the intricate mechanism by which the three peptidasic activities of 20S proteasome are regulated by fatty acids was made by Yamada and co-authors, who reported that the pattern of activation of the T-L peptidase is distinctly different from those of CT-L and PHPH-L; thus, linoleic and oleic acids strongly activated both CT-L and the C-L hydrolase-type activities in a biphasic activation pattern (Yamada et al., 1998). Conversely, the activation pattern of tryptic-type peptidase occurs in a tri-phasic manner through an inhibition over the low concentration range, activation in a middle concentration range and inhibition again over a higher concentration range. These apparently conflicting results were explained by hypothesizing the existence of two classes of binding sites, namely “latency sites” and “activation sites”, and the fatty acid activation or inhibition phenomena have been interpreted as the result of binding to these different sites.
Over last decades, the studies on isolated/purified proteasome have been replaced by investigations performed through cellular or clinical studies. Some of these reports focused on the role of polyunsaturated fatty acids on protein- breakdown in muscle mass of cachectic cancer patients. As an example, the neuroprotective effects of Long-Chain Polyunsaturated Fatty Acids (LCPUFA) have been ascribed to some modulatory effects on the UPS, albeit no evidences on direct interaction with 20S proteasome have been reported (Undurti, 2006). Docosahexaenoic acid, the most unsaturated omega-3 fatty acid, displaying pro-apoptotic activity against tumour cells, was reported to exert its anti-cancer activity acting on UPS, even though no evidence of a direct interaction with 20S has been reported (Jing et al., 2014). There is also evidence describing the inhibitory (Hamel, 2009) effect on proteasome by the saturated fatty acid palmitate; this fatty acid is believed to contribute to type-2 diabetes, blocking UPS activity with a consequent lipotoxic effect on pancreatic beta cells. Recently, fatty acid derivatives have been designed to obtain gate-opening 20S stimulators with drug-like properties (Coleman et al., 2019). Using an arachidonic acid derivative AM-404, a very potent but toxic molecule, a series of molecules containing the aminophenol head group linked to aliphatic chains of varying length and degree of unsaturation have been synthesized and characterized. Their effect on the 20S activity indicates that, beside the chain length, saturated chains are generally not able to stimulate the activity of the 20S, while the most important structural feature useful to induce the stimulator activity on 20S, is the cis-double bond in the carbon chain. Furthermore, a greatly diminished capacity to stimulate the 20S was observed when the phenolic amide was substituted by aryl groups. These derivatives still need a careful pharmacological evaluation, also taking into account that, beyond their role as a nutritional energy source, fatty acids have several molecular targets, such as enzymes, receptors, and they are increasingly considered as important signalling molecules that can induce several physiological and pathophysiological effects.
4.5.3. Repositioning “old drugs” to activate the proteasome: the case of aminopyrine
Although over the last decade our knowledge of human diseases has greatly increased, its translation into new drugs and therapeutic benefits has been much slower than expected (Ashburn & Thor, 2004; Scannell, Blanckley, Boldon, & Warrington, 2012) The reasons that may explain this apparent incongruity are multi-faceted and include the increased time needed to pipeline new drugs to the market, a high attrition rate of drug candidates in clinical trials (Pammolli, Magazzini, & Riccaboni, 2011), and rapidly changing regulatory requirements. Some reports estimate that, on average, for every dollar invested by the pharmaceutical industry in research and development (R&D) less than a dollar is returned, thus suggesting that investments on R&D will rapidly decline in the very next future (Pushpakom et al., 2019).
Therefore, drug repositioning (or repurposing), an approach to identify new medical applications for drugs, already approved for different therapeutic uses (Nosengo, 2016), offers a number of advantages over the development of entirely new drugs, such as (i) the reduced costs in the case of failure, and (ii) the shorter time interval for the transfer to the market because safety assessment has been already completed. Thus, given the urgent need to find a treatment for neurodegenerative diseases (such as AD, PD and HD), it is not surprising that an increasingly large number of existing drugs are tested for these disorders; in this respect, an important example is the repositioning of galantamine, one of the drugs now available on the market for the treatment of AD (Durães, Pinto, & Sousa, 2018).
Despite many examples of drug repositioning have been based on serendipity, a rational development of a repurposed drug implies a detailed knowledge of the pathways involved. As an example, there is evidence that AD development is associated to aggregation of the neurotoxic amyloid β (Aβ) peptide (Kang et al., 1987), being the consequence of a failure of proteasome function and a consequent accumulation of poly-ubiquitinated substrates, as detected in AD neuronal tissues (Perry, Friedman, Shaw, & Chau, 1987). In particular, in response to the increased oxidative and proteotoxic stress, the percentage of uncapped 20S proteasome is significantly increased in AD neuronal tissues in comparison to healthy cells (Wang, Yen, Kaiser, & Huang, 2010). Moreover, the Aβ peptide, as well as other intrinsically disordered proteins (IDPs), is a substrate of the 20S proteasome.
On these premises, it has been reported that pyrazolones, a class of small molecules extensively used in the past as painkillers and antipyretic drugs, induce proteasome activation in mice moels of Amyotrophic lateral Sclerosis (Trippier et al., 2014). In a recent report, it was demonstrated that some members of this class of molecules (i.e., aminopyrine, 4-aminoantiypirine and nifenazone) may enhance CT-L proteasome activity in tube tests (Santos et al., 2019). Proteasome activity assays, carried out in parallel in the presence of an excess of reducing agents (i.e., ascorbic acid or glutathione), underscored that proteasome activation by pyrazolones is not directly related to their antioxidant properties, thus suggesting that an alternative mechanism of action should be proposed. In the case of aminopyrine, the evidence that it is able to activate native 20S, but it is ineffective on a mutant (i.e., α3ΔN, which has a permanently opened gate, see Section 2.2.2), envisaged that its effect on proteasome activity is mainly related to enhanced dynamics of the outer α-rings, which is a common mechanism of proteasome activation by small molecules (see Section 2.2.2) (Njomen & Tepe, 2019). Furthermore, the effects on the different cellular proteasome forms (i.e., 20S, 26S and 30S, see 2.2.2, 2.3.2) were also assayed in neuroblastoma SH-SY5Y cells by separating proteasome assemblies in non-denaturing gels. It was observed that proteasome assemblies resulted significantly stimulated two hours after the treatment with the drug, even though this effect vanishes over 24 h after stimulation, reflecting the pharmacokinetic properties of aminopyrine (with a half-life in blood serum of approximately 2 h). Docking simulations, performed using aminopyrine, antipyrine, 4-aminoantipyrine and nifenazone as ligands of human 20S, have outlined that they interact with α-rings, involving the α1/α2 and α5/α6 binding pockets; the most active molecules display a binding free energy ~30 Kcal/mol more favourable than the less active ones. In particular, it was observed that aminopyrine bridges α1and α2 subunits since its phenyl ring is involved in hydrophobic interactions with residues L22, Y25, E26, A126, and A157 of the α1 subunit; the oxygen atom of aminopyrine is also H-bonded with Y159 and the N-methyl group with residue A32 of the same subunit. Moreover, residues G30, G31 and A32 of the α2 subunit are linked by non-polar interactions to the 4-(dimethylamino) group of aminopyrine. Next, T-shaped stacking interactions bridge the residue F162 of the α5 subunit with the phenyl ring of aminopyrine, and the residue Q60 of the α6 subunit turns out to be H-bonded to the oxygen atom of the ligand. It is important to remind here that the α1/α2 grooves are the preferential anchoring sites of the HbYX motif which binds the 20S with the Rpt3 subunit of the regulatory particle 19S (see 2.2.2, 2.3.2) (Smith et al., 2007); furthermore, small molecules, known to mobilize 20S gating dynamics, bind the α5/α6 grooves (Di Di Dato et al., 2017). Notably, the inactive compound antipyrine mainly interacts through T-shaped aromatic interactions and H-bonds with two residues of α2 subunit Y159 and Y160, respectively, but no bridging interactions with any other subunit of the α-ring are observed (see Figs. 5A and 6A in Santos et al., 2019). MALDI-MS experiments, performed using as a substrate Aβ1–28, a water-soluble fragment of the amyloid peptide, further demonstrated that aminopyrine may enhance the rate of peptide degradation. Cell viability assays, carried out on differentiated SH-SY5Y neuroblastoma cells, underscored the neuroprotective properties of aminopyrine, and further experiments, conducted in the presence of the proteasome inhibitor bortezomib, demonstrated that aminopyrine rescues neuron-like cells from Aβ proteotoxicity with a mechanism of protection mostly related to proteasome activation. These results are likely to stimulate further studies focusing on proteasome activation by repurposed drugs and, ultimately, to relaunch investments from pharmaceutical industries in this risky area.
5. Insights on additional potential applications of proteasome inhibition: a role in SARS-Covid19 therapy?
Although not deeply investigated yet, proteasome inhibitors discussed so far display a known anti-inflammatory activity, envisaging a therapeutic efficacy in combined regimen in subjects with acute severe inflammatory processes, such as in viral infections. This potentiality assumes a particularly updated importance nowadays during the recent pandemia due to SARS-Cov-2, which is likely to infect millions of people worldwide with a significant lethality rate. Besides the public health and victim tolls, which are by far the most urgent topics, disease spread is dramatically impacting on world social and economic activities, making even more urgent the identification of a specific therapy or a vaccine (Andersen, Rambaut, Lipkin, Holmes, & Garry, 2020; Sheeren et al., 2020; Baden & Rubin, 2020; Lipsitch, Swerdlow, & Finelli, 2020).
The limited clinical and laboratory data available so far suggest that in most cases SARS-Cov-2 infection evolves through symptoms overlapping those of canonical flu, even though a very large number of subjects do not develop symptoms (Baden & Rubin, 2020; Zhang et al., 2020). In a limited, but significant, especially for the health assistance burden, number of cases, infection progresses toward a clinical picture of interstitial pneumonia sustained by the massive stimulation of the immune system the virus appears to be able to elicit (Baden & Rubin, 2020; Zhang, Lin, et al., 2020).
The cytokine storm which underscores this disease progression often induces Acute Respiratory Distress Syndrome (ARDS) and diffuse thrombotic angiopathy which are, to date, the prevalent cause of death of ill patients.
Nonetheless, the massive inflammatory response rather than virus replication is gaining increasingly relevance as the real target of therapy. This is emphasized by the apparent efficacy of therapies based on biological drugs which target the pro-inflammatory cytokines (Baden & Rubin, 2020). In any case, specific trials will definitively address the therapeutic efficacy of these approaches, hopefully allowing to identify a therapeutic regimen which can be early undertaken to prevent disease complication and sanitarian costs.
The most studied models for SARS-CoV infection, spread in 2003; encompass: (i) original strains of SARS-CoV isolated from human subjects for infection of cell cultures in vitro; (ii) the Coronavirus Mouse Hepatitis virus (MHV) infection in murine models. MHV belongs to the same coronavirus genus of SARS-CoV and displays significant similarity concerning both structural features and pathogenesis, including the marked innate immune inflammatory cytokine release (Ma et al., 2010).
Among studies, reported to date, there is a compelling evidence that the UPS could regulate the virus infectious cycle at multiple levels, with the exception of virus internalization which occurs through endocytosis following recognition of Angiotensin Converting Enzyme-2 (ACE2) receptor on host cells by the spike protein (S) (Mathewson et al., 2008; Li et al., 2007; Li et al., 2003). Interestingly, chemical inhibitors of proteasome induced the virus particles to accumulate in late endosomes and lysosomes, suggesting a UPS role in virus release from endosomes (Yu & Lai, 2005). Once released in the cytoplasm, the nucleic acid of SARS-Cov (a positively single stranded RNA) encodes four structural proteins, nucleocapsid (N), envelope (E), membrane (M) and spike (S) proteins and about 16 non-structural proteins which are translated as a single poly-protein (Yu & Lai, 2005; Li, Sui, et al., 2007; Schneider et al., 2012). The highly antigenic protein N of SARS-CoV, which is an extensively glycosylated and positively charged protein of the nucleocapsid, forms a helical ribonucleoprotein complex with the viral RNA, and it was found to interact with ATPase 6 (i.e., a 19S subunit) in lung fibroblasts infected with the virus (Wang, Xu, et al., 2010). Therefore, although further studies are required to confirm this hypothesis, a direct down-regulation of proteasome complexes by SARS-CoV can be envisaged, since in the intracellular space the ATPase 6 subunit (because of its position in the multi-subunits complex, see Fig. 2 and Lander et al., 2012) could be accessible for binding also when assembled in 19S particles; as a consequence, proteasome inhibition would appear as a strategy to halt the antigenic processing of virus proteins. If so, this process should be properly balanced, since proteasome activity is further relevant for advancement of cell cycle, especially G1-S transition which allows the virus to replicate (Wang, Xu, et al., 2010).
Studies on non-structural proteins, which intervene either in the virus replication and in the interaction with the host machineries, besides confirming that the UPS does not affect virus internalization, envisaged that virus-UPS interaction may account for the mechanisms of immune system evasion (Wong et al., 2018; Liu et al., 2014; Hu, Yen, Singh, Kao, & Wu-Hsieh, 2012). In this respect, the SARS-CoV Papain-like proteasome (PLpro), which, along with the main protease Mpro (also called 3CLpro), is essential for the cleavage and processing of the viral poly-protein, was found to repress IFNγ synthesis and secretion in lung cell cultures by targeting IRF3 phosphorylation and nuclear translocation (Devaraj et al., 2007; Zhang et al., 2020). Interestingly, PLpro was reported to have DUB activity in human cells by recognizing the LXGG consensus de-ubiquitination motif and by directly binding to the proteasome through the N-terminal Ub-like domain which is present in its structure (Ma et al., 2010). Although the DUB activity was shown to be ineffective in altering the repertoire of host poly-Ub proteins, since Ub-conjugation is a post-translational mechanism of regulation of intracellular trafficking and receptor signalling, it would be relevant to uncover the link between IRF3 signalling and Ub-labelling by PLpro.
In this framework, viral accessory protein 3a was reported to promote IFN-typeI receptor ubiquitination and proteasome degradation (Minakshi et al., 2009). Along with this, accessory proteins 8a and 8b or 8ab, which were found to be expressed either as a single protein or spliced protein in different viruses at different stages of SARS-CoV infection, were found to bind to intracellular ubiquitinated proteins through an extensively glycosylated functional domain which account for ubiquitin binding and ubiquitin conjugation (Le et al., 2007; Keng et al., 2011; Li & Johnson, 2012). Interestingly, 8b and 8ab appear to stimulate the UPS-mediated degradation of IRF3 at later stages of virus replication than PLpro was supposed to do (Wong et al., 2018). Remarkably, proteasome inhibition was found to be ineffective in the assembly of virus (Raaben, Grinwis, Rottier, & de Haan, 2010; Raaben et al., 2010). Although proteasome inhibition appears ineffective in the assembly of virus (Raaben, Grinwis, et al., 2010; Raaben, Posthuma, et al., 2010), the role of the UPS in the virus infectious cycle is a relevant topic, and its putative role was suggested to rely in M-calpain rather than proteasome inhibition (Schneider et al., 2012). What looks more convincing is that the UPS plays a key role in handling the immune response to the pathogen, but also in the aberrant inflammatory response the virus may elicit. From the first point of view, bortezomib treatment increased disease progression in the liver of mice infected by MHV, underscoring the relevance of the protective role UPS in inflammatory response against the viral agent (Raaben, Posthuma, et al., 2010). Several independent investigations on different microbial pathogens reveal that a major role for the UPS is to drive antigenic processing and inflammation activation especially in monocytes/macrophages (Forget, Gregory, & Olivier, 2005; Horan et al., 2013; Silswal, Reis, Qureshi, Papasian, & Qureshi, 2017). Furthermore, proteasome dysfunction in alveolar type II epithelial cells is associated with ARDS in the alveolar space of Rpt3-KO mice (Sitaraman et al., 2019). From the second point of view, besides the known contribution in modulating the transcriptional activity of nuclear factors, such as NF-kB (see Section 3.2.1), additional tissue-specific mechanisms which should intervene in coronavirus infection are expected to take part to the pathogenesis. In this regard, the clearance of specific proteins, such as elafin by the exceedingly activated UPS might be a relevant factor in sustaining inflammation. In fact, elafin is a serine-proteasome inhibitor and its inhibitory activity resides within the C-terminal domain which has specificity for NE and proteinase 3. Notably, transglutaminase substrate binding motif (GQDPVK) is present at the N-terminus which allows it to cross-link extracellular matrix proteins (Kerrin et al., 2013).
In addition, clinical studies highlight that the 20S proteasome is released in the alveolar space during ARDS in an active configuration, and that immune proteasome subunits are increased in the alveolar space envisaging a prognostic relevance of its quantification (Sixt et al., 2009; Sixt et al., 2012; de Bruin et al., 2016). As a whole, the disastrous SARS-Cov-2 experience suggests that multi-faceted efforts by the scientific community are demanded to clarify the pathogenesis of coronaviruses especially in view of the concrete possibility that additional spill-overs in the next future might come up posing new pandemic threats. The central role of the UPS in regulating the complex dynamic of interactions between the pathogen and the host, along with the growing interest in the development of UPS modulators, could provide further clues for the identification of valid approaches which allow to limit the sanitarian, social and economic costs of similar pandemia.
Box 1: Elements in proteasome heterogeneity
Proteasome is a highly dynamic complex as demonstrated by the existence of alternative forms of proteasome which deal with specific biological roles. The immun-oproteasome is the most studied alternative form of proteasome and its proteolytic activity has been long linked only to generation of antigenic peptides for MHC class I presentation (Murata, Takahama, Kasahara, & Tanaka, 2018; Rousseau & Bertolotti, 2018). However, a number of studies have reported a role for immunoproteasome in B and T cell differentiation, monocytes and dendritic cells activation, in the maintenance of pluripotency of stem cells (Atkinson et al., 2012), in the differentiation of non-immune cells, such as skeletal muscle ones, and also in the homeostasis of nervous cells (Kaur & Batra, 2016; Kimura, Caturegli, Takahashi, & Suzuki, 2015). Along with this, immunoproteasome dys-regulation has also been associated with various human diseases, including cancer, immune and inflammatory disorders: in fact, either hyper-activation or hypo-activation may turn out into a hyper-immune or hypo-immune phenotype (Eskandari, Seelen, Lin, & Azzi, 2017). Therefore, there has been a great effort to develop specific immunoproteasome inhibitors, which showed minimal cross-reactivity with constitutive proteasome. From the structural point of view, the immunoproteasome differs from the canonical 20S for the replacement of catalytic subunits with its immune highly homolog counterparts β1i (LMP2), β2i (MECL1), and b5i (LMP7) (Sherman & Li, 2020; Sijts & Kloetzel, 2011). Remarkably, these immunoproteasome subunits are constitutively expressed in different tissues, such as thymus and spleen. Moreover, non-immune cells preferentially incorporate them during the assembly de novo of 20S particles following exposure to proinflammatory stimuli (e.g., IFN-γ, TNF-α, and lipopolysaccharide) or cytokine-independent stressors (e.g., oxidative stress) (Griffin et al., 1998; Heink, Ludwig, Kloetzel, & Krüger, 2005; Ferrington & Gregerson, 2012; Murata et al., 2018). Immune-subunits incorporation has been proposed to proceed cooperatively, since the direct binding of β5i to chaperone POMP is followed by the quick recruitment of b1i and b2i. Therefore, the rate of their assembly is about four times faster than that of canonical subunits in the forming 20S, a finding consistent with the primary biological role of immunoproteasome, which is demanded to cope with patho-physiological challenges in a dynamic and highly efficient manner (Griffin et al., 1998; Groettrup, Standera, Stohwasser, & Kloetzel, 1997; Murata et al., 2018; Murata et al., 2001). The subunit substitution accounts for a shift in the catalytic preferences and activity; in fact, immunoproteasome exhibits elevated level of CT-L and T-L activities which favour the production of peptides with terminal basic or hydrophobic residues that fit better into the cleft of the MHC class I molecule (Murata et al., 2018; Rousseau & Bertolotti, 2018).
IFN-γ also induces the expression of another important regulator, besides 19S, of 20S activity, the 11S regulator (PA28) (Sherman & Li, 2020; Cascio, Hilton, Kisselev, Rock, & Goldberg, 2001; Cascio, Call, Petre, Walz, & Goldberg, 2002; Cascio, 2014).
Mammalian cells express three different subunits of 11S regulator: PA28α, PA28β, and PA28γ. PA28α and PA28β assemble into a hetero-heptameric complex, primarily located in the cytoplasm, while homo-heptameric PA28γ is mainly present inside the nucleus (Wójcik, Tanaka, Paweletz, Naab, & Wilk, 1998; Cascio, 2014). Although the role of these regulators is not clear, it has been reported that both forms increase after oxidative stress, suggesting their involvement in the degradation of damaged proteins (Pickering et al., 2010; Pickering et al., 2012; Kors, Geijtenbeek, Reits, & Schipper-Krom, 2019; Thibaudeau and Smith, 2018). Accordingly, PA28α-β association with the 20S does not enhance the degradation of poly-ubiquitinated protein/peptide substrates in vitro (Li, Powell, & Wang, 2011; Cascio, 2014; Lobanova et al., 2018). In addition to its role in preserving cellular homeostasis after oxidative stress, the PA28 role in the regulation of the immune response has been extensively studied (Tanahashi et al., 1997; Früh & Yang, 1999; Preckel et al., 1999; Cascio, 2014). In fact, following INFγ stimulation, the level of PA28α-β binding to inducible 20S increases, enhancing its proteolytic activity and mediating the generation of antigenic peptides (Groettrup et al., 1996; Sijts et al., 2002; Sijts et al., 2011; Fort, Kajava, Delsuc, & Coux, 2015). Unlike PA28α-β, PA28γ is not induced by IFNγ, suggesting a different biological role for PA28γ-20S complex. Although the role of this complex remains elusive, a number of studies imply an involvement in the cell cycle progression (Kors et al., 2019).
In addition to immunoproteasome, other tissue-specialized forms of proteasome are thymo-proteasome and testis-proteasome (spermato-proteasome). The first one is expressed by cortical thymic epithelial cells and contains two immune catalytic subunits, β1i and β2i, and a thymus specific subunit (β5t), that, unlike β5 and β5i, is characterized by a number of hydrophilic amino acids in its catalytic pocket. Thus, thymo-proteasome produces a distinct spectrum of peptide fragments, and cells expressing it display a unique set of peptides associated with MHC-I molecules (Florea et al., 2010; Murata et al., 2007; Sasaki et al., 2015). Thymo-proteasome role accounts for the positive selection of developing T cells, since it is essential to optimize the release of the repertoire of peptides for CD8+ T cell (Murata et al., 2018; Nitta et al., 2010; Takada et al., 2015; Xing, Jameson, & Hogquist, 2013).
Spermato-proteasome is a testis-specific form of proteasome (described exclusively in spermatocytes, spermatids and sperm), and it is characterized by a chronologically defined expression. It contains a specific α4 subunit (α4s) (PMSA8 gene) in the place of corresponding constitutive α-subunit, whose incorporation into a newly formed 20S is mutually exclusive and does not alter the catalytic preferences of the constitutive 20S (Morozov & Karpov, 2019; Qian et al., 2013; Uechi et al., 2014). Elevated expression of PSMA8 has been identified in different tumours, such as large B-cell lymphoma, thymoma, and testicular germ cell tumours. However, the biological significance and the possibility to make α4s a therapeutic target candidate has not been elucidated yet (Bruggeman, Koster, Lodder, Repping, & Hamer, 2018; Morozov & Karpov, 2019). The incorporation of α4s in spermato-proteasome seems to favour the 20S association with another gate-activating RP, namely PA200 (Blm10 in yeast) (Qian et al., 2013). PA200 is a nuclear-specific proteasome activator and it is expressed in all mammalian tissues, but it is particularly abundant in the testis, where it plays a crucial role in spermatogenesis (Khor et al., 2006; Ustrell, Pratt, Gorbea, & Rechsteiner, 2005). Accordingly, PA200 deletion markedly reduces fertility of male mice (Khor et al., 2006), and the PA200/20S spermato-proteasome complex catalyses ubiquitin-independent degradation of acetylated core histones during DNA repair and spermatogenesis (Qian et al., 2013). PA200 also binds constitutive proteasome and the amount of PA200/constitutive 20S, as well as of PA28/20S complex, increases upon 26S inhibition, contributing to adapt the pool of different proteasome populations to the cell condition (Welk et al., 2016). The crystal structure of this complex has revealed that C-terminal HbYX motif in PA200 fits between α5 and α6 inter-subunit pocket, mediating 20S gate opening (Sadre-Bazzaz et al., 2010; Witkowska et al., 2017). Interestingly, it strongly stimulates the rate of C-L activity, although its biological significance is poorly clear yet (Ustrell, Hoffman, Pratt, & Rechsteiner, 2002). Recently, it has been reported the identification of a non-canonical variant of constitutive 20S in mammalian cells, previously identified in yeast. It is known as alternative ‘α4-α4’ proteasome, which assembles upon replacement of α3 with an additional α4 subunit in the position normally occupied by the former (Kusmierczyk, Kunjappu, Funakoshi, & Hochstrasser, 2008; Padmanabhan, Vuong, & Hochstrasser, 2016; Velichutina, Connerly, Arendt, Li, & Hochstrasser, 2004). Importantly, mammalian cells, primed to assemble these alternative proteasomes, exhibit enhanced resistance to cellular stress induced by metal ions (Padmanabhan et al., 2016).
The existence of interchangeable subunits, and thus, of alternative proteasome forms above described, as well as the identification of hybrid proteasome particles (i.e., 19S–20S-11Sα-β, 19S–20S-11Sγ,19S–20S-PA200) whose biological function is poorly known, underlie how cells modify proteasome repertoire in relation to its specific needs (Cascio et al., 2002; Morozov & Karpov, 2019; Thibaudeau & Smith, 2019).
Box 2: Inhibition of UPS targetting deubiquitinases: Usp14 at a glance
De-ubiquitinases (DUBs), which catalyse ubiquitin moieties removal from target proteins, are key components of the UPS, being involved in ubiquitin recycling and editing (Yuan et al., 2018). Three DUBs are associated with the proteasome: Rpn11, a Zn2+ metallo-protease, which is part of the lid, USP14 and Uch37 which are two cysteine proteases, extrinsically associated with the base (see Section 2.3.1) (D'Arcy, Wang, & Linder, 2015). Several studies have proposed that de-ubiquitination by Rpn11 stimulates the substrate degradation by removing bulky ubiquitin chains that otherwise might impair further substrate translocation into the proteasome (de Poot et al., 2017) on the other hand, the ATP/independent de-ubiquitination by Usp14 and Uch37 is envisaged to suppress substrate degradation, promoting its premature dissociation from the proteasome (Lam et al., 1997; Hanna et al., 2006; Lee et al., 2010; Lee et al., 2016b). Among the three DUBs, Usp14 is the most attractive way of intervention to regulate proteasome activity (Chakraborty et al., 2018; Wertz & Murray, 2019). Human USP14 consists of two domains, namely (i) a N-terminal ubiquitin-like domain (UBL) and (ii) a C-terminal DUB domain, which contains the catalytic triad, Cys114, His435, and Asp451. In the free unbound state, the catalytic domain of Usp14 is characterized by a low level of de-ubiquitinase activity, whereas when it binds proteasome its activity is increased by about 800-fold and shows a preference for substrates ubiquitinated at more than one site (Koulich, Li, & DeMartino, 2008; Hu et al., 2005; Lee et al., 2010; Lee et al., 2016b). It has been shown that Usp14 inhibition stimulates the degradation of some specific proteasome substrate in mammalian cells, such as cancer and neurodegeneration related proteins (Lee et al., 2010; Homma et al., 2015; Zhu, Zhang, et al., 2016a; McKinnon et al., 2016; Boselli et al., 2017; Liao et al., 2017; de Poot et al., 2017). Additionally, Usp14 activation through phosphorylation of Ser432 residue by AKT results in the suppression of the degradation of short-lived proteins, that may in turn promote tumour cell proliferation (Kim & Goldberg, 2017; Wei et al., 2017; Xu et al., 2015). Some evidences also suggest that Usp14 expression is closely related to the onset of different tumours, including breast, gastric and lung cancer (Wu et al., 2013; Zhu, Zhang, et al., 2016b; Zhang et al., 2017; Fu et al., 2018) and its activity is required for nervous system development and functioning (Chen, Retzlaff, et al., 2011; Kiprowska et al., 2017; Vaden et al., 2015; Wilson et al., 2002). Thus, considerable efforts have been dedicated to the discovery of small molecules that functionally inhibit Usp14 and several ones have been identified, such as b-AP15, auranofin, WP1130 and curcumin analogue AC17 (Kapuria et al., 2010; Zhou et al., 2013; Wang et al., 2014; Coughlin et al., 2014; Liu et al., 2014; Tian et al., 2014; D'Arcy et al., 2015; Wang et al., 2015; Wang et al., 2016; Wang et al., 2018; Xia et al., 2019; Wertz & Murray, 2019; Ma et al., 2020). A common feature of most of these compounds is the presence of α,β-unsaturated carbonyl groups which can form covalent adducts with free thiols in the active site by Michael addition (D'Arcy et al., 2015; Wang et al., 2015). Unfortunately, these compounds usually have poor selectivity across the DUB family, since most of them are cysteine enzymes which are easily “druggable” by compounds containing Michael acceptors (D'Arcy et al., 2015). The small molecule IU1 was the first specific inhibitor identified, exhibiting excellent selectivity for USP14 over other DUBs (Lee et al., 2010; Lee et al., 2016). Co-crystal studies reveal a unique mechanism of action of IU1 that exerts its inhibitory activity by binding to the thumb-palm cleft region of Usp14 catalytic domain, sterically preventing ubiquitin binding to the C-terminal of Usp14 (Wang et al., 2018). Interestingly, the prion protein shows accelerated degradation upon IU1 treatment, and a more powerful analogue (IU1–47) enhances tau degradation in neurons (Boselli et al., 2017; Homma et al., 2015; McKinnon et al., 2016), rendering it an intriguing target also for neurodegenerative diseases. VLX1570 is an analogue of b-AP15, being characterized by a higher potency and an improved solubility, which shows consistent anti-tumour activity in orthotopic and xenograft models of MM, lymphoma, Ewing's sarcoma, and other malignancies (D'Arcy et al., 2014; Chitta et al., 2015; Wang et al., 2016; Shukla et al., 2016). Moreover, VLX1570 retains prominent activity in bortezomib-resistant MM cells (Rowinsky et al., 2020; Shukla et al., 2016). These studies together with the good tolerability profile, reported in preclinical models, provided the rationale for investigating this drug in clinical trials on patients with RRMM (Rowinsky et al., 2020). Thus, in a phase 1 study fourteen patients with RRMM were enrolled and treated with escalating doses of intravenous infusion of VLX1570 ranging from 0.05 to 1.2 mg/kg and anti-myeloma effects were observed at dose of 0.6 mg/kg or more. Unfortunately, two patients treated with 1.2 mg/kg dose experienced severe and progressive respiratory insufficiency, culminating in death; thus, due to severity of the toxicity, the study was discontinued (Rowinsky et al., 2020). Beside this molecule, no other inhibitors targeting DUBs have entered into clinical trial so far. However, since they are abnormally expressed in a variety of tumours and/or in tumour microenvironment (Yuan et al., 2018), making them ideal anticancer target candidates, the identification of selective small-molecule inhibitors for Usp14 and in general for other specific DUBs remains an active and extremely challenging task.
Box 3: Metabolic control of proteasome function
The regulation of metabolic control of proteasome function is a challenging point in proteasome biology, which deserves particular attention, although many aspects are still obscures. The coordinated balance of the two post-translational modifications (i.e., O-linked N-acetylglucosamine (O-GlcNAc) and phosphorylation) appears to be crucial in this process (Rousseau & Bertolotti, 2018;Zhang et al., 2003 ; Zhang et al., 2007; Fardini et al., 2015). Recent studies have shown that different kinases, such as CaMKII, PKG, DYRK2 and PKA, induce the phosphorylation of proteasome subunits. In particular, the activation of PKA, which follows the increase of cAMP level, results in the phosphorylation of Ser14 of Rpn6, leading to proteasome activity enhancement (VerPlank and Goldberg, 2018, VerPlank and Goldberg, 2017). In vitro and in vivo studies have shown that a series of hormones with different biological functions, spanning from energy control (i.e., glucagon and epinephrine) to water resorption (i.e., antidiuretic hormone), stimulate adenylate cyclase, thus raising cAMP level and enhancing, through the Rpn6 phosphorylation, degradation by 26S proteasome of short-lived regulatory proteins and/or damaged proteins (VerPlank et al., 2019). These findings suggest that proteasome activation, mediated by CAMKII and cAMP-PKA, could be a common cellular mechanism to answer a series of endocrine stimuli which, inducing a quick destruction of regulatory and/or damaged proteins, should help cells to adapt their proteome to the novel conditions determined by the hormones exposure (VerPlank et al., 2019). O-GlcNAc is a post-translational modification that occurs at serine or threonine residues, which are also a target site of phosphorylation. It is catalyzed by the enzyme O-linked N-acetylglucosamine transferase, which modulates the addition of a GlcNAc moiety to target proteins, by using as substrate a product of hexosamine biosynthesis, the UDP-GlcNAc, whose availability is influenced by nutritional conditions. In fact, an increase in glucose availability raises UDP-GlcNAc levels and consequently promotes protein O-GlcNAc (Comer & Hart, 2000; Zachara & Hart, 2004). It has been reported that 26S proteasome function is inhibited by the addition of sugar moieties to the Rpt2 subunit (Zhang et al., 2003), a finding which provides a link between glucose metabolism and protein turnover (Zhang et al., 2007). In the liver and muscle tissues of mouse models, when blood glucose drops (e.g., under fasting conditions or during exercise) the PKA increase stimulates 26S activity selectively toward the clearance of short-lived regulatory proteins, with no alterations in proteasome content, confirming that proteasome activation occurs through post-synthetic modification of already existing particles (Rousseau & Bertolotti, 2018;VerPlank et al., 2019). The low level of glucose also reduces the entry of this mono-saccharide into the hexosamine pathway, limiting the availability of UDP-GlcNAc; as a consequence, the O-GlcNAc modification of Rpt2 decreases thus removing the signal that inhibits proteasome function (Zhang et al., 2003; Zhang et al., 2007). Moreover, it has been shown that lack of nutrient rapidly inhibits the stress and nutrient response of the mTOR complex. It results into the enhancement of autophagic process and of the rate of long-lived protein ubiquitination and, therefore, of their degradation by UPS (Zhao et al., 2015). All these events bring to a general activation of proteolysis, promoting cellular adaptation, facilitating damaged and potentially toxic protein clearance, and providing essential amino acids for the synthesis of proteins necessary for cell survival and energy production (Zhao et al., 2015; VerPlank and Goldberg, 2015; Rosseau and Bertolotti, 2016).
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgment
The authors would like to thank Prof. G. Manni for his fruitful discussion.
This work was supported by the Italian Ministery of University and Research (PRIN 2017SNRXH3), the Italian Association for Cancer Research (AIRC) under IG 2017 - ID. 20353 project-PI_Grazia Graziani, the Italian Ministry of Health RC18-2638151 to P.M.L. Special thanks are due to Omikron Italia SrL for a liberal donation.
Contributor Information
G.R. Tundo, Email: grazia.tundo@libero.it.
G. Graziani, Email: graziani@uniroma2.it.
M. Coletta, Email: coletta@med.uniroma2.it.
References
- Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nature Reviews. Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. The proteasome: structure, function, and role in the cell. Cancer Treatment Reviews. 2003;29(Suppl. 1):3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Adams J. The proteasome: a suitable antineoplastic target. Nature Reviews. Cancer. 2004;4(5):349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- Adams J., Behnke M., Chen S., Cruickshank A.A., Dick L.R., Grenier L.…Stein R.L. Potent and selective inhibitors of the proteasome: Dipeptidyl boronic acids. Bioorganic & Medicinal Chemistry Letters. 1998;8(4):333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- Adams J., Palombella V.J., Sausville E.A., Johnson J., Destree A., Lazarus D.D.…Elliott P.J. Proteasome inhibitors: A novel class of potent and effective antitumor agents. Cancer Research. 1999;59(11):2615–2622. [PubMed] [Google Scholar]
- Agarwal R., Gupta S.K., Agarwal P., Saxena R., Agrawal S.S. Current concepts in the pathophysiology of glaucoma. Indian Journal of Ophthalmology. 2009;57(4):257–266. doi: 10.4103/0301-4738.53049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous A., Rohatiner A., Rule S., Hunter H., Kerr J.P., Neeson S.M.…Lister A. Weekly versus twice weekly bortezomib given in conjunction with rituximab, in patients with recurrent follicular lymphoma, mantle cell lymphoma and Waldenström macroglobulinaemia. British Journal of Haematology. 2010;151(4):346–353. doi: 10.1111/j.1365-2141.2010.08340.x. [DOI] [PubMed] [Google Scholar]
- Aggarwal B.B. Nuclear factor-kappaB: The enemy within. Cancer Cell. 2004;6(3):203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Aghdam S.Y., Gurel Z., Ghaffarieh A., Sorenson C.M., Sheibani N. High glucose and diabetes modulate cellular proteasome function: Implications in the pathogenesis of diabetes complications. Biochemical and Biophysical Research Communications. 2013;432(2):339–344. doi: 10.1016/j.bbrc.2013.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agholme L., Nath S., Domert J., Marcusson J., Kågedal K., Hallbeck M. Proteasome inhibition induces stress kinase dependent transport deficits - implications for Alzheimer's disease. Molecular and Cellular Neuroscience. 2014;58:29–39. doi: 10.1016/j.mcn.2013.11.001. Scopus. [DOI] [PubMed] [Google Scholar]
- Aguileta M.A., Korac J., Durcan T.M., Trempe J.-F., Haber M., Gehring K.…Husnjak K. The E3 ubiquitin ligase parkin is recruited to the 26 S proteasome via the proteasomal ubiquitin receptor Rpn13. The Journal of Biological Chemistry. 2015;290(12):7492–7505. doi: 10.1074/jbc.M114.614925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian T.N., Kisselev A.F., Goldberg A.L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. The Journal of Biological Chemistry. 1997;272(3):1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- Al Mamun A., Uddin M.S., Kabir M.T., Khanum S., Sarwar M.S., Mathew B.…Ashraf G.M. Exploring the promise of targeting ubiquitin-proteasome system to combat Alzheimer's disease. Neurotoxicity Research. Scopus. 2020 doi: 10.1007/s12640-020-00185-1. [DOI] [PubMed] [Google Scholar]
- Albert M.-C., Brinkmann K., Kashkar H. Noxa and cancer therapy. Molecular & Cellular Oncology. 2014;1(1) doi: 10.4161/mco.29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsson-Lindblad A., Kolstad A., Laurell A., Räty R., Grønbæk K., Sundberg J.…Jerkeman M. Lenalidomide-bendamustine-rituximab in patients older than 65 years with untreated mantle cell lymphoma. Blood. 2016;128(14):1814–1820. doi: 10.1182/blood-2016-03-704023. [DOI] [PubMed] [Google Scholar]
- Alderden R.A., Hall M.D., Hambley T.W. The discovery and development of cisplatin. Journal of Chemical Education. 2006;83(5):728. doi: 10.1021/ed083p728. [DOI] [Google Scholar]
- Alexanian R., Haut A., Khan A.U., Lane M., McKelvey E.M., Migliore P.J.…Wilson H.E. Treatment for multiple myeloma. Combination chemotherapy with different melphalan dose regimens. JAMA. 1969;208(9):1680–1685. doi: 10.1001/jama.208.9.1680. [DOI] [PubMed] [Google Scholar]
- Ali R.E., Rattan S.I. Curcumin's biphasic hormetic response on proteasome activity and heat-shock protein synthesis in human keratinocytes. Annals of the New York Academy of Sciences. 2006;1067:394–399. doi: 10.1196/annals.1354.05. [DOI] [PubMed] [Google Scholar]
- Alkalay I., Yaron A., Hatzubai A., Orian A., Ciechanover A., Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(23):10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almond J.B., Cohen G.M. The proteasome: A novel target for cancer chemotherapy. Leukemia. 2002;16(4):433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- Alsina M., Trudel S., Furman R.R., Rosen P.J., O'Connor O.A., Comenzo R.L.…Goy A. A phase I single-agent study of twice-weekly consecutive-day dosing of the proteasome inhibitor carfilzomib in patients with relapsed or refractory multiple myeloma or lymphoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18(17):4830–4840. doi: 10.1158/1078-0432.CCR-11-3007. [DOI] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Amiri K.I., Horton L.W., LaFleur B.J., Sosman J.A., Richmond A. Augmenting chemosensitivity of malignant melanoma tumors via proteasome inhibition: implication for bortezomib (VELCADE, PS-341) as a therapeutic agent for malignant melanoma. Cancer Research. 2004;64(14):4912–4918. doi: 10.1158/0008-5472.CAN-04-0673. [DOI] [PubMed] [Google Scholar]
- Amor-Gutiérrez O., Costa-Rama E., Arce-Varas N., Martínez-Rodríguez C., Novelli A., Fernández-Sánchez M.T., Costa-García A. Competitive electrochemical immunosensor for the detection of unfolded p53 protein in blood as biomarker for Alzheimer's disease. Analytica Chimica Acta. 2020;1093:28–34. doi: 10.1016/j.aca.2019.09.042. [DOI] [PubMed] [Google Scholar]
- An W.G., Hwang S.-G., Trepel J.B., Blagosklonny M.V. Protease inhibitor-induced apoptosis: accumulation of wt p53, p21 WAF1/CIP1 , and induction of apoptosis are independent markers of proteasome inhibition. Leukemia. 2000;14(7):1276–1283. doi: 10.1038/sj.leu.2401812. [DOI] [PubMed] [Google Scholar]
- Anandhan A., Rodriguez-Rocha H., Bohovych I., Griggs A.M., Zavala-Flores L., Reyes-Reyes E.M.…Franco R. Overexpression of alpha-synuclein at non-toxic levels increases dopaminergic cell death induced by copper exposure via modulation of protein degradation pathways. Neurobiology of Disease. 2015;81:76–92. doi: 10.1016/j.nbd.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nature Medicine. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov S., Takahashi M., Kakihana T., Katsuragi Y., Kitaura H., Zhang L.…Fujii M. G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. Scientific Reports. 2019;9(1):12896. doi: 10.1038/s41598-019-46237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao L., Reichel D., Hu D., Jeong H., Kim K.B., Bae Y., Lee W. Polymer micelle formulations of proteasome inhibitor carfilzomib for improved metabolic stability and anticancer efficacy in human multiple myeloma and lung cancer cell lines. The Journal of Pharmacology and Experimental Therapeutics. 2015;355(2):168–173. doi: 10.1124/jpet.115.226993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao L., Wu Y., Kim D., Jang E.R., Kim K., Lee D.-M.…Lee W. Development of peptide-based reversing agents for p-glycoprotein-mediated resistance to carfilzomib. Molecular Pharmaceutics. 2012;9(8):2197–2205. doi: 10.1021/mp300044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Nishiyama T., Imahashi N., Kitamura K. Efficacy of continuous, daily, oral, ultra-low-dose 200 mg acyclovir to prevent herpes zoster events among bortezomib-treated patients: A report from retrospective study. Japanese Journal of Clinical Oncology. 2011;41(7):876–881. doi: 10.1093/jjco/hyr063. [DOI] [PubMed] [Google Scholar]
- Arastu-Kapur S., Anderl J.L., Kraus M., Parlati F., Shenk K.D., Lee S.J.…Kirk C.J. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: A link to clinical adverse events. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2011;17(9):2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- Arba, M., Nur-Hidayat, A., Ruslin, Yusuf, M., Sumarlin, Hertadi, R., Wahyudi, S. T., Surantaadmaja, S. I., & Tjahjono, D. H. (2018). Molecular modeling on porphyrin derivatives as β5 subunit inhibitor of 20S proteasome. Computational Biology and Chemistry, 74, 230–238. 10.1016/j.compbiolchem.2018.03.002. [DOI] [PubMed]
- Arendt C.S., Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. The EMBO Journal. 1999;18(13):3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt A., Bauer I., Schafmayer C., Tepel J., Müerköster S.S., Brosch M.…Schäfer H. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009;28(45):3983–3996. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- Armon T., Ganoth D., Hershko A. Assembly of the 26 S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. The Journal of Biological Chemistry. 1990;265(34):20723–20726. [PubMed] [Google Scholar]
- Arnsburg K., Kirstein-Miles J. Interrelation between protein synthesis, proteostasis and life span. Current Genomics. 2014;15(1):66–75. doi: 10.2174/1389202915666140210210542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnulf B., Pylypenko H., Grosicki S., Karamanesht I., Leleu X., van de Velde H.…Moreau P. Updated survival analysis of a randomized phase III study of subcutaneous versus intravenous bortezomib in patients with relapsed multiple myeloma. Haematologica. 2012;97(12):1925–1928. doi: 10.3324/haematol.2012.067793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo A.P., Tanaka K., Goldberg A.L., Welch W.J. Identity of the 19S “prosome” particle with the large multifunctional protease complex of mammalian cells (the proteasome) Nature. 1988;331(6152):192–194. doi: 10.1038/331192a0. [DOI] [PubMed] [Google Scholar]
- Asai M., Tsukamoto O., Minamino T., Asanuma H., Fujita M., Asano Y.…Kitakaze M. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. Journal of Molecular and Cellular Cardiology. 2009;46(4):452–462. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Asano S., Fukuda Y., Beck F., Aufderheide A., Förster F., Danev R., Baumeister W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science (New York, N.Y.) 2015;347(6220):439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nature Reviews Drug Discovery. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Asher G., Tsvetkov P., Kahana C., Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes & Development. 2005;19(3):316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaraf Y.G., Brozovic A., Gonçalves A.C., Jurkovicova D., Linē A., Machuqueiro M.…Vasconcelos M.H. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2019;46 doi: 10.1016/j.drup.2019.100645. [DOI] [PubMed] [Google Scholar]
- Atkinson S.P., Collin J., Irina N., Anyfantis G., Kyung B.K., Lako M., Armstrong L. A putative role for the immunoproteasome in the maintenance of pluripotency in human embryonic stem cells. Stem Cells (Dayton, Ohio) 2012;30(7):1373–1384. doi: 10.1002/stem.1113. [DOI] [PubMed] [Google Scholar]
- Attal M., Lauwers-Cances V., Hulin C., Leleu X., Caillot D., Escoffre M., Arnulf B., Macro M., Belhadj K., Garderet L., Roussel M., Payen C., Mathiot C., Fermand J.P., Meuleman N., Rollet S., Maglio M.E., Zeytoonjian A.A., Weller E.A.…Moreau P. Lenalidomide, bortezomib, and dexamethasone with transplantation for myeloma. New England Journal of Medicine. 2017;376(14):1311–1320. doi: 10.1056/NEJMoa1611750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Tam M.-E., Olivares A.O., Sauer R.T., Baker T.A., Lang M.J. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145(2):257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Loiseau H., Fonseca R., Siegel D., Dimopoulos M.A., Špička I., Masszi T., Hájek R., Rosiñol L., Goranova-Marinova V., Mihaylov G., Maisnar V., Mateos M.-V., Wang M., Niesvizky R., Oriol A., Jakubowiak A., Minarik J., Palumbo A., Bensinger W.…Moreau P. Carfilzomib significantly improves the progression-free survival of high-risk patients in multiple myeloma. Blood. 2016;128(9):1174–1180. doi: 10.1182/blood-2016-03-707596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., Rubin E.J. SARS-Cov-2 — The search for effective therapy. New England Journal of Medicine. 2020 doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badros A.Z., Vij R., Martin T., Zonder J.A., Kunkel L., Wang Z.…Niesvizky R. Carfilzomib in multiple myeloma patients with renal impairment: Pharmacokinetics and safety. Leukemia. 2013;27(8):1707–1714. doi: 10.1038/leu.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Finley D., Glickman M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Current Biology: CB. 2003;13(13):1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- Bajorek M., Glickman M.H. Keepers at the final gates: Regulatory complexes and gating of the proteasome channel. Cellular and Molecular Life Sciences: CMLS. 2004;61(13):1579–1588. doi: 10.1007/s00018-004-4131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science (New York, N.Y.) 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annual Review of Immunology. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. The Journal of Clinical Investigation. 2001;107(3):241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Ji C., Mayfield J.E., Goel A., Xiao J., Dixon J.E., Guo X. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(32):8155–8160. doi: 10.1073/pnas.1806797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks P.M., Chan J., Cleary M.L., Delsol G., De Wolf-Peeters C., Gatter K.…Jaffe E.S. Mantle cell lymphoma. A proposal for unification of morphologic, immunologic, and molecular data. The American Journal of Surgical Pathology. 1992;16(7):637–640. doi: 10.1097/00000478-199207000-00001. [DOI] [PubMed] [Google Scholar]
- Banno A., Garcia D.A., van Baarsel E.D., Metz P.J., Fisch K., Widjaja C.E.…Chang J.T. Downregulation of 26S proteasome catalytic activity promotes epithelial-mesenchymal transition. Oncotarget. 2016;7(16):21527–21541. doi: 10.18632/oncotarget.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., Martin A. Structure and function of the 26S proteasome. Annual Review of Biochemistry. 2018;87:697–724. doi: 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlogie B., Jagannath S., Vesole D.H., Naucke S., Cheson B., Mattox S.…Tricot G. Superiority of tandem autologous transplantation over standard therapy for previously untreated multiple myeloma. Blood. 1997;89(3):789–793. [PubMed] [Google Scholar]
- Barrault M.-B., Richet N., Godard C., Murciano B., Le Tallec B., Rousseau E.…Peyroche A. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(17):E1001–E1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio S., Stühmer T., Da-Viá M., Barrio-Garcia C., Lehners N., Besse A., Cuenca I., Garitano-Trojaola A., Fink S., Leich E., Chatterjee M., Driessen C., Martinez-Lopez J., Rosenwald A., Beckmann R., Bargou R.C., Braggio E., Stewart A.K., Raab M.S.…Kortüm K.M. Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia. 2019;33(2):447–456. doi: 10.1038/s41375-018-0216-8. [DOI] [PubMed] [Google Scholar]
- Basu Ray S. PNPLA3-I148M: A problem of plenty in non-alcoholic fatty liver disease. Adipocyte. 2019;8(1):201–208. doi: 10.1080/21623945.2019.1607423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh J.M., Viktorova E.G., Pilipenko E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. Journal of Molecular Biology. 2009;386(3):814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Dahlmann B., Hegerl R., Kopp F., Kuehn L., Pfeifer G. Electron microscopy and image analysis of the multicatalytic proteinase. FEBS Letters. 1988;241(1–2):239–245. doi: 10.1016/0014-5793(88)81069-x. [DOI] [PubMed] [Google Scholar]
- Beaumont V., Zhong S., Lin H., Xu W., Bradaia A., Steidl E., Gleyzes M., Wadel K., Buisson B., Padovan-Neto F.E., Chakroborty S., Ward K.M., Harms J.F., Beltran J., Kwan M., Ghavami A., Häggkvist J., Tóth M., Halldin C.…Munoz-Sanjuan I. Phosphodiesterase 10A inhibition improves cortico-basal ganglia function in Huntington's disease models. Neuron. 2016;92(6):1220–1237. doi: 10.1016/j.neuron.2016.10.064. [DOI] [PubMed] [Google Scholar]
- Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., Sakata E.…Förster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(37):14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith R., Estrin E., Worden E.J., Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nature Structural & Molecular Biology. 2013;20(10):1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L., Hay D., Devoy A., Paine S., Powe D.G., Seth R.…Mayer R.J. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(33):8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke S., Ley S.C. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. The Biochemical Journal. 2004;382:393–409. doi: 10.1042/BJ20040544. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belch A., Kouroukis C.T., Crump M., Sehn L., Gascoyne R.D., Klasa R.…Eisenhauer E.A. A phase II study of bortezomib in mantle cell lymphoma: The National Cancer Institute of Canada Clinical Trials Group trial IND.150. Annals of Oncology: Official Journal of the European Society for Medical Oncology. 2007;18(1):116–121. doi: 10.1093/annonc/mdl316. [DOI] [PubMed] [Google Scholar]
- Belenky M.A., Smeraski C.A., Provencio I., Sollars P.J., Pickard G.E. Melanopsin retinal ganglion cells receive bipolar and amacrine cell synapses. The Journal of Comparative Neurology. 2003;460(3):380–393. doi: 10.1002/cne.10652. [DOI] [PubMed] [Google Scholar]
- Bellia F., Lanza V., García-Viñuales S., Ahmed I.M.M., Pietropaolo A., Iacobucci C.…Milardi D. Ubiquitin binds the amyloid β peptide and interferes with its clearance pathways. Chemical Science. 2019;10(9):2732–2742. doi: 10.1039/c8sc03394c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook D.M., Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Experimental Oncology. 2012;34(3):286–297. [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science (New York, N.Y.) 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G., Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4(3):862–884. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentea E., Verbruggen L., Massie A. The proteasome inhibition model of Parkinson's disease. Journal of Parkinson's Disease. 2017;7(1):31–63. doi: 10.3233/JPD-160921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. Journal of Biological Chemistry. 1989;264(27):15949–15952. [PubMed] [Google Scholar]
- Berenson J.R., Cartmell A., Bessudo A., Lyons R.M., Harb W., Tzachanis D.…Berdeja J.G. CHAMPION-1: a phase 1/2 study of once-weekly carfilzomib and dexamethasone for relapsed or refractory multiple myeloma. Blood. 2016;127(26):3360–3368. doi: 10.1182/blood-2015-11-683854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenson J.R., Matous J., Swift R.A., Mapes R., Morrison B., Yeh H.S. A phase I/II study of arsenic trioxide/bortezomib/ascorbic acid combination therapy for the treatment of relapsed or refractory multiple myeloma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2007;13(6):1762–1768. doi: 10.1158/1078-0432.CCR-06-1812. [DOI] [PubMed] [Google Scholar]
- Berisha F., Feke G.T., Trempe C.L., McMeel J.W., Schepens C.L. Retinal abnormalities in early Alzheimer's disease. Investigative Ophthalmology & Visual Science. 2007;48(5):2285–2289. doi: 10.1167/iovs.06-1029. [DOI] [PubMed] [Google Scholar]
- Bersuker K., Olzmann J.A. Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochimica Et Biophysica Acta. Molecular and Cell Biology of Lipids. 2017;1862(10 Pt B):1166–1177. doi: 10.1016/j.bbalip.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse A., Besse L., Kraus M., Mendez-Lopez M., Bader J., Xin B.-T.…Driessen C. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell Chemical Biology. 2019;26(3):340–351.e3. doi: 10.1016/j.chembiol.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Besse A., Stolze S.C., Rasche L., Weinhold N., Morgan G.J., Kraus M.…Driessen C. Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma. Leukemia. 2018;32(2):391–401. doi: 10.1038/leu.2017.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.P., Yan S., Wang C.-E., Li S., Li X.-J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(15):5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow S., Hough R., Rechsteiner M. The selective degradation of injected proteins occurs principally in the cytosol rather than in lysosomes. Cell. 1981;25(1):83–93. doi: 10.1016/0092-8674(81)90233-6. [DOI] [PubMed] [Google Scholar]
- Bingol B., Schuman E.M. A proteasome-sensitive connection between PSD-95 and GluR1 endocytosis. Neuropharmacology. 2004;47(5):755–763. doi: 10.1016/j.neuropharm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Bingol B., Schuman E.M. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441(7097):1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- Bingol B., Wang C.-F., Arnott D., Cheng D., Peng J., Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140(4):567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Blasiak J., Pawlowska E., Szczepanska J., Kaarniranta K. Interplay between autophagy and the ubiquitin-proteasome system and its role in the pathogenesis of age-related macular degeneration. International Journal of Molecular Sciences. 2019;20(1) doi: 10.3390/ijms20010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. The proteasome. Annual Review of Biophysics and Biomolecular Structure. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Bogyo M., Shin S., McMaster J.S., Ploegh H.L. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chemistry & Biology. 1998;5(6):307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- Bohn S., Sakata E., Beck F., Pathare G.R., Schnitger J., Nágy I.…Förster F. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochemical and Biophysical Research Communications. 2013;435(2):250–254. doi: 10.1016/j.bbrc.2013.04.069. [DOI] [PubMed] [Google Scholar]
- Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E.…Millan M.J. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nature Reviews. Drug Discovery. 2018;17(9):660–688. doi: 10.1038/nrd.2018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfili L., Cecarini V., Amici M., Cuccioloni M., Angeletti M., Keller J.N., Eleuteri A.M. Natural polyphenols as proteasome modulators and their role as anti-cancer compounds. FEBS Journal. 2008;275(22):5512–5526. doi: 10.1111/j.1742-4658.2008.06696.x. [DOI] [PubMed] [Google Scholar]
- Bonnet A., Moreau P. Safety of ixazomib for the treatment of multiple myeloma. Expert Opinion on Drug Safety. 2017;16(8):973–980. doi: 10.1080/14740338.2017.1344212. [DOI] [PubMed] [Google Scholar]
- Borissenko L., Groll M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chemical Reviews. 2007;107(3):687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- Boselli M., Lee B.-H., Robert J., Prado M.A., Min S.-W., Cheng C.…Finley D. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. The Journal of Biological Chemistry. 2017;292(47):19209–19225. doi: 10.1074/jbc.M117.815126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan J.A., Dodson G., Duggleby H.J., Moody P.C., Smith J.L., Tomchick D.R., Murzin A.G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378(6555):416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- Braun T., Carvalho G., Fabre C., Grosjean J., Fenaux P., Kroemer G. Targeting NF-kappaB in hematologic malignancies. Cell Death and Differentiation. 2006;13(5):748–758. doi: 10.1038/sj.cdd.4401874. [DOI] [PubMed] [Google Scholar]
- Bredesen D.E., Rao R.V., Mehlen P. Cell death in the nervous system. Nature. 2006;443(7113):796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Liu Y., Sheikh A., Marrero B., Omoyinmi E., Zhou Q., Montealegre G., Biancotto A., Reinhardt A., Almeida de Jesus A., Pelletier M., Tsai W.L., Remmers E.F., Kardava L., Hill S., Kim H., Lachmann H.J., Megarbane A., Chae J.J.…Goldbach-Mansky R. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. The Journal of Clinical Investigation. 2015;125(11):4196–4211. doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J., Del Tredici K., Lee V.M.-Y., Trojanowski J.Q. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nature Reviews. Neuroscience. 2015;16(2):109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringhen S., De Wit E., Dimopoulos M.-A. New Agents in Multiple Myeloma: An Examination of Safety Profiles. Clinical Lymphoma, Myeloma & Leukemia. 2017;17(7):391–407.e5. doi: 10.1016/j.clml.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Lain S., Verma C.S., Fersht A.R., Lane D.P. Awakening guardian angels: drugging the p53 pathway. Nature Reviews. Cancer. 2009;9(12):862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Brugarolas J., Chandrasekaran C., Gordon J.I., Beach D., Jacks T., Hannon G.J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377(6549):552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Bruggeman J.W., Koster J., Lodder P., Repping S., Hamer G. Massive expression of germ cell-specific genes is a hallmark of cancer and a potential target for novel treatment development. Oncogene. 2018;37(42):5694–5700. doi: 10.1038/s41388-018-0357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin G., Xin B.T., Kraus M., van der Stelt M., van der Marel G.A., Kisselev A.F.…Overkleeft H.S. A set of activity-based probes to visualize human (immuno)proteasome activities. Angewandte Chemie (International Ed. in. English) 2016;55(13):4199–4203. doi: 10.1002/anie.201509092. [DOI] [PubMed] [Google Scholar]
- Buac D., Shen M., Schmitt S., Kona F.R., Deshmukh R., Zhang Z.…Dou Q.P. From bortezomib to other inhibitors of the proteasome and beyond. Current Pharmaceutical Design. 2013;19(22):4025–4038. doi: 10.2174/1381612811319220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budenholzer L., Cheng C.L., Li Y., Hochstrasser M. Proteasome structure and assembly. Journal of Molecular Biology. 2017;429(22):3500–3524. doi: 10.1016/j.jmb.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullova P., Cougnoux A., Marzouca G., Kopacek J., Pacak K. Bortezomib Alone and in Combination With Salinosporamid A Induces Apoptosis and Promotes Pheochromocytoma Cell Death In Vitro and in Female Nude Mice. Endocrinology. 2017;158(10):3097–3108. doi: 10.1210/en.2017-00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L., Höckendorff J., Boehm U., Klamp T., Dohmen R.J., Lévy F. Identification and characterization of a mammalian protein interacting with 20S proteasome precursors. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10348–10353. doi: 10.1073/pnas.190268597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante H.A., González A.E., Cerda-Troncoso C., Shaughnessy R., Otth C., Soza A., Burgos P.V. Interplay between the autophagy-lysosomal pathway and the ubiquitin-proteasome system: A target for therapeutic development in alzheimer's disease. Frontiers in Cellular Neuroscience. 2018;12 doi: 10.3389/fncel.2018.00126. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero M., Liton P.B., Challa P., Epstein D.L., Gonzalez P. Effects of donor age on proteasome activity and senescence in trabecular meshwork cells. Biochemical and Biophysical Research Communications. 2004;323(3):1048–1054. doi: 10.1016/j.bbrc.2004.08.195. [DOI] [PubMed] [Google Scholar]
- Caballero M., Liton P.B., Epstein D.L., Gonzalez P. Proteasome inhibition by chronic oxidative stress in human trabecular meshwork cells. Biochemical and Biophysical Research Communications. 2003;308(2):346–352. doi: 10.1016/s0006-291x(03)01385-8. [DOI] [PubMed] [Google Scholar]
- Calderwood S.K., Khaleque M.A., Sawyer D.B., Ciocca D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends in Biochemical Sciences. 2006;31(3):164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Campello L., Esteve-Rudd J., Cuenca N., Martín-Nieto J. The ubiquitin-proteasome system in retinal health and disease. Molecular Neurobiology. 2013;47(2):790–810. doi: 10.1007/s12035-012-8391-5. [DOI] [PubMed] [Google Scholar]
- Campo E., Rule S. Mantle cell lymphoma: Evolving management strategies. Blood. 2015;125(1):48–55. doi: 10.1182/blood-2014-05-521898. [DOI] [PubMed] [Google Scholar]
- Cao J., Zhong M.B., Toro C.A., Zhang L., Cai D. Endo-lysosomal pathway and ubiquitin-proteasome system dysfunction in Alzheimer's disease pathogenesis. Neuroscience Letters. 2019;703:68–78. doi: 10.1016/j.neulet.2019.03.016. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravita T., de Fabritiis P., Palumbo A., Amadori S., Boccadoro M. Bortezomib: Efficacy comparisons in solid tumors and hematologic malignancies. Nature Clinical Practice. Oncology. 2006;3(7):374–387. doi: 10.1038/ncponc0555. [DOI] [PubMed] [Google Scholar]
- Cardozo C., Vinitsky A., Michaud C., Orlowski M. Evidence that the nature of amino acid residues in the P3 position directs substrates to distinct catalytic sites of the pituitary multicatalytic proteinase complex (proteasome) Biochemistry. 1994;33(21):6483–6489. doi: 10.1021/bi00187a014. [DOI] [PubMed] [Google Scholar]
- Carmony K.C., Lee W., Kim K.B. High-resolution snapshots of proteasome inhibitors in action revise inhibition paradigms and inspire next-generation inhibitor design. Chembiochem: A European Journal of Chemical Biology. 2016 doi: 10.1002/cbic.201600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale C., Manni G., Roberti G., Micera A., Bruno L., Cacciamani A.…Oddone F. Human vitreous concentrations of citicoline following topical application of citicoline 2% ophthalmic solution. PLoS One. 2019;14(11) doi: 10.1371/journal.pone.0224982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell R.W., Lomas D.A. Conformational disease. The Lancet. 1997;350(9071):134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Carvalho A.S., Rodríguez M.S., Matthiesen R. Review and literature mining on proteostasis factors and cancer. Methods in Molecular Biology (Clifton, N.J.) 2016;1449:71–84. doi: 10.1007/978-1-4939-3756-1_2. [DOI] [PubMed] [Google Scholar]
- Cascio P. PA28αβ: The enigmatic magic ring of the proteasome? Biomolecules. 2014;4(2):566–584. doi: 10.3390/biom4020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio P., Call M., Petre B.M., Walz T., Goldberg A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. The EMBO Journal. 2002;21(11):2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio P., Hilton C., Kisselev A.F., Rock K.L., Goldberg A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. The EMBO Journal. 2001;20(10):2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavo M., Tacchetti P., Patriarca F., Petrucci M.T., Pantani L., Galli M., Di Raimondo F., Crippa C., Zamagni E., Palumbo A., Offidani M., Corradini P., Narni F., Spadano A., Pescosta N., Deliliers G.L., Ledda A., Cellini C., Caravita T.…GIMEMA Italian Myeloma Network Bortezomib with thalidomide plus dexamethasone compared with thalidomide plus dexamethasone as induction therapy before, and consolidation therapy after, double autologous stem-cell transplantation in newly diagnosed multiple myeloma: A randomised phase 3 study. Lancet (London, England) 2010;376(9758):2075–2085. doi: 10.1016/S0140-6736(10)61424-9. [DOI] [PubMed] [Google Scholar]
- Cecarini V., Bonfili L., Amici M., Angeletti M., Keller J.N., Eleuteri A.M. Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Research. 2008:1209. doi: 10.1016/j.brainres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Cengiz Seval G., Beksac M. The safety of bortezomib for the treatment of multiple myeloma. Expert Opinion on Drug Safety. 2018;17(9):953–962. doi: 10.1080/14740338.2018.1513487. [DOI] [PubMed] [Google Scholar]
- Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T.C., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science (New York, N.Y.) 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty J., von Stockum S., Marchesan E., Caicci F., Ferrari V., Rakovic A.…Ziviani E. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Molecular Medicine. 2018;10(11) doi: 10.15252/emmm.201809014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanan-Khan A., Sonneveld P., Schuster M.W., Stadtmauer E.A., Facon T., Harousseau J.-L.…Richardson P.G. Analysis of herpes zoster events among bortezomib-treated patients in the phase III APEX study. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2008;26(29):4784–4790. doi: 10.1200/JCO.2007.14.9641. [DOI] [PubMed] [Google Scholar]
- Chang S.C., Ding J.L. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochimica Et Biophysica Acta. Reviews on Cancer. 2018;1870(2):165–175. doi: 10.1016/j.bbcan.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Chari A., Hajje D. Case series discussion of cardiac and vascular events following carfilzomib treatment: Possible mechanism, screening, and monitoring. BMC Cancer. 2014;14:915. doi: 10.1186/1471-2407-14-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule V.K., Burchell L., Barber K.R., Sidhu A., Leslie S.J., Shaw G.S., Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. The EMBO Journal. 2011;30(14):2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M.…Anderson K.C. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from bortezomib. Cancer Cell. 2005;8(5):407–419. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Chauhan D., Hideshima T., Anderson K.C. Proteasome inhibition in multiple myeloma: therapeutic implication. Annual Review of Pharmacology and Toxicology. 2005;45:465–476. doi: 10.1146/annurev.pharmtox.45.120403.100037. [DOI] [PubMed] [Google Scholar]
- Chauhan D., Singh A.V., Aujay M., Kirk C.J., Bandi M., Ciccarelli B.…Anderson K.C. A novel orally active proteasome inhibitor ONX 0912 triggers in vitro and in vivo cytotoxicity in multiple myeloma. Blood. 2010;116(23):4906–4915. doi: 10.1182/blood-2010-04-276626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Singh A.V., Ciccarelli B., Richardson P.G., Palladino M.A., Anderson K.C. Combination of novel proteasome inhibitor NPI-0052 and lenalidomide trigger in vitro and in vivo synergistic cytotoxicity in multiple myeloma. Blood. 2010;115(4):834–845. doi: 10.1182/blood-2009-03-213009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Tian Z., Zhou B., Kuhn D., Orlowski R., Raje N.…Anderson K.C. In vitro and in vivo selective antitumor activity of a novel orally bioavailable proteasome inhibitor MLN9708 against multiple myeloma cells. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2011;17(16):5311–5321. doi: 10.1158/1078-0432.CCR-11-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Barton L.F., Chi Y., Clurman B.E., Roberts J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Molecular Cell. 2007;26(6):843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.-C., Bhattacharyya B.J., Hanna J., Minkel H., Wilson J.A., Finley D.…Wilson S.M. Ubiquitin homeostasis is critical for synaptic development and function. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2011;31(48):17505–17513. doi: 10.1523/JNEUROSCI.2922-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Cui Q.C., Yang H., Dou Q.P. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Research. 2006;66(21):10425–10433. doi: 10.1158/0008-5472.CAN-06-2126. Scopus. [DOI] [PubMed] [Google Scholar]
- Chen D., Daniel K.J., Chen M.S., Kuhn D.J., Landis-Piwowar K.R., Dou Q.P. Dietary flavonoids as proteasome inhibitors and apoptosis inducers in human leukemia cells. Biochemical Pharmacology. 2005;69(10):1421–1432. doi: 10.1016/j.bcp.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Chen D., Frezza M., Schmitt S., Kanwar J., Dou Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Current Cancer Drug Targets. 2011;11(3):239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hagler J., Palombella V.J., Melandri F., Scherer D., Ballard D., Maniatis T. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes & Development. 1995;9(13):1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Chen P., Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. The EMBO Journal. 1995;14(11):2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86(6):961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- Chen L., Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Research. 2005;65(13):5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- Chen B., Retzlaff M., Roos T., Frydman J. Cellular strategies of protein quality control. Cold Spring Harbor Perspectives in Biology. 2011;3(8):a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J., Wu H., Shen X.-Z. The ubiquitin–proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Letters. 2016;379(2):245–252. doi: 10.1016/j.canlet.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Chène P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nature Reviews. Cancer. 2003;3(2):102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- Cheungpasitporn W., Leung N., Rajkumar S.V., Cornell L.D., Sethi S., Angioi A., Fervenza F.C. Bortezomib-induced acute interstitial nephritis. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association - European Renal Association. 2015;30(7):1225–1229. doi: 10.1093/ndt/gfv222. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annual Review of Biochemistry. 2006;75(1):333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annual Review of Biochemistry. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- Chitta K., Paulus A., Akhtar S., Blake M.K.K., Caulfield T.R., Novak A.J.…Chanan-Khan A. Targeted inhibition of the deubiquitinating enzymes, USP14 and UCHL5, induces proteotoxic stress and apoptosis in Waldenström macroglobulinaemia tumour cells. British Journal of Haematology. 2015;169(3):377–390. doi: 10.1111/bjh.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.H., de Poot S.A.H., Lee J.H., Kim J.H., Han D.H., Kim Y.K.…Lee M.J. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nature Communications. 2016;7:10963. doi: 10.1038/ncomms10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.-H., Jo H.S., Lim S., Kim H.-T., Lee K.W., Moon K.H.…Kim J.W. mTORC1 accelerates retinal development via the immunoproteasome. Nature Communications. 2018;9(1):2502. doi: 10.1038/s41467-018-04774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrogianni N., Gonos E.S. Proteasome inhibition induces a senescence-like phenotype in primary human fibroblasts cultures. Biogerontology. 2004;5(1):55–61. doi: 10.1023/B:BGEN.0000017687.55667.42. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N., Gonos E.S. Overexpression of hUMP1/POMP proteasome accessory protein enhances proteasome-mediated antioxidant defence. Experimental Gerontology. 2007;42(9):899–903. doi: 10.1016/j.exger.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N., Gonos E.S. Proteasome function determines cellular homeostasis and the rate of aging. In: Tavernarakis N., editor. Protein Metabolism and Homeostasis in Aging. Springer US; 2010. pp. 38–46. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N., Kapeta S., Chinou I., Vassilatou K., Papassideri I., Gonos E.S. Anti-ageing and rejuvenating effects of quercetin. Experimental Gerontology. 2010;45(10):763–771. doi: 10.1016/j.exger.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Chu I.M., Hengst L., Slingerland J.M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature Reviews Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nature Reviews. Molecular Cell Biology. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: From a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorganic & Medicinal Chemistry. 2013;21(12):3400–3410. doi: 10.1016/j.bmc.2013.01.056. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron. 2003;40(2):427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Gonen H., Bercovich B., Cohen S., Fajerman I., Israël A.…Orian A. Mechanisms of ubiquitin-mediated, limited processing of the NF-kappaB1 precursor protein p105. Biochimie. 2001;83(3–4):341–349. doi: 10.1016/s0300-9084(01)01239-1. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Heller H., Elias S., Haas A.L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(3):1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Hod Y., Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. 1978. Biochemical and Biophysical Research Communications. 2012;425(3):565–570. doi: 10.1016/j.bbrc.2012.08.025. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Kwon Y.T. Protein quality control by molecular chaperones in neurodegeneration. Frontiers in Neuroscience. 2017;11:185. doi: 10.3389/fnins.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciehanover A., Hod Y., Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochemical and Biophysical Research Communications. 1978;81(4):1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- Cliffe R., Sang J.C., Kundel F., Finley D., Klenerman D., Ye Y. Filamentous aggregates are fragmented by the proteasome holoenzyme. Cell Reports. 2019;26(8):2140–2149. doi: 10.1016/j.celrep.2019.01.096. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos J., Roeten M.S., Franke N.E., van Meerloo J., Zweegman S., Kaspers G.J., Jansen G. (Immuno)proteasomes as therapeutic target in acute leukemia. Cancer Metastasis Reviews. 2017;36(4):599–615. doi: 10.1007/s10555-017-9699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho S.C., Almeida G.M., Santos-Silva F., Pereira M.C., Coelho M.A.N. Enhancing the efficiency of bortezomib conjugated to pegylated gold nanoparticles: An in vitro study on human pancreatic cancer cells and adenocarcinoma human lung alveolar basal epithelial cells. Expert Opinion on Drug Delivery. 2016;13(8):1075–1081. doi: 10.1080/17425247.2016.1178234. [DOI] [PubMed] [Google Scholar]
- Coleman R.A., Muli C.S., Zhao Y., Bhardwaj A., Newhouse T.R., Trader D.J. Analysis of chain length, substitution patterns, and unsaturation of AM-404 derivatives as 20S proteasome stimulators. Bioorganic and Medicinal Chemistry Letters. 2019;29(3):420–423. doi: 10.1016/j.bmcl.2018.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M., Snyder E.M., Crozier R.A., Soderling J.A., Jin Y., Langeberg L.K.…Scott J.D. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40(3):595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins G.A., Goldberg A.L. The logic of the 26S proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer F.I., Hart G.W. O-glycosylation of nuclear and cytosolic proteins dynamic interplay between O-GlcNAc ando-phosphate. Journal of Biological Chemistry. 2000;275(38):29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- Concannon C.G., Koehler B.F., Reimertz C., Murphy B.M., Bonner C., Thurow N.…Prehn J.H.M. Apoptosis induced by proteasome inhibition in cancer cells: predominant role of the p53/PUMA pathway. Oncogene. 2007;26(12):1681–1692. doi: 10.1038/sj.onc.1209974. [DOI] [PubMed] [Google Scholar]
- Corpas R., Griñán-Ferré C., Rodríguez-Farré E., Pallàs M., Sanfeliu C. Resveratrol induces brain resilience against Alzheimer neurodegeneration through proteostasis enhancement. Molecular Neurobiology. 2019;56(2):1502–1516. doi: 10.1007/s12035-018-1157-y. [DOI] [PubMed] [Google Scholar]
- Coughlin K., Anchoori R., Iizuka Y., Meints J., MacNeill L., Vogel R.I.…Bazzaro M. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2014;20(12):3174–3186. doi: 10.1158/1078-0432.CCR-13-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annual Review of Biochemistry. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Cree I.A., Charlton P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer. 2017;17(1) doi: 10.1186/s12885-016-2999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps D., Thomas S.N., Jeng Y., Yang F., Davies P., Yang A.J. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. The Journal of Biological Chemistry. 2006;281(16):10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- Cron K.R., Zhu K., Kushwaha D.S., Hsieh G., Merzon D., Rameseder J.…Kozono D. Proteasome inhibitors block DNA repair and radiosensitize non-small cell lung cancer. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuanalo-Contreras K., Moreno-Gonzalez I. Natural products as modulators of the proteostasis machinery: Implications in neurodegenerative diseases. International Journal of Molecular Sciences. 2019;20(19) doi: 10.3390/ijms20194666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundiff M.D., Hurley C.M., Wong J.D., Boscia J.A., Bashyal A., Rosenberg J.…Kraut D.A. Ubiquitin receptors are required for substrate-mediated activation of the proteasome's unfolding ability. Scientific Reports. 2019;9(1):14506. doi: 10.1038/s41598-019-50857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dächsel J.C., Lücking C.B., Deeg S., Schultz E., Lalowski M., Casademunt E.…Gasser T. Parkin interacts with the proteasome subunit alpha4. FEBS Letters. 2005;579(18):3913–3919. doi: 10.1016/j.febslet.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Dahlmann B. Mammalian proteasome subtypes: Their diversity in structure and function. Archives of Biochemistry and Biophysics. 2016;591:132–140. doi: 10.1016/j.abb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Becher B., Sobek A., Ehlers C., Kopp F., Kuehn L. In vitro activation of the 20S proteasome. Enzyme & Protein. 1993;47(4–6):274–284. doi: 10.1159/000468685. [DOI] [PubMed] [Google Scholar]
- Dahlmann B., Rutschmann M., Kuehn L., Reinauer H. Activation of the multicatalytic proteinase from rat skeletal muscle by fatty acids or sodium dodecyl sulphate. Biochemical Journal. 1985;228(1):171–177. doi: 10.1042/bj2280171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist J., Klar J., Tiwari N., Schuster J., Törmä H., Badhai J.…Dahl N. A single-nucleotide deletion in the POMP 5’ UTR causes a transcriptional switch and altered epidermal proteasome distribution in KLICK genodermatosis. American Journal of Human Genetics. 2010;86(4):596–603. doi: 10.1016/j.ajhg.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist J., Törmä H., Badhai J., Dahl N. siRNA silencing of proteasome maturation protein (POMP) activates the unfolded protein response and constitutes a model for KLICK genodermatosis. PLoS One. 2012;7(1):e29471. doi: 10.1371/journal.pone.0029471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Dai S., Cao J. Proteotoxic stress of cancer: Implication of the heat-shock response in oncogenesis. Journal of Cellular Physiology. 2012;227(8):2982–2987. doi: 10.1002/jcp.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Vechio F.H., Cerqueira F., Augusto O., Lopes R., Demasi M. Peptides that activate the 20S proteasome by gate opening increased oxidized protein removal and reduced protein aggregation. Free Radical Biology & Medicine. 2014;67:304–313. doi: 10.1016/j.freeradbiomed.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Dambacher C.M., Worden E.J., Herzik M.A., Martin A., Lander G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. ELife. 2016;5 doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K.G., Gupta P., Harbach R.H., Guida W.C., Dou Q.P. Organic copper complexes as a new class of proteasome inhibitors and apoptosis inducers in human cancer cells. Biochemical Pharmacology. 2004;67(6):1139–1151. doi: 10.1016/j.bcp.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Dantuma N.P., Bott L.C. The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Frontiers in Molecular Neuroscience. 2014;7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D.S., Ray A., Song Y., Richardson P., Trikha M., Chauhan D., Anderson K.C. Synergistic anti-myeloma activity of the proteasome inhibitor marizomib and the IMiD® immunomodulatory drug pomalidomide. British Journal of Haematology. 2015;171(5):798–812. doi: 10.1111/bjh.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy P., Wang X., Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacology & Therapeutics. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Dasgupta S., Fishman M.A., Mahallati H., Castro L.M., Tashima A.K., Ferro E.S., Fricker L.D. Reduced levels of proteasome products in a mouse striatal cell model of Huntington's disease. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Dato A.D., Cunsolo A., Persico M., Santoro A.M., D'Urso A., Milardi D.…Coletta M. Electrostatic map of proteasome α-rings encodes the design of allosteric porphyrin-based inhibitors able to affect 20S conformation by cooperative binding. Scientific Reports. 2017;7(1):17098. doi: 10.1038/s41598-017-17008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D.C., Layfield R., Serpell L., Narain Y., Goedert M., Spillantini M.G. Proteasomal degradation of tau protein. Journal of Neurochemistry. 2002;83(1):176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- Davies K.J. Protein modification by oxidants and the role of proteolytic enzymes. Biochemical Society Transactions. 1993;21(2):346–353. doi: 10.1042/bst0210346. [DOI] [PubMed] [Google Scholar]
- Davies K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83(3–4):301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- De Duve C., Gianetto R., Appelmans F., Wattiaux R. Enzymic content of the mitochondria fraction. Nature. 1953;172(4390):1143–1144. doi: 10.1038/1721143a0. [DOI] [PubMed] [Google Scholar]
- De Duve C., Wattiaux R. Functions of lysosomes. Annual Review of Physiology. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- De La Mota-Peynado A., Lee S.Y.-C., Pierce B.M., Wani P., Singh C.R., Roelofs J. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. The Journal of Biological Chemistry. 2013;288(41):29467–29481. doi: 10.1074/jbc.M113.491662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehabadi M.H., Davis B.M., Wong T.K., Cordeiro M.F. Retinal manifestations of Alzheimer's disease. Neurodegenerative Disease Management. 2014;4(3):241–252. doi: 10.2217/nmt.14.19. [DOI] [PubMed] [Google Scholar]
- Demo S.D., Kirk C.J., Aujay M.A., Buchholz T.J., Dajee M., Ho M.N.…Bennett M.K. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Research. 2007;67(13):6383–6391. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- Deng T., Yan G., Song X., Xie L., Zhou Y., Li J.…Ye M. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proceedings of the National Academy of Sciences. 2018;115(18):4678–4683. doi: 10.1073/pnas.1714938115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82(4):675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Deriziotis P., André R., Smith D.M., Goold R., Kinghorn K.J., Kristiansen M.…Tabrizi S.J. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. The EMBO Journal. 2011;30(15):3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes-Simard X., Lessard F., Gaumont-Leclerc M.-F., Bardeesy N., Ferbeyre G. Cellular senescence and protein degradation. Cell Cycle. 2014;13(12):1840–1858. doi: 10.4161/cc.29335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biology. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N.…Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. Journal of Biological Chemistry. 2007;282(44):32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. The Journal of Biological Chemistry. 1994;269(10):7059–7061. [PubMed] [Google Scholar]
- Devine T., Dai M.-S. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Current Pharmaceutical Design. 2013;19(18):3248–3262. doi: 10.2174/1381612811319180009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di K., Lloyd G.K., Abraham V., MacLaren A., Burrows F.J., Desjardins A.…Bota D.A. Marizomib activity as a single agent in malignant gliomas: Ability to cross the blood-brain barrier. Neuro-Oncology. 2016;18(6):840–848. doi: 10.1093/neuonc/nov299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Hernández M., Valera A.G., Morán M.A., Gómez-Ramos P., Alvarez-Castelao B., Castaño J.G.…Lucas J.J. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. Journal of Neurochemistry. 2006;98(5):1585–1596. doi: 10.1111/j.1471-4159.2006.03968.x. [DOI] [PubMed] [Google Scholar]
- Dick T.P., Nussbaum A.K., Deeg M., Heinemeyer W., Groll M., Schirle M.…Schild H. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. The Journal of Biological Chemistry. 1998;273(40):25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J.…Petrucelli L. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. The Journal of Clinical Investigation. 2007;117(3):648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J.A., Ponugoti B. Ubiquitin-dependent proteolysis in G1/S phase control and its relationship with tumor susceptibility. Genes & Cancer. 2010;1(7):717–724. doi: 10.1177/1947601910382902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science (New York, N.Y.) 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dimopoulos M.A., Goldschmidt H., Niesvizky R., Joshua D., Chng W.-J., Oriol A.…Moreau P. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): An interim overall survival analysis of an open-label, randomised, phase 3 trial. The Lancet. Oncology. 2017;18(10):1327–1337. doi: 10.1016/S1470-2045(17)30578-8. [DOI] [PubMed] [Google Scholar]
- Dimopoulos M.A., Moreau P., Palumbo A., Joshua D., Pour L., Hájek R., Facon T., Ludwig H., Oriol A., Goldschmidt H., Rosiñol L., Straub J., Suvorov A., Araujo C., Rimashevskaya E., Pika T., Gaidano G., Weisel K., Goranova-Marinova V.…Investigators, E.N.D.E.A.V.O.R Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): A randomised, phase 3, open-label, multicentre study. The Lancet. Oncology. 2016;17(1):27–38. doi: 10.1016/S1470-2045(15)00464-7. [DOI] [PubMed] [Google Scholar]
- Dimopoulos M.A., Roussou M., Gavriatopoulou M., Psimenou E., Ziogas D., Eleutherakis-Papaiakovou E.…Kastritis E. Cardiac and renal complications of carfilzomib in patients with multiple myeloma. Blood Advances. 2017;1(7):449–454. doi: 10.1182/bloodadvances.2016003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos M., Siegel D., White D.J., Boccia R., Iskander K.S., Yang Z.…Niesvizky R. Carfilzomib vs bortezomib in patients with multiple myeloma and renal failure: A subgroup analysis of ENDEAVOR. Blood. 2019;133(2):147–155. doi: 10.1182/blood-2018-06-860015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel L., Huber R., Mann K., Heinemeyer W., Wolf D.H., Groll M. Conformational constraints for protein self-cleavage in the proteasome. Journal of Molecular Biology. 1998;279(5):1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- Djakovic S.N., Marquez-Lona E.M., Jakawich S.K., Wright R., Chu C., Sutton M.A., Patrick G.N. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2012;32(15):5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic S.N., Schwarz L.A., Barylko B., DeMartino G.N., Patrick G.N. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. The Journal of Biological Chemistry. 2009;284(39):26655–26665. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S., Hartmann M.D., Habeck M., Ursinus A., Zwickl P., Martin J.…Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Molecular Cell. 2009;34(5):580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Dong Z., Cui H. The autophagy-lysosomal pathways and their emerging roles in modulating proteostasis in tumors. Cells. 2018;8(1) doi: 10.3390/cells8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Zhang S., Wu Z., Li X., Wang W.L., Zhu Y.…Mao Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2019;565(7737):49–55. doi: 10.1038/s41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduijn J.K., Zijlstra J.M., Lugtenburg P.J., Kersten M.J., Böhmer L.H., Minnema M.C.…Kluin-Nelemans H.C. Bortezomib maintenance after R-CHOP, cytarabine and autologous stem cell transplantation in newly diagnosed patients with mantle cell lymphoma, results of a randomised phase II HOVON trial. British Journal of Haematology. 2020 doi: 10.1111/bjh.16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey B.D., Iqbal M., Chatterjee S., Menta E., Bernardini R., Bernareggi A.…Mallamo J.P. Discovery of a potent, selective, and orally active proteasome inhibitor for the treatment of cancer. Journal of Medicinal Chemistry. 2008;51(4):1068–1072. doi: 10.1021/jm7010589. [DOI] [PubMed] [Google Scholar]
- Drach J., Huang H., Samoilova O., Belch A., Farber C., Bosly A.…Cavalli F. Efficacy and safety of frontline rituximab, cyclophosphamide, doxorubicin and prednisone plus bortezomib (VR-CAP) or vincristine (R-CHOP) in a subset of newly diagnosed mantle cell lymphoma patients medically eligible for transplantation in the randomized, phase 3 LYM-3002 study. Leukemia & Lymphoma. 2018;59(4):896–903. doi: 10.1080/10428194.2017.1365855. [DOI] [PubMed] [Google Scholar]
- Du H.-P., Yang Q.-Q., Zhang Y. Bortezomib-based chemotherapy to treat refractory angioimmunoblastic T-cell lymphoma: A case report and review of the literature. Oncology Letters. 2016;11(3):2310–2314. doi: 10.3892/ol.2016.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufey E., Urra H., Hetz C. ER proteostasis addiction in cancer biology: Novel concepts. Seminars in Cancer Biology. 2015;33:40–47. doi: 10.1016/j.semcancer.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Durães F., Pinto M., Sousa E. Old drugs as new treatments for neurodegenerative diseases. Pharmaceuticals. 2018;11(2):44. doi: 10.3390/ph11020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durie B.G.M., Hoering A., Abidi M.H., Rajkumar S.V., Epstein J., Kahanic S.P.…Dispenzieri A. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): A randomised, open-label, phase 3 trial. Lancet (London, England) 2017;389(10068):519–527. doi: 10.1016/S0140-6736(16)31594-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C., Pressman B.C., Gianetto R., Wattiaux R., Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochemical Journal. 1955;60(4):604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nature Reviews. Molecular Cell Biology. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Eastman A. Cell cycle checkpoints and their impact on anticancer therapeutic strategies. Journal of Cellular Biochemistry. 2004;91(2):223–231. doi: 10.1002/jcb.10699. [DOI] [PubMed] [Google Scholar]
- Eberhardt R., Birbamer G., Gerstenbrand F., Rainer E., Traegner H. Citicoline in the treatment of Parkinson's disease. Clinical Therapeutics. 1990;12(6):489–495. [PubMed] [Google Scholar]
- Ebstein F., Poli Harlowe M.C., Studencka-Turski M., Krüger E. Contribution of the unfolded protein response (UPR) to the pathogenesis of proteasome-associated autoinflammatory syndromes (PRAAS) Frontiers in Immunology. 2019;10:2756. doi: 10.3389/fimmu.2019.02756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlinger, A., Park, S., Fahmy, A., Lary, J. W., Cole, J. L., Finley, D., & Walters, K. J. (2013). Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding. Structure (London, England: 1993), 21(5), 753–765. 10.1016/j.str.2013.02.021. [DOI] [PMC free article] [PubMed]
- Eisele M.R., Reed R.G., Rudack T., Schweitzer A., Beck F., Nagy I.…Sakata E. Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Reports. 2018;24(5):1301–1315. doi: 10.1016/j.celrep.2018.07.004. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. The Journal of Biological Chemistry. 2004;279(26):26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Enrico O., Gabriele B., Nadia C., Sara G., Daniele V., Giulia C.…Mario P. Unexpected cardiotoxicity in haematological bortezomib treated patients. British Journal of Haematology. 2007;138(3):396–397. doi: 10.1111/j.1365-2141.2007.06659.x. [DOI] [PubMed] [Google Scholar]
- Erales J., Coffino P. Ubiquitin-independent proteasomal degradation. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843(1):216–221. doi: 10.1016/j.bbamcr.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erales J., Hoyt M.A., Troll F., Coffino P. Functional asymmetries of proteasome translocase pore. The Journal of Biological Chemistry. 2012;287(22):18535–18543. doi: 10.1074/jbc.M112.357327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S., Vonk W.I.M., Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nature Cell Biology. 2013;15(10):1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari S.K., Seelen M.a.J., Lin G., Azzi J.R. The immunoproteasome: An old player with a novel and emerging role in alloimmunity. American Journal of Transplantation : Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2017;17(12):3033–3039. doi: 10.1111/ajt.14435. [DOI] [PubMed] [Google Scholar]
- Estrin E., Lopez-Blanco J.R., Chacón P., Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure (London, England: 1993) 2013;21(9):1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Etlinger J.D., Goldberg A.L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etlinger J.D., Goldberg A.L. Control of protein degradation in reticulocytes and reticulocyte extracts by hemin. Journal of Biological Chemistry. 1980;255(10):4563–4568. [PubMed] [Google Scholar]
- Evan G.I., Vousden K.H. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- Eytan E., Ganoth D., Armon T., Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(20):7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre B., Lambour T., Garrigues L., Ducoux-Petit M., Amalric F., Monsarrat B.…Bousquet-Dubouch M.-P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. Journal of Proteome Research. 2014;13(6):3027–3037. doi: 10.1021/pr500193k. [DOI] [PubMed] [Google Scholar]
- Faiq M.A., Wollstein G., Schuman J.S., Chan K.C. Cholinergic nervous system and glaucoma: From basic science to clinical applications. Progress in Retinal and Eye Research. 2019;72:100767. doi: 10.1016/j.preteyeres.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.-Y., Agyekum B., Venkatesan A., Hall D.R., Keightley A., Bjes E.S.…Price J.L. Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron. 2013;80(4):984–996. doi: 10.1016/j.neuron.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.M., Maniatis T. Generation of p50 subunit of NF-kappa B by processing of p105 through an ATP-dependent pathway. Nature. 1991;354(6352):395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- Fedorowicz M.A., de Vries-Schneider R.L.A., Rüb C., Becker D., Huang Y., Zhou C.…Przedborski S. Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Reports. 2014;15(1):86–93. doi: 10.1002/embr.201337294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feling R.H., Buchanan G.O., Mincer T.J., Kauffman C.A., Jensen P.R., Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus salinospora. Angewandte Chemie (International Ed. in English) 2003;42(3):355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- Felszeghy S., Viiri J., Paterno J.J., Hyttinen J.M.T., Koskela A., Chen M., Leinonen H., Tanila H., Kivinen N., Koistinen A., Toropainen E., Amadio M., Smedowski A., Reinisalo M., Winiarczyk M., Mackiewicz J., Mutikainen M., Ruotsalainen A.-K., Kettunen M.…Kaarniranta K. Loss of NRF-2 and PGC-1α genes leads to retinal pigment epithelium damage resembling dry age-related macular degeneration. Redox Biology. 2019;20:1–12. doi: 10.1016/j.redox.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenical W., Jensen P.R., Palladino M.A., Lam K.S., Lloyd G.K., Potts B.C. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorganic & Medicinal Chemistry. 2009;17(6):2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Y., Verhaegen M., Miller T.P., Rush J.L., Steiner P., Opipari A.W.…Soengas M.S. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Research. 2005;65(14):6294–6304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- Fernandez-Godino R., Pierce E.A. C3a triggers formation of sub-retinal pigment epithelium deposits via the ubiquitin proteasome pathway. Scientific Reports. 2018;8(1):9679. doi: 10.1038/s41598-018-28143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor: Basic science and clinical progress. Endocrine Reviews. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Ferrington D.A., Gregerson D.S. Immunoproteasomes: Structure, function, and antigen presentation. Progress in Molecular Biology and Translational Science. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual Review of Biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Chen X., Walters K.J. Gates, channels, and switches: Elements of the proteasome machine. Trends in Biochemical Sciences. 2016;41(1):77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Prado M.A. The proteasome and its network: Engineering for adaptability. Cold Spring Harbor Perspectives in Biology. 2019:a033985. doi: 10.1101/cshperspect.a033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D.F., De Vos R.a.I., Van Dijk R., De Vrij F.M.S., Proper E.A., Sonnemans M.a.F.…Van Leeuwen F.W. Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. The FASEB Journal. 2003;17(14):2014–2024. doi: 10.1096/fj.03-0205com. [DOI] [PubMed] [Google Scholar]
- Fisher R.I., Bernstein S.H., Kahl B.S., Djulbegovic B., Robertson M.J., de Vos S.…Goy A. Multicenter phase II study of bortezomib in patients with relapsed or refractory mantle cell lymphoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2006;24(30):4867–4874. doi: 10.1200/JCO.2006.07.9665. [DOI] [PubMed] [Google Scholar]
- Fisher R.I., Dahlberg S., Nathwani B.N., Banks P.M., Miller T.P., Grogan T.M. A clinical analysis of two indolent lymphoma entities: Mantle cell lymphoma and marginal zone lymphoma (including the mucosa-associated lymphoid tissue and monocytoid B-cell subcategories): a Southwest Oncology Group study. Blood. 1995;85(4):1075–1082. [PubMed] [Google Scholar]
- Florea B.I., Verdoes M., Li N., van der Linden W.A., Geurink P.P., van den Elst H.…Overkleeft H.S. Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chemistry & Biology. 2010;17(8):795–801. doi: 10.1016/j.chembiol.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco E.J., Busconi L., Martone C.B., Sanchez J.J. Multicatalytic proteinase in fish muscle. Archives of Biochemistry and Biophysics. 1988;267(2):599–605. doi: 10.1016/0003-9861(88)90067-7. [DOI] [PubMed] [Google Scholar]
- Fonseca R., Vabulas R.M., Hartl F.U., Bonhoeffer T., Nägerl U.V. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52(2):239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Forget G., Gregory D.J., Olivier M. Proteasome-mediated degradation of STAT1alpha following infection of macrophages with Leishmania donovani. The Journal of Biological Chemistry. 2005;280(34):30542–30549. doi: 10.1074/jbc.M414126200. [DOI] [PubMed] [Google Scholar]
- Förster A., Whitby F.G., Hill C.P. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. The EMBO Journal. 2003;22(17):4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel A.M., Peña M.M.O., Xing Y.Y., Rafique Z., Berger F.G. Structural determinants for the intracellular degradation of human thymidylate synthase. Biochemistry. 2004;43(7):1972–1979. doi: 10.1021/bi035894p. [DOI] [PubMed] [Google Scholar]
- Fort P., Kajava A.V., Delsuc F., Coux O. Evolution of proteasome regulators in eukaryotes. Genome Biology and Evolution. 2015;7(5):1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosgerau K., Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discovery Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Fricke B., Heink S., Steffen J., Kloetzel P.-M., Krüger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Reports. 2007;8(12):1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker L.D. Proteasome Inhibitor Drugs. Annual Review of Pharmacology and Toxicology. 2020;60:457–476. doi: 10.1146/annurev-pharmtox-010919-023603. [DOI] [PubMed] [Google Scholar]
- Früh K., Yang Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Current Opinion in Immunology. 1999;11(1):76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- Fu Y., Ma G., Liu G., Li B., Li H., Hao X., Liu L. USP14 as a novel prognostic marker promotes cisplatin resistance via Akt/ERK signaling pathways in gastric cancer. Cancer Medicine. 2018;7(11):5577–5588. doi: 10.1002/cam4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara C., Dirden J.C., Tosini G. Photic regulation of melatonin in rat retina and the role of proteasomal proteolysis. Neuroreport. 2001;12(17):3833–3837. doi: 10.1097/00001756-200112040-00046. [DOI] [PubMed] [Google Scholar]
- Fukunaga K., Kudo T., Toh-e A., Tanaka K., Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochemical and Biophysical Research Communications. 2010;396(4):1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- Funakoshi M., Tomko R.J., Kobayashi H., Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137(5):887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado M., Johnson R., Kruger A., Turner D., Rule S. Addition of bortezomib to standard dose chop chemotherapy improves response and survival in relapsed mantle cell lymphoma. British Journal of Haematology. 2015;168(1):55–62. doi: 10.1111/bjh.13101. [DOI] [PubMed] [Google Scholar]
- Gacche R.N., Assaraf Y.G. Redundant angiogenic signaling and tumor drug resistance. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2018;36:47–76. doi: 10.1016/j.drup.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Gaczynska M., Osmulski P.A. Harnessing proteasome dynamics and allostery in drug design. Antioxidants & Redox Signaling. 2014;21(17):2286–2301. doi: 10.1089/ars.2013.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallerani E., Zucchetti M., Brunelli D., Marangon E., Noberasco C., Hess D.…Sessa C. A first in human phase I study of the proteasome inhibitor CEP-18770 in patients with advanced solid tumours and multiple myeloma. European Journal of Cancer. 2013;49(2):290–296. doi: 10.1016/j.ejca.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Gandolfi S., Laubach J.P., Hideshima T., Chauhan D., Anderson K.C., Richardson P.G. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Reviews. 2017;36(4):561–584. doi: 10.1007/s10555-017-9707-8. [DOI] [PubMed] [Google Scholar]
- Gao J., Li M., Qin S., Zhang T., Jiang S., Hu Y.…Jiang C. Cytosolic PINK1 promotes the targeting of ubiquitinated proteins to the aggresome-autophagy pathway during proteasomal stress. Autophagy. 2016;12(4):632–647. doi: 10.1080/15548627.2016.1147667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garderet L., Iacobelli S., Moreau P., Dib M., Lafon I., Niederwieser D., Masszi T., Fontan J., Michallet M., Gratwohl A., Milone G., Doyen C., Pegourie B., Hajek R., Casassus P., Kolb B., Chaleteix C., Hertenstein B., Onida F.…Gahrton G. Superiority of the triple combination of bortezomib-thalidomide-dexamethasone over the dual combination of thalidomide-dexamethasone in patients with multiple myeloma progressing or relapsing after autologous transplantation: The MMVAR/IFM 2005-04 Randomized Phase III Trial from the Chronic Leukemia Working Party of the European Group for Blood and Marrow Transplantation. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2012;30(20):2475–2482. doi: 10.1200/JCO.2011.37.4918. [DOI] [PubMed] [Google Scholar]
- Gatti L., Zuco V., Zaffaroni N., Perego P. Drug combinations with proteasome inhibitors in antitumor therapy. Current Pharmaceutical Design. 2013;19(22):4094–4114. doi: 10.2174/1381612811319220015. [DOI] [PubMed] [Google Scholar]
- Ghobrial I.M., Vij R., Siegel D., Badros A., Kaufman J., Raje N.…Berdeja J.G. A phase Ib/II study of oprozomib in patients with advanced multiple myeloma and Waldenström macroglobulinemia. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2019;25(16):4907–4916. doi: 10.1158/1078-0432.CCR-18-3728. [DOI] [PubMed] [Google Scholar]
- Giampà C., Laurenti D., Anzilotti S., Bernardi G., Menniti F.S., Fusco F.R. Inhibition of the striatal specific phosphodiesterase PDE10A ameliorates striatal and cortical pathology in R6/2 mouse model of Huntington's disease. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J., Lee J.W., Argiris A., Haigentz M., Feldman L.E., Jang M.…Forastiere A.A. Phase II 2-arm trial of the proteasome inhibitor, PS-341 (bortezomib) in combination with irinotecan or PS-341 alone followed by the addition of irinotecan at time of progression in patients with locally recurrent or metastatic squamous cell carcinoma of the head and neck (E1304): A trial of the Eastern Cooperative Oncology Group. Head & Neck. 2013;35(7):942–948. doi: 10.1002/hed.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani N., Storti P., Bolzoni M., Palma B.D., Bonomini S. Angiogenesis and multiple myeloma. Cancer Microenvironment. 2011;4(3):325–337. doi: 10.1007/s12307-011-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulivi C., Pacifici R.E., Davies K.J. Exposure of hydrophobic moieties promotes the selective degradation of hydrogen peroxide-modified hemoglobin by the multicatalytic proteinase complex, proteasome. Archives of Biochemistry and Biophysics. 1994;311(2):329–341. doi: 10.1006/abbi.1994.1245. [DOI] [PubMed] [Google Scholar]
- Giżyńska M., Witkowska J., Karpowicz P., Rostankowski R., Chocron E.S., Pickering A.M.…Jankowska E. Proline- and arginine-rich peptides as flexible allosteric modulators of human proteasome activity. Journal of Medicinal Chemistry. 2019;62(1):359–370. doi: 10.1021/acs.jmedchem.8b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner G.G., Wong C.W. Alzheimer's disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochemical and Biophysical Research Communications. 1984;120(3):885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goedert M., Clavaguera F., Tolnay M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends in Neurosciences. 2010;33(7):317–325. doi: 10.1016/j.tins.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Goldberg A.L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin) Proceedings of the National Academy of Sciences of the United States of America. 1972;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.L., St John A.C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annual Review of Biochemistry. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldberg A.L., Zhao J., Collins G.A. Blocking cancer growth with less POMP or proteasomes. Molecular Cell. 2015;59(2):143–145. doi: 10.1016/j.molcel.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Bougie P., Wuillème-Toumi S., Ménoret E., Trichet V., Robillard N., Philippe M.…Amiot M. Noxa up-regulation and Mcl-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer Research. 2007;67(11):5418–5424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- Gonen N., Assaraf Y.G. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2012;15(4):183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Gong B., Radulovic M., Figueiredo-Pereira M.E., Cardozo C. The ubiquitin-proteasome system: Potential therapeutic targets for alzheimer's disease and spinal cord injury. Frontiers in Molecular Neuroscience. 2016;9(JAN) doi: 10.3389/fnmol.2016.00004. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorusupudi A., Liu A., Hageman G.S., Bernstein P.S. Associations of human retinal very long-chain polyunsaturated fatty acids with dietary lipid biomarkers. Journal of Lipid Research. 2016;57(3):499–508. doi: 10.1194/jlr.P065540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy A., Bernstein S.H., Kahl B.S., Djulbegovic B., Robertson M.J., de Vos S.…Fisher R.I. Bortezomib in patients with relapsed or refractory mantle cell lymphoma: Updated time-to-event analyses of the multicenter phase 2 PINNACLE study. Annals of Oncology. 2009;20(3):520–525. doi: 10.1093/annonc/mdn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goy A., Younes A., McLaughlin P., Pro B., Romaguera J.E., Hagemeister F.…Rodriguez A.M. Phase II study of proteasome inhibitor bortezomib in relapsed or refractory B-cell non-Hodgkin's lymphoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2005;23(4):667–675. doi: 10.1200/JCO.2005.03.108. [DOI] [PubMed] [Google Scholar]
- Gozzetti A., Papini G., Candi V., Brambilla C.Z., Sirianni S., Bocchia M. Second generation proteasome inhibitors in multiple myeloma. Anti-Cancer Agents in Medicinal Chemistry. 2017;17(7):920–926. doi: 10.2174/1871520616666160902101622. [DOI] [PubMed] [Google Scholar]
- Grandin E.W., Ky B., Cornell R.F., Carver J., Lenihan D.J. Patterns of cardiac toxicity associated with irreversible proteasome inhibition in the treatment of multiple myeloma. Journal of Cardiac Failure. 2015;21(2):138–144. doi: 10.1016/j.cardfail.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Grasso G., Pietropaolo A., Spoto G., Pappalardo G., Tundo G.R., Ciaccio C.…Rizzarelli E. Copper(I) and copper(II) inhibit Aβ peptides proteolysis by insulin-degrading enzyme differently: Implications for metallostasis alteration in Alzheimer's disease. Chemistry - A European Journal. 2011;17(9):2752–2762. doi: 10.1002/chem.201002809. [DOI] [PubMed] [Google Scholar]
- Grasso G., Salomone F., Tundo G.R., Pappalardo G., Ciaccio C., Spoto G.…Rizzarelli E. Metal ions affect insulin-degrading enzyme activity. Journal of Inorganic Biochemistry. 2012;117:351–358. doi: 10.1016/j.jinorgbio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Grasso G., Santoro A.M., Lanza V., Sbardella D., Tundo G.R., Ciaccio C.…Milardi D. The double faced role of copper in A? Homeostasis: A survey on the interrelationship between metal dyshomeostasis, UPS functioning and autophagy in neurodegeneration. Coordination Chemistry Reviews. 2017;347:1–22. [Google Scholar]
- Greene A.W., Grenier K., Aguileta M.A., Muise S., Farazifard R., Haque M.E.…Fon E.A. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Reports. 2012;13(4):378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori L., Hainfeld J.F., Simon M.N., Goldgaber D. Binding of amyloid beta protein to the 20 S proteasome. The Journal of Biological Chemistry. 1997;272(1):58–62. doi: 10.1074/jbc.272.1.58. [DOI] [PubMed] [Google Scholar]
- Gressin R., Daguindau N., Tempescul A., Moreau A., Carras S., Tchernonog E., Schmitt A., Houot R., Dartigeas C., Pignon J.M., Corm S., Banos A., Mounier C., Dupuis J., Macro M., Fleury J., Jardin F., Sarkozy C., Damaj G.…Lymphoma Study Association A phase 2 study of rituximab, bendamustine, bortezomib and dexamethasone for first-line treatment of older patients with mantle cell lymphoma. Haematologica. 2019;104(1):138–146. doi: 10.3324/haematol.2018.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin T.A., Nandi D., Cruz M., Fehling H.J., Kaer L.V., Monaco J.J., Colbert R.A. Immunoproteasome assembly: Cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. The Journal of Experimental Medicine. 1998;187(1):97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoreva T.A., Tribulovich V.G., Garabadzhiu A.V., Melino G., Barlev N.A. The 26S proteasome is a multifaceted target for anti-cancer therapies. Oncotarget. 2015;6(28):24733–24749. doi: 10.18632/oncotarget.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M., Standera S., Stohwasser R., Kloetzel P.M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(17):8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Bajorek M., Köhler A., Moroder L., Rubin D.M., Huber R.…Finley D. A gated channel into the proteasome core particle. Nature Structural Biology. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Groll M., Berkers C.R., Ploegh H.L., Ovaa H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure (London, England: 1993) 2006;14(3):451–456. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H.D., Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Groll M., Heinemeyer W., Jäger S., Ullrich T., Bochtler M., Wolf D.H., Huber R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):10976–10983. doi: 10.1073/pnas.96.20.10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Huber R. Substrate access and processing by the 20S proteasome core particle. The International Journal of Biochemistry & Cell Biology. 2003;35(5):606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Groll M., Nazif T., Huber R., Bogyo M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chemistry & Biology. 2002;9(5):655–662. doi: 10.1016/s1074-5521(02)00144-8. [DOI] [PubMed] [Google Scholar]
- Grune T., Catalgol B., Licht A., Ermak G., Pickering A.M., Ngo J.K., Davies K.J.A. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radical Biology & Medicine. 2011;51(7):1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T., Reinheckel T., Davies K.J. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. The Journal of Biological Chemistry. 1996;271(26):15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Jordan R., Lim M., Catzavelos C., Mestan J., Slingerland J., Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro J.I., Sunde M., Jones J.A., Campbell I.D., Dobson C.M. Amyloid fibril formation by an SH3 domain. Proceedings of the National Academy of Sciences. 1998;95(8):4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., He X.-B., Li S., Le W. A Central Role for Phosphorylated p38α in Linking Proteasome Inhibition-Induced Apoptosis and Autophagy. Molecular Neurobiology. 2017;54(10):7597–7609. doi: 10.1007/s12035-016-0260-1. [DOI] [PubMed] [Google Scholar]
- Guo Q., Lehmer C., Martínez-Sánchez A., Rudack T., Beck F., Hartmann H.…Fernández-Busnadiego R. In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell. 2018;172(4):696–705.e12. doi: 10.1016/j.cell.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang Y. Glutamate stimulates glutamate receptor interacting protein 1 degradation by ubiquitin-proteasome system to regulate surface expression of GluR2. Neuroscience. 2007;145(1):100–109. doi: 10.1016/j.neuroscience.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Gupta N., Hanley M.J., Venkatakrishnan K., Bessudo A., Rasco D.W., Sharma S.…Nemunaitis J. Effects of strong CYP3A inhibition and induction on the pharmacokinetics of ixazomib, an oral proteasome inhibitor: Results of drug-drug interaction studies in patients with advanced solid tumors or lymphoma and a physiologically based pharmacokinetic analysis. Journal of Clinical Pharmacology. 2018;58(2):180–192. doi: 10.1002/jcph.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Hanley M.J., Venkatakrishnan K., Wang B., Sharma S., Bessudo A.…Nemunaitis J. The effect of a high-fat meal on the pharmacokinetics of ixazomib, an oral proteasome inhibitor, in patients with advanced solid tumors or lymphoma. The Journal of Clinical Pharmacology. 2016;56(10):1288–1295. doi: 10.1002/jcph.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Hanley M.J., Xia C., Labotka R., Harvey R.D., Venkatakrishnan K. Clinical pharmacology of ixazomib: The first oral proteasome inhibitor. Clinical Pharmacokinetics. 2019;58(4):431–449. doi: 10.1007/s40262-018-0702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.P., Herzlich A.A., Sauer T., Chan C.-C. Retinal anatomy and pathology. Developments in Ophthalmology. 2016;55:7–17. doi: 10.1159/000431128. [DOI] [PubMed] [Google Scholar]
- Gupta I., Singh K., Varshney N.K., Khan S. Delineating Crosstalk Mechanisms of the Ubiquitin Proteasome System That Regulate Apoptosis. Frontiers in Cell and Development Biology. 2018;6:11. doi: 10.3389/fcell.2018.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Zhang S., Pusalkar S., Plesescu M., Chowdhury S., Hanley M.J.…Shepard D.R. A phase I study to assess the mass balance, excretion, and pharmacokinetics of [14C]-ixazomib, an oral proteasome inhibitor, in patients with advanced solid tumors. Investigational New Drugs. 2018;36(3):407–415. doi: 10.1007/s10637-017-0509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Zhao Y., Hui A.-M., Esseltine D.-L., Venkatakrishnan K. Switching from body surface area-based to fixed dosing for the investigational proteasome inhibitor ixazomib: A population pharmacokinetic analysis. British Journal of Clinical Pharmacology. 2015;79(5):789–800. doi: 10.1111/bcp.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman A., Glickman M.H. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. The Journal of Biological Chemistry. 2004;279(3):1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- Haas A.L., Rose I.A. Hemin inhibits ATP-dependent ubiquitin-dependent proteolysis: Role of hemin in regulating ubiquitin conjugate degradation. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(11 II):6845–6848. doi: 10.1073/pnas.78.11.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hagen M., Piebes D.G.E., de Leeuw W.C., Vuist I.M., van Roon-Mom W.M.C., Moerland P.D., Verschure P.J. The dynamics of early-state transcriptional changes and aggregate formation in a Huntington's disease cell model. BMC Genomics. 2017;18(1):373. doi: 10.1186/s12864-017-3745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J., Hirayama S., Murata S. Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis. PLoS Genetics. 2015;11(7) doi: 10.1371/journal.pgen.1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J., Iemura S.-I., Natsume T., Yashiroda H., Tanaka K., Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. The EMBO Journal. 2006;25(19):4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel F.G. Preliminary report: Inhibition of cellular proteasome activity by free fatty acids. Metabolism, Clinical and Experimental. 2009;58(8):1047–1049. doi: 10.1016/j.metabol.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Hamilton A.M., Oh W.C., Vega-Ramirez H., Stein I.S., Hell J.W., Patrick G.N., Zito K. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74(6):1023–1030. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman J.H., Enslin G.M., Kotzé A.F. Oral delivery of peptide drugs: Barriers and developments. BioDrugs: Clinical Immunotherapeutics, Biopharmaceuticals and Gene Therapy. 2005;19(3):165–177. doi: 10.2165/00063030-200519030-00003. [DOI] [PubMed] [Google Scholar]
- Han D.H., Na H.-K., Choi W.H., Lee J.H., Kim Y.K., Won C.…Lee M.J. Direct cellular delivery of human proteasomes to delay tau aggregation. Nature Communications. 2014;5:5633. doi: 10.1038/ncomms6633. [DOI] [PubMed] [Google Scholar]
- Hanada M., Sugawara K., Kaneta K., Toda S., Nishiyama Y., Tomita K.…Oki T. Epoxomicin, a new antitumor agent of microbial origin. The Journal of Antibiotics. 1992;45(11):1746–1752. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanna J., Hathaway N.A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D.S.…Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hanna J., Meides A., Zhang D.P., Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129(4):747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hanssum A., Zhong Z., Rousseau A., Krzyzosiak A., Sigurdardottir A., Bertolotti A. An inducible chaperone adapts proteasome assembly to stress. Molecular Cell. 2014;55(4):566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Suzuki K., Kimura H. TAK-063, a novel phosphodiesterase 10A inhibitor, protects from striatal neurodegeneration and ameliorates behavioral deficits in the R6/2 mouse model of Huntington's disease. The Journal of Pharmacology and Experimental Therapeutics. 2017;360(1):75–83. doi: 10.1124/jpet.116.237388. [DOI] [PubMed] [Google Scholar]
- Harbour J.W., Luo R.X., Dei Santi A., Postigo A.A., Dean D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- Harding R.J., Tong Y.-F. Proteostasis in Huntington's disease: Disease mechanisms and therapeutic opportunities. Acta Pharmacologica Sinica. 2018;39(5):754–769. doi: 10.1038/aps.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harer S.L., Bhatia M.S., Bhatia N.M. Proteasome inhibitors mechanism; source for design of newer therapeutic agents. The Journal of Antibiotics. 2012;65(6):279–288. doi: 10.1038/ja.2011.84. [DOI] [PubMed] [Google Scholar]
- Hari P., Lin H.M., Zhu Y., Berg D., Richardson P.G., Moreau P. Healthcare resource utilization with ixazomib or placebo plus lenalidomide-dexamethasone in the randomized, double-blind, phase 3 TOURMALINE-MM1 study in relapsed/refractory multiple myeloma. Journal of Medical Economics. 2018;21(8):793–798. doi: 10.1080/13696998.2018.1474745. [DOI] [PubMed] [Google Scholar]
- Hari P., Matous J.V., Voorhees P.M., Shain K.H., Obreja M., Frye J.…Sonneveld P. Oprozomib in patients with newly diagnosed multiple myeloma. Blood Cancer Journal. 2019;9(9) doi: 10.1038/s41408-019-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari P., Paba-Prada C.E., Voorhees P.M., Frye J., Chang Y.-L., Moreau P.…Shain K.H. Efficacy and safety results from a phase 1b/2, multicenter, open-label study of oprozomib and dexamethasone in patients with relapsed and/or refractory multiple myeloma. Leukemia Research. 2019;83:106172. doi: 10.1016/j.leukres.2019.106172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harousseau J.-L., Attal M., Avet-Loiseau H., Marit G., Caillot D., Mohty M.…Moreau P. Bortezomib plus dexamethasone is superior to vincristine plus doxorubicin plus dexamethasone as induction treatment prior to autologous stem-cell transplantation in newly diagnosed multiple myeloma: Results of the IFM 2005-01 phase III trial. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2010;28(30):4621–4629. doi: 10.1200/JCO.2009.27.9158. [DOI] [PubMed] [Google Scholar]
- Harris J.R. Release of a macromolecular protein component from human erythrocyte ghosts. Biochimica et Biophysica Acta. 1968;150(3):534–537. doi: 10.1016/0005-2736(68)90157-0. [DOI] [PubMed] [Google Scholar]
- Harrison S.J., Mainwaring P., Price T., Millward M.J., Padrik P., Underhill C.R.…Spencer A. Phase I clinical trial of marizomib (NPI-0052) in patients with advanced malignancies including multiple myeloma: Study NPI-0052-102 final results. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2016;22(18):4559–4566. doi: 10.1158/1078-0432.CCR-15-2616. [DOI] [PubMed] [Google Scholar]
- Harshbarger W., Miller C., Diedrich C., Sacchettini J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure (London, England: 1993) 2015;23(2):418–424. doi: 10.1016/j.str.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature Structural & Molecular Biology. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- He J., Kulkarni K., da Fonseca P.C.A., Krutauz D., Glickman M.H., Barford D., Morris E.P. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric α-helical rings. Structure (London, England: 1993) 2012;20(3):513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Heckman P.R.A., Blokland A., Bollen E.P.P., Prickaerts J. Phosphodiesterase inhibition and modulation of corticostriatal and hippocampal circuits: Clinical overview and translational considerations. Neuroscience & Biobehavioral Reviews. 2018;87:233–254. doi: 10.1016/j.neubiorev.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Hegde A.N., Smith S.G., Duke L.M., Pourquoi A., Vaz S. Perturbations of ubiquitin-proteasome-mediated proteolysis in aging and Alzheimer's disease. Frontiers in Aging Neuroscience. 2019;11 doi: 10.3389/fnagi.2019.00324. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Fischer M., Krimmer T., Stachon U., Wolf D.H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. The Journal of Biological Chemistry. 1997;272(40):25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- Heink S., Ludwig D., Kloetzel P.-M., Krüger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(26):9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann A., Rule S., Walewski J., Shpilberg O., Feng H., van de Velde H.…Louw V.J. Effect of cytochrome P450 3A4 inducers on the pharmacokinetic, pharmacodynamic and safety profiles of bortezomib in patients with multiple myeloma or non-Hodgkin's lymphoma. Clinical Pharmacokinetics. 2011;50(12):781–791. doi: 10.2165/11594410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Hemmis C.W., Heard S.C., Hill C.P. Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13. The Journal of Biological Chemistry. 2019;294(25):9659–9665. doi: 10.1074/jbc.AC119.008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon T.M., Deisseroth A., Kaminskas E., Kane R.C., Koti K.M., Rothmann M.D.…Pazdur R. U.s. Food and Drug Administration approval: Carfilzomib for the treatment of multiple myeloma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2013;19(17):4559–4563. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Heller H., Haas A.L., Rose I.A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(4):1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Eytan E., Ciechanover A., Haas A.L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. The Journal of Biological Chemistry. 1982;257(23):13964–13970. [PubMed] [Google Scholar]
- Hideshima T., Chauhan D., Richardson P., Mitsiades C., Mitsiades N., Hayashi T.…Anderson K.C. NF-kappa B as a therapeutic target in multiple myeloma. The Journal of Biological Chemistry. 2002;277(19):16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- Hideshima T., Mitsiades C., Akiyama M., Hayashi T., Chauhan D., Richardson P.…Anderson K.C. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101(4):1530–1534. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- Hideshima T., Richardson P., Chauhan D., Palombella V.J., Elliott P.J., Adams J., Anderson K.C. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Research. 2001;61(7):3071–3076. [PubMed] [Google Scholar]
- Hindo S.S., Frezza M., Tomco D., Heeg M.J., Hryhorczuk L., McGarvey B.R.…Verani C.N. Metals in anticancer therapy: Copper(II) complexes as inhibitors of the 20S proteasome. European Journal of Medicinal Chemistry. 2009;44(11):4353–4361. doi: 10.1016/j.ejmech.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M.S., Park S.-H., Hartl F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends in Cell Biology. 2014;24(9):506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Hayashi H., Iemura S.-I., Hendil K.B., Niwa S.-I., Kishimoto T.…Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Molecular Cell. 2006;24(6):977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Hendil K.B., Yashiroda H., Iemura S., Nagane R., Hioki Y.…Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437(7063):1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Hirano H., Kimura Y., Kimura A. Biological significance of co- and post-translational modifications of the yeast 26S proteasome. Journal of Proteomics. 2016;134:37–46. doi: 10.1016/j.jprot.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Hoefer M.M., Boneberg E.-M., Grotegut S., Kusch J., Illges H. Possible tetramerisation of the proteasome maturation factor POMP/proteassemblin/hUmp1 and its subcellular localisation. International Journal of Biological Macromolecules. 2006;38(3–5):259–267. doi: 10.1016/j.ijbiomac.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Homma T., Ishibashi D., Nakagaki T., Fuse T., Mori T., Satoh K.…Nishida N. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Scientific Reports. 2015;5 doi: 10.1038/srep11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D.P., Han S., Fink A.L., Uversky V.N. Characterization of the non-fibrillar α-synuclein oligomers. Protein and Peptide Letters. 2011;18(3):230–240. doi: 10.2174/092986611794578332. [DOI] [PubMed] [Google Scholar]
- Horan K.A., Hansen K., Jakobsen M.R., Holm C.K., Søby S., Unterholzner L.…Paludan S.R. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. Journal of Immunology (Baltimore, MD.: 1950) 2013;190(5):2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Jin J., Xu Y., Wu D., Ke X., Zhou D., Lu J., Du X., Chen X., Li J., Liu J., Gupta N., Hanley M.J., Li H., Hua Z., Wang B., Zhang X., Wang H., van de Velde H.…Moreau P. Randomized, double-blind, placebo-controlled phase III study of ixazomib plus lenalidomide-dexamethasone in patients with relapsed/refractory multiple myeloma: China Continuation study. Journal of Hematology & Oncology. 2017;10(1):137. doi: 10.1186/s13045-017-0501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. The Journal of Biological Chemistry. 1986;261(5):2400–2408. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. The Journal of Biological Chemistry. 1987;262(17):8303–8313. [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D.…Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. The EMBO Journal. 2005;24(21):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yen Y.-T., Singh S., Kao C.-L., Wu-Hsieh B.A. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunology. 2012;25(4):277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Chen C. Proteasome regulators: Activators and inhibitors. Current Medicinal Chemistry. 2009;16(8):931–939. doi: 10.2174/092986709787581860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.T., Dhungel B., Shrestha R., Bridle K.R., Crawford D.H.G., Jayachandran A., Steel J.C. Spotlight on bortezomib: Potential in the treatment of hepatocellular carcinoma. Expert Opinion on Investigational Drugs. 2019;28(1):7–18. doi: 10.1080/13543784.2019.1551359. [DOI] [PubMed] [Google Scholar]
- Huang L., Ho P., Chen C.H. Activation and inhibition of the proteasome by betulinic acid and its derivatives. FEBS Letters. 2007;581(25):4955–4959. doi: 10.1016/j.febslet.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Liu N., Zhao K., Zhu C., Lu X., Li S.…Liu J. Sanggenon C decreases tumor cell viability associated with proteasome inhibition. Frontiers in Bioscience (Elite Edition) 2011;3:1315–1325. doi: 10.2741/e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Wu Y., Zhou X., Xu J., Zhu W., Shu Y., Liu P. Efficacy of therapy with bortezomib in solid tumors: A review based on 32 clinical trials. Future Oncology (London, England) 2014;10(10):1795–1807. doi: 10.2217/fon.14.30. [DOI] [PubMed] [Google Scholar]
- Huber E.M., Heinemeyer W., Li X., Arendt C.S., Hochstrasser M., Groll M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nature Communications. 2016;7(1):1–10. doi: 10.1038/ncomms10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y.…Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453(7194):481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong S.A., Kapphahn R.J., Phillips S.L., Maldonado M., Ferrington D.A. Immunoproteasome deficiency alters retinal proteasome's response to stress. Journal of Neurochemistry. 2010;113(6):1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong S.A., Roehrich H., Kapphahn R.J., Maldonado M., Pardue M.T., Ferrington D.A. A novel role for the immunoproteasome in retinal function. Investigative Ophthalmology & Visual Science. 2011;52(2):714–723. doi: 10.1167/iovs.10-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrskyluoto A., Bruelle C., Lundh S.H., Do H.T., Kivinen J., Rappou E.…Korhonen L. Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: Involvement of the proteasome and ER stress-activated kinase IRE1α. Human Molecular Genetics. 2014;23(22):5928–5939. doi: 10.1093/hmg/ddu317. [DOI] [PubMed] [Google Scholar]
- Imai J., Maruya M., Yashiroda H., Yahara I., Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. The EMBO Journal. 2003;22(14):3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante J.R., Mendelson D.S., Burris H.A., Bendell J.C., Tolcher A.W., Gordon M.S.…Papadopoulos K.P. A first-in-human dose-escalation study of the oral proteasome inhibitor oprozomib in patients with advanced solid tumors. Investigational New Drugs. 2016;34(2):216–224. doi: 10.1007/s10637-016-0327-x. [DOI] [PubMed] [Google Scholar]
- Irrcher I., Aleyasin H., Seifert E.L., Hewitt S.J., Chhabra S., Phillips M., Lutz A.K., Rousseaux M.W.C., Bevilacqua L., Jahani-Asl A., Callaghan S., MacLaurin J.G., Winklhofer K.F., Rizzu P., Rippstein P., Kim R.H., Chen C.X., Fon E.A., Slack R.S.…Park D.S. Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Human Molecular Genetics. 2010;19(19):3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Isasa M., Katz E.J., Kim W., Yugo V., González S., Kirkpatrick D.S.…Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Molecular Cell. 2010;38(5):733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseri P.K., Altinaş O., Tokay T., Yüksel N. Relationship between cognitive impairment and retinal morphological and visual functional abnormalities in Alzheimer disease. Journal of Neuro-Ophthalmology: The Official Journal of the North American Neuro-Ophthalmology Society. 2006;26(1):18–24. doi: 10.1097/01.wno.0000204645.56873.26. [DOI] [PubMed] [Google Scholar]
- Jagannath S., Barlogie B., Berenson J., Siegel D., Irwin D., Richardson P.G.…Anderson K.C. A phase 2 study of two doses of bortezomib in relapsed or refractory myeloma. British Journal of Haematology. 2004;127(2):165–172. doi: 10.1111/j.1365-2141.2004.05188.x. [DOI] [PubMed] [Google Scholar]
- Jagannath S., Durie B.G.M., Wolf J., Camacho E., Irwin D., Lutzky J.…Crowley J. Bortezomib therapy alone and in combination with dexamethasone for previously untreated symptomatic multiple myeloma. British Journal of Haematology. 2005;129(6):776–783. doi: 10.1111/j.1365-2141.2005.05540.x. [DOI] [PubMed] [Google Scholar]
- Jain D., Singh K., Chirumamilla S., Bibat G.M., Blue M.E., Naidu S.R., Eberhart C.G. Ocular MECP2 protein expression in patients with and without Rett syndrome. Pediatric Neurology. 2010;43(1):35–40. doi: 10.1016/j.pediatrneurol.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Zode G., Kasetti R.B., Ran F.A., Yan W., Sharma T.P.…Sheffield V.C. CRISPR-Cas9–based treatment of myocilin-associated glaucoma. Proceedings of the National Academy of Sciences. 2017;114(42):11199–11204. doi: 10.1073/pnas.1706193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowiak A.J., Dytfeld D., Griffith K.A., Jasielec J., McDonnell K., Lebovic D.…Kaminski M.S. Treatment outcome with the combination of carfilzomib, lenalidomide, and low-dose dexamethasone (CRd) for newly diagnosed multiple myeloma (NDMM) after extended follow-up. Journal of Clinical Oncology. 2013;31(15_suppl):8543. doi: 10.1200/jco.2013.31.15_suppl.8543. [DOI] [Google Scholar]
- Jakubowiak A., Offidani M., Pégourie B., De La Rubia J., Garderet L., Laribi K.…Palumbo A. Randomized phase 2 study: Elotuzumab plus bortezomib/dexamethasone vs bortezomib/dexamethasone for relapsed/refractory MM. Blood. 2016;127(23):2833–2840. doi: 10.1182/blood-2016-01-694604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda E., Isidoro C., Carresi C., Mollace V. Defective autophagy in Parkinson's disease: Role of oxidative stress. Molecular Neurobiology. 2012;46(3):639–661. doi: 10.1007/s12035-012-8318-1. [DOI] [PubMed] [Google Scholar]
- Jang H.H. Regulation of Protein Degradation by Proteasomes in Cancer. Journal of Cancer Prevention. 2018;23(4):153–161. doi: 10.15430/JCP.2018.23.4.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome T.J., Kwapis J.L., Ruenzel W.L., Helmstetter F.J. CaMKII, but not protein kinase A, regulates Rpt6 phosphorylation and proteasome activity during the formation of long-term memories. Frontiers in Behavioral Neuroscience. 2013;7 doi: 10.3389/fnbeh.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger K.A. Interaction between pathogenic proteins in neurodegenerative disorders. Journal of Cellular and Molecular Medicine. 2012;16(6):1166–1183. doi: 10.1111/j.1582-4934.2011.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.-X., Zhao M., Qiu X.-B. Substrate receptors of proteasomes. Biological Reviews of the Cambridge Philosophical Society. 2018;93(4):1765–1777. doi: 10.1111/brv.12419. [DOI] [PubMed] [Google Scholar]
- Jing K., Shin S., Jeong S., Kim S., Song K.S., Park J.H.…Lim K. Docosahexaenoic acid induces the degradation of HPV E6/E7 oncoproteins by activating the ubiquitin-proteasome system. Cell Death & Disease. 2014;5(11) doi: 10.1038/cddis.2014.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe M.K., Nakaya N., Abu-Asab M., Tomarev S.I. Mutated myocilin and heterozygous Sod2 deficiency act synergistically in a mouse model of open-angle glaucoma. Human Molecular Genetics. 2015;24(12):3322–3334. doi: 10.1093/hmg/ddv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E. The ubiquitin-proteasome system: Opportunities for therapeutic intervention in solid tumors. Endocrine-Related Cancer. 2015;22(1):T1–17. doi: 10.1530/ERC-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.L., Tepe J.J. Proteasome activation to combat proteotoxicity. Molecules. 2019;24(15) doi: 10.3390/molecules24152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju D., Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. Journal of Biological Chemistry. 2004;279(23):23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- Juenemann K., Jansen A.H.P., van Riel L., Merkx R., Mulder M.P.C., An H.…Reits E.A. Dynamic recruitment of ubiquitin to mutant huntingtin inclusion bodies. Scientific Reports. 2018;8 doi: 10.1038/s41598-018-19538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenemann K., Wiemhoefer A., Reits E.A. Detection of ubiquitinated huntingtin species in intracellular aggregates. Frontiers in Molecular Neuroscience. 2015:8. doi: 10.3389/fnmol.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabayama H., Tokushige N., Takeuchi M., Kabayama M., Fukuda M., Mikoshiba K. Parkin promotes proteasomal degradation of synaptotagmin IV by accelerating polyubiquitination. Molecular and Cellular Neurosciences. 2017;80:89–99. doi: 10.1016/j.mcn.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Kageyama M., Ota T., Sasaoka M., Katsuta O., Shinomiya K. Chemical proteasome inhibition as a novel animal model of inner retinal degeneration in rats. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0217945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C.…Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Lemaire H.G., Unterbeck A., Salbaum J.M., Masters C.L., Grzeschik K.H.…Müller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325(6106):733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kaplan G.S., Torcun C.C., Grune T., Ozer N.K., Karademir B. Proteasome inhibitors in cancer therapy: Treatment regimen and peripheral neuropathy as a side effect. Free Radical Biology & Medicine. 2017;103:1–13. doi: 10.1016/j.freeradbiomed.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Kapuria V., Peterson L.F., Fang D., Bornmann W.G., Talpaz M., Donato N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Research. 2010;70(22):9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- Karin M., Cao Y., Greten F.R., Li Z.-W. NF-kappaB in cancer: from innocent bystander to major culprit. Nature Reviews. Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Karin M., Greten F.R. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nature Reviews. Immunology. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Karpowicz P., Osmulski P.A., Witkowska J., Sikorska E., Giżyńska M., Belczyk-Ciesielska A.…Jankowska E. Interplay between structure and charge as a key to allosteric modulation of human 20S proteasome by the basic fragment of HIV-1 tat protein. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0143038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastritis E., Dialoupi I., Gavriatopoulou M., Roussou M., Kanellias N., Fotiou D., Ntanasis-Stathopoulos I., Papadopoulou E., Ziogas D.C., Stamatelopoulos K., Manios E., Ntalianis A., Eleutherakis-Papaiakovou E., Papanikolaou A., Migkou M., Papanota A.-M., Gakiopoulou H., Psimenou E., Tselegkidi M.I.…Dimopoulos M.A. Primary treatment of light-chain amyloidosis with bortezomib, lenalidomide, and dexamethasone. Blood Advances. 2019;3(20):3002–3009. doi: 10.1182/bloodadvances.2019000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiki M., Chondrogianni N., Chinou I., Rivett A.J., Gonos E.S. The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Research. 2007;10(2):157–172. doi: 10.1089/rej.2006.0513. [DOI] [PubMed] [Google Scholar]
- Kaundal M., Zameer S., Najmi A.K., Parvez S., Akhtar M. Betulinic acid, a natural PDE inhibitor restores hippocampal cAMP/cGMP and BDNF, improve cerebral blood flow and recover memory deficits in permanent BCCAO induced vascular dementia in rats. European Journal of Pharmacology. 2018;832:56–66. doi: 10.1016/j.ejphar.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Kaur G., Batra S. Emerging role of immunoproteasomes in pathophysiology. Immunology & Cell Biology. 2016;94(9):812–820. doi: 10.1038/icb.2016.50. [DOI] [PubMed] [Google Scholar]
- Keck S., Nitsch R., Grune T., Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. Journal of Neurochemistry. 2003;85(1):115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- Keller J.N., Hanni K.B., Markesbery W.R. Impaired proteasome function in Alzheimer's disease. Journal of Neurochemistry. 2000;75(1):436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Keren-Kaplan T., Zeev Peters L., Levin-Kravets O., Attali I., Kleifeld O., Shohat N.…Prag G. Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism. Nature Communications. 2016;7(1):1–12. doi: 10.1038/ncomms12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrin A., Weldon S., Chung A.H.-K., Craig T., Simpson A.J., O'Kane C.M.…Taggart C.C. Proteolytic cleavage of elafin by 20S proteasome may contribute to inflammation in acute lung injury. Thorax. 2013;68(4):315–321. doi: 10.1136/thoraxjnl-2012-202536. [DOI] [PubMed] [Google Scholar]
- Khor B., Bredemeyer A.L., Huang C.-Y., Turnbull I.R., Evans R., Maggi L.B.…Sleckman B.P. Proteasome activator PA200 is required for normal spermatogenesis. Molecular and Cellular Biology. 2006;26(8):2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis E.A., Gidalevitz T., Morimoto R.I. Protein homeostasis in models of aging and age-related conformational disease. Advances in Experimental Medicine and Biology. 2010;694:138–159. doi: 10.1007/978-1-4419-7002-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.B., Crews C.M. From epoxomicin to carfilzomib: Chemistry, biology, and medical outcomes. Natural Product Reports. 2013;30(5):600–604. doi: 10.1039/c3np20126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.T., Goldberg A.L. The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. The Journal of Biological Chemistry. 2017;292(23):9830–9839. doi: 10.1074/jbc.M116.763128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Hawke N., Baldwin A.S. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death and Differentiation. 2006;13(5):738–747. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- Kim B.S., Savinova O.V., Reedy M.V., Martin J., Lun Y., Gan L.…Johnson R.L. Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Molecular and Cellular Biology. 2001;21(22):7707–7713. doi: 10.1128/MCB.21.22.7707-7713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Zaret K.S. Reprogramming of human cancer cells to pluripotency for models of cancer progression. The EMBO Journal. 2015;34(6):739–747. doi: 10.15252/embj.201490736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Caturegli P., Takahashi M., Suzuki K. New insights into the function of the immunoproteasome in immune and nonimmune cells. Journal of Immunology Research. 2015;2015:541984. doi: 10.1155/2015/541984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.W., Deshaies R.J., Peters J.M., Kirschner M.W. How proteolysis drives the cell cycle. Science (New York, N.Y.) 1996;274(5293):1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kiprowska M.J., Stepanova A., Todaro D.R., Galkin A., Haas A., Wilson S.M., Figueiredo-Pereira M.E. Neurotoxic mechanisms by which the USP14 inhibitor IU1 depletes ubiquitinated proteins and Tau in rat cerebral cortical neurons: relevance to Alzheimer's disease. Biochimica et Biophysica Acta. 2017;1863(6):1157–1170. doi: 10.1016/j.bbadis.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kish-Trier E., Hill C.P. Structural biology of the proteasome. Annual Review of Biophysics. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev A.F., Akopian T.N., Castillo V., Goldberg A.L. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Molecular Cell. 1999;4(3):395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Goldberg A.L. Proteasome inhibitors: From research tools to drug candidates. Chemistry & Biology. 2001;8(8):739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Kaganovich D., Goldberg A.L. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. The Journal of Biological Chemistry. 2002;277(25):22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Songyang Z., Goldberg A.L. Why does threonine, and not serine, function as the active site nucleophile in proteasomes? The Journal of Biological Chemistry. 2000;275(20):14831–14837. doi: 10.1074/jbc.275.20.14831. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., van der Linden W.A., Overkleeft H.S. Proteasome inhibitors: An expanding army attacking a unique target. Chemistry & Biology. 2012;19(1):99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips C.L., Jayaraj G.G., Hartl F.U. Pathways of cellular proteostasis in aging and disease. The Journal of Cell Biology. 2018;217(1):51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinknecht A., Popova B., Lázaro D.F., Pinho R., Valerius O., Outeiro T.F., Braus G.H. C-terminal tyrosine residue modifications modulate the protective phosphorylation of serine 129 of α-synuclein in a yeast model of Parkinson's disease. PLoS Genetics. 2016;12(6) doi: 10.1371/journal.pgen.1006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkradt S., Naudé R.J., Muramoto K., Oelofsen W. Purification and characterization of proteasome from ostrich liver. International Journal of Biochemistry & Cell Biology. 1997;29(4):611–622. doi: 10.1016/s1357-2725(96)00143-4. [DOI] [PubMed] [Google Scholar]
- Kniepert A., Groettrup M. The unique functions of tissue-specific proteasomes. Trends in Biochemical Sciences. 2014;39(1):17–24. doi: 10.1016/j.tibs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Knowles A., Koh K., Wu J.-T., Chien C.-T., Chamovitz D.A., Blau J. The COP9 signalosome is required for light-dependent timeless degradation and Drosophila clock resetting. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2009;29(4):1152–1162. doi: 10.1523/JNEUROSCI.0429-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Bajorek M., Groll M., Moroder L., Rubin D.M., Huber R.…Finley D. The substrate translocation channel of the proteasome. Biochimie. 2001;83(3–4):325–332. doi: 10.1016/s0300-9084(01)01242-1. [DOI] [PubMed] [Google Scholar]
- Köhler A., Cascio P., Leggett D.S., Woo K.M., Goldberg A.L., Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Molecular Cell. 2001;7(6):1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Komura H., Kakio S., Sasahara T., Arai Y., Takino N., Sato M.…Hoshi M. Alzheimer Aβ assemblies accumulate in excitatory neurons upon proteasome inhibition and kill nearby NAKα3 neurons by secretion. IScience. 2019;13:452–477. doi: 10.1016/j.isci.2019.01.018. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde N., Roschewski M., Zingone A., Kwok M., Manasanch E.E., Bhutani M., Tageja N., Kazandjian D., Mailankody S., Wu P., Morrison C., Costello R., Zhang Y., Burton D., Mulquin M., Zuchlinski D., Lamping L., Carpenter A., Wall Y.…Landgren O. Treatment with carfilzomib-lenalidomide-dexamethasone with lenalidomide extension in patients with smoldering or newly diagnosed multiple myeloma. JAMA Oncology. 2015;1(6):746–754. doi: 10.1001/jamaoncol.2015.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kors S., Geijtenbeek K., Reits E., Schipper-Krom S. Regulation of proteasome activity by (post-)transcriptional mechanisms. Frontiers in Molecular Biosciences. 2019;6 doi: 10.3389/fmolb.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulich E., Li X., DeMartino G.N. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Molecular Biology of the Cell. 2008;19(3):1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouroukis C.T., Fernandez L.A.V., Crump M., Gascoyne R.D., Chua N.S., Buckstein R.…Eisenhauer E. A phase II study of bortezomib and gemcitabine in relapsed mantle cell lymphoma from the National Cancer Institute of Canada Clinical Trials Group (IND 172) Leukemia & Lymphoma. 2011;52(3):394–399. doi: 10.3109/10428194.2010.546015. [DOI] [PubMed] [Google Scholar]
- Koyuncu S., Saez I., Lee H.J., Gutierrez-Garcia R., Pokrzywa W., Fatima A.…Vilchez D. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington's disease patients. Nature Communications. 2018;9(1):2886. doi: 10.1038/s41467-018-05320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozai T., Sekiguchi T., Satoh T., Yagi H., Kato K., Uchihashi T. Two-step process for disassembly mechanism of proteasome α7 homo-tetradecamer by α6 revealed by high-speed atomic force microscopy. Scientific Reports. 2017;7(1):1–9. doi: 10.1038/s41598-017-15708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjærg-Hansen A.…Cohen J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nature Genetics. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus M., Bader J., Geurink P.P., Weyburne E.S., Mirabella A.C., Silzle T.…Driessen C. The novel β2-selective proteasome inhibitor LU-102 synergizes with bortezomib and carfilzomib to overcome proteasome inhibitor resistance of myeloma cells. Haematologica. 2015;100(10):1350–1360. doi: 10.3324/haematol.2014.109421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Molecular Cell. 2009;33(4):496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Krebiehl G., Ruckerbauer S., Burbulla L.F., Kieper N., Maurer B., Waak J.…Krüger R. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson's disease-associated protein DJ-1. PLoS One. 2010;5(2) doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger E., Kloetzel P.M., Enenkel C. 20S proteasome biogenesis. Biochimie. 2001;83(3–4):289–293. doi: 10.1016/s0300-9084(01)01241-x. [DOI] [PubMed] [Google Scholar]
- Kubiczkova L., Pour L., Sedlarikova L., Hajek R., Sevcikova S. Proteasome inhibitors - molecular basis and current perspectives in multiple myeloma. Journal of Cellular and Molecular Medicine. 2014;18(6):947–961. doi: 10.1111/jcmm.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudriaeva A.A., Belogurov A.A. Proteasome: A nanomachinery of creative destruction. Biochemistry. Biokhimiia. 2019;84(Suppl. 1):S159–S192. doi: 10.1134/S0006297919140104. [DOI] [PubMed] [Google Scholar]
- Kudriaeva A., Kuzina E.S., Zubenko O., Smirnov I.V., Belogurov A. Charge-mediated proteasome targeting. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2019;33(6):6852–6866. doi: 10.1096/fj.201802237R. [DOI] [PubMed] [Google Scholar]
- Kulak N.A., Geyer P.E., Mann M. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Molecular & Cellular Proteomics: MCP. 2017;16(4):694–705. doi: 10.1074/mcp.O116.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.K., Bensinger W.I., Zimmerman T.M., Reeder C.B., Berenson J.R., Berg D.…Niesvizky R. Phase 1 study of weekly dosing with the investigational oral proteasome inhibitor ixazomib in relapsed/refractory multiple myeloma. Blood. 2014;124(7):1047–1055. doi: 10.1182/blood-2014-01-548941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.K., Berdeja J.G., Niesvizky R., Lonial S., Laubach J.P., Hamadani M.…Richardson P.G. Safety and tolerability of ixazomib, an oral proteasome inhibitor, in combination with lenalidomide and dexamethasone in patients with previously untreated multiple myeloma: An open-label phase 1/2 study. The Lancet. Oncology. 2014;15(13):1503–1512. doi: 10.1016/S1470-2045(14)71125-8. [DOI] [PubMed] [Google Scholar]
- Kumar S.K., Berdeja J.G., Niesvizky R., Lonial S., Laubach J.P., Hamadani M.…Richardson P.G. Ixazomib, lenalidomide, and dexamethasone in patients with newly diagnosed multiple myeloma: Long-term follow-up including ixazomib maintenance. Leukemia. 2019;33(7):1736–1746. doi: 10.1038/s41375-019-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.K., LaPlant B.R., Reeder C.B., Roy V., Halvorson A.E., Buadi F., Gertz M.A., Bergsagel P.L., Dispenzieri A., Thompson M.A., Crawley J., Kapoor P., Mikhael J., Stewart K., Hayman S.R., Hwa Y.L., Gonsalves W., Witzig T.E., Ailawadhi S.…Lacy M.Q. Randomized phase 2 trial of ixazomib and dexamethasone in relapsed multiple myeloma not refractory to bortezomib. Blood. 2016;128(20):2415–2422. doi: 10.1182/blood-2016-05-717769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.K., LaPlant B., Roy V., Reeder C.B., Lacy M.Q., Gertz M.A.…Dispenzieri A. Phase 2 trial of ixazomib in patients with relapsed multiple myeloma not refractory to bortezomib. Blood Cancer Journal. 2015;5 doi: 10.1038/bcj.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Singh D., Singh B.K., Singh S., Mittra N., Jha R.R.…Singh C. Alpha-synuclein aggregation, ubiquitin proteasome system impairment, and L-Dopa response in zinc-induced Parkinsonism: Resemblance to sporadic Parkinson's disease. Molecular and Cellular Biochemistry. 2018;444(1–2):149–160. doi: 10.1007/s11010-017-3239-y. [DOI] [PubMed] [Google Scholar]
- Kumatori A., Tanaka K., Inamura N., Sone S., Ogura T., Matsumoto T.…Ichihara A. Abnormally high expression of proteasomes in human leukemic cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(18):7071–7075. doi: 10.1073/pnas.87.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjappu M.J., Hochstrasser M. Assembly of the 20S proteasome. Biochimica et Biophysica Acta. 2014;1843(1):2–12. doi: 10.1016/j.bbamcr.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupperman E., Lee E.C., Cao Y., Bannerman B., Fitzgerald M., Berger A.…Bolen J. Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Research. 2010;70(5):1970–1980. doi: 10.1158/0008-5472.CAN-09-2766. [DOI] [PubMed] [Google Scholar]
- Kurimoto E., Satoh T., Ito Y., Ishihara E., Okamoto K., Yagi-Utsumi M.…Kato K. Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation. Protein Science: A Publication of the Protein Society. 2017;26(5):1080–1085. doi: 10.1002/pro.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup P.K., Xu J., Videira R.A., Ononenyi C., Baltazar G., Lombroso P.J., Nairn A.C. STEP61 is a substrate of the E3 ligase parkin and is upregulated in Parkinson's disease. Proceedings of the National Academy of Sciences. 2015;112(4):1202–1207. doi: 10.1073/pnas.1417423112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmierczyk A.R., Kunjappu M.J., Funakoshi M., Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nature Structural & Molecular Biology. 2008;15(3):237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. Huntington's disease: Underlying molecular mechanisms and emerging concepts. Trends in Biochemical Sciences. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. The biology of proteostasis in aging and disease. Annual Review of Biochemistry. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal P.M., Graziani G. Therapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitors. Pharmacological Research. 2018;136:97–107. doi: 10.1016/j.phrs.2018.08.023. [DOI] [PubMed] [Google Scholar]
- Lam Y.A., Xu W., DeMartino G.N., Cohen R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385(6618):737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lamm W., Kaufmann H., Raderer M., Hoffmann M., Chott A., Zielinski C., Drach J. Bortezomib combined with rituximab and dexamethasone is an active regimen for patients with relapsed and chemotherapy-refractory mantle cell lymphoma. Haematologica. 2011;96(7):1008–1014. doi: 10.3324/haematol.2011.041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A.P., Chen J., Chai Z.F., Hu Y. The neurotoxicity of iron, copper and cobalt in Parkinson's disease through ROS-mediated mechanisms. BioMetals. 2016;29(4):665–678. doi: 10.1007/s10534-016-9942-4. [DOI] [PubMed] [Google Scholar]
- Lander G.C., Estrin E., Matyskiela M.E., Bashore C., Nogales E., Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis-Piwowar K.R., Milacic V., Chen D., Yang H., Zhao Y., Chan T.H.…Dou Q.P. The proteasome as a potential target for novel anticancer drugs and chemosensitizers. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2006;9(6):263–273. doi: 10.1016/j.drup.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Lau J.L., Dunn M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic & Medicinal Chemistry. 2018;26(10):2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Le W. Role of iron in UPS impairment model of Parkinson's disease. Parkinsonism & Related Disorders. 2014;20(Suppl. 1):S158–S161. doi: 10.1016/S1353-8020(13)70038-5. [DOI] [PubMed] [Google Scholar]
- Le Y.-Z. VEGF production and signaling in Müller glia are critical to modulating vascular function and neuronal integrity in diabetic retinopathy and hypoxic retinal vascular diseases. Vision Research. 2017;139:108–114. doi: 10.1016/j.visres.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B., Barrault M.-B., Courbeyrette R., Guérois R., Marsolier-Kergoat M.-C., Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Molecular Cell. 2007;27(4):660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Le T.M., Wong H.H., Tay F.P.L., Fang S., Keng C.-T., Tan Y.J., Liu D.X. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. The FEBS Journal. 2007;274(16):4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc R., Catley L.P., Hideshima T., Lentzsch S., Mitsiades C.S., Mitsiades N.…Anderson K.C. Proteasome inhibitor PS-341 inhibits human myeloma cell growth in vivo and prolongs survival in a murine model. Cancer Research. 2002;62(17):4996–5000. [PubMed] [Google Scholar]
- Lecker S.H., Goldberg A.L., Mitch W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. Journal of the American Society of Nephrology: JASN. 2006;17(7):1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Lee V.M., Balin B.J., Otvos L., Trojanowski J.Q. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science (New York, N.Y.) 1991;251(4994):675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- Lee S.Y.-C., De la Mota-Peynado A., Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. The Journal of Biological Chemistry. 2011;286(42):36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.C., Fitzgerald M., Bannerman B., Donelan J., Bano K., Terkelsen J., Bradley D.P., Subakan O., Silva M.D., Liu R., Pickard M., Li Z., Tayber O., Li P., Hales P., Carsillo M., Neppalli V.T., Berger A.J., Kupperman E.…Janz S. Antitumor activity of the investigational proteasome inhibitor MLN9708 in mouse models of B-cell and plasma cell malignancies. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2011;17(23):7313–7323. doi: 10.1158/1078-0432.CCR-11-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Kim Y., Park J., Shim S., Lee J., Hong S.…Jung Y.-K. iRhom1 regulates proteasome activity via PAC1/2 under ER stress. Scientific Reports. 2015;5:11559. doi: 10.1038/srep11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.H., Lee J.M., Lim C., Kim S., Kim S.G. Structural requirements within protoporphyrin IX in the inhibition of heat shock protein 90. Chemico-Biological Interactions. 2013;204(1):49–57. doi: 10.1016/j.cbi.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Lee B.-H., Lee M.J., Park S., Oh D.-C., Elsasser S., Chen P.-C.…Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Lee J.H., Rubinsztein D.C. Tau degradation: The ubiquitin-proteasome system versus the autophagy-lysosome system. Progress in Neurobiology. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Lee Y., Lim H.-S. Skp2 Inhibitors: Novel Anticancer Strategies. Current Medicinal Chemistry. 2016;23(22):2363–2379. doi: 10.2174/0929867323666160510122624. [DOI] [PubMed] [Google Scholar]
- Lee B.-H., Lu Y., Prado M.A., Shi Y., Tian G., Sun S.…Finley D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532(7599):398–401. doi: 10.1038/nature17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Miller Z., Park J.E., Bhattarai D., Lee W., Kim K.B. H727 cells are inherently resistant to the proteasome inhibitor carfilzomib, yet require proteasome activity for cell survival and growth. Scientific Reports. 2019;9(1):1–9. doi: 10.1038/s41598-019-40635-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leestemaker Y., de Jong A., Witting K.F., Penning R., Schuurman K., Rodenko B.…Ovaa H. Proteasome activation by small molecules. Cell Chemical Biology. 2017;24(6):725–736. doi: 10.1016/j.chembiol.2017.05.010. e7. [DOI] [PubMed] [Google Scholar]
- Leestemaker Y., Ovaa H. Tools to investigate the ubiquitin proteasome system. Drug Discovery Today: Technologies. 2017;26:25–31. doi: 10.1016/j.ddtec.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Leggett D.S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R.T.…Finley D. Multiple associated proteins regulate proteasome structure and function. Molecular Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C. Ecm29 fulfils quality control functions in proteasome assembly. Molecular Cell. 2010;38(6):879–888. doi: 10.1016/j.molcel.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Leleu X., Martin T.G., Einsele H., Lyons R.M., Durie B.G.M., Iskander K.S., Ailawadhi S. Role of proteasome inhibitors in relapsed and/or refractory multiple myeloma. Clinical Lymphoma, Myeloma & Leukemia. 2019;19(1):9–22. doi: 10.1016/j.clml.2018.08.016. [DOI] [PubMed] [Google Scholar]
- Leleu X., Masszi T., Bahlis N.J., Viterbo L., Baker B., Gimsing P.…Richardson P.G. Patient-reported health-related quality of life from the phase III TOURMALINE-MM1 study of ixazomib-lenalidomide-dexamethasone versus placebo-lenalidomide-dexamethasone in relapsed/refractory multiple myeloma. American Journal of Hematology. 2018 doi: 10.1002/ajh.25134. [DOI] [PubMed] [Google Scholar]
- Levêque D., Carvalho M.C.M., Maloisel F. Review. Clinical pharmacokinetics of bortezomib. In Vivo (Athens, Greece) 2007;21(2):273–278. [PubMed] [Google Scholar]
- Levin N., Spencer A., Harrison S.J., Chauhan D., Burrows F.J., Anderson K.C.…Trikha M. Marizomib irreversibly inhibits proteasome to overcome compensatory hyperactivation in multiple myeloma and solid tumour patients. British Journal of Haematology. 2016;174(5):711–720. doi: 10.1111/bjh.14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Amazit L., Long W., Lonard D.M., Monaco J.J., O'Malley B.W. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Molecular Cell. 2007;26(6):831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Li B., Fu J., Chen P., Ge X., Li Y., Kuiatse I.…Orlowski R.Z. The nuclear factor (erythroid-derived 2)-like 2 and proteasome maturation protein axis mediate bortezomib resistance in multiple myeloma. The Journal of Biological Chemistry. 2015;290(50):29854–29868. doi: 10.1074/jbc.M115.664953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Ho L., Piperdi B., Elrafei T., Camacho F.J., Rigas J.R.…Gucalp R. Phase II study of the proteasome inhibitor bortezomib (PS-341, Velcade) in chemotherapy-naïve patients with advanced stage non-small cell lung cancer (NSCLC) Lung Cancer (Amsterdam, Netherlands) 2010;68(1):89–93. doi: 10.1016/j.lungcan.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Li X., Huang Q., Long H., Zhang P., Su H., Liu J. A new gold(I) complex-au(PPh3)PT is a deubiquitinase inhibitor and inhibits tumor growth. EBioMedicine. 2019;39:159–172. doi: 10.1016/j.ebiom.2018.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang J., Zeng B., Yang D., Sun J., Yin X.…Ren G. PSMD2 regulates breast cancer cell proliferation and cell cycle progression by modulating p21 and p27 proteasomal degradation. Cancer Letters. 2018;430:109–122. doi: 10.1016/j.canlet.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Li C., Johnson D.E. Bortezomib induces autophagy in head and neck squamous cell carcinoma cells via JNK activation. Cancer Letters. 2012;314(1):102–107. doi: 10.1016/j.canlet.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Johnson D.E. Liberation of functional p53 by proteasome inhibition in human papilloma virus-positive head and neck squamous cell carcinoma cells promotes apoptosis and cell cycle arrest. Cell Cycle (Georgetown, Texas) 2013;12(6):923–934. doi: 10.4161/cc.23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li R., Grandis J.R., Johnson D.E. Bortezomib induces apoptosis via Bim and Bik up-regulation and synergizes with cisplatin in the killing of head and neck squamous cell carcinoma cells. Molecular Cancer Therapeutics. 2008;7(6):1647–1655. doi: 10.1158/1535-7163.MCT-07-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A.…Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Powell S.R., Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. The FASEB Journal. 2011;25(3):883–893. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sui J., Huang I.-C., Kuhn J.H., Radoshitzky S.R., Marasco W.A.…Farzan M. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367(2):367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Tian G., Langager D., Sokolova V., Finley D., Park S. Nucleotide-dependent switch in proteasome assembly mediated by the Nas6 chaperone. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(7):1548–1553. doi: 10.1073/pnas.1612922114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wu X., Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. Journal of Gynecologic Oncology. 2016;27(4):e43. doi: 10.3802/jgo.2016.27.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-W., Yang T.-C., Wan L., Lin Y.-J., Tsai F.-J., Lai C.-C., Lin C.-W. Correlation between TGF-β1 expression and proteomic profiling induced by severe acute respiratory syndrome coronavirus papain-like protease. Proteomics. 2012;12(21):3193–3205. doi: 10.1002/pmic.201200225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.C., Yu E., Ring S.C., Chovan J.P. Boronic acid-containing proteasome inhibitors: Alert to potential pharmaceutical bioactivation. Chemical Research in Toxicology. 2013;26(4):608–615. doi: 10.1021/tx400032n. [DOI] [PubMed] [Google Scholar]
- Li G., Yuan S., Pan Y., Liu Y., Huang G. Binding states of protein-metal complexes in cells. Analytical Chemistry. 2016;88(22):10860–10866. doi: 10.1021/acs.analchem.6b00032. [DOI] [PubMed] [Google Scholar]
- Li C., Zang Y., Sen M., Leeman-Neill R.J., Man D.S.K., Grandis J.R., Johnson D.E. Bortezomib up-regulates activated signal transducer and activator of transcription-3 and synergizes with inhibitors of signal transducer and activator of transcription-3 to promote head and neck squamous cell carcinoma cell death. Molecular Cancer Therapeutics. 2009;8(8):2211–2220. doi: 10.1158/1535-7163.MCT-09-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou Z., Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death and Differentiation. 2008;15(12):1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- Liao Y., Liu N., Hua X., Cai J., Xia X., Wang X.…Liu J. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death & Disease. 2017;8(2):e2585. doi: 10.1038/cddis.2016.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg M., Mansilla A., Zecchini V.R., Fleming A., Rubinsztein D.C. The Parkinson's disease protein LRRK2 impairs proteasome substrate clearance without affecting proteasome catalytic activity. Cell Death & Disease. 2011;2 doi: 10.1038/cddis.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Lachenmayer M.L., Wu S., Liu W., Kundu M., Wang R.…Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genetics. 2015;11(2) doi: 10.1371/journal.pgen.1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limanaqi F., Biagioni F., Gaglione A., Busceti C.L., Fornai F. A sentinel in the crosstalk between the nervous and immune system: The (Immuno)-proteasome. Frontiers in Immunology. 2019;10 doi: 10.3389/fimmu.2019.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-T., Chang W.-C., Chen H.-M., Lai H.-L., Chen C.-Y., Tao M.-H., Chern Y. Regulation of feedback between protein kinase A and the proteasome system worsens Huntington's disease. Molecular and Cellular Biology. 2013;33(5):1073–1084. doi: 10.1128/MCB.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-C., Chen K.-C., Chen C.-C., Cheng A.-L., Chen K.-F. CIP2A-mediated Akt activation plays a role in bortezomib-induced apoptosis in head and neck squamous cell carcinoma cells. Oral Oncology. 2012;48(7):585–593. doi: 10.1016/j.oraloncology.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Lin L., Jin Z., Tan H., Xu Q., Peng T., Li H. Atypical ubiquitination by E3 ligase WWP1 inhibits the proteasome-mediated degradation of mutant huntingtin. Brain Research. 2016;1643:103–112. doi: 10.1016/j.brainres.2016.03.027. [DOI] [PubMed] [Google Scholar]
- Lin X., Parisiadou L., Gu X.-L., Wang L., Shim H., Sun L.…Cai H. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64(6):807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of SARS-Cov-2 — Studies needed. New England Journal of Medicine. 2020;382(13):1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- Lipski D.A., Dewispelaere R., Foucart V., Caspers L.E., Defrance M., Bruyns C., Willermain F. MHC class II expression and potential antigen-presenting cells in the retina during experimental autoimmune uveitis. Journal of Neuroinflammation. 2017;14(1):136. doi: 10.1186/s12974-017-0915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Research. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hettinger C.L., Zhang D., Rezvani K., Wang X., Wang H. The proteasome function reporter GFPu accumulates in young brains of the APPswe/PS1dE9 Alzheimer's disease mouse models. Cellular and Molecular Neurobiology. 2014;34(3):315–322. doi: 10.1007/s10571-013-0022-9. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Li X., Huang H., Zhao C., Liao S., Yang C., Liu S., Song W., Lu X., Lan X., Chen X., Yi S., Xu L., Jiang L., Zhao C., Dong X., Zhou P., Li S., Wang S.…Liu J. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget. 2014;5(14):5453–5471. doi: 10.18632/oncotarget.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-W., Li X., Thompson D., Wooding K., Chang T., Tang Z.…DeMartino G.N. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Molecular Cell. 2006;24(1):39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Qiao F., Leiferman P.C., Ross A., Schlenker E.H., Wang H. FOXOs modulate proteasome activity in human-induced pluripotent stem cells of Huntington's disease and their derived neural cells. Human Molecular Genetics. 2017;26(22):4416–4428. doi: 10.1093/hmg/ddx327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanova E.S., Finkelstein S., Li J., Travis A.M., Hao Y., Klingeborn M.…Arshavsky V.Y. Increased proteasomal activity supports photoreceptor survival in inherited retinal degeneration. Nature Communications. 2018;9(1):1738. doi: 10.1038/s41467-018-04117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda M., Cukor B., Tam S.W., Lavin P., Fiorentino M., Draetta G.F.…Pagano M. Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nature Medicine. 1997;3(2):231–234. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- Lokireddy S., Kukushkin N.V., Goldberg A.L. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(52):E7176–E7185. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M., Parravano M., Serrao S., Ducoli P., Stirpe M., Lombardo G. Analysis of retinal capillaries in patients with type 1 diabetes and nonproliferative diabetic retinopathy using adaptive optics imaging. Retina (Philadelphia, PA.) 2013;33(8):1630–1639. doi: 10.1097/IAE.0b013e3182899326. [DOI] [PubMed] [Google Scholar]
- London A., Benhar I., Schwartz M. The retina as a window to the brain-from eye research to CNS disorders. Nature Reviews. Neurology. 2013;9(1):44–53. doi: 10.1038/nrneurol.2012.227. [DOI] [PubMed] [Google Scholar]
- Lopes U.G., Erhardt P., Yao R., Cooper G.M. p53-dependent induction of apoptosis by proteasome inhibitors. The Journal of Biological Chemistry. 1997;272(20):12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- Lopez Salon M., Pasquini L., Besio Moreno M., Pasquini J.M., Soto E. Relationship between beta-amyloid degradation and the 26S proteasome in neural cells. Experimental Neurology. 2003;180(2):131–143. doi: 10.1016/s0014-4886(02)00060-2. [DOI] [PubMed] [Google Scholar]
- Lopez-Salon M., Alonso M., Vianna M.R., Viola H., Mello e Souza, T., Izquierdo, I., Pasquini, J. M., & Medina, J. H. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. The European Journal of Neuroscience. 2001;14(11):1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- LoRusso P.M., Venkatakrishnan K., Ramanathan R.K., Sarantopoulos J., Mulkerin D., Shibata S.I.…Ivy P. Pharmacokinetics and safety of bortezomib in patients with advanced malignancies and varying degrees of liver dysfunction: Phase I NCI Organ Dysfunction Working Group Study NCI-6432. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18(10):2954–2963. doi: 10.1158/1078-0432.CCR-11-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell M.A., Robertson J.D., Teesdale W.J., Campbell J.L., Markesbery W.R. Copper, iron and zinc in Alzheimer's disease senile plaques. Journal of the Neurological Sciences. 1998;158(1):47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science (New York, N.Y.) 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lu H., Zong C., Wang Y., Young G.W., Deng N., Souda P.…Ping P. Revealing the dynamics of the 20 S proteasome phosphoproteome: A combined CID and electron transfer dissociation approach. Molecular & Cellular Proteomics: MCP. 2008;7(11):2073–2089. doi: 10.1074/mcp.M800064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łuczkowska K., Rogińska D., Ulańczyk Z., Machaliński B. Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells. International Journal of Molecular Sciences. 2020;21(3) doi: 10.3390/ijms21030751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H., Moreau P., Dimopoulos M.A., Mateos M.-V., Kaiser M., Hajek R.…Weisel K. Health-related quality of life in the ENDEAVOR study: Carfilzomib-dexamethasone vs bortezomib-dexamethasone in relapsed/refractory multiple myeloma. Blood Cancer Journal. 2019;9(3):23. doi: 10.1038/s41408-019-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Cao L., Liang X., Du A., Peng T., Li H. Herp promotes degradation of mutant Huntingtin: Involvement of the proteasome and molecular chaperones. Molecular Neurobiology. 2018;55(10):7652–7668. doi: 10.1007/s12035-018-0900-8. [DOI] [PubMed] [Google Scholar]
- Lv H., Wei G.-Y., Guo C.-S., Deng Y.-F., Jiang Y.-M., Gao C., Jian C.-D. 20S proteasome and glyoxalase 1 activities decrease in erythrocytes derived from Alzheimer's disease patients. Neural Regeneration Research. 2020;15(1):178–183. doi: 10.4103/1673-5374.264473. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.-Z., Bartczak A., Zhang J., Khattar R., Chen L., Liu M.F.…McGilvray I.D. Proteasome inhibition in vivo promotes survival in a lethal murine model of severe acute respiratory syndrome. Journal of Virology. 2010;84(23):12419–12428. doi: 10.1128/JVI.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.-S., Wang X.-F., Zhang Y.-J., Luo P., Long H.-D., Li L.…Fu D. Inhibition of USP14 Deubiquitinating Activity as a Potential Therapy for Tumors with p53 Deficiency. Molecular Therapy - Oncolytics. 2020;16:147–157. doi: 10.1016/j.omto.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.H., Yang H.H., Parker K., Manyak S., Friedman J.M., Altamirano C.…Berenson J.R. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2003;9(3):1136–1144. [PubMed] [Google Scholar]
- MacLaren A.P., Chapman R.S., Wyllie A.H., Watson C.J. p53-dependent apoptosis induced by proteasome inhibition in mammary epithelial cells. Cell Death and Differentiation. 2001;8(3):210–218. doi: 10.1038/sj.cdd.4400801. [DOI] [PubMed] [Google Scholar]
- Mahul-Mellier A.-L., Fauvet B., Gysbers A., Dikiy I., Oueslati A., Georgeon S.…Lashuel H.A. C-Abl phosphorylates α-synuclein and regulates its degradation: Implication for α-synuclein clearance and contribution to the pathogenesis of Parkinson's disease. Human Molecular Genetics. 2014;23(11):2858–2879. doi: 10.1093/hmg/ddt674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard R.A., Chistol G., Sen M., Righini M., Tan J., Kaiser C.M.…Bustamante C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145(3):459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P., Rudack T., Beck F., Danev R., Pfeifer G., Nagy I., Baumeister W. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(2):534–539. doi: 10.1073/pnas.1817752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasanch E.E., Orlowski R.Z. Proteasome inhibitors in cancer therapy. Nature Reviews. Clinical Oncology. 2017;14(7):417–433. doi: 10.1038/nrclinonc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton C.A., Johnson B., Singh M., Bailey C.P., Bouchier-Hayes L., Chandra J. Induction of cell death by the novel proteasome inhibitor marizomib in glioblastoma in vitro and in vivo. Scientific Reports. 2016;6:18953. doi: 10.1038/srep18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A.J., Palanimurugan R., Matias A.C., Ramos P.C., Dohmen R.J. Catalytic mechanism and assembly of the proteasome. Chemical Reviews. 2009;109(4):1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Vierstra R.D. Dynamic regulation of the 26S proteasome: From synthesis to degradation. Frontiers in Molecular Biosciences. 2019;6:40. doi: 10.3389/fmolb.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Baker T.A., Sauer R.T. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nature Structural & Molecular Biology. 2008;15(11):1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon, E. B., Molloy, S. A., Zimnicka, A. M., & Kaplan, J. H. (2007). Copper entry into human cells: Progress and unanswered questions. BioMetals, 20(3–4), 355–364. 10.1007/s10534-006-9066-3. [DOI] [PubMed]
- Marzano C., Pellei M., Tisato F., Santini C. Copper complexes as anticancer agents. Anti-Cancer Agents in Medicinal Chemistry. 2009;9(2):185–211. doi: 10.2174/187152009787313837. [DOI] [PubMed] [Google Scholar]
- Masamha C.P., Benbrook D.M. Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells. Cancer Research. 2009;69(16):6565–6572. doi: 10.1158/0008-5472.CAN-09-0913. [DOI] [PubMed] [Google Scholar]
- Mateos M.-V., Cavo M., Blade J., Dimopoulos M.A., Suzuki K., Jakubowiak A., Knop S., Doyen C., Lucio P., Nagy Z., Pour L., Cook M., Grosicki S., Crepaldi A., Liberati A.M., Campbell P., Shelekhova T., Yoon S.-S., Iosava G.…San-Miguel J. Overall survival with daratumumab, bortezomib, melphalan, and prednisone in newly diagnosed multiple myeloma (ALCYONE): a randomised, open-label, phase 3 trial. Lancet (London, England) 2020;395(10218):132–141. doi: 10.1016/S0140-6736(19)32956-3. [DOI] [PubMed] [Google Scholar]
- Mateos M.-V., Hernández J.-M., Hernández M.-T., Gutiérrez N.-C., Palomera L., Fuertes M., Díaz-Mediavilla J., Lahuerta J.-J., de la Rubia J., Terol M.-J., Sureda A., Bargay J., Ribas P., de Arriba F., Alegre A., Oriol A., Carrera D., García-Laraña J., García-Sanz R.…San Miguel J.-F. Bortezomib plus melphalan and prednisone in elderly untreated patients with multiple myeloma: Results of a multicenter phase 1/2 study. Blood. 2006;108(7):2165–2172. doi: 10.1182/blood-2006-04-019778. [DOI] [PubMed] [Google Scholar]
- Mateos M.-V., Hernández J.M., Hernández M.T., Gutiérrez N.C., Palomera L., Fuertes M., Garcia-Sanchez P., Lahuerta J.J., de la Rubia J., Terol M.-J., Sureda A., Bargay J., Ribas P., Alegre A., de Arriba F., Oriol A., Carrera D., García-Laraña J., García-Sanz R.…San Miguel J.F. Bortezomib plus melphalan and prednisone in elderly untreated patients with multiple myeloma: Updated time-to-events results and prognostic factors for time to progression. Haematologica. 2008;93(4):560–565. doi: 10.3324/haematol.12106. [DOI] [PubMed] [Google Scholar]
- Mateos M.-V., Masszi T., Grzasko N., Hansson M., Sandhu I., Pour L.…Moreau P. Impact of prior therapy on the efficacy and safety of oral ixazomib-lenalidomide-dexamethasone vs. placebo-lenalidomide-dexamethasone in patients with relapsed/refractory multiple myeloma in TOURMALINE-MM1. Haematologica. 2017;102(10):1767–1775. doi: 10.3324/haematol.2017.170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos M.-V., Richardson P.G., Schlag R., Khuageva N.K., Dimopoulos M.A., Shpilberg O.…San Miguel J.F. Bortezomib plus melphalan and prednisone compared with melphalan and prednisone in previously untreated multiple myeloma: Updated follow-up and impact of subsequent therapy in the phase III VISTA trial. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2010;28(13):2259–2266. doi: 10.1200/JCO.2009.26.0638. [DOI] [PubMed] [Google Scholar]
- Mathewson A.C., Bishop A., Yao Y., Kemp F., Ren J., Chen H.…Jones I.M. Interaction of severe acute respiratory syndrome-coronavirus and NL63 coronavirus spike proteins with angiotensin converting enzyme-2. The Journal of General Virology. 2008;89:2741–2745. doi: 10.1099/vir.0.2008/003962-0. Pt 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias A.C., Ramos P.C., Dohmen R.J. Chaperone-assisted assembly of the proteasome core particle. Biochemical Society Transactions. 2010;38(Pt 1):29–33. doi: 10.1042/BST0380029. [DOI] [PubMed] [Google Scholar]
- Matsumura K., Aketa K. Proteasome (multicatalytic proteinase) of sea urchin sperm and its possible participation in the acrosome reaction. Molecular Reproduction and Development. 1991;29(2):189–199. doi: 10.1002/mrd.1080290215. [DOI] [PubMed] [Google Scholar]
- Matteucci A., Gaddini L., Villa M., Varano M., Parravano M., Monteleone V.…Pricci F. Neuroprotection by rat Müller glia against high glucose-induced neurodegeneration through a mechanism involving ERK1/2 activation. Experimental Eye Research. 2014;125:20–29. doi: 10.1016/j.exer.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Magnus T. Aging and neuronal vulnerabilitys. Nature Review. Neuroscience. 2006;7(4):278–294. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela M.E., Lander G.C., Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nature Structural & Molecular Biology. 2013;20(7):781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayaux J.F., Bousseau A., Pauwels R., Huet T., Hénin Y., Dereu N.…Le Pecq J.B. Triterpene derivatives that block entry of human immunodeficiency virus type 1 into cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(9):3564–3568. doi: 10.1073/pnas.91.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlary L., Plotkin S.S., Cashman N.R. Emerging developments in targeting proteotoxicity in neurodegenerative diseases. CNS Drugs. 2019;33(9):883–904. doi: 10.1007/s40263-019-00657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon C., Goold R., Andre R., Devoy A., Ortega Z., Moonga J.…Tabrizi S.J. Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathologica. 2016;131(3):411–425. doi: 10.1007/s00401-015-1508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon C., Tabrizi S.J. The ubiquitin-proteasome system in neurodegeneration. Antioxidants & Redox Signaling. 2014;21(17):2302–2321. doi: 10.1089/ars.2013.5802. [DOI] [PubMed] [Google Scholar]
- McNaught K.S.P., Mytilineou C., Jnobaptiste R., Yabut J., Shashidharan P., Jennert P., Olanow C.W. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. Journal of Neurochemistry. 2002;81(2):301–306. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- McNaught K.S.P., Perl D.P., Brownell A.-L., Olanow C.W. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson's disease. Annals of Neurology. 2004;56(1):149–162. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- McPherson S.W., Heuss N.D., Pierson M.J., Gregerson D.S. Retinal antigen-specific regulatory T cells protect against spontaneous and induced autoimmunity and require local dendritic cells. Journal of Neuroinflammation. 2014:11. doi: 10.1186/s12974-014-0205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Mohan R., Kwok B.H., Elofsson M., Sin N., Crews C.M. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe M.J., Huang Q., Figueiredo-Pereira M.E. Coordination between proteasome impairment and caspase activation leading to TAU pathology: Neuroprotection by cAMP. Cell Death & Disease. 2012;3(6):e326. doi: 10.1038/cddis.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L., Gan N., Chung F.-L. Isothiocyanates inhibit proteasome activity and proliferation of multiple myeloma cells. Carcinogenesis. 2011;32(2):216–223. doi: 10.1093/carcin/bgq242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micale N., Schirmeister T., Ettari R., Cinellu M.A., Maiore L., Serratrice M.…Messori L. Selected cytotoxic gold compounds cause significant inhibition of 20S proteasome catalytic activities. Journal of Inorganic Biochemistry. 2014;141:79–82. doi: 10.1016/j.jinorgbio.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Micera A., Quaranta L., Esposito G., Floriani I., Pocobelli A., Saccà S.C.…Oddone F. Differential protein expression profiles in glaucomatous trabecular meshwork: An evaluation study on a small primary open angle glaucoma population. Advances in Therapy. 2016;33(2):252–267. doi: 10.1007/s12325-016-0285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhael J.R., Schuster S.R., Jimenez-Zepeda V.H., Bello N., Spong J., Reeder C.B.…Fonseca R. Cyclophosphamide-bortezomib-dexamethasone (CyBorD) produces rapid and complete hematologic response in patients with AL amyloidosis. Blood. 2012;119(19):4391–4394. doi: 10.1182/blood-2011-11-390930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.F., Drake J.C., Naylor B., Price J.C., Hamilton K.L. The measurement of protein synthesis for assessing proteostasis in studies of slowed aging. Ageing Research Reviews. 2014;18:106–111. doi: 10.1016/j.arr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4(12) doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Sheng X., Yoshitake K., Sergeev Y., Iejima D., Shibagaki Y.…Iwata T. CCT2 mutations evoke Leber congenital amaurosis due to chaperone complex instability. Scientific Reports. 2016;6:33742. doi: 10.1038/srep33742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitch W.E., Goldberg A.L. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. The New England Journal of Medicine. 1996;335(25):1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- Mitsiades N., Mitsiades C.S., Richardson P.G., Poulaki V., Tai Y.-T., Chauhan D.…Anderson K.C. The proteasome inhibitor PS-341 potentiates sensitivity of multiple myeloma cells to conventional chemotherapeutic agents: Therapeutic applications. Blood. 2003;101(6):2377–2380. doi: 10.1182/blood-2002-06-1768. [DOI] [PubMed] [Google Scholar]
- Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Momand J., Wu H.H., Dasgupta G. MDM2--master regulator of the p53 tumor suppressor protein. Gene. 2000;242(1–2):15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- Moreau P., Attal M., Hulin C., Arnulf B., Belhadj K., Benboubker L., Béné M.C., Broijl A., Caillon H.L., Caillot D., Corre J., Delforge M., Dejoie T., Doyen C., Facon T., Sonntag C.C., Fontan J., Garderet L., Jie K.-S.…Sonneveld P. Bortezomib, thalidomide, and dexamethasone with or without daratumumab before and after autologous stem-cell transplantation for newly diagnosed multiple myeloma (CASSIOPEIA): A randomised, open-label, phase 3 study. The Lancet. 2019;394(10192):29–38. doi: 10.1016/S0140-6736(19)31240-1. [DOI] [PubMed] [Google Scholar]
- Moreau P., Kolb B., Attal M., Caillot D., Benboubker L., Tiab M.…Facon T. Phase 1/2 study of carfilzomib plus melphalan and prednisone in patients aged over 65 years with newly diagnosed multiple myeloma. Blood. 2015;125(20):3100–3104. doi: 10.1182/blood-2015-02-626168. [DOI] [PubMed] [Google Scholar]
- Moreau P., Kumar S., Boccia R., Iida S., Goldschmidt H., Cocks K.…Dimopoulos M. Convenience, satisfaction, health-related quality of life of once-weekly 70 mg/m2 vs. twice-weekly 27 mg/m2 carfilzomib (randomized A.R.R.O.W. study) Leukemia. 2019;33(12):2934–2946. doi: 10.1038/s41375-019-0480-2. [DOI] [PubMed] [Google Scholar]
- Moreau P., Masszi T., Grzasko N., Bahlis N.J., Hansson M., Pour L., Sandhu I., Ganly P., Baker B.W., Jackson S.R., Stoppa A.-M., Simpson D.R., Gimsing P., Palumbo A., Garderet L., Cavo M., Kumar S., Touzeau C., Buadi F.K.…Richardson P.G. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. New England Journal of Medicine. 2016;374(17):1621–1634. doi: 10.1056/NEJMoa1516282. [DOI] [PubMed] [Google Scholar]
- Moreau P., Mateos M.-V., Berenson J.R., Weisel K., Lazzaro A., Song K.…Stewart A.K. Once weekly versus twice weekly carfilzomib dosing in patients with relapsed and refractory multiple myeloma (A.R.R.O.W.): Interim analysis results of a randomised, phase 3 study. The Lancet. Oncology. 2018;19(7):953–964. doi: 10.1016/S1470-2045(18)30354-1. [DOI] [PubMed] [Google Scholar]
- Moreau P., Pylypenko H., Grosicki S., Karamanesht I., Leleu X., Grishunina M.…Harousseau J.-L. Subcutaneous versus intravenous administration of bortezomib in patients with relapsed multiple myeloma: A randomised, phase 3, non-inferiority study. The Lancet. Oncology. 2011;12(5):431–440. doi: 10.1016/S1470-2045(11)70081-X. [DOI] [PubMed] [Google Scholar]
- Moreau P., Stewart K.A., Dimopoulos M., Siegel D., Facon T., Berenson J.…Mateos M.-V. Once-weekly (70 mg/m2) vs twice-weekly (56 mg/m2) dosing of carfilzomib in patients with relapsed or refractory multiple myeloma: A post hoc analysis of the ENDEAVOR, A.R.R.O.W., and CHAMPION-1 trials. Cancer Medicine. 2020 doi: 10.1002/cam4.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morice-Picard F., Jonca N., Pichery M., Mermin D., Leauté-Labrèze C., Taïeb A.…Boralevi F. KLICK syndrome: Recognizable phenotype and hot-spot POMP mutation. Journal of the European Academy of Dermatology and Venereology: JEADV. 2017;31(3):e154–e156. doi: 10.1111/jdv.13898. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & Development. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A., Kimura M., Itokawa Y. The effect of aging on the mineral status of female mice. Biological Trace Element Research. 1994;42(2):165–177. doi: 10.1007/BF02785387. [DOI] [PubMed] [Google Scholar]
- Morozov A.V., Karpov V.L. Proteasomes and several aspects of their heterogeneity relevant to cancer. Frontiers in Oncology. 2019;9:761. doi: 10.3389/fonc.2019.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M., Knudsen G.M., Maeda S., Trinidad J.C., Ioanoviciu A., Burlingame A.L., Mucke L. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nature Neuroscience. 2015;18(8):1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsi R.Z., Hage-Sleiman R., Kobeissy H., Dbaibo G. Noxa: Role in Cancer Pathogenesis and Treatment. Current Cancer Drug Targets. 2018;18(10):914–928. doi: 10.2174/1568009618666180308105048. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O., Schermelleh L., Walter J., Cardoso M.C., Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(25):8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovitz O., Ben-Nissan G., Fainer I., Pollack D., Mizrachi L., Sharon M. The Parkinson's-associated protein DJ-1 regulates the 20S proteasome. Nature Communications. 2015;6:6609. doi: 10.1038/ncomms7609. [DOI] [PubMed] [Google Scholar]
- Mullauer F.B., Kessler J.H., Medema J.P. Betulinic acid, a natural compound with potent anticancer effects. Anti-Cancer Drugs. 2010;21(3):215–227. doi: 10.1097/CAD.0b013e3283357c62. [DOI] [PubMed] [Google Scholar]
- Murakami A. Modulation of protein quality control systems by food phytochemicals. Journal of Clinical Biochemistry and Nutrition. 2013;52(3):215–227. doi: 10.3164/jcbn.12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T.…Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murata S., Sasaki K., Kishimoto T., Niwa S.-I., Hayashi H., Takahama Y., Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science (New York, N.Y.) 2007;316(5829):1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Murata S., Takahama Y., Kasahara M., Tanaka K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nature Immunology. 2018;19(9):923–931. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- Murata S., Udono H., Tanahashi N., Hamada N., Watanabe K., Adachi K.…Chiba T. Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. The EMBO Journal. 2001;20(21):5898–5907. doi: 10.1093/emboj/20.21.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muz B., Ghazarian R.N., Ou M., Luderer M.J., Kusdono H.D., Azab A.K. Spotlight on ixazomib: Potential in the treatment of multiple myeloma. Drug Design, Development and Therapy. 2016;10:217–226. doi: 10.2147/DDDT.S93602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku, N., Clelland, C. L., Emrani, S., Kukushkin, N. V, Yu, W. H., Goldberg, A. L., & Duff, K. E. (2016). Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nature Medicine, 22(1), 46–53. 10.1038/nm.4011. [DOI] [PMC free article] [PubMed]
- Myeku N., Duff K.E. Targeting the 26S proteasome to protect against proteotoxic diseases. Trends in Molecular Medicine. 2018;24(1):18–29. doi: 10.1016/j.molmed.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J., Kim K.B., Lindsten K., Dantuma N.P., Crews C.M. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Molecular Cell. 2001;7(2):411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- Naash M.I., Al-Ubaidi M.R., Anderson R.E. Light exposure induces ubiquitin conjugation and degradation activities in the rat retina. Investigative Ophthalmology & Visual Science. 1997;38(11):2344–2354. [PubMed] [Google Scholar]
- Nag T.C., Wadhwa S. Ultrastructure of the human retina in aging and various pathological states. Micron (Oxford, England: 1993) 2012;43(7):759–781. doi: 10.1016/j.micron.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Nakanishi C., Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nature Reviews. Cancer. 2005;5(4):297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- Narayanan S., Cai C.-Y., Assaraf Y.G., Guo H.-Q., Cui Q., Wei L.…Chen Z.-S. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resistance Updates. 2020;48:100663. doi: 10.1016/j.drup.2019.100663. [DOI] [PubMed] [Google Scholar]
- Niazi S., Purohit M., Niazi J.H. Role of p53 circuitry in tumorigenesis: A brief review. European Journal of Medicinal Chemistry. 2018;158:7–24. doi: 10.1016/j.ejmech.2018.08.099. [DOI] [PubMed] [Google Scholar]
- Niewerth D., Jansen G., Assaraf Y.G., Zweegman S., Kaspers G.J.L., Cloos J. Molecular basis of resistance to proteasome inhibitors in hematological malignancies. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2015;18:18–35. doi: 10.1016/j.drup.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Nijholt D.A.T., de Graaf T.R., van Haastert E.S., Oliveira A.O., Berkers C.R., Zwart R.…Scheper W. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: Implications for Alzheimer's disease. Cell Death and Differentiation. 2011;18(6):1071–1081. doi: 10.1038/cdd.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki T., Endo J., Takahashi-Niki K., Yasuda T., Okamoto A., Saito Y.…Iguchi-Ariga S.M.M. DJ-1-binding compound B enhances Nrf2 activity through the PI3-kinase-Akt pathway by DJ-1-dependent inactivation of PTEN. Brain Research. 2020;1729:146641. doi: 10.1016/j.brainres.2019.146641. [DOI] [PubMed] [Google Scholar]
- Nitta T., Murata S., Sasaki K., Fujii H., Ripen A.M., Ishimaru N.…Takahama Y. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32(1):29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Njomen E., Tepe J.J. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. Journal of Medicinal Chemistry. 2019 doi: 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- Nunes A.T., Annunziata C.M. Proteasome inhibitors: Structure and function. Seminars in Oncology. 2017;44(6):377–380. doi: 10.1053/j.seminoncol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytemans K., Theuns J., Cruts M., Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Human Mutation. 2010;31(7):763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor O.A., Moskowitz C., Portlock C., Hamlin P., Straus D., Dumitrescu O.…Winter J. Patients with chemotherapy-refractory mantle cell lymphoma experience high response rates and identical progression-free survivals compared with patients with relapsed disease following treatment with single agent bortezomib: Results of a multicentre phase 2 clinical trial. British Journal of Haematology. 2009;145(1):34–39. doi: 10.1111/j.1365-2141.2008.07466.x. [DOI] [PubMed] [Google Scholar]
- O'Connor O.A., Stewart A.K., Vallone M., Molineaux C.J., Kunkel L.A., Gerecitano J.F., Orlowski R.Z. A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2009;15(22):7085–7091. doi: 10.1158/1078-0432.CCR-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor O.A., Wright J., Moskowitz C., Muzzy J., MacGregor-Cortelli B., Stubblefield M.…Zelenetz A.D. Phase II clinical experience with the novel proteasome inhibitor bortezomib in patients with indolent non-Hodgkin's lymphoma and mantle cell lymphoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2005;23(4):676–684. doi: 10.1200/JCO.2005.02.050. [DOI] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T.…Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science (New York, N.Y.) 2000;288(5468):1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Oddone F., Lucenteforte E., Michelessi M., Rizzo S., Donati S., Parravano M., Virgili G. Macular versus retinal nerve fiber layer parameters for diagnosing manifest glaucoma: A systematic review of diagnostic accuracy studies. Ophthalmology. 2016;123(5):939–949. doi: 10.1016/j.ophtha.2015.12.041. [DOI] [PubMed] [Google Scholar]
- Oerlemans R., Franke N.E., Assaraf Y.G., Cloos J., van Zantwijk I., Berkers C.R.…Jansen G. Molecular basis of bortezomib resistance: Proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112(6):2489–2499. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- Offidani M., Corvatta L., Caraffa P., Gentili S., Maracci L., Leoni P. An evidence-based review of ixazomib citrate and its potential in the treatment of newly diagnosed multiple myeloma. Oncotargets and Therapy. 2014;7:1793–1800. doi: 10.2147/OTT.S49187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso Y., Tomida A., Kim H.D., Tsuruo T. Glucose starvation and hypoxia induce nuclear accumulation of proteasome in cancer cells. Biochemical and Biophysical Research Communications. 1999;258(2):448–452. doi: 10.1006/bbrc.1999.0635. [DOI] [PubMed] [Google Scholar]
- Ohsawa K., Yoshida M., Izumikawa M., Takagi M., Shin-Ya K., Goshima N.…Doi T. Synthesis and biological evaluation of thielocin B1 analogues as protein-protein interaction inhibitors of PAC3 homodimer. Bioorganic & Medicinal Chemistry. 2018;26(23–24):6023–6034. doi: 10.1016/j.bmc.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.…Matsuda N. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes to Cells. 2010;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazuka K., Ishida T. Proteasome inhibitors for multiple myeloma. Japanese Journal of Clinical Oncology. 2018;48(9):785–793. doi: 10.1093/jjco/hyy108. [DOI] [PubMed] [Google Scholar]
- Olzscha H. Posttranslational modifications and proteinopathies: How guardians of the proteome are defeated. Biological Chemistry. 2019;400(7):895–915. doi: 10.1515/hsz-2018-0458. [DOI] [PubMed] [Google Scholar]
- Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G.…Vabulas R.M. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144(1):67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Opazo C., Huang X., Cherny R.A., Moir R.D., Roher A.E., White A.R.…Bush A.I. Metalloenzyme-like activity of Alzheimer's disease beta-amyloid. Cu-dependent catalytic conversion of dopamine, cholesterol, and biological reducing agents to neurotoxic H(2)O(2) The Journal of Biological Chemistry. 2002;277(43):40302–40308. doi: 10.1074/jbc.M206428200. [DOI] [PubMed] [Google Scholar]
- Orciuolo E., Buda G., Pelosini M., Petrini M. Fludarabine, Bortezomib, Myocet and rituximab chemotherapy in relapsed and refractory mantle cell lymphoma. British Journal of Haematology. 2010;148(5):810–812. doi: 10.1111/j.1365-2141.2009.07998.x. [DOI] [PubMed] [Google Scholar]
- Orlowski M. Selective activation of the 20 S proteasome (multicatalytic proteinase complex) by histone h3. Biochemistry. 2001;40(50):15318–15326. doi: 10.1021/bi0116240. [DOI] [PubMed] [Google Scholar]
- Orlowski M., Cardozo C., Hidalgo M.C., Michaud C. Regulation of the peptidylglutamyl-peptide hydrolyzing activity of the pituitary multicatalytic proteinase complex. Biochemistry. 1991;30(24):5999–6005. doi: 10.1021/bi00238a025. [DOI] [PubMed] [Google Scholar]
- Orlowski R.Z., Nagler A., Sonneveld P., Bladé J., Hajek R., Spencer A.…San-Miguel J.F. Final overall survival results of a randomized trial comparing bortezomib plus pegylated liposomal doxorubicin with bortezomib alone in patients with relapsed or refractory multiple myeloma. Cancer. 2016;122(13):2050–2056. doi: 10.1002/cncr.30026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski R.Z., Nagler A., Sonneveld P., Bladé J., Hajek R., Spencer A.…Harousseau J.-L. Randomized phase III study of pegylated liposomal doxorubicin plus bortezomib compared with bortezomib alone in relapsed or refractory multiple myeloma: Combination therapy improves time to progression. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2007;25(25):3892–3901. doi: 10.1200/JCO.2006.10.5460. [DOI] [PubMed] [Google Scholar]
- Orlowski R.Z., Stinchcombe T.E., Mitchell B.S., Shea T.C., Baldwin A.S., Stahl S.…Soignet S.L. Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2002;20(22):4420–4427. doi: 10.1200/JCO.2002.01.133. [DOI] [PubMed] [Google Scholar]
- Orlowski M., Wilk S. A multicatalytic protease complex from pituitary that forms enkephalin and enkephalin containing peptides. Biochemical and Biophysical Research Communications. 1981;101(3):814–822. doi: 10.1016/0006-291x(81)91823-4. [DOI] [PubMed] [Google Scholar]
- Oromendia A.B., Amon A. Aneuploidy: Implications for protein homeostasis and disease. Disease Models & Mechanisms. 2014;7(1):15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z., Díaz-Hernández M., Lucas J.J. Is the ubiquitin-proteasome system impaired in Huntington's disease? Cellular and Molecular Life Sciences: CMLS. 2007;64(17):2245–2257. doi: 10.1007/s00018-007-7222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z., Lucas J.J. Ubiquitin-proteasome system involvement in Huntington's disease. Frontiers in Molecular Neuroscience. 2014;7:77. doi: 10.3389/fnmol.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmulski P.A., Karpowicz P., Jankowska E., Bohmann J., Pickering A.M., Gaczyńska M. New peptide-based pharmacophore activates 20S proteasome. Molecules (Basel, Switzerland) 2020;25(6) doi: 10.3390/molecules25061439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottobelli L. Citicoline oral solution in glaucoma: is there a role in slowing disease progression? Ophthalmologica. 2013 doi: 10.1159/000350496. PMID 23615390. [DOI] [PubMed] [Google Scholar]
- Pack C.-G., Yukii H., Toh-e A., Kudo T., Tsuchiya H., Kaiho A.…Saeki Y. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nature Communications. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
- Padmanabhan A., Vuong S.A.-T., Hochstrasser M. Assembly of an evolutionarily conserved alternative proteasome isoform in human cells. Cell Reports. 2016;14(12):2962–2974. doi: 10.1016/j.celrep.2016.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladini G., Perfetti V., Obici L., Caccialanza R., Semino A., Adami F.…Merlini G. Association of melphalan and high-dose dexamethasone is effective and well tolerated in patients with AL (primary) amyloidosis who are ineligible for stem cell transplantation. Blood. 2004;103(8):2936–2938. doi: 10.1182/blood-2003-08-2788. [DOI] [PubMed] [Google Scholar]
- Palumbo A., Bringhen S., Larocca A., Rossi D., Di Raimondo F., Magarotto V., Patriarca F., Levi A., Benevolo G., Vincelli I.D., Grasso M., Franceschini L., Gottardi D., Zambello R., Montefusco V., Falcone A.P., Omedé P., Marasca R., Morabito F.…Cavo M. Bortezomib-melphalan-prednisone-thalidomide followed by maintenance with bortezomib-thalidomide compared with bortezomib-melphalan-prednisone for initial treatment of multiple myeloma: Updated follow-up and improved survival. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2014;32(7):634–640. doi: 10.1200/JCO.2013.52.0023. [DOI] [PubMed] [Google Scholar]
- Palumbo A., Bringhen S., Rossi D., Cavalli M., Larocca A., Ria R., Offidani M., Patriarca F., Nozzoli C., Guglielmelli T., Benevolo G., Callea V., Baldini L., Morabito F., Grasso M., Leonardi G., Rizzo M., Falcone A.P., Gottardi D.…Boccadoro M. Bortezomib-melphalan-prednisone-thalidomide followed by maintenance with bortezomib-thalidomide compared with bortezomib-melphalan-prednisone for initial treatment of multiple myeloma: a randomized controlled trial. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2010;28(34):5101–5109. doi: 10.1200/JCO.2010.29.8216. [DOI] [PubMed] [Google Scholar]
- Palumbo A., Chanan-Khan A., Weisel K., Nooka A.K., Masszi T., Beksac M.…Sonneveld P. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. New England Journal of Medicine. 2016;375(8):754–766. doi: 10.1056/NEJMoa1606038. [DOI] [PubMed] [Google Scholar]
- Pammolli F., Magazzini L., Riccaboni M. The productivity crisis in pharmaceutical R&D. Nature Reviews. Drug Discovery. 2011;10(6):428–438. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- Panasenko O.O., Collart M.A. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Molecular and Cellular Biology. 2011;31(8):1610–1623. doi: 10.1128/MCB.01210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit B., Gartel A.L. Proteasome inhibitors induce p53-independent apoptosis in human cancer cells. The American Journal of Pathology. 2011;178(1):355–360. doi: 10.1016/j.ajpath.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H., Chen D., Cui Q.C., Ping Dou Q. Sodium diethyldithiocarbamate, an AIDS progression inhibitor and a copper-binding compound, has proteasome-inhibitory and apoptosis-inducing activities in cancer cells. International Journal of Molecular Medicine. 2007;19(5):809–816. Scopus. [PubMed] [Google Scholar]
- Pao K.-C., Wood N.T., Knebel A., Rafie K., Stanley M., Mabbitt P.D.…Virdee S. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature. 2018;556(7701):381–385. doi: 10.1038/s41586-018-0026-1. [DOI] [PubMed] [Google Scholar]
- Papadopoulos K.P., Burris H.A., Gordon M., Lee P., Sausville E.A., Rosen P.J.…Infante J.R. A phase I/II study of carfilzomib 2-10-min infusion in patients with advanced solid tumors. Cancer Chemotherapy and Pharmacology. 2013;72(4):861–868. doi: 10.1007/s00280-013-2267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos K.P., Siegel D.S., Vesole D.H., Lee P., Rosen S.T., Zojwalla N.…Badros A. Phase I study of 30-minute infusion of carfilzomib as single agent or in combination with low-dose dexamethasone in patients with relapsed and/or refractory multiple myeloma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2015;33(7):732–739. doi: 10.1200/JCO.2013.52.3522. [DOI] [PubMed] [Google Scholar]
- Papaevgeniou N., Sakellari M., Jha S., Tavernarakis N., Holmberg C.I., Gonos E.S., Chondrogianni N. 18α-Glycyrrhetinic acid proteasome activator decelerates aging and Alzheimer's disease progression in Caenorhabditis elegans and neuronal cultures. Antioxidants and Redox Signaling. 2016;25(16):855–869. doi: 10.1089/ars.2015.6494. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou C.N., Daliani D.D., Nix D., Yang H., Madden T., Wang X.…Logothetis C.J. Phase I trial of the proteasome inhibitor bortezomib in patients with advanced solid tumors with observations in androgen-independent prostate cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2004;22(11):2108–2121. doi: 10.1200/JCO.2004.02.106. [DOI] [PubMed] [Google Scholar]
- Parisi V., Coppola G., Centofanti M., Oddone F., Angrisani A.M., Ziccardi L.…Manni G. Evidence of the neuroprotective role of citicoline in glaucoma patients. Progress in Brain Research. 2008;173:541–554. doi: 10.1016/S0079-6123(08)01137-0. [DOI] [PubMed] [Google Scholar]
- Parisi V., Oddone F., Roberti G., Tanga L., Carnevale C., Ziccardi L., Manni G. Enhancement of retinal function and of neural conduction along the visual pathway induced by treatment with citicoline eye drops in liposomal formulation in open angle glaucoma: A pilot electrofunctional study. Advances in Therapy. 2019;36(4):987–996. doi: 10.1007/s12325-019-0897-z. [DOI] [PubMed] [Google Scholar]
- Parisi V., Oddone F., Ziccardi L., Roberti G., Coppola G., Manni G. Citicoline and retinal ganglion cells: Effects on morphology and function. Current Neuropharmacology. 2018;16(7):919–932. doi: 10.2174/1570159X15666170703111729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi V., Restuccia R., Fattapposta F., Mina C., Bucci M.G., Pierelli F. Morphological and functional retinal impairment in Alzheimer's disease patients. Clinical Neurophysiology: Official Journal of the International Federation of Clinical Neurophysiology. 2001;112(10):1860–1867. doi: 10.1016/s1388-2457(01)00620-4. [DOI] [PubMed] [Google Scholar]
- Parisi V et al. (2015) Treatment with citicoline eye drops enhances retinal function and neural conduction along the visual pathways in open angle glaucoma. Graefes Arch Clin Exp Ophthalmol. PMDI 26004075 [DOI] [PubMed]
- Park S., Kim W., Tian G., Gygi S.P., Finley D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. The Journal of Biological Chemistry. 2011;286(42):36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Li X., Kim H.M., Singh C.R., Tian G., Hoyt M.A.…Finley D. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497(7450):512–516. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Miller Z., Jun Y., Lee W., Kim K.B. Next-generation proteasome inhibitors for cancer therapy. Translational Research: The Journal of Laboratory and Clinical Medicine. 2018;198:1–16. doi: 10.1016/j.trsl.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., Lee S.J., Aujay M., Suzuki E., Levitsky K., Lorens J.B.…Bennett M.K. Carfilzomib can induce tumor cell death through selective inhibition of the chymotrypsin-like activity of the proteasome. Blood. 2009;114(16):3439–3447. doi: 10.1182/blood-2009-05-223677. [DOI] [PubMed] [Google Scholar]
- Parravano M., Oddone F., Boccassini B., Centofanti M., Tanga L., Cacciamani A.…Varano M. Functional retinal impairment in type 1 diabetic patients without any signs of retinopathy. Ophthalmic Research. 2013;50(2):108–112. doi: 10.1159/000350412. [DOI] [PubMed] [Google Scholar]
- Pathare G.R., Nagy I., Bohn S., Unverdorben P., Hubert A., Körner R.…Bracher A. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(1):149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare G.R., Nagy I., Śledź P., Anderson D.J., Zhou H.-J., Pardon E.…Baumeister W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X.-Y., Dai Y., Grant S. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2004;10(11):3839–3852. doi: 10.1158/1078-0432.CCR-03-0561. [DOI] [PubMed] [Google Scholar]
- Pekol T., Daniels J.S., Labutti J., Parsons I., Nix D., Baronas E.…Miwa G. Human metabolism of the proteasome inhibitor bortezomib: Identification of circulating metabolites. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2005;33(6):771–777. doi: 10.1124/dmd.104.002956. [DOI] [PubMed] [Google Scholar]
- de la Peña A.H., Goodall E.A., Gates S.N., Lander G.C., Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science (New York, N.Y.) 2018;362(6418) doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B., Chen H., Lou X. Bortezomib-induced syndrome of inappropriate antidiuresis in a patient with multiple myeloma: A case report and literature review. International Journal of Clinical Pharmacology and Therapeutics. 2017;55(12):910–914. doi: 10.5414/CP203109. [DOI] [PubMed] [Google Scholar]
- Pereira R.B., Evdokimov N.M., Lefranc F., Valentão P., Kornienko A., Pereira D.M.…Gomes N.G.M. Marine-Derived Anticancer Agents: Clinical Benefits, Innovative Mechanisms, and New Targets. Marine Drugs. 2019;17(6) doi: 10.3390/md17060329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins N.D. The diverse and complex roles of NF-κB subunits in cancer. Nature Reviews. Cancer. 2012;12(2):121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- Perry G., Friedman R., Shaw G., Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(9):3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A.…Hutton M. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Human Molecular Genetics. 2004;13(7):703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- Pham L.V., Tamayo A.T., Yoshimura L.C., Lo P., Ford R.J. Inhibition of constitutive NF-kappa B activation in mantle cell lymphoma B cells leads to induction of cell cycle arrest and apoptosis. Journal of Immunology (Baltimore, MD: 1950) 2003;171(1):88–95. doi: 10.4049/jimmunol.171.1.88. [DOI] [PubMed] [Google Scholar]
- Picconi F., Mataluni G., Ziccardi L., Parravano M., Di Renzo A., Ylli D.…Frontoni S. Association between early neuroretinal dysfunction and peripheral motor unit loss in patients with type 1 diabetes mellitus. Journal of Diabetes Research. 2018;2018:9763507. doi: 10.1155/2018/9763507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picconi F., Parravano M., Sciarretta F., Fulci C., Nali M., Frontoni S.…Caccuri A.M. Activation of retinal Müller cells in response to glucose variability. Endocrine. 2019;65(3):542–549. doi: 10.1007/s12020-019-02017-5. [DOI] [PubMed] [Google Scholar]
- Picconi F., Parravano M., Ylli D., Pasqualetti P., Coluzzi S., Giordani I.…Frontoni S. Retinal neurodegeneration in patients with type 1 diabetes mellitus: The role of glycemic variability. Acta Diabetologica. 2017;54(5):489–497. doi: 10.1007/s00592-017-0971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickering A.M., Davies K.J.A. Differential roles of proteasome and immunoproteasome regulators Pa28αβ, Pa28γ and Pa200 in the degradation of oxidized proteins. Archives of Biochemistry and Biophysics. 2012;523(2):181–190. doi: 10.1016/j.abb.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering A.M., Koop A.L., Teoh C.Y., Ermak G., Grune T., Davies K.J.A. The immunoproteasome, the 20S proteasome, and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. The Biochemical Journal. 2010;432(3):585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piippo N., Korhonen E., Hytti M., Kinnunen K., Kaarniranta K., Kauppinen A. Oxidative stress is the principal contributor to inflammasome activation in retinal pigment epithelium cells with defunct proteasomes and autophagy. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2018;49(1):359–367. doi: 10.1159/000492886. [DOI] [PubMed] [Google Scholar]
- Pisha E., Chai H., Lee I.S., Chagwedera T.E., Farnsworth N.R., Cordell G.A.…Brown D.M. Discovery of betulinic acid as a selective inhibitor of human melanoma that functions by induction of apoptosis. Nature Medicine. 1995;1(10):1046–1051. doi: 10.1038/nm1095-1046. [DOI] [PubMed] [Google Scholar]
- Piva R., Ruggeri B., Williams M., Costa G., Tamagno I., Ferrero D., Giai V., Coscia M., Peola S., Massaia M., Pezzoni G., Allievi C., Pescalli N., Cassin M., di Giovine S., Nicoli P., de Feudis P., Strepponi I., Roato I.…Inghirami G. CEP-18770: A novel, orally active proteasome inhibitor with a tumor-selective pharmacologic profile competitive with bortezomib. Blood. 2008;111(5):2765–2775. doi: 10.1182/blood-2007-07-100651. [DOI] [PubMed] [Google Scholar]
- Podar K., Tai Y.T., Davies F.E., Lentzsch S., Sattler M., Hideshima T.…Anderson K.C. Vascular endothelial growth factor triggers signaling cascades mediating multiple myeloma cell growth and migration. Blood. 2001;98(2):428–435. doi: 10.1182/blood.v98.2.428. [DOI] [PubMed] [Google Scholar]
- Poli M.C., Ebstein F., Nicholas S.K., de Guzman M.M., Forbes L.R., Chinn I.K., Mace E.M., Vogel T.P., Carisey A.F., Benavides F., Coban-Akdemir Z.H., Gibbs R.A., Jhangiani S.N., Muzny D.M., Carvalho C.M.B., Schady D.A., Jain M., Rosenfeld J.A., Emrick L.…Orange J.S. Heterozygous truncating variants in POMP escape nonsense-mediated decay and cause a unique immune dysregulatory syndrome. American Journal of Human Genetics. 2018;102(6):1126–1142. doi: 10.1016/j.ajhg.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot S.A.H., Tian G., Finley D. Meddling with fate: The proteasomal deubiquitinating enzymes. Journal of Molecular Biology. 2017;429(22):3525–3545. doi: 10.1016/j.jmb.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts B.C., Albitar M.X., Anderson K.C., Baritaki S., Berkers C., Bonavida B., Chandra J., Chauhan D., Cusack J.C., Fenical W., Ghobrial I.M., Groll M., Jensen P.R., Lam K.S., Lloyd G.K., McBride W., McConkey D.J., Miller C.P., Neuteboom S.T.C.…Palladino M.A. Marizomib, a proteasome inhibitor for all seasons: Preclinical profile and a framework for clinical trials. Current Cancer Drug Targets. 2011;11(3):254–284. doi: 10.2174/156800911794519716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts B.C., Lam K.S. Generating a generation of proteasome inhibitors: from microbial fermentation to total synthesis of salinosporamide a (marizomib) and other salinosporamides. Marine Drugs. 2010;8(4):835–880. doi: 10.3390/md8040835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annual Review of Biochemistry. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Poyurovsky M.V., Priest C., Kentsis A., Borden K.L.B., Pan Z.-Q., Pavletich N., Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. The EMBO Journal. 2007;26(1):90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preckel T., Fung-Leung W.P., Cai Z., Vitiello A., Salter-Cid L., Winqvist O.…Yang Y. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science (New York, N.Y.) 1999;286(5447):2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A.…Pirmohamed M. Drug repurposing: Progress, challenges and recommendations. Nature Reviews. Drug Discovery. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Qian M.-X., Pang Y., Liu C.H., Haratake K., Du B.-Y., Ji D.-Y., Wang G.-F., Zhu Q.-Q., Song W., Yu Y., Zhang X.-X., Huang H.-T., Miao S., Chen L.-B., Zhang Z.-H., Liang Y.-N., Liu S., Cha H., Yang D.…Qiu X.-B. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153(5):1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.-Z., Ziffra J., Stennett L., Bodner B., Bonish B.K., Chaturvedi V.…Nickoloff B.J. Proteasome Inhibitors Trigger NOXA-Mediated Apoptosis in Melanoma and Myeloma Cells. Cancer Research. 2005;65(14):6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- Qiu X.-B., Ouyang S.-Y., Li C.-J., Miao S., Wang L., Goldberg A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. The EMBO Journal. 2006;25(24):5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Shen X., Shyam R., Yue B.Y.J.T., Ying H. Cellular processing of myocilin. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0092845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach H., White D., Spencer A., Ho P.J., Bhutani D., White M.…Gyger M. Pharmacokinetics and safety of carfilzomib in patients with relapsed multiple myeloma and end-stage renal disease (ESRD): An open-label, single-arm, phase I study. Cancer Chemotherapy and Pharmacology. 2017;79(6):1067–1076. doi: 10.1007/s00280-017-3287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta L., Riva I., Gerardi C., Oddone F., Floriano I., Konstas A.G.P. Quality of life in glaucoma: A review of the literature. Advances in Therapy. 2016;33(6):959–981. doi: 10.1007/s12325-016-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnel B., Preudhomme C., Oscier D., Lepelley P., Hooghe M.C., Facon T.…Fenaux P. Over-expression of the MDM2 gene is found in some cases of haematological malignancies. British Journal of Haematology. 1994;88(2):415–418. doi: 10.1111/j.1365-2141.1994.tb05044.x. [DOI] [PubMed] [Google Scholar]
- Qureshi A.A., Guan X.Q., Reis J.C., Papasian C.J., Jabre S., Morrison D.C., Qureshi N. Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by resveratrol, a potent proteasome inhibitor. Lipids in Health and Disease. 2012;11(1):1. doi: 10.1186/1476-511X-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi A.A., Zuvanich E.G., Khan D.A., Mushtaq S., Silswal N., Qureshi N. Proteasome inhibitors modulate anticancer and anti-proliferative properties via NF-kB signaling, and ubiquitin-proteasome pathways in cancer cell lines of different organs. Lipids in Health and Disease. 2018;17(1) doi: 10.1186/s12944-018-0697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Grinwis G.C.M., Rottier P.J.M., de Haan C.A.M. The proteasome inhibitor Velcade enhances rather than reduces disease in mouse hepatitis coronavirus-infected mice. Journal of Virology. 2010;84(15):7880–7885. doi: 10.1128/JVI.00486-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Posthuma C.C., Verheije M.H., te Lintelo E.G., Kikkert M., Drijfhout J.W.…de Haan C.A.M. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. Journal of Virology. 2010;84(15):7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S., Varadan R., Fushman D., Pickart C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nature Structural & Molecular Biology. 2005;12(8):708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rabl J., Smith D.M., Yu Y., Chang S.-C., Goldberg A.L., Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Molecular Cell. 2008;30(3):360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi N. The ubiquitin-proteasome system meets angiogenesis. Molecular Cancer Therapeutics. 2012;11(3):538–548. doi: 10.1158/1535-7163.MCT-11-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiss C.C., Braun T.S., Konings I.B.M., Grabmayr H., Hassink G.C., Sidhu A.…Claessens M.M.A.E. Functionally different α-synuclein inclusions yield insight into Parkinson's disease pathology. Scientific Reports. 2016;6:23116. doi: 10.1038/srep23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A.M., Kumar S. New investigational drugs with single-agent activity in multiple myeloma. Blood Cancer Journal. 2016;6(7) doi: 10.1038/bcj.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran K.V., Margolis S.S. A mammalian nervous system-specific plasma membrane proteasome complex that modulates neuronal function. Nature Structural & Molecular Biology. 2017;24(4):419–430. doi: 10.1038/nsmb.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P.C., Dohmen R.J. PACemakers of proteasome core particle assembly. Structure (London, England: 1993) 2008;16(9):1296–1304. doi: 10.1016/j.str.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Rangel L.P., Costa D.C.F., Vieira T.C.R.G., Silva J.L. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8(1):75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel L.P., Ferretti G.D.S., Costa C.L., Andrade S.M.M.V., Carvalho R.S., Costa D.C.F., Silva J.L. p53 reactivation with induction of massive apoptosis-1 (PRIMA-1) inhibits amyloid aggregation of mutant p53 in cancer cells. The Journal of Biological Chemistry. 2019;294(10):3670–3682. doi: 10.1074/jbc.RA118.004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi N., Mishra D.P. Therapeutic targeting of cancer cell cycle using proteasome inhibitors. Cell Division. 2012;7(1):26. doi: 10.1186/1747-1028-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raule M., Cerruti F., Benaroudj N., Migotti R., Kikuchi J., Bachi A.…Cascio P. PA28αβ reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chemistry & Biology. 2014;21(4):470–480. doi: 10.1016/j.chembiol.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Raule M., Cerruti F., Cascio P. Enhanced rate of degradation of basic proteins by 26S immunoproteasomes. Biochimica et Biophysica Acta. 2014;1843(9):1942–1947. doi: 10.1016/j.bbamcr.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Raynes R., Pomatto L.C.D., Davies K.J.A. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Molecular Aspects of Medicine. 2016;50:41–55. doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M.A., Neish A.S., Luscinskas F.W., Palombella V.J., Maniatis T., Collins T. The proteasome pathway is required for cytokine-induced endothelial-leukocyte adhesion molecule expression. Immunity. 1995;2(5):493–506. doi: 10.1016/1074-7613(95)90030-6. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Hill C.P. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends in Cell Biology. 2005;15(1):27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Regitz C., Fitzenberger E., Mahn F.L., Dußling L.M., Wenzel U. Resveratrol reduces amyloid-beta (Aβ₁₋₄₂)-induced paralysis through targeting proteostasis in an Alzheimer model of Caenorhabditis elegans. European Journal of Nutrition. 2016;55(2):741–747. doi: 10.1007/s00394-015-0894-1. [DOI] [PubMed] [Google Scholar]
- Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K.J., Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. The Biochemical Journal. 1998;335(Pt 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T., Ullrich O., Sitte N., Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Archives of Biochemistry and Biophysics. 2000;377(1):65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- Resch Z.T., Fautsch M.P. Glaucoma-associated myocilin: A better understanding but much more to learn. Experimental Eye Research. 2009;88(4):704–712. doi: 10.1016/j.exer.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Barlogie B., Berenson J., Singhal S., Jagannath S., Irwin D., Rajkumar S.V., Srkalovic G., Alsina M., Alexanian R., Siegel D., Orlowski R.Z., Kuter D., Limentani S.A., Lee S., Hideshima T., Esseltine D.-L., Kauffman M., Adams J.…Anderson K.C. A phase 2 study of bortezomib in relapsed, refractory myeloma. The New England Journal of Medicine. 2003;348(26):2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Barlogie B., Berenson J., Singhal S., Jagannath S., Irwin D.H.…Anderson K.C. Extended follow-up of a phase II trial in relapsed, refractory multiple myeloma: Final time-to-event results from the SUMMIT trial. Cancer. 2006;106(6):1316–1319. doi: 10.1002/cncr.21740. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Baz R., Wang M., Jakubowiak A.J., Laubach J.P., Harvey R.D.…Lonial S. Phase 1 study of twice-weekly ixazomib, an oral proteasome inhibitor, in relapsed/refractory multiple myeloma patients. Blood. 2014;124(7):1038–1046. doi: 10.1182/blood-2014-01-548826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Briemberg H., Jagannath S., Wen P.Y., Barlogie B., Berenson J.…Amato A.A. Frequency, characteristics, and reversibility of peripheral neuropathy during treatment of advanced multiple myeloma with bortezomib. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2006;24(19):3113–3120. doi: 10.1200/JCO.2005.04.7779. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Delforge M., Beksac M., Wen P., Jongen J.L., Sezer O., Terpos E., Munshi N., Palumbo A., Rajkumar S.V., Harousseau J.L., Moreau P., Avet-Loiseau H., Lee J.H., Cavo M., Merlini G., Voorhees P., Chng W.J., Mazumder A.…Sonneveld P. Management of treatment-emergent peripheral neuropathy in multiple myeloma. Leukemia. 2012;26(4):595–608. doi: 10.1038/leu.2011.346. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Hofmeister C.C., Rosenbaum C.A., Htut M., Vesole D.H., Berdeja J.…Baz R. Twice-weekly Oral MLN9708 (ixazomib citrate), An investigational proteasome inhibitor, in combination with lenalidomide (Len) and dexamethasone (Dex) in patients (Pts) with newly diagnosed multiple myeloma (MM): Final phase 1 results and phase 2 data. Blood. 2013;122(21):535. doi: 10.1182/blood.V122.21.535.535. [DOI] [Google Scholar]
- Richardson P.G., Hofmeister C.C., Rosenbaum C.A., Htut M., Vesole D.H., Berdeja J.G.…Baz R. Twice-weekly ixazomib in combination with lenalidomide-dexamethasone in patients with newly diagnosed multiple myeloma. British Journal of Haematology. 2018;182(2):231–244. doi: 10.1111/bjh.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Mitsiades C., Hideshima T., Anderson K.C. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annual Review of Medicine. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Oriol A., Beksac M., Liberati A.M., Galli M., Schjesvold F., Lindsay J., Weisel K., White D., Facon T., Miguel J.S., Sunami K., O'Gorman P., Sonneveld P., Robak P., Semochkin S., Schey S., Yu X., Doerr T.…Yee A. Pomalidomide, bortezomib, and dexamethasone for patients with relapsed or refractory multiple myeloma previously treated with lenalidomide (OPTIMISMM): a randomised, open-label, phase 3 trial. The Lancet Oncology. 2019;20(6):781–794. doi: 10.1016/S1470-2045(19)30152-4. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Sonneveld P., Schuster M., Irwin D., Stadtmauer E., Facon T., Harousseau J.-L., Ben-Yehuda D., Lonial S., Goldschmidt H., Reece D., Miguel J.S., Bladé J., Boccadoro M., Cavenagh J., Alsina M., Rajkumar S.V., Lacy M., Jakubowiak A.…Anderson K.C. Extended follow-up of a phase 3 trial in relapsed multiple myeloma: Final time-to-event results of the APEX trial. Blood. 2007;110(10):3557–3560. doi: 10.1182/blood-2006-08-036947. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Sonneveld P., Schuster M.W., Irwin D., Stadtmauer E.A., Facon T., Harousseau J.-L., Ben-Yehuda D., Lonial S., Goldschmidt H., Reece D., San-Miguel J.F., Bladé J., Boccadoro M., Cavenagh J., Dalton W.S., Boral A.L., Esseltine D.L., Porter J.B.…Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. The New England Journal of Medicine. 2005;352(24):2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- Richardson P.G., Weller E., Lonial S., Jakubowiak A.J., Jagannath S., Raje N.S., Avigan D.E., Xie W., Ghobrial I.M., Schlossman R.L., Mazumder A., Munshi N.C., Vesole D.H., Joyce R., Kaufman J.L., Doss D., Warren D.L., Lunde L.E., Kaster S.…Anderson K.C. Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood. 2010;116(5):679–686. doi: 10.1182/blood-2010-02-268862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Xie W., Jagannath S., Jakubowiak A., Lonial S., Raje N.S., Alsina M., Ghobrial I.M., Schlossman R.L., Munshi N.C., Mazumder A., Vesole D.H., Kaufman J.L., Colson K., McKenney M., Lunde L.E., Feather J., Maglio M.E., Warren D.…Anderson K.C. A phase 2 trial of lenalidomide, bortezomib, and dexamethasone in patients with relapsed and relapsed/refractory myeloma. Blood. 2014;123(10):1461–1469. doi: 10.1182/blood-2013-07-517276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Zweegman S., O'Donnell E.K., Laubach J.P., Raje N., Voorhees P.…Lonial S. Ixazomib for the treatment of multiple myeloma. Expert Opinion on Pharmacotherapy. 2018;19(17):1949–1968. doi: 10.1080/14656566.2018.1528229. [DOI] [PubMed] [Google Scholar]
- Robak T., Huang H., Jin J., Zhu J., Liu T., Samoilova O.…Cavalli F. Bortezomib-based therapy for newly diagnosed mantle-cell lymphoma. New England Journal of Medicine. 2015;372(10):944–953. doi: 10.1056/NEJMoa1412096. [DOI] [PubMed] [Google Scholar]
- Robak T., Huang H., Jin J., Zhu J., Liu T., Samoilova O.…Cavalli F. Association between bortezomib dose intensity and overall survival in mantle cell lymphoma patients on frontline VR-CAP in the phase 3 LYM-3002 study. Leukemia & Lymphoma. 2019;60(1):172–179. doi: 10.1080/10428194.2017.1321750. [DOI] [PubMed] [Google Scholar]
- Robak T., Jin J., Pylypenko H., Verhoef G., Siritanaratkul N., Drach J.…LYM-3002 investigators Frontline bortezomib, rituximab, cyclophosphamide, doxorubicin, and prednisone (VR-CAP) versus rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) in transplantation-ineligible patients with newly diagnosed mantle cell lymphoma: final overall survival results of a randomised, open-label, phase 3 study. The Lancet. Oncology. 2018;19(11):1449–1458. doi: 10.1016/S1470-2045(18)30685-5. [DOI] [PubMed] [Google Scholar]
- Robak P., Robak T. Bortezomib for the treatment of hematologic malignancies: 15 years later. Drugs in R&D. 2019;19(2):73–92. doi: 10.1007/s40268-019-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti G., Oddone F., Agnifili L., Katsanos A., Michelessi M., Mastropasqua L.…Manni G. Steroid-induced glaucoma: Epidemiology, pathophysiology, and clinical management. Survey of Ophthalmology. 2020 doi: 10.1016/j.survophthal.2020.01.002. [DOI] [PubMed] [Google Scholar]
- Roberti G., Tanga L., Michelessi M., Quaranta L., Parisi V., Manni G., Oddone F. Cytidine 5′-diphosphocholine (citicoline) in glaucoma: Rationale of its use, current evidence and future perspectives. International Journal of Molecular Sciences. 2015;16(12):28401–28417. doi: 10.3390/ijms161226099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccaro A.M., Hideshima T., Raje N., Kumar S., Ishitsuka K., Yasui H.…Anderson K.C. Bortezomib mediates antiangiogenesis in multiple myeloma via direct and indirect effects on endothelial cells. Cancer Research. 2006;66(1):184–191. doi: 10.1158/0008-5472.CAN-05-1195. [DOI] [PubMed] [Google Scholar]
- Roelofs J., Park S., Haas W., Tian G., McAllister F.E., Huo Y.…Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459(7248):861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeten M.S.F., Cloos J., Jansen G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemotherapy and Pharmacology. 2018;81(2):227–243. doi: 10.1007/s00280-017-3489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson I.B. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Letters. 2002;179(1):1–14. doi: 10.1016/s0304-3835(01)00847-3. [DOI] [PubMed] [Google Scholar]
- Rose S.A., Wass S., Jankowski J.J., Feldman J.F., Djukic A. Impaired visual search in children with Rett syndrome. Pediatric Neurology. 2019;92:26–31. doi: 10.1016/j.pediatrneurol.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen K.M., Moussa C.E.-H., Lee H.-K., Kumar P., Kitada T., Qin G.…Querfurth H.W. Parkin reverses intracellular beta-amyloid accumulation and its negative effects on proteasome function. Journal of Neuroscience Research. 2010;88(1):167–178. doi: 10.1002/jnr.22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C.L., Wong E., Petty E.M., Bale A.E., Tsujimoto Y., Harris N.L., Arnold A. PRAD1, a candidate BCL1 oncogene: Mapping and expression in centrocytic lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(21):9638–9642. doi: 10.1073/pnas.88.21.9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y., Bercovich Z., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. European Journal of Biochemistry. 1989;185(2):469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- Rosiñol L., Oriol A., Rios R., Sureda A., Blanchard M.J., Hernández M.T., Martínez-Martínez R., Moraleda J.M., Jarque I., Bargay J., Gironella M., de Arriba F., Palomera L., González-Montes Y., Martí J.M., Krsnik I., Arguiñano J.M., González M.E., González A.P.…Bladé J. Bortezomib, lenalidomide, and dexamethasone as induction therapy prior to autologous transplant in multiple myeloma. Blood. 2019;134(16):1337–1345. doi: 10.1182/blood.2019000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosiñol L., Oriol A., Teruel A.I., Hernández D., López-Jiménez J., de la Rubia J., Granell M., Besalduch J., Palomera L., González Y., Etxebeste M.A., Díaz-Mediavilla J., Hernández M.T., de Arriba F., Gutiérrez N.C., Martín-Ramos M.L., Cibeira M.T., Mateos M.V., Martínez J.…Programa para el Estudio y la Terapéutica de las Hemopatías Malignas/Grupo Español de Mieloma (PETHEMA/GEM) group Superiority of bortezomib, thalidomide, and dexamethasone (VTD) as induction pretransplantation therapy in multiple myeloma: A randomized phase 3 PETHEMA/GEM study. Blood. 2012;120(8):1589–1596. doi: 10.1182/blood-2012-02-408922. [DOI] [PubMed] [Google Scholar]
- Rousseau A., Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536(7615):184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A., Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nature Reviews Molecular Cell Biology. 2018;19(11):697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- Roussel M., Lauwers-Cances V., Robillard N., Hulin C., Leleu X., Benboubker L., Marit G., Moreau P., Pegourie B., Caillot D., Fruchart C., Stoppa A.-M., Gentil C., Wuilleme S., Huynh A., Hebraud B., Corre J., Chretien M.-L., Facon T.…Attal M. Front-line transplantation program with lenalidomide, bortezomib, and dexamethasone combination as induction and consolidation followed by lenalidomide maintenance in patients with multiple myeloma: A phase II study by the Intergroupe francophone du Myélome. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2014;32(25):2712–2717. doi: 10.1200/JCO.2013.54.8164. [DOI] [PubMed] [Google Scholar]
- Rowinsky E.K., Paner A., Berdeja J.G., Paba-Prada C., Venugopal P., Porkka K.…Landgren O. Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Investigational New Drugs. 2020 doi: 10.1007/s10637-020-00915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Martin P., Furman R.R., Lee S.M., Cheung K., Vose J.M.…Leonard J.P. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2011;29(6):690–697. doi: 10.1200/JCO.2010.31.1142. [DOI] [PubMed] [Google Scholar]
- Rubin D.M., Glickman M.H., Larsen C.N., Dhruvakumar S., Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. The EMBO Journal. 1998;17(17):4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Mena I., Mahillo E., Arribas J., Castaño J.G. Kinetic mechanism of activation by cardiolipin (diphosphatidylglycerol) of the rat liver multicatalytic proteinase. Biochemical Journal. 1993;296(1):93–97. doi: 10.1042/bj2960093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Krupnik Y., Keating M., Chandra J., Palladino M., McConkey D. The proteasome inhibitor NPI-0052 is a more effective inducer of apoptosis than bortezomib in lymphocytes from patients with chronic lymphocytic leukemia. Molecular Cancer Therapeutics. 2006;5(7):1836–1843. doi: 10.1158/1535-7163.MCT-06-0066. [DOI] [PubMed] [Google Scholar]
- Ruschak A.M., Religa T.L., Breuer S., Witt S., Kay L.E. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467(7317):868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- Sadre-Bazzaz K., Whitby F.G., Robinson H., Formosa T., Hill C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Molecular Cell. 2010;37(5):728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M.E.M., Mahmoud N., Sugimoto Y., Efferth T., Abdel-Aziz H. Betulinic Acid Exerts Cytotoxic Activity Against Multidrug-Resistant Tumor Cells via Targeting Autocrine Motility Factor Receptor (AMFR) Frontiers in Pharmacology. 2018;9:481. doi: 10.3389/fphar.2018.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y., Saitoh A., Toh-e A., Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochemical and Biophysical Research Communications. 2002;293(3):986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Tanaka K. Assembly and function of the proteasome. Methods in Molecular Biology (Clifton, N.J.) 2012;832:315–337. doi: 10.1007/978-1-61779-474-2_22. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Toh-e A., Kudo T., Kawamura H., Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137(5):900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Safren N., El Ayadi A., Chang L., Terrillion C.E., Gould T.D., Boehning D.F., Monteiro M.J. Ubiquilin-1 overexpression increases the lifespan and delays accumulation of Huntingtin aggregates in the R6/2 mouse model of Huntington's disease. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0087513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala A.J., Bott L.C., Morimoto R.I. Shaping proteostasis at the cellular, tissue, and organismal level. The Journal of Cell Biology. 2017;216(5):1231–1241. doi: 10.1083/jcb.201612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel J.F., Schlag R., Khuageva N.K., Dimopoulos M.A., Shpilberg O., Kropff M., Spicka I., Petrucci M.T., Palumbo A., Samoilova O.S., Dmoszynska A., Abdulkadyrov K.M., Delforge M., Jiang B., Mateos M.-V., Anderson K.C., Esseltine D.-L., Liu K., Deraedt W.…Richardson P.G. Persistent overall survival benefit and no increased risk of second malignancies with bortezomib-melphalan-prednisone versus melphalan-prednisone in patients with previously untreated multiple myeloma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2013;31(4):448–455. doi: 10.1200/JCO.2012.41.6180. [DOI] [PubMed] [Google Scholar]
- San Miguel J.F., Schlag R., Khuageva N.K., Dimopoulos M.A., Shpilberg O., Kropff M., Spicka I., Petrucci M.T., Palumbo A., Samoilova O.S., Dmoszynska A., Abdulkadyrov K.M., Schots R., Jiang B., Mateos M.-V., Anderson K.C., Esseltine D.L., Liu K., Cakana A.…Trial Investigators V.I.S.T.A. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. The New England Journal of Medicine. 2008;359(9):906–917. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- Sanagawa A., Hotta Y., Kondo M., Nishikawa R., Tohkin M., Kimura K. Tumor lysis syndrome associated with bortezomib: A post-hoc analysis after signal detection using the US Food and Drug Administration Adverse Event Reporting System. Anti-Cancer Drugs. 2020;31(2):183–189. doi: 10.1097/CAD.0000000000000862. [DOI] [PubMed] [Google Scholar]
- Sanchez E., Li M., Li J., Wang C., Chen H., Jones-Bolin S.…Berenson J.R. CEP-18770 (delanzomib) in combination with dexamethasone and lenalidomide inhibits the growth of multiple myeloma. Leukemia Research. 2012;36(11):1422–1427. doi: 10.1016/j.leukres.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Sanchez E., Li M., Steinberg J.A., Wang C., Shen J., Bonavida B.…Berenson J.R. The proteasome inhibitor CEP-18770 enhances the anti-myeloma activity of bortezomib and melphalan. British Journal of Haematology. 2010;148(4):569–581. doi: 10.1111/j.1365-2141.2009.08008.x. [DOI] [PubMed] [Google Scholar]
- Sanchez E., Li M., Wang C.S., Tang G., Gillespie A., Chen H., Berenson J.R. Anti-angiogenic and anti-multiple myeloma effects of oprozomib (OPZ) alone and in combination with pomalidomide (Pom) and/or dexamethasone (Dex) Leukemia Research. 2017;57:45–54. doi: 10.1016/j.leukres.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Sánchez-Serrano I. Success in translational research: Lessons from the development of bortezomib. Nature Reviews. Drug Discovery. 2006;5(2):107–114. doi: 10.1038/nrd1959. [DOI] [PubMed] [Google Scholar]
- Sanchorawala V., Palladini G., Kukreti V., Zonder J.A., Cohen A.D., Seldin D.C.…Merlini G. A phase 1/2 study of the oral proteasome inhibitor ixazomib in relapsed or refractory AL amyloidosis. Blood. 2017;130(5):597–605. doi: 10.1182/blood-2017-03-771220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Miguel J., Bladé J., Shpilberg O., Grosicki S., Maloisel F., Min C.-K.…Z R. Phase 2 randomized study of bortezomib-melphalan-prednisone with or without siltuximab (anti-IL-6) in multiple myeloma. Blood. 2014;123(26):4136–4142. doi: 10.1182/blood-2013-12-546374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Miguel J.F., Echeveste Gutierrez M.-A., Špicka I., Mateos M.-V., Song K., Craig M.D.…Lonial S. A phase I/II dose-escalation study investigating all-oral ixazomib-melphalan-prednisone induction followed by single-agent ixazomib maintenance in transplant-ineligible newly diagnosed multiple myeloma. Haematologica. 2018;103(9):1518–1526. doi: 10.3324/haematol.2017.185991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Miguel J.F., Hungria V.T.M., Yoon S.-S., Beksac M., Dimopoulos M.A., Elghandour A., Jedrzejczak W.W., Günther A., Nakorn T.N., Siritanaratkul N., Corradini P., Chuncharunee S., Lee J.-J., Schlossman R.L., Shelekhova T., Yong K., Tan D., Numbenjapon T., Cavenagh J.D.…Richardson P.G. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: A multicentre, randomised, double-blind phase 3 trial. The Lancet. Oncology. 2014;15(11):1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- San-Miguel J.F., Hungria V.T.M., Yoon S.-S., Beksac M., Dimopoulos M.A., Elghandour A.…Richardson P.G. Overall survival of patients with relapsed multiple myeloma treated with panobinostat or placebo plus bortezomib and dexamethasone (the PANORAMA 1 trial): a randomised, placebo-controlled, phase 3 trial. The Lancet. Haematology. 2016;3(11):e506–e515. doi: 10.1016/S2352-3026(16)30147-8. [DOI] [PubMed] [Google Scholar]
- Santoro A.M., Cunsolo A., D'Urso A., Sbardella D., Tundo G.R., Ciaccio C.…Purrello R. Cationic porphyrins are tunable gatekeepers of the 20S proteasome. Chemical Science. 2016;7(2):1286–1297. doi: 10.1039/c5sc03312h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A.M., Lo Giudice M.C., D'Urso A., Lauceri R., Purrello R., Milardi D. Cationic porphyrins are reversible proteasome inhibitors. Journal of the American Chemical Society. 2012;134(25):10451–10457. doi: 10.1021/ja300781u. [DOI] [PubMed] [Google Scholar]
- Santoro A.M., Monaco I., Attanasio F., Lanza V., Pappalardo G., Tomasello M.F.…Milardi D. Copper(II) ions affect the gating dynamics of the 20S proteasome: A molecular and in cell study. Scientific Reports. 2016;6(1):1–10. doi: 10.1038/srep33444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., Morais S., Pereira C., Sequeiros J., Alonso I. Parkin truncating variants result in a loss-of-function phenotype. Scientific Reports. 2019;9(1):16150. doi: 10.1038/s41598-019-52534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Hamazaki J., Koike M., Hirano Y., Komatsu M., Uchiyama Y.…Murata S. PAC1 gene knockout reveals an essential role of chaperone-mediated 20S proteasome biogenesis and latent 20S proteasomes in cellular homeostasis. Molecular and Cellular Biology. 2010;30(15):3864–3874. doi: 10.1128/MCB.00216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Takada K., Ohte Y., Kondo H., Sorimachi H., Tanaka K.…Murata S. Thymoproteasomes produce unique peptide motifs for positive selection of CD8 + T cells. Nature Communications. 2015;6(1):1–10. doi: 10.1038/ncomms8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Yagi-Utsumi M., Okamoto K., Kurimoto E., Tanaka K., Kato K. Molecular and structural basis of the proteasome α subunit assembly mechanism mediated by the proteasome-assembling chaperone PAC3-PAC4 heterodimer. International Journal of Molecular Sciences. 2019;20(9) doi: 10.3390/ijms20092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satriano C., Sfrazzetto G.T., Amato M.E., Ballistreri F.P., Copani A., Giuffrida M.L.…Toscano R.M. A ratiometric naphthalimide sensor for live cell imaging of copper(i) Chemical Communications. 2013;49(49):5565–5567. doi: 10.1039/c3cc42069h. [DOI] [PubMed] [Google Scholar]
- Savolainen M.H., Albert K., Airavaara M., Myöhänen T.T. Nigral injection of a proteasomal inhibitor, lactacystin, induces widespread glial cell activation and shows various phenotypes of Parkinson's disease in young and adult mouse. Experimental Brain Research. 2017;235(7):2189–2202. doi: 10.1007/s00221-017-4962-z. [DOI] [PubMed] [Google Scholar]
- Sbardella D., Coletta A., Tundo G.R., Ahmed I.M.M., Bellia F., Oddone F.…Coletta M. Structural and functional evidence for citicoline binding and modulation of 20S proteasome activity: Novel insights into its pro-proteostatic effect. Biochemical Pharmacology. 2020;177:113977. doi: 10.1016/j.bcp.2020.113977. [DOI] [PubMed] [Google Scholar]
- Sbardella D., Tundo G.R., Coletta A., Marcoux J., Koufogeorgou E.I., Ciaccio C.…Coletta M. The insulin-degrading enzyme is an allosteric modulator of the 20S proteasome and a potential competitor of the 19S. Cellular and Molecular Life Sciences: CMLS. 2018;75(18):3441–3456. doi: 10.1007/s00018-018-2807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbardella D., Tundo G.R., Cunsolo V., Grasso G., Cascella R., Caputo V., Santoro A.M., Milardi D., Pecorelli A., Ciaccio C., Di Pierro D., Leoncini S., Campagnolo L., Pironi V., Oddone F., Manni P., Foti S., Giardina E., De Felice C.…Marini S. Defective proteasome biogenesis into skin fibroblasts isolated from Rett syndrome subjects with MeCP2 non-sense mutations. Biochimica Et Biophysica Acta. Molecular Basis of Disease. 2020;1866(7):165793. doi: 10.1016/j.bbadis.2020.165793. [DOI] [PubMed] [Google Scholar]
- Sbardella D., Tundo G.R., Campagnolo L., Valacchi G., Orlandi A., Curatolo P., Borsellino G., D'Esposito M., Ciaccio C., Di Cesare S., Di Pierro D., Galasso C., Santarone M.E., Hayek J., Coletta M., (2017). Stefano Marini Retention of Mitochondria in Mature Human Red Blood Cells as the Result of Autophagy Impairment in Rett Syndrome Sci Rep. 7(1):12297. http://doi.org/10.1038/s41598-017-12069-0. [DOI] [PMC free article] [PubMed]
- Sbardella D., Tundo G.R., Sciandra F., Bozzi M., Gioia M., Ciaccio C.…Marini S. Proteasome activity is affected by fluctuations in insulin-degrading enzyme distribution. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nature Reviews Drug Discovery. 2012;11(3):191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Schaler A.W., Myeku N. Cilostazol, a phosphodiesterase 3 inhibitor, activates proteasome-mediated proteolysis and attenuates tauopathy and cognitive decline. Translational Research: The Journal of Laboratory and Clinical Medicine. 2018;193:31–41. doi: 10.1016/j.trsl.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Nuber U., Huibregtse J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R.…Wanker E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/S0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Schipper-Krom S., Juenemann K., Reits E.A.J. The ubiquitin-proteasome system in Huntington's disease: Are proteasomes impaired, initiators of disease, or coming to the rescue? Biochemistry Research International. 2012;2012 doi: 10.1155/2012/837015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Finley D. Regulation of proteasome activity in health and disease. Biochimica et Biophysica Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke G., Kraft R., Kostka S., Henklein P., Frömmel C., Löwe J.…Schmidt M. Analysis of mammalian 20S proteasome biogenesis: The maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. The EMBO Journal. 1996;15(24):6887–6898. [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Ackermann K., Stuart M., Wex C., Protzer U., Schätzl H.M., Gilch S. Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. Journal of Virology. 2012;86(18):10112–10122. doi: 10.1128/JVI.01001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenheimer R. 2nd ed. Mass Harvard University Press; Cambridge: 1946. The Dynamic State of Body Constituents.https://trove.nla.gov.au/work/14374148 [Google Scholar]
- Schoenheimer R., Ratner S., Rittenberg D. Studies in protein metabolism vii. The metabolism of tyrosine. Journal of Biological Chemistry. 1939;127(1):333–344. [Google Scholar]
- Schrader J., Henneberg F., Mata R.A., Tittmann K., Schneider T.R., Stark H.…Chari A. The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science. 2016;353(6299):594–598. doi: 10.1126/science.aaf8993. [DOI] [PubMed] [Google Scholar]
- Schuld N.J., Hussong S.A., Kapphahn R.J., Lehmann U., Roehrich H., Rageh A.A.…Ferrington D.A. Immunoproteasome deficiency protects in the retina after optic nerve crush. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.A., Hernandez A., Mark Evers B. The role of NF-kappaB/IkappaB proteins in cancer: implications for novel treatment strategies. Surgical Oncology. 1999;8(3):143–153. doi: 10.1016/s0960-7404(00)00012-8. [DOI] [PubMed] [Google Scholar]
- Seemuller E., Lupas A., Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382(6590):468–471. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. Proteasome from Thermoplasma acidophilum: A threonine protease. Science (New York, N.Y.) 1995;268(5210):579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- Seki N., Toh U., Sayers T.J., Fujii T., Miyagi M., Akagi Y.…Yamana H. Bortezomib sensitizes human esophageal squamous cell carcinoma cells to TRAIL-mediated apoptosis via activation of both extrinsic and intrinsic apoptosis pathways. Molecular Cancer Therapeutics. 2010;9(6):1842–1851. doi: 10.1158/1535-7163.MCT-09-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatorov V., Malyukova I., Fariss R., Wawrousek E.F., Swaminathan S., Sharan S.K., Tomarev S. Expression of mutated mouse myocilin induces open-angle glaucoma in transgenic mice. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2006;26(46):11903–11914. doi: 10.1523/JNEUROSCI.3020-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabaneh T.B., Downey S.L., Goddard A.L., Screen M., Lucas M.M., Eastman A., Kisselev A.F. Molecular basis of differential sensitivity of myeloma cells to clinically relevant bolus treatment with bortezomib. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Herman-Bachinsky Y., Buchsbaum S., Lewinson O., Haj-Yahya M., Hejjaoui M.…Ciechanover A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Molecular Cell. 2012;48(1):87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Shagufta, Ahmad I. Transition metal complexes as proteasome inhibitors for cancer treatment. Inorganica Chimica Acta. 2020;506:119521. doi: 10.1016/j.ica.2020.119521. [DOI] [Google Scholar]
- Shah C., Bishnoi R., Wang Y., Zou F., Bejjanki H., Master S., Moreb J.S. Efficacy and safety of carfilzomib in relapsed and/or refractory multiple myeloma: Systematic review and meta-analysis of 14 trials. Oncotarget. 2018;9(34):23704–23717. doi: 10.18632/oncotarget.25281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Usmani S., Stadtmauer E.A., Rifkin R.M., Berenson J.R., Berdeja J.G.…Niesvizky R. Oprozomib, pomalidomide, and dexamethasone in patients with relapsed and/or refractory multiple myeloma. Clinical Lymphoma, Myeloma & Leukemia. 2019;19(9):570–578. doi: 10.1016/j.clml.2019.05.017. e1. [DOI] [PubMed] [Google Scholar]
- Shakya R., Peng F., Liu J., Heeg M.J., Verani C.N. Synthesis, structure, and anticancer activity of gallium(III) complexes with asymmetric tridentate ligands: Growth inhibition and apoptosis induction of cisplatin-resistant neuroblastoma cells. Inorganic Chemistry. 2006;45(16):6263–6268. doi: 10.1021/ic060106g. [DOI] [PubMed] [Google Scholar]
- Shang F., Taylor A. Role of the ubiquitin-proteasome in protein quality control and signaling: Implication in the pathogenesis of eye diseases. Progress in Molecular Biology and Translational Science. 2012;109:347–396. doi: 10.1016/B978-0-12-397863-9.00010-9. [DOI] [PubMed] [Google Scholar]
- Sharma S.K., Chorell E., Steneberg P., Vernersson-Lindahl E., Edlund H., Wittung-Stafshede P. Insulin-degrading enzyme prevents α-synuclein fibril formation in a nonproteolytical manner. Scientific Reports. 2015;5(1):1–10. doi: 10.1038/srep12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.K., Chorell E., Wittung-Stafshede P. Insulin-degrading enzyme is activated by the C-terminus of α-synuclein. Biochemical and Biophysical Research Communications. 2015;466(2):192–195. doi: 10.1016/j.bbrc.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Sharon M., Taverner T., Ambroggio X.I., Deshaies R.J., Robinson C.V. Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biology. 2006;4(8) doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff R.J., Singer J.D., Swanger J., Smitherman M., Roberts J.M., Clurman B.E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Molecular Cell. 2000;5(2):403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- Shen W.-C., Li H.-Y., Chen G.-C., Chern Y., Tu P.-H. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2015;11(4):685–700. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Luo X., Liu S., Shen Y., Nawy S., Shen Y. Rod bipolar cells dysfunction occurs before ganglion cells loss in excitotoxin-damaged mouse retina. Cell Death & Disease. 2019;10(12):905. doi: 10.1038/s41419-019-2140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Song G., An M., Li X., Wu N., Ruan K.…Hu R. The use of hollow mesoporous silica nanospheres to encapsulate bortezomib and improve efficacy for non-small cell lung cancer therapy. Biomaterials. 2014;35(1):316–326. doi: 10.1016/j.biomaterials.2013.09.098. [DOI] [PubMed] [Google Scholar]
- Shen X., Ying H., Qiu Y., Park J.-S., Shyam R., Chi Z.-L.…Yue B.Y.J.T. Processing of optineurin in neuronal cells. The Journal of Biological Chemistry. 2011;286(5):3618–3629. doi: 10.1074/jbc.M110.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman D.J., Li J. Proteasome inhibitors: Harnessing proteostasis to combat disease. Molecules. 2020;25(3):671. doi: 10.3390/molecules25030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & Development. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shi Y., Chen X., Elsasser S., Stocks B.B., Tian G., Lee B.-H.…Walters K.J. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science (New York, N.Y.) 2016;351(6275) doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Schlossmacher M.G., Hattori N., Frosch M.P., Trockenbacher A., Schneider R.…Selkoe D.J. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: Implications for Parkinson's disease. Science (New York, N.Y.) 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Shimura H., Schwartz D., Gygi S.P., Kosik K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. The Journal of Biological Chemistry. 2004;279(6):4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- Shringarpure R., Grune T., Mehlhase J., Davies K.J.A. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. The Journal of Biological Chemistry. 2003;278(1):311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- Shukla N., Somwar R., Smith R.S., Ambati S., Munoz S., Merchant M., D'Arcy P., Wang X., Kobos R., Antczak C., Bhinder B., Shum D., Radu C., Yang G., Taylor B.S., Ng C.K.Y., Weigelt B., Khodos I., de Stanchina E.…Ladanyi M. Proteasome Addiction Defined in Ewing Sarcoma Is Effectively Targeted by a Novel Class of 19S Proteasome Inhibitors. Cancer Research. 2016;76(15):4525–4534. doi: 10.1158/0008-5472.CAN-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Martin T., Nooka A., Harvey R.D., Vij R., Niesvizky R.…Lonial S. Integrated safety profile of single-agent carfilzomib: Experience from 526 patients enrolled in 4 phase II clinical studies. Haematologica. 2013;98(11):1753–1761. doi: 10.3324/haematol.2013.089334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D.S., Martin T., Wang M., Vij R., Jakubowiak A.J., Lonial S., Trudel S., Kukreti V., Bahlis N., Alsina M., Chanan-Khan A., Buadi F., Reu F.J., Somlo G., Zonder J., Song K., Stewart A.K., Stadtmauer E., Kunkel L.…Jagannath S. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 2012;120(14):2817–2825. doi: 10.1182/blood-2012-05-425934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijts E.J.A.M., Kloetzel P.M. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cellular and Molecular Life Sciences: CMLS. 2011;68(9):1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijts A., Sun Y., Janek K., Kral S., Paschen A., Schadendorf D., Kloetzel P.-M. The role of the proteasome activator PA28 in MHC class I antigen processing. Molecular Immunology. 2002;39(3–4):165–169. doi: 10.1016/s0161-5890(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Silswal N., Reis J., Qureshi A.A., Papasian C., Qureshi N. Of mice and men: Proteasome's role in LPS-induced inflammation and tolerance. Shock (Augusta, GA.) 2017;47(4):445–454. doi: 10.1097/SHK.0000000000000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.L., De Moura Gallo C.V., Costa D.C.F., Rangel L.P. Prion-like aggregation of mutant p53 in cancer. Trends in Biochemical Sciences. 2014;39(6):260–267. doi: 10.1016/j.tibs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Simons B.D., Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145(6):851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Simpson M.V. The release of labeled amino acids from the proteins of rat liver slices. The Journal of Biological Chemistry. 1953;201(1):143–154. [PubMed] [Google Scholar]
- Sin N., Kim K.B., Elofsson M., Meng L., Auth H., Kwok B.H., Crews C.M. Total synthesis of the potent proteasome inhibitor epoxomicin: A useful tool for understanding proteasome biology. Bioorganic & Medicinal Chemistry Letters. 1999;9(15):2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- Singh B.N., Malhotra B.K. Effects of food on the clinical pharmacokinetics of anticancer agents: Underlying mechanisms and implications for oral chemotherapy. Clinical Pharmacokinetics. 2004;43(15):1127–1156. doi: 10.2165/00003088-200443150-00005. [DOI] [PubMed] [Google Scholar]
- Singh A.V., Palladino M.A., Lloyd G.K., Potts B.C., Chauhan D., Anderson K.C. Pharmacodynamic and efficacy studies of the novel proteasome inhibitor NPI-0052 (marizomib) in a human plasmacytoma xenograft murine model. British Journal of Haematology. 2010;149(4):550–559. doi: 10.1111/j.1365-2141.2010.08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi K., Swarup G. Defects in autophagy caused by glaucoma-associated mutations in optineurin. Experimental Eye Research. 2016;144:54–63. doi: 10.1016/j.exer.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Sitaraman S., Na C.-L., Yang L., Filuta A., Bridges J.P., Weaver T.E. Proteasome dysfunction in alveolar type 2 epithelial cells is associated with acute respiratory distress syndrome. Scientific Reports. 2019;9(1):1–15. doi: 10.1038/s41598-019-49020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixt S.U., Adamzik M., Spyrka D., Saul B., Hakenbeck J., Wohlschlaeger J.…Peters J. Alveolar extracellular 20S proteasome in patients with acute respiratory distress syndrome. American Journal of Respiratory and Critical Care Medicine. 2009;179(12):1098–1106. doi: 10.1164/rccm.200802-199OC. [DOI] [PubMed] [Google Scholar]
- Sixt S.U., Alami R., Hakenbeck J., Adamzik M., Kloß A., Costabel U.…Peters J. Distinct proteasome subpopulations in the alveolar space of patients with the acute respiratory distress syndrome. Mediators of Inflammation. 2012;2012 doi: 10.1155/2012/204250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklirou A., Papanagnou E.-D., Fokialakis N., Trougakos I.P. Cancer chemoprevention via activation of proteostatic modules. Cancer Letters. 2018;413:110–121. doi: 10.1016/j.canlet.2017.10.034. [DOI] [PubMed] [Google Scholar]
- Śledź P., Unverdorben P., Beck F., Pfeifer G., Schweitzer A., Förster F., Baumeister W. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingerland J., Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. Journal of Cellular Physiology. 2000;183(1):10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Smith D.M. Could a common mechanism of protein degradation impairment underlie many neurodegenerative diseases? Journal of Experimental Neuroscience. 2018;12 doi: 10.1177/1179069518794675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.M., Chang S.-C., Park S., Finley D., Cheng Y., Goldberg A. Docking of the proteasomal ATPases' C-termini in the 20S proteasomes alpha ring opens the gate for substrate entry. Molecular Cell. 2007;27(5):731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.M., Kafri G., Cheng Y., Ng D., Walz T., Goldberg A.L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Molecular Cell. 2005;20(5):687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Smith D.C., Kalebic T., Infante J.R., Siu L.L., Sullivan D., Vlahovic G.…Thompson J.A. Phase 1 study of ixazomib, an investigational proteasome inhibitor, in advanced non-hematologic malignancies. Investigational New Drugs. 2015;33(3):652–663. doi: 10.1007/s10637-015-0230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.L., Pozueta J., Gong B., Arancio O., Shelanski M. Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proceedings of the National Academy of Sciences. 2009;106(39):16877–16882. doi: 10.1073/pnas.0908706106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolewski P., Rydygier D. Ixazomib: An investigational drug for the treatment of lymphoproliferative disorders. Expert Opinion on Investigational Drugs. 2019;28(5):421–433. doi: 10.1080/13543784.2019.1596258. [DOI] [PubMed] [Google Scholar]
- Soares T.R., Reis S.D., Pinho B.R., Duchen M.R., Oliveira J.M.A. Targeting the proteostasis network in Huntington's disease. Ageing Research Reviews. 2019;49:92–103. doi: 10.1016/j.arr.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave C.L., Guerin T., Liu J., Dou Q.P. Targeting the ubiquitin-proteasome system for cancer treatment: discovering novel inhibitors from nature and drug repurposing. Cancer Metastasis Reviews. 2017;36(4):717–736. doi: 10.1007/s10555-017-9705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V., Li F., Polovin G., Park S. Proteasome activation is mediated via a functional switch of the Rpt6 C-terminal tail following chaperone-dependent assembly. Scientific Reports. 2015;5(1):1–15. doi: 10.1038/srep14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T., Saeki Y., Toh-e A., Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. The Journal of Biological Chemistry. 2004;279(27):28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- Song P., Li S., Wu H., Gao R., Rao G., Wang D.…Chen Q. Parkin promotes proteasomal degradation of p62: Implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson's disease. Protein & Cell. 2016;7(2):114–129. doi: 10.1007/s13238-015-0230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneveld P., Goldschmidt H., Rosiñol L., Bladé J., Lahuerta J.J., Cavo M.…Moreau P. Bortezomib-based versus nonbortezomib-based induction treatment before autologous stem-cell transplantation in patients with previously untreated multiple myeloma: A meta-analysis of phase III randomized, controlled trials. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2013;31(26):3279–3287. doi: 10.1200/JCO.2012.48.4626. [DOI] [PubMed] [Google Scholar]
- Sontag E.M., Vonk W.I.M., Frydman J. Sorting out the trash: The spatial nature of eukaryotic protein quality control. Current Opinion in Cell Biology. 2014;26:139–146. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., Nalls M.A., Clark J.M., Mitchell B.D., Shuldiner A.R., Butler J.L., Tomas M., Hoffmann U., Hwang S.-J., Massaro J.M.…Consortium G.O.L.D. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genetics. 2011;7(3) doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A., Harrison S., Zonder J., Badros A., Laubach J., Bergin K.…Richardson P. A phase 1 clinical trial evaluating marizomib, pomalidomide and low-dose dexamethasone in relapsed and refractory multiple myeloma (NPI-0052-107): Final study results. British Journal of Haematology. 2018;180(1):41–51. doi: 10.1111/bjh.14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A., Lentzsch S., Weisel K., Avet-Loiseau H., Mark T.M., Spicka I., Masszi T., Lauri B., Levin M.-D., Bosi A., Hungria V., Cavo M., Lee J.-J., Nooka A.K., Quach H., Lee C., Barreto W., Corradini P., Min C.-K.…Mateos M.-V. Daratumumab plus bortezomib and dexamethasone versus bortezomib and dexamethasone in relapsed or refractory multiple myeloma: Updated analysis of CASTOR. Haematologica. 2018;103(12):2079–2087. doi: 10.3324/haematol.2018.194118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyracopoulos L. The proteasome: More than a means to an end. Structure (London, England: 1993) 2016;24(8):1221–1223. doi: 10.1016/j.str.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Stelzma R.A., Schnitzlein H.N., Murllagh F.R. VIEWPOINT An english translation of Alzheimer's 1907 Paper, "ijber eine eigenartige Erlranliung der Hirnrinde". Clinical Anatomy. 1995;8:429–443. doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- Sterz J., Jakob C., Kuckelkorn U., Heider U., Mieth M., Kleeberg L.…von Metzler I. BSc2118 is a novel proteasome inhibitor with activity against multiple myeloma. European Journal of Haematology. 2010;85(2):99–107. doi: 10.1111/j.1600-0609.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- Stewart A.K., Dimopoulos M.A., Masszi T., Špička I., Oriol A., Hájek R.…Palumbo A. Health-related quality-of-life results from the open-label, randomized, phase III ASPIRE trial evaluating carfilzomib, lenalidomide, and dexamethasone versus lenalidomide and dexamethasone in patients with relapsed multiple myeloma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2016;34(32):3921–3930. doi: 10.1200/JCO.2016.66.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A.K., Rajkumar S.V., Dimopoulos M.A., Masszi T., Špička I., Oriol A., Hájek R., Rosiñol L., Siegel D.S., Mihaylov G.G., Goranova-Marinova V., Rajnics P., Suvorov A., Niesvizky R., Jakubowiak A.J., San-Miguel J.F., Ludwig H., Wang M., Maisnar V.…ASPIRE Investigators Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. The New England Journal of Medicine. 2015;372(2):142–152. doi: 10.1056/NEJMoa1411321. [DOI] [PubMed] [Google Scholar]
- Strauss S.J., Higginbottom K., Jüliger S., Maharaj L., Allen P., Schenkein D.…Joel S.P. The proteasome inhibitor bortezomib acts independently of p53 and induces cell death via apoptosis and mitotic catastrophe in B-cell lymphoma cell lines. Cancer Research. 2007;67(6):2783–2790. doi: 10.1158/0008-5472.CAN-06-3254. [DOI] [PubMed] [Google Scholar]
- Striha A., Ashcroft A.J., Hockaday A., Cairns D.A., Boardman K., Jacques G.…Cook G. The role of ixazomib as an augmented conditioning therapy in salvage autologous stem cell transplant (ASCT) and as a post-ASCT consolidation and maintenance strategy in patients with relapsed multiple myeloma (ACCoRd [UK-MRA Myeloma XII] trial): study protocol for a Phase III randomised controlled trial. Trials. 2018;19(1):169. doi: 10.1186/s13063-018-2524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S.W., Jensen K.B., Jensen M.S., Silva D.S., Kesslak P.J., Danscher G., Frederickson C.J. Histochemically-reactive zinc in amyloid plaques, angiopathy, and degenerating neurons of Alzheimer's diseased brains. Brain Research. 2000;852(2):274–278. doi: 10.1016/s0006-8993(99)02096-x. [DOI] [PubMed] [Google Scholar]
- Sun L., Büeler H. Proteasome inhibition promotes mono-ubiquitination and nuclear translocation of mature (52 kDa) PINK1. Biochemical and Biophysical Research Communications. 2019;517(2):376–382. doi: 10.1016/j.bbrc.2019.07.051. [DOI] [PubMed] [Google Scholar]
- Sunwoo J.B., Chen Z., Dong G., Yeh N., Crowl Bancroft C., Sausville E.…Van Waes C. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2001;7(5):1419–1428. [PubMed] [Google Scholar]
- Swarup G., Sayyad Z. Altered functions and interactions of glaucoma-associated mutants of optineurin. Frontiers in Immunology. 2018:9. doi: 10.3389/fimmu.2018.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney P., Park H., Baumann M., Dunlop J., Frydman J., Kopito R.…Hodgson R. Protein misfolding in neurodegenerative diseases: Implications and strategies. Translational Neurodegeneration. 2017;6 doi: 10.1186/s40035-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szokalska A., Makowski M., Nowis D., Wilczyński G.M., Kujawa M., Wójcik C., Młynarczuk-Biały I., Salwa P., Bil J., Janowska S., Agostinis P., Verfaillie T., Bugajski M., Gietka J., Issat T., Głodkowska E., Mrówka P., Stoklosa T., Hamblin M.R.…Golab J. Proteasome inhibition potentiates antitumor effects of photodynamic therapy in mice through induction of endoplasmic reticulum stress and unfolded protein response. Cancer Research. 2009;69(10):4235–4243. doi: 10.1158/0008-5472.CAN-08-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H.-C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nature Reviews. Neuroscience. 2008;9(11):826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Tai H.-C., Serrano-Pozo A., Hashimoto T., Frosch M.P., Spires-Jones T.L., Hyman B.T. The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. The American Journal of Pathology. 2012;181(4):1426–1435. doi: 10.1016/j.ajpath.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Van Laethem F., Xing Y., Akane K., Suzuki H., Murata S.…Takahama Y. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells. Nature Immunology. 2015;16(10):1069–1076. doi: 10.1038/ni.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Kitaura H., Kakita A., Kakihana T., Katsuragi Y., Nameta M.…Fujii M. USP10 is a driver of ubiquitinated protein aggregation and aggresome formation to inhibit apoptosis. IScience. 2018;9:433–450. doi: 10.1016/j.isci.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi N., Yokota K., Ahn J.Y., Chung C.H., Fujiwara T., Takahashi E.…Tanaka K. Molecular properties of the proteasome activator PA28 family proteins and γ-interferon regulation. Genes to Cells. 1997;2(3):195–211. doi: 10.1046/j.1365-2443.1997.d01-308.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K. The proteasome: Overview of structure and functions. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 2009;85(1):12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Waxman L., Goldberg A.L. ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. The Journal of Cell Biology. 1983;96(6):1580–1585. doi: 10.1083/jcb.96.6.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Seredenina T., Coppola G., Kuhn A., Geschwind D.H., Luthi-Carter R., Thomas E.A. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lengths reveals genes associated with disease onset and progression in Huntington's disease. Neurobiology of Disease. 2011;42(3):459–467. doi: 10.1016/j.nbd.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji K., Mori F., Mimura J., Itoh K., Kakita A., Takahashi H., Wakabayashi K. Proteinase K-resistant alpha-synuclein is deposited in presynapses in human Lewy body disease and A53T alpha-synuclein transgenic mice. Acta Neuropathologica. 2010;120(2):145–154. doi: 10.1007/s00401-010-0676-z. [DOI] [PubMed] [Google Scholar]
- Tarohda T., Yamamoto M., Amamo R. Regional distribution of manganese, iron, copper, and zinc in the rat brain during development. Analytical and Bioanalytical Chemistry. 2004;380(2):240–246. doi: 10.1007/s00216-004-2697-8. [DOI] [PubMed] [Google Scholar]
- Taromi S., Lewens F., Arsenic R., Sedding D., Sänger J., Kunze A., Möbs M., Benecke J., Freitag H., Christen F., Kaemmerer D., Lupp A., Heilmann M., Lammert H., Schneider C.-P., Richter K., Hummel M., Siegmund B., Burger M.…Grabowski P. Proteasome inhibitor bortezomib enhances the effect of standard chemotherapy in small cell lung cancer. Oncotarget. 2017;8(57):97061–97078. doi: 10.18632/oncotarget.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr J.M., Kaul K., Chopra M., Kohner E.M., Chibber R. Pathophysiology of diabetic retinopathy. ISRN Ophthalmology. 2013;2013:343560. doi: 10.1155/2013/343560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp A.T.N., Green C., Xu G., Denovan-Wright E.M., Rising A.C., Fromholt S.E.…Borchelt D.R. Transgenic mice expressing caspase-6-derived N-terminal fragments of mutant huntingtin develop neurologic abnormalities with predominant cytoplasmic inclusion pathology composed largely of a smaller proteolytic derivative. Human Molecular Genetics. 2011;20(14):2770–2782. doi: 10.1093/hmg/ddr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh B.W., Harrison S.J., Worth L.J., Slavin M.A. Antiviral prophylaxis for varicella zoster virus infections in patients with myeloma in the era of novel therapies. Leukemia & Lymphoma. 2016;57(7):1719–1722. doi: 10.3109/10428194.2015.1106538. [DOI] [PubMed] [Google Scholar]
- Teicher B.A., Ara G., Herbst R., Palombella V.J., Adams J. The proteasome inhibitor PS-341 in cancer therapy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 1999;5(9):2638–2645. [PubMed] [Google Scholar]
- Teodorovic I., Pittaluga S., Kluin-Nelemans J.C., Meerwaldt J.H., Hagenbeek A., van Glabbeke M.…Peeters C.D. Efficacy of four different regimens in 64 mantle-cell lymphoma cases: clinicopathologic comparison with 498 other non-Hodgkin's lymphoma subtypes. European Organization for the Research and Treatment of Cancer Lymphoma Cooperative Group. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 1995;13(11):2819–2826. doi: 10.1200/JCO.1995.13.11.2819. [DOI] [PubMed] [Google Scholar]
- Thibaudeau T.A., Anderson R.T., Smith D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nature Communications. 2018;9(1):1097. doi: 10.1038/s41467-018-03509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaudeau T.A., Smith D.M. A practical review of proteasome pharmacology. Pharmacological Reviews. 2019;71(2):170–197. doi: 10.1124/pr.117.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.N., Cripps D., Yang A.J. Proteomic analysis of protein phosphorylation and ubiquitination in Alzheimer's disease. Methods in Molecular Biology (Clifton, N.J.) 2009;566:109–121. doi: 10.1007/978-1-59745-562-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., D'Arcy P., Wang X., Ray A., Tai Y.-T., Hu Y.…Anderson K.C. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123(5):706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Park S., Lee M.J., Huck B., McAllister F., Hill C.P.…Finley D. An asymmetric interface between the regulatory and core particles of the proteasome. Nature Structural & Molecular Biology. 2011;18(11):1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Zhao J., Tai Y.-T., Amin S.B., Hu Y., Berger A.J.…Anderson K.C. Investigational agent MLN9708/2238 targets tumor-suppressor miR33b in MM cells. Blood. 2012;120(19):3958–3967. doi: 10.1182/blood-2012-01-401794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B.G., Li H., Bernstein S.H., Fisher R.I., Burack W.R., Rimsza L.M.…Friedberg J.W. Phase II trial of R-CHOP plus bortezomib induction therapy followed by bortezomib maintenance for newly diagnosed mantle cell lymphoma: SWOG S0601. British Journal of Haematology. 2016;172(2):208–218. doi: 10.1111/bjh.13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofaris G.K., Layfield R., Spillantini M.G. Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Letters. 2001;509(1):22–26. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- Tomaru U., Takahashi S., Ishizu A., Miyatake Y., Gohda A., Suzuki S.…Kasahara M. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. The American Journal of Pathology. 2012;180(3):963–972. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Tomasello M.F., Nardon C., Lanza V., Di Natale G., Pettenuzzo N., Salmaso S.…Fregona D. New comprehensive studies of a gold(III) Dithiocarbamate complex with proven anticancer properties: Aqueous dissolution with cyclodextrins, pharmacokinetics and upstream inhibition of the ubiquitin-proteasome pathway. European Journal of Medicinal Chemistry. 2017;138:115–127. doi: 10.1016/j.ejmech.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Tomko R.J., Funakoshi M., Schneider K., Wang J., Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Molecular Cell. 2010;38(3):393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R.J., Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Molecular Cell. 2011;44(6):907–917. doi: 10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R.J., Hochstrasser M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Molecular Cell. 2014;53(3):433–443. doi: 10.1016/j.molcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R.J., Taylor D.W., Chen Z.A., Wang H.-W., Rappsilber J., Hochstrasser M. A single α helix drives extensive remodeling of the proteasome lid and completion of regulatory particle assembly. Cell. 2015;163(2):432–444. doi: 10.1016/j.cell.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E.M., Dephoure N., Panneerselvam A., Tucker C.M., Whittaker C.A., Gygi S.P.…Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143(1):71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E.M., Sokolsky T., Tucker C.M., Chan L.Y., Boselli M., Dunham M.J., Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science (New York, N.Y.) 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres E.M., Williams B.R., Amon A. Aneuploidy: Cells losing their balance. Genetics. 2008;179(2):737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M.J. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. The EMBO Journal. 2001;20(10):2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traenckner E.B., Pahl H.L., Henkel T., Schmidt K.N., Wilk S., Baeuerle P.A. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. The EMBO Journal. 1995;14(12):2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traenckner E.B., Wilk S., Baeuerle P.A. A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. The EMBO Journal. 1994;13(22):5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippier P.C., Zhao K.T., Fox S.G., Schiefer I.T., Benmohamed R., Moran J.…Silverman R.B. Proteasome activation is a mechanism for pyrazolone small molecules displaying therapeutic potential in amyotrophic lateral sclerosis. ACS Chemical Neuroscience. 2014;5(9):823–829. doi: 10.1021/cn500147v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.W., Tsai R.T., Liu S.P., Chen C.S., Tsai M.C., Chien S.H.…Fu R.H. Neuroprotective effects of betulin in pharmacological and transgenic Caenorhabditis elegans models of Parkinson's disease. Cell Transplantation. 2017;26(12):1903–1918. doi: 10.1177/0963689717738785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng B.P., Green K.N., Chan J.L., Blurton-Jones M., LaFerla F.M. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiology of Aging. 2008;29(11):1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsihlias J., Kapusta L., Slingerland J. The prognostic significance of altered cyclin-dependent kinase inhibitors in human cancer. Annual Review of Medicine. 1999;50:401–423. doi: 10.1146/annurev.med.50.1.401. [DOI] [PubMed] [Google Scholar]
- Tsolekile N., Nelana S., Oluwafemi O.S. Porphyrin as diagnostic and therapeutic agent. Molecules. 2019;24(14):2669. doi: 10.3390/molecules24142669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P., Mendillo M.L., Zhao J., Carette J.E., Merrill P.H., Cikes D.…Lindquist S. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. ELife. 2015;4 doi: 10.7554/eLife.08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P., Sokol E., Jin D., Brune Z., Thiru P., Ghandi M.…Lindquist S. Suppression of 19S proteasome subunits marks emergence of an altered cell state in diverse cancers. Proceedings of the National Academy of Sciences. 2017;114(2):382–387. doi: 10.1073/pnas.1619067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tullio M.B., Castelletto V., Hamley I.W., Martino Adami P.V., Morelli L., Castaño E.M. Proteolytically inactive insulin-degrading enzyme inhibits amyloid formation yielding non-neurotoxic Aβ peptide aggregates. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0059113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundo G.R., Sbardella D., Ciaccio C., Bianculli A., Orlandi A., Desimio M.G.…Marini S. Insulin-degrading enzyme (IDE): A novel heat shock-like protein. The Journal of Biological Chemistry. 2013;288(4):2281–2289. doi: 10.1074/jbc.M112.393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundo G.R., Sbardella D., Ciaccio C., Grasso G., Gioia M., Coletta A.…Coletta M. Multiple functions of insulin-degrading enzyme: A metabolic crosslight? Critical Reviews in Biochemistry and Molecular Biology. 2017;52(5):554–582. doi: 10.1080/10409238.2017.1337707. [DOI] [PubMed] [Google Scholar]
- Tundo G.R., Sbardella D., Coletta M. Insights into proteasome conformation dynamics and intersubunit communication. Trends in Biochemical Sciences. 2018;43(11):852–853. doi: 10.1016/j.tibs.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Tundo G.R., Sbardella D., Lacal P.M., Graziani G., Marini S. On the horizon: Targeting next-generation immune checkpoints for cancer treatment. Chemotherapy. 2019;64(2):62–80. doi: 10.1159/000500902. [DOI] [PubMed] [Google Scholar]
- Tzotzos S., Doig A.J. Amyloidogenic sequences in native protein structures. Protein Science: A Publication of the Protein Society. 2010;19(2):327–348. doi: 10.1002/pro.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi H., Hamazaki J., Murata S. Characterization of the testis-specific proteasome subunit α4s in mammals. The Journal of Biological Chemistry. 2014;289(18):12365–12374. doi: 10.1074/jbc.M114.558866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugras S., Daniels M.J., Fazelinia H., Gould N.S., Yocum A.K., Luk K.C.…Ischiropoulos H. Induction of the immunoproteasome subunit Lmp7 links proteostasis and immunity in α-synuclein aggregation disorders. EBioMedicine. 2018;31:307–319. doi: 10.1016/j.ebiom.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um J.W., Im E., Lee H.J., Min B., Yoo L., Yoo J.…Chung K.C. Parkin directly modulates 26S proteasome activity. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2010;30(35):11805–11814. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um J.W., Min D.S., Rhim H., Kim J., Paik S.R., Chung K.C. Parkin ubiquitinates and promotes the degradation of RanBP2. The Journal of Biological Chemistry. 2006;281(6):3595–3603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure (London, England: 1993) 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Unverdorben P., Beck F., Śledź P., Schweitzer A., Pfeifer G., Plitzko J.M.…Förster F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proceedings of the National Academy of Sciences. 2014;111(15):5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V., Hoffman L., Pratt G., Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. The EMBO Journal. 2002;21(13):3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V., Pratt G., Gorbea C., Rechsteiner M. Purification and assay of proteasome activator PA200. Methods in Enzymology. 2005;398:321–329. doi: 10.1016/S0076-6879(05)98026-9. [DOI] [PubMed] [Google Scholar]
- Uttamsingh V., Lu C., Miwa G., Gan L.-S. Relative contributions of the five major human cytochromes P450, 1A2, 2C9, 2C19, 2D6, and 3A4, to the hepatic metabolism of the proteasome inhibitor bortezomib. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2005;33(11):1723–1728. doi: 10.1124/dmd.105.005710. [DOI] [PubMed] [Google Scholar]
- Vaden J.H., Watson J.A., Howard A.D., Chen P.-C., Wilson J.A., Wilson S.M. Distinct effects of ubiquitin overexpression on NMJ structure and motor performance in mice expressing catalytically inactive USP14. Frontiers in Molecular Neuroscience. 2015;8 doi: 10.3389/fnmol.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahid S., Thaper D., Zoubeidi A. Chaperoning the cancer: The proteostatic functions of the heat shock proteins in cancer. Recent Patents on Anti-Cancer Drug Discovery. 2017;12(1):35–47. doi: 10.2174/1574892811666161102125252. [DOI] [PubMed] [Google Scholar]
- Vallelian F., Deuel J.W., Opitz L., Schaer C.A., Puglia M., Lönn M.…Schaer D.J. Proteasome inhibition and oxidative reactions disrupt cellular homeostasis during heme stress. Cell Death and Differentiation. 2015;22(4):597–611. doi: 10.1038/cdd.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2007;13(4):1076–1082. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- VanderLinden R.T., Hemmis C.W., Yao T., Robinson H., Hill C.P. Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism. The Journal of Biological Chemistry. 2017;292(23):9493–9504. doi: 10.1074/jbc.M117.785287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The early history of the ubiquitin field. Protein Science: A Publication of the Protein Society. 2006;15(3):647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I., Connerly P.L., Arendt C.S., Li X., Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. The EMBO Journal. 2004;23(3):500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan K., Rader M., Ramanathan R.K., Ramalingam S., Chen E., Riordan W.…Chatta G. Effect of the CYP3A inhibitor ketoconazole on the pharmacokinetics and pharmacodynamics of bortezomib in patients with advanced solid tumors: a prospective, multicenter, open-label, randomized, two-way crossover drug-drug interaction study. Clinical Therapeutics. 2009;31(Pt 2):2444–2458. doi: 10.1016/j.clinthera.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Venner C.P., Lane T., Foard D., Rannigan L., Gibbs S.D.J., Pinney J.H.…Wechalekar A.D. Cyclophosphamide, bortezomib, and dexamethasone therapy in AL amyloidosis is associated with high clonal response rates and prolonged progression-free survival. Blood. 2012;119(19):4387–4390. doi: 10.1182/blood-2011-10-388462. [DOI] [PubMed] [Google Scholar]
- Verbrugge S.E., Assaraf Y.G., Dijkmans B.A.C., Scheffer G.L., Al M., den Uyl D.…Jansen G. Inactivating PSMB5 mutations and P-glycoprotein (multidrug resistance-associated protein/ATP-binding cassette B1) mediate resistance to proteasome inhibitors: Ex vivo efficacy of (immuno)proteasome inhibitors in mononuclear blood cells from patients with rheumatoid arthritis. The Journal of Pharmacology and Experimental Therapeutics. 2012;341(1):174–182. doi: 10.1124/jpet.111.187542. [DOI] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., Koonin E.V., Deshaies R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science (New York, N.Y.) 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Vernazza S., Tirendi S., Scarfì S., Passalacqua M., Oddone F., Traverso C.E.…Saccà S.C. 2D- and 3D-cultures of human trabecular meshwork cells: A preliminary assessment of an in vitro model for glaucoma study. PLoS One. 2019;14(9) doi: 10.1371/journal.pone.0221942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank J.J.S., Goldberg A.L. Regulating protein breakdown through proteasome phosphorylation. The Biochemical Journal. 2017;474(19):3355–3371. doi: 10.1042/BCJ20160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank J.J.S., Goldberg A.L. Exploring the regulation of proteasome function by subunit phosphorylation. Methods in Molecular Biology (Clifton, N.J.) 2018;1844:309–319. doi: 10.1007/978-1-4939-8706-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank J.J.S., Lokireddy S., Zhao J., Goldberg A.L. 26S proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proceedings of the National Academy of Sciences. 2019;116(10):4228–4237. doi: 10.1073/pnas.1809254116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielberg M.-T., Bauer V.C., Groll M. On the trails of the proteasome fold: Structural and functional analysis of the ancestral β-subunit protein anbu. Journal of Molecular Biology. 2018;430(5):628–640. doi: 10.1016/j.jmb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D., Sullivan M.L. Hemin inhibits ubiquitin-dependent proteolysis in both a higher plant and yeast. Biochemistry. 1988;27(9):3290–3295. doi: 10.1021/bi00409a025. [DOI] [PubMed] [Google Scholar]
- Vigouroux S., Briand M., Briand Y. Linkage between the proteasome pathway and neurodegenerative diseases and aging. Molecular Neurobiology. 2004;30(2):201–221. doi: 10.1385/MN:30:2:201. [DOI] [PubMed] [Google Scholar]
- Vij R., Wang M., Kaufman J.L., Lonial S., Jakubowiak A.J., Stewart A.K., Kukreti V., Jagannath S., McDonagh K.T., Alsina M., Bahlis N.J., Reu F.J., Gabrail N.Y., Belch A., Matous J.V., Lee P., Rosen P., Sebag M., Vesole D.H.…Siegel D.S. An open-label, single-arm, phase 2 (PX-171-004) study of single-agent carfilzomib in bortezomib-naive patients with relapsed and/or refractory multiple myeloma. Blood. 2012;119(24):5661–5670. doi: 10.1182/blood-2012-03-414359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo O.V., Sant'Angelo A., Costanzo V., Battaglia F., Arancio O., Shelanski M. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: Reversibility by drugs that enhance cAMP signaling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science (New York, N.Y.) 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl D.T., Martin T.G., Vij R., Hari P., Mikhael J.R., Siegel D.…Gasparetto C. Phase I/II study of the novel proteasome inhibitor delanzomib (CEP-18770) for relapsed and refractory multiple myeloma. Leukemia & Lymphoma. 2017;58(8):1872–1879. doi: 10.1080/10428194.2016.1263842. [DOI] [PubMed] [Google Scholar]
- Voigt V., Wikstrom M.E., Kezic J.M., Schuster I.S., Fleming P., Makinen K.…Forrester J.V. Ocular antigen does not cause disease unless presented in the context of inflammation. Scientific Reports. 2017;7(1):14226. doi: 10.1038/s41598-017-14618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose J.M. Mantle cell lymphoma: 2017 update on diagnosis, risk-stratification, and clinical management. American Journal of Hematology. 2017;92(8):806–813. doi: 10.1002/ajh.24797. [DOI] [PubMed] [Google Scholar]
- Voutsadakis I.A. Proteasome expression and activity in cancer and cancer stem cells. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39(3) doi: 10.1177/1010428317692248. [DOI] [PubMed] [Google Scholar]
- de Vrij F.M.S., Fischer D.F., van Leeuwen F.W., Hol E.M. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Progress in Neurobiology. 2004;74(5):249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., Tanji K., Mori F., Takahashi H. The Lewy body in Parkinson's disease: Molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology: Official Journal of the Japanese Society of Neuropathology. 2007;27(5):494–506. doi: 10.1111/j.1440-1789.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., Tanji K., Odagiri S., Miki Y., Mori F., Takahashi H. The Lewy body in Parkinson's disease and related neurodegenerative disorders. Molecular Neurobiology. 2013;47(2):495–508. doi: 10.1007/s12035-012-8280-y. [DOI] [PubMed] [Google Scholar]
- Walerych D., Lisek K., Del Sal G. Mutant p53: One, No One, and One Hundred Thousand. Frontiers in Oncology. 2015;5:289. doi: 10.3389/fonc.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D., Lisek K., Sommaggio R., Piazza S., Ciani Y., Dalla E.…Del Sal G. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nature Cell Biology. 2016;18(8):897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- Wallington‐Beddoe C.T., Sobieraj‐Teague M., Kuss B.J., Pitson S.M. Resistance to proteasome inhibitors and other targeted therapies in myeloma. British Journal of Haematology. 2018;182(1):11–28. doi: 10.1111/bjh.15210. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Xu G.L. Cytosine methylation and DNA repair. Current Topics in Microbiology and Immunology. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- Wang X., Chemmama I.E., Yu C., Huszagh A., Xu Y., Viner R.…Huang L. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. The Journal of Biological Chemistry. 2017;292(39):16310–16320. doi: 10.1074/jbc.M117.803619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., D'Arcy P., Caulfield T.R., Paulus A., Chitta K., Mohanty C.…Linder S. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chemical Biology & Drug Design. 2015;86(5):1036–1048. doi: 10.1111/cbdd.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fang Y., Teague J., Wong H., Morisseau C., Hammock B.D.…Wang Z. In vitro metabolism of oprozomib, an oral proteasome inhibitor: Role of epoxide hydrolases and cytochrome P450s. Drug Metabolism and Disposition. 2017;45(7):712–720. doi: 10.1124/dmd.117.075226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang Y., Ding S., Li J., Song N., Ren Y.…Mei Z. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Research. 2018;28(12):1186–1194. doi: 10.1038/s41422-018-0091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li C., Zhang Q., Wang T., Li J., Guan W.…Li D. Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42. Virology Journal. 2010;7:99. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li M., Zhang Z., Xue H., Chen X., Ji Y. Physiological function of myocilin and its role in the pathogenesis of glaucoma in the trabecular meshwork (review) International Journal of Molecular Medicine. 2019;43(2):671–681. doi: 10.3892/ijmm.2018.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Martin T., Bensinger W., Alsina M., Siegel D.S., Kavalerchik E.…Niesvizky R. Phase 2 dose-expansion study (PX-171-006) of carfilzomib, lenalidomide, and low-dose dexamethasone in relapsed or progressive multiple myeloma. Blood. 2013;122(18):3122–3128. doi: 10.1182/blood-2013-07-511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Mazurkiewicz M., Hillert E.-K., Olofsson M.H., Pierrou S., Hillertz P.…D'Arcy P. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Scientific Reports. 2016;6 doi: 10.1038/srep26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Stafford W., Mazurkiewicz M., Fryknäs M., Brjnic S., Zhang X.…Linder S. The 19S Deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Molecular Pharmacology. 2014;85(6):932–945. doi: 10.1124/mol.113.091322. [DOI] [PubMed] [Google Scholar]
- Wang J., Xu X., Elliott M.H., Zhu M., Le Y.-Z. Müller cell-derived VEGF is essential for diabetes-induced retinal inflammation and vascular leakage. Diabetes. 2010;59(9):2297–2305. doi: 10.2337/db09-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang J., Kirk C., Fang Y., Alsina M., Badros A.…Infante J.R. Clinical pharmacokinetics, metabolism, and drug-drug interaction of carfilzomib. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2013;41(1):230–237. doi: 10.1124/dmd.112.047662. [DOI] [PubMed] [Google Scholar]
- Wang X., Yen J., Kaiser P., Huang L. Regulation of the 26S proteasome complex during oxidative stress. Science Signaling. 2010;3(151):ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman A.J., Clasen S., Hwang W.-T., Garfall A., Vogl D.T., Carver J.…Weiss B.M. Carfilzomib-associated cardiovascular adverse events: A systematic review and meta-analysis. JAMA Oncology. 2018;4(3) doi: 10.1001/jamaoncol.2017.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A.A., Cleveland D.W. Does aneuploidy cause cancer? Current Opinion in Cell Biology. 2006;18(6):658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Wehmer M., Rudack T., Beck F., Aufderheide A., Pfeifer G., Plitzko J.M.…Sakata E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proceedings of the National Academy of Sciences. 2017;114(6):1305–1310. doi: 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmer M., Sakata E. Recent advances in the structural biology of the 26S proteasome. The International Journal of Biochemistry & Cell Biology. 2016;79:437–442. doi: 10.1016/j.biocel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Wei J., Dong S., Bowser R.K., Khoo A., Zhang L., Jacko A.M.…Zhao J. Regulation of the ubiquitylation and deubiquitylation of CREB-binding protein modulates histone acetylation and lung inflammation. Science Signaling. 2017;10(483) doi: 10.1126/scisignal.aak9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert O., Weidmann E., Mueck R., Bentz M., von Schilling C., Rohrberg R.…Dreyling M. A novel regimen combining high dose cytarabine and bortezomib has activity in multiply relapsed and refractory mantle cell lymphoma - long-term results of a multicenter observation study. Leukemia & Lymphoma. 2009;50(5):716–722. doi: 10.1080/10428190902856790. [DOI] [PubMed] [Google Scholar]
- Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma. JAMA. 2014;311(18):1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenburger D.D., Vose J.M., Greiner T.C., Lynch J.C., Chan W.C., Bierman P.J.…Armitage J.O. Mantle cell lymphoma. A clinicopathologic study of 68 cases from the Nebraska Lymphoma Study Group. American Journal of Hematology. 2000;64(3):190–196. doi: 10.1002/1096-8652(200007)64:3<190::AID-AJH9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Welk V., Coux O., Kleene V., Abeza C., Trümbach D., Eickelberg O., Meiners S. Inhibition of proteasome activity induces formation of alternative proteasome complexes. The Journal of Biological Chemistry. 2016;291(25):13147–13159. doi: 10.1074/jbc.M116.717652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M., Le Rhun E., Preusser M., Tonn J.-C., Roth P. How we treat glioblastoma. ESMO Open. 2019;4(Suppl. 2) doi: 10.1136/esmoopen-2019-000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., Murray J.M. Structurally-defined deubiquitinase inhibitors provide opportunities to investigate disease mechanisms. Drug Discovery Today: Technologies. 2019;31:109–123. doi: 10.1016/j.ddtec.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Wester R., van der Holt B., Asselbergs E., Zweegman S., Kersten M.J., Vellenga E.…Sonneveld P. Phase 2 study of carfilzomib, thalidomide, and low-dose dexamethasone as induction and consolidation in newly diagnosed, transplant eligible patients with multiple myeloma, the carthadex trial. Haematologica. 2019 doi: 10.3324/haematol.2018.205476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven R.H., Pang B., Assaraf Y.G., Neefjes J. Old drugs, novel ways out: Drug resistance toward cytotoxic chemotherapeutics. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2016;28:65–81. doi: 10.1016/j.drup.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Wilk S., Chen W.E. Synthetic peptide-based activators of the proteasome. Molecular Biology Reports. 1997;24(1–2):119–124. doi: 10.1023/a:1006851428691. [DOI] [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Cation-sensitive neutral endopeptidase: Isolation and specificity of the bovine pituitary enzyme. Journal of Neurochemistry. 1980;35(5):1172–1182. doi: 10.1111/j.1471-4159.1980.tb07873.x. [DOI] [PubMed] [Google Scholar]
- Wilk S., Orlowski M. Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. Journal of Neurochemistry. 1983;40(3):842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D. The discovery of ubiquitin-dependent proteolysis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15280–15282. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.R., Amon A. Aneuploidy –Cancer's fatal flaw? Cancer Research. 2009;69(13):5289–5291. doi: 10.1158/0008-5472.CAN-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Willik K.D., Schagen S.B., Ikram M.A. Cancer and dementia: Two sides of the same coin? European Journal of Clinical Investigation. 2018;48(11) doi: 10.1111/eci.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Bhattacharyya B., Rachel R.A., Coppola V., Tessarollo L., Householder D.B.…Jenkins N.A. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nature Genetics. 2002;32(3):420–425. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- Windheim M., Peggie M., Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. The Biochemical Journal. 2008;409(3):723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- Witkowska J., Giżyńska M., Grudnik P., Golik P., Karpowicz P., Giełdoń A.…Jankowska E. Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome – Insights into the enzyme activation mechanism. Scientific Reports. 2017;7(1):1–11. doi: 10.1038/s41598-017-05997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt E., Zantopf D., Schmidt M., Kraft R., Kloetzel P.M., Krüger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. Journal of Molecular Biology. 2000;301(1):1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- Wójcik C., Tanaka K., Paweletz N., Naab U., Wilk S. Proteasome activator (PA28) subunits, alpha, beta and gamma (Ki antigen) in NT2 neuronal precursor cells and HeLa S3 cells. European Journal of Cell Biology. 1998;77(2):151–160. doi: 10.1016/s0171-9335(98)80083-6. [DOI] [PubMed] [Google Scholar]
- Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden E.J., Dong K.C., Martin A. An AAA motor-driven mechanical switch in Rpn11 controls deubiquitination at the 26S proteasome. Molecular Cell. 2017;67(5):799–811. doi: 10.1016/j.molcel.2017.07.023. e8. [DOI] [PubMed] [Google Scholar]
- Worden E.J., Padovani C., Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nature Structural & Molecular Biology. 2014;21(3):220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- Wu S., Hyrc K.L., Moulder K.L., Lin Y., Warmke T., Snider B.J. Cellular calcium deficiency plays a role in neuronal death caused by proteasome inhibitors. Journal of Neurochemistry. 2009;109(5):1225–1236. doi: 10.1111/j.1471-4159.2009.06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Liu C., Bai C., Han Y.-P., Cho W.C.S., Li Q. Over-Expression of Deubiquitinating Enzyme USP14 in Lung Adenocarcinoma Promotes Proliferation through the Accumulation of β-Catenin. International Journal of Molecular Sciences. 2013;14(6):10749–10760. doi: 10.3390/ijms140610749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Sahara K., Hirayama S., Zhao X., Watanabe A., Hamazaki J.…Murata S. PAC1-PAC2 proteasome assembly chaperone retains the core α4-α7 assembly intermediates in the cytoplasm. Genes to Cells: Devoted to Molecular & Cellular Mechanisms. 2018;23(10):839–848. doi: 10.1111/gtc.12631. [DOI] [PubMed] [Google Scholar]
- Wu Z.-H., Shi Y. When ubiquitin meets NF-κB: a trove for anti-cancer drug development. Current Pharmaceutical Design. 2013;19(18):3263–3275. doi: 10.2174/1381612811319180010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-X., Yang J.-H., Saitsu H. Bortezomib-resistance is associated with increased levels of proteasome subunits and apoptosis-avoidance. Oncotarget. 2016;7(47):77622–77634. doi: 10.18632/oncotarget.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich K., Golubnitschaja O., Pache M., Eberle A.-N., Flammer J. Increased plasma levels of 20S proteasome alpha-subunit in glaucoma patients: An observational pilot study. Molecular Vision. 2002;8:431–435. [PubMed] [Google Scholar]
- Xia X., Huang C., Liao Y., Liu Y., He J., Guo Z.…Huang H. Inhibition of USP14 enhances the sensitivity of breast cancer to enzalutamide. Journal of Experimental & Clinical Cancer Research. 2019;38(1):220. doi: 10.1186/s13046-019-1227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Jameson S.C., Hogquist K.A. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(17):6979–6984. doi: 10.1073/pnas.1222244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Shan B., Lee B.-H., Zhu K., Zhang T., Sun H.…Yuan J. Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. ELife. 2015;4 doi: 10.7554/eLife.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Barker N., Hoon S., He P., Thakur T., Abdeen S.R.…Lane D.P. Bortezomib Stabilizes and Activates p53 in Proliferative Compartments of Both Normal and Tumor Tissues In Vivo. Cancer Research. 2019;79(14):3595–3607. doi: 10.1158/0008-5472.CAN-18-3744. [DOI] [PubMed] [Google Scholar]
- Yam G.H.-F., Gaplovska-Kysela K., Zuber C., Roth J. Aggregated myocilin induces russell bodies and causes apoptosis. The American Journal of Pathology. 2007;170(1):100–109. doi: 10.2353/ajpath.2007.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Yamada J., Sato K., Tokumoto T., Yasutomi M., Ishikawa K. Irreversible potent activation and reversible inhibition of trypsin-like activity of 20S proteasome purified from Xenopus oocytes by fatty acid. Zoological Science. 1998;15(1):43–49. doi: 10.2108/zsj.15.43. [DOI] [PubMed] [Google Scholar]
- Yamano T., Mizukami S., Murata S., Chiba T., Tanaka K., Udono H. Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. The Journal of Biological Chemistry. 2008;283(42):28060–28065. doi: 10.1074/jbc.M803077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gehrke S., Haque M.E., Imai Y., Kosek J., Yang L.…Lu B. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(38):13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Gong N., Zhang L., Lu Y. Isostructurality among 5 solvatomorphs of betulin: X-ray structure and characterization. Journal of Pharmaceutical Science. 2016;105(6):1867–1873. doi: 10.1016/j.xphs.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Yang H., Landis-Piwowar K.R., Chen D., Milacic V., Dou Q.P. Natural compounds with proteasome inhibitory activity for cancer prevention and treatment. Current Protein & Peptide Science. 2008;9(3):227–239. doi: 10.2174/138920308784533998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang Z., Fang Y., Jiang J., Zhao F., Wong H.…Kirk C.J. Pharmacokinetics, pharmacodynamics, metabolism, distribution, and excretion of carfilzomib in rats. Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2011;39(10):1873–1882. doi: 10.1124/dmd.111.039164. [DOI] [PubMed] [Google Scholar]
- Yang F., Yang Y., Mao C., Liu L., Zheng H., Hu L., Liu C. Crosstalk between the proteasome system and autophagy in the clearance of α-synuclein. Acta Pharmacologica Sinica. 2013;34(5):674–680. doi: 10.1038/aps.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Cohen R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao J., Qiu Y., Frontera E., Jia L., Khan N.W., Klionsky D.J.…Zacks D.N. Inhibiting autophagy reduces retinal degeneration caused by protein misfolding. Autophagy. 2018;14(7):1226–1238. doi: 10.1080/15548627.2018.1463121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Song L., Xu W., DeMartino G.N., Florens L., Swanson S.K.…Cohen R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nature Cell Biology. 2006;8(9):994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Ye Y., Klenerman D., Finley D. N-terminal ubiquitination of amyloidogenic proteins triggers removal of their oligomers by the proteasome holoenzyme. Journal of Molecular Biology. 2020;432(2):585–596. doi: 10.1016/j.jmb.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo I.J., Lee M.J., Baek A., Miller Z., Bhattarai D., Baek Y.M.…Kim K.B. A dual inhibitor of the proteasome catalytic subunits LMP2 and Y attenuates disease progression in mouse models of Alzheimer's disease. Scientific Reports. 2019;9(1) doi: 10.1038/s41598-019-54846-z. Scopus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerlikaya A., Okur E., Ulukaya E. The p53-independent induction of apoptosis in breast cancer cells in response to proteasome inhibitor bortezomib. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33(5):1385–1392. doi: 10.1007/s13277-012-0386-3. [DOI] [PubMed] [Google Scholar]
- Ying H., Turturro S., Nguyen T., Shen X., Zelkha R., Johnson E.C.…Yue B.Y.J.T. Induction of autophagy in rats upon overexpression of wild-type and mutant optineurin gene. BMC Cell Biology. 2015;16:14. doi: 10.1186/s12860-015-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba S., Iwase M., Kurihara S., Uchida M., Kurihara Y., Watanabe H., Shintani S. Proteasome inhibitor sensitizes oral squamous cell carcinoma cells to TRAIL-mediated apoptosis. Oncology Reports. 2011;25(3):645–652. doi: 10.3892/or.2010.1127. [DOI] [PubMed] [Google Scholar]
- Yu D., Carroll M., Thomas-Tikhonenko A. p53 status dictates responses of B lymphomas to monotherapy with proteasome inhibitors. Blood. 2007;109(11):4936–4943. doi: 10.1182/blood-2006-10-050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.-Y., Lai M.M.C. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. Journal of Virology. 2005;79(1):644–648. doi: 10.1128/JVI.79.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Livnat-Levanon N., Kleifeld O., Mansour W., Nakasone M.A., Castaneda C.A.…Glickman M.H. Base-CP proteasome can serve as a platform for stepwise lid formation. Bioscience Reports. 2015;35(3) doi: 10.1042/BSR20140173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T., Yan F., Ying M., Cao J., He Q., Zhu H., Yang B. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Frontiers in Pharmacology. 2018;9 doi: 10.3389/fphar.2018.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel C., Nguyen H.P., Hin S.C., Hartl D., Mao L., Klose J. Proteasome and oxidative phoshorylation changes may explain why aging is a risk factor for neurodegenerative disorders. Journal of Proteomics. 2010;73(11):2230–2238. doi: 10.1016/j.jprot.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Zachara N.E., Hart G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochimica et Biophysica Acta (BBA) - General Subjects. 2004;1673(1):13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Zang Y., Kirk C.J., Johnson D.E. Carfilzomib and oprozomib synergize with histone deacetylase inhibitors in head and neck squamous cell carcinoma models of acquired resistance to proteasome inhibitors. Cancer Biology & Therapy. 2014;15(9):1142–1152. doi: 10.4161/cbt.29452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y., Thomas S.M., Chan E.T., Kirk C.J., Freilino M.L., DeLancey H.M.…Johnson D.E. Carfilzomib and ONX 0912 inhibit cell survival and tumor growth of head and neck cancer and their activities are enhanced by suppression of Mcl-1 or autophagy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18(20):5639–5649. doi: 10.1158/1078-0432.CCR-12-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Tallaksen-Greene S.J., Wang B., Albin R.L., Paulson H.L. The de-ubiquitinating enzyme ataxin-3 does not modulate disease progression in a knock-in mouse model of Huntington disease. Journal of Huntington's Disease. 2013;2(2):201–215. doi: 10.3233/JHD-130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Bi C., Buac D., Fan Y., Zhang X., Zuo J.…Dou Q.P. Organic cadmium complexes as proteasome inhibitors and apoptosis inducers in human breast cancer cells. Journal of Inorganic Biochemistry. 2013;123:1–10. doi: 10.1016/j.jinorgbio.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Bi C., Fan Y., Wang H., Bao Y. Cefepime, a fourth-generation cephalosporin, in complex with manganese, inhibits proteasome activity and induces the apoptosis of human breast cancer cells. International Journal of Molecular Medicine. 2015;36(4):1143–1150. doi: 10.3892/ijmm.2015.2297. [DOI] [PubMed] [Google Scholar]
- Zhang N., Fan Y., Huang G., Buac D., Bi C., Ma Y.…Dou Q.P. L-tryptophan Schiff base cadmium(II) complexes as a new class of proteasome inhibitors and apoptosis inducers in human breast cancer cells. Inorganica Chimica Acta. 2017;466:478–485. doi: 10.1016/j.ica.2017.07.006. [DOI] [Google Scholar]
- Zhang X., Frezza M., Milacic V., Ronconi L., Fan Y., Bi C.…Dou Q.P. Inhibition of tumor proteasome activity by gold–dithiocarbamato complexes via both redox-dependent and -independent processes. Journal of Cellular Biochemistry. 2010;109(1):162–172. doi: 10.1002/jcb.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Hu Y., Huang P., Toleman C.A., Paterson A.J., Kudlow J.E. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. The Journal of Biological Chemistry. 2007;282(31):22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P.D., Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Molecular Cell. 2009;34(4):473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Li M., Huang P., Guan X., Zhu Y. Overexpression of ubiquitin specific peptidase 14 predicts unfavorable prognosis in esophageal squamous cell carcinoma. Thoracic Cancer. 2017;8(4):344–349. doi: 10.1111/1759-7714.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L.…Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Sadler P.J. Redox-active metal complexes for anticancer therapy. European Journal of Inorganic Chemistry. 2017;2017(12):1541–1548. doi: 10.1002/ejic.201600908. [DOI] [Google Scholar]
- Zhang X., Schulz R., Edmunds S., Krüger E., Markert E., Gaedcke J.…Dobbelstein M. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Molecular Cell. 2015;59(2):243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- Zhang F., Su K., Yang X., Bowe D.B., Paterson A.J., Kudlow J.E. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115(6):715–725. doi: 10.1016/S0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wei P.-F., Song Y.-H., Dong L., Wu Y.-D., Hao Z.-Y.…Wen L.-P. MnFe2O4 nanoparticles accelerate the clearance of mutant huntingtin selectively through ubiquitin-proteasome system. Biomaterials. 2019;216:119248. doi: 10.1016/j.biomaterials.2019.119248. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W., Chen H., Ding X., Zhao H., Zhang H., Wang C., Zhao J., Sun X., Tian R., Wu W., Wu D., Ma J., Chen Y., Zhang D.…Zhang S. Coagulopathy and Antiphospholipid antibodies in patients with SARS-Cov-2. New England Journal of Medicine. 2020 doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Chen X., Zang D., Lan X., Liao S., Yang C.…Liu J. A novel nickel complex works as a proteasomal deubiquitinase inhibitor for cancer therapy. Oncogene. 2016;35(45):5916–5927. doi: 10.1038/onc.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Foster N.R., Meyers J.P., Thomas S.P., Northfelt D.W., Rowland K.M.…Adjei A.A. A phase I/II study of bortezomib in combination with paclitaxel, carboplatin, and concurrent thoracic radiation therapy for non-small-cell lung cancer: North Central Cancer Treatment Group (NCCTG)-N0321. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer. 2015;10(1):172–180. doi: 10.1097/JTO.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhai B., Gygi S.P., Goldberg A.L. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(52):15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Clabough E.B.D., Sarkar S., Futter M., Rubinsztein D.C., Zeitlin S.O. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genetics. 2010;6(2) doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Liu T., Zheng J., Hu J. Clarifying the molecular mechanism associated with carfilzomib resistance in human multiple myeloma using microarray gene expression profile and genetic interaction network. Oncotargets and Therapy. 2017;10:1327–1334. doi: 10.2147/OTT.S130742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitomirsky B., Assaraf Y.G. Lysosomes as mediators of drug resistance in cancer. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2016;24:23–33. doi: 10.1016/j.drup.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Zhou H.-J., Aujay M.A., Bennett M.K., Dajee M., Demo S.D., Fang Y., Ho M.N., Jiang J., Kirk C.J., Laidig G.J., Lewis E.R., Lu Y., Muchamuel T., Parlati F., Ring E., Shenk K.D., Shields J., Shwonek P.J., Stanton T.…Yang J. Design and synthesis of an orally bioavailable and selective peptide Epoxyketone proteasome inhibitor (PR-047) Journal of Medicinal Chemistry. 2009;52(9):3028–3038. doi: 10.1021/jm801329v. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Grinchuk O., Tomarev S.I. Transgenic mice expressing the Tyr437His mutant of human myocilin protein develop glaucoma. Investigative Ophthalmology & Visual Science. 2008;49(5):1932–1939. doi: 10.1167/iovs.07-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Zuo Y., Li B., Wang H., Liu H., Wang X.…Bu X. Deubiquitinase Inhibition of 19S Regulatory Particles by 4-Arylidene Curcumin Analog AC17 Causes NF-κB Inhibition and p53 Reactivation in Human Lung Cancer Cells. Molecular Cancer Therapeutics. 2013;12(8):1381–1392. doi: 10.1158/1535-7163.MCT-12-1057. [DOI] [PubMed] [Google Scholar]
- Zhu W., Liu J., Nie J., Sheng W., Cao H., Shen W.…Cao J. MG132 enhances the radiosensitivity of lung cancer cells in vitro and in vivo. Oncology Reports. 2015;34(4):2083–2089. doi: 10.3892/or.2015.4169. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Ramos J., Kampa K., Adimoolam S., Sirisawad M., Yu Z.…Lopez C.D. Control of ASPP2/53BP2L Protein Levels by Proteasomal Degradation Modulates p53 Apoptotic Function. Journal of Biological Chemistry. 2005;280(41):34473–34480. doi: 10.1074/jbc.M503736200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang T., Xin Z., Zhan Y., Gu G., Li X.…Liu C. An oral second-generation proteasome inhibitor oprozomib significantly inhibits lung cancer in a p53 independent manner in vitro. Acta Biochimica et Biophysica Sinica. 2019;51(10):1034–1040. doi: 10.1093/abbs/gmz093. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang W.L., Yu D., Ouyang Q., Lu Y., Mao Y. Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome. BioRxiv. 2017 doi: 10.1101/132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Zhang C., Gu C., Li Q., Wu N. Function of Deubiquitinating Enzyme USP14 as Oncogene in Different Types of Cancer. Cellular Physiology and Biochemistry. 2016;38(3):993–1002. doi: 10.1159/000443051. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Zhang Y., Sui Z., Zhang Y., Liu M., Tang H. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget. 2016;8(30):48725–48736. doi: 10.18632/oncotarget.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieba B.A., Henry L., Lacroix M., Jemaà M., Lavabre-Bertrand T., Meunier L.…Stoebner P.-E. The proteasome maturation protein POMP increases proteasome assembly and activity in psoriatic lesional skin. Journal of Dermatological Science. 2017;88(1):10–19. doi: 10.1016/j.jdermsci.2017.04.009. [DOI] [PubMed] [Google Scholar]
- Zondler L., Kostka M., Garidel P., Heinzelmann U., Hengerer B., Mayer B.…Danzer K.M. Proteasome impairment by α-synuclein. PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0184040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P., Kleinz J., Baumeister W. Critical elements in proteasome assembly. Nature Structural Biology. 1994;1(11):765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]